Abstract
Wobble GU pairs (or G•U) occur frequently within double-stranded RNA helices interspersed between standard G=C and A-U Watson–Crick pairs. Another type of G•U pair interacting via their Watson–Crick edges has been observed in the A site of ribosome structures between a modified U34 in the tRNA anticodon triplet and G + 3 in the mRNA. In such pairs, the electronic structure of the U is changed with a negative charge on N3(U), resulting in two H-bonds between N1(G)…O4(U) and N2(G)…N3(U). Here, we report that such pairs occur in other highly conserved positions in ribosomal RNAs of bacteria in the absence of U modification. An anionic cis Watson–Crick G•G pair is also observed and well conserved in the small subunit. These pairs are observed in tightly folded regions.
Keywords: G•U pair, rRNA, anticodon, codon, tRNA, mRNA, anionic
INTRODUCTION
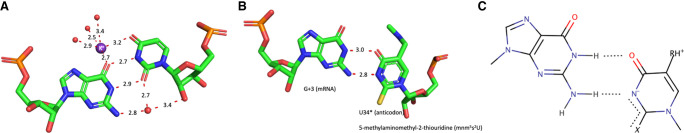
The diversity of interactions between Gs and Us has been previously analyzed and reviewed (see for example Masquida and Westhof 2000; Varani and McClain 2000), and predominantly consist of standard G•U wobble pairs. In a standard wobble G•U pair, the U moves into the deep major groove leaving a cavity on the shallow minor groove side frequently occupied by a water molecule that links the O2′(U), O2(U), and the N2(G) (Fig. 1A; Westhof 1988; Auffinger and Westhof 1998; Trikha et al. 1999). The displacement of the U also creates a binding site frequently occupied by a hydrated potassium ion with direct binding to O4(U) and O6(G), and via one hydration water molecule to N7(G) in the major groove (Fig. 1A; Klein et al. 2004; Leonarski et al. 2019). Furthermore, because of the movement of the U, a G•U pair is not isosteric (or superimposable) to a U•G pair, unlike the usual Watson–Crick G=C and A-U pairs (Crick 1966; Westhof 2014). A thorough analysis of G•U pairs in ribosomal structures is presented in Mokdad et al. (2006). Interestingly, two alternative G•U pairs (Ogle et al. 2002; Ogle and Ramakrishnan 2005; Weixlbaumer et al. 2007; Demeshkina et al. 2012, 2013; Rozov et al. 2015, 2016a,b,c, 2018) occur in codon–anticodon interactions within functional ribosomes, the tautomeric and the anionic forms, as reviewed in Westhof et al. (2019). With a modified U34 in the anticodon loop, the U34*•G3 pairs can adopt either a tautomeric Watson–Crick-like form with three H-bonds or an anionic pair with the U34* moved into the shallow groove (instead of the deep major groove seen with a standard U•G pair) (Fig. 1B,C). In both cases, the formed U34*•G3 pairs are isosteric either to a G34•U3 or a G34=C3 pair depending on the type of modification (Weixlbaumer et al. 2007; Kurata et al. 2008; Cantara et al. 2013; Rozov et al. 2016a; Westhof et al. 2019). It should be noted that in the anionic U34*•G3 pair, depending on the type of modifications, U34* is in a zwitterionic form with the positive charge on the C5 modification and the negative charge on the N3 of U (Fig. 1C; Sochacka et al. 2015, 2017).These observations show how the U34 modifications restrict the movements of U34*•G3 to be accommodated within the ribosomal grip during efficient mRNA decoding (see the local environment in Supplemental Fig. S1).
FIGURE 1.
(A) A standard wobble G•U pair (from PDB 1HQ1, Batey et al. 2001) with a water molecule in the minor groove (red sphere) and a hydrated potassium ion (purple sphere) in the major groove. All distances are in Å and between the heavy atoms. The distance between the two C1′ carbon atoms is 10.7 Å (values between 10.4 Å and 10.7 Å are commonly observed). (B) The observed G•U pair between G3 and modified U34* in the ternary complex between the ribosomal A site, the anticodon loop of the A-tRNA and the mRNA (from PDB 5E81, Rozov et al. 2016a). The tRNA U34* is modified in 5-methylaminomethyl-2-thioUridine. The distance between the two C1′ carbon atoms is 11.4 Å. (C) Possible electronic structure of the modified U34* where X at position 2 is S and R at position 5 is methylaminomethyl, probably charged and thus forming a zwitterion. The negative charge may be delocalized between the N3 and S2. For chemical data and a discussion, see Sochacka et al. (2015, 2017).
