Abstract
Neurogenesis in the developing human cerebral cortex occurs at a particularly slow rate owing in part to cortical neural progenitors preserving their progenitor state for a relatively long time, while generating neurons. How this balance between the progenitor and neurogenic state is regulated, and whether it contributes to species-specific brain temporal patterning, is poorly understood. Here, we show that the characteristic potential of human neural progenitor cells (NPCs) to remain in a progenitor state as they generate neurons for a prolonged amount of time requires the amyloid precursor protein (APP). In contrast, APP is dispensable in mouse NPCs, which undergo neurogenesis at a much faster rate. Mechanistically, APP cell-autonomously contributes to protracted neurogenesis through suppression of the proneurogenic activator protein–1 transcription factor and facilitation of canonical WNT signaling. We propose that the fine balance between self-renewal and differentiation is homeostatically regulated by APP, which may contribute to human-specific temporal patterns of neurogenesis.
The slow tempo of human cortical neurogenesis requires the Alzheimer's disease Amyloid Precursor Protein.
INTRODUCTION
The human brain has expanded over 15-fold since our divergence from old world monkeys and 3-fold since our divergence from a common ancestor with chimpanzees (1). The cerebral cortex (neocortex) accounts for two-thirds of brain size with enormous diversity in cell type, morphology, connectivity, and function. It is typically structured into six radial layers of cells with highly elaborate neuronal circuits and is responsible for complex cognitive behaviors including sensory perception and skilled motor planning, attention, language, emotion, and consciousness (2, 3). Human cortical neurogenesis begins at around 5 gestational weeks (GW) and is mostly completed by 28 GW (4, 5). An important factor underlying human cortical expansion is an intrinsic capacity of human cortical progenitors to generate very large numbers of neurons over extended periods of time relative to other mammals, while retaining a self-renewing progenitor state for a particularly long time, presumably through species-specific mechanisms that remain poorly understood (6, 7).
The highly conserved class I transmembrane amyloid precursor protein (APP) is broadly expressed throughout nervous system development and has been extensively studied because its proteolytic processing is linked to Alzheimer’s disease (AD) (8–10), yet its physiological function, especially in humans, is unclear. APP and its homologs in various species are involved in many biological processes such as axonal outgrowth after injury (11), endolysosomal pathway (12–14), stress response after hypoxia/ischemia (15, 16), cell signaling processes, and brain development and plasticity (17, 18). In the worm Caenorhabditis elegans, loss of the APP homolog Apl is lethal because of molting deficits, while causing mild and low penetrance defects in various aspects of neuronal differentiation, function, and survival in mouse and Drosophila (19). In humans, point mutations of even a single copy of APP cause familial AD (20). APP is highly expressed in human telencephalic neurospheres and during the differentiation and migration of cortical neurons (21–23), suggesting that it may be involved in neural stem cells proliferation, differentiation, and/or maturation (24, 25). In the adult rodent brain, APP is also abundantly expressed in progenitor cells found in the ventricular zone (VZ)–subventricular zone (SVZ) (26, 27), but its loss does not cause obvious deficits in neurogenesis. We generated APP-knockout human induced pluripotent stem cells (iPSCs) with two different genetic backgrounds and queried the potential effect on human cortical neurogenesis.
RESULTS
Increased generation of human cortical neurons in the absence of APP
To examine APP expression during human fetal cortical development, we queried single-cell RNA sequencing (scRNA-seq) data from the developing human cortex at GW6 to GW10 (28) (https://cells-test.gi.ucsc.edu/?ds=early-brain) and found that APP is present in all six types of cells, with expression detected in 23.50% of neuroepithelial cell (NEC), 35.45% of radial glial cell (RGC), 43.66% of intermediate progenitor cell (IPC) and 43.48% of neurons (Fig. 1, A to D) present in that dataset. Furthermore, this expression appears particularly dynamic as cells transition from NECs to RGCs and from RGCs to IPCs. We next queried a second scRNA-seq dataset from the human developing cortex at GW17 to GW18 (22) and found that APP is expressed in all 16 types of cells present in the human developing cortex (Fig. 1, E and F). We also confirmed APP expression in all the regions of human developing cortex from VZ to cortical plate (CP) at GW14 using RNA scope (Fig. 1, G and H). APP mRNA appears enriched on the apical/ventricular pole of RGCs (Fig. 1, I and J). Together, these data show that APP is expressed in human cortical progenitors throughout neurogenesis raising the question of what its function may be at that stage of human cortex development. To gain initial insight into whether APP is potentially required during human fetal development, we queried genetic loss of function data obtained from the human population by the Genome Aggregation Database project (29, 30). These analyses reveal that APP has a loss of function observed/expected (LOEUF) score of 0.42 (https://gnomad.broadinstitute.org/gene/ENSG00000142192?dataset=gnomad_r2_1), meaning that complete loss of APP causes ~60% developmental mortality, a rate unexpectedly higher than that of key neurodevelopmental genes such as Neurog2 (LOEUF = 0.54) and Ascl1 (LOEUF = 0.59) that show less than 50% developmental mortality (Fig. 1K). Together, these data suggest that APP may play an important role in human cortex development.
Fig. 1. APP expression in the developing human cortex.
(A to D) APP is expressed in all cell types detected by scRNAseq of the human fetal cortex during GW6 to GW10 (28) and (E and F) GW17 to GW18 of development (22). (G and H) APP is expressed in different regions of human developing cortex at GW14 from VZ to CP (scale bars, 1000 and 100 μm). (I and J) Enrichment of APP on apical pole of aRGCs. (K) APP loss of function results in significant developmental lethality in the human population with a lower observed/expected (LOEUF) score (0.42) compared to two key neurodevelopmental genes such as NEUROG2 (LOEUF = 0.54) and ASCL1 (LOEUF = 0.59). (L) APP presence in control iPSC-derived human NPCs was confirmed by Western blot.
To test this hypothesis, we generated APP-KO iPSCs by CRISPR-Cas9 (fig. S1, A to K; see methods for iPSC lines and experimental details) and chose three APP-KO clones and one APP isogenic control clone, which received Cas9 but was not mutated, for further analysis. We verified the karyotype and the APP locus in all clones to ensure isogenicity. Upon induction of cortical differentiation, the morphology and molecular identity of neural progenitor cells (NPCs) were confirmed with NESTIN, SOX2, SOX1, and PAX6 (fig. S2). Cortical identity was verified using FOXG1 and EMX2 (fig. S3, A to H). We first confirmed that APP protein is expressed in human cortical progenitors (Fig. 1L). Seven days after initiation of neuronal differentiation, we stained the cells for SOX2 (progenitors), doublecortin (DCX; intermediate progenitors and immature neurons), and Tubulin β-III (TUJ1; neurons) to determine cell type. We observed a notable increase in TUJ1 (fig. S3, I to P) and DCX (fig. S3, Q to X) expression 7 days post differentiation in all three APP-KO clones relative to the control clone. To examine whether this effect is general or brain-region specific, we generated motor neurons (31) from control and APP-KO iPSCs. While human cortical neurogenesis occurs over 3 to 4 months, spinal motor neurons are generated at a much faster rate in 2 to 3 weeks. We used a 17-day protocol with 10 days of neural induction and 7 days of neural differentiation. We chose two early time points (day 1 and day 4 after differentiation; i.e., days 11 and 14 of culture) that approximately correspond to the 7 days postdifferentiation time point in cortical neurons (fig. S4A). We first confirmed the expression of APP protein in motor neuron progenitor cells (day 10 of culture; fig. S4B) and then examined their differentiating daughters for expression of ISLET1 (MN marker) and TUJ1 (fig. S4, C to N). No significant differences were detected in the number of ISLET1+ cells in APP-KO2 and control (fig. S4O) at either time point. These data suggest that loss of APP preferentially affects the temporally slower cortical neurogenesis compared to the temporally faster motor neurogenesis.
To determine the neurogenic potential of control and APP-KO cortical NPCs, we sparsely labeled them (70 control NPCs and an average of 63.6 NPCs in the 3 APP-KO clones; table S1) with pLv794_pTrip_PromSynapsin1_GFP_DeltaU3, a green fluorescence protein (GFP)–expressing lenti-viral vector (32, 33) (Fig. 2A) and quantified the number of GFP+ cells progenitors (SOX2) and GFP+ neurons (TUJ1) at days 0, 7, and 30 after differentiation (Fig. 2, B to U). After 7 days, we observed a significant decrease in NPCs (Fig. 2V) paralleled by a significant increase in neurons over time (Fig. 2W), in all three APP-KO compared to control. Notably, while the proportion of neurons generated by APP-KO progenitors shows a plateau between days 7 and 30, the slope is still rising in controls (arrow in Fig. 2W), meaning that neurogenesis is still ongoing in the control background, consistent with previous reports (33). Quantitatively, we found that one control NPC produced on average eight NPCs and 3.6 neurons after 7 days of differentiation. In contrast, within the same time frame, each APP-KO NPC produced on average 1.25 NPCs and 22.9 neurons (Fig. 2X and table S1), indicating a major switch of cortical NPCs from self-amplifying state to neurogenic state. This means that, in the absence of APP, a starting pool of NPCs does not produce more progenitors but is nevertheless maintained to produce neurons.
Fig. 2. Following neurogenesis using sparse labeling of progenitors confirms increased neurogenesis in APP-KO clones.
(A) Schematic illustration of the sparse labeling approach using a lentiviral vector (pLv794_pTrip_PromSynapsin1_GFP_DeltaU3) to follow the fate of the progeny of labelled progenitors (icons adapted from BioRender.com). (B to U) Representative immunofluorescence images showing expression of SOX2 and TUJ1 within GFP+ cells in isogenic control and APP-KOs 7 days after differentiation (scale bar, 100 μm; N, neuron; P, progenitor). (V) Decrease in the number of SOX2+ cells normalized to GFP+ cells and (W) Increase in the number of TUJ1+ cells normalized to GFP+ cells in APP-KO clones compared to isogenic control within 7 days [two-way analysis of variance (ANOVA), ****P < 0.0001]. At 30 days after differentiation, the ratios of progenitors and neurons have plateaued in APP-KO clones. The ratios in isogenic control clone have reached almost similar levels as in KO clones, but they are still on a dropping (progenitors) and rising (neurons, gray arrow) trajectories, respectively. (X) Number of progenitors and neurons generated by a single progenitor in control and APP-KO at 7 days after differentiation shows that one control NPC produced in average eight NPCs and 3.6 neurons. In contrast, within the same time frame, each APP-KO NPC produced on average 1.25 NPCs and 22.9 neurons. See table S1 for the detail of the numbers. We used six coverslips per condition per time point with the total number of quantified GFP+ cells at DAY 0 (Control = 74; APP-KO1 = 65; APP-KO2 = 64; APP-KO3 = 90), day 7 (Control = 814; APP-KO1 = 860; APP-KO2 = 1892; APP-KO3 = 1868) and day 30 (Control = 657; APP-KO1 = 599; APP-KO2 = 524; APP-KO3 = 552).
Cortical neurons are generated on a shorter time scale in the absence of APP
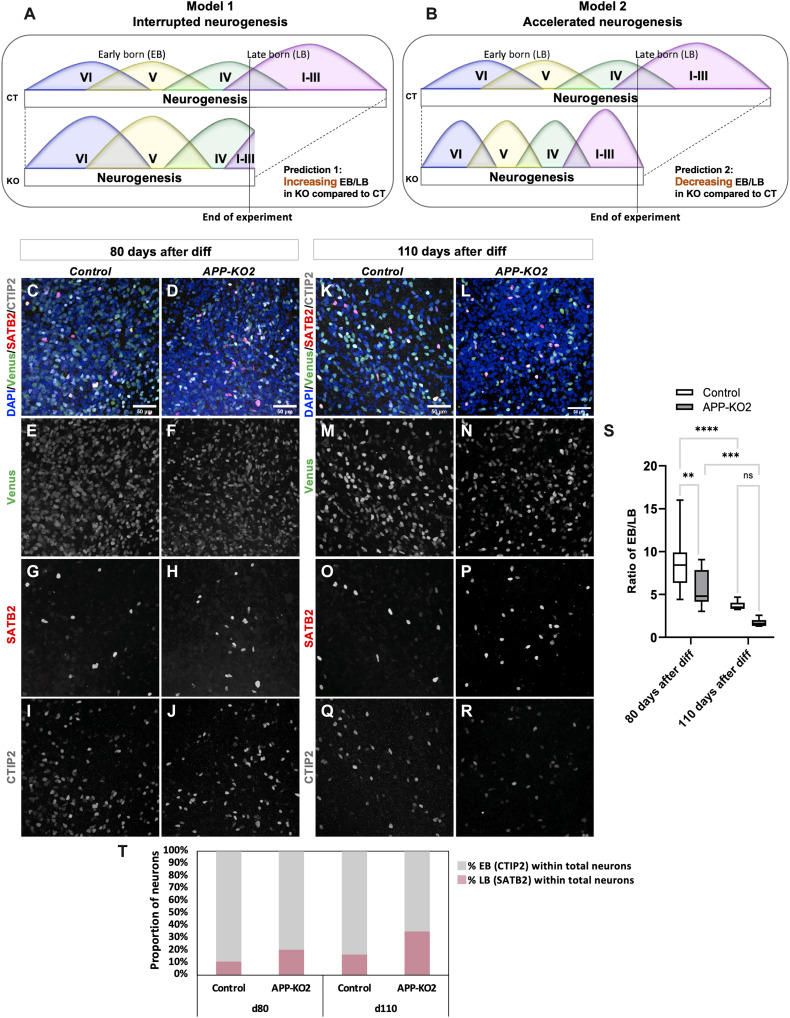
One of the key features of cortical development is the sequential generation of different neuronal subtypes that will occupy different layers of the neocortex. This feature is preserved in two-dimensional (2D) cultures, whereby early-born (EB; deep layer) neuronal fate markers appear before late-born (LB; upper layer) cell fate markers (7, 34). To assess whether loss of APP affects this order, differentiation was launched, and cells were tracked for 7, 15, and 30 days after differentiation. We first observed clear expression of EB neuronal marker CTIP2 at day 15 followed by SATB2 at day 30 in both genotypes.
