Abstract
Chemical analysis by analytical instrumentation has played a major role in diseases diagnosis, which is a necessary step for diseases treatment. While the treatment process often targets specific organs or compounds, the diagnostic step can occur through various means, including physical or chemical examination. Chemically, the genome may be evaluated to give information about potential genetic outcomes, the transcriptome to provide information about expression actively occurring, the proteome to offer insight on functions causing metabolite expression, or the metabolome to provide a picture of both past and ongoing physiological function in the body. Mass spectrometry (MS) has been elevated among other analytical instrumentation because it can be used to evaluate all four biological machineries of the body. In addition, MS provides enhanced sensitivity, selectivity, versatility, and speed for rapid turnaround time, qualities that are important for instance in clinical procedures involving the diagnosis of a pediatric patient in intensive care or a cancer patient undergoing surgery. In this review, we provide a summary of the use of MS to evaluate biomarkers for newborn screening and cancer diagnosis. As many reviews have recently appeared focusing on MS methods and instrumentation for metabolite analysis, we sought to describe the biological basis for many metabolomic and additional omics biomarkers used in newborn screening and how tandem MS methods have recently been applied, in comparison to traditional methods. Similar comparison is done for cancer screening, with emphasis on emerging MS approaches that allow biological fluids, tissues, and breath to be analyzed for the presence of diagnostic metabolites yielding insight for treatment options based on the understanding of prior and current physiological functions of the body.
Keywords: Mass spectrometry, metabolomics, newborn screening, cancer diagnosis
I. Introduction
Modern humans can expect to live longer, healthier lives, due in large part to advancements in chemical instrumentation that have improved the sensitivity, accuracy, speed, and accessibility of medical testing.(Wender et al., 2019) Since the 1900s, there has been a widely reported trend that has shown a ubiquitous increase in life expectancy and life span equality (Kinsella, 1992; Woolf and Schoomaker, 2019; Aburto et al., 2020). A precipitous drop in infantile deaths over the decades has led to a greater equality in life expectancy across the globe (Aburto et al., 2020).
While there are many factors that have influenced this improved quality of life, the current review will focus on diagnostic techniques, specifically those advancements in the field of mass spectrometry that have allowed for enhanced screening for various illnesses at the early stages of the disease. Early disease detection leads to early treatment, which has been shown to be one of the most important factors in affecting prognosis. Indeed, when accurate testing is readily available to patients at the onset of the illness, prognosis and long-term quality of life for patients suffering from cancer (Houssami et al., 2009; Jang et al., 2018; Hawkes, 2019), Alzheimer’s disease (Chu, 2012; Rasmussen and Langerman, 2019; Al-Chalabi, 2021), Diabetes (Herman et al., 2015; Thornton Snider et al., 2019), cystic fibrosis (Kerem et al., 1992; Dankert-Roelse and te Meerman, 1995; Dankert-Roelse and Mérelle, 2005; Breuer et al., 2018), among many others (Geelhoed et al., 2005; Howard, 2005; van der Plas et al., 2011; Lê et al., 2018; Sandler et al., 2018), starkly increase.
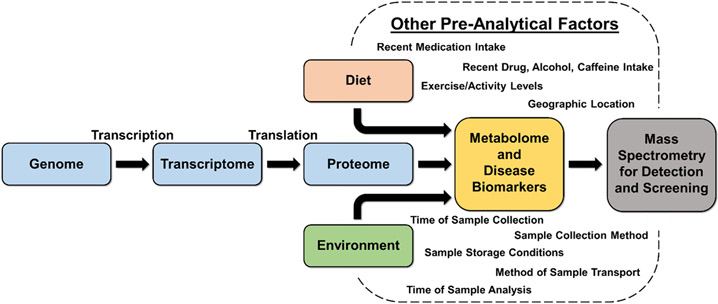
Some of the diseases that are fatal or are associated with lifelong, uncurable side effects are those that humans are genetically predisposed to (e.g., cystic fibrosis (Gadsby et al., 2006), phenylketonuria (Knappskog et al., 1995)), while others are predominantly influenced by dietary and environmental factors (e.g., emphysema (Taraseviciene-Stewart and Voelkel, 2008), obesity (Wright and Aronne, 2012), certain types of cancer (Godtfredsen, 2005)). Still, other diseases can arise from the combination of genetic, environmental, and dietary factors (e.g., diabetes (Riserus et al., 2009; Langenberg and Lotta, 2018) and asthma (Cockcroft, 2018; Morales and Duffy, 2019)). Genetic diseases are often diagnosed at-birth, during neonatal screening, whereas other diseases that onset later in life can be monitored and diagnosed with routine exams. Regardless of the cause of diseases, whether it be some combination of genetic, environmental, or dietary factors, each is characterized by an alteration of the body’s biology, which is driven by more fundamental biochemical deviations. These chemical abnormalities are the targets for current screening and diagnostic platforms. The targets for various analytical methods differ but must have direct link to any of the four body machineries, including the genome, transcriptome, proteome, and metabolome, as illustrated in Figure 1. The metabolome represents the most downstream stage in molecular biology and thus have higher significance in early disease detection. Given that each disease manifests itself differently, the associated small molecule metabolites are also expected to be different, being up or down regulated when compared with the absence of the diseases. Therefore, there is a great demand for a variety of highly sensitive and accurate analytical tools to provide rapid screening and accurate detection of metabolites in various biological samples (biofluids and tissues) for diagnostic purposes.
Figure 1:
Schematic illustration of factors leading to alteration of the body’s biology giving rise to metabolites and disease biomarkers which are the targets of current mass spectrometric screening and diagnostic platforms along with a collection of pre-analytical factors affecting disease screening and diagnosis, thus demanding highly sensitive and accurate methods.
In general, analytical techniques used for disease screening include visual methods that are based on chemical tests, assays based on molecular recognition, separation methods combined with various online spectroscopic techniques, nuclear magnetic resonance, and mass spectrometry. Perhaps the most common diagnostic tool is the use of colorimetric detection, including those used to differentiate tissues (specifically the differentiation of cancerous versus non-cancerous tissues). Recent advances have made great strides in digitization through the use of artificial intelligence and/or handheld devices, such as cellphones (Evered and Dudding, 2011; Holmström et al., 2021; Joh et al., 2021; Tang et al., 2021) that have transformed visual-based analytical methods. Tissue samples can be taken via biopsies, which may be warranted depending on the preliminary results of other screening tests, such as radiological tests (e.g., X-ray, ultrasound, magnetic resonance imaging), colonoscopies, or simple, non-invasive surface inspection (e.g., changes in skin pigmentation). While the collection of these methods is considered the standard for the diagnosis of cancer, there are instances where the visual diagnostic methods cannot be made. In some cases, speed (i.e., turnaround time) is of utmost importance requiring the use of a more rapid alternative method. Since changes to biology are expected at the onset of the disease, the analysis of biofluids can also provide a useful and less invasive approach to tissue sampling. The small changes in the biochemistry of the patient make it imperative to develop highly sensitive chemical instrumentation for diseases testing.
Optical density (via spectrophotometer) or fluorescence (via microplate fluorimeter) signal transduction methods have been developed for molecular assay. These assays are typically performed on a solid substrate where a “capture” molecule previously immobilized on the surface is used to react selectively with the analyte of interest. If the analyte of interest was present in the sample, then the optical density or fluorescence is monitored to yield a diagnostic signal indicating a positive test. Multiple commercial assays have been developed over the past couple of decades for the quantification of various classes of molecules including lipids (Li et al., 2018; Visnovitz et al., 2019) and proteins (most notably, antibodies via Enzyme-Linked Immunosorbent Assay, or ELISA) (Wu et al., 2019; Beavis et al., 2020; Van Elslande et al., 2020). While immunoassays are incredibly sensitive, highly selective, and generally low-cost, they often provide targeted analysis for specific molecules or classes of molecules, and are subject to both false positive and false negative results (O’Kennedy et al., 2017). While fluorescence is a good detection method for molecular recognition assays, it too can stand as an independent analytical technique for clinical purposes. For example, fluorimetry has been found useful in tissue pathology, specifically in oncology, cardiology, ophthalmology, and neurology (Shahzad et al., 2010; Marcu, 2012). Fluorimetry offers high sensitivity and selectivity but suffers from external factors such as temperature and pH that influence fluorescence measurements (Marcu, 2012), including often unavoidable background noise that tend to reduce signal-to-noise ratio.
Measurement approaches based on mass spectrometry (MS) have been found to provide a wealth of molecular information regarding biological samples in a single experiment and have recently proven to be a valuable tool in clinical settings. The increase in popularity of MS in recent years in disease detection can be largely attributed to the developments made in ion sources, mass analyzers, tandem MS (MS/MS) scan modes, and effective software development. Mass spectrometers can be easily coupled to separation techniques for the facile introduction of samples of complex mixtures, such as gas chromatography (GC-MS), liquid chromatography (LC-MS), and capillary electrophoresis (Carroll et al., 1975; Maxwell and Chen, 2008). The MS technique can also be used independently with a variety of ionization methods, such as matrix assisted laser desorption ionization (MALDI) (Karas and Hillenkamp, 1988), electrospray ionization (ESI) (Fenn et al., 1989), atmospheric pressure chemical ionization (APCI) (Carroll et al., 1974, 1975), or atmospheric pressure photoionization ionization (APPI) (Hanold et al., 2004). One of the most recent innovations in MS involves ambient ionization, which allows direct analysis of the complex samples without front-end separation or sample pre-treatment. With the availability of various ion sources and mass analyzers, MS can now be applied to analyze any type of biomarker for genome, transcriptome, proteome, and metabolome information.
In this review, we focus mainly on metabolomics analysis. We summarize the biochemistry of various genetic diseases and provide the fundamental basis for using the metabolome for diagnosis. We believe the collection of this information into a single review article will prove valuable to the MS community in terms of new method development and validation. When necessary, the genome and proteome are discussed for selected diseases. Most of the MS methods designed for genetic diseases are based on different tandem MS scan modes. Therefore, these are described in some detail prior to the discussion of the specific diseases. The review then shifts from genetic diseases to cancer diagnosis/screening, also based on metabolomic analysis. In this case, we focus on recent developments in ambient MS where biofluids and tissues can be analyzed in their native state without prior sample preparation. Lastly, we discuss recent advances in exhalome in which exhaled breath is used for disease diagnosis. We note that several review articles have recently appeared that described the chemical characteristics of metabolites such as lipids and their analysis by various MS methods (Heiles, 2021; Xia and Wan). Recent reviews by us ((Frey et al., 2020; Swiner et al., 2020; Lee et al., 2021)) and others ((Zang et al., 2019; Macklin et al., 2020; Ma and Fernández, 2022)) have also discussed advancements in clinical mass spectrometry that cover general methods such as microsampling and pre-analytical techniques, miniaturization, and data analysis for proteomics and metabolomics. By providing the biological relevance of metabolites to various diseases in this review, we hope to add a unique perspective to the existing literature.
II. Emerging Mass Spectrometry Methods for Newborn Disease Screening
One of the clinical applications of metabolomics has involved newborn screening. Perhaps this is not surprising giving that after birth, the neonate must transition from assured continuous transplacental supply of glucose to a variable fat-based fuel economy. The neonate must become accustomed to periodic feeding and fasting, a challenge that is met through well-controlled metabolic and hormonal adaptive changes that ensure a continuing supply of energy. This constitutes the neonatal metabolic adaptation, which when studied carefully can yield diagnostic information for different forms of disorders. This section of the review will focus on the application of MS to screen specific disorders in infants and how MS can be used to obtain molecular information from all levels of the human biochemical machinery.
Recently there has been an increase in the need for newborn clinical screening and diagnostic methodologies with high sensitivity, and specificity, with emphasis on the power of early detection. Early diagnosis is critical in newborn screening (NBS) where early intervention can dictate patient longevity and survival (Etzioni et al., 2003; Solomon et al., 2012). In the past decade, MS has grown to be a powerful tool in the multidisciplinary fields of disease screening– demonstrating superior analytical performance in terms of higher turnaround time (Wu et al., 2022; Zhang et al., 2022), enhanced sensitivity, and specificity (Annesley et al., 2016; Mussap et al., 2018; Adhikari et al., 2020). Easy automation and reduced sample pre-treatment enable MS experiments to be performed in a high throughput fashion (Chen et al., 2022; Davidovics et al., 2022). Modern instruments are capable of high-resolution mass-to-charge measurements as well as tandem MS (MS/MS) methodologies both of which offer unparalleled selectivity (Blevins et al., 2022; Varenina et al., 2022). Femtomolar sensitivity is easily achieved via MS/MS methods due to reduced noise during the mass selection process (Kisiala et al., 2019). Higher sensitivity is important in NBS where sample size is limited, and early detection can mitigate disease progression as discussed time and again within the reports discussed herein. These improvements in sensitivity can be gained with specialized separation methods at the front-end of the mass spectrometer to reduce noise due to coeluting species and ion suppression effects. Such separations-based MS/MS methods have been applied in the NBS of Krabbe disease, Sickle Cell disease, beta-Thalassemia Syndrome, newborn lysosomal disorders, mucopolysaccharidoses, Pompe disease, Fabry disease, metabolomic disorders, and muscular atrophy and will be discussed here.
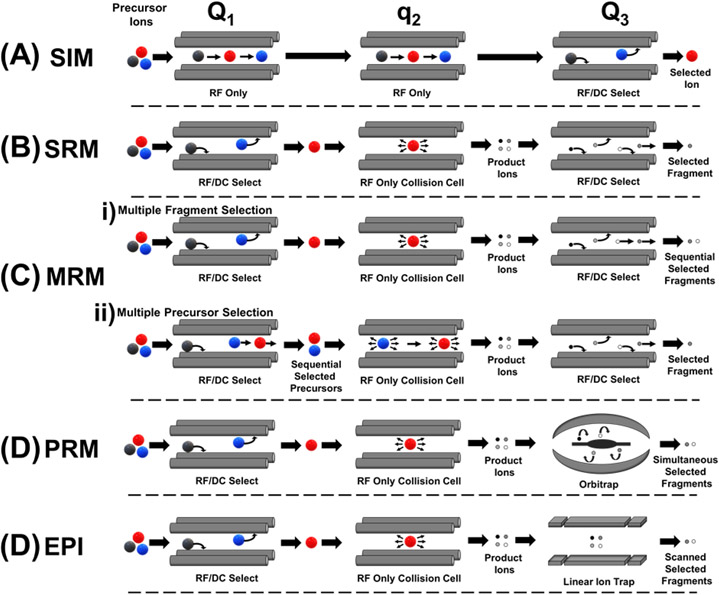
In more specific terms, the enhancements in sensitivity and specificity are due to the application of quantitative MS operational modes such as selected ion monitoring (SIM), single reaction monitoring (SRM), multiple reaction monitoring (MRM), parallel reaction monitoring (PRM), and enhanced product ion (EPI) scanning, as depicted in Figure 2, in conjunction with unique internal standards based on the system of interest. Quantitation of diagnostically useful mass-to-charge (m/z) ranges are afforded by the above methods as the instrument scans only in the desired mass range, omitting those not useful to the analysis which increases time of analysis. Due to the scanning nature of these modes, they are best performed in MS instruments with triple quadrupole mass analyzers where ion selection and fragmentation happens in separate quadrupole analyzers. Figure 2A describes SIM mode in which the quadrupole of the mass spectrometer selects only the ions of interest and omits all other ions and noise, thus increasing accumulation of only the ions of interest by a factor of 10 to 100 on the same analytical timescale (Middleditch and Desiderio, 1973), which inherently decreases analytical detection limit. However, SIM does not structurally target compounds of interest due to the lack of the added dimension of MS/MS. That is, although the ion of interest is selected, no collisional energy is applied. Figure 2B and 2C depict SRM and MRM respectively in which either one targeted ion (SRM) or multiple target ions (MRM) are selected in quadrupole one, fragmented in quadrupole two, and then specific product ions are selected in quadrupole three for detection to give the aforementioned detector time and sensitivity improvements along with the diagnostic MS/MS mode of operation for added compound of interest specificity. MRM may be performed as described above with multiple precursor ion selection (Figure 2Cii) or via selection of one precursor ion in quadrupole 1, collisional dissociation in quadrupole 2, and sequential selection of multiple fragment product ions in quadrupole three. Figure 2D describes PRM which is similar in principle to SRM and MRM techniques, however, this method replaces the third quadrupole with a high resolution orbitrap mass analyzer. As with SRM and MRM, ions of interest are selected and fragmented in quadrupoles one and two, respectively. In lieu of quadrupole three selecting one product ion at a time, the orbitrap analyzer scans all product ions with high resolution and mass accuracy which yields the high sensitivity targeted MS/MS approach of MRM along with decreasing interferences in complex mixtures where isobaric species and background interference can cause false positives – making PRM ideal for assay development (Bourmaud et al., 2016). EPI, depicted in Figure 2E, is an analogous configuration to PRM – however quadrupole three is replaced with a linear ion trap. An additional MS method of note for NBS is ion mobility-mass spectrometry (IMS), which has been shown recently to yield improved figures of merit (e.g., speed and selectivity) in the screening of potential biomarkers of interest (Dodds and Baker, 2021; Mukherjee et al., 2022). IMS separates analytes based on structural features such as size and shape. When coupled with MS/MS, excellent separation and elucidation of target NBS molecules have been shown (Dodds and Baker, 2021). The applications of various MS/MS methods in newborn screening for diagnosing different diseases are discussed below, including Krabbe disease, lysosomal storage disease, and mucopolysaccharidoses.
