Abstract
Plasmids are the primary vector for horizontal transfer of antimicrobial resistance (AMR) within bacterial populations. We applied the MOB-suite, a toolset for reconstructing and typing plasmids, to 150 767 publicly available Salmonella whole-genome sequencing samples covering 1204 distinct serovars to produce a large-scale population survey of plasmids based on the MOB-suite plasmid nomenclature. Reconstruction yielded 183 017 plasmids representing 1044 primary MOB-clusters and 830 potentially novel MOB-clusters. Replicon and relaxase typing were able to type 83.4 and 58 % of plasmids, respectively, compared to 99.9 % for MOB-clusters. Within this work, we developed an approach to characterize the horizonal transfer of MOB-clusters and AMR genes across different serotypes, as well as the diversity of MOB-cluster associations with AMR genes. Aggregating conjugative mobility predictions provided by the MOB-suite and their corresponding serovar entropy demonstrated that non-mobilizable plasmids were associated with fewer serotypes compared to mobilizable or conjugative MOB-clusters. The host-range predictions for MOB-clusters also showed differences between the mobility classes, with mobilizable MOB-clusters accounting for 88.3 % of the multi-phyla (broad-host-range) predictions compared to 3 and 8.6 % for conjugative and non-mobilizable, respectively. A total of 296 (22 %) of identified MOB-clusters were associated with at least one resistance gene, indicating that the majority of Salmonella plasmids are not involved in AMR dissemination. Shannon entropy analysis of horizontal transfer of AMR genes across serovars and MOB-clusters demonstrated that horizonal transfer of genes is higher between serovars compared to transfer between different MOB-clusters. In addition to the population structure characterization based on primary MOB-clusters, we characterized a multi-plasmid outbreak responsible for the global dissemination of bla CMY-2 across different serotypes using higher resolution MOB-suite secondary cluster codes. The plasmid characterization approach developed here can be applied to different organisms to identify plasmids and genes which pose high risks for horizontal transfer.
Keywords: antimicrobial resistance, horizontal transfer, mobile genetic elements, plasmids, Salmonella
Data Summary
Scripts used in this study can be accessed on GitHub at https://github.com/phac-nml/plasmid_analysis. Supplementary material can be accessed through Zenodo at https://doi.org/10.5281/zenodo.6617143. The MOB-suite database is accessible through Zenodo at https://doi.org/10.5281/zenodo.3786915. Supplementary Tables and Supplementary Data have been uploaded to Figshare: https://doi.org/10.6084/m9.figshare.22194235.v1 [1].
Impact Statement.
There is a resurgence of interest in plasmids due to their role in disseminating antimicrobial resistance (AMR), which has been facilitated by advances in long-read sequencing technology and tools such as MOB-suite. Publicly available whole-genome sequencing data for pathogens provides an incredible resource for examining the population-level dynamics of plasmids. A broad characterization of the plasmids circulating in Salmonella and their associations with known AMR genes is presented here. The analytical approach presented in this work represents a novel framework for categorizing plasmids into risk categories based on their associations with AMR genes and population distribution.
Introduction
Salmonella is a frequent foodborne pathogen that typically results in self-limiting enteric illness, but it can result in severe complications and death in vulnerable populations [2–4]. Antimicrobial resistance (AMR) in pathogens such as Salmonella is considered a serious concern to public health due to its increasing prevalence [5]. It is estimated that 16 % of Salmonella cases in the USA are resistant to at least one essential antibiotic [6]. Bacteria can become resistant to antibiotics through spontaneous mutations or by acquiring mobile genetic elements (MGEs) that contain genes that confer resistance to antimicrobials. MGEs such as plasmids and integrative conjugative elements (ICEs) are common vectors for the horizontal transfer of AMR genes [7, 8]. It is now possible to gain an unprecedented population view of the prevalence of AMR and compositions of different pathogen subtypes due to routine whole-genome sequencing by public-health laboratories around the world. However, population-level plasmid characterization efforts have been hampered by the lack of tools capable of extracting plasmid contigs from assemblies and reconstructing the individual plasmids in the sample [9]. Identification and typing of plasmid sequences are critical to understanding AMR transmission, since plasmids are the primary vectors for AMR dissemination in Enterobacteriaceae, including priority pathogens such as Salmonella , Escherichia coli and Klebsiella [10].
Plasmids are autonomously replicating MGEs that can provide advantageous traits that promote their host cells' survival and are widely distributed in diverse bacterial species. Plasmids employ multiple different replication mechanisms and are highly variable in gene content, and it is well known that there are no universal plasmid marker sequences [11–16]. Most plasmids are circular, but linear plasmids have been described within Gram-positive and some Gram-negative bacteria, such as Salmonella [11–14]. Multiple marker-based methods have been developed for plasmid classification, with replicon typing and relaxase typing being the two most established methods [11, 12, 17, 18]. These approaches provide valuable evolutionary insights into the distribution of important plasmid types within bacterial populations and associations with traits such as AMR and virulence [19]. Replicon and relaxase typing success rates range from 56–85 % and 43–65 %, depending on the analysed dataset [12, 20, 21]. Due to the limited resolution provided by both replicon and relaxase typing, higher resolution sub-typing methods such as multilocus sequence typing (MLST) have been developed for several important plasmid groups, including IncA/IncC [22], Inc11 [23] and IncN [24], to facilitate epidemiological investigations of plasmid horizontal dissemination in outbreak contexts. The considerable variability in both the complement and phylogenetic concordance of genes within plasmid types complicates the development of higher-resolution typing methods based on adding more loci [19, 25, 26]. An analysis of the shared gene content of plasmids belonging to the same relaxase type showed that there was poor conservation across the diversity of all plasmids within a single type, demonstrating that they are poorly related [15]. All of these marker-based approaches will miss novel plasmids and require experts to manually curate and designate new types, as well as requiring bespoke creation of new MLST schemes for each plasmid type of interest [19].
To address the limitations of the current marker-based typing systems, there has been development of novel and complementary approaches for plasmid classification systems using network [16, 27, 28] or tree-based analysis [9, 21]. An exhaustive overview of the analytical approaches to develop plasmid nomenclature is outside the scope of this work; however, it is important to highlight two recent large-scale network-based approaches that utilized large datasets of ~10 000 complete plasmids. The number of plasmids classified into groups varied significantly between the studies, which ranged from 3725 (35 %) [28] to 5371 (51 %) [27] of the dataset. Similarly, the number of groups identified ranged from 276 [28] to 561 [27], indicating that the methodological approach and parameters will significantly influence the classification outcome. Plasmids can be assigned into the known plasmid taxonomic units (pTU) identified by Redondo-Salvo et al. [28] using the tool copla [28]. In contrast, MOB-suite employs a tree-based, complete-linkage clustering approach to partition the complete plasmid sequences into taxonomic units termed MOB-clusters based on empirically derived Mash [29] distance thresholds [9, 21]. To facilitate short-term outbreak analysis and broader plasmid surveillance, the MOB-suite implements a multi-level nomenclature for plasmid typing consisting of two distance thresholds that cluster plasmids into groups based on empirically derived Mash distances [29] of the entire plasmid sequence [9, 21]. Mash distances correlate with average nucleotide identity (ANI) for genomes with >90 % ANI and simultaneously capture point mutations and gene content differences [29], and our previous work demonstrated that Mash distances provided higher overall cluster concordance with replicon and relaxase typing compared to ANI. MOB-clusters are formed by the selection of a distance threshold, and partitioning all of the input data into a unique cluster code and so, by design, 100 % of the 17 779 unique sequences used to establish a database were assigned a MOB-cluster code [9, 21]. The primary MOB-cluster threshold is designed for surveillance and reconstruction of plasmids, and was selected using a multi-optimization approach that ensures that no two plasmids with the same cluster designation will co-occur in the same cell, as well as to maximize congruences with existing replicon and relaxase typing approaches and favour larger cluster sizes. The secondary MOB-cluster threshold was selected to identify plasmids that are near duplicates, and was derived from an examination of the Mash distances between complete and draft assemblies of plasmids from the same sample [9, 21].
