Abstract
The complete sequence of the Bombyx mori fibroin gene has been determined by means of combining a shotgun sequencing strategy with physical map-based sequencing procedures. It consists of two exons (67 and 15 750 bp, respectively) and one intron (971 bp). The fibroin coding sequence presents a spectacular organization, with a highly repetitive and G-rich (~45%) core flanked by non-repetitive 5′ and 3′ ends. This repetitive core is composed of alternate arrays of 12 repetitive and 11 amorphous domains. The sequences of the amorphous domains are evolutionarily conserved and the repetitive domains differ from each other in length by a variety of tandem repeats of subdomains of ~208 bp which are reminiscent of the repetitive nucleosome organization. A typical composition of a subdomain is a cluster of repetitive units, Ua, followed by a cluster of units, Ub, (with a Ua:Ub ratio of 2:1) flanked by conserved boundary elements at the 3′ end. Moreover some repeats are also perfectly conserved at the peptide level indicating that the evolutionary pressure is not identical along the sequence. A tentative model for the constitution and evolution of this unusual gene is discussed.
INTRODUCTION
The silkworm Bombyx mori is a well studied model eukaryotic organism. The most significant aspect of this organism is the differentiation of the silkgland, a model system for chromatin organization, DNA replication, tissue-specific and efficient gene expression and regulation as well as packaging and secretion of mature silk proteins.
Silk fibroin consists of heavy (H) and light (L) chain polypeptides of ~350 and 25 kDa, respectively, linked by a disulfide bond at the C-terminus of the two subunits (1–3). In addition, another 25 kDa polypeptide, encoded by the P25 gene (4), associates with the H–L complex primarily by hydrophobic interactions (3). Genes encoding the three polypeptides are located on different chromosomes, but their expression seems to be coordinately regulated in the posterior silkgland (5–7). Moreover, interactions between H-chain and L-chain or P25 are essential for the secretion of fibroin (1,3).
Identification of the gene encoding the fibroin heavy chain (generally called ‘fibroin gene’) at the molecular level started with the isolation of its mRNA from the posterior silkgland by a novel method based on its high molecular weight and GC-rich content (8). A partial DNA sequence of the fibroin gene (from –550 to ~ +1474) including the 5′ flanking region, exon 1, intron and a small part of exon 2 has been determined (access code GenBank V00094) (9). The promoter region (from –240 to ~ +24) contains a cluster of homeodomain binding sites interacting with three silkgland factors which induce its tissue-specific expression (5). The intron also contributes to transcriptional regulation and contains multiple octamer-like AT-rich elements recognized by fibroin-modulator-binding proteins (10). The 3′ end and the flanking region (557 bp, including a 258-base sequence at the 3′ end and a 299-base downstream extension) as well as the stop codon were also determined (access code GenBank AB01736) (11). However, to date, the majority of the fibroin gene coding sequence was unknown, and even the precise length of such a well-known gene was still controversial. All understanding and predictions of the core region were based on restriction patterns (12,13) and fragments of cDNA sequence (11,14). The difficulty has been that the 15–16 kb core region of exon 2 is GC-rich and highly repetitive allowing the obtention of only short DNA sequences, while long overlaps are necessary to construct the real contigs.
In order to solve this problem, we combined currently applied shotgun procedures in genomic sequencing with a traditional physical map-directed sequencing strategy. Random sequencing of AlwNI fragments in a library subcloned from exon 2 produced enough raw data to construct primary contigs. These contigs were subsequently placed in the correct orientation by producing the forward and reverse sequences from AvaII and HinfI-completely digested subcloning libraries. Finally, a unique contig covering the intact exon 2 was built.
Determination of the complete sequence of the fibroin gene has allowed us to understand its global fine organization. The precise analysis of the basic repetitive units and boundary elements, defined for the first time in this paper, shed light on the evolution and duplication of this complex gene. Moreover, the deduced primary structure and properties of intact fibroin will facilitate the industrial application of this fibrous protein of great economic significance (15).
MATERIALS AND METHODS
The bacterial artificial chromosome (BAC) clone containing the fibroin gene, provided by Yuji Yasukochi (National Institute of Sericultural and Entomological Science of Japan), was screened from a BAC library of silkworm p50 (16).