Tautomeric and anionic pairs have been observed in solution using NMR spectroscopy (Kimsey et al. 2015). We wondered whether such alternative G•U pairs, without U modifications, are observed in other RNA structures, especially ribosome structures. Thanks to the progress in cryo-electron microscopy of biological macromolecules, structures of bacterial ribosomes are now available with sufficiently high resolution to be confident of the hydrogen bond distances and the relative positions of the nucleotides in G•U pairs. For E. coli ribosomes, high-resolution structures are now available at 2.0 Å (Watson et al. 2020) and 1.55 Å (Fromm et al. 2023).
RESULTS
The anionic pairs in the ribosome
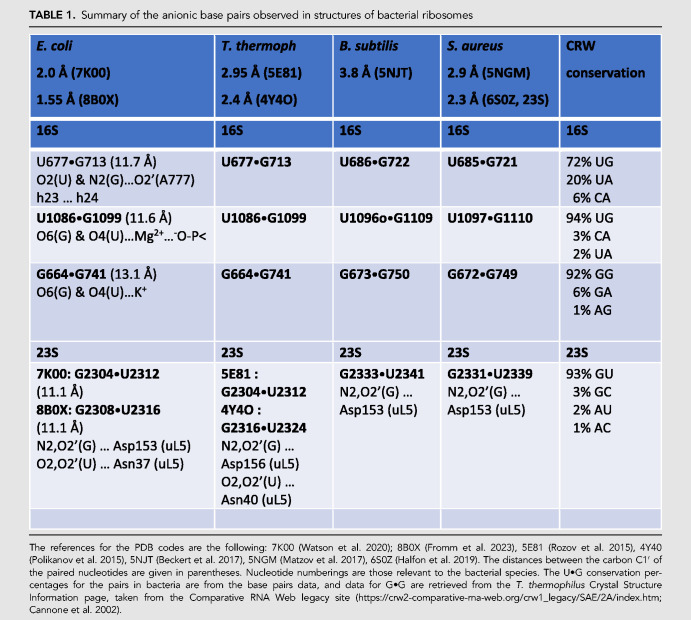
Table 1 gathers the three main anionic G•U pairs found in bacterial ribosomes (two in the 16S and one in the 23S rRNA) together with an anionic G•G pair in high or good resolution structures of four bacterial species. The identifications are based on the relative positions of the two interacting bases (toward the minor or the major groove) together with distances between the interacting heavy atoms (N or O). The identifications follow the calculations previously performed (Sochacka et al. 2015, 2017). Both the Gs and the Us are highly conserved based on structural alignments of 544 bacterial 16S rRNAs (Hosseini et al. 2018). Although a comparable structural alignment of 23S rRNAs is not available, we were able to retrieve the conservation in Bacteria for all four pairs from the legacy Comparative RNA Web site (Cannone et al. 2002). None of the residues in the 16S rRNA, discussed in the present paper, are universally conserved (i.e., throughout the three kingdoms of life) according to a recent survey (Noller et al. 2022).
TABLE 1.
Summary of the anionic base pairs observed in structures of bacterial ribosomes
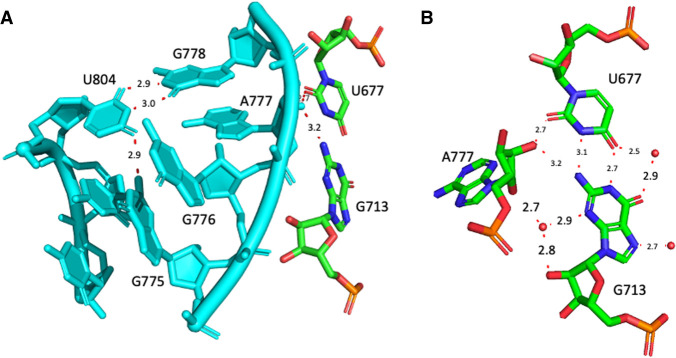
The anionic U677•G713 pair
The U is highly conserved (>90%) and, in some sequences, one can observe a switch from a U•G to a C•A opposition. The U677•G713 pair (Fig. 2A), in helix 23, interacts with the O2′ of A777, which is conserved at more than 99% and located in the bulge of helix h24 (Fig. 2B). Interestingly, the apical loops of h23 and h24 are involved, respectively, in the formation of the E-site and the P-site. Identical contacts are present in the other structures of bacterial ribosomes analyzed. In the high-resolution structure of an archaeal 30S subunit (PDB: 7ZHG, Kazan et al. 2022), the U•G pair is substituted with a C=G Watson–Crick pair C644=G680, however with the G680 2′-O-methylated. In Archaea, that pair is 87.5% C=G, 9.9% C/U, 2.2% U•G (Cannone et al. 2002). Despite the local differences in nucleotides, nts 701 to 741 of 7K00 and nts 668 to 708 of 7ZHG (Kazan et al. 2022) superimpose extremely well (RMSD less than 0.5 Å).
