Abstract
Purpose of review
The development of deep learning (DL) systems requires a large amount of data, which may be limited by costs, protection of patient information and low prevalence of some conditions. Recent developments in artificial intelligence techniques have provided an innovative alternative to this challenge via the synthesis of biomedical images within a DL framework known as generative adversarial networks (GANs). This paper aims to introduce how GANs can be deployed for image synthesis in ophthalmology and to discuss the potential applications of GANs-produced images.
Recent findings
Image synthesis is the most relevant function of GANs to the medical field, and it has been widely used for generating ‘new’ medical images of various modalities. In ophthalmology, GANs have mainly been utilized for augmenting classification and predictive tasks, by synthesizing fundus images and optical coherence tomography images with and without pathologies such as age-related macular degeneration and diabetic retinopathy. Despite their ability to generate high-resolution images, the development of GANs remains data intensive, and there is a lack of consensus on how best to evaluate the outputs produced by GANs.
Summary
Although the problem of artificial biomedical data generation is of great interest, image synthesis by GANs represents an innovation with yet unclear relevance for ophthalmology.
Keywords: artificial intelligence, deep learning, generative adversarial networks, medical image synthesis, ophthalmology
INTRODUCTION
In the field of ophthalmology over the last few years, clinically applicable deep learning (DL) systems have been developed to detect different eye diseases, such as diabetic retinopathy (DR) [1–4], glaucoma [3,5], age-related macular degeneration (AMD) [6,7], and retinopathy of prematurity (ROP) [8]. This has led to the real possibility that such DL systems may be implemented soon in appropriate clinical settings, such as in DR screening programs [9,10].
Despite the substantial promise of DL, the development of a robust DL algorithm or system is data intensive, meaning that a large amount of data exhibiting representative variability (i.e., disease and normal) is required for the training and validation process [11]. The availability of such large datasets is often limited by the lack of corresponding clinical cohorts, the high costs of starting a primary data collection from baseline, and the need to protect the privacy of patients. Personal information of patients must be protected under rigorously controlled conditions and in accordance with the best research practices [12,13]. Moreover, medical images are considered identifiable personal information and cannot be anonymized easily, and consent is difficult to obtain for large retrospective datasets [14–16]. In addition, annotated data of the more severe phenotypes of certain pathologies such as advanced glaucoma and neovascular or late AMD are often too uncommon in existing population studies to be useful for conducting DL analysis.
Recent developments in DL have provided an innovative alternative to these challenges, by using generative adversarial networks (GANs) to artificially create new images based on smaller real image datasets. There is significant potential to generate a large number of images required to train, develop, validate and test new DL algorithms and systems.
WHAT ARE GENERATIVE ADVERSARIAL NETWORKS?
GANs are a special type of neural network model based on a game theoretic approach, with the objective being to find Nash equilibrium between two networks: a generator and a discriminator (Fig. 1). The idea is to sample from a simple distribution, like Gaussian, and then learn to transform this noise to a targeted data distribution, using universal function approximators such as convolutional neural networks, by the adversarial training of generator and discriminator simultaneously. The generator model learns to capture the targeted data distribution, and the discriminator model estimates the probability that a sample comes from the targeted data distribution rather than the distribution generated by the generator. In other words, the task of generator is to generate natural looking images and the task of discriminator is to decide whether the image is fake or real. This can be thought of as a minimax two-player game, i.e., generator vs discriminator, where the performance of both the networks ideally improves over time iteratively. Although the generator tries to fool the discriminator by generating images that appear as real as possible, the discriminator tries to not get fooled by the generator by improving its discriminative capability [17].
FIGURE 1.