Put together, our data so far can be explained by two very different models. In a first model, which we term the “interrupted neurogenesis” model, APP-KO progenitors initiate neurogenesis prematurely causing an exhaustion of the progenitor pool while they are still in their early temporal phases. In a second model, which we term the “accelerated neurogenesis” model, APP-KO progenitors go through their entire temporal program but in a much shorter period. These two models make diametrically opposite predictions for the EB-to-LB neuron ratio (EB/LB ratio) at different stage of neurogenesis. The first model predicts that the EB/LB ratio would be higher in APP-KO and would increase over time compared to control (Fig. 3A) because the mutant progenitors would be consumed before enough LB neurons are produced. The second model predicts that the EB/LB ratio would be initially lower in APP-KO and would decrease over time compared to control (Fig. 3B) because APP-KO progenitors would have completed their temporal program while control progenitors are still in early phases of generating LB neurons. However, eventually the control progenitors would “catch up” and the ratio would be similar between the two conditions. To test these predictions, we followed the cultures for 50, 80, and 110 days after differentiation. We first stained 50 days postdifferentiation cultures for the neuronal fate markers FOXP2 (EB) and SATB2 (LB) and found that the ratio of FOXP2/SATB2 (EB/LB ratio) is lower in all three APP-KO clones compared to the control reaching significance in two of the three cases (fig. S5, A to N) consistent with the accelerated neurogenesis model. Next, we followed the ratios of neuronal fate at 80 (Fig. 3, C to J) and 110 days (Fig. 3, K to R) after differentiation by transducing progenitors of control and APP-KO2 with pLv794_pTrip_PromSynapsin1_Venus-NLS_DeltaU3 and staining their neuronal progeny for Venus, CTIP2 (EB), and SATB2 (LB). We found that the difference in EB/LB ratios (CTIP2/SATB2) within the labeled neurons decreases overtime and is not significant anymore at 110 days after differentiation (Fig. 3, S and T). These data suggest that APP contributes specifically to the absolute temporal scale of human neurogenesis but not the relative temporal birth order of cortical neurons. To have an insight on maturation of control and APP-KO cortical neurons, we stained them for synapse markers. At 60 days after differentiation, neither control nor APP-KO neurons do express synaptic markers. In contrast, at 120 days after differentiation, both control and APP-KO neurons express presynaptic (MUNC13) and postsynaptic (PSD95) markers (fig. S5, O to V), and no significant difference was observed in synapse density per micrometer in control and APP-KO2 (fig. S5W). To determine the functional properties of cortical neurons, we produced cortical organoids from control and APP-KO2 using previously published protocol (35), and extracellular recordings were performed from cortical organoids at 160 days of differentiation using the multielectrode array (MEA) system (fig. S6, A and B). Representative raster plots and heat maps of neuronal network activity displays spontaneous activity in both control and APP-KO2 cortical organoids (fig. S6, C to F). Most organoids displayed spontaneous spiking activity, even though the known variability between organoids was also observed. No significant difference was observed between control and APP-KO2 for either the number of active electrodes or the mean firing rate (fig. S6, G and H). Together, these data suggest that loss of APP does not significantly alter the density of synapses or functional activity of the neurons.
Fig. 3. Accelerated production of different types of cortical neuron in APP-KO clones.
(A) The “interrupted neurogenesis” model in which APP KO progenitors are prematurely depleted before producing sufficient LB neurons predicts increasing EB-to-LB neuron ratio compared to isogenic controls (B) Conversely, the “accelerated neurogenesis” model in which APP KO progenitors undergo the same temporal neurogenic series but with faster dynamics predicts decreasing EB/LB neuron ratio at the same time point during active neurogenesis. (C to R) 80 and 110 days postdifferentiation neurons stained for Venus, CTIP2, and SATB2 in isogenic control and APP-KO2 (scale bar, 50 μm for all the images). (S) Lower ratio of EB/LB neurons that would decrease over time in APP-KO2 compared to isogenic control supports the accelerated neurogenesis model in the absence of APP (n = 3, ordinary two-way ANOVA, **P = 0.0025 for 80 days after diff Control versus 80 days after diff APP-KO2, ****P < 0.0001 for 80 days versus 110 days after diff Control, ***P = 0.0006 for 80 days versus 110 days after diff APP-KO2). (T) Percentage of CTIP2 and SATB2 cells within total neurons at 80 and 110 days after differentiation.
Loss of APP drives human cortical progenitors into neurogenic state
Given the expression of APP in fetal human cortical progenitors (22, 28) (Fig. 1, A to J) and the acceleration of neurogenesis observed in APP-KO cells, we hypothesized that that APP is required in the progenitors themselves to maintain cortical NPCs in a prolonged self-amplifying progenitor state. To test this idea, we quantified the ratio of NPCs versus IPCs and EB neurons by staining pure NPC cultures for SOX2 and DCX (Fig. 4, A to O) before the onset of neuronal differentiation, where there should be very little spontaneous differentiation under control conditions. Notably, we observed a significant decrease in the ratio of SOX2+ cells (Fig. 4P) and a significant increase in the ratio of DCX+ cells (Fig. 4Q) in APP-KOs relative to the isogenic parental (see Materials and Methods) and guide RNA/Cas9-treated controls. To rule out genetic background effects, we generated an APP-KO clone from a different independent iPSC line (iPSC line 2; see Materials and Methods for details) (fig. S1, L to N) and observed the same results in those NPCs (fig. S7, A to K). We also stained cortical organoids from control and APP-KO clones of two different iPSC lines for SOX2 and DCX at day 15, when most cells are in the NPC state (iPSC line 1; fig. S7, L to S, and iPSC line2; fig. S7, U to Z″). We performed 3D reconstructions of the organoids and quantified DCX expression (iPSC line 1; movies S1 and S2 and iPSC line2; movies S3 and S4). We observed more total DCX in APP-KO organoids relative to control in both genetic backgrounds (fig. S7, T and Z‴).Next, we performed a rescue experiment by transfecting APP-KO2 NPCs with integrative (PiggyBac) expressing vectors containing either GFP alone (pPB-CAG-IRES-EGFP) or GFP and APP (pPB-CAG-hAPP-IRES-EGFP) and staining for SOX2 and DCX. Quantification of GFP+/SOX2+ and GFP+/DCX+ cells showed that APP-KO NPCs that received the APP-containing vector but not GFP alone were restored to a progenitor state and the number of their neuronal progeny was reduced back to control levels (Fig. 4, R1 to V). We noted that restoring APP only rescued NPCs expressing it, but not neighboring cells (arrows in Fig. 4T2), suggesting a cell-autonomous requirement. Together, these data show that APP is cell-autonomously required to repress premature neurogenesis in human cortical NPCs. Last, to test whether this effect on cortical neurogenesis was specific to human cortical NPCs, we tested for signs of premature neuronal differentiation in the APP-KO mouse cortex at embryonic day 10.5 (E10.5), a stage when most cells should still be progenitors. Previous studies of the role of APP and its paralogs APLP1 and APLP2 in mouse neurogenesis have consistently reported contradictory results, including increased neurogenesis, decreased neurogenesis, and absence of any effects (36–39). We examined the developing mouse cortex at E10.5 and observed no Tuj1+ cells in the progenitor layer and no difference in Pax6 (NPC) and Tuj1 expression between App-WT and App-KO brains (fig. S8, A to N). We then examined the number of progenitors (Pax6), EB (Ctip2), and LB (Satb2) neurons at E17.5 toward the end of neurogenesis. We observed no significant change in the number of Satb2-, Ctip2-, or Pax6-positive cells between App-WT and App-KO brains (fig. S8, O to Z′). These results suggest that APP’s requirement in cortical progenitors may not be as critical in the mouse cortex as it is in humans, perhaps because mouse cortical neurogenesis is much faster.
Fig. 4. Loss of APP drives premature differentiation of human NPCs.
(A to O) NPCs in proliferating medium from parental, isogenic control and APP-KOs stained for SOX2/DCX. (P) Significant decrease in ratio of SOX2+ cells (n = 3, one-way ANOVA, P < 0.0001 for all the conditions). (Q) Significant increase in ratio of DCX+ in APP-KOs background compared to parental and isogenic control (n = 3, one-way ANOVA, *P = 0.0235, ****P < 0.0001, **P = 0.0061 for parental versus APP-KO1, APP-KO2 and APP-KO3 respectively. *P = 0.0487, ****P < 0.0001, *P = 0.0151 for isogenic control versus APP-KO1, APP-KO2 and APP-KO3, respectively; scale bar, 50 μm). (R1 to R5) Isogenic control NPCs transfected with pPB-CAG-IRES-EGFP. (S1 to S5) APP-KO2 NPCs transfected with pPB-CAG-IRES-EGFP. (T1 to T5) APP-KO2 NPCs transfected with pPB-CAG-hAPP-IRES-EGFP and stained for GFP/SOX2/DCX. APP rescued the phenotype of transfected cells but not neighboring cells (arrows in T2). (U) Ratio of SOX2+ and (V) DCX+ cells in all three conditions (n = 3 independent biological repeat, ordinary one-way ANOVA, ****P < 0.0001; scale bar, 50 μm). We used four coverslips per condition in each biological repeats. In total, 1431, 1252, and 1540 cells were quantified for Control-EGFP, APP-KO2-EGFP, and APP-KO2-EGFP-hAPP, respectively.
To gain insight into how APP regulates the fate of human cortical NPCs, we performed scRNA-seq on control and APP-KO NPCs using a novel, nonmicrofluidics-based method (table S2, BioProject accession number PRJNA678443). First, we confirmed that both control and APP-KO NPCs mapped to progenitor cells in the human fetal brain by integrating them with recently reported single-cell transcriptomes (22) using Integrated Anchors analysis followed by clustering with Seurat (Fig. 5, A and B) (40). Unsupervised clustering identified eight clusters as shown by uniform manifold approximation and projection (UMAP) 2D representation (Fig. 5C). To more precisely identify these cells, we performed clustering analyses according to cell identity, following the cell-type annotation of Polioudakis et al. (22), which confirmed the presence of closely related control and APP-KO clusters (UMAP 2D representation; fig. S9A and heatmaps; fig. S9, B and C) of RGCs, cycling progenitors 1 and 2 (CP1 and CP2), intermediate progenitors (IP) and neurons (N), as well as a cluster that we refer to as the neurogenic progenitors (NP) cluster (see below). NPs represent 13% of the APP-KO population and together with the neurons appear at the expense of cycling progenitors, which show a reduction of ~20% in APP-KO cells compared to control (fig. S9D), suggesting premature cell cycle exit and increased differentiation of APP-KO cells. Consistent with this, we found a 20% reduction in cells expressing the cell cycle marker Ki67 in APP-KO NPCs compared to controls (fig. S9, E to G). Because APP is known as stress response gene due to its elevated expression in brain injury and ischemia (41), part of this reduction might be due to general loss of cells caused by cell death. We investigated cell death by immunostaining for cleaved caspase 3. Although we found a small but significant increase in cell death in APP-KO background compared to control (fig. S9, H to J), the percentage of dying cells was well below 5% and therefore does not explain the observed reduction in proliferation. The NP cluster that represents cluster 4 of the previous analysis (Fig. 5C) contained both control and APP-KO cells, with the majority being APP-KO (Fig. 5D). To verify the neurogenic state of these progenitors, we performed SOX2/NEUROG2/DCX triple staining (Fig. 5, E to L) on control and APP-KO NPCs. NEUROG2 expression marks commitment to differentiation of mammalian NPCs (42, 43). We observed a ~15-fold increase in the number of NEUROG2+ progenitors in APP-KO compared to control (Fig. 5M). This is close to the increased ratio of neurons to progenitors (~18-fold) we found to be produced by APP-KO NPCs (table S1).
Fig. 5. APP-KO NPCs are in a faster temporal trajectory toward neurogenesis.
