Abstract
Citrullination is a key post-translational modification (PTM) that affects protein structures and functions. Although it has been linked to various biological processes and disease pathogenesis, the underlying mechanism remains poorly understood due to a lack of effective tools to enrich, detect, and localize this PTM. Herein, we report the design and development of a biotin thiol tag that enables derivatization, enrichment, and confident identification of citrullination via mass spectrometry. We perform global mapping of the citrullination proteome of mouse tissues. In total, we identify 691 citrullination sites from 432 proteins which represents the largest dataset to date. We discover novel distribution and functions of this PTM. This study depicts a landscape of protein citrullination and lays the foundation for further deciphering their physiological and pathological roles.
Graphical Abstract
Introduction
Protein citrullination/deimination is an emerging post-translational modification (PTM) resulting from the conversion of peptidyl arginine to citrulline and is catalyzed by a calcium-regulated family of enzymes called protein arginine deiminases (PADs)1–4. This PTM leads to the loss of positive charges on the basic amino acid residues under physiological conditions, and therefore has a profound effect on protein conformations, protein-protein interactions and protein functions1,2,5.
The pathological involvement of this PTM was initially explored in rheumatoid arthritis in which pain in the joints is caused by PAD dysregulation. Proteins with aberrant citrullination also stimulate the generation of anti-citrullinated protein antibodies that are related to atypical autoimmune and inflammatory responses6–13. In another extensively studied disease multiple sclerosis, excessive citrullination of myelin basic protein (MBP) is considered to be a major driver of partial unfolding of myelin sheath and the resultant impaired neuronal signal transduction14–17. Moreover, recent accumulating evidence has revealed that citrullination is associated with the development of diverse pathological states including prion disease18, psoriasis19, Alzheimer’s disease (AD)20–22 and cancers23–28, which raises a fast-growing interest in studying this important PTM.
Despite the emerging interests, knowledge of the citrullination proteome is still limited primarily due to the lack of effective analytical tools. Antibody-based techniques such as Western blotting and immunohistochemistry are currently the most prevalent methods to detect this PTM29–31. However, these approaches are neither suitable for high-throughput analysis nor able to pinpoint exact sites of this PTM with confidence32,33. Mass spectrometry (MS)-based strategies, on the other hand, are gaining popularity as powerful tools for large-scale characterization and localization of various PTM. However, its application to mapping the citrullination proteome suffers from several challenges32–36. Firstly, signals of this low-abundance PTM can be largely suppressed by other molecules in the sample and effective enrichment methods are lacking. Secondly, the small mass shift induced by citrullination (+ 0.984 Da) is easily confused with deamidation (+ 0.984 Da) and 13C isotopic peaks (+ 1.0033 Da). These limitations contribute to the poor-quality tandem MS spectra, which pose challenges for confident identification and localization of this PTM. To combat these issues, significant effort has been devoted to improving aspects of the analytical workflow. However, none of the reported methods have overcome all the difficulties so far. For example, direct MS analysis is possible but often requires high mass accuracy of the instrument and time-consuming manual examination of the spectra13,37,38. Delicate searching algorithms and statistical modeling have also been developed to aid in the direct analysis39–43. Chemical derivatization of the PTM prior to MS analysis is an alternative to enlarge the mass shift but usually suffers from incomplete reaction44–46. The above-mentioned strategies did not address the challenge associated with intrinsic low abundance of this PTM either. Alternative studies have sought the means of using chemical probes for simultaneous introduction of mass shift and enrichment groups47. Nevertheless, previous designs led to unsatisfying fragmentation of the peptide backbones and thus limited the identified citrullination sites8,47,48.
Here, we design a novel biotin thiol tag that enables derivatization and enrichment of citrullinated peptides with high specificity and efficiency. We then develop a reliable and robust proteomics approach for large-scale characterization of this PTM from complex samples. The utility of this pipeline is demonstrated by comprehensive profiling of the landscape of protein citrullination from different mouse tissues.
Experimental Section
Full descriptions about chemicals and materials, synthesis of biotin thiol tag, peptide standard as well as the rest of the procedures can be found in the Supporting Information.
Protein extraction and digestion of mouse tissues.
For method optimization, brain tissue was collected from one mouse. For tissue-specific citrullination profiling, five brain regions and six body organs were collected: Bcortex (cerebral cortex), Scortex (hippocampus and thalamus), hypothalamus, cerebellum, medulla, spleen, pancreas, kidney, lung, heart, and liver. Each tissue was collected as triplicates from three mice. Tissues were dissolved in 150 μL of extraction buffer solution (4 % SDS, 50 mM Tris buffer) and sonicated using a probe sonicator (Thermo). Protein extracts were reduced with 10 mM dithiothreitol (DTT) for 30 min at room temperature and alkylated with 50 mM iodoacetamide for another 30 min in dark before quenched with DTT. Proteins were then precipitated with 80% (v/v) cold acetone (−20 °) overnight. Samples were centrifuged at 14,000 g for 15 min after which supernatant containing SDS (in the extraction buffer) was discarded. Pellets were rinsed with cold acetone again and air-dried at room temperature. Five moles of guanidine hydrochloride (GuHCl) were added to dissolve the pellets and 50 mM Tris buffer was used to dilute the samples to a GuHCl concentration <0.5 M. On-pellet digestion was performed with either trypsin, LysC or LysC/trypsin mixture (Promega) in a 50:1 ratio (protein:enzyme, w/w) at 37 ° overnight. The digestion was quenched with 1% TFA and samples were desalted with Sep-Pak C18 cartridges (Waters). Concentrations of peptide mixture were measured by peptide assay (Thermo). Four hundred microgram of peptide was aliquoted for each sample and dried in vacuo.
Derivatization and enrichment of citrullinated peptides in histone and mouse tissues.
Three hundred microgram of biotin thiol tag was added to each sample tube containing peptides from mouse tissues or histone and resuspended in 40 μL 12.5% TFA solution. Ten microliters of 2,3-butanedione solution prepared as mentioned before was added to initiate the reaction. The rest of derivatization, SCX and enrichment steps were the same as those for citrullinated peptide standard which was indicated in Supporting Information.
LC-MS/MS analysis.
Samples were analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo) coupled to a Dionex UltiMate 3000 UPLC system. Each sample was dissolved in 3% ACN, 0.1% FA in water before loaded onto a 75 μm inner diameter homemade microcapillary column which is packed with 15 cm of Bridged Ethylene Hybrid C18 particles (1.7 μm, 130 Å, Waters) and fabricated with an integrated emitter tip. Mobile phase A was composed of water and 0.1% FA while mobile phase B was composed of ACN and 0.1% FA. LC separation was achieved across a 100-min gradient elution of 3% to 30% mobile phase B at a flow rate of 300 nL/min. Survey scans of peptide precursors from 350 to 1500 m/z were performed at a resolving power of 60k (at m/z 200) with an AGC target of 2 × 105 and maximum injection time of 100 ms. For histone samples, survey scans of peptide precursors were performed from 200 to 1500 m/z.
Data analysis.
