Abstract
Circular RNA (circRNA) is a class of endogenous non-coding RNA, a type of single-stranded covalently closed RNA molecule formed by alternative splicing of exons or introns. Previous studies have demonstrated that circRNA participates in modulating biological processes such as cell proliferation, differentiation and apoptosis, and plays key roles in tumor occurrence and development. CircRNA nuclear receptor interacting protein 1 (circ_NRIP1), a form of circRNA, is abnormally expressed in certain human tumor types. It is present at a higher abundance compared with cognate linear transcripts and can regulate malignant biological behaviors such as tumor proliferation, invasion and migration, revealing a currently unexplored frontier in cancer progression. The present review presents a pattern of circ_NRIP1 expression in various malignant tumor types and highlights its significance in cancer development, in addition to its potential as a disease indicator or future therapeutic agent.
Keywords: circular RNA, circular RNA nuclear receptor interacting protein 1, tumor development, therapeutic potential
1. Introduction
Circular RNA (circRNA) is a recent addition to an expanding list of non-coding RNAs. CircRNA is produced through a non-canonical splicing mechanism called back splicing, which involves the covalent linkage of the downstream splice-donor site to its upstream counterpart (1,2). Despite the absence of polyadenylation and capping, circRNAs exhibit a remarkable tolerance to RNase and a greater stability compared with linear RNAs due to their covalently closed circular structure (3,4). Over the previous two decades, scientists have identified circRNAs in plant viroids, yeast mitochondrial RNAs and hepatitis D virus, and subsequently in the human Ets1 gene and the mouse Sry gene (5–9). A small number of circRNAs derived from eukaryotic genomes have been discovered and are thought to arise from abnormal splicing events, lacking any discernible regulatory functions (10,11). To date, the utilization of RNA-seq and newly developed bioinformatic approaches have facilitated the identification of >10,000 different circRNAs in various organisms (3). However, biological functions have only been investigated for a minor fraction of the circRNAs identified, the majority of which have been proposed to act as miRNA sponges (12–15). In addition, crosslinking immunoprecipitation data sets suggest that circRNAs interact with numerous different RNA-binding proteins to act as protein sponges (16–19). Furthermore, a subset of circRNAs may undergo cap-independent translation under specific conditions (20–22). Taken together, these findings suggest a close association between circRNAs and both biological and pathological processes.
CircRNA nuclear receptor interacting protein 1 (circ_NRIP1) is a 203 bp circRNA (circBase ID: hsa_circ_0004771) derived from exons 2 and 3 of the NRIP1 host gene which is located at human chromosome 21q11.2 (Fig. 1). Several studies in previous years have demonstrated that circ_NRIP1 can reinforce proliferation and extracellular matrix accumulation in keloid-derived fibroblasts and impede apoptosis (23). Additionally, circ_NRIP1 can also mediate alcohol dependence through the modulation of neuron projection and axon regeneration (24). Further research has revealed that abnormal expression of circ_NRIP1 is an independent predictor of poor prognosis in ovarian and gastric cancer (25–30). This manuscript offers a thorough review of research on the oncogenic functions, regulatory mechanisms and therapeutic potential of circ_NRIP1 in malignant tumors affecting humans (Tables I and II).
Figure 1.
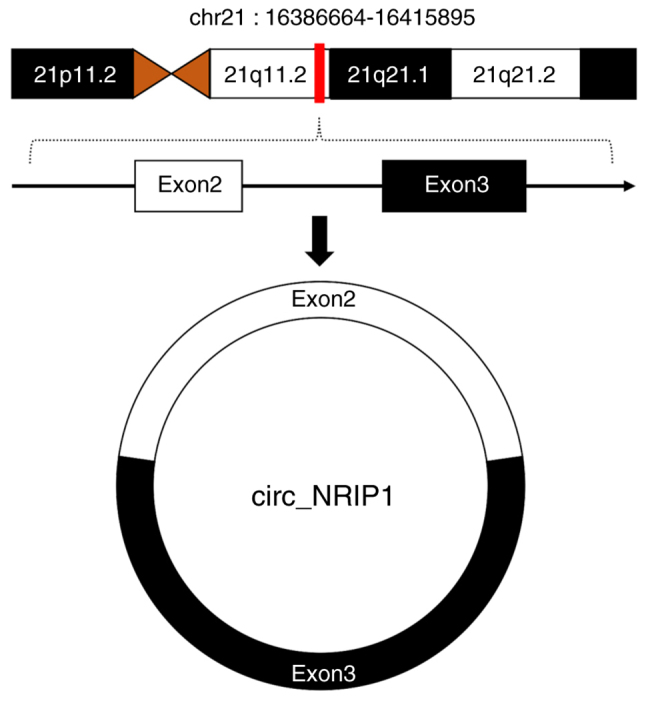
Chromosomal location of circ_NRIP1.
Table I.
Characterization of circ_NRIP1 in cancers.
| First author, year | Cancer type | circ_NRIP1 regulation | Role | Clinicopathologic features | Reported drug resistance | (Refs.) |
|---|---|---|---|---|---|---|
| Li, 2021 | OC | Increased | Onco- | - | Paclitaxel | (40) |
| Li, 2020 | CC | Increased | Onco- | Lymph node metastasis, advanced FIGO stage | - | (49) |
| Pan, 2019, Liu, 2020 | CRC | Increased | Onco- | Diagnostic biomarker, advanced TNM stage, poor overall survival | 5-Fluorouracil | (59,60) |
| Huang, 2020, Zhou, 2021 | ESCC | Increased | Onco- | Diagnostic biomarker | - | (61,63) |
| Zhang, 2019, Liu, 2020, Xu, 2020 | GC | Increased | Onco- | Advanced TNM stage, poor disease-free survival | - | (67,70,71) |
| Ding, 2021, Xie, 2019 | BC | Increased | Onco- | Advanced TNM stage, poor overall survival | - | (74,77) |
| Lin, 2021 | NPC | Increased | Onco- | - | Cisplatin | (85) |
| Li, 2021, Fu, 2022 | PTC | Increased | Onco- | Advanced TNM stage, poor overall survival | - | (86,90) |
| D'Ambrosi, 2021 | NSCLC | Decreased | - | Negative correlation with TNM, Diagnostic biomarker | - | (98) |
| Meng, 2021, Shi, 2021 | OS | Increased | Onco- | Advanced TNM stage, poor overall survival | - | (111,113) |
OC, ovarian cancer; CC, cervical cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell cancer; GC, gastric cancer; BC, breast cancer; NPC, nasopharyngeal carcinoma; PTC, papillary thyroid carcinoma; NSCLC, non-small cell lung cancer; OS, osteosarcoma; Onco-, oncogenic.
Table II.
Regulatory mechanism of circ_NRIP1 in certain cancer types.
