Abstract
Vaccination and natural infection both elicit potent humoral responses that provide protection from subsequent infections. The immune history of an individual following such exposures is in part encoded by antibodies. While there are multiple immunoassays for measuring antibody responses, the majority of these methods measure responses to a single antigen. A commonly used method for measuring antibody responses is ELISA—a semiquantitative assay that is simple to perform in research and clinical settings. Here, we present FLU‐LISA (fluorescence‐linked immunosorbent assay)—a novel antigen microarray‐based assay for rapid high‐throughput antibody profiling. The assay can be used for profiling immunoglobulin (Ig) G, IgA and IgM responses to multiple antigens simultaneously, requiring minimal amounts of sample and antigens. Using several influenza and severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) antigen microarrays, we demonstrated the specificity and sensitivity of our novel assay and compared it with the traditional ELISA, using samples from mice, chickens and humans. We also showed that our assay can be readily used with dried blood spots, which can be collected from humans and wild birds. FLU‐LISA can be readily used to profile hundreds of samples against dozens of antigens in a single day, and therefore offers an attractive alternative to the traditional ELISA.
Keywords: Antibody profiling, antigen microarrays, ELISA, FLU‐LISA, immune history
We developed a novel high‐throughput antibody binding assay for profiling immune history to previous infections and vaccines. We demonstrate its use using samples from mice, birds and humans, and compare it with the traditional ELISA.
INTRODUCTION
Immunoassays are a broad set of methods that can be used to detect the presence of immune responses to antigens such as pathogens and autoantigens. Antibodies encode the immune history of an individual following exposures to both infections and vaccines, and also offer protection from subsequent infections. A common method that has been widely used for antibody characterization is ELISA. 1 , 2 The ELISA method can be used to detect the presence of hormones, peptides, proteins and antibodies against a specific antigen of interest. The assay uses an enzyme–substrate reaction that can be measured and quantified by optical density. There are several common ELISA techniques, including direct, indirect, sandwich and competitive ELISAs. 3 ELISA is a high‐accuracy semiquantitative assay that is simple to perform in the laboratory using a variety of sample types and available reagents and equipment. 4 ELISA is widely used (1) to detect ligands in various biological samples including swabs, blood, sera and stool; (2) for clinical diagnosis of diseases such as HIV and malaria 5 (3) and for characterizing antibody responses to vaccines and natural infections. 6 Each ELISA can quantify antibodies to a single antigen. Therefore, characterizing binding of a single sample to multiple antigens using this assay is laborious and requires high volumes of sample and antigens.
Antigen microarrays (AMs) are a high‐throughput antibody‐binding assay that allow the quantification of antibody responses to hundreds or thousands of antigens simultaneously. 7 In essence, the AM provides an efficient and highly sensitive antibody binding screen. This platform can accommodate a variety of different antigens, including proteins, peptides, lipids and whole viruses, providing a comprehensive binding antibody profile. It has been extensively used to study antibody responses to both viral and bacterial infections, 8 , 9 , 10 as well as to identify cancer 11 and autoimmune 12 , 13 biomarkers.
Here we developed and optimized an AM‐based assay for rapid high‐throughput antibody profiling (Figure 1). We developed influenza‐specific AMs spotted with recombinant hemagglutinin (HA) and neuraminidase proteins of multiple influenza strains from the A/H1N1, A/H3N2, A/H5N1 and B subtypes. We demonstrated the specificity of our assay using subtype‐specific and cross‐reactive anti‐influenza monoclonal antibodies (mAbs), and characterized the immune history to previous influenza exposures using serum samples from mice and humans. We also showed that the assay can be used to profile antibodies from dried blood spots, which are easier to collect, especially in field studies of wild birds and in serological surveys. Finally, we developed “FLU‐LISA” (fluorescence‐linked immunosorbent assay), an ELISA‐like antibody‐binding assay based on AMs, in which each antigen is spotted in several serial concentrations. FLU‐LISA allows rapid semiquantitative characterization of binding profiles to a large panel of antigens simultaneously, using minimal sample and antigen volumes. Using human serum samples, we compared our novel assay with the traditional ELISA, and demonstrated its concordance.
Figure 1.
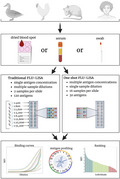
FLU‐LISA: a rapid high‐throughput antibody binding assay. FLU‐LISA is an antigen microarray‐based binding assay that can simultaneously quantify responses to multiple antigens using minimal sample volumes. The assay can be performed on multiple sample types from multiple species including humans, mice, chickens and wild birds. Two alternative approaches to perform FLU‐LISA are available: (1) traditional FLU‐LISA, an ELISA‐like assay, performed using multiple sample dilutions that are incubated on separate arrays, in which each antigen is spotted at a single concentration; (2) one‐shot FLU‐LISA, in which each sample is incubated once at a single dilution and antigens are spotted at multiple concentrations on each array. Both alternatives provide semiquantitative characterization of the antibody repertoire to multiple antigens simultaneously. FLU‐LISA, fluorescence‐linked immunosorbent assay; MFI, Median Fluorescent Intensity.
RESULTS
Monovalent influenza infection or immunization in mice generates a subtype‐specific antibody profile
To test the specificity and sensitivity of the AMs, mice (n = 9) were infected intranasally with a sublethal dose of A/Puerto Rico/8/1934 (PR8) (A/H1N1). Serum samples were collected at baseline and 28 days after the infection. Immunoglobulin (Ig) G antibody profiles from each mouse were generated at baseline and day 28, using the M1 influenza HA Ams spotted with recombinant HA proteins from 11 A/H1N1 (H1), 18 A/H3N2 (H3), 7 A/H5N1 (H5) and 5 influenza B strains (see the “Methods” section and Supplementary table 1). At baseline, mice displayed none, or very low levels of IgG binding to the PR8 HA protein. After the infection anti‐PR8 IgG levels were significantly higher than baseline levels (P = 0.007; Figure 2a). The anti‐PR8 antibody responses varied significantly between mice and one mouse failed to mount an immune response following sublethal infection (Figure 2a). Geometric mean magnitude titers were computed for antibody reactivity to each of the four influenza subtypes. Baseline titers were low to all four subtypes, while only H1 and H5 titers significantly increased after PR8 infection (P = 0.003 and P = 0.002, respectively; Figure 2b). Previous work reported that antibodies generated following H1N1 infection or vaccination can cross‐react with H5N1 strains. 14 , 15 , 16 , 17 , 18
Figure 2.
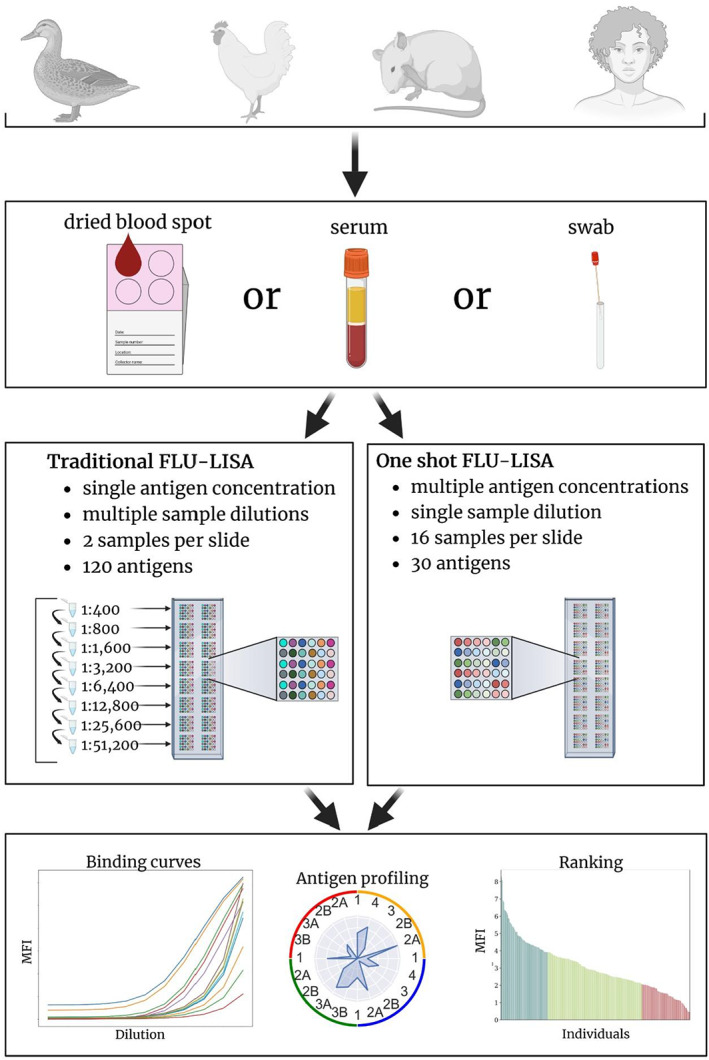
Magnitude and breadth of anti‐influenza immunoglobulin (Ig) G antibodies to hemagglutinin (HA) proteins. Nine C57BL/6 female mice were infected intranasally with A/Puerto Rico/8/1934 (PR8) H1N1 virus, as previously described. 37 Serum samples were collected before the infection (blue) and 28 days after the infection (orange). Antigen microarrays spotted with HA proteins from A/H1N1 (n = 11), A/H3N2 (n = 18), B (n = 5) and A/H5N1 (n = 7) influenza strains (microarray M1 in Supplementary table 1) were hybridized and the median fluorescent intensity (MFI) of IgG binding was measured. Data from one representative experiment of three experiments are shown. (a) IgG binding to the HA of the infection strain PR8. (b) IgG geometric mean titers (GMTs) to the panel of HA proteins from each subtype. In panels a and b, horizontal lines represent the median, boxes denote the 25th and 75th percentiles and the error bars represent 1.5 times the interquartile range. Statistical significance was assessed using the Wilcoxon signed‐rank test comparing baseline and postvaccination responses: *P < 0.05, **P < 0.005. (c) Serum IgG profiles of four representative mice at baseline (blue) and after PR8 infection (orange) across A/H1N1, A/H3N2 and B influenza strains. (d) A spider plot of the mean IgG‐binding profile across all nine mice at baseline (blue) and after vaccination (orange). In panels c and d, each vertex represents the normalized MFI to a single HA protein. The numbers listed around the inner circle denote the year each influenza strain was isolated (Supplementary table 1). Counterclockwise, HA proteins from H5N1 strains 1997–2008 (brown); H3N2 strains 1968–2014 (purple); H1N1 strains 1918–2015 (blue) and B strains 1988–2013 (green).
