Abstract
Acinetobacter baumannii is an important opportunistic pathogen known for its high levels of resistance to many antibiotics, particularly those considered last resorts such as colistin and carbapenems. Plasmids of this organism are increasingly associated with the spread of clinically important antibiotic resistance genes. Although A. baumannii is a ubiquitous organism, to date, most of the focus has been on studying strains recovered from clinical samples ignoring those isolated in the environment (soil, water, food, etc.). Here, we analysed the genetic structures of eight novel plasmids carried by an environmental colistin-resistant A. baumannii (strain E-072658) recovered in a recycled fibre pulp in a paper mill in Finland. It was shown that E-072658 carries a new variant of the mcr-4 colistin resistance gene (mcr-4.7) in a novel Tn3-family transposon (called Tn6926) carried by a novel plasmid p8E072658. E-072658 is also resistant to sulphonamide compounds; consistent with this, the sul2 sulphonamide resistance gene was found in a pdif module. E-072658 also carries six additional plasmids with no antibiotic resistance genes, but they contained several pdif modules shared with plasmids carried by clinical strains. Detailed analysis of the genetic structure of all eight plasmids carried by E-072658 showed a complex evolutionary history revealing genetic exchange events within the genus Acinetobacter beyond the clinical or environmental origin of the strains. This work provides evidence that environmental strains might act as a source for some of the clinically significant antibiotic resistance genes.
Keywords: Acinetobacter baumannii, colistin resistance, mcr-4.7, plasmid, environmental, Tn6926 , Transposon
Data Summary
Complete genome assemblies of Acinetobacter baumannii E-072658 including its chromosome and eight plasmids p1E072658, p2E072658, p3E072658, p4E072658, p5E072658, p6E072658, p7E072658 and p8E072658 are available in the GenBank non-redundant database under GenBank accession numbers CP061705, CP061713, CP061712, CP061711, CP061710, CP061709, CP061708, CP061707 and CP061706, respectively.
Impact Statement.
Acinetobacter baumannii has become a globally significant opportunistic pathogen that is difficult to treat given its high levels of resistance to the last line of antibiotic defence, such as colistin and carbapenems. Hence it is imperative to understand how resistance emerges, evolves and spreads through A. baumannii populations. However, understating the evolution of resistance has largely been hampered by the fact that most studies have focused on clinical strains, which are often highly clonal. This ignores the possibility that the source for some of the clinically significant resistance genes might be in the environment that ultimately accumulate in clinical strains. Here, we studied the evolution of eight plasmids carried by an environmental strain (E-072658) and showed that one of the plasmids carries a novel variant of the mcr-4 colistin resistance gene, which confers resistance to colistin, considered a last-resort antibiotic. This novel variant of the mcr-4 gene (mcr-4.7) might be the origin of the variants of the mcr-4 genes that are found in clinical strains. Moreover, this study showes several links of specific modules found in other plasmids of E-072658 to their closely related homologues of plasmids in clinical strains, providing further evidence of genetic exchange events between clinical and environmental strains. This study calls for further investigation of environmental strains to reveal the intricacies of the origin and evolution of antibiotic resistance.
Introduction
Acinetobacter baumannii is an opportunistic Gram-negative pathogen that has attracted attention given its ability to develop resistance to many antibiotics [1, 2]. A. baumannii causes a wide range of nosocomial infections, including pneumonia, bacteraemia, and urinary tract and wound infections [1]. Indeed, the spread of A. baumannii strains resistant to all commercially available antibiotics, including those considered last resort (e.g. colistin and carbapenems) [3, 4], has become a significant global problem [3, 5]. This is recognized by the World Health Organisation, deeming carbapenem-resistant A. baumannii as the number one priority for antibiotic development [6]. Although A. baumannii strains are mainly associated with clinical settings, they have also been routinely identified in a wide range of non-clinical environments, including food, water, sewage and soil, as well as from pig and cattle faecal samples [7–15]. Therefore, it has recently been suggested that A. baumannii is a One Health problem (with the environment, humans and animals considered interconnected), indicating a need for a transdisciplinary research approach to study A. baumannii strains isolated from both clinical environmental sources (soil, water, food, etc.) [16].
In A. baumannii , the spread of antibiotic resistance is known to occur mainly by the horizontal acquisition of antibiotic resistance genes (ARGs) primarily via mobile genetic elements (MGEs) such as transposons and plasmids [3, 17–19]. In A. baumannii, most antibiotic-resistance genes are often in chromosomal genomic islands [3, 17, 18]. However, plasmids are also increasingly recognized as a significant mode for disseminating antibiotic-resistance genes, such as those conferring resistance to the front-line carbapenems and colistin antibiotics [20–23]. A. baumannii has a unique repertoire of plasmids that capture and mobilize a wide range of genetic material involved in pathogenesis and antibiotic resistance [20, 21, 24] that has recently been classified [25]. Most comparative genomics studies have focused on nosocomial strains, leaving the evolution, origin and genetic structure of plasmids carried by environmental strains poorly understood. Here, we analysed the genetic structure of eight plasmids carried by E-072658, an environmental A. baumannii strain recovered in recycled fibre pulp in a paper mill in Finland. This study shows that all eight plasmids carried by E-072658 are novel, including one that carries a novel variant of the mcr-4 colistin resistance gene [4], and have a complex evolutionary history and share common origins with those carried by clinical strains. Furthermore, only recently characterized, pdif modules have been uncovered in Acinetobacter plasmids where they comprise pairs of Xer recombination sites called plasmid–dif (pdif) sites that flank a gene or genes [26, 27]. These pdif sites are remarkable for two reasons. First, pdif modules have been isolated from plasmids of bacteria in permafrost cores, and many gene types, including ARGs and heavy metal resistance genes, are located in pdif-modules in plasmids. These pdif-modules assist resistance genes that they carry to move between different plasmids. Second, pdif sites flank carbapenem resistance genes [e.g. tet39, mph-msr(E), oxa58 and oxa24] in major global clones of A. baumannii [26, 27]. However, little is known about the distribution of pdif modules in Acinetobacter plasmids from different environmental niches. Here, pdif modules found on E-072658 plasmids were identified, analysed and tracked.
Methods
Bacterial genome sequence
Sequences of eight novel plasmids previously reported in the genome sequence of A. baumannii strain E-072658 [28], recovered in recycled fibre pulp in a Finnish paper mill, and Hamidian et al. [29] were analysed in this study.
Antibiotic resistance profile
The antibiotic resistance profile against 23 antibiotics was previously determined [29] using the standard CDS (http://cdstest.net) disc diffusion method. Here, we measured the minimal inhibitory concentration (MIC) against colistin using the standard micro broth dilution method as previously described [30, 31] and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines for Acinetobacter spp. [32].
Bioinformatics and sequence analysis
The resistance genes and insertions sequences were manually annotated using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) and ISFinder (https://www-is.biotoul.fr/), respectively. Protein coding regions were characterized using blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins), Pfam (http://pfam.xfam.org/) and UniProt (https://www.uniprot.org/) searches. Standalone blast (www.ncbi.nlm.nih.gov/books/) was used to describe the structure of plasmids carried by E-072658. A combination of manual sequence analysis and a local database of known pdif sites [26, 27] that was curated here were used to identify pdif modules in E-072658 plasmids. Putative plasmid replication initiation proteins were typed (and named) using the Acinetobacter Plasmid Typing (APT) scheme publicly available at GitHub (https://github.com/MehradHamidian/AcinetobacterPlasmidTyping) [25]. The CRISPR (clustered regularly interspaced short palindromic repeats) sequences were explored using the CRISPRCasFinder webtool available at https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index. The SnapGene (V6.0.5) software manually annotates regions of interest and draws figures to scale using the Illustrator (v26.2.1) program.
