Abstract
Eukaryotic gene expression is regulated at multiple levels, from transcription to messenger RNA (mRNA) processing, transport, localization, turnover, and translation. RNA-binding proteins (RBPs) are critical regulators of gene expression and are involved in all aspects of mRNA processing, from splicing to translation. The RNS-binding protein Human antigen R (HuR) mainly regulates the mRNA transport from nucleus-to-cytoplasm, stability, and translation. Dysregulation of HuR is linked to diseases such as cancer, neurodegenerative disorders, and immune-related disorders. HuR targets mRNAs containing AU-rich elements at their 3’untranslated region, which encodes proteins involved in cell growth, tumorigenesis, angiogenesis, immune evasion, tumor inflammation, invasion, and metastasis. HuR overexpression has been reported in many tumor types, which led to a poor prognosis for patients. Hence, HuR is an attractive drug target for cancer therapy. Recently, there has been an intense effort to identify small molecule inhibitors for blocking HuR functions. This article reviews the current prospects of drugs that target HuR in multiple cancer types. Furthermore, we will summarize drugs that interfered with HuR-RNA interactions and established themselves as novel therapeutics. We will also highlight the significance of HuR overexpression in multiple cancers and discuss its role in immune functions. This review provides evidence of a new era of HuR-targeted small molecules that can be used for cancer therapeutics either as a monotherapy or combination with other cancer treatment modalities.
Keywords: Oral squamous cell carcinoma, post-transcriptional gene regulation, RNA-binding proteins, mRNA turnover, mRNA translation, WNT signaling, and oral cancer progression
Graphical Abstract:
Targeting HuR for anticancer activity. Left panel: Overexpressed HuR is phosphorylated by kinases that bind with ARE mRNAs and promote their protein expression to facilitate tumor growth and proliferation. Right panel: Inhibition of HuR phosphorylation by small-molecule BML-277 or disruption of HuR binding with ARE mRNA using CMLD2 reduced ARE mRNA encoding protein and promoted cell death.

1. Introduction
The significance of RNA-binding protein (RBP) mediated post-transcriptional gene regulation (PTR) in eukaryotes has emerged as a necessary regulatory mechanism in gene expression. This level of PTR is mainly controlled by RBPs, which play a significant role in all aspects of RNA maturation and the regulation of messenger RNA (mRNA) stability, localization, and translation (well-reviewed in ref. [1]). Recent emerging high-throughput genomics and mass spectrometry-based methods have identified hundreds of RBPs in mammalian cells [2–5], suggesting that the human genome may contain 1,542 or more RBP-encoding genes [6]. Due to vast differences in protein motifs, RBPs recognize different RNA sequences and are functionally distinct from each other. Thus, RBPs play critical roles in controlling the fate of the RNA molecules with which they associate.
Despite ongoing efforts in identifying RBPs and their RNA interactome, many questions remain unanswered. For example, there is a lack of understanding of how RBP-RNA binding specificities are defined and how this RBP-RNA interface can be used for drug targeting by chemical and biophysical methodologies. Recently, several attempts have been made to identify small molecule chemical probes that target RBPs and interfere with RNA binding, representing valuable tools for developing novel therapeutics to block the function of RBPs [7]. Although RBPs have been considered impossible molecules for drug-targeting due to the complexity of protein structure and domains, efforts have been made to discover small molecules that target specific RBPs have prompted considerable interest. Small molecules are often designed to target RBP’s RNA-binding domains, such as the RNA-recognition motif (RRM), hnRNP K homology (KH), and the zinc-finger domain. Other examples include small molecules that target RBPs such as spliceosomal proteins for treating muscular dystrophy [8, 9], cancer activity [10], EIF4 family proteins [11], TDP-43 for the treatment of Amyotrophic Lateral Sclerosis (ALS) [12], and Human antigen R (HuR) for anti-cancer activities [13–15]. We direct the readers to the following reviews to further read RBP probes and their medicinal uses [7, 16]. Here, we review the RBP HuR’s role in cancer and immune functions, detail its RNA-binding motifs, analyze its expression across different cancers, and highlight drug-targeting opportunities that will enable us to develop HuR-dependent therapeutics for future medicine.
Chemical and biophysical approaches such as fluorescence polarization (FP) and fluorescence resonance energy transfer (FRET) assays are the principal methods used to determine RBP interaction with small molecules for drug screening. FP is a well-known technique to study molecular interactions, particularly those between proteins and nucleic acids. FP utilizes the polarized light emission from a 3’ or 5’ or bodily labeled RNA and monitors its binding to RBPs. The ability of FP to screen drugs relies solely on the changes in the polarization of corresponding unbound RNA, which is indicative of the small molecule blockade of RNA-RBP interaction [7]. FRET is a photophysical event in which energy is transferred between two appropriate fluorophores called donor and acceptor, appropriately oriented and distanced to study their interactions. The donor and acceptor are individually attached to RNA and RBP, respectively, and the interference of small molecule binding will decrease the emission of the acceptor fluorophore [7]. For example, to determine the spliceosome complex, FRET was employed to measure dynamic protein-protein interactions between splicing factors [17–19]. In addition to FP and FRET, another proximity assay, Alphascreen (Amplified Luminescent Proximity Homogenous Assay), has been used to detect RBP inhibitors. Recently, Alphascreen was used to identify HuR inhibitor CMLD-2 by disturbing the interaction between HuR and AU-rich element (ARE) mRNA [20].
This review will identify FDA-approved drugs that target HuR or other pending therapies for cancer. We will also summarize examples of potential RNA-targeted therapies that inhibit HuR and RNA interactions and act as antitumor drugs. We will review the posttranslational modification of HuR carried out by signaling kinases, which are targets of small molecule inhibitors that interfere with HuR functions. Finally, we will highlight small animal models of HuR and their contribution to immune dysfunctions, HuR’s cancer traits in multiple cancers, and provide insight into the mRNA-HuR targeted therapies.
2. Biology of HuR
2.1. ELAV family of proteins:
HuR belongs to the ELAV (embryonic lethal abnormal visual system) family of RNA binding proteins, present in prokaryotes and eukaryotes [21]. In Drosophila, the ELAV protein was the first discovered member of the family, followed by its homologs [22]. The ELAV family of proteins contains three RRMs, which have characteristic ribonucleoprotein consensus sequences [23]. Among 1,542 human RBPs, approximately 692 proteins are mRNA binding proteins in which 225 proteins contain canonical RRM [6], indicating that RRM is the primary domain among all the other RBP motifs. Furthermore, a search of RRM in the Protein Data Bank (PDB) yields over 500 structures and is estimated to be present in 1% of all the human proteins [24].
HuR has canonical protein motifs to bind RNA. Protein ELAVL1, alias HuR, is composed of three highly conserved canonical RRMs. All three RRMs adopt a βαββαβ topology, where a four-stranded antiparallel β-sheet is packed against two α-helices [25]. The tandem RRM1 and 2 are separated from the C-terminal RRM (RRM3) by a ~50-residue basic hinge region containing the nucleocytoplasmic export signal [26]. Both RRM 1 and 2 are the significant domains that bind ARE-containing RNAs and stabilize them [27], but RRM3 recognizes AREs and is involved in dimerization and competes with both RRM 1 and 2 for mRNA degradation [28]. Further studies demonstrated that RRM3, with hinge region of HuR, is involved in protein-protein interactions [29], protein dimerization [30], and multimerization in cancer cells [31]. Both RRM3 and hinge regions counter miRNA-mediated repression and promote miRNA-induced silencing complex release from target mRNAs [32]. Multiple studies reported that structural details of HuR and its family members are essential for RNA recognition [33–35]. Based on these structural features of HuR, several investigators attempted to develop small-molecule ligands that modulate HuR functions and block RNA binding activity (reviewed in [36, 37]). In this review, we will update and summarize the significant role of HuR in cancer and immune regulation in conjunction with the potential impact of targeting HuR for cancer therapeutics. As our understanding of HuR protein-ligand interactions grows, it is becoming evident that structure-based computer-aided drug discovery (molecular docking) using bioinformatic approaches will further develop multiple therapeutic opportunities.
2.2. Posttranslational modifications (PTM) of HuR:
Many excellent reviews have covered the different posttranslational modifications (PTM) of HuR and their modes of RNA binding and biological functions [38–40]. Therefore, we will limit the depth of our discussion for each PTM and focus our attention on drug targeting of upstream protein kinase modifiers. Figure-1 illustrates the protein structure and posttranslational modification of HuR by signaling kinases, which can serve as small molecule drug targeting enzymes to block HuR localization and functions.
Figure 1:
Posttranslational modification of HuR by signaling kinases. Schematics illustrates the protein domains and amino acids of HuR modified at the posttranslational level by signaling kinases targeted using specified small molecules. References citing the relevant studies are listed.
2.2.1. HuR phosphorylation by kinases provides an opportunity to indirectly target HuR:
HuR is phosphorylated by several kinases, which modulate its RNA binding and turnover activities. The checkpoint kinase 2 (Chk2), activated and rapidly phosphorylated in response to DNA damage, phosphorylates HuR at S88, S100, and threonine (T)118 [41]. This phosphorylation affects HuR binding with SIRT1 mRNA and triggers its mRNA decay. In a subsequent study, the authors showed that ionizing radiation-induced CHK2 phosphorylates HuR, consequently promoting dissociation of HuR from its target mRNAs, including mRNAs that encoded proapoptotic and proliferative proteins [42]. These findings provide an opportunity to target Chk2 for the control of HuR phosphorylation at the pharmacological level. Accordingly, a small-molecule inhibitor BML-277 (Inhibitor II) of Chk2 dephosphorylate HuR showed decreased mRNA association in cancer cells [43].