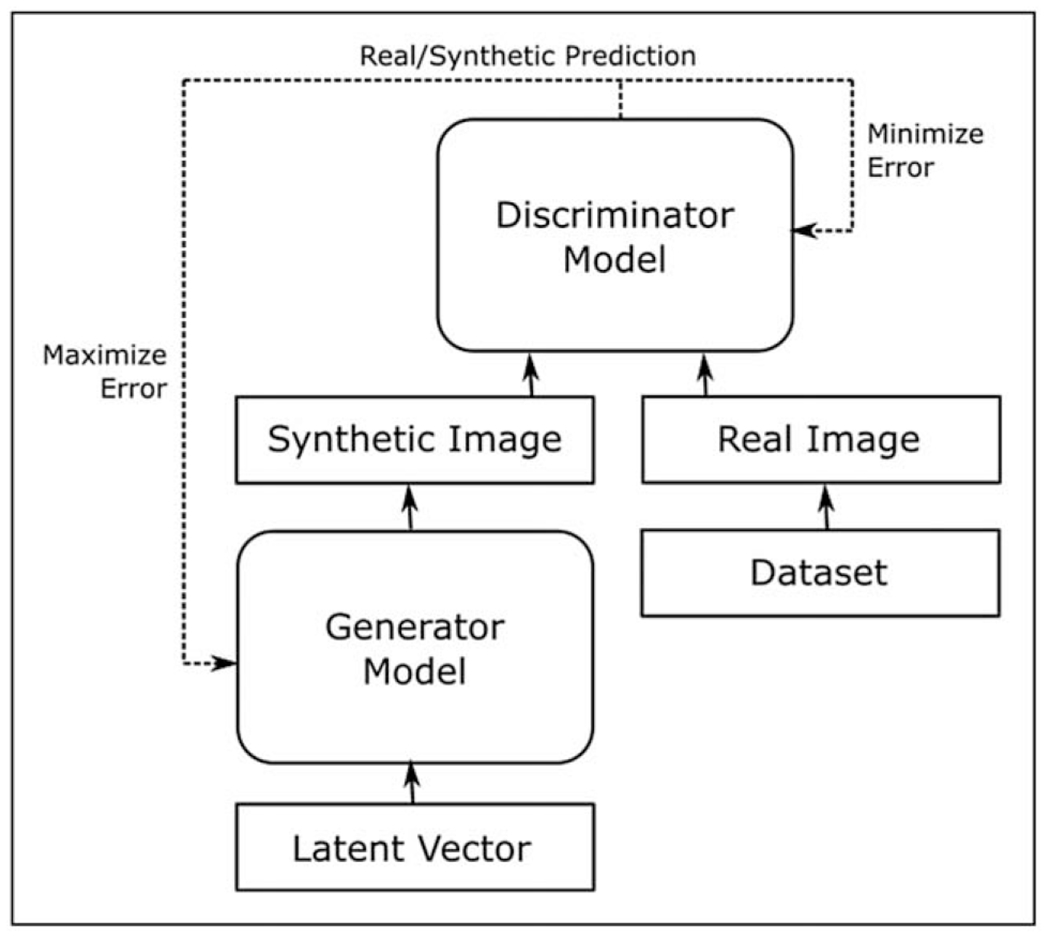
General architecture of a GAN. The generator model and discriminator model are in competition with each other, with the generator model’s objective being to produce increasingly realistic synthetic images, and the discriminator model’s objective being to distinguish these synthetic images from real images. GAN, generative adversarial network.
There are many ways to incorporate GANs in medical imaging tasks, such as segmentation [18], classification [19], detection [20], registration [21], image reconstruction [22] and image synthesis (Table 1) [23]. GANs have been used in research studies for generating medical images of various image modalities, including breast ultrasound [24], mammograms [25], computed tomography (CT) [26–29], magnetic resonance images (MRI) [30], cancer pathology images [31], and contrast agent-free ischemic heart disease images [32]. Moreover, GANs have been shown to be capable of cross-modality image synthesis, such as generating MRI based on ultrasound [33] or CT [34,35]. This paper focuses on the image synthesis aspect of GANs in ophthalmology via introducing different types of GANs, summarizing GAN models reported in the literature of ophthalmology (Table 2), discussing the outcome measures for GANs, and evaluating the advantages and disadvantages of GANs.
