Abstract
Decoding the gene regulatory mechanisms mediating self-renewal of hematopoietic stem cells (HSCs) during their amplification in the fetal liver (FL) is relevant for advancing therapeutic applications aiming to expand transplantable HSCs, a long-standing challenge. Here, to explore intrinsic and extrinsic regulation of self-renewal in FL-HSCs at the single cell level, we engineered a culture platform designed to recapitulate the FL endothelial niche, which supports the amplification of serially engraftable HSCs ex vivo. Leveraging this platform in combination with single cell index flow cytometry, serial transplantation assays, and single cell RNA-sequencing, we elucidated previously unrecognized heterogeneity in immunophenotypically defined FL-HSCs and demonstrated that differentiation latency and transcriptional signatures of biosynthetic dormancy are distinguishing properties of self-renewing FL-HSCs with capacity for serial, long-term multilineage hematopoietic reconstitution. Altogether, our findings provide key insights into HSC expansion and generate a novel resource for future exploration of the intrinsic and niche-derived signaling pathways that support FL-HSC self-renewal.
Introduction
Hematopoietic stem cells (HSCs) have the distinguishing properties of life-long self-renewal and the ability to reconstitute multilineage hematopoiesis upon transplantation. Understanding the biology of HSC development is essential to unlocking methods for de novo HSC generation and expansion to facilitate advances in hematopoietic stem cell transplantation, gene therapies, and disease modeling for blood and immune disorders.
Following their initial emergence in embryonic arteries, HSCs seed the fetal liver (FL), where they have conventionally been thought to both rapidly expand (generating sufficient HSCs to sustain life-long hematopoiesis in the adult) and differentiate (generating mature blood cells essential to support the developing embryo). To facilitate these processes, FL-HSCs have been postulated to exist in a state of proliferative and biosynthetic activation during prenatal development, followed by a transition to a state of quiescence upon their migration to the bone marrow (BM) perinatally (Bowie et al., 2007; Bowie et al., 2006).
Several elegant studies using methodologies to track the fate of nascent HSCs and progenitors through development have recently called for a revision of this classical paradigm of developmental hematopoiesis. These studies suggest that distinct waves of HSC-independent progenitors also seed the fetal liver and serve as the primary source of mature blood cells throughout prenatal development, with minimal contribution from HSCs, which are instead reserved to contribute primarily to adult hematopoiesis (Yokomizo et al., 2022). Further supporting this novel paradigm, we and others have uncovered distinct ontogenies for oligopotent and even multipotent hematopoietic progenitors that precede HSCs during various waves of endothelial to hematopoietic transition in embryonic blood vessels (Dignum et al., 2021; Ulloa et al., 2021; Yokomizo et al., 2022). While some HSC-independent progenitors possess properties that overlap with HSCs, such as multilineage hematopoietic activity, they are generally lacking in durable self-renewal capacity assayed by serial transplantation, the gold standard methodology for defining functional HSCs of relevance for translational applications.
Recent studies also suggest that the number of precursors emerging in the embryo that contribute to hematopoiesis in the adult when traced in vivo is substantially larger than the number of engraftable HSCs detected by direct transplantation assays (Ganuza et al., 2017; Pei et al., 2017) and that this compartment of long-lived hematopoietic precursors undergoes only limited expansion in the fetal liver (Ganuza et al., 2022). Lineage barcoding studies in vivo suggest significant heterogeneity in this pool of long-lived precursors in the FL, identifying a distinct population of embryonic multipotent progenitors (eMPPs) independent of HSC origin. Remarkably, eMPPs were found to contribute to mature blood cells in the adult when traced in situ, but lack long-term engraftment properties in transplantation assays essential to classify them as bona fide HSCs (Patel et al., 2022). In contrast, another study identified a subset of lymphoid-biased “developmentally-restricted HSCs” in the FL possessing long-term engraftment upon transplantation, yet failing to contribute to adult hematopoiesis in situ (Beaudin et al., 2016), in line with previous work identifying populations of FL-HSCs with distinct engraftment properties (Benz et al., 2012).
Altogether, these reports highlight substantial heterogeneity of the HSCs and HSCs-independent progenitors that seed the fetal liver in early gestation and contribute to various aspects of hematopoiesis during prenatal development and into adulthood. Though prior studies have identified surface markers that enable enrichment of engraftable HSCs by FACS to study their properties at a population level, uncovering the unique phenotypic, functional, and molecular properties of the rare subset with long-term, multilineage engraftment capacity requires studies at a single cell level. Furthermore, a better understanding of how functionally engraftable HSCs undergo maturation and self-renewal in the fetal liver niche could provide insight into engineering more robust protocols for generation of engraftable HSCs in vitro.
To address these issues at single cell level, we developed a serum-free culture platform that mimics the FL vascular niche, which enabled ex vivo amplification of serially engrafting HSCs from individual immunophenotypic FL-HSCs isolated by FACS at early and mid-gestation in the mouse embryo. We leveraged this platform to study the heterogeneity of the FL-HSC compartment by integrating flow cytometry, single cell RNA-sequencing (scRNAseq), and functional transplantation assays. Our studies elucidate previously unrecognized heterogeneity in immunophenotypic FL-HSCs, revealing that relative biosynthetic dormancy, propensity for symmetric self-renewal, and differentiation latency are key properties of serially engraftable FL-HSCs, which provide essential insight for future strategies for ex vivo amplification of HSCs.
Results
Establishment of a clonal HSC amplification platform reveals that serially engrafting FL-HSCs exhibit delayed colony formation, differentiation latency, and homogeneous expansion in vitro
Aiming to establish a platform to study FL-HSC properties at single cell resolution, we first set out to isolate the immunophenotypic population enriched in functional long-term engrafting HSC activity in the early (embryonic day (E) 13.5) and mid-gestation (E15.5/16.5) FL based on established HSC surface markers in AGM and FL (Che et al., 2022; Hadland et al., 2017; Iwasaki et al., 2010; McKinney-Freeman et al., 2009). Notably, SLAM marker CD150 is not a reliable marker of nascent HSCs in the early embryo until E14.5 FL stage(McKinney-Freeman et al., 2009); thus, we did not include this marker in our initial sorting strategy for E13.5 FL-HSCs. Using stringent gating for known FL-HSC markers, Endothelial Cell Protein C Receptor (EPCR) and Stem Cell Antigen-1 (SCA1), we found that CD45+DAPI−GR1−F4/80−SCA1highEPCRhigh immunophenotype (hereafter denoted SEhi) efficiently excluded cells expressing other lineage markers as well as CD48, allowing us to simplify the sorting methodology for immunophenotypic FL-HSCs (Fig. S1, A and B). As expected, this population provides long-term, multilineage hematopoietic engraftment following transplantation into congenic strain adult mice (Fig. S1C).
Next, to evaluate functional heterogeneity within the immunophenotypically defined FL-HSC fraction at the single cell level, we established a coculture platform recapitulating the FL endothelial niche (Fig. 1A). Briefly, FL-derived endothelial cells (ECs) were transduced with a lentivirus encoding constitutively active AKT1 (myristoylated AKT1; MyrAKT) (henceforth “FL-AKT-ECs”), which enables EC propagation in serum-free culture conditions while maintaining endogenous properties of the native EC of origin, as previously described (Hadland et al., 2015; Kobayashi et al., 2010). We first tested immunophenotypically defined HSCs isolated from E15.5/16.5 FL, a developmental stage when engrafting HSCs are relatively abundant in the FL based on limiting dilution transplantation assays and are enriched based on CD150 expression (Ema and Nakauchi, 2000; Kim et al., 2006). SEhiCD150+ FL-HSCs were individually index sorted into one well of a 96 well-plate plated with FL-AKT-ECs in serum-free media with limited cytokines (SCF and TPO). The formation of hematopoietic colonies in coculture was monitored visually over time by microscopy, and a portion of each colony was harvested by pipetting for flow cytometric analysis. Following coculture, most colonies contained a population of cells co-expressing EPCR and SCA1 (Fig. 1B), an in vitro immunophenotype previously shown to correlate with multilineage engraftment potential (Che et al., 2022; Hadland et al., 2017; Hadland et al., 2018), thus suggesting maintenance and expansion of HSCs during culture. The presence of the FL-AKT-EC niche is crucial for maintenance of immunophenotypic HSCs, as FL-HSCs cultured without FL-AKT-ECs generated colonies of cells largely lacking EPCR and SCA1 co-expression (not shown). Interestingly, we observed a spectrum of colonies containing various proportions of SCA1+EPCR+ cells following FL-AKT-EC coculture, some consisting nearly exclusively of SCA1+EPCR+ cells, suggesting symmetric self-renewal and delayed differentiation in the EC niche (Fig. 1B, left panel), and others containing increased proportions of more differentiated progeny lacking expression of EPCR and/or SCA1 (Fig. 1B, right panel). Interestingly, colonies emerging early in coculture (prior to day 12) were characterized by relatively lower percentages of SCA1+EPCR+ cells compared to those observed to emerge only after 12 days of coculture (Fig. S1D). These studies suggest significant heterogeneity even of immunophenotypically pure (CD150+SEhi) FL-HSCs when assayed at the single cell level in a developmentally relevant ex vivo niche, revealing intrinsic differences in proliferative potential and propensity for self-renewal verses differentiation in culture.
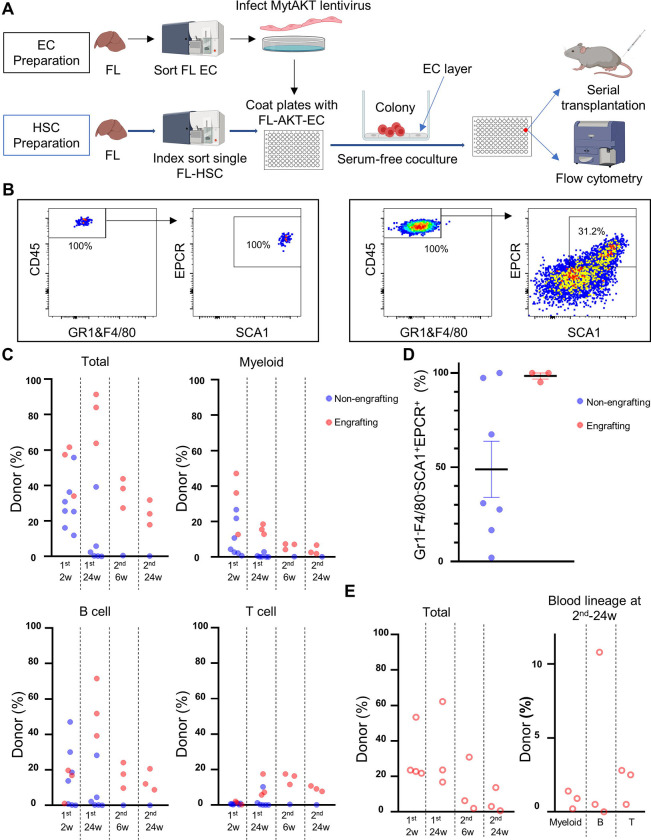
Figure 1: Establishment of a FL vascular niche platform supporting ex vivo amplification of clonal FL-HSCs.
(A) Overview of methodology. Sorted FL-derived ECs were infected with lentivirus encoding myristoylated AKT to generate FL-AKT-ECs. Single FL-HSCs were sorted into each well of a 96 well-plate coated with FL-AKT-EC in serum free media with hematopoietic cytokines (SCF, TPO). Following a period of coculture, emerging colonies were assessed by flow cytometry for immunophenotype and by serial transplantation into lethally irradiated mice to measure long-term hematopoietic engraftment (See Materials & Methods for details).
(B) Representative immunophenotypes of colonies emerging from single E16.5 SEhiCD150+ FL-HSC following coculture with FL-AKT-EC. Colonies were analyzed on day 15 by flow cytometry. Shown is expression of EPCR and SCA1 within cells gated as CD45+GR1−F4/80−, after exclusion of dead cells (DAPI+) and VECadherin+ FL-AKT-EC.
(C) Donor chimerism (total, myeloid, B cell, and T cell) in peripheral blood of primary and secondary recipients transplanted with the progeny of single E16.5 FL-HSCs following coculture with FL-AKT-ECs. 50% of cells from each colony was subject to flow cytometric analysis and a portion of the residual cells (25–50%) was used for transplantation (n=10).
(D) Frequency of GR1−F4/80−SCA1+EPCR+ cells (amongst total viable CD45+ cells) in individual colonies possessing serial multilineage engraftment (Engrafting, red) (n=3) or lacking serial multilineage engraftment (Non-engrafting, blue) (n=7).
(E) For one colony in Figure 1C/D, 25% of remaining cells were transplanted into multiple recipients (n=5, 5% per recipient) and serial multilineage engraftment was measured based on donor-derived peripheral blood.
Next, to characterize the engraftment properties of the progeny of FL-HSCs following coculture, we sorted single E15.5/16.5 SEhiCD150+ FL cells to FL-AKT-EC and harvested cells from individual colonies following coculture. Flow cytometry was performed using half of each culture well to assess immunophenotypic HSCs (Fig. 1B), and the remaining cells from each colony were transplanted into lethally irradiated mice. There was substantial heterogeneity in observed engraftment, with only a subset of colonies providing long-term, multilineage engraftment at 24 weeks after secondary transplantation (Fig. 1C). Notably, when plotted as a function of percentage of SCA1+EPCR+ cells, those colonies providing serial engraftment were exclusively from the subset that had near homogenous expression of SCA1+EPCR+ (Fig. 1D). Furthermore, when cells from a single SCA1+EPCR+-predominant colony were transplanted into multiple recipients, multilineage engraftment was observed in both primary and secondary recipients at 24 weeks post-transplant in all recipients (Fig. 1E), indicating the FL-AKT-EC niche was able to support expansion of serially engrafting HSCs from single FL-HSCs in vitro.
