Abstract
Hybridization is a key mechanism involved in lineage diversification and speciation, especially in ecosystems that experienced repeated environmental oscillations. Recently radiated plant groups, which have evolved in mountain ecosystems impacted by historical climate change provide an excellent model system for studying the impact of gene flow on speciation. We combined organellar (whole-plastome) and nuclear genomic data (RAD-seq) with a cytogenetic approach (rDNA FISH) to investigate the effects of hybridization and introgression on evolution and speciation in the genus Soldanella (snowbells, Primulaceae). Pervasive introgression has already occurred among ancestral lineages of snowbells and has persisted throughout the entire evolutionary history of the genus, regardless of the ecology, cytotype, or distribution range size of the affected species. The highest extent of introgression has been detected in the Carpathian species, which is also reflected in their extensive karyotype variation. Introgression occurred even between species with dysploid and euploid cytotypes, which were considered to be reproductively isolated. The magnitude of introgression detected in snowbells is unprecedented in other mountain genera of the European Alpine System investigated hitherto. Our study stresses the prominent evolutionary role of hybridization in facilitating speciation and diversification on the one hand, but also enriching previously isolated genetic pools. [chloroplast capture; diversification; dysploidy; European Alpine system; introgression; nuclear-cytoplasmic discordance; ribosomal DNA.]
Hybridization is a key evolutionary process shaping diversification and speciation processes (Soltis and Soltis 2009; Taylor and Larson 2019; Nieto Feliner et al. 2020). Recent methodological developments allowed uncovering numerous, often unexpected, cases of hybridization and introgression (Mallet 2007; Whitney et al. 2010; Abbott et al. 2013, 2016; Mallet et al. 2016; Taylor and Larson 2019). Identification of historical hybridization in various groups of organisms has opened avenues to investigate the long-term consequences of gene flow in evolution, including hybrid speciation (Greig et al. 2002; Harris and Nielsen 2016; vonHoldt et al. 2016; Taylor and Larson 2019), adaptive introgression (Choler et al. 2004; Suarez-Gonzalez et al. 2018), but also adaptive radiation (e.g., Meier et al. 2017). A direct consequence of speciation induced by gene exchange is genomic mosaicism (Jiggins et al. 2008). The substantially increased allelic diversity of introgressed populations can facilitate their adaptation to novel and challenging environments. Hence, this mechanism can directly trigger the emergence of new species and fuel adaptive radiations (Pease et al. 2016; Meier et al. 2017). On the other hand, excessive introgression can reintegrate previously separated gene pools into a single biological entity, and thus hampering diversification (Bog et al. 2017; Kearns et al. 2018).
Key extrinsic triggers facilitating hybridization are dynamic changes in the environment, which enable previously allopatric populations to shift their ranges and come into secondary contact (Stebbins 1985). Mountain landscapes are highly diverse, forming steep altitudinal gradients featuring mosaics of niches with specific microclimates and bedrock. Such fine-scale, ecologically-heterogeneous systems promote secondary contacts of originally allopatric or parapatric species and hence provide ample opportunities for gene exchange (e.g., Raven 1973; Körner and Spehn 2002; Emadzade et al. 2015; Antonelli et al. 2018; Pouchon et al. 2018; Kandziora et al. 2022). The Quaternary climatic oscillations led to pronounced range contractions and expansions, especially in the mountain regions of the Northern hemisphere (Stebbins 1985; Hewitt 1996, 2000, 2004; Flantua et al. 2019). Consequently, the extensive range shifts along vertical and horizontal gradients created opportunities for secondary contacts and gene flow among species and populations lacking substantial mating barriers. Hybridization events associated with Quaternary glaciations in the European Alpine System (EAS, hereafter; Ozenda 1985) have primarily been evidenced among closely related species (Paun et al. 2008; Boucher et al. 2016; Klein and Kadereit 2016; Olšavská et al. 2016; Melichárková et al. 2020). Although the impact of gene flow on plant speciation at generic level remains understudied in European mountains, a crucial role for hybridization has been recently demonstrated, for instance, in South American mountain plant groups (Cortés et al. 2018; Morales-Briones et al. 2018; Pouchon et al. 2018; Kandziora et al. 2022).
The present study investigates the role of hybridization in the diversification of the mountain-dwelling genus Soldanella L. (snowbells; Primulaceae). Based on morphological and molecular data, it was hypothesized that the evolutionary history of this genus has been affected by hybridization (Vierhapper 1904a, 1904b; Pawłowska 1972; Meyer 1985; Zhang et al. 2001; Zhang and Kadereit 2002; Steffen and Kadereit 2014). Classical phylogenetic reconstructions uncovered shallow and poorly resolved topologies that hampered explicit detection of hybridization and introgression (cf. Zhang et al. 2001; Zhang and Kadereit 2002; Steffen and Kadereit 2014; Bellino et al. 2015). The weak morphological and genetic divergence between snowbell species from the Carpathians and Balkans might indicate incomplete speciation or widespread hybridization-induced erosion of species boundaries (Zhang et al. 2001; Zhang and Kadereit 2002). Interestingly, a traditional hypothesis that the widespread S. alpina is of hybrid origin has not received support in a recent study (Steffen and Kadereit 2014). Nevertheless, the role of hybridization in snowbells evolution remains speculative and needs to be addressed by high-resolution genomic data.
Using an exhaustive species sampling and a combination of NGS-based restriction site-associated DNA sequencing (RAD-seq), whole-plastome sequencing, and cytogenetic data (rDNA FISH) we aim here to investigate the prevalence of hybridization across the history of an EAS-dwelling plant genus, given that such a system must have been exposed to significant Quaternary climatic fluctuations. Specifically, we aim to test the following two hypotheses: 1) predominantly Carpathian and Balkan Soldanella species, usually featuring large ecological amplitude, have been affected by hybridization more than alpine zone-dwelling snowbell species, and 2) hybridization contributed to diversification of S. alpina, but also triggered the breakdown of species boundaries in Carpathian species.
Materials and Methods
Study System
The genus Soldanella consists of 18 species, of which four include two subspecies and one includes three subspecies (Zhang et al. 2001; Zhang and Kadereit 2002; Niederle 2003, 2016; Bellino et al. 2015) and is distributed across EAS, including the Alps, Apennines, Balkan mountains, Carpathians, Cordillera Cantábrica, Bohemian Massif, and the Pyrenees. Most species are endemic to a single mountain range, while others, such as S. alpina are more widespread (Zhang et al. 2001; Zhang and Kadereit 2002). Snowbells are cespitose to clonally growing, evergreen perennials with campanulate or funnel-shaped flowers with laciniate lobes, and capsule forming fruits. The genus includes snow bed specialists (S. minima and S. pusilla) occupying arctic-alpine environments in the alpine or subnival zone (Supplementary Fig. S1—https://doi.org/10.5061/dryad.mcvdnck41; Zhang et al. 2001; Zhang and Kadereit 2002), but the majority of snowbells have large ecological amplitude and occur from sheltered, humid forests to open habitats in the alpine zone (Zhang and Kadereit 2002; Štubňová et al. 2017; Valachovič et al. 2019). From a karyological perspective, most of species have the same ploidy and chromosome number with 2n = 40. A dysploid cytotype with 2n = 38 was, however, reported in several species (Štubňová et al. 2017).
Sampling Design
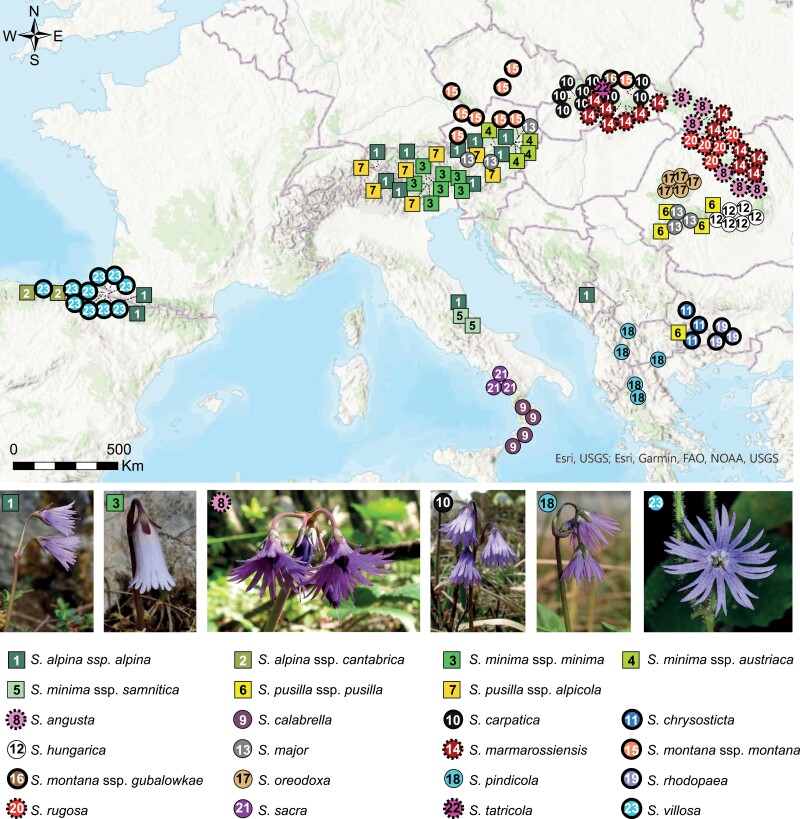
We sampled all members of the genus Soldanella, covering its entire distribution range (Fig. 1) including localities from which taxa were originally described (Supplementary Table S1). Fresh, intact leaves were conserved in silica gel. The RAD-seq based phylogenomic inference was performed with a minimum of four accessions per species, originating from different populations. Exceptions represented taxa with a narrow distribution (Supplementary Table S1). For RAD-seq analyses, we included 123 individuals from 118 populations. The plastome analyses were based on a selection of 42 Soldanella accessions, plus 47 outgroup accessions (Supplementary Table S1). For cytogenetic analyses, all taxa except S. minima subsp. samnitica and S. callabrella were used, representing 50 accessions from 42 localities (Supplementary Table S1). Vouchers labeled with the population and individual code number have been deposited in the SAV herbarium.