Both genetic and epidemiological data are necessary to link pathogens or plasmids to transmission events with the genetic data used to narrow the search space for epidemiological investigations rather than defining outbreaks. It is common practice in the analysis of pathogen epidemiology to establish thresholds of genetic relatedness where more distant samples can be ‘ruled out’ as part of a potential transmission chain [30–32]. Similarly, plasmids assigned to different primary MOB-clusters are sufficiently unrelated to not be considered part of an epidemiologically relevant transmission event. Plasmids that share the same primary cluster designation require higher resolution typing to determine whether they have sufficient similarity to be considered part of a recent transfer. The secondary MOB-cluster designation is higher resolution, and two plasmids assigned the same ID share high sequence similarity and are strong candidates for outbreak investigation. For example, nosocomial plasmid transmission may be suspected if two patients in the same ward of a hospital are colonized during their stay with two different pathogen species that both carry a plasmid assigned to the same secondary MOB-cluster, providing evidence for a potential plasmid outbreak within the hospital. Conversely, a plasmid outbreak can be ruled out if the plasmids belonged to different primary clusters.
The ability of a plasmid to establish itself in different hosts depends on a complex interplay between a variety of plasmid and host factors, such as restriction-modification systems [33–35]. In the context of this work, the replicative plasmid host range (host range) is defined as the taxonomic breadth in which a plasmid could potentially replicate. The MOB-suite uses three features to predict potential replicative host ranges of plasmids: the replicon and relaxase marker sequences, along with MOB-cluster codes in a database of closed plasmid sequences [21]. The host range of a plasmid is described in terms of the taxonomic rank and name of the lowest common ancestor for all three features. Previous analysis of closed plasmids showed that MOB-clusters were usually restricted to a single species, but is likely an underestimate the true plasmid host range [21]. Horizontal transmission of plasmids through bacterial populations often occurs through conjugation, where direct transfer of intact plasmids occurs between donor and recipient cells [36]; however, naturally competent bacteria can also be transformed through their uptake of free plasmid DNA. Plasmids can be classified into three mobility classes based on their conjugative potential: conjugative, mobilizable and non-mobilizable [9, 37]. A conjugative plasmid is considered to be the most transmissible type of plasmid since it encodes the complete set of features needed for transfer: an origin of transfer (oriT), a DNA relaxase, a type IV coupling protein (T4CP) and a type IV secretion system (T4SS) [37, 38]. Mobilizable plasmids are expected to be less transmissible due to missing one or more of these features, which must be provided externally. Non-transmissible plasmids are restricted in their transfer rate since they are missing the necessary oriT sequence needed to utilize the conjugative apparatus and, therefore, rely on transformation or transduction [39, 40] to propagate in new hosts.
There are a plethora of alpha-diversity measures to characterize genetic and taxonomic biodiversity with Shannon entropy being a commonly used measure in ecology that is based on the number of taxa and their relative abundance within a community that can provide insights into the diversity and dynamics of biological communities [41–43]. Within a biodiversity framework, Shannon entropy is used as a measure of the uncertainty in the identity of an individual drawn from a community at random [41–43]. The community composition with a high Shannon entropy will have a greater number of taxa with similar abundances [41–43]. In contrast, a community with a low Shannon entropy will have fewer distinct taxa and wider range of abundances [41–43]. Conceptually, Shannon entropy can be used to measure the distributions of genes and plasmids within a population to quantify the serovar diversity that they occur in. In a similar manner, Shannon entropy can be used to measure the diversity of plasmids that individual genes are associated with. Within this work, we utilize Shannon entropy as a novel measure of horizontal transfer of genes and plasmids as their occurrence in different genetic contexts indicate a violation of vertical inheritance.
We previously developed the MOB-suite, which performs scalable reconstruction (MOB-recon) and typing (MOB-typer) of plasmids from draft and complete whole-genome sequencing assemblies [9]. Using a large public dataset of Salmonella Illumina read sets, we characterize the plasmidome and resistome within a single pathogen at an unprecedented level by establishing the consistency of this approach with existing literature and highlighting novel findings. An analytical approached based on Shannon entropy is developed here to measure the degree of horizontal transfer of genes and plasmids within the Salmonella population. Shannon entropy captured signals reflecting the conjugation mobility predictions of the MOB-suite, which demonstrates that an understanding of the conjugative potential of a plasmid provides insights regarding their distribution within the Salmonella population. The analytical process presented here can be extended to other pathogens as a starting point toward a risk-based prioritization framework for AMR plasmids. Using known examples of important AMR plasmids, we have tuned parameters for identifying priority MOB-clusters for further investigation based on plasmids known to be important in AMR dissemination.
Methods
Sequence analysis of genomes
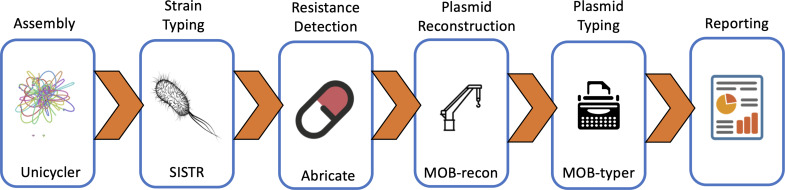
An overview of the entire workflow is presented in Fig. 1. Sample metadata of available Illumina paired-end Salmonella was retrieved from the National Center for Biotechnology Information (NCBI) (n=150 767) (download date October 18 2019) and the reads were downloaded from the SRA (Sequence Read Archive) and assembled with Unicycler v. 4.4 using the default parameters (normal mode) [44]. Unicycler includes detection of circular contigs and an iterative Pilon [45] based polishing step to remove variants in the assembly not supported by the original reads [44]. Serovar predictions were determined from the assemblies using sistr v. 1.1.0 with the default parameters, and genomes with fewer than 290 of the 330 core-genome loci used for serovar prediction were excluded from further analysis [46]. Resistance genes were identified in the assemblies using ABRicate v. 1.0.0 [47] and the NCBI resistance gene database v. 2020-01-06.1 [48] using a coverage and identity threshold of 95 %. MOB-suite v. 3.0.0 with database v. 2.0 [9] using the default parameters (80 % coverage and identity of sequence queries) was used to predict, reconstruct, type and predict the host range of plasmids from the draft Unicycler assemblies using a genome masking database of all closed Salmonella chromosomes (Table S1). This feature of the MOB-recon identifies reference genomes that match closely to the assembly using a Mash [29] distance of 0.02 and then labels any contigs with ≥80 % coverage and identity as chromosomal sequences. This results in selective depletion of plasmids that have integrated within specific lineages but are also autonomously replicating plasmids elsewhere. The sanitized and standardized sample metadata, along with read statistics for all assemblies that passed quality control, are available in Table S2.
Fig. 1.
Overall plasmid analytical workflow.
Identification of closed plasmids and MOB database update
From the 150 767 assemblies, the potentially complete plasmid contigs were identified from the first round of MOB-recon reports by selecting contigs flagged by Unicycler as circular and labelled as a plasmid by MOB-suite (i.e. passed the chromosome-masking step). This initial set was then filtered to remove any contigs with a coverage depth less than 0.5× of the chromosome, a length of less than 1.5 kb, or that were missing both a replicon and relaxase sequence. A total of 70 604 circular plasmid contigs that met these criteria and also contained either a replicon or relaxase sequence were identified (Table S3). Duplicate sequences were identified using the md5 hash of the sequences, and a single representative was retained for each set of duplicates. cd-hit-est v. 4.8.1 [49] (minimum identity 95%, minimum coverage of the smaller sequence 95 %) was used to identify near-duplicate sequences and variants that were simply truncated versions of other sequences. These deduplicated candidate sequences were queried against a custom blast v. 2.10.1 database of NCBI RefSeq prophage sequences. Any sequences that matched the database with ≥60 % coverage and ≥80 % identity were filtered out. Remaining sequences that passed all of these checks were added to the MOB-suite database, and the input assemblies were re-analysed in a second pass with the MOB-suite to reflect the newly added sequences.
