Abstract
Four new complexes prepared from 5-Methyl-2-carboxaldehyde-thiophene and 2,6-pyridinediamine with cobalt, nickel, copper and cadmium ions have been synthesized because of the significance of these complexes in pharmacological research and catalytic reduction. The prepared compounds were characterized using elemental analysis, mass, UV–visible, NMR and FTIR spectroscopy, as well as molar conductance, magnetic susceptibility measurements, fluorescence properties and TGA analysis. The stoichiometry mode was confirmed as 1:1 (metal: ligand) for Co, Ni and Cu complexes and 1:2 (metal: ligand) for Cd complex according to the results of the elemental and spectral studies. Furthermore, the thermal stability and luminescence properties of complexes have been studied. Thermal studies confirmed the presence of water molecules. The thermodynamic properties of the complexes were measured via the Coats-Redfern procedure. The geometric structures of the complexes were found to be octahedral around the metal ions. The optical energy gaps (Eopt) vary between 2.92 and 3.71 eV indicating that these compounds can be used as selective absorbing solar energy in photovoltaic applications. In the presence of NaBH4, the greatest reduction efficiency for the conversion of 2-NP to 2-AP was discovered to be 73–91% within 15–25 min. In vitro, high antifungal and antibacterial activity was shown by complexes than the ligand alone. The Cd(II) complex was shown to have greater activity than all of the examined microorganisms when compared to the reference drug in addition it had 4.94 μg/ml minimal inhibitory concentration against “S. aureus”, “B. subtilis”, and “E. coli”. The bond angles, bond lengths, and quantum chemical factors of the ligand and complexes were shown in the molecular modeling using the DFT approach. The studied compounds' binding modes were confirmed using the Gaussian 09 program.
Keywords: Optical, Catalytic reduction, Metal removal, DFT, Docking studies
1. Introduction
Schiff bases with donor atoms of nitrogen, sulfur, or oxygen can easily form stable complexes with different coordination sites [[1], [2], [3], [4], [5]]. Environmental pollution such as nitroaromatic compounds can be controlled to some extent by reducing to amino compounds with a reducing agent. These compounds are extensively applied in many industries and directly released into the environment without any treatment causing damage to the environment, soil, living organisms and human health [6,7]. As a result, there is growing interest in developing new Schiff base compounds, which could act as a catalytic reduction for these compounds. The volume of heavy metal pollution has increased due to an increase in various industries in many countries [8]. Accordingly, adsorption technology is considered one of the best ways to get rid of heavy metal ions because it is low-cost and simple [9]. These complexes possess various applications in many fields luminescent materials [10], anti-inflammatory action [11], and antioxidant [12,13], antitumor [14], microbial properties [[15], [16], [17], [18]]. Heterocyclic compounds were effective ligands such as pyridine and its derivatives, which have a nitrogen atom [19,20]. This paper describes the preparation of a new ligand (L) formed by the condensation of 5-methyl-2-carboxaldehyde-thiophene and pyridine-2,6-diamine, as well as its Co, Ni, Cu, and Cd complexes. To characterize the synthesized compounds, numerous spectroscopic methods were used, including C, H, N, S analysis, IR, NMR, molar conductivity, Uv–vis, TG and fluorescence studies. The thermal behavior, optical properties and catalytic reduction of the obtained complexes were investigated. All compounds were tested for biological studies. Furthermore, optimization and docking studies for the L and its compounds were performed to determine the receptor under the study’s active site binding affinity.
2. Experimental
2.1. Materials and physical measurements
The solvents were all extremely pure. From Merck/Sigma-Aldrich, the compounds were purchased. The IR spectra were measured by a spectrophotometer (model 1430). The NMR was recorded using a Bruker (400 MHz) spectrometer. The elemental analysis of the ligand and complexes (C, S, H and N) were evaluated through Perkin–Elmer (2400). The molar conductance was determined using Jenway conductivity meter 4010. A Shimadzu (3101) pc spectrophotometer was used to measure the spectra of solid reflectance. Thermal analysis was performed on a Shimadzu apparatus (TGA-50H). The spectra of fluorescence were investigated on Perkin Elmer LS 55 spectrofluorometer.
2.2. Synthesis
2.2.1. Preparation of ligand (L)
Pyridine-2,6-diamine (10 mmol) was added to a stirred mixture of 5-methyl-2-carboxaldehyde-thiophene (10 mmol) in 20 mL ethanol, and 7 h were spent stirring the reaction mixture. The solvent was evaporated to obtain solid residue product L (Fig. 1). After being washed with ethanol, the product was vacuum-dried.
Fig. 1.
The preparation of the ligand, L and its complexes, as well as the suggested structures.
2.2.2. Preparation of complexes
Metal complexes were synthesized by adding 20 ml of a hot methanolic solution of metal nitrate or chloride (1 mmol) to 20 mL of ligand (L) solution in THF (1 mmol). The mixture was then refluxed for 5 h. The product was separated by filtration, then washed numerous times with diethyl ether (Fig. 1). The precipitate was then dried in a vacuum.
2.3. Catalytic reduction of the synthesized complexes
Organic compounds such as nitroaromatic compounds cause various pollutants because they are directly released into the environment without any treatment [21]. Catalytic hydrogenation using a hydrogen donor e.g. sodium borohydride in the presence of catalysts is considered to be the most effective and eco-friendly route to reduce the nitro group to amino [22]. The progress of the reaction is monitored by UV–Vis. spectroscopy. UV–vis spectroscopy was used to calculate the percentage color reduction by observing changes in the absorbance values using the equation: % reduction = (A0 − At)/A0 × 100, where A0 is the absorbance of 2-nitrophenol (2-NP) at 0 min and At is the absorbance at time t. 4 ml of a 2-nitrophenol aqueous solution (1 mmol) was added with stirring to a beaker containing 30 ml of NaBH4 solution (50 mmol). Then, 0.01 g of Co, Ni, or Cu complex as a catalyst was added to the reaction and the absorbance was measured at 400 nm at different times.
2.4. Metal ion removal
The batch approach was employed for the metal ion uptake investigation. Ion adsorption was performed by adding 6 mg of ligand (L) to 30 ml aqueous solutions of each of the cobalt, nickel, copper or cadmium salts in a 100 ml flask at room temperature. Three concentrations of 1 × 102−, 1 × 103− and 1 × 104− M for the metal ions were done after various times of stirring 1, 2, and 5 h. The concentration of residual metal in the solution after filtration was estimated complexometrically by EDTA-Na [23]. The subsequent equation was applied to calculate the percentage of metal removal: % of removal is calculated as Co−Ce/Ce, where Ce is the metal ion concentration at time t (in mg/L) and Co is the starting metal ion concentration (in mg/L).
2.5. Computational studies
The Gaussian09 software [24] was used for the geometry optimization of the ligand and its complexes. DFT method-based functional B3LYP by LANL2DZ basis for metals and 6-311G++(dp) for C. H, N and Cl [25,26]. The various chemical global reactivity descriptors were computed. Additionally, the potential modes of binding for the produced compounds with the receptors of Bacillus subtilis bacteria (PDB ID: 1QD9) [27] and COVID-19 (PDB ID: 6lu7) [5] in the situations of (L) ligand and copper and cadmium compounds were studied using the program MOA2019 [28]. The crystal structures of the receptors were downloaded from the protein data bank (http://www.rcsb.org./pdb).
2.6. Antimicrobial analysis
The compounds were tested on a nutrient agar medium against four bacterial species. On sabouraud dextrose agar medium, antifungal activity against two fungal species was also investigated. The standard Gentamycin and Ketoconazole references were utilized as positive controls. The zone of inhibition against the test organisms was measured in the disc diffusion assay to assess the activity. Every assay was carried out twice [29].
2.7. MIC determination
NCCLS suggestions achieved the minimum amount of inhibitory effect [30]. The inhibitory zone was screened using the well diffusion approach [31,32]. Different test chemical concentrations (10, 5 and 2.5 mg/ml) in DMSO solvent were utilized. After 24 h, the vicinity of each well's inhibitory zone was identified at 37 °C with DMSO controls.
3. Results and discussion
3.1. Characterization using spectroscopy
There have been many failed attempts to make single crystals from the complexes because they are only weakly soluble in ordinary organic solvents. The amount of molar conductivity for the Cd complex was discovered to be 80 Ω−1 mol−1 cm2 suggesting a 1:1 nature electrolytic, while the remaining compounds observed non-electrolytic character (Table 1) [33]. Table 1 contains the synthesized compounds' analytical and physical data.
