Abstract
Background/purpose(s)
During a viral infection, the immune response is mediated by the toll-like receptors and myeloid differentiation Factor 88 (MyD88) that play an important role sensing infections such as SARS-CoV-2 which has claimed the lives of more than 6.8 million people around the world.
Methods
We carried out a cross-sectional with a population of 618 SARS-CoV-2-positive unvaccinated subjects and further classified based on severity: 22% were mild, 34% were severe, 26% were critical, and 18% were deceased. Toll Like Receptor 7 (TLR7) single-nucleotide polymorphisms (rs3853839, rs179008, rs179009, and rs2302267) and MyD88 (rs7744) were genotyped using TaqMan OpenArray. The association of polymorphisms with disease outcomes was performed by logistic regression analysis adjusted by covariates.
Results
A significant association of rs3853839 and rs7744 of the TLR7 and MyD88 genes, respectively, was found with COVID-19 severity. The G/G genotype of the rs3853839 TLR7 was associated with the critical outcome showing an Odd Ratio = 1.98 (95% IC = 1.04–3.77). The results highlighted an association of the G allele of MyD88 gene with severe, critical and deceased outcomes. Furthermore, in the dominant model (AG + GG vs. AA), we observed an Odd Ratio = 1.70 (95% CI = 1.02–2.86) with severe, Odd Ratio = 1.82 (95% CI = 1.04–3.21) with critical, and Odd Ratio = 2.44 (95% CI = 1.21–4.9) with deceased outcomes.
Conclusion
To our knowledge this work represents an innovative report that highlights the significant association of TLR7 and MyD88 gene polymorphisms with COVID-19 outcomes and the possible implication of the MyD88 variant with D-dimer and IFN-α concentrations.
Keywords: COVID-19, MyD88, Polymorphism, SARS-CoV-2 and TLR7
Introduction
During activation of the innate and adaptive immune responses, the participation of toll-like receptors (TLRs) is crucial in recognizing pathogen-associated molecular patterns (PAMPs).1 TLRs 3, 7, and 8 are endolysosomal receptors that recognize double-stranded RNA and single-stranded RNA (TLR 7 and 8).2 However, recognition of PAMPs by the TLR 7 and 8 is not sufficient to trigger an antiviral response since myeloid differentiation Factor 88 (MyD88) is required as an adaptor molecule to form the Myddosome complex, which initiates the signalling that leads to production of inflammatory cytokines such as TNF-α, IL-6, and Interleukin-1 (IL-1) family as well as type I IFNs like IFN-α.3 , 4 Activation of MyD88 mediated by TLR7 and TLR8 is responsible for sensing infections by single-stranded RNA viruses such as HIV, influenza viruses, HCV, Sendai virus, and CBVs, among others.5
Considering that the SARS-CoV-2 virus has infected more than 761 million individuals and caused the death of 6.8 million who have developed COVID-19, the scientific community has devoted vast resources to examining the immunopathogenesis of the virus and therapeutic targets. TLR7 is in the X-chromosome, and expressed on monocyte-macrophages and dendritic cells. Genetic variants of TLR7 are associated with COVID-19 progression and patient outcomes, suggesting a role for TLR7 in its pathogenesis.6 Fallerini et al. reported TLR7 loss-of-function variants that contribute to disease susceptibility in young males.7 In addition, in four young males without a history of chronic disease, loss-of-function TLR7 was found and associated with impaired type I and II IFN responses.8 In COVID-19 patients, TLR7 deficiency was reported in at least 1% of men under 60 years of age by Asano et al.9
A single nucleotide polymorphism (SNP) of the MyD88 gene in the 3′ untranslated region (3′UTR) has been reported to be associated with diverse pathologies, such as Buerger's disease,10 higher death risk at 90 days of septic shock,11 and cardiovascular artery disease.12 Some reports suggest the participation of MyD88 in COVID-19.13 , 14 Nevertheless, the efficacy of the antiviral response depends on both the molecular diversity of the pathogen and functional versatility and genetic variability of the myddosome.15 In this context, we investigated the association of TLR7 and MyD88 gene variants with COVID-19 outcomes.
Methods
Setting and participants
We carried out a cross-sectional study. From June 2020–March 2021, unvaccinated patients during the first wave of SARS-CoV-2 infection, were recruited from the following hospitals of the Mexican Governmental Health System: Instituto Nacional de Rehabilitación “Luis Guillermo Ibarra Ibarra”, Instituto Nacional de Cardiología “Ignacio Chávez”, Hospital Central Militar, Instituto Nacional de Ciencias Médicas y Nutrición “Salvador Zubirán”, Hospital General “Dr. Manuel Gea González”, Hospital General ISSSTE “Tláhuac”, and Hospital Central Norte Pemex.
Inclusion criteria were not familiar related, independent of gender, age ≥18 years, unvaccinated, and nonpregnant women with clinical manifestations of COVID-19 and positive qRT–PCR test. The exclusion criteria were incomplete clinical history. These individuals were classified according to previously described16 according to Gandhi criteria as: mild, those ambulatory subjects with symptoms such as fever, headache, fatigue, odynophagia, cough, rhinorrhea, diarrhea, anosmia or dysgeusia, with or without dyspnea or pneumonia, not requiring hospitalization; severe, those hospitalized individuals with any of the following symptoms: tachypnea (FR > 30 bpm), dyspnea for small efforts; and critical, those patients requiring invasive mechanical ventilation who could course with shock and multiorgan failure17 The bioethics and research committees of the participating institutions approved this study. Written informed consent was obtained from each participant.
Blood samples
Blood samples were collected for DNA extraction and serum was obtained by centrifugation. Serum samples were stored immediately at −80 °C until further use.
SNPs selection and genotyping
Genomic DNA was isolated from peripheral blood white cells using a commercial kit column-based method (QIAmp 250 DNA Blood Mini Kit, Qiagen, Hilden, Germany). Genomic DNA samples at 10 ng/uL were deposited into genotyping OpenArray plates previously loaded with the genotyping primers and probes using the AccuFill System (Thermo Fisher Scientific). Real-time PCR amplification was carried out following the manufacturer's protocol using OpenArray technology in a QuantStudio 12 K flex System (Thermo Fisher Scientific). The results were analyzed using TaqMan Genotyper v1.6 software.
Statistical methods
The normality of the variable distribution was evaluated. For continuous variables, the Kruskal–Wallis test was used to compare nonparametric distributions among the studied groups, and the results were described using the median and interquartile range (IQR). The chi-squared test was performed for categorical variables. For all tests, a value of p < 0.05 was considered statistically significant. Hardy-Weinberg Equilibrium (HWE) was assessed for all polymorphisms in the mild group. For the TLR7 polymorphisms the HWE was estimated in women of the mild group. Linkage disequilibrium (LD) among TLR7 gene variants was assessed using HaploView software V4.2. A logistic regression analysis was used to evaluate the association between genetic variants and outcomes of COVID-19, adjusted by age, stratified by < 60 years and ≥60 years old, sex, hypertension, type 2 diabetes, and obesity. The final models were evaluated using the Hosmer–Lemeshow goodness-of-fit test. The correction for multiple comparisons was 0.05/5(SNPs) = 0.001, which was considered statistically significant. The correlation between SNPs and clinical features was assessed by comparing their distribution among alleles and genotypes by the Kruskal–Wallis test and stratified by disease outcome. The analysis was performed using the STATA v.13 statistical package (StataCorp Texas, USA).
