Abstract
NeuroD-related factor (NDRF) is a basic helix–loop–helix (bHLH) protein whose expression is restricted to the central nervous system, and is considered to be responsible for maintenance of differentiated neurons as well as neurogenesis. NDRF structurally resembles NeuroD in the bHLH region and can induce neurogenesis ectopically in ectodermal cells of the Xenopus embryo. In this study, we delineated the functional domains of NDRF. Using GAL4/NDRF fusion proteins, we identified the C-terminal activation domain (C-AD) in NDRF between amino acid positions 294 and 383. This region was highly homologous to one part of the activation domain of NeuroD. We further investigated the transactivational function of C-AD in the mouse type 1 inositol 1,4,5-trisphosphate receptor promoter, which has an NDRF site. Truncation of C-AD resulted in reduction of the activation function, whereas the DNA-binding specificity was not affected. These results suggest that C-AD has a stimulatory function in the mammalian nervous system.
INTRODUCTION
In myogenesis, myogenic basic helix–loop–helix (bHLH) factors such as MyoD, myogenin, Myf5 and MRF4 are involved in cell fate determination (1–4). Similarly, formation of central and peripheral neurons of Drosophila have been shown to be controlled by bHLH proteins encoded by the achaete–scute complex (AS-C) (5) and atonal genes (6,7). Recently, mammalian homologues of Drosophila AS-C (Mash) and atonal (MATH, neurogenin, NeuroD) have been identified. In mammals, bHLH factors such as Mash1 (8,9), neurogenin (10) and MATH1 (11) are expressed in neural precursor cells, whereas NeuroD (12) and MATH2/NEX-1 (13,14) are expressed in postmitotic neural cells (15). These tissue-specific bHLH proteins (i.e. class B bHLH factors) form heterodimers with ubiquitous (class A) bHLH factors, including E2A-encoded E12 and E47 proteins (16).
Among various mammalian neurogenic bHLH factors, NeuroD and MATH2/NEX-1 are highly conserved in their bHLH regions and belong to a single subfamily. Since their expression persists in the adult central nervous system (CNS), they are believed to play a crucial role in the development and maintenance of the mammalian CNS. NeuroD was identified as a neural differentiation factor in Xenopus ectodermal cells undergoing neurogenesis (12). NeuroD also has been identified as BETA2 (17), which is involved in the expression of the hamster insulin gene in the pancreatic β cells. NeuroD/BETA2 knockout mice showed defective morphogenesis of pancreas (18). NeuroD/beta2 transgenes specifically expressed in the pancreas of the neuroD-null mice rescued them from neonatal lethality. These mice showed neural defects in the granule layers of the cerebellum and hippocampus (19). Yasunami et al. identified another bHLH factor, NeuroD-related factor (NDRF), that was also categorized as a NeuroD-type bHLH factor with respect to structure of its bHLH region (20). NDRF has also been referred to as NeuroD2 and KW8 (21,22). NDRF can convert ectodermal cells into neural cells in Xenopus embryos as can NeuroD, and its expression is restricted to the CNS of adult mice (21). In the mammalian embryonic and postnatal CNS, expression patterns of those multiple NeuroD-like factors are basically unique, though overlapping distribution is observed in various subsets of neurons (21,23). Although NeuroD-like proteins are important for the nervous system, their target genes and transcriptional activation mechanism have not been fully elucidated.
Inositol 1,4,5-trisphosphate receptor type 1 (IP3R1) is a major calcium ion channel protein that is abundantly expressed in the CNS, especially in the cerebellar Purkinje and hippocampal neurons (24–26). IP3R1 plays a crucial role in neural function via regulation of calcium ion concentration. Targeting disruption of the IP3R1 gene resulted in ataxia and epileptic seizures in mice (27). A transgenic mouse study revealed that the mouse IP3R1 promoter sequence from –528 to +169 was responsible for predominant gene expression in cerebellar Purkinje cells and the hippocampus (28). We have studied CNS-specific transcriptional regulation of the mouse IP3R1 gene using a cell-free in vitro transcription system, and demonstrated that a CNS-enriched bHLH factor positively functions in transcriptional regulation of the IP3R1 gene upon binding to its E-box-containing element designated as box-I (29). A subsequent study demonstrated that NDRF specifically binds to the box-I element through heterodimerisation with E47 and activates transcription of the mouse IP3R1 gene (30). In the postnatal mouse CNS, NDRF and IP3R1 mRNAs spatio-temporally coexisted in various subsets of neurons, suggesting that NDRF is involved in cell-specific transcriptional regulation of the IP3R1 gene in the CNS (30). In this study, we identified the activation domain within the C-terminal region of NDRF. Furthermore, we compared the structures and function for transcriptional stimulation of C-terminal regions between NDRF and NeuroD.
