Abstract
Accumulating data shows that altered metabolic activity contributes to glioma development. Recently, modulation of SSADH (succinic semialdehyde dehydrogenase) expression, implicated in the catabolism of GABA neurotransmitter, was shown to impact glioma cell properties, such as proliferation, self-renewal and tumorigenicity. The purpose of this study was to investigate the clinical significance of SSADH expression in human gliomas. Using public single-cell RNA-sequencing data from glioma surgical resections, we initially grouped cancer cells according to ALDH5A1 (Aldehyde dehydrogenase 5 family member A1) expression, which encodes SSADH. Gene ontology enrichment analysis of genes differentially expressed between cancer cells expressing high or low levels of ALDH5A1, highlighted enrichment in genes implicated in cell morphogenesis and motility. In glioblastoma cell lines, ALDH5A1 knockdown inhibited cell proliferation, induced apoptosis and reduced their migratory potential. This was accompanied by a reduction in the mRNA levels of the adherens junction molecule ADAM-15 and deregulation in the expression of EMT biomarkers, with increased CDH1 and decreased vimentin mRNA levels. Evaluation of SSADH expression in a cohort of 95 gliomas using immunohistochemistry showed that SSADH expression was significantly elevated in cancer tissues compared to normal brain tissues, without any significant correlation with clinicopathological characteristics. In summary, our data show that SSADH is upregulated in glioma tissues irrespective of the histological grade and its expression sustains glioma cell motility.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-023-01354-8.
Keywords: ALDH5A1, Cell motility, ADAM-15, CDH1, Vimentin, Gene silencing, Gliomas, Perinecrotic areas
Introduction
Gliomas are the most common primary malignant brain tumors, accounting for 81% of all central nervous system (CNS) neoplasms. Based on the latest WHO classification of CNS tumors, gliomas are classified into distinct families [1]. The first group comprises the adult-type diffuse gliomas which represent the majority of primary adult brain tumors and includes the Isocitrate Dehydrogenase (IDH)-wild type (WT) glioblastoma (GB). The second large group of tumors involves the circumscribed astrocytic gliomas which are further divided based on their histological type into different grades using Arabic numbers. The other four families of tumors include the pediatric high-grade and low-grade gliomas, glioneural and neuronal tumors and ependymomas [1].
Alteration of the expression and/or activity of metabolic enzymes leading to metabolic reprogramming is one of the major changes observed in glioma tissues. Metabolic reprogramming constitutes the crosstalk between glioma’s genetic alterations and oncogenic signaling with the tumor microenvironment. Mutations in IDH enzymes, which result in accumulation of the oncometabolite D-2-hydroxyglutarate, lead to drastic changes in epigenetic marks and redox homeostasis. Moreover, alterations of mitochondrial metabolic genes play a central role in malignant transformation and tumor growth [2].
Of interest, a dysregulation of aldehyde dehydrogenase (ALDH) family of metabolic enzymes has been observed in brain tumors. ALDHs have been detected aberrantly expressed in gliomas and associated to glioma progression, while ALDH inhibitors have been shown to downregulate glioma stem-like proliferation [3]. More specifically, ALDH1A1 and A2 have been referred to as glioma migration-related genes [4], while ALDH1A1 has also been implicated in GB temozolomide resistance [5]. ALDH1A3 has been proposed as a stem-like cell biomarker in gliomas, regulating glucose metabolism [6]. Its activity and glycolytic effects are more enhanced in mesenchymal-type cancer stem cells (CSCs) compared to proneural-type CSCs and have been significantly associated with tumor invasion [7]. Moreover, ALDH3B1 and ALDH16A1 have been associated with genes regulating cell cycle checkpoints and extracellular matrix disassembly pathways [8]. Silencing of ALDH3B1 and ALDH16A1 was shown to induce cell cycle arrest at the G2/M phase and inhibit the epithelial-mesenchymal transition (EMT) in glioma cells. They both affect EMT-related protein expression, as well as the expression of aurora A and cyclin B1, involved in G2/M cell cycle checkpoints and modulate glioma cell proliferation. An association of ALDHs with deregulated DNA repair has also been described in gliomas, possibly correlating with immune dysfunction and the poor prognosis of glioma patients [9]. They have been shown to affect immunocytes and the formation of the tumor immune environment by regulating the induction and action of regulatory T cells, as well as macrophage polarization [10].
ALDH5A1 is a less studied ALDH family member in human gliomas. It codes for succinic semialdehyde dehydrogenase (SSADH), a key enzyme of glutamate and gamma-aminobutyric acid (GABA) metabolism [11, 12]. GABA is metabolized into succinate semialdehyde (SSA) and converted into either γ-hydroxybutyrate (GHB) or succinic acid by SSADH. Succinic acid can then further fuel the TCA cycle, which is connected to the GABA shunt, thus recycling GABA [13].
Increased glutamate and GABA have been demonstrated to serve as energy substrates by glioma cells, most likely via the activity of SSADH. This may explain why long-term GABA treatment greatly increased the proliferation rates of U251 wild-type cells and less significantly of IDH1-mutated cells [12]. SSADH levels were increased in almost all types of glioma cells compared to normal brain tissue [12], with SSADH staining mainly prevailing in highly proliferative and non-differentiated tumor territories [11]. These findings suggest that at the tissue level, SSADH overexpression may demonstrate cancer cell growth, survival, and stem cell–favoring effects. This evidence could be explained by the fact that it allows glioma cells to use the GABA shunt to enable energy production, proliferation and metabolic adaptation [11, 12, 14]. In this way, SSADH activity may be implicated in glioma onset and progression as demonstrated in other cancer types [15].
Therefore, the aim of our study was to specifically investigate the biological role and clinical significance of ALDH5A1 in diffuse gliomas.
Materials and Methods
Public Data Acquisition
Three datasets comprising of single-cell transcriptomes from gliomas obtained using the SMART-seq2 technology were downloaded from the Single Cell Portal (https://portals.broadinstitute.org/single_cell): 4916 neoplastic cells from twenty IDH-wildtype glioblastoma patients from Neftel and colleagues [16], 4044 neoplastic cells from six IDH-mutant oligodendroglioma patients from Tirosh and colleagues [17] and 5097 neoplastic cells from ten IDH-mutant astrocytoma patients from Venteicher and colleagues [18]. The datasets are hereafter designated as IDHwt-GB, IDHmut-O and IDHmut-A, respectively. We chose datasets obtained using the SMART-seq2 technology since it generates deeper sequencing data compared to other technologies like 10X Genomics. For each dataset, neoplastic and normal cells were distinguished according to the cell annotations provided. Expression values used in the analyses were in log2(TPM + 1).
Analyzing Single-Cell RNA Sequencing Data
All bioinformatics analyses were performed using the R software version ≥ 3.6.1 (https://cran.r-project.org/). R scripts used in this study are provided in Datafile S1.
Identifying ALDH5A1low and ALDH5A1high Cell Groups
Neoplastic cells were split into two main groups based on the mean ALDH5A1 expression level in each dataset: one group of cells expressing the lowest ALDH5A1 levels (ALDH5A1low) and another cell group expressing the highest levels (ALDH5A1high). For this analysis, we considered only neoplastic cells in which ALDH5A1 was detected: 3139, 3095 and 3657 cells for the IDHwt-GB, IDHmut-O and IDHmut-A datasets, respectively. We calculated Normalized Mutual Information (NMI) values (ClusterR package version 1.2.5 [19]) to assess the contribution of each tumor to each cell group as previously described [20]. NMI values range from 0 to 1, a value of 0 indicating that each group is composed of cells from each tumor, and a value of 1 indicating that cell groups contain cells from a single tumor. Chord plots were used to visualize the contribution of each tumor to each cell group (circlize R package version 0.4.13 [21]).
Differential Gene Expression Analysis
Genes differentially expressed between differing cell groups were identified using the Mann–Whitney test [22]. The p-values were adjusted for multiple testing using Benjamini-Hochberg (BH) approach. Significance level was set at BH-adjusted p-value < 0.01. Fold change (FC) for gene i was calculated as follows: FCi = xi − yi, where xi and yi are the log2expression levels of gene i in conditions x and y, respectively. Only genes detected in at least 3% of neoplastic cells were considered for this analysis.
Functional Enrichment Analysis
Gene symbols were first updated based on gene metadata files downloaded from HGNC website (https://www.genenames.org/download/custom/) and NCBI website [downloaded using limma R package version 3.48.3 [23]. Genes absent from these metadata files, or with ambiguous symbols (e.g., genes whose current approved symbol is the previous symbol of another gene), or whose symbol was previously associated with more than one gene were excluded from the lists. Functional enrichment analysis was then carried out using the clusterProfiler R package (version 4.0.0) [24]. The human genome was used as background. Significance level was set at BH-adjusted p-value < 0.05. Redundant terms were excluded. Graphs were generated using the ggplot2 R package (version 3.3.5).
