Abstract
Microglia are brain-resident macrophages capable of long-term maintenance through self-renewal. Yet the mechanism governing the turnover and lifespan of microglia remains unknown. In zebrafish, microglia arise from two sources, rostral blood island (RBI) and aorta-gonad-mesonephros (AGM). The RBI-derived microglia are born early but have a short lifespan and diminish in adulthood, while the AGM-derived microglia emerge later and are capable of long-term maintenance in adulthood. Here, we show that the attenuation of RBI microglia is due to their less competitiveness for neuron-derived interleukin-34 (Il34) caused by age-dependent decline of colony-stimulating factor-1 receptor a (csf1ra). Alterations of Il34/Csf1ra levels and removal of AGM microglia revamp the proportion and lifespan of RBI microglia. The csf1ra/CSF1R expression in zebrafish AGM-derived microglia and murine adult microglia also undergo age-dependent decline, leading to the elimination of aged microglia. Our study reveals cell competition as a general mechanism controlling the turnover and lifespan of microglia.
Cell competition is a general mechanism to regulate microglia turnover and lifespan.
INTRODUCTION
Microglia are specialized tissue-resident macrophages in the central nervous system (CNS), and they play essential roles in the regulation of CNS development and homeostasis (1, 2). The importance of microglia in the CNS is further exemplified by the observations that the occurrence and progression of many neurodegenerative disorders are often accompanied by the aberrant function of microglia (3, 4). Thus, a comprehensive understanding of how microglia pools are established and maintained is essential for elucidating the versatile functions of microglia in physiological and pathological conditions.
In mice, microglial precursors begin colonizing the brain rudiment at around embryonic day 9.5 (5, 6). Fate mapping studies reveal that the early-colonizing microglial precursors are of yolk sac (YS) origin, and these YS-derived microglia rapidly expand and continue to populate the adult brain throughout the lifespan of the animals (5, 7). It is now well accepted that microglia in the brain are capable of maintaining its population through self-renewal (5, 8, 9). However, because of the heterogeneities (10, 11) and inconsistency of estimated turnover rates (9, 12–14) of microglia in adult mice, the cellular and molecular mechanisms dictating their turnover and lifespan remain largely unclear. Unlike mammals, a recent temporospatial-specific fate mapping study in zebrafish revealed that microglia in the brain arise from two distinct sources, the rostral blood island (RBI) and the aorta-gonad-mesonephros (AGM) (15). In contrast to the long-term maintenance of AGM-derived microglia (referred to as AGM microglia) in adult zebrafish, the RBI-derived embryonic microglia (referred to as RBI microglia) have a relatively short lifespan and diminish in adulthood. This dynamic shift of microglia pools in zebrafish provides an ideal system to explore the mechanism that determines the turnover and lifespan of microglia.
Cell competition is a process that selects fit cells (winners) to populate the tissue while eliminates less-fit but intrinsically viable cells (losers) from a heterogeneous cell population (16, 17). In general, three different models are applied to explain the mechanisms of cell competition (16, 17). The first model is competition for growth factors and/or nutrients, thus cells with lower capability to sense or transduce the signals are eliminated. For instance, comparing to the surrounding wild-type (WT) cells, Drosophila Minute mutant cells and dMyc overexpression cells were less competitive for the extracellular growth factor Dpp in the wing discs (18, 19). The second mechanism is direct fitness comparison via cell-cell contact, which is best described by the interaction of tyrosine phosphatase Ptp10D expressed on the surface of Drosophila scrib mutant cells with the Sas expressed on the surface of neighboring WT cells (20, 21). Last, mechanical force sensing is another mechanism for cells competing for space (22, 23). Although cell competition has been thought to be a highly conserved process that exerts fundamental functions in the quality control of cells, especially in the conditions of stress, aging, and tumorigenesis (16, 17, 24), its roles in the establishment and maintenance of immune cells, which are highly mobile and are capable of colonizing various tissues, have not been explored.
In this study, by combining fate mapping, genetic manipulation, and single molecular RNA expression analysis, we demonstrate that cell competition for neuron-secreted trophic factor interleukin-34 (IL34) in the brain controls the turnover and lifespan of microglia.
RESULTS
Generation of transgenic reporter line distinguishing microglia of different origins
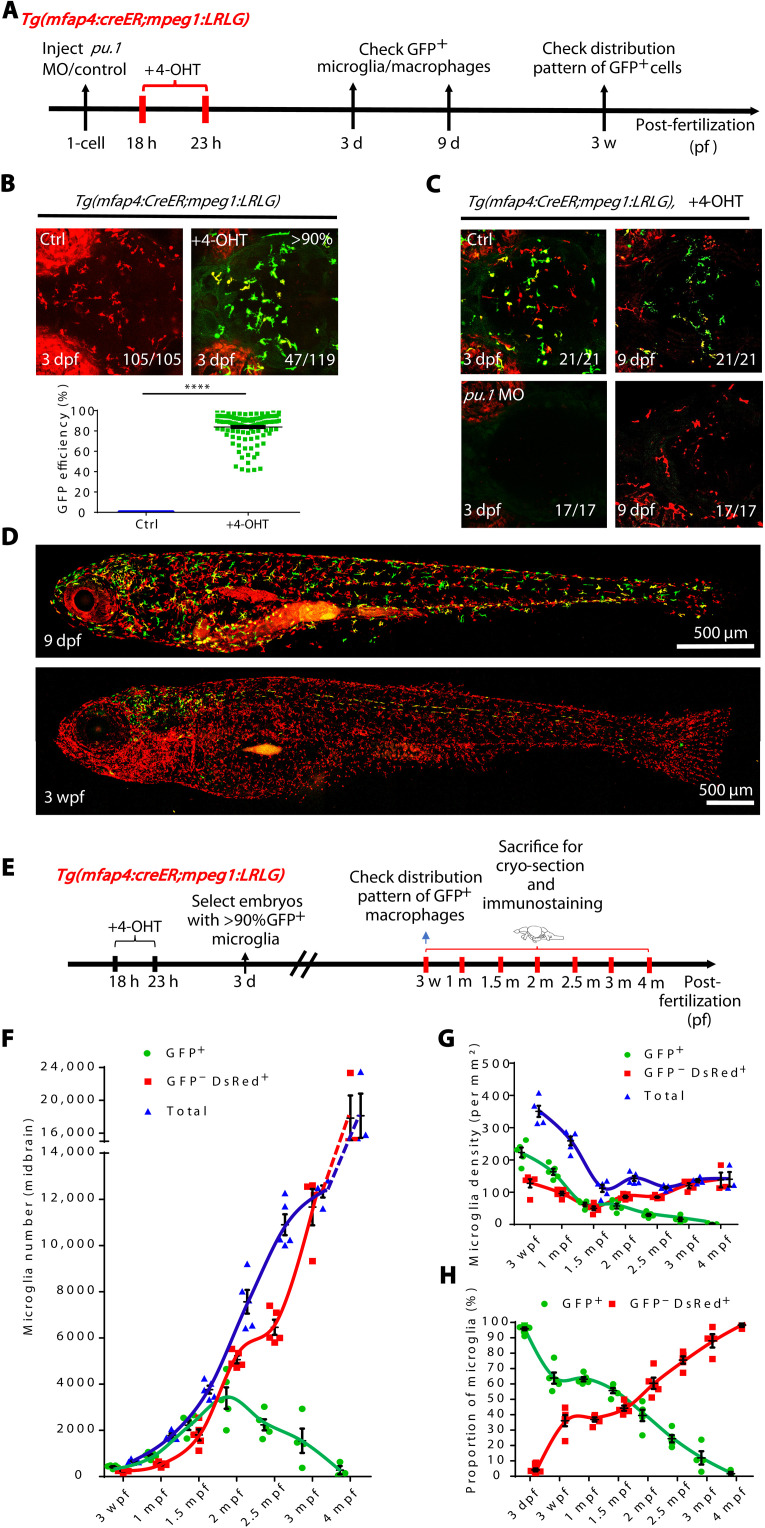
To dissect the cellular and molecular mechanisms underlying the attenuation of embryonic RBI microglia to adult AGM microglia in zebrafish, we generated a Tg(mfap4:CreER) transgenic line (fig. S1), in which the expression of CreER was under the control of macrophage-specific mfap4 promoter (25), and outcrossed this line with the Tg(mpeg1:Loxp-DsRedx-Loxp-GFP) [referred to as Tg(mpeg1:LRLG) hereafter] fish (26) to generate a Tg(mfap4:CreER;mpeg1:LRLG) double transgenic line where DsRed+ macrophages (including microglia) would switch to GFP+ upon 4-hydroxytamoxifen (4-OHT) treatment (fig. S1B). As the RBI-derived macrophages emerge at ~21 hours postfertilization (hpf) and the AGM-born macrophages are generated at around 4 to 5 days postfertilization (dpf) (26), we reasoned that in the Tg(mfap4:CreER;mpeg1:LRLG) fish, a short course of 4-OHT treatment at ~20 hpf would preferentially convert RBI microglia to GFP+ without affecting the AGM microglia population. We therefore treated the Tg(mfap4:CreER;mpeg1:LRLG) embryos with 4-OHT from 18 to 23 hpf and monitored the labeling efficiency of the RBI microglia by quantifying GFP+ microglia at 3 dpf (Fig. 1A), when the developing brain contains RBI microglia exclusively (27). We showed that, while GFP+ microglia were not detectable in the control embryos (n = 105/105), all the 4-OHT–treated embryos contained abundant GFP+ microglia in the brains (n = 119/119) (Fig. 1B), of which about 40% (n = 47/119) had >90% GFP+ microglia (Fig. 1B), indicating that a short course of 4-OHT treatment effectively labels RBI microglia in the Tg(mfap4:CreER;mpeg1:LRLG) fish. Morpholinos (MOs) are antisense oligonucleotides that transiently inhibit gene expression either by preventing the splicing of pre-mRNA or by blocking the translation of mRNA. Our previous study has revealed that injection of pu.1 MOs at one-cell stage specifically blocks the development of RBI microglia but has little effect on the formation of AGM microglia (28). We therefore used pu.1 MOs to selectively deplete the RBI microglia and then tested if the short course of 4-OHT treatment might accidentally label AGM microglia (Fig. 1A). Results showed that the pu.1 MO-injected Tg(mfap4:CreER;mpeg1:LRLG) embryos contained no or few GFP+ microglia after 4-OHT treatment (n = 17/17) (Fig. 1C), indicating that this short course of 4-OHT treatment indeed labels the RBI microglia with high specificity. To further support this conclusion, we analyzed the distribution pattern of GFP+ macrophages in the 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) fish at around 3 weeks postfertilization (wpf), when the RBI-derived macrophages are diminished in the periphery and the majority of them have colonized the brain (15). As anticipated, by 3 wpf, abundant GFP+ microglia were detected exclusively in the brain (spinal cord as well) but not in the periphery in the 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) fish (Fig. 1D). These observations show that the RBI and AGM microglia can be well-separated by a short course of 4-OHT treatment (18 to 23 hpf) in the Tg(mfap4:CreER;mpeg1:LRLG) fish.
