Abstract
Biomedical devices comprise a major component of modern medicine, however immune-mediated fibrosis and rejection can limit their function over time. Here, we describe a humanized mouse model that recapitulates fibrosis following biomaterial implantation. Cellular and cytokine responses to multiple biomaterials were evaluated across different implant sites. Human innate immune macrophages were verified as essential to biomaterial rejection in this model and were capable of cross-talk with mouse fibroblasts for collagen matrix deposition. Cytokine and cytokine receptor array analysis confirmed core signaling in the fibrotic cascade. Foreign body giant cell formation, often unobserved in mice, was also prominent. Last, high-resolution microscopy coupled with multiplexed antibody capture digital profiling analysis supplied spatial resolution of rejection responses. This model enables the study of human immune cell–mediated fibrosis and interactions with implanted biomaterials and devices.
After multiple attempts, a humanized (immune system) mouse variant capable of recapitulating biomaterial fibrosis was identified.
INTRODUCTION
Implanted biomedical devices, whether single biomaterial or multicomponent, are an integral part of modern medicine, essential for many therapies, and used in millions of surgeries every year (1, 2). Naturally derived alginate hydrogel has a long history of use as a multipurpose biomaterial for a range of biomedical device applications including biosensors, tissue regeneration, cell encapsulation, and drug delivery (3–8). Synthetic polymer polydimethylsiloxane (PDMS) is also used in a large number of biomaterial devices, with breast implants being a prominent example (9). Polystyrene (PS) is another synthetic polymer used in microshunt implants for glaucoma (10) and nanofibrous membrane patches for pelvic reconstruction (11). It is also a major component (30 wt %) of an U.S. Food and Drug Administration (FDA)–approved medical grade thermoplastic silicone elastomer SIBS30 (12) used for prosthetic heart valves (13) and drug-eluting stent/balloon coatings (14). However, regardless of their being naturally derived or synthetic, micron to macroscale biomaterial systems typically elicit immunogenic responses that lead to their fibrotic encapsulation as part of foreign body response (FBR) rejection (15, 16).
To better understand and find solutions to this interfering FBR, numerous groups have carried out biocompatibility studies in mice (17–20) and non-human primate (NHP) cynomolgus monkeys (18–21) that mimic FBR observed in humans (4–6). Notably, this investigation has indicated that innate immune myeloid populations, namely, monocyte/macrophages, are required to confer profibrotic ability. Macrophages are a key mediator of material recognition, adhering to the surface of many types of biomaterials (15, 22–24), fusing into foreign body giant cells (FBGCs) (15, 25), before myofibroblast induction and fibrous capsule formation (15, 16, 23, 26).
To date, most biocompatibility studies have been carried out in rodents due to limited accessibility to nonhuman primates and multiple barriers to working in humans. For instance, ethical and technical concerns may make in vivo testing in large animals and humans difficult. However, one must be mindful that rodent models may not always be predictive of the translational potential of biomaterial platforms in human patients as a result of immunologic differences between species (27). Further complicating the field over many decades, different mouse strains have varying levels of fibrotic response: a general lack of fibrosis in BALB/c mice (17, 28) versus much more substantial in C57BL/6 mice (18–21).
Despite these issues, a number of interesting biomaterial device modifications have been identified in preclinical models. While device architecture is often driven by design, a unique physical feature—specifically a larger size and spherical shape of implants—was found to abrogate the immune response to multiple classes of biomaterials commonly used in modern biomedical devices that were implanted in rodents and nonhuman primates (20). Furthermore, certain biomaterial surface chemistries (21) and drugs (18, 19) have also been identified to significantly reduce fibrosis in both models. However, without these innovations going through clinical trial testing, one cannot be sure of the level of their translatability into human patients.
Thankfully, humanized mouse models have been developed in an attempt to recapitulate human immune responses observed in patients (29). To do so, humanized mice may be created and modified using human tissues, cells, and/or genes. This is done to improve the faithfulness of immune responses in numerous disease models including cancer, tissue inflammation, infectious agents, and immune dysregulation (30). However, even humanized model variants can be limited in their ability to mimic the full range and complexity of human immune responses. Depending on how they are generated, numerous differences in immune cell development and maturation as well as lymph node and secondary lymphoid tissue development have been observed between humanized model variants (30, 31). Furthermore, while humanized mice have been under study for the past 30 years, needed improvements in human immune cell function occurred with the development of immunodeficient IL2rg−/− mice that have supported heightened levels of immune engraftment (32) and function over earlier humanized non-obese diabetic (NOD)-scid models (33). Moreover, certain hematopoietic stem cell (HSC)–engrafted humanized models often support adaptive immune B and T cell development, which is ideal for studies in pathogen and transplant response (29, 30, 34–37), but not for fibrosis.
Many recognize the additional utility that studying human immune system dynamics in a host mouse provides the ability to more frequently sample tissues and better dissect molecular mechanism (31). To this point, previous reports have described humanized mouse models to evaluate host responses to allogeneic and xenogeneic transplants (36–38) and various inflammation responses to biomaterials (39, 40). However, identification of a humanized model that is capable of recapitulating biomaterial-induced FBR and fibrosis to enable study of direct human immune cell interactions in foreign body biomaterial response has not been reported.
We hypothesized that further advances in engraftment, maintenance, and behavior of immune cell subsets were required to create humanized mouse models as predictive preclinical tools for fibrosis. Here, we describe a humanized mouse model that engrafts with a robust human immune system at both the level of innate and adaptive immunity. To broaden the strength of our findings, we used this model to evaluate the fibrotic response to both natural and synthetic biomaterials as well as across multiple [subcutaneous (SC) and intraperitoneal (IP)] implantation sites.
RESULTS
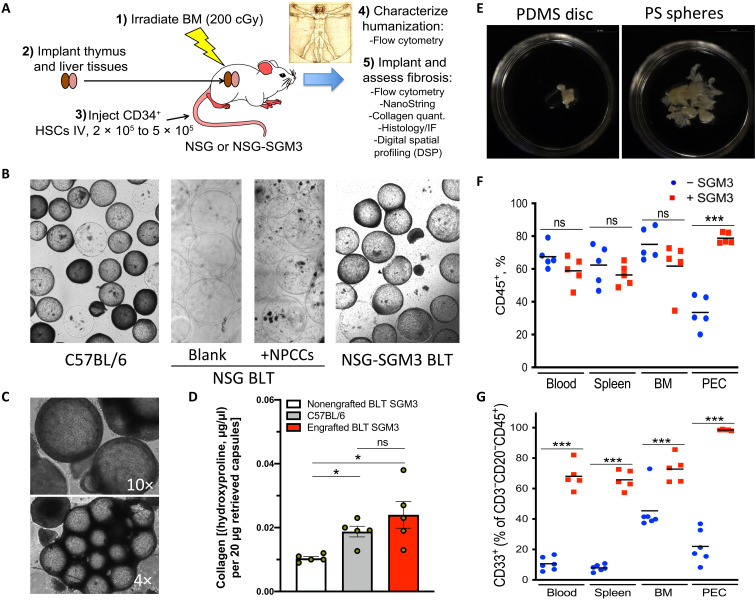
To identify an appropriate humanized model capable of allowing the investigation of the fibrotic response to implanted biomaterials, we studied NOD-scid mice bearing a targeted mutation in the interleukin 2 (IL2) receptor common gamma chain locus (IL2gnull), commonly abbreviated as NOD-scid-gamma (NSG) (41), as well as NSG mice that transgenically expressed human stem cell factor (SCF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and IL-3, abbreviated as NSG-SGM3. First, we implanted empty 500-μm-diameter hydrogel spheres (0.5 ml per mouse) and transplanted similar spherical capsules containing a high load (10,000 clusters per mouse) of foreign xenogeneic neonatal porcine cell clusters (NPCCs), used in an attempt to engage a more prominent host rejection response, into the IP cavity of both nonengrafted NSG mice as well as human fetal thymus and liver engrafted mice that were also injected with autologous human CD34+ HSCs, termed “NSG-BLT” mice (Fig. 1A). As expected, in nonengrafted NSG control mice, no fibrotic response was observed after 4 weeks. However, as compared to fibrosed NPCC-containing 500-μm SLG20 alginate capsule controls retrieved from fibrosis-positive control C57BL/6 mice (Fig. 1B, left), even in NSG-BLT mice engrafted with a functional human immune system, no fibrotic response was observed after 4 weeks with either empty capsules or capsules containing NPCCs (Fig. 1B, middle). However, upon inspection, we realized that while both models had appropriate levels of adaptive immune T and B cells, the innate immune arm of their immune systems was deficient.
Fig. 1. Identifying a humanized model capable of biomaterial fibrosis.
500-μm-diameter SLG20 alginate spheres (0.5-ml volume) were implanted into the IP space of C57BL/6 and various humanized mouse models for 28 days and then analyzed for the degree of fibrosis. (A) Graphic showing steps required to achieve fibrotic FBR in the human HSC–engrafted NSG-SGM3 BLT model. BM, bone marrow; IV, intravenous. (B) Bright-field images of retrieved spheres reveal significant overgrowth in control C57BL/6 mice (left), with no fibrosis in NSG BLT mice, without or with xenogeneic neonatal pig cell clusters (NPCCs) (middle images). FBR was only observed after transgenic expression of three human cytokines [SCF, GM-CSF, and IL-3 (SGM3)] for innate immune cell maintenance. (C) Bright-field images (4× and 10×) of fibrosed spheres retrieved from engrafted NSG-SGM3 BLT mice 28 days after implantation. (D) Collagen levels on retrieved capsules by hydroxyproline assay from nonengrafted NSG-SGM3 BLT (white), wild-type C57BL/6 (gray), or engrafted (humanized) NSG-SGM3 BLT mice (red). (E) Phase contrast images of fibrosed and epididymal fat-embedded 5-mm PDMS discs and 500-μm PS spheres retrieved 28 days after IP implantations (alginate in fig. S1B). (F) Flow cytometry analysis reflected similar CD45+ engraftment without (blue) versus with (red) SGM3 incorporation in the blood, spleen, and bone marrow; however, a significant increase occurred in the IP space. PEC, peritoneal exudate cell. (G) With SGM3 adoption, CD45+CD3−CD20−CD33+ innate myeloid lineages were significantly increased in all compartments. For statistical analysis, one-way analysis of variance (ANOVA) with Bonferroni multiple comparison correction was used. *P < 0.05 and ***P < 0.0001; ns, not significant. n = 5 (biologic replicates) per group. Experiments were repeated three times.
