Abstract
Many universally and conditionally important genes are genomically aggregated within clusters. Here, we introduce fai and zol, which together enable large-scale comparative analysis of different types of gene clusters and mobile-genetic elements (MGEs), such as biosynthetic gene clusters (BGCs) or viruses. Fundamentally, they overcome a current bottleneck to reliably perform comprehensive orthology inference at large scale across broad taxonomic contexts and thousands of genomes. First, fai allows the identification of orthologous or homologous instances of a query gene cluster of interest amongst a database of target genomes. Subsequently, zol enables reliable, context-specific inference of protein-encoding ortholog groups for individual genes across gene cluster instances. In addition, zol performs functional annotation and computes a variety of statistics for each inferred ortholog group. These programs are showcased through application to: (i) longitudinal tracking of a virus in metagenomes, (ii) discovering novel population-genetic insights of two common BGCs in a fungal species, and (iii) uncovering large-scale evolutionary trends of a virulence-associated gene cluster across thousands of genomes from a diverse bacterial genus.
Introduction
Within bacterial genomes, genes are often co-located within smaller genetic structures such as operons1,2, phages3, metabolic gene clusters4, biosynthetic gene clusters (BGCs)5, and pathogenicity islands6,7. Although less prevalent, eukaryotic genomes also contain genes aggregated within discrete clusters5,8.
Sometimes gene clusters are highly conserved, encoding for products essential to the survival of the organism9. In other cases, a single gene cluster can exhibit variability in gene carriage and order across different strains or species10–12. This is often the case for BGCs encoding specialized metabolites or virulence-associated gene clusters, where evolution of gene content and sequence divergence can influence fitness and contribute to adaptation within a changing ecosystem.
Bioinformatic toolkits to perform accurate pangenomic and comparative genomic analyses have been heavily developed over the past two decades13–18; however, tool development to aid the identification and comparative analysis of smaller homologous gene clusters has been more limited and largely designed for specific types of gene clusters19–22. In addition, while methods for comprehensive comparative genomics within species exist and are scalable17,23,24, methods for reliable, large-scale comparative genomics of thousands of genomes representing a greater breadth of taxonomic diversity are lacking and bear heavy computational costs25,26. Context-specific inference of orthologous genes within focal gene clusters offers a targeted and reliable solution to overcome challenges with scalability27,28. Such an approach was recently taken to infer orthologous genes between instances of homologous BGCs22.
Here, we introduce fai (find-additional-instances) and zol (zoom-on-locus), which are designed for the identification (fai) and in-depth evolutionary genomics investigations (zol) of a wide array of gene cluster types. We demonstrate the utility of these programs through application to three types of gene clusters within different genomic contexts including a novel bacteriophage within environmental metagenomes, a fungal secondary metabolite encoding biosynthetic gene clusters, and a conserved polysaccharide antigen locus within the diverse bacterial genus of Enterococcus.
Results
fai and zol allow for the rapid inference of gene cluster orthologs across diverse genomes
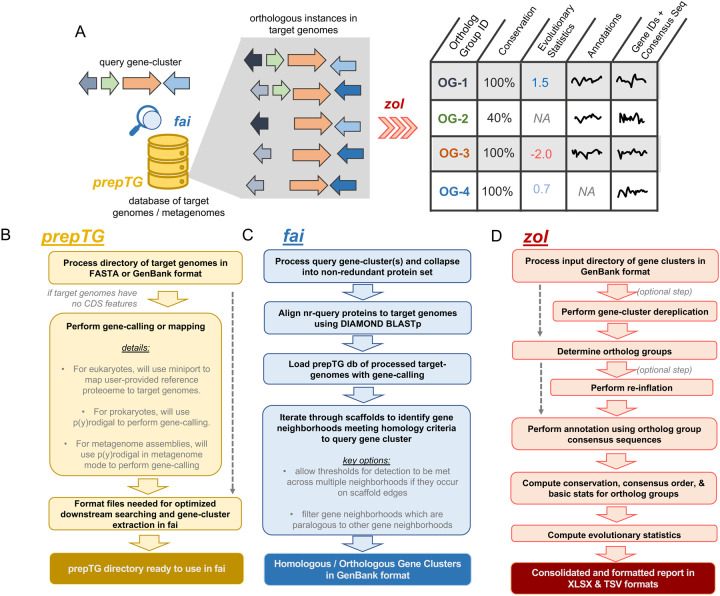
The two programs, fai and zol, build upon approaches we recently reported in lsaBGC29 that were developed to investigate evolutionary trends of BGCs in a single taxon. Within fai and zol, algorithmic adjustments have been implemented to broaden the application for searching any type of gene cluster across a diverse set of target genomes (Figure 1A). First, fai allows users to rapidly search for gene cluster instances in a target set of genomes. Then, zol can be used to compute evolutionary statistics and functional annotations of gene cluster content in table-based reports. Importantly, because fai has an option to filter secondary, potentially paralogous, instances of gene clusters found in target genomes, downstream ab initio clustering of proteins using a flexible, InParanoid-type algorithm14 by zol can be used to reliably infer ortholog groups.
Figure 1: Overviews of fai and zol.
A) A schematic of how prepTG, fai, and zol are integrated to perform evolutionary investigations by searching for gene-clusters. An overview of the prepTG (B), fai (C) and zol (D) algorithms and workflows.
In addition to filtering secondary instances of query gene clusters identified in target genomes, detection criteria in fai can be adjusted by assessing whether gene cluster homologs lie near scaffold edges in target genomic assemblies. This feature overcomes challenges inherent to the identification of full gene-clusters in metagenomic assemblies or metagenome-assembled genomes, which can be highly fragmented (Figure S1). fai can further accept query gene-clusters in different formats to ease searching for gene clusters and genomic islands cataloged in databases such as ICEberg30, MIBiG31, or IslandViewer32. In addition, to promote consistency in gene calling across target genomes, we have incorporated computationally lightweight dependencies for de novo gene prediction in prokaryotic genomes33,34 and genemapping in eukaryotic genomes35 within prepTG, to prepare and format target genomes for optimized gene-cluster searching in fai (Figure 1B). Together these unique features and options differentiate fai from other software with similar functionalities, such as cblaster21 (Figure 1C, S1; Table S1; Supplementary Text).
zol is differentiated from lsaBGC29, where ortholog groups are inferred across full genomes using OrthoFinder18, by delineating ortholog groups within the context of a homologous or orthologous set of gene clusters, similar to the approach taken within CORASON22 to visualize similarities between BGCs. While CORASON uses bidirectional besthits to identify direct orthologs, zol accounts for the presence of in-paralogs and comprehensively partitions proteins into ortholog groups. Similar to lsaBGC-PopGene29, zol will then construct a tabular report with information on conservation, evolutionary trends, and annotation for individual ortholog groups (Figure 1D). To make annotated reports generated by zol more broadly informative for a variety of gene clusters, several databases have been included, such as VOGs36, VFDB37, ISFinder38, and CARD39. In addition, zol incorporates HyPhy40 as a dependency and calculates evolutionary statistics not previously reported in lsaBGC-PopGene, such as sequence entropy in the 100 bp upstream of an ortholog group, where important regulatory differences could exist41. Ultimately, beyond high-throughput inference of ortholog groups across diverse genomic datasets, the rich tabular report produced by zol provides complementary information to figures generated by comparative visualization software such as clinker42, CORASON22, gggenomes43, and Easyfig44.
Another key feature in zol is the ability to dereplicate gene clusters directly using skani45, which was recently shown to be more reliable at estimating ANI between genomes of variable contiguity relative to comparative methods. Dereplication allows for more appropriate inference of evolutionary statistics to overcome availability or sampling biases in genomic databases46. Finally, zol allows for comparative investigations of gene-clusters based on taxonomic or ecological groupings47–49. For instance, users can designate a subset of gene clusters as belonging to a specific population to allow zol to calculate ortholog group conservation across just the focal set of gene clusters. In addition, if comparative investigations are requested, zol will also compute the fixation index50, FST, for each ortholog group to assess gene flow between the focal and complementary sets of gene clusters.