(A and B) scRNA-seq data from isogenic control and APP-KO NPCs both map to normal human progenitor cell types. (C and D) Cluster analysis without a priori cell identity identified eight clusters most of which are composed of similar ratios of isogenic control and APP-KO cells (see fig. S9) with the exception of cluster 4 where the majority of cells in cluster 4 are APP-KO NPCs. (E to L) Staining of isogenic control and APP-KO2 NPCs for NEUROG2/SOX2/DCX. (M) Ratio of NEUROG2+ neurogenic NPCs is 15-fold higher in APP-KO2 compare to isogenic control (unpaired t test, P < 0.0001; scale bar, 50 μm). (N) Cluster 4 cells map to oRG or a transitionary state from vRG to oRG in human fetal progenitors. (O) Pseudotime analysis of the scRNA-seq data shows a temporal shift toward a differentiated state in the whole population of APP-KO NPCs compared to isogenic control (****P = 8.418 × 10−13, Kolmogorov-Smirnov test).
To further understand the cell identity [ventricular radial glia (vRG), outer radial glia (oRG), and IPC] of NPs, we mapped them to human fetal brain cells and identified that most of them mapped to oRG or transitionary state from vRG to oRG (Fig. 5N) suggesting APP in the human fetal brain might influence the generation of oRG cells. This is supported by other evidence in our single cell data. First, some of oRG-enriched genes such as PTPRZ1 and FABP7 are markers of the NP cluster (table S2). Second, cell type analysis in control and APP-KO (fig. S9K) according to Polioudakis et al. (22), shows that while the majority of progenitors are vRGs under both conditions, there is a an increase in oRGs in APP mutants.
Our scRNA-seq dataset gave us the opportunity to further test the accelerated neurogenesis model by performing pseudotime analysis (44, 45) to determine the temporal trajectory of APP-KO NPCs relative to control. Pseudotime analysis shows a significant overall temporal shift toward a differentiated state in the APP-KO population compared to control (Fig. 5O) which was true for all classes of progenitors but not IPs and neurons (fig. S9L). It has been reported that lengthening of cell cycle duration increases neurogenesis at the expense of progenitors (46–48). We used the cumulative labeling assay (49) to measure cell cycle length. We treated control and APP-KO progenitors with 1 μM Edu and sampling was performed overtime from 2 to 55 hours with Edu continuously in the culture (fig. S10, A to I). While average cell cycle length in control is 36 hours, APP-KO progenitors shows protracted cell cycle length about 48 hours (fig. S10J). Together, our data show that loss of APP creates an accelerated drive toward neurogenesis in human cortical progenitors, suggesting that APP regulates downstream mechanisms required to maintain cortical NPCs in a progenitor state.
Loss of APP in human cortical progenitors triggers activator protein–1 activation to drive neurogenesis
To gain insight into the mechanisms of the gain of neurogenic state in the absence of APP, we performed bulk RNA sequencing in APP-KO and control NPCs. We obtained 763 differentially expressed genes (558 up; 205 down including APP as expected) between control and APP-KO2 NPC (table S3 and fig. S11, A and B). Consistent with the APP-KO phenotype, bulk RNA-seq showed down-regulation of primary progenitor’s markers PAX6 and CDON, and up-regulation of neurogenic genes NEUROG2, BCL6, ASCL1, NHLH1, and DCX. Moreover, CDKN2B, a well-known cell cycle inhibitor, and differentiation promoting factors DLL3, JAG2, and DNER were also up-regulated (fig. S11C). Consistent with the scRNA-seq data, we observed up-regulation of some oRG markers such as TNC, MOXD1, FABP7, and ACSBG1 in our bulk data (fig. S11D). Notably, TNC expression (4.2 log2FC in APP-KO compare to control) was also confirmed by Western blot showing approximately sixfold increase in protein level (fig. S11, E and F). One pathway whose activity is intimately linked to APP function in a variety of organisms and neuronal contexts is the JUN N-terminal kinase (JNK) pathway (50) whose effector proteins, JUN and FOS, combine to form the activator protein–1 (AP-1) transcription factor. Significant evidence supports APP-dependent transcriptional repression of c-Jun and reduced basal activity of c-Jun N-terminal kinase (JNK) in PC12 cells (51) with activation of the JNK signaling pathway reduced either by APP overexpression or treatment with sAPPα (51–53). Moreover, c-Fos expression is up-regulated in the prefrontal cortex of App-KO mice (54, 55). We observed increased expression of JUN and FOS mRNAs in APP-KO NPCs (1.3 and 2.7 log2FC, respectively; Fig. 6A), suggesting that APP also regulates their expression in human context. JUN is activated upon phosphorylation by JNK (56–58), and we observed a 1.5-fold increase in phospho-JUN in APP-KO NPCs compared to control (Fig. 6, B and C). Last, AP-1 regulates the expression of BCL6 in germinal-center B cells (59), and BCL6, which we found to be up-regulated in APP-KO NPCs, drives neurogenesis of cortical progenitors in the developing mouse cortex (60, 61). Analysis of genes characterizing the APP-KO NP cluster of our single-cell transcriptomic dataset showed enrichment for binding of JUN and JUND to their promoters by ChIP-X Enrichment Analysis (ChEA) (62); http://amp.pharm.mssm.edu/Enrichr/, Fig. 6D). Furthermore, we also found consensus AP-1 binding sites in the regulatory regions of genes up-regulated in APP-KO NPCs in the bulk RNA-seq data, including DCX (table S4). To test whether AP-1 activation might drive APP-KO NPCs into a neurogenic state, we treated control and APP-KO2 NPCs with a specific AP-1 inhibitor (SR11302) (63, 64) and stained them for SOX2, NEUROG2, and DCX (Fig. 6, E to T). While the ratio of SOX2+ cells remained unchanged after treatment (Fig. 6U), the number of NEUROG2+ and DCX+ cells was significantly rescued (Fig. 6, V and W). These results suggest that loss of APP causes premature differentiation through AP-1 activation independently of NPC self-renewal, indicating that there is a second APP-dependent mechanism for NPC amplification.
Fig. 6. Loss of APP increases differentiation of NPCs through activating the JNK pathway.
(A) Up-regulation of JUN and FOS in APP-KO2 NPCs in bulk RNA sequencing results (ordinary one-way ANOVA, P < 0.0001). (B and C) Western blot shows significant increase in phospho-JUN in APP-KO2 NPCs compared to isogenic control (n = 3 biologically independent repeats, unpaired t test, ****P < 0.0001). (D) JUN family members bind to marker genes of NPs in the ChEA database (http://amp.pharm.mssm.edu/Enrichr/). (E to T) NPCs treated with AP-1 inhibitor (AP-1 inh, SR11302) and stained for SOX2/DCX/NEUROG2 showed (U) no increase in ratio of SOX2+ cells in APP-KO2 treated with SR11302 compared to untreated APP-KO2 NPCs (n = 3, ordinary one-way ANOVA, ****P < 0.0001 for all the significant condition). (V) Significant decrease in NEUROG2+ cells in APP-KO2 treated with SR11302 compared to untreated APP-KO2 NPCs (n = 3, ordinary one-way ANOVA, ****P < 0.0001 for all the significant conditions). (W) Significant decrease in DCX+ cells in APP-KO2 treated with SR11302 compared to untreated APP-KO2 NPCs (n = 3, ordinary one-way ANOVA, ****P < 0.0001 for isogenic control-DMSO versus APP-KO2-DMSO, isogenic control-SR11302 versus APP-KO2-DMSO and APP-KO2-DMSO versus APP-KO2-SR11302 and ***P = 0.0008 for isogenic control-SR11302 versus APP-KO2-SR11302). Scale bar, 50 μm for all the images.
Loss of APP reduces human cortical progenitor amplification by attenuating the canonical WNT pathway
The canonical WNT contain different proteins including Frizzled family of proteins as main receptors, co-receptors LRP5/6 and downstream protein β-catenin and is required in a variety of contexts for stem cell maintenance (65, 66). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of down-regulated genes in our bulk RNA-seq data indicated that WNT signaling is reduced in APP-KO NPCs (Fig. 7A), with up-regulation of canonical WNT inhibitors DKK1, DKK3, KREMEN1, APCDD1, and ALPK2 (Fig. 7B). We also observed down-regulation of WNT7A and up-regulation of WNT7B in our dataset. We asked whether this reduction in WNT signaling might contribute to reduced NPC amplification in APP-KO NPCs. WNT3A is a prototypical example of a ligand that consistently activates the canonical WNT pathway (21, 22). We compared control and APP-KO NPCs treated with the canonical ligand WNT3A for 48 hours to untreated NPCs and stained them for SOX2 and DCX (Fig. 7, C to N) and proliferation marker Ki67+ (fig. S12, A to H). As expected, Wnt3A treatment induced increased proliferation in control progenitors. In APP-KO progenitors, we observed a complete rescue in proliferation (fig. S12I), a partial but significant rescue of the loss of SOX2+, but no reduction in DCX+ cells (Fig. 7, O and P). Thus, consistent with all our analyses, APP-KO progenitors lose their progenitor status more readily even when they are induced to proliferate but require suppression of JNK signaling to differentiate.
Fig. 7. Loss of APP decreases amplification of NPCs by attenuating the canonical WNT pathway.
(A) KEGG pathway analysis shows down-regulation of WNT signaling pathway in APP-KO background. (B) Up-regulation of canonical WNT inhibitors DKK1, DKK3, KREMEN1, APCDD1, and ALPK2 in APP-KO2 NPC compared to isogenic control (ordinary one-way ANOVA, ****P < 0.0001). (C to N) Isogenic control and APP-KO2 NPCs treated with canonical WNT ligand (WNT3A) and stained for SOX2 and DCX. (O) Ratio of SOX2+ was significantly rescued in APP-KO2 NPCs treated with WNT3A (n = 3, ordinary one-way ANOVA, ****P < 0.0001 for isogenic control + PBS/BSA versus APP-KO2 + PBS/BSA and APP-KO2 + PBS/BSA versus APP-KO2 + WNT3A, ****P = 0.0001 for isogenic control + PBS/BSA versus APP-KO2 + WNT3A, ***P = 0.0006 for isogenic control + WNT3A versus APP-KO2 + WNT3a). (P) Ratio of DCX+ cells was not rescued after treatment with WNT3a (n = 3, ordinary one-way ANOVA, ****P < 0.0001 for all the significant conditions).
DISCUSSION
Here, we report what is, to our knowledge, the first evidence for a genetic network that specifically regulates the absolute timing at which human cortical progenitors generate neurons. Our evidence suggests that this timing requires simultaneously sufficient levels of canonical WNT signaling to promote progenitor identity and proliferation as well as sufficient repression of AP-1 activation to inhibit neurogenesis. We find that these two processes are genetically coupled by the activity of the APP and can be uncoupled downstream of it. Our evidence supports a model in which this genetic network is required for the prolonged maintenance of human cortical NPCs in a progenitor state. In this model, loss of APP reduces WNT signaling and activates AP-1 thus lowering the threshold for NPCs to transition from a self-renewing progenitor state to a neurogenic state. Notably, NPCs lacking APP generate cortical neurons in the correct temporal order. This means that the absolute amount of time a human cortical progenitor takes to generate neurons can be genetically uncoupled from the relative temporal order of the generation of cortical neurons. Obviously, evolution has achieved such uncoupling in the timing of cortical development in different mammalian species, where the same subtypes of neurons are generated in the same relative temporal sequence despite massive differences in the total amount of time required for cortical neurogenesis. Our findings suggest either that loss of APP mimics this evolutionary uncoupling or, more excitingly, that regulation of genes in the APP–WNT–AP-1 network may be part of the evolutionary uncoupling mechanism. Last, it is noteworthy that the acceleration of neurogenesis upon loss of APP does not appear to alter the final ratio of EB to LB neurons and does not appear to result in significantly faster maturation of the neurons produced. This contrasts with accelerated neuronal production using cocktails of small-molecule inhibitors, including the Notch signaling inhibitor DAPT, which appears to accelerate both processes (67). This may indicate the clock of neuronal production and the clock of neuronal maturation are genetically separable and that APP is specifically required for the former. Nevertheless, several points remain to be clarified in future work. First, it will be important to determine whether the acceleration is proportional along the entire duration of cortical neurogenesis or whether the effect varies over time, especially during the generation of upper layer neurons. Second, it will be interesting to determine whether the neuronal maturation clock is completely normal or whether there are also subtle but critical, effects yet to be found, such as changes in axonal branching and/or dendritic spine complexity, as we have recently observed in mouse cortical neurons (68). Last, determining whether prematurely born APP-KO human cortical neurons can maintain a healthy differentiated state over time or whether they have a shorter life span will be very interesting to investigate.
We observed an increase in the percentage and molecular markers of outer/basal RGCs upon loss of APP. Outer/basal radial glia are a progenitor population that was massively increased during the evolution of the primate brain. Together with an increase in NPs, this may suggest that the APP-dependent temporal program regulates the proportion of different types of progenitors in the developing human cortex. An indirect consequence of this notion would be that the relative spatial organization of the VZ and outer SVZ of the human fetal cortex is, in part, a derivative of the temporal progression of progenitor fate. In addition, whether the greater requirement for APP in humans compared to rodents may offer a clue into fAD is worth further investigation. We noted the up-regulation of MAPT gene (1.36 log2FC, table S3), encoding the other key AD protein Tau, in APP mutant progenitors. It is tempting to speculate that fAD mutations in APP have subtle effects on early human cortical development that cause reduced cortical robustness and resistance to a lifetime of neuronal and glial stress partially contributing to premature neurodegeneration.
