Abstract
This study aimed to analyze the polymorphism of the vasoactive intestinal peptide receptor-1 (VIPR-1) gene and its association with growth traits in quail using the PCR-RFLP and sequencing techniques. Genomic DNA was extracted from blood samples of 36 female Savimalt (SV) quails and 49 female French Giant (FG) quails. Growth traits were measured and used for VIPR-1 gene analysis, as body weight (BW), tibia length (TL), chest width (CW), chest depth (CD), sternum length (SL), body length (BL), and tibia circumference (TC). The results showed that 2 SNPs (BsrD I and HpyCH4 IV) were detected in exon 4 to 5 and exon 6 to 7 of the VIPR-1 gene, respectively. The results of association showed that the BsrD I site was not significantly associated with growth traits at 3 or 5 wk of age in the SV strain (P < 0.05), while the BsrD I site was significantly associated with BL at 3 or 5 wk of age in FG (P < 0.05). The HpyCH4 IV site was significantly associated with TL, CW, CD, SL, and BL at 3 wk of age in the SV strain (P < 0.05), while the HpyCH4 IV site was significantly correlated with BW, CW, SL, and BL at 5 wk of age in SV (P < 0.05). The HpyCH4 IV site was significantly associated with TL and TC at 3 wk of age in FG (P < 0.05), while the HpyCH4 IV site was significantly associated with TC at 5 wk of age in FG (P < 0.05). Four haplotype combinations based on 2 SNPs showed significantly association with BW, CW, CD, SL, BL, and TC at 3 or 5 wk of age in SV (P < 0.05). There was not significant association between 3 haplotype combinations with growth trait at 3 or 5 wk of age in FG (P > 0.05). In conclusion, the VIPR-1 gene could be used as a molecular genetic marker to improve growth traits in quail.
Key words: VIPR-1, polymorphism, PCR-RFLP, growth traits, quail
INTRODUCTION
The vasoactive intestinal peptide receptor-1 (VIPR-1) gene is a member of the VIP protein receptor family, with a large hydrophilic extracellular N terminus, 7 highly conserved hydrophobic transmembrane helices, and a cytoplasmic C terminus (Gaudin et al., 1998). It is a partial basic protein, and its secondary structure is mainly composed of α-helix, β-fold, and β-turn (Kansaku et al., 2001). The VIPR-1 gene was expressed major in the pituitary, and only the differential mRNA expression of the VIPR-1 gene in the pituitary was associated with reproductive changes (Chaiseha et al., 2004). Furthermore, Zhou et al. (2008) have proved that polymorphism of the VIPR-1 gene has a significant impact on broodiness traits of chicken. Pu et al. (2016) purported that the polymorphism of the VIPR-1 gene was significantly associated with egg weight in quail. These studies have shown a close correlation between VIPR-1 gene and animals’ economic traits.
Quail is an economically important poultry species widely raised in China, which has characteristics such as a short feeding cycle, high reproductive ability, small investment scale, and high nutritional benefits (Bai et al., 2021). However, how to improve the growth trait to further increase economic efficiency remains a key issue in quail breeding. The growth traits such as body weight (BW), tibia length (TL), chest width (CW), chest depth (CD), sternum length (SL), body length (BL), and tibia circumference (TC) are influenced not only by environmental factors but also by genetic factors (Gaur et al., 2022). At present, there are few studies on VIPR-1 gene polymorphism and its association with growth traits in quails. In this study, the polymorphisms of BsrD I and HpyCH4 IV sites of the VIPR-1 gene were analyzed by PCR-RFLP and sequencing techniques, and their association with growth traits in French Giant meat quail and Savimalt meat quail was evaluated to provide a reference for molecular assisted breeding in the quail breeding industry.
MATERIALS AND METHODS
Ethics Statement
This research strictly compliance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China). All procedures were passed by the Animal Care and Use Committee of Henan University of Science and Technology (Luoyang, China).
Experimental Animals, Housing Condition, and Phenotypic Measurements
Thirty six female SV strain and 49 female FG strain quails were randomly selected from Henan University of Science and Technology Quail Breeding Co. Ltd., Luoyang, Henan, China. All quails were fed at the experimental farm at Henan University of Science and Technology under the same conditions. All samples were separated into 2 groups as to the population. During the whole investigation, all quails were allowed to feed and drink ad libitum. Supplemental heaters were provided first 2 wk of growth. The daily lighting schedule was 14L/10D until 35 d. The feeding schedule of these quails was 2,800 kcal of ME/kg and 20% CP for d 1 to 35 in the SV and FG strains. The growth traits of the SV and FG meat quails were recorded at 3 and 5 wk. The growth traits included BW, CD, CW, SL, BL, TL, and TC.