Figure 2:
Illustration of various MS/MS operational modes on a triple quadrupole and related hybrid instruments for enhanced sensitivity with (A) selected ion monitoring (SIM) in which all precursor ions are sequentially passed through the first two quadrupoles (e.g. Q1 and q2 respectively) with subsequent selection of analyte ion in quadrupole three (Q3) without inducing fragmentation of the selected ions, (B) single reaction monitoring (SRM) in which a single precursor ion of interest is selected in Q1 followed by collisional dissociation in q2 and selection of a single fragment product ion in Q3, (C) illustration of two types of multiple reaction monitoring (MRM) operation: (i) multiple fragment selection mode in which a single ion is selected and fragmented as in SRM and Q3 sequentially selects multiple fragment product ions of interest, and (ii) multiple precursor selection mode in which two or more precursor ions are sequentially selected in Q1, followed by collisional dissociation in q2 and selection of a single fragment product ion that the selected precursor ions have in common, (D) parallel reaction monitoring (PRM) in which Q1 and q2 operation is analogous to SRM but Q3 is replaced with a high resolution orbitrap mass analyzer that simultaneously selects fragment product ions, (E) enhanced product ion (EPI) in which Q1 and q2 have analogous operation to SRM and PRM but Q3 is replaced with a linear ion trap mass analyzer where fragment product ions of interest can be accumulated prior to detection to enhance sensitivity.
A. Krabbe Disease
Globoid cell leukodystrophy, known as Krabbe disease (KD), is a rare and often fatal lysosomal storage disease caused by galactocerebrosidase (GALC) lysosomal enzyme deficiency, which acts to degrade galactolipids in the myelin sheath (Suzuki, 2003). KD is an inherited autosomal recessive disorder, and rapidly progresses in patients who have onset of the disease early in life (Suzuki, 2003). Accelerated disease progression in early onset KD patients have given rise to two subsets generally used to describe infantile KD: early-infantile and late-infantile KD in which symptoms appear before 6 months of age and between 7 to 12 months of age, respectively. Both types of KD bring about similar symptoms but are associated with different survival rates (Escolar et al., 2016). Symptoms of infantile KD are progressive neurologic decline – including seizures, psychomotor regression, hearing loss, vision loss, and death within the first 2-3 years of life in the case of early-infantile KD (Escolar et al., 2016). Patients with late-infantile KD have longer survival rates, while onset of the disease later in life yield varying survival rates. Absence of GALC causes excessive galactolipid accumulation, triggering inflammatory response and demyelination of the nervous system (Escolar et al., 2016). GALC is a hydrolytic lysosome specific to glycolipid selection (Suzuki, 2003). The principal substrate of GALC is galactosylceramide, degradation of which yields demyelination (Suzuki, 2003). Early detection of the disease in newborns is critical as umbilical cord blood and bone marrow transplantation have been effective in suppressing onset of symptoms and increasing longevity (Escolar et al., 2005). Early detection of KD relies on measurement of GALC enzymatic activity, which requires high sensitivity for not only sufficient differentiation between symptomatic and asymptomatic patients, but also to allow pre-symptomatic KD screening which is of utmost importance for early detection and treatment of the disease.
NBS techniques for Krabbe Disease (KD) have shifted from flourimetry to the use of liquid chromatographic (LC)-MS/MS methods for enhanced analytical sensitivity (Liao et al., 2017b). Liao used LC-MS/MS to study the deficiency of GALC via a new assay measuring enzymatic activity. MRM was used to measure small residual amounts of GALC with high accuracy, which gave a 20-fold increase in dynamic range than conventional radiometric assay. The LC-MS/MS method differentiated cells with null GALC activity from those with traces of the enzyme to 0.3% of normal levels. The success of the MS/MS methodology rose from the use of a structurally identical internal standard (GALC-IS) to the GALC product (GALC-P), which was the ion produced via MRM reaction. The assay employed by the authors involved a cocktail of substrate glacatosylceraminde with a 7-carbon heptanoyl chain (GALC-S) and GALC-IS with sodium oleate in a citrate phosphate buffer addition to cell lysate. After incubation and centrifugation, the supernatant was applied to the LC-MS/MS method. The LC method employed a C18 stationary phase and a linear gradient from 50 to 100 % B over 1.5 minutes after which eluent was ionized in a heated ESI source with subsequent mass analysis via a Waters Xevo TQ in positive ion mode. MRM analysis was performed via the expected fragmentations on the GALC-P ion with precursor at m/z 412.38 and product at m/z 264.27, the GALC-IS ion with precursor m/z 417.41 and product m/z 264.27, as well as the GALC-S ion with precursor m/z 574.43 and product m/z 264.20. Analysis was performed using the GM13793 cell line as the high-GALC constituent and cell line GM06805 as the low-GALC constituent as it was from a patient devoid of the enzyme.
The authors performed control assays to validate this method for NBS application, the results of which are summarized in Figure 3. The High Lysate + GALC-S control used GM13793-HIGH as the high enzymatic quality control sample was performed to ensure the MRM response from the high GALC cell lysate indicated no isobaric or coeluting compounds with GALC-P. Enzymatic activity for this control gave 0.58 ± 0.04 nmol/h/mg. The second control labeled “No Lysate Control” in Figure 3 shows GALC-P with no lysate giving 3.6E-4 ± 1.2E-4 nmol/h/mg GALC activity which resulted in 0.07 % of signal observed with lysate demonstrating that GALC-S undergoes no nonenzymatic breakdown to GALC-P. The High Lysate – GALC-S control shows the GM13793-HIGH cell lysate MRM response in the absence of GALC-S. Lastly the Low Lysate + GALC-S control in Figure 3 shows the MRM response for the low GALC cell lysate, which was 10-fold higher than the no lysate control and 0.4 % of the signal attained from the high GALC lysate at 3.7E-3 ± 3.8E-4 nmol/h/mg. This was attributed to non-GALC enzymes in the lysate capable of breaking down GALC-S to GALC-P such as GLB1, β-galactosidase 1, deficiency of which causes two lysosomal storage disorders. In summary, the authors demonstrated that the high GALC cell lysate response was 150-fold higher than the low GALC cell lysate response indicating the importance of a method with high sensitivity and a large analytical range.
Figure 3:
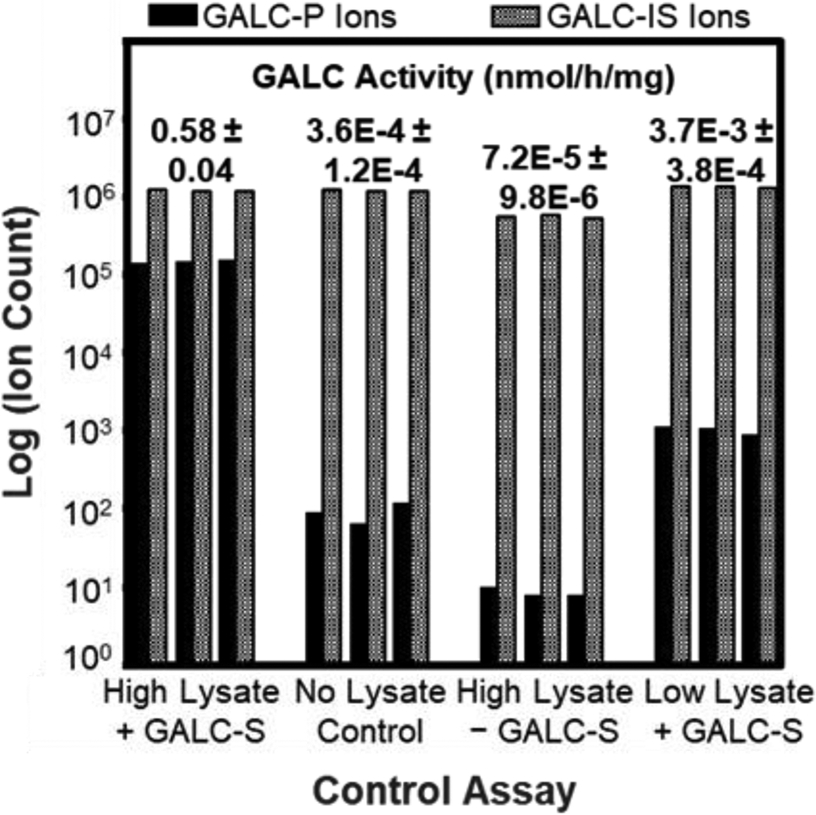
Graphical summary of findings from Liao et al (2017) showing GALC enzymatic activity levels across four control experiments obtained via MRM.
The authors compared this method to benchmark radiometric assay and found the analytical range to be 20 times lower than the LC-MS/MS assay. Also, of note the radiometric assay was performed with 50 μg of cell lysate which required 2-3 mL of patient blood which is undesirable in NBS whereas the LC-MS/MS assay used <5 μg of lysate, requiring only 0.5 mL of patient blood for >15 GALC assays further increasing confidence via replicates. The LC-MS/MS assay was also performed on samples from 3 early infantile KD patients and similar levels of GALC were seen as in the low GALC containing cell line along with asymptomatic patient samples which showed significantly elevated levels of GALC. Analogous analysis was performed using the radiometric assay with data from the New York Krabbe disease NBS program and the clear correlation between low GALC percentage and KD patient using the LC-MS/MS method was not apparent. Thus, radiometric assay alone fails to give the predictive power that the more sensitive LC-MS/MS assay provides and is not adequate to measure small differences in GALC enzyme activity when enzyme percentage is close to zero. The LC-MS/MS method indicates that severity of Krabbe disease and symptom onset may be controlled by the level of residual GALC activity, which gives more accurate predictive power in NBS for when and if patients will develop the disease.
B. Newborn Screening of Lysosomal Storage Disease
Lysosomal storage diseases (LSDs) are a broad class of diseases related to inborn errors in metabolism corresponding to lysosome impairment. LSDs that are caused by specific lysosomal transporter or enzyme deficiency or absence gives rise to non-metabolized macromolecule substrate build-up within lysosomes. These lead to a wide range of physiological complications in patients depending on the specific type (Burlina et al., 2019). In the following sections, the LSDs mucopolysaccharidoses, Pompe disease, and newborn Fabry disease will be discussed in detail with regards to recent advancements in MS/MS methodology for enhanced sensitivity NBS.
Tandem MS SRM and MRM method development have been widely applied to screen LSDs in infants. Burlina employed a multiplexed MS/MS method using the NeoLSD assay system – a commercialized kit from Perkin Elmer for assaying of Gaucher, Pompe, Fabry, and MPS-I LSDs in dried blood spot (DBS) samples (Burlina et al., 2018). Enzymatic cutoff values were used for NBS of 44,411 newborns of which 10 had pathogenic mutations (two Pompe, two Gaucher, five Fabry, one MPS-I). MRM MS/MS allowed for simultaneous determination of multiple enzyme activities via biochemical markers and allowed successful detection of LSDs in newborns. This MS/MS assay method was evaluated further via the recall of 138 newborn samples for collection and testing of a second DBS(Burlina et al., 2019). Low activity was confirmed in 62 samples, which underwent confirmatory testing yielding eight newborns with Pompe, seven with Gaucher, eight with Fabry, and two with Mucopolysaccharidosis type 1 disease. This illustrated that when NBS of LSDs is combined with second tier testing, the occurrence of false-positives can be greatly reduced in a rapid manner. In another study, Baerg et al. used MS/MS integrated with Collaborative Laboratory Integrated Reports (CLIR) - a multivariate pattern recognition software available to NBS programs – to select cases where a second-tier biochemical test is required (Minter Baerg et al., 2018). Use of this postanalytical interpretive tool decreased false-positives (0.0018 % from 55,161 samples) and yielded high predictive power for LSD detection.
1. Mucopolysaccharidoses
Mucopolysaccharidoses (MPS) are a series of inherited metabolic diseases that are caused by lysosomal enzyme deficiency, which are required for glycosaminoglycan (GAG) degradation in lysosomes (Dorfman and Matalon, 1976). MPS are separated into different categories based on the specific enzyme deficiency. Patients with MPS can exhibit a large range of symptoms depending on the type and severity of MPS. Symptoms include enlargement of organs, altered physical appearance, physical growth degradation, bone abnormalities, and neurological complications (Byers et al., 1998). Generally, MPS is divided into two groups based on the type of GAG that is stored. For MPS VI and MPS VIA, dermatan sulfate and keratan sulfate GAGs are stored, yielding abnormalities in patient skeletal structure. In MPSIII, heparan sulfate GAG is stored and exhibits central nervous system complications in addition to skeletal (Byers et al., 1998). GAGs are found throughout connective tissue where they bind to proteoglycans and have unique repeating disaccharide unit backbones (Byers et al., 1998). Proteoglycans control nutrient transfer through connective tissue, and GAGs bind to and regulate biologically active molecules which play a critical role in tissue growth and development (Byers et al., 1998). Thus, the lack of lysosomal enzymes necessary for GAG turnover cause these MPS disorders. GAGs that remain undegraded due to enzyme deficiency accumulate in cells and are expelled in urine when in excess in MPS affected patients. The early detection of MPS is crucial for the early initiation of enzyme replacement therapy, hematopoietic stem cell transplantation therapy, and gene therapy – all of which may prevent irreversible disease progression and improve long term quality of life for patients (Muenzer et al., 2017b, 2017a; Penati et al., 2017; Whiteman and Kimura, 2017).
Mucopolysaccharidoses NBS has been recently evaluated using enzymatic activities measured via tandem MS from DBS. Chan utilized a multiplexed LC-MS/MS based enzyme assay for MPS-I, MPS-II, and MPS-VI LSDs (Chan et al., 2019). In general, mass spectrometry analysis was performed via heated ESI with an applied DC voltage of 4.5 kV, 80 °C source, 250 °C source inlet, and 500 L/h desolvation gas on a tandem triple quadrupole mass spectrometer operating in positive-ion mode and MRM mode using collisional induced dissociation. Sample was injected in an automated manner with methanol and 0.2% formic acid spray solvent at 200 μL/min flow rate. Tandem data was analyzed within 45 seconds of infusion. Optimized cutoff values were combined with second tier testing to eradicate false-positive results. This pilot study indicated that NBS of MPS-I, MPS-II, and MPS-VI via MS/MS is possible and applicable to NBS at large due to the high throughput and specificity offered (Chan et al., 2019). Multiplexing of LC-MS/MS methods for NBS have been recently applied by Liu et al. for the detection of seven lysosomal enzymes in DBS (MPS-I, -II, IIIB, -IVA, -VI, and -VII) as well as type 1 neuronal ceroid lipofuscinosis (Liu et al., 2017). Isotopic labeled internal standards and MRM of ion fragmentations giving enzyme products allowed quantification of enzyme activities. Activity ratios showed a clear separation between healthy and affected newborns due to the large analytical range provided by the MS/MS method (1-2 orders of magnitude greater than traditional fluorometric assays). When compared with a glycosaminoglycan method for MPS screening of DBS, the multiplexed assay yielded a shorter overall time of analysis for 300 samples (28 hours versus 42 hours) illustrating the applicability of MS/MS method for high throughput and sensitivity NBS (Liu et al., 2017). A pilot study from Kubaski analyzed 2862 DBS from newborns for MPS-I, -II, and -III via glycosaminoglycan (GAG) digestion and LC-MS/MS analysis for quantification to determine cutoffs distinguishing affected patients from controls (Kubaski et al., 2017). When two or more different GAGs were combined as indicated in the flow chart in Figure 4, distinction between MPS patients and unaffected controls were improved indicating the potential for the use of GAG as a MPS biomarker in NBS. Polo et al. expanded on the aforementioned technique in the NBS of MPS-I from DBS (Polo et al., 2020). MPS-I screening in 125,000 newborns was conducted via enzyme activity levels alone with subsequent second-tier GAG quantification in DBS for the reduction of false positive rate (Polo et al., 2020).
Figure 4:
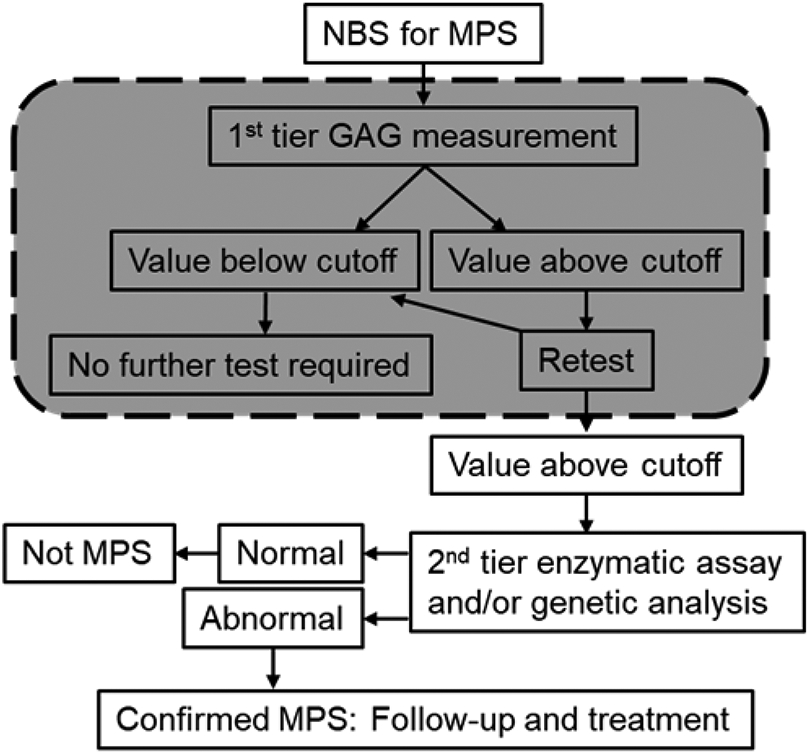
General flow chart for the two-tiered NBS patient testing for MPS involving a first-tier, high-throughput methodology performed via LC-MS/MS (shaded region) followed by a second-tier, more selective NBS technique. A first tier GAG screening could eliminate 99% of unaffected newborns giving rise to a valuable reduction in cost and time of more selective genetic tests. Figure adapted from Kubaski et al 2017.