Results
Patients
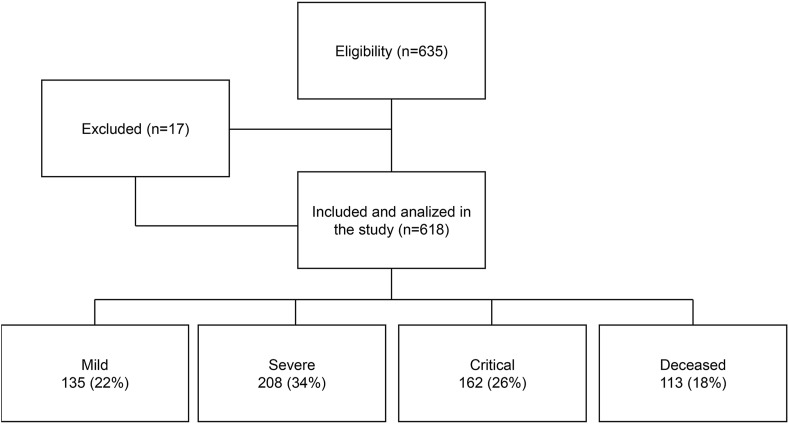
In this study, 618 COVID-19 patients were enrolled and classified based on disease severity. Of these, 22% were mild, 34% were severe, 26% were critical, and 18% were deceased (Fig. 1 ).
Figure 1.
Flow-chart of participants in the study.
The median age of patients was 52 years; however, in the mild group, the median age was 41 years, while the median age of the deceased group was 63 years. In this context, we found that 63% of the study population were males. Additionally, we showed the comorbidities and clinical symptoms in Table 1 .
Table 1.
Clinical and anthropometric characteristics of the study population.
| Total n = 618 | Mild n = 135 | Severe n = 208 | Critical n = 162 | Deceased n = 113 | P value | |
|---|---|---|---|---|---|---|
| Agea | 52 (43–63) | 41 (31–49) | 53 (43–64) | 52 (46–63) | 63 (54–70) | <0.001 |
| Gender | ||||||
| Male | 392 (63%) | 67 (49%) | 135 (65%) | 116 (72%) | 74 (65%) | 0.001 |
| Obesity | 195 (31%) | 20 (15%) | 70 (34%) | 67 (41%) | 38 (34%) | <0.001 |
| Type 2 Diabetes | 191 (31%) | 13 (10%) | 75 (37%) | 54 (33%) | 49 (43%) | <0.001 |
| Hypertension, n (%) | 189 (31%) | 14 (10%) | 65 (32%) | 61 (37%) | 49 (43%) | <0.001 |
| Heart rate, median (IQR), bpm+ | 93 (81–105) | 89 (78–100) | 93 (80–105) | 96 (87.5–110) | 92 (81–104) | 0.13 |
| Oxygen saturation % (IQR) | 87 (79–93) | 94 (92–96) | 87 (80–92) | 83 (72–88) | 81 (70–89) | <0.001 |
| Fever, n (%) | 274 (45%) | 46 (34%) | 101 (49%) | 85 (52%) | 42 (37%) | 0.003 |
| Cough, n (%) | 429 (70%) | 78 (58%) | 145 (71%) | 129 (80%) | 77 (69%) | 0.001 |
| Dyspnoea, n (%) | 379 (62%) | 29 (21%) | 150 (74%) | 125 (77%) | 75 (67%) | <0.001 |
| Headache, n (%) | 341 (56%) | 80 (59%) | 105 (52%) | 109 (68%) | 47 (42%) | <0.001 |
| Odynophagia, n (%) | 252 (41%) | 59 (44%) | 80 (39%) | 75 (46%) | 38 (34%) | 0.19 |
| Myalgia, n (%) | 336 (55%) | 68 (50%) | 111 (55%) | 105 (65%) | 52 (46%) | 0.013 |
| Vomiting, n (%) | 45 (7%) | 8 (6%) | 10 (5%) | 18 (11%) | 9 (8%) | 0.13 |
IQR= Interquartile Range.
Kruskal–Wallis test. Chi square test.
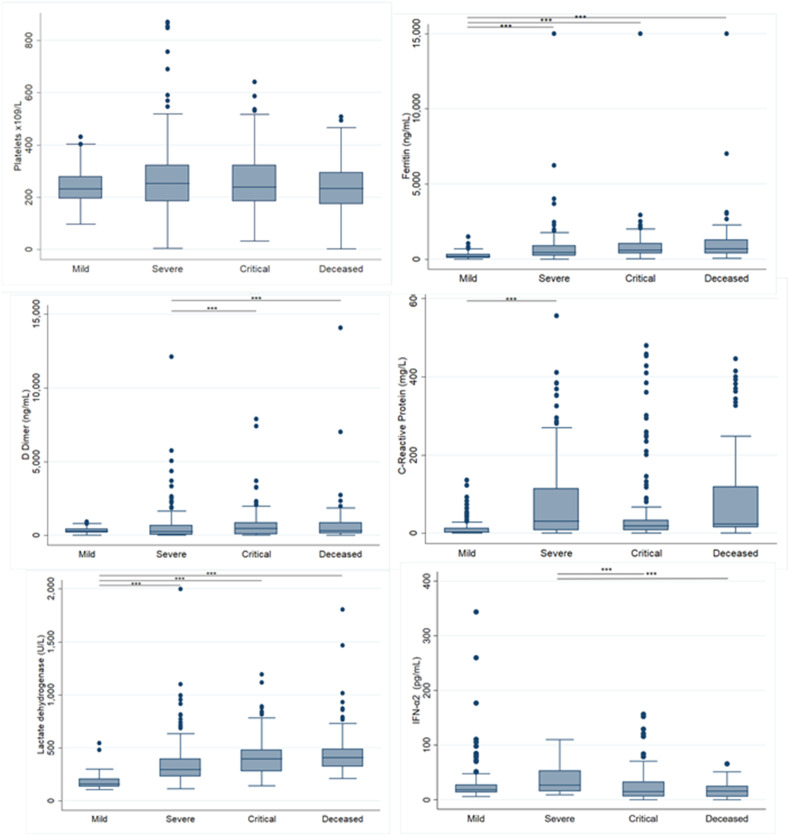
Clinical laboratory parameters are shown in Fig. 2 . We observed an increasing trend of ferritin and Lactate Dehydrogenase (LDH) as the COVID-19 outcome severity increased. The median ferritin level in the mild group was 138.5 (ng/mL) (Interquartile Range (IQR) = 27.2–312.6) and 694.3 (ng/mL) in the deceased group (IQR = 398.05–1286.3). The median LDH was 152.5 (IQR = 124.5–199) in the mild group and 407 (U/L) (IQR = 322–488.4) in the deceased group. We observed a decreased level in IFN-α in mild outcome with 18.14(IQR = 13.5–27.9) versus deceased group 15.9(IQR = 5.9–26.7).