DNA Extraction, PCR Amplification, Sequencing, and PCR-RFLP Analysis
Blood samples were taken from the wings of 85 quails into a syringe containing 2% EDTA used as an anticoagulant and stored at −80°C for further experiment. Genomic DNA was isolated from the venous blood samples using a poultry whole blood genomic DNA extraction kit (Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China). The primer pairs of the VIPR-1 gene were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA), were synthesized by Wuhan Aoke Biotechnology Co., Ltd. (Wuhan, China). The information of primer pairs was F-GCGTTCTATGGCACAGT and R-AAAGCAATGTTCGGGTT at BsrD I site, and HpyCH4 IV site was F-GCTGCTGGTGGAAGGGT and R-CCGTCCAAGCAGTGAT. The primer specificity was verified by the BLAST module at NCBI databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Then the quail genomic DNA from 85 quails was used as a template to synthesize the desired product by PCR. The PCR reaction systems were as follows: a total volume of 15 μL, including 7.5 μL the 2 × Taq PCR Master Mix, 1 μL genomic DNA template, 5.5 μL double-distilled water, 1 μL of each primer. The PCR reaction conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of 94°C for 35 s, annealing for 54°C and 58.7°C for 35 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The expected amplified segment size was 925 bp and 1,096 bp at BsrD I and HpyCH4 IV sites, respectively. Then, the amplified samples of the VIPR-1 gene were sent for sequencing (Tsingke Biotechnology Co., Ltd., Beijing, China). The restriction enzyme system was 10 μL: 1 μL 1× buffer, 7.5 μL PCR product, restriction enzyme 0.5 μL BsrD I (5 U/μL) or HpyCH4 IV (10 U/μL), 1 μL ddH2O. After centrifugation, the BsrD I system was digested overnight in an incubator at 37°C, while HpyCH4 IV system was digested in an incubator at 65°C for 1 h (Pu et al., 2016). The enzyme products were detected by 1% agarose gel electrophoresis.
Statistical Analysis
The SNPs were determined by imaging results, and genotypes and alleles were recorded 36 samples from SV and 49 samples from FG using Excel (version 2016; Microsoft, Redmond, WA). The Popgene version 1.32 was used to calculated the population genetic information including heterozygosity (He), homozygosity (Ho), Hardy-Weinberg equilibrium test (HWE), effective allele numbers (Ne), polymorphism information content (PIC) (Yeh and Boyle, 1997). Finally, association analysis of polymorphisms was accomplished with the measured growth traits using Duncan's multiple range test in SPSS (version 26.0; IBM Corp., Armonk, NY) and expressed as means ± standard error (SE). Differences were considered highly significant or significant at P ≤ 0.01 or P ≤ 0.05, respectively. The model of association analysis was as follows:
where Yi is the phenotype value, μ is the total mean value, Gi is the fixed effect of genotype, and eij is the random error.
