Abstract
There are very few small-molecule antivirals for SARS-CoV-2 that are either currently approved (or emergency authorized) in the US or globally, including remdesivir, molnupiravir, and paxlovid. The increasing number of SARS-CoV-2 variants that have appeared since the outbreak began over three years ago raises the need for continual development of updated vaccines and orally available antivirals in order to fully protect or treat the population. The viral main protease (Mpro) and the papain-like protease (PLpro) are key for viral replication; therefore, they represent valuable targets for antiviral therapy. We herein describe an in vitro screen performed using the 2560 compounds from the Microsource Spectrum library against Mpro and PLpro in an attempt to identify additional small-molecule hits that could be repurposed for SARS-CoV-2. We subsequently identified 2 hits for Mpro and 8 hits for PLpro. One of these hits was the quaternary ammonium compound cetylpyridinium chloride with dual activity (IC50 = 2.72 ± 0.09 μM for PLpro and IC50 = 7.25 ± 0.15 μM for Mpro). A second inhibitor of PLpro was the selective estrogen receptor modulator raloxifene (IC50 = 3.28 ± 0.29 μM for PLpro and IC50 = 42.8 ± 6.7 μM for Mpro). We additionally tested several kinase inhibitors and identified olmutinib (IC50 = 0.54 ± 0.04 μM), bosutinib (IC50 = 4.23 ± 0.28 μM), crizotinib (IC50 = 3.81 ± 0.04 μM), and dacominitinib (IC50 = IC50 3.33 ± 0.06 μM) as PLpro inhibitors for the first time. In some cases, these molecules have also been tested by others for antiviral activity for this virus, or we have used Calu-3 cells infected with SARS-CoV-2. The results suggest that approved drugs can be identified with promising activity against these proteases, and in several cases we or others have validated their antiviral activity. The additional identification of known kinase inhibitors as molecules targeting PLpro may provide new repurposing opportunities or starting points for chemical optimization.
Introduction
It has been over three years since the initial outbreak of SARS-CoV-2 in Wuhan, China, in November 2019 which caused the COVID-19 disease,1,2 and at the time of writing this paper there have been over 760 million cases and 6.8 million deaths3 caused by this enveloped, positive-sense, single-stranded RNA betacoronavirus. In the USA, currently, only remdesivir is FDA-approved as an antiviral for COVID-19, while molnupiravir and paxlovid have emergency use authorizations for this virus. The lower efficacy of molnupiravir and concerns that it may induce mutations in patient DNA4 have somewhat dampened enthusiasm for it. The continued emergence of new SARS-CoV-2 variants points to the need for the development of additional vaccines and oral antivirals if we are to overcome this virus globally.
Part of the challenge in SARS-CoV-2 and antiviral drug discovery, in general, is developing molecules against targets that may be less susceptible to resistance than the spike protein. There has been a long history of the development of protease inhibitors for human immunodeficiency virus (HIV) and hepatitis C virus (HCV).5−7 For SARS-CoV-2, there are two proteases: the main protease (Mpro also known as 3CLpro) and the papain-like protease (PLpro).8 The catalytic activity of these enzymes is key for viral replication, making their inhibition a compelling strategy for antiviral therapy for SARS-CoV-2.8 For example, Mpro is inhibited by many known cysteine protease inhibitors, primarily via covalent modification of the active site cysteine and represents an opportunity for repurposing.9−12
Paxlovid consists of a combination of nirmatrelvir (PF-07321332), which is an inhibitor of the main protease (Mpro), and ritonavir, which is commonly used to improve the half-life. Mutations have been observed in Mpro, yet these have proved susceptible to nirmatrelvir. This drug was developed from an earlier covalent active-site-directed inhibitor of the SARS Mpro inhibitor PF-00835231, which contains an indole and was proposed as a P-glycoprotein (P-gp) substrate in Vero cells and hence had low activity and poor bioavailability.13 It was recently determined that, in A549 + ACE2 cells, PF-00835231 is efficacious and suggests that P-gp may not be an issue after all.14
PLpro cleaves peptide bonds to produce non-structural proteins. PLpro also has an effect by countering the effect of viral infection on the immune response via deubiquitinating and deISGylating activities and releasing ubiquitin and ISG15, which suppresses the immune system.15 Recent detailed reviews of drug discovery for SARS-CoV-2 PLpro inhibitors have highlighted over 50 non-covalent and covalent molecules.15 What is apparent from this body of research for PLpro is the lack of an approved drug that displays both enzyme and antiviral activity. One potential issue with this target is the structural differences between the target in MERS-CoV and SARS-CoV that make a broad spectrum antiviral against this target difficult.16 Target selectivity is also challenging because of other deubiquitinating enzymes and the need for counter screens against these cysteine proteases. Like any target, PLpro is also susceptible to inhibitors that may be promiscuous, which might limit their value. While there have been some repurposing efforts for PLpro, most of the molecules have been natural products or not evaluated in counter screens or orthogonal assays.17 PLpro therefore represents a relatively untapped target for treating COVID. We now describe the hits derived from our initial repurposing screens with PLpro and Mpro and important observations relevant to ongoing studies with these molecules.
Results
PLpro and Mpro
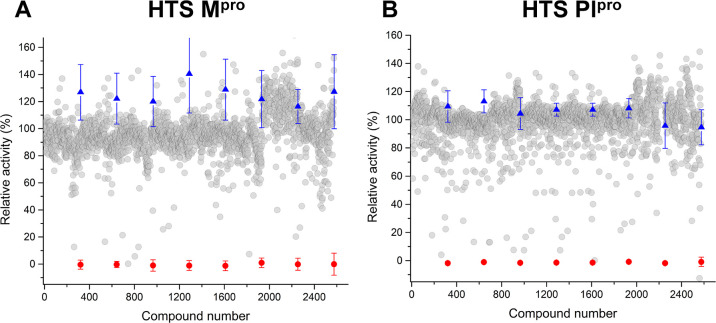
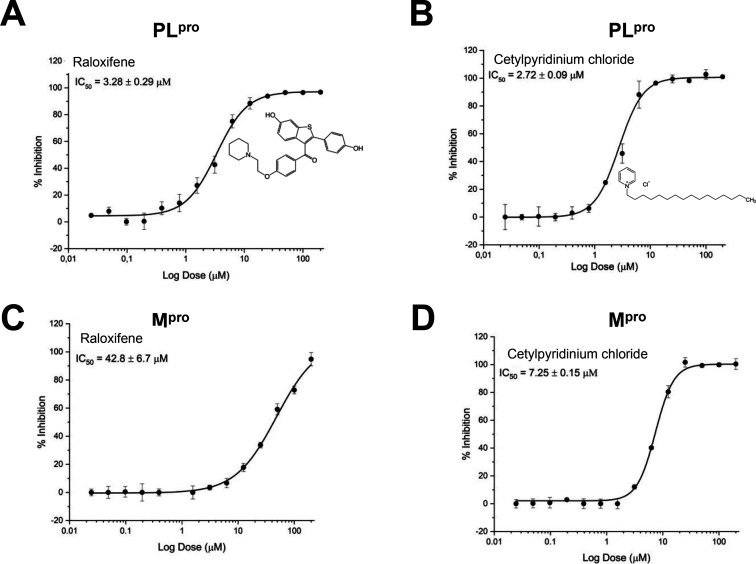
We developed high-throughput assays for PLpro and Mpro and used these to screen the Microsource Spectrum collection composed of 2560 compounds. The assays are suitable for screening (average Z = 0.76 ± 0.21 for Mpro and average Z = 0.67 ± 0.13 for PLpro) when compounds were screened at 10 μM, and compounds able to inhibit at least 80% of enzyme activity were selected (Figure 1). From the initial screening, we identified eight compounds with activity against PLpro and two compounds with activity against Mpro (Table 1). Surprisingly, we identified two compounds that inhibited both proteases, and we followed them up by generating dose–response curves. Raloxifene inhibits PLpro with an IC50 = 3.28 ± 0.29 μM and Mpro IC50 = 42.8 ± 6.7 μM, and cetylpyridinium chloride inhibits PLpro with an IC50 = 2.72 ± 0.09 μM and Mpro IC50 = 7.25 ± 0.15 μM (Figure 2).