Table 1.
Physical and elemental results of the compounds.
| Compound (molecular formula) | M.Wt. | Color (% yield) | M.P. (°C) | Λm (Ω−1mol−1 cm2) | Elemental analysis % Found (calc.) |
||||
|---|---|---|---|---|---|---|---|---|---|
| C | N | H | S | M | |||||
| L (C11H11N3S) | 217.28 | Brown (88) | 250 | – | 60.89 (60.80) | 19.03 (19.33) | 12.37 (12.10) | 14.75 (14.76) | – |
| [Co(C11H11N3S)(H2O)Cl2]·H2O | 383.13 | Pale brown (70) | >300 | 10 | 34.84 (34.48) | 10.78 (10.6) | 3.04 (3.94) | 8.18 (8.37) | 15.05 (15.38) |
| [Ni(C11H11N3S)(H2O)Cl2]·2H2O | 400.90 | Brown (78) | >300 | 16 | 32.54 (32.95) | 10.65 (10.48) | 4.03 (4.27) | 7.65 (7.99) | 14.32 (14.63) |
| [Cu(C11H11N3S)(H2O)Cl2]·4H2O | 441.77 | Black (60) | >300 | 7 | 29.45 (29.90) | 9.34 (9.51) | 2.23 (2.50) | 7.12 (7.25) | 14.11 (14.38) |
| [Cd(C11H11N3S)2(H2O)(NO3)]·(NO3)·2H2O | 725.00 | Light brown (80) | >300 | 80 | 36.51 (36.44) | 17.40 (17.38) | 3.12 (3.05) | 8.91 (8.83) | 15.21 (15.50) |
The FT-IR spectrum of the ligand displayed two peaks at 3464 and 3386 cm−1 which corresponds to asymmetric and symmetric N–H stretching of the amino group [34]. The complexes' spectra attributed to v(OH) showed a broad band between 3340 and 3442 cm−1 (Table 2) [35]. Additionally, the v(C=N) vibration shifted from 1613 cm−1 in the spectrum of the L to 1644–1630 cm−1 range in the spectra of complexes (Fig. S1) [36]. The peak attributed to δ(py) was moved to higher values after complexation, indicating coordination via the N atom of a pyridyl ring [37]. The coordinated υ(H2O) was seen in all IR spectra of complexes [38]. Also, The IR spectrum of the cadmium complex showed a band at 1384 cm−1 of ionic NO3 and three bands that were ascribed to the monodentate NO3 [39]. Finally, ν(M-S) vibrations were observed in the spectra of complexes except for that of the cadmium complex [40]. The bands at 530–535 cm−1 were assigned to M–N stretching [41].
Table 2.
IR spectra of the compounds.
| Assignments | υ(NH2) or υ(H2O) | υ(C=N) | Coord. υ(H2O) |
δ(py) | υ (M − N) | υ(M − S) | Additional bands |
|---|---|---|---|---|---|---|---|
| L | 3464 m, 3386 m | 1613s | – | 626w | – | – | – |
| Co-L | 3379b | 1644br | 987w | 631w | 532w | 468w | – |
| Ni-L | 3376b | 1630br | 926w | 630w | 530w | 457w | – |
| Cu-L | 3439b | 1631br | 979w | 631w | 532w | 450w | – |
| Cd-L2 | 3442b, 3340b | 1642br | 951w | 630w | 535w | – | 1384, 825 (ionic NO3) 453,1021,798 (Coord. NO3) |
s = strong; w = weak; br = broad; m = medium.
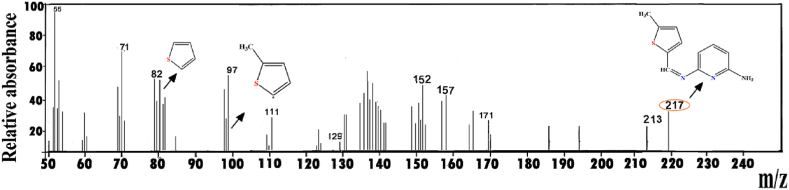
The ligand's mass spectrum showed a molecular ion peak (m/z = 217.00 amu), which agrees with its molecular formula (C11H11N3S) (Fig. 2). Fig. S2 depicts some notable fragmentation ions of the ligand. The molecular ion peaks at m/z = 383, 400, 441 and 752, which corresponds to the molecular weights of the relevant compounds, were seen in the mass spectra of the Co, Ni, Cu and Cd complexes, respectively.
Fig. 2.
Mass spectrum of the L ligand.
Three singlet signals at 4.32, 8.12, and 9.53 ppm were identified as CH3, –CH=N– and NH2 protons in the NMR spectrum of the ligand (Fig. S3) [42]. Aromatic protons were seen between δ6.23 and δ8.72 ppm [43]. The azomethine and aromatic protons in the NMR spectrum of complex cadmium displayed signals at δ 7.87 ppm and δ 6.12–7.45 ppm, respectively. This shift in peak values indicated the emergence of a complex [44].
3.2. Magnetic moments and electronic spectra
The electronic spectrum of L in DMSO revealed two absorbing peaks at 245 and 345 nm which were caused by π–π* and n–π* transition [45]. The wavelengths of π- π∗ were shifted to 237, 229, 239 and 238 nm for Co, Ni, Cd and Cu complexes, respectively [46]. The bands ascribed to n–π* for the Co, Ni, Cu and Cd complexes were noticed at 328, 330, 383 and 324 nm, respectively. The Co complex’s two absorption bands, which appeared at 526 and 698 nm in the spectra, were identified as (ν3) 4T1g(F) → 4T1g(P) and (ν2) 4T1g(F) → 4A2g(F), respectively [47]. The Ni complex diffuse reflectance spectrum observed two bands at 437 and 680 nm attributed to the d-d transitions (ν3) 3A2g(F) → 3T1g(P) and (ν2) 3A2g(F) → 3T1g(F) [48]. The octahedral shape of a Cd (II) (d10) compound has been proposed according to its empirical formula [49]. Co(II) (d7) and Ni(II) (d8) compounds were found to have magnetic moments of 4.6 and 2.64 BM, respectively [50,51]. This indicated that these compounds have an octahedral structure. The band is seen at 631 nm (2 Eg → 2T2g) in the diffuse reflection spectrum indicating an octahedral geometry and the magnetic moment of the Cu(II)(d9) complex was determined to be 1.76 BM [52,53].
3.3. Photoluminescence study
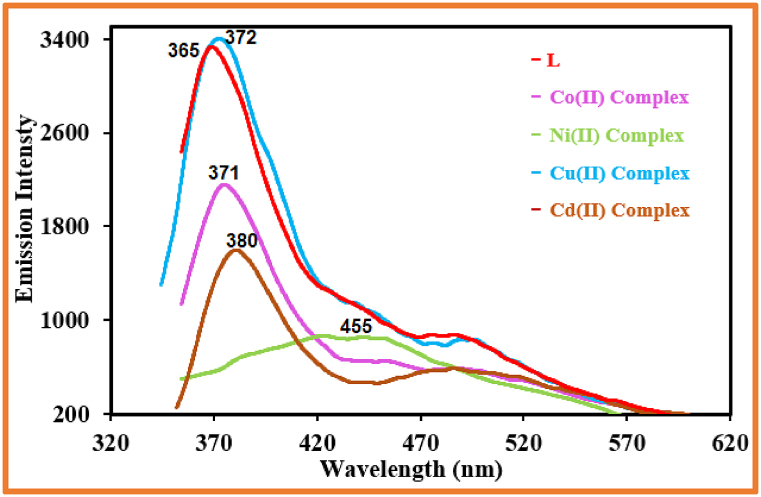
Fluorescence properties were seen in the synthesized compounds (Fig. 3). When excited at 309, 329, 340, 319, and 323 nm, the excitation spectra of the ligand and its Co, Ni, Cu, and Cd complexes revealed fluorescence emission bands at 365, 371, 455, 372, and 380 nm, respectively. This demonstrated that, due to their distinctive luminescence properties, all compounds have the potential to be photoactive substances [54].
Fig. 3.
Fluorescence spectra of the produced compounds.