RESULTS AND DISSCUSION
Polymorphism of the BsrD I and HpyCH4 IV Sites of the VIPR-1 Gene
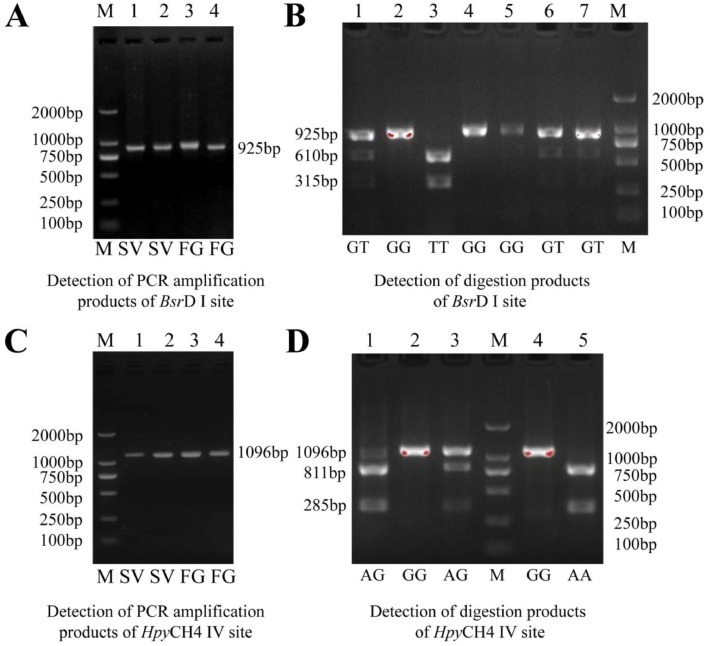
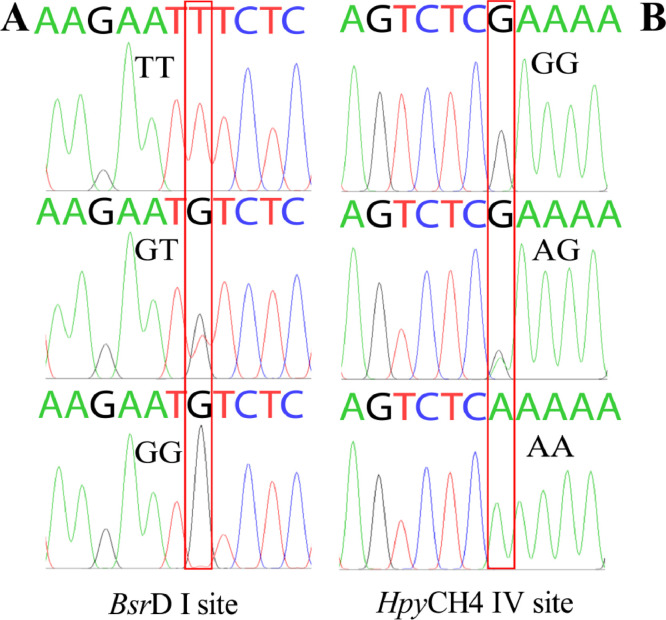
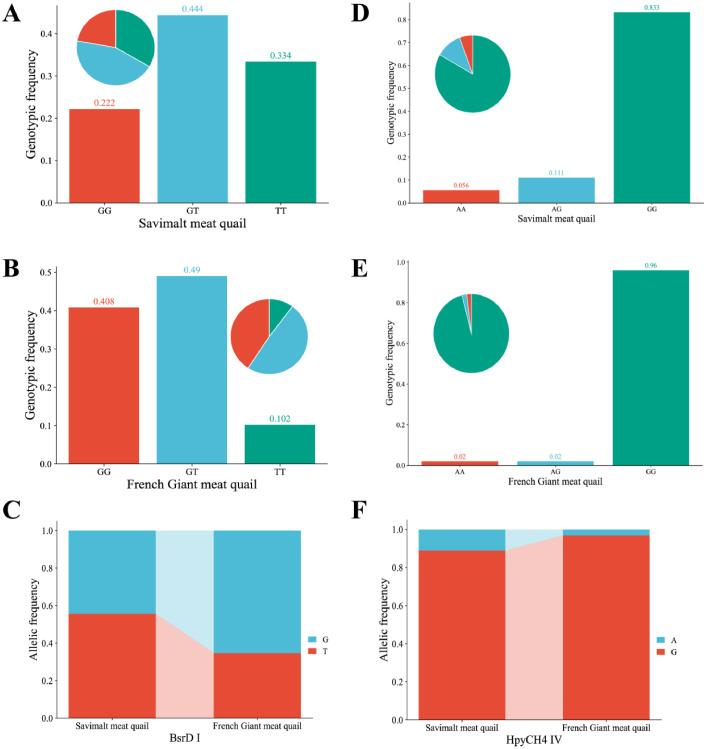
The advancement of molecular breeding techniques has enabled researchers to identify novel genetic markers and evaluate their association with economically important traits in poultry breeding (Bhattacharya et al., 2019; Zhao et al., 2023). This experiment aimed to explore the association between VIPR-1 gene polymorphisms and growth traits of quail by PCR-RFLP and sequencing technique. The electrophoretic patterns of PCR amplification products and digestion products at the BsrD I and HpyCH4 IV sites were shown in Figure 1. The PCR product in exon 6 to 7 of the VIPR-1 gene was digested by HpyCH4 IV to generate 3 genotypes: GG (1,096 bp), AA (285 bp/811 bp), and AG (285 bp/811 bp/1,096 bp). The PCR product in exon 4 to 5 of the VIPR-1 gene was digested by BsrD I to produce 3 genotypes: GG (925 bp), TT (315 bp/610 bp), and GT (315 bp/610 bp /925 bp). Sequencing results of the BsrD I and HpyCH4 IV sites of VIPR-1 gene were shown in Figure 2. The statistical results of genotype frequency, allele frequency, and population genetic information of BsrD I and HpyCH4 IV sites of VIPR-1 gene were shown in Figure 3 and Table 1. The frequencies of GG, GT, and TT genotypes were 22.2, 44.4, 33.4, 40.8, 49.0, and 10.2% at the BsrD I site in SV and FG, respectively. The GT genotype frequency was the highest in SV and FG (Figure 3A and B). Furthermore, allele T (55.6%) was the dominant gene at the BsrD I site in SV, allele G (65.3%) was the dominant gene at the BsrD I site in FG (Figure 3C). The frequencies of AA, AG, and GG genotypes were 5.6, 11.1, 83.3, 2.0, 2.0, 96.0% at the HpyCH4 IV site in SV and FG, respectively. The GG genotype frequency was the highest in SV and FG (Figure 3D and E). Furthermore, allele G (88.9%) was the dominant gene