Fig. 1. The developmental dynamics of RBI and AGM microglia.
(A) A timeline indicates 4-OHT treatment and fluorescent imaging of Tg(mfap4:CreER;mpeg1:LRLG) embryos. (B) Fluorescence imaging of microglia in the brain of 3 dpf Tg(mfap4:CreER;mpeg1:LRLG) embryos treated with 4-OHT from 18 to 23 hpf (top) and quantification of the label efficiency by percentage (GFP+ cells in total microglia in 3-dpf brain) (bottom) (Ctrl n = 105, +4-OHT n = 119). (C) Fluorescence imaging of microglia in the brain of 3 and 9 dpf control or pu.1 MO injected Tg(mfap4:CreER;mpeg1:LRLG) embryos treated with 4-OHT from 18 to 23 hpf. (D) Fluorescence imaging of 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) larva at 9 dpf and 3 wpf. Zebrafish larvae were in lateral view with anterior to the left to visualize the distribution of GFP+ cells throughout the body. (E) Schematic diagram shows the experimental setup for the treatment of embryos with 4-OHT and collection of samples for cryo-section. (F) Quantification of the absolute number of RBI and AGM microglia in the developing and adult midbrain. (G) Quantification of the density of RBI and AGM microglia in the developing and adult midbrain. (H) Quantification of the proportion of RBI and AGM microglia in the developing and adult midbrain. (F) to (H) Data were presented as mean ± SEM (3 dpf n = 12, 3 wpf n = 5, 1 mpf n = 5, 1.5 mpf n = 5, 2 mpf n = 5, 2.5 mpf n = 5, 3 mpf n = 4, and 4 mpf n = 3).
RBI and AGM microglia pools undergo dynamic changes during zebrafish development and maturation
To monitor the dynamic changes of the RBI and AGM microglia from early developmental stage to adulthood, we applied above labeling method to convert RBI microglia to GFP+ cells in the Tg(mfap4:CreER;mpeg1:LRLG) fish. Since only the embryos with >90% RBI microglia labeling efficiency were preselected for the experiments in this study, we therefore assumed that the majority of the GFP+ and GFP−DsRed+ cells in the brains represented the RBI microglia and AGM microglia, respectively. Because the unlabeled cells are less than 10%, they would not affect the overall observations. To monitor the dynamic changes of the RBI and AGM microglia, the number, density, and proportion of the RBI (GFP+) and AGM (GFP−DsRed+) microglia in the brains of the selected fish were then determined at 3 wpf, 1 month postfertilization (mpf), 1.5 mpf, 2 mpf, 2.5 mpf, 3 mpf, and 4 mpf. To characterize the attenuation of RBI microglia in details, we quantified the microglia in the midbrain, retina, and spinal cord, after cryo-section and immunostaining (Fig. 1E). We first focused on the midbrain where it contains most abundant microglia. Results showed that the total number of microglia (RBI + AGM microglia) in the midbrain increased steadily as fish grew and matured from 3 to 4 mpf (Fig. 1F, blue). The steady increase of microglia appeared to correlate well with the expansion of the brain volume (fig. S2A). We noticed that, despite a marked drop in early juvenile stage (from ~350/per mm2 in 3 wpf to ~120/per mm2 in 1.5 mpf) (Fig. 1G, blue) possibly due to the rapid increase of the brain volume (fig. S2A), the overall density of microglia (the total microglia number on each brain slice divided by the brain slice area) remained relatively constant (around ~120/per mm2) from late-juvenile stage onward (Fig. 1G, blue). In contrast, RBI and AGM microglia behaved very differently during zebrafish development (Fig. 1, F to H, and fig. S2B). In early and mid-juvenile stages (3 to 1.5 mpf), the RBI microglia pool expanded rapidly (Fig. 1F, green), presumably to accommodate the rapid expansion of the brain volume (fig. S2A). Intriguingly, from 1.5 to 2 mpf, the growth rate of the RBI microglia slowed down and from 2 mpf onward, the number of RBI microglia rapidly declined and became almost undetectable by 4 mpf (Fig. 1F, green). On the other hand, AGM microglia expanded steadily throughout all developmental stages to accommodate the growth of the brain volume (Fig. 1F, red). Likewise, in the retina and spinal cord, RBI microglia showed similar trends of decline as fish aged, whereas AGM microglia gradually became the dominant population (fig. S3). These results indicate that the number and proportion of RBI and AGM microglia are profoundly influenced by the extrinsic environment as well as by the intrinsic property of each population.
RBI and AGM microglia manifest different proliferation potential and death rate
To delineate the cellular basis underpinning the dynamic shift of the microglia pools in the developing and adult brain, we measured the proliferation and death rate of RBI and AGM microglial populations by 5-bromo-2′-deoxyuridine (BrdU) incorporation assay and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining, respectively. In this experiment, the 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) embryos were raised to desired developmental stages and injected with BrdU for five consecutive days (one dose, per day). At 24 hours postinjection, the fish were sacrificed, fixed, and subjected to anti-BrdU and TUNEL staining after cryo-section (Fig. 2, A, B, and D). BrdU incorporation assay showed that the proliferation rate of RBI microglia increased steadily from early juvenile stage and reached the peak at around 1 mpf (Fig. 2C, green). Notably, the death rate of RBI microglia was also found to increase from early juvenile stage and peak at around 1 mpf (Fig. 2E, green), which is consistent with the previous findings showing that the proliferation and death of microglia in adult mice are usually coupled together to maintain the homeostasis of the microglia pool (12). However, from 1 mpf onward, the proliferation rate of RBI microglia dropped rapidly and reduced to minimum level by 3 mpf (Fig. 2C, green), while the death rate decreased only moderately and remained relatively high in adulthood (Fig. 2E, green). In contrast, the proliferation rate of AGM microglia increased progressively and continued to maintain at a relatively high level in adulthood (Fig. 2C, red), whereas the death rate declined overtime (Fig. 2E, red). Together, these results indicate that the dynamic shift from embryonic RBI microglia to adult AGM microglia is largely attributed to the different proliferation and death rate of the two microglia pools.