To visualize the baseline and postvaccination IgG profiles of each individual mouse separately, we generated spider plots to display the normalized MFI staining for all H1, H3, H5 and B HA antigens. Representative spider plots for four mice are presented in Figure 2c. Most mice generated very narrow baseline responses to the HA proteins. Each mouse has a unique IgG profile that varies in both the overall breadth and the magnitude of responses, but also in the specificity to each subtype. For example, mouse number 4 (S4 in Figure 2c) had a very broad postinfection response, including high IgG binding to all the three influenza A subtypes, as well as to influenza B HA proteins. By contrast, mouse number 2 (S2) responded predominantly to H1. We computed the average IgG binding of all the nine mice with each HA protein at both time points (Figure 2d). We found that the highest IgG postinfection responses were against the infection strain (PR8; Figure 2d) and the H1N1 A/Solomon Islands/3/2006 strain. However, postinfection IgG levels were also observed for strains from all other influenza A subtypes.
To further examine the specificity of the AMs, we analyzed sera from mice exposed intramuscularly to inactivated viruses from different influenza A subtypes. C57BL/6 mice were given either H3N2 A/HKx31 (X31), mouse‐adapted H1N1 A/California/07/2009 (Cal09) or H5N1 A/Vietnam/1203/04 (Viet1203). Serum samples were collected 28 days later, and were incubated with M2 influenza HA AMs spotted with 8 H1, 11 H3 and 4 H5 proteins. Overall, we observed extensive heterogeneity in the antibody profiles of mice within each of the immunization groups (Figure 3), as observed in previous studies. 10 Mice immunized with Cal09 generated an IgG response to Has from the H1N1 subtype (Figure 3a). Moreover, IgG‐binding levels to the Cal09 HA protein were the highest in 9/10 Cal09 immunized mice. Importantly, none of these mice responded to H3N2 HAs (Figure 3b), and there were varying levels of cross‐reactive responses to H5N1 strains (Figure 3c). Only a single Cal09 injected mouse had broad and potent cross‐reactive antibody responses to both H1N1 and H5N1 HAs (compare Figure 3a, c). Significant levels of anti‐H3 antibodies were detected only in mice that were injected with the A/H3N2 X31 strain (Figure 3b). The X31‐injected mice generated none to very weak antibody responses to HA proteins from the H1N1 and H5N1 subtypes (Figure 3a, c). All the mice that were immunized with the H5N1 Viet1203 strain developed high levels of IgG antibodies to multiple H1 and H5 proteins, which belong to the same antigenic group (Figure 3a, c). 19 , 20 Two of the H5N1‐immunized mice generated very weak cross‐reactive responses to H3 HAs (Figure 3b). Overall, we find almost no cross‐reactive responses between H3 and H1 or H5 antibody responses and varying levels of cross‐reactivity between H5 and H1 antibody responses.
Figure 3.
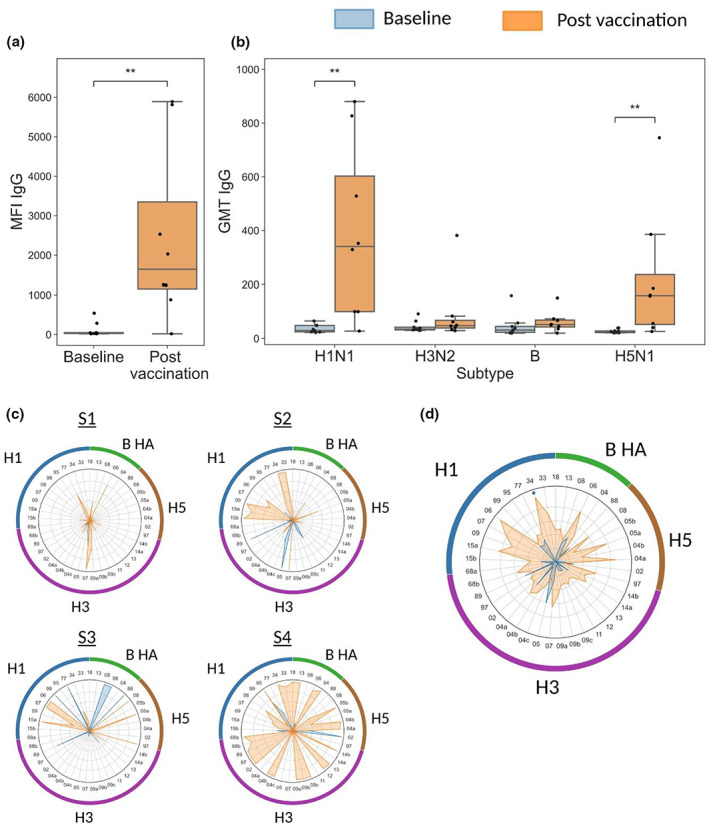
Subtype specificity of influenza hemagglutinin (HA) microarrays. Four groups of female mice (n = 10 per group) were injected intramuscularly with 40 μg of inactivated influenza A viruses from three different subtypes: A/California/07/2009 (Cal09, H1N1), A/HKx31 (X31, H3N2) and A/Vietnam/1203/2004 (Viet1203, H5N1). A negative control group was injected with phosphate‐buffered saline (PBS). Serum samples were collected at day 28 after immunization, and binding of immunoglobulin (Ig) G antibodies to influenza recombinant HA proteins was profiled using the M2 influenza antigen microarray (Supplementary table 1). Data from one representative experiment of two experiments are shown. (a) IgG responses to the HA protein of H1N1 strains (n = 8), (b) IgG responses to the HA protein of H3N2 strains (n = 11) and (c) IgG responses to the HA protein of H5N1 strains (n = 4). Each bar represents the cumulative MFI for a single mouse to all of the recombinant HA proteins listed. (d) The S1 and receptor‐binding domain (RBD) proteins of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) Wuhan strain were spotted on sCoV antigen microarrays (AMs) in six serial concentrations (2.03–65 μg mL−1; Supplementary table 3). Samples of 30 individuals collected prior to the SARS‐CoV‐2 pandemic (negative controls) were compared with 14 individuals that received three doses of the Pfizer‐BioNTech BNT162b2 vaccine (SARS‐CoV‐2 vaccinated). Responses of IgG antibodies were quantified using the area under the curve (AUC) statistic across all six antigen concentrations. ****P‐value < 0.0001 (Wilcoxon rank‐sum test).
As humans are continuously exposed to influenza viruses, and even newborns have maternal anti‐influenza antibodies, we further validated the specificity of our arrays using samples from a human cohort vaccinated three times with the Pfizer severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) messenger RNA vaccine, and samples from a human cohort collected prior to the SARS‐CoV‐2 pandemic. We used sCoV AMs spotted with recombinant spike and receptor‐binding domain (RBD) proteins of Wuhan vaccine strain (see the “Methods” section and Supplementary table 3). We found that none of the prepandemic samples generated responses to SARS‐CoV‐2 antigens, while all of the samples from vaccinated individuals generated varying levels of responses to these antigens (Figure 3d). Both pre‐ and postpandemic samples had antibodies to the RSV protein G which was spotted on the same array (Supplementary figure 1).
Antibody profiles generated from dried blood spots are comparable to serum antibody profiles
Collecting dried blood spots provides an attractive alternative to serum collection because it requires very small blood volumes (125–500 μL), does not require centrifugation and freezing and is minimally invasive, allowing collection of samples in field studies of wild birds, and from newborn babies and children. To compare the antibody repertoires obtained from serum and dried blood spots, we used paired samples from both chickens and humans.
We used serum and dried blood samples collected from 36 breeder chickens that were vaccinated two times with an H9N2 influenza vaccine to compare the anti‐influenza antibody repertoires obtained from the two sample types. Blood samples were collected from 40–41‐day‐old chicks, 34 days after the vaccination (n = 16), and from 2.5–3‐month‐old chickens 30–40 days after the boost (n = 20). Four blood spots (125–500 μL) from each sample were dropped on a Whatman FTA blood card and left to dry, while the rest of the sample was centrifuged for serum isolation. To compare the binding profiles of IgY from a single dried blood spot and the serum with influenza antigens, we used the pan‐influenza Ams (see the “Methods” section) spotted with 64 recombinant influenza proteins from both human and avian strains and 4 PR8 internal proteins: M1, NS1, NS2 and NP, which are relatively conserved. As only a small volume of serum could be collected, in particular from young chickens, serum samples were run at a dilution of 1:4000. By contrast, each dried blood spot was reconstituted in 2 mL buffer and further diluted only by 1:20 for microarray incubation. We found high correlations between IgY levels measured in the serum and from dried blood spots with an average correlation of 0.932 (P < 2 × 10−7, Pearson correlation; Figure 4a, b). Correlations were < 0.9 in 8/36 samples as a result of IgY responses that were detected in dried blood spots but not in sera (Supplementary figure 2). We also observed lower background (nonspecific binding) to our arrays when using dried blood spots.
Figure 4.