FIGURE 2.
(A) The internal loop of helix h24 in 16S rRNA in contact with the anionic U677•G713 pair is shown in cyan. Please note the stacking of the two G776 and G775 and the usual wobble pair at G778•U804. (B) The anionic U677•G713 pair with the contact to the O2′ of A777 in the 16S rRNA of E. coli (from 8B0X, Fromm et al. 2023). For comparison, the distances in PDB 7K00 (Watson et al. 2020) corresponding to N1(G)…O4(U), N2(G)…N3(U) are 2.7 Å and 3.0 Å and those for N2(G)…O2′(A777), O2(U)…O2′(A777) are 3.1 Å and 2.4 Å. The red spheres represent water molecules in close proximity.
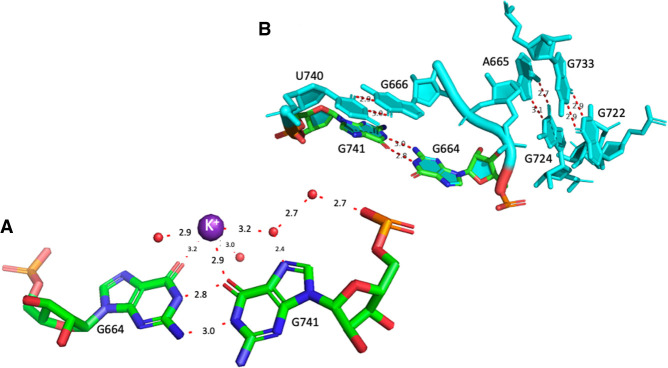
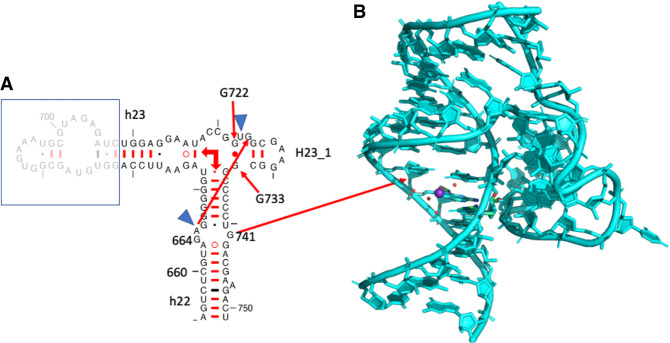
The anionic G664•G741 pair
This pair is not a G•U pair but it also involves a loss of a proton, in this case of one of the Gs (G741) and not of an U. Such a G•G pair is cis Watson–Crick/Watson–Crick and the only instance missing in that pair family Table (Leontis et al. 2002). In another possibility, less probable but that cannot be excluded (especially in the absence of other examples of such a pair in crystal structures), G664 would adopt a tautomeric form with O6H and G741 would remain neutral. The G664•G741 is conserved at 92% in Bacteria (Table 1). In a few sequences, one of the Gs is replaced by an A in which case the pair most probably adopts a wide and neutral cis Watson–Crick/Watson–Crick G•A pair (with a distance between C1′ atoms of 12.7 Å, Li et al. 2007).
The G664•G741 pair (Fig. 3A) is not far away in space from the preceding anionic G•U pair. It presents a cross-strand stacking with G666 (Fig. 3B), which forms a usual wobble pair with U740 forcing A665 (Fig. 3B) to bulge out for pairing with G724 (conserved at 96.7%). G724 stacks with G722 (conserved at 90%), which forms a common trans WC/WC GC pair with G733 (also conserved at 90%) (Fig. 3B). Figure 4A shows elements of the secondary structure with key nucleotides marked and Figure 4B illustrates the burying of the anionic G664•G741 within the hairpin h23_1. The complex interactions between G725 and C726 of h23.1 with G664 are shown in Supplemental Figure S2B. The G664•G741 pair is present in the complex of S15 with its rRNA fragment (PDB: 1G1X, Agalarov et al. 2000; see Supplemental Figure S2B). Interestingly, this G•G pair is conserved in the archaeal 30S ribosome structure (PDB: 7ZHG, G631•G708, Kazan et al. 2022). However, in Archaea, that pair is 61% G•U and 19.5% G=C or U•G (Cannone et al. 2002).