Figure 1.
Sampling map of Soldanella accessions used in the present study. Taxa specialized in alpine zone are represented by squares, while species that grow in forests or have a large ecological amplitude are represented by circles. The bold line on the symbol depicts species with a dysploid cytotype, whereas the dashed line denotes species revealed to be non-monophyletic through inferred tree analyses (Supplementary Figs. S2 and S3). Photos with their credits from left to right: S. alpina subsp. alpina (Jaromír Kučera—JK), S. minima subsp. minima (JK), S. angusta (Eliška Gbúrová Štubňová), S. carpatica (Marek Slovák), S. pindicola (JK) and S. villosa (JK).
DNA Isolation and RAD-seq Library Preparation and Sequencing
Genomic DNA was extracted using a DNeasy plant mini kit (Qiagen, Hilden, Germany) or a cetyltrimethylammonium bromide (CTAB) procedure (Doyle and Doyle 1987), with modifications proposed by Schönswetter et al. (2002), and purified using a NucleoSpin gDNA clean-up kit (Macherey-Nagel).
Preparation of single-digest RAD libraries followed a protocol adapted from Brandrud et al. (2017). The starting amount of genomic DNA was 120 ng per individual. Considering genome size (1C = 1.5–2.0 pg, Štubňová et al. 2017), 48 barcoded samples were pooled together in one library. Final libraries were sequenced on an Illumina HiSeq 2500 at the sequencing facility of the Vienna BioCenter Core Facilities (VBCF; https://www.viennabiocenter.org/) as 100 bp single-end reads. Individuals with less than 1.5 million high-quality reads were re-sequenced.
Filtering SNPs from RAD-seq Data
Raw Illumina reads were demultiplexed in two steps, first using BamIndexDecoder v.1.03 of the Picard Illumina2bam package (http://gq1.github.io/illumina2bam/index.html) and subsequently with process_radtags implemented in STACKS v.1.45 (Catchen et al. 2013), including quality filtering to retain only full-length sequences of 94 bp (without inline barcodes).
The filtered reads were further analyzed with denovo_map.pl from STACKS. To find the STACKS parameters that maximized the recovery of reliable polymorphic loci, whereas avoiding as much as possible over-splitting and collapsing of any paralogous loci, various settings were tested (Supplementary Table S2), following Heckenhauer et al. (2018). The optimal settings were m = 5, M = 1, n = 1, allowing for indels. These parameters resulted in the highest number of polymorphic loci with one to nine SNPs that were covered in at least 90% of individuals (Supplementary Table S2). To maximize the recovery of loci across the coverage and phylogenetic breadth in the data, we prepared a pseudo-reference based on consensus haplotypes of loci, following pipelines successfully applied in previous studies on species without reference genomes (e.g., Bateman et al. 2018; Heckenhauer et al. 2018; Brandrud et al. 2019).
The reads were further aligned against the pseudo-reference using BWA mem (v.0.7.17-r1188; Li and Durbin 2010), flagging shorter split hits as secondary. The aligned files were sorted by reference coordinates and read groups were added with Picard v.2.18.17 (https://broadinstitute.github.io/picard/). Indels were realigned in GATK v.3.7-1 (McKenna et al. 2010). Variants were finally called with the ref_map.pl script from STACKS with default parameters. Vcftools v.0.1.13 (Danecek et al. 2011) was used for SNPs filtration with various settings (Supplementary Table S3). The maximum allowed level of missing data was 5%.
RAD-seq based Phylogenetic Inference
The phylogenetic inference was performed using maximum likelihood (ML) and Bayesian inference (matrix RAD_A; Supplementary Table S3). The ML analyses were computed in RAxML v.8.1.3 (Stamatakis 2014), with the GTR + CAT approximation of rate heterogeneity, the standard Lewis acquisition bias correction (Lewis 2001) as recommended for concatenated SNPs, and 1000 rapid bootstrap replicates with a thorough ML search.
Bayesian phylogenomic trees were constructed in BEAST v.2.6.3 (Drummond and Bouckaert 2015), using the mutation model as selected by the in-built program, the relaxed lognormal molecular clock, and the Yule speciation model. Analyses were run for 300 million generations, sampling and logging every 15,000 generations. ESS values of parameters were checked for convergence in the software TRACER v.1.7 (Rambaut et. al 2018). Finally, TREEANNOTATOR v.2.6.3 (Bouckaert et al. 2014) was used to generate a maximum clade credibility tree discarding 25% of sampled trees as burn-in. The obtained trees were visualized with FIGTREE v.1.4.2 (available from http://tree.bio.ed.ac.uk/software/figtree/).
Detecting Genetic Admixture and Conflicting Signals
Neighbor-Net analysis (Bryant and Moulton 2004) was conducted in SPLITSTREE v.4.10 (Huson and Bryant 2006) using Matrix RAD_A. A co-ancestry heatmap based on genotype likelihoods was constructed with the genotype-free method of calling variants implemented in ANGSD v.0.921-26-g58a85440.910 (Korneliussen et al. 2014) on the indel-realigned mapping files. Only reads with base and mapping minimum qualities of 20 were used. Alleles were inferred from genotype likelihoods with a GATK-derived algorithm (i.e., GL 2). Only variable sites (P < 0.000001) that were detected in at least 50% of individuals and showed a minor allele frequency of 0.015 were kept. A covariance matrix was further calculated with PCangsd v.0.99 (Meisner and Albrechtsen 2018) and was plotted using the heatmap.2 function in gplots v.3.0.1.1 (Warnes et al. 2020). Population structure and recent admixture events were also inferred using NGSadmix v.32 (Skotte et al. 2013) based on data matrices containing a single SNP per locus. Namely, matrix RAD_C included all analyzed snowbell individuals, while Matrix RAD_D (79 individuals) was created to search for a finer genetic structure within taxa from the Carpathians, NE Alps, Balkan, and southern Italy. Clustering patterns were estimated for both datasets with K values from 1 to 15. Each analysis was run ten times for each K and started from a different seed.
Historical Hybridization and Introgression
To reconstruct the relationships among snowbell taxa in the face of historical gene flow, TREEMIX v.1.13 (Pickrell and Pritchard 2012) was used for Matrix RAD_B containing a single SNP per locus. To generate a TREEMIX plot, the input file from vcf and the population script from STACKS were utilized. TREEMIX was run with S. villosa as an outgroup and visualized in R. Migration edges were sequentially added to a ML tree of population ancestry, with 20 possible migrations tested. The contribution of each migration vector was identified using an f index representing the fraction of the variance in the sample covariance matrix (W ∂) accounted for by the model covariance matrix (W).
To identify candidate species involved in introgressive hybridization, we computed D-statistic (i.e., ABBA-BABA-derived test), f4-admixture ratio, and Fbranch (fb) for all trios using Dsuite v.0.4r43. These statistics have been specifically developed to infer introgression in the presence of ILS (Patterson et al. 2012; Malinsky et al. 2021). The fb statistic is a heuristic strategy for summing f4-admixture ratios over the entire tree topology to detect introgression events, in particular affecting internal branches (Malinsky et al. 2021). Matrix RAD_A was utilized for D-statistics computations. Soldanella villosa was selected as an out-group and species relationships were outlined on a pruned RAxML tree (Fig. 2), which was obtained with the ‘drop.tip’ command implemented in the R library ape (Paradise et al. 2004).
Figure 2.