Analysis of plasmid and resistance gene distributions
MOB-typer and MOB-recon reports were aggregated for the complete set of samples, and entropy analysis was performed using the Python code available at https://github.com/phac-nml/plasmid_analysis. The Shannon entropy calculations were performed using the SciPy stats library using the formula S = -Σ i Pi lnPi where Pi is the probability of a MOB-cluster or gene occurring in a serovar, or gene occurring in a MOB-cluster (Equation 1). Interactive visualizations of the results were produced using Plotly (https://github.com/plotly/plotly.py) with HTML versions of all figures available within the GitHub repository.
Results and Discussion
Plasmid distribution and diversity in Salmonella
To our knowledge, the current study is the largest population-level analysis of plasmids to date with 150 767 publicly available Salmonella isolates from 1204 distinct serovars predicted by sistr. From this data set, a total of 184 117 plasmids were identified representing 1044 MOB-clusters where sizes ranged from 0.9 to 379 kb with a mean of 50 kb. The MOB-suite database has good representation of plasmids that are circulating in Salmonella with only 1100 (0.6 %) reconstructed plasmids that could not be assigned to an existing MOB-cluster (Table S4). These unassigned plasmids were excluded from further analysis since long-read sequencing is required to validate them. Current estimates of plasmid diversity based on complete plasmids in the NCBI databases are serious underestimates with only 211 of the 7652 MOB-clusters previously known to occur in Salmonella . The power of population-level analysis is highlighted by our observed fivefold increase in known plasmid diversity within Salmonella where 1044 MOB-clusters were identified compared to the 211 observed in closed genomes. Most MOB-clusters (82 %) occur at a low frequency in the dataset with ≤100 members (Table S4) and in the absence of selection could represent ephemeral plasmid content that is destined to be lost, as has been observed in prolonged carriage of Salmonella enterica Typhimurium, and in laboratory conditions [21, 50–52].
Plasmid carriage was highly variable across serotypes with means ranging from 17–99 % within serotypes that had >100 members and a dataset mean of 65 % (Table S5). Similarly, the mean number of plasmids per isolate varied across serotypes ranging from 1.0 to 3.77 with a mean of 1.88. The dataset was heavily biased to a few highly predominant serovars, with the top five serovars accounting for 45 % of the entire set of samples (Table S5). Interestingly, there also appeared to be significant differences in carriage rates within a serovar since biphasic Typhimurium had a carriage rate of 80%, but the monophasic variant (I 1,4,[5],12:i-) had only a 60 % carriage rate (Table S5). These differences may be due to the adaptations of different lineages, stochastic effects based on habitat and opportunities for plasmid acquisition, or the differential presence of restriction-modification systems [53]. The observed plasmid carriage rates are consistent with previous work, which has shown that there are stable virulence plasmids that are inherited vertically with high prevalence in non-typhoidal serovars such as S . enterica Enteritidis, S . enterica Typhimurium and S . enterica Heidelberg [54–56]. The human restricted and closely related typhoidal serovars S . enterica Typhi and S . enterica Paratyphi A show similar carriage rates (32–35%), and this slightly lower than average plasmid prevalence may be due to their host specialization and selective pressure to maintain streamlined genomes [57, 58]. Establishing causative relationships between serovars and rates of plasmid carriage is beyond the scope of this work but differential presence of restriction modification systems [59] and CRISPR arrays [60, 61] across serotypes may yield insights into the reasons behind the observed patterns. These results are in agreement with previous work based on different analytical approaches and datasets, supporting the robustness and epidemiologically relevance of the plasmid distributions captured by our large-scale population-level results.
A total of 139 unique replicon (n=2686) and 237 relaxase (n=2050) query sequences from the MOB-suite database were identified in the reconstructed plasmids, indicating that only a small number of the query sequences are relevant within Salmonella . Consistent with previous studies of complete NCBI plasmids [19, 20], 16 % of reconstructed plasmids were missing a replicon, 42 % were missing a relaxase and 10 % were untypable by both methods (Fig. S1). With a total of 572 unique combinations, utilizing replicon typing information for epidemiological tracking of plasmids is complicated due to the presence of multi-replicon plasmids that comprise 45 % of reconstructed plasmids and subsequent ambiguity as to whether they should be treated as distinct plasmids. A total of 43 unique relaxase marker combinations was observed with 50 % of plasmids possessing a single relaxase. In general, there is usually one highly prevalent relaxase type associated with a given replicon, which is consistent with previous studies on closed plasmids [10, 12] (Fig. S1). Of the nine described relaxase types (MOBB, MOBC, MOBF, MOBH, MOBP, MOBL, MOBQ, MOBV, MOBT) [62], six were identified in the dataset with MOBV being the only relaxase type which was predominantly associated with MOB-clusters that did not have a known replicon associated with them (Table S6).
IncFII and IncFIB are the two most abundant replicon types in the dataset and a summary of the abundance of each of the replicon types and their associations with relaxase families is presented in Table S6. The other major IncF family replicons, IncFIA and IncFIC, were significantly rarer than IncFII and IncFIB (Table S6). Salmonella virulence plasmids belonging to the IncF replicon family are some of the best characterized and abundant plasmids within the genus, and it has been observed that their inheritance is primarily vertical [54, 55, 63]. The conjugative S . enterica Typhimurium virulence plasmid [54, 55] (AB460; IncFIB, IncFII, MOBP, conjugative) and the non-mobilizable S . enterica Enteritidis virulence plasmid [56] (AB461; IncFIB, IncFII, N/A, non-mobilizable) are the two most abundant plasmids in the dataset and significantly contribute to the prevalence of IncFII and IncFIB replicons in the dataset. With 99 % typing success, primary MOB-clusters provide more comprehensive coverage of plasmids and indicate that there is good representation of Salmonella plasmid diversity in the current MOB-suite database.
Identification of closed plasmid sequences from draft assemblies
Complete plasmid contigs were identified based on the circularity flag from Unicycler, and their sizes ranged from 1.5 to 279 kb with a median of 4.5 kb. It is known that short-reads can fully resolve plasmids as long as they do not have repeats that exceed the insert size of the library [44]; consequently, the complete plasmids identified from Illumina-only data tend to be small and without AMR genes as these genes are frequently associated with repeats [64]. The recovered complete plasmids in this study indeed tended to be small with 60 % ≤5 kb, but it was possible to have large plasmids >100 kb completely assembled with just Illumina data (Table S4). Exact sequence duplicates were removed to yield a dataset of 57 118 unique plasmids. Highly similar sequences and subsequences were deduplicated by cd-hit to produce 12 864 unique plasmids. We identified 751 distinct primary MOB-clusters and 16 novel primary MOB-clusters in the dataset of closed plasmids, with the vast majority being variations of members of existing primary MOB-clusters with novel secondary clusters. The set of closed plasmid sequences was used to expand the distribution of MOB-clusters in the database and the sequences were added to the sequence database where there was not an existing member assigned to the secondary MOB-cluster, and the full set of circular plasmid sequences identified in this work is available in the Supplementary Data. Our circular plasmid analysis demonstrates that there may be a large number of complete plasmids available in the public data that are not readily available to researchers since they would need to assemble large volumes of raw data, and use computational approaches to identify the circular plasmids from draft assemblies.