Mating assays
Recipients Escherichia coli K-12 (BW25113) and A. baumannii (ATCC 17978) were grown in cation adjusted Mueller Hinton (CAMH) broth without antibiotics. For conjugation experiments, A. baumannii strain E-072658 was grown in CAMH (Becton Dickinson) broth supplemented with 10 µg ml−1 colistin (Sigma-Aldrich) overnight at 37 °C with shaking at 200 r.p.m. Mating assays were performed by mixing the same volume of donor and recipient strains (a total of 100 µl of the overnight cultures) after washing with PBS followed by spotting on CAMH agar plates and incubating overnight at 37 °C. Cells were resuspended and diluted in PBS, and transconjugants were selected by plating on CAMH agar plates containing rifampicin (100 mg l−1) and colistin (5 mg l−1). Transconjugants were further tested for rifampicin resistance to which recipient cells were resistant to differentiate them from spontaneous rifampicin mutants of E-072658 by targeted PCR. For PCR tests, primer sets MH68 (5′-CAGATTTGCCCAAAGATGGT-3′) and MH69 (5′-CGATGATAAGCACGAGCAGA-3′), and mcr4.7F (5′-CCAGCATTGGTACGCTAGTT-3′) and mcr4.7R (5′-TCGTTGGCATTGGGATAGTC-3′) were used. The primer set MH68/69 was designed to target comM in E-072658, which is interrupted (by Tn6021) in the ATCC 17978 strain. The primer set mcr4.7F/R used for detecting the mcr4.7 gene in E-072658.
Results and discussion
A. baumannii E-072658 carries eight novel plasmids
We previously reported that the environmental A. baumannii strain E-072658 carries eight plasmids [29]. As also previously reported, E-072658 belongs to a novel sequence type (ST649) and it is not related to any other strains characterized to date (including all clinical and environmental strains). Its genome also contains over 440 insertion sequences, suggesting that it might have a defective CRISPR/cas system. The CRISPR/cas system is an adaptive immune system of bacteria and archaea against exogenous DNA. In fact, the CRISPR/cas system protects the bacteria from invaders, including MGEs or bacteriophages [33]. Here, our analysis showed that the E-072658 chromosome does not contain a CRISPR/cas locus, consistent with the accumulation of over hundreds of insertion sequences in its genome combined with eight plasmids.
Here, we dissected the genetic structures of all eight plasmids and examined their evolutionary relationship with other plasmids publicly available in GenBank. Analysis performed here showed all plasmids carried by E-072658 are novel given their differences to other known A. baumannii plasmid types. Specific modules found on the E-072658 plasmids were also tracked and revealed several potential exchange events with plasmids carried by clinical strains.
p8E072658, a novel large Rep_3 family plasmid carrying the mcr-4.7 colistin resistance gene, in a Tn3-family transposon
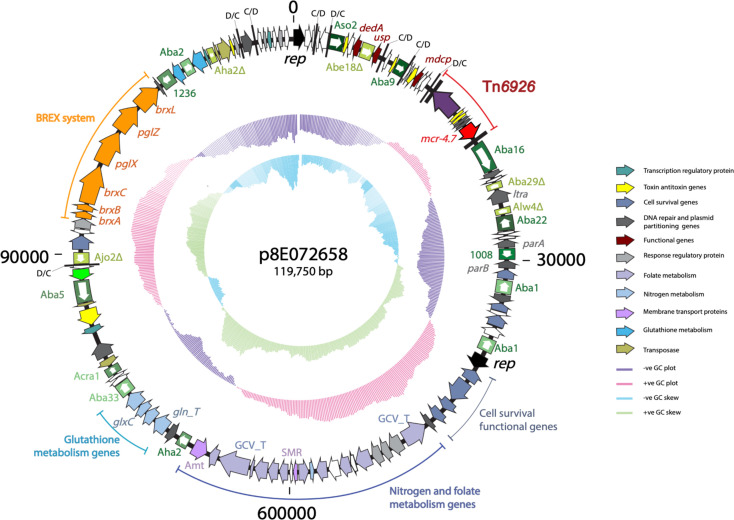
p8E072658 is 119 750 bp and the largest plasmid carried by E-072658 (Fig. 1). p8E072658 encodes two novel putative replication initiation proteins (Rep), which belong to the Rep_3 superfamily (Pfam01051), representing the most abundant Rep type in A. baumannii plasmids [25]. These Rep proteins were named R3-T73 (locus id: H2787_17600) and R3-T74 (locus id: H2787_17845) and included in the recently developed APT scheme [25]. The closest match to r3-T73 (locus id: H2787_17600) is r3-T45 (GenBank accession number CP044520.1) while H2787_17845 does not share any DNA identity with other known rep sequences.
Fig. 1.
Circular representation of p8E072658, showing the distribution of functional genes. The outer black ring represents the backbone of the plasmid. Arrows indicate the extent and orientation of genes and are colour coded as per their associated functions with the keys shown on the right. The boxed arrows are insertion sequences (IS) and are labelled along the IS elements. The two inner rings represent the GC plot and GC skew, respectively. Small vertical black bars labelled C/D or D/C represent the pdif recombination sites (XerC/D or XerD/C).
p8E072658 carries a novel allele of the mcr-4 colistin resistance gene named mcr-4.7 and represents one of the first reports of an mcr gene being isolated from an environmental isolate. The mcr-4.7 gene was registered in the NCBI’s Bacterial Antimicrobial Resistance Gene Database under https://www.ncbi.nlm.nih.gov/pathogens/refgene/#mcr-4.7. Colistin is a polymyxin antibiotic and is considered a last-resort antibiotic for treating multiple antibiotic-resistant A. baumannii infections. However, increasing resistance levels have become a significant concern given that colistin-resistant strains are also often resistant to several or all available antibiotics [34]. The mcr colistin resistance genes have mainly been reported in clinical strains; hence its presence in an environmental strain is significant as it also shows its presence in the environment. The mcr genes phosphoethanolamine transferases modify Lipid A of the cell’s lipopolysaccharide (LPS), rendering the cells colistin resistant due to the change of the overall charge of the outer membrane. To date, ten mcr genes (mcr-1 to mcr-10), each with several variants, have been identified worldwide in several Enterobacterales species (e.g. Escherichia coli , Klebsiella pneumoniae and Salmonella ) as well as A. baumannii with a wide range of clinical, environmental and animal origins [35]. The mcr-4.7 gene in p8E072658 differs from mcr-4.1, the first mcr-4 allele found in Escherichia coli and Salmonella spp. [36], by four base changes, two of which are also present in mcr-4.3 (Table 1). The four nucleotide changes cause the V179G, V236F, Q331R and V485I amino acid changes relative to Mcr-4.1 (Table 1; Fig. S1, available in the online version of this article). Resistance to colistin was tested by measuring the MIC. E-072658 was resistant to high levels of colistin (25 mg l−1), accounting for the presence of mcr-4.7. It has been shown that each mcr gene has different possible origins and genomic context as they can be plasmid-borne or located in the chromosome. For instance, the progenitors of mcr-1, mcr-3 and mcr-4 are Moracella sp., Aeromonas sp. and Shewanella sp., respectively. The mcr-1 gene is carried in a composite transposon (Tn6330), flanked by two copies of ISApl1, and was first identified on an Incl2 in E. coli [37] while mcr-4 was first found in an 8 kb ColE10 plasmid in Salmonella enterica recovered from a pig in Italy [35].