Therefore, to further enhance innate immune levels in the NSG-BLT model, we adopted triple transgenic NSG-SGM3 mice (also expressing three human cytokines: SCF, GM-CSF, and IL-3) to stimulate development and maintenance of human immune myeloid cell populations. Only following incorporation of the NSG-SGM3 BLT model (42) did we finally observe intact FBR against both NPCC-containing and blank (empty) alginate spheres [Fig. 1, B (right) and C, and fig. S1]. Establishing physiologically relevant magnitude response in this model, levels of collagen deposition were not significantly different between wild-type C57BL/6 and engrafted (humanized) NSG-SGM3 BLT mice (Fig. 1D). In addition, we also observed fibrosis of synthetic biomaterial polymer implants in this model with both PDMS 5-mm molded discs and 500-μm PS spheres after implantation for 4 weeks in the peritoneal cavity (Fig. 1E). When analyzing the human cell populations in the engrafted NSG-BLT mice, we observed that although human CD45+ immune cells were present throughout most of the mouse tissue compartments examined (blood, bone marrow, and spleen), few were present in the peritoneal cavity (~5 × 106 could be recovered from a peritoneal lavage) (Fig. 1F and fig. S2, A and B). Upon further inspection, NSG-SGM3 BLT mice exhibited human CD45+CD3−CD20−CD33+ innate myeloid cells in high numbers in circulating blood as well as the spleen, bone marrow, and within the peritoneal exudate cell (PEC; ~2 × 107) population (Fig. 1G and fig. S2, B and C).
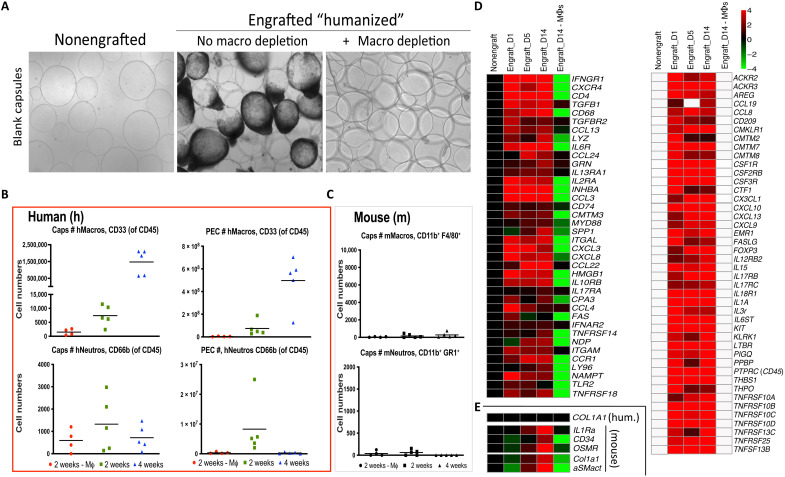
Previous work has demonstrated the important role of macrophages in fibrosis (15–17, 23, 25). Furthermore, study of fibrosis in wild-type C57BL/6 mice and nonhuman primate models has demonstrated the importance of macrophages in the fibrotic response in these animal systems (17–21). To study the role of human macrophages in the NSG-SGM3 profibrotic humanized mouse model, we performed clodrosome-based depletion (18). As expected, clodrosome depletion of phagocytic human monocyte/macrophages resulted in complete loss of FBR following 500-μm-diameter SLG20 alginate sphere 2-week peritoneal implantations in engrafted NSG-SGM3 BLT mice (Fig. 2A). Flow cytometry analysis of cells dissociated from the surface of retrieved alginate spheres at 2 and 4 weeks post-peritoneal cavity implantation, and nearby epididymal fat pads, or taken by peritoneal lavage (Fig. 2B, top, green and blue, respectively; and figs. S3 and S4) showed significantly increased levels of human CD45+CD3−CD20−CD33+ myeloid cells. These cells were also eliminated by clodrosome-maintained macrophage depletion throughout the 2-week implantation period (Fig. 2B, top, red). By comparison, human neutrophil abundance (CD45+CD66b+) did not significantly change in either case (Fig. 2B, bottom). The prior suppression of mouse immunity by irradiation was efficient, with absolute counts of residual mouse macrophages (F4/80+CD11b+) and neutrophils (Ly6g+CD11b+) being substantially reduced (Fig. 2C). NanoString array–based analysis of human physical cell markers, cytokines, and cytokine receptors was performed on RNA extracts taken from cells dissociated directly off of the surface of retrieved alginate spheres collected at 1, 5, and, 14 days after implantation in engrafted NSG-SGM3 BLT mice and compared to nonengrafted NSG-SGM3 controls and engrafted NSG-SGM3 BLT mice treated with macrophage-depleting clodrosomes (Fig. 2D). Last, no human collagen (COL1A1) expression was observed; instead, a slightly delayed increase of both mouse (host) fibroblast marker alpha-smooth muscle actin (αSMactin) and fibrotic extracellular matrix deposition marker collagen (Col1a1) was detected, indicating mouse myofibroblast cross-talk with the human immune system (Fig. 2E).
Fig. 2. FBR human macrophage dependence and mouse fibroblast cross-talk in NSG-SGM3 BLT humanized model.
(A) As compared to nonfibrosed (nonengrafted) and fibrosed (engrafted) controls, clodrosome depletion of human macrophages resulted in complete loss of fibrosis on 500-μm-diameter SLG20 alginate spheres following 2-week IP implantations in engrafted NSG-SGM3 BLT mice. (B) Flow cytometry analysis of human immune cell numbers (#) dissociated from capsule spheres (Caps) or PECs retrieved 2 or 4 weeks after IP implantation (green versus blue, respectively) showing significant increases of human (h)CD45+CD3−CD20−CD33+ myeloid cells, which were eliminated following clodrosome macrophage (Mϕ) depletion throughout 2-week implantations (red). By comparison, human neutrophils (CD45+CD66b+) did not significantly change. (C) As compared to very high absolute human immune cell numbers [in (B)], effective mouse immune cell elimination by irradiation was confirmed by low counts of residual mouse (m) macrophages (F4/80+CD11b+) and neutrophils (Ly6g+CD11b+), with no significant response by either population to material implantation or macrophage depletion. (D) NanoString analysis of cell and cytokine markers on alginate spheres at 1, 5, and 14 days after implantation in engrafted NSG-SGM3 BLT mice versus non-engrafted and engrafted but macrophage-depleted controls. Similar to wild-type FBR, macrophage and B cell markers were some of the largest dynamic responders. White, within background of the assay. (E) No human collagen was observed; instead, delayed mouse αSMactin fibroblast and collagen (Col1a1) expression indicated mouse myofibroblast engagement. For all, n = 5 (biologic replicates) per treatment. Flow cytometry was performed twice, and NanoString was performed once. For flow comparisons, one-way ANOVA (***P < 0.0001 versus clodrosome-depleted controls) was used. For NanoString, log2 scale was used; for statistical analysis, see Materials and Methods.
Flow cytometry analysis was extended to assay for potential adaptive immune responses in the engrafted NSG-SGM3 BLT model (fig. S5). While T and B cells were detected in both the blood and spleen compartments of engrafted NSG-SGM3 BLT mice (fig. S6), the majority of CD45+ immune cells on the surface of alginate capsules retrieved 2 weeks after implantation were CD33+ innate myeloid (CD3− and CD19−) cells (fig. S6A, right). Similar to what was observed in biomaterial-implanted wild-type C57BL/6 mice (18), CD19+ B cells were only present at ~5% and CD3+ T cells were not present at notable levels (<0.524 ± 0.11% of CD45+ cells) on the implanted biomaterial surface. Furthermore, the few CD3+ cells that were observed were also CD4− and CD8− (fig. S6B, right). However, the lack of T cell presence on the surface of retrieved biomaterial was not due to a lack of diverse T cell subsets in the NSG-SGM3 BLT model, with significantly positive staining for CD69, Human Leukocyte Antigen–DR isotype (HLA-DR), Programmed cell death protein 1 (PD-1), C-C chemokine receptor type 7 (CCR7), and CD45RA in the blood and spleen for both CD4+ and CD8+ T cell subsets, with naïve, activated, TEMRA, central memory, and effector/effector memory subsets detected (fig. S6, C to F). Demonstrating that these observed responses are faithful of wild-type response, comparable markers of T cell subsets were not only also observed systemically in implanted C57BL/6 mice (fig. S7) but, as seen in the NSG-SGM3 BLT model, were also absent from the surface of implanted alginate capsules.
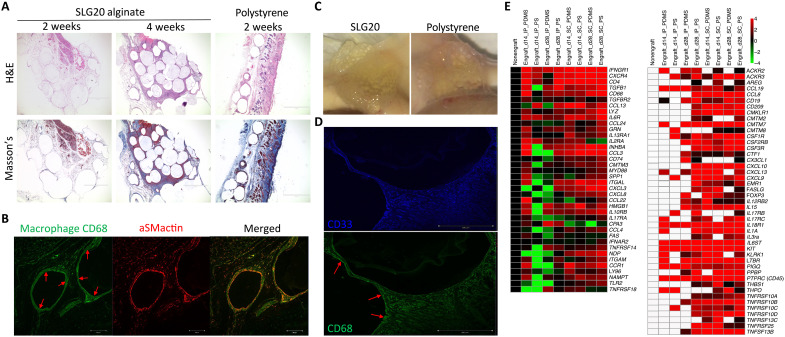
To confirm that fibrosis induction was not occurring solely in the peritoneal space of engrafted NSG-SGM3 BLT mice, we also implanted 500-μm SLG20 alginate and PS spheres into the SC space. Hematoxylin and eosin (H&E), Masson’s Trichrome, and αSMactin staining of sections of excised SC tissue after 14 and 28 days confirmed that induction of FBR was not an IP space-specific phenomenon (Fig. 3A and figs. S8 to S10). In addition to higher magnification histology (fig. S8, A to C), confocal microscopy also confirmed that FBGC formation (visualized by fused macrophage marker human CD68 staining, green) was intact in our humanized NSG-SGM3 BLT mouse model (Fig. 3, B and D, bottom, red arrows). In addition, FBGCs colocalized with an inner ring of myofibroblast marker αSMactin (Fig. 3B, red). Bright-field photos confirmed prominent fibrotic encapsulation of both alginate and PS spheres upon retrieval following 4-week SC implantations (Fig. 3C). Immunofluorescent staining of immune cell infiltration zones immediately around SC implants was also used to confirm that the majority of earlier identified human CD45+CD33+ myeloid cells coexpressed human CD68, indicating that they are largely human innate immune macrophages (Fig. 3D and figs. S11 and S12). Additional analysis of RNA collected from dissociated tissues (IP and SC) taken at either 14 or 28 days after implantation confirmed that many of the same cell and cytokine markers identified from naturally derived biomaterial alginate hydrogel IP implants (as shown in Fig. 2) were also significantly increased for both synthetic PDMS disc and PS sphere implants (Fig. 3E).
