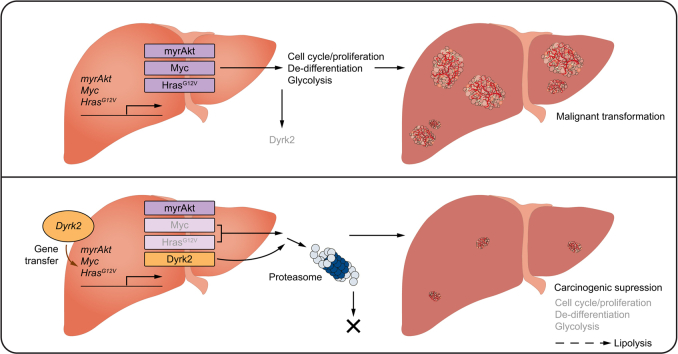
Abstract
Background & Aims
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide, and has a poor prognosis. However, the molecular mechanisms underlying hepatocarcinogenesis and progression remain unknown. In vitro gain- and loss-of-function analyses in cell lines and xenografts revealed that dual-specificity tyrosine-regulated kinase 2 (DYRK2) influences tumour growth in HCC.
Methods
To investigate the role of Dyrk2 during hepatocarcinogenesis, we developed liver-specific Dyrk2 conditional knockout mice and an in vivo gene delivery system with a hydrodynamic tail vein injection and the Sleeping Beauty transposon. The antitumour effects of Dyrk2 gene transfer were investigated in a murine autologous carcinogenesis model.
Results
Dyrk2 expression was reduced in tumours, and that its downregulation was induced before hepatocarcinogenesis. Dyrk2 gene transfer significantly suppressed carcinogenesis. It also suppresses Myc-induced de-differentiation and metabolic reprogramming, which favours proliferative, and malignant potential by altering gene profiles. Dyrk2 overexpression caused Myc and Hras degradation at the protein level rather than at the mRNA level, and this degradation mechanism was regulated by the proteasome. Immunohistochemical analyses revealed a negative correlation between DYRK2 expression and MYC and longer survival in patients with HCC with high-DYRK2 and low-MYC expressions.
Conclusions
Dyrk2 protects the liver from carcinogenesis by promoting Myc and Hras degradation. Our findings would pave the way for a novel therapeutic approach using DYRK2 gene transfer.
Impact and Implications
Hepatocellular carcinoma (HCC) is one of the most common cancers, with a poor prognosis. Hence, identifying molecules that can become promising targets for therapies is essential to improve mortality. No studies have clarified the association between DYRK2 and carcinogenesis, although DYRK2 is involved in tumour growth in various cancer cells. This is the first study to show that Dyrk2 expression decreases during hepatocarcinogenesis and that Dyrk2 gene transfer is an attractive approach with tumour suppressive activity against HCC by suppressing Myc-mediated de-differentiation and metabolic reprogramming that favours proliferative and malignant potential via Myc and Hras degradation.
Keywords: DYRK2, HCC, Liver cancer, Animal model, Carcinogenesis, HTVi, MYC, HRAS, De-differentiation, Metabolic reprogramming, Stemness, Gene transfer
Graphical abstract
Highlights
-
•
Some HCC models of mice decreased the expression of Dyrk2 mRNA and protein.
-
•
Dyrk2 gene transfer suppressed hepatocarcinogenesis in vivo.
-
•
DYRK2 overexpression induced degradation of Myc and Hras.
-
•
DYRK2 and MYC in HCC clinical specimens determine the prognosis for patient survival.
-
•
This study showed that DYRK2 gene transfer may be a future novel therapeutic strategy for HCC.
Introduction
The liver is known as a ‘silent organ’, and some patients with diseases such as hepatocellular carcinoma (HCC), have no subjective symptoms. The main causes of HCC include genomic alterations, toxin ingestion and metabolic stress caused by hepatitis B and C virus, non-alcoholic steatohepatitis (NASH), alcohol abuse, and aflatoxin B1 exposure.1 Surgical resection, radiofrequency ablation, and liver transplantation have been performed as curative therapies for early-stage HCC. However, these treatments are not recommended for advanced-stage HCC with multiple nodules, major vascular invasion, and/or lymph node and distant metastases. The underlying liver of HCC is frequently cirrhotic, which often recurs early after treatment and causes multicentric carcinogenesis, which makes the treatment difficult.2,3 Multi-kinase inhibitors, vascular endothelial growth factor (VEGF) inhibitors, and anti-programmed death-ligand 1 (anti-PD-L1) antibodies have been recently approved for clinical use in unresectable intermediated- and advanced-stage HCC, but their efficacy has been limited.[2], [3], [4] Furthermore, many of these treatments are not indicated for patients with HCC having decompensated cirrhosis and low hepatic functional reserve.2,3 Thus, liver cancer, including HCC, remains the fourth leading cause of cancer-related mortality worldwide in 2020, with a poor prognosis.5 Therefore, molecules that can become promising targets for future therapies should be identified.
Dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2 (DYRK2) is a member of the evolutionarily conserved DYRKs family that autophosphorylates its tyrosine and functions as serine/threonine kinases for its substrate. Accumulating evidence revealed the involvement of DYRK2 in organogenesis6,7 and tumour growth in various cancers, including breast,8,9 lung,10 stomach,11 blood cancers,12 and colon.9 We have previously reported that DYRK2 phosphorylates p53, cellular myelocytomatosis oncogene product (c-Myc, MYC), c-Jun, and SNAIL, leading to growth inhibition, apoptosis induction, and metastasis suppression in human cancer cell lines.8,[13], [14], [15] Our previous gain- and loss-of-function analyses in HCC cell lines in vitro and xenografts revealed that DYRK2 affects tumour growth by regulating cell proliferation and apoptosis, and low DYRK2 expression is associated with the aggressiveness and poor prognosis in patients with HCC.16 However, previous reports, by us, and others, regarding DYRK2 in cancers included experiments with culture in vitro or xenograft models using cell lines or retrospective studies analysing clinical and pathological data. Additionally, changes in DYRK2 expression during precancerous stages or the association between DYRK2 and carcinogenesis and have not been clarified.
This study aimed to evaluate the role of Dyrk2 in hepatocarcinogenesis in a murine autologous carcinogenesis model using newly developed liver-specific Dyrk2 knockout mice and an in vivo gene delivery system.
Materials and methods
Animals
B6.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J (Alb-cre, Dyrk2+/+) were purchased from Charles River Laboratories Japan (Yokohama, Japan). C57BL/Dyrk2tm1c were generated to mate C57BL/Dyrk2tm1a, which have loxP sequences sandwiching Dyrk2 exon 3 and flippase recognition target sites (FRTs) previously described6 and flippase (FLP) transgenic mice.17 Mice were maintained at 20–24 °C on a 12-h light–dark cycle. At autopsy, mice were weighed and euthanised under deep anaesthesia with isoflurane. Then, for use for the following experiments, the liver was removed after recirculation with PBS. All animal experiment protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Jikei University (No. 2018-078) and conformed to the Guidelines for the Proper Conduct of Animal Experiments of the Science Council of Japan (2006).
Human HCC specimens
This was a retrospective study on consecutive patients who were diagnosed with HCC at The Jikei University School of Medicine between 2009 and 2012. This study was conducted in accordance with the Declaration of Helsinki and ethical guidelines issued from administrative departments, and was approved by the Local Ethics Committee of The Jikei University School of Medicine (No. 29-038 [8654]) and carried out by the opt-out consent process. DYRK2 expression data were derived from immunohistochemical analysis of HCC specimens as described.16 Of these 67 data, 64 HCC specimens were used to assess c-Myc expression, excluding three specimens for which no specimens were left for staining. Paraffin-embedded blocks (4 μm) and the Discovery-ultra autostainer (Ventana Medical Systems, Tucson, AZ, USA) were used for all immunohistochemical staining reactions. After deparaffinisation, antigen retrieval was performed at 100 °C for 44 min in an autoclave with citrate buffer (pH 6.0), after which 3% hydrogen peroxidase was used for blocking. The sections were incubated with anti-c-Myc antibody (1:20; Abcam, Cambridge, UK) at 37 °C for 64 min. Slides were mounted in Permount (Vector Laboratories, Newark, NJ, USA), cover-slipped and evaluated by pathologists using a light microscope BX50 (Olympus, Tokyo, Japan). MYChigh-group was defined as more than 5% of HCC cells being positive.