| Cancer type | Biological function impacted by circ_NRIP1 | Targeted miRNA | Downstream molecules impacted | Year | (Refs.) |
|---|---|---|---|---|---|
| OC | P ↑ M ↑ I ↑ CR ↑ | miR-211-5p | HOXC8, cyclin D1, MMP2, MMP9 | 2020 | (40) |
| CC | P ↑ M ↑ I ↑ | miR-629-3p | PTP4A1, ERK1, ERK2 | 2020 | (49) |
| CRC | P ↑ M ↑ I ↑ CR↑ | miR-532-3p | - | 2021 | (60) |
| ESCC | P ↑ M ↑ I ↑ A ↑ EMT ↑ | miR-595 | SEMA4D, SNAI1 | 2021 | (63) |
| GC | P ↑ M ↑ I ↑ | miR-149-5p | AKT1 | 2019 | (67) |
| P ↑ M ↑ I ↑ G ↑ | miR-186-5p | MYH9 | 2020 | (70) | |
| P ↑ M ↑ I ↑ G ↑ | miR-138-5p | - | 2020 | (71) | |
| BC | P ↑ M ↑ I ↑ AG ↑ | miR-1253 | DDAH1 | 2021 | (74) |
| P ↑ M ↑ I ↑ EMT ↑ | miR-653 | ZEB2 | 2019 | (77) | |
| NPC | P ↑ M ↑ I ↑ CR ↑ | miR-515-5p | IL-25 | 2021 | (85) |
| PTC | P ↑ M ↑ I ↑ A ↑ | miR-195-5p | JAK2, STAT1, p38 | 2021 | (86) |
| miR-653-5p | PBX3 | 2022 | (90) | ||
| OS | P ↑ M ↑ I ↑ | miR-199a | FOXC2 | 2021 | (111) |
| miR-532-3p | AKT3 | 2021 | (113) |
OC, ovarian cancer; CC, cervical cancer, CRC, colorectal cancer; ESCC, esophageal squamous cell cancer; GC, gastric cancer; BC, breast cancer; NPC, nasopharyngeal carcinoma; PTC, papillary thyroid carcinoma; OS, osteosarcoma; P, proliferative capacity of tumor cells; M, migration capacity of tumor cells; I, invasive capacity of tumor cells; A, apoptosis of tumor cells; CR, chemoresistance; G, glycolysis; AG, Angiogenesis; EMT, epithelial-mesenchymal transition; HOXC8, homeobox C8; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; PTP4A1, protein tyrosine phosphatase 4A1; ERK1, extracellular regulated protein kinase1; ERK2, extracellular regulated protein kinase2; SEMA4D, semaphoring 4D; SNAI1, snail family transcriptional repressor 1; AKT1, AKT serine/threonine kinase 1; MYH9, myosin heavy chain 9; DDAH1, dimethylarginine dimethylaminohydrolase 1; ZEB2, zinc finger E-box binding homeobox 2; IL-25, interleukin 25; JAK2, Janus kinase 2; STAT1, signal transducer and activator of transcription 1; p38, p38 kinase; PBX3, pre-B-cell leukemia homeobox 3; FOXC2, forkhead box C2; AKT3, AKT serine/threonine kinase 3.
2. Circ_NRIP1 in malignant tumors
Ovarian cancer
Ovarian cancer is the most lethal gynecological malignancy, which is characterized by a rapid clinical course, high mortality rate and poor prognosis (31). Annually worldwide, 230,000 women will be diagnosed and 150,000 will succumb (32). Currently, chemotherapy is still the cornerstone of ovarian cancer treatment (33). Advanced stage ovarian cancers commonly present as sensitive to initial therapy; however, most (~75%) recur and develop resistance to chemotherapy (34,35). CircRNA has previously been identified as a primary regulator of initiation, progression and therapeutic response in certain cancers, including ovarian cancer (36). CircRNA is expected to be a potential target for overcoming resistance to cancer chemotherapy (37–39).
Li et al (40) demonstrated that circ_NRIP1 is significantly overexpressed in ovarian cancer tumor tissues. Similar results have also been observed in RT-qPCR detection in ovarian cancer cell lines (SKOV3). Notably, circ_NRIP1 expression levels in tumor tissues from paclitaxel (PTX)-resistant patients are markedly higher compared with those sensitive to PTX treatment. To investigate the roles of circ_NRIP1 in PTX-resistant ovarian cancer, the expression of circ_NRIP1 in SKOV3/PTX cells (PTX treated SKOV3 cells) was reduced by transfection with small interfering (siRNA). MTT, colony formation and Transwell assay results indicate that circ_NRIP1 knockdown prominently suppresses proliferation, migration and invasion of SKOV3/PTX cells. Western blot results demonstrate that circ_NRIP1-silencing suppresses the expression of proliferation marker cyclin D1 and metastasis markers MMP2 and MMP9, which is consistent with functional assay data (40).
To elucidate the impact of circ_NRIP1 on ovarian cancer, researchers have generated xenograft models in nude mice with either negative control or circ_NRIP1 knockdown cells. The results revealed that circ_NRIP1 suppression can impair xenograft tumor growth and augment the responsiveness of ovarian cancer to PTX in vivo (40). As the competitive endogenous RNA (ceRNA) regulatory network is frequently cited as the molecular mechanism of circRNA, researchers have postulated that circ_NRIP1 induces chemotherapy resistance in ovarian cancer through this competing ceRNA mechanism (41). LncBase has been used to predict the miRNA targets for circ_NRIP1, revealing the presence of miR-211-5p binding sites within circ_NRIP1 (40). The RT-qPCR detection and dual-luciferase reporter assay outcomes further verify that there is a targeted regulatory relationship between circ_NRIP1 and miR-211-5p. Emerging evidence has suggested that homeobox C8 (HOXC8) is aberrantly overexpressed in malignant tumors and is intimately associated with chemotherapy resistance (42,43). Through subsequent experiments, researchers have determined that HOXC8 gene is downstream of miR-211-5p. Functional assays indicate that overexpression of HOXC8 can promote malignant behaviors in SKOV3/PTX cells, which can be partially mitigated by silencing circ_NRIP1 or transfection with miR-211-5p mimics transfection. Therefore, circ_NRIP1 may modulate PTX resistance in ovarian cancer via the miR-211-5p/HOXC8 axis. Blocking this axis with suitable inhibitors or target molecules produced through bioengineering may be a promising future strategy to overcome chemotherapy resistance in ovarian cancer.
Cervical cancer
Despite being highly preventable, cervical cancer remains one of the leading causes of cancer-related mortality in women globally (44). Patients with advanced cervical cancer typically undergo treatment strategies that are associated with considerable side effects and a limited efficacy (45,46). Therefore, there is a pressing need for improved cervical cancer therapeutics such as targeted therapy and immunotherapy, which offer greater efficacy and suitability compared with current treatment strategies. Numerous studies have highlighted the importance of aberrant gene expression in tumor development, often coinciding with the altered expression of circRNAs (47,48). Notably, cervical cancer exhibits a dysregulated circRNA expression profile as revealed by microarray analysis, with circ_NRIP1 demonstrating a significant upregulation (49).
Successive studies have since been carried out to investigate the relationship between circ_NRIP1 and cervical cancer (49). Li et al (49) collected 40 cervical cancer tumor samples and compared these with matched healthy samples to evaluate circ_NRIP1 expression in cervical cancer. The results indicated that circ_NRIP1 is notably expressed in neoplasm tissues and its aberrant expression profiles are closely associated with lymph-vascular space invasion and advanced FIGO (International Federation of Gynecology and Obstetrics) stage (stage II). Functionally, siRNA-mediated silencing of circ_NRIP1 can markedly dampen proliferative, migration and invasive ability of cervical cancer cells Moreover, in instances where circ_NRIP1 is overexpressed in tumor cells, the malignant biological behavior of tumor cells, such as proliferation, migration and invasion, were reinforced, highlighting the numerous potential oncogenic roles of circ_NRIP1 (49). The crosstalk that occurs between circRNA and miRNA is an emerging research theme in cancer progression. Bioinformatic prediction data suggests that miR-629-3p is the downstream target of circ_NRIP1. Dual-luciferase reporter and RNA binding protein immunoprecipitation (RIP) assay results provide further evidence to verify this interaction (49). Additionally, dysfunction of the protein tyrosine phosphatase 4A1 (PTP4A1)/ERK signaling pathway has previously been implicated in human malignancies, such as breast cancer and colorectal cancer, where it plays a crucial role in the occurrence and development of tumors (50–53). Based on data presented in StarBase, PTP4A1 has been found to contain binding sites for miR-629-3p (49). Subsequently, western blot analysis and dual-luciferase reporter assays have been performed to confirm that circ_NRIP1 serves as a ceRNA by sequestering miR-629-3p through complementary binding, thus counteracting the suppressive effects of miR-629-3p on PTP4A1 (49). In vitro experiments also indicate that exogenous overexpression of PTP4A1 partially rescues the inhibition effects of si-circ_NRIP1 on tumor cells, via increasing the expression of ERK. Largely, the current treatments of cervical cancer remain inefficient, therefore targeted therapy based on the circ_NRIP1/miR-629-3p/PTP4A1 axis should be considered in future cervical cancer management.