Raw files were searched against the UniProt Mus musculus reviewed database (December 2018) using MaxQuant (version 1.5.2.8) with trypsin/P selected as the enzyme and three missed cleavages allowed. Histone data files were searched against the UniProt Homo sapiens reviewed database (February 2020). Carbamidomethylation of cysteine residues (+57.02146 Da) were chosen as fixed modifications and variable modifications included oxidation of methionine residues (+15.99492 Da), biotin tag-labeled citrullination of arginine (+354.10718 Da). A neutral loss of biotin tag (303.10752 Da) and two diagnostic ions of 227.08487 Da and 304.11479 Da were included in the search. Search results were filtered to 1% false discovery rate (FDR) at both peptide and protein levels. Peptides that were found as reverse or potential contaminant hits were filtered out and citrullination site localization probability threshold was set to 0.75. All other parameters were set as default. Bioinformatic analyses including Sankey diagram, arcplots and stacked bar graphs were performed using R packages. Heatmaps showing multi-tissue GO analyses were generated using Metascape49 (version 3.5). Sequence motif analyses were done using WebLogo50. For homology modeling, the 3D structure of mouse PKM2 (residues 14–531) was modeled according to the crystal structures of human PKM2, which delivered a sequence identity of 97.7% and represented the most similar crystal structures to mouse PKM2 retrievable from the Protein Data Bank. The homology model module of Discovery Studio 2016 was used for the multi-templates structure construction, and the ligands including PYR, SER, FBP were copied from the input templates. The output model with the lowest PDF total energy and DOPE score was adopted, and energy minimization was conducted on the adopted structure using CHARMm (version 40.1). PyMOL (version 2.4.0a0) was used to measure the Euclidean distances between the atoms of the selected arginine residues and atoms of annotated ligands (such as substrates and allosteric activator) in Å.
Data availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange51 Consortium via the PRIDE52 partner repository with the dataset identifier PXD023733. Public release of the data will be made on time for online publication of the paper. For anonymous access to peer-reviewers please use the following account information: Username: reviewer_pxd023733@ebi.ac.uk, Password: nGAPJjrm (https://www.ebi.ac.uk/pride/login). Mus musculus and Homo sapiens databases used for database searching were downloaded from UniProt (https://www.uniprot.org/).
Results and Discussion
Development of a novel biotin thiol tag for citrullination analysis.
Protein citrullination features a ureido group on the side chains that can be used for chemical derivatization as previously reported8,29,46–48. Here, we design a biotin thiol tag that can be easily synthesized with low cost (Figure S1) and can specifically react with citrulline residues together with 2,3-butanedione (Figure S1a). This derivatization not only increases the mass shift to allow more confident identification, but also introduces a biotin moiety that enables subsequent enrichment of the modified molecules.
We first performed a proof-of-principle test using a synthetic peptide standard containing one citrullination site and two arginine residues within the sequence (SAVRACitSSVPGVR) (Figure S2a). After 6 h, the reaction was complete without any observable side products (Figure S2b), suggesting high specificity towards ureido group. The low-abundance peak at m/z 1392 corresponds to the loss of biotin moiety caused by in-source fragmentation when using a matrix-assisted laser desorption/ionization (MALDI) source. We then tested an identical peptide containing an arginine at the modified position (SAVRARSSVPGVR) (Figure S2c) and no interfering reaction was observed on any arginine residues after incubation with biotin thiol tag (Figure S2d). To further verify the specificity and efficiency of this reaction, we repeated the experiment on six other peptide standards with varying lengths and sequences. The results proved that our biotin thiol tag-enabled reaction is highly selective towards only ureido groups on citrulline instead of other amino acid residues (Figure S3), which lays the foundation for further applications. The six peptide standard mixtures were also used as a sanity check to verify the specificity and efficiency of this reaction which again suggests its good specificity and efficiency as expected. Moreover, the reaction with multiple citrulline residues in the proximity also exhibited very good efficiency though with more complex in-source fragmentation pattern (Figure S5). We then evaluated the enrichment performance by spiking the derivatized peptide standard (SAVRACitSSVPGVR) into a complex peptide mixture at two different ratios (1:400 and 1:4000, w/w) (Figure S6a, S7a) followed by enrichment with streptavidin beads (Figure S6b, S7b). The results indicate that derivatized citrullinated peptides can be enriched with excellent specificity and released from streptavidin beads for MS analysis. The peak at m/z 1392 is still present after enrichment which further proves that it originates from in-source fragmentation instead of incomplete derivatization.
Previously reported chemical probes for citrullination analysis had bulky structures that negatively impacted the solubility of analytes. Upon derivatization, extensive yet uninformative fragments were generated from the tag, which severely impeded the peptide backbone fragmentation and therefore led to lower identification rates8. In contrast, our novel design of biotin thiol tag features a compact structure which only generates two fragment/diagnostic ions during higher-energy collisional dissociation (HCD) (Figure 1b and Figure S8a–c). Consequently, peptide backbones can preserve good fragmentation efficiency and produce rich b/y or c/z ion series during HCD or electron-transfer dissociation (ETD) (Figure S8a, d–g), respectively. The collected tandem MS spectra of the derivatized peptide standard delivered nearly full sequence coverage under HCD (Figure 1c), ETD (Figure S8h) or electron-transfer/higher-energy collision dissociation (EThcD) (Figure S8i) fragmentation. Our results indicate that the biotin thiol tag derivatized citrullinated peptides can generate high-quality tandem MS spectra for sequence annotation, which enhances the identification confidence of citrullination sites when coupled with various fragmentation techniques.
Figure 1. Design of biotin thiol tag for citrullination analysis.