Serovar distributions of plasmids based on in silico mobility predictions
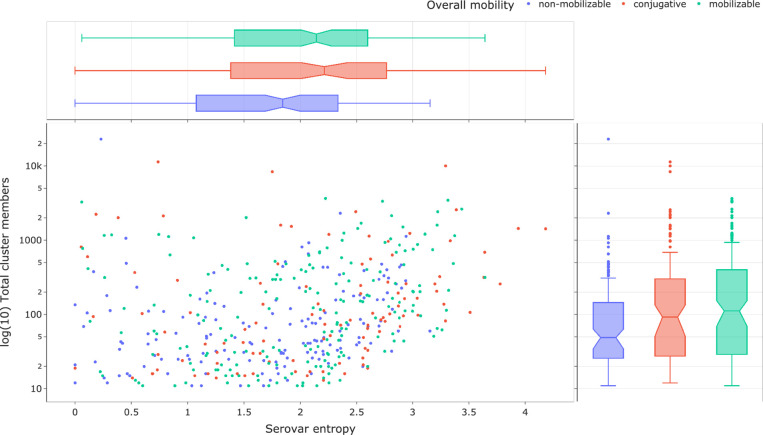
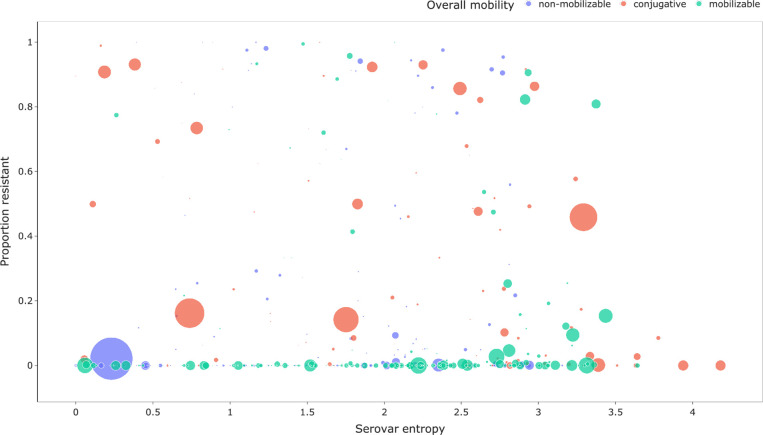
Plasmids have been studied extensively through an evolutionary perspective [61] but a novel aspect of this work is to apply ecological techniques to understand the horizontal transmission behaviour of genes and plasmids through a population using Shannon entropy, which is a commonly used measure of alpha-diversity [65–67]. To examine whether there is a relationship between conjugation mobility predictions and horizontal transfer between serotypes, we selected a total of 501 MOB-clusters that had ≥10 members and assigned them to their dominant mobility class prediction. Their corresponding serovar entropy was calculated according to Equation 1 (see Methods) where the counts of serotypes that the MOB-cluster occurred in were converted into probabilities and used as input, and resulting entropy scores were then plotted against the total frequency of the MOB-cluster (Fig. 2). MOB-suite utilizes a combination of genetic features to classify plasmids into three mobility classes: conjugative, mobilizable and non-mobilizable. Conjugative plasmids must contain both a relaxase combined with any number of mate-pair formation (MPF) proteins identified [9, 38]. Mobilizable plasmids contain a relaxase and/or a known oriT but lack a T4CP. Due to the reliance on MPF marker sequences and not evaluating a complete set of conjugative proteins, there may be some mobilizable plasmids that get classified as a conjugative. Non-mobilizable plasmids are missing both a relaxase and an oriT.
Fig. 2.
Scatter plot of individual MOB-clusters coloured based on their overall mobility class prediction. The y-axis is the log-scale number of occurrences of a MOB-cluster in the dataset and the x-axis is the serovar entropy (nats) based on sistr predictions. Box plots are presented for the marginal distributions for each axis.
Serovar entropy does not have a discernible relationship with the total number of members of a MOB-cluster, indicating that low-frequency plasmids do not have artificially low entropy due to sampling depth (Fig. 2). The median serovar entropy was determined for each of the three mobility classes, and the Kruskal–Wallis test revealed a significant difference in the entropy distribution in at least one sample (P=8.2×10−05). Post hoc Dunn analysis demonstrated a significant difference between non-mobilizable plasmids with a median of 1.8 nats to both conjugative (median=2.2 nats, P value=3.4×10−4) and mobilizable (median=2.1 nats, P value=1.2×10−3). The median entropies for conjugative and mobilizable plasmids were not significantly different, which could be the result of loss in precision based on picking the most abundant mobility classification in a MOB-cluster. However, this should only be a minor contribution since 85 % of the 501 MOB-clusters analysed had >86 % of their members assigned to a single mobility class. It is known that conjugative plasmids will frequently lose their capacity to conjugate [15] and the observed mobilizable plasmids may have lost parts of their transfer regions after conjugation into a new host to reduce fitness costs to the host. These results indicate that the conjugative mobility classifications contain information as to the behaviour of plasmids within a population. Taken together, the observed mobility class entropies fit the expectation that non-mobilizable plasmids would not undergo as much horizontal transfer as mobilizable or conjugative plasmids. Follow-up studies are necessary to understand mechanisms of transfer of high entropy non-mobilizable plasmids such as transformation and transduction [39, 40]. The obtained mobility results support the use of Shannon entropy as a measure of horizontal transfer due to its ability to capture differences in the conjugative mobility classes that fit with expectations.
Shannon entropy values are not readily interpretable in a biological context so we are using plasmids with known rates of conjugative transfer to guide interpretation. As stated previously, Salmonella virulence plasmids are known to be largely inherited vertically with little evidence for horizontal transmission [54]. The two highly abundant virulence plasmids represented by MOB-clusters AB461 (IncFIB, IncFII, N/A, non-mobilizable) (0.23 nats) and AB460 (IncFIB, IncFII, MOBP, conjugative) (0.74 nats) have low serovar entropy (≤1 nats) indicating that they predominantly found in one serovar (Fig. 2). The lower serovar entropy in the non-mobilizable MOB-cluster AB461 is consistent with experimental evidence that it is unable to conjugate due to the loss of critical elements of its transfer region [56]. In a similar manner, the slightly higher serovar entropy observed in the conjugative MOB-cluster AB460 is supported by experimental evidence that has demonstrated that it is able to undergo conjugation but at very low frequencies [55]. MOB-cluster AA474 (IncI-gamma/K1, MOBP) (3.29 nats) is an abundant conjugative plasmid with high serovar entropy (≥2.9 nats) and has been experimentally demonstrated to be conjugative and is associated with bla CMY-2 dissemination in multiple taxa [68–70]. Plasmids belonging to the IncN replicon family have been shown to have high conjugation rates [71, 72], but have also been shown to have a higher acquisition cost compared to other incompatibility groups [73], which may explain the intermediate entropy observed for AB159 (IncN, MOBF, conjugative) (1.95 nats). Interestingly, the small (~2.1 kb) MOB-cluster AC748 [Col(BS512), N/A, non-mobilizable] has high serovar entropy (≥2.9 nats) but lacks all known conjugative transfer elements and occurs in other species such as E. coli and Klebsiella in the MOB-suite database. AC748 plasmids may contain unidentified oriT sequences and be able to undergo conjugation by hijacking the machinery provided by another plasmid. Alternatively, due to the small size of the plasmid, it may be able to efficiently transfer to new hosts using extracellular vesicles [74]. The observed serovar entropies are the result of interactions between horizontal transfer rates, donor/host genetic background, fitness costs, clonal expansion, genetic drift and selection [60, 75–77].
Plasmid host range
The replicative host range of a given plasmid is the result of a complex interplay between the plasmid and its potential hosts, and depends on both the capacity for transfer and the ability to establish itself successfully. A summary of the host-range rank predicted for the reconstructed plasmids is presented in Fig. S2. MOB-suite uses a lowest-common ancestor approach based on replicon, relaxase and MOB-cluster distributions based on plasmid host information from its internal database of closed plasmids. A total of 155 172 (84 %) of the reconstructed plasmids representing 629 (74 %) of identified MOB-clusters were predicted to have a host range of Enterobacterales . This result is supported by a meta-analysis which demonstrated that plasmid transfer occurs at higher frequencies between genetically similar organisms [73]. Plasmids predicted to replicate in multiple phyla represented a much smaller number of plasmids with 12 775 (7 %) of reconstructed plasmids representing 191 (18 %) of the identified MOB-clusters. Intriguingly, 11 284 (88 %) of these broad-host-range plasmids were mobilizable which means that these plasmids would depend on trans-acting factors to transfer them or that they lost their conjugation ability after transfer [15]. A total of 198 (19 %) non-mobilizable MOB-clusters representing 35 440 (19 %) of reconstructed plasmids were predicted to have a host range restricted to Salmonella . It is not possible from these results to determine whether these plasmids truly can only replicate in Salmonella , or if they simply are restricted to the species due to lack of transfer capacity or in-depth sampling of other taxa. As mentioned previously, we identified 211 MOB-clusters where Salmonella was not previously known to be a host and of these MOB-clusters 139 of them were predicted to have host-range predictions restricted to a single genus. Many of these MOB-clusters have family-level host-range predictions of Enterobacteriaceae with the inclusion of Salmonella as a host. However, there are several MOB-clusters that are now assigned to multiple phyla with the inclusion of Salmonella as a host. The host-range analysis highlights that the majority of plasmids identified in Salmonella would have the capacity to replicate in other clinically relevant pathogens, and may also be transferred between commensal and pathogenic bacteria. Furthermore, the population-based analysis has increased knowledge of the potential host ranges of plasmids.