Colorectal cancer
Colorectal cancer (CRC) is the fourth most deadly cancer in the world, largely due to its non-specific symptoms and late-stage diagnosis, resulting in a 5-year relative survival rate of 14% (54). However, the 5-year survival rate for localized CRC is much higher at 90% (55,56). This factor has spurred great efforts to develop screening tools that allow for earlier detection of CRC. Previous studies have demonstrated that cancer cells may release exosomes containing circRNA into peripheral blood (57,58). Pan et al (59) collected serum samples from 135 patients with CRC and compared these with samples from 45 healthy controls (HCs). An upregulation in the expression of circ_NRIP1 in exosomes isolated from serum samples has been consistently observed in CRC patients compared with HCs, as demonstrated by RT-qPCR detection (59). Receiver operating characteristic (ROC) curve analysis data has demonstrated that the area under the curve to discriminate CRC patients from HCs is 0.92 with a sensitivity and specificity of 81.43 and 80%, respectively. These results suggest that circ_NRIP1 may be a promising new biomarker for early detection of CRC (59). In addition, Liu et al (60) have reported that circ_NRIP1 is significantly overexpressed in CRC tumor tissues compared with healthy specimens.
With respect to clinicopathological data, the abnormal expression of circ_NRIP1 is closely associated with lymph node invasion, distant metastasis and tumor, node and metastasis (TNM) stage. Moreover, Kaplan-Meier survival analysis data indicates that high expression levels of circ_NRIP1 are associated with poorer overall survival in CRC cases. To further investigate the roles of circ_NRIP1 in CRC, two specific siRNAs were synthesized to silence circ_NRIP1 expression. Subsequently, Cell Counting Kit-8 and Transwell assay results demonstrated that circ_NRIP1 knockdown significantly suppresses viability and motility of SW480 tumor cells. Collectively, these findings suggest that circ_NRIP1 functions as an oncogene in CRC (60).
Although 5-fluorouracil (5-FU) is a crucial component of systemic chemotherapy for CRC, its effectiveness is often limited by the development of chemoresistance. To determine whether circ_NRIP1 contributes to CRC 5-FU resistance, SW480 cells were transfected with si-circ_NRIP1 or si-NC, followed by treatment with varying concentrations of 5-FU. Functional assay data confirmed that siRNA mediates inhibition of circ_NRIP1 markedly increased SW480 cell sensitivity to 5-FU, meanwhile, a reduction in the IC50 values of 5-FU has been observed (60). Therefore, circ_NRIP1 serves as a ceRNA to sequester miR-532-3p through a complementary base pairing mechanism, thereby abrogating the miR-532-3p-mediated inhibitory effect on 5-FU resistant CRC.
Esophageal squamous cell cancer
Huang et al (61) demonstrated that circ_NRIP1 is highly upregulated in esophageal squamous cell cancer (ESCC) tumor specimens compared with non-tumor tissues. The aforementioned study also assessed the expression of circ_NRIP1 in peripheral blood, revealing an upregulation of circ_NRIP1 in plasma samples of patients with ESCC compared with matched healthy controls. Meanwhile, a marked decrease in circ_NRIP1 expression has been observed in post-operative samples from patients with ESCC (61). Notably, no significant change in plasma circ_NRIP1 expression has been detected after prolonged storage of plasma at 4°C or following repeated freeze-thaw cycles. This suggests that circ_NRIP1 is stable under these conditions and meets the basic requirements i) It must be produced by malignant tumor cells and can be detected in blood, tissue fluid, secretions or tumor tissue; ii) the content is low in normal tissues or benign tumors; iii) the tumor marker of a certain tumor can be detected in most patients with the tumor; iv) it can be detected before there is a clear clinical diagnosis of the tumor; v) the amount of tumor markers can reflect the size of the tumor; and vi) to some extent, it can help estimate the treatment effect and predict the recurrence and metastasis of the tumor.) to serve as a serum marker for ESCC (62). Moreover, ROC analysis further illustrates that circ_NRIP1 provides a less invasive diagnostic test for ESCC with an improved specificity and positive predictive value compared with serum CEA testing (61). These findings lay the foundation for the potential use of circ_NRIP1 as a plasma biomarker for ESCC. In addition, Zhou et al (63) confirm that si-circ_NRIP1 transfection markedly restrains proliferation, invasion and migration of ESCC cells. Mechanistically, circ_NRIP1 acts as a ceRNA to promote the expression of the oncogene semaphorin 4D (SEMA4D) by targeting miR-595 through base-pairing. Additionally, upregulation of SEMA4D can facilitate malignant behaviors in ESCC cells by activating the PI3K/AKT signaling pathway and inhibiting apoptosis and G0/G1 cell cycle arrest in tumor cells. Epithelial-mesenchymal transition (EMT) is a reversible biological process in which epithelial cells lose their polarity and tight junction contacts leading to a mesenchymal phenotype characterized by increased motility and invasiveness (63). EMT has previously been demonstrated to play a crucial role in the metastasis of cancer cells (64–66). Western blot analysis results verify that SEMA4D overexpression can increase snail family transcriptional repressor 1 expression, the master regulator of EMT (63). Broadly speaking, ESCC is a challenging and biologically heterogenous disease that requires the development of novel diagnostic, therapeutic and prognostic markers to improve current treatment options. Investigating the emerging role of circ_NRIP1 in these clinical facets may therefore aid in the development of future diagnostics and treatment options for ESCC.
Gastric cancer
Zhang et al (67) demonstrated that circ_NRIP1 in gastric cancer (GC) tumor tissues is increasingly expressed compared with healthy tissues. In addition, the dysregulated expression profiles of circ_NRIP1 correlate with GC tumor size and lymphatic invasion. Kaplan-Meier analysis also illustrates those patients in the circ_NRIP1 overexpression group demonstrate shortened overall and disease-free survival rates. Functionally, specific siRNA-mediated exogenous downregulation of circ_NRIP1 may repress the proliferation and metastasis of tumor cells and restrict the growth of xenograft tumors in immunodeficient mice. Additionally, experiments have been conducted to investigate the biological effects of circ_NRIP1 in gastric cancer. The results demonstrate that circ_NRIP1 acts as a molecular sponge by sequestering miR-149-5p, which attenuates miRNA suppression of the oncogene AKT serine/threonine kinase 1 (AKT1), thereby activating the AKT1/mTOR axis (67). This classic signaling pathway plays a positive role in promoting EMT. Quaking (QKI) is an RNA binding protein that has been reported to be a major regulator of circRNA biogenesis in EMT (68). To determine any potential post-transcriptional regulation of circ_NRIP1 formation by QKI in GC development, the flanking intron 1 and intron 3 of the host gene NRIP1 were aligned to the QKI binding motif, as circ_NRIP1 is derived from exon 2 and exon 3 of the host gene NRIP1 (67). A total of four canonical QKI binding sequences have been discovered. Furthermore, RIP and RT-qPCR assays confirm that QKI targets pre-mRNA to increase circ_NRIP1 expression in a post-transcriptional manner (67). In addition to epigenetic variations, altered energy metabolism is a distinct biochemical feature of cancer cells (69). It is becoming increasingly clear that glycolysis is tightly regulated in cancers and is coupled with tumor progression (66). Liu et al (70) reports that circ_NRIP1 can strengthen glycolysis by modulating the miR-186-5p/myosin heavy chain 9 axis. Similarly, Xu et al (71) has also demonstrated that circ_NRIP1 acts as a ceRNA to repress miR-138-5p, resulting in increased glycolysis in GC. In addition, regular follow-up checks are of great importance for patients with malignant tumors (72). However, there is a paucity of specific and effective biomarkers to screen for cancer recurrence (73). Xu et al (71) has demonstrated that circ_NRIP1 is abnormally higher expressed in the serum of primary patients with GC, which decreases to within the expected range after radical gastrectomy. Meanwhile, the expression of circ_NRIP1 remains upregulated in patients with postoperative recurrence, indicating a potential for dynamic monitoring of circ_NRIP1 in patients with GC. To summarize, circ_NRIP1 is a critical regulator in GC initiation, progression and dissemination, and ongoing efforts to uncover the mechanisms of action of circ_NRIP1 may lead to the development of an effective targeted therapy in the future.
