Abstract
Genetic deficiency for early components of the classical complement activation pathway (especially C1q, r, s, and C4) are the strongest monogenic causal factors for the prototypic autoimmune disease systemic lupus erythematosus (SLE), but their prevalence is extremely rare. In contrast, isotype genetic deficiency for the acidic C4A and acquired deficiency of C1q by autoantibodies are frequent among patients with SLE. Here we review the genetic basis of complement deficiencies in autoimmune disease, discuss the complex genetic diversity seen in complement C4 and its association with autoimmune disease, provide guidance as to when clinicians should suspect and test for complement deficiencies, and outline the current understanding of the mechanisms relating complement deficiencies to autoimmunity. We focus primarily on SLE, as the role of complement in SLE is well-established, but will also discuss other informative diseases such as inflammatory arthritis and myositis.
I. INTRODUCTION – Complement pathways
The complement system is an ancient form of immune defense that has existed since the emergence of thioester proteins with opsonic functions in insects and worms (1, 2). Increasing evidence implicates complement in diverse biological processes in mammals ranging from modulation of immunity and tolerance and inflammatory and autoimmune diseases to intracellular signaling involved in metabolism. In all cases, appropriate balance of complement activation and inhibition is required to maintain homeostasis and health (3). Over-activation may lead to excessive inflammation and tissue damage while under-activation may impair pathogen clearance, lead to rampant infection, and possibly predispose towards autoimmune responses against self-antigens. So, while this review will focus on what is known and future directions for research regarding complement’s role in autoimmune diseases with emphases on systemic lupus erythematosus (SLE) and the idiopathic inflammatory myopathies (IIM), it is written with circumspection and consideration for the fact that a small modification of the complement system can have far ranging ramifications in immunity and autoimmunity.
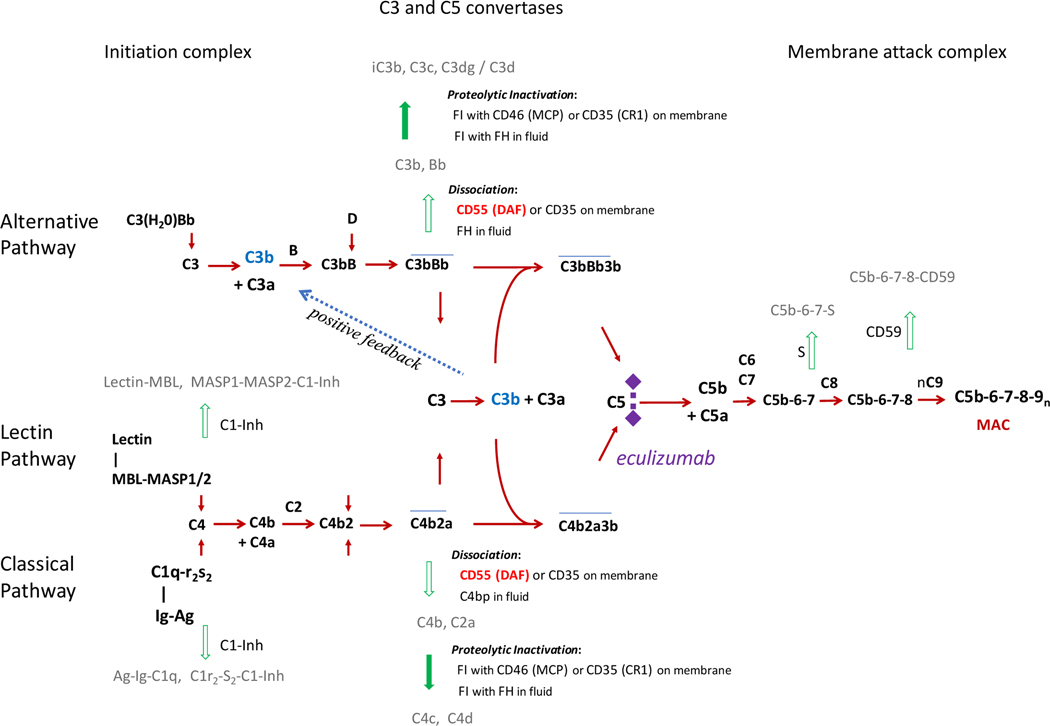
Complement was first described at the end of the 19th century as a soluble and heat-labile factor in the blood that efficiently lysed bacteria (4). Since then, the classical CP), alternative (AP), and lectin (LP) pathways of complement activation in immune defense have been elucidated (5–7). Figure 1 illustrates the three activation pathways of the complement system and positive feedback amplification leading to the release of anaphylatoxins, formation of complement C3 convertases, and the activation of a common terminal pathway (5, 8). Notably, there is a high degree of regulation at almost every step of the activation pathways to ensure rapid and robust destruction of foreign targets with minimal self-inflicted injury (3, 9, 10). Briefly, these pathways are respectively triggered by antigen/antibody complexes binding to C1q of the C1 complex (classical pathway), spontaneous hydrolysis of C3 (alternative pathway), or foreign carbohydrate moieties binding to mannose binding lectin (MBL) or ficolin (lectin pathway). Importantly, all three activation pathways lead to the generation of C3 convertases, C4b2a or C3bBb, and converge at the cleavage and activation of C3 and the amplification loop. In turn, C3b contributes to the formation of the C5 convertase C4b2a3b for the CP or LP, or (C3b)2Bb for the AP, which are essential for the activation of C5 and for the formation of the membrane attack complex (MAC) and direct lysis of target cells.
Figure 1. Activation and regulation of the human complement system.
There are 3 known arms of the complement cascade, including the classical, alternative, and lectin pathways (10). The ligands and sequence of activation are shown for each pathway. Note that all three pathways converge to cleave complement C3, which in turn allows for the generation of C5 convertases and formation of the membrane attack complex (MAC) composed of C5b, C6, C7, C8, and C9 multimers. This complex serves to perforate the outer membrane of invading microbes or aberrant cells necessitating clearance and can result in lysis or sub-lytic permeabilization of the target. Incorporated in the system are tight regulations that prevent or abort inadvertent activations on self or host cell membrane by dissociation of multimolecular complexes and proteolytic degradation of anchor proteins such as C3b and C4b. Eculizumab is a drug or biologic that blocks activation of complement C5. Activation of zymogens and progression of pathways are shown in red and regulatory steps in green. A positive feedback loop (shown in blue) leading to auto-amplification is common for all three activation pathways.
II. COMPLEMENT DEFICIENCIES AND IMMUNE DYSFUNCTION
A. Complement deficiency and infection susceptibility
Deficiencies of components of the complement classical pathway have been associated with increased susceptibility to infections caused by encapsulated bacteria (especially pneumonia and meningitis) and recurrent respiratory infections. Similar patterns of infection have been noted in patients with deficiencies of alternative pathway components. Reduced expression of mannose-binding lectin is associated with increased severity of infection by both bacteria and yeast. MBL deficiency has also been implicated in tuberculosis, malaria, and a variety of viral infections (11–13).
Frequent infections are also the predominant manifestation of deficiencies of C3, factors I and H, and the components of the membrane attack complex, C5-C8. Almost all reported patients with homozygous C3 deficiency have been infants or young children with severe bacterial infections (meningitis, pneumonitis, peritonitis, and osteomyelitis) (14). The reason for increased susceptibility to infection in cases of complement deficiency is easy to conceive: reduced complement availability impairs pathway activation, opsonization, and formation of MAC on the outer membranes of microbial invaders and leads to poor microbial killing. This phenomenon was demonstrated in a recent case report in which a patient with SLE, urticarial vasculitis, and acquired hypocomplementemia developed gonococcemia and septic shock (15). Reduced levels of complement components may also lead to the decreased generation of the anaphylatoxins C3a and C5a, which serve to recruit immune cells to the site of foreign pathogen invasion (16), and thus reduce immunocompetence. In addition to facilitating bacterial and fungal clearance, complement components including MBL are important in the innate immune response to viral infections (17), including upper respiratory infections such as the influenza (18, 19) and other viruses (20). It is known that viral infection temporarily weakens immunity and can open the door for bacterial superinfection (18, 21–23), and therefore complement deficiency may indirectly lead to increased susceptibility to bacterial infection in addition to the loss of its active role in pathogen killing.
B. Complement deficiency and autoimmunity
In addition to targeting invading pathogens with the MAC, the complement system is known to have a range of other immune and immunoregulatory roles, including: (1) the formation and release of the anaphylatoxins C3a and C5a, which serve to attract inflammatory cells to the site of complement activation, (2) opsonization and solubilization of native immune complexes (IC) composed of autoantigens and self-reactive antibodies, and (3) facilitating clearance of IC from the circulation by enabling the binding of complement receptor CR1 on erythrocytes or CR3 or CR4 on phagocytic myeloid cells to opsonized IC (24, 25). Thus, engagement of complement on IC helps reduce the potentially harmful sequelae associated with the presence of immunogenic autoantigen/antibody conglomerates and the subsequent generation of autoantibodies, which may then be deposited on and cause damage to otherwise healthy tissues (24, 26–31).
Deficiencies of various complement components therefore also have undeniable ramifications regarding autoimmunity. While C1q and C4 deficiency are the most strongly related to autoimmune disease (32–35), other complement components including C1r, C1s, C2, and C3 have also been implicated (31, 36, 37). The autoimmune phenotypes associated with the various complement deficiencies as well as literature pertaining to each is summarized in Table 1. The initial trigger for complement activation in various autoimmune diseases may be distinct: synovial proteins and C-reactive protein (CRP) in rheumatoid arthritis (RA) (38, 39); apoptotic cells, neutrophil extracellular traps containing DNA and modified DNA-binding proteins (40–44) and immune complexes in SLE (45); and possibly infection, malignancy, and certain medications in inflammatory myositis (46). In addition, regulation of the complement cascade has been shown to be altered in various autoimmune disorders including RA (47) and SLE (48).
Table 1.
Autoimmune diseases associated with complement deficiencies
| Component | Autoimmune phenotype | Sources |
|---|---|---|
| C1q | Severe lupus-like disease, including nephritis, discoid rash, oral ulcers and anti-Smith antibodies with an unusually negative anti-dsDNA and less frequent arthritis. Female to male ratio approximately 1:1. | (37, 87–95, 240, 241) |
| C1r | Glomerulonephritis, severe cutaneous lupus-like disease. May have increased interferon signaling. Female to male ratio approximately 1.5:1. | (112–114) |
| C1s | Glomerulonephritis, severe cutaneous lupus-like disease. May have increased interferon signaling. Female to male ratio approximately 1.5:1. | (112–115) |
| C2 | Childhood onset but more limited lupus-like symptoms with more severe skin manifestations but milder kidney disease; may include sunsensitive skin lesions, alopecia, febrile episodes, and arthritis. Anti-DNA antibody tests are usually negative, and severe kidney disease is rare. The penetrance of C2 deficiency on SLE is only about 10%. Anti-Ro often positive. Female predominance. | (120–124) |
| C3 | More likely to present with increased risk of severe infections. Autoimmune phenotype may be very mild or even asymptomatic. Very few known cases of autoimmune phenotype. | (125–127, 240) |
| C4 | Early disease onset, a severe photosensitive skin rash, presence of anti-Ro/SSA, and high ANA titers are common. May also present with vasculitis, lupus nephritis. Nearly 80% of homozygous deficient patients develop SLE. Complete deficiencies are associated with a female to male ratio of approximately 1:1. | (79, 132, 146–154, 159, 162, 168, 169, 240, 242, 243) |
| Other components: MBL, complement factor H, membrane cofactor protein, Factor I, Factor B | More commonly present with increased infection susceptibility. May develop atypical hemolytic uremic syndrome. MBL deficiency has been seen in SLE, and anti-MBL antibodies have been detected, though their clinical significance is unclear at this time. | (66, 175, 244–246) |
Similar to the mechanisms by which insufficient complement activation leads to impaired antimicrobial immunity, C1, C2 or C4 deficiency can increase susceptibility to autoimmune disease through impaired opsonization and clearance of autoantigens and IC as well as through decreased formation of MAC on membranes of apoptotic cells (49–53). Decreased complement compromises phagocytes’ response to immunogenic IC and apoptotic debris and increases the amount of time dendritic cells, T cells, and B cells are exposed to autoantigens (54–57). In addition, impairment of complement signaling may skew immunity towards more pro-inflammatory responses, thus contributing to the increased inflammation and immune reactivity seen in autoimmune disease (58, 59).
III. CLINICAL RELEVANCE OF COMPLEMENT IN AUTOIMMUNE DISEASE
This section will highlight the clinical signs and symptoms that suggest that clinicians should consider complement deficiency in their differential diagnosis. Complement deficiencies in general are under-diagnosed, in part because of the diversity in clinical presentation of these disorders, which varies based on the type of complement deficiency (60, 61).
A. Clinical findings suggestive of complement deficiency
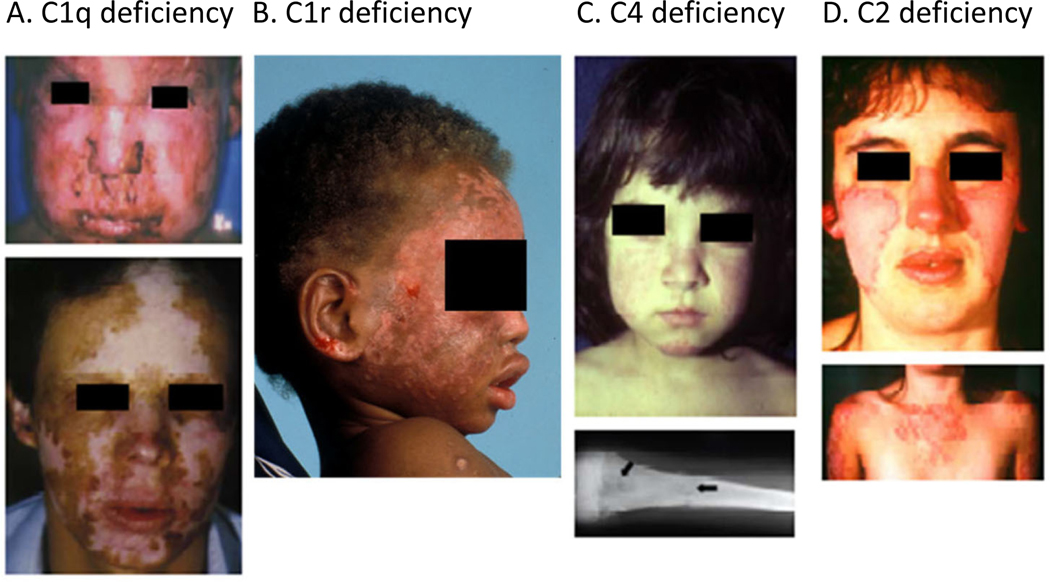
History and physical examination features suggestive of complement abnormalities include multi-generational autoimmune disease, including nephritis, early onset of skin lesions resembling lupus rash, alopecia, photosensitivity, increased susceptibility to infection with encapsulated bacteria such as Streptococcus pneumoniae and Neisseria meningitides, increased susceptibility to viral infections, and angioedema (62, 63). The presentation of complement deficiency may vary across a spectrum from asymptomatic to invasive infection to severe rheumatic diseases resembling SLE. Examples of lupus-like rashes seen in various complement deficiencies are shown in Figure 2.
Figure 2. Systemic lupus erythematous and homozygous deficiency of early components of the classical pathway of complement activation.
Severe cutaneous lesions are common clinical presentations in SLE patients with a complete complement deficiency. (A) A homozygous C1q-deficient male child with cutaneous infection (upper panel), and with discoid lupus erythematosus and scarring lesions on face when he was 22 years old (lower panel). (B) A male child with discoid lupus at 16 months of age with homozygous C1r-deficiency. (C) Complete C4-deficiency in a girl at age 3 with a butterfly rash and cheilitis. (D) A homozygous C2-deficient young woman with acute cutaneous lupus erythematosus. The upper panel shows the butterfly rash, and the lower panel shows photosensitive lesions on sun exposed area. (Source of photographs: (116, 249).)
Clinicians should consider complement deficiencies in a patient presenting with hemolytic anemia, acute kidney failure, and thrombocytopenia concerning for hemolytic uremic syndrome (HUS). While typical HUS is preceded by an episode of bloody diarrhea most commonly caused by a Shigella-like toxin producing E. coli (64), there is also a rare, chronic, and severe form of HUS known as atypical hemolytic uremic syndrome (aHUS) (65). Atypical HUS is caused by genetic defects with a variety of mutations in genes of the complement pathway including complement factor H (CFH), membrane cofactor protein (MCP), Factor I, Factor B, and C3 (66). Reduced serum levels of complement C3 with normal levels of C4 have been reported in patients with atypical HUS, likely reflecting complement consumption as a result of AP activation (67). Knowing the genetic defect underlying aHUS is important for treatment. Atypical HUS is associated with a mortality rate of 20–25% and a morbidity of 48%, as pediatric patients typically progress to end stage renal disease (68).
B. Laboratory testing in cases of suspected complement deficiency
Laboratory evaluation in suspected complement deficiency should include C3, C4, and CH50 to not only assess C3 and C4 protein levels but also complement hemolytic activity. Functional screening for the complement system includes tests for the CP (CH50), the AP (AH50), and the LP. Low CH50 and normal AH50 suggest early classical complement component (C1, C2, and C4) deficiency. Low AH50 with normal CH50 suggests a deficiency of early AP factors (factor B, factor D, and properdin). Low AH50 and low CH50 suggest common terminal complement (C3, C5, C6, C7, C8, or C9) deficiency. If CH50 and AH50 are both normal and the clinician still suspects complement deficiency, MBL functional assay is indicated (16). Furthermore, clinicians should complete a serological work up for SLE including complete blood count (CBC), comprehensive metabolic panel (CMP), antinuclear antibody (ANA), anti-double-stranded DNA (anti-dsDNA), anti-Smith antibodies, and urinalysis. As SLE is extremely rare in children less than 5 years of age, findings suggestive of SLE in young patients should always trigger a deeper investigation into possible genetic causes and/or immunodeficiency.
C. Factors to consider when evaluating complement levels
a. C4 GCN and serum protein levels.
Physiologically, serum protein levels of complement C3 and C4 are correlated with each other very strongly. Immune-complex mediated consumption can lower the serum levels of C4 and C3, particularly during active disease in SLE. Hypocomplementemia in SLE is one of the established criteria for diagnosis of SLE (69–71). Serum levels of C4 and C3 fluctuate with disease activity in SLE, with concurrently low levels of C4 and C3 during disease relapses. Thus, C3 and C4 are useful biomarkers for monitoring disease activity and response to therapy.
Among SLE patients, there exist distinct profiles of C4 serum protein levels over time (72), dependent on the C4 genetic background and disease activities. Patients with low C4 gene copy numbers (i.e., with only two or three copies of total C4) tend to have consistently low levels of serum C4 even during disease remission (72, 73). Patients with medium to high copy number of C4 genes are more likely to demonstrate fluctuating serum levels of C4 and C3 that return to normal range during disease remission. Although low complement levels are seen in active lupus, especially lupus nephritis, it can be difficult to ascertain whether low complement levels are due to consumption during inflammation or due to an inherent isotype deficiency. Even more obfuscating, the two scenarios may coexist in one individual (74). A return to normal C3 but not C4 would imply low gene copy number (GCN) of C4 or other possibilities such as acquired deficiency of C1-inhibitor leading to unchecked turnover (75). An important difference between patients with low C4 due to low GCN versus those with high turnover is that the latter is accompanied by the presence of high levels of serum C4a, and/or high levels of cell bound C4d on the membranes of red blood cells, reflecting ongoing activation and consumption as the cause of hypocomplementemia.
In other autoimmune rheumatic disease such as myositis, complement levels may be depressed to the low “normal” range, but this is often not clinically remarkable and may go undetected if baseline levels are not known. However, there are readily detectable complement activation products in the circulation and deposited on the membrane of circulating blood cells that can provide laboratory evidence of disease activity (76–79). The clinical utility and availability of this test is not universal, however, and is often limited to the research rather than the clinical arena.
b. Immune factors contributing to acquired deficiency of complement.
The presence of C3 or C4 nephritic factors or autoantibodies against C3bBb and C4b2a can impair dissociation of complement molecular complexes and thus affect regulation of the AP and CP C3 convertases, respectively, leading to low levels of C3. Clinically, genetic or acquired deficiency of C1 inhibitor would also lead to excessive turnover and very low levels of complement C4.
In addition, complement C3 is a strong acute phase protein whose expression may be stimulated by many cytokines, including TGFβ. Infections are known to stimulate immune cell function and cytokine secretion, and thus can cause fluctuations in C3 independent of autoimmune disease activity. The only known cytokine that stimulates the expression of complement C4 is IFNγ, which may also be stimulated by infection (especially viral) (80).
c. Non-immunologic sources of complement protein level variation.
The primary site of biosynthesis for most complement proteins in humans is the liver, but adipose tissues and tissue-resident myeloid cells including macrophages also synthesize complement proteins for local defense (81). Therefore, liver disease and obesity can affect complement levels in a manner independent of their correlation with autoimmune disease. The body mass index (BMI) of an individual strongly correlates with serum C3 level and oftentimes levels of C4, as well (82, 83). As male subjects tend to have higher BMI than female subjects, it is also natural to observe slightly higher serum complement C3 and C4 levels in males than females (82, 84).
Because of variations in GCN and gene size dichotomy plus the influence of body mass indices, circulating cytokines, and medications, the baseline levels of complement C4 and C3 vary greatly among different human subjects. It is prudent to establish complement profiles in each patient with data acquired during disease remissions and disease flares and to compare levels across time to accurately interpret the patient’s current complement levels and to apply appropriate therapies.
IV. COMPLEMENT IN HUMAN SYSTEMIC LUPUS ERYTHEMATOSUS
Human SLE is an autoimmune disease characterized by the generation of autoantibodies against nuclear and cytoplasmic antigens accompanied by complement activation with dramatic longitudinal fluctuations of serum C4 and C3 levels and immune-mediated tissue injury (85). The etiology and pathogenesis of SLE are complex and involve multiple genetic risk factors and environmental triggers for disease onset, progression, and response to therapy.
A. Complement genetic deficiency and SLE
Not surprisingly given complement’s role in immune complex formation and antigen clearance, homozygous or complete deficiencies for early components of the CP (i.e., C1q, r, s, C4A and C4B, and C2) are amongst the strongest genetic risk or causal factors for human SLE, although their incidences are extremely rare (33). As a result of the strength of the associations between complement deficiencies and lupus, the role of the complement system in autoimmune disease is the best studied in lupus. Importantly, cases of complement deficiencies provide important insights into the pathogenic mechanisms of human lupus (Table 1).
a. Genetic deficiency of C1q
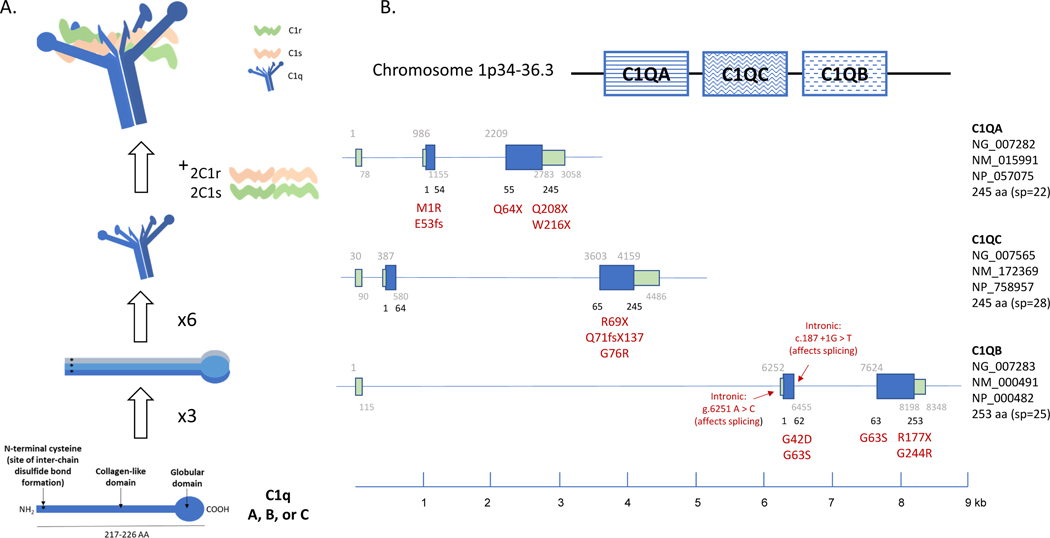
C1q consists of 18 polypeptides with hexamers of A, B and C chains intertwined together into a bouquet arrangement (Figure 3, panel A) (86). Missense or nonsense mutations in one of the A, B or C chains can disrupt the structure of C1q and render the protein non-functional. C1q also forms a complex with two C1r and two C1s subunits, and impaired interactions between these can cause functional deficiency (87).