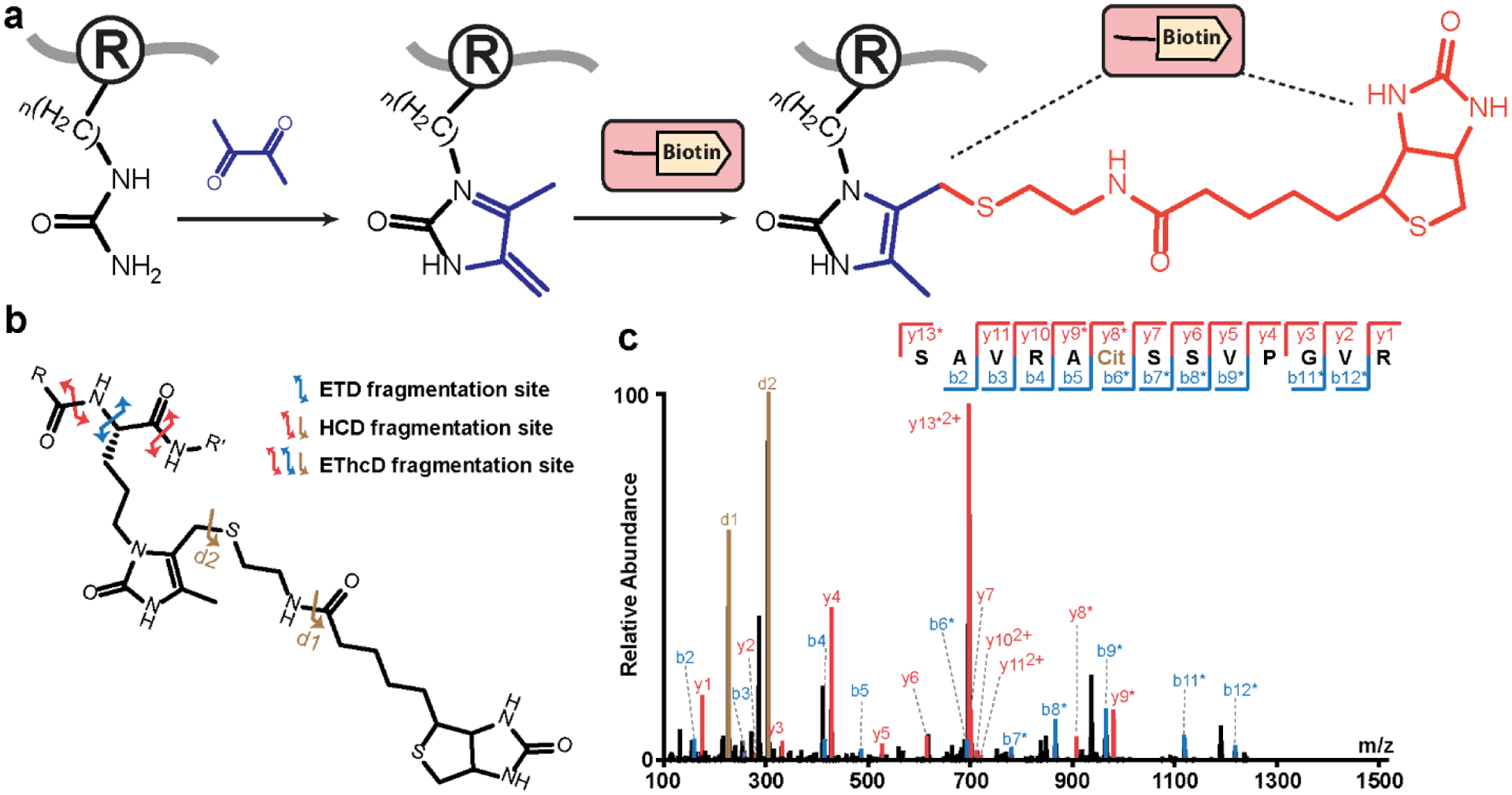
a, Derivatization of citrullinated peptides using biotin thiol tag and 2,3-butanedione. b, Fragmentation sites of biotin thiol tag-derivatized citrullinated peptides upon HCD, ETD, or EThcD fragmentations. c, Tandem MS spectrum of the biotin thiol tag-derivatized citrullinated peptide standard SAVRACitSSVPGVR upon HCD fragmentation.
Improved in vitro protein citrullination analysis with biotin thiol tag.
Following the initial experiments, we streamlined the citrullination analysis using our biotin thiol tag and MS-based bottom-up proteomics approach (Figure 2a). Proteins were extracted from biological samples and enzymatically digested to peptides. The biotin tag was incubated with the peptides under acidic conditions and reacted with citrulline residues. Excess tag was removed by strong cation exchange (SCX), and derivatized citrullinated peptides were enriched by streptavidin resin. The enriched peptides were then released for liquid chromatography coupled with tandem MS (LC-MS/MS) analysis and data processing. We tested this procedure using recombinant human histone H3 protein with or without in vitro PAD treatment and LysC was used during the digestion step. As a negative control, non-treated histone protein resulted in no peptides being identified as citrullinated, demonstrating again the high specificity of the biotin tag reaction (Figure 2b). After in vitro PAD treatment, we found many arginine residues were catalyzed to citrulline with abundant peptides confidently identified as citrullinated (Figure 2c), indicating the high efficacy of our method. A rich series of b/y ions were generated upon HCD fragmentation, allowing for unambiguous characterization of both singly (Figure 2d) and multiply (Figure 2e) citrullinated peptides.
Figure 2. Improved in vitro protein citrullination analysis with biotin thiol tag.
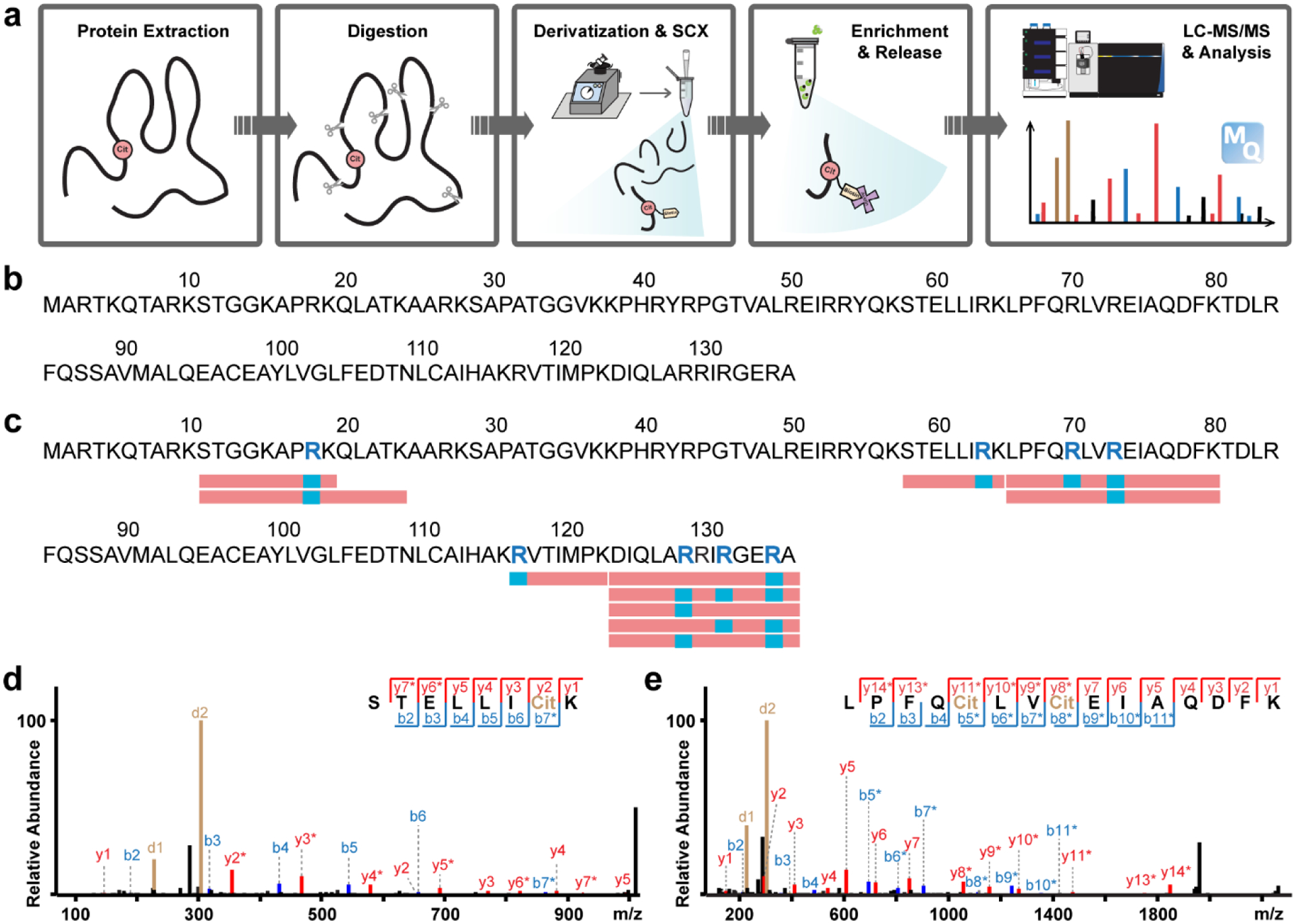
a, Experimental workflow of protein citrullination analysis with biotin thiol tag. b,c, Citrullination analysis of histone H3 protein before (b) and after (c) in vitro PAD treatment. Identified citrullination sites are highlighted as blue letters within the sequence. Red rectangles below the sequence indicate confidently identified citrullinated peptides while citrullination sites are shown in blue. d, Example tandem MS spectrum of an identified citrullinated peptide from PAD treated histone H3 (R64Cit). e, Example tandem MS spectrum of two citrullination sites (R70Cit and R73Cit) identified on the same peptide from PAD treated histone H3.
