Abstract
In recent years, highly pathogenic avian influenza H5 subtype (HPAI H5) viruses have been prevalent around the world in both avian and mammalian species, causing serious economic losses to farmers. HPAI H5 infections of zoonotic origin also pose a threat to human health. Upon evaluating the global distribution of HPAI H5 viruses from 2019 to 2022, we found that the dominant strain of HPAI H5 rapidly changed from H5N8 to H5N1. A comparison of HA sequences from human- and avian-derived HPAI H5 viruses indicated high homology within the same subtype of viruses. Moreover, amino acid residues 137A, 192I, and 193R in the receptor-binding domain of HA1 were the key mutation sites for human infection in the current HPAI H5 subtype viruses. The recent rapid transmission of H5N1 HPAI in minks may result in the further evolution of the virus in mammals, thereby causing cross-species transmission to humans in the near future. This potential cross-species transmission calls for the development of an H5-specific influenza vaccine, as well as a universal influenza vaccine able to provide protection against a broad range of influenza strains.
Subject terms: Immunology, Molecular biology
Introduction
Avian influenza is an infectious disease that affects poultry and wildfowl. It is caused by highly pathogenic avian influenza (HPAI) or low pathogenic avian influenza (LPAI) viruses, which belong to the Orthomyxoviridae family and have a single-stranded negative-sense RNA genome. Avian influenza viruses (AIVs) are mainly classified on the basis of their surface proteins, hemagglutinin (HA) and neuraminidase (NA). HA protein on the surface of the virion, the main antigenic site in vaccine design, causes erythrocyte agglutination in vitro and in vivo1. Over the years, outbreaks of HPAI H5 subtype viruses in poultry have caused huge economic losses to the farming industry. In 2022, more than 25 million poultry and wild birds were infected with HPAI H5 worldwide, resulting in 5.28 million deaths (https://wahis.woah.org/). Recently, HPAI H5 has caused more sporadic cases, or even outbreaks, in mammals, including minks, otters, foxes, and sea lions2–4. With possible further mutations in avian and mammalian species, HPAI H5 has a strong potential to cause human infection and trigger a global pandemic. Therefore, it is essential to develop an H5-specific vaccine, as well as a universal influenza vaccine, to fully cover a broad range of influenza strains.
Global distribution of HPAI H5 viruses
H5N1 was the first strain isolated among the HPAI H5 viruses in Scotland in 1959, and it was shown to infect a variety of avians5. In 1997, HPAI H5N1 (Gs/GD/96) emerged in China and it was first confirmed to infect humans6. In 2000, H5N1 broke out among poultry in several countries, including the Netherlands, Vietnam, Indonesia, and Thailand7. A few years later (after 2005), H5N1 further spread to poultry in Europe and Africa8,9. Owing to homologous recombination among influenza strains in poultry, other non-N1 recombinant AIVs strains, such as H5N2, H5N6, and H5N8, have emerged in many countries. To classify H5 subtype AIVs, the HA gene was selected by the WHO/OIE/FAO H5N1 Evolution Working Group to divide AIVs into diffident clades based on the similarity of HA nucleic acid sequences. Each distinct clade was determined to have an average distance > 1.5% from other clades10. From 2013 to 2019, HPAI viruses of subclades 2.3.2.1 and 2.3.4.4 began to spread around the world11–16. HPAI H5 subclade 2.3.4.4 was first detected in domestic ducks in China17,18 and was further divided into 8 subclades, 2.3.4.4a to 2.3.4.4h19.
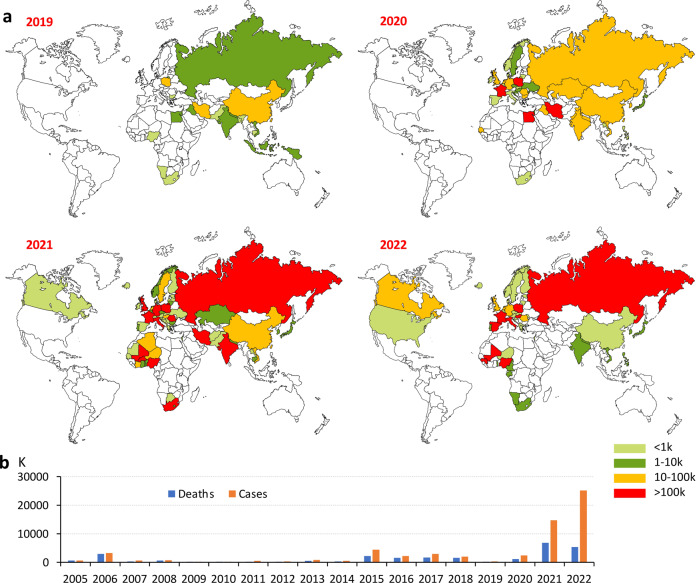
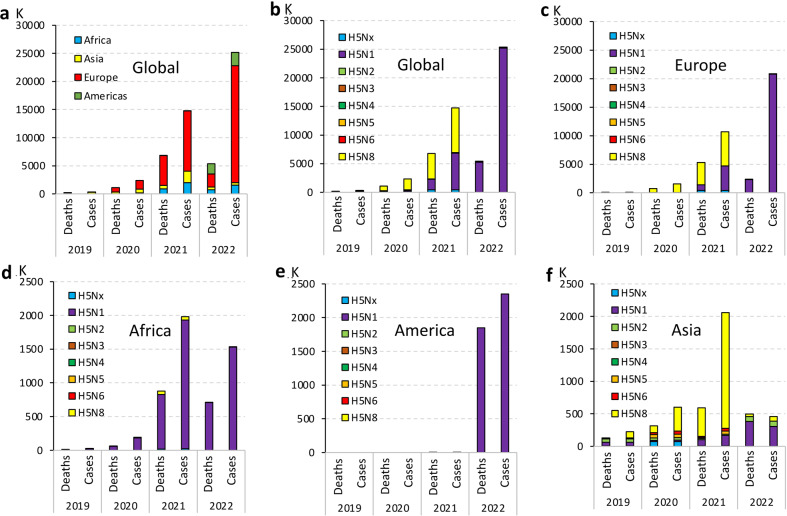
From 2019 to 2022, HPAI H5 viruses have been circulating among avian populations in Europe, Africa, and Asia20–24, resulting in a significant increase in global avian cases from 0.343 to 25.19 million (Fig. 1). Europe has become the primary site of spread accounting for 82.7% of avian cases and 43.9% of deaths globally in 2022 (Fig. 2a). Notably, the main HPAI subtype virus causing global epidemics gradually changed from H5N8 to H5N1 between 2019 and 2022 (Fig. 2b). For example, the epidemic of HPAI viruses in Europe was dominated by H5N8 from 2019 to 2021. However, it changed to H5N1 in 2022. During this year, infections and mortality rates caused by H5N1 accounted for ~99.9% among all HPAI H5 viruses in the same period (Fig. 2c). Since 2019, the H5N1 subtype has been dominant in Africa and the Americas, accounting for more than 99.9% of cases (Fig. 2d, e). Similar to Europe, H5N8 was the main HPAI subtype in Asia from 2019 to 2021, but it also changed to H5N1 in 2022. In 2022, the H5N1 subtype accounted for 67.4% of infections and 76.3% of mortalities among all H5 subtypes in Asia (Fig. 2f).
Fig. 1. Global distribution of HPAI H5 viruses is shown by avian cases and deaths from 2019 to 2022 (https://wahis.woah.org/#/dashboards/qd-dashboard).