Existence of the intact fibroin gene in this BAC was confirmed by restriction patterns. About 50 µg BAC DNA was prepared according to the procedure of She and colleagues (17). After NotI digestion and pulse-field gel electrophoresis, ~5 µg of insert DNA was recovered from the agarose gel by electroelution and then nebulized (mechanically sheared by formation of a fine spray, Systam-CPS, Systeme Assistance Medical, Villeneuve sur Lot, France) under a pressure of 1 bar for 12 s. Resultant fragments were repaired with Klenow enzyme and T7 DNA polymerase, and inserted into pUC18/SmaI/BAP. All clones in the subcloning library were checked by agarose gel electrophoresis and those with insert fragments longer than 1 kb (about 360 clones in total) were selected as templates for sequencing.
Subcloning libraries of the fibroin gene core region were constructed according to a novel stategy. Based on the restriction map of the 64-kb upstream region, the core of exon 2 (about 15~16 kb, Fig. 1A) was recovered from the BAC after complete digestion with three enzymes, EcoRI, HindIII and PstI. The core is flanked by PstI and EcoRI as shown in Figure 1B, and HindIII was added to digest the upstream region of the fibroin gene in the BAC into smaller fragments so that the 16 kb fibroin core DNA could be separated from the upstream fragments. Insert fragments were obtained by restriction digestion with either AlwNI, HinfI or AvaII.
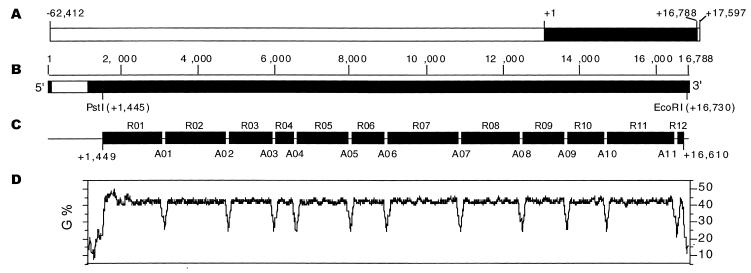
Figure 1.
Organization of fibroin gene in alternate arrays of repetitive domains and amorphous domains. (A) Location of fibroin gene (filled rectangle) on the BAC (80 009 bp), with the initiation point at position +1. (B) The fibroin gene consists of two exons (filled rectangles, 67 and 15 750 bp, respectively) and one intron (empty rectangle, 971 bp). (C) The central region of exon 2 contains 12 repetitive domains (R01–R12 shown by filled rectangles) separated by 11 amorphous domains (A01–A11, short lines to link the rectangles) in the central region of exon 2. (D) Percent G of exon 2 coding sequence identified by the Window program in GCG with a window setting of 100 bp and increments of 3 bp.
DNA sequencing was performed with the BigDye Terminator Kit (PE Applied Biosystems) on an ABI 373 Sequencer (Perkin-Elmer). The contigs were constructed from the raw sequence data by the Fragment Assembly program in SeqLab of the GCG sequence analysis software package version 10.0 (Genetics Computer Group, Madison, WI, USA). Restriction patterns and grouping of restriction fragments were carried out with Map and MapSort programs in GCG, respectively. The complete or partial sequences of the fibroin gene were compared with sequences in GenBank using the NetBlast program. Percent G was determined and plotted with the Window and StatPlot programs with a window setting of 100 bp and increments of 3 bp. To identify the occurrence and distribution of the putative motifs, the MapPlot program in GCG SeqLab was used. All patterns and their flanking bases were further checked by the Findpatterns program. The MultAlin program, developed by Corpet (18), was used for the multiple alignment of DNA sequences (http://www.toulouse.inra.fr/multalin.html ). The secondary structure of fibroin mRNA was predicted with the Stemloop program in GCG. The secondary structure of fibroin was predicted with the PHD program (Profile network prediction Heidelberg).