Fig. 2. BrdU and TUNEL assays reveal different proliferation and death rates of RBI and AGM microglia.
(A) Schematic diagram shows the experimental setup for BrdU and TUNEL assays. (B) Representative images of GFP+ RBI microglia and GFP−DsRed+ AGM microglia costained with BrdU and DAPI. (C) Quantification of the proliferation rate of RBI (BrdU+GFP+/GFP+) and AGM (BrdU+GFP−DsRed+/GFP−DsRed+) microglia. (D) Representative images of GFP+ RBI microglia and GFP−DsRed+ AGM microglia costained with TUNEL and DAPI. (E) Quantification of the death rate of RBI (TUNEL+GFP+/GFP+) and AGM (TUNEL+GFP−DsRed+/GFP−DsRed+) microglia. not significant (n.s.), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM (n = 5). i.p., intraperitoneal.
Depletion of AGM microglia leads to the expansion and prolonged lifespan of RBI microglia
The depletion of RBI microglia accompanied with the dominance of AGM microglia in adult brain prompted us to speculate that the attenuation of RBI microglia might be caused by the less competitiveness of RBI microglia in comparison with AGM microglia in the brain environment. To support this notion, we traced RBI microglia in runx1w84x mutants, in which the development of AGM-born hematopoietic cells, including microglia/macrophages, is largely abolished (29, 30), and asked whether the removal of AGM microglia would prolong the lifespan of RBI microglia (Fig. 3A). Contrary to the adult brains of control siblings where RBI microglia (GFP+) were rarely detected from 3 mpf onward (Fig. 3, B and C), runx1w84x adult mutants contained abundant amount of RBI microglia (GFP+) in the brains even at 5 mpf (Fig. 3B). Quantification analysis revealed that, while the overall number of microglia in adult runx1w84x mutants was comparable to that in control siblings (Fig. 3C), more than 60% of them were of RBI origin (Fig. 3D), indicating that the proportion and lifespan of RBI microglia are notably increased in runx1w84x mutants. The expansion and prolonged lifespan of RBI microglia observed in runx1w84x mutants was not caused by the direct effect of runx1 gene mutation on RBI microglia but rather due to the presence of the AGM microglia, as restoration of the AGM microglia by transplanting WT adult kidney marrow cells (which contain AGM-derived macrophages) into 2-dpf runx1w84x mutants resulted in the complete replenishment of the RBI microglia with AGM microglia in the adult brains in the recipients (26). From these results, we conclude that the attenuation of RBI microglia in adult fish is largely ascribed to the competition from AGM microglia.
Fig. 3. Expansion and prolonged lifespan of RBI microglia in AGM microglia-deficient runx1w84x mutants.
(A) Schematic diagram shows the experimental setup for the tracing of RBI microglia in siblings and runx1w84x mutants. (B) Representative images of the midbrain cross section of 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) siblings and runx1w84x;Tg(mfap4:CreER;mpeg1:LRLG) mutant fish at 5 mpf. GFP+ and GFP−DsRed+ cells represent RBI and AGM microglia, respectively. (C and D) Quantification of the number (C) and proportion (D) of RBI (GFP+) and AGM (GFP−DsRed+) microglia in (B). n.s., P > 0.05; **P < 0.01; ***P < 0.001 (Student’s t test). Data were presented as mean ± SEM (sibling n = 5, runx1w84x n = 3).
Neuron-derived Il34 is a key extrinsic factor controlling the proportion and lifespan of RBI microglia
In general, three possible mechanisms, i.e., competition for survival signals, direct fitness comparison, and mechanical force sensing can lead to cell competition (16, 17). Given that both direct fitness comparison and mechanical force sensing require direct cell-cell contact between the competing cells, we reasoned that competition for survival signals would likely be the mechanism mediating the competition between RBI and AGM microglia. We hence focused on the neuron-secreted trophic factor IL34 (fig. S4), a cytokine, together with its receptor CSF1R, which has been shown to be essential for microglia migration and survival in mammal and zebrafish (31–35). To test the role of the IL34 in the establishment and turnover of microglia, we crossed the Tg(mfap4:CreER;mpeg1:LRLG) transgenic reporter line with the Tg(Xla.Tubb:il34) transgenic fish (Fig. 4A), in which il34 is overexpressed under the control of the neuron-specific β-tubulin (Xla.Tubb) promoter (27), and asked whether overexpressing il34 in the brain, which presumably relieved the competition pressure, would alter the proportion and lifespan of RBI microglia. il34 overexpression led to a fourfold increase of total microglia in the adult brain of the transgenic fish (Fig. 4, B and C), and about half of the microglia in adult fish were of RBI origin and this ratio was maintained at least until 6 mpf (Fig. 4D). These data support the notion that the attenuation of RBI microglia in adult fish is due to their less competitiveness for brain-derived trophic factor Il34, and ectopically producing excessive amount of Il34 is sufficient to maintain the proportion and prolong the lifespan of RBI microglia. To further support this notion, we analyzed the microglia in il34-deficient mutants (Fig. 4E) (31) and asked whether the removal of Il34, which presumably leads to the increase of competition pressure, would have a more profound effect on RBI microglia maintenance. Results showed that, while AGM microglia remained comparable between mutants and siblings, RBI microglia had an about 50% decrease in il34-deficient mutants at 2 mpf (Fig. 4, F to H). This 50% reduction of RBI microglia in 2-mpf il34 mutants was not due to the colonization or developmental defect during zebrafish early development (31), as RBI microglia in the mutants gradually recovered from 6 dpf onward and displayed only ~20% reduction by 9 dpf (fig. S5). The moderate reduction of RBI microglia in il34-deficient mutants was likely due to the compensation of csf1a and csf1b (two other Csf1r ligands in zebrafish) (31), as RBI microglia was further decreased in il34;csf1a;csf1b triple mutants (fig. S6, A to D). Consistent with this hypothesis, ectopic overexpression of csf1a also significantly expanded the population of RBI microglia by 3 mpf, although the effect was much weaker than il34 overexpression (fig. S6, E to H). These observations demonstrate that the availability of Il34 and Csf1 in the brain is the key extrinsic factor regulating the proportion and lifespan of RBI microglia, and the attenuation of RBI microglia in adulthood is due to their less competitiveness for neuron-derived factors, predominately the Il34.
Fig. 4. Alteration of neuron-derived Il34 level changes the proportion and lifespan of RBI microglia.
(A) Schematic diagram shows the experimental setup for RBI microglia tracing in WT and Tg(Xla.Tubb:il34) fish. (B) Representative images of the midbrain cross section of 4-OHT–treated Tg(mfap4:creER;mpeg1:LRLG) and Tg(Xla.Tubb:il34;mfap4:creER;mpeg1:LRLG) fish at 6 mpf. GFP+ and GFP−DsRed+ cells represent RBI and AGM microglia, respectively. (C and D) Quantification of the number (C) and proportion (D) of RBI (GFP+) and AGM (GFP−DsRed+) microglia in (B) (n = 3 for each genotype). (E) Schematic diagram shows the experimental setup for RBI microglia tracing in siblings and il34−/− mutants. (F) Representative images of the midbrain cross section of 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) siblings and il34−/−;Tg(mfap4:CreER;mpeg1:LRLG) mutant fish at 2 mpf. GFP+ and GFP−DsRed+ cells represent RBI and AGM microglia, respectively. (G and H) Quantification of the number (G) and proportion (H) of RBI (GFP+) and AGM (GFP−DsRed+) microglia in (F) (sibling n = 6, il34−/− n = 5). n.s., P > 0.05; *P < 0.05; ***P < 0.001; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM.