Table 1.
Changes in nucleotide/amino acid in mcr-4 alleles
|
mcr-4 allele |
First identification |
Nucleotide differences compared to mcr-4.1 |
Amino acid differences compared to Mcr-4.1 |
References |
|---|---|---|---|---|
|
mcr-4.1 |
Escherichia coli, Salmonella spp. |
na |
na |
[36] |
|
mcr-4.2 |
Escherichia coli, Salmonella Typhimurium |
A992G |
Gln331Arg |
[47, 48] |
|
mcr-4.3 |
T536G, G706T |
Val179Gly, Val236Phe, |
[49] |
|
|
mcr-4.4 |
C613A, A992G |
Gln331Arg |
[47] |
|
|
mcr-4.5 |
C329T, A992G |
Gln331Arg |
[47] |
|
|
mcr-4.6 |
Salmonella keduugou |
G706T |
Val236Phe, |
[50] |
|
mcr-4.7 |
T536G, G706T, A992G, G1453A |
Val179Gly, Val236Phe, Gln331Arg, Val485Ile |
This study |
na, Not applicable.
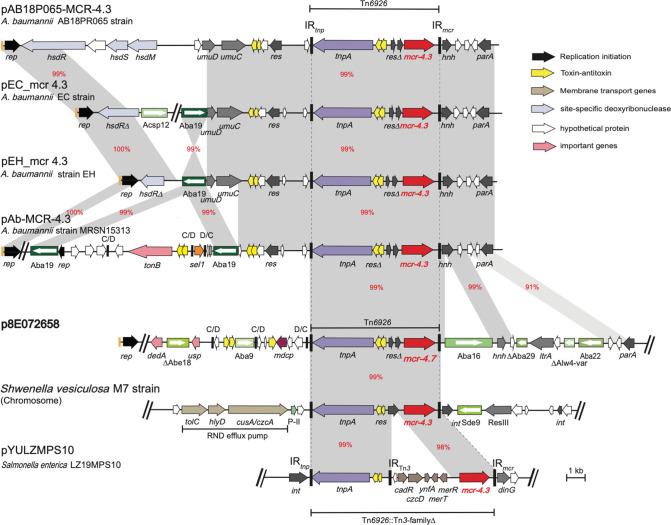
Later, two studies reported the presence of mcr-4.3 in A. baumannii strains, recovered in clinical and pig faeces, in transposons related to Tn3 [38, 39]. Here, analysis of the surrounding region of mcr-4.7 indicated that this gene is in a transposon, which we named here Tn6926. Tn6926 is a 6494 bp transposon with 38 bp inverted terminal repeats and encodes a TnpA and TnpR that are distantly related to the TnpAR of Tn1-3 (with 23 and 25% amino acid identity, respectively). This, combined with the presence of a 38 bp terminal repeat, suggests that Tn6926 also belongs to the Tn1-3 transposon family but is distantly related to Tn1-3. Tn6926 is not flanked by target site duplication. In addition to those previously described [38, 39], here we tracked and found several variants of Tn6926 in various A. baumannii plasmids as well as a Salmonella enterica plasmid (pYULZMPS10, GenBank accession number CP100354.1; Fig. 2). It has been suggested that the genus Shewanella is the origin of the mcr-4 gene [39]. A previous study showed that the mcr-4.3 gene is in a Tn3-family transposon that is interrupted by another transposon (called TnShfr1) in Shewanella frigidimarina NCIMB 400 (GenBank accession number CP000447) [39]. However, here, we found an intact form of a variant of Tn6926 in Shwenella vesiculosa strain M7 (GenBank accession number CP073588.1; Fig. 2) that confirms the genus Shwenella as an origin of mcr-4. It also showed a significant role for Tn6926 to spread the mcr-4 variants from Shwenella to Acinetobacter . Notably, Tn6926 does not appear to generate target site duplications while others (in Fig. 2) are flanked by various numbers (three to five) 3–5of duplications, suggesting they do not always generate an exact number of target site duplications.
Fig. 2.
Linearized partial map of p8E072658 encoding a new allele of mcr-4 within Tn6926. The horizontal central black lines represent the plasmid backbone. The arrows indicate gene orientation, and the boxed arrows represent IS. The grey shadings demonstrate significant percentage identities, marked in red. The genes/ORFs are labelled below according to their associated functions and the key is shown on the right. Iterons are shown in orange bars preceding the rep genes. Small vertical black bars labelled C/D or D/C represent the two pdif recombination sites (XerC/D or XerD/C). The long vertical bars (marked IRL and IRR) denote Tn6926. The scale bar is shown below and drawn to scale from GenBank accession numbers CP061706 (p8E072658), MK360916.1 (pAB18P065-MCR-4.3), CP038265.1 (pEC_mcr 4.3), CP038261.1 (pEH_mcr 4.3), CP033872.1 (pAb-MCR-4.3) and CP100354.1 (pYULZMPS10) and chromosome of Shewenella vesiculosa strain M7 from accession no. CP073588.1.
The emergence and spread of the mcr genes in A. baumannii strains is a concerning public health threat given that the wide dissemination of mcr from environmental to clinical samples has increased the risk of spread to humans via various routes. Prior to the emergence of mcr genes, colistin resistance was considered not transferable and intrinsic to the organism. Mating assays performed here did not result in conjugative transfer of p8E072658, suggesting that, under the conditions tested, p8E072658 is not conjugative, consistent with the absence of transfer functions on this plasmid. However, the finding of mcr-4.7 in a transposon on a plasmid in an environmental A. baumannii strain is remarkable in many ways. Colistin is considered a last-resort antibiotic, and the spread of mcr genes further limits treatment options. It also shows a significant role for the environment as a source for a clinically important resistance gene, highlighting that more attention should be given to study environmental strains as the potential source of many clinically important genes.