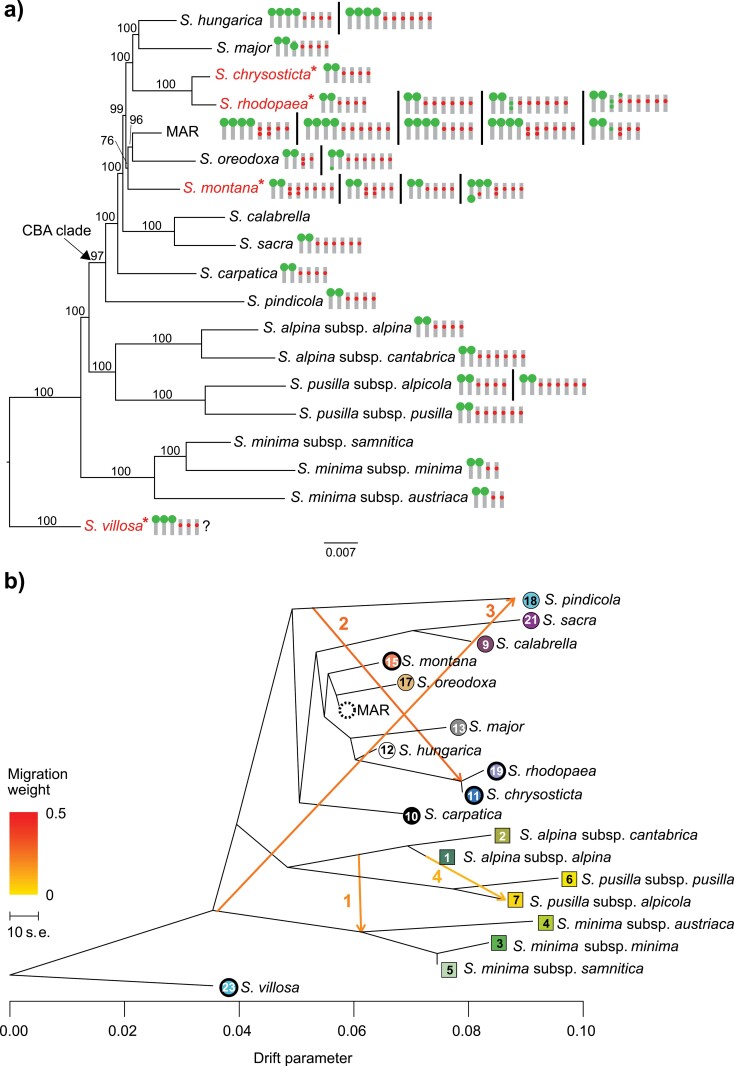
(a) Collapsed best-scoring ML tree of Soldanella based on 39,416 SNPs and 119 individuals (Matrix RAD_A). Bootstrap support is indicated above branches. Chromosomes carrying 5S (internal marks) and 35S (terminal bullets) ribosomal DNA loci detected by FISH analyses are shown on tree terminals. All detected karyotypes are shown for each taxon. The question mark in S. villosa indicates an approximate number of 5S loci. Species marked by asterisk have a dysploid cytotype with 2n = 38, while the rest have 2n = 40. The polyphyletic S. marmarossiensis, S. angusta, and S. rugosa are grouped into an arbitrary taxon “MAR”. (b) The phylogenetic network of relationships between Soldanella taxa as inferred by TREEMIX, based on 11,804 SNPs and 119 individuals. The first four migration edges between species are marked by arrows indicating the direction toward the recipient group and colored according to the ancestry percentage received from the donor. Symbols at the tips follow taxon identity from Fig. 1.
Plastome Data—Divergence Time Estimation and Phylogenetic Analyses of Plastome Data
For building a plastid phylogeny, we used genome skimming data generated by the sequencing project PhyloAlps, and recently published by Smyčka et al. (2022). For further details, see Supplementary data 1.
Annotated plastomes of Soldanella taxa together with outgroup taxa were processed in Geneious v.7.1 (Biomatters, Auckland, New Zealand). Divergence times were estimated in a Bayesian framework using BEAST v.2.5.2. (Drummond et al. 2012) and a combination of primary (fossil-based) and secondary calibration points from the studies of Magallón et al. (2015) and de Vos et al. (2014) (for more details see Supplementary data 1).
Matrix cp_B that included 42 Soldanella accessions and Hottonia as an outgroup (70 genes, 43 individuals, 103,787 bp) was used for computation of the ML analysis conducted in RAxML v.8.1.3 (Stamatakis 2014), with the GTR + CAT model and 1000 rapid bootstrap replicates, each with a subsequent thorough ML search.
Chromosome Preparation and Fluorescence In Situ Hybridization
Chromosome spreads were prepared for 50 individuals (Supplementary Table S1) as described by Mandáková and Lysak (2016a) and in Supplementary data 1.
The BAC clone T15P10 (AF167571) of Arabidopsis thaliana bearing 35S rRNA gene repeats was used for in situ localization of nucleolar organizer regions (NORs), and the Arabidopsis clone pCT4.2 (M65137), corresponding to a 500-bp 5S rDNA repeat, was used for localization of 5S rDNA loci. The rDNA probes were labeled with biotin-dUTP and digoxigenin-dUTP by nick translation as previously described (Mandáková and Lysak, 2016b; see also Supplementary data 1).
Results
RAD-seq Data
An average of 3,162,605 (± 1,253,129 SD) high-quality RAD-seq reads were obtained per individual after demultiplexing and filtering. Altogether 23,498 polymorphic loci (94‒112 nt long) were included in the synthetic reference after optimizing parameters and filtering. On average, 25.8% (± 2.2% SD) of the reads could be mapped to this reference, for a final mean coverage of 34.7×. The raw dataset contained 322,278 SNPs. Four samples (including S. chrysosticta subsp. serbica) were discarded due to high levels of missing data. Hence, the final dataset consisted of a total of 119 Soldanella individuals. The RAD-seq reads have been deposited in the NCBI Short Reads Archive (BioProject ID PRJNA820454, SRA: SRR18507145-SRR18507263).
Phylogenetic Tree Reconstruction
ML and BI analyses served to uncover overall phylogenetic relationships among snowbell taxa. Soldanella villosa appeared in a sister position to the rest of the snowbells after midpoint rooting, which is in line with outcomes of previous studies (e.g., Zhang et al. 2001; de Vos et al., 2014). Species occupying the alpine zone (S. alpina, S. minima and S. pusilla) branched off before the radiation of a clade containing species mostly from the Balkans and Carpathians (Fig. 2a, Supplementary Figs. S2 and S3). By contrast, S. villosa and S. minima were clustered in sister positions to the rest of the taxa in BI analyses, although with poor support (PP = 0.51; Supplementary Fig. S3). The rest of the snowbell taxa from the Carpathians, Balkans, and southern Apennines formed a clade, referred to as the CBA clade hereafter. Individuals belonging to the same taxon were grouped into strongly supported clades (BS 100%, PP 1.0), except for some Carpathian species that were reciprocally non-monophyletic (S. marmarossiensis, S. angusta, and S. rugosa). Interestingly, the dysploid cytotype (2n = 38) has very likely evolved independently in four species (Fig. 2a).
Genetic Admixture and Conflicting Signal Detection
Neighbor-Net results (Supplementary Fig. S4) displayed multiple reticulations across the network, with parallel splits connecting particularly S. alpina, S. minima, S. pusilla, and S. villosa reciprocally, but also with the rest of snowbell taxa.
The co-ancestry heatmap indicated the existence of two distinct species groups: alpine species + S. villosa, and the CBA clade (Supplementary Fig. S5). Soldanella pindicola appeared to share relatively equal co-ancestry with both groups. The most pronounced traces of genetic admixture were visible between S. alpina and S. minima, as well as between S. alpina and S. pusilla. Intensive mutual gene flow was also indicated for the Carpathian taxa, with a lack of taxon boundaries among S. angusta, S. marmarossiensis and S. rugosa.
The first run of admixture analysis including all taxa found higher ΔK peaks (Evanno et al. 2005) when individuals were clustered into 2, 4, 10, and 13 groups (Supplementary Fig. S7a–e). When clustered into two groups, S. pindicola appears admixed between two major species groups formed by S. minima, S. alpina, S. pusilla and S. villosa on one hand, and the rest of snowbells on the other hand (Supplementary Fig. S7a). In the case of four groups, S. rhodopaea, S. chrysosticta, S. calabrella, and S. sacra were genetically intermediate between S. pindicola-S. carpatica and the other Carpathian species (Supplementary Fig. S7b). The second run including only CBA taxa uncovered interpretable results at K = 2, 3, 5, 6, and 8 (Supplementary Fig. S8 a–e). The new insight with respect to the first run was provided by results with three and five clusters, where S. rhodopaea and S. chrysosticta were remarkably admixed with S. pindicola, and S. calabrella shares genetic material with S. carpatica, respectively (Supplementary Fig. S8b,c). Finally, S. angusta, S. hungarica, S. marmarossiensis, and S. rugosa share alleles from various gene pools especially for the analyses with high number of clusters (Supplementary Fig. S8c–e).
The TREEMIX graph rooted with S. villosa revealed nearly the same topology as the ML and BI trees (Fig. 2b). Residuals from the covariance matrix did not significantly improve after including four migration edges, as also indicated by the curve of variance (Supplementary Fig. S6a,b). The first migration edge showed a historical gene flow between lineages ancestral to S. alpina and S. minima (weight 19.3%), the second edge suggested a migration from S. pindicola to the ancestor of S. chrysosticta + S. rhodopaea (weight 27.6%), and the third edge indicated a shared ancestry of S. pindicola with the ancestor of S. minima (weight 24.4%). Finally, the last migration edge suggested a historical gene flow between populations of S. alpina subsp. alpina and S. pussila subsp. alpicola (weight 10.2%).
D and derived statistics uncovered a pervasive, but generally mild gene exchange, whereby several hybridization/introgression events have most likely occurred between ancestral lineages and others occurred between currently existing taxa (Fig. 3, Supplementary Table S4). Soldanella alpina, S. minima and S. pusilla appear to have interbred frequently in different phases of their evolutionary histories. Strong signals of likely historical gene flow and admixture were identified between S. alpina and S. minima, and also between S. alpina and S. pusilla subsp. alpicola (Fig. 3). Some level of gene flow is indicated also between species with the euploid and dysploid cytotypes (i.e., between the euploid S. pindicola and the dysploid S. chrysosticta-rhodopaea clade, as well as between the dysploid S. montana and the euploid MAR clade and S. hungaricaFig. 3b,c).
