Summary
Mouse models of colorectal cancer (CRC) have been crucial in the identification of the role of genes responsible for the full range of pathology of the human disease and have proved to be dependable for testing anti-cancer drugs. Recent research points toward the relevance of tumor, angiogenic, and immune microenvironments in CRC progression to late-stage disease, as well as the treatment of it. This study examines important mouse models in CRC, discussing inherent strengths and weaknesses disclosed during their construction. It endeavors to provide both a synopsis of previous work covering how investigators have defined various models and to evaluate critically how researchers are most likely to use them in the future. Accumulated evidence regarding the metastatic process and the hope of using checkpoint inhibitors and immunological inhibitor therapies points to the need for a genetically engineered mouse model that is both immunocompetent and autochthonous.
Subject areas: Cancer, Model organism
Graphical abstract

Cancer; Model organism
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide with more than 1.9 million new cases diagnosed in 2020.1 Almost half the population of developed countries will have at least one benign intestinal tumor during their lifetime.2 There has been a decrease in mortality attributed to better screening and removal of early lesions in people aged forty-five and older. However, clinicians increasingly see younger people with early CRC that may be attributable to age versus screening recommendations or diet.3,4,5,6,7 Overall survival frequency is 64% at five years;8 staging at diagnosis is the most significant indicator for survival. At diagnosis approximately 35% of CRC patients are stage IV metastatic; twenty to fifty percent of stage II and stage III patients progress to metastatic disease. Conventional therapeutic approaches include surgery, cytotoxic agents, targeted therapies, and immune checkpoint inhibitors that extend survival for 12 to 24 months. Almost all develop resistance, and expert opinion generally recognizes the inadequacy of the approaches. This has led to the suggestion that new pharmaceutical targets and immuno-oncologic options be identified. Target authentication ultimately depends on understanding the cause of the disease and the role different genetic and genomic changes play in its initiation and progression. In an appropriate, if somewhat ironic turn of events, the development of genetically engineered mouse models (GEMMs) that more closely mimic late-stage disease are being called for to help elucidate how mutations cooperate in the natural environment.9 Investigators are now using CRISPR-Cas9 technology to generate “autochthonous” mouse models where tumor initiation happens in a normal cell within the intact organism permitting identification of a gene’s pathophysiological relevance.
Researchers have used mouse and cell-based models for decades to study the molecular etiology of CRC and more recently to clinically identify drug and immune response to defined CRC types. Attributes that led to mice being a choice model have included short lifespan, ease in generation and maintenance, and relative lack of expense compared to larger mammals such as dogs, pigs, or chimpanzees. The anatomy and histology of the mouse are close to humans in structure and function in the adult animal and during development. Research on humans directly, prior to the development of next-generation sequencing (2003), was generally reasoned unethical, making the use of animal models a necessity. The public also questioned the ethics of animal research, leading scientists to replace, reduce, and refine mouse work in cancer research. The issue is, as many dichotomous issues are, debated without unequivocal resolution for or against. For many years written protocols designed for both humans and animals and approved by internal review boards have been required and have helped to establish better ethical standards in the treatment of humans and animals used for research. Mouse facilities at academic institutions funded by federal grants are staffed by veterinarians and animal welfare overseen routinely.
Human CRC subtypes (consensus molecular subtypes, CMS1-4)10 currently fall into four groups: CMS1 (microsatellite unstable and strong immune activation), CMS2 (epithelial, Wnt signaling activation (wingless-related integration site) and MYC activation (MYC proto-oncogene/BHLH transcription factor), CMS3 (epithelial, metabolic dysregulation), and CMS4 (mesenchymal, transforming growth factor β [TGF-β] activation, stromal invasion, and angiogenesis). Investigators are considering subtyping mouse models analogous to the human CMS1-4 phenotypes; mice seem to have a narrower phenotype when compared to human tumors suggesting they will need to be subtyped themselves before comparing with human subtypes.11 This is especially true as murine backgrounds are often congenic and artificially altered; modifiers exist that are not fully identified that affect tumor phenotype. Interestingly, studies of gut microbiota versus immune phenotype have led to examining naturalistic indoor housing system for mice closer to that of a farmyard as opposed to hygienic laboratory mice; these animals appear to have elevated levels of memory T cells and late-stage natural killer (NK) cells12,13
One of the most frequent problems mentioned in the literature is that no mouse model exactly recapitulates the progression of human disease from adenoma and adenocarcinoma to metastasis, or changes in microenvironment. While there were multiple reasons for making early GEMMs, a major ambition was to mimic human CRC to explore drug efficacy pre-clinically. Despite fundamental discoveries being made about stem cells, homeostasis, and transformation, the community was proclaimed to have “failed successfully”9 due to the lack of a reliable and consistent metastatic phenotype. At one time mouse models did not have significant penetrance of the metastatic phenotype, and tumors tended to form in the small intestine as opposed to the colon unless induced by laparoscopy.14 In 2013 the National Institute of Health officially terminated its Mouse Models in Human Cancer Consortium (MMHCC), and researchers, in addition to using GEMMs, began using other models such as patient-derived xenografts (PDXs) and patient-derived organoids (PDOs) to study the disease. The establishment of PDX15 in mice and the identification of intestinal stem cells by the leucine rich repeat containing G protein-coupled receptor 5 (LGR5) marker16 with the development of organoids17 led to the development of transplantation models for the study of conventional treatment response, response prediction, and various immunotherapies.
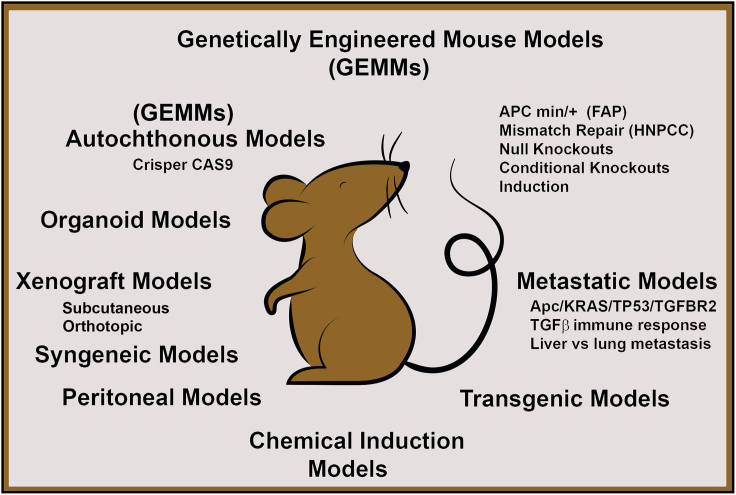
At present, as research focuses on tumor microenvironment, partially because of its relatively recent finding of importance in metastasis but also with the hope of using checkpoint inhibitors and other immunological and inhibitor therapies, the lack of a perfect model continues to be an issue, and the need for immunocompetent autochthonous model has become even more important. An interesting proposal for the use of such a mouse has been for single-cell RNA sequencing and its application in understanding mechanisms underlying immune-modulating therapies.18 However, despite inadequacies the abundant ways researchers have used mice, the flexibility of the system, and the ability to deductively create an image of CRC from the different models that exist are striking (Figure 1).19 The mouse selected by an investigator for a study can be made with some prior knowledge of its advantages and disadvantages relative to the study goals, whether they want to determine therapy for a patient or to elucidate the mechanism behind the disease and identify gene function.
Figure 1.
Schematic representation of mouse models in coloretal cancer
FAP, Familial adenomatous polyposis; HNPCC, Hereditary non-polyposis colon cancer.
It has been said that “history, like a badly constructed concert hall, has occasional dead spots where the music can’t be heard”.20 The metaphor is out of its context here, but it is oddly applicable to reviews, systematic or subjective, as all have vantage points, search terms, or simply lack of space that can lead to “unheard” work. This article presents an accounting of important mouse models in colon cancer and the technology behind them, discussing inherent strengths and weaknesses revealed in their making. Thousands of reports covering mouse models in colon cancer and their variations exist; not all could be examined in this study. The review does endeavor to provide both a synopsis of what experts in the field have thought in yearly chronology of the past and to evaluate critically and collectively on its own how mice are used at present and are likely to be used in the future.
Adenomatous polyposis coli (APC)
The earliest mouse models for CRC were the result of genetic engineering prior to the introduction of CRISPR-Cas9 techniques. Apc Min/+21 was the first model; it was identified and named by its phenotype of “multiple intestinal neoplasia”. The mouse was made by chemical mutagenesis, and at the time of its initial characterization the gene responsible for the phenotype was not known. It was later determined to be the result of a nonsense mutation at codon 850 of the adenomatous polyposis coli gene. Clinicians had identified the familial adenomatous phenotype (FAP) in 1947,22,23 but the disease entity, extraordinarily, was described centuries earlier in a publication dating to 1721.24 The region responsible for the phenotype was localized to human chromosome 5 in 1987,25 and the gene26,27 that would come to be considered the initiating event in CRC was identified shortly afterward through linkage analysis followed by sequencing of candidate genes in the region of interest.28,29
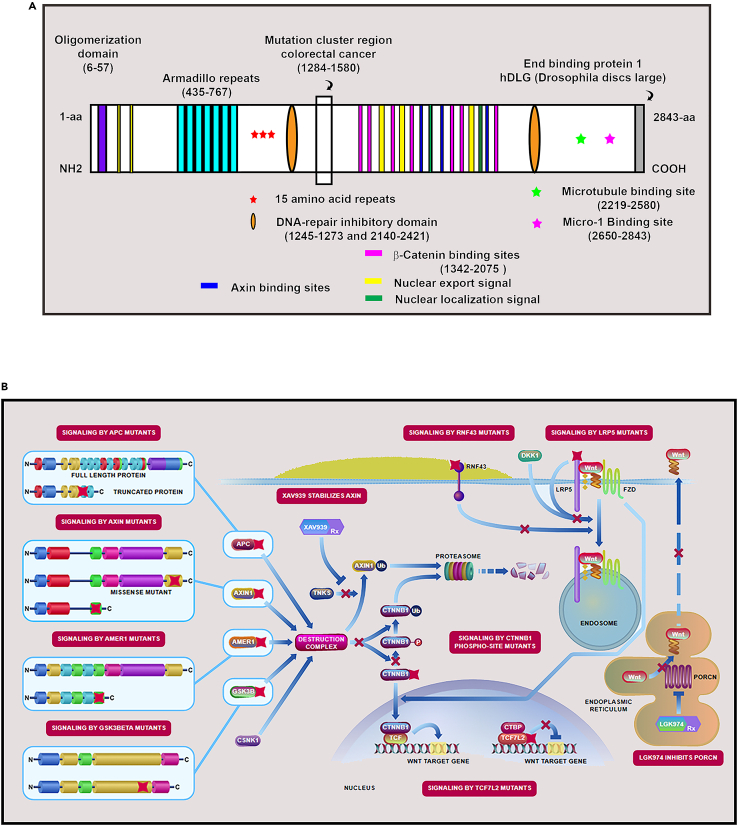
Apc was subsequently shown to control Wnt signaling through the beta catenin complex (cadherin association proteins [eg. CTNNB1] (Figure 2A -B). Despite the importance of proband analysis in gene identification, only three to four percent of all cases have been found attributable to inherited disease. Almost ninety-seven percent of CRC are the result of spontaneous mutations, seventy to eighty percent having truncation mutations to the APC gene. Data supporting this finding are from The Cancer Genome Atlas (TCGA) initiated in 2005 which provided a comprehensive understanding of genomic alterations that underlie all major human cancers; its first report on colon cancer was in 2012.30
Figure 2.
The adenomatous polyposis coli (APC) gene
(A) Important APC motifs.
(B)Wnt signaling controlled by APC (Pathway obtained from National Center for Biotechnology Information, NCBI31).
The Apc Min/+ mouse, made in 1990, was followed by the construction of Apc 1638N four years later which removed the 15th exon of the gene using technology that took advantage of cellular homologous recombination.32 These two genetic changes in the Apc gene led to different phenotypes, where the Apc Min/+ mice developed as many as 40 adenomas by four months of life; the Apc1638N mice developed fewer than five adenomas in nine months of time (Table 1). In the next ten to twelve years, Apc functional motifs and their contribution to phenotype were explored, and investigators began looking at the contribution of important secondary genes in double knockout mice. The general finding, despite small differences in phenotype, was that Apc alteration alone led to adenomas of the small intestine without progression to metastases. This unexpectedly included dual knockouts of Apc and TP53, found by multiple groups. Table 1 presents models in order of publication year (not strictly chronological for month and date), partially drawing from earlier reviews.33,34,35 Comparing genotype/phenotype information in Apc mouse models to human lesions has gained interest recently as many removed during screening colonoscopies are adenomas; it is thought the comparison may be useful in the elucidation of the basic pathways of carcinogenesis and chemoprevention in humans.33
Table 1.