Score Calculation
Astrocyte (AC), mesenchymal (MES), neural-progenitor (NPC) and oligodendrocyte-progenitor (OPC) scores provided by the authors were used to generate the 2D representation of malignant cellular states (Supplementary Fig. 3) as identified by Neftel and colleagues [16]. Cells were separated into AC-like, MES-like, NPC-like and OPC-like groups based on position on 2D representation (AC-like: x < 0 & y < 0; MES-like: x > 0 & y < 0; NPC-like: x > 0 & y > 0; OPC-like: x < 0 & y > 0).
Migration score was calculated per cell as the geometric mean of the expression of a motility signature defined by Saurty-Seerunghen and colleagues [25].
Glioma Cell Lines and Culturing
The T98G (human glioblastoma multiforme), 1321N1 (human astrocytoma) and GOS-3 (human glioblastoma) cell lines were purchased from ATCC and cultured in RPMI 1640 medium GlutaMAX (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies), 1% penicillin–streptomycin mixture (10,000 U/ml of penicillin and 10,000 lg/mL of streptomycin; Gibco, Life Technologies), and 0.1% Fungizone antimycotic (250 lg/mL; Gibco, Life Technologies). The CHLA-200 and SJ-GBM2 glioblastoma cell lines were kindly provided by the Texas Tech University Health Sciences Center, School of Medicine Cancer Center, COG Cell Line & Xenograft Repository (www.cccells.org). Cell lines were antibiotic-free, mycoplasma-free, authenticated and validated by short tandem repeat (STR) genotyping. Cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Biosera) supplemented with 1% penicillin–streptomycin (Gibco, Life Technologies), 20% fetal bovine serum FBS (Gibco, Life Technologies), 4 mM L-Glutamine (Gibco, Life Technologies) and 1X ITS (Millipore, MA, USA) (5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenous acid). Cell cultures were incubated at 37 °C in a humidified atmosphere containing 5% CO2–95% air.
Knockdown of ALDH5A1 by siRNA in GOS-3, SJ-GBM2 and T98G cells
Small-interfering RNA (siRNA) targeting ALDH5A1 (siALDH5A1, #sc-72480), as well as a non-targeting (nt)-siRNA (nt, #sc-37007) were purchased from Santa Cruz Biotechnology. ALDH5A1 siRNA (h) is a pool of 3 target-specific 19–25 nt siRNAs designed to knock down gene expression as described in the manufacturer’s datasheet.
GOS-3, SJ-GBM2 and T98G cells were seeded in 6-, 24- and 96-well plates to a confluence of 70–80%. Transfection with siRNA for ALDH5A1 knockdown was accomplished using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Final siALDH5A1 and nt-siRNA concentrations were 20 nM. Cells were divided into two groups: (1) siALDH5A1 cells transfected with a mixture of 20 nM siALDH5A1 and Lipofectamine 3000, each diluted in Opti-MEM (a reduced serum medium), and (2) nt-siRNA cells transfected with a mixture of 20 nM nt-siRNA and Lipofectamine 3000, again diluted in Opti-MEM.
After 6 h, RPMI and IMDM growth media were added to each cell line, respectively and the cells were incubated for 24–48 h at 37 °C in a CO2 incubator. Subsequently, the cells were collected for protein extraction and western blotting, which were performed as described below, in order to verify a successful gene knockdown.
XTT Proliferation Assay
At 24 h post transfection, when cells reached 70–80% confluency, the assessment of GOS-3 and SJ-GBM2 cell proliferation was performed with the sodium 3′-[1- (phenylaminocarbonyl)- 3,4- tetrazolium]-bis (4-methoxy6-nitro) benzene sulfonic acid hydrate) XTT Cell Proliferation Assay Kit (10,010,200, Cayman Chemical, USA). At 24 h post siRNA transfection, the media and complexes in the 96-well plate were replaced with 200 μL/well of XTT mixture diluted in serum-free, phenol red (PR)–free medium and mixed gently for 1 min on an orbital shaker. The XTT mixture was prepared based on the manufacturer’s assay protocol. The cells were then incubated for 2.5 h at 37 °C in a CO2 incubator. The absorbance of each sample was measured using a microplate reader at 450 nm. Each experiment was conducted in triplicates.
Real-Time PCR Analysis
Total RNA was extracted from GOS-3 and SJ-GBM2 cells, using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s recommendations. Samples included cells transfected with siRNA sequences specific for ALDH5A1 silencing and untreated cells, which served as control. The quantity and quality of total RNA were validated by UV spectroscopy, by calculating the ratio of absorbance at 260 and 280 nm. PrimeScript RT Reagent Kit-Perfect Real Time (Takara Bio, Japan) for RT-PCR was used for cDNA synthesis. Real-Time PCR was performed with SYBR Green using the KAPA SYBR® FAST qPCR Master Mix (2X) Kit (Sigma Aldrich) and specific primer pairs (Table 1), in order to assess the expression levels of the following genes: GAPDH, CDH1, Vimentin, ADAM-15, and BAX. Conditions for the PCR reaction were 3 min at 95 °C followed by two-step PCR for 40 cycles, each consisting of 3 s at 95 °C and 40 s at 60 °C. A melting curve analysis was performed to confirm the specificity of quantitative polymerase chain reaction (qPCR) products. The Fold value, which depicts the differential expression of each selected gene, resulted from the processing of the PCR data according to the 2−ΔΔct method, using GAPDH as a reference gene for normalization.
Table 1.
Primer pairs used in real-time PCR analysis
| Gene | Forward primer (5′- > 3′) | Reverse primer (5′- > 3′) |
|---|---|---|
| GAPDH | AGGGCTGCTTTTAACTCTGGT | CCCCACTTGATTTTGGAGGGA |
| CDH1 | ATTCTGATTCTGCTGCTCTTG | AGTAGTCATAGTCCTGGTCTT |
| Vimentin | TGTCCAAATCGATGTGGATGTTTC | TTGTACCATTCTTCTGCCTCCTG |
| ADAM-15 | GAGAAAGCCCTCCTGGATG | GGGCAGAATTCAGGCAAGT |
| BAX | CCGCCGTGGACACAGAC | CAGAAAACATGTCAGCTGCCA |
Cell Migration Assay
The cell migration assay was performed at 48 h post siRNA transfection in GOS-3 and SJ-GBM2 cells expressing high ALDH5A1 levels as well as in T98G cells expressing low ALDH5A1 levels to validate specificity (Supplementary Fig. 2).
Briefly, the cell monolayer was scraped with a sterile 500 μL pipette tip, marking the point of zero migration at 0 h. Fresh medium was added to the plates and the cells were incubated with RPMI or IMDM media for T98G, GOS-3 and SJ-GBM2, respectively. Each well was photographed at 20 × magnification using computer assisted microscopy. Phase contrast images of the cells were taken at 0 and 2 h, each time after washing for debris removal. The scratch and wound areas were measured for these timepoints, using the WimScratch Software (Wimasis image analysis platform). The results were expressed as percentages of the cell etched and covered area.
Patients’ Description
The study cohort included 95 adult patients with supratentorial diffuse infiltrating astrocytomas (grades II to IV) for which archival primary tumor material at diagnosis, prior to radio/chemotherapy, was available. Patients had been diagnosed in the First Department of Pathology, Laikon Hospital, National and Kapodistrian University of Athens, and treated as well as followed up in Evangelismos, Asklepeion and Metropolitan Hospitals between 1996 and 2009. In all cases, the diagnoses and grading were peer-reviewed according to the principles laid down in the World Health Organization (WHO) Classification. Distinction between primary (53 cases) and secondary (11 cases) glioblastomas was based on the WHO criteria 2016 and IDH1-R132H expression [26]. Written informed consent was obtained from all patients and the study was approved by the University of Athens Medical School Ethics Committee (protocol number 5105). Post-operative radiation consisted of a total dose of 60 Gy in 30 to 33 fractions. According to the existing treatment protocols, no chemotherapy was administered for cases diagnosed before 2007. Table 2 summarizes the demographic data of our patients.
Table 2.