Age-dependent reduction of csf1ra expression contributes to the less competitiveness and attenuation of RBI microglia
To dissect the molecular basis underlying the different competition capability of RBI and AGM microglia for the trophic factor Il34, we analyzed the expression of the Il34 receptor, csf1ra, in each microglia population at 2 mpf by RNAscope single-molecule fluorescent in situ hybridization (smFISH) (Fig. 5A) (36). To quantify the csf1ra smFISH signals in each microglia population, we performed multicolor staining consisting of RNAscope, anti-GFP (label RBI microglia), anti-Lcp1 (label both RBI and AGM microglia), and 4′,6-diamidino-2-phenylindole (DAPI) to distinguish the smFISH signals in RBI (GFP+Lcp1+DAPI+) and AGM (GFP−Lcp1+DAPI+) microglia. On the basis of the smFISH signals, we classified the csf1ra+ microglia into four categories (C1 to C4) based on the average smFISH signals: csf1rarare (1 to 9 dots per cell), csf1ralow (10 to 20 dots per cell), csf1ramed (21 to 30 dots per cell), and csf1rahi (>30 dots per cell) (Fig. 5, A and B). As we anticipated, the proportion of high csf1ra expression cells (20 to 30 dots per cell and >30 dots per cell) in AGM microglia (45.2%) was significantly higher than that in RBI microglia (36.9%) (Fig. 5B), and vice versa, a higher percentage of low csf1ra expression cells (1 to 9 dots per cell and 10 to 20 dots per cell) was detected in RBI microglia (63.1%) compared to that in AGM microglia (54.8%) (Fig. 5B). The H-score quantification further confirmed that there was a significant lower expression of csf1ra in RBI microglia compared to that in AGM microglia (Fig. 5C). Collectively, these data indicate that the less competitiveness of RBI microglia for neuron-derived Il34 is largely attributed to their lower expression of csf1ra.
Fig. 5. Age-dependent decline of csf1ra expression accounts for the less competitiveness of RBI microglia.
(A) Diagram shows the design for RNAscope smFISH (red), anti-GFP (green), anti-Lcp1 (white), and DAPI (blue) staining (top) and the representative images of RBI microglia with high and rare csf1ra expression (bottom). (B) Quantification of four categories of RBI and AGM microglia expressing different csf1ra levels at 2 mpf (n = 4). (C) H-score quantification of csf1ra expression levels in RBI and AGM microglia (n = 4). (D) Average number of csf1ra smFISH dots in 3-dpf, 3-wpf, and 2-mpf RBI microglia and in 2-mpf AGM microglia (3 dpf n = 6, 3 wpf n = 4, 2 mpf n = 4). (E) Quantification of four categories of RBI microglia expressing different csf1ra level (3 dpf n = 6, 3 wpf n = 4, 2 mpf n = 4). (F) Diagram shows the experimental setup for RNAscope smFISH (red), anti-GFP (green), EdU or TUNEL (white), and DAPI (blue) staining (top) at 2 mpf, and representative images show the expression of csf1ra in EdU+GFP+ and TUNEL+GFP+ microglia at 2 mpf (bottom). (G) Quantification of csf1ra smFISH dots in EdU+GFP+, TUNEL+GFP+, and GFP+ (total) microglia in (F) (total n = 1255, TUNEL+ n = 9, EdU+ n = 18). (H) Diagram shows the design for RBI microglia tracing in WT, csf1ra+/−, and csf1ra−/− mutants (top) and quantification of the number and proportion of RBI (GFP+) and AGM (GFP−DsRed+) microglia in WT and csf1ra+/− fish at 2 mpf (WT n = 3, csf1ra+/− n = 4). n.s., P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM.
Since RBI and AGM microglia originate from distinct sources and are generated at different time windows during zebrafish development (15), two possible reasons may explain the lower expression of csf1ra in RBI microglia than in AGM microglia at 2 mpf: RBI microglia are born with lower expression of csf1ra or the csf1ra expression declines in an age-dependent manner as RBI microglia are born much earlier than AGM microglia. To distinguish these two possibilities, we performed RNAscope smFISH to compare the csf1ra expression in nascent (3 dpf) RBI microglia with that in 2 mpf AGM microglia as well as with that in young (3 wpf) and aged (2 mpf) RBI microglia. Quantification of the csf1ra smFISH signals revealed that the average expression level of csf1ra in nascent RBI microglia was significantly higher than that in 2 mpf AGM microglia but steadily declined in young and aged RBI microglia (Fig. 5D). Accordingly, the proportions of high csf1ra expression cells (21 to 30 dots per cell and >30 dots per cell) were dominant in nascent RBI microglia (95.7%) but moderately reduced in young RBI microglia (51.8%) and further decreased in aged RBI microglia (36.9%) (Fig. 5E). These observations suggest that the less competitiveness of RBI microglia is closely associated with age-dependent decline of csf1ra expression. Moreover, when we costained csf1ra smFISH signals with 5-ethynyl-2-deoxyuridine (EdU) (labeling proliferation cells) and TUNEL (labeling apoptotic cells), respectively, at 2 mpf (Fig. 5F), we found that the expression levels of csf1ra were significantly higher in proliferating microglia (EdU+GFP+) than that in the dying microglia (TUNEL+GFP+) or overall (GFP+) microglia (Fig. 5G), indicating that the expression levels of csf1ra is directly associated with the fitness of RBI microglia. To prove that this is indeed the case, we analyzed the maintenance of RBI microglia in csf1ra heterozygous (csf1ra+/−) fish (Fig. 5H), in which the development of RBI microglia is largely unaffected (31, 37), and asked whether partial reduction of Csf1ra function would have more profound effects on the competition capability of RBI microglia. RBI microglia were significantly reduced in csf1ra+/− heterozygous fish by 2 mpf compared to that in WT control fish (Fig. 5H and fig. S7, B to D). Furthermore, when we generally overexpressed csf1ra in both RBI and AGM microglia in the csf1ra+/− fish using the macrophage-specific mpeg1 promoter, which presumably reduces the difference of csf1ra expression between RBI and AGM microglia, it significantly increased the number and proportion of RBI microglia (fig. S8), suggesting that reducing the difference of csf1ra expression level between RBI and AGM microglia indeed enhances the competitiveness and promote the maintenance of RBI microglia. Together, these results demonstrate that the attenuation of RBI microglia in adulthood is due to, at least in part, by the age-dependent decline of the csf1ra expression, reducing their competitiveness for neuron-derived trophic factor Il34.
IL34-CSF1R signaling is a general pathway regulating the fitness and lifespan of microglia
The above data indicate that the Il34-Csf1r signaling pathway facilitates the elimination of RBI microglia in an age-dependent manner. We speculated that this mechanism serves as a general principle to eliminate the aged microglia regardless the origin of microglia. To test this hypothesis, we turned to AGM microglia and asked whether a similar mechanism was used for the elimination of aged cells within AGM microglia pool. To do so, we first monitored the dynamic changes of the csf1ra expression and tested whether its expression level was down-regulated in aged-dependent manner in AGM microglia. Because the lifespan of AGM microglia is currently unknown and AGM microglia are presumably maintained via self-renewal, we therefore used a pulse-chase labeling strategy to define young and aged AGM microglia. In this experiment, WT Tg(mpeg1:eGFP) adult zebrafish (elder than 6 mpf), where microglia are exclusively of the AGM origin (Fig. 1, F to H) (15), were intraperitoneally injected with EdU for five or seven consecutive days (one dose per day) and collected at 1 and 30 days postinjection (dpi; Fig. 6A). In principle, the EdU+ microglia at 1 dpi are considered as young microglia (newly proliferated cells), whereas the EdU+ microglia at 30 dpi represent relatively aged microglia as these cells have experienced longer intervals from the last cell division. Consistent with our hypothesis, we found that the average expression levels of csf1ra in 30-dpi EdU+ AGM microglia (aged microglia) were significantly lower than that in 1-dpi EdU+ AGM microglia (young microglia) (Fig. 6B), suggesting that AGM microglia indeed undergo age-dependent reduction of csf1ra expression. Moreover, costaining of csf1ra smFISH signals with EdU and TUNEL (Fig. 6A) further revealed that the expression levels of csf1ra in the proliferating (EdU+GFP+) microglia were significantly higher than that in the dying microglia (TUNEL+GFP+) or overall (GFP+) microglia (Fig. 6, C and D). These data indicate that, like RBI microglia, the fitness and maintenance of AGM microglia are determined by the expression levels of csf1ra. This conclusion was further supported by the observations showing that the number of AGM microglia in 5 mpf csf1ra+/− heterozygous and csf1ra−/− homozygous fish was significantly lower than that in WT control (fig. S9). To further extend our findings from zebrafish to mammals, we analyzed the correlation of Csf1r expression levels and microglia fitness and maintenance by costaining of Csf1r RNAscope smFISH signals with EdU and TUNEL in 4-month-old male C57BL/6j mice (Fig. 6E). As expected, we found that the number of Csf1r smFISH signals in EdU+IBA1+ microglia was substantially higher than that in TUNEL+IBA1+ or total IBA1+ microglia (Fig. 6, F and G). Collectively, these data demonstrate that the IL34-CSF1R signaling is an evolutionary conserved pathway controlling the establishment, fitness, and lifespan of microglia from zebrafish to mammals.
