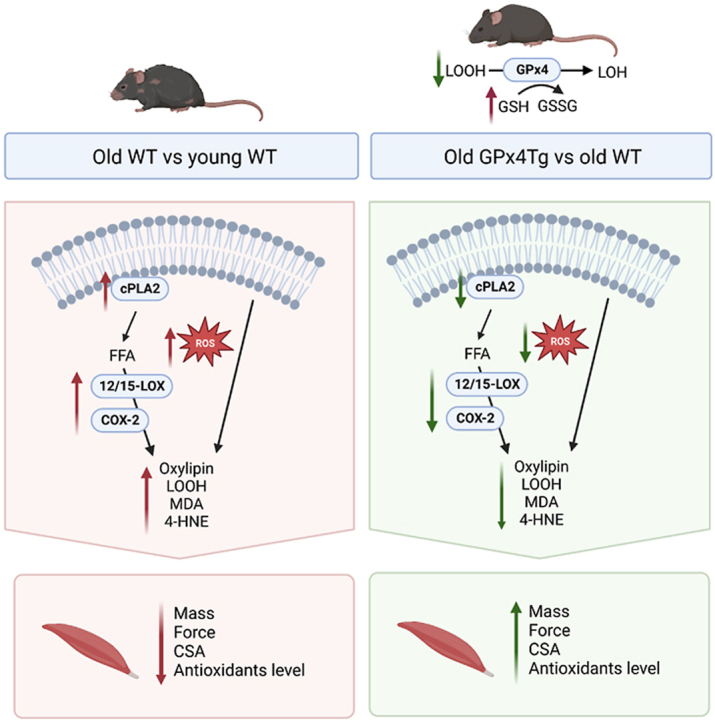
Abstract
Our previous studies support a key role for mitochondrial lipid hydroperoxides as important contributors to denervation-related muscle atrophy, including muscle atrophy associated with aging. Phospholipid hydroperoxide glutathione peroxidase 4 (GPX4) is an essential antioxidant enzyme that directly reduces phospholipid hydroperoxides and we previously reported that denervation-induced muscle atrophy is blunted in a mouse model of GPX4 overexpression. Therefore, the goal of the present study was to determine whether GPX4 overexpression can reduce the age-related increase in mitochondrial hydroperoxides in skeletal muscle and ameliorate age-related muscle atrophy and weakness (sarcopenia). Male C57Bl6 WT and GPX4 transgenic (GPX4Tg) mice were studied at 3 to 5 and 23–29 months of age. Basal mitochondrial peroxide generation was reduced by 34% in muscle fibers from aged GPX4Tg compared to old WT mice. GPX4 overexpression also reduced levels of lipid peroxidation products: 4-HNE, MDA, and LOOHs by 38%, 32%, and 84% respectively in aged GPX4Tg mice compared to aged WT mice. Muscle mass was preserved in old GPX4 Tg mice by 11% and specific force generation was 21% higher in old GPX4Tg versus age matched male WT mice. Oxylipins from lipoxygenases (LOX) and cyclooxygenase (COX), as well as less abundant non-enzymatically generated isomers, were significantly reduced by GPX4 overexpression. The expression of cPLA2, 12/15-LOX and COX-2 were 1.9-, 10.5- and 3.4-fold greater in old versus young WT muscle respectively, and 12/15-LOX and COX-2 levels were reduced by 37% and 35%, respectively in muscle from old GPX4Tg mice. Our study suggests that lipid peroxidation products may play an important role in the development of sarcopenia, and their detoxification might be an effective intervention in preventing muscle atrophy.
Graphical abstract
1. Introduction
Sarcopenia is characterized by a progressive degeneration in skeletal muscle, including loss of both mass and strength. Age-related loss of muscle function leads to falls, weakness and consequently secondary illnesses and increased mortality, which makes sarcopenia a major factor contributing to deterioration in the quality of life in the elderly [[1], [2], [3], [4]]. Effective prevention of sarcopenia still remains a challenge due to the limited understanding of the biochemical and molecular mechanisms underlying age-related muscle atrophy [1,5].
One of the hallmarks of aging cells, including skeletal muscle fibers, is an increase in oxidative stress and damage [[6], [7], [8], [9]]. Our previous studies have shown that increased mitochondrial production of hydroperoxides is associated with muscle atrophy in a variety of conditions related to neurogenic atrophy [8,10,11]. Hydroperoxides produced in response to denervation are predominately LOOHs rather than H2O2 generated by the electron transport chain, and are associated with activation of phospholipase A2 (cPLA2) [[12], [13], [14]]. Peroxide production is reduced in the presence of the cPLA2 inhibitor (arachidonyl trifluoromethyl ketone; AACOCF3) associated with a reduction in the amount of muscle loss due to denervation [12,13]. cPLA2 releases mainly arachidonic acid (AA), but also releases eicosapentaenoic (EPA), and linoleic acid (LA) from membrane phospholipids, if they are present at substantial levels. Free fatty acids can be further metabolized to generate many lipid oxidation products [15]. An excess and accumulation of these products in a cell disturb its normal function and can lead to a number of deleterious outcomes, including a form of cell death called ferroptosis [16].
Lipid hydroperoxides (LOOHs) are the primary products of peroxidation of polyunsaturated fatty acids (PUFA), cholesterol, and to a smaller extent, monounsaturated fatty acids (MUFA) [[17], [18], [19]]. Another important product of this process is oxylipins. These are signaling molecules generated from PUFA via activity of lipoxygenases (5-LOX, 12-LOX, and 12/15-LOXcyclooxygenases (COX-1 and COX-2), and cytochrome P450 (CYP) enzymes, as well as non-enzymatic pathways [20,21]. Oxylipins are elevated in various pathological conditions in a variety of cells and tissues [[21], [22], [23], [24]]. Chain propagation caused by uncontrolled lipid peroxidation also leads to formation of secondary products that might have negative biological effects. For example, PUFA fragmentation by-products such as malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) cause cytotoxicity including mitochondrial damage characterized by reduced respiration, ROS generation, reduced enzyme activity, and protein modification [[25], [26], [27]].
Phospholipid hydroperoxide glutathione peroxidase 4 (GPX4) is an intracellular antioxidant enzyme, that can directly reduce phospholipid, fatty acid, and cholesterol hydroperoxides [19,28]. We previously reported that GPX4 overexpression reduces denervation-induced muscle loss [12]. We hypothesize that lipid oxidation products such as LOOHs and oxylipins are key factors in the loss of muscle mass and strength in aging mice. Thus, the goal of the present study is to investigate whether the neutralization of oxidized lipids by GPX4 can inhibit sarcopenia in older mice. Here we used young and old transgenic mice overexpressing GPX4 to study the effects of detoxifying oxidized lipids in aging muscle as well as major pathways of lipid metabolism.
2. Materials and methods
2.1. Mice
GPX4 transgenic mice overexpressing GPX4 (GPX4Tg) were previously generated using a genomic clone containing the human GPX4 gene, and are described in detail by Ran et al. [29]. Male mice were kept on a 12:12 h light-dark cycle, and had access to standard rodent chow and water. 3–5 and 23–29 month old wild type (WT) and GPX4Tg male mice were used for all experiments. All procedures were approved by the Institutional Animal Care and Use Committees at Oklahoma Medical Research Foundation.
2.2. Tibialis anterior cross-sectional area
We sectioned Tibialis Anterior muscle after sacrifice, mounted in optimum cutting temperature compound (OCT), and flash frozen in liquid nitrogen-cooled isopentane. We cut sections (10 μm), mounted, and stained the sections with laminin. Briefly, we blocked sections with 10% normal goat serum and 2% bovine serum albumin in Phosphate Buffered Saline (PBS) for 1 h. We then stained sections overnight with laminin antibody (Sigma L9393) diluted in PBS with 1% BSA. The next day we washed the sections in PBS 3 times for 5 min followed by incubation with a FITC conjugated secondary antibody. We washed sections in PBS 3 times for 5 min. We visualized and captured images with Zeiss Axiovert 200 M microscope, Zeiss Axiocam MRC camera, and Zeiss AxioVision software V4.8.2.0 (Carl Zeiss AG, Oberkochen, Germany) at 10× magnification. We then measured the cross-sectional area of the muscle fibers using an ImageJ automated analysis as previously described [30].
2.3. Western blot
Proteins were isolated from gastrocnemius of wild-type and GPX4Tg male mice using RIPA buffer (50 mM Tris (pH 7.5), 140 mM NaCl, 1% IGEPAL (v/v; Sigma Aldrich, I8896), 1 mM EDTA (pH 8.0), 0.5 mM EGTA, 50 mM NAF, 1 mM NaO vandate) with 1% (v/v) protease inhibitor (Protease Inhibitor Cocktail Set III, EDTA-Free, Millipore, 539134). After centrifugation and collecting the supernatants, protein concentrations was determined using Bio-Rad protein assay dye reagent (Bio-Rad, 5000006) as stated in the manufacturer's instructions. Samples were prepared using Laemmli sample buffer (Bio-Rad, 1610737) and 20–30 μg of protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Bound proteins were blocked with 1% nonfat dry milk in Tris-buffered saline with 1% Tween 20 (TBS-T), incubated with specific primary antibodies (4HNE: Abcam, ab46545; cPLA2: Cell Signaling, 2832; iPLA2 (PLA2G6): Thermo Fisher, PA5-27945; 12/15-LOX: Santa Cruz, sc-133085; COX-2: Cell Signaling, 4842), washed with TBS-T and incubated with horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology). Protein bands were detected by ECL reagent using G-Box (Syngene), and the band intensity was measured using Image Studio™ Lite Quantification software.
2.4. Fiber permeabilization
Approximately 2–4 mg of red gastrocnemius muscle was dissected, and fibers were mechanically separated along their striations in cold buffer X (pH 7.1; 7.23 mM K2EGTA, 2.77 mM CaK2EGTA, 20 mM imidazole, 0.5 mM DTT, 20 mM taurine, 5.7 mM ATP, 14.3 mM PCr, 6.56 mM MgCl2–6H2O, 50 mM K-MES). The fiber bundle was permeabilized using saponin (30 μg/ml) in buffer Z (pH 7.1; 30 mM KCl, 10 mM KH2PO4, 0.6 mg/ml BSA, 5 mM MgCl2–6H2O, 1 mM EGTA, 105 mM K-MES) with 1 mM EGTA (wash buffer) for 30 min. Next, the fibers were washed 3 times using wash buffer.