Resistance gene associations
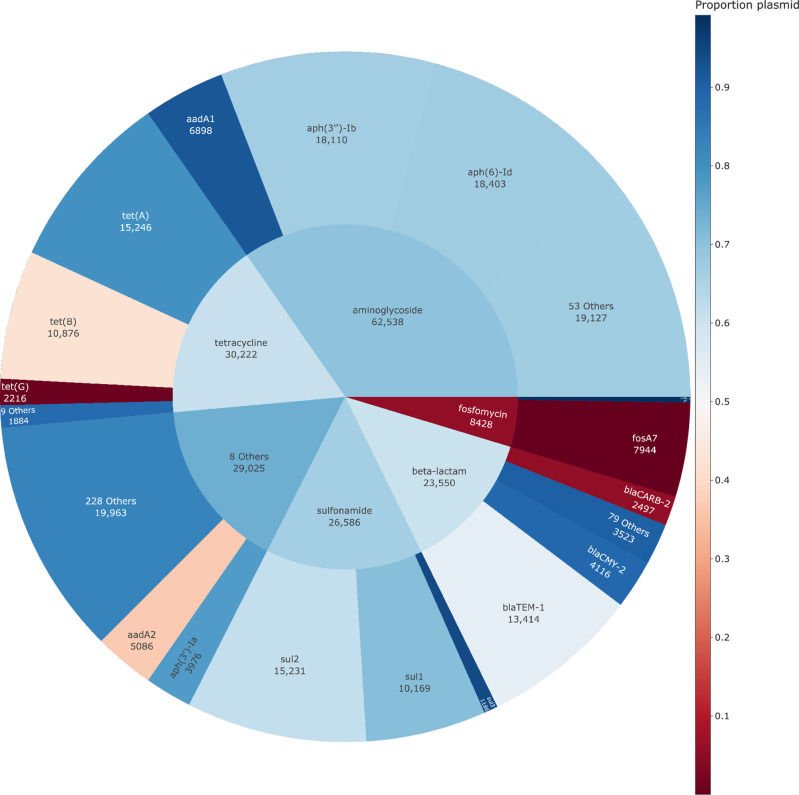
Surveillance of AMR focuses heavily on cataloguing the abundance and distribution of known AMR genes or mutations associated with resistance to specific pharmaceutical drug classes, but understanding their genomic context and associations with plasmids is critical to understanding their horizontal transmission risk. Using the NCBI AMR database of known acquired resistance genes, we examined the association of specific resistance genes with respect to the genomic context and aggregated them according to their drug classes. A sunburst plot is presented in (Fig. 3), where the prevalence of individual genes and their genomic context is presented. As expected, the majority of the acquired resistance genes from the database are predominantly associated with plasmid sequences (64 %) (Fig. 3). Aminoglycoside-resistance genes are highly abundant in Salmonella and the two most abundant genes, aph(6)-Id (67 %) and aph(3’’)-Ib (67 %), are primarily associated with plasmids (Fig. 3). Fosfomycin is the only drug class to be nearly exclusively associated with the chromosome, and this is in large part due to the fosA7 gene (Fig. 3), which is frequently identified in S . enterica Heidelberg where it was originally described [78]. Tetracycline (81 %) and β-lactam (60 %) resistance is mediated largely through plasmids but, notably, tet(G) and blaCarb-2 are heavily associated with the chromosome (Fig. 3) and these resistance elements are localized within the chromosomally integrated Salmonella genomic island 1 (SGI-1) [79]. The results presented here highlight that there is considerable heterogeneity in the genomic context of NCBI resistance genes and is the largest characterization of their genomic context within a single pathogen to date. Characterizing gene association with chromosomal or plasmid sequences provides an initial basis for informing the potential horizontal transmission risk for a given resistance element.
Fig. 3.
Sunburst plot of individual NCBI resistance genes grouped by drug class and coloured based on the proportion of occurrences of the gene associated with the chromosome or a plasmid.
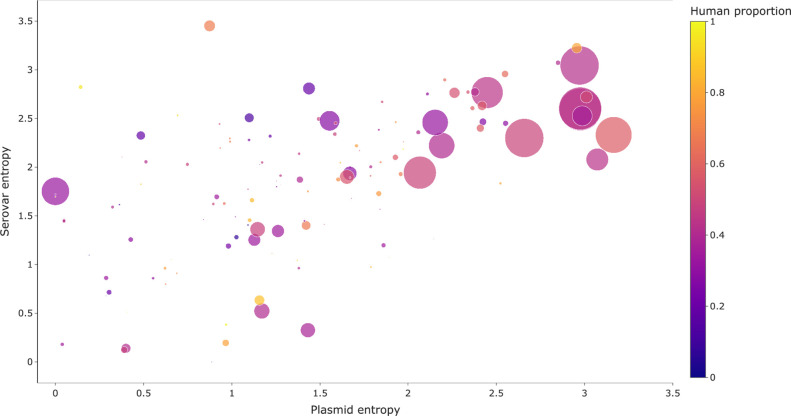
Resistance gene serovar and plasmid distributions
Out of the 183 017 reconstructed plasmids, 22 % contained at least one resistance gene. These resistance-gene-containing plasmids comprised 296 MOB-clusters (26 % of the total number of MOB-clusters observed in this study). This result shows that the majority of plasmids in Salmonella are not involved in AMR, which is notable since it means that addressing AMR transmission requires focusing on a much smaller subset of the circulating plasmids. Using a gene-centric approach, the MOB-cluster and serovar entropies were plotted to explore the relationship between these two entropies (Fig. 4). The examined resistance genes had a median MOB-cluster entropy of 1.38 nats and a median serovar entropy of 1.98 nats, which indicates there is greater uncertainty in the serovar identity compared to a MOB-cluster. This observation indicates that it is a rarer event for a gene to change its local genetic context through recombination or transposition compared to plasmid-mediated horizontal transfer. There is a positive relationship between serovar and MOB-cluster entropy and the more abundant genes have high entropies for both (Fig. 4). Genes present in a variety of MOB-clusters are likely associated with active cis-acting MGEs, such as insertion sequences or transposable elements, and so these genes are likely to have higher entropy since they can be mobilized through multiple mechanisms and are more likely spread to different serovars. For example, bla TEM-1 (n=13 423) has a MOB-cluster entropy of 3.16 nats and a serovar entropy of 2.33 nats and has been shown in the literature to be associated with IS26 [80]. The predominantly human-associated fluoroquinolone-resistance gene qnrSr1 (n=1057) has the highest combined entropies, with a MOB-cluster entropy of 2.96 nats and a serovar entropy of 3.22. The observed lack of non-human sources in the dataset for qnrS1 is consistent with the literature [6], and so further sampling of different commodities and sources may be necessary to better characterize the epidemiology of this gene.
Fig. 4.