Table 1.
Other applications of GANs in medical imaging besides image synthesis [23]
| Functions of GANs | Methods | Examples |
|---|---|---|
| Segmentation | Use discriminator to count the adversarial losses | Conditional GAN for segmenting myocardium and blood pool simultaneously on patients with congenital heart disease [18] |
| Classification | Use generator and discriminator as a feature extractor, or use discriminator as a classifier | Use semi-supervised GAN for cardiac abnormality classification in chest X-rays [19] |
| Detection | Train discriminator to detect the lesions | Use GAN to detect brain lesions on MRI [20] |
| Registration | Train generator to produce transformation parameters or the transformed images | Use spatial transformation network to perform prostate MR to transrectal ultrasound image registration [21] |
| Image reconstruction | Use cGANs to tackle low spatial resolution, noise contamination and under-sampling | Use pix2pix framework for low dose CT denoising [22] |
CT, computed tomography; GAN, generative adversarial network; MRI, magnetic resonance images.
Table 2.
the summary of studies using image synthesis function of GANs in ophthalmology
| GAN models | Year | Target imaging modality | Type of GANs | Training datasets | Outputs | ||
|---|---|---|---|---|---|---|---|
| Costa et al. [45] | 2018 | Fundus images | Adversarial autoencoder, image-to-image translation | 614 normal fundus images | An end-to-end retinal image synthesis system with corresponding vessel networks | ||
| Guibas et al. [46] | 2017 | Fundus images | Deep convolutional GAN | 40 pairs of retinal fundus images and vessel segmentation masks | - F1 accuracy rating of 0.8877 and 0.8988 for synthetically trained and real image trained u-net respectively - Kullback-Leibler (KL) divergence score of 4.759 comparing the synthetic and real images |
||
| Zhao et al. [47] | 2018 | Fundus images | Tub-GAN | 10 fundus images | Able to synthesize multiple images with diversity despite a small training set | ||
| Yu et al. [48] | 2019 | Fundus images | Cycle-GAN | 101 fundus images (70 glaucomatous eyes and 31 normal eyes) | Higher structural similarity index and peak signal-to-noise ratio than single vessel tree approach | ||
| Burlina et al. [49] | 2019 | AMD fundus images of AMD | ProGAN | 133,821 AREDS fundus images | - Accuracy of 53.67% and 59.50% in differentiating real from synthetic images - AUC of DLS trained on synthetic images alone: 0.9235 |
||
| Diaz-Pinto et al. [50] | 2019 | Glaucomatous fundus images | Deep convolutional GAN | 86,926 fundus images | - t-SNE plots showing that the synthesized images are closer to the real images than Costa’s method and semi-supervised DCGAN model - AUC of 0.9017 in detecting glaucomatous fundus images |
||
| Zhou et al. [51■] | 2020 | DR fundus images | DR-GAN | 1,842 images | - Accuracy of 65.8% in differentiating real from synthetic images - Accuracy of 90.46% in DR grading by the classifying model augmented with synthetic images |
||
| Beers et al. [52] | 2018 | Posterior pole retinal photographs of ROP | ProGAN | 5,550 images | AUC of 0.97 comparing to the segmentation maps from real images | ||
| Zheng et al. [53■] | 2020 | OCT images | ProGAN | 108,312 OCT images | - Accuracy of discriminating real vs synthetic OCT images: 59.50% and 53.67% - AUC of 0.905 in detecting urgent referrals by DL system trained on synthetic data |
||
| Liu et al. [56■] | 2019 | Posttreatment OCT images | Pix2pixHD | 476 pairs of pre and posttherapeutic OCT images | - sensitivity 84% and specificity 86% in predicting the posttreatment macular classification | ||
| Lee et al. [57] | 2021 | Posttreatment OCT images | cGAN | 15,183 paired OCT B-scans from 241 patients | Predicting | Sensitivity | Specificity |
| Intraretinal fluid | 33.3% | 95.1% | |||||
| Subretinal fluid | 21.2% | 95.1% | |||||
| Pigmented epithelial detachment | 70.4% | 94.6% | |||||
| Subretinal hyperreflective material | 76.5% | 94.1% | |||||
| Tavakkoli et al. [59■■] | 2020 | Generate FA from fundus images | cGAN | Pairs of FA and fundus images from 59 patients | Fréchet Inception Distance: 43 Structural similarity measures: 0.67 |
||
AMD, age-related macular degeneration; AREDS, the Age-Related Eye Disease Study; AUC, area under the receiver operating characteristic (ROC) curve; DR, diabetic retinopathy; DL, deep learning; GAN, generative adversarial network; cGAN, conditioning of GAN; OCT, optical coherence tomography; ProGAN, Progressive GAN; ROP, retinopathy of prematurity.
