Abstract
Photolyase is a DNA repair enzyme that reverses UV-induced photoproducts in DNA in a light-dependent manner. Recently, photolyase homologs were identified in higher eukaryotes. These homologs, termed cryptochromes, function as blue light photoreceptors or regulators of circadian rhythm. In contrast, most bacteria have only a single photolyase or photolyase-like gene. Unlike other microbes, the chromosome of the cyanobacterium Synechocystis sp. PCC6803 contains two ORFs (slr0854 and sll1629) with high similarities to photolyases. We have characterized both genes. The slr0854 gene product exhibited specific, light-dependent repair activity for a cyclobutane pyrimidine dimer (CPD), whereas the sll1629 gene product lacks measurable affinity for DNA in vitro. Disruption of either slr0854 or sll1629 had little or no effect on the growth rate of the cyanobacterium. A mutant lacking the slr0854 gene showed severe UV sensitivity, in contrast to a mutant lacking sll1629. Phylogenetic analysis showed that sll1629 is more closely related to the cryptochromes than photolyases. We conclude that sll1629 is a bacterial cryptochrome. To our knowledge, this is the first description of a bacterial cryptochrome.
INTRODUCTION
Sunlight is the most important energy source for life on Earth. The UV component of sunlight, however, threatens life by producing cytotoxic, mutagenic and carcinogenic lesions within DNA (1). UV-induced damage was probably even more severe before the formation of the ozone shield by photosynthetic organisms like cyanobacteria. Therefore, selective pressure existed to develop a self-defense system, such as the photolyases. Photolyases repair UV-damaged DNA by using light of the near-UV/blue light region (2). Thus, two types of photolyase have been identified: the well-characterized CPD photolyase and the (6–4) photolyase (2,3). CPD photolyases have been found in a wide range of organisms, including eubacteria, archaebacteria, plants, insects and vertebrates (2,4,5). CPD photolyase genes have been isolated from many sources (2,5). According to their amino acid sequences, CPD photolyases are classified into two classes, class I and class II (5,6). The amino acid sequences of the class I photolyases are highly diverged from those of the class II photolyases. Reflecting the sequence differences, the divergence between the classes is considered to be ancient. The differences in sequence between the class I and class II enzymes and the functional/structural implications were discussed by Kanai et al. (6). In contrast to the CPD photolyases, the (6–4) photolyases have only been found thus far in some higher eukaryotes (3,7–9). Interestingly, despite their substrate specificity, the (6–4) photolyases show strong similarities to the class I CPD photolyases and belong to a large cluster of the photolyase/cryptochrome family (8).
Photolyases contain stoichiometric amounts of two non-covalently bound chromophores, flavin adenine dinucleotide (FAD) and either methyltetrahydrofolate (MTHF) or 8-hydroxy-5-deazaflavin (8-HDF) as a second chromophore (2). The proposed repair mechanism is as follows: (i) the enzyme binds DNA and recognizes the UV-induced photoproduct; (ii) the second chromophore absorbs a photon in the visible spectral region and transfers the energy to reduced FAD by the Förster mechanism; (iii) the excited FAD transfers an electron to the photoproduct in the DNA strand; (iv) the photoproduct splits and the electron transfers back to FAD to restore the dipyrimidine and the functional form of FAD (2).
With the formation of the ozone shield, photolyases became less important and cells developed alternative DNA repair systems, e.g. excision repair, mismatch repair and recombination (1,4,10). In the lineage to higher eukaryotes, the photolyase genes have been subjected to gene duplication and subsequent functional divergence, giving rise to the cryptochrome/blue light photoreceptors (6,11,12). The amino acid sequences of the cryptochromes are more similar to those of the class I photolyases than the class II photolyases, although they lack any apparent DNA repair activity. Cryptochromes bind similar chromophores and absorb in the blue light region, like the photolyases (13–17). Cashmore et al. suggested that the appearance of animal cryptochromes is independent of that of plant cryptochromes during the course of evolution (11). Cryptochromes were originally identified from plants as blue light receptors (18). Subsequently, cryptochrome homologs, which regulate the circadian rhythm in a light-dependent or light-independent manner, were identified in animals (19–22). Microbial cryptochromes have not been found thus far. Here, we use the term ‘cryptochrome’ as a member of this protein family, which is involved in some type of signal transduction.
Genome projects on various species have revealed the diversity of organisms, whereas the functions of most of the gene products still remain unknown. Total sequencing of the Synechocystis sp. PCC6803 genome by the Kazusa Research Institute revealed two ORFs (slr0854 and sll1629) with similarities to photolyase (23). Hence, both slr0854 and sll1629 show similarities to the class I photolyases as well as the (6–4) photolyases and cryptochromes. The existence of two photolyase-like genes in one bacterium is unique and attracted our attention. Previously, we analyzed the photolyase/cryptochrome family genetically and reported that the divergence of cryptochromes from CPD photolyases occurred before the appearance of eukaryotes (6). In this study we found that slr0854 is a CPD photolyase, but sll1629 is probably a cryptochrome. The existence of cryptochrome-like genes in prokaryotes will be discussed.
MATERIALS AND METHODS
DNA repair assay
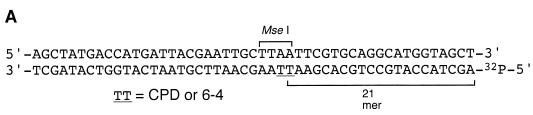
Isotopically labeled d(AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT) oligonucleotides, containing either a CPD or (6–4) photoproduct (TT in the center of the sequence), were prepared as described previously (24–26). One nanomole of oligonucleotide was labeled with [γ-32P]ATP (3000 Ci/mmol) by T4 polynucleotide kinase and was annealed with the complementary strand by heating at 75°C for 10 min and cooling to room temperature for 2–3 h. The labeled duplex DNA, containing a single photoproduct, was purified by agarose gel electrophoresis and used as substrate for the repair assay.
Photoreversal activity was tested by a coupled enzyme assay as described (26). In the assay, the MseI restriction enzyme recognition site, TTAA, which becomes uncleavable due to the photoproduct at the TT site, is restored by the photoreversal reaction. One picomole of oligonucleotides containing a single photoproduct at the MseI site was illuminated for 2 h by daylight fluorescent lamps in 20 µl of 100 mM Tris–HCl, pH 8.0, 1 mM DTT and either 2 µl of cell extract from Synechocystis sp. PCC6803 or 0.5 pM of protein, either slr0854, sll1629, Escherichia coli CPD or Xenopus (6–4) photolyase. The solutions were diluted to 150 µl with 100 mM Tris–HCl, pH 8.0, 1 mM DTT, treated with 150 µl phenol/chloroform (50% phenol, 50% chloroform v/v) and precipitated with ethanol. The oligonucleotides obtained were subjected to MseI digestion and subsequent electrophoresis on a 10% polyacrylamide gel containing 7 M urea.
Cell extracts were prepared by sonication of Synechocystis sp. PCC6803 cells in phosphate-buffered saline (PBS) and centrifugation at 12 000 g for 1 h at 4°C. The supernatant was used for the photoreversal assay. Escherichia coli CPD photolyase and Xenopus (6–4) photolyase were purified as described previously (7,27).