Three observations suggest that human brain development may be particularly sensitive to loss of APP. First, there is a strongly reduced tolerance to complete loss of APP in human genetic data (29, 69–71). Second, both in vivo and in vitro studies on mouse NPCs lacking APP have shown no consistent evidence for altered neurogenesis (8, 36, 37). Third, among the genes whose expression is altered in APP-KO NPCs, 33 have been reported to be located within human accelerated regions, which are conserved genomic loci with elevated divergence in humans (72, 73). We also did not observe any effect on human motor neurogenesis, suggesting that APP function may be more important during cortex development. Because APP is highly conserved in sequence and expression among mammalian species, we speculate that the notable requirement for APP in human cortical neurogenesis is less due to anything special about the human APP protein itself and more due to the special features of human cortical neurogenesis. In particular, we propose that the links between APP, WNT, and JNK pathways have been co-opted in human, and perhaps more generally primates, to prolong the ability of the cortical NPCs to retain a progenitor state while also generating more differentiated daughters. For example, human/primate cortical NPCs may express human/primate-specific regulators of APP activity or stability that allow them to suppress JNK signaling and enhance WNT signaling more efficiently than in other mammalian species.
We have recently shown that APP is a co-receptor for Wnt-3a and that Wnt-3a binding protects APP from degradation (68). We observed that WNT receptor FZD5 that mediates both canonical and noncanonical pathway (74, 75) is up-regulated (2.4 L2Fc) in our bulk RNA-seq data. It is possible that the rescue of progenitor proliferation defects observed in APP-KO NPCs by WNT3A is partially mediated by up-regulation of FZD5. Furthermore, it has recently been reported that there is a correlation between down-regulation of WNT signaling and emergence of oRG cells meaning cortical organoids generated using Triple-i (Dual-SMAD and WNT inhibition) specifically enrich for oRG cells compare to classical Dual-SMAD inhibition protocol (76). This is in line with our findings that WNT signaling pathway is down-regulated in APP-KO progenitors and APP-KO progenitors show a shift toward oRG fate. Together, these data suggest that APP regulates the balance between NPC proliferation and differentiation through partially parallel mechanisms, facilitating WNT signaling to promote NPC proliferation and repressing AP-1 levels to prevent premature NPC differentiation. In addition, human cortical NPCs may be particularly sensitive to cellular stress. Some studies suggest that increased stress drives premature neurogenesis in cortical progenitors (77). APP is a conserved stress response protein, as are JUN and FOS. Therefore, the absence of APP may mimic cellular stress, activate AP-1–mediated responses, and drive premature neurogenesis. We have previously shown that the coding sequence contributes to evolutionary changes in the neurogenic activity of proneural proteins (78). We cannot exclude that subtle changes to the otherwise highly conserved APP coding sequence may be involved in its acquisition of this function in human cortical NPCs.
Increasing evidence points to human-specific genetic alterations, such as new gene isoforms (33, 79–82), microRNAs (83), and regulatory sequences, creating human-specific transcriptional networks (72, 84) potentially resulting in human-specific brain features. A tantalizing speculation is that members of the genetic network we uncovered here may be regulated by one or more human-specific protein isoform or microRNA whose expression is enriched in the cortex. A clinical case of a homozygous nonsense truncating mutation in APP leading to microcephaly has been reported (70, 71). The reported patient carries mutations in three genes including homozygous mutations in APP, that strongly reduce its expression levels, and SETX as well as a heterozygous mutation in CLN8. Because CLN8 is associated with autosomal-recessive neuronal ceroid lipofuscinosis [Mendelian Inheritance in Man (MIM): 600143] and the patient is heterozygote for that, the authors conclude that the phenotype is not related to this gene. SETX is associated with autosomal-dominant juvenile amyotrophic lateral sclerosis type 4 (ALS; MIM: 602433) and autosomal recessive ataxia with ocular apraxia type 2 (AOA2; MIM: 606002). Because ALS and AOA2 usually have onset in adolescence, they conclude that it is more probable that the phenotype is related to the mutation of APP and not SETX. This supports the notion that human brain development is vulnerable to loss of APP and that perhaps the status of APP heterozygosity should be included in clinical genetic counseling.
MATERIALS AND METHODS
iPSC culture and maintenance
iPSC cell lines WTSIi002-A (https://cells.ebisc.org/WTSIi002-A/, iPSC line1) and WTSIi008-A (https://cells.ebisc.org/WTSIi008-A/, iPSC line2) were purchased from EBISC (European Bank for Induced pluripotent Stem Cells) and maintained on Geltrex LDEV-Free hESC-qualified Reduced Growth Factor Basement Membrane Matrix (Thermo Fisher Scientific) in Essential 8 Flex Media Kit (Thermo Fisher Scientific) with 0.1% penicillin/streptomycin. Cultures were fed every other day and passaged every 5 to 7 days by ReLeSR (STEMCELL Technologies).
Creating APP-KO clones by CRISPR-Cas9
iPSC line1
Four guide RNAs were designed by Crispor (http://crispor.tefor.net/) to target APP’s first exon and cloned in pCAG-CAS9-GFP (fig. S1, A and B). To evaluate the cleavage efficiencies of the guide RNAs, human embryonic kidney–293 cells were transfected with p-CAG-Cas9-GFP containing four guide RNAs (fig. S1C). DNA extraction and polymerase chain reaction (PCR) were performed 48 hours after transfection for the cleaved region. Sequenced PCR products were analyzed in TIDE (Tracking of Indels by Decomposition) to obtain cleavage efficiency. The sequence of each guide RNA, their cleavage efficiency, and off-target are listed in table S5. The best guide RNA with 8.5% total efficiency and lowest off-target was chosen for further experiments (fig. S1D). Karyotyping was performed on the iPSC line (which we refer to as “parental line” 1) before the CRISPR approach and the results showed a deletion in chromosome 18. The iPSC line was transfected with the best guide RNA using Lipofectamine stem reagent (Thermo Fisher Scientific). GFP-positive cells were isolated by fluorescence-activated cell sorting 48 hours after transfection and seeded at low density. A total of 118 clones were picked after 5 days and expanded, and 60 were screened for possible mutations (fig. S1, E to H). Sixteen of the 60 clones had homozygous mutations for APP. These mutations are classified as 1-, 2-, 3-, 20-, 22-, and 33-bp deletions and 1- and 2-bp insertions (table S6). The absence of expression of the APP protein was confirmed by Western blot in 14 clones (fig. S1I). The efficiency of producing APP knock-out clones was 23% (fig. S1J). Three APP knock-out clones were chosen for further experiments, and low APP mRNA levels were confirmed by quantitative PCR (fig. S1K). The control, which was chosen for further experiments, expressed guide RNA and CAS9, but the APP gene remained un-mutated. Karyotyping was performed for the APP-KO clones, control, and parental iPSC. The results showed that the same deletion in chromosome 18 in parental line was also confirmed in control and APP-KO clones. No other deletion or duplication was observed in APP-KO clones and control after CRISPR approach. The pluripotency of APP-KO cells and the control were confirmed by octamer binding transcription factor 4 (OCT4) staining (fig. S2, D to G).
iPSC line2
Karyotyping was performed on the iPSC line before the CRISPR approach (which we refer to as “parental line” 2), and the results showed a duplication in chromosome 20. The guide RNA 1 and the ribonucleoprotein approach were used to generate APP-KO. A total of 1 × 106 iPSCs were nucleofected with the ribonucleoprotein complex from Integrated DNA Technologies (225 pmol of each RNA and 120 pmol of Cas9 protein). iPSCs were plated 48 hours later at very low density (10 cells/cm2) on Laminin-521 with CloneR supplement (Stem Cell Technology) for clonal selection. One week later, iPSC clones were picked under a stereomicroscope and cultured on Laminin-521 in 96 well plates (Duscher). The resulting iPSC clones were duplicated after confluency and used for cryoconservation and DNA extraction. One base-pair deletion was observed in screening results and the absence of expression of APP protein was confirmed by Western blot. The APP knock-out clone and control were used for further experiments, and the low level of APP mRNA was confirmed by quantitative PCR (fig. S1, L to N). Karyotyping was also performed for the APP-KO clone, control, and parental iPSC. The results confirmed the same duplication in chromosome 22 in all the conditions. No other deletion or duplication was observed in APP-KO clones and control after CRISPR approach.
Cortical differentiation of human iPSC
The cortical differentiation protocol is based on Dual-SMAD inhibition (85, 86) with some modification. The protocol is divided into two main steps: neural induction and neural differentiation. At day 0, iPSCs were detached by Accutase (Thermo Fisher Scientific) and transferred to T75 ultralow attachment flasks (VWR) in Essential 8 medium with 0.1% penicillin/streptomycin and Stemgent hES Cell Cloning & Recovery Supplement (Ozyme, 01-0014-500) to form embryoid body. At day 1, the medium was switched to embryoid body medium containing Dulbecco’s modified Eagle’s medium/F12 Glutamax, 20% knockout serum replacement, 1% nonessential amino acids, 1% penicillin/streptomycin, and 0.55 mM 2-mercaptoethanol and supplemented with dorsomorphin (1 μM, Sigma-Aldrich, P5499), SB431542 (10 μM, Abcam, ab120163) for 8 days. At day 8, embryoid bodies were collected and seeded on Geltrex and maintained for 6 to 8 days in Neurobasal without vitamin A medium and B27 without vitamin A supplemented with human epidermal growth factor (EGF; 10 ng/ml) and human fibroblast growth factor 2 (FGF2) (10 ng/ml). Cells were fed every day from day 0 to day 16. Rosettes were manually picked at day 15 and dissociated with Accutase, seeded on poly-ornithine and laminin-coated dishes for expansion, and maintained with passage for two additional weeks to achieve a large pool of NPCs (fig. S2, A to C). Characterization for NPCs at passage numbers 3 to 5 was performed by staining for Nestin, SOX2, SOX1, and PAX6 (fig. S2, H1 to K6) and FOXG1 and EMX2 (fig. S3, A to H). NPC with passage numbers 4 to 9 was used for the experiments. We used control and APP-KO NPC with similar passage numbers in all experiment to avoid unknown effects of passage number. NPCs were seeded on 24-well plates with coverslips, and the day after seeding, the medium was switched to the B-27 Plus Neuronal Culture System (Thermo Fisher Scientific) supplemented with ascorbic acid. Cortical neurons were stained for EB neural markers and LB neural markers at the indicated early time points or kept for 3 to 4 months in culture and stained for synaptic markers at 120 days after differentiation.
Labeling and tracing of NPCs for short-term neuronal culture
Sparse labeling was performed using viral vector approach as described (32, 33). We used GFP-expressing lent viral vector (pLv794_pTrip_PromSynapsin1_GFP_DeltaU3). We tested the vector before performing any experiment with different concentration of virus [MOI (multiplicity of infection) = 1, MOI = 0.1, MOI = 0.05, and MOI = 0.01]. First, we found that the pLv794_pTrip_PromSynapsin1_GFP_DeltaU3 promoter does initiate GFP expression in SOX2+ progenitors and is thus an appropriate tool. Second, we found that MOI = 1 and MOI = 0.5 concentrations were toxic and thus chose MOI = 0.05 for the experiment. Next, APP-KO and control NPCs were transduced by pLv794_pTrip_PromSynapsin1_GFP_DeltaU3. The medium was changed to fresh Neurobasal without vitamin A and B27 without vitamin A supplemented with EGF and hFGF2 to wash out lentiviruses 1 day after initial infection. When the GFP+ cells appeared (4 days after transduction), it was considered day 0 and the medium was switched to differentiation medium (B-27 Plus Neuronal Culture System) and kept for 7 and 30 days. Lentiviral-expressing cells were detected by anti-GFP antibody, and cell type was determined by anti-SOX2 and anti-TUJ1 antibodies.
Labeling and tracing of NPCs for long-term neuronal culture
APP-KO and control NPCs were seeded on poly-l-ornithine/laminin-coated coverslips and transduced by lentiviral vector containing Venus-NLS (pLv794_pTrip_PromSynapsin1_Venus-NLS_DeltaU3). When the Venus+ cells appeared, it was considered as day 0 and the medium was switched to differentiation medium (B-27 Plus Neuronal Culture System) and kept for 80 and 110 days. Venus expressing cells were detected by anti-GFP antibody and EB and LB neurons were detected by the antibodies listed in table S7.
Motor neuron differentiation and staining
Control and mutant iPSC clones were differentiated into motor neurons as described (31). In summary, after dissociation of embryoid bodies containing motor neuron progenitors at D10, single cells were plated on poly ornithine-laminin–coated coverslips at 2 × 105 cells per coverslip for 1, 4, and 7 more days. Cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 10 min at room temperature (RT). For staining, cells were treated with 5% goat serum with 0.1% Triton X-100 (Thermo Fisher Scientific) for 1 hour at RT and then incubated overnight at 4°C with a mouse anti–βIII-Tubulin (TUJ1, Sigma-Aldrich, 1/500) and a rabbit anti-ISLET1 (Abcam, 1/100) antibodies in 5% goat serum. Secondary antibodies [goat anti-mouse IgG2a-Alexa 555 (1/2000) and goat anti-rabbit IgG Alexa 488 (1/2000) from Thermo Fisher Scientific] were incubated for 1 hour at RT with Hoechst33342 to stain nuclei.