MATERIALS AND METHODS
DNA constructs
pBIND (Promega, Madison, WI) was used as an expression vector for GAL4 chimerical constructs. This vector has a GAL4 DNA-binding domain (DBD) as a protein-coding sequence and expresses a fusion protein with the GAL4 DBD (1–147; amino acid positions from the N-terminus) under the control of the cytomegarovirus immediate early promoter. This vector also expresses the Renilla luciferase (31) driven by the SV40 early promoter within identical DNA. Mouse NDRF or NeuroD cDNA sequence was inserted into pBIND to produce in-frame GAL4-fusion proteins. The pG5luc reporter plasmid (Promega) contains five GAL4-binding sites upstream of the TATA box-containing adenovirus major late promoter (MLP) from –35 to +33, and expresses the firefly luciferase gene (32) dependent on a GAL4-fused activator. In assaying NDRF-dependent transcriptional activation, wild-type (1–383) and C-terminally truncated (1–315) NDRFs and the bHLH region of E47 (473–651) were expressed under control of the cytomegarovirus immediate early promoter using the pCI-neo expression vector (Promega). The mouse IP3R1 promoter from –524 to +165 is located upstream of the firefly luciferase gene and was used as a reporter plasmid (30).
Cell culture and transfection
PC12 cells were maintained in RPMI1640 (Sigma, St Louis, MO) supplemented with 10% foetal calf serum. Transfection was performed using Transfast reagent (Promega). Cells were cultured in 24-well plates at 3 × 104 cells/well 24 h prior to transfection. 400 ng of DNA, which was diluted in 200 µl of OPTI-MEM (Gibco BRL, Rockville, MD) containing 2.4 µl of Transfast reagent and then incubated for 15 min at room temperature, was changed to growth medium, and cells were incubated at 37°C for 1 h. Then, 400 µl of growth medium was added, and the cells were then incubated for 48 h for expression of the luciferase. The luciferase activity was analysed by the Dual-Luciferase Reporter Assay System (Promega). The reporter gene activities were normalised by the Renilla luciferase activities directed by the SV40 early promoter.
Western blotting
PC12 cells were suspended with phosphate-buffered saline 2 days after the transfection. One-tenth of the cells were used for the Renilla luciferase activity for normalisation of the transfection efficiency, and the remaining cells were boiled for 3 min in SDS loading buffer [50 mM Tris–HCl (pH 6.8), 100 mM DTT, 2% SDS, 0.1% BPB and 10% glycerol]. The amounts of extracts used for western blotting were normalised by the Renilla luciferase activity. Proteins separated by 10–15% SDS–polyacrylamide gel were transferred to a polyvinylidene difluoride membrane (Millipore, Waltham). Horseradish peroxidase-conjugated protein A (Amersham-Pharmacia, Bucks, UK) was used to detect the first antibody. Specific antibody interaction was detected by enhanced chemiluminescence plus western blotting detection reagent (Amersham).
Preparation of recombinant proteins and electrophoretic mobility shift assay (EMSA)
Recombinant NDRF and E47 carrying an oligo-histidine tag at their N-terminus were expressed in Escherichia coli BL21(DE3)pLysS by use of the pET vector system (33). Soluble proteins were purified using Ni2+-agarose (Qiagen, Hilden, Germany). EMSA was performed as previously described (30). For the Ebx16-1 probe, two oligonucleotides, 5′-GAATTCTAGTGACACCTGACTTG and 5′-GAATTCAAGTCAGGTGTCACTAG, were annealed. Ebx16M has the mutated E-box sequence CCAAGG instead of CACCTG of Ebx16-1 (underlined). NDRF and E47 were preincubated at 0°C for 1 h in a reaction mixture containing 50 mM NaCl, 0.1 mM EDTA, 20 mM HEPES–KOH (pH 7.5), 0.5 mM DTT and 10% glycerol, and then for an additional 30 min with probe DNA. Competitor DNA was added to the reaction mixture 15 min prior to the addition of probe DNA.
RESULTS
Transcriptional activation domain is located in the C-terminal region of NDRF
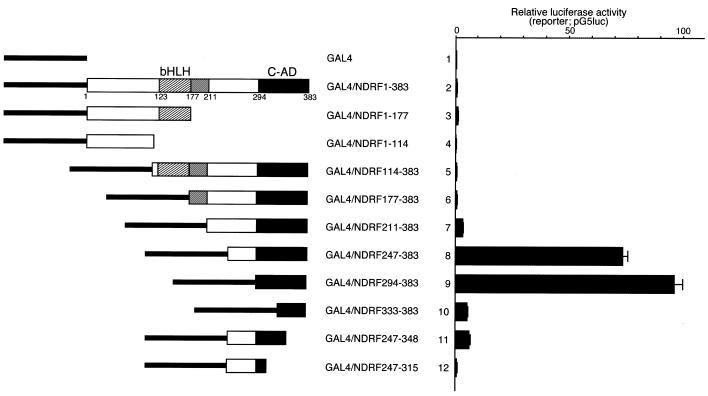
To identify the transcriptional activation domain in NDRF, we performed a transient reporter assay using various effector plasmids that express the GAL4 DBD/NDRF fusion proteins (Fig. 1). Experimental luciferase activities were normalised by the reference Renilla luciferase activity. The full-length (GAL4/NDRF1–383) and N-terminal region of NDRF (GAL4/NDRF1–177 and GAL4/NDRF1–114) displayed no significant transcriptional activity (Fig. 1, lanes 2–4). However, truncation of the N-terminus enhanced the transcription activity markedly (Fig. 1, lanes 7–9). The highest activity was seen for GAL4/NDRF294–383 carrying an NDRF sequence from 294 to 383. Further deletion reduced the activation function severely (Fig. 1, lanes 10–12). Western blotting of GAL4-fusion proteins revealed that the results shown in Figure 1 were not due to differences in the amounts of fusion proteins (Fig. 2A). For example, the amounts of GAL4/NDRF247–383 and GAL4/NDRF294–383 (Fig. 2A, lanes 7 and 8, respectively), both of which exhibited potent transcription stimulation activities, were comparable with those of other fusion proteins. GAL4/294–383 revealed the highest activity in PC12 and other cell lines such as Neuro2a, HeLa and NIH 3T3, while some cell specificity was observed in transcriptional activity (data not shown). From these results, we assigned the region from 294 to 383 as the C-terminal activation domain (C-AD) of NDRF.