A yearly chronological listing of selected apc mouse models and their phenotypes
| Year published | Alterations | Description of Genotypea | Phenotype | Reference |
|---|---|---|---|---|
| 1990 | Apc Min/+ | Nonsense mutation in codon 850 of the Apc gene (Armadillo repeats present). | Adenomas of small intestine upon loss of heterozygosity, high mortality with increasing age, is a result of intestinal obstruction and anemia, no progression to adenocarcinoma. | Moser et al.21 |
| 1994 | Apc 1638N | Truncation mutation at Exon 15 (1638 aa) made using cellular homologous recombination (Armadillo repeats present). | Adenomas of small intestine (fewer than Apc Min/+) often duodenal, occasionally in colon. Fibromatosis (desmoid tumors), gastric carcinoma, cutaneous cysts, occasional liver metastasis found. | Fodde et al.32 |
| 1995 | Apc Δ716/+ | Nonsense mutation in codon 716 of the Apc gene using cellular homologous recombination (Armadillo repeats disrupted, C57BL/6J background). | Adenomas of the small intestine, ten-fold increase over Apc Min/+. | Oshima et al.36 |
| 1995 | Apc Min/+ p53−/− |
Apc Min/+ mutation21 simultaneous with p53 mutation.37 | Adenomas of the small intestine, no difference to Apc Min/+ mutation alone. | Clarke et al.38 |
| 1996 | Apc Min/+/Msh2+/+ and Apc Min/+/Msh2+/− and Apc Min/+/Msh2−/− | Apc Min/+ mutation21 simultaneous with Msh2 null mutation.39 | Accelerated small intestinal tumor formation (MSH2-deficient mice develop lymphomas). | Reitmair et al.39 |
| 1997 | Apc 580S | Apc Exon 14 flanked by loxP, induction by Adenovirus-Cre (AxCANCre or AxSRaCre) causes exon fourteen deletion and introduces a frameshift mutation at codon 580 (Armadillo repeats disrupted). | Homozygotes develop adenomas of distal rectum attributed to the location of virus delivery. When mice were left to age over one year, approximately 50% progressed to invasive adenocarcinoma, no metastasis. | Shibata et al.40 |
| 1997 | Apc Min/+p53−/− and Apc Min/+ p53+/− | Apc Min mutation21 simultaneous with homozygous recessive and heterozygous p53 mutation.41 | Adenomas of the small intestine, no difference to Apc Min/+ mutation alone. | Fazeli et al.42 |
| 1998 | Apc 1309/+ | Nonsense mutation at codon 1309 (Armadillo repeats and beta-catenin sites present). | Adenomas of small intestine, fewer in number and more proximal than Apc Min/+. | Quesada et al.43 |
| 1998 | Apc 1638N p53−/− | Apc 1638N mutation32 simultaneous with p53 homozygous recessive mutation.41 | Seven-fold increase in desmoid multiplicity. | Smits et al.44 |
| 1998 | Apc +/Δ716/Dpc4/+ | Apc Δ716 mutation36 in combination with Dpc4 (known at present as SMAD4) inactivated in the study. | Dpc4 (SMAD4) and Apc D716 double heterozygous mice lead to adenomas that develop to adenocarcinomas in the small intestine and colon with submucosal infiltration. | Takaku et al.45 |
| 1999 | Apc 1638N/+/Mlh1 −/− | Apc 1638N32 mutation simultaneous with Mlh1 homozygous recessive mutation inactivated in the study. | Increased tumor incidence and multiplicity, 30% adenocarcinomas, significantly reduced lifespan, high number of tumors outside the intestine. | Edelmann et al.46 |
| 2000 | Apc Min/+ Mom1/+/ p53−/− |
Apc Min/+21 simultaneous with Mom1AKR,47 (known at present as PLA2G2A) heterozygotes and p53 homozygous recessive mutations.48 | In this study, contrary to earlier model implications, p53 deficiency increases intestinal adenoma multiplicity, and malignancy mutations were on congenic background (C57BL6/J). | Halberg et al.49 |
| 2000 | Apc Min/+ Msh2−/− Apc 1638N Msh2−/− | Apc Min/+21 or Apc1638N/+32 simultaneous with Msh2 null knockout (exon 7 deletion).50 | Rapid tumor formation in the small intestine, early death (2–3 months), more tumors in Apc Min/+ Msh2 −/− mice. | Smits et al.50 |
| 2000 | Apc Δ474 | Mutant form of mRNA encoding only 474 aa of the Apc gene, created by exon duplication using homologous recombination. | Adenomas of the small intestine are present in duodenum, stomach, colon, and mammary gland. Mice treated with COX-2 inhibitor reduced the number of polyps. | Sasai et al.51 |
| 2002 | Apc Min/+ treated with azoxymethane (AOM) | Apc Min/+21 simultaneous with random unidentified mutations. | Significantly increased incidence of colonic adenocarcinomas. | Møllersen, Suzui et al.52,53 |
| 2002 | Apc Δ716/+ SMAD2/+ | Nonsense mutation in codon 716 of the Apc gene36 and heterozygous mutation of SMAD2 inactivated in the study. | Combination of Apc mutation and loss of SMAD2 leads to no changes in tumor size or properties compared to Apc Δ716/+ mice. | Takaku et al.54 |
| 2002 | Apc Δ580/+ SMAD2/+ | Nonsense mutation in codon 580 of the Apc gene40 and heterozygous mutation of SMAD2 inactivated in study. | Combination of Apc mutation and loss of SMAD2 leads to larger tumors, higher incidence of malignant phenotype compared to Apc Δ580/+ mice. | Hamamoto et al.55 |
| 2002 | Apc 1638N/+ Fen1/+ | Apc 1638N/+32 and Fen1/+ double heterozygous mice inactivated in study. | Increased malignancy of intestinal tumors compared to Apc1638N mice, Fen1 heterozygosity led to microsatellite instability in tumors. | Kucherlapati et al.56 |
| 2002 | Apc Δ716/+ Smad4 Apc Δ716/+ PTGS2(COX-2) Apc Δ716/+ PTGER2 |
Nonsense mutation in codon 716 of the Apc gene36 with Smad4,45 PTGS2(COX-2),57 or PTGER258 alterations. | Micro-vessel density (MVD) is determined in tumors. Apc Δ716 alone produced polyps, with Smad4 polyps progress to adenocarcinomas, MVD was size dependent; with PTGS and PTGER2, adenoma growth was suppressed, MVD did not increase. | Seno et al.59 |
| 2003 | Apc Δ716/+ CDX2/+ | Apc Δ716/+36 and CDX2/+ inactivated in the study, double heterozygous mice. | Increased adenoma formation in the colon reported, reduced number of polyps in the small intestine. | Aoki et al.60 |
| 2004 | Apc Δ14/+ | Heterozygotes have Apc Exon 14 deletion (Armadillo repeats deleted), Cre-loxP strategy (MeuCre40 transgene). | Reported shift of tumors to distal colon and rectum associated with rectal prolapse. | Colnot et al.61 |
| 2004 | Apc Min/+ treated with PhIP | PHIP (2-amino-1-methyl-6 phenylimidazo[4,5-b] pyridine) treatment of Apc Min/+21 mice leads to APC LOH. | Increased tumor initiation two- to three-fold compared to Apc Min/+ mice. | Møllersen et al.62 |
| 2005 | Apc Min/+ BubR1/+ | Apc Min/+21 BubR1/+ double heterozygous mice. | Increased colonic tumor initiation compared to Apc Min/+ (ten-fold) and progression by chromosomal instability. | Rao et al.63 |
| 2005 | Apc Min/+ EphB2 −/+ Apc Min/+ EphB3 −/+ Apc Min/+ EphB3 −/− | Apc Min/+21 mutation simultaneous with homozygous recessive and heterozygous EphB364 and heterozygous EphB2 mutation65 (dominant negative, transgenic). | Reduced adenomas of the small intestine, increased adenocarcinomas of the colorectum. | Batlle et al.66 |
| 2005 | Apc Min/+ MIF −/− | Apc Min/+ mutations simultaneous with homozygous MIF (cytokine macrophage migratory inhibitory factor) | MIF expression and “tautomerase” activity were higher in Apc Min/+ tumors (epithelial and stromal cells) over normal mucosa, increasing with higher grade tumors. Loss of MIF associated with reduction in the number of adenomas and angiogenesis suggesting MIF drives tumorigenesis. | Wilson et al.67 |
| 2006 | Apc CKO/FRT | Exon 14 of Apc flanked by loxP and FRT sites, “FLP e-deleter” used to make Apc CKO/+ mice. | Hung et al. used this conditional knockout to create sporadic colon tumors with metastasis by laparoscopy. | Kuraguchi et al.68, Hung et al.14 |
| 2006 | Apc Min/+ treated with dextran sulfate sodium salt (DSS) | Apc Min/+ mutation21 in the presence of inflammation and disruption of the intestinal epithelial lining caused by DSS. | High incidence of well-differentiated colonic carcinomas. | Tanaka et al.69 |
| 2006 | Apc 580S AhCreT/+ Kras/LSLV12/+ | Apc Exon 14 flanked by loxP,40 induction and deletion by AhCre, and Kras V12 mutation. (Cre induction in liver and intestine by napthoflavone treatment). | Kras V12 mutation has no effect on intestinal epithelium, APC loss in the presence of activated Kras increases tumor progression, 17% are invasive adenocarcinomas. | Sansom et al.70 |
| 2006 | Apc 1638N/TGFBR2flx/flx Villin-Cre | Apc1638N/+32 simultaneous with TGFRB2 −/− predominantly in intestine and colon | Tumor incidence unchanged, tumor progression to invasive adenocarcinoma | Muñoz et al.71 |
| 2006 | Apc Min/+ SMAD3−/− | Apc Min/+ mutation21 simultaneous with SMAD3 homozygous recessive mutation | Tumor progression in distal colon with mixed histology reduces lifespan, no metastases | Sodir et al.72 |
| 2006 | Apc 1638N SMAD4/+ | Apc +/1638N32 mutation simultaneous with SMAD4/DPC4 (SMAD4 E6sad, single nucleotide deletion in the exon 6 splice acceptor site results in unstable mRNA). Genes map to the same chromosome and are therefore analyzed "in cis" and "intrans" to each other. | Increased tumor multiplicity for double heterozygotes both "incis" and "intrans" LOH of SMAD4 occurs at later stages of tumor progression | Alberici et al.73 |
| 2007 | Apc 1638N/+ Exo1 +/− Fen1+/− | Double and triple heterozygous mice combinations for Apc 1638N,32 Exonuclease 1,74 and Flap endonuclease 1.56 | Increased tumor incidence, higher progression to malignancy, high incidence of hematopoietic cancers. | Kucherlapati et al.75 |
| 2007 | Apc Min/+ MYD88 −/− |
Apc Min/+21 MYD88−/− (innate immune signal transduction adaptor) |
MYD88 reduced the number of polyps, mortality decreased. | Rakoff-Nahoum et al.76 |
| 2008 | Apc 2lox14/+ Kras LSL-G12D/+ Apc 2lox14/+ Nras LSL-G12D/+ Fapbl-Cre (or) Villin-Cre |
APC Exon 14 flanked by loxP Kras and Nras flanked by loxP. Induction and deletion by Fapbl-Cre77 (or) Villin-Cre78 |
Kras G12D, but not NrasG12D, drives colon cancer progression. | Haigis et al.79 |
| 2008 | Apc1638N Rb1−/− Villin-Cre |
Apc1638N32 Rbtm2Brn80 Rb1 exon 19(20) flanked by loxP Villin-Cre78 |
Tumors of the small and large intestine. The small intestinal tumors are a mixture of adenomas and adenocarcinomas. Tumors of the cecum and proximal colon are mostly adenomas. | Kucherlapati et al.81 |
| 2009 | Apc Min/+ (C57BL/6) Apc Min/+ (SWR/J) Apc Min/+ (C57BR/cdcJ Apc Min/+ (Nbs1DB) |
C57BL/6J Apc Min/+21 bred with mice of different backgrounds to make hybrids (SWR/J or C57BR/cdcJ); APC Min/+ also bred with Nbs1DB (Nijmegen breakage syndrome alteration) | Hybrid Apc Min/+ mice had decreased adenoma frequency, longer survival, increased adenocarcinomas with occasional metastasis to the lymph nodes. The presence Nbs1 DB alteration created MSI that did not affect tumor multiplicity, mouse lifespan, or tumor progression |
Halberg et al.82 |
| 2009 | cis-ApcΔ716/+Smad4/+ Mmp7−/− | Apc truncated at codon 716 heterozygote,36 Smad4 heterozygote,45 with metalloproteinase (MMP7−/−)83 homozygous recessive | MMP7 knockout reduces tumor frequency and size but not invasiveness | Kitamura et al.84 |
| 2009 | Apc 1638N/+ Notch1 | Apc 1638N32 mutation simultaneous with a constitutively activated NOTCH1 receptor | Notch1 activation reported to result in increased adenoma formation at an earlier age, decreased survival, dysplastic lesions in the colon | Fre et al.85 |
| 2009 | Apc 1322T | Nonsense mutation at codon 1322, transcript retain the first 20- armadillo repeats | High number of adenomas of the proximal small intestine, small number in the colon and stomach | Pollard et al.86 |
| 2010 | APC Δ242 | Gene trap cassette placed between exons 7 and 8 of Apc gives truncated fusion transcript without armadillo repeats | Adenomas found higher in numbers, smaller in size, histology same as for ApcMin/+ mice | Crist et al.87 |
| 2010 | Apc 15lox/+ | Exon 15 flanked by loxP sites, induction by Fabpl (Fatty acid binding protein) Cre | Increased survival due to small number of tumors, larger tumors found in the colon. Ninety-one percent of mice have low-grade adenoma, 50% adenocarcinoma | Robanus-Maandag et al.88 |
| 2010 | Apc 580S/+ | Nonsense mutation in codon 580 of the Apc gene40 (armadillo repeats present), induction by Cre transgene under control of Carbonic anhydrase 1 promoter (limited to large intestine) | Transgene expression limited to the large intestine, adenoma formation without malignancy. | Xue et al.89 |
| 2010 | Apc fle 1-15 | Complete deletion of the entire APC gene (exons 1–15) by Villin-Cre | More severe polyposis compared to Apc Min/+ mice | Cheung et al.90 |
| 2010 | APC CKO/FRT/Kras | Removal of exon 14 of APC68 by orthotopic application of Adenoviral-Cre, mated to mice with K-ras G12D activation mutation | Adenocarcinomas of the colon, 20% liver metastasis | Hung et al.,14 Roper et al.91 |
| 2010 | Apc Min/+ Fbw7ΔG | APC Min/+ mutation21 simultaneous with E3 ubiquitination ligase F box and WD repeat containing 7 (Fbw7) exon 5 deleted | APC Min+/− Fbw7 −/− mice had enhanced tumor numbers and tumor size, APC Min+/− Fbw7 +/− mice increased tumor number but not tumor size. | Sancho et al.92 |
| 2010 | Apc Min/+ IL17A −/− |
Apc Min/+21 IL17A (exons 1 and 2 deleted)93 |
Intestinal tumorigenesis reduced, recovery of thymic and splenic cellularity. | Chae et al.94 |
| 2010 | Apc Min/+ MYD88 −/− |
Apc Min/+21 mutation simultaneous with MYD8895 (innate immune signal transduction adaptor) removal. | Lower phospho-ERK levels, fewer and smaller intestinal epithelial cell tumors than Apc Min/+ mice. | Lee et al.96 |
| 2011 | Apc Min/+ IL-17F −/− |
Apc Min/+21 IL-17F (exons two deleted)97 |
Inhibition of spontaneous intestinal tumorigenesis, partially restores thymic atrophy, does not affect splenomegaly. | Chae et al.98 |
| 2012 | Apc Min/+ MCP1 −/− |
Apc Min/+21 MCP1/CCL2 −/− (Monocyte chemoattractant protein, purchased from Jackson Labs) |
MCP1 knockout decreased polyps 20–45%. | McClellan et al.99 |
| 2012 | Apc Min/+ IKKβ(transgene/overexpression) |
Apc Min/+21 IKKβ (inhibitor of NF-kB). |
More early lesions and visible small intestinal and colonic tumors than Apc Min/+. | Shaked et al.100 |
| 2013 | Apc 580S/+ CDX2P-CreERT2 | APC 580S40 conditional knockout by CDX2P-CreERT2-Cre regulation through tamoxifen added to chow. | Apc knockout led to changes in proliferation, apoptosis, morphology, mitotic spindle axis misorientation, b-catenin nuclear localization, induction of stem cell marker and Sox9, ectopic Paneth-like cell so observed. | Feng et al.101 |