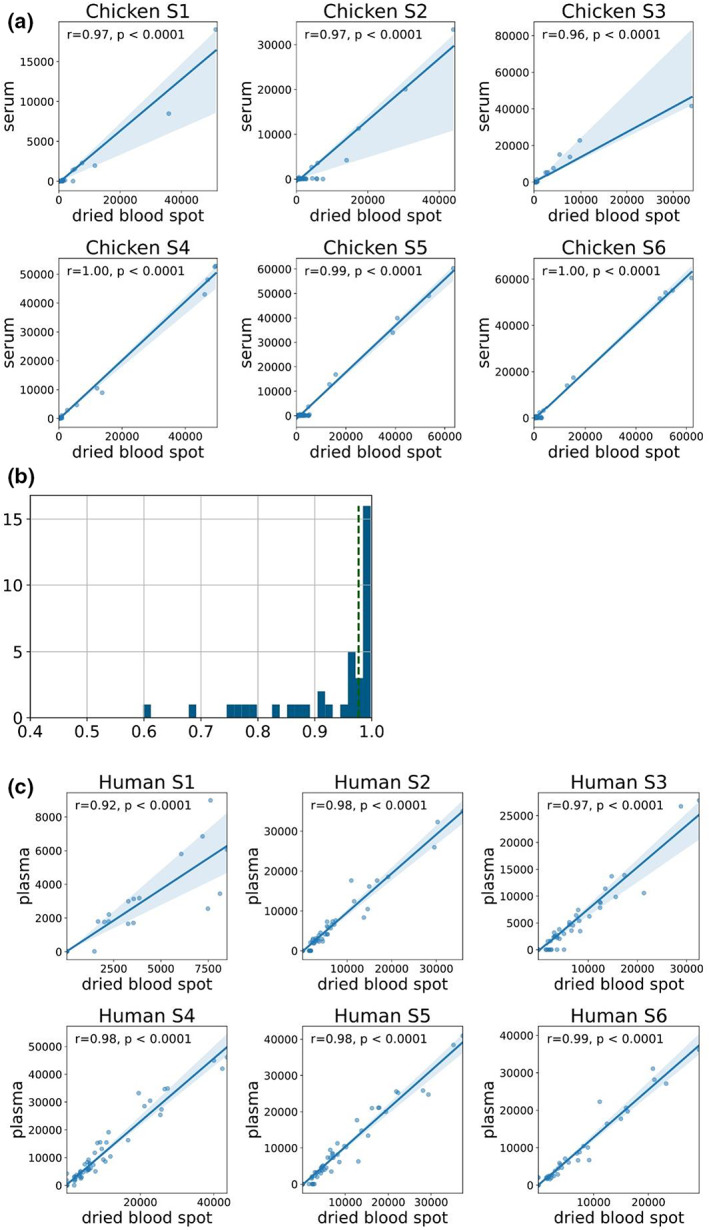
Antibody profiles of dried blood spots are highly correlated with those measured from serum or plasma. Chickens were vaccinated with an H9N2 influenza vaccine two times, and blood samples were collected 34 days after the first vaccination (n = 16) or 30 days following the second vaccination (n = 20). Serum and dried blood spots were collected from each blood sample. The immunoglobulin (Ig) Y response to vaccination was profiled using an antigen microarray spotted with recombinant influenza proteins from 15 influenza A and B subtypes (see Supplementary table 2). Data from one representative experiment of two experiments are shown. (a) Scatter plots of the median fluorescent intensity (MFI) for each serum–blood spot pair are presented for six chickens. Each dot represents the MFI to a single antigen as measured by dried blood spot (x‐axis) and serum (y‐axis). The Pearson correlation was computed for each pair. Samples S1 and S2 are from 41‐day‐old female chicks, samples S3 and S4 are from 2.5‐month‐old male chickens and samples S5 and S6 are from 3‐month‐old female chickens. (b) The distribution of Pearson correlation coefficients of the 36 serum–blood spot pairs. The dotted green line represents the median r value of 0.977. (c) Dried blood spots and plasma samples were also collected from six human adults that were previously vaccinated with the Pfizer severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) vaccine. The scatter plots of the IgG responses to a human SARS‐CoV‐2 antigen microarray (Supplementary table 3). Each dot represents the MFI to a single antigen as measured by dried blood spot (x‐axis) and plasma (y‐axis). The Pearson correlation coefficient and P‐value are presented for each individual.
We then used dried blood spots and plasma samples from six human blood samples to compare the antibody profiles generated from these paired samples. The samples were collected from human adults following three vaccinations with the Pfizer‐BioNTech BNT162b2 SARS‐CoV‐2 vaccine, and IgG response to human coronaviruses, including many SARS‐CoV‐2 variants, was profiled using the CoV Ams (Supplementary table 3). Similar to our findings with antibody profiles to avian influenza in the chicken samples, we found very high correlations between the IgG profiles to human coronaviruses generated from dried blood spots and plasma samples in humans (r > 0.92; Figure 4c). Interestingly, in human samples we also observed that some responses were detected in the dried blood spot samples but not in the serum samples (e.g. sample 3; Figure 4c).
Developing and optimizing FLU‐LISA
The accuracy of the traditional ELISA derives in part from testing each sample in several serial dilutions, which enables calculation of the area under the curve (AUC) statistic. When AMs are spotted with a single concentration for each antigen, as in the aforesaid experiments, the incubation of a set of microarrays with a set of serially diluted samples is not efficient, and sometimes impossible when sample volume is limited. An important advantage of the AM assay is that antigens can be spotted in multiple dilutions on each array, allowing us to also characterize antibody binding using a single dilution of the sample, while considering responses to each antigen across all of its dilutions. We therefore sought to develop “FLU‐LISA,” an alternative AM‐based assay in which each antigen is spotted in several serial concentrations such that incubation with a single dilution of the sample can allow computing an AUC across dilutions of each of the antigens spotted on the array. We spotted FLU‐LISA microarrays with four influenza HA proteins. Each protein was spotted in 11 serial twofold dilutions (5 ng mL−1 to 125 μg mL−1) in triplicates.
We compared the traditional ELISA with traditional FLU‐LISA, using serial dilutions of the sample and considering only one concentration of the spotted antigen (Figure 1). We used a mouse mAb against the spotted H3N2 A/Brisbane/10/2007 HA protein as a primary antibody for assay calibration. The same A/Brisbane/10/2007 HA protein was also used to coat 384‐well ELISA plates at a single concentration (4 μg mL−1) to compare the FLU‐LISA results with the traditional ELISA. Serial concentrations of the primary mAb (5 ng mL−1–80 μg mL−1) were incubated with the FLU‐LISA arrays and with the coated ELISA plates. A secondary anti‐mouse IgG antibody was used to detect the binding of the primary antibody to the antigen. For the traditional ELISA, the secondary antibody was bound to a horseradish peroxidase, and for the FLU‐LISA the secondary antibody was conjugated to a fluorescent dye (Alexa 635). A five‐parameter logistic regression model was used to fit curves to the median fluorescent intensity (MFI) measured by FLU‐LISA as a function of the antigen concentration, or to the relative light units as a function of the sample dilution in the traditional ELISA. We found that the Pearson correlation between the curves of the traditional ELISA and FLU‐LISA was r = 0.975 (P = ; Figure 5a).
Figure 5.
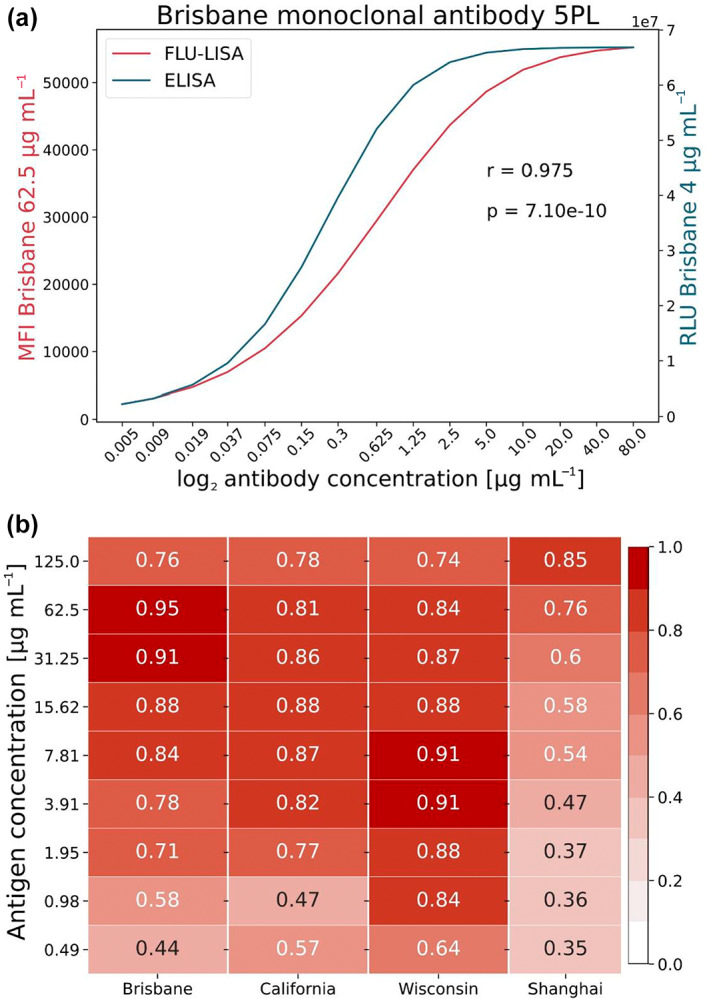
Comparison of the ELISA and FLU‐LISA titer curves for anti‐HA monoclonal antibody and human serum samples. (a) Binding of 15 serial twofold dilutions of anti‐A/Brisbane/10/2007 HA monoclonal antibody to a single concentration of the A/Brisbane/10/2007 HA protein, as measured by ELISA (blue; 4 μg mL−1 protein) or FLU‐LISA (red; 62.5 μg mL−1 protein). Curves were fitted using a five‐parameter logistic regression model. The Pearson correlation between the two curves was r = 0.975, P = . Data from one representative experiment of two experiments are shown. (b) ELISA and FLU‐LISA correlations using human serum samples. Immunoglobulin (Ig) G antibodies against HA proteins from four influenza strains were quantified in serum samples of 10 healthy adult individuals using both ELISA and FLU‐LISA. Both assays were performed over 15 twofold serial dilutions of the serum samples. ELISA used a single antigen concentration (4 μg mL−1), while FLU‐LISA was run against antigens spotted in 11 twofold serial dilutions. MFI or relative light unit (RLU) curves as a function of the sample dilution were fitted using a five‐parameter logistic regression model for each antigen concentration separately, and Pearson correlation coefficients were computed between the area under the curve (AUC) values of the two assays for each antigen concentration. Data from one representative experiment of two experiments are shown. FLU‐LISA, fluorescence‐linked immunosorbent assay; HA, hemagglutinin; MFI, Median Fluorescent Intensity.