Data up to January 2023 are included. a Distribution of HPAI H5 viruses between 2019 and 2022 based on the number of infections. b Number of avian infections and deaths caused by HPAI H5 viruses from 2005 to 2022.
Fig. 2. Global distribution of HPAI H5 avian infections from 2019 to 2022.
a Distribution of HPAI H5 in Asia, Europe, Africa, and the Americas. b Global distribution of each subtype of HPAI H5. c–f Continent-specific distribution of each subtype of HPAI H5. Raw data for avian cases and deaths were taken from WAHIS (https://wahis.woah.org/#/dashboards/qd-dashboard). Data are included up to January 2023.
Human infections with HPAI H5 viruses
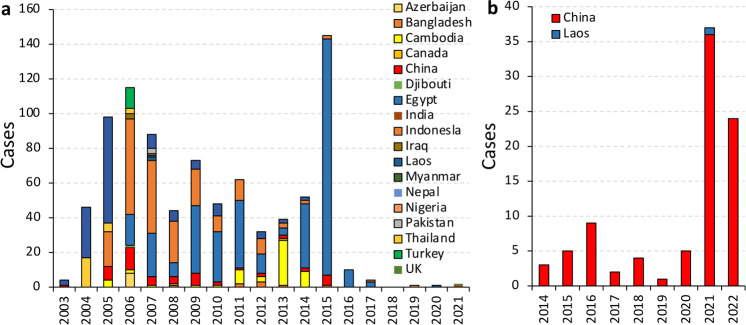
The current global epidemic of HPAI H5 mainly involves three subtypes: H5N1, H5N8, and H5N6. Indeed, the widespread epidemic of AIVs among wild birds increases the risk of infection for poultry and other avians. However, it is generally believed that human infection with HPAI H5 viruses is closely related to outbreaks in poultry and wild birds, and according to the WHO, 864 human cases of H5N1 infection have been reported worldwide, resulting in 456 deaths from 2014 to 2021(Fig. 3a). It was previously believed that only cumulative mutations of AIVs in avians could lead to spillover, causing mammalian (or human) infections and deaths. Currently, no evidence of HPAI H5 transmission has been observed among mammals. However, H5N1 was recently detected in mink farms in the United States and Spain, and more than 50,000 mink were killed to prevent further spread2–4. These events provide strong evidence that HPAI H5 viruses can spread rapidly among mammals and that minks may serve as a potential intermediate host to increase the possibility of the H5N1 epidemic in humans. In fact, human infections caused by the HPAI H5 viruses have recently been reported in Ecuador, Cambodia, and Chile (https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON434; https://www.cdc.gov/flu/avianflu/spotlights/2022-2023/chile-first-case-h5n1-addendum.htm).
Fig. 3. Global number of human cases of H5N1 and H5N8 infection.
a Global number of human cases of H5N1 infection from 2003 to 2021 (https://www.who.int/publications/m/item/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who-2003-2023-3-march-2023). b Global number of human cases of H5N6 infection from 2014 to 2022 (https://search.fresh.gov.hk/chp/sc/search_result.php?q=influenza+virus&fq_yr=2023&fq_ct=&fq_ft=&sort=&page=1).
More concerning evidence has been reported on human infection with the H5N6 subtype virus. From 2014 to 2022, 87 human cases of H5N6 infection were reported (86 in China and one in Laos), with most infections reported in 2021 and 2022 (Fig. 3b). The number of H5N6 infection cases in 2021 and 2022 accounted for 67% of the total number of infections from 2014 to 2022, suggesting that the Chinese government should strengthen protective measures to prevent further spread of the H5N6 virus. Additionally, the first case of H5N8 infection in humans was reported in Russia in 202025. The significant increase in human cases of H5N6 and the emergence of a new human case of H5N8 are alarming signs for human safety.
Analysis of HPAI H5 HA sequences and assessment of the risk for human infection based on the infection data from 2019 to 2022
In the process of viral infection, HA binds to sialic acid receptors on the cell surface and mediates the fusion of the viral membrane and the host endosomal membrane to deliver viral nucleic acid into the cytoplasm of host cells, thereby playing a key role in the process of infection. HA protein is hydrolyzed to produce HA1 and HA2. HA1 binds to cell receptors via the receptor-binding domain (RBD) and is prone to mutations, while HA2 mediates the membrane fusion process and is relatively conserved. The HA1 of AIVs binds to the α-2,3-sialic acid receptors in avian species, while it binds to the α-2,6-sialic acid receptors in humans. The difference in receptor usage partly prevents the transmission of AIVs from birds to humans. Therefore, we compared HA sequence, HA1 sequence, and RBD key sites of HPAI H5 viruses isolated from avians and humans in recent years to assess the potential risk of human infection.
The amino acid sequences of HA proteins were derived from GISAID (https://gisaid.org/). The major HPAI H5 HA sequences from avian and human (Fig. 4) belong to subclades 2.3.4.4b (H5N1, H5N6, H5N8), 2.3.4.4h (H5N6) and 2.3.2 (H5N1). Notably, human infection was closely related to the outbreak of AIVs in avians since strains that caused both human and avian infections showed a close evolutionary relationship in all subclades of HPAI viruses, except for 2.3.2.1c. In addition, subclades 2.3.4.4b and 2.3.4.4h mainly broke out in Europe, Africa, Asia, and the Americas, while subclade 2.3.2 has only recently appeared in Egypt, South Asia, and other countries. HA1 amino acid sequences of HPAI H5 were highly homologous with only individual mutations, or even no mutations, for strains from different hosts, subtypes, and regions separated by time (Table 1). The HA1 sequence of human-derived H5N8 strain A/Astrakhan/3212/2020 was used as a reference sequence for comparison with the HA1 sequences of strains derived from humans or birds. It was surprising to find that the HA1 sequence of A/Astrakhan/3212/2020 from humans was identical to that of A/chicken/Kosovo/22-2 22VIR3124-13/2022 from avian. Moreover, only one amino acid (T192I) separated it from another avian-derived HA1 (A/whooper swan/Shanxi/4-1/2020), suggesting that these avian-derived strains hold a high risk for human infection. Most of these site differences in HA1 are located in the RBD region (E130D, A144T, V152L R173Q, T192I, and V214A). Previous studies have performed key amino acid mutations in RBD to analyze the effects on α-2,3/6-sialic acid affinities26–33. We collected these key amino acid mutations and compared them with the recent sequences from avian and human sources of the H5N1, H5N6, and H5N8 subtypes (Table 2).
Fig. 4. Phylogenetic tree based on HA amino acid sequences of HPAI H5 viruses isolated between 2019 and 2022.
