Abstract
The study of eukaryotic tRNA processing has given rise to an explosion of new information and insights in the last several years. We now have unprecedented knowledge of each step in the tRNA processing pathway, revealing unexpected twists in biochemical pathways, multiple new connections with regulatory pathways, and numerous biological effects of defects in processing steps that have profound consequences throughout eukaryotes, leading to growth phenotypes in the yeast Saccharomyces cerevisiae and to neurological and other disorders in humans. This review highlights seminal new results within the pathways that comprise the life of a tRNA, from its birth after transcription until its death by decay. We focus on new findings and revelations in each step of the pathway including the end-processing and splicing steps, many of the numerous modifications throughout the main body and anticodon loop of tRNA that are so crucial for tRNA function, the intricate tRNA trafficking pathways, and the quality control decay pathways, as well as the biogenesis and biology of tRNA-derived fragments. We also describe the many interactions of these pathways with signaling and other pathways in the cell.
Keywords: decay, modification, splicing, tRNA-derived fragments, tRNA
INTRODUCTION
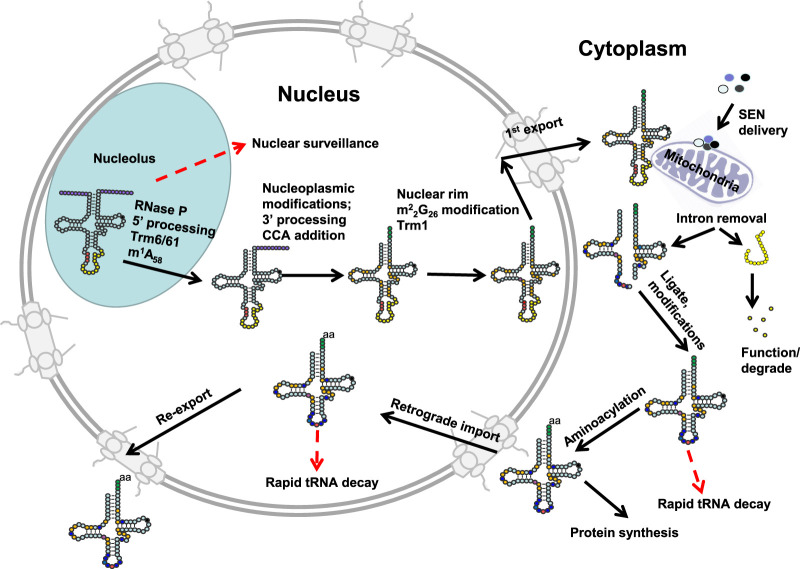
The elemental steps of eukaryotic tRNA biogenesis have been known for some time. After transcription by RNA polymerase III, pre-tRNA maturation involves a number of size-altering steps, including endonucleolytic removal of the 5′ leader, endonucleolytic and/or exonucleolytic removal of the 3′ trailer, untemplated CCA addition to the 3′ end, untemplated addition of a G−1 residue to the 5′ end of tRNAHis, and enzymatic splicing of the introns found between N37 and N38 in a subset of tRNAs. Each of these stages also involves the formation of modifications, ∼13 in the typical cytoplasmic tRNA from the budding yeast Saccharomyces cerevisiae, with each tRNA having its own specific combination of the 25 chemically distinct modifications that occur in 36 different locations in the tRNA. In addition, each tRNA is subject to a number of intracellular trafficking steps, which themselves may differ among different tRNAs (Fig. 1).
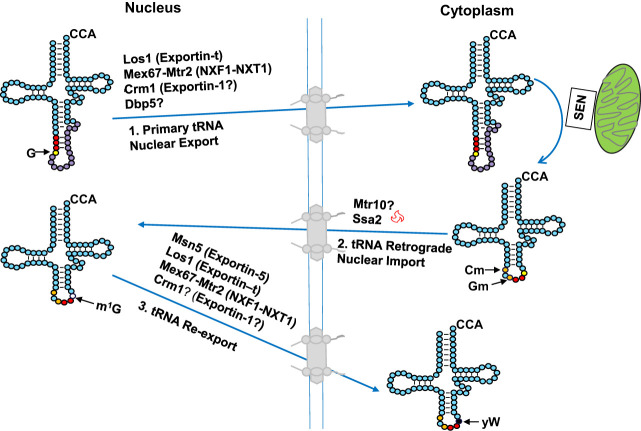
FIGURE 1.
Schematic of tRNA biogenesis, subcellular dynamics, and quality control turnover pathways in S. cerevisiae. tRNAs are transcribed in the nucleolus where the 5′ leader (left purple circles) of the initial transcript is removed by RNase P and likely where m1A58 is modified (black circle) by Trm6/61. About half of the known modifications (examples, orange circles) occur in the nucleoplasm where 3′ CCA nucleotides (green circles) are also added. Dimethylation of G26 (magenta circle) is catalyzed by Trm1, which is located on the inner nuclear membrane, prior to nuclear export of the end-matured, partially processed, intron-containing (yellow circles) pre-tRNAs; end-processed, partially modified tRNAs encoded by genes lacking introns are also exported to the cytoplasm. Introns are removed on the mitochondrial cytoplasmic surface. After/during splicing, additional modifications are added in the cytoplasm (examples, blue circles), and the freed introns are destroyed. Processed/modified cytoplasmic tRNAs return to the nucleoplasm via retrograde tRNA nuclear import and under stress conditions accumulate there; in favorable conditions the tRNAs return to the cytoplasm via reexport where they participate in protein synthesis. There are quality control steps, indicated by red dashed arrows, that destroy tRNAs that have not undergone the canonical (black arrows) steps appropriately. Further details of the cell biology and quality control pathways are provided in the text and Figures 7 and 8.
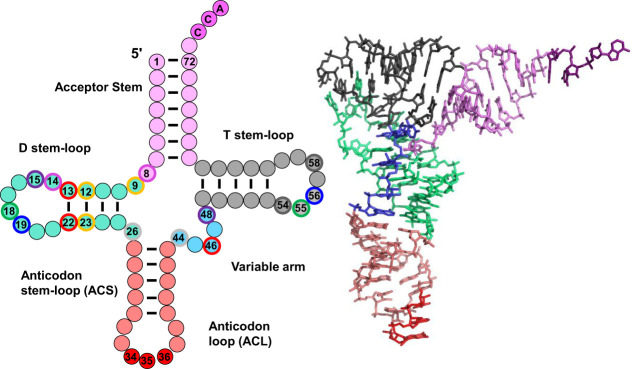
The tRNA that emerges after this processing pathway has the canonical cloverleaf secondary structure, which is folded into the classical L-shape by a combination of stacking interactions and conserved tertiary interactions (Fig. 2; Kim et al. 1974a,b; Giege et al. 2012). The resulting tRNA has its acceptor stem stacked on the T-stem to form an extended helix with the 3′-CCAOH end protruding from the acceptor stem, and at approximately right angles, the D-stem is weakly stacked on the anticodon stem (ACS), with the anticodon loop (ACL) protruding. Subsequently, the tRNA is charged at the CCA end by its cognate aminoacyl tRNA synthetase (aaRS) to form the corresponding aminoacyl-tRNA (aa-tRNA), which is now ready for its crucial role in translation.
FIGURE 2.
tRNA structure. A schematic of tRNA structure. tRNA is shown in its usual secondary structure, with colored circles representing nucleotides in and adjacent to the acceptor stem (pink), D stem–loop (green), anticodon stem–loop (red), variable arm (aqua) and T-stem–loop (gray), and lines representing base pairs. The 3′ CCA residues N74–N76 are shown in dark pink, and the anticodon residues N34–N36 are dark red. Outer disks of circles are colored to indicate common tertiary interactions, as first detailed for tRNAPhe from yeast (Kim et al. 1974b) (8–14, dark pink; 9–12–23, yellow; 13–22–46, red; 15–48, purple; 18–55, green; 19–56, blue; 26–44, light gray; 54–58, dark gray). Note that different tRNA species can have a D-stem with only 3 bp, a D-loop of variable length, a variable arm with 4 nt or a longer variable arm comprising a stem–loop. Note that tRNA residues are numbered so as to conserve constant numbering of major structural and functional elements, with the anticodon as N34–N36 and the CCA end as N74–N76 (Sprinzl et al. 1998). To this end, additional residues in the D-loop and variable arm have specialized names, and missing residues in some tRNA species are designated by gaps in the numbering for the appropriate residues. On the right is the corresponding crystal structure of tRNAPhe (1EHZ) (Shi and Moore 2000), with residues colored to match the schematic.
Research in the last several years has enormously increased our understanding of almost every step in the eukaryotic tRNA processing pathway in the budding yeast Saccharomyces cerevisiae, and in many cases in other eukaryotic systems, revealing a number of surprises and insights. It is now known that failure of any of a number of the processing steps can lead to tRNA with defects in charging, decoding, or stability, resulting in a number of distinct growth defects in S. cerevisiae and neurological and/or mitochondrial disorders in humans. It has also become apparent that there are multiple points at which tRNA processing intersects with regulatory pathways that respond to nutrients and other environmental factors, stress response pathways, and signaling pathways, to mediate cell growth and translation.
This review aims to capture some of the seminal findings in the biology of eukaryotic tRNA processing during the past several years, with a focus on cytoplasmic tRNAs of S. cerevisiae and other well-studied eukaryotic systems. In the review, we first discuss each tRNA processing step in end maturation and splicing, in their usual in vivo order. This is followed by a discussion of the biology of modifications in and around the ACL, and then the biology of modifications in the main tRNA body, after which there is a discussion of tRNA decay pathways, tRNA nuclear cytoplasmic subcellular dynamics, and tRNA fragments. Along the way, we discuss the intersection of all of these pathways with stress and regulatory pathways. We do not focus on the rich biology of tRNA transcription, aminoacyl tRNA synthetases, and mRNA decoding in the ribosome, as these are covered by numerous other reviews (for example, see Rozov et al. 2016; Graczyk et al. 2018; Rubio Gomez and Ibba 2020).
END PROCESSING AND SPLICING STEPS OF THE tRNA BIOGENESIS PATHWAY
Unexpected finding of frequent 5′ capping of pre-tRNAs
It is now known that Pol III transcription of tRNA is frequently followed by 5′ end capping of the pre-tRNA transcript in S. cerevisiae and human cells, albeit not as frequently as for mRNAs (Ohira and Suzuki 2016). The discovery of pre-tRNA capping was surprising because no interaction exists between the capping machinery and the Pol III transcription machinery, as is well established for the Pol II transcription machinery (for review, see Bentley 2014). Nonetheless, mass spectrometry analysis of pre-tRNAs shows that capping occurs between 5% and 22% of the time on different pre-tRNAs in wild-type (WT) cells, including each of several tRNAs examined from intron-containing and intronless genes. Furthermore, pre-tRNA capping appears to occur by the same mechanism as that for mRNA capping, based on genetic depletion experiments and analysis of intermediates. Moreover, capped pre-tRNAs accumulate to a greater extent when removal of the pre-tRNA 5′ leader by RNase P is inhibited, suggesting that pre-tRNA capping frequency is based on availability of the pre-tRNA (Ohira and Suzuki 2016).
5′ end removal catalyzed by RNAs of RNase P RNPs and protein-only RNase P (PRORP) enzymes
Following the paradigm-breaking discovery that endonucleolytic removal of the tRNA 5′ leader was catalyzed by the RNA component of bacterial RNase P ribonucleoprotein (RNP) (Guerrier-Takada et al. 1983), subsequent work extended RNA catalysis of 5′ leader removal to archaea (Pannucci et al. 1999) and eukaryotes (Kikovska et al. 2007), even as the number of protein subunits of the RNPs increased from one in bacteria to four to five in archaea, and nine to ten in eukaryotes (Supplemental Table S1; Chamberlain et al. 1998; for reviews, see Walker and Engelke 2006; Jarrous and Gopalan 2010). Although the protein subunits do not participate directly in catalysis, they are all essential in yeast (Chamberlain et al. 1998), and cryoEM structures of the human holoenzyme, and the yeast holoenzyme with and without bound pre-tRNA, revealed that the protein subunits stabilize the RNA subunit for catalysis and substrate recognition and participate in recognition of the tRNA 5′ leader (Lan et al. 2018; Wu et al. 2018; see Phan et al. 2021).
One intriguing aspect of RNase P biology is that many of its protein subunits are also part of other essential RNPs (for review, see Jarrous 2017). Indeed, all but one of the subunits of yeast RNase P are shared with the essential and highly conserved RNase MRP (Chamberlain et al. 1998), which has a role in maturation of rRNA and specific mRNAs, but remarkably, a recent cryoEM structure of yeast RNase MRP revealed that several of the shared protein subunits undergo remodeling driven by its distinct RNA subunit (Perederina et al. 2020). In addition, several RNase P subunits are implicated in different roles: three subunits are part of the telomerase complex, helping to stabilize the complex and promoting nuclear localization (Lemieux et al. 2016; Garcia et al. 2020); and another subunit is implicated in different organisms in female gametophyte development and sterility, piRNA synthesis, or fungal resistance (Wang et al. 2012; Molla-Herman et al. 2015; Li et al. 2021). Although these additional functions of RNase P subunits make it more difficult to untangle auxiliary roles of subunits from their specific roles in 5′ leader removal, reconstitution experiments may clarify this (Perederina et al. 2018).
Because of the well-established role of RNA catalysis in RNase P function, it was a distinct surprise to discover that removal of pre-tRNA 5′ leaders was catalyzed by a protein-only RNase P (PRORP) of three subunits in human mitochondria (Holzmann et al. 2008) and a single subunit PRORP in the plant Arabidopsis thaliana (Gobert et al. 2010). Indeed, Arabidopsis PRORPs likely catalyze all 5′ leader removal from tRNAs in vivo in each of the nuclear/cytoplasmic, mitochondrial, and chloroplast compartments (Gutmann et al. 2012). Remarkably, the yeast RNase P function can be replaced by the single subunit nuclear PRORP of Trypanosoma brucei or Arabidopsis, without an obvious growth defect in the latter case (Taschner et al. 2012; Weber et al. 2014).
Subsequent phylogenetic analysis indicates that PRORPs and RNase P RNAs are each widely found in distinct clades within the subgroups of eukaryotes, and in distinct nuclear, mitochondrial, or chloroplast compartments in subsets of these organisms (Lechner et al. 2015), as well as in a small number of bacterial and archaeal phyla (Nickel et al. 2017; Daniels et al. 2019). One unexplained curiosity is why in two cases, examined bacteria sometimes have both a functional PRORP and a functional RNase P RNA (Nickel et al. 2017; Daniels et al. 2019).
Recent structural analysis shows that the human three subunit PRORP binds and positions the pre-tRNA through a subcomplex of two subunits including the TRM10C tRNA methyltransferase, which then recruits the endonuclease PRORP catalytic subunit (Bhatta et al. 2021), and the bacterial single subunit PRORP binds the pre-tRNA with one subunit of the homodimer, to catalyze cleavage by the other subunit (Li et al. 2022).
3′ trailer removal catalyzed by different exonucleases and endonucleases
The processing machinery that removes the 3′ trailer from pre-tRNA in eukaryotes is now understood to result from a combination of nucleases. For most tRNAs, removal of the 3′ trailer sequence occurs after removal of the 5′ leader by RNase P (Fig. 1; Lee et al. 1991; O'Connor and Peebles 1991). As in E. coli (Li and Deutscher 1996), removal of the 3′ trailer sequence in eukaryotes is catalyzed by a combination of endonucleases and exonucleases. Trz1 catalyzes endonucleolytic removal of the 3′ trailer of a number of pre-tRNAs (Schiffer et al. 2002; Takaku et al. 2003), and is known to play a prominent role in 3′ trailer removal in vivo, based on northern analysis after siRNA depletion in Drosophila (Dubrovsky et al. 2004), temperature shift experiments in conditional mutants of the fission yeast Schizosaccharomyces pombe (Zhang et al. 2013), and promoter shut-off experiments in S. cerevisiae (Skowronek et al. 2014). In addition, the 3′ exonuclease Rex1 has a prominent role in 3′ trailer removal of pre-tRNAs in S. cerevisiae. Rex1 was initially implicated in tRNAArg maturation of the tandemly transcribed tRNAArg–Asp genes of S. cerevisiae (van Hoof et al. 2000). Subsequent northern analysis showed that Rex1 had a significant role in 3′ trailer removal in pre-tRNAs with slightly longer 3′ trailers, including two of the four pre-tRNAMeti species and two pre-tRNAVal(CAC) species (Ozanick et al. 2009). Additional experiments showed clear evidence for collaboration in 3′ trailer removal, with Trz1 playing a major role in conjunction with Rex1, with minor additional contributions from Rrp6 and Rex2 (Copela et al. 2008; Skowronek et al. 2014).
The La protein also has a major noncatalytic role in affecting the pathways of 3′ end formation of pre-tRNAs. La protein is an abundant nuclear protein, which binds pre-tRNAs (Rinke and Steitz 1982) at their 3′ oligo(U) ends (Stefano 1984; Teplova et al. 2006; for reviews, see Wolin and Cedervall 2002; Maraia and Bayfield 2006; Porat et al. 2021). La binding leads to endonucleolytic cleavage of the 3′ trailer sequence of the pre-tRNA, and protects the 3′ end of the pre-tRNA from exonucleases in S. cerevisiae (Yoo and Wolin 1997). Thus, in an S. cerevisiae strain lacking La protein (Lhp1), Rex1 acts in conjunction with the 3′ exonuclease Rrp6 to process the 3′ end of the tRNA (Copela et al. 2008) and mutations in La expose tRNAs to Rrp6 in S. pombe (Huang et al. 2006).
CCA addition and removal
The CCA sequence is found at the 3′ ends of all functional tRNAs in all organisms, comprising residues N74–N76, with one of the A76 ribose hydroxyls (2′ or 3′) serving as the covalent attachment site of the cognate amino acid during tRNA charging. The CCA sequence must be added during processing in all eukaryotes and most other organisms, as they lack encoded CCA in their tRNA genes, although some archaea and bacteria (such as E. coli) have encoded CCA in some or all of their tRNA genes. Remarkably, CCA addition is an untemplated addition reaction. In most organisms, CCA addition is catalyzed by a single tRNA nucleotidyl transferase (also known as the CCA-adding enzyme) (Supplemental Table S1), which catalyzes three successive nucleotide additions, although in some ancient bacteria such as Aquifex aeolicus, and in some eukaryotes such as S. pombe, there are separate C74C75-adding and A76-adding enzymes (Tomita and Weiner 2001; Preston et al. 2019). CCA adding enzymes are divided into two classes, each with a similar catalytic domain but with different sequences and overall structures, with class I CCA-adding enzymes in archaea, and class II enzymes in bacteria and eukaryotes (for review, see Xiong and Steitz 2006). Both S. cerevisiae and humans have a single CCA-adding enzyme acting on both nuclear-encoded and mitochondrial-encoded tRNAs (Wolfe et al. 1994; Sasarman et al. 2015).
Previous seminal work elucidated the biochemical gymnastics used by CCA-adding enzymes to precisely add CMP, CMP, and then AMP to the N73 residue of tRNAs without the aid of a template. Both class I and class II CCA-adding enzymes successively add the three NTPs in a single active site, by fixing the acceptor stem through a set of charge and shape interactions with the protein, followed by presentation of the incoming CTP or ATP at each step through interactions that exclude GTP or UTP (Tomita et al. 2004; Xiong and Steitz 2004, 2006). The class I A. fulgidus CCA enzyme features a refolded tRNA 3′ end at each step to position the growing 3′ end at the same location, and to position the incoming CTP or ATP identically, with size discrimination at steps 1 and 2 to exclude ATP, and selection against CTP during step 3 due to incorrect positioning of its α-phosphate (Xiong and Steitz 2004; Pan et al. 2010).
Prior work also revealed that the CCA-adding enzyme has a crucial function in repair of frayed CCA ends of tRNA, in addition to its de novo CCA-addition activity. Thus, although E. coli tRNA genes all have encoded CCA ends, mutants lacking CCA-adding enzyme have reduced growth due to partial removal of some of the ends by RNase T (Zhu and Deutscher 1987). Similarly, while S. cerevisiae cca1 mutants lacking the enzyme are inviable due to the lack of encoded CCA ends in its tRNA genes, mutants lacking Cca1 in the cytoplasmic compartment, but retaining Cca1 in the nucleus and mitochondria, have a similar growth defect and accumulate tRNAs with shortened ends (Wolfe et al. 1996).
It is now clear that the CCA end is implicated in at least four regulatory pathways. First, the CCA end has an important role in the stress response of cells. Thus, oxidative stress treatment of mammalian cells results in shortening of ∼30 of the tRNA 3′ CCA ends, ascribed to angiogenin (ANG), resulting in reduced cap-dependent translation before recovery (Czech et al. 2013) and the accumulation of the truncated tRNAs in nuclei (Schwenzer et al. 2019), discussed further below. Similarly, nutritional stress in T. brucei results in massive removal of ∼70% of the 3′ CCA ends of tRNAs by the conserved Ccr4 homolog LCCR4, which is rapidly reversed by the CCA-adding enzyme when the stress is removed (Cristodero et al. 2021). Second, the CCA end of the peptidyl tRNA has an important role in ribosome-associated quality control triggered by aberrantly stalled ribosomes. During the response, incomplete polypeptides are degraded after release of the peptidyl tRNA from the ribosomal P site by mammalian ANKZF1 (Vms1 in S. cerevisiae), which precisely cleaves the CCA end from the tRNA, leaving a tRNA ending in a 2′–3′ cyclic phosphate at N73. This tRNA is subsequently recycled by removal of the cyclic phosphate by the Trz1 homolog ELAC1, which is found primarily in vertebrates and plants, followed by CCA addition by the CCA-adding enzyme TRNT1 (Yip et al. 2019, 2020). Third, the CCA end of certain tRNAs is subject to a decay pathway triggered by addition of a second CCA repeat. Thus, the instability of the mouse MEN β tRNA-like small cytoplasmic RNA was found to be due to the combination of an unstable acceptor stem and a 5′ end starting with two successive G residues, which leads to aberrant CCACCA addition, and S. cerevisiae tRNASer(CGA) variants with reduced stability that are targeted for the rapid tRNA decay (RTD) pathway (discussed further below) are subject to similar CCACCA addition (Wilusz et al. 2011). Subsequent analysis showed that unstable tRNAs that elicited the aberrant CCACCA addition had refolded on the enzyme after the initial CCA addition so as to loop out three residues and pair C74 and C75 with G2 and G1, setting up a new round of CCA addition (Kuhn et al. 2015; for review, see Wilusz 2015). This pathway of tRNA quality control mediated by CCACCA addition is also found in E. coli cells, likely leading to decay mediated by RNase R (Wellner et al. 2018). Fourth, it is possible that CCA addition is itself regulatory, as initial evidence indicates that CCA addition becomes limiting in S. cerevisiae when tRNA expression is unchecked due to lack of the repressor Maf1 (Foretek et al. 2017), and wild-type E. coli cells have significant amounts of tRNAs with incomplete CCA ends during exponential growth (Czech 2020). In this regard, two independent likely hypomorphic mutations of the human homolog TRNT1 have been associated with multiple clinical manifestations and early death and defective CCA levels in the noncanonical mitochondrial tRNASer(AGY) (Sasarman et al. 2015).
G−1 addition to tRNAHis and reverse polymerization by Thg1 family proteins
The biology of the tRNAHis guanylyltransferase Thg1 and its related proteins continues to reveal surprises (for reviews, see Jackman et al. 2012; Chen et al. 2019).
Virtually all tRNAHis species have an additional G−1 residue (Fig. 3), which is a critical determinant for tRNAHis recognition and charging by HisRS (Rudinger et al. 1994; Nameki et al. 1995), and arises by two very different mechanisms. In bacteria, the G−1 residue is encoded in the genome, and remains at the 5′ end of tRNAHis due to noncanonical processing by RNase P (Orellana et al. 1986). In contrast, in eukaryotes the G−1 residue is added posttranscriptionally opposite A73 by the essential tRNAHis guanylyltransferase Thg1 (Fig. 3; Supplemental Table S1), which catalyzes an unusual 3′–5′ nt addition reaction involving adenylylation of the 5′-phosphate of tRNAHis to activate it; nucleophilic attack of the 3′-OH of GTP to add the G−1 residue to the 5′-phosphate while displacing the adenylate; and pyrophosphatase to generate the mature G−1 monophosphate 5′ end (Cooley et al. 1982; Jahn and Pande 1991; Gu et al. 2003). Thg1 recognizes the GUG anticodon of tRNAHis as a unique determinant (Jackman and Phizicky 2006a), and biochemical evidence suggests that during tRNAHis maturation CCA is added before G−1 addition (Pohler et al. 2019).
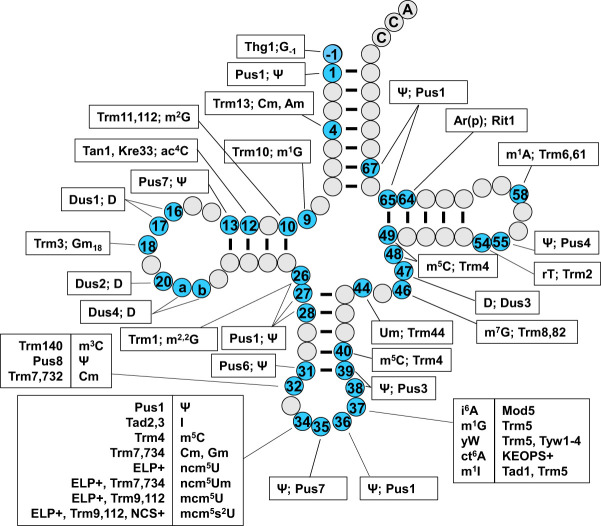
FIGURE 3.
A schematic of modifications and the corresponding genes found in cytoplasmic tRNA in S. cerevisiae. The tRNA secondary structure has gray circles indicating residues without known modifications and blue numbered circles indicating residues with modifications, for each of which the boxed text indicates the corresponding modification and the required gene products. a and b represent nucleotides N20a and N20b, which are found in some tRNAs. Names in all caps (ELP+, NCS+, KEOPS+) refer to the main text for the corresponding genes involved in modification. Conventional abbreviations are used; they are described in the Modomics database (https://genesilico.pl/modomics/) (Boccaletto et al. 2022). (Ψ) pseudouridine, (Am) 2′-O-methyladenosine, (Cm) 2′-O-methylcytidine, (m1G) 1-methylguanosine, (m2G) 2-methylguanosine, (ac4C) 4-acetylcytidine, (D) dihydrouridine, (Gm) 2′-O-methylguanosine, (m2,2G) N2,N2-dimethylguanosine, (m3C) 3-methylcytidine, (I) inosine, (m5C) 5-methylcytidine, (mcm5U) 5-methoxycarbonylmethyluridine, (mcm5s2U) 5-methoxycarbonylmethyl-2-thiouridine, (ncm5U) 5-carbamoylmethyluridine, (ncm5Um) 5-carbamoylmethyl-2′-O-methyluridine, (m1I) 1-methylinosine, (i6A) N6-isopentenyl adenosine, (yW) wybutosine, (t6A) N6-threonylcarbamoyladenosine, (ct6A) cyclic form of t6A, (Um) 2′-O-methyluridine, (m7G) 7-methylguanosine, (rT) ribothymidine, [Ar(p)] 2′-O-ribosyladenosine (phosphate).
Remarkably, Thg1 also catalyzes a true reverse polymerization reaction, involving the template-dependent addition of multiple nucleotides to the 5′ end of tRNAHis variants bearing C73, G73, or U73 instead of A73 (Jackman and Phizicky 2006b), and this reverse polymerization was readily detected in vivo on an S. cerevisiae tRNAHis variant bearing C73 (Preston and Phizicky 2010).
Structural analysis of Thg1 led to a surprise as, despite the lack of sequence similarity, Thg1 was structurally similar to canonical 5′–3′ DNA polymerases, with a palm domain, conserved carboxylates, and two Me++ ions in the active site, suggesting a canonical two-metal ion catalytic mechanism (Hyde et al. 2010; Nakamura et al. 2013). Additional mechanistic analysis showed critical roles for the two conserved aspartate residues that coordinate the Me++ ions for each of the three reaction steps (Smith and Jackman 2012), and showed that reduced pyrophosphatase activity was correlated with increased reverse polymerization, consistent with competition between the two reaction pathways (Smith and Jackman 2014; Desai et al. 2018).
After the unexpected discovery of organisms with tRNAHis species lacking G−1 in a clade within alphaproteobacteria (Wang et al. 2007), tRNAHis species lacking G−1 were also found in several eukaryotes. For example, T. brucei and A. castellanii were found to have tRNAHis lacking G−1 and multiple organisms have no recognizable Thg1 homolog, suggesting that this is much more general (Rao et al. 2013; Rao and Jackman 2015). Remarkably also, the lethality of an S. cerevisiae thg1Δ strain could be suppressed by expression of the corresponding noncanonical HisRS and companion tRNAHis species from T. brucei, A. castellanii, and C. elegans (Rao and Jackman 2015; Lee et al. 2019). Moreover, the virtually normal growth of the S. cerevisiae thg1Δ strain expressing A. castellanii HisRS and tRNAHis essentially proved that the only important role of the G−1 residue of tRNAHis in S. cerevisiae is as an identity element for charging by HisRS (Rao and Jackman 2015).
It is now known that Thg1 is part of the Thg1 superfamily, comprised of a clade of Thg1 orthologs that are widely found in eukaryotes, and a clade of Thg1-like proteins (TLPs) that are found in some archaea, bacteria, and eukaryotes (Heinemann et al. 2009, 2010; Jackman et al. 2012). The bacterial TLPs from Bacillus thuringiensis and Myxococcus xanthus and each of four archaeal TLPs tested all catalyzed templated addition of nucleotides to tRNAs in vitro, and expression of the B. thurigiensis and the four archaeal TLPs each complemented the lethality of an S. cerevisiae thg1Δ strain (Abad et al. 2010; Heinemann et al. 2010; Rao et al. 2011), through U−1 addition to tRNAHis across from A73 in the case of the B. thuringiensis TLP (Dodbele et al. 2019).
Nonetheless, the biochemical activity of TLPs suggests that their primary role is in tRNA editing, in which tRNAs missing one or more 5′ nt are 5′ end-repaired by templated reverse polymerization. This 5′ end repair activity was first inferred by comparison of the sequences of tRNAs and their corresponding genes in mitochondria of the eukaryotic microbe Acanthamoeiba castellanii (Lonergan and Gray 1993). In support of this editing function of TLPs, the B. thuringiensis TLP has increased kcat/KM values for addition of nucleotides to 5′ truncated tRNAs, compared to that for G−1 addition to the mature tRNAHis (Rao et al. 2011); two of the four Thg1/TLPs (TLP3 and TLP4) from Dictyostelium discoideum have substantial kcat/KM values for templated nucleotide addition to 5′ truncated tRNAs (Abad et al. 2011); and depletion of D. discoideum TLP3 results in a severe growth defect and decreased mitochondrial tRNA 5′ editing (Long et al. 2016).
Dictyostelium discoideum TLP4 has a critical but as yet unknown role (Long et al. 2016). Whereas depletion of D. discoideum Thg1 leads to the expected severe growth defect and cytoplasmic tRNAHis lacking G−1, and knockout of TLP2 leads to a minor but distinct growth defect and mitochondrial tRNAHis lacking G−1, depletion of TLP4 leads to a severe growth defect for unknown reasons. Although the function of TLP4 is not yet known, its cytoplasmic location suggests a nonorganellar role, and its biochemical activity on 5S RNAs and a ncRNA emphasizes the potential for TLP4 to act naturally on non-tRNA substrates (Long et al. 2016).
It is also not fully understood how Thg1 acts to regulate mitochondrial function. A V55A mutation in the human Thg1 ortholog THG1L (Supplemental Table S1) is associated with cerebellar ataxia and decreased mitochondrial fusion (Fig. 4; Edvardson et al. 2016). Furthermore, reduced expression of human THG1L (also called IHG-1, induced in high glucose-1) leads to reduced mitochondrial respiration and mitochondrial fusion, linked to reduced interaction with Mfn1 and Mfn2, which mediate mitochondrial fusion (Hickey et al. 2014). It is unknown how THG1L interacts with Mfn1 and Mfn2 and if this interaction is perturbed in the V55A variant as part of a moonlighting role of THG1L, or if the V55A variant has reduced G−1 addition activity on mitochondrial tRNAHis (Suzuki and Suzuki 2014; Nakamura et al. 2018). The THG1L–V55A variant has normal activity in vitro, but its expression in an S. cerevisiae thg1Δ strain results in a growth defect, unlike for the WT THG1L protein (Edvardson et al. 2016).
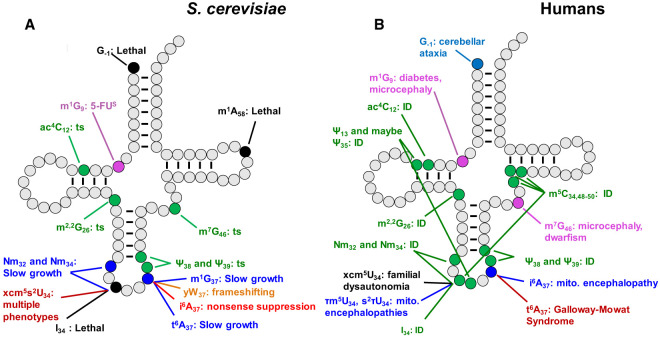
FIGURE 4.
Effect of lack of tRNA modifications in S. cerevisiae and humans. (A) Prominent phenotypes resulting from mutations in S. cerevisiae modification genes. 5-FUs, sensitivity to 5-fluorouracil; ts, temperature sensitivity. (B) Prominent diseases and disorders resulting from mutations in human modification genes. (ID) Intellectual disability.
Pre-tRNA splicing
The discovery of pre-tRNAs with transcribed introns in budding yeast and vertebrate cells (Hopper et al. 1978; Knapp et al. 1978; O'Farrell et al. 1978; De Robertis and Olson 1979) occurred nearly simultaneously with the discovery of mRNA introns in Drosophila and vertebrate cells (Berget et al. 1977; Chow et al. 1977; White and Hogness 1977). However, unlike pre-mRNA splicing, which involves two RNA catalyzed phosphoester transfer reactions occurring in a large RNP complex called the spliceosome to remove the intron in circular form, pre-tRNA splicing is catalyzed by a small endonuclease complex that generates two exons and a linear, or in some cases a circular (Lu et al. 2015), excised intron, followed by exon joining by a ligase enzyme and a small cast of additional proteins.
tRNA introns: characteristics and functions
In all eukaryotes examined (http://gtrnadb.ucsc.edu; Chan and Lowe 2016), a subset of tRNAs is encoded by intron-containing genes. Eukaryotic tRNA introns are located 1 nt 3′ of the anticodon, between N37 and N38, and are generally short, ranging from 14–60 nt in budding yeast, to as long as 133 nt for some introns in other organisms (Chan and Lowe 2016; for reviews, see Yoshihisa 2014; Schmidt and Matera 2020). The percentage of intron-containing tRNA genes differs among organisms, with ∼6% in mouse, rat, and humans and 24% in budding yeast, but their occurrence is clustered in specific gene families (Chan and Lowe 2016; for review, see Schmidt and Matera 2020). For example, every gene member in each of the 10 intron-containing gene families in budding yeast and the 16 intron-containing families in fission yeast contains an intron, and all eukaryotic tRNATyr genes in studied organisms have an intron. However, in mouse and humans, introns are not always found in all members of isoacceptor gene families with introns. For example, although in humans all 13 tRNATyr(GUA) genes and all five tRNAIle(UAU) genes contain an intron, only 5/6 of the tRNAArg(UCU), and 5/7 tRNALeu(CAA) gene family members contain introns (Chan and Lowe 2016). It is also interesting to note that in budding yeast, the intron sequences within each intron-containing gene family are either identical or very similar to each other, but for fission yeast and vertebrates the sequences of introns vary among the family members (Chan and Lowe 2016). A number of archaeal tRNA genes also have introns which, as in eukaryotes, are generally relatively short, occur mostly between N37 and N38, and are generally clustered in all the genes of each isoacceptor gene family member with introns (Yoshihisa 2014).
Although tRNA introns do not possess conserved sequence motifs at their termini, they generally have structure (Fig. 5). The classical archaeal exon–intron structure is a bulge-helix-bulge (BHB) RNA structure, in which nucleotides N32–N35 form a helix with corresponding residues in the intron, with cleavage sites in the adjacent 3 nt single-stranded bulges (Thompson and Daniels 1988, 1990; Yoshihisa 2014). Eukaryotic tRNA introns have a similar, but less well-defined structure, generally with nucleotide sequences that are complementary with N33–N35 or sometimes N34–N36 or other combinations of nucleotides within the ACL, extended by an additional base pair, called the anticodon–intron (A–I) base pair between C32/U32 of the ACL and the antepenultimate A/G of the intron. This results in a bulge-helix-loop (BHL) exon–intron structure (Fig. 5), with the cleavage sites in the single-stranded regions comprising the bulge and the loop (Lee and Knapp 1985; Baldi et al. 1992; Di Nicola Negri et al. 1997; Schmidt and Matera 2020).
FIGURE 5.
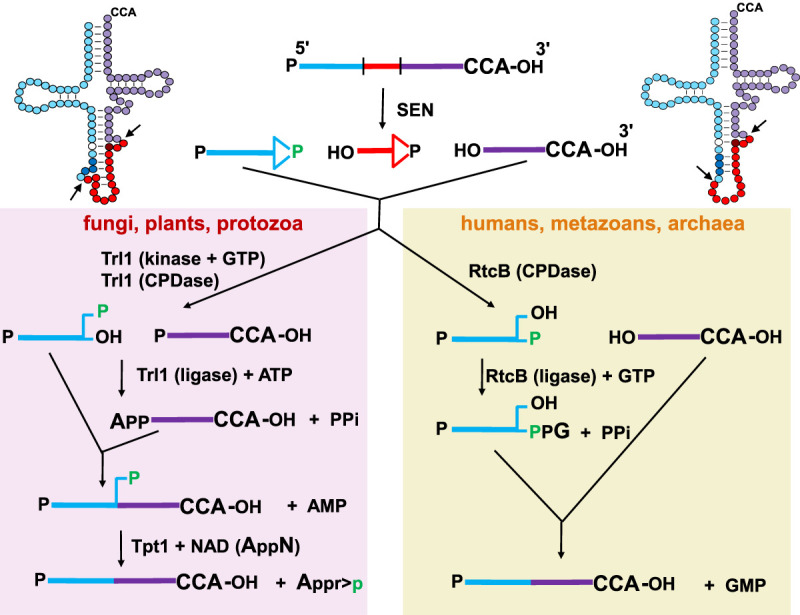
Schematic of tRNA splicing pathways in different eukaryotes. (Top left and right) A typical unspliced pre-tRNA is shown in its accepted secondary structure, with the intron residues indicated by red circles except for the antepenultimate intron residue (dark red); residues N1–N37 of the 5′ exon indicated by light blue circles, except for N32 (white) and anticodon residues N34–N36, (dark blue); and residues N38–N73 indicated by purple residues. The antepenultimate intron residue pairs with N32 in the pre-tRNA. Arrows indicate sites of endonucleolytic cleavage of the pre-tRNA by the SEN/TSEN splicing complex. (Top left) Canonical pre-tRNA with a well-defined BHL motif. (Top right) One of several pre-tRNAs with a slightly different BHL motif. (Top center) A typical unspliced pre-tRNA is shown in linear form with the 5′ exon in blue, the intron in red, and the 3′ exon in purple. Endonucleolytic cleavage of the pre-tRNA results in formation of a 2′–3′-cyclic phosphate at the 3′ end of both the 5′ exon and and the intron, leaving a 5′-OH at the 5′ end of both the 3′ exon and the intron. (Left panel) In fungi, plants, and protozoa, the RNA 5′-kinase activity of the ligase Trl1 phosphorylates the 5′-OH end of the 3′-half-molecule using GTP, and the cyclic phosphodiesterase (CPDase) activity of Trl1 opens the 2′–3′ cyclic phosphate to a 2′-phosphate (green). Then the ligase activity of Trl1 joins the half-molecules by adenylylation of the 5′-phosphate of the 3′ exon and ligation to the 3′-OH of the 5′ exon, leaving a 2′ phosphate (green) at the splice junction. This 2′-phosphate is subsequently transferred to NAD by the 2′-phosphotransferase (Tpt1). (Right panel) In humans and metazoans, as well as in some archaea, the CPDase activity of the ligase RtcB opens the 2′–3′ cyclic phosphate of the 5′ exon to form a 3′-phosphate (green). Then, the ligase activity of RtcB joins the half-molecules by guanylylation of the 3′-phosphate of the 5′ exon and ligation to the 5′-OH of the 3′ exon, releasing GMP.
Splicing is essential in all studied eukaryotes because in each organism for at least one intron-containing isoacceptor gene family, all of the genes possess introns. Intron-containing tRNAs cannot function in protein synthesis prior to splicing because tRNA introns disrupt the ACL and there is at least one report that documented intron-containing tRNAs cannot be aminoacylated (O'Farrell et al. 1978; for reviews, see Phizicky and Hopper 2010; Yoshihisa 2014; Chan and Lowe 2016; Schmidt and Matera 2020).