Immunocytochemistry
Cells were fixed in 4% PFA and incubated for 30 min in blocking buffer (PBS and 0.3% Triton X-100 and 5% horse serum). Primary antibodies (table S7) were diluted in antibody solution (PBS and 0.1% Triton X-100 and 1% horse serum) and applied overnight at RT. After three washes in PBS, secondary antibodies conjugated to Alexa fluorophores (Molecular Probes, Eugene, OR, USA) were diluted at 1:1000 in antibody solution and applied for 1 hour at RT. Cells were washed in PBS followed by nuclear staining by DAPI (Sigma-Aldrich) for 10 min. Cells were washed three more times and mounted by VECTASHIELD Mounting Medium-Vector Laboratories. Confocal image acquisition was performed using Olympus FV1200 and SP8 Leica DLS.
Generation of cortical organoids
Cortical organoids were generated from human iPSCs using a previously reported protocol (35), with some modifications. Human iPSCs were incubated with Accutase (Life Technologies, A1110501) at 37°C for 7 min and dissociated into single cells. To obtain uniformly sized spheroids, approximately 3 × 106 single cells were added per well in the AggreWell 800 plate (STEMCELL Technologies, 34815) with Essential 8 flex medium supplemented with Stemgent hES Cell Cloning & Recovery Supplement (1X, Ozyme, STE01-0014-500) and incubated at 37°C with 5% CO2. After 24 hours, spheroids from each microwell were collected by firmly pipetting medium in the well up and down and transferred into Corning nontreated culture dishes (Merck, CLS430591-500EA) in TeSR-E6 (StemCell Technologies, no. 05946) supplemented with two inhibitors of the SMAD signaling pathway, dorsomorphin (2.5 μM, Sigma-Aldrich, P5499) and SB-431542 (10 μM, Abcam, ab120163). From day 2 to day 5, TeSR-E6 supplemented with dorsomorphin and SB-431542 was changed every day. On the sixth day in suspension, neural spheroids were transferred to neural medium containing Neurobasal minus vitamin A (Life Technologies, 10888), B-27 supplement without vitamin A (Life Technologies, 12587), GlutaMax (1%, Life Technologies, 35050), Penicillin-Streptomycin (1%, Life technologies, 15140122) and 2-mercaptoethanol (5 mM, Life Technologies, 31350010). The neural medium was supplemented with EGF (10 ng/ml; PreproTech, AF-100-15) and basic FGF (10 ng/ml; R&D Systems, 234-FSE-025) and changed daily until day 12. From day 12 to day 24, medium was changed every other day. At day 25, the medium was switched to the B-27 Plus Neuronal Culture System (Thermo Fisher Scientific) supplemented with ascorbic acid. Cortical organoids were fed twice per week and kept for 3 to 4 months in culture. They were stained at day 15 for SOX2 and DCX.
Organoid collection, immunohistochemistry, and imaging
3D staining
At day 15, organoids were fixed with 4% PFA at 4°C for 6 hours and rinsed three times with PBS for 10 min. Following two washing with PBS + 2% Triton X-100 for 2 hours, spheroids were treated overnight at RT with PBS + 2% Triton X-100 + 2% Tween 20 + 20% dimethyl sulfoxide. Blocking/permeabilization was performed with PBS + 10% horse serum, 3% bovine serum albumin (BSA), and 2% Triton X-100 for 24 hours at RT. Primary antibodies (Sox2 1/500, Millipore AB5603 and DCX 1/2000, Millipore AB2253) were incubated in the same solution supplemented with 0.05% Azide for 3 days at 4°C. After multiple washing with PBS + 0.5% Tween 20 until the next day, organoids were then incubated for 24 hours with secondary antibodies. After 24 hours of washing with PBS + 0.5% Tween 20, samples were cleared in RapiClear 1.49 (SunJin lab) before mounting on cavity slides (Hecht Karl 42410010) and confocal image acquisition was performed using Nikon A1R HD25.
DCX quantification
Whole-mount day 15 organoids were imaged with a Nikon A1R HD25 confocal microscope at 1-μm intervals with a 10× objective (MRD71120). Analyses on organoids were performed using the NIS-Elements software (Nikon) and Imaris. Image stacks were reconstructed in 3D to measure volume of DCX clusters and total 4′,6-diamino-2-phenylindol (DAPI). Each measurement was normalized to the total volume of DAPI in the organoid. Each organoid is considered as n = 1.
Quantification of synapse markers
The quantification was performed using a semiautomatic method using two plugins in imageJ. First, Neurite tracing was drawn manually by NeuronJ plugin (www.imagescience.org/meijering/software/neuronj/) followed by quantification of synapses by SynapCountJ plugin (87).
MEA recording on organoids
Spike activity was recorded using a 64-channel MEA system (MED64, Alpha MED Scientific). Cortical organoids at day 160 were collected 48 to 72 hours after changing the culture medium and recordings were performed in this conditioned culture media. The organoids were carefully placed into a poly-l-ornithine/laminin–coated MEA well (MED-P515A) and immobilized by a small platinum anchor. Recordings started after a period of recovery of 2 min. Extracellular field potentials were filtered with a band-pass filter (100 Hz and 10 kHz as cutoff frequencies) and acquired at a sampling rate of 20 kHz. Recordings were performed at 37°C. Spikes were detected using MOBIUS software with a detection threshold set at six times the SDs of the estimated noise for each electrode. Data were analyzed using a home-made R script. Electrodes that detected at least 5 spikes per minute were classified as active electrodes.
RNA scope on human fetal brain samples
Formalin-fixed paraffin-embedded fetal brain sections at 14 weeks of gestation were obtained from the brain collection “Hôpitaux Universitaires de l’Est Parisien – Neuropathologie du développement” (Biobank identification number BB-0033-00082). For all cases studied, informed consent was obtained for autopsy of the brain and histological examination. After removal, brains were fixed with formalin for 5 to 12 weeks. Macroscopic analysis was performed allowing the selection and conditioning of samples (paraffin embedding, 7-μm slicing) of brain tissue for histological analysis. The RNAScope assay was performed according to the standard provider’s protocol (ACD Bio-Techne). In summary, slides were deparaffinized twice with xylene for 5 min, followed by ethanol treatment for three times, and dried at 60°C for 30 min. Peroxidase blocking with H2O2 performed at RT for 10 min. After washing with water, antigen retrieval was performed in a pressure cooker, followed by rinsing with water and ethanol for 3 min. Slides were air-dried for 10 min and baked at 60°C for 30 min, followed by rinsing with PBS. TrueView (Vector Laboratories, SP-8400-15) applied according to the provider’s recommendations to reduce sample autofluorescence, followed by three times washing with PBS. Protease Plus treatment was applied for 30 min. RNAscope Probe-Hs-APP (C1) (Bio-Techne 418321) hybridization was performed for 2.5 hours, followed by rinsing twice with washing buffer. Slides were kept ON in 4× Saline-Sodium Citrate and amplification steps were performed the day after. The fluorophore OPAL 650 used to label the APP probe. Sections were stained with DAPI (1:5000) and sealed with VECTASHIELD Vibrance Mounting Medium (Vector Laboratories H-1700) and allowed to polymerize O.N. in the dark. Confocal image acquisition was performed using SP8 Leica DLS.
Embryo collection, immunohistochemistry, and antibodies
E10.5 and E17.5 embryos were collected from App-WT or App-KO pregnant mice, whole embryos were fixed in 2% PFA in PBS at 4°C for 2 to 3 hours, then dehydrated in 30% sucrose in 1× PBS overnight (o/n). After all the samples sank into the bottom of the tube, they were embedded in OCT compound (TissueTek) and frozen at −20°C. Sagittal sections were performed by cryostat (Leica) at 20 μm and then slices stored at −80°C. For the immunostaining, sections were fixed with 4% PFA for 10 min at RT, then blocked with 10% normal donkey or goat serum in 1× PBS with 0.1% TritonX-100 (PBT) for 1 hour at RT followed by three washes in 1× PBT. Thereafter, these sections were incubated with primary antibodies diluted in 0.1% 1× PBT containing 1% normal donkey or goat serum o/n at 4°C or 3 to 4 hours at RT. After three washes with 1× PBT, samples were incubated with appropriate secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647 (1:500, Invitrogen) in 0.1% 1× PBT containing 1% normal donkey or goat serum for 1 to 2 hours at RT. After washing with 1× PBT three times and then counterstained with DAPI (1:2000, Sigma-Aldrich), the slides were mounted by using VECTASHIELD (Vector) after rinsing. After staining, images were obtained by using a confocal microscope, Olympus FV-1200.
Quantitative PCR
The cells were lysed directly in the wells by addition of 300 μl of Buffer RLT supplemented with 15 mM β-mercaptoethanol (Thermo Fisher Scientific) after a wash with Dulbecco’s PBS (DPBS, Life Technologies). Total RNA was isolated using the RNeasy Mini extraction kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s protocol. RNA levels and quality were quantified using a NanoDrop spectrophotometer. cDNA synthesis was performed by Thermo Fisher Scientific Verso cDNA Synthesis Kit and Quantitative PCR assay was performed by Sybergreen Gene Expression Assays in triplicate wells of 96-well plates. Primers are listed in table S8 and 2−(Cp GOI−Cp internal gene) was used for analysis. The housekeeping gene (glyceraldehyde-3-phosphate dehydrogenase) was selected to control for variation in cDNA amounts.
Bulk RNA sequencing and analysis
The cells were lysed directly in the wells by addition of 300 μl of Buffer RLT supplemented with 15 mM beta-mercaptoethanol (Thermo Fisher Scientific) after a wash with DPBS (Life Technologies). Total RNA was isolated using the RNeasy Mini extraction kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s protocol. RNA levels and quality were quantified using a NanoDrop spectrophotometer. RNA sample purity/integrity was assessed using an Agilent 2200 Tapestation. mRNA library preparation was completed following the manufacturer’s recommendations (KAPA mRNA hyperprep ROCHE). Final samples pooled library prep were sequenced on NextSeq 500 ILLUMINA with MidOutPut cartridge (2 × 130 million 75 base reads) with one run, corresponding to 2 × 20 million reads per sample after demultiplexing. The quality of raw data was evaluated with FastQC. Poor-quality sequences were trimmed or removed with fastp software to retain only good quality paired reads without adapters. Star v2.5.3a (88) was used to align reads on the hg19 reference genome using standard options. Quantification of gene and isoform abundances was carried out with rsem 1.2.28 (89), before normalization on library size with the edgeR (90) bioconductor package. Last, differential analysis was conducted with the glm framework likelihood ratio test from edgeR. Multiple hypotheses adjusted P values were calculated with the Benjamini-Hochberg (91) procedure to control FDR.
scRNA-seq and analysis
For each control and knockout cell line, single cell samples were prepared for 3′ mRNA sequence determinations using the Scipio bioscience protocol (92) with barcoding beads (NxSeq Single-cell RNA-seq Beads, LGC Biosearch Technologies). For beads that captured mRNA molecules, reverse transcription, PCR amplification of cDNA, and sequencing library preparation were performed according to published procedures (93). A total of 35,106,546 reads, 56,108,512 reads, and 54,702,467 reads were generated by NovaSeq from two replicates of APP-KO2 (KO2-1 and KO2-2) and one CT (control) sample, corresponding to 2000, 2000, and 1500 cells. The reads passed QC process using FASTQC v0.11.8. Sample analyses were performed using UMI-tools v1.0.0. Reads were aligned to GRCh38.94 using STAR v2.7 with default parameters. FeatureCounts v1.6.4 (94) (Ensembl GRCh38.94.GTF) was used to count the number of aligned reads per feature, followed by umi_tools dedup to collapse those reads belonging to a barcode and mapping the same position of a gene. To build the count table umi_tools count was applied. Seurat V3.1.4 (40) was used: (i) to filter cells having more than 200 genes and less than 20% mitochondrial genes, with 1326, 1594, and 1299 cells passing the QC filters for the KO2-1, KO2-2, and CT samples, respectively.; (ii) to pool both KO samples (2920 total); (iii) to cluster KO and CT cells using Seurat clustering with 10 PCA dimensions and resolution 0.5; (iv) to identify cluster markers using FindAllMarkers with default parameters. Visualizations were performed with Seurat and ggplot2 packages. DEGs in KO versus CT were analyzed for enrichment in gene ontology, biological and disease associations using Enrichr tools to further explore ChIP-seq datasets linking transcription regulators with DEGs (95). Integration with other early fetal cells (22) was performed using Integrated Anchors analysis followed by clustering with Seurat (40).