Figure 2.
Laboratory values of the population study. (a) Platelets ×109/L. (b) Serum ferritin concentrations (ng/ml). (c) D-dimer (ng/mL). (d) C-reactive protein (CRP) (mg/L) (e) Lactate dehydrogenase (LDH) (U/L).
Allelic, genotypes and linkage disequilibrium
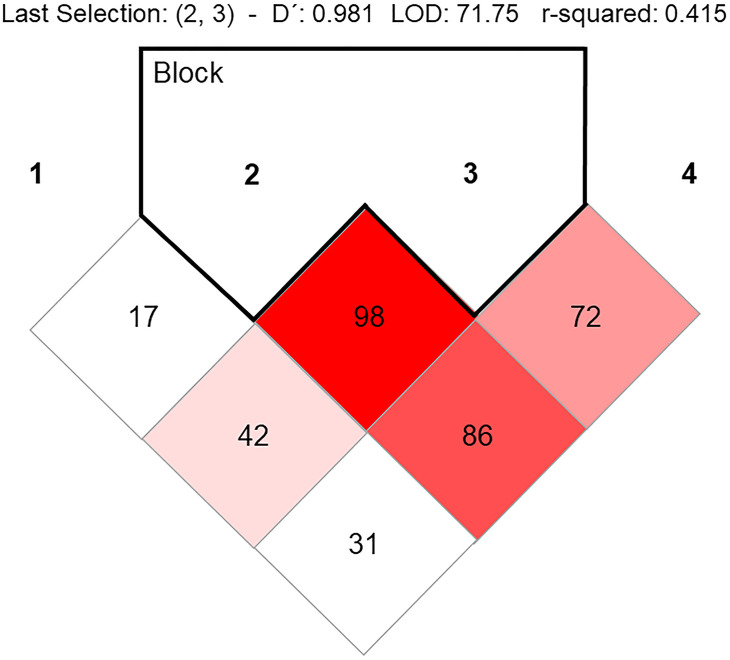
Table 2 shows the allelic and genotypic distribution of five SNPs on the TLR7 and MyD88 genes. We found statistically significant differences in the frequencies of SNPs in the TLR7 gene, all SNPs were in HWE. We observed a strong LD between rs179008 and rs179009 variants, showing D′0.98 (Fig. 3 ).
Table 2.
Allelic and genotype TLR7 and MyD88 gene frequencies in the study population.
| Frequencies (%) |
Pa | HWEb | |||||
|---|---|---|---|---|---|---|---|
| Total (n = 618) |
Mild (n = 135) |
Severe (n = 208) |
Critical (N = 162) |
Deceased (n = 113) |
|||
| TLR7 | |||||||
| rs179008 | |||||||
| A | 965 (78%) | 199 (74%) | 328 (79%) | 254 (78%) | 184 (81%) | 0.20 | |
| T | 271 (22%) | 71 (26%) | 88 (21%) | 70 (22%) | 42 (19%) | ||
| AA | 451 (73%) | 89 (66%) | 156 (75%) | 117 (72%) | 89 (78%) | 0.10 | 0.07c |
| AT | 63 (10%) | 21 (15%) | 16 (8%) | 20 (12%) | 6 (5%) | ||
| TT | 104 (17%) | 25 (18%) | 36 (17%) | 25 (15%) | 18 (16%) | ||
| rs179009 | |||||||
| A | 743 (60%) | 148 (55%) | 253 (61%) | 205 (63%) | 137 (61%) | 0.20 | |
| G | 493 (40%) | 122 (45%) | 163 (39%) | 119 (37%) | 89 (39%) | ||
| AA | 319 (52%) | 56 (41%) | 109 (52%) | 92 (57%) | 62 (55%) | 0.02 | 0.6c |
| AG | 105 (17%) | 36 (27%) | 35 (17%) | 21 (13%) | 13 (11%) | ||
| GG | 194 (31%) | 43 (32%) | 64 (31%) | 49 (30%) | 38 (34%) | ||
| rs3853839 | |||||||
| C | 793 (65%) | 158 (59%) | 265 (64%) | 230 (71%) | 140 (63%) | 0.01 | |
| G | 443 (35%) | 112 (41%) | 147 (36%) | 92 (29%) | 84 (37%) | ||
| CC | 344 (56%) | 62 (46%) | 118 (57%) | 105 (65%) | 59 (53%) | 0.02 | 1c |
| CG | 105 (17%) | 34 (25%) | 29 (14%) | 20 (12%) | 22 (20%) | ||
| GG | 169 (27%) | 39 (29%) | 61 (29%) | 37 (22%) | 32 (28%) | ||
| rs2302267 | |||||||
| T | 1085 (88%) | 241 (89%) | 364 (87%) | 281 (87%) | 199 (88%) | 0.87 | |
| G | 151 (12%) | 29 (11%) | 52 (13%) | 43 (13%) | 27 (12%) | ||
| TT | 521 (84%) | 115 (85%) | 174 (84%) | 136 (85%) | 96 (85%) | 0.89 |
0.17c |
| TG | 43 (7%) | 11 (8%) | 16 (8%) | 9 (6%) | 7 (6%) | ||
| GG |
54 (9%) |
9 (7%) |
18 (9%) |
17 (10%) |
10 (9%) |
||
|
MyD88 | |||||||
| rs7744 | |||||||
| A | 954 (77%) | 224 (83%) | 315 (76%) | 245 (76%) | 170 (75%) | 0.08 | |
| G | 282 (23%) | 46 (17%) | 101 (24%) | 79 (24%) | 56 (25%) | ||
| A/A | 370 (60%) | 92 (68%) | 120 (58%) | 93 (57%) | 65 (57%) | 0.64 | |
| A/G | 214 (35%) | 40 (30%) | 75 (36%) | 59 (36%) | 40 (35%) | 0.34 | |
| G/G | 34 (5%) | 3 (2%) | 13 (6%) | 10 (6%) | 8 (7%) | ||
Chi square test.
HWE (Hardy Weinberg Equilibrium).
HWE in women. Text in bold denotes statistical significance.
Figure 3.
Linkage disequilibrium of TLR7 variants.
Correlation of clinical biomarkers with polymorphisms of the TLR7 and MyD88 genes
In a subsequent analysis with complete clinical data, we explored the distribution of the genotypes of the TLR7 and MyD88 genes. In this sense, we observed significant differences in ferritin (ng/mL), C reactive protein (mg/L) and LDH levels among the genotypes of the rs3853839, rs179008, and rs179009 variants of the TLR7 gene; nevertheless, only ferritin and LDH showed significant differences for the rs2302267 variant (Table 3 ). Regarding rs7744 of the MyD88 gene, significant differences were observed only in D-dimer (ng/mL) (P = 0.03), with increasing levels among genotypes: for AA, a median of 281.3 (IQR 74.5–665.5); for AG, a median of 291 (ng/mL) (IQR = 69.5–683.5); and for GG, a median of 547.75 (ng/mL) (IQR = 260–845).