Longitudinal tracking of a virus within lake metagenomic assemblies
Viruses are important members of host and environmental microbiomes51–53, influencing the microbial composition and participating in several metabolic pathways. Targeted identification of a specific virus or bacteriophage within metagenomes can thus offer greater insight into their elusive functional roles in microbiomes.
Recently, changes in the composition and function of the metagenome at three different depths of a lake was reported using longitudinal shotgun metagenomics54. Using metagenome assemblies generated from this dataset, large (≥20kb) and predicted-circular phages were identified independently across a subset of metagenomes from the three different depths at the the earliest sampling date using VIBRANT55. Subsequent clustering based on the sequence and syntenic similarity of protein domains identified a ~36kb highly conserved virus in two metagenomes sampled from lower lake depths.
fai was then used to perform a rapid, targeted search for this ~36kb Caudovirales virus across the full set of 16 metagenomes to identify additional instances of the virus. fai completed its search of the metagenomes, featuring >20 million proteins and 10.7 million contigs, in less than seven minutes using 20 threads. Of the 16 total metagenomes, spanning five distinct sampling timepoints and four distinct sampling depths, nine metagenomes containing the virus were identified (Figure 2A) exclusively from anoxic conditions (p=8.7e-5; two-sided Fisher’s exact test). This suggests the viral host likely performs anaerobic respiration. Application of zol further revealed that 34 (64%) of the 53 total distinct ortholog groups were core to all instances of the virus across nine metagenomes and completely conserved in sequence over the course of 2.5 months (Figure 2B; Table S2). Furthermore, seven of the 53 ortholog groups were not observed in the query viruses from the earliest sampling date, demonstrating the ability of fai to identify new genes within additional instances of known gene clusters.
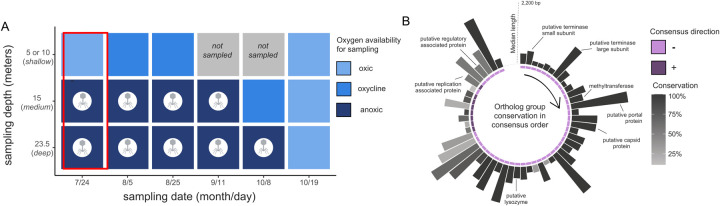
Figure 2: Targeted viral detection in metagenomes using fai.
A) Total metagenomes from a single site in Lake Mendota across multiple depths and timepoints from Tran et al. 2023 were investigated using fai for the presence of a virus found in two of the three earliest microbiome samplings (red box). The presence of the virus is indicated by a phage icon. Metagenome samples are colored according to whether they corresponded to oxic, oxycline, or anoxic. The most shallow sampling depths varied for different dates and consolidated as a single row corresponding to a sampling depth of either 5 or 10 meters. B) The pangenome of the virus is shown based on the consensus order and directionality of coding sequences inferred by zol. Bar heights correspond to the median length of coding sequences and are colored based on the percentages of the nine metagenomes the virus was detected in. BioRender was used in generation of this figure.
Investigating population-level and species-wide evolutionary trends of BGCs in the eukaryotic species Aspergillus flavus
The fungal genus of Aspergillus is a source of several natural products, including aflatoxins, a common and economically impactful contaminant of food. The genus also contains species that are model organisms for studying fungal secondary metabolism56–58. Examination of the secondary metabolome of A. flavus has revealed that different clades or populations comprising certain species can exhibit variability in their metabolite production despite high conservation of core BGC genes encoding enzymes for synthesis of these metabolites12,59,60. For instance, population B A. flavus were identified as producing a greater abundance of the insecticide leporin B relative to populations A and C12,61.
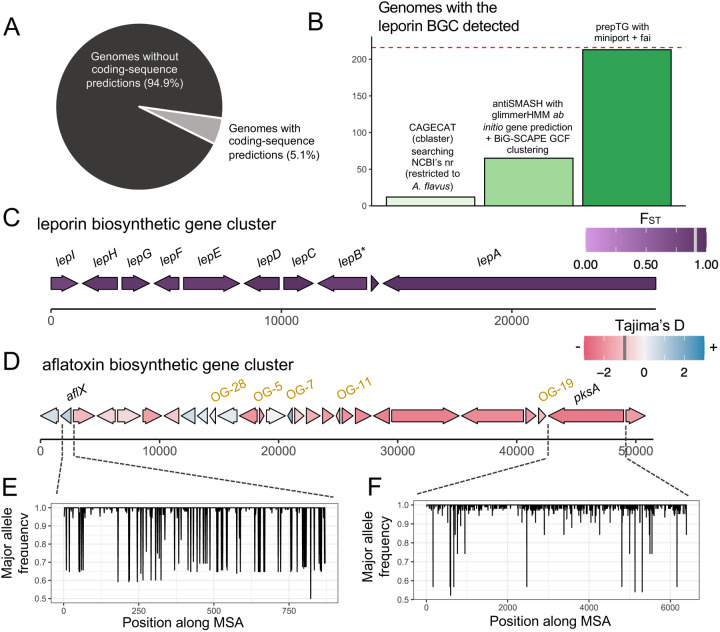
To further understand the genomic basis for differences in metabolite content between populations, we investigated the leporin BGC using fai and zol. While the leporin cluster was previously identified as a core component of the A. flavus genome12, a recent study suggested that the full BGC was specific to a single clade from the species60. Low sensitivity in direct assessment of gene cluster presence in eukaryotic genome assemblies can arise from their incompleteness, leading to gene clusters being fragmented across multiple scaffolds, and challenges in ab initio gene prediction62,63. Further deterring the direct prediction of gene clusters in eukaryotic assemblies is the lack of gene annotations, with only 11 (5.1%) of 216 A. flavus genomes in NCBI’s GenBank database having coding sequence predictions (Figure 3A). Therefore, we used miniprot35, which is integrated within prepTG, to directly map high-quality coding genes predictions based on transcriptomics data from the genome of strain A. flavus NRRL 335764 to the 216 genomes available for the species. Running fai in “draft mode” led to the identification of the leporin BGC within 212 (98.1%) assemblies, consistent with prior read mapping-based investigations12. This increase in sensitivity when fai is run with miniprot-based gene-mapping is substantial when compared to common alternate approaches for identifying homologous instances of BGCs across genomes (Figure 3B; Supplementary Text).
Figure 3: Evolutionary trends of common BGCs in A. flavus.
A) The proportion of 216 A. flavus genomes from NCBI’s GenBank database with coding-sequence predictions available. B) Comparison of the sensitivity of fai and alternate approaches based on assemblies for detecting the leporin BGC. The red-line indicates the total number of genomes (n=216) assessed. A schematic of the (C) leporin and (D) aflatoxin BGCs is shown with genes present in ≥ 10% of samples shown in consensus order and relative directionality. Coloring of genes in (C) corresponds to FST values and in (D) to Tajima’s D values, as calculated by zol. Grey bars in the legends, at (C) 0.92 and (D) −0.98, indicate the mean values for the statistics across genes in the BGC. *For the leporin BGC, lepB corresponds to an updated open-reading frame (ORF) prediction by Skerker et al. 2021 which was the combination of AFLA_066860 and AFLA_066870 ORFs in the MIBiG entry BGC0001445 used as the query for fai. For the aflatoxin BGC, ORFs which were not represented in the MIBiG entry BGC0000008 but predicted to be within the aflatoxin BGC by mapping of gene-calls from A. flavus NRRL 3357 by Skerker et al. 2021 are shown in gold. The major allele frequency distributions are shown for (E) aflX and (F) pksA, which depict opposite trends in sequence conservation according to their respective Tajima’s D calculations.