Pseudo time analysis
The pseudotime was performed on the basis of a machine learning ordinal regression algorithm (44, 45), using the annotated stages of differentiation present in the UMAP 1 component (CP1 > RGC > CP2 > IP > N), ordered from the most undifferentiated to most differentiated ones. Using the control cells (CT), the model adapts weights (that can be considered as importance) to genes so that when using the expression of the genes and their corresponding weights, the algorithm orders the cells along the differentiation axis to build a pseudotime axis, and the genes weights was used to predict the pseudotime distribution of the APP-KO cells (KO).
Rescue experiments
NPCs of Control and APP-KO2 were transfected with pPB-CAG-IRES-EGFP and APP-KO2 NPCs were transfected by pPB-CAG-hAPP- IRES -EGFP and kept for 4 days. Cells were fixed and stained for GFP/SOX2/DCX. The GFP+ cells were quantified, and cell type was determined by SOX2 and DCX as progenitor and differentiated cell, respectively.
Cell cycle length measurement
For measuring cell-cycle length, 1 μM EdU was added to the NPC culture medium. After 2, 4, 14, 24, 30, 36, 48, and 55 further hours in culture, cells were fixed with 4% PFA, and EdU incorporation was visualized using the Click-iT imaging kit (Life Technologies). Cell-cycle lengths were calculated from cumulative labeling as described (49).
Neural precursor treatment
Recombinant human WNT3A (R&D Systems) and SR11302 (TOCRIS) were used at final concentrations of 150 ng/ml and 10 μM, respectively. NPCs of control and APP-KO2 were treated for 48 hours and then stained for SOX2 and DCX.
Western blot
Cells were placed on ice, lysed directly in wells by adding RIPA buffer (Sigma-Aldrich) supplemented with Complete protease inhibitor cocktail (Roche), and agitated for 20 min. Thereafter, the samples were collected and centrifuged at >14000 g, 30 min at 4°C and the supernatant was transferred to a fresh tube. Protein determination was performed using the Pierce Detergent Compatible Bradford Assay Kit (Thermo Fisher Scientific). Sample buffer (BOLT LDS, Life technologies) was added to equal amounts of protein, and samples were loaded onto a BOLT 4 to 12% Bis-tris gel and transferred using MES buffer (all from Life technologies). Proteins were blotted onto a 0.2 μM nitrocellulose membrane (GE Healthcare) using semi-dry technique. Membranes were blocked in 5% Nonfat-Dried Milk bovine (Sigma-Aldrich) and incubated overnight at 4°C with primary antibodies listed in table S7. After washing, membranes were incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour at RT. Protein detection was performed by Pierce ECL Western Blotting substrate (Thermo Fisher Scientific) and bands were visualized using ChemiDoc Touch Imaging System (BioRad Laboratories). Band intensities were calculated using Image Lab software.
Statistical analysis
Statistical analysis was performed by GraphPad Prism. Data in figure panels reflect three independent experiments performed on different days. An estimate of variation within each group of data is indicated using SEM. We performed unpaired t test for assessing the significance of differences in the analyses containing two conditions, one-way analysis of variance (ANOVA) correction in the analyses containing more than three conditions and two-way ANOVA in the group analysis.
Acknowledgments
We thank N. Baumann and D. Jabaudon for sharing the pseudotime analysis code and B. De Strooper for sharing the anti-APP antibody. We especially thank A. Gelot (APHP Hôpital Trousseau) for providing human fetal brain sections. We are grateful to S. Nedelec, J. De Wit, D. Jabaudon, members of the Hasan laboratory as well as A. Graindorge, and S. Edelstein from Scipio biosciences for helpful discussions, and D. Héron for facilitating access to human brain samples. Scipio bioscience was supported by the Investissements d’Avenir program and the Région Île-de-France. This work benefited from equipment and services from the ICM’s genotyping and sequencing core facility (iGenSeq). Mouse breeding work was conducted at the PHENO-ICMice facility. We thank P. Ravassard for providing the lentiviral vector (pLv794_pTrip_PromSynapsin1_GFP_DeltaU3) and B. Bonnamy and C. Ripoll from iVECTOR core facility for technical assistance and production of the lentiviral vectors. Light microscopy was carried out at the ICM.Quant facility. We gratefully acknowledge C. Dalle from ePHYS (electrophysiology core facility) facility of ICM for Electrophysiology experiment on organoids and F.-X. Lejeune from the DAC (Data Analysis core) for developing the script for analysis. We also thank all core technical staff involved especially S. Bigou from CELIS for advice on iPSCs and CRISPR-Cas9 work and C. Lovo from ICM.Quant for help with image analysis.
Funding: This work was supported by the Investissements d’Avenir program (ANR-10-IAIHU-06), Paris Brain Institute–ICM core funding, a Sorbonne Université Emergence grant, a Neuro-Glia foundation grant, and the Roger De Spoelberch Prize (to B.A.H.). T.L. is funded by the Chinese Scholarship Council (CSC). The Core is supported by two Investissements d’Avenir grants (ANR-10- IAIHU-06 and ANR-11-INBS-0011-NeurATRIS) and the “Fondation pour la Recherche Médicale”. iPSC work was carried out at the CELIS core facility with support from Program Investissements d’Avenir (ANR-10-IAIHU-06).
Author contributions: K.S., C.P., and B.A.H. conceived the project, interpreted the data, and wrote the manuscript with input from all authors. K.S. and B.A.H. designed, and K.S. performed experiments and data analysis for most of the project. J.P. performed experiment on NPCs derived from the second iPSC line and 3D culture derived from both iPSC lines. M.B.T.Z. performed the viral approach experiment. T.L. performed mouse embryo work. J.K. performed sample preparation for single-cell RNA-seq. A.S. and C.P. performed single-cell data analyses. N.D. generated the vectors for the rescue experiment. E.L. and D.B. performed motor neuron culture and staining. R.L. helped optimize the iPSC and NPC culture and differentiation protocols.
Competing interests: A.S. and J.K. are employees of Scipio biosciences. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1, S4 to S8
Legends for tables S2 and S3
Legends for movies S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S2 and S3
Movies S1 to S4
Lentiviral vector sequence fasta file
REFERENCES AND NOTES
- 1.S. Herculano-Houzel, The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc. Natl. Acad. Sci. U.S.A. 109, 10661–10668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. Harris, G. S. Tomassy, P. Arlotta, Building blocks of the cerebral cortex: From development to the dish. Wiley Interdiscip. Rev. Dev. Biol. 4, 529–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. Mancinelli, S. Lodato, Decoding neuronal diversity in the developing cerebral cortex: From single cells to functional networks. Curr. Opin. Neurobiol. 53, 146–155 (2018). [DOI] [PubMed] [Google Scholar]
- 4.T. Rabinowicz, G. M. De Courten-Myers, J. M. C. Petetot, G. Xi, E. De Los Reyes, Human cortex development: Estimates of neuronal numbers indicate major loss late during gestation. J. Neuropathol. Exp. Neurol. 55, 320–328 (1996). [PubMed] [Google Scholar]
- 5.G. Meyer, J. P. Schaaps, L. Moreau, A. M. Goffinet, Embryonic and early fetal development of the human neocortex. J. Neurosci. 20, 1858–1868 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.I. Espuny-Camacho, K. A. Michelsen, D. Gall, D. Linaro, A. Hasche, J. Bonnefont, C. Bali, D. Orduz, A. Bilheu, A. Herpoel, N. Lambert, N. Gaspard, S. Péron, S. N. Schiffmann, M. Giugliano, A. Gaillard, P. Vanderhaeghen, Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 77, 440–456 (2013). [DOI] [PubMed] [Google Scholar]
- 7.N. Gaspard, T. Bouschet, R. Hourez, J. Dimidschstein, G. Naeije, J. Van Den Ameele, I. Espuny-Camacho, A. Herpoel, L. Passante, S. N. Schiffmann, A. Gaillard, P. Vanderhaeghen, An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357 (2008). [DOI] [PubMed] [Google Scholar]
- 8.P. Bergström, L. Agholme, F. H. Nazir, T. M. Satir, J. Toombs, H. Wellington, J. Strandberg, T. O. Bontell, H. Kvartsberg, M. Holmström, C. Boreström, S. Simonsson, T. Kunath, A. Lindahl, K. Blennow, E. Hanse, E. Portelius, S. Wray, H. Zetterberg, Amyloid precursor protein expression and processing are differentially regulated during cortical neuron differentiation. Sci. Rep. 6, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Nicolas, B. A. Hassan, Amyloid precursor protein and neural development. Development 141, 2543–2548 (2014). [DOI] [PubMed] [Google Scholar]
- 10.R. Coronel, C. Palmer, A. Bernabeu-Zornoza, M. Monteagudo, A. Rosca, A. Zambrano, I. Liste, Physiological effects of amyloid precursor protein and its derivatives on neural stem cell biology and signaling pathways involved. Neural Regen. Res. 14, 1661–1671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. Leyssen, D. Ayaz, S. S. Hébert, S. Reeve, B. De Strooper, B. A. Hassan, Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. EMBO J. 24, 2944–2955 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. O. Y. Hung, F. J. Livesey, Altered γ-secretase processing of APP disrupts lysosome and autophagosome function in monogenic Alzheimer’s disease. Cell Rep. 25, 3647–3660.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.I. Kessissoglou, D. Langui, A. Hasan, M. Maral, S. B. Dutta, P. R. Hiesinger, B. Hassan, The Drosophila amyloid precursor protein homologue mediates neuronal survival and neuro-glial interactions. PLOS Biol. 18, e3000703 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. Kwart, A. Gregg, C. Scheckel, E. Murphy, D. Paquet, M. Duffield, J. Fak, O. Olsen, R. Darnell, M. Tessier-Lavigne, A large panel of isogenic APP and PSEN1 mutant human iPSC neurons reveals shared endosomal abnormalities mediated by APP β-CTFs, not Aβ. Neuron 104, 256–270.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 15.A. Salminen, A. Kauppinen, K. Kaarniranta, Hypoxia/ischemia activate processing of amyloid precursor protein: Impact of vascular dysfunction in the pathogenesis of Alzheimer’s disease. J. Neurochem. 140, 536–549 (2017). [DOI] [PubMed] [Google Scholar]
- 16.R. Sayer, D. Robertson, D. J. K. Balfour, K. C. Breen, C. A. Stewart, The effect of stress on the expression of the amyloid precursor protein in rat brain. Neurosci. Lett. 431, 197–200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.G. F. Chen, T. H. Xu, Y. Yan, Y. R. Zhou, Y. Jiang, K. Melcher, H. E. Xu, Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 38, 1205–1235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Coronel, M. Lachgar, A. Bernabeu-Zornoza, C. Palmer, M. Domínguez-Alvaro, A. Revilla, I. Ocaña, A. Fernández, A. Martínez-Serrano, E. Cano, I. Liste, Neuronal and glial differentiation of human neural stem cells is regulated by Amyloid Precursor Protein (APP) levels. Mol. Neurobiol. 56, 1248–1261 (2019). [DOI] [PubMed] [Google Scholar]
- 19.U. C. Müller, T. Deller, M. Korte, Not just amyloid: Physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. 18, 281–298 (2017). [DOI] [PubMed] [Google Scholar]
- 20.R. J. O’Brien, P. C. Wong, Amyloid precursor protein processing and alzheimer’s disease. Annu. Rev. Neurosci. 34, 185–204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. Lorent, L. Overbergh, D. Moechars, B. de Strooper, F. van Leuven, H. van den Berghe, Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the alpha-2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, alpha-2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience 65, 1009–1025 (1995). [DOI] [PubMed] [Google Scholar]