Although tRNA splicing is essential, the reverse is not the case; that is, the presence of introns in any tRNA gene family is not essential. Early studies from the Abelson laboratory demonstrated that a budding yeast strain possessing a deletion of the intron from the single copy essential tRNASer(CGA) gene was viable, thereby documenting that the intron in this tRNA gene is unessential (Ho and Abelson 1988). Subsequent studies by the Yoshihisa group created 10 yeast strains, each missing the intron from every member of the corresponding intron-containing isoaccepter gene family; this tour-de-force report documented that introns for all budding yeast tRNA genes are unessential (Mori et al. 2011; Hayashi et al. 2019). In fact, each of the 10 yeast deletion strains had rather few growth defects (Hayashi et al. 2019).
That tRNA introns are unessential for life (at least for budding yeast) raises the very interesting question as to why the presence of tRNA introns has been conserved throughout eukaryotes. Surprisingly, multiple roles for tRNA introns have been documented, thereby providing selection pressure for their conservation. These roles include: the efficiency of tRNA genes in functioning as transcription barriers for local ORFs (Donze and Kamakaka 2001); posttranscriptional regulation of mature tRNA levels via the Met22-dependent pre-tRNA decay (MPD) turnover pathway (discussed below), which has specificity for intron-containing pre-tRNAs (Payea et al. 2020); the presence of particular tRNA modifications; and the altered modification pattern of tRNAs. It is well documented that particular tRNA modification enzymes have specificity for intron-containing pre-tRNAs. For example, if an intron is removed from one of the eight genes encoding tRNATyr(GUA) (SUP6), the resulting tRNATyr lacks Ψ at anticodon residue U35 and its function as a suppressor tRNA is reduced (Johnson and Abelson 1983). Similarly, intron removal from tRNALeu(CAA) genes results in lack of m5C at C34 in the anticodon (Strobel and Abelson 1986; Hayashi et al. 2019). In each of these cases, removal of the intron eliminates the specificity of the corresponding modification enzymes, Pus7 and Trm4, respectively, for the corresponding tRNAs (Behm-Ansmant et al. 2003; for review, see Grosjean et al. 1997). Introns in pre-tRNAs also have been shown to dictate modification fidelity; for example, intron deletions of the tRNAIle(UAU) genes in budding yeast result in a lack of Ψ34 and instead U34 is erroneously modified with 5-carbamoylmethyluridine (ncm5U) (Hayashi et al. 2019).
Eukaryotic tRNA splicing endonucleases
Intron removal from eukaryotic pre-tRNAs is catalyzed by a heterotetramic protein endonuclease complex called tRNA splicing endonuclease (SEN in budding yeast or TSEN vertebrate cells, Supplemental Table S1; Peebles et al. 1979, 1983; Trotta et al. 1997; Paushkin et al. 2004; Hayne et al. 2022). Two of the four subunits (Sen2 and Sen34) of the SEN/TSEN complexes are conserved and possess catalytic activity, while the remaining two subunits (Sen15 and Sen54) are not conserved (Paushkin et al. 2004). Genes that encode proteins similar to the SEN catalytic subunits that function in pre-tRNA splicing have been discovered in plants, trypanosomes, and Drosophila (Akama et al. 2000; Rubio et al. 2013; Schmidt et al. 2019; for reviews, see Fabbri et al. 1998; Phizicky and Hopper 2010; Yoshihisa 2014; Schmidt and Matera 2020). Since pre-tRNA splicing is necessary to generate the entire cadre of tRNAs required to decode the genome, it is not surprising that each of the SEN and TSEN subunits is essential for life in budding and fission yeast and in human cell lines (Giaever et al. 2002; Kim et al. 2010a; Wang et al. 2015). Interestingly, autosomal recessive mutations in each of the TSEN subunits cause subclasses of Pontocerebellar Hypoplasia (PCH), congenital neurodegenerative diseases (Budde et al. 2008; Cassandrini et al. 2010; Breuss et al. 2016; for review, see Sekulovski and Trowitzsch 2022). It is unclear why the TSEN mutations preferentially affect neuronal tissues, but this is a common phenomenon in tRNA processing biology in humans, as mutations in human modification genes are often linked to neurological disorders (Fig. 4).
Prior results described crucial similarities and differences between the structure and substrate recognition properties of TSEN and the equivalent archaeal tRNA splicing endonuclease. Whereas the archaeal tRNA splicing endonuclease from H. volcanii recognizes an isolated BHB RNA structure (Thompson and Daniels 1988, 1990), the S. cerevisiae SEN complex recognizes a combination of features, including the mature domain of the intron-containing pre-tRNA, the distance from the mature domain to the splice sites, and the A–I base pair in the context of the BHL structure found in eukaryotic pre-tRNAs (Reyes and Abelson 1988; Baldi et al. 1992; Di Nicola Negri et al. 1997). Structural analysis of the archaeal endonuclease from Methanococcus jannaschii and a co-crystal structure of the Archaeoglobus fulgidus enzyme with a BHB RNA substrate revealed an active site His-Tyr-Lys triad that is conserved between eukaryotes and archaea, with substrate bulge recognition aided by two nearby arginines, which originate in the other subunit of the homodimer, and form a cation-π sandwich with one of the substrate adenine residues in the bulge (Li et al. 1998; Xue et al. 2006; Calvin et al. 2008). Remarkably this cross-subunit interaction is functionally conserved for the corresponding arginine and tryptophan residues of the S. cerevisiae Sen34 subunit, as these residues are required for cleavage of the 5′ splice site by the Sen2 subunit of TSEN (Trotta et al. 2006).
Additional work has added substantially to our understanding of how the different recognition properties of the archaeal and eukaryotic endonucleases evolved. It was initially found that the A. fulgidus endonuclease recognized an isolated BHB motif RNA substrate, but could only recognize the more relaxed BHL motif in the context of a pre-tRNA containing the mature domain (Tocchini-Valentini et al. 2007). Subsequently, it was found that the crenarchaeal endonuclease from Aeropyrum pernix had specificity for both the BHB and the BHL structural motifs, and that this was due to a crenarchaeal specific loop (CSL) which, when inserted into the A. fulgidus enzyme, converted it to an enzyme that recognized the BHL structural motif (Hirata et al. 2011; Kaneta et al. 2018; for review, see Hirata 2019).
Until recently, it was not possible to understand the biochemical functions of the two eukaryote-specific noncatalytic SEN15 and SEN54 subunits of the heterotetrameric SENs, because efforts to reconstitute functional SEN or TSEN complexes from purified recombinant subunits had failed for decades. However, in vitro reconstitutions have now succeeded (Hayne et al. 2020, 2022; Sekulovski et al. 2021). Hayne et al. obtained functional human endonuclease expressed in E. coli or HEK cells when all four TSEN (2, 15, 34, and 54) subunits were coexpressed, whereas Sekulovski et al. were able to reconstitute human endonuclease activity from recombinant TSEN15–34 and TSEN2–54 heterodimers expressed in insect or mammalian cells (Hayne et al. 2020, 2022; Sekulovski et al. 2021). Success in reconstitution of TSEN provided the opportunity for structural and biochemical analysis.
Recently, three groups (Hayne et al. 2023; Sekulovski et al. 2023; Zhang et al. 2023) obtained high resolution (2.9–3.9 Å) cryo-EM structures of the human TSEN heterotetramer enzyme in complex with intron-containing pre-tRNAs. The enzyme-substrate complexes were trapped by either modifying the RNA cleavage sites and/or by utilizing enzyme with alterations of catalytic amino acids. Overall, the resolved heterotetrameric enzyme-tRNA complexes are structurally similar to the archaeal enzymes, documenting their evolutionary relationship. However, the human TSEN subunits contain extensions and insertions that provide additional enzyme-substrate interactions. Importantly, although the reconstituted human TSEN complex can utilize short RNA sequences containing just the intron and the anticodon stem–loop as substrates (albeit with low kinetic activity), the structural analyses show that TSEN54 has extensive interactions with the mature tRNA domain, supporting the earlier model that TSEN54 acts as a molecular ruler to regulate cutting at the appropriate splice sites (Trotta et al. 1997). TSEN15 interactions with tRNAs were not resolved, but it is predicted that TSEN15 “mediates interactions with the intron surrounding the 5′ splice site” (Hayne et al. 2023).
The high-resolution TSEN structures provide further information regarding how the TSEN mutations that cause PCH may affect TSEN structure/function. None of the causative alterations lie within catalytically important locations, but rather they disrupt subunit interactions (Hayne et al. 2023; Sekulovski et al. 2023). Thus, the TSEN mutations likely affect the SEN complex stoichiometry, thermostability of the heterotetramer, and/or efficiency of tRNA splicing (Breuss et al. 2016; Sekulovski et al. 2021).
Clp1 and TSEN
The role of the human RNA kinase hsClp1 in tRNA splicing continues to be an enigma. Clp1 functions in mRNA 3′ end processing (de Vries et al. 2000), but also copurifies with the TSEN isolated from human 293 cell lines (Paushkin et al. 2004) and coexpressed recombinant hsClp1 copurifies with TSEN (Hayne et al. 2020; Sekulovski et al. 2021). Similarly to mutations of the TSEN subunits, autosomal recessive mutations in CLP1 genes are linked to PCH-like disorders in human patients, as well as in zebrafish and mouse models (Schaffer et al. 2014). Moreover, the CLP1 mutations were reported to affect the endonuclease subunit stoichiometry and tRNA splicing in vitro activity (Hanada et al. 2013; Karaca et al. 2014; Schaffer et al. 2014). Therefore, it was surprising that Clp1 is not required for either TSEN complex in vitro assembly or for pre-tRNA splicing by the reconstituted human complex from either E. coli or mammalian cells (Hayne et al. 2020; Sekulovski et al. 2021). Moreover, in vivo studies of the Drosophila Clp1 ortholog provided evidence that Clp1 may instead function to negatively regulate the ligation step of pre-tRNA splicing (Hayne et al. 2020). Further, a recent study that created mouse models with the PCH relevant CLP1 mutations documented changes in tRNA processing intermediates, but these tRNA processing alterations did not correlate with pathogenicity; rather, the pathogenicity correlated with alterations of 3′ poly(A) site selection of particular RNAs; thus, the authors suggest that PCH due to CLP1 mutations may result from defects in RNA 3′ processing instead of tRNA biology (Monaghan et al. 2021). Nevertheless, cryo-EM structures of TSEN in complex with hsClp1 document that TSEN54 interacts with Clp1 (Hayne et al. 2023; Sekulovski et al. 2023). Future studies are required to resolve the functional relationship of TSEN and Clp1.
Subcellular location for tRNA splicing
The subcellular location of pre-tRNA splicing differs among organisms. Early studies using Xenopus oocytes reported that pre-tRNA splicing occurs in the nucleus (Melton et al. 1980; De Robertis et al. 1981). Later studies verified this nuclear location in human cells (Paushkin et al. 2004). In contrast, the budding and fission yeast SEN complexes are not located in the nucleus, but rather are peripherally associated on the cytoplasmic surface of mitochondria (Fig. 1; Yoshihisa et al. 2003, 2007; Wan and Hopper 2018). For both budding and fission yeast, a conserved mitochondrial membrane protein component of the mitochondrial import machinery, Tom70, is important for tethering of the SEN complexes to mitochondria (Wan and Hopper 2018), documenting conservation for the location of, and the mechanism of, achieving SEN location to mitochondria for at least 600 million years (Parfrey et al. 2011). tRNA splicing in the protozoan Trypanosome brucei (Lopes et al. 2016) and, likely, in the plant Arabidopsis thaliana (Park et al. 2005) also occurs after pre-tRNAs are exported from the nucleus to the cytoplasm; however, there is no evidence that either the Trypanosome or Arabidopsis TSEN localize at the mitochondrial surface (Englert et al. 2007; Lopes et al. 2016). It would be very interesting to discern the subcellular location of TSEN in other eukaryotic organisms to learn when and why the split from cytoplasmic to nuclear pre-tRNA splicing occurred.
Additional SEN RNA substrates
Since the preponderance of studies of eukaryotic SEN indicated that it interacted with the mature tRNA anticodon stem–loop rather than the splice junctions or intron sequences (Reyes and Abelson 1988; Sekulovski et al. 2021, 2023; Hayne et al. 2023), and that accurate pre-tRNA cleavage proceeds by a mechanism measuring the length of the anticodon stem (Reyes and Abelson 1988), it was not anticipated that there would be SEN substrates in addition to tRNAs. However, the Xenopus, budding yeast, and reconstituted human TSEN can cleave mini substrates in vitro that contain tRNA stem–loop structures (Fabbri et al. 1998; Hayne et al. 2020), and there is in vivo evidence suggesting that the budding yeast SEN complex has substrates in addition to intron-containing pre-tRNAs. Two studies generated yeast strains that were able to bypass the requirement for SEN to generate mature tRNAs and the results demonstrated that even though cells possessed normal levels of mature, spliced tRNAs they nevertheless required all four functional SEN subunits for viability (Dhungel and Hopper 2012; Cherry et al. 2018), and cells with nuclear SEN and defective mitochondrially located SEN have defects in an unessential step in pre-rRNA processing (Volta et al. 2005; Dhungel and Hopper 2012). These data support the hypothesis that there are essential cytoplasmic non-tRNA substrates for the SEN complex.
Additional SEN substrates have been identified. Budding yeast SEN complex functions in cleavage/turnover of mRNAs encoding proteins that are imported into mitochondria such as CBP1 mRNA, encoding an unessential mitochondrial protein, at the boundary of a stem–loop structure (Tsuboi et al. 2015). Most recently, the van Hoof laboratory, using an unbiased bioinformatics approach, identified several budding yeast mRNAs that encode additional essential and unessential mitochondrial proteins, which are cleaved by SEN (Hurtig et al. 2021). Interestingly, although there is no known sequence specificity of SEN required for removal of tRNA introns, mRNA cleavage by the SEN complex appears to require an A nucleotide located at the −1 position of the mRNA cleavage sites (Hurtig et al. 2021). This newly discovered tRNA endonuclease-initiated decay (TED) role for the SEN complex likely functions in quality control of mRNAs encoding mitochondrial proteins that are located at the mitochondrial cytoplasmic surface (Hurtig et al. 2021).
The two eukaryotic pathways for ligation of tRNA exons
The ligation step of eukaryotic tRNA splicing proceeds by two very different mechanisms to join the 5′ exon, which terminates with a 2′–3′ cyclic phosphate, to the 3′ exon, which initiates with a 5′-OH end (Fig. 5).
In S. cerevisiae, the single subunit ligase Trl1 (also called Rlg1) first heals the ends, using its cyclic phosphodiesterase (CPDase) activity to open the 2′–3′ cyclic phosphate to a 2′-phosphate and its polynucleotide kinase (PNK) to phosphorylate the 5′-OH (Fig. 5; Supplemental Table S1). Then Trl1 joins the ends with its ligase activity, first activating the 5′-phosphate by formation of an adenylylated intermediate (Greer et al. 1983; Phizicky et al. 1986). The resulting ligated RNA has a 2′-phosphate at the splice junction, which is transferred to NAD by the 2′-phosphotransferase Tpt1 to form ADP-ribose 1″–2″-cyclic phosphate (McCraith and Phizicky 1991; Culver et al. 1993, 1997; Spinelli et al. 1997). This ligation mechanism is conserved in fungi (Remus et al. 2017; Banerjee et al. 2019a; Peschek and Walter 2019), protozoa (Lopes et al. 2016), and plants (Englert and Beier 2005; Wang et al. 2006), and also in several metazoan species, albeit through a separate ligase and PNK/CPDase and/or Clp1 (Englert et al. 2010).
In contrast, in most metazoans and archaea, the RNA ligase joins the two exons by incorporating the phosphate from the 2′–3′ cyclic phosphate of the 5′ exon in the junction (Nishikura and De Robertis 1981; Filipowicz and Shatkin 1983; Laski et al. 1983; Zofallova et al. 2000), a reaction that is catalyzed by RtcB (Supplemental Table S1; Englert et al. 2011; Popow et al. 2011; Tanaka and Shuman 2011; for review, see Popow et al. 2012). The biochemical reaction of RtcB is unusual (Fig. 5). Although joining of the 2′–3′ cyclic phosphate to a 5′-OH should in principle be isoenergetic, the RtcB reaction uses a more circuitous route to ligation. RtcB first uses its CPDase activity to generate a 3′-phosphate, which is then followed by guanylylation of RtcB using GTP, transfer of the guanylate to the RNA-3′-p to generate the activated RNA-p-pG intermediate, and then ligation by attack by the 5′-OH of the 3′ exon to generate the products RNA and GMP (Tanaka et al. 2011a; Chakravarty and Shuman 2012; Chakravarty et al. 2012; Englert et al. 2012; Desai et al. 2013; Banerjee et al. 2021).
RtcB has additional partners that affect its activity. Although E. coli RtcB can function independently to replace Trl1 function in S. cerevisiae for both tRNA splicing and HAC1 mRNA splicing (Tanaka et al. 2011b), the activity of Pyrococcus horikoshii RtcB is stimulated by Archease, a member of the same operon (Desai et al. 2014), and remarkably, Archease stimulates some RtcB orthologs from single turnover to multiple turnover enzymes (Desai et al. 2015). Moreover, in humans the Archease ortholog ARCH (ZBTB8OS) interacts with RTCB and is crucial for tRNA splicing in vivo and in vitro, and the RTCB guanylation step is stimulated in vitro and in vivo by the DEAD box helicase DDX1 (Popow et al. 2014). In contrast, overexpression of the 2′–3′-cyclic phosphatase activity of ANGEL2 (which completely removes the phosphate) can compete with tRNA ligase and inhibit mammalian tRNA splicing (Pinto et al. 2020).
In both S. cerevisiae and mammalian cells, the respective ligase pathways also participate in the ligation step of the noncanonical mRNA splicing of HAC1/XBP1, encoding a crucial transcription factor in the unfolded protein response (UPR) pathway, after endonucleolytic excision of the HAC1/XBP1 intron by Ire1 (Sidrauski et al. 1996; Sidrauski and Walter 1997; Jurkin et al. 2014; Lu et al. 2014).
Mechanistic studies of the Trl1 ligation step and the Tpt1 2′-phosphotransferase step
A series of elegant papers have illuminated the unique biochemical properties of the single subunit funga/plant Trl1 ligase. The modular nature of the Trl1 domains has been well documented by showing functional complementation of S. cerevisiae trl1 mutants lacking complete Trl1 function by individual kinase (Ramirez et al. 2008) and CPDase subunits (Schwer et al. 2008). The unique GTP specificity of the Trl1 kinase activity in Trl1 is accounted for by a unique G-loop and extensive guanine-specific interactions with residues in the G-loop (Remus et al. 2017), and the unique 2′-phosphate specificity of the Trl1 ligase activity (Remus and Shuman 2013) is plausibly explained by a sulfate binding site in the structure of the ligase domain (Banerjee et al. 2019a).
Much has been learned about the unusual mechanism by which Tpt1 catalyzes transfer of the 2′-phosphate from the splice junction of ligated tRNA to NAD to form ADP-ribose 1″–2″-cyclic phosphate. Prior biochemical analysis showed that Tpt1 catalyzes nucleophilic attack by the RNA-2′-phosphate oxygen at the 1″-position of NAD+ to displace nicotinamide and form an ADP-ribosylated RNA covalent intermediate, followed by cyclization catalyzed by the neighboring 2′-OH to form the product ADP-ribose 1″–2″-cyclic phosphate, with concomitant release of the dephosphorylated RNA (Spinelli et al. 1999; Sawaya et al. 2005; Steiger et al. 2005). Subsequent kinetic analysis of variants in Runella slithyformis Tpt1 revealed that the R68A variant was completely unaffected in the rate of formation of the covalent intermediate (step 1) but had a severe 200-fold reduction in the rate of step 2, in which the dephosphorylated RNA was released during cyclization (Munir et al. 2018a). The crystal structure of the Clostridium thermocellum Tpt1 showed four critical residues in the active site (Banerjee et al. 2019b), which had been previously implicated in catalysis (Steiger et al. 2005; Munir et al. 2018a), and revealed two highly informative bound ligands: ADP-ribose 1″-phosphate, mimicking the ADP-ribose 1″–2″-cyclic phosphate product of step 2, after subsequent CPDase activity in the crystal; and acetyl-coA, with its adenosine 3′,5′ bis-phosphate (pAp) moiety mimicking the substrate RNA after dephosphorylation of the 2′-phosphate (Banerjee et al. 2019b).
The puzzle of Tpt1 in other organisms
One major unanswered question regarding the Tpt1 protein family is why its members are found widely in bacterial, archaeal, and eukaryotic organisms that do not apparently require its enzymatic activity. For example, mouse, E. coli, and R. slithyformis each have a functional Tpt1 ortholog that complements the lethality of an S. cerevisiae tpt1Δ mutant (Spinelli et al. 1998; Munir et al. 2018a), although mammals use the metazoan/archaeal RtcB pathway for splicing of both tRNA and HAC1/XBP1, which does not generate a 2′-phosphate (Popow et al. 2011; Jurkin et al. 2014), and bacteria such as E. coli do not undergo tRNA splicing or have a known pathway that generates an RNA with internal 2′-phosphate. Indeed, neither the E. coli nor the mouse Tpt1 ortholog is essential in their respective organisms (Spinelli et al. 1998; Harding et al. 2008).
Recent experiments suggest that Tpt1 could have other biochemical functions in some of these and other organisms. Remarkably, a subset of Tpt1 enzymes can catalyze NAD-dependent ADP-ribosylation of the 5′-phosphate of RNA or DNA, with an oxygen of the 5′-phosphate of the oligonucleotide acting as nucleophile (like the 2′-phosphate of ligated tRNA during the canonical Tpt1 reaction), forming an ADP-ribosyl cap on the nucleotide (Munir et al. 2018b). This RNA and DNA ADP-ribosylation activity extends to the human TRPT1 ortholog and, although the product 5′ capped oligonucleotide cannot be resolved by the Tpt1 reaction to release the RNA or DNA, the product can be reversed by a number of ADP-ribosylhydrolases as well as by some macrodomains (Munnur et al. 2019). Other results show that C. thermocellum and A. pernix Tpt1 proteins can catalyze removal of terminal 2′-phosphates, and to some extent 3′-phosphates, from RNA (Munir et al. 2019). It thus seems likely that one or more noncanonical Tpt1 reactions like these could explain the evolutionarily widespread occurrence of Tpt1 in organisms that do not generate RNA with an internal 2′-phosphate, or that have a functional RtcB. One of these noncanonical Tpt1 activities might also explain the puzzling result that overexpression of S. cerevisiae TPT1 rescues the synthetic lethality of S. cerevisiae elg1Δ srs2Δ mutants, which are defective in the repair of DNA damage, as does overexpression of the CPDase Cpd1 (Gazy et al. 2013), which generates Appr-1″-p from ADP-ribose 1″–2″-cyclic phosphate (Martzen et al. 1999; Nasr and Filipowicz 2000).
tRNA intron turnover
A quantitatively important by-product of tRNA splicing is the excised introns, which are produced in equimolar amounts to spliced tRNA, at the rate of ∼600,000 times a generation in budding yeast (Wu and Hopper 2014). Even though eukaryotic cells generate these enormous quantities of freed linear introns during tRNA splicing, these introns are rarely detected under normal physiological conditions, because the excised introns are either converted to more stable molecules or are rapidly and efficiently destroyed.
The excised introns derived from budding yeast pre-tRNAs remain as linear RNAs (Knapp et al. 1979; Wu and Hopper 2014), which are subject to decay. One pathway by which yeast linear tRNA introns are degraded is the kinase-mediated decay pathway, in which the RNA kinase activity of the Trl1/Rlg1 ligase phosphorylates the 5′ terminus of the linear excised intron, rendering the intron as a substrate for the cytoplasmic 5′ to 3′ exoribonuclease, Xrn1 (Wu and Hopper 2014). Curiously, the kinase-mediated decay pathway functions in the turnover of only a subset of the budding yeast tRNA introns (Wu and Hopper 2014); the gene products involved in the turnover of other excised tRNA introns have not yet been delineated. In addition, the kinase-mediated decay pathway acts at two points during the UPR pathway: to degrade the 3′ exon and therefore inhibit HAC1 ligation by competition with Trl1 (Cherry et al. 2019; Peschek and Walter 2019), and to degrade the HAC1 intron and activate HAC1 translation by relieving an inhibiting interaction with the HAC1 mRNA (Mori et al. 2010; Cherry et al. 2019).
In contrast, the excised tRNA introns in Drosophila exist as circular molecules (Lu et al. 2015; for review, see Schmidt and Matera 2020), arising from RtcB-mediated direct ligation of the 5′ and 3′ termini (Schmidt et al. 2019), as introduction of RtcB to yeast also efficiently converts the introns to circular molecules (Schmidt et al. 2019). How these circular introns are turned over remains unknown.
tRNA MODIFICATIONS
It is well known that tRNAs are by far the most modified class of RNAs in the cell. A total of 155 nucleoside or base modifications are currently listed in the Modomics database (Boccaletto et al. 2022), the vast majority of which are found in tRNAs (Grosjean 2015). These modifications (Fig. 3; Supplemental Table S1) provide substantial chemical diversity to tRNAs (Helm and Alfonzo 2014), and their lack frequently leads to growth defects in S. cerevisiae and neurological or mitochondrial disorders in humans (Fig. 4; for reviews, see Hopper 2013; Ramos and Fu 2019; Suzuki 2021). Previous analysis of a database of 561 sequenced tRNAs from bacteria, archaea, eukaryotes, mitochondria, and chloroplasts (Sprinzl and Vassilenko 2005, now Juhling et al. 2009) found chemical modifications on 11.9% of tRNA residues, with a median of eight modifications per tRNA species (Phizicky and Alfonzo 2010; Phizicky and Hopper 2010), and a range of average modification frequencies from 6.5% to 16.5% in different subgroups of species and from 8.6% to 10.2% in organelles (Machnicka et al. 2014). Among 34 S. cerevisiae cytoplasmic sequenced tRNA species, there are 25 chemically distinct modifications, which are found at 36 different residues, with an average of 12.6 modifications per species, ranging from 7 to 17 modifications per tRNA; and for 17 sequenced S. cerevisiae mitochondrial tRNAs, 9.5% of the residues have modifications, with six to nine modifications per tRNA (Phizicky and Alfonzo 2010; Phizicky and Hopper 2010).
Modifications within the main tRNA body and outside the ACL region (i.e., N1–N30 and N40–N76, and not N31–N39) comprise the majority of tRNA modifications found in tRNAs (Figs. 2, 3). For example, of the ∼12.6 modifications found in a typical cytoplasmic tRNA in S. cerevisiae, 10 are body modifications, and comprise 14 of the 25 different modifications (Fig. 3; Supplemental Table S1). As described in detail later in this review, the body modifications have important roles in tRNA folding and/or stability, and their biology intersects several other important cellular pathways.
ACL modifications play major roles in decoding mRNA at the A-site of the ribosome. Most of the diversity in tRNA modifications occurs within the ACL region (Machnicka et al. 2014). Indeed, the ACL region contains 15 of the 25 distinct modifications in S. cerevisiae, 17 of the 28 in humans, and 21 of the 28 in E. coli, and on average ∼30% of the residues in this region are modified in eukaryotes (Han and Phizicky 2018).
Below, we provide highlights in the biology of modifications, starting with modifications in the ACL region.
MODIFICATIONS IN THE ANTICODON LOOP REGION
Of the modifications in the ACL region of tRNAs, the N34 and N37 modifications are by far the most commonly modified and have the most variety. Among ∼600 completely analyzed tRNAs, N34 modifications are found in 255 tRNAs and N37 modifications occur 426 times, and remarkably, the 29 chemically distinct modifications at N34 and 13 at N37 together comprise 70% of the chemically different modifications in this data set (Machnicka et al. 2014).
The major driving forces for N34 and N37 modifications are to stabilize codon–anticodon interactions that have multiple A–U pairs, and to properly discriminate pairing between the wobble nucleotide (N34) and the third nucleotide of the codon (B3) at the A-site of the ribosome (for review, see Grosjean and Westhof 2016). The N34–B3 interactions at the A-site require structural accommodation within the codon–anticodon helix (Demeshkina et al. 2012). N34 modifications help achieve this accommodation by the changes in chemical properties of N34, which can alter the population of tautomeric forms of the base, orientation of the base about the glycosidic bond, hydrogen bonding, or the sugar pucker, to allow a proper fit of N34–B3 within the decoding site of the ribosome (Murphy and Ramakrishnan 2004; Murphy et al. 2004; Weixlbaumer et al. 2007; Kurata et al. 2008; Vendeix et al. 2012; Grosjean and Westhof 2016). N37 modifications improve stacking. For example, t6A37 in E. coli tRNALys(UUU) promotes stacking interactions with A38 in the ACL, and a cross-strand stacking interaction with the B1 base of the codon in the decoding center of the ribosome A site (Murphy et al. 2004), and the ms2 moiety of ms2t6A37 found in human tRNALys(UUU) enhances the stacking interactions with the B1 base (Vendeix et al. 2012).
In the sections below, we elaborate on specific examples of the biology of ACL region modifications (Fig. 3; Supplemental Table S1), with emphasis on relatively recent discoveries.
The essential deamination of adenosine to form I34 by the Tad2:Tad3 (ADAT2:ADAT3) complex
In all eukaryotes, tRNAs with A34 are deaminated to form I34, allowing decoding of codons ending in U, C, or A. This reaction is catalyzed by the essential Tad2:Tad3 complex in S. cerevisiae, with Tad2 as the catalytic subunit, requiring formation of the complex with Tad3 for activity (Auxilien et al. 1996; Gerber and Keller 1999; Liu et al. 2020), and by the homologous ADAT2:ADAT3 complex in mouse (Ramos-Morales et al. 2021). Although lack of I34 is lethal due to lack of proper decoding by tRNAs with A34, the ADAT3–V144M mutation is linked to severe intellectual disability (Fig. 4) and strabismus in patients from multiple families (Alazami et al. 2013). Analysis of tRNAs from lymphoblastoid cell lines (LCLs) derived from patients showed reduced levels of I34 modification for each of several tRNAs, which was associated with an increased frequency of aggregates in the corresponding ADAT2:ADAT3–V144M complex, relative to the WT complex (Ramos et al. 2019).
Modification of U34 by elongator and other proteins to form mcm5U34, mcm5s2U34, ncm5U34, and ncm5Um34
Of the 29 chemically distinct N34 modifications in different organisms (Machnicka et al. 2014), nine are found in S. cerevisiae tRNAs, four of which contain the carboxymethyluridine moiety (xcm5U34), with x representing an attached methyl (m) or amino (n) group (Karlsborn et al. 2014b). These xcm5U modifications are found in S cerevisiae on 11 of the 13 tRNA species with U34, including five with ncm5U, one with ncm5Um, two with mcm5U, and three with mcm5s2U (Fig. 6). As elaborated below, these modifications are subject to complex biochemistry, and have major regulatory roles.
FIGURE 6.
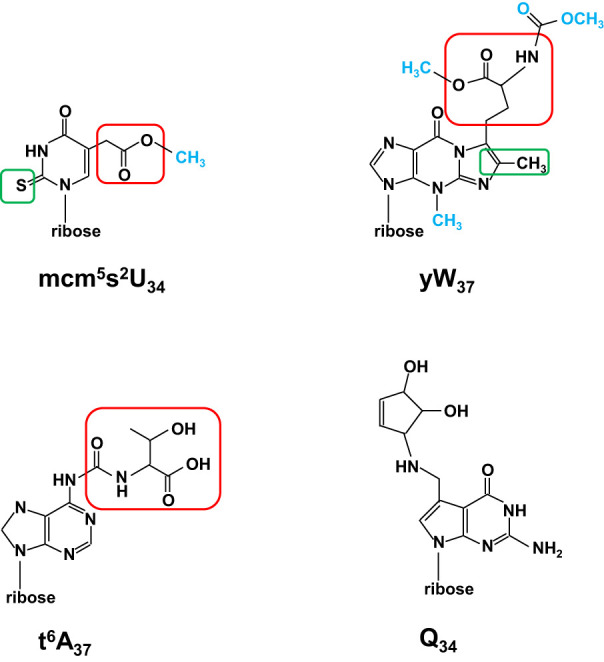
Schematic of complex modifications. All modifications are shown as nucleosides. (Top left) The mcm5s2U34 modification. The schematic is shown with the 2-thio moiety s2 boxed in green, the 5-carboxymethyl moiety cm5 boxed in red, and the terminal methyl group colored blue. In ncm5U, the terminal methyl group would be an amino group, and the sulfur in the 2-thio moiety would be an oxygen. (Top right) The yW37 modification. The schematic is shown with the methyl/methylene residues added to m1G to form the additional ring of imG14 boxed in green, the α-amino-α-carboxypropyl group added from S-adenosylmethionine boxed in red, and other added groups colored blue. (Bottom left) The t6A37 modification. The schematic is shown with the threonylcarbamoyl group boxed in red. (Bottom right) The Q34 modification.
The apparatus required for xcm5U34 modification in S. cerevisiae is enormous, as 15 genes are required for formation of mcm5U, 11 genes are required for formation of the s2U group of mcm5s2U (for review, see Karlsborn et al. 2014b), and two are required for formation of the Um moiety of ncm5Um (Pintard et al. 2002; Guy et al. 2012). Three findings led the way in identifying the components required for these modifications. First, Bystrom and coworkers cloned the S. pombe sin3+ gene (Huang et al. 2005) by screening for complementation of a sin3 mutant, previously shown to have reduced nonsense suppression and reduced mcm5s2U in tRNAs (Heyer et al. 1984), and sequencing revealed that it encoded the S. cerevisiae ortholog of Elp3, a subunit of the elongator complex, previously implicated in other functions. Consistent with a requirement for the elongator complex in xcm5U34 modification, mutations in each of the six elongator genes eliminated the modification and reduced nonsense suppression in S. cerevisiae (Huang et al. 2005). Second, the connection was made between resistance to γ-toxin from Kluyveronmyces lactis and lack of mcm5s2U34 in tRNAs. Thus, S. cerevisiae kti11, kti12, and kti13 mutants, like elp mutants, were each resistant to K. lactis γ-toxin produced by the zymocin complex (Frohloff et al. 2001; Jablonowski et al. 2001; Fichtner et al. 2002), and had reduced xcm5U in tRNAs and reduced nonsense suppression (Huang et al. 2005). As K. lactis γ-toxin was shown to encode an endonuclease that cleaved S. cerevisiae tRNAs with mcm5s2U34 at U34 (Lu et al. 2005), other genes important for mcm5s2U34 formation were identified by screening for γ-toxin resistance (Mehlgarten and Schaffrath 2003; Huang et al. 2008). Third, several laboratories uncovered the biochemical pathway by which the s2U tRNA modification was made in a sulfur relay from cysteine to tRNA, via Nfs1, Tum1, Uba4, and then Urm1, followed by thiolation of tRNA by Ncs2–Ncs6 using the Urm1-activated thiol (Nakai et al. 2008; Leidel et al. 2009; Noma et al. 2009).
The biochemical function/activity of some of these proteins has become clearer. Elp3 is known to be the catalytic component for carboxymethylation of U34, generating a 5′-deoxyadenosyl radical from S-adenosylmethionine that is used to generate an acetyl coA radical, to catalyze formation of cm5U34 (Selvadurai et al. 2014). The external methyl group of mcm5U and mcm5s2U is known to be attached by a complex of Trm9 (human ALKBH8) and Trm112 in yeast and humans (Kalhor and Clarke 2003; Fu et al. 2010; Mazauric et al. 2010; Songe-Moller et al. 2010). Surprisingly, however, yeast trm9Δ and trm112Δ mutants accumulate the corresponding ncm5U and ncm5s2U modifications, rather than the anticipated cm5U and cm5s2U modifications, suggesting either that ncm5U (ncm5s2U) is the precursor for mcm5U (mcm5s2U), rather than cm5U, or that ncm5U (ncm5s2U) is a default modification in the absence of the corresponding methyl modification (Mazauric et al. 2010; Chen et al. 2011a). An additional surprise was the discovery of mammalian tRNAs with a hydroxylated version of mcm5U, with tRNAGly(UCC) bearing (S)-mchm5U, catalyzed in vivo and in vitro by the AlkB domain of ALKBH8, and tRNAArg(UCG) bearing the (R)-diastereomer (van den Born et al. 2011).
Remarkably, the multiple phenotypic consequences of elongator mutants in S. cerevisiae (Fig. 4) are almost all due to reduced function of some combination of the three tRNAs with the mcm5s2U modification (tRNAGln(UUG), tRNALys(UUU), and tRNAGlu(UUC)) (for review, see Karlsborn et al. 2014b). Although the elongator complex was initially implicated in Pol II transcription elongation due to chromatin remodeling by histone acetylation (Otero et al. 1999; Wittschieben et al. 1999; Kim et al. 2002; Winkler et al. 2002), and in polarized transport of secretory vesicles to the bud tip (Rahl et al. 2005), these phenotypes are all due to reduced function of tRNAGln(UUG) and tRNALys(UUU), as they were completely rescued by overexpression of the two tRNA species (Esberg et al. 2006). Moreover, many of the phenotypes of elongator mutants were also found in an ncs2Δ mutant strain, which lacks the s2U moiety of mcm5s2U34 (Esberg et al. 2006). Similarly, defects in telomeric gene silencing and the DNA damage response that were ascribed to elongator mutants (Li et al. 2009) were rescued by overexpression of all three tRNAs with mcm5s2U, and tuc2Δ (ncs2Δ) mutants, lacking the s2U moiety, had the same phenotypes as the elongator mutants (Chen et al. 2011b).
The xcm5U and mcm5s2U modifications are found widely in eukaryotes (Karlsborn et al. 2014b), in which mutants invariably have significant defects. In S. pombe, lack of the conserved Ctu1–Ctu2 (Ncs6–Ncs2) complex resulted in loss of s2U from mcm5s2U in tRNAs, associated with temperature sensitivity and a septation defect leading to aberrant ploidy, and the temperature sensitivity was rescued by overproduction of tRNALys(UUU) and tRNAGlu(UUC) (Dewez et al. 2008). Similarly, S. pombe elp3 mutants, which have U34 instead of mcm5s2U34, are sensitive to H2O2 stress due to reduced function of tRNALys(UUU) (Fernandez-Vazquez et al. 2013). In metazoans and plants, mutations in the mcm5s2U modification components have distinct phenotypes, but the corresponding tRNA rescue experiments to directly link the effects to tRNA biology have not been reported. In C. elegans, mcm5s2U is implicated in neurological and developmental defects, based on analysis of five mutants with reduced levels of the mcm5U or s2U moieties (Dewez et al. 2008; Chen et al. 2009a; Kim et al. 2010b), accompanied by a temperature sensitive germ-line maturation defect for three tested mutants (Dewez et al. 2008; Chen et al. 2009a), and a temperature sensitive defect in a chemotaxis learning assay for two elongator mutants (Chen et al. 2009a). In humans, an intronic mutation in the ELP1 ortholog IKBKAP has been linked to the recessive neurodegenerative genetic disease familial dysautonomia (Fig. 4; Slaugenhaupt et al. 2001) and is associated with reduced mcm5s2U (Karlsborn et al. 2014a). In Drosophila and mouse, the corresponding elp3 mutants and null IKBKAP (ELP1) mutants are embryonic lethal, with vascular and neural development defects in the mouse (Chen et al. 2009b; Walker et al. 2011), and germ-line-specific conditional mutants cause male infertility, associated with defective chromosome synapsis and meiotic recombination, and reduced xcm5U modification (Lin et al. 2013).
There has been substantial progress in understanding the precise translation defect due to lack of mcm5U or s2U in S. cerevisiae. It was noted previously that the rescue of xcm5U mutant phenotypes by overexpression of unmodified tRNAs implied reduced tRNA function at or before the A-site decoding step, rather than a miscoding defect arising from the lack of modifications (Bjork et al. 2007). A subsequent seminal study measured decoding potential by examining growth of mutants lacking xcm5U or s2U groups, in combination with deletions of selected nonessential tRNA genes with C34. As C34-containing tRNAs can only decode G ending codons, whereas U34-containing tRNAs can in principle decode both A-ending and G-ending codons, this strategy allowed for decoding analysis in codon boxes that are decoded with both U34-containing tRNAs and nonessential C34-containing tRNAs (Johansson et al. 2008). For example, it was found that deletion of both copies of tRNAGly(CCC) resulted in little growth defect, but was nearly lethal in combination with an elp3Δ mutation, providing strong evidence that mcm5U34 in tRNAGly(UCC) was important for reading GGG proline codons. This and other similar experiments suggested that mcm5U and ncm5U improve decoding of G-ending codons, and that in tRNAs with mcm5s2U34, both mcm5U and s2U cooperate to improve decoding of G-ending codons (Johansson et al. 2008), although in this study only a few of the tRNAs with xcm5U or mcm5s2U could be examined.
Subsequent examination of mcm5s2U and xcm5U modifications by mass spectrometry and ribosome footprint profiling techniques substantially enhanced understanding of their translation roles. A mass spectrometry study in S. cerevisiae showed that lack of either xcm5U or s2U led to underrepresentation of proteins with a high abundance of AAA, CAA, and GAA codons, decoded by tRNALys(UUU), tRNAGln(UUG), and tRNAGlu(UUC), respectively, the three tRNAs with mcm5s2U (Rezgui et al. 2013). Ribosome profiling analysis extended these results. Ribosome profiling of elp3Δ mutants lacking mcm5U showed a minor but distinct increased occupancy of CAA and GAA codons, and several mutants lacking s2U had increased occupancy of CAA and AAA, codons, with little effect on the corresponding G-ending codons (Zinshteyn and Gilbert 2013). In addition, elp3Δ and mutants lacking s2U had low level induction of GCN4 translation that was independent of Gcn2 (the eIF2α kinase), suggesting constitutive activation of the general amino acid control (GAAC) signaling pathway (Natarajan et al. 2001; Hinnebusch 2005; Castilho et al. 2014; Wu et al. 2020) as a consequence of a lack of mcm5s2U (Zinshteyn and Gilbert 2013).