Of the 212 genomes with the leporin BGC, 202 contain instances that were not near scaffold edges. This set of 202 instances of the gene cluster were further investigated using zol, with comparative investigation of BGC instances from A. flavus population B genomes to instances from other populations requested. High sequence conservation was observed for all genes in the leporin gene cluster as previously reported12 (Table S3). Further, alleles for genes in the BGC from population B genomes were generally more similar to each other than to alleles from outside the population as indicated by high FST values (>0.85 for 9 of 10 genes) (Figure 3C; Table S3). While regulation of secondary metabolites in Aspergillus is complex65, zol analysis showed that the three essential genes for leporin production61 also had the lowest variation in the 100 bps upstream their exonic coordinates (Figure S2). This suggests higher variability is occurring in the transcription of the accessory lep genes within the species. This supports experimental evidence that has shown gene knockouts depleting certain leporin species will still permit the production of others61.
fai and zol were also applied to the BGC encoding aflatoxin across A. flavus66 (Table S4). Similar to the leporin BGC, the aflatoxin BGC was highly prevalent in the species and found in 71.8% of genomes. However, in contrast to the leporin BGC, the aflatoxin BGC contains several genes with positive Tajima’s D values, indicating greater sequence variability for these coding regions across the species (Figure 3D). One of the genes with a positive Tajima’s D value is aflX, which has been shown to influence conversion of the precursor veriscolorin A to downstream intermediates in the aflatoxin biosynthesis pathway67 (Figure 3E). An abundance of sites with mid-frequency alleles in the oxidoreductase encoding gene could represent granular control for the amount of aflatoxin relative to intermediates produced. The polyketide synthase gene pksA had the lowest Tajima’s D value of −2.4, which suggests it is either highly conserved or under purifying selection (Figure 3F). In addition, because a recent predicted reference proteome was used to infer genomic coding regions, fai and zol detected several highly conserved genes within the aflatoxin BGC that are not represented in the original reference gene cluster input for fai31. This includes a gene annotated as a noranthrone monooxygenase recently characterized as contributing to aflatoxin biosynthesis68,69 (Figure 3D).
Large-scale identification of the Enterococcal polysaccharide antigen and assessment of context restricted orthology inference
The Enterococcal polysaccharide antigen (Epa) is a signature component of the cellular envelope of multiple species within Enterococcus70–73, which has mostly been characterized in the species Enterococcus faecalis70,74–77. While molecular studies have provided evidence that the locus contributes to enterococcal host colonization76, evasion of immune systems78, and sensitivity to antibiotics79 and phages79,80, it was only recently that the structure of Epa was resolved and a model for its biosynthesis and localization formally proposed77. A homologous instance of the epa locus was identified in the other prominent pathogenic species from the genus, Enterococcus faecium71,73,81; however, the prevalence and conservation of epa across the diverse genus of Enterococcus82–84 remains poorly studied.
fai was used to search for homologous instances of epa across 5,291 Enterococcus, genomes estimated by GTDB to represent 92 species85, using a sensitive searching criterium and coordinates of the locus along the E. faecalis V583 genome as a reference75,77 (Supplementary Text). For detection of epa orthologous regions, co-location of at least seven of the 14 epa genes previously identified as conserved in both E. faecalis and E. faecium was required. The default threshold for syntenic conservation of homologous instances to the query gene cluster was also disregarded to increase sensitivity for the detection of epa in more distantly related enterococcal species to E. faecalis. To allow for capture and downstream analysis of auxiliary genes which might be species or strain-specific but related to Epa production or decoration, 20 kb flanking contexts of the core epa genes identified in each target genome were extracted.
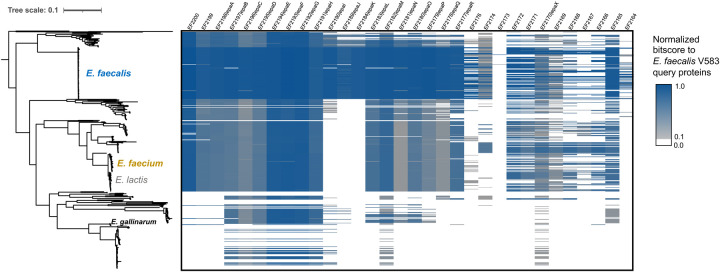
Using these criteria, 5,085 (96.1%) genomes from across the genus were found to possess an epa locus, confirming the locus as nearly core to the genus. Visual inspection of the epa genes among 463 representative Enterococcus genomes revealed that the core genes epaA-epaR are highly conserved in three of four major clades (Figure 4; Supplementary Text). Based on the detection criteria in fai, the epa locus in the fourth clade, previously referred to as the Enterococcus columbae group82, was either missing or encoded for highly divergent homologs of these genes. This clade includes Enterococcus gallinarum, one of the only other species in the genus, besides E. faecalis and E. faecium, reported to cause nosocomial outbreaks86,87.
Figure 4: The epa locus is conserved across most enterococcal species.
The distribution of the epa locus and associated genes, based on criteria used for running fai, is shown across 463 representative genomes across Enterococcus. Coloring of the heatmap corresponds to the normalized bitscore of the best alignment to coding sequences from E. faecalis V583.
Evolutionary trends and sequence diversity for individual genes with the epa locus, were next computed using zol after assessing zol’s reliability for gene cluster context-limited inference of orthology and the impact of dereplication on the calculation of evolutionary statistics by zol.
Gene-context specific orthology inference using fai and zol are concordant with genome-wide ortholog group predictions
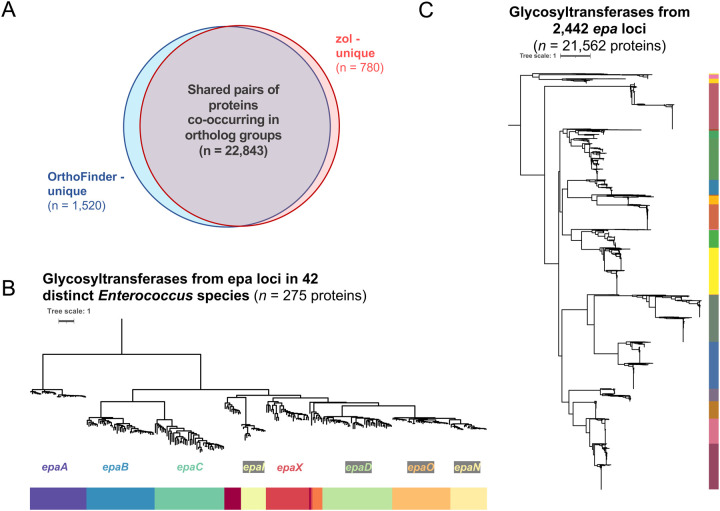
Genome-wide orthology inference is currently difficult to scale to hundreds or thousands of genomes belonging to multiple species. However, orthology inference can be made more accessible if larger loci are first identified as orthologous between genomes, through leveraging syntenic support23,27. To assess whether ortholog group inference was reliable when zol is applied on orthologous gene clusters identified across multiple species, we ran zol on high-quality instances of the epa locus from 42 different species (Figure 5C). Ortholog group predictions by zol were then compared to genome-wide orthology predictions by OrthoFinder18, which has been shown to yield highly accurate predictions in benchmarking experiments involving genomes from multiple species88. Orthology predictions were highly concordant between zol and OrthoFinder for proteins from diverse instances of the epa locus. zol identified 23,623 pairs of proteins within ortholog groups, of which 22,843 (96.70%) were also grouped together by OrthoFinder. Only 1,520 (6.24%) pairs of epa-associated proteins which were identified by OrthoFinder to belong to the same ortholog group were missed by zol.
Figure 5: Assessment of gene-cluster restricted ortholog grouping by fai and zol.