Figure 1.
Scatter plots showing the data from the high-throughput screening (HTS) against Mpro (A) and PLpro (B). Single-shot inhibition data are shown for compounds from the Microsource Spectrum collection containing 2560 compounds that were screened at 10 μM. Average Z = 0.76 ± 0.21 for Mpro and average Z = 0.67 ± 0.13 for PLpro. Compounds’ individual results are represented as gray spheres. Positive control averages are indicated as red spheres. Negative controls are indicated as blue triangles. Error bars are ± standard deviation from relative controls.
Table 1. Compounds Identified against Plpro and Mpro from the Microsource Spectrum Collection Library Screena.
| compound | PLpro IC50 (μM) | Mpro IC50 (μM) |
|---|---|---|
| raloxifene | 3.28 ± 0.29 | 42.8 ± 6.7 |
| cetylpyridinium chloride | 2.72 ± 0.09 | 7.25 ± 0.15 |
| cefonicid sodium | 2.17 ± 1.2 | |
| citicoline | 1.35 ± 0.27 | |
| colistin sulfate | 2.94 ± 0.54 | |
| β-lapachone | 5.30 ± 0.29 | |
| tanshinone iia sulfonate sodium | 2.76 ± 0.28 | |
| lobaric acid | 10 ± 1.11 |
It should be noted that compounds such as β-lapachone and tanshinone may be potential false positives involved in redox cycling.18
Figure 2.
Dose–response curves for raloxifene and cetylpyridinium chloride tested against PLpro (A,B) and Mpro (C,D). Both compounds were identified by screening the Microsource Spectrum collection. All experiments were performed in triplicate, and all data are expressed as the mean ± standard deviation.
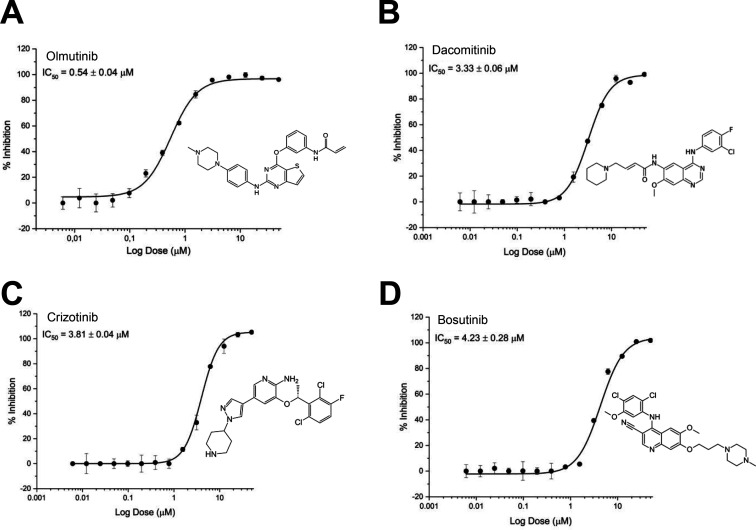
We previously identified vandetanib, a vascular endothelial growth factor (VGFR) inhibitor, with activity against SARS-CoV-2 in A549 – ACE2 cells, which blocked the cytokine storm in infected mice.19 We then evaluated other kinase inhibitors against PLpro and Mpro and showed that olmutinib inhibits PLpro with an IC50 = 0.54 ± 0.04 μM (Figure 3A), dacomitinib with an IC50 = 3.33 ± 0.06 μM (Figure 3B), crizotinib with an IC50 = 3.81 ± 0.04 μM (Figure 3C), and bosutinib with an IC50 = 4.23 ± 0.28 μM (Figure 3D) were similarly active. None of the kinase inhibitors demonstrated inhibition with Mpro.
Figure 3.
Dose–response curves of kinase inhibitors tested against PLpro. (A) Olmutinib, IC50 0.54 ± 0.04 μM, (B) Dacomitinib, IC50 3.30 ± 0.06 μM, (C) Crizotinib 3.80 ± 0.04 μM, and (D) Bosutinib IC50 4.20 ± 0.28 μM. All experiments were performed in triplicate, and all data are expressed as the mean ± standard deviation.
Cell Assays
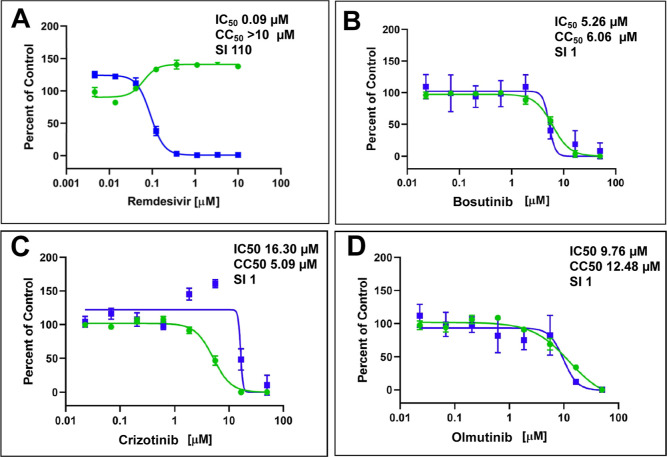
Raloxifene20−22 and cetylpyridium chloride23,24 were previously described to have activity against SARS-CoV-2 in various cell types. We tested activity of 3 of the 4 kinase inhibitors due to limited resources such that bosutinib, crizotinib, and olmutinib were tested in Calu-3 cells available via the NIAID. Remdesivir was used as a general control for in vitro inhibition. Bosutinib showed an IC50 = 5.26 μM and CC50 = 6.06 μM, crizotinib showed an IC50 = 16.30 μM and CC50 = 5.09 μM, and olmutinib showed an IC50 = 9.76 μM and CC50 = 12.48 μM (Figure 4). All the kinase inhibitors tested showed substantial cell toxicity in Calu-3 cells.
Figure 4.
Kinase inhibitors tested against SARS-CoV-2 in Calu-3 cells. (A) Control: remdesivir, (B) bosutinib, (C) crizotinib, and (D) olmutinib. Sample well data was normalized to DMSO control wells and plotted versus drug concentration to determine the IC50 (infection: blue) and CC50 (toxicity: green). The x axis shows concentrations in a logarithmic scale.
