Summary
Just as mammals have coevolved with the intestinal bacterial communities that are part of the microbiota, intestinal helminths represent an important selective force on their mammalian host. The complex interaction between helminth, microbes and their mammalian host is likely an important determinant of mutual fitness. The host immune system in particular is a critical interface with both helminths and the microbiota, and this crosstalk often determines the balance between resistance and tolerance against these widespread parasites. Hence, there are many examples of how both helminths and the microbiota can influence tissue homeostasis and homeostatic immunity. Understanding these processes at a cellular and molecular level is an exciting area of research that we seek to highlight in this review and that will potentially guide future treatment approaches.
Keywords: Helminth, Microbiota, Parasites, Intestine, Inflammatory disease
In Brief
Both the intestinal microbiota and helminths represent important selective forces on their mammalian hosts. Loke and Harris examine trans-kingdom interactions between the microbiota and helminths that occur across a large range of parasite and host species as well as their impacts on parasite fitness and host health and disease.
Introduction
The intestine is a particularly competitive ecological environment. Intestinal inhabitants have found ways to utilize the host to achieve a competitive advantage in this nutrient rich environment. In this continuous arms race, evolution brings an equilibrium that enables the survival of complex eco-cultures. For example, earlier studies examined whether helminth infections would alter the composition of bacterial populations in the gut of their mammalian host, a topic that has been extensively reviewed elsewhere 1–3. However, because such equilibrium states are highly context dependent, with every human population having unique dietary and lifestyle properties, the contribution of helminths into the intestinal ecoculture is highly variable. Furthermore, trans-kingdom interactions with other organisms (e.g. protozoans, fungi and viruses) in this system are largely unexplored.
Undoubtedly, the consequences of helminths to the social network of microbes will have physiological consequences on homeostasis, health and disease in their mammalian host 4. The biochemical communication system underlying such physiology is gradually being uncovered at a molecular and cellular level. Fitness of the helminths themselves are also highly dependent on the relationships with bacterial communities 5,6. With newer technological developments, gaps in our knowledge are being filled, with potential consequences of improving health in countries whereby the burden of these parasites remains substantial – and of understanding how the absence of these parasites alter intestinal ecocultures in those countries and regions where exposure to helminths has been adequately controlled.
Coevolution of helminth and bacterial populations in the mammalian gut.
What shapes the gut microbiota depends on a myriad of factors that differ considerably between industrialized and non-industrialized nations. In an industrialized society, cohabitation is one of the biggest determinants of an individual’s microbiome profile 7, and helminth infections are likely to have different effects from one family to the next. Indeed, by conducting a fecal metagenomic study in Malaysia, we have found that the study village is the largest determinant of fecal microbiome profiles and there is a significant interaction between village and helminths on the fecal microbiome 8. These relationships are probably established in early life. While all infants may start with a predominantly Bifidobacteria community, vertical transmission of specific taxa can establish long term trajectories in future communities 9. By the time children encounter and become infected with intestinal helminths as toddlers, major components of pre-existing communities are already well established. Hence, any effects of helminth infections would be dependent on the pre-existing microbial populations in the gut. Indeed, how helminth infections during pregnancy shape the microbiota of the infants and the immune consequence of this interaction remains poorly understood 10. Interestingly, exposure to Nippostrongylus brasiliensis infection in mice prior to pregnancy can alter the breastmilk microbiota and offspring CD4+ T cells along with their microbial communities 11. Hence, the microbial network from early life will imprint the immune system of the mammalian host for the future. Another complication lies in the finding that helminths impact microbial communities even at sites distant to the worm. Experimental infection with the small intestinal parasite Heligmosomoides polygyrus in mice revealed impacts on mucous and luminal bacterial communities throughout the small and large intestine 12, whilst a recent human study reported helminth-induced alterations to microbial communities in the feces and salvia 13.
Specific mechanisms by which helminths affect bacterial communities are being elucidated (Figure 1). Recently, Hu et. al., 13 identified in mice a new antimicrobial protein (AMP), small proline-rich protein 2A (SPRR2A), that is phylogenetically distinct from previously known AMPs. The authors demonstrated that SPRR2A was produced in response to H. polygyrus infection in mice, was dependent on type 2 cytokines, and specifically targeted gram-positive bacteria (Figure 1). In contrast, resistin-like molecule β (RELMβ) is another intestinal bactericidal protein that specifically targets Gram-negative bacteria 14. Similar to SPRR2A, RELMβ is induced during intestinal helminth infections by type 2 cytokines and may also have direct anti-nematode effects by affecting their chemosensory system 15. Increased mucus production in the intestine is another key feature of type 2 responses to helminth infections 16 and forms a key interface with bacterial populations. Studies with Retnlb−/− mice find that RELMβ is particularly important in limiting the abundance of Gram-negative bacteria in the mucus layer 14(Figure 1). Additionally, this mucus rich environment may favor the expansion of Clostridiales bacteria that can have anti-inflammatory properties during helminth infections 17(Figure 1). Hence, the host type 2 response plays an important role in maintaining the social context of helminth-microbe interactions. Other examples of these physiological interactions include gut motility 18.
Figure 1: Predicted mechanisms of helminth-bacterial interactions.