RESULTS
A novel strategy to sequence fibroin gene exon 2
With the transcriptional start point of fibroin gene at position +1 determined by Tsuda and Suzuki (19), the BAC (80 009 bp) ranges from positions –62 412 to +17 597 (Fig. 1A). By using the procedure of a shotgun sequencing strategy, the sequence of a 64 kb upstream region of the fibroin gene, including its exon 1, intron and a part of exon 2 as well as the 3′ end region (~1 kb) of the BAC insert, was determined (Zhou et al., manuscript in preparation). We failed to obtain a contig covering the complete fibroin gene located at the 3′ proximal region of the BAC (Fig. 1A) since only five sequenced clones were found in exon 2. The GC-rich core of the fibroin gene (~60%) is more resistant to nebulization than the upstream regions. The longer nebulized fragments in this region result in lower efficiency of ligation and transformation, explaining why most fragments in exon 2 were excluded from the subcloning library. Therefore, we have developed a novel strategy based on digestion with restriction enzymes to sequence this region. Three libraries were constructed from the exon 2 core. One contains insert fragments enriched in 1–2 kb lengths after partial digestion with AlwNI (CAGNNN↓CTG) to obtain random fragments from the core region. The other two carry subfragments of exon 2 after complete digestion with either AvaII (G↓GWCC) (W = A or T) or HinfI (G↓ANTC) which have been previously identified or predicted in the amorphous regions (12) (Fig. 1C). About 200 recombinant plasmids (100 from AlwNI-, 50 from AvaII- and 50 from HinfI-library) were prepared and purified for sequencing, and the insert size of each plasmid was determined by electrophoresis. Short primary contigs were constructed by using the GCG Fragment Assembly program with stringent settings for GelMerge (word size for overlap determination of 30 bp, minimum fraction of matching words in overlap of 0.99, minimum overlap length of 300 bp, minimum gap size of 2 bp). Then the orientations and positioning of these short contigs on exon 2 were determined by comparison with pairs of forward and reverse sequences from the AvaII or HinfI libraries combined with the sizes of insert fragments. Finally, the results of sequence-based GCG MapSort of exon 2 agree with the electrophoresis-based restriction patterns (data not shown).
An overview of fibroin gene organization
The fibroin gene (+1 ~ +16 788) consists of one intron and two exons (Fig. 1B). Exon 1 is 67 bp long, including a 25 bp untranslated domain (+1 ~ +25) and a 42-base coding region for 14 amino acid residues (+26 ~ +67). The intron (971 bp, +68 ~ +1038) contains a truncated sequence of B.mori repetitive element L1Bm (158 bp, from +565 to +722, identity of 83.4%, accession number AB002270) and multiple octamer-like AT-rich fibroin-modulator for the transcriptional regulation (10). The region from +1039 to +16 788 is exon 2 (15 750 bp), comprising the majority of the fibroin gene. The stop codon (TAA) is located 709 bp upstream to the 3′ end of the BAC insert.
The 5′ (+1 ~ +1449) and 3′ (+16 396 ~ +16 788) ends are identical at 99% or more to the previously reported sequences (GenBank V00094, AB01736), but the sequence next to the 3′ end region (~1900–400 bp upstream from the stop codon) is quite different from the sequences of Mita et al. (14). The discrepancies may result from polymorphism between different strains (12). The one used here is genomic DNA of p50 whereas Mita and colleagues used cDNA fragments from the F1 hybrid Kinshu×Showa (14). Both the 5′ (from +1 to +1450) and 3′ (from +16 612 to +16 788) non-repetitive regions of the fibroin gene have been thoroughly analyzed in previous papers (9,14), and in this paper we will mainly focus on the central repetitive region (from +1449 to +16 610) (Fig. 1C).
This repetitive core is composed of 12 repetitive domains (R01~R12) separated by 11 amorphous regions (A01~A11). A distinct property of the repetitive domains is their high GC content (~63%), especially the considerably high percentage of G (~45%) in the coding strand (Fig. 1D). At the 3′ end of repetitive domains (R01–R10), we found a 36-base sequence, (GGTGCT)(GGTGCC)(GGTGCT)(GGAGCT)(GGTGCA)(G-GAACA), functioning as an R-A border, which encodes the peptide (GlyAla)5GlyThr. The last one located at the end of R11 is the only exception with a deletion of the second 6-base stretch, GGTGCC.
Fine organization of repetitive domains
In each repetitive domain (except for R12), there are two types of repeat units, termed Ua and Ub. Ua is characterized by (GGNGCN)nGGNTCW (n = 0~6, W = A or T, N = A, G, T or C). In group Ua, the dominant repeat unit (~70%) is an 18-base sequence, (GGTGCT)2GGTTCA, termed Ua0 which encodes the well-known fibroin repeat peptide GlyAlaGlyAlaGlySer (20). Other Uas either have single/double changes especially at the third position of codons except for the last one (TCW), or share different lengths, due to various repeats of the dicodon GGNGCN. In fact, only 6 out of 368 Uas use the serine codon TCT, so that the predominant motif of Ua elements is the serine codon TCA. Ub can be described as (GGNGYN)nGGNTAY (n = 1~8, Y = T or C). About 75% of Ub units encode peptides of GlyAla repeats followed by GlyTyr, and the remaining 25% are those interrupted by irregular substitution of Ala with Val resulting from a C-or-T transition at the second position of codon GYN. The signature of Ub elements is the last codon for tyrosine (TAY).