Discussion
Since the outbreak of SARS-CoV-2, there has been an enormous global effort to identify and develop potential antivirals using a vast number of techniques. Much of the early work included extensive use of computational approaches25,26 as well as high-throughput screens, which has had admittedly mixed success, as briefly described here. Extensive effort has been applied toward developing Mpro inhibitors using a range of experimental approaches. For example, a ligand-based virtual screen of 790,000 compounds followed by structure-based screens for Mpro led to the testing of 30 compounds, of which 8 had high 10’s μM IC50 and antiviral activity.27 A docking and molecular dynamics approach was used to develop a covalent inhibitor of Mpro from a fragment hit, although the antiviral activity or selectivity was not assessed.28 An in silico approach to screening over a billion compounds led to over 400 being synthesized, of which only 5 were active and ultimately resulted in a non-covalent inhibitor with a Mpro IC50 = 1 μM. Virtual pharmacophore and docking-based screening of over 8000 drugs against Mpro identified the kinase inhibitor nilotinib as a low μM antiviral in Vero cells but did not test inhibition of Mpro or PLpro.10 An ensemble docking approach was used with a Mpro to screen around 2000 natural products and identify 5 with high μM Ki values. Antiviral activity was not determined for these hits either.29 A virtual repurposing screen of 8700 compounds against Mpro led to the identification of the preclinical molecule MG-132 with sub μM antiviral activity in Vero cells.11 Docking FDA drugs in the Mpro structure identified dipyridamole (IC50 = 0.53 μM), which was active in Vero cells (EC50 ∼ 0.1 μM).30 An early screen of Mpro used 10,000 compounds, finding 7 hits including ebselen (IC50 = 0.67 μM) that was active in Vero cells (EC50 = 4.67 μM).31 A screen of protease inhibitors against Mpro identified boceprevir (IC50 = 4.13 μM), which was similarly active in Vero cells (EC50 = 1.31 μM).32 A high-throughput repurposing screen of over 6000 drugs against Mpro identified 50 hits and 8 with IC50 < 50 μM, and this data did not correlate with docking in the protein. Several of the best hits were hepatitis C inhibitors including boceprevir, but the antiviral activity was not determined.9 The COVID moonshot initiative, which is a consortium that uses open science and open data to rapidly develop patent-free antivirals, reported the discovery of novel chemical scaffolds for Mpro active in biochemical and live virus assays, which were synthesized with model-generated routes.33 Generative approaches have also been proposed for use with Mpro but to date this appears to have not been acted upon.34
These represent a small snapshot of the massive number of virtual or high-throughput screens undertaken to date. Others have used boceprevir for crystallography and structure-based design to identify further analogues with antiviral activity and in vivo efficacy in a mouse model of infection.35 The flavonoid natural product baicalein is a sub μM inhibitor of Mpro and has been crystallized with this target from SARS-CoV-2.36 Following crystallography of covalent inhibitors, low nM antiviral inhibitors selective (sub μM) for Mpro have been developed that reduced viral replication and viral load, increasing survival in SARS-CoV-2-infected mice.37 The protease inhibitor 13b is less potent versus SARS-CoV-2 Mpro (IC50 = 0.67 μM) but shows activity in Calu-3 cells (EC50 = 4–5 μM).38 Additional indole and indoline compounds were developed as Mpro inhibitors with antiviral activity against SARS-CoV-2.39 There has also been extensive development of peptidic Mpro nM inhibitors such as tripeptides,40 tripeptide mimics41 and boceprevir analogues with antiviral activity IC50 = 1 μM.42 The covalent inhibitor halicin was also shown to be a nM Mpro inhibitor but was not tested against other proteases or for its antiviral activity against SARS-CoV-2.43 An orally available low nM Mpro α-ketoamide containing inhibitor Y180 improved survival in a mouse model of SARS-CoV-2 infection.44 Several early in vitro hits for Mpro when tested in the mouse model ultimately had modest efficacy. For example, GC37614 is a potent inhibitor of SARS-CoV-2 Mpro (Ki = 12 nM)45 with sub μM antiviral activity in A549+ACE2 cells and in Vero cells45 but performed poorly in vivo.46
Compared to Mpro, there have been far fewer publications describing studies attempting to find PLpro inhibitors. For example, one study identified disulfiram and analogues47 as covalent inhibitors and another described the natural product celastrol,48 which binds similarly to PLpro, Mpro, and cathepsin L. Yet, another natural product, anarcardic acid, is a weak inhibitor of PLpro and Mpro.49 An initial hit for PLpro came from the naphthalene derivative GRL-0617, which had previously been identified for SARS-CoV and demonstrated activity against SARS-CoV-2.50 A study identified analogues of GRL-0617, but these did not have desirable antiviral activity versus SARS-CoV-2.51 Another study described an analogue with sub μM PLpro inhibition and antiviral activity.52 Several virtual repurposing screens have used docking against PLpro to identify hits such as the antimalarial mefloquine.53 A combination of pharmacophores and docking led to 4 low μM hits against PLpro although no antiviral activity was assessed.54 A MedChemExpress library high throughput screen of 9791 compounds against PLpro led to three hits including the clinical candidate FXR agonist tropifexor55 which was a low μM hit active in Calu-3 cells. A docking and structure-based design approach was used to identify indole dual inhibitors for PLpro and Mpro with antiviral activity.56 We have previously described the in vitro (A549-ACE2 IC50 = 0.23 μM) and in vivo efficacy of pyronaridine tetraphosphate against SARS-CoV-2,57 while more recently, it has been demonstrated to also inhibit PLpro (IC50 = 1.8 μM).58 We additionally described several pyronaridine analogues with similar activity as well as analogues that lacked activity altogether. Pyronaridine is an antimalarial drug that is approved in Europe and used as an antimalarial. It may hold promise as it has showed increased IFN-1β levels and decreased IL-6, CXCL1, and CCL4 while also decreasing viral load and improving lung histopathology in mice infected witih SARS-CoV-2.58
Herein, we have now described the screening of the Microsource Spectrum collection that led to the identification of cetylpyridinium chloride and raloxifene as PLpro and Mpro inhibitors. We had earlier used a text mining approach to identify molecules with antiviral effects against coronaviruses59 and this uncovered cetylpyridinium chloride, a quaternary ammonium compound which is widely used in mouthwashes, toothpastes, lozenges, throat sprays, breath sprays, and nasal sprays.59 The target for this molecule against SARS-CoV-2 was previously unknown. Others have described the SARS-CoV-2 virucidal activity of this molecule in Vero cells upon short exposure and that it interfered with the spike and ACE2 interaction.60 This antiviral efficacy seems also to be observed across different viral strains and in saliva, while the mechanism is occurring without disruption of the viral particles and that the denaturing effect of the spike protein may have a role.61 Cetylpyridinium chloride has also been tested in Caco-2 cells (IC50 = 0.62 μM).23 Several clinical trials have also indicated efficacy against SARS-CoV-2.62,63 A lipidomics study has compared host and viral cell composition showing that the cholesterol/phospholipid ratio is comparable to lysosomes, as well as evaluated cetylyridinium chloride containing mouthwashes to show a viral reduction in vitro. This was followed by a clinical study, which showed that mouthwash containing cetylpyridinium chloride reduced the viral load.24
In contrast, raloxifene is a selective estrogen receptor modulator approved in the US and Europe for osteoporosis in post-menopausal women and reduces the potential risk of breast cancer.64 A pseudovirus screen against MERS identified raloxifene, which also had activity against SARS-CoV-2 in Vero cells and in the hamster model reduced the virus in the lung.21 Raloxifene was tested in Vero (IC50 = 5.9 μM) and Calu-3 cells (IC50 = 9 μM and SI 2.7) and was similarly active across different variants.20 A screen of Sendai virus in human iPSCs also identified raloxifene, which was subsequently tested against SARS-CoV-2 in Vero cells (IC50 = 3.9 μM).22 Raloxifene has also completed a Phase 2 randomized, double blinded, placebo-controlled trial in patients with early mild to moderate COVID-19 and showed evidence of effect in the primary endpoint and shortened the time for viral shedding.65
There have been relatively few studies that have to date described molecules that inhibit both Mpro and PLpro; these include the covalent inhibitor disulfiram66,67 and non-covalent HCV protease inhibitors grazoprevir, simeprevir, and vaniprevir.68 A recent screen of over 1.8 million compounds found only six molecules that inhibited both Mpro and PLpro.69
FDA-approved small-molecule kinase inhibitors could potentially be repurposed as antivirals because many kinase host targets are necessary or required for the viral life cycle, replication, and infection of multiple virus types.70 We recently demonstrated that vandetanib blocks the cytokine storm in mice infected by SARS-CoV-2,19 which prompted us to evaluate other kinase inhibitors against PLpro and Mpro. Here, we have demonstrated for the first time that olmutinib, bosutinib, dacomitinib, and crizotinib are PLpro inhibitors, and this may contribute alongside any host effects the compounds may have. Olmutinib showed the lowest IC50 = 0.54 ± 0.04 μM against PLpro in this study, and it is also an investigational anti-cancer drug for non-small cell lung cancer (NSCLC), which inhibits the epidermal growth factor receptor (EGFR) by binding covalently to a cysteine residue near the kinase domain.71 Whether olmutinib binds covalently remains to be investigated. Dacomitinib is a selective and irreversible inhibitor of EGFR, approved for the treatment of NSCLC with EGFR gene mutation,72 which demonstrated potent antiviral activity against SARS-CoV-2 in Calu-3 cells with IC50 = 0.04 μM and CC50 = 9 μM.73 EGFR is an essential pathway for SARS-CoV-2 replication74 and many viruses such as hepatitis C, Epstein-Barr virus, and influenza, have been shown to use the EGFR as an entry receptor.75−77 Bosutinib is a BCR-ABL and proto-oncogene tyrosine-protein kinase Src inhibitor used for the treatment of chronic myelogenous leukemia78 and it was previously demonstrated to have anti-SARS-CoV-2 entry activity with an EC50 = 2.45 μM and an SI = 7.08 in Vero cells.79 Crizotinib is an anaplastic lymphoma kinase (ALK) and an inhibitor used for the treatment of NSCLC.80,81 Previously, it was shown that (S)-crizotinib inhibited SARS CoV-2 in three cell lines and variants of concern such as delta and omicron, while crizotinib hydrochloride showed activity in one cell line Vero-TMPRSS2.82 Olmutinib, bosutinib, and crizotinib were tested by us in Calu-3 cells and demonstrated cell toxicity. The activity of these compounds in other cell lines such as A549-ACE2, Huh-7, and Caco-2 needs to be determined, as we and others have previously demonstrated cell-type specific activities of compounds against SARS-CoV-2.57,73
In conclusion, we have described the discovery of several new repurposed inhibitors for PLpro and Mpro using high-throughput screening of a small library of drugs and the specific selection of kinase inhibitors that had previously demonstrated in vitro activity in cell lines, suggesting a direct antiviral effect. All these molecules are clinically accessible and could be further evaluated in animal models or clinical studies for SARS-CoV-2. Perhaps the molecule of most interest to us is cetylpyridinium chloride. While it does not represent an oral drug, its application as a mouthwash or nasal spray may have therapeutic applications, as demonstrated in several clinical studies, and this was also proposed by us at the very outset of the pandemic.59 We have now described for the first time how PLpro and Mpro may represent the viral targets for this readily available molecule that may have already been in widespread use during the pandemic in various consumer products.