3.4. Thermogravimetric analyses
The TGA curves of the all-new metal complexes for the thiophene ligand were collected in Fig. 4a–d. The thermal decomposition steps and assignment of the evolved species were presented in Table S1. TG curves of [Co(C11H11N3S)(H2O)Cl2]·H2O and [Ni(C11H11N3S)(H2O)Cl2]·2H2O were thermally decomposed in three stages. The first and second degradation stages in the Co complex led to weight losses of 5.11% (calc. 4.72%) and 48.72% (calc. 48.55%) due to the release of 2H2O, Cl2, and C5H9N2 fragments, respectively. The evolution of the C6H2NS fragment with the final remnant Co metal at 538–750 °C demonstrated the end process. The initial breakdown stage in the Ni complex occurred between 47 and 213 °C and involved the loss of 3H2O. Between 213 and 429 °C, the other part of a complex (½Cl2 and C2H2) were decomposed with a mass of 15.37% (calc. 14.63%). At temperatures between 429 and 726 °C, the last breakdown stage took place, leaving Ni metal as the byproduct. The TG and DTG curves of [Cu(C11H11N3S)(H2O)Cl2]·4H2O and [Cd(C11H11N3S)2 (NO3)(H2O)](NO3)·2H2O were decomposed into three and four main steps, respectively. In Cu(II) complex, the loss of 5H2O was consistent with the first stage between 50 and 149 °C. The complex was completely decomposed in the temperature range 149–741 °C at the last two steps leaving copper metal with a mass of 14.48% (calc. 14.38%) as residue. In Cd(II) complex, the loss of 2H2O hydrated and H2O coordinated occurred at 48–123 °C and 123–170 °C temperature ranges in the first and second stages, respectively. In the third step, C11H11N4O3S species was lost with a mass of 38.61% (calculated as 38.55%). The 4th step of decomposition entailed losing C11H11N4O2S between 513 and 708 °C, leaving CdO as the end product.
Fig. 4.
TG/DTG thermograms for (a) [CoL(H2O)Cl2]·H2O, (b) [NiL(H2O)Cl2]·2H2O, (c) [CuL(H2O)Cl2]·4H2O and (d) [CdL2(H2O)(NO3)](NO3)·2H2O.
3.5. Kinetic studies
The equation below visually determined kinetic parameters, including E* (energy of activation) and A (pre-exponential factor) [55]:
A straight line was produced by plotting log(log(W∞/(W∞-W))/T2 (log M) against 1/T (Fig. S4), where R, θ, W and W∞ represent the gas constant, the heating rate, the mass loss up to temperature T, and the final mass loss, respectively. Eyring equations can also be used to compute the additional parameters, including S* (entropy of activation), H* (enthalpy of activation), and G* (free energy change of activation) (Table S2): H* = E* - RT, G* = H* − TS*, S* = 2.3.3[log (Ah/kBTs)]R.
The Planck and Boltzmann constants, respectively, are h and kB, and Ts is the lowest temperature measured by the DTG. The findings revealed that all substances had negative activation of entropy, which suggested that breakdown processes proceed more slowly than usual.
3.6. Optical studies
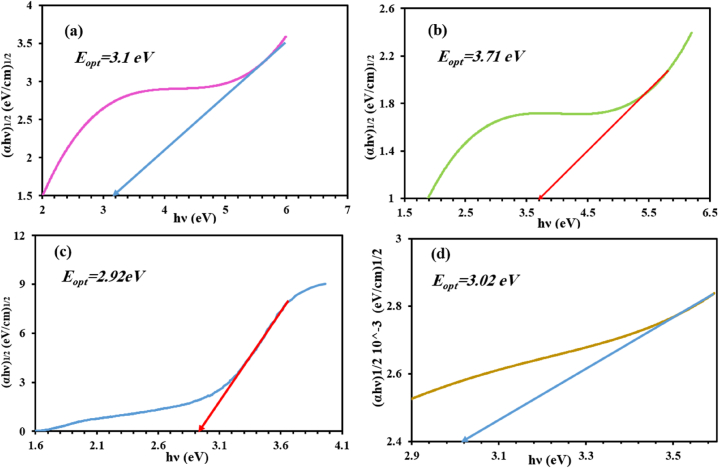
The optical energy gap (Eopt) for the studied complexes is determinable by considering that these complexes have indirect transitions during the occurrence of optical transition between the valence and conduction bands. According to the Tauc relation, the optical band energy gap (Eopt) value was evaluated [56]. The results showed that the values of Eopt for Co, Ni, Cu and Cd complexes were determined to be 3.1, 3.71, 2.92, and 3.02 eV, respectively (Fig. 5a–d). These findings suggested that these compounds may be employed as semiconductors in solar cell projects [57]. An Urbach energy Eu, is a parameter that indicated the effects of any defects or disorder degree in the structure of the complexes. From the equation: α = α0 exp(hυ/Eu), the value of Urbach energy Eu was discovered to be 0.22, 0.17, 0.19 and 0.23 eV for Co, Ni, Cu and Cd compounds, respectively (Fig. S5). These values are in agreement with previous work in the range Eu (0.045–0.66) eV for semiconductors [58].
Fig. 5.
Optical band gap for (a) [CoL(H2O)Cl2]·H2O, (b) [NiL(H2O)Cl2]·2H2O, (c) [CuL(H2O)Cl2]·4H2O and (d) [CdL2(H2O)(NO3)](NO3)·2H2O.
3.7. Catalytic reduction results
The most effective and eco-friendly application area to remove/degrade organic compounds such as nitroaromatic compounds is the catalytic reduction process [21]. In this process, the catalytic proficiency of synthesized compounds was examined by reducing 2-nitrophenol (2-NP) using NaBH4 at room temp [22]. The UV–vis absorption spectrum of 2-nitrophenol displayed peaks at 265 and 350 nm and with NaBH4 these peaks were shifted to 400 nm due to the production of 2-nitrophenolate ions via deprotonation [59] and a new peak due to the generation of 2-AP emerged around 270–280 nm (Fig. 6a). The catalytic reduction 2-NP in the presence of Co, Ni and Cu complexes as a catalyst can be monitored with time by measuring a decrease in absorbance at 400 nm and an increase at 270–280 nm due to the formation of 2-aminophenol (Fig. 6–d). The degradation of 2-NP occurred at 83.66%, 91.38% and 73.41% in the presence of Co, Ni and Cu complexes at 20, 20, and 25 min, respectively (Table S3 and Fig. 7). The reduction rate constant (k) was determined by applying pseudo-first-order kinetics concerning the concentration of 2-NP and the constant concentration of BH4− throughout the reaction [60]. A linear plot of ln(At/A0) vs reaction time t of the 2-NP reduction was used to get the K value (Fig. 6e). Co, Ni, and Cu metal complexes were found to have rate constant values of 12.06 × 10−1, 19.7 × 10−1 and 4.91 × 10−2 min−1, respectively. Fig. 8 illustrated the suggested mechanism for the reduction of 2-NP on the catalyst's surface. The mechanism was based on electron transfer (ET) from the BH4− donor to the 2-NP acceptor on the catalyst's surface via π-π interactions. The catalyst receives an electron from BH4− ion which reacts with the proton (H+) producing hydrogen molecules (H2). On the catalyst’s surface, the H2 molecules adsorb and reduce 2-NP to 2-AP.
Fig. 6.
Electronic spectra of reduction of 2-NP without catalyst and with complexes as catalyst (a–d) and (e) plot of reaction time (t) against ln(At/A0) versus.
Fig. 7.
The % of 2-NP reduction utilizing metal complexes as the catalyst changes with time.
Fig. 8.
The mechanism of 2-NP conversion to 2-AP.
3.8. Metal removal results
The adsorption of the studied metal ions with varied concentrations utilizing L was determined at various points in time. The findings showed that over time, the percentage of metal ion elimination increased (Fig. 9a). The maximum removal percentages of Co, Ni, Cu, and Cd ions were found to be 45.6, 45.7, 29.7, and 68.9% at 0.01 M, 55.4, 64.8, 53.2, and 70.5% at 0.001 M, and 65.8, 77.2, 68.6 and 87.7% at 0.0001 M, respectively. Furthermore, the percentage of metal ion removal appeared to rise as metal ion concentration fell (Fig. 9b) [61].
Fig. 9.
The percentage of Co, Ni, Cu, and Cd ions removed from solution.