| 2013 | APC CKO/FRT/BrafVE | Removal of exon fourteen of APC68 and exons 15–17 of Braf (BrafV600E) by orthotopic application of Adenoviral-Cre leads to focal inactivation of Apc and activation of Braf. | Sessile serrated adenoma with dysplasia, adenocarcinoma, and invasive carcinoma. | Coffee et al.102 |
| 2013 | Apc Min/+ IL-8 human transgene/overexpression |
Apc Min/+21 IL-8 (interleukin 8/initiator of inflammation) transgene not found naturally in mice. | Increased tumorigenesis with transgene. | Asfaha et al.103 |
| 2014 | Apc Min/+ on hybrid background | Apc Min/+21 model described earlier82 on different backgrounds, treated with DSS | Histology reassessed, tumors develop in the distill colon, with occasional progression to adenocarcinoma. | Paul Olson et al.104 |
| 2014 | Apc 580S/+ Kras LSL-G12D/+ CAC+ | CAC-Cre under the control of Carbonic anhydrase promoter inactivates APC40 and Kras | Adenomas formed. | Byun et al.105 |
| 2014 | Apc 580S Pten fl/fl Kras LSL/+ Villin-Cre (ERT) |
APC 580S: conditional knockout40 PTEN: Exons 4–5 flanked by LoxP106 Kras (LoxP/Stop/LoxP)107 gives G12V inducible oncogenic mutation. Villin-Cre ERT78 is under the control of the Estrogen receptor promoter. |
Survival reduction, adenomas, occasional adenocarcinomas of the small intestine, no metastasis. | Davies et al.108 |
| 2015 | Sh Apc Kras G12D P53 fl/fl Lgr5 +/+ |
Short hairpin RNA regulated by doxycycline responsive promoter/Cre suppresses Apc expression in colonic epithelium. The effects of compound mutations to Kras (activation) and p53 (deactivation) and reversible Apc loss are examined. | The histological and molecular features of colon cancer are recapitulated and are reversed upon Apc re-expression. Apc loss increases MYC-dependent proliferation. | Dow et al.109 |
| 2015 | Apc Δ 716 TGFBR2 fl/fl Villin-Cre (ERT) |
Apc36 and TGFBR2 are conditionally knocked down using Villin-Cre (ERT) | Only invasive adenocarcinomas found to have inflammatory microenvironment; treatment with DSS caused invasive colon cancer. TGFΒ signaling disruption in the presence of inflammation is sufficient to produce colon cancer | Oshima et al.110 |
| 2015 | Apc fl/fl KrasG12D/+ PIK3CA+ |
Apc fl/fl68 Kras G12D PIK3CA Cre-Adenovirus (orthotopic injection) |
Additional mutations to Apc cause progression to adenocarcinoma and metastasis. | Hadac et al.111 |
| 2016 | A/J Apc Min/+ | Apc Min/+21 on A/J background | Increased tumor initiation in intestine, increased progression (50% adenocarcinoma). Carcinoma of the colon (21%). | Sødring et al.112 |
| 2016 | Apc CKO/FRT LSL Kras G12D CDX2P9.5-G22Cre |
Apc68 inactivation, Kras activation by CDX2P9.5-G22Cre (“CDX2” is caudal type homeobox protein, normally expressed in the proximal colon). | Moderately differentiated adenocarcinomas of the proximal colon, Kras mutation did not increase malignancy. | Kawaguchi et al.113 |
| 2016 | Apc CKO/FRT Kras LSL G12D/+ P53KO Smad4 fl/fl Car1CreER |
Apc68 inactivation, Kras activation, p53 knockout, Smad4 inactivation using Car1CreERT2 (Carbonic anhydrase promoter enhancer under the control of Estrogen receptor 2 controls Cre recombinase upon tamoxifen induction). | No adenomas of the small intestine. Apc and Kras led to colonic adenomas. Compound mutations Apc, Kras, p53, Smad4 led to aggressive colonic carcinomas with occasional lymph-node invasion. | Tetteh et al.114 |
| 2017 | Apc Δ716 TP53 +/LSL-R270H Apc Δ716 TP53 LSL-R270H/LSL-R270H Villin-Cre ER |
Apc36 inactivation by Villin-Cre, TP53 mutant allele TP53R270H/R270H generated after removal of floxed TP53 by Villin-Cre ER. | Apc Δ716 with homozygous mutation to p53 die of lymphoma, heterozygotes have invasive adenocarcinomas with evidence of epithelial to mesenchyme transition (EMT). | Nakayama et al.115 |
| 2017 | Apc CKO/FRT | Orthotopic injection of PGK::Cre lentivirus, Ad5CMV::Cre, Villin-Cre ER, or U6::sgApc-EFS::Cas9-P2A-GFP into “mucosa bubble” of Apc CKO/FRT68 mice inactivates Apc. | Rapid tumor formation with histological features of adenomatous change and CTNNB1 nuclear localization using PGK::Cre lentivirus, Villin-Cre ER. U6::sgAPC-efs::Cas9-P2A-GFP delivered the Crispr-Cas9 system and inactivated Apc produced tumors in 34% of the mice. (The study also looked at tumor-associated genes TP52 and Kras in combination with Apc). |
Roper et al.116 |
| 2017 | Apc Min/+ TDG fl/fl Fabpl:Cre |
Apc Min/+21 mutation with Thymine DNA Glycosylase (TDG) exons 3–6 are deleted by Fabpl::Cre | TDG knockout in Apc Min/+ mice led to a two-fold increase in adenomas in female mice. | Xu et al.117 |
| 2017 | Apc Min/+ PLD1 −/− AOM/DSS |
Apc Min/+21 PLD −/− exons 13–14 deleted. |
Decreased number of adenomas. | Kang et al.118 |
| 2018 | Apc Δ716 Kras +/LSL-G12D TGFBR2 fl/fl TRP53 +/LSL-R270H FBXW7fl/fl Villin-CreER |
Various combinations examined: Apc Δ71636 Kras +/LSL-G12D119 TGFBR2 fl/fl120 TRP53 +/LSL-R270H121 FBXW7fl/fl122 Villin-CreER78 |
Apc alteration led to adenomas, addition of TRP53R270H or TGFBR2 deletion induced submucosal invasion. KrasG12D activation led to epithelial-mesenchymal transition (EMT) and lymph vessel intravasation. Apc alteration with Kras G12D and FBXW7 insufficient for submucosal invasion but not EMT histology. Apc, Kras, TGFBR2 were sufficient for metastasis. |
Sakai et al.123 |
| 2018 | Apc Min/+ | Apc Min/+21 | Computer learning-based algorithm gives quantitative analysis of immunofluorescence staining on whole colon sections, used to characterize the immune environment. Total leukocytes and T cell sub-populations are counted in colonic mucosa, lymphoid follicles, and tumors. T cells quantified in lymphoid follicles. | Cassé et al.124 |
| 2018 | Apc Min/+ Tob1 AOM/DSS treated |
Apc Min/+21 Tob1 −/− |
Reduced tumorigenesis | Li et al.125 |
| 2019 | Apc Min/+ Apc 580S Villin-Cre ERT2 Rosa |
Apc Min/+21 Apc 580S40 Villin-Cre ERT2 Rosa (reference not given) Vaccine to MYB (overexpressed transcription factor in polyps) given to both models with anti PD-1 antibody. |
The TetMYB vaccine is shown to be effective in both prophylactic and therapeutic animal models, the vaccine will be used in a future clinical trial. | Pham et al.126 |
| 2019 | Apc580S Cdx2-CreERT2 Mcl1 fl/fl LysMCre AOM/DSS |
Apc580S40 Mcl1−/− (neutrophil deficient)127 Microbiota assessed |
Neutrophils slowed colon tumor growth and progression over controls by restricting numbers of bacteria and tumor-associated inflammatory response. | Triner et al.128 |
| 2019 | Apc fle 1-15 | Apc fle 1–1590 Villin-Cre TFAMfl/fl129 |
TFAM deficiency diminishes mitochondrial formation and oxidative phosphorylation, site-specific removal prevented tumorigenesis in Apc-mutant mouse intestines. | Wen et al.130 |
| 2020 | Apc Min/+ Mucin 2 −/−(Winnie) |
Apc Min/+21 Mucin 2 −/−(Winnie)131 |
Winnie-ApcMin/+ mice combine an inflammatory background with a genetic predisposition to CRC. Aberrant crypt foci form in the colon recapitulating human CRC. | De Santis et al.132 |
| 2021 | Apc Min/+ | Apc Min/+21 pretreated with rapamycin | Rapamycin extended lifespan of Apc Min/+ mice. | Parihar et al.133 |
| 2021 | Apc Min/+ | Apc Min/+21 inoculated with Fusobacterium nucleatum, treated with aspirin. | Aspirin-supplemented chow is sufficient to inhibit F. nucleatum-potentiated colonic tumorigenesis. | Brennan et al.134 |
| 2021 | Apc fle 1-15 Cdx2P-CreERT2 |
Apc fle 1–1590 or Apc Min/+ animals treated with NSAID (sulindac) and rexinoid (bexarotene). | Significant polyp reduction. | Bowen et al.135 |
In Table 1, the first time an Apc mutation is mentioned the alteration is described under “Description of Genotype” and the reference for its creation given under “Reference”. When mentioned in a second study, references for all alterations are given under “Description of Genotype”, and the study examining compound gene alterations is under “Reference”.
By the mid-2000s the drawbacks of null knockouts, described below, would lead to the development of conditional knockout technology and its application to the Apc gene. Around the same time and until the present, observations exploring the effects of inflammation and immunity in intestinal cancer were undertaken. Conclusions from Apc dual knockout models regarding inflammation are complex as deficiency in some factors (e.g., interleukin 17A [IL17A], interleukin 17F [IL17F], prostaglandin-endoperoxide synthase 2 [PTGS2/COX-2], C-C motif chemokine ligand 2 [CCL2/MCP1], macrophage migration inhibitory factor [MIF], myeloid differentiation primary response protein/ MYD88 innate immune signal transduction adaptor [MYD88]) alters or blocks inflammation inhibiting intestinal tumor development in Apc Min/+ mice, whereas overexpression in others (e.g., nuclear factor kappa B subunit 1 [NFKB1], C-X-C motif chemokine ligand 8/interleukin 8 [CXCL8/IL8], and interleukin 6 [IL-6]) promotes it.35 Apc mouse models despite their drawbacks (no metastases and tumor location) are considered autochthonous and widely used at present for innate and adaptive immunological study.
Hereditary non-polyposis colon cancer (HNPCC)
After APC was shown responsible for familial adenomatous polyposis, a second inherited type of colon cancer, HNPCC, was found to result from germline mutations in genes in DNA mismatch repair (MMR, 1993-6).136,137,138,139,140 MutL homologue 1 [MLH1]141 and MutS homologue 2 [MSH2]142 were the first two MMR genes implicated in colon cancer; other components of the MMR system were later identified and examined. The relevance of mammalian MMR to HNPCC was possible because of earlier work on the Mut HLS complex (methyl directed mismatch repair system comprised of mutH/mutL/mutS proteins) in Escherichia coli143,144 and in yeast. Mouse models were used later to explore MMR’s relationship to colon cancer.145,146 Mice with a null knockout of the Mlh1 gene (1996) developed intestinal tumors and additionally resulted in sterility of both male and female mice caused by failure to progress beyond meiotic prophase and arrest at pachytene.141 Null knockout of Msh2 alone in mice (1995) led to the development of lymphomas,142 in combination with the Apc Min/+ mouse Msh2 could accelerate intestinal tumor formation.39 Conditional knockout of Msh2 alone (2010) under the control of a constitutive Villin-Cre transgene (cre, cyclization recombinase of bacteriophage P1) permitted routine development of adenocarcinomas of the small intestine147 without lymphoma.
The ability to identify and understand mammalian orthologues of Mut HLS led to understanding MMR contribution to intestinal cancer. This colon cancer genotype has turned out to be the first and one of the most successfully treated tumor types with immune checkpoint inhibitor therapies148 when knockout mouse model studies in 2004 showed that the programmed cell death 1 ligand 1 gene [CD274/PD-L1] negatively regulated T cells.149
Chemical induction of CRC
The creation of the APC min/+ mouse was achieved using ethyl nitrosourea (ENU), followed by screening for the adenomatous phenotype; chemical induction was the first method used to generate CRC in mice, reports being published as early as the 1940s.150 An accounting of reagents and their use has been recently made.151 Chemically induced models are not invasive and do not have metastases as a rule; tumors initiated in this manner reflect progression from aberrant crypt foci and adenoma to adenocarcinoma. Currently, researchers use these animals to examine the effects of diet, colitis-associated carcinogenesis, chemoprevention, and gut microbiota.152,153,154 Mouse strain,155 housing conditions, and route of administration are known to have effect on carcinogenic potential. Routes of administration include feeding, oral gavage, injection (intraperitoneal, subcutaneous, or intramuscular), or enema.
Systematized approaches for the use of azoxymethane (AOM) and/or dextran sulfate sodium salt (DSS) in C57BL/6J and A/J mice have been generated.156,157 These animals turn out to be good models for colitis-associated CRC;158 however, because they generate an inflammatory response, they are not considered applicable for the study of normal tumor microenvironment and its immune response. Additionally, intestinal tumors produced by AOM appear to have higher numbers of mutations to Kras proto-oncogene GTPase [Kras] and Ctnnb1 than A and tumor protein p53 [Tp53].159,160 This mutation profile deviates from human CRC where APC was found to be mutated in seventy-two percent of TCGA cases, and TP53, fifty-nine percent. The underlying mechanism of AOM/DSS is a result of the formation of O6 and N7 methylguanine in the DNA leading to carcinogenesis, with DSS physically damaging the colonic epithelium by altering mucosal permeability leading to inflammation and a shortened length of time to tumor development. Tosti et al.154 have examined MMR-deficient mouse models with compound transforming growth factor beta receptor 2 [Tgfbr2] deficiency, often found accompanying MMR alteration, along with AOS/DSS chemical induction of tumorigenesis. TGFBR2, as mentioned below, is associated in non-MMR CRC with metastasis; in CRC MMR-deficient mice Tgfbr2 was found responsible for microbiota alterations161 and induction of the equivalent of human inflammatory bowel disease (IBD).
GEMMs
Most early GEMMs were null knockouts made by using the cellular homologous recombination system.162,163 When it was found that homozygous knockouts often had embryonic lethality as a developmental phenotype, the loxP/cre system164 was developed and used mostly to generate gene knockdown in the adult animal rather than the zygote. Cre, a bacterial recombinase that specifically cleaved loxP sites, was placed under the control of tissue-specific promoters and introduced into animals by transgenic injection. These strains were then mated to mice where the gene of interest had been flanked by loxP sites,165 giving progeny with gene deletions where the promoter/cre was active. Investigators thus had some ability to stop specific gene expression in a tissue-specific manner and to generate site-specific tumors.