at the HpyCH4 IV site in SV, allele G (96.9%) was also the dominant gene at the HpyCH4 IV site in FG (Figure 3F). The PIC of BsrD I site was in moderate polymorphism (0.25 < PIC < 0.5) in SV and FG. The PIC of HpyCH4 IV site was in low polymorphism (0 < PIC < 0.25) in SV and FG. The BsrD I site was consistent with HWE in FG and SV (P > 0.05). The HpyCH4 IV site was significantly deviated from HWE (P < 0.05) in SV, and was consistent with HWE (P > 0.05) in FG. Pu et al. (2016) reported that the detection of 2 SNPs (G373T and A313G) in the VIPR-1 gene of the 3 quail strains was similar to the results of this study.
Figure 1.
Detection of PCR amplification products and digestion products at the BsrD I and HpyCH4 IV sites of VIPR-1 gene.
Figure 2.
Sequencing results at the BsrD I and HpyCH4 IV sites of VIPR-1 gene.
Figure 3.
Genotype frequency and allele frequency at the BsrD I and HpyCH4 IV sites of VIPR-1 gene.
Table 1.
Genotype frequency, allele frequency, and Hardy-Weinberg's law date at the BsrD I and HpyCH4 IV sites of VIPR-1 gene in meat quail.
| SNP | S | Genotypic frequency | Allelic frequency |
HWE2 |
Ho | He | PIC | Ne | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | χ2 | P | |||||||||
| BsrD I | SV (36) | GG (0.222) | GT (0.444) | TT (0.334) | 0.556 | 0.444 | 0.360 | 0.549 | 0.506 | 0.494 | 0.372 | 1.976 |
| FG (49) | GG (0.408) | GT (0.490) | TT (0.102) | 0.653 | 0.347 | 0.321 | 0.571 | 0.547 | 0.453 | 0.350 | 1.829 | |
| HpyCH4 IV | SV (36) | AA (0.056) | AG (0.111) | GG (0.833) | 0.889 | 0.111 | 6.891 | 0.009 | 0.802 | 0.198 | 0.178 | 0.246 |
| FG (49) | AA (0.020) | AG (0.020) | GG (0.960) | 0.969 | 0.031 | 21.095 | 1.000 | 0.941 | 0.059 | 0.058 | 1.063 | |
Abbreviations: FG, French Giant meat quail; He, heterozygosity; Ho, homozygosity; HWE, Hardy-Weinberg equilibrium test; Ne, effective allele numbers; PIC, polymorphism information content; S, strain; SNP, single nucleotide polymorphism; SV, Savimalt meat quail.