Table 3.
Genotypes of TLR7 and MyD88 variant genes with clinical characteristics.
| TLR7 | ||||
|---|---|---|---|---|
| rs3853839 | CC | CG | GG | Pa |
| Oxygen saturation % | 88 (80–93) | 89 (82–95) | 86.5 (78–93) | 0.09 |
| Platelets ×109/L | 230 (167–295) | 260 (204–321) | 240 (186–308) | 0.18 |
| Ferritin (ng/mL) | 493.7(259.8–904.3) | 230.6(96.6–489.3) | 511.5(257–987.9) | <0.001 |
| D Dimer (ng/mL) | 278.5 (61.4–690) | 348.5 (120–677) | 319 (90.3–754) | 0.76 |
| C-Reactive Protein (mg/L) | 20.05(6.8–90) | 13.03(3.1–31.73) | 19.67(6.11–61.73) | 0.04 |
| DHL (U/L) | 279.5(202–436.5) | 251.9(159.2–354) | 327.1(228–453) | <0.001 |
| IFNα (pg/mL) | 17.9 (9.7–34.3) | 15.5 (9.6–23.6) | 18.2 (11.4–40.9) | 0.21 |
| rs179008 | AA | AG | GG | |
| Oxygen saturation % | 87 (78–93) | 90 (83–94) | 89 (80–94) | 0.15 |
| Platelets ×109/L | 238.5 (185–310) | 250 (193–289) | 231 (164–300) | 0.41 |
| Ferritin (ng/mL) | 463(219–888.3) | 159.9(42.1–503.2) | 529.1(259.8–1000) | <0.001 |
| D Dimer (ng/mL) | 313 (74.7–672.4) | 263 (77–616) | 294.35 (84–828) | 0.62 |
| C-Reactive Protein (mg/L) | 18.98(5.30–66.96) | 10.2(3.53–24.81) | 20.83(5.5–123.49) | 0.01 |
| DHL (U/L) | 309.5(214–448) | 233.95(147–323) | 313.9(221.5–442) | 0.001 |
| IFNα (pg/mL) | 18.12 (10.8–34.7) | 19.8 (9.8–31.8) | 16.05 (9.8–26.1) | 0.66 |
| rs179009 | AA | AG | GG | |
| Oxygen saturation % | 87 (77–92) | 90 (82–94) | 88 (79–93) | 0.09 |
| Platelets ×109/L | 237 (184–305) | 252 (198–320) | 226 (174–300) | 0.13 |
| Ferritin (ng/mL) | 486.5(237–894.1) | 170.35(46–433.9) | 580.3(259.8–1021.6) | <0.001 |
| D Dimer (ng/mL) | 297 (73–627) | 286 (77–666) | 322.7 (87–793.43) | 0.46 |
| C-Reactive Protein (mg/L) | 18.03(5.39–62.32) | 11.05(2.6–28.42) | 22.35(7.1–90) | 0.006 |
| DHL (U/L) | 308(217–443.7) | 219.5(154–323) | 334.2(238–456.5) | <0.001 |
| IFNα (pg/mL) | 17.6 (10.8–34.3) | 20.4 (12.3–31.8) | 17.6 (10.4–33.8) | 0.6 |
| rs2302267 | TT | TG | GG | |
| Oxygen saturation % | 87 (79–93) | 90.5 (83–95) | 85 (71–93) | 0.05 |
| Platelets ×109/L | 235 (180–307) | 250 (204–336) | 230 (185–295) | 0.30 |
| Ferritin (ng/mL) | 457.3(205.1–885.9) | 212.25(61–390.6) | 584.4(387.7–1042.1) | <0.001 |
| D Dimer (ng/mL) | 325.5 (83.15–751) | 260 (77–564) | 162.05 (44.1–465) | 0.07 |
| C-Reactive Protein (mg/L) | 18.34 (5.3–62.9) | 9.7 (5–71.55) | 20.24 (5.72–163.72) | 0.32 |
| DHL (U/L) | 306.30(211–438) | 209.15(158.5–269.7) | 360.75(250–480) | <0.001 |
| IFNα (pg/mL) | 18.2 (10.7–33.8) | 14.3 (9.9–19.7) | 17.6 (11.9–50.4) | 0.31 |
| MyD88 | ||||
| rs7744 | AA | AG | GG | |
| Oxygen saturation % | 88 (80–93) | 87 (77–93) | 83 (71–92) | 0.09 |
| Platelets ×109/L | 237 (178–303) | 233 (184–304.5) | 271 (204–331) | 0.39 |
| Ferritin (ng/mL) | 487 (184.6–888.3) | 438.3 (225–865.4) | 359.3 (208.4–558.9) | 0.81 |
| D Dimer (ng/mL) | 281.3(74.5–665.5) | 291(69.5–683.5) | 547.75(260–845) | 0.03 |
| C-Reactive Protein (mg/L) | 17.12 (4.65–82.9) | 17.65 (5.42–51.74) | 24.5 (18.31–38.51) | 0.30 |
| DHL (U/L) | 304 (202–438.5) | 293.6 (212.1–436.5) | 324 (276–389) | 0.62 |
| IFNα (pg/mL) | 18.16 (11.8–37.2) | 17.6 (9.7–27.4) | 17.02 (6.14–22.6) | 0.16 |
IQR = Interquartile Range.
Kruskal–Wallis test. Chi square test.
We performed the effect of TLR7 alleles separately in males and females, we observed for the MAF allele of rs3853839 significant differences between C Reactive Protein (P = 0.009) observing a decreased level in the Minor Allele Frequency (MAF) with a median of 19.9 mg/L (IQR 6.01–79.7) and LDH (P = 0.01) with increasing levels in the MAF, median of 349 U/L (IQR 252–470) in men group. In this sense, we found significant differences in D-dimer (ng/mL) between women group (P = 0.01).
Logistic regression analysis
In the logistic regression analysis adjusted by age, sex, hypertension, type 2 diabetes, and obesity, we found a statistically significant association of rs7744 (1244 A > G) of the MyD88 gene with outcome severity. Additionally, we observed an increase in the magnitude of the association according to COVID-19 outcome progression. It was shown a statistically significant association of the A/G genotype with an OR = 8.83 (95% CI = 1.82–42.23; P = 0.007) with fatal COVID-19 outcome. For the dominant model (AG + GG vs. AA), we observed an OR = 2.44 (95% CI = 1.21–4.9; P = 0.01) with deceased outcome. For the recessive model we found and OR = 6.75 (95% CI = 1.45–31.33; P = 0.01) with deceased outcome (Table 4 ).
Table 4.