Demographic data of patients
| Variable | Patients’ cohort (n = 95) |
|---|---|
| Median (range) | |
| Age | 58.5 (19–83) years |
| Number of cases (%) | |
| Gender: | |
| Male | 52 (54.7%) |
| Female | 43 (45.3%) |
| Grade: | |
| II | 17 (17. 9%) |
| III | 14 (14.7%) |
| IV | 64 (67.4%) |
| Events: | |
| Death | 70 follow-up period: median 8 (range 1–86) months |
| Censored | 25 follow-up period: median 18 (range 3–48) months |
| Surgery: | |
| Partial | 36 (37.9%) |
| Complete | 53 (55.8%) |
| NA | 6 (6.3%) |
| Radiotherapy: | |
| Yesa | 44 (46.3%) |
| No | 11 (11.6%) |
| NA | 40 (42.1%) |
| Chemotherapy (Temozolomide): | |
| Yesb | 7 (7.4%) |
| No | 54 (56.8%) |
| NA | 34 (35.8%) |
NA not available
a post-operative radiotherapy (a total dose of 60 Gy in 30 to 33 fractions)
b According to the Protocols existing between 1999 and 2006, no chemotherapy was admitted to these tumors
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections from 95 human brain diffuse astrocytoma tissues of grades II–IV and normal brain tissues were sliced at 4 μm thickness. Tissues were deparaffinized at 59 °C for 30 min and 3 consecutive xylene buffers for 3 min each. They were immersed for hydration in ethanol buffers of 100, 96, 80, and 70% concentration consecutively and washed with distilled H2O (dH2O). Νext, tissues were treated in the microwave for 20 min with Target Retrieval Solution, pH 9.0 (Dako, Glostrup, Denmark) for antigen unmasking and, subsequently, with 3% H2O2 (v/v) in dH2O for 30 min at room temperature, in order to block endogenous peroxidase activity. After washing with Tris-buffered saline (TBS; 0.1 M Tris–HCl, 0.15 M NaCl, pH 7.6), sections were incubated with polyclonal primary antibody ALDH5A1 at 4 °C overnight (1:50, ABGENT #AP1472d). Tissues were washed in TBS, incubated for 30 min with the Dako Real Envision, Peroxidase/DAB + , Rabbit/Mouse (Dako, Glostruo, Denmark) and labeling was visualized with diaminobenzidine for 3 min. Hematoxylin solution was used for counterstaining (Sigma-Aldrich, St. Louis, USA), and sections, after dehydration in ethanol buffers of 70, 80, 96, and 100% concentration consecutively, were mounted with mounting medium onto glass coverslips. Immunohistochemical evaluation was performed by three pathologists (PK, GL, AS), without knowledge of the clinical information. A labeling index (LI) based on the percentage of stained neoplastic cells was calculated. Staining intensity was also evaluated in three subcategories (low, moderate, intense). Moreover, 28 of our cases were also stained for nestin from our previous investigation.
Western Immunoblotting
Protein extraction from four diffuse astrocytoma tissues (1 Grade II, 2 Grade III, 1 Grade IV), normal brain tissues and glioma cells lines, was performed using ice-cold RIPA buffer (Thermo Scientific, Rockford, IL, USA) and a protease inhibitor cocktail (Thermo Scientific). Bradford assay (Bio-Rad) was used to assess protein concentration in the extracts. Approximately 30 μg of total protein was separated on a 12% gel using the sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) system. The resulting protein pattern was transferred onto a nitrocellulose membrane at 300 mA for 2 h. Protein membrane was blocked for 1 h with PBST (Tween 0.01%, 5% milk) at room temperature to prevent non-specific binding of the primary antibody. After an overnight incubation with ALDH5A1 antibody (ABGENT, #AP1472d, 1:1000) or cleaved caspase-3 (#9661, Cell Signaling Technology, Danvers, MA, USA) or actin (sc-8035, Santa Cruz Biotechnology, Dallas, TX, USA), the membrane was washed four times (10 min each) with PBST (Tween 0.01%, 1% milk) and incubated for 1 h at room temperature with goat anti-rabbit secondary antibody (Millipore, MA, 1:2000, #12–348). The membrane was further washed three times (10 min each) with PBST (Tween 0.01%, 1% milk) and two more times (10 min each) with PBS. After the washes, the membrane was incubated for 1 min in horseradish peroxidase (HRP) substrate for enhanced chemiluminescence (Clarity Western ECL Substrate (Bio-Rad), #1,705,061) and films were developed in the dark room. Actin was used as loading control.
Statistical Analysis
Statistical analysis in tissue samples was performed by a M.Sc. Biostatistician (GL). In the basic statistical analysis, ALDH5A1 expression was treated as continuous variable. Associations of the molecules under study with clinicopathological parameters were tested using non-parametric tests with correction for multiple comparisons (Kruskal–Wallis, ANOVA, Mann–Whitney U-test), as appropriate. Numerical variables were categorized on the basis of cut-off values provided by ROC curves. Statistical calculations were performed using the statistical package STATA 11.0 for Windows. All results with a two-sided p-value level < 0.05 were considered statistically significant.
Results
Glioma Cells Expressing High Levels of ALDH5A1 Exhibit Enhanced Migration in Patients’ Tumors
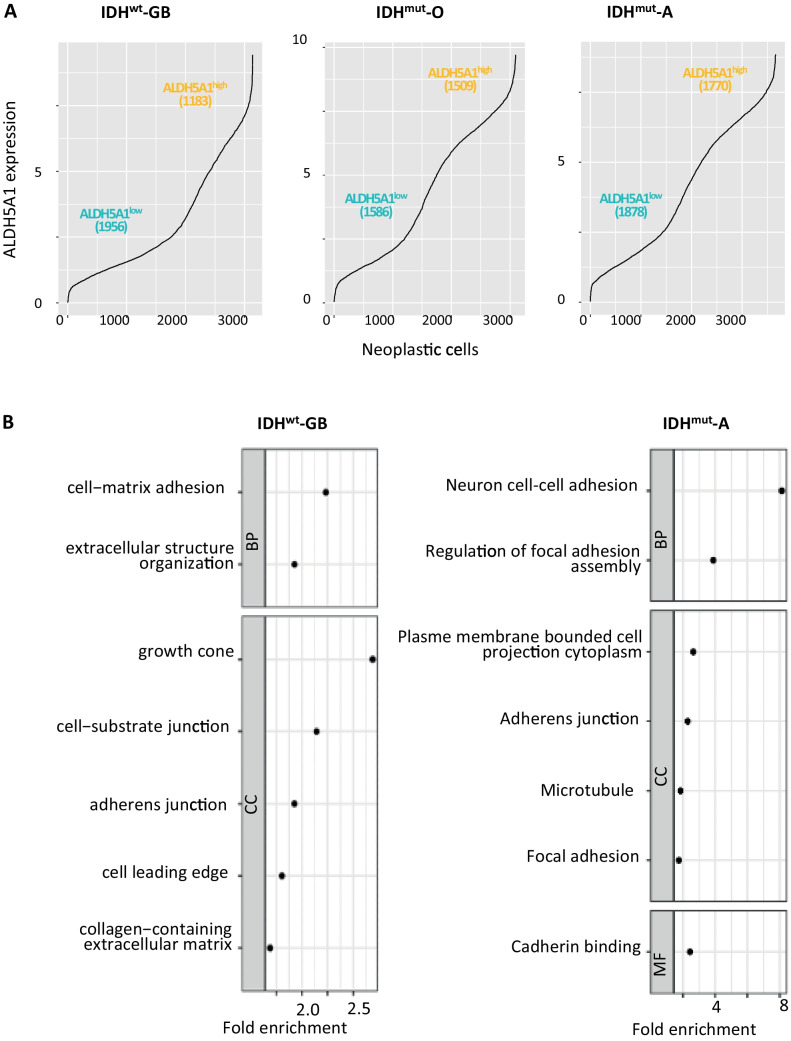
We sought for the molecular pathways sustaining cancer cells expressing high levels of ALDH5A1, focusing on glioma patients’ tumors. To this aim, we took advantage of transcriptomes from 3139 single cancer cells obtained directly from surgical resections of adult patients bearing IDH-wild type glioblastoma [16], hereafter designated as IDHwt-GB. Neoplastic cells, in which ALDH5A1 was detected, were split into two main groups, one containing 1956 cells with the lowest ALDH5A1 expression levels (ALDH5A1low) and one containing 1183 cells with the highest levels (ALDH5A1high) (Fig. 1A). Of note, ALDH5A1low and ALDH5A1high cells were isolated from each of the different patient tumors, as confirmed by the low NMI value (Supplementary Fig. 1). We identified 1255 genes differentially expressed between ALDH5A1low and ALDH5A1high cell groups (Mann–Whitney test, BH-adjusted p-value < 0.01), with 71% of them being overexpressed in ALDH5A1high cells compared to ALDH5A1low cells (891/1255, Supplementary Table 1-Sheet2). Gene ontology and KEGG pathway analyses of the overexpressed genes highlight an enrichment in several motility-related terms (Fig. 1B, Supplementary Table 2-Sheet2). Coherently, we observed a higher score for a GB-specific motility signature defined by Saurty-Seerunghen and colleagues [25] in ALDH5A1high cells compared to ALDH5A1low cells (Supplementary Fig. 2A). ALDH5A1 expression was also found to be higher in glioblastoma cells with high versus low motile potential in three of the four datasets analyzed [25] (see example in Supplementary Fig. 2B). These results suggest that ALDH5A1 may play a role in glioblastoma cell motility. Of note, ALDH5A1 expression was not restricted to a specific cellular state as identified by Neftel and colleagues [16] (Supplementary Fig. 3).
Fig. 1.