Figure 3. C1 complex: structure and genetics.
(A) The structure of hexameric C1q in a complex with 2 subunits each of C1r and C1s (C1r2s2) is shown in a cartoon. The structural domains of each C1q subunit are outlined in the bottom panel, including the globular and collagen-like regions. Each C1q subunit is composed of C1qA, C1qB, and C1qC. Six of these subunits then assemble into a bouquet conformation as depicted. Note the N terminal cysteine residues, which are critical for disulfide bond formation between C1qA, B, and C. (B) The configuration of C1qA, C1qB, and C1qC on chromosome 1 is shown in the top panel with each gene structure represented diagrammatically below. Known mutations and their relative location in the various introns and exons of C1qA, C1qB, and C1qC are listed (89–95). GenBank accession numbers for C1q genes, mRNA and proteins are shown.
Clinical presentations of C1q deficiency.
Genetic deficiency of C1q is rare, with only 74 known cases (88, 89). A total of 17 nonsense mutations have been identified to cause C1q genetic deficiency (87, 90–94). The mutations that have been associated with disease are listed under their genetic location in C1qA, C1qB, and C1qC (Figure 3, panel B). Many patients described were of European or Middle Eastern ancestry. Homozygosity was almost exclusively the result of consanguineous marriages. While clinical presentations among C1q-deficient patients varied considerably, two common symptoms are overwhelmingly associated with a genetic deficiency of C1q: (a) SLE or lupus-like disease occurred in 88% of patients, and (b) recurrent bacterial infections in 41% of patients (87, 89–91, 95, 96).
As for SLE and lupus-like disorders, many patients had disease onset in early childhood. The range was 6 months to 42 years (37). The female to male sex ratio was close to 1:1. Many patients with C1q-deficiency died at a young age secondary to septicemia or renal failure. Among the C1q-deficient patients with SLE or lupus-like disease, cutaneous disorders, especially photosensitivity, were prominent with a frequency of 84%. Glomerulonephritis and neurologic disease affected about 30% and 19% of patients, respectively.
C1q deficiency leads to poorer clearance of apoptotic cells and increased exposure to potentially immunogenic autoantigens (97–100). Cai et al showed that UV damage-induced apoptosis caused C1q binding to nucleolar DNA and that C1q allowed C1r/s to degrade the nucleosomes, presumably decreasing their availability to trigger autoimmune responses (100). Furthermore, impairments in this pathway have been directly observed in vitro using cells from patients with SLE, with poorer C1q binding and impaired clearance of apoptotic cell debris by phagocytes (101). C1q was not sequenced, so it was not known if these results were due to C1q genetic deficiency, but the logical conclusion from this work is that C1q-mediated clearance of apoptotic debris is critical to prevent autoimmunity from developing and that this process can be disrupted by a lack of C1q or poor function of C1q in vivo, which might be inherited or acquired.
C1q deficiency is also associated with heightened levels of IFNα in the blood and cerebrospinal fluid and affects whether immune complexes are preferentially taken up by phagocytes rather than dendritic cells in vitro (102), which may be why plasmacytoid dendritic cells (pDC) from patients with C1q deficiency demonstrate higher IFNα production (59). As pDC are primary antigen presenting cells, increased uptake of immune complexes and stimulation of IFNα signaling is a plausible mechanism for increased adaptive immune activation and autoimmune responses seen in patients with C1q deficiency. Indeed, work by Hosszu et al suggests that locally synthesized C1q regulates dendritic cell (DC) differentiation and function in a way that negatively impacts self-tolerance (97, 103, 104). As IC clearance is decreased in a dose-dependent manner in C1q deficiency (55), it is also plausible that low C1q not only results in prolonged exposure to IC, but also to C1q itself, which may explain in part why patients with SLE are noted to frequently test positive for antibodies against C1q and other complement components (105, 106).
Therapy and potential cure of SLE with C1q-deficiency.
Plasmapheresis or infusion of fresh frozen plasma (FFP) can restore C1q activity in C1q-deficient patients temporarily and ameliorate lupus disease symptoms. However, complement activity drops off rapidly and frequent treatments are necessary (90, 107). Furthermore, FFP infusions bring with them their own risk of infection and thrombotic complications and are therefore a far from ideal therapy.
Unlike most other complement components, the primary site of biosynthesis for C1q is not in the liver but rather in myeloid cells including macrophages, monocytes, and dendritic cells, which originate from the bone marrow. Proof of concept that complement function can be reconstituted through bone marrow transplant has been shown in mouse models. Bone marrow transplantation (BMT) of hematopoietic stem cells from wild-type mice has been shown to be effective in treating C1q deficient animals (108, 109). BMT of allogenic hematopoietic stem cells therapy (HSCT) has been performed in three C1q deficient patients to date, two of which were successful and one of which resulted in the patient’s death (110, 111). The two successes demonstrate that allogenic hematopoietic stem cell transplant in humans can potentially restore complement function and eliminate an important factor contributing to lupus disease. However, there is a risk of considerable side effects, such as post-transplant lymphoproliferative disease, reactivation of latent viruses, and graft versus host disease. The two cases with excellent outcomes received bone marrow from siblings with matched HLA, which minimized graft versus host disease. Unfortunately, HLA-matched related donors are not always available. However, with the advent of CRISPR/cas9 technology for gene editing, correction of C1q deficiency using engineered autologous hematopoietic stem cells to cure SLE could become a reality.
b. Genetic deficiency of C1r or C1s
Deficiencies in sub-components of the C1 complex, C1r and C1s, were among the earliest reported linking complement deficiency with human glomerulonephritis or a lupus-like disease (112–115). A total of 20 cases of C1r and/or C1s deficiency have been reported, which includes 12 cases of C1r deficiency from eight families, and 8 cases of C1s deficiency from five families.
Clinical presentation of C1r or C1s deficiency.
Among the C1r/C1s deficient subjects studied, all but three had recurrent bacterial, viral, or fungal infections (85%). Many patients died at young ages due to severe infections. Thirteen subjects developed SLE or a lupus-like disease (65%). The female to male ratio among C1r/C1s deficient subjects with SLE was 1.5 to 1. Mortality at a young age due to fulminant infections likely explains the slightly lower frequency of lupus disease association. Most patients had severe cutaneous lesions. Eight patients had renal disease due to lupus nephritis (40%). The prevalence of anti-nuclear antibodies (ANA) among patients with SLE was only about 60% (116).
Whole exome sequencing of a multiplex family with SLE identified monogenic lupus with homozygous deficiency of C1r. A homozygous, single T-nucleotide deletion at coding sequence position 1332 of complement C1R (c1332delT) was found in all four SLE patients studied (117). This deletion resulted in the synthesis of a truncated protein without the serine proteinase domain in C1r. Of note, the same homozygous mutation was also detected in a 9-year-old female sibling who remained asymptomatic at the time of study. A heterozygous mutation was present in each of the four parents plus two other healthy siblings. Extensive screening of 300 patients with non-familial SLE, plus 1706 healthy subjects and 1618 patients with Behçet’s disease, all from Turkey, did not detect this mutation. Thus, the c1332delT in C1R of this Turkish family was a private, recessive mutation that caused SLE with high penetrance. Gene expression profiling using PAXgene RNA revealed enhanced type I interferon stimulated gene expression signature in patients with SLE but not the healthy subjects or carriers. Such IFN-I gene expression signature persisted among patients even without active disease (117), which is in contrast to findings in C4 deficiency will be discussed below (118).
Two patients with C1r deficiency, a female with disease onset at 13 years old and a male with disease onset at 7 years old, were further studied at the National Institutes of Health in the US (117). The female patient had neurologic and cognitive impairments and proteinuria with renal biopsy showing mesangial proliferative nephritis. Laboratory results showed that she had a strong neutrophil signature as defined by enhanced neutrophil extracellular trap (NET) formation when induced by bacterial lipopolysaccharide (LPS), and marked increase of low-density granulocytes (CD10+, CD14-low, CD15+) in PBMC. Her brother presented with proteinuria and a renal biopsy showed acute proliferative glomerulonephritis. Clinical laboratory tests revealed a significant increase in serum levels of cytokines and chemokines such as IL-2, IL-7, IL-10, IL-13, MCP-1, and MIP-1α and spontaneous and LPS-induced NET formation.
c. C2 or C3 deficiency
C2 deficiency.
Among individuals of European descent, C2 deficiency occurs with an estimated prevalence of 1/20,000, which probably accounts for <1% of SLE patients. There are two types of C2 deficiency (119, 120), caused either by nonsense mutations leading to the absence of protein biosynthesis (type 1, the predominant cause of deficiency) or by missense mutations C111Y, S189F, and G444R (121, 122) (type 2, only ~10% of C2 deficiency). One example of type 1 deficiency is a 28-bp deletion that generates a premature stop codon (123). This 28-bp deletion is present in the HLA haplotype with A10 (A25) and B18 in the class I region; BF-S, C2Q0, C4A4, and C4B2 in the class III region; and DRB1*15 (DR2) in the class II region. A second type 1 C2 deficiency is associated with the haplotype HLA A3, B35, DR4, BF-F, C2Q0, C4A3, and C4A2 (124).
Unlike C1 or C4 deficiency, the penetrance of C2 deficiency on SLE is only about 10%. Like other risk factors for SLE, there is a female predominance. C2-deficient SLE patients tend to have early childhood onset but a milder disease course with prominent photosensitive dermatologic manifestations, speckled ANA [a pattern common for the Ro (SSA) antigen], and a family history of SLE. Anti-DNA antibody tests are usually negative, and severe kidney disease is rare.
C3 deficiency.
C3 deficiency is generally associated with severe infections (125). Despite its prominent role in the complement functions of opsonization, phagocytosis, and clearance of immune complexes, C3 deficiency has not been found to associate with increased SLE predisposition except in Japanese subjects, with five out of six patients identified as C3 deficient being diagnosed with SLE (125–127).
d. Genetic diversity and deficiency of C4.
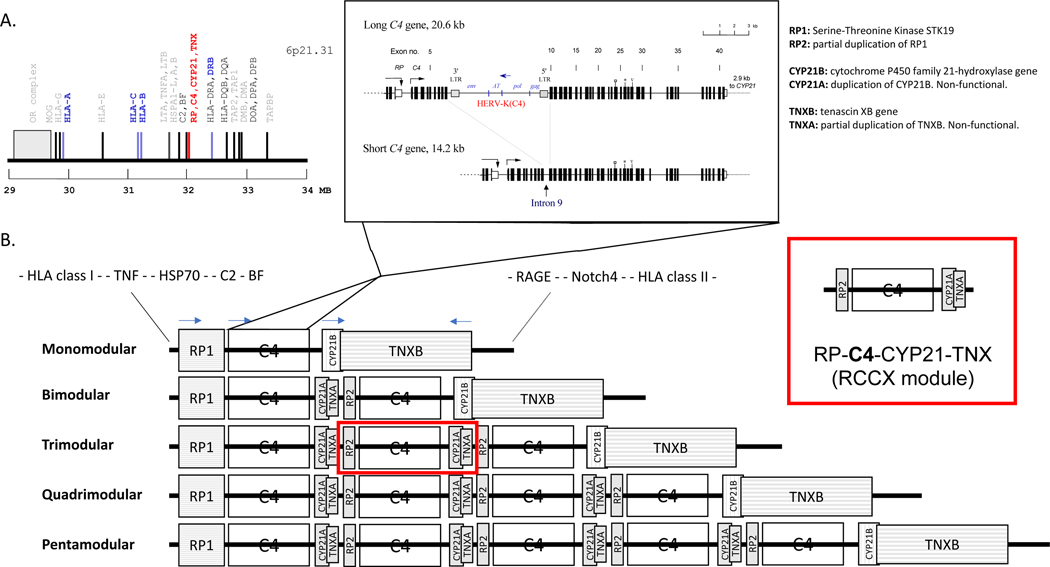
Figure 4 summarizes the unusual diversity seen in human C4 genetics. Panel A depicts the genetic locations for constituents of the C3 convertases—C4 and the enzymatic components C2 and factor B—in the class III region of the human major histocompatibility complex (MHC or HLA) on the short arm of the chromosome 6 (128–130). Panel B of Figure 4 illustrates the segmental duplications with one to five modules of the RP-C4-CYP21-TNX (RCCX) in haplotypes of the HLA (83, 131–136). Each duplicated segment, which includes C4 as well as CYP21, can be 30.6 or 24.2 kb in size. Panel B further depicts the exon-intron structures and the highly unusual gene size dichotomy of human C4 genes explaining this size variation (137). While each C4 gene consists of 41 exons (138), the long gene contains a 6.4 kb endogenous retrovirus, HERV-K(C4), in the ninth intron (137, 139, 140). In most ethnic groups, the majority (approximately 75%) of C4 genes are long genes. The single exception is in African American populations, in which only 60% of all C4 genes are long (141). The benefit or detriment of long versus short genes and the physiologic relevance of the coexistence of both isoforms of C4 genes are not yet well understood, though it has been proposed that the retrovirus may be a source of anti-sense mRNA that may be useful for antiviral defense and regulation of gene expression (139, 142).
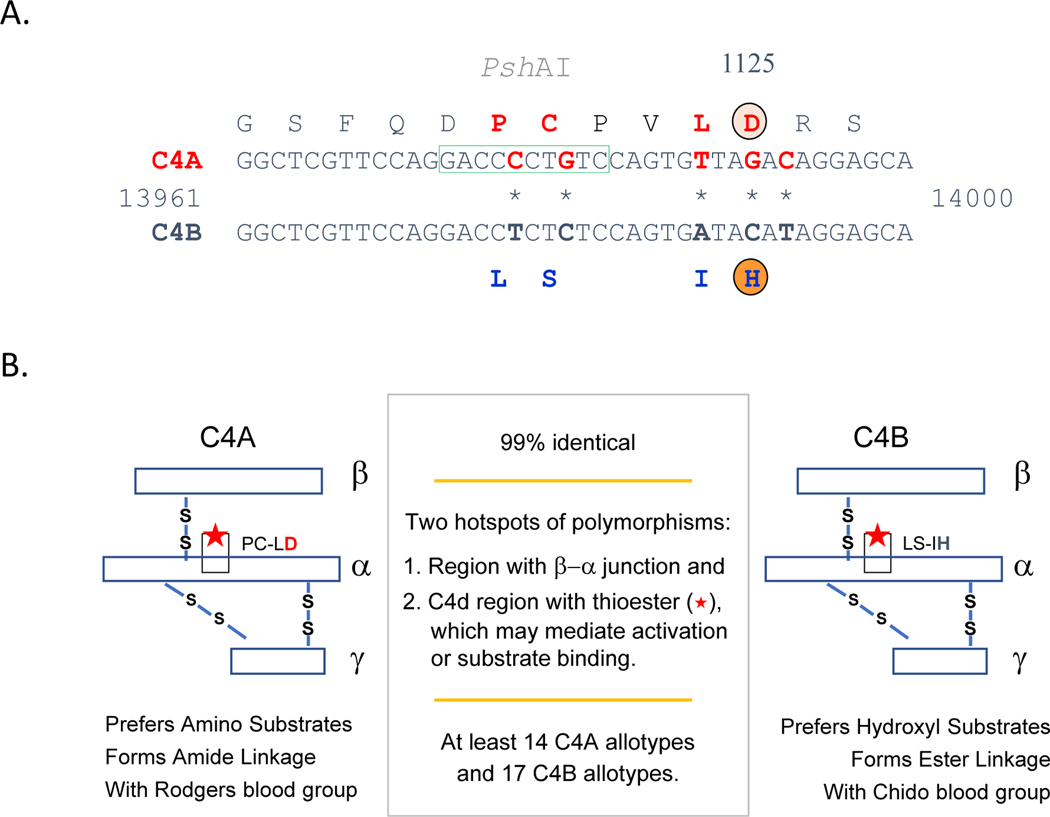
Figure 4. C4 genetics.
(A) Genetic locations for constituents of the C3 convertases for classical and alternative pathways. (B) Segmental duplications with one to five modules of the RP-C4-CYP21-TNX (RCCX) in haplotypes are located in the class III region of the HLA. Panel B inset: Dichotomy of human C4 gene size with the long gene containing endogenous retrovirus HERV-K(C4) in the ninth intron and the short gene without the endogenous retrovirus. Note the otherwise complex gene structure, with 41 exons and intervening introns (133, 134, 137, 138, 250).
Polymorphisms of C4A and C4B genes and proteins.
Figure 5 shows the molecular basis of isotypic changes for the acidic C4A and the basic C4B proteins (138, 140, 143). The boxed sequence in Panel A is specific for PshAI restriction enzyme cleavage, which has been used as a tool to distinguish the A and B isotypes of C4 through restriction fragment length polymorphisms (RFLP) (135, 144, 145). Panel B describes the functional implications of the biochemical differences between the two isotypes. In short, C4A is thought to be superior at binding amino groups, including those present in antibodies that contribute to immune complexes, while C4B tends to bind hydroxyl groups more readily, which may explain the finding that it is superior at binding to and triggering the lysis of red blood cells in vitro [70].
Figure 5. C4A and C4B genetic and functional differences.
(A) Gene sequence differences between C4A and C4B are shown with the corresponding amino acid differences noted above and below. (B) Graphical representation of C4A and C4B, including amino acid substitutions. Biochemical properties and bioreactivity for C4A and C4B are included (140, 143, 154–156). Note that despite high homology, there are dozens of different allotypes of C4A and C4B, allowing for incredible diversity.
Complete C4A and C4B deficiency.
A total of 28 individuals with a complete genetic deficiency of both C4A and C4B have been reported (146–150). These C4-deficient subjects come from 19 families, characterized by 16 different HLA haplotypes and with European, African, and Asian ancestries. Among those subjects with complete C4 deficiency, 22 (78.6%) were diagnosed with SLE or a lupus-like disease, and four others had renal disease such as glomerulonephritis. The female to male ratio was 1:1. Early disease onset, a severe photosensitive skin rash, presence of anti-Ro/SSA, and high ANA titers were common. The frequency of sepsis or severe recurrent infections was 29%.
Almost all completely C4 deficient patients were homozygous with HLA class I and class II markers for both copies of chromosome 6, suggesting consanguinity. The molecular basis of complete C4 deficiency has been determined in 15 cases (146–150). All nonsense mutations except one were private mutations that were only observed in the patient’s family or local community. The exception was a 2-bp insertion in exon 29 (codon 1232) of C4A that has an allelic frequency of 1 to 3% among Europeans (151–153).
In a European family from Austria and Northern Italy with multiplex lupus-related mortality and low serum C4 levels, the culprit was a heterozygous, recurrent haplotype with HLA-A30, B18 and DR7 that segregated with two defective and short C4B genes (C4B-C4B/SS) with identical mutations at the donor splice site of intron 28 (gt→at) (154). Notably such duplicated splice-junction mutations were found previously in two other families with complete (homozygous) C4 deficiency, vasculitis, and lupus nephritis who resided in the same geographic area (150). Such phenomena suggest that C4 hypocomplementemia caused by heterozygous deficiencies could increase the risk of SLE.
C4A or C4B isotype deficiency.
Similarly, in complement C4 isotype deficiency—especially that of C4A, which binds amino groups (and thus immune complexes) better than does C4B (155, 156)—reduced ability to opsonize immune complexes has been suggested as a putative trigger for autoimmune responses and disease (157, 158). In addition, in comparison with other complement components, C4 exhibits substantial heterogeneity in gene copy number (GCN) variation among different individuals, gene size dichotomy with long and short genes, DNA sequence and protein polymorphism, biochemical and serological properties, and expression levels, all of which may contribute to C4’s association with autoimmune conditions such as systemic lupus erythematous (SLE). Importantly, this layered system of diversity in innate immunity has not been fully appreciated in the past and is distinct to complement C4, suggesting critical and varied roles for this molecule in immune defense. This complex diversity will be discussed in depth in later sections of this review.
A retrospective chart review of medical histories for 32 patients with homozygous C4A deficiency, 87 patients with homozygous C4B deficiencies, and 120 patients without C4A or C4B deficiencies was performed in Helsinki University Hospital in Finland (159). Besides the increased prevalence of autoimmune disease such as SLE, it was found that patients with homozygous C4A deficiency had increased frequencies of lymphoma, celiac disease, and sarcoidosis. Moreover, patients with homozygous C4A or C4B deficiency tended to have a higher incidence of adverse drug reactions, especially to antimicrobials (159).
While complete or heterozygous genetic deficiency of C1 or both C4A and C4B are likely causal factors for SLE pathogenesis, the prevalence of these anomalies is extremely rare and not seen regularly in rheumatology or nephrology clinics. However, there are two common complement C4-associated risk factors that modulate susceptibility to and the clinical presentation of SLE. The first is the low copy numbers of total C4 genes or C4A genetic deficiency. The second is the generation of autoantibodies against neo-epitopes of complement proteins, which are readily observable in patients with lupus nephritis.
Interferon stimulated gene expression signatures in complement C4 isotype deficiency.
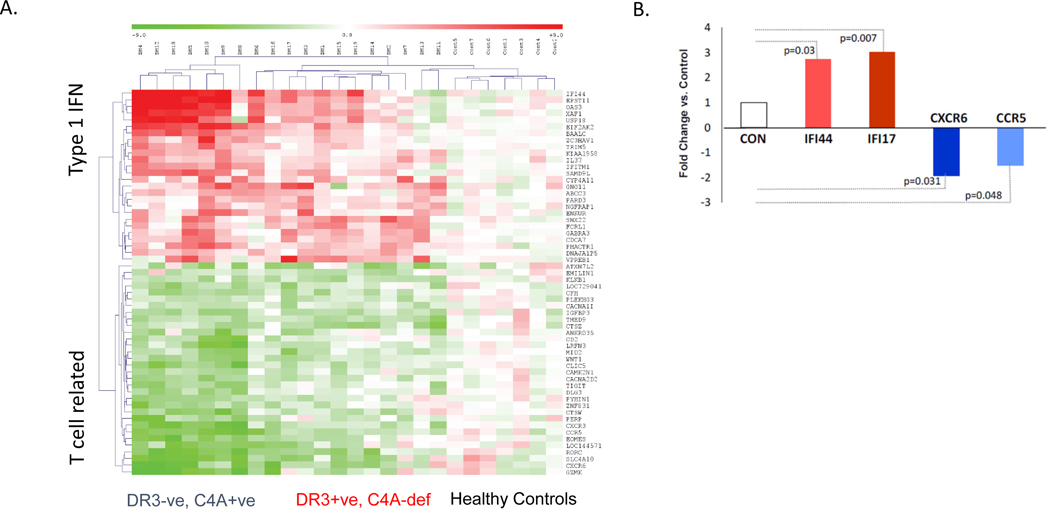
Type 1 interferon stimulated gene (ISG) expression signatures in the absence of infection have become recognized as a hallmark of many autoimmune diseases (160, 161). However, they are not conspicuous among patients with C4 (C4A) deficiency (or mice with C3 deficiency). In a study of SLE, ten patients with a C4 protein deficiency but normal C3 did not present with interferon α-induced gene expression signature (118). In this study, it was reasonably argued that patients with SLE who had persistently low C4 but normal C3 were likely to have a genetic deficiency (or low GCN) of C4, although this was not proven by a parallel genetic analysis. This observation was consistent with another study on patients with juvenile dermatomyositis (JDM), in which gene expression analyses using RNA microarray and real time PCR experiments of cDNA from peripheral blood transcripts revealed that patients who exhibited strong type I interferon-induced gene expression signatures had neither complement C4A genetic deficiency nor HLA-DR3 (Figure 6) (77).
