Abstract
LncRNAs comprise a heterogeneous class of RNA-encoding genes typified by low expression, nuclear enrichment, high tissue-specificity, and functional diversity, but the vast majority remain uncharacterized. Here, we assembled the mouse liver noncoding transcriptome from >2000 bulk RNA-seq samples and discovered 48,261 liver-expressed lncRNAs, a majority novel. Using these lncRNAs as a single-cell transcriptomic reference set, we elucidated lncRNA dysregulation in mouse models of high fat diet-induced nonalcoholic steatohepatitis and carbon tetrachloride-induced liver fibrosis. Trajectory inference analysis revealed lncRNA zonation patterns across the liver lobule in each major liver cell population. Perturbations in lncRNA expression and zonation were common in several disease-associated liver cell types, including nonalcoholic steatohepatitis-associated macrophages, a hallmark of fatty liver disease progression, and collagen-producing myofibroblasts, a central feature of liver fibrosis. Single-cell-based gene regulatory network analysis using bigSCale2 linked individual lncRNAs to specific biological pathways, and network-essential regulatory lncRNAs with disease-associated functions were identified by their high network centrality metrics. For a subset of these lncRNAs, promoter sequences of the network-defined lncRNA target genes were significantly enriched for lncRNA triplex formation, providing independent mechanistic support for the lncRNA–target gene linkages predicted by the gene regulatory networks. These findings elucidate liver lncRNA cell-type specificities, spatial zonation patterns, associated regulatory networks, and temporal patterns of dysregulation during hepatic disease progression. A subset of the liver disease-associated regulatory lncRNAs identified have human orthologs and are promising candidates for biomarkers and therapeutic targets.
Keywords: lncRNA, high fat diet, carbon tetrachloride, liver fibrosis, BigSCale2 networks, liver disease marker genes
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is rapidly becoming the most common chronic liver disease, affecting 25% of the world's adult population, most notably individuals with obesity, type II diabetes and metabolic syndrome (Younossi et al. 2018). NAFLD comprises a continuum of liver pathologies, ranging from fat accumulation, known as simple steatosis or nonalcoholic fatty liver, to nonalcoholic steatohepatitis (NASH), a more severe disease subtype characterized by excessive lipid accumulation, chronic inflammation, hepatocyte ballooning, and varying degrees of fibrosis (Friedman et al. 2018; Sheka et al. 2020). Liver fibrosis is characterized by excessive accumulation of collagens and other extracellular matrix proteins (Kisseleva and Brenner 2021) and can be induced in NASH or by environmental chemical-induced injury and alcohol abuse (Roehlen et al. 2020). The underlying mechanisms of development and progression of these liver diseases are still poorly understood. Moreover, there are no approved therapeutics for advanced NASH and liver fibrosis (Raza et al. 2021), which all too frequently advance to liver cirrhosis and hepatocellular carcinoma (Anstee et al. 2019; Gines et al. 2021).
Long noncoding RNAs (lncRNAs) comprise a heterogeneous class of RNA-encoding genes, primarily defined by their low protein coding potential/low translational activity and by a minimum RNA length of 200 nt. LncRNAs often have low expression, show strong nuclear enrichment (Guo et al. 2020), have high tissue-specificity (Gloss and Dinger 2016), and may regulate gene expression through effects on chromatin states and transcriptional regulation or at the post-transcriptional level (Zhang et al. 2019; Statello et al. 2021). Thousands of liver-expressed lncRNAs have been identified, a subset of which are responsive to endogenous hormones (Melia et al. 2016; Melia and Waxman 2019; Goldfarb et al. 2022) or exposure to xenobiotics (Lodato et al. 2017; Dempsey and Cui 2019; Karri and Waxman 2020; Goldfarb and Waxman 2021), many of which can promote NASH, cirrhosis and other liver pathologies (Jin et al. 2020; Massart et al. 2022; Rajak et al. 2022). Prior studies identified individual lncRNAs that impact liver disease (He et al. 2020; Huang et al. 2021; Unfried et al. 2021); examples include SRA, which promotes hepatic steatosis by repressing adipose triglyceride lipase expression (Chen et al. 2016a), GAS5, which attenuates carbon tetrachloride (CCl4)-induced liver fibrosis by acting as a sponge for miRNA-23a (Dong et al. 2019), and HULC, which inhibits liver fibrosis associated with NAFLD (Shen et al. 2019). Thousands of other liver-expressed lncRNAs are uncharacterized or even unidentified, many of which are likely to impact liver pathophysiology.
Hepatocytes account for 60%–70% of all cells in the liver, with the balance largely comprised of three major nonparenchymal cell types: endothelial cells, hepatic stellate cells, and Kupffer cells (liver resident macrophages). Liver cell type-specific gene expression patterns and their zonated regulation across the liver lobule have been elucidated in both healthy liver (Halpern et al. 2017; Kietzmann 2017; Cunningham and Porat-Shliom 2021) and in high fat diet-induced liver disease (Xiong et al. 2019; Loft et al. 2021; Su et al. 2021) by using single-cell (sc)RNA-sequencing technologies. scRNA-seq has also elucidated the role of hepatic mesenchymal cells, including hepatic stellate cells (HSCs), in liver fibrosis induced by hepatotoxins such as CCl4 (Fig. 1A; Dobie et al. 2019; Ramachandran et al. 2019; Yang et al. 2021; Zhang et al. 2021). While these studies have determined the roles of liver cell subpopulations and individual protein coding genes (PCGs) (Ramachandran et al. 2020), prior studies of lncRNAs have largely been limited to lncRNAs with RefSeq gene or other annotations, which comprise only a small fraction of the tens of thousands of lncRNAs thought to be encoded by the genome. Many lncRNAs are expressed in bulk tissue at much lower levels than PCGs, but often exhibit high tissue specificity, raising the possibility that detection sensitivity may actually be increased by using scRNA-seq technology to characterize lncRNAs whose expression is restricted to a specific subpopulation of cells in the liver.
FIGURE 1.
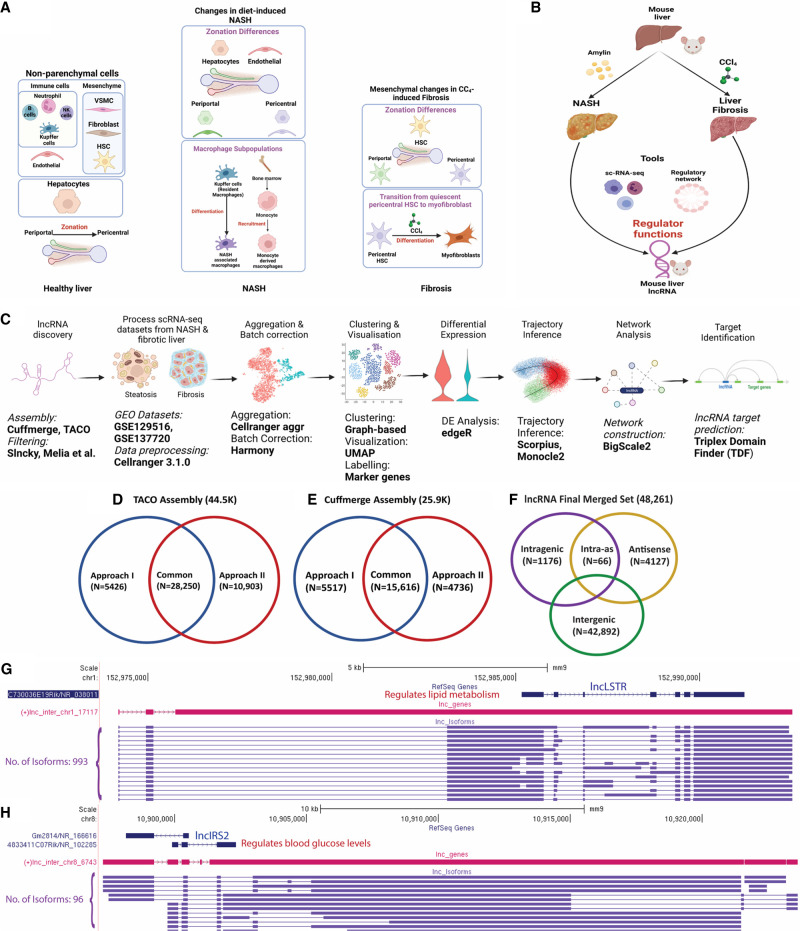
LncRNA discovery and role in NASH and liver fibrosis analyzed using single-cell technology. (A) Healthy liver is comprised of hepatocytes and nonparenchymal cells, notably endothelial cells, mesenchymal cells, and immune cell populations. With the emergence of NASH, changes in gene expression and zonation occur in hepatocytes and endothelial cells and new macrophage subpopulations emerge. In CCl4-induced liver fibrosis, changes in hepatic mesenchymal cells include zonation differences in hepatic stellate cells (HSC) and the transition of pericentral stellate cells to collagen-producing myofibroblasts. (B) Discovery of regulatory roles of lncRNAs in two liver disease models: Amylin diet-induced NASH and CCl4-induced liver fibrosis. (C) Computational workflow for characterization of functional roles of liver-expressed lncRNAs used in this study. (D,E) Numbers of mouse (mm9) liver lncRNAs discovered using TACO assembly (D) and Cuffmerge assembly (E), with two different filtering approaches, I and II. (F) Final set of 48,261 mouse lncRNAs, classified based on their location with respect to PCGs after conversion to mouse mm10 genomic coordinates. (G,H) Two liver-expressed lncRNAs, both found to be comprised of many novel isoforms, a subset of which is shown.
Here, we sought to elucidate on a global scale the roles of liver-expressed lncRNAs in biological pathways related to liver disease development. We used an integrative approach to assemble the hepatic transcriptome from >2000 bulk murine liver RNA-seq samples and discovered more than 48,000 liver-expressed lncRNAs, a majority novel and previously uncharacterized, including many lncRNAs that share orthology with corresponding human sequences. We integrated multiple public scRNA-seq data sets for healthy mouse liver to create a single-cell transcriptomic reference atlas, which enabled us to characterize the liver cell-type specificities of thousands of novel lncRNAs and identify more than 100 liver cell type-specific lncRNA marker genes. We elucidated liver lncRNA zonation profiles using trajectory inference algorithms for five liver cell populations, complementing those reported earlier for PCGs (Fig. 1A; Halpern et al. 2018; Ben-Moshe et al. 2019; Dobie et al. 2019; Kalucka et al. 2020). Further, to elucidate lncRNA dysregulation in liver disease, we analyzed liver scRNA-seq data from mice fed a high fat, high fructose diet (HFHFD, also known as AMLN diet) (Su et al. 2021), which induces disease progression from healthy liver to NAFLD (simple steatosis) and then NASH, revealing lncRNA transcriptomic dysregulation patterns during disease progression. We also analyzed the role of lncRNAs in the hepatic mesenchyme from healthy and CCl4-induced fibrotic liver (Fig. 1B; Dobie et al. 2019). Finally, we constructed gene regulatory networks for both healthy and diseased liver to discover key regulatory lncRNAs based on gene network centrality metrics, and we identified a subset of regulatory lncRNAs whose PCG target gene promoters are significantly enriched for direct interactions via triplex-based lncRNA binding (Fig. 1C). Overall, our findings reveal an unanticipated complexity of hepatic lncRNA biology. The data sets obtained are expected to serve as a rich resource for discovery of lncRNA biomarkers and in studies targeting lncRNAs implicated in development of NASH and foreign chemical-induced liver fibrosis.
RESULTS
Global discovery of liver-expressed lncRNAs
We reconstructed the mouse liver transcriptome from 2089 bulk RNA-seq samples representing a wide range of biological conditions (Supplemental Table S1A). We integrated results from two different transcriptome assembly methods, TACO (Niknafs et al. 2017) and Cuffmerge (Trapnell et al. 2012), and used two approaches to analyze the output and identify liver-expressed lncRNA genes and isoform structures (Chen et al. 2016b; Melia et al. 2016). The transcriptomes generated by each method were processed and analyzed separately using two discovery pipelines (Fig. 1D,E), which after integrating with a prior set of 15,558 mouse liver lncRNAs based on a much smaller number of bulk RNA-seq samples (Melia and Waxman 2019) yielded a global set of 48,261 liver-expressed lncRNAs, including 5656 multiexonic genes (Supplemental Table S1C) and a total of 150,280 isoforms (Supplemental Table S1D). Eighty-nine percent of the 48,261 lncRNAs are intergenic (Fig. 1F). Further, 9543 (19.8%) of the lncRNAs have orthologous sequences in the human genome (hg38), and 1722 were orthologous to an incomplete set comprised of 5795 lncRNAs that we previously identified in rat liver (Supplemental Table S1E; Karri and Waxman 2020). The final lncRNA data set includes many novel isoforms of some well-characterized liver lncRNAs. For example, we identified hundreds of isoforms of LncLSTR (lnc17117), a liver-enriched RNA that regulates lipid metabolism in mice (Fig. 1G; Li et al. 2015) and 96 isoforms of LincIRS2 (lnc6743), which protects against diabetes and whose knockdown increases blood glucose, insulin resistance and aberrant glucose output (Fig. 1H; Pradas-Juni et al. 2020). However, the vast majority of the 48,261 liver lncRNAs are novel and of unknown function.
LncRNA expression in liver cell subpopulations
The low expression of many lncRNAs makes it difficult to reliably characterize their expression using bulk tissue RNA-seq, where expression patterns are dominated by hepatocytes, which comprise 60%–70% of all cells in adult liver (Si-Tayeb et al. 2010; Braet et al. 2018). Moreover, the high frequency of scRNA-seq drop out of low abundance transcripts limits the ability of scRNA-seq to detect and characterize the many thousands of lowly expressed lncRNAs. To increase the sensitivity for lncRNA detection, we pooled, integrated, and harmonized data from four public mouse liver scRNA-seq data sets comprising 39,878 liver cells (Supplemental Table S1G,H) to give a single, uniform single-cell landscape for healthy mouse liver. The resulting UMAP, which is enriched in nonparenchymal cells (Fig. 2A), contains 13 major cell clusters identified by their marker gene expression patterns (Supplemental Fig. S1A): hepatocytes, endothelial cells, Kupffer cells (liver macrophages), mesenchymal subpopulations comprised of hepatic stellate cells (HSCs), vascular smooth muscle cells (VSMCs) and fibroblasts, as well as dendritic cells, cholangiocytes, natural killer and T cells, B cells, B plasma cells, neutrophils, and dividing cells. A total of 30,092 distinct lncRNAs were detectable in this healthy liver data set, of which 24,961 (83%) are novel genes (Supplemental Table S2B). A total of 1352 of these lncRNAs were detectable in ≥5% of cells from at least one cell cluster (Fig. 2B; Supplemental Table S2A,B). Fourteen lncRNAs (11 with human orthologs, indicated by *) were detected in all liver cell types at >5% cells/cluster (Supplemental Table S2C). Examples of such widely expressed liver lncRNAs with established roles in liver biology and pathophysiology include: Malat1 (lnc31752*) and Norad/LINC00657 (lnc1906*), which promote hepatocellular carcinoma (Toraih et al. 2018; Yang et al. 2019), as do Snhg8 (lnc2803*) (Dong et al. 2018) and Dleu2 (lnc25736*) (Guo et al. 2019); Gas5 (lnc733*), which alleviates collagen accumulation in fibrotic liver (Yu et al. 2015); Neat1 (lnc14746*), which promotes NAFLD by facilitating hepatic lipid accumulation (Chen et al. 2019) and promotes liver fibrosis (He et al. 2020); Cyrano/Oip5os1 (lnc34166*), an essential developmental lncRNA (Smith et al. 2018); and Pint (lnc4993), which interacts with the Polycomb repressive complex 2 and is required to target specific genes for histone-H3 K27 trimethylation and gene repression (Marin-Bejar et al. 2013).
FIGURE 2.
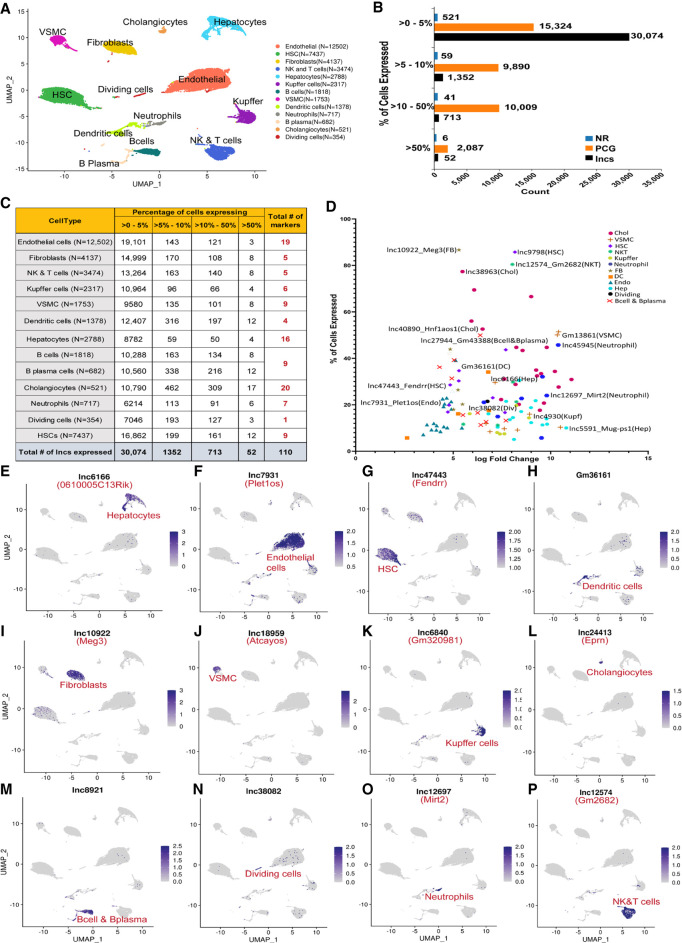
LncRNA detection in hepatic subpopulations and cell type-specific markers. (A) UMAP of liver cell clusters based on 39,878 single-cell transcriptomes integrated across four scRNA-seq data sets. Cell counts for each cell type are indicated in parentheses and in Supplemental Table S1H. (B) Bar plots indicating number of genes in the indicated gene classes (lncRNAs, PCGs, other noncoding RNAs [NR]) that are detectably expressed in one or more liver cell types in up to 5% of cells in a cluster, in >5%–10% of cells in a cluster, in >10%–50% of cells in a cluster, or in >50% of cells in a cluster, based on scRNA-seq data sets from healthy (control) adult male mouse liver. (C) Number of lncRNAs detected in each liver cell type, presented as a percentage of cells that express the lncRNA at ≥1 UMI/cell. Thus, 19,101 lncRNAs were detectably expressed in up to 5% of endothelial cells, and three lncRNAs were detected in >50% of endothelial cells. Cell type-specific marker genes for each cell cluster (last column) are based on Supplemental Table S2D. (D) Plot depicting 110 liver cell type-specific marker lncRNAs, from C. X-axis, log2 fold-change value for differential expression of the cluster marker gene lncRNA compared to its expression across all other clusters; y-axis, percentage of cells that express the lncRNA marker (see Supplemental Table S2D). (E–P) Feature plots with examples of lncRNA marker genes for each liver cell cluster. Shown are UMAPs as in A, with color intensity indicating expression level of the indicated marker lncRNA in individual cells.