A major breakthrough in understanding of the translation defect due to lack of xcm5U and/or s2U modifications came from further ribosome profiling, combined with gene expression analysis using RNA seq, which revealed a prominent proteotoxic stress defect associated with lack of the modifications (Nedialkova and Leidel 2015). Thus, ribosomes in S. cerevisiae ncs2Δ and elp6Δ mutants (lacking s2U and mcm5U, respectively) had increased occupancy of CAA and AAA codons accompanied by the accumulation of protein aggregates, which was linked to poor clearance of stress induced protein aggregates, and both phenotypes were suppressed by overexpression of the three tRNA species with mcm5s2U. Remarkably, these functions of mcm5s2U are conserved, as ribosomes in C. elegans ncs2−/− mutants also had increased occupancy of CAA and GAA codons and a similar protein aggregation phenotype (Nedialkova and Leidel 2015).
Several studies have described how the xcm5U and s2U modifications contribute to decoding interactions in the context of human tRNALys(UUU) anticodon stem–loops with mcm5s2U34, ms2t6A37, and Ψ39. Physical and NMR analysis showed that mcm5U34 has virtually no effect on the structure of U34 or of the ACL, whereas the s2U modification promotes stacking of U34 and U35 and modestly increases the 3′ endo conformation of U34 (Durant et al. 2005). Further studies of the human tRNALys(UUU) ASL at the ribosome A-site showed that mcm5s2U34 has normal Watson–Crick pairing with the corresponding B3 nucleotide, with mcm5s2U34 shifting to the enol form (Vendeix et al. 2012). Analysis of translation showed that lack of s2U in yeast tRNALys(UUU) with mcm5U results in reduced tRNA binding at the A-site due to increased off-rate, a reduced rate of conformational changes necessary for the translation cycle, and a greater rate of rejection before peptide formation (Ranjan and Rodnina 2017).
The xcm5U modifications are subject to multiple levels of regulation. Intriguingly, it appears that proper elongator modification function requires intermediate levels of Elp1 phosphorylation. Thus, zymocin resistance (reduced mcm5s2U modification) in S. cerevisiae is associated with mutations in the protein kinase Kti14 (Hrr25) that result in reduced Elp1 phosphorylation, as well as with a sit4Δ mutation, which is associated with increased Elp1 phosphorylation; and related experiments show that a sit4Δ mutation in an hrr25-3 mutant restores the normal moderate Elp1 phosphorylation levels of WT cells and normal zymocin sensitivity (Mehlgarten et al. 2009). In support of a direct effect of phosphorylation, subsequent experiments provided evidence that Hrr25 directly phosphorylated Elp1, and showed that the Elp1 phosphorylation state was important for interactions with Kti12 (Abdel-Fattah et al. 2015). Kti12 is structurally similar to O-phosphoseryl tRNA kinase (PSTK), a protein involved in biosynthesis of tRNASec, and like PSTK, binds efficiently to bulk tRNAs, has a tRNA-dependent ATPase activity that is essential for xcm5U modification, and binds directly to Elp1 and stimulates the ATPase (Krutyholowa et al. 2019). However, it is not known how this ATPase activity is linked to elongator modification function.
Furthermore, it is now known that elongator modifications reciprocally regulate TOR signaling in S. pombe. Thus, elongator mutants up-regulate TORC1 by down-regulating inhibitors such as Tsc2, and down-regulate TORC2 by inhibiting expression of positive effectors such as Ste20, resulting in increased TORC1 signaling, unbalanced TORC signaling, and sensitivity to the TORC1 regulator rapamycin (Candiracci et al. 2019). Conversely, TOR also regulates elongator function, as overexpression of the TORC1 kinase Tor2 leads to reduced levels of xcm5U and xcm5Um modifications, whereas rapamycin treatment of WT cells (inhibiting TORC1), or overexpression of the TORC2 kinase Tor1, increased the amount of xcm5U and xcm5Um. Activation of elongator by the TORC2 pathway in this set of experiments appeared to occur by down-regulation of glycogen synthase kinase (Gsk3), which phosphorylates and inhibits Elp4 function in the elongator complex (Candiracci et al. 2019).
Levels of s2U34 are also subject to environmental regulation. In WT S. cerevisiae strains, the s2U modification is significantly reduced at temperatures higher than 30°C, due to lack of formation of the modification at higher temperatures (Alings et al. 2015; Damon et al. 2015; Han et al. 2015). Study of the effects of growth conditions showed that s2U modification is significantly reduced in synthetic medium containing glucose (fermenting sugar) or lactate (nonfermenting), due to reduced intracellular methionine and cysteine, and that sulfur starvation leads to reduced expression of Uba4 in the s2U pathway and reduced s2U in tRNA, resulting in increased expression of genes involved in biosynthesis of methionine and cysteine (Laxman et al. 2013). Subsequent metabolic analysis showed that tRNA thiolation mutants reroute carbon flux as if the cells are starved, down-regulating the pentose phosphate pathway and nucleotide biosynthesis pathway and up-regulating pathways leading to storage carbohydrates trehalose and glycogen, and that this is due to down-regulation of phosphate homeostasis and reduced intracellular phosphate (Gupta et al. 2019; for review, see Gupta and Laxman 2020).
Trm7/FTSJ1, partners Trm732/THADA and Trm734/WDR6, and Nm32 and Nm34 modification
The importance of ribose 2′-O-methylation (Nm) in the ACL of eukaryotes has been evident since the identification of S. cerevisiae Trm7 as the 2′-O-methyltransferase responsible for Nm32 and Nm34 formation of its three substrate tRNAs (tRNAPhe, tRNATrp, and tRNALeu(UAA)), and the finding that mutants had a severe growth defect (Fig. 4) linked to poor translation (Pintard et al. 2002).
It is now known that S. cerevisiae Trm7 requires interaction partners Trm732 and Trm734 for formation of Nm32 and Nm34, respectively, on each Trm7 substrate tRNA, and that the critical tRNA substrate is tRNAPhe, as overproduction of tRNAPhe almost completely suppresses the S. cerevisiae trm7Δ growth defect (Guy et al. 2012). Moreover, the Cm32 and Gm34 modifications of tRNAPhe(GAA) each help direct formation of wybutosine (yW) from 1-methylguanosine at G37 (m1G37) (discussed further below), as tRNAPhe from trm7Δ mutants has m1G37 instead of yW37, and tRNAPhe from trm732Δ and trm734Δ mutants each has only partial yW37 modification. Thus, in a trm7Δ mutant the entire ACL of tRNAPhe is undermodified at C32, G32, and G37. In addition, the severe growth defect of trm7Δ mutants is known to be due to loss of both the Cm32 and Gm34 modifications, as trm732Δ trm734Δ mutants phenocopy the severe growth defect of trm7Δ mutants and completely lack yW37, whereas trm732Δ and trm734Δ mutants each grow normally, as do trm732Δ tyw1Δ and trm734Δ tyw1Δ double mutants (Guy et al. 2012), which also have m1G37 instead of yW37 due to lack of Tyw1 (Waas et al. 2005; Noma et al. 2006).
This modification circuitry for Nm32 and Nm34 formation in tRNA substrates and yW formation in tRNAPhe is conserved widely through eukaryotes. Thus, tRNAPhe from S. pombe trm7Δ mutants and from patients with null mutations in the human TRM7 ortholog FTSJ1 each lack detectable Cm32 and Gm34, and have m1G37 instead of yW37 or the human yW derivative peroxywybutosine (Guy and Phizicky 2015; Guy et al. 2015). Furthermore, S. pombe trm732Δ and trm734Δ mutants each lack the corresponding Cm32 and Gm34 modifications, and expression of S. pombe Trm732 or its human ortholog THADA complements the growth defect of an S. cerevisiae trm732Δ trm734Δ mutant and restores Cm32 formation in tRNAPhe (Guy and Phizicky 2015; Guy et al. 2015). As Trm7, Trm732, and Trm734 orthologs are found in diverse eukaryotes, as is yW37 or its derivatives, it seems likely that the Trm7 modification circuitry is widely conserved in eukaryotes (Guy and Phizicky 2015).
Intriguingly, there are two Trm7 paralogs in D. melanogaster and related genus members, one of which (CG5220, dTrm7_32) is required for Nm32 modification, and the other (CG7009, dTrm7_34) for Nm34 modification (Angelova et al. 2020). Nonetheless, it seems likely that the Trm732 ortholog (CG15618, DmTHADA) and Trm734 ortholog WDR6 (CG33172) are also required in Drosophila for formation of Nm32 and Nm34, respectively, as knockdowns of these genes each have a similar phenotype as knockdowns of Trm7 orthologs, and the dTrm7_34 protein physically interacts with the Drosophila Trm734/WDR6 (Angelova et al. 2020).
TRM7/FTSJ1 is important in all eukaryotes examined, although the biological manifestations of mutations differ in different eukaryotes. In both S. cerevisiae and S. pombe, trm7Δ mutants have a severe growth defect due to reduced tRNAPhe function (Pintard et al. 2002; Guy et al. 2012; Guy and Phizicky 2015). Moreover, both S. cerevisiae and S. pombe trm7Δ mutants constitutively and robustly activate the GAAC pathway in the absence of an apparent charging defect, presumably due to increased ribosome collisions (Chou et al. 2017; Han et al. 2018). Remarkably, the constitutive GAAC activation is itself part of the reason for the severe growth defect of S. cerevisiae trm7Δ mutants, as their severe growth defect is partially alleviated by mutation of the GAAC pathway (Han et al. 2018).
In multicellular organisms, lack of TRM7 is manifested by distinct phenotypes. In Drosophila, homozygous null mutants of either dTrm7_34 or dTrm7_32 do not have a noticeable growth defect but each mutant (or dsRNA knockdown), as well as the corresponding DmTHADA or WDR6 knockdowns, inhibit Ago2-dependent silencing by the siRNA pathway and piRNA-mediated silencing (Angelova et al. 2020). Furthermore, homozygous double mutant flies lacking both dTrm7_32 and dTrm7_34 have modestly reduced size and weight, reduced life spans, and some locomotion defects (Angelova et al. 2020). In A. thaliana, a trm7 mutant (scs9) has reduced resistance to a bacterial infection and a mild growth defect (Ramirez et al. 2018). In humans, FTSJ1 mutations lead to nonsyndromic X-linked intellectual disability (Fig. 4, NSXLID; Freude et al. 2004; Ramser et al. 2004; Froyen et al. 2007; Takano et al. 2008; Guy et al. 2015), suggesting little other obvious abnormality, whereas mouse Ftsj1−/− males have impaired learning as well as several phenotypes related to metabolism (Jensen et al. 2019).
As speculated by Carré and colleagues (Angelova et al. 2020), it is intriguing to note the possible connection between Trm7 biology and transposable elements in Drosophila and S. cerevisiae. Lack of either Drosophila TRM7 ortholog, or their partner proteins DmTHADA or WDR6 results in derepression of expression of the retrotransponon gypsy in ovarian follicle cells, while lack of TRM7 or TRM734 in S. cerevisiae results in expression of the Ty1 transposable element (Nyswaner et al. 2008). These phenomena might reflect a common theme.
It is not known in multicellular animals and plants if a single tRNA is responsible for the different phenotypes of TRM7/FTSJ1 mutants, as in fungi for tRNAPhe(GAA). In this connection, it is intriguing to note that 2,6-diaminopurine binds to and inhibits FTSJ1 in human Calu-6 cancer cells, resulting in reduced Cm34 modification of tRNATrp(CCA) and increased readthrough of UGA stop codons (Trzaska et al. 2020). It is also not known if other phenotypes attributed to lack of TRM732/THADA or TRM734/WDR6 are related to tRNA biology. S. cerevisiae Trm734 was previously identified as Ere2, a protein that interacts with Ere1 in the retromer-mediated pathway to recycle cell membrane proteins back to the cell surface after internalization (Shi et al. 2011). In addition, Drosophila THADA mutants are cold sensitive and obese, with elevated triglycerides, and THADA was shown to interact with the sarco/ER Ca2+ ATPase as an uncoupler (Moraru et al. 2017). As both TRM732/THADA and TRM734/WDR6 are large proteins with relatively small highly conserved domains (Guy and Phizicky 2015; Hirata et al. 2019; Funk et al. 2022), it is possible that these functions of Trm732 and Trm734 are unrelated to tRNA biology.
The crucial and universal m1G37 modification and its tRNAPhe derivative wybutosine, yW37
The highly conserved m1G37 modification is crucially important for tRNA function in organisms in all domains of life. An early biochemical study showed that m1G37 on tRNAAsp protects the tRNA from mischarging by yeast arginyl-tRNA synthetase (Putz et al. 1994). Subsequent seminal studies showed that lack of m1G37 due to null or near null mutations in S. cerevisiae TRM5 or S. typhimurium trmD, respectively, leads to a severe growth defect, consistent with the widespread occurrence of m1G37 in tRNAs in organisms (Bjork et al. 2001) and showed a prominent role of m1G37 in preventing +1 frameshifting (Bjork et al. 1989; Urbonavicius et al. 2001, 2003).
Recent results have substantially increased our knowledge of the role of m1G37. Biochemical analysis with ASL's based on tRNAPro(CGG) show that m1G37 improves the binding constant for RNA binding to a cognate CCG codon by threefold (relative to the ASL with G37) and weakens the binding constant by ninefold to a +1 CCC-U frameshifting codon (Nguyen et al. 2019). While it is not known how lack of m1G37 affects eukaryotic translation, ribosome profiling in E. coli shows that lack of m1G37 leads to ribosome stalling at the A-site of a subset of codons (Pro CCN, Arg CGG, and Leu CUA codons), showing a direct role for m1G37 in decoding that is distinct from frameshifting, attributed to reduced charging of tRNAPro and tRNAArg(CCG), and reduced peptide bond formation for some of the tRNAs (Masuda et al. 2021).
Wybutosine, yW37 (Fig. 6), and its various derivatives are found ubiquitously on tRNAPhe in eukaryotes and archaea and is formed from m1G37 (Droogmans and Grosjean 1987). Although the biogenesis pathway of wybutosine and derivatives varies in different organisms (discussed in Sample et al. 2015), these pathways always involve formation of wyosine (imG-14) by methylation of m1G37 and ring closure on the WC face of G37 (Fig. 6), catalyzed by Tyw1/Taw1, followed by various further maturation steps, which in S. cerevisiae involves addition of the main body of methionine from S-adenosylmethionine to the Hoogsteen side of the third ring by Tyw2, methylation at N3 of the guanosine moiety by Tyw3/Taw3, and esterification and amidation of the carboxyl and amino groups of the methionine moiety by Tyw4 (Kalhor et al. 2005; Waas et al. 2005; Noma et al. 2006). tRNAPhe with m1G37 instead of yW37 stimulated frameshifting in vitro (Carlson et al. 2001), and each of the successive maturation steps in wybutosine in S. cerevisiae further reduced −1 frameshifting of a test sequence in a reporter in vivo (Waas et al. 2007).
Recent developments have emphasized the crucial importance of m1G37 and yW37 in mitochondrial function. Prior work in S. cerevisiae showed that m1G37 is found on at least eight mitochondrially encoded tRNA species, including tRNAfMet (Canaday et al. 1980), that Trm5 is localized to mitochondria in addition to the nuclear/cytoplasmic compartment, and that specific loss of mitochondrial Trm5 significantly reduces oxygen consumption, albeit with little evident growth phenotype on media containing the nonfermentable carbon source glycerol (Lee et al. 2007). Curiously, although T. brucei TRM5 is also localized to both the mitochondrial and the nuclear/cytoplasmic compartments, and down-regulation of TRM5 leads to reduced m1G37 in both mitochondrial and cytoplasmic tRNAs, translation is reduced in mitochondria but not in the cytoplasm (Paris et al. 2013). The importance of m1G37 in T. brucei mitochondrial tRNAs also extends to the imG-14 derivatives unexpectedly found in T. brucei mitochondrial tRNAPhe (Sample et al. 2015). T brucei has a nuclear Tyw1 paralog that is responsible for imG-14 formation in cytoplasmic tRNAPhe, some of which is imported into the mitochondria, and a mitochondrial Tyw1 paralog that is responsible for imG-14 formation of mitochondrial tRNAPhe, which is imported before modification. Down-regulation of either paralog resulted in little growth defect in normal growth media but resulted in reduced growth in low-glucose media, in which cells need full mitochondrial function. This result suggests an important unexpected role for the imG-14 or derivative modification on mitochondrial tRNAPhe. Although other explanations are possible, one attractive explanation advanced by the authors was that the extensive U-rich mRNAs that arise from pan-editing in kinetoplasmid mitochondria might impose strict requirements that all tRNAs are modified as completely as possible to prevent frameshifting (Sample et al. 2015).
A similar apparent mitochondrial bias for TRM5 function has also been found in humans. Thus, each of two unrelated patients with compound heterozygous TRMT5 mutations had lactic acidosis and multiple deficiencies in mitochondrial function in skeletal muscle, accompanied by reduced m1G37 in mitochondrial tRNA. Moreover, analysis in yeast showed that the mutations were functionally hypomorphic as expression of the corresponding trm5 variants as the only source of mitochondrial TRM5 led to partially reduced oxygen consumption and respiratory function (Powell et al. 2015).
The biology of the universally important N6-threonylcarbamoyladenosine modification, t6A37
Much has been learned about the occurrence, biosynthesis, and function of t6A since its original discovery in tRNA (Schweizer et al. 1969). The t6A modification, or derivatives of it, is found in all organisms examined in all domains of life, and is invariably found at residue A37, immediately 3′ of U36 of tRNAs with NNU anticodons. Indeed, only a few tRNAs with the NNU anticodon do not have the t6A37 modification including, most prominently, initiator tRNA in prokaryotes, organelles, and archaea, which often harbor unmodified A37 (summarized in Morin et al. 1998). Initial studies showed that formation of t6A required ATP to incorporate a one-carbon group and threonine at N6 of A37 (Elkins and Keller 1974; Korner and Soll 1974), and physical studies showed that t6A37 of the tRNALys(UUU) ASL unexpectedly decreased stacking in the ACL, bulging out the adjacent U36 residue, and forming a cross-strand stack with N1 of the codon (Murphy et al. 2004; Durant et al. 2005).
Discovery of the genes involved in t6A biosynthesis facilitated study of its biology. Bioinformatic analysis, coupled with biochemical analysis, showed that the highly conserved Sua5 (S. cerevisiae)/YrdC (E. coli) family of proteins is directly involved in t6A biosynthesis (El Yacoubi et al. 2009), and genetic analysis established that lack of the gene results in a severe slow growth phenotype in S. cerevisiae (Na et al. 1992), and lethality in E. coli (El Yacoubi et al. 2009). Subsequent experiments in S. cerevisiae showed that Kae1, Bud32, and Pcc1 of the highly conserved KEOPS complex are involved in t6A biosynthesis, and that mutants have very similar slow growth phenotypes; in contrast, the KEOPS complex member Cgi121 is not involved in t6A biosynthesis and mutants grow nearly normally (El Yacoubi et al. 2011; Srinivasan et al. 2011).
Biochemical experiments with purified B. subtilis proteins established a synthetic route (for review, see Thiaville et al. 2014) involving direct carboxylation of threonine with bicarbonate (or CO2) and transfer to ATP, displacing PPi to form the intermediate threonylcarbamoyl AMP (TC-AMP), which was then transferred to N6 of A37 to form t6A (Fig. 6; Lauhon 2012). Subsequent experiments with purified yeast components likewise showed that Sua5 catalyzes TC-AMP formation, and that Bud32, Kae1, and Pcc1 of the KEOPS complex catalyzes its transfer to N6A37 of substrate tRNAs, requiring the Bud32 (TsaE in bacteria) ATPase (Perrochia et al. 2013a,b). Remarkably, the Kae1 paralog Qri7 of S. cerevisiae mitochondria can replace the entire KEOPS complex in vivo and in vitro (Wan et al. 2013). Additional structural and functional analysis showed that the TC-transfer step is catalyzed by the TsaD (Kae1) subunit in complex with TsaB, and elaborated the role of TsaE (Bud32) in regulation of the TsaD catalytic activity, allowing multiple turnover reactions through its ATPase activity (Luthra et al. 2018, 2019; Missoury et al. 2018).
Unexpectedly, t6A37 is not the final modification product in E. coli and S. cerevisiae, and based on phylogenetic analysis, likely also in most proteobacteria and bacteroidetes, most fungi, and several protists and plants. In these organisms, the normal t6A modification is converted to the cyclic derivative ct6A by TcdA dehydratase and related family members, in which one of the hydroxyls from the terminal carboxyl group of the threonine moiety is lost during a condensation reaction that results in an oxazolone ring instead of the more customary linear threonine adduct (Miyauchi et al. 2013). Although the corresponding E. coli tcdA mutant grows at a normal rate, it does not compete with WT, and the corresponding S. cerevisiae tcd1Δ and tcd2Δ mutants each lack ct6A and grow poorly on glycerol-containing media, which require respiration. Consistent with a mild translation defect, tRNALys(UUU) lacking ct6A in E. coli tcdA mutants have reduced ability to decode near cognate AGA and noncognate UAG codons (Miyauchi et al. 2013). Intriguingly, echinoderm mitochondria decode AAA as asparagine instead of lysine, and their tRNALys(CUU) has hydroxy-t6A (ht6A) instead of t6A and binds more poorly to ribosomes with AAA codons, in principle allowing for AAA decoding by tRNAAsn(GUU) (Nagao et al. 2017).
Consistent with early analysis, t6A is generally essential in bacteria and archaea, whereas mutants in eukaryotes are slow growing but viable (Fig. 4; El Yacoubi et al. 2009, 2011; Srinivasan et al. 2011; Naor et al. 2012); however, the source of this discrepancy is unknown (for review, see Thiaville et al. 2015).
In S. cerevisiae, there are a number of consequences of lack of t6A modification. Temperature sensitive mutants in the KEOPS complex components trigger translation of GCN4, the transcriptional activator of the GAAC pathway, attributed to defective recognition of the AUG codons of the normally inhibitory upstream open reading frames of GCN4 by tRNAMet(CAU)i lacking t6A (Daugeron et al. 2011). Additional experiments showed that null mutants in the t6A pathway accumulate aggregates indicative of proteotoxic stress (Thiaville et al. 2016), particularly in the absence of both mcm5U and t6A (Pollo-Oliveira et al. 2020), similar to the accumulation of aggregates previously observed in mutants lacking mcm5U and/or s2U (Nedialkova and Leidel 2015). In addition, null mutants in the t6A pathway were sensitive to stresses such as temperature, ethanol, and rapamycin, and had ribosome occupancy profiles suggesting that t6A acts to homogenize translation rates across codons and to prevent increased translation initiation at non-AUG codons (Thiaville et al. 2016).
In metazoans, reduced levels of t6A also have dramatic effects. In Drosophila, hemizygous kae1 larvae have reduced t6A that is correlated with a characteristic mass called a Black spot phenotype, an extended larval period, defective imaginal discs, and reduced proliferation of mitotic vs. nonproliferating tissues (Lin et al. 2015). In humans, pedigree analysis linked a homozygous kae1 missense mutation to a global developmental delay and renal defects, and the corresponding yeast mutant had reduced t6A (Edvardson et al. 2017). Furthermore, mutations in each of the gene products of the KEOPS complex are associated with Galloway–Mowat syndrome (GAMOS) (Fig. 4; Braun et al. 2017). A patient with early onset nephrotic syndrome associated with microcephaly and developmental delays had a mutation in OSGEP (KAE1, TsaD), the catalytic subunit of the TC-AMP transfer step, and whole exome sequencing of a panel of 907 patients with nephrotic syndrome, including 91 with GAMOS, revealed 32 familes with mutations in this or other subunits of the KEOPS complex (LAGE3, TP53RK, and TPRKB; orthologs of yeast Pcc1, Bud32, and Cgi121) (Braun et al. 2017). Moreover, corresponding knockouts (OSGEP and TPRKB) in zebrafish recapitulated the microcephaly with marked apoptosis in the brain; mouse knockouts (OSGEP, TPRKB, and LAGE3) reproduced the microcephaly; and human podocyte cell lines expressing shRNAs directed against two of these genes had reduced t6A, accompanied by increased apoptosis and decreased cell survival (Braun et al. 2017). More recently, examination of 14 GAMOS-affected patients revealed mutations in the remaining two genes associated with t6A biosynthesis, human GON7 (also called C14orf142) and YRDC (ortholog of yeast SUA5). Consistent with their yeast growth phenotypes, the patients with GON7 mutations had a milder disease presentation than those with YRDC mutations (Arrondel et al. 2019).
The intriguing biology of N6-isopentenyl-adenosine, i6A37, and Mod5/TRIT1
The N6-isopentenyl-adenosine (i6A) modification was first linked to tRNA function by the isolation of an S. cerevisiae mod5 mutant that reduced nonsense suppression by the tyrosine-inserting nonsense suppressor SUP7 and had tRNA with reduced amounts of i6A37 (Laten et al. 1978). The i6A37 modification occurs widely in tRNA from bacteria and eukaryotes, and its formation is catalyzed by the isopentenyl transferase (dimethylallyl transferase), called Mod5 in S. cerevisiae, miaA in E. coli, Tit1 in S. pombe, GRO-1 in C. elegans, and TRIT1 in humans (Dihanich et al. 1987; Lemieux et al. 2001; Soderberg and Poulter 2001; Spinola et al. 2005). Mod5 catalyzes isopentenylation at the N6 position of A37 of tRNAs with A36–A37–A38 in their ACLs (Motorin et al. 1997; Soderberg and Poulter 2000), with additional recognition of N34 for some orthologs (Lamichhane et al. 2011). In S. cerevisiae, the single MOD5 gene is responsible for the modification of both cytoplasmic and mitochondrial tRNA substrates (Dihanich et al. 1987) due to separate translation starts (Gillman et al. 1991; Slusher et al. 1991), and Mod5 is found in the nucleus, cytoplasm, and mitochondria (Boguta et al. 1994).
Mutants lacking i6A37 have several additional phenotypes. S. pombe tit1Δ mutants, like S. cerevisiae SUP7 mod5 mutants, have reduced decoding, as shown for each of two tested tRNAs on a reporter, suggesting a common role for i6A37 in decoding efficiency (Lamichhane et al. 2013). In addition, tit1Δ mutants have a growth defect in media containing the TOR inhibitor rapamycin that is almost completely rescued by overexpression of cytoplasmic tRNATyr(GUA) and tRNATrp(CCA), and a growth defect in media containing the nonfermentable carbon source glycerol that is unexpectedly partially rescued by these same two tRNAs, suggesting that the growth defect is not entirely due to a mitochondrial defect (Lamichhane et al. 2013, 2016). In contrast, a human TRIT1-R323Q mutation, in a highly conserved residue near the active site (Zhou and Huang 2008), was linked to encephalopathy and myoclinc epilepsy, and to multiple defects in oxidative phosphorylation that were attributed to reduced i6A in mitochondrial tRNASer (Yarham et al. 2014). Likewise, a pleiotropic C. elegans gro-1 mutant had developmental, behavioral, and reproductive defects, and increased life span, and these were all rescued by expression of a mitochondrial GRO-1 but not cytoplasmic GRO-1 (Lemieux et al. 2001).
Intriguingly, Mod5 is also required in S. cerevisiae for tRNA-mediated gene silencing of neighboring genes due to the nucleolar localization of tRNA genes being transcribed, and expression of human TRIT1 in S. cerevisiae confers this same property (Thompson et al. 2003; Wang et al. 2005; Pratt-Hyatt et al. 2013). As this silencing function is not due to the catalytic activity of Mod5, it is presumably a conserved moonlighting function of Mod5/TRIT1, which may impact human biology (Pratt-Hyatt et al. 2013).
The most intriguing aspect of i6A biology is its prion-related functions in S. cerevisiae, linked to Mod5 aggregation using a novel aggregation domain (Suzuki et al. 2012). Like other genetically characterized prions such as [PSI+], [MOD5+] cells have reduced function, which is mitotically stable over multiple generations, but can be reversed by chemical or genetic treatment (in this case by inhibition of the Hsp104 chaperone); is genetically dominant; and can be transmitted to a cell that is not [MOD5+] by introduction of the [MOD5+] protein. Moreover, consistent with previous results with S. cerevisiae mod5 hypomorphic mutants (Benko et al. 2000), [MOD5+] cells had reduced i6A in their tRNA and increased ergosterol due to increased activity of Erg20, which competes with Mod5 for the same dimethylallyl pyrophosphate substrate. Of special note, [MOD5+] yeast were resistant to several antifungal agents, likely due to increased ergosterol, and treatment with antifungal agents in wild-type cells triggered the generation of [MOD5+] cells, which was reversible upon removal of the drugs (Suzuki et al. 2012).
Although there is currently no detailed structural information of the effect of i6A on individual translation steps, the ASL structures show that i6A37 disrupts C32–A+38 base pairing and U33–A37 base–base interactions and increases dynamics in the loop (Denmon et al. 2011).
The biology of Dnmt2 and m5C38 and queueosine at N34 (Q34)
The m5C38 modification, catalyzed by Dnmt2 (Goll et al. 2006), and the Q34 modification (Fig. 6) are considered together here because of the partial overlap of their biology in eukaryotes (for review, see Ehrenhofer-Murray 2017). Dnmt2 enzymes and m5C38 are widely found in eukaryotes, but not in S. cerevisiae. Q34 is widely found in bacteria and eukaryotes (but not in S. cerevisiae), where it is found in tRNAs with GUN anticodons (tRNATyr(GUA), tRNAHis(GUG), tRNAAsn(GUU), and tRNAAsp(GUC)). Whereas bacteria form Q34 in a complicated pathway involving biosynthesis of pre-Q1, exchange of pre-Q1 for G34 by a tRNA-guanine transglycosylase (TGT), and further processing, eukaryotes form Q34 by direct transfer of free queuine found in cells (derived from bacteria) to the tRNA by a eukaryotic TGT, displacing guanosine at G34 (for review, see El Yacoubi et al. 2012).
There has been intensive study of the role of Dnmt2 in tRNA since the discovery that Dnmt2 from mouse, Arabidopsis, Drosophila, and humans was not a DNA methyltransferase, as anticipated based on phylogenetic analysis (Goll and Bestor 2005), but was instead a tRNA methyltransferase that catalyzed formation of m5C38 on tRNAAsp (Goll et al. 2006; Jurkowski et al. 2008; for review, see Jeltsch et al. 2017). Mapping of m5C sites in WT and mutant strains (Schaefer et al. 2009) showed that Drosophila and mouse Dnmt1 modifies tRNAAsp, tRNAVal(AAC), and tRNAGly(GCC) (Schaefer et al. 2010; Tuorto et al. 2012), whereas the S. pombe Dnmt2 ortholog Pmt1 primarily targets tRNAAsp, with partial modification of tRNAGlu (Becker et al. 2012). Moreover, m5C38 modification of tRNAAsp in S. pombe, Dictyostelium, and mouse is strongly dependent on prior Q34 modification in vivo, and in vitro for S. pombe Pmt1, thus linking cellular queuine and Q34 modification to m5C38 modification of tRNAAsp (Muller et al. 2015; Tuorto et al. 2018).
Although S. pombe pmt1Δ (dnmt2) mutants have no obvious growth or stress phenotype (Becker et al. 2012), several important Dnmt2 roles have emerged from study in other organisms. Thus, Drosophila Dnmt2−/− mutants are sensitive to growth at high temperature and to oxidative stress, which is correlated with stress-induced tRNA cleavage due to lack of m5C38 (Schaefer et al. 2010), which in turn leads to significant changes in dsRNAs, siRNAs and viral sensitivity, as discussed further below (Durdevic et al. 2013a). In mouse, Dnmt2−/− mutants have delayed endochondral ossification in newborns and reduced populations of hematopoietic stem cells and progenitor cells, accompanied by increased mistranslation of Asp codons by tRNAGlu and of Glu codons by tRNAAsp (Tuorto et al. 2015). In addition, the mouse Dnmt2 gene is required for epigenetic regulation of the Kit gene, responsible for altered fur color, and of Sox9, resulting in excess growth of the embryo and adult body (Kiani et al. 2013).
All of these phenotypes are highly likely to be due to tRNA m5C38 modification, as no other RNA substrates have been validated, and reported DNA substrates have not been supported by more rigorous analysis (Schaefer and Lyko 2010; Raddatz et al. 2013). However, it is possible that some of the phenotypes of mutants can be explained by other properties of the Dnmt2 protein, such as chaperone effects or binding effects (Jeltsch et al. 2017).
Intriguingly, Q34 has slightly different effects on translation in mouse and S. pombe. In mammalian cells, lack of Q34 in tRNA resulted in increased ribosome occupancy and reduced translation of codons for all four tRNAs that normally have the queuosine modification, with the U-ending codons reduced more than the C-ending codons (except for tRNAAsp(GUC), which also lack m5C38). The reduced translation through these codons was reflected in reduced amounts of proteins richer in these codons, an increase in unfolded proteins, and activation of the UPR (Tuorto et al. 2018). However, in S. pombe strains lacking queuosine, ribosome profiling showed a reduced decoding rate for C-ending codons for tRNAAsp and tRNAHis, and faster decoding of U-ending codons for tRNAAsn and tRNATyr, the effect of which was to even translation rates of the synonymous codons with Q34 (Muller et al. 2019a).
Recent results also show that Q34 levels are regulated by oxidative stress in HepG2 cells, altering translation and gene expression (Huber et al. 2022). Thus, each of three different oxidative stress treatments, including arsenite treatment, resulted in increased Q34 modification of tRNAs in HepG2 cells, resulting in codon bias-linked up-regulation of proteins involved in glycolysis, and down-regulation of oxidative phosphorylation. In contrast, queuine limitation resulted in increased arsenite sensitivity and increased levels of reactive oxygen species, linked to mitochondrial dysfunction.
m3C32 modification by Trm140 family members
The m3C modification is frequently found in tRNAs at three locations in cytoplasmic tRNAs: at C32 in the ACL of eukaryotic tRNAThr and tRNASer isoacceptors, and mammalian tRNAArg(CCU) and tRNAArg(UCU) isoacceptors; at either e1 or e4 within the variable arm of metazoan tRNASer isoacceptors; and at N20 of human tRNAMet(CAU)e (Clark et al. 2016; Boccaletto et al. 2018). In addition, m3C32 is found in Bos taurus mitochondrial tRNASer(UGA) (Boccaletto et al. 2018), and has been found in mRNA in mammalian cells (Xu et al. 2017). These m3C modifications are catalyzed by Trm140 family members. In S. cerevisiae, Trm140 protein has m3C32 methyltransferase activity and is required in vivo for m3C32 modification of all three tRNAThr isoacceptors and all three tRNASer isoacceptors with C32 (D'Silva et al. 2011; Noma et al. 2011).
Trm140 family members have very different mechanisms for substrate recognition and m3C modification in different fungi and metazoans. For example, in S. pombe, Trm140 is required for m3C32 modification of tRNAThr substrates and Trm141 for m3C32 modification of tRNASer substrates (Arimbasseri et al. 2016). Furthermore, m3C modification of S. pombe tRNASer substrates requires prior i6A37 formation by Tit1/Mod5 (Arimbasseri et al. 2016), suggesting a similar modification circuitry to that observed for other ACL modifications (Guy et al. 2012; Guy and Phizicky 2015; Muller et al. 2015; for review, see Han and Phizicky 2018). Moreover, phylogenetic analysis showed that the TRM140 (METTL2) and TRM141 (METTL6) family members are widely distributed in fission yeasts and metazoans, and extend to a third family member (METTL8) in vertebrates (Arimbasseri et al. 2016).
In contrast, S. cerevisiae Trm140 has two seemingly distinct recognition modes, enabling modification of both tRNAThr and tRNASer substrates (Han et al. 2017). For tRNAThr isoacceptors, the ACL residues G35–U36–t6A37 are both necessary and sufficient for Trm140 recognition and m3C32 modification; whereas for tRNASer, m3C modification is stimulated in vivo and in vitro by seryl-tRNA synthetase and the distinctive tRNASer variable arm that SerRS recognizes, as well as by t6A37 or i6A37. As i6A and t6A are not chemically related, it is not clear why both modifications stimulate m3C formation, although it seems plausible that they each expose C32 for modification. The presence of a single Trm140 family member is conserved through the Saccharomycotina and Pezizomycotina subdivisions of the phylum Ascomycota, and to a more limited extent, in Basidiomycota, implying that this dual tRNAThr and tRNASer recognition mechanism is retained in these organisms (Han et al. 2017).
In other eukaryotes, the theme of interacting proteins required for m3C modification recurs. Thus, METTL2 is required in humans and mouse for m3C32 modification of both tRNAThr(UGU) and tRNAArg(CCU) (Noma et al. 2011; Xu et al. 2017), and subsequent experiments show that m3C32 modification of tRNAArg substrates in humans requires interaction of METTL2 with DALRD3, based on complex formation in lysates, copurification of tRNAArg(CCU) and tRNAArg(UCU) with the complex, and the loss of the m3C modification in tRNAArg species in a human DALRD3 knockout cell line (Lentini et al. 2020). Similarly, METTL6 targets tRNASer, likely due to its interaction with seryl-tRNA synthetase (Xu et al. 2017). Furthermore, T. brucei TRM140 forms a complex with ADAT2/ADAT3, which normally deaminates A34 to I34, and all three proteins are required to catalyze m3C32 modification of tRNAThr(IGU) and for subsequent m3C32 deamination to form m3U32 (Rubio et al. 2017; McKenney et al. 2018). Intriguingly, the human METTL8 Trm140 paralog targets mitochondrial tRNAs (tRNASer(UGA) and tRNAThr(UGU) for m3C32 modification (Lentini et al. 2022), and may also target mRNA (Xu et al. 2017).
The biological role of m3C modification and Trm140 family members is not clear. There is no obvious growth defect in S. cerevisiae trm140Δ mutants and in S. pombe trm140Δ trm141Δ mutants in a variety of conditions (D'Silva et al. 2011; Arimbasseri et al. 2016), although S. cerevisiae trm140Δ trm1Δ strains, lacking both m3C32 and m2,2G26, have a mild, but distinct, growth defect in low concentrations of cycloheximide, suggesting a translation defect (D'Silva et al. 2011). In contrast, a homozygous human mutation in DALRD3 is associated with a developmental delay and infantile epilepsy in patients and lack of m3C32 in tRNAArg(CCU) and tRNAArg(UCU) (Lentini et al. 2020), and in T. brucei, METTL6 depletion results in reduced ribosome stability and a cytokinesis defect, although it is not known which RNAs are modified by T. brucei METTL6 (Fleming et al. 2016).
It is also not clear how m3C32 affects tRNA function. The pKa of 8.7 for 3-methylcytidine (Brookes and Lawley 1962; Ueda and Fox 1963) suggests a positive charge in the tRNA ACL, which likely also affects noncanonical N32–N38 interactions that are commonly found in tRNAs (Auffinger and Westhof 1999). As N32–N38 interactions are known to modulate the binding of tRNA to the A-site (Olejniczak and Uhlenbeck 2006; Olejniczak et al. 2005) and the fidelity of translation (Ledoux et al. 2009; Pernod et al. 2020), it is possible that m3C has similar or related effects.
Ψ38,39,40 and Pus3
The pseudouridine (Ψ) modification of U38 and U39 in tRNAs is found in all domains of life, including in the streamlined bacterial genome of Mycoplasma capricolum (Andachi et al. 1989), and the TruA/Pus3 family of pseudouridylases that catalyzes formation of Ψ38 Ψ39 is similarly highly conserved (Koonin 1996; Mueller and Ferre-D'Amare 2009; de Crecy-Lagard et al. 2012). Bacterial TruA from E. coli and Salmonella typhimurium catalyze formation of Ψ40 in addition to Ψ38 and Ψ39 (Singer et al. 1972; Hur and Stroud 2007), whereas the eukaryotic Pus3 (Deg1) ortholog from S. cerevisiae catalyzes only Ψ38 and Ψ39 modification (Lecointe et al. 1998).
Lack of the TruA/Pus3 pseudouridylases results in several defined phenotypes (Fig. 4). E. coli and S. typhimurium truA (hisT) mutants de-repress the histidine operon, and have a modest-to-severe reduction in growth rate that depends on growth supplements (Chang et al. 1971; Tsui et al. 1991), and S. cerevisiae pus3Δ mutants grow slowly and are temperature sensitive (Carbone et al. 1991; Lecointe et al. 2002; Han et al. 2015). In contrast, in humans PUS3 mutations have been linked to intellectual disability, associated with loss of Ψ in a representative tRNA with Ψ39 (Shaheen et al. 2016; Abdelrahman et al. 2018).