Plasmids
Sleeping Beauty transposase-, myristoylated Akt-, Myc-, and HrasG12V-expressing cassette vectors (pT2-cLuc/SB13-PGK, pT3-EF1a-myr-AKT-HA, pT3-EF1a-MYC, and pT3-EF1a-FLAG-HRASG12V), in which the two loxP sites in the original plasmid were removed, were described previously18,19 Dyrk2, Dyrk2K247R, and human influenza hemagglutinin (HA)-expressing cassette vector (pT3-EF1a-mDYRK2-HA, pT3-EF1a-mDYRK2K247R-HA, and pT3-EF1a-HA) were generated from pT3-EF1a-myr-AKT-HA and pcDNA3-HAC-mDYRK2, pcDNA3-HAC-mDYRK2K247R or HA sequences to use NEBuilder HiFi DNA Assembly (New England BioLabs Japan Inc., Tokyo, Japan). These plasmids were duplicated by ECOS™ Competent E. coli DH5α (Nippon Gene Co. Ltd., Tokyo, Japan) and extracted to use EndoFree Plasmid Maxi Kit (QIAGEN, Hilden, Germany). pFLAG-MYC, pEGFP-Empty, pEGFP-hDYRK2, and pEGFP-hDYRK2KR were generated and used in a previous report.15
The murine hepatocarcinogenesis models
For the model of hepatocarcinogenesis by introducing oncogenes into hepatocytes in vivo, the Sleeping Beauty transposon system and hydrodynamic tail vein injection (HTVi) were performed according to previous reports.18,20,21 Ringer’s solution (Otsuka Pharmaceutical Factory, Tokushima, Japan) was mixed with four basic plasmids and rapidly injected within 8 s via the lateral tail vein of male mice at 8–12 weeks old. To express Dyrk2, Dyrk2KR, or HA, the either of pT3-EF1a-mDYRK2-HA, pT3-EF1a-mDYRK2K247R-HA, or pT3-EF1a-HA was also mixed at the same time. The volume of the solution was 10% body weight (max 2.5 ml), and the molars of pT2-cLuc/SB13-PGK and the other mixed plasmids were 0.74 and 1.12 pmol/ml, respectively. The mice were housed under observation for 2 weeks.
For the NASH-related carcinogenesis model, mice were intraperitoneally injected with diethylnitrosamine (DEN) at 3–5 weeks old; 24 h later, the mice had been fed either a choline-deficient l-amino acid-defined high-fat diet (CDAHFD) (A06071302, Research Diets Inc., New Brunswick, USA) or a choline-deficiency high-fat diet (HFCD) (HFD-60 without choline bitartrate, Oriental Yeast Co., Ltd., Tokyo, Japan) for 24 weeks.19,[22], [23], [24]
Cell cultures and transfections
The human liver cancer cell lines HuH7 and PLC/PRF/5, were obtained from RIKEN and JCRB Cell Bank, respectively. Cells were cultured in DMEM (Nacalai Tesque Inc., Kyoto, Japan) with 10% foetal bovine serum (Biowest, Nuaillé, France) and maintained at 37 °C in a humidified atmosphere of 5% CO2. All transfection assays were performed with Lipofectamine 3000 transfection reagent (Invitrogen, Massachusetts, USA), according to the manufacturer’s instructions. After transfection, cells were treated with 10 μM MG132 (Sigma-Aldrich, St Louis, USA) or DMSO for 4 h to inhibit the effects of proteases (for Western blots).
Quantitative real-time reverse transcriptase-polymerase chain reaction
Liver tissues were stored in RNAlater™ Stabilization Solution (ThermoFisher, entrusted to Cell Innovator Co., Fukuoka, Japan) until analysis. Total RNA in cells or tissue was extracted using Sepasol-RNA I Super G (Nacalai Tesque Inc., Kyoto, Japan) and RNeasy kit (QIAGEN, Hilden, Germany). cDNA was synthesised using PrimeScript 1st strand cDNA Synthesis Kit (Takara Bio Inc., Shiga, Japan). qRT-PCR was performed using the ΔΔCt method with KAPA SYBR FAST Master Mix (Nippon genetics Co., Tokyo, Japan). The mRNA expressions were normalised against Gapdh. The sequences of the specific primers are listed in Table S2.
Western immunoblotting
Cells and tissues were lysed by radioimmunoprecipitation (RIPA) buffer without sodium deoxycholate (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40) with various inhibitors (1 mM Na3VO4, 1 mM AEBSF, 1 mM DTT, 10 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mM NaF, 1 μg/ml pepstatin A) and cOmplete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Protein concentrations in the supernatants were detected by the DC™ protein assay kit (Bio-Rad Laboratories, Hercules, USA). The equal amounts of proteins were separated by SDS-PAGE. Proteins were detected by the following antibodies diluted by CanGetSignal Solution 1 (Takara Bio Inc.); anti-GAPDH (Bethyl Laboratories, Montgomery, TX, USA), anti-DYRK2 (Sigma-Aldrich), anti-phospho-Akt (Ser473) (Cell Signaling Technology, Danvers, MA, USA), anti-Myc, anti-Myc-Ser62 (Abcam), anti-FLAG (Sigma-Aldrich), anti-Hras, anti-cyclin E, anti-cyclin D1, and anti-cyclin D2 (Santa Cruz Biotechnology, Dallas, TX, USA). Membranes were developed by ImmunoStar LD (Fujifilm Wako Pure Chemical Co., Osaka, Tokyo) and imaged using FUSION SOLO 4 M, and analysed using Fusion Capt Advance software (M&S Instruments Inc., Osaka, Tokyo).
Microarray
For microarray analyses, extracted RNA samples with an RNA integrity number >9.0 were used by Cariom™ D Assay, mouse (ThermoFisher, entrusted to Cell Innovator Co., Fukuoka, Japan). To create the heat map, the number of signals for each gene was converted to a logarithm with a base of 2. The results are available in the Gene Expression Omnibus database under accession no. GSE214053.
Statistical analysis
Experimental results were shown as mean and SEM. Group comparisons were performed using the non-paired t test, Welch’s t test, Mann–Whitney U test, Kruskal–Wallis test, Fisher’s exact test and Χ2 test, as appropriate. Cumulative survival rates for each variable were calculated using the Kaplan–Meier method and were compared between groups using the log-rank test. A p-value of <0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism 9.4.0 (GraphPad Software, San Diego, CA, USA).
Other materials and methods can be found in the Supplementary information.
Results
Generation of liver-specific Dyrk2 knockout mice
We have previously reported that systemic Dyrk2 knockout mice were lethal after birth as a result of fatal congenital malformations with lung hypoplasia.7 Thus, the precise functions of Dyrk2 in the liver have not been elucidated as systemic Dyrk2 knockout mice are lethal and exhibit no liver malformations. To explore the functions of Dyrk2 in the liver, we generated the liver-specific Dyrk2 conditional knockout mice (Dyrk2Δhep). Dyrk2 flox mice, which have two loxP sequences sandwiching Dyrk2 exon 3 as previously used,6 were mated with Alb-Cre mice (Dyrk2+/+) (Fig. S1).
Dyrk2Δhep foetuses were born healthy and not lethal. Their survival was comparable to that of Dyrk2+/+, and they did not spontaneously develop liver cancer by 1.5 years of age (Kamioka, Oikawa and Yoshida, unpublished data). Hence, Dyrk2 deficiency has no direct effect on liver organogenesis and cancer development, and additional hits are needed for hepatocarcinogenesis in Dyrk2Δhep.
Dyrk2 expression was reduced in hepatocarcinogenesis models
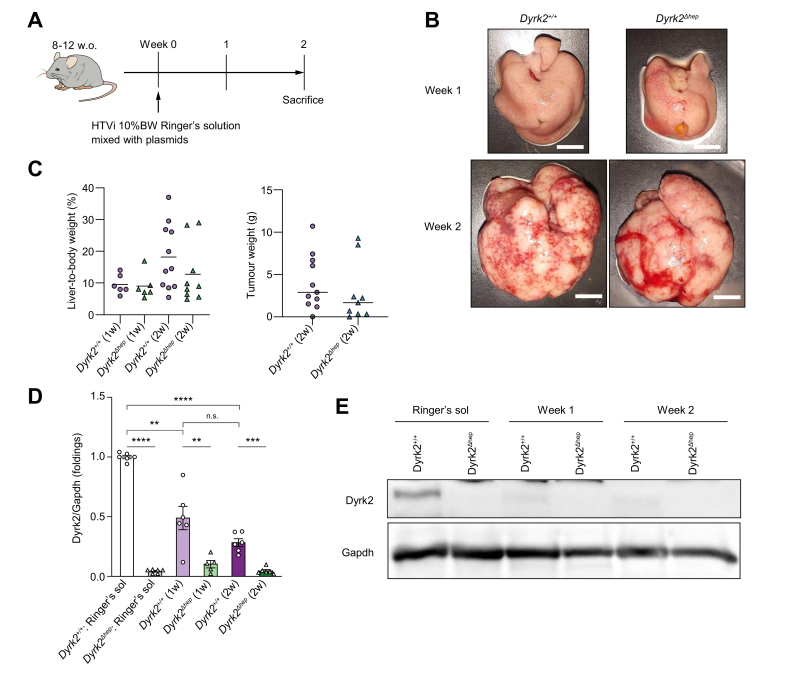
We hypothesised that liver-specific Dyrk2 deficiency may affect hepatocarcinogenesis, considering the previous findings that Dyrk2 acts as a tumour suppressor in several cancers, including liver cancers, and therefore a low DYRK2 expression in patients with HCC is associated with a poor prognosis,16 Hence, a murine model of hepatocarcinogenesis, with a hydrodynamic tail vein injection (HTVi), and the Sleeping Beauty transposon were used, which does not require a special diet and induces stable oncogenes expression in hepatocytes in vivo, leading to HCC development after 2 weeks18,25 (Fig. 1A). Plasmids consisting of the Sleeping Beauty transposase-expressing gene and three oncogenes, myristoylated Akt, Myc, and HrasG12V,18,25 were intravenously injected into Dyrk2+/+ and Dyrk2Δhep. The liver relative to the body weight ratio and tumour weight in Dyrk2Δhep were not significantly different from those of Dyrk2+/+ (18.2 ± 3.1 vs. 12.8 ± 3.1 %, p = 0.242; 4079 ± 976.4 vs. 2831 ± 1182.0 mg, p = 0.484, respectively), although both Dyrk2+/+ and Dyrk2Δhep developed and progressed to HCC 2 weeks after HTVi (Fig. 1B and C). Quantitative real-time PCR (qRT-PCR) after the development of HCC was used to verify Dyrk2 expression. Surprisingly, Dyrk2 mRNA expression was progressively decreased by half at 1 week and one-third at 2 weeks in Dyrk2+/+ tumours introduced with Sleeping Beauty transposase and three oncogenes against those injected with the Ringer’s solution alone as controls, but not as much as in Dyrk2Δhep (p = 0.002; Fig. 1D). Western blotting detected no protein Dyrk2 levels in HCC of both mice (Fig. 1E).