DIFFERENT TYPES OF GENERATIVE ADVERSARIAL NETWORKS
There are many different formulations of GANs [36], which might firstly be categorized according to their objectives. Although GANs have been employed to generate sequential data such as text, their most common usage has been in imaging tasks, including video. Within image processing, GANs have been employed to perform texture synthesis, super-resolution, object detection and image synthesis, all of which could have potential applications in medicine. GANs can be further defined by their features, the more prominent of which may be their neural architecture for generators and discriminators, their objective function, and their training procedure. Each of these components has undergone significant development since the introduction of GANs. For example, the conditioning of GANs referred as cGAN, on both the generator and discriminator would be demonstrated soon after their inception [37], through the addition of a prior as input. Various objective functions have been proposed to address issues such as instability in training, some of the most prominent amongst of which include Wasserstein GAN [38], which seeks to maintain a continuous distance when real and generated data distributions are disjoint, and LS-GAN [39], which encourages the generated data distribution to be closer to the real distribution through the implementation of mean squared loss instead of log loss. PatchGAN [40] is proposed to run discriminators on patches or on images at different scales in order to improve the quality of image synthesis such as angiography image synthesis. As for training procedure, individual learning rates for the discriminator and generator have been shown to converge to a local equilibrium [41].
Improving the quality of synthesized images at higher resolutions has been of particular interest for medical applications, which are often especially sensitive to subtle details within images. This has been enabled by innovations such as the progressive growing of the generator and discriminator with Progressive GAN (ProGAN) [42], orthogonal regularization with BigGAN [43], and the extension of ProGAN with a number of incremental improvements such as a mapping network bilinear upsampling and block noise into StyleGAN [44]. It should be noted that many individual features of various GANs are possibly compatible, and may thus be combined into custom GAN architectures towards specific applications. If the desired objective is to improve the performance of existing classification models, the realism of the generated images may not be the top priority, since it is possible that the generated images may nevertheless augment the training data in a useful way, particularly for classes where data is sparse. Achieving the optimal mix of real and synthetic data in such cases remains an area of active research.
GENERATIVE ADVERSARIAL NETWORKS FOR IMAGE SYNTHESIS IN OPHTHALMOLOGY
Fundus images
Using generated vessel trees as an intermediate stage, Costa et al. were among the first few groups to deploy adversarial learning for building an end-to-end system for synthesizing retinal fundus images. This system was trained on a small data set of 614 normal fundus images and tends to generate fixed outputs that are lack of diversity and pathological features [45]. Following this, various methods have been attempted to improve the quality and diversity of the synthetic fundus photos. Guibas et al. proposed a two-stage GAN pipeline by first generating synthetic retinal blood vessel trees and then translating these masks to photorealistic images. The synthetic images were used to train a U-net segmentation network, achieving similar F1-score as the network trained on real images [46]. Using direct mapping from manual tubular structured annotation back to a raw image, the model developed by Zhao et al. can synthesize multiple images with diversity using a dataset as small as 10 training examples [47]. Yu et al. built a multiple-channels-multiple-landmarks pipeline using a combination of vessel tree, optic disc and optic cup images to generate colour fundus photos, which produced superior images than single-vessel-based approach [48].