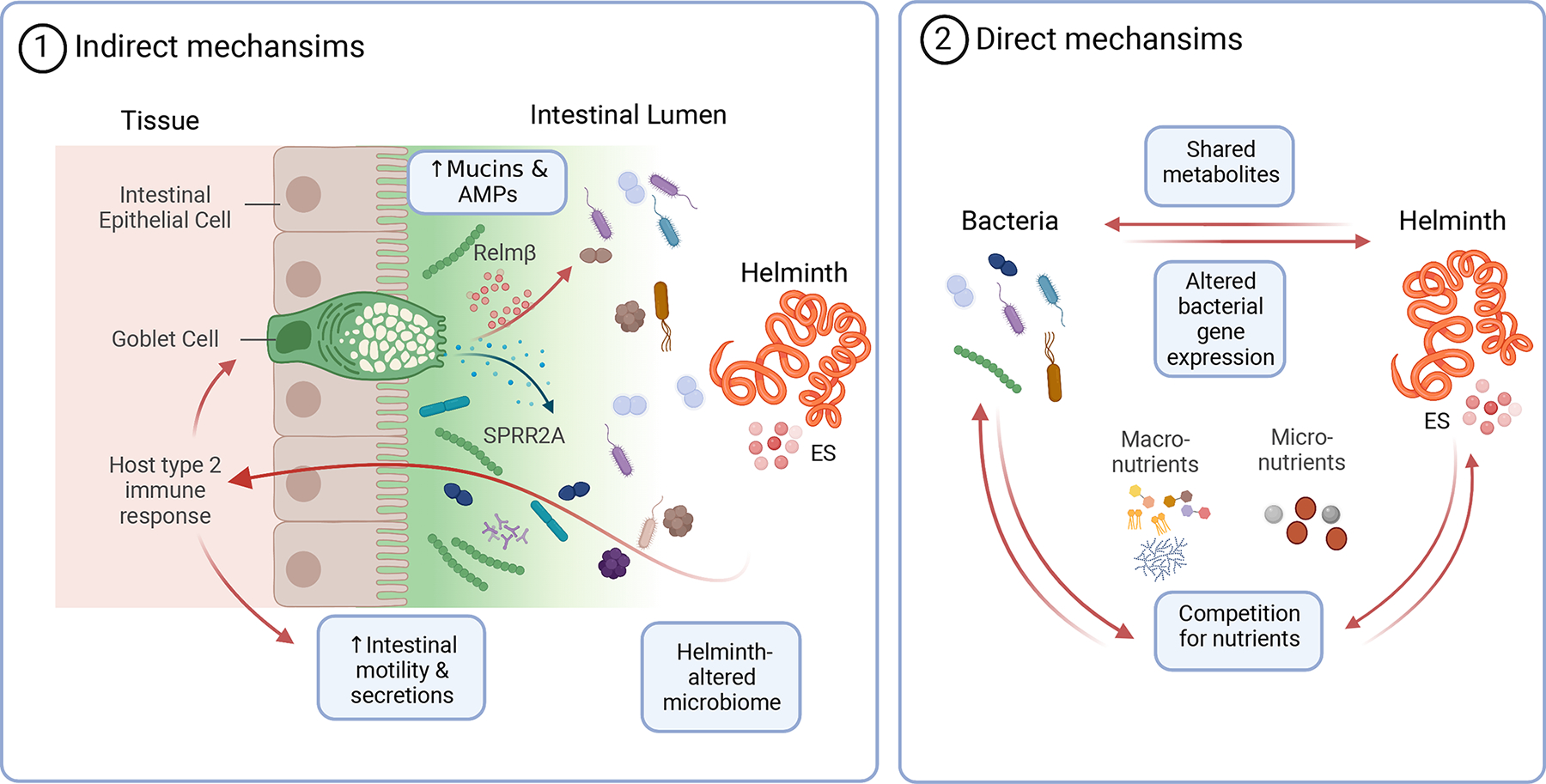
The mechanisms by which helminth infection results in altered intestinal bacterial communities are many and include indirect and direct interactions. Indirect effects result from helminth-induced type two immune response in the mammalian host. This response has been shown to alter intestinal bacterial communities via a number of mechanisms including; i) increased production of SPRR2A and Relmβ proteins that exert antimicrobial activity against gram-positive and gram-negative bacterial spp, respectively and ii) increased and altered production of mucins. It is also likely that type 2 cytokine mediated “weep and sweep’ response, characterised by increased intestinal motility and secretions, impacts microbial communities along the entire intestinal tract. A direct impact of helminth on bacterial communities is likely to occur through multiple mechanisms including, but not restricted to, helminth-induced modulation of bacterial gene expression, the production of shared metabolites and/or toxic metabolic by-products, or competition for essential nutrients. Nutrients could be derived from the host diet, the helminth or bacteria and include; i) macronutrients such as carbohydrates (CHO), lipids and proteins, ii) micronutrients (minerals and vitamins). “Created with BioRender.com”.
The relationship between helminths and microbiota, shaped by evolution, is bidirectional. For example, the Trichuris genus are intestinal parasites that live in the large intestines of diverse mammalian hosts. The eggs embryonate in the soil but will only hatch in the cecum when they encounter the presence of bacteria 19, which means germ-free mice cannot be colonized by Trichuris muris 6. Egg hatching can be triggered by binding to bacterial fimbriae 19. Hatching is but the first step, as the worms will also acquire some of its intestinal microbiota from the mammalian host and cannot fully mature and develop if this microbial acquisition is disrupted 6. Additionally, the worms may be giving themselves an advantage by altering the composition of the microbiota to inhibit other worms from infecting the same host 6. Hence, the dynamically altering bacterial landscape can have substantial impact on the colonization efficiencies of incoming Trichuris worms. Indeed, when laboratory mice are reintroduced into an outdoor enclosure, they become more susceptible to Trichuris muris infection 20, as their microbiota becomes more diverse and their intestinal immune response becomes more biased towards Type 1 cytokines such as IFNγ 20. Whether the increased worm burden is a direct effect of the microbiota changes, or the resulting intestinal immune responses is unclear, but certainly it is not simply a result of enhanced egg hatching because the individual worms themselves have a greater biomass and are healthier in the outdoor environment. Notably, an IFNγ signature characterized by expression of innate immune effector Isg15 is also a feature of the early events of Trichuris larval infection as they form syncytial tunnels in the epithelial cells 21. Perhaps worms exploit the host IFNγ response to improve their ability to colonize and grow in the intestine.