We next used the FL-AKT-EC coculture assay established above to examine the functional heterogeneity of SEhi FL-HSCs at E13.5. At this early stage, the FL has recently been seeded with nascent HSCs and HSC-independent progenitors emerging from hemogenic endothelium in embryonic vessels, and long-term engrafting HSCs are still relatively infrequent (Ema and Nakauchi, 2000). Similar to colonies derived from E15.5/16.5 CD150+SEhi FL-HSCs, only a minor subset of colonies from E13.5 SEhi FL-HSCs provided long-term donor engraftment in secondary transplants (Fig. 2A). In our series of transplants following single cell SEhi FL-HSC coculture with FL-AKT-EC, we retrospectively observed that colonies possessing serial multilineage engraftment consisted exclusively of those with high level (greater than 80%) SCA1+EPCR+ cells, although not all colonies meeting this criterion provided long-term engraftment (Fig. 1, C and D and Fig. 2, A and B). Based on this correlation between colony immunophenotype and engraftment properties, we classified “HSC colonies” to include only those with >80% of SCA1+EPCR+ cells and compared frequency of each colony type and the number of cells per colony type across multiple experiments at E13.5 and E15.5/16.5. HSC colonies had significantly lower number of cells following coculture at both developmental stages and were of relatively lower frequency amongst SEhi FL-HSCs examined at E13.5 compared with E15.5/16.5 (Fig. 2, C and D). Retrospectively examining the immunophenotype of index sorted SEhi FL-HSCs across experiments for expression of CD150 and ESAM (another established marker of FL and adult HSCs) (Yokota et al., 2009), we also observed the majority of HSC colonies originate from SEhi cells co-expressing ESAM and CD150 in the E15.5/16.5 FL, consistent with previous reports (Kim et al., 2006; McKinney-Freeman et al., 2009). However, E13.5 SEhi FL-HSCs generating HSC colonies in our assay uniformly expressed ESAM but were heterogeneous for CD150 expression, suggesting CD150 does not reliably mark functional FL-HSCs at this earlier stage (Fig. 2E and Fig. S1E). Transplantation of the progeny of HSC colonies in a subset of experiments confirmed these findings (Fig. S1F). Furthermore, at both stages, SEhi FL-HSCs that failed to generate HSC colonies also heterogeneously expressed CD150 and uniformly expressed ESAM, suggesting neither is a specific marker for FL-HSCs with serial engraftment potential in this assay.
Figure 2: SEhi FL-HSCs demonstrate heterogeneity in functional engraftment, proliferation, and immunophenotype following ex vivo coculture.
(A) Donor chimerism (total, myeloid, B cell, and T cell) in peripheral blood of primary and secondary recipients transplanted with the progeny of single E13.5 FL-HSCs following coculture with FL-AKT-ECs. 50% of cells from each colony were subject to flow cytometric analysis and the residual cells were used for transplantation (n=22 total colonies transplanted to n=24 total recipients; results pooled from 3 independent experiments). Two colonies (open circles) were each transplanted to 2 recipients.
(B) Frequency of GR1−F4/80−SCA1+EPCR+ cells (amongst total viable CD45+ cells) in individual colonies possessing serial multilineage engraftment (Engrafting, red) (n=4) or lacking serial multilineage engraftment (Non-engrafting, blue) (n=18).
(C) Frequency of HSC colonies, other colony types, or no colonies, observed following coculture of single E13.5 or E15.5/16.5 SEhi FL-HSCs with FL-AKT-ECs. (HSC colonies were defined as containing >80% GR1−F4/80−SCA1+EPCR+ cells amongst total viable CD45+ cells after exclusion of VECadherin+ FL-AKT-EC). (D) Total viable CD45+ cells per colony determined by flow cytometry following coculture of single E13.5 or E15.5/16.5 SEhi FL-HSCs with FL-AKT-ECs. Cell numbers were normalized to HSC colonies for each experiment. (E13.5: n=32 HSC colonies, n=119 other colonies, from 3 independent experiments; E15.5/16.5: n=95 HSC colonies, n=80 other colonies, from 5 independent experiments). ****P<0.0001 (Mann-Whitney test).
(E) Expression of ESAM and CD150 by index analysis of E13.5 (left) and E15.5 (right) SEhi FL-HSCs giving rise to immunophenotypic HSC colonies (red) or other colonies (gray) following FL-AKT-EC coculture.
Altogether, these studies establish a novel assay to assess the immunophenotypic properties and engraftment potential of FL-HSCs at single cell resolution in a developmentally relevant ex vivo niche. Specifically, this approach revealed functional heterogeneity in both the immunophenotypic E13.5 SEhi and E15.5/16.5 SEhiCD150+ FL-HSC populations at the single cell level based on their differential capacity to generate serially engrafting HSCs during FL-AKT-EC coculture. Furthermore, these results suggest that long-term engrafting FL-HSCs are uniquely characterized by delayed colony formation concomitant with latency to differentiate toward progenitors and propensity to symmetrically self-renew (based on both homogenous HSC immunophenotype and clonal expansion of serially engrafting HSCs) during FL-AKT-EC coculture ex vivo. Remarkably, this differentiation latency and self-renewal behavior in vitro is consistent with recent studies that have tracked FL-HSC fates in the native FL niche in vivo (Patel et al., 2022; Yokomizo et al., 2022).
Single cell RNA sequencing uncovers transcriptional heterogeneity in immunophenotypically defined FL-HSCs
In light of the functional heterogeneity of SEhi FL-HSCs revealed by our ex vivo single cell assays above, we next sought to explore the corresponding transcriptional heterogeneity of the immunophenotypic SEhi FL-HSC population in the early-stage FL, which may provide insight into the gene regulatory programs responsible for observed differences in self-renewal and engraftment potential of cells in this immunophenotypically homogenous population. To this end, we sorted SEhi cells from freshly isolated FL at E13.5 for scRNAseq (Fig. 3A and Fig. S2A). After filtering out poor quality cells, we obtained genome-wide transcriptome data for 338 cells. Using the Monocle3 toolkit (Cao et al., 2019), we performed dimensionality reduction by uniform manifold approximation and projection (UMAP) and unsupervised clustering by the Louvain algorithm to identify distinct cell clusters (Fig. 3B). E13.5 immunophenotypic SEhi FL-HSCs globally express Procr (EPCR), as expected, as well as other genes associated with HSCs, such as Hlf, Esam, Mecom, Fgd5, H19, and Cdkn1c, and mostly lack expression of genes associated with differentiating progenitors and mature blood cells (Fig. S2B and C). Interestingly, however, the expression levels of HSC-associated transcripts varied among cell clusters across UMAP space, suggesting underlying heterogeneity in the SEhi population. Moreover, although this population is negative for CD48 by flow cytometry (Fig. S1A), we observed heterogenous expression of Cd48 transcript (Fig. S2C). Flt3 expression was also heterogeneously observed in some of the cells (Fig. S2C), which may imply presence of previously described eMPP (Patel et al., 2022) or developmentally-restricted HSCs (Beaudin et al., 2016) (known to express Flt3) in the SEhi population. Furthermore, while FL-HSCs are generally thought to be proliferative (Bowie et al., 2006; Nakamura-Ishizu et al., 2020), we observed substantial variability in the expressions of genes associated with cell cycle activity within the immunophenotypically defined SEhi FL-HSCs, including a distinct population with low expression of cell cycle genes and low proliferation index, a transcriptional measure of cell cycle activity (Fig. S2D).
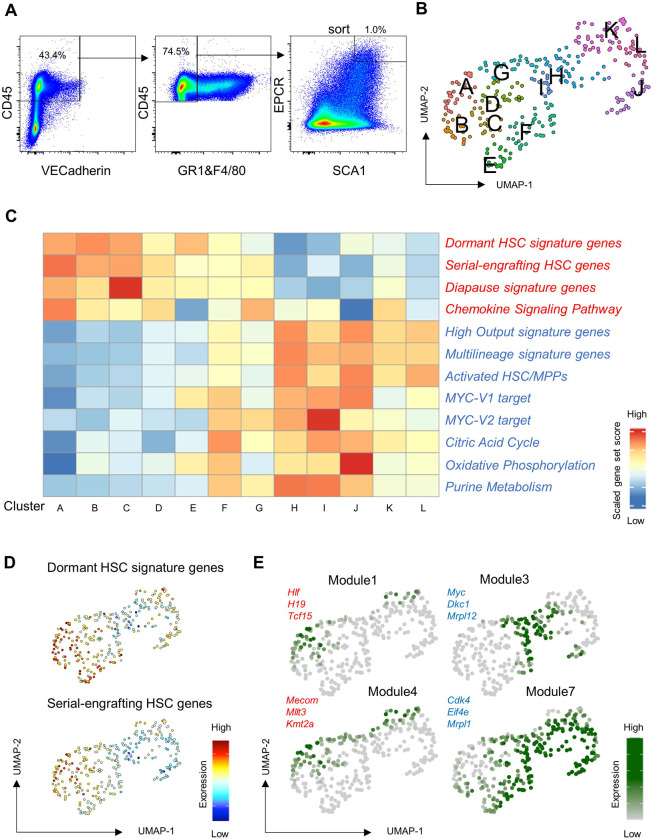
Figure 3: Single cell RNA sequencing reveals transcriptional heterogeneity amongst immunophenotypically-defined SEhi FL-HSCs.
(A) Sorting strategy for SEhi FL-HSCs from freshly isolated E13.5 FL used for scRNAseq.
(B) Unsupervised clustering of E13.5 SEhi FL-HSC scRNAseq data in UMAP.
(C) Heatmap of gene-set scores by cluster for genes associated with HSC dormancy (Cabezas-Wallscheid et al., 2017), serial engrafting HSCs (Rodriguez-Fraticelli et al., 2020), diapause (Boroviak et al., 2015) (Duy et al., 2021), and chemokine signaling (WP_CHEMOKINE_SIGNALING_PATHWAY) (red font) or genes associated with activated HSC/MPP states including high output and multilineage signatures (Cabezas-Wallscheid et al., 2017; Rodriguez-Fraticelli et al., 2020; Schönberger et al., 2021), Myc pathway activation (Hallmark Myc Target Genes V1, V2), and metabolic activity (WP_TCA_CYCLE, HALLMARK_OXIDATIVE_PHOSPHORYLATION, WP_PURINE_METABOLISM) (blue font).
(D) Gene-set expression heatmaps for dormant HSC signature genes and serial-engrafting HSC genes (Cabezas-Wallscheid et al., 2017; Rodriguez-Fraticelli et al., 2020).
(E) Heatmaps for expression scores of gene modules. Gene modules 1, 3, 4 and 7 are shown with representative genes identified in each module.
We next examined expression of gene-sets that characterize dormant HSCs in adult BM across cell clusters in the SEhi FL-HSC scRNAseq data (Fig. 3C) (Boroviak et al., 2015; Cabezas-Wallscheid et al., 2017; Rodriguez-Fraticelli et al., 2020; Wilson et al., 2015). We observed a marked variation in the expression levels of the adult HSC and dormancy-related gene-sets across the population of SEhi cells, with clusters exhibiting the highest dormancy-associated gene set scores predominantly localized to regions of UMAP with the lowest expression of genes associated with active cell cycle state and low proliferation index (Fig. 3, C and D and Fig. S2D). In contrast, genes characterizing activated HSCs/MMPs in adult BM and MYC target genes that promote biosynthetic and cell cycle activation were lower in this subset (Fig. 3C) (Cabezas-Wallscheid et al., 2017; Liberzon et al., 2015; Rodriguez-Fraticelli et al., 2020). Interestingly, the expression pattern of genes associated with diapause (Boroviak et al., 2015; Duy et al., 2021), a reversible embryonic state of dormancy unrelated to hematopoiesis, showed a similar pattern to the HSC dormancy gene set, as did genes associated with chemokine signaling characteristic of dormant HSCs in adult BM (Cabezas-Wallscheid et al., 2017) (Fig. 3C).
To further characterize transcriptional heterogeneity of the immunophenotypic population of FL-HSCs in an unbiased manner, we performed gene module analysis across cell clusters to determine co-regulated genes (Fig. 3E, Fig. S2E and Table S1). We identified two modules (#1 and #4) of genes with cluster-specific expression patterns that are similar to dormant and serial-engrafting HSC and diapause gene-sets (compare Fig. S2E and Fig. 3C). Consistent with this observation, genes expressed in these modules include known HSC-specific genes (e.g. Hlf, Mecom, Mllt3) (Table S1). Gene Ontogeny (GO) analysis of genes in module 3 and module 7, which have non-overlapping expression patterns to modules 1 and 4, identified terms associated with activated metabolic and cell cycle states, such as DNA replication initiation (Alvarez et al., 2015; Flach et al., 2014), ATP biosynthetic process (Inoue et al., 2010; Simsek et al., 2010), oxidative phosphorylation (Abbas et al., 2010; Maryanovich et al., 2015; Tothova et al., 2007) and translation (Keyvani Chahi et al., 2022) (Kruta et al., 2021) (Table S1). Furthermore, gene-sets specifically related to the cellular metabolism of activated HSCs and MPPs (Cabezas-Wallscheid et al., 2017; Karigane et al., 2016; Takubo et al., 2013) exhibit similar cluster-specific expression patterns to these gene modules (compare Fig. S2E and Fig. 3C).