Exploring different fragmentation techniques and enzymatic digestion methods for optimized citrullination analysis from complex biological samples.
We moved forward to evaluate our method with complex biological samples. We first compared three MS fragmentation methods, including stepped HCD, HCD product ion-triggered ETD (HCD-pd-ETD) and HCD product ion-triggered EThcD (HCD-pd-EThcD), using mouse brain digest. All three methods were able to achieve in-depth citrullination analysis with decent numbers of identifications (Figure S9a) while stepped HCD method slightly outperformed the other two, likely due to shorter cycle time. Different methods show certain overlaps but are also complementary to one another (Figure S9b, c), suggesting the importance of choosing an appropriate one depending on specific applications. When comparing the same citrullination site identified with various fragmentation techniques, we observed that they all produced high-quality spectra though EThcD showed even better sequence coverage as expected (Figure S9d–f). Thus, we conclude that stepped HCD confers optimal performance for citrullination analysis of complex samples due to its faster acquisition rate and shorter duty cycle while EThcD shines in providing more informative fragment ions and hence is more beneficial for in-depth characterization of citrullinated peptides in relatively simple systems.
We then sought to optimize the enzymatic digestion methods to see if various enzymes will significantly improve the total citrullination identification numbers and may also suggest possible direction of choosing an appropriate one depending on specific targets. Lower identification numbers were observed when only using LysC to digest the samples (Figure S10a) probably because LysC digestion produces longer peptides, which results in lower fragmentation efficiency. Though trypsin digestion produced higher identification rate, different procedures generated complementary results (Figure S10b), suggesting the potential benefits to use a combination of multiple digestion methods. Interestingly, some peptides were identified with citrullination sites located at peptide C-termini, indicating that citrulline residues could potentially be cleaved by trypsin. When searching the results of LysC digestion with tryptic peptide parameters, we found almost all of the citrullination sites were still identified in the middle of the peptide sequence (Figure S10c), which demonstrated that no artificial cleavage of citrulline residues happened after enzymatic digestion. Some citrullination sites were confidently identified with different digestion protocols (Figure S10d–f), which further supported our observations of trypsin cleavable C-terminal citrullinated arginine. However, it remains controversial whether trypsin is able to cleave after citrulline residues. While some researchers believe citrulline is resistant to trypsin digestion due to its neutral-charge property and even use it as a rule to exclude their identifications37,53, others have reported the presence of numerous C-terminal citrullination sites though manual inspection of the spectra is usually required23,40,54. These authors also suggested that the cleavage might be protein-specific and/or at a lower rate35,54. Although some previous studies as well as ours provided evidence to support this observation, we believe that future investigations into the mechanisms are highly desirable and essential before including the peptide C-terminal citrullines as true hits, especially considering that Bennike et al. have reported the inability of trypsin to cleave citrulline in vitro using synthetic peptides53.
Taking consideration of citrullination identification rate and economic cost, we determined that using LysC/trypsin digestion and stepped HCD fragmentation technique would be the optimal strategy for interrogating the citrullination proteome. For our later applications, we remove the C-terminal citrullination identifications to ensure our results are as confident and reliable as possible. One can also utilize LysC digestion to avoid the dilemma but with a cost of lower identification rate and higher experimental expense. In addition, we evaluated the reproducibility by analyzing three biological replicates and the good overlap among these replicates indicates the reliability and robustness of our optimized methods (Figure S11).
Large-scale citrullinome profiling of different mouse tissues.
Next, we ask whether the developed method can delineate the citrullination landscape from biological samples and offers potential to elucidate the regulatory mechanisms of citrullination in cells. We performed an in-depth citrullinome analysis of six body organs and five brain regions in mice, generating a first tissue-specific atlas of mouse citrullinome. In total, we identified 691 citrullination sites from 432 citrullinated proteins with high confidence (Figure 3a and Table S1). More importantly, about 60% of these proteins were not reported to be proteins with PTMs retrievable from the UniProt database (Figure S12), which suggests that our results greatly expand the knowledge and understanding of citrullination and how these substrate proteins are subjected to modulation via PTM. We then compared the citrullination sites identified in our study with a recent study by Fert-Bober et al. who mapped citrullination sites in multiple mouse organs using hypercitrullinated library generated by in vitro citrullination experiment (Figure S13a and Table S2). About 24% of in vivo citrullination sites identified in our study were overlapped with the hypercitrullinated library reported by Fert-Bober et al. This result confirms the validity of our method. We also aligned the citrullinated proteins identified in two studies from mice with another deep citrullinome analysis reported by Lee, C.Y., et al. who mined MS-based proteomics data of 30 human tissues to identify citrullination sites (Figure S13b and Table S2). Because the PAD enzyme could non-selectively catalyze arginine to citrulline in vitro to generate hyper-citrullinated spectral library, it is expected that only a small percentage of the citrullination sites would overlap with the in vivo citrullination sites from mouse samples. Similarly, 28% of citrullinated proteins identified in Lee et al., were also observed in our study. The uniquely identified in vivo citrullination sites and citrullinated proteins in our study further highlight the advantage of our approach with enrichment and high-confidence identification, revealing novel citrullination sites and citrullinated proteins with deeper coverage.
Figure 3. Large-scale citrullinome profiling of different mouse tissues.