Fig. 1.
Introducing Sleeping Beauty transposase and oncogenes, myristoylated Akt, HrasG12V and Myc into Dyrk2+/+or Dyrk2Δhep rapidly induced liver tumours and reduced Dyrk2 expression.
(A) Schematic representation of the protocol. (B) Gross appearances of livers in Dyrk2+/+ and Dyrk2Δhep at 1 and 2 weeks after the introduction of Sleeping Beauty transposase and oncogenes (scale bar, 1 cm) and (C) the percentages of liver-to-body weight and tumour weights (unpaired t test, no significance). (D) qRT-PCR analysis of Dyrk2 mRNA expression in Akt/Myc/Hras-induced tumours of Dyrk2+/+ and Dyrk2Δhep at 1 and 2 weeks compared with that of Dyrk2+/+ injected with Ringer’s solution without plasmids as control (Welch’s t test, n.s.; no significance, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001). Data are expressed as mean ± SEM. (E) Western immunoblotting of Dyrk2. Gapdh was used as an internal control. Dyrk2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; Hras, harvey rat sarcoma viral oncogene homologue; HTVi, hydrodynamics tail vein injection; Myc, myelocytomatosis oncogene.
Next, we used another murine model induced by DEN and CDAHFD (lacks choline, and is supplemented with 0.1 weight by weight % methionine). Dyrk2Δhep was intraperitoneally injected with DEN at 4 weeks old and then fed with CDAHFD for 24 weeks22,23 (Fig. S2A). Although the mice developed liver cancer, the ratio of liver relative to the body weight, the liver weight, and the tumour weight in Dyrk2Δhep was not significantly different from those of Dyrk2+/+ (9.5 ± 0.5 % vs. 9.1 ± 0.3 %, p = 0.655; 2097.0 ± 102.8 mg vs. 2050.0 ± 81.6 mg, P = 0.869; 692.6 ± 136.1 mg vs. 663.1 ± 93.0 mg, p = 0.943, respectively) (Fig. S2A and C). A significant difference was found in Dyrk2 mRNA expression between Dyrk2+/+ and Dyrk2Δhep 24 weeks after DEN administration and CDAHFD feeding, but Dyrk2 mRNA levels were significantly reduced compared with Dyrk2+/+ fed a normal chow diet (NCD). Dyrk2 protein was not detected, similar to the other above-mentioned murine model (Fig. S2D and E).
These findings led us to explore special feeds could affect Dyrk2 expression. We compared Dyrk2 mRNA and protein expressions in livers of Dyrk2+/+ induced with DEN and fed either an NCD, a CDAHFD, or a HFCD (rich in methionine compared with CDAHFD) (Fig. S2F). Liver cancer only observed in the DEN + CDAHFD group and not in the HFCD or NCD groups (Fig. S2G). DYRK2 mRNA was reduced only in CDAHFD-fed mice and was not detected at the protein level, whereas its expression was not affected in HFCD-fed mice that did not develop liver cancer (Fig. S2H and I). Importantly, Dyrk2 expression was reduced at the early stage of the fatty liver after only 2 weeks of CDAHFD feeding without DEN and was not restored after sequential switching to NCD (Fig. S2J–L). Thus, even a short-term CDAHFD feeding can induce an irreversible decrease in Dyrk2 before hepatocarcinogenesis.
Therefore, Dyrk2 expression was reduced by DEN + CDAHFD and HTVi transduction of the proto-oncogene, but not by DEN + HFCD, in a mouse model of hepatocarcinogenesis.
Dyrk2 suppressed carcinogenesis in the hepatocarcinogenic model
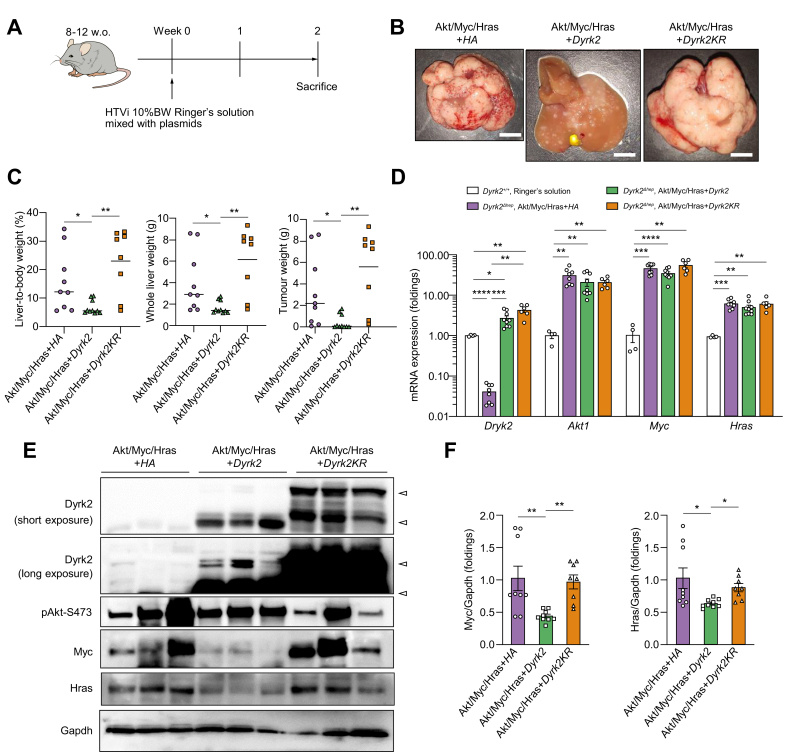
Next, an in vivo Dyrk2 over-expression study was conducted with Dyrk2 gene transfer in the murine hepatocarcinogenesis model with a combination of the Sleeping Beauty transposon system and HTVi. We constructed plasmids expressing Dyrk2-HA, Dyrk2K274R-HA (murine kinase-dead Dyrk2 mutant; Dyrk2KR), and HA-tag with transposase ITRs to transpose, and one of which was introduced into Dyrk2Δhep in addition to Sleeping Beauty transposase and three oncogenes, with HTVi (Fig. 2A). Numerous tumours replaced the whole liver in Dyrk2Δhep introduced with Sleeping Beauty transposase, three oncogenes, and HA-tag (Akt/Myc/Hras+HA). Notably, the introduction of Sleeping Beauty transposase, three oncogenes, and Dyrk2-HA (Akt/Myc/Hras+Dyrk2) by HTVi significantly suppressed tumorigenesis after 2 weeks, but not Sleeping Beauty transposase, three oncogenes, and Dyrk2KR (Akt/Myc/Hras+Dyrk2KR) (Fig. 2B and C). Additionally, this suppressed tumour formation by Dyrk2 gene transfer in vivo was observed when Dyrk2+/+ was used in this model (Fig. S3). Intratumoural mRNA and protein expressions of Dyrk2, Akt, Myc, and Hras were confirmed using qRT-PCR and Western blots (Fig. 2D and E). Increased Dyrk2 expression in Dyrk2Δhep liver tumours introduced with Akt/Myc/Hras+Dyrk2 or Dyrk2KR by HTVi was observed not only at mRNA but also at protein levels (Fig. 2E, arrows indicated long-, and short-variants Dyrk2 protein in the long and short Western blot exposures, respectively). HTVi-mediated proto-oncogene introduction upregulated mRNA expressions of Akt1, Myc, and Hras in this murine hepatocarcinogenesis model. Interestingly, Western blot analyses revealed that Akt/Myc/Hras+Dyrk2 introduction reduced Myc and Hras protein levels, but with no corresponding protein degradation with Dyrk2KR introduction (Fig. 2E and F). Akt phosphorylated at Ser473 was not affected in Dyrk2-overexpressing tumours. This finding was further supported by immunohistochemical analyses (Fig. 2, Fig. 3A).
Fig. 2.
Dyrk2 gene transfer suppressed carcinogenesis in Akt/Myc/Hras-induced tumours of Dyrk2Δhep.
Dyrk2Δhep were introduced with either HA, Dyrk2, or Dyrk2KR in addition to Sleeping Beauty transposase and oncogenes (n = 9, 10, and 8, respectively). (A) Schematic representation of the protocol. (B) Gross appearances of livers in Dyrk2Δhep introduced HA, Dyrk2, and Dyrk2KR (scale bar, 1 cm). (C) The percentages of liver-to-body weight, whole liver weights, and tumour weights. (D) qRT-PCR analysis of relative Dyrk2, Akt1, Myc, and Hras mRNA expressions in Akt/Myc/Hras-induced tumours of Dyrk2Δhep with expressing HA, or Dyrk2, or Dyrk2KR compared with those of Dyrk2+/+ injected with Ringer’s solution without plasmids. (E) Western immunoblotting of proteins in tumours (arrowhead; target protein) and (F) Quantitative analysis of relative protein levels of Myc and Hras in all mice. Data are expressed as mean ± SEM (Kruskal-Wallis test, ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001). Dyrk2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; Hras, harvey rat sarcoma viral oncogene homologue; HTVi, hydrodynamics tail vein injection; Myc, myelocytomatosis oncogene.