2. Pompe Disease
Glycogen storage disease type II, also known as Pompe Disease (PD) is a rare autosomal recessive lysosomal storage disease that causes accumulation of glycogen derived glucose polymers in skeletal muscle, among many other tissues, due to α-glucosidase (GAA) deficiency (Kishnani et al., 2014). The most severe form is early-onset PD, which can manifest within the first weeks to months of life and results in progressing symptoms, cardiomyopathy, and eventual cardiorespiratory failure resulting in death before the second year of life (van den Hout et al., 2003; Kishnani et al., 2006). Late-onset PD manifests after 12 months and is associated with progressive respiratory muscle weakness that gives rise to high mortality rate (Kishnani and Howell, 2004). General PD symptoms from excess accumulation of glycogen include organ failure, clinical debilitation, and death. Deficiency of the GAA enzyme remains the standard for PD screening and diagnosis, and high sensitivity methods are necessary as early detection of PD can allow enzyme treatment therapy which can improve the prognosis for patients (Kishnani et al., 2014).
Liao et al. demonstrated enzymatic activity of GAA in DBS from newborn screening using a MS/MS methodology (Liao et al., 2017a). MRM analysis in a triple quadrupole instrument was used in conjunction with GAA internal standards to quantify GAA activities via peak ratio. The analytical range of the MS/MS method was calculated and compared with that of the fluorimetric assay of GAA in DBS. The fluorimetric assay yielded a range 15 times smaller than that of the MS/MS assay. The realization that MS/MS GAA activity is more powerful than standard fluorimetric assays for the distinction of Pompe affected patients speaks to the ability of MS/MS to effectively screen for Pompe disease and reduce the number of patients referred for follow up. Lin et al. utilized a similar LC-MS/MS method for the first distinction between infantile onset Pompe disease (IOPD) and late onset Pompe disease (LOPD) via enzyme assay of GAA using blood samples (Lin et al., 2017). This is particularly important due to the different prognoses and treatment paths for IOPD and LOPD. Data between the MS/MS method and benchmark fluorometry method was compared (Figure 5), again showing the predictive power of the MS method over the fluorimetric assay as indicated by the high precision measurements in tight error bars.
Figure 5:
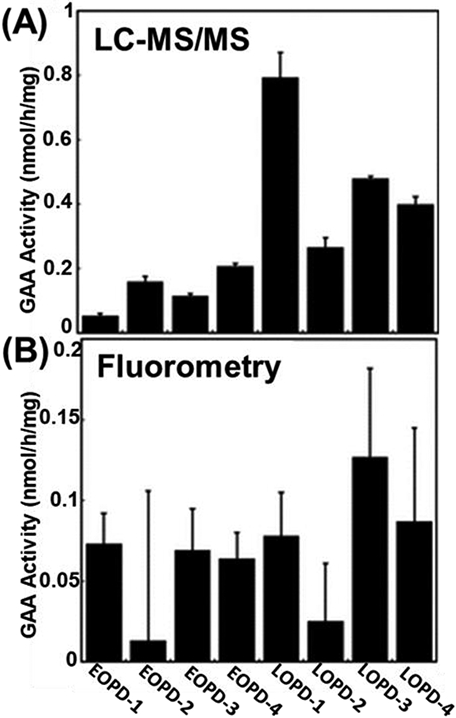
Comparison of LC-MS/MS method applied by Lin et al (2017) with conventional fluorometry assay method for the NBS of Pompe disease with (A) LC-MS/MS analysis showing EOPD with lower activities than LOPD with statistical contrast as seen in the non-overlapping error bars, and (B) fluorometry assay analysis showing no correlation between GAA activity and severity of disease in EOPD and LOPD (reprinted with permission from Lin et al., 2017, copyright 2017 Oxford University Press).
3. Newborn Fabry Disease
Fabry’s disease is a lysosomal storage disorder that is X-chromosome linked and resultant from α-galactosidase A enzyme deficiency (Fabry, 2007). Symptoms typically manifest early in life with life threatening problems arising by middle age (Zarate and Hopkin, 2008). Symptoms of Fabry’s disease resultant of α-galactosidase A enzyme deficiency include inflammation, fibrosis, organ dysfunction, and irreversible tissue damage, α-galactosidase A acts to break down globotriaosylceramide and related glycosphingolipids (Zarate and Hopkin, 2008). The monitoring of enzyme activity related to Fabry disease has not given desirable differentiation between normal and symptomatic levels (Lu et al., 2018). To overcome such false negative results in NBS of Fabry disease, the use of molecular genetic high-throughput mutation detection has been developed to identify genetic variations associated with Fabry mutation (Lu et al., 2018).
Lu et al. used high throughput, cost effective DNA MS for improved sensitivity in NBS of Fabry disease (Lu et al., 2018). An advancement upon the Agena iPLEX MassARRAY platform was used to detect 21 pathogenic mutations in one assay. Briefly, the MassARRAY platform utilizes an allele-specific primer for amplification of the extension products, and has been proven as a powerful tool for detection of thousands of gene variation in hundreds of individuals concurrently (Calvo et al., 2010). The limitation of this method rests in its ability to only detect known mutations, thus novel mutations will be missed. The authors customized the iPLEX PCR primers and extension primers to fit the 21 pathogenic mutations observed in Taiwanese newborn Fabry disease.
C. Newborn Screening of Metabolism Disorders
Inborn errors of metabolism (IEMs) are an extensive group of diseases caused by the deficiency of an enzyme, co-enzyme, or a transporter leading to substrate accumulation (Wang et al., 2019). IEMs are associated with abnormal physical and neurological growth during all stages of life and can result in irreversible mental degradation, disability, physical complications and death. IEMs are inherited in an autosomal recessive manner, meaning variation in genes are inherited mostly from parents. Additionally, clinical and symptomatic expression of IEMs are varying and multifaceted (Yang et al., 2019). Interpretation of IEM screening results includes multiple indicators and ratios when relying on enzyme, co-enzyme, or subsequent transporter deficiency, alone. Thus there has been a recent shift to genetic diagnosis for early detection and treatment of IEMs (Yang et al., 2019).
Tandem MS methods have begun to be applied to NBS for inborn errors of metabolism (IEMs) due to the recent display of the technique to provide sensitive, accurate, and specific results in a high throughput manner in NBS laboratories. Yang et al. and Liu et al. followed a MS/MS protocol using a non-derivatized MS/MS kit from PerkinElmer (Turku, Finland) to explore the effectiveness of NBS by MS/MS and the use of next-generation sequencing (NGS) for gene diagnosis (Liu et al., 2019; Yang et al., 2019). DBS analysis via their MS/MS method was used in conjunction with cut-off values to determine abnormalities in NBS samples. NGS was used as a diagnostic platform after high-risk infants were screened via the MS/MS method. MS/MS was performed after assay with a NeoBase non-derivatized MS/MS Kit (PerkinElmer). If results fell outside of a cutoff value, repeat analysis was performed after which the newborn would be examined further if results remained abnormal. Wang et al. developed an expanded NBS protocol for IEMs that involved the following steps (Wang et al., 2019). Newborns were initially screened via MS/MS, if the result fell outside of the cut-off value the newborn was recalled for another DBS sample after which MS/MS would be performed with the new sample. Newborns with a second abnormal test result would be referred for diagnostic testing. Those with one of the targeted IEMs were referred for genetic analysis. The method yielded 89 reported genetic mutations and 51 novel mutations in 25 IMEs out of 138 patients. All mutations reported were likely pathogenic, however, they expand the mutational spectrum of IEMs which has value in diagnosis and treatment plans for IEMs.
D. Newborn Muscular Atrophy
Spinal muscular atrophy (SMA) is a neurodegenerative disorder characterized by alpha motor neuron degeneration in spinal cord anterior horn cells that is inherited in an autosomal recessive manner, and is the main cause of infant death with a 1 in 10,000 live birth incidence (Verhaart et al., 2017a, 2017b). Symptoms of SMA are wide ranging depending on the type and related age of onset. SMA type I accounts for 60% of affected patients with motor neuron loss, respiratory failure, and death within the first 2 years of life (Kolb et al., 2017). In ~95% of patients, SMA is caused by homozygous deletion or mutation in the survival motor neuron 1 (SMN1) that results in SMN protein deficiency (Lefebvre et al., 1997). A homologue of the SMN1 gene is the SMN2 gene which differs by five nucleotides. The presence of SMN2 is inversely correlated to disease severity as more SMN2 copies ensures higher SMN protein quantities, and SMN2 defects do not seemingly cause SMA (Wirth et al., 2013). Early intervention to prevent neurodegeneration onset is crucial for pre-symptomatic SMA patients (Mendell et al., 2017) yet diagnosis is often delayed (Lin et al., 2015), thus there is an ongoing need for sensitive, cost-effective, and high throughput SMA NBS method.
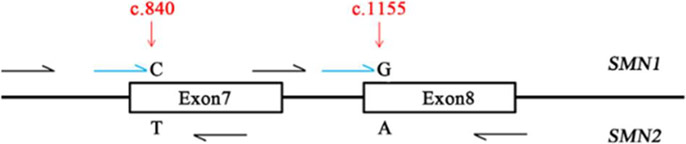
Lin et al. screened for spinal muscular atrophy (SMA) in newborns via DBS using the the Agena iPLEX MassARRAY as a SMA assay (Lin et al., 2019). Early detection and treatment of SMA is important due to the high infant mortality rate associated with this disease. SMA does not have a specific biochemical analyte therefore DNA testing is necessary. For application to NBS the method must be fast, sensitive, and specific all of which are fulfilled via the MS/MS MassARRAY method. The authors expanded this array for the detection of homozygous SMN1 deletion (Figure 6). The similarity of SMN1 and SMN2 genes requires the two well-known positions (c.840 and c.1155) to be used as targets for PCR and single base extension. MALDI-TOF-MS allowed detection of mass of the extended primer identifying the allele. PCR and single base extension primers were designed for two variants of the SMN1 and SMN2 genes. 29,364 newborns were screened with three of them being identified with SMA having homozygous SMN1 deletions at exons 7 and 8. Patients 1, 2, and 3 had one peak at 6710.3 Da and one peak at 4665.9 Da indicating SMN1 deletion at exons 7 and 8, respectively. The healthy individual control had just one peak at 6630.3 Da and one peak at 4586.0 Da which indicated a normal copy number of SMN1.
Figure 6:
Schematic description of the Agena iPLEX SMA assay design consisting of a target specific PCR reaction, then a single base extension with molecular weight modified dideoxynucleotide terminators of the extension primer, annealing directly upstream of the subject polymorphic site. Here, the two well defined positions c.840 and c.1155 of SMN1 and SMN2 genes were utilized as targets for PCR and single base extension primer design. Black arrows represent multiplex PCR while blue arrows represent multiplex extension primers (reprinted with permission from Lin et al., 2019, copyright 2019 Frontiers Media S.A.).
E. Newborn Sickle Cell Disease
Sickle cell disease (SCD) is a severe genetic disorder involving clinical complications associated with the sickling of red blood cells and has high occurrence and mortality during early childhood (Serjeant, 1997). Patients with SCD are subject to life threatening complications with encapsulated bacteria such as pneumococci that causes overwhelming post-splenectomy infection, complications with anemia, splenic sequestration, and aplastic crisis associated with erythrotropic viral infection (Rees et al., 2010; Piel et al., 2017; Ware et al., 2017). Effective preventative treatments are available in the form of antibiotics and vaccination to avoid these life-threatening pathologies, however early diagnosis and awareness of the disease is required. The biochemical analytes of interest in SCD are adult hemoglobin (HbA) and sickling hemoglobin (HbS). In newborns with SCD HbS will exceed HbA due to the HBB:c.20A > T point mutation, which is the mutation that causes the variant HbS structural beta globin, and will be vastly exceeded by fetal hemoglobin (HbF) which is a hemoglobin that is present early in life (Wild et al., 2004; Lobitz et al., 2019). Homozygous presence of this HBB:c.20A > T mutation results in complete substitution of HbA for HbS resulting in severe disease phenotype, while heterozygosity for HbS with another β globin variant results in less severe SCD in comparison(Lobitz et al., 2019). SCD diagnosis is based on the presence of HbS due to its substitution of HbA, which is particularly challenging in neonates where HbF remains the majority of hemoglobin constituent and considering the wide array of additional abnormal hemoglobin traits not specific to SCD, thus requiring a highly sensitive and specific analytical technique for effective NBS (Wild et al., 2004).
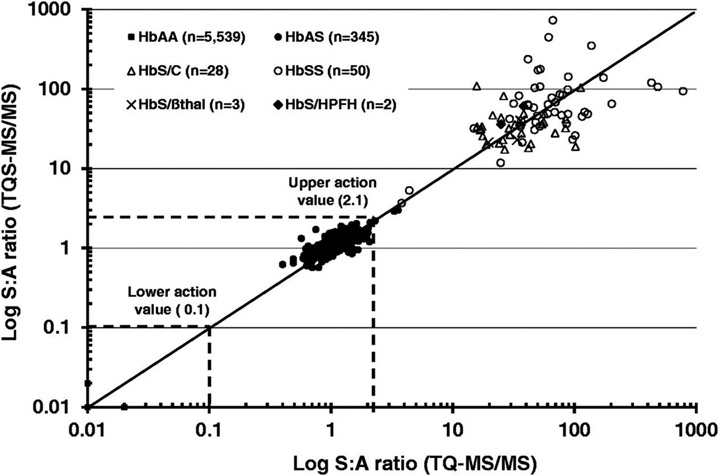
Recent advances in NBS of Sickle Cell Disease (SCD) have also been in the regime of improved MS/MS methodology. Initial attempts at using MS for screening of SCD were limited by analytical resolution. ESI-MS of undigested hemoglobin (Hb) gave a wide array of multiply charged ions requiring software deconvolution to yield α, β, and γ Hb wild-type peaks along with variant Hb peaks shifted 30 Da lower than wild-type β chains. However, adequate resolution to deconvolute the variant Hb peak region was not achievable, leading to the conclusion that ESI-MS alone allowed for detection of SCD but confirmatory methods would be needed for adequate screening (Wild et al., 2004). This narrative was shifted by MS/MS methodology as displayed in two recent studies that have utilized similar electrospray ionization (ESI) MS/MS workflows with no prior chromatography step and demonstrated that MS/MS methods are not only a sensitive and specific tool for NBS of SCD, but they also utilize MS hardware and expertise already available in NBS laboratories whereas traditional screening methods for SCD via capillary electrophoresis (CE) and high performance liquid chromatography (HPLC) require additional instrumentation (Moat et al., 2017; Lobitz et al., 2019). The MS/MS method of interest employed DBS sampling from newborns with a tryptic digestion protocol that has recently been widely accepted in NBS laboratories for SCD (Moat et al., 2017). The DBS samples were prepared by punching and extraction, followed by internal standard Hb and trypsin addition. After incubation and solvent addition to stop the reaction and dilute the solution, the sample was directly injected to the triple quadrupole MS via ESI for subsequent SRM and MRM in which the tryptic peptide ion is first selected followed by CID. The ESI source was operated in positive-ion mode for the detection of target peptides and internal standards.
Moat et al. used this method to address NBS for SCD in premature infants, infants post-transfusion, and older infants – all for whom may have altered Hb β chain expression thus affecting interpretation of results and subsequent disease diagnosis via benchmark clinical methods (Moat et al., 2017). The authors developed action values determined by the ratio of wild-type Hb peptide to variant Hb peptide to screen premature and transfused infant DBS samples. Table 1 shows MS/MS acquisition for relevant Hb peptides using their protocol along with ratios used for screening. The method was also evaluated across two instruments to assess the ability to transfer action values onto different MS/MS instruments and excellent ratio agreement was observed (Figure 6). Results from a sample size of 100,456 newborns screened for SCD yielded 10 true positive, 6 false positives, and no false negatives as validated by traditional methods. The protocol prevented the misidentification of an estimated 810 Hb variant carrier infants as the method gave the specificity to detect the disease states of SCD only. The study indicates the MS/MS method is a sensitive and specific tool for detection of Hb peptides with clinical relevance and demonstrates that the method may be used as an optimized NBS method for SCD as it utilizes technology and expertise already available in NBS laboratories.
Table 1:
Summary of MS/MS acquisition precursor/product ions for Hb peptides and internal standards used for SCD NBS protocol for identification of clinically relevant beta-chain hemoglobinopathies (reprinted with permission from Moat et al., 2017, copyright 2017 SAGE Publications).
| Target peptide |
Tryptic peptide and ion |
Parent/daughter ions (m/z ratios) |
Hb variant peptide ratios |
|---|---|---|---|
| HbS | βT1 y4 | 461.66/472.15 | SβT1 y4/wt βT1y4 (S:A) |
| HbC | βT1 b4 | 694.30/451.15 | CβT1 b4/wt βT1y4 (C:A) |
| HbDPunjab | βT13 b3 | 689.22/377.08 | DβT13 b3/wt βT13 b3 (DPunjab:A) |
| HbOArab | βT13 y9 | 625.25/1001.45 | OβT13 y9/wt βT13 y9 (OArab:A) |
| HbE | βT3 y6 | 458.65/604.15 | EβT3 y6/wt βT3 y9 (E:A) |
| HbLepore | δT2y6 | 480.16/688.20 | δT2 y6/wt βT2 y6 (Lepore:A) |
| HbF | γT2 y6 | 488.3/691.30 | |
| HbA | wt βT1 y4 | 476.67/502.15 | wt βT1 y4/HbFγT2 y6 (A(βT1):F) |
| wt βT2 y6 | 466.76/675.38 | HbFγT2 y6/wt βT2y6 (F:A (βT2)) | |
| wt βT3 y9 | 657.70/887.25 | wt βT3 y9/HbFγT2 y6 (A(βT3):F) | |
| wt βT13 b3 | 689.72/378.05 | ||
| wt βT13 y9 | 689.85/1001.35 | ||
| Internal Standard | 465.65/480.15 |
Lobitz et al. used an analogous ESI-MS/MS method to directly compare it with the traditional CE screening method via DBS analysis of 29,079 German newborns. Both CE and ESI-MS/MS methods gave 100 % agreement in results – observing 7 newborns with Hb peptide patterns consistent with SCD (Lobitz et al., 2019). The authors state that MS/MS is an adequate replacement for CE in NBS of SCD.