Another striking feature of p8E072658 is the presence of 18 insertion sequences (IS) including multiple partial IS indicating a complex evolutionary history (Fig. 1). Moreover, p8E072658 encodes a BREX system (Bacteriophage Exclusion system; Fig. 1). The BREX system is a phage resistance system and has been added to the previously known mechanisms (e.g. CRISPR-Cas and restriction-modification systems) in the arsenal of bacterial defence against phages [38]. A related BREX system was found in pTol5, a 117 kb plasmid of an Acinetobacter sp. (GenBank accession number AP024709.1) that belongs to the diverse plasmid family of A. baumannii conjugative plasmids represented by pA297-3 (GenBank accession number KU744946) [23, 40]. BREX determinants were found in another Acinetobacter plasmid type, as recently described [41]. However, as p8E072658 is a different plasmid type this indicates that various Acinetobacter plasmid types carry BREX systems. p8E072658 also has a few additional regions that encode metabolic and cell survival functions (Fig. 1), suggesting possible environmental co-selection and important roles of these functions in adapting E-072658 in the environment.
p7E072658 carries the sul2 sulphonamide resistance gene in a pdif module
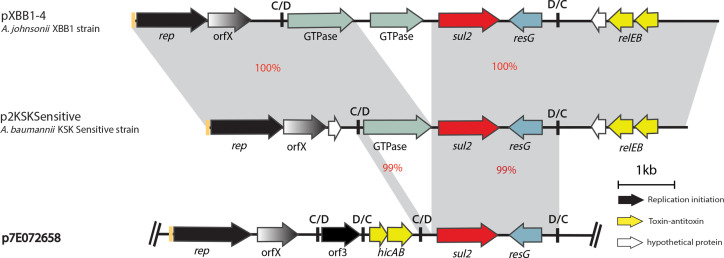
p7E072658 (GenBank accession number CP061707) is a 19 661 bp plasmid that encodes two novel Rep proteins belonging to the Rep_3 superfamily (Pfam01051), which were named R3-T75 (locus id H2787_18135) and R3-T76 (locus id H2787_18195). Closet matches to H2787_18135 and H2787_18195 are r3-T17 (GenBank accession number CP015365) and r3-T10 (GenBank accession number LR026972), respectively. p7E072658 carries a sul2 sulfonamide resistance gene and the relEB genes encoding toxin–antitoxin functions. Like most plasmids that encode Rep_3 family Rep proteins [25], p7E072658 contains multiple pdif modules (Fig. 3). The sul2 gene is often associated with the GIsul2 genomic island [42]. Here, analysis of the sul2 context showed that a 1222 bp segment, including sul2 and 100 bp and 297 bp upstream and downstream of this gene, was identical to GIsul2, indicating a common origin and that this segment has derived from GIsul2. However, further analysis showed that the sul2 gene is part of a 2699 bp long pdif module (Fig. 3), suggesting that it can move via spread of this module. To date, several antibiotic resistance genes have been found in pdif modules, but to the best of our knowledge this is the first time sul2 has been found in this context. Here, we tracked this module and found two variants in pXBB1-4 (GenBank accession number CP061707.1) carried by Acinetobacter johnsoni XBB1-4 isolated in hospital sewage and p2KSKSensitive (GenBank accession number CP061707.1) carried by A. baumannii p2KSKSensitive recovered in a respiratory specimen, indicating spread of the sul2 pdif modules across species in various sources (hospital sewage and clinical samples).
Fig. 3.
Schematic of the linearized partial map of p7E072658 containing the sul2 gene within a pdif module compared to similar pdif modules found in unrelated plasmids. The horizontal central black lines represent the plasmid backbone. The arrows indicate gene orientation, and the boxed arrows represent IS. The grey shadings demonstrate significant percentage identities, coloured red. The genes/ORFs are labelled below according to their associated functions and the key is shown on the right. Iterons are shown in orange bars preceding the rep genes. Small vertical black bars labelled C/D or D/C represent the two pdif recombination sites (XerC/D or XerD/C). The scale bar is shown below and drawn to scale from GenBank accession numbers CP061707 (p7E072658), CP061707.1 (p2KSKSensitive) and CP061707.1 (pXBB1-4).
E-072658 carries six novel cryptic plasmids containing various pdif modules shared with other clinical and environmental Acinetobacter plasmids
The remaining plasmids (p1E072658–p6E072658; Fig. S2) were cryptic. However, they encode important proteins involved in various functions such as usher protein, ABC transporter, inorganic ion transporter and toxin–antitoxin systems.
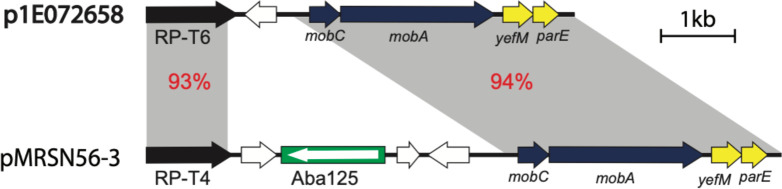
p1E072658 is 4483 bp in length and the only plasmid that encodes a novel putative replication protein (Locus id H2787_18615) belonging to the Rep_PriCT1 superfamily (pfam03090) (Fig. 4). The closest match to H2787_18615 is rP-T4 (GenBank accession number CM009039). This novel Rep was named RP-T78 (Table 2). Notably, most plasmids that belong to this family are large and involved in the spread of aminoglycoside and carbapenem resistance genes [25]. However, unlike other members of this diverse family, p1E072658 is cryptic. p1E072658 is a novel plasmid; however, it shares approximately 3.5 kb with pMRSN56-2 (GenBank accession number CP080455) with 93–94 % DNA identity, indicating these plasmids belong to a diverse family. Moreover, pMRSN56-2 is carried by A. baumannii MRSN56 recovered in 2010 from a hip infection, indicating that plasmids related to pMRSN56-2 and p1E072658 are spread across clinical and environmental samples.
Fig. 4.
Comparison of the genetic structure of p1E072658 and pMRSN56-3. The horizontal central black lines represent the plasmid backbone. The arrows indicate gene orientation, and the boxed arrow represents IS. The grey shadings demonstrate significant percentage identities, marked in red. The genes/ORFs are labelled below according to their associated functions. The scale bar is shown below and drawn to scale from GenBank accession numbers CP061713 (p1E072658) and CP080455 (pMRSN56-2).
Table 2.
Properties of plasmids carried by A. baumannii E-072658
|
Plasmid name |
Size (bp) |
Rep type* |
Locus_id |
Rep family |
Pfam |
Mob |
Pfam |
pdif modules |
IS |
Important functions |
GenBank Acc. no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
p8E072658 |
119 750 |
R3-T73 R3-T74 |
H2787_17600 H2787_17845 |
Rep-3 Rep-3 |
01051 |
– |
– |
8 |
ISAba1, IS1008, ISAba22, ISAlw4Δ (92%), ISAba9, ISAba29Δ, ISAba16, ISAba8, ISAbe18Δ, ISAba27Δ, IS1236, ISAba5Δ (92%), ISAcra1, ISAba33, ISAha2 |
Colistin resistance |
|
|
p7E072658 |
19 661 |
R3-T75 R3-T76 |
H2787_18135 H2787_18195 |
Rep-3 Rep-3 |
01051 |
MobA/L |
03389 |
7 |
IS17∆ ISA1w14 |
Sulfonamide resistance |
|
|
p6E072658 |
18 806 |
R3-T36 R3-T77 |
H2787_18240 H2787_18280 |
Rep-3 Rep-3 |
01051 |
– |
– |
7 |
– |
Usher protein |
|
|
p5E072658 |
15 545 |
– |
na |
– |
– |
– |
– |
– |
ISAlw34, ISAba125 ISAha3Δ, ISaba31 ISAba27Δ, ISAba7Δ |
ABC transporter |
|
|
p4E072658 |
7989 |
R3-T2 |
H2787_18430 |
Rep-3 |
01051 |
– |
– |
3 |
– |
Inorganic ion transporter |
|
|
p3E07265 |
6763 |
R3-T49 |
H2787_18495 |
Rep-3 |
01051 |
MobA/L |
03389 |
1 |
– |
Toxin- antitoxin |
|
|
p2E072658 |
6018 |
R3-T15 |
H2787_18560 |
Rep-3 |
01051 |
MobA/L |
03389 |
1 |
– |
Toxin- antitoxin |
|
|
p1E072658 |
4483 |
RP-T6 |
H2787_18615 |
RepPriCT_1 |
03090 |
MobC MbeA |
05713 03432 |
– |
– |
Toxin- antitoxin |
*Letters in bold type indicate novel Reps identified in this study. na, Not applicable.