In contrast to T. muris, H. polygyrus enters the host as an infective larvae and is able to complete its lifecycle within the small intestine even in the complete absence of bacteria 18. The presence or absence of a microbiota does however impact worm colonization with both larvae and adult worms being located in the more proximal (towards the duodenum) habitat in antibiotic-treated or germ-free animals 18. This bacterial-induced alteration to worm positioning stems from reduced host production of the excitatory neurotransmitter acetylcholine, resulting in a consequent reduction in intestinal motility. Increased intestinal motility plays a critical role in the protective host ‘weep and sweep’ response that expels adult worms from the intestine 22 and mice lacking a complex microbiota were consequently unable to expel H. polygyrus in a timely manner. Whether or how the microbiota impacts host resistance to helminth species other than T. muris and H. polygyrus remains to be determined. However, alterations to bacterial genes in response to human helminth infection, included those associated with arachidonic acid metabolism and arachidonic acid, is closely linked to host resistance against helminths 23(Table 1),
Table 1:
Known and predicted impacts of helminth infection on the production of microbial-derived metabolites.
| Helminth | Host | Bacterial metabolite or metabolic pathway | Consequence for the host | Reference |
|---|---|---|---|---|
| Ascaris lumbricoides, Necator americanus, Trichuris trichiura | Homo sapiens | ↑Arachidonic acid metabolism | Predicted to alter host resistance to intestinal helminths | 23 |
| T. muris | Mus musculus | ↑Ethanolamine utilization | ↑ T. muris egg hatching | 5 |
| H. polygyrus | Mus musculus | ↑SCFA | ↓Allergic airway inflammation | 29 |
| H. diminuta | Mus musculus | ↑SCFA | ↓Chemical-induced colitis | 30 |
| ES-62 secreted by A. vitea | Mus musculus | ↑SCFA | ↓Collagen-induced arthritis | 32 |
| H. polygyrus | Mus musculus | ↑SCFA | ↓HFD-induced weight gain and glucose intolerance | 33 |
| H. polygyrus | Mus musculus | ↑NE | ↑Adipose tissue beiging and ↓weight gain with HFD | 34 |
| H. polygyrus | Mus musculus | ↓Undefined protective metabolites | ↑Invasion of intestinal epithelial cells by Salmonella Typhimurium | 36 |
Helminths and bacterial communities share a common goal of establishing a long-term residence in the intestine without eliciting inflammation. To avoid bacterial dissemination and ongoing inflammation, it is imperative the host rapidly repairs any tissue damage the helminth inflicts, a process that seems especially important for those helminths that penetrate the mucosa or epithelial barrier as part of their lifecycle. One notable recent discovery is that in mice intestinal epithelial cells residing in the crypts adjacent to tissue dwelling H. polygryus larvae adopt a genetic program reminiscent of fetal development. Re-initiation of this developmental genetic program was required for efficient tissue repair and was driven by IFNγ 24. Interestingly a separate study reported that IFNγ production elicited by H. polygyrus infection peaks during larval tissue invasion and IFNγ, but not type 2 cytokines, required the presence of a complex microbiome 18. This raises the intriguing question as to whether the tissue protective response reported by Nusse et. al., 24 is microbiota dependent. Type 2 cytokine production following H. polygyrus infection drives the production of SPRR2A with increased SPRR2A acting to protect the host against helminth-induced bacterial invasion of intestinal tissues 13. These studies suggest a strong evolutionary drive for mammalian hosts to reinforce the intestinal epithelial barrier following helminth infection – with programs directed at both tissue repair and anti-bacterial defense.
Another important component of the helminth-bacterial interaction could be nutritional (Figure 1). E. coli mutants that reduce Caenorhabditis elegans fertility because of nutritional defects in fatty acid biosynthesis and ethanolamine utilization will also affect the proper development of Trichuris muris parasites in gnotobiotic mice colonized with these E. coli mutants 5. T. muris infection increased the expression of bacterial genes involved in ethanolamine utilization presumably in an attempt to promote its own fitness 5(Table 1). Iron is also an essential resource that can be regulated by bacteria to provide appropriate developmental cues to C. elegans 25 and is likely to affect helminth-microbiota relationships with the host 26. Other trace metals such as manganese, copper and zinc are also important for the survival and metabolism of microbes, helminths and their mammalian hosts. Understanding the tight regulation of these processes remains poor in the complex social network between microbes and host. Certainly, microbial by-products and metabolites are critical component of these interactions, and helminths themselves secrete a variety of factors called excretory secretory (ES) products including proteins, extracellular vesicles and metabolites 19(Figure 1)
In summary, the biological network between helminth-microbiota-host is important for competition, as well as mutual benefits in regulating tissue repair and homeostasis as well as inflammation and nutrition.
Helminth-microbiome interactions can modulate host health and disease
Helminths are amongst the most common and long-lived of all pathogens, an attribute often ascribed to their potent ability to interfere with host immune responses. This also corresponds well with the “hygiene hypothesis” which states that reduced infections are responsible for the increased prevalence of allergic, autoimmune, inflammatory and potentially metabolic diseases observed in urbanised populations 27,28. An increasing number of studies point to a central role of helminth-microbiota interactions in this phenomenon.
Inflammatory diseases
Early reports detailed the impact of helminth-altered microbial communities on experimental disease models. The first reported that H. polygyrus infection increased the availability of bacterial-derived short-chain fatty acids (SCFA), which in turn attenuated experimental allergic airway inflammation 29(Table 1). The second reported that T. muris promoted the expansion of mutualistic Clostridium spps that inhibited the outgrowth of a pathogenic bacterium, Bacteriodes vulgatus, and thus prevent intestinal inflammation 17. Since this time, reports that helminths can alter host health via the microbiome have grown. Shute et al., 30 demonstrated that the tapeworm Hymenolepis diminuta was able to protect mice against chemical-induced colitis with disease modulation requiring helminth-induced alterations to the microbiome as evidenced using both antibiotic depletion and transfer of fecal filtrates. Metabolomic analysis of the filtrate revealed increased levels of SCFAs and disease protection was required the host SCFA receptor GPR43 30(Table 1). Infection of mice with adult male Schistosoma mansoni worms was also reported to suppress chemical-induced colitis, although the presence of both male and female worms exacerbated disease. In the latter case worms are able to mate and eggs shed from the female worms transit from the mucosal vasculature through the intestinal wall causing tissue damage. Close inspection of the microbiome revealed that male only, or both male and female, worm infections altered the microbiome. However the presence of male worms uniquely prevented the emergence of a colitogenic microbiota, possibly reflecting an active attempt by the parasite to mitigate the colonic tissue damage caused by transiting eggs 31. Given that male worms reside in the vasculature rather than intestine, these findings indicate that the worm infection can alter distant microbial communities, likely via soluble products released by the host or worm. In line with this Doonan et. al., investigated the impact of a protein, ES-62, secreted by the tissue dwelling helminth Acanthocheilonema vitea 32. Subcutaneous delivery of ES-62 was sufficient to alter the intestinal microbiota and attenuate the severity of collagen-induced arthritis (CIA). Disease modulation was ablated by antibiotic treatment and the involvement of SCFAs was postulated by the finding that ES-62 treatment maintained the presence of butyrate producing Butyrivibrio bacterial spp in diseased mice (Table 1). It remains unclear however whether ES-62 treatment prevents CIA by altering the gut microbiota, or whether it simply prevents inflammation-induced dysbiosis. Further studies, investigating helminth-microbiome interactions in autoimmune diseases would be of great interest.
