Abstract
Neuroendocrine (NE) cells comprise ~1% of epithelial cells in benign prostate and prostatic adenocarcinoma (PCa). However, they become enriched in hormonally-treated and castration-resistant PCa (CRPC). In addition, close to 20% of hormonally-treated tumors recur as small cell neuroendocrine carcinoma (SCNC), composed entirely of NE cells, which may be the result of clonal expansion or lineage plasticity. Since NE cells do not express androgen receptor (AR), they are resistant to hormonal therapy and contribute to therapy failure. Here, we describe the identification of glypican-3 (GPC3) as an oncofetal cell surface protein specific to NE cells in prostate cancer. Functional studies revealed that GPC3 is critical to the viability of NE tumor cells and tumors displaying NE differentiation and that it regulates calcium homeostasis and signaling. Since our results demonstrate that GPC3 is specifically expressed by NE cells, patients with confirmed SCNC may qualify for GPC3-targeted therapy which has been developed in the context of liver cancer and displays minimal toxicity due to its tumor-specific expression.
Keywords: Prostate Cancer, Genitourinary Oncology, Neuroendocrine, Precision Oncology, Biomarkers, Immunotherapy, Cellular Heterogeneity
Introduction
Prostate Cancer (PCa) is the most common non-cutaneous malignancy in men and the second most common cause of cancer-related mortality[1]. Although most men diagnosed with with primary prostate cancer have adenocarcinomas, treatable by radical prostatectomy or radiation therapy [2,3], a subset of men develop biochemical recurrence followed by metastasis requiring systemic therapy to suppress tumor growth[4]. Since the majority of tumor cells of adenocarcinoma glands are luminal-type cells and androgen receptor (AR)-dependent[5], a commonly used treatment is hormone therapy to suppress androgen production or inhibit AR activity [6]. Although this treatment is initially successful, all hormonally-treated tumors eventually recur as castration-resistant PCa (CRPC) which responds temporarily to newer drugs such as enzalutamide and abiraterone that further inhibit AR signaling, but therapy failure is inevitable, eventually leading to death [6].
The vast majority of cells that comprise adenocarcinomas are luminal-type cells, expressing AR and prostate-specific antigen (PSA), although ~1% of the cells are neuroendocrine (NE) cells, which do not express AR and are resistant to hormone therapy [7–9]. Following hormone therapy, NE cells are observed to become enriched in the tumor cell population which may be due to lineage plasticity or clonal expansion [10]. Furthermore, after AR-targeted therapy, ~17–30% of patients develop small cell neuroendocrine carcinoma (SCNC), comprised entirely of NE cells [11]. Effective, durable therapy should target NE cells as well as luminal tumor cells which is an urgent unmet clinical need. As a result, it is important to identify molecular targets on NE cells.
Our laboratory has previously shown that CXCR2 is expressed on NE cells of the prostate, and that this is a suitable therapeutic target for PCa displaying NE differentiation [12,13]. Although targeting CXCR2 can result in tumor regression, there is significant risk for neutropenia as this protein is also highly expressed by circulating neutrophils[14]. We therefore wanted to identify other proteins specific to NE cells that are both critical to disease progression and whose targeting carries less side effects.
We have successfully identified glypican-3 (GPC3) as a cell-surface protein specifically expressed by NE cells of the prostate. GPC3 is a heparan sulfate proteoglycan (HSPG) that is only expressed during embryogenesis but not in normal adult tissue [15]. Interestingly, it is aberrantly expressed by some tumors including hepatocellular carcinoma (HCC) and yolk sac tumor of the testis and ovary [16]. Targeting GPC3 has been studied in the context of liver cancer [17–21], where clinical trials show minimal toxicity [22]. Herein we have thoroughly characterized the expression of GPC3 in PCa using multiple molecular-based methods and demonstrate its critical function to tumor viability. Mechanistically, GPC3 regulates intracellular calcium levels which is critical to NE cell function. The results of this study set the premise for targeting GPC3 as a novel therapeutic strategy to treat advanced, therapy-resistant PCa.
Materials and methods
Study design
The objective of this study was to demonstrate another useful biomarker and therapeutic target for NE cells of the prostate. Liquid chromatography-mass spectrometry (LC/MS) was used to study the metabolic signature of the previously described LNCaP/CXCR2 cell line. Potential targets and findings were then validated in vitro and in human tissue data using multiple molecular methods. Previously published and publicly deposited mRNA-sequencing and single-cell sequencing data [23, 4] were used to determine GPC3 expression status in human NE cells. Human tissue slides representing hormone-sensitive adenocarcinoma and CRPC-adenocarcinoma were obtained from Duke University Department of Pathology. SCNC cases were obtained as a gift from Dr. Liang Cheng, MD of the Department of Pathology and Laboratory Medicine, Indiana University. All human tissues were de-identified and the use of anatomic materials was approved by Duke University’s Institutional Review Board.
Various functional studies were carried out in vitro and in vivo to demonstrate GPC3 function and mechanism of action in NE cells of the prostate. Experimental mice were maintained on a regular diet in a pathogen-free setting on a 12-h light/dark cycle with continuous access to food and water. The number of animals used for the xenograft study was determined by a power calculation and all animal work was conducted in accordance with the NIH Guidelines of Care and Use of Laboratory Animals [25] and approved by the Duke Institutional Animal Care and Use Committee (Protocol #: A055–22-03)
Cell culture
LNCaP, C4–2, 22RV1, DU-145, and PC-3 cells were cultured in RPMI 1640 medium (ThermoFisher (Waltham, MA, USA)) supplemented with 10% fetal bovine serum (Corning (Glendale, AZ, USA), and 1% penicillin-streptomycin stock solution (ThermoFisher). LAPC4 cells were maintained in IMDM medium (ThermoFisher), also supplemented with fetal bovine serum and penicillin-streptomycin. NCI-H660 cells were maintained in Advanced DMEM/F-12 (ThermoFisher) supplemented with 1X B-27 supplement (ThermoFisher), 10 ng/ml FGF2 (Peprotech (Cranbury, NJ, USA)), 10 ng/ml EGF (Peprotech), 1X Glutamax (ThermoFisher) and 1X penicillin-streptomycin (ThermoFisher). LNCaP-CXCR2 cells were generated as previously described[13] and maintained in RPMI 1640 medium, supplemented with 400 μg/ml Geneticin (ThermoFisher), 10% fetal bovine serum, and 1% penicillin-streptomycin stock solution.
Metabolite extraction and liquid chromatography-mass spectrometry (LC-MS)
Metabolite extraction was carried out according to a protocol established by the Locasale Laboratory at Duke University. In brief, 1×106 cells/well were plated in a standard 6-well plate (3 wells per group) with 2 ml culture medium per well. After 24 h, plates were placed on dry ice, medium was aspirated and 80% HPLC grade methanol/water (extraction solvent) was added to each well. Plates were then transferred to –80 °C for 15 min for enzyme deactivation. Subsequently, cells were scraped into the extraction solvent and centrifuged for 10 min. Supernatants were collected and vacuum concentrator was used at room temperature for 4 h. Dry pellets were obtained and sent for LC-MS analysis.
Single cell sequencing and data analysis
Six patient samples were collected at Duke University Hospital representing the spectrum of PCa (hormone-sensitive adenocarcinoma (4), localized CRPC (1), metastatic CRPC (1), and SCNC (1)), as previously published[24, 26]. Single cells were encapsulated into single droplets using Chromium Controller (10X Genomics (Pleasanton, CA, USA)) following the manufacturer’s protocol. Libraries were sequenced on a NovaSeq 6000 S2 as 150-bp paired-end reads by NantOmics LLC, Culver City CA, USA; or on a NovaSeq 6000 S1 as 100-bp paired-end reads by the GCB Sequencing and Genomic Technologies Core at Duke University. The CellRanger (10X Genomics) analysis pipeline was used for sample demultiplexing, barcode processing, and single cell 3’-gene counting to generate a digital gene-cell matrix. The gene expression matrix was then processed and analyzed using Seurat under the R software environment [27]. Mitochondrial and ribosomal genes were removed from the gene expression matrix. Violin plots were used to visualize clusters of cells that passed quality control. Clusters were associated with cell types based on canonical marker genes as described previously [24].