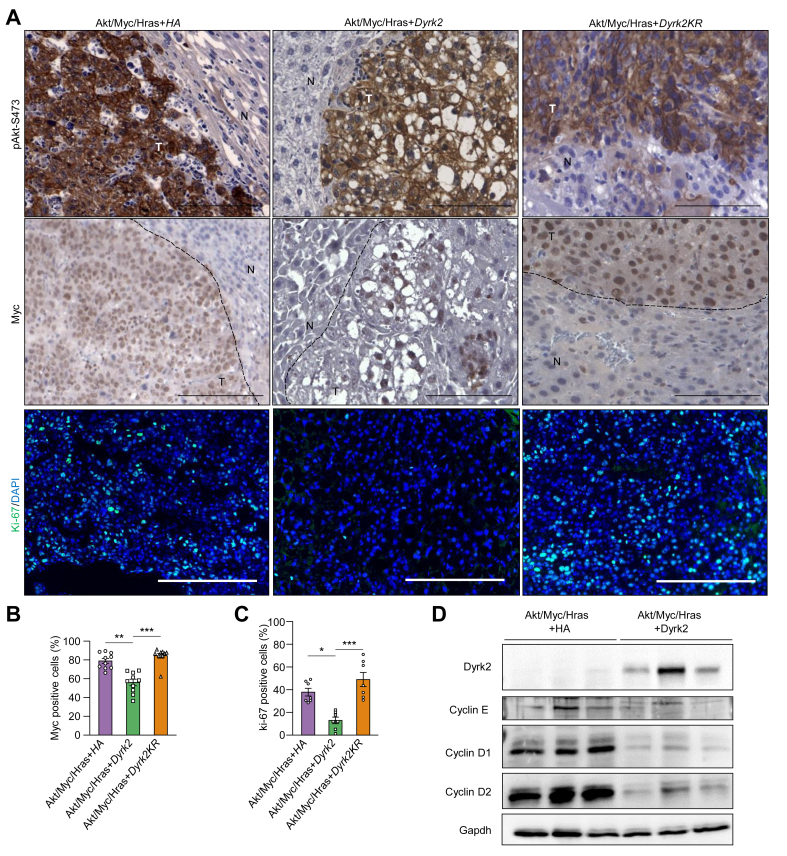
Fig. 3.
Dyrk2-overexpression led to the degradation of protein levels of Myc and Hras.
(A) Immunohistochemistry for phosphorylated Akt at Ser473 (pAkt-S473) and Myc (T, tumour area; N, normal liver area), and immunofluorescence staining for Ki-67 (scale bar, 100 μm). (B) Quantitative analysis of the percentages of Myc-positive cells in the tumour area. (C) Quantitative analysis of the percentages of Ki-67-positive cells in the tumour area. (D) Western immunoblotting of proteins including cyclin subtypes in liver cancers. Data are expressed as mean ± SEM (Kruskal-Wallis test, ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001). Dyrk2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; Hras, harvey rat sarcoma viral oncogene homologue; Myc, myelocytomatosis oncogene.
We have previously reported that DYRK2 forced expression inhibits proliferation and tumour growth of HCC cell lines in vitro and xenografts in vivo via the delayed G1 phase of the cell cycle along with decreased cyclins D1 and D2 expressions.16 Consistent with previous findings, tumour suppression was observed in Akt/Myc/Hras+Dyrk2-induced tumours accompanied by decreased cyclins (D1, D2, and E) and Ki67-positive cells (Fig. 3A, C, and D). These results suggest that Dyrk2 gene transfer leads to tumorigenesis and tumour growth suppression through delayed G1-S transition and Myc and Hras protein degradation in hepatocarcinogenesis.
Dyrk2 reduced malignant potential by inhibiting de-differentiation and metabolic reprogramming
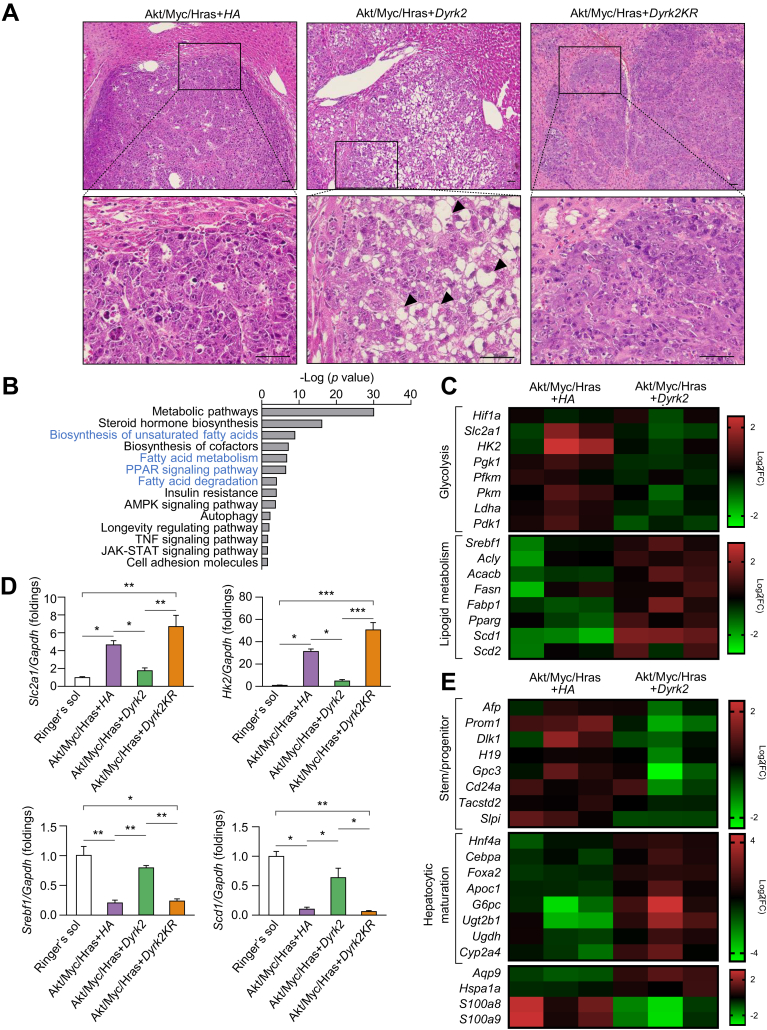
Given that Dyrk2 gene transfer suppressed the tumorigenesis in a murine hepatocarcinogenesis model, we hypothesised that Dyrk2 could affect the histological grade of liver tumours. All tumours showed histological features of moderately to poorly differentiated HCC with severe nuclear atypia, prominent nucleoli, frequent mitoses, neutrophilic and lymphocytic infiltration, and abundant apoptotic tumour cells induced by proto-oncogenes, myrAkt, Myc, and HrasG12V, as previously reported.18 Notably, well to moderately differentiated HCC histologically observed, including mild nuclear atypia, inconspicuous nucleoli cytoplasmic, and intracellular lipid accumulation, in tumours of Dyrk2Δhep introduced with Akt/Myc/Hras+Dyrk2 (Fig. 4A).
Fig. 4.
Dyrk2-overexpression led to the reduction of malignant potential by inhibiting de-differentiation and metabolic reprogramming.
(A) H&E staining in tumours (scale bar, 50 μm). Tumours introduced with Akt/Myc/Hras+Dyrk2 were well to moderately differentiated HCC with lipid accumulation (arrowheads), whilst both expressing Akt/Myc/Hras+HA- and Akt/Myc/Hras+Dyrk2KR-expressing tumours were moderately to poorly differentiated HCC with lack of lipid accumulation. (B) Microarray analysis of Akt/Myc/Hras+HA- and Akt/Myc/Hras+Dyrk2-expressing tumours in Dyrk2Δhep (n = 3 in each group). DAVID analysis of upregulated signalling pathways. The pathways related to lipid metabolism are shown in blue. (C) The heat maps of genes related to glycolysis and lipid metabolism. (D) mRNA expression of Scl2a1, Hk2, Srebf1, and Scd1 in the livers not introduced with plasmids or tumours introduced with Akt/Myc/Hras+HA, Akt/Myc/Hras+Dyrk2, or Akt/Myc/Hras+Dyrk2KR (n = 4, 9, 10, and 8, respectively). (E) The heat maps of genes related to stem/progenitor, hepatocytic maturation. Data are expressed as mean ± SEM (Kruskal-Wallis test, ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001). Dyrk2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; Hras, harvey rat sarcoma viral oncogene homologue.