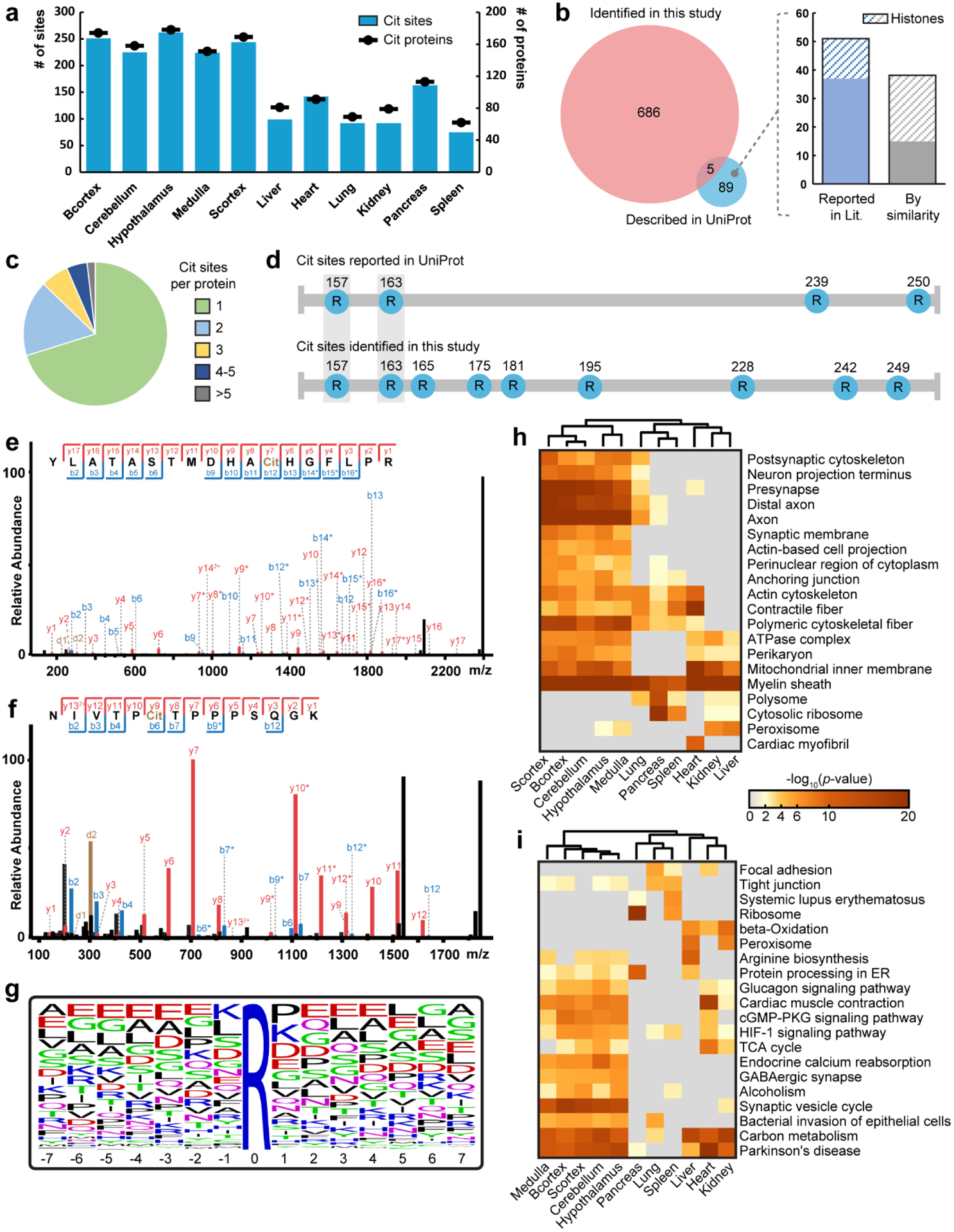
a, Number of identified citrullinated proteins and citrullination sites in different mouse tissues. Identification numbers are presented as a total from three mice. Cit, citrullination; Bcortex, cerebral cortex; Scortex, hippocampus and thalamus. b, Overlap of citrullination sites identified in this study with those reported in the UniProt database. Many of the sites only reported in UniProt have that description based on similarity prediction or location on histone proteins. c, Distribution of the number of citrullination sites per citrullinated proteins identified. d, Comparison of citrullination sites identified in this study and those reported in the UniProt database on myelin basic protein. e,f, Example tandem MS spectra of two citrullination sites identified on myelin basic protein, R157Cit (e) and R228Cit (f). g, Sequence motif of identified citrullinated peptides. Citrullination sites are centered in the middle as “0” position. The height of letters indicates the relative frequency of each amino acid at certain positions. h,i, Heatmaps generated using Metascape showing the significantly enriched (p value < 0.01 in at least one tissue region) cellular components (h) and Kyoto encyclopedia of genes and genomes (KEGG) pathways (i) in different mouse tissues. The most significant 20 terms are shown in each heatmap. Color coding indicates −log10 (p values). Columns are clustered based on their profile similarity.
Intriguingly, we found each examined brain region doubles in the number of identifications compared to other organs (Figure 3a); however, the total number of citrullinated proteins in the brain is lower than that in the body (Figure S12). To investigate the seemingly contradictory results, we generated two arcplots where the width of ribbons connecting two tissues is proportional to the number of overlapping proteins or sites between them (Figure S14a, b). We observed a higher degree of overlap between brain regions with many more shared proteins and sites in between (Figure S14c–f). This could indicate protein citrullination functions importantly and similarly across multiple brain regions, while in body organs it is involved in diverse biological processes. Our results greatly expand the knowledge of the substrate proteome for citrullination although the overlapped fraction with UniProt repository is negligible (Figure 3b). This is likely because nearly 40% of the citrullination sites described in UniProt are based on similarity extrapolation without experimental evidence which are inconsistent with the identified in vivo citrullination proteome. In addition, many of those reported sites are located on histone proteins especially at protein termini that may escape detection with our bottom-up strategies (Figure 3b). Figure 3c captures the prevalence of singly- and multiply-citrullinated proteins where 70% of the identified proteins were observed with only one citrullination site.
The newly discovered citrullination proteome provides a valuable resource to conjecture the regulatory mechanisms of this PTM. For instance, we identified nine citrullination sites on MBP while there are only four reported in the UniProt database (Figure 3d). Our results provided high-quality tandem MS spectra, which not only confirmed the presence of known modification sites (Figure 3e), but also identified unknown sites with confidence (Figure 3f). These findings may partially explain why MBP is more susceptible to hypercitrullination when PADs are dysregulated under pathological conditions and thus can help better understand the underlying mechanisms in related diseases. Two citrullination sites described in UniProt were not detected in our study which could result from the complementarity of various analytical tools. But again, these sites from UniProt are all based on similarity extrapolation from human and our results might indeed indicate a species-specific molecular profile of protein citrullination. Another interesting example is glial fibrillary acidic protein (GFAP), which is an astrocyte-specific protein marker and is involved in astrocyte-neuron interactions. Increased expression of citrullinated GFAP was also observed in brains from patients with AD20,22. In this study, we identified nine citrullination sites on GFAP compared to four described in the UniProt database (Figure S15), which underscored the importance of citrullination in regulating GFAP functions and understanding the pathology of AD and possible other astrocyte disorders. In addition, we detected many novel citrullinated proteins for the first time. For example, we identified one citrullination site on apolipoprotein E (Figure S16a) and microtubule-associated protein tau (Figure S16b). These two proteins have been proven to be closely associated with the initiation and progression of AD55–57 and our results suggest the possible roles of their citrullinated forms in the pathogenesis of these neurodegenerative diseases.