The first available cre/promoters acted constitutively which could lead to cre expression in unwanted places. cre/promoters were later designed to have inducible expression using tamoxifen166 or tetracycline167 permitting site-specific cre expression during adulthood; this was accomplished specifically in the small intestine and colonic epithelium of mice.77 The system was also later creatively modified to “knockin” activation mutations using “loxP/Stop/loxP (LSL)” cassettes that removed inhibitory sequences adjacent to gene sequences of choice by cre recombinase, permitting gene expression; another important advancement saw short hairpin RNA (2002168,169,170) under the control of an LSL cassette permitting expression to be turned off and then returned to normal levels using an inducible cre recombinase.
The drawbacks and complexities of the loxP system are aptly illustrated by studies of the retinoblastoma (Rb1) tumor suppressor and cell-cycle regulator in intestinal tissues. Rb1 conditional knockouts80,171 were put under the control recombinases, constitutive Villin-cre81,172 or Fabpl-cre171 (fatty acid-binding protein of the liver). Both “Villin” and “Fabpl” promoters, function in the mouse intestine; both promoters have known activity in extraintestinal tissues. Several Villin models exist that make use of different promoter fragments,78,173 with varied activity in extraintestinal tissues174 (including the tamoxifen inducible promoter). Initial studies of Rb1−/− Villin-cre animals showed they died of extraintestinal tumors by 17 months. By comparison, this was not seen with analogous Msh2−/− Villin-cre animals,147 which developed invasive adenocarcinomas of the small intestine, dying at 18 months. Dual knockout of Rb1−/− by Villin-cre and Apc1638N (Table 1) led to tumors of the cecum and proximal colon81 with all animals dying at 12 months. In Parisi et al.,171 Rb1−/− Fabpl-cre mice were found to live almost a full year longer (28 months), leading to invasive, poorly differentiated adenocarcinomas of the colon at an old age. Researchers examined Apc expression by immunohistochemistry in this interesting report, and it raised the question if truncation mutations occurred. APC is thought to be expressed upstream of G1/S in human CRC preventing RB1 phosphorylation,175 likely during G2/M of the cell cycle in colon adenocarcinoma.176 TCGA found “C to T”, the predominant mutational signature of Apc in these tumors, a signature frequently associated with transcriptional mutation, suggesting transcription sometimes is not turned off fast enough in G2/M phase and when chromosomes begin to condense uracil gets incorporated (by R loops) resulting in nonsense mutations. This could place cell cycle or basic transcriptional machinery as the initial “hit(s)” in colon tumor initiation.
Conditional knockouts are best characterized as capable of giving great insight and unexpected results, leading to further questions and hypotheses.
Metastasis and tumor microenvironment
By 2017 it was well known that intestinal tumor development required multiple genetic changes. Examination of human CRC revealed that activation mutations in KRAS, loss-of-function mutations in SMAD (supressor of mothers against decapentaplegic transcriptional factors) genes, and loss-of-function mutations in TP53 were common in CRC; genetic and genomic analysis revealed the involvement of many other genes. Many mouse models with modifications in these genes were developed and studied. With the development of the “iKAP” mouse model, Boutin et al.177 identified the progressive genomic changes necessary for a metastatic phenotype and were able to increase the number of tumors with correct location in the colon. In iKAP mice, Apc and Tp53 were knocked out and the Kras G12D activation mutation knocked into colonic intestinal epithelial cells engineered with appropriate transgenes to accomplish the process in a multi-step manner. Prior observations in multiple studies supported the involvement of KRAS in the formation of metastases; these included high mutation rate to KRAS and mutation association with highly invasive stage CRC and liver metastasis. Investigators suggested the limited success in KRAS-targeting strategies could be explained by a role for the gene in tumor progression rather than CRC maintenance; the TGF-β pathway was observed to be one of the major effectors of the process.
Subsequently, in a model also examining quadruple mutations to Apc, Kras, Tp53, and Tgfbr2 in intestinal stem cells, Tauriello et al.178 found Tgf-β able to cause metastasis by its ability to drive immune evasion in MMR stabile CRC. Inhibition of Cd274/Pd-l1 gave only a limited response in these tumors, and inhibition of Tgf-β created a cytotoxic T cell response against them that prevented metastasis. Tumor microenvironment through TGF-β, long thought to be involved in the relationship of tumor and host,179 was thus identified as a means of immune evasion that promoted T cell exclusion and blocked acquisition of the TH1-effector (CD4/Type 1 T helper) phenotype. The study implied that therapies against TGF-β signaling180 would have broad application in treating patients with advanced CRC. Upon examining NOTCH signaling activation (neurogenic locus homologue signaling) in the Kras G12D mice, researchers found TGF-β-dependent neutrophil recruitment drove poor-prognosis tumor subtypes, suggesting neutrophil recruitment as a potential therapeutic approach.181 Sakai et al. also demonstrated Apc, Kras, and Tgfbr2 were sufficient to drive metastasis in CRC.123
In the studies mentioned above177,178 metastases were exclusively found in the liver; human CRC metastasizes to both the liver and the lung. A recent report finds reliable lung metastases arise when Rag-1 knockout mice, another immune-deficient mouse negative for B, T, and NK cells, were orthotopically transplanted during colonoscopy with the human HCT116 cell line (HCT, human colorectal tumor).182 Investigators suggested that Rag-1 mice differ from the nude mice used previously because leakage of B and T cells does not happen as the mice age. While the study is encouraging, liver metastases are absent in the Rag-1 model; the report itself is based on a very few numbers of mice, and the investigators do not mention the genetic background of the animals. However, the involvement of the immune system and tumor microenvironment in the metastasis observed is not likely incidental. Other mouse models containing different combinations of genetic changes have been studied. These include combinations of different Apc-mutant mice with those containing Kras, Tp53, and others. Overall, combinations of different mutations including tumor microenvironment generally result in acceleration of the development of intestinal tumors.
Transgenic mouse models
GEMMs permitted researchers to remove a gene and examine the change in phenotype; transgenic mice permitted overexpression of genes for the same purpose. First constructed in the Ruddle laboratory in the 1980s,183 transgenics specific for the study of colon cancer were made in the early 1990s.184,185 Initially recombinant vectors containing the gene of interest adjacent to promoter/enhancer elements were injected into the pronuclei of fertilized eggs, and the DNA transmitted to progeny by randomly inserting into the mouse genome. Later with the development of CRISPR/Cas9 technology, the desired gene could be “knocked in” at a specific locus. The technology is still being used to understand gene function in CRC.185
Peritoneum models
CRC coexists with peritoneal carcinomatosis (PC) in approximately 3% of patients without metastases and 25–30% of patients with recurrent metastatic disease.186 It is not entirely clear if PC is part of metastasis or represents its own separate phenotypic and molecular entity. This has limited their use in studying CRC, and researchers use them more often now to examine nonsolid tumors or to assess pharmacologic responsiveness.
In a recent report, investigators demonstrated an orthotopic xenograft from a patient with PC could be labeled with red-fluorescent protein (RFP) simply by stromal invasion of the graft. Human stroma in PDX becomes replaced very quickly by murine stroma, despite the histological characteristics of the tumor remaining unchanged.187,188 When placed in a transgenic mouse expressing RFP, enough stroma from the transgenic animal invaded the tumor that it could be followed by imaging when those tumors were repositioned into a second non-transgenic mouse.189 The study anticipates the process might be useful in the identification of effective treatments using a personalized approach for PC patients.
At present, there are no spontaneous models (GEMMs) for PC. Syngeneic and xenografted mice model the condition, with mouse cell lines injected into the intraperitoneal cavity and human colon cancer cells xenografted subcutaneously or orthotopically. A few transgenic animals for genes found associated with PC have been reported.190 The use of CRISPR-Cas9 technology is expected to lead to the development of genetically engineered models soon.
Syngeneic (allograft) models
In syngeneic mouse models, investigators graft tumor tissues into mice with the same genetic background; grafts are not rejected as the immune response is not triggered. The transplanted tissue is often from cell lines induced originally by chemical carcinogenesis. These animals have an intact immune system and are considered useful in studies of immunotherapy; the drawbacks are that cell lines are not the original tumor genotype and human material cannot be used.191 The first syngeneic transplants were done in the 1960s;192 the system is still being used in treatment response studies in CRC193 and the immune response to colon cancer.194
Xenografted mice
Xenografts refer to the transfer of tissue from one species into another; the earliest work was done in the 1940s.195 Today the term refers to the transfer of fresh human tumor, intact or single cell, into mice that are necessarily immuno-incompetent. Implantation of tumor fragments has been shown preferable to cell suspensions in orthotopic models.196 Immuno-incompetency may include genetic deficiency of an entire branch of the immune system (athymic nude mice), combined immunodeficiency, or animals that have had their immune systems humanized. Transplantation routes include subcutaneous, intrasplenic, or orthotopic,197,198 with orthotopic transplantation being the delivery of cancer cells to the anatomic location or tissue in mice from which a tumor type was derived.15
Subcutaneous xenografts generally do not result in metastases, and injection into the spleen, portal vein, and liver to create them artificially does not permit identification of the molecular changes necessary for their development.199,200 Patient-derived orthotopic xenografts better mimic metastasis than subcutaneous xenografts.201 In 1991, the Hoffman laboratory showed it is possible to construct a CRC model using orthotopic transplantation with a variety of clinical behaviors reflecting human pathology, including local growth, abdominal metastasis, abdominal carcinomatosis with extensive peritoneal seeding, lymph-node metastasis, liver metastasis, and colonic obstruction, with a high rate of success, and then went on to use adaptations of the model for drug discovery.202 Advantages to xenograft models that led to their original development and their continued use include that cancer alterations growing in the graft are close to the patients’ original tumor, and they are far less expensive to make than most other models. Investigators can make cell lines with human tumor material and use early passages to measure various biological properties. Xenografted mice have long been considered the most reliable model for research and testing of anti-cancer drugs, the idea being that one had to examine human as opposed to mouse tumors to accurately model chemotherapy.203
A serious drawback using subcutaneous or orthotopic PDX in modeling CRC is that the microenvironment in an immuno-deficient mouse is significantly different from that of a tumor growing in an otherwise normal gastrointestinal tract.197,204 Immune-deficient mice are prone to metastasis, and this has been taken advantage of for the testing of combinations of many drugs, but a major aim of personalized medicine has been to develop chemotherapeutic regimens such as checkpoint inhibitors that are less deleterious to the patient. To test the effectiveness of an immune checkpoint inhibitor in a mouse, there must be an immune system to release.
Both patient-derived tumors and organoids205 are xenografted subcutaneously and orthotopically; 3D cultures are made of both. Researchers are increasingly culturing PDX onto sponge-matrixes (Gelfoam)206 when doing “histoculture drug-response assays” (HDRA), giving the ability to discern drug sensitivity between individual patients with similarly classified tumors.207 By comparison, organoids are grown on a laminar-rich extracellular matrix known as Matrigel.208,209 Murine tumors and organoids, syngeneic and non-syngeneic, have also been transplanted in the same manner as human xenografts.210 RNA interference of genes of interest is made in cell lines, and the resulting altered cell lines are xenografted into mice for studies on chemosensitivity in colon cancer.211 In a recent comparison of PDXs to patient-derived organoids (PDOs) upon orthotopic transplantation into mice,212 analysis showed good in vivo reproduction by both.
Organoids
The development of organoids permitted another approach to the creation and utilization of mouse models. Over the last four years organoid banks have been assembled, becoming publicly available for almost all human tissues and patient-derived tumors, as well as mouse tumors. Researchers defined them as three-dimensional structures embedded in an extracellular matrix, which originate from embryonic, pluripotent, or adult stem cells (ASCs). These structures develop important characteristics of the expected organ through self-organization, sorting, and spatially restricted lineage commitment of cells.17 ASC differs from pluripotent organoids (iPSCs, induced pluripotent stem cells) by being lower in complexity and having an absence of stroma nerves and vasculature. They were originally developed to study intestinal cancer,213,214 when LGR5-positive stem cells were cultured into crypt-villus-like organoids. A recent systematic literature review215 reports overall acceptance that organoids have potential use in both personalized medicine and in understanding disease progression. Progress in endoscopic capability, transplantation techniques,198 and the identification of ASC by marker has permitted organoid use in mouse models to predict therapeutic drug response in patients.216,116 Accepted limitations in their use in mouse models include unwanted genomic alterations during culture, an inability to model the human tumor immune environment completely, lack of stroma when derived from ASC, and reliance on Matrigel for extracellular matrix. Major strengths in studies using organoids include greater ability to establish the organoid compared to establishment of a 2D cell line, the ability to determine drug response to patient-derived tumors, and the ability to compare matched pairs of cancer versus normal tissues.
Discussion
Inferences and implications
Data, primarily from the Human Genome Project (1990–2003), estimated the total number of coding genes in one nucleus at about 20,000. The mouse genome was later found to have 24,408, with 17,094 human orthologs. Interestingly, an organism’s size was not directly proportional to the number of its genes; “Daphnia magna” (the water flea) has 31,000. Researchers for many years altered genes in multiple species to determine function and considered mutations in human disease genes as nature’s experiments. The development of “next-generation sequencing”, the initiation of TCGA, and xenograft and organoid transplantation all dramatically changed how mouse models were made and used in cancer research. An argument was made that one did not have to understand gene function; it was best to identify compounds that were effective against tumor pathways, and human tumors transplanted into mice would be the most informative. The argument had weight; transplanted organoids and xenografts have been effectively used to determine treatment response, and this type of modeling is being further developed. But it is also found to be an over-simplification as time has shown that the immune response can be altered to treat some tumors successfully, angiogenesis is altered, and that micro-environment plays an important role in metastasis and cancer progression (Table 2). These observations are responsible for investigators suggesting the development of an immunocompetent autochthonous mouse that models the full spectrum of CRC. GEMMs in the past were criticized as expensive endeavors, which will likely be true in future; the adage suggesting we are too poor to afford bargain commodities may well be deemed practicable. This direction implies that the Herculean but finite task of understanding how most of 20,000–24,000 human and mouse genes function in ontogeny may be necessary to fully understand alterations in cancer and treat them successfully. It infers the numerous pathways and processes we have uncovered in any one nucleus are components of a jigsaw puzzle responsible for multiple cell fates that interact to form an organism and/or a cancer cell, and we are over time required to assemble the full picture.
Table 2.