A) zol gene-cluster constricted ortholog group predictions for epa locus proteins from 42 distinct representative enterococcal species were compared to genome-wide predictions of ortholog groups by OrthoFinder. A phylogeny based on gap-filtered protein alignments of ortholog groups with domains featuring “glycosyl” and “transferase” as key words is shown from (B) epa loci in the 42 representative genomes and (C) a more comprehensive set of 2,442 epa loci. Each node represents a specific protein and coloring of the track corresponds to their ortholog group designations by zol. Note, (B) 2 (0.07%) and (C) 79 proteins (0.4%) were removed prior to phylogeny construction due to an abundance of gaps in the trimmed alignment.
Because the epa locus encodes multiple characterized and putative glycosyltransferases89, we used phylogenetics to examine the relationship between proteins belonging to ortholog groups with glycosyltransferase domains to confirm that major clades correspond to distinct ortholog group designations (Figure 5B). zol also has an option to “re-inflate” ortholog groups, expanding them to include proteins from gene clusters which were deemed redundant during dereplication. To demonstrate the scalability of zol, this “re-inflation”-based approach was next applied on the full set of high-quality and contiguous epa instances and a comprehensive phylogeny of ortholog groups corresponding to glycosyltransferases was constructed. In concordance with our analysis of the 42 representative genomes, distinct phylogenetic clades for glycosyltransferases corresponded to different ortholog groups identified by zol (Figure 5C).
Dereplication can impact taxa-wide inferences of selection-informative statistics
Dereplication, or removal of redundant gene cluster instances, is important to consider when working with highly sequenced bacterial taxa, including E. faecalis, where certain lineages, such as those commonly isolated at clinics, can be overrepresented in genomic databases. Over-representation of select lineages will skew estimates for some evolutionary statistics, such as those informative of selective pressures, complicating evaluation of evolutionary trends across the entire taxonomic group. We thus assessed the impact of dereplication on the calculation of evolutionary statistics for instances of epa in E. faecalis using two different approaches: (i) genome-wide dereplication with dRep90,91 and (ii) gene cluster specific dereplication with skani45. Dereplication at the gene cluster level with skani was performed directly in zol. The “re-inflation” option was also used to simulate comprehensive processing and calculation of evolutionary statistics while avoiding excessive computation.
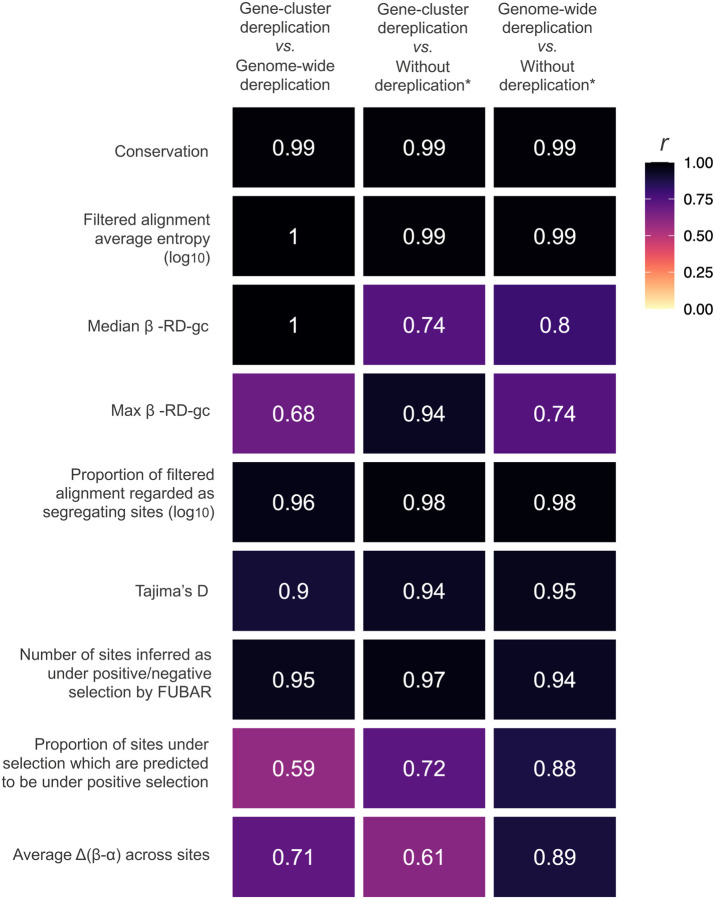
Regardless of the approach for dereplication, genome-wide or gene cluster-specific, the estimates of evolutionary and genomic statistics for analogous ortholog groups were highly concordant (Figure 6, S3). However, gene cluster based dereplication can overestimate or underestimate selection informative statistics, such as Tajima’s D or FUBAR-based inference of the number of sites under selection, relative to genome-wide dereplication performed using similar thresholds. This is likely because the core epa locus is highly conserved across E. faecalis which led to fewer representative gene clusters following dereplication and a lower weight being placed on conserved alleles when estimating such statistics. In contrast, more simplistic statistics, such as average sequence entropy and the proportion of total alignment sites regarded as segregating sites, were closely estimated for genes regardless of the dereplication method used. In addition, using the “reinflation” option in zol to infer orthology relationships across a comprehensive set of 1,232 high-quality and contiguous epa locus instances from the species produced concordant values for selection informative statistics to values generated using genome-wide based dereplication.
Figure 6: Effects of dereplication on the calculation of evolutionary statistics by zol.
The heatmap shows the correlation of values for analogous ortholog groups for various evolutionary statistics computed by zol when different approaches to dereplication are used. See Methods for further details. *To simulate no dereplication, gene-cluster dereplication with re-inflation parameters were used in zol.
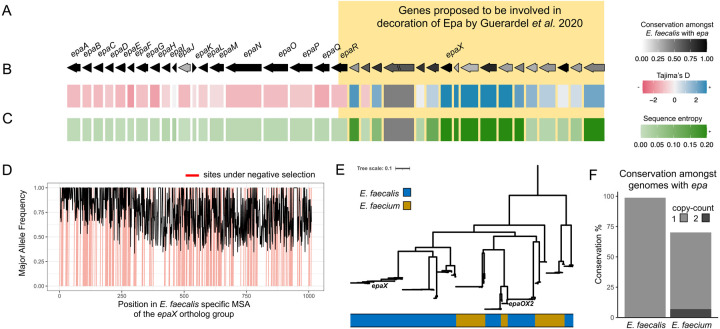
zol identifies genetic diversity of epaX-like glycosyltransferases
Because Epa biosynthesis and its conditional importance has mostly been investigated in E. faecalis70,74,75,77, we first examined evolutionary trends for proteins across instances of the epa locus from 75 E. faecalis representative genomes following genome-wide dereplication. In accordance with prior studies71,77, zol reported that one end of the locus corresponds to genes which are highly conserved and core to E. faecalis (epaA-epaR) whereas the other end contains strain-specific genes (Figure 7A; Table S5). Using zol, we further found that variably conserved genes exhibit high sequence dissimilarity, as measured using both Tajima’s D and average sequence entropy, in comparison to the core genes of the locus (Figure 7BC). Comparative and multi-species analysis of the epa locus between and across E. faecalis and E. faecium was next performed using gene cluster based dereplication with re-inflation using zol (Table S6). zol reported conservation statistics were consistently in agreement with previous studies71,73.
Figure 7: Distribution of the epa locus and associated genes across the genus of Enterococcus.
A) A schematic is shown for the epa locus in E. faecalis for genes which were found in ≥ 25% of 83 representative genomes for the species presented in consensus order with consensus directionality as inferred by zol. The coloring corresponds to the conservation of individual genes. Genes upstream and/or including epaR were recently proposed to be involved in decoration of Epa by Guerardel et al. 2020. “//” indicates that the ortholog group was not single-copy in the context of the gene-cluster. The tracks below the gene showcase their sequence similarity across the E. faecalis genomes measured using (B) Tajima’s D and (C) the average sequence alignment entropy. D) The major allele frequency is depicted across the alignment for the ortholog group featuring epaX. Sites predicted to be under negative selection by FUBAR, Prob(α>β) ≥ 0.9, are marked in red. E) An approximate maximum-likelihood phylogeny based on gap-filtered codon alignments for the ortholog group corresponding to epaX and epaX-like proteins in the joint E. faecalis and E. faecium investigation of the epa locus using zol. F) Conservation of epaX is shown amongst E. faecalis and E. faecium genomes with a high-quality representation of the epa locus available. Coloring of the bars corresponds to the proportion of genomes with a certain copy-count of the epaX-like ortholog group.