F. Beta-Thalassemia Syndrome
Beta-thalassemia (β-thalassemia) syndromes are a group of recessive inherited blood disorders classified by reduced hemoglobin in red blood cells from reduced beta globin chain synthesis (Thein, 2013). β-thalassemia syndromes can be classified broadly as β-thalassemia major, β-thalassemia intermedia, and β-thalassemia minor. In patients with β-thalassemia major, symptoms onset between 6 to 24 months old and consist of fever and progressive abdomen enlargement. When blood transfusions are not performed, complications can include growth hindrance, musculature problems, and skeletal issues from bone marrow expansion. Patients with β-thalassemia intermedia show symptoms at the age of two to six years old and can survive with regular transfusion. Individuals that carry β-thalassemia minor are typically asymptomatic but can present mild anemia symptoms, β-thalassemias are caused by a deficiency in β-globin chain synthesis that, if detected early, can allow transplantation of stem cells which is the only effective treatment (Gambari et al., 2015). Thus, a high sensitivity NBS method is desirable.
Traditional methods for β-thalassemia screening are time consuming and prone to failure due to degradation of cell Hb in stored blood samples. These methods include mean corpuscular hemoglobin (MCH) and mean corpuscular volume (MCV), erythrocyte osmotic fragility test (EOFT), Hb electrophoresis, isoelectric focusing (IEF), and HPLC. They depend on the structural integrity of tetramers which can be affected by degradation of whole blood and hemolysis. MCH and MCV are two common screening methods but lack specificity. Direct Hb electrophoresis via CE has been employed as this method yields different electrophoretic behavior of Hb tetramers compared to normal Hb (Oleske et al., 2014). This method is prone to fault due to the degradation of whole blood samples in storage (Hoppe, 2009). IEF and HPLC in conjunction offers screening for most hemoglobinpathies but is time consuming and requires expertise not typically available in screening laboratories making the method not applicable to large scale screening efforts.
As discussed previously, MS/MS methods have proven robust and valuable for SCD hemoglobinpathy screening, thus the technique should be amenable to β-thalassemia syndrome as well. Yu et al. have achieved this goal by developing a simplified MS/MS protocol that is low cost and high-throughput for detection of thalassemia and subsequent clinical application to DBS analysis for disease screening (Yu et al., 2017). The authors used stable isotopic labeled peptide internal standards for quantification as well as the calculation of α:β globin ratio, which showed statistical difference between healthy patient samples and patients with β-thalassemia. Four of the most informative peptides and their stable isotopic labeled internal standard counterparts were chosen for analysis and yielded high accuracy. Proteospecific peptides produced via tryptic digestion of each globin were used to evaluate α:β globin ratios and the highest intensity peptides for α and β globin were commercially obtained to determine the sensitivity of the technique. When applied to DBS samples, cutoff ratios were used to differentiate between the healthy individuals and those with thalassemia.
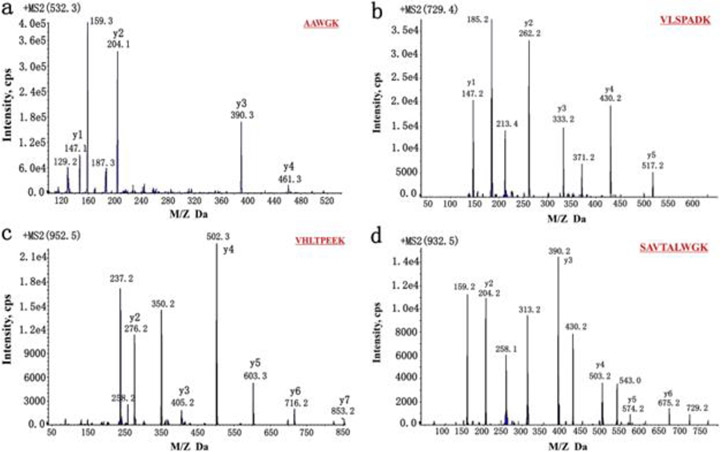
The simplified, low cost, high throughput method of detecting thalassemia in DBS is as follows. DBS constituting 3.2 μL of blood were treated with acetonitrile and formic acid for denaturing followed by a tryptic digest. The sample was diluted in a solution containing the stable isotope labeled internal standard, after which it was introduced to the LC-ESI-MS/MS method. MRM transitions of interest were first identified and optimized via direct infusion tandem MS experiments, and the two mass transitions of highest intensity were selected for the quantitative MRM acquisition method (Figure 8). MS/MS was performed in a QTRAP triple quadrupole instrument using an ESI source operating in positive ion mode. For each peptide of interest, two MRM reaction were simultaneously monitored via a scheduled MRM acquisition method. Peak ratio of internal standard and analyte led to both quantification and α:β globin ratios of interest.
Figure 8:
Product ion scanning MS/MS of selected proteospecific peptides. (a) T3 of alpha-globin product ion spectrum, (b) T1 of alpha-globin product ion spectrum, (c) T1 of beta-globin product ion spectrum, and (d) T2 of beta-globin product ion spectrum all obtained to identify fragment ions of highest value for MRM analysis (reprinted with permission from Yu et al., 2017, copyright 2017 Elsevier).
The α:β globin ratio cutoffs were used to differentiate between healthy and thalassemia samples and the method was applied to 781 patients and 300 healthy individuals, and the method showed significant differences in globin ratios between the two groups with 99 % confidence (Yu et al., 2017). The data show cutoff values for different subtypes of thalassemia alongside the mean with 1st and 99th percentile ratios in healthy controls and thalassemia patients indicating significant difference. A parallel study screened 600 DBS and confirmed results via genotyping indicating the accuracy of this method. This method also displayed insignificant matrix effects and carryover indicating its amenability to high throughput screening. When combined with the aforementioned MS/MS approach for SCD screening, tandem MS methodology is a powerful tool for the screening of both structural abnormality of Hb (SCD) and abnormalities in Hb expression (β-thalassemia), which could significantly increase the efficiency, sensitivity, and specificity of Hb type diseases.
III. Emerging Mass Spectrometry Methods for Cancer Screening
As with newborn screening, tandem MS methodologies have also recently been applied to cancer diagnosis. Deng et al. aimed to develop a low cost, high sensitivity, and high throughput MS based technique for colon cancer screening based on a urine metabolomic test that facilitated detection of adenomatous polyps (Deng et al., 2017). A targeted LC-MS/MS method for quantification of metabolites in 685 urine samples was developed and validated via NMR. The area under curve (AUC) of this method outperformed NMR test, and sensitivity and specificity values were compared against commercially available fecal tests in which the method showed enhanced sensitivity (86 %) versus that of current fecal tests (<18 %) (Deng et al., 2017). Similar metabolomic analysis of plasma set from 282 stage 0-II colorectal cancer patients and 291 healthy individuals was performed by Nishiumi et al. via GC-MS/MS methodology in an attempt to identify biomarkers of stage 0-II colorectal cancer (Nishiumi et al., 2017). GC-MS/MS using a triple quadrupole instrument and MRM analysis was used to distinguish single metabolite peaks from coeluted peaks and background noise. The method yielded 99.3 % sensitivity and 93.8 % specificity once again demonstrating the analytical power of MS/MS methods.
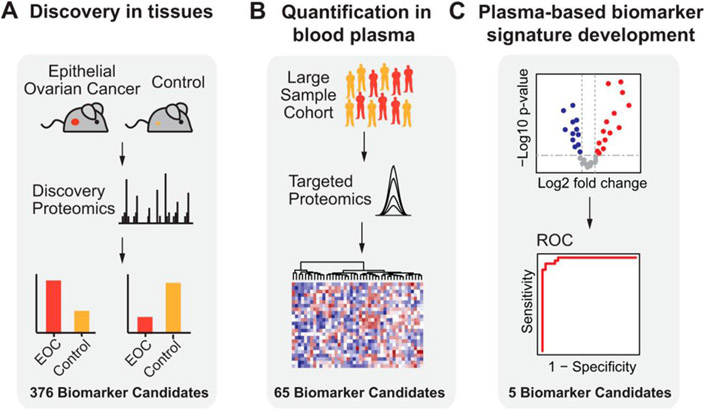
Epithelial ovarian cancer (EOC) screening has been widely improved upon by recent advancements in MS based analysis. Lipid and proteome metabolites from EOC have been evaluated via MS/MS methods. MALDI-MS imaging has been applied with top-down proteomics for distinction of tumor and benign tissue and for EOC stage specific metabolite analysis. Li (Li et al., 2017) used MS to determine patients with EOC could be distinguished clearly based on lipid profiles. LysoPG was shown as a predictive biomarker, and decreased levels of triglycerides were discovered to be a metabolic feature that foreshadowed EOC relapse. Huttenhain (Hüttenhain et al., 2019) used a SRM MS/MS based strategy to develop protein biomarkers from plasma for EOC prediction. This strategy, illustrated in Figure 9, involves first identifying proteome metabolites of interest via discovery MS, followed by quantification of the biomarker candidates using SRM analysis, and finally accompanied by a validation of the protein biomarkers for EOC in patient tissues. This discovery dependent LC-MS/MS protocol was used to identify five protein signatures for the distinction of healthy individuals from those with EOC. When used in conjunction with CA125 ELISA based assay, the sensitivity of biomarker identification increased.
Figure 9:
Overview of study used with permission from Huttenhain et al (2019) with (A) discovery phase in which EOC biomarker candidates were found using a proteomics-based study with EOC tissue sample. (B) Biomarker candidates, which are the plasma-detectable, orthologous human proteins detected to be differentially abundant in (A) were then quantified in plasma samples from a large group of EOC patients along with healthy controls via SRM. (C) The biomarker candidates for EOC detection that were most predictive were chosen, combined in a protein biomarker signature, and submitted to further evaluation in an independent validation set (reprinted with permission from Hüttenhain et al., 2019, copyright 2019 Elsevier).
IV. Ambient Ionization Platforms for Rapid Cancer Screening
Although traditional MS methods can offer several advantages, such as sensitivity and selectivity, there remain barriers to overcome, including lengthy sample preparation, large volume consumption, and complex and expensive instrumentation. These limitations can create unnecessary delays in analysis time and create difficulties in the creation of high throughput testing with MS. To overcome some of these barriers, several ambient ionization MS techniques have been developed that maintain the selectivity and sensitivity of the traditional MS techniques while offering additional advantages. Ambient ionization (AI) allows for the direct ionization and analysis of analytes in the raw sample (urine, sweat, blood, serum, plasma, etc.), eliminating the need for sample preparation and separation/extraction, allowing for a rapid analysis time and a uniquely convenient pairing in the clinical realm. Although ambient ionization methods have been applied to newborn screening, they have made significant contribution in cancer diagnosis. The application of ambient ionization methods in cancer screening has come in the form of direct biofluid analysis and tissue imaging and profiling. Some of the most common methods discussed here fall within two major ambient ionization methodologies: substrate-based platforms and spray-based platforms. The common methods in these categories are paper spray (Wang et al., 2010; Jackson et al., 2018; Swiner et al., 2019) and desorption electrospray ionization (DESI) (Takáts et al., 2005).
A. Direct Biofluid Analysis by Spray-Based Ambient Ionization Platforms
MS is particularly adept at tackling relevant, real-world issues due to the versatility of the instrument and the ability to be mobilized outside of the traditional lab space. Indeed, recent advancements relating to instrument design and ion source have permitted the analysis to be performed in the field or in the operating room. The ability to take the analysis to the field is due in large part to advancements in ambient ionization methods. For an ionization platform to be considered ambient, it must fulfill specific criteria: that the ionization process occurs in the ambient environment, that analysis can be performed directly without prior sample preparation steps (e.g. extraction), that there is a capacity to interface the ionization source with a mass spectrometer with an appropriate outfitting of an atmospheric pressure interface, and that it generates ions softly (Monge et al., 2013). These criteria work well to specifically address concerns regarding the analysis of biological samples, where turnaround time and sensitivity are paramount. Ambient ionization is particularly well-suited for in-situ analysis of biological samples due to its ability to be brought directly to patients, its ability to ionize analytes directly from tissues or biofluids, and its ability to softly generate ions to preserve important molecular information. Consequently, many ambient ionization platforms have been specifically developed for the purpose of providing a targeted approach to analyze analytes to advance fields such as forensics and clinical chemistry. As such, it is no wonder that many researchers seek to either use ambient ionization platforms to advance disease screening or to directly implement ambient ionization techniques in a clinical setting.
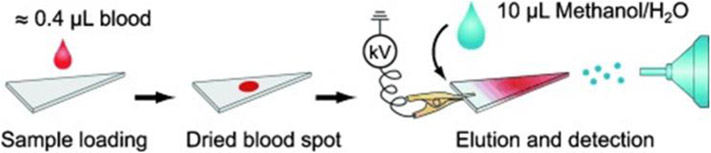
Among ambient ionization methods, substrate-based spray techniques have emerged as particularly valuable direct MS analysis methods as they offer on-line extraction of analytes for rapid, targeted analysis. Compatible substrates for such spray-based techniques are numerous but cellulose paper is most commonly used in the form of paper spray MS (Figure 10) (Wang et al., 2010). In paper spray, the sample (biofluid or tissue) is deposited onto a paper substrate cut to a triangular sharp tip. Then, an appropriate solvent is applied to selectively extract the analyte from the complex mixture. The application of direct current (DC) high voltage to the wet paper substrate releases charged droplets from the tip of the paper triangle via an electrospray mechanism. These charged droplets contain the extracted analytes, which subsequently ionizes and transfers them to the proximal mass spectrometer. Aside from paper, other substrates such as thread, (Jackson et al., 2018; Swiner et al., 2019) metal blade, (Gómez-Ríos et al., 2017) leaf, (Liu et al., 2011; Malaj et al., 2012) and wooden toothpick (Yang et al., 2014). Variants of these methods have also been implemented where chemical modification of the substrate (e.g., salinization to create hydrophobic layer) has been used to improve performance (Manicke et al., 2011; Wang et al., 2011; Gómez-Ríos and Pawliszyn, 2014; Damon et al., 2016).
Figure 10:
Analysis of DBS via paper spray-MS in which whole blood is placed directly on chromatography paper cut into a triangle (reprinted with permission from Wang et al., 2010, copyright 2010 John Wiley and Sons).
Recent advancements in paper spray have shown that it can be used as a tool for the screening of cancer biomarkers (Mendes et al., 2020; Mahmud et al., 2021). To show the potential of paper spray-MS to screen for cervical cancer, Mendes (Mendes et al., 2020) employed an unmodified paper substrate, typical of this kind of ionization, wherein blood plasma from participants was able to be deposited for subsequent analysis. Willing participants were of healthy (n=49) and diseased (n=37) populations, as determined by the absence or presence of intraepithelial lesions/malignancies, respectively. Of these 86 total samples, they were divided into a training set and test set, where the training set was used for model optimization and the test set was used for model evaluation. The screening metabolites chosen were various lipid species that were extracted and detected in positive ion mode, given their widely understood relationship to cancer diagnostics. The lipid profiles for the healthy and diseased samples were compared and it was found that there was a significant difference, as determined by the interval-successive projection algorithm linear discriminant analysis performed. The results from the training set established a clear differentiation between healthy and diseased samples and the confirmation of the model showed agreement.
The established system provided an accuracy of 77% with a sensitivity of 86% whilst using a low-resolution mass spectrometer and analysis took merely 90 seconds total (Mendes et al., 2020). Mahmud (Mahmud et al., 2021) used paper spray-MS to diagnose prostate cancer progression. Using an untargeted approach, they looked at urine liquid samples from the healthy participants (n=10) and of prostate cancer patients whose Gleason scores were GS7, GS8 and GS9 (n=10 each) and performed the paper spray-MS analysis using paper spray cartridges. Mass spectra for the different stages of prostate cancer were then compared to a metabolite library and the tumor stage-specific metabolic features were determined. Using these features, they were able to rapidly discriminate among tumor stages of prostate cancer within one minute noninvasively using urine samples (Mahmud et al., 2021).