3.9. Antimicrobial activities
Investigations on the biological effects of each compound were conducted on two fungi and four bacteria (Fig. 10). Table 3 provides a summary of the inhibition zone diameter (mm) and percent activity index data (Fig. S6). The ligand, L, is shown negligible activity against E. coli, A. flavus, and C. albicans and moderate activity against S. aureus, B. subtilis, and P. vulgaris. The most effective compound against the reference medication, all tested bacteria, and all tested fungi was Cd(II) complex. The Co, Ni, and Cu complexes had greater activity than the ligand, L.
Fig. 10.
Antimicrobial properties of synthetic substances and their effects.
Table 3.
Antimicrobial activity of the compounds.
| Gram- + ve bacteria |
Gram--ve bacteria |
Fungi |
||||
|---|---|---|---|---|---|---|
| S. aureus | B.subtilis | P.vulgaris | E.coli | A. flavus | C.albicans | |
| L | 11(46) | 15 (58) | 13(52) | NA | NA | NA |
| Co-L | 17 (71) | 18 (69) | 16 (64) | 20 (67) | 8 (50) | 12 (60) |
| Ni-L | 11 (46) | 15 (58) | 14 (56) | 16 (53) | 13 (81) | 13 (65) |
| Cu-L | 22 (92) | 19 (73) | 14 (56) | 12 (40) | 15 (94) | 14 (70) |
| Cd-L2 | 25 (104) | 40 (153.8) | 20 (80) | 34 (113) | 21 (131) | 20 (100) |
| Gentamycin | 24 | 26 | 25 | 30 | – | – |
| Ketoconazole | – | – | – | – | 16 | 20 |
| DMF | – | – | – | – | – | – |
% Activity index are in parentheses.
NA: No activity.
3.10. Minimum inhibitory concentration (MIC)
The MIC for the highly active Cd(II) complex against “S. aureus”, “B. subtilis”, and “E. coli” was calculated using the agar dilution method. A 4.94 μg/ml was found to be the complex's minimal inhibitory concentration (MIC).
3.11. Computational studies
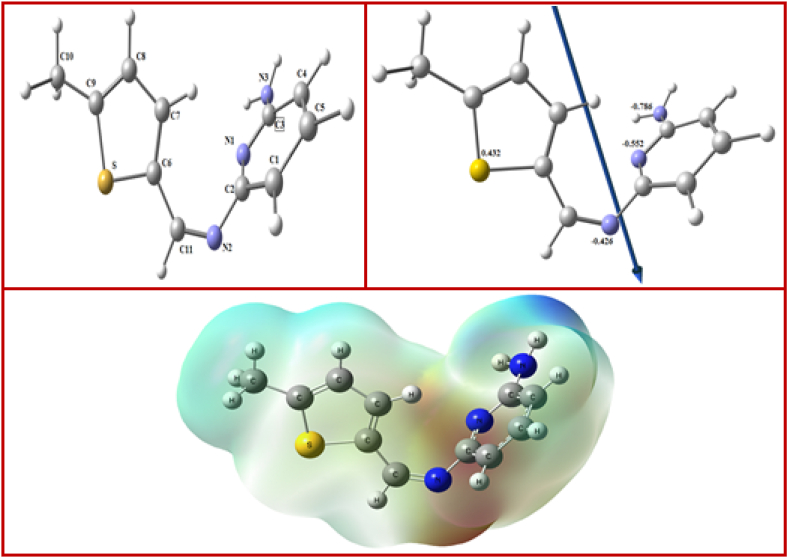
The optimized structure and surface of molecular electrostatic potential (MEP) of the ligand and the complexes were displayed in Fig. 11, Fig. 12. The octahedral geometry of the metal atoms had six coordinates. The atoms “N1, N2, S, and O” were discovered in the same plane but with a −2.060°, −0.312° and −2.543° deviation in Co, Ni, and Cu octahedral complexes field, respectively (Table 4). In Cd complex, the atoms N1, O1, N4 and O4 were deviated by +0.442° (Table S4). In complexation, the distances in the ligand between N2- - - –S, N1- - - –S and N1- - - -N2 were longer than those in the metal complexes Also, the N2-M-S bond angle was in the range 84.79–88.96 lower than 90° indicating a distorted octahedral structure. The bond angles N1-M-S, N2-M-O, Cl1-M-Cl2, N1–Cd–N4, N3–Cd–N6, O1–Cd–N4 showed a deviation from 180° (Tables 4 and S4). The ligand's natural bond orbital analysis and those of its complexes were investigated. The more negatively charged active positions were N1 (−0.552), N2 (−0.426), N3 (−0.786), and S (0.452), according to the natural charges (NBO). In the instance of the Co-L, NBO data on the atoms revealed Co (+0.114), N1(−0.695), O(−0.677), N2(−0.765), Cl1(−0.478), S(0.671), and Cl2(−0.442), while in the case of the Ni-L, NBO data revealed Ni (+0.037), N1(−0.526), O(−0.763), N2(−0.395), S(0.468), Cl2(−0.404) and Cl1(−(−0.365). Additionally, the charge on the atoms in the Cu-L was Cu (+1.354), N1(−0.677), O2(−0.783), N4(−0.617), N2(−0.741) Cl (−0.719) and N6(−0.626. Also, the atoms in Cd-L2 have the following charges: Cd (+1.395), O4(−0.664), O1(−0.905), N3(−0.580), N1(−0.623), N6 (−0.568) and N4(−0.634) (Fig. 12).
Fig. 11.
The ligand's vector of the dipole moment and molecule electrostatic potential surface, as well as their optimal structures.
Fig. 12.
Compounds' vector of the dipole moment and molecular electrostatic potential (MEP) surface, as well as their optimal structure.
Table 4.
Significant optimized bond lengths and angles.
| Type of bond | Bond length(Å) L |
Bond length (Å) Co-L |
Bond length (Å) Ni-L |
Bond length (Å) Cu-L |
Type of Angle | Angle (°) Co-L |
Angle (°) Ni-L |
Angle (°) Cu-L |
|---|---|---|---|---|---|---|---|---|
| M-N1 | – | 1.896 | 1.821 | 2.110 | N1-M-N2 | 70.79 | 71.28 | 66.09 |
| M-N2 | – | 1.840 | 1.837 | 1.969 | N1-M-O | 100.5 | 104.3 | 103.7 |
| M-S | – | 2.091 | 2.151 | 2.394 | N2-M-S | 87.26 | 88.96 | 84.79 |
| M-O | – | 2.028 | 1.795 | 2.114 | O-M-S | 101.4 | 95.45 | 105.3 |
| M-Cl1 | – | 2.335 | 2.157 | 2.433 | Cl1-M-N1 | 87.10 | 91.29 | 89.61 |
| M-Cl2 | – | 2.312 | 2.172 | 2.417 | Cl1-M-N2 | 102.2 | 91.91 | 101.1 |
| N1- - - - –S | 5.151 | 3.914 | 3.914 | 4.359 | Cl1-M-O | 78.94 | 88.75 | 80.72 |
| N2- - - - –S | 4.000 | 2.719 | 2.804 | 2.958 | Cl1-M-S | 96.98 | 89.40 | 95.17 |
| N1- - - - -N2 | 2.338 | 2.164 | 2.132 | 2.227 | Cl2-M-N1 | 89.22 | 89.63 | 91.08 |
| Cl2-M-N2 | 94.91 | 88.33 | 95.01 | |||||
| Cl2-M-O | 82.65 | 91.08 | 82.29 | |||||
| Cl2-M-S | 93.68 | 89.72 | 92.65 | |||||
| N1-M-S | 158.0 | 160.2 | 150.9 | |||||
| N2-M-O | 171.1 | 175.6 | 169.5 | |||||
| Cl1-M-Cl2 | 160.2 | 179.1 | 162.6 | |||||
| N1–N2–S–O | −2.060∗ | −0.312∗ | −2.543∗ |
Dihedral angle.
The frontier molecular orbital of all coordination compounds and the ligand are shown in Fig. 13. Due to the stability of the complexes, their overall energy was more negative than that of the free ligand. The energy gap in the L ligand was smaller than in complexes indicating complex formation (Fig. 13). The different reactivity descriptors (I, A, χ, μ, η, S and ω) are displayed in Table 5. Smaller values of η for the compounds represent the ability to transfer charges. As a result, the sequence of increasing its values within the investigated complexes was: Co complex > Ni complex > Cu complex > Cd complex > L.
Fig. 13.
The synthesized compounds' HOMO and LUMO charge density maps.
Table 5.