Obesity
A convincing role for helminth-altered microbiomes in modulating the severity of diet-induced obesity has emerged in recent years (reviewed in 28), and two recent reports indicate disease modulation involves helminth-microbiota interactions 33,34. Both studies reported that H. polygyrus infection reduced weight gain and increased adiposity in mice fed a high fat diet (HFD), correlating with increased expression of uncoupling protein 1 (UCP1) in the visceral adipose tissue. UCP1 is an integral membrane protein expressed in the mitochondria of brown adipocytes, that can also be upregulated in white adipocytes in a phenomenon referred to as ‘beiging’. UCP1 activation protects against adiposity by increasing the utilization of lipids for heat production in response to norepinephrine (NE). Shimokawa et. al., 34 reported increased concentrations of NE in both the circulation and feces of H. polygyrus infected mice and reasoned that the microbiota may contribute to increased NE that drives UCP1 expression (Table 1). Infection of antibiotic treated mice fed a HFD did not result in increased fecal NE levels or UCP-1 expression in the adipose tissue and mice were not protected against weight gain. Additionally, the increased fecal NE was positivity correlated with the abundance of bacteria belonging to the Bacillus and Esherichia genera, known NE producers. Su et. al., 33, also utilized H. polygyrus in mice to invoke protection against HFD-induced obesity and noted that protection required an intact host type 2 immune responses. A role for the microbiome was demonstrated through fecal microbiome transfer (FMT) experiments and disease protection correlated with increased SCFA availability (Table 1). Notably, the microbiome of H. polygyrus infected mice also afford protection against HFD-induced glucose intolerance. Together these reports provide tantalizing evidence for a role of helminth-altered microbial communities in modulating host responses to high fat diet.
Pathogens
In contrast to chemically-induced colitis, the H. polygyrus-modified microbiome can promote the expansion of a pathogenic bacterium Citrobacter rodentium, and worsen bacterial-driven colitis 35. Similarly, Reynolds et. al., 36 observed increased colonisation of Salmonella Typhimurium in mice harboring H. polygyrus. H. polygyrus infection was associated with alterations to the intestinal metabolome and this resulted in the loss of metabolites able to afford protection against bacterial invasion of intestinal epithelial cells, a process key to the ability of S. Typhimurium to establish infection 36(Table 1). Lastly, analysis of fecal microbiomes in sheep experimentally infected with the agriculturally important sheep helminth Teladorsagia circumcincta or ‘brown stomach worm’ highlighted an association between the worm infection and the presence of known pathobionts (normally harmless bacterial species that are associated with chronic inflammation under specific circumstances). These pathobionts included members of Prevotella and Porhyromonas genera and their presence was notable given that T. circumcincta is a leading cause of gastroenteritis in sheep 37.
Helminths also alter host susceptibility to viruses with H. polygyrus infection promoting immunity to respiratory syncytial virus (RSV) through its ability to induce production of type 1 interferon 38. Of note, protection required the presence of a complex microbiome as H. polygyrus infection of germ-free mice did not induce type 1 interferon production or reduce viral titers following RSV infection. By contrast, infection with Trichinella spirilas or H. polygyrus enhances host susceptibility to murine norovirus as a result of their ability to elicit type 2 immune responses, and this occurs independently of helminth-induced changes to the microbiota 39. How and whether the microbiota impacts on the outcome of other helminth-microbe co-infections has yet to be determined.
Technological developments in studying helminth – bacterial interactions
Advances in high dimensional assay technologies and computational approaches have been major driving forces in understanding the influences of the intestinal microbiota. One of the primary drivers has been sequencing technology. While initial efforts centered around 16S rRNA sequencing to provide an overview of bacterial populations, more recent studies have used shotgun metagenomic sequencing to provide a more complete picture of microbiome content. An advantage of this strategy is the ability to assemble full genomes, as well as to infer functional gene profiles of from the genomic sequences (Figure 2). However, microbiome sequencing data is still underrepresented among human populations for which the helminth infection status is known. Hence, mapping metagenomic sequences obtained from rural populations where helminth infections are prevalent remains challenging. Recently, metagenomic data linked with helminth infection status has been compiled as an online resource to increase data accessibility 40. Also, larger scale metagenomic studies whereby helminth infection status is known is also now available from Malaysia 8 and Cameroon 41. The availability of more data from rural, indigenous and non-industrialized societies will facilitate the assembly of better genome databases for organisms that are missing in industrialized countries.
Figure 2: Technological advances in studying network interactions of helminths, microbes, and host.