2.5. Measurement of respiration and hydroperoxide production
Permeabilized red gastrocnemius muscle fiber bundles were used for simultaneous measure of oxygen consumption rates (OCR) and hydroperoxide generation using the Oroboros Oxygraph-2k machine (O2k, OROBOROS Instruments, Innsbruck, Austria). OCR was determined using an oxygen probe, while the hydroperoxide production rate was determined using O2k-Fluo LED2-Module Fluorescence-Sensor Green. Measurements were performed at 37 °C in buffer Z containing 1 mM EGTA, 22.7 mM creatine monohydrate, 10 μM Amplex® UltraRed (Thermofisher, A36006), 1 U/ml horseradish peroxidase (HRP), and 25 μM blebbistatin. The assay chambers were hyper-oxygenated and the substrate–inhibitor titration protocol was used as follows: 10 mM glutamate and 2 mM malate, 5 mM ADP, 10 mM succinate, 1 μM rotenone, and 1 μM Antimycin A. OCR measurements were normalized to levels obtained with antimycin A addition to account for non-mitochondrial oxygen consumption. Hydroperoxide generation was determined using calibration curve for H2O2. All the results were normalized to wet tissue weights.
2.6. Force generation
Force generation was measured as previously described [31,32]. Briefly, EDL muscle was suspended on a dual‐mode muscle lever system (300C‐LR, Aurora Scientific Inc, Aurora, Canada) and a hook in Krebs buffer (137 mM NaCl, 5 mM KCl, 1 mM NaH2PO4, 1 mM MgSO4, 24 mM NaHCO3, 2 mM CaCl2). Muscles were placed at optimal length and allowed 10 min of thermoequilibration at 32 °C. A supramaximal current (600–800 mA) of 0.25 ms pulse duration was delivered through a stimulator (701 C, Aurora Scientific Inc.), while train duration for isometric contractions was 300 ms. Data were recorded and analyzed using commercial software (DMC and DMA, Aurora Scientific). Specific force (N/cm2) of EDL muscles were multiplied to the ratio of fiber length to muscle length published previously [33].
2.7. Reduced (GSH) and oxidized (GSSG) glutathione levels
GSH and GSSG were separated by Thermoscientific Dianox Ultimate 3000 HPLC-ECD system using a reverse-phase Accucore RP-MS (150*2.1 mm) column obtained from Thermoscientific (Waltham, MA). LC conditions include mobile phase of 25 mM Sodium Phosphate, 0.5 mM 1-octanesulfonic acid and 4% acetonitrile adjusted to pH 7 using 85% phosphoric acid, temperature at 35° and flow rate 0.5 ml/min. Gastrocnemius samples were homogenized in ice-cold antioxidant buffer (50 mM potassium phosphate buffer, pH 7.4, containing 2 mM EDTA and 0.1 mM butylated hydroxytolune in tissue lyzer (Rebrin and Sohal., 2004). The samples were briefly centrifuged at 1000 g for 1min at 4°. An aliquot in duplicate was taken to estimate total protein by BCA method according to manufacturer's instructions (Pierce, Rockford, IL, USA). 10% ice-cold MPA was added to the crude homogenized samples, incubated for 30min on ice. Then, the homogenates were centrifuged for 20min at 14000 g at 4°. The acidified supernatant was then transferred to HPLC autosampler vials for separation of GSH and GSSG. The system was equilibrated well enough to maintain the baseline stability before the samples were injected.
ECD detection: Clean cell potential of 1900 mV for 10s was applied after every run. In order to filter the electrochemically active compounds that could possibly interfere with GSH and GSSG, we used a conditioning coulometric OMNI cell (6020RS, Thermoscientific) before the BDD (Boron doped diamond) electrode. The GSH and GSSG peak were eluted at 1.26min and 2.25min at 500 mV and 1500 mV for OMNI and BDD respectively. The oxidation potential for GSH and GSSG was performed every time the electrode is replaced to avoid the changes due to cell difference. The peaks were quantified using Thermo Scientific Chromeleon CDS Chromatography software.
2.8. Cytosolic phospholipase A2 (cPLA2) activity
cPLA2 activity was assessed using the Cytosolic Phospholipase A2 Assay Kit (Abcam; ab133090) according to the manufacturer's instructions. Proteins were isolated from gastrocnemius muscle using 500 μL homogenization buffer (pH 7.4; 50 mM HEPES, 1 mM EDTA). Samples were centrifuged at 10,000×g for 15 min, then the supernatant was filtered using membrane filter with a molecular weight cut-off of 30 kDa (Amicon; UFC503024) at 14,000×g for 10 min to remove sPLA2. Protein concentration was estimated using Bio-Rad protein assay dye reagent (Bio-Rad, 5000006) as stated in the manufacturer's instructions. Samples were incubated with 5 μM bromoenol lactone for 15 min at room temperature to inhibit iPLA2. 100 μg protein (15 μL) and 200 μL of Arachidonoyl Thio-PC (as a substrate) were added to wells. After 60 min at RT, 10 μL mixture of 25 mM DTNB and 475 mM EGTA in 0.5 M Tris-HCl was added to stop enzyme catalysis. Absorbance was read at 414 nm and converted to enzyme activity using the manufacturer's calculations.
2.9. Malondialdehyde (MDA) level
20 mg of gastrocnemius was homogenized in 250 μL of 7.5% trichloroacetic acid (TCA). After centrifugation and filtration, supernatants were added to equal volume of the mixture containing 10% TCA and 0.5% TBA. Samples were boiled for 30 min in a dry thermoblock and cooled down. The absorbance for TBA–MDA complex was read at 532 nm and corrected for non-specific absorbance at 600 nm. Concentration of MDA was calculated using the molar extinction coefficient of MDA equal to 155 mM−1 cm−1.
2.10. Lipid hydroperoxide level
The concentration of lipid hydroperoxides was assessed using the Lipid Hydroperoxide Assay Kit (Abcam; ab133085) according to the manufacturer's instructions. Lipids were isolated from gastrocnemius muscle using 500 μL of ultra-pure water. 500 μL of methanol solution of extract R and 1 ml of cold chloroform were added to the homogenate. Samples were mixed and centrifuged at 3000×g for 5 min. The bottom chloroform layer was collected and used for the assay. 450 μL of chloroform-methanol solvent (2:1) and 50 μL of chromogen were added to 500 μL of chloroform extract. After 5 min at RT, absorbance was read at 500 nm and the amount of LOOHs was calculated on the basis of a calibration curve.
2.11. Lipid extraction and oxylipin quantitation using LC/MS/MS
Lipids were extracted from gastrocnemius muscle and processed as previously described in Brown et al. [30]. Tissue samples were homogenized with ceramic beads in 1 ml anti-oxidation buffer containing 100 μM diethylenetriaminepentaacetic acid (DTPA) and 100 μM butylated hydroxytoluene (BHT) in phosphate buffered saline using a Bead Ruptor Elite for 1 × 30 s interval at 6 m/s, under cooled nitrogen gas (4 °C). Samples were spiked with 2.1–2.9 ng of 12-HETE-d8, 15-HETE-d8, 13-HODE-d4, standards (Cayman Chemical) and 10 ng of 15:0–18:1(d7) PE (Avanti) prior to homogenization. Lipids were extracted from the lysates by adding a 2.5 ml solvent mixture (1 M acetic acid/isopropanol/hexane; 2:20:30, v/v/v) to 1 ml homogenate in a glass extraction vial and vortexed for 60 sec. 2.5ml hexane was then added to samples and after vortexing for 60 s, tubes were centrifuged (500 g for 5 min at 4 °C) to recover lipids in the upper hexane layer (aqueous phase), which was transferred to a clean tube. Aqueous samples were re-extracted as above by addition of 2.5 ml hexane, and upper layers were combined. Lipid extraction from the lower aqueous layer was then completed according to the Bligh and Dyer technique. Specifically, 3.75 ml of a 2:1 ratio of methanol:chloroform was added followed by vortexing for 60 s. Subsequent additions of 1.25 ml chloroform and 1.25 ml water were followed with a vortexing step for 60 s, and the lower layer was recovered following centrifugation as above and combined with the upper layers from the first stage of extraction. Solvent was dried under vacuum and lipid extract was reconstituted in 100 μl HPLC grade methanol. For oxylipin analysis, lipids were separated by liquid chromatography (LC) using a gradient of 30–100% B over 20 min (A: Water:Mob B 95:5 + 0.1% Acetic Acid, B: Acetonitrile: Methanol – 80:15 + 0.1% Acetic Acid) on an Eclipse Plus C18 Column (Agilent), and analyzed on a Sciex QTRAP® 6500 LC-MS/MS system. Source conditions: TEM 475 °C, IS -4500, GS1 60, GS2 60, CUR 35. Lipids were detected using MRM monitoring with the following parent to daughter ion transitions: 12-HETE [M − H]- 319.2/179.1, 15-HETE [M − H]- 319.2/219.1, 13-HODE [M − H]- 295.2/195.1, 15-HETrE [M − H]- 321.2/221.1, 15-HEPE [M − H]- 317.2/219.1, 14-HDOHE [M − H]- 343.2/205.1, 17-HDOHE [M − H]- 343.2/201.1, 13-HOTrE [M − H]- 293.2/195.1. Deuterated internal standards were monitored using parent to daughter ions transitions of: 12-HETE-d8 [M − H]- 327.2/184.1, 15-HETE-d8 [M − H]- 327.2/226.1 and 13-HODE-d4 [M − H]- 299.2/198.1. Chromatographic peaks were integrated using Multiquant 3.0.2 software (Sciex). The criteria for assigning a peak was signal:noise of at least 5:1 and with at least 7 points across a peak. The ratio of analyte peak areas to internal standard was taken and lipids quantified using a standard curve made up and run at the same time as the samples. Each oxylipin was then standardized per mg of tissue. For oxPL measurements, lipids were separated by liquid chromatography (LC) using a gradient of 50–100% B over 10 min, followed by 30 min at 100% B (A: methanol:acetonitrile:water, 1 mM ammonium acetate, 60:20:20. B: methanol, 1 mM ammonium acetate), with a flow rate of 0.2 ml/min on a Luna C18 column (15 cm × 2 mm, 3 μm) (Phenomenex), and analyzed on a Sciex QTRAP® 6500 LC-MS/MS system. Source conditions: TEM 500 °C, IS -4500, GS1 40, GS2 30, CUR 35. Lipid were detecting using MRM monitoring with the following parent to daughter ion transitions: 18:0a/20:4(O)-PE and 16:0a/20:4(O)-PC [M − H]- 782.6/319.2, 18:0p/20:4(O)-PE [M − H]- 766.6/319.2, 18:1p/20:4(O)-PE [M − H]- 764.6/319.2, 16:0p/20:4(O)-PE [M − H]- 738.6/319.2 and 18:0a/20:4(O)-PC [M − H]- 810.7/319.2.15:0–18:1(d7) PE internal standard was monitored using parent to daughter ion transition of: [M − H]- 709.5/288.2. Chromatographic peaks were integrated using Analyst 1.7 software (Sciex). The criteria for assigning a peak was signal:noise of at least 5:1 and with at least 7 points across a peak. These MRM transitions gave rise to multiple peaks which were integrated as a group. This method was used since MS/MS data showed these peaks to be a mixture of oxidized PLs (not isolated to HETE-PL's, but also other oxidized FA containing PL's), however it was not possible to assign individual IDs to each peak. The ratio of analyte peak areas to internal standard was taken and displayed as relative values.