Overexpression and purification of the slr0854 and sll1629 gene products
The slr0854 and sll1629 genes were amplified from a genomic library of Synechocystis sp. PCC6803 by PCR using the following primers with tagged BamHI and SalI restriction sites: 0854N, 5′-GGAAGGATCCATGGGGAGGCAGTTGATT-3′; 0854C, 5′-CTCTGTCGACTTATTTGACCAATTGATAACG-3′; 1629N1, 5′-CGATGTCCATAGCTCTTG-3′; 1629N2, 5′-GAGAGGATCCATGAAACACGTCCCCCCC-3′; 1629C1, 5′-TGGCGTTGATAGCATTGC-3′; 1629C2, 5′-GAGAGTCGACCTAAGCAATAACACCCA-3′.
The resultant fragments were digested with BamHI and SalI and cloned into the BamHI and SalI sites of the pGEX-4T-2 vector (Amersham Pharmacia Biotech) behind the gene for glutathione S-transferase (GST), yielding the plasmids pGEX/slr0854 and pGEX/sll1629, respectively. Escherichia coli SY2 (uvrA–, recA–, phr –) was transformed with either pGEX/slr0854 or pGEX/sll1629 (28) and was grown at 37°C in 3 l of LB medium containing 150 mg/l ampicillin until an OD600 of 0.9–1.0 was reached. Isopropyl β-d-thiogalactopyranoside (IPTG) (0.1 mM) was added to induce expression of the chimeric genes. After incubation at 37°C for 12 h, the cells were spun down and resuspended in 45 ml of PBS. Cell extracts were prepared by sonication of the cells in PBS, followed by centrifugation to remove debris. Cell free extracts were loaded onto a glutathione–Sepharose column. The slr0854 and sll1629 fusion proteins were identified by SDS–PAGE and Coomassie staining. Fractions containing the recombinants were pooled and concentrated with a Centriprep 50 concentrator (Amicon). GST was removed from the sll1629 protein by cleavage with thrombin according to the manufacturer’s instructions (Amersham Pharmacia Biotech). The sll1629 protein was purified further with a heparin–Sepharose column (Amersham Pharmacia Biotech). To detect DNA-binding activity, band shift assays for the sll1629 protein were performed as described previously (26).
The purified proteins were denatured by heating at 75°C for 15 min and the precipitated apoenzymes were removed by centrifugation. The respective supernatants were analyzed by HPLC using a Waters µ Bondasphere 5 µ C18 300 Å column with a 7–13% gradient of acetonitrile in 0.1 M TEAA over 20 min. Authentic FAD was purchased from Tokyo Chemical Industry Co. Ltd.
Photoreversal of UV-induced DNA damage
Escherichia coli strain SY2 (uvrA–, recA–, phr –) was transformed with pGEX/sll1629 or pGEX-4T-2. The transformed cells were grown overnight in LB medium with 150 mg/l ampicillin at 37°C. Expression of pGEX-4T-2 and pGEX/sll1629 was induced by adding IPTG to the medium to a final concentration of 0.1 mM and shaking for 1 h at 37°C. The cells were plated on LB agar and first UV-irradiated with intensities of 0.2, 0.3 and 0.4 J/m2 and subsequently with daylight fluorescence lamps for 1 h as previously described (29). Samples were incubated at 37°C overnight. Surviving colonies were counted the next day. All experiments, except photoreactivation treatments, were performed under yellow light.
Disruption of the slr0854 and sll1629 genes
Based on the Ω cassette plasmid pHP45Ω (30), we constructed two cassette plasmids with additional restriction sites by replacing the spectinomycin resistance gene of pHP45Ω with the chloramphenicol or kanamycin resistance genes. The plasmids were named pCCm and pCKm for the chloramphenicol resistance and kanamycin resistance genes, respectively. The construction of pCCm and pCKm will be described in detail elsewhere.
To disrupt the sll1629 gene, we constructed a plasmid carrying part of the sll1629 gene with the chloramphenicol resistance gene. A 3076 bp StuI–NheI fragment containing the sll1629 gene was isolated from a Is0009 λ phage library (Kazusa DNA Research Institute) and was ligated with the NruI–NheI fragment of pBR322 (TaKaRa). We replaced the XbaI–HpaI fragment of sll1629 in the resultant plasmid with the chloramphenicol resistance gene of pCCm, yielding the plasmid pDeltaSll1629.
To disrupt the slr0854 gene, we constructed a plasmid containing the slr0854 gene in which the kanamycin resistance gene was inserted. The upstream region of slr0854 was amplified by PCR with the primers UpR0854U (5′-CTTGATGGTTTCGCTTC-3′) and UpR0854L (5′-GGATCCGGTAATAACGTTTTCTGC-3′) (underlining indicates the BamHI recognition site) and was ligated with the pGem-T vector (Promega). The downstream region of slr0854 was amplified with the primers LoR0854U (5′-GGATCCAATCAGCAACAACGGGAA-3′) and LoR0854L (5′-TCTGGATTTACATTGGCA-3′) and was ligated with pGem-T along with the upstream region. Then, we inserted both the SpeI–BamHI fragment of the slr0854 downstream region and the kanamycin resistance gene digested from pCKm with BglII into the site between the BamHI and SpeI sites of pGem-T containing the upstream region of slr0854. The resultant plasmid was named pDeltaSlr0854.
Synechocystis sp. PCC6803 is naturally transformable by exogenous DNA (31). By using pDeltaSll1629 and pDeltaSlr0854, we disrupted the slr0854 and sll1629 genes by insertion of the described antibiotic resistance gene by homologous recombination, as described previously (32). Disruption of the sll1629 gene was confirmed by PCR using primers UpL1129U (5′-CTGGCAAAATCGGGGTAG-3′) and LoL1129L (5′-CAACGACAGCAAAGTGGT-3′) and subsequent digestion of the PCR products with NheI. For the slr0854 disruption, the described primers, UpR0854U and LoR0854L, were used and the PCR products were digested with ClaI.
Wild-type and mutant Synechocystis sp. PCC6803 strains were grown at 30°C in BG-11 medium under illumination with white light at an intensity of ~66 µE/m2/s (33).
Determination of the growth rates of Synechocystis sp. PCC6803 mutants
Synechocystis sp. PCC6803 wild-type and mutants were grown in BG-11 medium at 30°C under continuous illumination with white light and shaking until an OD730 of 0.5 was reached. Six- hundred milliliters of BG-11 medium in a Roux flask was then inoculated with ~3 × 107 cells and cultured at 30°C under aerobic conditions and illumination with white light (66 µE/m2/s). The growth rate was measured as the optical density at 730 nm.
UV sensitivity tests of Synechocystis sp. PCC6803
Synechocystis sp. PCC6803 wild-type and mutants were cultured in BG-11 medium under aeration with air and continuous illumination with white light. When an OD730 of ~0.5 was reached, each culture was placed in the dark for 12 h and was then illuminated with red light (27.1 µE/m2/s) for 6 h. Synechocystis cells were washed with BG-11 and resuspended in BG-11 to a density of 2 × 107 cells/ml. All manipulations were carried out under red light conditions to prevent photoreactivation. Seven milliliters of cells in a Petri dish were irradiated with UV light (14.6 W/m2). After UV irradiation, several samples were irradiated with blue light (11.4 µE/m2/s) for 60 min, while control samples were put in the dark. For estimation of cell viability, each sample was diluted, spread on a BG-11 agar plate and cultured under white light (blue light irradiated samples) or red light (dark samples).