A previous paper had revealed that the core region of the fibroin gene is composed of alternate arrays of two elements, a and b (14), which are included in the two repetitive units Ua and Ub, defined in the present paper, respectively. After comparing the various codon patterns in this region, we found two kinds of boundary elements, B1 and B2 (Fig. 2). B1 is an 18-base sequence GGAGCAGGAGCAGGAAGC, also coding for the same hexapeptide (GlyAlaGlyAlaGlySer) as Ua0, but with a distinct codon pattern. There are 60 B1 elements and 26 of those possess a single mismatch, predominantly at the 12th base, ‘A’. B2 encodes a tetrapeptide GlyAlaAlaSer with another distinctive codon pattern GGAGCTGCCTCT (with one mismatch except for the last serine codon TCT). Mita and colleagues have pointed out the existence of a boundary element similar to B2 (14). As shown in Figure 2A, with B2 as the only boundary element, the repetitive domains R03, R05, R07 and R08 would not be able to be well organized, which does not seem like a reasonable explanation of the fine organization of the fibroin gene. In fact 39 out of 45 B2 elements follow B1 elements, except for the first five B2 at the 5′ half of repetitive domain R01 and the last B2 in R12. Another set of 21 B1 elements follows a kind of B2-like element, GGAGCTGGCTCA that encodes GlyAlaGlySer (not shown in Fig. 2A). This modified B2 may be derived from a B2 element after double mutations, C-to-G and T-to-A. With two types of boundary elements described above, each repetitive domain is further separated into subdomains of ~208 bp on average (Table 1). A distinguishable feature of boundary elements compared with repetitive units is that the former use rare codons compared with the preferred codons in the repetitive regions.
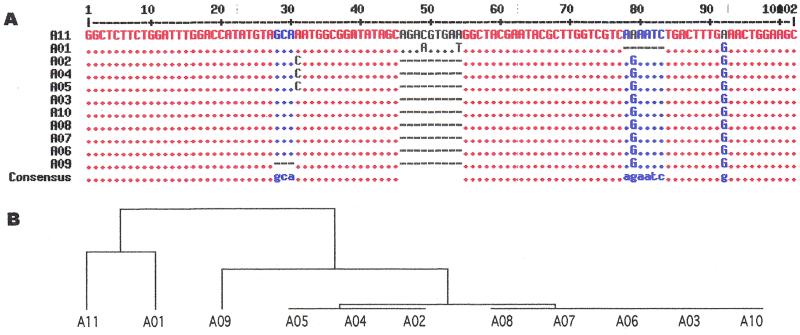
Figure 2.
Fine organization of repetitive domains with Ua and Ub as basic units, and B1 and B2 as boundary elements. The patterns used here are defined as those in GCG FindPatterns: Ua (red), (GGNGCN){0,}GGNTCW (no mismatch); Ub (blue), (GGNGCN){0,}GGNTAY (no mismatch); B1 (black), GGAGCAGGAGCAGGAAGC (with one mismatch); B2 (green), GGAGCTGCCTCT (with one mismatch). (A) Distribution of Ua, Ub, B1 and B2 in the fibroin gene repetitive domains, R01–R12. (B) Enlarged figures of R01, R06 and R12. (C) Sequences of the first subdomain in R01, a typical subdomain (the third) in R06, and the last repetitive domain R12 (the 3′ end of the truncated B1-like sequence is not shown).
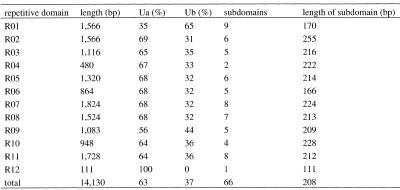
Table 1. Composition of repetitive domains.
Ua (Ub) (%) represents the percentage of the total length of Ua (Ub) to the length of correlated repetitive domain excluding the boundary elements. The figures in the Subdomain column represent the numbers of subdomains in each repetitive domain as defined in the text. Length of subdomain is the average length of subdomains in each repetitive domain separated by boundary elements.