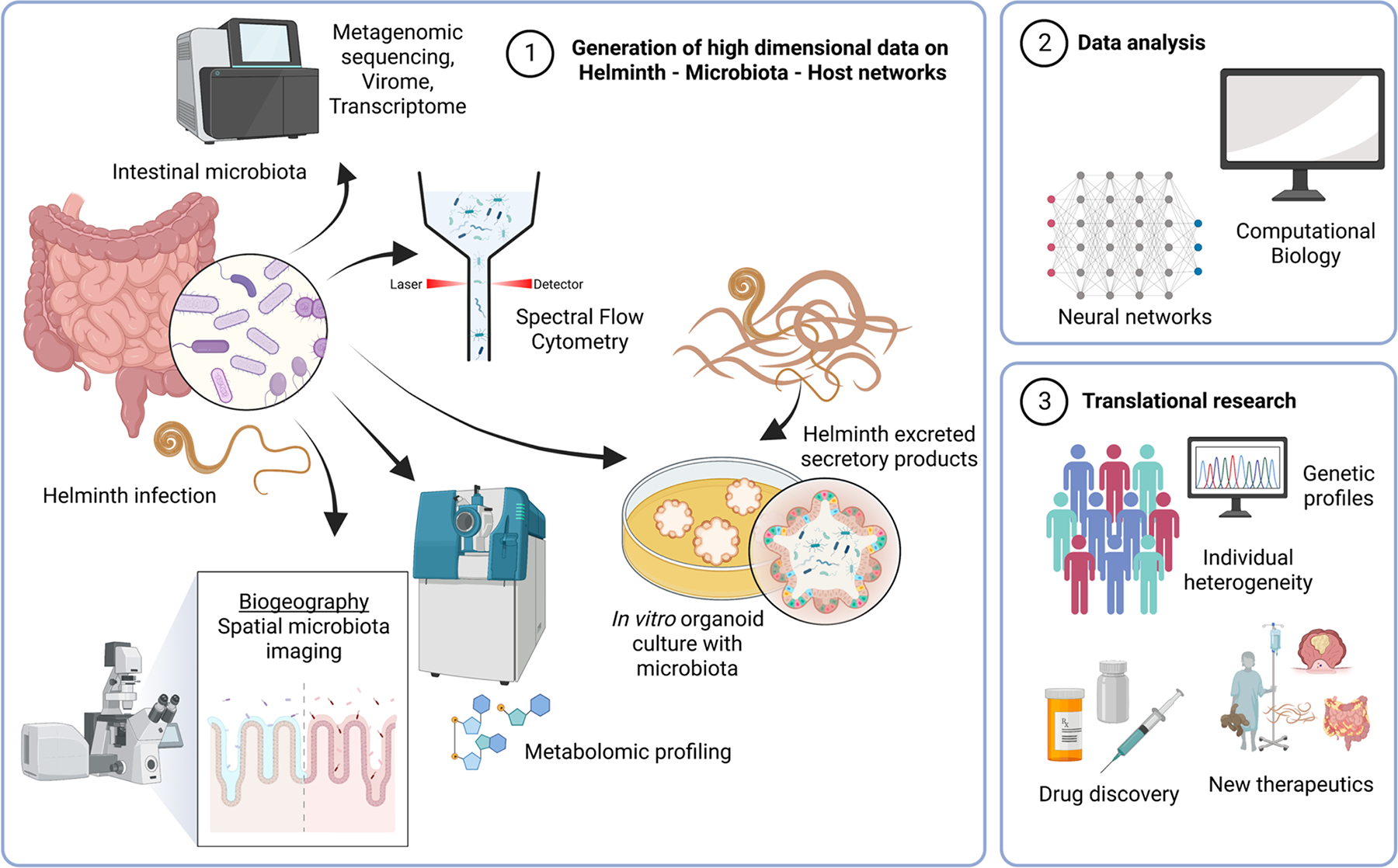
Advances in high dimensional technologies and computational approaches are driving forces in studying the interactions of microbes and helminths with the host immune system. As intestinal helminths and the gut microbiota occupy the same ecological niche, the host intestines have been the major focus of this work. Sequencing technology is a primary driver of many analytical approaches, with newer strategies providing a comprehensive picture of the metagenome, virome and transcriptome of the intestinal microbiota. Other developments include the use of flow cytometry, which can provide absolute count data, as well as sort bacterial populations of interest. Metabolomic profiling provide insights into the biochemical language exchange between host, bacteria and helminths. New imaging and sequencing approaches could enable a better understanding of the biogeography of helminth-bacterial interactions in the intestine. Intestinal biopsies can be used to generate organoid cultures, providing in vitro assay systems to study interactions between helminth secreted molecules and epithelial cells. Bacteria can also be introduced into the organoids in this reductionist system. These high dimensional datasets will then have to undergo extensive data analysis with modern computational biology approaches, including neural network strategies to discern interaction networks. Finally, this information needs to be translated to the clinical setting where there is greater heterogeneity at the host level with many different genetic profiles but has the promise of developing better therapeutic strategies against helminth infections, as well as inflammatory diseases. “Created with BioRender.com”.
Recent advancements in organoid technology may also provide new insights into helminth-microbe interactions with their mammalian host (Figure 2). Whereas these reductionist in vitro systems have been used to study virus, bacteria and helminth interactions independently, investigators have yet to report using organoids to study helminth-microbe interactions. For example, excretory-secreted (ES) products from H. polygyrus can induce epithelial cells in organoid cultures to adopt a fetal-like state spheroid state, which blocks the ability of these cells to differentiate into tuft cells necessary for parasite expulsion 42,43. Although this system showed that succinate induced tuft cell expansion is inhibited by H. polygyrus ES 42, other relationships with microbial metabolites or bacteria are unexplored. These in vitro systems have also been used to examine early events in Trichuris muris larval infection of caecal epithelial cells, including the intracellular formation of syncytial tunnels by burrowing through rows of cells 21, but have yet to be combined with bacteria that trigger the hatching of the eggs to release the larvae. Nonetheless, we are many steps closer towards recapitulating key events in helminth life cycles in vitro, with these advances. Concurrently, technology for co-culturing organoids with bacteria and other microbes are also providing new insights into host-microbe interactions, reviewed elsewhere 44. For example, the relationship between mutations in colorectal cancer and specific bacteria 45,46, could be combined with helminth larvae, or ES products, to investigate relationships between intestinal helminths and colorectal cancer 47 or with viral infections 48. These complex relationships may begin to be further dissected through this reductionist approach. Addition of immune cells to organoid cultures 49 could then add greater realism these models.