p2E072658 is a 6018 bp novel plasmid. It encodes the R3-T15 replication protein (belonging to the Rep_3 superfamily, pfam01051) (locus id H2787_18560; GenBank accession number CP061712; Table 2). p2E072658 also encodes the MobA/L mobilization proteins (belonging to pfam03389) and a BrnTA toxin–antitoxin system that is widely spread in Acinetobacter plasmids. p2E072658 contains one pdif module that encodes two hypothetical proteins (ORF module). We found several plasmids that contained segments of p2E072658 with 60–83% coverage (and DNA identities ranging from 92 to 97 %). These plasmids were from different Acinetobacter species and various environmental sources, and included p2_010062 (GenBank accession number CP033122.1) carried by Acinetobacter wuhouensis WCHAW010062 recovered in sewage (with 72 % coverage and 97 % DNA identity), p9_010034 (GenBank accession number CP032277.1) found in Acinetobacter sp. WCHAc010034 isolated in hospital sewage (66 % coverage and ~92 % DNA identity), p2014S01-097-4 (GenBank accession number CP033554.1) in Acinetobacter nosocomialis strain 2014S01-097 isolated in a clinical sample (50 % coverage and 83 % identity), pXMC5x702–5k (GenBank accession number CP084309.1) in Acinetobacter pseudolwoffii strain XMC5x702 recovered in chicken faeces (50 % coverage and ~91 % identity) and pAS80-5 (GenBank accession number CP061548.1) in Acinetobacter seifertii strain AS80 isolated in human blood. Notably, most sequence homologies indicated were around the segments encoding mobilization and replication functions – again indicating a wide dissemination of these modules beyond the species and source boundaries. The ORF pdif module with 99 % DNA identity was also found in two Acinetobacter plasmids – pApW20-3 (GenBank accession number CP027661.1) and pOCUAc18-3 (GenBank accession number AP024805.1) carried by Acinetobacter pittii strain Ap-W20 isolated from fish and A. baumannii strain OCU_Ac18 isolated from a patient’s blood in Japan, respectively [43].
p3E072658 is an 6763 bp plasmid that encodes a Rep3 family (R3-T49) replication protein, a MobA mobilization protein and a toxin–antitoxin protein required for usual plasmid stabilization. p3E072658 contains a pdif module that encodes three hypothetical proteins, one toxin and antitoxin system and a macrodomain-containing protein. Macrodomains are highly evolutionarily conserved proteins in all three domains of life, highlighting their fundamental roles in diverse biological functions. They are known to interact with ADP-ribose metabolites, but their specific functional role in bacteria is not fully understood. However, one study showed that macrodomain proteins recognize ADP-ribosylated protein targets by bacterial exotoxin in host cells [44]. An identical module to that in p3E072658 was detected only in A. towneri strain SCLZS30 in plasmid p1_SCLZS30 (GenBank accession number CP090385), isolated from wastewater in China. p3E072658 and p1_SCLZS30 shared no other region, indicating that this pdif module can move between plasmids.
p4E072658 (7989 bp) is an R3-T2 Rep protein (formerly known as RepAci2) with no mobilization function. Plasmids encoding R3-T2 are common in ST2 strains representing global clone 2, the most widespread clinical strains globally. p4E072658 contains three pdif modules, one module encoding a RelE/ParE toxin–antitoxin system, flanked by two other modules encoding a SulP on the left and three hypothetical proteins on the other side.
NCBI blast searches revealed several plasmids of clinical strains with segments identical to p4E072658: for instance, the SulP transporter region of several A. seifertii strains isolated from human clinical samples (e.g. pAS47-2 from A. seifertii AS47 recovered from a blood sample in Taiwan; GenBank accession number CP061631), indicating a link between these plasmids.
To date, several Acinetobacter plasmids have been described that do not encode a rep gene, including the small plasmid pRAY and the large conjugative plasmid pA297-3 [25]. p5E072658 (15545 bp) does not encode an identifiable rep gene but contains several genes encoding ABC membrane transporter genes. p5E072658 adds yet another plasmid type that needs further investigation to reveal its replication mode.
p6E072658 is an 18806 bp plasmid that encodes two Rep3 family Rep proteins, including an R3-T36 and a novel Rep called R3-T77. The r3-T77 rep gene (locus id: H2787_18280) does not share significant DNA identity with any known rep sequence typed to date. No pdif module was found. p6E072658 carries the fimD gene, fimC gene and a spore coat protein U domain-containing genes. These genes (with ~98% DNA identity) were found in pXM9F202-2-186k and pAV_175–3, two plasmids in Acinetobacter variabilis strain XM9F202-2 (CP060812), and in plasmid AV_175 (CP078029), respectively. The A. variabilis strains were isolated from hospital alcohol dispensers in China.
Together, analysis of cryptic plasmids of E-072658 show the presence of several distinct pdif modules, and other segments that are not part of a pdif, shared with several baumannii and non-baumannii Acinetobacter strains recovered in clinical sources and a wide range of environmental sources. This analysis provides clear evidence for the significant role of Acinetobacter plasmids in spreading genetic material, beyond antibiotic resistance determinants, involved in various biological and physiological processes.
A. baumannii genomes carrying multiple plasmids
To draw an overal picture of the number of plasmids carried by A. baumannii genomes, all complete genomes of this organism (n=423 as of mid-August 2022) were analysed. Analysis of these genomes showed that of the 423 genomes, 113 carried no plasmids, 273 carried three or fewer plasmids, and 35 complete A. baumannii genomes carried four or more plasmids (genomes with five or more in Table 3 and genomes with four plasmids in Table S1).
Table 3.