We then used both traditional FLU‐LISA and traditional ELISA methods to measure the levels of serum IgG antibodies to HA proteins of four influenza strains in serum samples from 10 healthy adults that were collected in a clinical study in Israel during the 2018 winter season. Each serum sample was run at 15 serial dilutions in both assays. While ELISA was performed with a single antigen concentration (4 μg mL−1), the FLU‐LISA results were calculated for each spotted antigen concentration. We used the same five‐parameter logistic regression model to fit curves measured by FLU‐LISA for each antigen concentration as a function of the sample dilution, and the AUC was calculated. Similarly, the AUC of ELISA curves (relative light units as a function of the sample dilution) was calculated using the same model. We computed the Pearson correlations between the FLU‐LISA AUCs and the ELISA AUCs. The FLU‐LISA AUC was calculated for each antigen concentration spotted on the array. We found that the correlations were high for the three human influenza HA proteins for all relevant concentrations (0.71 ≤ r ≤ 0.95, P < 0.0001; Figure 5b), and lower for the H7N9 Shanghai strain, an avian strain to which the individuals were not exposed (Figure 5b).
Generating binding curves using antigen dilutions
We then compared the traditional FLU‐LISA (multiple sample dilutions and a single antigen concentration; Figure 1) with the one‐shot FLU‐LISA (single sample concentration across multiple antigen concentrations; Figure 1) using an anti H3N2 Brisbane 2007 mouse mAb tested at different concentrations. We found that the assay was able to distinguish between the different concentrations of the same mAb (Figure 6a). To further compare the two assays, we plotted antibody levels as a function of both mAb and antigen concentrations. We found that the signal decay across antigen dilutions or mAb dilutions was similar (Figure 6b).
Figure 6.
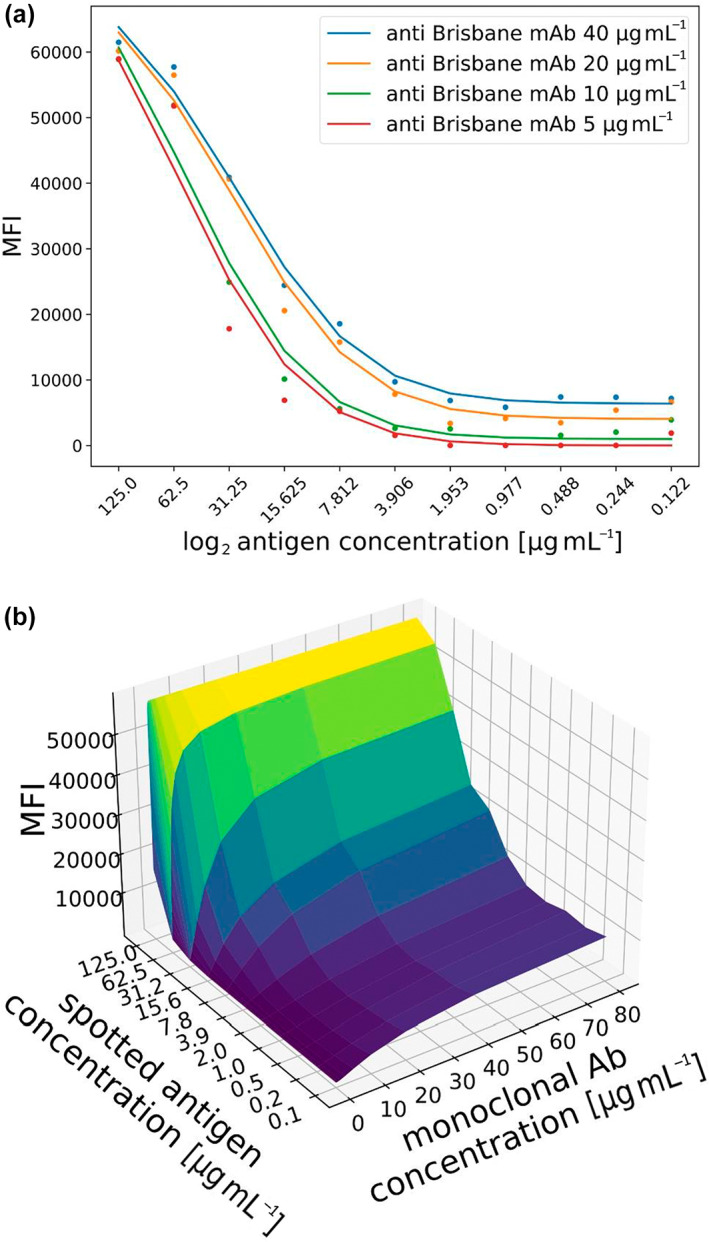
Comparison of one‐shot FLU‐LISA and traditional FLU‐LISA binding curves. (a) Comparison of the binding curves of an anti‐H3N2 Brisbane 2007 monoclonal antibody (mAb) using one‐shot FLU‐LISA. The microarray included 11 serial twofold dilutions of the A/Brisbane/10/2007 hemagglutinin antigen. The mAb was run at four different dilutions, and each dilution was incubated on a separate microarray. (b) Comparison of one‐shot FLU‐LISA with traditional FLU‐LISA. The same anti‐H3N2 mAb was run at 15 multiple dilutions on FLU‐LISA microarrays in which the antigen was spotted at 11 dilutions. Binding curves were generated across antigen dilutions (y‐axis) and across mAb dilutions (x‐axis). FLU‐LISA, fluorescence‐linked immunosorbent assay.
To compare the quantitative values of the traditional ELISA with the one‐shot FLU‐LISA, we computed the AUC statistic for 10 human serum samples according to both assays for the four Has spotted on the FLU‐LISA arrays: H3N2 A/Wisconsin/67/2005 (Wisconsin), H3N2 A/Brisbane/10/2007 (Brisbane), H1N1 A/California/07/2009 (California) and H7N9 A/Shanghai/1/2013 (Shanghai). The FLU‐LISA AUC values were computed using a single sample concentration (1:3200) across all antigen concentrations, whereas the ELISA AUC values were computed for a single antigen concentration (4 μg mL−1) across all sample dilutions. We found that the Spearman correlation coefficients () of the two assays were 0.89, 0.92, 0.75 and 0.72 for Brisbane, California, Wisconsin and Shanghai.
To compare the specificity of the one‐shot FLU‐LISA and traditional ELISA, we also compared Spearman correlations between AUC values of IgG response in the 10 human serum samples with the four antigens in each assay. We found that overall, there were higher correlations between the IgG levels to different antigens according to ELISA, even for strains from different subtypes, as compared with FLU‐LISA (Figure 7; Supplementary tables 4 and 5). In particular, the ELISA results included a moderate correlation between IgG responses to H3N2 Brisbane and H7N9 Shanghai proteins, whereas such a correlation was not detected by FLU‐LISA. Higher correlations were found also between the IgG levels to the two H3N2 HA proteins by ELISA compared with FLU‐LISA.
Figure 7.
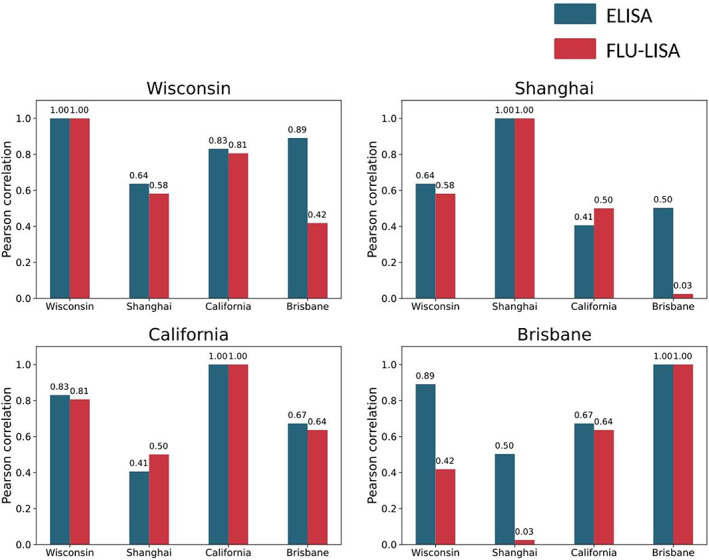
Pearson correlations between ELISA and one‐shot FLU‐LISA for four influenza antigens. We compared the correlations between the immunoglobulin (Ig) G area under the curve (AUC) for serum samples of 10 healthy adult individuals against four influenza strains: A/Wisconsin/67/2005 (H3N2), A/Brisbane/10/2007 (H3N2), A/California/07/2009 (H1N1) and the A/Shanghai/1/2013 (H7N9). ELISA was run using 15 serial dilutions (1:25–1:409 600) of each serum and a single antigen concentration, thus ELISA AUC was calculated across serum dilutions. FLU‐LISA microarrays were incubated once with each serum sample diluted 1:3200, and AUC was calculated across antigen dilutions. Pairwise correlations between FLU‐LISA AUC values with the different antigens are shown in red, and correlations between ELISA AUC values are shown in blue. In each of the four panels the correlation values between a single strain (Wisconsin, Shanghai, California or Brisbane) with all four strains (including itself) are visualized for each assay separately. We found that overall correlations between ELISA AUC values were higher. FLU‐LISA, fluorescence‐linked immunosorbent assay.