- 22.D. Polioudakis, L. de la Torre-Ubieta, J. Langerman, A. G. Elkins, X. Shi, J. L. Stein, C. K. Vuong, S. Nichterwitz, M. Gevorgian, C. K. Opland, D. Lu, W. Connell, E. K. Ruzzo, J. K. Lowe, T. Hadzic, F. I. Hinz, S. Sabri, W. E. Lowry, M. B. Gerstein, K. Plath, D. H. Geschwind, A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103, 785–801.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.T. L. Young-Pearse, J. Bai, R. Chang, J. B. Zheng, J. J. Loturco, D. J. Selkoe, A critical function for β-amyloid precursor protein in neuronal migration revealed by In Utero RNA interference. J. Neurosci. 27, 14459–14469 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R. Coronel, A. Bernabeu-Zornoza, C. Palmer, M. Muñiz-Moreno, A. Zambrano, E. Cano, I. Liste, Role of Amyloid Precursor Protein (APP) and its derivatives in the biology and cell fate specification of neural stem cells. Mol. Neurobiol. 55, 7107–7117 (2018). [DOI] [PubMed] [Google Scholar]
- 25.D. H. Small, Y. Hu, M. Bolós, E. Dawkins, L. Foa, K. M. Young, β-Amyloid precursor protein: Function in stem cell development and alzheimer’s disease brain. Neurodegener Dis. 13, 96–98 (2014). [DOI] [PubMed] [Google Scholar]
- 26.I. Caillé, B. Allinquant, E. Dupont, C. Bouillot, A. Langer, U. Müller, A. Prochiantz, Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development 131, 2173–2181 (2004). [DOI] [PubMed] [Google Scholar]
- 27.S. Wang, M. Bolós, R. Clark, C. L. Cullen, K. A. Southam, L. Foa, T. C. Dickson, K. M. Young, Amyloid β precursor protein regulates neuron survival and maturation in the adult mouse brain. Mol. Cell. Neurosci. 77, 21–33 (2016). [DOI] [PubMed] [Google Scholar]
- 28.U. C. Eze, A. Bhaduri, M. Haeussler, T. J. Nowakowski, A. R. Kriegstein, Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 24, 584–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Lek, K. J. Karczewski, E. V. Minikel, K. E. Samocha, E. Banks, T. Fennell, A. H. O’Donnell-Luria, J. S. Ware, A. J. Hill, B. B. Cummings, T. Tukiainen, D. P. Birnbaum, J. A. Kosmicki, L. E. Duncan, K. Estrada, F. Zhao, J. Zou, E. Pierce-Hoffman, J. Berghout, D. N. Cooper, N. Deflaux, M. DePristo, R. Do, J. Flannick, M. Fromer, L. Gauthier, J. Goldstein, N. Gupta, D. Howrigan, A. Kiezun, M. I. Kurki, A. L. Moonshine, P. Natarajan, L. Orozco, G. M. Peloso, R. Poplin, M. A. Rivas, V. Ruano-Rubio, S. A. Rose, D. M. Ruderfer, K. Shakir, P. D. Stenson, C. Stevens, B. P. Thomas, G. Tiao, M. T. Tusie-Luna, B. Weisburd, H. H. Won, D. Yu, D. M. Altshuler, D. Ardissino, M. Boehnke, J. Danesh, S. Donnelly, R. Elosua, J. C. Florez, S. B. Gabriel, G. Getz, S. J. Glatt, C. M. Hultman, S. Kathiresan, M. Laakso, S. McCarroll, M. I. McCarthy, D. McGovern, R. McPherson, B. M. Neale, A. Palotie, S. M. Purcell, D. Saleheen, J. M. Scharf, P. Sklar, P. F. Sullivan, J. Tuomilehto, M. T. Tsuang, H. C. Watkins, J. G. Wilson, M. J. Daly, D. G. MacArthur, H. E. Abboud, G. Abecasis, C. A. Aguilar-Salinas, O. Arellano-Campos, G. Atzmon, I. Aukrust, C. L. Barr, G. I. Bell, S. Bergen, L. Bjørkhaug, J. Blangero, D. W. Bowden, C. L. Budman, N. P. Burtt, F. Centeno-Cruz, J. C. Chambers, K. Chambert, R. Clarke, R. Collins, G. Coppola, E. J. Córdova, M. L. Cortes, N. J. Cox, R. Duggirala, M. Farrall, J. C. Fernandez-Lopez, P. Fontanillas, T. M. Frayling, N. B. Freimer, C. Fuchsberger, H. García-Ortiz, A. Goel, M. J. Gómez-Vázquez, M. E. González-Villalpando, C. González-Villalpando, M. A. Grados, L. Groop, C. A. Haiman, C. L. Hanis, A. T. Hattersley, B. E. Henderson, J. C. Hopewell, A. Huerta-Chagoya, S. Islas-Andrade, S. B. Jacobs, S. Jalilzadeh, C. P. Jenkinson, J. Moran, S. Jiménez-Morale, A. Kähler, R. A. King, G. Kirov, J. S. Kooner, T. Kyriakou, J. Y. Lee, D. M. Lehman, G. Lyon, W. MacMahon, P. K. Magnusson, A. Mahajan, J. Marrugat, A. Martínez-Hernández, C. A. Mathews, G. McVean, J. B. Meigs, T. Meitinger, E. Mendoza-Caamal, J. M. Mercader, K. L. Mohlke, H. Moreno-Macías, A. P. Morris, L. A. Najmi, P. R. Njølstad, M. C. O’Donovan, M. L. Ordóñez-Sánchez, M. J. Owen, T. Park, D. L. Pauls, D. Posthuma, C. Revilla-Monsalve, L. Riba, S. Ripke, R. Rodríguez-Guillén, M. Rodríguez-Torres, P. Sandor, M. Seielstad, R. Sladek, X. Soberón, T. D. Spector, S. E. Tai, T. M. Teslovich, G. Walford, L. R. Wilkens, A. L. Williams, Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.K. J. Karczewski, L. C. Francioli, G. Tiao, B. B. Cummings, J. Alföldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O’Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski, C. A. A. Salinas, T. Ahmad, C. M. Albert, D. Ardissino, G. Atzmon, J. Barnard, L. Beaugerie, E. J. Benjamin, M. Boehnke, L. L. Bonnycastle, E. P. Bottinger, D. W. Bowden, M. J. Bown, J. C. Chambers, J. C. Chan, D. Chasman, J. Cho, M. K. Chung, B. Cohen, A. Correa, D. Dabelea, M. J. Daly, D. Darbar, R. Duggirala, J. Dupuis, P. T. Ellinor, R. Elosua, J. Erdmann, T. Esko, M. Färkkilä, J. Florez, A. Franke, G. Getz, B. Glaser, S. J. Glatt, D. Goldstein, C. Gonzalez, L. Groop, C. Haiman, C. Hanis, M. Harms, M. Hiltunen, M. M. Holi, C. M. Hultman, M. Kallela, J. Kaprio, S. Kathiresan, B. J. Kim, Y. J. Kim, G. Kirov, J. Kooner, S. Koskinen, H. M. Krumholz, S. Kugathasan, S. H. Kwak, M. Laakso, T. Lehtimäki, R. J. F. Loos, S. A. Lubitz, R. C. W. Ma, D. G. MacArthur, J. Marrugat, K. M. Mattila, S. McCarroll, M. I. McCarthy, D. McGovern, R. McPherson, J. B. Meigs, O. Melander, A. Metspalu, B. M. Neale, P. M. Nilsson, M. C. O’Donovan, D. Ongur, L. Orozco, M. J. Owen, C. N. A. Palmer, A. Palotie, K. S. Park, C. Pato, A. E. Pulver, N. Rahman, A. M. Remes, J. D. Rioux, S. Ripatti, D. M. Roden, D. Saleheen, V. Salomaa, N. J. Samani, J. Scharf, H. Schunkert, M. B. Shoemaker, P. Sklar, H. Soininen, H. Sokol, T. Spector, P. F. Sullivan, J. Suvisaari, E. S. Tai, Y. Y. Teo, T. Tiinamaija, M. Tsuang, D. Turner, T. Tusie-Luna, E. Vartiainen, M. P. Vawter, H. Watkins, R. K. Weersma, M. Wessman, J. G. Wilson, R. J. Xavier, B. M. Neale, M. J. Daly, D. G. MacArthur; Genome Aggregation Database Consortium , Author Correction: The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 590, E53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.E. C. Genin, B. Madji Hounoum, S. Bannwarth, K. Fragaki, S. Lacas-Gervais, A. Mauri-Crouzet, F. Lespinasse, J. Neveu, B. Ropert, G. Augé, C. Cochaud, C. Lefebvre-Omar, S. Bigou, A. Chiot, F. Mochel, S. Boillée, C. S. Lobsiger, D. Bohl, J. E. Ricci, V. Paquis-Flucklinger, Mitochondrial defect in muscle precedes neuromuscular junction degeneration and motor neuron death in CHCHD10S59L/+ mouse. Acta Neuropathol. 138, 123–145 (2019). [DOI] [PubMed] [Google Scholar]
- 32.T. Otani, M. C. Marchetto, F. H. Gage, B. D. Simons, F. J. Livesey, 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell 18, 467–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.I. K. Suzuki, D. Gacquer, R. Van Heurck, D. Kumar, M. Wojno, A. Bilheu, A. Herpoel, N. Lambert, J. Cheron, F. Polleux, V. Detours, P. Vanderhaeghen, Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.B. J. Molyneaux, P. Arlotta, J. R. L. Menezes, J. D. Macklis, Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437 (2007). [DOI] [PubMed] [Google Scholar]
- 35.S. A. Sloan, J. Andersen, A. M. Pașca, F. Birey, S. P. Pașca, Generation and assembly of human brain region–Specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Y. Hu, A. C. Hung, H. Cui, E. Dawkins, M. Bolós, L. Foa, K. M. Young, D. H. Small, Role of cystatin C in amyloid precursor protein-induced proliferation of neural stem/progenitor cells. J. Biol. Chem. 288, 18853–18862 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Q. H. Ma, T. Futagawa, W. L. Yang, X. D. Jiang, L. Zeng, Y. Takeda, R. X. Xu, D. Bagnard, M. Schachner, A. J. Furley, D. Karagogeos, K. Watanabe, G. S. Dawe, Z. C. Xiao, A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat. Cell Biol. 10, 283–294 (2008). [DOI] [PubMed] [Google Scholar]
- 38.B. A. Bergmans, S. A. M. Shariati, R. L. P. Habets, P. Verstreken, L. Schoonjans, U. Müller, C. G. Dotti, B. De Strooper, Neurons generated from APP/APLP1/APLP2 triple knockout embryonic stem cells behave normally in vitro and in vivo: Lack of evidence for a cell autonomous role of the amyloid precursor protein in neuronal differentiation. Stem Cells 28, 399–406 (2010). [DOI] [PubMed] [Google Scholar]
- 39.S. A. M. Shariati, P. Lau, B. A. Hassan, U. Müller, C. G. Dotti, B. De Strooper, A. Gärtner, APLP2 regulates neuronal stem cell differentiation during cortical development. J. Cell Sci. 126, 1268–1277 (2013). [DOI] [PubMed] [Google Scholar]
- 40.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck, Y. Hao, M. Stoeckius, P. Smibert, R. Satija, Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.K. R. Pennypacker, C. A. Kassed, S. Eidizadeh, J. P. O’Callaghan, Brain injury: Prolonged induction of transcription factors. Acta Neurobiol. Exp. (Wars) 60, 515–530 (2000). [DOI] [PubMed] [Google Scholar]
- 42.M. B. Johnson, P. P. Wang, K. D. Atabay, E. A. Murphy, R. N. Doan, J. L. Hecht, C. A. Walsh, Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 18, 637–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.N. Urbán, F. Guillemot, Neurogenesis in the embryonic and adult brain: Same regulators, different roles. Front. Cell. Neurosci. 8, 396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L. Telley, G. Agirman, J. Prados, N. Amberg, S. Fièvre, P. Oberst, G. Bartolini, I. Vitali, C. Cadilhac, S. Hippenmeyer, L. Nguyen, A. Dayer, D. Jabaudon, Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364, eaav2522 (2019). [DOI] [PubMed] [Google Scholar]
- 45.C. Mayer, C. Hafemeister, R. C. Bandler, R. Machold, R. Batista Brito, X. Jaglin, K. Allaway, A. Butler, G. Fishell, R. Satija, Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. Lange, W. B. Huttner, F. Calegari, Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5, 320–331 (2009). [DOI] [PubMed] [Google Scholar]
- 47.L. J. Pilaz, J. J. McMahon, E. E. Miller, A. L. Lennox, A. Suzuki, E. Salmon, D. L. Silver, Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron 89, 83–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Mitchell-Dick, A. Chalem, L. J. Pilaz, L. J. Pilaz, L. J. Pilaz, D. L. Silver, D. L. Silver, D. L. Silver, D. L. Silver, D. L. Silver, Acute lengthening of progenitor mitosis influences progeny fate during cortical development in vivo. Dev. Neurosci. 41, 300–317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. S. Nowakowski, S. B. Lewin, M. W. Miller, Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 18, 311–318 (1989). [DOI] [PubMed] [Google Scholar]
- 50.C. Liu, C. W. Zhang, Y. Zhou, W. Q. Wong, L. C. Lee, W. Y. Ong, S. O. Yoon, W. Hong, X. Y. Fu, T. W. Soong, E. H. Koo, L. W. Stanton, K. L. Lim, Z. C. Xiao, G. S. Dawe, APP upregulation contributes to retinal ganglion cell degeneration via JNK3. Cell Death Differ. 25, 663–678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. Kögel, R. Schomburg, E. Copanaki, J. H. M. Prehn, Regulation of gene expression by the amyloid precursor protein: Inhibition of the JNK/c-Jun pathway. Cell Death Differ. 12, 1–9 (2005). [DOI] [PubMed] [Google Scholar]
- 52.E. Copanaki, S. Chang, A. Vlachos, J. A. Tschäpe, U. C. Müller, D. Kögel, T. Deller, SAPPα antagonizes dendritic degeneration and neuron death triggered by proteasomal stress. Mol. Cell. Neurosci. 44, 386–393 (2010). [DOI] [PubMed] [Google Scholar]
- 53.G. P. Eckert, S. Chang, J. Eckmann, E. Copanaki, S. Hagl, U. Hener, W. E. Müller, D. Kögel, Liposome-incorporated DHA increases neuronal survival by enhancing non-amyloidogenic APP processing. Biochim. Biophys. Acta Biomembr. 1808, 236–243 (2011). [DOI] [PubMed] [Google Scholar]