The kinase Cyclin-dependent kinase-1 (CDK1) dependent phosphorylation of HuR at S202 facilitates its interaction with protein 14–3-3 for nuclear retention [44]. However, when CDK1 is inactive and amino acid S202 is dephosphorylated, HuR translocates from the nucleus to the cytoplasm. Interestingly, inhibition of CDK1 by pharmacologic agent (Cdk1-specific inhibitor, CGP74514A) promotes HuR translocation to the cytoplasm and stabilizes CDKN1A mRNA, a tumor suppressor gene [45]. Thus, pharmacological targeting of kinases that phosphorylate HuR is an alternative strategy to target HuR in cancer cells at the therapeutic level.
The protein kinase C (PKC) family members phosphorylate HuR and promote its cytoplasmic export and control of mRNA stability [46–48] of encoding proteins involved in various cancer pathways. Among many PKC isoforms, PKCα phosphorylates S158 and S221 residues of HuR to promote its cytoplasmic localization to enhance its binding to COX2 mRNA leading to increased mRNA stability, COX2 protein, and prostaglandin E2 biosynthesis [48]. However, the authors demonstrated that inhibition of PKCα by small molecules such as gö6976, CGP 41251, or rottlerin prevents cytoplasmic HuR in an ATP-dependent manner, suggesting that targeting PKCα may indirectly control HuR-mediated gene expression [48]. Consequently, the same group demonstrated that HuR phosphorylation at serines 221 and 318 confers binding to COX2 mRNA, which PKCδ differentially regulates in human mesangial cells [47]. Since both PKC isoforms phosphorylate HuR at different sites, it is most likely that those serine residues induce oligomerization of HuR and regulate its RNA-binding capacity [49]. However, pharmacological inhibition of PKCδ by rottlerin abolished the S318 phosphorylation of HuR, and its binding to target mRNAs in colon cancer cells demonstrated that targeting PKC isoforms may alter HuR functions in multiple cancer cells [50, 51]. The p38MAPK also participates in HuR phosphorylation at Thr-118 and promotes its translocation to the cytoplasm to stabilize p21 mRNA [52]. Further studies also supported that p38 MAPK-mediated Thr-118 phosphorylation of HuR plays a crucial role in regulating the COX-2 expression during inflammation [53]. Concurrently, blocking p38MAPK by small-molecule with SB203580 abolished the effect of HuR phosphorylation and target COX-2 expression [53]. HuR phosphorylation by other kinases and their coordinated regulation of HuR target mRNAs are well-reviewed (please see ref. [38]).
2.2.2. Proteolytic cleavage of HuR:
We [54–56] and others [57–60] have shown that apoptotic stress stimuli promote HuR translocation to the cytoplasm and proteolytic cleavage of HuR at the aspartate D226 by caspases 3 and 7, subsequently generating two active HuR cleavage products, HuR-CP1 and HuR-CP2. Furthermore, our group showed that HuR-CP1 alone facilitates cell death, whereas the non-cleavable isoform of HuR enhances cancer cell proliferation [56]. We further showed that specific inhibition of caspase-3 by a small molecule compound, NSC321205, blocks proteolytic degradation of HuR and promotes the oral epithelial membrane integrity after ionizing radiation treatment [55]. These findings demonstrated that the use of caspase inhibitor NSC321205 indirectly supports HuR functions by establishing HuR’s role in epithelial homeostasis. However, more specific epithelial-specific HuR gain- or loss-of-function studies are required to fully understand the biology of HuR and its cleavage products in epithelial homeostasis.
2.2.3. Ubiquitinylation (protein degradation) of HuR:
HuR undergoes Lys-182 residue-dependent proteolytic degradation by proteosomes during heat shock [61]. However, this proteolytic degradation is opposed by phosphorylation of HuR by chk2 [61]. Further studies demonstrated that the tumor suppressor esophageal cancer-related gene 2 (ECRG2) was shown to increase in response to DNA damage, in turn favoring ubiquitination of HuR at K182 and HuR degradation [62]. HuR is also noted for the enzyme ubiquitin E3 ligase βTrCP1 mediated degradation by glycolytic stress in prostate cancer cells [51]. Hence, an agonist approach to activating ECRG2 or βTrCP1 may promote HuR ubiquitinylation, which may provide a therapeutic window of opportunity to target HuR in cancer cells. Interestingly, studies demonstrated that ubiquitination of HuR with a short K29 chain attached to Lys-313 and Lys-326 did not affect protein stability but promoted the disassociation of HuR from its target mRNAs [63]. Thus, altering ubiquitinylation and de-ubiquitinylation may regulate HuR’s expression and RNA-binding activity in cancer cells.
2.2.4. Methylation:
The protein coactivator-associated arginine methyltransferase 1 (CARM1/PRMT4) methylates HuR at the arginine residue R217 in macrophages [64], human cervical carcinoma cells [65], and human embryonic stem cells (hESCs) [66] and led HuR to promote mRNA stabilization of its targets. As CARM1 and other protein methyltransferase inhibitors are used to treat a variety of cancers [67, 68], inhibition of CARM1 with small molecules may have a significant impact on HuR functions.
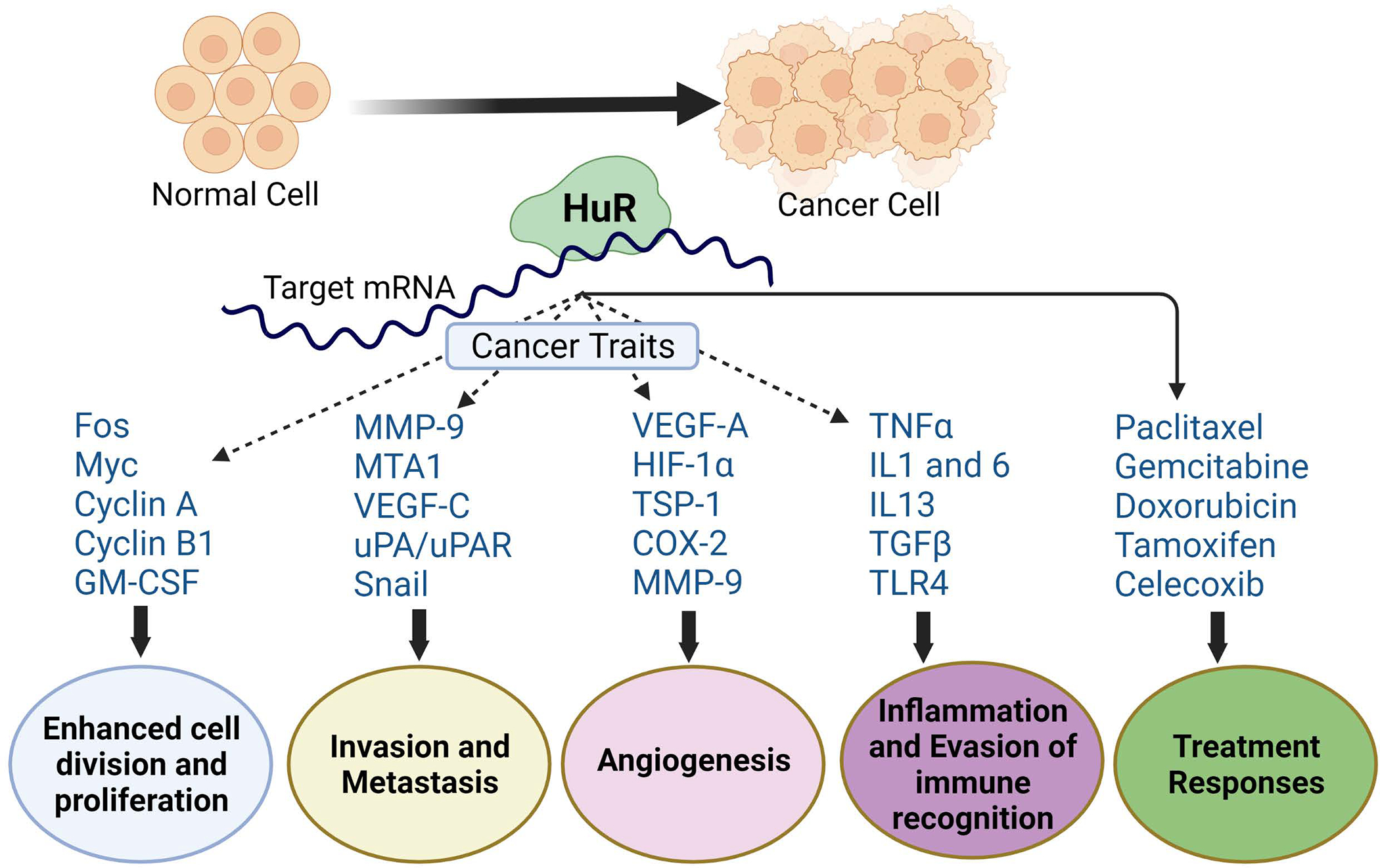
3. HuR in cancers:
While increased expression of HuR is reported in many cancers and provides a poor prognosis, the role and function of HuR in cancers have been well-reviewed by many publications [69–72]. Hence, we limit the detailed review of the HuR’s role in cancers. For the past 20 years, more than a hundred papers have detailed the role of HuR in cancers using in vitro cell culture methods with fewer in vivo studies. We have summarized the essential cancer types in which HuR and its target mRNAs encoding proteins play a critical role in carcinogenesis (Table 1).