The roles of Pus3 pseudouridylation in tRNA function are not yet clear. As Pus3 targets both U38 in the ACL and U39 in the closing base pair of the anticodon stem, lack of Pus modifications could in principle have different roles in each capacity. In S. cerevisiae, loss of Ψ38 impaired function of one tRNA examined, loss of Ψ39 impaired function of one of three tRNAs examined, and the growth defect of pus3Δ trm10Δ double mutants, lacking Ψ38, Ψ39, and m1G9, was primarily due to reduced function of a single tRNA with Ψ39 and m1G9, although three other tRNAs have Ψ39 and m1G9, and four others have Ψ38 and m1G9 (Han et al. 2015). Curiously, in addition, a higher frequency of −1 frameshifting in test sequences in S. cerevisiae was correlated with tRNAs with Ψ39 and found to be reduced in pus3Δ mutants (Bekaert and Rousset 2005). As biochemical and structural analysis has shown that Ψ stabilizes both duplex and single-stranded RNA due to coordination of a water molecule and to enhanced stacking of its favored 3′ endo conformation (Arnez and Steitz 1994; Davis 1995; Durant and Davis 1999; Charette and Gray 2000), it seems plausible that the selective Ψ39 effects will be explained by similar physical or structural effects. The effects of Ψ38 may be due to related stabilization effects or may also be due in part to alteration of the noncanonical 32–38 interactions found in many tRNAs (Auffinger and Westhof 1999).
m5C34,40,48,49,50 and Trm4/NSUN2
Understanding the biology of m5C and the corresponding Trm4/NSUN2 methyltransferase family is complicated by its presence at different locations in tRNA, as well as in other RNAs. In S. cerevisiae and in mouse, Trm4/NSUN2 catalyzes m5C formation in tRNA substrates at C34 in the ACL, at C40 in the anticodon stem, at C48 in the variable loop, and at C49 and C50 in the T-stem (Motorin and Grosjean 1999; Brzezicha et al. 2006; Tuorto et al. 2012). In contrast, S. pombe has two Trm4 paralogs, with Trm4a methylating C34 and C48 and Trm4b methylating C49 and C50 modification (Muller et al. 2019b). Furthermore, multiple m5C-modified sites have been found within mRNAs and other RNAs in human cells by bisulfite sequencing methods (Squires et al. 2012; Amort et al. 2017; David et al. 2017). As m5C modifications can contribute differently to tRNA function in the tRNA ACL, the tRNA body, and in other RNA substrates, it has been difficult to sort these out to ascribe known biological phenotypes to specific m5C modification sites.
Lack of m5C modification in fungi results in relatively mild phenotypes. S cerevisiae trm4Δ mutants have little growth defect in a variety of normal media conditions, but have increased sensitivity to paromomycin (Wu et al. 1998) and are sensitive to oxidative stress (Chan et al. 2010), which has been linked in WT cells to a 70% increase in m5C34 in tRNALeu(CAA), improved tRNALeu(CAA) decoding, and increased translation of proteins rich in UUG codons, including a specific ribosome subunit that is important for survival of peroxide stress (Chan et al. 2012). Curiously also in S. cerevisiae, m5C levels in tRNAHis (but not in two other tRNAs), are increased after amino acid starvation, rapamycin treatment, growth to stationary phase, and growth of temperature sensitive strains at nonpermissive temperature, although it is not clear why this occurs (Preston et al. 2013). In S. pombe, lack of either or both of Trm4a and Trm4b results in little or no growth defect under a variety of conditions, including oxidative stress conditions and paromomycin sensitivity, although trm4aΔ mutants are mildly resistant to CaCl2, suggesting some type of mitochondrial function (Muller et al. 2019b).
Lack of NSUN2 and m5C modifications has been linked to numerous phenotypes in mammals. In humans, NSUN2 mutations are linked to autosomal recessive intellectual disability (Fig. 4; Abbasi-Moheb et al. 2012; Khan et al. 2012; Martinez et al. 2012), and mouse Nsun2−/− mutants have aberrant stem cell differentiation in hair follicle stem cells (Blanco et al. 2011), blocked meiotic progression into pachytene in testis germ cells (Hussain et al. 2013), reduced survival of neurons, and reduced spatial working memory (Blanco et al. 2014). Furthermore, lack of NSUN2 in mouse and human skin cells results in increased sensitivity to UVB radiation and oxidative stress, resulting in accumulation of tRNA-derived fragments and downstream consequences, as discussed further below (Blanco et al. 2014; Gkatza et al. 2019). However, mRNA levels could also contribute to phenotypes of NSUN2 mutants, as m5C-modified mRNAs are recognized by ALYREF/THOC4 in mammals to promote mRNA export (Yang et al. 2017), and in zebrafish m5C-modified maternal mRNAs have higher stability during the maternal-to-zygotic transition, mediated by interaction with the m5C-RNA binding protein Ybx1 (Yang et al. 2019).
There is also a report that lack of NSUN2/TRM4 might result in reduced tRNA stability. A. thaliana trm4b mutants have shorter primary roots and are sensitive to different oxidative stresses (David et al. 2017), and have reduced levels of tRNAAsp(GTC), a representative tRNA substrate with m5C at C48, C49, and C50 (David et al. 2017). Although mouse Nsun2−/− mutants are not known to result in reduced levels of tRNAs in mouse (Blanco et al. 2014), it is known that Nsun2−/− Dnmt2−/− mouse mutants, lacking both sources of m5C in tRNA, are mostly inviable, and survivors have reduced levels of tRNAs with both modifications (Tuorto et al. 2012).
MODIFICATIONS IN THE tRNA BODY
m1A9 and m1G9 and the catalytically versatile Trm10 family
The m1A9 and m1G9 (collectively m1R9) modifications are frequently found in tRNAs from eukaryotes and archaea. Seminal prior work provided compelling evidence that the m1A9 modification prevents misfolding of mitochondrial tRNALys(UUU), as the unmodified tRNA could adopt an alternative structure with an elongated acceptor stem, which was prevented by m1A9 (Helm et al. 1998, 1999; Helm and Attardi 2004). Subsequently, it was shown that S. cerevisiae Trm10 is responsible for m1G9 modification of all nine known substrate tRNAs in vivo, and that the Trm10 family is widespread in eukaryotes and archaea, with two or three paralogs in different metazoans (Jackman et al. 2003). Although S. cerevisiae trm10Δ mutants do not have any obvious growth phenotype in rich or minimal media over a range of temperatures (Jackman et al. 2003), the mutants are unusually sensitive to the anticancer drug 5-fluorouracil (Fig. 4; Gustavsson and Ronne 2008).
Further investigation has revealed several surprises in the biology of the Trm10 family and m1R9 modifications. Because guanosine and adenosine are chemically very different, it was a distinct surprise to discover that the Crenarchaean Trm10 ortholog from Sulfolobus acidocaldarius catalyzes formation of m1A9, rather than m1G9, on a tRNAMeti transcript, and that Trm10 from the Euryarchaean Thermococcus kodakaraensis catalyzes both m1A9 and m1G9 formation on different tRNAs (Kempenaers et al. 2010). Remarkably also, the human TRMT10C ortholog and the short chain dehydrogenase/reductase SDR5C1 comprise two of the three different subunits of the human protein-only RNase P trimer, and moonlight as an m1R9 methyltransferase subcomplex that catalyzes formation of both m1A9 and m1G9 on mitochondrial substrate tRNAs (Vilardo et al. 2012), all of which bear m1R9 in bovine mitochondrial tRNAs with a purine at this residue and canonical cloverleaf structure (Suzuki and Suzuki 2014). Moreover, it is now known that whereas human TRMT10A catalyzes m1G9 modification on all tested cytoplasmic tRNAs known to have the modification, the human TRMT10B ortholog is specific for m1A9 modification of tRNAAsp(GUC) (Howell et al. 2019), the sole human cytoplasmic tRNA with m1A9 (Clark et al. 2016), and knockout cell lines prove the mutually exclusive specificity of TRMT10A for m1G9 and TRMT10B for m1A9 in vivo (Vilardo et al. 2020).
Structural and biochemical analyses have clarified aspects of the mechanism of the Trm10 protein family (for review, see Krishnamohan and Jackman 2019). The structure of Trm10 from S. cerevisiae and S. pombe shows the typical fold of the SPOUT family of methyltransferases, but acting as a monomer (Shao et al. 2014), and biochemical analysis has revealed a noncanonical methyltransferase mechanism. Thus, the mechanism of S. cerevisiae Trm10 involves a collaborative catalytic role for two highly conserved aspartate residues near the active site of S. cerevisiae Trm10, with added contributions from a nearby glutamate residue (Krishnamohan and Jackman 2017). Consistent with these findings, catalysis by the dual specificity T. kodakareinsis Trm10 is also synergistically inhibited in the corresponding double carboxylate variants (Singh et al. 2018).
Nonetheless, it remains to be determined exactly how specificity for m1A9 and/or m1G9 modification are established within Trm10 family members. Whatever the mechanism, the catalytic difference between TRMT10A acting on m1G9 and TRMT10B on m1A9 is not reflected in binding affinity, as each protein bound substrate and nonsubstrate tRNAs with comparable affinity (Howell et al. 2019). It also remains to be determined why S. cerevisiae Trm10 modifies certain tRNAs with equal efficiency in vitro but is more selective in vivo (Swinehart et al. 2013).
Although human cell lines lacking TRMT10A, TRMT10B, or both have no obvious growth defect accompanying the lack of m1G9 and/or m1A9 in their cytoplasmic tRNAs (Vilardo et al. 2020), mutations in TRMT10A result in human disorders (Fig. 4). Thus, early onset diabetes and microcephaly was linked to a TRMT10A-R127* nonsense mutation (Igoillo-Esteve et al. 2013), and a homozygous TRMT10A–G206R mutation was associated with defective glucose metabolism, microcephaly, and intellectual disability (Gillis et al. 2014), and in each case the corresponding TRMT10A variant was catalytically inactive (Gillis et al. 2014) or the corresponding patient cell lines had no detectable m1G9 in tRNAs examined (Cosentino et al. 2018). Initial study showed that TRMT10A knockdown is associated with apoptosis in rat and human β-cells, as well as in a rat pancreatic β-cell line, in this case after treatment with fatty acids, high concentration of glucose, or any of several ER-stressors (Igoillo-Esteve et al. 2013) and, as described further below, these have been linked to tRNA-derived fragments (Cosentino et al. 2018).
Three other phenomena related to TRM10 biology have appeared, each of which adds new dimensions to its roles. First, TRMT10A appears to functionally interact with the mRNA m6A demethylase FTO to regulate m6A levels in mRNA, as endogenous TRMT10A and FTO reciprocally coimmunopreciptate, TRMT10A knockdown or knockout results in increased m6A levels in poly(A) mRNAs, TRMT10A stimulates FTO m6A-demethylation activity in vitro, and there is significant overlap among mRNAs with increased m6A-modification in TRMT10A knockdowns and mRNAs identified by CLIP-seq of TRMT10A and FTO (Ontiveros et al. 2020). The consequences of this potential coregulation of tRNA m1G9 modification and mRNA m6A modification are not yet known. Second, natural S. cerevisiae TRM10 variants have been reported to affect UGA stop codon readthrough efficiency, and SUP45 (encoding the polypeptide release factor eRF1) and TRM10 variants are found in distinct linkage disequilibrium, suggesting evolutionary pressure to moderate termination readthrough efficiency (Torabi and Kruglyak 2011). Third, the biology of S. cerevisiae TRM10 is even more intricate than expected due to the 18-mer ncRNA derived from TRM10 that down-regulates translation (Pircher et al. 2014).
m2,2G26 and Trm1/TRMT1
The m2,2G26 modification occurs frequently in tRNAs from eukaryotes and archaea. Among a set of characterized tRNAs with an encoded G26, the m2,2G26 modification is found in 127 of 160 eukaryotic cytosolic tRNAs (including 21 of 22 in S. cerevisiae and 8 of 10 in humans) and 14 of 32 mitochondrial tRNAs (three of nine in S. cerevisiae and one in humans) (Juhling et al. 2009). In S. cerevisiae and humans, the methyltransferase Trm1/TRMT1 modifies both cytoplasmic and mitochondrial tRNAs to m2,2G26 (Phillips and Kjellin-Straby 1967; Hopper et al. 1982; Ellis et al. 1986; Liu and Straby 2000; Dewe et al. 2017), but it should be noted that the m2G26 modification often found at G26 is also likely catalyzed by Trm1 family members (Edqvist et al. 1994; Urbonavicius et al. 2006). S. cerevisiae Trm1 is localized to the inner nuclear rim (Li et al. 1989) and trm1Δ mutants are temperature sensitive due (Fig. 4) to decay of substrate tRNAs by the RTD pathway (Dewe et al. 2012; discussed further below). In humans, TRMT1 mutations have been linked to intellectual disability (Fig. 4; Najmabadi et al. 2011; Davarniya et al. 2015; Blaesius et al. 2018; Zhang et al. 2020), associated with reduced m2,2G26 in patient-derived lymphoblastoid cell lines (Zhang et al. 2020) and with reduced m2,2G26, reduced proliferation, and sensitivity to oxidative stress in HEK 293T TRMT1 knockout cells (Dewe et al. 2017).
4-acetylcytidine at C12, ac4C12, and the Tan1/THUMPD1:Kre33/NAT10 complex
The ac4C modification is typically found at C12 in eukaryotic tRNAs with a long variable arm, including all but one of the 26 characterized cytoplasmic tRNALeu species, 15 of 19 cytoplasmic tRNASer species, and five mitochondrial tRNALeu species with these properties (Juhling et al. 2009). The ac4C modification is typically found at the middle cytidine of a CCG motif, and remarkably, in archaea, ac4C is found in numerous tRNAs at numerous positions, as well as in other RNAs (except for mRNA), and its levels are dramatically increased at high temperature (Sas-Chen et al. 2020).
Biochemical and genetic analysis has shown that the ac4C12 modification of tRNA requires a complex of Tan1/THUMPD1 and Kre33/NAT10. A prior genetic screen revealed that S. cerevisiae Tan1 was required for ac4C12 formation (Johansson and Bystrom 2004). Subsequent work showed that ac4C12 formation is catalyzed in yeast and humans by Kre33/NAT10 in complex with Tan1/THUMPD1, and surprisingly, that Kre33/NAT10 also acts independently of Tan1 to catalyze acetylation of two cytidine residues in 18S rRNAs in S. cerevisiae and humans (Sharma et al. 2015). This requirement for Tan1 to direct Kre33/NAT10 to tRNA for acetylation is consistent with the tRNA binding activity of Tan1 (Johansson and Bystrom 2004). Kre33/THUMPD1 has a helicase domain that is important for function (Sharma et al. 2015), like that of the related E. coli TmcA protein that acetylates C34 of elongator tRNAMet (Ikeuchi et al. 2008; Chimnaronk et al. 2009), but its role is not yet known.
Tan1 is biologically important in both S. cerevisiae and humans (Fig. 4). In S. cerevisiae, tan1Δ mutants are temperature sensitive due to degradation of substrate tRNAs by the RTD pathway (Chernyakov et al. 2008; Kotelawala et al. 2008; Dewe et al. 2012, discussed further below). In humans, mutations in THUMPD1 are associated with developmental delay, intellectual disability, and behavioral abnormalities, associated with loss of ac4C12 modification of tRNA (Broly et al. 2022).
m7G46 and the Trm8/METTL1:Trm82/WDR4 complex
The m7G46 modification is widely found in tRNAs from prokaryotes and eukaryotes, as well as in some mitochondrial and plastid tRNAs, when G46 is the middle residue of a tRNA with a 5 nt variable loop (Okamoto et al. 2004; Matsumoto et al. 2007; Juhling et al. 2009), comprising the RAGGU motif in yeast and humans (Lin et al. 2018). The corresponding tRNA m7G46 methyltransferase is a two subunit complex in S. cerevisiae and humans, encoded by TRM8/METTL1 and TRM82/WDR4, both components of which are highly conserved in eukaryotes (Alexandrov et al. 2002), although they can be functionally replaced in S. cerevisiae by single subunit bacterial Trm8 homologs from E. coli or Thermotoga maritima (Alexandrov et al. 2005; for review see Tomikawa 2018). Structural analysis has revealed a substantial interaction surface between yeast Trm8 and Trm82 including two salt bridges whose residues are highly conserved (Leulliot et al. 2008). A slightly different interaction surface is found in the human METTL1:WDR4 complex (Li et al. 2023; Ruiz-Arroyo et al. 2023), stabilized by two different salt bridges as well as three hydrogen bonding interactions, and biochemical experiments show that each of three mutations that disrupt these intersubunit interactions eliminates activity (Li et al. 2023). Intriguingly, comparison of crystal and cryo-EM structures with and without tRNA and cofactor (S-adenosylmethionine or S-adenosylhomocysteine) show that tRNA and cofactor binding is accompanied by substantial changes in METTL1 and bending of the tRNA, resulting in local unwinding of the variable loop and base flipping of G46 into the active site, with a prominent role for the previously disordered METTL1 amino terminus (Li et al. 2023; Ruiz-Arroyo et al. 2023).
Trm8 and Trm82 are biologically important in eukaryotes (Fig. 4). Lack of TRM8 and/or TRM82 is associated with temperature sensitivity in S. cerevisiae and S. pombe, linked to decay of substrate tRNAs by the RTD pathway (Alexandrov et al. 2005; Chernyakov et al. 2008; Dewe et al. 2012; De Zoysa and Phizicky 2020, discussed further below). In humans, mutations in WDR4/TRM82 are linked to microcephaly and primordial dwarfism (Fig. 4), and in each of two unrelated families, the same conserved R170 residue is mutated to leucine or glutamine (Shaheen et al. 2015; Trimouille et al. 2018). Consistent with this clinical manifestation, the WDR4-R170L mutation results in reduced tRNA m7G modification activity in vitro and reduced or eliminated activity in vivo (Shaheen et al. 2015; Li et al. 2023), and the WDR4-R170Q mutation results in the elimination of activity in vitro and in vivo (Li et al. 2023). In addition, human METTL1 is subject to phosphorylation at S27 by Akt (protein kinase B) and RSK in vitro and in vivo, which resulted in loss of m7G modification activity in vitro (Cartlidge et al. 2005), consistent with the lack of function of the corresponding yeast and human phosphomimetic mutants (Cartlidge et al. 2005; Li et al. 2023; Ruiz-Arroyo et al. 2023). The consequences of this regulation are not yet known.
Recent results in mammals have revealed profound biological effects associated with lack of m7G46, linked to reduced levels of tRNAs. Mouse embryonic stem cells (mESCs) lacking METTL1 have substantially reduced levels of six of the 22 tRNA species normally bearing m7G46, associated with increased ribosome pausing at the corresponding codons, reduced translation efficiency of mRNAs rich in these codons, and defective self-renewal and neural differentiation (Lin et al. 2018).
Remarkably, recent experiments show that m7G modification of one specific tRNA by METTL1/WDR4 drives oncogenic transformation. In support of the causal link between METTL1/WDR4 expression and oncogenic transformation, METTL1 and WDR4 expression and m7G46 modification are up-regulated in certain cancers, associated with poor prognosis (Dai et al. 2021; Orellana et al. 2021); knockout of METTL1 or WDR4 results in a reduction in cell proliferation, oncogenicity, and tumor growth, which is associated with reduced m7G46 modification and reduced levels of several m7G46-modified tRNAs; and overexpression of METTL1/WDR4 (but not catalytic dead variants) results in increased oncogenicity and increased translation of a subset of genes associated with cell cycle regulation (Dai et al. 2021; Orellana et al. 2021). Furthermore, tRNAArg(TCT) accounts for most of this biology, as translation is most affected for genes particularly enriched in AGA codons, which is decoded by tRNAArg(TCT); tRNAArg(TCT) expression correlates with METL1/WDR4 expression in tumors and is associated with poor survival; and overexpression of tRNAArg(TCT) is oncogenic and phenocopies many of the properties of METTL1/WDR4 overexpression (Orellana et al. 2021).
m1A58, Trm6:Trm61, and tRNAMeti
The m1A58 modification is ubiquitous in tRNA from eukaryotes, occurring in 33 of the 55 tRNA isodecoders of cytosolic tRNA from S. cerevisiae and in the majority of those from human cytosolic tRNAs (Saikia et al. 2010; Cozen et al. 2015; Boccaletto et al. 2022), as well as in a limited number of tRNAs in prokaryotes, archaea, and mitochondria of animals and plants (Juhling et al. 2009). Formation of m1A58 is catalyzed by the essential Trm6:Trm61 (Gcd10:Gcd14) complex in S. cerevisiae (Anderson et al. 1998, 2000), which is widely conserved in eukaryotes (Bujnicki 2001; Ozanick et al. 2005). This complex is comprised of a dimer of Trm6:Trm61 heterodimers in which the noncatalytic Trm6 subunit positions the tRNA for Trm61 to modify at A58, which is exposed by separation of the T-loop and D-loop (Finer-Moore et al. 2015).
The m1A58 modification is crucial for structure and function of initiator tRNA, tRNAMet(CAU)i (for review, see Kolitz and Lorsch 2010). Thus, although m1A58 is found on numerous S. cerevisiae tRNAs, the lethality of a trm6Δ mutation is suppressed by overexpression of tRNAMet(CAU)i, suggesting that this is the only biologically important substrate (Anderson et al. 1998). This finding is consistent with the unique structure of initiator tRNAMeti, which features a tRNA substructure involving hydrogen bonding interactions between A58 and residues A54 and A60 in the T-loop, and between A20 of the D-loop and G57, A59, and A60 of the T-loop (Basavappa and Sigler 1991). It seems highly likely that this substructure is uniquely common to all eukaryotic initiator tRNA species, as the residues A20, A54, and A60 (and the lack of N17) are normally found in initiator tRNA, but are only rarely found among elongator tRNAs (particularly not in combination), and m1A58 is found in all but one characterized eukaryotic tRNAMeti (Marck and Grosjean 2002; Kolitz and Lorsch 2010; Boccaletto et al. 2022).
The lack of m1A58 in S. cerevisiae leads to reduced levels of tRNAMet(CAU)i, due to decay (Anderson et al. 1998), by both the nuclear surveillance pathway (Kadaba et al. 2004) and the RTD pathway (Tasak and Phizicky 2022), as discussed further below. The m1A58 modification is also likely important for cell health and tRNAMeti stability in other eukaryotes. In mammalian cells, knockdown of either TRM6 or TRM61 in a rat glioma cell line was reported to result in a slow growth phenotype and reduced levels of tRNAMeti, which could be partially rescued by overexpression of tRNAMeti, and overexpression of TRM6–TRM61 resulted in increased levels of tRNAMeti, as well as of tRNAMete (Macari et al. 2016). In Arabidopsis, lack of either the TRM6 or the TRM61 ortholog leads to embryo arrest and seed abortion and reduced TRM61 expression is associated with reduced levels of tRNAMeti (Tang et al. 2020). It remains to be determined how the levels of tRNAMet(CAU)i are reduced in these and other multicellular organisms.
Lack of m1A58 can also affect the function of at least one other tRNA. Thus, cells from patients with MERFF (myoclonus epilepsy, ragged-red fibers) due to an A54G mutation in mitochondrial tRNALys lack m1A58 as well as the taurine modification normally associated with this disease, and the lack of m1A58 was directly linked to reduced translation by mitochondrial tRNALys (Richter et al. 2018).
It is now known that m1A58 levels in tRNAs are subject to regulation by members of the AlkB family of dioxygenases with tRNA m1A demethylase activity. AlkBH1 was documented to have tRNA m1A demethylase activity based on CLIP-Seq experiments showing binding to mature tRNAs, in vitro assays that documented tRNA m1A demethylation activity, accompanied by ALKBH1 knockdown experiments that resulted in increased m1A levels in specific tRNAs, and ALKBH1 overexpression experiments that resulted in reduced levels of m1A in these tRNAs (Liu et al. 2016). Strikingly, it was also shown that transient ALKBH1 knockdown led to a threefold increase in levels of tRNAMeti, associated with increased cellular proliferation, and increased translation, and that glucose starvation led to increased ALKBH1 expression, reduced m1A levels in tRNA targets, and reduced tRNAMeti levels and reduced translation (Liu et al. 2016). In addition, m1A levels in tRNAs may also be regulated by two other members of the AlkB protein family. Thus, ALKBH3 has tRNA m1A demethylation activity in vitro and is highly expressed in tumor cells, and its knockdown in tumor cells reduces proliferation (Ueda et al. 2017), and FTO catalyzes m1A demethylase activity in vitro in addition to its known m6Am and m6A demethylation activity, and FTO knockdown in cell lines and in Fto−/− MEFs resulted in increased m1A levels in specific tRNAs (Wei et al. 2018).
In addition, as dicussed further below, it is now known that increased TRMT6:TRMT61-dependent m1A levels in some tRNA-derived fragments leads to their reduced gene silencing activity (Su et al. 2022).
tRNA TURNOVER PATHWAYS
The half-life of typical tRNAs is extraordinarily long, estimated to be 44 and 50 h in Euglena gracilis and chicken muscle, respectively, similar to that of rRNAs (Nwagwu and Nana 1980; Karnahl and Wasternack 1992) and ∼9 h in S. cerevisiae (Gudipati et al. 2012). However, although tRNAs are stable, lack of any of several tRNA body modifications in S. cerevisiae and S. pombe leads to decay of a specific subset of the hypomodified tRNA species by either of two decay pathways. The nuclear surveillance pathway targets pre-tRNAs for 3′–5′ exonucleolytic decay shortly after transcription, and the RTD pathway targets mature tRNAs for 5′–3′ exonucleolytic decay after maturation (Figs. 1, 7; Supplemental Table S1). These pathways are described in more detail below.
FIGURE 7.
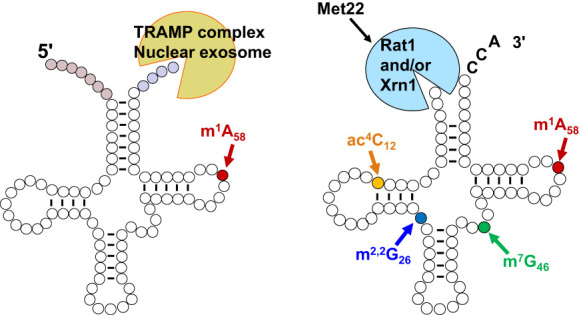
Two different tRNA decay pathways in S. cerevisiae. (Left) A pre-tRNAMeti molecule is depicted in the typical secondary structure shortly after transcription, with uncolored circles representing tRNA residues, pale red circles representing the 5′ leader nucleotides, pale blue circles representing the 3′ trailer nucleotides, and a bright red circle indicating the site for m1A58 modification. A pre-tRNAMeti lacking m1A58 is targeted for decay by the nucelar surveillance pathway in S. cerevisiae, involving oligoadenylation of the pre-tRNA by Trf4 of the TRAMP complex, and then 3′–5′ exonucleolytic degradation of the pre-tRNA by Rrp6 and Rrp44 of the nuclear exosome. Spliced leader-containing pre-tRNAs are also targeted for decay by the nuclear surveillance pathway (Kramer and Hopper 2013; Chatterjee et al. 2022). (Right) A mature tRNA with a CCA end is depicted in its typical secondary structure, with residues that are normally modified to form ac4C12 in yellow, m2,2G26 in blue, m7G46 in green, and m1A58 in red. Specific mature tRNAs lacking one of these modifications are targeted for decay by the rapid tRNA decay pathway in S. cerevisiae, involving 5′–3′ exonucleolytic decay of the tRNA by Rat1 and Xrn1 in the nucleus and cytoplasm, respectively, both of which are inhibited by pAp, which accumulates in met22Δ mutants.
The nuclear surveillance pathway
Earlier groundbreaking work in S. cerevisiae defined the nuclear surveillance pathway by identification and characterization of spontaneous suppressors of the temperature sensitivity of trm6-504 mutants (Kadaba et al. 2004), which was known to be due to reduced levels of tRNAMeti (Anderson et al. 1998). Thus, the isolation of suppressing mutants in TRF4 and in RRP44, encoding, respectively, a protein with poly(A) polymerase activity and a 3′–5′ exonuclease in the exosome, led to the definition of a pathway (Fig. 7) in which pre-tRNAMeti lacking m1A58 was targeted for 3′–5′ exonucleolytic decay by Rrp6 and the nuclear exosome after polyadenylation by Trf4 (Kadaba et al. 2004). Biochemical analysis showed that Trf4 was part of the TRAMP complex, along with Air1 or Air2 and the RNA helicase Mtr4 (LaCava et al. 2005; Vanacova et al. 2005), and that the TRAMP complex and nuclear exosome could degrade a mature tRNAMeti transcript but not native tRNAMeti, and a tRNAAla(GGC) transcript with a destabilizing D-stem mutation, but not the corresponding WT tRNAAla(GGC) transcript (Vanacova et al. 2005). Further, in vitro analysis showed that the TRAMP complex and recombinant Rrp44, the sole nuclease of the core exosome (Dziembowski et al. 2007), specifically acted on mature tRNAMeti lacking m1A58, but not on any of several other tRNAs examined, in a preparation of bulk RNA (Schneider et al. 2007). This in vitro specificity for tRNAMeti lacking m1A58 recapitulated the specificity observed in vivo and was consistent with the known unique involvement of residue A58 of initiator tRNAMeti in tertiary interactions with A54 and with A60 as part of the unique substructure of tRNAMeti (Basavappa and Sigler 1991). Although there is a similar TRAMP5 complex containing the highly related Trf4 homolog Trf5 (Houseley and Tollervey 2006), its role in tRNA decay is less clear.
Further in vivo analysis in S. cerevisiae provided evidence that the nuclear surveillance pathway also targets a large portion of newly transcribed pre-tRNAs in WT cells (Gudipati et al. 2012). Thus, tiling arrays revealed a global increase in steady state tRNA levels in mutants conditionally lacking Dis3/Rrp44 or lacking Rrp6, and a synergistic increase in tRNA levels in rrp6Δ dis3− double mutants. This data, combined with pulse chase experiments, showed that more than 50% of the global population of transcribed tRNAs is degraded by the nuclear surveillance pathway as pre-tRNAs, while also revealing that the half-life of mature tRNAs in S. cerevisiae is ∼9 h, and is independent of Dis3. The cause of this strikingly high level of pre-tRNA decay is unknown, but was speculated to be due to some combination of pre-tRNA misfolding after transcription, pre-tRNA instability, stochastic mutations arising during transcription, and competition between the maturation machinery and the nuclear surveillance pathway for normally folded pre-tRNAs (Gudipati et al. 2012).
The nuclear surveillance pathway in S. cerevisiae is known to compete with early steps of tRNA processing and 3′ end formation. Failure of proper 3′ trailer removal by Trz1, Rex1, and Rrp6 can lead to polyadenylation and pre-tRNA decay by the nuclear surveillance pathway, as documented for two pre-tRNAs with longer structured 3′ trailers that are targeted by Trz1 (Skowronek et al. 2014), and for three pre-tRNAs with longer 3′ ends, including two pre-tRNAMeti species lacking m1A58 that are normally processed by Rex1 (Ozanick et al. 2009). Consistent with direct competition between the nuclear surveillance pathway and the tRNA 3′ end formation machinery, overexpression of the La protein (Lhp1) prevents decay of pre-tRNAMeti lacking m1A58 by the nuclear surveillance pathway (Anderson et al. 1998).
It also appears that the nuclear surveillance pathway can target pre-tRNAs at different points in the biogenesis pathway in S. cerevisiae. An early analysis showed that the species of pre-tRNAMeti lacking m1A58 that was targeted by the nuclear surveillance pathway had complete 5′ leaders and a portion of their 3′ trailers, implying targeting shortly after initial transcription (Kadaba et al. 2004, 2006; Ozanick et al. 2009). However, the nuclear surveillance pathway also appears to target end-matured unspliced pre-tRNAs that are 3′ trimmed by Rex1, and competition also occurs at this stage, as overexpression of La prevents access of Rex1 to the 3′ ends and the ensuing decay (Copela et al. 2008). Given the very different stages in biogenesis of these pre-tRNA targets, it seems likely that the nuclear surveillance pathway targets pre-tRNAs at all nuclear steps of biogenesis, in competition with components of the maturation machinery.
It seems likely that the nuclear surveillance pathway will target pre-tRNAs for decay widely throughout eukaryotes. The nuclear exosome is widely conserved across eukaryotes (Houseley and Tollervey 2009; Januszyk and Lima 2014), as are the components of the TRAMP complex (Win et al. 2006; Schmidt and Butler 2013), and it is known in S. pombe that pre-tRNAs lacking La protein are targeted by Rrp6 of the nuclear exosome (Huang et al. 2006). Although there is both a nucleolar and a nuclear TRAMP complex in S. cerevisiae (Wolin et al. 2012), and an analogous nucleolar TRAMP complex in S. pombe to recruit RNAs (Win et al. 2006), in humans there is both a nucleolar TRAMP complex and a nuclear exosome targeting (NEXT) complex that recruits RNA substrates (Lubas et al. 2011, 2015; Schmidt and Butler 2013). Although tRNA transcription and early tRNA processing events are nucleolar in yeast (Thompson et al. 2003), and other tRNA processing steps take place in the nucleoplasm or at the inner nuclear membrane (Rose et al. 1995; Murthi and Hopper 2005), it is not known where in the nucleus tRNA biogenesis takes place in other organisms. Thus, although the nuclear exosome has a wide swath of other RNA substrates (Houseley et al. 2006; Wolin et al. 2012), it is plausible that in other organisms the TRAMP and/or the NEXT complexes have roles in targeting pre-tRNAs for decay by the nuclear exosome. Global analysis of the effects of the nuclear exosome on tRNA levels or stability has not been examined in organisms other than S. cerevisiae.
The rapid tRNA decay pathway
In S. cerevisiae, lack of m7G46, ac4C12, or m2,2G26, alone or in combination with other modifications, is associated with temperature sensitivity due to 5′–3′ exonucleolytic decay of a subset of the mature hypomodified tRNAs by Xrn1 and/or Rat1 of the RTD pathway (Fig. 7). Thus, the temperature sensitivity of trm8Δ trm4Δ mutants, lacking m7G46 and m5C49, is due to decay of tRNAVal(AAC), as levels of tRNAVal(AAC) but not other hypomodified tRNAs were reduced at high temperatures, the temperature sensitivity was suppressed by overexpression of tRNAVal(AAC), and both decay and temperature sensitivity were suppressed by mutation of RAT1, XRN1, or MET22 (Alexandrov et al. 2006; Chernyakov et al. 2008). Mutation of MET22 suppresses decay by the RTD pathway (Fig. 7) because it leads to increased levels of the metabolite pAp, an inhibitor of Rat1 and Xrn1 (Murguia et al. 1996; Dichtl et al. 1997; Yun et al. 2018). A similar set of experiments showed that the temperature sensitivity of tan1Δ trm44Δ mutants (lacking ac4C12 and Um44) and of trm1Δ trm4Δ mutants (lacking m2,2G26 and m5C) was due to RTD of tRNASer(CGA) and tRNASer(UGA), and not other tRNAs with the corresponding modifications (Chernyakov et al. 2008; Whipple et al. 2011; Dewe et al. 2012), and showed that this decay occurred at the level of mature tRNAs, rather than pre-tRNAs (Alexandrov et al. 2006; Chernyakov et al. 2008). Additional experiments expanded the scope of the RTD pathway to single mutants lacking m7G46, m2,2G26, or ac4C12, as the temperature sensitivity of each single mutant was suppressed by a met22Δ mutation and associated with tRNA decay at the restrictive temperature (Dewe et al. 2012).
Mechanistic studies suggest that the RTD pathway targets tRNAs that expose their 5′ end due to reduced stability, which can arise from lack of stabilizing body modifications or from destabilizing mutations in the tRNA body, and which is amplified by growth at higher temperatures. Temperature is a prominent feature of the RTD pathway as each of the hypomodified strains implicated in RTD is temperature sensitive due to the RTD pathway, although decay is still evident at lower temperatures (Chernyakov et al. 2008; Whipple et al. 2011; Dewe et al. 2012). To analyze the importance of destabilizing mutations and lack of body modifications in triggering RTD, the growth properties of strains expressing variants of the essential tRNASer(CGA) gene SUP61 were compared in WT and tan1Δ trm44Δ backgrounds (with and without a met22Δ mutation to inhibit RTD), and then compared to the predicted stabilities of the variants in the combined acceptor and T-stem, which are normally stacked in mature tRNA (Whipple et al. 2011). This analysis revealed that lack of ac4C12 and Um44 in tan1Δ trm44Δ strains acts as if it destabilizes the tRNA by 1.0–1.5 kcal/mol, and that met22Δ strains (in which RTD is inhibited) could tolerate ∼1.5–2 kcal/mol more destabilization than WT strains in the acceptor and T-stems. In addition, biochemical analysis of 5′ end accessibility of purified tRNAs showed that the 5′ end was more sensitive to purified Xrn1 or calf intestinal phosphatase in tRNAs lacking modifications that triggered RTD than in WT tRNAs, and in variants with destabilizing mutations in the acceptor and T-stem than in WT tRNAs (Whipple et al. 2011).
As ac4C12 and Um44 are both located in residues known to participate in tertiary interactions (Fig. 2; Kim et al. 1974a,b; Giege et al. 2012), this data supports a model in which lack of these modifications destabilize the tertiary structure of the tRNA, which is known to be the initial step in the overall melting of tRNAs (Shelton et al. 2001; Wilkinson et al. 2005), making it more likely for the subsequent helix unwinding to expose the 5′ end to exonucleases. In support of this argument, the other modification mutants implicated in the RTD pathway lack m7G46 or m2,2G26, and now also m1A58 (see below), and all of these residues are known to participate in tertiary interactions in some tRNAs (Kim et al. 1974a,b; Basavappa and Sigler 1991; Giege et al. 2012). This tertiary structure destabilization model also explains why a number of variants of fully modified SUP4oc (tRNATyr(GUA) with a UUA anticodon) that trigger RTD at low temperature (28°C) have mutations in the D-stem–loop, the V-loop or the T-loop, as well as in the acceptor and T-stems (Guy et al. 2014).
Additional studies emphasize the importance of both reduced overall tRNA stability and of higher temperature in increased susceptibility to RTD. Thus, high-throughput analysis of S. cerevisiae SUP4oc variants reveals a correlation between RTD at 28°C and a reduction in the predicted ΔΔG° of variants (Guy et al. 2014), and shows that the pervasive temperature sensitivity of SUP4oc variants observed between growth at 28°C and 37°C is highly correlated with susceptibility to RTD, suggesting that temperature sensitivity is frequently due to RTD (Payea et al. 2018).
Recent results show that the RTD pathway is conserved in the phylogenetically distant fission yeast S. pombe, and extend the use of the RTD pathway to another body modification mutant in both S. pombe and S. cerevisiae. Thus, S. pombe trm8Δ mutants are now known to be temperature sensitive due to decay of tRNATyr and to some extent tRNAPro(AGG) by the RTD pathway, as levels of these tRNAs were reduced at high temperature, overexpression of these tRNAs suppressed the temperature sensitivity, and each of four spontaneous suppressors of the temperature sensitivity had mutations in the Rat1 ortholog DHP1 and prevented decay of these tRNAs (De Zoysa and Phizicky 2020).
Similarly, S. pombe trm6Δ mutants are now known to be temperature sensitive due to decay of tRNAMeti by the RTD pathway, as tRNAMeti levels were reduced at high temperature, overexpression of tRNAMeti suppressed the temperature sensitivity, each of three spontaneous suppressors of the temperature sensitivity and tRNA decay had mutations in DHP1 or the MET22 ortholog TOL1, and each of nine suppressors of the exacerbated growth defect of trm6Δ imt06Δ mutants (also lacking one of four copies of the tRNAMeti gene) had mutations in DHP1 or TOL1 (Tasak and Phizicky 2022). Furthermore, the TRAMP complex had little role in quality control of tRNAMeti in S. pombe trm6Δ mutants, as deletion of the TRF4 ortholog CID14 in trm6Δ mutants had little effect on growth or in preventing tRNAMeti decay (Tasak and Phizicky 2022). Moreover, reexamination of S. cerevisiae trm6 mutants showed a prominent role of the RTD pathway in preventing tRNAMeti decay, in addition to the known role of the nuclear surveillance pathway. Thus, both the temperature sensitivity and the tRNAMeti decay observed in S. cerevisiae trm6-504 mutants were suppressed by mutation of any of the components of the RTD pathway (RAT1, XRN1, and MET22), and the lethality of S. cerevisiae trm6Δ mutants could be suppressed by mutation of both the nuclear surveillance pathway (trf4Δ) and the RTD pathway (met22Δ) but not by either alone (Tasak and Phizicky 2022).
As S. pombe and S. cerevisiae diverged ∼600 Mya (Parfrey et al. 2011), these results fuel speculation that the RTD pathway will target decay of specific hypomodified tRNAs throughout eukaryotes in these and other body modification mutants. Thus, it seems plausible that the RTD pathway is responsible for the reduced levels of specific hypomodified tRNAs in mammals lacking m7G46 (Lin et al. 2018; Dai et al. 2021), and in mouse strains lacking m5C in their tRNAs (Tuorto et al. 2012; Hussain et al. 2013). Indeed, there is evidence that the RTD pathway acts in humans, as heat stress at 43°C in HeLa cells resulted in loss of tRNAMeti levels which was prevented by knockdown of XRN1 and the human RAT1 ortholog XRN2 (Watanabe et al. 2013).
Consistent with its targeting of mature tRNAs, the RTD pathway competes with elements of the translation pathway. Thus, RTD is prevented by overexpression of elongation factor 1A (EF-1A), which normally binds charged tRNA to escort the tRNA to the ribosome A-site, and is enhanced by reduced expression of EF-1A (Dewe et al. 2012; Turowski et al. 2012). Similarly, overexpression of ValRS suppresses RTD of tRNAVal(AAC) in trm8Δ trm4Δ mutants (Turowski et al. 2012). Competition might also explain the apparent paradox of why reduced Pol III transcription, resulting from overexpression of the negative regulator Maf1 or from Pol III mutants, protects against RTD, as the reduced numbers of tRNAs would be more easily protected by the available EF-1A (Turowski et al. 2012).