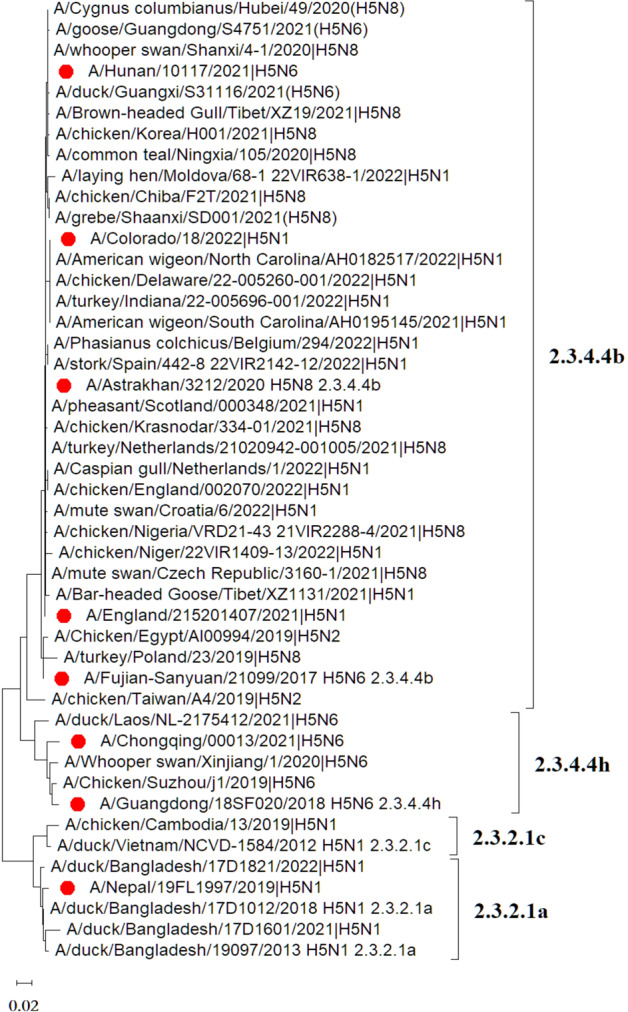
These sequences were obtained from GISAID. The human-derived HPAI H5 sequences are marked in red. The phylogenetic tree was drawn with MEGA 11 obtained from https://megasoftware.net/.
Table 1.
Differences in HA1 amino acid sequences between HPAI H5 virus derived from humans and avians.
| Host | Isolation | Mutation |
|---|---|---|
| Human | A/Astrakhan/3212/2020 | H5N8 | Reference sequence |
| Human | A/England/215201407/2021 | H5N1 | A144T |
| A/Hunan/10117/2021 | H5N6 | T192I | |
| A/Colorado/18/2022 | H5N1 | L108M, V214A | |
| Avian | A/Brown-headed Gull/Tibet/XZ19/2021 | H5N8 | H277N |
| A/chicken/Korea/H001/2021 | H5N8 | Q19R | |
| A/common teal/Ningxia/105/2020 | H5N8 | V214I | |
| A/whooper swan/Shanxi/4-1/2020 | H5N8 | T192I | |
| A/Cygnus columbianus/Hubei/49/2020 | H5N8 | T192I | |
| A/goose/Guangdong/S4751/2021 | H5N6 | E130D | |
| A/American wigeon/North Carolina/AH0182517/2022 | H5N1 | L108M, V214A | |
| A/Phasianus colchicus/Belgium/294/2022 | H5N1 | Q19K | |
| A/stork/Spain/442-8 22VIR2142-12/2022 | H5N1 | Q19K | |
| A/mute swan/Croatia/6/2022 | H5N1 | S124N | |
| A/Caspian gull/Netherlands/1/2022 | H5N1 | V152L | |
| A/chicken/England/002070/2022 | H5N1 | V152L | |
| A/turkey/Netherlands/21020942-001005/2021 | H5N8 | No mutation | |
| A/chicken/Krasnodar/334-01/2021 | H5N8 | No mutation | |
| A/pheasant/Scotland/000348/2021 | H5N1 | No mutation | |
| A/chicken/Nigeria/VRD21-43 21VIR2288-4/2021 | H5N8 | R173Q |
Table 2.
Comparison of key amino acid sites in the RBD region of HPAI H5 viruses.
| Key amino acid site | 133 | 136 | 137 | 138 | 153 | 158 | 160 | 183 | 186 | 190 | 192 | 193 | 196 | 216 | 221 | 225 | 226 | 227 | 228 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aviana | L | T/A | S | A | A | N | T | F | N | E | T | L | Q | R | S | G | Q | S | G |
| Humana | V | S | A | V | W | S | A | H | K | D | I | R | H | E | P | D | L | N | S |
| H5N8 HA1 from avian and human | |||||||||||||||||||
| A/chicken/Kosovo/22-2 22VIR3124-13/2022b | L | S | A | A | W | N | A | H | N | E | T | N | K | K | S | G | Q | R | G |
| A/whooper swan/Shanxi/4-1/2020H5N8 | . | . | . | . | . | . | . | . | . | . | I | . | . | . | . | . | . | . | . |
| A/chicken/Egypt/Army/1201/2022 | R | . | . | . | . | . | . | . | . | . | I | . | . | . | . | . | . | . | . |
| A/Astrakhan/3212/2020c | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| H5N6 HA1 from avian and human | |||||||||||||||||||
| A/duck/Hunan/S40199/2021 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| A/duck/Yunnan/S4318/2021 | . | . | . | . | . | . | . | . | . | . | I | . | . | . | . | . | . | . | . |
| A/duck/Zhejiang/S4854/2021 | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . |
| A/Hunan/09285/2021 | . | . | . | . | . | . | . | . | . | . | I | . | . | . | . | . | L | . | . |
| A/Chongqing/02/2021 | . | . | . | . | . | . | . | . | . | . | I | . | . | . | . | . | X | . | . |
| A/Guangdong/18SF020/2018 | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . |
| A/Jiangsu/1/2020 | . | . | . | . | . | . | . | . | . | . | . | K | . | . | . | . | . | G | . |
| A/Chongqing/00013/2021 | . | . | . | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . |
| A/Fujian-Sanyuan/21099/2017 | . | . | T | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . |
| H5N1 HA1 from avian and human | |||||||||||||||||||
| A/duck/Guizhou/S1321/2022 | . | . | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . |
| A/laying hen/Moldova/68-2 22VIR638-2/2022 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| A/duck/Bangladesh/17D1821/2022 | . | . | . | . | . | D | . | . | . | . | . | R | Q | . | . | . | . | S | . |
| A/chicken/Hong Kong/AP156/2008 | . | . | . | . | . | . | T | . | . | . | . | R | Q | . | . | . | . | S | . |
| A/duck/Egypt/D1Br12/2007 | S | . | S | . | . | . | T | . | . | . | . | M | Q | . | . | . | . | S | . |
| A/Nepal/19FL1997/2019 | . | . | . | . | . | . | . | . | . | . | . | R | Q | . | . | . | . | N | . |
| A/Laos/2121/2020 | . | . | . | . | . | D | . | . | X | . | . | R | Q | R | P | . | . | S | . |
| A/England/215201407/2021 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| A/Anhui/1/2005 | S | . | S | . | . | . | T | . | . | . | . | K | Q | . | . | . | . | S | . |
| A/Thailand/1(KAN-1)/2004 | . | . | S | . | . | . | T | . | . | . | . | K | Q | R | . | . | . | S | . |
| A/Vietnam/1203/2004 | S | . | S | . | . | D | . | . | . | . | . | R | Q | . | . | . | . | S | . |
| A/Indonesia/05/2005 | . | . | S | . | . | . | T | . | . | . | . | K | Q | R | . | . | . | S | . |
Sequence data were downloaded from https://legacy.fludb.org/brc/home.spg?decorator=influenza.
aTypical amino acids at the indicated positions in avian- or human-susceptible H5 viruses.
bReference sequence.
cHPAI viruses isolated from humans are underlined.