Association of TLR7 and MyD88 polymorphisms with COVID-19 outcomes.
| Polymorphisms | Severe |
Critical |
Deceased |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ORa |
95% CI |
P |
ORa |
95% CI |
P |
ORa |
95% CI |
P |
|
| TLR7 | |||||||||
| rs179008 | |||||||||
| A | Reference | Reference | Reference | ||||||
| T | 0.86 | 0.56–1.30 | 0.48 | 0.94 | 0.60–1.47 | 0.79 | 0.67 | 0.37–1.19 | 0.17 |
| AA | Reference | Reference | Reference | ||||||
| AT | 0.54 | 0.21–1.37 | 0.19 | 1.40 | 0.54–3.61 | 0.48 | 0.91 | 0.25–3.24 | 0.89 |
| TTc | 0.93 | 0.48–1.81 | 0.84 | 0.85 | 0.41–1.77 | 0.67 | 0.61 | 0.24–1.52 | 0.28 |
| AT + TTd | 0.74 | 0.42–1.32 | 0.32 | 1.26 | 0.66–2.40 | 0.47 | 0.75 | 0.32–1.78 | 0.52 |
| TTr | 0.94 | 0.47–1.87 | 0.87 | 0.97 | 0.45–2.11 | 0.95 | 0.66 | 0.22–1.17 | 0.45 |
| Log additive | 0.89 | 0.64–1.24 | 0.51 | 0.85 | 0.54–1.34 | 0.50 | 0.82 | 0.49–1.37 | 0.46 |
| rs179009 | |||||||||
| A | Reference | Reference | Reference | ||||||
| G | 0.74 | 0.52–1.06 | 0.10 | 0.79 | 0.54–1.17 | 0.25 | 0.86 | 0.53–1.38 | 0.54 |
| AA | Reference | Reference | Reference | ||||||
| AG | 0.65 | 0.29–1.46 | 0.30 | 0.38 | 0.15–0.99 | 0.05 | 0.35 | 0.11–1.14 | 0.08 |
| GGc | 0.70 | 0.39–1.26 | 0.24 | 0.81 | 0.44–1.4 | 0.50 | 0.91 | 0.43–1.93 | 0.82 |
| AG + GGd | 0.69 | 0.41–1.14 | 0.15 | 0.67 | 0.38–1.17 | 0.16 | 0.71 | 0.36–1.41 | 0.33 |
| GGr | 1.08 | 0.64–1.82 | 0.75 | 1.19 | 0.67–2.09 | 0.54 | 1.42 | 0.71–2.83 | 0.31 |
| Log additive | 0.82 | 0.62–1.09 | 0.19 | 0.87 | 0.64–1.17 | 0.37 | 0.91 | 0.63–1.32 | 0.63 |
| rs3853839 | |||||||||
| C | Reference | Reference | Reference | ||||||
| G | 1.09 | 0.75–1.56 | 0.64 | 1.83 | 1.21–2.75 | 0.004 | 1.36 | 0.83–2.23 | 0.21 |
| CC | Reference | Reference | Reference | ||||||
| CG | 0.83 | 0.34–1.99 | 0.68 | 1.16 | 0.41–3.25 | 0.78 | 2.02 | 0.56–7.20 | 0.27 |
| GGc | 1.09 | 0.61–1.95 | 0.75 | 1.98 | 1.04–3.77 | 0.03 | 1.57 | 0.70–3.52 | 0.26 |
| CG + GGd | 0.86 | 0.50–1.48 | 0.59 | 1.85 | 0.98–3.49 | 0.05 | 1.17 | 0.56–2.43 | 0.67 |
| GGr | 1.49 | 0.91–2.44 | 0.10 | 1.91 | 1.08–3.39 | 0.03 | 1.28 | 0.63–2.58 | 0.49 |
| Log additive | 1.09 | 0.82–1.45 | 0.51 | 1.42 | 1.03–1.96 | 0.03 | 1.22 | 0.83–1.81 | 0.30 |
| rs2302267 | |||||||||
| T | Reference | Reference | Reference | ||||||
| G | 0.72 | 0.40–1.28 | 0.26 | 1.10 | 0.61–1.99 | 0.73 | 0.64 | 0.29–1.40 | 0.26 |
| TT | Reference | Reference | Reference | ||||||
| TG | 1.34 | 0.48–3.72 | 0.56 | 1.24 | 0.38–4.02 | 0.72 | 1.23 | 0.42–3.55 | 0.70 |
| GGc | 0.52 | 0.19–1.41 | 0.20 | 1.14 | 0.43–2.98 | 0.88 | 1.01 | 0.40–2.51 | 0.98 |
| TG + GGd | 0.71 | 0.35–1.44 | 0.34 | 1.03 | 0.49–2.16 | 0.93 | 0.71 | 0.25–1.97 | 0.51 |
| GGr | 0.76 | 0.29–2.00 | 0.58 | 1.53 | 0.58–4.02 | 0.38 | 0.53 | 0.15–1.85 | 0.32 |
| Log additive |
0.82 |
0.53–1.28 |
0.40 |
1.11 |
0.70–1.74 |
0.64 |
0.77 |
0.42–1.40 |
0.39 |
|
MyD88 | |||||||||
| Rs7744 | |||||||||
| A | Reference | Reference | Reference | ||||||
| G | 1.58 | 1.01–2.45 | 0.04 | 1.76 | 1.10–2.82 | 0.02 | 2.45 | 1.38–4.34 | 0.002 |
| A/A | Reference | Reference | Reference | ||||||
| A/G | 1.66 | 0.96–2.84 | 0.06 | 1.69 | 0.94–3.05 | 0.08 | 2.03 | 0.97–4.25 | 0.06 |
| G/Gc | 2.34 | 0.58–9.40 | 0.23 | 3.51 | 0.86–14.24 | 0.07 | 8.83 | 1.82–42.23 | 0.007 |
| A/G + G/Gd | 1.70 | 1.02–2.86 | 0.04 | 1.82 | 1.04–3.21 | 0.03 | 2.44 | 1.21–4.9 | 0.01 |
| G/Gr | 1.96 | 0.49–7.77 | 0.33 | 2.91 | 0.73–11.64 | 0.13 | 6.75 | 1.45–31.33 | 0.01 |
| Log additive | 1.61 | 1.02–2.54 | 0.04 | 1.75 | 1.09–2.84 | 0.02 | 2.41 | 1.35–4.32 | 0.003 |
Adjusted for age, sex, hypertension, type 2 diabetes, and obesity. d: dominant inheritance model, the reference group is formed by the major allele homozygote genotype; r: recessive inheritance model, the reference group is formed by the major allele homozygote and heterozygote genotypes. The text in bold denotes statistical significant.
Interestingly, for rs3853839 of the TLR7 gene for the GG genotype under the codominant model we found an OR = 1.98 (95% CI = 1.04–3.77) with critical outcome, while for the recessive model (GG), we observed an OR = 1.91 (95% CI = 1.08–3.39) with critical outcome. In the log additive model, the OR was 1.42 (95% CI = 1.03–1.96) (Table 4). However, with the correction for multiple comparisons the allele G was associated with critical outcome (OR = 1.83; 95% IC = 1.21–2.75; P = 0.004). In a special analysis restricted only to young men (<60 years) without hypertension, type 2 diabetes, and obesity, the results showed an OR = 4.3 (95% CI = 1.11–16.52; P = 0.002) with critical outcome.
