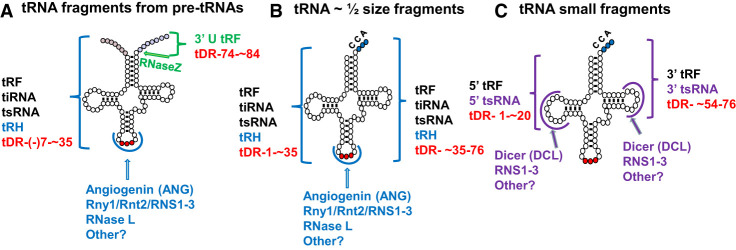
FIGURE 9.
Biogenesis of tRNA fragments. (A) tRNA fragments generated from pre-tRNAs prior to 5′ leader and 3′ trailer removal. Green font and bracket indicate tRNA fragments derived from 3′ trailers upon endonucleolytic cleavage by RNase Z. Blue bracket demarcates region of fragments resulting from cleavage of 5′ leader-containing pre-tRNAs in the ACL. (B) 5′ (left blue bracket) or 3′ (right blue bracket) ∼ half size tRNA fragments generated upon cleavage of mature tRNAs in the ACL. (C) 5′ (left puple bracket) or 3′ (right purple bracket) ∼ ¼ size tRNA framents resulting from endonucleolytic cleavage of mature tRNAs in the D- or T-loops, respectively. Black, blue, and purple fonts near brackets indicate the various names of the tRNA fragments; blue font nomenclature is used in this review. Red font refers to the proposed future systematic nomenclature for tRNA fragments. Arcs indicate the possible locations of loop cleavages. Names below each arc refer to the various endonucleases implicated in cleavages. Angiogenin (also refered to as ANG) is a vertebrate RNase A-like enzyme, and RNase L is an interferon induced 2′–5′ oligoadenylate synthetase-dependent RNase. Rny1 is a yeast T2-like endonuclease; Rnt2 and RNS1, RNS2, and RNS3 are plant T2-like endonucleases. Metazoan Dicer and plant Dicer-like DCLs are RNase III-like enzymes also functioning in pre-miRNA biogenesis.