Breast cancer
Ding et al (74) has reported that circ_NRIP1 expression is markedly elevated in breast cancer (BC) tumor tissues compared with adjacent benign tissues. Moreover, analysis of the correlation between circ_NRIP1 expression and clinicopathological traits of patients with BC demonstrates that circ_NRIP1 upregulation is closely associated with lymph node metastasis and advanced TNM stage (74). Kaplan-Meier analysis results also confirms that cases of BC with overexpressed circ_NRIP1 demonstrate worse overall patient survival, which may potentially guide clinical decision-making. Furthermore, functional experiments have been conducted to investigate the impact of circ_NRIP1 on oncogenic phenotypes, and the results demonstrate that circ_NRIP1 knockdown depresses proliferation, migration and invasion abilities of tumor cells (74). In addition, the western blot analysis data have demonstrated that silencing circ_NRIP1 results in restricted expression of Vimentin, the mesenchymal marker associated with cell polarity (75). These results suggest that circ_NRIP1 plays numerous roles in maintaining the BC phenotype. While normal angiogenesis is critical for development and tissue growth, pathological angiogenesis is important for the growth and spread of cancers by fueling cancer progression and providing a conduit for distant metastasis (76). Dimethylarginine dimethylaminohydrolase 1 (DDAH1) is the cysteine hydrolase enzyme that metabolizes asymmetric dimethylarginine and N-monomethyl L-arginine, which are inhibitors of endogenous nitric oxide synthase (76). By doing so, DDAH1 enhances the angiogenesis of prostate cancer (76). Circ_NRIP1 has been demonstrated to function as a ceRNA to sequester miR-1253, resulting in increased DDAH1 expression (74). Rescue experiments confirm that DDAH1 overexpression can reverse the impaired proliferative, migratory and invasive potential of tumor cells caused by circ_NRIP1 silencing (74). In addition, Xie et al (77) has also demonstrated that circ_NRIP1 is prominently overexpressed in pathological tissues of BC, which is associated with advanced histological grade and positively correlates with poor prognosis. Zinc finger E-box binding homeobox 2 (ZEB2) is a transcription factor composed of two zinc finger clusters and a central repression region. ZEB2 is primarily involved in executing EMT processes during cancer development (78,79). Circ_NRIP1 has been reported to act as pivotal regulator in BC, which enhances the transcriptional activity of ZEB2 by sponging miR-653 (77). In summary, circ_NRIP1 has the potential to change the conventional approach to BC management through use as an effective method for cancer screening. Moreover, novel drug delivery technologies such as light-activated siRNA endosomal release can also provide a possibility for circNRIP1-based targeted therapy, which may ultimately lead to increased BC control.
Nasopharyngeal carcinoma (NPC)
NPC is an epithelial-derived malignant tumor that arises in the nasopharynx. It is characterized by its unique geographical distribution and a marked propensity to invade and metastasize (80,81). Currently, the extensive application of intensity-modulated radiotherapy and chemotherapy regimens has resulted in enhanced survival outcomes for individuals with NPC. However, despite these advancements in treatment, Chinese patients with NPC still have a poor prognosis with a 5-year survival rate of 50% (82). This is largely attributed to the fact that advanced NPC is initially responsive to concurrent radiotherapy and chemotherapy using cisplatin, resulting in complete disease control and remission, but relapses with metastasis can occur (83,84). Therefore, it is essential to clarify the mechanistic basis for cisplatin resistance in NPC. In a cohort of 138 patients with NPC, circ_NRIP1 expression in serum isolated from 72 cisplatin resistant cases was significantly elevated in comparison with the serum from cisplatin sensitive patients (85). To explore the possible role of circ_NRIP1 in the development of cisplatin chemoresistance, Lin et al (85) subjected NPC tumor cells (HK-1) to successive passages in varying concentrations of cisplatin. This ultimately led to the emergence of a subpopulation of HK-1 cells that were able to withstand cisplatin treatment, known as cisplatin-resistant HK-1 cells (HK-1/CDDP). Notably, RT-qPCR analysis revealed that circ_NRIP1 expression is significantly upregulated in HK-1/CDDP cells despite its increased baseline expression in parental HK-1 cells (85). To investigate whether the upregulation of circ_NRIP1 in HK-1/CDDP cells was functionally remarkable, researchers further assessed the proliferative capacity of these cells (85). It was demonstrated that circ_NRIP1 knockdown in HK-1/CDDP cells led to the loss of their baseline proliferative advantage when cultured in cisplatin, exhibiting a proliferation profile similar to parental HK-1 cells (85). Therefore, researchers concluded that circ_NRIP1 augments cisplatin resistance in NPC by promoting interleukin 25 (IL-25) expression by targeting miR-515-5p (85). Additionally, rescue studies have also confirmed that the proliferative advantage of HK-1/CDDP, conferred by circ_NRIP1 overexpression, is partially reversed by upregulation of miR-515-5p or silencing of IL-25 (85). Broadly speaking, novel mediators of tumor aggression, such as circ_NRIP1, can provide insight into the mechanism of cisplatin chemotherapy resistance. As bioengineering and pharmaceutical technology advances, this increased understanding may help to develop more targeted and efficient strategies to further improve the management of NPC.
Papillary thyroid carcinoma
Li et al (86) demonstrated that circ_NRIP1 is overexpressed in papillary thyroid carcinoma (PTC) tumor tissues, and that this increased expression is associated with advanced TNM stage. The relationship between circ_NRIP1 expression and clinicopathological features in patients with PTC reveals a strong association between circ_NRIP1 upregulation and poor overall survival, highlighting the potential of circ_NRIP1 as a prognostic biomarker (86). Additionally, as determined by the RT-qPCR detection results, circ_NRIP1 expression has been observed to be more highly expressed in PTC tumor cells (TPC-1) compared with healthy human thyroid follicular epithelial cells (86). To investigate how circ_NRIP1 expression impacts PTC, researchers performed gain and loss of function assays (86). Subsequently, MTT and Transwell assays were used to demonstrate that knocking down circ_NRIP1 can significantly attenuate the viability and mobility of TPC-1 cells (86). In parallel, flow cytometry detection results also revealed that circ_NRIP1 depletion expedites TPC-1 cell apoptosis (86). A growing body of evidence has demonstrated that circRNA can serve as a natural miRNA sponge and regulate functions of miRNAs (41). Circ_NRIP1 has been demonstrated to competitively bind miR-195-5p to promote PTC progression (86). In addition, previous studies have demonstrated that the JAK/STAT and MARK signaling pathways regulate cell proliferation and PTC development (87–89). Thus, researchers measured the expression of genes involved in these pathways in TPC-1 cells (86). The results indicate that knockdown of circ_NRIP1 suppresses the expression of Janus kinase 2, signal transducer and activator of transcription 1 and mitogen-activated protein kinase 14, while silencing of miR-195-5p increased expression of these molecules, confirming that circ_NRIP1 activates the JAK/STAT and MARK signaling pathways by targeting miR-195-5p (86). In summary, these results demonstrate that overexpression of circ_NRIP1 promotes PTC progression by activating the JAK/STAT and MARK signaling pathway in a ceRNA-dependent manner. Researchers also found that increased circ_NRIP1 expression promotes PTC cell proliferation, migration and invasion via the miR-653-5p/PBX3 axis (90). In conclusion, understanding the important roles of circ_NRIP1 and aberrant activation of the associated signaling pathway will increase our knowledge of the biological basis of PTC and may open the door for future novel therapeutic strategies.