2.12. Targeted quantitative mass spectrometry
Targeted quantitative mass spectrometry was utilized to measure protein abundance as previously described [30,[34], [35], [36]]. Briefly, gastrocnemius samples were homogenized in a radioimmunoprecipitation assay buffer containing 10 mM Tris‐Cl (pH 8.0), 1 mM EDTA, 1% Triton X‐100 (v/v), 0.1% sodium deoxycholate (w/v), 0.1% sodium dodecyl sulfate (w/v), 140 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride, with protease inhibitor cocktail (Calbiochem Set III, EDTA‐free; EMD Millipore; Billerica, MA, USA), and Bradford assay was used to determine protein concentration. Targeted proteomic analysis with LC-tandem mass spectrometry was done in the Oklahoma Nathan Shock Center Multiplexed Protein Analysis Laboratory as previously described [30]. Briefly, 60 μg protein was taken for analysis, mixed with 1 μg of BSA as an internal standard, and processed to reduce, alkylate and digest with trypsin. The digest was analyzed by selected reaction monitoring (SRM) on a Thermo Scientific TSQ Quantiva using methods that were developed and validated previously for each protein target. Data was analyzed using Skyline [37] to find and integrate the appropriate chromatographic peak.
2.13. Statistical analyses
Prior to statistical analyses, the results were tested for equal variances and normal distribution to establish the appropriate statistical test. All data were presented as mean values ± SEM, and comparisons among different groups were performed with ordinary one-way or two-way ANOVA, with post hoc multiple comparisons test, or t-test (for oxylipins), taking p < 0.05 using GraphPad Prism 9.3.0 (GraphPad Sofware, La Jolla, CA).
3. Results
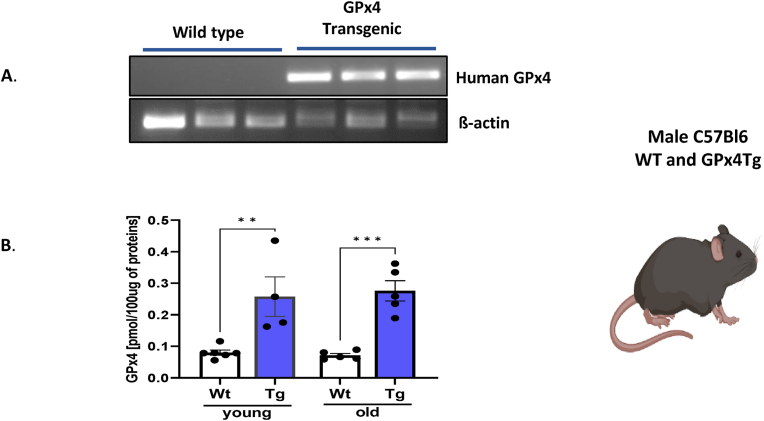
Mouse model. Young (3–5 month) and old (23–29 month old male C57Bl6/J wildtype (WT) and transgenic (Tg) mice were used to study the effect of GPX4 overexpression on mitochondrial function, oxylipin production, metabolism, and muscle mass and function. The presence of the human GPX4 transgene in the GPX4Tg mice was verified using a PCR method (Fig. 1A). Further, mass spectrometry-based protein analysis confirmed elevated expression of this enzyme in muscle from GPX4Tg mice (3.2 -fold, p = 0.0038 in young mice and 3.9 fold change (p = 0.0009) in old transgenic mice compared to aged matched WT mice (Fig. 1B).
Fig. 1.
Model characterization. (A) Representative RT-PCR gel showing the presence of human GPX4 mRNA in gastrocnemius tissue from GPX4Tg mice but not in tissues from wild type mice. (B) Murine and human GPX4 protein content. Measure by mass spectrometry *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 4–5. Data are presented as mean value ± SEM.
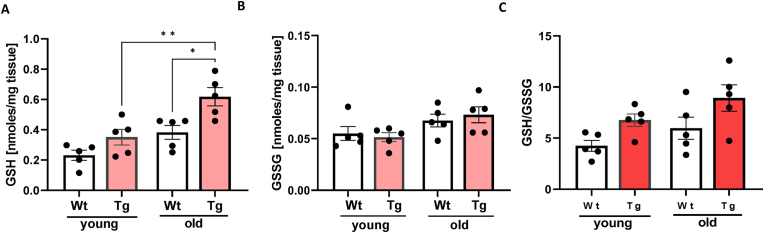
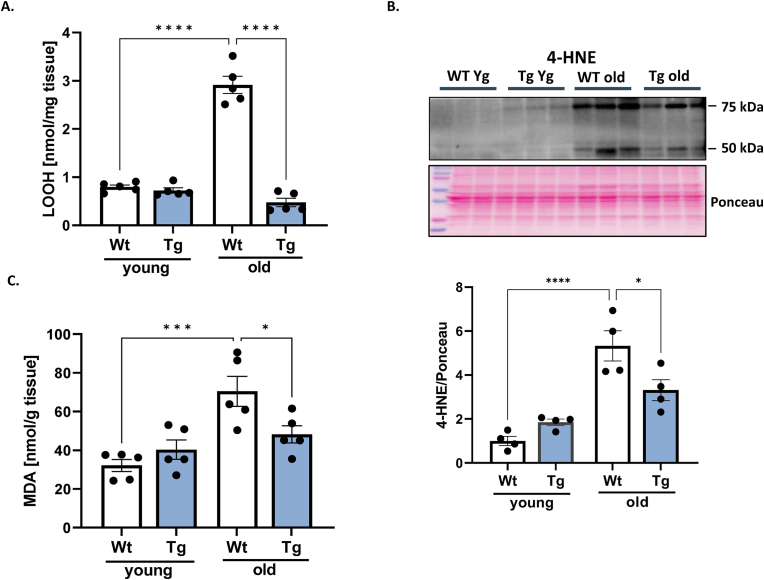
Lipid peroxidation products and redox status. GPX4 is a GSH-dependent enzyme. LOOHs reduction generates not only a lipid hydroxide but also selenic acid, which has to be reduced by two glutathione molecules [38]. Therefore, both the GSH level and the ratio of GSH to its oxidized form GSSG were measured in gastrocnemius muscle. As shown in Fig. 2A, GSH levels were 52% higher in muscle from young GPX4Tg mice and 62% higher in old GPX4Tg mice versus muscle from age matched wildtype (WT) mice. Oxidized glutathione levels (GSSG) were not different among the groups (Fig. 2B). The GSH/GSSG ratio showed changes similar to GSH, although these results did not reach statistical significance (Fig. 2C). Because LOOHs are an important component of hydroperoxides in denervation-related loss of muscle mass [12,13], we measured lipid peroxidation products in muscle from young and old WT and GPX4 Tg mice. LOOH levels in muscle from old WT mice are 3.7-fold higher (p < 0.0001) than LOOH production in muscle from young WT mice. GPX4 overexpression blunted this increase by 84%; the LOOH concentration was 2.915 nM/mg of tissue in old WT mice vs 0.473 nM/mg of tissue in old GPX4Tg mice, p < 0.0001 (Fig. 3A). The concentration of by-products of lipid oxidation also increased with age. For example, the level of 4-HNE and MDA (commonly used as markers of lipid peroxidation) increased 5.3 fold (p < 0,0001) and 2.2 fold (p = 0.0006) in WT animals, respectively. The amounts of these substances were reduced in the muscles from GPX4 mice by 38% (p = 0.0288) for 4-HNE and 32% (p = 0.0430) for MDA (Fig. 3B and C).
Fig. 2.
Skeletal muscle glutathione status. (A) Glutathione (GSH) level, (B) oxidized glutathione (GSSG) level and (C) ratio of GSH to its oxidized form-glutathione disulfide (GSSG) measured in gastrocnemius muscle tissue. *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 5. Data are presented as mean value ± SEM.
Fig. 3.
Lipid peroxidation in gastrocnemius muscle. (A) Lipid hydroperoxide (LOOHs) concentration measured in muscle homogenate. (B) Representative Western blot and the quantified data for 4-hydroxynonenal (4-HNE) level, and (C) malondialdehyde (MDA) concentration. *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 3–5. Data are presented as mean value ± SEM.
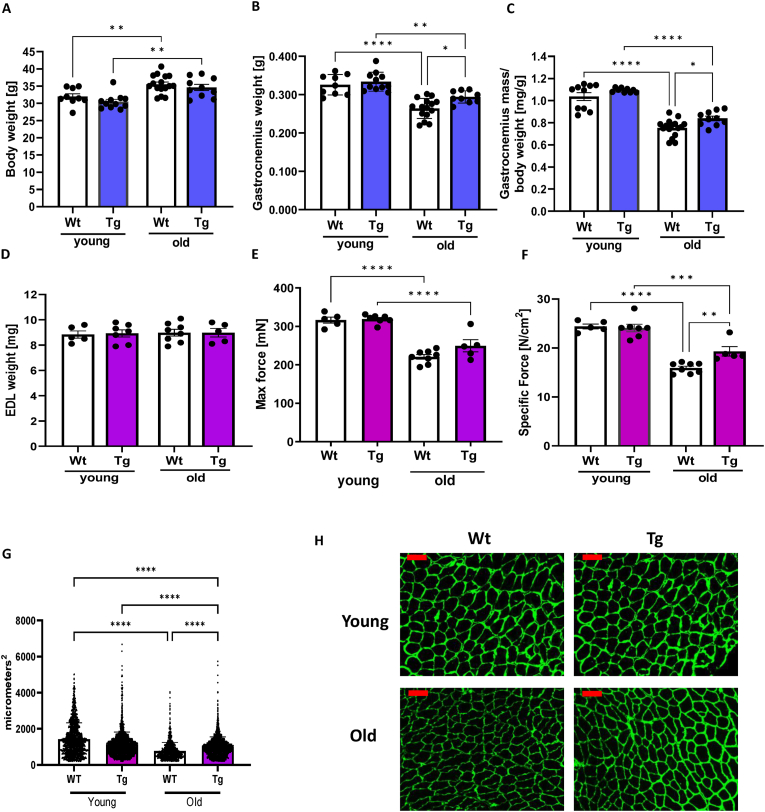
Muscle mass and function. To investigate the effect of GPX4 overexpression on age-related loss of muscle function, we measured body and muscle weights, as well as contractile function in the extensor digitorum longus (EDL) muscle. As shown in Fig. 4, despite approximately 10% higher bodyweight in old WT and GPX4Tg mice compared to the young mice in each group, the gastrocnemius muscle weight decreased by an average of 26% (Fig. 4B and C). GPX4 overexpression does not affect the bodyweight (Fig. 4A) but increases both absolute and relative gastrocnemius mass in old mice by 11%; 0.264 vs 0.294 mg, p = 0.0287 (Figs. 4B) and 0.753 vs 0.839 mg/bwt, p = 0.0454 (Fig. 4C). EDL mass was not different between groups (Fig. 4D). However, EDL maximal force was reduced by 30% while specific force by 35% in old WT versus young WT mice (Fig. 4E and F). Specific force (force expressed relative to muscle mass) was rescued by GPX4 overexpression by 21%; 15.913 vs 19.315 N/cm2, p = 0.008 (Fig. 4F). Fiber cross sectional area measured in tibialis anterior (TA) muscle was 50% lower in muscle from aged versus young wildtype mice; 1548.71 vs 767.05, p < 0.0001 (Fig. 4G). In addition, fiber cross sectional area was 30% greater in TA muscle from aged GPX4Tg mice when compared to aged wildtype mice; 1001.29 vs 767.05, p < 0.0001 (Fig. 4G). Representative CSA images are in Fig. 4H.