FIGURE 3.
(A) The anionic G664•G741 pair with some water molecules and a hydrated K+ ion in the deep major groove. (B) The highly conserved organization around the anionic G664•G741 in E. coli (PDB 8B0X, Fromm et al. 2023). The residues G666•U740 form a usual wobble G•U pair. A665/G724 form a cis Hoogsteen/Watson–Crick pair and G722•G733 a trans Watson–Crick/Watson–Crick pair with G722 in the unusual syn conformation. For additional contacts to G664, see Supplemental Figure S2A.
FIGURE 4.
(A) Part of the secondary structure of the 16S rRNA showing helices h22, h23, h23_1. The blue triangles indicate that those two residues bulge out so that A665 and G724 can form a cis Hoogsteen/Watson–Crick pair. The right-angle arrows indicate the coaxial stacking between the helices in the three-way junction. (B) The figure shows how the shallow minor groove side of the anionic G664•G741 binds tightly to the hairpin h23_1. Drawing based on E. coli (PDB 8B0X, Fromm et al. 2023).
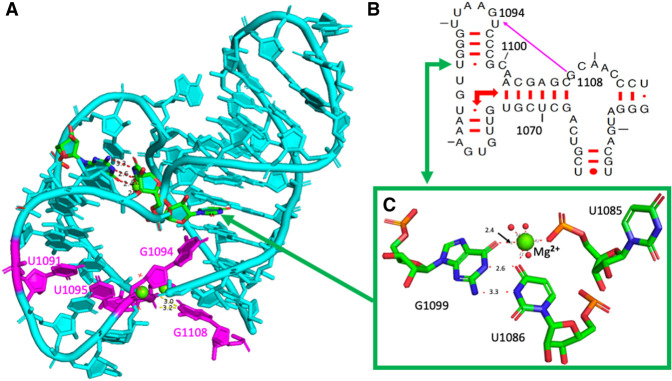
The anionic U1086•G1099 pair
This unusual U•G pair is within a three-way junction and important for positioning the hairpin h37 so that G1108 can interact directly with the 5′ phosphate of U1095 forcing G1094 to bulge out (Fig. 5A,B). The U1086•G1099 pair is conserved at 94% in bacterial ribosomes. In the major groove, O6(G) and O4(U) directly bind a hydrated Mg ion that contacts also an anionic phosphate oxygen of U1085 (Fig. 5C) which is 100% conserved in bacteria. It is also near the carboxyl terminus of ribosomal protein bS21, which packs against h37 containing the U•G pair and forms a stacking interaction with A1167 in the closing loop of helix h40 (See Fig. 3A,B in Watson et al. 2020).
FIGURE 5.
(A) The fold of the three-way junction with the anionic G1099•U1086 pair with, in purple, the contacts between G1108 and the 5′-phosphate of U1095 forcing G1094 to bulge out. (B) Elements of the secondary structure of the three-way junction. The right-angle arrows indicate the coaxial stacking between the helices in the three-way junction. (C) The anionic G1099•U1086 pair with a hydrated Mg2+ ion (green sphere) with its hydration sphere (red spheres) and a direct contact with an anionic phosphate oxygen of U1085. The distances to the Mg2+ ion are between 1.9 Å and 2.1 Å, except the one to the O6(G1099). Drawings based on E. coli (PDB 8B0X, Fromm et al. 2023).