Immunohistochemistry and immunofluorescence
All tissues were stained using a BioCare Medical (Concord, CA, USA) Intellipath autostainer. Sections were deparaffinized, rehydrated, and boiled for 40 mins in citrate buffer pH 6 (Azer Scientific, Cat # ES35535) followed by cooling for 20 min. An antibody to GPC3 (Diagnostic Biosystems (Pleasanton, CA, USA), Cat # Mob561) was incubated at a dilution of 1:2000 for 60 min at room temperature (RT). Detection was accomplished with the DAKO (Glostrup, Denmark) Universal Polymer System for 30 min followed by a hematoxylin counterstain. For immunofluorescence, antibodies to GPC3 (Diagnostic Systems, Cat # Mob561) and CHGA (Novus Biologicals (Centennial, CO, USA), Cat # NB120–15160) were incubated at RT for 60 min at dilutions of 1:2000 and 1:20,000, respectively. Conjugated secondary antibodies (AlexaFluor 488, Thermo Fisher, Cat #: A11001 and AlexaFluor594, ThermoFisher, Cat #: A11037) were incubated at RT for 30 min at dilutions of 1:500. Fluorescent antibody-labeled slides were counterstained with DAPI, and images were obtained in a single plane on a Zeiss upright 180 confocal microscope.
Flow cytometry
1×106 cells from each cell line (LNCaP, LNCaP/CXCR2, LAPC4, C4–2, 22RV1, PC-3, DU-145, and NCI-H660) were harvested, washed twice with PBS, re-suspended in staining buffer (PBS + 0.2% FBS) and blocked (FcR Blocking Reagent, Miltenyi Biotec (Bergisch, Germany), Cat #: 130–059-901). Cells were then incubated with an antibody to GPC3 (Diagnostic Biosystems, Cat # Mob561) at a concentration of 1 μg/ml for 30 min at 4 °C. Following incubation, cells were washed twice with PBS, re-suspended in staining buffer, and incubated with PE-conjugated secondary antibody (BD Biosciences (Franklin Lakes, NJ, USA), Cat # 550083) at a concentration of 1 μg/ml for 30 min at 4 °C. Following two washes with PBS, cells were re-suspended in staining buffer and 30,000 events were recorded for each tube using a BD LSRII Cell Analyzer. All data was further processed using FlowJo software.
RT-qPCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen (Germantown, MD, USA)) following the manufacturer’s instructions. In brief, cells were lysed in Trizol reagent followed by chloroform extraction and ethanol precipitation. Reverse-transcription and qPCR was performed using a qScript One-Step SYBR Green RT-qPCR kit (QuantaBio (Beverly, MA, USA)) following the manufacturer’s instructions. All primer sequences are listed in supplementary materials, Table S1. Data was collected and analyzed using a ViiA 7 Real-Time PCR instrument (Applied Biosystems (Waltham, MA, USA)).
Western blotting
Total protein was extracted from cells using RIPA buffer (Sigma Aldrich (St. Louis, MO, USA)) supplemented with phosphatase and protease inhibitor cocktail (ThermoFisher). Lysates were cleared by centrifugation and protein concentration was determined using a BCA Assay Kit (ThermoFisher, Cat #: 23227). Equal amounts of total protein were resolved using 12% SDS-polyacrylamide gels, transferred to PVDF membranes, and blocked with 5% non-fat milk powder. Primary antibodies against GPC3 (Diagnostic Biosystems, Cat # Mob-561, 1:200), SYP (Santa Cruz Biotechnology (Dallas, TX, USA), Cat # sc-17750, 1:100), p-CAMKII (Cell Signaling Technology (Danvers, MA, USA), Cat # 12716, 1:500), CAMKII (Cell Signaling Technology, Cat # 3362, 1:500), HSP90 (BD Biosciences, Cat #: 610418, 1:2000), and β-actin (Cell Signaling Technology, Cat # 4970, 1:1000) were used. HRP-conjugated secondary antibody (BioRad (Hercules, CA, USA), Cat #172–1011, Cat #170–6515, 1:3000) were then used and the antigen-antibody reaction was visualized by enhanced chemiluminescence assay (Thermo Fisher, Cat #: 34096).
Gene silencing and overexpression
GPC3 knockdown was achieved by stable transduction with an shRNA lentivirus. Lentiviral plasmids expressing shRNAs were purchased from Sigma Aldrich (shGPC3 1: TRCN0000078561, shGPC3 2: TRCN0000315782, shCAMK2D: TRCN0000000475, shCTRL: SHC001) and infected into cells. After 48 h, cells were grown with 1 μg/ml puromycin until only resistant cells remained (0.3 μg/ml puromycin selection used for NCI-H660 cells). GPC3 overexpression was also achieved by stable transduction with lentivirus and a corresponding empty vector was used as negative control. In brief, a GPC3 human tagged ORF clone was purchased from Origene (Rockville, MD, USA) (Cat #: RG205911). A stop codon was introduced by site-directed mutagenesis and GPC3 was cloned into a Gateway entry vector using restriction enzymes. Using clonase recombination, the GPC3 ORF was inserted into a CMV-driven lentiviral expression plasmid containing a puromycin-resistant gene. All lentiviral transduction vectors were generated by transfection of 293T cells with helper plasmids using the Mirus (Madison, WI) TransIT System (Cat #: MIR6000), following the manufacturer’s protocol, with collection of medium at 48 h (for GPC3 overexpression, medium was collected at 24, 48, and 72 h followed by ultracentrifugation). Knockdown or overexpression was confirmed using RT-qPCR and western blotting.
Cell proliferation/Viability assays
For LNCaP/CXCR2, cells were seeded in 0.2 ml medium in a 96-well plate (3,000 cells/well, 4 wells/group). After 24 h, cell confluency was measured daily for 7 days using an Incucyte Live Cell Imaging System. For BrdU incorporation, cells were seeded in a 6-well plate (1×105 cells/well, 3 wells/group) and incubated for 72 h. Following incubation, cells were pulsed with BrdU and further incubated for 1 h. Cells were then permeabilized and stained using a FITC-BrdU kit (BD Biosciences, Cat #: 559619), following the manufacturer’s protocol. Data was collected using a BD LSRII Cell Analyzer. All data was further processed using FlowJo software. For NCI-H660, cells were trypsinized to dissociate cells for 1 min and then plated in 0.2 ml medium in a 96-well, round-bottom plate (2,000 cells/well, 6 wells/group). The plate was then immediately centrifuged for 5 min (300 x g, RT) to allow single cells to aggregate at the center of each well. The area (μm2) of the aggregates was monitored and calculated with daily measurements over a period of 13 days using an Incucyte Live Cell System (Sartorius (Göttingen, Germany)). For the Calcein AM viability assay, cells were plated in a 6-well plate (1×106/well, 3 wells/group) and incubated for 72 h. Following incubation, cells were harvested and stained with Calcein AM dye according to manufacturer’s protocol (ThermoFisher, Cat #: C1430). Data was collected using a BD LSRII Cell Analyzer and all data was further processed using FlowJo software.
Xenograft study
Ten immunocompromised male NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice were obtained from the Division of Animal Laboratory Resources at Duke University. 6 ×104 of NCI-H660/shCTRL or NCI-H660/shGPC3 cells were suspended in media containing 50% Matrigel (Corning) and inoculated subcutaneously bilaterally into the flanks of six-week old NSG mice, as described previously[13]. Once tumors were palpable, they were measured twice a week using a caliper and the volume was calculated over time (volume (mm3) = (length x height2)/2). At 54 days post-implantation, mice were euthanized using CO2 and xenografts were harvested and weighed.
mRNA sequencing and analysis
Total RNA from LNCaP/CXCR2/shCTRL and LNCaP/CXCR2/shGPC3 was extracted as described above and library preparation/subsequent sequencing was performed by Novogene (Sacramento, CA, USA) In brief, sequencing libraries were generated using an NEBNext Ultra RNA Library Prep Kit for Illumina (NEB (Ipswich, MA, USA)) following the manufacturer’s protocol, and index codes were added to attribute sequences to each sample. The clustering of index-coded samples was performed on a CBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina (San Diego, CA, USA)), following the manufacturer’s protocol. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated. Raw data (reads) in FASTQ format were processed through fastp and clean reads were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. After mapping to reference genome, differential expression analysis was performed using DESeq2 R package. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling false discovery rate (FDR). Genes with an adjusted P value < 0.05 found by DESeq2 were assigned as differentially expressed. Enrichment analysis with differentially expressed genes was performed using Enrichr [28].