Table 1:
A representative list of HuR associated cancer types and their role on prognosis.
| Cancer Type | Association between High HuR Expression and Clinicopathological Features | Pathology and Prognosis | References |
|---|---|---|---|
| Breast (DCIS) | ↑ tumor grade; ↑ aggressiveness; More frequently observed in PR(-ve) cases | Elevated HuR expression observed in atypical ductal hyperplasia (ADH) and ductal in situ carcinomas (DCIS) compared to normal controls, associated with high grade, progesterone receptor negativity, and microinvasion and/or tumor-positive sentinel nodes of the DCIS. | [137] |
| Breast (DIC) | ↑ histological grade; ↑ ductal tumor grade; More frequently observed in PR(-ve), ER(-ve) cases; ↑ p53; ↓ OS | In non-BRCA1/2 breast cancer patients, cytoplasmic HuR protein expression is present in 39.4% of the BRCA1/2 negative patients and associated with high tumor grade and ductal type of the tumor. However, in patients with BRCA1 or BRCA2 mutations, cytoplasmic HuR expression is more frequent (62.7% for BRCA1 and 61.7% for BRCA2) and associated with poor survival. | [138] |
| Esophageal squamous | 46.6% of specimens had positive cytoplasmic staining for HuR. | Cytoplasmic HuR expression was positively associated with lymph node metastasis, depth of tumor invasion, advanced tumor stage, and exhibits a low 5-year survival rate. Patients positive for cytoplasmic HuR expression had a cumulative 5-year survival rate of 25.3 %, whereas it was 43.8 % for patients negative for cytoplasmic HuR expression. | [139] |
| Gallbladder carcinoma | ↑ advanced tumor stage, histological grade, vascular and perineurial invasion, tumor necrosis, Ki-67 labeling index; ↑ cyclin A expression; ↓ DSS & DFS | Nuclear HuR expression correlated with poor disease-free survival, but cytoplasmic HuR expression was strongly associated with inferior disease-specific survival and disease-free survival. | [140] |
| Gastointestinal stromal tumors (GISTs) | Epithelioid histology, larger tumor size, NIH risk category and nuclear expression of Ki67 and cyclin A. Cytoplasmic HuR and cyclin A overexpression were strongly associated with worse DFS. |
Both HuR cytoplasmic expression (P < 0.001) and cyclin A overexpression (P < 0.001) are strongly associated with worse disease-free survival in GISTs patients. | [141] |
| Lung carcinoma | ↑ lymph-node involvement at presentation; ↑ risk of death and metastasis | High cytoplasmic HuR expression is directly associated with the risk of death and metastasis. The levels of nuclear and cytoplasmic HuR were allowed the authors to discriminate between patients with the highest risk of metastasis and death with lung adenocarcinomas. | [142] |
| Meningioma | ↑ tumor grade; ↓ PFS & OS; HuR knockdown decreases meningioma cell growth and hypoxia-mediated resistance | High HuR cytoplasmic expression correlated with tumor grade and negatively with progression-free and overall survival of meningioma patients. | [143] |
| Oral squamous cell carcinoma | ↑ IAP2; ↓ OS | Positive cytoplasmic HuR expression was significantly associated with positive cellular inhibitors of apoptosis protein-2 (cIAP2) in high-grade tumors, which highly correlated with poor patient survival. | [144] |
| Oral squamous cell carcinoma | ↑ Lymph node metastasis | The expression of cytoplasmic HuR is correlated positively with IMP3 and negatively with p53, significantly associated with the risk of death and poor patient survival. | [145] |
| Ovarian cancer (metastatic highgrade serous carcinoma) | ↑ HuR mRNA and protein level | Higher HuR protein expression was associated with higher serum Cancer Antigen 125 levels at diagnosis. HuR protein as detected in the nucleus and cytoplasm of tumor cells in 98% and 58% respectively, of the effusion samples. Higher HuR mRNA levels were significantly associated with poor OS. | [146] |
| Pancreatic cancer | In pre-treatment tumor samples, Deoxycytidine kinase (dCK) and HuR cytoplasmic expression were strongly correlated. | The dCK levels were prognostic and had predictive value for sensitivity to 5-FU. However, cytoplasmic HuR expression was not associated with either prognostic or predictive to 5-FU treatment. | [147] |
| Urinary tract urothelial carcinoma | Cytoplasmic HuR and nuclear cyclin A expressions were correlated with disease-specific survival (DSS), metastasis-free survival (MeFS), urinary bladder recurrence-free survival (UBRFS), and treatment response. | Increased Tumor grade, lymph node metastasis, histological grade, vascular and perineurial invasion and cyclin A expression; decreased DSS, MeFS, UBRFS; High-risk patients (pT3 or pT4 with/without nodal metastasis) with high cytoplasmic HuR had better DSS if given adjuvant chemotherapy. | [148] |
4. HuR as an immune regulator:
Given the critical roles played by HuR in tumor progression in a variety of cancers, understanding its role in the tumor microenvironment (TME) and the immune response is essential to investigate the potential for HuR as a target for cancer therapeutics. HuR is the most often studied RBP in the immune system. Conditional deletion of HuR impairs the B-cell population in the bone marrow and the periphery leading to reduced serum immunoglobulins (IgG) [73, 74]. HuR is also critical for T-cell development, and mice lacking HuR develop egress from the thymus, which results in lymphopenia in the periphery [75]. HuR expression and functions are cell-specific [76], as it controls both pro-inflammatory and anti-inflammatory cytokines. For example, myeloid cell-specific deletion of HuR resulted in an inflammatory phenotype with an enhanced inflammatory response in chemically induced colitis [77]. Similarly, heterozygous HuR mice showed steadily decreased mRNA levels of Gata3, Il4, and Il13 without affecting protein expression; however, HuR KO homozygous mice showed increased expression of Il2, Il4, and Il13 mRNA and protein [78]. The distinct differences observed between these studies indicate that intrinsic differences in immune cell types determine HuR immune cell functions.
Another example of HuR expression is correlated with increased Tnf mRNA stability and TNF synthesis in macrophages [79, 80]. However, in B-cells, HuR controls the oxidative response induced upon mitogen activation, which allows the cells for the metabolic switch and cell growth [74]. When HuR is absent in B-lymphocyte development and differentiation (HuRfl/fl Mb1Cre mice), the germinal center (GC) reaction is not formed, and affinity maturation is severely compromised [73, 74]. HuR, in association with other RBPs like PCBP1, promotes the production of pro-inflammatory cytokines in T cells and contributes to the development of T-cell mediated inflammation [81, 82]. Thus, these collective findings indicate that HuR is critical for immunosurveillance and promotes a hijack of an immune response in cancer cells.
5. Targeting HuR through RNA interference:
Although HuR is overexpressed in various cancers, miRNAs can control its expression at post-transcriptional gene regulation. The miRNAs miR-519, miR-125a, miR-16, miR-34a suppress HuR levels either by RNA stability or translational repression [83–86]. Hence, a miRNA-mediated manipulation of HuR is feasible using an antisense oligonucleotide targeted approach. We have observed that smoking-induced downregulation of miR-133a directly controls the expression of its targets HuR and EGFR in human papillomavirus-infected patient samples and oral pharyngeal carcinoma cells [87]. In addition to miRNAs, HuR silencing is carried out by delivering siRNAs targeting HuR using a nanoparticle formulation. For example, a folate or transferrin receptor-targeted nanoparticle formulated with siRNAs against HuR reduced lung cancer cell growth, migration, and invasion [88, 89]. Another study demonstrated that siRNA conjugated with cholesterol was loaded onto exosomal vesicles for the delivery and silencing of HuR [90]. HuR is silenced in the retina of diabetic retinopathy rats, in which cationic liposomes loaded with siRNAs target HuR and result in downregulation of VEGF and retinal thickness [91]. In another study, siRNAs targeting HuR were conjugated with Cy3-labeled DNA dendrimer nanocarrier delivered into ovarian cancer xenografts and showed antitumor activity [92]. Additional findings have demonstrated that siRNAs against HuR inhibit the expression of HuR and its targets, including Bcl-2, to repress the survival of colorectal cancer cells [93]. In other settings, an antisense oligonucleotide (ASO) knockdown of HuR reduces neuropathic pain through changes in target gene expression to inhibit microglia-mediated spinal neuroinflammation and promote an anti-inflammatory and neuroprotectant response [94]. Further, multiple sclerosis (MS) model ASO silencing of HuR exhibited improved experimental autoimmune encephalomyelitis in mice-related motor dysfunction, pain hypersensitivity, and body weight loss. Hence, targeting HuR using ASO is a promising approach to improve neurological disturbances in MS patients [95]. Together, these findings demonstrated that the possibility of siRNA or ASO therapeutics has the favorable effect of targeting HuR in multiple cell systems.
6. Small molecules targeting HuR:
The first report to develop inhibitors for HuR used a confocal fluctuation spectroscopy assay with recombinant HuR (both RRM1 and RRM2), and a tetramethylrhodamine (TMR) labeled ARE-RNA using a 50,000-compound library from Novartis [15]. In a competitive binding assay between HuR and ARE-RNA complex, the investigators identified MS-444, okicenone, and dehydromutactin as small molecules that block HuR binding with ARE-RNA. Out of the three compounds, MS-444 has been widely used as an inhibitor of HuR by many laboratories for anti-cancer activity [96–98], including melanoma [99], colorectal [100], pancreatic carcinoma [101], and glioma [102] by in vitro and other xenograft studies. However, the failure of in vivo tumor models to establish changes in HuR-mediated TME studies minimizes the promise of MS-444 as a potent HuR inhibitor. It is uncertain whether Novartis will continue the studies related to MS-444. Using a confocal nanoscanning screening method, a selective inhibitor compound, H1N, was identified against HuR RRM3. The compound H1N belongs to the dicarboxylic acid scaffold sub-library, which provides an opportunity to identify an ATP-binding pocket associated with 3′-terminal adenosyltransferase activity in HuR RRM3 [103].