In the central repetitive domains, R02–R11, the ratio of Ua:Ub is ~2:1 on average (Table 1). Each subdomain is composed of a cluster of Uas followed by a cluster of Ubs, and flanked by combined B1&B2/B2-like at the 3′ end (Fig. 2B). The sequence of a typical subdomain in R06 is shown in Figure 2C.
The compositions of repetitive units in the first and last domains are quite different. R01 is rich in Ub units (65% in general), especially in the first half ahead of the first B1 boundary element (~90%) (Fig. 2B). The first repetitive unit in R01, as well as the first one in the repetitive core, is a minor Ua with a serine codon TCT (Fig. 2C). The second half of R01 also shares the same Ua:Ub ratio of 2:1 as that in the central repetitive domains (Fig. 2B). In contrast, R12 is composed of only Uas (Fig. 2B), flanked by B2 at the 5′ end and a shortened B1-like GGTAGC (not shown in Fig. 2C) at the 3′ end.
Multiple comparison of amorphous domains
Compared with the repetitive domains sharing variable lengths and different composition, the amorphous domains and their upstream flanking regions, where R–A borders are located, are relatively well conserved. The sequences of A01–A11 were multiple-aligned in Figure 3A. From the distance tree (Fig. 3B), clearly two clusters emerge from this unrooted tree. One cluster includes two sequences, the first (A01) and the last (A11) amorphous domains. The other amorphous domains are grouped in the second cluster. A previous study found that the amorphous domains share a novel MobII–AvaII–HinfI pattern, except for the missing HinfI site in the first (A01) and last (A11) domains (13). One possibility is that the ancestral sequence of amorphous domains was probably similar to the sequence of A11. Originally there was no HinfI site in the ancestral sequence and an ‘A-to-G’ transition at position 79 generated the HinfI sites in the central amorphous domains. A01 could have been derived from A11 by a deletion of a 6-base sequence (AAAATC) at the potential HinfI site, thus losing that site, and three single-base mutations at positions 49, 54 and 92 (Fig. 3A). A03, A06~A08 and A10 could be derived from A11 by a 9-base deletion (positions 46–54) and two A-to-G transitions at positions 79 and 92. From the sequence A03, an A-to-C transversion at position 31 could generate A02, A04 and A05, whereas a 3-base deletion (positions 28–30) could generate A09. A02, A04 and A05 are identical and share an additional A-to-C transversion, whereas A09 has a further 3-base deletion from A03.
Figure 3.
(A) Multi-alignment of 11 amorphous domains. Consensus levels: high = 90% (red), low = 50% (blue), and neutral (grey). An asterisk at any position represents the same nucleotide as in A11. Deletions are signified by dashes. (B) The distance tree.
Specific codon usage pattern and secondary structure of fibroin mRNA
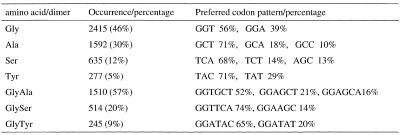
Besides the GC-rich content and repetitive features, another characteristic distinguishing repetitive domains from amorphous domains is the specific codon pattern. The first and second position GC bias are up to ~90 and 80%, respectively, due to the enrichment of glycine (GGN) and alanine (GCN) residues in fibroin. The third position AT bias is ~90% on average, resulting from the iso-codons used in fibroin mRNA. The enriched amino acid residues (dipeptides) and their unique codon patterns are shown in Table 2. An extreme example of codon pattern bias is that five out of six serine codons are used in amorphous regions, except for that preferred in Ua0 (TCA).
Table 2. The preferential codon patterns of enriched amino acid residuals and dipeptides in the fibroin protein.
Percentages calculated as the ratio of the occurrence to the total length of the protein (5263 residues).
With the GCG Stemloop program, we find that the alternate GGU-GCU dicodons for Gly-Ala repeats lead to the abundance of inverted repeats forming strong stem–loop structures in the subdomains of fibroin mRNA (data not shown). Conversely, the base-paired stems are destroyed by different codon patterns in boundary elements, and stems formed in amorphous domains are very weak. This specific codon choice is thought to play an essential role in sustaining the stability of the secondary structure of fibroin mRNA (14), which explains the translational pauses during the synthesis of fibroin (21, and references cited therein).