We then performed a motif analysis and found there was no strongly conserved amino acid sequence patterns flanking identified citrullination sites (Figure 3g), though glutamic acid residues are present more frequently in the proximity especially on the N-terminal side, which is consistent with the mouse citrullinome analysis reported by Fert-Bober et al.39. On the other hand, Tanikawa et al. found that approximately one-fifth of the PAD4 substrates contained an RG/RGG motif in human tissue and plasma58. Similarly, Lee et al. and other researchers observed that aspartate and glycine residues were overrepresented at the +1 position37,59,60. Notably, these latter studies also used human cell lines or tissues which might lead to discrepancies, and our results could indicate the sequence patterns surrounding citrullination sites are different among species. To better discern the general functions that citrullinated proteins are involved in, we generated heatmaps showing multi-organ gene ontology (GO) analyses. Twenty most significantly enriched cellular components (Figure 3h) or Kyoto encyclopedia of genes and genomes (KEGG) pathways (Figure 3i) are shown where the color coding indicates the p values of a certain term in different tissues. We found that there are clear disparities between brain and body while citrullinated proteins are more involved in brain functions. Specifically, citrullinated proteins are concentrated in axon, myelin sheath, perikaryon and synapse, and consequently function importantly in the central nervous system. Furthermore, they also participate in many critical metabolic processes including respiration and are observed to be enriched in mitochondria. In accordance with this, we also identified four citrullination sites on an essential glycolytic enzyme pyruvate kinase (PKM). Interestingly, many of these sites on PKM are located at critical positions (Figure S17a), which raises the likelihood that citrullination can influence the kinase activity and supports a recent study concluding that citrullination regulates glycolysis61. For instance, R455 is located in the proximity of one allosteric center (Figure S17b), which was known to regulate enzyme activity62. Another position R399 which we found to be citrullinated was also shown to be very important in stabilizing the highly active tetrameric form (Figure S17c, d)62,63. Our results greatly expand current understandings of protein citrullination by demonstrating its widespread distribution (Figure S18) and involvement in many other biological processes (Figure S19), molecular functions (Figure S20) and KEGG pathways (Figure S21).
Additionally, we noticed that 24 citrullination sites are colocalized with other arginine modifications especially omega-n-methylarginine (Figure S22a). For example, we identified five citrullination sites on heterogeneous nuclear ribonucleoproteins A2/B1 (Hnrnpa2b1) and four of them were also reported as arginine methylation sites (Figure S22b). Hnrnpa2b1 was shown to influence RNA metabolism and transport, and arginine methylation could regulate the nucleocytoplasmic distribution of this protein64. Our results raise the possibility that citrullination indirectly participates in biological processes through an interplay with other protein modifications such as arginine methylation. Future studies will examine potential PTM crosstalk involving citrullination and their role in protein function and biological processes.
Conclusions
Herein, we report the design and development of a biotin thiol tag that specifically reacts with citrulline and allows for enrichment of target molecules. After demonstrating its efficacy using standard peptide and recombinant protein, we streamline the workflow to detect citrullination from complex biological samples. We then apply this protocol to profile protein citrullination of five brain regions and six body organs in mice. In total, we identify 691 citrullination sites from 432 proteins, which is the largest dataset to date. Our study reveals the critical roles of this PTM which may play in the nervous system and indicate they also have important functions in many metabolic processes including respiration and glycolysis. Collectively, our results expand the current understanding of protein citrullination by mapping their widespread distribution in different tissues and participation in various biological processes than hitherto anticipated. More importantly, we envision our method can provide a simple yet powerful tool for unambiguous identification of this intriguing modification, which will also inspire and benefit future investigations into its functional role under physiological and pathological conditions.
Supplementary Material
Acknowledgements
This study was supported in part by grant funding from the NIH (R21AG060242, U01CA231081, RF1AG052324, R21AG065728, and R01DK071801). The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. The MALDI TOF/TOF RapifleX mass spectrometer was purchased through the support of an NIH shared instrument grant S10OD025084. L.L. acknowledges a Vilas Distinguished Achievement Professorship and the Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Supporting information
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Additional experimental details, materials, and methods, including synthesis of biotin thiol tag, derivatization of citrullinated peptide standard using biotin thiol tag, enrichment of derivatized citrullinated peptide standard, MALDI-MS analysis of citrullinated peptide standard and its derivatized form, fragmentation of derivatized citrullinated peptide standard, PAD treatment and digestion of histone H3, fragmentation parameters. Synthesis and characterization of biotin thiol tag. Proof-of-principle test of biotin thiol tag using citrullinated peptide standard. Verification of reaction specificity and efficiency of biotin thiol tag. Reaction of multiple citrulline residues in the proximity. Evaluation of enrichment performance using citrullinated peptide standard. Fragmentation behavior of biotin thiol tag-derivatized citrullinated peptides. Optimization of different fragmentation methods in detecting protein citrullination. Optimization of different enzymatic digestion methods in detecting protein citrullination. Evaluation of method reproducibility in citrullination analysis. Tissue-specific distribution of identified citrullinated proteins in this study. Comparison of citrullinome in this study with previous large-scale citrullinome studies. Overlap of identified citrullinated proteins and citrullination sites in different tissues. Examples of novel citrullination sites identified on glial fibrillary acidic protein in this study. Examples of novel citrullinated proteins identified in this study. Homology modeling of mouse pyruvate kinase isozymes M2 (PKM2). Enrichment of cellular component in different mouse tissues. Enrichment of biological processes in different mouse tissues. Enrichment of molecular function in different mouse tissues. Enrichment of KEGG pathway in different mouse tissues. Colocalization of citrullination with arginine methylation. List of all identified citrullinated proteins and sites from five brain regions and six body organs in mice. Comparison of citrullinome in this study with previous large-scale citrullinome studies.
The authors declare the following competing financial interests: The University of Wisconsin–Madison is in the process of filing a patent application P190302US01 based on this work. L.L., Y.S. and Z.L. are named as inventors on this provisional patent application.