Figure 1.
Transcriptional activation domain of mouse NDRF is located within the C-terminus. NDRF deletion mutants linked to the GAL4 DBD (1–147) are schematically indicated. The bHLH region (hatched boxes), the highly conserved region among NeuroD-like factors (grey boxes) and the C-terminal activation domain (C-AD) (black boxes) are indicated. PC12 cells were transfected with pG5luc reporter plasmid and individual experimental GAL4/NDRF-expressing vectors. Normalised luciferase activities are shown. Data are mean + S.E. (bars) values obtained from five independent experiments.
Figure 2.
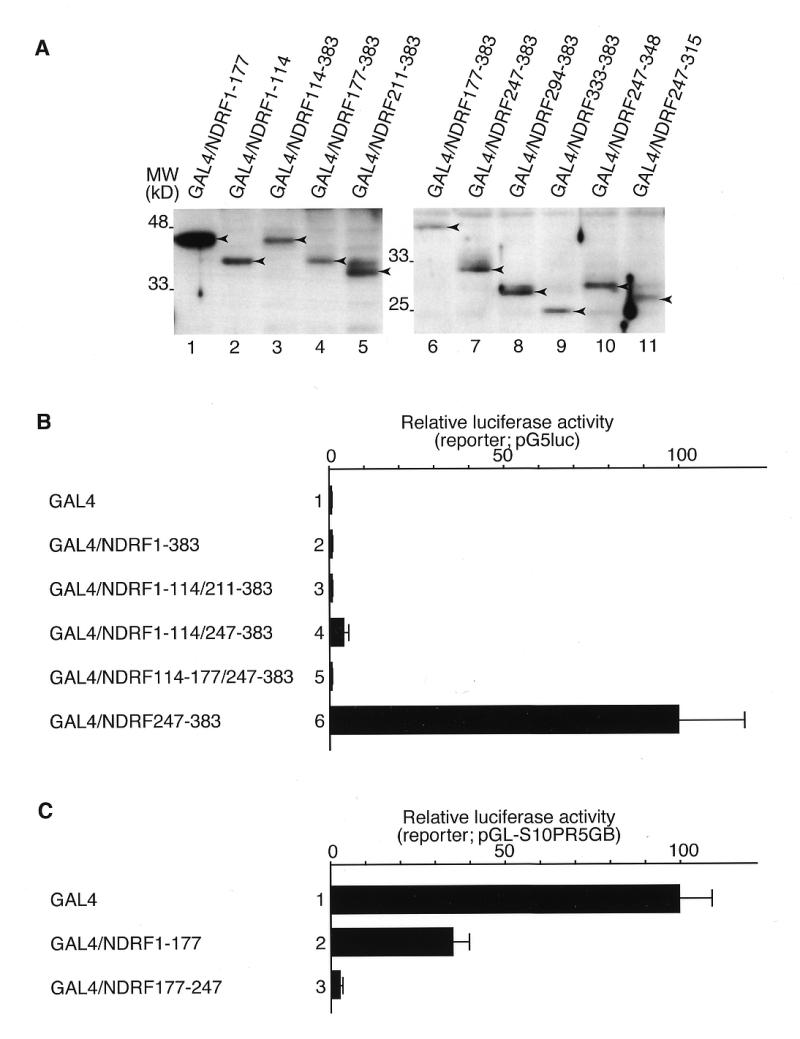
Identification of repressive function within NDRF. (A) Extracts of PC12 cells expressed GAL4/NDRF fusion proteins were analysed by western blotting using the antibody against the GAL4 DBD. Arrowheads indicate positions of each GAL4 fusion protein. (B) Transcriptional activation function of GAL4/NDRF mutants were measured as Figure 1. Data were obtained from four independent experiments. (C) Expression vectors of GAL4/NDRF1–177 and GAL4/NDRF177–247 were co-transfected to PC12 cells together with the reporter plasmid pGL-S10PR5GB, containing the SCG10 promoter flanked by five copies of the GAL4-binding site linked to a luciferase gene (34). Results were obtained from four independent experiments.