Association Analysis Between VIPR-1 Gene and Growth Traits
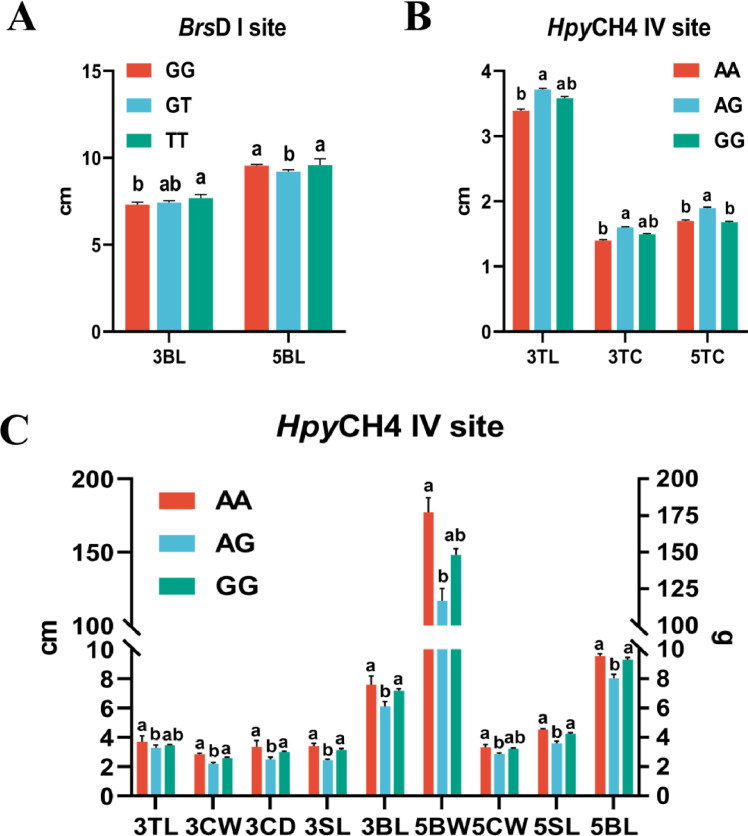
Similarly to the present study, a previous investigation conducted in 3 strains of laying quail demonstrated a significant association between egg production traits and 2 SNPs (G373T and A313G) located in the VIPR-1 gene. Additionally, 7 diplotypes (GGGG, GGGT, GGTA, GGGA, GTTA, TTAA, and GTAA) based on 4 haplotypes (GG, GT, TA, and GA) were found to be significantly correlated with age at first laying, egg weight, and egg number at 20 wk of age (Pu et al., 2016). It is reported that a mutation (C1704887T) of the VIPR-1 gene was correlated with chicken egg number at 300 d of age (Xu et al., 2011). Zhou et al. (2008) have shown that 7 sites (A-284G, A+457G, C+598T, D+19820I, C+37454T, C+42913T, and C+53327T) of VIPR-1 gene might be associated with broodiness. Hosseinpour et al. (2020) have shown that 2 SNPs (C17687T and A17690T) of VIPR-1 gene were associated with egg production traits, egg number, total egg weight, and laying period in turkey hens. The present study was conducted to determine if the polymorphisms of the VIPR-1 gene were correlated with the growth traits at 3 and 5 wk of age in quails (Figure 4). The results showed that the BsrD I site was significantly associated with the BL at 3 wk of age in FG (P < 0.05), and individuals with the TT genotype had significantly higher BL than that of GG genotype (P < 0.05). The BsrD I site was significantly associated with the BL at 5 wk of age in FG (P < 0.05), and individuals with the GT genotype had significantly lower BL (P < 0.05, Figure 4A). The HpyCH4 IV site was significantly associated with TL and TC at 3 wk of age in FG (P < 0.05), and individuals with of AG genotype had a significantly higher TL and TC (P < 0.05). The HpyCH4 IV site was significantly associated with TC at 5 wk of age in FG (P < 0.05), and individuals with of AG genotype had significantly higher TC (P < 0.05, Figure 4B). The HpyCH4 IV site was significantly associated with TL, CW, CD, SL and BL at 3 wk of age in SV (P < 0.05), and individuals with the AA genotype had a significantly higher TL, CW, CD, SL, and BL (P < 0.05). The HpyCH4 IV site was significantly associated with BW, CW, SL, and BL at 5 wk of age in SV (P < 0.05), and individuals with the AG genotype had significantly lower BL, CW, SL, and BL (P < 0.05, Figure 4C). No significant differences were observed of 2 sites with other growth traits (P > 0.05, Table S1). This finding indicates that 2 SNPs of the VIPR-1 gene may affect growth traits, and the VIPR-1 gene could be used as a novel molecular marker in SV and FG quail strains.
Figure 4.