Relationship between serovar and MOB-cluster entropy (nats) for individual NCBI-curated resistance genes. The size of each circle represents the abundance of the gene in the dataset and colours indicate the percentage of samples associated with human sources.
bla CMY-2 multi-plasmid outbreak analysis
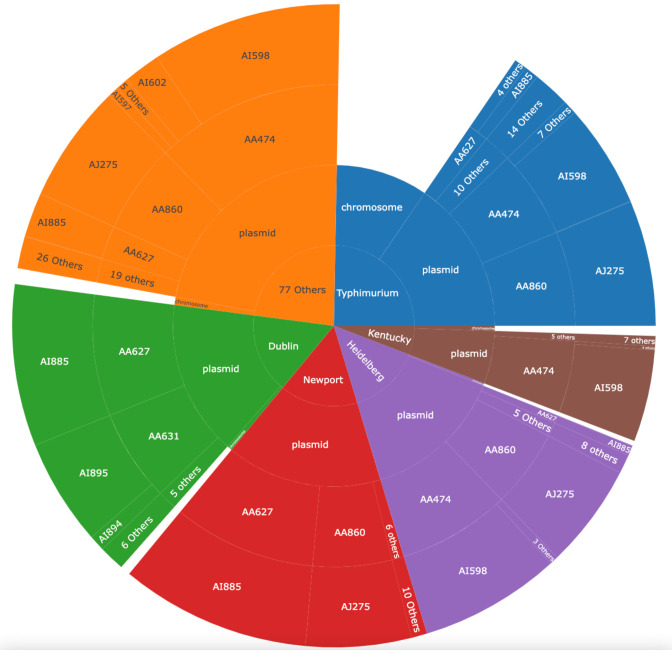
The β-lactamase gene bla CMY-2 provides resistance to third-generation cephalosporins and is one of the best-characterized examples where the removal of the antibiotic from animal production in Canada resulted in a dramatic reduction in resistance [7]. A total of 73 % of observed bla CMY-2 (n=4233) are found in non-human sources across 84 serovars (2.5 nats) and associated with 10 primary MOB-clusters (1.6 nats) (Fig. 5). Five serovars account for 77 % of all bla CMY-2 samples, with S . enterica Typhimurium being the most abundant (Fig. 5). There is also evidence for transfer and clonal expansion of bla CMY-2 within S . enterica Typhimurium due to it accounting for 82 % of all chromosomally associated bla CMY-2 (Fig. 5). Mechanistically, it is known that bla CMY-2 is associated with functional insertion sequence elements, which can explain the transfer to the chromosome [69, 80, 81]. Despite this observed mobilization of bla CMY-2 to the chromosome, its plasmid entropy is lower than its serovar entropy due to three primary MOB-clusters AA474 (IncI-gamma/K1, MOBP, conjugative), AA627 (IncC, MOBH, conjugative), AA860 (IncC, MOBH, conjugative) accounting for 77 % of all bla CMY-2 genes (Fig. 5). From these results, it is clear that bla CMY-2 is capable of mobilization into different genetic contexts but the horizontal dissemination of the gene is largely due to the conjugal transfer of these three MOB-clusters, which are discussed in further detail below.
Fig. 5.
Sunburst plot of the bla CMY-2 (n=4116) multi-plasmid outbreak with the inner ring showing association between chromosome or plasmid contig sequences. The middle ring shows the primary MOB-cluster and outer ring shows the secondary MOB-cluster associated with the gene.
As mentioned previously, MOB-suite secondary clusters provide higher resolution subtyping information, which is suitable for performing outbreak analysis of plasmids. If two plasmids are assigned to the same secondary MOB-cluster, they can be considered sufficiently related to be considered potentially part of an outbreak. Within Salmonella there is an ongoing multi-plasmid outbreak of bla CMY-2 due to the presence of the same secondary MOB-clusters in different serotypes. With the notable exception of S . enterica Dublin, most serotypes contain multi-phyla host-range plasmids assigned to the secondary MOB-cluster AJ275 belonging to AA860 (IncC, MOBH, conjugative), which could be the major bla CMY-2 vector to distantly related taxa. S . enterica Dublin contains the secondary MOB-cluster AA627, which is part of AA627 (IncC, MOBH, conjugative) and is found in a variety of serotypes but most notably in S . enterica Dublin and S . enterica Newport. IncI plasmids are known to be poultry associated with demonstrated inter-species transfer occurring, and within the dataset we observe that two poultry associated serotypes, S . enterica Heidelberg and S . enterica Kentucky, primarily contain members of AI598 as part of AA474 (IncI-gamma/K1, MOBP, conjugative), which due to their shared ecological niche provide opportunity for transfer to occur [82–84]. It is not possible from this analysis to establish the frequency of transfer or directionality of transfer as it would require ancestral state reconstruction across different lineages. The presence of highly similar plasmids distributed in different serotypes provides direct evidence of horizontal transfer of these plasmids and subsequent clonal expansion.
MOB-cluster informed AMR transmission risk
A goal of this work is to develop a risk-based prioritization method for addressing horizontal transmission of AMR genes and their plasmid vectors. Abundance of plasmids in the population, associations with AMR and their frequency of horizontal transmission are major factors to consider when examining the potential risk a given MOB-cluster presents for the dissemination of AMR. MOB-cluster members variably contain AMR genes, which indicates that there are dynamics within a cluster, which demonstrates that members of that group are susceptible to AMR acquisition. MOB-clusters that rarely contain AMR and do not readily undergo horizontal transfer can be considered low risk compared to those with a high prevalence of AMR and high serovar entropy. The majority of MOB-clusters in Salmonella are low frequency, with 82 % of MOB-clusters found in fewer than 100 samples, which gives a useful threshold for delineating plasmids that are contributing broadly to AMR dissemination. All MOB-clusters were analysed according to the percentage of members containing at least one resistance gene against its corresponding serovar entropy (Fig. 6). The virulence plasmid AB461 (IncFIB, IncFII, N/A, non-mobilizable) from S . enterica Enteritidis represents a low-risk MOB-cluster as it has low serovar entropy and fewer than 2 % of its members contain at least one resistance gene. The abundant bla CMY-2 outbreak plasmids AA627 (IncC, MOBH, conjugative) and AA860 (IncC, MOBH, conjugative) represent high-risk MOB-clusters as roughly 90 % of their members contain ≥1 resistance gene and have moderate serovar entropies, 2.3 nats and 1.92 nats, respectively. However, the third bla CMY-2 outbreak MOB-cluster AA474 (IncI-gamma/K1, MOBP, conjugative) represents a more moderate risk since only 44 % of its members carry AMR genes. Using the examples of the virulence and bla CMY-2 outbreak MOB-clusters, other clusters with similar entropy and resistance prevalence can be identified. Out of 1044 known MOB-clusters detected in Salmonella , there are only 41 MOB-clusters that have at least 100 members, and at least 40 % of their members contain at least one resistance gene. This significantly reduces the scope of plasmids that merit more detailed analysis of their epidemiology and risk for AMR dissemination in different situations such as food production and clinical settings.
Fig. 6.
Relationship between serovar entropy (nats) and proportion of members containing at least one NCBI resistance gene for individual MOB-clusters. The size of the circles represents the abundance of the MOB-cluster and overall mobility prediction is used to colour the circles.
Conclusions
To our knowledge, this study provides the largest to-date characterization of plasmids and their associations with AMR genes within a single species based on publicly available Illumina sequencing data. We have presented a novel application of the alpha-diversity measure Shannon entropy to characterize horizontal transfer behaviour of plasmids across serotypes, as well as transfer of genes between plasmids and serotypes. As long reads become more prevalent, the need for plasmid reconstruction methods will decrease, but short-read samples will remain important due to the huge number of historical strains that will not be re-sequenced using long-read technologies. The ability to identify complete and circular plasmids from Illumina-only data is underappreciated and, based on our results, it is likely that there is a large number of plasmids in other pathogens that could likewise be completely assembled using only Illumina reads. Using the analytical approach developed here, we characterized an ongoing multi-plasmid outbreak responsible for the horizontal dissemination of bla CMY-2 between different serovars. The analytical approached developed here can be applied to other pathogens with large amounts of publicly available sequence data. Due to the large scope of this study, it is not feasible to go into great detail for specific serovars, MOB-clusters or genes, but a major goal for this work is to foster follow-up studies that can provide targeted long-read sequencing of samples with novel plasmids, or examine MOB-cluster or resistance gene horizontal transfer in more detail.