Benefits and disadvantages of different mouse model types
| Type | Models early stage | Late stage | Used for drug response | Immune status | Benefits | Disadvantages |
|---|---|---|---|---|---|---|
| GEMMS | ||||||
| Null | Yes | No | Yes | Normal | Can identify phenotype of single genetic alteration on congenic backgrounds. | Phenotypes may differ between mice and humans. |
| Can identify genes involved in tumor initiation and progression. | Tumor genotypes may differ between mice and humans. | |||||
| Can assess tumor response to drugs. | Drug response not to human tumor. | |||||
| Can identify developmental genes and haploinsufficient phenotypes. | Embryonic lethality may prevent identification of function in adult tissues. | |||||
| Technology adapted to knockout by CRISPR-Cas9. | Technology considered expensive and time consuming. | |||||
| Conditional | Yes | Potentially | Yes | Normal unless by design | Bypasses embryonic lethality phenotype. | Optimally, Cre recombinase should be expressed solely in the desired tissue; promoters driving Cre recombinase in the colon almost all have some extraintestinal activity. |
| Can relocate Apc-initiated tumor from small intestine to colon. | ||||||
| Laparotomy | Yes | No | Yes | Altered inflammation | Can relocate Apc-initiated tumor from small intestine to colon. | Procedure creates atopic inflammation. |
| Metastatic | Yes | Yes | Yes | Altered through TGF-β | Can reproduce metastasis by changing immune response (APC, KRAS, TP53, TGFBR2). | No model gives both liver and lung metastasis routinely. |
| TRANSGENIC | ||||||
| Random | No | No | No | Normal | Can observe phenotype when gene is overexpressed. | Random insertion affects genes other than gene of interest. |
| Site specific | No | Potentially | No | Normal | RNA guide can eliminate random insertion. | |
| CHEMICAL INDUCTION | Yes | No | Yes | Altered inflammation | Tumor progression from adenoma to adenocarcinoma similar with human histology. | Tumors rarely proceed to metastasis. |
| Tumors have high KRAS/CTNNB1 and low APC/TP53 mutation frequency. | Human tumors have high APC/TP53 and lower KRAS mutation frequency. | |||||
| AOS/DSS good model for inflammatory bowel disease. | Not applicable for the study of the normal immune response, requires chemical injury to colon. | |||||
| Technically simple, inexpensive. | Low tumor burden, tumors take a long time to develop. | |||||
| PERITONEUM | No | Potentially | Yes | Normal | Technically simple. | Dependent on syngeneic, xenografted, or transgenic mice to model. |
| Spontaneous GEMMs models do not yet exist | ||||||
| SYNGENEIC | Yes | No | Yes | Normal | No tumor rejection. | Cannot use human tumor material. |
| Cell lines often used for grafts are not an original tumor genotype. | ||||||
| XENOGRAFT | ||||||
| Subcutaneous | No | No | Yes | Altered deficient | Tests human cancer directly. | Immuno-deficient. |
| Technically simple, rapid tumor formation. | Tumors are not present in the correct microenvironment. | |||||
| Can assess tumor response to drugs. | Not always easy to identify antitumoral drug response. | |||||
| Mouse stroma replaces human. | ||||||
| No insight into tumor initiation events. | ||||||
| Mostly non-metastatic, rarely models late-stage disease. | ||||||
| Orthotopic | No | Potentially | Yes | Altered deficient | Tests human cancer directly. | Immuno-deficient. |
| Histology similar. | Mouse stroma replaces human. | |||||
| Potential to generate liver metastasis. | Tumor burden is lower than subcutaneous xenografts. | |||||
| Genetically engineered mice are not necessary. | No insight into tumor initiation events. | |||||
| Can assess tumor response to drugs. | Not always easy to identify antitumoral drug response. | |||||
| ORGANOID | ||||||
| Subcutaneous | No | No | Yes | Altered deficient | Technically simple, rapid tumor formation. | Tumors are not present in the correct microenvironment. |
| Immuno-deficient. | ||||||
| No insight into tumor initiation events. | ||||||
| Mostly non-metastatic, rarely models late-stage disease. | ||||||
| Orthotopic Transplantation and in situ gene editing | Yes | Yes | Yes | Potentially altered deficient | Mouse and human organoids are both transplantable into the colon. | Requires colonoscopy system and special equipment. |
| Can obtain metastasis. | If using human tissue for transplantation, mice must be immuno-deficient. | |||||
| It is not necessary to generate genetically engineered mice. | ||||||
| Use of CRISPR-Cas9 | Can get tumor formation in selected sites in distal colon. | |||||
| Tumors monitored with colonoscopy. | ||||||
Conclusion
Many types of mouse models have been used to understand the biology of CRC. Xenografting human cell lines into immune-deficient mice or transplantation of mouse cell lines into immune-competent mice have been very popular in evaluating drug toxicity and efficacy; however, they do not provide basic information about tumor development and maintenance processes, nor do they adequately describe innate and adaptive immunity to human tumors. Newer approaches using PDOs have become useful tools in developing models for human CRC. GEMMs in the past permitted recreation of multiple alterations seen in CRC providing novel insights into the process of tumor development. Other methods of developing mutant mice such as the CRISPR-CAS9 system are now being used in the development of new GEMMs that are both immunocompetent and autochthonous. All are expected to give further insight into the oncological process and to be used in evaluating new drugs with the abiding hope of providing effective therapeutic opportunities for patients.
Acknowledgments
The author would like to thank Dr. Raju Kucherlapati for the idea to write the review and for reading the manuscript and providing comments.
Author contributions
MK was solely responsible for data analysis and interpretation and manuscript drafting of important intellectual content. The author gives approval of the final version for publication and takes responsibility for the content.
Declaration of interests
The author declares there are no competing financial and/or non-financial interests in relation to the work described.
References
- 1.International, W.C.R.F. https://www.wcrf.org/cancer-trends/colorectal-cancer-statistics/
- 2.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA A Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Brenner D.R., Heer E., Sutherland R.L., Ruan Y., Tinmouth J., Heitman S.J., Hilsden R.J. National trends in colorectal cancer incidence among older and younger adults in Canada. JAMA Netw. Open. 2019;2:e198090. doi: 10.1001/jamanetworkopen.2019.8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ullah M.F., Fleming C.A., Mealy K. Changing trends in age and stage of colorectal cancer presentation in Ireland - from the nineties to noughties and beyond. Surgeon. 2018;16:350–354. doi: 10.1016/j.surge.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Vuik F.E., Nieuwenburg S.A., Bardou M., Lansdorp-Vogelaar I., Dinis-Ribeiro M., Bento M.J., Zadnik V., Pellisé M., Esteban L., Kaminski M.F., et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut. 2019;68:1820–1826. doi: 10.1136/gutjnl-2018-317592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu P.H., Wu K., Ng K., Zauber A.G., Nguyen L.H., Song M., He X., Fuchs C.S., Ogino S., Willett W.C., et al. Association of obesity with risk of early-onset colorectal cancer among women. JAMA Oncol. 2019;5:37–44. doi: 10.1001/jamaoncol.2018.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hales C.M., Fryar C.D., Carroll M.D., Freedman D.S., Ogden C.L. Trends in obesity and severe obesity prevalence in US youth and adults by sex and age, 2007-2008 to 2015-2016. JAMA. 2018;319:1723–1725. doi: 10.1001/jama.2018.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crutcher M.M., Baybutt T.R., Kopenhaver J.S., Snook A.E., Waldman S.A. Emerging drug targets for colon cancer: a preclinical assessment. Expert Opin. Ther. Targets. 2022;26:207–216. doi: 10.1080/14728222.2022.2039119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackstadt R., Sansom O.J. Mouse models of intestinal cancer. J. Pathol. 2016;238:141–151. doi: 10.1002/path.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guinney J., Dienstmann R., Wang X., de Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lannagan T.R., Jackstadt R., Leedham S.J., Sansom O.J. Advances in colon cancer research: in vitro and animal models. Curr. Opin. Genet. Dev. 2021;66:50–56. doi: 10.1016/j.gde.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnesen H., Knutsen L.E., Hognestad B.W., Johansen G.M., Bemark M., Pabst O., Storset A.K., Boysen P. A model system for feralizing laboratory mice in large farmyard-like pens. Front. Microbiol. 2020;11:615661. doi: 10.3389/fmicb.2020.615661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnesen H., Hitch T.C.A., Steppeler C., Müller M.H.B., Knutsen L.E., Gunnes G., Angell I.L., Ormaasen I., Rudi K., Paulsen J.E., et al. Naturalizing laboratory mice by housing in a farmyard-type habitat confers protection against colorectal carcinogenesis. Gut Microb. 2021;13:1993581. doi: 10.1080/19490976.2021.1993581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung K.E., Maricevich M.A., Richard L.G., Chen W.Y., Richardson M.P., Kunin A., Bronson R.T., Mahmood U., Kucherlapati R. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. USA. 2010;107:1565–1570. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu X.Y., Besterman J.M., Monosov A., Hoffman R.M. Models of human metastatic colon cancer in nude mice orthotopically constructed by using histologically intact patient specimens. Proc. Natl. Acad. Sci. USA. 1991;88:9345–9349. doi: 10.1073/pnas.88.20.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 17.Schutgens F., Clevers H. Human organoids: tools for understanding biology and treating diseases. Annu. Rev. Pathol. 2020;15:211–234. doi: 10.1146/annurev-pathmechdis-012419-032611. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O'Brien S.A., He Y., Wang L., Zhang Q., Kim A., et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 19.Vectorstock. https://www.vectorstock.com/royalty-free-vector/isolated-mouse-cartoon-design-vector-27980414
- 20.MacLeish A. The seat behind the pillar. The New York times, January 21, 1967. 1967. file://cifs2.partners.org/colony1$/Mouse%20models%20in%20Colon%20Cancer%20review/Archibald%20MacLeish_The%20seat%20behind%20the%20Pillar.pdf
- 21.Moser A.R., Pitot H.C., Dove W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 22.Estes W.L., Jr. Familial polyposis and carcinoma of the colon. Trans. South Surg. Assoc. 1947;59:325–335. [PubMed] [Google Scholar]
- 23.Guptill P. Familial polyposis of the colon: two families, five cases. Surgery. 1947;22:286–304. [PubMed] [Google Scholar]
- 24.Menezelio D. De excrescentals verrucosa cristois in intestininis crassis dysenteriam passi observatis. Acta Medicorum Berolinensium. 1721;4:68–71. [Google Scholar]
- 25.Bodmer W.F., Bailey C.J., Bodmer J., Bussey H.J., Ellis A., Gorman P., Lucibello F.C., Murday V.A., Rider S.H., Scambler P., et al. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature. 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 26.Groden J., Thliveris A., Samowitz W., Carlson M., Gelbert L., Albertsen H., Joslyn G., Stevens J., Spirio L., Robertson M., et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 27.Kinzler K.W., Nilbert M.C., Su L.K., Vogelstein B., Bryan T.M., Levy D.B., Smith K.J., Preisinger A.C., Hedge P., McKechnie D., et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 28.Meera Khan P., Tops C.M., vd Broek M., Breukel C., Wijnen J.T., Oldenburg M., vd Bos J., van Leeuwen-Cornelisse I.S., Vasen H.F., Griffioen G., et al. Close linkage of a highly polymorphic marker (D5S37) to familial adenomatous polyposis (FAP) and confirmation of FAP localization on chromosome 5q21-q22. Hum. Genet. 1988;79:183–185. doi: 10.1007/BF00280563. [DOI] [PubMed] [Google Scholar]
- 29.Leppert M., Burt R., Hughes J.P., Samowitz W., Nakamura Y., Woodward S., Gardner E., Lalouel J.M., White R. Genetic analysis of an inherited predisposition to colon cancer in a family with a variable number of adenomatous polyps. N. Engl. J. Med. 1990;322:904–908. doi: 10.1056/NEJM199003293221306. [DOI] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Center for Biotechnology, N. https://reactome.org/content/detail/R-HSA-4791275
- 32.Fodde R., Edelmann W., Yang K., van Leeuwen C., Carlson C., Renault B., Breukel C., Alt E., Lipkin M., Khan P.M., et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc. Natl. Acad. Sci. USA. 1994;91:8969–8973. doi: 10.1073/pnas.91.19.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Washington K. Apc-related models of intestinal neoplasia: a brief review for pathologists. BMC Surgical and Experimental Pathology. 2019 doi: 10.1186/s42047-019-0036-9. [DOI] [Google Scholar]
- 34.Bürtin F., Mullins C.S., Linnebacher M. Mouse models of colorectal cancer: past, present and future perspectives. World J. Gastroenterol. 2020;26:1394–1426. doi: 10.3748/wjg.v26.i13.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu S., Yin Y., Wang Q., Wang L. Dual gene deficient models of Apc(Min/+) mouse in assessing molecular mechanisms of intestinal carcinogenesis. Biomed. Pharmacother. 2018;108:600–609. doi: 10.1016/j.biopha.2018.09.056. [DOI] [PubMed] [Google Scholar]
- 36.Oshima M., Oshima H., Kitagawa K., Kobayashi M., Itakura C., Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc. Natl. Acad. Sci. USA. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke A.R., Purdie C.A., Harrison D.J., Morris R.G., Bird C.C., Hooper M.L., Wyllie A.H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 38.Clarke A.R., Cummings M.C., Harrison D.J. Interaction between murine germline mutations in p53 and APC predisposes to pancreatic neoplasia but not to increased intestinal malignancy. Oncogene. 1995;11:1913–1920. [PubMed] [Google Scholar]
- 39.Reitmair A.H., Cai J.C., Bjerknes M., Redston M., Cheng H., Pind M.T., Hay K., Mitri A., Bapat B.V., Mak T.W., Gallinger S. MSH2 deficiency contributes to accelerated APC-mediated intestinal tumorigenesis. Cancer Res. 1996;56:2922–2926. [PubMed] [Google Scholar]