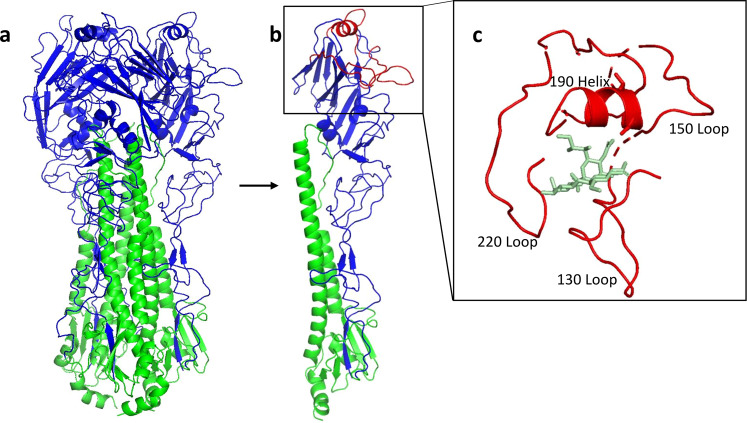
RBD is located in the head of HA1 and contains 190-helix, 130-loop, 150-loop, 220-loop, and other amino acid residues26,34,35 (Fig. 5). Yang et al.36 found that the introduction of S137A and T192I mutations in the RBD of A/Thailand/KAN 1/2004 endowed this Avian strain with the ability to bind with α-2,6-sialic acid receptors present in humans. In our selected sequences, the 137A and 192I sites were found to be present in both human- and avian-derived H5N8 and H5N6 strains, indicating that they are key sites for cross-species transmission. In addition, although 192T exists in A/Astrakhan/3212/2020 (H5N8) and A/Fujian-Sanyuan/21099/2017 (H5N6), these strains still retain the ability to infect humans. This indicates that a single-site mutation (S137A/T) may also change the receptor-binding ability of AIVs. It has been reported that some mutation sites, such as the K193R mutation in the A/Vietnam/1203/2004 strain37, the Q196H mutation in the A/duck/Egypt/ D1Br12/2007 strain38, and the Q226L, S227N, and G228S mutations in the A/Indonesia/05/2005 strain39, can enhance the ability of stains to utilize the α-2,6-sialic acid receptor. In our selected sequences, the 193R site is present in both human- and avian-derived H5N1 strains, indicating that these H5N1 strains may have already gained the ability to utilize α-2,6-sialic acid receptors.
Fig. 5. Structure of HA protein derived from HPAI H5 virus.
a Trimeric HA is shown in cartoon representation with HA1 displayed in blue and HA2 in green. b Critical domains, including three loops and one helix, in RBD are shown in red in a single HA molecule. c Red domains in RBD (130-loop, 150-loop, 190-helix, and 220-loop) from b were enlarged and displayed with a sialic acid molecule (light green) in the groove. HA sequence of A/Astrakhan/3212/2020 was modeled by SWISS-MODEL and drawn by PyMOL software obtained from https://pymol.org/2/. The PDB number for the α-2,6-sialic acid molecule is 5E35.
Antiviral therapy for influenza
Some small-molecule compounds have been developed for the treatment of influenza viruses. These compounds target various stages of the viral life cycle, e.g., virus adsorption, fusion, nucleic acid release, nucleic acid replication, and virus budding. HA protein inhibitors block virus adsorption or fusion, which can be divided into HA1 and HA2 inhibitors. HA1 inhibitors, such as Dextran sulfate and DSA18140, block the binding of HA1 to receptors on the cell surface. Meanwhile, HA2 inhibitors like arbidol41 and BMY-2770942 block virus entry by preventing HA2-mediated membrane fusion. In addition, Basu et al. identified two small-molecule compounds, MBX2329 and MBX2546, which were able to bind to the stem region of the HA trimer and inhibit HA-mediated fusion43. The fusion process of the influenza virus also depends on endosomal acidification and a series of host enzymes, like proteases. Therefore, inhibitors of these host enzymes have also been developed as anti-influenza drugs, such as bafilomycin A144 and aprotinin45. After membrane fusion, viral RNA enters the host cell through the M2 ion channel. M2 inhibitors like amantadine and rimantadine, which block ion channel activity, were developed to prevent the release of the viral genome into the cytoplasm. M2 inhibitors are effective for the influenza A virus but not for the influenza B virus because of its lack of M2 protein. It has been reported that S31N mutation is the main culprit causing resistance to M2 inhibitors, thus accounting for 92% of drug-resistant strains in the United States46. Consequently, M2 inhibitors are currently not recommended for treatment. NA protein is related to the maturation and release of viruses, and it plays an important role in regulating receptor binding and virus budding. NA inhibitors, such as oseltamivir, zanamivir, and peramivir, can effectively inhibit the release of progeny viruses from infected cells47,48. However, amino acid mutations in the NA protein, e.g., E119A, H274Y, and N294S, usually lead to resistance to NA inhibitors49,50. Furthermore, viral nucleic acid replication inhibitors include PB2 inhibitors (VX78751), PA inhibitors (flutamide52 and Baloxavir53), RNA-dependent RNA polymerase (RdRp) inhibitors (Favipiravir54), and NP inhibitors (nucleolin55).
In addition, some monoclonal antibodies (Mab) have been developed and are highly anticipated for post-exposure prophylaxis and clinical treatment. For example, a novel humanized Mab 8A56 neutralized H5N1 by binding to two types of epitopes on HA. Li et al. described a chimeric Mab, termed C12H5, which could neutralize representative strains of H1N1 circulating from 1991 to the present; it could even cross-neutralize H5N157. It has been reported that neutralizing antibodies against HA, isolated from volunteers vaccinated with seasonal influenza vaccines, could protect mice from H1N1 and H3N2 viruses in vivo58. Recently, the FDA confirmed that humanized polyclonal antibody SAB-176 could recognize multiple epitopes and provide protection against multiple influenza virus strains (https://ir.sab.bio/static-files/b332c893-5795-4d05-af96-a9ebcd917f24). A phase 2b clinical trial is about to be launched in patients with high-risk severe diseases. Some polypeptide drugs, such as EB-peptide59, iHA60, FluPep61, NDFRSKT62, P163, and P9R64, have also been developed against influenza viruses. However, the accumulation of mutations in AIVs still increases the probability of immune evasion65,66. Therefore, updating existing antiviral drugs cannot keep pace with the continuous variation of AIVs. This calls for new antiviral strategies, such as drugs and therapeutic Mabs targeting more conserved viral epitopes or cytokines, or immunomodulatory drugs, in response to emerging strains with epidemic potential43,67.
Development of H5-specific influenza vaccines
Currently, the main types of avian influenza vaccines include inactivated recombinant vaccines, subunit vaccines, viral vector vaccines, and DNA vaccines. Inactivated vaccines were previously the primary means of preventing influenza and were mainly prepared from low pathogenicity strains isolated from farms68. However, traditional inactivated vaccines are not conducive to vaccine production owing to such defects as dependence on embryo culture and low virus titer. At present, the vaccines used for H5 and H7 avian influenza in China are mainly recombinant inactivated vaccines. These vaccines are prepared by co-transfecting Vero cells with the viral RNA expression plasmid of HA and NA genes of the current epidemic strains and six internal genes (PB2, PB1, PA, NP, M, and NS) of PR8 (A/Puerto Rico/8/1934), together with four PR8 protein expression plasmids (PB2, PB1, PA, and NP)69. The basic terminal sequence R/KRRKR of HA from the HPAI virus was modified to RETR, endowing the recombinant virus with both the epitope of the pandemic strain and the high-titer characteristics of PR8 chicken embryo adaptation. The team led by Dr. Hualan Chen in China has developed a series of recombinant vaccines for the prevention of HPAI H5, among which Re-13 (A/duck/Fujian/S1424/2020 H5N6 2.3.4.4h) and Re-14 (A/whooper_ swan/Shanxi/4-1/2020 H5N8 2.3.4.4b) were developed in 202270. According to Chen et al.71, the H5N1 AIV strain bearing the subclade 2.3.4.4b HA gene was isolated from China in 2021–2022 and exhibited antigenic sites similar to those of H5-Re14. Since this type of recombinant vaccine is widely used in China, it plays a crucial role in the prevention and control of AIVs. WHO updated its AIV strain recommendations in 2022 and selected A/Astrakhan/3212/2020 H5N8 2.3.4.4b, A/Guangdong/18SF020/2018-like H5N6 2.3.4.4h and A/Fujian-Sanyuan /21099/2017-like H5N6 2.3.4.4b as candidate vaccine strains (https://www.who.int/teams/global-influenza-programme/vaccines/who-recommendations).