Figure 6. Interferon signature in juvenile dermatomyositis.
(A) A heat map showing a comparison of blood transcripts between patients with JDM and healthy subjects. Type 1 interferon stimulated gene expression was prominent among seven patients without HLA-DRB1*03 who were C4A proficient (DR3-ve, C4A+ve, left columns). T-cell related transcripts were markedly reduced (green), suggesting migration of T lymphocytes away from blood to tissues (possibly to muscles and skin). Those with HLA-DRB1*03 and C4A deficiency (DR3+ve, C4A-def) demonstrated moderate IFN-stimulated gene expression and the reduction of T cell specific transcripts was also less remarkable (middle columns). Healthy control results are shown in the right most columns. (B) These results were further confirmed by SYBR-green qPCR assays for IFN-stimulated gene expression (red) and reduced expression of T-cell related genes (blue) (77).
One possible explanation for these findings is that lupus secondary to genetic C4 deficiency leads to activation of a distinct pathway not related to type I IFN signaling. In a murine model of lupus, mice exhibited strong type 1 interferon gene expression signature except when the mice had deficiencies of IgM, C3, or CD18. CD18 is the common chain for complement receptors CR3 and CR4. Thus, a pathway leading to elevated expression of interferon α or the presence of type 1 interferon induced gene expression signature in autoimmune rheumatic disease appears to require the presence of C4 (or C4A) in human, or C3, complement receptors, and natural IgM in mice (118). In the absence of C4, this pathway is less active and alternative mechanisms to promote inflammation and autoimmune disease must therefore be at the forefront of disease pathogenesis.
B. Gene copy number (GCN) variation of total C4 and C4A in SLE among different racial groups
a. C4 and C4A gene copy number among patients with SLE of European ancestry.
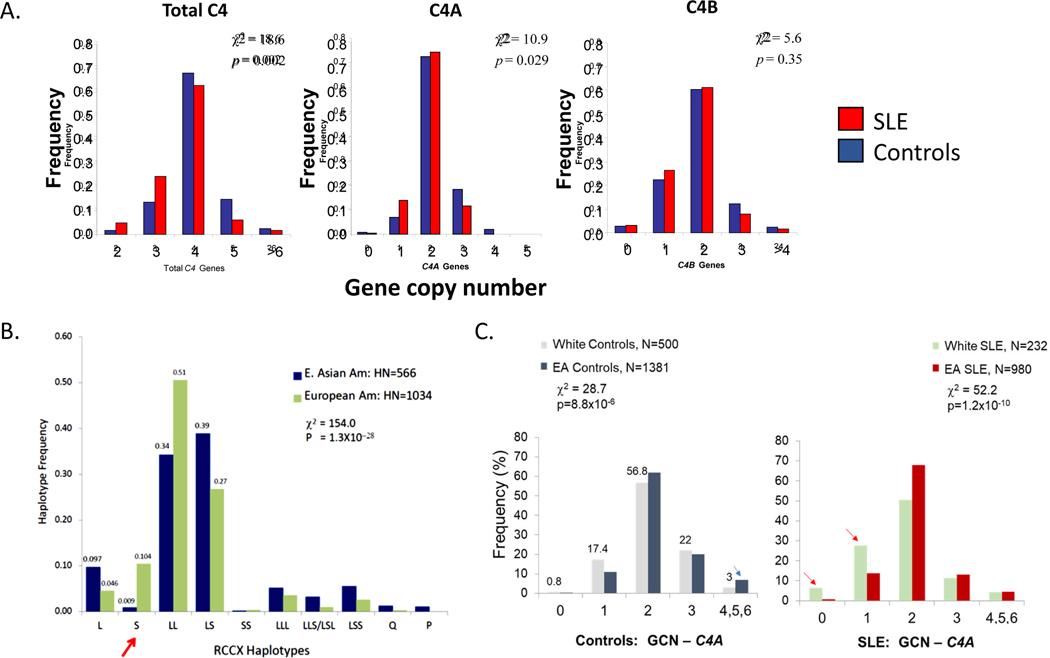
The first comprehensive study to investigate the complement C4 genetic diversities in SLE of European decent and race-matched controls was published in 2007 (153). The investigators employed (a) regular genomic Southern blot analyses of TaqI and PshAI-PvuII digested DNA resolved by regular agarose gel electrophoresis to define the relative dosage of C4A and C4B genes, (b) long-range mapping by PmeI digested genomic DNA fragments resolved by pulsed field gel electrophoresis, and (c) C4A and C4B protein phenotyping by immunofixation to define protein polymorphisms (144, 145, 153). The primary study population included 1241 European Americans with 233 SLE patients and 356 first degree relatives, plus 517 unrelated healthy controls (Figure 7, panel A; Table 2). In comparison to healthy controls, the distribution of total C4 and C4A, but not C4B, GCN was left-shifted in SLE patients. The risk associated with low C4 GCN was particularly evident among female SLE patients: 9.3% had only two copies of total C4 genes, and 6.5% had a homozygous deficiency of C4A, compared to 1.5% and 1.3%, respectively, in healthy controls. The risk of SLE (odds ratio, OR) for a subject with only two copies of total C4 genes in a diploid genome was 6.5 times greater (OR=6.5) than for those with three or more copies of C4 genes. As for subjects with homozygous deficiency of C4A genes, the risk of SLE was nearly 6 times greater than those with one or more copies of C4A genes (OR=5.7) (153).
Figure 7. Copy number variation of C4 genes in healthy subjects and patients with systemic lupus erythematous.
(A) Comparisons of gene copy number (GCN) groups for total C4, C4A, and C4B between healthy American subjects and patients with systemic lupus erythematous (SLE). Both groups were of European ancestry. SLE patients show significantly higher frequencies for lower copy numbers of total C4 and C4A but not C4B. More than one-third of patients with SLE had only 0 or 1 copies of the C4A gene, compared with one-fifth in healthy White control subjects. Lower panels: (B) Comparisons of C4 GCN groups in healthy subjects of either European or East Asian (EA) descent. (C) Comparison of C4A GCN groups in SLE patients versus healthy subjects (132, 153). White subjects are of European American descent.
Table 2.
Seven independent studies on associations of complement C4 gene copy number (GCN) variations with human autoimmune diseases
| Studies | Parameters | Disease Cases | Healthy Controls | P | OR (95% CI) |
|---|---|---|---|---|---|
| 1. European SLE: Ohio, USA; Southern blots, phenotyping (153) | |||||
| N | 232 | 500 | |||
| Sex: F, M | 216, 16 | 389, 111 | |||
| C4T, GCN ± SD | 3.56±.78 | 3.83±.69 | 3.56E-06 | ||
| C4T=2, % | 9.05 | 1.40 | 1.55E-06 | 7.00 (2.93–16.7) | |
| C4T=2+3, % | 42.2 | 29.2 | 0.0006 | 1.77 (1.28–2.45) | |
| C4A, GCN ± SD | 1.80± .90 | 2.09±.75 | 3.38E-06 | ||
| C4A=0, % | 6.47 | 0.80 | 1.70E-05 | 8.57 (2.81–26.1) | |
| C4A=0+1, % | 34.1 | 18.2 | 3.67E-06 | 2.30 (1.63–3.31) | |
| C4B, GCN ± SD | 1.78 ±.59 | 1.73±.63 | 0.40, ns | ||
| C4B=0, % | 3.02 | 3.00 | 0.99, ns | ||
| C4B=0+1, % | 22.2 | 28.50 | 0.077, ns | 0.72 (0.50–1.04) | |
| C4L, GCN ± SD | 2.64±1.16 | 2.95±.98 | 0.0002 | ||
| C4L=0, % | 5.17 | 0.60 | 9.71E-05 | 9.04 (2.52–32.3) | |
| C4L-0+1, % | 14.2 | 6.2 | 0.0005 | 2.51 (1.50–4.21) | |
| C4L=0+1+2, % | 45.3 | 32.4 | 0.0008 | 1.72 (1.25–2.37) | |
| C4S, GCN ± SD | 0.98±.85 | 0.89±.79 | 0.17, ns | ||
| C4S=0, % | 31.5 | 34.4 | 0.43, ns | ||
| C4A/C4T | 0.48±.20 | 0.54±.17 | 2.40E-05 | ||
| C4B/C4T | 0.52±.20 | 0.46±.16 | 6.90E-06 | ||
| C4L/C4T | 0.72±.27 | 0.76±.21 | 0.012 | ||
| C4S/C4T | 0.30±.28 | 0.24±.21 | 0.0006 | ||
| 2. East Asian SLE: Taiwan, Hong Kong, Ohio, Southern blots, real time PCR, phenotyping (132) | |||||
| N | 999 | 1347 | |||
| Sex: F, M | 920, 79 | 700, 674 | |||
| C4T, GCN±SD | 3.95±.97 | 4.14±.92 | 3.70E-07 | ||
| C4T=2 | 2.1 | 1.55 | 0.33, ns | ||
| C4T=2+3 | 27.0 | 20.3 | 0.0002 | 1.45 (1.20–1.77) | |
| C4A, GCN± SD | 2.09±.79 | 2.25±.82 | 3.40E-06 | ||
| C4A=0, % | 0.9 | 0.07 | 0.0015 | 12.4 (1.57–97.9) | |
| C4A=0+1, % | 14.7 | 11.2 | |||
| C4B, GCN±SD | 1.85±.68 | 1.88±.71 | 0.24 | ||
| C4B=0, % | 1.14 | 2.28 | 0.038 | 0.50 (0.24–0.99) | |
| C4B=0+1, % | 27.9 | 25.8 | 0.25, ns | ||
| C4L, GCN±SD | 2.89±.79 | 2.93±.82 | 0.24, ns | ||
| C4L=0 | 0 | 0 | |||
| C4L=1 | 0.88 | 0.26 | 0.062, NS | 3.40 (0.88–13.2) | |
| C4L=1+2 | 33.2 | 32.7 | 0.82 | NS | |
| C4S, GCN±SD | 1.04±1.00 | 1.19±.96 | 0.0008 | ||
| C4S=0 | 31.4 | 22.3 | 6.50E-06 | 1.60 (1.30–1.96) | |
| C4S=0+1 | 75.3 | 68.9 | 0.0022 | 1.37 (1.12–1.68) | |
| C4A/C4T | 0.532±.151 | 0.543±.157 | 0.10 | ||
| C4B/C4T | 0.466±.151 | 0.450±.155 | 0.01 | ||
| C4L/C4T | 0.625±.338 | 0.615±.315 | 0.47 | ||
| C4S/C4T | 0.202±.202 | 0.230±.197 | 0.0009 | ||
| 3. Hispanic SLE: Sao Paolo, Brazil; Real-time PCR (168, 169) | |||||
| N | 427 | 301 | |||
| Sex: F, M | 406, 21 | 286, 15 | |||
| C4T (GCN ± SD) | 3.87±.96 | 4.17±.83 | <0.001 | ||
| C4T=2, % | 7.0 | 1.0 | <0.001 | ||
| C4T=2+3, % | 29.0 | 13.6 | <0.001 | 2.62 (1.77–3.87) | |
| C4A (GCN ± SD) | 2.06 ±.86 | 2.24±.73 | 0.001 | ||
| C4A=0, % | 2.1 | 0.3 | 0.053 | ||
| C4A=0+1, % | 20.4 | 6.6 | <0.001 | 3.59 (2.15–5.99) | |
| C4B (GCN± SD) | 1.87 ±.77 | 1.95±.71 | 0.19 | ||
| C4B=0, % | 3.3 | 1.7 | 0.24 | ||
| C4B=0+1, % | 27.1 | 20.2 | 0.033 | 1.46 (1.03–2.08) | |
| 4. European SLE: Germany, Real-time PCR (162) | |||||
| N | 169 | 520 | |||
| Sex: F, M | 154, 15 | 471,49 | |||
| C4T, GCN | na | na | |||
| C4T=2, % | 6.9 | 1.9 | 0.0068 | 3.70 (1.50–9.10) | |
| C4T=2+3, % | 43.8 | 31.3 | na | ||
| C4A GCN | na | na | |||
| C4A=0, % | 4.3 | 0.8 | 0.0077 | 5.33 (1.54–18.4) | |
| C4A=0+1 | 37.4 | 21.9 | na | ||
| C4B=0 | 0 | 2.3 | na | ||
| C4B=0+1 | 17.1 | 25.3 | na | ||
| 5a. European SLE: London, UK, Paralog ratio test (PRT)* (242) | |||||
| N | 501 | 719 | |||
| C4T, GCN ± SD | 3.79±.98 | 3.89±.76 | 0.046 | ||
| C4T=2, % | 7.18 | 3.47 | |||
| C4T=2+3, % | 38.7 | 26.8 | |||
| C4A, GCN ± SD | 1.82±.93 | 2.08±.83 | <0.001 | ||
| C4A=0, % | 5.18 | 1.25 | |||
| C4A=0+1, % | 38.1 | 22 | |||
| C4B, GCN ± SD | 1.96±.82 | 1.81±.72 | <0.001 | ||
| C4B=0, % | 2.6 | 3.6 | |||
| C4B=0+1, % | 27.5 | 29.3 | |||
| 5b. South European SLE: Barcelona, Spain; Paralog ratio test (PRT)* (242) | |||||
| N | 527 | 460 | |||
| C4T, GCN ± SD | 3.80 ± .92 | 4.13 ± 1.02 | <0.001 | ||
| C4T=2, % | 6.03 | 4.23 | |||
| C4T=2+3, % | 36.9 | 23.4 | |||
| C4A, GCN ± SD | 1.82 ± .93 | 2.08 ± .83 | |||
| C4A=0, % | 5.18 | 1.25 | |||
| C4A=0+1, % | 25.0 | 13.1 | |||
| C4B, GCN ± SD | 1.96±.82 | 1.81±.72 | <0.001 | ||
| C4B=0, % | 3.00 | 2.70 | |||
| C4B=0+1, % | 39.6 | 29.7 | |||
| 6. Myositis: US, UK, Czech Republic, Sweden, Belgium; real time PCR, Southern blots, phenotyping (79) | |||||
| N | 1632 | 3276 | |||
| Sex: F, M | 1091, 541 | 2390, 886 | |||
| C4T, GCN ± SD | 3.50±.78 | 3.83±.76 | 1.40E-46 | ||
| C4T=2, % | 7.14 | 3.29 | 2.03E-09 | 2.26 (1.74–2.95) | |
| C4T=2+3, % | 52.63 | 29.8 | 2.81E-55 | 2.62 (2.32–2.95) | |
| C4A, GCN ± SD | 1.74± .88 | 2.10± .84 | 6.00E-46 | ||
| C4A=0, % | 4.24 | 1.74 | 1.90E-03 | 2.49 (1.76–3.54) | |
| C4A=0+1, % | 42.87 | 21.01 | 8.70E-22 | 2.82 (2.48–3.21) | |
| C4B, GCN ± SD | 1.74± .59 | 1.73± .62 | 0.45 | ||
| C4B=0, % | 2.09 | 3.15 | 0.03 | 0.659 (.446-.972) | |
| C4B=0+1, % | 29.21 | 30.43 | 0.11 | ns | |
| C4L, GCN ± SD | 2.41± 1.13 | 2.94± 1.08 | 1.70E-54 | ||
| C4L=0, % | 3.67 | 1.38 | 5.09E-07 | 2.72 (1.85–4.02) | |
| C4L-0+1, % | |||||
| C4L=0+1+2, % | 51.62 | 33.4 | 2.74E-49 | 2.13 (1.77–2.56) | |
| C4S, GCN ± SD | 1.06±.72 | 0.90±.77 | 3.00E-12 | ||
| C4S=0, % | 21.1 | 32.3 | 8.80E-17 | 0.561 (.486-.645) | |
| C4S=0+1 | 74.59 | 80.05 | 1.44E-05 | 0.732 (.636-.842) | |
| 7. Graves’ Disease: Taiwan; real time PCR, phenotyping (243) | |||||
| N | 624 | 160 | |||
| F, M | 491, 133 | 128, 32 | |||
| C4T=2+3, % | 21.5 | 32.5 | 0.003 | 0.51 (0.34–0.78) | |
| C4A=0+1, % | 12.7 | 20.6 | 0.01 | 0.58 (0.36–0.95) | |
| C4B=0+1, % | 22.9 | 33.1 | 0.008 | 0.49 (0.32–0.74) | |
Abbreviations: GCN, gene copy number (mean); C4T, total C4; C4L, long C4; C4S, short C4; C4A, acidic C4; C4B, basic C4; F, female; M, male; SD, standard deviation
Note: Two recent publications not included in this list are Lundtoft et al. (247) and Kamitaki et al. (248) that engaged data from read-depths of next-generation sequencing and imputation of single nucleotide polymorphisms.
Results for entry Number 5 on British and Spanish subjects have been discussed in reference (33).
Parallel increases in the frequency of heterozygous C4A deficiency (GCN=1), and moderately low copy number of total C4 (GCN=3) were also observed amongst patients with SLE, but these yielded smaller effect sizes or impact on disease risk (153). On the other hand, human subjects with high copy-numbers of total C4 or C4A genes were protected against SLE disease risk. The frequency of subjects with C4T≥5 was 6.0% in patients with SLE but 12.0% in controls (OR=0.47). Similarly, 15.3% of subjects with SLE had C4A≥3 in comparison with 23.8% of controls (OR=0.57). In the same study, family-based association tests further revealed that monomodular-short (mono-S) RCCX haplotypes with single C4B gene and the absence of C4A (i.e., deficiency) were a risk factor for SLE (Figure 7, panel B and red arrow). The mono-S haplotype with C4A deficiency is in strong linkage disequilibrium with HLA-DRB1*03:01 (also known as HLA-DR3) among Caucasian subjects (153).
The natural conclusion from these results is that low gene copy number of total C4 or C4A is a risk factor for, and high copy-number of total C4 or C4A is a protective factor against, SLE disease susceptibility in European-Americans (153). A replication study including 128 independent patients of Caucasian descent with SLE yielded similar results. It must be acknowledged, however, that linkage disequilibrium with HLA-DR3 confounds a causal analysis.
Nevertheless, additional studies have yielded similar results. TaqMan-based real time PCR was employed to interrogate copy number variation (CNV) of C4A and C4B in 169 patients with SLE and 520 healthy controls in Kiel, Germany (162, 163). C4T GCN was calculated by summing GCNs of C4A and C4B. Low C4 GCN (C4T=2) was more common in subjects with SLE compared with controls: 6.9% versus 1.9%, respectively (OR=3.7, p=6.8×10−3). In addition, 4.3% of subjects with SLE demonstrated homozygous C4A deficiency (C4A=0), compared with only 0.8% of controls (OR=5.33, p=7.7×10−3). Intragroup analyses of German patients with SLE revealed that patients with homozygous C4A deficiency experienced a more severe disease course based on their requirement of cyclophosphamide therapy (OR=5.8, p=0.034). Patients with homozygous C4A deficiency were also diagnosed with SLE about ten years earlier than those without C4A deficiency (24 versus 34 years of age).
b. Low copy number of C4 and C4A deficiency in East Asians.
SLE affects all racial groups, but patients of East Asian and African ancestries and those of Hispanic ethnicity have more severe disease including a higher prevalence of renal complications (164). Although risk factors for SLE identified in European populations, such as C4A deficiency, are germane to other racial groups (165), there are substantial differences between races with respect to the frequency and effect sizes of risk factors. In a study including 999 patients with SLE and 1,347 healthy subjects of East Asian ancestry from Taiwan, Hong Kong, and the US, gene copy number variations of total C4, C4A, and C4B and their polymorphisms were determined by either TaqMan-based real time PCR (163) or by genomic Southern blot analyses and protein phenotyping (132, 144). In this study population, high GCN of total C4 (GCN≥5) and C4A (GCN≥3) were protective factors against SLE, whereas medium and low copy numbers of total C4 and C4A were risk factors for SLE (Table 2).
Homozygous deficiency of C4A is infrequent among East Asian subjects but has a remarkable odds ratio of 12.4 for SLE disease susceptibility (p= 0.0015). The haplotype including HLA-DR3 (or DRB1*03:01) and mono-S C4 (with one copy of C4B and total C4A deficiency) is basically absent among East Asians. Instead, East Asian subjects with C4A deficiency usually present with bimodular LS (long-short) RCCX with two C4B genes (i.e., C4B-C4B, but no C4A), a haplotype that is linked to HLA DRB1*15:01 (or DR2), which is common amongst Chinese patients with SLE (132).
Comparisons of C4A GCN variations between European and East Asian healthy subjects and between patients with SLE revealed highly significant differences in the distributions of gene copy number groups for C4A (Figure 7, panel B). Notably, European subjects were more likely to have low copy numbers of C4A, which was especially evident when considering only subjects with SLE (Figure 7, right side of panel C). It is notable that mono-modular short-RCCX with a single short C4B gene (i.e., S or mono-S) is quite prevalent among White subjects but infrequent among East Asian subjects (red arrow in Figure 7, panel B). Other differences are seen, as well. For example, the most common haplotype for White subjects is bimodular LL with a frequency of 51% and for East Asian subjects is bimodular LS with a frequency of 39%.
In two independent studies of East Asian SLE in Han-Chinese and South Koreans, C4A and C4B gene copy numbers were also interrogated by TaqMan-based real time qPCR methods (163, 166, 167). Significantly lower GCN of C4A and total C4 were observed in SLE compared with race-matched controls. Again, a notable feature in those two East Asian populations was the low frequency of homozygous C4A deficiency in both SLE (≤1%) and healthy controls.
c. Low GCNs of complement C4 in SLE of Hispanic populations.
A slightly different version of TaqMan real time PCR methodology was used to determine C4 GCN for 427 patients with SLE and 301 matched healthy controls from São Paulo, Brazil (168). Subjects’ ethnicity was not described, but Brazil is known to be a melting pot for European, African, indigenous, and Asian ethnicities. Mean copy numbers for total C4 (C4T) and C4A were statistically lower in subjects with SLE compared with controls. Low C4T GCN (C4T<4) and C4A deficiency (C4A GCN<2) more common in subjects with SLE compared to controls with odds ratios of 2.62 and 3.59, respectively. Homozygous deficiency of C4A was more common in subjects with SLE: 2.1% compared with 0.3% of controls (OR=6.49; p=0.053). It was noted that low gene copy number of C4B also appeared to be a moderate risk factor for SLE among the Brazilian subjects, which was not found in studies of either Europeans/European Americans or East Asians/Asian Americans outlined above. Furthermore, low GCN of C4A was associated with higher permanent disease damage indices and higher frequencies of serositis in subjects with SLE. Similarly, patients with lower GCN of C4B had a higher frequency of arthritis. An earlier publication from the same team found that juvenile onset patients with SLE had lower GCN of total C4, C4A, and C4B than adult-onset patients (169).