Phylogenetic analysis
The amino acid sequence alignment was performed with the program CLUSTALW (34) and the positions of the gaps were slightly modified to avoid their introduction into regions corresponding to α-helix or β-strand, according to the alignment constructed previously (6). Information about the secondary structures was obtained from the X-ray crystal structures of the CPD photolyases (35,36). The genetic distance of every pair of aligned sequences was calculated by the maximum likelihood method (37). Using the distances, an unrooted phylogenetic tree was constructed by the neighbor joining method (38). The reliability of each node of the tree was evaluated by a bootstrap analysis (39) with 1000 tree reconstructions. The software packages PHYLIP (40) and MOLPHY (41) were used for the molecular phylogenetic analysis. TREEVIEW was used for the graphical representation (42).
RESULTS
Photoreversal activity of Synechocystis sp. PCC6803
We detected the photoreversal activity of CPD but not the (6–4) photoproducts in Synechocystis sp. PCC6803 cell extracts by an enzyme coupled repair assay, as shown in Figure 1. This assay is based on light-dependent restoration of the UV-damaged recognition site for the restriction enzyme MseI, as described in Materials and Methods.
Figure 1.
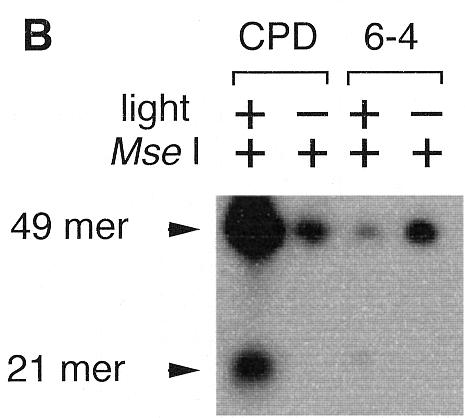
Photoreversal activity of Synechocystis sp. PCC6803. (A) Oligonucleotide sequence containing a photoproduct. (B) Photoenzymatic repair of the CPD and (6–4) photoproducts in the 49mer DNA by a Synechocystis sp. PCC6803 cell extract.
Characterization of the recombinant slr0854 and sll1629 gene products
Stable folded proteins were produced in E.coli by recombinant overexpression of each of the two photolyase-like ORFs of Synechocystis sp. PCC6803 (slr0854 and sll1629) as GST fusion proteins. The recombinant proteins were purified by affinity chromatography on glutathione–Sepharose (Fig. 2A and B). The growth rate of E.coli transformed with pGEX/slr0854 was poor as compared with that of pGEX/slr1629 and the slr0854 recombinant fused with GST was unstable during the incubation with thrombin at room temperature for GST removal. Therefore, the GST-fused slr0854 protein was used for in vitro activity tests, assuming that the fused GST does not prevent photolyase activity. In contrast, sll1629 was efficiently expressed in E.coli. After GST cleavage with thrombin, the protein was purified further with a heparin column and yielded a band on a polyacrylamide gel with an apparent molecular weight of 50 kDa (Fig. 2B).
Figure 2.
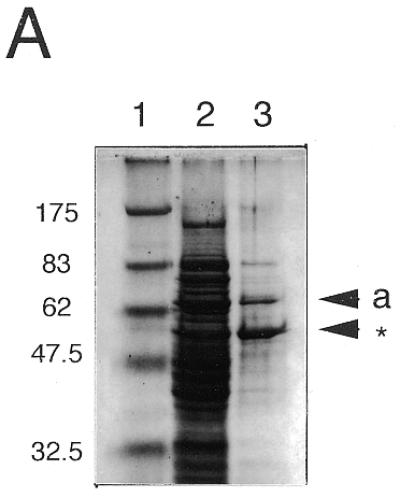
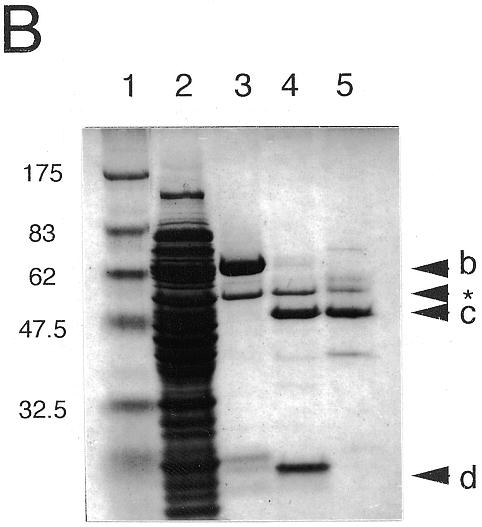
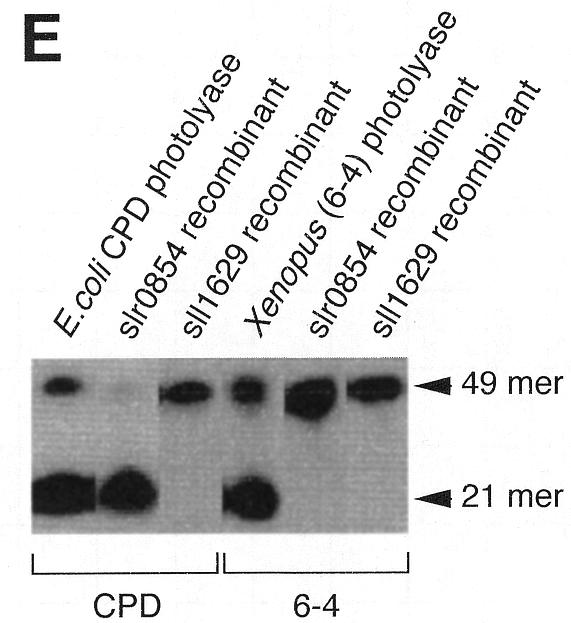
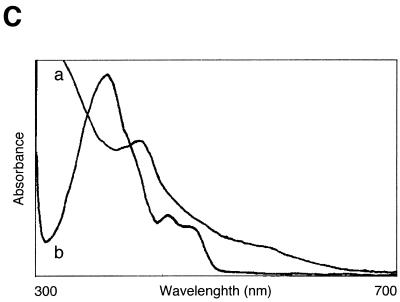
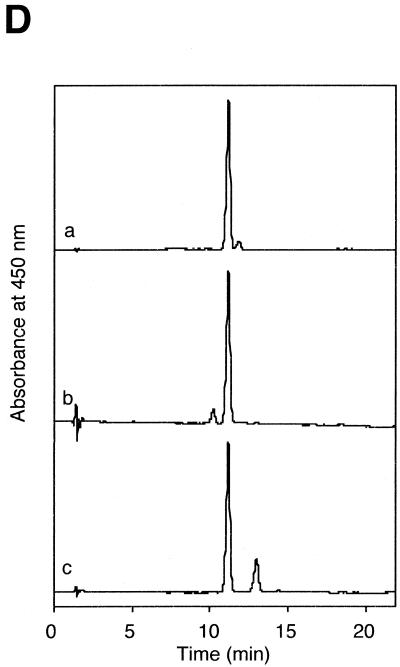
Characterization of the recombinant slr0854 and sll1629 gene products. (A) SDS–polyacrylamide gel (4–20% gradient). Lane 1, molecular weight markers; lane 2, an extract of E.coli pGEX/slr0854; lane 3, semi-purified slr0854 fusion protein from a glutathione–Sepharose column. a, GST-fused slr0854 recombinant. The asterisk shows an unknown protein. (B) SDS–polyacrylamide gel (4–20% gradient). Lane 1, molecular weight markers; lane 2, an E.coli pGEX/sll1629 extract; lane 3, the eluate from a glutathione–Sepharose column; lane 4, cleavage of GST by thrombin; lane 5, purified sll1629 protein after purification on a heparin–Sepharose column. b, GST-fused sll1629 recombinant; c, sll1629 recombinant; d, GST. The asterisk shows an unknown protein. (C) Absorption spectra of the slr0854 fusion protein (a) and the sll1629 protein (b). (D) Analysis of the prosthetic groups of the slr0854 and sll1629 fusion proteins by HPLC. a, standard FAD; b, the slr0854 protein; c, the sll1629 protein. (E) Photoenzymatic repair of photoproducts in a 49mer DNA.