Properties of publicly available strains that contain five or more plasmids
|
Strain |
Date |
Source |
Country |
ST |
Plasmid name |
Size (kb) |
Rep |
Tra |
Mob |
Resistance genes |
Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
OCU_Ac18 |
2016 |
Blood |
Australia |
Novel |
pOCU_Ac18-1 |
90 090 |
R3-T44 |
– |
– |
– |
|
|
|
|
pOCU_Ac18-2 |
12 817 |
R3-T6 |
– |
MobAL |
– |
||||
|
|
|
pOCU_Ac18-3 |
10 831 |
R3-T67 |
– |
MobAL |
– |
||||
|
|
|
pOCU_Ac18-4 |
8948 |
R3-T24 |
– |
– |
– |
||||
|
|
|
pOCU_Ac18-5 |
7104 |
R3-T2 |
– |
MobAL |
– |
||||
|
|
|
pOCU_Ac18-6 |
5049 |
R3-T70 |
– |
MobA |
– |
||||
|
|
|
pOCU_Ac18-7 |
4293 |
R3-T79 |
– |
MobAL |
– |
||||
|
|
|
pOCU_Ac18-8 |
3948 |
R3-T71 |
– |
MobAL |
– |
||||
|
|
|
pOCU_Ac18-9 |
3496 |
R1-T8 |
– |
– |
– |
||||
|
|
|
pOCU_Ac18-10 |
2444 |
– |
– |
– |
– |
||||
|
|
|
pOCU_Ac18-11 |
2287 |
R3-T15 |
– |
– |
– |
||||
|
E47 |
2013 |
na |
Australia |
1547 |
pE47_001 |
327 867 |
– |
– |
– |
mph/msr(E), aac(3)-lld, cmlB1, sul1, catB3, aac(6')-lb4, bla lMP-4, dfrA19, sul1, bla CARB-2, aphA1 |
|
|
|
|
pE47_002 |
59 744 |
R3-T7 |
– |
– |
– |
||||
|
|
|
pE47_003 |
8795 |
R3-T66 |
– |
MobL |
– |
||||
|
|
|
pE47_004 |
7703 |
R3-T2 |
– |
MobAL |
– |
||||
|
|
|
pE47_005 |
5234 |
R3-T18 |
– |
MobL |
– |
||||
|
|
|
pE47_006 |
5039 |
– |
– |
MobAC |
– |
||||
|
|
|
pE47_007 |
4715 |
RP-T5 |
– |
MobC |
– |
||||
|
|
|
pE47_008 |
3065 |
R3-T55 |
– |
– |
– |
||||
|
|
|
pE47_009 |
2427 |
R3-T47 |
– |
– |
– |
||||
|
ACN21a |
2018 |
Blood |
India |
85 |
pACN-1 |
116 047 |
R3-T3 |
– |
– |
mph/msr(E), armA, sul1, blaCARB-2, blaOXA-420 |
|
|
|
|
pACN-2 |
57 333 |
R3-T60 |
– |
– |
aadB |
||||
|
|
|
pACN-3 |
9909 |
– |
– |
MobC |
– |
||||
|
|
|
pACN-4 |
9205 |
R3-T6 |
– |
MobAL |
aadB |
||||
|
|
|
pACN-5 |
7396 |
– |
– |
MobC |
aadB |
||||
|
|
|
pACN-6 |
6944 |
– |
– |
MobC |
aadB |
||||
|
|
|
pACN-7 |
5844 |
– |
– |
MobC |
– |
||||
|
|
|
pACN-8 |
5734 |
– |
– |
MobC |
mph/msr(E), armA, sul1, blaCARB-2, bla OXA-420 |
||||
|
E-072658 |
2009 |
Paper pulp mill |
Finland |
649 |
p8E072658 |
119 750 |
R3-T73 R3-T74 |
– |
– |
mcr-4.7 |
|
|
|
|
p7E072658 |
19 661 |
R3-T75 R3-T76 |
– |
MobAL |
sul2 |
||||
|
|
|
p6E072658 |
18 806 |
R3-T5 R3-T77 |
– |
– |
– |
||||
|
|
|
p5E072658 |
15 545 |
– |
– |
– |
– |
||||
|
|
|
p4E072658 |
7989 |
R3-T2 |
– |
– |
– |
||||
|
|
|
p3E072658 |
6763 |
R3-T49 |
– |
MobAL |
– |
||||
|
|
|
p2E072658 |
6018 |
R3-T15 |
– |
MobAL |
– |
||||
|
|
|
p1E072658 |
4483 |
RP-T6 |
– |
MobC |
– |
||||
|
VB2486 |
2019 |
Sputum |
India |
1 |
pVB2486_1 |
99 090 |
RP-T1 |
TraLEKBVCWUNFHG |
– |
bla OXA-23 |
|
|
|
|
pVB2486_2 |
14 906 |
R3-T11 |
– |
– |
– |
||||
|
|
|
pVB2486_3 |
12 574 |
R3-T23 |
– |
MobAL |
mph/msr(E) |
||||
|
|
|
pVB2486_4 |
5679 |
– |
TraD |
MobAL |
aphA6 |
||||
|
|
|
pVB2486_5 |
5432 |
R3-T56 |
TrbL |
MobAL |
aadA1 |
||||
|
|
|
pVB2486_6 |
2457 |
– |
– |
MobAL |
– |
||||
|
PM194188 |
2019 |
BAL |
India |
10 |
pPM194122_1 |
150 385 |
R3-T60 |
TraHY |
MobL |
mph/msr(E), armA, sul1, cmlA5, arr-2, sul2, strB, aph(3'')-lb, ble-MBL, bla NDM-1, sul2, tet(B) |
|
|
|
|
pPM194122_2 |
18 783 |
R3-T11 |
– |
– |
– |
||||
|
|
|
pPM194122_3 |
7695 |
R3-T17 |
– |
– |
– |
||||
|
|
|
pPM194122_4 |
7540 |
R3-T1 |
– |
– |
– |
||||
|
|
|
pPM194122_5 |
2762 |
R3-T30 |
– |
– |
– |
||||
|
|
|
pPM194122_6 |
2419 |
– |
– |
– |
– |
||||
|
VB958a |
2019 |
Blood |
India |
Novel |
pVB958-1 |
561 419 |
– |
– |
– |
– |
|
|
|
|
pVB958-2 |
538 712 |
– |
– |
– |
bla OXA-23, carO |
||||
|
|
|
pVB958-3 |
82 500 |
RP-T1 |
TraLEKBVCW NFHG, TrbC |
– |
blaOXA-23 |
||||
|
|
|
pVB958-4 |
47 143 |
– |
– |
– |
– |
||||
|
|
|
pVB958-5 |
16 490 |
R3-T11 |
– |
– |
– |
||||
|
|
|
pVB958-6 |
16 485 |
R3-T11 |
– |
– |
– |
||||
|
Ab-D10a-a |
2016 |
Spinal Fluid |
Ghana |
103 |
pAb-D10a-a_1 |
48 239 |
R3-T20 |
– |
MobC |
dfrA1, mph/msr(E), tet39, aacC2d |
|
|
|
|
pAb-D10a-a_2 |
8495 |
R3-T10 |
– |
– |
– |
||||
|
|
|
pAb-D10a-a_3 |
8215 |
– |
TraD |
MobAL |
bla OXA-23 |
||||
|
|
|
pAb-D10a-a_4 |
6619 |
Novel |
– |
– |
– |
||||
|
|
|
pAb-D10a-a_5 |
2697 |
R1-T3 |
– |
– |
– |
||||
|
A85 |
2003 |
Sputum |
Australia |
1 |
pA85-3 |
86 334 |
RP-T1 |
TraALEKBVCWUNFHG, TrwBC,TrbC |
– |
bla OXA23 |
|
|
|
|
pA85-2 |
8731 |
R3-T1 |
– |
– |
– |
||||
|
|
|
pA85-1b |
4484 |
– |
TraD |
MobA |
dfrA44 |
||||
|
|
|
pA85-1a |
2726 |
R1-T2 |
– |
– |
– |
||||
|
|
|
pA85-1 |
2343 |
R1-T1 |
– |
– |
– |
||||
|
PM193665 |
2019 |
Wound |
India |
10 |
pPM193665_1 |
150 385 |
R3-T60 |
TraHY |
MobL |
mph/msr(E), armA, sul1, cmlA5, arr-2, sul2, strB, aph(3'')-lb, ble-MBL, bla NDM-1, sul2, tet(B) |
|
|
|
|
pPM193665_2 |
52 509 |
– |
– |
– |
– |
||||
|
|
|
pPM193665_3 |
18 783 |
R3-T11 |
– |
– |
– |
||||
|
|
|
pPM193665_4 |
7540 |
R3-T1 |
– |
– |
– |
||||
|
|
|
pPM193665_5 |
2762 |
R3-T30 |
– |
– |
– |
||||
|
A52 |
2015 |
Sputum |
China |
77 |
pA52-1 |
110 713 |
R3-T3 |
– |
– |
– |
|
|
|
|
pA52-2 |
27 452 |
R3-T27 |
TraA |
– |
– |
||||
|
|
|
pA52-5 |
8493 |
R3-T24 |
– |
– |
bla OXA72 |
||||
|
|
|
pA52-3 |
8420 |
– |
– |
– |
– |
||||
|
|
|
pA52-4 |
3610 |
R3-T58 |
– |
MobL |
– |
||||
|
DS002 |
2015 |
Soil |
India |
novel |
pTS134338 |
134 338 |
R3-T7 |
– |
– |
– |
|
|
|
|
pTS37365 |
37 365 |
– |
– |
– |
– |
||||
|
|
|
pTS11291 |
11 291 |
R3-T2 |
– |
MobA |
– |
||||
|
|
|
pTS9900 |
9900 |
R3-T6 |
– |
MobA |
– |
||||
|
|
|
pTS4586 |
4586 |
– |
– |
MobA |
– |
*Named here. na, Not applicable.