Intracellular calcium measurements
NCI-H660/shCTRL, NCI-H660/shGPC3, LNCaP/CXCR2/shCTRL, LNCaP/CXCR2/shGPC3, LNCaP, and LNCaP/GPC3 cells for [Ca2+]i measurements were plated in a 6-well plate (1 × 106 cells/well, 3 wells/group) and incubated for 48 h. Following incubation, cells were loaded with a Cal-520 AM dye and further incubated for 30 min, following the manufacturer’s protocol (Abcam (Cambridge, UK), Cat #: ab233472). Cells were then harvested, and data was collected using a BD LSR11 Cell Analyzer. All data was further processed using FlowJo software.
IP3 competitive ELISA
Equal number of cells were harvested from NCI-H660/shCTRL, NCI-H660/shGPC3, LNCaP/CXCR2/shCTRL, LNCaP/CXCR2/shGPC3, LNCaP, and LNCaP/GPC3 cultures and protein lysates prepared as described above. Protein yield for each group of cells was quantified using BCA Assay Kit (ThermoFisher, Cat #: 23227) and quantitative competitive ELISA was then performed using an IP3 ELISA Kit (MyBioSource (San Diego, CA, USA), Cat #: MBS2515875) following the manufacturer’s protocol. Data was collected on a micro-plate reader set to 450 nm and final IP3 concentration was normalized to total protein.
Quantitation and statistical analyses
T tests were used to test differences between means of continuous variables for two groups. All p values <0.05 were considered statistically significant (p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****). For IHC analysis of human tissue, whole-slide tissue cases were quantified by H-score according to the following formula: 3 x percentage of strongly staining cells, 2 x percentage of moderately staining cells, and 1 x percentage of weakly staining cells. Neurite length quantification in LNCaP/GPC3 model was performed using the single neurite tracer (SNT) function of ImageJ. All statistical analyses were conducted using GraphPad.
Results
PCa cell lines with NE phenotype express glypican-3
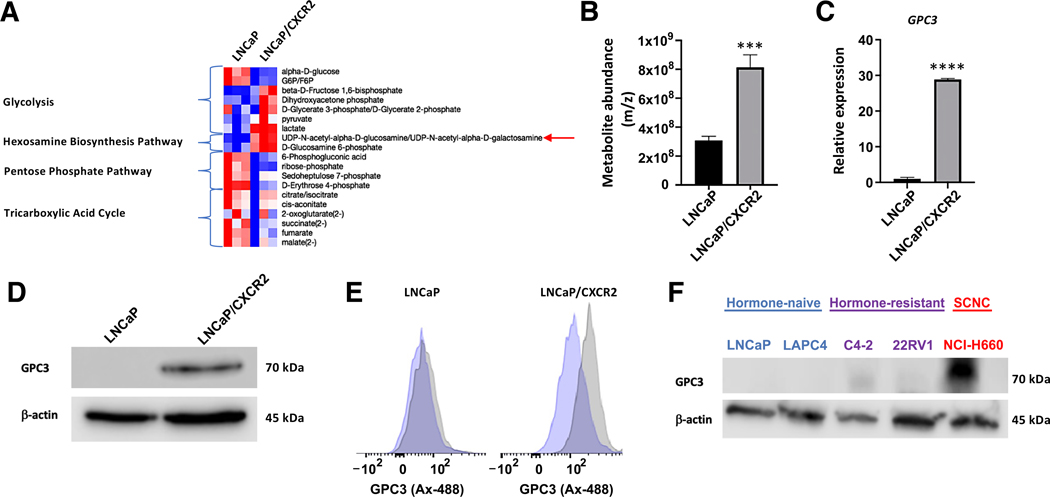
We have previously published that CXCR2 expression in LNCaP cells induces an NE phenotype, including resistance to ADT and up-regulation of NE markers[13]. To understand the molecular mechanisms underlying this lineage switch, we conducted a metabolite profiling study comparing LNCaP (luminal phenotype) to LNCaP/CXCR2 (NE phenotype) to study metabolic changes that occur intracellularly. Our results showed alterations in the levels of many metabolites comprising a number of different pathways including glycolysis, pentose phosphate pathway (PPP), and the tricarboxylic acid (TCA) cycle (Figure 1A). Although the levels of most metabolites were comparatively decreased, the levels of metabolites involved in the hexosamine biosynthesis pathway (HBP) were significantly up-regulated (p < 0.001) (Figure 1A,B).
Figure 1.
Cell lines with NE phenotype display up-regulated GPC3. (A) Heat map showing metabolite changes between LNCaP and LNCaP/CXCR2. (B) LC/MS results showing metabolite abundance of UDP-N-acetylglucosamine between LNCaP and LNCaP/CXCR2. (C) RT-qPCR results showing GPC3 relative expression between LNCaP and LNCaP/CXCR2. (D) Western blot analysis shows upregulation of GPC3 in LNCaP/CXCR2 relative to LNCaP (E) Flow cytometric analysis showing GPC3 surface expression between LNCaP and LNCaP/CXCR2. (F) Western blot analysis of GPC3 expression across PCa cell lines of different phenotype. Statistics: t-test with p < 0.05 considered significant. p < 0.0001 (***), p < 0.00001 (****).
The HBP represents a minor pathway where ~2–5% of consumed glucose is converted to UDP-N-acetylglucosamine rather than continuing on through glycolysis to be converted to pyruvate[29]. These nucleotide sugars are then commonly utilized to donate N-acetylglucosamine to processes such as O-GlcNAcylation and N-linked glycosylation, as well as in the synthesis of glycosaminoglycans (GAGs), such as heparan sulfates. To determine whether O-GlcNAcylation was up-regulated, we studied the mRNA levels of the rate-limiting enzyme, O-GlcNAc transferase (OGT), and our results showed no significant difference between LNCaP and LNCaP/CXCR2 cells (supplementary material, Figure S1A). We also observed no significant difference in the expression of oligosaccharyltransferase (STT3A, STT3B), the enzyme central for co-translationally attaching N-glycans to a growing peptide. (supplementary material, Figure S1A). We next determined whether proteins that utilize acetylglucosamine (heparan sulfate proteoglycans) in their GAG structures were up-regulated. We performed an RT-qPCR screen for all HSPG genes and observed that GPC3 was significantly up-regulated (relative fold-change >20, p < 0.001), providing explanation for increased HBP activity (Figure 1C,D, supplementary material, Figure S1B). As glypican-3 (GPC3) is a cell-surface oncofetal antigen which is not expressed in normal adult tissue[16], we confirmed this finding using flow cytometry to demonstrate its expression on the cell surface, and demonstrated that LNCaP/CXCR2 cells were positive for GPC3 while LNCaP cells were largely negative (Figure 1D,E).
We next characterized GPC3 expression in PCa cell lines representing different disease stages (LNCaP, LAPC4, C4–2, 22RV1, and NCI-H660). By western blot analysis, we found an absence of protein expression in cell lines representing hormone-sensitive adenocarcinoma (LNCaP and LAPC4) and weakly detectable protein expression in CRPC-adenocarcinoma (CRPC-adeno) cell lines (C4–2 and 22RV1). NCI-H660, the PCa cell line which models SCNC, displayed high expression of GPC3 (Figure 1F). Flow cytometric analysis of these cell lines (supplementary material, Figure S1C) showed that hormone-naïve adenocarcinoma cell lines contained very small proportions of GPC3+ cells (LNCaP = 0.63%, LAPC4 = 0.21%), while the proportions were higher in CRPC cell lines (C4–2 = 4.54%, 22RV1 = 8.61%). SCNC cell line, NCI-H660, uniformly expressed GPC3 (100%). It is well known that LNCaP cells develop NE phenotype in vitro following 7–14 days ADT or enzalutamide treatment [30]. To determine whether cells up-regulate GPC3 during this process, we treated LNCaP cells with 10 μM enzalutamide or vehicle (DMSO) for 1 and 2 weeks and looked for cell-surface GPC3 expression by flow cytometry. We found that LNCaP cells gradually become positive for GPC3 (vehicle treatment = 0.9%, enzalutamide treatment, 1 week = 17.6%, enzalutamide treatment, 2 weeks = 34.5%), further supporting that GPC3 is associated with NE phenotype (supplementary material, Figure S1D). Overall, the expression pattern of GPC3 was similar to that of CXCR2 and represents an oncofetal protein specifically expressed by NE cells of the prostate.