Development of universal influenza vaccines
As AIV is a single-stranded RNA virus, its nucleic acid sequence is prone to mutation, thereby reducing the protective efficacy of the vaccine over time. Although it is possible to predict the next dominant strain for vaccine strain selection, production, and distribution, the circulating strain may further mutate, resulting in a decrease in vaccine protection efficiency. Therefore, it is necessary to develop universal influenza vaccines that target more conservative epitopes to counter potential antigenic drift or shift in AIVs. Accordingly, scientists have focused on several common targets for the development of universal influenza vaccines, including the conserved stalk domain of HA protein, the conserved regions of NA protein, the ectodomain of M2 ion channel (M2e), and the internal proteins, nucleoprotein (NP) and matrix protein 1 (M1). The aim is to expand existing immune memory response by multiple immunizations in order to produce the widest range of protective antibodies against different subtypes of influenza virus72.
Effective humoral heterosubtypic immunity is rare, mainly based on antibodies targeting the HA stalk domain73,74. As mentioned, the RBD region in the head of HA protein is prone to mutations leading to viral immune evasion, while the HA stalk domain is rarely exposed to neutralizing antibodies, thus facing less selection pressure from the host immune system. As a result, the HA stalk domain is highly conserved in AIVs, making it an attractive target for universal vaccine design. A strategy for inducing high levels of stalk-reactive antibodies is based on chimeric HAs (cHAs), which combine exogenous head domains with conserved stalk domains. The cHAs with different head domains have been used in sequential vaccination programs to break the immunodominance of the head domain of HA and induce high titers of stalk-reactive antibodies75. However, the vaccine targeting the conserved stalk domain of HA can only produce cross-protection that occurs between strains within the same subtype or multiple subtypes of the same group, making it difficult to induce broadly reactive antibodies against influenza viruses across different groups. However, the construction of chimeric HA stalk domain with other conserved antigens, such as M2e, could improve cross-protection against multiple AIVs from different groups and improve the broad protection of universal influenza vaccines76. Chen et al. 77 have reported that influenza virus infection induces high titers of NA-reactive antibodies, which effectively inhibit the enzymatic activity of NA and provide robust prophylactic protection against avian H5N1 viruses in vivo. This observation suggests that some conserved regions in NA recognized by NA-reactive antibodies could be incorporated into influenza vaccines to elicit durable and broad protection against divergent influenza strains.
Emerging vaccine platforms can help trigger a better immune response than that induced by traditional influenza vaccines. For example, virus-like particles (VLPs) can present natural conformational antigens, stimulate the immune system through a virus-like pathway, and efficiently induce immune protection. An H5N1 VLP-based vaccine, designed with computationally optimized broadly cross-reactive antigen (COBRA), elicits broadly reactive antibodies in mice and ferrets. Therefore, this strategy is potentially paradigm-shifting for H5 universal influenza vaccines78. A candidate universal influenza vaccine, which uses M2e-based VLP to present M2e, has been shown to protect mice from homosubtypic and heterosubtypic AIVs79. In addition, nanoparticle platforms have been used to develop universal influenza vaccines owing to their dominance in expressing antigens at high densities and providing adjuvant-like functions. For example, the OVX836 vaccine is based on oligomerized nanoparticles (NPs) that can induce humoral and cellular immunity in mice and ferrets, thereby providing protection against influenza A and B80,81. A ‘mosaic’ quadrivalent influenza vaccine based on two-component nanoparticle immunogens not only showed better protective antibody response than the 2017–2018 quadrivalent influenza vaccine (QIV) but also triggered heterosubtypic antibody response and protective immunity in several animal models82. Viral vector-based vaccines can be delivered through both systemic or mucosal routes to trigger strong humoral and cellular immunity. An adenovirus vector-based H5N1 conserved multi-epitope influenza vaccine showed broad immune protection against H5, H7, and H9 influenza viruses in mice83. The nucleic acid platform includes DNA- and mRNA-based vaccines, which can respond quickly to emerging outbreaks. Based on their novel contribution to the coronavirus disease 2019 (COVID-19) pandemic, mRNA-based vaccines have become the focus of new vaccine technologies. Freyn et al. 84 demonstrated the broad protective effect of nucleoside-modified mRNA-LNP vaccines based on conserved antigens (HA, NA, NP, and M2) against influenza virus challenge in mice. Koen et al. 85 evaluated heterosubtypic protection from a nucleoside-modified mRNA vaccine that encodes the conserved NP, M1, and PB1 (polymerase basic protein 1) of one H1N1 strain. This vaccine induced a broadly reactive T-cell response in ferrets. Recently, Arevalo et al.86 developed an mRNA-LNP vaccine encoding HA from 20 known influenza A and B virus subtypes, and it triggered high levels of cross-reactivity and subtype-specific antibodies in mice and ferrets. This is a new antigen design concept for developing a universal influenza vaccine.
In addition, new vaccine adjuvants support ideas for universal vaccine design. Appropriate vaccine adjuvants can improve immunogenicity, regulate immune response types, and even enhance the universality of vaccine protection87,88. Only 6 new adjuvants have been approved by the FDA in the past century, including MF59, AS04, AS03, AS01, CpG1018, and Matrix-M adjuvants for emergency use in COVID-19. MF59 and AS03 have improved the protective efficiency of influenza vaccines73,89. More prominently, the quadrivalent influenza nanoparticle vaccine (qNIV) with Matrix-M has been shown to enhance antigen presentation, expand the antibody epitope library, boost cross-neutralizing antibody responses, and improve the induction of potent CD4+ and CD8+ T cell responses in a variety of cells90. It has now successfully completed key phase III trials. Compared with adjuvant-free vaccines, influenza vaccines with adjuvants have shown higher immunogenicity and effects on heterologous strains. The 2′,3′-cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) is an effective natural agonist of stimulator of interferon genes (STING), which induces type I interferon (IFN-I) response and proinflammatory cytokine production by activating STING91,92. Wang et al.93 demonstrated intranasal immunization with the PS-cGAMP-adjuvanted inactivated H1N1 vaccine, which triggered a strong protective effect against different subtypes of influenza (H3N2, H5N1, and H7N9) in mice. During the COVID-19 pandemic, the emerging non-nucleotide small-molecule STING agonist CF501 showed higher protective efficacy compared to the cGAMP-adjuvanted vaccine, suggesting that CF501 can also be used as an adjuvant to boost the original vaccine for effective, extensive and long-term immune protection94.