C. APPLE clinical trial investigating complement in SLE
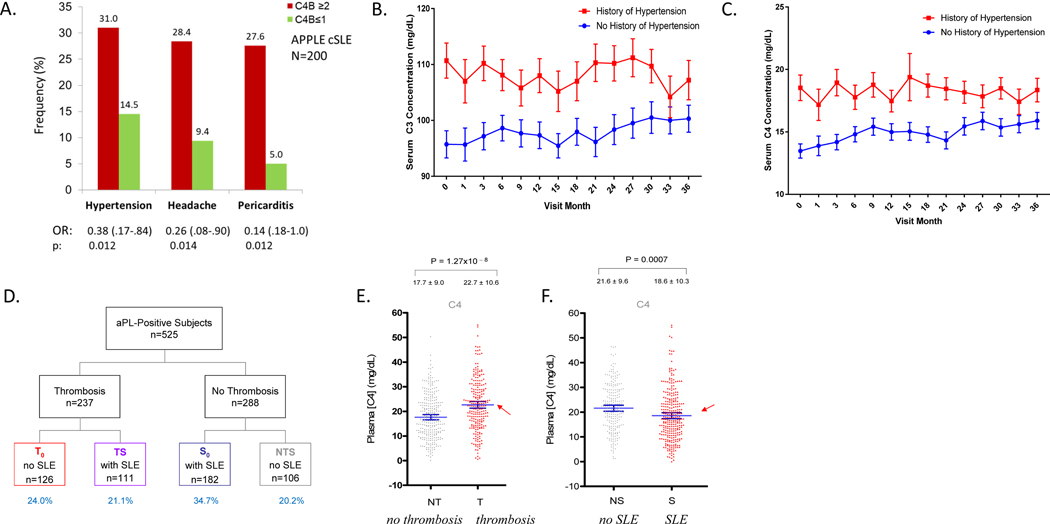
Leveraging biorepository samples and data from the clinical trial APPLE (atherosclerosis prevention in pediatric lupus erythematosus), Mulvihill and colleagues determined CNV of C4 genes in 200 children with SLE (cSLE) (170, 171). This three-year trial recruited patients between 10 and 21 years old for randomized atorvastatin or placebo-controlled therapy. Patients with ≥2 copies of C4B genes had 2.5 times the odds of having hypertension and higher diastolic blood pressure (Figure 8, panel A). C4B GCN≥2 was also associated with headaches and pericarditis, though potential mechanisms relating complement with these problems were not elucidated. Patients with ≥2 copies of C4B genes also appeared to respond more effectively to atorvastatin treatment with reduced progression of atherosclerosis as shown by slower increase in carotid intima media thickness, a surrogate marker of atherosclerosis, when compared with those treated with placebo (170).
Figure 8. Complement, systemic lupus erythematous, and anti-phospholipid syndrome.
(A) Clinical differences in the frequency of reported hypertension, headache, and pericarditis between patients with more than two copies of C4B genes (red columns) and those with less than two copies of C4B genes (green columns) (170). Patients with hypertension consistently presented with higher C3 (panel B) and C4 (panel C) serum protein levels (red curves) than those without (blue curves). (D) Study design of patients with anti-phospholipid (aPL) antibodies. Subjects were divided into groups based on the presence or absence of thrombosis and systemic lupus erythematous (SLE). T0= patients with thrombosis only. TS= patients with both thrombosis and SLE. S0= patients with SLE only. NTS= patients with neither thrombosis nor SLE. (E) Remarkably, patients with aPL antibodies and thromboses had significantly higher serum C4 protein levels than those without thromboses. (F) As expected, patients who were diagnosed with SLE who also had aPL antibodies had lower levels of serum C4 than those who did not present with SLE (right bottom panel) (175).
In the APPLE trial, serum complement protein levels also varied with the severity of disease sequelae. Longitudinal studies of serum complement protein levels in SLE patients have revealed three distinct serum complement profiles, as discussed previously (72, 73, 172, 173). As serum complement protein levels fluctuate, so do levels of complement degradation products like C4d. C4d can be found in the fluid phase or attached to membranes of red blood cells (RBC-C4d ) and other cell types in the circulation (76, 78, 174). The deposition of C4d on cell surfaces can be monitored by flow cytometry using antibodies against C4d. It is suggested that levels of RBC-C4d reflect complement activation in the past 60 days, the levels of reticulocyte-C4d the past 2–3 days, and platelet-C4d the past 5 to 10 days. In the APPLE trial, patients with hypertension were found to have significantly higher serum levels of complement C3 and C4 than normotensive patients at the baseline and throughout the trial study period (Figure 8, panels B and C). The relatively higher complement C4 protein levels among patients with hypertension, as well as amongst patients with antiphospholipid (aPL) antibodies and thrombosis or recurrent pregnancy loss (RPL) (175) reveal the pro-inflammatory aspect of complement. Normal range or elevated levels of complement in these patients likely aggravated complement-mediated tissue damage.
D. Opposite profiles of complement in SLE and antiphospholipid syndrome
Antiphospholipid syndrome (APS) is defined by the persistence of aPL on two laboratory tests separated by 6–8 weeks and clinically by thrombosis or recurrent pregnancy loss (176, 177). Antiphospholipid antibodies are heterogeneous autoantibodies whose targets include β2 glycoprotein I (β2GPI), prothrombin, and phospholipids found in the cell membrane, including cardiolipin. Some of these antibodies can interfere with clotting in vitro assays and therefore have been dubbed lupus anticoagulants, though in vivo they predispose patients towards thrombosis rather than bleeding events. β2GPI is an enigmatic plasma protein that consists of five short consensus repeats (SCR), which are characteristic of complement control proteins (178–180). Similar to ANA, aPL antibodies form immune complexes and fix complement (181, 182). Anti-phospholipid antibodies are detectable in ~30% of SLE patients, and 10–15% of SLE patients are diagnosed with APS (183–186).
Concurrence of thrombosis and SLE imposes a very high risk of recurrent pregnancy loss; 40% of aPL-positive patients with recurrent pregnancy loss had thrombotic SLE in one study (175). Remarkably, all aPL antibody-positive female patients with homozygous C4A deficiency experienced recurrent pregnancy loss. In contrast, female patients with homozygous C4B deficiency were protected from recurrent pregnancy loss (187). This phenomenon underscores the importance of C4A in the protection against the onset of autoimmune disease and the engagement of C4B in complement-mediated tissue injury. Similarly, genetic ablation of C1q, C4, C3, or C5 in a rodent model protected against aPL antibody-induced clinical symptoms such as fetal resorption (181, 188–193). While complement activation is important in the pathology of both APS and SLE, the genetic and immunological profiles of complement in these related diseases are very different (187). For example, the thrombo-inflammatory process in APS seems to drive not only complement activation and consumption (as is seen in SLE), but also higher production of component proteins, particularly C4 (187, 194).
In a cross-sectional study of 525 human subjects with aPL (187), 45% developed thrombosis, 56% had SLE, and 24% of female patients experienced recurrent pregnancy loss (Figure 8, panel D). Meticulous analyses of complement proteins among these aPL antibody-positive patients revealed significantly higher levels of C4 and C3 proteins among patients with thrombosis only, and lower levels of these proteins among patients with SLE only (Figure 8, panels E and F). Amongst all aPL antibody-positive patients, those with concurrent SLE but not thrombosis had significantly higher factor H and functional MBL levels (187, 195, 196). Patients with APS also demonstrated elevated levels of complement activation products such as the anaphylatoxins C5a and C3a and the MAC (181, 195). aPL antibody-positive patients with both SLE and thrombosis and subjects without either disease had intermediate C4 and C3 protein levels.
E. Autoantibodies to complement in SLE
There are many autoantibodies to complement proteins in SLE (197), which are mostly directed against neo-epitopes exposed after activation or inactivation of complement proteins or against multi-molecular complexes formed during the activation process. The binding of autoantibodies to complement can lead to a state of acquired complement deficiency and contribute to disease pathogenesis.
a. Autoantibodies to C1q
About 30% of SLE patients synthesize autoantibodies to C1q. The presence of C1q autoantibodies correlates with anti-dsDNA, nephritis, and low complement levels in about 75% of such patients (60, 198, 199). The development of these autoantibodies may be a secondary phenomenon that occurs when immune complexes (IC) bind C1q and activate the classical pathway (CP). Following CP activation, the C1 inhibitor strips the C1r and C1s proteases from the C1 complex, which is bound to Ab. C1q remains attached to the IC at the site of inflammation. Local proteases will next degrade IgG, C1q, and the autoantigen and generate multiple proteolytic fragments and neoantigens, to which novel autoantibodies may be generated. A large study was undertaken to assess the specificity of anti-C1q antibodies and their potential association with SLE manifestations and diagnostic tests (200). The authors confirmed an association between anti-C1q antibodies, low complement (C4 and C3), and anti-dsDNA antibodies. This combination also had the strongest serological association with the clinical development of renal disease. Anti-C1q antibodies were seen in 28% of all SLE patients but 68% of patients with renal disease. By contrast, the absence of anti-C1q antibodies makes a nephritis flare less likely (199).
b. Autoantibodies to C3 or C4
Immunoconglutinins were among the earliest complement-directed autoantibodies discovered (201). Immunoconglutinins are autoantibodies against solid-bound or processed fragments of C3. The IgG isotype of immunoconglutinins is strongly correlated with SLE disease exacerbation (202), increased anti-C1q IgG level and anti-dsDNA, and lower levels of serum C3 and/or C4 in ~30% patients with lupus nephritis (203, 204). Binding of such autoantibodies to C3b interfere with the interaction between C3b and regulatory molecules such as factor H or CR1 and thus exacerbates the activation and consumption of serum C3. Increased C3b autoantibodies have been observed to be more specific but less sensitive than anti-C1q for lupus nephritis flares (205).
C3 and C4 nephritic factors bind to neoepitopes present in the AP C3 convertase (C3bBb or C3bBbP) and the CP C3 convertase (C4b2a), respectively. They prolong the half-life of the convertases from minutes to hours and enhance the consumption of serum C3 and variably C5 as well. Both C3 and C4 nephritic factors are associated with membranoproliferative glomerulonephritis (206, 207). C3 nephritic factors are associated with acquired partial lipodystrophy (206, 207). C4 nephritic factors are associated with SLE and post-infectious acute glomerulonephritis (208, 209).
V. COMPLEMENT IN THE IDIOPATHIC INFLAMMATORY MYOPATHIES
IIM are a group of autoimmune diseases characterized by myositis-related autoantibodies, infiltration of leukocytes into muscles and/or the skin, and destruction of blood vessels and muscle fibers, which together cause chronic weakness and fatigue. While complement-mediated destruction of capillary endothelia is implicated in pediatric and adult dermatomyositis, the complex role of C4 in IIM pathology is largely unknown.
A. C4 genetic diversity and IIM
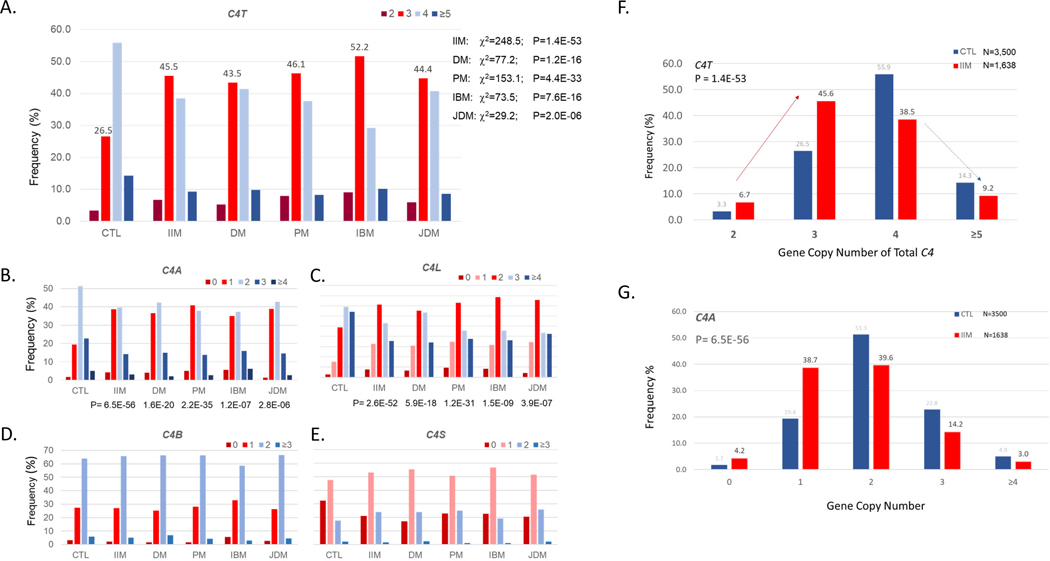
C4 GCN were quantified for 1,644 Caucasian patients with IIM plus 3,526 matched healthy controls using real-time PCR or Southern blot analyses, as summarized in Figure 9 (79). C4T GCN (C4T<4) and C4A deficiency were correlated with increased risk of IIM with odds ratios of 2.58 (2.28–2.91, p=5.0×10−53) for C4T; and 2.82 (2.48–3.21, p=7.0×10−57) for C4A. Contingency analyses showed that amongst patients with C4A deficiency, the presence of HLA-DRB1*03 (DR3) became insignificant as a risk factor in IIM except for inclusion body myositis (IBM), in which 98.2% of patients had HLA-DR3 with an OR of 11.02 (1.44–84.4). This study revealed that C4A deficiency is a relevant risk factor in both juvenile-onset and adult-onset dermatomyositis, that both C4A deficiency and HLA-DRB1*03 contribute interactively to the risk of polymyositis, and that HLA-DRB1*03 is singularly important in IBM (79).
Figure 9. Complement C4 gene copy number variation in the idiopathic inflammatory myopathies.
(A through E) Gene copy number (GCN) variation of total C4 (C4T), C4A, C4 long (C4L), C4B, and C4 short (C4S) between healthy control subjects (CTL) and patients with idiopathic inflammatory myopathy (IIM) and its major subgroups: dermatomyositis (DM), polymyositis (PM), inclusion body myositis (IBM), and juvenile dermatomyositis (JDM). Note that more than half (52.3%) of patients with IIM had C4T GCN<4 (panel F); C4A deficiency (C4A GCN<2) was present in 42.9% of the IIM patients (panel G) (77, 79).
Further analyses revealed that low GCNs of total C4, i.e., C4T=2 [OR=2.26 (1.74–2.95)] and C4T=2+3 [OR=2.62 (2.32–2.95)], had similar magnitude of effects on the genetic risk of IIM. In the case of C4A deficiency, 4.2% and 38.6% of subjects with IIM had 0 and 1 copies of C4A, respectively, with odds ratios of 2.49 (1.76–3.54; p=3.6×10−7) for C4A=0 and 2.82 (2.48–3.21; p=2.9 ×10−57) for C4A=0+1. Of note, this finding is analogous to an autosomal dominant phenotype in Mendelian genetics and stands in stark contrast with results in SLE, in which the risk associated with C4A deficiency exhibits gene dosage effects (OR= 5.27 for C4A=0, OR=1.61 for C4A=1).
B. Serum complement levels and their activation products in IIM
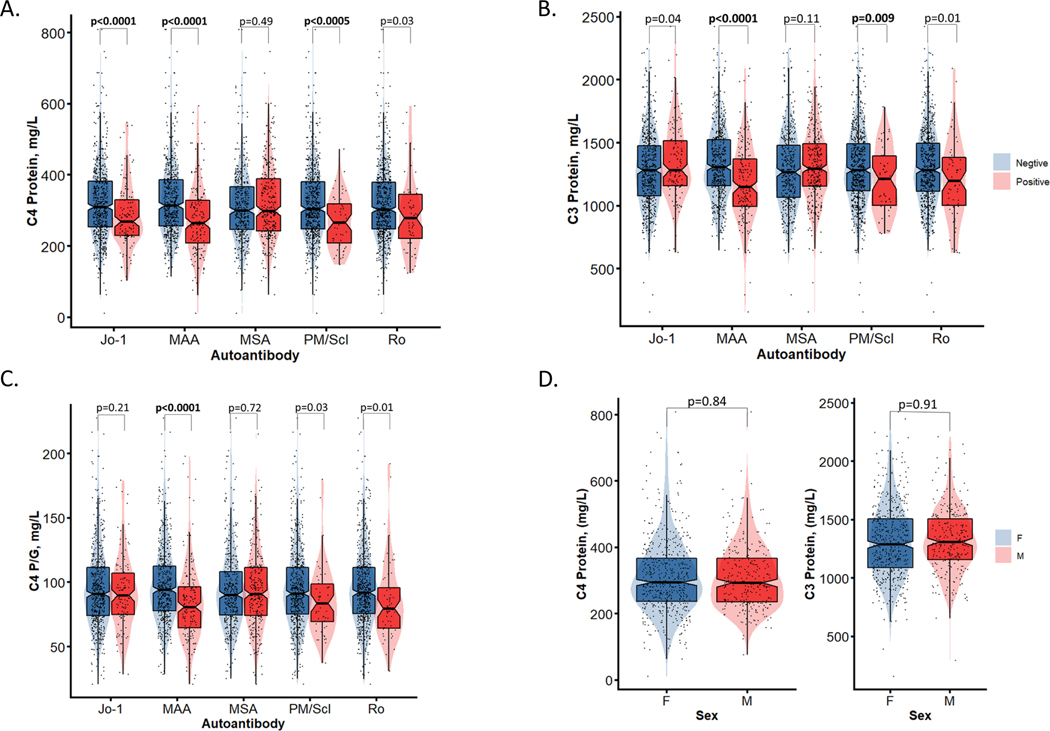
Phenotypic studies support a pathological relationship between circulating complement protein levels and IIM. Our work has shown that in addition to differences in circulating complement protein levels between healthy subjects and patients with IIM, there are also differences between subgroups of IIM patients, especially when comparing those with or without specific myositis-specific (MSA) and myositis-associated (MAA) antibodies, as shown in Figure 10 (210). These data suggest that MSA and MAA may modulate complement protein levels and activity, possibly through formation of immune complexes that in turn activate and consume complement.
Figure 10. Comparisons of plasma protein levels among patients with idiopathic inflammatory myopathies. Plasma complement levels were determined by single radial immunodiffusion assays.
(A and B) Serum C4 and C3 protein levels were measured in patients with inflammatory idiopathic myopathies (IIM) who also had myositis-specific (MSA) and myositis-associated (MAA) antibodies. Specific MSA including Jo-1 are shown, as are the MAA PM/Scl and Ro. (C) C4 protein levels were corrected for gene copy number (GCN) variation, and C4 protein yield per copy of C4 gene was plotted for patients with MSA, MAA, Jo-1, PM-Scl, and Ro antibodies. (D) C4 (left) and C3 (right) protein levels were compared between males and females (79).
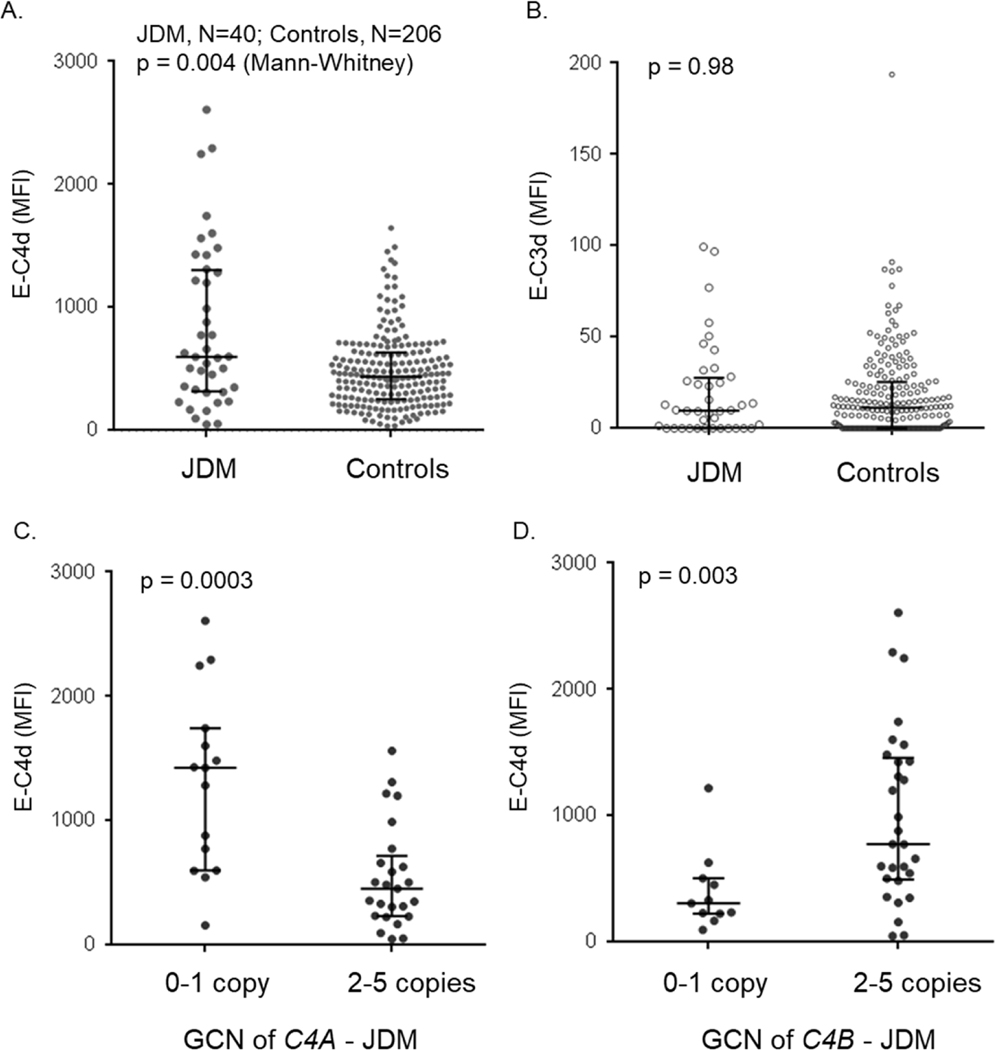
Additional work has supported the relationship between autoimmune myopathy and complement, including JDM. Our group took advantage of complement biology in which complement C4 byproducts, such as C4d, are deposited on cell surfaces (including on erythrocytes) upon activation and cleavage of C4. This deposition of the byproduct C4d (erythrocyte-C4d or E-C4d) creates record of complement activation (76, 174). Figure 11 shows mean fluorescence intensity, a measure of antigen density on target cells, of E-C4d in patients with JDM versus controls (left upper panel) and compares E-C4d levels based on C4A and C4B deficiency (bottom panels). E-C3d levels are also shown, though patients with JDM do not demonstrate any appreciable differences in C3d deposition than controls. The results demonstrate that E-C4d levels are higher in JDM compared with controls, likely reflecting chronic complement activation and inflammation. However, higher E-C4d levels were also seen in patients with C4A deficiency and in patients with higher GCN of C4B. This remarkable relationship suggests that activated C4B may be the primary isotype of C4 involved in complement activation and deposition on red blood cells, which can lead to complement-mediated cellular or tissue destruction. Alternatively, C4A may be less efficient at or protect against this process. In either case, these findings offer a hint of the mechanisms connecting complement genetics and autoimmune diseases such as JDM (77–79).
Figure 11. Erythrocyte-bound C4d (E-C4d) and E-C3d in patients with juvenile dermatomyositis and healthy controls.
(A and B) A comparison of E-C4d and E-C3d in juvenile dermatomyositis (JDM) and controls. (C) A comparison of E-C4d in C4A-deficient (C4A gene copy number <2) and C4A-proficient (C4A GCN≥2) JDM patients. (D) A comparison of E-C4d in C4B-deficient (C4B GCN <2) and C4B-proficient (C4B GCN≥2) JDM patients. The median for each group is indicated by a horizontal bar, while the shorter bars represent interquartile ranges; the p-value for Mann Whitney test is indicated.
VI. COMPLEMENT IN OTHER AUTOIMMUNE DISEASES
A. Rheumatoid arthritis
Autoimmune arthritis is one of the most common rheumatological disorders and includes diseases such as rheumatoid arthritis (RA) in adults and juvenile idiopathic arthritis (JIA) in children. Autoimmune arthritides are thought to be largely T cell-mediated diseases, but complement has been implicated in both the initiation and ongoing pathogenesis of RA (211–216), psoriatic arthritis (214, 217), and JIA (218–220).
A study published very recently by Banda et al examined the role of complement in early RA. This work showed that gene expression of complement components and receptors such as C2, FCN1, FCN3, CFB, CFP, C3AR1, C5AR1, and CR1 positively correlated with disease activity as measured by the DAS28-ESR (disease activity score in 28 joints for erythrocyte sedimentation rate) (215). Conversely, expression of other complement genes such as Colec12, C5, C6, MASP-1, CFH, and MCP was inversely correlated with disease activity. Synovial samples underwent immunohistochemical staining that showed local alterations in complement and complement-activating and inhibitory proteins. The authors interpreted these results as evidence that an imbalance between complement activation and regulatory activity contributes to early pathogenesis in RA.