Twenty genes determined to be present in the majority (>95%) of epa clusters across both species, including epaABCDEFGH, epaLM, and epaOPQR. In addition, default parameters for orthologous clustering of proteins in zol detected a known truncated variant of the glycosyltransferase epaN in E. faecium.
The gene epaX, encoding a glycosyltransferase, was identified as one ortholog group with the greatest sequence variation in E. faecalis (Figure 7BD, S4). epaX was previously shown to be critical for E. faecalis host-gut colonization and proposed to be involved in the decoration of the rhamnan backbone structure of Epa with galactose and N-acetyl glucosamine76. Comparative analysis using E. faecium as the focal taxa further showed that the epaX-containing ortholog group has a low FST value, indicating alleles from E. faecalis and E. faecium species are phylogenetically interspersed. This was confirmed through phylogenetic assessment of the ortholog group (Figure 7E). In addition, although some allelic clades encode sequences from both species, genes remained sub-partitioned by species. This phylogenetic structure for the ortholog group, together with our prior observation that the epaX-containing ortholog group in E. faecalis has greater sequence variability relative to other glycosyltransferases from the locus, suggests extensive and ancestral sequence evolution of epaX-like glycosyltransferases. Further, while only 70% of E. faecium found to carry epa possess an epaX-like ortholog group, approximately 7% of them encode the ortholog in multi-copy (Figure 7F), suggesting the occurrence of intra-locus gene duplication.
Discussion
Here fai and zol are introduced to enable large scale evolutionary investigations of gene clusters in diverse taxa. Together these tools overcome current bottlenecks in computational biology to infer orthologous sets of genes at scale across thousands of diverse genomes.
Both fai and cblaster21 can be used to identify additional gene clusters within target genomes and extract them as GenBanks for downstream investigations using zol. For those lacking computational resources needed for fai analysis, cblaster offers remote searching of BGCs using NCBI’s BLAST infrastructure and non-redundant databases. More recently, CAGECAT92, a highly accessible web-application for running cblaster, was also developed and can similarly be used to identify and extract gene cluster instances from genomes represented in NCBI databases. In contrast to these tools, fai contains algorithms and options for users interested in: (i) identifying gene clusters across a comprehensive or redundant set of genomic assemblies, (ii) improved sensitivity for gene cluster detection in draft-quality assemblies, and (iii) automated filtering of secondary, or paralogous, matches to query gene clusters. In addition, users can apply zol to further investigate homologous sets of gene clusters identified from IslandCompare93, BiG-SCAPE22, or vConTACT294 analyses, which perform comprehensive clustering of predicted genomic islands, BGCs, or viruses.
The utility of fai is demonstrated here through rapid, targeted detection of a virus directly from lake metagenomic assemblies. Targeted detection of specific viruses longitudinally presents an efficient and tractable approach to understand how viral pangenomes evolve over time. In addition, by permitting fragmented detection of gene clusters and detection of proximity to scaffold edges, users can assess whether phages or other gene clusters corresponding to MGEs are present in their metagenomes. fai and zol will continue to compliment metagenomic applications as long-read sequencing becomes more economical and commonly used to profile microbial communities. For example, their application could be useful for assessing the presence of concerning MGEs conferring antimicrobial resistance traits95–97 and identifying novel auxiliary genes within known BGCs which may tailor the resulting specialized metabolites and expand chemical diversity98,99.
Reidentifying gene-clusters in eukaryotic genomes remains difficult due to technical challenges in gene prediction owing to the presence of alternative splicing. The ability of fai and zol to perform population-level genetics on common BGCs from the eukaryotic species A. flavus was demonstrated. While there are over 200 genomes of A. flavus on NCBI, only 5.1% have coding-sequence information readily available. We used miniprot35 to map high quality gene coordinate predictions from a representative genome in the species64 to the remainder of genomic assemblies within prepTG which enabled high sensitivity detection of BGCs with fai. Our analysis provides additional support that the leporin BGC is conserved in full across the species12 using an assembly-based approach.
Application of fai and zol to exopolysaccharide encoding gene clusters from pathogens of interest allows a better understanding of their conservation and evolutionary trends. This information can then aid the identification of potential genes to target for antivirulence efforts103,104 or genes underlying host-pathogen interactions76,105. fai was used to identify orthologous instances of the epa locus, encoding for an extracellular polysaccharide antigen, across thousands of diverse genomes from the genus of Enterococcus. Subsequently, application of zol reliably produced comparable orthology predictions to OrthoFinder, a highly dependable genome-wide orthology inference software18,88. While zol missed a small percentage of orthologous instances identified by OrthoFinder in our testing, this could be due to threshold settings for percent identity and coverage between pairs of proteins set in zol. Such thresholds are not enforced in OrthoFinder. However, parameters controlling these thresholds are adjustable in zol and allow users to increase or decrease orthology sensitivity at the expense of incurring false positives as they deem appropriate for their research objective.
Using zol, it was determined that an ortholog group containing epaX-like glycosyltransferases possess high sequence divergence relative to other glycosyltransferases within the epa locus in E. faecalis. In addition to influencing the ability of E. faecalis to colonize hosts76, mutations in epaX and other genes from the ortholog group have also been shown to impact susceptibility to phage predation100–102. Thus, because similar epaX-like glycosyltransferases are found in both E. faecalis and E. faecium, we hypothesize that extensive ancestral evolution of the epaX-containing ortholog group may have occurred to support evasion from phages and confer colonization of new hosts. In this study, we further found that the E. columbae group might lack or possess highly divergent versions of core epa genes found in E. faecalis and E. faecium, suggesting that development of anti-virulence approaches to broadly target Epa in all pathogenic enterococci might be difficult to achieve. Similar investigations with fai and zol can readily be performed for other exopolysaccharide encoding gene clusters of pathogens to better understand their conservation, evolutionary trends, identify appropriate genes to target for antivirulence efforts103,104, and infer whether certain genes underlie host-pathogen interactions76,105.
Options for dereplication and re-inflation provided within zol enable scalability to thousands of gene cluster instances. The usage of these options can further aid in performing more accurate evolutionary investigations for genes broadly across focal taxa or between clades, by overcoming biases due to overrepresentation of certain lineages in genomic databases12,47. Depending on the underlying origin of input gene clusters, zol can also be used to assess temporal48,106 or spatial49 evolutionary trends.
Practically, zol presents a comprehensive analysis tool for comparative genetics of related gene clusters to facilitate detection of evolutionary patterns that might be less apparent from visual analysis. Fundamentally, the algorithms presented within fai and zol enable the reliable detection of orthologous gene clusters, and subsequently orthologous proteins, across multi-species datasets spanning thousands of genomes and help overcome a key barrier in scalability for comparative genomics.
Methods
Software availability
zol is provided as an open-source software suite, developed primarily in Python3 on GitHub at: https://github.com/Kalan-Lab/zol. Docker and Bioconda107 based installations of the suite are supported. For the analyses presented in this paper, we used v1.2.0 of the zol software package. Minor patches, since incorporated into the software since v1.25, were added retrospectively to this version pertaining to safer acquisition of stored statistics when generating the final report. Version information for major dependencies of the zol suite33,35,40,45,108–115 or software generally used22,55,116 for analyses in this study is provided in Supplementary Table S7.