In this article, we have analyzed the global epidemic of HPAI H5 and revealed that the number of infections has risen significantly in recent years. Furthermore, it has been observed that the dominant HPAI virus worldwide rapidly changed from H5N8 to H5N1 in 2022. According to the sequence alignment analysis of HA1, we found that the HA1 sequences of strains isolated from avians and humans were highly homologous or even identical, suggesting that the existing AIVs strains circulating in birds may infect humans. Amino acids 137A, 192I, and 193R in the RBD of HA are key sites that exist in both avian and human source sequences. These sites enable the current HPAI H5 strains to bind α-2,6-sialic acid receptors in humans, indicating that the mutated HPAI H5 viruses may have jumped from birds to mammals and that such spillover may cause human infection.
It should be mentioned that receptor affinity is not the only factor affecting the transmission of AIVs in humans. In the process of viral infection, HA mediates membrane fusion between the virus and host cells95. Next, nucleic acid is released with the assistance of the M protein and enters the nucleus to complete viral replication in the presence of viral polymerases PA, PB1, and PB2. Finally, the progeny virus is released from infected cells with the assistance of the NA protein. Many HPAI H5 viruses can enter host cells, but they cannot replicate successfully owing to the difference in amino acids at position 627 of PB2 protein, namely glutamic acid in AIVs and lysine in human influenza virus96. Hence, mutations in the RBD domain may only affect receptor binding and cell entry of AIVs, while replication efficiency of the virus in cells must be assisted by other viral proteins, such as PA, PB1, PB2, and NA, to gain successful cross-species transmission97. Therefore, mutations in these proteins and homologous recombination between strains deserve more attention.
Nowadays, the HPAI H5 virus belonging to the 2.3.4.4b subclade is widespread among wild birds and poultry worldwide, resulting in significant economic losses. The prevention and control strategy for the HPAI H5 virus in Europe and North America mainly relies on culling, while the strategy in China is “vaccine and culling”. The latter strategy did reduce HPAI H5 virus infections in avians in China (Fig. 1a)25. In addition, after vaccination of the H5/H7 vaccine in poultry, the isolation of H7N9 strains in China decreased by 93.3%, which largely prevented the prevalence of H7N9 among poultry98. The transmission modes of HPAI H5 among wild birds, poultry, and mammals also deserve more attention. Wildfowl is the natural host of the HPAI H5 virus, and the virus usually replicates in their intestines and respiratory tract. Nine major routes have been identified for migration across the world, increasing the likelihood of AIV infection in poultry and mammals99,100. Therefore, understanding the temporospatial characteristics and as well as environmental factors of HPAI H5 outbreaks is helpful for establishing an effective prevention and control system101. It is widely accepted that AIVs only infect mammals through avian transmission and that no reports have so far indicated its spread among mammals. Nonetheless, the recent spread of the H5N1 virus in mink has sounded the alarm for human safety2–4. Prevention should be emphasized in virus-susceptible areas, and measures should be taken to reduce human exposure to birds and mammals in order to minimize the risk of zoonotic infections. Protective measures and preventive vaccination should be taken seriously for populations susceptible to occupational hazards. Finally, real-time virus monitoring and rapid data sharing are crucial for assessing the risk of cross-species transmission of HPAI H5 and implementing effective prevention and control measures. The antigenic drift of the current epidemic strains should be monitored, and it should be determined whether the existing vaccines still have protective effects. Furthermore, H5-specific vaccines need to be developed, and the team led by Hualan Chen in China, whose work was noted above, serves as a model in this regard71,102,103.
While small-molecule compounds, peptides, and antibodies have been developed for influenza antiviral therapy, the constant mutation of the virus and its ability to evade immune response confound these efforts. Therefore, drugs and vaccines must be regularly updated to address the emergence of new strains. In response, scientists are trying different methods to develop universal vaccines against multiple subtypes of influenza viruses. HA is the main immunogen for vaccine design and mainly induces antibodies against the RBD region at the spherical head of HA, which is also highly prone to mutation. However, some cross-protective antibodies against highly conserved HA stalk may also be induced104. Emerging vaccine platforms and new vaccine adjuvants also provide pathways toward improving vaccine efficacy. Although not emphasized in this review, the potential of cross-reactive T cell-based responses for influenza vaccine design cannot be ignored. Currently, avian influenza vaccines are mandatory for poultry immunization. However, they are not included in routine human immunization but are only used as a preventive vaccination strategy during emergencies. HPAI H5 viruses are circulating in birds and have even caused outbreaks in mammals in recent years, thus raising concerns about HPAI H5 infections in humans. Heterologous prime–boost immunization strategies against H5N1 could induce broader cross-clade antibody responses. It is also worth considering priming with a universal vaccine and boosting with a specific vaccine against the current pandemic strain.
Acknowledgements
The authors thank Mr. Chuxiong Zheng at Guangdong Haid Group Co., Ltd. for his valuable suggestions in the data analysis. This study was supported by grants from the National Natural Science Foundation of China (92169112 to S.J.) and Guangzhou High-level Research Institute in Enterprises independent projects (202205110006 to C.P.).
Author contributions
C.P. and S.J. conceived the idea and planned the study. P.H., L.S., Q.W., and J.L. collected and analyzed the data, as well as prepared the figures and tables. P.H. and L.S. drafted the manuscript, while C.P., N.R., and S.J. revised and finalized the manuscript. All authors have read and approved the final version of the manuscript for submission.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pan Huang, Lujia Sun
Contributor Information
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
Chungen Pan, Email: pancg01@haid.com.cn.
References
- 1.Tonegawa K, et al. Analysis of epitope recognition of antibodies induced by DNA immunization against hemagglutinin protein of influenza A virus. Vaccine. 2003;21:3118–3125. doi: 10.1016/S0264-410X(03)00257-3. [DOI] [PubMed] [Google Scholar]
- 2.Kupferschmidt K. Bird flu spread between mink is a ‘warning bell’. Science. 2023;379:316–317. doi: 10.1126/science.adg8342. [DOI] [PubMed] [Google Scholar]
- 3.Sidik SM. Bird flu outbreak in mink sparks concern about spread in people. Nature. 2023;614:17. doi: 10.1038/d41586-023-00201-2. [DOI] [PubMed] [Google Scholar]
- 4.Agüero M, et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Euro. Surveill. 2023;28:2300001. doi: 10.2807/1560-7917.ES.2023.28.3.2300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira HG, Tůmová B, Law VG. Avian influenza A viruses. Bull. World Health Organ. 1965;32:855–860. [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC). Isolation of avian influenza A(H5N1) viruses from humans-Hong Kong, May-December 1997. MMWR Morb. Mortal. Wkly. Rep. 46, 1204–1207 (1997).. [PubMed]
- 7.World Health Organization Global Influenza Program Surveillance Network. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 2005;11:1515–1521. doi: 10.3201/eid1110.050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonassen CM, Handeland K. Avian influenza virus screening in wild waterfowl in Norway, 2005. Avian Dis. 2007;51:425–428. doi: 10.1637/7555-033106R1.1. [DOI] [PubMed] [Google Scholar]