Cell type-specific lncRNA marker genes
A total of 110 lncRNAs showed high specificity for expression in a single liver cell type; only 44 of these lncRNAs were previously known (i.e., have RefSeq or Ensembl annotations) (Fig. 2E–P; Supplemental Table S2D; see Materials and Methods). One example is Fendrr (lnc47443*), a strong marker for HSCs (Fig. 2G). Fendrr inhibits pulmonary fibrosis (Huang et al. 2020) and its overexpression inhibits hepatocellular carcinoma growth (Wang et al. 2019). Another example, Meg3 (lnc10922*), has liver antifibrotic activity (He et al. 2020) and is a marker for liver fibroblasts (Fig. 2I). Atcayos (lnc18959) is a marker for liver vascular smooth muscle cells (Fig. 2J) that regulates myogenic differentiation of satellite cells during skeletal muscle development (Qi et al. 2020), and Ephemeron (Eprn; lnc24413) fine-tunes the dynamics of the cell state transition in mouse embryonic stem cells (Li et al. 2017a) and in liver is a cholangiocyte marker (Fig. 2L). Finally, Mirt2 (lnc12697) is a neutrophil marker (Fig. 2O) that negatively regulates inflammation (Du et al. 2017).
LncRNA zonation across the healthy liver lobule
The liver is divided into small functional units called lobules fed by the hepatic artery and the portal vein, which drain to the central vein via sinusoidal capillaries. Gradients of oxygen, nutrients and hormones are established across the lobule, leading to spatial zonation of liver function and gene expression, as is seen in hepatocytes (Ben-Moshe and Itzkovitz 2019), endothelial cells (Halpern et al. 2018), and HSCs (Dobie et al. 2019). Here, we elucidated the zonation patterns for both lncRNAs and PCGs in healthy liver (chow diet-fed mice) in hepatocytes and several major nonparenchymal cell populations.
LncRNA zonation in hepatocytes
We used established spatial zonation markers (Halpern et al. 2017) for periportal, midlobular and pericentral hepatocytes in combination with trajectory inference analysis using Monocle2 (Qiu et al. 2017; Trapnell et al. 2017) to identify 111 lncRNAs showing significant zonation (q-val <0.001) across the liver lobule (Fig. 3A; Supplemental Table S3A). For example, lnc-LFAR1, a liver-enriched lncRNA that promotes liver fibrosis by activating TGFβ and notch signaling (Zhang et al. 2017b; He et al. 2020), was preferentially expressed in periportal hepatocytes, while lnc13605 and lnc14942 showed midlobular zonation, and Gas5 (lnc733*) and lnc6236* exemplify pericentrally zonated lncRNAs (Fig. 3A). PCGs expressed in periportal hepatocytes showed functional enrichment for specialized liver functions, such as lipid and steroid metabolism, acute inflammatory response and complement cascades, whereas pericentral hepatocytes were enriched for functions related to carboxylic acid metabolic process, oxidoreductase, peroxisome and response to xenobiotic stimulus (Supplemental Table S3B), consistent with prior work (Halpern et al. 2017).
FIGURE 3.
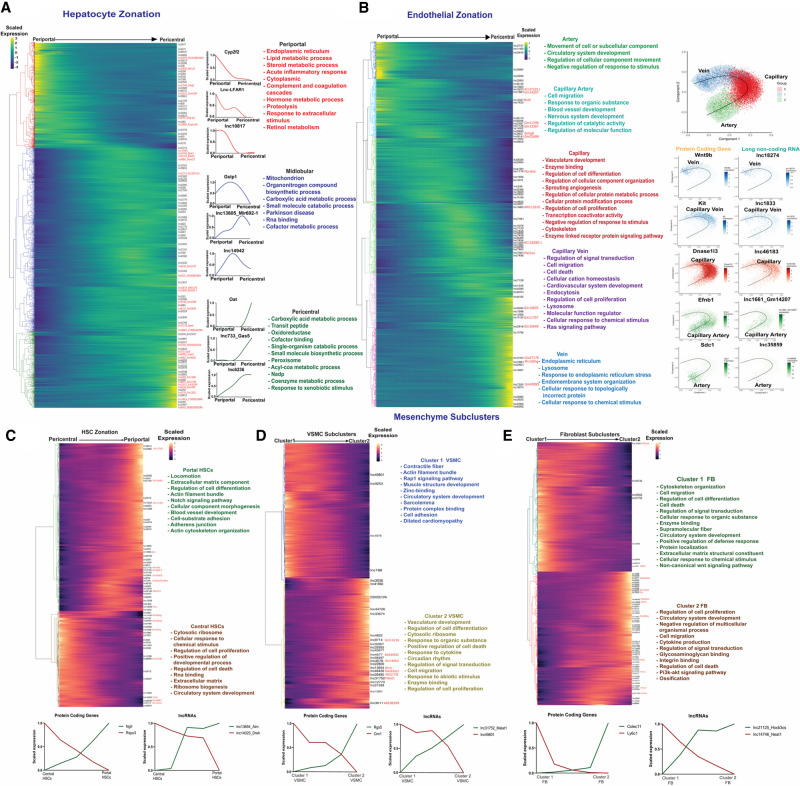
LncRNA zonation across healthy liver lobule. (A) Heatmap showing relative expression of PCGs and lncRNAs that are zonated in hepatocytes, ordered from periportal (left) to pericentral (right), with each row corresponding to one gene, as marked at the right for lncRNAs. Right of heatmap: Zonation profiles for select genes, along with top enriched terms for each of the three main hepatocyte zones. Font color for enriched terms matches the dendrogram color at the left of heatmap. (B) Heatmap showing zonated expression profiles for PCGs and lncRNAs that are zonated in endothelial cells, with clusters matching gene trajectories sequentially, from artery to capillary artery, capillary, capillary vein, and finally vein, with cluster assignments based on known spatial marker genes (see text). Top functional enrichment terms of each cluster (colored) are at the right. Further to the right are Scorpius pseudotime trajectories showing endothelial cell phenotypes along the artery-capillary-vein axis, with trajectory plots for select marker genes and mouse liver lncRNAs displayed across the pseudotime trajectory. (C–E) Heatmaps showing expression trajectories for PCGs and lncRNAs that are apparently zonated in the hepatic mesenchyme, which is comprised of HSCs (C), VSMCs (D) and fibroblasts (E), with top enriched terms and select marker genes as shown.
LncRNA zonation in endothelial cells
Five zonated endothelial cell clusters were obtained and identified using established spatial marker genes for arteries (marker gene: Sdc1), capillary arteries (arterioles; Efnb1), capillaries (Dnase1I3), capillary veins (venules; Kit) and veins (Wnt9b) (Kalucka et al. 2020). Overall, we identified 1163 zonated endothelial cell transcripts, including 71 zonated lncRNAs (q-val <0.001) (Supplemental Table S3C). Furthermore, we determined the enriched biological functions for the genes expressed in each cell cluster (Fig. 3B; Supplemental Table S3D). For example, capillary endothelial cells were enriched for vasculature development and sprouting angiogenesis; while capillary vein endothelial cells were enriched for cell migration and cell death gene expression, and for Ras signaling genes, which regulate hepatocyte zonation (Braeuning et al. 2007). LncRNAs preferentially expressed in capillary arteries of the endothelium include Bvht (lnc14557), an epigenetic regulator of cardiovascular lineage commitment (Xue et al. 2016), and Snhg8 (lnc2803*), which promotes tumorigenesis and predicts tumor recurrence in hepatocellular carcinoma (Dong et al. 2018), while the venous cell-enriched Mir100hg (lnc7851*) activates Wnt signaling via its embedded miRNAs (Lu et al. 2017). Other top zonation markers across the endothelium trajectory, derived using SCORPIUS (Cannoodt et al. 2016), are shown in Figure 3B (right).
lncRNA zonation in mesenchymal cells
We investigated gene expression zonation patterns in three mesenchymal cell populations (Fig. 3C–E): HSCs, fibroblasts, which are found in the mesenchyme surrounding the bile duct (Wells 2014), and VSMCs, which comprise a cell layer beneath endothelial cells lining the blood vessel (Bomzon and Ljubuncic 2001). Trajectory analysis partitioned HSCs into two subclusters: portal vein-associated HSCs and central vein-associated HSCs, whose identities we verified using the established HSC marker genes Rspo3 (pericentral) and Ngfr (periportal) (Fig. 3C; Dobie et al. 2019). We identified 58 HSC zonated lncRNAs, including Pvt1 (lnc12608), Airn (lnc13654) and Lnc-Dreh (lnc14025) (Fig. 3C; Supplemental Table S3E). Pvt1 activates HSCs under hypoxia and promotes liver fibrosis (Zheng et al. 2016; Yu et al. 2020) and cozonates with periportal HSC genes, as does Airn. Pathways associated with the PCGs in this HSC cluster include locomotion, extracellular matrix, Notch signaling, and blood vessel development. In contrast, lnc-Dreh, a tumor suppressor gene for hepatocellular carcinoma (Lv et al. 2017), was preferentially expressed in central vein-associated HSCs, which were most highly enriched for cytosolic ribosome genes and for regulation of cell proliferation (Fig. 3C; Supplemental Table S3F).
The spatial zonation of smooth muscle cells has been established in brain but has not been investigated for liver. Trajectory analysis identified two distinct clusters of liver VSMCs, with 381 genes (355 PCGs, 26 lncRNAs) showing significant differential expression between clusters (Supplemental Table S3G). VSMC cluster 1 was characterized by high expression of Rgs5, a marker for brain pericytes (Shen et al. 2016; Vanlandewijck et al. 2018), and was enriched in functions related to contractile fiber and muscle structure development. In contrast, VSMC cluster 2 was characterized by high expression of Cnn1, a marker for arterial smooth muscle cells in mouse brain (Vanlandewijck et al. 2018), and was enriched for a distinct set of biological processes, including vasculature development, cell differentiation, cytosolic ribosome, cell death, and response to cytokine (Fig. 3D; Supplemental Table S3H).
Fibroblast heterogeneity associated with discrete anatomical positions is seen in multiple tissues (Muhl et al. 2020) but has not been characterized for liver. Trajectory inference identified two distinct fibroblast clusters in mouse liver, which may correspond to distinct, zonated cell populations, and which encompassed 1335 PCGs and 69 lncRNAs (Supplemental Table S3I). Cluster 1 liver fibroblasts were enriched for cytoskeleton organization, cell migration, cell differentiation, and cell death, while cluster 2 fibroblasts were enriched for cell proliferation, circulatory system development, and extracellular matrix (Fig. 3E; Supplemental Table S3J). Neat1 (lnc14746*), which promotes liver fibrosis (He et al. 2020), was enriched in cluster 1 fibroblasts. Cluster 2 fibroblasts were enriched for Dnm3os (lnc17273*), which promotes hepatocellular carcinoma via an epigenetic mechanism (Wang et al. 2021), and for Carmn/Mir143HG (lnc14558*), whose loss in human VSMCs is associated with enhanced proliferation and migration (Wang and Sallam 2021).
LncRNAs dysregulation in diet-induced NASH
To elucidate the role of lncRNAs in NASH progression and the development of pathogenic cell states, we analyzed scRNA-seq data (Su et al. 2021) obtained from a combined total of 74,718 liver cells isolated from healthy mice (chow diet) and from mice fed HFHFD (high fat high fructose diet) for either 15 wk, to induce simple steatosis (NAFLD), or for 30–34 wk, to induce NASH (Supplemental Table S1H). In livers from 15 wk high-fat-diet fed mice (NAFLD livers), 1477 genes, including 308 lncRNAs, were differentially expressed (>2 fold-change at FDR < 0.05) in one or more of 12 major cell clusters (Fig. 4A; Supplemental Table S4A). These genes showed strong functional enrichment for inflammatory response, lipid metabolic process, leukocyte migration, cytokine production, and innate immune response (Supplemental Table S4B). After 30 wk of high-fat diet feeding (NASH livers), the number of differentially expressed genes increased to 2216, including 332 lncRNAs; and after 34 wk, it increased to 3090 genes, including 459 lncRNAs (Supplemental Table S4C,D). Overall, a total of 677 lncRNAs were dysregulated across the time course of NAFLD to NASH progression. The top enriched pathways for the NASH-induced and NASH-repressed genes were very similar to those seen in the NAFLD (15 wk) livers (Supplemental Table S4B).
FIGURE 4.
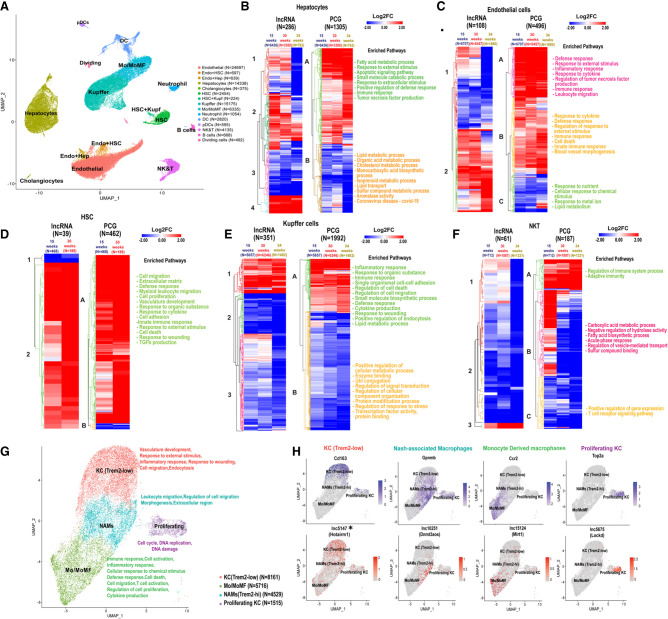
LncRNAs and PCGs perturbed across different cell types in NAFLD and NASH liver. (A) UMAP showing liver cell clusters for a total of 74,718 cells. Cells from healthy liver (19,364 cells) were aggregated with cells from livers of mice fed HFHFD for 15 wk (NAFLD liver; 23,961 cells), 30 wk (24,106 cells), or 34 wk (NASH livers; 7287 cells) (Supplemental Table S1H). Cell numbers in each cluster are shown in parentheses. Mo/MoMF, monocytes/monocyte-derived macrophages. (B–F) Heat maps showing differentially expressed lncRNAs and PCGs in the indicated cells clusters from 15-, 30-, and 34-wk HFHFD-fed livers compared to chow diet livers, in hepatocytes (B), endothelial cells (C), HSCs (D), Kupffer cells (E), and NK & T cells (F). Functional enrichment terms are shown at the right of each heatmap for each PCG heatmap subcluster (marked A–C) from each cell type. See Supplemental Table S4 for full data sets. (G) UMAP of Kupffer (macrophage) cell subpopulations, identified based on PCG markers in each cell subtype. Kupffer cells and Mo/MoMf cell clusters shown in A were reclustered to generate the four clusters shown at higher resolution, after filtering, to remove cells with high mitochondrial contamination. Functional enrichment terms of each cluster are shown at the right. (H) Feature plots showing expression of select PCG and lncRNAs marker genes in Kupffer cell subpopulations.
Genes differentially expressed between HFHFD and chow-fed mice at one or more of these three time points were identified for each major liver cell type (Supplemental Table S4E) and then clustered to give the heatmaps and enriched pathways shown in Figure 4B–F. Genes induced across the time course of NAFLD and NASH development were enriched for functions such as fatty acid metabolism and innate immune response in both hepatocytes and endothelial cells (Fig. 4B,C). In HSCs, genes responding to HFHFD were mostly up-regulated at both time points and were enriched for cell migration, extracellular matrix, defense response and innate immunity (Fig. 4D), consistent with the activation of NASH-associated fibrosis. Finally, in both Kupffer cells and in NK and T cells, more genes were down-regulated than were up-regulated, with distinct enriched functions (Fig. 4E,F).
Several of the lncRNAs identified here as dysregulated with HFHFD feeding have known functions relevant to NAFLD and NASH. These include Pvt1 (lnc12608), which regulates HSC activation to promote liver fibrosis (Zheng et al. 2016; Yu et al. 2020) and was induced twofold in endothelial cells from NAFLD liver; and Dnmt3aos (lnc10251), which regulates macrophage polarization (Li et al. 2020) and was induced 16-fold in Kupffer cells (Supplemental Table S4A). Airn (lnc13654), which promotes hepatocellular carcinoma progression (Oliva et al. 2009), was induced five- to sixfold in both Kupffer cells and hepatocytes from NASH liver (30 wk). Further, Hnf4aos (lnc1966*), a lncRNA that is antisense to the major hepatocyte transcription factor Hnf4a (Guo and Lu 2019), was repressed threefold in hepatocytes, while lnc-Plet1os (lnc7931), which is antisense to a marker gene for epithelial progenitor cells with liver regeneration capacity (Zhang et al. 2017a), was repressed threefold in both endothelial cells and hepatocytes (Supplemental Table S4C). Finally, the oncogenic Snhg8 (lnc2803*) (Dong et al. 2018) was down-regulated threefold in both hepatocytes and Kupffer cells from 34 wk NASH liver (Supplemental Table S4D).
