Abstract
Throughout history, humans have relied on plants as a source of medication, flavoring, and food. Plants synthesize large chemical libraries and release many of these compounds into the rhizosphere and atmosphere where they affect animal and microbe behavior. To survive, nematodes must have evolved the sensory capacity to distinguish plant-made small molecules (SMs) that are harmful and must be avoided from those that are beneficial and should be sought. This ability to classify chemical cues as a function of their value is fundamental to olfaction, and represents a capacity shared by many animals, including humans. Here, we present an efficient platform based on multi-well plates, liquid handling instrumentation, low-cost optical scanners, and bespoke software that can efficiently determine the chemotaxis valence of single SMs in the model nematode, Caenorhabditis elegans. Using this integrated hardware-wetware-software platform, we screened 90 plant SMs and identified 37 that attracted or repelled wild-type animals, but had no effect on mutants defective in chemosensory transduction. Genetic dissection indicates that for at least 10 of these SMs, response valence emerges from the integration of opposing signals, arguing that olfactory valence is often determined by integrating chemosensory signals over multiple lines of information. This study establishes that C. elegans is an effective discovery engine for determining chemotaxis valence and for identifying natural products detected by the chemosensory nervous system.
Introduction
Odors and other chemical cues shape behaviors like feeding, mating and the avoidance of predators and other hazards. Humans and other animals, including invertebrates, perceive attractive odors as pleasant and repellent ones as foul and reliably classify chemical cues according to this single dimension of valence [1–3]. This process starts when odor molecules bind to receptors expressed by specialized chemosensory neurons (CSNs). In mammalian and nematode CSNs, odors and pheromones are typically detected by G-protein coupled receptors (GPCRs) and GPCR activation is transduced into electrical signals via activation of adenylate cyclase and cyclic-nucleotide gated ion channels or phospholipase C and transient receptor potential (TRP) channels. How these molecular and cellular events culminate in similar integrated behaviors (approach or withdrawal) across phyla is an incompletely understood and fundamental problem in neuroscience.
The roundworm Caenorhabitis elegans has provided compelling insights into the genetic, molecular, and neural basis of chemosensation for five decades (reviewed by Refs. [4,5]). The primary strategy worms use to accumulate near attractants is to suppress turns (pirouettes) and to increase forward run duration when moving up a chemical gradient [6]. The converse strategy underpins the avoidance of repellents [7]. Both strategies make it possible to monitor chemotaxis by observing the position of groups of animals following timed exposure to spatial chemical gradients. Chemotaxis behavior depends on signaling by one or more of the worm’s 32 chemosensory neurons [4], organized into 16 classes of paired neurons. Roughly three dozen organic chemicals and salts are thus far known to elicit chemotaxis. Some individual classes of CSNs are associated with promoting attraction or repulsion [e.g. Ref. [8]], mirroring the single dimension of valence. While ample evidence links specific odorants to particular CSNs and the receptors they express, how the broader chemical space of odorants that a worm might encounter could interact with one or more receptors to produce either attraction or repulsion is incompletely understood.
With a genome encoding 1532 GPCRs, including more than 1000 proposed chemosensory receptors [9], C. elegans has substantial capacity for chemical sensing. Each class of chemosensory neurons expresses a distinct ensemble of hundreds of GPCRs [10,11], a level of multiplexing that is also found in mammalian cells [12]. Chemosensory transduction by hundreds of GPCRs expressed in C. elegans CSNs is thought to converge on either TAX-4-dependent cyclic nucleotide-gated ion channels or OSM-9-dependent TRP channels. At least ten pairs of chemosensory neurons express TAX-4 [13], including six that also express OSM-9 [14]. Six pairs of neurons appear to express OSM-9 alone [14]. These expression patterns divide the 16 classes of chemosensory neurons into three groups (TAX-4 only, OSM-9 only, and TAX-4 and OSM-9), all of which depend on conserved chemosensory transduction pathways converging on ion channels.
Like other nematode species [15], C. elegans are found below ground throughout the soil matrix across the globe [16,17] and must therefore navigate complex environments that contain a wealth of plant-derived secondary metabolites and other small molecules (SMs). It is estimated plants make at least 200,000 chemically-distinct SMs and that many of these compounds are released into the soil-root interface where they affect animal and microbial behavior [18]. Thus, plant SMs are an important component of the natural environment of C. elegans and are very likely to be ethologically relevant chemotactic cues. In the laboratory, it is common to monitor C. elegans chemotaxis by observing the position of groups of animals following timed exposure to spatial chemical gradients [19,20]. This artisanal method is not well-suited for screening chemical libraries, however. Inspired by efforts to create semi-automated methods for measuring C. elegans lifespan [21,22] and feeding behaviors [23,24], we developed a chemotaxis platform and integrated analytic workflow compatible with testing chemical libraries for their ability to attract or repel C. elegans. Our approach integrates hardware, wetware, and software and supports performing chemotaxis assays at scale. As this platform is compatible with any chemical library, we used it to screen plant SMs for their ability to evoke C. elegans chemotaxis. This evolution-inspired approach to natural product screening may also deepen understanding of the link between chemical cues and behavior.
By screening a curated library of 90 plant SMs and 6 reference conditions, we found a total of 37 SMs that evoked chemotaxis in wild-type C. elegans, but not anosmic mutants lacking tax-4 and osm-9. Most of these chemoactive compounds (26 of 37) were attractants and only 11 were repellents. A similar enrichment of attractants is also seen in prior studies of C. elegans chemotaxis [25]. Taking advantage of the scale of our approach, we dissected the dependence of these responses on perturbations of tax-4 or osm-9 and discovered that while a handful of odorants were dependent on a single transduction pathway, most were dependent on both. Strikingly, loss of either tax-4 or osm-9 function reversed the response valence of 10 compounds. This finding implies that the response valence exhibited in wild-type animals reflects integration of signaling from multiple CSNs and/or receptors. More broadly, these results suggest that many SMs engage receptors expressed in multiple sensory neuron types and that behavioral valence emerges from integration of signals across multiple CSNs. These data demonstrate the value of our high-throughput behavioral screening approach for characterizing diverse chemical libraries, reveals that plant derived SMs are salient chemical cues for C. elegans, and sets the stage for using phenotypic assays to discover novel actuators of the nervous system and their cognate receptors.
Methods
C. elegans strains
We used four Caenorhabditis elegans strains in this study:
wild-type [N2 (Bristol), [RRID:WB-STRAIN:WBStrain00000001];
PR678 tax-4(pr678) III [RRID:WB-STRAIN:WBStrain00030785];
CX10 osm-9(ky10) IV [RRID:WB-STRAIN:WBStrain00005214];
GN1077 tax-4(pr678) III; osm-9(ky10) IV.
For the purposes of this study, N2 (Bristol) was the wild type, tax-4(pr678) and osm-9(ky10) are null alleles, and were derived in the N2 background. We made GN1077 by crossing GN1065 osm-9(ky10) IV; pat-2(pg125[pat-2::wrmScarlet) III with GN1076 tax-4(p678) III; oxTi915 [eft-3p::GFP::2xNLS] IV and selecting non-fluorescent progeny as candidate tax-4;osm-9 double mutants. The final double mutant was verified by PCR and sequencing using the following primers for osm-9 (Forward -GCAGAAGAGAAACTCCTCAC; Reverse -CCACCTTCATAATCTCCAGC) and tax-4 (Forward -CCAATGGAATTGGCTCTCCTC; Reverse -CATCCCAAGTCAGGATACTG).
C. elegans husbandry
We maintained C. elegans in 10 cm diameter plates (Fisher Scientific, 229695) on nematode growth medium (NGM) seeded with OP50 E. coli and generated age-synchronized cohorts of young adults suitable for behavioral testing, using standard methods [26]. We thawed animals from frozen stocks prior to each round of screening and maintained them on OP50-seeded 10-cm NGM growth plates. The procedure for age-synchronization was as follows: 1) Using sterile, filtered, osmotically-purified water, wash worms from growth plates into 15-mL conical tube; 2) Concentrate worms by centrifugation (1 minute, 4000-RPM, Thermo Scientific Sorvall Legend X1R), discard the supernatant, and distribute the pellet in ~250μL pellets into 15-mL tubes; 3) Resuspend pellets in water (4 mL) and add household bleach (1 mL, Clorox) and 5M KOH (0.5 mL), vortex, and incubate until adult worms disintegrate and eggs are released (5–10 minutes); 4) Concentrate eggs by centrifugation (1 minute, 4000 RPM) and discard the supernatant; 5) Wash in water (10 mL) and concentrate by centrifugation (1 minute, 4000 RPM), four times; 6) Resuspend egg pellets in water (2 mL) and deliver 1200–1800 embryos onto OP50-seeded, 10-cm NGM growth plates. Animals were incubated at 20°C, reached adulthood in ~3 days, and were used for behavioral testing..
Custom chemical library curation
We assembled a custom library of 94 compounds and 2 null reference conditions (DMSO:DMSO and DMSO:H2O). Four compounds known to attract (isoamyl alcohol, diacetyl) or repel (2-nonanone, 1-octanol) wild-type C. elegans [4] were included to provide a direct comparison between the findings emerging from our screening platform and prior studies. The other 90 compounds were small molecules synthesized by plants, soluble in DMSO, and purchased from commercial suppliers (Table S1). They were selected based upon a search of the published literature for SMs that attract or repel animals that consume plants and/or are known to induce physical effects on animals. We expanded the set by searching for SMs that were chemically similar to an initial set of compounds or synthesized in the same biosynthetic pathway as these SMs. The library includes SMs made by plants used in medicine, human foods, or human rituals, such as camphor [27], salvinorinA and its propionate analog [28], and sinomenine hydrochloride [29]. The library also includes three SM pairs that map to the same compound according to the CAS registration number but have different common names and were purchased from different suppliers. For this reason, the SM pairs provide a potential window in reproducibility. The SM pairs are: CAS No. 496-16-2—2,3-dihydrobenzofuran and coumarin; CAS No. 106-22-9—citronellol and β-citronellol; CAS No. 474-58-8—daucosterol and sitogluside.
Chemicals reagents
The chemical library was sourced as indicated in Table S1. Other chemical reagents were obtained from Sigma-Aldrich.
Chemotaxis assays
We conducted end-point assays of populations of synchronized, young adult wild-type and mutant C. elegans. Our implementation involves novel behavioral arenas, methods for linking the chemical library format to assay plates, strategies for dispensing worms onto assay arenas using automated liquid handling equipment, and humidity-controlled environments for running assays (schematized in Figure S1). We pooled results across at least three independent biological replicates and masked the identity of test compounds and C. elegans genotype during all experiments, which were performed by a team of two investigators.
Chemical gradient set-up
We arrayed our chemical library into 96-well microplates at a uniform concentration of 20 mM in dimethyl sulfoxide (DMSO) for all compounds except the reference set. Attractive reference compounds (isoamyl alcohol, diacetyl) were diluted serially in DMSO to 1:1000, 2-nonanone was diluted to 1:10, 1-octanol and DMSO were added directly to the plates. These concentrations were drawn from the literature and take into account the observation that a single compound can elicit attraction or repulsion, depending on concentration [30–32]. For all assays, compound identity was masked until after screening was completed. We used a variable-spacing multichannel pipette (Thermo E1-ClipTip 2-125 uL) to transfer 3.5 μL of each compound from the chemical library plate into assay plates (Nunc™ 4-well plates, Thermo Fisher, Cat # 267061). We used each of the lanes of 4-well multiwell assay plates to create four two-dimensional behavioral arenas consisting of solid media and a custom-fabricated foam corral in each multiwell plate (see below). Test compounds were dispensed into one apex and the solvent, DMSO, was dispensed into the opposite apex. Assay orientation was standardized by delivering test compounds to the notched side of each arena (Figure 1). Once loaded with test compounds and the solvent vehicle, we held assay plates at room temperature for 1 hour to establish a chemical gradient.
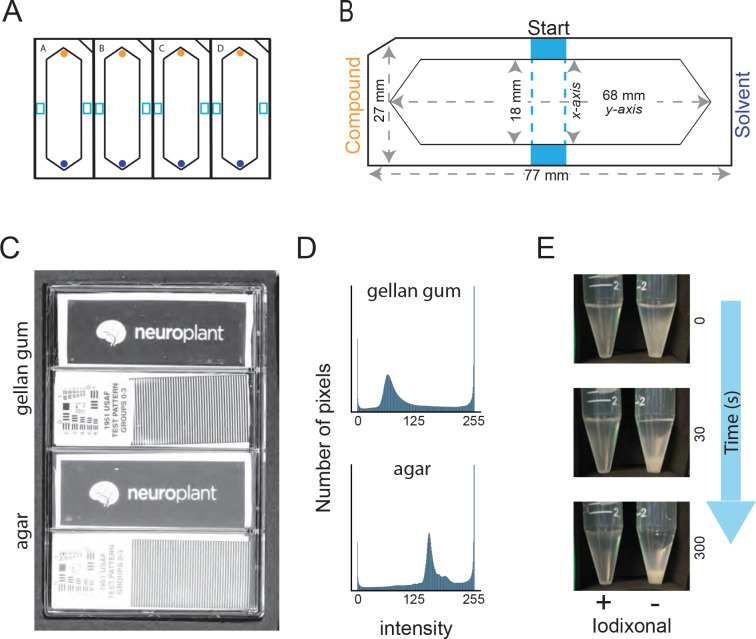
Figure 1: Measures that enable increased throughput of population-based C. elegans chemotaxis assays.