ALDH5A1 high glioma cells from patient tumors appear to exhibit enhanced motility. A Splitting cells into groups with high (ALDH5A1high) or low (ALDH5A1low) expression levels of ALDH5A1. Expression distribution across neoplastic cells. From left to right, datasets: IDH-wildtype glioblastoma (IDHwt-GB), IDH-mutant oligodendroglioma (IDHmut-O) and IDH-mutant astrocytoma (IDHmut-A). B Motility-related terms highlighted by enrichment analyses of genes overexpressed in ALDH5A1high neoplastic cells compared to ALDH5A1low cells. Dot plot representation. IDH-wildtype glioblastoma (left panel) and IDH-mutant astrocytoma (right panel). See Supplementary Fig. 1B for results corresponding to IDH-mutant oligodendroglioma
We next determined whether these observations could be extended to other adult diffuse gliomas. We applied the same analytical approach on single-cell transcriptomes obtained from surgical resections of adult patients bearing IDH-mutant oligodendroglioma [17] and IDH-mutant astrocytoma [18], hereafter designated as IDHmut-O and IDHmut-A, respectively. We identified 1586 ALDH5A1low and 1509 ALDH5A1high cells in the IDHmut-O dataset, and 1878 ALDH5A1low and 1770 ALDH5A1high cells in the IDHmut-A dataset (Fig. 1A). In both datasets, each patient tumor contributed to each cell group (Supplementary Fig. 1). As for the IDHwt-GB dataset, we observed an enrichment in numerous motility-related terms (Fig. 1B, Supplementary Fig. 2C, Supplementary Table 2-Sheets 3–4) when enrichment analyses were performed on genes overexpressed in ALDH5A1high cells compared to ALDH5A1low cells (Supplementary Table 1-Sheets 3–4).
Taken together, these results point towards a potential role for ALDH5A1 in glioma cell motility within patient tumors.
ALDH5A1 Gene Knockdown Reduces Glioblastoma Cell Viability
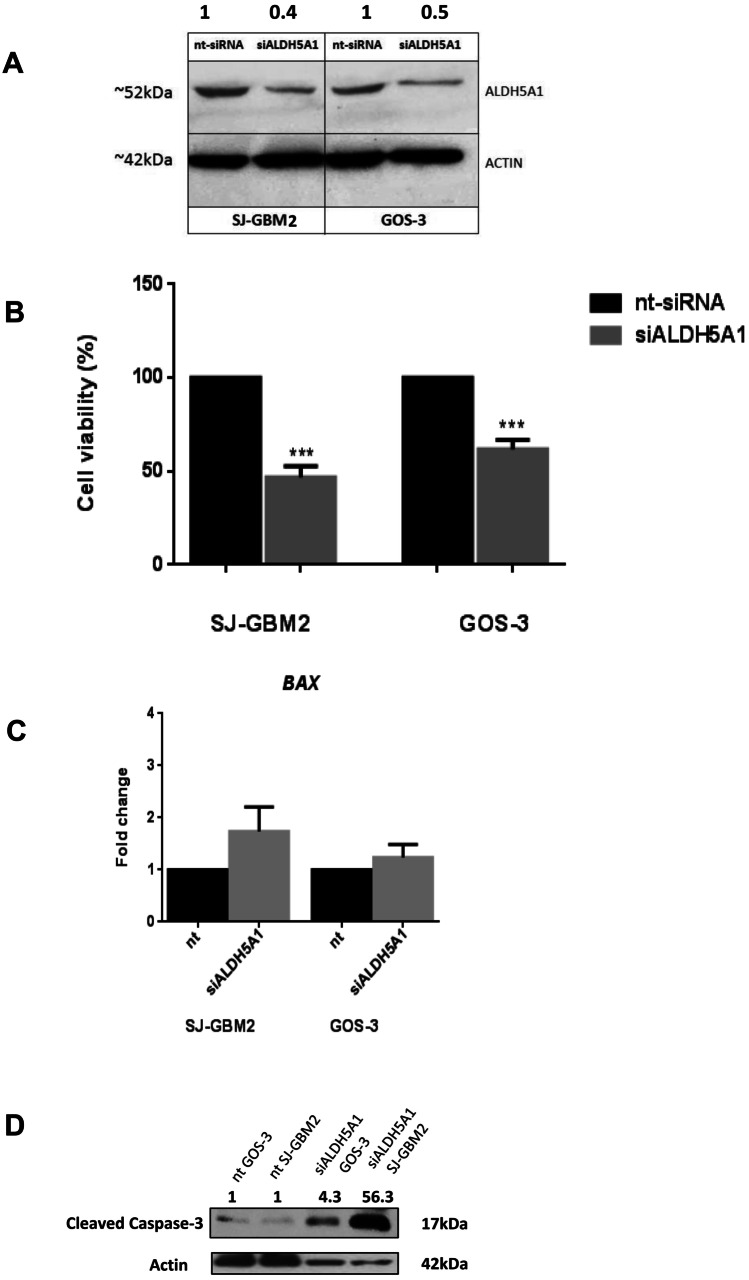
In order to examine the functional significance of ALDH5Α1 in glioma progression, we proceeded to ALDH5A1 gene knockdown using the siRNA approach. We selected two glioblastoma cell lines, GOS-3 and SJ-GBM2, that expressed high ALDH5A1 levels (Supplementary Fig. 4) and transiently transfected them with ALDH5A1-siRNA or control siRNA (nt-siRNA). Forty-eight hours post-siRNA transfection, we verified gene silencing by Western blot analysis. Blotting for the ALDH5A1 indicated siRNA specificity and ALDH5A1 expression was reduced by 61% SJ-GBM2 and by 49% in GOS-3-siRNA-transfected cells compared to nt-siRNA cells (Fig. 2A).
Fig. 2.
Glioblastoma cells exhibit reduced cell proliferation following siALDH5A1 transfection, increased BAX mRNA levels and induced caspase-3 cleavage. A Decreased ALDH5A1 protein expression after cell transfection with siRNA of GOS-3 and SJ-GBM2 cells by western immunoblotting and quantification of protein levels. B Reduced cellular viability after transfection with siALDH5A1 of GOS-3 and SJ-GBM2 cells. All experiments were repeated at least 3 times. Mean values of the three experiments are shown, t-test, ***: p-value < 0.001, *: p-value < 0.05. C Fold change in BAX mRNA expression between nt-control and siALDH5A1-transfected SJ-GBM2 and GOS-3 cells by real-time PCR. All experiments were repeated at least 3 times. Mean values of the three experiments are shown, t-test. D Protein levels of cleaved caspase-3 between nt-control and siALDH5A1-transfected GOS-3 and SJ-GBM2 cells by western immunoblotting. All experiments were repeated at least 3 times and representative image is shown
For assessing glioma cell viability after ALDH5A1 knockdown, XTT assays were performed at 24 h of culture. The measured absorbance rates were lower in siALDH5A1-transfected cells, compared to the nt-siRNA cells. The cell viability calculated using the ratio: (ODsiALDH5A1/ODnt-siRNA)*100% was significantly reduced in SJ-GBM2 cells by 53.1% and in GOS-3 cells by 38.2% (Fig. 2B). In order to explore the mechanisms by which ALDH5A1 knockdown affects cell survival, we investigated the pro-apoptotic gene BAX mRNA levels in both cell lines. BAX was elevated in SJ-GBM2 (Fold change = 1.7) and in GOS-3 cells (Fold change = 1.2) after ALDH5A1 gene silencing, however without reaching statistical significance (Fig. 2C). We then proceeded to investigate the cleaved caspase-3 protein levels after siALDH5A1 transfection. Increased cleavage of caspase-3 was detected in both siALDH5A1 transfected GOS-3 and SJ-GBM2 cells compared to nt controls (Fig. 2D).
These results show that ALDH5A1 down-regulation affects glioma cell viability and stimulates apoptotic pathways.