Figure 3.
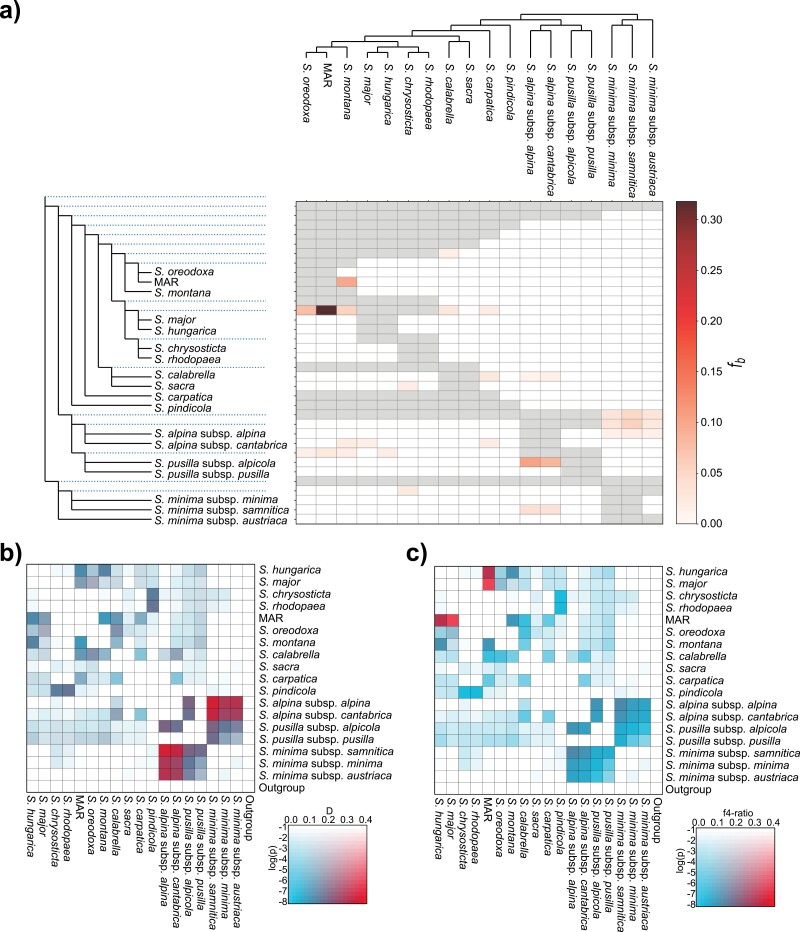
Results of D-suite statistics. (a) f-branch statistics showing deeper introgressions among taxa and lineages. Gray shadings indicate untestable pairs, given our sampling and the phylogenetic tree. (b) D-statistics showing that most taxa have been significantly affected by introgression. (c) f4 statistics showing the proportion of the genome affected by admixture. All analyses were based on Matrix_RAD_A (39,416 SNPs and 119 individuals). The polyphyletic S. marmarossiensis, S. angusta, and S. rugosa are grouped into an arbitrary taxon named “MAR” (Supplementary Figs. S2 and S3).
As determined by D-statistics, the most extensive hybridization occurred between the following pairs: S. alpina and S. minima, S. alpina and S. pusilla subsp. alpicola, S. pindicola and the S. chrysosticta-rhodopaea clade (Fig. 3b). The f4 statistics revealed that S. major and S. hungarica, or more likely their ancestor, hybridized with the MAR clade, which has affected a substantial portion of their genomes (i.e., around 30%). Genetic admixture, hybridization, and introgression in snowbells as resolved by various analytical approaches are summarized in Supplementary Table S5.
Plastid Phylogeny and Molecular Dating
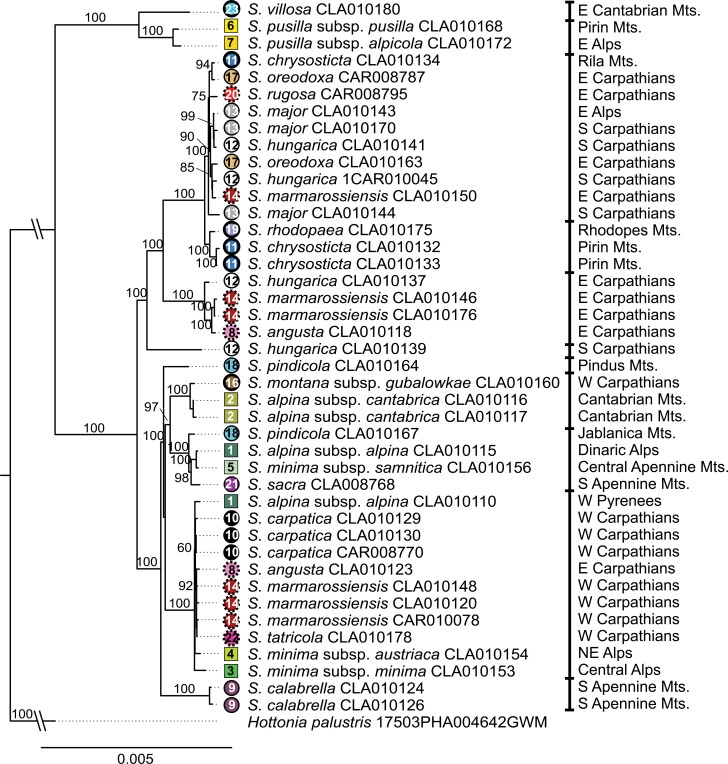
The topology of the inferred plastome tree contrasts strongly with topologies obtained from RAD-seq data (Fig. 4, Supplementary Fig. S9). Plastome haplotypes of the vast majority of species reflected neither the current taxonomic concept nor the morphology or ecology of the taxa studied. However, individual haplotype subclades were clustered based on narrower or wider geographical units. Plastome-based molecular divergence time estimation indicates that the genus Soldanella is of a Middle Tertiary origin. Its MRCA split from Hottonia in the early Miocene (21.2 Ma, Supplementary Fig. S9). After a prolonged diversification stasis in the late Miocene (6.3 Ma), the MRCA lineage was divided into two major sister clades. Ancestral lineages of recent taxa branched off at the end of the Pliocene (3.1 Ma) and the beginning of the Pleistocene (2.3 Ma). All recent species are of a Pleistocene origin, mostly being younger than 1 Ma.
Figure 4.
Best-scoring ML tree of Soldanella based on plastome data including 43 accessions and 103,787 bp (Matrix cp_B). Bootstrap support is indicated above branches. The branch lengths to the outgroup and ingroup are interrupted and not to scale. Symbols at the tips follow Fig. 1.
Variation in Chromosome Number and rDNA Loci
The euploid chromosome number (2n = 40) dominated in the genus, whereas the dysploid chromosome number (2n = 38) was proved in S. chrysosticta, S. montana, S. rhodopaea, and S. villosa. A high variation in the number and localization of 35S and 5S rDNA loci was observed (Fig. 2a; Supplementary Fig. S10). The rDNA FISH revealed 16 unique and clearly interpretable patterns. Probes corresponding to 35S rDNA provided 2–5 hybridization signals in chromosome complements. Interstitial 35S rDNA loci were identified in S. major and the MAR clade (Supplementary Figs. S10b,g,h,k). NORs on both termini of a single chromosome were observed in S. oreodoxa and S. montana (Supplementary Fig. S10i and n).
All 5S rDNA loci were observed in the interstitial position on chromosomes. However, the number of 5S rDNA hybridization sites varied extensively from two in S. minima to eight in the MAR clade (Supplementary Fig. S10p and d). In S. montana, S. oreodoxa and the MAR clade, duplicated 5S rDNA loci were observed within one or two chromosomes (Supplementary Fig. S10d,f,j,k,m–o). Two evolutionary trends are obvious: 1) a single NOR-bearing chromosome pair and two 5S-bearing chromosome pairs represent the most frequent rDNA pattern (Fig. 2a and Supplementary Fig. S10c) and 2) the highest intraspecific rDNA variation occurs only in the young Carpathian species (Fig. 2a). No correlation in the rDNA chromosome positioning was identified between the euploid and the dysploid cytotype, except for the sister species pair of S. chrysosticta and S. rhodopaea (Fig. 2a; Supplementary Fig. S10c).