The RTD pathway may also compete with the cellular retrograde tRNA transport pathway in which tRNAs are imported from the cytoplasm to the nucleus (Shaheen and Hopper 2005; Takano et al. 2005), as discussed further below. Thus, tRNATyr(GUA) and tRNALys(UUU) lacking m2,2G26 accumulate in the cytoplasm if retrograde transport is inhibited genetically by either of two mechanisms, as well as in cells lacking the RTD exonuclease Xrn1 (Kramer and Hopper 2013). The parsimonious explanation of these results is that the RTD pathway and the retrograde transport pathway are in competition for the same substrate tRNAs lacking m2,2G26, to degrade them or give them a second chance to be modified (Kramer and Hopper 2013).
It also seems likely that 5′ end capping of pre-tRNA transcripts competes with 5′–3′ decay by the RTD pathway, as the preferential accumulation of 5′ capped pre-tRNA (relative to uncapped pre-tRNA) that occurs when RNase P is inhibited is exacerbated in a met22Δ derivative strain, and in some cases in an xrn1Δ-derivative strain, indicating decay of uncapped pre-tRNA by an RTD-like mechanism (Ohira and Suzuki 2016).
In addition, recent experiments establish that the onset of the RTD pathway in both S. pombe and S. cerevisiae triggers activation of the GAAC pathway (the integrated stress response pathway in humans), which reprograms transcription and translation after stress treatments leading to uncharged tRNA and/or ribosome collisions (Hinnebusch 2005; Udagawa et al. 2008; Castilho et al. 2014; Duncan et al. 2018; Wu et al. 2020; Yan and Zaher 2021; Kim and Zaher 2022). Thus, in S. pombe, mutations in any of four genes of the GAAC pathway fully suppressed the temperature sensitivity of trm8Δ mutants and partially restored tRNATyr(GUA) levels, and temperature shift experiments showed that the growth defect, tRNATyr(GUA) decay, and GAAC induction start at exactly the same temperature, and are due to the tRNA decay and not the temperature shift itself (De Zoysa and Phizicky 2020). Furthermore, S. cerevisiae modification mutants subject to RTD also activate the GAAC pathway, but with the opposite effect on growth (De Zoysa and Phizicky 2020). Thus, for the well-studied S. cerevisiae trm8Δ trm4Δ mutant, GAAC activation occurs at the lowest temperature at which the growth defect and decay of tRNAVal(AAC) is observed, and deletion of any of several genes in the GAAC pathway exacerbates the temperature sensitivity and the loss of tRNAVal(AAC). As a gcn2Δ mutation exacerbates the temperature sensitivity of each of four other S. cerevisiae modification mutants that undergo RTD, it seems likely that activation of the GAAC pathway is a general consequence of onset of the RTD pathway in S. cerevisiae (De Zoysa and Phizicky 2020).
Intriguingly, the TOR pathway is also possibly linked to RTD in HeLa cells, as the XRN1 and XRN2/RAT1-mediated decay of tRNAMeti that occurs under heat stress is inhibited by rapamycin, concomitant with increased nucleolar and reduced nucleoplasmic localization of XRN2/RAT1 (Watanabe et al. 2014).
The Met22-dependent pre-tRNA decay pathway in S. cerevisiae
Recent results document the existence of an additional tRNA decay pathway in S. cerevisiae that is related to RTD by its Met22-dependence, but different due to the nature of its tRNA substrates. Whereas the RTD pathway typically targets mature tRNAs (Alexandrov et al. 2006; Chernyakov et al. 2008), this MPD pathway acts specifically on end-matured intron-containing pre-tRNAs, which accumulate due to mutations in the anticodon stem–loop (ASL) or the introns that perturb ASL-intron structure (Payea et al. 2020). As described above, eukaryotic tRNA introns form a characteristic BHB or BHL structure with the ASL that is recognized by the endonuclease (Thompson and Daniels 1988, 1990; Xue et al. 2006; Yoshihisa 2014; Schmidt and Matera 2020), and tRNA variants that disrupt this structure trigger MPD (Payea et al. 2020). Moreover, MPD is quantitatively comparable to that observed for classical RTD substrates with acceptor stem mutations, and removal of the intron eliminates most of the observed decay (Payea et al. 2020). It remains to be determined which Met22-dependent exonucleases or endonucleases act in MPD, although presumably the nucleases are inhibited by the pAp that builds up in met22Δ mutants (Dichtl et al. 1997; Yun et al. 2018). It also remains to be determined if the MPD pathway extends to intron-containing tRNAs in other eukaryotes (Chan and Lowe 2016; Schmidt and Matera 2020).
tRNA NUCLEAR-CYTOPLASMIC SUBCELLULAR DYNAMICS
The tRNA retrograde process
tRNAs surprisingly travel bidirectionally between the nucleus and cytoplasm in both budding yeast (Shaheen and Hopper 2005; Takano et al. 2005) and vertebrate cells (Zaitseva et al. 2006; Shaheen et al. 2007). The distribution of tRNAs between the nucleus and the cytoplasm results from the balance between: (1) tRNA nuclear export to the cytoplasm after transcription (primary tRNA nuclear export); (2) retrograde import of cytoplasmic tRNA into the nucleus (tRNA retrograde nuclear import); and (3) return of tRNAs that have been imported into the nucleus to the cytoplasm (tRNA reexport) (Fig. 8).
FIGURE 8.
Bidirectional tRNA trafficking between the nucleus and cytoplasm and generation of tRNAPhe yW37 in S. cerevisiae. Step 1. Upon 5′ and 3′ processing and addition of several nucleoside modifications to newly transcribed intron-containing tRNAs, Los1, Mex67–Mtr2, and Crm1 escort the end-processed, partially modified intron-containing tRNAs to the cytoplasm via the primary tRNA nuclear export step. The tRNAs are then spliced on the mitochondrial outer membrane. Numerous additional nucleoside modifications also occur in the cytoplasm after splicing. Cm32 and Gm34 (orange circles) modifications added in the cytoplasm are important for yW biogenesis. Step 2. Spliced, modified tRNAs are returned to the nucleus via the tRNA retrograde nuclear import step. Mtr10 functions indirectly in tRNA nuclear import both constitutively and upon amino acid deprivation (red symbol), whereas Ssa2 functions only upon amino acid deprivation. tRNAPhe imported into the nucleus is further modified at G37 (yellow circle) to m1G37 (empty colored circle). Step 3. Msn5, Los1, Mex67–Mtr2, and pehaps also Crm1, escort the imported tRNAs back to the cytoplasm via the tRNA reexport step. Once reexported to the cytoplasm, tRNAPhe m1G37 is further modified to yW (black circle). Red circles indicate anticodon nucleotides 34, 35, and 36.
Initial studies of tRNA nuclear/cytoplasmic distribution were conducted by using Xenopus oocyte injections (Arts et al. 1998a; Kutay et al. 1998; Lund and Dahlberg 1998), reconstituted nuclear import assays using vertebrate cells (Zaitseva et al. 2006), and tRNA fluorescence in situ hybridization, FISH (Hellmuth et al. 1998; Sarkar and Hopper 1998; Shaheen and Hopper 2005; Takano et al. 2005; Shaheen et al. 2007). More recently, organelle fractionation and RNA-seq have been used (Schwenzer et al. 2019). Furthermore, analysis of the kinetics of tRNA nuclear export vs. nuclear import has become possible by using microinjection of tagged tRNAs and confocal imaging of the tagged tRNA in single vertebrate cells (Dhakal et al. 2019).
For vertebrate cells, the ability to distinguish primary tRNA nuclear export from tRNA reexport generally relies upon employment of transcription inhibitors (Shaheen et al. 2007; Schwenzer et al. 2019). In contrast, in budding and fission yeast and Trypanosoma brucei, it is possible to distinguish primary tRNA nuclear export from the reexport process because splicing of pre-tRNAs occurs in the cytoplasm; thus, those tRNAs encoded by genes possessing introns leave the nucleus in the primary export process with their introns, whereas tRNAs in the nucleus that have been spliced have undergone retrograde nuclear import, and exit the nucleus by reexport as spliced tRNAs (Yoshihisa et al. 2003; Murthi et al. 2010; Lopes et al. 2016; Kessler et al. 2018; Wan and Hopper 2018).
The distribution of tRNAs between the nucleus and the cytoplasm responds to nutrient status and environmental stresses. In budding yeast, cytoplasmic tRNAs accumulate in the nucleus upon amino acid, phosphate, and glucose deprivation (Shaheen and Hopper 2005; Hurto et al. 2007; Whitney et al. 2007). In vertebrate cells, cytoplasmic tRNAs accumulate in nuclei upon amino acid and glucose deprivation and in response to H202-induced oxidative stress (Shaheen et al. 2007; Dhakal et al. 2019; Schwenzer et al. 2019). Moreover, in vertebrate cells cytoplasmic tRNAMeti accumulates in nuclear granules upon temperature stress (Miyagawa et al. 2012; Watanabe et al. 2013). Furthermore, the tRNA nucleus–cytoplasm trafficking is relatively fast. Upon various stress impositions, for both budding yeast and vertebrate cells, cytoplasmic tRNAs rapidly redistribute to the nucleus and, likewise, rapidly return to the cytoplasm upon stress relief (Shaheen and Hopper 2005; Shaheen et al. 2007; Whitney et al. 2007; Dhakal et al. 2019; Schwenzer et al. 2019).
tRNA nuclear exporters
The first tRNA nuclear exporter identified was the conserved GTPase, Ran binding β-importin member, Los1 (budding yeast)/Xpo-t (fission yeast)/Exportin-t (vertebrates)/PAUSED (plants). This member of the β-importin family was identified decades ago (Hopper et al. 1980; Arts et al. 1998a; Hellmuth et al. 1998; Kutay et al. 1998; Lund and Dahlberg 1998; Sarkar and Hopper 1998; Hunter et al. 2003; Li and Chen 2003), and its interaction with tRNA substrates and RanGTP to form nuclear export complexes was described in a 3.2 Å resolution structure for the S. pombe Xpot in complex with a partial tRNA and RanGTP (Cook et al. 2009). However, insects lack a Los1/Exportin-t homolog (Lippai et al. 2000) and Los1 appears not to function in tRNA nuclear export in the kinetoplastid, T. brucei (Hegedusova et al. 2019). Furthermore, Los1 and its orthologs are unessential in every organism from which it has been deleted, including budding yeast, fission yeast, the plant A. thaliana, and haploid human cell lines (Hurt et al. 1987; Hunter et al. 2003; Li and Chen 2003; Cherkasova et al. 2012; Blomen et al. 2015; Hart et al. 2015; Wang et al. 2015; Azizi et al. 2020). Therefore, since tRNA nuclear export is essential for translation, eukaryotes must possess tRNA nuclear exporters that are independent of Los1 homologs.
For budding yeast there are at least three additional tRNA nuclear exporters that function in parallel to Los1: (1) the β-importin family member Msn5 which, for intron-containing tRNAs, appears to function solely in the tRNA nuclear reexport step (Murthi et al. 2010; Huang and Hopper 2015); (2) the mRNA nuclear exporter Mex67–Mtr2 heterodimer, which functions in both the tRNA primary and nuclear reexport steps (Wu et al. 2015; Chatterjee et al. 2017); and (3) the β-importin protein nuclear exporter, Crm1, which also functions in primary nuclear export (Fig. 8; Wu et al. 2015; Chatterjee et al. 2022). The RNA helicase, Dbp5, also functions in tRNA nuclear export (Lari et al. 2019), but it likely serves as an adapter/scaffold for Mex67–Mtr2 and/or Crm1.
Prior studies in vertebrate cells showed that, although unessential, Exportin-t appeared to be the dominant tRNA nuclear exporter (Arts et al. 1998a; Kutay et al. 1998), and that the Msn5 homolog Exportin-5, had a minor role in tRNA nuclear export (Bohnsack et al. 2002; Calado et al. 2002). However, a recent study documented that the human Mex67 homolog, NXF1, is a tRNA nuclear exporter (Chen et al. 2021), and the Mtr2 homolog, NXT1, has been reported to bind tRNA (Ossareh-Nazari et al. 2000). Further, in Arabidopsis deletions of both PAUSED and the exportin-5 homolog HASTY have a more severe phenotype than either individual deletion (Hunter et al. 2003), indicating that HASTY may function in tRNA nuclear export. Finally, in T. brucei, the Los1 and Msn5 homologs apparently have no important role in tRNA nuclear export; instead, Mex67 and Mtr2 function in tRNA nuclear export. Interestingly, as determined by RNA FISH studies, it appears that T. brucei Mex67 exports different families of tRNAs than does Mtr2 (Hegedusova et al. 2019). Unlike for budding yeast, the human Crm1 homolog, Xpo-1, appears not to play a role in tRNA nuclear export (Lund et al. 2004; Chen et al. 2021).
tRNA family preferences of the various tRNA nuclear exporters appears to be a theme, based on results from several approaches. In budding yeast, accumulation of end-processed, intron-containing tRNAs as assessed by northern analyses has served as a proxy for tRNA nuclear export due to the cytoplasmic location of SEN; thus, tRNAs retained in the nucleus are unspliced. More recently, in vivo copurification of nuclear exporters in complex with pre-tRNA cargo has served to assess tRNA nuclear export complexes (Huang and Hopper 2015). Both of these methodologies have documented that yeast Los1, Mex67–Mtr2, and Crm1 possess different preferences for each of the 10 different tRNA families encoded by intron-containing genes (Chatterjee et al. 2017, 2022). Moreover, vertebrate Exportin-t also has been reported to bind various tRNAs with different affinities (Li and Sprinzl 2006). tRNA family preferences for Los1/Exportin-t are surprising since this exporter is dedicated to tRNA nucleus–cytoplasm traffic and it interacts with the tertiary structure of mature tRNAs that is shared by all tRNAs (Arts et al. 1998b; Lipowsky et al. 1999; Cook et al. 2009).
The Los1/Exportin-t-independent tRNA nuclear exporters Mex67–Mtr2/NXF1–NXT1 and Crm1, which bind numerous RNAs and protein adapters (for reviews, see Kelly and Corbett 2009; Chatterjee et al. 2018), would not necessarily have been expected to recognize all tRNA families equally well. As predicted, Mex67–Mtr2 and Crm1 have been documented to possess tRNA family preferences and these preferences differ from each other and from Los1 (Chatterjee et al. 2022).
How tRNA family-specific tRNA nuclear export is achieved is not understood. Mex67–Mtr2 and NXF1–NXT1 function in mRNA nuclear export generally by binding RNA substrates via various protein adapters (for reviews, see Kelly and Corbett 2009; Chatterjee et al. 2018). Likewise, Crm1/Exportin-1 functions to export proteins from the nucleus to the cytoplasm via its interaction with proteins harboring leucine-rich nuclear export sequences (NES) (Fischer et al. 1995; Wen et al. 1995; Fornerod et al. 1997). Therefore, such proteins could preferentially bind subsets of nuclear tRNAs to function in preferential family-specific tRNA nuclear export. Other possible means for tRNA recognition include tRNA modifications and/or subnuclear locations of the nuclear exporters or the tRNAs.
In sum, there are multiple tRNA nuclear exporters in nature and their employment for efficient tRNA nuclear export differs among eukaryotic organisms. The implications of the individual nuclear exporters having tRNA family preferences could be far-reaching, as translation of the proteome could be affected by alteration of the cellular balance/activities of individual tRNA exporters. To fully understand tRNA family preferences for tRNA nuclear export, it will be important to understand how the various tRNA exporters interact with specific tRNA cargoes and to identify the protein adapters and possibly the RNA competitors.
tRNA nuclear importers
The gene products involved in retrograde tRNA nuclear import are not well defined. To date, two budding yeast proteins, the β-importin family member Mtr10 (Shaheen and Hopper 2005; Murthi et al. 2010) and the Ssa2 member of the chaperonin family (Takano et al. 2015) were shown to affect the levels of cytoplasmic tRNAs that accumulate in nuclei upon nutrient deprivation. Recently an assay has been developed that allows analysis of both constitutive and stress-induced tRNA nuclear import and reexport in budding yeast. This assay assesses modification of G37 of tRNAPhe to wybutosine (yW) (Fig. 6). yW modification of tRNAPhe requires all three steps of the tRNA retrograde pathway (Ohira and Suzuki 2011). The first step of the yW biogenesis is acquisition of m1G37 catalyzed by the nucleus-localized tRNA methyltransferase, Trm5. Trm5 can only modify spliced tRNAs; so, intron-containing pre-tRNAPhe is first exported via the primary tRNA nuclear export step to the cytoplasm where it is spliced and further modified by Trm7. Upon nuclear import of the spliced tRNA, Cm32 and Gm34 modified tRNAPhe G37 becomes a Trm5 substrate and G37 is thus modified to m1G37. Then, the m1G37 bearing tRNAPhe returns to the cytoplasm where m1G37 is further converted to yW via catalysis by the four cytoplasmic enzymes, Tyw1, Tyw2, Tyw3, and Tyw4. Thus, completion of yW modification of tRNAPhe requires primary nuclear export, tRNA nuclear retrograde import, and tRNA reexport (Ohira and Suzuki 2011). tRNAPhe possessing yW37 can be cleaved at position 37 upon treatment with HCl followed by aniline treatment (Thiebe and Zachau 1968; Ladner and Schweizer 1974) to generate tRNA halves, which are easily detected by northern analysis (Nostramo and Hopper 2020). Using this HCl-aniline assay, Mtr10 was shown to function in tRNA retrograde nuclear import both constitutively and upon amino acid deprivation, whereas Ssa2 functions in amino acid deprivation-induced tRNA nuclear import but not detectably in constitutive tRNA nuclear import (Nostramo and Hopper 2020). Ssa2, binds tRNAs and the nuclear pore protein, Nup116, and therefore, Ssa2 likely functions directly in tRNA retrograde nuclear import (Takano et al. 2015). In contrast, in vivo pull-down studies failed to document physical interactions between Mtr10 and tRNA (Huang and Hopper 2015) and therefore, it is not clear whether Mtr10 directly functions in tRNA nuclear import (Fig. 8). Whether there are additional budding yeast tRNA nuclear importers remains unknown but seems likely. The putative human Mtr10 homolog, Transportin 3, does not appear to affect tRNA nuclear import (Zhou et al. 2011) and, to date, no vertebrate tRNA nuclear importers have been reported.
Environmental stresses and tRNA nuclear/cytoplasmic dynamics
How environmental stresses result in redistribution of tRNAs between the nucleus and the cytoplasm is not well understood. As documented in budding yeast (Shaheen and Hopper 2005; Whitney et al. 2007; Takano et al. 2015; Lari et al. 2019; for reviews, see Huang and Hopper 2016; Chatterjee et al. 2018) as well as in vertebrate cells (Shaheen et al. 2007 PMCID: PMC1183567; Barhoom et al. 2011; Miyagawa et al. 2012; Watanabe et al. 2013; Dhakal et al. 2019; Schwenzer et al. 2019) environmental stresses affect both tRNA nuclear export and retrograde nuclear import steps. Recent studies using injected fluorescently tagged functional tRNAs measured the kinetics of tRNA subcellular movements and demonstrated that in response to nutrient deprivation, RNA nuclear import is down-regulated but tRNA nuclear export is nearly completely blocked, resulting in a net retrograde nuclear accumulation of tRNA (Dhakal et al. 2019).
There are different mechanisms that could cause altered nuclear vs. cytoplasmic pools of tRNAs upon stress (Huang and Hopper 2014). First, part of the pool of a given tRNA isoacceptor could be altered in response to stress. For example, budding yeast Ssa2, which is important for nuclear import of cytoplasmic tRNAs upon amino acid deprivation, preferentially binds tRNAs with destabilized aminoacyl acceptor stems (Takano et al. 2015). Likewise, using reconstituted nuclear import assays, the Fassati group reported that tRNAs deleted for 3′ nt, sometimes extending into the tRNA body, were preferentially imported into nuclei (Zaitseva et al. 2006). More recent tRNA sequencing studies of the tRNAs imported upon oxidative stress documented preferential nuclear accumulation of tRNA with 3′ deletions/truncations (Schwenzer et al. 2019). One possibility is that aberrant tRNAs might have preferential access to the tRNA nuclear importers because the truncated tRNAs are unable to interact with the translation machinery and thus are not otherwise engaged. Another possibility is that there are proteins that are able to recognize particular tRNAs and monitor integrity of the tRNA 3′ ends.
Alternatively, as documented by RNA sequencing of the tRNA cytoplasmic vs. nuclear pools upon stress in HeLa cells, there is tRNA family-specific tRNA nuclear accumulation upon stress (Schwenzer et al. 2019), possibly giving rise to changes of the proteome upon stress. tRNA family-specific nuclear accumulation could result from alteration of the levels of individual tRNA exporters and importers and/or the putative adaptors that may change in response to stresses, or the subcellular distributions of the exporters and importers themselves may be altered upon various stresses. Regarding the latter possibility, upon glucose deprivation and oxidative stress the steady state nuclear vs. cytoplasmic localization of the budding yeast tRNA nuclear exporters, Los1, Msn5, and Crm1, are inverted; normally, the steady state distribution of these proteins is nucleoplasmic or nuclear rim located, but under stress conditions the proteins appear to be predominately cytoplasmic (Quan et al. 2007; Huang and Hopper 2014). Likewise, glucose deprivation results in the inversion of the steady state nuclear/cytoplasmic distribution from primarily cytoplasmic to nucleoplasmic for the putative tRNA nuclear importer, Mtr10 (Huang and Hopper 2014). However, upon amino acid deprivation in budding yeast, Los1, Msn5, and Crm1 did not display altered nuclear vs. cytoplasmic distributions (Huang and Hopper 2014), whereas the level of the nucleus-located subpool of Ssa2 was reported to increase (Takano et al. 2015). To date, there have been no reports of similar studies of the subcellular locations of the budding yeast Mex67–Mtr2 tRNA nuclear exporter under conditions that alter tRNA subcellular dynamics; nor is it known whether there are altered levels or subcellular distributions of the various exporters in response to stresses in vertebrate cells.
The tRNA retrograde pathway and constitutive tRNA biogenesis/quality control
The tRNA retrograde pathway also serves important constitutive functions. One such function concerns tRNA modifications. As detailed above, yW modification of tRNAPhe requires tRNA nuclear import from the cytoplasm followed by reexport back to the cytoplasm. Similarly, queuosine (Q) modification of G34 of tRNATyr in T. brucei requires that pre-tRNATyr first be exported to the cytoplasm where tRNA splicing occurs (Lopes et al. 2016; Kessler et al. 2018). Upon tRNA retrograde nuclear import, the nucleus-localized tRNA-guanine transglycosylase, which has specificity for spliced tRNATyr as substrate, converts G34 to Q34. Then, the Q-modified tRNA exits the nucleus via Mex67–Mtr2 mediated tRNA reexport for appropriate function in translation in the cytoplasm (Kessler et al. 2018; Hegedusova et al. 2019). It is unknown whether other tRNA modifications in organisms that have cytoplasmic tRNA splicing also require tRNA retrograde nuclear import for tRNA modifications.
The tRNA retrograde pathway also serves an important role in tRNA quality control as demonstrated by studies in budding yeast (Kramer and Hopper 2013; Chatterjee et al. 2022). Errors sometimes occur such that aberrant tRNAs that are unprocessed at the 5′ and 3′ termini or that are hypomodified are prematurely exported to the cytoplasm. The levels of these inappropriate tRNAs increase upon deletion of the putative tRNA nuclear importer, Mtr10 (Kramer and Hopper 2013). Thus, tRNA retrograde nuclear import appears to remove aberrant tRNAs from the cytoplasm, returning them to the nucleus where they may be repaired and/or destroyed by the 5′ to 3′ nuclease Rat1 of the RTD pathway, or by the 3′ to 5′ nuclear surveillance pathway (Kramer and Hopper 2013; Chatterjee et al. 2022). As described above, the retrograde pathway is also in apparent competition with the cytoplasmic RTD quality control pathway that destroys hypomodified tRNAs or tRNAs with destabilizing mutations (Whipple et al. 2011; Kramer and Hopper 2013; Guy et al. 2014). Further, although Los1 and Crm1 have high fidelity in nuclear export of only those tRNAs with mature 5′ termini, Mex67 is able to export 5′ leader containing tRNAs to the cytoplasm, where they are spliced. Upon tRNA retrograde nuclear import these aberrant 5′ leader-containing spliced tRNAs can be destroyed by the 3′ to 5′ nuclear surveillance pathway, but not by the nuclear RTD process (Chatterjee et al. 2022). Protection from Rat1 turnover may be due to possession of the triphosphate of the initial tRNA transcripts or to caps at the 5′ temini (Ohira and Suzuki 2016; Chatterjee et al. 2022).
tRNA FRAGMENTS AND REGULATION OF GENE EXPRESSION
Although the canonical function of tRNAs as adaptors in gene expression, delivering amino acids to the translation machinery, has been appreciated since 1958 (Hoagland et al. 1958), numerous additional noncanonical functions for tRNAs have been described in the subsequent decades (for reviews, see Raina and Ibba 2014; Schimmel 2018). Most recently, this list of noncanonical functions has expanded due to the discoveries of tRNA fragments that serve numerous unanticipated roles in biology. The rapid pace of the discoveries and the important roles of the tRNA fragments in gene expression, development, and health have been summarized in numerous excellent review articles (Anderson and Ivanov 2014; Kumar et al. 2016; Oberbauer and Schaefer 2018; Guzzi and Bellodi 2020; Kim et al. 2020; Xie et al. 2020; Chu et al. 2022; George et al. 2022; Hou et al. 2022; Pekarsky et al. 2022). We direct the reader to these reviews regarding the roles of tRNA fragments in development, health and disease. Here, we highlight the various means by which the myriad of tRNA fragments are generated, the roles of nucleotide modifications in their production and functions, and the varying mechanisms by which tRNA fragments regulate gene expression in eukaryotic cells.
RNases involved in tRNA fragment production
The first discovered nucleases that cleave tRNAs were the E5 and D subsets of the bacterial colicins (Ogawa et al. 1999; Tomita et al. 2000; for review, see Ogawa 2016) and the fungal killer toxins from K. lactis and Pichia acaciae (Lu et al. 2005; Klassen et al. 2008). These secreted RNases cleave specific mature tRNAs in the ACL to generate half molecules, which reduces environmental competition by recipient cells via depletion of the recipient's active tRNA pools and therefore inhibition of their protein synthesis. Similarly, in bacterial and archaeal organisms the toxin–antitoxin systems that regulate cell growth upon various stresses can act via tRNA endonucleolytic cleavage. For example, the Type II MazF and VapB/C toxin–antitoxins cleave various tRNAs in the ACL to inhibit translation (Cintron et al. 2019; for review, see Walling and Butler 2019).
In contrast, the more recently discovered mechanisms that generate tRNA fragments in eukaryotic cells generally do not cause significant reduction of tRNA pools; rather, they generate novel noncoding RNAs that possess various activities able to regulate gene expression. These tRNA fragments have several different nomenclatures. Some RNases cleave specific mature tRNAs in or near the ACL and generate tRNA ∼half molecules (30–40 oligonucleotides) that are variously dubbed tRNA fragments (tRFs), tiRNAs (tRNA stress-induced RNAs), tsRNAs (tRNA-derived small RNAs), tdRs (tRNA-derived RNAs) or tRHs (tRNA halves); hereafter these tRNA fragments are referred to as tRHs (Fig. 9). RNases also cleave mature tRNAs in or near the D-loop or the T-loop to generate smaller, 13–26 nt fragments, named 5′- or 3′-tsRNAs or 5′- or 3′-tRFs (hereafter referred to as 5′- or 3′-tsRNAs). tRNA fragments can also be derived from pre-tRNAs; the 3′U tRFs are derived from the 3′ trailer of pre-tRNAs and the 5′ leader exon fragments are derived from initial tRNA transcripts containing the 5′ leader that have been cleaved in the ACL (Fig. 9; for reviews, see Anderson and Ivanov 2014; Raina and Ibba 2014). A single system for naming the myriad of tRNA fragments has been proposed (Holmes et al. 2023). Accordingly, the fragments will be referred to as tDRs (tRNA-derived RNAs) with numbers denoting the starting and ending positions of the mature tRNAs, according to conventional tRNA numbering (Sprinzl et al. 1998) (e.g., tDR-1:15); the specific tRNA from which the fragments are derived will also be designated (e.g., tDR-1:15-Val-AAC-1) and, finally the nomenclature will also contain information to link the particular tRNA fragments to tRNAs in the genomic database (http://gtrnadb.ucsc.edu) (see Fig. 9). This proposed nomenclature promises to eliminate future confusion; however, because the discoveries summarized here unfortunately generally do not have sufficient information to utilize the new systematic nomenclature, we will utilize the terms tRHs, 5′ or 3′-tsRNAs, and 3′U tRFs (Fig. 9).
FIGURE 9.
Biogenesis of tRNA fragments. (A) tRNA fragments generated from pre-tRNAs prior to 5′ leader and 3′ trailer removal. Green font and bracket indicate tRNA fragments derived from 3′ trailers upon endonucleolytic cleavage by RNase Z. Blue bracket demarcates region of fragments resulting from cleavage of 5′ leader-containing pre-tRNAs in the ACL. (B) 5′ (left blue bracket) or 3′ (right blue bracket) ∼ half size tRNA fragments generated upon cleavage of mature tRNAs in the ACL. (C) 5′ (left puple bracket) or 3′ (right purple bracket) ∼ ¼ size tRNA framents resulting from endonucleolytic cleavage of mature tRNAs in the D- or T-loops, respectively. Black, blue, and purple fonts near brackets indicate the various names of the tRNA fragments; blue font nomenclature is used in this review. Red font refers to the proposed future systematic nomenclature for tRNA fragments. Arcs indicate the possible locations of loop cleavages. Names below each arc refer to the various endonucleases implicated in cleavages. Angiogenin (also refered to as ANG) is a vertebrate RNase A-like enzyme, and RNase L is an interferon induced 2′–5′ oligoadenylate synthetase-dependent RNase. Rny1 is a yeast T2-like endonuclease; Rnt2 and RNS1, RNS2, and RNS3 are plant T2-like endonucleases. Metazoan Dicer and plant Dicer-like DCLs are RNase III-like enzymes also functioning in pre-miRNA biogenesis.
Generation of tRNA halves
Numerous RNases function in the production of tRHs, 5′ or 3′-tsRNAs, and 3′U tRFs (Fig. 9), with some of the RNases functioning in tRNA cutting primarily under stress conditions. The discovery of amino acid starvation-induced cleavage of tRNAs in their ACLs in the protozoan Tetrahymena thermophila (Lee and Collins 2005) was followed by definition of other stress-induced tRNA cleavage events in other organisms, and definition of their mechanisms. Budding yeast Rny1 (vertebrate RNASET2) is an RNase T2-like endonuclease that, upon stress treatment, catalyzes cleavage of substrate tRNAs, rRNAs, and snRNAs. Rny1 cleaves some mature tRNAs in the ACL after exposure of cells to oxidative stress or high culture density/stationary phase (Thompson and Parker 2009). Rny1 is a resident of the yeast vacuole (lysosome in vertebrates); it is unknown whether tRNA cleavage results from release of Rny1 to the cytoplasm upon stress or, instead, whether tRNAs access the vacuole upon stress via autophagy (Luhtala and Parker 2012). Cleavages in the tRNA anticodon in Tetrahymena and the plant, Arabidopsis are catalyzed by combinations of multiple Rny1 orthologs, Rnt2 A, B, and C and RNS1, RNS2, and RNS3, respectively (Andersen and Collins 2012; Megel et al. 2019). In contrast, for vertebrate cells, generation of tRNA halves is generally catalyzed by ANG, an RNase A-like ribonuclease. Under normal environmental conditions, ANG is primarily localized in the nucleus; cytoplasmic pools exist in complex with an inhibitor, RNH1. Upon stress, nuclear ANG cleaves specific cytoplasmic tRNAs (for review, see Anderson and Ivanov 2014). Interestingly, the T. brucei protozoan genome does not encode either ANG-like or Rny1-like endonucleases (Fricker et al. 2019); so, it is unknown how the stress-induced tRNA half molecules are derived in this organism. Finally, a specialized mammalian endonuclease, RNase L, that is dependent upon 2′,5′ oligoadenylate for dimerization and activity, cleaves particular tRNAs in the ACL (Donovan et al. 2017 and references therein).
After cleavage in the ACL, the 5′ and 3′ halves may not be separated due to the base pairs that form the cloverleaf secondary structure. Indeed, it has recently been reported that tRNAs nicked in the ACL can be repaired (Chen and Wolin 2023; Costa et al. 2023). Identification of helicases that may aid separation of the nicked halves are a subject of current investigation (Drino et al. 2023).
Generation of 13–26 nt tRNA fragments
The 3′ U tRF species are comprised of the 3′ trailers of pre-tRNA transcripts, and result from the cleavage of these pre-tRNAs at the 3′ mature border by the endonuclease RNase Z (Trz1 in yeast) (Haussecker et al. 2010; Su et al. 2019; for reviews, see Anderson and Ivanov 2014; Keam and Hutvagner 2015; Xie et al. 2020). However, less is known regarding the enzymes required for generating the fragments from mature tRNAs that are smaller than tRNA halves (5′-tsRNAs and 3′-tsRNAs). The endonuclease, Dicer, which functions in the biogenesis of miRNAs, has been implicated in the generation of both the 5′-tsRNAs and 3′-tsRNAs in several biological systems (Cole et al. 2009; Haussecker et al. 2010; Durdevic et al. 2013b; Maute et al. 2013; Martinez et al. 2017; Luo et al. 2018). In contrast, other studies have shown that production of 5′- and 3′-tsRNAs can be independent of Dicer (Li et al. 2012; Kumar et al. 2014; for review, see Keam and Hutvagner 2015). For example, even though in Arabidopsis pollen cells a member of the Dicer family, DCL1, was reported to function in the generation of 5′-tsRNAs that target transposable element RNAs (Martinez et al. 2017), Arabidopsis missing all three of the unessential Dicer genes (DCL2, 3, and 4) and possessing a hypomorphic allele of the essential DCL1 gene (dcl1234) exhibited no differences in the tRNA cleavage products compared to the wild-type plants. Rather, deletion of the Rny1-like genes, RNS1, RNS2, and RNS3, affected the production of short tRNA fragments in a tissue-specific manner (Alves et al. 2017; Megel et al. 2019). Production of 5′- and 3′-tsRNAs also appears to be independent of ANG; for example, studies of stressed and unstressed human cells overexpressing ANG or possessing an ANG knockout reported comparable levels of 3′-tsRNAs (Su et al. 2019). A recent report documented that the tRNA substrate generating 3′-tsRNAs are mature aminoacylated tRNAs (Liu et al. 2021). A future challenge will be to detail the biogenesis pathways of the various small tRNA fragments. This may be complex given the exceedingly large number of tRNA fragments that can be generated from the various isoacceptor and isodecoder tRNAs encoded by eukaryote genomes and the numerous RNases with differing specificities.
tRNA modifications and tRNA fragments
RNA modifications play surprisingly important roles in tRNA fragment production and/or function (for review, see Lyons et al. 2018). Some tRNA modifications enhance tRNA cleavage. The fungal zymocin γ toxin subunit provides a eukaryotic example of this. K. lactis produces γ toxin that is toxic to S. cerevisiae, because when introduced into S. cerevisiae the γ toxin cleaves three different tRNAs, all of which possess the mcm5s2U34 modification; the modification is important for substrate cleavage, but additional surrounding nucleotides affect cleavage efficiency (Lu et al. 2005; Huang et al. 2008). There are examples of the requirements of tRNA modifications for the production of vertebrate tRNA fragments. For example, pseudouridylation functions in production of tsRNAs, as in human ECS cells pseudouridylation at U8 by Pus7 enhances production (or stability) and the activities of short 5′-tsRNAs that possess a terminal oligo guanosine (TOG) motif that are derived from tRNAAla(AGC/CGC/TGC), tRNACys(GCA), and tRNAVal(AAC) (Guzzi et al. 2018; for review, see Guzzi and Bellodi 2020).
Other modifications protect tRNAs from cleavage; there are many reports of such protection from several organisms/tissues involving several different modifications including Q34, m5C38, m5C48.49, C34m, m1G9. For example, in HEK293T and HeLa cells queuosine (Q34) modification of tRNAHis(GTC) and tRNAAsn(GTT/GTC) protected these tRNAs from cleavage by ANG in vitro and in vivo (Wang et al. 2018). Modification of m5C38 by Drosophila Dnmt2 protected several tRNAs from stress-induced cleavage; Dnmt2−/− mutants lacking m5C38, are sensitive to growth at high temperature and to oxidative stress (Schaefer et al. 2010). Similarily, the presence of m5C38 in mouse sperm inhibited fragmentation of tRNAGly into 5′ and 3′ tsRNAs (Zhang et al. 2018). m5C modifications catalyzed by Trm4/NSUN2 are also important for production of 5′ tRHs in mouse and human skin cells. Lack of m5C48,49 resulted in an ANG-dependent accumulation of 5′ tRHs from a subset of tRNA species (Blanco et al. 2014). Moreover, in human cell lines, C34 2′-O-methylation of tRNAMete (C34m) is generated by small guide RNAs, SNOD97 and SCARNA97; this Cm34 modification protects tRNAMete from stress-induced cleavage by ANG (Vitali and Kiss 2019). Further, m1G9 protects tRNAs from fragment production. Thus, lymphoblast cell lines derived from TRMT10A deficient patients accumulated tRNAGln 5′ tRHs as well as 5′ fragments of ∼22 nt. TRMT10A knockdown in a rat pancreatic β-cell line resulted in increased reactive oxidative species that led to apoptosis, and apoptosis was also caused by transfection of tRNAGln 5′ fragments into TRMT10A-competent EndoC-βH1 cells (Cosentino et al. 2018). Finally, in human cell lines, 5′ monophosphate methylation of tRNAHis(GTG) by BCDIN3D is reported to protect this mature tRNA from cleavage, resulting in reduced levels of 3′-tsRNAHisGTG (Reinsborough et al. 2019). As many of the discoveries of the roles of modifications in the biogenesis/function of tRNA fragments are recent, it is likely that future studies will uncover other such examples.
Diverse mechanisms of action of tRNA fragments
tRNA fragments can participate in some of the same noncanonical functions that mature tRNAs participate in. For example, both mature cytoplasmic tRNAs and tRHs are able to bind cytochrome C released from mitochondria and, in doing so, activate caspase and thereby inhibit apoptosis (Mei et al. 2010; Saikia et al. 2014). In another example, mature tRNAs prime retrotranscription by base pairing with the primer binding site (PBS) in retroviruses and endogenous LTR retroelements; likewise, the 5′ tRNAMeti derived tRH serves as the primer for Drosophila copia retroviral replication (Kikuchi et al. 1986), and 3′-tsRNAs that are complementary to human T-cell leukemia virus (HTLV) PBS serve as primers for reverse transcription in vitro (Ruggero et al. 2014). In contrast, in mouse 3′-tsRNAs with perfect complementarity to the PBS of retroelements compete with mature tRNA for PBS binding and thereby inhibit retrotranscription (Schorn et al. 2017; for review, see Schorn and Martienssen 2018).
tRNA fragments also serve unique functions that are unrelated to activities of full length tRNAs. These novel functions result from either binding of tRNA fragments to proteins or protein complexes or from complementary base pairing (often dependent upon Argonaute proteins) to target RNAs, thereby affecting the structure, stability, or activities of the target RNAs. Through these various mechanisms tRNA halves and tRNA small fragments can affect RNA transcription and epigenetic inheritance, RNA processing, RNA stability, RNA structure, or translation. Here, we provide a few examples of the various mechanisms of action by tRNA fragments.
tRNA fragments functioning via protein interaction
A well-described function of tRNA fragments is to inhibit protein synthesis initiation (for reviews, see Anderson and Ivanov 2014; Guzzi and Bellodi 2020). Ivanov et al. (2011) and Anderson and Ivanov (2014) reported that specific 5′ halves of tRNAAla and tRNACys, in combination with the YB-1 translational repressor, inhibit translation via displacement of initiation factor eIF4F from capped mRNAs, thereby globally inhibiting translation in response to stress. These tRHs contain the 4–5 5′ G nucleotides comprising the TOG motif that is important to generate the RNP complex. Similarly, in human ESC cells, TOG motif-containing small 5′-tsRNAs derived from tRNAAla(AGC/CGC/TGC), tRNACys(GCA), and tRNAVal(AAC), modified with Ψ8 (see above), bind the poly (A) binding protein, PABPC1, which is required for translation initiation (Guzzi et al. 2018). tRNA fragments have also been reported to affect translation by binding ribosomes. In the archaeon, Haloferax volcanii, stress-induced 5′-tsRNAs derived from tRNAVal bind to the small ribosome subunits and compete with mRNA binding, thereby inhibiting translation initiation (Gebetsberger et al. 2017). In contrast, in the protozoan, T. brucei, binding of a stress-induced tRNA 3′ tRH, derived from tRNAThr(AGU), to ribosomes or polysomes resulted in enhanced translation (Fricker et al. 2019). Interestingly, studies of differentiating mouse embryonic stem cells reported different modes of action of particular 5′ tRHs during differentiation: in stem and retinoic acid induced differentiating states, particular tRHs interact with ribosomes and ribosomal subunits, globally modulating translation; however, a set of tRHs also interact with and sequester the insulin growth factor-like mRNA binding protein, Igf2bp1, resulting in c-Myc mRNA instability (Krishna et al. 2019).
tRNA fragment interactions with proteins can also affect RNA processing. For example, 3′-tsRNAs from Tetrahymena affect pre-rRNA processing (Couvillion et al. 2012). These 3′-tsRNAs interact with a Piwi protein, Twi12, as well as other proteins to form an RNP complex that contains the 5′ to 3′ exonuclease Xrn2. The complex forms in the cytoplasm and it is required for Xrn2′s nuclear import/stability and its role in pre-rRNA processing.
tRNA fragments that function via RNA–RNA complementarity
There are numerous examples of tRNA fragments that cause down-regulation of specific mRNA targets via RNP complexes consisting of Argonaute proteins and tRNA fragments with limited complementarity to the target mRNAs (for review, see Kumar et al. 2014). This mechanism to regulate gene expression resembles the manner in which miRNAs and piRNAs affect gene expression. In fact, some small noncoding regulatory RNAs that were originally identified as miRNAs are actually 3′-tsRNA molecules derived from mature tRNAs (e.g., Haussecker et al. 2010; Maute et al 2013; Reinsborough et al. 2019) or 3′U tRFs derived from the 3′ trailers of pre-tRNAs (e.g., Haussecker et al. 2010; Pekarsky et al. 2016). However, the tRNA-derived fragments differ in important ways from miRNAs. First, they are transcribed by RNA polymerase III, rather than RNA polymerase II. Second, tRNA fragments generally have different biogenesis pathways than miRNAs (for review, see Ha and Kim 2014) or piRNAs (for review, see Han and Zamore 2014). Third, although complementary base pairing for some tRNA-derived fragments is similar to the mechanism by which miRNAs and piRNAs interact with target RNAs via short 7 nt 5′ seed sequences that base pair with the mRNA 3′ UTR (e.g., Kuscu et al. 2018), other tRNA fragments appear to interact with target RNAs differently. These tRNA fragments have been proposed to have seed sequences located in the 5′ ends, the middle, and/or 3′ ends of the tRNA fragments that are complementary with the 5′ UTR, the coding sequence, or the 3′ UTR of the target mRNAs (e.g., Luo et al. 2018). Finally, there are examples in which tRNA fragments affect target mRNAs via complementary base pairing, but independently of Argonaute proteins (Jehn et al. 2020).