At a different scale in the in vivo setting, the spatial 3-dimensional relationships between organisms are in early stages of exploration. The spatial patterns of organisms in the tissues will determine how they interact at local or more systemic levels. This biogeography will influence interactions with host tissues and the balance between pathogenesis and homeostasis 50. Technological advances in microscopy, sequencing and spatial transcriptomics are beginning to combine high dimensional data with high resolution spatial localization 51. Such approaches could provide spatial maps of intestinal helminth interactions with the microbiota in a high dimensional scale (Figure 2). Flow cytometry is another promising new strategy for bacterial microbiota analysis, with the main advantage of providing absolute counts 52 and being able to sort for culture 53. Combined with spectral cytometry that could provide an auto-fluorescent signature of bacterial populations, we have begun to utilize this approach to characterize changes during helminth infections (Figure 2).
Conclusions and Future perspectives
There is now no doubt that helminth infections can alter the microbiome, the microbiome can affect helminths directly and that these interactions both depend on the host response and affect host physiology, homeostasis and disease. However, while there is a conserved type 2 immune response mounted by the host against almost all helminth infections, identification of a conserved type 2 bacterial signature in response to helminth infections is lacking. The type 2 immune response is elicited regardless of the tissues colonized by different helminths, yet the differential effects of a worm that lives in the colon, compared to the small intestine or other tissue, on the bacterial communities in the gut appears substantial. Additionally, the context of the bacterial communities prior to the helminth infection, shaped by early life, diet and other lifestyle determinants also influences the downstream consequences of the worm colonization.
The efficacy of deworming treatment may also be influenced by helminth-bacterial interactions although this remains poorly understood with clear practical implications. Unlike hookworm and Ascaris, Trichuris infection in humans can have low cure rates after anthelminthic treatment 54). Interestingly, intestinal bacterial enterotypes pre-treatment can be associated with the efficacy of deworming treatment in some individuals 55. More specifically, pre-treatment bacterial populations are correlated with treatment efficacy of ivermectin based therapy for both T. trichiura and hookworms 55. This variability in drug efficacy could be associated with xenobiotic degradation properties that different bacterial encoded enzymes may exert on the metabolism of anthelminthic drugs. A major focus of the potential of helminth-microbiota interactions to modulate human disease is evident in the literature. Such work, currently in its infancy, needs to be expanded to human settings before its utility can be judged. Yet the possibility that helminth-altered microbiomes could represent a ‘treasure trove’ for the identification of novel probiotics or microbe-based therapeutic modalities is clear and further work is warranted. However, the complexity of helminth-microbiota interactions on various inflammatory and metabolic diseases indicates that there can be both protective, as well as injurious properties of helminth infection. We suggest that helminth-induced microbiomes can worsen rather than improve important health outcomes in some settings and believe it would be important to test this hypothesis for those conditions associated with naturally occurring helminth infection, such as impaired physical and cognitive development in children.
The biological networks described here are also critical determinants of pathogenesis for infection with other important pathogens. Little is known about helminth-microbiota effects on infection with SARS-CoV-2, HIV, TB, malaria and other important pathogens. Helminths and the microbiota should be considered important environmental contributors towards the inter-individual heterogeneity of immune responses against other pathogens, as well as inflammatory diseases prevalent in industrialized societies. Combined with genetic heterogeneity of the mammalian host, an important direction for the future is to better understand how the response of individuals are shaped by genetic-environmental interactions (Figure 2), particularly with regard to infectious challenges and inflammatory diseases. High dimensional data generated through technological advances in microbial profiling, genomics and immunology combined with computational innovations will play a vital part in this future work.
Acknowledgements
P. L. is supported by the Division of Intramural Research, NIAID, NIH, USA and N.L.H. is supported by a National Health and Medical Research Council (NHMRC) of Australia SRF-B fellowship.
Footnotes
Declaration of interest: N.L. Harris is a member of the Cell Host and Microbe advisory board
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cortes A, Peachey L, Scotti R, Jenkins TP, and Cantacessi C (2019). Helminth-microbiota cross-talk - A journey through the vertebrate digestive system. Mol Biochem Parasitol 233, 111222. 10.1016/j.molbiopara.2019.111222. [DOI] [PubMed] [Google Scholar]
- 2.Leung JM, Graham AL, and Knowles SCL (2018). Parasite-Microbiota Interactions With the Vertebrate Gut: Synthesis Through an Ecological Lens. Front Microbiol 9, 843. 10.3389/fmicb.2018.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapin A, and Harris NL (2018). Helminth-Bacterial Interactions: Cause and Consequence. Trends Immunol 39, 724–733. 10.1016/j.it.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Gause WC, and Maizels RM (2016). Macrobiota - helminths as active participants and partners of the microbiota in host intestinal homeostasis. Curr Opin Microbiol 32, 14–18. 10.1016/j.mib.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venzon M, Das R, Luciano DJ, Burnett J, Park HS, Devlin JC, Kool ET, Belasco JG, Hubbard EJA, and Cadwell K (2022). Microbial byproducts determine reproductive fitness of free-living and parasitic nematodes. Cell Host Microbe 30, 786–797 e788. 10.1016/j.chom.2022.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White EC, Houlden A, Bancroft AJ, Hayes KS, Goldrick M, Grencis RK, and Roberts IS (2018). Manipulation of host and parasite microbiotas: Survival strategies during chronic nematode infection. Sci Adv 4, eaap7399. 10.1126/sciadv.aap7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gacesa R, Kurilshikov A, Vich Vila A, Sinha T, Klaassen MAY, Bolte LA, Andreu-Sanchez S, Chen L, Collij V, Hu S, et al. (2022). Environmental factors shaping the gut microbiome in a Dutch population. Nature 604, 732–739. 10.1038/s41586-022-04567-7. [DOI] [PubMed] [Google Scholar]