Discussion
Pervasive Introgression in Snowbells
Recurring waves of hybridization and introgression have played a vital role in the evolutionary history and speciation processes in the genus Soldanella. Hybridization did not affect only forest species, primarily from the CBA clade as we hypothesized (cf. Zhang et al. 2001), but it was even more extensive in species inhabiting alpine habitats. Even though we employed a variety of methods to infer gene flow, the detected patterns and intensity of gene flow were not fully consistent across methods. This might be explained by extremely complicated patterns caused by recurrent hybridization across various time horizons. In addition, the employed methods are sensitive to gene flow at different time horizons. This could result in various degrees of masking of ancestral introgression across analytical approaches (e.g., Dsuite or TREEMIX versus ADMIXTURE, cf. Lawson et al. 2018). Extensive introgression was, apart from the genetic admixture, suggested also by the exceptional diversity in the number of 35S and 5S loci and their positions on chromosomes (Fig. 2a; Supplementary Fig. S10). Speciation-related chromosomal re-patterning further increases/decreases the number of rDNA sites or their chromosomal repositioning. The dynamics of rDNA loci may be thus regarded as a strong indicator for significant hybridization processes (Raskina et al. 2008; Waminal et al. 2021). The genus underwent several descending dysploidies leading to the recurrent formation of dysploid cytotypes (from 2n = 40 to 38) and speciation (cf. Blöch et al. 2009; Moraes et al. 2016; Singhal et al. 2017; Melichárková et al. 2020). Although descending dysploidy can act as an instant reproduction barrier between the newly formed dysploid and its euploid parental cytotype (Mandáková and Lysak 2018; Winterfeld et al. 2018), hybridization events between dysploid and euploid Soldanella species were revealed by our analyses (Fig. 2b,3, and Supplementary Figs. S5, S7b, and S8b).
To our best knowledge, such a high level of within-genus hybridization has never been observed in mountain plants from the EAS. Snowbells lack specific dispersal mechanisms (e.g., anemophily, anemochory, or extensive clonality), which could otherwise promote secondary contacts among distinct populations and thus explain the extensive introgression detected. Indeed, it has been suggested that the reduced floral morphology of alpine-dwelling snowbells indicates a shift from insect pollination towards self-pollination (Steffen and Kadereit 2014), which would rather hamper such a pervasive gene exchange.
During the rapid diversification of snowbells, however, there may not have been enough time to build strong reproductive barriers between species, which might have been further weakened by repeated episodes of introgression. Importantly, the majority of snowbell taxa, especially from the CBA clade, share large ecological amplitudes that range from foothill sheltered and humid forest understory to exposed alpine meadows and snow bed communities (Štubňová et al. 2017; Valachovič et al. 2019). Recurrent elevation shifts triggered by the glacial-interglacial range contraction most likely facilitated secondary contacts of the mountain and alpine populations both within and between species (cf. Smyčka et al. 2022). In combination with the absence of inter-specific reproductive barriers, these frequent secondary interactions significantly boosted the probability of effective hybridization between snowbell populations. Nevertheless, historical range shifts did not need to play a substantial role in promoting gene flow in alpine species; at least at present they frequently occur in parapatry or sympatry (Zhang et al. 2001; Zhang and Kadereit 2002), which enhances the frequency of secondary contacts among them.
Strong Cytonuclear Discordance Reflects Introgression and Chloroplast Capture
The most blatant case of reticulation visible at first sight in the snowbell phylogeny is the cytonuclear discordance documented by the incongruent plastome (Fig. 4 and Supplementary Fig. S9) and nuclear SNP-based phylogenies (Fig. 2a, Supplementary Figs. S2, S3). Cytonuclear discordance is mostly attributed to ILS and/or hybridization (e.g., Folk et al. 2017; García et al. 2017; Pouchon et al. 2018; Heckenhauer et al. 2019; Rose et al. 2020). ILS is often prominent in rapidly radiating lineages, and affects phylogenetic inferences by a lack of hierarchical phylogenetic structuring, accumulation of speciation events in short periods, and sharing haplotypes across clades without a geographic pattern (Wendel and Doyle 1998; Acosta and Premoli 2010; Xu et al. 2012; Pouchon et al. 2018). Nevertheless, given the present results, we suggest that the major source of the cytonuclear discordance during the evolutionary history of snowbells could rather be attributed to extensive hybridization and introgression than to ILS (see results of Dsuite statistics, Fig 3; see Malinsky et al. 2021). However, a certain level of ILS is expected, especially in young taxa. Extensive introgression among S. alpina, S. pusilla, and S. minima (Figs. 2b and 3, Supplementary Figs. S4 and S5) finally resulted in distinct cytonuclear discordances and their altering reciprocal positions in the nuclear and plastome phylogenies (Figs. 2a and 4, Supplementary Figs. S2, S3, and S9). Significant genetic admixture, as would be expected in nuclear signals of recent hybrid entities, was, however, not revealed in these taxa using admixture analysis (Supplementary Figs. S7a–e; cf. Acosta and Premoli 2010; Yi et al. 2015; Folk et al. 2017; Pouchon et al. 2018; Rose et al. 2020). In reality, ADMIXTURE parsimoniously describes variation between individuals, but it does not implement a historical model and assumes that all populations are descended from a single ancestral population. Under such a relatively simple scenario, identified admixture can be misleadingly interpreted or even omitted. Moreover, if all members of a certain taxon are equally admixed or affected by genetic drift, there is no variation for the model to explain, and such admixture might be missed (see Lawson et al. 2018). Furthermore, introgression traces might have vanished from the biparentally inherited nuclear genome via long-term unidirectional backcrossing, leaving signals only in the plastid genomes as a result of chloroplast capture (Soltis and Soltis 1995; Fehrer et al. 2007). More specifically, the ancient hybridization between parental species gave rise to F1 hybrids that consequently might have undergone asymmetrical, repetitive backcrossing towards only one parental species, causing the nuclear material of the other parental species to be essentially eradicated. Backcrossed populations thus maintained the nuclear material from one parent while the chloroplast was obtained from another parent via chloroplast capture (Acosta and Premoli 2010; Yi et al. 2015; Folk et al. 2017; Pouchon et al. 2018; Rose et al. 2020).
Extensive and Recurrent Introgression Facilitates but also Prevents Speciation
Hybridization can accelerate diversification via the formation of hybrid species (Seehausen 2004; Litsios and Salamin 2014; Stankowski and Streisfeld 2015; Sefc et al. 2017; White et al. 2020), but it can also slow down diversification via the breakdown of species reproductive barriers followed by extensive introgression. This could further lead to swamping of previously separated lineages and extinction of parental taxa via genetic swamping (Huxel 1999; Gómez et al. 2015; Todesco et al. 2016). We did not identify key hybridization events that would directly trigger the evolution of some snowbell clades as shown in other organisms (Meier et al. 2017; Irisarri et al. 2018; Svardal et al. 2020). Nevertheless, we uncovered several introgression events which most likely contributed to the evolution of numerous snowbell species, especially within the CBA clade. The cascade of diversification and speciation events facilitated by ancestral introgressions might thus have significantly contributed to the emergence of most Carpathian and Balkan taxa.
The widespread S. alpina was traditionally considered a hybrid species, but this status has been left unsupported by previous genetic data (Zhang et al. 2001; Steffen and Kadereit 2014). The present analyses showed that this species underwent recurrent introgression with the alpine-dwelling snowbed specialists S. pusilla and S. minima (Fig. 3, Supplementary Figs. S3 and S5). Furthermore, polyphyly of S. alpina plastomes across the geographically delimited sub-clades implies that S. alpina might have hybridized and captured plastids recurrently not only from the parapatric S. minima and S. pusilla, but also from a more distant and recently allopatric species (Fig. 4, Supplementary Fig. S9). This could imply local extinctions of one of the parental taxa after hybridization events followed by long-term unidirectional introgression of the hybrid offspring with S. alpina, a phenomenon coined “ghost introgression” (Ottenburghs 2020). Given the foregoing, piecing together a puzzle of hybridization episodes involving S. alpina is still exceedingly difficult, which precludes solid conclusions as to whether or not it is a hybridogeneous entity.
Unexpected and exciting is the genetic mosaicism found in S. pindicola and the S. chrysosticta–S. rhodopaea clade, occupying mountains in the SE and E Balkans, respectively. Soldanella pindicola was likely the recipient of the genetic material from an ancestor close to S. minima (Supplementary Fig. S2b) and, at the same time, served as a donor of a significant proportion of the genetic material to the ancestor of the S. chrysosticta-S. rhodopaea clade (Figs. 2b and 3, Supplementary Figs. S5, S7b and S8b). Indeed, the extensive admixture found in S. pindicola and the S. chrysosticta-S. rhodopaea clade may be indicated also by their increased absolute genome sizes (Štubňová et al. 2017). Given the significant genetic admixture present in the genomes of all three Balkan species (see Figs 2b and 3b,c, as well as Supplementary Figs S7a,b and S8b), we can conclude that they can be considered to be of hybridogeneous origin.
Substantial introgression has been observed among the young Carpathian species, namely, among S. major and S. hungarica from the southern parts of the Carpathians, and the S. marmarossiensis-angusta-rugosa group occurring across almost the entire Carpathian arc (Fig. 3; Supplementary Fig. S5). Apart from the genetic evidence, extensive gene flow among these taxa and their lineages is also reflected by the astonishing diversity of their 35S and 5S karyotype patterns (Fig. 2a; Supplementary Fig. S10). The extensive gene flow resulted in pronounced weakening of their species boundaries, which finally hampered and/or slowed down their speciation processes. From an evolutionary standpoint, it appears that S. angusta, S. marmarossiensis, and S. rugosa, which are currently recognized as separate species (Zhang et al. 2001; Zhang and Kadereit 2002), might represent a single polymorphic biological entity. Additional taxonomically focused research based on comprehensive population sampling is required to determine whether certain Carpathian taxa are hybridogeneous or merely highly affected by more recent introgression.