Fig. 6. Csf1ra/CSF1R level determines the fitness of adult microglia in zebrafish and mice.
(A) Schematic diagram shows the experimental setup for RNAscope smFISH (red), anti-GFP/Lcp1 (green), EdU or TUNEL (white), and DAPI (blue) multicolor staining in adult zebrafish. (B) Quantification of csf1ra smFISH dots in EdU+GFP+ at 1 dpi and 30 dpi (1 dpi n = 3, 30 dpi n = 4). (C) Representative images show the expression of csf1ra in EdU+GFP+ and TUNEL+GFP+ microglia in adult zebrafish. (D) Quantification of csf1ra smFISH dots in EdU+GFP+, TUNEL+GFP+, and GFP+ (total) microglia at 1 dpi (total n = 312, TUNEL+ n = 7, EdU+ n = 18). (E) Schematic diagram shows the experimental setup for RNAscope smFISH (red), anti-IBA1 (green), EdU or TUNEL (white), and DAPI (blue) multicolor staining in adult mice. (F) Representative images show the expression of Csf1r in EdU+IBA1+ and TUNEL+IBA1+ microglia in 4-month-old C57BL/6j mice. (G) Quantification of csf1r smFISH dots in EdU+IBA1+, TUNEL+IBA1+, and IBA1+ (total) microglia shown in (F) (total n = 1348, TUNEL+ n = 18, EdU+ n = 32). **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM.
DISCUSSION
It is well documented that both YS-derived macrophages and fetal liver–derived monocytes (FL-Mos) are capable of colonizing various tissues during early mouse embryo development (7, 38). However, in adult stage, the tissue-resident macrophage populations in different mouse tissues are quite heterogeneous. For example, microglia in the brain are largely of YS origin (5, 7), while alveolar macrophages in the lung and Kupffer cells in the liver predominantly arise from FL-Mos (38–41). Yet the mechanisms underlying the heterogeneity of tissue-resident macrophages in different organs remain poorly defined. Our findings suggest that the heterogeneity and lifespan of tissue-resident macrophages in various tissues are determined, at least in part, by the ability of each population to compete for the survival signals produced by the microenvironments. Consistent with this notion, previous studies in mammals showed that granulocyte-macrophage colony-stimulating factor secreted by lung epithelial cells can induce FL-Mos into long-lived alveolar macrophages (39, 42), and YS-derived primitive macrophages, which are not normally present in the alveolar niche of adult mouse, can develop into functional alveolar macrophages in the absence of FL-Mo-derived macrophages (43). These observations, together with our findings, indicate that competition for survival factors produced by the niches is a general mechanism governing the establishment, composition, and lifespan of tissue-resident macrophages across different species.
Our results pinpoint Il34-Csf1r axis as a key cascade in regulating the proportion and lifespan of RBI and AGM microglia. We speculate that the Il34-Csf1r axis must be tightly controlled in steady state to maintain the fitness and number of microglia, and dysregulation of this pathway may lead to aberrant activity of microglia and neurons. Chronic activation of microglia by IL34 has been shown to cause aberrant microglia proliferation and promote neuroinflammation and neurodegeneration in the prion disease mouse model (44) as well as to exacerbate mutant Huntingtin (mHTTx1)–induced degeneration of striatal medium spiny neurons in a rat brain tissue slice model (45). Yet the mechanism controlling Il34-Csf1r signaling remains undefined. In mammals, IL34 has been shown to be mainly produced by neurons in the regions of the cortex, hippocampus, and striatum (32, 33, 46). Intriguingly, the mHTTx1 aggregates and the excitotoxin NMDA have been shown to elicit robust induction of IL34 transcripts in human dopaminergic neurons (45), indicating that neuronal stress and aberrant excitation of neurons may modulate the expression of IL34. As microglia in steady state constantly extrude/retract their process to surveil the surrounding environment and frequently contact neurons (47), it is reasonable to speculate that the interplay of microglia and neurons may have a role in maintaining the homeostasis of microglia through IL34-CSF1R pathway. This notion is supported by quantitative reverse transcription polymerase chain reaction data showing that a transient depletion of microglia in adult zebrafish brain leads to a significant increase of il34 expression (fig. S10). Similarly, although the transcriptional regulation and chromatin architecture of Csf1r locus have been discussed in previous reviews (48, 49), little is known about the mechanism underlying the differential expression levels of csf1ra in RBI and AGM microglia. Considering RBI microglia are born and colonize the brain earlier than AGM microglia, we believe that the relatively low csf1ra expression in RBI microglia is possibly because they age earlier than AGM microglia. We found that the expression level of csf1ra in RBI microglia is clearly lower in late stage comparing to that in early stage (Fig. 5D). Likewise, we also noticed a significant decrease of csf1ra expression in aged microglia in comparison with young cells in AGM microglia population (Fig. 6B). It will be of great interest to set up interdisciplinary methods to explore the upstream regulators or epigenetic modifiers that control the expression of csf1ra during the aging of microglia.
Another unsolved question is whether the dynamic shift of microglia pool from embryo to adult has any physiological relevance. A recent study by Li et al. (50) has shown that microglia in neonatal mice, but not adult mice, mediate scar-free wound healing after spinal cord crush injury by transiently secreting fibronectin for bridge formation of the crushed site and peptidase inhibitors for inflammation resolving. Although the phenotypical differences between neonatal and adult microglia can be ascribed to age-dependent phenotype transition shaped by the CNS niches, the possibility of distinct origins of neonatal and adult microglia cannot be ruled out, as dual origins of microglia in mice has also been suggested (51, 52). It would be interesting to investigate the role of RBI and AGM microglia during axon regeneration in zebrafish. Our data show that after 1.5-mpf RBI microglia manifest low proliferation ability and high apoptotic rate, which resemble the features of senescent/aged microglia (53, 54). In this regard, the elimination of RBI microglia may be beneficial to the homeostasis of the CNS, as senescent microglia have been suggested to elicit robust expression of senescent-associated secretory phenotype, such as proinflammatory cytokines like tumor necrosis factor–α, IL-1β, IL-6, and IL-8 etc., which are detrimental to neurons and have been linked to the onset and progression of several neurodegenerative diseases (54, 55). Hence, understanding the functional differences between RBI and AGM microglia may provide previously unknown cues about the physiological relevance of the heterogeneity of microglia in general.
MATERIALS AND METHODS
Zebrafish husbandry and lines used
Zebrafish were maintained according to the standard protocol (56). runx1w84x (30), Tg(Xla.Tubb:il34)hkz19Tg (31), Tg(Xla.Tubb:csf1a)hkz17Tg (31), il34hkz11 (31), csf1ahkz9 (31), csf1bhkz10 (31), csf1raj4e1 (37), Tg(mpeg1:LRLG) short for Tg(mpeg1:loxP-DsRedx-loxP-eGFP)hkz015Tg (27), Tg(mpeg1:loxP-eGFP)hkz032Tg (57), Tg(mfap4:CreER)hkz036Tg, and Tg(mpeg1:csf1ra)hkz037Tg were used in this study. For all the experiments, sibling or mutant/transgenic fish were raised under the same condition or in the same tank, and they were genotyped and chosen randomly without gender bias.
Mouse strains
Four-month-old male C57BL/6j WT mice were used and purchased from Jiangsu Wukong Biotechnology Co. Ltd. (China). All animal experiments and procedures were proved by Ethics Committee for The Welfare of Laboratory Animals in Shenzhen PKU-HKUST Medical Center (reference number for approval: 2022-014).