Fig. 3. FBR and foreign body giant cell formation is observed in the SC site in engrafted NSG-SGM3 BLT humanized mice.
500-μm SLG20 alginate or PS spheres (0.5 ml) and 5-mm PDMS discs were implanted for 14 or 28 days into the SC space of NSG-SGM3 BLT humanized mice and analyzed for degree of fibrosis upon retrieval. (A) H&E- and Masson’s Trichrome–stained histological sections of excised SC tissue 14 and 28 days after implantation for alginate and 14 days for PS (additional photos in figs. S8 and S9) (4×). Scale bars, 500 μm. (B) Confocal microscopy showing prominent FBGC formation (by macrophage marker CD68, green) colocalized with an inner ring of myofibroblast marker αSMactin (red) at day 28. (C) Photos showing prominent encapsulation of alginate and PS spheres upon retrieval following 4-week SC implantations. (D) Confocal microscopy confirming that the majority of earlier identified (Figs. 1 and 2) CD33+ myeloid cells coexpress macrophage marker CD68. CD68 and CD33 antibodies are anti-human; our αSMactin Ab is pan-species and detecting mouse fibroblasts here. (E) NanoString analysis of physical cell and cytokine markers analyzed from deposited cell RNA extracts from PS spheres and 5-mm PDMS discs collected at 14 and 28 days after IP or SC implantations in engrafted NSG-SGM3 BLT mice as compared to nonengrafted controls. White, within assay background. NanoString analysis was performed once (log base-2 scale); for statistical analyses, see Materials and Methods. n = 5 (biologic replicate) mice per group for all assays. Experiments were repeated three times.
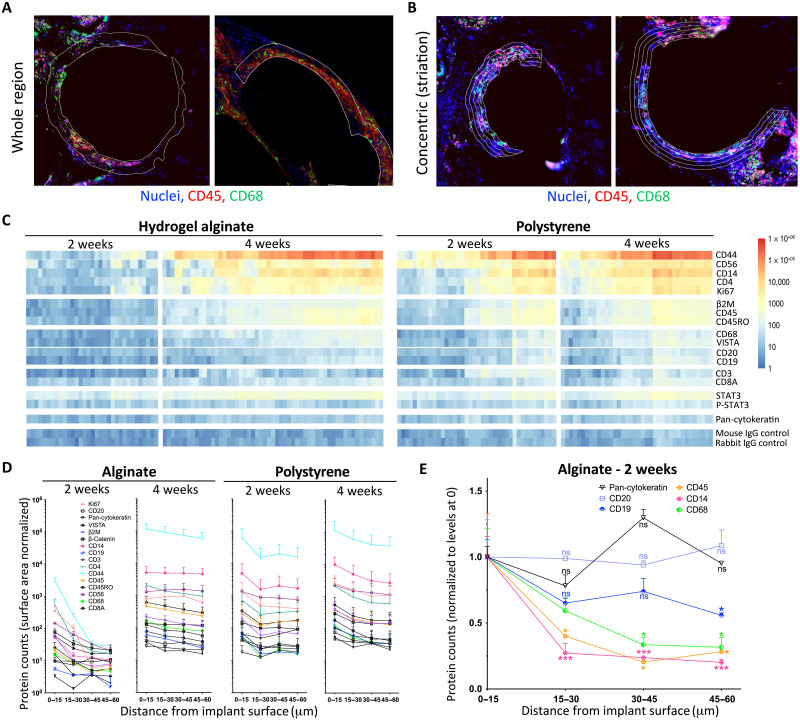
To further discern whether immune cells were differentially localizing in the immunologic infiltration around our biomaterial implants, we used a NanoString protein-level three-dimensional digital spatial profiling (DSP) analysis platform of human immune cell–mediated FBR in engrafted NSG-SGM3 BLT mice. 500-μm-diameter SLG20 alginate or PS spheres were implanted into the SC space of NSG-SGM3 BLT mice for 14 and 28 days. Per NanoString DSP protocols, immunofluorescence microscopy using 4′,6-diamidino-2-phenylindole (DAPI) nuclear (blue), human leukocyte marker CD45 (red), and human macrophage marker CD68 (green) staining was used to identify (whole) regions of interest (ROIs) (white lined areas) for subsequent ultraviolet (UV) laser ablation to decouple linked nucleotide probes to measure binding of a 30-plex human antibody (Ab) panel (Fig. 4A and figs. S11 and S12, A to C). In discussion with NanoString, we determined that ROIs could be performed around the same implant site but with successive rounds of ablation where the UV laser was incrementally increased 15 μm at a time, thereby capturing concentric rings of Ab binding and immune localization data as a function of distance (0 to 15, 15 to 30, 30 to 45, and 45 to 60 μm) moving away from the implant surface (Fig. 4B and fig. S12, D and E). Heatmap plots and log-scale line graph depictions of changing Ab data as a function of distance were produced for the entire DSP NanoString human 30-plex Ab panel, with data for both earlier 2-week and later 4-week alginate and PS implants (Fig. 4, C and D, and fig. S13). While alginate had a more muted human immune response at 2 weeks (similar to reduced immunogenicity at early timepoints in wild-type C57BL/6 mice), the response was as robust as against PS by 4 weeks (Fig. 4D, left versus right). Furthermore, we determined that macrophage marker for human (h)CD68 was largely localized directly at the implant surface and dropped off exponentially as distance from the implant increased, while B cell markers hCD19 and hCD20 decreased more slowly in a linear fashion, moving away from the implant surface at early times for less immunogenic hydrogel alginate spheres (Fig. 4E), with hCD19 localizing more toward the implant surface (fig. S13). While the NanoString DSP platform was utilizable on human/humanized tissues, it was not the case with previously collected, in-house nonhuman primate samples (fig. S14).
Fig. 4. DSP of human immune presence on and around implanted biomaterials in engrafted NSG-SGM3 BLT mice.
500-μm-diameter SLG20 alginate or PS spheres (0.5 ml) were implanted into the SC space of humanized NSG-SGM3 BLT mice for 14 and 28 days. (A) Per NanoString digital spatial profiling (DSP) protocols, immunofluorescence microscopy with DAPI nuclear (blue), leukocyte CD45 (red), and macrophage CD68 (green) staining was used to identify (whole) ROIs (white lines) for UV laser ablation to decouple probes from a 30-plex (anti-human, h) Ab panel. (B) With the DSP team, we found that concentric ring ROIs could be collected around the same implant with successive rounds of ablation where the UV laser was incrementally increased to 15 μm, thereby capturing data as a function of distance from its surface. (C) Heatmaps showing the ROI data for all 30 antibodies for both 2-week and 4-week alginate and PS implants. (D) Log-scale line graphs of data [as reflected in (C)]. Muted at 2 weeks, alginate FBR is as robust as PS’s by 4 weeks. (E) On the basis of our concentric ring DSP method [in (B)], macrophage CD68 was largely localized at the implant surface, while B cell markers CD19 and CD20 decreased, slowly moving away. One-way ANOVA (Bonferroni comparison) was used (*P < 0.05 and ***P < 0.0001, versus 0-μm distance). Data are presented as means ± SE. n = 5 (biologic replicate) mice per group. DSP analysis was run once across n = 2 biologic replicates per treatment group (SLG20 and PS) per time point (2 and 4 weeks), with 20 areas of interest per sample. STAT3, signal transducers and activators of transcription 3; P-STAT3, phosphorylated STAT3; IgG, immunoglobulin G.
DISCUSSION
Humanized mice are typically created from highly immunodeficient mice that are then irradiated and engrafted with functional human immune systems following transplantation with human HSCs and/or tissues/organoids (30). Their highly immune compromised state allows for significant reduction of inflammation-related complications following irradiation of residual bone marrow and pre-existing mouse immunity. Following subsequent human immune system engraftment, these models have been used by many groups to investigate human immune cell dynamics in response to various disease settings (e.g., autoimmunity, cancer, pathogen, and transplantation) (30). Generation of humanized systems was desired to recapitulate immune responses observed in humans (29) while enabling more extensive exploration of microscopic phenomena and mechanism that is often too difficult to carry out in patients. However, it must also be noted that there are many humanized mouse model variants (>40 to 50) (31), and depending on how they are created, this many models result in a wide array of human immune cell as well as lymphatic development and behaviorial permutations (30, 31).
While humanized mice have been under investigation for 30+ years, needed improvements in human immune cell function still remained and did not occur until the development of further immunodeficient IL2rg−/− mice. Notably, this strain has supported heightened levels of immune engraftment (32), due in part to improved major histocompatibility with human cells, as well as function over earlier humanized NOD-scid models (33). These NSG humanized variants have been used for improved studies in human hematopoiesis, studying individual immune cell functions, and regenerative medicine in addition to the aforementioned applications in disease. Furthermore, reports with the NSG strain have also described humanized models to evaluate host responses to allogeneic and xenogeneic transplants (36–38) as well as various inflammation responses to biomaterials (39, 40). However, identification of a humanized model (NSG or otherwise) that was capable of recapitulating biomaterial-induced FBR and fibrosis remained elusive. The lack of fibrosis was observed for samples retrieved from another humanized variant, HSC-engrafted NRG-Akita mice (35), as well.