Conclusions
Despite the extensive research on the evolutionary history of plants in the EAS, there is still a gap in our understanding of how introgression has impacted the diversity and speciation of mountain plants. An unprecedented degree of introgression in comparison with other plants from the EAS was detected in almost all snowbell species. While the detected introgression potentially bolstered the formation of new snowbell species, in other cases it likely promoted reintegration of previously isolated gene pools and slowed down speciation in the young Carpathian taxa. The wide ecological amplitude of multiple snowbell taxa, growing in habitats from foothill forests to snow-beds in the alpine zone, was most likely one of the key factors facilitating allopatric lineage interconnection and gene flow.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.mcvdnck41
Acknowledgments
Authors are thankful to Juliane Baar for valuable help with RAD sequencing library preparations and to thank Michal Melichárek for his generous assistance with bioinformatic data processing. We are deeply thankful to Anna Berešová, Clara Bertel, Božo Frajman, Iva Hodálová, Judita Kochjarová, Monika Kolářová, Patrik Mráz, Jana Pilátová, Doubravka Požárová, Katarína Skokanová, Marie Smyčková, Peter Schönswetter, Stanislav Španiel, Milan Štech, Andreas Tribsch and Milan Valachovič for help with sampling of analyzed material. In addition, selected data analyses were computed in the CIPRES Science Gateway. We are also thankful to Lukáš Demovič for help with computation of some specific analyses.
Contributor Information
Marek Slovák, Department of Evolution and Systematics, Plant Science and Biodiversity Centre, Slovak Academy of Sciences, Institute of Botany, Bratislava, Slovakia; Department of Botany, Charles University, Prague, Czech Republic.
Andrea Melichárková, Department of Evolution and Systematics, Plant Science and Biodiversity Centre, Slovak Academy of Sciences, Institute of Botany, Bratislava, Slovakia.
Eliška Gbúrová Štubňová, Department of Evolution and Systematics, Plant Science and Biodiversity Centre, Slovak Academy of Sciences, Institute of Botany, Bratislava, Slovakia; Slovak National Museum, Natural History Museum, Bratislava, Slovakia.
Jaromír Kučera, Department of Evolution and Systematics, Plant Science and Biodiversity Centre, Slovak Academy of Sciences, Institute of Botany, Bratislava, Slovakia.
Terezie Mandáková, Central European Institute of Technology, Department of Experimental Biology, Faculty of Science, Masaryk University, Kamenice 753/5, CZ-625 00 Brno, Czech Republic.
Jan Smyčka, Department of Botany, Charles University, Prague, Czech Republic; Center for Theoretical Study, Charles University and the Academy of Sciences of the Czech Republic, Jilská 1, 110 00 Praha, Czech Republic; Université Grenoble Alpes, University of Savoie Mont Blanc, CNRS, Grenoble, France.
Sébastien Lavergne, Université Grenoble Alpes, University of Savoie Mont Blanc, CNRS, Grenoble, France.
Nicodemo Giuseppe Passalacqua, Department of Biology, Ecology and Earth Sciences, University of Calabria, Rende, Cosenza, Italy.
Peter Vďačný, Department of Zoology, Comenius University in Bratislava, Bratislava, Slovakia.
Ovidiu Paun, Department of Botany and Biodiversity Research, University of Vienna, Vienna, Austria.
Funding
This study was financially supported by the Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and Slovak Academy of Sciences, VEGA 2/0041/19 (to Marek Slovák, Bratislava), by the project “Conserving the endemic flora of the Carpathian Region” coordinated by RBG Kew’s Millennium Seed Bank (United Kingdom), and by the mobility grant 2015-05-15-001 of the Action Austria-Slovakia (OeAD). Sequencing and fieldwork related to the PhyloAlps project were supported from France Génomique (ANR-10-INBS-09-08) and an ANR-SNF project (ANR-16-CE93-0004, SNF-549 310030L_170059). Terezie Mandáková was supported by the Czech Science Foundation (project no. 22-16826S to Terezie Mandáková) and Eliška Gbúrová Štubňová was supported by the Operational Programme Research and Innovations and co-financed by the European Fund for Regional Development (EFRD) ITMS2014 + 313021W683: DNA barcoding of Slovakia (SK-BOL), as a part of the International Barcode of Life (iBOL).
References
- Abbott R., Albach D., Ansell S., Arntzen J.W., Baird S.J., Bierne N., Boughman J., Brelsford A., Buerkle C.A., Buggs R., Butlin R.K., Dieckmann U., Eroukhmanoff F., Grill A., Cahan S.H., Hermansen J.S., Hewitt G., Hudson A.G., Jiggins C., Jones J., Keller B., Marczewski T., Mallet J., Martinez-Rodriguez P., Möst M., Mullen S., Nichols R., Nolte A.W., Parisod C., Pfennig K., Rice A.M., Ritchie M.G., Seifert B., Smadja C.M., Stelkens R., Szymura J.M., Väinölä R., Wolf J.B., Zinner D.. 2013. Hybridization and speciation. J. Evol. Biol. 26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Abbott R., Barton N.H., Good J.. 2016. Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 25(11):2325–2332. doi: 10.1111/mec.13685. [DOI] [PubMed] [Google Scholar]
- Acosta M.C., Premoli A.C.. 2010. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Biol. Evol. 54:235–242. doi: 10.1016/j.ympev.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Antonelli A., Kissling W.D., Flantua S.G.A., Bermúdez M.A., Mulch A., Muellner-Riehl A.N., Kreft H., Linder H.P., Badgley C., Fjeldsa J., Fritz S.A., Rahbek C., Herman F., Hooghiemstra H., Hoorn C.. 2018. Geological and climatic influences on mountain biodiversity. Nat. Geosci. 11:718–725. doi: 10.1038/s41561-018-0236-z. [DOI] [Google Scholar]
- Bateman R., Sramkó G., Paun O.. 2018. Integrating RAD-seq with morphological cladistic analysis clarifies evolutionary relationships among species groups of bee orchids. Ann. Bot. 121:85–105. doi: 10.1093/aob/mcx129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellino A., Bellino L., Baldantoni D., Saracino A.. 2015. Evolution, ecology and systematics of Soldanella (Primulaceae) in the southern Apennines (Italy). BMC Evol. Biol. 15(1):158. doi: 10.1186/s12862-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blöch C., Weiss-Schneeweiss H., Schneeweiss G.M., Barfuss M.H., Rebernig C.A., Villaseñor J.L., Stuessy T.F.. 2009. Molecular phylogenetic analyses of nuclear and plastid DNA sequences support dysploid and polyploid chromosome number changes and reticulate evolution in the diversification of Melampodium (Millerieae, Asteraceae). Mol. Phylogenet. Evol. 53(1):220–233. doi: 10.1016/j.ympev.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bog M., Bässler C., Oberprieler C.. 2017. Lost in the hybridisation vortex: high-elevation Senecio hercynicus (Compositae, Senecioneae) is genetically swamped by its congener S. ovatus in the Bavarian Forest National Park (SE Germany). Evol. Ecol. 31:401–420. doi: 10.1007/s10682-017-9890-7. [DOI] [Google Scholar]
- Boucher F.C., Casazza G., Szövényi P., Conti E.. 2016. Sequence capture using RAD probes clarifies phylogenetic relationships and species boundaries in Primula sect. Auricula. Mol. Phylogenet. Evol. 104:60–72. doi: 10.1016/j.ympev.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C.-H., Xie D., Suchard M.A., Rambaut A., Drummond A.J.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandrud M.K., Paun O., Lorenz R., Baar J., Hedrén M.. 2019. Restriction-site associated DNA sequencing supports a sister group relationship of Nigritella and Gymnadenia (Orchidaceae). Mol. Phylogenet. Evol. 136:21–28. doi: 10.1016/j.ympev.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandrud M.K., Paun O., Lorenzo M.T., Nordal I., Brysting A.K.. 2017. RADseq provides evidence for parallel ecotypic divergence in the autotetraploid Cochlearia officinalis in Northern Norway. Sci. Rep. 7(1):5573. doi: 10.1038/s41598-017-05794-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D., Moulton V., Moulton V.. 2004. NeighborNet: an agglomerative algorithm for the construction of planar phylogenetic networks. Mol. Biol. Evol. 21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Catchen J., Hohenlohe P.A., Bassham S., Amores A., Cresko W.A.. 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choler P., Erschbamer B., Tribsch A., Gielly L., Taberlet P.. 2004. Genetic introgression as a potential to widen a species’ niche: insights from alpine Carex curvula. PNAS 101(1):171–176. doi: 10.1073/pnas.2237235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A.J., Garzón L.N., Valencia J.B., Madriñán S.. 2018. On the causes of rapid diversification in the Páramos: isolation by ecology and genomic divergence in Espeletia. Front. Plant Sci. 9:1700. doi: 10.3389/fpls.2018.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R., Lunter G., Marth G., Sherry S.T., McVean G., Durbin R., 1000 Genomes Project Analysis Group. 2011. The Variant Call Format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos J.M., Hughes C.E., Schneeweiss G.M., Moore B.R., Conti E.. 2014. Heterostyly accelerates diversification via reduced extinction in primroses. Proc. R. Soc. B 281:20140075. doi: 10.1098/rspb.2014.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. Bot. Soc. Am. 19:11–15. [Google Scholar]