A) Schematic of a 4-lane assay plate (standard microtiter plate footprint) showing foam inserts.
B) Dimensions of a single foam insert. Panels A and B show the starting zone for animals (light blue), position of the test compound (side with notched corner, orange), and the reference or solvent (opposite, dark blue).
C) Image collected on a flatbed scanner of a single 4-well assay plate (left) containing Gelrite™ gellan gum (top two lanes) and agar (bottom two lanes). Transparent test patterns (Neuroplant logo, 1951 USAF test pattern) placed on the surface of the solid media demonstrate improved clarity for gellan gum compared to agar.
D) Intensity histogram drawn from the image of the test pattern imaged through gellan gum (top) and agar (bottom).
E) Still images of a time lapse observation of worms suspended in chemotaxis buffer with (+, left) and without (−, right) Optiprep™ solution of iodixonal (7:3 chemotaxis buffer: Optiprep™).
Behavioral arenas
We used thin foam to define assay arenas because it is hydrophobic, non-absorbent, and easy to cut with precision and reproducibility [33] with a computer-controlled cutting machine (Cricut Maker® and Cricut Maker3, Cricut, Inc.). The material we used is thin sheets of EVA foam (Cleverbrand, Inc., 9”×12”×1/16” or BetterOfficeProducts, 9”×12”×1/12”). The precise dimensions of each insert is shown in Figure 1B and we cut twenty inserts from a single 9”×12” foam sheet.
We filled assay lanes with gellan gum (Gelrite®, Research Products International, G35020-100.0) instead of agar, floating pre-cut foam inserts on top of the molten media so that it formed a worm-proof seal as the media solidified at room temperature. We sealed assay plates in plastic wrap and stored them at 4°C for up to 14 days prior to use. We selected gellan gum because of its superior optical clarity (Figure 1C) and settled on 2.5% (w/v) concentration as a practical balance between cost, stiffness, and clarity. We dissolved gellan gum (2.5% w/v) in ddH2O and heated it above 75°C by autoclaving. Chemotaxis buffer [aka CB: 5 mM KPO4, pH6) supplemented with MgCl2 (1 mM) and CaCl2 (1 mM)], prepared as described in Ref. [34], was added when the media cooled 60°C. Using serological pipettes, we added buffered, molten gellan gum (10 mL) to each assay lane and floated pre-cut foam inserts (see below) on top of the molten media.
Preparing worms for large-scale behavioral assays
We generated synchronized populations of worms and collected them for behavioral assays as follows. First, we examined NGM agar growth plates for signs of starvation or microbial contamination and discarded plates with starved animals or visible contaminants. Next, we collected young adult worms by dispensing 2.5 mL of sterile ddH2O and gently swirling the plate to dislodge worms from the agar surface. We transferred the worm slurry to a 15-mL conical tube, concentrated the animals in a centrifuge for one minute at 3000 RPM, and washed the worm pellet three times with sterile ddH20 to remove trace E. coli OP50. Washed pellets were resuspended in a 7:3 ratio of chemotaxis buffer (see above) and Optiprep (60% iodixanol in sterile H2O w/v, Sigma-Aldrich, D-1556), resulting in a net dilution to 18% iodixanol w/v. As shown in Figure 1D, worms remain suspended in chemotaxis buffer + iodixanol (CBI), an effect that reduces the variation in the number of worms delivered using liquid handling instruments or by manual pipetting. Iodixanol is a non-toxic polymer used to tune the index of refraction for live imaging applications [35]. It is also used in density gradient centrifugation applications [36,37], making it an ideal chemical tool to improve consistency of dispensing C. elegans in liquid. We have previously added iodixanol to physiological buffers and used this solution with C. elegans to reduce optical reflections inside microfluidic chips [38]. Finally, we resuspended a ~0.5 mL worm pellet in 3.5 mL of CBI to deliver ~250 worms/assay arena onto 12 assay plates.
Liquid handling and worm dispensing
To increase throughput, and reduce trial-to-trial variation of the number of worms dispensed into each assay arena, we adapted a multimode reagent dispenser (Biotek, Multiflo) and plate stacker (Biotek Biostack 3) to automatically dispense worms suspended in CBI. In brief, we separated a single line near the center of an 8-channel cassette (10 uL #423526) and adjusted the Liquid Handling Control software (LHC 2.22.) to deliver worm-laden drops in the center of the assay arena. To achieve this goal with sufficient precision, we used the 1536-well preset configuration in the LHC software to deliver a single droplet at the center of each of the four wells. Finally, we adjusted the flow rate and dispensing volumes to minimize splatter during dispensing events and droplet spread while the plates were in motion on the working surface of the liquid handler and plate stacker. Once the dispense cycle was completed, we flushed the line of any remaining worms by flowing 100% ethanol for 10 seconds, followed by ddH2O for 20 seconds. Using this approach, we dispensed 100–450 worms into each arena (~20 s per plate) and processed 12 plates in parallel for a total elapsed run time of ~250 seconds.
Running the chemotaxis assay
Once dispensed onto the assay plate, worms were retained in the liquid droplet. Thus, excess liquid needed to be removed to disperse animals and enable free movement. To achieve this goal, we placed absorbent PVA eye spears (BVI Ultracell -40400-8) on the center of the liquid droplet to withdraw as much liquid as possible by capillary action and used the fine point of the eye spear to disperse animals across the width of the assay arena, disrupting clumps of animals. Finally, assay plates prepared with chemical gradients and animals were transferred to a dry cabinet (Forspark, Cat.# FSDCBLK30) set at 31% relative humidity and allowed to move freely for 1-hour at room temperature (20 to 24°C).
Image capture
To efficiently capture images of the distribution of worms at the end of each chemotaxis assay, we used flatbed scanners (Epson, Perfection V600 Photo). We captured 8-bit grayscale images at 1200 dpi, with both brightness and contrast set at 50, choosing these settings to maximize contrast and resolution of the worms. The scan-bed on this instrument was large enough to simultaneously scan four assay plates positioned on the scanner surface using a frame cut from a sheet of black foam (9”×12”×1/6”, Cleverbrand Fun Foam, Black). The frame helped to map the four plates captured in a single image to their respective metadata and increased image contrast by setting consistent black levels. In addition, we adjusted the position of the scanner’s camera lens to achieve a sharp image at the surface of the gellan gel media that formed the assay arena. To achieve this goal, we modified consumer-grade flatbed scanners [21]. Specifically, we used standardized, transparent resolution patterns (USAF, 1951 Test Patterns, Edmund Scientific, #38-710) placed in position mimicking the assay surface, and adjusted the lens position to maximize pattern sharpness.
Image processing to locate worms
We transformed endpoint images of assay plates into arrays of worm position in each assay arena using a custom Python (v 3.7.4) code base. The software, which we call OWL (Our Worm Locator) locates the centroid of animals in scanned images and is built upon scikit-image [39]. Raw 8-bit 1200 dpi grayscale Tagged Image File Format (*.tiff) files of the chemotaxis endpoint were read and converted into Numpy (v 1.16.4) arrays. We used Otsu’s method [40] to determine a global thresholding value and generated a binary matrix of pixels for each image. All pixels with a pixel intensity greater than the thresholding value were set to white and all pixels less than the thresholding value were set to black. We then used the close function to repair defects that occurred during binarization. Contiguous groups of white pixels were labeled and all labeled objects were stored in a Pandas dataframe. These data included: centroid location , object area and bounding box values. The white foam inserts are the largest detectable objects within the image and were used to sort the data frame and assign well IDs based on their area and position in the image. Using the coordinates of the foam insert, we generated a mask which allowed us to dynamically divide image scans into images of each well. Objects in the cropped image were then relabeled and filtered to retain only those with an area greater than 50 pixels and less than 2500 pixels. This range of values excluded small objects that were not worms (eggs, dust, etc.) as well as large clumps of worms, but included small clumps of worms that were counted as single objects. Instead of attempting to estimate the number of worms in clumps [41], we sought to reduce their occurrence by manually dispersing animals across the width of the assay arena in the starting zone. The centroid coordinates of each identified worm-like object were exported as a comma separated values (*.csv) file for each well and used to evaluate chemotaxis. To support users, we used PySimpleGUI to create a graphical user interface for OWL.
Metadata and digital data management
For each round of screening, we established and maintained two types of data files (location, summary) and one metadata (plate ID, strain ID, compound ID), connecting each assay to the conditions in that particular trial using Python scripts (see Code Availability). Each assay arena is associated with a location file and a summary file. The location file contains the coordinates (in pixel units) of all the worms detected in the arena. We linked each location file to its assay conditions using an automated file naming convention in which the file name contained the image ID, scanner slot number (location of the plate in the scanned image), and the well ID (location of the well within the plate). The summary file contains the total number of worms counted in the assay arena, the calculated chemotaxis index, and the distance between apices (in pixels) (3041 ± 20, mean±s.d., arenas), test compound, worm strain, image ID and plate ID. All raw and processed data files are stored in open-source file formats (*.tiff, *.csv) or as Google Sheets. Each image is assigned a unique image ID, linking the image to its respective metadata and image analysis results. Metadata are stored as Google Sheets and include assay date, experimenter, image ID, plate ID, scanner slot number, compound ID, strain ID, relative humidity, and temperature.
Assessing the accuracy of image-based measures of chemotaxis behavior
We assessed OWL’s accuracy by comparing human and machine analyzed images. First, we identified three cropped endpoint images for the reference conditions [isoamyl alcohol, 2-nonanone, 1-octanol, symmetric DMSO (DMSO:DMSO) and asymmetric DMSO (DMSO:H20)] and four cropped endpoint images for diacetyl. In total, 19 images were identified for human scoring. Next, two people were assigned to score each cropped image using the same manual scoring protocol, described as follows. Each image was loaded into FIJI (ImageJ2) [42]. Next, human counters logged the location of individual worms using the ‘“multi-point” selection tool. Once all worms were located and logged in an image, the human counter used the “Measure” function to return the coordinates (pixel) of all counted worms in the image and exported these data as a *.csv file.
We used two metrics to analyze the OWL’s performance: 1) total number of worms counted in an assay arena and 2) the mean position of worms within an assay arena. For both metrics, we used Pearson’s correlation coefficient (computed by linregress in scipy.stats (version 1.7.1)) to evaluate the similarity between human scorers and between each human and the OWL software. Mean worm positions were calculated using the mean module in the Python statistics package (v 3.7.4). Residuals were calculated and plotted for both analyses using the Seaborn (v. 0.9.0) residplot package. Finally, we generated kernel density estimation plots to compare the worm locations in each well identified by both human scorers and OWL using the Seaborn kdeplot package (v 0.9.0).
Data and statistical analysis
Each assay arena is associated with a *.csv file of the pixel positions (in units of dots per inch or DPI) of worms detected in the endpoint image of the experimental arena. In this coordinate system, the -axis extends along the chemical gradient and the -axis indicates position across the width of the arena. We collected images at a resolution of 1200 DPI (pixels/inch). For data visualization, we transformed units from pixels to millimeters and repositioned the origin of the -axis to the center of the arena as follows:
Where position along the -coordinate in mm, position along the -coordinate in pixels, and between the arena apices in pixels. Positive values of indicate positions closer to the test compound and negative values for indicate positions closer to the solvent reference. The total range for is −35 to +35 mm.
We established and maintained metadata sheets to link these datasets to the conditions of each assay (see below) and used these datasets to evaluate trial to trial variation, pool results across trials, and to determine the effect of test compounds on chemotaxis. Our analysis approach used the distribution of animals along the axis of the chemical gradient, which we designated as the -axis in our coordinate system, to determine chemotactic responses. Conditions that resulted in roughly equal numbers of animals migrating toward each apex in the arena and an average worm position indistinguishable from zero were considered evidence of indifference to the chemical conditions in the arena. On the other hand, distributions biased toward or away from the test compound were classified as positive and negative chemotaxis, respectively. We also refer to these outcomes as attraction and repulsion, respectively.