Both the sll1629 protein and the slr0854 fusion protein have absorbance spectra with near-UV/blue light peaks characteristic of flavoproteins (Fig. 2C). Particularly, the sll1629 recombinant has a peak at 450 nm and a shoulder at 470 nm due to fully oxidized FAD, indicating that the FAD is partially oxidized (~7%) in the purified protein. This is in agreement with the fact that the FAD in photolyases, which is reduced in vivo, becomes partially oxidized after purification and shows a peak at 450 nm and a shoulder at 470 nm (7,9). The isolation of prosthetic groups in the supernatant after denaturation indicated that the cofactors are non-covalently bound to the slr0854 and sll1629 fusion proteins. Since the isolated prosthetic groups of the slr0854 and sll1629 proteins were identical to authentic FAD by HPLC analysis, we concluded that both the slr0854 and sll1629 proteins contain FAD as a cofactor (Fig. 2D).
The photoreversal activity of the two recombinant proteins was tested with the same assay as shown in Figure 1. Escherichia coli CPD photolyase and Xenopus (6–4) photolyase were used as controls. The appearance of a 21mer, due to the elimination of CPD, strongly suggested that slr0854 encodes a CPD photolyase (Fig. 2E). In contrast, the sll1629 protein showed no photoreversal activity for either CPD or (6–4) photoproducts (Fig. 2E). The sll1629 recombinant lacks a measurable affinity for DNA containing a CPD or a (6–4) photoproduct, while E.coli CPD photolyase and Xenopus (6–4) photolyase bind to their substrates specifically (data not shown).
UV sensitivity of Synechocystis sp. PCC6803 mutants
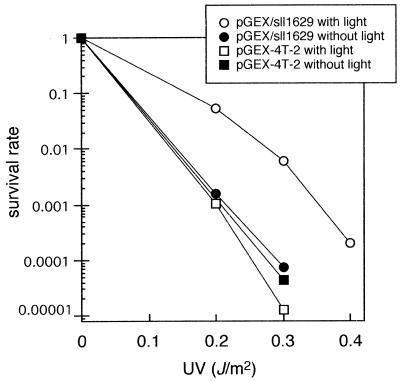
The presence of pGEX/sll1629 in the uvrA–, recA–, phr – host strain E.coli SY2, which is deficient in photoreversal activity for CPDs, resulted in an increase in the UV resistance after photoreactivation, relative to SY2 cells carrying the vector pGEX-4T-2 (Fig. 3). This suggested that sll1629 shows photorepair activity for CPD in vivo.
Figure 3.
Photoreactivation of UV-induced damage in the repair-defective E.coli SY2 (uvrA–, recA–, phr –) strain with pGEX-4T-2 (squares) or pGEX/sll1629 (circles). After UV irradiation the E.coli cells were either kept in the dark (closed symbols) or illuminated with white light (open symbols).
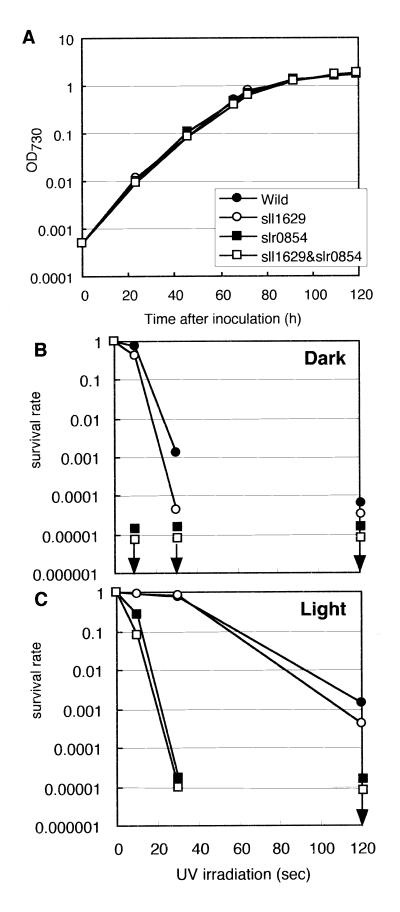
We disrupted slr0854 and sll1629, either alone or together, in order to test the abilities of the slr0854 and sll1629 proteins to repair DNA in situ (Fig. 4). The disruption had no effect on growth rate (Fig. 5A). It has been suggested that both DNA repair systems, excision repair and photoreactivation, exist in Synechocystis sp. PCC6803 (43). Under dark conditions, not only the mutants lacking the photolyase-like gene but also the wild-type were rather UV sensitive at a relatively low dose (30 J/m2), as compared with the wild-type in the light (Fig. 5B). This indicates that photorepair is more important than excision repair for UV-induced DNA lesions in Synechocystis. Mutants lacking the slr0854 gene showed high UV sensitivity. Disruption of the sll1629 gene had little or no effect on UV sensitivity (Fig. 5C). This result is consistent with the photoreversal activity in vitro and suggests that sll1629 is also irrelevant for DNA repair in situ.
Figure 4.