Next, we performed a microarray analysis in tumours induced with Akt/Myc/Hras+HA or Akt/Myc/Hras+Dyrk2, each from three different mice. Microarray analysis revealed that the genes and the pathways in glycolysis and lipid metabolism were most significantly altered in Akt/Myc/Hras+Dyrk2 overexpressing tumours relative to those induced with Akt/Myc/Hras+HA control (Fig. 4B and C). Akt/Myc/Hras+Dyrk2 tumours have unique molecular features of low expression of glycolysis-related genes, including Slc2a1 and Hk2, and high expression of lipid metabolism-related genes, such as Srebf1 and Scd1 (Fig. 4D), consistent with previous reports of the association between MYC with glycolysis26 and lipid metabolism18,20 via Hif1a. Furthermore, Dyrk2 introduction suppressed the mRNA expression of alpha-foetoprotein (Afp), prominin-1 (Prom1, also known as Cd133), delta-like 1 (Dlk1), h19 imprinted maternally expressed transcript (H19), glypican-3 (Gpc3), Cd24a, tumour associated calcium signal transducer 2 (Tacstd2, also known as Trop2), and secretory leukocyte peptidase inhibitor (Slpi), which are expressed in hepatic stem/progenitor cells and/or foetal liver, as well as liver cancer stem cells, whereas the mRNA expression of hepatocyte nuclear factor 4 alpha (Hnf4a), CCAAT enhancer binding protein alpha (Cebpa), forkhead box a2 (Foxa2), apolipoprotein c1 (Apoc1), glucose-6-phosphatase catalytic (G6pc), uridine 5′-diphosphate glucuronosyltransferase family 2 member b17 (Ugt2b1), uridine 5′-diphosphate-glucose 6-dehydrogenase (Ugdh), and cytochrome p450 family 2 subfamily a member 4 (Cyp2a4), which are previously identified as hepatocytic maturation markers,[27], [28], [29], [30], [31], [32] was increased (Fig. 4E). Notably, aquaporin 9 (Aqp9), which has been linked to tumour suppressive activity by suppressing the stemness of liver cancer stem cells and the deceased expression associated with poorly differentiated HCC and poor prognosis,33 is highly expressed in Dyrk2-overexpressing tumours, but lowly expressed in control tumours (Akt/Myc/Hras-HA). Hsp1a1 (also known as Hsp70) was previously identified as one of the molecules highly expressed in well differentiated HCC,34 and Dyrk2 overexpression increased Hsp1a1 expression. Additionally, mRNA expression of s100 calcium binding protein a9 (S100a9), and s100 calcium binding protein a8 (S100a8), which encode calprotectin, known as a heterodimeric protumorigenic protein, was decreased in Dyrk2-overexpressing tumours. Moreover, Dyrk2 gene transfer inhibited mRNA expression of the G1-S phase in cell cycle-related genes, such as cyclin d1 (Ccnd1), cyclin d2 (Ccnd2), cyclin e1 (Ccne1), cyclin e2 (Ccne2), cyclin dependent kinase 4 (Cdk4), and cyclin dependent kinase 6 (Cdk6) (Fig. S4). These gene signatures support the abovementioned histological findings. Taken together, these findings suggest that Dyrk2 forced expression suppresses carcinogenesis by degrading Myc and Hras, inhibiting de-differentiation with the acquisition of stemness and glycolytic and lipid metabolism reprogramming in favour of cancer development and progression, inducing cell differentiation to less aggressive phenotypes, and reducing proliferative, and malignant potential.
DYKR2 downregulated MYC and HRAS expression in a kinase activity-dependent manner
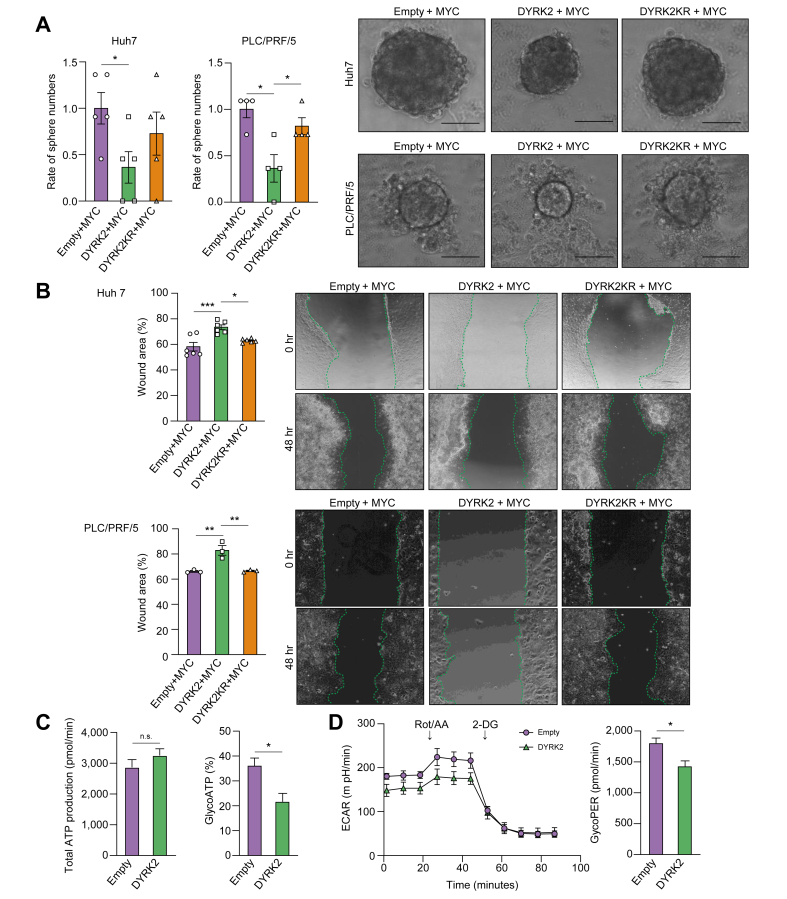
Our in vitro culture and in vivo xenograft studies of human HCC cell lines previously demonstrated the inhibitory effect of DYRK2 on cell growth using gain- and loss-of-function analyses.16 Sphere formation assays, migration assays, and seahorse analysis were performed in HCC cell lines Huh7 and PLC/PRF/5 to confirm our concept obtained in a murine hepatocarcinogenesis model that DYRK2 regulates malignant potential by controlling stemness acquisition and metabolic reprogramming in favour of tumour cell growth and functional analyses by DYRK2 forced expression. HCC cells were co-transfected with pFLAG-MYC and either pEGFP, pEGFP-DYRK2, or pEGFP-DYRK2K239R (human kinase-dead DYRK2 mutant). The number of spheres derived from DYRK2-overexpressing HCC cells was significantly decreased compared to those of controls and DYRK2 mutants lacking the active kinase (Fig. 5A). Furthermore, HCC cells with DYRK2 forced expression revealed significantly reduced cell migration-induced wound recovery (Fig. 5B).
Fig. 5.
DYRK2-overexpression suppressed the malignant potential of human HCC cells.
HCC cells were co-transfected with the combination of FLAG-MYC and either EGFP, EGFP-DYRK2, or EGFP-DYRK2KR. (A) Sphere formation assays. Representative sphere images and the relative number of spheres between the three groups are shown (scale bar: 100 μm). (B) Migration assays. Wound recoveries induced by cell migration were analysed (Kruskal-Wallis test). (C) Quantification of basal total ATP production rate and the percentages of glycolytic ATP production in HCC cells transfected with Empty or DYRK2. (D) Representative extracellular acidification rate (ECAR) glycolytic rate assay profile and quantification of compensatory glycolysis in HCC cells transfected Empty or DYRK2 (Mann-Whitney U test). Data are expressed as mean ± SEM. ns; no significance, ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001. DYRK2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; HCC, hepatocellular carcinoma; MYC, myelocytomatosis oncogene.
Generally, tumour cells require a lot of energy for survival and ATP production by glycolysis is enhanced as an energy source in the Warburg effect.35 Seahorse analysis revealed a reduced rate of ATP production by glycolysis in DYRK2-overexpressing HCC cells under normal culture conditions compared with Empty plasmid-expressing cells (control), although with no significant difference in the basal total ATP production between both cells (Fig. 5C). Additionally, HCC cells with DYRK2 overexpression revealed reduced compensatory glycolysis under anaerobic conditions treated with rotenone/Antimycin a (Fig. 5D). Thus, DYRK2 forced expression in human HCC cells revealed a significant decrease in the glycolytic activity. These results support our findings on gene expression profiles (e.g. stemness and glycolysis) in tumours of a murine hepatocarcinogenesis model by Dyrk2 gene transfer (Fig. 4C–E).
Therefore, our data indicate that DYRK2 directly regulated malignant properties through stemness acquisition and metabolic reprogramming in HCC cells.