B. Direct Tissue Imaging with Desorption Electrospray Ionization
Mass spectrometry imaging (MSI) has recently increased in prevalence in clinical omics approaches. MSI relies on the use of an ionization source with sufficient spatial resolution for mapping of analyte distribution in a given sample based on the m/z detected within regions of the sample. Broadly, MSI may be broken into vacuum-based imaging techniques such as matrix-assisted laser desorption ionization (MALDI) and atmospheric pressure (AP) methods such as DESI and AP-MALDI. For direct tissue imaging, (Wiseman et al., 2006) clinical ambient ionization rely on the principles of desorption electrospray ionization (Takáts et al., 2005). DESI imaging has been applied to the extraction and subsequent analysis of small metabolites and lipids from tissue samples for the diagnosis and screening of various cancers. For example, in 2017, Banerjee et al used DESI imaging platform to diagnose prostate cancer (Banerjee et al., 2017). In this work, a label-free DESI imaging protocol is employed to differentiate cancerous and normal tissues by using known metabolic changes related to prostate cancer, which is particularly pertinent for the removal of tissues, as it can assist in understanding where tissues need to be excised and where it can remain. To capture this idea, various prostate biopsies were taken and analyzed (n=54) using DESI imaging, looking specifically for metabolites and lipids that have been previously associated with cancerous tissue (e.g., Krebs cycle metabolites, fatty acids, various phospholipids), relative to healthy tissue. From these specific metabolites, they found that there was greater accuracy in tissue differentiation among smaller metabolites, when compared to the larger lipids. This finding agrees with a current-standing hypothesis that malignant prostate cells undergo a more bioenergetically efficient Krebs cycle than normal prostate cells which supports the claim that smaller metabolites of the Krebs cycle could be particularly good at differentiating tissues. Specifically, it was determined that the ratio between glucose and citrate was to be one of the more profound indicators for normal versus cancerous tissues (Figure 11). Cancerous tissues demonstrate higher concentrations of glucose and lower concentrations of citrate, relative to the non-cancerous tissues. Specifically, it was found that the ratio of glucose to citrate was greater than 1 for cancerous tissues and was less than 0.5 for normal tissues (Banerjee et al., 2017).
Figure 11:
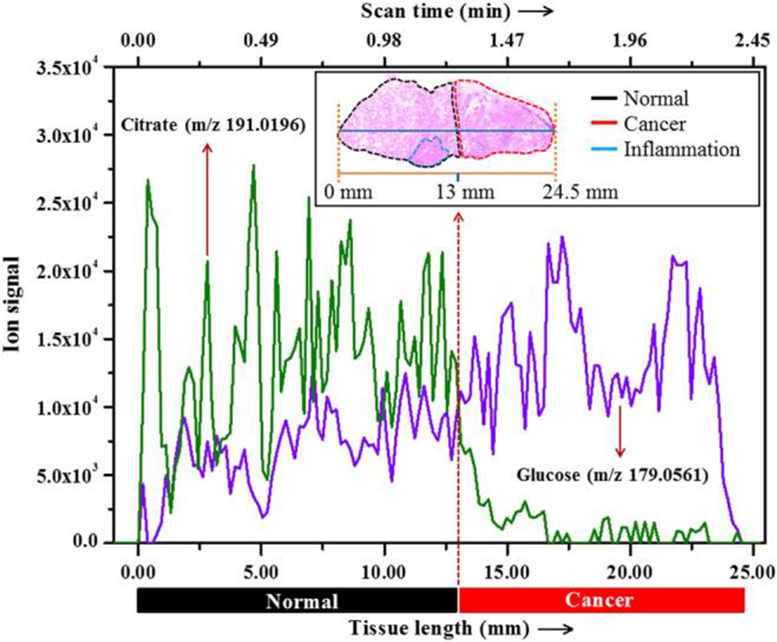
Extracted ion chromatogram of glucose and citrate over a line scan of a typical prostate tissue specimen that contains both normal (inset, left side) and cancerous (inset, right side) areas. Inset shows staining of the tissue and position of the line scan (horizontal line) (reprinted with permission from Banerjee et al., 2017, copyright 2017 National Academy of Science).
Another similar DESI imaging technique has been used to predict aggressiveness of ovarian cancer (Sans et al., 2017) performing analysis on normal ovarian samples (n=15), borderline ovarian tumors (n=15), and high-grade serous carcinoma (n=48). Sans et al selected various lipids to investigate to see how lipid concentration is dependent on cell type. Of particular note are the changes in fatty acid and lipid metabolism, given the differences in tumor biology, which can be particularly helpful in identifying proliferation and aggressiveness of cancers. Specifically, higher degrees of unsaturation have a higher correspondence to high-proliferating tumor cells with polyunsaturated fatty acids (specifically noted were FA 20:3 and FA 22:4) being in higher concentrations for high-grade serous carcinoma than any other type of tissue. Different lipid classes are also associated with different degrees of cellular aggressiveness and proliferation, with ceramides being noted as particularly abundant in the borderline ovarian tumors and phosphatidylinositol and phosphatidylglycerol being characteristic of high-grade serous carcinoma (Sans et al., 2017). Another such imaging technique has been able to distinguish benign from malignant skin abnormalities using DESI imaging, described by Margulis (Margulis et al., 2018). The researchers discuss the imaging of very small, excised samples of tissue wherein DESI imaging was performed and analysis was performed on a variety of analytes to determine those most closely associated with normal versus cancerous tissues. Similar to previous studies that used DESI imaging to differentiate healthy versus cancerous tissues, abnormalities can be traced to differences in metabolism in the Krebs cycle. Using skin tissue from willing participants (n=86), they were able to establish the efficacy of the platform to differentiate tissue types on the physical scale of less than 200 μm in size.
Unlike previous discussion regarding imaging, MALDI imaging is not an ambient technique but the principle is similar to that established by DESI imaging (Cornett et al., 2007; Walch et al., 2008). MALDI imaging uses a laser to oblate a sample and the released molecules are then analyzed and detected via MS. These analytes, like DESI imaging, can be used to create fingerprints or profiles for typical tissue samples for healthy and cancerous patients. Specifically, many recent works have shown the abilities of MALDI imaging to investigate lung cancer, ovarian cancer, and cervical cancer, among others (Delcourt et al., 2017; Ros-Mazurczyk et al., 2017; Briggs et al., 2019; Shu et al., 2020).
It turns out that the interaction of lasers with tissues can be performed outside of the vacuum environment of the mass spectrometer and still produce ion imaging information. When a matrix is used, the process is called AP-MALDI. By changing the wavelength of the laser source, endogenous compounds in the tissue (e.g., water when using IR laser) can serve as a matrix to facilitate the desorption of analytes from the tissue sample without applying any external matrix. This experiment becomes an ambient ionization since no matrix is applied and the analysis is performed in the open environment without transferring sample into vacuum. However, in the absence of a matrix, neutral species are desorbed from the surface. Ion yield is enhanced when the neutral plume is intercepted with electrospray charged droplets. This experiment, in which mid-IR laser is used for desorption and subsequent ionization, is achieved with ESI called laser ablation electrospray ionization (LAESI) MS.(Nemes and Vertes, 2007) When UV photons are used for analyte desorption, the process is called electrospray-assisted laser desorption/ionization (ELDI) MS.(Huang et al., 2006) Both LEASI-MS and ELDI-MS offer higher spatial resolution than DESI.
C. Real-Time Tissue Profiling with Handheld Ambient Ionization Devices
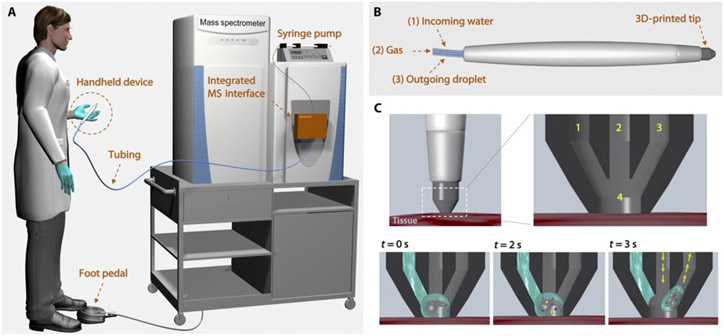
While the analysis of tissues by DESI requires biopsied samples, there is great interest in the differentiation of tissue samples in-vivo and in real-time for application in the operating room. Such endeavors have led to the development of handheld ionization sources such as the MasSpec Pen (Zhang et al., 2017) and iKnife (Balog et al., 2013). The MasSpec Pen (Figure 12) was first discussed by Zhang (Zhang et al., 2017) and describes a powerful, handheld tool for profile analysis of tissues for diagnosis.
Figure 12:
Schematic of MasSpec Pen operation with (A) complete visualization of operating room use of the handheld device with integrated MS interface, (B) view of the handheld device alone showing (1) incoming water, (2) gas input, and (3) outgoing droplet capillary, (C) schematic of tissue sampling with the device showing the 3-D printed tip at three different times of sampling: t = 0 s showing initial solvent interaction, t = 2 s showing solvation of sample on the surface, and t = 3 s sequestering of sample into the outgoing droplet capillary (reprinted with permission from Zhang et al., 2017, copyright 2017 The American Association for the Advancement of Science).
The MasSpec Pen connects the handheld device to the mass spectrometer directly via PTFE tubing. The handheld device itself has three conduits which provide solvent through one conduit and gas through a second conduit and these exit through a third conduit. The pen is placed upon a tissue surface and solvent flow is initiated with a foot pedal after which the solvent interacts with the surface of the tissue to extract molecules, which are then drawn into the mass spectrometer via pressure differential for analysis. Given the various chemical profiles noted, tissue type and characteristic can be identified without the need for biopsy or lengthy preparation time (Zhang et al., 2017). Since the establishment of the MasSpec Pen, the diagnosis of ovarian cancer using the device has been investigated (Sans et al., 2019). This work, conducted by Sans et al, does not show in-vivo analysis but it does provide validation of the method through high degrees of accuracy and sensitivity (>86% and >81%, respectively, for all analyses performed) in the differentiation of normal tissue samples, low-grade serous carcinoma samples, and high-grade serous carcinoma samples. Three different sample sets, with analyses performed across multiple years, involving more than 100 samples using both a relatively low resolution mass spectrometer (linear ion trap) and a relatively high resolution mass spectrometer (orbitrap) helped to elucidate typical profiles for healthy and cancerous tissues at both low-grade and high-grade severities. Specifically, Sans et al selected small metabolites (e.g., ascorbate, taurine, glutamate) and phospholipid species (e.g., glycerophosphoethanolamine, glycerophosphoinositol) as the ions most typical of the representative spectra and would be the ions to aid in differentiation of tissue type. Through machine learning, it was found that differentiation between not only healthy and cancerous tissues was possible, but also a differentiation between low-grade and high-grade serous carcinoma, even on the relatively low-resolution mass spectrometer.
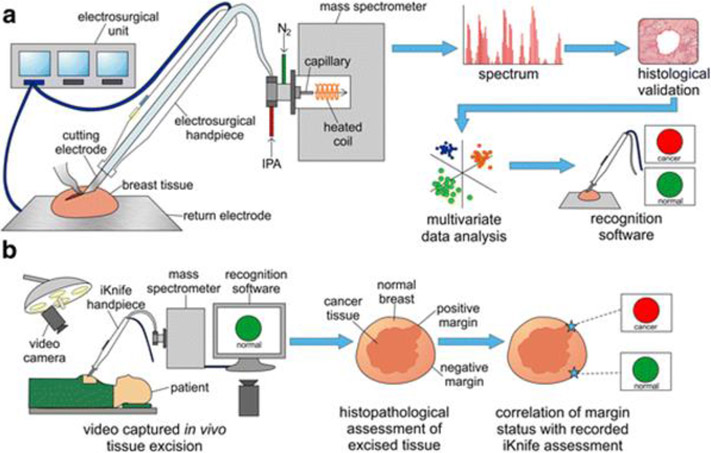
Another notable example of a handheld ambient ionization device is the intelligent knife (iKnife). The iKnife was first described by Balog (Balog et al., 2013) and relies on rapid evaporative ionization mass spectrometry (REIMS) to produce ions from tissues (Schäfer et al., 2009; St John et al., 2017) (Figure 13).
Figure 13:
Schematic description of iKnife operation with (A) description of REIMS operation on a breast tissue sample with subsequent histological validation, multivariate data analysis, and application to recognition software and (B) Surgery room operation of the iKnife with recognition software identifying cancerous versus normal tissue (reprinted with permission from St. John et al., 2017, copyright 2017 Springer Nature).
REIMS allows for analysis to be performed directly upon the various surfaces, such as tissues, by analyzing the aerosols released during electrosurgical dissection. Upon the application of electrosurgical activation, aerosols will be given off (dependent on tissue size and type) which will be transferred through a plastic tube to the mass spectrometer. Recent work specifically has shown that the iKnife is a powerful tool for the identification of breast pathology and can be a powerful tool during breast cancer surgery, as noted in work by St John (St John et al., 2017). In these works, typical spectra for normal samples (n=253) and tumor samples (n=106) were established to create fingerprint spectra. It was noted that certain spectral features and intensities were dependent on the tissue type, as well as the setting used for the device itself. Notably, there was a high intensity of peaks for phospholipids and triglycerides for normal tissue but for cancerous tissues the intensities of phospholipid peaks were substantially higher, while triglyceride was substantially lower. Application of the iKnife device yielded sensitivity of 91% and specificity of 99% (St John et al., 2017).
V. Exhaled Breath Analysis
Sinues, et al (Martinez-Lozano Sinues et al., 2013) suggested that the volatile organic compounds exhaled can be used to monitor and probe the health of a patient, particularly as it pertains to lung health. Given the potential breadth of the exhalome, the original capabilities of exhaled breath analysis seemed vast and it seemed a promising technique, as it allowed for a noninvasive means for diagnosis of lung diseases. Since volatile organic compounds (VOC) in exhaled breath are the analytes of interest, it is easily coupled to techniques such as gas chromatography (GC).
A. Diagnosis of SARS-CoV-2 (COVID-19) via Exhaled Breath
The SARS-CoV-2 (COVID-19) pandemic swept across the globe in late 2019 and early 2020 and demonstrated just how valuable it was for information to be gathered quickly to tailor public health initiatives and patient care as effectively as possible. To gather this information, much research was also given to investigating the implications of COVID-19 infections on a patient’s biochemistry through a variety of means. Widespread COVID-19 screening has been performed mainly via real time reverse-transcriptase polymerase chain reaction (RT-PCR), which involves amplification of viral genetic material (Liu et al., 2020), and antigen-detection rapid diagnostic tests (Ag-RDTs), which involves replication of viral proteins (Agulló et al., 2021). MS-based methods have utilized exhaled breath to assist in characterizing the metabolic profile of exhaled breath by critically ill patients, such as the works performed by Liangou et al (Liangou et al., 2021) and Grassin-Delyle et al (Grassin-Delyle et al., 2021) in 2021. In the work by Grassin-Delyle et al (Grassin-Delyle et al., 2021) the metabolic profile of patients afflicted with respiratory diseases were studied with a time-of-flight (TOF) MS. In this work, expired air was introduced into a proton transfer quadrupole TOF MS via a heated transfer tube that could be connected directly to the patient’s endotracheal tube. These data were compiled for patients with acute respiratory distress syndrome (ARDS) who were COVID-19 positive and those that were COVID-19 negative. Principle component analysis (PCA) was performed to establish a “breathprint” for VOCs associated with the disease, to establish commonalities between the COVID-19 positive patients (n=18) versus COVID-19 negative patients (n=10). Through three different machine learning algorithms, the group was able to assign four specific VOCs (methylpent-2-enal, 2,4-octadiene, 1-chloroheptane, and nonanal) that can be used to differentiate between patients with ARDS that have COVID-19 and those that do not, as seen in Figure 14 (Grassin-Delyle et al., 2021). For the work performed by both Liangou and Grassin-Delyle, the accuracy in assignment was consistently above 80%, for COVID-19 positive and COVID-19 negative patients, when accounting for variables such as smoking status and age.
Figure 14:
Longitudinal analysis of VOCs in expired breath with the four features at m/z 99.08, 111.12, 135.09, and 143.15 which contributed the most to models and were assessed in (A) the first available sample for each patient and (B) over time during intubated, mechanically ventilated patients in ICU with COVID-19 ARDs (n=28, red) or non-COVID-19 ARDs (n=12, blue) (reprinted with permission from Grassin-Delyle et al., 2021 copyright 2021 Elsevier).
Other researcher groups have sought to couple separations-based GC to the exhaled breath analysis via MS to enhance the capabilities for untargeted metabolite coverage to assist with diagnosis of COVID-19 Many of the works that use orthogonal GC-MS techniques were inspired by preliminary work performed by Ruszkiewicz et al that employs the analysis of exhaled breath by gas chromatography-ion mobility spectrometry (IMS) (Ruszkiewicz et al., 2020). While IMS is a powerful tool for the analysis of various metabolites, there are certainly advantages in using mass spectrometry. Ibrahim et al (Ibrahim et al., 2021) and Barberis et al (Barberis et al., 2021) both utilize tandem GC coupled to MS to accomplish analysis of molecules from exhaled breath. Barberis et al utilized orthogonal two-dimensional gas chromatography mass spectrometry (GCxGC-MS) to investigate exhaled breath condensate, as opposed to the exhaled breath itself. Exhaled breath condensate is the liquid phase of exhaled air and contains diluted, nonvolatile molecules which can serve as a source for biomarkers for various respiratory diseases.