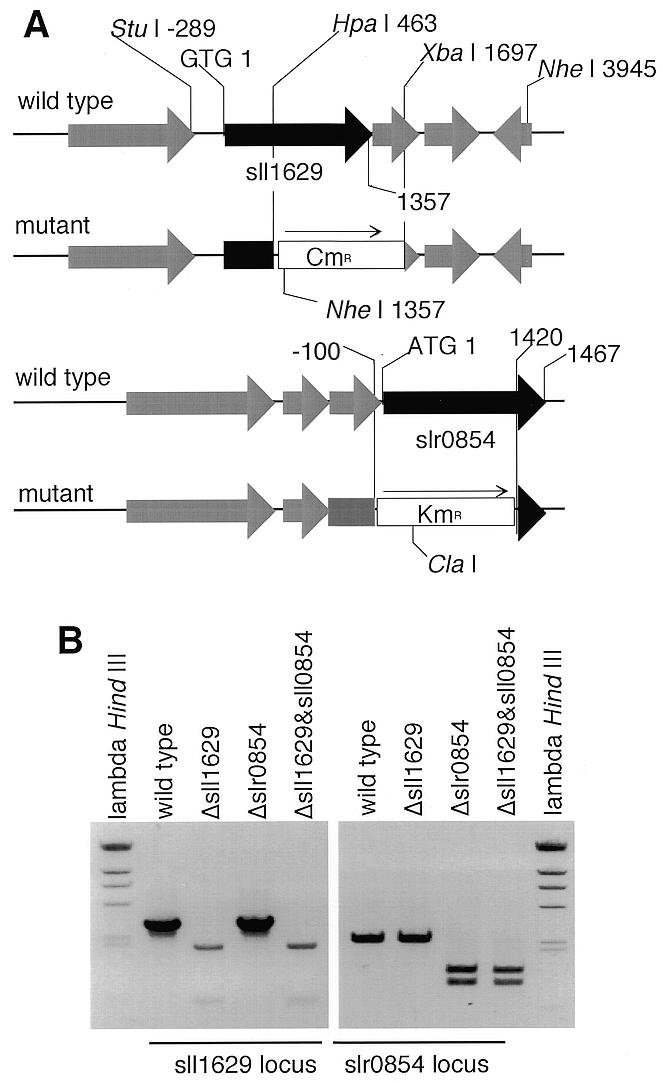
Disruption of the sll1629 and slr0854 genes. (A) The sll1629 and slr0854 loci in wild-type and mutant genomic DNA of the Synechocystis sp. PCC6803 strain. CmR and KmR stand for the chloramphenicol and kanamycin resistance genes, respectively. (B) Agarose gel analysis of PCR products. The region amplified by PCR to check for disruption of sll1629 was –520 to 2430, digested with NheI. To confirm the slr0854 disruption, the region from –505 to 1900 was amplified by PCR and digested with ClaI.
Figure 5.
(A) Growth rate of Synechocystis sp. PCC6803 wild-type and mutants with disrupted slr0829 and sll1629 genes in liquid culture under white light. Cell density was measured as optical density at 730 nm. (B and C) Viability of each strain after UV irradiation. After UV irradiation (14.6 W/m2), samples were incubated in the dark for 60 min and then cultured under red (B) or white light (C). Arrowheads indicate that viability is less than 0.0001 and cannot be measured at the specified irradiation time.
Evolutionary relationship of slr0854 and sll1629 to other members of the photolyase/cryptochrome family
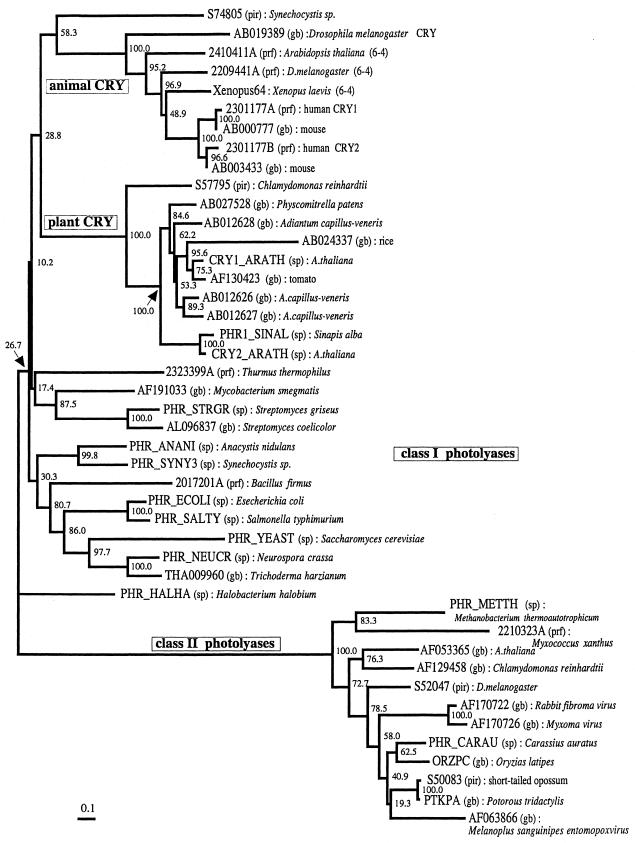
To investigate the evolutionary positions of slr0854 and sll1629 within the photolyase/cryptochrome family, the amino acid sequences of 44 members of the family, including the two homologs from Synechocystis, were aligned (http://www.beri.co.jp/~protein ). Almost the entire regions of the two bacterial homologs showed significant sequence similarity to those of the (6–4) photolyases, the cryptochromes and the class I photolyases, although the bacterial homologs lacked the C-terminal extended regions, which are specific for cryptochromes derived from plants. They also showed sequence similarity to the class II photolyases over the entire primary structure, although the N-terminal regions were highly diverged between them. Seven residues of E.coli CPD photolyase, with side chains that interact with FAD, Tyr222, Thr234, Ser235, Asn341, Arg344, Asp372 and Asn378, were identified through X-ray crystallographic studies (35). It is known that the sites are highly conserved among the members of the photolyase/cryptochrome family (6). Actually, all of the corresponding sites of the two homologs from Synechocystis sp. PCC6803 were either identical or replaced with physicochemically similar residues (Thr→Ser in the sll1629 protein). Based on the alignment, an unrooted phylogenetic tree of 44 proteins was constructed (Fig. 6). The result is quite similar to previously published trees and shows two clusters, one for the class II CPD photolyases and a second one comprising the remaining members (6,11). The latter was further divided into three subclusters. One of them consists of the animal cryptochromes and also includes the (6–4) photolyases. The figure suggests that the (6–4) photolyases have evolved from the cryptochromes. The tree topology is consistent with that constructed by Cashmore et al., although the details differ slightly (11). The second subcluster contains the plant cryptochromes. The third comprises the class I CPD photolyases. These photolyases, however, did not form a single subcluster. Therefore, the third subcluster is defined merely as the set of proteins remaining after definition of the first and second subclusters. It is difficult to discuss the relationships among the three subclusters, because the bootstrap probabilities of the nodes associated with divergence of the three subclusters were too small. However, the bootstrap probabilities for the subclusters of the animal and plant cryptochromes were both 100.0%. Therefore, division into the three subclusters is considered to have statisitcal significance. The tree shows that sll1629 occupies a position relatively close to the animal cryptochromes, while slr0854 belongs to the subcluster of classical class I photolyases, like other microbial photolyases. As shown in Figure 6, slr0854 is closely related to a class I CPD photolyase homolog from Aspergillus nidulans with 100.0% bootstrap probability. In contrast, the position of sll1629 has low bootstrap probability (57.8%). To confirm the location of sll1629 in the tree, we tried a quartet test by the maximum likelihood method. In the test, sll1629 was used as the first operational taxonomic unit (OTU) and the second OTU was selected from the subcluster of animal cryptochromes. The third OTU was selected from the subcluster of plant cryptochromes and the fourth OTU from the subcluster of class I CPD photolyases. These results also suggest that sll1629 is closely related to the animal cryptochromes and the (6–4) photolyases, rather than the plant cryptochromes and the class I CPD photolyases (data not shown). This observation is consistent with the biological properties of the slr0854 and sll1629 proteins in vitro and in vivo.