The computed quantum chemical parameters.
| Property | L | Co-L | Ni-L | Cu-L | Cd-L2 |
|---|---|---|---|---|---|
| The total energy E (a.u.) | −988.40 | −2130.19 | −2154.325 | −2181.29 | −2381.05 |
| HOMO (eV) | −5.74 | −5.02 | −4.42 | −6.24 | −8.16 |
| LUMO (eV) | −1.84 | −3.11 | −2.41 | −3.61 | −5.47 |
| Eg = ELUMO- EHOMO (eV) | 3.90 | 1.90 | 2.01 | 2.62 | 2.69 |
| Dipole moment (Debye) | 4.59 | 3.47 | 2.44 | 3.65 | 11.29 |
| Ionization potential I = -EHOMO |
5.74 | 5.02 | 4.42 | 6.24 | 8.16 |
| electron affinity A = -ELUMO (eV) |
1.84 | 3.11 | 2.41 | 3.61 | 5.47 |
| Electronegativity χ=(I + A)/2(eV) |
3.79 | 4.06 | 3.42 | 4.92 | 6.81 |
| chemical hardness η=(I -A)/2(eV) |
1.95 | 0.95 | 1.00 | 1.31 | 1.34 |
| chemical softness S = 1/2η(eV−1) |
0.25 | 0.52 | 0.49 | 0.38 | 0.37 |
| chemical potential μ = -χ |
−3.79 | −4.06 | −3.42 | −4.92 | −6.81 |
| Electrophilicity ω = μ2/2η |
3.68 | 8.68 | 5.82 | 9.23 | 17.26 |
3.12. Molecular docking studies
Molecular docking was carried out using the MOA2019 program to ascertain the many ways that the investigated chemicals bind to the active position of the “Bacillus subtilis” bacterium receptor (PDB ID:1QD9) [27,28]. Using the protein bank data, the bacterial receptor's crystal structure was obtained. The L and Co, Ni, Cu, and Cd complexes were found to have binding energies of −4.0, −42.7, −32.3, −59.5, and −67.8 kcal/mol, respectively (Table 6). The order of the interaction was [Cd(L)2(H2O)(NO3)]+ > [CuL(H2O)Cl2] > [CoL(H2O)Cl2] > [NiL(H2O)Cl2] > L according to the increase in the -ve binding energy. Fig. 14 depicts 2 and 3-dimensional plots showing the interaction of ligand and complexes with the Bacillus receptor. Additionally, molecular docking of the interaction between the ligand, copper, and cadmium complexes and the COVID-19 receptor (PDB ID: 6lu7) was examined (Fig. 15). Their structures were built in PDB format file from Gaussian09 program. From the bank website, the structures of the COVID-19 receptor of the main viral protease protein (PDB ID: 6lu7) were obtained. Table 7 showed the interaction results of the docking and binding free energy data. [Cd(L)2(H2O)(NO3)]+ showed stronger interaction than both [CuL(H2O)Cl2] and ligand, L, since it has more negative binding energy.
Table 6.
Docking calculations data of interaction L and its complexes with the active sites of the receptor of B. subtilis gene regulation (PDB ID: 1QD9).
| Receptor | Interaction | Distance(Å)∗ | E (kcal/mol) | |
|---|---|---|---|---|
| L | ||||
| O 23 | OD1 ASN 25 (A) | H-donor | 2.77 (1.81) | −1.9 |
| N 17 | ND2 ASN 24 (C) | H-acceptor | 3.08 (2.15) | −2.1 |
| [CoL(H2O)Cl2] | ||||
| N 15 | OD2 ASP 80 (A) | H-donor | 2.87 (1.75) | −3.7 |
| O 30 | OD2 ASP 80 (A) | H-donor | 2.60 (2.11) | −15.2 |
| N 4 | OD2 ASP 80 (A) | Ionic | 3.02 | −4.3 |
| S 13 | OD1 ASP 80 (A) | Ionic | 3.11 | −3.8 |
| S 13 | OD2 ASP 80(A) | Ionic | 3.35 | −2.5 |
| N 15 | OD2 ASP 80 (A) | Ionic | 2.87 | −5.4 |
| O 30 | OD2 ASP 80 (A) | Ionic | 2.60 | −7.8 |
| [NiL(H2O)Cl2] | ||||
| O 30 | OE2 GLU 119 (C) | H-donor | 2.71 (1.81) | −16.6 |
| N 4 | OE2 GLU 119 (A) | Ionic | 3.38 | −2.4 |
| S 13 | OE2 GLU 119 (B) | Ionic | 2.88 | −5.4 |
| N 14 | OE2 GLU 119 (B) | Ionic | 3.73 | −1.1 |
| O 30 | OE2 GLU 119 (C) | Ionic | 2.71 | −6.8 |
| [CuL(H2O)Cl2] | ||||
| O 30 | OD1 ASP 80 (A) | H-donor | 2.63 (1.63) | −13.8 |
| O 30 | OE1 GLU 106(A) | H-donor | 2.71 (1.76) | −18.6 |
| N 4 | OE2 GLU 106(A) | Ionic | 3.86 | −0.8 |
| S 13 | OD1 ASP 80 (A) | Ionic | 3.36 | −2.5 |
| S 13 | OD2 ASP 80 (A) | Ionic | 3.44 | −2.1 |
| N 15 | OE2 LU 106 (A) | Ionic | 2.82 | −5.8 |
| O 30 | OD1 ASP 80 (A) | Ionic | 2.63 | −7.5 |
| O 30 | OE1 GLU 106 (A) | Ionic | 2.71 | −6.7 |
| O 30 | OE2 GLU 106 (A) | Ionic | 3.77 | −1.0 |
| 6-ring | CD ARG 109 (B) | pi-H | 3.69 | −0.7 |
| [Cd(L)2(H2O)(NO3)]+ | ||||
| O 23 | O GLU 82 (A) | H-donor | 2.64 (1.72) | −12.3 |
| O 23 | OE2 GLU 86 (A) | H-donor | 2.69 (1.69) | −25.0 |
| S 43 | OE1 GLU 82 (A) | H-donor | 2.98 (1.95) | −6.6 |
| N 4 | OE2 GLU 86 (A) | Ionic | 3.87 | −0.8 |
| N 10 | OE2 GLU 86 (A) | Ionic | 2.95 | −4.8 |
| O 23 | OE1 GLU 86 (A) | Ionic | 2.77 | −6.2 |
| O 23 | OE2 GLU 86 (A) | Ionic | 2.69 | −7.0 |
| S 43 | OE1 GLU 82 (A) | Ionic | 2.98 | −4.6 |
| 6-ring | CB GLU 82 (A) | pi-H | 3.69 | −0.5 |
The lengths of H-bonds are in brackets.
Fig. 14.
2- and 3-dimension plots of the interactions between studied compounds with the receptor of B. subtilis.
Fig. 15.
L, Cu, and Cd complexes engage in a molecular docking interaction with the COVID-19 receptor’s active site.
Table 7.
Docking calculations data of interaction L and its Cu and Cd complexes with the active sites of the receptor of COVID-19.
| Receptor | Interaction | Distance(Å)∗ | E (kcal/mol) | |
|---|---|---|---|---|
| L | ||||
| N 13 | O PHE 140 | H-donor | 3.17 (2.15) | −1.2 |
| N 12 | N GLU 166 | H-acceptor | 3.32 (2.47) | −0.5 |
| 5-ring | NE2 GLN 189 | pi-H | 4.38 | −0.6 |
| [CuL(H2O)Cl2] | ||||
| O 30 | OD1 ASP 153 | H-donor | 2.66 (1.67) | −20.0 |
| O 30 | OD2 ASP 153 | H-donor | 2.96 (2.18) | −2.6 |
| N 14 | OD1 ASP 153 | Ionic | 3.08 | −3.9 |
| O 30 | OD1 ASP 153 | Ionic | 2.66 | −7.2 |
| O 30 | OD2 ASP 153 | Ionic | 2.96 | −4.8 |
| 6-ring | 6-ring PHE 294 | pi-pi | 3.64 | −0.1 |
| [Cd(L)2(H2O)(NO3)]+ | ||||
| O 23 | OD1 ASP 33 | H-donor | 2.58 (1.64) | −24.3 |
| O 23 | OD2 ASP 33 | H-donor | 2.99 (2.33) | −2.7 |
| C 34 | OD1 ASP 33 | H-donor | 3.39 (2.6) | −0.5 |
| N 4 | OD1 ASP 33 | Ionic | 2.95 | −4.8 |
| N 4 | OD2 ASP 33 | Ionic | 3.85 | −0.8 |
| N 10 | OD1 ASP 33 | Ionic | 3.41 | −2.3 |
| N 10 | OD2 ASP 33 | Ionic | 2.99 | −4.6 |
| O 23 | OD1 ASP 33 | Ionic | 2.58 | −8.0 |
| O 23 | OD2 ASP 33 | Ionic | 2.99 | −4.5 |
| N 33 | OD1 ASP 33 | Ionic | 3.74 | −1.1 |
The lengths of H-bonds are in brackets.