Two independent studies revealed that the bioactive flavonoid quercetin disrupts the HuR binding with TNFα and IL-6 mRNA using electrophoretic mobility shift assays (EMSA), suggesting that both mRNAs are destabilized in the presence of quercetin [104, 105]. Further screening by PerkinElmer demonstrated that cetylpyridinium chloride and mitoxantrone prevented the formation of HuR-TNFα ARE complex [106]. In addition, mitoxantrone disrupts the interaction between HuR and SOX2 mRNA, which results in translational activation of SOX2 in mesenchymal stem cells [107]. Further studies from the same screening approach established the inhibitor DHTS (15, 16-dihydrotanshinone-I), which disrupts the binding between HuR and TNFα mRNA in the nanomolar range. Furthermore, treatment with DHTS exhibited anti-cancer activity in breast cancer cells [108], reduced colon cancer cell growth and proliferation [109], and cytotoxicity towards glioma cells [31]. Based on the structure of DHTS, treatment of its derivatives tanshinone mimic 6a and 6n reduced the cell growth of breast and pancreatic cancer cells. However, they are less potent when compared to DHTS [110]. In another study, using an actin-depolymerizing macrolide latrunculin A, or blebbistatin, an inhibitor of myosin II ATPase activity, the authors showed reduction in the cytoplasmic HuR content of HepG2 and Huh7 hepatocellular carcinoma (HCC) cells, which concomitantly changes the intracellular HuR localization, and markedly decreased the half-lives of COX-2, cyclin A and cyclin D1 resulting in a significant reduction in their expression levels in HepG2 cells [111].
Wu et al. used an FP assay optimized for high throughput screening (HTS) to screen 6000 compounds for HuR/ARE-mRNA disruptors [112]. The HTS analysis yielded six compounds of coumarin derivatives, CMLD1–6, in which CMLD-2 was the most potent HuR/ARE disruptor identified. CMLD-2 showed promising antitumor activity in lung, breast, colon, and thyroid cancers [20, 113, 114], in addition to triggering apoptosis and autophagy through deregulation of HuR targets Bcl-2 and XIAP [112]. Further efforts using FP-based assays identified small molecule azaphilones, derivatives of the fungal natural product asperbenzaldehyde, which disrupt the binding between HuR and c-Fos ARE mRNA [115]. An FDA-approved drug, suramin, an anti-trypanosomal drug, competitively binds with HuR and exerts antitumor activity in oral cancer cells [116]. Trichostatin A (TSA) and 5-Aza 2’deoxycytidine (AZA), two well-characterized pharmacologic inhibitors of histone deacetylation and DNA methylation, affect HuR translocation in ER-negative breast cancer cell lines, modulate HuR dependent ER mRNA [117] to reduce tumor burden [117].
Table 2 illustrates the list of HuR inhibitors demonstrating the antitumor activity, and promising, potent drugs are listed in the order of their discovery. Figure 2 shows small molecules that are either interrupting HuR binding with RNA or inhibiting HuR activity.
Table 2:
Small molecules targeting HuR show anticancer activity.
| Small-molecules | Drug action on HuR | Method of identification | References |
|---|---|---|---|
| Mitoxantrone and 15,16- dihydritanshinone-1 | Blocks HuR association with target mRNAs | Amplified luminescent proximity homogeneous assay (AL-PHA Screen) | [123] |
| Tanshinone and derivatives | Blocks HuR association with target mRNAs | ALPHA Screen | [127] |
| MS-444 | HuR translocation | In vitro/ in vivo | [13, 91] |
| 5-aza 2” deoxycytidine/ trichostatin A | HuR translocation | In vitro/ in vivo | [134] |
| Blebbistatin | HuR trafficking | In vitro/ in vivo | [128] |
| CMLD2 | HuR target mRNA binding inhibitor | In vitro only | [105] |
| Tanshinone II | HuR target mRNA binding inhibitor | In vitro only | [125] |
| Suramin | HuR target mRNA binding inhibitor | In vitro/ in vivo | [133] |
| NSC# 651084 | HuR target mRNA binding inhibitor | In vitro | [12] |
Figure 2:

Small molecule inhibitors of HuR. Small molecules interrupting RNA binding or protein localization of HuR are displayed.
7. Small animal models of HuR provide a therapeutic window of opportunity to treat human diseases:
Recent efforts from multiple groups showing a conditional knockout of HuR have a significant impact on cell survival. Timothy Hla’s group established the first reported HuR flox mice [118] and showed deletion of HuR induced atrophy of hematopoietic organs, extensive loss of intestinal villi, obstructive enterocolitis, and lethality within ten days. The same HuR flox mice (Jax mice stock number # 021431) model was used to generate multiple tissue-specific HuR silencing, such as adipose and vascular smooth muscle [119, 120], intestinal epithelium [121], skeletal muscle [122], hepatic tissue [123], cardiac-specific [124], and mouse neurons [125]. For example, fat-specific knockout of HuR greatly enhances adipogenic gene program in fatty tissues, accompanied by a systemic glucose intolerance and insulin resistance [126]. In another study, neuron-specific HuR-deficient mice developed a phenotype consisting of poor balance, decreased movement, and strength [127]. These findings demonstrated that deletion of HuR will have a complex differential post-transcriptional regulation and biological functions in a tissue-specific manner. A complete gene knockout differs primarily from the inhibition of protein activity because the phenotypes obtained from gene knockouts are likely to be different from those obtained after inhibitor treatment. It is evident that genetic silencing of HuR in murine is embryonically lethal and disturbs cellular development and normal homeostasis [118]. However, these effects were not observed when mice were treated with a HuR inhibitor MS-444 [100]. Therefore, drug targeting of HuR is more efficient than genetic silencing in cancer cells.
As HuR belongs to the ELAV family of RBP, its complete knockout is embryonically lethal. Hence, tissue-specific HuR deletion is critical to determine its functions. For example, heterozygous HuR conditional (OX40-Cre HuRfl/+) KO in T cells had decreased steady-state levels of Gata3, Il4, and Il13 mRNAs with minor changes at the protein level. However, homozygous HuR conditional (OX40-Cre HuRfl/fl) KO mice showed increased Il2, Il4, and Il13 mRNA and protein via different mechanisms [128]. These findings demonstrate that the dosage of HuR is critical for the management of cytokines in the immune system. In another study, vascular HuRSMKO (smooth muscle-specific HuR knockout) mice showed hypertension and cardiac hypertrophy [129]. The functional abnormalities in HuRSMKO mice were attributed to decreased mRNA and protein levels of RGS (regulator of G-protein signaling) protein(s) RGS2, RGS4, and RGS5, which increased intracellular calcium levels and elevated blood pressure. Consistently, the degree of intracellular calcium ion increase and blood pressure in HuR-deficient smooth muscle cells was reduced by overexpression of RGS2, RGS4, or RGS5 [129]. Since HuR has an alleged role in the post-transcriptional activation of inflammatory mRNAs, an inducible increase of HuR in murine innate compartments suppresses inflammatory responses in vivo [130]. In macrophages, HuR overexpression induced the translational silencing of specific cytokine mRNAs despite positive or nominal effects on their corresponding turnover. The study demonstrated that HuR acts in a pleiotropic fashion in inflammation through its functional interactions with specific mRNA subsets and negative post-transcriptional modules [130]. Taken together, knowledge gained from HuR KO mice models further supports the use of HuR inhibitors in clinical settings. However, the lack of studies related to HuR’s role in the TME, and immune crosstalk provides a positive framework for us to block HuR activity and study its role in antitumor functions in vivo using mouse models.
8. Conclusions and future perspectives:
HuR functions in a wide range of biological activity in a tissue-specific manner, both in mRNA stability and translation. HuR overexpression or dysregulation, including altered ARE mRNA interactions, has been implicated in many diseases, including neurodegenerative disorders, immune disorders, and cancers [76]. Hence, HuR is attractive to chemically target and manipulate its binding to ARE mRNAs for the control of gene expression. As highlighted here and recently reviewed comprehensively [36], several FDA-approved drugs have been identified that target HuR. Despite these advances in HuR targeting, it is still unanswered how HuR targeting drugs can control tumors in an in vivo TME and immune response. The major obstacle for this nonresponsive action is the absence of suitable high throughput assays for HuR small-molecule probe discovery and the heterogeneous population of cancer cells. As we described here, the recent advancement of single-cell RNA sequencing combined with HuR manipulation may reveal which cancer cell types respond to HuR inhibition.
Targeting RBPs with specific inhibitors is still an emerging field compared to targeting other proteins like kinases and transcription factors [131, 132]. In addition to targeting HuR and altering its biological activity, small molecule inhibitors that indirectly modulate its interaction with ARE mRNAs can serve as indirect modulators of HuR (see graphical illustration). RNA is negatively charged, and RBPs are positively charged, making their interacting surface have many RNA-proteins in the interface. The disturbance of the interface makes it harder for small molecules that effectively inhibit RBPs through competitive binding studies with RNAs. Although the RNA-competitive RBP inhibitors have weaknesses, the allosteric RBP inhibitors might have low off-target effects against other RBPs and will likely attract more attention in the coming years [16]. Both FRET and FP are conventional screening methods used to detect HuR inhibitors. However, in looking beyond these assays, established methods like proximity-based labeling strategies and photo-crosslinking capture methods will support the identification of additional novel inhibitors for HuR.
The recent emergence of reports linking RBPs to the formation of phase-separated posttranscriptional condensates [133, 134] logically raises questions about how the physicochemical properties of condensates could influence the action of RBP-targeting small-molecules. Studies have indicated that drugs that target proteins have significant preferences for entering posttranscriptional condensates over other types of condensates [135]. For example, HuR, along with other RBPs, can phase separate in microtubules, which can be used as a platform to understand the mechanisms underlying liquid-liquid phase separation and their deregulation in human diseases [136]. It has also been demonstrated that certain small molecules preferentially interact with specific condensates based on their physicochemical properties [135]. Therefore, it is appropriate to evaluate the physicochemical properties associated with partitioning into condensates, which could significantly affect a small molecule’s ability to reach its target. Moreover, it will be imperative to understand the circumstances where entering a condensate is required for drug action. At present, there are many more questions than answers regarding the relationship between condensates and drug action. Still, future insights may have a significant impact on the ways we approach RBP-targeting and small-molecule design.
In conclusion, research into HuR inhibition using small molecules or siRNAs has evolved rapidly in the past decade, providing an opportunity for us to develop a monotherapy or combination therapy with anti-cancer drugs. However, many HuR inhibitors were identified and tested in vitro cell culture approaches. HuR’s critical role in the TME remains an understudied area as well as how the inhibition of HuR changes the TME using in vivo mouse models.