Supplementary Data
Funding information
This work was funded by the Public Health Agency of Canada and the Government of Canada’s Genomics R and D Initiative Phase VI Shared Priority Project Management Plan on Antimicrobial Resistance.
Acknowledgements
We thank the Bioinformatics core facility of the National Microbiology Laboratory of the Public Health Agency of Canada for computational infrastructure.
Author contributions
J. R.: conceptualization, software, methodology, data curation, formal analysis, investigation, writing – original draft, visualization. J. S.: writing – review and editing. K. B.: software, writing – review and editing. P. B.: writing – review and editing. J. H.E N.: funding acquisition, project administration, resources, supervision, writing – original draft.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ANI, average nucleotide identity; MGE, mobile genetic element; MLST, multilocus sequence typing; NCBI, National Center for Biotechnology Information.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary figures, six supplementary tables and supplementary data are available with the online version of this article or via Figshare.
References
- 1.Robertson J, Schonfeld J, Bessonov K, Bastedo P, Nash JHE. 2023. A global survey of Salmonella Plasmids and their associations with antimicrobial resistance. Microbiology Society. Dataset. [DOI] [PMC free article] [PubMed]
- 2.Balasubramanian R, Im J, Lee J-S, Jeon HJ, Mogeni OD, et al. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum Vaccin Immunother. 2019;15:1421–1426. doi: 10.1080/21645515.2018.1504717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. The global burden of nontyphoidal Salmonella gastroenteritis . Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 4.Pires SM, Desta BN, Mughini-Gras L, Mmbaga BT, Fayemi OE, et al. Burden of foodborne diseases: think global, act local. Curr Opin Food Sci. 2021;39:152–159. doi: 10.1016/j.cofs.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Antimicrobial Resistance: Global Report on Surveillance 2014. Geneva: World Health Organization; 2014. [Google Scholar]
- 6.US Food and Drug Administration NARMS Update: Integrated Report Summary. Apr 19, 2022. [ June 2; 2022 ]. https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2019-narms-update-integrated-report-summary accessed.
- 7.Botelho J, Schulenburg H. The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 2021;29:8–18. doi: 10.1016/j.tim.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Johnson CM, Grossman AD. Integrative and Conjugative Elements (ICEs): what they do and how they work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. 2018;4:e000206. doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 11.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlek A, Phan H, Sheppard AE, Doumith M, Ellington M, et al. Ordering the MOB: insights into replicon and MOB typing schemes from analysis of a curated dataset of publicly available plasmids. Plasmid. 2017;91:42–52. doi: 10.1016/j.plasmid.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker S, Hardy J, Sanderson KE, Quail M, Goodhead I, et al. A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 2007;3:e59. doi: 10.1371/journal.ppat.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson J, Lin J, Wren-Hedgus A, Arya G, Carrillo C, et al. Development of a multi-locus typing scheme for an Enterobacteriaceae linear plasmid that mediates inter-species transfer of flagella. PLoS One. 2019;14:e0218638. doi: 10.1371/journal.pone.0218638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coluzzi C, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. Evolution of plasmid mobility: origin and fate of conjugative and nonconjugative plasmids. Mol Biol Evol. 2022;39:msac115. doi: 10.1093/molbev/msac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat Commun. 2020;11:3602. doi: 10.1038/s41467-020-17278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Alvarado A, Garcillán-Barcia MP, de la Cruz F, Cloeckaert A. A Degenerate Primer MOB Typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One. 2012;7:e40438. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlek A, Stoesser N, Anjum MF, Doumith M, Ellington MJ, et al. Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front Microbiol. 2017;8:182. doi: 10.3389/fmicb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douarre PE, Mallet L, Radomski N, Felten A, Mistou MY. Analysis of COMPASS, a new comprehensive plasmid database revealed prevalence of multireplicon and extensive diversity of IncF plasmids. Front Microbiol. 2020;11:483. doi: 10.3389/fmicb.2020.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson J, Bessonov K, Schonfeld J, Nash JHE. Universal whole-sequence-based plasmid typing and its utility to prediction of host range and epidemiological surveillance. Microb Genom. 2020;6:000435. doi: 10.1099/mgen.0.000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock SJ, Phan M-D, Peters KM, Forde BM, Chong TM, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother. 2017;61:e01740-16. doi: 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008;61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 24.García-Fernández A, Villa L, Moodley A, Hasman H, Miriagou V, et al. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother. 2011;66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- 25.Fondi M, Bacci G, Brilli M, Papaleo MC, Mengoni A, et al. Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol Biol. 2010;10:59. doi: 10.1186/1471-2148-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tazzyman SJ, Bonhoeffer S. Why there are no essential genes on plasmids. Mol Biol Evol. 2015;32:3079–3088. doi: 10.1093/molbev/msu293. [DOI] [PubMed] [Google Scholar]
- 27.Acman M, van Dorp L, Santini JM, Balloux F. Large-scale network analysis captures biological features of bacterial plasmids. Nat Commun. 2020;11:2452. doi: 10.1038/s41467-020-16282-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redondo-Salvo S, Bartomeus-Peñalver R, Vielva L, Tagg KA, Webb HE, et al. COPLA, a taxonomic classifier of plasmids. BMC Bioinformatics. 2021;22:390. doi: 10.1186/s12859-021-04299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stimson J, Gardy J, Mathema B, Crudu V, Cohen T, et al. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol Biol Evol. 2019;36:587–603. doi: 10.1093/molbev/msy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, et al. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes . J Clin Microbiol. 2015;53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H, Liu W, Qin T, Liu C, Ren H. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Klebsiella pneumoniae . Front Microbiol. 2017;8:371. doi: 10.3389/fmicb.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks LE, Kaze M, Sistrom M. Where the plasmids roam: large-scale sequence analysis reveals plasmids with large host ranges. Microb Genom. 2019;5:e000244. doi: 10.1099/mgen.0.000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain A, Srivastava P. Broad host range plasmids. FEMS Microbiol Lett. 2013;348:87–96. doi: 10.1111/1574-6968.12241. [DOI] [PubMed] [Google Scholar]
- 35.Popowska M, Krawczyk-Balska A. Broad-host-range IncP-1 plasmids and their resistance potential. Front Microbiol. 2013;4:44. doi: 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsay JP, Kwong SM, Murphy RJT, Eto KY, Price KJ, et al. An updated view of plasmid conjugation and mobilization in Staphylococcus . Mob Genet Elements. 2016;6:e1208317. doi: 10.1080/2159256X.2016.1208317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcillán-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev. 2011;35:936–956. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 38.Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol. 2015;6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey S, San Millán Á, Toll-Riera M, Connolly J, Flor-Duro A, et al. Staphylococcal phages and pathogenicity islands drive plasmid evolution. Nat Commun. 2021;12:5845. doi: 10.1038/s41467-021-26101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeifer E, Moura de Sousa JA, Touchon M, Rocha EPC. Bacteria have numerous distinctive groups of phage-plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res. 2021;49:2655–2673. doi: 10.1093/nar/gkab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beck J, Schwanghart W. Comparing measures of species diversity from incomplete inventories: an update. Methods Ecol Evol. 2010;1:38–44. doi: 10.1111/j.2041-210X.2009.00003.x. [DOI] [Google Scholar]
- 42.Chao A, Shen TJ. Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ Ecol Stat. 2003;10:429–443. doi: 10.1023/A:1026096204727. [DOI] [Google Scholar]