- 40.Shibata H., Toyama K., Shioya H., Ito M., Hirota M., Hasegawa S., Matsumoto H., Takano H., Akiyama T., Toyoshima K., et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 41.Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 42.Fazeli A., Steen R.G., Dickinson S.L., Bautista D., Dietrich W.F., Bronson R.T., Bresalier R.S., Lander E.S., Costa J., Weinberg R.A. Effects of p53 mutations on apoptosis in mouse intestinal and human colonic adenomas. Proc. Natl. Acad. Sci. USA. 1997;94:10199–10204. doi: 10.1073/pnas.94.19.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quesada C.F., Kimata H., Mori M., Nishimura M., Tsuneyoshi T., Baba S. Piroxicam and acarbose as chemopreventive agents for spontaneous intestinal adenomas in APC gene 1309 knockout mice. Jpn. J. Cancer Res. 1998;89:392–396. doi: 10.1111/j.1349-7006.1998.tb00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smits R., van der Houven van Oordt W., Luz A., Zurcher C., Jagmohan-Changur S., Breukel C., Khan P.M., Fodde R. Apc1638N: a mouse model for familial adenomatous polyposis-associated desmoid tumors and cutaneous cysts. Gastroenterology. 1998;114:275–283. doi: 10.1016/s0016-5085(98)70478-0. [DOI] [PubMed] [Google Scholar]
- 45.Takaku K., Oshima M., Miyoshi H., Matsui M., Seldin M.F., Taketo M.M. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 46.Edelmann W., Yang K., Kuraguchi M., Heyer J., Lia M., Kneitz B., Fan K., Brown A.M., Lipkin M., Kucherlapati R. Tumorigenesis in Mlh1 and mlh1/apc1638N mutant mice. Cancer Res. 1999;59:1301–1307. [PubMed] [Google Scholar]
- 47.Gould K.A., Dietrich W.F., Borenstein N., Lander E.S., Dove W.F. Mom1 is a semi-dominant modifier of intestinal adenoma size and multiplicity in Min/+ mice. Genetics. 1996;144:1769–1776. doi: 10.1093/genetics/144.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr., Butel J.S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 49.Halberg R.B., Katzung D.S., Hoff P.D., Moser A.R., Cole C.E., Lubet R.A., Donehower L.A., Jacoby R.F., Dove W.F. Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proc. Natl. Acad. Sci. USA. 2000;97:3461–3466. doi: 10.1073/pnas.97.7.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smits R., Hofland N., Edelmann W., Geugien M., Jagmohan-Changur S., Albuquerque C., Breukel C., Kucherlapati R., Kielman M.F., Fodde R. Somatic Apc mutations are selected upon their capacity to inactivate the beta-catenin downregulating activity. Genes Chromosomes Cancer. 2000;29:229–239. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1033>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 51.Sasai H., Masaki M., Wakitani K. Suppression of polypogenesis in a new mouse strain with a truncated Apc(Delta474) by a novel COX-2 inhibitor, JTE-522. Carcinogenesis. 2000;21:953–958. doi: 10.1093/carcin/21.5.953. [DOI] [PubMed] [Google Scholar]
- 52.Møllersen L., Paulsen J.E., Alexander J. Loss of heterozygosity and nonsense mutation in Apc in azoxymethane-induced colonic tumours in min mice. Anticancer Res. 2004;24:2595–2599. [PubMed] [Google Scholar]
- 53.Suzui M., Okuno M., Tanaka T., Nakagama H., Moriwaki H. Enhanced colon carcinogenesis induced by azoxymethane in min mice occurs via a mechanism independent of beta-catenin mutation. Cancer Lett. 2002;183:31–41. doi: 10.1016/s0304-3835(02)00114-3. [DOI] [PubMed] [Google Scholar]
- 54.Takaku K., Wrana J.L., Robertson E.J., Taketo M.M. No effects of Smad2 (madh2) null mutation on malignant progression of intestinal polyps in Apc(delta716) knockout mice. Cancer Res. 2002;62:4558–4561. [PubMed] [Google Scholar]
- 55.Hamamoto T., Beppu H., Okada H., Kawabata M., Kitamura T., Miyazono K., Kato M. Compound disruption of smad2 accelerates malignant progression of intestinal tumors in apc knockout mice. Cancer Res. 2002;62:5955–5961. [PubMed] [Google Scholar]
- 56.Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M.F., Fan K., Russell R., Brown A.M.C., et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oshima M., Dinchuk J.E., Kargman S.L., Oshima H., Hancock B., Kwong E., Trzaskos J.M., Evans J.F., Taketo M.M. Suppression of intestinal polyposis in Apc delta716 knockout mice by inhibition of cyclooxygenase 2 (COX-2) Cell. 1996;87:803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 58.Ushikubi F., Segi E., Sugimoto Y., Murata T., Matsuoka T., Kobayashi T., Hizaki H., Tuboi K., Katsuyama M., Ichikawa A., et al. Impaired febrile response in mice lacking the prostaglandin E receptor subtype EP3. Nature. 1998;395:281–284. doi: 10.1038/26233. [DOI] [PubMed] [Google Scholar]
- 59.Seno H., Oshima M., Ishikawa T.O., Oshima H., Takaku K., Chiba T., Narumiya S., Taketo M.M. Cyclooxygenase 2- and prostaglandin E(2) receptor EP(2)-dependent angiogenesis in Apc(Delta716) mouse intestinal polyps. Cancer Res. 2002;62:506–511. [PubMed] [Google Scholar]
- 60.Aoki K., Tamai Y., Horiike S., Oshima M., Taketo M.M. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/- compound mutant mice. Nat. Genet. 2003;35:323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- 61.Colnot S., Niwa-Kawakita M., Hamard G., Godard C., Le Plenier S., Houbron C., Romagnolo B., Berrebi D., Giovannini M., Perret C. Colorectal cancers in a new mouse model of familial adenomatous polyposis: influence of genetic and environmental modifiers. Lab. Invest. 2004;84:1619–1630. doi: 10.1038/labinvest.3700180. [DOI] [PubMed] [Google Scholar]
- 62.Møllersen L., Vikse R., Andreassen A., Steffensen I.L., Mikalsen A., Paulsen J.E., Alexander J. Adenomatous polyposis coli truncation mutations in 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-induced intestinal tumours of multiple intestinal neoplasia mice. Mutat. Res. 2004;557:29–40. doi: 10.1016/j.mrgentox.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Rao C.V., Yang Y.M., Swamy M.V., Liu T., Fang Y., Mahmood R., Jhanwar-Uniyal M., Dai W. Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc. Natl. Acad. Sci. USA. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orioli D., Henkemeyer M., Lemke G., Klein R., Pawson T. Sek4 and Nuk receptors cooperate in guidance of commissural axons and in palate formation. EMBO J. 1996;15:6035–6049. [PMC free article] [PubMed] [Google Scholar]
- 65.Batlle E., Henderson J.T., Beghtel H., van den Born M.M.W., Sancho E., Huls G., Meeldijk J., Robertson J., van de Wetering M., Pawson T., Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 66.Batlle E., Bacani J., Begthel H., Jonkheer S., Gregorieff A., van de Born M., Malats N., Sancho E., Boon E., Pawson T., et al. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435:1126–1130. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]
- 67.Wilson J.M., Coletta P.L., Cuthbert R.J., Scott N., MacLennan K., Hawcroft G., Leng L., Lubetsky J.B., Jin K.K., Lolis E., et al. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology. 2005;129:1485–1503. doi: 10.1053/j.gastro.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 68.Kuraguchi M., Wang X.P., Bronson R.T., Rothenberg R., Ohene-Baah N.Y., Lund J.J., Kucherlapati M., Maas R.L., Kucherlapati R. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka T., Kohno H., Suzuki R., Hata K., Sugie S., Niho N., Sakano K., Takahashi M., Wakabayashi K. Dextran sodium sulfate strongly promotes colorectal carcinogenesis in Apc(Min/+) mice: inflammatory stimuli by dextran sodium sulfate results in development of multiple colonic neoplasms. Int. J. Cancer. 2006;118:25–34. doi: 10.1002/ijc.21282. [DOI] [PubMed] [Google Scholar]
- 70.Sansom O.J., Meniel V., Wilkins J.A., Cole A.M., Oien K.A., Marsh V., Jamieson T.J., Guerra C., Ashton G.H., Barbacid M., Clarke A.R. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muñoz N.M., Upton M., Rojas A., Washington M.K., Lin L., Chytil A., Sozmen E.G., Madison B.B., Pozzi A., Moon R.T., et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 72.Sodir N.M., Chen X., Park R., Nickel A.E., Conti P.S., Moats R., Bading J.R., Shibata D., Laird P.W. Smad3 deficiency promotes tumorigenesis in the distal colon of ApcMin/+ mice. Cancer Res. 2006;66:8430–8438. doi: 10.1158/0008-5472.CAN-06-1437. [DOI] [PubMed] [Google Scholar]
- 73.Alberici P., Jagmohan-Changur S., De Pater E., Van Der Valk M., Smits R., Hohenstein P., Fodde R. Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene. 2006;25:1841–1851. doi: 10.1038/sj.onc.1209226. [DOI] [PubMed] [Google Scholar]
- 74.Wei K., Clark A.B., Wong E., Kane M.F., Mazur D.J., Parris T., Kolas N.K., Russell R., Hou H., Jr., Kneitz B., et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kucherlapati M., Nguyen A., Kuraguchi M., Yang K., Fan K., Bronson R., Wei K., Lipkin M., Edelmann W., Kucherlapati R. Tumor progression in apc(1638N) mice with Exo1 and Fen1 deficiencies. Oncogene. 2007;26:6297–6306. doi: 10.1038/sj.onc.1210453. [DOI] [PubMed] [Google Scholar]
- 76.Rakoff-Nahoum S., Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 77.Saam J.R., Gordon J.I. Inducible gene knockouts in the small intestinal and colonic epithelium. J. Biol. Chem. 1999;274:38071–38082. doi: 10.1074/jbc.274.53.38071. [DOI] [PubMed] [Google Scholar]
- 78.el Marjou F., Janssen K.P., Chang B.H.J., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 79.Haigis K.M., Kendall K.R., Wang Y., Cheung A., Haigis M.C., Glickman J.N., Niwa-Kawakita M., Sweet-Cordero A., Sebolt-Leopold J., Shannon K.M., et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 2008;40:600–608. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vooijs M., van der Valk M., te Riele H., Berns A. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene. 1998;17:1–12. doi: 10.1038/sj.onc.1202169. [DOI] [PubMed] [Google Scholar]
- 81.Kucherlapati M.H., Yang K., Fan K., Kuraguchi M., Sonkin D., Rosulek A., Lipkin M., Bronson R.T., Aronow B.J., Kucherlapati R. Loss of Rb1 in the gastrointestinal tract of Apc1638N mice promotes tumors of the cecum and proximal colon. Proc. Natl. Acad. Sci. USA. 2008;105:15493–15498. doi: 10.1073/pnas.0802933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Halberg R.B., Waggoner J., Rasmussen K., White A., Clipson L., Prunuske A.J., Bacher J.W., Sullivan R., Washington M.K., Pitot H.C., et al. Long-lived Min mice develop advanced intestinal cancers through a genetically conservative pathway. Cancer Res. 2009;69:5768–5775. doi: 10.1158/0008-5472.CAN-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson C.L., Heppner K.J., Labosky P.A., Hogan B.L., Matrisian L.M. Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin. Proc. Natl. Acad. Sci. USA. 1997;94:1402–1407. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kitamura T., Biyajima K., Aoki M., Oshima M., Taketo M.M. Matrix metalloproteinase 7 is required for tumor formation, but dispensable for invasion and fibrosis in SMAD4-deficient intestinal adenocarcinomas. Lab. Invest. 2009;89:98–105. doi: 10.1038/labinvest.2008.107. [DOI] [PubMed] [Google Scholar]
- 85.Fre S., Pallavi S.K., Huyghe M., Laé M., Janssen K.P., Robine S., Artavanis-Tsakonas S., Louvard D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc. Natl. Acad. Sci. USA. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pollard P., Deheragoda M., Segditsas S., Lewis A., Rowan A., Howarth K., Willis L., Nye E., McCart A., Mandir N., et al. The Apc 1322T mouse develops severe polyposis associated with submaximal nuclear beta-catenin expression. Gastroenterology. 2009;136:2204–2213.e1-13. doi: 10.1053/j.gastro.2009.02.058. [DOI] [PubMed] [Google Scholar]
- 87.Crist R.C., Roth J.J., Baran A.A., McEntee B.J., Siracusa L.D., Buchberg A.M. The armadillo repeat domain of Apc suppresses intestinal tumorigenesis. Mamm. Genome. 2010;21:450–457. doi: 10.1007/s00335-010-9288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robanus-Maandag E.C., Koelink P.J., Breukel C., Salvatori D.C.F., Jagmohan-Changur S.C., Bosch C.A.J., Verspaget H.W., Devilee P., Fodde R., Smits R. A new conditional Apc-mutant mouse model for colorectal cancer. Carcinogenesis. 2010;31:946–952. doi: 10.1093/carcin/bgq046. [DOI] [PubMed] [Google Scholar]
- 89.Xue Y., Johnson R., Desmet M., Snyder P.W., Fleet J.C. Generation of a transgenic mouse for colorectal cancer research with intestinal cre expression limited to the large intestine. Mol. Cancer Res. 2010;8:1095–1104. doi: 10.1158/1541-7786.MCR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheung A.F., Carter A.M., Kostova K.K., Woodruff J.F., Crowley D., Bronson R.T., Haigis K.M., Jacks T. Complete deletion of Apc results in severe polyposis in mice. Oncogene. 2010;29:1857–1864. doi: 10.1038/onc.2009.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roper J., Richardson M.P., Wang W.V., Richard L.G., Chen W., Coffee E.M., Sinnamon M.J., Lee L., Chen P.C., Bronson R.T., et al. The dual PI3K/mTOR inhibitor NVP-BEZ235 induces tumor regression in a genetically engineered mouse model of PIK3CA wild-type colorectal cancer. PLoS One. 2011;6:e25132. doi: 10.1371/journal.pone.0025132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sancho R., Jandke A., Davis H., Diefenbacher M.E., Tomlinson I., Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 93.Nakae S., Komiyama Y., Nambu A., Sudo K., Iwase M., Homma I., Sekikawa K., Asano M., Iwakura Y. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 2002;17:375–387. doi: 10.1016/s1074-7613(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 94.Chae W.J., Gibson T.F., Zelterman D., Hao L., Henegariu O., Bothwell A.L.M. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc. Natl. Acad. Sci. USA. 2010;107:5540–5544. doi: 10.1073/pnas.0912675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akira S., Hoshino K., Kaisho T. The role of Toll-like receptors and MyD88 in innate immune responses. J. Endotoxin Res. 2000;6:383–387. [PubMed] [Google Scholar]