From the results of Figure 1, some negative transcriptional regulatory function appeared to be localised downstream from 247 in the N-terminus. We constructed several internal deletions to investigate the repressive function of individual regions. As shown in Figure 2B, transcriptional activation function within 247–383 was repressed when fused to each sequence upstream of 247. We further studied whether this sequence functions as a bona fide repressor when fused to the DBD. pGL-S10PR5GB (34) was used in this assay, in which the minimal adenovirus major late promoter pG5luc was replaced with the SCG10 promoter, since basal promoter activity of pG5luc was faint. GAL4/NDRF1–177 showed weak (~3-fold) but significant repressive function for SCG10 promoter (Fig. 2C, lane 2). Moreover, GAL4/NDRF177–247 alone remarkably repressed the SCG10 promoter activity (~40-fold; Fig. 2C, lane 3).
Role of the C-AD in the IP3R1 promoter function
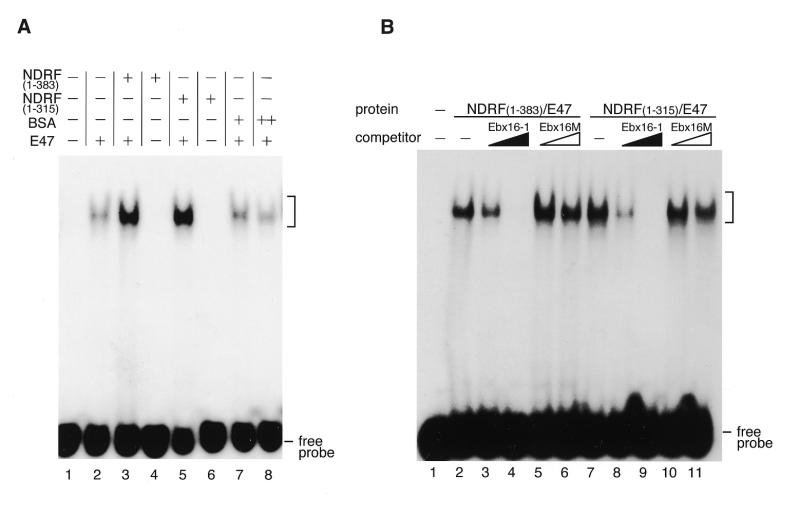
We investigated whether the C-AD mediates E-box-directed transcriptional stimulation by using C-AD-truncated NDRF. At first, to examine the DNA-binding activity of both wild-type and C-AD truncated NDRF, we performed EMSA. For probe sequence, we used the box-I element of the IP3R1 promoter, which contains an E-box sequence and was suggested to be a target of NDRF (30). Recombinant proteins of wild-type NDRF (1–383), C-AD truncated NDRF (1–315) and E47 (473–651) were used for the assay together with box-I sequence. Consistent with a previous report (30), E47 per se bound to the box-I sequence weakly (Fig. 3A, lane 2). Addition of the wild-type NDRF resulted in a marked enhancement of the DNA-binding of E47, whereas NDRF alone did not bind to the probe DNA (Fig. 3A, lanes 3 and 4). The C-AD-truncated NDRF (1–315) enhanced DNA-binding ability of E47 to the same degree as did the wild-type NDRF (Fig. 3A, lanes 5 and 6). We also performed competition EMSA to examine how DNA-binding specificity is altered in accordance with the structured NDRF. The specific competitor (Ebx16-1) reduced the shifted band generated with the wild-type NDRF and C-AD-truncated NDRF (Fig. 3B, lanes 3–4 and 8–9). On the other hand, the shifted band was not affected by the addition of E-box-mutated competitor (Fig. 3B, lanes 5–6 and 10–11). Thus, no difference in DNA-binding specificity was observed between wild-type NDRF and its mutant.
Figure 3.
Comparison of DNA-binding functions between wild-type NDRF and its C-terminal truncation. (A) EMSA for wild-type and C-AD-truncated NDRFs. Probe, box-I sequence in the IP3R1 promoter from –334 to –318 containing an E-box. Each recombinant protein was used as indicated. (+) and (++) indicate 1 ng and 2 ng of proteins, respectively. Positions of the nucleoprotein complexes are shown by brackets. (B) Elimination of C-AD does not affect the DNA-binding specificity. DNA-binding specificity of wild-type and C-AD truncated NDRFs was examined using box-I competition (Ebx16-1) and its E-box mutant (Ebx16M).