Modifications also are implicated in tRNA fragment function via base pairing. For example, TRMT6/61A-dependent m1A modification in the seed region of particular 3′ts RNAs inhibits miRNA function. Inhibition is due to reduced base pairing with target mRNAs rather than to interaction with Argonaute. Over production of TRMT6/61A and fragment modification is correlated with bladder cancer (Su et al. 2022).
Although most known small RNAs that base pair with target mRNAs cause decreased gene expression, either due to increased turnover or to decreased translation, a tRNA fragment that enhances translation upon complementary base pairing with its target mRNA has been reported. Following up on the observations that a 3′-tsRNA derived from tRNALeu(CAG) in HeLa and HCT-116 cells is important for cell growth and efficiency of translation, the Kay group learned that this 3′-tsRNA possesses conserved complementarity with a region in the RPS28 coding sequence in mouse and human cells. It has been proposed that base pairing of the tRNA fragment with RPS28 mRNA alters mRNA structure, unfolding the mRNA to allow efficient translation at a step after initiation (Kim et al. 2017, 2019). There is no evidence for the interaction of this 3′-tsRNA with Argonaute proteins (Kim et al. 2017).
CONCLUDING REMARKS AND PERSPECTIVES FOR THE FUTURE
As documented above, the last several years have witnessed an explosion in our understanding of the biology of tRNA processing, tRNA modification, tRNA decay, and tRNA fragments. These advances set the stage for significant discoveries in the future, aided by ever more powerful new technologies. Four particularly interesting future topics are elucidated below.
First, there is great promise for breakthroughs in our understanding of the mechanisms of the numerous neurological, mitochondrial, and other disorders due to defects in tRNA processing. Multiple studies cited here and elsewhere (Suzuki 2021) have documented examples in which mutations leading to reduced function or to lack of different tRNA processing or modification components result in neurological or other disorders (Fig. 4). It seems highly likely that future studies will unravel why so many of these mutations selectively target the neurological system, why the mutations have different manifestations, and how they exert their effects at a mechanistic level. It also seems likely that some of the different manifestations will be due to tissue-specific differences in expression of isodecoders (Ishimura et al. 2014) or of different tRNA species.
Second, it seems likely that there will be significant new insights regarding the regulation of modifications. We described above a number of examples highlighting the variability of modifications in response to different stress or environmental conditions (Chan et al. 2010, 2012; Czech et al. 2013; Laxman et al. 2013; Preston et al. 2013; Alings et al. 2015; Damon et al. 2015; Han et al. 2015; Gupta et al. 2019; Cristodero et al. 2021; Huber et al. 2022), and in several cases there is significant understanding of the consequences of the altered modifications on translation (Chan et al. 2012; Czech et al. 2013), signaling pathway regulation (Damon et al. 2015), and metabolic regulation (Laxman et al. 2013; Gupta et al. 2019; Huber et al. 2022). Future analysis will undoubtedly reveal a more complete description of the pervasiveness of modification regulation, aided in part by the continued development of technology to facilitate collection of modification profiles of individual tRNAs (Liu et al. 2019; Furlan et al. 2021). The importance of modification regulation seems likely also to be extended by additional findings that tRNA modifications are removed in vivo in response to stress or other conditions, as shown for AlkBH1 demethylase (Liu et al. 2016), or findings that modification levels have tissue-specific differences due to variability in expression of the modification enzymes.
Third, it seems likely that there will be new surprises revealed about the interplay between tRNA biology and different regulatory or stress response pathways. Previous analysis has documented interactions between the Mod5 i6A modification enzyme and a central enzyme of sterol biosynthesis (Benko et al. 2000), reciprocal interactions between elongator function in xcm5U34 modification and the TORC1 and TORC2 signaling pathways (Candiracci et al. 2019), between xcm5U34 and the proteotoxic stress pathway (Nedialkova and Leidel 2015), several different interactions between the biology of different modifications and the GAAC pathway (Zinshteyn and Gilbert 2013; Chou et al. 2017; Han et al. 2018; De Zoysa and Phizicky 2020), and interactions between modifications such as s2U34 and queuine and metabolic pathways (Laxman et al. 2013; Gupta et al. 2019; Huber et al. 2022). It is likely that more such cross-pathway interactions will be discovered using the sophisticated modern arsenal of methodologies for analysis of transcription, translation, and the proteome.
Fourth, it is highly likely that there will be a huge increase in our knowledge of the biology of tRNA fragments. We cited above several well-studied examples in which tRNA fragments have been shown to inhibit apoptosis (Mei et al. 2010; Saikia et al. 2014), stimulate translation (Fricker et al. 2019), prime retroviral replication (Kikuchi et al. 1986), inhibit protein synthesis (Ivanov et al. 2011; Anderson and Ivanov 2014; Gebetsberger et al. 2017), impair production of siRNAs (Durdevic et al. 2013b), and affect pre-rRNA processing (Couvillion et al. 2012). Based on the large number of tRHs that continue to be found using modern sequencing methods, it is virtually certain that there will be additional insights into their different modes of regulation.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elizabeth Grayhack and members of the Phizicky and Hopper laboratories for valuable discussions during the course of this work. This research was supported by National Institutes of Health (NIH) grants GM052347 to E.M.P. and GM122884 to A.K.H.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079620.123.
Freely available online through the RNA Open Access option.
REFERENCES
- Abad MG, Rao BS, Jackman JE. 2010. Template-dependent 3′–5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc Natl Acad Sci 107: 674–679. 10.1073/pnas.0910961107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad MG, Long Y, Willcox A, Gott JM, Gray MW, Jackman JE. 2011. A role for tRNAHis guanylyltransferase (Thg1)-like proteins from Dictyostelium discoideum in mitochondrial 5′-tRNA editing. RNA 17: 613–623. 10.1261/rna.2517111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K, et al. 2012. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet 90: 847–855. 10.1016/j.ajhg.2012.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Fattah W, Jablonowski D, Di Santo R, Thuring KL, Scheidt V, Hammermeister A, Ten Have S, Helm M, Schaffrath R, Stark MJ. 2015. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet 11: e1004931. 10.1371/journal.pgen.1004931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrahman HA, Al-Shamsi AM, Ali BR, Al-Gazali L. 2018. A null variant in PUS3 confirms its involvement in intellectual disability and further delineates the associated neurodevelopmental disease. Clin Genet 94: 586–587. 10.1111/cge.13443 [DOI] [PubMed] [Google Scholar]
- Akama K, Junker V, Beier H. 2000. Identification of two catalytic subunits of tRNA splicing endonuclease from Arabidopsis thaliana. Gene 257: 177–185. 10.1016/S0378-1119(00)00408-X [DOI] [PubMed] [Google Scholar]
- Alazami AM, Hijazi H, Al-Dosari MS, Shaheen R, Hashem A, Aldahmesh MA, Mohamed JY, Kentab A, Salih MA, Awaji A, et al. 2013. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J Med Genet 50: 425–430. 10.1136/jmedgenet-2012-101378 [DOI] [PubMed] [Google Scholar]
- Alexandrov A, Martzen MR, Phizicky EM. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266. 10.1017/S1355838202024019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Grayhack EJ, Phizicky EM. 2005. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11: 821–830. 10.1261/rna.2030705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96. 10.1016/j.molcel.2005.10.036 [DOI] [PubMed] [Google Scholar]
- Alings F, Sarin LP, Fufezan C, Drexler HC, Leidel SA. 2015. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 21: 202–212. 10.1261/rna.048199.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CS, Vicentini R, Duarte GT, Pinoti VF, Vincentz M, Nogueira FT. 2017. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol Biol 93: 35–48. 10.1007/s11103-016-0545-9 [DOI] [PubMed] [Google Scholar]
- Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, Jia XY, Micura R, Lusser A. 2017. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol 18: 1. 10.1186/s13059-016-1139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y, Yamao F, Muto A, Osawa S. 1989. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol 209: 37–54. 10.1016/0022-2836(89)90168-X [DOI] [PubMed] [Google Scholar]
- Andersen KL, Collins K. 2012. Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol Biol Cell 23: 36–44. 10.1091/mbc.e11-08-0689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Ivanov P. 2014. tRNA fragments in human health and disease. FEBS Lett 588: 4297–4304. 10.1016/j.febslet.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. 1998. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Gene Dev 12: 3650–3662. 10.1101/gad.12.23.3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG. 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 5173–5178. 10.1073/pnas.090102597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova MT, Dimitrova DG, Da Silva B, Marchand V, Jacquier C, Achour C, Brazane M, Goyenvalle C, Bourguignon-Igel V, Shehzada S, et al. 2020. tRNA 2′-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res 48: 2050–2072. 10.1093/nar/gkaa002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Iben J, Wei FY, Rijal K, Tomizawa K, Hafner M, Maraia RJ. 2016. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA 22: 1400–1410. 10.1261/rna.056259.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez JG, Steitz TA. 1994. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. 10.1021/bi00190a008 [DOI] [PubMed] [Google Scholar]
- Arrondel C, Missoury S, Snoek R, Patat J, Menara G, Collinet B, Liger D, Durand D, Gribouval O, Boyer O, et al. 2019. Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat Commun 10: 3967. 10.1038/s41467-019-11951-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW. 1998a. Identification of a nuclear export receptor for tRNA. Curr Biol 8: 305–314. 10.1016/S0960-9822(98)70130-7 [DOI] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW. 1998b. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17: 7430–7441. 10.1093/emboj/17.24.7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 1999. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J Mol Biol 292: 467–483. 10.1006/jmbi.1999.3080 [DOI] [PubMed] [Google Scholar]
- Auxilien S, Crain PF, Trewyn RW, Grosjean H. 1996. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol 262: 437–458. 10.1006/jmbi.1996.0527 [DOI] [PubMed] [Google Scholar]
- Azizi A, SharifiRad A, Enayati S, Azizi M, Bayat M, Khalaj V. 2020. Absence of AfuXpot, the yeast Los1 homologue, limits Aspergillus fumigatus growth under amino acid deprived condition. World J Microbiol Biotechnol 36: 28. 10.1007/s11274-020-2805-8 [DOI] [PubMed] [Google Scholar]
- Baldi MI, Mattoccia E, Bufardeci E, Fabbri S, Tocchini-Valentini GP. 1992. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 255: 1404–1408. 10.1126/science.1542788 [DOI] [PubMed] [Google Scholar]
- Banerjee A, Ghosh S, Goldgur Y, Shuman S. 2019a. Structure and two-metal mechanism of fungal tRNA ligase. Nucleic Acids Res 47: 1428–1439. 10.1093/nar/gky1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Munir A, Abdullahu L, Damha MJ, Goldgur Y, Shuman S. 2019b. Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2′-PO4 recognition and ADP-ribosylation. Nat Commun 10: 218. 10.1038/s41467-018-08211-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Goldgur Y, Shuman S. 2021. Structure of 3′-PO4/5′-OH RNA ligase RtcB in complex with a 5′-OH oligonucleotide. RNA 27: 584–590. 10.1261/rna.078692.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhoom S, Kaur J, Cooperman BS, Smorodinsky NI, Smilansky Z, Ehrlich M, Elroy-Stein O. 2011. Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA. Nucleic Acids Res 39: e129. 10.1093/nar/gkr601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa R, Sigler PB. 1991. The 3 Å crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J 10: 3105–3111. 10.1002/j.1460-2075.1991.tb07864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Muller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. 2012. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res 40: 11648–11658. 10.1093/nar/gks956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. 2003. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite–multisubstrate RNA:ψ-synthase also acting on tRNAs. RNA 9: 1371–1382. 10.1261/rna.5520403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Rousset JP. 2005. An extended signal involved in eukaryotic −1 frameshifting operates through modification of the E site tRNA. Mol Cell 17: 61–68. 10.1016/j.molcel.2004.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko AL, Vaduva G, Martin NC, Hopper AK. 2000. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc Natl Acad Sci 97: 61–66. 10.1073/pnas.97.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA. 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci 74: 3171–3175. 10.1073/pnas.74.8.3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatta A, Dienemann C, Cramer P, Hillen HS. 2021. Structural basis of RNA processing by human mitochondrial RNase P. Nat Struct Mol Biol 28: 713–723. 10.1038/s41594-021-00637-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Wikstrom PM, Bystrom AS. 1989. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244: 986–989. 10.1126/science.2471265 [DOI] [PubMed] [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239. 10.1093/emboj/20.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Huang B, Persson OP, Bystrom AS. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13: 1245–1255. 10.1261/rna.558707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaesius K, Abbasi AA, Tahir TH, Tietze A, Picker-Minh S, Ali G, Farooq S, Hu H, Latif Z, Khan MN, et al. 2018. Mutations in the tRNA methyltransferase 1 gene TRMT1 cause congenital microcephaly, isolated inferior vermian hypoplasia and cystic leukomalacia in addition to intellectual disability. Am J Med Genet A 176: 2517–2521. 10.1002/ajmg.a.38631 [DOI] [PubMed] [Google Scholar]
- Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. 2011. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 7: e1002403. 10.1371/journal.pgen.1002403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. 2014. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33: 2020–2039. 10.15252/embj.201489282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 350: 1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, Kurkowska M, Shirvanizadeh N, Destefanis E, Groza P, et al. 2022. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res 50: D231–D235. 10.1093/nar/gkab1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. 1994. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol 14: 2298–2306. 10.1128/mcb.14.4.2298-2306.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215. 10.1093/emboj/cdf613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Rao J, Mollet G, Schapiro D, Daugeron MC, Tan W, Gribouval O, Boyer O, Revy P, Jobst-Schwan T, et al. 2017. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat Genet 49: 1529–1538. 10.1038/ng.3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuss MW, Sultan T, James KN, Rosti RO, Scott E, Musaev D, Furia B, Reis A, Sticht H, Al-Owain M, et al. 2016. Autosomal-recessive mutations in the tRNA splicing endonuclease subunit TSEN15 cause pontocerebellar hypoplasia and progressive microcephaly. Am J Hum Genet 99: 228–235. 10.1016/j.ajhg.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broly M, Polevoda BV, Awayda KM, Tong N, Lentini J, Besnard T, Deb W, O'Rourke D, Baptista J, Ellard S, et al. 2022. THUMPD1 bi-allelic variants cause loss of tRNA acetylation and a syndromic neurodevelopmental disorder. Am J Hum Genet 109: 587–600. 10.1016/j.ajhg.2022.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes P, Lawley PD. 1962. Methylation of cytosine and cytidine. J Chem Soc 1348–1351. 10.1039/JR9620001348 [DOI] [Google Scholar]
- Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. 2006. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNALeu(CAA). Nucleic Acids Res 34: 6034–6043. 10.1093/nar/gkl765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, et al. 2008. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nature Genet 40: 1113–1118. 10.1038/ng.204 [DOI] [PubMed] [Google Scholar]
- Bujnicki JM. 2001. In silico analysis of the tRNA:m1A58 methyltransferase family: homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett 507: 123–127. 10.1016/S0014-5793(01)02962-3 [DOI] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller EC, Otto A, Kutay U. 2002. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21: 6216–6224. 10.1093/emboj/cdf620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvin K, Xue S, Ellis C, Mitchell MH, Li H. 2008. Probing the catalytic triad of an archaeal RNA splicing endonuclease. Biochemistry 47: 13659–13665. 10.1021/bi801141q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaday J, Dirheimer G, Martin RP. 1980. Yeast mitochondrial methionine initiator tRNA: characterization and nucleotide sequence. Nucleic Acids Res 8: 1445–1457. 10.1093/nar/8.7.1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiracci J, Migeot V, Chionh YH, Bauer F, Brochier T, Russell B, Shiozaki K, Dedon P, Hermand D. 2019. Reciprocal regulation of TORC signaling and tRNA modifications by Elongator enforces nutrient-dependent cell fate. Sci Adv 5: eaav0184. 10.1126/sciadv.aav0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone ML, Solinas M, Sora S, Panzeri L. 1991. A gene tightly linked to CEN6 is important for growth of Saccharomyces cerevisiae. Curr Genet 19: 1–8. 10.1007/BF00362080 [DOI] [PubMed] [Google Scholar]
- Carlson BA, Mushinski JF, Henderson DW, Kwon SY, Crain PF, Lee BJ, Hatfield DL. 2001. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology 279: 130–135. 10.1006/viro.2000.0692 [DOI] [PubMed] [Google Scholar]
- Cartlidge RA, Knebel A, Peggie M, Alexandrov A, Phizicky EM, Cohen P. 2005. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J 24: 1696–1705. 10.1038/sj.emboj.7600648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandrini D, Biancheri R, Tessa A, Di Rocco M, Di Capua M, Bruno C, Denora PS, Sartori S, Rossi A, Nozza P, et al. 2010. Pontocerebellar hypoplasia: clinical, pathologic, and genetic studies. Neurology 75: 1459–1464. 10.1212/WNL.0b013e3181f88173 [DOI] [PubMed] [Google Scholar]
- Castilho BA, Shanmugam R, Silva RC, Ramesh R, Himme BM, Sattlegger E. 2014. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim Biophys Acta 1843: 1948–1968. 10.1016/j.bbamcr.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S. 2012. The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res 40: 8558–8567. 10.1093/nar/gks558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Subbotin R, Chait BT, Shuman S. 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci 109: 6072–6077. 10.1073/pnas.1201207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690. 10.1101/gad.12.11.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. 2010. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247. 10.1371/journal.pgen.1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3: 937. 10.1038/ncomms1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GW, Roth JR, Ames BN. 1971. Histidine regulation in Salmonella typhimurium. 8. Mutations of the hisT gene. J Bacteriol 108: 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49: 341–351. 10.1080/152165400410182 [DOI] [PubMed] [Google Scholar]
- Chatterjee K, Majumder S, Wan Y, Shah V, Wu J, Huang HY, Hopper AK. 2017. Sharing the load: Mex67–Mtr2 cofunctions with Los1 in primary tRNA nuclear export. Genes Dev 31: 2186–2198. 10.1101/gad.305904.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee K, Nostramo RT, Wan Y, Hopper AK. 2018. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: location, location, location. Biochim Biophys Acta Gene Regul Mech 1861: 373–386. 10.1016/j.bbagrm.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee K, Marshall WA, Hopper AK. 2022. Three tRNA nuclear exporters in S. cerevisiae: parallel pathways, preferences, and precision. Nucleic Acids Res 50: 10140–10152. 10.1093/nar/gkac754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wolin SL. 2023. Transfer RNA halves are found as nicked tRNAs in cells: evidence that nicked tRNAs regulate expression of an RNA repair operon. RNA 29: 620–629. 10.1261/rna.079575.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tuck S, Bystrom AS. 2009a. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561. 10.1371/journal.pgen.1000561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Hims MM, Shetty RS, Mull J, Liu L, Leyne M, Slaugenhaupt SA. 2009b. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol Cell Biol 29: 736–744. 10.1128/MCB.01313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Anderson JT, Bystrom AS. 2011a. Unexpected accumulation of ncm5U and ncm5S2U in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5S2U. PLoS One 6: e20783. 10.1371/journal.pone.0020783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Eliasson M, Ryden P, Bystrom AS. 2011b. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet 7: e1002258. 10.1371/journal.pgen.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AW, Jayasinghe MI, Chung CZ, Rao BS, Kenana R, Heinemann IU, Jackman JE. 2019. The role of 3′ to 5′ reverse RNA polymerization in tRNA fidelity and repair. Genes (Basel) 10: 250. 10.3390/genes10030250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Long Q, Borrie MS, Sun H, Zhang C, Yang H, Shi D, Gartenberg MR, Deng W. 2021. Nucleoporin TPR promotes tRNA nuclear export and protein synthesis in lung cancer cells. PLoS Genet 17: e1009899. 10.1371/journal.pgen.1009899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V, Maury LL, Bacikova D, Pridham K, Bahler J, Maraia RJ. 2012. Altered nuclear tRNA metabolism in La-deleted Schizosaccharomyces pombe is accompanied by a nutritional stress response involving Atf1p and Pcr1p that is suppressible by Xpo-t/Los1p. Mol Biol Cell 23: 480–491. 10.1091/mbc.e11-08-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′–3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380. 10.1101/gad.1654308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry PD, White LK, York K, Hesselberth JR. 2018. Genetic bypass of essential RNA repair enzymes in budding yeast. RNA 24: 313–323. 10.1261/rna.061788.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry PD, Peach SE, Hesselberth JR. 2019. Multiple decay events target HAC1 mRNA during splicing to regulate the unfolded protein response. Elife 8: e42262. 10.7554/eLife.42262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, Tanaka I. 2009. RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J 28: 1362–1373. 10.1038/emboj.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ. 2017. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol Cell 68: 978–992.e974. 10.1016/j.molcel.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8. 10.1016/0092-8674(77)90180-5 [DOI] [PubMed] [Google Scholar]
- Chu X, He C, Sang B, Yang C, Yin C, Ji M, Qian A, Tian Y. 2022. Transfer RNAs-derived small RNAs and their application potential in multiple diseases. Front Cell Dev Biol 10: 954431. 10.3389/fcell.2022.954431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintron M, Zeng JM, Barth VC, Cruz JW, Husson RN, Woychik NA. 2019. Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci Rep 9: 5949. 10.1038/s41598-019-41548-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. 2016. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22: 1771–1784. 10.1261/rna.056531.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160. 10.1261/rna.1738409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E. 2009. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65. 10.1038/nature08394 [DOI] [PubMed] [Google Scholar]
- Cooley L, Appel B, Soll D. 1982. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc Natl Acad Sci 79: 6475–6479. 10.1073/pnas.79.21.6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL. 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227. 10.1261/rna.1050408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino C, Toivonen S, Diaz Villamil E, Atta M, Ravanat JL, Demine S, Schiavo AA, Pachera N, Deglasse JP, Jonas JC, et al. 2018. Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res 46: 10302–10318. 10.1093/nar/gky839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa B, Li Calzi M, Castellano M, Blanco V, Cuevasanta E, Litvan I, Ivanov P, Witwer K, Cayota A, Tosar JP. 2023. Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc Natl Acad Sci 120: e2216330120. 10.1073/pnas.2216330120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. 2012. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 48: 509–520. 10.1016/j.molcel.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. 2015. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. 10.1038/nmeth.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristodero M, Brogli R, Joss O, Schimanski B, Schneider A, Polacek N. 2021. tRNA 3′ shortening by LCCR4 as a response to stress in Trypanosoma brucei. Nucleic Acids Res 49: 1647–1661. 10.1093/nar/gkaa1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Zillmann M, Kierzek R, Michaud N, LaReau RD, Turner DH, Phizicky EM. 1993. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1″–2″ cyclic phosphate. Science 261: 206–208. 10.1126/science.8392224 [DOI] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM. 1997. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem 272: 13203–13210. 10.1074/jbc.272.20.13203 [DOI] [PubMed] [Google Scholar]
- Czech A. 2020. Deep sequencing of tRNA's 3′-termini sheds light on CCA-tail integrity and maturation. RNA 26: 199–208. 10.1261/rna.072330.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech A, Wende S, Morl M, Pan T, Ignatova Z. 2013. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet 9: e1003767. 10.1371/journal.pgen.1003767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W, Zhu S, Peng B, Li S, Lai J, et al. 2021. N7-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell 81: 3339–3355.e8. 10.1016/j.molcel.2021.07.003 [DOI] [PubMed] [Google Scholar]
- Damon JR, Pincus D, Ploegh HL. 2015. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol Biol Cell 26: 270–282. 10.1091/mbc.E14-06-1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CJ, Lai LB, Chen TH, Gopalan V. 2019. Both kinds of RNase P in all domains of life: surprises galore. RNA 25: 286–291. 10.1261/rna.068379.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L, Saveanu C, Jacquier A, et al. 2011. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res 39: 6148–6160. 10.1093/nar/gkr178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davarniya B, Hu H, Kahrizi K, Musante L, Fattahi Z, Hosseini M, Maqsoud F, Farajollahi R, Wienker TF, Ropers HH, et al. 2015. The role of a novel TRMT1 gene mutation and rare GRM1 gene defect in intellectual disability in two Azeri families. PLoS One 10: e0129631. 10.1371/journal.pone.0129631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR. 2017. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29: 445–460. 10.1105/tpc.16.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026. 10.1093/nar/23.24.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crecy-Lagard V, Marck C, Grosjean H. 2012. Decoding in Candidatus Riesia pediculicola, close to a minimal tRNA modification set? Trends Cell Mol Biol 7: 11–34. [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259. 10.1038/nature10913 [DOI] [PubMed] [Google Scholar]
- Denmon AP, Wang J, Nikonowicz EP. 2011. Conformation effects of base modification on the anticodon stem-loop of Bacillus subtilis tRNATyr. J Mol Biol 412: 285–303. 10.1016/j.jmb.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Olson MV. 1979. Transcription and processing of cloned yeast tyrosine tRNA genes microinjected into frog oocytes. Nature 278: 137–143. 10.1038/278137a0 [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Black P, Nishikura K. 1981. Intranuclear location of the tRNA splicing enzymes. Cell 23: 89–93. 10.1016/0092-8674(81)90273-7 [DOI] [PubMed] [Google Scholar]
- Desai KK, Bingman CA, Phillips GN Jr, Raines RT. 2013. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry 52: 2518–2525. 10.1021/bi4002375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KK, Cheng CL, Bingman CA, Phillips GN Jr, Raines RT. 2014. A tRNA splicing operon: archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res 42: 3931–3942. 10.1093/nar/gkt1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KK, Beltrame AL, Raines RT. 2015. Coevolution of RtcB and Archease created a multiple-turnover RNA ligase. RNA 21: 1866–1872. 10.1261/rna.052639.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Kim K, Buchsenschutz HC, Chen AW, Bi Y, Mann MR, Turk MA, Chung CZ, Heinemann IU. 2018. Minimal requirements for reverse polymerization and tRNA repair by tRNAHis guanylyltransferase. RNA Biol 15: 614–622. 10.1080/15476286.2017.1372076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. 2000. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904. 10.1093/emboj/19.21.5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Whipple JM, Chernyakov I, Jaramillo LN, Phizicky EM. 2012. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 18: 1886–1896. 10.1261/rna.033654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D. 2017. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol Cell Biol 37: e00214-17. 10.1128/MCB.00214-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. 2008. The conserved Wobble uridine tRNA thiolase Ctu1–Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci 105: 5459–5464. 10.1073/pnas.0709404105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zoysa T, Phizicky EM. 2020. Hypomodified tRNA in evolutionarily distant yeasts can trigger rapid tRNA decay to activate the general amino acid control response, but with different consequences. PLoS Genet 16: e1008893. 10.1371/journal.pgen.1008893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal R, Tong C, Anderson S, Kashina AS, Cooperman B, Bau HH. 2019. Dynamics of intracellular stress-induced tRNA trafficking. Nucleic Acids Res 47: 2002–2010. 10.1093/nar/gky1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhungel N, Hopper AK. 2012. Beyond tRNA cleavage: novel essential function for yeast tRNA splicing endonuclease unrelated to tRNA processing. Genes Dev 26: 503–514. 10.1101/gad.183004.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Stevens A, Tollervey D. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J 16: 7184–7195. 10.1093/emboj/16.23.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. 1987. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol 7: 177–184. 10.1128/MCB.7.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nicola Negri E, Fabbri S, Bufardeci E, Baldi MI, Gandini Attardi D, Mattoccia E, Tocchini-Valentini GP. 1997. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89: 859–866. 10.1016/S0092-8674(00)80271-8 [DOI] [PubMed] [Google Scholar]
- Dodbele S, Moreland B, Gardner SM, Bundschuh R, Jackman JE. 2019. 5′-end sequencing in Saccharomyces cerevisiae offers new insights into 5′-ends of tRNAHis and snoRNAs. FEBS Lett 593: 971–981. 10.1002/1873-3468.13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Rath S, Kolet-Mandrikov D, Korennykh A. 2017. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23: 1660–1671. 10.1261/rna.062000.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J 20: 520–531. 10.1093/emboj/20.3.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drino A, Konig L, Capitanchik C, Sanadgol N, Janisiw E, Rappol T, Vilardo E, Schaefer MR. 2023. Identification of RNA helicases with unwinding activity on angiogenin-processed tRNAs. Nucleic Acids Res 51: 1326–1352. 10.1093/nar/gkad033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans L, Grosjean H. 1987. Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: dependence on the anticodon sequence. EMBO J 6: 477–483. 10.1002/j.1460-2075.1987.tb04778.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva S, Haider SJ, Phizicky EM. 2011. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA 17: 1100–1110. 10.1261/rna.2652611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. 2004. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res 32: 255–262. 10.1093/nar/gkh182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CDS, Rodriguez-Lopez M, Ruis P, Bahler J, Mata J. 2018. General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc Natl Acad Sci 115: E1829–E1838. 10.1073/pnas.1713991115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Davis DR. 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol 285: 115–131. 10.1006/jmbi.1998.2297 [DOI] [PubMed] [Google Scholar]
- Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. 2005. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44: 8078–8089. 10.1021/bi050343f [DOI] [PubMed] [Google Scholar]
- Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, Schaefer M. 2013a. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep 14: 269–275. 10.1038/embor.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. 2013b. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep 4: 931–937. 10.1016/j.celrep.2013.07.046 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22. 10.1038/nsmb1184 [DOI] [PubMed] [Google Scholar]
- Edqvist J, Blomqvist K, Straby KB. 1994. Structural elements in yeast tRNAs required for homologous modification of guanosine-26 into dimethylguanosine-26 by the yeast Trm1 tRNA- modifying enzyme. Biochemistry 33: 9546–9551. 10.1021/bi00198a021 [DOI] [PubMed] [Google Scholar]
- Edvardson S, Elbaz-Alon Y, Jalas C, Matlock A, Patel K, Labbe K, Shaag A, Jackman JE, Elpeleg O. 2016. A mutation in the THG1L gene in a family with cerebellar ataxia and developmental delay. Neurogenetics 17: 219–225. 10.1007/s10048-016-0487-z [DOI] [PubMed] [Google Scholar]
- Edvardson S, Prunetti L, Arraf A, Haas D, Bacusmo JM, Hu JF, Ta-Shma A, Dedon PC, de Crecy-Lagard V, Elpeleg O. 2017. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur J Hum Genet 25: 545–551. 10.1038/ejhg.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE. 2017. Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules 7: 14. 10.3390/biom7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins BN, Keller EB. 1974. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry 13: 4622–4628. 10.1021/bi00719a024 [DOI] [PubMed] [Google Scholar]
- Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC. 1986. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem 261: 9703–9709. 10.1016/S0021-9258(18)67571-4 [DOI] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. 2009. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909. 10.1093/nar/gkp152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D AGM, de Crecy-Lagard V. 2011. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30: 882–893. 10.1038/emboj.2010.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crecy-Lagard V. 2012. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 46: 69–95. 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- Englert M, Beier H. 2005. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res 33: 388–399. 10.1093/nar/gki174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Latz A, Becker D, Gimple O, Beier H, Akama K. 2007. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 89: 1351–1365. 10.1016/j.biochi.2007.06.014 [DOI] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Gundllapalli S, Beier H, Soll D. 2010. Branchiostoma floridae has separate healing and sealing enzymes for 5′-phosphate RNA ligation. Proc Natl Acad Sci 107: 16834–16839. 10.1073/pnas.1011703107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR III, Soll D. 2011. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci 108: 1290–1295. 10.1073/pnas.1018307108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yao M, Eom SH, Konigsberg WH, Soll D, Wang J. 2012. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc Natl Acad Sci 109: 15235–15240. 10.1073/pnas.1213795109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Bystrom AS. 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24: 139–148. 10.1016/j.molcel.2006.07.031 [DOI] [PubMed] [Google Scholar]
- Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola Negri E, Baldi MI, Attardi DG, Mattoccia E, Tocchini-Valentini GP. 1998. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280: 284–286. 10.1126/science.280.5361.284 [DOI] [PubMed] [Google Scholar]
- Fernandez-Vazquez J, Vargas-Perez I, Sanso M, Buhne K, Carmona M, Paulo E, Hermand D, Rodriguez-Gabriel M, Ayte J, Leidel S, et al. 2013. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 9: e1003647. 10.1371/journal.pgen.1003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner L, Frohloff F, Burkner K, Larsen M, Breunig KD, Schaffrath R. 2002. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol 43: 783–791. 10.1046/j.1365-2958.2002.02794.x [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ. 1983. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell 32: 547–557. 10.1016/0092-8674(83)90474-9 [DOI] [PubMed] [Google Scholar]
- Finer-Moore J, Czudnochowski N, O'Connell JD III, Wang AL, Stroud RM. 2015. Crystal structure of the human tRNA m1A58 methyltransferase-tRNA3Lys complex: refolding of substrate tRNA allows access to the methylation target. J Mol Biol 427: 3862–3876. 10.1016/j.jmb.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483. 10.1016/0092-8674(95)90436-0 [DOI] [PubMed] [Google Scholar]
- Fleming IM, Paris Z, Gaston KW, Balakrishnan R, Fredrick K, Rubio MA, Alfonzo JD. 2016. A tRNA methyltransferase paralog is important for ribosome stability and cell division in Trypanosoma brucei. Sci Rep 6: 21438. 10.1038/srep21438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretek D, Nuc P, Zywicki M, Karlowski WM, Kudla G, Boguta M. 2017. Maf1-mediated regulation of yeast RNA polymerase III is correlated with CCA addition at the 3′ end of tRNA precursors. Gene 612: 12–18. 10.1016/j.gene.2016.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060. 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Freude K, Hoffmann K, Jensen LR, Delatycki MB, des Portes V, Moser B, Hamel B, van Bokhoven H, Moraine C, Fryns JP, et al. 2004. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet 75: 305–309. 10.1086/422507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker R, Brogli R, Luidalepp H, Wyss L, Fasnacht M, Joss O, Zywicki M, Helm M, Schneider A, Cristodero M, et al. 2019. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun 10: 118. 10.1038/s41467-018-07949-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. 2001. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J 20: 1993–2003. 10.1093/emboj/20.8.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Bauters M, Boyle J, Van Esch H, Govaerts K, van Bokhoven H, Ropers HH, Moraine C, Chelly J, Fryns JP, et al. 2007. Loss of SLC38A5 and FTSJ1 at Xp11.23 in three brothers with non-syndromic mental retardation due to a microdeletion in an unstable genomic region. Hum Genet 121: 539–547. 10.1007/s00439-007-0343-1 [DOI] [PubMed] [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. 2010. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30: 2449–2459. 10.1128/MCB.01604-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk HM, DiVita DJ, Sizemore HE, Wehrle K, Miller CLW, Fraley ME, Mullins AK, Guy AR, Phizicky EM, Guy MP. 2022. Identification of a Trm732 motif required for 2'-O-methylation of the tRNA anticodon loop by Trm7. ACS Omega 7: 13667–13675. 10.1021/acsomega.1c07231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan M, Delgado-Tejedor A, Mulroney L, Pelizzola M, Novoa EM, Leonardi T. 2021. Computational methods for RNA modification detection from nanopore direct RNA sequencing data. RNA Biol 18: 31–40. 10.1080/15476286.2021.1978215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PD, Leach RW, Wadsworth GM, Choudhary K, Li H, Aviran S, Kim HD, Zakian VA. 2020. Stability and nuclear localization of yeast telomerase depend on protein components of RNase P/MRP. Nat Commun 11: 2173. 10.1038/s41467-020-15875-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazy I, Liefshitz B, Bronstein A, Parnas O, Atias N, Sharan R, Kupiec M. 2013. A genetic screen for high copy number suppressors of the synthetic lethality between elg1Δ and srs2Δ in yeast. G3 (Bethesda) 3: 917–926. 10.1534/g3.113.005561 [DOI] [Google Scholar]
- Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. 2017. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14: 1364–1373. 10.1080/15476286.2016.1257470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S, Rafi M, Aldarmaki M, ElSiddig M, Al Nuaimi M, Amiri KMA. 2022. tRNA derived small RNAs—small players with big roles. Front Genet 13: 997780. 10.3389/fgene.2022.997780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W. 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286: 1146–1149. 10.1126/science.286.5442.1146 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Giege R, Juhling F, Putz J, Stadler P, Sauter C, Florentz C. 2012. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA 3: 37–61. 10.1002/wrna.103 [DOI] [PubMed] [Google Scholar]
- Gillis D, Krishnamohan A, Yaacov B, Shaag A, Jackman JE, Elpeleg O. 2014. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J Med Genet 51: 581–586. 10.1136/jmedgenet-2014-102282 [DOI] [PubMed] [Google Scholar]
- Gillman EC, Slusher LB, Martin NC, Hopper AK. 1991. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol Cell Biol 11: 2382–2390. 10.1128/mcb.11.5.2382-2390.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkatza NA, Castro C, Harvey RF, Heiss M, Popis MC, Blanco S, Bornelov S, Sajini AA, Gleeson JG, Griffin JL, et al. 2019. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol 17: e3000297. 10.1371/journal.pbio.3000297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P. 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744. 10.1038/nsmb.1812 [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514. 10.1146/annurev.biochem.74.010904.153721 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398. 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- Graczyk D, Ciesla M, Boguta M. 2018. Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein. Biochim Biophys Acta Gene Regul Mech 1861: 320–329. 10.1016/j.bbagrm.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, Abelson J. 1983. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell 32: 537–546. 10.1016/0092-8674(83)90473-7 [DOI] [PubMed] [Google Scholar]
- Grosjean H. 2015. RNA modification: the Golden Period 1995–2015. RNA 21: 625–626. 10.1261/rna.049866.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Westhof E. 2016. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 44: 8020–8040. 10.1093/nar/gkw608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Szweykowska-Kulinska Z, Motorin Y, Fasiolo F, Simos G. 1997. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie 79: 293–302. 10.1016/S0300-9084(97)83517-1 [DOI] [PubMed] [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. 2003. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 17: 2889–2901. 10.1101/gad.1148603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Seraphin B, Steinmetz LM, Jacquier A, Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48: 409–421. 10.1016/j.molcel.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. 10.1016/0092-8674(83)90117-4 [DOI] [PubMed] [Google Scholar]
- Gupta R, Laxman S. 2020. tRNA wobble-uridine modifications as amino acid sensors and regulators of cellular metabolic state. Curr Genet 66: 475–480. 10.1007/s00294-019-01045-y [DOI] [PubMed] [Google Scholar]
- Gupta R, Walvekar AS, Liang S, Rashida Z, Shah P, Laxman S. 2019. A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife 8: e44795. 10.7554/eLife.44795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Ronne H. 2008. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 14: 666–674. 10.1261/rna.966208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann B, Gobert A, Giege P. 2012. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev 26: 1022–1027. 10.1101/gad.189514.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Phizicky EM. 2015. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 21: 61–74. 10.1261/rna.047639.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Podyma BM, Preston MA, Shaheen HH, Krivos KL, Limbach PA, Hopper AK, Phizicky EM. 2012. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 18: 1921–1933. 10.1261/rna.035287.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Young DL, Payea MJ, Zhang X, Kon Y, Dean KM, Grayhack EJ, Mathews DH, Fields S, Phizicky EM. 2014. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev 28: 1721–1732. 10.1101/gad.245936.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Shaw M, Weiner CL, Hobson L, Stark Z, Rose K, Kalscheuer VM, Gecz J, Phizicky EM. 2015. Defects in tRNA anticodon loop 2′-O-methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum Mutat 36: 1176–1187. 10.1002/humu.22897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Bellodi C. 2020. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol 17: 1214–1222. 10.1080/15476286.2020.1732694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M, et al. 2018. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173: 1204–1216.e1226. 10.1016/j.cell.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Han L, Phizicky EM. 2018. A rationale for tRNA modification circuits in the anticodon loop. RNA 24: 1277–1284. 10.1261/rna.067736.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Zamore PD. 2014. piRNAs. Curr Biol 24: R730–R733. 10.1016/j.cub.2014.07.037 [DOI] [PubMed] [Google Scholar]
- Han L, Kon Y, Phizicky EM. 2015. Functional importance of ψ38 and ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 21: 188–201. 10.1261/rna.048173.114 [DOI] [Google Scholar]