Most studies have examined serum complement levels, not genetic diversity in RA. A notable exception is a study in which C4 gene copy number was elucidated for 160 patients with RA and both healthy control subjects (n=51) and patients with rheumatological diseases besides RA (221). Unlike SLE, there was no increased risk of RA associated with C4A deficiency; in fact, C4B deficiency was most strongly associated with RA, especially in patients with autoantibodies such as rheumatoid factor (RF) or anti-citrullinated protein antibodies (ACPA).
Other groups have examined how single nucleotide polymorphisms (SNP) such as those identified in genome wide association studies impact disease activity and complement activation in RA (222–226). One group specifically interrogated the SNP rs17611 in complement C5 and found a dose-dependent effect on circulating C5 levels and activation. Variable susceptibility to cleavage by proteases such as elastase was hypothesized to underlie differences in C5 activation and disease activity observed in association with the SNP (227).This association with C5 is perplexing, as C5a receptor blockade does not reduce markers of joint inflammation in RA (228) and especially because C5 knock out mice have been shown to be resistant to collagen-induced antibody-mediated arthritis (229). Another group found that the SNP rs292001 in C1q was associated with reduced circulating C1q levels, though no functional assays were performed (225). In a study of Brazilian patients with RA and their healthy family members, alleles associated with lower production and relative deficiency of mannose-binding lectin (MBL) were associated with increased risk of disease (230), though other studies have not corroborated that finding and have instead showed that MBL deficiency is associated with increased disease severity in RA (231, 232). Mechanistic studies to understand the underlying pathophysiology connecting SNP of MBL with disease in RA are notably lacking.
B. Juvenile idiopathic arthritis
Juvenile idiopathic arthritis (JIA) is one of the most common pediatric autoimmune disorders and therefore is of high clinical significance. Much like our work in lupus and JDM, our group has investigated the relationship between complement C4 GCN and disease in patients with JIA versus controls (233). We interrogated total C4, C4A, C4B, C4L, and C4S gene copy number in 103 patients with oligoarticular JIA and over 3,500 race-matched controls. In contrast to the phenomenon observed in adult RA, described above, we did not detect significant differences in C4B GCN in patients with JIA (221). While there were no differences in the patterns of total C4, C4B, or long C4 genes, we observed significantly different distributions of C4A and C4S GCN. More than two-thirds of patients with oligoarticular JIA had two copies of C4A (68.9%), compared to 51.3% in controls; nearly half (46.6%) of patients with oligoarticular JIA had zero copies of short C4 genes (controls: 32.3%). Further clinical studies are necessary to understand the physiologic implications of such variations and whether GCN variations in oligoarticular JIA are secondary to HLA-class II gene polymorphisms (221).
C. Type I diabetes mellitus
Type I diabetes mellitus (T1DM) is an autoimmune disease of the young in which the immune system attacks and destroys β islet cells in the pancreas that are responsible for producing insulin. While the direct damage to the pancreas is thought to be mediated by cytotoxic T cells, increased deposition of complement C4 has been observed in T1DM (234, 235). Genetic analyses have implicated various haplotypes of the major histocompatibility complex (MHC) on chromosome 6 as risk factors for T1DM (236–238), but several complement genes (including C2, C4A, and C4B) are also located nearby between the human leukocyte antigen (HLA) class I and class II regions. Due to extensive linkage dysequilibrium, it remains relevant to critically analyze which genetic variants confer the greatest risk and if multiple variants contribute to increased risk or disease progression from onset to complications of T1D.
As a part of The Environmental Determinants of Diabetes in the Young (TEDDY) study, 15 known SNPs within complement genes were studied for possible associations with the development of autoantibodies associated with T1DM, including those against insulin, GAD65, or IA2 and/or association with the onset of clinical T1DM disease (239). Only one SNP, rs2230199, remained significant after correction; this variant was strongly associated with the development of T1DM in patients who carried two copies of HLA-DRB*04:01 (DR4/4), with a hazard ratio of 3.2 (239). While these results offer somewhat underwhelming evidence of complement’s role in T1DM, it is important to note that the group did not study gene copy number variation, which others have shown to correlate with T1DM risk or progression to full-blown disease after partial remission (235). Specifically, Kingery et al showed that higher C4A GCN and plasma levels of C4A protein were associated with greater preservation of insulin production (as measured by C peptide levels) 1 month after diagnosis (235). Conversely, lower C4B GCN and protein concentration were protective at 9 months after diagnosis. This apprarent dichotomy between C4A and C4B is consistent with effects we have observed in other autoimmune diseases, such as JDM, pediatric lupus (170), and recurrent pregnancy loss in antiphospholipid syndrome (187).
VII. CONCLUDING REMARKS
Although extremely rare in prevalence, complete deficiencies of C1q, r, or s and C4 have provided strong evidence for the role of early components of the classical complement activation pathway in the pathogenesis of systemic autoimmune disease such as SLE. Acquired deficiency of C1q by autoantibodies and genetic deficiency of the C4A isotype secondary to gene copy number variation of C4 genes are frequent complement abnormalities that can be present in 30–50% of SLE patients, particularly those of European ancestry. While likely often missed clinically, complement deficiency should be included in the differential diagnosis of patients presenting with recurrent infections and/or signs of SLE-like autoimmune disease, especially when similar problems exist in other family members and when patients present with symptoms at a young age.
We are gaining an increasing understanding of complement genetics and the role of complement polymorphisms and deficiency in various diseases. While some complement components are essential and lead to high penetrance immune disorders, the effects of deficiencies of other components are more subtle. Complement biology continues to teach us much about how the immune system functions and offers an avenue to further explore the interface of antimicrobial immunity, autoimmunity, and immunodeficiency.
This review has summarized what is known regarding complement C1q, r, and s; C2; C3; and C4 deficiencies and their role in autoimmune disease. C1 and C4 deficiencies in particular present with severe infections and highly penetrant autoimmune disorders. In contrast, disease severity in C2 deficiency is generally lower, and C3 deficiency may present without autoimmune symptoms at all. The body of evidence strongly suggests, however, that complement is involved in the pathogenesis of various autoimmune conditions.
Complement C4 in particular is genetically extraordinary with inter-individual gene copy number variations, gene size dichotomy, and polymorphic protein isotypes with distinct biochemical properties. It is that diversity that allows for much of the flexibility and efficacy of the classical complement pathway. Associations with complement components including C4 may vary between disorders, however, which to us suggests a nuanced model of complement’s role in autoimmunity: one in which the balance of various complement components and other immune factors determines disease phenotype. For example, our work has shown that there is a clear C4A gene dosage effect on the risk of disease susceptibility in SLE and other conditions such as inflammatory myositis. In contrast, C4B shows no such strong associations for most diseases. An exception to this rule is rheumatoid arthritis, in which low C4B GCN increases the odds of disease. Low gene copy number or deficiency of C4A remains a common risk factor for SLE across different racial groups despite divergent HLA haplotypes. In SLE and patients with antiphospholipid antibodies, high copy numbers of C4B are associated with greater incidence of cardiovascular complications including a history of hypertension, higher incidence of thrombosis, and recurrent pregnancy loss in female patients. Additional evidence implicates complement C4A in autoimmune arthritis and type I diabetes mellitus, but the determination of whether associations suggest a protective or risk role for various complement components is incomplete.
Additional subtleties exist regarding the downstream effects of complement deficiencies in autoimmune disorders. Patients with SLE and myositis both present with type I interferon stimulated gene expression signatures in peripheral blood samples and local tissues. However, this phenomenon is less conspicuous among patients with C4A deficiency or with HLA-DR3 haplotypes. The mechanistic relevance of C4A deficiency and HLA-DR3 in type I interferon induced gene expression requires further inquiry, but these findings suggest that complement signaling may contribute to autoimmunity in a way that is distinct from type I interferon pathways.
Despite ongoing advancements in our understanding of complement biology, there remain many puzzles to solve. Examples include the relative contribution of various complement deficiencies or polymorphisms and HLA haplotype in various autoimmune disorders, the relevance of the long versus short C4 genes, as well as the mechanisms by which complement contributes to autoimmune phenotypes. Furthermore, technical and scientific challenges remain on the path to understanding the pathophysiology of complement in autoimmune disease. Recent advances in whole-genome or whole-exome next-generation sequencing offer alternative methodologies to impute copy number variations for some innate immunity genes. However, validation is essential in genes with copy number variation and can be cumbersome, time-consuming, and expensive.
Finally, recent utilization of bone marrow transplantation and hematopoietic stem cell transfer has enabled complementation with intact complement C1q in C1q-deficient patients. Additional prospects for future gene therapy offer encouraging and innovative approaches to treat subjects with complement deficiency. Naturally, there remain many hurdles to overcome before this idea can become a reality, including graft versus host disease and viral reactivation under immunosuppressive conditions. Nevertheless, our increasing understanding of complement biology and its implications in immunity and autoimmunity is opening the door to more rational therapeutic design and offers hope for patients with a variety of immune-mediated conditions, from immunodeficiency to autoimmune disease.
Complement and Autoimmunity: Highlights.
While complement’s role in host defense against infection is well known, this article will focus on the role that various complement components play in autoimmune diseases.
The over-arching purpose of this review is to provide a comprehensive summary of the association of complement with various autoimmune diseases for the clinician and researcher alike. Importantly, putative mechanisms by which complement contributes to autoimmunity will be discussed.
To facilitate clinicians’ ability to address possible complement deficiencies, the clinical history and physical exam features seen in patients with complement deficiencies will be reviewed. In addition, a suggested laboratory work-up in cases of suspected complement deficiency will be provided.
The complex role of complement in systemic lupus erythematous, including a point-by-point discussion of each complement component and known clinical cases of their deficiencies, will be reviewed. Particular attention will be paid to complement C1 and C4, which are most strongly associated with autoimmune disease.
Complement C1q in particular is strongly associated with lupus-like autoimmune diseases. Treatment options for C1 deficiency will be discussed.
Complement C4 is also strongly associated with systemic lupus erythematous, as well as other autoimmune disorders.
C4 is a unique molecule. Not only does it boast multiple layers of genetic polymorphisms, but there also exists a wide range of gene copy number that contributes an additional dimension of complexity to C4’s role in autoimmune disease. Our group has studied complement extensively, and what is known regarding the genetics of complement C4 in autoimmune disease is briefly summarized.
Complement’s role in autoimmune disease is best studied in systemic lupus erythematous, but this review will also briefly address what is known regarding the role of complement in other autoimmune disorders such as the idiopathic inflammatory myopathies, autoimmune arthritis, and type I diabetes mellitus.
ACKNOWLEDGEMENTS
We are indebted to volunteers and donors for human blood samples. We are grateful to students, fellows, colleagues and collaborators who contributed to original data reviewed in this article. Our work are supported by NIH grants R01 AR073311, R01 AR054459, R21 AR070509, and P01 DK55546 from the NIAMS, NIDDK, NIAID and NICHS, and by a research grants from the CureJM Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nonaka M. Evolution of the complement system. Subcell Biochem. 2014;80:31–43. Epub 2014/05/07. doi: 10.1007/978-94-017-8881-6_3. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Thangamani S, Ho B, Ding JL. The ancient origin of the complement system. EMBO J. 2005;24(2):382–94. Epub 2004/12/24. doi: 10.1038/sj.emboj.7600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter RR. Complement polymorphism, the major histocompatibility complex and associated diseases: a speculation. Mol Biol Med. 1983;1(1):161–8. Epub 1983/07/01. [PubMed] [Google Scholar]
- 4.Nesargikar PN, Spiller B, Chavez R. The complement system: history, pathways, cascade and inhibitors. Eur J Microbiol Immunol (Bp). 2012;2(2):103–11. Epub 2012/06/01. doi: 10.1556/EuJMI.2.2012.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344(14):1058–66. Epub 2001/04/05. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 6.Walport MJ. Complement. Second of two parts. N Engl J Med. 2001;344(15):1140–4. Epub 2001/04/12. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 7.Reid KB, Porter RR. The proteolytic activation systems of complement. Annu Rev Biochem. 1981;50:433–64. Epub 1981/01/01. doi: 10.1146/annurev.bi.50.070181.002245. [DOI] [PubMed] [Google Scholar]
- 8.Physiology Peters K. and pathology of the C3 amplification cycle: A retrospective. Immunol Rev. 2022. Epub 2022/11/22. doi: 10.1111/imr.13165. [DOI]
- 9.Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S, Martinez P, Kulski J. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275–304. Epub 1999/05/13. doi: 10.1111/j.1600065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 10.Yu CY, Ardoin SP. Complement inhibitor for therapy of CHAPLE. Nat Immunol. 2021;22(2):106–8. Epub 2021/01/06. doi: 10.1038/s41590-020-00842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ram S, Lewis LA, Rice PA. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin Microbiol Rev. 2010;23(4):740–80. Epub 2010/10/12. doi: 10.1128/cmr.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skattum L, van Deuren M, van der Poll T, Truedsson L. Complement deficiency states and associated infections. Mol Immunol. 2011;48(14):1643–55. Epub 2011/06/01. doi: 10.1016/j.molimm.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Liu M. Complement C4, Infections, and Autoimmune Diseases. Front Immunol. 2021;12:694928. Epub 2021/08/03. doi: 10.3389/fimmu.2021.694928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roord JJ, Daha M, Kuis W, Verbrugh HA, Verhoef J, Zegers BJ, Stoop JW. Inherited deficiency of the third component of complement associated with recurrent pyogenic infections, circulating immune complexes, and vasculitis in a Dutch family. Pediatrics. 1983;71(1):81–7. Epub 1983/01/01. [PubMed] [Google Scholar]
- 15.Belfeki N, Zayet S, Hamrouni S, Diamantis S, Boutboul D. Extreme gonococcal susceptibility associated with acquired complement deficiency secondary to hypocomplementemic urticarial vasculitis and systemic lupus erythematosus. J Infect Chemother. 2022;28(2):308–10. Epub 2021/11/06. doi: 10.1016/j.jiac.2021.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Ekdahl KN, Persson B, Mohlin C, Sandholm K, Skattum L, Nilsson B. Interpretation of Serological Complement Biomarkers in Disease. Front Immunol. 2018;9:2237. Epub 2018/11/09. doi: 10.3389/fimmu.2018.02237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellors J, Tipton T, Longet S, Carroll M. Viral Evasion of the Complement System and Its Importance for Vaccines and Therapeutics. Front Immunol. 2020;11:1450. Epub 2020/08/01. doi: 10.3389/fimmu.2020.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kase T, Suzuki Y, Kawai T, Sakamoto T, Ohtani K, Eda S, Maeda A, Okuno Y, Kurimura T, Wakamiya N. Human mannan-binding lectin inhibits the infection of influenza A virus without complement. Immunology. 1999;97(3):385–92. Epub 1999/08/14. doi: 10.1046/j.1365-2567.1999.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang WC, White MR, Moyo P, McClear S, Thiel S, Hartshorn KL, Takahashi K. Lack of the pattern recognition molecule mannose-binding lectin increases susceptibility to influenza A virus infection. BMC Immunol. 2010;11:64. Epub 2010/12/25. doi: 10.1186/1471-2172-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friborg JT, Jarrett RF, Koch A, Garred P, Freeland JM, Andersen A, Melbye M. Mannose-binding lectin genotypes and susceptibility to epstein-barr virus infection in infancy. Clin Vaccine Immunol. 2010;17(9):1484–7. Epub 2010/07/09. doi: 10.1128/CVI.00527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small CL, Shaler CR, McCormick S, Jeyanathan M, Damjanovic D, Brown EG, Arck P, Jordana M, Kaushic C, Ashkar AA, Xing Z. Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J Immunol. 2010;184(4):2048–56. Epub 2010/01/20. doi: 10.4049/jimmunol.0902772. [DOI] [PubMed] [Google Scholar]
- 22.Hament JM, Kimpen JL, Fleer A, Wolfs TF. Respiratory viral infection predisposing for bacterial disease: a concise review. FEMS Immunol Med Microbiol. 1999;26(3–4):18995. Epub 1999/11/27. doi: 10.1111/j.1574-695X.1999.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 23.Oliva J, Terrier O. Viral and Bacterial Co-Infections in the Lungs: Dangerous Liaisons. Viruses. 2021;13(9). Epub 2021/09/29. doi: 10.3390/v13091725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornacoff JB, Hebert LA, Smead WL, VanAman ME, Birmingham DJ, Waxman FJ. Primate erythrocyte-immune complex-clearing mechanism. J Clin Invest. 1983;71(2):236–47. Epub 1983/02/01. doi: 10.1172/jci110764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schifferli JA, Ng YC, Peters DK. The role of complement and its receptor in the elimination of immune complexes. N Engl J Med. 1986;315(8):488–95. Epub 1986/08/21. doi: 10.1056/NEJM198608213150805. [DOI] [PubMed] [Google Scholar]
- 26.Elkon KB, Santer DM. Complement, interferon and lupus. Curr Opin Immunol. 2012;24(6):665–70. Epub 2012/09/25. doi: 10.1016/j.coi.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Horgan C, Taylor RP. Studies on the kinetics of binding of complement-fixing dsDNA/anti-dsDNA immune complexes to the red blood cells of normal individuals and patients with systemic lupus erythematosus. Arthritis Rheum. 1984;27(3):320–9. Epub 1984/03/01. doi: 10.1002/art.1780270312. [DOI] [PubMed] [Google Scholar]
- 28.Kavai M. Immune complex clearance by complement receptor type 1 in SLE. Autoimmun Rev. 2008;8(2):160–4. Epub 2008/07/08. doi: 10.1016/j.autrev.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Petersen I, Baatrup G, Jepsen HH, Svehag SE. Complement-mediated solubilization of immune complexes and their interaction with complement C3 receptors. Complement. 1985;2(2–3):97–110. Epub 1985/01/01. doi: 10.1159/000467850. [DOI] [PubMed] [Google Scholar]
- 30.Schena FP, Pastore A, Manno C, Varvara B. Effects of plasma-exchange on complement-mediated solubilization of circulating immune complexes in patients with systemic lupus erythematosus. Int J Artif Organs. 1985;8 Suppl 2:3–6. Epub 1985/07/01. [PubMed] [Google Scholar]
- 31.Arason GJ, Jorgensen GH, Ludviksson BR. Primary immunodeficiency and autoimmunity: lessons from human diseases. Scand J Immunol. 2010;71(5):317–28. Epub 2010/05/27. doi: 10.1111/j.1365-3083.2010.02386.x. [DOI] [PubMed] [Google Scholar]
- 32.Macedo AC, Isaac L. Systemic Lupus Erythematosus and Deficiencies of Early Components of the Complement Classical Pathway. Front Immunol. 2016;7:55. Epub 2016/03/05. doi: 10.3389/fimmu.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lintner KE, Wu YL, Yang Y, Spencer CH, Hauptmann G, Hebert LA, Atkinson JP, Yu CY. Early Components of the Complement Classical Activation Pathway in Human Systemic Autoimmune Diseases. Front Immunol. 2016;7:36. Epub 2016/02/26. doi: 10.3389/fimmu.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sontheimer RD, Racila E, Racila DM. C1q: its functions within the innate and adaptive immune responses and its role in lupus autoimmunity. J Invest Dermatol. 2005;125(1):14–23. Epub 2005/06/29. doi: 10.1111/j.0022-202X.2005.23673.x. [DOI] [PubMed] [Google Scholar]
- 35.Carneiro-Sampaio M, Coutinho A. Early-onset autoimmune disease as a manifestation of primary immunodeficiency. Front Immunol. 2015;6:185. Epub 2015/05/23. doi: 10.3389/fimmu.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leffler J, Bengtsson AA, Blom AM. The complement system in systemic lupus erythematosus: an update. Ann Rheum Dis. 2014;73(9):1601–6. Epub 2014/05/23. doi: 10.1136/annrheumdis-2014-205287. [DOI] [PubMed] [Google Scholar]