The inhibition constants for interactions were obtained from the equation (Ki) = exp(ΔG × 1000)/(RT), where ΔG is the docking binding energy kCal/mol, R gas constant = 1.9872 cal, and T = 298.15 K [3]. The inhibition constants are 2.004E-44, 1.238E-38, 5.017E-26, and 2.107E-18 μM for Cd, Cu, Co, Ni complexes, respectively and 1169.388 for the free ligand. The more negative binding energy the lower inhibition constant the more stable the interaction between the complex and the protein i.e, the Cd complex has the largest affinity of interaction.
4. Conclusion
This work involved the synthesis and use of several spectral techniques (IH NMR, IR, mass spectra, UV–Visible, and thermogravimetric analysis) to describe the Schiff base ligand produced from 5-Methyl-2-carboxaldehyde-thiophene and 2,6-pyridinediamine and its four complexes. The low conductance of the cobalt, copper and nickel complexes supports the non-electrolytic nature of the complexes. The fluorescence spectra of all the synthesized compounds have been recorded. The presence of water in or out of the coordination sphere was found with the help of thermogravimetric analysis. The kinetic parameters were calculated with the help of Coats-Redfern equations. Based on elemental and spectral studies, six coordinated geometry has been assigned for all the metal complexes. The study of the optical properties of complexes showed that these complexes can be employed as semiconductors in solar cells. The catalytic activity of the synthesized compounds in the reduction of 2-nitrophenol was investigated. It has a variable degree of catalytic efficiency. The complexes displayed stronger antibacterial activity than the ligand, L. The Cd complex was also discovered to have the highest measured activity. The geometry of the substances under study was examined using the DFT method. The determination of the free energy binding of the compounds included a study of their simulated docking. At the active sites, the complexes demonstrated better binding than the ligand.
Author contribution statement
Doaa A. Nassar: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Omyma A. M. Ali: Conceived and designed the experiments.
Mohamed R. Shehata: Contributed reagents, materials, analysis tools or data.
Abeer S.S. Sayed: Performed the experiments.
Data availability statement
Data will be made available on request.
Ethical approval
The manuscript does not include any studies on humans or animals.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e16973.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Abu-Dief M., Mohamed I.M.A. A review on versatile applications of transition metal complexes incorporating Schiff bases. Beni-Suef Univ. J. Basic and Appl. Sci. 2015;4:119–133. doi: 10.1016/j.bjbas.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu-Dief A.M., El-Metwaly N.M., Alzahrani S.O., Bawazeer A.M., Shaaban S., Adam M.S.S. Targeting ctDNA binding and elaborated in-vitro assessments concerning novel Schiff base complexes: synthesis, characterization, DFT and detailed in-silico confirmation. J. Mol. Liq. 2021;322 doi: 10.1016/j.molliq.2020.114977. [DOI] [Google Scholar]
- 3.Koca I., Yiğitcan S., Gümüş M., Gökce H., Sert Y. A new series of sulfa drugs containing pyrazolyl acylthiourea moiety: synthesis, experimental and theoretical spectral characterization and molecular docking studies. J. Mol. Struct. 2020;1204 doi: 10.1016/j.molstruc.2019.127479. . [DOI] [Google Scholar]
- 4.Gümüş M., Babacan Ş.N., Demir Y., Sert Y., Koca I., Gülçin I. Discovery of sulfadrug–pyrrole conjugates as carbonic anhydrase and acetylcholinesterase inhibitors. Arch. Pharm. 2022;355(1):1–14. doi: 10.1002/ardp.202100242. . [DOI] [PubMed] [Google Scholar]
- 5.Almalki S.A., Bawazeer T.M., Asghar B., Alharbi A., Aljohani M.M., Khalifa M.E., El-Metwaly N. Synthesis and characterization of new thiazole-based Co (II) and Cu (II) complexes; therapeutic function of thiazole towards COVID-19 in comparing to current antivirals in treatment protocol. J. Mol. Struct. 2021;1244:130961–130973. doi: 10.1016/j.molstruc.2021.130961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Gammal O.A., Mohamed F.Sh., Rezk G.N., El-Bindary A.A. Structural characterization and biological activity of a new metal complexes based of Schiff base. J. Mol. Liq. 2021;330 doi: 10.1016/j.molliq.2021.115522. [DOI] [Google Scholar]
- 7.Suneetha G., Ayodhya D., Manjari P.S. Schiff base stabilized gold nanoparticles: synthesis, characterization, catalytic reduction of nitroaromatic compounds, fluorometric sensing, and biological activities. Results Chem. 2023;5:100688–100699. doi: 10.1016/j.rechem.2022.100688. [DOI] [Google Scholar]
- 8.Eltaweil A.S., Hashem O.A., Abdel-Hamid H., Abd El-Monaem E.M., Ayoup M.S. Synthesis of a new magnetic Sulfacetamide-Ethylacetoacetate hydrazone-chitosan Schiff-base for Cr(VI) removal. Int. J. Biol. Macromol. 2022;222:1465–1475. doi: 10.1016/j.ijbiomac.2022.09.081. [DOI] [PubMed] [Google Scholar]
- 9.Betiha M.A., Moustafa Y.M., El-Shahat M.F., Rafik E. Polyvinylpyrrolidone-Aminopropyl-SBA-15 schiff Base hybrid for efficient removal of divalent heavy metal cations from wastewater. J. Hazard Mater. 2020;397 doi: 10.1016/j.jhazmat.2020.122675. [DOI] [PubMed] [Google Scholar]
- 10.Dailey S., Feast J.W., Peace R.J., Sage I.C., Till S., Wood E.L. Synthesis and device characterisation of side-chain polymer electron transport materials for organic semiconductor applications, Metal-organic conductors special issue: editorial. J. Mater. Chem. 2001;11:2238–2243. doi: 10.1039/b104674h. [DOI] [Google Scholar]
- 11.Leung C.H., Lin S., Zhong H.J., Ma D.L. Metal complexes as potential modulators of inflammatory and autoimmune responses. Chem. Sci. 2015;6:871–884. doi: 10.1039/C4SC03094J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turan N., Buldurun K., Bursal E., Mahmoudi G. Pd(II)-Schiff base complexes: synthesis, characterization, Suzuki–Miyaura and Mizoroki–Heck cross-coupling reactions, enzyme inhibition and antioxidant activities. J. Organomet. Chem. 2022;970–971 doi: 10.1016/j.jorganchem.2022.122370. [DOI] [Google Scholar]
- 13.Kirubavathy S.J., Chitra S. Synthesis, characterization, DFT, In-vitro anti-microbial, cytotoxicity evaluation, and DNA binding interactions of transition metal complexes of quinoxaline Schiff base ligand. Mater. Today: Proc. 2020;33:2331–2350. doi: 10.1016/j.matpr.2020.04.699. [DOI] [Google Scholar]
- 14.Li X., Li X.-.Q., Liu H.-.M., Zhou X.-.Z., Shao Z.-.H. Synthesis and evaluation of antitumor activities of novel chiral 1,2,4-triazole Schiff bases bearing γ-butenolide moiety. Org. Med. Chem. Lett. 2012;2(26):1–5. doi: 10.1186/2191-2858-2-26. http://www.orgmedchemlett.com/content/2/1/26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keshtkar N., Zamanpour A., Esmaielzadeh S. Bioactive Ni(II), Cu(II) and Zn(II) complexes with an N3 functionalized Schiff base ligand: synthesis, structural elucidation, thermodynamic and DFT calculation studies. Inorg. Chim. Acta. 2022;541(15) doi: 10.1016/j.ica.2022.121083. [DOI] [Google Scholar]