- 8.Tee MZ, Er YX, Easton AV, Yap NJ, Lee IL, Devlin J, Chen Z, Ng KS, Subramanian P, Angelova A, et al. (2022). Gut Microbiome of Helminth Infected Indigenous Malaysians Is Context Dependent. bioRxiv, 2022.2001.2021.477162. 10.1101/2022.01.21.477162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olm MR, Dahan D, Carter MM, Merrill BD, Yu FB, Jain S, Meng X, Tripathi S, Wastyk H, Neff N, et al. (2022). Robust variation in infant gut microbiome assembly across a spectrum of lifestyles. Science 376, 1220–1223. 10.1126/science.abj2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor M, Pillaye J, and Horsnell WGC (2021). Inherent maternal type 2 immunity: Consequences for maternal and offspring health. Semin Immunol 53, 101527. 10.1016/j.smim.2021.101527. [DOI] [PubMed] [Google Scholar]
- 11.Nyangahu DD, Darby M, Havyarimana E, Brown BP, Horsnell W, and Jaspan HB (2020). Preconception helminth infection alters offspring microbiota and immune subsets in a mouse model. Parasite Immunol 42, e12721. 10.1111/pim.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rapin A, Chuat A, Lebon L, Zaiss MM, Marsland BJ, and Harris NL (2020). Infection with a small intestinal helminth, Heligmosomoides polygyrus bakeri, consistently alters microbial communities throughout the murine small and large intestine. Int J Parasitol 50, 35–46. 10.1016/j.ijpara.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Zhang C, Sifuentes-Dominguez L, Zarek CM, Propheter DC, Kuang Z, Wang Y, Pendse M, Ruhn KA, Hassell B, et al. (2021). Small proline-rich protein 2A is a gut bactericidal protein deployed during helminth infection. Science 374, eabe6723. 10.1126/science.abe6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Propheter DC, Chara AL, Harris TA, Ruhn KA, and Hooper LV (2017). Resistin-like molecule beta is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc Natl Acad Sci U S A 114, 11027–11033. 10.1073/pnas.1711395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, Swain GP, Knight PA, Donaldson DD, Lazar MA, Miller HR, et al. (2004). RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci U S A 101, 13596–13600. 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharpe C, Thornton DJ, and Grencis RK (2018). A sticky end for gastrointestinal helminths; the role of the mucus barrier. Parasite Immunol 40, e12517. 10.1111/pim.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, Honda K, Gause WC, Blaser MJ, Bonneau RA, et al. (2016). Helminth infection promotes colonization resistance via type 2 immunity. Science 352, 608–612. 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyat M, Lebon L, Perdijk O, Wickramasinghe LC, Zaiss MM, Mosconi I, Volpe B, Guenat N, Shah K, Coakley G, et al. (2022). Microbial regulation of intestinal motility provides resistance against helminth infection. Mucosal Immunol. 10.1038/s41385-022-00498-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson MAE, Roberts IS, and Grencis RK (2021). The interplay between Trichuris and the microbiota. Parasitology, 1–8. 10.1017/S0031182021000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung JM, Budischak SA, Chung The H, Hansen C, Bowcutt R, Neill R, Shellman M, Loke P, and Graham AL (2018). Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol 16, e2004108. 10.1371/journal.pbio.2004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duque-Correa MA, Goulding D, Rodgers FH, Gillis JA, Cormie C, Rawlinson KA, Bancroft AJ, Bennett HM, Lotkowska ME, Reid AJ, et al. (2022). Defining the early stages of intestinal colonisation by whipworms. Nat Commun 13, 1725. 10.1038/s41467-022-29334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris NL, and Loke P (2017). Recent Advances in Type-2-Cell-Mediated Immunity: Insights from Helminth Infection. Immunity 47, 1024–1036. 10.1016/j.immuni.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Rosa BA, Supali T, Gankpala L, Djuardi Y, Sartono E, Zhou Y, Fischer K, Martin J, Tyagi R, Bolay FK, et al. (2018). Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 6, 33. 10.1186/s40168-018-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113. 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Li X, Olmedo M, Holdorf AD, Shang Y, Artal-Sanz M, Yilmaz LS, and Walhout AJM (2019). A Delicate Balance between Bacterial Iron and Reactive Oxygen Species Supports Optimal C. elegans Development. Cell Host Microbe 26, 400–411 e403. 10.1016/j.chom.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SC, Tang MS, Easton AV, Devlin JC, Chua LL, Cho I, Moy FM, Khang TF, Lim YAL, and Loke P (2019). Linking the effects of helminth infection, diet and the gut microbiota with human whole-blood signatures. PLoS Pathog 15, e1008066. 10.1371/journal.ppat.1008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maizels RM (2020). Regulation of immunity and allergy by helminth parasites. Allergy 75, 524–534. 10.1111/all.13944. [DOI] [PubMed] [Google Scholar]
- 28.van der Zande HJP, Zawistowska-Deniziak A, and Guigas B (2019). Immune Regulation of Metabolic Homeostasis by Helminths and Their Molecules. Trends Parasitol 35, 795–808. 10.1016/j.pt.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Zaiss MM, Rapin A, Lebon L, Dubey LK, Mosconi I, Sarter K, Piersigilli A, Menin L, Walker AW, Rougemont J, et al. (2015). The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity 43, 998–1010. 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shute A, Callejas BE, Li S, Wang A, Jayme TS, Ohland C, Lewis IA, Layden BT, Buret AG, and McKay DM (2021). Cooperation between host immunity and the gut bacteria is essential for helminth-evoked suppression of colitis. Microbiome 9, 186. 10.1186/s40168-021-01146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floudas A, Aviello G, Schwartz C, Jeffery IB, O’Toole PW, and Fallon PG (2019). Schistosoma mansoni Worm Infection Regulates the Intestinal Microbiota and Susceptibility to Colitis. Infect Immun 87. 