Data availability
Genomes and metagenomes used to showcase the application of fai and zol are listed with GenBank accession identifiers in Supplementary Table S8. Total metagenomes and their associated information from Lake Mendota microbiome samplings were originally described in Tran et al. 202354 and deposited in NCBI under BioProject PRJNA758276. Genomic assemblies available for A. flavus in NCBI’s GenBank database on Jan 31st, 2023 were downloaded in GenBank format using ncbi-genome-download (https://github.com/kblin/ncbi-genomedownload). Genomic assemblies for Enterococcus that met quality and taxonomic criteria for belonging to the genus or related genera (e.g. Enterococcus_A, Enterococcus_B, etc.) in GTDB85 release R207 were similarly downloaded from NCBI’s GenBank database using ncbi-genome-download in FASTA format.
Application of fai and zol to identify phages within metagenomes
VIBRANT was used to identify viral contigs or sub-contigs in the three total metagenomes from Tran et al. 202354 sampled on the earliest date of 07/24. Afterwards, predicted circular contigs were clustered using BiG-SCAPE22 which revealed a ~36 kb virus was found in two of the three metagenomes.
prepTG was run on all 16 total metagenomic assemblies from the Tran et al. 2023 study, performing gene calling with pyrodigal in metagenomics mode33 to prepare for comprehensive targeted searching of the virus. Afterwards, fai was run with default settings, with filtering of paralogous (or secondary) instances of the phage requested to retain only the best matching scaffold or scaffold segment resembling the queries.
Microevolutionary investigations of leporin and aflatoxin BGCs in Aspergillus flavus
Genomic assemblies downloaded from NCBI GenBank were processed using prepTG. Of the 217 genomic assemblies downloaded, one, GCA_000006275.3, was dropped from the analysis because the original GenBank had multiple CDS features with the same name, leading to difficulties in performing BGC prediction with antiSMASH116, and because alternate assemblies were available for the isolate. prepTG was run on all assemblies with miniprot35 based gene-mapping of the high-quality gene coordinate predictions available for A. flavus NRRL 3357 (GCA_009017415.1)64 requested. Target genomes were then searched for the leporin (BGC0001445) and aflatoxin (BGC0000008) BGCs using GenBanks provided on MIBiGv331. For leporin, AFLA_066840, as represented in the MIBiG database, was treated as a key protein required for detection of the BGC. Similarly, for aflatoxin, PksA (AAS90022.1), as represented in the MIBiG database, was treated as a key protein required for detection of the BGC. Draft-mode and filtering of paralogous segments was requested but turned off by default.
We reidentified population B as previously delineated12 using k-mer based ANI estimation117 and neighbor-joining tree construction118. A discrete clade (n=81) in the tree was validated to feature all isolates previously determined as part of population B12 and thus regarded as such.
For comprehensive and de novo BGC prediction, antiSMASH was run on the 216 genomic assemblies with ‘glimmerhmm’ requested for the option ‘--genefinding-tool’. BGCs were clustered using default settings in BiG-SCAPE with MIBiG reference BGC integration requested and a PKS-NRPS hybrid GCF was found to feature the leporin B BGC representative (BGC0001445). Only 65 (30.1%) of the 216 genomic assemblies featured this GCF, likely resulting from the use of distant gene models based on Cryptococcus genomes with glimmerhmm119. For remote clinker analysis, CAGECAT92 was used to search NCBI’s nr database with proteins from the leporin BGC representative (BGC0001445) provided as a query. Only 13 scaffolds, belonging to 12 assemblies (including GCA_000006275.3), were identified.
Evolutionary investigations of the epa locus across Enterococcus
All Enterococcus genomes represented in GTDB R20785 (n=5,291) were downloaded using ncbi-genome-download and processed in prepTG with gene-calling performed using pyrodigal33. Coordinates extending from 2,071,671 to 2,115,174 along the E. faecalis V583 chromosome, corresponding to genes EF2164 to EF2200. When using direct coordinates along a reference, fai reperforms gene-calling along the reference and extracts a local GenBank corresponding to the region between the coordinates. Gene calling is performed using pyrodigal. Because prior comparative analyses had shown that gene-conservation and gene-order can be slightly variable between epa loci from E. faecalis and E. faecium71, we relaxed the syntenic similar to query in fai from 0.6 to 0.0 and minimum percentage of query proteins needed to report a homologous instance of the epa locus to 10%. Instead, we required the presence of 50% of key epa proteins found in both E. faecalis and E. faecium, epaABCDEFGHLMOPQR, for the identification of valid homologous instances of the epa locus. To gather auxiliary genes flanking the core epa regions detected, we further requested the inclusion of CDS features found within 20 kb of the boundary core epa genes.
Genome selection for comparing ortholog grouping of proteins by zol with OrthoFinder:
Genome-wide dereplication of all Enterococcus genomes using dRep90 with fastANI91 and a secondary ANI clustering threshold of 99.0% led to the identification of 463 distinct genomes, including 101 E. faecalis genomes. Of these 101 genomes, 75 had high-quality epa instances which were not located near scaffold edges. zol was run on the 75 high-quality epa instances using default ortholog grouping parameters and similarly OrthoFinder v2.5.4 was run using default settings on the full, genome-wide set of 75 proteomes. To assess the concordance between OrthoFinder and zol for more diverse gene-clusters, gathered from multiple species, dRep was applied a second time on the set of 463 Enterococcus genomes using an ANI threshold of 95.0% to approximate selection of one representative genome per species120. This secondary dereplication identified 89 genomes, of which 42 featured highly-quality instances of the epa locus.
Phylogenetic analysis of glycosyltransferases found in or near the epa locus:
Ortholog groups from the zol analysis on the 42 representative and 2,442 comprehensive multi-species epa instances (Figure 5BC), as well as the 75 representative E. faecalis epa instances (Figure S4), were identified as glycosyltransferases if they featured the key words: “glycosyl” and “transferase” in Pfam protein domain annotations121. For each gene cluster set, protein sequences belonging to the ortholog groups were extracted, retaining association information with particular ortholog groups, and subsequently aligned using MUSCLE115. Alignment filtering was next performed using trimal with options “-keepseqs -gt 0.9”, sequences with greater than 25% of sites being gaps were filtered, and an approximate maximum-likelihood phylogeny was finally constructed using FastTree2110, midpoint rooted, and visualized using iTol122. Ortholog groups were assigned to specific epa gene designations based on sequence alignment of E. faecalis V583 proteins.
Assessing the impact of dereplication on the calculation of evolutionary statistics computed by zol:
To assess the impact of dereplication on the estimation of evolutionary statistics using zol, we focused on high-quality instances (<10% of bases ambiguous) of the epa locus that were not near scaffold edges from E. faecalis genomes. We ran dereplication at the genome scale using dRep90 with fastANI91 and a secondary ANI clustering threshold of 99.0% and dereplication at the gene-cluster scale using skani45 at 99.0% identity and 99.0% coverage with single-linkage clustering. We additionally simulated comprehensive processing of all high-quality gene-clusters distant from scaffold edges using the re-inflation option in zol, which allows expansion of ortholog groups determined in the dereplicated gene cluster set to the full listing of gene-clusters. Comparisons of estimates for various evolutionary statistics by zol between the different dereplication approaches were performed by first identifying the best matching ortholog groups from the three distinct analyses to each epa-associated gene from EF2164 to EF2200 in the E. faeacalis V583 reference genome based on E-value. Only ortholog groups which were found in single-copy within the epa context were considered.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health awarded to L.R.K (NIAID U19AI142720 and NIGMS R35GM137828) and the Broad Institute (U19AI110818). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors are grateful to James Kosmopoulos, Dr. Caitlin Sande, and Mary Hannah Swaney for feedback and assistance with data acquisition as well as Dr. Devon Ryan and Dr. Robert A. Petit III for assistance with incorporation of the suite into Bioconda.