Figure 6.
An unrooted phylogenetic tree of the photolyase/cryptochrome family. The tree was constructed according to the amino acid sequence alignment of 44 members of the family. The number associated with each node indicates the bootstrap probability of the cluster at the node. Instead of the citations of sequence data, the ID codes and the corresponding databases for the sequence data are shown as the leaves of the tree. The abbreviated names for the databases are shown in the figure, where pir, prf, sp and gb indicate PIR, PRF, SwissProt and GenBank, respectively. The animal CRY cluster comprises not only cryptochromes but also (6–4) photolyases. To distinguish them in the cluster, (6–4) or CRY is attached to the names of the species only when the data have been identified as (6–4) photolyase or cryptochrome. The scale bar under the tree indicates the branch length, corresponding to 0.1 amino acid substitutions per site.
The phylogenetic tree shown in Figure 6 includes three photolyase homologs derived from viruses. This is the first report on the phylogenetic positions of the viral members of the photolyase/cryptochrome family. The family Poxviridae contains two subfamilies; the Entomopoxvirinae, pox viruses of insects, and the Chrodopoxvirinae, poxviruses of vertebrates. One of the three viral homologs is derived from an entomopoxvirus that infects the North American grasshopper, Melanoplus sanguinipes, and other important orthopteran pests (44). The other two are derived from closely related chrodopoxviruses, Shope fibroma virus and myxome virus (45,46). Both viruses infect rabbits. As shown in Figure 6, the homologs derived from poxviruses belong to class II and are distantly related to the two homologs from Synechocystis. Here, we will not enter into the details of the evolution of viral homologs and their functions, because information on the viral proteins is too scant for further discussion.
DISCUSSION
In order to determine whether Synechocystis sp. PCC6803 can repair CPD, (6–4) photoproducts or both, we tested the photoreactivity of a cell-free extract. As shown in Figure 1, only photoreactivity for CPD was detected and the (6–4) photoproduct was not repaired light-dependently. This result suggested that at least one of the products of slr0854 and sll1629 functions as a CPD photolyase. To clarify this, we overexpressed the slr0854 and sll1629 genes in E.coli and characterized their products. Since the slr0854 and sll1629 proteins show no remarkable absorption of visible light, we conclude that neither the slr0854 nor the sll1629 recombinant protein has a stoichiometric second chromophore, like MTHF or 8-HDF, which is not essential for photolyase function. On the contrary, the absorption spectrum with bands in the blue light region near 400 nm and the HPLC analysis indicated that both slr0854 and sll1629 contain FAD as a cofactor, like photolyases and cryptochromes (Fig. 2C and D). Hence, we could demonstrate that the slr0854 protein photoreversed CPD specifically. In contrast, the sll1629 recombinant did not show photoreversal activty in vitro (Fig. 2E) and lacked measurable binding affinity for photodamaged DNA.
On the other hand, expression of the sll1629 gene in E.coli SY2 cells, which lack the gene for CPD photolyase, conveyed slight protection against UV irradiation, unlike other cryptochromes (Fig. 3; 13–17). The gene product of sll1629 does not seem to lack DNA repair capability completely but retains slight photoreversal activity for CPDs in vivo. To investigate the photoreactivity in situ, we disrupted the slr0854 and sll1629 genes (Fig. 4). Only a mutant lacking the slr0854 gene exhibited UV sensitivity, whereas disruption of sll1629 had little or no effect on UV sensitivity (Fig. 5C). The results were consistent with the photoreactivity in vitro, as shown in Figure 2D, indicating that the repair activity of the sll1629 protein is negligible in situ, despite the slight photoreversal activity in vivo. These observations suggest that sll1629 is different from the other class I CPD photolyases in microbes, but that slr0854 is a CPD photolyase.
A phylogenetic tree obtained from an amino acid sequence comparison showed that sll1629 is relatively close to the cryptchromes, rather than class I CPD photolyase, while slr0854 belongs to the subcluster of class I CPD photolyases (Fig. 6). The (6–4) photolyases are also involved in the subcluster of animal cryptochromes. It is known that a certain consensus sequence is required for (6–4) photolyase in order to arrange a hydroxyl or amino group on the 5′-side of the (6–4) photoproduct (Hitomi et al., unpublished data). The sll1629 protein lacks this sequence. In addition, the sll1629 protein lacks affinity and photoreversal activity for the (6–4) photoproduct, as described above. Our experimental results and the phylogenetic analysis suggest that sll1629 may encode a bacterial cryptochrome, although its function as a blue light receptor has yet to be demonstrated.
Plant cryptochromes regulate plant growth and development, while animal cryptochromes participate in circadian clocks (18–22). Nevertheless, all cryptochromes exhibit similarities to class I CPD photolyase. Thus far, two crystal structures of class I CPD photolyases have been solved, but there is no crystal structure of a cryptochrome at present (35,36). However, cryptochromes have 20–25% sequence identity with class I CPD photolyases and the two crystal structures of the CPD photolyases reveal that the FAD binding sites are well conserved, not only in photolyases but also in cryptochromes. Hence, the FAD binding sites identified from the crystal structure of the CPD photolyases were also conserved in both slr0854 and sll1629, which is consistent with the observation that both the slr0854 and sll1629 recombinants are flavoproteins, as shown in Figure 2C and D.
The primary structural features that distinguish cryptochromes from photolyases remain unknown. Therefore, it is difficult to predict whether the sll1629 product is a photolyase or a cryptochrome from the amino acid sequence. As shown in Figure 6, however, our phylogenetic study demonstrated that the sll1629 product is closer to the animal cryptochromes and their relatives than any other members of the family. It is known that the functional mechanism is diverse, even among the members of the animal cryptochromes. Recently it was reported that mammalian cryptochromes do not function as light receptors in the circadian clock (47,48), while the Drosophila cryptochrome sequesters TIMELESS light-dependently (49). The Drosophila TIMELESS protein is involved in the circadian clock, consisting of negative feedback loops of gene regulation that facilitate adaptation to cycles of light and darkness (49). Cyanobacteria are the simplest organisms known to have a circadian clock (50). Unlike insects and vertebrates, cyanobacteria have a gene cluster, kaiABC, as a circadian feedback element (50). The negative feedback control of kaiC expression by KaiC generates a circadian oscillation in cyanobacteria, and KaiA sustains the oscillation by enhancing kaiC expression. However, disruption of either the slr0854 or sll1629 gene did not affect the circadian rhythm (data not shown). Given the similarities between Synechocystis and plants as photosynthetic organisms, sll1629 may have a function similar to that of the plant cryptochromes. In higher plants, phytochromes and cryptochromes are known to be involved in entrainment (51). In fact, the Synechocystis sp. PCC6803 chromosome sequence includes an ORF that appears to encode a protein with similarities to a plant phytochrome (52–54). Phytochrome photoreceptors are almost certainly ubiquitous in the plant kingdom, as well as cryptochromes, and regulate numerous developmental processes throughout their life cycle (Table 1; 55). Interestingly, Wilde et al. isolated a gene designated plpA (phytochrome-like protein) from Synechocystis sp. PCC6803 and reported that the interruption or partial deletion of plpA yielded mutants unable to grow under blue light (54). Two different types of interactions between cryptochromes and phytochromes have been reported. An Arabidopsis cryptochrome conferring responsivity to the blue light receptor interacts with a phytochrome (56). As described above, plant cryptochromes specifically possess additional amino acid sequences in the C-terminal region. The C-terminal extended region of the Arabidopsis cryptochrome is phosphorylated by a phytochrome-associated kinase activity. On the other hand, expression of a cryptochrome from the fern Adiatum capillus-veneris, which lacks the C-terminal extended region, is regulated by a phytochrome in a light-dependent manner (57). Unlike the plant cryptochromes, the sll1629 product lacks the C-terminal region, while it is known that several genes encoding phytochrome-like proteins are present within the Synechocystis genome and that some of them work in bacteria, as discussed above. The interaction between the sll1629 product and the phytochrome-like proteins encoded by the Synechocystis genome may be similar to that observed in the fern. However, our knowledge about the interaction between cryptochromes and phytochromes in cyanobacteria is too sparse to make a definite statement on this problem at this stage.