Generation of transgenic zebrafish line
The 1.6-kb promoter of mfap4 gene (25) and coding sequence of CreER were cloned into the modified pBluescript II SK(+) vector containing two arms of Tol2 sequence. Similarly, the 4-kb promoter of mpeg1 gene (42) and coding sequence of csf1ra were cloned into the modified pBluescript II SK(+) vector containing two arms of Tol2 sequence. The resulting mfap4-CreER or mpeg1-csf1ra construct, together with the mRNA of transposase, was injected into one-cell stage–fertilized embryos (58). The injected embryos were raised to adult for germline transmission screening.
Tamoxifen treatment
Tg(mfap4:CreER) transgenic fish were outcrossed with Tg(mpeg1:LRLG) reporter line to generate Tg(mfap4:CreER;mpeg1:LRLG) double transgenic fish. After de-chorion, the Tg(mfap4:CreER;mpeg1:LRLG) embryos were incubated with 5 μM (Z)–4-OHT (H7904, Sigma-Aldrich) from 18 to 23 hpf, washed intensively with fresh egg water for at least five times, and changed to a new dish for further incubation.
Morpholino
pu.1 sp3 MO oligonucleotides (5′-AATAACTGATACAAACTCACCGTTC-3′) (59) were used in this study. Two nanoliters of pu.1 sp3 MO (0.5 mM) was injected into one-cell stage embryos.
Cryo-section
Three-wpf- and 1-mpf-old juvenile fish were anesthetized on ice and fixed in 4% paraformaldehyde (PFA) at 4°C overnight. Fish older than 1.5 mpf were anesthetized on ice, and their brains were dissected and fixed in 4% PFA at 4°C overnight. After washing with phosphate-buffered saline (PBS) at room temperature for 1 day and dehydration with 30% sucrose in PBS at 4°C overnight, the samples were soaked in coagulating solution [optimal cutting temperature (OCT) compound] for further cryo-section in transverse planes with 30-μm thickness.
BrdU incorporation, TUNEL staining, and immunostaining
BrdU (B5002, Sigma-Aldrich) was dissolved in PBS to a stock concentration of 10 mg/ml. A total of 166.7 μg BrdU per gram of body weight was intraperitoneally injected for five consecutive days at different desired stages. At 6 dpi, injected fish were sacrificed, and the brains were dissected for further cryo-section. For each fish, two adjacent brain section slices were collected separately on two slides and were subjected to anti-BrdU and TUNEL staining, respectively.
BrdU staining was performed as previously reported (60). TUNEL assay was performed using Click-iT Plus TUNEL assay (Invitrogen, C10619). Subsequent immunostaining was performed as previously described (61). Primary antibodies used in this study are anti-GFP antibody (ab6658, Abcam), anti-DsRed antibody (632496, Clontech), anti-BrdU antibody (11170376001, Roche), anti–caspase 3 antibody (559565, BD Biosciences), and anti-Lcp1 antibody (30). The secondary antibodies used in this study are Alexa 488 anti-goat antibody (A11055, Invitrogen), Alexa 555 anti-rabbit antibody (A31572, Invitrogen), and Alexa 647 anti-mouse antibody (A31571, Invitrogen).
Imaging
Fluorescent signals were imaged under a Zeiss LSM980 confocal microscope equipped with Plan-Apochromat 10×/0.45 M27 and Plan-Apochromat 20×/0.8 M27 objectives. The images were processed using ZEN3.1 software (maximum z-projection).
Quantification of microglia number, proportion, and density in tracing experiments
For general microglia counting and analysis, each group contained at least three fish, with the body length within ±2 mm of the average and at least three representative midbrain slices were chosen for each fish. Rare cases with GFP+ macrophages in periphery, which were nonspecifically labeled AGM microglia due to leaky CreER activity, were excluded in the experiments. The number of representative midbrain slices for microglia quantification in BrdU incorporation and TUNEL staining are as follows: 3 wpf/1 mpf fish, 4 slices; 1.5 mpf/2 mpf fish, 6 slices, 2.5 mpf/3 mpf/4 mpf fish, 8 slices. GFP and DsRed single-positive cells represent RBI and AGM microglia, respectively. The proportion is calculated by RBI/AGM microglial number divided by total microglial number. The absolute number is calculated by multiplying the average microglia number on slices with the total number of brain slices. The density of microglia is calculated by dividing the microglial number with the brain area.
RNAscope smFISH and immunostaining
Expression and localization of csf1ra and il34 was evaluated by RNAscope smFISH technology [Advanced Cell Diagnostics (ACD)] according to the manufacturer’s instructions. The multiplexed assay was performed on cryo-section at 14-μm thickness in transverse planes. Probes were hybridized and amplified using the RNAscope Multiplex Fluorescent Reagent Kit (323100, ACD) according to the instructions. Three-plex Negative Control Probe (ACD) was used in negative control group under the same conditions. After in situ hybridization, sequential GFP and Lcp1 (for csf1ra smFISH) or Elavl3 (HuC) (for il34 smFISH) immunostaining was performed as previously described (61). The primary antibodies used here are anti-GFP antibody (ab6658, Abcam), anti-Lcp1 antibody (30), and HuC/HuD monoclonal antibody (A21271, Invitrogen). The secondary antibodies used here are Alexa 488 anti-goat antibody (A11055, Invitrogen), Alexa 647 anti-rabbit antibody (A31573, Invitrogen), and Alexa 647 anti-mouse antibody (A31571, Invitrogen). Last, the slices were counterstained with DAPI and analyzed.
RNAscope smFISH, EdU incorporation/TUNEL staining, and immunostaining
EdU (A10044, Invitrogen) was dissolved in PBS to make a stock concentration of 10 mg/ml. For zebrafish, 2-mpf fish were intraperitoneally injected with 2.5 μl of EdU, and adult fish were injected 5 μl of EdU for five consecutive days. For mice, 4-month-old mice were intraperitoneally injected with 100 μl of EdU for five consecutive days. The injected fish and mice were sacrificed, and the brains were dissected for cryo-section. After RNAscope in situ hybridization, sequential EdU (Click-iT EdU Imaging Kit, Invitrogen, C10340) and TUNEL (Vazyme, A112-02 and A113-02) staining was performed according to the manufacturer’s instructions. Anti-GFP or anti-Lcp1 (fish) and anti-IBA1 (mice) immunostaining was used to identify microglia in the brain. Last, the slices were counterstained with DAPI. The primary antibodies used here are anti-GFP antibody (ab6658, Abcam), anti-Lcp1 antibody (30), and anti-IBA1 antibody (019-19741, FUJIFILM Wako). The secondary antibodies used here are Alexa 488 anti-goat antibody (A11055, Invitrogen), Alexa 488 anti-rabbit antibody (A21206, Invitrogen), Alexa 647 anti-goat antibody (ab150131, Abcam), and Alexa 647 anti-rabbit antibody (A31573, Invitrogen).
Image analysis and quantification of smFISH
smFISH signals were imaged under a Zeiss Celldiscoverer7/LSM900 microscope with Airyscan2 mode and with a Plan-Apochromat 20×/0.95 objective. Z-stack images with a 0.32-μm interval were used. Three representative midbrain sections per fish were selected, and at least half of brain of each section were imaged for analysis. Each mouse was imaged in three different areas: cortex, hippocampus, and thalamus. For EdU/TUNEL costaining slices, a Zeiss LSM980 microscope with Airyscan2 mode and with a LD C-Apochromat 40×/1.1 objective was used to image EdU+ or TUNEL+ microglia. The image processing was done by ZEN3.1/3.2 software.
For csf1ra/Csf1r smFISH signals analysis, Imaris 9.5.1 software was used. Briefly, Imaris Surface function was used to classify and contour RBI (GFP+Lcp1+DAPI+), AGM (GFP−Lcp1+DAPI+) microglia, and mouse adult microglia (IBA1+DAPI+). The Spots function was used to distinguish and calculate the number of csf1ra signals on the three-dimensional images. Then, the number of csf1ra/Csf1r signals in each microglia was acquired manually. On the basis of the csf1ra signals, the csf1ra+ RBI and AGM microglia were classified into four categories (C1 to C4): C1, csf1rarare (1to 9 dots per cell); C2, csf1ralow (9 to 20 dots per cell); C3, csf1ramed (20 to 30 dots per cell); and C4, csf1rahi (>30 dots per cell). The dynamic range of csf1ra expression is determined by generating the H-score for each individual. H-score was calculated following the guideline recommended by ACD for RNAscope data analysis: H-score = (1 × C1 cells %) + (2× C2 cells %) + (3× C3 cells %) + (4× C4 cells %).