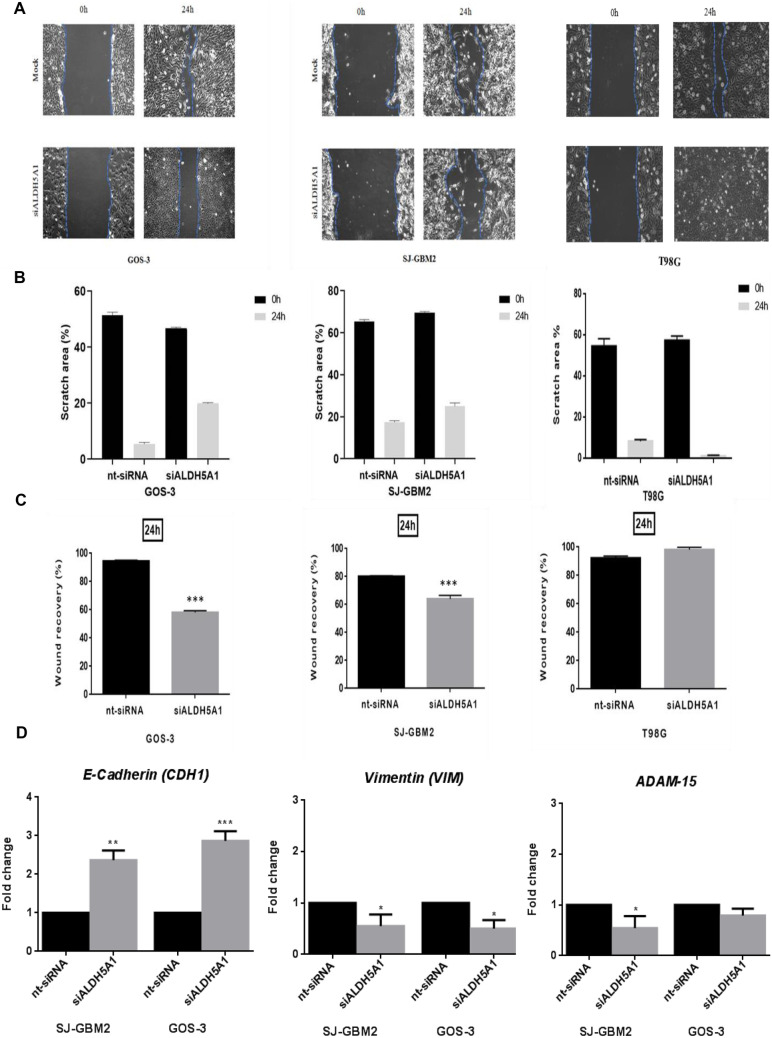
ALDH5A1 Knockdown Reduces Migration of GOS-3 and SJ-GBM2 Cells
To investigate the effects of ALDH5A1 on glioma cell migratory ability, monolayer scratch migration assays were performed at 24 h after their 48 h siRNA transfection in GOS-3 and SJ-GBM2 cells as well as in T98G cell line expressing low levels of ALDH5A1 (Supplementary Fig. 4). Cell migration in the scratch area was reduced by 58.1% in GOS-3 ALDH5A1-knockdown cells and 64.2% in SJ-GBM2 ALDH5A1-knockdown cells at 24 h. Scratch area was fully covered in siALDH5A1-transfected T98G cells at 24 h (Fig. 3A–B). The wound recovery was significantly decreased in both cell lines expressing high ALDH5A1 levels at 24 h. Specifically, wound recovery was reduced by 58.17% in siALDH5A1-transfected GOS-3 cells compared to 94.58% in nt-control (p < 0.001) and by 64% in siALDH5A1-transfected SJ-GBM2 compared to 80.17% in nt-control (p < 0.001). On the contrary, siALDH5A1-transfected T98G cells exhibited similar levels of wound recovery to nt-control cells (92.26% in nt-control and 98.1% in siALDH5A1-transfected T98G cells, Fig. 3C).
Fig. 3.
Glioblastoma cells exhibit reduced migratory abilities following siALDH5A1 transfection. A Reduced migration in the scratch area following siALDH5A1 transfection. Microphotographs depict examples of results of the scratch test-migration assay at 0 h and 24 h of GOS-3 and SJ-GBM2 cells expressing high ALDH5A1 levels and T98G cells expressing low ALDH5A1 levels. B Cell migration in the scratch area was reduced by 58.1% in GOS-3 ALDH5A1-knockdown cells and 64.2% in SJ-GBM2 ALDH5A1-knockdown cells compared to nt-controls at 24 h (A–B). Scratch area was fully covered in siALDH5A1-transfected T98G cells compared to nt-control at 24 h (A–B). C The wound recovery was significantly decreased in both cell lines expressing high ALDH5A1 levels by 58.17% in siALDH5A1-transfected GOS-3 cells compared to 94.58% in nt-control (p < 0.001) and by 64% in siALDH5A1-transfected SJ-GBM2 compared to 80.17% in nt-control (p < 0.001) at 24 h. On the contrary, siALDH5A1-transfected T98G cells exhibited similar levels of wound recovery to nt-control cells (92.26% in nt-control and 98.1% in siALDH5A1-transfected T98G cells, C). D Altered mRNA levels of epithelio-mesenchymal transition (EMT) biomarkers, CDH1, and vimentin, as well as of the adherens junction molecule ADAM-15 in GOS-3 and SJ-GBM2 cells by real-time PCR. Fold change in CDH1, Vimentin and ADAM-15 mRNA expression between nt-control and siALDH5A1-transfected SJ-GBM2 and GOS-3 cells. All experiments were repeated at least 3 times and mean values of the three experiments are shown. t-test, ***: p-value < 0.001, **: p-value < 0.01, *: p-value < 0.05
To further investigate the role of ALDH5A1 in glioma cell migration, we examined the expression of epithelio-mesenchymal transition (EMT) biomarkers, CDH1 and vimentin mRNA levels at 24 h after ALDH5A1 knockdown. Compared with nt-siRNA, CDH1 expression was significantly increased, while vimentin expression was significantly downregulated in ALDH5A1-siRNA cells (Fold change CDH1 = 2.4, p < 0.01 in SJ-GBM2 and 2.9, p < 0.001 in GOS-3 cells, respectively; Fold change Vimentin = 0.6, p < 0.05 in SJ-GBM2 and 0.5, p < 0.05 in GOS-3) (Fig. 3D). Moreover, we observed a reduction of the expression of the adherens junction molecule ADAM-15 in siALDH5A1 cells compared to nt-siRNA cells (Fold change = 0.5, p < 0.05 in SJ-GBM2 cells and 0.8 in GOS-3, respectively) (Fig. 3D). This is in coherence with our in silico results, showing an upregulation of ADAM-15 gene in ALDH5A1high GB cells compared to ALDH5A1low cells and a downregulation of CDH1 gene encoding E-cadherin in ALDH5A1high GB cells (BH-adjusted p-value < 0.01 and < 0.05, respectively) (Supplementary Table 1—Sheet 2). ADAM-15 is also upregulated in ALDH5A1high IDH-mutant glioma cells compared to ALDH5A1low cells (BH-adjusted p-value < 0.05) (Supplementary Table 1—Sheets 3–4).
These results show that ALDH5A1 down-regulation affects glioma migration via regulation of EMT and the adherens junction molecule ADAM-15.
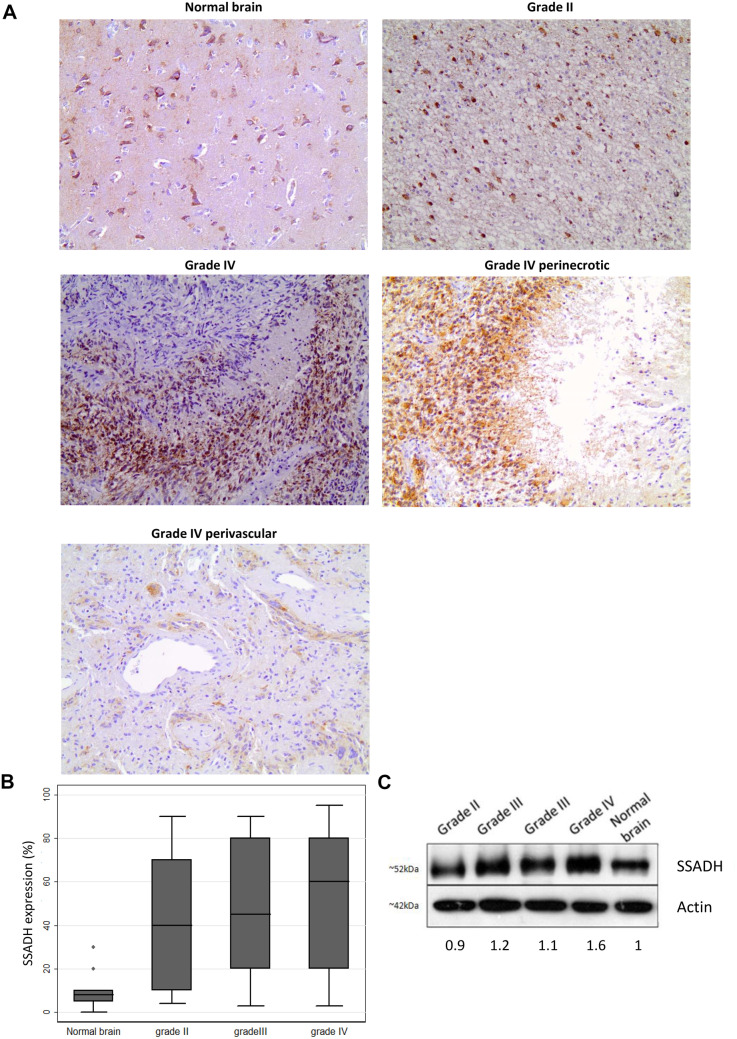
SSADH Expression is Upregulated in Human Glioma Tumors
Next, we attempted to evaluate the clinical significance of SSADH expression in human glioma tumors. Our results show that SSADH immunoreactivity was cytoplasmic and was observed in all the examined cases (95/95, 100%), ranging from 3 to 95% (median value 50%) (Table 3, Fig. 4A). SSADH showed a striking propensity for gemistocytic astrocytic tumors. It was not expressed in mitotic figures or endothelial cells. Staining intensity did not vary significantly among the examined cases, the vast majority of them (78/95, 82%) showing a moderate intensity of staining, and was therefore not included in our statistical analysis.