- Han L, Marcus E, D′Silva S, Phizicky EM. 2017. S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. RNA 23: 406–419. 10.1261/rna.059667.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Guy MP, Kon Y, Phizicky EM. 2018. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet 14: e1007288. 10.1371/journal.pgen.1007288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. 2013. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495: 474–480. 10.1038/nature11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D. 2008. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA 14: 225–232. 10.1261/rna.859908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163: 1515–1526. 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695. 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Mori S, Suzuki T, Suzuki T, Yoshihisa T. 2019. Impact of intron removal from tRNA genes on Saccharomyces cerevisiae. Nucleic Acids Res 47: 5936–5949. 10.1093/nar/gkz270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayne CK, Schmidt CA, Haque MI, Matera AG, Stanley RE. 2020. Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage. Nucleic Acids Res 48: 7609–7622. 10.1093/nar/gkaa438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayne CK, Lewis TA, Stanley RE. 2022. Recent insights into the structure, function, and regulation of the eukaryotic transfer RNA splicing endonuclease complex. Wiley Interdiscip Rev RNA 13: e1717. 10.1002/wrna.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayne CK, Butay KJU, Stewart ZD, Krahn JM, Perera L, Williams JG, Petrovitch RM, Deterding LJ, Matera AG, Borgnia MJ, et al. 2023. Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex. Nat Struct Mol Biol 10.1038/s41594-023-00991-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedusova E, Kulkarni S, Burgman B, Alfonzo JD, Paris Z. 2019. The general mRNA exporters Mex67 and Mtr2 play distinct roles in nuclear export of tRNAs in Trypanosoma brucei. Nucleic Acids Res 47: 8620–8631. 10.1093/nar/gkz671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Soll D. 2009. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci 106: 21103–21108. 10.1073/pnas.0912072106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, Randau L, Tomko RJ Jr, Soll D. 2010. 3′–5′ tRNAHis guanylyltransferase in bacteria. FEBS Lett 584: 3567–3572. 10.1016/j.febslet.2010.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386. 10.1128/MCB.18.11.6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Alfonzo JD. 2014. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 21: 174–185. 10.1016/j.chembiol.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Attardi G. 2004. Nuclear control of cloverleaf structure of human mitochondrial tRNALys. J Mol Biol 337: 545–560. 10.1016/j.jmb.2004.01.036 [DOI] [PubMed] [Google Scholar]
- Helm M, Brule H, Degoul F, Cepanec C, Leroux JP, Giege R, Florentz C. 1998. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res 26: 1636–1643. 10.1093/nar/26.7.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Giege R, Florentz C. 1999. A Watson–Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38: 13338–13346. 10.1021/bi991061g [DOI] [PubMed] [Google Scholar]
- Heyer WD, Thuriaux P, Kohli J, Ebert P, Kersten H, Gehrke C, Kuo KC, Agris PF. 1984. An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the “wobble” base in the anticodon of tRNAs. J Biol Chem 259: 2856–2862. 10.1016/S0021-9258(17)43226-1 [DOI] [PubMed] [Google Scholar]
- Hickey FB, Corcoran JB, Griffin B, Bhreathnach U, Mortiboys H, Reid HM, Andrews D, Byrne S, Furlong F, Martin F, et al. 2014. IHG-1 increases mitochondrial fusion and bioenergetic function. Diabetes 63: 4314–4325. 10.2337/db13-1256 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- Hirata A. 2019. Recent insights into the structure, function, and evolution of the RNA-splicing endonucleases. Front Genet 10: 103. 10.3389/fgene.2019.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A, Kitajima T, Hori H. 2011. Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res 39: 9376–9389. 10.1093/nar/gkr615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A, Okada K, Yoshii K, Shiraishi H, Saijo S, Yonezawa K, Shimizu N, Hori H. 2019. Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition. Nucleic Acids Res 47: 10942–10955. 10.1093/nar/gkz856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Abelson J. 1988. Testing for intron function in the essential Saccharomyces cerevisiae tRNASerUCG gene. J Mol Biol 202: 667–672. 10.1016/0022-2836(88)90295-1 [DOI] [PubMed] [Google Scholar]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. 1958. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231: 241–257. 10.1016/S0021-9258(19)77302-5 [DOI] [PubMed] [Google Scholar]
- Holmes AD, Chan PP, Chen Q, Ivanov P, Drouard L, Polacek N, Kay MA, Lowe TM. 2023. A standardized ontology for naming tRNA-derived RNAs based on molecular origin. Nat Methods 20: 627–628. 10.1038/s41592-023-01813-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. 2008. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474. 10.1016/j.cell.2008.09.013 [DOI] [PubMed] [Google Scholar]
- Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Banks F, Evangelides V. 1978. A yeast mutant which accumulates precursor tRNAs. Cell 14: 211–219. 10.1016/0092-8674(78)90108-3 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA. 1980. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751. 10.1016/S0092-8674(80)80050-X [DOI] [PubMed] [Google Scholar]
- Hopper AK, Furukawa AH, Pham HD, Martin NC. 1982. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell 28: 543–550. 10.1016/0092-8674(82)90209-4 [DOI] [PubMed] [Google Scholar]
- Hou J, Li Q, Wang J, Lu W. 2022. tRFs and tRNA halves: novel cellular defenders in multiple biological processes. Curr Issues Mol Biol 44: 5949–5962. 10.3390/cimb44120405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep 7: 205–211. 10.1038/sj.embor.7400612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. 10.1016/j.cell.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539. 10.1038/nrm1964 [DOI] [PubMed] [Google Scholar]
- Howell NW, Jora M, Jepson BF, Limbach PA, Jackman JE. 2019. Distinct substrate specificities of the human tRNA methyltransferases TRMT10A and TRMT10B. RNA 25: 1366–1376. 10.1261/rna.072090.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Hopper AK. 2014. Separate responses of karyopherins to glucose and amino acid availability regulate nucleocytoplasmic transport. Mol Biol Cell 25: 2840–2852. 10.1091/mbc.e14-04-0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Hopper AK. 2015. In vivo biochemical analyses reveal distinct roles of β-importins and eEF1A in tRNA subcellular traffic. Genes Dev 29: 772–783. 10.1101/gad.258293.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Hopper AK. 2016. Multiple layers of stress-induced regulation in tRNA biology. Life (Basel) 6: 16. 10.3390/life6020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS. 2005. An early step in wobble uridine tRNA modification requires the elongator complex. RNA 11: 424–436. 10.1261/rna.7247705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Bayfield MA, Intine RV, Maraia RJ. 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol 13: 611–618. 10.1038/nsmb1110 [DOI] [PubMed] [Google Scholar]
- Huang B, Lu J, Bystrom AS. 2008. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14: 2183–2194. 10.1261/rna.1184108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SM, Begley U, Sarkar A, Gasperi W, Davis ET, Surampudi V, Lee M, Melendez JA, Dedon PC, Begley TJ. 2022. Arsenite toxicity is regulated by queuine availability and oxidation-induced reprogramming of the human tRNA epitranscriptome. Proc Natl Acad Sci 119: e2123529119. 10.1073/pnas.2123529119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS. 2003. PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol 132: 2135–2143. 10.1104/pp.103.023309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S, Stroud RM. 2007. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell 26: 189–203. 10.1016/j.molcel.2007.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt DJ, Wang SS, Lin YH, Hopper AK. 1987. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol 7: 1208–1216. 10.1128/mcb.7.3.1208-1216.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtig JE, Steiger MA, Nagarajan VK, Li T, Chao TC, Tsai KL, van Hoof A. 2021. Comparative parallel analysis of RNA ends identifies mRNA substrates of a tRNA splicing endonuclease-initiated mRNA decay pathway. Proc Natl Acad Sci 118: e2020429118. 10.1073/pnas.2020429118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurto RL, Tong AH, Boone C, Hopper AK. 2007. Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics 176: 841–852. 10.1534/genetics.106.069732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M. 2013. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 33: 1561–1570. 10.1128/MCB.01523-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. 2010. tRNAHis guanylyltransferase (THG1), a unique 3′–5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′–3′ DNA polymerases. Proc Natl Acad Sci 107: 20305–20310. 10.1073/pnas.1010436107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoillo-Esteve M, Genin A, Lambert N, Desir J, Pirson I, Abdulkarim B, Simonis N, Drielsma A, Marselli L, Marchetti P, et al. 2013. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet 9: e1003888. 10.1371/journal.pgen.1003888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, Kitahara K, Suzuki T. 2008. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J 27: 2194–2203. 10.1038/emboj.2008.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. 2014. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345: 455–459. 10.1126/science.1249749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D, Frohloff F, Fichtner L, Stark MJ, Schaffrath R. 2001. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol Microbiol 42: 1095–1105. 10.1046/j.1365-2958.2001.02705.x [DOI] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM. 2006a. tRNAHis guanylyltransferase adds G–1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12: 1007–1014. 10.1261/rna.54706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM. 2006b. tRNAHis guanylyltransferase catalyzes a 3′–5′ polymerization reaction that is distinct from G–1 addition. Proc Natl Acad Sci 103: 8640–8645. 10.1073/pnas.0603068103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Montange RK, Malik HS, Phizicky EM. 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9: 574–585. 10.1261/rna.5070303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Gott JM, Gray MW. 2012. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA 18: 886–899. 10.1261/rna.032300.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D, Pande S. 1991. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. II. Catalytic mechanism. J Biol Chem 266: 22832–22836. 10.1016/S0021-9258(18)54429-X [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. 2014. The eukaryotic RNA exosome. Curr Op Struct Biol 24: 132–140. 10.1016/j.sbi.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N. 2017. Roles of RNase P and its subunits. Trends Genet 33: 594–603. 10.1016/j.tig.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Jarrous N, Gopalan V. 2010. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res 38: 7885–7894. 10.1093/nar/gkq701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn J, Treml J, Wulsch S, Ottum B, Erb V, Hewel C, Kooijmans RN, Wester L, Fast I, Rosenkranz D. 2020. 5′ tRNA halves are highly expressed in the primate hippocampus and might sequence-specifically regulate gene expression. RNA 26: 694–707. 10.1261/rna.073395.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A, Ehrenhofer-Murray A, Jurkowski TP, Lyko F, Reuter G, Ankri S, Nellen W, Schaefer M, Helm M. 2017. Mechanism and biological role of Dnmt2 in nucleic acid methylation. RNA Biol 14: 1108–1123. 10.1080/15476286.2016.1191737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LR, Garrett L, Holter SM, Rathkolb B, Racz I, Adler T, Prehn C, Hans W, Rozman J, Becker L, et al. 2019. A mouse model for intellectual disability caused by mutations in the X-linked 2′-O-methyltransferase Ftsj1 gene. Biochim Biophys Acta Mol Basis Dis 1865: 2083–2093. 10.1016/j.bbadis.2018.12.011 [DOI] [PubMed] [Google Scholar]
- Johansson MJ, Bystrom AS. 2004. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10: 712–719. 10.1261/rna.5198204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. 2008. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28: 3301–3312. 10.1128/MCB.01542-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PF, Abelson J. 1983. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature 302: 681–687. 10.1038/302681a0 [DOI] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, Martinez J. 2014. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 33: 2922–2936. 10.15252/embj.201490332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. 2008. Human DNMT2 methylates tRNAAsp molecules using a DNA methyltransferase-like catalytic mechanism. RNA 14: 1663–1670. 10.1261/rna.970408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240. 10.1101/gad.1183804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521. 10.1261/rna.2305406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor HR, Clarke S. 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 23: 9283–9292. 10.1128/MCB.23.24.9283-9292.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor HR, Penjwini M, Clarke S. 2005. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem Biophys Res Comm 334: 433–440. 10.1016/j.bbrc.2005.06.111 [DOI] [PubMed] [Google Scholar]
- Kaneta A, Fujishima K, Morikazu W, Hori H, Hirata A. 2018. The RNA-splicing endonuclease from the euryarchaeaon Methanopyrus kandleri is a heterotetramer with constrained substrate specificity. Nucleic Acids Res 46: 1958–1972. 10.1093/nar/gky003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, et al. 2014. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157: 636–650. 10.1016/j.cell.2014.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsborn T, Tukenmez H, Chen C, Bystrom AS. 2014a. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem Biophys Res Comm 454: 441–445. 10.1016/j.bbrc.2014.10.116 [DOI] [PubMed] [Google Scholar]
- Karlsborn T, Tukenmez H, Mahmud AK, Xu F, Xu H, Bystrom AS. 2014b. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol 11: 1519–1528. 10.4161/15476286.2014.992276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnahl U, Wasternack C. 1992. Half-life of cytoplasmic rRNA and tRNA, of plastid rRNA and of uridine nucleotides in heterotrophically and photoorganotrophically grown cells of Euglena gracilis and its apoplastic mutant W3BUL. Int J Biochem 24: 493–497. 10.1016/0020-711X(92)90044-2 [DOI] [PubMed] [Google Scholar]
- Keam SP, Hutvagner G. 2015. tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel) 5: 1638–1651. 10.3390/life5041638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Corbett AH. 2009. Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 10: 1199–1208. 10.1111/j.1600-0854.2009.00944.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempenaers M, Roovers M, Oudjama Y, Tkaczuk KL, Bujnicki JM, Droogmans L. 2010. New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res 38: 6533–6543. 10.1093/nar/gkq451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler AC, Kulkarni SS, Paulines MJ, Rubio MAT, Limbach PA, Paris Z, Alfonzo JD. 2018. Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. brucei. RNA Biol 15: 528–536. 10.1080/15476286.2017.1377878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, et al. 2012. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet 90: 856–863. 10.1016/j.ajhg.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. 2013. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet 9: e1003498. 10.1371/journal.pgen.1003498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikovska E, Svard SG, Kirsebom LA. 2007. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci 104: 2062–2067. 10.1073/pnas.0607326104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Ando Y, Shiba T. 1986. Unusual priming mechanism of RNA-directed DNA synthesis in copia retrovirus-like particles of Drosophila. Nature 323: 824–826. 10.1038/323824a0 [DOI] [PubMed] [Google Scholar]
- Kim KQ, Zaher HS. 2022. Canary in a coal mine: collided ribosomes as sensors of cellular conditions. Trends Biochem Sci 47: 82–97. 10.1016/j.tibs.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. 1974a. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435–440. 10.1126/science.185.4149.435 [DOI] [PubMed] [Google Scholar]
- Kim SH, Sussman JL, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC, Rich A. 1974b. The general structure of transfer RNA molecules. Proc Natl Acad Sci 71: 4970–4974. 10.1073/pnas.71.12.4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lane WS, Reinberg D. 2002. Human elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci 99: 1241–1246. 10.1073/pnas.251672198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. 2010a. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623. 10.1038/nbt.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Johnson W, Chen C, Sewell AK, Bystrom AS, Han M. 2010b. Allele-specific suppressors of lin-1(R175Opal) identify functions of MOC-3 and DPH-3 in tRNA modification complexes in Caenorhabditis elegans. Genetics 185: 1235–1247. 10.1534/genetics.110.118406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. 2017. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552: 57–62. 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Xu J, Chu K, Park H, Jang H, Li P, Valdmanis PN, Zhang QC, Kay MA. 2019. A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice. Cell Rep 29: 3816–3824.e3814. 10.1016/j.celrep.2019.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Yeom JH, Kay MA. 2020. Transfer RNA-derived small RNAs: another layer of gene regulation and novel targets for disease therapeutics. Mol Ther 28: 2340–2357. 10.1016/j.ymthe.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Paluszynski JP, Wemhoff S, Pfeiffer A, Fricke J, Meinhardt F. 2008. The primary target of the killer toxin from Pichia acaciae is tRNAGln. Mol Microbiol 69: 681–697. 10.1111/j.1365-2958.2008.06319.x [DOI] [PubMed] [Google Scholar]
- Knapp G, Beckmann JS, Johnson PF, Fuhrman SA, Abelson J. 1978. Transcription and processing of intervening sequences in yeast tRNA genes. Cell 14: 221–236. 10.1016/0092-8674(78)90109-5 [DOI] [PubMed] [Google Scholar]
- Knapp G, Ogden RC, Peebles CL, Abelson J. 1979. Splicing of yeast tRNA precursors: structure of the reaction intermediates. Cell 18: 37–45. 10.1016/0092-8674(79)90351-9 [DOI] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. 2010. Eukaryotic initiator tRNA: finely tuned and ready for action. FEBS Lett 584: 396–404. 10.1016/j.febslet.2009.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24: 2411–2415. 10.1093/nar/24.12.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner A, Soll D. 1974. N-(purin-6-ylcarbamoyl)threonine: biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett 39: 301–306. 10.1016/0014-5793(74)80135-3 [DOI] [PubMed] [Google Scholar]
- Kotelawala L, Grayhack EJ, Phizicky EM. 2008. Identification of yeast tRNA Um44 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA 14: 158–169. 10.1261/rna.811008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Hopper AK. 2013. Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc Natl Acad Sci 110: 21042–21047. 10.1073/pnas.1316579110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, Yim DG, Lakshmanan V, Tirumalai V, Koh JL, Park JE, Cheong JK, Low JL, Lim MJ, Sze SK, et al. 2019. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep 20: e47789. 10.15252/embr.201947789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamohan A, Jackman JE. 2017. Mechanistic features of the atypical tRNA m1G9 SPOUT methyltransferase, Trm10. Nucleic Acids Res 45: 9019–9029. 10.1093/nar/gkx620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamohan A, Jackman JE. 2019. A family divided: distinct structural and mechanistic features of the SpoU-TrmD (SPOUT) methyltransferase superfamily. Biochemistry 58: 336–345. 10.1021/acs.biochem.8b01047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutyholowa R, Hammermeister A, Zabel R, Abdel-Fattah W, Reinhardt-Tews A, Helm M, Stark MJR, Breunig KD, Schaffrath R, Glatt S. 2019. Kti12, a PSTK-like tRNA dependent ATPase essential for tRNA modification by Elongator. Nucleic Acids Res 47: 4814–4830. 10.1093/nar/gkz190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, Wilusz JE, Zheng Y, Beal PA, Joshua-Tor L. 2015. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 160: 644–658. 10.1016/j.cell.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A. 2014. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12: 78. 10.1186/s12915-014-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, Dutta A. 2016. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci 41: 679–689. 10.1016/j.tibs.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. 2008. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U•G wobble pairing during decoding. J Biol Chem 283: 18801–18811. 10.1074/jbc.M800233200 [DOI] [PubMed] [Google Scholar]
- Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. 2018. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24: 1093–1105. 10.1261/rna.066126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D. 1998. Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369. 10.1016/S1097-2765(00)80036-2 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Ladner JE, Schweizer MP. 1974. Effects of dilute HCl on yeast tRNAPhe and E. coli tRNA1fMet. Nucleic Acids Res 1: 183–192. 10.1093/nar/1.2.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Maraia RJ. 2011. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA 17: 1846–1857. 10.1261/rna.2628611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Crawford AK, Cherkasova VA, Iben JR, Begley TJ, Farabaugh PJ, Maraia RJ. 2013. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol 33: 2918–2929. 10.1128/MCB.00278-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Arimbasseri AG, Rijal K, Iben JR, Wei FY, Tomizawa K, Maraia RJ. 2016. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA 22: 583–596. 10.1261/rna.054064.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Tan M, Zhang Y, Niu S, Chen J, Shi S, Qiu S, Wang X, Peng X, Cai G, et al. 2018. Structural insight into precursor tRNA processing by yeast ribonuclease P. Science 362: eaat6678. 10.1126/science.aat6678 [DOI] [PubMed] [Google Scholar]
- Lari A, Arul Nambi Rajan A, Sandhu R, Reiter T, Montpetit R, Young BP, Loewen CJ, Montpetit B. 2019. A nuclear role for the DEAD-box protein Dbp5 in tRNA export. Elife 8: e48410. 10.7554/eLife.48410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Fire AZ, RajBhandary UL, Sharp PA. 1983. Characterization of tRNA precursor splicing in mammalian extracts. J Biol Chem 258: 11974–11980. 10.1016/S0021-9258(17)44327-4 [DOI] [PubMed] [Google Scholar]
- Laten H, Gorman J, Bock RM. 1978. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res 5: 4329–4342. 10.1093/nar/5.11.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauhon CT. 2012. Mechanism of N6-threonylcarbamoyladenonsine (t6A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry 51: 8950–8963. 10.1021/bi301233d [DOI] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. 2013. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154: 416–429. 10.1016/j.cell.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Rossmanith W, Hartmann RK, Tholken C, Gutmann B, Giege P, Gobert A. 2015. Distribution of ribonucleoprotein and protein-only RNase P in eukarya. Mol Biol Evol 32: 3186–3193. 10.1093/molbev/msv187 [DOI] [PubMed] [Google Scholar]
- Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H. 1998. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of ψ38 and ψ39 in tRNA anticodon loop. J Biol Chem 273: 1316–1323. 10.1074/jbc.273.3.1316 [DOI] [Google Scholar]
- Lecointe F, Namy O, Hatin I, Simos G, Rousset JP, Grosjean H. 2002. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem 277: 30445–30453. 10.1074/jbc.M203456200 [DOI] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC. 2009. A sequence element that tunes Escherichia coli tRNAAlaGGC to ensure accurate decoding. Nat Struct Mol Biol 16: 359–364. 10.1038/nsmb.1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749. 10.1074/jbc.M510356200 [DOI] [PubMed] [Google Scholar]
- Lee MC, Knapp G. 1985. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem 260: 3108–3115. 10.1016/S0021-9258(18)89479-0 [DOI] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR. 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol 11: 721–730. 10.1128/mcb.11.2.721-730.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Kramer G, Graham DE, Appling DR. 2007. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J Biol Chem 282: 27744–27753. 10.1074/jbc.M704572200 [DOI] [PubMed] [Google Scholar]
- Lee YH, Lo YT, Chang CP, Yeh CS, Chang TH, Chen YW, Tseng YK, Wang CC. 2019. Naturally occurring dual recognition of tRNAHis substrates with and without a universal identity element. RNA Biol 16: 1275–1285. 10.1080/15476286.2019.1626663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458: 228–232. 10.1038/nature07643 [DOI] [PubMed] [Google Scholar]
- Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, Barnes T, Hekimi S. 2001. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics 159: 147–157. 10.1093/genetics/159.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, Krasilnikov AS, Wellinger RJ. 2016. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell 165: 1171–1181. 10.1016/j.cell.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini JM, Alsaif HS, Faqeih E, Alkuraya FS, Fu D. 2020. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat Commun 11: 2510. 10.1038/s41467-020-16321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini JM, Bargabos R, Chen C, Fu D. 2022. Methyltransferase METTL8 is required for 3-methylcytosine modification in human mitochondrial tRNAs. J Biol Chem 298: 101788. 10.1016/j.jbc.2022.101788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. 2008. Structure of the yeast tRNA m7G methylation complex. Structure 16: 52–61. 10.1016/j.str.2007.10.025 [DOI] [PubMed] [Google Scholar]
- Li J, Chen X. 2003. PAUSED, a putative exportin-t, acts pleiotropically in Arabidopsis development but is dispensable for viability. Plant Physiol 132: 1913–1924. 10.1104/pp.103.023291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. 1996. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86: 503–512. 10.1016/S0092-8674(00)80123-3 [DOI] [PubMed] [Google Scholar]
- Li S, Sprinzl M. 2006. Interaction of immobilized human exportin-t with calf liver tRNA. RNA Biol 3: 145–149. 10.4161/rna.3.4.3679 [DOI] [PubMed] [Google Scholar]
- Li JM, Hopper AK, Martin NC. 1989. N2,N2-dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J Cell Biol 109: 1411–1419. 10.1083/jcb.109.4.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Trotta CR, Abelson J. 1998. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280: 279–284. 10.1126/science.280.5361.279 [DOI] [PubMed] [Google Scholar]
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. 2009. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet 5: e1000684. 10.1371/journal.pgen.1000684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. 2012. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 40: 6787–6799. 10.1093/nar/gks307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xiong Y, Lai LB, Zhang K, Li Z, Kang H, Dai L, Gopalan V, Wang GL, Liu W. 2021. The rice RNase P protein subunit Rpp30 confers broad-spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol J 19: 1988–1999. 10.1111/pbi.13612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Su S, Gao Y, Lu G, Liu H, Chen X, Shao Z, Zhang Y, Shao Q, Zhao X, et al. 2022. Crystal structures and insights into precursor tRNA 5′-end processing by prokaryotic minimal protein-only RNase P. Nature Comm 13: 2290. 10.1038/s41467-022-30072-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang L, Hahn Q, Nowak RP, Viennet T, Orellana EA, Roy Burman SS, Yue H, Hunkeler M, Fontana P, et al. 2023. Structural basis of regulated m7G tRNA modification by METTL1-WDR4. Nature 613: 391–397. 10.1038/s41586-022-05566-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Shen L, Jang CW, Falnes PO, Zhang Y. 2013. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet 9: e1003516. 10.1371/journal.pgen.1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Smibert P, Zhao X, Hu JF, Ramroop J, Kellner SM, Benton MA, Govind S, Dedon PC, Sternglanz R, et al. 2015. An extensive allelic series of Drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA 21: 2103–2118. 10.1261/rna.053934.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. 2018. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell 71: 244–255.e245. 10.1016/j.molcel.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H, Gorlich D. 1999. Coordination of tRNA nuclear export with processing of tRNA. RNA 5: 539–549. 10.1017/S1355838299982134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippai M, Tirian L, Boros I, Mihaly J, Erdelyi M, Belecz I, Mathe E, Posfai J, Nagy A, Udvardy A, et al. 2000. The Ketel gene encodes a Drosophila homologue of importin-β. Genetics 156: 1889–1900. 10.1093/genetics/156.4.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Straby KB. 2000. The human tRNA(m22G26)dimethyltransferase: functional expression and characterization of a cloned hTRM1 gene. Nucleic Acids Res 28: 3445–3451. 10.1093/nar/28.18.3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. 2016. ALKBH1-mediated tRNA demethylation regulates translation. Cell 167: 816–828.e816. 10.1016/j.cell.2016.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. 2019. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10: 4079. 10.1038/s41467-019-11713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen R, Sun Y, Chen R, Zhou J, Tian Q, Tao X, Zhang Z, Luo GZ, Xie W. 2020. Crystal structure of the yeast heterodimeric ADAT2/3 deaminase. BMC Biol 18: 189. 10.1186/s12915-020-00920-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Kim HK, Xu J, Jing Y, Kay MA. 2021. The 3′tsRNAs are aminoacylated: implications for their biogenesis. PLoS Genet 17: e1009675. 10.1371/journal.pgen.1009675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW. 1993. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science 259: 812–816. 10.1126/science.8430334 [DOI] [PubMed] [Google Scholar]
- Long Y, Abad MG, Olson ED, Carrillo EY, Jackman JE. 2016. Identification of distinct biological functions for four 3′–5′ RNA polymerases. Nucleic Acids Res 44: 8395–8406. 10.1093/nar/gkw681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RR, Silveira Gde O, Eitler R, Vidal RS, Kessler A, Hinger S, Paris Z, Alfonzo JD, Polycarpo C. 2016. The essential function of the Trypanosoma brucei Trl1 homolog in procyclic cells is maturation of the intron-containing tRNATyr. RNA 22: 1190–1199. 10.1261/rna.056242.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11: 1648–1654. 10.1261/rna.2172105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liang FX, Wang X. 2014. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell 55: 758–770. 10.1016/j.molcel.2014.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. 2015. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 21: 1554–1565. 10.1261/rna.052944.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. 10.1016/j.molcel.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Lubas M, Andersen PR, Schein A, Dziembowski A, Kudla G, Jensen TH. 2015. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep 10: 178–192. 10.1016/j.celrep.2014.12.026 [DOI] [PubMed] [Google Scholar]
- Luhtala N, Parker R. 2012. Structure-function analysis of Rny1 in tRNA cleavage and growth inhibition. PLoS One 7: e41111. 10.1371/journal.pone.0041111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085. 10.1126/science.282.5396.2082 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. 2004. Nuclear export of microRNA precursors. Science 303: 95–98. 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- Luo S, He F, Luo J, Dou S, Wang Y, Guo A, Lu J. 2018. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res 46: 5250–5268. 10.1093/nar/gky189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra A, Swinehart W, Bayooz S, Phan P, Stec B, Iwata-Reuyl D, Swairjo MA. 2018. Structure and mechanism of a bacterial t6A biosynthesis system. Nucleic Acids Res 46: 1395–1411. 10.1093/nar/gkx1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra A, Paranagama N, Swinehart W, Bayooz S, Phan P, Quach V, Schiffer JM, Stec B, Iwata-Reuyl D, Swairjo MA. 2019. Conformational communication mediates the reset step in t6A biosynthesis. Nucleic Acids Res 47: 6551–6567. 10.1093/nar/gkz439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Fay MM, Ivanov P. 2018. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett 592: 2828–2844. 10.1002/1873-3468.13205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macari F, El-Houfi Y, Boldina G, Xu H, Khoury-Hanna S, Ollier J, Yazdani L, Zheng G, Bieche I, Legrand N, et al. 2016. TRM6/61 connects PKC α with translational control through tRNAiMet stabilization: impact on tumorigenesis. Oncogene 35: 1785–1796. 10.1038/onc.2015.244 [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. 2014. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol 11: 1619–1629. 10.4161/15476286.2014.992273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Bayfield MA. 2006. The La protein-RNA complex surfaces. Mol Cell 21: 149–152. 10.1016/j.molcel.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Marck C, Grosjean H. 2002. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189–1232. 10.1017/S1355838202022021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al-Gazali L, Gleeson JG. 2012. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet 49: 380–385. 10.1136/jmedgenet-2011-100686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Choudury SG, Slotkin RK. 2017. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45: 5142–5152. 10.1093/nar/gkx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286: 1153–1155. 10.1126/science.286.5442.1153 [DOI] [PubMed] [Google Scholar]
- Masuda I, Hwang JY, Christian T, Maharjan S, Mohammad F, Gamper H, Buskirk AR, Hou YM. 2021. Loss of N1-methylation of G37 in tRNA induces ribosome stalling and reprograms gene expression. Elife 10: e70619. 10.7554/eLife.70619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Toyooka T, Tomikawa C, Ochi A, Takano Y, Takayanagi N, Endo Y, Hori H. 2007. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8–Trm82 complex). FEBS Lett 581: 1599–1604. 10.1016/j.febslet.2007.03.023 [DOI] [PubMed] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci 110: 1404–1409. 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazauric MH, Dirick L, Purushothaman SK, Bjork GR, Lapeyre B. 2010. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 285: 18505–18515. 10.1074/jbc.M110.113100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith SM, Phizicky EM. 1991. An enzyme from Saccharomyces cerevisiae uses NAD+ to transfer the splice junction 2′-phosphate from ligated tRNA to an acceptor molecule. J Biol Chem 266: 11986–11992. 10.1016/S0021-9258(18)99054-X [DOI] [PubMed] [Google Scholar]
- McKenney KM, Rubio MAT, Alfonzo JD. 2018. Binding synergy as an essential step for tRNA editing and modification enzyme codependence in Trypanosoma brucei. RNA 24: 56–66. 10.1261/rna.062893.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megel C, Hummel G, Lalande S, Ubrig E, Cognat V, Morelle G, Salinas-Giege T, Duchene AM, Marechal-Drouard L. 2019. Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res 47: 941–952. 10.1093/nar/gky1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Schaffrath R. 2003. Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactis zymocin in budding yeast. Mol Genet Genomics 269: 188–196. 10.1007/s00438-003-0807-5 [DOI] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. 2009. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol 73: 869–881. 10.1111/j.1365-2958.2009.06811.x [DOI] [PubMed] [Google Scholar]
- Mei Y, Yong J, Stonestrom A, Yang X. 2010. tRNA and cytochrome c in cell death and beyond. Cell Cycle 9: 2936–2939. 10.4161/cc.9.15.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA, De Robertis EM, Cortese R. 1980. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature 284: 143–148. 10.1038/284143a0 [DOI] [PubMed] [Google Scholar]
- Missoury S, Plancqueel S, de la Sierra-Gallay IL, Zhang W, Liger D, Durand D, Dammak R, Collinet B, van Tilbeurgh H. 2018. The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification. Nucleic Acids Res 46: 5850–5860. 10.1093/nar/gky323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa R, Mizuno R, Watanabe K, Ijiri K. 2012. Formation of tRNA granules in the nucleus of heat-induced human cells. Biochem Biophys Res Comm 418: 149–155. 10.1016/j.bbrc.2011.12.150 [DOI] [PubMed] [Google Scholar]
- Miyauchi K, Kimura S, Suzuki T. 2013. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 9: 105–111. 10.1038/nchembio.1137 [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, Valles AM, Ganem-Elbaz C, Antoniewski C, Huynh JR. 2015. tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription. EMBO J 34: 3009–3027. 10.15252/embj.201591006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan CE, Adamson SI, Kapur M, Chuang JH, Ackerman SL. 2021. The Clp1 R140H mutation alters tRNA metabolism and mRNA 3′ processing in mouse models of pontocerebellar hypoplasia. Proc Natl Acad Sci 118: e2110730118. 10.1073/pnas.2110730118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru A, Cakan-Akdogan G, Strassburger K, Males M, Mueller S, Jabs M, Muelleder M, Frejno M, Braeckman BP, Ralser M, et al. 2017. THADA regulates the organismal balance between energy storage and heat production. Dev Cell 41: 72–81.e76. 10.1016/j.devcel.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Ogasawara C, Inada T, Englert M, Beier H, Takezawa M, Endo T, Yoshihisa T. 2010. Dual functions of yeast tRNA ligase in the unfolded protein response: unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol Biol Cell 21: 3722–3734. 10.1091/mbc.e10-08-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Kajita T, Endo T, Yoshihisa T. 2011. The intron of tRNA-TrpCCA is dispensable for growth and translation of Saccharomyces cerevisiae. RNA 17: 1760–1769. 10.1261/rna.2851411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Auxilien S, Senger B, Tewari R, Grosjean H. 1998. Structural requirements for enzymatic formation of threonylcarbamoyladenosine (t6A) in tRNA: an in vivo study with Xenopus laevis oocytes. RNA 4: 24–37. [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Grosjean H. 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118. 10.1017/S1355838299982201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Bec G, Tewari R, Grosjean H. 1997. Transfer RNA recognition by the Escherichia coli isopentenyl-pyrophosphate:tRNA-isopentenyl transferase: dependence on the anticodon arm structure. RNA 3: 721–733. [PMC free article] [PubMed] [Google Scholar]
- Mueller EG, Ferre-D'Amare AR. 2009. Pseudouridine formation, the most common transglycosylation in RNA. In DNA and RNA modification enzymes: structure mechanism, function and evolution (ed. Grosjean H), pp. 363–376. Landes Bioscience, Austin, TX. [Google Scholar]
- Muller M, Hartmann M, Schuster I, Bender S, Thuring KL, Helm M, Katze JR, Nellen W, Lyko F, Ehrenhofer-Murray AE. 2015. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res 43: 10952–10962. 10.1093/nar/gkv980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Legrand C, Tuorto F, Kelly VP, Atlasi Y, Lyko F, Ehrenhofer-Murray AE. 2019a. Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res 47: 3711–3727. 10.1093/nar/gkz063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Samel-Pommerencke A, Legrand C, Tuorto F, Lyko F, Ehrenhofer-Murray AE. 2019b. Division of labour: tRNA methylation by the NSun2 tRNA methyltransferases Trm4a and Trm4b in fission yeast. RNA Biol 16: 249–256. 10.1080/15476286.2019.1568819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Abdullahu L, Damha MJ, Shuman S. 2018a. Two-step mechanism and step-arrest mutants of Runella slithyformis NAD+-dependent tRNA 2′-phosphotransferase Tpt1. RNA 24: 1144–1157. 10.1261/rna.067165.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Banerjee A, Shuman S. 2018b. NAD+-dependent synthesis of a 5′-phospho-ADP-ribosylated RNA/DNA cap by RNA 2′-phosphotransferase Tpt1. Nucleic Acids Res 46: 9617–9624. 10.1093/nar/gky792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Abdullahu L, Banerjee A, Damha MJ, Shuman S. 2019. NAD+-dependent RNA terminal 2′ and 3′ phosphomonoesterase activity of a subset of Tpt1 enzymes. RNA 25: 783–792. 10.1261/rna.071142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D, Bartlett E, Mikolcevic P, Kirby IT, Rack JGM, Mikoc A, Cohen MS, Ahel I. 2019. Reversible ADP-ribosylation of RNA. Nucleic Acids Res 47: 5658–5669. 10.1093/nar/gkz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murguia JR, Belles JM, Serrano R. 1996. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J Biol Chem 271: 29029–29033. 10.1074/jbc.271.46.29029 [DOI] [PubMed] [Google Scholar]
- Murphy FV, Ramakrishnan V. 2004. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11: 1251–1252. 10.1038/nsmb866 [DOI] [PubMed] [Google Scholar]
- Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. 2004. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186–1191. 10.1038/nsmb861 [DOI] [PubMed] [Google Scholar]
- Murthi A, Hopper AK. 2005. Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: roles for N-acetylation and an integral membrane protein. Genetics 170: 1553–1560. 10.1534/genetics.105.043620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK. 2010. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell 21: 639–649. 10.1091/mbc.e09-07-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na JG, Pinto I, Hampsey M. 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131: 791–801. 10.1093/genetics/131.4.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Ohara M, Miyauchi K, Yokobori SI, Yamagishi A, Watanabe K, Suzuki T. 2017. Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria. Nat Struct Mol Biol 24: 778–782. 10.1038/nsmb.3449 [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. 2011. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478: 57–63. 10.1038/nature10423 [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Hayashi H. 2008. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 283: 27469–27476. 10.1074/jbc.M804043200 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nemoto T, Heinemann IU, Yamashita K, Sonoda T, Komoda K, Tanaka I, Soll D, Yao M. 2013. Structural basis of reverse nucleotide polymerization. Proc Natl Acad Sci 110: 20970–20975. 10.1073/pnas.1321312111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Wang D, Komatsu Y. 2018. Biochemical analysis of human tRNAHis guanylyltransferase in mitochondrial tRNAHis maturation. Biochem Biophys Res Commun 503: 2015–2021. 10.1016/j.bbrc.2018.07.150 [DOI] [PubMed] [Google Scholar]
- Nameki N, Asahara H, Shimizu M, Okada N, Himeno H. 1995. Identity elements of Saccharomyces cerevisiae tRNAHis. Nucleic Acids Res 23: 389–394. 10.1093/nar/23.3.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor A, Thiaville PC, Altman-Price N, Cohen-Or I, Allers T, de Crecy-Lagard V, Gophna U. 2012. A genetic investigation of the KEOPS complex in halophilic Archaea. PLoS One 7: e43013. 10.1371/journal.pone.0043013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr F, Filipowicz W. 2000. Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1″,2″-cyclic phosphate. Nucleic Acids Res 28: 1676–1683. 10.1093/nar/28.8.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21: 4347–4368. 10.1128/MCB.21.13.4347-4368.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161: 1606–1618. 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HA, Hoffer ED, Dunham CM. 2019. Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGPro for decoding. J Biol Chem 294: 5281–5291. 10.1074/jbc.RA119.007410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel AI, Waber NB, Gossringer M, Lechner M, Linne U, Toth U, Rossmanith W, Hartmann RK. 2017. Minimal and RNA-free RNase P in Aquifex aeolicus. Proc Natl Acad Sci 114: 11121–11126. 10.1073/pnas.1707862114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, De Robertis EM. 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol 145: 405–420. 10.1016/0022-2836(81)90212-6 [DOI] [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, Suzuki T. 2006. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 25: 2142–2154. 10.1038/sj.emboj.7601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T. 2009. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37: 1335–1352. 10.1093/nar/gkn1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Yi S, Katoh T, Takai Y, Suzuki T, Suzuki T. 2011. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA 17: 1111–1119. 10.1261/rna.2653411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostramo RT, Hopper AK. 2020. A novel assay provides insight into tRNAPhe retrograde nuclear import and re-export in S. cerevisiae. Nucleic Acids Res 48: 11577–11588. 10.1093/nar/gkaa879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwagwu M, Nana M. 1980. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J Embryol Exp Morphol 56: 253–267. 10.1242/dev.56.1.253 [DOI] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178: 197–214. 10.1534/genetics.107.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbauer V, Schaefer MR. 2018. tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes (Basel) 9: 607. 10.3390/genes9120607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor JP, Peebles CL. 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol 11: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PZ, Cordell B, Valenzuela P, Rutter WJ, Goodman HM. 1978. Structure and processing of yeast precursor tRNAs containing intervening sequences. Nature 274: 438–445. 10.1038/274438a0 [DOI] [PubMed] [Google Scholar]
- Ogawa T. 2016. tRNA-targeting ribonucleases: molecular mechanisms and insights into their physiological roles. Biosci Biotechnol Biochem 80: 1037–1045. 10.1080/09168451.2016.1148579 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. 1999. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 283: 2097–2100. 10.1126/science.283.5410.2097 [DOI] [PubMed] [Google Scholar]
- Ohira T, Suzuki T. 2011. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc Natl Acad Sci 108: 10502–10507. 10.1073/pnas.1105645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira T, Suzuki T. 2016. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat Chem Biol 12: 648–655. 10.1038/nchembio.2117 [DOI] [PubMed] [Google Scholar]
- Okamoto H, Watanabe K, Ikeuchi Y, Suzuki T, Endo Y, Hori H. 2004. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J Biol Chem 279: 49151–49159. 10.1074/jbc.M408209200 [DOI] [PubMed] [Google Scholar]
- Olejniczak M, Uhlenbeck OC. 2006. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88: 943–950. 10.1016/j.biochi.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. 2005. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat Struct Mol Biol 12: 788–793. 10.1038/nsmb978 [DOI] [PubMed] [Google Scholar]