Besides generating normal fundus images, GANs have been used to synthesize fundus photos of specific eye diseases, such as AMD, glaucoma, DR and ROP (Fig. 2). Using over 100,000 colour fundus photographs from the Age-Related Eye Disease Study (AREDS), Burlina et al. built two ProGAN models to synthesize referable (intermediate and/or advanced AMD) and nonreferable AMD images respectively [49]. The outputs were realistic enough that two retinal specialists could not distinguish real images from the synthetic ones, with accuracy of 53.67% and 59.50%, respectively. Furthermore, the DL system trained on synthetic data alone showed comparable diagnostic performance to the algorithm trained on real images in detecting referable AMD [area under the receiver operating characteristic (ROC) curve (AUC) of 0.92 vs. AUC of 0.97, respectively]. The question of how best to incorporate synthetic data with available real data for training remains open, to the best of our knowledge. Diaz-Pinto et al. built a retinal image synthesizer and semi-supervised classifier for glaucoma detection using GANs on over 80,000 images. Their system was capable of generating realistic synthetic images with features of glaucoma and labelling glaucomatous images automatically with an AUC of 0.902 [50]. Furthermore, Zhou et al. report a DR fundus photo generator that can be directly deployed to train DR classifier, via modification with arbitrary grading and lesion information to synthesize high-quality images [51■]. Beers et al. trained a ProGAN model with 5,550 posterior pole retinal photographs of ROP, which could produce realistic fundus images of ROP. They evaluated the performance of a segmentation algorithm trained on synthetic images, reporting an AUC of 0.97 comparing to the segmentation maps from real images [52].
FIGURE 2.
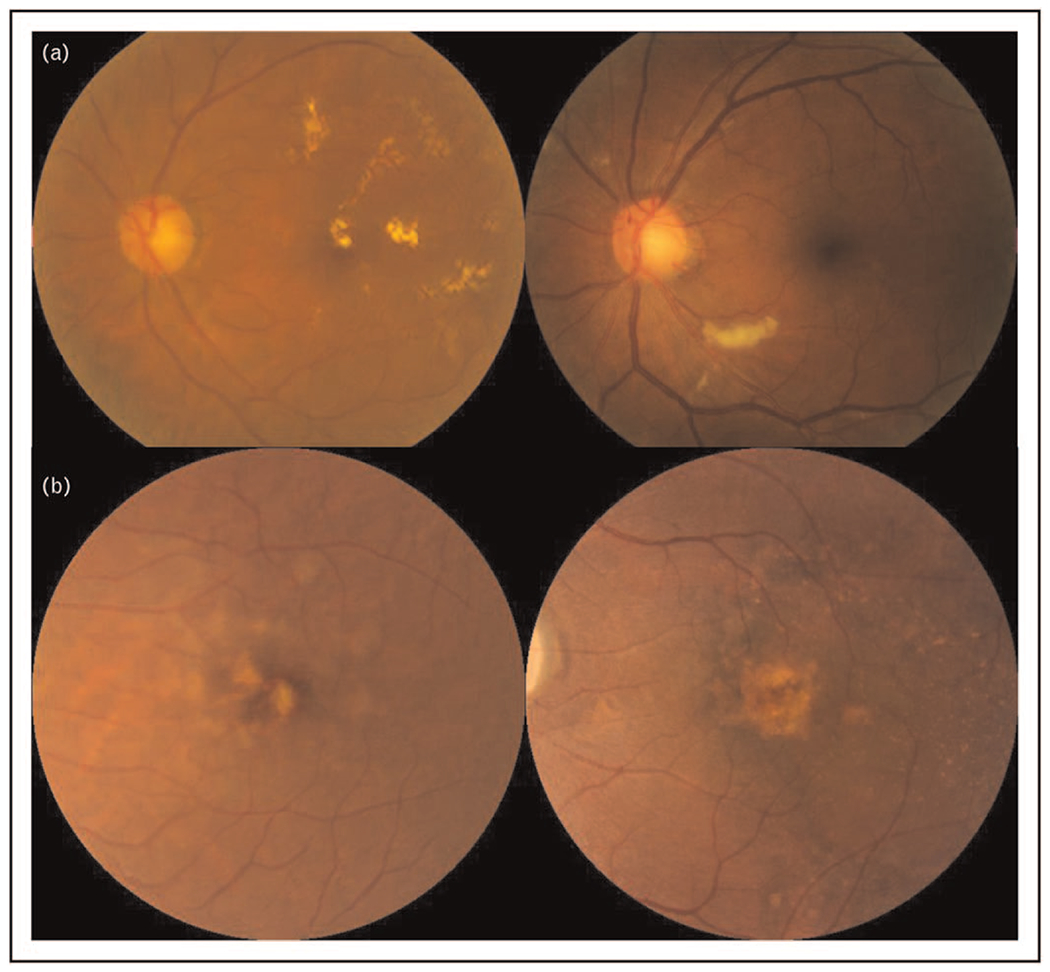
Examples of GAN synthesized images for DR and AMD. a. GAN synthesized images (left) compared to real images (right) of DR, b. GAN synthesized images (left) compared to real images (right) of and AMD. The AMD images were preprocessed with macular segmentation. AMD, age-related macular degeneration; DR, diabetic retinopathy; GAN, generative adversarial network.
Optical coherence tomography