Rapid profiling of a panel of human monoclonal antibodies using FLU‐LISA
Recent advances in the ability to isolate human mAbs (hmAbs) following vaccination or infection 21 , 22 , 23 have highlighted the importance of rapid profiling of their binding specificities. We therefore selected a representative set of eight hmAbs, which were isolated from human individuals following vaccination with monovalent or quadrivalent influenza vaccines 24 , 25 (summarized in Supplementary table 6). Each of the eight hmAbs were previously tested against specific influenza antigens to classify their binding to influenza A/H1N1, A/H3N2 and (in some cases) B subtypes, as well as their binding to the HA head or stalk regions. 24 , 25 , 26 , 27 , 28 We profiled each of the eight hmAbs using the “hmAbs” AMs—influenza AMs spotted with a panel of 28 HA antigens (Figure 8). Each protein was spotted in a single concentration of 32.5 μg mL−1. All antibodies were profiled in three dilutions (6, 1.5 and 0.375 μg mL−1), and the AUC was computed for each hmAb across the three dilutions. We found that overall, antibodies bound to HA antigens from the specific subtypes that they were previously reported to bind, that is, hmAb 047‐1G05 bound to several H1N1 strains from multiple lineages as previously reported, whereas 030–09 2B03 bound H1N1 strains from the H1N1 2009 pandemic (pH1N1) and onward. However, in some cases, hmAbs also bound influenza antigens from additional subtypes, against which they were not previously tested, or failed to bind antigens from a subtype that they were previously reported to bind. For example, the 030‐09 1E05 hmAb was previously reported to bind an influenza B strain that was not detected here, and the FI6 hmAb bound several influenza B antigens, although it was not previously reported to cross‐react with subtype B HAs.
Figure 8.
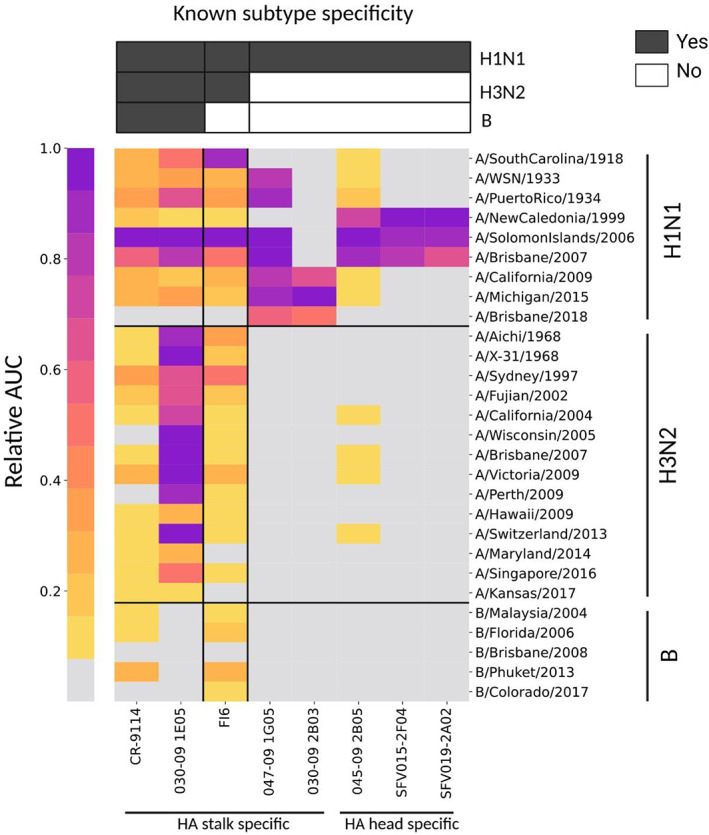
Rapid profiling of a panel of human monoclonal antibodies (hmAbs) using FLU‐LISA. A panel of eight representative influenza hemagglutinin (HA) hmAbs (Supplementary table 6) were profiled using traditional FLU‐LISA against a panel of A/H1N1, A/H3N2 and B influenza recombinant HA proteins (hmAb antigen microarrays; Supplementary table 1). The hmAbs included five HA stalk‐specific antibodies and three HA head‐specific antibodies. Each hmAb was run at three dilutions: 6, 1.5 and 0.375 μg mL−1, and the area under the curve (AUC) was computed for each antigen separately. Responses of each hmAb were normalized to its maximal AUC value across all antigens. Colors denote the normalized AUC scores for each hmAb to each of the antigens, with darker colors representing higher normalized AUC values. Gray represents normalized AUC values < 0.05. Each of the eight hmAbs was previously characterized using ELISA against specific H1, H3 and B HA antigens, as summarized on the top bars (dark gray cells). FLU‐LISA, fluorescence‐linked immunosorbent assay.
DISCUSSION
Here we presented FLU‐LISA, a novel AM binding assay that can be used as a high‐throughput alternative to ELISA for semiquantitative profiling of antibody levels to many antigens concurrently. As proof of concept, we used several types of influenza AMs spotted with recombinant HA proteins, as well as human coronavirus microarrays spotted with spike and RBD recombinant proteins. Using serum samples from mice with known influenza exposure history, we showed that our influenza arrays can be used for identification of the influenza subtype. We also showed that sublethal PR8 (H1N1) infection generates a broad cross‐reactive response to H1N1 strains, as well as to H5N1 strains, which are both group 1 influenza strains. 19 , 20 Comparing the antibody profiles of individual mice, we found extensive heterogeneity in the breadth and magnitude of the response following sublethal influenza infection or immunization, showcasing the ability of the influenza AMs to profile influenza immune history.
We showed that the AMs can also be used to generate antibody profiles from dried blood spots taken from chickens and humans, and that these profiles were highly similar to those measured from the serum or plasma of the same individual. The antibody profiles generated from both the serum and dried blood spots required minimal sample volumes. Taken together, these data suggest that the FLU‐LISA AMs provide a useful alternative to ELISA, especially for profiling antibody responses in small wild animals, as well as from newborns and young children from which very limited sample volumes can be obtained. Another advantage of dried blood spots is that they do not require refrigeration or centrifugation, making them highly amenable for field studies. In the FLU‐LISA AMs each antigen is spotted in serial dilutions, allowing us to calculate an AUC statistic for each antigen and sample. We found that this was a more robust measure of the antibody levels as compared with measuring antibody levels from a single antigen concentration. While the traditional ELISA uses a single concentration of the antigen and requires running each sample in serial dilutions for antibody quantitation when a standard control does not exist, FLU‐LISA can estimate antibody levels using a single sample dilution. We demonstrated that AUC values obtained by FLU‐LISA are comparable to those obtained by ELISA, as evidenced by the significant correlations between the AUC values of the two assays. However, our assay can also be used as a multiplex ELISA by running the traditional FLU‐LISA version. Furthermore, when both antigens are spotted at multiple dilutions and samples are tested in serial dilutions, we can obtain three‐dimensional ELISA‐like curves as demonstrated in Figure 6b. We demonstrated that one‐shot FLU‐LISA can be used to quantify binding of mouse mAbs, and generates binding curves highly similar to those obtained using ELISA. Finally, we showed that FLU‐LISA can be used for rapidly characterizing the specificity and cross‐reactivity profiles of a panel of hmAbs.
Using mice immunized with specific influenza strains from the H1N1 and H3N2 subtypes, we showed that FLU‐LISA can detect serum cross‐reactive antibody responses within subtype, and that mice infected with H1N1 do not generate an H3N2 response and vice versa. However, we also found that the assay can capture cross reactivity to other subtypes. In particular, we found that mice with H1N1 also generated weaker antibody responses to strains from the H5N1 subtype, which belongs to the same antigenic group. 20 , 29 These data suggest that influenza antibody profiles may discriminate between animals exposed to an H1N1 natural infection and an H5N1 natural infection. As such, this may be used as an effective tool for monitoring influenza exposures of wild migrating birds.
Interestingly, we found a difference in the cross‐reactivity profile of IgG antibodies following intranasal infection or intramuscular immunization with H1N1 viruses. Intranasal infection with PR8 induced heterosubtypic antibodies that bound to H3 proteins (influenza group 2), while this was not observed in mice immunized intramuscularly with Cal09 (compare Figures 2 and 3). These results coincide with previous reports that intranasal influenza infection or immunization in mice induces a more heterosubtypic neutralizing antibody response compared with intramuscular immunization (e.g. Lorenzo et al. 30 and Budimir et al. 31 ). Moreover, female C57BL/6 mice develop higher levels of heterosubtypic neutralizing antibodies compared with male C57BL/6 mice following sublethal intranasal infection. 30
The analysis of the hmAb profiles generated using FLU‐LISA yielded results similar to those obtained in previous studies. 24 , 25 , 26 , 27 , 28 This showcases the feasibility of using FLU‐LISA for rapid mapping of large panels of mAbs. By increasing the initial set of mAbs characterized using this platform, focusing on mAbs with known solved structures and binding properties, this platform may in the future be further developed for inferring the binding footprints of novel mAbs. Some of the mAbs we tested were more responsive at lower dilutions than others, and this is in agreement with the fact that different antibodies have different binding affinities to their target. 32 To overcome this limitation, we tested each mAb using the traditional FLU‐LISA (i.e. using several serial dilutions of the mAb and computing the AUC statistic). Some of the mAbs exhibited moderate binding for their target antigens (i.e. 030–09 2B03), and bound more strongly to other strains, against which they were never previously tested. For example, mAb 051‐09 4A03 strongly bound influenza B strains and 217‐1A02 had weak binding to H1N1 and H3N2 strains. Some of the H1 mAbs, especially the HA head‐specific mAbs, had very weak binding to the California 2009 pH1N1 HA (e.g. SFV019‐2A02 and SFV015‐2F04). We conducted additional serum binding experiments using this antigen and have concluded that responses to this antigen are lower overall.
A clear advantage of FLU‐LISA compared with ELISA is its ability to measure antibody binding to multiple antigens in parallel. For example, spotting 16 microarrays on a single 2.5 × 7.5‐cm2 microarray slide (yielding arrays of 6400 × 6400 μm2 in size), allows parallel quantification of antibody levels of 40 antigens spotted at three dilutions in triplicates. A limitation of AMs is batch‐to‐batch variability, which has been widely studied. 33 , 34 , 35 This can be addressed using a variety of standard normalization methods for array data. 36 Furthermore, we note that a single batch of FLU‐LISA slides with 40 antigens per array generates 2240 arrays, which is a sufficiently large batch for most studies.