Collectively, we here observed that immunophenotypically defined E13.5 SEhi FL-HSCs are transcriptionally heterogeneous in their biosynthetic and cell cycle activation states, and we unexpectedly identified a subset exhibiting gene signatures characteristic of dormant adult BM HSCs. These results are consistent with our observation that the same immunophenotypic SEhi FL-HSCs exhibit heterogeneous self-renewal, differentiation, and engraftment properties revealed by single cell coculture in the FL-AKT-EC niche. Furthermore, the distinct in vitro behavior of the subset of E13.5 SEhi FL-HSCs with serial engraftment potential (i.e. delayed colony formation, lower cell numbers per colony, latency to differentiate, and homogenous HSC immunophenotype) suggests that this subset corresponds to cells in our scRNAseq data characterized by a transcriptional state of relative biosynthetic dormancy and decreased cell cycle activity.
Integrating single cell transcriptomics and transplantation assays from clonal progeny of FL-HSCs identifies transcriptional signatures of self-renewing HSCs with serial multilineage engraftment
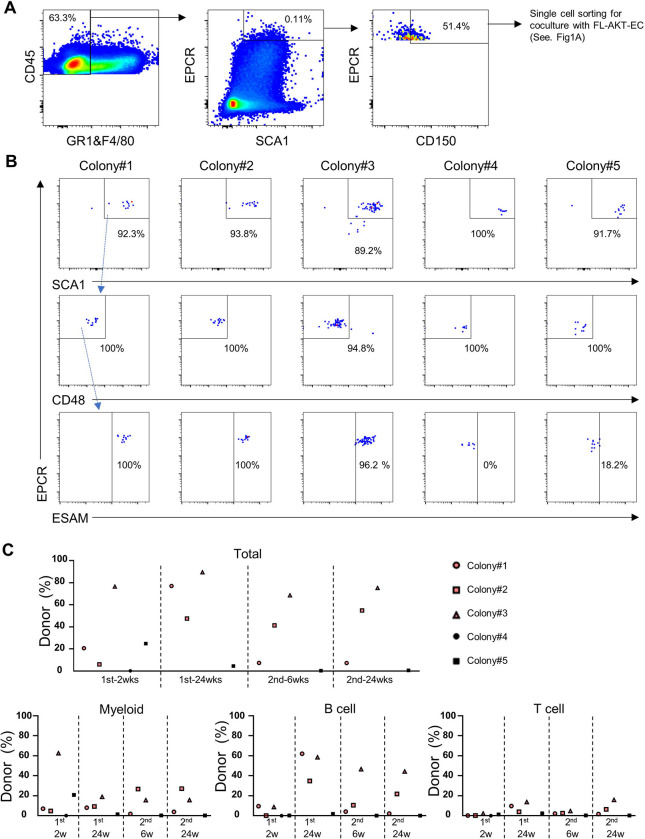
Based on our single cell transcriptomic analysis of freshly isolated E13.5 SEhi FL-HSCs above, we hypothesized that the subset of HSC colonies arising in vitro from E13.5 SEhi FL-HSCs possessing serial engraftment potential could be distinguished by transcriptional signatures of biosynthetic dormancy and decreased cell cycle activity. To test this hypothesis, we integrated scRNA-seq into our ex vivo platform for clonal analysis of FL-HSCs (Fig. 4A). Briefly, we index sorted single E13.5 SEhi FL-HSCs into a 96-well plate containing FL-AKT-EC. Following coculture a portion of each colony (15%) was used for immunophenotyping by flow cytometry to identify HSC colonies (>80% SCA1+EPCR+) for further analysis (Fig. S3A). A portion of the remaining cells from each of these colonies was used for transplantation (10% per recipient, to three recipients per colony) and for scRNAseq (55%). To minimize batch effects between colonies, we individually labelled each colony with oligonucleotide-barcode-conjugated antibodies to ubiquitous murine cell surface antigens such that cells from each colony could be pooled for downstream steps of scRNAseq (Stoeckius et al., 2018). This approach allows us to correlate the functional engraftment properties of each HSC colony to the unique transcriptomes of cells in the colony, as well as to the prospectively collected immunophenotype of the individually sorted E13.5 SEhi FL-HSCs from which each HSC colony was derived. Using this approach, we found that most of the identified HSC colonies contributed to short-term engraftment following transplantation; however, only colony #2 contained functional long-term, multilineage engrafting HSCs by serial transplantation (Fig. 4B). Notably, this HSC colony was derived from an SEhiCD150−ESAM+ FL-HSC (Fig. S3B). After filtering for poor quality cells, we obtained scRNAseq data for 1,030 cells, which were distinguished by colony of origin based on detection of barcoded oligonucleotides. Analysis of the single cell transcriptomic data was performed using dimensionality reduction by UMAP (Fig. 4C). A subset of cells expressed genes associated with HSC state, including Hlf, Procr, and Mecom, though expression levels of these genes varied significantly in UMAP space, as did expression of Cd48 and other genes associated with differentiation of HSCs to multipotent progenitors. Cells from HSC colonies generally lacked expression of genes associated with more lineage-restricted progenitors and mature blood cells (Fig. S3C), though a small cluster of cells expressing markers of granulocyte differentiation (Elane) was observed in cells coming primarily from colony #1 (Fig. S3C and Fig. 4C), which was notably more proliferative than other HSC colonies and contained a larger proportion of cells lacking SCA1+EPCR+ co-expression (Fig. S3A).
Figure 4: Analysis of clonal progeny of single FL-HSCs by integrating scRNAseq, flow cytometry, and transplantation reveals transcriptional and functional heterogeneity of HSC colonies.
(A) Overview of the experiment design. 15% of each well was used for flow cytometric analysis, 55% for scRNAseq, and 30% for serial transplantation (10% to each of 3 recipient mice).
(B) Donor chimerism in peripheral blood of primary and secondary recipients transplanted with the progeny of single E13.5 FL-HSCs following coculture with FL-AKT-ECs. Left: Total donor chimerism at indicated timepoints following primary and secondary transplant. Right: Donor myeloid, B cell, and T cell contribution at 24 weeks in secondary recipients. (n=3 mice transplanted for each colony).
(C) UMAP analysis with cells labeled based on colony of origin (colony #1: 513 cells, colony #2: 16 cells, colony #3: 202 cells, colony #4: 44 cells, colony #5: 102 cells, colony #6: 153 cells).
(D) Distribution of colony #2 (serially engrafting colony) verses all other colonies.
(E) Heatmap of gene-set scores characterizing dormant HSCs (Cabezas-Wallscheid et al., 2017). Violin plots of gene-set scores by colony type. p values indicate Wilcoxon rank-sum test.
(F) Heatmap of gene-set scores characterizing serial-engrafting HSCs (Rodriguez-Fraticelli et al., 2020). Violin plots of gene-set scores by colony type. p values indicate Wilcoxon rank-sum test.
(G) Heatmap of gene-set scores for Myc target genes (HALLMARK_MYC_TARGETS_V2). Violin plots of gene-set scores by colony type. p values indicate Wilcoxon rank-sum test.
(H) Heatmap showing the expression of gene module 1 in UMAP with representative genes (left). Heatmap of expression of gene modules by colony type (right)
Interestingly, the majority of cells from colony #2 clustered together in regions of UMAP space with the highest expression of HSC-associated genes and absence of genes associated with differentiated progenitors (Fig. 4D). Furthermore, when we examined published adult HSC-specific gene signatures associated with dormancy and serial engraftment (Cabezas-Wallscheid et al., 2017; Rodriguez-Fraticelli et al., 2020; Wilson et al., 2015), we found expression to be significantly higher in cells from colony #2 compared to cells from other types of colonies (Fig. 4,E and F and Fig. S3D). Interestingly, cells in colony #2 also exhibited significantly higher expression of genes recently associated with long-term, multilineage engraftment following in vitro expansion of adult HSCs (Che et al., 2022), genes associated with diapause (Boroviak et al., 2015; Duy et al., 2021) and chemokine signaling (Fig. S3D). In contrast, MYC target genes that promote biosynthetic and cell cycle activation, as well as other gene-sets characterizing differentiated progenitors, were significantly lower in cells from colony #2 (Fig. 4G and Fig. S3E). Differences in biosynthetic signatures were also confirmed by the comparison of cellular metabolism gene-sets which are reportedly higher in activated HSC/MPPs (Cabezas-Wallscheid et al., 2017; Karigane et al., 2016; Takubo et al., 2013) (Fig. S4A).
To further characterize the transcriptional heterogeneity of HSC colonies in an unbiased manner, we used unsupervised clustering (Fig. S4B) to identify distinct cell clusters and performed gene module analysis across cell clusters to determine co-regulated genes (Fig. 4H and Fig. S4C). One of the identified gene modules (module 1) which specifically marks cluster 1 (containing the majority of cells from colony #2) includes known HSC-specific genes such as Hlf, Mecom, and Mllt3, while non-overlapping gene modules feature active cell cycle genes and genes related to biosynthetic activity (module 2–4), including ribosome biogenesis, translation, and ATP biosynthetic process based on Gene Ontogeny (GO) analysis (Table S2).
Next, to identify the genes potentially related to the unique engraftment properties of colony #2, we conducted regression analysis to determine significantly differentially expressed genes between cells in colony #2 and cells from other HSC colonies lacking serial engraftment (Table S3). Cross-referencing these genes with genes expressed in module 1 (Table S2), we identified 123 common genes representing a gene-set that we termed “Serially engrafting FL-HSC genes” (Table S5). Remarkably 40% of the genes identified in our novel gene-set were shared with those in published gene-sets either associated with dormant adult BM HSCs or serially engrafting adult HSCs (Cabezas-Wallscheid et al., 2017; Rodriguez-Fraticelli et al., 2020), and these gene-sets marked similar subsets of cells in both our cultured and freshly isolated FL-HSC scRNAseq data (Fig. S, 4D and E). Furthermore, common genes identified include a number of key regulators of adult BM HSC dormancy and self-renewal, including Angpt1, Mecom, Mllt3, Cd63, and Ndn (Arai et al., 2004; Asai et al., 2012; Calvanese et al., 2019; Hu et al., 2022; Kataoka et al., 2011; Kustikova et al., 2013; Yokomizo et al., 2022). Interestingly, the Cxcr4-expressing subset of hemogenic endothelium (HE) in the E9-9.5 mouse embryo, which we recently demonstrated to contain the majority of HSC-competent HE prior to the emergence of the first functionally transplantable HSCs (Dignum et al., 2021), also showed significantly higher expression of this gene-set compared with HE lacking Cxcr4 expression, which can give rise to HSC-independent MPP but largely lack HSC potential (Fig. S4F).
Taken together, these studies reveal substantial transcriptional heterogeneity amongst otherwise immunophenotypically similar HSC colonies that predicts their functional capacity for serial engraftment in transplantation assays. Specifically, serially engrafting HSC colonies are characterized by gene expression patterns associated with dormancy in adult BM HSCs, including lower biosynthetic/Myc pathway activation and decreased cell cycle activity (Cabezas-Wallscheid et al., 2017). The finding that this gene expression profile also uniquely characterizes a subset of freshly isolated E13.5 SEhi FL-HSCs as well as HSC-competent HE in the early embryo (Dignum et al., 2021) suggests an HSC-specific gene expression program of relative biosynthetic dormancy that is conserved across embryonic and prenatal development into adulthood.
ESAM expression identifies HSC colonies with long-term engraftment potential derived from CD63-expressing SEhi FL-HSCs
Given heterogeneity in functional engraftment properties of HSC colonies that appear immunophenotypically similar, we next sought to further explore the scRNAseq data to identify candidate surface proteins that may serve as prospective markers of long-term engraftment of ex vivo expanded FL-HSCs. To this end, we identified genes encoding surface markers (Bausch-Fluck et al., 2015) specific to module #1 compared with other modules (#2–4) (Fig. 4H, Table S2). Amongst genes identified, we focused on Cd48 as a negative marker and Esam as a positive marker. ESAM is a known surface marker of FL-HSCs (Yokota et al., 2009), is highly expressed in HSCs with durable reconstitution ability after 5-FU treatment (Sudo et al., 2012), and was also recently reported as a marker of transplantable adult BM-derived HSCs following in vitro expansion (Che et al., 2022). Interestingly, while Esam was ubiquitously expressed in freshly isolated E13.5 SEhi FL-HSC by scRNAseq (Fig. S2B) and surface expression (Fig. 2E), its expression after coculture was significantly enriched in cells from HSC colony #2 (which uniquely demonstrating serial engraftment) compared to other HSC colonies (Fig. S4G).
To determine the utility of ESAM as a marker of serial engraftment in FL-HSC derived colonies, we sorted single E15.5 SEhiCD150+ FL-HSCs for coculture with FL-AKT-EC. Following coculture, amongst colonies classified as “HSC colonies”, we observed heterogeneity of ESAM expression, while CD48 expression was largely absent (>90% CD48 negative) (Fig. 5B). When we transplanted the remaining cells from each of these HSC colonies, we observed long-term, serial multilineage engraftment in HSC colonies containing predominantly ESAM+ cells, but not from HSC colonies containing predominantly ESAM− cells (Fig. 5, B and C). In an independent experiment from E15.5 SEhiCD150+ FL-HSCs, ESAM expression in HSC colonies also predicted high level donor engraftment of BM HSCs (LSK-SLAM: Lin−SCA1+KIT+CD150+CD48−) at 24 weeks post-transplant (Fig. S5).
Figure 5: ESAM-expressing HSC colonies exhibit serial hematopoietic reconstitution capacity.
(A) Single cell sorting strategy of E15.5 SEhiCD150+ FL-HSCs for coculture with FL-AKT-EC.
(B) Expression of CD48 and ESAM in 5 HSC colonies analyzed by flow cytometry following 15 days of coculture. 50% of the cells from each colony were used for flow cytometry and the remaining cells from each of the 5 HSC colonies shown were used for serial transplantation. (HSC colonies were defined as containing >80% CD48−SCA1+EPCR+ cells amongst total viable CD45+ cells).
