TO THE EDITOR:
Molecular analysis of pediatric and adult acute myeloid leukemia (AML) is used routinely to identify subtype-defining driver structural variants and mutations which may provide important information for risk stratification. For example, the core-binding factor (CBF) AML subgroup defined by t(8;21) RUNX1::RUNX1T1 or inv(16)/t(16;16) CBFB::MYH11 are associated with favorable outcomes.1,2 Despite the extensive genomic characterization of pediatric and adult AML, there remains an important proportion of previously unclassified cases where new driver lesions are still being identified, including those that can influence patient management owing to their association with outcomes. This includes the recent identification of tandem duplications in UBTF in pediatric AML3,4 and structural variants that dysregulate BCL11B in lineage-ambiguous acute leukemia.5 Herein we describe an additional new subtype of AML characterized by a recurrent insertion mutation in CBFB.
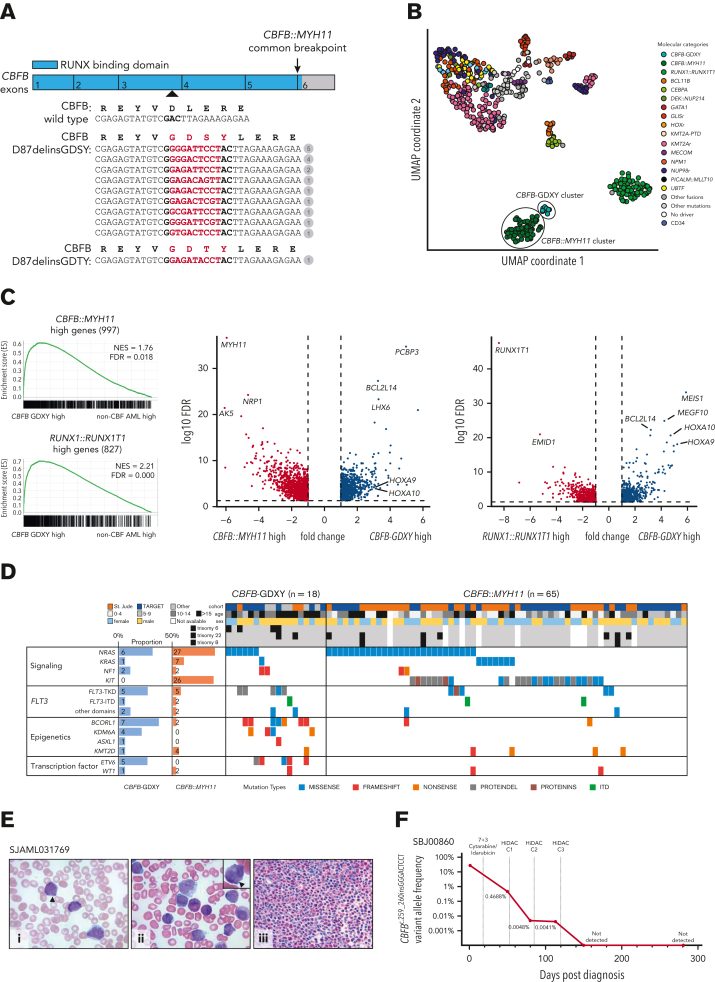
We initially reanalyzed a cohort of 553 pediatric AML transcriptomes from our previous study3 and identified 2 patients (PARANT:SJAML040573, PARUTH:SJAML040605) with similar gene expression profiles to CBFB::MYH11 AML, but without a CBFB::MYH11 fusion or other known leukemic driver by conventional testing, and without finding a CBFB::MYH11 fusion by manual inspection of RNA sequence data (supplemental Figure 1, available on the Blood website; Table 1). However, in both of these patients, we identified a somatic 9-base pair insertion in exon 3 of CBFB (NM_022845.3). The CBFβ protein encoded by CBFB forms the non–DNA-binding regulatory subunit of a heterodimeric transcription factor complex with a DNA-binding CBFα subunit (RUNX1, RUNX2, or RUNX3). Interestingly, both CBFB mutations were predicted to lead to the same amino acid change, substituting aspartic acid at position 87 (D87) for glycine, aspartic acid, serine, and tyrosine [p.(Asp87delinsGlyAspSerTyr); GDSY] within the N-terminal RUNX-binding domain6 (Figure 1A).
Table 1.