While both ELISA and FLU‐LISA are semiquantitative antibody‐binding assays, they differ in multiple parameters, including (1) amplification mode, enzyme‐catalyzed colorimetric reaction versus fluorescent labeling, which in turn also affects their sensitivity and their dynamic range; (2) FLU‐LISA allows multiplexed testing, while by ELISA each antigen is tested individually; (3) sample volume requirements—FLU‐LISA requires significantly lower sample volume as compared with running multiple ELISA because of its multiplex nature; (4) antigen quantity—because of the small spot sizes used on FLU‐LISA, the amount of antigen (and cost) required per sample is significantly lower than that in ELISA; (5) testing capacity—the FLU‐LISA method can readily be used to generate antibody profiles for up to 192 samples per day (12 slides), which would require significantly more time using the traditional ELISA even with proper liquid dispensing automation; (6) fabrication—a clear advantage of ELISA is that plates can be coated manually without the need to sophisticated array‐spotting equipment required for fabricating AMs; (7) assay readout—ELISA requires a plate reader which is widely available in multiple laboratories, while scanning arrays require dedicated laser scanners that are less prevalent.
To further illustrate the advantages of FLU‐LISA over ELISA, we compared their use for testing 90 samples in two settings: (1) running each sample at a dilution curve using four serial dilutions—in this setting we compared ELISA using four serial dilutions of the sample with traditional FLU‐LISA using four serial dilutions of the antigen spotted on the microarrays (Table 1; Figure 7b) and (2) running each sample at a single sample dilution and a single concentration of antigen coated on the ELISA plates or spotted on the FLU‐LISA microarrays (Table 2).
Table 1.
Comparison of one‐shot FLU‐LISA and traditional ELISA for profiling antibodies to 30 antigens in 90 samples using four serial dilutions.
| FLU‐LISA single antigen | FLU‐LISA 30 antigens | ELISA single antigen | ELISA 30 antigens | |
|---|---|---|---|---|
| Number of samples | 90 | 90 | 90 | 90 |
| Number of slides/plates | 6 a | 6 | 3 b | 90 |
| Antigen quantity (μg) c | 2.08 (triplicate) d | — | 74.4 (triplicate) | — |
| Antigen concentration (μg mL−1) e | 8–65 | 4 | ||
| Sample volume (μL) | 1 | 1 | 2 (triplicate) | 60 |
| Serum dilution (IgG) f | 1:100 | 1:100–1:800 | ||
| Work time (h) | 6 | 6 | 6 | 120 g |
Traditional ELISA is run at four sample dilutions using a single antigen concentration, while FLU‐LISA is run using a single sample dilution across four antigen concentrations spotted on the AM.
AM, antigen microarray; FLU‐LISA, fluorescence‐linked immunosorbent assay; Ig, immunoglobulin.
Using microarray slides with 16 arrays per slide, each including 30 antigens spotted at four concentrations in triplicates.
Using the 384‐well format ELISA, where each sample is tested at four dilutions using triplicate wells; 30 individuals can be tested on a single 384‐well plate.
cAntigen quantity per single antigen assuming triplicate spots (FLU‐LISA) or wells (ELISA).
2.08 μg will be sufficient to spot 140 microarray slides—sufficient for testing 2100 samples.
Antigens are spotted on the microarray at four serial concentrations (8.12, 16.25, 32.5 and 65 μg mL−1).
Minimal serum dilution used for IgG profiling.
Assuming six 384‐well ELISA plates can be run on a single work day.
Table 2.
Comparison of the FLU‐LISA and traditional ELISA methods for profiling antibodies to 120 antigens in 90 samples using a single sample dilution.
| FLU‐LISA single antigen | FLU‐LISA 120 antigens | ELISA single antigen | ELISA 120 antigens | |
|---|---|---|---|---|
| Number of samples | 90 | 90 | 90 | 90 |
| Number of slides/plates | 6 a | 6 | 1 b | 90 c |
| Antigen quantity d | 0.52 μg (triplicate) | — | 18.6 μg (triplicate) | — |
| Antigen concentration | 32.5 μg mL−1 | 32.5 μg mL−1 | 4 μg mL−1 | 4 μg mL−1 |
| Sample volume | 1 μL | 1 μL | 1 μL (triplicate) | 108 μL |
| Serum dilution (IgG) e | 1:100–1:3200 | 1:100–1:3200 | ||
| Work time (h) | 6 h | 6 h | 4 h | 90 h f |
In both assays each sample is run at a single dilution and each antigen is spotted/coated at a single concentration.
FLU‐LISA, fluorescence‐linked immunosorbent assay; Ig, immunoglobulin.
Using microarray slides with 16 arrays per slide, each including 120 antigens spotted at a single concentration in triplicates.
To test 90 individuals in triplicates, 75% of a single 384‐well plate is required.
Using the 384‐well format ELISA, where each sample is tested using triplicate wells; 90 samples can be run using 75% of a single plate.
Antigen quantity per single antigen assuming triplicate spots (FLU‐LISA) or wells (ELISA).
Minimal serum dilution used for IgG profiling.
Assuming six 384‐well ELISA plates can be run on a single work day.
While ELISA uses an enzyme substrate reaction that significantly amplifies signals allowing increased sensitivity, the AM assay uses fluorescence detection and may therefore have a reduced dynamic range of detection. This can be partially mitigated by increasing the concentration of antigens used for array spotting. As antigens are spotted in micrometer spots using nanodrops (300–360 pL), antigens can be cost‐effectively spotted at much higher concentrations than those used to coat ELISA plates. Nevertheless, our results demonstrate that FLU‐LISA produces qualitatively similar results to ELISA. The high‐throughput nature of FLU‐LISA, as well as the low volume of biological material required per assay, makes FLU‐LISA an alternative antibody‐binding method for large‐scale screening. A clear advantage of FLU‐LISA over the traditional ELISA is the ability to test multiple antigens simultaneously, allowing to rapidly generate antibody‐binding profiles to hundreds of antigens simultaneously using low volume samples, which is particularly important in studies of newborns and young children.
Microarrays incubated with serum samples may display some background staining that is sample specific (data not shown), and typically longitudinal serum samples collected from the same individual display similar levels of background staining. Interestingly, when we compared serum samples that displayed a relatively higher background on microarrays with dried blood spots that were collected from the same individual and reconstituted in phosphate‐buffered saline (PBS)–Tween buffer, the reconstituted blood spots displayed a significantly lower level of background staining. This phenomenon was also observed with the chicken blood samples described here. These data suggest that dried blood spots may be a viable alternative to serum for antibody profiling, and have reduced signal‐to‐noise ratios.
In summary, here we presented FLU‐LISA, a novel AM‐based assay and compared it with the traditional ELISA. We demonstrated several antibody profiling applications in which the traditional ELISA is not feasible. The ability to perform high‐throughput antibody profiling using minimal sample volumes allows to rapidly and cost‐effectively screen large data sets, and can be used as a filtering step to identify important samples and antigens that should be further studied using functional antibody assays.
METHODS
Antigens
Influenza antigens (influenza recombinant proteins were used as antigens): Antigens were purchased from Sino Biological Inc (Beijing, China), The Native Antigen Company (Kidlington, UK) or were obtained as a gift from the International Reagent Resource (CDC, USA) or from BEI Resources (NIAID, USA), as described in Supplementary tables 1 and 2.
SARS‐CoV‐2 antigens: SARS‐CoV‐2 recombinant spike, S1 subunit and RBD proteins from multiple SARS‐CoV‐2 strains, as well as other human coronaviruses, were purchased from Sino Biological Inc (Beijing, China) or obtained from BEI Resources (NIAID, USA), as described in Supplementary table 3.
Antigen microarray spotting
Recombinant proteins were spotted onto N‐hydroxysuccinimide ester–derivatized hydrogel slides (H slides type B) using a Scienion sciFLEXARRAYER SX noncontact array spotter. Spot volumes ranged between 300 and 360 pL. Antigens at each concentration were spotted in triplicates. Sixteen identical microarrays were spotted on each microarray slide. Each printing batch included up to 140 microarray slides, yielding a total of up to 2240 arrays per batch.
Antigen microarray design
We used the following AMs for antibody profiling:
Influenza recombinant HA AMs, which included 28–44 recombinant influenza HA proteins or HA1 subunits from A/H1N1, A/H3N2, A/H5N1 and B influenza strains. Three different arrays were spotted: “hmAbs,” “M1” and “M2” (Supplementary table 1). All proteins were spotted at a single concentration (32.5 μg mL−1) in sciSPOT D1 spotting buffer (Scienion, Germany). These AMs were used for screening anti‐influenza hmAbs, and for profiling serum samples from mice that were exposed to sublethal doses of influenza viruses or immunized with inactivated influenza viruses.
Pan‐influenza AMs, which included 46 recombinant HA proteins and 14 recombinant neuraminidase proteins from human and avian influenza A subtypes and influenza B strains. The arrays also included four influenza internal proteins from the H1N1 A/Puerto Rico/8/1934 (PR8) strain: M1, NS1, NS2 and NP (Supplementary table 2). All antigens were spotted at a single concentration of 16.25 μg mL−1 in 0.01% Triton X‐100. These arrays were used for profiling chicken IgY anti‐influenza antibodies.
FLU‐LISA influenza AMs: To compare the FLU‐LISA AM with the standard ELISA, we used recombinant HA proteins from four influenza strains that were spotted in 11 serial concentrations in the range of 122 ng mL−1–125 μg mL−1 in sciSPOT D1 spotting buffer (Scienion, Germany). These included three seasonal vaccine strains (north hemisphere): H3N2 A/Wisconsin/67/2005; H3N2 A/Brisbane/10/2007 and H1N1 A/California/07/2009 and the avian influenza H7N9 A/Shanghai/1/2013 strain (FLU‐LISA AM in Supplementary table 1).