- 43.Brose U, D. Martinez N. Estimating the richness of species with variable mobility. Oikos. 2004;105:292–300. doi: 10.1111/j.0030-1299.2004.12884.x. [DOI] [Google Scholar]
- 44.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, et al. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeman T. GitHub; 2020. https://github.com/tseemann/abricate [Google Scholar]
- 48.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, et al. Validating the amrfinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Godzik A. CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 50.Octavia S, Wang Q, Tanaka MM, Sintchenko V, Lan R. Genomic variability of serial human isolates of Salmonella enterica serovar Typhimurium associated with prolonged carriage. J Clin Microbiol. 2015;53:3507–3514. doi: 10.1128/JCM.01733-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wein T, Hülter NF, Mizrahi I, Dagan T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat Commun. 2019;10:2595. doi: 10.1038/s41467-019-10600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, Larsson M, Robinson RC, Chen SL. Direct and convenient measurement of plasmid stability in lab and clinical isolates of E. coli . Sci Rep. 2017;7:4788. doi: 10.1038/s41598-017-05219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dimitriu T, Marchant L, Buckling A, Raymond B. Bacteria from natural populations transfer plasmids mostly towards their kin. Proc Biol Sci. 2019;286:20191110. doi: 10.1098/rspb.2019.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng Y, Liu J, Li Y-G, Cao F-L, Johnston RN, et al. Inheritance of the Salmonella virulence plasmids: mostly vertical and rarely horizontal. Infect Genet Evol. 2012;12:1058–1063. doi: 10.1016/j.meegid.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Ahmer BMM, Tran M, Heffron F. The virulence plasmid of Salmonella typhimurium is self-transmissible. J Bacteriol. 1999;181:1364–1368. doi: 10.1128/JB.181.4.1364-1368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu C, Feng Y, Chien AC, Hu S, Chu CH, et al. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics. 2008;92:339–343. doi: 10.1016/j.ygeno.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 57.Baddam R, Kumar N, Shaik S, Lankapalli AK, Ahmed N. Genome dynamics and evolution of Salmonella Typhi strains from the typhoid-endemic zones. Sci Rep. 2014;4:7457. doi: 10.1038/srep07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holt KE, Thomson NR, Wain J, Langridge GC, Hasan R, et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics. 2009;10:36. doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveira PH, Touchon M, Rocha EPC. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mamontov V, Martynov A, Morozova N, Bukatin A, Staroverov DB, et al. Persistence of plasmids targeted by CRISPR interference in bacterial populations. Proc Natl Acad Sci U S A. 2022;119:e2114905119. doi: 10.1073/pnas.2114905119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lanza VF, Tedim AP, Martínez JL, Baquero F, Coque TM. The plasmidome of Firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol Spectr. 2015;3:PLAS-0039-2014. doi: 10.1128/microbiolspec.PLAS-0039-2014. [DOI] [PubMed] [Google Scholar]
- 62.Soler N, Robert E, Chauvot de Beauchêne I, Monteiro P, Libante V, et al. Characterization of a relaxase belonging to the MOBT family, a widespread family in Firmicutes mediating the transfer of ICEs. Mob DNA. 2019;10:18. doi: 10.1186/s13100-019-0160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu C, Chiu CH. Evolution of the virulence plasmids of non-typhoid Salmonella and its association with antimicrobial resistance. Microbes Infect. 2006;8:1931–1936. doi: 10.1016/j.micinf.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 64.Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, et al. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother. 2016;60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorelick R. Combining richness and abundance into a single diversity index using matrix analogues of Shannon’s and Simpson’s indices. Ecography. 2006;29:525–530. doi: 10.1111/j.0906-7590.2006.04601.x. [DOI] [Google Scholar]
- 66.Grabchak M, Marcon E, Lang G, Zhang Z. The generalized Simpson’s entropy is a measure of biodiversity. PLoS One. 2017;12:e0173305. doi: 10.1371/journal.pone.0173305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medina O, Manian V, Chinea JD. Biodiversity assessment using hierarchical agglomerative clustering and spectral unmixing over hyperspectral images. Sensors. 2013;13:13949–13959. doi: 10.3390/s131013949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Folster JP, Tolar B, Pecic G, Sheehan D, Rickert R, et al. Characterization of blaCMY plasmids and their possible role in source attribution of Salmonella enterica serotype Typhimurium infections. Foodborne Pathog Dis. 2014;11:301–306. doi: 10.1089/fpd.2013.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castellanos LR, van der Graaf-van Bloois L, Donado-Godoy P, Mevius DJ, Wagenaar JA, et al. Phylogenomic investigation of INCI1-Iγ plasmids harboring blaCMY-2 and blaSHV-12 in Salmonella enterica and Escherichia coli in multiple countries. Antimicrob Agents Chemother. 2019;63:e02546-18. doi: 10.1128/AAC.02546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM. Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog Dis. 2011;8:1289–1294. doi: 10.1089/fpd.2011.0944. [DOI] [PubMed] [Google Scholar]
- 71.Guzman-Otazo J, Joffré E, Agramont J, Mamani N, Jutkina J, et al. Conjugative transfer of multi-drug resistance IncN plasmids from environmental waterborne bacteria to Escherichia coli. . Front Microbiol. 2022;13:997849. doi: 10.3389/fmicb.2022.997849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dorr M, Silver A, Smurlick D, Arukha A, Kariyawasam S, et al. Transferability of ESBL-encoding IncN and IncI1 plasmids among field strains of different Salmonella serovars and Escherichia coli . J Glob Antimicrob Resist. 2022;30:88–95. doi: 10.1016/j.jgar.2022.04.015. [DOI] [PubMed] [Google Scholar]
- 73.Alderliesten JB, Duxbury SJN, Zwart MP, de Visser JAGM, Stegeman A, et al. Effect of donor-recipient relatedness on the plasmid conjugation frequency: a meta-analysis. BMC Microbiol. 2020;20:135. doi: 10.1186/s12866-020-01825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran F, Boedicker JQ. Plasmid characteristics modulate the propensity of gene exchange in bacterial vesicles. J Bacteriol. 2019;201:e00430-18. doi: 10.1128/JB.00430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slater FR, Bailey MJ, Tett AJ, Turner SL. Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol Ecol. 2008;66:3–13. doi: 10.1111/j.1574-6941.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 76.Hernández-Beltrán JCR, San Millán A, Fuentes-Hernández A, Peña-Miller R. Mathematical models of plasmid population dynamics. Front Microbiol. 2021;12:606396. doi: 10.3389/fmicb.2021.606396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harrison E, Dytham C, Hall JPJ, Guymer D, Spiers AJ, et al. Rapid compensatory evolution promotes the survival of conjugative plasmids. Mob Genet Elements. 2016;6:e1179074. doi: 10.1080/2159256X.2016.1179074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother. 2017;61:e00410-17. doi: 10.1128/AAC.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol. 2005;55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 80.Singh NS, Singhal N, Virdi JS. Genetic environment of blaTEM-1, blaCTX-M-15, blaCMY-42 and characterization of integrons of Escherichia coli isolated from an Indian urban aquatic environment. Front Microbiol. 2018;9:00382. doi: 10.3389/fmicb.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Su L-H, Chen H-L, Chia J-H, Liu S-Y, Chu C, et al. Distribution of a transposon-like element carrying bla(CMY-2) among Salmonella and other Enterobacteriaceae . J Antimicrob Chemother. 2006;57:424–429. doi: 10.1093/jac/dki478. [DOI] [PubMed] [Google Scholar]
- 82.Abraham S, Kirkwood RN, Laird T, Saputra S, Mitchell T, et al. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J. 2018;12:2352–2362. doi: 10.1038/s41396-018-0200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carattoli A, Villa L, Fortini D, García-Fernández A. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid. 2021;118:102392. doi: 10.1016/j.plasmid.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 84.Castellanos LR, Donado-Godoy P, León M, Clavijo V, Arevalo A, et al. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS One. 2017;12:e0170777. doi: 10.1371/journal.pone.0170777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Robertson J, Schonfeld J, Bessonov K, Bastedo P, Nash JHE. 2023. A global survey of Salmonella Plasmids and their associations with antimicrobial resistance. Microbiology Society. Dataset. [DOI] [PMC free article] [PubMed]