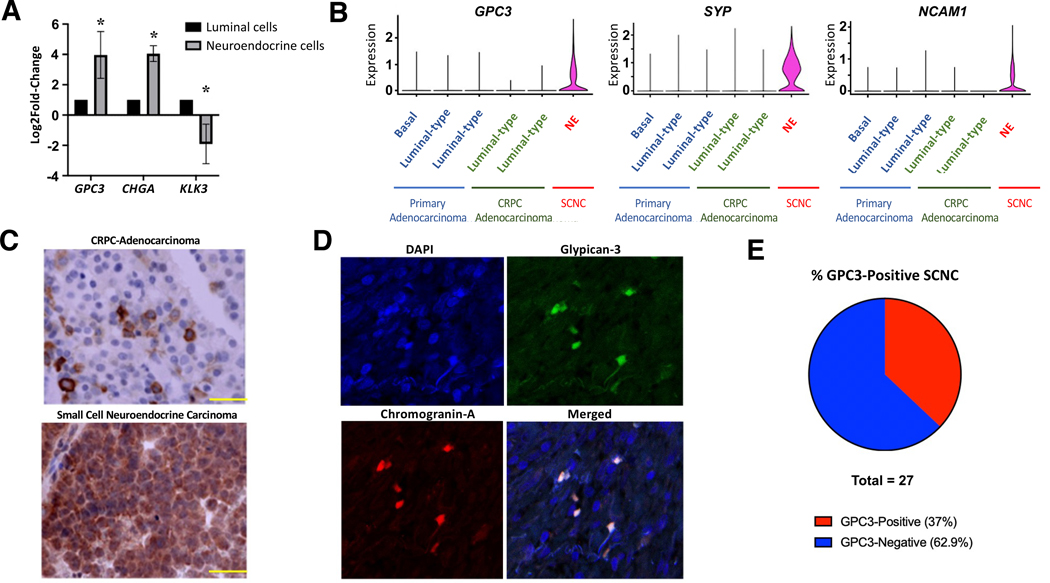
Glypican-3 is expressed specifically by NE cells in human PCa tissue
The above results demonstrate that cell lines displaying NE signature (LNCaP/CXCR2 and NCI-H660) uniformly express GPC3. To determine whether NE cells of human PCa also express GPC3, we interrogated our previously published mRNA-sequencing dataset for FACS-sorted human luminal and NE cells derived from fresh, human prostatic adenocarcinoma tissue which contain a minor component of NE cells[13]. Our data showed that GPC3 expression in purified NE cells is much higher (log2FC >3, p < 0.05) than in luminal-type tumor cells, with a similar expression pattern to chromogranin A (CHGA), a commonly used NE marker (Figure 2A). To further corroborate our findings, we performed a single-cell sequencing study [24] where we collected 6 PCa cases including primary hormone-naïve adenocarcinoma, locally recurrent CRPC with adenocarcinoma morphology (CRPC-adeno), metastatic CRPC with adenocarcinoma morphology (mCRPC), and treatment-emergent SCNC with a total of 24,385 single epithelial cells[24]. We found that GPC3 expression was restricted to NE tumor cells (SYP-Positive, NCAM1-Positive), whereas it was undetectable in the luminal clusters or basal cluster (defined in [24]) (Figure 2B). To further corroborate our findings, we interrogated a recently published single-cell sequencing dataset deposited by Dong, et al[31] which includes 3 patients with a diagnosis of SCNC. Here, 1 of 3 patients displayed significant GPC3 expression, suggesting a subset of SCNC tumors express GPC3 (supplementary material, Figure S2). Bulk RNA-sequencing datasets[32, 33] have not yet demonstrated significant GPC3 expression (supplementary material, Figure S3A,B) which we reason is due to mixed benign and adenocarcinoma components because PCa tissue is never pure due to its unique infiltrative growth. Interestingly, in lung cancer, bulk RNA sequencing[34] demonstrates significant upregulation of GPC3 (supplementary material, Figure S3C) in SCNC relative to non-small cell. This potentially indicates GPC3 may be a good target for NE tumors outside of the prostate as well and is deserving of further study.
Figure 2.
GPC3 is expressed on human NE cells of the prostate. (A) mRNA-seq results of FACS-sorted luminal and NE cells from prostatic adenocarcinoma (n = 3) demonstrating GPC3, CHGA, and KLK3 expression. (B) sc-RNA seq results from human PCa cases (n = 6) showing GPC3, SYP, and NCAM1 expression in different cell clusters throughout the spectrum of PCa. (C) Representative IHC image showing GPC3 expression in CRPC-adeno and SCNC (scale bar, 50 μm). (D) IF images showing GPC3 and CgA co-expression in CRPC-adeno tissue. (E) Circle chart demonstrating percentage of SCNC cases that were positive for GPC3 by IHC. Statistics: t-test with p < 0.05 considered significant. p < 0.05 (*).
We studied paraffin tissue of CRPC-adeno histology and SCNC, respectively, using immunohistochemistry (IHC). This demonstrated scattered GPC3+ tumor cells among the more abundant GPC3– cells in CRPC (characteristic of NE and luminal tumor cells, respectively) while SCNC was highly positive (Figure 2C). Subsequent immunofluorescence (IF) staining of CRPC-adeno tissue demonstrated perfectly concordant expression of GPC3 with the NE marker CHGA, further proving that GPC3 is expressed by NE cells of the prostate (Figure 2D).
SCNC, comprised entirely of NE tumor cells, represents the most lethal histologic variant of PCa, with no effective therapy. As GPC3 is only expressed during embryonic development and an ideal therapy target, we wished to determine whether a significant subset of human SCNC cases are positive and could therefore qualify for GPC3-targeted therapy to treat their tumors. We stained 27 whole-slide tissue cases of treatment-emergent SCNC and demonstrated that 10/27 (37%) of the SCNC cases were positive for GPC3 while 17/27 (62.9%) were negative (Figure 2E, supplementary material, Figure S4A,B)). In contrast, whole slide hormone-naïve or castration resistant adenocarcinomas contained rare, scattered GPC3 positive cells (supplementary material, Figure S4A), consistent with the presence of a minor component of NE cells in such tumors. Finally, we explored two patient-derived xenograft (PDX) tumor lines that represent SCNC (LTL-545 and LTL-352) where it was observed that one of the two lines had significant GPC3-positivity while the other was entirely negative (supplementary material, Figure S4C).
At this time, it is unknown exactly why some NE cells express GPC3 while others do not. Using our sc-RNA seq data, we further classified our NE cells into 8 clusters and found that GPC3 expression was not unique to any specific NE cluster in our dataset (supplementary material, Figure S5). However, GPC3 expression was observed to be extremely low in one of the clusters (C7). Pathway analysis demonstrates that the GPC3-high clusters are particularly enriched for pathways involved in DNA repair, as well oncogenic pathways (mTOR, IL-6/JAK/STAT3, E2F, MYC) and N-glycosylation relative to the GPC3-low cluster. However, future studies would be needed to confirm these findings as well as determine the exact role of GPC3 in these processes.
Taken together, our data indicate that a significant subset of patients with prostatic SCNC express this oncofetal protein and could potentially qualify for GPC3-targeted therapy, which has already been developed for liver cancer[22]. However, this finding could allow for the first time the development of a low toxicity, molecularly-targeted approach for patients with SCNC.