Acknowledgments
This work was supported by the National Institutes of Health (NIH Grant R01 DE030013). NIH Grant Number UL1 TR001450 partly funds this study. We want to thank Dr. Carl E. Heltzel at Hollings Cancer Center for proofreading the manuscript.
Abbreviations:
- HuR
Human antigen R
- ELAVL1
Embryonic Lethal, Abnormal Vision, Drosophila-Like 1
- PTR
Posttranscriptional gene regulation
- RBP
RNA-binding proteins
- mRNP
messenger RNA nucleoproteins
- ARE
AU-rich elements
- UTR
Untranslated region
- ALS
Amyotrophic Lateral Sclerosis
- ARE
AU-rich elements
- Chk2
Checkpoint Kinase 2
- DHTS
15,16-dihydrotanshinone-I
- EMSA
electrophoretic mobility shift assays
- ELAVL1
Embryonic Lethal, Abnormal Vision, Drosophila-Like 1
- FP
Fluorescence Polarization
- FRET
fluorescence resonance energy transfer
- GC
germinal center
- HTS
High throughput screening
- HuR
Human antigen R
- mRNP
messenger RNA nucleoproteins
- PDB
Protein Data Bank
- PTR
Posttranscriptional gene regulation
- RBP
RNA-binding proteins
- RRM
RNA-recognition motif
- TME
tumor microenvironment
Footnotes
Conflict of interest: The author declares no conflict of interest.
References
- [1].Siomi H, Dreyfuss G, RNA-binding proteins as regulators of gene expression, Curr Opin Genet Dev, 7 (1997) 345–353. [DOI] [PubMed] [Google Scholar]
- [2].Brannan KW, Jin W, Huelga SC, Banks CA, Gilmore JM, Florens L, Washburn MP, Van Nostrand EL, Pratt GA, Schwinn MK, Daniels DL, Yeo GW, SONAR Discovers RNA-Binding Proteins from Analysis of Large-Scale Protein-Protein Interactomes, Mol Cell, 64 (2016) 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN, The RNA-binding protein repertoire of embryonic stem cells, Nat Struct Mol Biol, 20 (2013) 1122–1130. [DOI] [PubMed] [Google Scholar]
- [4].Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M, The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts, Mol Cell, 46 (2012) 674–690. [DOI] [PubMed] [Google Scholar]
- [5].Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW, Insights into RNA biology from an atlas of mammalian mRNA-binding proteins, Cell, 149 (2012) 1393–1406. [DOI] [PubMed] [Google Scholar]
- [6].Gerstberger S, Hafner M, Tuschl T, A census of human RNA-binding proteins, Nat Rev Genet, 15 (2014) 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Julio AR, Backus KM, New approaches to target RNA binding proteins, Curr Opin Chem Biol, 62 (2021) 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cheung AK, Hurley B, Kerrigan R, Shu L, Chin DN, Shen Y, O’Brien G, Sung MJ, Hou Y, Axford J, Cody E, Sun R, Fazal A, Fridrich C, Sanchez CC, Tomlinson RC, Jain M, Deng L, Hoffmaster K, Song C, Van Hoosear M, Shin Y, Servais R, Towler C, Hild M, Curtis D, Dietrich WF, Hamann LG, Briner K, Chen KS, Kobayashi D, Sivasankaran R, Dales NA, Discovery of Small Molecule Splicing Modulators of Survival Motor Neuron-2 (SMN2) for the Treatment of Spinal Muscular Atrophy (SMA), J Med Chem, 61 (2018) 11021–11036. [DOI] [PubMed] [Google Scholar]
- [9].Ratni H, Ebeling M, Baird J, Bendels S, Bylund J, Chen KS, Denk N, Feng Z, Green L, Guerard M, Jablonski P, Jacobsen B, Khwaja O, Kletzl H, Ko CP, Kustermann S, Marquet A, Metzger F, Mueller B, Naryshkin NA, Paushkin SV, Pinard E, Poirier A, Reutlinger M, Weetall M, Zeller A, Zhao X, Mueller L, Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 ( SMN2) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA), J Med Chem, 61 (2018) 6501–6517. [DOI] [PubMed] [Google Scholar]
- [10].Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, Xie Y, Williams NS, Nijhawan D, Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15, Science, 356 (2017). [DOI] [PubMed] [Google Scholar]
- [11].Chen X, Kopecky DJ, Mihalic J, Jeffries S, Min X, Heath J, Deignan J, Lai S, Fu Z, Guimaraes C, Shen S, Li S, Johnstone S, Thibault S, Xu H, Cardozo M, Shen W, Walker N, Kayser F, Wang Z, Structure-guided design, synthesis, and evaluation of guanine-derived inhibitors of the eIF4E mRNA-cap interaction, J Med Chem, 55 (2012) 3837–3851. [DOI] [PubMed] [Google Scholar]
- [12].Francois-Moutal L, Felemban R, Scott DD, Sayegh MR, Miranda VG, Perez-Miller S, Khanna R, Gokhale V, Zarnescu DC, Khanna M, Small Molecule Targeting TDP-43’s RNA Recognition Motifs Reduces Locomotor Defects in a Drosophila Model of Amyotrophic Lateral Sclerosis (ALS), ACS Chem Biol, 14 (2019) 2006–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu X, Gardashova G, Lan L, Han S, Zhong C, Marquez RT, Wei L, Wood S, Roy S, Gowthaman R, Karanicolas J, Gao FP, Dixon DA, Welch DR, Li L, Ji M, Aube J, Xu L, Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis, Commun Biol, 3 (2020) 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang Z, Bhattacharya A, Ivanov DN, Identification of Small-Molecule Inhibitors of the HuR/RNA Interaction Using a Fluorescence Polarization Screening Assay Followed by NMR Validation, PLoS One, 10 (2015) e0138780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S, Harrer N, Woisetschlaeger M, Buehler C, Uhl V, Auer M, Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR, Nat Chem Biol, 3 (2007) 508–515. [DOI] [PubMed] [Google Scholar]
- [16].Wu P, Inhibition of RNA-binding proteins with small molecules, Nature Reviews Chemistry, 4 (2020) 441–458. [DOI] [PubMed] [Google Scholar]
- [17].Stanek D, Neugebauer KM, Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer, J Cell Biol, 166 (2004) 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chusainow J, Ajuh PM, Trinkle-Mulcahy L, Sleeman JE, Ellenberg J, Lamond AI, FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35, RNA, 11 (2005) 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ellis JD, Lleres D, Denegri M, Lamond AI, Caceres JF, Spatial mapping of splicing factor complexes involved in exon and intron definition, J Cell Biol, 181 (2008) 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Allegri L, Baldan F, Roy S, Aube J, Russo D, Filetti S, Damante G, The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells, Sci Rep, 9 (2019) 7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Good PJ, A conserved family of elav-like genes in vertebrates, Proc Natl Acad Sci U S A, 92 (1995) 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yao KM, Samson ML, Reeves R, White K, Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans, J Neurobiol, 24 (1993) 723–739. [DOI] [PubMed] [Google Scholar]
- [23].Keene JD, Why is Hu where? Shuttling of early-response-gene messenger RNA subsets, Proc Natl Acad Sci U S A, 96 (1999) 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Corley M, Burns MC, Yeo GW, How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms, Mol Cell, 78 (2020) 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maris C, Dominguez C, Allain FH, The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression, FEBS J, 272 (2005) 2118–2131. [DOI] [PubMed] [Google Scholar]
- [26].Fan XC, Steitz JA, HNS, a nuclear-cytoplasmic shuttling sequence in HuR, Proc Natl Acad Sci U S A, 95 (1998) 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen CY, Xu N, Shyu AB, Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization, Mol Cell Biol, 22 (2002) 7268–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ripin N, Boudet J, Duszczyk MM, Hinniger A, Faller M, Krepl M, Gadi A, Schneider RJ, Sponer J, Meisner-Kober NC, Allain FH, Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM, Proc Natl Acad Sci U S A, 116 (2019) 2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brennan CM, Gallouzi IE, Steitz JA, Protein ligands to HuR modulate its interaction with target mRNAs in vivo, J Cell Biol, 151 (2000) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Scheiba RM, de Opakua AI, Diaz-Quintana A, Cruz-Gallardo I, Martinez-Cruz LA, Martinez-Chantar ML, Blanco FJ, Diaz-Moreno I, The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets, RNA biology, 11 (2014) 1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Filippova N, Yang X, Ananthan S, Sorochinsky A, Hackney JR, Gentry Z, Bae S, King P, Nabors LB, Hu antigen R (HuR) multimerization contributes to glioma disease progression, J Biol Chem, 292 (2017) 16999–17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W, HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA, Nucleic Acids Res, 40 (2012) 5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang H, Zeng F, Liu Q, Liu H, Liu Z, Niu L, Teng M, Li X, The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding, Acta Crystallogr D Biol Crystallogr, 69 (2013) 373–380. [DOI] [PubMed] [Google Scholar]
- [34].Wang X, Tanaka Hall TM, Structural basis for recognition of AU-rich element RNA by the HuD protein, Nat Struct Biol, 8 (2001) 141–145. [DOI] [PubMed] [Google Scholar]
- [35].Inoue M, Muto Y, Sakamoto H, Yokoyama S, NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C, Nucleic Acids Res, 28 (2000) 1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wu M, Tong CWS, Yan W, To KKW, Cho WCS, The RNA Binding Protein HuR: A Promising Drug Target for Anticancer Therapy, Curr Cancer Drug Targets, 19 (2019) 382–399. [DOI] [PubMed] [Google Scholar]