Methods
Chemicals and Reagents
The Microsource Spectrum Collection compound library was used for screening. Bosutinib, crizotinib, dacomitinib, cetylpyridinium chloride, and raloxifene were purchased from MedChemExpress (MCE). The purity of these compounds is greater than 95%. Compound 15c83 was purchased from Sigma-Aldrich.
SARS-CoV-2 High-Throughput Screening against PLpro and Mpro
SARS-Cov-2 PLpro and Mpro expression, purification, and activity assays were described previously.58 Briefly, the Mpro inhibition assay was performed using an fluorescence resonance energy transfer (FRET)-based fluorescent peptide substrate DABCYL-KTSAVLQ↓SGFRKM-E(EDANS)-NH2 (purchased from Genscript), with an enzyme concentration of 140 nM and a 30 μM fluorescent substrate in assay buffer (20 mM Tris pH 7.3, 1 mM EDTA, and 1 mM DTT) at 37 °C for 30 min. Activity was detected in the spectrofluorometer system Spectramax Gemini EM (Molecular Devices), with λex = 360 nm and λem = 460 nm. For the PLpro, a FRET-based fluorescent peptide substrate, Abz-TLKGG↓APIKEDDPS-EDDnp (kindly provided by Dr. Maria Aparecida Juliano, Federal University of São Paulo, Brazil), was used. For the reaction, compounds were incubated with 70 nM enzyme and 27 μM fluorescent substrate in PLpro assay buffer (50 mM HEPES pH 7.5, 0.01% Triton X-100, and 5 mM DTT) at 37 °C for 30 min. Activity was measured in the plate reader system, Spectramax Gemini EM (Molecular Devices), with λex = 320 nm and λem = 420 nm. For both, controls in reactions without enzyme (negative control) and without inhibitor (positive control).
The Microsource Spectrum collection library containing 2560 compounds was tested against PLpro and Mpro in a high-throughput screening format (HTS) using 384 well-plates.
Proteins in the described assay conditions58 were incubated with 1% DMSO and compounds at 10 μM for 30 min at 37° C in an end-point format. The activity of compound reaction was normalized based on the relative activity of the enzymes in the presence of 1% DMSO. Compounds that inhibited each protease activity in more than 80% were assayed in a dose-dependent manner to determine their half-inhibitory concentrations, IC50. The results were analyzed using OriginPro 9.0 Software (Origin Lab), and the IC50 for each compound was estimated using the Hill1 function fitting. The dose response assays were carried out in triplicate of the same point in the experiment.
Calu-3 Cells
Compounds were incubated for 2 h with Calu-3 (ATCC, HTB-55) cells, followed by infection with SARS-CoV-2 (isolate USA WA1/2020) at a MOI = 0.5. 48 h post-infection, cells were fixed, immunostained, and imaged by automated microscopy for infection (dsRNA + cells/total cell number) and cell number. Sample well data was normalized to aggregated DMSO control wells and plotted versus drug concentration to determine the IC50 (infection: blue) and CC50 (toxicity: green).
Acknowledgments
The authors would like to kindly acknowledge their many collaborators around the world who have assisted in our various COVID-19 projects since 2020. Dr. Mindy Davis and colleagues are gratefully acknowledged for assistance with the NIAID virus screening capabilities. Dr. Ethan Perlstein is kindly acknowledged for providing the Microsource library.
Glossary
Abbreviations USED
- ALK
anaplastic lymphoma kinase
- COVID-19
coronavirus disease
- EGFR
epidermal growth factor receptor
- HIV
human immunodeficiency virus
- HCV
hepatitis C virus
- Mpro
main protease
- MERS-CoV
middle east respiratory syndrome coronavirus
- NSCLC
non-small cell lung cancer
- PLpro
papain-like protease
- P-gp
P-glycoprotein
- ROS1
c-ros oncogene 1
- SARS-CoV-2
severe acute respiratory coronavirus 2
- VGFR
vascular endothelial growth factor
Author Contributions
All authors read and accept the manuscript. A.S.G., A.C.P., R.S.F., G.O., and S.E. conceived and codirected the study. A.C.P., A.S.G., and R.S.F. designed the experiments. A.S.G., G.D.N., A.M.N., and V.O.G. performed in vitro experiments. A.C.P. and S.E. drafted the manuscript.
SE kindly acknowledges NIH funding R44GM122196-02A1 from NIH NIGMS 1R43AT010585-01 from NIH/NCCAM. Collaborations Pharmaceuticals, Inc. has utilized the non-clinical and pre-clinical services program offered by the National Institute of Allergy and Infectious Diseases. This project was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES—Project 88887.516153/2020-00, ASG) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP projects 2013/07600-3, 2015/16811-3, and 2016/19712-9, GO).
The authors declare the following competing financial interest(s): SE is CEO of Collaborations Pharmaceuticals, Inc. ACP is an employee at Collaborations Pharmaceuticals, Inc. Other authors have no conflicts.
References
- Hui D. S.; I Azhar E.; Madani T. A.; Ntoumi F.; Kock R.; Dar O.; Ippolito G.; Mchugh T. D.; Memish Z. A.; Drosten C.; Zumla A.; Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.; Zhao S.; Yu B.; Chen Y. M.; Wang W.; Song Z. G.; Hu Y.; Tao Z. W.; Tian J. H.; Pei Y. Y.; Yuan M. L.; Zhang Y. L.; Dai F. H.; Liu Y.; Wang Q. M.; Zheng J. J.; Xu L.; Holmes E. C.; Zhang Y. Z. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO; WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int (accessed May 23 2023).