We hypothesized that a remaining yet unestablished advance in engraftment, maintenance, and behavior of immune cell subsets was therefore required to create a humanized mouse model for biomaterial fibrosis. This suspicion was driven by the fact that multiple earlier humanized model attempts failed to demonstrate biomaterial-induced FBR and fibrosis (Fig. 1B, middle) (35, 39, 40) in conjunction with our beginning to understand which immune cell types were not only required but also necessary for driving FBR in our wild-type mouse and non-human primate models (18, 19). Upon closer inspection of our earlier NSG-BLT model attempts, we realized that while the mice had appropriate levels of adaptive immune T and B cells, the innate immune arm of their immune systems (including macrophages, required for intact FBR) was deficient (i.e., opposite to SCID or Rag1 or Rag2 knockout mice, which have intact innate immunity but lack T and B cells) (Fig. 1G and fig. S2). Therefore, to produce appropriate innate immune levels in the NSG-BLT model, we adopted triple transgenic NSG-SGM3 mice, which are also designed to stably express three human cytokines (SCF, GM-CSF, and IL-3) to stimulate the development and maintenance of human immune myeloid cell populations following engraftment.
Following adoption of this triple transgenic model, not only did we finally see the formation of stable foreign body and biomaterial-induced fibrotic response (Fig. 1), but we also detected immune responses similar to those seen in our wild-type animal models (18)—namely, macrophages (e.g., CD68, CSF1R, and EMR1), B cells (e.g., CD19), and macrophage-expressed B cell chemoattractant CXCL13—as some of the strongest dynamic cell and cytokine responders (Figs. 2, B and D, and 3, B, D, and E). Furthermore, while we detected significant CD4 increases both at the RNA (Fig. 3D) and protein (Fig. 4C) levels, the lack of observed T cell response on implanted capsules by flow cytometry (fig. S6B, right) suggests that CD4 expression is likely, instead originating from, responding macrophages. Notably, not only has CD4 expression been observed on monocyte-derived macrophages (43), but also CD4 ligation on human macrophages triggers their differentiation (44). Similar to the need for macrophages in wild-type fibrotic response (18, 19), we confirmed that macrophages are required to confer fibrosis in this humanized system, as fibrosis was lost upon their depletion (Fig. 2). Our data in the NSG-SGM3 BLT model (Fig. 1G) confirms that the vast majority (90+ %) of CD45+ cells in the peritoneum (PEC) are CD3−CD20−CD33+ and that the vast majority of cells on 500-μm alginate spheres retrieved 2 weeks after implantation are CD33+ (~40+ %) (fig. S6A). This percentage mirrors magnitude response of CD68+ macrophages (~40 to 50%) on alginate spheres in wild-type C57BL/6 mice (18). Furthermore, we used immunofluorescent staining to show that the majority of CD33+ cells are co-positive CD68+ and therefore macrophages (Fig. 3D). While we cannot make claims regarding CD33+ cell phenotypes or subtypes at this time, we plan to use the NSG-SGM3 BLT model to investigate this in the future. Furthermore, we believe that the SGM3 modification resulted in such a large expansion in both CD45+ and CD45+CD3−CD20−CD33+ fractions in the PEC as a consequence of the SGM3 modification supporting myeloid/macrophage immune cell proliferation and maintenance. Notably, macrophages make up the vast majority (~50 to 60%) of immune cells in the PEC in wild-type C57BL/6 mice (18); therefore, without the SGM3 cytokines supporting myeloid and macrophage maintenance, functional depletion of these cells in the peritoneum is observed in the non-SGM3 variant humanized mouse model (Fig. 1, F and G). Last, while CD33 is largely recognized as a myeloid cell marker, it can be present on alloantigen-activated T and natural killer (NK) cells (45). However, these responses should not be relevant in this model due to autologous matching of our human tissues and HSCs for engraftment. Furthermore, our gating demonstrated little to no co-positivity of our CD33 cells with CD3 or NKp46 (<1% in both cases). Last, the vast majority of the CD45+CD3−CD20−CD33+ cells in our model were also depleted from both the surface of retrieved capsules and from the PEC following clodrosome treatment (Fig. 2B, top, red), suggesting that these cells are predominantly monocytes/macrophages rather than lymphocytes.
Furthermore, B cells have previously been implicated in potentiating fibrosis (18), perhaps due to their ability to regulate macrophage phenotype and activation (46). The chemokine CXCL13 has also been reported to be responsible for B cell recruitment in a model of lymphoid neogenesis (47) and recently identified as a biomarker for idiopathic pulmonary fibrosis (48), perhaps suggesting further uses of this humanized variant for other model systems beyond biomaterial FBR. Adding to the utility of this model for FBR, FBGC formation, which has historically been less apparent in mice as opposed to easily visible in NHPs (18) and humans, was not only intact but also easily visible in our humanized NSG-SGM3 BLT mouse model (Fig. 3, B and D, bottom, red arrows).
For the purposes of this study, we wanted to ensure that fibrosis in our model would be observed regardless of biomaterial or implant architecture. To this point, gene expression analysis of IP and SC tissues taken at either 14 or 28 days after implantation confirmed that many of the same core—essential cell and cytokine markers, established by prior functional knockout (18) and identified on naturally derived biomaterial alginate 500-μm sphere hydrogel implants (as shown in Fig. 2)—were also significantly increased for both synthetic PDMS (5 mm wide–by–0.5 mm thick discs) and PS (also 500-μm sphere) implants (Fig. 3E). These results, together with intact fibrotic response in both IP and SC compartments, collectively suggest that human immune cell population(s) and signaling factors important in the fibrotic cascade are intact in this humanized mouse model for multiple biomaterial classes and across different anatomical sites. With that said, there were also some observed genes that showed differential responses over time (14 days versus 28 days) across our PS and PDMS implants as well as IP and SC implant sites. While some of the differences may be explained by differences in biomaterial and implant geometry, which have shown in the past affect host response kinetics (20), we also expect there to be implant site-specific phenomena as well. As an example, in wild-type C57BL/6 mice, more encompassing fibrotic overgrowth often takes longer in the SC compartment. There, an outer layer (around all spheres) forms first with ingrowth and individual sphere separation by fibrotic response following (18). This is in contrast to fibrosis seen on free-floating biomaterials spheres in the IP space, which are often shown to be individually fibrosed by 1 to 2 weeks after implantation, with subsequent fusion of fibrotic overgrowth and material clumping over more time. Therefore, further exploration in follow-on studies is needed to more deeply explore such biomaterial, architecture, and implant site-specific phenomena observed in wild-type animals in this humanized system.
The ability to compare similarities in FBR across different species (e.g., rodents, nonhuman primates, and human patients), while powerful, may sometimes be limited due to advanced technology toolsets that have been explicitly optimized for use with human specimens. Notably, many of the antibodies in the 30-plex NanoString DSP system used in this study displayed poor signal detection of the same respective biomarkers in nonhuman primate model tissues (fig. S14), likely due to the need for further Ab clone (titration and choice) optimization. With that said, this assay, which was originally developed for human tissue use (with no options for rodent tissue testing), worked incredibly well on specimens retrieved from humanized mice, suggesting that this model may provide a better surrogate model for use in conjunction with tools optimized for human specimens. This is especially poignant in cases where either no or limited human tissue might be available. Thus far, early DSP usage has been presented, showing traditional use of singular, lump-sum analysis zones (e.g., singular contiguous circles usually for analyzing tumor infiltration) (49). From early discussion with the NanoString DSP team, in the present study, we demonstrate modification of the ablation (probe decoupling) UV laser in a diameter-specific manner [e.g., smaller to get the first inner 15 μm area around the implant and then successively larger concentric circles in 15-μm increments to show staining quantification as a function of distance (0–15, 15–30, 30–45, and 45–60 um, respectively] away from the implant surface (Fig. 4A versus Fig. 4B). Notably, we observed some immune markers decreased slowly (more linearly) and other signals deteriorated quickly as a function of distance away from the implanted biomaterial surface (Fig. 4E and fig. S14). This suggests that certain immune cell responses (i.e., macrophage CD68) are highly localized to the implant surface, whereas others (i.e., B cell CD19) are more evenly spread throughout the surrounding infiltration zone.
Last, regarding the fibroblastic response, hydroxyproline proline (general collagen) quantification was used to directly compare collagen deposition at the same retrieval time point (day 28) following implantation of 500-μm-diameter alginate spheres in humanized NSG-SGM3 BLT mice and wild-type C57BL/6 mice (Fig. 1D). As shown (gray versus red), there were no statistically significant differences in magnitude of response. Furthermore, regarding fibroblast response kinetics and migration to the implantation site, the marker αSMactin used throughout the study at both RNA and protein levels indicated similar presence and magnitude of fibroblast response between both wild-type and humanized NSG-SGM3 BLT mice over multiple time points (days 1, 5, and 14) after implantation. As a commonly used marker for myofibroblast response during FBR, αSMactin increased in a delayed time point at 5 days after implantation in the humanized BLT SGM3 model, increasing further at days 14 and 28 (Figs. 2E and 3B, and fig. S10). These kinetics mirror wild-type fibroblast response to 500-μm-diameter alginate spheres in C57BL/6 mice, where initial response (~5% compositionally) was noted by day 7 and increasing further to ~20% into days 14 and 28 after implantation (18). While the present study was more focused on earlier events of human immune cell engagement required for recapitulating fibrosis, we feel that this humanized NSG-SGM3 BLT model system will be important for looking more into functional characterization of responding fibroblasts in the future. With that said, one interesting gene subset that was identified within our dataset mirroring αSMactin response kinetics includes IL1Ra, CD34, Oncostatin M receptor (OSMR), and Col1a1 (Fig. 2E). Notably, IL-1 signaling as well as CD34, OSMR, and Col1a1 have all been implicated in fibroblastic responses, with all highlighted within at least one of five distinct subsets that have emerged within arthritis and cardiac human single-cell atlas datasets (50, 51).