Non-small cell lung cancer (NSCLC)
Lung cancer is the leading cause of cancer-associated mortalities worldwide. Studies show that ~85% of patients with lung cancer have NSCLC, a group of histological subtypes of cancer with a five-year survival rate of 15%, which has barely improved in the past two decades (91,92). Currently, the lack of a routine screening method for the early detection of this disease is a major contributor to its high mortality rate. However, the advent of large databases characterizing the molecular features of human tumors have improved our understanding of NSCLC, shifting the focus from traditional histopathological descriptions to precise molecular and genetic identification (93,94). CircRNAs are emerging as promising molecular biomarkers for cancer diagnosis due to their high stability, abundance, conservation and accessibility in certain bodily fluids including bile, saliva and gastric juice (95). Several studies have demonstrated that blood platelets can serve as platforms for detecting circRNAs, as platelets can carry biomolecules from their environment, including tumor-derived circRNAs (96,97). Therefore, analyzing the alterations in platelet circRNA profiles can be employed as a biomarker for liquid biopsy diagnosis. D'Ambrosi et al (98) performed RNA sequencing of 12 platelet samples derived from patients with NSCLC and healthy controls, which were age and gender matched. RNA sequencing analysis demonstrated 411 aberrantly expressed circRNAs, of which only circ_NRIP1, showed increased upregulation in the NSCLC samples with a false discovery rate value of <0.05. To investigate whether circ_NRIP1 could potentially serve as a biomarker for NSCLC, researchers investigated circ_NRIP1 expression in an independent cohort of platelet samples isolated from 23 patients with NSCLC and 24 asymptomatic individuals, and the results demonstrated that circ_NRIP1 is overexpressed in the NSCLC group (98), consistent with previous RNA sequencing data (98). Subsequent statistical analysis confirmed a strong correlation between circ_NRIP1 expression, lymph node invasion and advanced TNM stage, indicating that circ_NRIP1 can serve as a novel biomarker for NSCLC (98). Previous studies have demonstrated that circRNAs, such as circ_USP7, circ_SATB2 and circ_PTK2, are differentially expressed in NSCLC and play vital roles in tumor initiation and progression (99–101). Similarly, circ_NRIP1 may also have the potential to promote cancer development. Circ_NRIP1 has also been demonstrated to be highly abundant in platelet-derived exosomes (102). This may indicate that circ_NRIP1 can promote cancer progression by being released from platelets via exosomes. Tumors exhibit a high heterogeneity, with remarkable biological differences between certain tumor types and even within subtypes of the same tumor. The differential roles of circNRIP1 in different tumors are not unique, as other circRNAs, such as circRHOBTB3, circFNDC3B, circGRAMD1B and circOXCT1, can also act as oncogenes or tumor suppressors depending on the tumor context (103–110). This suggests that when designing targeted drugs for circRNA, drug carriers and administration routes should be comprehensively considered. Otherwise, the off-target effects of drugs may bring unexpected side effects to patients, violating the basic ethical principle of ‘do no harm’. Further studies on circ_NRIP1 have the potential to improve management strategies for NSCLC, in addition to improving its early detection.
Osteosarcoma
Meng et al (111) demonstrated that circ_NRIP1 expression is markedly elevated in osteosarcoma tumor tissues compared with adjacent healthy tissues. When combined with clinicopathological parameters of patients with osteosarcoma, researchers demonstrated that the abnormal expression of circ_NRIP1 is positively associated with distant metastasis of the tumor and TNM stage (111). Kaplan-Meier analysis results also demonstrated that patients with osteosarcoma who overexpressed circ_NRIP1 have a worse overall survival rate compared to low expression group (70–50%) (111). In vitro experiments that knocked down circ_NRIP1 with siRNA effectively decreased tumor cell proliferation, migration and invasion. In parallel, flow cytometry data revealed that circ_NRIP1-silencing induces cell cycle arrest (111). Therefore, it can be concluded that miR-199a contains a complementary sequence (UGUGACCA) to circ_NRIP1, as identified through a bioinformatics database (StarBase; http://starbase.sysu.edu.cn). Furthermore, downregulation of circ_NRIP1 expression leads to a significant increase in miR-199a expression, while the overexpression of circ_NRIP1 exhibits the opposite effect (111). Subsequently, the biotin-labeled pull-down analysis demonstrated that circ_NRIP1 can negatively regulate miR-199a via a ceRNA mechanism. Forkhead box C2 (FOXC2) is a well-characterized oncogene that is targeted by miR-199a in certain tumors such as breast cancer (112). Notably, knockdown of circ_NRIP1 leads to a significant decrease in FOXC2 expression, while transfection with a miR-143 inhibitor upregulates circ_NRIP1 expression. Function assays further demonstrated that upregulation of FOXC2 increased tumor cell malignancy. These results demonstrated that circ_NRIP1 promotes tumor cell growth and metastasis via the miR-199a/FOXC2 axis (111). Similarly, Shi et al (113) reported that circ_NRIP1 can also be transferred between cells via extracellular vesicles, which can contribute to the formation of the tumor microenvironment. Shi et al also revealed that the regulatory mechanism of circ_NRIP1, as a molecular sponge of miR-532-3p, leads to elevated expression of AKT3 and PI3K/AKT signaling pathway activation, thereby facilitating osteosarcoma progression. Previous advances in RNA modification research have shed light on the pivotal role of N6-methyladenosine (m6A) in circRNA metabolism and tumor biology (113). Deposition of m6A on circRNA is catalyzed by the methyltransferase-like 3 (METTL3), which acts as the main catalytic subunit in RNA methylation (114–116). Methylated RNA immunoprecipitation analysis has indicated that circ_NRIP1 contains an m6A modification site. Additionally, western blot and RT-qPCR analysis results demonstrate that METTL3 is significantly upregulated in osteosarcoma tumor tissues and is positively associated with circ_NRIP1 expression. This suggests that the aberrant expression of circ_NRIP1 may be regulated by methylation modifications of the NRIP1 parent gene (113). Therefore, circ_NRIP1 can promote osteosarcoma progression by sponging miR-532-3p and activating the PI3K/AKT signaling pathway, indicating its potential use in osteosarcoma targeted therapy. Moreover, circ_NRIP1 m6A modification demonstrates a new layer of epigenetic regulation in cancer and opens up new possibilities in osteosarcoma research. This discovery may also contribute to the future development of novel therapeutics that are more specific and effective at treating osteosarcoma.
3. Conclusion and future perspectives
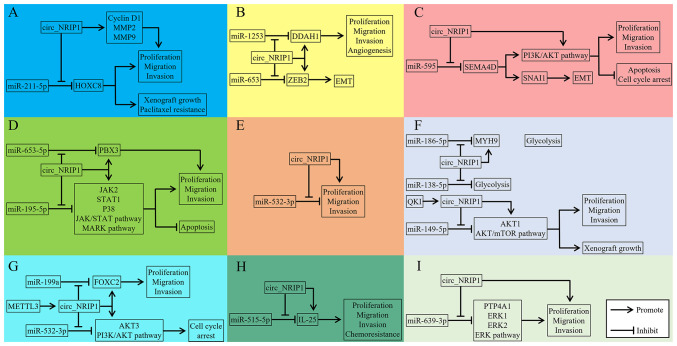
Emerging evidence has continued to recognize circRNA as a tightly regulated transcript that is involved in numerous versatile biological functions in addition to being a byproduct in pre-mRNA splicing. Circ_NRIP1, one of the circRNA families, has been found to interfere with crucial steps during cancer initiation and progression thereby promoting proliferation, metastasis and adaptation to the cellular microenvironment. Circ_NRIP1 acts as a vital regulator in certain cancers through various mechanisms such as molecular sponging and activation of signaling pathways (Fig. 2). Although the ceRNA function of circ_NRIP1 is well documented, exploration of the upstream and downstream regulators that may modulate circ_NRIP1 expression and activity is still a work in progress. Additionally, the discovery of circ_NRIP1 regulatory mechanisms could contribute not only to furthering the field of genomics research but also to improving the diagnosis and targeted therapeutic applications for cancers. It is important to validate circ_NRIP1 expression in diverse populations with cancer to realize its diagnostic potential. Notably, circ_NRIP1 based targeted therapy also faces major challenges, such as off-target effects and immune system activation, which can induce an uncontrolled hyperinflammatory response in patients. Future research will be marked by the further mechanisms of circRNA which may contribute to successful translation of circRNA based targeted therapy from the bench to bedside. With a clearer understanding of circRNA biological functions, future cancer treatments may present an exciting facet to personalized medicine.
Figure 2.