To investigate the role of C-AD in E-box-directed transcriptional activation, we performed luciferase reporter gene assay. PC12 cells were transfected with an expression vector producing wild-type NDRF (1–383) or its C-AD-truncated version (1–315) (Fig. 4A and B). The IP3R1 promoter was stimulated ~5-fold by co-transfection of the wild-type NDRF (1–383), while it was activated only 2-fold by the C-AD-truncated NDRF (Fig. 4C, lanes 4 and 7). Since NDRF enhanced DNA-binding ability of E47 (Fig. 3), we examined the effects of the transcriptional activation function of class A proteins using a mutant E47 (473–651) that lacks its activation domain(s) (Fig. 4A). Transcriptional activation functions of both wild-type (1–383) and mutant (1–315) NDRF were repressed by co-expression of E47 (473–651) (Fig. 4C, lanes 6 and 9), while no repression was observed by co-expression of wild-type E47 (1–651) (Fig. 4C, lanes 5 and 8). However, though significant transcriptional promotion function was still retained in NDRF (1–383) (Fig. 4C, lane 6), no significant activation was detected for NDRF (1–315) (Fig. 4C, lane 9). Effect of co-transfected E47 expression vectors on the NDRF expression was only slight (Fig. 4D). Thus, it is indicated that the C-AD is a unique E-box-mediated activation domain in NDRF, and no activation function appears to exist in the N-terminal and bHLH region. A mutant E47 alone (473–651) had no significant effect on the reporter activity (Fig. 4C, lane 3). This phenomenon is thought to be due to the absence of NDRF and its functionally related factors in the PC12 cells. Therefore, we could not detect NDRF protein by western blotting and no significant reduction in IP3R1 promoter activity in PC12 cells was observed by the deletion of the box-I element (data not shown).
Figure 4.
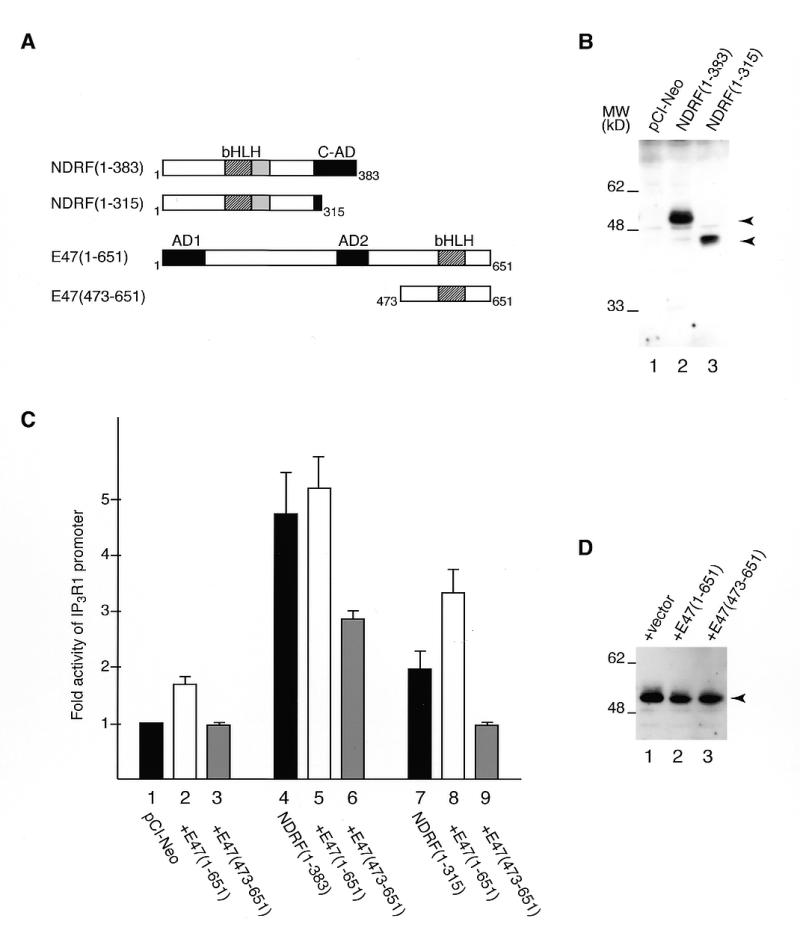
Effect of C-AD on the stimulation of the IP3R1 promoter. (A) Schematic representation of NDRF, E47 and their mutants. (B) Detection of full-length and C-AD-truncated NDRF proteins expressed in PC12 cells. Proteins were detected by western blotting using an antibody that recognises the bHLH region of NDRF. Specific bands are indicated by arrowheads. (C) Luciferase activity directed by NDRF and its truncation mutant in the presence of control vector (black bars), wild-type E47 (open bars) or mutant E47 (grey bars). Reporter plasmid containing the IP3R1 promoter upstream of a luciferase gene (100 ng) was co-transfected with 200 ng of NDRF vector, 100 ng of E47 vector and 0.4 ng of control vector express the Renilla luciferase under the direction of the SV40 early promoter. Data are mean values obtained from five independent assays. (D) Analysis of NDRF expression level in the condition of (C) (lanes 4–6). The arrowhead indicates specific signals of NDRF (1–383) detected by western blotting.