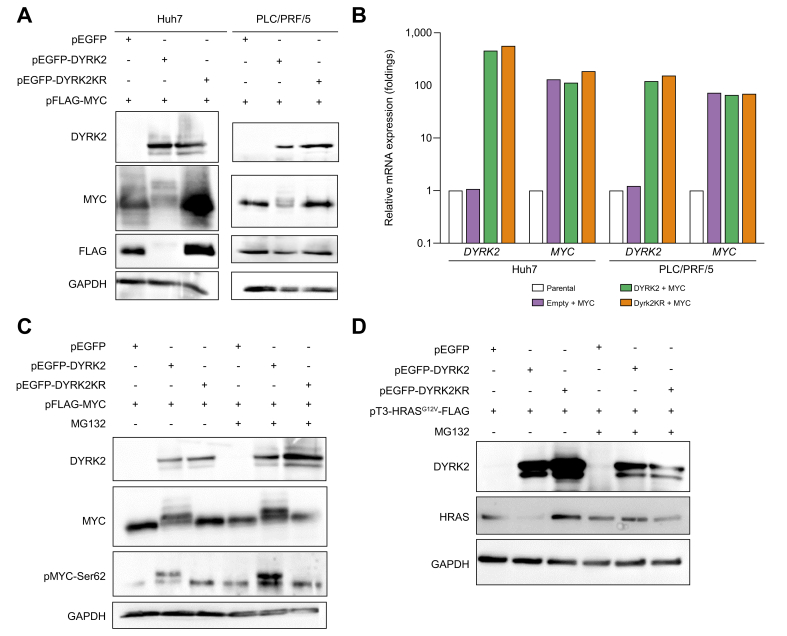
Next, we investigate the mechanism of MYC and HRAS degradation induced by DYRK2 overexpression. MYC and DYRK2 co-transfection resulted in a shift in MYC bands and a kinase-dependent decrease in MYC protein levels in both lines, although MYC mRNA levels were comparable (Fig. 6A and B). MG132 treatment, which is a proteasome inhibitor, restored MYC protein expression, and its phosphorylation at Ser62, especially in the shifted band (Fig. 6C). HRASG12V and DYRK2 co-transfection decreased HRAS expression, similar to MYC expression, and HRAS degradation was suppressed by MG132 (Fig. 6D). In contrast, MG132 treatment did not inhibit HRAS degradation in cells co-transfected with HRASG12V and DYRK2KR. These results suggest that gene transfer of DYRK2 degrades MYC and HRAS proteins in a kinase-dependent manner via the proteasome in human HCC cells.
Fig. 6.
DYRK2 overexpression induced the degradation of MYC and HRAS via a proteasome in human HCC cells.
HCC cells were co-transfected with the combination of FLAG-MYC or FLAG-HRASG12V, and either EGFP, EGFP-DYRK2, or EGFP-DYRK2KR. (A) Western immunoblot of DYRK2, MYC, and HRAS proteins in MYC-expressing HCC cells with overexpression of either EGFP, EGFP-DYRK2, or EGFP-DYRK2KR (kinase-dead mutant). (B) qRT-PCR of relative DYRK2 and MYC mRNA expressions (n = 3 in each group). Data are expressed as mean ± SEM. (C, D) Effects of the proteasome inhibitor, MG132 in DYRK2-overexpressing HCC cells. DMSO was used as a control. GAPDH was used as an internal control. DYRK2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCC, hepatocellular carcinoma; HRAS, harvey rat sarcoma viral oncogene homologue; MYC, myelocytomatosis oncogene.
DYRK2 and MYC expressions in HCC clinical specimens are prognostic of patients’ survival
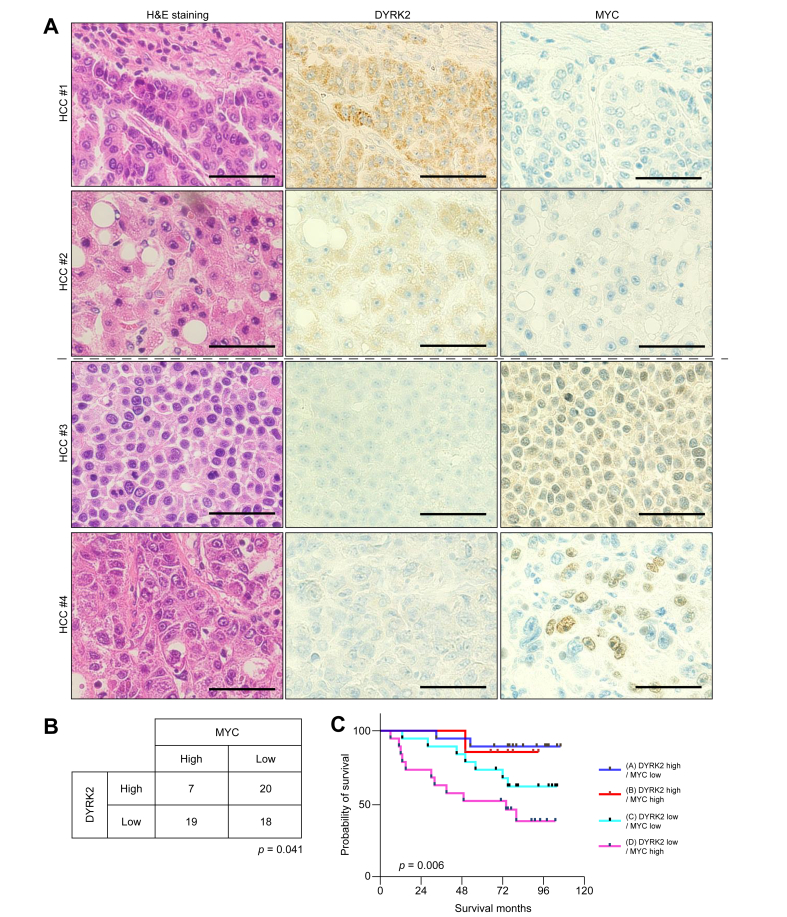
We analysed the association of DYRK2 with MYC expression in 64 surgical specimens of human HCC. Immunohistochemical analyses revealed an inverse correlation between DYRK2 and MYC expression (Fig. 7A and B; p = 0.041). We have previously reported the poor prognosis in patients with HCC with low DYRK2 expression.16 Shorter survival was observed in patients with HCC with high-MYC expression in this cohort (Fig. S5; p = 0.033). We assessed the overall survival of patients with HCC classified into four groups according to DYRK2 and MYC expression levels. Interestingly, patients with HCC with low-DYRK2 and high-MYC expressions (indicated as pink; group-D) were associated with shorter survival, whereas longer survival was correlated with those with high-DYRK2 and low-MYC expressions (blue; group-A) (Fig. 7C; p = 0.006 for all groups, p = 0.002 for group-A vs. group-D). The clinicopathological features of HCC with low-DYRK2 and high-MYC HCC tended to be associated with cirrhosis, older age, and pathologically poorly differentiated carcinoma (Table S1). These results suggest that DYRK2 gene transfer might improve HCC prognosis through proteasome-mediated kinase-dependent MYC and HRAS protein degradation.
Fig. 7.
Relationship between DYRK2 and MYC expression in HCC patients.
(A) H&E staining and immunohistochemistry for DYRK2 (cytoplasmic pattern), and MYC (nuclear pattern) in human HCC specimens (scale bar, 50 μm). (B) Correlation of DYRK2 with MYC expression in HCC patients (Χ2 test). (C) Overall survival of HCC patients categorised into four groups according to expression levels of DYRK2 and MYC. Kaplan–Meier analysis indicated that HCC patients with low-DYRK2 and high-MYC had a poor prognosis (log-rank test). DYRK2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; HCC, hepatocellular carcinoma; MYC, myelocytomatosis oncogene.
Discussion
This study is the first to reveal Dyrk2 gene transfer as an attractive approach with tumour suppressive activity in a murine model of autologous carcinogenesis. Dyrk2 was decreased at both mRNA and protein levels during hepatocarcinogenesis. Dyrk2 gene transfer led to tumorigenesis suppression via proteasome-mediated Myc and Hras degradation in a murine hepatocarcinogenesis model-induced Sleeping Beauty transposon system and HTVi (Fig. 2, Fig. 3, Fig. 4). Furthermore, low-DYRK2 and high-Myc expressions were indicative of aggressiveness and poor prognosis in human liver cancers because DYRK2 negatively correlated with MYC (Fig. 7). Therefore, we propose DYRK2 as a future therapeutic candidate against liver cancer.
We have previously reported reduced DYRK2 expression in HCC using cell lines and human surgical specimens.16 However, the time of decreased DYRK2 expression is decreased in the process of carcinogenesis in any organ is unclear. We hereby report, for the first time, that reduced Dyrk2 expression can be already observed in pre-carcinogenetic liver tissues using murine hepatocarcinogenesis models (both DEN + CDAHFD diet and Sleeping Beauty transposon + HTVi models). This suggests that Dyrk2 expression downregulation is induced before hepatocarcinogenesis and needs to be regulated at an early stage of pre-carcinogenesis to eliminate HCC. Thus, the clear effects of Dyrk2 deficiency on tumour growth were not assessed when Dyrk2Δhep was used. Therefore, we decided to conduct in vivo gain of function studies.
The modification of the Sleeping Beauty transposon system previously developed by HTVi used the ‘gene therapeutic model’ to introduce Dyrk2 into a murine model of autologous hepatocarcinogenesis.18,21,25,36 Cell lines and xenografts were used to analyse the effect of a specific gene on cancer progression, but not on the process of carcinogenesis.36 Our technique has the advantage of continuously expressing a specific gene in vivo, allowing functional analyses not only during cancer development, but also during the carcinogenic process. Furthermore, this system is characterised by simplicity, requiring no toxic or infectious vectors or reagents, and continuously induced gene expression for at least 2 weeks after a single administration (Fig. 2D). However, the effects of in vivo forced expression by the Dyrk2 gene transfer is unclear to persist for ≥2 weeks, and further validation is required. Thus, our developed in vivo gene delivery system is a reliable tool for elucidating functional mechanisms of hepatocarcinogenesis.
DYRK2 is a serine/threonine kinase with its various substrates being reported.37 Our prior investigations with cancer cell lines revealed that DYRK2 phosphorylates MYC at Ser62 as priming phosphorylation of glycogen synthase kinase 3 beta (GSK3β) and subsequent ubiquitination, and Dyrk2 knockdown promotes tumour cell proliferation through cell cycle dysregulation.15 Hence, Myc degradation promoted by direct phosphorylation of Dyrk2 may lead to tumour growth inhibition in animal carcinogenesis models in vivo. The suppressed carcinogenesis and tumour growth in Dyrk2-overexpressing tumours with reduced Myc and cyclin D expression in a murine hepatocarcinogenesis model (Fig. 2, Fig. 3C).