- Zhou S.; Hill C. S.; Sarkar S.; Tse L. V.; Woodburn B. M. D.; Schinazi R. F.; Sheahan T. P.; Baric R. S.; Heise M. T.; Swanstrom R. β-d-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells. J. Infect. Dis. 2021, 224, 415–419. 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T.; Wang Y. F.; Harrison R. W. HIV Protease: Historical Perspective and Current Research. Viruses 2021, 13, 839. 10.3390/v13050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meewan I.; Zhang X.; Roy S.; Ballatore C.; O’Donoghue A. J.; Schooley R. T.; Abagyan R. Discovery of New Inhibitors of Hepatitis C Virus NS3/4A Protease and Its D168A Mutant. ACS Omega 2019, 4, 16999–17008. 10.1021/acsomega.9b02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leuw P.; Stephan C. Protease inhibitors for the treatment of hepatitis C virus infection. GMS Infect. Dis. 2017, 5, Doc08. 10.3205/id000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A.; Banerjee S.; Ghosh K.; Gayen S.; Jha T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021, 29, 115860. 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. D.; Uhrich R. L.; Kraemer G. C.; Love J. E.; Kraemer B. C. A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease. PLoS One 2021, 16, e0245962 10.1371/journal.pone.0245962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S.; Yadav S.; Banerjee S.; Fakayode S. O.; Parvathareddy J.; Reichard W.; Surendranathan S.; Mahmud F.; Whatcott R.; Thammathong J.; Meibohm B.; Miller D. D.; Jonsson C. B.; Dubey K. D. Drug Repurposing to Identify Nilotinib as a Potential SARS-CoV-2 Main Protease Inhibitor: Insights from a Computational and In Vitro Study. J. Chem. Inf. Model. 2021, 61, 5469–5483. 10.1021/acs.jcim.1c00524. [DOI] [PubMed] [Google Scholar]
- Kuzikov M.; Costanzi E.; Reinshagen J.; Esposito F.; Vangeel L.; Wolf M.; Ellinger B.; Claussen C.; Geisslinger G.; Corona A.; Iaconis D.; Talarico C.; Manelfi C.; Cannalire R.; Rossetti G.; Gossen J.; Albani S.; Musiani F.; Herzog K.; Ye Y.; Giabbai B.; Demitri N.; Jochmans D.; Jonghe S.; Rymenants J.; Summa V.; Tramontano E.; Beccari A. R.; Leyssen P.; Storici P.; Neyts J.; Gribbon P.; Zaliani A. Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen. ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina F.; Puhl A. C.; Ekins S. Recent advances in drug repurposing using machine learning. Curr. Opin. Chem. Biol. 2021, 65, 74–84. 10.1016/j.cbpa.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. R.; Allerton C. M. N.; Anderson A. S.; Aschenbrenner L.; Avery M.; Berritt S.; Boras B.; Cardin R. D.; Carlo A.; Coffman K. J.; Dantonio A.; Di L.; Eng H.; Ferre R.; Gajiwala K. S.; Gibson S. A.; Greasley S. E.; Hurst B. L.; Kadar E. P.; Kalgutkar A. S.; Lee J. C.; Lee J.; Liu W.; Mason S. W.; Noell S.; Novak J. J.; Obach R. S.; Ogilvie K.; Patel N. C.; Pettersson M.; Rai D. K.; Reese M. R.; Sammons M. F.; Sathish J. G.; Singh R. S. P.; Steppan C. M.; Stewart A. E.; Tuttle J. B.; Updyke L.; Verhoest P. R.; Wei L.; Yang Q.; Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586. 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- de Vries M.; Mohamed A. S.; Prescott R. A.; Valero-Jimenez A. M.; Desvignes L.; O’Connor R.; Steppan C.; Devlin J. C.; Ivanova E.; Herrera A.; Schinlever A.; Loose P.; Ruggles K.; Koralov S. B.; Anderson A. S.; Binder J.; Dittmann M. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CL(pro) inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 2021, 95, e01819 10.1128/jvi.01819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.; Hu Y.; Jadhav P.; Tan B.; Wang J. Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease. J. Med. Chem. 2022, 65, 7561–7580. 10.1021/acs.jmedchem.2c00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K.; Kim J.; Chang J.; Hong S.; Kim I.; Oh S.; Jeon S.; Lee J. C.; Park H. J.; Kim S.; Lee W. Chemical screen uncovers novel structural classes of inhibitors of the papain-like protease of coronaviruses. iScience 2022, 25, 105254. 10.1016/j.isci.2022.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja D. J.; Lessene G.; Komander D. Inhibitors of SARS-CoV-2 PLpro. Front. Chem. 2022, 10, 876212. 10.3389/fchem.2022.876212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares K. M.; Blackmon N.; Shun T. Y.; Shinde S. N.; Takyi H. K.; Wipf P.; Lazo J. S.; Johnston P. A. Profiling the NIH Small Molecule Repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev. Technol. 2010, 8, 152–174. 10.1089/adt.2009.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C.; Gomes G. F.; Damasceno S.; Fritch E. J.; Levi J. A.; Johnson N. J.; Scholle F.; Premkumar L.; Hurst B. L.; Lee-Montiel F.; Veras F. P.; Batah S. S.; Fabro A. T.; Moorman N. J.; Yount B. L.; Dickmander R. J.; Baric R. S.; Pearce K. H.; Cunha F. Q.; Alves-Filho J. C.; Cunha T. M.; Ekins S. Vandetanib Blocks the Cytokine Storm in SARS-CoV-2-Infected Mice. ACS Omega 2022, 7, 31935–31944. 10.1021/acsomega.2c02794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconis D.; Bordi L.; Matusali G.; Talarico C.; Manelfi C.; Cesta M. C.; Zippoli M.; Caccuri F.; Bugatti A.; Zani A.; Filippini F.; Scorzolini L.; Gobbi M.; Beeg M.; Piotti A.; Montopoli M.; Cocetta V.; Bressan S.; Bucci E. M.; Caruso A.; Nicastri E.; Allegretti M.; Beccari A. R. Characterization of raloxifene as a potential pharmacological agent against SARS-CoV-2 and its variants. Cell Death Dis. 2022, 13, 498. 10.1038/s41419-022-04961-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K.; Chang J.; Park S. M.; Kim J.; Jeon S.; Kim D. H.; Kim Y. E.; Lee J. C.; Im S.; Jo Y.; Min J. Y.; Lee H.; Yeom M.; Seok S. H.; On D. I.; Noh H.; Yun J. W.; Park J. W.; Song D.; Seong J. K.; Kim K. C.; Lee J. Y.; Park H. J.; Kim S.; Nam T. G.; Lee W. Rapid discovery and classification of inhibitors of coronavirus infection by pseudovirus screen and amplified luminescence proximity homogeneous assay. Antiviral Res. 2023, 209, 105473. 10.1016/j.antiviral.2022.105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K.; Sakurai Y.; Enami T.; Shibukawa R.; Nishi Y.; Ohta A.; Shu T.; Kawaguchi J.; Okada S.; Hoenen T.; Yasuda J.; Inoue H. iPSC screening for drug repurposing identifies anti-RNA virus agents modulating host cell susceptibility. FEBS Open Bio 2021, 11, 1452–1464. 10.1002/2211-5463.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger B.; Bojkova D.; Zaliani A.; Cinatl J.; Claussen C.; Westhaus S.; Keminer O.; Reinshagen J.; Kuzikov M.; Wolf M.; Geisslinger G.; Gribbon P.; Ciesek S. A SARS-CoV-2 cytopathicity dataset generated by high-content screening of a large drug repurposing collection. Sci. Data 2021, 8, 70. 