Deduced amino acid sequence of fibroin heavy chain
The deduced fibroin amino acid sequence is 5263 residues long, with a molecular weight of 391 367 Da and an isoelectric point of 4.22 which was calculated with the Isoelectric program in GCG (charged groups, 14 Arg, 12 Lys, 5 His, 30 Glu and 25 Asp, 277 Tyr and 5 Cys). It is made of a low-complexity region bordered by short N- and C-terminal segments of more standard amino acid composition. The bulk of the low-complexity region (4754 residues corresponding to ~90% of the fibroin) is made of repeats of a GX dipeptide motif where X is Ala in 65%, Ser in 23% and Tyr in 10% of the repeats compared with all residues in fibroin. Val and Thr also occur as X, but at a lower abundance (~2%). All 14 other amino acid types are absent with the exception of one Phe, two Ile and one Gln residues. In 1% the first position is Ala instead of Gly, where the translation of B2, GlyAlaAlaSer, is located. The GX are distributed in 12 domains, designated GX-domains, of between 39 and 612 amino acids separated by 11 nearly identical copies of a boundary sequence (Table 3). The boundary sequences, translated from the amorphous domain, contain the only tryptophan residues found in fibroin, and 11 of 14 prolines. They also contain charged residues, which are absent from the GX-domains. Acidic Asp/Glu residues are present in all 11 boundary sequences, basic Arg/Lys residues in the first and the last ones. Within the GX-domains, the GX alternance is strictly conserved with the exception of GX9 (residues 3819–4183) where a single residue insertion changes the phase at position 4108. Most of the GX dipeptide units are present as part of the two hexapeptides GAGAGS (432 copies) and GAGAGY (120 copies) which, together, account for 72% of the low-complexity region. As can be seen in the partial (324 residues) sequence of Mita et al. (14), these hexapeptides often occur in stretches, but whereas the stretches may contain up to 10 consecutive copies, none of the GX-domains is a simple repeat of hexapeptides.
Table 3. GX-domains and boundary sequences in fibroin.
$Region of the N-terminal domain with homology to the boundary sequence.
*All GX-domains start and end with dipeptide GS except for GX1 starting with dipeptide GA.
Beyond 5205, the C-terminal sequence is still low-complexity, but with no obvious feature. In contrast, N-terminal residues 1–151 have a regular amino acid composition and may form a globular domain. This sequence was suggested as a signal peptide for the secretion of the fibroin because the N-terminal residue of the mature fibroin is Gly instead of Met (22). Residues 127–147 may be related to the boundary sequences between GX-domains (Table 3). A BLAST search against the PRODOM database (23) indicates homology (score 94, 44% identity) of residues 1–104 to a sequence of 84 residues at the N-terminus of the Chinese oak silkworm Anthereae pernyi fibroin (accession number EMBL O76786). The rest of this fibroin sequence is low-complexity, but it is of a different class and unrelated to the B.mori sequence, containing polyalanine stretches instead of GX repeats.
We found that the H chain has five Cys residues, all being located near the N- or C-terminus. The single disulfide bond between fibroin H and L chains was determined recently (3). The cysteine residue involved in the disulfide linkage is Cys-5244 in our fibroin sequence. Two Cys-containing peptides have been identified chemically and sequenced. The RALPCNVC peptide of Earland and Robins (24) is identical to our C-terminal sequence. In contrast, the GAGAGC(D,N)SAVC peptide sequenced by Robson et al. (25) has no counterpart.
The silk structure of B.mori
X-ray diffraction patterns of silk fibres indicate that the bulk of the protein forms regular β-sheets similar to those of poly(Ala-Gly). In the alternating co-polymer, all the glycines are on one face of the β-sheets, all the alanine side-chains on the other. The β-sheets pack upside-down, so that short vertical spacings (~3.6 Å) for glycine-glycine packing alternate with longer ones (~5 Å) for alanine-alanine packing (26). With the GX-domains, we observe a strict alternance of Gly residues with either Ala, Ser or Tyr. Thus, one face of the β-sheet is only glycines, permitting the same tight glycine-glycine packing as in poly(Ala-Gly). The other face is shaped less regularly, containing large side chains (Tyr) and an abundance of hydroxyl groups (Ser, Tyr). The assembly of two such surfaces must position these groups correctly for hydrogen bonding.