- 96.Lee S.H., Hu L.L., Gonzalez-Navajas J., Seo G.S., Shen C., Brick J., Herdman S., Varki N., Corr M., Lee J., Raz E. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat. Med. 2010;16:665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang X.O., Chang S.H., Park H., Nurieva R., Shah B., Acero L., Wang Y.H., Schluns K.S., Broaddus R.R., Zhu Z., Dong C. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chae W.J., Bothwell A.L.M. IL-17F deficiency inhibits small intestinal tumorigenesis in ApcMin/+ mice. Biochem. Biophys. Res. Commun. 2011;414:31–36. doi: 10.1016/j.bbrc.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 99.McClellan J.L., Davis J.M., Steiner J.L., Enos R.T., Jung S.H., Carson J.A., Pena M.M., Carnevale K.A., Berger F.G., Murphy E.A. Linking tumor-associated macrophages, inflammation, and intestinal tumorigenesis: role of MCP-1. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G1087–G1095. doi: 10.1152/ajpgi.00252.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shaked H., Hofseth L.J., Chumanevich A., Chumanevich A.A., Wang J., Wang Y., Taniguchi K., Guma M., Shenouda S., Clevers H., et al. Chronic epithelial NF-kappaB activation accelerates APC loss and intestinal tumor initiation through iNOS up-regulation. Proc. Natl. Acad. Sci. USA. 2012;109:14007–14012. doi: 10.1073/pnas.1211509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng Y., Sentani K., Wiese A., Sands E., Green M., Bommer G.T., Cho K.R., Fearon E.R. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. Am. J. Pathol. 2013;183:493–503. doi: 10.1016/j.ajpath.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coffee E.M., Faber A.C., Roper J., Sinnamon M.J., Goel G., Keung L., Wang W.V., Vecchione L., de Vriendt V., Weinstein B.J., et al. Concomitant BRAF and PI3K/mTOR blockade is required for effective treatment of BRAF(V600E) colorectal cancer. Clin. Cancer Res. 2013;19:2688–2698. doi: 10.1158/1078-0432.CCR-12-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Asfaha S., Dubeykovskiy A.N., Tomita H., Yang X., Stokes S., Shibata W., Friedman R.A., Ariyama H., Dubeykovskaya Z.A., Muthupalani S., et al. Mice that express human interleukin-8 have increased mobilization of immature myeloid cells, which exacerbates inflammation and accelerates colon carcinogenesis. Gastroenterology. 2013;144:155–166. doi: 10.1053/j.gastro.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paul Olson T.J., Hadac J.N., Sievers C.K., Leystra A.A., Deming D.A., Zahm C.D., Albrecht D.M., Nomura A., Nettekoven L.A., Plesh L.K., et al. Dynamic tumor growth patterns in a novel murine model of colorectal cancer. Cancer Prev. Res. 2014;7:105–113. doi: 10.1158/1940-6207.CAPR-13-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Byun A.J., Hung K.E., Fleet J.C., Bronson R.T., Mason J.B., Garcia P.E., Crott J.W. Colon-specific tumorigenesis in mice driven by Cre-mediated inactivation of Apc and activation of mutant Kras. Cancer Lett. 2014;347:191–195. doi: 10.1016/j.canlet.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Suzuki A., Yamaguchi M.T., Ohteki T., Sasaki T., Kaisho T., Kimura Y., Yoshida R., Wakeham A., Higuchi T., Fukumoto M., et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14:523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 107.Guerra C., Mijimolle N., Dhawahir A., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 108.Davies E.J., Marsh Durban V., Meniel V., Williams G.T., Clarke A.R. PTEN loss and KRAS activation leads to the formation of serrated adenomas and metastatic carcinoma in the mouse intestine. J. Pathol. 2014;233:27–38. doi: 10.1002/path.4312. [DOI] [PubMed] [Google Scholar]
- 109.Dow L.E., O'Rourke K.P., Simon J., Tschaharganeh D.F., van Es J.H., Clevers H., Lowe S.W. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161:1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oshima H., Nakayama M., Han T.S., Naoi K., Ju X., Maeda Y., Robine S., Tsuchiya K., Sato T., Sato H., et al. Suppressing TGFbeta signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res. 2015;75:766–776. doi: 10.1158/0008-5472.CAN-14-2036. [DOI] [PubMed] [Google Scholar]
- 111.Hadac J.N., Leystra A.A., Paul Olson T.J., Maher M.E., Payne S.N., Yueh A.E., Schwartz A.R., Albrecht D.M., Clipson L., Pasch C.A., et al. Colon tumors with the simultaneous induction of driver mutations in APC, KRAS, and PIK3CA still progress through the adenoma-to-carcinoma sequence. Cancer Prev. Res. 2015;8:952–961. doi: 10.1158/1940-6207.CAPR-15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sødring M., Gunnes G., Paulsen J.E. Spontaneous initiation, promotion and progression of colorectal cancer in the novel A/J Min/+ mouse. Int. J. Cancer. 2016;138:1936–1946. doi: 10.1002/ijc.29928. [DOI] [PubMed] [Google Scholar]
- 113.Kawaguchi Y., Hinoi T., Saito Y., Adachi T., Miguchi M., Niitsu H., Sasada T., Shimomura M., Egi H., Oka S., et al. Mouse model of proximal colon-specific tumorigenesis driven by microsatellite instability-induced Cre-mediated inactivation of Apc and activation of Kras. J. Gastroenterol. 2016;51:447–457. doi: 10.1007/s00535-015-1121-9. [DOI] [PubMed] [Google Scholar]
- 114.Tetteh P.W., Kretzschmar K., Begthel H., van den Born M., Korving J., Morsink F., Farin H., van Es J.H., Offerhaus G.J.A., Clevers H. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc. Natl. Acad. Sci. USA. 2016;113:11859–11864. doi: 10.1073/pnas.1614057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nakayama M., Sakai E., Echizen K., Yamada Y., Oshima H., Han T.S., Ohki R., Fujii S., Ochiai A., Robine S., et al. Intestinal cancer progression by mutant p53 through the acquisition of invasiveness associated with complex glandular formation. Oncogene. 2017;36:5885–5896. doi: 10.1038/onc.2017.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roper J., Tammela T., Cetinbas N.M., Akkad A., Roghanian A., Rickelt S., Almeqdadi M., Wu K., Oberli M.A., Sánchez-Rivera F.J., et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 2017;35:569–576. doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu J., Cortellino S., Tricarico R., Chang W.C., Scher G., Devarajan K., Slifker M., Moore R., Bassi M.R., Caretti E., et al. Thymine DNA Glycosylase (TDG) is involved in the pathogenesis of intestinal tumors with reduced APC expression. Oncotarget. 2017;8:89988–89997. doi: 10.18632/oncotarget.21219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kang D.W., Lee S.W., Hwang W.C., Lee B.H., Choi Y.S., Suh Y.A., Choi K.Y., Min D.S. Phospholipase D1 acts through akt/TopBP1 and RB1 to regulate the E2F1-dependent apoptotic program in cancer cells. Cancer Res. 2017;77:142–152. doi: 10.1158/0008-5472.CAN-15-3032. [DOI] [PubMed] [Google Scholar]
- 119.Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chytil A., Magnuson M.A., Wright C.V.E., Moses H.L. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 121.Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 122.Onoyama I., Tsunematsu R., Matsumoto A., Kimura T., de Alborán I.M., Nakayama K., Nakayama K.I. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sakai E., Nakayama M., Oshima H., Kouyama Y., Niida A., Fujii S., Ochiai A., Nakayama K.I., Mimori K., Suzuki Y., et al. Combined mutation of apc, Kras, and Tgfbr2 effectively drives metastasis of intestinal cancer. Cancer Res. 2018;78:1334–1346. doi: 10.1158/0008-5472.CAN-17-3303. [DOI] [PubMed] [Google Scholar]
- 124.Lopès A., Cassé A.H., Billard E., Boulcourt-Sambou E., Roche G., Larois C., Barnich N., Naimi S., Bonnet M., Dumas B. Deciphering the immune microenvironment of a tissue by digital imaging and cognition network. Sci. Rep. 2018;8:16692. doi: 10.1038/s41598-018-34731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li D., Xiao L., Ge Y., Fu Y., Zhang W., Cao H., Chen B., Wang H., Zhan Y.Y., Hu T. High expression of Tob1 indicates poor survival outcome and promotes tumour progression via a Wnt positive feedback loop in colon cancer. Mol. Cancer. 2018;17:159. doi: 10.1186/s12943-018-0907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pham T., Carpinteri S., Sampurno S., Pereira L., Roth S., Narasimhan V., Darcy P., Desai J., Heriot A.G., Ramsay R.G. Novel vaccine targeting colonic adenoma: a pre-clinical model. J. Gastrointest. Surg. 2019;23:626–633. doi: 10.1007/s11605-018-4060-y. [DOI] [PubMed] [Google Scholar]
- 127.Dzhagalov I., St John A., He Y.W. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood. 2007;109:1620–1626. doi: 10.1182/blood-2006-03-013771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Triner D., Devenport S.N., Ramakrishnan S.K., Ma X., Frieler R.A., Greenson J.K., Inohara N., Nunez G., Colacino J.A., Mortensen R.M., Shah Y.M. Neutrophils restrict tumor-associated microbiota to reduce growth and invasion of colon tumors in mice. Gastroenterology. 2019;156:1467–1482. doi: 10.1053/j.gastro.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 130.Wen Y.A., Xiong X., Scott T., Li A.T., Wang C., Weiss H.L., Tan L., Bradford E., Fan T.W.M., Chandel N.S., et al. The mitochondrial retrograde signaling regulates Wnt signaling to promote tumorigenesis in colon cancer. Cell Death Differ. 2019;26:1955–1969. doi: 10.1038/s41418-018-0265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heazlewood C.K., Cook M.C., Eri R., Price G.R., Tauro S.B., Taupin D., Thornton D.J., Png C.W., Crockford T.L., Cornall R.J., et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.De Santis S., Verna G., Serino G., Armentano R., Cavalcanti E., Liso M., Dicarlo M., Coletta S., Mastronardi M., Lippolis A., et al. Winnie-APC(Min/+) mice: a spontaneous model of colitis-associated colorectal cancer combining genetics and inflammation. Int. J. Mol. Sci. 2020;21:2972. doi: 10.3390/ijms21082972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Parihar M., Dodds S.G., Hubbard G., Javors M.A., Strong R., Hasty P., Sharp Z.D. Rapamycin extends life span in apc(min/+) colon cancer FAP model. Clin. Colorectal Cancer. 2021;20:e61–e70. doi: 10.1016/j.clcc.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brennan C.A., Nakatsu G., Gallini Comeau C.A., Drew D.A., Glickman J.N., Schoen R.E., Chan A.T., Garrett W.S. Aspirin Modulation of the Colorectal Cancer-Associated Microbe Fusobacterium Nucleatum. mBio. 2021;12 doi: 10.1128/mBio.00547-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bowen C.M., Walter L., Borras E., Wu W., Ozcan Z., Chang K., Bommi P.V., Taggart M.W., Thirumurthi S., Lynch P.M., et al. Combination of sulindac and bexarotene for prevention of intestinal carcinogenesis in familial adenomatous polyposis. Cancer Prev. Res. 2021;14:851–862. doi: 10.1158/1940-6207.CAPR-20-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 137.Kolodner R.D., Hall N.R., Lipford J., Kane M.F., Rao M.R., Morrison P., Wirth L., Finan P.J., Burn J., Chapman P. Structure of the human MSH2 locus and analysis of two Muir-Torre kindreds for msh2 mutations. Genomics. 1994;24:516–526. doi: 10.1006/geno.1994.1661. [DOI] [PubMed] [Google Scholar]
- 138.Lindblom A., Tannergård P., Werelius B., Nordenskjöld M. Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat. Genet. 1993;5:279–282. doi: 10.1038/ng1193-279. [DOI] [PubMed] [Google Scholar]
- 139.Bronner C.E., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 140.Kolodner R.D., Hall N.R., Lipford J., Kane M.F., Morrison P.T., Finan P.J., Burn J., Chapman P., Earabino C., Merchant E., et al. Structure of the human MLH1 locus and analysis of a large hereditary nonpolyposis colorectal carcinoma kindred for mlh1 mutations. Cancer Res. 1995;55:242–248. [PubMed] [Google Scholar]
- 141.Edelmann W., Cohen P.E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R., et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 142.Reitmair A.H., Schmits R., Ewel A., Bapat B., Redston M., Mitri A., Waterhouse P., Mittrücker H.W., Wakeham A., Liu B., et al. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat. Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 143.Siegel E.C., Bryson V. Mutator gene of Escherichia coli B. J. Bacteriol. 1967;94:38–47. doi: 10.1128/jb.94.1.38-47.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wildenberg J., Meselson M. Mismatch repair in heteroduplex DNA. Proc. Natl. Acad. Sci. USA. 1975;72:2202–2206. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kolodner R.D. A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair. 2016;38:3–13. doi: 10.1016/j.dnarep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee K., Tosti E., Edelmann W. Mouse models of DNA mismatch repair in cancer research. DNA Repair. 2016;38:140–146. doi: 10.1016/j.dnarep.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kucherlapati M.H., Lee K., Nguyen A.A., Clark A.B., Hou H., Jr., Rosulek A., Li H., Yang K., Fan K., Lipkin M., et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology. 2010;138:993–1002.e1. doi: 10.1053/j.gastro.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chan A.T., Baba Y., Shima K., Nosho K., Chung D.C., Hung K.E., Mahmood U., Madden K., Poss K., Ranieri A., et al. Cathepsin B expression and survival in colon cancer: implications for molecular detection of neoplasia. Cancer Epidemiol. Biomarkers Prev. 2010;19:2777–2785. doi: 10.1158/1055-9965.EPI-10-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Latchman Y.E., Liang S.C., Wu Y., Chernova T., Sobel R.A., Klemm M., Kuchroo V.K., Freeman G.J., Sharpe A.H. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA. 2004;101:10691–10696. doi: 10.1073/pnas.0307252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lorenz E., Stewart H.L. Tumors of the alimentary tract induced in mice by feeding olive-oil emulsions containing carcinogenic hydrocarbons. J. Natl. Cancer Inst. 1947;7:227–238. [PubMed] [Google Scholar]
- 151.Nascimento-Gonçalves E., Mendes B.A.L., Silva-Reis R., Faustino-Rocha A.I., Gama A., Oliveira P.A. Animal models of colorectal cancer: from spontaneous to genetically engineered models and their applications. Vet. Sci. 2021;8:59. doi: 10.3390/vetsci8040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rausch P., Basic M., Batra A., Bischoff S.C., Blaut M., Clavel T., Gläsner J., Gopalakrishnan S., Grassl G.A., Günther C., et al. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol. 2016;306:343–355. doi: 10.1016/j.ijmm.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 153.Franklin C.L., Ericsson A.C. Microbiota and reproducibility of rodent models. Lab Anim. 2017;46:114–122. doi: 10.1038/laban.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tosti E., Almeida A.S., Tran T.T.T., Barbachan E Silva M., Broin P.Ó., Dubin R., Chen K., Beck A.P., McLellan A.S., Vilar E., et al. Loss of MMR and TGFBR2 increases the susceptibility to microbiota-dependent inflammation-associated colon cancer. Cell. Mol. Gastroenterol. Hepatol. 2022;14:693–717. doi: 10.1016/j.jcmgh.2022.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Suzuki R., Kohno H., Sugie S., Nakagama H., Tanaka T. Strain differences in the susceptibility to azoxymethane and dextran sodium sulfate-induced colon carcinogenesis in mice. Carcinogenesis. 2006;27:162–169. doi: 10.1093/carcin/bgi205. [DOI] [PubMed] [Google Scholar]