Table 1. Cryptochrome and phytochrome homologs of photosynthetic organisms.
| Cryptochrome | References | Phytochrome | References | |
|---|---|---|---|---|
| Synechocystis sp. PCC6803 | sll1629 | This paper | slr0473 | 53 |
| plpA (sll1124) | 54 | |||
| Adiantum capillus-veneris | CRY 1–5 | 57,58 | PHY 1–3 | 60–62 |
| Arabidopsis thaliana | HY 4 (CRY 1) | 18 | PHY A–E | 63 |
| CRY 2 | 59 |
We must await further experiments to decipher the actual function of sll1629 and to conclude that the product is a cryptochrome. However, we previously reported that divergence between the photolyases and the cryptochromes occurred before the appearance of eukaryotes (6). Therefore, the existence of a gene like sll1629 in a prokaryote is plausible, although we cannot exclude the possibility of the introduction of sll1629 by horizontal gene transfer. Considering that the mechanism of conversion of the light signal via FAD by cryptochromes still remains unknown, the discovery of a cryptochrome in a simple prokaryote such as Synechocystis is quite significant and may help to decipher the still unknown reaction mechanism of the cryptochromes.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs Ronald Brudler and Elizabeth D. Getzoff for insightful discussions and critical reading of the manuscript and Dr Sang-Tae Kim, Yoshie Fujiwara, Yuri Kobayashi and Jun Hirayama for their invaluable help. M.I. thanks Dr Kazuo Yamamoto (Tohoku University) for helpful discussion. This study was supported by a grant from the Japanese Ministry of Education, Science and Culture (No. 11304055) and from the BRAIN (SEIKINKIKO).
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis, American Society for Microbiology, Washington, DC.
- 2.Sancar A. (1994) Biochemistry, 33, 2–9. [DOI] [PubMed] [Google Scholar]
- 3.Todo T., Takemori,H., Ryo,H., Ihara,M., Matsunaga,T., Nikaido,O., Sato,K. and Nomura,T. (1993) Nature, 361, 371–374. [DOI] [PubMed] [Google Scholar]
- 4.Kato T. Jr, Todo,T., Ayaki,H., Ishizaki,K., Morita,T., Mitra,S. and Ikenaga,M. (1994) Nucleic Acids Res., 22, 4119–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasui A., Eker,A.P., Yasuhira,S., Yajima,H., Kobayashi,T., Takao,M. and Oikawa,A. (1994) EMBO J., 13, 6143–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanai S., Kikuno,R., Toh,H., Ryo,H. and Todo,T. (1997) J. Mol. Evol., 45, 535–548. [DOI] [PubMed] [Google Scholar]
- 7.Todo T., Kim,S.T., Hitomi,K., Otoshi,E., Inui,T., Morioka,H., Kobayashi,H., Ohtsuka,E., Toh,H. and Ikenaga,M. (1997) Nucleic Acids Res., 25, 764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todo T., Ryo,H., Yamamoto,K., Toh,H., Inui,T., Ayaki,H., Nomura,T. and Ikenaga,M. (1996) Science, 272, 109–112. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima S., Sugiyama,M., Iwai,S., Hitomi,K., Otoshi,E., Kim,S.T., Jiang,C.Z., Todo,T., Britt,A.B. and Yamamoto,K. (1998) Nucleic Acids Res., 15, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y.F., Kim,S.T. and Sancar,A. (1993) Proc. Natl Acad. Sci. USA, 90, 4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cashmore A.R., Jarillo,J.A., Wu,Y.J. and Liu,D. (1999) Science, 284, 760–765. [DOI] [PubMed] [Google Scholar]
- 12.Todo T. (1999) Mutat. Res., 434, 89–97. [DOI] [PubMed] [Google Scholar]
- 13.Lin C., Robertson,D.E., Ahmad,M., Raibekas,A.A., Jorns,M.S., Dutton,P.L. and Cashmore,A.R. (1995) Science, 269, 968–970. [DOI] [PubMed] [Google Scholar]
- 14.Malhotra K., Kim,S.T., Batschauer,A., Dawut,L. and Sancar,A. (1995) Biochemistry, 34, 6892–6899. [DOI] [PubMed] [Google Scholar]
- 15.Todo T., Tsuji,H., Otoshi,E., Hitomi,K., Kim,S.T. and Ikenaga,M. (1997) Mutat. Res., 384, 195–204. [DOI] [PubMed] [Google Scholar]
- 16.Selby C.P. and Sancar,A. (1999) Photochem. Photobiol., 69, 105–107. [PubMed] [Google Scholar]
- 17.Okano S., Kanno,S., Takao,M., Eker,A.P., Isono,K., Tsukahara,Y. and Yasui,A. (1999) Photochem. Photobiol., 69, 108–113. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad M. and Cashmore,A.R. (1993) Nature, 366, 162–166. [DOI] [PubMed] [Google Scholar]
- 19.Thresher R.J., Vitaterna,M.H., Miyamoto,Y., Kazantsev,A., Hsu,D.S., Petit,C., Selby,C.P., Dawut,L., Smithies,O., Takahashi,J.S. and Sancar,A. (1998) Science, 282, 1490–1494. [DOI] [PubMed] [Google Scholar]
- 20.Emery P., So,W.V., Kaneko,M., Hall,J.C. and Rosbash,M. (1998) Cell, 95, 669–679. [DOI] [PubMed] [Google Scholar]
- 21.Stanewsky R., Kaneko,M., Emery,P., Beretta,B., Wager-Smith,K., Kay,S.A., Rosbash,M. and Hall,J.C. (1998) Cell, 95, 681–692. [DOI] [PubMed] [Google Scholar]
- 22.van der Horst G.T., Muijtjens,M., Kobayashi,K., Takano,R., Kanno,S., Takao,M., de Wit,J., Verkerk,A., Eker,A.P., van Leenen,D., Buijs,R., Bootsma,D., Hoeijmakers,J.H. and Yasui,A. (1999) Nature, 398, 627–630. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko T., Sato,S., Kotani,H., Tanaka,A., Asamizu,E., Nakamura,Y., Miyajima,N., Hirosawa,M., Sugiura,M., Sasamoto,S., Kimura,T., Hosouchi,T., Matsuno,A., Muraki,A., Nakazaki,N., Naruo,K., Okumura,S., Shimpo,S., Takeuchi,C., Wada,T., Watanabe,A., Yamada,M., Yasuda,M. and Tabata,S. (1996) DNA Res., 3, 109–136. [DOI] [PubMed] [Google Scholar]