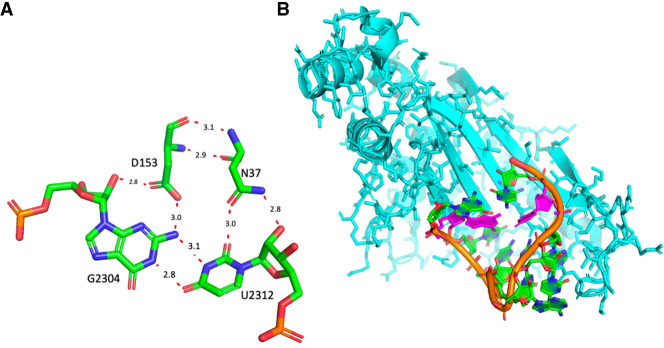
The anionic G2304•U2312 of the 23S rRNA
This is the only anionic G•U pair that we identified systematically within the large ribosomal subunit. This pair occurs in the central protuberance of the 50S ribosomal subunit and interacts with the side chains of two amino acids in ribosomal protein uL5 (Asn37 and Asp153 in E. coli) (Fig. 6A). It is the last pair of a hairpin and the capping loop is buried within uL5 (Fig. 6B). This interaction is conserved in all structures analyzed. Depending on the resolution, the contact with Asn37 (or equivalent) is not observed and the pair appears bent. Although uL5 is a universal ribosomal protein, the rRNA pair changes to a G=C Watson–Crick pair in the rabbit 60S ribosomal subunit (PDB 7O7Y, Bhatt et al. 2021). In Eukaryotes, that pair is G•U only in 4% of the sequences and otherwise Watson–Crick (Cannone et al. 2002). However, the superimposition of nts 2301 to 2310 from 7K00 with nts 3993 to 4002 from 7O7Y gives a RMSD of less than 0.5 Å. Further, the rabbit uL5 residues Asp129 and Asn23 form similar contacts with G3996=C4004. Similarly, in the structure of the complex between tRNA_Ala and its cognate aaRS (Naganuma et al. 2014; Chong et al. 2018), an Asp residue interacts with N2(G3) (and O2 of U70) while an Asn residues interacts in the major groove (instead of the minor groove) with O4(U70) (see Supplemental Fig. S3).
FIGURE 6.
(A) The anionic G2304•U2312 pair in the 23S rRNA and its contacts with two conserved residues of protein uL5. (B) The drawing illustrates how the capping loop following the anionic pair (in purple) is buried within uL5. Drawings based on E. coli (PDB 8B0X, Fromm et al. 2023). Please note that in 8B0X that pair is numbered G2308•U2316 (see Table 1).
Occurrence of anionic G•U pairs in other RNA structures
An anionic G•U pair is also detected in a Mg2+-sensing riboswitch (Dann et al. 2007), although the resolution of the structure is 2.6 Å. The anionic G76•U65 pair binds in the major groove a hydration water from a hydrated Mg ion and is stacked between a G=C pair and a one H-bond C•A pair (PDB 2QBZ, (Dann et al. 2007; see the local environment in Supplemental Fig. S4). Mapping G76•U65 of 2QBZ to Rfam family RF00380 gives the following sequence variability: C=G 389, U•G 130, A-U 84, U-A 62. So, there is some support for that pair to stay U•G, but more support for it becoming C=G.
DISCUSSION
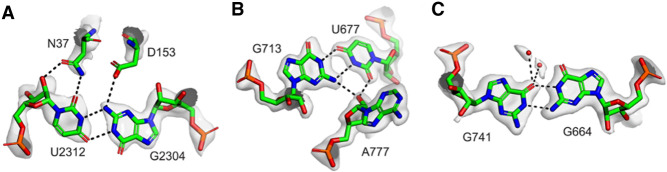
Compared with the standard wobble pair, the anionic G•U pairs display the U displaced into the shallow minor groove with a distance between the sugar carbon C1′ larger by about 1 Å (see Table 1). The high quality of the recent structures (Watson et al. 2020; Fromm et al. 2023), together with the residue conservations in sequence alignments and their systematic occurrences in previous ribosomal structures, exclude a systematic error in molecular fitting in the observed anionic G•U pairs. The quality of the fits is illustrated by the cryo-EM maps of two anionic G•U pairs (Fig. 7A,B) and the anionic G664•G741 pair (Fig. 7C) are displayed. Besides, functionally, they are not scattered throughout the ribosome but gather near the platform and the neck of the 30S ribosomal subunit (Fig. 8A,B). In the large ribosomal subunit, protein uL5 has a major role in the assembly of the central protuberance (Korepanov et al. 2012) and is involved in the formation of the intersubunit bridge 1b (Yusupov et al. 2001). Ribosomal protein uL5 also binds the 5S rRNA.
FIGURE 7.
Views of the cryo-EM maps around some anionic pairs based on the E. coli ribosome structure (7K00, Watson et al. 2020). (A) Map around the anionic G2304•U2312 pair in the 23S rRNA and its contacts with two conserved residues of protein uL5, N37, and D153. The nature of the amino acid side chain atoms in contact with G2304 and U2312 cannot be ascertained at this stage. The distance between the nitrogen atom of N37 and one of the oxygen atoms of D153 is 3.4 Å. Note also that the density for the carboxylate group is weak, which could be due to radiation damage (Marques et al. 2019). (B) Map around the anionic U677•G713 pair and A777 in the 16S rRNA. (C) Map around the anionic G664•G741 in E. coli. The water molecules bound to O4(G741) are above and below the plane of the base.