- 16.Ahmed Y.M., Mohamed G.G. Synthesis, spectral characterization, antimicrobial evaluation and molecular docking studies on new metal complexes of novel Schiff base derived from 4,6-dihydroxy-1,3-phenylene-diethanone. J. Mol. Struct. 2022;1256 doi: 10.1016/j.molstruc.2022.132496. [DOI] [Google Scholar]
- 17.Alyaninezhad Z., Bekhradnia A., Gorgi R.Z., Ghanbarimasir Z., Fouladpour M. Synthesis, characterization and biological activity of Mn(II), Co(II), Ni(II), Cu(II) and Zn(II) complexes derived from Schiff base ligand quinoxaline-2-carboxaldehyde and 4-aminoantipyrine. J. Mol. Struct. 2022;1262 doi: 10.1016/j.molstruc.2022.132990. [DOI] [Google Scholar]
- 18.Hasan Md.R., Hossain M.A., Salam Md.A., Uddin M.N. Nickel complexes of Schiff bases derived from mono/diketone with anthranilic acid: synthesis, characterization and microbial evaluation. J. Taibah Univ. Sci. 2016;10:766–773. doi: 10.1016/j.jtusci.2015.11.007. [DOI] [Google Scholar]
- 19.Coombs R.R., Westcott S.A., Decken A., Baerlocher F. Palladium(II) Schiff base complexes derived from sulfanilamides and aminobenzothiazoles. J. Trans. Met. Chem. 2005;30:411–418. doi: 10.1007/s11243-004-7625-4. [DOI] [Google Scholar]
- 20.Omar M.M., Mohammed G.G. Potentiometric, spectroscopic and thermal studies on the metal chelates of 1-(2-thiazolylazo)-2-naphthalenol. Spectrochim. Acta. 2005;61:929–936. doi: 10.1016/j.saa.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 21.Vaidya M.J., Kulkarni S.M., Chaudhari R.V. Synthesis of p-aminophenol by catalytic hydrogenation of p-nitrophenol. Org. Process Res. Dev. 2003;27:202–208. doi: 10.1021/op025589w. [DOI] [Google Scholar]
- 22.Abbar A.H., Sulaymon A.H., Jalhoom M.G. Scale-up of a fixed bed electrochemical reactor consisting of parallel screen electrode used for p-aminophenol production. Electrochim. Acta. 2007;53:1671–1679. doi: 10.1016/j.electacta.2007.07.075. [DOI] [Google Scholar]
- 23.Vogel A.I. A text book of quantitative chemical Analysis. 5th ed. Longman’s; London: 1989. [Google Scholar]
- 24.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B.G., Gomperts R., Mennucci B., Hratchian H.P., Ortiz J.V., Izmaylov A.F., Sonnenberg J.L., Williams-Young D., Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V.G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Keith T., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Millam J.M., Klene M., Adamo C., Cammi R., Ochterski J.W., Martin R.L., Morokuma K., Farkas O., Foresman J.B., Fox D.J. Gaussian, Inc.; Wallingford CT): 2016. Gaussian 09, Revision A.02.https://gaussian.com/g09citation/ [Google Scholar]
- 25.Aljohani E.T., Shehata M.R., Alkhatib F., Alzahrani S.O., Abu Dief A.M. Development and structure elucidation of new VO2+, Mn2+, Zn2+, and Pd2+ complexes based on azomethine ferrocenyl ligand: DNA interaction, antimicrobial, antioxidant, anticancer activities, and molecular docking. Appl. Organomet. Chem. 2021;35 e6154:1–24. doi: 10.1002/aoc.6154. [DOI] [Google Scholar]
- 26.Mahmudov I., Demir Y., Sert Y., Abdullayev Y., Sujayev A., Alwasel S.H., Gulcin I. Synthesis and inhibition profiles of N-benzyl-and N-allyl aniline derivatives against carbonic anhydrase and acetylcholinesterase–A molecular docking study. Arab. J. Chem. 2022;15(3) doi: 10.1016/j.arabjc.2021.103645. . [DOI] [Google Scholar]
- 27.Sinha S., Rappu P., Lange S.C., Mäntsälä P., Zalkin H., Smith J.L. Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved. YjgF Family. 1999;96(23):13074–13079. doi: 10.1073/pnas.96.23.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molecular Operating Environment (MOE) 2019.01; Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2021. https://www.chemcomp.com/Research-Citing_MOE.htm
- 29.Abou-Hussein A.A., Linert W. Synthesis, spectroscopic studies and inhibitory activity against bacteria and fungi of acyclic and macrocyclic transition metal complexes containing a triamine coumarin Schiff base ligand. Spectrochim. Acta. 2015;141:223–232. doi: 10.1016/j.saa.2015.01.063. [DOI] [PubMed] [Google Scholar]
- 30.Souza L.C., Rodrigues N.S., Cunha D.A., Feitosa V.P., Santiago S.L., Reis A., Loguercio A.D., Perdigão J., Saboia V.P.A. Two-year clinical evaluation of a proanthocyanidins-based primer in non-carious cervical lesions: a double-blind randomized clinical, trial. J. Dent. 2020;96:103325–103359. doi: 10.1016/j.jdent.2020.103325. [DOI] [PubMed] [Google Scholar]
- 31.Jesus R.S., Piana M., Freitas R.B., Brum T.F., Alves C.F.S., Belke B.V., Mossmann N.J., Cruz R.C., Santos R.C.V., Dalmolin T.V., Bianchini B.V., Campos M.M.A., Bauermann L.F. In vitro antimicrobial and antimycobacterial activity and HPLC–DAD screening of phenolics from Chenopodium ambrosioides. L. Braz. J. Microbiol. 2018;49:296–302. doi: 10.1016/j.bjm.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong K.Y., Vikram P., Chiruvella K.K., Mohammed A. Phytochemical screening and antimicrobial potentials of Borreria sps (Rubiaceae) J. King Saud Univ. Sci. 2015;27:302–311. doi: 10.1016/j.jksus.2014.12.001. [DOI] [Google Scholar]
- 33.Ayoub M.A., Abd-Elnasser E.H., Ahmed M.A., Rizk M.G. Some new metal (II) complexes based on bis-Schiff base ligand derived from 2-acetylethiophine and 2,6-diaminopyridine: syntheses, structural investigation, thermal, fluorescence and catalytic activity studies. J. Mol. Struct. 2018;1163:379–387. doi: 10.1016/j.molstruc.2018.03.006. [DOI] [Google Scholar]
- 34.Ali O.A.M., El-Medani S.M., Ahmed D.A., Nassar D.A. Synthesis, characterization, fluorescence and catalytic activity of some new complexes of unsymmetrical Schiff base of 2-pyridinecarboxaldehyde with 2,6-diaminopyridine. Spectrochim. Acta. 2015;144:99–106. doi: 10.1016/j.saa.2015.02.078. [DOI] [PubMed] [Google Scholar]
- 35.Ali O.A.M., El-Medani S.M., Ahmed D.A., Nassar D.A. Metal carbonyl complexes with Schiff bases derived from 2-pyridinecarboxaldehyde: syntheses, spectral, catalytic activity and antimicrobial activity studies. J. Mol. Struct. 2014;1074(25):713–722. doi: 10.1016/j.molstruc.2014.05.035. [DOI] [Google Scholar]
- 36.Li X., Li Ch.-H., Jiang J.-H., Gu H.-W., Wei D.-L., Ye L.-J., Hu J.-L., Xiao S.-X., Guo D.-C., Li X., Zhang H., Li Q.-G. Synthesis and microcalorimetric determination of the bioactivities of a new Schiff base and its bismuth(III) complex derived from o-vanillin and 2,6-pyridinediamine. J. Therm. Anal. Calorim. 2017;127:1767–1776. doi: 10.1007/s10973-016-5892-x. [DOI] [Google Scholar]
- 37.Tofiq D.I., Hassan H.Q., Abdalkarim K.A. Preparation of a novel Mixed-Ligand divalent metal complexes from solvent free Synthesized Schiff base derived from 2,6-Diaminopyridine with cinnamaldehyde and 2,20-Bipyridine: characterization and antibacterial activities. Arab. J. Chem. 2021;14 doi: 10.1016/j.arabjc.2021.103429. [DOI] [Google Scholar]