Clinical characteristics of the patients with acute leukemia harboring a CBFB-GDXY mutation
| Identifier | CBFB-GDXY mutation (CBFB NM_022845.3) | Age/Sex | Diagnosis | FAB category | Karyotype | Cooperating gene mutations | Risk group | Treatment protocol | Outcome | References |
|---|---|---|---|---|---|---|---|---|---|---|
| PARANT:SJAML040573 | c.259_260insGAGATTCCT p.(Asp87delinsGlyAspSerTyr) |
17M | AML | M2 | 46,XY | ETV6, KRAS, NF1 | Standard | AAML0531 | Alive/CR | 9,10 |
| PARCEC | c.259_260insGAGACTCCT p.(Asp87delinsGlyAspSerTyr) |
23F | AML | Unknown | 46,XX | NRAS | Standard | AAML0531 | Refractory/dead | 9,10 |
| PARUTH:SJAML040605 | c.259_260insGGGACTCCT p.(Asp87delinsGlyAspSerTyr) |
9M | AML | Unknown | 46,XY.nuc ish CBFB×3 | FLT3, NRAS, BCORL1 | Standard | AAML0531 | Relapse/dead | 9 |
| PAVLBB | c.259_260insGTGACTCCT p.(Asp87delinsGlyAspSerTyr) |
15M | AML | Unknown | 46,XY | Standard | AAML1031 | Refractory/dead | Unpublished | |
| PAWIHN | c.259_260insGGGATTCCT p.(Asp87delinsGlyAspSerTyr) |
17F | AML | Unknown | 46,XX,i(7)(p10) | KDM6A | Standard | AAML1031 | Alive/CR | Unpublished |
| PAWZIX | c.259_260insGGGACTCCT p.(Asp87delinsGlyAspSerTyr) |
13F | AML | Unknown | 46,XX | ETV6, NRAS | Standard | AAML1031 | Relapse/alive/CR2 | Unpublished |
| PAXCCW | c.259_260insGGGACTCCT p.(Asp87delinsGlyAspSerTyr) |
12F | AML | Unknown | 47,XX,+6 | NRAS | Standard | AAML1031 | Alive/CR | Unpublished |
| PAXDVZ | c.259_260insGGGATTCCT p.(Asp87delinsGlyAspSerTyr) |
20M | AML | Unknown | 46,XY | BCORL1, KDM6A, NRAS | Standard | AAML1031 | Refractory/dead | Unpublished |
| SJMPAL017975 | c.259_260insGAGACAGTT p.(Asp87delinsGlyAspSerTyr) |
18M | B/M MPAL | Unknown | 51,XY,+Y,+4,+6,+13,+22 | FLT3, ASXL1, BCORL1 | Unknown | Unknown | Alive/CR | 11 |
| SJAML016545 | c.259_260insGAGACTCGT p.(Asp87delinsGlyAspSerTyr) |
16M | AML | M2 | 47,XY,+22 | FLT3 | Intermediate | AML02 | Alive/CR | 12 |
| SJAML031769 | c.259_260insGGGATTCCT p.(Asp87delinsGlyAspSerTyr) |
12M | AML | M2 | 47,XY,+6 | NRAS, FLT3 | Intermediate | AML16 | Alive/CR | Unpublished |
| SJAML033048 | c.259_260insGGGATTCCT p.(Asp87delinsGlyAspSerTyr) |
14F | AML | M2 | 46,XX | BCORL1 | Intermediate | AML16 | Alive/CR | Unpublished |
| SBJ00860 | c.259_260insGGGACTCCT p.(Asp87delinsGlyAspSerTyr) |
25M | AML | M1 | BCORL1, ETV6, KMT2D | Intermediate | Induction: 7 + 3 Cytarabine/idarubicin Consolidation: HiDAC ×3 cycles |
Alive/CR | Unpublished | |
| AML075 | c.259_260insGGGATTCGT p.(Asp87delinsGlyAspSerTyr) |
10M | AML | M0-NOS | 46,XY,inv(9)(q11q12) | NF1, KDM6A, WT1 | Intermediate | NOPHO-AML-93 | Relapse/dead | 4,13 |
| AMLNOS004 | c.259_260insGCGATTCCT p.(Asp87delinsGlyAspSerTyr) |
15M | AML | M1 | 47,XY,+6 | FLT3, BCORL1, KDM6A | Standard | NOPHO AML 2004 | Alive/CR | Unpublished |
| ALG201115 | c.259_260insGAGATTCCT p.(Asp87delinsGlyAspSerTyr) |
27M | AML | M1 | 46,XY | Intermediate | VP2010-2012 | Relapse/alive/CR2 | Unpublished | |
| MLL_75644 | c.259_260insGGGATTCCT p.(Asp87delinsGlyAspSerTyr) |
17M | AML | M1 | 46,XY | FLT3, BCORL1, ETV6 | Intermediate | Unknown | Unknown | Unpublished |
| 115225 | c.259_260insGAGATACCT p.(Asp87delinsGlyAspThrTyr) |
22M | AML | Indeterminate | 46,XY | FLT3, ETV6, WT1, DNMT3A | Unknown | Induction: 7 + 3 cytarabine/daunorubicin, with concurrent midostaurin (vs placebo) consolidation: high-dose cytarabine ×3 cycles with concurrent midostaurin (vs placebo) + 12 mth maintenance midostaurin (vs placebo) |
Relapse/alive/CR2 | 14 |
AML, acute myeloid leukemia; CR, complete response; MPAL, mixed phenotype acute leukemia.