Coronavirus AMs
To compare antibody profiles from human plasma and dried blood spots stored from the same human blood sample, CoV AMs were spotted with recombinant spike, S1 or RBD proteins from all the seven human coronaviruses, including many SARS‐CoV‐2 variants (Supplementary table 3). The proteins were spotted in three serial concentrations (32.5, 16.25 and 8 μg mL−1 in 0.0025% Triton X‐100).
sCoV FLU‐LISA AMs
To test the specificity of FLU‐LISA also to SARS‐CoV‐2 antigens, sCoV microarrays were spotted with spike and RBD antigens of many SARS‐CoV‐2 variants (Supplementary table 3). Each protein was spotted in six serial concentrations (2.03–65 μg mL−1) in 0.0025% Triton X‐100.
Mouse serum samples
Two mice experiments were performed using female C57BL/6 mice. In the first experiment, approved by the Ben Gurion University Committee for the Ethical Care and Use of Animals in Experiments, nine mice were infected intranasally with a sublethal dose (100 pfu) of A/Puerto Rico/8/1934 (PR8) H1N1 influenza virus, as previously described. 37 All mice were symptomatically infected, lost weight and recovered. In the second experiment, approved by the Animal Ethics Committee at St. Jude Children's Research Hospital, forty 8‐week‐old C57BL/6 mice were injected intramuscularly with 40 μg of either one of three whole inactivated viruses: A/X31/1968 (X31, H3N2), mouse‐adapted A/California/07/2009 (Cal09, H1N1), A/Vietnam/1203/04 (Viet1203, H5N1) viruses or PBS. Each immunization group included 10 mice. Serum samples were collected 28 days after the infection in both experiments.
Chicken serum and dried blood spot samples
Blood samples were obtained from 36 breeder chickens that were vaccinated two times with two avian H9N2 2018 influenza vaccines: the first vaccination with strain 215 (Biovac, Israel) was injected into 12–17‐day‐old chicks, and the second vaccination with strain 947 (Phibro, Israel) was injected into 40–41‐day‐old chickens. Blood samples were collected from 40–41‐day‐old female chicks, 34 days following the first vaccination (n = 16); and from 2.5–3‐month‐old male and female chickens about 40 days following the second dose (n = 20). Four blood spots from each sample were dropped on Whatman FTA blood cards (125–500 μL drops), and the rest of the sample was centrifuged for serum isolation. The dried blood spots and sera were stored frozen at −20°C. For microarray experiments, each blood spot was incubated in 2 mL of 0.05% PBS‐T (0.05% Tween‐20 in PBS) overnight on a shaker, and the liquid was collected and stored frozen.
Human serum samples
The serum samples used in this study were collected from healthy adults by two separate clinical studies:
IDF (Israel Defense Forces) study: a serological study of soldiers conducted in 2018. The study was approved by the Institutional Review Board of the Israeli Defense Force (IRB approval number 1854–2017, IDF IRB). Samples from 40 individuals collected in January 2018 were used in this study for (a) comparing FLU‐LISA with the traditional ELISA (n = 10); and (b) comparing pre‐ and post‐SARS‐CoV‐2 pandemic samples using a coronavirus AM (n = 30).
Booster study: A study of health care workers that were previously vaccinated three times with the Pfizer‐BioNTech SARS‐CoV‐2 vaccine. The study was approved by the Institutional Review Board of the Soroka University Medical Center (IRB approval number 0404‐21‐SOR‐C). Samples from 14 individuals were used for comparing pre‐ and post‐SARS‐CoV‐2 pandemic samples using a coronavirus AM.
Human plasma and blood spot samples
Blood samples were obtained from six healthy individuals from the booster study described above (IRB approval number 0404‐21‐SOR‐C). Four blood spots from each sample were dropped on Whatman FTA blood cards (125–500 μL drops), and the rest of the sample was centrifuged for plasma isolation. The dried blood spots and sera were stored frozen at –20°C. For microarray experiments, each blood spot was incubated in 1 mL of 0.05% PBS‐T (0.05% Tween‐20 in PBS) for 4 h on a shaker, and the liquid was collected and stored frozen. The clinical study was approved by the Institutional Review Board of the Soroka University Medical Center (approval number 0404‐21‐SOR‐C).
Mouse monoclonal antibody
To compare our FLU‐LISA with the traditional ELISA we used a mouse mAb against the HA protein of the H3N2 A/Brisbane/10/2007 influenza strain (Sino Biologicals, catalog number 11056‐MM01, China).
Human monoclonal antibodies
A set of eight hmAbs isolated from individuals vaccinated with various influenza vaccines 24 , 25 was kindly obtained as a gift from Patrick Wilson (Weill Cornell Medicine, New York, NY, USA), and is described in Supplementary table 6.
ELISA
To run efficient ELISA with reduced sample volumes and antigens, we optimized our ELISA for 384‐well plate format, using a liquid dispensing robot (EzMate 601; Arise Biotech Corp.). The 384‐well white MaxiSorp‐coated plates (120 μL wells; catalog number 460372; Thermo Fisher, USA) were coated with 17 μL of 4 μg mL−1 recombinant HA protein per well (diluted in PBS) and incubated overnight at 4°C. Plates were washed five times with PBS‐T washing buffer (0.1% Tween‐20 in PBS, 60 μL per well) using a plate washer (ELx405 Select Deep‐Well Microplate Washer; BioTek, USA). Plates were then blocked with 100 μL of 10% skim milk powder (Sigma, Germany) in PBS‐T and incubated for 1 h at 37°C. Following five PBS‐T washes, human serum samples were diluted in twofold serial dilutions (1:25–1:409 600) in 2% skimmed milk in PBS‐T, and added to the plates in triplicates (30 μL per well) for 1‐h incubation at 37°C in the incubator, and then washed five times with PBS‐T. The secondary antibody, Peroxidase‐AffiniPure Goat Anti‐Human IgG (H + L) (catalog number 109–035‐088, Jackson ImmunoResearch, USA) or Anti‐Mouse IgG (H + L), horseradish peroxidase conjugate (catalog number W4021, Promega), was diluted 1:10 000 or 1:2500, respectively, in 2% skimmed milk in PBS‐T and added to the plates (30 μL per well). After incubation for 1 h at 37°C and five additional PBS‐T washes, equal volumes of peroxide and luminol were mixed and added (30 μL stable peroxide +30 μL luminol/enhancer per well, SuperSignal West Pico Chemiluminescent Substrate, catalog number 34578, Thermo Fisher). Following 1‐min incubation, the luminescence was measured by an ELISA reader (Infinite 200 PRO, TECAN, Switzerland) at 600‐nm wavelength.
ELISA
The median optical density values of each triplicate were calculated and negative sample control of 2% skimmed milk was subtracted to get the relative light units. ELISA curves were fitted using a five‐parameter logistic model:
where a is the minimal value obtained (lower asymptote); b is the slope of the curve; c is the inflection point; d is the maximal value (upper asymptote) and g is the asymmetric factor. The model was fitted using the curve fit function of the scipy.optimize library in Python.
Antigen microarray assay
AM slides were blocked with 4 mL of chemical blocking solution (50 mM ethanolamine, 50 mM borate, pH 9.0) per slide for 1 h at room temperature (RT) on a shaker in rectangular 4‐well plates (catalog number 267061; NUNC). After blocking, the liquid was vacuumed and the slide was washed two times for 3 min in a washing buffer (0.05% Tween‐20 in PBS), two times for 3 min in PBS and an additional 3‐min wash in double‐deionized water. Every wash was performed with 3 mL liquid per slide on a shaker at RT. Samples were diluted in an incubation buffer (1% bovine serum albumin/0.025% Tween‐20 in PBS). Human serum samples were diluted 1:1000 or 1:3200, as specified in the results and figure legends; mice serum samples were diluted 1:100; chicken serum samples were diluted 1:4000. Chicken dried blood spots (approximately 500 μL) were reconstituted in 2 mL washing buffer and diluted 1:20, and human dried blood spots (approximately 500 μL) were reconstituted in 1 mL washing buffer and diluted 1:100. hmAbs were incubated in three serial concentrations: 6, 1.5 and 0.375 μg mL−1, and the mouse mAb was incubated in 15 twofold serial concentrations ranging between 5 ng mL−1 and 125 μg mL−1. Following a 2‐h incubation, the slides were dried by centrifugation at RT for 5 min at speed 800 relative centrifugal force in a slide holder padded with Kim wipes, loaded on divided incubation trays (PEPperCHIP; PEPperPRINT, Germany) and then the samples were added and incubated with the arrays for 2 h at RT on a shaker. After incubation, the samples were discarded and the slides were washed two times with a washing buffer and two times with PBS, as described earlier. After washes, the slides were incubated for 45 min on the shaker at RT with a fluorescently labeled polyclonal secondary antibody diluted in the incubation buffer. The secondary antibody for human samples and hmAbs was Alexa Fluor 647–conjugated AffiniPure Donkey Anti‐Human IgG (H + L) (catalog number 709–605‐149; Jackson ImmunoResearch, USA), diluted 1:1000. The secondary antibody for mouse serum samples was Alexa Fluor 647–conjugated AffiniPure Goat Anti‐Mouse IgG Fcγ Fragment Specific (catalog number 115–605‐008; Jackson ImmunoResearch), diluted 1:3000 (PR8 infection) or 1:4000 (H1N1, H5N1 or H3N2 infections). The secondary antibody for chicken serum and dried blood spot samples was Alexa Fluor 647–conjugated AffiniPure Goat Anti‐Chicken IgY (IgG) (H + L) (catalog number 103–605‐155; Jackson ImmunoResearch, USA), used at a dilution of 1:1000 for serum samples and at a dilution of 1:2000 for dried blood spot samples. To detect bound Igs, slides were scanned on a three‐laser GenePix 4400A scanner. Images were analyzed using GenePix Pro version 7 to obtain the median fluorescent intensity (MFI) of each spot after subtracting the mean local background fluorescence intensity (0 ≤ MFI ≤ 65 000).