Glypican-3 is critical to the viability of NE cells and SCNC
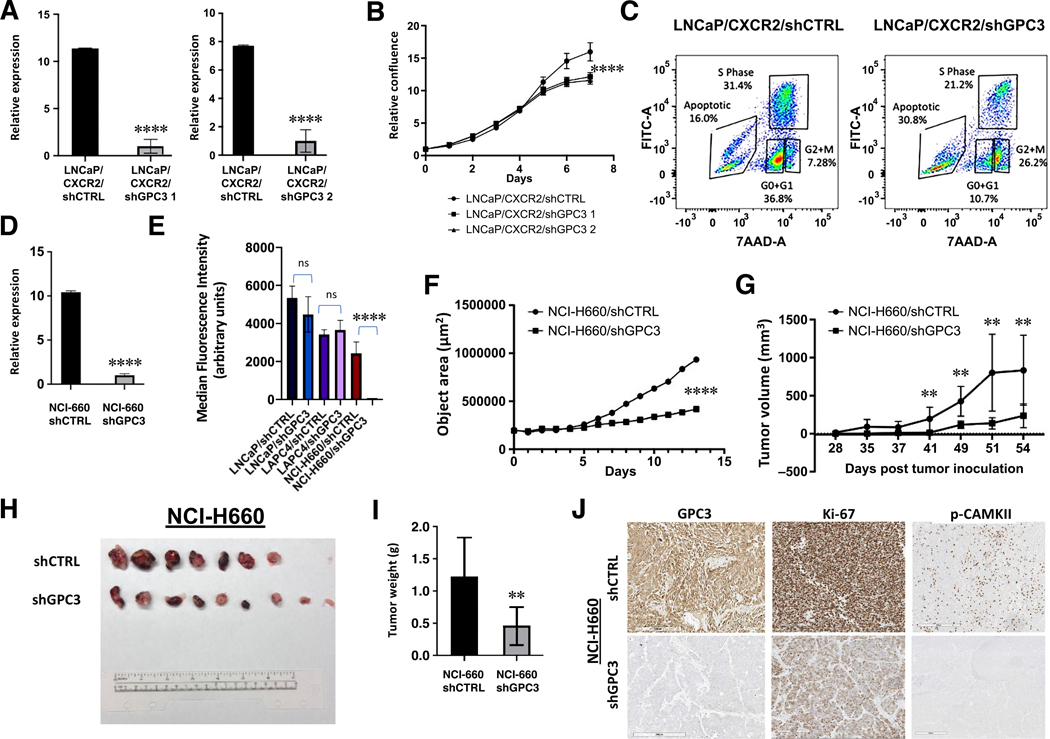
GPC3 is a cell surface HSPG which can sequester growth factors and ions to regulate a variety of signaling processes[35]. Due to this function, we hypothesized that GPC3 could have significant effects on NE cell viability. To examine this, we stably knocked down GPC3 expression in LNCaP/CXCR2 cells using two lentiviruses expressing a short hairpin RNA (shGPC3 1 and shGPC3 2. shGPC3 1 was chosen for remainder of studies) (Figure 3A). This cell line, engineered by our lab, displays NE phenotype[13] and expresses GPC3 (Figure 1C–E). Although no changes in lineage marker expression (CHGA, SYP, NSE, AR, PSA) were observed, we observed a significant (p < 0.01) decrease in cell proliferation as determined by daily measurements of confluency (Figure 3B). We performed a BrdU incorporation study, which showed a reduction of cells in the S-phase (LNCaP/CXCR2/shCTRL = 31.4%, LNCaP/CXCR2/shGPC3 = 21.2%) and a higher degree of apoptosis (LNCaP/CXCR2/shCTRL = 16.0%. LNCaP/CXCR2/shGPC3 = 30.8%) as determined through 7-AAD staining for total DNA (Figure 3C). Since LNCaP cells are well established to develop NE phenotype after 14 days of AR signaling inhibition[30] and demonstrate upregulated GPC3 (supplementary material, Figure S1D) we further knocked down GPC3 in long-term (>1 month) androgen deprived cells and observed a significant growth suppressive effect (supplementary material, Figure S6). These results suggest that GPC3 may have a role in maintaining NE cell viability.
Figure 3.
GPC3 is critical to the viability of NE cells and SCNC. (A) RT-qPCR demonstrating GPC3 knockdown in LNCaP/CXCR2. (B) Growth curve comparing LNCaP/CXCR2/shCTRL versus LNCaP/CXCR2 /shGPC3. (C) BrdU Incorporation of LNCaP/CXCR2/shCTRL versus LNCaP/CXCR2/shGPC3. (D) RT-qPCR demonstrating GPC3 knockdown in NCI-H660 cells. (E) Quantification of MFI values from Calcein AM viability assay comparing LNCaP/shCTRL versus LNCaP/shGPC3, LAPC4/shCTRL versus LAPC4/shGPC3, and NCI-H660/shCTRL versus NCI-H660/shGPC3. (F) 3D aggregate growth curve comparing NCI-H660/shCTRL versus NCI-H660/shGPC3. (G) Tumor growth rate of NCI-H660/shCTRL and NCI-H660/shGPC3 xenografts. (H) Images of resected xenograft tissue from mice inoculated with NCI-H660/shCTRL versus NCI-H660/shGPC3 cells. (I) Final tumor weights of resected NCI-H660 xenograft tissue. (J) Representative image demonstrating GPC3, Ki-67, and p-CAMKII expression in resected xenograft tissue (scale bar, 200 μm). Statistics: t-test with p < 0.05 considered significant. p < 0.01 (**), p < 0.00001 (****).
As SCNC represents the most lethal form of PCa, we explored whether knockdown of GPC3 in NCI-H660 cells (Figure 3D) can result in decreased viability. Here, we performed a Calcein AM flow cytometric assay, where the fluorescence of the dye taken up correlates positively with cell viability, and our results demonstrate a significant (p < 0.001) decrease in cell viability in NCI-H660/shGPC3 relative to control (Figure 3E). Since these are suspension cells which grow in aggregates, we further performed a 3D aggregate growth assay by plating an equal number of cells of either NCI-H660/shCTRL or NCI-H660/shGPC3 (2,000 cells/well, 4 replicates per group) and inducing aggregate formation through centrifugation. Aggregate growth over time was significantly (p < 0.001) suppressed in NCI-H660/shGPC3 relative to cells expressing non-targeting control, consistent with our results from the Calcein AM viability assay (Figure 3F).
A major question with any gene associated with NE phenotype is whether or not it can induce lineage plasticity (i.e. cause luminal cells to transdifferentiate to NE cells). As LNCaP is the classic cell line for modeling NE transdifferentiation[30], we stably overexpressed GPC3 in LNCaP cells (supplementary material, Figure S7A) and tested for NE marker expression as well as studied morphology. Although there was no significant change in the expression of luminal or NE-defining markers, we observed a significant morphology change, consistent with neurite outgrowth (supplementary material, Figure S7B,C), which has been reported previously in other studies[36–39]. Since PC-3 cells represent a cell line with NE characteristics but is not true SCNC-derived, we overexpressed GPC3 to observe whether its expression is able to enhance NE marker expression. Interestingly, in this cell line, we observed significant upregulation of both SYP and ENO2 (NSE), two classic NE markers (supplementary material, Figure S7D). Overall, our results indicate that GPC3 may have a role in promoting NE transdifferentiation; however, its specific effect likely depends on the genetic background of the cells and the expression of collaborating factors which have yet to be identified.
To explore whether GPC3 knockdown can inhibit SCNC tumor growth in vivo, we performed a xenograft study where we subcutaneously injected both flanks of 10 mice with NCI-H660 cells (60,000 cells/injection) expressing either non-targeting shCTRL or shGPC3. Our results demonstrated a significant (p < 0.01) decrease in tumor growth rate over the experimental period (54 days) (Figure 3G) and resected tissue showed a significant (p < 0.01) decrease in average tumor weight (shCTRL = 1.22 g, shGPC3 = 0.460 g) and Ki-67 positivity (Figure 3H–J). Collectively, these results indicate that GPC3 is vital to NE tumor cell viability and may be an important contributor to SCNC.