The strength of each putative chemotaxis response was determined using estimation plots [43,44] comparing distributions evoked by exposure to test compounds with those found for two null conditions: symmetric solvent (DMSO:DMSO) and DMSO opposite ddH2O (DMSO:ddH2O). Effect sizes were calculated using a statistical bootstrapping approach implemented via the Dabest software library (v 0.3.1) [43]. Additional statistical tests were performed using scipy.stats packages (v 1.7.1) including the Shapiro-Wilk test for normality [45], the Kruskal-Wallis H test (non-parametric ANOVA) [46] and the Mann-Whitney U test [47] for post hoc analysis. We report bootstrapped effect sizes for each test condition as 95% confidence intervals along with their respective mean differences and the results of a Mann-Whitney U test to determine statistical significance relative to the reference condition (Table S2).
Structured literature review
To evaluate the novelty of SMs in our chemical library as either attractants or repellants of C. elegans or other nematodes, we designed and performed a structured search of the PubMed and Web of Science (WOS) databases on the subset of SMs we identified as either attractants or repellants. The search terms consisted of compound name, CAS No., and species name (C. elegans or Caenorhabditis elegans) or compound name, CAS No., and “nematode NOT elegans” together with “chemotax*”. Next, we excluded studies that used plant extracts or complex mixtures, studies in which worms were used as pathogen vectors, or transformed with human peptides. Finally, we eliminated duplicates, generating a set of 61 unique publications.
Code Availability
We developed OWL and the OWL GUI software in Python version 3.7.4 and used Anaconda (v 2020.02) to set up a virtual environment that contains all of the Python packages and versions necessary to run these tools. The full codebase is publicly available in a Github repository, https://github.com/Neuroplant-Resources, and includes a *.yml file to define package and version information (NP_conda_env.yml file).
Results
This work harnesses chemical communication between plants and nematodes [48–50] to identify small molecules that are detected by the chemosensory nervous system. Our approach relies on testing small molecules synthesized by plants for their ability to either attract or repel the model roundworm, Caenorhabditis elegans. This behavior, known as chemotaxis, has at least two advantages for the purposes of identifying chemical cues detected by neurons. First, because animals are not immersed in test chemicals, there is little, if any, risk of lethality. Second, all putative receptors expressed by the 32 chemosensory neurons are tested in parallel. Each class of chemosensory neurons expresses a distinct ensemble of ion channels and receptors [9]. The data available from neuron-specific and single-neuron RNASeq datasets [11] and promoter fusions [10,51] indicate that a single chemosensory neuron expresses ~100 GPCRs and 3–5 receptor guanylate cyclases and that no two classes of chemosensory neurons express identical subsets of either class of membrane receptors. Thus, by working with a defined sensorimotor behavior, we test as many as 1000 putative receptors for plant SMs without building libraries of cells expressing putative receptors or establishing in vitro assays of their function.
A four-lane highway for nematode chemotaxis assays
We followed a rapid prototyping, design-build-test approach to retool classical laboratory assays for C. elegans chemotaxis. Our prototyping cycles were guided by these design rules: 1) minimize manual handling; 2) use uniform behavioral arenas; 3) use common scientific or consumer equipment; 4) automate analysis; and 5) integrate data acquisition and management. Classical C. elegans chemotaxis assays are often performed on round (6-cm or 10-cm diameter) agar plates bearing a chemical gradient created by a small volume of test compound at the edge of one side of the plate and the relevant vehicle on the opposite side [4,52]. Animals are dispensed into the center of the assay plate and allowed to move freely for a defined time. Following the assay, the number of animals on the compound and solvent sides are counted manually and these counts are used to compute a chemotaxis index that has a value of +1 for ideal attractants, −1 for ideal repellents, and 0 for compounds that are not chemoactive. This chemotaxis assay is simple, widely-used and reduces a complex behavior (chemotaxis) to a single endpoint metric, but its throughput is limited to a single assay condition per plate.
Based on our goals and design rules, we selected standard multiwell plates with 4 lanes for behavioral arenas (Figure 1A). To further standardize assay arenas, we fabricated foam inserts and floated them on top of optically-clear solid media (gellan gum) deposited in each lane (Methods). The foam’s hydrophobic surface retains animals within the arena and its shape standardizes the placement of both animals and compounds on the arena surface (Figure 1A, 1B). These choices allow for a workflow based on standard instrumentation compatible with multiwell plates. We exploited this feature by using a liquid handler and plate stacker to dispense worms onto assay plates. The liquid handler not only dispenses worms onto 12 plates (48 assay arenas) in ~4 minutes, but also dramatically increases the repeatability and accuracy of the number of worms dispensed (coefficient of variation, ) compared to manual pipetting .
Worms do not stay suspended in conventional buffers, leading to systematic variations in the number of worms dispensed in liquid. We counteracted this effect using iodixanol, a non-toxic polymer, to adjust buffer density so that C. elegans are neutrally buoyant in solution. After 30 seconds, C. elegans animals in standard buffer form a visible pellet, but animals in iodixanol buffer remain suspended (Figure 1D). This effect reduces variability in dispensing animals and could be extended to other workflows, including those that rely on manual pipetting. The dispensing liquid must be dispersed before animals crawl freely on the gel surface. At present, this step is performed manually using lint-free, absorbent eye spears to withdraw excess liquid and to disperse animals across the width of the behavioral arena. Collectively, these maneuvers accelerate and improve chemotaxis assay reliability.
Iterative improvements in imaging and automated chemotaxis measurements
We adapted a consumer flatbed scanner to rapidly image assay plates at high contrast, and developed an image processing pipeline, Our Worm Locator (OWL), for detecting worm positions. Our prototyping cycle identified four modifications that were instrumental in reaching this goal. First, we replaced agar with gellan gum because agar lacks the optical clarity needed to achieve high contrast images (Figure 1C). Gellan gum is a natural heteropolysaccharide purified from the bacterium Sphingomonas elodea [53,54], which can be cast into stable, solid gels similar to agar. Second, we modified the flatbed scanner to achieve sharp focus at the gellan gum surface (Methods). Third, we used custom foam inserts to standardize behavioral arenas, to improve image contrast, and to simplify downstream image processing. We programmed the cutting machine to mark the worm starting zone equidistant from the apices of the arena (Figure 1A, 1B). The apices define locations for spotting compounds and solvent controls, while the hydrophobic surface repels worms, retaining animals in the main arena. Fourth, we cut black craft foam to generate guides for consistent positioning of four assay plates on the scan bed. The scanner captures a full-field image of four assay plates in ~2 minutes, yielding a single image at near-uniform time point. Because of the sharp contrast and consistent positioning, our codebase de-multiplexes scanner images into images of single assay plates and each assay plate is de-multiplexed into single assay arenas (Figure S2). Compared to the initial iteration of the design-build-test cycle, these actions generated a 16-fold increase in data collection efficiency and a 40-fold increase in image capture efficiency.
Imaging processing pipeline to determine worm position
Borrowing imaging principles from software for tracking worm movement [55] and similar to other reports [56,57], OWL locates and logs the centroid position of all worms in our assay arenas. OWL removed multiple, significant barriers to scaling up chemotaxis assays that depend on manual counting, which is time-consuming and error-prone. The OWL software, by contrast, determines the locations of hundreds of worms from images collected at a single time and generates large, digital datasets that can be efficiently analyzed at any time. As part of our design-build-test cycle, we pooled data across 16 assays in which animals were exposed to solvent (DMSO) on both sides of the arena (Figure 2A) and used bootstrapping techniques to determine how the number of assayed in a given arena affects the chemotaxis index. Across four C. elegans genotypes (wild-type, tax-4, osm-9, tax-4;osm-9), we observed the mean chemotaxis index, but not the variance was independent of this metric. As expected for a random or pseudo-random process, variance was inversely proportional to the total number of animals (Figure 2B). Because the variance reaches a steady minimum near 150 worms, we used this value as a quality control threshold — including assays with at least this many worms and excluding those with fewer worms.
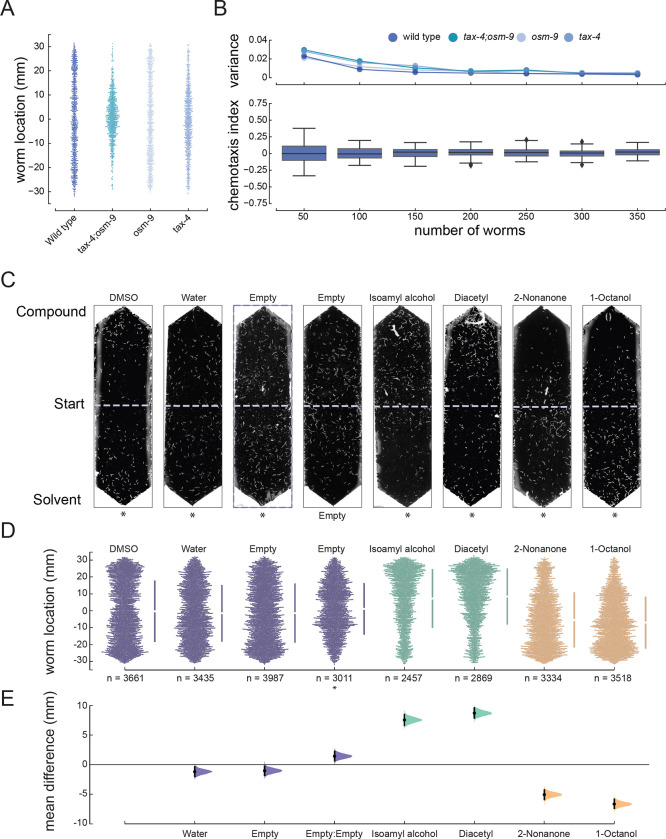
Figure 2: Optimization and validation of chemotaxis performance and derivation of average position as a robust chemotaxis metric.
A) Distribution of animals following exposure to symmetric DMSO. Each dot represents the y coordinate of a single animal of the indicated genotype, pooled across three biological replicates: wild-type (N2), tax-4(pr678);osm-9(ky10), osm-9(ky10), and tax-4(pr678).
B) Average (± s.d.) chemotaxis index for wild-type animals (bottom) and variance for the indicated genotypes (top) as a function of the number worms in an assay arena. The data are a bootstrap analysis of the data in panel A for sample sizes from 50 to 350 (increments of 50) animals.
C) Representative images of assay arenas following exposure to (left to right): four null conditions, two known attractants, and two known repellents. Except for the empty condition, DMSO is on the solvent (bottom) side, denoted by an asterisk (*).
D) Swarm plots pooled across 16 technical replicates for each condition shown in panel C. Bars to the right of each swarm show the ± one standard deviation, with the gap between the bars indicating the mean worm position.
E) Effect size relative to the DMSO:DMSO null condition. Black bars and shaded areas show the difference of the mean values and the 95% confidence intervals for this value, bootstrapped from the data for each test condition. Instances that exclude a mean difference of zero are considered bona fide responses compared to the null condition. Positive values indicate attraction (positive chemotaxis) and negative values indicate repulsion (negative chemotaxis).
Platform performance and validation
To assess the performance of our chemotaxis pipeline, we determined the response of the standard laboratory C. elegans strain (N2, Bristol) to four compounds with well established chemotaxis phenotypes and four null conditions predicted to result in indifference. We selected two attractants, isoamyl alcohol and diacetyl [58], and two repellants, 2-nonanone, and 1-octanol [58]. The four null conditions were: DMSO vs. DMSO (DMSO:DMSO or symmetric DMSO); DMSO vs water (DMSO:ddH2O); DMSO vs. empty (DMSO:null); and no compound added (null:null). We selected these conditions based on the use of DMSO as the solvent for all of our test SMs and to determine if animals were sensitive to this solvent. Figure 2C shows images of single assay arenas for each of the four null conditions and the four reference compounds. We pooled worm position along the chemical gradient across 16 replicate assays and used estimation statistics [43,44] to determine the effect size (Figure 2D, 2E). From these data, we infer that DMSO is a weak attractant and confirm that isoamyl alcohol and diacetyl are strong attractants and that 2-nonanone and 1-octananol are strong repellents.