Whole-mount in situ hybridization
Antisense digoxin (DIG)-labeled mfap4 RNA probe was synthesized in vitro. The embryos at desired stages were fixed by 4% PFA. Whole-mount in situ hybridization was performed following the standard protocol (56).
Statistical analysis
At least two independent replicated experiments were performed in this study except RNAscope smFISH costaining assays. Unpaired two-tailed Student’s t tests were used for statistical analysis. All the statistical analysis was performed using GraphPad Prism version 6. All values represent means with SEM. Statistical significance is shown as if not significant (n.s.), P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001.
Supplementary Material
Acknowledgments
We thank W. Chen and J. Pan in Shenzhen Peking University-Hong Kong University of Science and Technology Medical Center for assistance in the mouse experiment. We thank J. Xu from South China University of Technology for the discussion and suggestions in our manuscript preparation.
Funding: This work was supported by Shenzhen Science and Technology Innovation Committee Grant JCYJ20180223181227269 (T.Y.), National Natural Science Foundation of China Grant 31801211 (T.Y.), Major Program of Shenzhen Bay Laboratory (S201101002), the National Key Research and Development Program of China (2018YFA0800200), Research Grants Council of the Hong Kong Special Administrative Region Grants (16103920, AoE/M-09/12, T13-605/18-W, and T13-602/21-N), and Innovation and Technology Commission of the Hong Kong Special Administrative Region (ITCPD/17-9).
Author contributions: Conceptualization: T.Y. and Z.W. Methodology: T.Y. and H.K. Investigation: T.Y., H.K., X.W., Y.H., and J.W. Visualization: T.Y. and H.K. Supervision: Z.W. Writing—original draft: T.Y. Writing—review and editing: T.Y. and Z.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Correction (27 July 2023): The original caption for Fig. 1 swapped the descriptions of panels F and G. The figure caption has been corrected. The original Fig. 4A was incorrectly labeled as Tg(mfap4:CreER;mpeg1:LRLG) X Tg(Xla.Tubb:csf1a;mpeg1:LRLG). The figure has been corrected. The PDF and HTML have been updated.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Table S1
REFERENCES AND NOTES
- 1.D. Gomez-Nicola, V. H. Perry, Microglial dynamics and role in the healthy and diseased brain: A paradigm of functional plasticity. Neuroscientist 21, 169–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.I. Hristovska, O. Pascual, Deciphering resting microglial morphology and process motility from a synaptic prospect. Front. Integr. Neurosci. 9, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. K. Glass, K. Saijo, B. Winner, M. C. Marchetto, F. H. Gage, Mechanisms underlying inflammation in neurodegeneration. Cell 140, 918–934 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. Prinz, J. Priller, S. S. Sisodia, R. M. Ransohoff, Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat. Neurosci. 14, 1227–1235 (2011). [DOI] [PubMed] [Google Scholar]
- 5.F. Ginhoux, M. Greter, M. Leboeuf, S. Nandi, P. See, S. Gokhan, M. F. Mehler, S. J. Conway, L. G. Ng, E. R. Stanley, I. M. Samokhvalov, M. Merad, Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K. Kierdorf, D. Erny, T. Goldmann, V. Sander, C. Schulz, E. G. Perdiguero, P. Wieghofer, A. Heinrich, P. Riemke, C. Hölscher, D. N. Müller, B. Luckow, T. Brocker, K. Debowski, G. Fritz, G. Opdenakker, A. Diefenbach, K. Biber, M. Heikenwalder, F. Geissmann, F. Rosenbauer, M. Prinz, Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013). [DOI] [PubMed] [Google Scholar]
- 7.C. Schulz, E. G. Perdiguero, L. Chorro, H. Szabo-Rogers, N. Cagnard, K. Kierdorf, M. Prinz, B. Wu, S. E. W. Jacobsen, J. W. Pollard, J. Frampton, K. J. Liu, F. Geissmann, A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). [DOI] [PubMed] [Google Scholar]
- 8.B. Ajami, J. L. Bennett, C. Krieger, W. Tetzlaff, F. M. V. Rossi, Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10, 1538–1543 (2007). [DOI] [PubMed] [Google Scholar]
- 9.T. L. Tay, D. Mai, J. Dautzenberg, F. Fernández-Klett, G. Lin, Sagar, M. Datta, A. Drougard, T. Stempfl, A. Ardura-Fabregat, O. Staszewski, A. Margineanu, A. Sporbert, L. M. Steinmetz, J. A. Pospisilik, S. Jung, J. Priller, D. Grün, O. Ronneberger, M. Prinz, A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20, 793–803 (2017). [DOI] [PubMed] [Google Scholar]
- 10.V. Stratoulias, J. L. Venero, M.-È. Tremblay, B. Joseph, Microglial subtypes: Diversity within the microglial community. EMBO J. 38, e101997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.F. Provenzano, M. J. Pérez, M. Deleidi, Redefining microglial identity in health and disease at single-cell resolution. Trends Mol. Med. 27, 47–59 (2021). [DOI] [PubMed] [Google Scholar]
- 12.K. Askew, K. Li, A. Olmos-Alonso, F. Garcia-Moreno, Y. Liang, P. Richardson, T. Tipton, M. A. Chapman, K. Riecken, S. Beccari, A. Sierra, Z. Molnár, M. S. Cragg, O. Garaschuk, V. H. Perry, D. Gomez-Nicola, Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 18, 391–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.L. J. Lawson, V. H. Perry, S. Gordon, Turnover of resident microglia in the normal adult mouse brain. Neuroscience 48, 405–415 (1992). [DOI] [PubMed] [Google Scholar]
- 14.A. A. Babcock, M. Wirenfeldt, B. Finsen, in Microglia: Methods and Protocols, B. Joseph, J. L. Venero, Eds. (Humana Press, 2013), pp. 129–145. [Google Scholar]
- 15.J. Xu, L. Zhu, S. He, Y. Wu, W. Jin, T. Yu, J. Y. Qu, Z. Wen, Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in Zebrafish. Dev. Cell 34, 632–641 (2015). [DOI] [PubMed] [Google Scholar]
- 16.N. E. Baker, Emerging mechanisms of cell competition. Nat. Rev. Genet. 21, 683–697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Bowling, K. Lawlor, T. A. Rodríguez, Cell competition: The winners and losers of fitness selection. Development 146, dev167486 (2019). [DOI] [PubMed] [Google Scholar]
- 18.E. Moreno, K. Basler, G. Morata, Cells compete for Decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 416, 755–759 (2002). [DOI] [PubMed] [Google Scholar]
- 19.E. Moreno, K. Basler, dMyc transforms cells into super-competitors. Cell 117, 117–129 (2004). [DOI] [PubMed] [Google Scholar]
- 20.M. Yamamoto, S. Ohsawa, K. Kunimasa, T. Igaki, The ligand Sas and its receptor PTP10D drive tumour-suppressive cell competition. Nature 542, 246–250 (2017). [DOI] [PubMed] [Google Scholar]
- 21.J. Vaughen, T. Igaki, Slit-robo repulsive signaling extrudes tumorigenic cells from epithelia. Dev. Cell 39, 683–695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.G. T. Eisenhoffer, P. D. Loftus, M. Yoshigi, H. Otsuna, C. B. Chien, P. A. Morcos, J. Rosenblatt, Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E. Marinari, A. Mehonic, S. Curran, J. Gale, T. Duke, B. Baum, Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature 484, 542–545 (2012). [DOI] [PubMed] [Google Scholar]
- 24.M. Marques-Reis, E. Moreno, Role of cell competition in ageing. Dev. Biol. 476, 79–87 (2021). [DOI] [PubMed] [Google Scholar]
- 25.E. M. Walton, M. R. Cronan, R. W. Beerman, D. M. Tobin, The macrophage-specific promoter mfap4 allows live, long-term analysis of macrophage behavior during mycobacterial infection in Zebrafish. PLOS ONE 10, e0138949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. He, J. Chen, Y. Jiang, Y. Wu, L. Zhu, W. Jin, C. Zhao, T. Yu, T. Wang, S. Wu, X. Lin, J. Y. Qu, Z. Wen, W. Zhang, J. Xu, Adult zebrafish Langerhans cells arise from hematopoietic stem/progenitor cells. eLife 7, e36131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. Xu, T. Wang, Y. Wu, W. Jin, Z. Wen, Microglia colonization of developing zebrafish midbrain is promoted by apoptotic neuron and lysophosphatidylcholine. Dev. Cell 38, 214–222 (2016). [DOI] [PubMed] [Google Scholar]