In summary, here, we show that the triple transgenic NSG-SGM3 BLT humanized mouse model has utility for the study of human immune fibrosis response and validate its use as a faithful surrogate model for biomaterial FBR. We established the utility of the human immune cell–engrafted NSG-SGM3 BLT model for use with the multiplexed Ab DSP array, optimized for human protein detection, thereby enabling the determination of human immune cell localization as a function of distance from implant surfaces. The fibrotic response to PDMS and related materials is intact in this model, with immunologic events comparable to previous findings in wild-type immune competent, profibrotic C57BL/6, New Zealand White Rabbit, and NHP models and in previously analyzed human clinical specimens (9, 18–21). While humanized mouse models are time-consuming and relatively expensive, efforts continue for further optimization to improve the ease, speed, and costs associated with their generation. Despite these needed optimizations, our findings suggest that the NSG-SGM3 BLT humanized mouse model identified here may enable useful preclinical modeling of human immune system and fibrotic responses to implanted materials and medical devices.
MATERIALS AND METHODS
Materials/reagents
In vitro reagents were obtained from Life Technologies (Carlsbad, CA), unless otherwise noted. Antibodies such as Alexa Fluor–conjugated anti-human CD45, CD14, CD33, and CD66b (described below) as well as anti-mouse F4/80 and Ly6g/Gr1 were purchased from BD Biosciences Inc. (San Jose, CA, USA), eBioscience (San Diego, CA, USA), BioLegend (San Diego, CA, USA), or Miltenyi Biotec (San Diego, CA, USA), as denoted below. For human/primate immunostaining, anti-human CD68 Alexa Fluor–conjugated (488 or 647) Ab was purchased from Santa Cruz Biotechnology (Dallas, TX). The same CD11b (anti-mouse/human) Ab (BioLegend) was used for both human and mouse staining. Cy3-conjugated anti-mouse αSMactin Ab was purchased from Sigma-Aldrich (St. Louis, MO). Medium PS spheres (mean, 526.6 ± 53.3 μm) (catalog no. 136, lot no. 30055) were purchased from Phosphorex (Hopkinton, MA). Material aliquots used in this study were submitted for endotoxin testing by a commercial vendor (Charles River Laboratories, Wilmington, MA), with results showing that spheres contained endotoxin levels of <0.05 EU/ml (below detectable limits) (table S1).
Fabrication of alginate hydrogel spheres
Alginate hydrogel spheres were made with an in-house customized electro-jetting system, as previously described (18). Spheres were made with a 1.4% solution of sterile alginate (PRONOVA SLG20, NovaMatrix, Sandvika, Norway), dissolved in 0.9% saline, and crosslinked with 250 ml of sterile 20 mM BaCl2 solution. 500-μm-diameter alginate hydrogel spheres were generated with a 25-gauge blunt needle, a voltage of 5 kV, and a flow rate of 200 μl/min. After, alginate spheres were washed with Hepes buffer four times and stored overnight at 4°C. Immediately before implantation, spheres were washed an additional two times with 0.9% saline.
Implantation surgeries
All protocols were approved by the UMass Medical School and Massachusetts Institute of Technology (MIT) Committees on Animal Care, with all surgical procedures and post-operative care supervised by UMass and MIT veterinary staff. All wild-type male immune-competent C57BL/6 mice or immunodeficient strains used for producing humanized models (described just below) were ordered pathogen-free from the Jackson Laboratory (Bar Harbor, ME). Implanted mice were anesthetized with 3% isoflurane in oxygen, and their abdomens were shaved and sterilized using betadine and isopropanol. Preoperatively, all mice received buprenorphine (0.05 mg/kg) and 0.2 ml of 0.9% saline SC for presurgery analgesia and hydration. A midline incision (0.5 mm) was made, and the peritoneal lining was exposed using blunt dissection. The peritoneal wall was then grasped with forceps, and an incision of 0.5 to 1 mm was made along the linea alba. Spheres (500 μl) were then loaded into a sterile pipette and implanted into the IP space. The incision was then closed using 5-0 PDS II absorbable sutures, and the skin was closed using wound clips and VetBond tissue glue. For SC implantation, ~200 to 300 μl of 500-μm SLG20 or PS spheres were injected SC following anesthesia with isofluorane. For macrophage depletion, clodrosomes (200 μl per mouse) (Encapsula NanoSciences, Nashville, TN) were injected intraperitoneally starting at −3 days (before implant day 0) and for every 7 days thereafter (on days −3, 4, and 11).
Mice used for humanized models
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NOD-scid IL2rgnull, NSG) mice and NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSG-SGM3) mice (all males) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). All mice were housed in a specific pathogen–free facility in microisolator cages, provided autoclaved food, and maintained on sulphamethoxazole-trimethoprim medicated water (Goldline Laboratories, Ft Lauderdale, FL, USA) and acidified autoclaved water on alternate weeks. All experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School and the recommendations in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council, National Academy of Sciences, 1996).
Engraftment of mice with human HSCs and generation of BLT mice
Human fetal tissues (gestational age 16 to 20 weeks) were obtained from Advanced Bioscience Resources (Alameda, CA, USA). The tissues were washed with RPMI 1640 supplemented with penicillin G (100 U/ml), streptomycin (100 mg/ml), fungizone (0.25 μg/ml), and gentamycin (5 μg/ml), and fragments (1 mm3) of the fetal thymus and liver were implanted in the renal subcapsular space of 8- to 12-week-old NSG and NSG-SGM3 mice that were previously irradiated with 200 and 100 centigray, respectively. Implanted mice were subsequently injected intravenously with 1 × 105 CD34+ autologous fetal liver-derived HSCs (determined by flow cytometry with a CD34-specific Ab; clone 581, BD Biosciences) between 4 to 6 hours after irradiation, as described previously (52). Mice were injected subcutaneously with gentamycin (0.2 mg) and cefazolin (0.83 mg) after surgery. After 12 weeks, flow cytometry analyses of the peripheral blood of HSC recipients determined the engraftment of the human immune system.
Retrieval of cells, tissues, and materials
As done previously (18), at desired time points after implantation, as specified in figures, mice were euthanized by CO2 administration, followed by cervical dislocation. In certain instances, 5 ml of ice-cold phosphate-buffered saline (PBS) was first injected intraperitoneally to rinse out and collect free-floating IP immune cells. An incision was then made using forceps and scissors along the abdomen, and IP lavage volumes were pipetted out into fresh 15-ml Falcon tubes (each prepared with 5 ml of RPMI 1640 cell culture media). Next, a wash bottle tip was inserted into the abdominal cavity. Krebs buffer was then used to wash out all material spheres into petri dishes for collection. After ensuring all the spheres were washed out or manually retrieved (if fibrosed directly to IP tissues, particularly epididymal and mesentery fat pads), they were transferred into 50-ml conical tubes for downstream processing and imaging. In certain instances, after IP lavage, portions of fibrosed IP tissues and material spheres were also excised for downstream flow cytometry and expression analyses.
Imaging of the retrieved material spheres
As done previously (18), for phase contrast imaging, retrieved materials were gently washed using Krebs buffer and transferred into 35-mm petri dishes for phase contrast microscopy using an EVOS XL microscope (Advanced Microscopy Group). For bright-field imaging of retrieved materials, samples were gently washed using Krebs buffer and transferred into 35-mm petri dishes for bright-field imaging using a Leica stereoscopic microscope.
Hydroxyproline assay
A similar volume (~250 μl from 500 μl total) of retrieved capsules per sample were snap frozen in a cryotube and stored at −80°C for long-term storage. Then, after placing the samples on ice, 200 μl of NP-40 buffer (Invitrogen, catalog no. FNN0021) was added to each tube and sonicated with a GE 130 PB probe sonicator at an amplitude of 20 for one to three rounds of 30- to 45-s pulses. Additional NP-40 buffer up to 200 μl was added if the capsules were not seen to break down (e.g., sample solution was too viscous) with the minimum amount of NP-40 buffer. The protein concentration was quantified using the microplate procedure in the BCA protocol (Pierce, catalog no. 23225) with samples run in duplicate.
Once the protein concentration was known, a hydroxyproline assay (Sigma-Aldrich, catalog no. MAK008) was performed according to the manufacturer’s instructions. Briefly, 20 μg of each lysate sample was transferred into a pressure-sealed screw cap polypropylene tube with 100 μl of concentrated hydrochloric acid (12 M HCl) and placed on a heat block set to 120°C for 3 hours. Samples were then mixed and centrifuged at 10,000g for 3 min. Then, 10 to 50 μl of supernatant from each tube were transferred into a 96-well plate and placed in a vacuum chamber overnight. Afterward, 10 μl of a standard solution (1 mg/ml) was diluted with 90 μl of deionized water to get a standard solution of 0.1 mg/ml. The new standard was used to make standards of 0, 0.2, 0.4, 0.6, 0.8, and 1.0 μg per well by loading 0, 2, 4, 6, 8, and 10 μl into the 96-well plate containing the dried samples. Then, a volume of 100 μl of the chloramine T/oxidation buffer (6 μl of chloramine T concentrate to 94 μl of oxidation buffer) was added and mixed to each protein sample and standard well and incubated for 5 min at room temperature. After, 100 μl of diluted 4-(dimethylamino)benzaldehyde (DMAB) solution (50 μl of DMAB concentrate to 50 μl of perchloric acid/isopropanol solution) was added to each sample and standard well and incubated for 90 min in an oven at 60°C. Last, the absorbance was measured at 560 nm using a plate reader, and a standard curve was generated with background noise subtracted from each sample. Using this standard curve, the concentration of hydroxyproline for each sample was generated.
Confocal immunofluorescence
Immunofluorescent imaging was used to determine immune populations attached to spheres. Retrieved SC tissues were fixed using 4% paraformaldehyde at 4°C. Samples were then dehydrated and embedded in paraffin, sectioned, and then permeabilized with 0.1% Triton X-100, and blocked with 1% bovine serum albumin (BSA) for 1 hour. Next, spheres were incubated for 1 hour in an Ab cocktail solution consisting of DAPI (500 nM) and specific marker probes (1:200 dilution) in BSA. After staining, samples were washed and cover-slipped following addition of ProLong Gold Antifade Mountant (Thermo Fisher Scientific, catalog no. P10144) and then imaged using a LSM 700 point scanning confocal microscope (Carl Zeiss Microscopy, Jena Germany) equipped with 5× and 10× objectives. For nonhuman primates (cynomolgus macaques), SC sphere-embedded samples were sectioned and stained according to traditional antigen retrieval and immunofluorescent methods, specifically looking at cellular nuclei (DAPI), macrophage marker CD68-AF488 (Santa Cruz Biotechnology, CA) and Cy3-conjugated anti-mouse αSMactin (fibrosis-associated myofibroblast marker) (Sigma-Aldrich, St. Louis, MO).