Association analysis of the BsrD I and HpyCH4 IV sites of VIPR-1 gene with growth trait in quail. (A) Association analysis of the BsrD I site of VIPR-1 gene with growth trait in French Giant meat quail; 3BL, body length at 3 wk of age; 5BL, body length at 5 wk of age. (B) Association analysis of the HpyCH4 IV site of VIPR-1 gene with growth trait in French Giant meat quail; 3TL, tibia length at 3 wk of age; 3TC, tibia circumference at 3 wk of age; 5TC, tibia circumference at 5 wk of age. (C) Association analysis of the HpyCH4 IV site of VIPR-1 gene with growth trait in Savimalt meat quail; 3TL, tibia length at 3 wk of age; 3CW, chest width at 3 wk of age; 3CD, chest depth at 3 wk of age; 3SL, sternum length at 3 wk of age; 3BL, body length at 3 wk of age; 5BW, body weight at 5 wk of age; 5CW, chest width at 5 wk of age; 5SL, sternum length at 5 wk of age; 5BL, body length at 5 wk of age. abThe difference between genotypes with different lowercase letters was significant (P < 0.05).
Linkage Disequilibrium Analysis and Haplotype Analysis of SNPs of VIPR-1 Gene
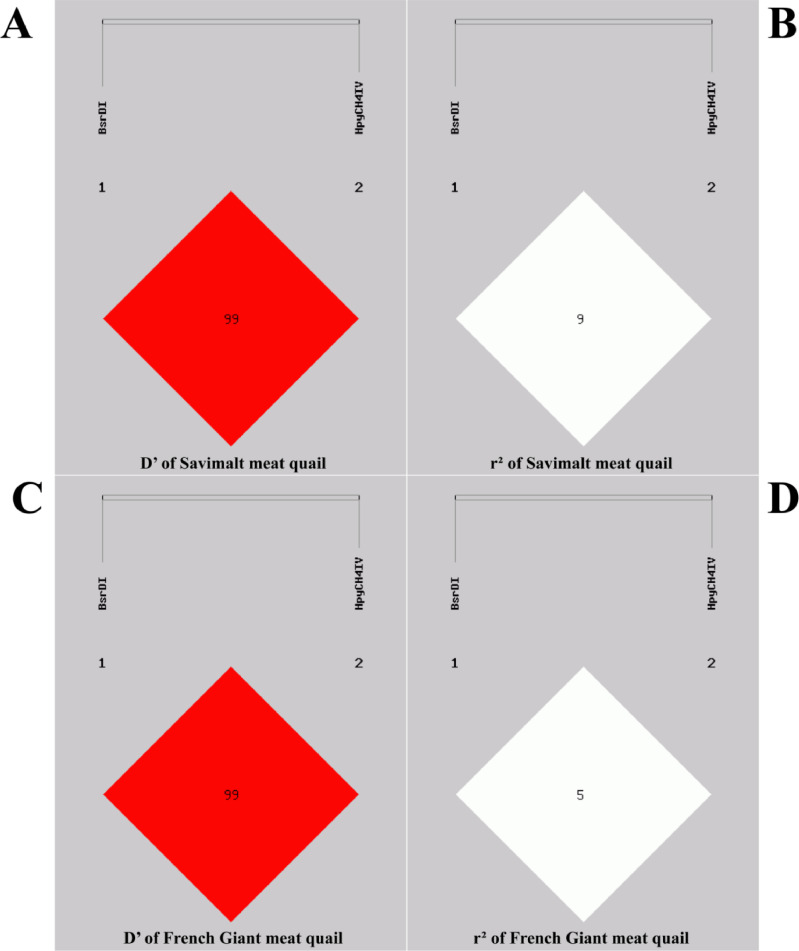
The linkage disequilibrium (LD) analysis of 2 SNPs showed there was no significant LD (D' > 0.8 and r2 > 0.33) at the BsrD I and HpyCH4 IV sites in SV and FG, indicating that they tend to be genetically independent of each other (Figure 5). Haplotypes based on the 2 SNPs showed that a total of 3 (A1, A2, A3) and 3 (B1, B2, B3) haplotypes (frequencies greater than 3%) were identified in SV and FG.
Figure 5.
Linkage disequilibrium coefficient between SNPs (D’ and r2) in quail of the Savimalt strain and French Giant strain, the numbers are the D’ and r2 value (%).