LncRNA markers in NASH-associated macrophages
Macrophages play a critical role in NASH pathogenesis; they are strongly linked to disease progression and highly responsive to therapeutic interventions (Kazankov et al. 2019; Oates et al. 2019). We reclustered the Kupffer cells and monocyte/macrophage cells aggregated from both control and HFHFD livers to identify four macrophage subpopulations (Fig. 4G; Supplemental Table S4G). A total of 96 lncRNA markers were discovered for the individual macrophage subpopulations, of which 27 have human orthologs (Supplemental Table S4H). The four subpopulations were characterized as Trem2 (low) macrophages, NASH-associated macrophages (NAMs; Trem2-high, a hallmark of mouse and human NASH) (Ramachandran et al. 2019), monocyte/monocyte-derived macrophages, and proliferating cells, very similar to those described earlier using a nonparenchymal liver cell-enriched population (Xiong et al. 2019). In NASH liver, Trem2 (low) macrophages decreased from 81% to 30%–35% of the overall macrophage population (Supplemental Table S1H); these cells preferentially express Hotairm1 (lnc5147*), a tumor-associated lncRNA that participates in cell proliferation, migration, and apoptosis (Luo et al. 2019), and the Kupffer cell activation marker and scavenger receptor CD163 (Fig. 4H; Nielsen et al. 2020). In contrast, the NAM cell cluster (Trem2-high; Supplemental Fig. S2) expanded from 8% to 24% of the overall macrophage population in NASH liver and was marked by high expression of Gpnmb (Fig. 4H) and of Dnmt3aos (lnc10251), which regulates macrophage polarization via its effects on the expression of Dnmt3 (Li et al. 2020). Monocyte-derived macrophages, which infiltrate and can replace resident Kupffer cells under inflammatory conditions (Daemen et al. 2021), increased from 5% to 34%–40% of the macrophage population (Supplemental Table S1H), and were marked by Ccr2 and by Mirt1 (lnc15124), an inhibitor of NF-κB signaling that can decrease expression of inflammatory factors (Li et al. 2017b). The proliferating macrophage population was marked by Top2a, which is associated with poor prognosis for hepatocellular carcinoma (Cai et al. 2020), and by Lockd (lnc5675) (Fig. 4H), which acts as an enhancer of the mitotic cell cycle factor Cdkn1b (Paralkar et al. 2016) and is decreased in offspring liver in response to maternal high fat diet-induced obesity (Chen et al. 2022). Other lncRNA markers of macrophage subpopulations are shown in Supplemental Figure S2. Analysis of the distinct sets of marker genes for each macrophage subpopulation (Supplemental Table S4G) revealed unique functions for each cell cluster: Trem2 (low) macrophages were enriched for vasculature development, inflammatory response, response to wounding and cell migration; NAMs were associated with regulation of cell migration/leukocyte migration; monocyte-derived macrophages were enriched for functions related to immune response, inflammatory response, cell death, T cell activation and cytokine production; and proliferating cells were enriched for functions related to DNA replication and cell cycle.
Zonation dysregulation during NAFLD and NASH pathogenesis
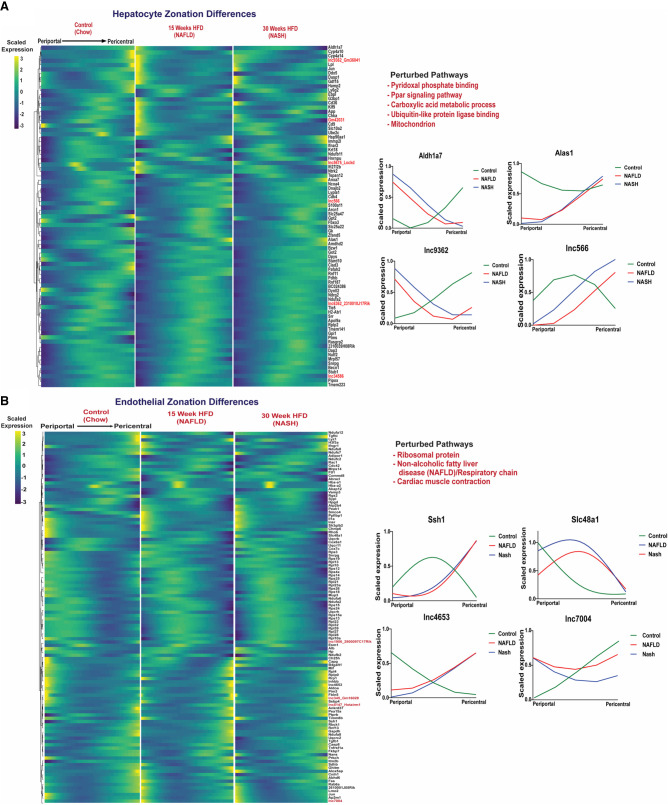
The distinctive liver lobule zone-dependent gene expression seen in healthy liver (Fig. 3) is important for liver function and can be perturbed in disease states (Kietzmann 2019). We found significant zonal perturbations in NAFLD and NASH liver compared to chow diet liver for 74 PCGs and six lncRNAs in hepatocytes, and for 107 PCGs and five lncRNAs in endothelial cells (Fig. 5A,B; Supplemental Table S4I). The zonally perturbed hepatocyte genes were enriched for pyridoxal phosphate binding and PPAR signaling, while the endothelial cell zonation perturbed genes showed strong enrichment for ribosomal proteins and for NAFLD/respiratory chain (Supplemental Table S4J). Specific examples include Aldh1a7, which can protect hepatocytes by catabolism of reactive aldehydes formed during oxidative stress and was shifted from pericentral to periportal hepatocytes during NAFLD and NASH development, while the heme biosynthetic enzyme Alas1 shifted from periportal to pericentral hepatocytes (Fig. 5A). In liver endothelial cells, Ssh1 shifted from midlobular expression in chow diet livers to pericentral expression in NAFLD and NASH livers (Fig. 5B). This actin remodeling gene is involved in endothelial cell inflammatory signaling and has vascular antifibrotic activity (Williams et al. 2019).
FIGURE 5.
Perturbation of zonation in NAFLD and NASH liver. Matched heatmaps of genes (PCGs and lncRNAs) that are differentially zonated between control, NAFLD, and NASH livers at FDR < 0.001, in hepatocytes (A), and in endothelial cells (B). Pathways perturbed were identified by DAVID functional enrichment analysis of the differential PCGs (Supplemental Table S4I,J). Shown at the right are zonation profiles for select genes for each cell type across three conditions (chow diet, NAFLD, NASH).
Global discovery of network-essential regulatory lncRNAs in healthy, NAFLD and NASH liver
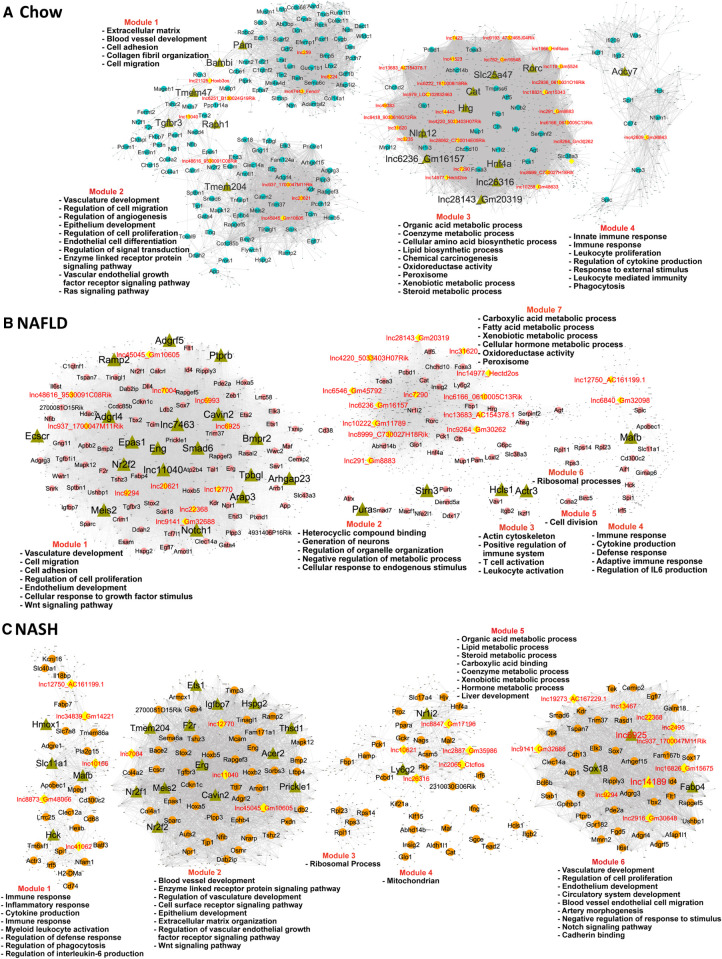
We implemented gene coexpression network analysis using bigSCale2 (Iacono et al. 2019) to develop gene regulatory networks and associate individual lncRNAs with specific biological functions in healthy liver, in NAFLD liver (15 wk HFHFD) and in NASH liver (30 wk HFHFD) (Fig. 6; Supplemental Table S5A,B). We inferred the identities of key network-essential regulatory genes based on four network centrality metrics extracted from each network, namely, Betweenness, PageRank centrality, Closeness, and Degree (Iacono et al. 2019), which serve as proxies for a gene's influence on the network (see Fig. 6, where labeled nodes are the inferred regulatory genes; Supplemental Table S5C). A total of 65 such network-essential lncRNAs were identified across the three liver networks. Functional annotation clusters associated with the gene targets of each of these network-essential regulatory lncRNAs (Supplemental Table S5D) revealed many common enriched functional annotations across the set of lncRNAs. This is consistent with the high gene densities of all three bigSCale2 networks (Supplemental Table S5E) and the sharing of gene targets between regulatory lncRNAs within a network. Thus, organic acid metabolic process described the top enriched annotation cluster (Benjamini-corrected P-value: 10−28 to 10−87) for 27 of the 65 regulatory lncRNAs, in either the healthy liver (chow diet) network or the NAFLD network, but none in the NASH network. In contrast, vascular development or cardiovascular system development described the top enriched annotation cluster of 17 other regulatory lncRNAs in the NAFLD and NASH networks (Benjamini P-value: 10−16 to 10−34) but for none in the healthy liver network (at P < 10−10), consistent with the role of angiogenesis in progression from liver fibrosis to more advanced liver disease (Elpek 2015; Li 2021).
FIGURE 6.
Gene regulatory networks for healthy, NAFLD, and NASH liver. BigSCale2 networks based on scRNA-seq data for chow-fed (A), NAFLD (B), and NASH (C) mouse livers, where PCGs and lncRNAs are nodes, and the edges between genes are correlation values based on an adaptive threshold. Nodes displayed here represent network-essential/regulatory genes (circular and diamond-shaped nodes, with lncRNAs nodes colored yellow), as predicted based on top network metrics. Triangular nodes represent master regulators (predicted to regulate the network-essential regulators) and are defined as nodes (genes) with high network centrality metrics calculated for subnetworks extracted from all top 100 ranked PCG nodes plus all top 50 ranked lncRNA nodes. The networks are subdivided into gene modules that are enriched for the biological functions listed. Also see Supplemental Table S5.
Forty-five genes were identified as network-essential for all three liver networks (Supplemental Fig. S3; Supplemental Table S5C, column AH), including three lncRNAs, two with human orthologs (lnc937*, lnc11040, lnc45045*). Other network-essential regulatory lncRNAs of interest include: Hnf4aos (lnc1966*), which is antisense to the major liver transcription factor HNF4A and showed high connectivity to genes involved in various metabolic processes in the healthy liver network (Supplemental Table S5D); Gm45792 (lnc6546), which is antisense to acyl-CoA synthetase medium chain family member 1 (Acsm1) and is an essential node in the NAFLD liver network; and Ctcflos (lnc2065), which regulates transcription of hepatic Pck1 by modulating glucocorticoid receptor function (Yoon et al. 2023) and was an essential node in the NASH network, where it makes a second-degree connection with Pck1 via lnc10621 (Supplemental Fig. S4).
Master regulators in healthy and NAFLD/NASH liver
The sets of network-essential regulatory genes identified in each network (both lncRNAs and PCGs) were extracted and used to construct subnetworks comprised exclusively of the putative regulatory genes themselves. We ranked the subnetwork genes using a modified scoring approach (see Materials and Methods) to identify n = 16–23 master regulators for each liver network (Fig. 6, nodes marked with green triangles; Supplemental Table S5C, columns AP-BU). Seven of the master regulators were lncRNAs (healthy liver network: Gm16157 [lnc6236*], lnc26316, and Gm20319 [lnc28143]; NAFLD network: lnc7463, lnc11040; NASH network: lnc6925, lnc14189*). Validating this approach to discovery of bonafide liver network regulatory genes, several of the master regulators are liver transcription factors with well-established regulatory functions, and many of the master regulators have regulatory functions specifically related to liver disease (Supplemental Table S5F, columns F and L). Specific examples include: Hnf4a, a master regulator of hepatocyte gene expression that protects against NASH development (Xu et al. 2021), in the healthy liver network; Nr2f2 (COUP-TFII), whose loss inhibits HSC/myofibroblast activation in liver injury (Ceni et al. 2017), in the NAFLD liver network; and Meis2, which promotes hepatocellular carcinoma (Guan et al. 2019), and Ets1, whose loss decreases diet-induced hepatocyte apoptosis, fibrosis and NASH (Liu et al. 2019), in the NASH liver network. We further validated the functionality of the regulatory gene networks by directly comparing the known biological activities of each master regulator to the sets of highly enriched functional annotations of its network target genes (Supplemental Table S5G), and in many cases found good agreement (Supplemental Table S5F). For example: Notch1, a master regulator in the NAFLD network, promotes the migration and invasion of hepatocellular carcinoma cells (Lu et al. 2022), and its target genes were enriched for regulation of cell migration (Benjamini P = 6.46 × 10−17); while the master regulator Cavin2 regulates endothelial nitric-oxide synthase in angiogenesis, and its NAFLD network target genes were strongly enriched for regulation of angiogenesis (P = 2.96 × 10−15). Finally, two master regulators from the chow diet liver network that are DNA-binding proteins were identified as upstream regulators of their liver network-predicted target genes by IPA Upstream Regulator analysis: Hnf4a, at P = 5.80 × 10−19; and Rorc, at P = 9.71 × 10−08 (Supplemental Table S5F, column I).
Cell type-specific responses and lncRNAs dysregulated in CCl4-induced liver fibrosis
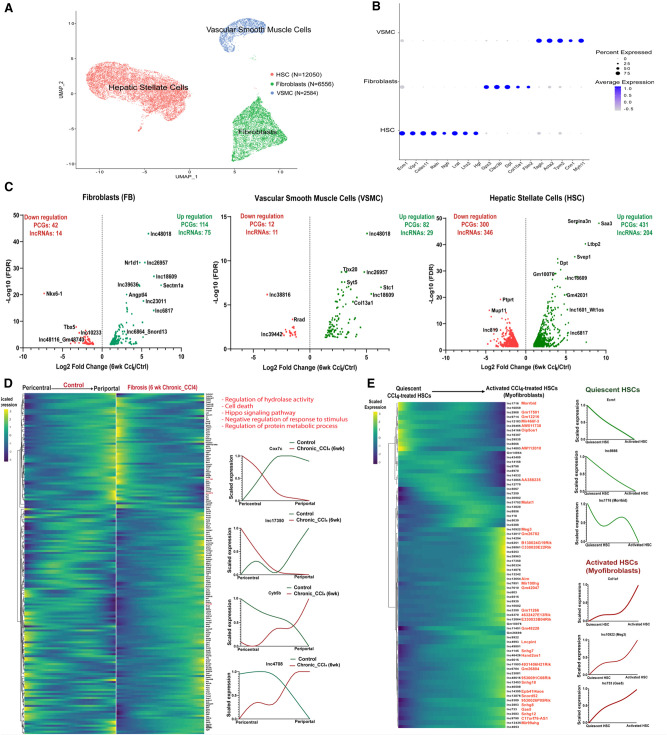
We examined the utility of the computational framework described above to identify disease-relevant liver lncRNAs in a second disease model. Thus, we analyzed an scRNA-seq data set (Dobie et al. 2019) comprised of mesenchymal cells from healthy mouse liver and from livers of mice following chronic (6 wk) exposure to CCl4, which induces advanced liver fibrosis. Three distinct cell populations were identified by clustering mesenchymal cells aggregated from the healthy and CCl4-treated livers, namely, HSCs, fibroblasts and VSMCs (Fig. 7A,B), consistent with prior findings (Dobie et al. 2019). Differential expression analysis across these three subpopulations identified 1550 genes, including 631 lncRNAs, that were dysregulated by CCl4 treatment (Fig. 7C; Supplemental Table S6A), a subset of which was dysregulated in multiple subpopulations (Supplemental Fig. S6). Genes preferentially or specifically dysregulated by CCl4 in the fibroblast subpopulation include a 174-fold induction of Sectm1a, which stabilizes tissue resident macrophages in response to acute inflammation (Mu et al. 2021), and a 160-fold suppression of Nkx6-1, an unfavorable prognostic marker for human hepatocellular carcinoma (Huang et al. 2015). Genes specifically dysregulated in VSMCs include Stc1, a classic inflammation marker in fibrotic disease (Chan et al. 2022), as well as lnc24481*, which was up-regulated 16-fold, and lnc43318, which was fivefold down-regulated by CCl4 exposure. HSCs showed the most extensive dysregulation, impacting 1296 genes (743 PCGs, 553 lncRNAs), >90% of which were specifically dysregulated in HSCs (Supplemental Table S6A). Examples include: Ltbp2 (172-fold increase), a marker of cardiac fibrosis (Park et al. 2018); Gpx3 (16-fold increase), an antioxidant enzyme induced by oxidative stress (Kim et al. 2018); and Fcna (fourfold decrease), a marker for resting HSCs (Krenkel et al. 2019). Wt1, which moderates fibrogenesis after injury (Kendall et al. 2019), and the divergently transcribed Wt1os (lnc1601), were both strongly induced (>20-fold) in HSCs. Finally, we identified 172 genes, including three lncRNAs, whose expression along the trajectory was significantly different between healthy and chronic CCl4-exposed HSCs (Fig. 7D; Supplemental Table S6B). This finding is consistent with the dynamic changes in zonation seen in HSCs during CCl4-induced fibrotic liver injury (Dobie et al. 2019). The top enriched term was cell death (Supplemental Table S6C). In many cases, CCl4 exposure substantially reversed the HSC zonation pattern seen in control liver.