For the X-linked inheritance of the TLR7 variants we performed a logistic regression for alleles stratified by gender and we found for G allele in the rs3853839 an OR = 2.49 (95% CI = 1.43–4.32; P = 0.001) with critical outcome in men (Table 5 ).
Table 5.
Association of TLR7 polymorphisms with COVID-19 outcomes in men.
| Polymorphisms | Severe |
Critical |
Deceased |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ORa |
95% CI |
P |
ORa |
95% CI |
P |
ORa |
95% CI |
P |
|
| TLR7 | |||||||||
| rs179008 | |||||||||
| A | Reference | Reference | Reference | ||||||
| T | 1.06 | 0.63–1.82 | 0.82 | 0.66 | 0.36–1.20 | 0.17 | 0.77 | 0.37–1.59 | 0.48 |
| rs179009 | |||||||||
| A | Reference | Reference | Reference | ||||||
| G | 0.78 | 0.49–1.22 | 0.28 | 0.59 | 0.35–98 | 0.04 | 0.99 | 0.54–1.82 | 0.99 |
| rs3853839 | |||||||||
| C | Reference | Reference | Reference | ||||||
| G | 1.22 | 0.76–1.98 | 0.4 | 2.49 | 1.43–4.32 | 0.001 | 1.42 | 0.74–2.7 | 0.28 |
| rs2302267 | |||||||||
| T | Reference | Reference | Reference | ||||||
| G | 0.79 | 0.37–1.68 | 0.55 | 1.72 | 0.80–3.68 | 0.15 | 0.88 | 0.33–2.34 | 0.82 |
Adjusted for age, hypertension, type 2 diabetes, and obesity.
Discussion
Since COVID-19 pandemic emerged, multiple studies of susceptibility have been published showing different epidemiologic risks, such as non-communicable diseases.18 , 19 In the present study, we found obesity, type 2 diabetes, and hypertension to be the most common comorbidities with a 31% frequency. In Mexico, the prevalence of obesity has been reported to be 36%, for type 2 diabetes 15.7%, and 30.2% for hypertension.20 According to age and sex, Martínez-Martínez et al. reported a median age of 43.6 ± 17.07 years and an increased incidence of severe COVID-19 in men.21 De la Cruz-Cano et al. reported a mean age of 59.62 years with a frequency of 60.52% for males.22 In our study, the median age was 52 years (IQR, 43–63), and men accounted for a higher proportion of COVID-19 cases (63%).18
The main symptoms in our study population were cough (70%), dyspnoea (62%), and headache (56%); however, in other reports, the three main symptoms were headache (50%), arthralgia or myalgia (38%), and sore throat (36%).18
The susceptibility to COVID-19 has been described in multiple studies.18 , 19 However, the genetic and molecular mechanisms are unclear. Previous studies on host genetics23, 24, 25 have reported some loci that could affect the loss-of-function of immune molecules implicated in the response to infectious diseases. Reports have suggested that host genome variations play a role in COVID-19 outcomes.26 , 27 In this sense, TLR7 and MyD88 gene variants could influence the susceptibility to infectious diseases.28
In our study, gene frequencies are similar to those reported in other populations. We found statistically significant differences in frequencies of TLR7 gene SNPs, located on the X chromosome. The allele frequencies of rs3853839, located in the 3′UTR, were similar to the reported in HapMap for Mexican population29. This work represents the first report proposing the rs3853839 to increase the risk of developing severe outcomes in COVID-19.
Recently, rare variants of the TLR7 gene were associated with COVID-19.7 , 9 , 30 , 31 In this sense, Fallerini et al. (2021) identified these variants associated with COVID-19, especially in young males (<60 years old) hospitalized with supplemental oxygen (CPAP/BiPAP and intubated). This suggests that variants in the TLR7 gene are responsible for severely affecting young males with COVID-19.7 Moreover, van de Veerdonk FL and Netea MG (2021) suggested that screening TLR7 variants in patients and their relatives could be a potential therapeutic strategy during interferon gamma treatment.32
A recent work showed that TLR7 is responsible for IFN-α production in response to SARS-CoV-2,33 that could explain why some variants of the TLR7 gene are implicated in immune activation during COVID-19. The type I IFN response is associated with severe disease that has been demonstrated in patients with inborn errors of type I IFN.34 A deficiency in the signaling pathway would result in abrogated innate and adaptive immune responses, like we can observe, the IFN-α decreased in the deceased group suggesting that the study of these SNPs could be associated with a deficiency of signaling pathway in the antiviral immune responses of COVID-19.
On the other hand, the MyD88 gene is located on chromosome 3p22 locus. Variants in this gene are associated with diverse pathologies, such as ulcerative colitis and Buerger's disease.10 , 35 The rs7744 variant is located at the 3′UTR of the MyD88 gene and might play a key role in the severity of COVID-19. In the present work, we found a significant association of rs7744 with the severity of COVID-19, increasing the odds ratio of severity in the codominant and log additive models. MyD88 is implicated in TLR/interleukin-1 receptor (IL-1R) family signalling in response to pathogens and injury.36 In a review of MyD88 as a therapeutic target for inflammatory lung diseases, the authors reported that mice deficient in this gene have inflammatory responses in models such as endotoxin-induced acute respiratory distress, allergic asthma, tobacco smoke inflammation,37 bronchitis and lung fibrosis.38
Matsunga et al.35 proposed that this variant could be implicated with high levels of MyD88 as binding site of miRNA is closely located to rs7744 resulting in a non-regulated cleavage of mRNA and impair protein synthesis. However, to understand the function of rs7744, further studies are necessary. Given the impact of the association seen between rs7744 MyD88 gene variant and fatal outcomes of COVID-19, as well as the locus of this polymorphism, we wanted to predict its functional impact on miRNA transcriptional regulation of MyD88 gene. We explored the PolymiRTS database to search for miRNA that recognize the 3′UTR sequence where the variant falls.39 In this sense, two miRNA (miR-6866-5p and miR-877-5p) can bind to the MyD88 mRNA of rs7744 variant ancestral allele; while with the minor allele these miRNAs lose their target. Then, the minor allele generates a target sequence for two miRNA (miR-520g-5p and miR-6837-3p). This points out the functional impact over MyD88 gene expression of the rs7744 variant, and further functional studies are needed to corroborate which miRNA is involved in the high risk of developing fatal outcomes for COVID-19.