Comparison of the activation domain of NDRF with that of NeuroD
Recently, Sharma et al. reported that the transcriptional activation domain of NeuroD is located in the C-terminal region (35). Since there has been little study on functional comparisons between the NeuroD-like factors, we extensively compared activation domains of NDRF and NeuroD using GAL4/NeuroD-expression vectors (Fig. 5A). The activation domain of mouse NeuroD (190–357) corresponds to a region from 211 to 383 of mouse NDRF (Figs 5A and 6). We compared luciferase activities directed by GAL4/NDRF and GAL4/NeuroD in PC12 cells. Western blot analysis showed that GAL4-fusion proteins were expressed to similar levels in PC12 cells (Fig. 5B). GAL4/NDRF211–383 showed only one-thirtieth of the activity of GAL4/NDRF294–383 (Fig. 5C, lanes 1 and 2). In contrast, the activity of GAL4/NeuroD190–357 was three times higher than that of GAL4/NeuroD267–357 (Fig. 5C, lanes 3 and 4). These observations suggest that there are repressive and stimulatory functions in 211–293 of NDRF and 190–266 of NeuroD, respectively. Although GAL4/NDRF294–383 and GAL4/NeuroD267–357 exhibited strong stimulatory ability (Fig. 1, lane 9; Fig. 5C, lane 4), further deletions resulted in loss of activation function (Fig. 1, lane 10; Fig. 5C, lane 6), suggesting that both 294–383 of NDRF and 267–357 of NeuroD contribute to the transcriptional activation.
Figure 5.
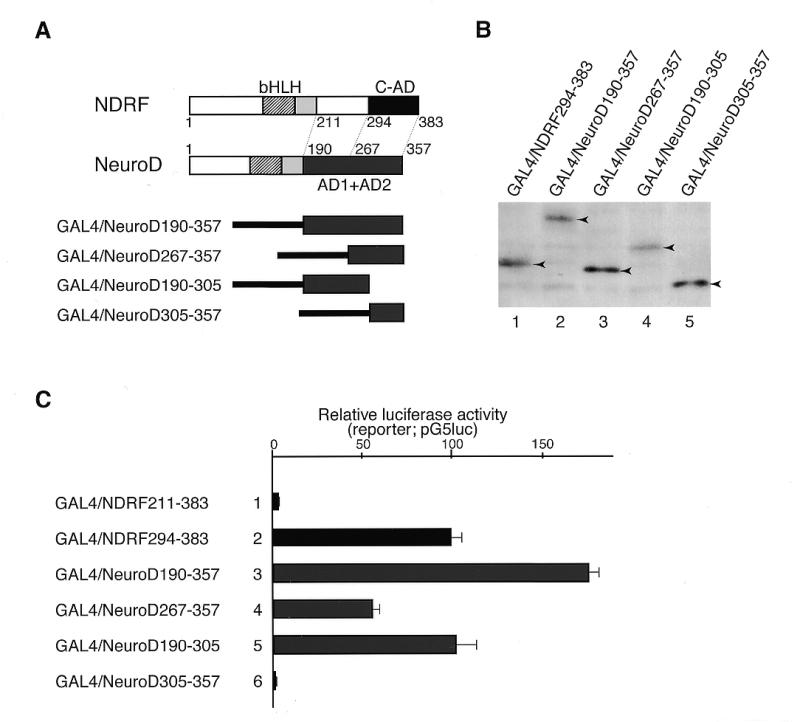
Functional comparison of C-terminal activation domains of NDRF and NeuroD. (A) Schematic representation of deletion mutants of NeuroD linked to the GAL4 DBD. The activation domain of NeuroD comprises AD1 and AD2, which have comparable activities among cell lines (35). (B) Western blot analysis of GAL4/NeuroD fusion proteins expressed in PC12 cells using the antibody against the GAL4 DBD. Specific signals are indicated by arrowheads. (C) Luciferase activity directed by various GAL4/NDRF and GAL4/NeuroD constructs in PC12 cells. Data are mean values obtained from five independent assays.
Figure 6.
Comparison of C-terminal regions of mouse NDRF and NeuroD. Amino acid sequences were aligned by the CLUSTALW program (36). Identical and similar amino acids between two proteins are indicated by asterisks and dots, respectively. C-AD of NDRF and its corresponding region of NeuroD are indicated by the solid line and dotted line, respectively.
Figure 6 shows an alignment of C-terminal regions of mouse NDRF and NeuroD. Interestingly, homology between 294–383 of NDRF and 267–357 of NeuroD was considerably higher than 211–293 of NDRF and 190–266 of NeuroD (Fig. 6 and Table 1). A structurally homologous region of NeuroD was found in a sequence indicated by the dotted line in Figure 6. The corresponding region in Xenopus NeuroD for the mouse sequence was involved in the induction of ectopic neurogenesis in Xenopus embryos (35).
Table 1. Comparison between transcriptional activation domains of NDRF and NeuroDa.
| NDRF vs NeuroDb | Identity | Positive | Gap |
|---|---|---|---|
| 211–293:190–266 | 22/85 (24%) | 33/85 (36%) | 10/85 (12%) |
| 294–383:267–357 | 41/91 (45%) | 62/91 (68%) | 3/91 (3%) |
aThe calculation is based on the results of Figure 6.
bAmino acid positions are indicated.