FIGURE 7.
Mesenchymal cell lncRNAs and PCGs perturbed in CCl4-induced fibrotic mouse liver. (A) UMAP showing 21,190 cells comprised of three hepatic mesenchyme subpopulations aggregated from healthy liver (11,710 cells) and fibrotic mouse liver (6 wk CCl4 exposure; 9480 cells), with total cell numbers indicated for each cluster (Supplemental Table S1H). (B) Dot plot showing average expression of mesenchymal cell subpopulation marker genes (Dobie et al. 2019). Dot size is proportional to the percentage of cells expressing each marker gene. (C) Volcano plots showing differentially expressed genes in each cell cluster at log2 |fold-change| > 1 and FDR < 0.05, displayed as −log10 value on y-axis. (D) Matched heatmaps of genes expressed in HSCs that are differentially zonated between control and fibrotic mouse liver (at FDR < 0.001), with perturbed pathways based on DAVID functional enrichment analysis. (Right) Zonation profiles for select genes whose zonation pattern is perturbed by CCl4 exposure. (E) Heatmap showing relative expression of lncRNAs that are differentially expressed between quiescent pericentral HSCs and myofibroblasts from CCl4-exposed mouse liver mesenchymal cells. See Supplemental Figure S5B. (Right) Expression patterns for select genes. Cell identities were verified using the uninjured HSC marker gene Ecm1 and the profibrogenic marker gene Col1a1 (right).
LncRNAs expressed in collagen-producing myofibroblasts
The pericentral HSC population of CCl4-treated liver includes activated HSCs (also known as myofibroblasts), which produce collagen and promote fibrosis progression (Dobie et al. 2019; Krenkel et al. 2019). We investigated this activated HSC subpopulation to discover lncRNAs associated with pathogenic collagen production. The activation of pericentral HSCs was validated by the expression patterns of profibrogenic marker genes (Col1a1, Col1a2) and by the reduced expression of established marker genes for quiescent pericentral HSCs (Ecm1, Rgs5) (Fig. 7E; Supplemental Fig. S5A; Dobie et al. 2019). Overall, 73 lncRNAs showed differential expression between quiescent and activated pericentral HSCs (i.e., myofibroblasts) from CCl4-induced fibrotic liver (Supplemental Table S6D). For example, the lncRNA Morrbid (lnc1716), a critical regulator of immune cell prosurvival cytokine responses (Kotzin et al. 2016), was more highly expressed in quiescent than in activated HSCs.
Meg3 (lnc10922*), a marker for pulmonary fibroblasts (Xie et al. 2018) and an inhibitor of liver fibrosis (He et al. 2020), and Gas5 (lnc733*), which suppresses HSC activation and counters liver fibrosis (Yu et al. 2015), were more highly expressed in activated HSCs. Overall, a total of 1420 PCGs were differentially expressed between these two pericentral HSC subpopulations (Supplemental Fig. S5B; Supplemental Table S6D). PCGs expressed in the quiescent HSCs were strongly enriched for cellular response to chemical stimulus and cell migration (Supplemental Table S6E), consistent with quiescent HSC migration being linked to cell proliferation (Ikeda et al. 1999), while PCGs in the activated, myofibroblast subpopulation showed strong enrichment for terms such as extracellular matrix, circulatory system development and connective tissue development (Supplemental Table S6E).
Network-essential lncRNA regulators in CCl4-exposed liver
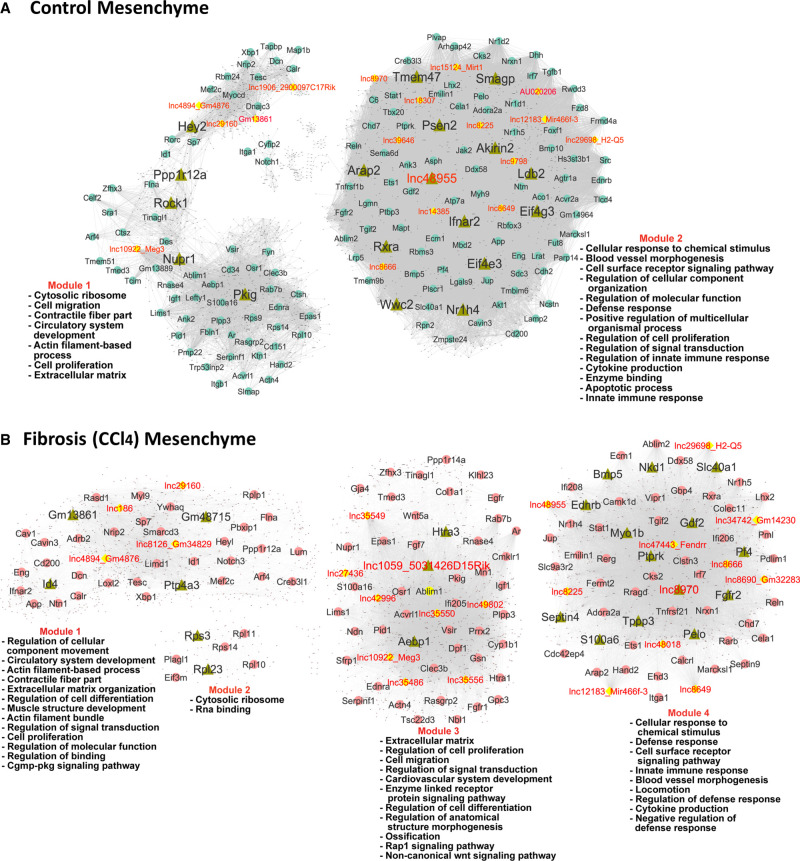
Analysis of the mesenchymal single-cell populations using bigSCale2 yielded functional gene coexpression networks for both healthy (control) and CCl4-exposed liver (Fig. 8). Using network centrality metrics, we identified a total of 34 network-essential regulatory lncRNAs: 19 for the healthy mesenchymal cell network and 26 for the CCl4-exposed liver fibrosis network; of these, eight network-essential regulatory lncRNAs were specific to the control liver network, 15 to the CCl4 network, and 11 were common to both networks (Supplemental Table S5C; Supplemental Fig. S7). One of the network-common lncRNAs, Meg3 (lnc10922*), inhibits HSC activation and accelerates the reversal of CCl4-induced fibrosis (Wu et al. 2021). Network-essential PCG regulators common to both networks include: Ifnar2, which mediates many antiviral immune responses and has a genetic polymorphism linked to hepatocellular carcinoma (Ma et al. 2018); Ets1, which regulates HSC activation in NASH liver and is a profibrogenic marker in CCl4-induced fibrosis (Marcher et al. 2019), where it is a critical mediator of extracellular matrix remodeling (Trojanowska 2000); Tesc, which had the highest PageRank centrality score in the CCl4 network and negatively regulates cell proliferation and survival in hepatocellular carcinoma (Zhou et al. 2020); and Nr1h4 (FXR), which regulates lipid and glucose homeostasis and is involved in innate immune responses (Fiorucci et al. 2018).
FIGURE 8.
Gene regulatory networks for healthy liver (A) and CCl4-induced fibrotic liver (B). Shown are BigSCale2 networks, as in Figure 6, with nodes representing network-essential regulatory genes (circular nodes, with lncRNAs shown as yellow nodes) predicted based on top network metrics. Triangular nodes indicate master regulators. The networks are subdivided into gene modules, which were enriched for the biological functions listed. See Supplemental Table S5.
Many of the gene targets of the 34 mesenchymal cell network-essential regulatory lncRNAs were enriched for common functional annotations, with notable differences between control and CCl4-exposed liver (Supplemental Table S5D). For example, response to cytokine/response to interferon-β was the top enriched term for six regulatory lncRNAs in CCl4-exposed mesenchymal cells (Benjamini P-value: 10−13 to 10−22) versus no lncRNAs in control liver. Similarly, extracellular matrix was among the top three terms for nine regulatory lncRNAs from CCl4 liver versus one regulatory lncRNAs in mesenchymal cells from healthy liver (Benjamini P-value: 10−09 to 10−15). These findings are consistent with the central roles of each of these biological processes in liver fibrosis. In contrast, top two enriched terms more frequently found for gene targets of mesenchymal cell regulatory lncRNAs from control liver included response to organic substance (seven in regulatory lncRNAs in control liver vs. 0 lncRNAs in CCl4 liver; Benjamini P-value: 10−10 to 10−17) and blood vessel/cardiovascular system development and closely related terms (12 regulatory lncRNAs in control liver vs. six lncRNAs in CCl4 liver; Benjamini P-value: 10−09 to 10−17) (Supplemental Table S5D).
Master regulators in the mesenchymal cell networks
Analysis of the sets of mesenchymal cell network regulatory genes as described above for healthy and NAFLD/NASH identified n = 18–23 master regulators for each mesenchymal cell network along with enriched functional annotations of their target genes (Fig. 8, nodes shown as green triangles; Supplemental Table S5C,F). Four lncRNAs were identified as master regulators of the CCl4 mesenchymal cell network: Gm13861 (target genes enriched in cytoskeletal protein binding, at Benjamini P = 7.12 × 10−10); Gm48715 (muscle system process, P = 9.39 × 10−09), lnc1059* (extracellular matrix, P = 7.64 × 10−11); and lnc8970 (blood vessel morphogenesis, P = 1.66 × 10−11). One master regulatory lncRNA was identified in the control mesenchymal cell network (lnc48955, targets enriched in locomotion, at P = 4.62 × 10−22) (Fig. 8; Supplemental Table S5C,F,G). Validating our approach to discovery of liver network-essential regulatory genes, many of the master regulators from the CCl4-exposed mesenchymal cell network have regulatory roles in liver fibrosis, a hallmark of CCl4-exposed liver, and the network target genes of the master regulators often showed highly enriched functional annotations that match the known biological properties of their master regulators (Supplemental Table S5F).
Functional clustering of regulatory lncRNAs
PCGs that were targets of the network-essential regulatory lncRNAs from each of the above five liver networks (Figs. 6, 8) were input to Metascape (Zhou et al. 2019) to facilitate comparisons across the target gene lists of each lncRNA, and thereby cluster the regulatory lncRNAs based on commonality of function. Regulatory lncRNAs from chow fed liver comprised three major functional clusters (Supplemental Fig. S8), two of which were variously enriched for diverse metabolic processes, while a third lncRNA cluster was functionally enriched for angiogenesis and vasculature development, extracellular matrix, and cell–cell adhesion. Similarly, we identified three clusters of regulatory lncRNAs from the NAFLD network (Supplemental Fig. S9), and four clusters with distinct enrichment patterns for the regulatory lncRNAs from NASH liver (Supplemental Fig. S10). Overall, the NAFLD and NASH network-derived regulatory lncRNAs showed more extensive enrichment for vasculature development and angiogenic processes than those from control liver (Supplemental Fig. S11), whereas the NAFLD and control network regulatory lncRNAs both showed enrichment for diverse metabolic processes.
Similarly, we identified two distinct regulatory lncRNA clusters from the healthy liver mesenchymal cell network (Supplemental Fig. S12), with one cluster of five lncRNAs enriched for terms related to muscle contraction, notch signaling, and ion homeostasis, and a separate cluster comprised of 14 lncRNAs enriched for diverse biological processes, including cell morphogenesis, regulation of defense response, vascular development, and interferon responses. Finally, regulatory lncRNAs from the CCl4-exposed mesenchymal cell network yielded three clusters with strongest enrichments and highest specificities for muscle contraction and notch signaling, extracellular matrix organization, and response to interferon-β, respectively (Supplemental Fig. S13). Pathways that were either common or specific to healthy versus CCl4-exposed mesenchymal cell lncRNA gene targets are shown in Supplemental Figure S14.
Triplex potential of liver disease-associated regulatory lncRNAs
We evaluated the potential of each of the above network-essential regulatory lncRNAs to form a triple helix with the promoter sequence of its putative protein coding target genes in the gene coexpression network, as determined by Triplex Domain Finder (Kuo et al. 2019). For each lncRNA, we computed the enrichment of sequence-specific lncRNA–protein coding gene promoter triplex interactions in the lncRNA's network-predicted target gene set, compared to a background set comprised of protein coding gene promoters. First, we used Triplex Domain Finder to define a set of DNA-binding domains within each lncRNA of interest, that is, lncRNA sequences with high triplex formation oligonucleotide activity. We then determined, for each such DNA-binding domain, whether the set of target gene promoters was significantly enriched for containing at least one Triplex Target Site (TTS) as compared to that for the set of nontarget gene promoters. For the chow-fed liver network, we determined that seven of 39 lncRNAs identified as key regulatory genes based on network metrics (Supplemental Table S5C) showed significant enrichment for lncRNA–promoter DNA triplexes at FDR < 0.05 (Fisher's exact test) (Supplemental Table S7A). Four of these seven lncRNAs formed triplexes with genes enriched for metabolic processes such as oxidoreductase, peroxisomes and lipid metabolism (Supplemental Fig. S15A; 5033403H07Rik [lnc4220], lnc7423, 0610031O16Rik [lnc2936], Prox1os [lnc979*]) and three lncRNAs formed triplexes with genes enriched for extracellular matrix (Supplemental Table S7A; lnc259, Fendrr [lnc47443*], lnc8224). Furthermore, nine out of 28 regulatory lncRNAs in the NAFLD network were significantly enriched for triplex formation with their gene targets, as were five out of 29 regulatory lncRNAs in the NASH network. Enriched gene functions associated with these regulatory lncRNA gene targets include vasculature development and angiogenesis (Supplemental Fig. S15B,C).
In healthy liver mesenchymal cells, eight out of 19 network-essential regulatory lncRNAs were significantly enriched for triplex formation with their target gene promotors; the associated enriched functions included response to chemical stimulus, vasculature development, cell migration, immune response, and extracellular matrix (Supplemental Fig. S16A). In the CCl4-induced liver mesenchymal network, 11 out of 26 regulatory lncRNAs were enriched for triplex formation with their target gene promoters for pathways linked to extracellular matrix, response to cytokine and interferons (β and γ), blood morphogenesis, and defense response (Supplemental Fig. S16B). Six network-essential regulatory lncRNAs were common to both the control and the CCl4-exposed liver mesenchymal cell network, but new regulatory interactions emerged in the CCl4 network. For instance, Meg3 (lnc10922*) formed an isolated subnetwork in control mesenchymal cells (Supplemental Fig. S16A) but had extensive shared binding targets with lnc35550*, lnc35556*, and 5031426D15Rik (lnc1059*) in the CCl4-induced liver fibrosis network (Supplemental Fig. S16B). We identified specific PCG targets for each lncRNA, as well as many shared targets between regulatory lncRNAs, which is indicative of their complex regulatory crosstalk (Supplemental Table S7C–G).
DISCUSSION
The overall goal of this study was to elucidate on a global scale the roles of liver-expressed lncRNAs in biological pathways related to liver disease development. Single-cell RNA-seq was used to characterize the long noncoding transcriptional landscape of mouse liver using a reference catalog of 48,261 liver-expressed lncRNAs, a majority of them novel, which we discovered by transcriptome reconstruction from >2000 bulk public mouse liver RNA-seq data sets, a major update to our earlier reference set of 15,558 mouse liver lncRNAs (Melia and Waxman 2019). Prior studies identified several hundred mouse liver-expressed lncRNAs that show hormone-regulated, sex-biased expression (Melia and Waxman 2019; Lau-Corona et al. 2022) or respond to xenobiotic exposures (Lodato et al. 2017; Dempsey and Cui 2019; Goldfarb and Waxman 2021). However, a global analysis of lncRNAs dysregulated in liver diseased states, most notably, lncRNAs expressed in specific cell types and zonal subpopulations across the liver lobule, was lacking. Here, we characterized liver cell type-specific expression patterns for healthy mouse liver and for two disease models for a total of 76,011 genes, including 48,261 liver-expressed lncRNAs plus 4700 other lncRNAs from RefSeq and Ensemble databases. Importantly, we showed that single-cell RNA-seq technology is sufficiently sensitive to detect and characterize more than 30,000 liver lncRNAs, including 25,000 novel liver-expressed lncRNA genes, 110 of which we identified as cell type-specific marker genes for the 13 major cell types identified in healthy adult mouse liver (see overall summary in Supplemental Table S8A). Using public scRNA-seq data sets (Dobie et al. 2019; Su et al. 2021) we uncovered striking liver cell type-dependent perturbations in the expression of 677 lncRNAs in NAFLD compared to healthy mouse liver, and in the transition from NAFLD to NASH liver. Furthermore, we identified a largely nonoverlapping set of 631 lncRNAs dysregulated in liver mesenchymal cells from livers that develop extensive fibrosis following chronic exposure to CCl4. Given that thousands of liver-expressed lncRNAs are nuclear, tightly bound to chromatin and polyadenylated (Goldfarb and Waxman 2021), we can anticipate an even greater sensitivity for lncRNA detection when using single nucleus RNA-seq in place of scRNA-seq (Zeng et al. 2016; Goldfarb et al. 2022). Single nucleus RNA-seq may also provide a more representative picture of the relative abundance of each cell type in mouse liver, something that is lacking in the single-cell populations from healthy (normal chow diet) mouse liver used in our analysis (Fig. 2), which were aggregated and harmonized across four data sets variously enriched for mouse nonparenchymal cells.