Fig. 4.
Muscle mass and Function. (A) Body mass, (B) absolute gastrocnemius weight, and (C) gastrocnemius wet mass normalized to body weight in young and old wildtype and GPX4 transgenic mice. *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 5–16. (D) Mass of extensor digitorum longus (EDL) used for contractile function experiments mass. (E) Maximum and (F) specific force measured for EDL muscle. (G)Tibialis anterior muscle cross sectional area analysis. (H) Representative images for tibialis anterior muscle cross sectional area analysis. The scale bar in the images displayed in (H) is 100 μm *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 5–8.4 mice per group was used for tibialis anterior cross sectional area analysis. Data are presented as mean value ± SEM.
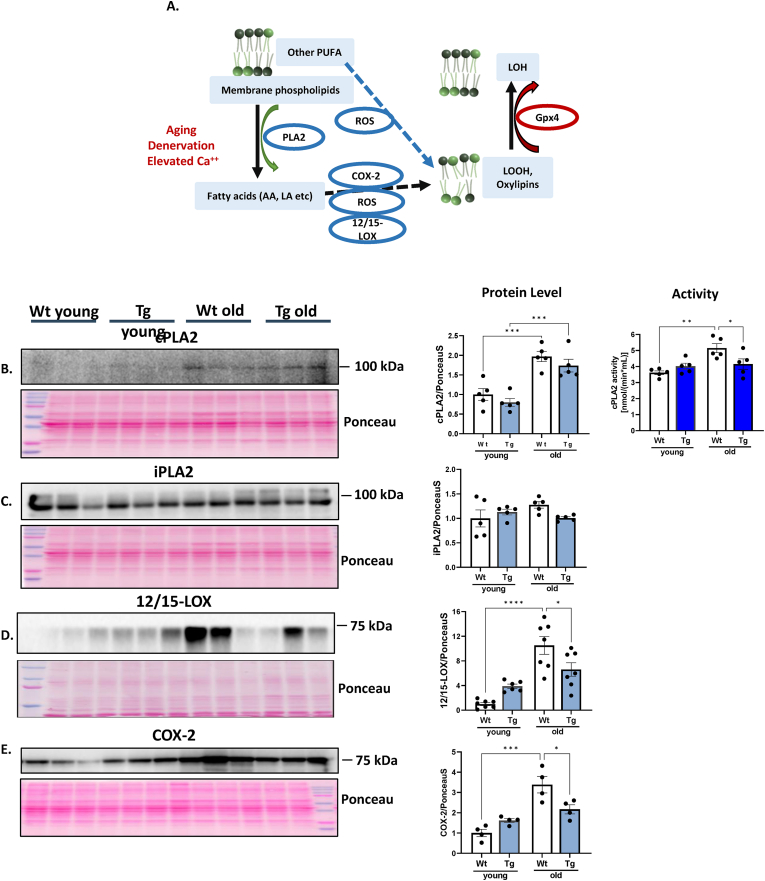
Mediators of oxylipin generation induced by denervation. Our previous work has shown induction of the activity of cPLA2 and 12/15 lipoxygenase (12/15 LOX) enzymes in response to denervation and in aging muscle. As shown in Fig. 5A, fatty acids, including arachidonic acid, (AA) are released by the activity of phospholipase A2 (cPLA2). AA released by cPLA2 and other PUFAs are oxidized by COX, LOX and CYP family members as well as non-enzymatically to LOOHs and oxylipins. GPX4 reduces LOOHs (including some oxylipins) to the chemically less reactive lipid hydroxide (LOH) [21,28] (Fig. 5A). In order to determine the enzymatic pathways associated with lipid peroxidation in sarcopenia, we measured several key enzymes involved in generating LOOHs and oxylipin formation using whole gastrocnemius muscle homogenates by western blotting. Expression of cPLA2 (95 kDa) was approximately 2-fold higher in muscle from old mice. There was no difference between WT and GPX4Tg mice in the expression of this protein, but cPLA2 activity was 26% lower for old GPX4Tg when compared with age matched WT (Fig. 5B). There were no changes between groups in the amount of the mitochondrial iPLA2 (Fig. 5C). The expression of both 12/15-LOX (∼75 kDa) and cyclooxygenase-2 (COX-2) (∼69 kDa) was increased 10.5-fold (p < 0.0001) and 3.4-fold (p = 0.0001) in old versus young WT muscle and reduced by 37% (p = 0.0355, Figs. 5D) and 35% (p = 0.0237, Fig. 5E), respectively in muscle from old GPX4Tg mice.
Fig. 5.
Oxylipin generation pathways. A) Simplified diagram of the action of the tested enzymes: Fatty acids are released from membrane phospholipids by the activity of PLA enzymes. The released fatty acids and other PUFAs are oxidized by the action of the 12/15-LOX and COX-2 enzymes and non-enzymatically to LOOHs. GPX4 reduces LOOHs to the chemically less reactive lipid hydroxide (LOH). Representative Western blot and pooled data for: (B) phospholipase A2 (PLA2), (C) Calcium-independent phospholipase A2 (iPLA2), (D) 12/15-lipoxygenase (12/15-LOX), and (E) cyclooxygenase-2 (COX-2) measured in gastrocnemius muscle homogenates from young and old wildtype and GPX4 transgenic mice *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 4–7. Data are presented as mean value ± SEM.
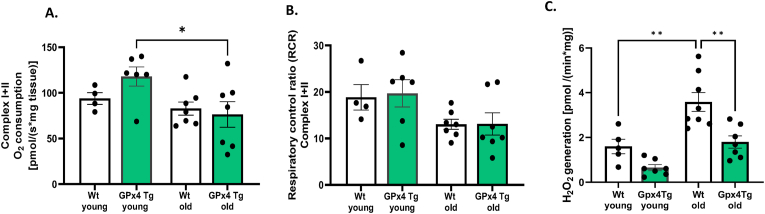
Mitochondrial function. Mitochondrial dysfunction is commonly associated with muscle atrophy conditions [39]. To assess the effect of GPX4 overexpression on mitochondrial function in young and old skeletal muscle, we measured mitochondrial respiration (OCR), respiratory control ratio (RCR) and hydroperoxide production in permeabilized muscle fibers from the red portion of the gastrocnemius muscle. We measured respiration from electron transport chain from complex I + II (in response to glutamate malate, ADP, and succinate). Maximally stimulated OCR and RCR are reduced in fibers from old WT mice when compared to young WT (Fig. 6A and B). There were no differences between WT and GPX4Tg mice. State 1 (mitochondria respiring without addition of external substrate) hydroperoxide production was 2.2 times higher in muscle fibers from old WT mice versus young WT (p = 0.0019). This was reduced by GPX4 overexpression by 50%; 3.587 vs 1.795 pM/(min*mg), p = 0.002 (Fig. 6C). We noticed the same trend for the production of hydroperoxides with complex I, and complex I + II stimulated respiration: 2 and 2.2 fold increased for old WT group compared to young and reduction by 26 and 42% in old GPX4Tg muscle, however, the differences were not statistically significant (data not shown).
Fig. 6.
Mitochondrial function in permeabilized gastrocnemius fibers. (A) Respiratory rate with appropriate substrates and inhibitors for the complex I + II, (B) respiratory control ratio (RCR) for complex I + II, (C) Reactive oxygen species (ROS) production rate with no substrate (basal). *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 5–8. Data are presented as mean value ± SEM.