(C) Donor chimerism (total, myeloid, B cell, and T cell) in peripheral blood of primary and secondary recipients transplanted with cells from each of the HSC colonies shown in B (n=5).
Having validated ESAM expression as a predictive marker of HSC colonies with long-term, serial engraftment, we next set out to identify surface markers expressed by FL-HSCs that would prospectively predict their ability to give rise to ESAM+ HSC colonies. To this end, we identified genes encoding surface antigens that were enriched in the serially engrafting HSC colony in our scRNAseq studies (Table S5), and focused on Cd63 since it has previously been shown to be enriched in serially engrafting adult HSCs based on both transcript and surface protein expression, and is implicated in HSC self-renewal and dormancy through interactions with the TGF-β pathway (Rodriguez-Fraticelli et al., 2020) (Hu et al., 2022). Supporting CD63 surface expression as a potential marker of FL-HSCs, we observed that the frequency of CD63+ cells and the median intensity of CD63 expression were higher in the EPCRhi fraction compared to the EPCRmedium and EPCR−/low fractions of CD45+SCA1+GR1−F4/80− cells from freshly isolated E13.5 FL (Fig. 6, A and B). Furthermore, we observed that ESAM+ HSC colonies were primarily derived from the CD63+ fraction of E13.5 SEhi FL-HSCs based on index flow cytometry analysis (Fig. 6C). Altogether, these results suggest that SEhi FL-HSCs with serial engraftment potential are further characterized by CD63 expression and by their unique propensity for symmetric self-renewal behavior, clonally giving rise to a relatively homogenous population of SEhiESAM+ HSCs in our ex vivo assay.
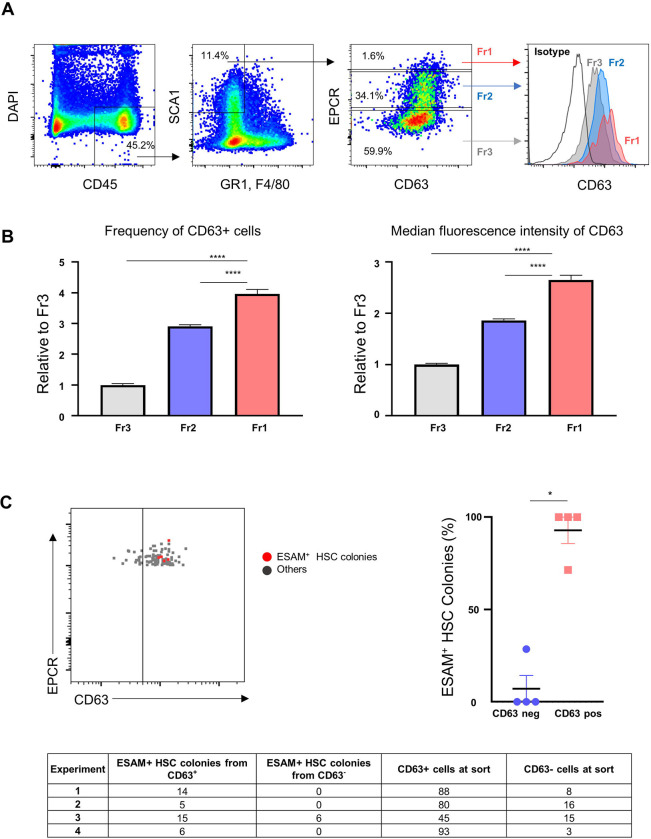
Figure 6: ESAM-expressing HSC colonies originate from CD63+ SEhi FL-HSCs.
(A) Expression of CD63 in CD45+GR1−F4/80−SCA1+ cells from E13.5 FL divided by EPCR expression. Fr1: EPCR high (red), Fr2: EPCR medium (blue), Fr3: EPCR low (gray). Fr: Fraction.
(B) Frequency of CD63 positive cells (left) and median fluorescence intensity of CD63 (right) in each fraction. 3 pooled FL samples from 4 different litters were used for analysis (total n=12) in 2 independent experiments. Values are normalized to Fr3 in each experiment. One-Way ANNOVA with Dunnett’s multiple comparisons test was used for statistical comparison. **** P<0.0001
(C) Expression of CD63 by index analysis of E13.5 SEhi FL-HSCs giving rise to immunophenotypic ESAM-expressing (>60% ESAM+) HSC colonies (red) or other colony types (gray) following FL-AKT-EC coculture (left: representative index profile; right: frequency of ESAM+ HSC colonies arising from CD63 negative vs CD63 positive fraction of SEhi FL-HSCs; bottom: summary of distribution of sorted SEhi FL-HSCs and ESAM+ HSC colonies based on CD63 expression at the time of index sorting). Total n=348 colonies from 4 independent experiments. Mann-Whitney testing was used for statistical analysis. *P<0.05.
Single cell transcriptomic analysis identifies candidate signaling interactions supporting FL-HSC self-renewal in the FL-AKT-EC niche
FL-AKT-ECs provide a niche sufficient to support expansion of FL-HSCs, revealing that the subset of immunophenotypic SEhi FL-HSCs with serial engraftment potential are intrinsically primed for differentiation latency and symmetric self-renewal behavior. To elucidate potential extrinsic signaling interactions regulating the process of FL-HSC self-renewal, we first integrated scRNAseq data from FL-AKT-ECs and serially engrafting HSCs (from HSC colony #2, Fig. 4B), and broadly identified candidate ligand-receptor interactions using a comprehensive database of curated pairs of ligands and receptors (Hadland et al., 2022; Ramilowski et al., 2015) (Table S4). This unbiased analysis identified an extensive list of potential signaling interactions, many of which have been previously shown to regulate various aspects of HSC self-renewal in the context of embryonic development and/or the adult BM, including those involving Notch, TGF-β, Wnt/β-catenin, integrins, and cytokines/chemokines (SCF, CXCL12, IGF1).
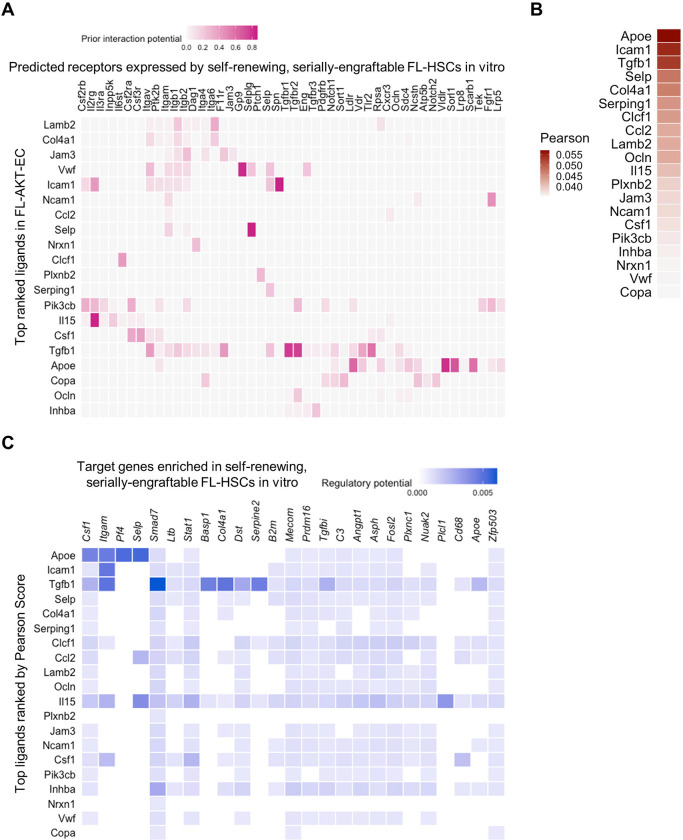
To further define the pathways that may be specifically required for HSC self-renewal in the FL-AKT-EC niche, we applied NicheNet (Browaeys et al., 2020), a computational algorithm that identifies ligand-receptor pairs from scRNAseq data based on existing knowledge of downstream signaling pathways and gene regulatory networks. Integrating scRNAseq data from FL-AKT-EC and HSC colonies, NicheNet identified candidate ligand-receptor signaling interactions prioritized based on likelihood of accounting for genes significantly enriched in serially engrafting HSCs (from colony #2) compared with cells from other HSC colonies in our scRNAseq data (Fig. 7, A–C). Multiple ligand-receptor pairs and downstream pathways were identified, including tgfb1 (TGFβ1), a well-established regulator of HSC quiescence in adult BM (Yamazaki et al., 2011; Yamazaki et al., 2009), transmembrane and extracellular cell adhesion molecules (egs. Icam1, Col4a1, Lamb2) that bind and activate integrin receptors (Itgb1, Itgav) essential for colonization of HSCs to the FL (Potocnik et al., 2000) and for regulating the size of the FL-HSC pool (Biswas et al., 2020), Selp (P-selectin), implicated in HSC expansion in the zebrafish caudal hematopoietic territory (CHT, equivalent to the mammalian FL) (Esain et al., 2015), and Jam3, encoding a cell surface adhesion protein implicated in HSC maintenance in the adult BM (Arcangeli et al., 2011). Numerous ligand-receptor signals not previously implicated in HSC self-renewal or fate determination were also identified. Notably, a large proportion of identified interactions were predicted to contribute to regulating downstream genes that are essential to HSC dormancy and self-renewal, particularly Mecom, Prdm16, and Angpt1 (Arai et al., 2004; Gudmundsson et al., 2020; Kataoka et al., 2011; Yokomizo et al., 2022). These results suggest that combinatorial ligand-receptor interactions between niche ECs and FL-HSCs involving numerous signaling pathways are likely to contribute to the observed self-renewal of functional HSCs in vitro (Fig. 8). Altogether, our ex vivo clonal FL-HSC expansion platform and correlative single cell transcriptomic data provide a valuable resource for future studies to further examine how these complex receptor-ligand interactions in the FL vascular niche coordinately regulate downstream transcriptional programs essential to FL-HSC self-renewal.
Figure 7: Complementary analysis of scRNAseq data identifies candidate receptor-ligand interactions regulating self-renewal of serially engrafting FL-HSCs in the FL-AKT-EC niche.
(A) Heatmap showing candidate ligands (expressed by FL-AKT-EC) interacting with receptors (expressed by cells in the serially-engrafting HSC colony #2) inferred by NicheNet (Browaeys et al., 2020).
(B) Heatmap showing candidate ligands (expressed by FL-AKT-EC) ranked by Pearson correlation.
(C) Heatmap showing the regulatory potential between the top ranked ligands (expressed by FL-AKT-EC) and downstream genes whose expression is enriched in cells from the serially engrafting HSC colony (#2) compared to cells from other HSC colonies lacking serial engraftment.
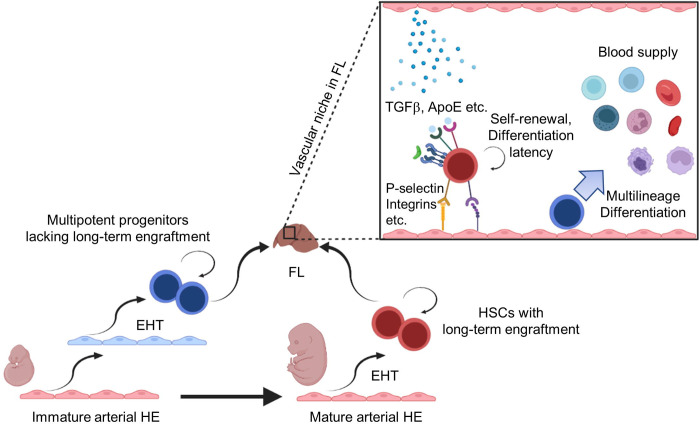
Figure 8: Proposed model for the independent origins of self-renewing, differentiation-latent FL-HSCs and multipotent hematopoietic progenitors contributing to heterogeneity in the FL hematopoietic compartment.
The FL hematopoietic compartment is seeded by heterogeneous populations of HSCs and HSC-independent multipotent progenitors derived from distinct populations of hemogenic endothelium (HE) (Dignum et al., 2021; Patel et al., 2022; Yokomizo et al., 2022). Multipotent progenitors lack long-term engraftment by transplantation assays but natively differentiate to produce mature blood cells, whereas self-renewing FL-HSCs possess long-term, serial engraftment potential and exhibit differentiation latency. Self-renewal of FL-HSCs may be supported, at least in part, by complex interactions with numerous ligands derived from the FL vascular niche.
Discussion
Here we establish a novel ex vivo platform that supports the expansion of functional, serially engrafting HSCs from individually sorted FL-HSCs. This platform provides a powerful tool to analyze the intrinsic properties of functionally validated FL-HSCs at single cell resolution, as well as the extrinsic niche-derived signals enabling HSC self-renewal in vitro. To this end, we leveraged this platform to resolve heterogeneity in immunophenotypically defined FL-HSCs by simultaneously integrating index flow cytometry, single cell transcriptomics, and transplantation assays, identifying a rare subset of FL-HSCs with serial engraftment potential that are characterized by propensity for symmetric self-renewal, differentiation latency, and transcriptional signatures of biosynthetic dormancy.