- 28.T. Yu, W. Guo, Y. Tian, J. Xu, J. Chen, L. Li, Z. Wen, Distinct regulatory networks control the development of macrophages of different origins in zebrafish. Blood 129, 509–519 (2017). [DOI] [PubMed] [Google Scholar]
- 29.C. E. Burns, D. Traver, E. Mayhall, J. L. Shepard, L. I. Zon, Hematopoietic stem cell fate is established by the Notch–Runx pathway. Genes Dev. 19, 2331–2342 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.H. Jin, R. Sood, J. Xu, F. Zhen, M. A. English, P. P. Liu, Z. Wen, Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development 136, 647–654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S. Wu, R. Xue, S. Hassan, T. M. L. Nguyen, T. Wang, H. Pan, J. Xu, Q. Liu, W. Zhang, Z. Wen, Il34-Csf1r pathway regulates the migration and colonization of microglial precursors. Dev. Cell 46, 552–563.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Y. Wang, K. J. Szretter, W. Vermi, S. Gilfillan, C. Rossini, M. Cella, A. D. Barrow, M. S. Diamond, M. Colonna, IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13, 753–760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Greter, I. Lelios, P. Pelczar, G. Hoeffel, J. Price, M. Leboeuf, T. M. Kündig, K. Frei, F. Ginhoux, M. Merad, B. Becher, Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. R. P. Elmore, A. R. Najafi, M. A. Koike, N. N. Dagher, E. E. Spangenberg, R. A. Rice, M. Kitazawa, B. Matusow, H. Nguyen, B. L. West, K. N. Green, Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.B. Erblich, L. Zhu, A. M. Etgen, K. Dobrenis, J. W. Pollard, Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLOS ONE 6, e26317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.F. Wang, J. Flanagan, N. Su, L. C. Wang, S. Bui, A. Nielson, X. Wu, H. T. Vo, X. J. Ma, Y. Luo, RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D. M. Parichy, D. G. Ransom, B. Paw, L. I. Zon, S. L. Johnson, An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 127, 3031–3044 (2000). [DOI] [PubMed] [Google Scholar]
- 38.G. Hoeffel, J. Chen, Y. Lavin, D. Low, F. F. Almeida, P. See, A. E. Beaudin, J. Lum, I. Low, E. C. Forsberg, M. Poidinger, F. Zolezzi, A. Larbi, L. G. Ng, J. K. Y. Chan, M. Greter, B. Becher, I. M. Samokhvalov, M. Merad, F. Ginhoux, C-Myb+ erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Guilliams, I. de Kleer, S. Henri, S. Post, L. Vanhoutte, S. de Prijck, K. Deswarte, B. Malissen, H. Hammad, B. N. Lambrecht, Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. Gomez Perdiguero, K. Klapproth, C. Schulz, K. Busch, E. Azzoni, L. Crozet, H. Garner, C. Trouillet, M. F. de Bruijn, F. Geissmann, H. R. Rodewald, Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Sheng, C. Ruedl, K. Karjalainen, most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity 43, 382–393 (2015). [DOI] [PubMed] [Google Scholar]
- 42.C. Schneider, S. P. Nobs, M. Kurrer, H. Rehrauer, C. Thiele, M. Kopf, Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15, 1026–1037 (2014). [DOI] [PubMed] [Google Scholar]
- 43.L. van de Laar, W. Saelens, S. de Prijck, L. Martens, C. L. Scott, G. van Isterdael, E. Hoffmann, R. Beyaert, Y. Saeys, B. N. Lambrecht, M. Guilliams, Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44, 755–768 (2016). [DOI] [PubMed] [Google Scholar]
- 44.D. Gómez-Nicola, N. L. Fransen, S. Suzzi, V. H. Perry, Regulation of microglial proliferation during chronic neurodegeneration. J. Neurosci. 33, 2481–2493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. Khoshnan, A. Sabbaugh, B. Calamini, S. A. Marinero, D. E. Dunn, J. H. Yoo, J. Ko, D. C. Lo, P. H. Patterson, IKKβ and mutant huntingtin interactions regulate the expression of IL-34: Implications for microglial-mediated neurodegeneration in HD. Hum. Mol. Genet. 26, 4267–4277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. Wei, S. Nandi, V. Chitu, Y. G. Yeung, W. Yu, M. Huang, L. T. Williams, H. Lin, E. R. Stanley, Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J. Leukoc. Biol. 88, 495–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. Nimmerjahn, F. Kirchhoff, F. Helmchen, Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). [DOI] [PubMed] [Google Scholar]
- 48.R. Rojo, C. Pridans, D. Langlais, D. A. Hume, Transcriptional mechanisms that control expression of the macrophage colony-stimulating factor receptor locus. Clin. Sci. 131, 2161–2182 (2017). [DOI] [PubMed] [Google Scholar]
- 49.E. R. Stanley, V. Chitu, CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 6, ea021857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Y. Li, X. He, R. Kawaguchi, Y. Zhang, Q. Wang, A. Monavarfeshani, Z. Yang, B. Chen, Z. Shi, H. Meng, S. Zhou, J. Zhu, A. Jacobi, V. Swarup, P. G. Popovich, D. H. Geschwind, Z. He, Microglia-organized scar-free spinal cord repair in neonatal mice. Nature 587, 613–618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. De, D. Van Deren, E. Peden, M. Hockin, A. Boulet, S. Titen, M. R. Capecchi, Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development 145, dev152306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H.-R. Chen, Y. Y. Sun, C. W. Chen, Y. M. Kuo, I. S. Kuan, Z. R. Tiger Li, J. C. Short-Miller, M. R. Smucker, C. Y. Kuan, Fate mapping via CCR2-CreER mice reveals monocyte-to-microglia transition in development and neonatal stroke. Sci. Adv. 6, eabb2119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.B. Spittau, Aging Microglia—Phenotypes, functions and implications for age-related neurodegenerative diseases. Front. Aging Neurosci. 9, 194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. R. Stojiljkovic, Q. Ain, T. Bondeva, R. Heller, C. Schmeer, O. W. Witte, Phenotypic and functional differences between senescent and aged murine microglia. Neurobiol. Aging 74, 56–69 (2019). [DOI] [PubMed] [Google Scholar]
- 55.D. M. Angelova, D. R. Brown, Microglia and the aging brain: Are senescent microglia the key to neurodegeneration? J. Neurochem. 151, 676–688 (2019). [DOI] [PubMed] [Google Scholar]
- 56.M. Westerfield, The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio), ed. 4, (Univ. Oregon Press, Eugene. 2000). [Google Scholar]
- 57.T. Yu, H. Kuang, J. Chen, X. Lin, Y. Wu, K. Chen, M. Zhang, W. Zhang, Z. Wen, Tripartite-motif family protein 35-28 regulates microglia development by preventing necrotic death of microglial precursors in zebrafish. J. Biol. Chem. 295, 8846–8856 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. Kawakami, A. Shima, N. Kawakami, Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. U.S.A. 97, 11403–11408 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.H. Jin, L. Li, J. Xu, F. Zhen, L. Zhu, P. P. Liu, M. Zhang, W. Zhang, Z. Wen, Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 119, 5239–5249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.X. Li, Y. Lan, J. Xu, W. Zhang, Z. Wen, SUMO1-activating enzyme subunit 1 is essential for the survival of hematopoietic stem/progenitor cells in zebrafish. Development 139, 4321–4329 (2012). [DOI] [PubMed] [Google Scholar]
- 61.T. Wendl, K. Lun, M. Mione, J. Favor, M. Brand, S. W. Wilson, K. B. Rohr, pax2.1 is required for the development of thyroid follicles in zebrafish. Development 129, 3751–3760 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Table S1