Full information is found in Supplemental Table 1.
Figure 1.
A novel subtype of AML characterized by CBFB-GDXY mutations. (A) Graphical representation of CBFB exons 1 to 6 (NM_022845.3) showing the location (arrowhead) and sequence of the 9 bp insertion mutations (highlighted in red) relative to the wild-type complementary DNA sequence. The number of patients with each mutation is shown in the circles. The predicted protein sequence of the CBFB mutations is also shown in red. (B) Uniform manifold approximation and projection (UMAP) of expression profiles of the pediatric AML cohort (AML, n = 561; cord blood CD34+ control, n = 5) performed with the top 133 most variably expressed genes. Dots are colored by the molecular feature of the sample. (C) Gene set enrichment analysis (GSEA) between AML with CBFB insertions and non-CBF AML (left) using gene sets derived from differentially expressed genes in CBFB::MYH11 AML or RUNX1::RUNX1T1 AML against non-CBF AML. Volcano plots (right) of genes differentially expressed between AML with CBFB insertions and CBFB::MYH11 or RUNX1::RUNX1T1. (D) Mutational landscape of CBFB-GDXY AML (n = 18) in this study (mutations detected at diagnosis are shown) and CBFB::MYH11 AML (n = 65) collected in the previous study.3 Seventy-five preselected genes frequently mutated in AML were subjected to mutation calling from RNA sequencing data. Eight FLT3 mutations were detected in seven patients and are categorized as internal tandem duplications (ITD), mutations in the tyrosine kinase domain (TKD), or mutations outside the TKD (other domains). (E) (i) Giemsa-stained peripheral blood showed blasts with myeloid and monoblastic features; arrowhead marks single slender Auer rod (original magnification ×1000). (ii) Giemsa-stained bone marrow aspirate smears showed immature myeloid elements with granules, blasts, and eosinophils (original magnification ×1000); arrowhead marks salmon-colored granules in the cytoplasm of the myeloid cell (inset). (iii) Hematoxylin and eosin–stained bone marrow biopsy (original magnification ×500) showed a hypercellular marrow almost completely replaced by a diffuse infiltrate of medium-sized blasts with increased eosinophils in the background. (F) Measurable residual disease assessment of the CBFB c.259_260insGGGACTCCT mutation by ultradeep next-generation sequencing in SBJ00860.
Given these findings, we hypothesized that CBFB mutations may be recurrent in AML and a defining feature of a novel subtype. Through a combination of published data, clinical sequencing, and screening driver-negative AML cohorts from independent sources (supplemental Methods), we identified an additional 16 cases with CBFB insertions involving D87, including 15 AML and 1 B/myeloid mixed phenotype acute leukemia, for a total of 18 cases (Table 1). These additional cases also lacked a CBFB::MYH11 fusion or other known leukemic driver alterations (supplemental Table 1). CBFB mutations were confirmed in both DNA and RNA sequencing when available, or Sanger sequencing, and were confirmed to be somatic in 11 of 11 cases where matched germline data was available (supplemental Table 1; supplemental Methods; supplemental Figure 2). Remarkably, we identified 10 different nucleotide insertions at codon 87 in these 18 cases; 9 out of 10 were predicted to encode for the same in-frame GDSY amino acid change (p.(Asp87delinsGlyAspSerTyr)), with the other nucleotide insertion leading to a GDTY amino acid change (p.(Asp87delinsGlyAspThrTyr)) (Figure 1A). This highly stereotyped change at the protein level (ie, GDXY) strongly implies a functional relevance.