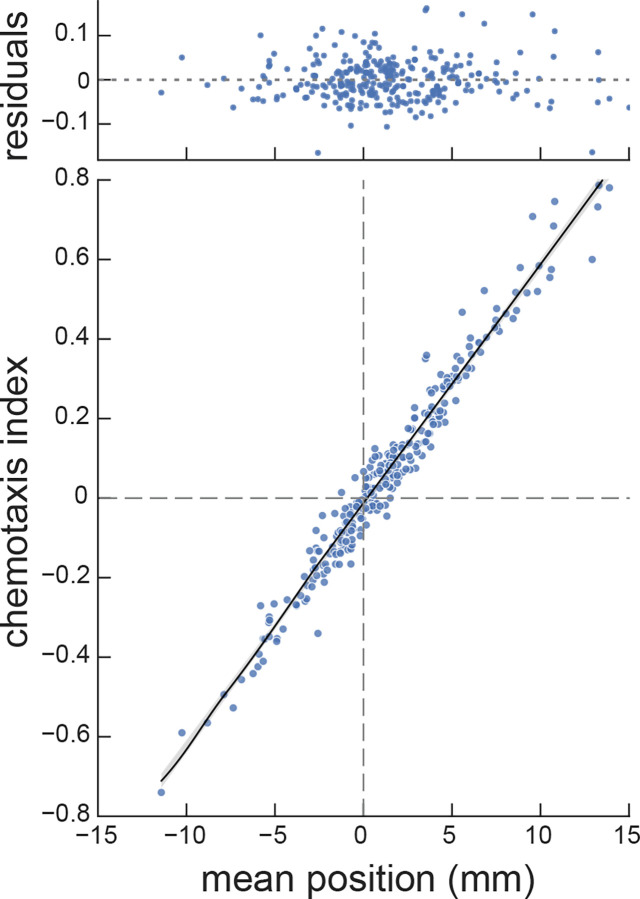
To validate the mean position as an indicator of chemosensitivity, we compared it to the chemotaxis index. Classically, researchers have reported the results of chemotaxis assays using a chemotaxis index [52,58,59]: where is the number of animals on the side of the test chemical and is the number on the opposite or control side. Consistent with prior practice and to minimize the impact of variation in movement ability, animals in the starting zone were excluded from analysis. We defined as the total number of worms on the side of the test compound and as the total number of worms in the four regions on the opposite side. Comparing three biological replicates testing wild-type against 96 conditions consisting of 90 plant SMs, two null reference conditions, two attractants (isoamyl alcohol, diacetyl), and two repellents (2-nonanone, 1-octanol), we found that chemotaxis index and average worm position were tightly correlated with one another (Figure 3, ), indicating that our analytical approach is consistent with classical studies. Thus, the average worm position is correlated with and essentially equivalent to the chemotaxis index. Evaluating the distribution of worm positions, however, enables us to pool responses across replicates and this step increases the robustness and statistical power of the assay. For these reasons, we rely on worm position for subsequent analyses.
Figure 3: Chemotaxis index and mean worm position are similar across a range of values and test conditions.
Each point represents the chemotaxis index and mean worm position computed from a single assay. The dataset represents 288 assays of the response of wild-type worms to 96 compounds ( biological replicates). Black line is a least-squares fit to the data with a slope of .061 , the shaded area shows the 95% confidence interval for the fit.
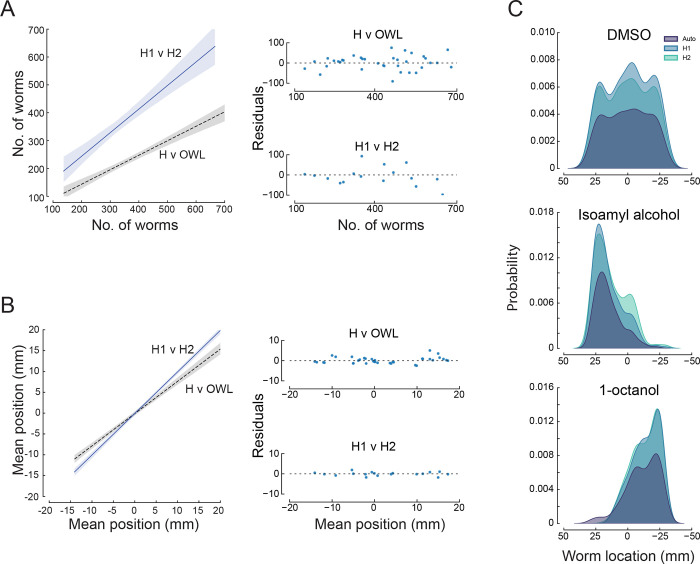
Next, to assess OWL’s performance, we benchmarked the software against human scorers. To do this, we generated a test dataset and recruited two team members to manually tag the location of worms in each arena using FIJI [42]. The test dataset included 19 images of assays performed with two attractants (diacetyl, isoamyl alcohol), two repellents (2-nonanone, 1-octanol), symmetric solvent (DMSO:DMSO), and solvent (DMSO) opposite ddH2O (DMSO:ddH2O). To assess the agreement between the human observers and OWL, we compared the total number of worms (Figure 4A) (Pearson’s correlation coefficient = 0.901) and mean worm positions (Figure 4B) (Pearson’s correlation coefficient = 0.978).
Figure 4: Performance of human scorers (H1,H2) and OWL software.
A) Relationship (left) between the total number of worms detected by H1 and H2 (solid blue line, slope = .846 ) and by the average human and OWL software (dashed black line, slope = 0.519; ). Shaded areas show the 95% confidence intervals of the fit. The fit residuals (right) indicate no systematic effect of the number of worms.
B) Relationship (left) between the mean worm position detected by H1 and H2 (solid blue line, slope = 0.993; ) and by the average human and OWL software (dashed black line, slope = 0.774; ). Shaded areas show the 95% confidence intervals of the fit. The fit residuals (right) indicate no systematic effect of the mean position.
C) Density as function of distance along the chemical gradient for three conditions (top to bottom): null condition, DMSO:DMSO, known attractant, isoamyl alcohol, known repellent, 1-octanol. Distributions scored by humans (light blue and aqua) and determined by OWL software (dark blue) are similar. Each image in the test dataset was scored by two human experimenters and by the OWL software, as described in Methods. The test dataset included 19 assay images (4 of diacetyl assays and 3 each for all other conditions).
The strong agreement between automated worm location and manual counting is similar to the findings of Crombie, et al. [56] who paired large-particle sorting hardware (COPAS biosorter) with custom software to automate nematode chemotaxis assays performed on round Petri dishes. While OWL undercounted worms relative to human observers, human observers were also discordant. Importantly, the average worm position measured by human observers was similar to that extracted by OWL. We suspect that the primary difference in worm counts resides in imperfect attempts by human observers to count aggregated animals. OWL excludes such aggregates (based on their size), a factor likely to account for the fact that humans find more worms. These effects are independent of position in the arena, however, since the distribution of worms as a function of position along the -axis is similar when measured by human observers and by OWL (Figure 4C). Thus, similar to the parallel worm tracker [55], the concordance between human observers resembles that found between a single human observer and OWL. In summary, the OWL image processing pipeline reliably determines average worm position, does not compromise reproducibility compared to pairs of human observers, and dramatically increases experimental throughput.
Dozens of plant-derived small molecules attract or repel C. elegans
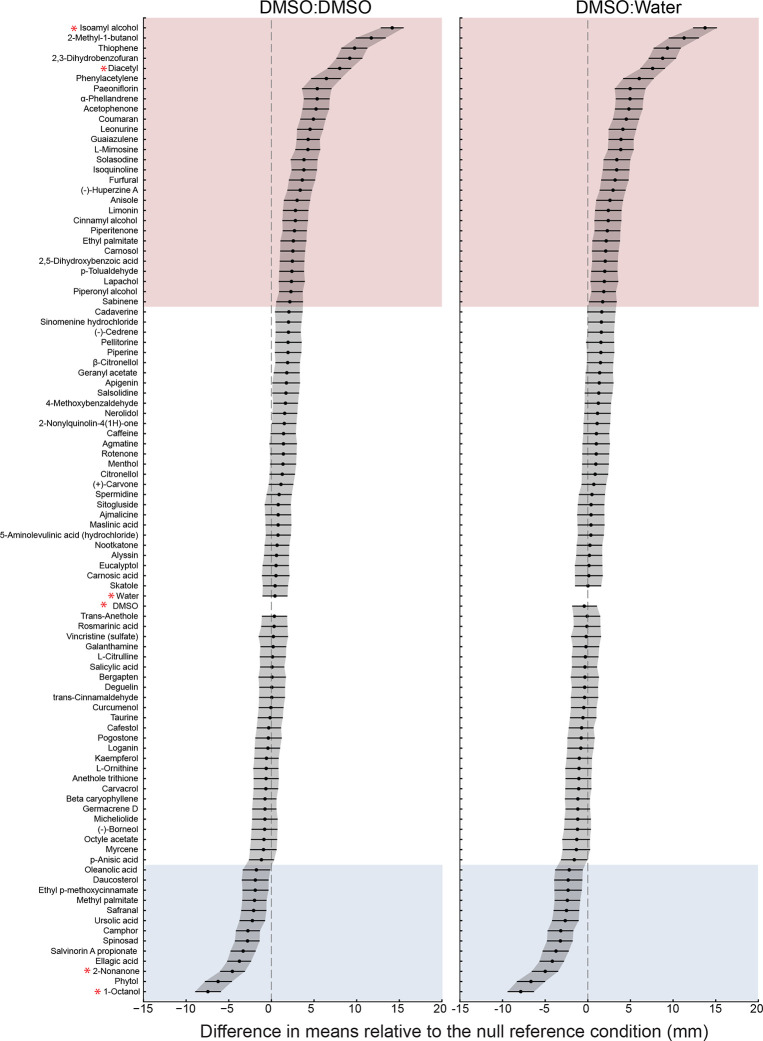
We applied our platform and integrated data handling workflow to screen 90 plant SMs and 6 reference conditions (isoamyl alcohol, diacetyl, 2-nonanone, 1-octanol, DMSO:DMSO, DMSO:ddH2O). A given compound was considered chemoactive if the mean worm position observed in arenas containing that compound differed from our two null reference conditions (DMSO:DMSO and DMSO:ddH2O). Using estimation statistics and bootstrapping [43,44], we computed the difference of the mean position for each compound relative to each of the null reference conditions. Figure 5 plots the distributions of mean differences and arranges the results by magnitude and valence such that the strongest attractants are at the top and the strongest repellents are at the bottom. In addition to null reference conditions, the library contained three pairs of SMs that were nominally identical compounds obtained from different suppliers (Methods). For two of the three SM pairs, the response of wild-type worms to compounds were distinct from one another: CAS No. 496-16-2—2,3-dihydrobenzofuran and coumarin, , Student’s t-test; CAS No. 474-58-8: daucosterol and sitogluside, , Student’s t-test). These findings could reflect a true difference in the chemicals we tested. For the third pair, citronellol and β-citronellol (CAS. No. 106-22-9), the responses were indistinguishable (Student’s t-test, ). Forty-one compounds in total (including 4 reference compounds) induced a response different from both null conditions. Excluding references, 27 compounds attracted wild-type worms and 11 repelled them. One compound, safranal, was omitted from further consideration because it elicited weak responses that were poorly distinguished from the DMSO:water null condition. Thus, our screening platform uncovers SMs that attract or repel wild-type C. elegans with high confidence and efficiency.
Figure 5: A screen of 96 conditions reveals 37 plant SMs that are chemoactive in wild-type C. elegans, evoking either attraction (pink) or repulsion (blue).
The chemical panel contained 90 plant SMs and 6 reference conditions (red asterisks: isoamyl alcohol, diacetyl, 2-nonanone, 1-octanol, DMSO, and water). Results are sorted (top to bottom) according to the difference in mean position relative to two null reference conditions: symmetric DMSO:DMSO (left) and asymmetric DMSO:water (right). We used a bootstrapping approach [43] to compute differences in mean position. Positive values correspond to attraction or positive chemotaxis and negative values correspond to repulsion or negative chemotaxis. Black points and lines are, respectively, the difference of the mean position in each test condition relative to the reference condition and the 95% confidence intervals of these values. independent biological replicates, at least 150 worms per assay. Table S2 contains the following for all screening conditions: difference of the mean position (in mm) in experimental vs. reference conditions (±95% confidence intervals), and statistical testing against the reference condition (Mann-Whitney U test).
We next sought to determine which of these 37 plant SMs had been tested previously for their ability to attract or repel C. elegans or other nematodes. To achieve this goal with similar coverage for all compounds, we used a defined key-word search of a standard bibliographic database (Methods). With the exception of two attractive compounds, furfural [58] and 2-methyl-1-butanol [60–62], we found that very few of these plant SMs had been tested for their activity in chemotaxis assays in C. elegans or any other nematode. We also searched for studies applying these SMs to C. elegans or other nematodes for any other purpose. Six compounds (phytol, ellagic acid, camphor, ursolic acid, furfural, and 2-methyl-1-butanol) have been tested for effects on lifespan, oxidative stress, fecundity, or as nematicides [63–67]. Three compounds, furfural, solasodine, and phytol, have been tested as tools for managing root-knot nematodes that parasitize plants, including important crops [68,69][70–73]. This raises the possibility that additional compounds in this dataset may prove relevant to agriculture. More broadly, our systematic review buttresses the idea that combining an evolution-inspired screen design with an efficient phenotypic screening platform is a highly effective tool for discovering novel chemoactive compounds made by plants.