Histological processing for H&E and Masson’s Trichrome staining
Retrieved material-containing tissue (IP and/or SC) was fixed overnight using 4% paraformaldehyde at 4°C. After fixation, alginate sphere or retrieved tissue samples were washed using 70% alcohol. The materials were then paraffin embedded, sectioned at 5 μm thickness, and stained according to standard histological (H&E or Masson’s Trichrome) methods.
Flow cytometry analysis
Tissue samples for flow cytometry were prepared from mice as described below. Single-cell suspensions were prepared from the bone marrow and spleen. PECs were recovered from mice by flushing the peritoneal cavity using 5 ml of PBS. Whole blood was collected by submandibular bleed or cardiac puncture into tubes containing 1000 U of heparin sodium injection, USP (Pfizer Injectables, New York, NY). Single-cell suspensions of 1 × 106 cells in 100 μl or 100 μl of whole blood were washed with fluorescence-activated cell sorter (FACS) buffer (PBS supplemented with 2% fetal bovine serum) and then incubated with rat anti-mouse CD16/CD32 (clone 2.4G2, BD Biosciences) to block Fc binding. Fluorescently conjugated antibodies were then added to the samples and incubated for 30 min at 4°C in the dark. Stained cell suspensions were washed and fixed with 2% paraformaldehyde. Alternatively, blood or single-cell suspensions were treated with BD FACS Lysing Solution (BD Biosciences). Fixed samples were then suspended in FACS buffer and analyzed using a flow cytometry. At minimum of 50,000 events were acquired on LSRII or FACSCalibur instruments (BD Biosciences). Data analysis was performed with FlowJo (Tree Star Inc., Ashland, OR, USA) software.
Single-cell suspensions of freshly excised tissues were prepared using a gentleMACS Dissociator (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s protocol and as previously described (9, 18–21). Single-cell suspensions were prepared in a passive PEB dissociation buffer [1× PBS (pH 7.2), 0.5% BSA, and 2 mM EDTA], and suspensions were passed through 70-μm filters (catalog no. 22363548, Thermo Fisher Scientific, Pittsburgh, PA). All tissue and material sample-derived, single-cell populations were then subjected to red blood cell lysis with 5 ml of 1× red blood cell lysis buffer (catalog no. 00-4333, eBioscience, San Diego, CA, USA) for 5 min at 4°C. The reaction was terminated by the addition of 20 ml of sterile 1× PBS. The cells remaining were centrifuged at 300 to 400g at 4°C and resuspended in a minimal volume (~50 μl) of eBioscience staining buffer (catalog no. 00-4222) for Ab incubation. All samples were then costained in the dark for 25 min at 4°C with fluorescently tagged monoclonal antibodies (1:100 dilution). For human/primate immunostaining, anti-human CD68 (clone KP1) Alexa Fluor–conjugated (488 or 647) Ab was purchased from Santa Cruz Biotechnology (Dallas, TX). Antibodies for flow cytometry were purchased from BD Biosciences Inc. (San Jose, CA, USA), eBioscience (San Diego, CA, USA), BioLegend (San Diego, CA, USA), or Miltenyi Biotec (San Diego, CA, USA). For mouse, antibodies specific for the cell markers CD68 [1 μl (0.5 μg) per sample; CD68-Alexa647, clone FA-11, catalog no. 11-5931, BioLegend], F4/80 [1 μl (0.5 μg) per sample; F4/80-Alexa647, clone BM8, catalog no. 123122, BioLegend], Ly-6G (Gr-1) [1 μl (0.5 μg) per sample; Ly-6G-Alexa647, clone RB6-8C5, catalog no. 108418, BioLegend], and/or CD11b [1 μl (0.2 μg) per sample; CD11b-Alexa488, clone M1/70, catalog no. 101217, BioLegend] were used. For advanced multiparameter analysis of human and mouse cell populations by flow cytometry, monoclonal antibodies specific for human antigens such as CD14-BV650 (1 μl; clone M5E2, catalog no. 301836, BioLegend), CD33-AF488 (2 μl; clone WM53, catalog no. 564588, BD Bioscience), CD45–allophycocyanin (APC)–H7 (0.5 μl; clone 2D1, catalog no. 560274, BD Bioscience), CD66b-phycoerythrin (PE) (2 μl; clone G10F5, catalog no. 305106, BioLegend), CD19–fluorescein isothiocyanate (FITC) (4 μl; clone 4G7, catalog no. 392508, BioLegend), CD3-BUV395 (1 μl; clone UCHT1, catalog no. 563546, BD Bioscience), CD4-AmCyan (3 μl; RPA-T4, catalog no. 300546, BioLegend), CD8-BV421 (1 μl; clone HIT8a, catalog no. 300928, BioLegend), CD45RA-Alexa700 (1 μl; clone HI100, catalog no. 304120, BioLegend), CCR7-PE-Cy7 (2 μl; clone G043H7, catalog no. 353226, BioLegend), CD69-PE-TexasRed (2 μl; clone FN50, catalog no. 310942, BioLegend), HLA-DR–APC (1 μl; clone G4-6, catalog no. 559866, BD Bioscience), or PD1-BV786 (1 μl; clone EH12.2H7, catalog no. 329930, BioLegend) as well as mouse CD45-APC (clone 30-F11, catalog no. 103132, BioLegend), CD3–peridinin-chlorophyll-protein (clone 145-2C11, catalog no. 551163, BD Bioscience), CD19-BV421 (clone 6D5, catalog no. 115538, BioLegend), CD11b-PE-Cy7 (clone M1/70, catalog no. 101216, BioLegend), CD4-AF700 (clone RM4-5, catalog no. 557956, BD Bioscience), CD8-APC-Cy7 (clone 53-6.7, catalog no. 560182, BD Bioscience), CD44-FITC (clone IM7, catalog no. 553133, BD Bioscience), CD62L-PE (clone MEL-14, catalog no. 553151, BD Bioscience), CD69-PE-Dzl594 (clone H1.2F3, catalog no. 104535, BioLegend), and PD1-BV786 (clone 19F.1A12, catalog no. 135225, BioLegend) were used (volumes indicated used are for 106 cells in 100-μl staining volume). In general, 2 ml of eBioscience flow cytometry staining buffer (catalog no. 00-4222, eBioscience) was then added, and the samples were centrifuged at 400 to 500g for 5 min at 4°C. Supernatants were aspirated, and this wash step was repeated two more times with staining buffer. Following the third wash, each sample was resuspended in 300 μl of flow cytometry staining buffer and run through a 40-μm filter (catalog no. 22363547, Thermo Fisher Scientific) for eventual FACS analysis using an LSRII (BD Biosciences, San Jose, CA, USA). For proper background and laser intensity settings, unstained, single Ab, and immunoglobulin G (labeled with either Alexa-488 or Alexa-647, BioLegend) controls were also run.
NanoString gene expression analysis
RNAs for mock-implanted (saline)–treated controls or for 500-μm alginate or PS sphere-bearing humanized mouse strains (n = 5 per group) were isolated from tissue samples taken at various time points after implantation, as described. In general, respective RNAs were quantified and normalized to the appropriate loading concentration (100 ng/μl), and then 500 ng of each sample was processed according to NanoString’s manufacturer protocols for expression analysis via our customized multiplexed human or mouse cytokine and cytokine receptor expression panels (also including major immune marker probes), used for both comparisons between implant sites (IP and SC), early and late time points (2 and 4 weeks), and alginate versus PS sphere implantation. RNA levels (absolute copy numbers) were obtained following nCounter (NanoString Technologies Inc., Seattle, WA) quantification, and group samples were analyzed using nSolver analysis software (NanoString Technologies Inc., Seattle, WA).
NanoString DSP analysis
An automated imaging and sample collection system was developed by NanoString, modified from a standard microscope setup. A multiplexed primary Ab cocktail (with each Ab containing a unique UV photocleavable indexing oligo and two fluorescent marker Abs) was applied to Formalin-Fixed Paraffin-Embedded (FFPE) tissue sections. Tissue slides were placed on a stage of an inverted microscope, and a custom gasket was used to allow the tissue to be submerged in 1.5 ml of buffer. A microcapillary tip, connected to a syringe-pump primed with buffer, allowed for accurate small volume aspiration (e.g., <2.5 μl). Wide-field fluorescence imaging was then performed to locate tissue areas of interest. Photos (20×) were taken and stitched together to form a high-resolution image. ROIs were selected on the basis of the fluorescence information (using the above-mentioned two color-based Abs) and then sequentially processed by automation. Processing steps for each ROI were as follows: (i) The microcapillary tip was washed by dispensing clean buffer; (ii) the tissue slide was bulk-washed with buffer solution via the gasket clamp inlet and outlet ports; (iii) the microcapillary tip was then moved directly over the ROI (50 μm from the tissue); (iv) regions of tissue around ROIs were rinsed with 100 μl of buffer solution; (v) ROIs were then illuminated with UV light to release indexing oligos in those regions; (vi) following each UV exposure, released oligos were collected via microcapillary aspiration and transferred to individual wells of a plate; and (vii) once all ROIs were processed, collected oligos were hybridized to NanoString fluorescent barcodes for digital counting (nCounter) and subsequent analysis (nSolver). In instances of concentric UV light ablation (as noted), the diameter of the UV light was incrementally increased to enable capture of released oligo probes in 15-μm-thick rings moving outward from the surface of material implants (e.g., 0 to 15, 15 to 30, 30 to 45, and 45 to 60 μm). Final data shown were normalized by ROI surface area to directly compare between those that were different in size.