10.1038/s41597-021-00848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saud Z.; Tyrrell V. J.; Zaragkoulias A.; Protty M. B.; Statkute E.; Rubina A.; Bentley K.; White D. A.; Rodrigues P. D. S.; Murphy R. C.; Kofeler H.; Griffiths W. J.; Alvarez-Jarreta J.; Brown R. W.; Newcombe R. G.; Heyman J.; Pritchard M.; McLeod R. W.; Arya A.; Lynch C. A.; Owens D.; Jenkins P. V.; Buurma N. J.; O’Donnell V. B.; Thomas D. W.; Stanton R. J. The SARS-CoV2 envelope differs from host cells, exposes procoagulant lipids, and is disrupted in vivo by oral rinses. J. Lipid Res. 2022, 63, 100208. 10.1016/j.jlr.2022.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratov E. N.; Amaro R.; Andrade C. H.; Brown N.; Ekins S.; Fourches D.; Isayev O.; Kozakov D.; Medina-Franco J. L.; Merz K. M.; Oprea T. I.; Poroikov V.; Schneider G.; Todd M. H.; Varnek A.; Winkler D. A.; Zakharov A. V.; Cherkasov A.; Tropsha A. A critical overview of computational approaches employed for COVID-19 drug discovery. Chem. Soc. Rev. 2021, 50, 9121–9151. 10.1039/d0cs01065k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Mottin M.; Ramos P.; Sousa B. K. P.; Neves B. J.; Foil D. H.; Zorn K. M.; Braga R. C.; Coffee M.; Southan C.; Puhl A. C.; Andrade C. H. Deja vu: Stimulating open drug discovery for SARS-CoV-2. Drug Discovery Today 2020, 25, 928–941. 10.1016/j.drudis.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercorelli B.; Desantis J.; Celegato M.; Bazzacco A.; Siragusa L.; Benedetti P.; Eleuteri M.; Croci F.; Cruciani G.; Goracci L.; Loregian A. Discovery of novel SARS-CoV-2 inhibitors targeting the main protease M(pro) by virtual screenings and hit optimization. Antiviral Res. 2022, 204, 105350. 10.1016/j.antiviral.2022.105350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury L.; Jing Z.; Cuzzolin A.; Deplano A.; Loco D.; Sattarov B.; Hedin F.; Wendeborn S.; Ho C.; El Ahdab D.; Jaffrelot Inizan T.; Sturlese M.; Sosic A.; Volpiana M.; Lugato A.; Barone M.; Gatto B.; Macchia M. L.; Bellanda M.; Battistutta R.; Salata C.; Kondratov I.; Iminov R.; Khairulin A.; Mykhalonok Y.; Pochepko A.; Chashka-Ratushnyi V.; Kos I.; Moro S.; Montes M.; Ren P.; Ponder J. W.; Lagardere L.; Piquemal J. P.; Sabbadin D. Computationally driven discovery of SARS-CoV-2 M(pro) inhibitors: from design to experimental validation. Chem. Sci. 2022, 13, 3674–3687. 10.1039/d1sc05892d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Martinez J.; Jimenez-Alesanco A.; Ceballos-Laita L.; Ortega-Alarcon D.; Vega S.; Calvo C.; Benitez C.; Abian O.; Velazquez-Campoy A.; Thomson T. M.; Granadino-Roldan J. M.; Gomez-Gutierrez P.; Perez J. J. Discovery of Diverse Natural Products as Inhibitors of SARS-CoV-2 M(pro) Protease through Virtual Screening. J. Chem. Inf. Model. 2021, 61, 6094–6106. 10.1021/acs.jcim.1c00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Li Z.; Liu S.; Sun J.; Chen Z.; Jiang M.; Zhang Q.; Wei Y.; Wang X.; Huang Y. Y.; Shi Y.; Xu Y.; Xian H.; Bai F.; Ou C.; Xiong B.; Lew A. M.; Cui J.; Fang R.; Huang H.; Zhao J.; Hong X.; Zhang Y.; Zhou F.; Luo H. B. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B 2020, 10, 1205–1215. 10.1016/j.apsb.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Ma C.; Sacco M. D.; Hurst B.; Townsend J. A.; Hu Y.; Szeto T.; Zhang X.; Tarbet B.; Marty M. T.; Chen Y.; Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.; McCorkindale W.; Consortium T. C. M.; Drayman N.; Chodera J. D.; Tay S.; London N.; Lee A. A. Discovery of SARS-CoV-2 main protease inhibitors using a synthesis-directed de novo design model. Chem. Commun. 2021, 57, 5909–5912. 10.1039/d1cc00050k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A. T.; Pandey M.; Smith J. R.; Ban F.; Fernandez M.; Cherkasov A. Targeting SARS-CoV-2 papain-like protease in the postvaccine era. Trends Pharmacol. Sci. 2022, 43, 906–919. 10.1016/j.tips.2022.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J.; Li Y. S.; Zeng R.; Liu F. L.; Luo R. H.; Huang C.; Wang Y. F.; Zhang J.; Quan B.; Shen C.; Mao X.; Liu X.; Sun W.; Yang W.; Ni X.; Wang K.; Xu L.; Duan Z. L.; Zou Q. C.; Zhang H. L.; Qu W.; Long Y. H.; Li M. H.; Yang R. C.; Liu X.; You J.; Zhou Y.; Yao R.; Li W. P.; Liu J. M.; Chen P.; Liu Y.; Lin G. F.; Yang X.; Zou J.; Li L.; Hu Y.; Lu G. W.; Li W. M.; Wei Y. Q.; Zheng Y. T.; Lei J.; Yang S. SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science 2021, 371, 1374–1378. 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.; Li D.; Zhang J.; Yin X.; Li J. Crystal structure of SARS-CoV 3C-like protease with baicalein. Biochem. Biophys. Res. Commun. 2022, 611, 190–194. 10.1016/j.bbrc.2022.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S.; Chen Y.; Lockbaum G. J.; Sen S.; Chaudhuri S.; Reyes A. C.; Lee J. M.; Kaur A. N.; Sultana N.; Cameron M. D.; Shaffer S. A.; Schiffer C. A.; Fitzgerald K. A.; Thompson P. R. Dual Inhibitors of Main Protease (M(Pro)) and Cathepsin L as Potent Antivirals against SARS-CoV2. J. Am. Chem. Soc. 2022, 144, 21035–21045. 10.1021/jacs.2c04626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Lin D.; Sun X.; Curth U.; Drosten C.; Sauerhering L.; Becker S.; Rox K.; Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 2020, 368, 409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S. I.; Higashi-Kuwata N.; Hayashi H.; Allu S. R.; Raghavaiah J.; Bulut H.; Das D.; Anson B. J.; Lendy E. K.; Takamatsu Y.; Takamune N.; Kishimoto N.; Murayama K.; Hasegawa K.; Li M.; Davis D. A.; Kodama E. N.; Yarchoan R.; Wlodawer A.; Misumi S.; Mesecar A. D.; Ghosh A. K.; Mitsuya H. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021, 12, 668. 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Yang K. S.; Geng Z. Z.; Alugubelli Y. R.; Shaabani N.; Vatansever E. C.; Ma X. R.; Cho C. C.; Khatua K.; Xiao J.; Blankenship L. R.; Yu G.; Sankaran B.; Li P.; Allen R.; Ji H.; Xu S.; Liu W. R. A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med. Chem. 2022, 240, 114570. 10.1016/j.ejmech.2022.114570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K.; Ishii T.; Kobayakawa T.; Higashi-Kuwata N.; Azuma C.; Nakayama M.; Onishi T.; Nakano H.; Wada N.; Hori M.; Shinohara K.; Miura Y.; Kawada T.; Hayashi H.; Hattori S. I.; Bulut H.; Das D.; Takamune N.; Kishimoto N.; Saruwatari J.; Okamura T.; Nakano K.; Misumi S.; Mitsuya H.; Tamamura H. Potent and biostable inhibitors of the main protease of SARS-CoV-2. iScience 2022, 25, 105365. 10.1016/j.isci.2022.105365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alugubelli Y. R.; Geng Z. Z.; Yang K. S.; Shaabani N.; Khatua K.; Ma X. R.; Vatansever E. C.; Cho C. C.; Ma Y.; Xiao J.; Blankenship L. R.; Yu G.; Sankaran B.; Li P.; Allen R.; Ji H.; Xu S.; Liu W. R. A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med. Chem. 2022, 240, 114596. 10.1016/j.ejmech.2022.114596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. S.; Alex Kuo S. T.; Blankenship L. R.; Geng Z. Z.; Li S. G.; Russell D. H.; Yan X.; Xu S.; Liu W. R. Repurposing Halicin as a potent covalent inhibitor for the SARS-CoV-2 main protease. Curr. Res. Chem. Biol. 2022, 2, 100025. 10.1016/j.crchbi.2022.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan B. X.; Shuai H.; Xia A. J.; Hou Y.; Zeng R.; Liu X. L.; Lin G. F.; Qiao J. X.; Li W. P.; Wang F. L.; Wang K.; Zhou R. J.; Yuen T. T.; Chen M. X.; Yoon C.; Wu M.; Zhang S. Y.; Huang C.; Wang Y. F.; Yang W.; Tian C.; Li W. M.; Wei Y. Q.; Yuen K. Y.; Chan J. F.; Lei J.; Chu H.; Yang S. An orally available M(pro) inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron. Nat. Microbiol. 2022, 7, 716–725. 10.1038/s41564-022-01119-7. [DOI] [PubMed] [Google Scholar]