Table 3.
Distribution of SSADH labeling index (LI) according to histological grade
| SSADH LI | |
|---|---|
| Mean (range) | |
| Total | 50% (3–95%) |
| Histological grade | |
| Grade II | 40% (4–90%) |
| Grade III | 45% (3–90%) |
| Grade IV | 60% (3–95%) |
Fig. 4.
ALDH5A1 protein expression in different grades of gliomas. A ALDH5A1 protein levels in normal human brain tissue and glioma tumors by immunohistochemical (IHC) analysis. In Grade IV, perinecrotic and perivascular expression was also observed. B Increased ALDH5A1 expression was observed in Grade II, Grade III and Grade IV tumors, compared to normal brain tissues. C Quantification of ALDH5A1 protein levels in normal human brain tissues and glioma tumors, assessed by western immunoblotting analysis. All experiments were repeated at least 3 times and representative data are shown. t-test, ***: p-value < 0.001, *: p-value < 0.05
SSADH expression was significantly elevated in astrocytoma tissues compared to adjacent normal brain (Fig. 4B). More specifically, SSADH was expressed at higher levels in Grade II compared to normal brain (Mann–Whitney U test for multiple comparisons, median values 40% vs 8%, p-value = 0.066), in Grade III (median values 45% vs 8%, p-value = 0.0010) and in Grade IV gliomas (median values 60% vs 8%, p-value = 0.0001).
SSAADH expression was, in some cases, more pronounced around highly vascular areas (‘perivascular niche’) or areas of necrosis (‘hypoxic niche’) in glioblastomas. It should be noted that only 3 glioblastomas (in 64 analyzed) showed only perinecrotic/perivascular distribution, 20 showed perinecrotic/perivascular staining along with the presence of staining in the remaining neoplastic tissue and the majority of glioblastomas (41) showed no perinecrotic/perivascular staining. There was not any significant correlation between SSADH expression and histologic grade (Kruskal–Wallis ANOVA, p-value > 0.10). SSADH expression was also not correlated with nestin expression or IDH1-R132H positivity, irrespective of any potential grade effect.
Western blot analysis was performed in 4 normal brain tissues, 4 glioma tissues (Grade II, III, IV). SSADH expression was lower in normal brain tissues compared to astrocytomas and was increasing progressively with histological grade, being higher in Grade IV (Fig. 4C).
Discussion
Accumulating evidence underlines the implication of metabolic alterations and defective mitochondrial metabolism in glioma pathobiology. Metabolic reprogramming constitutes the crosstalk between the glioma’s genetic alterations and oncogenic signaling with the tumor microenvironment and metabolic alterations being of high significance for tumor survival [27].
Previous studies have demonstrated that ALDH5A1 (SSADH) oxidizes SSA to succinic acid and then enters the TCA cycle, affecting mitochondrial metabolic function [28] which is essential for tumorigenesis, implicated in cell survival and growth. At the same time, ALDH5A1 plays a crucial role in normal brain homeostasis, being involved in GABA recycling and control of GABA and GHB levels in the CNS. SSADH deficiency (SSADH) leads to mental retardation, autism, ataxia, and epileptic seizures [29] while ALDH5A1 mutations and polymorphisms of the ALDH5A1 C allele promoter, disrupt GABA metabolism and result in accumulation of toxic substances, such as GHB in neurons.
The present investigation was undertaken in an attempt to shed light upon the biological significance and clinical relevance of ALDH5A1 in gliomagenesis. It had been demonstrated by our previous study that gamma-hydroxybutarate (GABA byproduct) accumulation and ALDH5A1 downregulation in glioblastomas resulted in a less aggressive tumor phenotype and increased differentiation of stem-like cancer cells [11].
In this study, the analysis of publicly available single-cell transcriptomes datasets from adult patients bearing IDH-wildtype glioblastoma [16], IDH-mutant oligodendroglioma [17] and IDH-mutant astrocytoma [18] for ALDH5A1low and ALDH5A1high expression, revealed that ALDH5A1high cell groups were characterized by overexpression of several genes, with a predominant enrichment in several motility-related terms, when compared to ALDH5A1low cell groups. This finding, together with the upregulation of ALDH5A1 in patient-derived glioblastoma cells with high motile potential [25], indicates that increased ALDH5A1 expression and activity, possibly affects glioma cell motility. The role of ALDH5A1 in cancer cell motility has been previously shown in papillary thyroid cancer cells, where suppression of ALDH5A1 using siRNA in those cells caused a decrease in their migratory and invasive abilities [15].
In order to further explore the functional relevance of ALDH5A1 in glioblastomas, we investigated its expression in several patient-derived glioblastoma cell lines. Of note, heterogeneity in ALDH5A1 expression levels was obtained among the studied cell lines, reflecting differential respiratory activities and metabolic pathways for energy production as well as different cell proliferation rates. Since these cell lines are patient-derived, they also indicate the variable expression levels of ALDH5A1 among astrocytoma patients.
Upon silencing of ALDH5A1 gene using siRNA in GOS-3 and SJ-GBM2 glioblastoma cell lines with high ALDH5A1 expression, a significant reduction was observed in ALDH5A1 expression and function that lowered cell proliferation capacity, increased the pro-apoptotic BAX gene marker, and induced caspase-3 cleavage, indicating its important functional role in intracellular metabolism and maintenance of cell viability and growth. Investigation of high ALDH5A1 effect in cell migration revealed a reduced migratory potential of siALDH5A1-transfected glioblastoma cells in scratch assay and wound healing analysis, possibly due to reduced mitochondrial activity and oxidative phosphorylation. Of importance, no reduction in cell migration was observed in siALDH5A1-transfected T98G cells expressing low ALDH5A1 levels.
In addition, our results indicate that ALDH5A1 depletion affects the expression of EMT markers, leading to increased CDH1 and decreased vimentin expression. This finding is consistent with the reversal of the EMT process and a shift towards a physiologic cellular phenotype [30, 31]. Therefore, a role for ALDH5A1 as an EMT driver is suggested which points out its potential for therapeutic targeting. The EMT-promoting role of ALDH5A1 has been also previously demonstrated in thyroid cancer cells [15]. Moreover, in accordance with the single-cell RNA-seq data analysis, ALDH5A1 silencing induced a decrease in ADAM-15 expression, a transmembrane protein involved in cell signalling, cell adhesion and protein ectodomain shedding. Since E-cadherin and N-cadherin are known substrates of ADAM-15 [32], it is possible that elevated activity of ALDH5A1 affects glioma cell migration through this interaction. Furthermore, ADAM-15 upregulation has been correlated with increased tumor aggressiveness and metastatic disease in breast and prostate cancer through regulation of cell motility and adhesion [33], suggesting a similar role in gliomas.
In our study, a significant upregulation of ALDH5A1 expression was observed in diffuse infiltrating astrocytomas of grades II–IV compared to adjacent normal brain. ALDH5A1 immunoreactivity was more pronounced in grade IV astrocytomas. However, no significant correlations were detected between ALDH5A1 expression and histological grade, clinicopathological parameters or IDH status.
Our findings are in accordance with the study of Hujber et al. [12] where ALDH5A1 overexpression was notable in all histological subtypes and stages of gliomas, without however presenting any significant associations with the disease’s clinicopathological parameters, such as age, gender and IDH mutation status, suggesting the presence of a metabolic compensatory mechanism in IDH1-mutant cells that uses oxidative phosphorylation and alternative substrates to glucose [12]. In the same study, glioma cells demonstrated high levels of glutamine, glutamate, and GABA oxidation and ALDH5A1 expression was positively correlated with GABA oxidation. Moreover, GABA addition to glioma cells led to an increased proliferation rates while GABA oxidation inhibition through D-2-hydroxyglutarate (2-HG) was demonstrated to contribute to a less aggressive glioma phenotype [12]. On the other hand, the study of Lenting et al. [34] detected a higher expression of ALDH5A1 in IDH1-mutated gliomas, suggesting that these cells are able to use the GABA shunt to replenish the TCA cycle. Although the underlying mechanism remains unknown, the presence of TP53 mutation was associated with low ALDH5A1 expression [35]. Additionally, reduced expression of ALDH5A1 in high-grade serous ovarian cancer correlated with poor survival and the presence of EMT gene signature, implying a role in cancer cell migration and invasion [36].