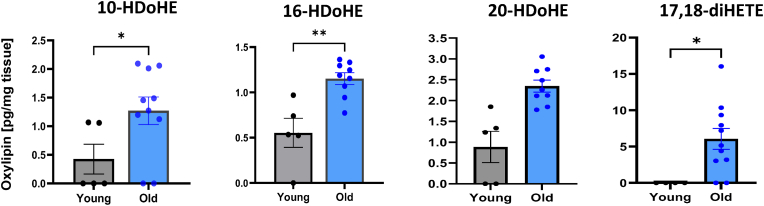
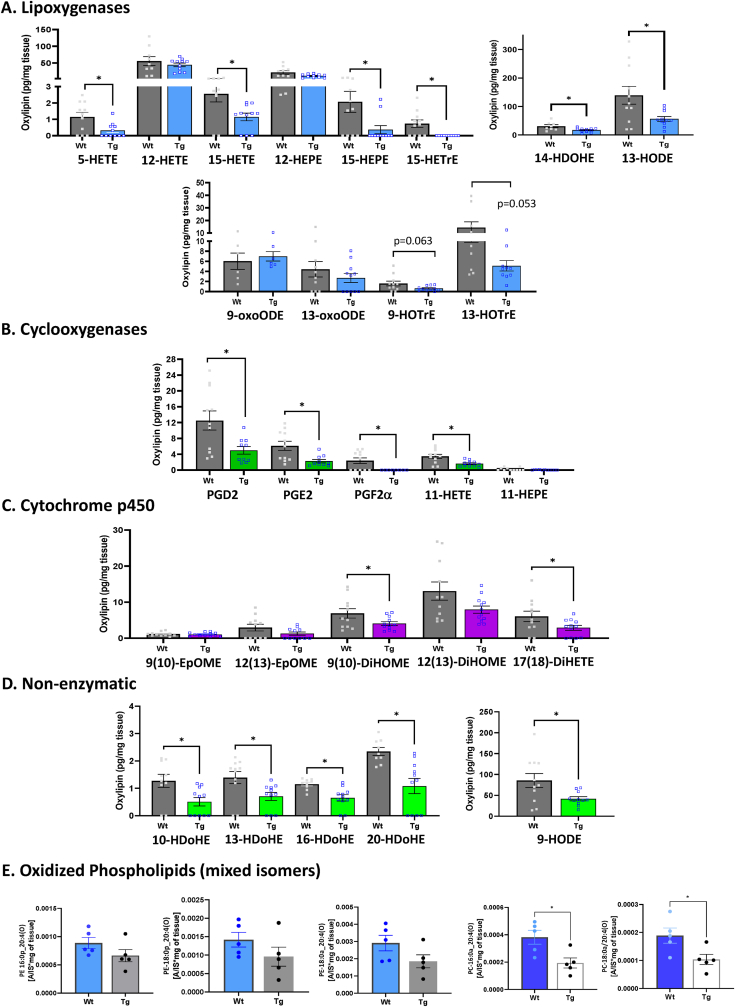
Oxylipin and oxidized phospholipids (oxPL) levels. The concentration of oxylipins and oxPLs were measured in gastrocnemius muscle of young WT, as well as old WT and GPX4Tg mice. Four of the 27 identified oxylipin species were significantly elevated with age, i.e., 10-, 16- and 20-HDoHE and 17,18 -diHETE (Fig. 7). Conversely, 16 out of 27 identified oxylipins were significantly reduced (p = 0.0023–0.0484) in the muscles from old GPX4Tg mice when compared to old WT mice (Fig. 8). Oxylipin levels (pg/mg tissue) were reduced more than 50% for 7 out of 12 detected species generated enzymatically by the activity of lipoxygenase from arachidonic acid (AA): 5- and 15-HETE [40,41], linoleic acid (LA): 13-HODE [40], alpha linoleic acid (ALA): 9- and 13-HOTrE [42,43], dihomo gamma linoleic acid (DGLA): 15-HETrE [44], eicosapentaenoic acid (EPA): 15-HEPE [45], and docosahexaenoic acid (DHA): 14-HDoHE [46]. Likewise, 11 detected oxylipins generated by COX or CYP activity or non-enzymatically, including 11-HETE, PGD2, PGE2, PGF2α (AA metabolites) [41], 9,10-DiHOME (LA metabolite) [47], 17,18-DiHETE (EPA metabolite) [40], 10-, 13-, 14-16-, 20-HDoHE (DHA metabolites) [46], and 9-HODE (LA metabolite) [48], were also reduced by GPX4 overexpression in aging muscle. A number of other oxylipins followed the same pattern but the changes were not statistically significant. Levels of oxidized phosphatidylcholines (PC) 16:0a_20:4(O), and 18:0a_20:4(O) were decreased in muscle from old GPX4Tg mice by 49% (p = 0.0173), and 45% (p = 0.0230) respectively, when comparing to old WT muscle. Oxidized phosphatidylethanolamines (PE) 16:0p_20:4(O), 18:0p_20:4(O), and 18:0a_20:4(O) were reduced by 25, 32, and 36%, respectively (changes not statistically significant).
Fig. 7.
Oxylipins elevated in muscle from aging wildtype mice. Four of 28 detected oxylipins were significantly elevated in aging muscle: 10-hydroxydocosa-4Z,7Z,11Z,13Z,16Z,19Z-hexaenoic acid (10-HDoHE), 16-Hydroxydocosa-4Z,7Z,10Z,13Z,17Z,19Z-hexaenoate (16-HDoHE), 20-Hydroxydocosa-4Z,7Z,10Z,13Z,16Z,18Z-hexaenoate (20-HDoHE), and 17,18-dihydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid (17,18-DiHETE). *Significant difference between the groups (p < 0.05, t-test), n = 9–12.
Fig. 8.
Oxylipins detected in skeletal muscle from old wildtype and old GPX4 transgenic mice with their precursor lipids and the most probable way of formation in the presented model. Note that some oxylipins may be produced by multiple enzymes and we have done only a general categorization. Oxylipins delivered from lipoxygenases thru various fatty acid precursors are shown in Panel (A). Arachidonic acid (AA): 5-hydroxyeicosatetraenoic acid (5-HETE), 12-Hydroxyeicosatetraenoic acid (12-HETE), 15-Hydroxyeicosatetraenoic acid (15-HETE); Linoleic acid (LA): 13-Hydroxyoctadecadienoic acid (13-HODE), 9-oxo-10E,12Z-octadecadienoic acid (9-oxoODE), 13-Oxo-9E,11E-octadecadienoic acid (13-oxoODE); dihomo-γ-linolenic acid (DGLA): 15-Hydroxy-(8Z,11Z,13E)-eicosatrienoic acid (15-HETrE); eicosapentaenoic acid (EPA): 12-hydroxyeicosapentaenoic acid (12-HEPE), 15-hydroxyeicosapentaenoic acid (15-HEPE); docosahexaenoic acid (DHA): 14-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14-HDoHE); α-linolenic acid (ALA): 9-Hydroxy-(10E,12Z,15Z)-octadecatrienoic acid (9-HOTrE), 13-hydroxy-(9Z,11E,15Z)-octadecatrienoic acid (13-HOTrE). Panel (B)- oxylipins generated by cyclooxygenase from: arachidonic acid (AA):11-Hydroxy-5,8,12,14-eicosatetraenoic acid (11-HETE), Prostaglandin D2 (PGD2), Prostaglandin E2 (PGE2), prostaglandin F2α (PGF2α), and from EPA: 11-hydroxy-(5Z,8Z,12E,14Z,17Z)-eicosapentaenoic acid (11-HEPE). Panel (C)- oxylipins generated through cytochrome p450 from: linoleic acid (LA): 9,10-Dihydroxy-12Z-octadecenoic acid (9,10-DiHOME), 12,13-dihydroxy-9Z-octadecenoic acid (12,13-DiHOME), 9,10-epoxy-(12Z)-octadecenoic acid (9(10)-EpOME), 12(13)epoxy-(9Z)-octadecenoic acid (12(13)-EpOME); and from eicosapentaenoic acid (EPA): 17,18-dihydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid (17,18-DiHETE). Panel (D)- oxylipins generated nonenzymatically from: docosahexaenoic acid (DHA): 10-hydroxydocosa-4Z,7Z,11Z,13Z,16Z,19Z-hexaenoic acid (10-HDoHE), 13-hydroxy-4Z,7Z,10Z,14E,16Z,19Z-docosahexaenoic acid (13-HDoHE), 16-Hydroxydocosa-4Z,7Z,10Z,13Z,17Z,19Z-hexaenoate (16-HDoHE), 20-Hydroxydocosa-4Z,7Z,10Z,13Z,16Z,18Z-hexaenoate (20-HDoHE), and from linoleic acid (LA): 9-hydroxy-10E,12Z-octadecadienoic acid (9-HODE). Panel (E)- Oxidized phospholipids (OxPLs): phosphatidylethanolamines (PE) 16:0p_20:4(O), 18:0p_20:4(O), and 18:0a_20:4(O); phosphatidylcholines (PC) 16:0a_20:4(O), and 18:0a_20:4(O). *Significant difference between the groups (p < 0.05, t-test), n = 5–12.
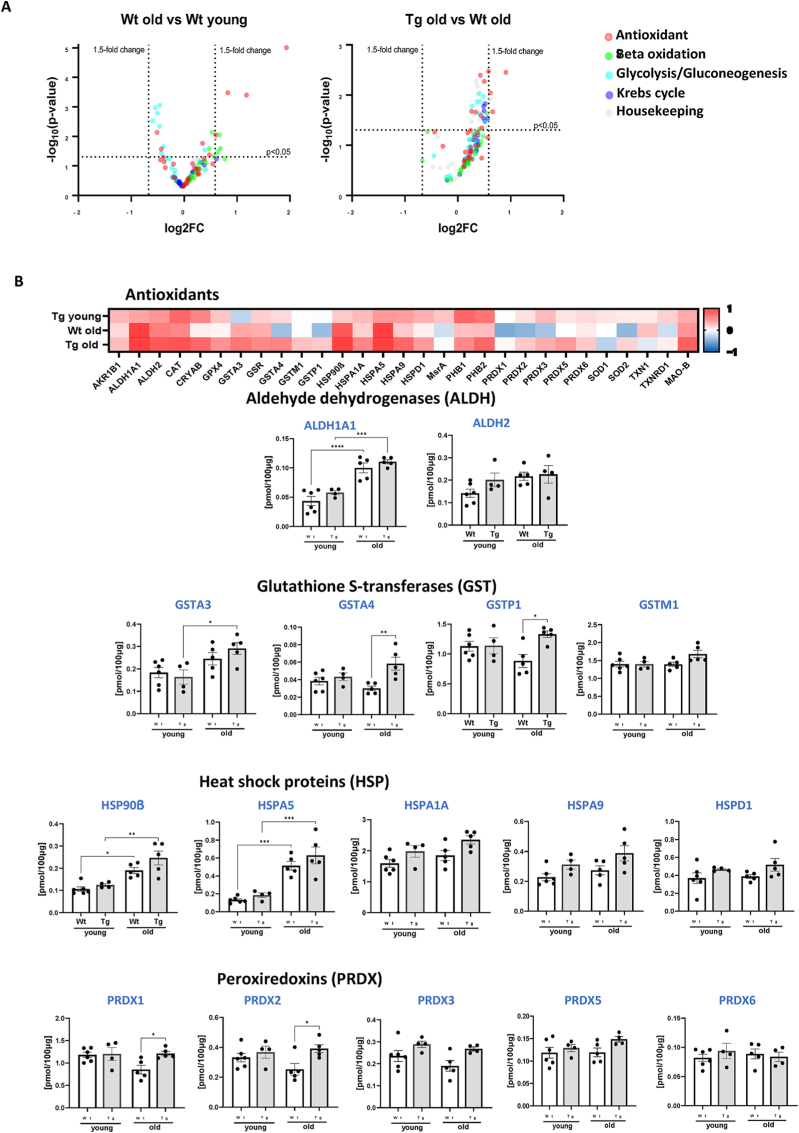
Metabolic Alterations. To determine whether there were changes in the metabolic profile in old WT and GPX4Tg gastrocnemius muscle, we measured key metabolic proteins involved in the TCA cycle, beta oxidation and glycolysis, as well as antioxidant related proteins. The greatest up-regulation was evident in proteins involved in the oxidative stress response and beta oxidation, while the levels of proteins involved in glucose metabolism tended to be reduced. Transgenic mice showed an up-regulation of most proteins when compared to WT animals (Fig. 9A). The significantly upregulated antioxidants (p = 0.0001–0.0181, Fig. 9B) for both old groups were: ALDH1A1 (∼2 fold change), and two members of heat shock proteins family: HSP90β (∼2 fold increase), and HSPA5 (3.4 fold change). There was a tendency for age-related down-regulation in the expression of some of the glutathione S-transferases and peroxiredoxins (Fig. 9B) for WT mice: GSTA4 and GSTP1 by 22%, PRDX1 by 28%, PRDX2 by 23%, and PRDX3 by 19% (statistically insignificant). Their expression was rescued in GPX4Tg by 94% (p = 0.0053), 50% (p = 0.0198), 30% (p = 0.0419), 42% (p = 0.0405) and 40% (statistically insignificant), respectively. As shown in Fig. 9C, among the proteins involved in beta oxidation, only LONP1 expression was slightly decreased (by 9%, non-significant), GPX4 overexpression increased it by 67% (p = 0.0081). The largest increase for old WT group was noted for two fatty-acid-binding proteins: FABP3 by 72% (p = 0.0673) and FABP4 by 46% (p = 0.0165), it was reduced in muscle from GPX4Tg mice by 50% (p = 0.0433) and 41% (p = 0.005), respectively. The amount of many proteins involved in glycolysis and gluconeogenesis was significantly reduced in aged WT compared to young mice (Fig. 9D). Proteins the most affected by GPX4 overexpression were ENO3 (increase by 41% in old GPX4Tg versus old WT, p = 0.0315) and PFKM (increase by 35%, p = 0.0384). There were no statistically significant differences between the groups in the Krebs cycle proteins (Fig. 9E).