- Hung H. C.; Ke Y. Y.; Huang S. Y.; Huang P. N.; Kung Y. A.; Chang T. Y.; Yen K. J.; Peng T. T.; Chang S. E.; Huang C. T.; Tsai Y. R.; Wu S. H.; Lee S. J.; Lin J. H.; Liu B. S.; Sung W. C.; Shih S. R.; Chen C. T.; Hsu J. T. Discovery of M Protease Inhibitors Encoded by SARS-CoV-2. Antimicrob. Agents Chemother. 2020, 64, e00872 10.1128/aac.00872-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres C. J.; Cardenas-Garcia S.; Carnaccini S.; Seibert B.; Rajao D. S.; Wang J.; Perez D. R. Efficacy of GC-376 against SARS-CoV-2 virus infection in the K18 hACE2 transgenic mouse model. Sci. Rep. 2021, 11, 9609. 10.1038/s41598-021-89013-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meewan I.; Kattoula J.; Kattoula J. Y.; Skinner D.; Fajtova P.; Giardini M. A.; Woodworth B.; McKerrow J. H.; Lage de Siqueira-Neto J.; O’Donoghue A. J.; Abagyan R. Discovery of Triple Inhibitors of Both SARS-CoV-2 Proteases and Human Cathepsin L. Pharmaceuticals 2022, 15, 744. 10.3390/ph15060744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzo C. A.; Martins R. B.; Fraga-Silva T. F. C.; Amstalden M. K.; Canassa De Leo T.; Souza J. P.; Lima T. M.; Faccioli L. H.; Okamoto D. N.; Juliano M. A.; Franca S. C.; Juliano L.; Bonato V. L. D.; Arruda E.; Dias-Baruffi M. Celastrol: A lead compound that inhibits SARS-CoV-2 replication, the activity of viral and human cysteine proteases, and virus-induced IL-6 secretion. Drug Dev. Res. 2022, 83, 1623–1640. 10.1002/ddr.21982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.; Liu Z.; Yan G.; Liu X.; Liu X.; Wang Y.; Chen Y. A robust high-throughput fluorescence polarization assay for rapid screening of SARS-CoV-2 papain-like protease inhibitors. Virology 2022, 574, 18–24. 10.1016/j.virol.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.; Huang B.; Tang J.; Liu S.; Liu M.; Ye Y.; Liu Z.; Xiong Y.; Zhu W.; Cao D.; Li J.; Niu X.; Zhou H.; Zhao Y. J.; Zhang G.; Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021, 12, 488. 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas B. T.; Ahiadorme D. A.; Bagul R. S.; Durie I. A.; Ghosh S.; Hill J.; Kramer N. E.; Murray J.; O’Boyle B. M.; Onobun E.; Pirrone M. G.; Shepard J. D.; Enos S.; Subedi Y. P.; Upadhyaya K.; Tripp R. A.; Cummings B. S.; Crich D.; Pegan S. D. Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses. ACS Infect. Dis 2022, 8, 596–611. 10.1021/acsinfecdis.1c00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H.; Liu J.; Shen J.; Dai J.; Xu G.; Lu K.; Han C.; Wang Y.; Xu X.; Tong Y.; Xiang H.; Ai Z.; Zhuang G.; Hu J.; Zhang Z.; Li Y.; Pan L.; Tan L. Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2. Cell Chem. Biol. 2021, 28, 855–865 e9. 10.1016/j.chembiol.2021.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulandaisamy R.; Kushwaha T.; Dalal A.; Kumar V.; Singh D.; Baswal K.; Sharma P.; Praneeth K.; Jorwal P.; Kayampeta S. R.; Sharma T.; Maddur S.; Kumar M.; Kumar S.; Polamarasetty A.; Singh A.; Sehgal D.; Gholap S. L.; Appaiahgari M. B.; Katika M. R.; Inampudi K. K. Repurposing of FDA Approved Drugs Against SARS-CoV-2 Papain-Like Protease: Computational, Biochemical, and in vitro Studies. Front. Microbiol. 2022, 13, 877813. 10.3389/fmicb.2022.877813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.; Zhao Q.; Chen X.; Peng Z.; Tan X.; Wang Q.; Chen L.; Yang Y. Discovery of Novel and Highly Potent Inhibitors of SARS CoV-2 Papain-Like Protease Through Structure-Based Pharmacophore Modeling, Virtual Screening, Molecular Docking, Molecular Dynamics Simulations, and Biological Evaluation. Front. Pharmacol. 2022, 13, 817715. 10.3389/fphar.2022.817715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Hu Y.; Wang Y.; Choza J.; Wang J. Drug-Repurposing Screening Identified Tropifexor as a SARS-CoV-2 Papain-like Protease Inhibitor. ACS Infect. Dis 2022, 8, 1022–1030. 10.1021/acsinfecdis.1c00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sarno V.; Lauro G.; Musella S.; Ciaglia T.; Vestuto V.; Sala M.; Scala M. C.; Smaldone G.; Di Matteo F.; Novi S.; Tecce M. F.; Moltedo O.; Bifulco G.; Campiglia P.; Gomez-Monterrey I. M.; Snoeck R.; Andrei G.; Ostacolo C.; Bertamino A. Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization. Eur. J. Med. Chem. 2021, 226, 113863. 10.1016/j.ejmech.2021.113863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C.; Fritch E. J.; Lane T. R.; Tse L. V.; Yount B. L.; Sacramento C. Q.; Fintelman-Rodrigues N.; Tavella T. A.; Maranhão Costa F. T.; Weston S.; Logue J.; Frieman M.; Premkumar L.; Pearce K. H.; Hurst B. L.; Andrade C. H.; Levi J. A.; Johnson N. J.; Kisthardt S. C.; Scholle F.; Souza T. M. L.; Moorman N. J.; Baric R. S.; Madrid P. B.; Ekins S. Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine, and Pyronaridine: In Vitro Activity against SARS-CoV-2 and Potential Mechanisms. ACS Omega 2021, 6, 7454–7468. 10.1021/acsomega.0c05996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C.; Gomes G. F.; Damasceno S.; Godoy A. S.; Noske G. D.; Nakamura A. M.; Gawriljuk V. O.; Fernandes R. S.; Monakhova N.; Riabova O.; Lane T. R.; Makarov V.; Veras F. P.; Batah S. S.; Fabro A. T.; Oliva G.; Cunha F. Q.; Alves-Filho J. C.; Cunha T. M.; Ekins S. Pyronaridine Protects against SARS-CoV-2 Infection in Mouse. ACS Infect. Dis 2022, 8, 1147–1160. 10.1021/acsinfecdis.2c00091. [DOI] [PubMed] [Google Scholar]