Response valence reflects integration of chemosensory signaling
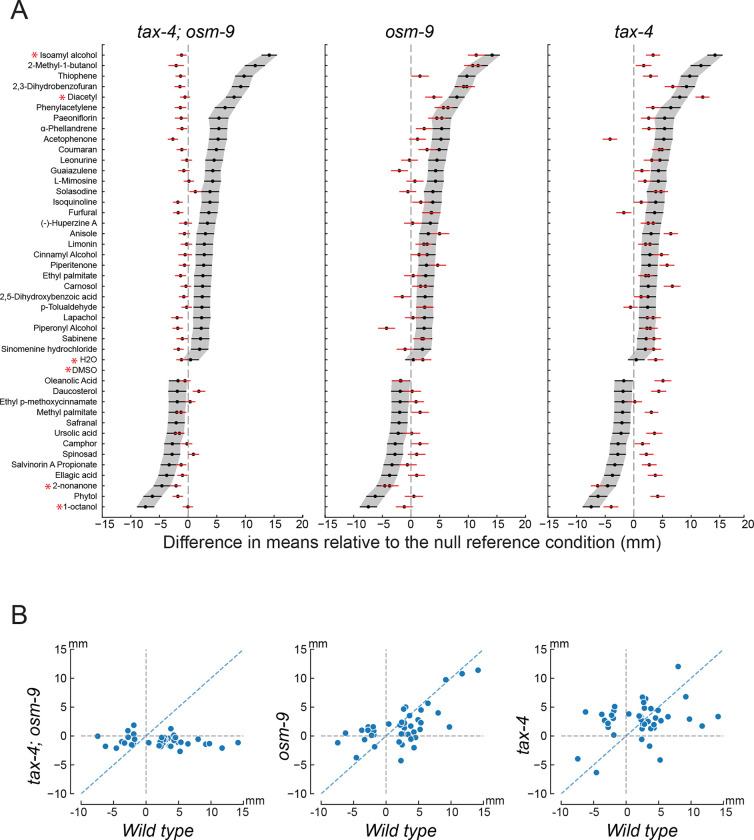
To learn more about the genetic basis of chemotaxis valence, we tested the 37 compounds against mutants lacking one or both of the two ion channel effectors, TAX-4 and OSM-9, required for chemosensory transduction (reviewed in [4]). To do this, we relied on two previously isolated null mutants, tax-4(p678) [74] and osm-9(ky10) [14,75], and used them to generate an anosmic tax-4;osm-9 double mutant. We first analyzed the response of tax-4;osm-9 double mutants to 41 SMs (37 test and 4 reference compounds) and two null conditions. For all attractants and repellents, tax-4;osm-9 mutants were either indifferent or weakly repelled (Figure 6A, 6B, left). These findings indicate that the responses we observed in wild-type animals depend on chemosensory signaling and are unlikely to reflect direct modulation of locomotion. Collectively, ~40% of the compounds in our curated library of plant SMs are biologically active chemical cues that activate one or more C. elegans chemosensory neurons. These findings suggest that the C. elegans chemosensing repertoire is likely to be larger than previously appreciated.
Figure 6: Chemoactive plant SMs evoke approach or avoidance based on signaling by CNG channels, TRPV channels, or both chemosensory ion channels.
A) Bootstrapped difference in the mean position (± 95% confidence interval) for each plant SM tested in mutants lacking the TAX-4 CNG channel, the OSM-9 TRP channel, or both: tax-4(pr678);osm-9(ky10) (left), osm-9(ky10) (middle), and tax-4(pr678) (right). The red points and lines represent the difference in bootstrapped mean position (± 95% confidence intervals) for responses in mutant animals; black points and lines are reproduced from Figure 5 for ease of comparison. Confidence intervals and Mann Whitney U test statistics comparing wild-type and mutant responses are reported in Table S3.
B) Mutant vs. wild-type responses across the 37 chemoactive plant SMs (mutant genotype, left to right): tax-4(pr678);osm-9(ky10), osm-9(ky10), and tax-4(pr678). Points are results for a single compound and overlap in some instances. Dashed black lines indicate values consistent with indifference in each mutant line; dashed blue lines at indicate the expected value if the mutant and wild-type phenotypes are indistinguishable.
Next, we sought to map chemoactive compounds onto chemosensory transduction signaling pathways based on the ion channel effectors, TAX-4 and OSM-9. Following the analytic approach described above, we classified responses as tax-4-dependent or osm-9-dependent as follows: 1) the mutant response was distinct from that of wild-type worms; and 2) similar to the response to the symmetric DMSO condition. Applying these criteria, four compounds evoked responses in wild-type that were different in tax-4, but not osm-9 single mutants. These compounds are: isoamyl alcohol, 2-methyl-1-butanol, furfural, and -tolualdehyde. This result reinforces prior work showing that tax-4 expressing neurons play an integral role in detecting isoamyl alcohol [14] and that furfural functions as chemoattractant [58]. Using the same framework, we classified five compounds as osm-9 dependent and tax-4-independent: leonurine, L-mimosine, solasodine, piperonyl alcohol and 1-octanol. One of these compounds, 1-octanol, is a strong repellant (95% CI [−.94, .85], ) and one of our reference compounds.
Notably, behavioral sensitivity to two reference compounds (diacetyl, 2-nonanone) was impaired in tax-4 and osm-9 single mutants and eliminated in tax-4;osm-9 mutants. From this finding, we infer that the ability to classify these compounds as desirable or toxic emerges from multiple signaling cascades. Consistent with this idea, diacetyl evokes calcium transients in five classes of chemosensory neurons: ASE, ASH, AWA, AWB, and AWC [76]. The ASE, ASG, ASJ, and AWC neurons express both tax-4 and osm-9, ASH and AWA are seen to express osm-9, but not tax-4, and AWB expresses tax-4, but not osm-9 [13,14]. The present studies of chemotaxis (Figure 6) and complementary calcium imaging datasets [76] suggest that the valence of a given chemical cue is driven by integration of multiple chemical signaling pathways.
Additional support for the general model in which chemical valence is the result of integrated signaling pathways is evident in our genetic dissection study of the 37 plant SMs (Figure 6). Responses to more than half (20) of chemoactive plant SMs were diminished, but not eliminated in osm-9 and tax-4 single null mutants (Figure 6A). For example, wild-type worms are strongly attracted to thiophene and this response was diminished, but not abolished in tax-4 mutants. Attraction to thiophene was abolished in osm-9 single mutants and in tax-4;osm-9 double mutants, however. Collectively, these results demonstrate that attraction to thiophene depends on multiple signaling pathways and they suggest that tax-4-dependent signaling contributes to response valence in an osm-9-dependent manner. More broadly, responses to eleven (11) plant SMs share a pattern in which chemotaxis strength is reduced in single mutants of either tax-4 or osm-9 but absent in tax-4;osm-9 double mutants.
For the remaining nine compounds that require both signaling pathways, we observed a valence change in the tax-4 null mutant compared to wild-type animals: acetophenone, oleanolic acid, daucesterol, methyl palmitate, ursolic acid, salvinorin A propionate, ellagic acid, spinosad and phytol. With the exception of acetophenone, all of these compounds were repellent to wild-type worms (Figure 6A). Ursolic acid and oleanolic acid are isomers and responses to these compounds were similar behaviors in all genotypes. Wild-type animals were strongly attracted to acetophenone, whereas tax-4;osm-9 double mutants and tax-4 mutants were repelled; osm-9 single mutants were essentially indifferent. If acetophenone attraction were exclusively dependent on osm-9 signaling, then tax-4 mutants should be attracted to acetophenone. This prediction did not hold, however, as acetophenone repelled tax-4 mutants. The other eight compounds in this group were repellent to wild-type worms and attractive to tax-4 mutants. These compounds evoked little or no response in osm-9 single mutants and tax-4;osm-9 mutant lines (Figure 6A). In the most extreme case, phytol was found to be strongly repellent to wild-type worms (95% CI [−7.80, −4.62 ] Mann Whitney U Test, ), indifferent to osm-9 (95% CI [−1.09, 2.10], Mann Whitney U Test, ), weakly repellent to osm-9;tax-4 (95% CI [−2.72, −0.88], Mann Whitney U Test, ) and attractive to tax-4 worms (95% CI [2.89, 5.41] Mann Whitney U Test, ). Phytol is an acyclic diterpene that is a component of chlorophyll and is found in all photosynthetic organisms. Interestingly, salvinorin A, a psychotropic compound produced by Salvia divinorum, and its analog, salvinorin A propionate are also acyclic diterpenes [28]. Salvinorin A propionate was also found to induce this valence change in the tax-4 mutant line. These findings suggest that chemosensory valence emerges as a consequence of the signals generated by the ensemble of chemosensory neurons sensitive to a given compound.
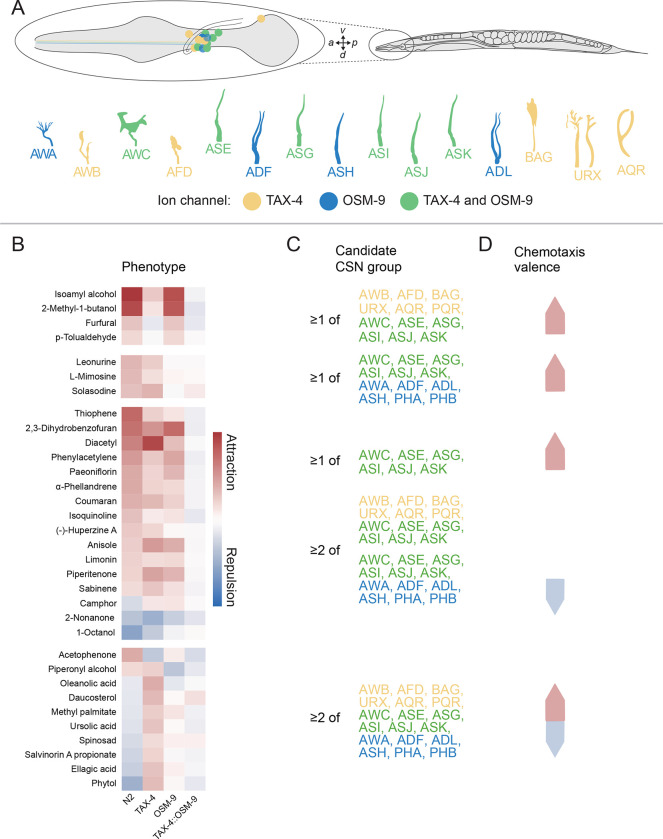
Based upon the pattern of phenotypes evident in the four genotypes we tested and the cellular expression patterns of the tax-4 and osm-9 ion channel genes, we draw inferences regarding the chemosensory neurons likely to detect the chemoactive compounds. As illustrated in Figure 7A, the tax-4 and osm-9 genes are co-expressed in six anterior chemosensory neurons (CSNs): AWC, ASE, ASG, ASI, ASJ, ASK. The AWB, AFD, BAG, URX, and AQR chemosensory neurons express tax-4, but do not appear to express osm-9. And, the AWA, ADF, ASH, and ADL neurons express osm-9, but do not appear to express tax-4 [13,14]. Considering only compounds that generated responses distinct from the wild-type response and exceeding each genotype’s response to symmetric DMSO, we infer that 4 and 3 compounds are detected by at least one CSN using TAX-4 and OSM-9, respectively, as an effector. Further, we propose that 26 compounds are detected by at least two CSNs using TAX-4, OSM-9, or both ion channels as effectors. Additional experimental work is needed to link individual plant SMs to chemosensory neurons and to their membrane protein receptors.
Figure 7: Graphical summary of behavioral responses to chemoactive compounds and proposed links to candidate chemosensory neurons (CSNs).
A) Schematic showing the position of C. elegans amphid chemosensory neurons (CSNs) on the right side of an adult animal (top). With the exception of AQR, CSNs are bilaterally symmetric. CSNs have distinctive cilia, shown schematically (bottom). Color indicates expression of chenosensory transduction ion channels in each CSN.
B) Mean difference in worm position for test vs. reference conditions shown as a heatmap. Rows are plant SMs, columns are C. elegans genotypes. Positive and negative values indicate attraction (red) and repulsion (blue) and values near zero indicate neutral responses (white). The four groups divide responses into those that depend primarily on a single ion channel signaling pathway (i: tax-4, ii: osm-9), depend on both tax-4 and osm-9 (iii), and responses that change valence in either the tax-4 or osm-9 single mutants (iv).
C) Candidate CSNs proposed to detect SMs in each grouping.
D) Chemosensory valence associated with each group: attraction (red), repulsion (blue).
Discussion
To spur efforts toward obtaining a general understanding of how chemical cues are encoded according to valence, we built an efficient platform for testing compounds for the ability to attract or repel nematodes. Compared to classical C. elegans chemotaxis assays, which depend on manual assays and hand-counting worms [4,59,77], our platform features liquid handling hardware for worm dispensing, wetware to optimize image quality and enable image de-multiplexing, and software to count animals, determine their position, and determine the strength and direction of chemotaxis. The workflow presented here makes it possible to screen hundreds of compounds in a single week with improved rigor and reproducibility. Across >250 assays, we demonstrate that mean worm position is equivalent to the classical chemotaxis index (Figure 3C). Recording worm position in a standardized, open-source digital data format opens the door to pooling results across replicates. This tactic also generates improved statistical power and is amenable to using estimation statistics to determine effect size relative to reference compounds and null conditions [43,44].