Association Analysis of Haplotype Combinations With Growth Traits
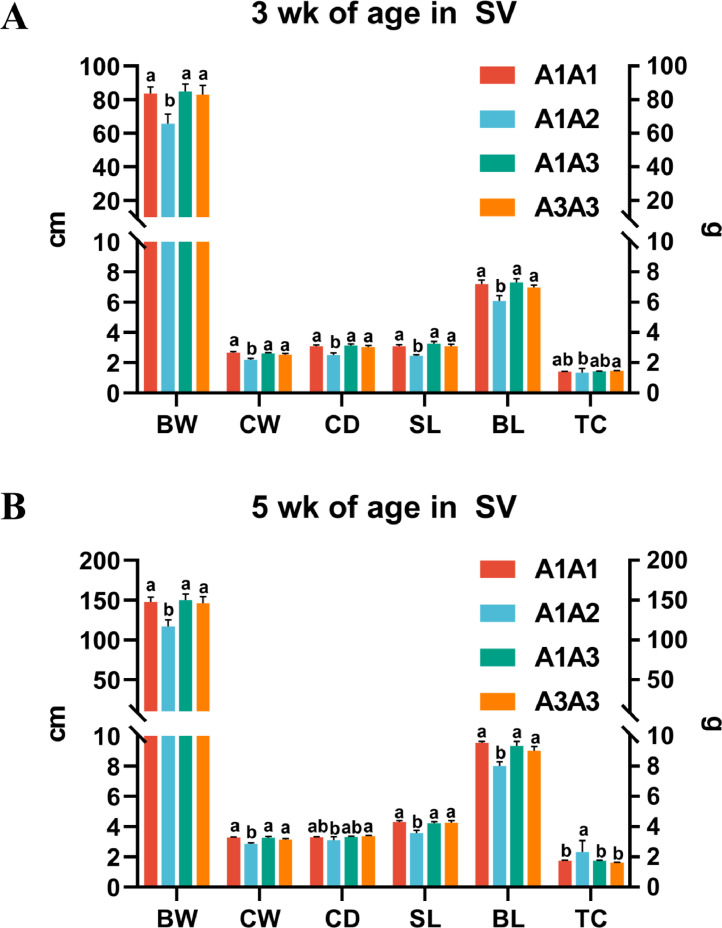
In the linkage between 2 SNPs, 4 and 3 haplotype combinations (combinations with the number of individuals higher than or equal to 3) were formed in SV and FG, respectively (Table S2). Haplotype combination A1A1, A1A3, and A3A3 had significantly higher BW, CW, CD, SL, and BL than that of A1A2 at 3 wk of age in SV (P < 0.05), A3A3 had significantly higher TC than of A1A2 in SV (P < 0.05, Figure 6A). A1A1, A1A3, and A3A3 combinations had significantly higher BW, CW, SL, and BL than that of A1A2 at 5 wk of age in SV (P < 0.05), A3A3 combination had a significantly higher CD than that of A1A2 in SV (P < 0.05), A1A2 combination had significantly higher TC than those of A1A1, A1A3, and A3A3 in SV (P < 0.05, Figure 6B). Three haplotype combinations were not significantly associated with growth traits at 3 or 5 wk of age in FG (P > 0.05, Table S2). This haplotype association analysis was consistent with the significant effect detected by the SNP association analysis, which was similarly reported in laying quails (Pu et al., 2016). This finding indicates that 4 diplotypes (A1A1, A1A2, A1A3, and A3A3) based on 3 haplotypes of the VIPR-1 gene may affect growth traits, and the VIPR-1 gene could be used as a novel molecular marker in meat type Japanese quail.
Figure 6.
Association analysis of the VIPR-1 gene haplotype combinations with growth trait in quail. (A) Association analysis of the VIPR-1 gene haplotype combinations with growth trait at 3 wk of age in Savimalt meat quail; BW, body weight; CW, chest width; CD, chest depth; SL, sternum length; BL, body length; TC, tibia circumference. (B) Association analysis of the VIPR-1 gene haplotype combinations with growth trait at 5 wk of age in Savimalt meat quail; BW, body weight; CW, chest width; CD, chest depth; SL, sternum length; BL, body length; TC, tibia circumference. abThe difference between genotypes with different lowercase letters was significant (P < 0.05).
In conclusion, our results showed that 2 SNPs (BsrD I and HpyCH4 IV) were detected in exon 4 to 5 and exon 6 to 7 of the VIPR-1 gene which were significantly correlated with growth traits in 2 meat type Japanese quail strains. The VIPR-1 gene could be used by quail breeders to improve growth traits. However, due to the limitation of the number of quail population, we have only carried out preliminary studies on these 2 strains of quail, further studies in large populations with different quail strains are required to further assess the associations of the polymorphisms of VIPR-1 gene with growth traits. Moreover, more studies on the polymorphism of VIPR-1 gene are required to improve quail carcass, meat quality, egg quality and egg production traits.