Cyclin D1, which is one of the major regulators of the G1-S phase transition in the cell cycle, is involved in carcinogenesis and cancer progression by regulating cell proliferation.38 Moreover, a strong correlation between tumorigenesis and self-renewal through Ras-Cyclin D2 activation has been demonstrated.39 MYC expression is known to promote carcinogenesis in the liver when mutant HRAS is expressed.40,41 These reports support our findings that Dyrk2 suppresses carcinogenesis and tumour growth either by degrading Myc and Hras proteins or by controlling the G1-S transition through direct cyclin D1 and D2 regulation.
Cancer development and growth, including HCC, requires large amounts of ATP, which is produced by the TCA cycle in normal cells under adequate oxygen and by glycolysis under hypoxic conditions. Conversely, cancer cells produce ATP from glycolysis in the presence or absence of oxygen, which is known as the Warburg effect.35 Hypoxia-inducible factor (HIF1) is a key player in glycolysis regulating glucose transporters, aldolase A, enolase 1, phosphoglycerate kinase 1, pyruvate kinase M, and lactate dehydrogenase.42 MYC and RAS regulate glycolysis expression through transcription activity, such as HIF1.26,43 AKT increases the transport of glucose and its metabolism.44 Thus, changes in ATP-generating metabolism caused by proto-oncogenes allow cancer cells to proliferate. We found that DYRK2 overexpression induced a dramatically reduced glycolytic activity of HCC cells in a seahorse analysis (Fig. 5). Hence, DYRK2 directly regulates cancer development and growth by altering metabolic reprogramming.
Recently reported findings corroborate our own because MYC overexpression in AKT/HRAS-induced HCC has shown highly proliferative activities and altered gene profiles for fatty acid synthesis and degradation and no intracytoplasmic lipid accumulation in the mouse hepatocarcinogenesis model.18 Thus, Nishikawa, and colleagues demonstrated that Myc-induced metabolic reprogramming favours tumour cell growth. Our finding revealed that Dyrk2 overexpression in AKT/HRAS/Myc-mediated HCC suppresses hepatocarcinogenesis by reducing proliferative activity and promoting a less aggressive histological phenotype and intracellular lipid accumulation through Myc and Hras degradation. Indeed, we found that tumours with Dyrk2 forced expression exhibit pathological differentiation (well to moderately differentiated) and gene expression patterns (Fig. 4), similar to the Akt/Hras-induced HCC (without Myc transduction).18 Furthermore, cancer stem cells are presumed factors in tumour initiation, progression, metastasis, and recurrence after conventional therapies, and have been associated with aggressiveness phenotype and poor prognosis. Aggressive phenotypic traits of HCC, such as self-renewal, tumorigenicity, invasiveness, and resistance to chemo- and radiation-therapy are assumed dependent on stemness acquisition.30,45,46 We have previously reported that loss of Dyrk2 augments stem cell-traits by promoting Krüpple-like factor 4 (KLF4) expression in breast cancer.9 Our functional analyses of human HCC cells revealed that DYRK2 forced expression suppressed sphere formation and cell migration (Fig. 5). Furthermore, Dyrk2 gene transfer degrades Myc, inhibits Myc-mediated de-differentiation with stemness acquisition and metabolic reprogramming, and promotes hepatocytic differentiation in a murine hepatocarcinogenesis model, resulting in less aggressive HCC phenotypes (Fig. 4A).
Recent studies have also reported that HRAS is ubiquitinated following GSK3β phosphorylation,47,48 and its activity varies depending on whether it binds GDP or GTP.49 This background makes our findings interpretable that Dyrk2 is likely the indirect regulator of Hras ubiquitination (Fig. 2, Fig. 4E), although HRAS does not have consensus sequences that bind to DYRK2.50 Functional analysis using cell lines has revealed that this degradation is attributable to proteasome-mediated regulation, considering the degradation mechanisms of MYC and HRAS in HCC. Therefore, our findings suggest that DYRK2 protects the liver from carcinogenesis by promoting Myc and mutant Hras degradation.
Our study revealed the irreversible drastic reduction in Dyrk2 expression in mice fed with CDAHFD for a short period and then switched to NCD, indicating that its downregulation is induced in the early stages of hepatocarcinogenesis and liver injury. However, the mechanistic details and upstream of Dyrk2 remain unknown, and further studies are necessary to explore the cellular/molecular mechanisms for DYRK2 expression during hepatocarcinogenesis and hepatocellular injury.
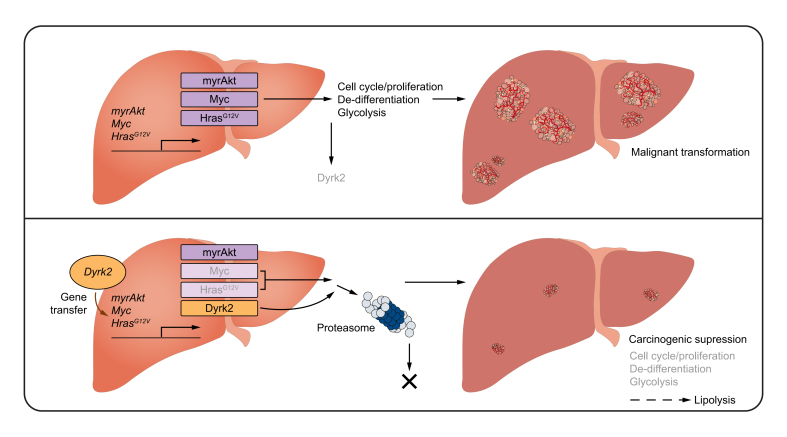
In summary, the loss of Dyrk2 plays an important role in hepatocarcinogenesis and tumour growth and is associated with a poor prognosis. In contrast, Dyrk2 gene transfer provided for proliferative and malignant potential inhibition, cell differentiation promotion with altered gene profiles related to stemness, hepatocytic maturation, glycolytic and lipid metabolism, and subsequently, carcinogenesis suppression through Myc and Hras expression regulation (Fig. 8).
Fig. 8.
Schematic diagram of the tumour suppressive role of Dyrk2 during hepatocarcinogenesis.
Dyrk2, dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2; Hras, harvey rat sarcoma viral oncogene homologue; Myc, myelocytomatosis oncogene.
Therefore, DYRK2 gene transfer may improve the prognosis and be a novel future therapeutic strategy against HCC.
Financial support
This work was supported by JSPS KAKENHI Grant Numbers JP19K08429 to TO, JP17H03584 and JP20H03519 to KY, by The Jikei University Research Fund for Graduate Students to HK and by Bristol-Myers Squibb Foundation Grant to TO.
Conflicts of interest
The authors have declared that no competing interests exist.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
Project conception and design: HK, SY, TO, KY. Designing the protocols: HK, SY. Advising on the research: SY. Conducting the research: HK. Instructing HK on generating mice: SY. Staining and pathological analysis of human HCC: DA, MS. Provision of materials: SY, TO. Contributed materials including plasmids for murine hepatocarcinogenesis models and pathological analysis of mouse tumours: YN. Analysing data: HK Analysis of microarray data: TO. Acquisition and analysis of patient data: TO, KU, CS, KH, TI. Interpretation of results: HK, SY, TO, KY, MS. Drafting the manuscript: HK. Drafting and editing of the manuscript: TO. Supervised the study: MS, KY. All of the authors have read and approved the final manuscript.
Data availability statement
Data presented in this manuscript are available through the corresponding author upon reasonable request.
Acknowledgements
We thank staff members at the Laboratory Animal Facility of the Jikei University School of Medicine for animal care and Dr Hiroshi Manya (Molecular Glycobiology, Research Team for Mechanism of Aging, Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology, Tokyo, Japan) and Dr Shushi Nagamori (Department of Laboratory Medicine, The Jikei University School of Medicine, Tokyo, Japan) for technical assistance in the Seahorse XFe24 Analyzer.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100759.
Contributor Information
Tsunekazu Oikawa, Email: oitsune@jikei.ac.jp.
Kiyotsugu Yoshida, Email: kyoshida@jikei.ac.jp.