- [37].Zhu S, Rooney S, Michlewski G, RNA-Targeted Therapies and High-Throughput Screening Methods, Int J Mol Sci, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Grammatikakis I, Abdelmohsen K, Gorospe M, Posttranslational control of HuR function, Wiley interdisciplinary reviews. RNA, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Eberhardt W, Doller A, Pfeilschifter J, Regulation of the mRNA-binding protein HuR by posttranslational modification: spotlight on phosphorylation, Curr Protein Pept Sci, 13 (2012) 380–390. [DOI] [PubMed] [Google Scholar]
- [40].Doller A, Pfeilschifter J, Eberhardt W, Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR, Cell Signal, 20 (2008) 2165–2173. [DOI] [PubMed] [Google Scholar]
- [41].Abdelmohsen K, Pullmann R Jr., Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M, Phosphorylation of HuR by Chk2 regulates SIRT1 expression, Mol Cell, 25 (2007) 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Masuda K, Abdelmohsen K, Kim MM, Srikantan S, Lee EK, Tominaga K, Selimyan R, Martindale JL, Yang X, Lehrmann E, Zhang Y, Becker KG, Wang JY, Kim HH, Gorospe M, Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation, Embo J, 30 (2011) 1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mazan-Mamczarz K, Hagner PR, Zhang Y, Dai B, Lehrmann E, Becker KG, Keene JD, Gorospe M, Liu Z, Gartenhaus RB, ATM regulates a DNA damage response posttranscriptional RNA operon in lymphocytes, Blood, 117 (2011) 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr., Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, Gorospe M, Nuclear HuR accumulation through phosphorylation by Cdk1, Genes Dev, 22 (2008) 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Al-Khalaf HH, Aboussekhra A, ATR controls the UV-related upregulation of the CDKN1A mRNA in a Cdk1/HuR-dependent manner, Mol Carcinog, 53 (2014) 979–987. [DOI] [PubMed] [Google Scholar]
- [46].Amadio M, Bucolo C, Leggio GM, Drago F, Govoni S, Pascale A, The PKCbeta/HuR/VEGF pathway in diabetic retinopathy, Biochem Pharmacol, 80 (2010) 1230–1237. [DOI] [PubMed] [Google Scholar]
- [47].Doller A, Akool el S, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W, Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA, Mol Cell Biol, 28 (2008) 2608–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W, Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2, Mol Biol Cell, 18 (2007) 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM, Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences, J Biol Chem, 282 (2007) 20948–20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Latorre E, Castiglioni I, Gatto P, Carelli S, Quattrone A, Provenzani A, Loss of protein kinase Cdelta/HuR interaction is necessary to doxorubicin resistance in breast cancer cell lines, J Pharmacol Exp Ther, 349 (2014) 99–106. [DOI] [PubMed] [Google Scholar]
- [51].Chu PC, Chuang HC, Kulp SK, Chen CS, The mRNA-stabilizing factor HuR protein is targeted by beta-TrCP protein for degradation in response to glycolysis inhibition, J Biol Chem, 287 (2012) 43639–43650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lafarga V, Cuadrado A, Lopez de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR, p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint, Mol Cell Biol, 29 (2009) 4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liao WL, Wang WC, Chang WC, Tseng JT, The RNA-binding protein HuR stabilizes cytosolic phospholipase A2alpha mRNA under interleukin-1beta treatment in non-small cell lung cancer A549 Cells, J Biol Chem, 286 (2011) 35499–35508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Janakiraman H, House RP, Talwar S, Courtney SM, Hazard ES, Hardiman G, Mehrotra S, Howe PH, Gangaraju V, Palanisamy V, Repression of caspase-3 and RNA-binding protein HuR cleavage by cyclooxygenase-2 promotes drug resistance in oral squamous cell carcinoma, Oncogene, 36 (2017) 3137–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Talwar S, House R, Sundaramurthy S, Balasubramanian S, Yu H, Palanisamy V, Inhibition of caspases protects mice from radiation-induced oral mucositis and abolishes the cleavage of RNA-binding protein HuR, J Biol Chem, 289 (2014) 3487–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Talwar S, Jin J, Carroll B, Liu A, Gillespie MB, Palanisamy V, Caspase-mediated cleavage of RNA-binding protein HuR regulates c-Myc protein expression after hypoxic stress, J Biol Chem, 286 (2011) 32333–32343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Beauchamp P, Nassif C, Hillock S, van der Giessen K, von Roretz C, Jasmin BJ, Gallouzi IE, The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation, Cell Death Differ, (2010). [DOI] [PubMed] [Google Scholar]
- [58].Kuwano Y GI-E, & Gorospe M (2010) Role of the RNA-Binding Protein HuR in Apoptosis and Apoptosome Function. Apoptosome, eds Cecconi F & D’Amelio M (Springer Netherlands; ), pp 203–220. [Google Scholar]; Yuki Kuwano, Gallouzi I-E, Gorospe M, Role of the RNA-Binding Protein HuR in Apoptosis and Apoptosome Function, in: Cecconi F, D’Amelio M (Eds.) Apoptosome, Springer Netherlands; 2010, pp. 203–220. [Google Scholar]
- [59].Mazroui R, Di Marco S, Clair E, von Roretz C, Tenenbaum SA, Keene JD, Saleh M, Gallouzi IE, Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis, J Cell Biol, 180 (2008) 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].von Roretz C, Lian XJ, Macri AM, Punjani N, Clair E, Drouin O, Dormoy-Raclet V, Ma JF, Gallouzi IE, Apoptotic-induced cleavage shifts HuR from being a promoter of survival to an activator of caspase-mediated apoptosis, Cell Death Differ, 20 (2013) 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, Gorospe M, Ubiquitin-mediated proteolysis of HuR by heat shock, Embo J, 28 (2009) 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lucchesi C, Sheikh MS, Huang Y, Negative regulation of RNA-binding protein HuR by tumor-suppressor ECRG2, Oncogene, 35 (2016) 2565–2573. [DOI] [PubMed] [Google Scholar]
- [63].Zhou HL, Geng C, Luo G, Lou H, The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP, Genes Dev, 27 (2013) 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA, Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase, J Biol Chem, 277 (2002) 44623–44630. [DOI] [PubMed] [Google Scholar]
- [65].Pang L, Tian H, Chang N, Yi J, Xue L, Jiang B, Gorospe M, Zhang X, Wang W, Loss of CARM1 is linked to reduced HuR function in replicative senescence, BMC Mol Biol, 14 (2013) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Calvanese V, Lara E, Suarez-Alvarez B, Abu Dawud R, Vazquez-Chantada M, Martinez-Chantar ML, Embade N, Lopez-Nieva P, Horrillo A, Hmadcha A, Soria B, Piazzolla D, Herranz D, Serrano M, Mato JM, Andrews PW, Lopez-Larrea C, Esteller M, Fraga MF, Sirtuin 1 regulation of developmental genes during differentiation of stem cells, Proc Natl Acad Sci U S A, 107 (2010) 13736–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xu J, Richard S, Cellular pathways influenced by protein arginine methylation: Implications for cancer, Mol Cell, 81 (2021) 4357–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kaniskan HU, Martini ML, Jin J, Inhibitors of Protein Methyltransferases and Demethylases, Chem Rev, 118 (2018) 989–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou F, Zhang F, Zhou C, Liang M, Cai Z, Lv H, Li W, Wei X, Human antigen R and drug resistance in tumors, Invest New Drugs, 37 (2019) 1107–1116. [DOI] [PubMed] [Google Scholar]
- [70].Abdelmohsen K, Gorospe M, Posttranscriptional regulation of cancer traits by HuR, Wiley interdisciplinary reviews. RNA, 1 (2010) 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR, Understanding and targeting the disease-related RNA binding protein human antigen R (HuR), Wiley interdisciplinary reviews. RNA, 11 (2020) e1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B, Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis, Int J Mol Sci, 14 (2013) 10015–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].DeMicco A, Naradikian MS, Sindhava VJ, Yoon JH, Gorospe M, Wertheim GB, Cancro MP, Bassing CH, B Cell-Intrinsic Expression of the HuR RNA-Binding Protein Is Required for the T Cell-Dependent Immune Response In Vivo, J Immunol, 195 (2015) 3449–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Diaz-Munoz MD, Bell SE, Fairfax K, Monzon-Casanova E, Cunningham AF, Gonzalez-Porta M, Andrews SR, Bunik VI, Zarnack K, Curk T, Heggermont WA, Heymans S, Gibson GE, Kontoyiannis DL, Ule J, Turner M, The RNA-binding protein HuR is essential for the B cell antibody response, Nat Immunol, 16 (2015) 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL, Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR, J Immunol, 182 (2009) 6779–6788. [DOI] [PubMed] [Google Scholar]
- [76].Srikantan S, Gorospe M, HuR function in disease, Front Biosci (Landmark Ed), 17 (2012) 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, Kontoyiannis DL, Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis, J Clin Invest, 122 (2012) 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR, HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities, J Biol Chem, 276 (2001) 47958–47965. [DOI] [PubMed] [Google Scholar]