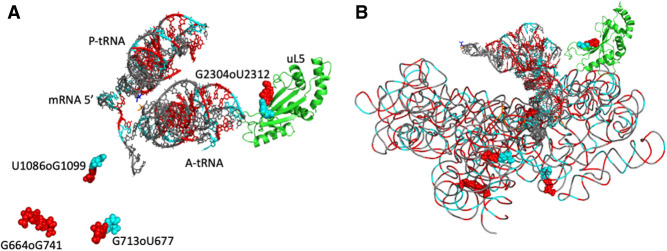
FIGURE 8.
Two views of the molecular environment around the anionic pairs and their proximity within the ribosome structure. The guanines are colored red and shown as van der Waals spheres with the uracils colored in cyan. (A) The A-tRNA and P-tRNA bound to the mRNA (with van der Waals spheres at the right) are shown. The protein uL5 in the 23S rRNA is colored green. (B) The backbone of the whole 16S rRNA is superimposed (with the same color code). The drawings are from 7K00 (Watson et al. 2020).
These observations show that anionic G•U pairs do occur with unmodified nucleotides, as dynamically observed by NMR (Kimsey et al. 2015). Here they were observed in crowded and tight molecular environments in specific regions of the bacterial ribosomes. The anionic G•U pairs have been observed in tight contacts with Asp and Asn side chains (G2304•U2312) or with the sugar-phosphate backbone of a nucleotide (U677•G713) in the minor groove. In other instances, the anionic pairs interact with partially hydrated potassium or magnesium ions, like the standard wobble pair, in the major groove. The diversity in pairing geometries in nucleic acids originates from the various combinations of contacts between the nucleotide base edges (Leontis et al. 2002). Both the tautomeric form of G•U pairs (Demeshkina et al. 2012) and the anionic G•U pair (Sochacka et al. 2015, 2017; Rozov et al. 2016a) involve a change in electronic structure (respectively, of either the G or the U, or only the U) that leads to a pairing diversity between the two Watson–Crick edges. Interestingly, among the four neutral bases, G is the most electropositive at its N1 and N2 edge and that edge is often observed interacting with anions like chloride or sulfate (Auffinger et al. 2004), anionic phosphate oxygens (Zirbel et al. 2009), or the negatively charged side chains of Asp or Glu (Treger and Westhof 2001). In the anionic G•U and G•G pairs described here, one neutral G interacts with an anionic base (U or another G). One may thus consider that the tautomeric and anionic pairs involving G or U contribute to molecular accommodation and stabilization during the evolution of RNA sequences.
MATERIALS AND METHODS
First, a manual inspection and analysis of PDB entry 7K00 (Watson et al. 2020) was performed together with map fitting. Unless otherwise indicated, nucleotide numbers refer to 7K00. This analysis was extended to other ribosome structures manually and then after a search using FR3D (Petrov et al. 2011). The diagrams for the secondary structures are from Cannone et al. (2002). The drawings were made using PyMOL (The PyMOL Molecular Graphics System, version 1.2r3pre, Schrödinger, LLC.). For the conservation of nucleotides in the 16S rRNA, we used the alignment present in the supplemental material of Hosseini et al. (2018). For the 23S rRNA, we retrieved base pair and nucleotide frequency data from the Comparative RNA Web legacy site (https://crw2-comparative-rna-web.org/crw1_legacy/SAE/2A/nt_Frequency/index.htm; Cannone et al. 2002). The latter values are given in Table 1. Throughout, we use the description of base pairs by Leontis and Westhof (2001) with the generic “=” for G=C pairs, “-“ for A-U pairs, and “•” for wobble G•U pairs or other bases interacting through their Watson–Crick edges. We use in the text the numbering of 7K00 (Watson et al. 2020); in Table 1 the nucleotide correspondences for the other PDB files are indicated.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robin Gutell (University of Texas at Austin) for help to retrieve the G•G sequence data. This work was supported by the LABEX: ANR-10-LABX-0036_NETRNA managed by the French National Research Agency as part of the Investments for the future program. Z.L.W. and J.H.D.C. were supported by the National Science Foundation Center for Genetically Encoded Materials (C-GEM), CHE-2002182. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM085328 to C.L.Z. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions: E.W., Z.L.W., C.L.Z., and J.H.D.C. participated in the research and contributed to the writing of the paper.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079583.123.