Plasmid incompatibility is known mainly due to two plasmids using the same replication machinery [45]. In fact, incompatibility generally occurs when copy number control elements from closely related plasmids act in trans to interfere with one another’s stable maintenance [46]. Almost all genomes with multiple plasmids were found to carry multiple R3-type (Rep3; Pfam01051) plasmids, suggesting they were compatible. The shared properties (the same Rep type and 122 bp iteron ~50 bp upstream of rep) of the Rep3 family plasmids and that there are many genomes with several Rep3 plasmid types raise the question of whether Acinetobacter plasmids, and in particular diverged Rep3-type plasmids, are compatible. Interestingly, no genome with more than one RP-type (encoding RepPriCT_1 type; Pfam03090) plasmid was found. While this might be coincidental, it does indicate that multiple R3-type plasmids can be stably maintained in the same cell or with other R1- or RP-type plasmids. Notably, only one strain was found (A85 recovered in Australia in 2003) that contained two R1-type (encoding R1 replication protein; Pfam01446) plasmids (GenBank accession numbers CP021783 and CP021784) (Tables 1 and S1). Although this provides clues to the incompatibility of A. baumannii plasmids, it does warrant extensive experimental work to establish the ability or inability of various plasmid types to be stably maintained in the same cell. Analysis of complete plasmids also showed many plasmids with no identifiable rep gene (Tables 1 and S1), raising the need to establish their replication mode. In addition, many small plasmids, mainly R3 type, were found to encode mobilization functions (Mob), suggesting they might be mobilizable.
Conclusion
Resistance to last-resort antibiotics such as colistin leaves limited to no option to treat A. baumannii infections, given that strains causing infections in hospitals are frequently resistant to several additional antibiotics. To date, most studies have focused on analysing clinical strains, while the origin of many clinically significant resistance genes remains poorly understood. Here, we described an environmental A. baumannii strain recovered in recycled fibre pulp in a Finnish paper mill and showed that it carries eight novel plasmids, one containing a novel colistin resistance gene (mcr-4.7), which is associated with high levels of the colistin resistance phenotype. Moreover, the strain analysed here contained seven additional novel plasmids, analysis of which revealed several shared modules with Acinetobacter plasmids carried by clinical strains, indicating the exchange of genetic material beyond species and environmental niches. This study highlights the role of the environment, and environmental strains, as potential reservoirs for clinically significant antibiotic resistance genes and that to better understand the emergence of resistance, more attention should be given to studying and analysing strains recovered in non-clinical niches.
Supplementary Data
Funding information
A.K.C. is supported by an Australian Research Council Future Fellowship (FT220100152). M.H. is supported by an Australian Research Council Discovery Early Career Research Award (DE200100111). I.T.P. is supported by an ARC Laureate Fellowship (FL140100021).
Author contributions
F.P., R.M. and L.T.: methodology, formal analysis. A.K.C., I.P. and M.H.: conceptualization, funding, review and editing, funding, methodology, and writing – review and editing. M.H.: writing – original draft.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Abbreviations: ARG, antibiotic resistance gene; BREX, bacteriophage exclusion system; IS, insertion sequence; LPS, lipopolysaccharide; MGE, mobile genetic element; MIC, minimum inhibitory concentration.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary figures and one supplementary table are available with the online version of this article.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roca I, Espinal P, Vila-Farrés X, Vila J. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol. 2012;3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii . Microb Genom. 2019;5:10. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling Z, Yin W, Shen Z, Wang Y, Shen J, et al. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother. 2020;75:3087–3095. doi: 10.1093/jac/dkaa205. [DOI] [PubMed] [Google Scholar]
- 5.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii . J Bacteriol. 2008;190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO World Health Organisation (WHO); 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. [Google Scholar]
- 7.Hamouda A, Findlay J, Al Hassan L, Amyes SGB. Epidemiology of Acinetobacter baumannii of animal origin. Int J Antimicrob Agents. 2011;38:314–318. doi: 10.1016/j.ijantimicag.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Adewoyin MA, Okoh AI. The natural environment as a reservoir of pathogenic and non-pathogenic Acinetobacter species. Rev Environ Health. 2018;33:265–272. doi: 10.1515/reveh-2017-0034. [DOI] [PubMed] [Google Scholar]
- 9.Al Atrouni A, Hamze M, Rafei R, Eveillard M, Joly-Guillou ML, et al. Diversity of Acinetobacter species isolated from different environments in Lebanon: a nationwide study. Future Microbiol. 2016;11:1147–1156. doi: 10.2217/fmb-2016-0082. [DOI] [PubMed] [Google Scholar]
- 10.Dekić S, Klobučar G, Ivanković T, Zanella D, Vucić M, et al. Emerging human pathogen Acinetobacter baumannii in the natural aquatic environment: a public health risk? Int J Environ Health Res. 2018;28:315–322. doi: 10.1080/09603123.2018.1472746. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlén J, Morris WF. Predicting changes in the distribution and abundance of species under environmental change. Ecol Lett. 2015;18:303–314. doi: 10.1111/ele.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furlan JPR, de Almeida OGG, De Martinis ECP, Stehling EG. Characterization of an environmental multidrug-resistant Acinetobacter seifertii and comparative genomic analysis reveals co-occurrence of antimicrobial resistance and metal tolerance determinants. Front Microbiol. 2019;10:2151. doi: 10.3389/fmicb.2019.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Garcera M, Touchon M, Brisse S, Rocha EPC. Metagenomic assessment of the interplay between the environment and the genetic diversification of Acinetobacter . Environ Microbiol. 2017;19:5010–5024. doi: 10.1111/1462-2920.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrenovic J, Durn G, Goic-Barisic I, Kovacic A. Occurrence of an environmental Acinetobacter baumannii strain similar to a clinical isolate in paleosol from Croatia. Appl Environ Microbiol. 2014;80:2860–2866. doi: 10.1128/AEM.00312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Repizo GD, Viale AM, Borges V, Cameranesi MM, Taib N, et al. The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and Niche range of a predominant nosocomial pathogen. Genome Biol Evol. 2017;9:2292–2307. doi: 10.1093/gbe/evx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernández-González IL, Castillo-Ramírez S. Antibiotic-resistant Acinetobacter baumannii is a one health problem. Lancet Microbe. 2020;1:e279. doi: 10.1016/S2666-5247(20)30167-1. [DOI] [PubMed] [Google Scholar]