- 37.Lipsker D, Hauptmann G. Cutaneous manifestations of complement deficiencies. Lupus. 2010;19(9):1096–106. Epub 2010/08/10. doi: 10.1177/0961203310373370. [DOI] [PubMed] [Google Scholar]
- 38.Happonen KE, Saxne T, Aspberg A, Mörgelin M, Heinegård D, Blom AM. Regulation of complement by cartilage oligomeric matrix protein allows for a novel molecular diagnostic principle in rheumatoid arthritis. Arthritis Rheum. 2010;62(12):3574–83. Epub 2010/08/26. doi: 10.1002/art.27720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molenaar ET, Voskuyl AE, Familian A, van Mierlo GJ, Dijkmans BA, Hack CE. Complement activation in patients with rheumatoid arthritis mediated in part by C-reactive protein. Arthritis Rheum. 2001;44(5):997–1002. Epub 2001/05/16. doi: . [DOI] [PubMed] [Google Scholar]
- 40.Courtney PA, Crockard AD, Williamson K, Irvine AE, Kennedy RJ, Bell AL. Increased apoptotic peripheral blood neutrophils in systemic lupus erythematosus: relations with disease activity, antibodies to double stranded DNA, and neutropenia. Ann Rheum Dis. 1999;58(5):309–14. Epub 1999/05/04. doi: 10.1136/ard.58.5.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren Y, Tang J, Mok MY, Chan AW, Wu A, Lau CS. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis Rheum. 2003;48(10):2888–97. Epub 2003/10/15. doi: 10.1002/art.11237. [DOI] [PubMed] [Google Scholar]
- 42.Bouts YM, Wolthuis DF, Dirkx MF, Pieterse E, Simons EM, van Boekel AM, Dieker JW, van der Vlag J. Apoptosis and NET formation in the pathogenesis of SLE. Autoimmunity. 2012;45(8):597–601. Epub 2012/08/24. doi: 10.3109/08916934.2012.719953. [DOI] [PubMed] [Google Scholar]
- 43.Knight JS, Carmona-Rivera C, Kaplan MJ. Proteins derived from neutrophil extracellular traps may serve as self-antigens and mediate organ damage in autoimmune diseases. Front Immunol. 2012;3:380. Epub 2012/12/19. doi: 10.3389/fimmu.2012.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz LE, van Bavel C, Franz S, Berden J, Herrmann M, van der Vlag J. Apoptosis in the pathogenesis of systemic lupus erythematosus. Lupus. 2008;17(5):371–5. Epub 2008/05/21. doi: 10.1177/0961203308089990. [DOI] [PubMed] [Google Scholar]
- 45.Tsokos GC, Lo MS, Costa Reis P, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol. 2016;12(12):716–30. Epub 2016/11/23. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- 46.Adler BL, Christopher-Stine L. Triggers of inflammatory myopathy: insights into pathogenesis. Discov Med. 2018;25(136):75–83. Epub 2018/03/27. [PMC free article] [PubMed] [Google Scholar]
- 47.Jones J, Laffafian I, Cooper AM, Williams BD, Morgan BP. Expression of complement regulatory molecules and other surface markers on neutrophils from synovial fluid and blood of patients with rheumatoid arthritis. Br J Rheumatol. 1994;33(8):707–12. Epub 1994/08/01. doi: 10.1093/rheumatology/33.8.707. [DOI] [PubMed] [Google Scholar]
- 48.Schanzenbacher J, Köhl J, Karsten CM. Anaphylatoxins spark the flame in early autoimmunity. Front Immunol. 2022;13:958392. Epub 2022/08/13. doi: 10.3389/fimmu.2022.958392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coutinho A, Carneiro-Sampaio M. Primary immunodeficiencies unravel critical aspects of the pathophysiology of autoimmunity and of the genetics of autoimmune disease. J Clin Immunol. 2008;28 Suppl 1:S4–10. Epub 2008/02/23. doi: 10.1007/s10875-007-9167-y. [DOI] [PubMed] [Google Scholar]
- 50.Pabón-Porras MA, Molina-Ríos S, Flórez-Suárez JB, Coral-Alvarado PX, Méndez-Patarroyo P, Quintana-López G. Rheumatoid arthritis and systemic lupus erythematosus: Pathophysiological mechanisms related to innate immune system. SAGE Open Med. 2019;7:2050312119876146. Epub 2019/09/13. doi: 10.1177/2050312119876146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kusner LL, Kaminski HJ. The role of complement in experimental autoimmune myasthenia gravis. Ann N Y Acad Sci. 2012;1274(1):127–32. Epub 2012/12/21. doi: 10.1111/j.1749-6632.2012.06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carneiro-Sampaio M, Liphaus BL, Jesus AA, Silva CA, Oliveira JB, Kiss MH. Understanding systemic lupus erythematosus physiopathology in the light of primary immunodeficiencies. J Clin Immunol. 2008;28 Suppl 1:S34–41. Epub 2008/04/12. doi: 10.1007/s10875-008-9187-2. [DOI] [PubMed] [Google Scholar]
- 53.Ogden CA, Elkon KB. Role of complement and other innate immune mechanisms in the removal of apoptotic cells. Curr Dir Autoimmun. 2006;9:120–42. Epub 2006/01/06. doi: 10.1159/000090776. [DOI] [PubMed] [Google Scholar]
- 54.Bijl M, Reefman E, Horst G, Limburg PC, Kallenberg CG. Reduced uptake of apoptotic cells by macrophages in systemic lupus erythematosus: correlates with decreased serum levels of complement. Ann Rheum Dis. 2006;65(1):57–63. Epub 2005/05/28. doi: 10.1136/ard.2005.035733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser DA, Laust AK, Nelson EL, Tenner AJ. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J Immunol. 2009;183(10):6175–85. Epub 2009/10/30. doi: 10.4049/jimmunol.0902232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Defendi F, Thielens NM, Clavarino G, Cesbron JY, Dumestre-Pérard C. The Immunopathology of Complement Proteins and Innate Immunity in Autoimmune Disease. Clin Rev Allergy Immunol. 2020;58(2):229–51. Epub 2019/12/14. doi: 10.1007/s12016-019-08774-5. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Y, Wei W, Liu ML. Extracellular vesicles and lupus nephritis - New insights into pathophysiology and clinical implications. J Autoimmun. 2020;115:102540. Epub 2020/09/08. doi: 10.1016/j.jaut.2020.102540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohlson SS, O’Conner SD, Hulsebus HJ, Ho MM, Fraser DA. Complement, c1q, and c1q-related molecules regulate macrophage polarization. Front Immunol. 2014;5:402. Epub 2014/09/06. doi: 10.3389/fimmu.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lood C, Gullstrand B, Truedsson L, Olin AI, Alm GV, Rönnblom L, Sturfelt G, Eloranta ML, Bengtsson AA. C1q inhibits immune complex-induced interferon-alpha production in plasmacytoid dendritic cells: a novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum. 2009;60(10):3081–90. Epub 2009/10/01. doi: 10.1002/art.24852. [DOI] [PubMed] [Google Scholar]
- 60.Walport MJ. Complement and systemic lupus erythematosus. Arthritis Res. 2002;4 Suppl 3:S279–93. Epub 2002/07/12. doi: 10.1186/ar586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grumach AS, Kirschfink M. Are complement deficiencies really rare? Overview on prevalence, clinical importance and modern diagnostic approach. Mol Immunol. 2014;61(2):110–7. Epub 2014/07/20. doi: 10.1016/j.molimm.2014.06.030. [DOI] [PubMed] [Google Scholar]
- 62.Sjoholm AG, Jonsson G, Braconier JH, Sturfelt G, Truedsson L. Complement deficiency and disease: an update. Mol Immunol. 2006;43(1–2):78–85. Epub 2005/07/20. doi: 10.1016/j.molimm.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 63.Ross SC, Densen P. Complement deficiency states and infection: epidemiology, pathogenesis and consequences of neisserial and other infections in an immune deficiency. Medicine (Baltimore). 1984;63(5):243–73. Epub 1984/09/01. [PubMed] [Google Scholar]
- 64.Corrigan JJ Jr., Boineau FG. Hemolytic-uremic syndrome. Pediatr Rev. 2001;22(11):365–9. Epub 2001/11/03. [PubMed] [Google Scholar]
- 65.Noris M, Remuzzi G. Atypical hemolytic-uremic syndrome. N Engl J Med. 2009;361(17):1676–87. Epub 2009/10/23. doi: 10.1056/NEJMra0902814. [DOI] [PubMed] [Google Scholar]
- 66.Caprioli J, Noris M, Brioschi S, Pianetti G, Castelletti F, Bettinaglio P, Mele C, Bresin E, Cassis L, Gamba S, Porrati F, Bucchioni S, Monteferrante G, Fang CJ, Liszewski MK, Kavanagh D, Atkinson JP, Remuzzi G, International Registry of R, Familial HT. Genetics of HUS: the impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood. 2006;108(4):1267–79. Epub 2006/04/20. doi: 10.1182/blood-2005-10-007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noris M, Ruggenenti P, Perna A, Orisio S, Caprioli J, Skerka C, Vasile B, Zipfel PF, Remuzzi G. Hypocomplementemia discloses genetic predisposition to hemolytic uremic syndrome and thrombotic thrombocytopenic purpura: role of factor H abnormalities. Italian Registry of Familial and Recurrent Hemolytic Uremic Syndrome/Thrombotic Thrombocytopenic Purpura. J Am Soc Nephrol. 1999;10(2):281–93. [DOI] [PubMed] [Google Scholar]
- 68.Lapeyraque AL, Bitzan M, Al-Dakkak I, Francis M, Huang SS, Kaprielian R, Larratt L, Pavenski K, Ribic C, Tosikyan A, Licht C, Philibert D. Clinical Characteristics and Outcome of Canadian Patients Diagnosed With Atypical Hemolytic Uremic Syndrome. Can J Kidney Health Dis. 2020;7:2054358119897229. Epub 2020/02/13. doi: 10.1177/2054358119897229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, Fortin PR, Bruce IN, Isenberg D, Wallace DJ, Nived O, Sturfelt G, Ramsey-Goldman R, Bae SC, Hanly JG, Sanchez-Guerrero J, Clarke A, Aranow C, Manzi S, Urowitz M, Gladman D, Kalunian K, Costner M, Werth VP, Zoma A, Bernatsky S, Ruiz-Irastorza G, Khamashta MA, Jacobsen S, Buyon JP, Maddison P, Dooley MA, van Vollenhoven RF, Ginzler E, Stoll T, Peschken C, Jorizzo JL, Callen JP, Lim SS, Fessler BJ, Inanc M, Kamen DL, Rahman A, Steinsson K, Franks AG Jr., Sigler L, Hameed S, Fang H, Pham N, Brey R, Weisman MH, McGwin G Jr., Magder LS. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64(8):2677–86. Epub 2012/05/04. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hebert LA, Cosio FG, Neff JC. Diagnostic significance of hypocomplementemia. Kidney Int. 1991;39(5):811–21. Epub 1991/05/01. doi: 10.1038/ki.1991.102. [DOI] [PubMed] [Google Scholar]
- 71.Schur PH. Complement and lupus erythematosus. Arthritis Rheum. 1982;25(7):793–8. Epub 1982/07/01. [DOI] [PubMed] [Google Scholar]
- 72.Wu YL, Higgins GC, Rennebohm RM, Chung EK, Yang Y, Zhou B, Nagaraja HN, Birmingham DJ, Rovin BH, Hebert LA, Yu CY. Three distinct profiles of serum complement C4 proteins in pediatric systemic lupus erythematosus (SLE) patients: tight associations of complement C4 and C3 protein levels in SLE but not in healthy subjects. Adv Exp Med Biol. 2006;586:227–47. Epub 2006/08/09. doi: 10.1007/0-387-34134-X_16. [DOI] [PubMed] [Google Scholar]
- 73.Birmingham DJ, Irshaid F, Nagaraja HN, Zou X, Tsao BP, Wu H, Yu CY, Hebert LA, Rovin BH. The complex nature of serum C3 and C4 as biomarkers of lupus renal flare. Lupus. 2010;19(11):1272–80. Epub 2010/07/08. doi: 10.1177/0961203310371154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandhu V, Quan M. SLE and Serum Complement: Causative, Concomitant or Coincidental? Open Rheumatol J. 2018;12:171. Epub 2018/10/06. doi: 10.2174/1874312901812010171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Longhurst H, Cicardi M. Hereditary angio-oedema. Lancet. 2012;379(9814):474–81. Epub 2012/02/07. doi: 10.1016/S0140-6736(11)60935-5. [DOI] [PubMed] [Google Scholar]
- 76.Manzi S, Navratil JS, Ruffing MJ, Liu CC, Danchenko N, Nilson SE, Krishnaswami S, King DE, Kao AH, Ahearn JM. Measurement of erythrocyte C4d and complement receptor 1 in systemic lupus erythematosus. Arthritis Rheum. 2004;50(11):3596–604. Epub 2004/11/06. doi: 10.1002/art.20561. [DOI] [PubMed] [Google Scholar]
- 77.Lintner KE, Patwardhan A, Rider LG, Abdul-Aziz R, Wu YL, Lundstrom E, Padyukov L, Zhou B, Alhomosh A, Newsom D, White P, Jones KB, O’Hanlon TP, Miller FW, Spencer CH, Yu CY. Gene copy-number variations (CNVs) of complement C4 and C4A deficiency in genetic risk and pathogenesis of juvenile dermatomyositis. Ann Rheum Dis. 2016;75(9):1599–606. Epub 2015/10/24. doi: 10.1136/annrheumdis-2015-207762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Putterman C, Furie R, Ramsey-Goldman R, Askanase A, Buyon J, Kalunian K, Chatham WW, Massarotti E, Kirou K, Jordan N, Blanco I, Weinstein A, Chitkara P, Manzi S, Ahearn J, O’Malley T, Conklin J, Ibarra C, Barken D, Dervieux T. Cell-bound complement activation products in systemic lupus erythematosus: comparison with anti-double-stranded DNA and standard complement measurements. Lupus Sci Med. 2014;1(1):e000056. Epub 2014/11/15. doi: 10.1136/lupus-2014-000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou D, King EH, Rothwell S, Krystufkova O, Notarnicola A, Coss S, Abdul-Aziz R, Miller KE, Dang A, Yu GR, Drew J, Lundstrom E, Pachman LM, Mamyrova G, Curiel RV, De Paepe B, De Bleecker JL, Payton A, Ollier W, O’Hanlon TP, Targoff IN, Flegel WA, Sivaraman V, Oberle E, Akoghlanian S, Driest K, Spencer CH, Wu YL, Nagaraja HN, Ardoin SP, Chinoy H, Rider LG, Miller FW, Lundberg IE, Padyukov L, Vencovsky J, Lamb JA, Yu CY, for MI. Low copy numbers of complement C4 and C4A deficiency are risk factors for myositis, its subgroups and autoantibodies. Ann Rheum Dis. 2022. Epub 2022/09/29. doi: 10.1136/ard-2022-222935. [DOI] [PMC free article] [PubMed]
- 80.Mitchell TJ, Naughton M, Norsworthy P, Davies KA, Walport MJ, Morley BJ. IFN-gamma up-regulates expression of the complement components C3 and C4 by stabilization of mRNA. J Immunol. 1996;156(11):4429–34. Epub 1996/06/01. [PubMed] [Google Scholar]
- 81.Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, Zhou B, Moulds JM, Yu CY. Genetic, structural and functional diversities of human complement components C4A and C4B and their mouse homologues, Slp and C4. Int Immunopharmacol. 2001;1(3):365–92. Epub 2001/05/23. doi: 10.1016/s1567-5769(01)00019-4. [DOI] [PubMed] [Google Scholar]
- 82.Yang Y, Chung EK, Zhou B, Blanchong CA, Yu CY, Fust G, Kovacs M, Vatay A, Szalai C, Karadi I, Varga L. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J Immunol. 2003;171(5):2734–45. Epub 2003/08/21. [DOI] [PubMed] [Google Scholar]
- 83.Saxena K, Kitzmiller KJ, Wu YL, Zhou B, Esack N, Hiremath L, Chung EK, Yang Y, Yu CY. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: a comparison of Asian-Indian and European American populations. Mol Immunol. 2009;46(7):1289–303. Epub 2009/01/13. doi: 10.1016/j.molimm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Copenhaver M, Yu CY, Hoffman RP. Complement Components, C3 and C4, and the Metabolic Syndrome. Curr Diabetes Rev. 2019;15(1):44–8. Epub 2018/04/18. doi: 10.2174/1573399814666180417122030. [DOI] [PubMed] [Google Scholar]
- 85.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–21. Epub 2011/12/02. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 86.Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR. Structure and activation of C1, the complex initiating the classical pathway of the complement cascade. Proc Natl Acad Sci U S A. 2017;114(5):986–91. Epub 2017/01/21. doi: 10.1073/pnas.1616998114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roumenina LT, Sene D, Radanova M, Blouin J, Halbwachs-Mecarelli L, Dragon-Durey MA, Fridman WH, Fremeaux-Bacchi V. Functional complement C1q abnormality leads to impaired immune complexes and apoptotic cell clearance. J Immunol. 2011;187(8):4369–73. Epub 2011/09/21. doi: 10.4049/jimmunol.1101749. [DOI] [PubMed] [Google Scholar]
- 88.Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv Immunol. 2000;76:227–324. Epub 2000/11/18. [DOI] [PubMed] [Google Scholar]
- 89.Schejbel L, Skattum L, Hagelberg S, Ahlin A, Schiller B, Berg S, Genel F, Truedsson L, Garred P. Molecular basis of hereditary C1q deficiency--revisited: identification of several novel disease-causing mutations. Genes Immun. 2011;12(8):626–34. Epub 2011/06/10. doi: 10.1038/gene.2011.39. [DOI] [PubMed] [Google Scholar]
- 90.Higuchi Y, Shimizu J, Hatanaka M, Kitano E, Kitamura H, Takada H, Ishimura M, Hara T, Ohara O, Asagoe K, Kubo T. The identification of a novel splicing mutation in C1qB in a Japanese family with C1q deficiency: a case report. Pediatr Rheumatol Online J. 2013;11(1):41. Epub 2013/10/29. doi: 10.1186/1546-0096-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jlajla H, Sellami MK, Sfar I, Laadhar L, Zerzeri Y, Abdelmoula MS, Gorgi Y, Dridi MF, Makni S. New C1q mutation in a Tunisian family. Immunobiology. 2014;219(3):241–6. Epub 2013/12/18. doi: 10.1016/j.imbio.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 92.Marquart HV, Schejbel L, Sjoholm A, Martensson U, Nielsen S, Koch A, Svejgaard A, Garred P. C1q deficiency in an Inuit family: identification of a new class of C1q disease-causing mutations. Clin Immunol. 2007;124(1):33–40. Epub 2007/05/22. doi: 10.1016/j.clim.2007.03.547. [DOI] [PubMed] [Google Scholar]
- 93.McAdam RA, Goundis D, Reid KB. A homozygous point mutation results in a stop codon in the C1q B-chain of a C1q-deficient individual. Immunogenetics. 1988;27(4):259–64. Epub 1988/01/01. [DOI] [PubMed] [Google Scholar]
- 94.van Schaarenburg RA, Daha NA, Schonkeren JJ, Nivine Levarht EW, van Gijlswijk-Janssen DJ, Kurreeman FA, Roos A, van Kooten C, Koelman CA, Ernst-Kruis MR, Toes RE, Huizinga TW, Lankester AC, Trouw LA. Identification of a novel non-coding mutation in C1qB in a Dutch child with C1q deficiency associated with recurrent infections. Immunobiology. 2015;220(3):422–7. Epub 2014/12/03. doi: 10.1016/j.imbio.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 95.Namjou B, Keddache M, Fletcher D, Dillon S, Kottyan L, Wiley G, Gaffney PM, Wakeland BE, Liang C, Wakeland EK, Scofield RH, Kaufman K, Harley JB. Identification of novel coding mutation in C1qA gene in an African-American pedigree with lupus and C1q deficiency. Lupus. 2012;21(10):1113–8. Epub 2012/04/05. doi: 10.1177/0961203312443993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mehta P, Norsworthy PJ, Hall AE, Kelly SJ, Walport MJ, Botto M, Pickering MC. SLE with C1q deficiency treated with fresh frozen plasma: a 10-year experience. Rheumatology. 2009;49(4):823–4. doi: 10.1093/rheumatology/kep387. [DOI] [PubMed] [Google Scholar]
- 97.Hosszu KK, Valentino A, Peerschke EI, Ghebrehiwet B. SLE: Novel Postulates for Therapeutic Options. Front Immunol. 2020;11:583853. Epub 2020/10/30. doi: 10.3389/fimmu.2020.583853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ogden CA, deCathelineau A, Hoffmann PR, Bratton D, Ghebrehiwet B, Fadok VA, Henson PM. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med. 2001;194(6):781–95. Epub 2001/09/19. doi: 10.1084/jem.194.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Païdassi H, Tacnet-Delorme P, Garlatti V, Darnault C, Ghebrehiwet B, Gaboriaud C, Arlaud GJ, Frachet P. C1q binds phosphatidylserine and likely acts as a multiligandbridging molecule in apoptotic cell recognition. J Immunol. 2008;180(4):2329–38. Epub 2008/02/06. doi: 10.4049/jimmunol.180.4.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vandivier RW, Ogden CA, Fadok VA, Hoffmann PR, Brown KK, Botto M, Walport MJ, Fisher JH, Henson PM, Greene KE. Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro: calreticulin and CD91 as a common collectin receptor complex. J Immunol. 2002;169(7):3978–86. Epub 2002/09/24. doi: 10.4049/jimmunol.169.7.3978. [DOI] [PubMed] [Google Scholar]
- 101.Donnelly S, Roake W, Brown S, Young P, Naik H, Wordsworth P, Isenberg DA, Reid KB, Eggleton P. Impaired recognition of apoptotic neutrophils by the C1q/calreticulin and CD91 pathway in systemic lupus erythematosus. Arthritis Rheum. 2006;54(5):1543–56. Epub 2006/04/29. doi: 10.1002/art.21783. [DOI] [PubMed] [Google Scholar]
- 102.Santer DM, Hall BE, George TC, Tangsombatvisit S, Liu CL, Arkwright PD, Elkon KB. C1q deficiency leads to the defective suppression of IFN-alpha in response to nucleoprotein containing immune complexes. J Immunol. 2010;185(8):4738–49. Epub 2010/09/17. doi: 10.4049/jimmunol.1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hosszu KK, Santiago-Schwarz F, Peerschke EI, Ghebrehiwet B. C1q is a molecular switch dictating the monocyte to dendritic cell (DC) transition and arrests DCs in an immature phenotype. The FASEB Journal. 2008;22(S1):673.1-.1. doi: 10.1096/fasebj.22.1_supplement.673.1. [DOI] [Google Scholar]
- 104.Hosszu KK, Santiago-Schwarz F, Peerschke EI, Ghebrehiwet B. Evidence that a C1q/C1qR system regulates monocyte-derived dendritic cell differentiation at the interface of innate and acquired immunity. Innate Immun. 2010;16(2):115–27. Epub 2009/08/28. doi: 10.1177/1753425909339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bigler C, Schaller M, Perahud I, Osthoff M, Trendelenburg M. Autoantibodies against complement C1q specifically target C1q bound on early apoptotic cells. J Immunol. 2009;183(5):3512–21. Epub 2009/08/04. doi: 10.4049/jimmunol.0803573. [DOI] [PubMed] [Google Scholar]
- 106.Shoenfeld Y, Szyper-Kravitz M, Witte T, Doria A, Tsutsumi A, Tatsuya A, Dayer JM, Roux-Lombard P, Fontao L, Kallenberg CG, Bijl M, Matthias T, Fraser A, ZandmanGoddard G, Blank M, Gilburd B, Meroni PL. Autoantibodies against protective molecules--C1q, C-reactive protein, serum amyloid P, mannose-binding lectin, and apolipoprotein A1: prevalence in systemic lupus erythematosus. Ann N Y Acad Sci. 2007;1108:227–39. Epub 2007/09/28. doi: 10.1196/annals.1422.025. [DOI] [PubMed] [Google Scholar]
- 107.Ekinci Z, Ozturk K. Systemic lupus erythematosus with C1q deficiency: treatment with fresh frozen plasma. Lupus. 2018;27(1):134–8. Epub 2017/11/09. doi: 10.1177/0961203317741565. [DOI] [PubMed] [Google Scholar]
- 108.Cortes-Hernandez J, Fossati-Jimack L, Petry F, Loos M, Izui S, Walport MJ, Cook HT, Botto M. Restoration of C1q levels by bone marrow transplantation attenuates autoimmune disease associated with C1q deficiency in mice. Eur J Immunol. 2004;34(12):3713–22. Epub 2004/11/02. doi: 10.1002/eji.200425616. [DOI] [PubMed] [Google Scholar]
- 109.Petry F, Botto M, Holtappels R, Walport MJ, Loos M. Reconstitution of the complement function in C1q-deficient (C1qa−/−) mice with wild-type bone marrow cells. J Immunol. 2001;167(7):4033–7. Epub 2001/09/21. doi: 10.4049/jimmunol.167.7.4033. [DOI] [PubMed] [Google Scholar]