- 9.Ducatez MF, et al. Molecular and antigenic evolution and geographical spread of H5N1 highly pathogenic avian influenza viruses in western Africa. J. Gen. Virol. 2007;88:2297–2306. doi: 10.1099/vir.0.82939-0. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir. Viruses. 2014;8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao K, et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet. Microbiol. 2013;163:351–357. doi: 10.1016/j.vetmic.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Wu H, et al. Novel reassortant influenza A(H5N8) viruses in domestic ducks, eastern China. Emerg. Infect. Dis. 2014;20:1315–1318. doi: 10.3201/eid2008.140339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Outbreaks of highly pathogenic avian influenza (H5N6) virus subclade 2.3.4.4h in swans, Xinjiang, Western China, 2020. Emerg. Infect. Dis. 2020;26:2956–2960. doi: 10.3201/eid2612.201201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat S, et al. Genetic and antigenic characterization of H5N1 viruses of clade 2.3.2.1 isolated in India. Microb. Pathog. 2015;88:87–93. doi: 10.1016/j.micpath.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Naguib MM, et al. Outbreaks of highly pathogenic avian influenza H5N1 clade 2.3.2.1c in hunting falcons and kept wild birds in Dubai implicate intercontinental virus spread. J. Gen. Virol. 2015;96:3212–3222. doi: 10.1099/jgv.0.000274. [DOI] [PubMed] [Google Scholar]
- 16.Arafa AS, et al. Emergence of a novel cluster of influenza A(H5N1) virus clade 2.2.1.2 with putative human health impact in Egypt, 2014/15. Euro. Surveill. 2015;20:2–8. doi: 10.2807/1560-7917.ES2015.20.13.21085. [DOI] [PubMed] [Google Scholar]
- 17.Bi Y, et al. Highly pathogenic avian influenza H5N1 Clade 2.3.2.1c virus in migratory birds, 2014-2015. Virol. Sin. 2016;31:300–305. doi: 10.1007/s12250-016-3750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchenko VY, et al. Highly pathogenic influenza H5N1 virus of clade 2.3.2.1c in Western Siberia. Arch. Virol. 2016;161:1645–1649. doi: 10.1007/s00705-016-2800-4. [DOI] [PubMed] [Google Scholar]
- 19.Antigua KJC, Choi WS, Baek YH, Song MS. The emergence and decennary distribution of clade 2.3.4.4 HPAI H5Nx. Microorganisms. 2019;7:156. doi: 10.3390/microorganisms7060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo FT, et al. Intercontinental spread of Eurasian highly pathogenic avian influenza A(H5N1) to Senegal. Emerg. Infect. Dis. 2022;28:234–237. doi: 10.3201/eid2801.211401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lean FZX, et al. Gross pathology associated with highly pathogenic avian influenza H5N8 and H5N1 in naturally infected birds in the UK (2020-2021) Vet. Rec. 2022;190:e731. doi: 10.1002/vetr.731. [DOI] [PubMed] [Google Scholar]
- 22.Banyard AC, et al. Detection of highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b in Great Skuas: a species of conservation concern in Great Britain. Viruses. 2022;14:212. doi: 10.3390/v14020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali M, et al. Genetic characterization of highly pathogenic avian influenza A(H5N8) virus in Pakistani live bird markets reveals rapid diversification of clade 2.3.4.4b viruses. Viruses. 2021;13:1633. doi: 10.3390/v13081633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong J, et al. Emerging highly pathogenic avian influenza (H5N8) virus in migratory birds in Central China, 2020. Emerg. Microbes Infect. 2021;10:1503–1506. doi: 10.1080/22221751.2021.1956372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, et al. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microbes Infect. 2023;12:2155072. doi: 10.1080/22221751.2022.2155072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 27.Martín J, et al. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241:101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- 28.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 29.Yamada S, et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 30.Auewarakul P, et al. An avian influenza H5N1 virus that binds to a human-type receptor. J. Virol. 2007;81:9950–9955. doi: 10.1128/JVI.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M, et al. Residue Y161 of influenza virus hemagglutinin is involved in viral recognition of sialylated complexes from different hosts. J. Virol. 2012;86:4455–4462. doi: 10.1128/JVI.07187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilyushina NA, et al. Human-like receptor specificity does not affect the neuraminidase-inhibitor susceptibility of H5N1 influenza viruses. PLoS Pathog. 2008;4:e1000043. doi: 10.1371/journal.ppat.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens J, et al. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J. Mol. Biol. 2008;381:1382–1394. doi: 10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 35.DuBois RM, et al. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog. 2011;7:e1002398. doi: 10.1371/journal.ppat.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang ZY, et al. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007;317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, et al. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J. Virol. 2010;84:6570–6577. doi: 10.1128/JVI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe Y, et al. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 2011;7:e1002068. doi: 10.1371/journal.ppat.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chutinimitkul S, et al. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J. Virol. 2010;84:6825–6833. doi: 10.1128/JVI.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belser JA, et al. DAS181, a novel sialidase fusion protein, protects mice from lethal avian influenza H5N1 virus infection. J. Infect. Dis. 2007;196:1493–1499. doi: 10.1086/522609. [DOI] [PubMed] [Google Scholar]
- 41.Boonma T, et al. Insights into binding molecular mechanism of hemagglutinin H3N2 of influenza virus complexed with arbidol and its derivative: a molecular dynamics simulation perspective. Comput. Biol. Chem. 2022;101:107764. doi: 10.1016/j.compbiolchem.2022.107764. [DOI] [PubMed] [Google Scholar]
- 42.Luo G, et al. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J. Virol. 1997;71:4062–4070. doi: 10.1128/jvi.71.5.4062-4070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu A, et al. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza a virus fusion. J. Virol. 2014;88:1447–1460. doi: 10.1128/JVI.01225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochiai H, et al. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antiviral Res. 1995;27:425–430. doi: 10.1016/0166-3542(95)00040-S. [DOI] [PubMed] [Google Scholar]
- 45.Zhirnov OP, Klenk HD, Wright PF. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res. 2011;92:27–36. doi: 10.1016/j.antiviral.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Bright RA, et al. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. J. Am. Med. Assoc. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 47.De Clercq E. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Clercq E, Neyts J. Avian influenza A (H5N1) infection: targets and strategies for chemotherapeutic intervention. Trends Pharmacol. Sci. 2007;28:280–285. doi: 10.1016/j.tips.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Li Y, Zhang L, Hou T. Theoretical studies on the susceptibility of oseltamivir against variants of 2009 A/H1N1 influenza neuraminidase. J. Chem. Inf. Model. 2012;52:2715–2729. doi: 10.1021/ci300375k. [DOI] [PubMed] [Google Scholar]
- 50.Burnham AJ, et al. Fitness costs for Influenza B viruses carrying neuraminidase inhibitor-resistant substitutions: underscoring the importance of E119A and H274Y. Antimicrob. Agents Chemother. 2014;58:2718–2730. doi: 10.1128/AAC.02628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, et al. Design, synthesis, biological evaluation, and molecular dynamics simulation of influenza polymerase PB2 inhibitors. Molecules. 2023;28:1849. doi: 10.3390/molecules28041849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomassini JE, et al. A novel antiviral agent which inhibits the endonuclease of influenza viruses. Antimicrob. Agents Chemother. 1996;40:1189–1193. doi: 10.1128/AAC.40.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Govorkova EA, et al. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2018-2020. Antiviral Res. 2022;200:105281. doi: 10.1016/j.antiviral.2022.105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Correa-Padilla E, et al. Modifications in the piperazine ring of nucleozin affect anti-influenza activity. PLoS ONE. 2023;18:e0277073. doi: 10.1371/journal.pone.0277073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan Y, et al. A novel humanized antibody neutralizes H5N1 influenza virus via two different mechanisms. J. Virol. 2015;89:3712–3722. doi: 10.1128/JVI.03014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T, et al. Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses. Nat. Commun. 2022;13:5182. doi: 10.1038/s41467-022-32926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun X, et al. Unique binding pattern for a lineage of human antibodies with broad reactivity against influenza A virus. Nat. Commun. 2022;13:2378. doi: 10.1038/s41467-022-29950-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones JC, et al. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J. Virol. 2006;80:11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito M, et al. Macrocyclic peptides exhibit antiviral effects against influenza virus HA and prevent pneumonia in animal models. Nat. Commun. 2021;12:2654. doi: 10.1038/s41467-021-22964-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicol MQ, et al. A novel family of peptides with potent activity against influenza A viruses. J. Gen. Virol. 2012;93:980–986. doi: 10.1099/vir.0.038679-0. [DOI] [PubMed] [Google Scholar]