Regulatory network of circ_NRIP1 in human malignant tumors. (A) Ovarian cancer, (B) breast cancer, (C) esophageal squamous cell cancer, (D) papillary thyroid carcinoma, (E) colorectal cancer, (F) gastric cancer, (G) osteosarcoma, (H) nasopharyngeal carcinoma and (I) cervical cancer. HOXC8, homeobox C8; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; PTP4A1, protein tyrosine phosphatase 4A1; ERK1, extracellular regulated protein kinase1; ERK2, extracellular regulated protein kinase2; FOXC2, forkhead box C2; AKT3, AKT serine/threonine kinase 3; PI3K, phosphatidylinositol 3-kinase; DDAH1, dimethylarginine dimethylaminohydrolase 1; ZEB2, zinc finger E-box binding homeobox 2; EMT, epithelial-mesenchymal transition; MYH9, myosin heavy chain 9; AKT1, AKT serine/threonine kinase 1; SNAI1, snail family transcriptional repressor 1; SEMA4D, semaphoring 4D; IL-25, interleukin 25; PBX3, pre-B-cell leukemia homeobox 3; JAK2, Janus kinase 2; STAT1, signal transducer and activator of transcription 1; p38, p38 kinase.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Heilongjiang Postdoctoral Science Foundation (grant no. LBH-Q21023), the National Natural Science Foundation of Heilongjiang Province (grant no. LH2020H058) and the Chen Xiaoping Foundation for the Development of Science and Technology of Hubei Province (grant no. CXPJJH12000002-2020015).
Availability of data and materials
Not applicable.
Authors' contributions
XG, YY and HW prepared the manuscript. GL and XS shortlisted research articles for inclusion in this review and helped to revise the manuscript. ZW and XJ edited the manuscript. All authors read and approved the final version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Mol Aspects Med. 2019;70:141–152. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler B, Hayashi M, Hayashi MN, Spiegelman S. Circularity of the replicating form of a single-stranded DNA virus. Science. 1964;143:47–49. doi: 10.1126/science.143.3601.47. [DOI] [PubMed] [Google Scholar]
- 6.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313–319. doi: 10.1016/0092-8674(80)90505-X. [DOI] [PubMed] [Google Scholar]
- 7.Kos A, Dijkema R, Arnberg AC, Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 8.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 9.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 10.Diener TO. Circular RNAs: Relics of precellular evolution? Proc Natl Acad Sci USA. 1989;86:9370–9374. doi: 10.1073/pnas.86.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 12.Guan C, Liu L, Zhao Y, Zhang X, Liu G, Wang H, Gao X, Zhong X, Jiang X. YY1 and eIF4A3 are mediators of the cell proliferation, migration and invasion in cholangiocarcinoma promoted by circ-ZNF609 by targeting miR-432-5p to regulate LRRC1. Aging (Albany NY) 2021;13:25195–25212. doi: 10.18632/aging.203735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Azhati B, Wang W, Rexiati M, Xing C, Wang Y. Circular RNA UBAP2 promotes the proliferation of prostate cancer cells via the miR-1244/MAP3K2 axis. Oncol Lett. 2021;21:486. doi: 10.3892/ol.2021.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, Huang L, Wang W, Guan C, Zhao Y, Liu L, Jiang X. Long non-coding RNA FOXD2-AS1 promotes proliferation, migration, and invasion in cholangiocarcinoma through regulating miR-760/E2F3 axis. Dig Dis Sci. 2022;67:546–558. doi: 10.1007/s10620-021-06876-9. [DOI] [PubMed] [Google Scholar]
- 15.Peng F, Gong W, Li S, Yin B, Zhao C, Liu W, Chen X, Luo C, Huang Q, Chen T, et al. CircRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 2021;70:603–615. doi: 10.2337/db20-0203. [DOI] [PubMed] [Google Scholar]
- 16.Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Wang H, Yu S, Gao X, Liu G, Sun D, Jiang X. An update on the roles of circRNA-ZFR in human malignant tumors. Front Cell Dev Biol. 2022;9:806181. doi: 10.3389/fcell.2021.806181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Deng Z, Yang H, Pan X, Wei Z, Shen HB, Choi KS, Wang L, Wang S, Wu J. CircRNA-binding protein site prediction based on multi-view deep learning, subspace learning and multi-view classifier. Brief Bioinform. 2022;23:bbab394. doi: 10.1093/bib/bbab394. [DOI] [PubMed] [Google Scholar]
- 20.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. Translation of circRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CK, Cheng R, Demeter J, Chen J, Weingarten-Gabbay S, Jiang L, Snyder MP, Weissman JS, Segal E, Jackson PK, Chang HY. Structured elements drive extensive circular RNA translation. Mol Cell. 2021;81:4300–4318. doi: 10.1016/j.molcel.2021.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, Xiong F, Guo C, Wu X, Li Y, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B, Yin H, Zhang H, Wang T. CircNRIP1 facilitates keloid progression via FXR1-mediated upregulation of miR-503-3p and miR-503-5p. Int J Mol Med. 2021;47:70. doi: 10.3892/ijmm.2021.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Li J, Bu H, Wang H, Zhang Y, Shen Q, Li M, Lu Z, Rong X, Zheng D, Peng Y. Circular RNA expression alteration identifies a novel circulating biomarker in serum exosomal for detection of alcohol dependence. Addict Biol. 2021;26:e13031. doi: 10.1111/adb.13031. [DOI] [PubMed] [Google Scholar]
- 25.Tao X, Shao Y, Yan J, Yang L, Ye Q, Wang Q, Lu R, Guo J. Biological roles and potential clinical values of circular RNAs in gastrointestinal malignancies. Cancer Biol Med. 2021;18:437–457. doi: 10.20892/j.issn.2095-3941.2020.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Samsami M. Emerging role of circular RNAs in the pathogenesis of ovarian cancer. Cancer Cell Int. 2022;22:172. doi: 10.1186/s12935-022-02602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira AL, Magalhães L, Pantoja RP, Araújo G, Ribeiro-Dos-Santos Â, Vidal AF. The biological role of sponge circular RNAs in gastric cancer: Main players or coadjuvants? Cancers (Basel) 2020;12:1982. doi: 10.3390/cancers12071982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Li Z, Xu S, Guo J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal. 2020;34:e23359. doi: 10.1002/jcla.23359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med. 2018;96:85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 30.Reis-das-Mercês L, Vinasco-Sandoval T, Pompeu R, Ramos AC, Anaissi AKM, Demachki S, Assumpção PP, Vidal AF, Ribeiro-Dos-Santos Â, Magalhães L. CircRNAs as potential blood biomarkers and key elements in regulatory networks in gastric cancer. Int J Mol Sci. 2022;23:650. doi: 10.3390/ijms23020650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurwitz LM, Pinsky PF, Trabert B. General population screening for ovarian cancer. Lancet. 2021;397:2128–2130. doi: 10.1016/S0140-6736(21)01061-8. [DOI] [PubMed] [Google Scholar]
- 32.Killock D. Viral gene therapy active in ovarian cancer. Nat Rev Clin Oncol. 2020;17:391. doi: 10.1038/s41571-020-0371-5. [DOI] [PubMed] [Google Scholar]
- 33.Barber E, Matei D. Immunotherapy in ovarian cancer: We are not there yet. Lancet Oncol. 2021;22:903–905. doi: 10.1016/S1470-2045(21)00303-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bairi EK, Singh S, Page LC. Revisiting platinum-resistant ovarian cancer: Advances in therapy, molecular biomarkers, and clinical outcomes. Semin Cancer Biol. 2021;77:1–2. doi: 10.1016/j.semcancer.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Luo F, Jiang X, Zhang WJ, Xiang T, Pan QZ, Cai L, Zhao J, Weng D, Li Y, et al. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J Immunother Cancer. 2022;10:e004029. doi: 10.1136/jitc-2021-004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, Cai JB, Yang X, Fan J, Ke AW, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19:92. doi: 10.1186/s12943-020-01213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, Zhang P, Zhao Y, He S, Wang Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19:33. doi: 10.1186/s12943-020-01149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Y, Gui R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J Gynecol Oncol. 2020;31:e75. doi: 10.3802/jgo.2020.31.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Cai J, Han X, Ren Y. Downregulation of circNRIP1 suppresses the paclitaxel resistance of ovarian cancer via regulating the miR-211-5p/HOXC8 axis. Cancer Manag Res. 2020;12:9159–9171. doi: 10.2147/CMAR.S268872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 42.Xu P, Zhang X, Ni W, Fan H, Xu J, Chen Y, Zhu J, Gu X, Yang L, Ni R, et al. Upregulated HOXC8 expression is associated with poor prognosis and oxaliplatin resistance in hepatocellular carcinoma. Dig Dis Sci. 2015;60:3351–3363. doi: 10.1007/s10620-015-3774-x. [DOI] [PubMed] [Google Scholar]