- 17.Hamidian M, Hall RM. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 - structure, origin and evolution. Drug Resist Updat. 2018;41:26–39. doi: 10.1016/j.drup.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Holt K, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, et al. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom. 2016;2:e000052. doi: 10.1099/mgen.0.000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Salgado-Camargo AD, Castro-Jaimes S, Gutierrez-Rios R-M, Lozano LF, Altamirano-Pacheco L, et al. Structure and evolution of Acinetobacter baumannii plasmids. Front Microbiol. 2020;11:1283. doi: 10.3389/fmicb.2020.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brovedan MA, Cameranesi MM, Limansky AS, Morán-Barrio J, Marchiaro P, et al. What do we know about plasmids carried by members of the Acinetobacter genus? World J Microbiol Biotechnol. 2020;36:109. doi: 10.1007/s11274-020-02890-7. [DOI] [PubMed] [Google Scholar]
- 22.Hamidian M, Hall RM. Genetic structure of four plasmids found in Acinetobacter baumannii isolate D36 belonging to lineage 2 of global clone 1. PLoS One. 2018;13:e0204357. doi: 10.1371/journal.pone.0204357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamidian M, Ambrose SJ, Hall RM. A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid. 2016;87–88:43–50. doi: 10.1016/j.plasmid.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Lean SS, Yeo CC. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: Good, Bad, Who Knows? Front Microbiol. 2017;8:1547. doi: 10.3389/fmicb.2017.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam MMC, Koong J, Holt KE, Hall RM, Hamidian M. Detection and typing of plasmids in Acinetobacter baumannii using rep genes encoding replication initiation proteins. Microbiology. 2022;2022 doi: 10.1101/2022.08.26.505409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balalovski P, Grainge I. Mobilization of pdif modules in Acinetobacter: a novel mechanism for antibiotic resistance gene shuffling? Mol Microbiol. 2020;114:699–709. doi: 10.1111/mmi.14563. [DOI] [PubMed] [Google Scholar]
- 27.Blackwell GA, Hall RM. The tet39 determinant and the msre-mphe genes in acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (xerc-xerd) sites. Antimicrob Agents Chemother. 2017;61:e00780–17. doi: 10.1128/AAC.00780-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suihko M-L, Skyttä E. Characterisation of aerobically grown non-spore-forming bacteria from paper mill pulps containing recycled fibres. J Ind Microbiol Biotechnol. 2009;36:53–64. doi: 10.1007/s10295-008-0472-0. [DOI] [PubMed] [Google Scholar]
- 29.Hamidian M, Maharjan RP, Farrugia DN, Delgado NN, Dinh H, et al. Genomic and phenotypic analyses of diverse non-clinical Acinetobacter baumannii strains reveals strain-specific virulence and resistance capacity. Microb Genom. 2022;8:000765. doi: 10.1099/mgen.0.000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell SM, Smith DD. The CDS disc method of antibiotic sensitivity testing (Calibrated dichotomous sensitivity test) Pathology. 1975;7:1–48. doi: 10.3109/00313027509082602. [DOI] [PubMed] [Google Scholar]
- 31.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 32.CLSI Performance Standards for Antimicrobial Susceptibility Testing; 29th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 33.Shabbir MAB, Shabbir MZ, Wu Q, Mahmood S, Sajid A, et al. CRISPR-cas system: biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann Clin Microbiol Antimicrob. 2019;18:21. doi: 10.1186/s12941-019-0317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffatt JH, Harper M, Adler B, Nation RL, Li J, et al. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii . Antimicrob Agents Chemother. 2011;55:3022–3024. doi: 10.1128/AAC.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mmatli M, Mbelle NM, Osei Sekyere J. Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: a current and emerging update. Front Cell Infect Microbiol. 2022;12:941358. doi: 10.3389/fcimb.2022.941358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poirel L, Kieffer N, Nordmann P. In vitro study of isapl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cameranesi MM, Kurth D, Repizo GD. Acinetobacter defence mechanisms against biological aggressors and their use as alternative therapeutic applications. Crit Rev Microbiol. 2022;48:21–41. doi: 10.1080/1040841X.2021.1939266. [DOI] [PubMed] [Google Scholar]
- 39.Martins-Sorenson N, Snesrud E, Xavier DE, Cacci LC, Iavarone AT, et al. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J Antimicrob Chemother. 2020;75:60–64. doi: 10.1093/jac/dkz413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafei R, Koong J, Osman M, Al Atrouni A, Hamze M, et al. Analysis of pCl107 a large plasmid carried by an ST25 Acinetobacter baumannii strain reveals a complex evolutionary history and links to multiple antibiotic resistance and metabolic pathways. FEMS Microbes. 2022;3:xtac027. doi: 10.1093/femsmc/xtac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran RA, Liu H, Doughty EL, Hua X, Cummins EA, et al. GR13-type plasmids in Acinetobacter potentiate the accumulation and horizontal transfer of diverse accessory genes. Microb Genom. 2022;8:mgen000840. doi: 10.1099/mgen.0.000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nigro SJ, Hall RM. GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J Antimicrob Chemother. 2011;66:2175–2176. doi: 10.1093/jac/dkr230. [DOI] [PubMed] [Google Scholar]
- 43.Sakiyama A, Matsumoto Y, Tsubouchi T, Suzuki M, Niki M, et al. Complete genome sequence of a clinical isolate of Acinetobacter baumannii harboring 11 plasmids. Microbiol Resour Announc. 2021;10:39. doi: 10.1128/MRA.00695-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han W, Li X, Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/MMBR.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carretto E, Brovarone F, Nardini P, Russello G, Barbarini D, et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. 2018;23:17–00821. doi: 10.2807/1560-7917.ES.2018.23.2.17-00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kostyanev T, Xavier BB, García-Castillo M, Lammens C, Bravo-Ferrer Acosta J, et al. Phenotypic and molecular characterizations of carbapenem-resistant Acinetobacter baumannii isolates collected within the EURECA study. Int J Antimicrob Agents. 2021;57:106345. doi: 10.1016/j.ijantimicag.2021.106345. [DOI] [PubMed] [Google Scholar]
- 49.Teo JWP, Kalisvar M, Venkatachalam I, Ng OT, Lin RTP, et al. mcr-3 and mcr-4 variants in carbapenemase-producing clinical enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol. 2018;56:e01562-17. doi: 10.1128/JCM.01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.