- 62.Rajik M, et al. Identification and characterisation of a novel anti-viral peptide against avian influenza virus H9N2. Virol. J. 2009;6:74. doi: 10.1186/1743-422X-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajik M, et al. A novel peptide inhibits the influenza virus replication by preventing the viral attachment to the host cells. Int. J. Biol. Sci. 2009;5:543–548. doi: 10.7150/ijbs.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao H, et al. A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020;11:4252. doi: 10.1038/s41467-020-17986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Vries E, Stelma FF, Boucher CA. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N. Engl. J. Med. 2010;363:1381–1382. doi: 10.1056/NEJMc1003749. [DOI] [PubMed] [Google Scholar]
- 66.Baz M, et al. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J. Infect. Dis. 2010;201:740–745. doi: 10.1086/650464. [DOI] [PubMed] [Google Scholar]
- 67.Lu L, Su S, Yang H, Jiang S. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184:1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 68.Swayne DE. Avian influenza vaccines and therapies for poultry. Comp. Immunol. Microbiol. Infect. Dis. 2009;32:351–363. doi: 10.1016/j.cimid.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 69.Li C, Bu Z, Chen H. Avian influenza vaccines against H5N1 ‘bird flu’. Trends Biotechnol. 2014;32:147–156. doi: 10.1016/j.tibtech.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Zeng X-y, et al. Protective efficacy of an H5/H7 trivalent inactivated vaccine produced from Re-11, Re-12, and H7-Re2 strains against challenge with different H5 and H7 viruses in chickens. J. Integr. Agric. 2020;19:2294–2300. doi: 10.1016/S2095-3119(20)63301-9. [DOI] [Google Scholar]
- 71.Cui P, et al. Global dissemination of H5N1 influenza viruses bearing the clade 2.3.4.4b HA gene and biologic analysis of the ones detected in China. Emerg. Microbes Infect. 2022;11:1693–1704. doi: 10.1080/22221751.2022.2088407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017;23:222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 74.Wei CJ, et al. Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discov. 2020;19:239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nachbagauer R, et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021;27:106–114. doi: 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- 76.Subbiah J, et al. A chimeric thermostable M2e and H3 stalk-based universal influenza A virus vaccine. NPJ Vaccines. 2022;7:68. doi: 10.1038/s41541-022-00498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen YQ, et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. 2018;173:417–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine. 2011;29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schotsaert M, et al. Long-lasting cross-protection against influenza A by neuraminidase and M2e-based immunization strategies. Sci. Rep. 2016;6:24402. doi: 10.1038/srep24402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Del Campo J, et al. OVX836 a recombinant nucleoprotein vaccine inducing cellular responses and protective efficacy against multiple influenza A subtypes. NPJ Vaccines. 2019;4:4. doi: 10.1038/s41541-019-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leroux-Roels I, et al. Randomized, double-blind, reference-controlled, phase 2a study evaluating the immunogenicity and safety of OVX836, a nucleoprotein-based influenza vaccine. Front. Immunol. 2022;13:852904. doi: 10.3389/fimmu.2022.852904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyoglu-Barnum S, et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature. 2021;592:623–628. doi: 10.1038/s41586-021-03365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hassan AO, et al. Adenovirus vector-based multi-epitope vaccine provides partial protection against H5, H7, and H9 avian influenza viruses. PLoS ONE. 2017;12:e0186244. doi: 10.1371/journal.pone.0186244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Freyn AW, et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van de Ven K, et al. A universal influenza mRNA vaccine candidate boosts T cell responses and reduces zoonotic influenza virus disease in ferrets. Sci. Adv. 2022;8:eadc9937. doi: 10.1126/sciadv.adc9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arevalo CP, et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science. 2022;378:899–904. doi: 10.1126/science.abm0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tregoning JS, Russell RF, Kinnear E. Adjuvanted influenza vaccines. Hum. Vaccin. Immunother. 2018;14:550–564. doi: 10.1080/21645515.2017.1415684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J, Xie X, Jiang S, Lu L. Immunoengineered adjuvants for universal vaccines against respiratory viruses. Fundam. Res. 2021;1:189–192. doi: 10.1016/j.fmre.2021.01.010. [DOI] [Google Scholar]
- 89.Del Giudice G, Rappuoli R, Didierlaurent AM. Correlates of adjuvanticity: a review on adjuvants in licensed vaccines. Semin. Immunol. 2018;39:14–21. doi: 10.1016/j.smim.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Shinde V, et al. Comparison of the safety and immunogenicity of a novel Matrix-M-adjuvanted nanoparticle influenza vaccine with a quadrivalent seasonal influenza vaccine in older adults: a phase 3 randomised controlled trial. Lancet Infect. Dis. 2022;22:73–84. doi: 10.1016/S1473-3099(21)00192-4. [DOI] [PubMed] [Google Scholar]
- 91.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kato K, Omura H, Ishitani R, Nureki O. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 2017;86:541–566. doi: 10.1146/annurev-biochem-061516-044813. [DOI] [PubMed] [Google Scholar]
- 93.Wang J, et al. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science. 2020;367:eaau0810. doi: 10.1126/science.aau0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Z, et al. A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res. 2022;32:269–287. doi: 10.1038/s41422-022-00612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang C, et al. Comparative review of respiratory diseases caused by coronaviruses and influenza A viruses during epidemic season. Microbes Infect. 2020;22:236–244. doi: 10.1016/j.micinf.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, et al. Multiple amino acid mutations in viral RNA polymerase may synergistically enhance the transmissibility and/or virulence of the 2009 pandemic influenza (H1N1) virus. Acta Virol. 2013;57:35–40. doi: 10.4149/av_2013_01_35. [DOI] [PubMed] [Google Scholar]
- 98.Shi J, et al. Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe. 2018;24:558–568.e7. doi: 10.1016/j.chom.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee DH, et al. Transmission dynamics of highly pathogenic avian influenza virus A(H5Nx) clade 2.3.4.4, North America, 2014-2015. Emerg. Infect. Dis. 2018;24:1840–1848. doi: 10.3201/eid2410.171891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tian H, et al. Avian influenza H5N1 viral and bird migration networks in Asia. Proc. Natl. Acad. Sci. USA. 2015;112:172–177. doi: 10.1073/pnas.1405216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen W, et al. Environmental factors and spatiotemporal distribution characteristics of the global outbreaks of the highly pathogenic avian influenza H5N1. Environ. Sci. Pollut. Res. Int. 2022;29:44175–44185. doi: 10.1007/s11356-022-19016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gu W, et al. Novel H5N6 reassortants bearing the clade 2.3.4.4b HA gene of H5N8 virus have been detected in poultry and caused multiple human infections in China. Emerg. Microbes Infect. 2022;11:1174–1185. doi: 10.1080/22221751.2022.2063076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cui P, et al. Genetic and biological characteristics of the globally circulating H5N8 avian influenza viruses and the protective efficacy offered by the poultry vaccine currently used in China. Sci. China Life Sci. 2022;65:795–808. doi: 10.1007/s11427-021-2025-y. [DOI] [PubMed] [Google Scholar]
- 104.Pardi N, et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]