- [79].Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M, The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation, PLoS Genet, 8 (2012) e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J, The 3’ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR, Mol Cell Biol, 21 (2001) 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chen J, Martindale JL, Abdelmohsen K, Kumar G, Fortina PM, Gorospe M, Rostami A, Yu S, RNA-Binding Protein HuR Promotes Th17 Cell Differentiation and Can Be Targeted to Reduce Autoimmune Neuroinflammation, J Immunol, 204 (2020) 2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang Z, Yin W, Zhu L, Li J, Yao Y, Chen F, Sun M, Zhang J, Shen N, Song Y, Chang X, Iron Drives T Helper Cell Pathogenicity by Promoting RNA-Binding Protein PCBP1-Mediated Proinflammatory Cytokine Production, Immunity, 49 (2018) 80–92 e87. [DOI] [PubMed] [Google Scholar]
- [83].Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M, miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels, Proc Natl Acad Sci U S A, 105 (2008) 20297–20302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Guo X, Wu Y, Hartley RS, MicroRNA-125a represses cell growth by targeting HuR in breast cancer, RNA biology, 6 (2009) 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Xu F, Zhang X, Lei Y, Liu X, Liu Z, Tong T, Wang W, Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma, Journal of cellular biochemistry, 111 (2010) 727–734. [DOI] [PubMed] [Google Scholar]
- [86].Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M, MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms, Prostate, 70 (2010) 1501–1512. [DOI] [PubMed] [Google Scholar]
- [87].House R, Majumder M, Janakiraman H, Ogretmen B, Kato M, Erkul E, Hill E, Atkinson C, Barth J, Day TA, Palanisamy V, Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer, PLoS One, 13 (2018) e0205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Muralidharan R, Babu A, Amreddy N, Srivastava A, Chen A, Zhao YD, Kompella UB, Munshi A, Ramesh R, Tumor-targeted Nanoparticle Delivery of HuR siRNA Inhibits Lung Tumor Growth In Vitro and In Vivo By Disrupting the Oncogenic Activity of the RNA-binding Protein HuR, Mol Cancer Ther, 16 (2017) 1470–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Muralidharan R, Babu A, Amreddy N, Basalingappa K, Mehta M, Chen A, Zhao YD, Kompella UB, Munshi A, Ramesh R, Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration, J Nanobiotechnology, 14 (2016) 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].O’Loughlin AJ, Mager I, de Jong OG, Varela MA, Schiffelers RM, El Andaloussi S, Wood MJA, Vader P, Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles, Mol Ther, 25 (2017) 1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Amadio M, Pascale A, Cupri S, Pignatello R, Osera C, D.A. V, D.A. AG, G.M. Leggio, B. Ruozi, S. Govoni, F. Drago, C. Bucolo, Nanosystems based on siRNA silencing HuR expression counteract diabetic retinopathy in rat, Pharmacol Res, 111 (2016) 713–720. [DOI] [PubMed] [Google Scholar]
- [92].Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, Jimbo M, Gonye GE, Brody JR, Getts RC, Sawicki JA, Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth, Cancer Res, 76 (2016) 1549–1559. [DOI] [PubMed] [Google Scholar]
- [93].Lin GL, Ting HJ, Tseng TC, Juang V, Lo YL, Modulation of the mRNA-binding protein HuR as a novel reversal mechanism of epirubicin-triggered multidrug resistance in colorectal cancer cells, PLoS One, 12 (2017) e0185625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Borgonetti V, Galeotti N, Intranasal delivery of an antisense oligonucleotide to the RNA-binding protein HuR relieves nerve injury-induced neuropathic pain, Pain, 162 (2021) 1500–1510. [DOI] [PubMed] [Google Scholar]
- [95].Borgonetti V, Sanna MD, Lucarini L, Galeotti N, Targeting the RNA-Binding Protein HuR Alleviates Neuroinflammation in Experimental Autoimmune Encephalomyelitis: Potential Therapy for Multiple Sclerosis, Neurotherapeutics, 18 (2021) 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner-Kober N, Londin E, Rigoutsos I, Sawicki JA, Risbud MV, Witkiewicz AK, McCue PA, Jiang W, Rui H, Yeo CJ, Petricoin E, Winter JM, Brody JR, The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells, Oncogene, 35 (2016) 2529–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Herdy B, Karonitsch T, Vladimer GI, Tan CS, Stukalov A, Trefzer C, Bigenzahn JW, Theil T, Holinka J, Kiener HP, Colinge J, Bennett KL, Superti-Furga G, The RNA-binding protein HuR/ELAVL1 regulates IFN-beta mRNA abundance and the type I IFN response, Eur J Immunol, 45 (2015) 1500–1511. [DOI] [PubMed] [Google Scholar]
- [98].Lu L, Zheng L, Si Y, Luo W, Dujardin G, Kwan T, Potochick NR, Thompson SR, Schneider DA, King PH, Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis, J Biol Chem, 289 (2014) 31792–31804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Moradi F, Berglund P, Linnskog R, Leandersson K, Andersson T, Prasad CP, Dual mechanisms of action of the RNA-binding protein human antigen R explains its regulatory effect on melanoma cell migration, Transl Res, 172 (2016) 45–60. [DOI] [PubMed] [Google Scholar]
- [100].Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, Sedlyarov V, Evstatiev R, Dammann K, Loy A, Kuzyk O, Kovarik P, Khare V, Beibel M, Roma G, Meisner-Kober N, Gasche C, HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis, Cancer Res, 77 (2017) 2424–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, Winter JM, Sawicki JA, Sachs JN, Meisner-Kober N, Yeo CJ, Vadigepalli R, Tykocinski ML, Brody JR, HuR Contributes to TRAIL Resistance by Restricting Death Receptor 4 Expression in Pancreatic Cancer Cells, Mol Cancer Res, 14 (2016) 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang J, Hjelmeland AB, Nabors LB, King PH, Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells, Cancer Biol Ther, 20 (2019) 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Meisner NC, Hintersteiner M, Seifert JM, Bauer R, Benoit RM, Widmer A, Schindler T, Uhl V, Lang M, Gstach H, Auer M, Terminal adenosyl transferase activity of posttranscriptional regulator HuR revealed by confocal on-bead screening, J Mol Biol, 386 (2009) 435–450. [DOI] [PubMed] [Google Scholar]
- [104].Ouhara K, Munenaga S, Kajiya M, Takeda K, Matsuda S, Sato Y, Hamamoto Y, Iwata T, Yamasaki S, Akutagawa K, Mizuno N, Fujita T, Sugiyama E, Kurihara H, The induced RNA-binding protein, HuR, targets 3’-UTR region of IL-6 mRNA and enhances its stabilization in periodontitis, Clin Exp Immunol, 192 (2018) 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH, Kim S, Park WY, Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA, Exp Mol Med, 41 (2009) 824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].D’Agostino VG, Adami V, Provenzani A, A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation, PLoS One, 8 (2013) e72426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Latorre E, Carelli S, Caremoli F, Giallongo T, Colli M, Canazza A, Provenzani A, Di Giulio AM, Gorio A, Human Antigen R Binding and Regulation of SOX2 mRNA in Human Mesenchymal Stem Cells, Mol Pharmacol, 89 (2016) 243–252. [DOI] [PubMed] [Google Scholar]
- [108].D’Agostino VG, Lal P, Mantelli B, Tiedje C, Zucal C, Thongon N, Gaestel M, Latorre E, Marinelli L, Seneci P, Amadio M, Provenzani A, Dihydrotanshinone-I interferes with the RNA-binding activity of HuR affecting its post-transcriptional function, Sci Rep, 5 (2015) 16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lal P, Cerofolini L, D’Agostino VG, Zucal C, Fuccio C, Bonomo I, Dassi E, Giuntini S, Di Maio D, Vishwakarma V, Preet R, Williams SN, Fairlamb MS, Munk R, Lehrmann E, Abdelmohsen K, Elezgarai SR, Luchinat C, Novellino E, Quattrone A, Biasini E, Manzoni L, Gorospe M, Dixon DA, Seneci P, Marinelli L, Fragai M, Provenzani A, Regulation of HuR structure and function by dihydrotanshinone-I, Nucleic Acids Res, 45 (2017) 9514–9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Manzoni L, Zucal C, Maio DD, D’Agostino VG, Thongon N, Bonomo I, Lal P, Miceli M, Baj V, Brambilla M, Cerofolini L, Elezgarai S, Biasini E, Luchinat C, Novellino E, Fragai M, Marinelli L, Provenzani A, Seneci P, Interfering with HuR-RNA Interaction: Design, Synthesis and Biological Characterization of Tanshinone Mimics as Novel, Effective HuR Inhibitors, J Med Chem, 61 (2018) 1483–1498. [DOI] [PubMed] [Google Scholar]
- [111].Doller A, Badawi A, Schmid T, Brauss T, Pleli T, zu Heringdorf DM, Piiper A, Pfeilschifter J, Eberhardt W, The cytoskeletal inhibitors latrunculin A and blebbistatin exert antitumorigenic properties in human hepatocellular carcinoma cells by interfering with intracellular HuR trafficking, Exp Cell Res, 330 (2015) 66–80. [DOI] [PubMed] [Google Scholar]
- [112].Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, Roy A, Turner BA, McDonald P, Tunge JA, Rogers SA, Dixon DA, Aube J, Xu L, Identification and validation of novel small molecule disruptors of HuR-mRNA interaction, ACS Chem Biol, 10 (2015) 1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Andrade D, Mehta M, Griffith J, Oh S, Corbin J, Babu A, De S, Chen A, Zhao YD, Husain S, Roy S, Xu L, Aube J, Janknecht R, Gorospe M, Herman T, Ramesh R, Munshi A, HuR Reduces Radiation-Induced DNA Damage by Enhancing Expression of ARID1A, Cancers (Basel), 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Muralidharan R, Mehta M, Ahmed R, Roy S, Xu L, Aube J, Chen A, Zhao YD, Herman T, Ramesh R, Munshi A, HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells, Sci Rep, 7 (2017) 9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kaur K, Wu X, Fields JK, Johnson DK, Lan L, Pratt M, Somoza AD, Wang CCC, Karanicolas J, Oakley BR, Xu L, De Guzman RN, The fungal natural product azaphilone-9 binds to HuR and inhibits HuR-RNA interaction in vitro, PLoS One, 12 (2017) e0175471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kakuguchi W, Nomura T, Kitamura T, Otsuguro S, Matsushita K, Sakaitani M, Maenaka K, Tei K, Suramin, screened from an approved drug library, inhibits HuR functions and attenuates malignant phenotype of oral cancer cells, Cancer Med, 7 (2018) 6269–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Pryzbylkowski P, Obajimi O, Keen JC, Trichostatin A and 5 Aza-2’ deoxycytidine decrease estrogen receptor mRNA stability in ER positive MCF7 cells through modulation of HuR, Breast Cancer Res Treat, 111 (2008) 15–25. [DOI] [PubMed] [Google Scholar]