- 24.Taylor J.-S., Brockie,I.R. and O’Day,C.L. (1987) J. Am. Chem. Soc., 109, 6735–6742. [Google Scholar]
- 25.Iwai S., Shimizu,M., Kamiya,H. and Ohtsuka,E. (1996) J. Am. Chem. Soc., 118, 7642–7643. [Google Scholar]
- 26.Hitomi K., Kim,S.T., Iwai,S., Harima,N., Otoshi,E., Ikenaga,M. and Todo,T. (1997) J. Biol. Chem., 272, 32591–32598. [DOI] [PubMed] [Google Scholar]
- 27.Sancar A., Smith,F.W. and Sancar,G.B. (1984) J. Biol. Chem., 259, 6028–6032. [PubMed] [Google Scholar]
- 28.Yasuhira S. and Yasui,A. (1992) J. Biol. Chem., 267, 25644–25647. [PubMed] [Google Scholar]
- 29.Yamamoto K. (1985) Mol. Gen. Genet., 201, 141–145. [DOI] [PubMed] [Google Scholar]
- 30.Prentki P. and Krisch,H.M. (1984) Gene, 29, 303–313. [DOI] [PubMed] [Google Scholar]
- 31.Grigorieva G. and Shestakov,S.V. (1982) FEMS Microbiol. Lett., 13, 367–370. [Google Scholar]
- 32.Williams J.G.K., (1988) Methods Enzymol., 167, 766–778. [Google Scholar]
- 33.Rippka R., Deruellus,J., Waterbury,J.B., Herdman,M. and Stanier,R.Y. (1979) J. Gen. Microbiol., 111, 1–16. [Google Scholar]
- 34.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H.W., Kim,S.T., Sancar,A. and Deisenhofer,J. (1995) Science, 268, 1866–1872. [DOI] [PubMed] [Google Scholar]
- 36.Tamada T., Kitadokoro,K., Higuchi,Y., Inaka,K., Yasui,A., de Ruiter,P.E., Eker,A.P. and Miki,K. (1997) Nature Struct. Biol., 4, 887–891. [DOI] [PubMed] [Google Scholar]
- 37.Felsenstein J. (1996) Methods Enzymol., 266, 418–427. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N. and Nei,M. (1987) Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. (1985) Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. (1993) PHYLIP (phylogeny inference package) v.3.5c. Department of Genetics, University of Washington, Seattle, WA.
- 41.Adachi J. and Hasegawa,M. (1996) MOLPHY (programs for molecular phylogenetics) v.2.3b3. Institute of Statistical Mathematics, Tokyo, Japan.
- 42.Page R.D. (1996) Comp. Appl. Biosci., 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien P.A. and Houghton,J.A. (1982) Photochem. Photobiol., 35, 359–364. [DOI] [PubMed] [Google Scholar]
- 44.Afonso C.L., Tulman,E.R., Lu,Z., Oma,E., Kutish,G.F. and Rock,D.L. (1999) J. Virol., 73, 533–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer D.O., McFadden,G. and Evans,D.H. (1999) Virology, 264, 319–343. [DOI] [PubMed] [Google Scholar]
- 46.Cameron C., Hota-Mitchell,S., Chen,L., Barrett,J., Cao,J.X., Macaulay,C., Willer,D., Evans,D. and McFadden,G. (1999) Virology, 264, 298–318. [DOI] [PubMed] [Google Scholar]
- 47.Griffin E.A.,Jr, Staknis,D. and Weitz,C.J. (1999) Science, 286, 768–771. [DOI] [PubMed] [Google Scholar]
- 48.Okamura H., Miyake,S., Sumi. Y., Yamaguchi,S., Yasui,A., Muijtjens,M., Hoeijmakers,J.H. and van der Horst,G.T. (1999) Science, 286, 2531–2534. [DOI] [PubMed] [Google Scholar]
- 49.Ceriani M.F., Darlington,T.K., Staknis,D., Mas,P., Petti,A.A., Weitz,C.J. and Kay,S.A. (1999) Science, 285, 553–556. [DOI] [PubMed] [Google Scholar]
- 50.Ishiura M., Kutsuna,S., Aoki,S., Iwasaki,H., Andersson,C.R., Tanabe,A., Golden,S.S., Johnson,C.H. and Kondo,T. (1998) Science, 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- 51.Somers D.E., Devlin,P.E. and Kay,S.A. (1998) Science, 282, 1488–1490. [DOI] [PubMed] [Google Scholar]
- 52.Hughes J., Lamparter,T., Mittmann,F., Hartmann,E., Gartner,W., Wilde,A. and Borner,T. (1997) Nature, 386, 663. [DOI] [PubMed] [Google Scholar]
- 53.Lamparter T., Mittmann,F., Gartner,W., Borner,T., Hartmann,E. and Hughes,J. (1997) Proc. Natl Acad. Sci. USA, 94, 11792–11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilde A., Churin,Y., Schubert,H. and Börner,T. (1997) FEBS Lett., 406, 89–92. [DOI] [PubMed] [Google Scholar]
- 55.Mathews S. and Donoghue,M.J. (1999) Science, 286, 947–950. [DOI] [PubMed] [Google Scholar]
- 56.Ahmad M., Jarillo,J.A., Smirnova,O. and Cashmore A.R. (1998) Mol. Cell, 1, 939–948. [DOI] [PubMed] [Google Scholar]
- 57.Imaizumi T., Kanegae,T. and Wada,M. (2000) Plant Cell, 12, 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanegae T. and Wada,M. (1998) Mol. Gen. Genet., 259, 345–353. [DOI] [PubMed] [Google Scholar]
- 59.Lin C., Yang,H., Guo,H., Mockler,T., Chen,J. and Cashmore,A.R. (1998) Proc. Natl Acad. Sci. USA, 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okamoto H., Hirano,Y., Abe,H., Tomizawa,K., Furuya,M. and Wada,M. (1993) Plant Cell Physiol., 34, 1329–1334. [Google Scholar]
- 61.Nozue K., Kanegae,T., Imaizumi,T., Fukuda,S., Okamoto,H., Yeh,K.C., Lagarias,J.C. and Wada,M. (1998) Proc. Natl Acad. Sci. USA, 95, 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nozue K., Fukuda,S., Kanegae,T. and Wada,M. (1998) Plant Physiol., 118, 712. [Google Scholar]
- 63.Elich T.D. and Chory,J. (1997) Cell, 91, 713–716. [DOI] [PubMed] [Google Scholar]