Glypican-3 is associated with Ca2+-regulated kinases
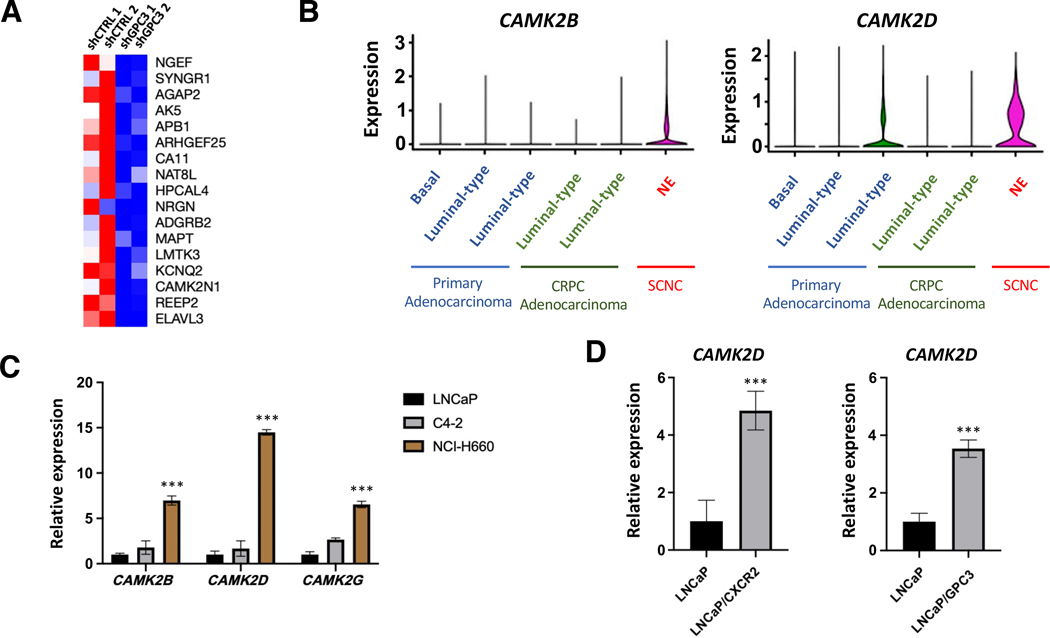
Our results showed that GPC3 is expressed specifically by NE cells and is critical to cell viability. To explore mechanisms for how GPC3 contributes to NE cell function, we took an unbiased approach and performed mRNA-sequencing on LNCaP/CXCR2/shCTRL and LNCaP/CXCR2/shGPC3 cells. Analysis of the list of all statistically significant (p < 0.05) differentially expressed genes, revealed that several genes associated with calcium regulated/dependent kinases were downregulated in the LNCaP/CXCR2/shGPC3 group relative to control (Figure 4A). We then analyzed sc-RNA seq data to determine if any calcium-regulated/dependent kinases were associated with SCNC. Here, we found that two CAMKII isozymes, encoded by CAMK2B and CAMK2D, were associated with the NE cell cluster. CAMK2B demonstrates NE-cell specific expression and CAMK2D has its highest expression in NE cells (Figure 4B). In analyzing our NE clusters, we observed CAMK2B and CAMK2D correlate with GPC3-high status of NE cells. Negligible expression was found in the GPC3-low NE cluster (C7) (supplementary material, Figure S8). Furthermore, in the dataset deposited by Dong, et al [31] both CAMK2B and CAMK2D were highest in the GPC3-positive SCNC case (P2) with very low/negligible expression in the GPC3-negative cases (P5 and P6) (supplementary material, Figure S8). Considered together, this data indicates that GPC3 correlates with CAMK2 in human PCa.
Figure 4.
GPC3 is associated with CAMKII. (A) Heat map showing genes commonly overexpressed with Ca2+-regulated kinases (ARCHS4 Kinases Coexpression Database) are significantly (p < 0.05) down-regulated in LNCaP/CXCR2/shGPC3 relative to LNCaP/CXCR2/shCTRL. (B) sc-RNA seq results from human PCa cases (n = 6) showing CAMK2B and CAMK2D expression in different cell clusters. (C) RT-qPCR showing CAMKII isozyme expression across PCa cell lines of different phenotype. (D) RT-qPCR showing CAMK2D expression in LNCaP/CXCR2 and LNCaP/GPC3 relative to LNCaP. Statistics: t-test with p < 0.05 considered significant. p < 0.0001 (***).
CAMKII is a calcium-dependent kinase which autophosphorylates to become constitutively active upon binding to a Ca2+/Calmodulin complex[40]. There are four isozymes: CAMK2A (alpha), CAMK2B (beta), CAMK2D (delta), and CAMK2G (gamma). The alpha isozyme is neuron-specific while the beta, delta, and gamma forms are expressed and active in various peripheral tissue types and disease settings[41]. To explore whether our findings in human tissue can be observed in PCa cell lines, we studied the expression of the various CAMKII isozymes across three major PCa cell lines representing hormone-sensitive adenocarcinoma (LNCaP), CRPC-adeno (C4–2), and SCNC (NCI-H660) by RT-qPCR. Although the alpha isozyme was undetectable across all cell lines tested, the beta, delta, and gamma isozymes all showed highest expression in NCI-H660 with lower expression in the adenocarcinoma lines (LNCaP and C4–2), an expression pattern similar to that of GPC3 (Figure 4D). We further tested LNCaP/CXCR2 and LNCaP/GPC3 and found that the delta isozyme in particular (CAMK2D) was up-regulated in LNCaP/CXCR2 and LNCaP/GPC3, relative to LNCaP (Figure 4E). However, all isozymes were high in NCI-H660 (Figure 4D), indicating that CAMKII may be an important kinase in the progression of SCNC.
Glypican-3 regulates [Ca2+]i to affect CAMKII activation in SCNC
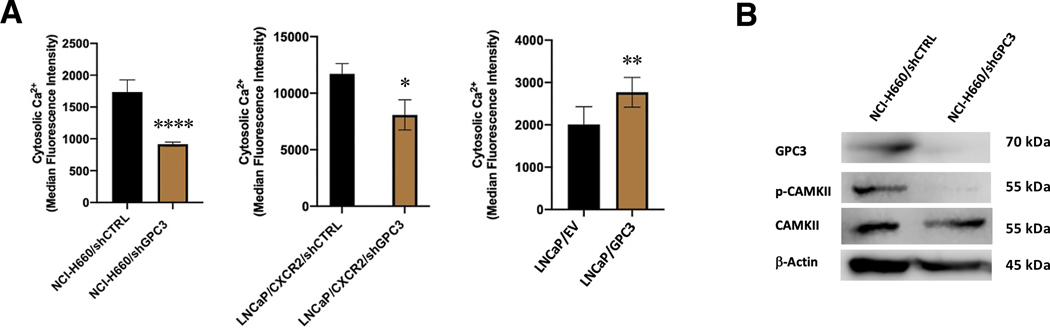
Structurally, GPC3 has negatively charged (anionic) heparan sulfate chains which may be involved in the recruitment of positively charged cations, such as Ca2+, to the plasma membrane. We sought therefore to determine whether GPC3 regulates cytosolic calcium which may then affect substrate availability to activate Ca2+-regulated/dependent kinases, such as CAMKII. To do this, we performed a flow cytometric assay where we loaded cells with a Cal-520 AM dye, which becomes highly fluorescent when complexed to cytosolic calcium. Quantification of the resulting fluorescence (median fluorescence intensity, MFI) revealed that knockdown of GPC3 significantly decreased cytosolic Ca2+ in both NCI-H660 and LNCaP/CXCR2 cells but overexpression increased Ca2+ in GPC3-negative LNCaP cells (Figure 5A). To determine if GPC3 is recruiting Ca2+ to the plasma membrane or mediating Ca2+ release from intracellular stores, we performed a competitive ELISA experiment to measure whether IP3, the product of PLC hydrolysis, was affected by GPC3 modulation. PLC signaling is the major mechanism for release of Ca2+ from intracellular stores, driven by IP3 binding to IP3-receptor (IP3R) on the ER surface[42, 43]. Our results show no significant difference in IP3 levels in all of our models (supplementary material, Figure S9), suggesting that GPC3 mediates [Ca2+]i through increased recruitment to the plasma membrane rather than increased release from the ER. Importantly, calcium has a vital role in maintaining neuronal morphology/function[44] and this may be a critical mechanism to maintaining the viability of NE cells.
Figure 5.
GPC3 regulates [Ca2+]i to control CAMKII activation in SCNC. (A) Quantification of MFI values from Cal-520 AM assays demonstrating [Ca2+]i in NCI-H660/shCTRL versus NCI-H660/shGPC3, LNCaP/CXCR2/shCTRL versus LNCaP/CXCR2/shGPC3 and LNCaP/EV versus LNCaP/GPC3. (B) Western blot analysis showing CAMKII activation status in NCI-H660/shCTRL versus NCI-H660/shGPC3. Statistics: t-test with p < 0.05 considered significant. p < 0.05 (*), p < 0.01 (**), p < 0.0001 (***).