We next integrated 8 of these additional cases into our transcriptome cohort and observed a tight cluster of cases with CBFB insertions adjacent to the CBFB::MYH11 cluster (Figure 1B). Gene set enrichment analysis confirmed broad similarities between AML with CBFB insertions and CBF AMLs (Figure 1C; supplemental Figure 3; supplemental Table 2). However, CBFB insertion cases showed uniquely high expression of BCL2L14, MEIS1, and HOXA cluster genes, demonstrating a more stem-like signature compared with CBFB::MYH11 AML.
We also examined the cooperating mutations in 10 CBFB insertion cases from RNA sequencing data and integrated these findings with mutational information from other studies (Figure 1D; supplemental Tables 1 and 3). Recurrent mutations were detected in BCORL1 (7/18 [39%]), FLT3 (7/18 [39%]), NRAS (6/18 [33%]), ETV6 (5/18 [28%]), KDM6A (4/18 [22%]), and NF1 (2/18 [11%]). FLT3 tyrosine kinase domain (TKD) mutations were most common, although internal tandem duplications and mutations outside the TKD were also observed. Overall, these mutations are different from the mutational spectrum previously reported for CBF leukemias,7 most notably the absence of KIT mutations and a higher frequency of FLT3-TKD and BCORL1 mutations. Recurrent chromosomal alterations in the CBFB insertion group included trisomy 6 and trisomy 22, whereas trisomy 8 was not observed (Figure 1D; supplemental Table 1). Additionally, the CBFB mutation was conserved at both diagnosis and relapse in 2 cases profiled at both time points (AML075 and 115225). Collectively, these data suggest that CBFB insertions are a subtype-defining lesion, and we have provisionally termed this group CBFB-GDXY.
Like CBF AML, AMLs with CBFB-GDXY mutations were observed in both children and adults, but were enriched in adolescent and young adult age groups (median age, 16.5 years; range, 9-27 years). Overall, this mutation in AML cohorts was rare, including 3 of 188 in the TARGET pediatric AML cohort, 5 of 1048 in the pediatric AAML1031 cohort and 1 of 350 in the Clinseq-AML Swedish adult cohort, whereas more than 2000 cases from multiple large cohorts composed primarily of adult AMLs did not harbor a CBFB insertion (supplemental Data for a description of cohorts screened). Additionally, unlike the typical myelomonocytic morphology with abnormal eosinophils (FAB M4 Eo) observed for CBFB::MYH11 AML, CBFB-GDXY AML had fewer mature morphologies (FAB M0, n = 1; FAB M1, n = 4; or FAB M2, n = 4), consistent with the stem-related expression profiles, where morphologic reports were available. However, an increase in eosinophils was still observed (Figure 1E). Like RUNX1::RUNX1T1 AML,8 we noted that CBFB-GDXY AML may express CD19 (supplemental Figure 4; supplemental Table 1). Further supporting this observation is the identification of the GDXY insertion in 1 case of B/myeloid mixed phenotype acute leukemia.
This cohort is small and collected from different sources with varied treatment protocols, precluding a definitive assessment of the impact of this mutation on outcomes. However, 8 of 17 patients (where data were available) had either relapsed or refractory disease after initial treatment, whereas patients with RUNX1::RUNX1T1 or CBFB::MYH11 AMLs commonly have a good outcome and these AMLs are considered favorable risks. To investigate the treatment response in one patient, we designed a CBFB mutation-specific ultradeep next-generation sequencing assay (supplemental Methods) for longitudinal tracking of measurable residual disease. This 25-year-old patient (SBJ00860) was treated with cytarabine and idarubicin induction (7 + 3), followed by high-dose cytarabine consolidation. Measurable residual disease assessment after each cycle of chemotherapy showed detectable but decreasing CBFB insertion variant allele frequency, becoming undetectable after the last cycle of therapy and remaining undetectable at 9 months of follow-up (Figure 1F).
In summary, we have reported a novel subtype of AML characterized by recurrent in-frame insertion mutations in CBFB, leading to a GDXY amino acid sequence change at position D87. Molecular characterization demonstrated transcriptional similarity to CBF AML, while also highlighting an enrichment of FLT3-TKD mutations, lack of KIT mutations, and stemness-related gene expression signature. Recognition of this subtype and further study in clinical trials, as well as investigation of the underlying leukemogenic mechanism of the CBFB insertion, will be important to understand the full clinical relevance of this novel entity.
Conflict-of-interest disclosure: P.G.E. receives an annual payment related to the Walter and Eliza Hall Institute distribution of royalties scheme. P.G.E. consults for Illumina. The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank all patients and their families from all participating institutions for their contribution of the biological specimens used in this study.