B. Chronic Disease Diagnosis with Exhaled Breath Analysis
A recent work performed by Markar et al (Markar et al., 2019) investigated the ability to use volatile organic compounds in exhaled breath to probe potential colorectal cancer metabolites. In this work Markar et al sought an alternative to fecal tests for the detection and diagnosis of colorectal cancer through the use of exhaled breath testing for the analysis of VOCs through selected ion flow tube mass spectrometry (SIFT-MS). SIFT-MS allows the researchers to overcome limitations associated with traditional GC-MS analyses, such as the semi-quantitative nature and the relatively long analysis time. SIFT-MS is based in the foundations of chemical ionization MS wherein VOCs interact with precursor ions that are injected into the carrier gas to cause ionization of the VOCs, which produce characteristic product ions. Given this workflow, SIFT-MS allows for the real-time quantification of VOCs without prior sample preparation. To determine the efficacy of the system in the diagnosis of colorectal cancer, samples were taken from patients with colorectal cancer (n=50), samples from patients with other, non-cancerous gastrointestinal conditions (positive control, n=50), and patients who had a healthy gastrointestinal tract (negative control, n=50). SIFT-MS and GC-MS analyses were performed simultaneously, and results were compared. These results showed that one such VOC, propanal, is an acceptable single breath biomarker for the diagnosis of colorectal cancer with accuracies >79% and sensitivities of >71%, which are similar to conventional fecal occult blood tests and fecal immunochemical tests. Similar techniques have also been used to diagnose tuberculosis, as well. In these studies, by Beccaria et al (Beccaria et al., 2018), patient exhaled breath was collected in a Tedlar bag and subsequently passed through a filter to remove potential pathogens into a thermal desorption tube which desorbed into a GCxGC-TOF MS. Patient samples (n=50) were taken and used for analysis. With these analyses, more than 2000 features were detected, only 52 of which were established as being discriminatory. The accuracy of the platform was found to be >80%, which serves as a testament to the importance of the VOCs found in exhaled breath (Beccaria et al., 2018).
VI. Prospect of Mass Spectrometry Based Omics
The unique building blocks of a mass spectrometer – including the ionization source, the mass analyzer, and the detector – allow ample pathways for innovation, creativity, and problem solving in the ever-growing omics field for disease screening and diagnosis. This includes the genome, transcriptome, proteome and metabolome as has been reflected in the recent advances described in this review for NBS, cancer, and chronic disease diagnosis. While we focus mainly on the advantages of tandem MS methodologies for NBS and ambient ionization MS for cancer and chronic disease diagnosis, the authors recognize that there are many studies utilizing tandem MS methods and additional ambient methods for cancer screening and diagnosis that yielded important insights into this field not discussed at large within this text. We briefly summarize these additional contributions in Table 2 with recent work from Eberlin (Eberlin et al., 2013), Paraskevaidi (Paraskevaidi et al., 2020), Sarbu (Sarbu et al., 2020), Fan (Fan et al., 2018), Wang (Wang et al., 2018), Li (Li et al., 2021), Principe (Principe et al., 2013), Øverbye (Øverbye et al., 2015), Sequeiros (Sequeiros et al., 2016), Fujita (Fujita et al., 2017), Andreu (Andreu et al., 2017), Raimondo (Raimondo et al., 2013), Zhang (Zhang et al., 2018), Luo (Luo et al., 2018), and Chi (Chi et al., 2020). The multitude of studies utilizing MS as an analytical tool for disease screening and diagnosis, as well as comparison and validation to standard methods relay the exciting prospect of this field – especially when considering the MS based methods perform comparatively or outperform standard methods in important figures of merit including sensitivity, specificity, speed, ease-of-use, and cost. However, challenges remain to be overcome for instance in the case of patient biofluid sampling and storage, which the authors have identified as a key step in biomarker stability and preservation in samples (Frey et al., 2022). Challenges associated with DBS sampling, utilized here both in LC-MS methods for NBS and ambient methods for cancer screening, include susceptibility to atmospheric degradation resulting in lowered analyte sensitivity and accuracy. Overcoming these challenges will be particularly important in the screening of rural and underserved populations where temperature, humidity, and time between sample collection and analysis play a nontrivial role (Frey et al., 2022; Lee et al., 2022). Increasing world population levels and inherent increase in cancer, chronic disease, and the need for NBS necessitates the field have robust methods for accurate measure of low concentration biomarkers. In the cases discussed herein, where early detection and diagnosis yield improved patient prognosis and quality of life, we opine that mass spectrometry-based methods will continue to play a critical role in omics fields. This will include the establishment of effective analytical laboratories based on portable instrumentation in developing countries (Lee et al., 2021).
Table 2:
Additional recent literature reporting advances in mass spectrometry-based cancer screening and diagnosis.
| Report | Disease Type | Method | Accuracy |
|---|---|---|---|
| Eberlin et al (2013) | Brain Cancer | DESI-MS | Histopathology Agreement |
| Paraskevaidi et al (2020) | Cervical Cancer | LA-REIMS | 94% Sensitivity and 83% Specificity |
| Sarbu, et al (2020) | Central Nervous System Disorders | IMS-MS/MS | Picomolar Sensitivity |
| Fan et al (2018) | Lung Cancer | Ultra high Resolution FT-MS | AUC of 0.51 – 0.85 |
| Wang et al (2018) | Lung Cancer | LC-MS/MS | 83.1% Sensitivity and 67% Specificity |
| Li et al (2021) | Lung cancer | PS-MS | 0.1 ppt sensitivity |
| Principe et al (2013) | Prostate Cancer | LC-MS/MS | Literature Validation |
| Øverbye et al (2015) | Prostate Cancer | LC-MS/MS | 100% Specificity and Sensitivity >60% |
| Sequeiros et al (2016) | Prostate Cancer | MS/MS (SRM) | Higher Sensitivity and Specificity than standard |
| Fujita et al (2017) | Prostate Cancer | LC-MS/MS (SRM/MRM) | AUC of 0.856 |
| Andreu et al (2017) | Bladder Cancer | LC-MS/MS | High Specificity, PCR comparison |
| Raimondo et al (2013) | Kidney Cancer | LC-MS/MS | AUC of 73 – 89% |
| Zhang et al (2018) | Pulmonary, Lung, Cervical, Colorectal, Bladder, Esophageal, Gastric Cancer | LC-MS/MS | AUC Ranging from 0.8747 – 0.9853 |
| Luo et al (2018) | Liver Cancer | LC-MS/MS | AUC Range of 0.807 – 0.901 and Sensitivity Range of 70-80.3% |
| Chi et al (2020) | Oral Cancer | Multiplexed LC-MS/MS (MRM) | AUC Range of 0.753 – 0.914 |
VII. Conclusion
Recent advances in mass spectrometry take advantage of the minute changes in a patient’s biochemistry to provide powerful, rapid, and targeted analyses to help provide detailed screening or diagnostic information. Work continues to be done on traditional methods, such as conventional GC-MS and LC-MS platforms to enhance the analytical capabilities, but much work is also being done on non-traditional platforms, such as those characterized by ambient ionization techniques. With recent developments in non-invasive and significantly more rapid techniques, such as the exhaled breath analysis, iKnife, and MasSpec Pen, it is possible to reduce barriers to access valuable healthcare for patients, while providing exceptional information quickly and easily for healthcare providers. In this review, we have discussed the biological basis for many genetic diseases and how various of types of MS/MS methods have been applied in newborn screening. We believe such understanding can guide the proper development of new analytical methods by targeting the correct chemical systems, while also improving sensitivity and turnaround time. Similar MS/MS methods have also been applied to cancer screening. However, the given the magnitude of cancer incidence and mortality, the effort to develop new analytical methods for this disease has received far more attention. We have summarized how direct ambient mass spectrometry can be used for metabolomics analysis targeting cancer screening. This can be achieved using biofluids or tissue samples. Cancer screening via tissue analysis can be achieved though profiling (iKnife and MasSpec Pen) or imaging (DESI) methods, with (MALDI) or without (DESI, MasSpec Pen, and iKnife) sample pre-treatment. Though invasive tissue analysis by profiling methods is often integrated with surgical operation procedures and do not significantly add to the needed sources. Biofluid analysis is far more simple and minimally invasive when compared with tissue analysis. A completely non-invasive method is also discussed in the form of exhaled breath analysis. Collectively, mass spectrometry provides a unique opportunity to analyze any type of sample to evaluate the biological machinery of the body, including the metabolome.
Figure 7:
Statistical analysis results to test instrument bias of SCD NBS from bloodspots with normal Hb infants, HbS carrier infants, HbS/S infants, HbS/C infants, and HbS/beta-thalassemia infants, and HbS/HPFH infants (reprinted with permission from Moat et al., 2017, copyright 2017 SAGE Publications).
References
- Aburto JM, Villavicencio F, Basellini U, Kjærgaard S, and Vaupel JW 2020. Dynamics of life expectancy and life span equality. Proc Natl Acad Sci. 117:5250–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari AN, Gallagher RC, Wang Y, Currier RJ, Amatuni G, Bassaganyas L, Chen F, Kundu K, Kvale M, Mooney SD, et al. 2020. The role of exome sequencing in newborn screening for inborn errors of metabolism. Nat Med. 26:1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulló V, Fernández-González M, de la Tabla VO, Gonzalo-Jiménez N, García JA, Masiá M, and Gutiérrez F 2021. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J Infect. 82:186–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A 2021. Preventing neurodegenerative disease. Brain. 144:1279–1280. [DOI] [PubMed] [Google Scholar]
- Andreu Z, Otta Oshiro R, Redruello A, López-Martín S, Gutiérrez-Vázquez C, Morato E, Marina AI, Olivier Gómez C, and Yáñez-Mó M 2017. Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur J Pharm Sci. 98:70–79. [DOI] [PubMed] [Google Scholar]
- Annesley T, Diamandis E, Bachmann L, Hanash S, Hart B, Javahery R, Singh R, and Smith R 2016. A Spectrum of Views on Clinical Mass Spectrometry. Clin Chem. 62:30–36. [DOI] [PubMed] [Google Scholar]
- Balog J, Sasi-Szabó L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, Mirnezami R, Dezső B, Damjanovich L, Darzi A, et al. 2013. Intraoperative Tissue Identification Using Rapid Evaporative Ionization Mass Spectrometry. Sci Transl Med. 5. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Zare RN, Tibshirani RJ, Kunder CA, Nolley R, Fan R, Brooks JD, and Sonn GA 2017. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc Natl Acad Sci. 114:3334–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis E, Amede E, Khoso S, Castello L, Sainaghi PP, Bellan M, Balbo PE, Patti G, Brustia D, Giordano M, et al. 2021. Metabolomics Diagnosis of COVID-19 from Exhaled Breath Condensate. Metabolites. 11:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis KG, Matushek SM, Abeleda APF, Bethel C, Hunt C, Gillen S, Moran A, and Tesic V 2020. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J Clin Virol. 129:104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccaria M, Bobak C, Maitshotlo B, Mellors TR, Purcaro G, Franchina FA, Rees CA, Nasir M, Black A, and Hill JE 2018. Exhaled human breath analysis in active pulmonary tuberculosis diagnostics by comprehensive gas chromatography-mass spectrometry and chemometric techniques. J Breath Res. 13:016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins MS, Shields SWJ, Cui W, Fallatah W, Moser AB, Braverman NE, and Brodbelt JS 2022. Structural Characterization and Quantitation of Ether-Linked Glycerophospholipids in Peroxisome Biogenesis Disorder Tissue by Ultraviolet Photodissociation Mass Spectrometry. Anal Chem. 94:12621–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourmaud A, Gallien S, and Domon B 2016. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications. PROTEOMICS. 16:2146–2159. [DOI] [PubMed] [Google Scholar]
- Breuer O, Caudri D, Stick S, and Turkovic L 2018. Predicting disease progression in cystic fibrosis. Expert Rev Respir Med. 12:905–917. [DOI] [PubMed] [Google Scholar]
- Briggs MT, Condina MR, Ho YY, Everest-Dass AV, Mittal P, Kaur G, Oehler MK, Packer NH, and Hoffmann P 2019. MALDI Mass Spectrometry Imaging of Early- and Late-Stage Serous Ovarian Cancer Tissue Reveals Stage-Specific N- Glycans. PROTEOMICS. 19:1800482. [DOI] [PubMed] [Google Scholar]
- Burlina AB, Polo G, Rubert L, Gueraldi D, Cazzorla C, Duro G, Salviati L, and Burlina AP 2019. Implementation of Second-Tier Tests in Newborn Screening for Lysosomal Disorders in North Eastern Italy. Int J Neonatal Screen. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlina AB, Polo G, Salviati L, Duro G, Zizzo C, Dardis A, Bembi B, Cazzorla C, Rubert L, Zordan R, et al. 2018. Newborn screening for lysosomal storage disorders by tandem mass spectrometry in North East Italy. J Inherit Metab Dis. 41:209–219. [DOI] [PubMed] [Google Scholar]
- Byers S, Rozaklis T, Brumfield LK, Ranieri E, and Hopwood JJ 1998. Glycosaminoglycan Accumulation and Excretion in the Mucopolysaccharidoses: Characterization and Basis of a Diagnostic Test for MPS. Mol Genet Metab. 65:282–290. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, et al. 2010. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet. 42:851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll DI, Dzidic I, Stillwell RN, Haegele KD, and Horning EC 1975. Atmospheric pressure ionization mass spectrometry. Corona discharge ion source for use in a liquid chromatograph-mass spectrometer-computer analytical system. Anal Chem. 47:2369–2373. [Google Scholar]
- Carroll DI, Dzidic I, Stillwell RN, Horning MG, and Horning EC 1974. Subpicogram detection system for gas phase analysis based upon atmospheric pressure ionization (API) mass spectrometry. Anal Chem. 46:706–710. [Google Scholar]
- Chan M-J, Liao H-C, Gelb MH, Chuang C-K, Liu M-Y, Chen H-J, Kao S-M, Lin H-Y, Huang Y-H, Kumar AB, et al. 2019. Taiwan National Newborn Screening Program by Tandem Mass Spectrometry for Mucopolysaccharidoses Types I, II, and VI. J Pediatr. 205:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wang Z-H, Cui W-Q, Zhang J-X, Zhang J-W, Wu D-Q, Wang Z-Y, Yu X-R, Luo Y-B, Hussain D, et al. 2022. High throughput and very specific screening of anabolic-androgenic steroid adulterants in healthy foods based on stable isotope labelling and flow injection analysis-tandem mass spectrometry with simultaneous monitoring proton adduct ions and chloride adduct ions. J Chromatogr A. 1667:462891. [DOI] [PubMed] [Google Scholar]
- Chi L-M, Hsiao Y-C, Chien K-Y, Chen S-F, Chuang Y-N, Lin S-Y, Wang W-S, Chang IY-F, Yang C, Chu LJ, et al. 2020. Assessment of candidate biomarkers in paired saliva and plasma samples from oral cancer patients by targeted mass spectrometry. J Proteomics. 211:103571. [DOI] [PubMed] [Google Scholar]
- Chu LW 2012. Alzheimer’s disease: early diagnosis and treatment. Hong Kong Med J Xianggang Yi Xue Za Zhi. 18:228–237. [PubMed] [Google Scholar]
- Cockcroft D 2018. Environmental Causes of Asthma. Semin Respir Crit Care Med. 39:012–018. [DOI] [PubMed] [Google Scholar]
- Cornett DS, Reyzer ML, Chaurand P, and Caprioli RM 2007. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 4:828–833. [DOI] [PubMed] [Google Scholar]
- Damon DE, Davis KM, Moreira CR, Capone P, Cruttenden R, and Badu-Tawiah AK 2016. Direct Biofluid Analysis Using Hydrophobic Paper Spray Mass Spectrometry. Anal Chem. 88:1878–1884. [DOI] [PubMed] [Google Scholar]
- Dankert-Roelse JE, and te Meerman GJ. 1995. Long term prognosis of patients with cystic fibrosis in relation to early detection by neonatal screening and treatment in a cystic fibrosis centre. Thorax. 50:712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankert-Roelse JE, and Mérelle ME 2005. Review of outcomes of neonatal screening for cystic fibrosis versus non-screening in Europe. J Pediatr. 147:S15–S20. [DOI] [PubMed] [Google Scholar]
- Davidovics R, Saw YL, Brown CO, Prinz M, McKiernan HE, Danielson PB, and Legg KM 2022. High-throughput seminal fluid identification by automated immunoaffinity mass spectrometry. J Forensic Sci. 67:1184–1190. [DOI] [PubMed] [Google Scholar]