- [118].Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T, Essential role of the RNA-binding protein HuR in progenitor cell survival in mice, J Clin Invest, 119 (2009) 3530–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Anthony SR, Guarnieri A, Lanzillotta L, Gozdiff A, Green LC, O’Grady K, Helsley RN, Owens Iii AP, Tranter M, HuR expression in adipose tissue mediates energy expenditure and acute thermogenesis independent of UCP1 expression, Adipocyte, 9 (2020) 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Li J, Gong L, Liu S, Zhang Y, Zhang C, Tian M, Lu H, Bu P, Yang J, Ouyang C, Jiang X, Wu J, Zhang Y, Min Q, Zhang C, Zhang W, Adipose HuR protects against diet-induced obesity and insulin resistance, Nat Commun, 10 (2019) 2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, Xu Y, Gorospe M, Wang JY, H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR, Mol Cell Biol, 36 (2016) 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Mynatt RL, Noland RC, Elks CM, Vandanmagsar B, Bayless DS, Stone AC, Ghosh S, Ravussin E, Warfel JD, The RNA binding protein HuR influences skeletal muscle metabolic flexibility in rodents and humans, Metabolism, 97 (2019) 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhang Z, Zong C, Jiang M, Hu H, Cheng X, Ni J, Yi X, Jiang B, Tian F, Chang MW, Su W, Zhu L, Li J, Xiang X, Miao C, Gorospe M, de Cabo R, Dou Y, Ju Z, Yang J, Jiang C, Yang Z, Wang W, Hepatic HuR modulates lipid homeostasis in response to high-fat diet, Nat Commun, 11 (2020) 3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Green LC, Anthony SR, Slone S, Lanzillotta L, Nieman ML, Wu X, Robbins N, Jones SM, Roy S, Owens AP 3rd, Aube J, Xu L, Lorenz JN, Blaxall BC, Rubinstein J, Benoit JB, Tranter M, Human antigen R as a therapeutic target in pathological cardiac hypertrophy, JCI Insight, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhao YF, He XX, Song ZF, Guo Y, Zhang YN, Yu HL, He ZX, Xiong WC, Guo W, Zhu XJ, Human antigen R-regulated mRNA metabolism promotes the cell motility of migrating mouse neurons, Development, 147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Siang DTC, Lim YC, Kyaw AMM, Win KN, Chia SY, Degirmenci U, Hu X, Tan BC, Walet ACE, Sun L, Xu D, The RNA-binding protein HuR is a negative regulator in adipogenesis, Nat Commun, 11 (2020) 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Sun K, Li X, Chen X, Bai Y, Zhou G, Kokiko-Cochran ON, Lamb B, Hamilton TA, Lin CY, Lee YS, Herjan T, Neuron-Specific HuR-Deficient Mice Spontaneously Develop Motor Neuron Disease, J Immunol, 201 (2018) 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Gubin MM, Techasintana P, Magee JD, Dahm GM, Calaluce R, Martindale JL, Whitney MS, Franklin CL, Besch-Williford C, Hollingsworth JW, Abdelmohsen K, Gorospe M, Atasoy U, Conditional knockout of the RNA-binding protein HuR in CD4(+) T cells reveals a gene dosage effect on cytokine production, Mol Med, 20 (2014) 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Liu S, Jiang X, Lu H, Xing M, Qiao Y, Zhang C, Zhang W, HuR (Human Antigen R) Regulates the Contraction of Vascular Smooth Muscle and Maintains Blood Pressure, Arterioscler Thromb Vasc Biol, 40 (2020) 943–957. [DOI] [PubMed] [Google Scholar]
- [130].Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL, HuR as a negative posttranscriptional modulator in inflammation, Mol Cell, 19 (2005) 777–789. [DOI] [PubMed] [Google Scholar]
- [131].Wu P, Nielsen TE, Clausen MH, FDA-approved small-molecule kinase inhibitors, Trends Pharmacol Sci, 36 (2015) 422–439. [DOI] [PubMed] [Google Scholar]
- [132].Henley MJ, Koehler AN, Advances in targeting ‘undruggable’ transcription factors with small molecules, Nat Rev Drug Discov, 20 (2021) 669–688. [DOI] [PubMed] [Google Scholar]
- [133].Shorter J, Phase separation of RNA-binding proteins in physiology and disease: An introduction to the JBC Reviews thematic series, J Biol Chem, 294 (2019) 7113–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wiedner HJ, Giudice J, It’s not just a phase: function and characteristics of RNA-binding proteins in phase separation, Nat Struct Mol Biol, 28 (2021) 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall’Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, Sagi I, Clark VE, Platt JM, Kar M, McCall PM, Zamudio AV, Manteiga JC, Coffey EL, Li CH, Hannett NM, Guo YE, Decker TM, Lee TI, Zhang T, Weng JK, Taatjes DJ, Chakraborty A, Sharp PA, Chang YT, Hyman AA, Gray NS, Young RA, Partitioning of cancer therapeutics in nuclear condensates, Science, 368 (2020) 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Maucuer A, Desforges B, Joshi V, Boca M, Kretov DA, Hamon L, Bouhss A, Curmi PA, Pastre D, Microtubules as platforms for probing liquid-liquid phase separation in cells - application to RNA-binding proteins, Journal of cell science, 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Heinonen M, Hemmes A, Salmenkivi K, Abdelmohsen K, Vilen ST, Laakso M, Leidenius M, Salo T, Hautaniemi S, Gorospe M, Heikkila P, Haglund C, Ristimaki A, Role of RNA binding protein HuR in ductal carcinoma in situ of the breast, J Pathol, 224 (2011) 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Heinonen M, Fagerholm R, Aaltonen K, Kilpivaara O, Aittomaki K, Blomqvist C, Heikkila P, Haglund C, Nevanlinna H, Ristimaki A, Prognostic role of HuR in hereditary breast cancer, Clin Cancer Res, 13 (2007) 6959–6963. [DOI] [PubMed] [Google Scholar]
- [139].Zhang C, Xue G, Bi J, Geng M, Chu H, Guan Y, Wang J, Wang B, Cytoplasmic expression of the ELAV-like protein HuR as a potential prognostic marker in esophageal squamous cell carcinoma, Tumour Biol, 35 (2014) 73–80. [DOI] [PubMed] [Google Scholar]
- [140].Sun DP, Lin CY, Tian YF, Chen LT, Lin LC, Lee SW, Hsing CH, Lee HH, Shiue YL, Huang HY, Li CF, Liang PI, Clinicopathological significance of HuR expression in gallbladder carcinoma: with special emphasis on the implications of its nuclear and cytoplasmic expression, Tumour Biol, 34 (2013) 3059–3069. [DOI] [PubMed] [Google Scholar]
- [141].Wei YC, Chou FF, Li CF, Li WM, Chen YY, Lan J, Li SH, Fang FM, Hu TH, Yu SC, Eng HL, Uen YH, Tian YF, Wang JC, Huang HY, HuR cytoplasmic expression is associated with increased cyclin A expression and inferior disease-free survival in patients with gastrointestinal stromal tumours (GISTs), Histopathology, 63 (2013) 445–454. [DOI] [PubMed] [Google Scholar]
- [142].Lauriola L, Granone P, Ramella S, Lanza P, Ranelletti FO, Expression of the RNA-binding protein HuR and its clinical significance in human stage I and II lung adenocarcinoma, Histol Histopathol, 27 (2012) 617–626. [DOI] [PubMed] [Google Scholar]
- [143].Gauchotte G, Hergalant S, Vigouroux C, Casse JM, Houlgatte R, Kaoma T, Helle D, Brochin L, Rech F, Peyre M, Labrousse F, Vallar L, Gueant JL, Vignaud JM, Battaglia-Hsu SF, Cytoplasmic overexpression of RNA-binding protein HuR is a marker of poor prognosis in meningioma, and HuR knockdown decreases meningioma cell growth and resistance to hypoxia, J Pathol, 242 (2017) 421–434. [DOI] [PubMed] [Google Scholar]
- [144].Cha JD, Kim HK, Cha IH, Cytoplasmic HuR expression: correlation with cellular inhibitors of apoptosis protein-2 expression and clinicopathologic factors in oral squamous cell carcinoma cells, Head Neck, 36 (2014) 1168–1175. [DOI] [PubMed] [Google Scholar]
- [145].Kim KY, Li S, Cha JD, Zhang X, Cha IH, Significance of molecular markers in survival prediction of oral squamous cell carcinoma, Head Neck, 34 (2012) 929–936. [DOI] [PubMed] [Google Scholar]
- [146].Davidson B, Holth A, Hellesylt E, Hadar R, Katz B, Trope CG, Reich R, HUR mRNA expression in ovarian high-grade serous carcinoma effusions is associated with poor survival, Hum Pathol, 48 (2016) 95–101. [DOI] [PubMed] [Google Scholar]
- [147].McAllister F, Pineda DM, Jimbo M, Lal S, Burkhart RA, Moughan J, Winter KA, Abdelmohsen K, Gorospe M, Acosta Ade J, Lankapalli RH, Winter JM, Yeo CJ, Witkiewicz AK, Iacobuzio-Donahue CA, Laheru D, Brody JR, dCK expression correlates with 5-fluorouracil efficacy and HuR cytoplasmic expression in pancreatic cancer: a dual-institutional follow-up with the RTOG 9704 trial, Cancer Biol Ther, 15 (2014) 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Liang PI, Li WM, Wang YH, Wu TF, Wu WR, Liao AC, Shen KH, Wei YC, Hsing CH, Shiue YL, Huang HY, Hsu HP, Chen LT, Lin CY, Tai C, Lin CM, Li CF, HuR cytoplasmic expression is associated with increased cyclin A expression and poor outcome with upper urinary tract urothelial carcinoma, BMC Cancer, 12 (2012) 611. [DOI] [PMC free article] [PubMed] [Google Scholar]