- Baker N.; Williams A. J.; Tropsha A.; Ekins S. Repurposing quaternary ammonium compounds as potential treatments for COVID-19. Pharm. Res. 2020, 37, 104. 10.1007/s11095-020-02842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N.; Saito A.; Okabayashi T.; Komine A. Virucidal activity and mechanism of action of cetylpyridinium chloride against SARS-CoV-2. J. Oral Maxillofac. Pathol. 2022, 34, 800–804. 10.1016/j.ajoms.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda R.; Sawa H.; Sasaki M.; Orba Y.; Maishi N.; Tsumita T.; Ushijima N.; Hida Y.; Sano H.; Kitagawa Y.; Hida K. Antiviral effect of cetylpyridinium chloride in mouthwash on SARS-CoV-2. Sci. Rep. 2022, 12, 14050. 10.1038/s41598-022-18367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne C. J.; Balan P.; Ko K. K. K.; Udawatte N. S.; Lai D.; Ng D. H. L.; Venkatachalam I.; Lim K. S.; Ling M. L.; Oon L.; Goh B. T.; Sim X. Y. J. Efficacy of commercial mouth-rinses on SARS-CoV-2 viral load in saliva: randomized control trial in Singapore. Infection 2021, 49, 305–311. 10.1007/s15010-020-01563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eduardo F. d. P.; Correa L.; Heller D.; Daep C. A.; Benitez C.; Malheiros Z.; Stewart B.; Ryan M.; Machado C. M.; Hamerschlak N.; Rebello Pinho J. R.; Bezinelli L. M. Salivary SARS-CoV-2 load reduction with mouthwash use: A randomized pilot clinical trial. Heliyon 2021, 7, e07346 10.1016/j.heliyon.2021.e07346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti M.; Cesta M. C.; Zippoli M.; Beccari A.; Talarico C.; Mantelli F.; Bucci E. M.; Scorzolini L.; Nicastri E. Repurposing the estrogen receptor modulator raloxifene to treat SARS-CoV-2 infection. Cell Death Differ. 2022, 29, 156–166. 10.1038/s41418-021-00844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastri E.; Marinangeli F.; Pivetta E.; Torri E.; Reggiani F.; Fiorentino G.; Scorzolini L.; Vettori S.; Marsiglia C.; Gavioli E. M.; Beccari A. R.; Terpolilli G.; De Pizzol M.; Goisis G.; Mantelli F.; Vaia F.; Allegretti M.; Raloxifene Territorial Health C. S. G. A phase 2 randomized, double-blinded, placebo-controlled, multicenter trial evaluating the efficacy and safety of raloxifene for patients with mild to moderate COVID-19. EClinicalMedicine 2022, 48, 101450. 10.1016/j.eclinm.2022.101450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Hu Y.; Townsend J. A.; Lagarias P. I.; Marty M. T.; Kolocouris A.; Wang J. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 2020, 3, 1265–1277. 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargsyan K.; Lin C. C.; Chen T.; Grauffel C.; Chen Y. P.; Yang W. Z.; Yuan H. S.; Lim C. Multi-targeting of functional cysteines in multiple conserved SARS-CoV-2 domains by clinically safe Zn-ejectors. Chem. Sci. 2020, 11, 9904–9909. 10.1039/d0sc02646h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafna K.; White K.; Harish B.; Rosales R.; Ramelot T. A.; Acton T. B.; Moreno E.; Kehrer T.; Miorin L.; Royer C. A.; Garcia-Sastre A.; Krug R. M.; Montelione G. T. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep. 2021, 35, 109133. 10.1016/j.celrep.2021.109133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y.; Su M.; Wang Q.; Cheng X.; Zhang W.; Zhao Y.; Chen T.; Jiang Y.; Shen Q.; Du J.; Tan Q.; Wang P.; Gao L.; Jin Z.; Zhang M.; Li C.; Zhu Y.; Feng B.; Tang B.; Xie H.; Wang M. W.; Zheng M.; Pan X.; Yang H.; Xu Y.; Wu B.; Zhang L.; Rao Z.; Yang X.; Jiang H.; Xiao G.; Zhao Q.; Li J. High-throughput screening of SARS-CoV-2 main and papain-like protease inhibitors. Protein Cell 2023, 14, 17–27. 10.1093/procel/pwac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E.; Parent A.; Yang P. L.; Sattler M.; Liu Q.; Liu Q.; Wang J.; Meng C.; Buhrlage S. J.; Gray N.; et al. Repurposing of Kinase Inhibitors for Treatment of COVID-19. Pharm. Res. 2020, 37, 167. 10.1007/s11095-020-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B. C.; Lin C. C.; Lee J. H.; Yang J. C. Update on recent preclinical and clinical studies of T790M mutant-specific irreversible epidermal growth factor receptor tyrosine kinase inhibitors. J. Biomed. Sci. 2016, 23, 86. 10.1186/s12929-016-0305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. C. M.; Batra U.; Mok T. S. K.; Loong H. H. Dacomitinib in the Management of Advanced Non-Small-Cell Lung Cancer. Drugs 2019, 79, 823–831. 10.1007/s40265-019-01115-y. [DOI] [PubMed] [Google Scholar]
- Dittmar M.; Lee J. S.; Whig K.; Segrist E.; Li M.; Kamalia B.; Castellana L.; Ayyanathan K.; Cardenas-Diaz F. L.; Morrisey E. E.; Truitt R.; Yang W.; Jurado K.; Samby K.; Ramage H.; Schultz D. C.; Cherry S. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021, 35, 108959. 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann K.; Bojkova D.; Tascher G.; Ciesek S.; Münch C.; Cinatl J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol. Cell 2020, 80, 164–174.e4. 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eierhoff T.; Hrincius E. R.; Rescher U.; Ludwig S.; Ehrhardt C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010, 6, e1001099 10.1371/journal.ppat.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C. P.; Meckes D. G.; Raab-Traub N. Epstein-Barr Virus LMP1 Activates EGFR, STAT3, and ERK through Effects on PKCδ. J. Virol. 2011, 85, 4399–4408. 10.1128/jvi.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupberger J.; Zeisel M. B.; Xiao F.; Thumann C.; Fofana I.; Zona L.; Davis C.; Mee C. J.; Turek M.; Gorke S.; Royer C.; Fischer B.; Zahid M. N.; Lavillette D.; Fresquet J.; Cosset F. L.; Rothenberg S. M.; Pietschmann T.; Patel A. H.; Pessaux P.; Doffoël M.; Raffelsberger W.; Poch O.; McKeating J. A.; Brino L.; Baumert T. F. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011, 17, 589–595. 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud A. I.; Krishnamurthi S. S.; Saleh M. N.; Gitlitz B. J.; Borad M. J.; Gold P. J.; Chiorean E. G.; Springett G. M.; Abbas R.; Agarwal S.; Bardy-Bouxin N.; Hsyu P. H.; Leip E.; Turnbull K.; Zacharchuk C.; Messersmith W. A. Phase I study of bosutinib, a src/abl tyrosine kinase inhibitor, administered to patients with advanced solid tumors. Clin. Cancer Res. 2012, 18, 1092–1100. 10.1158/1078-0432.ccr-11-2378. [DOI] [PubMed] [Google Scholar]
- Yang L.; Pei R. J.; Li H.; Ma X. N.; Zhou Y.; Zhu F. H.; He P. L.; Tang W.; Zhang Y. C.; Xiong J.; Xiao S. Q.; Tong X. K.; Zhang B.; Zuo J. P. Identification of SARS-CoV-2 entry inhibitors among already approved drugs. Acta Pharmacol. Sin. 2021, 42, 1347–1353. 10.1038/s41401-020-00556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P. J. Clinical use of crizotinib for the treatment of non-small cell lung cancer. Biologics 2013, 7, 91–101. 10.2147/btt.s29026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhash K.; Noronha V.; Joshi A.; Desai S.; Sahu A. Crizotinib: A comprehensive review. South Asian J. Cancer 2013, 2, 91–97. 10.4103/2278-330x.110506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.; Mathayan M.; Smieszek S. P.; Przychodzen B. P.; Koprivica V.; Birznieks G.; Polymeropoulos M. H.; Prabhakar B. S. Identification of potential COVID-19 treatment compounds which inhibit SARS Cov2 prototypic, Delta and Omicron variant infection. Virology 2022, 572, 64–71. 10.1016/j.virol.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Takayama J.; Rao K. V.; Ratia K.; Chaudhuri R.; Mulhearn D. C.; Lee H.; Nichols D. B.; Baliji S.; Baker S. C.; Johnson M. E.; Mesecar A. D. Severe Acute Respiratory Syndrome Coronavirus Papain-like Novel Protease Inhibitors: Design, Synthesis, Protein–Ligand X-ray Structure and Biological Evaluation. J. Med. Chem. 2010, 53, 4968–4979. 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]