MyD88 plays a critical role in protecting hosts against different pathogens, such as viruses, any dysfunction of this protein adaptor might result in abrogated innate and adaptive immune responses. In the context of immunity against severe acute respiratory syndrome coronaviruses, Sheahan et al. reported that MyD88 knockout mice showed enhanced pulmonary pathology and a higher mortality rate after infection with the novel human SARS-CoV from 2003,40 highlighting the pivotal role of this adaptor protein in providing immune protection against respiratory viruses. In a different study, Seo et al. reported that mice deficient in MyD88 showed significant susceptibility to primary influenza infection compared to their wild type counterparts. In the same study, the authors suggested that the absence of MyD88 could be correlated with a decreased production of pro-inflammatory cytokines, particularly Th1 cytokines, which could result in impaired T-cell mediated antiviral responses.41
Despite the key role of MyD88 in protecting hosts against infections, it has been reported that this adaptor protein can induce excessive inflammation and accelerate diseases,42 , 43 a phenomenon that is very relevant in the context of SARS-CoV-2. It has been reported that a large number of patients have cytokine release syndrome that triggers pathology progression. Therefore, it is relevant to take these findings further and understand whether mutations in the MyD88 gene could be correlated with disease progression caused by a lack of pro-inflammatory cytokines or excessive systemic inflammation.
The present study has some limitations, such as the inability to access all laboratory information from the study population. As well as, it is important to look for data on the cytokine and chemokine status of the patients included in this study. Another limitation is that we do not know patients smoking status, and the association could be modified. Another limitation that we hope to address soon is evaluating the role of TRIF in the observed correlations in this study, given that TRIF is an important adaptor protein that plays a significant role triggering immune responses against single-stranded RNA viruses mediated by TLR7/8.
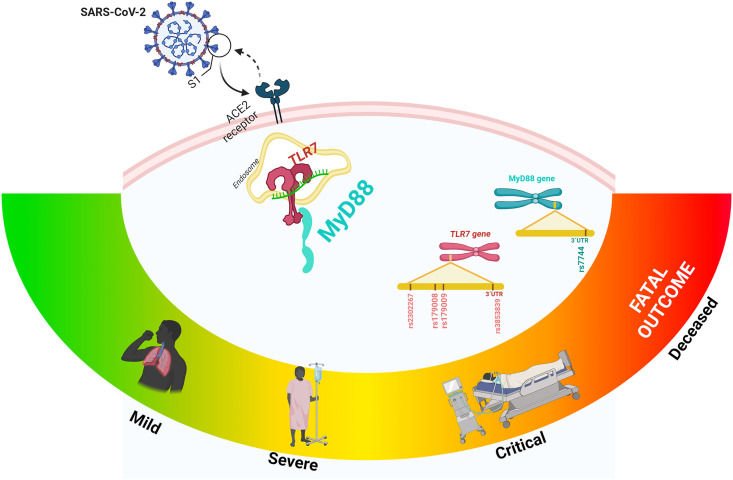
In conclusion, our results suggest that the MyD88 rs7744 variant and TLR7 rs3853839 are involved in COVID-19 progression. The identification of susceptibility variants to COVID-19 may lead to develop a personalized treatment (Fig. 4 ).
Figure 4.
The MyD88 rs7744 variant and TLR7 rs3853839 are involved in COVID-19 progression.
Author contributions
ALR, LEMG, GAMN and CP were the main contributors in the design, acquisition, management and interpretation of the data and writing the article. ALR acquisition of the financial support for the project and leadership responsibility for the research. PVC, JMRP, ALR, GAMN performed the formulation of overarching research. TTL, RPVV, GVA, FJMR, MMMM, EBG, JFMV, JG, RCZ and JMRP conducting a research and investigation evidence and process. JPRH, RPS, JMF, MLM, DMZA, GJVZ, AMC, LRT, RFC, PMMM and LELJ liaised with patients and provided access to samples, laboratory and clinical information. BHL, SOP and YSK performance the DNA extraction. OSML, LEMG, AHB performed the genotyping. CMA, DML, MCCR verification the replication/reproducibility of the results/experiments. JJM, MVF, CSA, MVA and LEMG maintain research data. LEMG performed the statistical analysis in STATA. GEJG creation of images. LEMG and DML drafted the manuscript. CP, GAMN and ALR reviewed and edited the manuscript. All authors reviewed and approved the final manuscript.
Funding
This study was funded by the Consejo Nacional de Ciencia y Tecnología; CONACYT 312513 SARS-COV 2.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully thank to PhD Margarita Valdés-Flores and the extraordinary effort of health-care workers who sacrificed their lives while saving patients. In memorial to PhD Margarita Valdés-Flores.
References
- 1.El-Zayat S.R., Sibaii H., Mannaa F.A. Toll-like receptors activation, signaling, and targeting: an overview. Bull Natl Res Cent. 2019;43(1):187. [Google Scholar]
- 2.Dalpke A., Helm M. RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol. 2012;9(6):828–842. doi: 10.4161/rna.20206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saikh K.U. MyD88 and beyond: a perspective on MyD88-targeted therapeutic approach for modulation of host immunity. Immunol Res. 2021;69(2):117–128. doi: 10.1007/s12026-021-09188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balka K.R., De Nardo D. Understanding early TLR signaling through the Myddosome. J Leukoc Biol. 2019;105(2):339–351. doi: 10.1002/JLB.MR0318-096R. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Espinoza I., Guerrero-Plata A. The relevance of TLR8 in viral infections. Pathogens. 2022;11(2) doi: 10.3390/pathogens11020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mabrey F.L., Morrell E.D., Wurfel M.M. TLRs in COVID-19: how they drive immunopathology and the rationale for modulation. Innate Immun. 2021;27(7–8):503–513. doi: 10.1177/17534259211051364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. Elife. 2021;10 doi: 10.7554/eLife.67569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324(7):663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci Immunol. 2021;6(62) doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z., Nakajima T., Inoue Y., Kudo T., Jibiki M., Iwai T., et al. A single nucleotide polymorphism in the 3'-untranslated region of MyD88 gene is associated with Buerger disease but not with Takayasu arteritis in Japanese. J Hum Genet. 2011;56(7):545–547. doi: 10.1038/jhg.2011.44. [DOI] [PubMed] [Google Scholar]
- 11.Jiménez-Sousa M., Fadrique A., Liu P., Fernández-Rodríguez A., Lorenzo-López M., Gómez-Sánchez E., et al. TNFAIP3, TNIP1, and MyD88 polymorphisms predict septic-shock-related death in patients who underwent major surgery. J Clin Med. 2019;8(3) doi: 10.3390/jcm8030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun D., Sun L., Xu Q., Gong Y., Wang H., Yang J., et al. SNP-SNP interaction between TLR4 and MyD88 in susceptibility to coronary artery disease in the Chinese han population. Int J Environ Res Publ Health. 2016;13(3) doi: 10.3390/ijerph13030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruscitti P., Berardicurti O., Di Benedetto P., Cipriani P., Iagnocco A., Shoenfeld Y., et al. Severe COVID-19, another piece in the puzzle of the hyperferritinemic syndrome. An immunomodulatory perspective to alleviate the storm. Front Immunol. 2020;11:1130. doi: 10.3389/fimmu.2020.