- 110.Arkwright PD, Riley P, Hughes SM, Alachkar H, Wynn RF. Successful cure of C1q deficiency in human subjects treated with hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2014;133(1):265–7. Epub 2013/09/17. doi: 10.1016/j.jaci.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 111.Olsson RF, Hagelberg S, Schiller B, Ringden O, Truedsson L, Ahlin A. Allogeneic Hematopoietic Stem Cell Transplantation in the Treatment of Human C1q Deficiency: The Karolinska Experience. Transplantation. 2016;100(6):1356–62. Epub 2015/10/31. doi: 10.1097/TP.0000000000000975. [DOI] [PubMed] [Google Scholar]
- 112.Day NK, Geiger H, Stroud R, DeBracco M, Mancaido B, Windhorst D, Good RA. C1r deficiency: an inborn error associated with cutaneous and renal disease. J Clin Invest. 1972;51(5):1102–8. Epub 1972/05/01. doi: 10.1172/JCI106902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moncada B, Day NK, Good RA, Windhorst DB. Lupus-erythematosus-like syndrome with a familial defect of complement. N Engl J Med. 1972;286(13):689–93. Epub 1972/03/30. doi: 10.1056/NEJM197203302861304. [DOI] [PubMed] [Google Scholar]
- 114.Pickering RJ, Naff GB, Stroud RM, Good RA, Gewurz H. Deficiency of C1r in human serum. Effects on the structure and function of macromolecular C1. J Exp Med. 1970;131(4):803–15. Epub 1970/04/01. doi: 10.1084/jem.131.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dragon-Durey MA, Quartier P, Fremeaux-Bacchi V, Blouin J, de Barace C, Prieur AM, Weiss L, Fridman WH. Molecular basis of a selective C1s deficiency associated with early onset multiple autoimmune diseases. J Immunol. 2001;166(12):7612–6. Epub 2001/06/08. doi: 10.4049/jimmunol.166.12.7612. [DOI] [PubMed] [Google Scholar]
- 116.Wu YL, Brookshire BP, Verani RR, Arnett FC, Yu CY. Clinical presentations and molecular basis of complement C1r deficiency in a male African-American patient with systemic lupus erythematosus. Lupus. 2011;20(11):1126–34. Epub 2011/07/26. doi: 10.1177/0961203311404914. [DOI] [PubMed] [Google Scholar]
- 117.Demirkaya E, Zhou Q, Smith CK, Ombrello MJ, Deuitch N, Tsai WL, Hoffmann P, Remmers EF, Takeuchi M, Park YH, Chae J, Barut K, Simsek D, Adrovic A, Sahin S, Caliskan S, Chandrasekharappa SC, Hasni SA, Ombrello AK, Gadina M, Kastner DL, Kaplan MJ, Kasapcopur O, Aksentijevich I. Brief Report: Deficiency of Complement 1r Subcomponent in Early-Onset Systemic Lupus Erythematosus: The Role of DiseaseModifying Alleles in a Monogenic Disease. Arthritis Rheumatol. 2017;69(9):1832–9. Epub 2017/05/26. doi: 10.1002/art.40158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhuang H, Han S, Li Y, Kienhofer D, Lee P, Shumyak S, Meyerholz R, Rosadzinski K, Rosner D, Chan A, Xu Y, Segal M, Sobel E, Yang LJ, Hoffmann MH, Reeves WH. A Novel Mechanism for Generating the Interferon Signature in Lupus: Opsonization of Dead Cells by Complement and IgM. Arthritis Rheumatol. 2016;68(12):2917–28. Epub 2016/06/09. doi: 10.1002/art.39781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johnson CA, Densen P, Wetsel RA, Cole FS, Goeken NE, Colten HR. Molecular heterogeneity of C2 deficiency. N Engl J Med. 1992;326(13):871–4. Epub 1992/03/26. doi: 10.1056/NEJM199203263261306. [DOI] [PubMed] [Google Scholar]
- 120.Yu CY, Hauptmann G, Yang Y, Wu YL, Birmingham DJ, Rovin BH, Hebert LA. Complement deficiencies in human systemic lupus erythematosus (SLE) and SLE nephritis: epidemiology and pathogenesis. In: Tsokos GC, Gordon C, Smolen JS, editors. Systemic Lupus Erythematosus, a companion to rheumatology. Philadelphia, PA: Mosby Elsevier; 2007. p. 183–93. [Google Scholar]
- 121.Wetsel RA, Kulics J, Lokki ML, Kiepiela P, Akama H, Johnson CA, Densen P, Colten HR. Type II human complement C2 deficiency. Allele-specific amino acid substitutions (Ser189 --> Phe; Gly444 --> Arg) cause impaired C2 secretion. J Biol Chem. 1996;271(10):5824–31. Epub 1996/03/08. doi: 10.1074/jbc.271.10.5824. [DOI] [PubMed] [Google Scholar]
- 122.Zhu ZB, Atkinson TP, Volanakis JE. A novel type II complement C2 deficiency allele in an African-American family. J Immunol. 1998;161(2):578–84. Epub 1998/07/22. [PubMed] [Google Scholar]
- 123.Johnson CA, Densen P, Hurford RK Jr., Colten HR, Wetsel RA. Type I human complement C2 deficiency. A 28-base pair gene deletion causes skipping of exon 6 during RNA splicing. J Biol Chem. 1992;267(13):9347–53. Epub 1992/05/15. [PubMed] [Google Scholar]
- 124.Wang X, Circolo A, Lokki ML, Shackelford PG, Wetsel RA, Colten HR. Molecular heterogeneity in deficiency of complement protein C2 type I. Immunology. 1998;93(2):184–91. Epub 1998/06/09. doi: 10.1046/j.1365-2567.1998.00392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Botto M, Fong KY, So AK, Rudge A, Walport MJ. Molecular basis of hereditary C3 deficiency. J Clin Invest. 1990;86(4):1158–63. Epub 1990/10/01. doi: 10.1172/JCI114821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.E SR, Falcao DA, Isaac L. Clinical aspects and molecular basis of primary deficiencies of complement component C3 and its regulatory proteins factor I and factor H. Scand J Immunol. 2006;63(3):155–68. Epub 2006/02/28. doi: 10.1111/j.1365-3083.2006.01729.x. [DOI] [PubMed] [Google Scholar]
- 127.Tsukamoto H, Horiuchi T, Kokuba H, Nagae S, Nishizaka H, Sawabe T, Harashima S, Himeji D, Koyama T, Otsuka J, Mitoma H, Kimoto Y, Hashimura C, Kitano E, Kitamura H, Furue M, Harada M. Molecular analysis of a novel hereditary C3 deficiency with systemic lupus erythematosus. Biochem Biophys Res Commun. 2005;330(1):298–304. Epub 2005/03/23. doi: 10.1016/j.bbrc.2005.02.159. [DOI] [PubMed] [Google Scholar]
- 128.Carroll MC, Campbell RD, Bentley DR, Porter RR. A molecular map of the human major histocompatibility complex class III region linking complement genes C4, C2 and factor B. Nature. 1984;307(5948):237–41. Epub 1984/01/19. doi: 10.1038/307237a0. [DOI] [PubMed] [Google Scholar]
- 129.Yang Z, Qu X, Yu CY. Features of the two gene pairs RD-SKI2W and DOM3Z-RP1 located between complement component genes factor B and C4 at the MHC class III region. Front Biosci. 2001;6:D927–35. Epub 2001/08/07. doi: 10.2741/yang. [DOI] [PubMed] [Google Scholar]
- 130.Yu CY, Yang Z, Blanchong CA, Miller W. The human and mouse MHC class III region: a parade of 21 genes at the centromeric segment. Immunol Today. 2000;21(7):320–8. Epub 2000/06/29. doi: 10.1016/s0167-5699(00)01664-9. [DOI] [PubMed] [Google Scholar]
- 131.Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, Sotos JF, Zipf WB, Rennebohm RM, Yung Yu C. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in caucasians. The load of RCCX genetic diversity on major histocompatibility complexassociated disease. J Exp Med. 2000;191(12):2183–96. Epub 2000/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen JY, Wu YL, Mok MY, Wu YJ, Lintner KE, Wang CM, Chung EK, Yang Y, Zhou B, Wang H, Yu D, Alhomosh A, Jones K, Spencer CH, Nagaraja HN, Lau YL, Lau CS, Yu CY. Effects of Complement C4 Gene Copy Number Variations, Size Dichotomy, and C4A Deficiency on Genetic Risk and Clinical Presentation of Systemic Lupus Erythematosus in East Asian Populations. Arthritis Rheumatol. 2016;68(6):1442–53. Epub 2016/01/28. doi: 10.1002/art.39589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chung EK, Yang Y, Rennebohm RM, Lokki ML, Higgins GC, Jones KN, Zhou B, Blanchong CA, Yu CY. Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am J Hum Genet. 2002;71(4):823–37. Epub 2002/09/13. doi: 10.1086/342777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem. 1999;274(17):12147–56. Epub 1999/04/17. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- 135.Yu CY, Campbell RD. Definitive RFLPs to distinguish between the human complement C4A/C4B isotypes and the major Rodgers/Chido determinants: application to the study of C4 null alleles. Immunogenetics. 1987;25(6):383–90. Epub 1987/01/01. [DOI] [PubMed] [Google Scholar]
- 136.Zhou D, Lai M, Luo A, Yu CY. An RNA Metabolism and Surveillance Quartet in the Major Histocompatibility Complex. Cells. 2019;8(9). Epub 2019/09/05. doi: 10.3390/cells8091008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu LC, Yu CY. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics. 1994;40(6):425–36. Epub 1994/01/01. [DOI] [PubMed] [Google Scholar]
- 138.Yu CY. The complete exon-intron structure of a human complement component C4A gene. DNA sequences, polymorphism, and linkage to the 21-hydroxylase gene. J Immunol. 1991;146(3):1057–66. Epub 1991/02/01. [PubMed] [Google Scholar]
- 139.Mack M, Bender K, Schneider PM. Detection of retroviral antisense transcripts and promoter activity of the HERV-K(C4) insertion in the MHC class III region. Immunogenetics. 2004;56(5):321–32. Epub 2004/08/17. doi: 10.1007/s00251-004-0705-y. [DOI] [PubMed] [Google Scholar]
- 140.Yu CY, Belt KT, Giles CM, Campbell RD, Porter RR. Structural basis of the polymorphism of human complement components C4A and C4B: gene size, reactivity and antigenicity. EMBO J. 1986;5(11):2873–81. Epub 1986/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yu CY, Whitacre CC. Sex, MHC and complement C4 in autoimmune diseases. Trends Immunol. 2004;25(12):694–9. Epub 2004/11/09. doi: 10.1016/j.it.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 142.Schneider PM, Witzel-Schlömp K, Rittner C, Zhang L. The endogenous retroviral insertion in the human complement C4 gene modulates the expression of homologous genes by antisense inhibition. Immunogenetics. 2001;53(1):1–9. Epub 2001/03/23. doi: 10.1007/s002510000288. [DOI] [PubMed] [Google Scholar]
- 143.Yu CY, Campbell RD, Porter RR. A structural model for the location of the Rodgers and the Chido antigenic determinants and their correlation with the human complement component C4A/C4B isotypes. Immunogenetics. 1988;27(6):399–405. Epub 1988/01/01. doi: 10.1007/bf00364425. [DOI] [PubMed] [Google Scholar]
- 144.Chung EK, Wu YL, Yang Y, Zhou B, Yu CY. Human complement components C4A and C4B genetic diversities: complex genotypes and phenotypes. Curr Protoc Immunol. 2005;Chapter 13:Unit 13.8. Epub 2008/04/25. doi: 10.1002/0471142735.im1308s68. [DOI] [PubMed]
- 145.Chung EK, Yang Y, Rupert KL, Jones KN, Rennebohm RM, Blanchong CA, Yu CY. Determining the one, two, three, or four long and short loci of human complement C4 in a major histocompatibility complex haplotype encoding C4A or C4B proteins. Am J Hum Genet. 2002;71(4):810–22. Epub 2002/09/12. doi: 10.1086/342778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fredrikson GN, Gullstrand B, Schneider PM, Witzel-Schlomp K, Sjoholm AG, Alper CA, Awdeh Z, Truedsson L. Characterization of non-expressed C4 genes in a case of complete C4 deficiency: identification of a novel point mutation leading to a premature stop codon. Hum Immunol. 1998;59(11):713–9. Epub 1998/10/31. [DOI] [PubMed] [Google Scholar]
- 147.Lokki ML, Circolo A, Ahokas P, Rupert KL, Yu CY, Colten HR. Deficiency of human complement protein C4 due to identical frameshift mutations in the C4A and C4B genes. J Immunol. 1999;162(6):3687–93. Epub 1999/03/27. [PubMed] [Google Scholar]
- 148.Rupert KL, Moulds JM, Yang Y, Arnett FC, Warren RW, Reveille JD, Myones BL, Blanchong CA, Yu CY. The molecular basis of complete complement C4A and C4B deficiencies in a systemic lupus erythematosus patient with homozygous C4A and C4B mutant genes. J Immunol. 2002;169(3):1570–8. Epub 2002/07/23. doi: 10.4049/jimmunol.169.3.1570. [DOI] [PubMed] [Google Scholar]
- 149.Wu YL, Hauptmann G, Viguier M, Yu CY. Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun. 2009;10(5):433–45. Epub 2009/03/13. doi: 10.1038/gene.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang Y, Lhotta K, Chung EK, Eder P, Neumair F, Yu CY. Complete complement components C4A and C4B deficiencies in human kidney diseases and systemic lupus erythematosus. J Immunol. 2004;173(4):2803–14. Epub 2004/08/06. [DOI] [PubMed] [Google Scholar]
- 151.Barba G, Rittner C, Schneider PM. Genetic basis of human complement C4A deficiency. Detection of a point mutation leading to nonexpression. J Clin Invest. 1993;91(4):1681–6. Epub 1993/04/01. doi: 10.1172/JCI116377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sullivan KE, Kim NA, Goldman D, Petri MA. C4A deficiency due to a 2 bp insertion is increased in patients with systemic lupus erythematosus. J Rheumatol. 1999;26(10):2144–7. Epub 1999/10/21. [PubMed] [Google Scholar]
- 153.Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, Hebert M, Jones KN, Shu Y, Kitzmiller K, Blanchong CA, McBride KL, Higgins GC, Rennebohm RM, Rice RR, Hackshaw KV, Roubey RA, Grossman JM, Tsao BP, Birmingham DJ, Rovin BH, Hebert LA, Yu CY. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80(6):1037–54. Epub 2007/05/16. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou D, Rudnicki M, Chua GT, Lawrance SK, Zhou B, Drew JL, Barbar-Smiley F, Armstrong TK, Hilt ME, Birmingham DJ, Passler W, Auletta JJ, Bowden SA, Hoffman RP, Wu YL, Jarjour WN, Mok CC, Ardoin SP, Lau YL, Yu CY. Human Complement C4B Allotypes and Deficiencies in Selected Cases With Autoimmune Diseases. Front Immunol. 2021;12:739430. Epub 2021/11/13. doi: 10.3389/fimmu.2021.739430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Law SK, Dodds AW, Porter RR. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 1984;3(8):1819–23. Epub 1984/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Isenman DE, Young JR. The molecular basis for the difference in immune hemolysis activity of the Chido and Rodgers isotypes of human complement component C4. J Immunol. 1984;132(6):3019–27. Epub 1984/06/01. [PubMed] [Google Scholar]
- 157.Taylor PR, Carugati A, Fadok VA, Cook HT, Andrews M, Carroll MC, Savill JS, Henson PM, Botto M, Walport MJ. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J Exp Med. 2000;192(3):359–66. Epub 2000/08/10. doi: 10.1084/jem.192.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nauta AJ, Daha MR, van Kooten C, Roos A. Recognition and clearance of apoptotic cells: a role for complement and pentraxins. Trends Immunol. 2003;24(3):148–54. Epub 2003/03/05. doi: 10.1016/s1471-4906(03)00030-9. [DOI] [PubMed] [Google Scholar]
- 159.Liesmaa I, Paakkanen R, Järvinen A, Valtonen V, Lokki ML. Clinical features of patients with homozygous complement C4A or C4B deficiency. PLoS One. 2018;13(6):e0199305. Epub 2018/06/22. doi: 10.1371/journal.pone.0199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–92. Epub 2006/09/19. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 161.Niewold TB, Kariuki SN, Morgan GA, Shrestha S, Pachman LM. Elevated serum interferon-alpha activity in juvenile dermatomyositis: associations with disease activity at diagnosis and after thirty-six months of therapy. Arthritis Rheum. 2009;60(6):1815–24. Epub 2009/05/30. doi: 10.1002/art.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Juptner M, Flachsbart F, Caliebe A, Lieb W, Schreiber S, Zeuner R, Franke A, Schroder JO. Low copy numbers of complement C4 and homozygous deficiency of C4A may predispose to severe disease and earlier disease onset in patients with systemic lupus erythematosus. Lupus. 2018;27(4):600–9. Epub 2017/10/21. doi: 10.1177/0961203317735187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu YL, Savelli SL, Yang Y, Zhou B, Rovin BH, Birmingham DJ, Nagaraja HN, Hebert LA, Yu CY. Sensitive and specific real-time polymerase chain reaction assays to accurately determine copy number variations (CNVs) of human complement C4A, C4B, C4-long, C4-short, and RCCX modules: elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J Immunol. 2007;179(5):3012–25. Epub 2007/08/22. [DOI] [PubMed] [Google Scholar]
- 164.Lau CS, Yin G, Mok MY. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus. 2006;15(11):715–9. Epub 2006/12/13. doi: 10.1177/0961203306072311. [DOI] [PubMed] [Google Scholar]
- 165.Dunckley H, Gatenby PA, Hawkins B, Naito S, Serjeantson SW. Deficiency of C4A is a genetic determinant of systemic lupus erythematosus in three ethnic groups. J Immunogenet. 1987;14(4–5):209–18. Epub 1987/08/01. doi: 10.1111/j.1744313x.1987.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 166.Kim JH, Jung SH, Bae JS, Lee HS, Yim SH, Park SY, Bang SY, Hu HJ, Shin HD, Bae SC, Chung YJ. Deletion variants of RABGAP1L, 10q21.3, and C4 are associated with the risk of systemic lupus erythematosus in Korean women. Arthritis Rheum. 2013;65(4):1055–63. Epub 2013/01/22. doi: 10.1002/art.37854. [DOI] [PubMed] [Google Scholar]
- 167.Lv Y, He S, Zhang Z, Li Y, Hu D, Zhu K, Cheng H, Zhou F, Chen G, Zheng X, Li P, Ren Y, Yin X, Cui Y, Sun L, Yang S, Zhang X. Confirmation of C4 gene copy number variation and the association with systemic lupus erythematosus in Chinese Han population. Rheumatol Int. 2012;32(10):3047–53. Epub 2011/09/10. doi: 10.1007/s00296-011-2023-7. [DOI] [PubMed] [Google Scholar]
- 168.Pereira KMC, Perazzio S, Faria AGA, Moreira ES, Santos VC, Grecco M, da Silva NP, Andrade LEC. Impact of C4, C4A and C4B gene copy number variation in the susceptibility, phenotype and progression of systemic lupus erythematosus. Adv Rheumatol. 2019;59(1):36. Epub 2019/08/08. doi: 10.1186/s42358-019-0076-6. [DOI] [PubMed] [Google Scholar]
- 169.Pereira KM, Faria AG, Liphaus BL, Jesus AA, Silva CA, Carneiro-Sampaio M, Andrade LE. Low C4, C4A and C4B gene copy numbers are stronger risk factors for juvenile-onset than for adult-onset systemic lupus erythematosus. Rheumatology (Oxford). 2016;55(5):869–73. Epub 2016/01/24. doi: 10.1093/rheumatology/kev436. [DOI] [PubMed] [Google Scholar]
- 170.Mulvihill E, Ardoin S, Thompson SD, Zhou B, Yu GR, King E, Singer N, Levy DM, Brunner H, Wu YL, Nagaraja HN, Schanberg LE, Yu CY. Elevated serum complement levels and higher gene copy number of complement C4B are associated with hypertension and effective response to statin therapy in childhood-onset systemic lupus erythematosus (SLE). Lupus Sci Med. 2019;6(1):e000333. Epub 2019/08/27. doi: 10.1136/lupus-2019-000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schanberg LE, Sandborg C, Barnhart HX, Ardoin SP, Yow E, Evans GW, Mieszkalski KL, Ilowite NT, Eberhard A, Imundo LF, Kimura Y, von Scheven E, Silverman E, Bowyer SL, Punaro M, Singer NG, Sherry DD, McCurdy D, Klein-Gitelman M, Wallace C, Silver R, Wagner-Weiner L, Higgins GC, Brunner HI, Jung L, Soep JB, Reed AM, Provenzale J, Thompson SD, Atherosclerosis Prevention in Pediatric Lupus Erythematosus I. Use of atorvastatin in systemic lupus erythematosus in children and adolescents. Arthritis Rheum. 2012;64(1):285–96. Epub 2011/10/28. doi: 10.1002/art.30645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Elliott JA Jr., Mathieson DR. Complement in disseminated (systemic) lupus erythematosus. AMA Arch Derm Syphilol. 1953;68(2):119–28. Epub 1953/08/01. [DOI] [PubMed] [Google Scholar]
- 173.Lewis EJ, Carpenter CB, Schur PH. Serum complement component levels in human glomerulonephritis. Ann Intern Med. 1971;75(4):555–60. Epub 1971/10/01. [DOI] [PubMed] [Google Scholar]
- 174.Liu CC, Manzi S, Kao AH, Navratil JS, Ruffing MJ, Ahearn JM. Reticulocytes bearing C4d as biomarkers of disease activity for systemic lupus erythematosus. Arthritis Rheum. 2005;52(10):3087–99. Epub 2005/10/04. doi: 10.1002/art.21305. [DOI] [PubMed] [Google Scholar]
- 175.Savelli SL, Roubey RAS, Kitzmiller KJ, Zhou D, Nagaraja HN, Mulvihill E, Barbar-Smiley F, Ardoin SP, Wu YL, Yu CY. Opposite Profiles of Complement in Antiphospholipid Syndrome (APS) and Systemic Lupus Erythematosus (SLE) Among Patients With Antiphospholipid Antibodies (aPL). Front Immunol. 2019;10:885. Epub 2019/05/28. doi: 10.3389/fimmu.2019.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Miyakis S, Lockshin MD, Atsumi T, Branch DW, Brey RL, Cervera R, Derksen RH, PG DEG, Koike T, Meroni PL, Reber G, Shoenfeld Y, Tincani A, Vlachoyiannopoulos PG, Krilis SA. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J Thromb Haemost. 2006;4(2):295–306. Epub 2006/01/20. doi: 10.1111/j.1538-7836.2006.01753.x. [DOI] [PubMed] [Google Scholar]
- 177.Wilson WA, Gharavi AE, Koike T, Lockshin MD, Branch DW, Piette JC, Brey R, Derksen R, Harris EN, Hughes GR, Triplett DA, Khamashta MA. International consensus statement on preliminary classification criteria for definite antiphospholipid syndrome: report of an international workshop. Arthritis Rheum. 1999;42(7):1309–11. Epub 1999/07/14. doi: . [DOI] [PubMed] [Google Scholar]
- 178.Koike T. Antiphospholipid syndrome: 30 years and our contribution. Int J Rheum Dis. 2015;18(2):233–41. Epub 2014/12/20. doi: 10.1111/1756-185X.12438. [DOI] [PubMed] [Google Scholar]
- 179.Steinkasserer A, Estaller C, Weiss EH, Sim RB, Day AJ. Complete nucleotide and deduced amino acid sequence of human beta 2-glycoprotein I. Biochem J. 1991;277 ( Pt 2):387–91. Epub 1991/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Weaver JC, Krilis SA, Giannakopoulos B. Oxidative post-translational modification of betaeta 2-glycoprotein I in the pathophysiology of the anti-phospholipid syndrome. Free Radic Biol Med. 2018;125:98–103. Epub 2018/04/01. doi: 10.1016/j.freeradbiomed.2018.03.048. [DOI] [PubMed] [Google Scholar]
- 181.Kim MY, Guerra MM, Kaplowitz E, Laskin CA, Petri M, Branch DW, Lockshin MD, Sammaritano LR, Merrill JT, Porter TF, Sawitzke A, Lynch AM, Buyon JP, Salmon JE. Complement activation predicts adverse pregnancy outcome in patients with systemic lupus erythematosus and/or antiphospholipid antibodies. Ann Rheum Dis. 2018;77(4):549–55. Epub 2018/01/27. doi: 10.1136/annrheumdis-2017-212224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shamonki JM, Salmon JE, Hyjek E, Baergen RN. Excessive complement activation is associated with placental injury in patients with antiphospholipid antibodies. Am J Obstet Gynecol. 2007;196(2):167 e1–5. Epub 2007/02/20. doi: 10.1016/j.ajog.2006.10.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cervera R. Antiphospholipid syndrome. Thromb Res. 2017;151 Suppl 1:S43–S7. Epub 2017/03/07. doi: 10.1016/S0049-3848(17)30066-X. [DOI] [PubMed] [Google Scholar]