References
- 1.Snyder L., Henkin T. M., Peters J. E. & Champness W. Molecular Genetics of Bacteria, 4th Edition. Preprint at 10.1128/9781555817169 (2013). [DOI] [Google Scholar]
- 2.Price M. N., Arkin A. P. & Alm E. J. The life-cycle of operons. PLoS Genet. 2, e96 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ptashne M. A genetic switch: Gene control and phage. lambda. (Palo Alto, CA (US); Blackwell Scientific Publications, 1986). [Google Scholar]
- 4.Andreu V. P. et al. gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota. Nature Biotechnology Preprint at 10.1038/s41587023-01675-1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischbach M. A., Walsh C. T. & Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. U. S. A. 105, 4601–4608 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gal-Mor O. & Finlay B. B. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell. Microbiol. 8, 1707–1719 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Kaper J. B., Nataro J. P. & Mobley H. L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Bolwell G. P. & Paul Bolwell G. Biochemistry & Molecular Biology of Plants. Phytochemistry vol. 58 185 Preprint at 10.1016/s0031-9422(01)00095-4 (2001). [DOI] [Google Scholar]
- 9.Lindahl L. & Zengel J. M. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 76, 6542–6546 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordero O. X. & Polz M. F. Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Salamzade R. et al. lsaBGC provides a comprehensive framework for evolutionary analysis of biosynthetic gene clusters within focal taxa. bioRxiv 2022.04.20.488953 (2022) doi: 10.1101/2022.04.20.488953. [DOI] [Google Scholar]
- 12.Drott M. T. et al. Microevolution in the pansecondary metabolome of Aspergillus flavus and its potential macroevolutionary implications for filamentous fungi. Proc. Natl. Acad. Sci. U. S. A. 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov R. L., Galperin M. Y., Natale D. A. & Koonin E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remm M., Storm C. E. & Sonnhammer E. L. Automatic clustering of orthologs and inparalogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Li L., Stoeckert C. J., Jr & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards D. J. & Holt K. E. Beginner’s guide to comparative bacterial genome analysis using next-generation sequence data. Microb. Inform. Exp. 3, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emms D. M. & Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medema M. H., Takano E. & Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abby S. S., Néron B., Ménager H., Touchon M. & Rocha E. P. C. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One 9, e110726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilchrist C. L. M. et al. Cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinformatics Advances 1, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro-Muñoz J. C. et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgescu C. H. et al. SynerClust: a highly scalable, synteny-aware orthologue clustering tool. Microb Genom 4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonkin-Hill G. et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21, 180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosentino S. & Iwasaki W. SonicParanoid: fast, accurate and easy orthology inference. Bioinformatics 35, 149–151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X. & Friedberg I. SwiftOrtho: A fast, memory-efficient, multiple genome orthology classifier. Gigascience 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallenet D. et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34, 53–65 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stam M. et al. NetSyn: genomic context exploration of protein families. bioRxiv (2023) doi: 10.1101/2023.02.15.528638. [DOI] [Google Scholar]
- 29.Salamzade R. et al. Evolutionary investigations of the biosynthetic diversity in the skin microbiome using lsaBGC. Microb Genom 9, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M. et al. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terlouw B. R. et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 51, D603–D610 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertelli C. et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larralde M. Pyrodigal: Python bindings and interface to Prodigal, an efficient method for gene prediction in prokaryotes. J. Open Source Softw. 7, 4296 (2022). [Google Scholar]
- 34.Hyatt D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. Protein-to-genome alignment with miniprot. Bioinformatics 39, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grazziotin A. L., Koonin E. V. & Kristensen D. M. Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res. 45, D491–D498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu B., Zheng D., Jin Q., Chen L. & Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siguier P., Perochon J., Lestrade L., Mahillon J. & Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcock B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond S. L. et al. HyPhy 2.5—A Customizable Platform for Evolutionary Hypothesis Testing Using Phylogenies. Mol. Biol. Evol. 37, 295–299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorpe H. A., Bayliss S. C., Sheppard S. K. & Feil E. J. Piggy: a rapid, large-scale pangenome analysis tool for intergenic regions in bacteria. Gigascience 7, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilchrist C. L. M. & Chooi Y.-H. clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Hackl T., Duponchel S., Barenhoff K., Weinmann A. & Fischer M. G. Virophages and retrotransposons colonize the genomes of a heterotrophic flagellate. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan M. J., Petty N. K. & Beatson S. A. Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw J. & Yu Y. W. Fast and robust metagenomic sequence comparison through sparse chaining with skani. bioRxiv 2023.01.18.524587 (2023) doi: 10.1101/2023.01.18.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blackwell G. et al. Exploring bacterial diversity via a curated and searchable snapshot of archived DNA. Access Microbiol. 4, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebreton F. et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lieberman T. D. et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat. Genet. 46, 82–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crits-Christoph A., Olm M. R., Diamond S., Bouma-Gregson K. & Banfield J. F. Soil bacterial populations are shaped by recombination and gene-specific selection across a grassland meadow. ISME J. 14, 1834–1846 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson R. R., Slatkin M. & Maddison W. P. Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran P. Q. & Anantharaman K. Biogeochemistry Goes Viral: towards a Multifaceted Approach To Study Viruses and Biogeochemical Cycling. mSystems 6, e0113821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barr J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U. S. A. 110, 10771–10776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lefeuvre P. et al. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 17, 632–644 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Tran P. Q. et al. Viral impacts on microbial activity and biogeochemical cycling in a seasonally anoxic freshwater lake. bioRxiv 2023.04.19.537559 (2023) doi: 10.1101/2023.04.19.537559. [DOI] [Google Scholar]
- 55.Kieft K., Zhou Z. & Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bok J. W. et al. Genomic mining for Aspergillus natural products. Chem. Biol. 13, 31–37 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Vadlapudi V. et al. Aspergillus Secondary Metabolite Database, a resource to understand the Secondary metabolome of Aspergillus genus. Sci. Rep. 7, 7325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robey M. T., Caesar L. K., Drott M. T., Keller N. P. & Kelleher N. L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. U. S. A. 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatmaker E. A. et al. Genomic and Phenotypic Trait Variation of the Opportunistic Human Pathogen Aspergillus flavus and Its Close Relatives. Microbiol Spectr 10, e0306922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie H. et al. Global multi-omics profiling reveals evolutionary drivers of phylogeographic diversity of fungal specialized metabolism. (2023) doi: 10.21203/rs.3.rs-2471999/v1. [DOI] [Google Scholar]
- 61.Cary J. W. et al. An Aspergillus flavus secondary metabolic gene cluster containing a hybrid PKS-NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genet. Biol. 81, 88–97 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Drăgan M.-A., Moghul I., Priyam A., Bustos C. & Wurm Y. GeneValidator: identify problems with protein-coding gene predictions. Bioinformatics 32, 1559–1561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scalzitti N., Jeannin-Girardon A., Collet P., Poch O. & Thompson J. D. A benchmark study of ab initio gene prediction methods in diverse eukaryotic organisms. BMC Genomics 21, 293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skerker J. M. et al. Chromosome assembled and annotated genome sequence of Aspergillus flavus NRRL 3357. G3 11, jkab213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang K., Tian J. & Keller N. P. Post-translational modifications drive secondary metabolite biosynthesis in Aspergillus: a review. Environ. Microbiol. 24, 2857–2881 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klich M. A. Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 8, 713–722 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Cary J. W., Ehrlich K. C., Bland J. M. & Montalbano B. G. The Aflatoxin Biosynthesis Cluster Gene, aflX, Encodes an Oxidoreductase Involved in Conversion of Versicolorin A to Demethylsterigmatocystin. Applied and Environmental Microbiology vol. 72 1096–1101 Preprint at 10.1128/aem.72.2.1096-1101.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cleveland T. E. et al. Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 27, 151–157 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Ehrlich K. C., Li P., Scharfenstein L. & Chang P.-K. HypC, the anthrone oxidase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 76, 3374–3377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y., Murray B. E. & Weinstock G. M. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 66, 4313–4323 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palmer K. L. et al. Comparative Genomics of Enterococci: Variation in Enterococcus faecalis, Clade Structure in E. faecium, and Defining Characteristics of E. gallinarum and E.casseliflavus. mBio vol. 3 Preprint at 10.1128/mbio.00318-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hancock L. E., Murray B. E. & Sillanpää J. Enterococcal Cell Wall Components and Structures. in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (eds. Gilmore M. S., Clewell D. B., Ike Y. & Shankar N.) (Massachusetts Eye and Ear Infirmary, 2014). [PubMed] [Google Scholar]