Single-cell analysis has enabled the functional characterization of the periportal to pericentral zonation gradients of PCG expression across the liver lobule, as was shown for hepatocytes (Halpern et al. 2017), endothelial cells (Halpern et al. 2018), and HSCs (Dobie et al. 2019). Here, we used spatial inference methods to elucidate zonation patterns for large numbers of lncRNAs, and PCGs, in each of these major liver cell types. We also identified novel subclusters for both VSMC and fibroblasts. Prior reports of VSMC and fibroblast zonation were limited to extrahepatic tissues, such as brain (Vanlandewijck et al. 2018; Muhl et al. 2020), whose zonated markers we used to tentatively annotate the zonation of the liver VSMC clusters we identified. Liver fibroblasts were subclustered to give two novel subtypes of unknown function, one of which was uniquely marked by Colec11, which plays an important role in innate immunity (Gajek et al. 2020), and the other by Ly6c1, an antigen expressed by monocytes/macrophages that ingest stressed erythrocytes and deliver iron to hepatocytes (Theurl et al. 2016). Further study, including experimental validation by single-molecule fluorescence in situ hybridization, will be needed to verify these lncRNA expression patterns and tentative zonation assignments.
Single-cell analysis can dissect the milieu of hepatic immune cells (Xiong et al. 2019), which are critical for hepatic immune surveillance and immune tolerance (Jenne and Kubes 2013) and protect against pathogens and dietary antigens (Bogdanos et al. 2013). Kupffer cells (liver resident macrophages) are essential for tissue repair and clearance of toxins (Wen et al. 2021) and can differentiate into the more highly pathogenic NASH-associated macrophages (NAMs) (Xiong et al. 2019). We identified a total of 96 lncRNA markers for four macrophage subpopulations from NAFLD and NASH liver, some of which may be useful as clinical indicators for disease diagnosis or progression and perhaps serve as functional therapeutic targets (Huang et al. 2021). These four cell subpopulations are consistent with prior findings (Xiong et al. 2019) and include Trem2-low macrophages, which are involved in innate immunity, NAMs (Trem2-high cells), which are an indicator of disease progression and provide opportunities for therapeutic intervention (van der Heide et al. 2019), and monocyte-derived macrophages, which are responsible for chemokine-mediated signaling and leukocyte migration and may replace the depleted Kupffer cells (Wen et al. 2021). Finally, proliferating Kupffer cells were characterized by high expression of cell division and cell proliferation genes (e.g., Top2a, Stmn1; Supplemental Fig. S17), reflecting the increase in macrophage proliferation in response to damage induced by NAFLD and NASH.
We explored lncRNA responses in chronic CCl4-exposed liver, a widely used model for chemical-induced liver fibrosis with properties that resemble human fibrosis, including widespread inflammation and collagen formation (Scholten et al. 2015; Bao et al. 2021). Using this model, we identified 631 lncRNAs dysregulated in one or more hepatic mesenchymal cell populations. Validating this model, we observed strong up-regulation of many HSC-expressed lncRNAs in association with the up-regulation of PCGs involved in extracellular matrix and collagen processes, a key feature of HSC activation and liver fibrosis (Pellicano et al. 2021). Further, spatial inference analysis revealed apparent zonation differences between control and fibrotic HSCs. Importantly, we identified 73 lncRNA marker genes, many of them novel, for the transition from the quiescent HSC state to the activated state of collagen-producing HSCs (myofibroblasts) following CCl4-induced centrilobular injury. Future studies may investigate these lncRNAs as potential fibrosis biomarkers and therapeutic targets and may expand the analysis reported here to include effects of CCl4-induced hepatotoxicity on the full repertoire of liver cell types, which were absent from the mesenchymal cell population we analyzed (Dobie et al. 2019). It will also be important to validate key findings using mouse models that may better mimic the pathological causes of liver fibrosis in humans (Bao et al. 2021), such as NASH-induced liver fibrosis (Ipsen et al. 2020).
Gene regulatory network inference is a promising approach to discover regulatory mechanisms based on transcription factor-target gene interactions in both healthy tissue and in disease phenotypes. We utilized the rich single-cell data sets analyzed here to construct gene regulatory networks, with three specific goals: (1) to deduce lncRNA gene functions based on membership in a functional gene module; (2) to identify network-essential regulatory lncRNAs, that is, lncRNAs that are expected to be critical for specific linked biological pathways; and (3) to discover new regulators that emerge in the rewiring of networks in diseased states. We analyzed bigSCale2-derived networks obtained for both healthy and diseased liver (NAFLD, NASH, and fibrotic liver) and used four distinct network centrality metrics to discover network-essential regulatory genes, including many lncRNAs. The four network metrics we used capture different types of network regulators. Thus, Betweenness is crucial for information flow between network modules, Closeness identifies genes that can rapidly spread information across the network, PageRank centrality marks genes that are highly influential on the network, and Degree, a measure of a node's centrality, identifies key network hubs (Iacono et al. 2019). Master regulators, both lncRNAs and PCGs, were also identified for each network based on their central roles in subnetworks comprised exclusively of the network-essential regulatory lncRNAs and PCG genes themselves. Many of the regulatory lncRNA gene targets from the diseased liver networks were enriched for common sets of functional annotations that are characteristic of the diseased states. For instance, clusters of lncRNAs in the NAFLD network were associated with cell migration and regulation of angiogenesis, while the sets of NAFLD and the NASH network regulatory lncRNA target genes were both associated with vascular development, and in CCl4-exposed mesenchymal cells, with cytokine and innate immune response and extracellular matrix. Validating this approach to the discovery of network regulatory genes, many of the PCG master regulators that we identified have well-established regulatory functions specifically related to liver disease, and many of their downstream network target genes were highly enriched for functional annotations that match the biological functions of their master regulators.
LncRNAs can act as scaffolds that promote interactions between proteins, RNA, and DNA (Luo et al. 2021). Furthermore, there is increasing evidence that some lncRNAs interact with genomic DNA in a sequence-specific manner via triple helix (triplex) formation, which may enable lncRNAs to assemble protein complexes at specific genomic regions and regulate gene expression (Kuo et al. 2019; Soibam and Zhamangaraeva 2021). We used the sequence-based algorithm Triplex Domain Finder to predict RNA–DNA triple helix binding between regulatory lncRNAs identified in our bigSCale2 networks and the DNA promoter sequences of the genes that make direct connections with those lncRNAs in the gene regulatory network. We found significant enrichments of triplex interactions in the set of target genes for 20%–32% of the network-essential regulatory lncRNAs identified in the NAFLD and NASH networks and for 42% of those from the CCl4-induced liver fibrosis network (Supplemental Table S7A). Little overlap was found between the NAFLD and NASH network-essential regulatory lncRNAs that formed significant triplexes with PCG promoters and those from the CCl4-induced fibrosis network. This is not unexpected as the fibrosis network input was limited to single-cell expression data from hepatic mesenchymal cell populations. Within each network, we observed many shared gene targets between regulatory lncRNAs, which indicates substantial regulatory crosstalk (Supplemental Figs. S15, S16).
Finally, supporting the human relevance of our findings, we identified 259 mouse liver lncRNAs with orthologous human sequences that were differentially expressed in liver disease (either NAFLD, NASH or CCl4-induced fibrosis) compared to healthy liver (Supplemental Table S8B). One example, Neat1 (lnc14746*), was up-regulated in NAFLD in our mouse liver data set and has a human ortholog that promotes NAFLD, liver fibrosis and hepatocellular carcinoma (Bu et al. 2020). Other human lncRNA orthologs associated with human liver disease include: SNHG18 (lnc12453*), a tumor suppressor and prognostic biomarker that is down-regulated in human hepatocellular carcinoma (Liu et al. 2018); LINC00657 (lnc1906*_2900097C17Rik), whose knockdown suppresses hepatocellular carcinoma progression (Cao et al. 2020); and LINC00862 (lnc590*_Gm19705), whose expression is elevated in hepatocellular carcinoma (Yu et al. 2021). In addition, 32 of the mouse lncRNAs with human orthologs were essential nodes in at least one of the five liver regulatory networks that we presented (Supplemental Table S8C). One of these lncRNAs, Meg3 (lnc10922*), is a marker for activated myofibroblasts and an essential node in both healthy and CCl4-induced mouse liver mesenchymal networks. In human liver, Meg3 has an antifibrotic effect in patients with liver fibrosis (Yu et al. 2018; Qin et al. 2022). Fendrr (lnc47443*) is an HSC-specific marker that serves as an essential node in both the chow-fed and fibrotic mouse liver mesenchymal networks; and in human liver, Fendrr inhibits proliferation and invasion of human hepatocellular carcinoma (Wang et al. 2019).
In conclusion, we present a comprehensive characterization of thousands of novel mouse liver lncRNAs including gene and isoform structures, cell-type expression patterns, inferred spatial location across the liver lobule, and dysregulation in liver disease. Many novel lncRNAs were identified as markers for pathogenic cell types in NASH and in liver fibrosis. A computational framework implemented to discover network-essential regulatory lncRNAs and their predicted gene targets was validated by multiple examples drawn from network-essential regulatory PCGs of known function. Finally, for a subset of the putative regulatory lncRNAs that we identified, promoter sequences upstream of the network-defined lncRNA target genes were shown to be significantly enriched for lncRNA triplex formation, a finding that gives independent mechanistic support for the lncRNA–target genes linkages predicted by our gene regulatory networks.
Limitations of this study
As this study is based on scRNA-seq data, there are technical limitations related to the sparse nature of such data sets, in particular when it comes to characterization of lncRNAs whose overall expression in the cell is often much lower than that of PCGs; greater sensitivity for lncRNA detection would likely have been achieved by using single nucleus RNA-seq. Moreover, lncRNA transcripts that are not polyadenylated would not be captured by the poly(A) capture-based scRNA-seq technology used to generate the data sets analyzed here; however, this is not a major concern, as a large majority of liver-expressed lncRNAs are polyadenylated, as shown in our prior studies (Goldfarb and Waxman 2021). Further, our analyses were limited to the specific set of 48,261 liver-expressed lncRNAs that we identified from our analysis of more than 2000 bulk mouse liver RNA samples, plus 4700 other lncRNA genes that we collected from established databases. Our inability to include other, presently unknown lncRNAs that may be important for liver function and pathophysiology is another limitation. Several of our conclusions are based on inferences drawn from analyses that are, at their core, correlative in nature. These include our analysis of pseudotime trajectories to infer zonation patterns for PCGs and lncRNAs across the liver lobule, as well as changes in zonation in liver disease. While such changes in zonation were, in some cases, experimentally verified by others for certain PCGs, the lack of experimental verification of the lncRNA zonation patterns reported here is a limitation, as is the lack of experimental verification of proposed marker genes and disease state-specific lncRNAs. Furthermore, while we were able to validate our use of single-cell-based gene regulatory networks to identify network-essential genes, as evidenced by multiple examples of network-essential regulatory PCGs whose known biological functions align with the enriched functions of their gene targets in the regulatory networks, the inferred nature of the gene regulatory networks is nevertheless a limitation. Experimental validation of the network-essential lncRNAs described here using lncRNA knockout or knockdown mouse models will ultimately be required to address this limitation.
MATERIALS AND METHODS
Transcriptome reconstruction
We downloaded from GEO, ENA, and Array express a total of 2089 bulk mouse liver RNA-seq samples extracted from 68 different studies (Supplemental Table S1A), all sequenced by paired-end, stranded Illumina RNA sequencing. Sequence reads were mapped to the mouse mm9 reference genome using TopHat2, and Cufflinks was used to assemble mapped reads into transcripts for each individual RNA-seq sample. We then used two algorithms for transcriptome assembly: TACO (Niknafs et al. 2017) and Cuffmerge (Trapnell et al. 2012), each of which produced a different assembly comprised of both coding and noncoding transcripts. The TACO-derived transcriptome was evaluated for read-through transcripts (He et al. 2018; Morgan et al. 2022), a product of transcriptional read-through of two adjacent genes on the same strand, which in many cases represent spurious transcriptional noise. A total of 23,346 out of 1,491,436 TACO-assembled transcripts were identified as potentially read-through based on their overlap with multiple PCGs or lncRNAs. For each of these putative read-through transcripts, we calculated isoform_sum_abs_frac, that is, the absolute fraction of all transcript isoforms of a given gene that is represented by the sum of all the read-through isoforms for that gene. Transcripts whose isoform_sum_abs_frac was >0.1 were retained, and the associated genes concatenated to give a new, longer gene structure. For transcripts whose isoform_sum_abs_frac was ≤0.1, we discarded the entire set of read-through transcripts for that gene, as they likely represent transcript assembly artifacts. Overall, 19,219 (82%) of the 23,346 read-through transcripts initially identified were discarded. In parallel, we assembled the transcriptome of each of the 2089 RNA-seq samples with Cufflinks (v2.1.1) (Trapnell et al. 2012) and discovered 547,300 transcripts that were used for downstream filtering and lncRNA discovery.
lncRNA discovery
The filtered transcriptome obtained from each assembly method (TACO and Cufflinks) was further processed using two different lncRNA discovery pipelines, as we described previously for rat liver lncRNAs (Karri and Waxman 2020). Method 1: LncRNA transcripts were identified based on transcript length >200 nt, low or no coding potential, and absence of overlap with known PCGs (Melia et al. 2016). Method 2: the lncRNA discovery tool Slncky (Chen et al. 2016b) was used to remove transcripts that overlap PCGs, and to assess the coding potential for small peptides and novel proteins. A filter based on synteny was used to remove transcripts that align to syntenic coding transcripts in other, related species. A nonsynonymous to synonymous (dN/dS) ratio was then calculated and used to evaluate the coding potential of each transcript. The TACO assembly yielded 109,937 sequences that passed our filters to qualify as lncRNAs but were found to be mono-exonic fragments that overlap a PCG intronic region. These were filtered out as likely derived from spurious transcription, and consequently, no mono-exonic intragenic lncRNAs were included in the final TACO assembled lncRNA gene list. Bedtools (v2.17.0) was used to perform overlap analysis between the set of 44,579 lncRNA gene structures obtained by TACO assembly, the 25,869 lncRNA gene structures generated by Cuffmerge, and the original set of 15,558 liver-expressed lncRNAs that we discovered previously using a much smaller set of input RNA-seq samples (Melia and Waxman 2019). These lncRNA genes and their isoforms were merged using the Bedtools overlap command. The gene structures and isoforms in the original set of 15,558 liver-expressed lncRNAs were updated based on the TACO and Cuffmerge assemblies while retaining the original lncRNA gene numbers (Melia and Waxman 2019). In many cases, the new lncRNA gene structure represents a merging of two or more adjacent lncRNAs from the original 15,558 lncRNA gene list; those lncRNAs were renamed by concatenating the old lncRNA gene numbers while using the lowest number to represent the new lncRNA structure. For example, the new lnc_inter_chr8_7423 (chr8:116,533,047-116,655,614) was obtained by merging the prior set of tandem minus-strand lncRNAs, numbered lnc7423 and lncs 7425–7430, and consequently, there are no lncRNAs numbered 7425 through 7430 in the final set of 48,261 liver-expressed lncRNAs. Lnc_inter_chr8_7424 (chr8:116,533,910–116,537,738) is a plus-strand lncRNA that is intragenic and antisense to the new lnc_inter_chr8_7423 and was therefore not included in the merger that formed the new lnc_inter_chr8_7423 (Supplemental Table S1B). Newly discovered lncRNAs that did not overlap the prior set of 15,558 lncRNAs were assigned new lncRNAs (Fig. 1F). gene numbers, beginning with lncRNA number 15,559 on Chr1.
Final set of 48,261 liver-expressed lncRNAs (mm10)
The final set of 48,361 lncRNAs and their total of 150,280 isoforms, defined for mouse genome mm9 (Supplemental Table S1B,D), was reduced to 48,261 lncRNAs after lift-over to mouse genome mm10 (Supplemental Table S1C), and was comprised of the following (based on mm10 mapping): 1176 intragenic lncRNAs, which overlap one or more PCG intronic regions on the same strand, and whose exons do not overlap any PCG exonic regions on the same strand; 4127 antisense lncRNAs, which overlap a PCG on the opposite strand; 42,892 intergenic lncRNAs, which do not overlap any PCG on either strand; and 66 intra-antisense lncRNAs, which have attributes of both antisense and intragenic lncRNAs (Fig. 1F). A total of 42,605 of the 48,261 lncRNAs are mono-exonic genes and 5656 are spliced, multiexonic genes. The designation “mono-exonic” is used here to describe a lncRNA whose union of all exons across all isoforms (“exon collapsed” sequence) covers the entire gene body without leaving any intronic gaps. However, 7754 of the 42,605 mono-exonic lncRNAs have multiple isoforms, which in many cases include intronic sequences (Supplemental Table S1D), and thus could be considered multiexonic.
The number of lncRNAs referred to in this study, for example, the number of lncRNAs expressed in a given liver single-cell cluster, or the number of lncRNAs that are differentially expressed under a given biological condition, is based on a total lncRNA count of 52,961, corresponding to the subset of the 48,261 liver-expressed lncRNAs that meet the specified condition, plus the subset of 4700 other long-noncoding genes that met the same condition: 4697 Ensembl noncoding lncRNAs assigned the gene biotype “lincRNA” (N = 3176) or “antisense” (N = 1521), plus three other lncRNAs not included in the above lists (lnc-LFAR1, LeXis, Lnclgr) (genes listed in Supplemental Table S2A).
Ortholog conservation
We used Slncky (Chen et al. 2016b) to assess primary sequence conservation and syntenic conservation to discover lncRNA orthologs in the human and rat genomes for the full set of 52,961 mouse liver lncRNAs described above. Two metrics were used to identify orthologs, as implemented by Slncky: transcript-genome identity (TGI; percent of identical, aligning sequences between a mouse lncRNA and a nontranscribed locus in a syntenic region in the rat or human genome); and transcript-transcript identity (TTI; percentage of identical, aligning sequences present in the exonic regions of lncRNAs in both species) (Chen et al. 2016b). A threshold of 30% conservation based on either the TTI metric or the TGI metric was used to define an orthologous human or rat sequence. To identify human orthologs, we input to Slncky a set of 18,151 human lncRNAs from GENCODE (version 23) (Frankish et al. 2019), 96,308 human lncRNAs from NONCODE (v5) (Zhao et al. 2016), and 662 human lncRNAs involved in cancer (onco-lncs) that we curated from PubMed. To identify rat orthologs, we input our published set of 5795 rat liver lncRNAs (Karri and Waxman 2020). As this set of rat lncRNAs is far from complete, there are likely to be many more than the 1805 rat lncRNA orthologs identified here (Supplemental Table S1E,F). We found human orthologs for 9543 of the 48,261 mouse liver-expressed lncRNAs (Supplemental Table S1E) and for 1166 of the 4700 other noncoding lncRNAs described above (Supplemental Table S1F).