10.1128/IAI.00275-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doonan J, Tarafdar A, Pineda MA, Lumb FE, Crowe J, Khan AM, Hoskisson PA, Harnett MM, and Harnett W (2019). The parasitic worm product ES-62 normalises the gut microbiota bone marrow axis in inflammatory arthritis. Nat Commun 10, 1554. 10.1038/s41467-019-09361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su CW, Chen CY, Jiao L, Long SR, Mao T, Ji Q, O’Donnell S, Stanton C, Zheng S, Walker WA, et al. (2020). Helminth-Induced and Th2-Dependent Alterations of the Gut Microbiota Attenuate Obesity Caused by High-Fat Diet. Cell Mol Gastroenterol Hepatol 10, 763–778. 10.1016/j.jcmgh.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimokawa C, Obi S, Shibata M, Olia A, Imai T, Suzue K, and Hisaeda H (2019). Suppression of Obesity by an Intestinal Helminth through Interactions with Intestinal Microbiota. Infect Immun 87. 10.1128/IAI.00042-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su C, Su L, Li Y, Long SR, Chang J, Zhang W, Walker WA, Xavier RJ, Cherayil BJ, and Shi HN (2018). Helminth-induced alterations of the gut microbiota exacerbate bacterial colitis. Mucosal Immunol 11, 144–157. 10.1038/mi.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds LA, Redpath SA, Yurist-Doutsch S, Gill N, Brown EM, van der Heijden J, Brosschot TP, Han J, Marshall NC, Woodward SE, et al. (2017). Enteric Helminths Promote Salmonella Coinfection by Altering the Intestinal Metabolome. J Infect Dis 215, 1245–1254. 10.1093/infdis/jix141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes A, Wills J, Su X, Hewitt RE, Robertson J, Scotti R, Price DRG, Bartley Y, McNeilly TN, Krause L, et al. (2020). Infection with the sheep gastrointestinal nematode Teladorsagia circumcincta increases luminal pathobionts. Microbiome 8, 60. 10.1186/s40168-020-00818-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFarlane AJ, McSorley HJ, Davidson DJ, Fitch PM, Errington C, Mackenzie KJ, Gollwitzer ES, Johnston CJC, MacDonald AS, Edwards MR, et al. (2017). Enteric helminth-induced type I interferon signaling protects against pulmonary virus infection through interaction with the microbiota. J Allergy Clin Immunol 140, 1068–1078 e1066. 10.1016/j.jaci.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, Tomov VT, Kobuley D, Tran SV, Bittinger K, et al. (2014). Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 345, 578–582. 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scotti R, Southern S, Boinett C, Jenkins TP, Cortes A, and Cantacessi C (2020). MICHELINdb: a web-based tool for mining of helminth-microbiota interaction datasets, and a meta-analysis of current research. Microbiome 8, 10. 10.1186/s40168-019-0782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubel MA, Abbas A, Taylor LJ, Connell A, Tanes C, Bittinger K, Ndze VN, Fonsah JY, Ngwang E, Essiane A, et al. (2020). Lifestyle and the presence of helminths is associated with gut microbiome composition in Cameroonians. Genome Biol 21, 122. 10.1186/s13059-020-02020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drurey C, Lindholm HT, Coakley G, Poveda MC, Loser S, Doolan R, Gerbe F, Jay P, Harris N, Oudhoff MJ, and Maizels RM (2022). Intestinal epithelial tuft cell induction is negated by a murine helminth and its secreted products. J Exp Med 219. 10.1084/jem.20211140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karo-Atar D, Ouladan S, Javkar T, Joumier L, Matheson MK, Merritt S, Westfall S, Rochette A, Gentile ME, Fontes G, et al. (2022). Helminth-induced reprogramming of the stem cell compartment inhibits type 2 immunity. J Exp Med 219. 10.1084/jem.20212311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puschhof J, Pleguezuelos-Manzano C, and Clevers H (2021). Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 29, 867–878. 10.1016/j.chom.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Iftekhar A, Berger H, Bouznad N, Heuberger J, Boccellato F, Dobrindt U, Hermeking H, Sigal M, and Meyer TF (2021). Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat Commun 12, 1003. 10.1038/s41467-021-21162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Y, Oh J, Xue M, Huh WJ, Wang J, Gonzalez-Hernandez JA, Rice TA, Martin AL, Song D, Crawford JM, et al. (2022). Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 378, eabm3233. 10.1126/science.abm3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayes KS, Cliffe LJ, Bancroft AJ, Forman SP, Thompson S, Booth C, and Grencis RK (2017). Chronic Trichuris muris infection causes neoplastic change in the intestine and exacerbates tumour formation in APC min/+ mice. PLoS Negl Trop Dis 11, e0005708. 10.1371/journal.pntd.0005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, et al. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393. 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, et al. (2018). Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 174, 1586–1598 e1512. 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azimi S, Lewin GR, and Whiteley M (2022). The biogeography of infection revisited. Nat Rev Microbiol 20, 579–592. 10.1038/s41579-022-00683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H, Grodner B, and De Vlaminck I (2021). Recent advances in tools to map the microbiome. Curr Opin Biomed Eng 19. 10.1016/j.cobme.2021.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. (2017). Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511. 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 53.Bellais S, Nehlich M, Ania M, Duquenoy A, Mazier W, van den Engh G, Baijer J, Treichel NS, Clavel T, Belotserkovsky I, and Thomas V (2022). Species-targeted sorting and cultivation of commensal bacteria from the gut microbiome using flow cytometry under anaerobic conditions. Microbiome 10, 24. 10.1186/s40168-021-01206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Else KJ, Keiser J, Holland CV, Grencis RK, Sattelle DB, Fujiwara RT, Bueno LL, Asaolu SO, Sowemimo OA, and Cooper PJ (2020). Whipworm and roundworm infections. Nat Rev Dis Primers 6, 44. 10.1038/s41572-020-0171-3. [DOI] [PubMed] [Google Scholar]
- 55.Schneeberger PHH, Gueuning M, Welsche S, Hurlimann E, Dommann J, Haberli C, Frey JE, Sayasone S, and Keiser J (2022). Different gut microbial communities correlate with efficacy of albendazole-ivermectin against soil-transmitted helminthiases. Nat Commun 13, 1063. 10.1038/s41467-022-28658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