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D., Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Targeted Ther. 2021;6(1):291. doi: 10.1038/s41392-021-00687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Gómez L.E., Ibarra-González I., Fernández-Lainez C., Tusie T., Moreno-Macías H., Martinez-Armenta C., et al. Metabolic reprogramming in SARS-CoV-2 infection impacts the outcome of COVID-19 patients. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi R.T., Lynch J.B., Del Rio C. Mild or moderate Covid-19. N Engl J Med. 2020;383(18):1757–1766. doi: 10.1056/NEJMcp2009249. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Rojas M.A., Luna-Ruiz Esparza M.A., Campos-Romero A., Calva-Espinosa D.Y., Moreno-Camacho J.L., Langle-Martínez A.P., et al. Epidemiology of COVID-19 in Mexico: symptomatic profiles and presymptomatic people. Int J Infect Dis. 2021;104:572–579. doi: 10.1016/j.ijid.2020.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mesta F., Coll A.M., Ramírez M., Delgado-Roche L. Predictors of mortality in hospitalized COVID-19 patients: a Mexican population-based cohort study. Biomedicine. 2021;11(2):1–4. doi: 10.37796/2211-8039.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamah-Levy T., Romero-Martinez M., Barrientos-Gutierrez T., Cuevas-Nasu L., Bautista-Arredondo S., Colchero M., et al. Resultados Nacionales; 2021. Encuesta nacional de Salud y nutricion 2020 Sobre COVID-19. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Martínez M.U., Alpízar-Rodríguez D., Flores-Ramírez R., Portales-Pérez D.P., Soria-Guerra R.E., Pérez-Vázquez F., et al. An analysis COVID-19 in Mexico: a prediction of severity. J Gen Intern Med. 2022;37(3):624–631. doi: 10.1007/s11606-021-07235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De la Cruz-Cano E., Jiménez-González C.D.C., Díaz-Gandarilla J.A., López-Victorio C.J., Escobar-Ramírez A., Uribe-López S.A., et al. Comorbidities and laboratory parameters associated with SARS-CoV-2 infection severity in patients from the southeast of Mexico: a cross-sectional study. F1000Research. 2022;11:10. doi: 10.12688/f1000research.74023.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mapping the human genetic architecture of COVID-19. Nature. 2021;600(7889):472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delorey T.M., Ziegler C.G.K., Heimberg G., Normand R., Yang Y., Segerstolpe Å., et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595(7865):107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Gómez L.E., Herrera-López B., Martinez-Armenta C., Ortega-Peña S., Camacho-Rea M.D.C., Suarez-Ahedo C., et al. ACE and ACE2 gene variants are associated with severe outcomes of COVID-19 in men. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.812940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas-Alarcón G., Ramírez-Bello J., Posadas-Sánchez R., Rojas-Velasco G., López-Reyes A., Martínez-Gómez L., et al. The rs8176740 T/A and rs512770 T/C genetic variants of the ABO gene increased the risk of COVID-19, as well as the plasma concentration Platelets. Biomolecules. 2022;12(4) doi: 10.3390/biom12040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deguine J., Barton G.M. MyD88: a central player in innate immune signaling. F1000Prime Rep. 2014;6:97. doi: 10.12703/P6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ensembl Genotypes for 1000GENOMES:phase_3:MXL. 2022. https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=X:12889039-12890039;v=rs3853839;vdb=variation;vf=93272713#373531_tablePanel
- 30.Bortolotti D., Gentili V., Rizzo S., Schiuma G., Beltrami S., Strazzabosco G., et al. TLR3 and TLR7 RNA sensor activation during SARS-CoV-2 infection. Microorganisms. 2021;9(9) doi: 10.3390/microorganisms9091820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solanich X., Vargas-Parra G., van der Made C.I., Simons A., Schuurs-Hoeijmakers J., Antolí A., et al. Genetic screening for TLR7 variants in young and previously healthy men with severe COVID-19. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van de Veerdonk F.L., Netea M.G. Rare variants increase the risk of severe COVID-19. Elife. 2021:10. doi: 10.7554/eLife.67860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Sluis R.M., Cham L.B., Gris-Oliver A., Gammelgaard K.R., Pedersen J.G., Idorn M., et al. TLR2 and TLR7 mediate distinct immunopathological and antiviral plasmacytoid dendritic cell responses to SARS-CoV-2 infection. EMBO J. 2022 doi: 10.15252/embj.2021109622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker J., Kalinke U. Toll-like receptors matter: plasmacytoid dendritic cells in COVID-19. EMBO J. 2022 doi: 10.15252/embj.2022111208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsunaga K., Tahara T., Shiroeda H., Otsuka T., Nakamura M., Shimasaki T., et al. The ∗1244 A>G polymorphism of MyD88 (rs7744) is closely associated with susceptibility to ulcerative colitis. Mol Med Rep. 2014;9(1):28–32. doi: 10.3892/mmr.2013.1769. [DOI] [PubMed] [Google Scholar]
- 36.Chen L., Zheng L., Chen P., Liang G. Myeloid differentiation primary response protein 88 (MyD88): the central hub of TLR/IL-1R signaling. J Med Chem. 2020;63(22):13316–13329. doi: 10.1021/acs.jmedchem.0c00884. [DOI] [PubMed] [Google Scholar]
- 37.Doz E., Noulin N., Boichot E., Guénon I., Fick L., Le Bert M., et al. Cigarette smoke-induced pulmonary inflammation is TLR4/MyD88 and IL-1R1/MyD88 signaling dependent. J Immunol. 2008;180(2):1169–1178. doi: 10.4049/jimmunol.180.2.1169. [DOI] [PubMed] [Google Scholar]
- 38.Agoro R., Piotet-Morin J., Palomo J., Michaudel C., Vigne S., Maillet I., et al. IL-1R1-MyD88 axis elicits papain-induced lung inflammation. Eur J Immunol. 2016;46(11):2531–2541. doi: 10.1002/eji.201646366. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya A., Ziebarth J.D., Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42(Database issue):D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., et al. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4(12) doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seo S.U., Kwon H.J., Song J.H., Byun Y.H., Seong B.L., Kawai T., et al. MyD88 signaling is indispensable for primary influenza A virus infection but dispensable for secondary infection. J Virol. 2010;84(24):12713–12722. doi: 10.1128/JVI.01675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura G.M., Balloy V., Ramphal R., Khun H., Huerre M., Ryffel B., et al. Lack of MyD88 protects the immunodeficient host against fatal lung inflammation triggered by the opportunistic bacteria Burkholderia cenocepacia. J Immunol. 2009;183(1):670–676. doi: 10.4049/jimmunol.0801497. [DOI] [PubMed] [Google Scholar]
- 43.Weighardt H., Kaiser-Moore S., Vabulas R.M., Kirschning C.J., Wagner H., Holzmann B. Cutting edge: myeloid differentiation factor 88 deficiency improves resistance against sepsis caused by polymicrobial infection. J Immunol. 2002;169(6):2823–2827. doi: 10.4049/jimmunol.169.6.2823. [DOI] [PubMed] [Google Scholar]