- 156.Arnesen H., Müller M.H.B., Aleksandersen M., Østby G.C., Carlsen H., Paulsen J.E., Boysen P. Induction of colorectal carcinogenesis in the C57BL/6J and A/J mouse strains with a reduced DSS dose in the AOM/DSS model. Lab. Anim. Res. 2021;37:19. doi: 10.1186/s42826-021-00096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Neufert C., Heichler C., Brabletz T., Scheibe K., Boonsanay V., Greten F.R., Neurath M.F. Inducible mouse models of colon cancer for the analysis of sporadic and inflammation-driven tumor progression and lymph node metastasis. Nat. Protoc. 2021;16:61–85. doi: 10.1038/s41596-020-00412-1. [DOI] [PubMed] [Google Scholar]
- 158.Okayasu I., Hatakeyama S., Yamada M., Ohkusa T., Inagaki Y., Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 159.Pan Q., Lou X., Zhang J., Zhu Y., Li F., Shan Q., Chen X., Xie Y., Su S., Wei H., et al. Erratum: genomic variants in mouse model induced by azoxymethane and dextran sodium sulfate improperly mimic human colorectal cancer. Sci. Rep. 2017;7:2784. doi: 10.1038/s41598-017-00076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pan Q., Lou X., Zhang J., Zhu Y., Li F., Shan Q., Chen X., Xie Y., Su S., Wei H., et al. Genomic variants in mouse model induced by azoxymethane and dextran sodium sulfate improperly mimic human colorectal cancer. Sci. Rep. 2017;7:25. doi: 10.1038/s41598-017-00057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gu S., Zaidi S., Hassan M.I., Mohammad T., Malta T.M., Noushmehr H., Nguyen B., Crandall K.A., Srivastav J., Obias V., et al. Mutated CEACAMs disrupt transforming growth factor beta signaling and alter the intestinal microbiome to promote colorectal carcinogenesis. Gastroenterology. 2020;158:238–252. doi: 10.1053/j.gastro.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Smithies O., Koralewski M.A., Song K.Y., Kucherlapati R.S. Homologous recombination with DNA introduced into mammalian cells. Cold Spring Harbor Symp. Quant. Biol. 1984;49:161–170. doi: 10.1101/sqb.1984.049.01.019. [DOI] [PubMed] [Google Scholar]
- 163.Mak T.W. Gene targeting in embryonic stem cells scores a knockout in Stockholm. Cell. 2007;131:1027–1031. doi: 10.1016/j.cell.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 164.Gu H., Marth J.D., Orban P.C., Mossmann H., Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 165.Lieber A., He C.Y., Kirillova I., Kay M.A. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J. Virol. 1996;70:8944–8960. doi: 10.1128/JVI.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Metzger D., Clifford J., Chiba H., Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hennighausen L., Wall R.J., Tillmann U., Li M., Furth P.A. Conditional gene expression in secretory tissues and skin of transgenic mice using the MMTV-LTR and the tetracycline responsive system. J. Cell. Biochem. 1995;59:463–472. doi: 10.1002/jcb.240590407. [DOI] [PubMed] [Google Scholar]
- 168.Paddison P.J., Hannon G.J. RNA interference: the new somatic cell genetics? Cancer Cell. 2002;2:17–23. doi: 10.1016/s1535-6108(02)00092-2. [DOI] [PubMed] [Google Scholar]
- 169.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Paddison P.J., Caudy A.A., Hannon G.J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Parisi T., Bronson R.T., Lees J.A. Inactivation of the retinoblastoma gene yields a mouse model of malignant colorectal cancer. Oncogene. 2015;34:5890–5899. doi: 10.1038/onc.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kucherlapati M.H., Nguyen A.A., Bronson R.T., Kucherlapati R.S. Inactivation of conditional Rb by Villin-Cre leads to aggressive tumors outside the gastrointestinal tract. Cancer Res. 2006;66:3576–3583. doi: 10.1158/0008-5472.CAN-05-2699. [DOI] [PubMed] [Google Scholar]
- 173.Madison B.B., Dunbar L., Qiao X.T., Braunstein K., Braunstein E., Gumucio D.L. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 174.Rutlin M., Rastelli D., Kuo W.T., Estep J.A., Louis A., Riccomagno M.M., Turner J.R., Rao M. The Villin1 gene promoter drives cre recombinase expression in extraintestinal tissues. Cell. Mol. Gastroenterol. Hepatol. 2020;10:864–867.e5. doi: 10.1016/j.jcmgh.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Heinen C.D., Goss K.H., Cornelius J.R., Babcock G.F., Knudsen E.S., Kowalik T., Groden J. The APC tumor suppressor controls entry into S-phase through its ability to regulate the cyclin D/RB pathway. Gastroenterology. 2002;123:751–763. doi: 10.1053/gast.2002.35382. [DOI] [PubMed] [Google Scholar]
- 176.Kucherlapati M.H. Co-expression patterns explain how a basic transcriptional role for MYC modulates Wnt and MAPK pathways in colon and lung adenocarcinomas. Cell Cycle. 2022;21:1619–1638. doi: 10.1080/15384101.2022.2060454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Boutin A.T., Liao W.T., Wang M., Hwang S.S., Karpinets T.V., Cheung H., Chu G.C., Jiang S., Hu J., Chang K., et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017;31:370–382. doi: 10.1101/gad.293449.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tauriello D.V.F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., Sevillano M., Ibiza S., Cañellas A., Hernando-Momblona X., et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 179.Holmes E.C. Immunology of tumor infiltrating lymphocytes. Ann. Surg. 1985;201:158–163. doi: 10.1097/00000658-198502000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kim B.G., Malek E., Choi S.H., Ignatz-Hoover J.J., Driscoll J.J. Novel therapies emerging in oncology to target the TGF-beta pathway. J. Hematol. Oncol. 2021;14:55. doi: 10.1186/s13045-021-01053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jackstadt R., van Hooff S.R., Leach J.D., Cortes-Lavaud X., Lohuis J.O., Ridgway R.A., Wouters V.M., Roper J., Kendall T.J., Roxburgh C.S., et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36:319–336.e7. doi: 10.1016/j.ccell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ahmad R., Kumar B., Tamang R.L., Xu W., Talmon G.A., Mohs A.M., Dhawan P., Singh A.B. Colonoscopy-based intramucosal transplantation of cancer cells for mouse modeling of colon cancer and lung metastasis. Biotechniques. 2021;71:456–464. doi: 10.2144/btn-2020-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gordon J.W., Ruddle F.H. Germ line transmission in transgenic mice. Prog. Clin. Biol. Res. 1982;85 Pt B:111–124. [PubMed] [Google Scholar]
- 184.Carroll S.L., Roth K.A., Gordon J.I. Liver fatty acid-binding protein: a marker for studying cellular differentiation in gut epithelial neoplasms. Gastroenterology. 1990;99:1727–1735. doi: 10.1016/0016-5085(90)90480-o. [DOI] [PubMed] [Google Scholar]
- 185.Hasegawa T., Isobe K., Tsuchiya Y., Oikawa S., Nakazato H., Ikezawa H., Nakashima I., Shimokata K. Establishment and characterisation of human carcinoembryonic antigen transgenic mice. Br. J. Cancer. 1991;64:710–714. doi: 10.1038/bjc.1991.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gremonprez F., Willaert W., Ceelen W. Animal models of colorectal peritoneal metastasis. Pleura Peritoneum. 2016;1:23–43. doi: 10.1515/pp-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Isella C., Terrasi A., Bellomo S.E., Petti C., Galatola G., Muratore A., Mellano A., Senetta R., Cassenti A., Sonetto C., et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 188.Isella C., Terrasi A., Bellomo S.E., Petti C., Galatola G., Muratore A., Mellano A., Senetta R., Cassenti A., Sonetto C., et al. Corrigendum: stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2016;48:1296. doi: 10.1038/ng1016-1296d. [DOI] [PubMed] [Google Scholar]
- 189.Park J.H., Zhao M., Oshiro H., Miyake K., Higuchi T., Reynoso J., Razmjooei S., Bouvet M., Clary B., Zhang Z., et al. Peritoneal metastases in a patient-derived orthotopic xenograft (PDOX) model of colon cancer imaged non-invasively via red fluorescent protein labeled stromal cells. Anticancer Res. 2019;39:3463–3467. doi: 10.21873/anticanres.13492. [DOI] [PubMed] [Google Scholar]
- 190.Nishino T., Ni J., Devuyst O. Transgenic mouse models. Perit. Dial. Int. 2007;27:625–633. [PubMed] [Google Scholar]
- 191.Kobaek-Larsen M., Thorup I., Diederichsen A., Fenger C., Hoitinga M.R. Review of colorectal cancer and its metastases in rodent models: comparative aspects with those in humans. Comp. Med. 2000;50:16–26. [PubMed] [Google Scholar]
- 192.Bruce W.R., Meeker B.E., Valeriote F.A. Comparison of the sensitivity of normal hematopoietic and transplanted lymphoma colony-forming cells to chemotherapeutic agents administered in vivo. J. Natl. Cancer Inst. 1966;37:233–245. [PubMed] [Google Scholar]
- 193.Delgado-Hernández R., Hernández-Balmaseda I., Rodeiro-Guerra I., Cesar Rodriguez Gonzalez J., De Wever O., Logie E., Declerck K., Pérez-Novo C., Vanden Berghe W. Anti-angiogenic effects of mangiferin and mechanism of action in metastatic melanoma. Melanoma Res. 2020;30:39–51. doi: 10.1097/CMR.0000000000000647. [DOI] [PubMed] [Google Scholar]
- 194.Huynh J., Baloyan D., Chisanga D., Shi W., O'Brien M., Afshar-Sterle S., Alorro M., Pang L., Williams D.S., Parslow A.C., et al. Host IL11 signaling suppresses CD4(+) T cell-mediated antitumor responses to colon cancer in mice. Cancer Immunol. Res. 2021;9:735–747. doi: 10.1158/2326-6066.CIR-19-1023. [DOI] [PubMed] [Google Scholar]
- 195.Greene H.S. The heterologous transplantation of mouse tumors induced in vitro. Cancer Res. 1946;6:396–402. [PubMed] [Google Scholar]
- 196.Hoffman R.M. Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest. N. Drugs. 1999;17:343–359. doi: 10.1023/a:1006326203858. [DOI] [PubMed] [Google Scholar]
- 197.Yang Y.S., Liu C.Y., Wen D., Gao D.Z., Lin S., He H.F., Zhao X.F. Recent advances in the development of transplanted colorectal cancer mouse models. Transl. Res. 2022;249:128–143. doi: 10.1016/j.trsl.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 198.Roper J., Tammela T., Akkad A., Almeqdadi M., Santos S.B., Jacks T., Yilmaz Ö.H. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat. Protoc. 2018;13:217–234. doi: 10.1038/nprot.2017.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Frampas E., Maurel C., Thedrez P., Remaud-Le Saëc P., Faivre-Chauvet A., Barbet J. The intraportal injection model for liver metastasis: advantages of associated bioluminescence to assess tumor growth and influences on tumor uptake of radiolabeled anti-carcinoembryonic antigen antibody. Nucl. Med. Commun. 2011;32:147–154. doi: 10.1097/MNM.0b013e328341b268. [DOI] [PubMed] [Google Scholar]
- 200.Schuh J.C.L. Trials, tribulations, and trends in tumor modeling in mice. Toxicol. Pathol. 2004;32(Suppl 1):53–66. doi: 10.1080/01926230490424770. [DOI] [PubMed] [Google Scholar]
- 201.Hoffman R.M. Patient-derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer. 2015;15:451–452. doi: 10.1038/nrc3972. [DOI] [PubMed] [Google Scholar]
- 202.Zhang Z., Hu K., Miyake K., Kiyuna T., Oshiro H., Wangsiricharoen S., Kawaguchi K., Higuchi T., Razmjooei S., Miyake M., et al. A novel patient-derived orthotopic xenograft (PDOX) mouse model of highly-aggressive liver metastasis for identification of candidate effective drug-combinations. Sci. Rep. 2020;10:20105. doi: 10.1038/s41598-020-76708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Richmond A., Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis. Model. Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhao X., Li L., Starr T.K., Subramanian S. Tumor location impacts immune response in mouse models of colon cancer. Oncotarget. 2017;8:54775–54787. doi: 10.18632/oncotarget.18423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Barbáchano A., Fernández-Barral A., Bustamante-Madrid P., Prieto I., Rodríguez-Salas N., Larriba M.J., Muñoz A. Organoids and colorectal cancer. Cancers. 2021;13:2657. doi: 10.3390/cancers13112657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yamamoto J., Sugisawa N., Hamada K., Nishino H., Miyake K., Matsuyama R., Inubushi S., Tanino H., Bouvet M., Endo I., Hoffman R.M. A universal gelfoam 3-D histoculture method to establish patient-derived cancer cells (3D-PDCC) without fibroblasts from patient-derived xenografts. Anticancer Res. 2020;40:6765–6768. doi: 10.21873/anticanres.14699. [DOI] [PubMed] [Google Scholar]
- 207.Hoffman R.M. 3D sponge-matrix histoculture: an overview. Methods Mol. Biol. 2018;1760:11–17. doi: 10.1007/978-1-4939-7745-1_2. [DOI] [PubMed] [Google Scholar]
- 208.Orkin R.W., Gehron P., McGoodwin E.B., Martin G.R., Valentine T., Swarm R. A murine tumor producing a matrix of basement membrane. J. Exp. Med. 1977;145:204–220. doi: 10.1084/jem.145.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Weaver V.M., Petersen O.W., Wang F., Larabell C.A., Briand P., Damsky C., Bissell M.J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Klemke L., Blume J.P., De Oliveira T., Schulz-Heddergott R. Preparation and cultivation of colonic and small intestinal murine organoids including analysis of gene expression and organoid viability. Bio. Protoc. 2022;12:e4298. doi: 10.21769/BioProtoc.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Guo J., Verma U.N., Gaynor R.B., Frenkel E.P., Becerra C.R. Enhanced chemosensitivity to irinotecan by RNA interference-mediated down-regulation of the nuclear factor-kappaB p65 subunit. Clin. Cancer Res. 2004;10:3333–3341. doi: 10.1158/1078-0432.CCR-03-0366. [DOI] [PubMed] [Google Scholar]
- 212.De Angelis M.L., Francescangeli F., Nicolazzo C., Xhelili E., La Torre F., Colace L., Bruselles A., Macchia D., Vitale S., Gazzaniga P., et al. An orthotopic patient-derived xenograft (PDX) model allows the analysis of metastasis-associated features in colorectal cancer. Front. Oncol. 2022;12:869485. doi: 10.3389/fonc.2022.869485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 214.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 215.DeHaan R.K., Sarvestani S.K., Huang E.H. Organoid models of colorectal pathology: do they hold the key to personalized medicine? A systematic review. Dis. Colon Rectum. 2020;63:1559–1569. doi: 10.1097/DCR.0000000000001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Roper J., Tammela T., Cetinbas N.M., Akkad A., Roghanian A., Rickelt S., Almeqdadi M., Wu K., Oberli M.A., Sánchez-Rivera F., et al. Corrigendum: in vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 2017;35:1211. doi: 10.1038/nbt1217-1211a. [DOI] [PubMed] [Google Scholar]