In principle, each GX-domain could constitute a single β-sheet which, in the case of GX7 (613 residues), would reach 200 nm in length. The boundary sequences contain a proline and are likely to break the β-strand allowing the polypeptide chain to change direction and, possibly, turn back on itself into an anti-parallel β-sheet. This β-sheet would be intra- rather than inter-chain.
DISCUSSION
In the present work, we determined the complete sequence of a famous eukaryotic gene, the B.mori fibroin gene. Analysis of the complete sequence reveals two basic repetitive units, Ua and Ub, and two boundary elements, B1 and B2, which form the subdomain in that order. The standard hierarchical organization of the fibroin gene repetitive core from the basic units (Ua, Ub, B1 and B2) to subdomains, and then repetitive domains can be deduced from Figure 2. The repetitive domain is followed by an amorphous domain, to form the high-order unit of the fibroin gene (Fig. 1C). This fine organization of the fibroin gene may be generated by duplication and unequal crossover (14,27). In addition, data shown in this paper reveal some new clues to the duplication of subdomains and evolution of the fibroin gene.
Origin and duplication of the repetitive units
The enrichment of Ub in R01 (Fig. 2A and Table 1), especially in the 5′ half, suggests that Ub should originate from R01. In other words, it seems there is a gradient of variation of Ub from the 5′ to the 3′ end. The last repetitive domain (R12) is the shortest one of all, the most proximal domain to one end of the fibroin gene, and flanked by the potential ancestor of the amorphous domains (A11). R12 consists of only Uas and is enriched for Ua0, the dominant repetitive unit. We thus conclude that Ua0 originated from R12, located at the 3′ end. Other units in group Ua were derived from Ua0 by single/double mutations and/or unequal crossing over.
Length polymorphism of repetitive domains in the core region
Kondo and colleagues have reported that the inactive forms of fibroin heavy chain gene chromatin are constructed by folding of the chromatin fiber on a regular array of nucleosomes (28). The average length of subdomains is 208 bp (Table 1), close to the repeat length of DNA in the nucleosome. This strongly suggests a chromatin organization of the fibroin gene based on the nucleosome surrounded by individual subdomains. This special organization may also be responsible for a simple mechanism of subdomain duplication during the course of evolution, allowing incremental or deletion in the central repetitive domains.
Predicted primordial sequence of the fibroin gene
After comparing the translation of amorphous domain A11 with the complete sequence of fibroin itself, we found that it is homologous to a domain in the N-terminal non-repetitive region (residues 127–147), near the start point of repetitive domain GX1 (Table 3). Hence, the amorphous domains probably derived from the 5′ end non-repetitive region. Ua could be originated from the 3′ end, whereas Ub could be from the 5′ end. Therefore, the primordial sequence of the fibroin gene could be simplified to an 838-bp sequence. It was composed of the current exon 1 (67 bp), 5′ non-repetitive domain of exon 2 (411 bp), the first subdomain in R01 (72 bp) and the 3′ ends region including R12 (111 bp) and the 3′ non-repetitive domain (177 bp). The intron was perhaps integrated between exon 1 and exon 2 mediated by the repetitive element L1Bm, a type of repetitive long interspersed element in the silkworm (29), because a 101 bp fragment in the intron is homologous to the truncated L1Bm. The first amorphous domain (A11) was derived from the 5′ non-repetitive domain of exon 2. After recombination between the 5′ and 3′ ends, mediated by A11 and A11-like elements at the 5′ end region (the potential ancestor of A11), the typical subdomain was generated. Based on this basic unit defined by the nucleosome, the repetitive domain was formed through duplication of the subdomain accomplished with unequal crossing over.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Yuji Yasukochi (National Institute of Sericultural and Entomological Science of Japan) for providing the BAC clone containing the fibroin gene. We also thank Hélène Fouché and Laurent Mallet for the help during experiments, and Mark Blight for critical reading of the manuscript. This project is supported by CNRS, Université Paris-Sud and Association Franco-Chinoise pour la Recherche Scientifique et Technique (AFCRST). C.-Z.Z. is funded by a postdoctoral fellowship from Ministère Français des Affaires Etrangères.