Overview of DSP workflow
Overview of in situ protein profiling workflow (Molecular profiling of antibodies with conjugated photocleavable oligos and the nCounter system): (i) For processing, FFPE slide-mounted tissue is incubated with a cocktail of primary antibodies conjugated with DNA oligos via a photocleavable linker together with visible wavelength-imaging reagents. (ii) For viewing, ROIs are identified with visible light-based imaging reagents at low plex to establish overall “architecture” of sphere slice (e.g., image nuclei and perhaps one or two key immune biomarkers). (iii) For profiling, select ROIs are chosen for high-resolution multiplex profiling, and oligos from the selected region are released upon exposure to UV light. (iv) For plating, free photocleaved oligos are then collected via a microcapillary tube and stored in a microplate well for subsequent quantitation. (v) For digital counting, photocleaved oligos from the spatially-resolved ROIs in the microplate are hybridized to four-color, six-spot optical barcodes, enabling up to ~1 million digital counts of the protein targets (distributed across all targets) in a single ROI using standard NanoString nCounter instruments.
Statistical analysis
Data are expressed as means ± SEM, and n = 5 mice per time point and per treatment group. These sample sizes were chosen on the basis of previous literature and for required power analysis to ascertain statistical significance while reducing mouse numbers as much as possible. All animals were included in analyses except in instances of unforeseen sickness or morbidity. Animal cohorts were randomly selected. Investigators were not blind to performed experiments. For FACS, data were analyzed for statistical significance by unpaired, two-tailed t test, or one-way analysis of variance (ANOVA) with Bonferroni multiple comparison correction, unless indicated otherwise, as implemented in GraphPad Prism 8 (*P < 0.05 and ***P < 0.0001). For NanoString, data were normalized using the geometric means of the NanoString positive controls and background levels were established using the means of the negative controls. Variance was similar between all compared groups. Housekeeping genes Tubb5, Hprt1, Bact, and Cltc were used to normalize between samples. Data were then log-transformed.
Acknowledgments
We would like to acknowledge the use of resources at W.M. Keck Biological Imaging Facility (Whitehead Institute) and Microscopy, Histology, Whole Animal Imaging, and Flow Cytometry Core Facilities (Swanson Biotechnology Centre, David H. Koch Institute for Integrative Cancer Research at MIT). We would also like to thank L. Covassin, E. Newcomb, and D. Reil for technical assistance.
Funding: This work was supported by the Juvenile Diabetes Research Foundation (JDRF) (grants 17-2007-1063 and 1-PNF-2019-782-S-B) and National Institutes of Health (NIH) (grants EB000244, EB000351, DE013023, and CA151884). J.C.D. was supported by JDRF postdoctoral fellowship (grant 3-PDF-2015-91-A-N), and J.C.D. and V.M.Q. are supported by an FDA award FDA-21-RFQ-1245838. A.R. was supported by the NSF Graduate Research Fellowship Program (DGE-1746891). D.L.G., L.D.S., and M.A.B. are supported by the NIH UC4 DK104218, R24OD0426640, and R01AI132963 (M.A.B and L.D.S.).
Author contributions: J.C.D., D.L.G., and D.G.A. designed the experiments, analyzed the data, and wrote the manuscript. J.C.D., M.M., A.S., H.H.T., S.F., J.H.-L., A.J.V., O.V., V.M.Q., A.R., S.A.-D., A.R.B., M.G., and M.A.B. performed the experiments. J.C.D., H.H.T., and M.A.B. performed the statistical analyses of datasets and aided in the preparation of displays communicating datasets. G.C.W., L.D.S., and D.L.G. provided conceptual advice and technical support. J.C.D., R.L., D.L.G., and D.G.A. supervised the study. All authors discussed the results and assisted in the preparation of the manuscript.
Competing interests: J.C.D., M.A.B., L.D.S., D.L.G., and D.G.A. are inventors on a pending patent filed by the Jackson Laboratory (D21-013-WO, filed 16 December 2022). The authors declare that they have no other competing interests. For a complete list of R.L.’s general competing interest disclosures or with which R.L. is or has been recently involved, compensated, or uncompensated, please visit www.dropbox.com/s/yc3xqb5s8s94v7x/Rev%20Langer%20COI.pdf?dl=0.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Table S1
REFERENCES AND NOTES
- 1.S. Kurtz, K. Ong, E. Lau, F. Mowat, M. Halpern, Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J. Bone Joint Surg. Am. 89, 780–785 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Institute of Medicine, Board on Population Health and Public Health Practice, Committee on the Public Health Effectiveness of the FDA 510(k) Clearance Process, Medical Devices and the Public's Health: The FDA 510(k) Clearance Process at 35 Years (National Academies Press, 2011), pp. 1–298. [Google Scholar]
- 3.P. de Vos, M. M. Faas, B. Strand, R. Calafiore, Alginate-based microcapsules for immunoisolation of pancreatic islets. Biomaterials 27, 5603–5617 (2006). [DOI] [PubMed] [Google Scholar]
- 4.D. Jacobs-Tulleneers-Thevissen, M. Chintinne, Z. Ling, P. Gillard, L. Schoonjans, G. Delvaux, B. L. Strand, F. Gorus, B. Keymeulen, D. Pipeleers; Beta Cell Therapy Consortium EU-FP7 , Sustained function of alginate-encapsulated human islet cell implants in the peritoneal cavity of mice leading to a pilot study in a type 1 diabetic patient. Diabetologia 56, 1605–1614 (2013). [DOI] [PubMed] [Google Scholar]
- 5.B. E. Tuch, G. W. Keogh, L. J. Williams, W. Wu, J. L. Foster, V. Vaithilingam, R. Philips, Safety and viability of microencapsulated human islets transplanted into diabetic humans. Diabetes Care 32, 1887–1889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.G. C. Weir, Islet encapsulation: Advances and obstacles. Diabetologia 56, 1458–1461 (2013). [DOI] [PubMed] [Google Scholar]
- 7.C. J. Kearney, D. J. Mooney, Macroscale delivery systems for molecular and cellular payloads. Nat. Mater. 12, 1004–1017 (2013). [DOI] [PubMed] [Google Scholar]
- 8.K. Y. Lee, D. J. Mooney, Alginate: Properties and biomedical applications. Prog. Polym. Sci. 37, 106–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. C. Doloff, O. Veiseh, R. de Mezerville, M. Sforza, T. A. Perry, J. Haupt, M. Jamiel, C. Chambers, A. Nash, S. Aghlara-Fotovat, J. L. Stelzel, S. J. Bauer, S. Y. Neshat, J. Hancock, N. A. Romero, Y. E. Hidalgo, I. M. Leiva, A. M. Munhoz, A. Bayat, B. M. Kinney, H. C. Hodges, R. N. Miranda, M. W. Clemens, R. Langer, The surface topography of silicone breast implants mediates the foreign body response in mice, rabbits and humans. Biomed. Eng. 5, 1115–1130 (2021). [DOI] [PubMed] [Google Scholar]
- 10.M. Ibarz Barbera, L. Morales-Fernandez, R. Gomez de Liano, P. Tana Rivero, M. A. Teus, Changes to corneal topography and biometrics after PRESERFLO microshunt surgery for glaucoma. J. Glaucoma 30, 921–931 (2021). [DOI] [PubMed] [Google Scholar]
- 11.L. Ge, Q. Li, J. Jiang, X. You, Z. Liu, W. Zhong, Y. Huang, M. M. Xing, Integration of nondegradable polystyrene and degradable gelatin in a core-sheath nanofibrous patch for pelvic reconstruction. Int. J. Nanomedicine 10, 3193–3201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. El Fray, P. Prowans, J. E. Puskas, V. Altstadt, Biocompatibility and fatigue properties of polystyrene-polyisobutylene-polystyrene, an emerging thermoplastic elastomeric biomaterial. Biomacromolecules 7, 844–850 (2006). [DOI] [PubMed] [Google Scholar]
- 13.J. Brubert, S. Krajewski, H. P. Wendel, S. Nair, J. Stasiak, G. D. Moggridge, Hemocompatibility of styrenic block copolymers for use in prosthetic heart valves. J. Mater. Sci. Mater. Med. 27, 32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. K. McDermott, C. S. Kim, D. M. Saylor, D. V. Patwardhan, Impact of copolymer ratio on drug distribution in styrene-isobutylene-styrene block copolymers. J. Biomed. Mater. Res. B Appl. Biomater. 101, 1191–1199 (2013). [DOI] [PubMed] [Google Scholar]
- 15.J. M. Anderson, A. Rodriguez, D. T. Chang, Foreign body reaction to biomaterials. Semin. Immunol. 20, 86–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. A. Wynn, T. R. Ramalingam, Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. King, S. Sandler, A. Andersson, The effect of host factors and capsule composition on the cellular overgrowth on implanted alginate capsules. J. Biomed. Mater. Res. 57, 374–383 (2001). [DOI] [PubMed] [Google Scholar]