Fig. 9.
Targeted proteomic analysis of key metabolic and antioxidant proteins. (A) Volcano plot of the proteins showing the Log2 fold changes plotted against corresponding –log10 p-value for wildtype old mice versus young, and GPX4Tg versus wildtype old. Data points in the upper right (ratio >1.5) and upper left (ratio <0.67, 1.5- fold decrease) sections with P > 0.05 represent proteins that are significantly changed. (B) Heatmap representing the log2 fold change in the levels of antioxidant proteins and theconcentration of selected proteins from the antioxidant panel: Aldehyde dehydrogenases (ALDH): ALDH1A1 and ALDH2; Glutathione S-transferases (GST): GSTA3, GSTA4, GSTP1, and GSTM1; Heat shock proteins (HSP): HSP90β, HSPA5, HSPA1A, HSP9, HSPD1; Peroxiredoxins (PRDX): PRDX1, PRDX2, PRDX3, PRDX4, PRDX5. (C) Heatmap representing the log2 fold change values of proteins level and the concentration of selected proteins from the (C) beta oxidation panel: fatty acid binding protein 3 (FABP3), fatty acid binding protein 4 (FABP4) and lon peptidase 1 (LONP1); (D) glycolysis/gluconeogenesis panel: enolase 3 (ENO3) and phosphofructokinase muscle-type (PFKM) and (D) heatmap for krebs cycle panel. *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 4–6.
4. Discussion
The mechanisms underlying oxidative stress and the age-related decline in muscle quantity and quality are still unclear. Based on our previous results tying LOOH generation to denervation induced atrophy, we hypothesized that an increase in LOOH production likely contributes to the phenotypes associated with sarcopenia. Further, and by extension, reducing their level may have a positive effect on maintaining muscle function in elderly. To test this hypothesis, we utilized a transgenic mouse with global overexpression of GPX4 to improve the ability of muscle cells to reduce LOOHs. We measured changes in muscle mass and force generation, the amount of lipid oxidation products (LOOHs, oxylipins, 4-HNE, and MDA) and proteins in major metabolic pathways, as well as mitochondrial function. The novel findings presented in this study show that reducing lipid hydroxides by GPX4 overexpression ameliorates the majority of muscle atrophy and weakness phenotypes present in mouse models of sarcopenia.
We have previously shown that LOOH production is higher in surgically denervated muscle when compared to sham control intact muscle [12,13]. Spendiff et al. also reported elevated production of LOOHs in skeletal muscle from humans older than 75 years compared to that of younger muscle tissue [49]. However, a causative role for LOOHs and other lipid oxidation products in triggering age-related loss of muscle strength and mass and the mechanisms by which this occurs have not been defined. In agreement with previous studies, here we showed that lipid oxidation and its products (LOOHs, 4-HNE, and MDA) are greater in muscles from old wildtype mice when compared to young wildtype mice.
The biological effect of many lipid peroxidation products depends on both their concentration and the conditions of the intracellular and extracellular environment. For example, oxylipins may be toxic or protective, anti-inflammatory or pro-inflammatory [50,51]. However, the accumulation of most oxylipins disrupts the proper functioning of cells at various levels. Specifically, lipid peroxidation can modify the structure of cell membranes and proteins, disrupting their proper functioning, and is involved in the pathogenesis of many various diseases such as degenerative disease of the brain, liver cirrhosis, atherosclerosis [[52], [53], [54], [55], [56], [57], [58]]. Aldehydes (such as MDA and 4-HNE) formed as secondary or decomposition products of lipid peroxidation are highly reactive and easily interact with proteins, DNA, and phospholipids. 4-HNE, the level of which is elevated over 5 times in muscle from old WT mice, reacts directly with thiol groups of amino acids generating pathogenic adducts and induces apoptosis of various cell types [51,59]. Furthermore, accumulation of LOOHs can be fatal and induce ferroptosis: iron-dependent, non-apoptotic cell death [60,61]. In support of this, a recent report by Eshima et al. [62] demonstrated that elevated LOOH and their reactive lipid aldehyde byproducts are effectors of muscle atrophy and weakness in a disuse atrophy model. It is reasonable to conclude that the toxic effects of lipid peroxidation products might contribute to the observed decrease in muscle mass and force generation in old WT mice.
GPX4 is the only mammalian enzyme able to reduce esterified phospholipids and cholesterol hydroperoxides directly in cell membranes [60]. Its overexpression reduced the amount of LOOHs by 84% and the amount of total hydroperoxides by 50% (basal state) in the muscles from old mice. We also noticed a decrease in secondary lipid peroxidation products: 4-HNE and MDA by 38 and 32%, respectively. This effect may contribute to the reduction of oxidative modification of proteins as well as prevent structural and functional modification of biological membranes and thus protect skeletal muscles mass and functions.
GPX4 overexpression lowers the concentration of many oxylipins present in aged muscle. The oxylipin literature in muscle is less extensive than other tissues. The concentration of individual oxylipins increases in muscles after exercise and injury, which plays an important role in the inflammatory response, repair, and muscle growth [[63], [64], [65], [66], [67]]. However, the function of oxylipins in skeletal muscle aging is unknown. In tissues other than muscle, elevated levels of oxylipins in age-related pathologies is associated with accelerated senescence [[68], [69], [70], [71], [72]]. More research is needed to understand how oxylipins may trigger muscle atrophy and weakness. In the current study, we have shown that overexpression of GPX4 reduces oxylipin content in aged muscle which is associated with improved contractile function, mitochondrial function, and increased muscle mass. It is interesting to note that the pattern of oxylipins detected in skeletal muscle is highly represented by oxylipin species generated by 12/15 lipoxygenase and prostaglandins generated by cyclooxygenase. Both of these enzymes are highly elevated with age, yet this is not reflected by an increase in these oxylipins with age. The potential significance of this discrepancy is uncertain at this point.
In our previous studies, we have shown that denervation-related hydroperoxides were produced downstream of cPLA2 and that 12/15-LOX knockout mice were protected from muscle mass loss after denervation [12,13,73]. We hypothesize that enzymes downstream of cPLA2 may be involved in oxylipin and other LOOHs generation during aging, and may contribute to age-related decline in muscle mass and function. Muscle from aged mice had increased level of cPLA2 but not iPLA2 (Ca2+-independent PLA2) proteins. We also noticed a significant increase in the amount of enzymes responsible for the metabolism of PUFAs in aged mice such as: 12/15-LOX and COX-2. GPX4 overexpression does not decrease the amount of cPLA2 protein but does decrease its activity in old mice. Many oxylipins are hydroperoxides formed by the action of LOX, COX and CYP, which are further converted into their reduced forms; for example by enzymes from the GPx family [58]. A decrease in reduced forms of oxylipins, as well as 12/15-LOX and COX-2 enzymes in GPX4Tg mice may indicate a reduced amount of initial substrates, including fatty acids released by cPLA2. LOX and COX enzymatic activity requires oxidation, thus, the effect of GPX4 in reducing redox tone may also contribute to suppressing LOX and COX activities directly [74]. Given the relative amounts of individual lipids in our analysis, we see that the most abundant are oxylipins that are likely to be generated by leukocyte 12/15-LOX (i.e., 12-HETE, 12-HEPE, 13-HODE). The levels of these oxylipins in particular are greatly reduced suggesting a lower activity of this enzyme. This could be due to a decrease in expression of 12/15-LOX or lower activity of either 12/15-LOX or cPLA2. We also see significantly reduced amounts of prostaglandins, which suggests that there may be reduced amount or activity of cyclooxygenase (either COX-1 or COX-2). Another possibility is an overall reduction of oxidative stress through the actions of GPX4 on other pathways of lipid peroxidation and consequently reduction of a need for some of the lipid signaling molecules.
It is well known that both LOX and COX enzymes leak off small amounts of lipid radicals from their active sites which can in turn mediate chain propagation that appears as “non-enzymatic” oxidation. Overall, the levels of non-enzymatically generated oxylipins in our study are very small and may just reflect the lower levels of LOX and COX found in the GPX4 transgenic mice. as a side product of the enzymatic activities. The levels of oxPL are also lower, and although they were mixed isomers, they could also be formed as a side product of LOX (e.g. through chain propagation of leaked radicals), which would account for the reduced levels in the GPX4 Tg mice.
We have shown that mitochondria are the primary source of LOOHs; therefore, we checked the mitochondrial function by measuring oxygen consumption and calculating the respiratory control ratio. Old mice exhibit a reduction in mitochondrial respiration and it does not change in GPX4Tg mice. Targeted proteomics was used to better understand the changes the content of metabolic and antioxidant proteins occurring in muscle during aging and the effect of GPX4 overexpression on these changes. There is a global change in the content of many proteins content of antioxidant, beta oxidation, glycolysis and gluconeogenesis enzymes. GPX4 overexpression has no effect on upregulated antioxidants but rescues downregulated proteins. It can improve the overall oxidative status of old muscle cells and specifically the ability of old muscles to detoxify active aldehydes by GSTs and LOOHs by PRDXs enzymes. Maintenance of the antioxidant status can be effective in preventing the degradation of muscle cells due to oxidative stress.