DISCUSSION
In this study, we identified the functional domain of the mouse NDRF. Using GAL4/NDRF fusion proteins, we detected an activation domain within the C-terminal region (Fig. 1). We performed a reporter assay in PC12 cells because significant activation of the IP3R1 promoter by NDRF has been observed in these cells (30). While no significant activation function was found in GAL4/NDRF1–383, elimination from the N-terminus resulted in strong transcriptional activation function (Fig. 1). Thus, it is suggested that some negative function is localised in certain regions within NDRF. Obviously, the negative function of 177–247 is significant. Because this region is directly linked to the C-AD, activation function of C-AD was revealed in accordance with the deletion of this region (Fig. 1, lanes 6–9). This region per se has potent repressive function (Fig. 2C, lane 3). Biological significance of these negative sequences is still unclear. We suppose that the negative region represses the C-AD function when NDRF does not bind properly to the E-box containing the target sequence.
To elucidate the role of C-AD in a physiological situation, full-length and C-AD-truncated NDRFs were analysed in the mouse IP3R1 promoter. The IP3R1 promoter activity was significantly reduced by elimination of C-AD, leaving its dimerisation and DNA-binding activity of NDRF unaltered (Figs 3 and 4C). We assumed that the weak but significant activation function of C-AD-truncated NDRF is elicited by a class A bHLH protein that forms a heterodimer with NDRF. In co-expression of full-length and C-AD-truncated NDRFs with E47 (473–651) that lacks its activation domains, NDRF (1–315) showed no significant activity, whereas NDRF (1–383) still maintained significant activation properties (Fig. 4C, lanes 6 and 9). These observations were thought to be a result of a dominant negative effect of the E47 mutant because reduced transcriptional activity was not provided by co-expression of wild-type E47 (Fig. 4C, lanes 5 and 8). Thus, we speculate that C-AD is a unique region in NDRF that acts as an activation domain. Consequently, two mechanisms of NDRF-directed neural gene expression are thought to be involved concomitantly or independently. (i) NDRF promotes the class A bHLH proteins to bind to the E-box-containing target sequence, and enhances the gene expression by the activation domains of class A bHLH factors. (ii) Furthermore, NDRF stimulates gene expression by its C-terminal activation domain.
Recently, Sharma et al. identified transcriptional activation domains within the C-terminal region of hamster and Xenopus NeuroD proteins (35). We compared the functions of activation domains between NDRF and NeuroD. Elimination of 211–293 from the C-terminal region of NDRF resulted in a significant increase in the stimulation capacity (Fig. 5C, lanes 1 and 2), suggesting that there is a negative function in the eliminated sequences. Interestingly, the corresponding region of NeuroD (i.e. from 190 to 266) had, on the contrary, a positive function (Fig. 5C, lanes 3 and 4). This region has a potent activating potential in cell lines that originate from pancreatic β cells and is thought to play a role in insulin gene transcription. It is noteworthy that this region of NeuroD is not critical for neurogenesis (35). These observations are thought to be relevant for tissue distribution of NeuroD and NDRF because NeuroD is expressed both in the CNS and pancreatic β-cells, but NDRF is expressed exclusively in the CNS in adult tissues (18). It therefore seems reasonable that NDRF lacks the activation domain for the pancreatic gene expression. On the contrary, both 294–383 of NDRF and 267–357 of NeuroD act as activation domains (Fig. 5C). As shown in Figure 6 and Table 1, 294–383 of NDRF and 267–357 of NeuroD showed high sequence similarity. Moreover, these regions are included in the functional domains that are involved in the induction of ectopic neurogenesis (Fig. 6, dotted line) (35). From these observations, 294–383 (C-AD) of NDRF and 267–357 of NeuroD are thought to have common functions and to use a common mediatory machinery in neural gene regulation.