- 18.J. C. Doloff, O. Veiseh, A. J. Vegas, H. H. Tam, S. Farah, M. Ma, J. Li, A. Bader, A. Chiu, A. Sadraei, S. Aresta-Dasilva, M. Griffin, S. Jhunjhunwala, M. Webber, S. Siebert, K. Tang, M. Chen, E. Langan, N. Dholokia, R. Thakrar, M. Qi, J. Oberholzer, D. L. Greiner, R. Langer, D. G. Anderson, Colony stimulating factor-1 receptor is a central component of the foreign body response to biomaterial implants in rodents and non-human primates. Nat. Mater. 16, 671–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Farah, J. C. Doloff, P. Muller, A. Sadraei, H. J. Han, K. Olafson, K. Vyas, H. H. Tam, J. Hollister-Lock, P. S. Kowalski, M. Griffin, A. Meng, M. McAvoy, A. C. Graham, J. McGarrigle, J. Oberholzer, G. C. Weir, D. L. Greiner, R. Langer, D. G. Anderson, Long-term implant fibrosis prevention in rodents and non-human primates using crystallized drug formulations. Nat. Mater. 18, 892–904 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O. Veiseh, J. C. Doloff, M. Ma, A. J. Vegas, H. H. Tam, A. R. Bader, J. Li, E. Langan, J. Wyckoff, W. S. Loo, S. Jhunjhunwala, A. Chiu, S. Siebert, K. Tang, J. Hollister-Lock, S. Aresta-Dasilva, M. Bochenek, J. Mendoza-Elias, Y. Wang, M. Qi, D. M. Lavin, M. Chen, N. Dholakia, R. Thakrar, I. Lacik, G. C. Weir, J. Oberholzer, D. L. Greiner, R. Langer, D. G. Anderson, Size- and shape-dependent foreign body immune response to materials implanted in rodents and non-human primates. Nat. Mater. 14, 643–651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. J. Vegas, O. Veiseh, J. C. Doloff, M. Ma, H. H. Tam, K. Bratlie, J. Li, A. R. Bader, E. Langan, K. Olejnik, P. Fenton, J. W. Kang, J. Hollister-Locke, M. A. Bochenek, A. Chiu, S. Siebert, K. Tang, S. Jhunjhunwala, S. Aresta-Dasilva, N. Dholakia, R. Thakrar, T. Vietti, M. Chen, J. Cohen, K. Siniakowicz, M. Qi, J. McGarrigle, S. Lyle, D. M. Harlan, D. L. Greiner, J. Oberholzer, G. C. Weir, R. Langer, D. G. Anderson, Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol. 34, 345–352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D. W. Grainger, All charged up about implanted biomaterials. Nat. Biotechnol. 31, 507–509 (2013). [DOI] [PubMed] [Google Scholar]
- 23.W. Kenneth Ward, A review of the foreign-body response to subcutaneously-implanted devices: The role of macrophages and cytokines in biofouling and fibrosis. J. Diabetes Sci. Technol. 2, 768–777 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.E. M. Sussman, M. C. Halpin, J. Muster, R. T. Moon, B. D. Ratner, Porous implants modulate healing and induce shifts in local macrophage polarization in the foreign body reaction. Ann. Biomed. Eng. 42, 1508–1516 (2014). [DOI] [PubMed] [Google Scholar]
- 25.T. R. Kyriakides, M. J. Foster, G. E. Keeney, A. Tsai, C. M. Giachelli, I. Clark-Lewis, B. J. Rollins, P. Bornstein, The CC chemokine ligand, CCL2/MCP1, participates in macrophage fusion and foreign body giant cell formation. Am. J. Pathol. 165, 2157–2166 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. Rodriguez, H. Meyerson, J. M. Anderson, Quantitative in vivo cytokine analysis at synthetic biomaterial implant sites. J. Biomed. Mater. Res. A 89, 152–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. Mestas, C. C. Hughes, Of mice and not men: Differences between mouse and human immunology. J. Immunol. 172, 2731–2738 (2004). [DOI] [PubMed] [Google Scholar]
- 28.B. Manoury, S. Caulet-Maugendre, I. Guenon, V. Lagente, E. Boichot, TIMP-1 is a key factor of fibrogenic response to bleomycin in mouse lung. Int. J. Immunopathol. Pharmacol. 19, 471–487 (2006). [DOI] [PubMed] [Google Scholar]
- 29.M. A. Brehm, L. D. Shultz, D. L. Greiner, Humanized mouse models to study human diseases. Curr. Opin. Endocrinol. Diabetes Obes. 17, 120–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. D. Shultz, M. A. Brehm, J. V. Garcia-Martinez, D. L. Greiner, Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 12, 786–798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. M. Allen, M. A. Brehm, S. Bridges, S. Ferguson, P. Kumar, O. Mirochnitchenko, K. Palucka, R. Pelanda, B. Sanders-Beer, L. D. Shultz, L. Su, M. PrabhuDas, Humanized immune system mouse models: Progress, challenges and opportunities. Nat. Immunol. 20, 770–774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. D. Shultz, F. Ishikawa, D. L. Greiner, Humanized mice in translational biomedical research. Nat. Rev. Immunol. 7, 118–130 (2007). [DOI] [PubMed] [Google Scholar]
- 33.T. Pearson, D. L. Greiner, L. D. Shultz, Humanized SCID mouse models for biomedical research. Humanized Mice 324, 25–51 (2008). [DOI] [PubMed] [Google Scholar]
- 34.M. W. Melkus, J. D. Estes, A. Padgett-Thomas, J. Gatlin, P. W. Denton, F. A. Othieno, A. K. Wege, A. T. Haase, J. V. Garcia, Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 12, 1316–1322 (2006). [DOI] [PubMed] [Google Scholar]
- 35.M. A. Brehm, R. Bortell, P. Diiorio, J. Leif, J. Laning, A. Cuthbert, C. Yang, M. Herlihy, L. Burzenski, B. Gott, O. Foreman, A. C. Powers, D. L. Greiner, L. D. Shultz, Human immune system development and rejection of human islet allografts in spontaneously diabetic NOD-Rag1null IL2rgammanull Ins2Akita mice. Diabetes 59, 2265–2270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.W. J. Racki, L. Covassin, M. Brehm, S. Pino, R. Ignotz, R. Dunn, J. Laning, S. K. Graves, A. A. Rossini, L. D. Shultz, D. L. Greiner, NOD-scid IL2rγnull mouse model of human skin transplantation and allograft rejection. Transplantation 89, 527–536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.N. Tonomura, A. Shimizu, S. Wang, K. Yamada, V. Tchipashvili, G. C. Weir, Y.-G. Yang, Pig islet xenograft rejection in a mouse model with an established human immune system. Xenotransplantation 15, 129–135 (2008). [DOI] [PubMed] [Google Scholar]
- 38.E. Perrier-Groult, E. Peres, M. Pasdeloup, L. Gazzolo, M. Duc Dodon, F. Mallein-Gerin, Evaluation of the biocompatibility and stability of allogeneic tissue-engineered cartilage in humanized mice. PLOS ONE 14, e0217183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.S. Chen, L. Ge, H. Wang, Y. Cheng, S. Gorantla, L. Y. Poluektova, A. F. Gombart, J. Xie, Eluted 25-hydroxyvitamin D3 from radially aligned nanofiber scaffolds enhances cathelicidin production while reducing inflammatory response in human immune system-engrafted mice. Acta Biomater. 97, 187–199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R. M. Wang, T. D. Johnson, J. He, Z. Rong, M. Wong, V. Nigam, A. Behfar, Y. Xu, K. L. Christman, Humanized mouse model for assessing the human immune response to xenogeneic and allogeneic decellularized biomaterials. Biomaterials 129, 98–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. D. Shultz, B. L. Lyons, L. M. Burzenski, B. Gott, X. Chen, S. Chaleff, M. Kotb, S. D. Gillies, M. King, J. Mangada, D. L. Greiner, R. Handgretinger, Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005). [DOI] [PubMed] [Google Scholar]
- 42.E. Billerbeck, W. T. Barry, K. Mu, M. Dorner, C. M. Rice, A. Ploss, Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rγnull humanized mice. Blood 117, 3076–3086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.B. Lee, M. Sharron, L. J. Montaner, D. Weissman, R. W. Doms, Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U.S.A. 96, 5215–5220 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. Zhen, S. R. Krutzik, B. R. Levin, S. Kasparian, J. A. Zack, S. G. Kitchen, CD4 ligation on human blood monocytes triggers macrophage differentiation and enhances HIV infection. J. Virol. 88, 9934–9946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.T. Hernández-Caselles, M. Martínez-Esparza, A. B. Pérez-Oliva, A. M. Quintanilla-Cecconi, A. García-Alonso, D. M. R. Alvarez-López, P. García-Peñarrubia, A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: Two isoforms of CD33 are generated by alternative splicing. J. Leukoc. Biol. 79, 46–58 (2006). [DOI] [PubMed] [Google Scholar]
- 46.N. I. Affara, B. Ruffell, T. R. Medler, A. J. Gunderson, M. Johansson, S. Bornstein, E. Bergsland, M. Steinhoff, Y. Li, Q. Gong, Y. Ma, J. F. Wiesen, M. H. Wong, M. Kulesz-Martin, B. Irving, L. M. Coussens, B cells regulate macrophage phenotype and response to chemotherapy in squamous carcinomas. Cancer Cell 25, 809–821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.H. S. Carlsen, E. S. Baekkevold, H. C. Morton, G. Haraldsen, P. Brandtzaeg, Monocyte-like and mature macrophages produce CXCL13 (B cell-attracting chemokine 1) in inflammatory lesions with lymphoid neogenesis. Blood 104, 3021–3027 (2004). [DOI] [PubMed] [Google Scholar]
- 48.L. J. Vuga, J. R. Tedrow, K. V. Pandit, J. Tan, D. J. Kass, J. Xue, D. Chandra, J. K. Leader, K. F. Gibson, N. Kaminski, F. C. Sciurba, S. R. Duncan, C-X-C motif chemokine 13 (CXCL13) is a prognostic biomarker of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 189, 966–974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.T. W. Fan, R. M. Higashi, H. Song, S. Daneshmandi, A. L. Mahan, M. S. Purdom, T. J. Bocklage, T. A. Pittman, D. He, C. Wang, A. N. Lane, Innate immune activation by checkpoint inhibition in human patient-derived lung cancer tissues. eLife 10, e69578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. P. Croft, J. Campos, K. Jansen, J. D. Turner, J. Marshall, M. Attar, L. Savary, C. Wehmeyer, A. J. Naylor, S. Kemble, J. Begum, K. Durholz, H. Perlman, F. Barone, H. M. McGettrick, D. T. Fearon, K. Wei, S. Raychaudhuri, I. Korsunsky, M. B. Brenner, M. Coles, S. N. Sansom, A. Filer, C. D. Buckley, Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. Litvinukova, C. Talavera-Lopez, H. Maatz, D. Reichart, C. L. Worth, E. L. Lindberg, M. Kanda, K. Polanski, M. Heinig, M. Lee, E. R. Nadelmann, K. Roberts, L. Tuck, E. S. Fasouli, D. M. DeLaughter, B. McDonough, H. Wakimoto, J. M. Gorham, S. Samari, K. T. Mahbubani, K. Saeb-Parsy, G. Patone, J. J. Boyle, H. Zhang, H. Zhang, A. Viveiros, G. Y. Oudit, O. A. Bayraktar, J. G. Seidman, C. E. Seidman, M. Noseda, N. Hubner, S. A. Teichmann, Cells of the adult human heart. Nature 588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.K. E. Aryee, L. D. Shultz, M. A. Brehm, Immunodeficient mouse model for human hematopoietic stem cell engraftment and immune system development. Methods Mol. Biol. 1185, 267–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Table S1