Among the changes in the metabolic proteins content, fatty acid-binding protein 3 and 4 content increases in the muscle of old mice, while overexpression of GPX4 in aged muscle prevents the increase of FABP3 and 4 in aged muscle. FABP3 expression can lead to pathological lipid remodeling and disrupt muscle homeostasis [75,76]. Furthermore, old mice with FABP3 knockdown exhibited increased protein synthesis, improved muscle recovery and retained a lipid composition similar to that of young muscle [75]. Reduced FABP3 content in aged muscle with GPX4 overexpression may be one of the reasons for the improvements in muscle mass and force generation.
In summary, our study shows that lipid hydroperoxides may play an important role in the development of sarcopenia. Reducing LOOHs by GPX4 overexpression ameliorates aged-related loss of both skeletal muscle mass and strength. We have shown that mice with increased content of GPX4 are characterized by a reduced amount of lipid peroxidation products including LOOHs, oxylipins, 4-HNE, and MDA. Further, GPX4 overexpression alters the content of many proteins involved in the metabolism of lipid oxidation products towards the young-like content. Collectively, we suggest that reduction of lipid hydroperoxides and GPX4 modulation can be a valuable target for intervention of sarcopenia.
Declaration of competing interest
None
Acknowledgements
Support for this work has been provided by a VA Merit I01BX004453 to Dr. Van Remmen as well as P01AG051442 and R01AG077812 from the National Institute on Aging. Dr. Van Remmen is also the recipient of a VA Senior Research Career Scientist award (1 IK6 BX005234-01). Contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Dr. Jacob Brown is currently supported by VA Career Development Award 1 IK2 BX005620-01A1. The authors would like to thank the members of the Van Remmen Laboratory for their contributions to the experiments presented here. We would also like to extend our gratitude to the numerous other faculties, staff and other researchers at the Oklahoma Medical Research Foundation and OUHSC for helpful discussions. Graphical Abstract was generated using BioRender.com.
Data availability
Data will be made available on request.
References
- 1.Christian C.J., Benian G.M. Animal models of sarcopenia. Aging Cell. 2020;19 doi: 10.1111/acel.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morley J.E., Abbatecola A.M., Argiles J.M., Baracos V., Bauer J., Bhasin S., et al. Sarcopenia with limited mobility: an international consensus. J. Am. Med. Dir. Assoc. 2011;12:403–409. doi: 10.1016/j.jamda.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sataranatarajan K., Pharaoh G., Brown J.L., Ranjit R., Piekarz K.M., Street K., et al. Molecular changes in transcription and metabolic pathways underlying muscle atrophy in the CuZnSOD null mouse model of sarcopenia. Geroscience. 2020;42:1101–1118. doi: 10.1007/s11357-020-00189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedmer P., Jung T., Castro J.P., Pomatto L.C.D., Sun P.Y., Davies K.J.A., et al. Sarcopenia - molecular mechanisms and open questions. Ageing Res. Rev. 2021;65 doi: 10.1016/j.arr.2020.101200. [DOI] [PubMed] [Google Scholar]
- 5.Zembron-Lacny A., Dziubek W., Rogowski L., Skorupka E., Dabrowska G. Sarcopenia: monitoring, molecular mechanisms, and physical intervention. Physiol. Res. 2014;63:683–691. doi: 10.33549/physiolres.932692. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton R.T., Walsh M.E., Van Remmen H. Mouse models of oxidative stress indicate a role for modulating healthy aging. J. Clin. Exp. Pathol. 2012 doi: 10.4172/2161-0681.S4-005. Suppl 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellanti F., Romano A.D., Lo Buglio A., Castriotta V., Guglielmi G., Greco A., et al. Oxidative stress is increased in sarcopenia and associated with cardiovascular disease risk in sarcopenic obesity. Maturitas. 2018;109:6–12. doi: 10.1016/j.maturitas.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Jang Y.C., Lustgarten M.S., Liu Y., Muller F.L., Bhattacharya A., Liang H., et al. Increased superoxide in vivo accelerates age-associated muscle atrophy through mitochondrial dysfunction and neuromuscular junction degeneration. Faseb. J. 2010;24:1376–1390. doi: 10.1096/fj.09-146308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur P.G., Grounds M.D., Shavlakadze T. Oxidative stress as a therapeutic target during muscle wasting: considering the complex interactions. Curr. Opin. Clin. Nutr. Metab. Care. 2008;11:408–416. doi: 10.1097/MCO.0b013e328302f3fe. [DOI] [PubMed] [Google Scholar]
- 10.Muller F.L., Song W., Jang Y.C., Liu Y., Sabia M., Richardson A., et al. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293:R1159–R1168. doi: 10.1152/ajpregu.00767.2006. [DOI] [PubMed] [Google Scholar]
- 11.Muller F.L., Song W., Liu Y., Chaudhuri A., Pieke-Dahl S., Strong R., et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic. Biol. Med. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharya A., Muller F.L., Liu Y., Sabia M., Liang H., Song W., et al. Denervation induces cytosolic phospholipase A2-mediated fatty acid hydroperoxide generation by muscle mitochondria. J. Biol. Chem. 2009;284:46–55. doi: 10.1074/jbc.M806311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pharaoh G., Brown J.L., Sataranatarajan K., Kneis P., Bian J., Ranjit R., et al. Targeting cPLA2 derived lipid hydroperoxides as a potential intervention for sarcopenia. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharya A., Lustgarten M., Shi Y., Liu Y., Jang Y.C., Pulliam D., et al. Increased mitochondrial matrix-directed superoxide production by fatty acid hydroperoxides in skeletal muscle mitochondria. Free Radic. Biol. Med. 2011;50:592–601. doi: 10.1016/j.freeradbiomed.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami M., Sato H., Taketomi Y. Updating phospholipase A2 biology. Biomolecules. 2020;10 doi: 10.3390/biom10101457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su L.J., Zhang J.H., Gomez H., Murugan R., Hong X., Xu D., et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/5080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayala A., Munoz M.F., Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014;2014 doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazawa T. Lipid hydroperoxides in nutrition, health, and diseases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021;97:161–196. doi: 10.2183/pjab.97.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forcina G.C., Dixon S.J. GPX4 at the crossroads of lipid homeostasis and ferroptosis. Proteomics. 2019;19 doi: 10.1002/pmic.201800311. [DOI] [PubMed] [Google Scholar]
- 20.Markworth J.F., Vella L., Lingard B.S., Tull D.L., Rupasinghe T.W., Sinclair A.J., et al. Human inflammatory and resolving lipid mediator responses to resistance exercise and ibuprofen treatment. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;305:R1281–R1296. doi: 10.1152/ajpregu.00128.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Signini E.F., Nieman D.C., Silva C.D., Sakaguchi C.A., Catai A.M. Oxylipin response to acute and chronic exercise: a systematic review. Metabolites. 2020;10 doi: 10.3390/metabo10060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nayeem M.A. Role of oxylipins in cardiovascular diseases. Acta Pharmacol. Sin. 2018;39:1142–1154. doi: 10.1038/aps.2018.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q., Rempel J.D., Ball T.B., Aukema H., Minuk G.Y. Plasma oxylipins levels in nonalcoholic fatty liver disease. Dig. Dis. Sci. 2020;65:3605–3613. doi: 10.1007/s10620-020-06095-8. [DOI] [PubMed] [Google Scholar]
- 24.Jurado-Fasoli L., Di X., Kohler I., Osuna-Prieto F.J., Hankemeier T., Krekels E., et al. Omega-6 and omega-3 oxylipins as potential markers of cardiometabolic risk in young adults. Obesity. 2022;30:50–61. doi: 10.1002/oby.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gegotek A., Skrzydlewska E. Biological effect of protein modifications by lipid peroxidation products. Chem. Phys. Lipids. 2019;221:46–52. doi: 10.1016/j.chemphyslip.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Del Rio D., Stewart A.J., Pellegrini N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metabol. Cardiovasc. Dis. 2005;15:316–328. doi: 10.1016/j.numecd.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Long J., Liu C., Sun L., Gao H., Liu J. Neuronal mitochondrial toxicity of malondialdehyde: inhibitory effects on respiratory function and enzyme activities in rat brain mitochondria. Neurochem. Res. 2009;34:786–794. doi: 10.1007/s11064-008-9882-7. [DOI] [PubMed] [Google Scholar]
- 28.Seibt T.M., Proneth B., Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019;133:144–152. doi: 10.1016/j.freeradbiomed.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Ran Q., Liang H., Gu M., Qi W., Walter C.A., Roberts L.J., 2nd, et al. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 30.Brown J.L., Peelor F.F., 3rd, Georgescu C., Wren J.D., Kinter M., Tyrrell V.J., et al. Lipid hydroperoxides and oxylipins are mediators of denervation induced muscle atrophy. Redox Biol. 2022;57 doi: 10.1016/j.redox.2022.102518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown J.L., Lawrence M.M., Ahn B., Kneis P., Piekarz K.M., Qaisar R., et al. Cancer cachexia in a mouse model of oxidative stress. J Cachexia Sarcopenia Muscle. 2020;11:1688–1704. doi: 10.1002/jcsm.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qaisar R., Bhaskaran S., Premkumar P., Ranjit R., Natarajan K.S., Ahn B., et al. Oxidative stress-induced dysregulation of excitation-contraction coupling contributes to muscle weakness. J Cachexia Sarcopenia Muscle. 2018;9:1003–1017. doi: 10.1002/jcsm.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks S.V., Faulkner J.A. Contractile properties of skeletal muscles from young, adult and aged mice. J. Physiol. 1988;404:71–82. doi: 10.1113/jphysiol.1988.sp017279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown J.L., Lawrence M.M., Ahn B., Kneis P., Piekarz K.M., Qaisar R., et al. Cancer cachexia in a mouse model of oxidative stress. J Cachexia Sarcopenia Muscle. 2020 doi: 10.1002/jcsm.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn B., Pharaoh G., Premkumar P., Huseman K., Ranjit R., Kinter M., et al. Nrf2 deficiency exacerbates age-related contractile dysfunction and loss of skeletal muscle mass. Redox Biol. 2018;17:47–58. doi: 10.1016/j.redox.