- 184.Cervera R, Piette JC, Font J, Khamashta MA, Shoenfeld Y, Camps MT, Jacobsen S, Lakos G, Tincani A, Kontopoulou-Griva I, Galeazzi M, Meroni PL, Derksen RH, de Groot PG, Gromnica-Ihle E, Baleva M, Mosca M, Bombardieri S, Houssiau F, Gris JC, Quere I, Hachulla E, Vasconcelos C, Roch B, Fernandez-Nebro A, Boffa MC, Hughes GR, Ingelmo M, Euro-Phospholipid Project G. Antiphospholipid syndrome: clinical and immunologic manifestations and patterns of disease expression in a cohort of 1,000 patients. Arthritis Rheum. 2002;46(4):1019–27. Epub 2002/04/16. [DOI] [PubMed] [Google Scholar]
- 185.Love PE, Santoro SA. Antiphospholipid antibodies: anticardiolipin and the lupus anticoagulant in systemic lupus erythematosus (SLE) and in non-SLE disorders. Prevalence and clinical significance. Ann Intern Med. 1990;112(9):682–98. Epub 1990/05/01. [DOI] [PubMed] [Google Scholar]
- 186.Pons-Estel GJ, Andreoli L, Scanzi F, Cervera R, Tincani A. The antiphospholipid syndrome in patients with systemic lupus erythematosus. J Autoimmun. 2017;76:10–20. Epub 2016/10/26. doi: 10.1016/j.jaut.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 187.Savelli SL, Roubey RAS, Kitzmiller KJ, Zhou D, Nagaraja HN, Mulvihill E, Barbar-Smiley F, Ardoin SP, Wu YL, Yu CY. Opposite Profiles of Complement in Antiphospholipid Syndrome (APS) and Systemic Lupus Erythematosus (SLE) among Patients with Antiphospholipid Antibodies. Front Immunol. 2019;10:885. doi: 10.3389/fimmu.2019.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Atkinson JP. Complement system on the attack in autoimmunity. J Clin Invest. 2003;112(11):1639–41. Epub 2003/12/09. doi: 10.1172/JCI20309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Branch DW, Dudley DJ, Mitchell MD, Creighton KA, Abbott TM, Hammond EH, Daynes RA. Immunoglobulin G fractions from patients with antiphospholipid antibodies cause fetal death in BALB/c mice: a model for autoimmune fetal loss. Am J Obstet Gynecol. 1990;163(1 Pt 1):210–6. Epub 1990/07/01. [DOI] [PubMed] [Google Scholar]
- 190.Cohen D, Buurma A, Goemaere NN, Girardi G, le Cessie S, Scherjon S, Bloemenkamp KW, de Heer E, Bruijn JA, Bajema IM. Classical complement activation as a footprint for murine and human antiphospholipid antibody-induced fetal loss. J Pathol. 2011;225(4):502–11. Epub 2011/06/21. doi: 10.1002/path.2893. [DOI] [PubMed] [Google Scholar]
- 191.Girardi G, Berman J, Redecha P, Spruce L, Thurman JM, Kraus D, Hollmann TJ, Casali P, Caroll MC, Wetsel RA, Lambris JD, Holers VM, Salmon JE. Complement C5a receptors and neutrophils mediate fetal injury in the antiphospholipid syndrome. J Clin Invest. 2003;112(11):1644–54. Epub 2003/12/09. doi: 10.1172/JCI18817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Holers VM, Girardi G, Mo L, Guthridge JM, Molina H, Pierangeli SS, Espinola R, Xiaowei LE, Mao D, Vialpando CG, Salmon JE. Complement C3 activation is required for antiphospholipid antibody-induced fetal loss. J Exp Med. 2002;195(2):211–20. Epub 2002/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tedesco F, Borghi MO, Gerosa M, Chighizola CB, Macor P, Lonati PA, Gulino A, Belmonte B, Meroni PL. Pathogenic Role of Complement in Antiphospholipid Syndrome and Therapeutic Implications. Front Immunol. 2018;9:1388. Epub 2018/07/05. doi: 10.3389/fimmu.2018.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ekdahl KN, Teramura Y, Hamad OA, Asif S, Duehrkop C, Fromell K, Gustafson E, Hong J, Kozarcanin H, Magnusson PU, Huber-Lang M, Garred P, Nilsson B. Dangerous liaisons: complement, coagulation, and kallikrein/kinin cross-talk act as a linchpin in the events leading to thromboinflammation. Immunol Rev. 2016;274(1):245–69. Epub 2016/10/27. doi: 10.1111/imr.12471. [DOI] [PubMed] [Google Scholar]
- 195.Oku K, Atsumi T, Bohgaki M, Amengual O, Kataoka H, Horita T, Yasuda S, Koike T. Complement activation in patients with primary antiphospholipid syndrome. Ann Rheum Dis. 2009;68(6):1030–5. Epub 2008/07/16. doi: 10.1136/ard.2008.090670. [DOI] [PubMed] [Google Scholar]
- 196.Oku K, Nakamura H, Kono M, Ohmura K, Kato M, Bohgaki T, Horita T, Yasuda S, Amengual O, Atsumi T. Complement and thrombosis in the antiphospholipid syndrome. Autoimmun Rev. 2016;15(10):1001–4. Epub 2016/08/04. doi: 10.1016/j.autrev.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 197.Dragon-Durey MA, Blanc C, Marinozzi MC, van Schaarenburg RA, Trouw LA. Autoantibodies against complement components and functional consequences. Mol Immunol. 2013;56(3):213–21. Epub 2013/06/25. doi: 10.1016/j.molimm.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 198.Fremeaux-Bacchi V, Weiss L, Demouchy C, Blouin J, Kazatchkine MD. Autoantibodies to the collagen-like region of C1q are strongly associated with classical pathway-mediated hypocomplementemia in systemic lupus erythematosus. Lupus. 1996;5(3):216–20. Epub 1996/06/01. doi: 10.1177/096120339600500309. [DOI] [PubMed] [Google Scholar]
- 199.Mahler M, van Schaarenburg RA, Trouw LA. Anti-C1q autoantibodies, novel tests, and clinical consequences. Front Immunol. 2013;4:117. Epub 2013/05/30. doi: 10.3389/fimmu.2013.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Orbai AM, Truedsson L, Sturfelt G, Nived O, Fang H, Alarcon GS, Gordon C, Merrill J, Fortin PR, Bruce IN, Isenberg DA, Wallace DJ, Ramsey-Goldman R, Bae SC, Hanly JG, Sanchez-Guerrero J, Clarke AE, Aranow CB, Manzi S, Urowitz MB, Gladman DD, Kalunian KC, Costner MI, Werth VP, Zoma A, Bernatsky S, Ruiz-Irastorza G, Khamashta MA, Jacobsen S, Buyon JP, Maddison P, Dooley MA, Van Vollenhoven RF, Ginzler E, Stoll T, Peschken C, Jorizzo JL, Callen JP, Lim SS, Fessler BJ, Inanc M, Kamen DL, Rahman A, Steinsson K, Franks AG Jr., Sigler L, Hameed S, Pham N, Brey R, Weisman MH, McGwin G Jr., Magder LS, Petri M. Anti-C1q antibodies in systemic lupus erythematosus. Lupus. 2015;24(1):42–9. Epub 2014/08/16. doi: 10.1177/0961203314547791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lachmann PJ. Conglutinin and immunoconglutinins. Adv Immunol. 1967;6:479–527. Epub 1967/01/01. [DOI] [PubMed] [Google Scholar]
- 202.Nilsson B, Ekdahl KN, Sjoholm A, Nilsson UR, Sturfelt G. Detection and characterization of immunoconglutinins in patients with systemic lupus erythematosus (SLE): serial analysis in relation to disease course. Clin Exp Immunol. 1992;90(2):251–5. Epub 1992/11/01. doi: 10.1111/j.1365-2249.1992.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ronnelid J, Gunnarsson I, Nilsson-Ekdahl K, Nilsson B. Correlation between anti-C1q and immune conglutinin levels, but not between levels of antibodies to the structurally related autoantigens C1q and type II collagen in SLE or RA. J Autoimmun. 1997;10(4):415–23. Epub 1997/08/01. doi: 10.1006/jaut.1997.0147. [DOI] [PubMed] [Google Scholar]
- 204.Vasilev VV, Noe R, Dragon-Durey MA, Chauvet S, Lazarov VJ, Deliyska BP, Fremeaux-Bacchi V, Dimitrov JD, Roumenina LT. Functional Characterization of Autoantibodies against Complement Component C3 in Patients with Lupus Nephritis. J Biol Chem. 2015;290(42):25343–55. Epub 2015/08/08. doi: 10.1074/jbc.M115.647008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Birmingham DJ, Bitter JE, Ndukwe EG, Dials S, Gullo TR, Conroy S, Nagaraja HN, Rovin BH, Hebert LA. Relationship of Circulating Anti-C3b and Anti-C1q IgG to Lupus Nephritis and Its Flare. Clin J Am Soc Nephrol. 2016;11(1):47–53. Epub 2015/12/25. doi: 10.2215/CJN.03990415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ito S, Tamura N, Fujita T. Effect of decay-accelerating factor on the assembly of the classical and alternative pathway C3 convertases in the presence of C4 or C3 nephritic factor. Immunology. 1989;68(4):449–52. Epub 1989/12/01. [PMC free article] [PubMed] [Google Scholar]
- 207.Jozsi M, Reuter S, Nozal P, Lopez-Trascasa M, Sanchez-Corral P, Prohaszka Z, Uzonyi B. Autoantibodies to complement components in C3 glomerulopathy and atypical hemolytic uremic syndrome. Immunol Lett. 2014;160(2):163–71. Epub 2014/02/05. doi: 10.1016/j.imlet.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 208.Daha MR, van Es LA. Relative resistance of the F-42-stabilized classical pathway C3 convertase to inactivation by C4-binding protein. J Immunol. 1980;125(5):2051–4. Epub 1980/11/01. [PubMed] [Google Scholar]
- 209.Miller EC, Chase NM, Densen P, Hintermeyer MK, Casper JT, Atkinson JP. Autoantibody stabilization of the classical pathway C3 convertase leading to C3 deficiency and Neisserial sepsis: C4 nephritic factor revisited. Clin Immunol. 2012;145(3):241–50. Epub 2012/11/03. doi: 10.1016/j.clim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhou D, King EH, Rothwell S, Krystufkova O, Notarnicola A, Coss S, Abdul-Aziz R, Miller KE, Dang A, Yu GR, Drew J, Lundström E, Pachman LM, Mamyrova G, Curiel RV, De Paepe B, De Bleecker JL, Payton A, Ollier W, O’Hanlon TP, Targoff IN, Flegel WA, Sivaraman V, Oberle E, Akoghlanian S, Driest K, Spencer CH, Wu YL, Nagaraja HN, Ardoin SP, Chinoy H, Rider LG, Miller FW, Lundberg IE, Padyukov L, Vencovský J, Lamb JA, Yu CY. Low copy numbers of complement C4 and C4A deficiency are risk factors for myositis, its subgroups and autoantibodies. Ann Rheum Dis. 2022. Epub 2022/09/29. doi: 10.1136/ard-2022-222935. [DOI] [PMC free article] [PubMed]
- 211.Holers VM, Banda NK. Complement in the Initiation and Evolution of Rheumatoid Arthritis. Front Immunol. 2018;9:1057. Epub 2018/06/13. doi: 10.3389/fimmu.2018.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cooke TD, Hurd ER, Jasin HE, Bienenstock J, Ziff M. Identification of immunoglobulins and complement in rheumatoid articular collagenous tissues. Arthritis Rheum. 1975;18(6):541–51. Epub 1975/11/01. doi: 10.1002/art.1780180603. [DOI] [PubMed] [Google Scholar]
- 213.Nakagawa K, Sakiyama H, Tsuchida T, Yamaguchi K, Toyoguchi T, Masuda R, Moriya H. Complement C1s activation in degenerating articular cartilage of rheumatoid arthritis patients: immunohistochemical studies with an active form specific antibody. Ann Rheum Dis. 1999;58(3):175–81. Epub 1999/06/12. doi: 10.1136/ard.58.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ballanti E, Perricone C, di Muzio G, Kroegler B, Chimenti MS, Graceffa D, Perricone R. Role of the complement system in rheumatoid arthritis and psoriatic arthritis: relationship with anti-TNF inhibitors. Autoimmun Rev. 2011;10(10):617–23. Epub 2011/05/10. doi: 10.1016/j.autrev.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 215.Banda NK, Deane KD, Bemis EA, Strickland C, Seifert J, Jordan K, Goldman K, Morgan BP, Moreland LW, Lewis MJ, Pitzalis C, Holers VM. Analysis of Complement Gene Expression, Clinical Associations, and Biodistribution of Complement Proteins in the Synovium of Early Rheumatoid Arthritis Patients Reveals Unique Pathophysiologic Features. J Immunol. 2022;208(11):2482–96. Epub 2022/05/03. doi: 10.4049/jimmunol.2101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mizuno M. A review of current knowledge of the complement system and the therapeutic opportunities in inflammatory arthritis. Curr Med Chem. 2006;13(14):1707–17. Epub 2006/06/22. doi: 10.2174/092986706777441959. [DOI] [PubMed] [Google Scholar]
- 217.Chimenti MS, Perricone C, Graceffa D, Di Muzio G, Ballanti E, Guarino MD, Conigliaro P, Greco E, Kroegler B, Perricone R. Complement system in psoriatic arthritis: a useful marker in response prediction and monitoring of anti-TNF treatment. Clin Exp Rheumatol. 2012;30(1):23–30. Epub 2012/01/21. [PubMed] [Google Scholar]
- 218.Aggarwal A, Bhardwaj A, Alam S, Misra R. Evidence for activation of the alternate complement pathway in patients with juvenile rheumatoid arthritis. Rheumatology. 2000;39(2):189–92. doi: 10.1093/rheumatology/39.2.189. [DOI] [PubMed] [Google Scholar]
- 219.Moore TL. Immune Complexes in Juvenile Idiopathic Arthritis. Front Immunol. 2016;7:177. Epub 2016/06/01. doi: 10.3389/fimmu.2016.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mollnes TE, Paus A. Complement activation in synovial fluid and tissue from patients with juvenile rheumatoid arthritis. Arthritis Rheum. 1986;29(11):1359–64. Epub 1986/11/01. doi: 10.1002/art.1780291108. [DOI] [PubMed] [Google Scholar]
- 221.Rigby WF, Wu YL, Zan M, Zhou B, Rosengren S, Carlson C, Hilton W, Yu CY. Increased frequency of complement C4B deficiency in rheumatoid arthritis. Arthritis Rheum. 2012;64(5):1338–44. Epub 2011/11/15. doi: 10.1002/art.33472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, Plant D, Gibbons LJ, Wilson AG, Bax DE, Morgan AW, Emery P, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Worthington J. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40(10):1156–9. Epub 2008/09/17. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kurreeman FA, Padyukov L, Marques RB, Schrodi SJ, Seddighzadeh M, Stoeken-Rijsbergen G, van der Helm-van Mil AH, Allaart CF, Verduyn W, Houwing-Duistermaat J, Alfredsson L, Begovich AB, Klareskog L, Huizinga TW, Toes RE. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4(9):e278. Epub 2007/09/21. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, Donn R, Symmons D, Hider S, Bruce IN, Wilson AG, Marinou I, Morgan A, Emery P, Carter A, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Strachan D, Worthington J. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–3. Epub 2007/11/06. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Trouw LA, Daha N, Kurreeman FAS, Böhringer S, Goulielmos GN, Westra HJ, Zhernakova A, Franke L, Stahl EA, Levarht EWN, Stoeken-Rijsbergen G, Verduijn W, Roos A, Li Y, Houwing-Duistermaat JJ, Huizinga TWJ, Toes REM. Genetic variants in the region of the C1q genes are associated with rheumatoid arthritis. Clinical and Experimental Immunology. 2013;173(1):76–83. doi: 10.1111/cei.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.van de Geijn FE, Hazes JMW, Geleijns K, Emonts M, Jacobs BC, Dufour-van den Goorbergh BCM, Dolhain RJEM. Mannose-binding lectin polymorphisms are not associated with rheumatoid arthritis—confirmation in two large cohorts. Rheumatology. 2008;47(8):1168–71. doi: 10.1093/rheumatology/ken226. [DOI] [PubMed] [Google Scholar]
- 227.Giles JL, Choy E, van den Berg C, Morgan BP, Harris CL. Functional analysis of a complement polymorphism (rs17611) associated with rheumatoid arthritis. J Immunol. 2015;194(7):3029–34. Epub 2015/03/01. doi: 10.4049/jimmunol.1402956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vergunst CE, Gerlag DM, Dinant H, Schulz L, Vinkenoog M, Smeets TJM, Sanders ME, Reedquist KA, Tak PP. Blocking the receptor for C5a in patients with rheumatoid arthritis does not reduce synovial inflammation. Rheumatology. 2007;46(12):1773–8. doi: 10.1093/rheumatology/kem222. [DOI] [PubMed] [Google Scholar]
- 229.Wang Y, Kristan J, Hao L, Lenkoski CS, Shen Y, Matis LA. A Role for Complement in Antibody-Mediated Inflammation: C5-Deficient DBA/1 Mice Are Resistant to Collagen-Induced Arthritis. The Journal of Immunology. 2000;164(8):4340–7. doi: 10.4049/jimmunol.164.8.4340. [DOI] [PubMed] [Google Scholar]
- 230.Goeldner I, Skare TL, Utiyama SR, Nisihara RM, Tong H, Messias-Reason IJ, Velavan TP. Mannose binding lectin and susceptibility to rheumatoid arthritis in Brazilian patients and their relatives. PLoS One. 2014;9(4):e95519. Epub 2014/04/23. doi: 10.1371/journal.pone.0095519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Graudal NA, Madsen HO, Tarp U, Svejgaard A, Jurik G, Graudal HK, Garred P. The association of variant mannose-binding lectin genotypes with radiographic outcome in rheumatoid arthritis. Arthritis Rheum. 2000;43(3):515–21. Epub 2000/03/23. doi: . [DOI] [PubMed] [Google Scholar]
- 232.Garred P, Madsen HO, Marquart H, Hansen TM, Sørensen SF, Petersen J, Volck B, Svejgaard A, Graudal NA, Rudd PM, Dwek RA, Sim RB, Andersen V. Two edged role of mannose binding lectin in rheumatoid arthritis: a cross sectional study. J Rheumatol. 2000;27(1):26–34. Epub 2000/01/27. [PubMed] [Google Scholar]
- 233.Rupert KL, Rennebohm RM, Yu CY. An unequal crossover between the RCCX modules of the human MHC leading to the presence of a CYP21B gene and a tenascin TNXB/TNXA-RP2 recombinant between C4A and C4B genes in a patient with juvenile rheumatoid arthritis. Exp Clin Immunogenet. 1999;16(2):81–97. Epub 1999/05/27. doi: 10.1159/000019099. [DOI] [PubMed] [Google Scholar]
- 234.Rowe P, Wasserfall C, Croker B, Campbell-Thompson M, Pugliese A, Atkinson M, Schatz D. Increased complement activation in human type 1 diabetes pancreata. Diabetes Care. 2013;36(11):3815–7. Epub 2013/09/18. doi: 10.2337/dc13-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kingery SE, Wu YL, Zhou B, Hoffman RP, Yu CY. Gene CNVs and protein levels of complement C4A and C4B as novel biomarkers for partial disease remissions in new-onset type 1 diabetes patients. Pediatr Diabetes. 2012;13(5):408–18. Epub 2011/12/14. doi: 10.1111/j.1399-5448.2011.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zavattari P, Lampis R, Motzo C, Loddo M, Mulargia A, Whalen M, Maioli M, Angius E, Todd JA, Cucca F. Conditional linkage disequilibrium analysis of a complex disease superlocus, IDDM1 in the HLA region, reveals the presence of independent modifying gene effects influencing the type 1 diabetes risk encoded by the major HLA-DQB1,- DRB1 disease loci. Human Molecular Genetics. 2001;10(8):881–9. doi: 10.1093/hmg/10.8.881. [DOI] [PubMed] [Google Scholar]
- 237.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, Todd JA. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450(7171):887–92. Epub 2007/11/16. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Noble JA, Valdes AM, Varney MD, Carlson JA, Moonsamy P, Fear AL, Lane JA, Lavant E, Rappner R, Louey A, Concannon P, Mychaleckyj JC, Erlich HA. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59(11):2972–9. Epub 2010/08/28. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Törn C, Liu X, Hagopian W, Lernmark Å, Simell O, Rewers M, Ziegler AG, Schatz D, Akolkar B, Onengut-Gumuscu S, Chen WM, Toppari J, Mykkänen J, Ilonen J, Rich SS, She JX, Sharma A, Steck A, Krischer J. Complement gene variants in relation to autoantibodies to beta cell specific antigens and type 1 diabetes in the TEDDY Study. Sci Rep. 2016;6:27887. Epub 2016/06/17. doi: 10.1038/srep27887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Al-Mayouf SM, Alreefi HA, Alsinan TA, AlSalmi G, AlRowais A, Al-Herz W, Alazami AM, Alsonbul A, Al-Mousa H. Lupus manifestations in children with primary immunodeficiency diseases: Comprehensive phenotypic and genetic features and outcome. Mod Rheumatol. 2021;31(6):1171–8. Epub 2021/02/11. doi: 10.1080/14397595.2021.1886627. [DOI] [PubMed] [Google Scholar]
- 241.Almlöf JC, Nystedt S, Leonard D, Eloranta ML, Grosso G, Sjöwall C, Bengtsson AA, Jönsen A, Gunnarsson I, Svenungsson E, Rönnblom L, Sandling JK, Syvänen AC. Whole-genome sequencing identifies complex contributions to genetic risk by variants in genes causing monogenic systemic lupus erythematosus. Hum Genet. 2019;138(2):141–50. Epub 2019/02/02. doi: 10.1007/s00439-018-01966-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Boteva L, Morris DL, Cortés-Hernández J, Martin J, Vyse TJ, Fernando MM. Genetically determined partial complement C4 deficiency states are not independent risk factors for SLE in UK and Spanish populations. Am J Hum Genet. 2012;90(3):445–56. Epub 2012/03/06. doi: 10.1016/j.ajhg.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Liu YH, Wan L, Chang CT, Liao WL, Chen WC, Tsai Y, Tsai CH, Tsai FJ. Association between copy number variation of complement component C4 and Graves’ disease. J Biomed Sci. 2011;18:71. Epub 2011/09/29. doi: 10.1186/1423-0127-18-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Garred P, Voss A, Madsen HO, Junker P. Association of mannose-binding lectin gene variation with disease severity and infections in a population-based cohort of systemic lupus erythematosus patients. Genes Immun. 2001;2(8):442–50. Epub 2002/01/10. doi: 10.1038/sj.gene.6363804. [DOI] [PubMed] [Google Scholar]
- 245.Ferluga J, Kouser L, Murugaiah V, Sim RB, Kishore U. Potential influences of complement factor H in autoimmune inflammatory and thrombotic disorders. Mol Immunol. 2017;84:84–106. Epub 2017/02/22. doi: 10.1016/j.molimm.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 246.Tsutsumi A, Sasaki K, Wakamiya N, Ichikawa K, Atsumi T, Ohtani K, Suzuki Y, Koike T, Sumida T. Mannose-binding lectin gene: polymorphisms in Japanese patients with systemic lupus erythematosus, rheumatoid arthritis and Sjögren’s syndrome. Genes Immun. 2001;2(2):99–104. Epub 2001/06/08. doi: 10.1038/sj.gene.6363744. [DOI] [PubMed] [Google Scholar]
- 247.Lundtoft C, Pucholt P, Martin M, Bianchi M, Lundstrom E, Eloranta ML, Sandling JK, Sjowall C, Jonsen A, Gunnarsson I, Rantapaa-Dahlqvist S, Bengtsson AA, Leonard D, Baecklund E, Jonsson R, Hammenfors D, Forsblad-d’Elia H, Eriksson P, Mandl T, Bucher SM, Norheim KB, Johnsen SJA, Omdal R, Kvarnstrom M, Wahren-Herlenius M, Notarnicola A, Andersson H, Molberg O, Diederichsen LP, Almlof J, Syvanen AC, Kozyrev SV, Lindblad-Toh K, consortium D, ImmunoArray C, Nilsson B, Blom AM, Lundberg IE, Nordmark G, Diaz-Gallo LM, Svenungsson E, Ronnblom L. Complement C4 copy number variation is linked to SSA/Ro and SSB/La autoantibodies in systemic inflammatory autoimmune diseases. Arthritis Rheumatol. 2022. Epub 2022/03/23. doi: 10.1002/art.42122. [DOI] [PMC free article] [PubMed]
- 248.Kamitaki N, Sekar A, Handsaker RE, de Rivera H, Tooley K, Morris DL, Taylor KE, Whelan CW, Tombleson P, Loohuis LMO, Schizophrenia Working Group of the Psychiatric Genomics C, Boehnke M, Kimberly RP, Kaufman KM, Harley JB, Langefeld CD, Seidman CE, Pato MT, Pato CN, Ophoff RA, Graham RR, Criswell LA, Vyse TJ, McCarroll SA. Complement genes contribute sex-biased vulnerability in diverse disorders. Nature. 2020;582(7813):577–81. Epub 2020/06/06. doi: 10.1038/s41586-020-2277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Atkinson JP, Yu CY. Genetic Susceptibility and Class III Complement Genes. In: Lahita RG, Tsokos G, Buyon J, Koike T, editors. Systemic Lupus Erythematosus. 5th ed. London: Elsevier Academic Press; 2011. p. 21–45. [Google Scholar]
- 250.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269(11):8466–76. Epub 1994/03/18. [PubMed] [Google Scholar]