As NCI-H660 represents SCNC and has the highest levels of GPC3 and CAMKII, we performed western blot analysis to determine if autophosphorylation of CAMKII is decreased with GPC3 knockdown. Our results demonstrate a significant decrease in phospho-CAMKII relative to cells expressing non-targeting control (Figure 5B, Figure 3J), indicating that GPC3 affects the activity of CAMKII. To confirm that CAMKII is functionally important in SCNC, we knocked down the expression of CAMK2D, the isozyme with strongest correlation with GPC3 in cell lines, and observed a marked decrease in cell viability (supplementary material, Figure S10A,B). To determine whether NCI-H660 is vulnerable to CAMKII inhibition, we treated cells with CAMKII inhibitor (KN-93 phosphate) or control (KN-92 phosphate) and performed a 3D aggregate growth assay to observe growth over time. Here, we observed marked suppression of tumor cell growth (supplementary material, Figure S10C), providing further evidence that CAMKII is important for SCNC growth and viability. Collectively, our results show that GPC3 controls CAMKII activation in SCNC, likely due to its ability to regulate cytosolic Ca2+, and represents an important signaling axis for SCNC.
Discussion
NE tumor cells are present throughout the spectrum of PCa but have not been well studied due to their rarity in most clinical settings. Current therapeutic strategies focus on targeting AR, which is not expressed by NE cells, resulting in enrichment of NE cells in the tumor population and the development of SCNC in a significant number of patients[9]. As such, there is an urgent, unmet need to identify targets for NE cells so that they can be targeted early in the disease process before significant expansion occurs leading to therapy resistance and disease progression.
Our laboratory has developed a robust system to isolate NE cells from fresh human PCa tissue[13], allowing us to successfully identify GPC3 as a novel biomarker for prostatic NE cells and SCNC. Furthermore, we have demonstrated that GPC3 is critical for NE cell viability and SCNC growth and have identified an important signaling axis for SCNC (supplementary material, Figure S9B), which has not been previously reported. Although GPC3 overexpression in LNCaP cells was unable to induce lineage plasticity alone, a significant morphology change consistent with neurite outgrowth was observed. Interestingly, in PC-3 cells, overexpression of GPC3 induced significant upregulation of SYP and ENO2 (NSE), two classic NE markers. Future studies would be needed to determine which specific factors in combination with GPC3 can induce lineage plasticity as well as the mechanism of action.
Since GPC3 is not expressed in normal adult tissue[15], its targeting is safe which has been demonstrated in several ongoing clinical trials of HCC [22]. Useful modalities include therapeutic antibodies, peptide vaccines, and CAR-T cells[22]. Due to the success of many of these earlier trials, GPC3-targeting drugs continue to be improved. For example, a bispecific antibody targeting GPC3 and CD47 demonstrates a long serum half-life and was highly effective at preventing HCC growth in xenograft models[45, 46]. An immunotoxin shown to be highly effective at targeting GPC3 in HCC (HN3-T20) has been modified to have an albumin-binding domain (HN3-ABD-T20), resulting in a 45-fold extended serum retention time and effective tumor regression at one-tenth the dose of the original form[46, 47]. GPC3 CAR-T cells co-expressing IL-15 and IL-21[46, 48] or IL-17 and PH-20[46, 49] were highly efficacious against HCC growth with minimal toxicity in vivo. Although several of these newer agents are still in the preclinical stage, it expected that these developments will continue and that several of these will be translated to clinic. Since SCNC is the most lethal form of PCa without any effective therapy, our findings make GPC3-targeting using these many agents a promising future direction. Our mechanistic studies show that intracellular calcium is regulated by GPC3 and that SCNC has high levels of CAMKII expression and activation. Although there have been early reports attempting to elucidate the role of CAMKII in PCa[50–52], it has not been previously linked to NE cells or SCNC as all of these studies have been carried out in models representing luminal phenotype. As calcium and CAMKII activity are critically important for maintaining neuronal phenotype and morphology, the results of our study highlight the close resemblance of NE cells to neurons in regards to intracellular signaling. In addition to targeting GPC3, these results show that SCNC is also vulnerable to CAMKII inhibition, which may offer another novel therapeutic direction.
Supplementary Material
Supplementary materials and methods
Figure S1. Cell lines with NE phenotype display upregulated GPC3
Figure S2. sc-RNA sequencing demonstrates a subset of SCNC patients express GPC3
Figure S3. Bulk RNA-Sequencing results for GPC3 in PCa datasets
Figure S4. IHC demonstrates that a subset of SCNC cases express GPC3
Figure S5. GPC3 expression in NE cell and pathway analysis of GPC3-high and -low clusters
Figure S6. Knockdown of GPC3 in androgen-resistant LNCaP cells
Figure S7. Potential role of GPC3 in lineage plasticity
Figure S8. GPC3-positive NE cells express high levels of CAMK2
Figure S9. GPC3 functions independently of IP3 in regulation of [Ca2+]i
Figure S10. CAMKII is critical to NE cell viability
Table S1. Primer sequences used for RT-qPCR
Table S2. shRNA sequences used for gene knockdown
Acknowledgements
We thank the Biorepository and Precision Pathology Core of Duke University for tissue processing, Jeff Groth for IHC technical support, Duke Cancer Institute Flow Cytometry Shared Resource for FACS, Duke Human Vaccine Institute for flow cytometric support, Duke Center for Genomic and Computational Biology for single-cell sequencing, and Novogene USA for mRNA-sequencing and analysis. The work was supported by 2018 Movember Foundation- PCF Valor Challenge Award (PI: Steven Patierno, PhD).
JH is a consultant for or owns shares in the following companies: Amgen, Artera, Kingmed Diagnostics, Teddy Clinical Research Laboratories, MoreHealth, OptraScan, Genetron, Omnitura, Vetonco, York Biotechnology, Genecode, VIVA Biotech, and Sisu Pharma, and received grants from Zenith Epigenetics, BioXcel Therapeutics, Inc., Dracen Pharmaceuticals, and Fortis Therapeutics. AJA is a consultant or advisor for the following companies: Astellas, Epic Sciences, Pfizer, Bayer, Janssen, Dendreon, BMS, AstraZeneca, Merck, Forma, Celgene, Clovis, Exact Sciences, Myovant, Exelixis, GoodRx, and Novartis. AJA has received research support/grants from NIH/NCI, PCF/Movember, DOD, Astellas, Pfizer, Bayer, Janssen, Dendreon, BMS, AstraZeneca, Merck, Forma, Celgene, Amgen, and Novartis.
Footnotes
Conflicts of interests
No other conflicts of interest were declared.
Data availability statement
The sc-RNA seq data has been deposited publicly in NCBI’s Genome Sequencing and Assembly and is accessible through accession number (PRJNA699369). All other data associated with this study is in the manuscript or supplementary materials.