- 38.Mohamed G.G., Attia H.A.E., Ibrahim N.A., Synthesis Characterization and fungicidal potentialities of some transition metal complexes of benzimidaz-oledithiocarbamate based ligand, Egypt. J. Agric. Res. 2018;96(2):527–543. doi: 10.21608/ejar.2018.135726. [DOI] [Google Scholar]
- 39.El-Boraey H.A., Serag El-Din A.A.S. Transition metal complexes of a new 15-membered [N5] penta-azamacrocyclic ligand with their spectral and anticancer studies. Spectrochim. Acta. 2014;132:663–671. doi: 10.1016/j.saa.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Shakira M., Abbasia A., Azama M., Khanb A.U. Synthesis, spectroscopic studies and crystal structure of the Schiff base ligand L derived from condensation of 2-thiophenecarboxaldehyde and 3,3-diaminobenzidine and its complexes with Co(II), Ni(II), Cu(II), Cd(II) and Hg(II): comparative DNA binding studies of L and its Co(II), Ni(II) and Cu(II) complexes. Spectrochim. Acta. 2011;79:1866–1875. doi: 10.1016/j.saa.2011.05.077. [DOI] [PubMed] [Google Scholar]
- 41.Mishra A.P., Tiwari A., Gupta S.K., Jain R., Synthesis Spectral and antimicrobial studies of some Co(II), Ni(II) and Cu(II) complexes containing 2-thiophenecarboxaldehyde moiety. E-J. Chem. 2012;9(3):1113–1121. doi: 10.1155/2012/585827. [DOI] [Google Scholar]
- 42.Mohammed L.W., Irzoqi A.A. New 3- hydrazonoindolin-2-one Cd(II) complexes with amino pyridine ligands, Synthesis, Characterization and biological activity evaluation. Tikrit J. Pure Sci. 2020;25(2):38–46. doi: 10.25130/tjps.25.2020.028. [DOI] [Google Scholar]
- 43.Daoud A.M., Mahdi H.A. Synthesis, characterization and evaluation biological activity of new Schiff base compounds and their complexes with some transition elements. J.Thi-Qar Sci. 2017;6(2):81–89. https://www.jsci.utq.edu.iq/index.php/main/article/view/7 [Google Scholar]
- 44.Mishra N., Gound S.S., Mondal R., Yadav R., Pandey Synthesis R. Characterization and antimicrobial activities of benzothiazoleimino-benzoic acid ligands and their Co(II), Ni(II), Cu(II), Zn(II) and Cd(II) complexes. Results Chem. 2019;1 doi: 10.1016/j.rechem.2019.100006. [DOI] [Google Scholar]
- 45.Ayoub M.A., Abd-Elnasser E.H., Abedel-aziz M.A., Rizk M.G. Synthesis, physicochemical, thermal and fluorescence studies of cobalt(II) complex with tridentate (ONS) schiff base ligand derived from 2-aminophenol and 2-acetylthiophene. J. Sci. Res. Sci. Article. 2016;33(1):434–448. doi: 10.21608/JSRS.2016.18341. [DOI] [Google Scholar]
- 46.Ali O.A.M., Abd El Wahab Z.H., Ismail B.A. Synthesis, structural characterization and evaluation of catalytic and antimicrobial properties of new mononuclear Ag(I), Mn(II), Cu(II) and Pt(IV) complexes. J. Mol. Struct. 2017;1139:175–195. doi: 10.1016/j.molstruc.2017.03.025. [DOI] [Google Scholar]
- 47.Abd El-Wahab Z.H., Marshaly M.M., Salman A.A., El-Shetary B.A., Faheim A.A. Co(II), Ce(III) and UO2(VI) bis-salicylatothiosemicarbazide complexes: binary and ternary complexes, thermal studies and antimicrobial activity. Spectrochim. Acta. 2004;60:2861–2873. doi: 10.1016/j.saa.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Megahed A.S., Al-Amoudi M.S., Refat M.S. A modern technique for preparation of zinc(II) and nickel(II) nanometric oxides using Schiff base compounds: synthesis, characterization, and antibacterial properties. Res. Chem. Intermed. 2014;40:1425–1439. doi: 10.1007/s11164-013-1049-8. [DOI] [Google Scholar]
- 49.Bharty M.K., Dani R.K., Kushawaha S.K., Singh N.K., Kharwar R.N., Butcher R.J. Mn(II), Ni(II), Cu(II), Zn(II), Cd(II), Hg(II) and Co(II) complexes of 1-phenyl-1Htetrazole-5-thiol: synthesis, spectral, structural characterization and thermal studies. Polyhedron. 2015;88:208–221. https://doi:10.1016/j.poly.2014.12.013 [Google Scholar]
- 50.Jha N.N., Ray I.P. Magnetic studies of Co(II) and Ni(II) complexes of hydroxamic acid. Asian J. Chem. 2000;12(3):703–706. https://asianjournalofchemistry.co.in/User/ViewFreeArticle.aspx?ArticleID=12_3_15 [Google Scholar]
- 51.Singh N.P., Anu, Singh J.V. Magnetic and spectroscopic studies of the synthesized metal complexes of bis(pyridine-2-carbo) hydrazide and their antimicrobial studies. E-J. Chem. 2012;9(4):1835–1842. doi: 10.1155/2012/521345. [DOI] [Google Scholar]
- 52.AL-Jibouri M.N., Jawad W.A., Balakit A.A., Obies M. Synthesis, characterization and theoretical study of some transition metal complexes with new schiff base derived from 1,2,4-triazole, Egypt. J. Chem. 2021;64:5227–5239. doi: 10.21608/EJCHEM.2021.71885.3580. [DOI] [Google Scholar]
- 53.Patil S.A., Unki Sh.N., Kulkarni A.D., Naik V.H., Kamble U., Badami P.S. Spectroscopic, in vitro antibacterial, and antifungal studies of Co(II), Ni(II), and Cu(II) complexes with 4-chloro-3-coumarinaldehyde Schiff bases. J. Coord. Chem. 2010;64(2):323–336. doi: 10.1080/00958972.2010.541240. [DOI] [Google Scholar]
- 54.Ashraf A., Islam M., Khalid M., Davis A.P., Ahsan M.T., Yaqub M., Syed A., Elgorban A.M., Bahkali A.H., Shafq Z. Naphthyridine derived colorimetric and fluorescent turn of sensors for Ni2+ in aqueous media. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-98400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Firuzabadi F.D., Asadi Z., Yousefi R. Synthesis of new nano Schiff base complexes: X-ray crystallography, thermal, electrochemical and anticancer studies of nano uranyl Schiff base complexes. Bull. Chem. Soc. Ethiop. 2018;32(1):89–100. doi: 10.4314/bcse.v32i1.8. [DOI] [Google Scholar]
- 56.Karipcin F., Dede B., Caglar Y., Hur D., Ilican S., Caglar M., Sahin Y. A new dioxime ligand and its trinuclear copper(II) complex: synthesis, characterization and optical properties. Opt Commun. 2007;272:131–137. doi: 10.1016/j.optcom.2006.10.079. [DOI] [Google Scholar]
- 57.Hosny N.M., Othman E., El Dossoki F.I. [Cd(Anthranilate)2]H2O as a precursor of CdO nanoparticles. J. Mol. Struct. 2019;1195:723–732. doi: 10.1016/j.molstruc.2019.06.041. [DOI] [Google Scholar]
- 58.Sumalatha B., Omkaram I., Rajavardhana Rao T., Linga Raju Ch. The effect of V2O5 on alkaline earth zinc borate glasses studied by EPR and optical absorption. J. Mol. Struct. 2011;1006:96–103. doi: 10.1016/j.molstruc.2011.08.025. [DOI] [Google Scholar]
- 59.Jeong W.K., Jiulong L., Akira S., Kazuyuki T., Weon B.K. Reduction of 2-nitrophenol using a hybrid C–Ni nanocomposite as a catalyst. Micro Nano Lett. 2018;13(9):1310–1314. doi: 10.1049/mnl.2018.0171. [DOI] [Google Scholar]
- 60.Suneetha G., Ayodhya D., Manjari P.S. Schiff base stabilized gold nanoparticles: synthesis, characterization, catalytic reduction of nitroaromatic compounds, fluorometric sensing, and biological activities. Results Chem. 2023;5 doi: 10.1016/j.rechem.2022.100688. [DOI] [Google Scholar]
- 61.Mahmoud W.H., Mohamed G.G., Elsawy H.A., Radwan M.A. Metal complexes of novel Schiff base derived from the condensation of 2-quinoline carboxaldehyde and ambroxol drug with some transition metal ions. Appl. Organomet. Chem. 2018;32(7):4392–4406. doi: 10.1002/aoc.4392. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.