DDBJ/EMBL/GenBank accession no. AF226688
REFERENCES
- 1.Takei F., Kikuchi,Y., Kikuchi,A., Mizuno,S. and Shimura,K. (1987) J. Cell Biol., 105, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka K., Mori,K. and Mizuno,S. (1993) J. Biochem. (Tokyo), 114, 1–4. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka K., Kajiyama,N., Ishikura,K., Waga,S., Kikuchi,A., Ohtomo,K., Takagi,T. and Mizuno,S.(1999) Biochim. Biophys. Acta, 1432, 92–103. [DOI] [PubMed] [Google Scholar]
- 4.Couble P., Chevillard,M., Moine,A., Ravel-Chapuis,P. and Prudhomme,J.C. (1985) Nucleic Acids Res., 13, 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui C.C., Matsuno,K. and Suzuki,Y. (1990) J. Mol. Biol., 213, 651–670. [DOI] [PubMed] [Google Scholar]
- 6.Hui C.C., Suzuki,Y., Kikuchi,Y. and Mizuno,S. (1990) J. Mol. Biol., 213, 395–398. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi Y., Mori,K., Suzuki,S., Yamaguchi,K. and Mizuno,S. (1992) Gene, 110, 151–158. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y. and Brown,D.D. (1972) J. Mol. Biol., 63, 409–429. [DOI] [PubMed] [Google Scholar]
- 9.Tsujimoto Y. and Suzuki,Y. (1979) Cell, 18, 591–600. [DOI] [PubMed] [Google Scholar]
- 10.Takiya S., Kokubo,H. and Suzuki,Y. (1997) Biochem. J., 321, 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mita K., Ichimura,S. and James,T.C. (1994) J. Mol. Evol., 38, 583–592. [DOI] [PubMed] [Google Scholar]
- 12.Manning R.F. and Gage,L.P. (1980) J. Biol. Chem., 255, 9451–9457. [PubMed] [Google Scholar]
- 13.Gage L.P. and Manning,R.F. (1980) J. Biol. Chem., 255, 9444–9450. [PubMed] [Google Scholar]
- 14.Mita K., Ichimura,S., Zama,M. and James,T.C. (1988) J. Mol. Biol., 203, 917–925. [DOI] [PubMed] [Google Scholar]
- 15.Heslot H. (1998) Biochimie, 80, 19–31. [DOI] [PubMed] [Google Scholar]
- 16.Wu C., Asakawa,S., Shimizu,N., Kawasaki,S. and Yasukochi,Y. (1999) Mol. Gen. Genet., 261, 698–706. [DOI] [PubMed] [Google Scholar]
- 17.She Q., Confalonieri,F., Zivanovic,Y., Medina,N., Billault,A., Awayez,M.J., Thi-Hgoc,H.P., Pham,B.-T.T., Van Der Oost,J., Duguet,M. and Garrett R. (2000) DNA Res., in press. [DOI] [PubMed] [Google Scholar]
- 18.Corpet F. (1988) Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuda M. and Suzuki,Y. (1981) Cell, 27, 175–182. [DOI] [PubMed] [Google Scholar]
- 20.Lucas F., Shan,J.T.B. and Smith,S.G. (1958) Adv. Protein Chem., 13, 107–242. [DOI] [PubMed] [Google Scholar]
- 21.Zama M. (1997) Nucleic Acids Symp. Ser., 37, 179–180. [PubMed] [Google Scholar]
- 22.Sasaki T and Noda,H. (1974) J. Biochem. (Tokyo), 76, 493–502. [DOI] [PubMed] [Google Scholar]
- 23.Corpet F., Gouzy,J. and Kahn,D. (1998) Nucleic Acids Res., 26, 323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Earland C. and Robins,S.P. (1973) Int. J. Pept. Protein Res., 5, 327–335. [DOI] [PubMed] [Google Scholar]
- 25.Robson A., Woodhouse,J.M. and Zaidi,Z.H. (1970) Int. J. Pept. Protein Res., 2, 181–189. [Google Scholar]
- 26.Fraser R.D., MacRae,T.P. and Miller,A. (1965) J. Mol. Biol., 14, 432–442. [DOI] [PubMed] [Google Scholar]
- 27.Ueda H., Mizuno,S. and Shimura,K. (1985) Gene, 34, 351–355. [DOI] [PubMed] [Google Scholar]
- 28.Kondo K., Aoshima,Y., Hagiwara,T., Ueda,H. and Mizuno,S. (1987) J. Biol. Chem., 262, 5271–5279. [PubMed] [Google Scholar]
- 29.Ichimura S., Mita,K. and Sugaya,K. (1997) J. Mol. Evol., 45, 253–264. [DOI] [PubMed] [Google Scholar]