- 73.Qin X. et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 12, 135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teng F., Jacques-Palaz K. D., Weinstock G. M. & Murray B. E. Evidence that the Enterococcal Polysaccharide Antigen Gene ( epa ) Cluster Is Widespread in Enterococcus faecalis and Influences Resistance to Phagocytic Killing of E. faecalis. Infection and Immunity vol. 70 2010–2015 Preprint at 10.1128/iai.70.4.2010-2015.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teng F., Singh K. V., Bourgogne A., Zeng J. & Murray B. E. Further Characterization of the epa Gene Cluster and Epa Polysaccharides of Enterococcus faecalis. Infection and Immunity vol. 77 3759–3767 Preprint at 10.1128/iai.00149-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rigottier-Gois L. et al. The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. J. Infect. Dis. 211, 62–71 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Guerardel Y. et al. Complete structure of the enterococcal polysaccharide antigen (EPA) of vancomycin-resistant Enterococcus faecalis V583 reveals that EPA decorations are teichoic acids covalently linked to a rhamnopolysaccharide backbone. MBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith R. E. et al. Decoration of the enterococcal polysaccharide antigen EPA is essential for virulence, cell surface charge and interaction with effectors of the innate immune system. PLoS Pathog. 15, e1007730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh K. V. & Murray B. E. Loss of a Major Enterococcal Polysaccharide Antigen (Epa) by Enterococcus faecalis Is Associated with Increased Resistance to Ceftriaxone and Carbapenems. Antimicrob. Agents Chemother. 63, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ho K., Huo W., Pas S., Dao R. & Palmer K. L. Loss-of-Function Mutations in epaR Confer Resistance to NPV1 Infection in Enterococcus faecalis OG1RF. Antimicrobial Agents and Chemotherapy vol. 62 Preprint at 10.1128/aac.00758-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fiore E., Van Tyne D. & Gilmore M. S. Pathogenicity of Enterococci. Microbiol Spectr 7, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lebreton F., Willems R. J. L. & Gilmore M. S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (eds. Gilmore M. S., Clewell D. B., Ike Y. & Shankar N.) (Massachusetts Eye and Ear Infirmary, 2014). [PubMed] [Google Scholar]
- 83.Lebreton F. et al. Tracing the Enterococci from Paleozoic Origins to the Hospital. Cell 169, 849–861.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwartzman J. A. et al. Global diversity of enterococci and description of 18 novel species. bioRxiv 2023.05.18.540996 (2023) doi: 10.1101/2023.05.18.540996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parks D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. (2021) doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reid K. C., Cockerill F. R. III & Patel R. Clinical and epidemiological features of Enterococcus casseliflavus/flavescens and Enterococcus gallinarum bacteremia: a report of 20 cases. Clin. Infect. Dis. 32, 1540–1546 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Monticelli J., Knezevich A., Luzzati R. & Di Bella S. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarum and Enterococcus casseliflavus/flavescens. J. Infect. Chemother. 24, 237–246 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Nevers Y. et al. The Quest for Orthologs orthology benchmark service in 2022. Nucleic Acids Res. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dale J. L., Cagnazzo J., Phan C. Q., Barnes A. M. T. & Dunny G. M. Multiple Roles for Enterococcus faecalis Glycosyltransferases in Biofilm-Associated Antibiotic Resistance, Cell Envelope Integrity, and Conjugative Transfer. Antimicrobial Agents and Chemotherapy vol. 59 4094–4105 Preprint at 10.1128/aac.00344-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olm M. R., Brown C. T., Brooks B. & Banfield J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T. & Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van den Belt M. et al. CAGECAT: The CompArative GEne Cluster Analysis Toolbox for rapid search and visualisation of homologous gene clusters. BMC Bioinformatics 24, 181 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bertelli C. et al. Enabling genomic island prediction and comparison in multiple genomes to investigate bacterial evolution and outbreaks. Microb. Genom. 8, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zablocki O., Jang H. B., Bolduc B. & Sullivan M. B. VConTACT 2: A tool to automate genome-based prokaryotic viral taxonomy. in Plant and Animal Genome XXVII Conference (January 12– 16, 2019) (PAG, 2019). [Google Scholar]
- 95.Salamzade R. et al. Inter-species geographic signatures for tracing horizontal gene transfer and long-term persistence of carbapenem resistance. Genome Med. 14, 37 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheppard A. E. et al. Nested Russian Doll-Like Genetic Mobility Drives Rapid Dissemination of the Carbapenem Resistance Gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Groussin M. et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053–2067.e18 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Crits-Christoph A., Diamond S., Butterfield C. N., Thomas B. C. & Banfield J. F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Bickhart D. M. et al. Generation of lineage-resolved complete metagenome-assembled genomes by precision phasing. bioRxiv 2021.05.04.442591 (2021) doi: 10.1101/2021.05.04.442591. [DOI] [Google Scholar]
- 100.Chatterjee A. et al. Bacteriophage Resistance Alters Antibiotic-Mediated Intestinal Expansion of Enterococci. Infect. Immun. 87, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chatterjee A. et al. Parallel genomics uncover novel enterococcal-bacteriophage interactions. Preprint at 10.1101/858506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Canfield G. S. et al. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Preprint at 10.1101/2020.09.22.309401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Tilburg Bernardes E., Charron-Mazenod L., Reading D. J., Reckseidler-Zenteno S. L. & Lewenza S. Exopolysaccharide-repressing small molecules with antibiofilm and antivirulence activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 61, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shen Y. & Loessner M. J. Beyond antibacterials - exploring bacteriophages as antivirulence agents. Curr. Opin. Biotechnol. 68, 166–173 (2021). [DOI] [PubMed] [Google Scholar]
- 105.Shankar-Sinha S. et al. The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect. Immun. 72, 1423–1430 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao S. et al. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25, 656–667.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grüning B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 15, 475–476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cock P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Capella-Gutiérrez S., Silla-Martínez J. M. & Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price M. N., Dehal P. S. & Arkin A. P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang Y., Niu B., Gao Y., Fu L. & Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eddy S. R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Buchfink B., Xie C. & Huson D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2014). [DOI] [PubMed] [Google Scholar]
- 114.Schreiber J. Pomegranate: fast and flexible probabilistic modeling in python. J. Mach. Learn. Res. (2017). [Google Scholar]
- 115.Edgar R. C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 13, 6968 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blin K. et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ondov B. D. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paradis E., Claude J. & Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Majoros W. H., Pertea M. & Salzberg S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 120.Olm M. R. et al. Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries. mSystems 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mistry J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Letunic I. & Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomes and metagenomes used to showcase the application of fai and zol are listed with GenBank accession identifiers in Supplementary Table S8. Total metagenomes and their associated information from Lake Mendota microbiome samplings were originally described in Tran et al. 202354 and deposited in NCBI under BioProject PRJNA758276. Genomic assemblies available for A. flavus in NCBI’s GenBank database on Jan 31st, 2023 were downloaded in GenBank format using ncbi-genome-download (https://github.com/kblin/ncbi-genomedownload). Genomic assemblies for Enterococcus that met quality and taxonomic criteria for belonging to the genus or related genera (e.g. Enterococcus_A, Enterococcus_B, etc.) in GTDB85 release R207 were similarly downloaded from NCBI’s GenBank database using ncbi-genome-download in FASTA format.