References
- (1).Fuhrmann J; Clancy KW; Thompson PR Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev 2015, 115 (11), 5413–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Fuhrmann J; Thompson PR Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem Biol 2016, 11 (3), 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Witalison E; Thompson P; Hofseth L Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Current Drug Targets 2015, 16 (7), 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mondal S; Thompson PR Protein Arginine Deiminases (PADs): Biochemistry and Chemical Biology of Protein Citrullination. Acc Chem Res 2019, 52 (3), 818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gyorgy B; Toth E; Tarcsa E; Falus A; Buzas EI Citrullination: a posttranslational modification in health and disease. The international journal of biochemistry & cell biology 2006, 38 (10), 1662–1677. [DOI] [PubMed] [Google Scholar]
- (6).Schellekens GA; de Jong BA; van den Hoogen FH; van de Putte LB; van Venrooij WJ Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest 1998, 101 (1), 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Elkon KB Poking holes in rheumatoid joints. Sci Transl Med 2013, 5 (209), 209fs239. [DOI] [PubMed] [Google Scholar]
- (8).Tutturen AE; Fleckenstein B; de Souza GA Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res 2014, 13 (6), 2867–2873. [DOI] [PubMed] [Google Scholar]
- (9).Pruijn GJ Citrullination and carbamylation in the pathophysiology of rheumatoid arthritis. Front Immunol 2015, 6, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Turunen S; Huhtakangas J; Nousiainen T; Valkealahti M; Melkko J; Risteli J; Lehenkari P Rheumatoid arthritis antigens homocitrulline and citrulline are generated by local myeloperoxidase and peptidyl arginine deiminases 2, 3 and 4 in rheumatoid nodule and synovial tissue. Arthritis Res Ther 2016, 18 (1), 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tilvawala R; Nguyen SH; Maurais AJ; Nemmara VV; Nagar M; Salinger AJ; Nagpal S; Weerapana E; Thompson PR The Rheumatoid Arthritis-Associated Citrullinome. Cell Chem Biol 2018, 25 (6), 691–704 e696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fert-Bober J; Darrah E; Andrade F Insights into the study and origin of the citrullinome in rheumatoid arthritis. Immunol Rev 2020, 294 (1), 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Raijmakers R; van Beers JJ; El-Azzouny M; Visser NF; Bozic B; Pruijn GJ; Heck AJ Elevated levels of fibrinogen-derived endogenous citrullinated peptides in synovial fluid of rheumatoid arthritis patients. Arthritis Res Ther 2012, 14 (3), R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Moscarello MA; Mastronardi FG; Wood DD The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem Res 2007, 32 (2), 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Bradford CM; Ramos I; Cross AK; Haddock G; McQuaid S; Nicholas AP; Woodroofe MN Localisation of citrullinated proteins in normal appearing white matter and lesions in the central nervous system in multiple sclerosis. J Neuroimmunol 2014, 273 (1–2), 85–95. [DOI] [PubMed] [Google Scholar]
- (16).Yang L; Tan D; Piao H Myelin Basic Protein Citrullination in Multiple Sclerosis: A Potential Therapeutic Target for the Pathology. Neurochem Res 2016, 41 (8), 1845–1856. [DOI] [PubMed] [Google Scholar]
- (17).Gs Chirivi R Citrullination: A Target for Disease Intervention in Multiple Sclerosis and other Inflammatory Diseases? Journal of Clinical & Cellular Immunology 2013, 04 (03), 1000146. [Google Scholar]
- (18).Jang B; Kim E; Choi JK; Jin JK; Kim JI; Ishigami A; Maruyama N; Carp RI; Kim YS; Choi EK Accumulation of citrullinated proteins by up-regulated peptidylarginine deiminase 2 in brains of scrapie-infected mice: a possible role in pathogenesis. The American journal of pathology 2008, 173 (4), 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ishida-Yamamoto A; Senshu T; Takahashi H; Akiyama K; Nomura K; Iizuka H Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Invest Dermatol 2000, 114 (4), 701–705. [DOI] [PubMed] [Google Scholar]
- (20).Ishigami A; Ohsawa T; Hiratsuka M; Taguchi H; Kobayashi S; Saito Y; Murayama S; Asaga H; Toda T; Kimura N; Maruyama N Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer’s disease. J Neurosci Res 2005, 80 (1), 120–128. [DOI] [PubMed] [Google Scholar]
- (21).Acharya NK; Nagele EP; Han M; Coretti NJ; DeMarshall C; Kosciuk MC; Boulos PA; Nagele RG Neuronal PAD4 expression and protein citrullination: possible role in production of autoantibodies associated with neurodegenerative disease. J Autoimmun 2012, 38 (4), 369–380. [DOI] [PubMed] [Google Scholar]
- (22).Ishigami A; Masutomi H; Handa S; Nakamura M; Nakaya S; Uchida Y; Saito Y; Murayama S; Jang B; Jeon YC; Choi EK; Kim YS; Kasahara Y; Maruyama N; Toda T Mass spectrometric identification of citrullination sites and immunohistochemical detection of citrullinated glial fibrillary acidic protein in Alzheimer’s disease brains. J Neurosci Res 2015, 93 (11), 1664–1674. [DOI] [PubMed] [Google Scholar]
- (23).Yuzhalin AE; Gordon-Weeks AN; Tognoli ML; Jones K; Markelc B; Konietzny R; Fischer R; Muth A; O’Neill E; Thompson PR; Venables PJ; Kessler BM; Lim SY; Muschel RJ Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat Commun 2018, 9 (1), 4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yuzhalin AE Citrullination in Cancer. Cancer Res 2019, 79 (7), 1274–1284. [DOI] [PubMed] [Google Scholar]
- (25).Stadler SC; Vincent CT; Fedorov VD; Patsialou A; Cherrington BD; Wakshlag JJ; Mohanan S; Zee BM; Zhang X; Garcia BA; Condeelis JS; Brown AM; Coonrod SA; Allis CD Dysregulation of PAD4-mediated citrullination of nuclear GSK3beta activates TGF-beta signaling and induces epithelial-to-mesenchymal transition in breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America 2013, 110 (29), 11851–11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chang X; Han J; Pang L; Zhao Y; Yang Y; Shen Z Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 2009, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang L; Song G; Zhang X; Feng T; Pan J; Chen W; Yang M; Bai X; Pang Y; Yu J; Han J; Han B PADI2-Mediated Citrullination Promotes Prostate Cancer Progression. Cancer Res 2017, 77 (21), 5755–5768. [DOI] [PubMed] [Google Scholar]
- (28).Thalin C; Lundstrom S; Seignez C; Daleskog M; Lundstrom A; Henriksson P; Helleday T; Phillipson M; Wallen H; Demers M Citrullinated histone H3 as a novel prognostic blood marker in patients with advanced cancer. PLoS One 2018, 13 (1), e0191231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Senshu T; Sato T; Inoue T; Akiyama K; Asaga H Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal. Biochem 1992, 203 (1), 94–100. [DOI] [PubMed] [Google Scholar]
- (30).Nicholas AP; King JL; Sambandam T; Echols JD; Gupta KB; McInnis C; Whitaker JN Immunohistochemical localization of citrullinated proteins in adult rat brain. The Journal of comparative neurology 2003, 459 (3), 251–266. [DOI] [PubMed] [Google Scholar]
- (31).Moelants EA; Van Damme J; Proost P Detection and quantification of citrullinated chemokines. PLoS One 2011, 6 (12), e28976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Verheul MK; van Veelen PA; van Delft MAM; de Ru A; Janssen GMC; Rispens T; Toes REM; Trouw LA Pitfalls in the detection of citrullination and carbamylation. Autoimmun Rev 2018, 17 (2), 136–141. [DOI] [PubMed] [Google Scholar]