This work was supported by grants from the Wilson Centre for Blood Cancer Genomics and the Snowdome Foundation, as well as the American Lebanese and Syrian Associated Charities of St. Jude Children's Research Hospital. The authors also gratefully acknowledge the Haematology Tissue Bank (Peter MacCallum Cancer Centre/Royal Melbourne Hospital) for assistance with sample collection and the Genomics Core Facility and Genomics Platform Group (University of Melbourne Centre for Cancer Research) for sequencing and analysis support, as well as the Biorepository and the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Center (P30 CA021765, Cancer Center Support Grant). Tumor samples and coded data were supplied by the Children’s Cancer Centre Biobank at the Murdoch Children’s Research Institute and The Royal Children’s Hospital (mcri.edu.au/research/projects/childrens-cancer-centre-biobank). The establishment and running of the Children’s Cancer Centre is made possible through generous support by Cancer In Kids @ RCH (www.cika.org.au), The Royal Children’s Hospital Foundation, and the Murdoch Children’s Research Institute. P.G.E. acknowledges the support of the Samuel Nissen Charitable Foundation and the Leukemia and Lymphoma Society USA Specialized Center of Research (SCOR) Project Grant “Apoptosis Controllers.” J.M.K. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is a previous recipient of the V Foundation Scholar Award (Pediatric).
This research content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: G.R., M.U., P.B., and J.M.K. conceived of the project; G.R., M.U., L.H., S.L., M.K.H., J.M., M.K., J.E.R., H.J.K., P.G.E., H.G., I.S.T., S.M.G., C.H., R.B.D., T.J.L., S.M., X.M., P.B., and J.M.K. identified cases and provided genomic data and clinical information; G.R., M.U., J.M., and X.M. analyzed genomic data and collated clinical information; G.R., M.U., P.B., and J.M.K. wrote the first version of the manuscript; all authors reviewed and approved the final version of the manuscript.
Footnotes
Data will be provided and shared upon request to Jeffery M. Klco, jeffery.klco@stjude.org; and Piers Blombery, piers.blombery@petermac.org.
The online version of this article contains a data supplement.
Contributor Information
Piers Blombery, Email: piers.blombery@petermac.org.
Jeffery M. Klco, Email: jeffery.klco@stjude.org.
Supplementary Material
References
- 1.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100(13):4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 2.Schlenk RF, Benner A, Krauter J, et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2004;22(18):3741–3750. doi: 10.1200/JCO.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Umeda M, Ma J, Huang BJ, et al. Integrated genomic analysis identifies UBTF tandem duplications as a recurrent lesion in pediatric acute myeloid leukemia. Blood Cancer Discov. 2022;3(3):194–207. doi: 10.1158/2643-3230.BCD-21-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stratmann S, Yones SA, Mayrhofer M, et al. Genomic characterization of relapsed acute myeloid leukemia reveals novel putative therapeutic targets. Blood Adv. 2021;5(3):900–912. doi: 10.1182/bloodadvances.2020003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montefiori LE, Bendig S, Gu Z, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage-ambiguous stem cell leukemia. Cancer Discov. 2021;11(11):2846–2867. doi: 10.1158/2159-8290.CD-21-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruijn M.F.T.R., Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23(24):4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 7.Faber ZJ, Chen X, Gedman AL, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48(12):1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter K, Cockerill PN, Barlow R, et al. Aberrant expression of CD19 in AML with t(8;21) involves a poised chromatin structure and PAX5. Oncogene. 2010;29(20):2927–2937. doi: 10.1038/onc.2010.56. [DOI] [PubMed] [Google Scholar]
- 9.Bolouri H, Farrar JE, Triche T, Jr., et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555(7696):371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander TB, Gu Z, Iacobucci I, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562(7727):373–379. doi: 10.1038/s41586-018-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornerod M, Ma J, Noort S, et al. Integrative genomic analysis of pediatric myeloid-related acute leukemias identifies novel subtypes and prognostic indicators. Blood Cancer Discov. 2021;2(6):586–599. doi: 10.1158/2643-3230.BCD-21-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratmann S, Yones SA, Garbulowski M, et al. Transcriptomic analysis reveals proinflammatory signatures associated with acute myeloid leukemia progression. Blood Adv. 2022;6(1):152–164. doi: 10.1182/bloodadvances.2021004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petti AA, Khan SM, Xu Z, et al. Genetic and transcriptional contributions to relapse in normal karyotype acute myeloid leukemia. Blood Cancer Discov. 2022;3(1):32–49. doi: 10.1158/2643-3230.BCD-21-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.