Supplementary data
The following are the supplementary data to this article:
References
- 1.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Heimbach J.K., Kulik L.M., Finn R.S., Sirlin C.B., Abecassis M.M., Roberts L.R., et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 3.European Association for the Study of the Liver EASL clinical Practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Finn R.S., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y., et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 5.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida S., Aoki K., Fujiwara K., Nakakura T., Kawamura A., Yamada K., et al. The novel ciliogenesis regulator DYRK2 governs Hedgehog signaling during mouse embryogenesis. Elife. 2020;9 doi: 10.7554/eLife.57381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yogosawa S., Ohkido M., Horii T., Okazaki Y., Nakayama J., Yoshida S., et al. Mice lacking DYRK2 exhibit congenital malformations with lung hypoplasia and altered Foxf1 expression gradient. Commun Biol. 2021;4:1204. doi: 10.1038/s42003-021-02734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mimoto R., Taira N., Takahashi H., Yamaguchi T., Okabe M., Uchida K., et al. DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail. Cancer Lett. 2013;339:214–225. doi: 10.1016/j.canlet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Ito D., Yogosawa S., Mimoto R., Hirooka S., Horiuchi T., Eto K., et al. Dual-specificity tyrosine-regulated kinase 2 is a suppressor and potential prognostic marker for liver metastasis of colorectal cancer. Cancer Sci. 2017;108:1565–1573. doi: 10.1111/cas.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita S., Chujo M., Tokuishi K., Anami K., Miyawaki M., Yamamoto S., et al. Expression of dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2 (DYRK2) can be a favorable prognostic marker in pulmonary adenocarcinoma. J Thorac Cardiovasc Surg. 2009;138:1303–1308. doi: 10.1016/j.jtcvs.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., Xiao R., Lu B., Wu H., Jiang C., Li P., et al. Kinase DYRK2 acts as a regulator of autophagy and an indicator of favorable prognosis in gastric carcinoma. Colloids Surf B Biointerfaces. 2022;209 doi: 10.1016/j.colsurfb.2021.112182. [DOI] [PubMed] [Google Scholar]
- 12.Park C.S., Lewis A.H., Chen T.J., Bridges C.S., Shen Y., Suppipat K., et al. A KLF4-DYRK2-mediated pathway regulating self-renewal in CML stem cells. Blood. 2019;134:1960–1972. doi: 10.1182/blood.2018875922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimoto R., Nihira N.T., Hirooka S., Takeyama H., Yoshida K. Diminished DYRK2 sensitizes hormone receptor-positive breast cancer to everolimus by the escape from degrading mTOR. Cancer Lett. 2017;384:27–38. doi: 10.1016/j.canlet.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Taira N., Nihira K., Yamaguchi T., Miki Y., Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol. 2007;25:725–738. doi: 10.1016/j.molcel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Taira N., Mimoto R., Kurata M., Yamaguchi T., Kitagawa M., Miki Y., et al. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J Clin Invest. 2012;122:859–872. doi: 10.1172/JCI60818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama-Mashima S., Yogosawa S., Kanegae Y., Hirooka S., Yoshida S., Horiuchi T., et al. Forced expression of DYRK2 exerts anti-tumor effects via apoptotic induction in liver cancer. Cancer Lett. 2019;451:100–109. doi: 10.1016/j.canlet.2019.02.046. [DOI] [PubMed] [Google Scholar]
- 17.Kanki H., Suzuki H., Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- 18.Xin B., Yamamoto M., Fujii K., Ooshio T., Chen X., Okada Y., et al. Critical role of Myc activation in mouse hepatocarcinogenesis induced by the activation of AKT and RAS pathways. Oncogene. 2017;36:5087–5097. doi: 10.1038/onc.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Xin B., Yamamoto M., Goto M., Ooshio T., Kamikokura Y., et al. Generation of combined hepatocellular-cholangiocarcinoma through transdifferentiation and dedifferentiation in p53-knockout mice. Cancer Sci. 2021;112:3111–3124. doi: 10.1111/cas.14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe K., Yamamoto M., Xin B., Ooshio T., Goto M., Fujii K., et al. Emergence of the dedifferentiated phenotype in hepatocyte-derived tumors in mice: roles of oncogene-induced epigenetic alterations. Hepatol Commun. 2019;3:697–715. doi: 10.1002/hep4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell J.B., Podetz-Pedersen K.M., Aronovich E.L., Belur L.R., McIvor R.S., Hackett P.B. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushida M., Kamendulis L.M., Peat T.J., Klaunig J.E. Dose-related induction of hepatic preneoplastic lesions by diethylnitrosamine in C57BL/6 mice. Toxicol Pathol. 2011;39:776–786. doi: 10.1177/0192623311409596. [DOI] [PubMed] [Google Scholar]
- 23.Kishida N., Matsuda S., Itano O., Shinoda M., Kitago M., Yagi H., et al. Development of a novel mouse model of hepatocellular carcinoma with nonalcoholic steatohepatitis using a high-fat, choline-deficient diet and intraperitoneal injection of diethylnitrosamine. BMC Gastroenterol. 2016;16:61. doi: 10.1186/s12876-016-0477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raubenheimer P.J., Nyirenda M.J., Walker B.R. A choline-deficient diet exacerbates fatty liver but attenuates insulin resistance and glucose intolerance in mice fed a high-fat diet. Diabetes. 2006;55:2015–2020. doi: 10.2337/db06-0097. [DOI] [PubMed] [Google Scholar]
- 25.Suda T., Gao X., Stolz D.B., Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129–137. doi: 10.1038/sj.gt.3302865. [DOI] [PubMed] [Google Scholar]
- 26.Dang C.V., Kim J.W., Gao P., Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 27.Oikawa T., Kamiya A., Kakinuma S., Zeniya M., Nishinakamura R., Tajiri H., et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136:1000–1011. doi: 10.1053/j.gastro.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita T., Ji J., Budhu A., Forgues M., Yang W., Wang H.Y., et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner R., Lozoya O., Wang Y., Cardinale V., Gaudio E., Alpini G., et al. Human hepatic stem cell and maturational liver lineage biology. Hepatology. 2011;53:1035–1045. doi: 10.1002/hep.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikawa T., Kamiya A., Zeniya M., Chikada H., Hyuck A.D., Yamazaki Y., et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyajima A., Tanaka M., Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Aizarani N., Saviano A., Sagar, Mailly L., Durand S., Herman J.S., et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X., Li C., Yu K., Shi S., Chen H., Qian Y., et al. Aquaporin-9, mediated by IGF2, suppresses liver cancer stem cell properties via augmenting ROS/beta-catenin/FOXO3a signaling. Mol Cancer Res. 2020;18:992–1003. doi: 10.1158/1541-7786.MCR-19-1180. [DOI] [PubMed] [Google Scholar]
- 34.Chuma M., Sakamoto M., Yamazaki K., Ohta T., Ohki M., Asaka M., et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198–207. doi: 10.1053/jhep.2003.50022. [DOI] [PubMed] [Google Scholar]
- 35.Racker E., Spector M. Warburg effect revisited: merger of biochemistry and molecular biology. Science. 1981;213:303–307. doi: 10.1126/science.6264596. [DOI] [PubMed] [Google Scholar]
- 36.He C.X., Shi D., Wu W.J., Ding Y.F., Feng D.M., Lu B., et al. Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J Gastroenterol. 2004;10:567–572. doi: 10.3748/wjg.v10.i4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Correa-Saez A., Jimenez-Izquierdo R., Garrido-Rodriguez M., Morrugares R., Munoz E., Calzado M.A. Updating dual-specificity tyrosine-phosphorylation-regulated kinase 2 (DYRK2): molecular basis, functions and role in diseases. Cell Mol Life Sci. 2020;77:4747–4763. doi: 10.1007/s00018-020-03556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musgrove E.A., Caldon C.E., Barraclough J., Stone A., Sutherland R.L. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 39.Lee J., Kanatsu-Shinohara M., Morimoto H., Kazuki Y., Takashima S., Oshimura M., et al. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation. Cell Stem Cell. 2009;5:76–86. doi: 10.1016/j.stem.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 40.Shachaf C.M., Kopelman A.M., Arvanitis C., Karlsson A., Beer S., Mandl S., et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 41.Stein T.J., Bowden M., Sandgren E.P. Minimal cooperation between mutant Hras and c-myc or TGFalpha in the regulation of mouse hepatocyte growth or transformation in vivo. Liver Int. 2011;31:1298–1305. doi: 10.1111/j.1478-3231.2011.02596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldman R.D., Kaplan N.O., Hall T.C. Lactic dehydrogenase in human neoplastic tissues. Cancer Res. 1964;24:389–399. [PubMed] [Google Scholar]
- 43.Dang C.V., Semenza G.L. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 44.Elstrom R.L., Bauer D.E., Buzzai M., Karnauskas R., Harris M.H., Plas D.R., et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 45.Oikawa T., Wauthier E., Dinh T.A., Selitsky S.R., Reyna-Neyra A., Carpino G., et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat Commun. 2015;6:8070. doi: 10.1038/ncomms9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oikawa T. Cancer stem cells and their cellular origins in primary liver and biliary tract cancers. Hepatology. 2016;64:645–651. doi: 10.1002/hep.28485. [DOI] [PubMed] [Google Scholar]
- 47.Jeong W.J., Yoon J., Park J.C., Lee S.H., Lee S.H., Kaduwal S., et al. Ras stabilization through aberrant activation of Wnt/beta-catenin signaling promotes intestinal tumorigenesis. Sci Signal. 2012;5:ra30. doi: 10.1126/scisignal.2002242. [DOI] [PubMed] [Google Scholar]
- 48.Lee S.K., Jeong W.J., Cho Y.H., Cha P.H., Yoon J.S., Ro E.J., et al. beta-Catenin-RAS interaction serves as a molecular switch for RAS degradation via GSK3beta. EMBO Rep. 2018;19 doi: 10.15252/embr.201846060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahearn I.M., Haigis K., Bar-Sagi D., Philips M.R. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cel Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soundararajan M., Roos A.K., Savitsky P., Filippakopoulos P., Kettenbach A.N., Olsen J.V., et al. Structures of Down syndrome kinases, DYRKs, reveal mechanisms of kinase activation and substrate recognition. Structure. 2013;21:986–996. doi: 10.1016/j.str.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this manuscript are available through the corresponding author upon reasonable request.