ACKNOWLEDGMENTS
This research was supported by grants from National Natural Science Foundation of China (No. 31201777).
I declare that research on live animals is in line with the guidelines approved by the institutional animal care and use Committee (IACUC) through the use of appropriate management and laboratory techniques to avoid unnecessary discomfort of animals.
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102781.
Appendix. Supplementary materials
REFERENCES
- Bai J.Y., Dong Z.H., Lei Y., Yang Y.B., Jia X.P., Li J.Y. Association analysis between polymorphism of gonadotrophin releasing hormone genes and growth traits of quail (Coturnix Coturnix) Braz. J. Poult. Sci. 2021;23 eRBCA-2020-1314. [Google Scholar]
- Bhattacharya T.K., Chatterjee R.N., Dange M., Bhanja S.K. Polymorphisms in GnRHI and GnRHII genes and their association with egg production and egg quality traits in chicken. Br. Poult. Sci. 2019;60:187–194. doi: 10.1080/00071668.2019.1575505. [DOI] [PubMed] [Google Scholar]
- Chaiseha Y., Youngren O.M., El Halawani M.E. Expression of vasoactive intestinal peptide receptor messenger RNA in the hypothalamus and pituitary throughout the turkey reproductive cycle. Biol. Reprod. 2004;70:593–599. doi: 10.1095/biolreprod.103.022715. [DOI] [PubMed] [Google Scholar]
- Gaudin P., Maoret J.-J., Couvineau A., Rouyer-Fessard C., Laburthe M. Constitutive activation of the human vasoactive intestinal peptide 1 receptor, a member of the new class II family of G protein-coupled receptors. J. Biol. Chem. 1998;273:4990–4996. doi: 10.1074/jbc.273.9.4990. [DOI] [PubMed] [Google Scholar]
- Gaur P., Malik Z.S., Bangar Y.C., Magotra A., Yadav A.S. Genetic and non-genetic effects associated with ewe productivity in Harnali sheep. Zygote. 2022;30:386–390. doi: 10.1017/S0967199421000897. [DOI] [PubMed] [Google Scholar]
- Hosseinpour L., Nikbin S., Hedayat-Evrigh N., Elyasi-Zarringhabaie G. Association of polymorphisms of vasoactive intestinal peptide and its receptor with reproductive traits of turkey hens. S. Afr. J. Anim. Sci. 2020;50:345–352. [Google Scholar]
- Kansaku N., Shimada K., Ohkubo T., Saito N., Suzuki T., Matsuda Y., Zadworny D. Molecular cloning of chicken vasoactive intestinal polypeptide receptor complementary DNA, tissue distribution and chromosomal localization1. Biol. Reprod. 2001;64:1575–1581. doi: 10.1095/biolreprod64.5.1575. [DOI] [PubMed] [Google Scholar]
- Pu Y.J., Wu Y., Xu X.J., Du J.P., Gong Y.Z. Association of VIPR-1 gene polymorphisms and haplotypes with egg production in laying quails. J. Zhejiang Univ. Sci. B. 2016;17:591–596. doi: 10.1631/jzus.B1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.P., Zeng H., Zhang D.X., Jia X.L., Luo C.L., Fang M.X., Nie Q.H., Zhang X.Q. Polymorphisms associated with egg number at 300 days of age in chickens. Genet. Mol. Res. 2011;10:2279–2289. doi: 10.4238/2011.October.3.5. [DOI] [PubMed] [Google Scholar]
- Yeh F.C., Boyle T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997;129:157. [Google Scholar]
- Zhao C.B., Hu B.W., Zhang Z., Luo Q.B., Nie Q.H., Zhang X.Q., Li H.M. Detection of CD36 gene polymorphism associated with chicken carcass traits and skin yellowness. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2023.102691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Lei M., Rao Y., Nie Q., Zeng H., Xia M., Liang F., Zhang D., Zhang X. Polymorphisms of vasoactive intestinal peptide receptor-1 gene and their genetic effects on broodiness in chickens. Poult. Sci. 2008;87:893–903. doi: 10.3382/ps.2007-00495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.