Acknowledgments
ACKNOWLEDGEMENTS
Y.K. and N.O. are research fellows of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Davis R.L., Weintraub,H. and Lassar,A.B. (1987) Cell, 51, 987–1000. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes S.J. and Konieczny,S.F. (1989) Genes Dev., 3, 2050–2061. [DOI] [PubMed] [Google Scholar]
- 3.Braun T., Winter,B., Bober,E. and Arnold,H.H. (1990) Nature, 346, 663–665. [DOI] [PubMed] [Google Scholar]
- 4.Weintraub H. (1993) Cell, 75, 1241–1244. [DOI] [PubMed] [Google Scholar]
- 5.Villares R. and Cabrera,C.V. (1987) Cell, 50, 415–424. [DOI] [PubMed] [Google Scholar]
- 6.Jarman A.P., Grau,Y., Jan,L.Y. and Jan,Y.N. (1993) Cell, 73, 1307–1321. [DOI] [PubMed] [Google Scholar]
- 7.Jan Y.N. and Jan,L.Y. (1993) Cell, 75, 827–830. [DOI] [PubMed] [Google Scholar]
- 8.Johnson J.E., Birren,S.J. and Anderson,D.J. (1990) Nature, 346, 858–861. [DOI] [PubMed] [Google Scholar]
- 9.Cau E., Grandwohl,G., Fode,C. and Guillemot,F. (1997) Development, 124, 1611–1621. [DOI] [PubMed] [Google Scholar]
- 10.Ma Q., Kintner,C. and Anderson,D.J. (1996) Cell, 87, 43–52. [DOI] [PubMed] [Google Scholar]
- 11.Akazawa C., Ishibashi,M., Shimizu,C., Nakanishi,S. and Kageyama,R. (1995) J. Biol. Chem., 270, 8730–8738. [DOI] [PubMed] [Google Scholar]
- 12.Lee J.E., Hollenberg,S.M., Snider,L., Turner,D.L., Lipnick,N. and Weintraub,H. (1995) Science, 268, 836–844. [DOI] [PubMed] [Google Scholar]
- 13.Bartholoma A. and Nave,K.A. (1994) Mech. Dev., 48, 217–228. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu C., Akazawa,C., Nakanishi,S. and Kageyama,R. (1995) Eur. J. Biochem., 229, 239–248. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama R. and Nakanishi,S. (1997) Curr. Opin. Genet. Dev., 7, 659–665. [DOI] [PubMed] [Google Scholar]
- 16.Roberts V.J., Steenbergen,R. and Murre,C. (1993) Proc. Natl Acad. Sci. USA, 90, 7583–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naya F.J., Strellrecht,C.M. and Tsai,M.J. (1995) Genes Dev., 9, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 18.Naya F.J., Huang,H.P., Qiu,Y., Mutoh,H., DeMayo,F.J., Leiter,A.B. and Tsai,M.J. (1997) Genes Dev., 11, 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyata T., Maeda,T. and Lee,J.E. (1999) Genes Dev., 13, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasunami M., Suzuki,K., Maruyama,H., Kawakami,H., Nagai,Y., Hagiwara,M. and Ohkubo,H. (1996) Biochem. Biophys. Res. Commun., 220, 754–758. [DOI] [PubMed] [Google Scholar]
- 21.McCormick M.B., Tamimi,R.M., Snider,L., Asakura,A., Bergstrom,D. and Tapscott,S.J. (1996) Mol. Cell. Biol., 16, 5792–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kume H., Maruyama,K., Tomita,T., Iwatsubo,T., Saido,T.C. and Obata,K. (1996) Biochem. Biophys. Res. Commun., 219, 526–530. [DOI] [PubMed] [Google Scholar]
- 23.Schwab M.H., Druffel-Augustin,S., Gass,P., Jung,M., Klugmann,M., Bartholomae,A., Rossner,M.J. and Nave,K.A. (1998) J. Neurosci., 18, 1408–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuichi T., Yoshikawa,S., Miyawaki,A., Wada,K., Maeda,N. and Mikoshiba,K. (1989) Nature, 342, 32–38. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi S., Maeda,N. and Mikoshiba,K. (1991) J. Neurosci., 11, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuichi T., Simon-Chazottes,D., Fujino,I., Yamada,N., Hasegawa,M., Miyawaki,A., Yoshikawa,S., Guenet,J.-L. and Mikoshiba,K. (1993) Recept. Channels, 1, 11–24. [PubMed] [Google Scholar]
- 27.Matsumoto M., Nakagawa,T., Inoue,T., Nagata,E., Tanaka,K., Takano,H., Minowa,O., Kuno,J., Sakakibara,S., Yamada,M., Yoneshima,H., Miyawaki,A., Fukuuchi,Y., Furuichi,T., Okano,H., Mikoshiba,K. and Noda,T. (1996) Nature, 379, 168–171. [DOI] [PubMed] [Google Scholar]
- 28.Furutama D., Shimoda,K., Yoshikawa,S., Miyawaki,A., Furuichi,T. and Mikoshiba,K. (1996) J. Neurochem., 66, 1793–1801. [DOI] [PubMed] [Google Scholar]
- 29.Konishi Y., Kobayashi,Y., Kishimoto,T., Makino,Y., Miyawaki,A., Furuichi,T., Okano,H., Mikoshiba,K. and Tamura,T. (1997) J. Neurochem., 69, 476–484. [DOI] [PubMed] [Google Scholar]
- 30.Konishi Y., Ohkawa,N., Makino,Y., Ohkubo,H., Kageyama,R., Furuichi,T., Mikoshiba,K. and Tamura,T. (1999) J. Neurochem., 72, 1717–1724. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz W.W., McCann,R.O., Longiaru,M. and Cormier,M.J. (1991) Proc. Natl Acad. Sci. USA, 88, 4438–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Wet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S. (1987) Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dudendorff,J.W. (1990) Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- 34.Naruse Y., Aoki,T., Kojima,T. and Mori,N. (1999) Proc. Natl Acad. Sci. USA, 96, 13691–13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma A., Moore,M., Marcora,E., Lee,J.E., Qiu,Y., Samaras,S. and Stein,R. (1999) Mol. Cell. Biol., 19, 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]