- Drummond A.J., Bouckaert R.R.. 2015. Bayesian evolutionary analysis with BEAST. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29(8):1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emadzade K., Lebmann M.J., Hoffmann M.H., Tkach N., Lone F.A., Hörandl E.. 2015. Phylogenetic relationships and evolution of high mountain buttercups (Ranunculus) in North America and Central Asia. Perspect. Plant Ecol. Evol. Syst. 17:131–141. doi: 10.1016/j.ppees.2015.02.001. [DOI] [Google Scholar]
- Evanno G., Regnaut S., Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fehrer J., Gemeinholzer B., ChrtekBräutigam J.S. Jr.. 2007. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol. Phylogenet. Evol. 42(2):347–361. doi: 10.1016/j.ympev.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Flantua S.G., O’Dea A., Onstein R.E., Giraldo C., Hooghiemstra H.. 2019. The flickering connectivity system of the north Andean páramos. J. Biogeogr. 46(8):1808–1825. doi: 10.1111/jbi.13607. [DOI] [Google Scholar]
- Folk R.A., Mandel J.R., Freudenstein J.V.. 2017. Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Syst. Biol. 66:320–337. doi: 10.1093/sysbio/syw083. [DOI] [PubMed] [Google Scholar]
- García N., Folk R.A., Meerow A.W., Chamala S., Gitzendanner M.A., Oliveira R.S., Soltis D.E., Soltis P.S.. 2017. Deep reticulation and incomplete lineage sorting obscure the diploid phylogeny of rain-lilies and allies (Amaryllidaceae tribe Hippeastreae). Mol. Phylogenet. Evol. 111:231–247. doi: 10.1016/j.ympev.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Gómez J.M., González-Megías A., Lorite J., Abdelaziz M., Perfectti F.. 2015. The silent extinction: climate change and the potential hybridization-mediated extinction of endemic high-mountain plants. Biodivers. Conserv. 24(8):1843–1857. doi: 10.1007/s10531-015-0909-5. [DOI] [Google Scholar]
- Greig D., Louis E.J., Borts R.H., Travisano M.. 2002. Hybrid speciation in experimental populations of yeast. Science 298:1773–1775. doi: 10.1126/science.1076374. [DOI] [PubMed] [Google Scholar]
- Harris K., Nielsen R., Nielsen R.. 2016. The genetic cost of Neanderthal introgression. Genetics 203(2):881–891. doi: 10.1534/genetics.116.186890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckenhauer J., Paun O., Chase M.W., Ashton P.S., Kamariah A.S., Samuel R.. 2019. Molecular phylogenomics of the tribe Shoreeae (Dipterocarpaceae) using whole plastid genomes. Ann. Bot. (Lond.) 123:857–865. doi: 10.1093/aob/mcy220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckenhauer J., Samuel R., Aston P.S., Salim K.A., Paun O.. 2018. Phylogenomics resolves evolutionary relationships and provides insights into floral evolution in the tribe Shoreeae. Mol. Phylogenet. Evol. 127:1–13. doi: 10.1016/j.ympev.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58(3):247–276. doi: 10.1006/bijl.1996.0035. [DOI] [Google Scholar]
- Hewitt G.M. 2000. The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B: Biol. Sci. 359(1442):183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huxel G.R. 1999. Rapid displacement of native species by invasive species: effects of hybridization. Biol. Conserv. 89(2):143–152. doi: 10.1016/S0006-3207(98)00153-0. [DOI] [Google Scholar]
- Irisarri I., Singh P., Koblmüller S., Torres-Dowdall J., Henning F., Franchini P., Fischer C., Lemmon A.R., Lemmon E.M., Thallinger G.G., Sturmbauer C., Meyer A.. 2018. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat. Commun. 9:3159. doi: 10.1038/s41467-018-05479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins C.D., Salazar C., Linares M., Mavarez J.. 2008. Hybrid trait speciation and Heliconius butterflies. Philos. Trans. R. Soc. Lond. B Biol. Sci 363(1506):3047–3054. doi: 10.1098/rstb.2008.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandziora M., Sklenář P., Kolář F., Schmickl R.. 2022. How to tackle phylogenetic discordance in recent and rapidly radiating groups? Developing a workflow using Loricaria (Asteraceae) as an example. Front. Plant Sci. 12:765719. doi: 10.3389/fpls.2021.765719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns A.M., Restani M., Szabo I., Schrøder-Nielsen A., Kim J.A., Richardson H.M., Marzluff J.M., Fleischer R.C., Johnsen A., Omland K.E.. 2018. Genomic evidence of speciation reversal in ravens. Nat. Commun. 9:906. doi: 10.1038/s41467-018-03294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J.T., Kadereit J.W.. 2016. Allopatric hybrids as evidence for past range dynamics in Sempervivum (Crassulaceae), a western Eurasian high mountain oreophyte. Alp. Bot. 126(2):119–133. doi: 10.1007/s00035-016-0164-8. [DOI] [Google Scholar]
- Korneliussen T.S., Albrechtsen A., Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinform. 15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C., Spehn E.M.. 2002. Mountain biodiversity: A global assessment. Parthenon Pub. Group, Boca Raton, 14, 336. [Google Scholar]
- Lawson D.J., van Dorp L., Falush, D.A.. 2018. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 9:3258. doi: 10.1038/s41467-018-05257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.O. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios G., Salamin N., Salamin N.. 2014. Hybridisation and diversification in the adaptive radiation of clownfishes. BMC Evol. Biol. 14:245. doi: 10.1186/s12862-014-0245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S., Gómez-Acevedo S., Sánchez-Reyes L.L., Hernández-Hernández T.. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207(2):437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- Malinsky M., Matschiner M., Svardal H.. 2021. Dsuite—Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 21:584–595. doi: 10.1111/1755-0998.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. 2007. Hybrid speciation. Nature 446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Mallet J., Besansky N., Hahn M.W.. 2016. How reticulated are species? Bioessays 38(2):140–149. doi: 10.1002/bies.201500149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A.. 2018. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 42:55–65. doi: 10.1016/j.pbi.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A., Lysak M.A.. 2016a. Chromosome preparation for cytogenetic analyses in Arabidopsis. Curr. Protoc. Plant Biol 1:43–51. doi: 10.1002/cppb.20009. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A., Lysak M.A.. 2016b. Painting of Arabidopsis chromosomes with chromosome-specific BAC clones. Curr. Protoc. Plant Biol 1(2):359–371. doi: 10.1002/cppb.20022. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J.I., Marques D.A., Mwaiko S., Wagner C.E., Excoffier L., Seehausen O.. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8:14363. doi: 10.1038/ncomms14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J., Albrechtsen A., Albrechtsen A.. 2018. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichárková A., Šlenker M., Zozomová-Lihová J., Skokanová K., Šingliarová B., Kačmárová T., Caboňová M., Kempa M., Šrámková G., Mandáková T., Lysák M.A., Svitok M., Mártonfiová L., Marhold K.. 2020. So closely related and yet so different: strong contrasts between the evolutionary histories of species of the Cardamine pratensis. Polyploid complex in central Europe. Front. Plant Sci. 11:588856. doi: 10.3389/fpls.2020.588856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F.K. 1985. Beitrag zur Kenntniss ost und südosteuropäischer Soldanella-Arten. Haussknechtia 2:7–41. [Google Scholar]
- Moraes A.P., Olmos Simões A., Ojeda Alayon D.I., de Barros F., Forni-Martins E.R.. 2016. Detecting mechanisms of karyotype evolution in Heterotaxis (Orchidaceae). PLoS One 11:e0165960. doi: 10.1371/journal.pone.0165960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Briones D.F., Liston A., Tank D.C.. 2018. Phylogenomic analyses reveal a deep history of hybridization and polyploidy in the Neotropical genus Lachemilla (Rosaceae). New Phytol. 218(4):1668–1684. doi: 10.1111/nph.15099. [DOI] [PubMed] [Google Scholar]
- Niederle J. 2003. Remarks on the genus Soldanella L. in the West Carpathians. Acta Mus. Richnoviensis, Sect. Natur. 10:171–174. [Google Scholar]
- Niederle J. 2016. Dřípatky ze Srbska. Skalničkářův rok 73:16. [Google Scholar]
- Nieto Feliner G., Casacuberta J., Wendel J.F.. 2020. Genomics of evolutionary novelty in hybrids and polyploids. Front. Genet. 11:792. doi: 10.3389/fgene.2020.00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olšavská K., Slovák M., Marhold K., Štubňová E., Kučera J.. 2016. On the origins of Balkan endemics: the complex evolutionary history of the Cyanus napulifer group (Asteraceae). Ann. Bot. 118(6):1071–1088. doi: 10.1093/aob/mcw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenburghs J. 2020. Ghost introgression: spooky gene flow in the distant past. Bioessays 42(6):2000012. doi: 10.1002/bies.202000012. [DOI] [PubMed] [Google Scholar]
- Ozenda P. 1985. La végétation de la chaîne alpine dans l’espace montagnard européen. Paris, Masson. [Google Scholar]