Our chemotaxis assay platform and integrated OWL software are versatile and compatible with any desired chemical library. Based on the long and continuing co-habitation of nematodes and plants, we reasoned that screening a library of plant-synthesized SMs would be especially productive and we screened a modest custom library of 90 plant SMs. Consistent with this evolution-inspired idea, we found that 37 of 90 or 41% of our curated plant SM library evoked chemotaxis in wild-type C. elegans. This group included 27 attractants and 10 repellents (Figure 5). The preponderance of attractants could reflect an unintended bias in our library, masking of repulsion by the weak attraction induced by DMSO, or be a true reflection of the bias in chemical communication between plants and nematodes. Regardless of its origin, a similar bias in favor of attractants was noted previously [58]. Most of the SMs identified in this study had not been tested previously in C. elegans chemotaxis assays and sensitivity to all 37 SMs was eliminated in taste- and smell-blind tax-4;osm-9 double mutants. Thus, these 37 SMs significantly expand the set of chemical cues known to evoke either positive or negative chemotaxis based on sensing by one or more C. elegans chemosensory neurons.
Valence depends on the integration of multiple signaling pathways
To learn more about how worms encode the valence of chemical cues, we analyzed responses in single mutants lacking either TAX-4 or OSM-9, the ion channel effectors responsible for chemosensory transduction (reviewed in [4]). Responses to more than half of the tested SMs were disrupted in both single of the mutants, indicating that behavioral valence most often reflects the integration of multiple chemosensory transduction pathways. Consistent with this inference, well-characterized attractants and repellents modulate calcium signaling in multiple chemosensory neurons [78]. Notably, response valence was inverted in single tax-4 or osm-9 mutants compared to wild-type in more than one-third (10 of 27) of the tested SMs. This findings indicate that the integrated behavior depends on both ion channel effectors (Figure 7). For instance, we found that phytol repels wild-type C. elegans but attracts tax-4 single mutants. Phytol has no detectable effect on either osm-9 mutants or tax-4;osm-9 double mutants. We observed an analogous response pattern for acetophenone, which attracts wild-type C. elegans, repels tax-4 single mutants, and has little or no effect on osm-9 single mutants and tax-4;osm-9 double mutants. Thus, with respect to the response valence to phytol and acetophenone, osm-9 is epistatic to tax-4. In other words, wild-type avoidance of phytol (or attraction to acetophenone) depends on an osm-9-dependent avoidance (or attraction) signal that supersedes a tax-4-dependent attraction (avoidance) signal. Such a complex mechanism for molecular encoding of chemosensory valence could enable plasticity based on the worm’s physiological or developmental state or the presence of these chemicals in complex mixtures. The scope of our screen reveals that complex encoding of behavioral valence is not rare, results that are aligned with calcium imaging studies of the responses to chemical cues [79] and suggest that studies examining panels of chemical cues will be needed to fully decipher how behavioral valence is encoded.
Some plant SMs detected by C. elegans are chemical cues for other nematodes and arthropods
Several of the 37 SMs are synthesized by additional organisms or known to affect other nematode species. For instance, 2-methyl-1-butanol is produced by bacteria, yeast, and a variety of plants [80]. It is also used by the nematode-eating fungus, Arthobotrys oligospora [60], to attract nematodes and as a sex pheromone in longhorn beetles [62]. Thus, this simple compound is a multifunctional chemical cue in nature and likely functions as a ligand for receptors present in multiple phyla. Whether or not the receptors themselves are conserved is an open question. Spinosad, a mixture of two complex macrocyclic lactones, is also produced by bacteria and is approved for use as an insecticide in purified form [81]. Our findings indicate that C. elegans is attracted to spinosad, although whether or not it is toxic to nematodes remains to be determined. Nevertheless, this finding suggests that the use of spinosad as an insecticide may have unintended consequences for nematode communities. Furfural, which attracts wild-type C. elegans, has been tested as a tool for managing Meloidogyne incognita [68,69], a root knot parasitic nematode that is a serious threat to agriculture. Phytol and methyl palmitate are other SMs in our collection that repel both C. elegans and root knot nematodes [73,82]. Camphor repels C. elegans (Figure 5), but attracts root knot nematodes [83]. Thus, sensitivity to some plant SMs is conserved among nematodes and might be exploited by their predators or hosts in nature. These findings also highlight the potential using C. elegans as a tool to screen for natural products that may aid in managing parasitic nematodes.
Twelve plant SMs detected by C. elegans are ligands for human GPCRs or ion channels
Numerous precedents suggest that plant SMs include ligands for G-protein coupled receptors (GPCRs) in C. elegans and humans. For instance, morphine, which is synthesized by the opium poppy, activates GPCRs in humans [84] and in C. elegans [85]. Consistent with this precedent and in spite of the limited sequence conservation within the repertoire of GPCR genes in C. elegans and humans [86], eight plant SMs that evoke C. elegans chemotaxis are also listed as ligands for human GPCRs in on-line databases [87,88]: acetophenone, anisole, camphor, cinnamyl alcohol, ellagic acid, methyl palmitate, oleanolic acid, ursolic acid. Acetophenone activates 11 human olfactory GPCRs [89–93] and 78 mouse olfactory GPCRs [94–97]. These GPCRs share a set of residues predicted to form the orthosteric binding pocket for acetophenone, but the proteins themselves are not otherwise considered orthologs or paralogs [98]. Our finding that acetophenone attracts wild-type C. elegans and repels tax-4 mutants (Figures 6, 7) indicates that there are also at least two acetophenone receptors in C. elegans. It is tempting to speculate that, regardless of the animals producing them, binding pockets in acetophenone receptors are structurally conserved. Three other plant SMs, anisole, camphor, and cinnamyl alcohol, also activate human olfactory GPCRs [89,99,100]. The repellents, ellagic acid and methyl palmitate, activate human GPR35 and the CB1/2 receptors [101,102], respectively, and oleanolic acid and ursolic acid both activate GPBAR1 [103,104]. Thus, the ability to detect and respond to individual plant SMs is conserved among animals as distantly related as nematodes, rodents, and humans. It is tempting to speculate that this shared ability depends upon receptors bearing structurally similar ligand binding pockets.
It remains to be determined if plant SM-evoked nematode attraction and repulsion is mediated primarily or exclusively by GPCRs, although at least one well-characterized attractant, diacetyl, has been linked to two GPCR genes [31,105] and responses to several other chemical cues depend on one or more G proteins expressed in chemosensory neurons [106]. GPCRs are not the only potential receptors for plant SMs, however. Several plant SMs that evoke attraction or repulsion also modulate ion channel function directly. Spinosad, which repels C. elegans (Figure 5) and is emerging as an agricultural insecticide [81,107]. Huperazine A, which is a C. elegans attractant, also modulates nAChR as well as ionotropic glutamate receptors [108]. Camphor, a weak repellant, is a well-characterized agonist for TRPV3 channels [109] and limonin, a weak attractant, blocks the human TMEM16A calcium-activated chloride channel [110]. Thus, nearly one-third of the plant SMs identified here as either C. elegans attractants or repellents also bind to one or more membrane proteins in mammals, including humans. These compounds comprise more than 10% of the library that we screened and these findings suggest that further screening is likely to yield additional ligands for membrane proteins in C. elegans and humans.
Limitations and future research
Chemical cues are widespread in nature and used by most, if not all animals to locate food and avoid harm. The platform we developed enables researchers to reliably and efficiently determine and quantify the valence of a library of chemical compounds presented to C. elegans nematodes. The platform is reliable, as evidenced in our ability to reproduce results from prior studies (Figure 2). The platform is simple, delivering all test compounds at a single concentration. This design choice limits the inferences that we might draw regarding response strength and might result in a failure to detect some bona fide responses. It might also affect response valence, since some chemical cues are attractive to wild-type C. elegans at low concentrations and repulsive at higher ones [30–32]. Thus, it is possible that our screen design omits some chemical cues or inverts responses to others. However, the large proportion of chemoactive compounds (41%) and the preponderance of attractants (73%) among chemoactive compounds suggest that these scenarios rarely occur in practice.
How does response valence emerge from chemosensory transduction? For many chemical cues studied here and elsewhere, chemotaxis behavior engages overlapping sets of chemosensory neurons and depends on dual chemosensory transduction pathways. For instance, the classical attractants isoamyl alcohol and diacetyl activate ASG and ASJ, respectively, and both chemicals activate AWA, AWB, AWC, ASE, and ASH [78]. Loss of tax-4 impairs attraction to isoamyl alcohol and enhances attraction to diacetyl (Figure 6). Conversely, loss of osm-9 has little impact on attraction to isoamyl alcohol and reduces attraction to diacetyl (Figure 6). From these findings, we infer that these two attractants are detected by distinct molecular signaling pathways and their presence is transformed into action based on signals generated by distinct, but overlapping sets of chemosensory neurons — despite their shared valence. Notably, these sets of neurons are not uniquely activated by attractants. Indeed, all of the chemosensory neurons activated by isoamyl alcohol and diacetyl are also activated by the classical repellent, 1-octanol [76]. Avoidance of 1-octanol depends primarily on osm-9-dependent signaling (Figure 6), even though osm-9 expression is not evident in all of the 1-octanol-sensitive chemosensory neurons. Derived from analyzing behavior (this study) and neural signaling across multiple chemical cues [78], these observations strongly imply that response valence is encoded at the ensemble level across chemosensory neurons and suggest an important role for signal integration within individual neurons co-expressing the key ion channel effectors.
The platform design is compatible with any chemical library and with most, if not all nematode species. Applicable nematodes include both lab-reared and wild C. elegans strains and other free-living nematodes, as well as parasites of plants and animals. Thus, this platform could be adapted to support discovery of chemical tools for control of parasitic nematodes or chemical actuators of the nervous system. Indeed, five of the chemoactive compounds studied here (~13%) are also listed in the Inxight Drugs portal [111] as relevant to neurological disease: carnosol, huperizine A, leonurine, l-mimosine, acetophenone, paeoniflorin. Whereas this study and many others primarily evaluate responses to pure compounds, natural chemical cues are present in complex mixtures. However, this experimental workflow is readily extended to experimenter-defined mixtures, to extracts of natural products obtained from plants, fungi, and bacteria, or even to small colonies of microorganisms. Given that it can be used to efficiently determine valence and response strength, the control of parasitic nematodes may be an especially valuable future application. With advanced liquid handling, it would become practical to determine the chemical valence exhibited by several nematode species or a collection of C. elegans strains in parallel. These tools would enable the simultaneous evaluation of responses of divergent nematode strains to a common chemical library, and make it possible to evaluate the co-variance of valence encoding and genetic variation. Combining this approach with advances in high-throughput tracking of freely-moving animals and imaging chemosensory neuron responses would deepen understanding of whether or not similarities in response valence reflect the engagement of a common sensorimotor encoding.
Supplementary Material
Table S1: Screening library. List of small molecules comprising the curated screening library, including CAS registry number (aka CAS No.), common name used in this study, vendor, and catalog number. Vendors are (alphabetical order): Ambeed, Inc. Arlington Heights, IL; Aobious, Inc., Gloucester, MA; Cayman Chemical, Ann Arbor, MI; Chem-Impex, Woodale, IL; MCE = MedChemExpress, Monmouth Junction, NJ; Sigma-Aldrich, St. Louis, MO; TargetMol, Boston, MA; TCI = TCI America, Portland, OR; VWR International, Radnor, PA.
Table S2: Responses of wild-type C. elegans to a curated chemical library. Tabulated list of the difference of the mean position between each test condition and a reference condition (aka “mean difference”), 95% confidence intervals for the mean difference, and statistical testing. Mean differences and confidence intervals obtained by bootstrapping using the Dabest statistical package ( Ho, et al., Nat Methods 16, 565–566 (2019). https://doi.org/10.1038/s41592-019-0470-3). All data are from assays conducted with wild-type (N2, Bristol) adult worms.
Table S3: Responses to chemoactive compounds as a function of genotype. Tabulated list of the difference of the mean position between each test condition and a reference condition (aka “mean difference”), 95% confidence intervals for the mean difference, and statistical testing. Mean differences and confidence intervals obtained by bootstrapping via the Dabest statistical package ( Ho, et al., Nat Methods 16, 565–566 (2019). https://doi.org/10.1038/s41592-019-0470-3). Results are grouped by the strain tested (top to bottom): peach, PR678 tax-4(p678); blue, CX10, osm-9(ky10); gold, GN1077 tax-4(p678);osm-9(ky10); green, N2 Bristol.
Figure S1: Schematic workflow for high-throughput C. elegans chemotaxis screening platform. Graphical timeline (minutes) illustrating time points (encircled) in assay workflow. Assay handling steps are described on the left side, worm handling steps are on the right side.