- Delcourt V, Franck J, Leblanc E, Narducci F, Robin Y-M, Gimeno J-P, Quanico J, Wisztorski M, Kobeissy F, Jacques J-F, et al. 2017. Combined Mass Spectrometry Imaging and Top-down Microproteomics Reveals Evidence of a Hidden Proteome in Ovarian Cancer. EBioMedicine. 21:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Chang D, Foshaug R, Eisner R, Tso V, Wishart D, and Fedorak R 2017. Development and Validation of a High-Throughput Mass Spectrometry Based Urine Metabolomic Test for the Detection of Colonic Adenomatous Polyps. Metabolites. 7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds JN, and Baker ES 2021. Improving the Speed and Selectivity of Newborn Screening Using Ion Mobility Spectrometry–Mass Spectrometry. Anal Chem. 93:17094–17102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman A, and Matalon R 1976. The mucopolysaccharidoses (a review). Proc Natl Acad Sci. 73:630–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Norton I, Orringer D, Dunn IF, Liu X, Ide JL, Jarmusch AK, Ligon KL, Jolesz FA, Golby AJ, et al. 2013. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci. 110:1611–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar ML, Richards KC, Wenger DA, Champagne M, and Kurtzberg J 2005. Transplantation of Umbilical-Cord Blood in Babies with Infantile Krabbe’s Disease. N Engl J Med. 13. [DOI] [PubMed] [Google Scholar]
- Escolar ML, West T, Dallavecchia A, Poe MD, and LaPoint K 2016. Clinical management of Krabbe disease: Clinical Management of Krabbe Disease. J Neurosci Res. 94:1118–1125. [DOI] [PubMed] [Google Scholar]
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, and Hartwell L 2003. The case for early detection. Nat Rev Cancer. 3:243–252. [DOI] [PubMed] [Google Scholar]
- Evered A, and Dudding N 2011. Accuracy and perceptions of virtual microscopy compared with glass slide microscopy in cervical cytology: Accuracy and perceptions of virtual microscopy. Cytopathology. 22:82–87. [DOI] [PubMed] [Google Scholar]
- Fabry H 2007. Angiokeratoma corporis diffusum - Fabry disease: historical review from the original description to the introduction of enzyme replacement therapy. Acta Paediatr. 91:3–5. [DOI] [PubMed] [Google Scholar]
- Fan TWM, Zhang X, Wang C, Yang Y, Kang W-Y, Arnold S, Higashi RM, Liu J, and Lane AN 2018. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal Chim Acta. 1037:256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn JB, Mann M, Meng CK, Wong SF, and Whitehouse CM 1989. Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science. 246:64–71. [DOI] [PubMed] [Google Scholar]
- Frey BS, Damon DE, and Badu-Tawiah AK 2020. Emerging trends in paper spray mass spectrometry: Microsampling, storage, direct analysis, and applications. Mass Spectrom Rev. 39:336–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BS, Damon DE, and Badu-Tawiah AK 2022. The Effect of the Physical Morphology of Dried Biofluids on the Chemical Stability of Analytes Stored in Paper and Direct Analysis by Mass Spectrometry. Anal Chem. 94:9618–9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Kume H, Matsuzaki K, Kawashima A, Ujike T, Nagahara A, Uemura M, Miyagawa Y, Tomonaga T, and Nonomura N 2017. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci Rep. 7:42961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby DC, Vergani P, and Csanady L 2006. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 440:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambari R, Finotti A, Breda L, Lederer C, Bianchi N, Zuccato C, Klenathous M, and Rivella S 2015. Recent trends in the gene therapy of β-thalassemia. J Blood Med. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelhoed E, Lewis B, Hounsome D, and O’Leary P 2005. Economic evaluation of neonatal screening for phenylketonuria and congenital hypothyroidism. J Paediatr Child Health. 41:575–579. [DOI] [PubMed] [Google Scholar]
- Godtfredsen NS 2005. Effect of Smoking Reduction on Lung Cancer Risk. JAMA. 294:1505. [DOI] [PubMed] [Google Scholar]
- Gómez-Ríos GA, and Pawliszyn J 2014. Development of Coated Blade Spray Ionization Mass Spectrometry for the Quantitation of Target Analytes Present in Complex Matrices. Angew Chem. 126:14731–14735. [DOI] [PubMed] [Google Scholar]
- Gómez-Ríos GA, Tascon M, Reyes-Garcés N, Boyacı E, Poole J, and Pawliszyn J 2017. Quantitative analysis of biofluid spots by coated blade spray mass spectrometry, a new approach to rapid screening. Sci Rep. 7:16104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassin-Delyle S, Roquencourt C, Moine P, Saffroy G, Carn S, Heming N, Fleuriet J, Salvator H, Naline E, Couderc L-J, et al. 2021. Metabolomics of exhaled breath in critically ill COVID-19 patients: A pilot study. EBioMedicine. 63:103154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanold KA, Fischer SM, Cormia PH, Miller CE, and Syage JA 2004. Atmospheric Pressure Photoionization. 1. General Properties for LC/MS. Anal Chem. 76:2842–2851. [DOI] [PubMed] [Google Scholar]
- Hawkes N 2019. Cancer survival data emphasise importance of early diagnosis. BMJ. 1408. [DOI] [PubMed] [Google Scholar]
- Heiles S 2021. Advanced tandem mass spectrometry in metabolomics and lipidomics—methods and applications. Anal Bioanal Chem. 413:5927–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman WH, Ye W, Griffin SJ, Simmons RK, Davies MJ, Khunti K, Rutten GEHM, Sandbaek A, Lauritzen T, Borch-Johnsen K, et al. 2015. Early Detection and Treatment of Type 2 Diabetes Reduce Cardiovascular Morbidity and Mortality: A Simulation of the Results of the Anglo-Danish-Dutch Study of Intensive Treatment in People With Screen-Detected Diabetes in Primary Care (ADDITION-Europe). Diabetes Care. 38:1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström O, Linder N, Kaingu H, Mbuuko N, Mbete J, Kinyua F, Törnquist S, Muinde M, Krogerus L, Lundin M, et al. 2021. Point-of-Care Digital Cytology With Artificial Intelligence for Cervical Cancer Screening in a Resource-Limited Setting. JAMA Netw Open. 4:e211740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe CC 2009. Newborn screening for non-sickling hemoglobinopathies. Hematology. 2009:19–25. [DOI] [PubMed] [Google Scholar]
- Houssami N, Ciatto S, Martinelli F, Bonardi R, and Duffy SW 2009. Early detection of second breast cancers improves prognosis in breast cancer survivors. Ann Oncol. 20:1505–1510. [DOI] [PubMed] [Google Scholar]
- van den Hout HMP, Hop W, van Diggelen OP, Smeitink JAM, Smit GPA, Poll-The B-TT, Bakker HD, Loonen MCB, de Klerk JBC, Reuser AJJ, et al. 2003. The Natural Course of Infantile Pompe’s Disease: 20 Original Cases Compared With 133 Cases From the Literature. Pediatrics. 112:332–340. [DOI] [PubMed] [Google Scholar]
- Howard DH 2005. Life expectancy and the value of early detection. J Health Econ. 24:891–906. [DOI] [PubMed] [Google Scholar]
- Huang M-Z, Hsu H-J, Lee J-Y, Jeng J, and Shiea J 2006. Direct Protein Detection from Biological Media through Electrospray-Assisted Laser Desorption lonization/Mass Spectrometry. J Proteome Res. 5:1107–1116. [DOI] [PubMed] [Google Scholar]
- Hüttenhain R, Choi M, Martin de la Fuente L, Oehl K, Chang C-Y, Zimmermann A-K, Malander S, Olsson H, Surinova S, Clough T, et al. 2019. A Targeted Mass Spectrometry Strategy for Developing Proteomic Biomarkers: A Case Study of Epithelial Ovarian Cancer. Mol Cell Proteomics. 18:1836–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim W, Cordell RL, Wilde MJ, Richardson M, Carr L, Sundari Devi Dasi A, Hargadon B, Free RC, Monks PS, Brightling CE, et al. 2021. Diagnosis of COVID-19 by exhaled breath analysis using gas chromatography–mass spectrometry. ERJ Open Res. 7:00139–02021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Swiner DJ, Capone PC, and Badu-Tawiah AK 2018. Thread spray mass spectrometry for direct analysis of capsaicinoids in pepper products. Anal Chim Acta. 1023:81–88. [DOI] [PubMed] [Google Scholar]
- Jang BG, Kim HS, Chang WY, Bae JM, Kim WH, and Kang GH 2018. Expression Profile of LGR5 and Its Prognostic Significance in Colorectal Cancer Progression. Am J Pathol. 188:2236–2250. [DOI] [PubMed] [Google Scholar]
- Joh DY, Heggestad JT, Zhang S, Anderson GR, Bhattacharyya J, Wardell SE, Wall SA, Cheng AB, Albarghouthi F, Liu J, et al. 2021. Cellphone enabled point-of-care assessment of breast tumor cytology and molecular HER2 expression from fine-needle aspirates. Npj Breast Cancer. 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas Michael., and Hillenkamp Franz. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 60:2299–2301. [DOI] [PubMed] [Google Scholar]
- Kerem E, Reisman J, Corey M, Canny GJ, and Levison H 1992. Prediction of Mortality in Patients with Cystic Fibrosis. N Engl J Med. 326:1187–1191. [DOI] [PubMed] [Google Scholar]
- Kinsella KG 1992. Changes in life expectancy 1900–1990. Am J Clin Nutr. 55:1196S–1202S. [DOI] [PubMed] [Google Scholar]
- Kishnani PS, Amartino HM, Lindberg C, Miller TM, Wilson A, and Keutzer J 2014. Methods of diagnosis of patients with Pompe disease: Data from the Pompe Registry. Mol Genet Metab. 113:84–91. [DOI] [PubMed] [Google Scholar]
- Kishnani PS, and Howell RR 2004. Pompe disease in infants and children. J Pediatr. 144:S35–S43. [DOI] [PubMed] [Google Scholar]
- Kishnani PS, Hwu W-L, Mandel H, Nicolino M, Yong F, and Corzo D 2006. A retrospective, multinational, multicenter study on the natural history of infantile-onset Pompe disease. J Pediatr. 148:671–676.e2. [DOI] [PubMed] [Google Scholar]
- Kisiala A, Kambhampati S, Stock NL, Aoki M, and Emery RJN 2019. Quantification of Cytokinins Using High-Resolution Accurate-Mass Orbitrap Mass Spectrometry and Parallel Reaction Monitoring (PRM). Anal Chem. 91:15049–15056. [DOI] [PubMed] [Google Scholar]
- Knappskog PM, Eiken HG, Martinez A, Flatmark T, and Apold J 1995. The PKU mutation S349P causes complete loss of catalytic activity in the recombinant phenylalanine hydroxylase enzyme. Hum Genet. 95:171–173. [DOI] [PubMed] [Google Scholar]
- Kolb SJ, Coffey CS, Yankey JW, Krosschell K, Arnold WD, Rutkove SB, Swoboda KJ, Reyna SP, Sakonju A, Darras BT, et al. 2017. Natural history of infantile-onset spinal muscular atrophy. Ann Neurol. 82:883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubaski F, Mason RW, Nakatomi A, Shintaku H, Xie L, van Vlies NN, Church H, Giugliani R, Kobayashi H, Yamaguchi S, et al. 2017. Newborn screening for mucopolysaccharidoses: a pilot study of measurement of glycosaminoglycans by tandem mass spectrometry. J Inherit Metab Dis. 40:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberg C, and Lotta LA 2018. Genomic insights into the causes of type 2 diabetes. The Lancet. 391:2463–2474. [DOI] [PubMed] [Google Scholar]
- Lê P-Q, Ferster A, Dedeken L, Vermylen C, Vanderfaeillie A, Rozen L, Heijmans C, Huybrechts S, Devalck C, Cotton F, et al. 2018. Neonatal screening improves sickle cell disease clinical outcome in Belgium. J Med Screen. 25:57–63. [DOI] [PubMed] [Google Scholar]
- Lee S, Chintalapudi K, and Badu-Tawiah AK 2021. Clinical Chemistry for Developing Countries: Mass Spectrometry. Annu Rev Anal Chem Palo Alto Calif. 14:437–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kulyk DS, Afriyie SO, Badu K, and Badu-Tawiah AK 2022. Malaria Diagnosis Using Paper-Based Immunoassay for Clinical Blood Sampling and Analysis by a Miniature Mass Spectrometer. Anal Chem. 94:14377–14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, and Melki J 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 16:265–269. [DOI] [PubMed] [Google Scholar]
- Li J, Xie H, Li A, Cheng J, Yang K, Wang J, Wang W, Zhang F, Li Z, Dhillon HS, et al. 2017. Distinct plasma lipids profiles of recurrent ovarian cancer by liquid chromatography-mass spectrometry. Oncotarget. 8:46834–46845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Huang Z, Ye Z, Zhang X, Chen L, and Xiao Y 2018. Total membrane lipid assay (MLA): simple and practical quantification of exosomes based on efficient membrane-specific dyes unaffected by proteins. Mater Chem Front. 2:2130–2139. [Google Scholar]
- Li Z, Li Y, Zhan L, Meng L, Huang X, Wang T, Li Y, and Nie Z 2021. Point-of-Care Test Paper for Exhaled Breath Aldehyde Analysis via Mass Spectrometry. Anal Chem. 93:9158–9165. [DOI] [PubMed] [Google Scholar]
- Liangou A, Tasoglou A, Huber HJ, Wistrom C, Brody K, Menon PG, Bebekoski T, Menschel K, Davidson-Fiedler M, DeMarco K, et al. 2021. A method for the identification of COVID-19 biomarkers in human breath using Proton Transfer Reaction Time-of-Flight Mass Spectrometry. EClinicalMedicine. 42:101207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-C, Chan M-J, Yang C-F, Chiang C-C, Niu D-M, Huang C-K, and Gelb MH 2017a. Mass Spectrometry but Not Fluorimetry Distinguishes Affected and Pseudodeficiency Patients in Newborn Screening for Pompe Disease. Clin Chem. 63:1271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-C, Spacil Z, Ghomashchi F, Escolar ML, Kurtzberg J, Orsini JJ, Turecek F, Scott CR, and Gelb MH 2017b. Lymphocyte Galactocerebrosidase Activity by LC-MS/MS for Post–Newborn Screening Evaluation of Krabbe Disease. Clin Chem. 63:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-W, Kalb SJ, and Yeh W-S 2015. Delay in Diagnosis of Spinal Muscular Atrophy: A Systematic Literature Review. Pediatr Neurol. 53:293–300. [DOI] [PubMed] [Google Scholar]
- Lin N, Huang J, Violante S, Orsini JJ, Caggana M, Hughes EE, Stevens C, DiAntonio L, Chieh Liao H, Hong X, et al. 2017. Liquid Chromatography–Tandem Mass Spectrometry Assay of Leukocyte Acid α-Glucosidase for Post-Newborn Screening Evaluation of Pompe Disease. Clin Chem. 63:842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lin C-H, Yin X, Zhu L, Yang J, Shen Y, Yang C, Chen X, Hu H, Ma Q, et al. 2019. Newborn Screening for Spinal Muscular Atrophy in China Using DNA Mass Spectrometry. Front Genet. 10:1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Cooks RG, and Ouyang Z 2011. Leaf Spray: Direct Chemical Analysis of Plant Material and Living Plants by Mass Spectrometry. Anal Chem. 83:7608–7613. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wu H, Liu Y, Li C, and Li L 2019. Application of tandem mass spectrometry in newborn genetic metabolic disease screening. Chin J Lab Med. 403–406. [Google Scholar]
- Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, and Zhu C 2020. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 505:172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yi F, Kumar AB, Kumar Chennamaneni N, Hong X, Scott CR, Gelb MH, and Turecek F 2017. Multiplex Tandem Mass Spectrometry Enzymatic Activity Assay for Newborn Screening of the Mucopolysaccharidoses and Type 2 Neuronal Ceroid Lipofuscinosis. Clin Chem. 63:1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobitz S, Klein J, Brose A, Blankenstein O, and Frömmel C 2019. Newborn screening by tandem mass spectrometry confirms the high prevalence of sickle cell disease among German newborns. Ann Hematol. 98:47–53. [DOI] [PubMed] [Google Scholar]
- Lu Y-H, Huang P-H, Wang L-Y, Hsu T-R, Li H-Y, Lee P-C, Hsieh Y-P, Hung S-C, Wang Y-C, Chang S-Κ, et al. 2018. Improvement in the sensitivity of newborn screening for Fabry disease among females through the use of a high-throughput and cost-effective method, DNA mass spectrometry. J Hum Genet. 63:1–8. [DOI] [PubMed] [Google Scholar]
- Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, Yin Z, Xie X, Wang X, Chen W, et al. 2018. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 67:662–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, and Fernández FM 2022. Advances in mass spectrometry imaging for spatial cancer metabolomics. Mass Spectrom Rev. n/a:e21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin A, Khan S, and Kislinger T 2020. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research. Clin Proteomics. 17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud I, Pinto FG, Rubio VY, Lee B, Pavlovich CP, Perera RJ, and Garrett TJ 2021. Rapid Diagnosis of Prostate Cancer Disease Progression Using Paper Spray Ionization Mass Spectrometry. Anal Chem. 93:7774–7780. [DOI] [PubMed] [Google Scholar]
- Malaj N, Ouyang Z, Sindona G, and Cooks RG 2012. Analysis of pesticide residues by leaf spray mass spectrometry. Anal Methods. 4:1913. [Google Scholar]
- Manicke NE, Abu-Rabie P, Spooner N, Ouyang Z, and Cooks RG 2011. Quantitative Analysis of Therapeutic Drugs in Dried Blood Spot Samples by Paper Spray Mass Spectrometry: An Avenue to Therapeutic Drug Monitoring. J Am Soc Mass Spectrom. 22:1501–1507. [DOI] [PubMed] [Google Scholar]
- Marcu L 2012. Fluorescence Lifetime Techniques in Medical Applications. Ann Biomed Eng. 40:304–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis K, Chiou AS, Aasi SZ, Tibshirani RJ, Tang JY, and Zare RN 2018. Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc Natl Acad Sci. 115:6347–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markar SR, Chin S-T, Romano A, Wiggins T, Antonowicz S, Paraskeva P, Ziprin P, Darzi A, and Hanna GB 2019. Breath Volatile Organic Compound Profiling of Colorectal Cancer Using Selected Ion Flow-tube Mass Spectrometry. Ann Surg. 269:903–910. [DOI] [PubMed] [Google Scholar]
- Martinez-Lozano Sinues P, Zenobi R, and Kohler M 2013. Analysis of the Exhalome. Chest. 144:746–749. [DOI] [PubMed] [Google Scholar]
- Maxwell EJ, and Chen DDY 2008. Twenty years of interface development for capillary electrophoresis–electrospray ionization–mass spectrometry. Anal Chim Acta. 627:25–33. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, et al. 2017. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N Engl J Med. 377:1713–1722. [DOI] [PubMed] [Google Scholar]