Upregulation of ALDH5A1 has also been detected in papillary thyroid cancer, where it correlated with tumor stage, metastasis, lymph node stage and poor disease-free survival [15]. Moreover, overexpression of ALDH5A1 on breast ductal carcinoma in situ models indicates its implication in premalignant progression [37]. In addition, breast cancer patients carrying the ALDH5A1rs1054899 AG/AA polymorphism exhibited worse response to the FAC regime (doxorubicin, 5′-fluorouracil and cyclophosphamide) chemotherapy, which was attributed to the detoxification role of ALDH5A1 in the cyclophosphamide metabolic pathway [38].
Altogether, our findings demonstrate the increased expression of ALDH5A1 in diffuse gliomas and a potential functional role in cell proliferation and motility. Although the underlying mechanism remains under investigation, SSADH gene knockdown in glioblastoma cell lines decreased their viability and migratory potential, via regulation of EMT and the adherens junction molecule ADAM-15, further indicating ALDH5A1 as a promising molecular target. Future studies are warranted to validate the potential of ALDH5A1 as a novel biomarker for glioma diagnosis and prognosis. Moreover, ALDH5A1 involvement in the metabolism of the chemotherapeutic agent cyclophosphamide in previous studies as well as the enhanced detoxification of aldophosphamide to carboxyphosphamide, indicates its therapeutic potential [39] that needs to be further exploited.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1 related to Figure 1. ALDH5A1high glioma cells come from distinct patient tumors. Chord plot representation of contribution of each tumor to each cluster. NMI: Normalized Mutual Information. IDH-wildtype glioblastoma (left), IDH-mutant oligodendroglioma (middle), IDH-mutant astrocytoma (right) datasets. Supplementary file1 (PDF 118 kb)
Supplementary Figure 2 related to Figure 1. ALDH5A1high glioma cells overexpress genes related to cell motility. A. Higher migration score in ALDH5A1high neoplastic cells from IDH-wildtype glioblastoma compared to ALDH5A1low cells. Box plot representation. Wilcoxon rank sum test with continuity correction. B. Higher ALDH5A1 expression in IDH-wildtype glioblastoma cells with high motile potential compared to cells with low motile potential. Neftel-S dataset. Wilcoxon rank sum test with continuity correction. Benjamini-Hochberg adjusted p-value (as in Supplementary Table 2 from Saurty-Seerunghen et al. [25]). Box plot representation. C. Motility-related terms highlighted by enrichment analyses of genes overexpressed in ALDH5A1high neoplastic cells from IDH-mutant oligodendroglioma compared to ALDH5A1low cells. Dot plot representation. Supplementary file2 (PDF 153 kb)
Supplementary Figure 3 related to Figure 1. ALDH5A1 expression is not restricted to a specific cellular state as identified by Neftel and colleagues [16]. Highlighting ALDH5A1 expression in malignant cellular states as identified by Neftel and colleagues [16]. Left panel: 2D representation of malignant cellular states as identified by Neftel and colleagues [16]. Cells coloured by ALDH5A1 expression. Right panel: Boxplot showing ALDH5A1 expression in each malignant cellular state. MeanAC = 2.17, MeanMES = 1.78, MeanNPC = 1.73, MeanOPC =2.22. Each group mean is different from one another, except AC-like vs OPC-like groups and MES-like vs NPC-like groups. Kruskal-Wallis test, pairwise comparisons using Mann-Whitney test (Benjamini Hochberg-adjusted p-value < 0.05). Supplementary file3 (PDF 2239 kb)
Supplementary Figure 4 related to Figure 2. Variable ALDH5A1 protein expression among different glioma cell lines, being highest in lines GOS-3, CHLA-200 and SJ-GBM2 cell lines. Western immunoblotting analysis. Supplementary file4 (PDF 18 kb)
Supplementary Table 1 related to Figure 1: List of genes differentially expressed between ALDH5A1high and ALDH5A1low cell groups in distinct glioma datasets. Sheet 1: Description of the contents. Sheets 2-4: List of genes differentially expressed between ALDH5A1high and ALDH5A1low cell groups from the IDH-wildtype glioblastoma (Sheet 2), the IDH-mutant oligodendroglioma (Sheet 3) and the IDH-mutant astrocytoma (Sheet 4) datasets. Supplementary file5 (XLSX 7.37 MB)
Supplementary Table 2 related to Figure 1: Results of the enrichment analyses done on genes overexpressed in ALDH5A1high cells compared to ALDH5A1low cells. Sheet 1: Description of the contents. Sheets 2-4: Results of enrichment analyses done on genes overexpressed in ALDH5A1high cells compared to ALDH5A1low cells from the IDH-wildtype glioblastoma (Sheet 2), the IDH-mutant oligodendroglioma (Sheet 3) and the IDH-mutant astrocytoma (Sheet 4) datasets. Supplementary file6 (XLSX 1.80 MB)
Datafile S1. Rscripts used in this study. Supplementary file10 (TXT 22 kb)
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Author Contribution
PK, CP, EAE: Conceptualization. MSS, AS, EAT, AK, MM, DS, EAK: Methodology, Investigation, Software. EL, EB, EAK, PK, CP: Data curation, Writing—Original draft preparation. AK, MM, AS, DS: Visualization, Investigation. CP, EAE PK: Supervision. DSK, GL: Software, Validation. CP, MM, EAE, PK: Writing—Reviewing and Editing.
Data Availability
The tree datasets comprising of single-cell transcriptomes from gliomas obtained using the SMART-seq2 technology were downloaded from the Single Cell Portal (https://portals.broadinstitute.org/single_cell).
Declarations
Ethics Approval
The research has been given ethical approval by the University of Athens Medical School Ethics Committee (Protocol number 5105).
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors Christina Piperi and Mirca S. Saurty-Seerunghen contributed equally.
The authors Elias A. El-Habr and Penelope Korkolopoulou contributed equally.
Contributor Information
Dimitrios S. Kanakoglou, Email: kanakogloud@biol.uoa.gr
Penelope Korkolopoulou, Email: pkorkol@med.uoa.gr.
References
- 1.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thakur S, Daley B, Gaskins K, et al. Metformin targets mitochondrial glycerophosphate dehydrogenase to control rate of oxidative phosphorylation and growth of thyroid cancer in vitro and in vivo. Clin Cancer Res Off J Am Assoc Cancer Res. 2018;24:4030–4043. doi: 10.1158/1078-0432.CCR-17-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park HH, Park J, Cho HJ, et al. Combinatorial therapeutic effect of inhibitors of aldehyde dehydrogenase and mitochondrial complex I, and the chemotherapeutic drug, temozolomide against glioblastoma tumorspheres. Molecules. 2021;26:282. doi: 10.3390/molecules26020282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Zhang H, Xu S, Liu Z, Cheng Q. The adaptive transition of glioblastoma stem cells and its implications on treatments. Signal Transduct Target Ther. 2021;6:124. doi: 10.1038/s41392-021-00491-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu S-L, Liu S, Cui W, et al. Aldehyde dehydrogenase 1A1 circumscribes high invasive glioma cells and predicts poor prognosis. Am J Cancer Res. 2015;5:1471–1483. [PMC free article] [PubMed] [Google Scholar]
- 6.Ni W, Xia Y, Luo L, et al. High expression of ALDH1A3 might independently influence poor progression-free and overall survival in patients with glioma via maintaining glucose uptake and lactate production. Cell Biol Int. 2020;44:569–582. doi: 10.1002/cbin.11257. [DOI] [PubMed] [Google Scholar]
- 7.Vasilogiannakopoulou T, Piperi C, Papavassiliou AG. Impact of aldehyde dehydrogenase activity on gliomas. Trends Pharmacol Sci. 2018;39:605–609. doi: 10.1016/j.tips.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Pan C-M, Chan K-H, Chen C-H, et al. MicroRNA-7 targets T-Box 2 to inhibit epithelial-mesenchymal transition and invasiveness in glioblastoma multiforme. Cancer Lett. 2020;493:133–142. doi: 10.1016/j.canlet.2020.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Jiang H, Wang Z, et al. Functional clustering analysis identifies specific subtypes of aldehyde dehydrogenase associated with glioma immunity. Transl Cancer Res. 2021;10:5052–64. 10.21037/tcr-21-1160. [DOI] [PMC free article] [PubMed]
- 10.Wang Z, Mo Y, Tan Y, et al. The ALDH family contributes to immunocyte infiltration, proliferation and epithelial-mesenchymal transformation in glioma. Front Immunol. 2022;12:756606. doi: 10.3389/fimmu.2021.756606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Habr EA, Dubois LG, Burel-Vandenbos F, et al. A driver role for GABA metabolism in controlling stem and proliferative cell state through GHB production in glioma. Acta Neuropathol. 2017;133:645–660. doi: 10.1007/s00401-016-1659-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hujber Z, Horváth G, Petővári G, et al. GABA, glutamine, glutamate oxidation and succinic semialdehyde dehydrogenase expression in human gliomas. J Exp Clin Cancer Res. 2018;37:271. doi: 10.1186/s13046-018-0946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Q, Yang Y, Feng D, et al. MAGI3 negatively regulates Wnt/β-catenin signaling and suppresses malignant phenotypes of glioma cells. Oncotarget. 2015;6:35851–65. 10.18632/oncotarget.5323. [DOI] [PMC free article] [PubMed]
- 14.Knerr I, Pearl PL, Bottiglieri T, et al. Therapeutic concepts in succinate semialdehyde dehydrogenase (SSADH; ALDH5a1) deficiency (gamma-hydroxybutyric aciduria). Hypotheses evolved from 25 years of patient evaluation, studies in Aldh5a1-/- mice and characterization of gamma-hydroxybutyric acid. J Inherit Metab Dis. 2007;30:279–294. doi: 10.1007/s10545-007-0574-2. [DOI] [PubMed] [Google Scholar]
- 15.Deng X-Y, Gan X-X, Feng J-H, et al. ALDH5A1 acts as a tumour promoter and has a prognostic impact in papillary thyroid carcinoma. Cell Biochem Funct. 2020 doi: 10.1002/cbf.3584. [DOI] [PubMed] [Google Scholar]
- 16.Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirosh I, Venteicher AS, Hebert C, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venteicher AS, Tirosh I, Hebert C, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017 doi: 10.1126/science.aai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning CD, Raghavan P, Schütze H. Introduction to information retrieval. Cambridge: Cambridge University Press; 2008. 10.1017/CBO9780511809071.