Figure S2: Schematic showing integrated data management and image analysis. Data management (left) and image analysis (right) for the screens occur simultaneously, reducing data processing time, reducing data processing errors, and increasing reproducibility.
Acknowledgements
We thank J. Casar, A. Das, and L. O’Connell for contributing to the prototyping team; C. Jaisinghani for assistance with genetics; S. R. Lockery for suggesting foam sheets to define behavioral arenas; J. A. Franco and L. Seyahi for assistance with data management and visualization, and Z. Liao for research & safety management. We also thank the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for C. elegans strains.
Funding
Wu Tsai Neuroscience Institutes ‘Big Ideas’ (MBG, TRC, SYR)
Wu Tsai Neuroscience Institutes Research Accelerator (MBG, TRC, SYR)
Chan-Zuckerberg BioHub Investigatorship (TRC)
National Institutes of Health grant R35NS10502-03S1 (MBG)
National Science Foundation grant IOS-1546838 (SYR)
National Institutes of Health fellowship, F31NS100318 (ALN)
Stanford Rise (LR, SG)
Stanford BioX Interdisciplinary Fellowship (LR)
Molecular Pharmacology Training Grant, T32GM113854 (LR)
Stanford Neuroscience Undergraduate Research Opportunity (NeURO) fellowship (HF)
Footnotes
Competing interests
Authors declare that they have no competing interests.
Data and materials availability
All processed data are available in the main text or the supplementary materials. Code used for all analyses and plots are publicly available on GitHub at https://github.com/wormsenseLab.
References
- 1.Manoel D, Makhlouf M, Arayata CJ, Sathappan A, Da’as S, Abdelrahman D, et al. Deconstructing the mouse olfactory percept through an ethological atlas. Curr Biol. 2021;31: 2809–2818.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haddad R, Weiss T, Khan R, Nadler B, Mandairon N, Bensafi M, et al. Global Features of Neural Activity in the Olfactory System Form a Parallel Code That Predicts Olfactory Behavior and Perception. Journal of Neuroscience. 2010. pp. 9017–9026. doi: 10.1523/jneurosci.0398-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeshurun Y, Sobel N. An odor is not worth a thousand words: from multidimensional odors to unidimensional odor objects. Annu Rev Psychol. 2010;61: 219–41, C1–5. [DOI] [PubMed] [Google Scholar]
- 4.Ferkey DM, Sengupta P, L’Etoile ND. Chemosensory signal transduction in Caenorhabditis elegans. Genetics. 2021;217. doi: 10.1093/genetics/iyab004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bargmann CI. Chemosensation in C. elegans. WormBook. 2006; 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19: 9557–9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanimoto Y, Yamazoe-Umemoto A, Fujita K, Kawazoe Y, Miyanishi Y, Yamazaki SJ, et al. Calcium dynamics regulating the timing of decision-making in C. elegans. Elife. 2017;6. doi: 10.7554/eLife.21629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91: 161–169. [DOI] [PubMed] [Google Scholar]
- 9.Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidal B, Aghayeva U, Sun H, Wang C, Glenwinkel L, Bayer EA, et al. An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 2018;16: e2004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, et al. Molecular topography of an entire nervous system. Cell. 2021;184: 4329–4347.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Insel PA, Sriram K, Gorr MW, Wiley SZ, Michkov A, Salmerón C, et al. GPCRomics: An Approach to Discover GPCR Drug Targets. Trends Pharmacol Sci. 2019;40: 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17: 707–718. [DOI] [PubMed] [Google Scholar]
- 14.Colbert HA, Smith TL, Bargmann CI. OSM-9, A Novel Protein with Structural Similarity to Channels, Is Required for Olfaction, Mechanosensation, and Olfactory Adaptation inCaenorhabditis elegans. J Neurosci. 1997;17: 8259–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Hoogen J, Geisen S, Routh D, Ferris H, Traunspurger W, Wardle DA, et al. Soil nematode abundance and functional group composition at a global scale. Nature. 2019;572: 194–198. [DOI] [PubMed] [Google Scholar]
- 16.Cook DC, Zdraljevic S, Tanny RE, Seo B, Riccardi DD, Noble LM, et al. The Genetic Basis of Natural Variation in Caenorhabditis elegans Telomere Length. Genetics. 2016; genetics.116.191148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmann T. From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry. 2007;68: 2831–2846. [DOI] [PubMed] [Google Scholar]
- 19.Margie O, Palmer C, Chin-Sang I. C. elegans chemotaxis assay. J Vis Exp. 2013; e50069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behavior Hart A.. WormBook. 2006. Available: http://www.wormbook.org/chapters/www_behavior/behavior.html [Google Scholar]
- 21.Stroustrup N, Ulmschneider BE, Nash ZM, López-Moyado IF, Apfeld J, Fontana W. The Caenorhabditis elegans Lifespan Machine. Nat Methods. 2013;10: 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Churgin MA, Jung S-K, Yu C-C, Chen X, Raizen DM, Fang-Yen C. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife. 2017;6. doi: 10.7554/eLife.26652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockery SR, Hulme SE, Roberts WM, Robinson KJ, Laromaine A, Lindsay TH, et al. A microfluidic device for whole-animal drug screening using electrophysiological measures in the nematode C. elegans. Lab Chip. 2012;12: 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahnel SR, Roberts WM, Heisler I, Kulke D, Weeks JC. Comparison of electrophysiological and motility assays to study anthelmintic effects in Caenorhabditis elegans. Int J Parasitol Drugs Drug Resist. 2021;16: 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7: 729–742. [DOI] [PubMed] [Google Scholar]
- 26.Stiernagle T. Maintenance of C. elegans. In: The C. elegans Research Community, editor. Wormbook. 2006. Available: doi/ 10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [Google Scholar]
- 27.Lee S-H, Kim D-S, Park S-H, Phytochemistry Park H. and Applications of Cinnamomum camphora Essential Oils. Molecules. 2022;27. doi: 10.3390/molecules27092695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eksi G, Kurbanoglu S, Erdem SA. Chapter 9 - Analysis of diterpenes and diterpenoids. In: Sanches Silva A, Nabavi SF, Saeedi M, Nabavi SM, editors. Recent Advances in Natural Products Analysis. Elsevier; 2020. pp. 313–345. [Google Scholar]
- 29.Qian L, Xu Z, Zhang W, Wilson B, Hong J-S, Flood PM. Sinomenine, a natural dextrorotatory morphinan analog, is anti-inflammatory and neuroprotective through inhibition of microglial NADPH oxidase. J Neuroinflammation. 2007;4: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng H, Liu Y, Xue Y, Shao J, Tan Z, Liu S, et al. Molecular Strategies for Intensity-Dependent Olfactory Processing in. Front Mol Neurosci. 2021;14: 748214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi G, Uozumi T, Kiriyama K, Kamizaki T, Hirotsu T. Screening of odor-receptor pairs in Caenorhabditis elegans reveals different receptors for high and low odor concentrations. Sci Signal. 2014;7: ra39. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida K, Hirotsu T, Tagawa T, Oda S, Wakabayashi T, Iino Y, et al. Odour concentration-dependent olfactory preference change in C. elegans. Nat Commun. 2012;3: 739. [DOI] [PubMed] [Google Scholar]
- 33.Katzen A, Chung H-K, Harbaugh WT, Della Iacono C, Jackson N, Yu SK, et al. The nematode worm C. elegans chooses between bacterial foods exactly as if maximizing economic utility. bioRxiv. 2021. p. 2021.April.25.441352. doi: 10.1101/2021.04.25.441352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim JP, Fehlauer H, Das A, Saro G, Glauser DA, Brunet A, et al. Loss of CaMKI Function Disrupts Salt Aversive Learning in C. elegans. J Neurosci. 2018;38: 6114–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boothe T, Hilbert L, Heide M, Berninger L, Huttner WB, Zaburdaev V, et al. A tunable refractive index matching medium for live imaging cells, tissues and model organisms. Elife. 2017;6. doi: 10.7554/eLife.27240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ford T, Graham J, Rickwood D. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal Biochem. 1994;220: 360–366. [DOI] [PubMed] [Google Scholar]
- 37.Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6: 973–985. [DOI] [PubMed] [Google Scholar]
- 38.Nekimken AL, Pruitt BL, Goodman MB. Touch-induced mechanical strain in somatosensory neurons is independent of extracellular matrix mutations in Caenorhabditis elegans. Mol Biol Cell. 2020;31: 1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, et al. scikit-image: image processing in Python. PeerJ. 2014;2: e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern. 1979;9: 62–66. [Google Scholar]
- 41.Wählby C, Kamentsky L, Liu ZH, Riklin-Raviv T, Conery AL, O’Rourke EJ, et al. An image analysis toolbox for high-throughput C. elegans assays. Nat Methods. 2012;9: 714–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nat Methods. 2019;16: 565–566. [DOI] [PubMed] [Google Scholar]
- 44.Calin-Jageman RJ, Cumming G. Estimation for Better Inference in Neuroscience. eNeuro. 2019. doi: 10.1523/ENEURO.0205-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro SS, Wilk MB. An Analysis of Variance Test for Normality (Complete Samples). Biometrika. 1965. p. 591. doi: 10.2307/2333709 [DOI] [Google Scholar]
- 46.Kruskal WH, Wallis WA. Use of Ranks in One-Criterion Variance Analysis. J Am Stat Assoc. 1952;47: 583–621. [Google Scholar]
- 47.Mann HB, Whitney DR. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann Math Stat. 1947;18: 50–60. [Google Scholar]
- 48.van Dam NM, Bouwmeester HJ. Metabolomics in the Rhizosphere: Tapping into Belowground Chemical Communication. Trends Plant Sci. 2016;21: 256–265. [DOI] [PubMed] [Google Scholar]
- 49.Rasmann S, Ali JG, Helder J, van der Putten WH. Ecology and evolution of soil nematode chemotaxis. J Chem Ecol. 2012;38: 615–628. [DOI] [PubMed] [Google Scholar]
- 50.Čepulytė R, Buda V. Toward Chemical Ecology of Plant-Parasitic Nematodes: Kairomones, Pheromones, and Other Behaviorally Active Chemical Compounds. J Agric Food Chem. 2022;70: 1367–1390. [DOI] [PubMed] [Google Scholar]
- 51.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94: 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bargmann CI. Chemosensation in C. elegans. WormBook. 2006; 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shungu D, Valiant M, Tutlane V, Weinberg E, Weissberger B, Koupal L, et al. GELRITE as an Agar Substitute in Bacteriological Media. Appl Environ Microbiol. 1983;46: 840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veramendi J, Villafranca MJ, Sota V, Mingo-Castel AM. Gelrite as an alternative to agar for micropropagation and microtuberization of Solanum tuberosum L. cv. Baraka. In Vitro Cellular & Developmental Biology - Plant. 1997;33: 195–199. [Google Scholar]
- 55.Ramot D, Johnson BE, Berry TL, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One. 2008;3: e2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crombie TA, Chikuturudzi C, Cook DE, Andersen EC. An automated approach to quantify chemotaxis index in C. elegans. MicroPubl Biol. 2022;2022. doi: 10.17912/micropub.biology.000567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Halloran DM. NemaCount: quantification of nematode chemotaxis behavior in a browser. Invert Neurosci. 2016;16: 5. [DOI] [PubMed] [Google Scholar]
- 58.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74: 515–527. [DOI] [PubMed] [Google Scholar]
- 59.Behavior Hart A.. WormBook. 2006. doi: 10.1895/wormbook.1.87.1 [DOI] [Google Scholar]
- 60.Hsueh Y-P, Gronquist MR, Schwarz EM, Nath RD, Lee C-H, Gharib S, et al. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. Elife. 2017;6. doi: 10.7554/eLife.20023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Worthy SE, Haynes L, Chambers M, Bethune D, Kan E, Chung K, et al. Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PLoS One. 2018;13: e0201158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molander MA, Larsson MC. Identification of the Aggregation-sex Pheromone of the Cerambycid Beetle Phymatodes pusillus ssp. pusillus and Evidence of a Synergistic Effect from a Heterospecific Pheromone Component. J Chem Ecol. 2018;44: 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sathya S, Shanmuganathan B, Balasubramaniam B, Balamurugan K, Devi KP. Phytol loaded PLGA nanoparticles regulate the expression of Alzheimer’s related genes and neuronal apoptosis against amyloid-β induced toxicity in Neuro-2a cells and transgenic Caenorhabditis elegans. Food Chem Toxicol. 2020;136: 110962. [DOI] [PubMed] [Google Scholar]