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pharaoh G., Sataranatarajan K., Street K., Hill S., Gregston J., Ahn B., et al. Metabolic and stress response changes precede disease onset in the spinal cord of mutant SOD1 ALS mice. Front. Neurosci. 2019;13:487. doi: 10.3389/fnins.2019.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conrad M., Friedmann Angeli J.P. Glutathione peroxidase 4 (GPX4) and ferroptosis: what's so special about it? Mol Cell Oncol. 2015;2 doi: 10.4161/23723556.2014.995047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyatt H., Deminice R., Yoshihara T., Powers S.K. Mitochondrial dysfunction induces muscle atrophy during prolonged inactivity: a review of the causes and effects. Arch. Biochem. Biophys. 2019;662:49–60. doi: 10.1016/j.abb.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misheva M., Kotzamanis K., Davies L.C., Tyrrell V.J., Rodrigues P.R.S., Benavides G.A., et al. Oxylipin metabolism is controlled by mitochondrial beta-oxidation during bacterial inflammation. Nat. Commun. 2022;13:139. doi: 10.1038/s41467-021-27766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajeyah A.A., Griffiths W.J., Wang Y., Finch A.J., O'Donnell V.B. The biosynthesis of enzymatically oxidized lipids. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.591819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cambiaggi L., Chakravarty A., Noureddine N., Hersberger M. The role of alpha-linolenic acid and its oxylipins in human cardiovascular diseases. Int. J. Mol. Sci. 2023:24. doi: 10.3390/ijms24076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galliard T., Phillips D.R. Lipoxygenase from potato tubers. Partial purification and properties of an enzyme that specifically oxygenates the 9-position of linoleic acid. Biochem. J. 1971;124:431–438. doi: 10.1042/bj1240431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iversen L., Fogh K., Kragballe K. Effect of dihomogammalinolenic acid and its 15-lipoxygenase metabolite on eicosanoid metabolism by human mononuclear leukocytes in vitro: selective inhibition of the 5-lipoxygenase pathway. Arch. Dermatol. Res. 1992;284:222–226. doi: 10.1007/BF00375798. [DOI] [PubMed] [Google Scholar]
- 45.Sawane K., Nagatake T., Hosomi K., Hirata S.I., Adachi J., Abe Y., et al. Dietary omega-3 fatty acid dampens allergic rhinitis via eosinophilic production of the anti-allergic lipid mediator 15-hydroxyeicosapentaenoic acid in mice. Nutrients. 2019;11 doi: 10.3390/nu11122868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derogis P.B., Freitas F.P., Marques A.S., Cunha D., Appolinario P.P., de Paula F., et al. The development of a specific and sensitive LC-MS-based method for the detection and quantification of hydroperoxy- and hydroxydocosahexaenoic acids as a tool for lipidomic analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hildreth K., Kodani S.D., Hammock B.D., Zhao L. Cytochrome P450-derived linoleic acid metabolites EpOMEs and DiHOMEs: a review of recent studies. J. Nutr. Biochem. 2020;86 doi: 10.1016/j.jnutbio.2020.108484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vangaveti V., Baune B.T., Kennedy R.L. Hydroxyoctadecadienoic acids: novel regulators of macrophage differentiation and atherogenesis. Ther Adv Endocrinol Metab. 2010;1:51–60. doi: 10.1177/2042018810375656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spendiff S., Vuda M., Gouspillou G., Aare S., Perez A., Morais J.A., et al. Denervation drives mitochondrial dysfunction in skeletal muscle of octogenarians. J. Physiol. 2016;594:7361–7379. doi: 10.1113/JP272487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parthasarathy S., Santanam N., Ramachandran S., Meilhac O. Potential role of oxidized lipids and lipoproteins in antioxidant defense. Free Radic. Res. 2000;33:197–215. doi: 10.1080/10715760000301381. [DOI] [PubMed] [Google Scholar]
- 51.Niki E. Lipid peroxidation: physiological levels and dual biological effects. Free Radic. Biol. Med. 2009;47:469–484. doi: 10.1016/j.freeradbiomed.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 52.Dalle-Donne I., Rossi R., Colombo R., Giustarini D., Milzani A. Biomarkers of oxidative damage in human disease. Clin. Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- 53.Chen C.M., Wu Y.R., Cheng M.L., Liu J.L., Lee Y.M., Lee P.W., et al. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington's disease patients. Biochem. Biophys. Res. Commun. 2007;359:335–340. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- 54.Basu S. F2-isoprostanes in human health and diseases: from molecular mechanisms to clinical implications. Antioxidants Redox Signal. 2008;10:1405–1434. doi: 10.1089/ars.2007.1956. [DOI] [PubMed] [Google Scholar]
- 55.Pratico D. Evidence of oxidative stress in Alzheimer's disease brain and antioxidant therapy: lights and shadows. Ann. N. Y. Acad. Sci. 2008;1147:70–78. doi: 10.1196/annals.1427.010. [DOI] [PubMed] [Google Scholar]
- 56.Gianazza E., Brioschi M., Martinez Fernandez A., Casalnuovo F., Altomare A., Aldini G., et al. Lipid peroxidation in atherosclerotic cardiovascular diseases. Antioxidants Redox Signal. 2021;34:49–98. doi: 10.1089/ars.2019.7955. [DOI] [PubMed] [Google Scholar]
- 57.Chen X., Li X., Xu X., Li L., Liang N., Zhang L., et al. Ferroptosis and cardiovascular disease: role of free radical-induced lipid peroxidation. Free Radic. Res. 2021;55:405–415. doi: 10.1080/10715762.2021.1876856. [DOI] [PubMed] [Google Scholar]
- 58.Negre-Salvayre A., Auge N., Ayala V., Basaga H., Boada J., Brenke R., et al. Pathological aspects of lipid peroxidation. Free Radic. Res. 2010;44:1125–1171. doi: 10.3109/10715762.2010.498478. [DOI] [PubMed] [Google Scholar]
- 59.Yang L.L., Chen H., Wang J., Xia T., Sun H., Yuan C.H., et al. 4-HNE induces apoptosis of human retinal pigment epithelial cells by modifying HSP70. Curr Med Sci. 2019;39:442–448. doi: 10.1007/s11596-019-2057-8. [DOI] [PubMed] [Google Scholar]
- 60.Liu H., Forouhar F., Seibt T., Saneto R., Wigby K., Friedman J., et al. Characterization of a patient-derived variant of GPX4 for precision therapy. Nat. Chem. Biol. 2022;18:91–100. doi: 10.1038/s41589-021-00915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Cao F., Yin H.L., Huang Z.J., Lin Z.T., Mao N., et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eshima H., Shahtout J.L., Siripoksup P., Pearson M.J., Mahmassani Z.S., Ferrara P.J., et al. Lipid hydroperoxides promote sarcopenia through carbonyl stress. Elife. 2023;12 doi: 10.7554/eLife.85289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanford K.I., Lynes M.D., Takahashi H., Baer L.A., Arts P.J., May F.J., et al. 12,13-diHOME: an exercise-induced lipokine that increases skeletal muscle fatty acid uptake. Cell Metabol. 2018;27:1111–11120 e3. doi: 10.1016/j.cmet.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Markworth J.F., Cameron-Smith D. Arachidonic acid supplementation enhances in vitro skeletal muscle cell growth via a COX-2-dependent pathway. Am. J. Physiol. Cell Physiol. 2013;304:C56–C67. doi: 10.1152/ajpcell.00038.2012. [DOI] [PubMed] [Google Scholar]
- 65.Ho A.T.V., Palla A.R., Blake M.R., Yucel N.D., Wang Y.X., Magnusson K.E.G., et al. Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc. Natl. Acad. Sci. U. S. A. 2017;114:6675–6684. doi: 10.1073/pnas.1705420114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giannakis N., Sansbury B.E., Patsalos A., Hays T.T., Riley C.O., Han X., et al. Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration. Nat. Immunol. 2019;20:626–636. doi: 10.1038/s41590-019-0356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vella L., Markworth J.F., Farnfield M.M., Maddipati K.R., Russell A.P., Cameron-Smith D. Intramuscular inflammatory and resolving lipid profile responses to an acute bout of resistance exercise in men. Phys. Rep. 2019;7 doi: 10.14814/phy2.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Currais A., Goldberg J., Farrokhi C., Chang M., Prior M., Dargusch R., et al. A comprehensive multiomics approach toward understanding the relationship between aging and dementia. Aging (Albany NY) 2015;7:937–955. doi: 10.18632/aging.100838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keenan A.H., Pedersen T.L., Fillaus K., Larson M.K., Shearer G.C., Newman J.W. Basal omega-3 fatty acid status affects fatty acid and oxylipin responses to high-dose n3-HUFA in healthy volunteers. J. Lipid Res. 2012;53:1662–1669. doi: 10.1194/jlr.P025577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caligiuri S.P., Aukema H.M., Ravandi A., Pierce G.N. Elevated levels of pro-inflammatory oxylipins in older subjects are normalized by flaxseed consumption. Exp. Gerontol. 2014;59:51–57. doi: 10.1016/j.exger.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 71.Khalesi S., Irwin C., Schubert M. Flaxseed consumption may reduce blood pressure: a systematic review and meta-analysis of controlled trials. J. Nutr. 2015;145:758–765. doi: 10.3945/jn.114.205302. [DOI] [PubMed] [Google Scholar]
- 72.Manev H., Uz T., Sugaya K., Qu T. Putative role of neuronal 5-lipoxygenase in an aging brain. Faseb. J. 2000;14:1464–1469. doi: 10.1096/fj.14.10.1464. [DOI] [PubMed] [Google Scholar]
- 73.Bhattacharya A., Hamilton R., Jernigan A., Zhang Y., Sabia M., Rahman M.M., et al. Genetic ablation of 12/15-lipoxygenase but not 5-lipoxygenase protects against denervation-induced muscle atrophy. Free Radic. Biol. Med. 2014;67:30–40. doi: 10.1016/j.freeradbiomed.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 74.O'Donnell V.B. Free radicals and lipid signaling in endothelial cells. Antioxidants Redox Signal. 2003;5:195–203. doi: 10.1089/152308603764816550. [DOI] [PubMed] [Google Scholar]
- 75.Lee S.M., Lee S.H., Jung Y., Lee Y., Yoon J.H., Choi J.Y., et al. FABP3-mediated membrane lipid saturation alters fluidity and induces ER stress in skeletal muscle with aging. Nat. Commun. 2020;11:5661. doi: 10.1038/s41467-020-19501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hwang C.Y., Kim K., Choi J.Y., Bahn Y.J., Lee S.M., Kim Y.K., et al. Quantitative proteome analysis of age-related changes in mouse gastrocnemius muscle using mTRAQ. Proteomics. 2014;14:121–132. doi: 10.1002/pmic.201200497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.