- 20.Saurty-Seerunghen MS, Bellenger L, El-Habr EA, et al. Capture at the single cell level of metabolic modules distinguishing aggressive and indolent glioblastoma cells. Acta Neuropathol Commun. 2019;7:155. doi: 10.1186/s40478-019-0819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 22.Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saurty-Seerunghen MS, Daubon T, Bellenger L, Delaunay V, Castro G, Guyon J, Rezk A, Fabrega S, Idbaih A, Almairac F, Burel-Vandenbos F, Turchi L, Duplus E, Virolle T, Peyrin JM, Antoniewski C, Chneiweiss H, El-Habr EA, Junier MP. Glioblastoma cell motility depends on enhanced oxidative stress coupled with mobilization of a sulfurtransferase. Cell Death Dis. 2022;13(10):913. doi: 10.1038/s41419-022-05358-8.PMID:36310164;PMCID:PMC9618559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 27.Bi J, Chowdhry S, Wu S, et al. Altered cellular metabolism in gliomas — an emerging landscape of actionable co-dependency targets. Nat Rev Cancer. 2020;20:57–70. doi: 10.1038/s41568-019-0226-5. [DOI] [PubMed] [Google Scholar]
- 28.Marquez J, Flores J, Kim AH, et al. Rescue of TCA cycle dysfunction for cancer therapy. J Clin Med. 2019 doi: 10.3390/jcm8122161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Didiášová M, Banning A, Brennenstuhl H, et al. Succinic semialdehyde dehydrogenase deficiency: an update. Cells. 2020;9:477. doi: 10.3390/cells9020477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onder TT, Gupta PB, Mani SA, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 31.Liu C-Y, Lin H-H, Tang M-J, Wang Y-K. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6:15966–83. 10.18632/oncotarget.3862. [DOI] [PMC free article] [PubMed]
- 32.Lorenzatti Hiles G, Bucheit A, Rubin JR, et al. ADAM15 is functionally associated with the metastatic progression of human bladder cancer. PLoS ONE. 2016;11:e0150138–e0150138. doi: 10.1371/journal.pone.0150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu JH, Guan YJ, Zhang YC, et al. ADAM15 correlates with prognosis, immune infiltration and apoptosis in hepatocellular carcinoma. Aging (Albany NY). 2021;13:20395–417. 10.18632/aging.203425. [DOI] [PMC free article] [PubMed]
- 34.Lenting K, Khurshed M, Peeters TH, et al. Isocitrate dehydrogenase 1–mutated human gliomas depend on lactate and glutamate to alleviate metabolic stress. FASEB J. 2019;33:557–571. doi: 10.1096/fj.201800907RR. [DOI] [PubMed] [Google Scholar]
- 35.Tian X, Han Y, Yu L, et al. Decreased expression of ALDH5A1 predicts prognosis in patients with ovarian cancer. Cancer Biol Ther. 2017;18:245–251. doi: 10.1080/15384047.2017.1295175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilvo M, de Santiago I, Gopalacharyulu P, et al. Accumulated metabolites of hydroxybutyric acid serve as diagnostic and prognostic biomarkers of ovarian high-grade serous carcinomas. Cancer Res. 2016;76:796–804. doi: 10.1158/0008-5472.CAN-15-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur H, Mao S, Li Q, et al. RNA-Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS ONE. 2012;7:e50249. doi: 10.1371/journal.pone.0050249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pamuła-Piłat J, Tęcza K, Kalinowska-Herok M, Grzybowska E. Genetic 3’UTR variations and clinical factors significantly contribute to survival prediction and clinical response in breast cancer patients. Sci Rep. 2020;10:5736. doi: 10.1038/s41598-020-62662-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Wang H. Oxazaphosphorine bioactivation and detoxification The role of xenobiotic receptors. Acta Pharm Sin B. 2012 doi: 10.1016/j.apsb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 related to Figure 1. ALDH5A1high glioma cells come from distinct patient tumors. Chord plot representation of contribution of each tumor to each cluster. NMI: Normalized Mutual Information. IDH-wildtype glioblastoma (left), IDH-mutant oligodendroglioma (middle), IDH-mutant astrocytoma (right) datasets. Supplementary file1 (PDF 118 kb)
Supplementary Figure 2 related to Figure 1. ALDH5A1high glioma cells overexpress genes related to cell motility. A. Higher migration score in ALDH5A1high neoplastic cells from IDH-wildtype glioblastoma compared to ALDH5A1low cells. Box plot representation. Wilcoxon rank sum test with continuity correction. B. Higher ALDH5A1 expression in IDH-wildtype glioblastoma cells with high motile potential compared to cells with low motile potential. Neftel-S dataset. Wilcoxon rank sum test with continuity correction. Benjamini-Hochberg adjusted p-value (as in Supplementary Table 2 from Saurty-Seerunghen et al. [25]). Box plot representation. C. Motility-related terms highlighted by enrichment analyses of genes overexpressed in ALDH5A1high neoplastic cells from IDH-mutant oligodendroglioma compared to ALDH5A1low cells. Dot plot representation. Supplementary file2 (PDF 153 kb)
Supplementary Figure 3 related to Figure 1. ALDH5A1 expression is not restricted to a specific cellular state as identified by Neftel and colleagues [16]. Highlighting ALDH5A1 expression in malignant cellular states as identified by Neftel and colleagues [16]. Left panel: 2D representation of malignant cellular states as identified by Neftel and colleagues [16]. Cells coloured by ALDH5A1 expression. Right panel: Boxplot showing ALDH5A1 expression in each malignant cellular state. MeanAC = 2.17, MeanMES = 1.78, MeanNPC = 1.73, MeanOPC =2.22. Each group mean is different from one another, except AC-like vs OPC-like groups and MES-like vs NPC-like groups. Kruskal-Wallis test, pairwise comparisons using Mann-Whitney test (Benjamini Hochberg-adjusted p-value < 0.05). Supplementary file3 (PDF 2239 kb)
Supplementary Figure 4 related to Figure 2. Variable ALDH5A1 protein expression among different glioma cell lines, being highest in lines GOS-3, CHLA-200 and SJ-GBM2 cell lines. Western immunoblotting analysis. Supplementary file4 (PDF 18 kb)
Supplementary Table 1 related to Figure 1: List of genes differentially expressed between ALDH5A1high and ALDH5A1low cell groups in distinct glioma datasets. Sheet 1: Description of the contents. Sheets 2-4: List of genes differentially expressed between ALDH5A1high and ALDH5A1low cell groups from the IDH-wildtype glioblastoma (Sheet 2), the IDH-mutant oligodendroglioma (Sheet 3) and the IDH-mutant astrocytoma (Sheet 4) datasets. Supplementary file5 (XLSX 7.37 MB)
Supplementary Table 2 related to Figure 1: Results of the enrichment analyses done on genes overexpressed in ALDH5A1high cells compared to ALDH5A1low cells. Sheet 1: Description of the contents. Sheets 2-4: Results of enrichment analyses done on genes overexpressed in ALDH5A1high cells compared to ALDH5A1low cells from the IDH-wildtype glioblastoma (Sheet 2), the IDH-mutant oligodendroglioma (Sheet 3) and the IDH-mutant astrocytoma (Sheet 4) datasets. Supplementary file6 (XLSX 1.80 MB)
Datafile S1. Rscripts used in this study. Supplementary file10 (TXT 22 kb)
Data Availability Statement
The tree datasets comprising of single-cell transcriptomes from gliomas obtained using the SMART-seq2 technology were downloaded from the Single Cell Portal (https://portals.broadinstitute.org/single_cell).