- 64.Zheng J, Heber D, Wang M, Gao C, Heymsfield SB, Martin RJ, et al. Pomegranate juice and extract extended lifespan and reduced intestinal fat deposition in Caenorhabditis elegans. Int J Vitam Nutr Res. 2017;87: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu F-L, Li W-H, Yu C-W, Hsieh Y-C, Yang Y-F, Liu J-T, et al. In vivo antioxidant activities of essential oils and their constituents from leaves of the Taiwanese Cinnamomum osmophloeum. J Agric Food Chem. 2012;60: 3092–3097. [DOI] [PubMed] [Google Scholar]
- 66.Sanadhya P, Bucki P, Liarzi O, Ezra D, Gamliel A, Braun Miyara S. Caenorhabditis elegans susceptibility to Daldinia cf. concentrica bioactive volatiles is coupled with expression activation of the stress-response transcription factor daf-16, a part of distinct nematicidal action. PLoS One. 2018;13: e0196870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chakrabarti S, Dicke C, Kalderis D, Kern J. Rice husks and their hydrochars cause unexpected stress response in the nematode Caenorhabditis elegans: reduced transcription of stress-related genes. Environ Sci Pollut Res Int. 2015;22: 12092–12103. [DOI] [PubMed] [Google Scholar]
- 68.Cheng W, Yang X, Xue H, Huang D, Cai M, Huang F, et al. Reproductive Toxicity of Furfural Acetone in Meloidogyne incognita and Caenorhabditis elegans. Cells. 2022;11. doi: 10.3390/cells11030401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Impact of direct and indirect application of rising furfural concentrations on viability, infectivity and reproduction of the root-knot nematode, Meloidogyne incognita in Pisum sativum. Microb Pathog. 2016;96: 26–34. [DOI] [PubMed] [Google Scholar]
- 70.Sivasankara Pillai S, Dandurand L-M. Effect of Steroidal Glycoalkaloids on Hatch and Reproduction of the Potato Cyst Nematode Globodera pallida. Plant Dis. 2021;105: 2975–2980. [DOI] [PubMed] [Google Scholar]
- 71.Kirwa HK, Murungi LK, Beck JJ, Torto B. Elicitation of Differential Responses in the Root-Knot Nematode Meloidogyne incognita to Tomato Root Exudate Cytokinin, Flavonoids, and Alkaloids. J Agric Food Chem. 2018;66: 11291–11300. [DOI] [PubMed] [Google Scholar]
- 72.Ochola J, Cortada L, Ng’ang’a M, Hassanali A, Coyne D, Torto B. Mediation of Potato-Potato Cyst Nematode, G. rostochiensis Interaction by Specific Root Exudate Compounds. Front Plant Sci. 2020;11: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujimoto T, Abe H, Mizukubo T, Seo S. Phytol, a Constituent of Chlorophyll, Induces Root-Knot Nematode Resistance in Arabidopsis via the Ethylene Signaling Pathway. Mol Plant Microbe Interact. 2021;34: 279–285. [DOI] [PubMed] [Google Scholar]
- 74.Dusenbery DB, Sheridan RE, Russell RL. Chemotaxis-defective mutants of the nematode Caenorhabditis elegans. Genetics. 1975;80: 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14: 803–812. [DOI] [PubMed] [Google Scholar]
- 76.Lin A, Qin S, Casademunt H, Wu M, Hung W, Cain G, et al. Functional imaging and quantification of multi-neuronal olfactory responses in C. elegans. bioRxiv. 2022. p. 2022.May.27.493772. doi: 10.1101/2022.05.27.493772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Queirós L, Marques C, Pereira JL, Gonçalves FJM, Aschner M, Pereira P. Overview of Chemotaxis Behavior Assays in Caenorhabditis elegans. Curr Protoc. 2021;1: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin A, Qin S, Casademunt H, Wu M, Hung W, Cain G, et al. Functional imaging and quantification of multineuronal olfactory responses in C. elegans. Sci Adv. 2023;9: eade1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin A, Qin S, Casademunt H, Wu M, Hung W, Cain G, et al. Functional imaging and quantification of multi-neuronal olfactory responses in C. elegans. Sci Adv. 2022. p. 2022.May.27.493772. doi: 10.1101/2022.05.27.493772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.PubChem. [No title]. [cited 7 Dec 2022]. Available: https://pubchem.ncbi.nlm.nih.gov/compound/2-Methyl-1-butanol
- 81.Kirst HA. The spinosyn family of insecticides: realizing the potential of natural products research. J Antibiot. 2010;63: 101–111. [DOI] [PubMed] [Google Scholar]
- 82.Lu Q, Liu T, Wang N, Dou Z, Wang K, Zuo Y. Nematicidal Effect of Methyl Palmitate and Methyl Stearate against Meloidogyne incognita in Bananas. J Agric Food Chem. 2020;68: 6502–6510. [DOI] [PubMed] [Google Scholar]
- 83.Mwamba S, Kihika-Opanda R, Murungi LK, Losenge T, Beck JJ, Torto B. Identification of Repellents from Four Non-Host Asteraceae Plants for the Root Knot Nematode, Meloidogyne incognita. J Agric Food Chem. 2021;69: 15145–15156. [DOI] [PubMed] [Google Scholar]
- 84.Chen Y, Mestek A, Liu J, Hurley JA, Yu L. Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Mol Pharmacol. 1993;44: 8–12. [PubMed] [Google Scholar]
- 85.Cheong MC, Artyukhin AB, You Y-J, Avery L. An opioid-like system regulating feeding behavior in C. elegans. Elife. 2015;4. doi: 10.7554/eLife.06683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robertson HM, Thomas JH. The putative chemoreceptor families of C. elegans. WormBook. 2006; 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma A, Saha BK, Kumar R, Varadwaj PK. OlfactionBase: a repository to explore odors, odorants, olfactory receptors and odorant-receptor interactions. Nucleic Acids Res. 2022;50: D678–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan WKB, Zhang H, Yang J, Brender JR, Hur J, Özgür A, et al. GLASS: a comprehensive database for experimentally validated GPCR-ligand associations. Bioinformatics. 2015;31: 3035–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanz G, Schlegel C, Pernollet J-C, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem Senses. 2005;30: 69–80. [DOI] [PubMed] [Google Scholar]
- 90.Matarazzo V, Clot-Faybesse O, Marcet B, Guiraudie-Capraz G, Atanasova B, Devauchelle G, et al. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses. 2005;30: 195–207. [DOI] [PubMed] [Google Scholar]
- 91.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2: ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mainland JD, Li YR, Zhou T, Liu WLL, Matsunami H. Human olfactory receptor responses to odorants. Sci Data. 2015;2: 150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Audouze K, Tromelin A, Le Bon AM, Belloir C, Petersen RK, Kristiansen K, et al. Identification of odorant-receptor interactions by global mapping of the human odorome. PLoS One. 2014;9: e93037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunami H. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat Neurosci. 2015;18: 1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kida H, Fukutani Y, Mainland JD, de March CA, Vihani A, Li YR, et al. Vapor detection and discrimination with a panel of odorant receptors. Nat Commun. 2018;9: 4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu Y, de March CA, Ni MJ, Adipietro KA, Golebiowski J, Matsunami H, et al. Responsiveness of G protein-coupled odorant receptors is partially attributed to the activation mechanism. Proc Natl Acad Sci U S A. 2015;112: 14966–14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones EM, Jajoo R, Cancilla D, Lubock NB, Wang J, Satyadi M, et al. A Scalable, Multiplexed Assay for Decoding GPCR-Ligand Interactions with RNA Sequencing. Cell Syst. 2019;8: 254–260.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cong X, Ren W, Pacalon J, Xu R, Xu L, Li X, et al. Large-Scale G Protein-Coupled Olfactory Receptor-Ligand Pairing. ACS Cent Sci. 2022;8: 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ieki T, Yamanaka Y, Yoshikawa K. Functional analysis of human olfactory receptors with a high basal activity using LNCaP cell line. PLoS One. 2022;17: e0267356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Veithen A, Wilin F, Philippeau M, Chatelain P. OR1D2 is a broadly tuned human olfactory receptor. Chemical Senses. OXFORD UNIV PRESS GREAT CLARENDON ST, OXFORD OX2 6DP, ENGLAND; 2015. pp. 262–263. [Google Scholar]
- 101.Deng H, Hu H, Ling S, Ferrie AM, Fang Y. Discovery of Natural Phenols as G Protein-Coupled Receptor-35 (GPR35) Agonists. ACS Med Chem Lett. 2012;3: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vandevoorde S, Tsuboi K, Ueda N, Jonsson K-O, Fowler CJ, Lambert DM. Esters, retroesters, and a retroamide of palmitic acid: pool for the first selective inhibitors of N-palmitoylethanolamine-selective acid amidase. J Med Chem. 2003;46: 4373–4376. [DOI] [PubMed] [Google Scholar]
- 103.Genet C, Strehle A, Schmidt C, Boudjelal G, Lobstein A, Schoonjans K, et al. Structure-activity relationship study of betulinic acid, a novel and selective TGR5 agonist, and its synthetic derivatives: potential impact in diabetes. J Med Chem. 2010;53: 178–190. [DOI] [PubMed] [Google Scholar]
- 104.Sato H, Genet C, Strehle A, Thomas C, Lobstein A, Wagner A, et al. Anti-hyperglycemic activity of a TGR5 agonist isolated from Olea europaea. Biochem Biophys Res Commun. 2007;362: 793–798. [DOI] [PubMed] [Google Scholar]
- 105.Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84: 899–909. [DOI] [PubMed] [Google Scholar]
- 106.Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21: 414–419. [DOI] [PubMed] [Google Scholar]
- 107.Sparks TC, Crouse GD, Benko Z, Demeter D, Giampietro NC, Lambert W, et al. The spinosyns, spinosad, spinetoram, and synthetic spinosyn mimics - discovery, exploration, and evolution of a natural product chemistry and the impact of computational tools. Pest Manag Sci. 2021;77: 3637–3649. [DOI] [PubMed] [Google Scholar]
- 108.Gordon RK, Nigam SV, Weitz JA, Dave JR, Doctor BP, Ved HS. The NMDA receptor ion channel: a site for binding of Huperzine A. J Appl Toxicol. 2001;21 Suppl 1: S47–51. [DOI] [PubMed] [Google Scholar]
- 109.Moqrich A, Hwang SW, Earley TJ, Petrus MJ, Murray AN, Spencer KSR, et al. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science. 2005;307: 1468–1472. [DOI] [PubMed] [Google Scholar]
- 110.Chang L, Chang R, Shen J, Wang Y, Song H, Kang X, et al. Self-healing pectin/cellulose hydrogel loaded with limonin as TMEM16A inhibitor for lung adenocarcinoma treatment. Int J Biol Macromol. 2022;219: 754–766. [DOI] [PubMed] [Google Scholar]
- 111.Siramshetty VB, Grishagin I, Nguyễn Ð-T, Peryea T, Skovpen Y, Stroganov O, et al. NCATS Inxight Drugs: a comprehensive and curated portal for translational research. Nucleic Acids Res. 2022;50: D1307–D1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Screening library. List of small molecules comprising the curated screening library, including CAS registry number (aka CAS No.), common name used in this study, vendor, and catalog number. Vendors are (alphabetical order): Ambeed, Inc. Arlington Heights, IL; Aobious, Inc., Gloucester, MA; Cayman Chemical, Ann Arbor, MI; Chem-Impex, Woodale, IL; MCE = MedChemExpress, Monmouth Junction, NJ; Sigma-Aldrich, St. Louis, MO; TargetMol, Boston, MA; TCI = TCI America, Portland, OR; VWR International, Radnor, PA.
Table S2: Responses of wild-type C. elegans to a curated chemical library. Tabulated list of the difference of the mean position between each test condition and a reference condition (aka “mean difference”), 95% confidence intervals for the mean difference, and statistical testing. Mean differences and confidence intervals obtained by bootstrapping using the Dabest statistical package ( Ho, et al., Nat Methods 16, 565–566 (2019). https://doi.org/10.1038/s41592-019-0470-3). All data are from assays conducted with wild-type (N2, Bristol) adult worms.
Table S3: Responses to chemoactive compounds as a function of genotype. Tabulated list of the difference of the mean position between each test condition and a reference condition (aka “mean difference”), 95% confidence intervals for the mean difference, and statistical testing. Mean differences and confidence intervals obtained by bootstrapping via the Dabest statistical package ( Ho, et al., Nat Methods 16, 565–566 (2019). https://doi.org/10.1038/s41592-019-0470-3). Results are grouped by the strain tested (top to bottom): peach, PR678 tax-4(p678); blue, CX10, osm-9(ky10); gold, GN1077 tax-4(p678);osm-9(ky10); green, N2 Bristol.
Figure S1: Schematic workflow for high-throughput C. elegans chemotaxis screening platform. Graphical timeline (minutes) illustrating time points (encircled) in assay workflow. Assay handling steps are described on the left side, worm handling steps are on the right side.
Figure S2: Schematic showing integrated data management and image analysis. Data management (left) and image analysis (right) for the screens occur simultaneously, reducing data processing time, reducing data processing errors, and increasing reproducibility.
Data Availability Statement
All processed data are available in the main text or the supplementary materials. Code used for all analyses and plots are publicly available on GitHub at https://github.com/wormsenseLab.