Notably, the observed behavior and transcriptional profiles of serially engrafting FL-HSCs in our study contrast with previous paradigms proposing that FL-HSCs reside in a state of biosynthetic and proliferative activation necessary to simultaneously expand their total numbers (providing sufficient HSCs for the adult) and differentiate to progenitors (providing functional blood cells for fetal development). Instead, our findings support a shift in this paradigm in line with several recent studies that have traced the origin and evolution of HSCs and HSC-independent progenitors from early embryonic development to adulthood by various clonal assays and lineage tracing methodologies in vitro and in vivo (Dignum et al., 2021; Kobayashi et al., 2023; Patel et al., 2022; Ulloa et al., 2021; Yokomizo et al., 2022). Together, these studies suggest that functional, long-term engrafting HSCs and HSC-independent embryonic multipotent progenitors (eMPP) emerge from distinct populations of hemogenic endothelium, that HSCs contribute minimally to differentiating progenitors and mature blood cells prenatally and even in early adulthood, and that eMPP contribute prenatally as well as postnatally to lifelong multilineage hematopoiesis natively in vivo but fail to provide long-term, multilineage hematopoietic engraftment as measured by transplantation assays (Fig. 8). Remarkably, our platform revealed intrinsic behaviors of individual FL-HSCs in vitro that recapitulated their predicted behaviors in vivo based on recent cellular barcoding and lineage tracing studies (Kobayashi et al., 2023; Patel et al., 2022; Yokomizo et al., 2022). Specifically, the rare subset of immunophenotypic (SEhi) FL-HSCs at early and mid-gestation with serial engraftment potential gave rise to colonies that emerged later in coculture, consisted of fewer total cell numbers, homogenously expressed immunophenotypic markers of HSCs (i.e. EPCR, SCA1, ESAM), and supported serial engraftment in multiple mice transplanted from a single colony, suggesting symmetric self-renewal events and restricted proliferative activity in vitro. Moreover, serially engrafting FL-HSC colonies demonstrated differentiation latency in vitro, failing to generate differentiated progenitors or mature blood cells based on immunophenotypic and transcriptomic analysis after extended coculture (greater than 12 days), which would temporally correspond to the postnatal period in vivo. Together, our data suggest the immunophenotypic (SEhi) FL-HSC compartment is heterogeneous, with only a minor subset representing functional, serially engrafting HSCs. Based on their in vitro behavior and clonal engraftment properties, we postulate that the remaining cells in this compartment may include a spectrum of separately emerging, HSC-independent lineages with multilineage hematopoietic potential but with minimal or restricted (short-term or lineage biased) engraftment potential, including recently described eMPP (which lack engraftment potential based on transplantation assays) (Patel et al., 2022) and “developmentally restricted” HSCs (that reconstitute multilineage hematopoiesis upon transplantation but with lymphoid biased engraftment) (Beaudin et al., 2016).
By integrating single cell transcriptomics into our platform, we also determined the unique transcriptional properties of serially engrafting FL-HSCs during the process of self-renewal in the ex vivo FL-AKT-EC niche. Surprisingly, this revealed a transcriptional signature of relative biosynthetic dormancy that overlaps with signatures of highly dormant and serially engrafting HSCs in the adult BM (Ganuza et al., 2022), as well as HSC-competent HE in the early embryo (Dignum et al., 2021). Together, these studies support a novel paradigm in which HSC fate across developmental stages from the early embryo to adult is linked to a state of relative dormancy that enables limited symmetric stem cell self-renewal while preventing excessive proliferation and differentiation to multilineage blood cells. Crucially, this mechanism would serve to minimize metabolic, proteotoxic, and genotoxic stress during HSC development that could contribute to hematopoietic dysfunction later in adult life. Furthermore, these cell intrinsic dormancy-related programs may synergize with extrinsic mechanisms in the FL, for example niche-derived antioxidant systems that minimize oxidative stress (Cacialli et al., 2021) and exogenous bile acids that alleviate endoplasmic reticulum stress (Sigurdsson et al., 2016), to protect self-renewing HSCs during their regulated expansion in the FL.
Studies of HSC development in the FL niche in vivo have been challenging due to the relative scarcity of functional, long-term engrafting HSCs amongst a sea of differentiating progenitors in the FL and the lack of markers that can prospectively identify FL-HSCs at single cell resolution with high specificity. By providing a culture system to functionally validate and characterize single FL-HSCs in vitro, our platform offers a complementary approach that provides several advantages and overcomes many challenges to studying FL-HSCs in vivo. We defined a combination of immunophenotypic markers in vitro (SCA1, EPCR, ESAM) sufficient to predict FL-HSCs expanded in vitro with serial engraftment potential in vivo, in line with previous studies that have identified these surface proteins as reliable markers of HSC potential following in vitro culture (Che et al., 2022; Fares et al., 2017; Hadland et al., 2018). Our platform thus provides a means to screen more rapidly for functional HSC potential at the single cell level while reducing the need for resource-intensive serial transplantation in live mouse models, which remains the gold standard to validate the engraftment potential of HSCs. Incorporating single cell index sorting into the platform, we also provide a means to retrospectively screen for immunophenotypic markers or other properties captured by FACS (such as fluorescent probes that measure metabolic activity, ROS, etc) that may predict functional HSC engraftment potential of each sorted cell. In the present study, for example, we demonstrated that functional FL-HSCs from the primary FL at E13.5 are enriched by CD63 surface expression, consistent with previous studies demonstrating CD63 expression in adult HSCs (Hu et al., 2022). Our platform also facilitates direct visualization of HSCs actively self-renewing in a developmentally relevant ex vivo niche, enabling longitudinal observations of their behavior and interactions with niche cells, which is not currently possible in vivo due to technical limitations and lack of markers that can reliably identify functional FL-HSCs at single cell resolution in situ. Future efforts to build on this platform, for example by incorporating advances in live imaging techniques and live fluorescence probes, will advance studies of the dynamic behavior and self-renewal mechanisms of FL-HSCs in real time at unprecedented resolution.
Since the FL-AKT-EC niche was sufficient to support the in vitro self-renewal of serial engrafting FL-HSCs, we also leveraged the platform to identify potential niche signaling interactions that regulate the process of FL-HSC self-renewal, using complementary scRNAseq data from the FL-AKT-EC and FL-HSCs expanded in vitro. To this end, NicheNet, a scRNAseq package which can infer and prioritize receptor-ligand pairs based on downstream gene expression changes in receiving cells, enabled identification of the signaling interactions between self-renewing HSCs and our engineered vascular environment that could account for genes identified as significantly enriched in engrafting HSC colonies verses immunophenotypically similar HSC colonies lacking long-term engraftment. Interestingly, TGFβ1, a key signaling molecule regulating dormancy in adult BM HSCs (Termini et al., 2022; Yamazaki et al., 2011; Yamazaki et al., 2009), was identified as one of the top 3 ligands in this analysis, with measurable regulatory potential for a large portion of differentially expressed genes in self-renewing HSCs in vitro (Fig. 7, B and C). This finding is consistent with the observation that serial engrafting FL-HSCs in our study were specifically enriched in transcriptional signatures associated with biosynthetic dormancy that are shared with adult BM HSCs. Thus, it is tempting to speculate a similar role for TGFβ signaling in the FL by regulating a relative state of biosynthetic and mitotic dormancy required to promote differentiation latency and limited, symmetric self-renewal of FL-HSCs. However, NicheNet also predicted complex regulatory interactions between multiple pathways that interact with TGFβ, including both positive and negative regulators and Smad7 (which can act via negative feedback on TGFβ/Activin pathways), suggesting fine tuning of TGFβ-mediated signals may be essential to achieve the appropriate balance of dormancy, allowing for FL-HSC expansion through controlled, symmetric self-renewal, while inhibiting excessive proliferation and differentiation. In addition to regulators of the TGFβ pathway, other signaling interactions and pathways identified by NicheNet have also been specifically implicated in HSC maintenance and self-renewal, including several intercellular and ECM adhesive interactions involving integrins, P-selectin, and Jam3 (Arcangeli et al., 2011; Biswas et al., 2020; Esain et al., 2015). Interestingly, the top ligand identified by NicheNet analysis, ApoE (whose transcript was also enriched in scRNAseq data from serially engrafting FL-HSCs in vitro) has been postulated to regulate HSC quiescence via modulation of cholesterol efflux and oxidative stress (Aires et al., 2021; Murphy et al., 2011; Tie et al., 2014). Indeed, ex vivo maintenance of engrafting HSCs has been shown to be dependent on exogenous cholesterol/fatty acids that may serve as a source of eicosanoid signaling molecules and other lipid-derivatives essential to HSC function (Kobayashi et al., 2019). Altogether, these results predict that complex, combinatorial interactions involving both niche-derived signals and cell intrinsic properties of FL-HSCs synergistically contribute to regulating the relative biosynthetic dormancy and cell cycle activity required for HSC self-renewal (Fig. 8). Our ex vivo system provides a resource for future exploration of the intricate interactions between intrinsic and extrinsic signaling networks regulating FL-HSC self-renewal at unprecedented resolution.
While the approach provided here offers advantages to studying the properties of FL-HSCs and how the EC niche supports their self-renewal in vitro, there are several limitations to our platform. Although we here focused on role of ECs from the FL, other FL niche components likely also contribute to self-renewal of FL-HSCs in the physiological FL environment in vivo. For example, macrophages play an important role in recycling glutathione that critically support amplification of developing HSPCs in zebrafish (Cacialli et al., 2021), and periportal Nestin+NG2+ arteriolar pericytes in the FL secrete HSC supportive factors facilitating FL-HSC expansion prenatally (Khan et al., 2016). Thus, future studies incorporating additional FL niche populations may provide additional insight into the complex regulation of FL-HSC self-renewal.
In summary, using a novel in vitro assay to assess for functional FL-HSCs at single cell resolution, we report here previously unrecognized heterogeneity of immunophenotypic FL-HSCs, revealing a subset of serially engraftable FL-HSCs characterized by intrinsic propensity for symmetric self-renewal, differentiation latency, and biosynthetic dormancy that is concomitantly dependent on critical extrinsic signals from the FL vascular niche. These findings support a paradigm shift in line with other recent studies that suggest prenatal hematopoiesis is largely supplied by HSC-independent progenitors whereas FL-HSCs are preserved to undergo limited self-renewal without differentiation during development to generate the adult HSC pool. By defining the developmental events necessary and sufficient to generate functional, engrafting HSCs ex vivo at single cell resolution, our approach delivers novel insights relevant to translational applications in regenerative medicine which aim to engineer and expand engrafting HSCs in vitro for transplantation and cellular therapies, and offers a powerful platform for future exploration of mechanisms of HSC self-renewal ex vivo.
Materials and methods
Mice
Wild type C57Bl6/J7 (CD45.2) and congenic C57BL/6.SJL-Ly5.1-Pep3b (CD45.1) mice were bred at the Fred Hutchinson Cancer Research Center. Male and female C57Bl6/J7 CD45.2 mice at 8–12 weeks of age were used for timed matings and transplantation experiments. All animal studies were conducted in accordance with the NIH guidelines for humane treatment of animals and were approved by the Institutional Animal Care and Use Committee at the Fred Hutchinson Cancer Center.
Dissection of embryos and cell sorting
Embryos were harvested from pregnant females and washed with PBS containing 2% FBS to minimize contaminating maternal tissues. Embryo age was determined by the date of observed maternal plugging and further confirmed by the following morphologic criteria. E13.5: Four lobes in the liver and retinal pigmentation were observed, digits were not completely separated but showed indentations. E15.5: Four lobes in the liver and retinal pigmentation is observed, digits were separated clearly and appeared unparallel. E16.5: Four lobes in the liver and retinal pigmentation were observed, digits were separated clearly and appeared parallel. Fetal livers were dissected under an inverted microscope by forceps, pooled in 10 ml conical tubes, and dissociated to a single cell suspension in 2% or 10% FBS with PBS by vigorous pipetting followed by passage through a 70um strainer to isolate single cells. Samples were then washed in PBS and resuspended in red blood cell lysis buffer (4L water, 33.2g Ammonium chloride (Fisher #6613), 4g sodium bicarbonate (Sigma Aldrich, S5761), and EDTA (0.1488g, Acros Organics, 147855000) or 200 uL/L of 0.5 M EDTA (Invitrogen, 15575-038)) for 5minutes at room temperature. Cells were washed once more, pre-incubated with anti-mouse CD16/32 (FcRII blocker, BD Biosciences Cat#553141), and stained with the following monoclonal anti-mouse antibodies as described in results: GR1 FITC (clone RB6-8C5, BD Biosciences, RRID: AB_394643), F4/80 FITC (clone BM8, Biolegend, RRID: AB_893502), CD201/EPCR PE (clone eBio1560, eBioscience, RRID: AB_914317), CD45 Peridinin-Chlorophyll-Protein (PerCP) Cyanine 5.5 (clone 30-F11, Invitrogen, RRID: AB_906233), SCA1 APC (clone D7, eBioscience, RRID: AB_469488), CD144 PE-cyanine7 (PE-Cy7) (clone eBioBV13, eBioscience, RRID: AB_2573402), CD150 biotin (clone TC15-12F12.2, Biolegend, RRID: AB_345278), Streptavidin APC-eFluor780(eBioscience, RRID: AB_10366688) or Streptavidin APC-cyanine7(APC-Cy7) (BD Biosciences, RRID: AB_10054651), GR1 APCeFluor780 (clone RB6-8C5, eBioscience, RRID: AB_1518804), F4/80 APCeFluor780 (clone BM8, eBioscience, RRID: AB_2735036), CD150 PerCP-Cyanine 5.5 (clone TC5-12F12.2, Biolegend, RRID: AB_2303663), CD45 PE-Cy7 (clone 30-F11, BD Biosciences, RRID: AB_394489), ESAM FITC (clone 1GB/ESAM, Biolegend, RRID: AB_2044017). CD201 PerCP-eFluor710 (clone eBio1560, Invitrogen, RRID: AB_10718383), CD63 PE (clone NVG-2, Biolegend, RRID: AB_11203532). DAPI (Millipore, Cat#268298) was used to exclude dead cells. Cells were sorted by either BD FACSymphony6 or Aria II equipped with BD FACS Diva Software with index sorting capability (Becton Dickinson). For index-sorted single cells, sorting was performed in single cell mode with maximum purity mask settings to minimize contaminating cells.