Antigen microarray analysis
The microarray results were analyzed using an in‐house pipeline developed in python. As each antigen at each concentration was spotted in triplicate, the median MFI intensity of each triplicate was calculated. During each experiment, a negative control array was incubated with the incubation buffer only. The background staining of each triplicate of spots in the negative control microarray was subtracted from all other microarrays. For antigens that were spotted in serial concentrations (FLU‐LISA influenza AMs), a five‐parameter logistic regression model was used to fit curves to the measured median fluorescent intensity (MFI) versus the antigen concentration, and the AUC was calculated. For the influenza microarrays, we sorted the HA and neuraminidase proteins into groups according to their subtype. The magnitude of antibody response to a group of antigens was defined as the sum of MFI levels to all the proteins included in this group. To compare groups with different numbers of proteins, the geometric mean magnitude was calculated.
Statistical analysis
Comparisons between experimental groups, and between the traditional ELISA and FLU‐LISA methods were performed using the Wilcoxon rank‐sum test. Correlations were computed using Pearson correlation, or Spearman correlation for comparison of different antigens binding in the same assay. All analysis was performed in Python.
CONFLICT OF INTEREST
The authors declare no nonfinancial interests but declare a competing financial interest. A patent application related to the microarrays used in this study has been filed by Tomer Hertz and Lilach M Friedman.
AUTHOR CONTRIBUTIONS
Shlomia Levy: Data curation; formal analysis; investigation; methodology; software; validation; visualization; writing – original draft; writing – review and editing. Marwa Abd Alhadi: Data curation; investigation; project administration; validation; visualization; writing – original draft; writing – review and editing. Asaf Azulay: Data curation; formal analysis; investigation; methodology; software; visualization; writing – original draft; writing – review and editing. Amit Kahana: Data curation; investigation; validation. Nir Bujanover: Investigation; resources. Roi Gazit: Investigation; resources; writing – review and editing. Maureen Ann McGargill: Investigation; resources; writing – review and editing. Lilach M Friedman: Data curation; formal analysis; investigation; methodology; project administration; software; supervision; validation; visualization; writing – original draft; writing – review and editing. Tomer Hertz: Conceptualization; formal analysis; funding acquisition; investigation; methodology; supervision; writing – original draft; writing – review and editing.
Supporting information
ACKNOWLEDGMENTS
This study was funded by the Israel Science Foundation (ISF) grant no. 882/17; NIH award no. 5R01AI114728 subaward 112 079 050–7 867 958 and by The National Institute for Biotechnology in Negev, Israel. The research reported in this publication was also supported by the Israel Defense Forces (IDF) Medical Corps and Directorate of Defense Research and Development, Israeli Ministry of Defense (IMOD DDR&D).
DATA AVAILABILITY STATEMENT
All raw data and code used in the analysis are available from the Hertz Lab website: https://www.hertz‐lab.org/.
REFERENCES
- 1. Engvall E, Jonsson K, Perlmann P. Enzyme‐linked immunosorbent assay. II. Quantitative assay of protein antigen, immunoglobulin G, by means of enzyme‐labelled antigen and antibody‐coated tubes. Biochim Biophys Acta 1971; 251: 427–434. [DOI] [PubMed] [Google Scholar]
- 2. Engvall E, Perlmann P. Enzyme‐linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 1971; 8: 871–874. [DOI] [PubMed] [Google Scholar]
- 3. Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015; 72: 4–15. [DOI] [PubMed] [Google Scholar]
- 4. Mikulskis A, Yeung D, Subramanyam M, Amaravadi L. Solution ELISA as a platform of choice for development of robust, drug tolerant immunogenicity assays in support of drug development. J Immunol Methods 2011; 365: 38–49. [DOI] [PubMed] [Google Scholar]
- 5. Lai S, Wang S, Luo J, Lee LJ, Yang ST, Madou MJ. Design of a compact disk‐like microfluidic platform for enzyme‐linked immunosorbent assay. Anal Chem 2004; 76: 1832–1837. [DOI] [PubMed] [Google Scholar]
- 6. Ng S, Nachbagauer R, Balmaseda A, et al. Novel correlates of protection against pandemic H1N1 influenza a virus infection. Nat Med 2019; 25: 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Legutki JB, Magee DM, Stafford P, Johnston SA. A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine 2010; 28: 4529–4537. [DOI] [PubMed] [Google Scholar]
- 8. Price JV, Jarrell JA, Furman D, et al. Characterization of influenza vaccine immunogenicity using influenza antigen microarrays. PLoS One 2013; 8: e64555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kunnath‐Velayudhan S, Davidow AL, Wang HY, et al. Proteome‐scale antibody responses and outcome of mycobacterium tuberculosis infection in nonhuman primates and in tuberculosis patients. J Infect Dis 2012; 206: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keating R, Hertz T, Wehenkel M, et al. The kinase mTOR modulates the antibody response to provide cross‐protective immunity to lethal infection with influenza virus. Nat Immunol 2013; 14: 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhong L, Hidalgo GE, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Using protein microarray as a diagnostic assay for non‐small cell lung cancer. Am J Respir Crit Care Med 2005; 172: 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quintana FJ, Farez MF, Viglietta V, et al. Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc Natl Acad Sci USA 2008; 105: 18889–18894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merbl Y, Zucker‐Toledano M, Quintana FJ, Cohen IR. Newborn humans manifest autoantibodies to defined self molecules detected by antigen microarray informatics. J Clin Invest 2007; 117: 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nachbagauer R, Shore D, Yang H, et al. Broadly reactive human monoclonal antibodies elicited following pandemic H1N1 influenza virus exposure protect mice against highly pathogenic H5N1 challenge. J Virol 2018; 92: e00949‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mahallawi WH, Kasbekar AV, McCormick MS, et al. Infection with 2009 H1N1 influenza virus primes for immunological memory in human nose‐associated lymphoid tissue, offering cross‐reactive immunity to H1N1 and avian H5N1 viruses. J Virol 2013; 87: 5331–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmed MS, Jacques LC, Mahallawi W, et al. Cross‐reactive immunity against influenza viruses in children and adults following 2009 pandemic H1N1 infection. Antiviral Res 2015; 114: 106–112. [DOI] [PubMed] [Google Scholar]
- 17. Jegaskanda S, Job ER, Kramski M, et al. Cross‐reactive influenza‐specific antibody‐dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 2013; 190: 1837–1848. [DOI] [PubMed] [Google Scholar]
- 18. Carreno JM, Strohmeier S, Kirkpatrick Roubidoux E, et al. H1 hemagglutinin priming provides long‐lasting heterosubtypic immunity against H5N1 challenge in the mouse model. MBio 2020; 11: e02090‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gostic KM, Ambrose M, Worobey M, Lloyd‐Smith JO. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 2016; 354: 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arevalo CP, Le Sage V, Bolton MJ, et al. Original antigenic sin priming of influenza virus hemagglutinin stalk antibodies. Proc Natl Acad Sci USA 2020; 117: 17221–17227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zost SJ, Gilchuk P, Chen RE, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS‐CoV‐2 spike protein. Nat Med 2020; 26: 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS‐CoV‐2 neutralizing antibodies and protection from disease in a small animal model. Science 2020; 369: 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pedrioli A, Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol 2021; 42: 1143–1158. [DOI] [PubMed] [Google Scholar]
- 24. Guthmiller JJ, Utset HA, Henry C, et al. An egg‐derived sulfated N‐Acetyllactosamine glycan is an antigenic decoy of influenza virus vaccines. MBio 2021; 12: e0083821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrews SF, Huang Y, Kaur K, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 2015; 7: 316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ekiert DC, Friesen RH, Bhabha G, et al. A highly conserved neutralizing epitope on group 2 influenza a viruses. Science 2011; 333: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science 2012; 337: 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza a hemagglutinins. Science 2011; 333: 850–856. [DOI] [PubMed] [Google Scholar]
- 29. Knight M, Changrob S, Li L, Wilson PC. Imprinting, immunodominance, and other impediments to generating broad influenza immunity. Immunol Rev 2020; 296: 191–204. [DOI] [PubMed] [Google Scholar]
- 30. Lorenzo ME, Hodgson A, Robinson DP, Kaplan JB, Pekosz A, Klein SL. Antibody responses and cross protection against lethal influenza a viruses differ between the sexes in C57BL/6 mice. Vaccine 2011; 29: 9246–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Budimir N, de Haan A, Meijerhof T, et al. Heterosubtypic cross‐protection induced by whole inactivated influenza virus vaccine in mice: influence of the route of vaccine administration. Influenza Other Respi Viruses 2013; 7: 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barrette RW, Urbonas J, Silbart LK. Quantifying specific antibody concentrations by enzyme‐linked immunosorbent assay using slope correction. Clin Vaccine Immunol 2006; 13: 802–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu H, Luo H, Yan M, Zuo X, Li QZ. Autoantigen microarray for high‐throughput autoantibody profiling in systemic lupus erythematosus. Genom Proteom Bioinform 2015; 13: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ayoglu B, Schwenk JM, Nilsson P. Antigen arrays for profiling autoantibody repertoires. Bioanalysis 2016; 8: 1105–1126. [DOI] [PubMed] [Google Scholar]
- 35. Brezina S, Soldo R, Kreuzhuber R, Hofer P, Gsur A, Weinhaeusel A. Immune‐signatures for lung cancer diagnostics: evaluation of protein microarray data normalization strategies. Microarrays 2015; 4: 162–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamelinck D, Zhou H, Li L, et al. Optimized normalization for antibody microarrays and application to serum‐protein profiling. Mol Cell Proteom 2005; 4: 773–784. [DOI] [PubMed] [Google Scholar]
- 37. Gazit R, Gruda R, Elboim M, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol 2006; 7: 517–523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data and code used in the analysis are available from the Hertz Lab website: https://www.hertz‐lab.org/.