- 54.A. Hendrickx, N. Pierrot, B. Tasiaux, O. Schakman, J. P. Brion, P. Kienlen-Campard, C. De Smet, J. N. Octave, Epigenetic induction of EGR-1 expression by the amyloid precursor protein during exposure to novelty. PLOS ONE 8, e74305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. Hendrickx, N. Pierrot, B. Tasiaux, O. Schakman, P. Kienlen-Campard, C. De Smet, J. N. Octave, Epigenetic regulations of immediate early genes expression involved in memory formation by the amyloid precursor protein of alzheimer disease. PLOS ONE 9, e99467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D. Barilá, R. Mangano, S. Gonfloni, J. Kretzschmar, M. Moro, D. Bohmann, G. Superti-Furga, A nuclear tyrosine phosphorylation circuit: c-Jun as an activator and substrate of c-Abl and JNK. EMBO J. 19, 273–281 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.T. Kallunki, T. Deng, M. Hibi, M. Karin, c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87, 929–939 (1996). [DOI] [PubMed] [Google Scholar]
- 58.B. J. Pulverer, J. M. Kyriakis, J. Avruch, E. Nikolakaki, J. R. Woodgett, Phosphorylation of c-jun mediated by MAP kinases. Nature 353, 670–674 (1991). [DOI] [PubMed] [Google Scholar]
- 59.E. Arguni, M. Arima, N. Tsuruoka, A. Sakamoto, M. Hatano, T. Tokuhisa, JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int. Immunol. 18, 1079–1089 (2006). [DOI] [PubMed] [Google Scholar]
- 60.L. Tiberi, J. Van Den Ameele, J. Dimidschstein, J. Piccirilli, D. Gall, A. Herpoel, A. Bilheu, J. Bonnefont, M. Iacovino, M. Kyba, T. Bouschet, P. Vanderhaeghen, BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat. Neurosci. 15, 1627–1635 (2012). [DOI] [PubMed] [Google Scholar]
- 61.J. Bonnefont, L. Tiberi, J. van den Ameele, D. Potier, Z. B. Gaber, X. Lin, A. Bilheu, A. Herpoel, F. D. Velez Bravo, F. Guillemot, S. Aerts, P. Vanderhaeghen, Cortical neurogenesis requires Bcl6-mediated transcriptional repression of multiple self-renewal-promoting extrinsic pathways. Neuron 103, 1096–1108.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. Lachmann, H. Xu, J. Krishnan, S. I. Berger, A. R. Mazloom, A. Ma’ayan, ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. P. Keasey, S. S. Kang, C. Lovins, T. Hagg, Inhibition of a novel specific neuroglial integrin signaling pathway increases STAT3-mediated CNTF expression. Cell Commun. Signal 11, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D. K. Mishra, M. P. Kim, SR 11302, an AP-1 inhibitor, reduces metastatic lesion formation in ex vivo 4D lung cancer model. Cancer Microenviron. 10, 95–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.C. A. Mutch, J. D. Schulte, E. Olson, A. Chenn, Beta-catenin signaling negatively regulates intermediate progenitor population numbers in the developing cortex. PLOS ONE 5, e12376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.N. Bengoa-Vergniory, R. M. Kypta, Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell. Mol. Life Sci. 72, 4157–4172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Y. Qi, X.-J. Zhang, N. Renier, Z. Wu, T. Atkin, Z. Sun, M. Z. Ozair, J. Tchieu, B. Zimmer, F. Fattahi, Y. Ganat, R. Azevedo, N. Zeltner, A. H. Brivanlou, M. Karayiorgou, J. Gogos, M. Tomishima, M. Tessier-Lavigne, S.-H. Shi, L. Studer, Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotechnol. 35, 154–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.T. Liu, T. Zhang, M. Nicolas, L. Boussicault, H. Rice, A. Soldano, A. Claeys, I. Petrova, L. Fradkin, B. De Strooper, M.-C. Potier, B. A. Hassan, The amyloid precursor protein is a conserved Wnt receptor. eLife 10, 1–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.K. J. Karczewski, L. C. Francioli, G. Tiao, B. B. Cummings, J. Alföldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O’Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski, C. A. A. Salinas, T. Ahmad, C. M. Albert, D. Ardissino, G. Atzmon, J. Barnard, L. Beaugerie, E. J. Benjamin, M. Boehnke, L. L. Bonnycastle, E. P. Bottinger, D. W. Bowden, M. J. Bown, J. C. Chambers, J. C. Chan, D. Chasman, J. Cho, M. K. Chung, B. Cohen, A. Correa, D. Dabelea, M. J. Daly, D. Darbar, R. Duggirala, J. Dupuis, P. T. Ellinor, R. Elosua, J. Erdmann, T. Esko, M. Färkkilä, J. Florez, A. Franke, G. Getz, B. Glaser, S. J. Glatt, D. Goldstein, C. Gonzalez, L. Groop, C. Haiman, C. Hanis, M. Harms, M. Hiltunen, M. M. Holi, C. M. Hultman, M. Kallela, J. Kaprio, S. Kathiresan, B. J. Kim, Y. J. Kim, G. Kirov, J. Kooner, S. Koskinen, H. M. Krumholz, S. Kugathasan, S. H. Kwak, M. Laakso, T. Lehtimäki, R. J. F. Loos, S. A. Lubitz, R. C. W. Ma, D. G. MacArthur, J. Marrugat, K. M. Mattila, S. McCarroll, M. I. McCarthy, D. McGovern, R. McPherson, J. B. Meigs, O. Melander, A. Metspalu, B. M. Neale, P. M. Nilsson, M. C. O’Donovan, D. Ongur, L. Orozco, M. J. Owen, C. N. A. Palmer, A. Palotie, K. S. Park, C. Pato, A. E. Pulver, N. Rahman, A. M. Remes, J. D. Rioux, S. Ripatti, D. M. Roden, D. Saleheen, V. Salomaa, N. J. Samani, J. Scharf, H. Schunkert, M. B. Shoemaker, P. Sklar, H. Soininen, H. Sokol, T. Spector, P. F. Sullivan, J. Suvisaari, E. S. Tai, Y. Y. Teo, T. Tiinamaija, M. Tsuang, D. Turner, T. Tusie-Luna, E. Vartiainen, J. S. Ware, H. Watkins, R. K. Weersma, M. Wessman, J. G. Wilson, R. J. Xavier, B. M. Neale, M. J. Daly, D. G. MacArthur, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. Klein, A. Goldman, H. Lee, S. Ghahremani, V. Bhakta, S. F. Nelson, J. A. Martinez-Agosto, Truncating mutations in APP cause a distinct neurological phenotype. Ann. Neurol. 80, 456–460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.K. V. Nguyen, K. Leydiker, R. Wang, J. Abdenur, W. L. Nyhan, A neurodevelopmental disorder with a nonsense mutation in the Ox-2 antigen domain of the amyloid precursor protein (APP) gene. Nucleosides Nucleotides Nucleic Acids 36, 317–327 (2017). [DOI] [PubMed] [Google Scholar]
- 72.R. N. Doan, B. Il Bae, B. Cubelos, C. Chang, A. A. Hossain, S. Al-Saad, N. M. Mukaddes, O. Oner, M. Al-Saffar, S. Balkhy, G. G. Gascon, M. Nieto, C. A. Walsh, Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167, 341–354.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. Levchenko, A. Kanapin, A. Samsonova, R. R. Gainetdinov, Human accelerated regions and other human-specific sequence variations in the context of evolution and their relevance for brain development. Genome Biol. Evol. 10, 166–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.F. Cavodeassi, F. Carreira-Barbosa, R. M. Young, M. L. Concha, M. L. Allende, C. Houart, M. Tada, S. W. Wilson, Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/β-catenin pathway. Neuron 47, 43–56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.C. Liu, J. Nathans, An essential role for frizzled 5 in mammalian ocular development. Development 135, 3567–3576 (2008). [DOI] [PubMed] [Google Scholar]
- 76.D. Rosebrock, S. Arora, N. Mutukula, R. Volkman, E. Gralinska, A. Balaskas, A. Aragonés Hernández, R. Buschow, B. Brändl, F. J. Müller, P. F. Arndt, M. Vingron, Y. Elkabetz, Enhanced cortical neural stem cell identity through short SMAD and WNT inhibition in human cerebral organoids facilitates emergence of outer radial glial cells. Nat. Cell Biol. 24, 981–995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C. Pérez Estrada, R. Covacu, S. R. Sankavaram, M. Svensson, L. Brundin, Oxidative stress increases neurogenesis and oligodendrogenesis in adult neural progenitor cells. Stem Cells Dev. 23, 2311–2327 (2014). [DOI] [PubMed] [Google Scholar]
- 78.S. Weinberger, M. P. Topping, J. Yan, A. Claeys, N. De Geest, D. Ozbay, T. Hassan, X. He, J. T. Albert, B. A. Hassan, A. Ramaekers, Evolutionary changes in transcription factor coding sequence quantitatively alter sensory organ development and function. eLife 6, 1–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. Florio, M. Albert, E. Taverna, T. Namba, H. Brandl, E. Lewitus, C. Haffner, A. Sykes, F. K. Wong, J. Peters, E. Guhr, S. Klemroth, K. Prüfer, J. Kelso, R. Naumann, I. Nüsslein, A. Dahl, R. Lachmann, S. Pääbo, W. B. Huttner, Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015). [DOI] [PubMed] [Google Scholar]
- 80.I. T. Fiddes, G. A. Lodewijk, M. Mooring, C. M. Bosworth, A. D. Ewing, G. L. Mantalas, A. M. Novak, A. van den Bout, A. Bishara, J. L. Rosenkrantz, R. Lorig-Roach, A. R. Field, M. Haeussler, L. Russo, A. Bhaduri, T. J. Nowakowski, A. A. Pollen, M. L. Dougherty, X. Nuttle, M. C. Addor, S. Zwolinski, S. Katzman, A. Kriegstein, E. E. Eichler, S. R. Salama, F. M. J. Jacobs, D. Haussler, Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.E. R. E. Schmidt, J. V. Kupferman, M. Stackmann, F. Polleux, The human-specific paralogs SRGAP2B and SRGAP2C differentially modulate SRGAP2A-dependent synaptic development. Sci. Rep. 9, 18692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.C. Y. Li, Y. Zhang, Z. Wang, Y. Zhang, C. Cao, P. W. Zhang, S. J. Lu, X. M. Li, Q. Yu, X. Zheng, Q. Du, G. R. Uhl, Q. R. Liu, L. Wei, A human-specific de novo protein-coding gene associated with human brain functions. PLOS Comput. Biol. 6, e1000734 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.K. Prodromidou, R. Matsas, Species-specific miRNAs in human brain development and disease. Front. Cell. Neurosci. 13, 559 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.G. Konopka, T. Friedrich, J. Davis-Turak, K. Winden, M. C. Oldham, F. Gao, L. Chen, G. Z. Wang, R. Luo, T. M. Preuss, D. H. Geschwind, Human-specific transcriptional networks in the brain. Neuron 75, 601–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.S. M. Chambers, C. A. Fasano, E. P. Papapetrou, M. Tomishima, M. Sadelain, L. Studer, Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.X. J. Li, S. C. Zhang, In vitro differentiation of neural precursors from human embryonic stem cells. Methods Mol. Biol. 331, 169–177 (2006). [DOI] [PubMed] [Google Scholar]
- 87.G. Mata, J. Heras, M. Morales, A. Romero, J. Rubio, SynapCountJ: A tool for analyzing synaptic densities in neurons. BIOIMAGING 2016 - 3rd Int. Conf. Bioimaging, Proceedings; Part 9th Int. Jt. Conf. Biomed. Eng. Syst. Technol. BIOSTEC 2016 2, 25–31 (2016).
- 88.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.B. Li, C. N. Dewey, RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. Bioinforma. Impact Accurate Quantif. Proteomic Genet. Anal. Res., 41–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.M. D. Robinson, D. J. McCarthy, G. K. Smyth, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Y. Benjamini, Y. Hochberg, Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
- 92.J. Komatsu, A. Cico, R. Poncin, M. Le Bohec, J. Morf, S. Lipin, A. Graindorge, H. Eckert, A. Saffarian, L. Cathaly, F. Guérin, S. Majello, D. Ulveling, A. Vayaboury, N. Fernandez, D. Dimitrova, X. Bussell, Y. Fourne, P. Chaumat, B. André, E. Baldivia, U. Godet, M. Guinin, V. Moretto, J. Ismail, O. Caille, N. Roblot, C. Beaupère, A. Liboz, G. Guillemain, B. Blondeau, P. Walrafen, S. Edelstein, RevGel-seq: instrument-free single-cell RNA sequencing using a reversible hydrogel for cell-specific barcoding. Scientific Reports 13 (2023). 10.1038/s41598-023-31915-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev, S. A. McCarroll, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Y. Liao, G. K. Smyth, W. Shi, FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 95.E. Y. Chen, C. M. Tan, Y. Kou, Q. Duan, Z. Wang, G. V. Meirelles, N. R. Clark, A. Ma’ayan, Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1, S4 to S8
Legends for tables S2 and S3
Legends for movies S1 to S4
Tables S2 and S3
Movies S1 to S4
Lentiviral vector sequence fasta file