GTF file for single-cell and single nucleus RNA-seq analysis
We created a custom reference GTF file for the mouse mm10 genome build for use with the CellRanger (v3.1.0) mkref command (Zheng et al. 2017). This GTF file is comprised of gene coordinates for 76,011 genes (Supplemental Table S2A) plus 91 ERCC spike-in controls: RefSeq PCGs (N = 20,973; 19,801 genes with NM accession numbers, 1159 genes with both NM and NR accession numbers, and 13 mitochondrial genes), the above-described set of mouse liver lncRNAs with mm10 genome coordinates (N = 48,261), RefSeq noncoding genes defined by NR accession numbers that do not overlap the set of 48,261 lncRNAs (N = 2077), Ensembl noncoding lncRNAs that do not overlap RefSeq NR genes or the set of 48,261 lncRNAs (N = 4697; 3176 genes designated “lincRNA” and 1521 genes designated “antisense” in Supplemental Table S2A), and three other lncRNAs (lnc-LFAR1, LeXis, Lnclgr) absent from the above lists. The full set of mouse liver lncRNAs identified in our analysis, originally discovered by mapping bulk RNA-seq fastq files to the mm9 reference genome, was converted to mm10 genomic coordinates using the UCSC genome browser LiftOver tool (Hinrichs et al. 2006). The GTF file used for scRNA-seq analysis (gtf76k_mm10_Liver_scRNAseq.gtf, available in Supplemental Material) contains exon region gene coordinates for all 76,011 genes and was used by 10× Genomics CellRanger software (v3.1.0) (Zheng et al. 2017) to count sequence reads mapping to exons for each scRNA-seq sample.
Data sets and data processing for single-cell analysis
Single-cell RNA-seq data for mouse liver was downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/). Raw sequencing data for hepatocytes (GSE109774) (Tabula Muris Consortium et al. 2018) and for the liver nonparenchymal cell clusters shown in Figure 2A (GSE137720, E-MTAB-8077, and GSE129516) (Dobie et al. 2019; Xiong et al. 2019; Kalucka et al. 2020) was obtained from healthy (untreated, normal chow diet) mouse livers (Supplemental Table S1G). We also analyzed 10× Genomics scRNA-seq data sets downloaded from GSE166504 (Su et al. 2021) to study the effects of high fat diet-induced NAFLD and NASH, and from GSE137720 (Dobie et al. 2019) to study CCl4-induced liver fibrosis (Supplemental Table S1G). Raw fastq files were processed and aligned to the mm10 mouse reference genome and processed using the single-cell analysis 76,011 gene custom GTF file described above. Feature-barcode matrices were generated using CellRanger (v3.1.0). Multiple scRNA-seq data sets were combined using the CellRanger aggr command and count data were then processed using Seurat v3 (Stuart et al. 2019).
Batch correction
As detailed further below, we used CellRanger aggr to merge and normalize the merged data sets for differences in sequencing depth. Next, we used Harmony to correct for differences in conditions, batches, chemistries, and studies. Cells with high mitochondrial contamination (cells with >5% gene reads mapping to the 13 mitochondrial genome genes) were excluded from all downstream analyses. Furthermore, we used the doublet finder method implemented in scDblFinder to remove doublets from the data. All of the published scRNA-seq data sets analyzed in this report were high quality scRNA-seq data sets with no indication of impactful levels of ambient RNA; accordingly, we did not use any algorithmic approaches to correct for ambient RNA.
Integration and clustering
Using Seurat v3, we normalized the integrated CellRanger count matrix data by dividing the UMI count per gene by the total UMI count in the corresponding cell followed by log-transformation. Cells with fewer than 200 genes detected, fewer than 400 UMI per cell, or cells with >5% gene reads mapping to the 13 mitochondrial genome genes were excluded from all downstream analyses. Highly variable genes were identified using the FindVariableGenes function of Seurat with default parameters. Variable genes were projected onto a low-dimensional space using principal component (PC) analysis, with the number of PCs set to 10 based on an inspection of elbow plots of the variance explained for each data set. We used Harmony (v1.0) (Korsunsky et al. 2019) to remove from the embedding the influence of data set-of-origin factors (e.g., the effects of varying batches or technologies) and to integrate shared cell types across different data sets. We input to Harmony normalized gene matrix files saved as a Seurat object along with a precalculated PC analysis embedding based on 10 PCs using default parameters. Graph-based clustering was performed to cluster the cells based on their gene expression profiles using the FindClusters function in Seurat. The clusters obtained were visualized using the UMAP function of Seurat v3 (resolution: 0.15 and PC = 10), and cell identities were assigned to each cluster based on the expression of established marker genes (Supplemental Fig. S1). scDblFinder (Germain et al. 2021), a doublet detection algorithm, was applied (with default parameters) to identify and remove doublets based on artificial doublet generation, parameter optimization and thresholding.
Detection of marker genes
Cell type-specific marker genes that drive the separation between single-cell clusters were identified by pairwise differential gene expression analysis (Yu et al. 2013), comparing each cell cluster against all other cell clusters using the Global Distinguishing function of Loupe Browser (10× Genomics, Loupe Browser 5.0 [https://www.10xgenomics.com/]). Genes expressed in >5% of the cells in a cluster were identified as putative marker genes for that cluster if they showed differential expression at log2|fold-change| >2 and FDR (Benjamini–Hochberg) <0.05 when compared to the set of all cells comprising all other clusters. We then removed from the list of putative marker genes all genes that were identified as putative marker genes for two or more clusters, resulting in the final set of cluster-specific marker genes (Supplemental Table S2D). Marker genes distinguishing the four macrophage clusters shown in Figure 4G were identified using a relaxed fold-change filter of log2|fold-change| >1 because of the close relationship between the four cell populations. Three Loupe Browser files (.cloupe format) containing the single-cell data sets analyzed in this study are available in Supplemental Material.
Inference of liver zonation
Pseudotemporal trajectories were inferred from the hepatocyte population obtained from chow-fed mouse liver (Su et al. 2021) and for each of four major nonparenchymal cell clusters (endothelial cells, HSCs, fibroblasts, VMSC; Fig. 2A) using the R package Monocle2 (v2.6.419) (Trapnell et al. 2014) with default parameters. Cells were ordered based on the inference pseudotime trajectory in Monocle2. PCGs and lncRNAs showing significant changes in expression along the pseudotime trajectory were identified using generalized linear models (differentialGeneTest function in Monocle2) with a threshold of Q-value <10−3 for significance. Hierarchical clustering of the genes that were cozonated across pseudotime was implemented using the pheatmap function of Monocle2. The zonation clusters were annotated as periportal versus pericentral for hepatocytes (Ben-Moshe et al. 2019), endothelial cells (Kalucka et al. 2020), and HSCs (Dobie et al. 2019) using established marker genes, and the resulting pseudotime values were scaled between 0 and 1. Cell trajectories were also determined for HSCs from healthy and from CCl4-treated livers. Hierarchical clustering in Monocle2 divided zonation heatmaps into clusters, which were assigned labels based on established zonation marker genes in each major cluster. SCORPIUS (Cannoodt et al. 2016) was used to further validate the endothelial cell zonation patterns (Fig. 3B). We included traditional endothelial cell phenotypes from the liver (Kalucka et al. 2020) (artery, capillary artery, capillary, capillary vein, vein), and performed the analysis using the parameters k = 3 and number of PC = 8.
Perturbations of zonation in disease states
Monocle2 was used to derive a common zonation trajectory across biological conditions within each cell type. This enabled us to compare the conditions along the trajectory and detect large scale gene expression changes indicative of differential progression. Differentially expressed genes along the trajectory between control (chow diet) and either NAFLD or NASH liver groups, and between the control and CCl4-exposed mesenchymal cell groups, were identified using the conditionTest function of the tradeSeq package (Van den Berge et al. 2020). Significantly perturbed PCGs and lncRNAs (FDR < 0.001) were extracted and used to prepare matched heatmaps for control liver and from diseased liver cells using the pheatmap function of Monocle2. In the case of NAFLD and NASH livers, we identified PCGs and lncRNAs where a change in zonation was significant for either NAFLD versus chow diet or for NASH versus chow diet. We then clustered heatmaps individually from each condition to assign the zonation labels shown in Supplemental Table S4I.
Functional enrichment analysis
Sets of PCGs obtained in various analyses were used as input for functional enrichment analysis using DAVID (Sherman et al. 2022) with default parameters, except that GO FAT terms were used in place of GO DIRECT terms to include a broader range of enrichment terms that were excluded by the default GO DIRECT option. We used a custom script prepared by Dr. Alan Downey–Wall of this laboratory (https://github.com/adowneywall/Davidaggregator) to combine and reformat DAVID output files from multiple gene lists, with each row presenting top enriched annotation clusters and individual columns presenting enrichment score, P-value, FDR and other such data. Top terms from each DAVID annotation cluster with a cluster enrichment score >3 and top FDR < 0.05 were used in downstream analysis.
Differential expression analysis between biological conditions
Differential gene expression comparisons between single-cell clusters and between biological conditions were performed using the Locally Distinguishing function of the 10× Genomics Loupe Browser (v.6.0). This method implements the negative binomial test based on the sSeq method (Yu et al. 2013) with Benjamini–Hochberg correction for multiple tests. Further, the method uses log-normalized average expression values and the distribution of UMIs across the two specific cell populations being compared to calculate fold-change and FDR values for each pair-wise comparison between cell clusters. Genes were identified as showing significant differential expression between control liver and diseased liver (NAFLD, NASH, or CCl4-induced liver fibrosis) if they met the thresholds of log2|fold-change| >1 and FDR < 0.05.
Transition from quiescent HSCs to activated HSCs (myofibroblasts)
Central vein-associated HSCs were extracted from livers of mice treated with CCl4 (6 wk treatment) to investigate the transition of gene expression from resting HSCs to collagen-producing myofibroblasts. The central vein-associated HSCs were spatially resolved into resting HSCs and myofibroblasts using Monocle2 (v2.6.419), and the two cell subtypes then characterized using established signature genes (Dobie et al. 2019). PCGs and lncRNAs showing zonation along the trajectory were identified using generalized linear models and a heatmap was prepared using Monocle2, as described above.
Construction of gene regulatory networks
Regulatory networks were generated using the R package bigSCale2 (Iacono et al. 2019) for scRNA-seq data sets comprised of all cells that passed the gene expression filters described above. Three separate bigSCale2 networks were generated from scRNA-seq data (GSE166504) from livers of mice fed chow diet (healthy liver) or mice fed HFHFD for either 15 wk (NAFLD liver) or 30 wk (NASH liver). Two other bigSCale2 networks were generated from all qualified mesenchymal cells from the scRNA-seq data sets for control and for CCl4-exposed mouse liver (GSE137720) (Supplemental Table S1G). Cells from each of the five data sets were split into individual Seurat objects and then input to bigSCale2. Each of the five networks was constructed using default clustering parameters, with granularity set to the highest setting and applying an edge cutoff of the top 99.9 quantile used for correlation coefficient. BigSCale2 retains edges that are expected to represent actual regulatory links by removing genes that did not have a direct edge with a known GO regulator (GO subsetting step). This was implemented using a list of GO regulators comprised of the union of genes in GO:0006355 “regulation of transcription, DNA-templated” (N = 2776) and genes in GO:000370 “DNA-binding transcription factor activity” (N = 1001). We also included the above list of 52,961 lncRNAs as potential regulators in the GO subsetting step. Network specifications are shown in Supplemental Table S5E. The BigSCale2 gene regulatory networks obtained were converted into json files for visualization by Cytoscape (Shannon et al. 2003) using the toCytoscape function of the package iGraph (v1.3.5) (https://igraph.org/r) in R. Specifically, the network json file was imported into Cytoscape using “Import Network from File.” Next, we used “forced-directed” layout in Cytoscape, where each network's layout was derived from 10,000 iterations of the Fruchterman–Reingold algorithm (Gajdoš et al. 2016) and using the Nogrid parameter (seed = 7). The resultant networks and derived subnetworks shown in the various figures are available at Ndexbio.org at https://tinyurl.com/scLiverNetworksKarriWaxman.
Analysis of gene regulatory networks
Gene modules in each network were discovered using the Glay community cluster algorithm (Su et al. 2010) in Cytoscape, whereby the overall network topology was used to subdivide the networks generated by bigSCale2 into functional modules. Genes in each module were input to DAVID (Sherman et al. 2022) for gene ontology enrichment analysis. Node rank was calculated for each of four key bigSCale2 network metrics (Betweenness, Closeness, Degree, PAGERANK). PCGs that ranked within the top 100 nodes for any one of the above four network metrics were deemed to be network-essential nodes that correspond to regulatory PCGs; and lncRNAs that ranked within the top 50 nodes for any one of the above four network metrics were deemed to be network-essential nodes identifying regulatory lncRNAs (Supplemental Table S5C).
Master regulators
We extracted subnetworks comprised of the network-essential regulatory PCGs and regulatory lncRNAs defined above, that is, all top-100 ranked PCG nodes and all top-50 ranked lncRNA nodes. We then recalculated the network metrics to identify critical nodes that are deemed network-essential regulators with regards to expression of the original set of regulators; these were designated master regulators. For this analysis, we ranked the nodes of the extracted networks using five different network metrics: Stress, Degree, Betweenness, Closeness, and Neighborhood Connectivity, as calculated using the “Analyze Network” option in Cytoscape. We then designated as master regulators the top 5 ranked nodes for each of the five metrics, which were considered independently in the ranking (Supplemental Table S5C, columns AP-BU). We excluded from the master regulator analysis the PageRank centrality metric (used in the original ranking, above), as it tended to select nodes with low Degree ranking due to the high degree of disconnectedness in these master regulator networks.
IPA analysis
We used Qiagen's Ingenuity Pathway Analysis (IPA) software (https://digitalinsights.qiagen.com) to validate the regulatory role of a total of the 20 master regulators derived from all five bigSCale2 networks that were identified as DNA-binding proteins. Putative gene targets of these 20 DNA-binding master regulators were provided as input to IPA. IPA's core analysis of these input gene lists was used to identify Upstream Regulators, along with the top enriched canonical pathways, diseases, and toxicological functions. The resulting IPA output for all 20 master regulators is included in Supplemental Table S5F.
Enrichment heatmaps
PCGs that made direct connections to a network-essential regulatory lncRNA in any of the bigSCale2 networks were used as input to Metascape with default parameters (Zhou et al. 2019). To identify pathways that are shared between, or are specific to, a lncRNA's gene targets, we performed multilist comparative analysis within and across the networks. The resulting clustered heatmaps depict −log10 P-values of the top enriched pathways across multiple lncRNAs target lists.
Triplex Domain Finder analysis
Regulatory lncRNAs identified for each gene network using bigSCale2 metrics (see above) were analyzed using the algorithm Triplex Domain Finder (TDF) (Kuo et al. 2019) to determine their capacity to form lncRNA-genomic DNA triplexes with the promoter sequences of their target genes. Target genes were defined as all PCGs that make a direct connection to the lncRNA in the corresponding bigSCale2 network. A background gene set was defined as the set of PCGs expressed in at least 0.5% of the cells used to develop the network, excluding all genes that were part of a modified bigSCale2 network derived from the same cell population used to identify the regulatory lncRNA being evaluated for triplex formation, but constructed using an edge cutoff at the 95% quantile (instead of the 99.9% quantile) correlation coefficient threshold. This relaxed threshold was implemented to obtain a modified bigSCale2 network specifically for the purpose of background gene identification, and with the goal of excluding from the background gene set any gene that might be a target of the network-essential regulatory lncRNA when using a less stringent correlation threshold of >95% quantile. Fisher exact test was used to assess the significance (at FDR < 0.05) of the observed number of target gene promoters predicted by Triplex Domain Finder to form lncRNA–DNA triplexes, compared to the number of background gene set promoters predicted to form triplexes with the same lncRNA. The Triplex Domain Finder-predicted gene binding targets of each regulatory lncRNA were used to derive lncRNA–gene directed subnetworks for each condition (Supplemental Figs. S15, S16) using Cytoscape (Shannon et al. 2003), and are available at https://tinyurl.com/scLiverNetworksKarriWaxman.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health (NIH) grant ES024421 (to D.J.W.).
Author contributions: K.K. and D.J.W. jointly conceptualized the study and its major aims. K.K. carried out all of the computational analysis, including methodology development, script writing, and figure preparation. Downstream data analysis and Supplemental Table preparation were carried out by K.K. with the assistance of D.J.W. K.K. prepared a draft manuscript and D.J.W. revised and edited the manuscript and finalized the text and Supplemental Tables. D.J.W. was responsible for project funding acquisition, project supervision, and administration.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079580.123.
MEET THE FIRST AUTHOR
Kritika Karri.

Meet the First Author(s) is an editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Kritika Karri is the first author of the paper, “Dysregulation of murine long noncoding single-cell transcriptome in nonalcoholic steatohepatitis and liver fibrosis.” She holds a Master's degree in Biomedical Informatics from the University of Nebraska and earned her PhD from Boston University under the guidance of Dr. David Waxman. Kritika's doctoral research focused on developing computational methodologies for the identification of lncRNAs and predicting their roles in liver diseases using bulk and single-cell RNA-seq data sets. Currently, she serves as a computational scientist at Celsius Therapeutics, where she contributes to the Data Science team's efforts in identifying clinical targets for inflammatory bowel disease (IBD).
What are the major results described in your paper and how do they impact this branch of the field?
In this study, we set out to uncover and characterize novel mouse liver long noncoding RNAs (lncRNAs) implicated in nonalcoholic steatohepatitis (NASH) and liver fibrosis using an extensive array of bulk and single-cell RNA-seq data sets. Our findings serve as a comprehensive reference for promising lncRNA candidates in these hepatic diseases. By leveraging single cell RNA-seq data from mouse liver samples, we shed light on the cell-type specificity, spatial zonation patterns, associated regulatory networks, and dysregulation patterns of these lncRNAs during hepatic disease progression.