Generation of FL-derived Akt-EC (FL-AKT-EC) and coculture with HSCs
FL-AKT-EC were generated as previously described for similar EC lines derived from murine AGM (Hadland et al., 2015; Kobayashi et al., 2010), which is further detailed in a protocol available at Nature Protocol Exchange (https://protocolexchange.researchsquare.com/). Briefly, FL tissues were dissected from pooled E12 embryos and VECadherin+CD45−CD41−Ter119− cells were isolated by FACS. Sorted cells (>50,000 cells) were cultured on 48-well tissue culture plates coated with RetroNectin (r-fibronectin CH-296; Takara Bio Inc.) in EC media; Iscove’s Modified Dulbecco’s Medium (Gibco), 20% FBS (HyClone, fisher scientific), 1%Penicillin/streptomycin (Gibco), 1%L-glutamine (STEMCELL Technologies), heparin 0.1 mg/ml (Sigma-Aldrich), endothelial mitogen 100 μg/ml (Biomedical Technologies)*, VEGF (50 ng/ml; PeproTech), CHIR009921 (5 μM; StemGent), and SB431542 (10 μM; R&D Systems). We are happy to provide FL-AKT-ECs upon request. *EC mitogen no longer available (see below for substitutes). Following 1–2 days culture, colonies of ECs were infected by lentivirus with MyrAKT construct as reported (Kobayashi et al., 2010). Cells were serially split, expanded in EC media and then frozen down for future use.
FL-AKT-EC coculture with FL-HSCs
For coculture experiments, FL-AKT-ECs at passage 12 or less were plated at a density of 1 × 104 cells per well on 96-well plates 1–2 days prior to use. Prior to starting coculture, FL-AKT-EC layers were washed with serum-free media (X-VIVO20, LONZA) to remove as much serum contamination as possible. For single cell index co-culture, FL-HSCs (from donor CD45.2 mice) were individually index sorted to each well of 96-well containing FL-AKT-EC in serum-free coculture media consisting of X-VIVO 20 (Lonza) with recombinant cytokines (PeproTech): murine stem cell factor (SCF) at 100 ng/ml and thrombopoietin (TPO) at 20 ng/ml. Formation of hematopoietic colonies in coculture was monitored visually over time by microscopy and following various periods of coculture as indicated, a portion of cells (as indicated for each experiment) were harvested by pipetting for phenotypic analysis by flow cytometry, and in some experiments, remaining cells were used for confirmatory transplantation assays and/or scRNAseq (described below). Based on initial observations of the kinetics of colony emergence (Fig. S1D), we assayed the immunophenotype of colonies after day 12 to ensure capture of the entire array of colony types simultaneously for most of the remaining experiments. When the number of viable CD45+ cell recorded by flow cytometry was <2, the well was classified as “no colony.” Based on initial experiments correlating colony immunophenotype with engraftment properties, colonies with greater than 80% GR1−F4/80−SCA1+EPCR+ (Fig. 1,2,4 and 6) or CD48−SCA1+EPCR+ (Fig. 5 and Fig. S5) cells amongst total viable CD45+ cells were retrospectively classified as “HSC colonies”. Hematopoietic colonies with all other immunophenotypes were classified as “other colonies.”
Flow cytometric analysis of colonies
Following co-culture, a fraction of the generated hematopoietic progeny in each 96-well was harvested by pipetting from the EC layer for analysis of surface phenotype by flow cytometry (unless otherwise indicated, 50% of cells generated following single cell index culture on FL-AKT-EC were used for flow cytometry). Cells were spun and re-suspended in PBS with 2% FBS, pre-incubated with anti-mouse CD16/CD32 (FcRII block) and then stained with the following anti-mouse monoclonal antibodies: GR1 FITC (clone RB6-8C5, BD Biosciences, RRID: AB_394643), F4/80 FITC (clone BM8, Biolegend, RRID: AB_893502), CD201 PE (clone eBio1560, eBioscience, RRID: AB_914317), CD45 PerCP-Cyanine5.5 (clone 30-F11, Invitrogen, RRID: AB_906233), SCA1 APC (clone D7, eBioscience, RRID: AB_469488), CD144 PE-Cy7 (clone eBioBV13, eBioscience, RRID: AB_2573402), CD45 PE-Cy7 (clone 30-F11, BD Biosciences, RRID: AB_394489), ESAM FITC(clone 1GB/ESAM, Biolegend, RRID: AB_2044017), SCA1 Alexa Fluor700 (clone D7, eBioscience, RRID: AB_657836), CD201 PerCPeFluor710(clone eBio1560, Invitrogen, RRID: AB_10718383), CD48 APC (clone HM48-1, Invitrogen, RRID: AB_469408), GR1 APCeFluor780 (clone RB6-8C5, eBioscience, RRID: AB_1518804), F4/80 APCeFluor780 (clone BM8, eBioscience, RRID: AB_2735036). DAPI was used to exclude dead cells. Cells were analyzed by either BD FACS Canto II or BD FACS LSR Fortessa equipped with BD FACS Diva Software (Becton Dickinson) and further analyzed by Flow Jo ver.10 software. For index sort analysis, Flow Jo Plugin v3.0.6 and R4.2.2 were used.
Flow cytometric analysis of fresh FL cells
For the phenotyping of freshly isolated FL, harvested FL cells were subjected to red blood cell lysis (as described above), pre-incubated with anti-mouse CD16/32 (FcRII blocker), and stained with the following monoclonal anti-mouse antibodies: GR1 APCeFluor780 (clone RB6-8C5, eBioscience, RRID: AB_1518804), F4/80 APCeFluor780 (clone BM8, eBioscience, RRID: AB_2735036), CD201 PE (clone eBio1560, eBioscience, RRID: AB_914317), SCA1 APC (clone D7, eBioscience, RRID: AB_469488), CD45 PE-Cy7 (clone 30-F11, BD Biosciences, RRID: AB_394489), TER119 FITC (clone TER119, eBiosceicne, RRID: AB_465311), CD2 FITC (clone RM2-5, eBioscience, RRID: AB_464874), CD5 FITC (clone 53-7.3, eBioscience, RRID: AB_464909), CD8a FITC (clone 53-6.7, eBioscience, RRID: AB_469897), B220 FITC (clone RA3-6B2, BD Biosciences, RRID: AB_394618), CD48 FITC (clone HM48-1, eBioscience, RRID: AB_465078). DAPI was used to exclude dead cells. Cells were analyzed by BD FACSymphony S6 equipped with BD FACS Diva Software (Becton Dickinson) and further analyzed by Flow Jo software.
Transplantation assays
Recipient C57BL/6.SJL-Ly5.1-Pep3b (CD45.1) mice (6–12 weeks) were lethally irradiated on day −1 or 0 with 1,000cGy using a Cesium source, and transplanted via tail vein injection. Bone marrow 1×105 cells from C57BL/6.SJL-Ly5.1-Pep3b (CD45.1) was used as hematopoietic rescue. For single cell index assays, a fraction of the colonies harvested by pipetting was used for flow cytometric analysis (described above) and the remaining cells (or a fraction indicated in each experiment) were subject to transplantation. For some experiments, a portion of the cells (as indicated for each experiment) were also used for scRNAseq studies. Secondary transplants were performed after 24 weeks of primary transplant by using 2×106 whole bone marrow cells collected from primary recipients to lethally irradiated C57BL/6.SJL-Ly5.1-Pep3b (CD45.1) secondary recipients via the tail vein. Serial, long-term multilineage engraftment was strictly defined as donor (CD45.2) contribution to the peripheral blood with detectable contribution (>0.1%) to each lineage of donor myeloid (Gr-1 and F4/80), B cells (CD19) and T-cells (CD3) at 24 weeks post-transplant in both primary and secondary recipients. Transplant data is summarized in Table S6. We observed in initial experiments that recipients without detectable multilineage donor engraftment at 24 weeks post-transplant in primary recipients failed to provide multilineage engraftment in secondary recipients; thus, for remaining experiments, mice failing to demonstrate multilineage engraftment in primary recipients at 24 weeks were not subject to secondary transplantation. Recipient mice that died before the final timepoint analyzed for engraftment were censored for analysis of long-term serial engraftment potential but peripheral blood engraftment for these mice is shown at the ultimate timepoint analyzed before death in relevant figures (as shown in Table S6 for all transplant data).
Analysis of donor chimerism in recipient mice
Leukocytes from peripheral blood samples collected by retro-orbital bleeding were analyzed at the indicated time points. Lineage-specific staining for donor (CD45.2) and recipient/hematopoietic rescue (CD45.1) cells from peripheral blood was performed as previously described (Hadland et al., 2015), using anti-mouse monoclonal antibodies: CD3 FITC (clone 17A2, BD Pharmingen, RRID: AB_395698), F4/80 PE (clone BM8, Invitrogen, RRID: AB_465923), GR1 PerCP-Cyanine 5.5 (clone RB6-8C5, BD Phrmingen, RRID: AB_394334), CD45.1 PE-Cy7 (clone A20, eBioscience, RRID:AB_469629), CD19 APC (clone 1D3/CD19, Biolegend, RRID: AB_2629839), CD45.2 APCeFluor780 (clone 104, Invitrogen, RRID: AB_1272175). Bone marrow was collected from primary recipient femur, fibula and tibiae and treated with red blood cell lysis buffer for 5 minutes at room temperature. Cells were then washed and stained using anti-mouse monoclonal antibodies: CD45.1 Brilliant Violet 510 (clone A20, BD Pharmingen, RRID: AB_2739150), CD45.2 FITC (clone 104, BD Pharmingen, RRID: AB_395041), CD2 PE (clone RM2-5, BD Pharmingen, RRID: AB_2073810), CD5 PE (clone 53-7.3, Biolegend, RRID: AB_312737), CD8a PE (clone 53-6.7, BD Pharmingen, RRID: AB_394571), GR1 PE (clone RB6-8C5, BD Pharmingen, RRID: AB_394644), TER119 PE (clone TER-119, eBioscience, RRID: AB_466043), CD11b PE (clone M1/70 BD Biosciences, RRID: AB_394775), B220 PE (clone RA3-6 B2, BD Pharmingen, RRID: AB_394620), CD150 PerCP-Cyanine 5.5 (clone TC5-12F12.2, Biolegend, RRID: AB_2303663), SCA1 APC (clone D7, eBioscience, RRID: AB_469488), CD117 APCeFluor780 (clone 2B8, eBioscience, RRID: AB_1272177), CD48 PE-Cy7 (clone HM48-1, Biolegend, RRID: AB_2075049). DAPI was used to exclude dead cells. Cells were analyzed by BD FACS Canto II equipped with BD FACS Diva Software (Becton Dickinson) and further analyzed by Flow Jo software.
Single cell RNA sequence experiments
Freshly isolated FL-HSC and FL-AKT-EC
For single cell RNA sequencing (scRNA-seq) studies, freshly sorted DAPI−VECadherin−/lowCD45+Gra1−F4/80−Sca1highEPCRhigh cells derived from E13.5 murine fetal liver samples were subject to scRNA-seq experiment. Following the sort (3,050 cells from E13.5), cells were washed and resuspended with PBS containing 0.04% ultrapure BSA (Invitrogen) on ice.
For FL-AKT-EC, confluent ECs in a 6-well plate were cultured in serum-free coculture media (described above) overnight, harvested by treatment with TRypLE Express (Gibco), then resuspended in PBS/10% FBS at 4°C, washed twice and resuspended PBS with 0.04% ultrapure BSA in on ice. A portion of cells (targeting 3,500 cells to load) were subject to downstream scRNAseq assay.
Cell suspensions were loaded into the Chromium Single Cell Chip G (10X Genomics) and processed in the Chromium single cell controller (10X Genomics). The 10X Genomics Version 3.1 single cell 3’ kit was used to prepare single cell mRNA libraries with the Index Kit T Set A, according to manufacturer protocols. Sequencing was performed for pooled libraries from each sample on an Illumina NextSeq 500 using the 75 cycle, high output kit, targeting a minimum of 100,000 reads per cell.
Cocultured FL-HSC colonies
For scRNAseq of cultured HSC colonies, 15% of each colony was used for flow cytometry, 30% for transplantation, and the remaining 55% for scRNAseq. TotalSeq™-B0301–306 anti-mouse Hashtag Antibodies (clone M1/42; 30-F11;, Biolegend, RRID: AB_ 2814067–2814072) were used to label individual colonies for multiplexing (Stoeckius et al., 2018), at a concentration of 125 mg/ml (determined by titration of PE-conjugated antibodies of the identical clones). Surface protein labeling was performed according to 10X protocol (Cell Surface Protein Labeling for Single Cell RNA Sequencing Protocols with Feature Barcoding technology). Cells labeled with hashing antibodies were pooled, loaded into the Chromium Single Cell Chip G (10X Genomics), and processed in the Chromium single cell controller (10X Genomics). Single cell mRNA libraries were prepared using the 10X Chromium Next GEM Single Cell3’ Reagent Kits version 3.1 with Feature Barcoding technology for Cell Surface Protein, according to manufacturer protocols. Sequencing was performed for pooled libraries from each sample on an Illumina NextSeq 500 using the 75 cycle, high output kit, targeting a minimum of 35,000 reads per cell.