- 43.Jiang Y, Wang Z, Ying C, Hu J, Zeng T, Gao L. FMR1/circCHAF1A/miR-211-5p/HOXC8 feedback loop regulates proliferation and tumorigenesis via MDM2-dependent p53 signaling in GSCs. Oncogene. 2021;40:4094–4110. doi: 10.1038/s41388-021-01833-2. [DOI] [PubMed] [Google Scholar]
- 44.Castle PE, Einstein MH, Sahasrabuddhe VV. Cervical cancer prevention and control in women living with human immunodeficiency virus. CA Cancer J Clin. 2021;71:505–526. doi: 10.3322/caac.21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrall L, Lin KY, Roden RBS, Hung CF, Wu TC. Cervical cancer immunotherapy: Facts and hopes. Clin Cancer Res. 2021;27:4953–4973. doi: 10.1158/1078-0432.CCR-20-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundström K, Elfström KM. Advances in cervical cancer prevention: Efficacy, effectiveness, elimination? PLoS Med. 2020;17:e1003035. doi: 10.1371/journal.pmed.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, Liu G, Li X, Yang Y, Chen R. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19:13. doi: 10.1186/s12943-020-1139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J, Du WW, Zeng K, Wu N, Fang L, Lyu J, Yee AJ, Yang BB. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther. 2021;29:2754–2768. doi: 10.1016/j.ymthe.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang X, Li X, Wu D, Wang D, Zhang S. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis. 2020;11:399. doi: 10.1038/s41419-020-2607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu JY, Yi W, Wei X, Zhang MY, Xu R, Zeng LS, Huang ZJ, Chen JS. MiR-601 is a prognostic marker and suppresses cell growth and invasion by targeting PTP4A1 in breast cancer. Biomed Pharmacother. 2016;79:247–253. doi: 10.1016/j.biopha.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, Kung HF, Zeng YX, Xie D. MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann Oncol. 2014;25:2196–2204. doi: 10.1093/annonc/mdu439. [DOI] [PubMed] [Google Scholar]
- 52.Flores-Pérez A, Marchat LA, Rodríguez-Cuevas S, Bautista VP, Fuentes-Mera L, Romero-Zamora D, Maciel-Dominguez A, Cruz OH, Fonseca-Sánchez M, Ruíz-García E, et al. Suppression of cell migration is promoted by miR-944 through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC Cancer. 2016;16:379. doi: 10.1186/s12885-016-2470-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai Y, Luo Y, Liu S, Zhang L, Shen K, Dong Y, Walls CD, Quilliam LA, Wells CD, Cao Y, Zhang ZY. PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein. J Biol Chem. 2011;286:42316–42324. doi: 10.1074/jbc.M111.286302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai Q, Wang M, Hu C, Tang Y, Li Y, Hao S. Circular RNA regulates the onset and progression of cancer through the mitogen-activated protein kinase signaling pathway. Oncol Lett. 2021;22:817. doi: 10.3892/ol.2021.13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 56.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA. 2021;325:669–685. doi: 10.1001/jama.2021.6027. [DOI] [PubMed] [Google Scholar]
- 57.Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu F, Li R, Zhang R, He M, Zhang Y. Knockdown of circNRIP1 sensitizes colorectal cancer to 5-FU via sponging miR-532-3p. Oncol Rep. 2021;46:218. doi: 10.3892/or.2021.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang E, Fu J, Yu Q, Xie P, Yang Z, Ji H, Wang L, Luo G, Zhang Y, Li K. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics. 2020;12:587–603. doi: 10.2217/epi-2019-0404. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Zhu C, Lu X. Advances in serum biomarkers for early diagnosis of gastric cancer. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2019;48:326–333. doi: 10.3785/j.issn.1008-9292.2019.06.14. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou S, Guo Z, Zhou C, Zhang Y, Wang S. Circ_NRIP1 is oncogenic in malignant development of esophageal squamous cell carcinoma (ESCC) via miR-595/SEMA4D axis and PI3K/AKT pathway. Cancer Cell Int. 2021;21:250. doi: 10.1186/s12935-021-01907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCabe EM, Rasmussen TP. LncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol. 2021;75:38–48. doi: 10.1016/j.semcancer.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van KA, Brown D, Moers V, Lemaire S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 67.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shingu T, Ho AL, Yuan L, Zhou X, Dai C, Zheng S, Wang Q, Zhong Y, Chang Q, Horner JW, et al. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat Genet. 2017;49:75–86. doi: 10.1038/ng.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Payne KK. Cellular stress responses and metabolic reprogramming in cancer progression and dormancy. Semin Cancer Biol. 2021;78:45–48. doi: 10.1016/j.semcancer.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, Jiang Y, Xu L, Qu C, Zhang L, Xiao X, Chen W, Li K, Liang Q, Wu H. Circ-NRIP1 promotes glycolysis and tumor progression by regulating miR-186-5p/MYH9 axis in gastric cancer. Cancer Manag Res. 2020;12:5945–5956. doi: 10.2147/CMAR.S245941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu G, Li M, Wu J, Qin C, Tao Y, He H. Circular RNA circNRIP1 sponges microRNA-138-5p to maintain hypoxia-induced resistance to 5-Fluorouracil through HIF-1α-dependent glucose metabolism in gastric carcinoma. Cancer Manag Res. 2020;12:2789–2802. doi: 10.2147/CMAR.S246272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernandes E, Sores J, Cotton S, Peixoto A, Ferreira D, Freitas R, Reis CA, Santos LL, Ferreira JA. Esophageal, gastric and colorectal cancers: Looking beyond classical serological biomarkers towards glycoproteomics-assisted precision oncology. Theranostics. 2020;10:4903–4928. doi: 10.7150/thno.42480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shieh Y, Eklund M, Sawaya GF, Black WC, Kramer BS, Esserman LJ. Population-based screening for cancer: Hope and hype. Nat Rev Clin Oncol. 2016;13:550–565. doi: 10.1038/nrclinonc.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding X, Zheng J, Cao M. Circ_0004771 accelerates cell carcinogenic phenotypes via suppressing miR-1253-mediated DDAH1 inhibition in breast cancer. Cancer Manag Res. 2021;13:1–11. doi: 10.2147/CMAR.S273783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soukup V, Babjuk M, Bellmunt J, Dalbagni G, Giannarini G, Hakenberg OW, Herr H, Lechevallier E, Ribal MJ. Follow-up after surgical treatment of bladder cancer: A critical analysis of the literature. Eur Urol. 2012;62:290–302. doi: 10.1016/j.eururo.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 76.Reddy KRK, Dasari C, Duscharla D, Supriya B, Ram NS, Surekha MV, Kumar JM, Ummanni R. Dimethylarginine dimethylaminohydrolase-1 (DDAH1) is frequently upregulated in prostate cancer, and its overexpression conveys tumor growth and angiogenesis by metabolizing asymmetric dimethylarginine (ADMA) Angiogenesis. 2018;21:79–94. doi: 10.1007/s10456-017-9587-0. [DOI] [PubMed] [Google Scholar]