What led you to study RNA or this aspect of RNA science?
LncRNAs are crucial due to their diverse and essential roles in gene regulation and cellular processes. Dysregulation of lncRNAs has been linked to numerous human diseases, making them potential therapeutic targets and diagnostic biomarkers. Furthermore, their tissue and cell-type specificity can offer valuable insights into their functions, advancing our understanding of the molecular mechanisms underlying various biological and pathological processes. Despite the growing research on lncRNAs, much remains to be discovered about their functional roles and mechanisms of action. This presents a fascinating area of study with the potential to reveal novel regulatory processes, deepen our understanding of complex biological systems, and ultimately contribute to the development of innovative therapeutic strategies and diagnostic tools for various human diseases.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
My fascination with the intricacy and complexity of human biology has always captivated me, sparking my curiosity to explore bioinformatics, where one can examine biological systems at the molecular level through genomic sequences. My formative internship experience at IIT Delhi ignited my passion for research, which continued to develop during my time as a Master's student at the University of Nebraska. There, I matured as a scientist and became inspired to utilize next-generation sequencing (NGS) data to investigate various diseases. At Boston University, under the guidance of Dr. Waxman, I had the opportunity to work with various NGS data sets to unravel diverse aspects of lncRNA which proved to be pivotal for my scientific growth, as it allowed me to expand my knowledge and refine my research skills in the field of lncRNA biology.
If you were able to give one piece of advice to your younger self, what would that be?
I would counsel my younger self to appreciate the importance of perseverance in research and the capacity to envision the broader context. In research, the most important step is to articulate a compelling biological question and ability to see the “Big Picture” before considering the most suitable tools, data, and methodologies to address that inquiry effectively.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
I attribute much of my scientific development to my advisor, Dr. David Waxman, who profoundly influenced my approach to research. He instilled in me the importance of examining problems from multiple perspectives and exhaustively investigating every aspect before drawing scientific conclusions. Additionally, he emphasized the value of effective science communication, both orally and in writing, which has been instrumental in my maturation as a scientist.
REFERENCES
- Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. 2019. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol 16: 411–428. 10.1038/s41575-019-0145-7 [DOI] [PubMed] [Google Scholar]
- Bao YL, Wang L, Pan HT, Zhang TR, Chen YH, Xu SJ, Mao XL, Li SW. 2021. Animal and organoid models of liver fibrosis. Front Physiol 12: 666138. 10.3389/fphys.2021.666138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Moshe S, Itzkovitz S. 2019. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol 16: 395–410. 10.1038/s41575-019-0134-x [DOI] [PubMed] [Google Scholar]
- Ben-Moshe S, Shapira Y, Moor AE, Manco R, Veg T, Bahar Halpern K, Itzkovitz S. 2019. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab 1: 899–911. 10.1038/s42255-019-0109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanos DP, Gao B, Gershwin ME. 2013. Liver immunology. Compr Physiol 3: 567–598. 10.1002/cphy.c120011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomzon A, Ljubuncic P. 2001. Oxidative stress and vascular smooth muscle cell function in liver disease. Pharmacol Ther 89: 295–308. 10.1016/S0163-7258(01)00129-2 [DOI] [PubMed] [Google Scholar]
- Braet F, Taatjes DJ, Wisse E. 2018. Probing the unseen structure and function of liver cells through atomic force microscopy. Semin Cell Dev Biol 73: 13–30. 10.1016/j.semcdb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Braeuning A, Menzel M, Kleinschnitz EM, Harada N, Tamai Y, Kohle C, Buchmann A, Schwarz M. 2007. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by β-catenin signaling in mouse hepatocytes. FEBS J 274: 4766–4777. 10.1111/j.1742-4658.2007.06002.x [DOI] [PubMed] [Google Scholar]
- Bu FT, Wang A, Zhu Y, You HM, Zhang YF, Meng XM, Huang C, Li J. 2020. LncRNA NEAT1: shedding light on mechanisms and opportunities in liver diseases. Liver Int 40: 2612–2626. 10.1111/liv.14629 [DOI] [PubMed] [Google Scholar]
- Cai H, Shao B, Zhou Y, Chen Z. 2020. High expression of TOP2A in hepatocellular carcinoma is associated with disease progression and poor prognosis. Oncol Lett 20: 232. 10.3892/ol.2020.12095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannoodt R, Saelens W, Sichien D, Tavernier S, Janssens S, Guilliams M, Lambrecht B, De Preter K, Saeys Y. 2016. SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. bioRxiv 10.1101/079509 [DOI] [Google Scholar]
- Cao X, Zhang G, Li T, Zhou C, Bai L, Zhao J, Tursun T. 2020. LINC00657 knockdown suppresses hepatocellular carcinoma progression by sponging miR-424 to regulate PD-L1 expression. Genes Genomics 42: 1361–1368. 10.1007/s13258-020-01001-y [DOI] [PubMed] [Google Scholar]
- Ceni E, Mello T, Polvani S, Vasseur-Cognet M, Tarocchi M, Tempesti S, Cavalieri D, Beltrame L, Marroncini G, Pinzani M, et al. 2017. The orphan nuclear receptor COUP-TFII coordinates hypoxia-independent proangiogenic responses in hepatic stellate cells. J Hepatol 66: 754–764. 10.1016/j.jhep.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Chan KK, Hon TC, Au KY, Choi HL, Wong DK, Chan AC, Yuen MF, Lai CL, Lo RC. 2022. Stanniocalcin 1 is a serum biomarker and potential therapeutic target for HBV-associated liver fibrosis. J Pathol 257: 227–238. 10.1002/path.5880 [DOI] [PubMed] [Google Scholar]
- Chen G, Yu D, Nian X, Liu J, Koenig RJ, Xu B, Sheng L. 2016a. LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose triglyceride lipase (ATGL). Sci Rep 6: 35531. 10.1038/srep35531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shishkin AA, Zhu X, Kadri S, Maza I, Guttman M, Hanna JH, Regev A, Garber M. 2016b. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol 17: 19. 10.1186/s13059-016-0880-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tan XR, Li SJ, Zhang XX. 2019. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci 235: 116829. 10.1016/j.lfs.2019.116829 [DOI] [PubMed] [Google Scholar]
- Chen R, Yang H, Song Y, Yu H, Zhang M, Rao W, Wang Y, Xiao X, Chen Q, He Q. 2022. Maternal obesity induces liver lipid accumulation of offspring through the lncRNA Lockd/mTOR autophagy pathway. Mol Genet Genomics 297: 1277–1287. 10.1007/s00438-022-01916-z [DOI] [PubMed] [Google Scholar]
- Cunningham RP, Porat-Shliom N. 2021. Liver zonation: revisiting old questions with new technologies. Front Physiol 12: 732929. 10.3389/fphys.2021.732929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daemen S, Gainullina A, Kalugotla G, He L, Chan MM, Beals JW, Liss KH, Klein S, Feldstein AE, Finck BN, et al. 2021. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep 34: 108626. 10.1016/j.celrep.2020.108626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JL, Cui JY. 2019. Regulation of hepatic long noncoding RNAs by pregnane X receptor and constitutive androstane receptor agonists in mouse liver. Drug Metab Dispos 47: 329–339. 10.1124/dmd.118.085142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, Wallenborg K, Picelli S, Zagorska A, Pendem SV, et al. 2019. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep 29: 1832–1847.e1838. 10.1016/j.celrep.2019.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. 2018. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem 51: 2262–2274. 10.1159/000495871 [DOI] [PubMed] [Google Scholar]
- Dong Z, Li S, Wang X, Si L, Ma R, Bao L, Bo A. 2019. lncRNA GAS5 restrains CCl4-induced hepatic fibrosis by targeting miR-23a through the PTEN/PI3K/Akt signaling pathway. Am J Physiol Gastrointest Liver Physiol 316: G539–G550. 10.1152/ajpgi.00249.2018 [DOI] [PubMed] [Google Scholar]
- Du M, Yuan L, Tan X, Huang D, Wang X, Zheng Z, Mao X, Li X, Yang L, Huang K, et al. 2017. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun 8: 2049. 10.1038/s41467-017-02229-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elpek G. 2015. Angiogenesis and liver fibrosis. World J Hepatol 7: 377–391. 10.4254/wjh.v7.i3.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Biagioli M, Zampella A, Distrutti E. 2018. Bile acids activated receptors regulate innate immunity. Front Immunol 9: 1853. 10.3389/fimmu.2018.01853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. 2018. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 24: 908–922. 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdoš P, Ježowicz T, Uher V, Dohnálek P. 2016. A parallel Fruchterman–Reingold algorithm optimized for fast visualization of large graphs and swarms of data. Swarm Evol Comput 26: 56–63. 10.1016/j.swevo.2015.07.006 [DOI] [Google Scholar]
- Gajek G, Swierzko AS, Cedzynski M. 2020. Association of polymorphisms of MASP1/3, COLEC10, and COLEC11 genes with 3MC syndrome. Int J Mol Sci 21: 5483. 10.3390/ijms21155483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain PL, Lun A, Garcia Meixide C, Macnair W, Robinson MD. 2021. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10: 979. 10.12688/f1000research.73600.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gines P, Krag A, Abraldes JG, Sola E, Fabrellas N, Kamath PS. 2021. Liver cirrhosis. Lancet 398: 1359–1376. 10.1016/S0140-6736(21)01374-X [DOI] [PubMed] [Google Scholar]
- Gloss BS, Dinger ME. 2016. The specificity of long noncoding RNA expression. Biochim Biophys Acta 1859: 16–22. 10.1016/j.bbagrm.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Goldfarb CN, Waxman DJ. 2021. Global analysis of expression, maturation and subcellular localization of mouse liver transcriptome identifies novel sex-biased and TCPOBOP-responsive long non-coding RNAs. BMC Genomics 22: 212. 10.1186/s12864-021-07478-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb CN, Karri K, Pyatkov M, Waxman DJ. 2022. Interplay between GH-regulated, sex-biased liver transcriptome and hepatic zonation revealed by single-nucleus RNA sequencing. Endocrinology 163: bqac059. 10.1210/endocr/bqac059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Li T, Ai N, Wang W, He B, Bai Y, Yu Z, Li M, Dong S, Zhu Q, et al. 2019. MEIS2C and MEIS2D promote tumor progression via Wnt/β-catenin and hippo/YAP signaling in hepatocellular carcinoma. J Exp Clin Cancer Res 38: 417. 10.1186/s13046-019-1417-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Lu H. 2019. Novel mechanisms of regulation of the expression and transcriptional activity of hepatocyte nuclear factor 4α. J Cell Biochem 120: 519–532. 10.1002/jcb.27407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Bai M, Lin L, Huang J, An Y, Liang L, Liu Y, Huang W. 2019. LncRNA DLEU2 aggravates the progression of hepatocellular carcinoma through binding to EZH2. Biomed Pharmacother 118: 109272. 10.1016/j.biopha.2019.109272 [DOI] [PubMed] [Google Scholar]
- Guo CJ, Xu G, Chen LL. 2020. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci 45: 947–960. 10.1016/j.tibs.2020.07.001 [DOI] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE. 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542: 352–356. 10.1038/nature21065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Massalha H, Toth B, Egozi A, Massasa EE, Medgalia C, David E, Giladi A, Moor AE, et al. 2018. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol 36: 962–970. 10.1038/nbt.4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yuan C, Chen L, Lei M, Zellmer L, Huang H, Liao DJ. 2018. Transcriptional-readthrough RNAs reflect the phenomenon of “a gene contains gene(s)” or “gene(s) within a gene” in the human genome, and thus are not chimeric RNAs. Genes (Basel) 9: 40. 10.3390/genes9010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Yang D, Fan X, Zhang M, Li Y, Gu X, Yang M. 2020. The roles and mechanisms of lncRNAs in liver fibrosis. Int J Mol Sci 21: 1482. 10.3390/ijms21041482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LL, Zhang Y, Zhang JX, He LJ, Lai YR, Liao YJ, Tian XP, Deng HX, Liang YJ, Kung HF, et al. 2015. Overexpression of NKX6.1 is closely associated with progressive features and predicts unfavorable prognosis in human primary hepatocellular carcinoma. Tumour Biol 36: 4405–4415. 10.1007/s13277-015-3080-4 [DOI] [PubMed] [Google Scholar]
- Huang C, Liang Y, Zeng X, Yang X, Xu D, Gou X, Sathiaseelan R, Senavirathna LK, Wang P, Liu L. 2020. Long noncoding RNA FENDRR exhibits antifibrotic activity in pulmonary fibrosis. Am J Respir Cell Mol Biol 62: 440–453. 10.1165/rcmb.2018-0293OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SF, Peng XF, Jiang L, Hu CY, Ye WC. 2021. LncRNAs as therapeutic targets and potential biomarkers for lipid-related diseases. Front Pharmacol 12: 729745. 10.3389/fphar.2021.729745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono G, Massoni-Badosa R, Heyn H. 2019. Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biol 20: 110. 10.1186/s13059-019-1713-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Wakahara T, Wang YQ, Kadoya H, Kawada N, Kaneda K. 1999. In vitro migratory potential of rat quiescent hepatic stellate cells and its augmentation by cell activation. Hepatology 29: 1760–1767. 10.1002/hep.510290640 [DOI] [PubMed] [Google Scholar]
- Ipsen DH, Lykkesfeldt J, Tveden-Nyborg P. 2020. Animal models of fibrosis in nonalcoholic steatohepatitis: Do they reflect human disease? Adv Nutr 11: 1696–1711. 10.1093/advances/nmaa081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne CN, Kubes P. 2013. Immune surveillance by the liver. Nat Immunol 14: 996–1006. 10.1038/ni.2691 [DOI] [PubMed] [Google Scholar]
- Jin J, Wahlang B, Shi H, Hardesty JE, Falkner KC, Head KZ, Srivastava S, Merchant ML, Rai SN, Cave MC, et al. 2020. Dioxin-like and non-dioxin-like PCBs differentially regulate the hepatic proteome and modify diet-induced nonalcoholic fatty liver disease severity. Med Chem Res 29: 1247–1263. 10.1007/s00044-020-02581-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. 2020. Single-cell transcriptome atlas of murine endothelial cells. Cell 180: 764–779.e720. 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- Karri K, Waxman DJ. 2020. Widespread dysregulation of long noncoding genes associated with fatty acid metabolism, cell division, and immune response gene networks in xenobiotic-exposed rat liver. Toxicol Sci 174: 291–310. 10.1093/toxsci/kfaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazankov K, Jorgensen SMD, Thomsen KL, Moller HJ, Vilstrup H, George J, Schuppan D, Gronbaek H. 2019. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol 16: 145–159. 10.1038/s41575-018-0082-x [DOI] [PubMed] [Google Scholar]
- Kendall TJ, Duff CM, Boulter L, Wilson DH, Freyer E, Aitken S, Forbes SJ, Iredale JP, Hastie ND. 2019. Embryonic mesothelial-derived hepatic lineage of quiescent and heterogenous scar-orchestrating cells defined but suppressed by WT1. Nat Commun 10: 4688. 10.1038/s41467-019-12701-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzmann T. 2017. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol 11: 622–630. 10.1016/j.redox.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzmann T. 2019. Liver zonation in health and disease: hypoxia and hypoxia-inducible transcription factors as concert masters. Int J Mol Sci 20: 2347. 10.3390/ijms20092347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kim HG, Son CG. 2018. Tissue-specific profiling of oxidative stress-associated transcriptome in a healthy mouse model. Int J Mol Sci 19: 3174. 10.3390/ijms19103174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Brenner D. 2021. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol 18: 151–166. 10.1038/s41575-020-00372-7 [DOI] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16: 1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, et al. 2016. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537: 239–243. 10.1038/nature19346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenkel O, Hundertmark J, Ritz TP, Weiskirchen R, Tacke F. 2019. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells 8: 503. 10.3390/cells8050503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CC, Hanzelmann S, Senturk Cetin N, Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt I, et al. 2019. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res 47: e32. 10.1093/nar/gkz037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau-Corona D, Ma H, Vergato C, Sarmento-Cabral A, Del Rio-Moreno M, Kineman RD, Waxman DJ. 2022. Constitutively active STAT5b feminizes mouse liver gene expression. Endocrinology 163: bqac046. 10.1210/endocr/bqac046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2021. Angiogenesis in the progression from liver fibrosis to cirrhosis and hepatocelluar carcinoma. Expert Rev Gastroenterol Hepatol 15: 217–233. 10.1080/17474124.2021.1842732 [DOI] [PubMed] [Google Scholar]
- Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H, Chen Y, Gucek M, Zhu J, Cao H. 2015. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 21: 455–467. 10.1016/j.cmet.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MA, Amaral PP, Cheung P, Bergmann JH, Kinoshita M, Kalkan T, Ralser M, Robson S, von Meyenn F, Paramor M, et al. 2017a. A lncRNA fine tunes the dynamics of a cell state transition involving Lin28, let-7 and de novo DNA methylation. Elife 6: e23468. 10.7554/eLife.23468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou J, Huang K. 2017b. Inhibition of the lncRNA Mirt1 attenuates acute myocardial infarction by suppressing NF-κB activation. Cell Physiol Biochem 42: 1153–1164. 10.1159/000478870 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Pei W, Zhang M, Yang H, Zhong M, Kong X, Xu Y, Zhu X, Chen T, et al. 2020. LncRNA Dnmt3aos regulates Dnmt3a expression leading to aberrant DNA methylation in macrophage polarization. FASEB J 34: 5077–5091. 10.1096/fj.201902379R [DOI] [PubMed] [Google Scholar]