- Ontiveros RJ, Shen H, Stoute J, Yanas A, Cui Y, Zhang Y, Liu KF. 2020. Coordination of mRNA and tRNA methylations by TRMT10A. Proc Natl Acad Sci 117: 7782–7791. 10.1073/pnas.1913448117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana O, Cooley L, Soll D. 1986. The additional guanylate at the 5′ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol Cell Biol 6: 525–529. 10.1128/mcb.6.2.525-529.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W, Lim J, Aspris D, Sendinc E, Garyfallos DA, et al. 2021. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell 81: 3323–3338.e3314. 10.1016/j.molcel.2021.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Maison C, Black BE, Levesque L, Paschal BM, Dargemont C. 2000. RanGTP-binding protein NXT1 facilitates nuclear export of different classes of RNA in vitro. Mol Cell Biol 20: 4562–4571. 10.1128/MCB.20.13.4562-4571.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 3: 109–118. 10.1016/S1097-2765(00)80179-3 [DOI] [PubMed] [Google Scholar]
- Ozanick S, Krecic A, Andersland J, Anderson JT. 2005. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11: 1281–1290. 10.1261/rna.5040605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res 37: 298–308. 10.1093/nar/gkn925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Xiong Y, Steitz TA. 2010. How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA. Science 330: 937–940. 10.1126/science.1194985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. 1999. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci 96: 7803–7808. 10.1073/pnas.96.14.7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci 108: 13624–13629. 10.1073/pnas.1110633108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris Z, Horakova E, Rubio MA, Sample P, Fleming IM, Armocida S, Lukes J, Alfonzo JD. 2013. The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 19: 649–658. 10.1261/rna.036665.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. 2005. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci 102: 3691–3696. 10.1073/pnas.0405570102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321. 10.1016/S0092-8674(04)00342-3 [DOI] [PubMed] [Google Scholar]
- Payea MJ, Sloma MF, Kon Y, Young DL, Guy MP, Zhang X, De Zoysa T, Fields S, Mathews DH, Phizicky EM. 2018. Widespread temperature sensitivity and tRNA decay due to mutations in a yeast tRNA. RNA 24: 410–422. 10.1261/rna.064642.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payea MJ, Hauke AC, De Zoysa T, Phizicky EM. 2020. Mutations in the anticodon stem of tRNA cause accumulation and Met22-dependent decay of pre-tRNA in yeast. RNA 26: 29–43. 10.1261/rna.073155.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles CL, Ogden RC, Knapp G, Abelson J. 1979. Splicing of yeast tRNA precursors: a two-stage reaction. Cell 18: 27–35. 10.1016/0092-8674(79)90350-7 [DOI] [PubMed] [Google Scholar]
- Peebles CL, Gegenheimer P, Abelson J. 1983. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32: 525–536. 10.1016/0092-8674(83)90472-5 [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, Rassenti LZ, Pass HI, Kipps TJ, Liu CG, et al. 2016. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci 113: 5071–5076. 10.1073/pnas.1604266113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Balatti V, Croce CM. 2022. tRNA-derived fragments (tRFs) in cancer. J Cell Commun Signal 17: 47–54. 10.1007/s12079-022-00690-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Berezin I, Krasilnikov AS. 2018. In vitro reconstitution and analysis of eukaryotic RNase P RNPs. Nucleic Acids Res 46: 6857–6868. 10.1093/nar/gky333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Li D, Lee H, Bator C, Berezin I, Hafenstein SL, Krasilnikov AS. 2020. Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP. Nat Commun 11: 3474. 10.1038/s41467-020-17308-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernod K, Schaeffer L, Chicher J, Hok E, Rick C, Geslain R, Eriani G, Westhof E, Ryckelynck M, Martin F. 2020. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res 48: 6170–6183. 10.1093/nar/gkaa221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrochia L, Crozat E, Hecker A, Zhang W, Bareille J, Collinet B, van Tilbeurgh H, Forterre P, Basta T. 2013a. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res 41: 1953–1964. 10.1093/nar/gks1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrochia L, Guetta D, Hecker A, Forterre P, Basta T. 2013b. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res 41: 9484–9499. 10.1093/nar/gkt720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J, Walter P. 2019. tRNA ligase structure reveals kinetic competition between non-conventional mRNA splicing and mRNA decay. Elife 8: e44199. 10.7554/eLife.44199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HD, Lai LB, Zahurancik WJ, Gopalan V. 2021. The many faces of RNA-based RNase P, an RNA-world relic. Trends Biochem Sci 46: 976–991. 10.1016/j.tibs.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JH, Kjellin-Straby K. 1967. Studies on microbial ribonucleic acid: IV. Two mutants of Saccharomyces cerevisiae lacking N2-dimethylguanine in soluble ribonucleic acid. J Mol Biol 26: 509–518. 10.1016/0022-2836(67)90318-X [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Alfonzo JD. 2010. Do all modifications benefit all tRNAs? FEBS Lett 584: 265–271. 10.1016/j.febslet.2009.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Schwartz RC, Abelson J. 1986. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem 261: 2978–2986. 10.1016/S0021-9258(17)35882-9 [DOI] [PubMed] [Google Scholar]
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J 21: 1811–1820. 10.1093/emboj/21.7.1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto PH, Kroupova A, Schleiffer A, Mechtler K, Jinek M, Weitzer S, Martinez J. 2020. ANGEL2 is a member of the CCR4 family of deadenylases with 2′,3′-cyclic phosphatase activity. Science 369: 524–530. 10.1126/science.aba9763 [DOI] [PubMed] [Google Scholar]
- Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N. 2014. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell 54: 147–155. 10.1016/j.molcel.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohler MT, Roach TM, Betat H, Jackman JE, Morl M. 2019. A temporal order in 5′- and 3′-processing of eukaryotic tRNAHis. Int J Mol Sci 20: 1384. 10.3390/ijms20061384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollo-Oliveira L, Klassen R, Davis N, Ciftci A, Bacusmo JM, Martinelli M, DeMott MS, Begley TJ, Dedon PC, Schaffrath R, et al. 2020. Loss of elongator- and KEOPS-dependent tRNA modifications leads to severe growth phenotypes and protein aggregation in yeast. Biomolecules 10: 322. 10.3390/biom10020322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, et al. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331: 760–764. 10.1126/science.1197847 [DOI] [PubMed] [Google Scholar]
- Popow J, Schleiffer A, Martinez J. 2012. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci 69: 2657–2670. 10.1007/s00018-012-0944-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Jurkin J, Schleiffer A, Martinez J. 2014. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511: 104–107. 10.1038/nature13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat J, Kothe U, Bayfield MA. 2021. Revisiting tRNA chaperones: new players in an ancient game. RNA 27: 543–559. 10.1261/rna.078428.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CA, Kopajtich R, D'Souza AR, Rorbach J, Kremer LS, Husain RA, Dallabona C, Donnini C, Alston CL, Griffin H, et al. 2015. TRMT5 mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am J Hum Genet 97: 319–328. 10.1016/j.ajhg.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, McLeod IX, Yates JR III, Hopper AK, Engelke DR. 2013. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci 110: E3081–E3089. 10.1073/pnas.1219946110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Phizicky EM. 2010. The requirement for the highly conserved G–1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 16: 1068–1077. 10.1261/rna.2087510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, D'Silva S, Kon Y, Phizicky EM. 2013. tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae. RNA 19: 243–256. 10.1261/rna.035808.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Porter DF, Chen F, Buter N, Lapointe CP, Keles S, Kimble J, Wickens M. 2019. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat Methods 16: 437–445. 10.1038/s41592-019-0370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz J, Florentz C, Benseler F, Giege R. 1994. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol 1: 580–582. 10.1038/nsb0994-580 [DOI] [PubMed] [Google Scholar]
- Quan X, Yu J, Bussey H, Stochaj U. 2007. The localization of nuclear exporters of the importin-β family is regulated by Snf1 kinase, nutrient supply and stress. Biochim Biophys Acta 1773: 1052–1061. 10.1016/j.bbamcr.2007.04.014 [DOI] [PubMed] [Google Scholar]
- Raddatz G, Guzzardo PM, Olova N, Fantappie MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F. 2013. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci 110: 8627–8631. 10.1073/pnas.1306723110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ, Collins RN. 2005. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell 17: 841–853. 10.1016/j.molcel.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Raina M, Ibba M. 2014. tRNAs as regulators of biological processes. Front Genet 5: 171. 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Shuman S, Schwer B. 2008. Human RNA 5′-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA 14: 1737–1745. 10.1261/rna.1142908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez V, Gonzalez B, Lopez A, Castello MJ, Gil MJ, Zheng B, Chen P, Vera P. 2018. A 2′-O-methyltransferase responsible for transfer RNA anticodon modification is pivotal for resistance to Pseudomonas syringae DC3000 in Arabidopsis. Mol Plant Microbe Interact 31: 1323–1336. 10.1094/MPMI-06-18-0148-R [DOI] [PubMed] [Google Scholar]
- Ramos J, Fu D. 2019. The emerging impact of tRNA modifications in the brain and nervous system. Biochim Biophys Acta Gene Regul Mech 1862: 412–428. 10.1016/j.bbagrm.2018.11.007 [DOI] [PubMed] [Google Scholar]
- Ramos J, Han L, Li Y, Hagelskamp F, Kellner SM, Alkuraya FS, Phizicky EM, Fu D. 2019. Formation of tRNA wobble inosine in humans is disrupted by a millennia-old mutation causing intellectual disability. Mol Cell Biol 39: e00203–19. 10.1128/MCB.00203-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Morales E, Bayam E, Del-Pozo-Rodriguez J, Salinas-Giege T, Marek M, Tilly P, Wolff P, Troesch E, Ennifar E, Drouard L, et al. 2021. The structure of the mouse ADAT2/ADAT3 complex reveals the molecular basis for mammalian tRNA wobble adenosine-to-inosine deamination. Nucleic Acids Res 49: 6529–6548. 10.1093/nar/gkab436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramser J, Winnepenninckx B, Lenski C, Errijgers V, Platzer M, Schwartz CE, Meindl A, Kooy RF. 2004. A splice site mutation in the methyltransferase gene FTSJ1 in Xp11.23 is associated with non-syndromic mental retardation in a large Belgian family (MRX9). J Med Genet 41: 679–683. 10.1136/jmg.2004.019000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N, Rodnina MV. 2017. Thio-modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J Am Chem Soc 139: 5857–5864. 10.1021/jacs.7b00727 [DOI] [PubMed] [Google Scholar]
- Rao BS, Jackman JE. 2015. Life without post-transcriptional addition of G–1: two alternatives for tRNAHis identity in Eukarya. RNA 21: 243–253. 10.1261/rna.048389.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, Maris EL, Jackman JE. 2011. tRNA 5′-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucleic Acids Res 39: 1833–1842. 10.1093/nar/gkq976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, Mohammad F, Gray MW, Jackman JE. 2013. Absence of a universal element for tRNAHis identity in Acanthamoeba castellanii. Nucleic Acids Res 41: 1885–1894. 10.1093/nar/gks1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsborough CW, Ipas H, Abell NS, Nottingham RM, Yao J, Devanathan SK, Shelton SB, Lambowitz AM, Xhemalce B. 2019. BCDIN3D regulates tRNAHis 3′ fragment processing. PLoS Genet 15: e1008273. 10.1371/journal.pgen.1008273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus BS, Shuman S. 2013. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA 19: 659–669. 10.1261/rna.038406.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus BS, Goldgur Y, Shuman S. 2017. Structural basis for the GTP specificity of the RNA kinase domain of fungal tRNA ligase. Nucleic Acids Res 45: 12945–12953. 10.1093/nar/gkx1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes VM, Abelson J. 1988. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55: 719–730. 10.1016/0092-8674(88)90230-9 [DOI] [PubMed] [Google Scholar]
- Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. 2013. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci 110: 12289–12294. 10.1073/pnas.1300781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Evans ME, Clark WC, Marttinen P, Shoubridge EA, Suomalainen A, Wredenberg A, Wedell A, Pan T, Battersby BJ. 2018. RNA modification landscape of the human mitochondrial tRNALys regulates protein synthesis. Nat Commun 9: 3966. 10.1038/s41467-018-06471-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. 1982. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell 29: 149–159. 10.1016/0092-8674(82)90099-X [DOI] [PubMed] [Google Scholar]
- Rose AM, Belford HG, Shen WC, Greer CL, Hopper AK, Martin NC. 1995. Location of N2,N2-dimethylguanosine-specific tRNA methyltransferase. Biochimie 77: 45–53. 10.1016/0300-9084(96)88103-X [DOI] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2016. New structural insights into translational miscoding. Trends Biochem Sci 41: 798–814. 10.1016/j.tibs.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Rubio MA, Paris Z, Gaston KW, Fleming IM, Sample P, Trotta CR, Alfonzo JD. 2013. Unusual noncanonical intron editing is important for tRNA splicing in Trypanosoma brucei. Mol Cell 52: 184–192. 10.1016/j.molcel.2013.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MA, Gaston KW, McKenney KM, Fleming IM, Paris Z, Limbach PA, Alfonzo JD. 2017. Editing and methylation at a single site by functionally interdependent activities. Nature 542: 494–497. 10.1038/nature21396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio Gomez MA, Ibba M. 2020. Aminoacyl-tRNA synthetases. RNA 26: 910–936. 10.1261/rna.071720.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Florentz C, Giege R. 1994. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues -1 and 73 but is dependent on the RNA context. Nucleic Acids Res 22: 5031–5037. 10.1093/nar/22.23.5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, Grassi A, Zanovello P, Bronte V, Ciminale V, et al. 2014. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol 88: 3612–3622. 10.1128/JVI.02823-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arroyo VM, Raj R, Babu K, Onolbaatar O, Roberts PH, Nam Y. 2023. Structures and mechanisms of tRNA methylation by METTL1-WDR4. Nature 613: 383–390. 10.1038/s41586-022-05565-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. 2010. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16: 1317–1327. 10.1261/rna.2057810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, et al. 2014. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34: 2450–2463. 10.1128/MCB.00136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample PJ, Koreny L, Paris Z, Gaston KW, Rubio MA, Fleming IM, Hinger S, Horakova E, Limbach PA, Lukes J, et al. 2015. A common tRNA modification at an unusual location: the discovery of wyosine biosynthesis in mitochondria. Nucleic Acids Res 43: 4262–4273. 10.1093/nar/gkv286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK. 1998. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 9: 3041–3055. 10.1091/mbc.9.11.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasarman F, Thiffault I, Weraarpachai W, Salomon S, Maftei C, Gauthier J, Ellazam B, Webb N, Antonicka H, Janer A, et al. 2015. The 3′ addition of CCA to mitochondrial tRNASer(AGY) is specifically impaired in patients with mutations in the tRNA nucleotidyl transferase TRNT1. Hum Mol Genet 24: 2841–2847. 10.1093/hmg/ddv044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, et al. 2020. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583: 638–643. 10.1038/s41586-020-2418-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Schwer B, Shuman S. 2005. Structure-function analysis of the yeast NAD+-dependent tRNA 2′-phosphotransferase Tpt1. RNA 11: 107–113. 10.1261/rna.7193705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Lyko F. 2010. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet 42: 920–921; author reply 921. 10.1038/ng1110-920 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Lyko F. 2009. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37: e12. 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595. 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, et al. 2014. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157: 651–663. 10.1016/j.cell.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S, Rosch S, Marchfelder A. 2002. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J 21: 2769–2777. 10.1093/emboj/21.11.2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. 2018. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol 19: 45–58. 10.1038/nrm.2017.77 [DOI] [PubMed] [Google Scholar]
- Schmidt K, Butler JS. 2013. Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip Rev RNA 4: 217–231. 10.1002/wrna.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, Matera AG. 2020. tRNA introns: presence, processing, and purpose. Wiley Interdiscip Rev RNA 11: e1583. 10.1002/wrna.1583 [DOI] [PubMed] [Google Scholar]
- Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. 2019. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47: 6452–6465. 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. 2007. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 27: 324–331. 10.1016/j.molcel.2007.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Martienssen R. 2018. Tie-break: host and retrotransposons play tRNA. Trends Cell Biol 28: 793–806. 10.1016/j.tcb.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. 2017. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170: 61–71.e11. 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer MP, Chheda GB, Baczynskyj L, Hall RH. 1969. Aminoacyl nucleosides. VII. N-(Purin-6-ylcarbamoyl)threonine. A new component of transfer ribonucleic acid. Biochemistry 8: 3283–3289. 10.1021/bi00836a023 [DOI] [PubMed] [Google Scholar]
- Schwenzer H, Juhling F, Chu A, Pallett LJ, Baumert TF, Maini M, Fassati A. 2019. Oxidative stress triggers selective tRNA retrograde transport in human cells during the integrated stress response. Cell Rep 26: 3416–3428.e3415. 10.1016/j.celrep.2019.02.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Aronova A, Ramirez A, Braun P, Shuman S. 2008. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA 14: 204–210. 10.1261/rna.858108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovski S, Trowitzsch S. 2022. Transfer RNA processing: from a structural and disease perspective. Biol Chem 403: 749–763. 10.1515/hsz-2021-0406 [DOI] [PubMed] [Google Scholar]
- Sekulovski S, Devant P, Panizza S, Gogakos T, Pitiriciu A, Heitmeier K, Ramsay EP, Barth M, Schmidt C, Tuschl T, et al. 2021. Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia. Nat Commun 12: 5610. 10.1038/s41467-021-25870-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovski S, Sušac L, Stelzl LS, Tampé R, Trowitzsch S. 2023. Structural basis of substrate recognition by human tRNA splicing endonuclease TSEN. Nat Struct Mol Biol doi:10.1038/s41594-023-00992-y [DOI] [PubMed] [Google Scholar]
- Selvadurai K, Wang P, Seimetz J, Huang RH. 2014. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat Chem Biol 10: 810–812. 10.1038/nchembio.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK. 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295. 10.1073/pnas.0503836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. 2007. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci 104: 8845–8850. 10.1073/pnas.0700765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Abdel-Salam GM, Guy MP, Alomar R, Abdel-Hamid MS, Afifi HH, Ismail SI, Emam BA, Phizicky EM, Alkuraya FS. 2015. Mutation in WDR4 impairs tRNA m7G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol 16: 210. 10.1186/s13059-015-0779-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, Alkuraya FS. 2016. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet 135: 707–713. 10.1007/s00439-016-1665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Yan W, Peng J, Zuo X, Zou Y, Li F, Gong D, Ma R, Wu J, Shi Y, et al. 2014. Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Res 42: 509–525. 10.1093/nar/gkt869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DL. 2015. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43: 2242–2258. 10.1093/nar/gkv075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton VM, Sosnick TR, Pan T. 2001. Altering the intermediate in the equilibrium folding of unmodified yeast tRNAPhe with monovalent and divalent cations. Biochemistry 40: 3629–3638. 10.1021/bi002646+ [DOI] [PubMed] [Google Scholar]
- Shi H, Moore PB. 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6: 1091–1105. 10.1017/S1355838200000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Stefan CJ, Rue SM, Teis D, Emr SD. 2011. Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol Biol Cell 22: 4093–4107. 10.1091/mbc.e11-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039. 10.1016/S0092-8674(00)80369-4 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87: 405–413. 10.1016/S0092-8674(00)81361-6 [DOI] [PubMed] [Google Scholar]
- Singer EE, Smith GR, Cortese R, Ames BN. 1972. Mutant tRNAHis ineffective in repression and lacking two pseudouridine modifications. Nat New Biol 238: 72–74. 10.1038/newbio238072a0 [DOI] [PubMed] [Google Scholar]
- Singh RK, Feller A, Roovers M, Van Elder D, Wauters L, Droogmans L, Versees W. 2018. Structural and biochemical analysis of the dual-specificity Trm10 enzyme from Thermococcus kodakaraensis prompts reconsideration of its catalytic mechanism. RNA 24: 1080–1092. 10.1261/rna.064345.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek E, Grzechnik P, Spath B, Marchfelder A, Kufel J. 2014. tRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA 20: 115–130. 10.1261/rna.041467.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, et al. 2001. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 68: 598–605. 10.1086/318810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusher LB, Gillman EC, Martin NC, Hopper AK. 1991. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci 88: 9789–9793. 10.1073/pnas.88.21.9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Jackman JE. 2012. Kinetic analysis of 3′–5′ nucleotide addition catalyzed by eukaryotic tRNAHis guanylyltransferase. Biochemistry 51: 453–465. 10.1021/bi201397f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Jackman JE. 2014. Saccharomyces cerevisiae Thg1 uses 5′-pyrophosphate removal to control addition of nucleotides to tRNAHis. Biochemistry 53: 1380–1391. 10.1021/bi4014648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg T, Poulter CD. 2000. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry 39: 6546–6553. 10.1021/bi992775u [DOI] [PubMed] [Google Scholar]
- Soderberg T, Poulter CD. 2001. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: site-directed mutagenesis of highly conserved residues. Biochemistry 40: 1734–1740. 10.1021/bi002149t [DOI] [PubMed] [Google Scholar]
- Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A. 2010. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 30: 1814–1827. 10.1128/MCB.01602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Consaul SA, Phizicky EM. 1997. A conditional lethal yeast phosphotransferase (tpt1) mutant accumulates tRNAs with a 2′-phosphate and an undermodified base at the splice junction. RNA 3: 1388–1400. [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Malik HS, Consaul SA, Phizicky EM. 1998. A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli. Proc Natl Acad Sci 95: 14136–14141. 10.1073/pnas.95.24.14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Kierzek R, Turner DH, Phizicky EM. 1999. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J Biol Chem 274: 2637–2644. 10.1074/jbc.274.5.2637 [DOI] [PubMed] [Google Scholar]
- Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA. 2005. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 24: 5502–5509. 10.1038/sj.onc.1208687 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS. 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140. 10.1093/nar/gki012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26: 148–153. 10.1093/nar/26.1.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. 2011. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30: 873–881. 10.1038/emboj.2010.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154. 10.1016/0092-8674(84)90083-7 [DOI] [PubMed] [Google Scholar]
- Steiger MA, Jackman JE, Phizicky EM. 2005. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: evidence for a conserved two-step reaction mechanism. RNA 11: 99–106. 10.1261/rna.7194605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel MC, Abelson J. 1986. Intron mutations affect splicing of Saccharomyces cerevisiae SUP53 precursor tRNA. Mol Cell Biol 6: 2674–2683. 10.1128/mcb.6.7.2674-2683.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Kuscu C, Malik A, Shibata E, Dutta A. 2019. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem 294: 16930–16941. 10.1074/jbc.RA119.009272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Monshaugen I, Wilson B, Wang F, Klungland A, Ougland R, Dutta A. 2022. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat Commun 13: 2165. 10.1038/s41467-022-29790-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. 2021. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol 22: 375–392. 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T. 2014. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357. 10.1093/nar/gku390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. 2012. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336: 355–359. 10.1126/science.1219491 [DOI] [PubMed] [Google Scholar]
- Swinehart WE, Henderson JC, Jackman JE. 2013. Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA 19: 1137–1146. 10.1261/rna.039651.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M. 2003. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res 31: 2272–2278. 10.1093/nar/gkg337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T. 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142. 10.1126/science.1113346 [DOI] [PubMed] [Google Scholar]
- Takano K, Nakagawa E, Inoue K, Kamada F, Kure S, Goto Y, Japanese Mental Retardation Consortium. 2008. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic X-linked mental retardation in a Japanese family. Am J Med Genet B Neuropsychiatr Genet 147B: 479–484. 10.1002/ajmg.b.30638 [DOI] [PubMed] [Google Scholar]
- Takano A, Kajita T, Mochizuki M, Endo T, Yoshihisa T. 2015. Cytosolic Hsp70 and co-chaperones constitute a novel system for tRNA import into the nucleus. Elife 4: e04659. 10.7554/eLife.04659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286: 7727–7731. 10.1074/jbc.C111.219022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Chakravarty AK, Maughan B, Shuman S. 2011a. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286: 43134–43143. 10.1074/jbc.M111.302133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S. 2011b. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem 286: 30253–30257. 10.1074/jbc.C111.274597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Jia P, Xin P, Chu J, Shi DQ, Yang WC. 2020. The Arabidopsis TRM61/TRM6 complex is a bona fide tRNA N1-methyladenosine methyltransferase. J Exp Bot 71: 3024–3036. 10.1093/jxb/eraa100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasak M, Phizicky EM. 2022. Initiator tRNA lacking 1-methyladenosine is targeted by the rapid tRNA decay pathway in evolutionarily distant yeast species. PLoS Genet 18: e1010215. 10.1371/journal.pgen.1010215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner A, Weber C, Buzet A, Hartmann RK, Hartig A, Rossmanith W. 2012. Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multicomponent RNA-protein complex. Cell Rep 2: 19–25. 10.1016/j.celrep.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ. 2006. Structural basis for recognition and sequestration of UUUOH 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell 21: 75–85. 10.1016/j.molcel.2005.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, Iwata-Reuyl D, de Crecy-Lagard V. 2014. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol 11: 1529–1539. 10.4161/15476286.2014.992277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, El Yacoubi B, Kohrer C, Thiaville JJ, Deutsch C, Iwata-Reuyl D, Bacusmo JM, Armengaud J, Bessho Y, Wetzel C, et al. 2015. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol Microbiol 98: 1199–1221. 10.1111/mmi.13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, Legendre R, Rojas-Benitez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crecy-Lagard V. 2016. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb Cell 3: 29–45. 10.15698/mic2016.01.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebe R, Zachau HG. 1968. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem 5: 546–555. 10.1111/j.1432-1033.1968.tb00404.x [DOI] [PubMed] [Google Scholar]
- Thompson LD, Daniels CJ. 1988. A tRNATrp intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J Biol Chem 263: 17951–17959. 10.1016/S0021-9258(19)81308-X [DOI] [PubMed] [Google Scholar]
- Thompson LD, Daniels CJ. 1990. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J Biol Chem 265: 18104–18111. 10.1016/S0021-9258(17)44723-5 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50. 10.1083/jcb.200811119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401. 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. 2007. The dawn of dominance by the mature domain in tRNA splicing. Proc Natl Acad Sci 104: 12300–12305. 10.1073/pnas.0705537104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C. 2018. 7-Methylguanosine modifications in transfer RNA (tRNA). Int J Mol Sci 19: 4080. 10.3390/ijms19124080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Weiner AM. 2001. Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science 294: 1334–1336. 10.1126/science.1063816 [DOI] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. 2000. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci 97: 8278–8283. 10.1073/pnas.140213797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O. 2004. Structural basis for template-independent RNA polymerization. Nature 430: 700–704. 10.1038/nature02712 [DOI] [PubMed] [Google Scholar]
- Torabi N, Kruglyak L. 2011. Variants in SUP45 and TRM10 underlie natural variation in translation termination efficiency in Saccharomyces cerevisiae. PLoS Genet 7: e1002211. 10.1371/journal.pgen.1002211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimouille A, Lasseaux E, Barat P, Deiller C, Drunat S, Rooryck C, Arveiler B, Lacombe D. 2018. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin Genet 93: 374–377. 10.1111/cge.13074 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. 1997. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. 10.1016/S0092-8674(00)80270-6 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Paushkin SV, Patel M, Li H, Peltz SW. 2006. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441: 375–377. 10.1038/nature04741 [DOI] [PubMed] [Google Scholar]
- Trzaska C, Amand S, Bailly C, Leroy C, Marchand V, Duvernois-Berthet E, Saliou JM, Benhabiles H, Werkmeister E, Chassat T, et al. 2020. 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat Commun 11: 1509. 10.1038/s41467-020-15140-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Yamazaki R, Nobuta R, Ikeuchi K, Makino S, Ohtaki A, Suzuki Y, Yoshihisa T, Trotta C, Inada T. 2015. The tRNA splicing endonuclease complex cleaves the mitochondria-localized CBP1 mRNA. J Biol Chem 290: 16021–16030. 10.1074/jbc.M114.634592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Arps PJ, Connolly DM, Winkler ME. 1991. Absence of hisT-mediated tRNA pseudouridylation results in a uracil requirement that interferes with Escherichia coli K-12 cell division. J Bacteriol 173: 7395–7400. 10.1128/jb.173.22.7395-7400.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Grone HJ, et al. 2015. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 34: 2350–2362. 10.15252/embj.201591382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Legrand C, Cirzi C, Federico G, Liebers R, Muller M, Ehrenhofer-Murray AE, Dittmar G, Grone HJ, Lyko F. 2018. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J 37: e99777. 10.15252/embj.201899777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Karkusiewicz I, Kowal J, Boguta M. 2012. Maf1-mediated repression of RNA polymerase III transcription inhibits tRNA degradation via RTD pathway. RNA 18: 1823–1832. 10.1261/rna.033597.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Nemoto N, Wilkinson CR, Narashimhan J, Jiang L, Watt S, Zook A, Jones N, Wek RC, Bahler J, et al. 2008. Int6/eIF3e promotes general translation and Atf1 abundance to modulate Sty1 MAPK-dependent stress response in fission yeast. J Biol Chem 283: 22063–22075. 10.1074/jbc.M710017200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Fox JJ. 1963. Spectrophotometric studies of nucleic acid derivatives and related compounds. V. On the structure of 3-methylcytosine. J Am Chem Soc 85: 4024–4028. 10.1021/ja00907a026 [DOI] [Google Scholar]
- Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K, Tsujikawa K. 2017. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep 7: 42271. 10.1038/srep42271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Qian O, Durand JMB, Hagervall TG, Bjork GR. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20: 4863–4873. 10.1093/emboj/20.17.4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Bjork GR. 2003. Transfer RNA modifications that alter +1 frameshifting in general fail to affect –1 frameshifting. RNA 9: 760–768. 10.1261/rna.5210803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Armengaud J, Grosjean H. 2006. Identity elements required for enzymatic formation of N2,N2-dimethylguanosine from N2-monomethylated derivative and its possible role in avoiding alternative conformations in archaeal tRNA. J Mol Biol 357: 387–399. 10.1016/j.jmb.2005.12.087 [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. 2011. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun 2: 172. 10.1038/ncomms1173 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365. 10.1093/emboj/19.6.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeix FA, Murphy FV, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. 2012. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto–enol tautomerism. J Mol Biol 416: 467–485. 10.1016/j.jmb.2011.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. 2012. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res 40: 11583–11593. 10.1093/nar/gks910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Amman F, Toth U, Kotter A, Helm M, Rossmanith W. 2020. Functional characterization of the human tRNA methyltransferases TRMT10A and TRMT10B. Nucleic Acids Res 48: 6157–6169. 10.1093/nar/gkaa353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Kiss T. 2019. Cooperative 2′-O-methylation of the wobble cytidine of human elongator tRNAMet(CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes Dev 33: 741–746. 10.1101/gad.326363.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta V, Ceci M, Emery B, Bachi A, Petfalski E, Tollervey D, Linder P, Marchisio PC, Piatti S, Biffo S. 2005. Sen34p depletion blocks tRNA splicing in vivo and delays rRNA processing. Biochem Biophys Res Comm 337: 89–94. 10.1016/j.bbrc.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Waas WF, de Crecy-Lagard V, Schimmel P. 2005. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J Biol Chem 280: 37616–37622. 10.1074/jbc.M506939200 [DOI] [PubMed] [Google Scholar]
- Waas WF, Druzina Z, Hanan M, Schimmel P. 2007. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem 282: 26026–26034. 10.1074/jbc.M703391200 [DOI] [PubMed] [Google Scholar]
- Walker SC, Engelke DR. 2006. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol 41: 77–102. 10.1080/10409230600602634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, Kwon SY, Badenhorst P, East P, McNeill H, Svejstrup JQ. 2011. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 187: 1067–1075. 10.1534/genetics.110.123893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LR, Butler JS. 2019. Toxins targeting transfer RNAs: translation inhibition by bacterial toxin-antitoxin systems. Wilet Interdiscip Rev RNA 10: e1506. 10.1002/wrna.1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Hopper AK. 2018. From powerhouse to processing plant: conserved roles of mitochondrial outer membrane proteins in tRNA splicing. Genes Dev 32: 1309–1314. 10.1101/gad.316257.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LC, Mao DY, Neculai D, Strecker J, Chiovitti D, Kurinov I, Poda G, Thevakumaran N, Yuan F, Szilard RK, et al. 2013. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res 41: 6332–6346. 10.1093/nar/gkt322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. 2005. Silencing near tRNA genes requires nucleolar localization. J Biol Chem 280: 8637–8639. 10.1074/jbc.C500017200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Schwer B, Englert M, Beier H, Shuman S. 2006. Structure–function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res 34: 517–527. 10.1093/nar/gkj441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sobral BW, Williams KP. 2007. Loss of a universal tRNA feature. J Bacteriol 189: 1954–1962. 10.1128/JB.01203-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SQ, Shi DQ, Long YP, Liu J, Yang WC. 2012. GAMETOPHYTE DEFECTIVE 1, a putative subunit of RNases P/MRP, is essential for female gametogenesis and male competence in Arabidopsis. PLoS One 7: e33595. 10.1371/journal.pone.0033595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W, Schwartz MH, Pan T. 2018. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24: 1305–1313. 10.1261/rna.067033.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Miyagawa R, Tomikawa C, Mizuno R, Takahashi A, Hori H, Ijiri K. 2013. Degradation of initiator tRNAMet by Xrn1/2 via its accumulation in the nucleus of heat-treated HeLa cells. Nucleic Acids Res 41: 4671–4685. 10.1093/nar/gkt153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ijiri K, Ohtsuki T. 2014. mTOR regulates the nucleoplasmic diffusion of Xrn2 under conditions of heat stress. FEBS Lett 588: 3454–3460. 10.1016/j.febslet.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Weber C, Hartig A, Hartmann RK, Rossmanith W. 2014. Playing RNase P evolution: swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms. PLoS Genet 10: e1004506. 10.1371/journal.pgen.1004506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. 2018. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 71: 973–985.e975. 10.1016/j.molcel.2018.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. 2007. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol 14: 498–502. 10.1038/nsmb1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner K, Czech A, Ignatova Z, Betat H, Morl M. 2018. Examining tRNA 3′-ends in Escherichia coli: teamwork between CCA-adding enzyme, RNase T, and RNase R. RNA 24: 361–370. 10.1261/rna.064436.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473. 10.1016/0092-8674(95)90435-2 [DOI] [PubMed] [Google Scholar]
- Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev 25: 1173–1184. 10.1101/gad.2050711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RL, Hogness DS. 1977. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell 10: 177–192. 10.1016/0092-8674(77)90213-6 [DOI] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK. 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686. 10.1091/mbc.e07-01-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc 127: 4659–4667. 10.1021/ja0436749 [DOI] [PubMed] [Google Scholar]
- Wilusz JE. 2015. Controlling translation via modulation of tRNA levels. WIRES RNA 6: 453–470. 10.1002/wrna.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. 2011. tRNAs marked with CCACCA are targeted for degradation. Science 334: 817–821. 10.1126/science.1213671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW. 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol 26: 1710–1721. 10.1128/MCB.26.5.1710-1721.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci 99: 3517–3522. 10.1073/pnas.022042899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 4: 123–128. 10.1016/S1097-2765(00)80194-X [DOI] [PubMed] [Google Scholar]
- Wolfe CL, Lou YC, Hopper AK, Martin NC. 1994. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J Biol Chem 269: 13361–13366. 10.1016/S0021-9258(17)36841-2 [DOI] [PubMed] [Google Scholar]
- Wolfe CL, Hopper AK, Martin NC. 1996. Mechanisms leading to and the consequences of altering the normal distribution of ATP(CTP):tRNA nucleotidyltransferase in yeast. J Biol Chem 271: 4679–4686. 10.1074/jbc.271.9.4679 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T. 2002. The La protein. Ann Rev Biochem 71: 375–403. 10.1146/annurev.biochem.71.090501.150003 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Sim S, Chen X. 2012. Nuclear noncoding RNA surveillance: is the end in sight? Trends Genet 28: 306–313. 10.1016/j.tig.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hopper AK. 2014. Healing for destruction: tRNA intron degradation in yeast is a two-step cytoplasmic process catalyzed by tRNA ligase Rlg1 and 5′-to-3′ exonuclease Xrn1. Genes Dev 28: 1556–1561. 10.1101/gad.244673.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Brockenbrough JS, Paddy MR, Aris JP. 1998. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene 220: 109–117. 10.1016/S0378-1119(98)00330-8 [DOI] [PubMed] [Google Scholar]
- Wu J, Bao A, Chatterjee K, Wan Y, Hopper AK. 2015. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear-cytoplasmic dynamics. Genes Dev 29: 2633–2644. 10.1101/gad.269803.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Niu S, Tan M, Huang C, Li M, Song Y, Wang Q, Chen J, Shi S, Lan P, et al. 2018. Cryo-EM structure of the human ribonuclease P holoenzyme. Cell 175: 1393–1404.e1311. 10.1016/j.cell.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Wu CC, Peterson A, Zinshteyn B, Regot S, Green R. 2020. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182: 404–416.e414. 10.1016/j.cell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yao L, Yu X, Ruan Y, Li Z, Guo J. 2020. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct Target Ther 5: 109. 10.1038/s41392-020-00217-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. 2004. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 430: 640–645. 10.1038/nature02711 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. 2006. A story with a good ending: tRNA 3′-end maturation by CCA-adding enzymes. Curr Opin Struct Biol 16: 12–17. 10.1016/j.sbi.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, et al. 2017. Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem 292: 14695–14703. 10.1074/jbc.M117.798298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H. 2006. RNA recognition and cleavage by a splicing endonuclease. Science 312: 906–910. 10.1126/science.1126629 [DOI] [PubMed] [Google Scholar]
- Yan LL, Zaher HS. 2021. Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol Cell 81: 614–628.e614. 10.1016/j.molcel.2020.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. 2017. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 27: 606–625. 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, Sun BF, Li A, Xia J, Chen J, et al. 2019. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell 75: 1188–1202.e1111. 10.1016/j.molcel.2019.06.033 [DOI] [PubMed] [Google Scholar]
- Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, et al. 2014. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet 10: e1004424. 10.1371/journal.pgen.1004424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip MCJ, Keszei AFA, Feng Q, Chu V, McKenna MJ, Shao S. 2019. Mechanism for recycling tRNAs on stalled ribosomes. Nat Struct Mol Biol 26: 343–349. 10.1038/s41594-019-0211-4 [DOI] [PubMed] [Google Scholar]
- Yip MCJ, Savickas S, Gygi SP, Shao S. 2020. ELAC1 repairs tRNAs cleaved during ribosome-associated quality control. Cell Rep 30: 2106–2114.e2105. 10.1016/j.celrep.2020.01.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402. 10.1016/S0092-8674(00)80220-2 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T. 2014. Handling tRNA introns, archaeal way and eukaryotic way. Front Genet 5: 213. 10.3389/fgene.2014.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279. 10.1091/mbc.e02-11-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. 2007. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 12: 285–297. 10.1111/j.1365-2443.2007.01056.x [DOI] [PubMed] [Google Scholar]
- Yun JS, Yoon JH, Choi YJ, Son YJ, Kim S, Tong L, Chang JH. 2018. Molecular mechanism for the inhibition of DXO by adenosine 3′,5′-bisphosphate. Biochem Biophys Res Commun 504: 89–95. 10.1016/j.bbrc.2018.08.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva L, Myers R, Fassati A. 2006. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol 4: e332. 10.1371/journal.pbio.0040332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Q, Huang Y. 2013. Partitioning of the nuclear and mitochondrial tRNA 3′-end processing activities between two different proteins in Schizosaccharomyces pombe. J Biol Chem 288: 27415–27422. 10.1074/jbc.M113.501569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J, et al. 2018. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 20: 535–540. 10.1038/s41556-018-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Lentini JM, Prevost CT, Hashem MO, Alkuraya FS, Fu D. 2020. An intellectual disability-associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity. Hum Mutat 41: 600–607. 10.1002/humu.23976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang F, Zhan X, Bian T, Xing Z, Lu Y, Shi Y. 2023. Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease. Mol Cell 83: 1328–1339. 10.1016/j.molcel.2023.03.015 [DOI] [PubMed] [Google Scholar]
- Zhou C, Huang RH. 2008. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc Natl Acad Sci 105: 16142–16147. 10.1073/pnas.0805680105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, Fassati A. 2011. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog 7: e1002194. 10.1371/journal.ppat.1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Deutscher MP. 1987. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J 6: 2473–2477. 10.1002/j.1460-2075.1987.tb02528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV. 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 9: e1003675. 10.1371/journal.pgen.1003675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofallova L, Guo Y, Gupta R. 2000. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA 6: 1019–1030. 10.1017/S1355838200000613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.