References
- 1.Miller KD, et al. , Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin, 2019. 69(5): p. 363–385. [DOI] [PubMed] [Google Scholar]
- 2.Barry MJ and Nelson JB, Patients Present with More Advanced Prostate Cancer since the USPSTF Screening Recommendations. J Urol, 2015. 194(6): p. 1534–6. [DOI] [PubMed] [Google Scholar]
- 3.Bekelman JE, Rumble RB, and Freedland SJ, Clinically Localized Prostate Cancer: ASCO Clinical Practice Guideline Endorsement of an AUA/ASTRO/SUO Guideline Summary. J Oncol Pract, 2018. 14(10): p. 618–624. [DOI] [PubMed] [Google Scholar]
- 4.Hussain A. and Dawson N, Management of advanced/metastatic prostate cancer: 2000 update. Oncology (Williston Park), 2000. 14(12): p. 1677–88; discussion 1688, 1691–4. [PubMed] [Google Scholar]
- 5.Culig Z. and Santer FR, Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev, 2014. 33(2–3): p. 413–27. [DOI] [PubMed] [Google Scholar]
- 6.Teo MY, Rathkopf DE, and Kantoff P, Treatment of Advanced Prostate Cancer. Annu Rev Med, 2019. 70: p. 479–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, et al. , Function and molecular mechanisms of neuroendocrine cells in prostate cancer. Anal Quant Cytol Histol, 2007. 29(3): p. 128–38. [PubMed] [Google Scholar]
- 8.Huang J, et al. , Immunohistochemical characterization of neuroendocrine cells in prostate cancer. Prostate, 2006. 66(13): p. 1399–406. [DOI] [PubMed] [Google Scholar]
- 9.Butler W. and Huang J, Neuroendocrine cells of the prostate: Histology, biological functions, and molecular mechanisms. Precis Clin Med, 2021. 4(1): p. 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kollermann J. and Helpap B, Neuroendocrine differentiation and short-term neoadjuvant hormonal treatment of prostatic carcinoma with special regard to tumor regression. Eur Urol, 2001. 40(3): p. 313–7. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal R, et al. , Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol, 2018. 36(24): p. 2492–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, et al. , Differential expression of interleukin-8 and its receptors in the neuroendocrine and non-neuroendocrine compartments of prostate cancer. Am J Pathol, 2005. 166(6): p. 1807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. , Targeting cellular heterogeneity with CXCR2 blockade for the treatment of therapy-resistant prostate cancer. Sci Transl Med, 2019. 11(521). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ocana A, et al. , Neutrophils in cancer: prognostic role and therapeutic strategies. Mol Cancer, 2017. 16(1): p. 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida T. and Kataoka H, Glypican 3-Targeted Therapy in Hepatocellular Carcinoma. Cancers (Basel), 2019. 11(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang SK, et al. , Discovery and diagnostic value of a novel oncofetal protein: glypican 3. Adv Anat Pathol, 2014. 21(6): p. 450–60. [DOI] [PubMed] [Google Scholar]
- 17.Ho M, Advances in liver cancer antibody therapies: a focus on glypican-3 and mesothelin. BioDrugs, 2011. 25(5): p. 275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng M, et al. , Therapeutically targeting glypican-3 via a conformation-specific single-domain antibody in hepatocellular carcinoma. Proc Natl Acad Sci U S A, 2013. 110(12): p. E1083–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng M. and Ho M, Glypican-3 antibodies: a new therapeutic target for liver cancer. FEBS Lett, 2014. 588(2): p. 377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng M, et al. , Recombinant soluble glypican 3 protein inhibits the growth of hepatocellular carcinoma in vitro. Int J Cancer, 2011. 128(9): p. 2246–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, et al. , Glypican-3-Specific Antibody Drug Conjugates Targeting Hepatocellular Carcinoma. Hepatology, 2019. 70(2): p. 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu Y, et al. , Next-Generation Cancer Immunotherapy Targeting Glypican-3. Front Oncol, 2019. 9: p. 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Human Prostate Cancer Single-cell RNA Sequencing.
- 24.Cheng Q, et al. , Pre-existing Castration-resistant Prostate Cancer-like Cells in Primary Prostate Cancer Promote Resistance to Hormonal Therapy. Eur Urol, 2022. 81(5): p. 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.in Guide for the Care and Use of Laboratory Animals. 2011: Washington (DC). [Google Scholar]
- 26.Goldstein AS, et al. , Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc, 2011. 6(5): p. 656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler A, et al. , Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol, 2018. 36(5): p. 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen EY, et al. , Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 2013. 14: p. 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akella NM, Ciraku L, and Reginato MJ, Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol, 2019. 17(1): p. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burchardt T, et al. , Transdifferentiation of prostate cancer cells to a neuroendocrine cell phenotype in vitro and in vivo. J Urol, 1999. 162(5): p. 1800–5. [PubMed] [Google Scholar]
- 31.Dong B, et al. , Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun Biol, 2020. 3(1): p. 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen WS, et al. , Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. Eur Urol, 2019. 76(5): p. 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beltran H, et al. , Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med, 2016. 22(3): p. 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghandi M, et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature, 2019. 569(7757): p. 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolluri A. and Ho M, The Role of Glypican-3 in Regulating Wnt, YAP, and Hedgehog in Liver Cancer. Front Oncol, 2019. 9: p. 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver EM, et al. , Posttranscriptional regulation of T-type Ca(2+) channel expression by interleukin-6 in prostate cancer cells. Cytokine, 2015. 76(2): p. 309–320. [DOI] [PubMed] [Google Scholar]
- 37.Palmer J, et al. , Constitutive activation of gp130 leads to neuroendocrine differentiation in vitro and in vivo. Prostate, 2005. 62(3): p. 282–9. [DOI] [PubMed] [Google Scholar]
- 38.Juarranz MG, et al. , Neuroendocrine differentiation of the LNCaP prostate cancer cell line maintains the expression and function of VIP and PACAP receptors. Cell Signal, 2001. 13(12): p. 887–94. [DOI] [PubMed] [Google Scholar]
- 39.Albrecht M, et al. , Proliferation of prostate cancer cells and activity of neutral endopeptidase is regulated by bombesin and IL-1beta with IL-1beta acting as a modulator of cellular differentiation. Prostate, 2004. 58(1): p. 82–94. [DOI] [PubMed] [Google Scholar]
- 40.Chang JY, et al. , CaMKII Autophosphorylation Is Necessary for Optimal Integration of Ca(2+) Signals during LTP Induction, but Not Maintenance. Neuron, 2017. 94(4): p. 800–808 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tombes RM, Faison MO, and Turbeville JM, Organization and evolution of multifunctional Ca(2+)/CaM-dependent protein kinase genes. Gene, 2003. 322: p. 17–31. [DOI] [PubMed] [Google Scholar]
- 42.Gill DL, Ghosh TK, and Mullaney JM, Calcium signalling mechanisms in endoplasmic reticulum activated by inositol 1,4,5-trisphosphate and GTP. Cell Calcium, 1989. 10(5): p. 363–74. [DOI] [PubMed] [Google Scholar]
- 43.Kania E, et al. , IP3 Receptor-Mediated Calcium Signaling and Its Role in Autophagy in Cancer. Front Oncol, 2017. 7: p. 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brini M, et al. , Neuronal calcium signaling: function and dysfunction. Cell Mol Life Sci, 2014. 71(15): p. 2787–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du K, et al. , A bispecific antibody targeting GPC3 and CD47 induced enhanced antitumor efficacy against dual antigen-expressing HCC. Mol Ther, 2021. 29(4): p. 1572–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X, et al. , Glypican-3: A Novel and Promising Target for the Treatment of Hepatocellular Carcinoma. Front Oncol, 2022. 12: p. 824208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleming BD, et al. , Engineered Anti-GPC3 Immunotoxin, HN3-ABD-T20, Produces Regression in Mouse Liver Cancer Xenografts Through Prolonged Serum Retention. Hepatology, 2020. 71(5): p. 1696–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batra SA, et al. , Glypican-3-Specific CAR T Cells Coexpressing IL15 and IL21 Have Superior Expansion and Antitumor Activity against Hepatocellular Carcinoma. Cancer Immunol Res, 2020. 8(3): p. 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong X, et al. , Co-expression of IL-7 and PH20 promote anti-GPC3 CAR-T tumour suppressor activity in vivo and in vitro. Liver Int, 2021. 41(5): p. 1033–1043. [DOI] [PubMed] [Google Scholar]
- 50.Wang T, et al. , The tumor suppressive role of CAMK2N1 in castration-resistant prostate cancer. Oncotarget, 2014. 5(11): p. 3611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rokhlin OW, et al. , Calcium/calmodulin-dependent kinase II plays an important role in prostate cancer cell survival. Cancer Biol Ther, 2007. 6(5): p. 732–42. [DOI] [PubMed] [Google Scholar]
- 52.Wang T, et al. , CAMK2N1 inhibits prostate cancer progression through androgen receptor-dependent signaling. Oncotarget, 2014. 5(21): p. 10293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Figure S1. Cell lines with NE phenotype display upregulated GPC3
Figure S2. sc-RNA sequencing demonstrates a subset of SCNC patients express GPC3
Figure S3. Bulk RNA-Sequencing results for GPC3 in PCa datasets
Figure S4. IHC demonstrates that a subset of SCNC cases express GPC3
Figure S5. GPC3 expression in NE cell and pathway analysis of GPC3-high and -low clusters
Figure S6. Knockdown of GPC3 in androgen-resistant LNCaP cells
Figure S7. Potential role of GPC3 in lineage plasticity
Figure S8. GPC3-positive NE cells express high levels of CAMK2
Figure S9. GPC3 functions independently of IP3 in regulation of [Ca2+]i
Figure S10. CAMKII is critical to NE cell viability
Table S1. Primer sequences used for RT-qPCR
Table S2. shRNA sequences used for gene knockdown
Data Availability Statement
The sc-RNA seq data has been deposited publicly in NCBI’s Genome Sequencing and Assembly and is accessible through accession number (PRJNA699369). All other data associated with this study is in the manuscript or supplementary materials.