Freely available online through the RNA Open Access option.
REFERENCES
- Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. 2000. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science 288: 107–113. 10.1126/science.288.5463.107 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 1998. Hydration of RNA base pairs. J Biomol Struct Dyn 16: 693–707. 10.1080/07391102.1998.10508281 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Bielecki L, Westhof E. 2004. Anion binding to nucleic acids. Structure 12: 379–388. 10.1016/j.str.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Batey RT, Sagar MB, Doudna JA. 2001. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J Mol Biol 307: 229–246. 10.1006/jmbi.2000.4454 [DOI] [PubMed] [Google Scholar]
- Beckert B, Abdelshahid M, Schafer H, Steinchen W, Arenz S, Berninghausen O, Beckmann R, Bange G, Turgay K, Wilson DN. 2017. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J 36: 2061–2072. 10.15252/embj.201696189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt PR, Scaiola A, Loughran G, Leibundgut M, Kratzel A, Meurs R, Dreos R, O'Connor KM, McMillan A, Bode JW, et al. 2021. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372: 1306–1313. 10.1126/science.abf3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantara WA, Murphy F, Demirci H, Agris PF. 2013. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci 110: 10964–10969. 10.1073/pnas.1222641110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YE, Guo M, Yang XL, Kuhle B, Naganuma M, Sekine SI, Yokoyama S, Schimmel P. 2018. Distinct ways of G:U recognition by conserved tRNA binding motifs. Proc Natl Acad Sci 115: 7527–7532. 10.1073/pnas.1807109115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. 1966. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 19: 548–555. 10.1016/S0022-2836(66)80022-0 [DOI] [PubMed] [Google Scholar]
- Dann CE III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130: 878–892. 10.1016/j.cell.2007.06.051 [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259. 10.1038/nature10913 [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2013. New structural insights into the decoding mechanism: translation infidelity via a G•U pair with Watson-Crick geometry. FEBS Lett 587: 1848–1857. 10.1016/j.febslet.2013.05.009 [DOI] [PubMed] [Google Scholar]
- Fromm SA, O'Connor KM, Purdy M, Bhatt PR, Loughran G, Atkins JF, Jomaa A, Mattei S. 2023. The translating bacterial ribosome at 1.55 A resolution generated by cryo-EM imaging services. Nat Commun 14: 1095. 10.1038/s41467-023-36742-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon Y, Matzov D, Eyal Z, Bashan A, Zimmerman E, Kjeldgaard J, Ingmer H, Yonath A. 2019. Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant. Sci Rep 9: 11460. 10.1038/s41598-019-48019-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M, Roy P, Sissler M, Zirbel CL, Westhof E, Leontis N. 2018. How to fold and protect mitochondrial ribosomal RNA with fewer guanines. Nucleic Acids Res 46: 10946–10968. 10.1093/nar/gky762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan R, Bourgeois G, Lazennec-Schurdevin C, Larquet E, Mechulam Y, Coureux PD, Schmitt E. 2022. Role of aIF5B in archaeal translation initiation. Nucleic Acids Res 50: 6532–6548. 10.1093/nar/gkac490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-Hashimi HM. 2015. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 519: 315–320. 10.1038/nature14227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10: 1366–1379. 10.1261/rna.7390804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korepanov AP, Korobeinikova AV, Shestakov SA, Garber MB, Gongadze GM. 2012. Protein L5 is crucial for in vivo assembly of the bacterial 50S ribosomal subunit central protuberance. Nucleic Acids Res 40: 9153–9159. 10.1093/nar/gks676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. 2008. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U•G wobble pairing during decoding. J Biol Chem 283: 18801–18811. 10.1074/jbc.M800233200 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531. 10.1093/nar/gkf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarski F, D'Ascenzo L, Auffinger P. 2019. Nucleobase carbonyl groups are poor Mg2+ inner-sphere binders but excellent monovalent ion binders—a critical PDB survey. RNA 25: 173–192. 10.1261/rna.068437.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. 10.1017/S1355838201002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, et al. 2007. Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res 35: 6424–6438. 10.1093/nar/gkm664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques MA, Purdy MD, Yeager M. 2019. CryoEM maps are full of potential. Curr Opin Struct Biol 58: 214–223. 10.1016/j.sbi.2019.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B, Westhof E. 2000. On the wobble GoU and related pairs. RNA 6: 9–15. 