- (33).Hensen SM; Pruijn GJ Methods for the detection of peptidylarginine deiminase (PAD) activity and protein citrullination. Mol Cell Proteomics 2014, 13 (2), 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Clancy KW; Weerapana E; Thompson PR Detection and identification of protein citrullination in complex biological systems. Curr Opin Chem Biol 2016, 30, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Vitorino R; Guedes S; Vitorino C; Ferreira R; Amado F; Van Eyk JE Elucidating Citrullination by Mass Spectrometry and Its Role in Disease Pathogenesis. J Proteome Res 2021, 20, (1), 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).De Ceuleneer M; Van Steendam K; Dhaenens M; Deforce D In vivo relevance of citrullinated proteins and the challenges in their detection. Proteomics 2012, 12 (6), 752–760. [DOI] [PubMed] [Google Scholar]
- (37).Lee CY; Wang D; Wilhelm M; Zolg DP; Schmidt T; Schnatbaum K; Reimer U; Ponten F; Uhlen M; Hahne H; Kuster B Mining the Human Tissue Proteome for Protein Citrullination. Mol Cell Proteomics 2018, 17 (7), 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Stobernack T; Glasner C; Junker S; Gabarrini G; de Smit M; de Jong A; Otto A; Becher D; van Winkelhoff AJ; van Dijl JM Extracellular Proteome and Citrullinome of the Oral Pathogen Porphyromonas gingivalis. Journal of proteome research 2016, 15 (12), 4532–4543. [DOI] [PubMed] [Google Scholar]
- (39).Fert-Bober J; Venkatraman V; Hunter CL; Liu R; Crowgey EL; Pandey R; Holewinski RJ; Stotland A; Berman BP; Van Eyk JE Mapping Citrullinated Sites in Multiple Organs of Mice Using Hypercitrullinated Library. J Proteome Res 2019, 18, (5), 2270–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wang X; Swensen AC; Zhang T; Piehowski PD; Gaffrey MJ; Monroe ME; Zhu Y; Dong H; Qian WJ Accurate Identification of Deamidation and Citrullination from Global Shotgun Proteomics Data Using a Dual-Search Delta Score Strategy. J Proteome Res 2020, 19 (4), 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Villacres C; Spicer V; Krokhin OV Confident Identification of Citrullination and Carbamylation Assisted by Peptide Retention Time Prediction. J Proteome Res 2021, 20, (3), 1571–1581. [DOI] [PubMed] [Google Scholar]
- (42).Huh S; Hwang D; Kim MS Statistical Modeling for Enhancing the Discovery Power of Citrullination from Tandem Mass Spectrometry Data. Anal Chem 2020, 92 (19), 12975–12986. [DOI] [PubMed] [Google Scholar]
- (43).De Ceuleneer M; Van Steendam K; Dhaenens M; Elewaut D; Deforce D Quantification of citrullination by means of skewed isotope distribution pattern. J Proteome Res 2012, 11 (11), 5245–5251. [DOI] [PubMed] [Google Scholar]
- (44).Stensland M; Holm A; Kiehne A; Fleckenstein B Targeted analysis of protein citrullination using chemical modification and tandem mass spectrometry. Rapid Commun Mass Spectrom 2009, 23 (17), 2754–2762. [DOI] [PubMed] [Google Scholar]
- (45).De Ceuleneer M; De Wit V; Van Steendam K; Van Nieuwerburgh F; Tilleman K; Deforce D Modification of citrulline residues with 2,3-butanedione facilitates their detection by liquid chromatography/mass spectrometry. Rapid Commun Mass Spectrom 2011, 25 (11), 1536–1542. [DOI] [PubMed] [Google Scholar]
- (46).Choi M; Song JS; Kim HJ; Cha S; Lee EY Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of peptide citrullination site using Br signature. Analytical biochemistry 2013, 437 (1), 62–67. [DOI] [PubMed] [Google Scholar]
- (47).Lewallen DM; Bicker KL; Subramanian V; Clancy KW; Slade DJ; Martell J; Dreyton CJ; Sokolove J; Weerapana E; Thompson PR Chemical Proteomic Platform To Identify Citrullinated Proteins. ACS Chem Biol 2015, 10 (11), 2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Tutturen AE; Holm A; Fleckenstein B Specific biotinylation and sensitive enrichment of citrullinated peptides. Anal Bioanal Chem 2013, 405 (29), 9321–9331. [DOI] [PubMed] [Google Scholar]
- (49).Zhou Y; Zhou B; Pache L; Chang M; Khodabakhshi AH; Tanaseichuk O; Benner C; Chanda SK Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 2019, 10 (1), 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Crooks GE; Hon G; Chandonia JM; Brenner SE WebLogo: a sequence logo generator. Genome Res 2004, 14 (6), 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Deutsch EW; et al. The ProteomeXchange consortium in 2020: enabling ‘big data’ approaches in proteomics. Nucleic acids research 2020, 48 (D1), D1145–D1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Perez-Riverol Y; et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic acids research 2019, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Bennike T; Lauridsen KB; Olesen MK; Andersen V; Birkelund S; Stensballe A Optimizing the Identification of Citrullinated Peptides by Mass Spectrometry: Utilizing the Inability of Trypsin to Cleave after Citrullinated Amino Acids. Journal of Proteomics & Bioinformatics 2013, 6 (12), 1000293. [Google Scholar]
- (54).Jin Z; Fu Z; Yang J; Troncosco J; Everett AD; Van Eyk JE Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics 2013, 13 (17), 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Yamazaki Y; Zhao N; Caulfield TR; Liu C-C; Bu G Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nature Reviews Neurology 2019, 15 (9), 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Liu CC; Liu CC; Kanekiyo T; Xu H; Bu G Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 2013, 9 (2), 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Iqbal K; Liu F; Gong CX; Grundke-Iqbal I Tau in Alzheimer disease and related tauopathies. Curr Alzheimer Res 2010, 7 (8), 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tanikawa C; Ueda K; Suzuki A; Iida A; Nakamura R; Atsuta N; Tohnai G; Sobue G; Saichi N; Momozawa Y; Kamatani Y; Kubo M; Yamamoto K; Nakamura Y; Matsuda K Citrullination of RGG Motifs in FET Proteins by PAD4 Regulates Protein Aggregation and ALS Susceptibility. Cell Rep 2018, 22 (6), 1473–1483. [DOI] [PubMed] [Google Scholar]
- (59).Stensland ME; Pollmann S; Molberg O; Sollid LM; Fleckenstein B Primary sequence, together with other factors, influence peptide deimination by peptidylarginine deiminase-4. Biol. Chem 2009, 390 (2), 99–107. [DOI] [PubMed] [Google Scholar]
- (60).Assohou-Luty C; Raijmakers R; Benckhuijsen WE; Stammen-Vogelzangs J; de Ru A; van Veelen PA; Franken KL; Drijfhout JW; Pruijn GJ The human peptidylarginine deiminases type 2 and type 4 have distinct substrate specificities. Biochimica et biophysica acta 2014, 1844 (4), 829–836. [DOI] [PubMed] [Google Scholar]
- (61).Coassolo S; Davidson G; Negroni L; Gambi G; Daujat S; Romier C; Davidson I Citrullination of pyruvate kinase by PADI1 and PADI3 regulates glycolysis and cancer cell proliferation. bioRxiv 2020, 718486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhang Z; Deng X; Liu Y; Liu Y; Sun L; Chen F PKM2, function and expression and regulation. Cell Biosci 2019, 9, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Gao X; Wang H; Yang JJ; Liu X; Liu ZR Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012, 45 (5), 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Friend LR; Landsberg MJ; Nouwens AS; Wei Y; Rothnagel JA; Smith R Arginine methylation of hnRNP A2 does not directly govern its subcellular localization. PloS one 2013, 8 (9), e75669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange51 Consortium via the PRIDE52 partner repository with the dataset identifier PXD023733. Public release of the data will be made on time for online publication of the paper. For anonymous access to peer-reviewers please use the following account information: Username: reviewer_pxd023733@ebi.ac.uk, Password: nGAPJjrm (https://www.ebi.ac.uk/pride/login). Mus musculus and Homo sapiens databases used for database searching were downloaded from UniProt (https://www.uniprot.org/).