- Paradise E., Claude J., Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D.. 2012. Ancient admixture in human history. Genetics 192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O., Schönswetter P., Winkler M., Tribsch A.. 2008. Evolutionary history of the Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Mol. Ecol. 14(19):4263–4275. doi: 10.1111/j.1365-294X.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawłowska S. 1972. Soldanella. In: Tutin T.G., Heywood V.H., Burges N.A., Moore D.M., Valentine D.H., Walters S.M., Webb D.A., editors. Flora Europaea, vol. 3, Cambridge University Press, UK, p. 23–24. [Google Scholar]
- Pease J.B., Haak D.C., Hahn M.W., Moyle L.C.. 2016. Phylogenomics reveals three sources of adaptive variation during rapid radiation. PLoS Biol. 14:e1002379. doi: 10.1371/journal.pbio.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J.K., Pritchard J.K.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouchon C., Fernández A., Nassar J.M., Boyer F., Aubert S., Lavergne S., Mavárez J.. 2018. Phylogenomic analysis of the explosive adaptive radiation of the Espeletia complex (Asteraceae) in the tropical Andes. Syst. Biol. 67(6):1041–1060. doi: 10.1093/sysbio/syy022. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A.. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskina O., Barber J.C., Nevo E., Belyayev A.. 2008. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytog. Gen. Res. 120:351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Raven P.H. 1973. Evolution of subalpine and alpine plant groups in New Zealand. N. Z. J. Bot. 11:177–200. doi: 10.1080/0028825X.1973.10430272. [DOI] [Google Scholar]
- Rose J.P., Toledo C.A., Lemmon E.M., Lemmon A.R., Sytsma K.J.. 2020. Out of sight, out of mind: widespread nuclear and plastid-nuclear discordance in the flowering plant genus Polemonium (Polemoniaceae) suggests widespread historical gene flow despite limited nuclear signal. Syst. Biol. 70:162–180. doi: 10.1093/sysbio/syaa049. [DOI] [PubMed] [Google Scholar]
- Schönswetter P., Tribsch A., Barfuss M., Niklfeld H.. 2002. Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. and Hoppe (Campanulaceae) in the European Alps. Mol. Ecol. 11:2637–2647. doi: 10.1046/j.1365-294x.2002.01651.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O. 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Sefc K.M., Mattersdorfer K., Ziegelbecker A., Neuhüttler N., Steiner O., Goessler W., Koblmüller S.. 2017. Shifting barriers and phenotypic diversification by hybridisation. Ecol. Lett. 20(5):651–662. doi: 10.1111/ele.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal V.K., Kumar R., Singhal H., Kumar P., Kaur D., Kaur M., Rana P.K., Gupta R.C.. 2017. A profile of male meiosis, chromosomal variation and status in species of Impatiens from North-West Himalaya in India. Caryologia 70:258–269. doi: 10.1080/00087114.2017.1344084. [DOI] [Google Scholar]
- Skotte L., Korneliussen T.S., Albrechtsen A.. 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyčka J., Roquet C., Boleda M., Alberti A., Boyer F., Douzet R., Perrier CH., Rome M., Valay J.G., Denoeud F., Šemberová K., Zimmermann N.E., Thuiller W., Wincker P., Alsos I.E., Coissac E.The PhyloAlps consortium, Lavergne S.. 2022. Tempo and drivers of plant diversification in the European mountain system. Nat. Commun. (preprint). doi: 10.21203/rs.3.rs-959411/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S.. 1995. The dynamic nature of polyploid genomes. Proc. Natl. Acad. Sci. U.S.A. 92(18):8089–8091. doi: 10.1073/pnas.92.18.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E.. 2009. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankowski S., Streisfeld M.A.. 2015. Introgressive hybridization facilitates adaptive divergence in a recent radiation of monkeyflowers. Proc. R. Soc. B 282:20151666. doi: 10.1098/rspb.2015.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G.L. 1985. Polyploidy, hybridization, and the invasion of new habitats. Ann. Mo. Bot. Gard. 72(4):824–832. doi: 10.2307/2399224. [DOI] [Google Scholar]
- Steffen S., Kadereit J.W.. 2014. Parallel evolution of flower reduction in two alpine Soldanella species (Primulaceae). Bot. J. Linn. 175:409–422. doi: 10.1111/boj.12174. [DOI] [Google Scholar]
- Štubňová E., Hodálová I., Kučera J., Mártonfiová L., Svitok M., Slovák M.. 2017. Karyological patterns in the European endemic genus Soldanella L.: absolute genome size variation uncorrelated with cytotype chromosome numbers. Am. J. Bot. 104:1241–1253. doi: 10.3732/ajb.1700153. [DOI] [PubMed] [Google Scholar]
- Suarez-Gonzalez A., Lexer C., Cronk Q.C.. 2018. Adaptive introgression: a plant perspective. Biol. Lett. 14(3):20170688. doi: 10.1098/rsbl.2017.0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svardal H., Quah F.X., Malinsky M., Ngatunga B.P., Miska E.A., Salzburger W., Genner M.J., Turner G.F., Durbin R.. 2020. Ancestral hybridization facilitated species diversification in the Lake Malawi Cichlid Fish Adaptive Radiation. Mol. Biol. Evol. 37(4):1100–1113. doi: 10.1093/molbev/msz294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.A., Larson E.L.. 2019. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 3(2):170–177. doi: 10.1038/s41559-018-0777-y. [DOI] [PubMed] [Google Scholar]
- Todesco M., Pascual M.A., Owens G.L., Ostevik K.L., Moyers B.T., Hübner S., Heredia S.M., Hahn M.A., Caseys C., Bock D.G., Rieseberg L.H.. 2016. Hybridization and extinction. Evol. Appl. 9(7):892–908. doi: 10.1111/eva.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valachovič M., Štubňová E., Senko D., Kochjarová J., Coldea G.. 2019. Ecology and species distribution pattern of Soldanella sect. Soldanella (Primulaceae) within vegetation types in the Carpathians and the adjacent mountains. Biologia 74:733–750. doi: 10.2478/s11756-019-00200-7. [DOI] [Google Scholar]
- Vierhapper F. 1904b. Neue Pflanzen-Hybriden. Österr. Bot. Z. 54:349–350. [Google Scholar]
- Vierhapper F. 1904a. Übersicht über die Arten und Hybriden der Gattung Soldanella. In: Urban I., Graebner P., editors. Festschrift zur Feier des siebzigsten Geburtstages des Herrn Prof. Dr. Paul Ascherson. Verlag Gebrüder Borntraeger, Leipzig, p. 500–508. [Google Scholar]
- vonHoldt B.M., Cahill J.A., Fan Z., Gronau I., Robinson J., Pollinger J.P., Shapiro B., Wall J., Wayne R.K.. 2016. Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Sci. Adv. 2:e1501714. doi: 10.1126/sciadv.1501714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waminal N.E., Pellerin R.J., Kang S.-H., Kim H.H.. 2021. Chromosomal mapping of tandem repeats revealed massive chromosomal rearrangements and insights into Senna tora dysploidy. Front. Plant Sci. 2021(12):629898. doi: 10.3389/fpls.2021.629898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M., Venables B., Galili T.. 2020. gplots: various R programming tools for plotting data. R package version 3. Accessed on Jan 01, 2022. https://github.com/talgalili/gplots. [Google Scholar]
- Wendel J.F., Doyle J.J.. 1998. Phylogenetic Incongruence: Window into Genome History and Molecular Evolution. In: Soltis D.E., Soltis P.S., Doyle J.J., editors. Molecular systematics of plants II. Springer, Boston, MA, p. 265–296 doi: 10.1007/978-1-4615-5419-6_10. [DOI] [Google Scholar]
- White O.W., Reyes-Betancort J.A., Chapman M.A., Carine M.A.. 2020. Geographical isolation, habitat shifts and hybridisation in the diversification of the Macaronesian endemic genus Argyranthemum (Asteraceae). New Phytol. 228(6):1953–1971. doi: 10.1111/nph.16980. [DOI] [PubMed] [Google Scholar]
- Whitney K.D., Ahern J.R., Campbell L.G., Albert L.P., King M.S.. 2010. Patterns of hybridization in plants. Perspect. Plant Ecol. Evol. Syst. 12(3):175–182. doi: 10.1016/j.ppees.2010.02.002. [DOI] [Google Scholar]
- Winterfeld G., Becher H., Voshell S., Hilu K., Röser M.. 2018. Karyotype evolution in Phalaris (Poaceae): the role of reductional dysploidy, polyploidy and chromosome alteration in a wide-spread and diverse genus. PLoS One 13:e0192869. doi: 10.1371/journal.pone.0192869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Wu N., Gao X.-F., Zhang L.-B.. 2012. Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Mol. Phylogenet. Evol. 62:346–358. doi: 10.1016/j.ympev.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Yi T.-S., Jin G.-H., Wen J.. 2015. Chloroplast capture and intra- and intercontinental biogeographic diversification in the Asian-New World disjunct plant genus Osmorhiza (Apiaceae). Mol. Phylogenet. Evol. 85:10–21. [DOI] [PubMed] [Google Scholar]
- Zhang L.-B., Comes H.P., Kadereit J.W.. 2001. Phylogeny and quaternary history of the European montane/alpine endemic Soldanella (Primulaceae) based on ITS and AFLP variation. Am. J. Bot. 88:2331–2345. doi: 10.2307/3558393. [DOI] [PubMed] [Google Scholar]
- Zhang L.-B., Kadereit J.W.. 2002. The systematics of Soldanella (Primulaceae) based on morphological and molecular (ITS, AFLPs) evidence. Nord. J. Bot. 22:129–169. doi: 10.1111/j.1756-1051.2002.tb01360.x. [DOI] [Google Scholar]