10.1017/S1355838200992082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzov D, Aibara S, Basu A, Zimmerman E, Bashan A, Yap MF, Amunts A, Yonath AE. 2017. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat Commun 8: 723. 10.1038/s41467-017-00753-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokdad A, Krasovska MV, Sponer J, Leontis NB. 2006. Structural and evolutionary classification of G/U wobble basepairs in the ribosome. Nucleic Acids Res 34: 1326–1341. 10.1093/nar/gkl025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma M, Sekine S, Chong YE, Guo M, Yang XL, Gamper H, Hou YM, Schimmel P, Yokoyama S. 2014. The selective tRNA aminoacylation mechanism based on a single G*U pair. Nature 510: 507–511. 10.1038/nature13440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Donohue JP, Gutell RR. 2022. The universally conserved nucleotides of the small subunit ribosomal RNAs. RNA 28: 623–644. 10.1261/rna.079019.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177. 10.1146/annurev.biochem.74.061903.155440 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. 10.1016/S0092-8674(02)01086-3 [DOI] [PubMed] [Google Scholar]
- Petrov AI, Zirbel CL, Leontis NB. 2011. WebFR3D—a server for finding, aligning and analyzing recurrent RNA 3D motifs. Nucleic Acids Res 39: W50–W55. 10.1093/nar/gkr249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov SV, Soll D, Steitz TA. 2015. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 22: 342–344. 10.1038/nsmb.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2015. Structural insights into the translational infidelity mechanism. Nat Commun 6: 7251. 10.1038/ncomms8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. 2016a. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun 7: 10457. 10.1038/ncomms10457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2016b. New structural insights into translational miscoding. Trends Biochem Sci 41: 798–814. 10.1016/j.tibs.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Rozov A, Westhof E, Yusupov M, Yusupova G. 2016c. The ribosome prohibits the G*U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res 44: 6434–6441. 10.1093/nar/gkw431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Wolff P, Grosjean H, Yusupov M, Yusupova G, Westhof E. 2018. Tautomeric G*U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria. Nucleic Acids Res 46: 7425–7435. 10.1093/nar/gky547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacka E, Szczepanowski RH, Cypryk M, Sobczak M, Janicka M, Kraszewska K, Bartos P, Chwialkowska A, Nawrot B. 2015. 2-Thiouracil deprived of thiocarbonyl function preferentially base pairs with guanine rather than adenine in RNA and DNA duplexes. Nucleic Acids Res 43: 2499–2512. 10.1093/nar/gkv109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacka E, Lodyga-Chruscinska E, Pawlak J, Cypryk M, Bartos P, Ebenryter-Olbinska K, Leszczynska G, Nawrot B. 2017. C5-substituents of uridines and 2-thiouridines present at the wobble position of tRNA determine the formation of their keto-enol or zwitterionic forms—a factor important for accuracy of reading of guanosine at the 3′-end of the mRNA codons. Nucleic Acids Res 45: 4825–4836. 10.1093/nar/gkw1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger M, Westhof E. 2001. Statistical analysis of atomic contacts at RNA-protein interfaces. J Mol Recognit 14: 199–214. 10.1002/jmr.534 [DOI] [PubMed] [Google Scholar]
- Trikha J, Filman DJ, Hogle JM. 1999. Crystal structure of a 14 bp RNA duplex with non-symmetrical tandem GxU wobble base pairs. Nucleic Acids Res 27: 1728–1739. 10.1093/nar/27.7.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, McClain WH. 2000. The G•U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1: 18–23. 10.1093/embo-reports/kvd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JH. 2020. Structure of the bacterial ribosome at 2 Å resolution. Elife 9: e60482. 10.7554/eLife.60482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Murphy F, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. 2007. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol 14: 498–502. 10.1038/nsmb1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E. 1988. Water: an integral part of nucleic acid structure. Annu Rev Biophys Biophys Chem 17: 125–144. 10.1146/annurev.bb.17.060188.001013 [DOI] [PubMed] [Google Scholar]
- Westhof E. 2014. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett 588: 2464–2469. 10.1016/j.febslet.2014.06.031 [DOI] [PubMed] [Google Scholar]
- Westhof E, Yusupov M, Yusupova G. 2019. The multiple flavors of GoU pairs in RNA. J Mol Recognit 32: e2782. 10.1002/jmr.2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. 10.1126/science.1060089 [DOI] [PubMed] [Google Scholar]
- Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB. 2009. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res 37: 4898–4918. 10.1093/nar/gkp468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.