- Liu XF, Thin KZ, Ming XL, Shuo L, Ping L, Man Z, Li ND, Tu JC. 2018. Small nucleolar RNA host gene 18 acts as a tumor suppressor and a diagnostic indicator in hepatocellular carcinoma. Technol Cancer Res Treat 17: 1533033818794494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wang K, Li K, Xu R, Chang X, Zhu Y, Sun P, Han X. 2019. Ets-1 deficiency alleviates nonalcoholic steatohepatitis via weakening TGF-β1 signaling-mediated hepatocyte apoptosis. Cell Death Dis 10: 458. 10.1038/s41419-019-1672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato NJ, Melia T, Rampersaud A, Waxman DJ. 2017. Sex-differential responses of tumor promotion-associated genes and dysregulation of novel long noncoding RNAs in constitutive androstane receptor-activated mouse liver. Toxicol Sci 159: 25–41. 10.1093/toxsci/kfx114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loft A, Alfaro AJ, Schmidt SF, Pedersen FB, Terkelsen MK, Puglia M, Chow KK, Feuchtinger A, Troullinaki M, Maida A, et al. 2021. Liver-fibrosis-activated transcriptional networks govern hepatocyte reprogramming and intra-hepatic communication. Cell Metab 33: 1685–1700.e1689. 10.1016/j.cmet.2021.06.005 [DOI] [PubMed] [Google Scholar]
- Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al. 2017. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med 23: 1331–1341. 10.1038/nm.4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ge G, Ji F, Wang J. 2022. LncRNA MORT overexpression inhibits cancer cell migration and invasion in hepatocellular carcinoma by downregulating NOTCH1. Cancer Biother Radiopharm 37: 537–543. 10.1089/cbr.2020.4020 [DOI] [PubMed] [Google Scholar]
- Luo Y, He Y, Ye X, Song J, Wang Q, Li Y, Xie X. 2019. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol Oncol Res 25: 1567–1577. 10.1007/s12253-018-00570-4 [DOI] [PubMed] [Google Scholar]
- Luo J, Qu L, Gao F, Lin J, Liu J, Lin A. 2021. LncRNAs: architectural scaffolds or more potential roles in phase separation. Front Genet 12: 626234. 10.3389/fgene.2021.626234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D, Wang Y, Zhang Y, Cui P, Xu Y. 2017. Downregulated long non-coding RNA DREH promotes cell proliferation in hepatitis B virus-associated hepatocellular carcinoma. Oncol Lett 14: 2025–2032. 10.3892/ol.2017.6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Zhang X, Yang L, Zhou J, Liu W, Gao X, Yu F, Zheng W, Ding S, Gao P, et al. 2018. Role of functional IFNL4, IFNLR1, IFNA, IFNAR2 polymorphisms in hepatitis B virus-related liver disease in Han Chinese population. J Viral Hepat 25: 306–313. 10.1111/jvh.12817 [DOI] [PubMed] [Google Scholar]
- Marcher AB, Bendixen SM, Terkelsen MK, Hohmann SS, Hansen MH, Larsen BD, Mandrup S, Dimke H, Detlefsen S, Ravnskjaer K. 2019. Transcriptional regulation of hepatic stellate cell activation in NASH. Sci Rep 9: 2324. 10.1038/s41598-019-39112-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, Huang L, Moreno I, Navarro A, Monzo M, et al. 2013. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 14: R104. 10.1186/gb-2013-14-9-r104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart J, Begriche K, Corlu A, Fromenty B. 2022. Xenobiotic-induced aggravation of metabolic-associated fatty liver disease. Int J Mol Sci 23: 1062. 10.3390/ijms23031062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T, Waxman DJ. 2019. Sex-biased lncRNAs inversely correlate with sex-opposite gene coexpression networks in diversity outbred mouse liver. Endocrinology 160: 989–1007. 10.1210/en.2018-00949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T, Hao P, Yilmaz F, Waxman DJ. 2016. Hepatic long intergenic noncoding RNAs: high promoter conservation and dynamic, sex-dependent transcriptional regulation by growth hormone. Mol Cell Biol 36: 50–69. 10.1128/MCB.00861-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Shiekhattar R, Shilatifard A, Lauberth SM. 2022. It's a DoG-eat-DoG world-altered transcriptional mechanisms drive downstream-of-gene (DoG) transcript production. Mol Cell 82: 1981–1991. 10.1016/j.molcel.2022.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fan H, Wang P, Li Y, Domenico K, Li Q, Wang X, Essandoh K, Chen J, Peng T, et al. 2021. Sectm1a facilitates protection against inflammation-induced organ damage through promoting TRM self-renewal. Mol Ther 29: 1294–1311. 10.1016/j.ymthe.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl L, Genové G, Leptidis S, Liu J, He L, Mocci G, Sun Y, Gustafsson S, Buyandelger B, Chivukula IV. 2020. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 11: 1–18. 10.1038/s41467-019-13993-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MC, Hvidbjerg Gantzel R, Claria J, Trebicka J, Moller HJ, Gronbaek H. 2020. Macrophage activation markers, CD163 and CD206, in acute-on-chronic liver failure. Cells 9: 1175. 10.3390/cells9051175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknafs YS, Pandian B, Iyer HK, Chinnaiyan AM, Iyer MK. 2017. TACO produces robust multisample transcriptome assemblies from RNA-seq. Nat Methods 14: 68–70. 10.1038/nmeth.4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates JR, McKell MC, Moreno-Fernandez ME, Damen M, Deepe GS Jr, Qualls JE, Divanovic S. 2019. Macrophage function in the pathogenesis of non-alcoholic fatty liver disease: the mac attack. Front Immunol 10: 2893. 10.3389/fimmu.2019.02893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, French SW. 2009. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol 87: 12–19. 10.1016/j.yexmp.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, Luan J, Davies JO, Hughes JR, Hardison RC, et al. 2016. Unlinking an lncRNA from its associated cis element. Mol Cell 62: 104–110. 10.1016/j.molcel.2016.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Ranjbarvaziri S, Lay FD, Zhao P, Miller MJ, Dhaliwal JS, Huertas-Vazquez A, Wu X, Qiao R, Soffer JM, et al. 2018. Genetic regulation of fibroblast activation and proliferation in cardiac fibrosis. Circulation 138: 1224–1235. 10.1161/CIRCULATIONAHA.118.035420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicano AJ, Spahn K, Zhou P, Goldberg ID, Narayan P. 2021. Collagen characterization in a model of nonalcoholic steatohepatitis with fibrosis; a call for development of targeted therapeutics. Molecules 26: 3316. 10.3390/molecules26113316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradas-Juni M, Hansmeier NR, Link JC, Schmidt E, Larsen BD, Klemm P, Meola N, Topel H, Loureiro R, Dhaouadi I, et al. 2020. A MAFG-lncRNA axis links systemic nutrient abundance to hepatic glucose metabolism. Nat Commun 11: 644. 10.1038/s41467-020-14323-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Hu M, Xiang Y, Wang D, Xu Y, Hou Y, Zhou H, Luan Y, Wang Z, Zhang W, et al. 2020. LncRNAs are regulated by chromatin states and affect the skeletal muscle cell differentiation. Cell Prolif 53: e12879. 10.1111/cpr.12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R, Huang W, Huang Y, Zhang Z, Su Y, Chen S, Wang H. 2022. lncRNA MEG3 modulates hepatic stellate cell activation by sponging miR‐145 to regulate PPARγ. Mol Med Rep 25: 3. 10.3892/mmr.2021.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14: 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajak S, Raza S, Tewari A, Sinha RA. 2022. Environmental toxicants and NAFLD: a neglected yet significant relationship. Dig Dis Sci 67: 3497–3507. 10.1007/s10620-021-07203-y [DOI] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, et al. 2019. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575: 512–518. 10.1038/s41586-019-1631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Matchett KP, Dobie R, Wilson-Kanamori JR, Henderson NC. 2020. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat Rev Gastroenterol Hepatol 17: 457–472. 10.1038/s41575-020-0304-x [DOI] [PubMed] [Google Scholar]
- Raza S, Rajak S, Upadhyay A, Tewari A, Anthony Sinha R. 2021. Current treatment paradigms and emerging therapies for NAFLD/NASH. Front Biosci (Landmark Ed) 26: 206–237. 10.2741/4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehlen N, Crouchet E, Baumert TF. 2020. Liver fibrosis: mechanistic concepts and therapeutic perspectives. Cells 9: 875. 10.3390/cells9040875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten D, Trebicka J, Liedtke C, Weiskirchen R. 2015. The carbon tetrachloride model in mice. Lab Anim 49: 4–11. 10.1177/0023677215571192 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheka AC, Adeyi O, Thompson J, Hameed B, Crawford PA, Ikramuddin S. 2020. Nonalcoholic steatohepatitis: a review. JAMA 323: 1175–1183. 10.1001/jama.2020.2298 [DOI] [PubMed] [Google Scholar]
- Shen J, Shrestha S, Yen YH, Scott MA, Soo C, Ting K, Peault B, Dry SM, James AW. 2016. The pericyte antigen RGS5 in perivascular soft tissue tumors. Hum Pathol 47: 121–131. 10.1016/j.humpath.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Guo H, Xu J, Wang J. 2019. Inhibition of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis by inhibiting the MAPK signaling pathway in rats with nonalcoholic fatty liver disease. J Cell Physiol 234: 18169–18179. 10.1002/jcp.28450 [DOI] [PubMed] [Google Scholar]
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. 2022. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50: W216–W221. 10.1093/nar/gkac194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, Duncan SA. 2010. Organogenesis and development of the liver. Dev Cell 18: 175–189. 10.1016/j.devcel.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Smith KN, Starmer J, Magnuson T. 2018. Interactome determination of a long noncoding RNA implicated in embryonic stem cell self-renewal. Sci Rep 8: 17568. 10.1038/s41598-018-34864-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soibam B, Zhamangaraeva A. 2021. LncRNA:DNA triplex-forming sites are positioned at specific areas of genome organization and are predictors for topologically associated domains. BMC Genomics 22: 397. 10.1186/s12864-021-07727-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M. 2021. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22: 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e1821. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G, Kuchinsky A, Morris JH, States DJ, Meng F. 2010. GLay: community structure analysis of biological networks. Bioinformatics 26: 3135–3137. 10.1093/bioinformatics/btq596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Kim SY, Adewale F, Zhou Y, Aldler C, Ni M, Wei Y, Burczynski ME, Atwal GS, Sleeman MW, et al. 2021. Single-cell RNA transcriptome landscape of hepatocytes and non-parenchymal cells in healthy and NAFLD mouse liver. iScience 24: 103233. 10.1016/j.isci.2021.103233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562: 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurl I, Hilgendorf I, Nairz M, Tymoszuk P, Haschka D, Asshoff M, He S, Gerhardt LM, Holderried TA, Seifert M, et al. 2016. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med 22: 945–951. 10.1038/nm.4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS, Hosny S. 2018. Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother 102: 653–669. 10.1016/j.biopha.2018.03.105 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single-cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Qiu X. 2017. Monocle: cell counting, differential expression, and trajectory analysis for single-cell RNA-Seq experiments. Bioconductor. https://www.bioconductor.org/packages/release/bioc/html/monocle.html
- Trojanowska M. 2000. Ets factors and regulation of the extracellular matrix. Oncogene 19: 6464–6471. 10.1038/sj.onc.1204043 [DOI] [PubMed] [Google Scholar]
- Unfried JP, Sangro P, Prats-Mari L, Sangro B, Fortes P. 2021. The landscape of lncRNAs in hepatocellular carcinoma: a translational perspective. Cancers (Basel) 13: 2651. 10.3390/cancers13112651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K, Roux de Bezieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L. 2020. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 11: 1201. 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heide D, Weiskirchen R, Bansal R. 2019. Therapeutic targeting of hepatic macrophages for the treatment of liver diseases. Front Immunol 10: 2852. 10.3389/fimmu.2019.02852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L, et al. 2018. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554: 475–480. 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- Wang D, Sallam T. 2021. Where in the (lncRNA) world is CARMN?: safeguarding against vascular dysfunction. Circ Res 128: 1276–1278. 10.1161/CIRCRESAHA.121.319150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Xian J, Zang J, Xiao L, Li Y, Sha M, Shen M. 2019. Long non-coding RNA FENDRR inhibits proliferation and invasion of hepatocellular carcinoma by down-regulating glypican-3 expression. Biochem Biophys Res Commun 509: 143–147. 10.1016/j.bbrc.2018.12.091 [DOI] [PubMed] [Google Scholar]
- Wang W, Wang Q, Huang DB, Sun QK, Wu SS, Zhao YJ, Jia W, Hu DS, He YF. 2021. Tumor-associated mesenchymal stem cells promote hepatocellular carcinoma metastasis via a DNM3OS/KDM6B/TIAM1 axis. Cancer Lett 503: 19–31. 10.1016/j.canlet.2021.01.011 [DOI] [PubMed] [Google Scholar]
- Wells RG. 2014. Portal fibroblasts in biliary fibrosis. Curr Pathobiol Rep 2: 185–190. 10.1007/s40139-014-0054-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Lambrecht J, Ju C, Tacke F. 2021. Hepatic macrophages in liver homeostasis and diseases-diversity, plasticity and therapeutic opportunities. Cell Mol Immunol 18: 45–56. 10.1038/s41423-020-00558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HC, Ma J, Weiss D, Lassegue B, Sutliff RL, San Martin A. 2019. The cofilin phosphatase slingshot homolog 1 restrains angiotensin II-induced vascular hypertrophy and fibrosis in vivo. Lab Invest 99: 399–410. 10.1038/s41374-018-0116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YY, Wu S, Li XF, Luo S, Wang A, Yin SQ, Huang C, Li J. 2021. LncRNA MEG3 reverses CCl4-induced liver fibrosis by targeting NLRC5. Eur J Pharmacol 911: 174462. 10.1016/j.ejphar.2021.174462 [DOI] [PubMed] [Google Scholar]
- Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, et al. 2018. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 22: 3625–3640. 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, Ji Y, Li C, Guo L, et al. 2019. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol Cell 75: 644–660.e645. 10.1016/j.molcel.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhu Y, Hu S, Xu Y, Stroup D, Pan X, Bawa FC, Chen S, Gopoju R, Yin L, et al. 2021. Hepatocyte nuclear factor 4α prevents the steatosis-to-NASH progression by regulating p53 and bile acid signaling (in mice). Hepatology 73: 2251–2265. 10.1002/hep.31604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, Boyer LA. 2016. A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell 64: 37–50. 10.1016/j.molcel.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Cai JB, Peng R, Wei CY, Lu JC, Gao C, Shen ZZ, Zhang PF, Huang XY, Ke AW, et al. 2019. The long noncoding RNA NORAD enhances the TGF-β pathway to promote hepatocellular carcinoma progression by targeting miR-202-5p. J Cell Physiol 234: 12051–12060. 10.1002/jcp.27869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, He H, Wang T, Su N, Zhang F, Jiang K, Zhu J, Zhang C, Niu K, Wang L, et al. 2021. Single-cell transcriptomic analysis reveals a hepatic stellate cell-activation roadmap and myofibroblast origin during liver fibrosis in mice. Hepatology 74: 2774–2790. 10.1002/hep.31987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-Y, Kim J-Y, Kim H-J, Ka N-L, Lee S-H, Lee M-O. 2023. LncRNA Ctcflos modulates glucocorticoid receptor-mediated induction of hepatic phosphoenolpyruvate carboxykinase in mice. Life Sci 312: 121254. 10.1016/j.lfs.2022.121254 [DOI] [PubMed] [Google Scholar]
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. 2018. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 15: 11–20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- Yu D, Huber W, Vitek O. 2013. Shrinkage estimation of dispersion in negative binomial models for RNA-seq experiments with small sample size. Bioinformatics 29: 1275–1282. 10.1093/bioinformatics/btt143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, Guo C, Liu Z, Fan X. 2015. Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem 290: 28286–28298. 10.1074/jbc.M115.683813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Geng W, Dong P, Huang Z, Zheng J. 2018. LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212. Cell Death Dis 9: 1014. 10.1038/s41419-018-1068-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Dong B, Dong P, He Y, Zheng J, Xu P. 2020. Hypoxia induces the activation of hepatic stellate cells through the PVT1-miR-152-ATG14 signaling pathway. Mol Cell Biochem 465: 115–123. 10.1007/s11010-019-03672-y [DOI] [PubMed] [Google Scholar]
- Yu AT, Berasain C, Bhatia S, Rivera K, Liu B, Rigo F, Pappin DJ, Spector DL. 2021. PHAROH lncRNA regulates Myc translation in hepatocellular carcinoma via sequestering TIAR. Elife 10: e68263. 10.7554/eLife.68263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Jiang S, Kong X, El-Ali N, Ball AR Jr, Ma CI, Hashimoto N, Yokomori K, Mortazavi A. 2016. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res 44: e158. 10.1093/nar/gkw739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li Y, Zhang XY, Liu L, Tong HZ, Han TL, Li WD, Jin XL, Yin NB, Song T, et al. 2017a. Therapeutic potential of human minor salivary gland epithelial progenitor cells in liver regeneration. Sci Rep 7: 12707. 10.1038/s41598-017-11880-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Han X, Zhang Z, Zheng L, Hu Z, Yao Q, Cui H, Shu G, Si M, Li C, et al. 2017b. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch pathways. Nat Commun 8: 144. 10.1038/s41467-017-00204-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. 2019. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci 20: 5573. 10.3390/ijms20225573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Conway SJ, Liu Y, Snider P, Chen H, Gao H, Liu Y, Isidan K, Lopez KJ, Campana G, et al. 2021. Heterogeneity of hepatic stellate cells in fibrogenesis of the liver: insights from single-cell transcriptomic analysis in liver injury. Cells 10: 2129. 10.3390/cells10082129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, et al. 2016. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44: D203–D208. 10.1093/nar/gkv1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yu F, Dong P, Wu L, Zhang Y, Hu Y, Zheng L. 2016. Long non-coding RNA PVT1 activates hepatic stellate cells through competitively binding microRNA-152. Oncotarget 7: 62886–62897. 10.18632/oncotarget.11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZG, Chen JB, Zhang RX, Ye L, Wang JC, Pan YX, Wang XH, Li WX, Zhang YJ, Xu L, et al. 2020. Tescalcin is an unfavorable prognosis factor that regulates cell proliferation and survival in hepatocellular carcinoma patients. Cancer Commun (Lond) 40: 355–369. 10.1002/cac2.12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.