- Mendes TPP, Pereira I, Lima L.A.S. de, Morais CLM, Neves ACON, Martin FL, Lima KMG, and Vaz BG 2020. Paper Spray Ionization Mass Spectrometry as a Potential Tool for Early Diagnosis of Cervical Cancer. J Am Soc Mass Spectrom. 31:1665–1672. [DOI] [PubMed] [Google Scholar]
- Middleditch BS, and Desiderio DM 1973. Comparison of selective ion monitoring and repetitive scanning during gas chromatography-mass spectrometry. Anal Chem. 45:806–808. [Google Scholar]
- Minter Baerg MM, Stoway SD, Hart J, Mott L, Peck DS, Nett SL, Eckerman JS, Lacey JM, Turgeon CT, Gavrilov D, et al. 2018. Precision newborn screening for lysosomal disorders. Genet Med. 20:847–854. [DOI] [PubMed] [Google Scholar]
- Moat SJ, Rees D, George RS, King L, Dodd A, Ifederu A, Ramgoolam T, and Hillier S 2017. Newborn screening for sickle cell disorders using tandem mass spectrometry: three years’ experience of using a protocol to detect only the disease states. Ann Clin Biochem Int J Lab Med. 54:601–611. [DOI] [PubMed] [Google Scholar]
- Monge ME, Harris GA, Dwivedi P, and Fernández FM 2013. Mass Spectrometry: Recent Advances in Direct Open Air Surface Sampling/Ionization. Chem Rev. 113:2269–2308. [DOI] [PubMed] [Google Scholar]
- Morales E, and Duffy D 2019. Genetics and Gene-Environment Interactions in Childhood and Adult Onset Asthma. Front Pediatr. 7:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenzer J, Giugliani R, Scarpa M, Tylki-Szymańska A, Jego V, and Beck M 2017a. Clinical outcomes in idursulfase-treated patients with mucopolysaccharidosis type II: 3-year data from the hunter outcome survey (HOS). Orphanet J Rare Dis. 12:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenzer J, Jones SA, Tylki-Szymańska A, Harmatz P, Mendelsohn NJ, Guffon N, Giugliani R, Burton BK, Scarpa M, Beck M, et al. 2017b. Ten years of the Hunter Outcome Survey (HOS): insights, achievements, and lessons learned from a global patient registry. Orphanet J Rare Dis. 12:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Fjeldsted JC, Masters CL, and Roberts BR 2022. Enhanced ion mobility resolution of Abeta isomers from human brain using high-resolution demultiplexing software. Anal Bioanal Chem. 414:5683–5693. [DOI] [PubMed] [Google Scholar]
- Mussap M, Zaffanello M, and Fanos V 2018. Metabolomics: a challenge for detecting and monitoring inborn errors of metabolism. Ann Transl Med. 6:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes P, and Vertes A 2007. Laser Ablation Electrospray Ionization for Atmospheric Pressure, in Vivo, and Imaging Mass Spectrometry. Anal Chem. 79:8098–8106. [DOI] [PubMed] [Google Scholar]
- Nishiumi S, Kobayashi T, Kawana S, Unno Y, Sakai T, Okamoto K, Yamada Y, Sudo K, Yamaji T, Saito Y, et al. 2017. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget. 8:17115–17126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kennedy R, Fitzgerald S, and Murphy C 2017. Don’t blame it all on antibodies – The need for exhaustive characterisation, appropriate handling, and addressing the issues that affect specificity. TrAC Trends Anal Chem. 89:53–59. [Google Scholar]
- Oleske DA, Huang RSP, Dasgupta A, Nguyen A, and Wahed A 2014. Higher Sensitivity of Capillary Electrophoresis in Detecting Hemoglobin A2′Compared to Traditional Gel Electrophoresis. Ann Clin Lab Sci. 44:291–293. [PubMed] [Google Scholar]
- Øverbye A, Skotland T, Koehler CJ, Thiede B, Seierstad T, Berge V, Sandvig K, and Llorente A 2015. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. 6:30357–30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevaidi M, Cameron SJS, Whelan E, Bowden S, Tzafetas M, Mitra A, Semertzidou A, Athanasiou A, Bennett PR, MacIntyre DA, et al. 2020. Laser-assisted rapid evaporative ionisation mass spectrometry (LA-REIMS) as a metabolomics platform in cervical cancer screening. EBioMedicine. 60:103017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penati R, Fumagalli F, Calbi V, Bernardo ME, and Aiuti A 2017. Gene therapy for lysosomal storage disorders: recent advances for metachromatic leukodystrophy and mucopolysaccaridosis I. J Inherit Metab Dis. 40:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel FB, Steinberg MH, and Rees DC 2017. Sickle Cell Disease. N Engl J Med. 376:1561–1573. [DOI] [PubMed] [Google Scholar]
- van der Plas EM, van den Tweel XW, Geskus RB, Heijboer H, Biemond BJ, Peters M, and Fijnvandraat K 2011. Mortality and causes of death in children with sickle cell disease in the Netherlands, before the introduction of neonatal screening: Mortality and Causes of Death in Children with SCD. Br J Haematol. 155:106–110. [DOI] [PubMed] [Google Scholar]
- Polo G, Gueraldi D, Giuliani A, Rubert L, Cazzorla C, Salviati L, Marzollo A, Biffi A, Burlina AP, and Burlina AB 2020. The combined use of enzyme activity and metabolite assays as a strategy for newborn screening of mucopolysaccharidosis type I. Clin Chem Lab Med CCLM. 58:2063–2072. [DOI] [PubMed] [Google Scholar]
- Principe S, Jones EE, Kim Y, Sinha A, Nyalwidhe JO, Brooks J, Semmes OJ, Troyer DA, Lance RS, Kislinger T, et al. 2013. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. PROTEOMICS. 13:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo F, Morosi L, Corbetta S, Chinello C, Brambilla P, Mina PD, Villa A, Albo G, Battaglia C, Bosari S, et al. 2013. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst. 9:1220–1233. [DOI] [PubMed] [Google Scholar]
- Rasmussen J, and Langerman H 2019. Alzheimer’s Disease – Why We Need Early Diagnosis. Degener Neurol Neuromuscul Dis. Volume 9:123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Williams TN, and Gladwin MT 2010. Sickle-cell disease. The Lancet. 376:2018–2031. [DOI] [PubMed] [Google Scholar]
- Riserus U, Willett W, and Hu F 2009. Dietary fats and prevention of type 2 diabetes. Prog Lipid Res. 48:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros-Mazurczyk M, Jelonek K, Marczyk M, Binczyk F, Pietrowska M, Polanska J, Dziadziuszko R, Jassem J, Rzyman W, and Widlak P 2017. Serum lipid profile discriminates patients with early lung cancer from healthy controls. Lung Cancer. 112:69–74. [DOI] [PubMed] [Google Scholar]
- Ruszkiewicz DM, Sanders D, O’Brien R, Hempel F, Reed MJ, Riepe AC, Bailie K, Brodrick E, Darnley K, Ellerkmann R, et al. 2020. Diagnosis of COVID-19 by analysis of breath with gas chromatography-ion mobility spectrometry - a feasibility study. EClinicalMedicine. 29–30:100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler S, Alfino L, and Saleem M 2018. The importance of preventative medicine in conjunction with modern day genetic studies. Genes Dis. 5:107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans M, Gharpure K, Tibshirani R, Zhang J, Liang L, Liu J, Young JH, Dood RL, Sood AK, and Eberlin LS 2017. Metabolic Markers and Statistical Prediction of Serous Ovarian Cancer Aggressiveness by Ambient Ionization Mass Spectrometry Imaging. Cancer Res. 77:2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans M, Zhang J, Lin JQ, Feider CL, Giese N, Breen MT, Sebastian K, Liu J, Sood AK, and Eberlin LS 2019. Performance of the MasSpec Pen for Rapid Diagnosis of Ovarian Cancer. Clin Chem. 65:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbu M, Raab S, Henderson L, Fabris D, Vukelić Ž, Clemmer DE, and Zamfir AD 2020. Cerebrospinal fluid: Profiling and fragmentation of gangliosides by ion mobility mass spectrometry. Biochimie. 170:36–48. [DOI] [PubMed] [Google Scholar]
- Schäfer K-C, Dénes J, Albrecht K, Szaniszló T, Balog J, Skoumal R, Katona M, Tóth M, Balogh L, and Takáts Z 2009. In Vivo, In Situ Tissue Analysis Using Rapid Evaporative Ionization Mass Spectrometry. Angew Chem Int Ed. 48:8240–8242. [DOI] [PubMed] [Google Scholar]
- Sequeiros T, Rigau M, Chiva C, Montes M, Garcia-Grau I, Garcia M, Diaz S, Celma A, Bijnsdorp I, Campos A, et al. 2016. Targeted proteomics in urinary extracellular vesicles identifies biomarkers for diagnosis and prognosis of prostate cancer. Oncotarget. 8:4960–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serjeant GR 1997. Sickle-cell disease. The Lancet. 350:725–730. [DOI] [PubMed] [Google Scholar]
- Shahzad A, Edetsberger M, and Koehler G 2010. Fluorescence Spectroscopy: An Emerging Excellent Diagnostic Tool in Medical Sciences. Appl Spectrosc Rev. 45:1–11. [Google Scholar]
- Shu W, Wang Y, Liu C, Li R, Pei C, Lou W, Lin S, Di W, and Wan J 2020. Construction of a Plasmonic Chip for Metabolic Analysis in Cervical Cancer Screening and Evaluation. Small Methods. 4:1900469. [Google Scholar]
- Solomon BD, Pineda-Alvarez DE, Bear KA, Mullikin JC, and Evans JP 2012. Applying Genomic Analysis to Newborn Screening. Mol Syndromol. 3:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John ER, Balog J, McKenzie JS, Rossi M, Covington A, Muirhead L, Bodai Z, Rosini F, Speller AVM, Shousha S, et al. 2017. Rapid evaporative ionisation mass spectrometry of electrosurgical vapours for the identification of breast pathology: towards an intelligent knife for breast cancer surgery. Breast Cancer Res. 19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K 2003. Globoid Cell Leukodystrophy (Krabbe’s Disease): Update. J Child Neurol. 18:595–603. [DOI] [PubMed] [Google Scholar]
- Swiner DJ, Jackson S, Burris BJ, and Badu-Tawiah AK 2020. Applications of Mass Spectrometry for Clinical Diagnostics: The Influence of Turnaround Time. Anal Chem. 92:183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiner DJ, Jackson S, Durisek GR, Walsh BK, Kouatli Y, and Badu-Tawiah AK 2019. Microsampling with cotton thread: Storage and ultra-sensitive analysis by thread spray mass Spectrometry. Anal Chim Acta. 1082:98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts Z, Wiseman JM, and Cooks RG 2005. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J Mass Spectrom. 40:1261–1275. [DOI] [PubMed] [Google Scholar]
- Tang H, Cai D, Kong Y, Ye H, Ma Z, Lv H, Tuo L, Pan Q, Liu Z, and Han X 2021. Cervical cytology screening facilitated by an artificial intelligence microscope: A preliminary study. Cancer Cytopathol. 129:693–700. [DOI] [PubMed] [Google Scholar]
- Taraseviciene-Stewart L, and Voelkel NF 2008. Molecular pathogenesis of emphysema. J Clin Invest. 118:394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein SL 2013. The Molecular Basis of -Thalassemia. Cold Spring Harb Perspect Med. 3:a011700–a011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton Snider J, Sullivan J, van Eijndhoven E, Hansen MK, Bellosillo N, Neslusan C, O’Brien E, Riley R, Seabury S, and Kasiske BL 2019. Lifetime benefits of early detection and treatment of diabetic kidney disease. PLOS ONE. 14:e0217487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J, Houben E, Depypere M, Brackenier A, Desmet S, André E, Van Ranst M, Lagrou K, and Vermeersch P 2020. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 26:1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenina I, Bilandžić N, Luburić ĐB, Kolanović BS, and Varga I 2022. High resolution mass spectrometry method for the determination of 13 antibiotic groups in bovine, swine, poultry and fish meat: An effective screening and confirmation analysis approach for routine laboratories. Food Control. 133:108576. [Google Scholar]
- Verhaart IEC, Robertson A, Leary R, McMacken G, König K, Kirschner J, Jones CC, Cook SF, and Lochmüller H 2017a. A multi-source approach to determine SMA incidence and research ready population. J Neurol. 264:1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaart IEC, Robertson A, Wilson IJ, Aartsma-Rus A, Cameron S, Jones CC, Cook SF, and Lochmüller H 2017b. Prevalence, incidence and carrier frequency of 5q–linked spinal muscular atrophy – a literature review. Orphanet J Rare Dis. 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnovitz T, Osteikoetxea X, Sódar BW, Mihály J, Lőrincz P, Vukman KV, Tóth EÁ, Koncz A, Székács I, Horváth R, et al. 2019. An improved 96 well plate format lipid quantification assay for standardisation of experiments with extracellular vesicles. J Extracell Vesicles. 8:1565263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch A, Rauser S, Deininger S-O, and Höfler H 2008. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem Cell Biol. 130:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu J, Cooks RG, and Ouyang Z 2010. Paper Spray for Direct Analysis of Complex Mixtures Using Mass Spectrometry. Angew Chem Int Ed. 49:877–880. [DOI] [PubMed] [Google Scholar]
- Wang H, Manicke NE, Yang Q, Zheng L, Shi R, Cooks RG, and Ouyang Z 2011. Direct Analysis of Biological Tissue by Paper Spray Mass Spectrometry. Anal Chem. 83:1197–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Song X, Liu L, Niu L, Wang X, Song X, and Xie L 2018. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 109:1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ma J, Zhang Q, Gao A, Wang Q, Li H, Xiang J, and Wang B 2019. Expanded Newborn Screening for Inborn Errors of Metabolism by Tandem Mass Spectrometry in Suzhou, China: Disease Spectrum, Prevalence, Genetic Characteristics in a Chinese Population. Front Genet. 10:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware RE, Montalembert M. de, Tshilolo L, and Abboud MR 2017. Sickle cell disease. The Lancet. 390:311–323. [DOI] [PubMed] [Google Scholar]
- Wender RC, Brawley OW, Fedewa SA, Gansler T, and Smith RA 2019. A blueprint for cancer screening and early detection: Advancing screening’s contribution to cancer control. CA Cancer J Clin. 69:50–79. [DOI] [PubMed] [Google Scholar]
- Whiteman D, and Kimura A 2017. Development of idursulfase therapy for mucopolysaccharidosis type II (Hunter syndrome): the past, the present and the future. Drug Des Devel Ther. Volume 11:2467–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild BJ, Green BN, and Stephens AD 2004. The potential of electrospray ionization mass spectrometry for the diagnosis of hemoglobin variants found in newborn screening. Blood Cells Mol Dis. 33:308–317. [DOI] [PubMed] [Google Scholar]
- Wirth B, Garbes L, and Riessland M 2013. How genetic modifiers influence the phenotype of spinal muscular atrophy and suggest future therapeutic approaches. Curr Opin Genet Dev. 23:330–338. [DOI] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Song Q, and Cooks RG 2006. Tissue Imaging at Atmospheric Pressure Using Desorption Electrospray Ionization (DESI) Mass Spectrometry. Angew Chem Int Ed. 45:7188–7192. [DOI] [PubMed] [Google Scholar]
- Woolf SH, and Schoomaker H 2019. Life Expectancy and Mortality Rates in the United States, 1959-2017. JAMA. 322:1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SM, and Aronne LJ 2012. Causes of obesity. Abdom Radiol. 37:730–732. [DOI] [PubMed] [Google Scholar]
- Wu L, Li G, Xu X, Zhu L, Huang R, and Chen X 2019. Application of nano-ELISA in food analysis: Recent advances and challenges. TrAC Trends Anal Chem. 113:140–156. [Google Scholar]
- Wu T, Sun G, Ma M, Pan X, Zhang S, and Zhang X 2022. Rapid quantitative analysis of hormones in serum by multilayer paper spray MS: Free MS from HPLC. Talanta. 237:122900. [DOI] [PubMed] [Google Scholar]
- Xia F, and Wan J-B Chemical derivatization strategy for mass spectrometry-based lipidomics. Mass Spectrom Rev. e21729. [DOI] [PubMed] [Google Scholar]
- Yang Y, Deng J, and Yao Z-P 2014. Pharmaceutical Analysis by Solid-Substrate Electrospray Ionization Mass Spectrometry with Wooden Tips. J Am Soc Mass Spectrom. 25:37–47. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang L, Wang B, Liu S, Yu B, and Wang T 2019. Application of Next-Generation Sequencing Following Tandem Mass Spectrometry to Expand Newborn Screening for Inborn Errors of Metabolism: A Multicenter Study. Front Genet. 10:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Huang S, Wang M, Zhang J, Liu H, Yuan Z, Wang X, He X, Wang J, and Zou L 2017. A novel tandem mass spectrometry method for first-line screening of mainly beta-thalassemia from dried blood spots. J Proteomics. 154:78–84. [DOI] [PubMed] [Google Scholar]
- Zang X, Monge ME, and Fernández FM 2019. Mass spectrometry-based non-targeted metabolic profiling for disease detection: Recent developments. TrAC Trends Anal Chem. 118:158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate YA, and Hopkin RJ 2008. Lysosomal Storage Disease 3 Fabry’s disease. 372:9. [DOI] [PubMed] [Google Scholar]
- Zhang C, Leng W, Sun C, Lu T, Chen Z, Men X, Wang Y, Wang G, Zhen B, and Qin J 2018. Urine Proteome Profiling Predicts Lung Cancer from Control Cases and Other Tumors. EBioMedicine. 30:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ji B, Yan X-H, Lv S, Fang F, Zhao S, Guo X-L, and Wu Z-Y 2022. Paper-based sample processing for the fast and direct MS analysis of multiple analytes from serum samples. Analyst. 147:4895–4902. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rector J, Lin JQ, Young JH, Sans M, Katta N, Giese N, Yu W, Nagi C, Suliburk J, et al. 2017. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci Transl Med. 9:eaan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]