- 77.Xie R, Tang J, Zhu X, Jiang H. Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR-653 by targeting ZEB2 signaling pathway. Biosci Rep. 2019;39:BSR20181919. doi: 10.1042/BSR20181919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Si W, Huang W, Zheng Y, Yang Y, Liu X, Shan L, Zhou X, Wang Y, Su D, Gao J, et al. Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27:822–836. doi: 10.1016/j.ccell.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 79.Li H, Xu L, Li C, Zhao L, Ma Y, Zheng H, Li Z, Zhang Y, Wang R, Liu Y, Qu X. Ubiquitin ligase Cbl-b represses IGF-I-induced epithelial mesenchymal transition via ZEB2 and microRNA-200c regulation in gastric cancer cells. Mol Cancer. 2014;13:136. doi: 10.1186/1476-4598-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 81.Tang LL, Chen WQ, Xue WQ, He YQ, Zheng RS, Zeng YX, Jia WH. Global trends in incidence and mortality of nasopharyngeal carcinoma. Cancer Lett. 2016;374:22–30. doi: 10.1016/j.canlet.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 82.Lei Y, Li YQ, Jiang W, Hong XH, Ge WX, Zhang Y, Hu WH, Wang YQ, Liang YL, Li JY, et al. A gene-expression predictor for efficacy of induction chemotherapy in locoregionally advanced nasopharyngeal carcinoma. J Natl Cancer Inst. 2021;113:471–480. doi: 10.1093/jnci/djaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang LQ, Li CF, Li J, Chen WH, Chen QY, Yuan LX, Lai XP, He Y, Xu YX, Hu DP, et al. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J Natl Cancer Inst. 2015;108:djv291. doi: 10.1093/jnci/djv291. [DOI] [PubMed] [Google Scholar]
- 84.Gatta G, Botta L, Sánchez MJ, Anderson LA, Pierannunzio D, Licitra L. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur J Cancer. 2015;51:2130–2143. doi: 10.1016/j.ejca.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 85.Lin J, Qin H, Han Y, Li X, Zhao Y, Zhai G. CircNRIP1 modulates the miR-515-5p/IL-25 axis to control 5-Fu and cisplatin resistance in nasopharyngeal carcinoma. Drug Des Devel Ther. 2021;15:323–330. doi: 10.2147/DDDT.S292180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li C, Zhu L, Fu L, Han M, Li Y, Meng Z, Qiu X. CircRNA NRIP1 promotes papillary thyroid carcinoma progression by sponging mir-195-5p and modulating the P38 MAPK and JAK/STAT pathways. Diagn Pathol. 2021;16:93. doi: 10.1186/s13000-021-01153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baldini C, Moriconi FR, Galimberti S, Libby P, Caterina DR. The JAK-STAT pathway: An emerging target for cardiovascular disease in rheumatoid arthritis and myeloproliferative neoplasms. Eur Heart J. 2021;42:4389–4400. doi: 10.1093/eurheartj/ehab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu ZK, Li C, Zhang RY, Wei D, Shang YK, Yong YL, Kong LM, Zheng NS, Liu K, Lu M, et al. EYA2 suppresses the progression of hepatocellular carcinoma via SOCS3-mediated blockade of JAK/STAT signaling. Mol Cancer. 2021;20:79. doi: 10.1186/s12943-021-01377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blaj C, Schmidt EM, Lamprecht S, Hermeking H, Jung A, Kirchner T, Horst D. Oncogenic effects of high MAPK activity in colorectal cancer mark progenitor cells and persist irrespective of RAS mutations. Cancer Res. 2017;77:1763–1774. doi: 10.1158/0008-5472.CAN-16-2821. [DOI] [PubMed] [Google Scholar]
- 90.Fu L, Huo J, Fitrat H, Kong Y, Zhang L, Shang C, Li G, Ji F, Fu X, Qiu X. CircNRIP1 exerts oncogenic functions in papillary thyroid carcinoma by sponging miR-653-5p and regulating PBX3 expression. J Oncol. 2022;2022:2081501. doi: 10.1155/2022/2081501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 93.Cagney DN, Martin AM, Catalano PJ, Brown PD, Alexander BM, Lin NU, Aizer AA. Implications of screening for brain metastases in patients with breast cancer and non-small cell lung cancer. JAMA Oncol. 2018;4:1001–1003. doi: 10.1001/jamaoncol.2018.0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.White RW, Horvitz E. Evaluation of the feasibility of screening patients for early signs of lung carcinoma in Web Search Logs. JAMA Oncol. 2017;3:398–401. doi: 10.1001/jamaoncol.2016.4911. [DOI] [PubMed] [Google Scholar]
- 95.Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, Xu X, Liang Q, Christiani DC, Wang M, et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol Cancer. 2021;20:13. doi: 10.1186/s12943-020-01298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lieben L. Diagnosis: RNA-seq for blood-based pan-cancer diagnostics. Nat Rev Cancer. 2015;15:696–697. doi: 10.1038/nrc4048. [DOI] [PubMed] [Google Scholar]
- 97.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.D'Ambrosi S, Visser A, Antunes-Ferreira M, Poutsma A, Giannoukakos S, Sol N, Sabrkhany S, Bahce I, Kuijpers MJE, Oude Egbrink MGA, et al. The analysis of platelet-derived circRNA repertoire as potential diagnostic biomarker for non-small cell lung cancer. Cancers. 2021;13:4644. doi: 10.3390/cancers13184644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, Lin K, Lu F, Xu JJ, Wu YB. Cancer cell-derived exosomal circUSP7 induces CD8+ T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 2021;20:144. doi: 10.1186/s12943-021-01448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, Dai X, Zhou H, Zhu J, Zhang H, Jiang Y. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101. doi: 10.1186/s12943-020-1131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Preußer C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, Santoso S, Bindereif A. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7:1424473. doi: 10.1080/20013078.2018.1424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang T, Shen P, Chen Q, Wu P, Yuan H, Ge W, Meng L, Huang X, Fu Y, Zhang Y, et al. FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2021;40:261. doi: 10.1186/s13046-021-02063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen C, Yu H, Han FY, Lai X, Ye KH, Lei S, Mai M, Lai M, Zhang H. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol Cancer. 2022;21:46. doi: 10.1186/s12943-022-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, Lin T, Huang J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tang B, Zhang QF, Liu K, Huang Y. Exosomal circRNA FNDC3B promotes the progression of esophageal squamous cell carcinoma by sponging miR-490-5p and regulating thioredoxin reductase 1 expression. Bioengineered. 2022;13:13829–13848. doi: 10.1080/21655979.2022.2084484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dai X, Guo X, Liu J, Cheng A, Peng X, Zha L, Wang Z. Circular RNA circGRAMD1B inhibits gastric cancer progression by sponging miR-130a-3p and regulating PTEN and p21 expression. Aging (Albany NY) 2019;11:9689–9708. doi: 10.18632/aging.102414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu X, Wang Y, Zhou G, Zhou JB, Tian Z, Xu J. circGRAMD1B contributes to migration, invasion and epithelial-mesenchymal transition of lung adenocarcinoma cells via modulating the expression of SOX4. Funct Integr Genomics. 2023;23:75. doi: 10.1007/s10142-023-00972-x. [DOI] [PubMed] [Google Scholar]
- 109.Luo H, Peng J, Yuan Y. CircRNA OXCT1 promotes the malignant progression and glutamine metabolism of non-small cell lung cancer by absorbing miR-516b-5p and upregulating SLC1A5. Cell Cycle. 2023;22:1182–1195. doi: 10.1080/15384101.2022.2071565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu J, Dai X, Guo X, Cheng A, Mac SM, Wang Z. Circ-OXCT1 suppresses gastric cancer EMT and metastasis by attenuating TGF-β pathway through the circ-OXCT1/miR-136/SMAD4 axis. Onco Targets Ther. 2020;13:3987–3998. doi: 10.2147/OTT.S239789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meng Y, Hao D, Huang Y, Jia S, Zhang J, He X, Liu D, Sun L. Circular RNA circNRIP1 plays oncogenic roles in the progression of osteosarcoma. Mamm Genome. 2021;32:448–456. doi: 10.1007/s00335-021-09891-3. [DOI] [PubMed] [Google Scholar]
- 112.Hollier BG, Tinnirello AA, Werden SJ, Evans KW, Taube JH, Sarkar TR, Sphyris N, Shariati M, Kumar SV, Battula VL, et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73:1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shi Z, Wang K, Xing Y, Yang X. CircNRIP1 encapsulated by bone marrow mesenchymal stem cell-derived extracellular vesicles aggravates osteosarcoma by modulating the miR-532-3p/AKT3/PI3K/AKT axis. Front Oncol. 2021;11:658139. doi: 10.3389/fonc.2021.658139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19:105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.