Single cell transcriptome computational analysis and quality control
The Cell Ranger 2.1.1 pipeline (10X Genomics) was used to align reads to the mm10 reference genome and to generate the feature barcode matrix, filtering low-quality cells using default parameters. Monocle3 (v.3.1.2.9) was used for downstream analysis, combining read-depth normalized data from freshly isolated E13.5, FL-AKT-EC, and cultured E13.5 colonies (Cao et al., 2019; Trapnell et al., 2014). Uniform Manifold Approximation (UMAP) was used for dimensionality reduction (Armstrong et al., 2021). The data was mapped onto the top principal components (default settings). The alignCDS() function was applied to remove batch effects between samples using a “mutual nearest neighbor” algorithm, Batchelor (v.1.2.4) (Haghverdi et al., 2018). Clustering was performed using the Louvain method implemented in Monocle3 (Levine et al., 2015). set.seed was used to specify the number of seeds to avoid variability in output due to a random number generator in the function. Cells with high mitochondrial genes (ENSMUSG00000064351, ENSMUSG00000064354, ENSMUSG00000064370, ENSMUSG00000064357) were excluded (>10% in HSC colonies and FL-AKT-EC or >5% in fresh FL-HSCs), as were cells with low genes per cell (<1,000) and cells with low UMI per cell (UMI<10,000 for scRNAseq data in fresh FL-HSCs or UMI<3,000 for scRNAseq data in HSC colonies). Outlying clusters of cells identified as non-hematopoietic populations (lacking expression of Ptprc, a pan-hematopoietic gene, and expressing either Cdh5, an EC-specific gene, or Hbb-bt, a globin gene expressed by red blood cells) were removed for downstream analysis. R scripts used for analysis are available on Github. The m3addon software was used to demultiplex the hashed samples in conjunction with Monocle3. Cells unique to each hashtag-oligo were assigned to the corresponding original colony names. All sequencing data is available at NCBI GEO (Accession number GSE233031).
Gene module analysis
The gene module methods provided in the Monocle3 package (the graph_test function and the find_gene_modules function) were used to find genes that vary across clusters and cell types, and group genes that have similar patterns of expressions into modules. Module genes with q-value<0.05 were selected.
Differential gene expression analysis
A quasipoisson distribution was used to evaluate the differential expression with the fit_model() and coefficient_table() functions in Monocle3. A q-value for multiple hypothesis testing was calculated by the Benjamini and Hochberg correction method, and q<0.05 was considered as statistically significant.
Gene-set scores
Gene set scores were calculated as the log-transformed sum of the size factor-normalized expression for each gene (Saunders et al., 2019) from published signature gene sets (Table S5) as well as the Molecular Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) including: Dormant HSC signature genes (Cabezas-Wallscheid et al., 2017), Serial-engrafting HSC signature genes (Rodriguez-Fraticelli et al., 2020), Diapause signature genes (Boroviak et al., 2015; Duy et al., 2021), Low output signature genes (Rodriguez-Fraticelli et al., 2020), MolO genes (Wilson et al., 2015), REPOPSIG genes (Che et al., 2022), High output signature genes (Rodriguez-Fraticelli et al., 2020), Multilineage signature genes (Rodriguez-Fraticelli et al., 2020), Hallmark Myc target genes (https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/HALLMARK_MYC_TARGETS_V1.html, https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/HALLMARK_MYC_TARGETS_V2.html), Activated HSC/MPPs (Cabezas-Wallscheid et al., 2017; Schönberger et al., 2021), WP_PURINE_METABOLISM (https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/WP_PURINE_METABOLISM.html), WP_TCA_CYCLE (https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/WP_TCA_CYCLE.html), HALLMARK_OXIDATIVE_PHOSPHORYLATION (https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/HALLMARK_OXIDATIVE_PHOSPHORYLATION.html), WP_MRNA_PROCESSING (https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/WP_MRNA_PROCESSING.html), REACTOME_TRANSLATION (https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/REACTOME_TRANSLATION.html). Wilcoxon Rank Sum Test (ggupbr package v0.4.0) was used to calculate p values.
Identification of genes encoding surface proteins
To identify genes encoding putative surface proteins in genes identified by our scRNAseq analysis, we cross-referenced identified genes with genes listed in CPSA validated surface protein (Bausch-Fluck et al., 2015) (http://wlab.ethz.ch/cspa/#downloads). Only those with CSPA category defined as 1-High confidence were included.
Gene Ontology (GO) analysis
Gene enrichment analysis was performed using an online functional annotations tool, The AmiGO v2.5.13 (Ashburner et al., 2000; Carbon et al., 2009; Gene Ontology, 2021) (http://geneontology.org/docs/go-citation-policy/), with the ‘GO biological process complete’ algorithm.
NicheNet
The nichenetr package (Browaeys et al., 2020) (https://github.com/saeyslab/nichenetr/blob/master/vignettes/ligand_activity_geneset.md) was applied to scRNAseq data to identify receptor-ligand interaction between FL-AKT-EC and cells in the serially engrafting HSC colony (#2). All genes expressed in cells from HSC colonies (colonies #1–6) were used as the background gene set, and the genes that were significantly enriched in serially-engrafting HSC colony #2 (Table S3) were used as the gene-set of interest (i.e., genes in the receiver cell population that are potentially affected by ligands).
Statistics
Wilcoxon signed-rank testing was used for scRNAseq studies. Mann-Whitney testing was used where indicated. One-way-ANOVA with Dunnett’s multiple comparisons test was used in statistical analysis for the comparison between multiple cohorts. A p-value < 0.05 was considered statistically significant. GraphPad Prism (GraphPad Software, La Jolla, CA) was used for all statistical analysis except for scRNAseq studies.
Supplementary Material
Acknowledgments
Authors declare no conflicting financial interest related to studies reported in this manuscript.
This work was supported by the American Society of Hematology Scholar Award (to B.H.), the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) (K08HL140143 to B.H.), and by the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH (RC2DK114777 to I.D.B., C.T., S.R., and B.H.). This work was also supported by the Flow Cytometry shared resource and the Genomics & Bioinformatics Shared Resource of the Fred Hutch/University of Washington/Seattle Children’s Cancer Consortium (NCI grant P30 CA015704).) T.I is supported for overseas medical research by Takeda Science Foundation, The Nakatomi Foundation and Kitasato University School of Medicine Alumni Association. The graphical abstract was created with BioRender.
Footnotes
Graphics
Some figures were created using BioRender.com.
Data and Code Availability
Raw sequencing data and Monocle 3 cell data sets have been deposited at NCBI GEO (accession number GSE233031) and are publicly available as of 5/24/2023.
All original code generated during this study has been deposited at Github and is publicly available as of the date of publication. Github repository: https://github.com/FredHutch/Ishida-etal-2023.
References
- Abbas H.A., Maccio D.R., Coskun S., Jackson J.G., Hazen A.L., Sills T.M., You M.J., Hirschi K.K., and Lozano G.. 2010. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell stem cell 7:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires R., Porto M.L., de Assis L.M., Pereira P.A.N., Carvalho G.R., Côco L.Z., Vasquez E.C., Pereira T.M.C., Campagnaro B.P., and Meyrelles S.S.. 2021. DNA damage and aging on hematopoietic stem cells: Impact of oxidative stress in ApoE. Exp Gerontol 156:111607. [DOI] [PubMed] [Google Scholar]
- Alvarez S., Díaz M., Flach J., Rodriguez-Acebes S., López-Contreras A.J., Martínez D., Cañamero M., Fernández-Capetillo O., Isern J., Passegué E., and Méndez J.. 2015. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nature Communications 6:8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai F., Hirao A., Ohmura M., Sato H., Matsuoka S., Takubo K., Ito K., Koh G.Y., and Suda T.. 2004. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118:149–161. [DOI] [PubMed] [Google Scholar]
- Arcangeli M.L., Frontera V., Bardin F., Obrados E., Adams S., Chabannon C., Schiff C., Mancini S.J., Adams R.H., and Aurrand-Lions M.. 2011. JAM-B regulates maintenance of hematopoietic stem cells in the bone marrow. Blood 118:4609–4619. [DOI] [PubMed] [Google Scholar]
- Armstrong G., Martino C., Rahman G., Gonzalez A., Vázquez-Baeza Y., Mishne G., and Knight R.. 2021. Uniform Manifold Approximation and Projection (UMAP) Reveals Composite Patterns and Resolves Visualization Artifacts in Microbiome Data. mSystems e0069121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Liu Y., Di Giandomenico S., Bae N., Ndiaye-Lobry D., Deblasio A., Menendez S., Antipin Y., Reva B., Wevrick R., and Nimer S.D.. 2012. Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood 120:1601–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., and Sherlock G.. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch-Fluck D., Hofmann A., Bock T., Frei A.P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R.L., Yoon C., Schiess R., Schmidt A., Mirkowska P., Härtlová A., Van Eyk J.E., Bourquin J.P., Aebersold R., Boheler K.R., Zandstra P., and Wollscheid B.. 2015. A mass spectrometric-derived cell surface protein atlas. PLoS One 10:e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin A.E., Boyer S.W., Perez-Cunningham J., Hernandez G.E., Derderian S.C., Jujjavarapu C., Aaserude E., MacKenzie T., and Forsberg E.C.. 2016. A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell stem cell 19:768–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C., Copley M.R., Kent D.G., Wohrer S., Cortes A., Aghaeepour N., Ma E., Mader H., Rowe K., Day C., Treloar D., Brinkman R.R., and Eaves C.J.. 2012. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell stem cell 10:273–283. [DOI] [PubMed] [Google Scholar]
- Biswas A., Roy I.M., Babu P.C., Manesia J., Schouteden S., Vijayakurup V., Anto R.J., Huelsken J., Lacy-Hulbert A., Verfaillie C.M., and Khurana S.. 2020. The Periostin/Integrin-αv Axis Regulates the Size of Hematopoietic Stem Cell Pool in the Fetal Liver. Stem Cell Reports 15:340–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A., and Bertone P.. 2015. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev Cell 35:366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie M.B., Kent D.G., Dykstra B., McKnight K.D., McCaffrey L., Hoodless P.A., and Eaves C.J.. 2007. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proceedings of the National Academy of Sciences of the United States of America 104:5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie M.B., McKnight K.D., Kent D.G., McCaffrey L., Hoodless P.A., and Eaves C.J.. 2006. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. The Journal of clinical investigation 116:2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R., Saelens W., and Saeys Y.. 2020. NicheNet: modeling intercellular communication by linking ligands to target genes. Nature Methods 17:159–162. [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Buettner F., Sommerkamp P., Klimmeck D., Ladel L., Thalheimer F.B., Pastor-Flores D., Roma L.P., Renders S., Zeisberger P., Przybylla A., Schönberger K., Scognamiglio R., Altamura S., Florian C.M., Fawaz M., Vonficht D., Tesio M., Collier P., Pavlinic D., Geiger H., Schroeder T., Benes V., Dick T.P., Rieger M.A., Stegle O., and Trumpp A.. 2017. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 169:807–823.e819. [DOI] [PubMed] [Google Scholar]
- Cacialli P., Mahony C.B., Petzold T., Bordignon P., Rougemont A.-L., and Bertrand J.Y.. 2021. A connexin/ifi30 pathway bridges HSCs with their niche to dampen oxidative stress. Nature Communications 12:4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V., Nguyen A.T., Bolan T.J., Vavilina A., Su T., Lee L.K., Wang Y., Lay F.D., Magnusson M., Crooks G.M., Kurdistani S.K., and Mikkola H.K.A.. 2019. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 576:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., Trapnell C., and Shendure J.. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., and Lewis S.. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J.L.C., Bode D., Kucinski I., Cull A.H., Bain F., Becker H.J., Jassinskaja M., Barile M., Boyd G., Belmonte M., Zeng A.G.X., Igarashi K.J., Rubio-Lara J., Shepherd M.S., Clay A., Dick J.E., Wilkinson A.C., Nakauchi H., Yamazaki S., Göttgens B., and Kent D.G.. 2022. Identification and characterization of in vitro expanded hematopoietic stem cells. 23:e55502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignum T., Varnum-Finney B., Srivatsan S.R., Dozono S., Waltner O., Heck A.M., Ishida T., Nourigat-McKay C., Jackson D.L., Rafii S., Trapnell C., Bernstein I.D., and Hadland B.. 2021a. Multipotent progenitors and hematopoietic stem cells arise independently from hemogenic endothelium in the mouse embryo. Cell Rep 36:109675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C., Li M., Teater M., Meydan C., Garrett-Bakelman F.E., Lee T.C., Chin C.R., Durmaz C., Kawabata K.C., Dhimolea E., Mitsiades C.S., Doehner H., D’Andrea R.J., Becker M.W., Paietta E.M., Mason C.E., Carroll M., and Melnick A.M.. 2021. Chemotherapy Induces Senescence-Like Resilient Cells Capable of Initiating AML Recurrence. Cancer Discovery 11:1542–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H., and Nakauchi H.. 2000. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95:2284–2288. [PubMed] [Google Scholar]
- Esain V., Kwan W., Carroll K.J., Cortes M., Liu S.Y., Frechette G.M., Sheward L.M., Nissim S., Goessling W., and North T.E.. 2015. Cannabinoid Receptor-2 Regulates Embryonic Hematopoietic Stem Cell Development via Prostaglandin E2 and P-Selectin Activity. Stem Cells 33:2596–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares I., Chagraoui J., Lehnertz B., MacRae T., Mayotte N., Tomellini E., Aubert L., Roux P.P., and Sauvageau G.. 2017. EPCR expression marks UM171-expanded CD34(+) cord blood stem cells. Blood 129:3344–3351. [DOI] [PubMed] [Google Scholar]
- Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., Le Beau M.M., Stohr B.A., Méndez J., Morrison C.G., and Passegué E.. 2014. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganuza M., Hall T., Finkelstein D., Chabot A., Kang G., and McKinney-Freeman S.. 2017. Lifelong haematopoiesis is established by hundreds of precursors throughout mammalian ontogeny. Nature cell biology 19:1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganuza M., Hall T., Myers J., Nevitt C., Sánchez-Lanzas R., Chabot A., Ding J., Kooienga E., Caprio C., Finkelstein D., Kang G., Obeng E., and McKinney-Freeman S.. 2022. Murine foetal liver supports limited detectable expansion of life-long haematopoietic progenitors. Nature cell biology 24:1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology C. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson K.O., Nguyen N., Oakley K., Han Y., Gudmundsdottir B., Liu P., Tessarollo L., Jenkins N.A., Copeland N.G., and Du Y.. 2020. Prdm16 is a critical regulator of adult long-term hematopoietic stem cell quiescence. Proc Natl Acad Sci U S A 117:31945–31953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B., Varnum-Finney B., Dozono S., Dignum T., Nourigat-McKay C., Heck A.M., Ishida T., Jackson D.L., Itkin T., Butler J.M., Rafii S., Trapnell C., and Bernstein I.D.. 2022. Engineering a niche supporting hematopoietic stem cell development using integrated single-cell transcriptomics. Nat Commun 13:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B.K., Varnum-Finney B., Mandal P.K., Rossi D.J., Poulos M.G., Butler J.M., Rafii S., Yoder M.C., Yoshimoto M., and Bernstein I.D.. 2017. A Common Origin for B-1a and B-2 Lymphocytes in Clonal Pre- Hematopoietic Stem Cells. Stem Cell Reports 8:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B.K., Varnum-Finney B., Nourigat-Mckay C., Flowers D., and Bernstein I.D.. 2018. Clonal Analysis of Embryonic Hematopoietic Stem Cell Precursors Using Single Cell Index Sorting Combined with Endothelial Cell Niche Co-culture. JoVE e56973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B.K., Varnum-Finney B., Poulos M.G., Moon R.T., Butler J.M., Rafii S., and Bernstein I.D.. 2015. Endothelium and NOTCH specify and amplify aorta-gonad-mesonephros-derived hematopoietic stem cells. J Clin Invest 125:2032–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Lun A.T.L., Morgan M.D., and Marioni J.C.. 2018. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Lu Y., Wang S., Zhang Z., Qi Y., Chen N., Shen M., Chen F., Chen M., Yang L., Chen S., Zeng D., Wang F., Su Y., Xu Y., and Wang J.. 2022. CD63 acts as a functional marker in maintaining hematopoietic stem cell quiescence through supporting TGFβ signaling in mice. Cell Death & Differentiation 29:178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S.-I., Noda S., Kashima K., Nakada K., Hayashi J.-I., and Miyoshi H.. 2010. Mitochondrial respiration defects modulate differentiation but not proliferation of hematopoietic stem and progenitor cells. FEBS Letters 584:3402–3409. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Arai F., Kubota Y., Dahl M., and Suda T.. 2010. Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood 116:544–553. [DOI] [PubMed] [Google Scholar]
- Karigane D., Kobayashi H., Morikawa T., Ootomo Y., Sakai M., Nagamatsu G., Kubota Y., Goda N., Matsumoto M., Nishimura Emi K., Soga T., Otsu K., Suematsu M., Okamoto S., Suda T., and Takubo K.. 2016. p38α Activates Purine Metabolism to Initiate Hematopoietic Stem/Progenitor Cell Cycling in Response to Stress. Cell stem cell 19:192–204. [DOI] [PubMed] [Google Scholar]
- Kataoka K., Sato T., Yoshimi A., Goyama S., Tsuruta T., Kobayashi H., Shimabe M., Arai S., Nakagawa M., Imai Y., Kumano K., Kumagai K., Kubota N., Kadowaki T., and Kurokawa M.. 2011. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med 208:2403–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyvani Chahi A., Belew M.S., Xu J., Chen H.T.T., Rentas S., Voisin V., Krivdova G., Lechman E., Marhon S.A., De Carvalho D.D., Dick J.E., Bader G.D., and Hope K.J.. 2022. PLAG1 dampens protein synthesis to promote human hematopoietic stem cell self-renewal. Blood 140:992–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J.A., Mendelson A., Kunisaki Y., Birbrair A., Kou Y., Arnal-Estapé A., Pinho S., Ciero P., Nakahara F., Ma’ayan A., Bergman A., Merad M., and Frenette P.S.. 2016. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 351:176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., He S., Yilmaz O.H., Kiel M.J., and Morrison S.J.. 2006. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood 108:737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Butler J.M., O’Donnell R., Kobayashi M., Ding B.S., Bonner B., Chiu V.K., Nolan D.J., Shido K., Benjamin L., and Rafii S.. 2010. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nature cell biology 12:1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Morikawa T., Okinaga A., Hamano F., Hashidate-Yoshida T., Watanuki S., Hishikawa D., Shindou H., Arai F., Kabe Y., Suematsu M., Shimizu T., and Takubo K.. 2019. Environmental Optimization Enables Maintenance of Quiescent Hematopoietic Stem Cells Ex Vivo. Cell Rep 28:145–158.e149. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Wei H., Yamanashi T., Azevedo Portilho N., Cornelius S., Valiente N., Nishida C., Cheng H., Latorre A., Zheng W.J., Kang J., Seita J., Shih D.J., Wu J.Q., and Yoshimoto M.. 2023. HSC-independent definitive hematopoiesis persists into adult life. Cell Rep 42:112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruta M., Sunshine M.J., Chua B.A., Fu Y., Chawla A., Dillingham C.H., Hidalgo San Jose L., De Jong B., Zhou F.J., and Signer R.A.J.. 2021. Hsf1 promotes hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. Cell stem cell 28:1950–1965.e1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova O.S., Schwarzer A., Stahlhut M., Brugman M.H., Neumann T., Yang M., Li Z., Schambach A., Heinz N., Gerdes S., Roeder I., Ha T.C., Steinemann D., Schlegelberger B., and Baum C.. 2013. Activation of Evi1 inhibits cell cycle progression and differentiation of hematopoietic progenitor cells. Leukemia 27:1127–1138. [DOI] [PubMed] [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir el A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., Finck R., Gedman A.L., Radtke I., Downing J.R., Pe’er D., and Nolan G.P.. 2015. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., and Tamayo P.. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanovich M., Zaltsman Y., Ruggiero A., Goldman A., Shachnai L., Zaidman S.L., Porat Z., Golan K., Lapidot T., and Gross A.. 2015. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nature Communications 6:7901. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman S.L., Naveiras O., Yates F., Loewer S., Philitas M., Curran M., Park P.J., and Daley G.Q.. 2009. Surface antigen phenotypes of hematopoietic stem cells from embryos and murine embryonic stem cells. Blood 114:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.J., Akhtari M., Tolani S., Pagler T., Bijl N., Kuo C.L., Wang M., Sanson M., Abramowicz S., Welch C., Bochem A.E., Kuivenhoven J.A., Yvan-Charvet L., and Tall A.R.. 2011. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J Clin Invest 121:4138–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Ishizu A., Ito K., and Suda T.. 2020. Hematopoietic Stem Cell Metabolism during Development and Aging. Developmental Cell 54:239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.H., Christodoulou C., Weinreb C., Yu Q., da Rocha E.L., Pepe-Mooney B.J., Bowling S., Li L., Osorio F.G., Daley G.Q., and Camargo F.D.. 2022. Lifelong multilineage contribution by embryonic-born blood progenitors. Nature 606:747–753. [DOI] [PubMed] [Google Scholar]
- Pei W., Feyerabend T.B., Rössler J., Wang X., Postrach D., Busch K., Rode I., Klapproth K., Dietlein N., Quedenau C., Chen W., Sauer S., Wolf S., Höfer T., and Rodewald H.-R.. 2017. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548:456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocnik A.J., Brakebusch C., and Fässler R.. 2000. Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity 12:653–663. [DOI] [PubMed] [Google Scholar]
- Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., and Forrest A.R.. 2015. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6:7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli A.E., Weinreb C., Wang S.W., Migueles R.P., Jankovic M., Usart M., Klein A.M., Lowell S., and Camargo F.D.. 2020. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature 583:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders L.M., Mishra A.K., Aman A.J., Lewis V.M., Toomey M.B., Packer J.S., Qiu X., McFaline-Figueroa J.L., Corbo J.C., Trapnell C., and Parichy D.M.. 2019. Thyroid hormone regulates distinct paths to maturation in pigment cell lineages. Elife 8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönberger K., Obier N., Romero-Mulero M.C., Cauchy P., Mess J., Pavlovich P.V., Zhang Y.W., Mitterer M., Rettkowski J., Lalioti M.E., Jäcklein K., Curtis J.D., Féret B., Sommerkamp P., Morganti C., Ito K., Ghyselinck N.B., Trompouki E., Buescher J.M., Pearce E.L., and Cabezas-Wallscheid N.. 2021. Multilayer omics analysis reveals a non-classical retinoic acid signaling axis that regulates hematopoietic stem cell identity. Cell Stem Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson V., Takei H., Soboleva S., Radulovic V., Galeev R., Siva K., Leeb-Lundberg L.M.F., Iida T., Nittono H., and Miharada K.. 2016. Bile Acids Protect Expanding Hematopoietic Stem Cells from Unfolded Protein Stress in Fetal Liver. Cell Stem Cell 18:522–532. [DOI] [PubMed] [Google Scholar]
- Simsek T., Kocabas F., Zheng J., DeBerardinis R.J., Mahmoud A.I., Olson E.N., Schneider J.W., Zhang C.C., and Sadek H.A.. 2010. The Distinct Metabolic Profile of Hematopoietic Stem Cells Reflects Their Location in a Hypoxic Niche. Cell stem cell 7:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan S.R., McFaline-Figueroa J.L., Ramani V., Saunders L., Cao J., Packer J., Pliner H.A., Jackson D.L., Daza R.M., Christiansen L., Zhang F., Steemers F., Shendure J., and Trapnell C.. 2020. Massively multiplex chemical transcriptomics at single-cell resolution. Science 367:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M., Smibert P., and Satija R.. 2018. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biology 19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo T., Yokota T., Oritani K., Satoh Y., Sugiyama T., Ishida T., Shibayama H., Ezoe S., Fujita N., Tanaka H., Maeda T., Nagasawa T., and Kanakura Y.. 2012. The endothelial antigen ESAM monitors hematopoietic stem cell status between quiescence and self-renewal. Journal of immunology (Baltimore, Md. : 1950) 189:200–210. [DOI] [PubMed] [Google Scholar]
- Takubo K., Nagamatsu G., Chiharu I. Kobayashi, Nakamura-Ishizu A., Kobayashi H., Ikeda E., Goda N., Rahimi Y., Randall S. Johnson, Soga T., Hirao A., Suematsu M., and Suda T.. 2013. Regulation of Glycolysis by Pdk Functions as a Metabolic Checkpoint for Cell Cycle Quiescence in Hematopoietic Stem Cells. Cell stem cell 12:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini C.M., Pang A., Li M., Fang T., Chang V.Y., and Chute J.P.. 2022. Syndecan-2 enriches for hematopoietic stem cells and regulates stem cell repopulating capacity. Blood 139:188–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie G., Messina K.E., Yan J., Messina J.A., and Messina L.M.. 2014. Hypercholesterolemia induces oxidant stress that accelerates the ageing of hematopoietic stem cells. J Am Heart Assoc 3:e000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H. 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J.R., Cohen O., Shah P., Lu D., Genshaft A.S., Hughes T.K., Ziegler C.G., Kazer S.W., Gaillard A., Kolb K.E., Villani A.C., Johannessen C.M., Andreev A.Y., Van Allen E.M., Bertagnolli M., Sorger P.K., Sullivan R.J., Flaherty K.T., Frederick D.T., Jané-Valbuena J., Yoon C.H., Rozenblatt-Rosen O., Shalek A.K., Regev A., and Garraway L.A.. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., Armstrong S.A., Passegué E., DePinho R.A., and Gilliland D.G.. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., and Rinn J.L.. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa B.A., Habbsa S.S., Potts K.S., Lewis A., McKinstry M., Payne S.G., Flores J.C., Nizhnik A., Feliz Norberto M., Mosimann C., and Bowman T.V.. 2021. Definitive hematopoietic stem cells minimally contribute to embryonic hematopoiesis. Cell Reports 36:109703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.K., Kent D.G., Buettner F., Shehata M., Macaulay I.C., Calero-Nieto F.J., Sánchez Castillo M., Oedekoven C.A., Diamanti E., Schulte R., Ponting C.P., Voet T., Caldas C., Stingl J., Green A.R., Theis F.J., and Göttgens B.. 2015. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell 16:712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Ema H., Karlsson G., Yamaguchi T., Miyoshi H., Shioda S., Makoto M. Taketo, Karlsson S., Iwama A., and Nakauchi H.. 2011. Nonmyelinating Schwann Cells Maintain Hematopoietic Stem Cell Hibernation in the Bone Marrow Niche. Cell 147:1146–1158. [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Iwama A., Takayanagi S., Eto K., Ema H., and Nakauchi H.. 2009. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood 113:1250–1256. [DOI] [PubMed] [Google Scholar]
- Yokomizo T., Ideue T., Morino-Koga S., Tham C.Y., Sato T., Takeda N., Kubota Y., Kurokawa M., Komatsu N., Ogawa M., Araki K., Osato M., and Suda T.. 2022. Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature 609:779–784. [DOI] [PubMed] [Google Scholar]
- Yokota T., Oritani K., Butz S., Kokame K., Kincade P.W., Miyata T., Vestweber D., and Kanakura Y.. 2009. The endothelial antigen ESAM marks primitive hematopoietic progenitors throughout life in mice. Blood 113:2914–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data and Monocle 3 cell data sets have been deposited at NCBI GEO (accession number GSE233031) and are publicly available as of 5/24/2023.
All original code generated during this study has been deposited at Github and is publicly available as of the date of publication. Github repository: https://github.com/FredHutch/Ishida-etal-2023.