Utilizing over 100,000 optical coherence tomography (OCT) images from eyes with balanced distribution of urgent referrals (choroidal neovascularization, diabetic macular oedema) and nonurgent referrals (drusen and normal eyes), Zheng et al. built a ProGAN model to synthesize OCT images that are realistic to retinal specialists. The DL framework trained on synthetic OCT images achieved an AUC of 0.905 in classifying urgent and nonurgent referrals, which was noninferior to the performance of the model trained on real OCT images (AUC = 0.937) [53■]. Apart from synthesizing OCT images from scratch, GAN has been used to enhance the image quality of OCT scans via denoising. Using noisy images and corresponding high-quality images from one normal eye, Ma et al. built an image-to-image cGAN, enabling the competition of the generator and the discriminator to learn the underlying structure of the retina layers and to reduce OCT speckle noise. Despite the small training dataset, their model was capable of generalizing to images with low signal-to-noise (LSTN) ratio from pathological eyes and from different OCT scanners [54]. Similarly, using a small set of OCT images with high signal-to-noise (HSTN) ratio and LSTN ratio from the same eye of 28 patients, Kande et al. equipped a GAN model with Siamese network to generate denoised spectra-domain OCT images that are closer to the ground truth images with HSTN ratio. The discriminator was designed to fool the generator to produce a denoised image via enabling extraction of the discriminative features from the HSNR patch and denoised patch by passing them through a twin network [55].
The image synthesis function of GANs could also be applied for predicting the posttreatment OCT images of patients receiving antivascular endothelial growth factor (anti-VEGF). Utilizing 476 pairs of pre and posttherapeutic OCT images of patients with neovascular AMD (nAMD) who received anti-VEGF treatment, Liu et al. proposed an image-to-image GAN model to generate predicted posttherapeutic OCT images based on their pretherapeutic ones. Their GAN model achieved a sensitivity of 84% and specificity of 86% in predicting the posttreatment macular classification (wet or dry macula) [56■]. Following this, Lee et al. trained a cGAN model to predict posttreatment OCT images in patients with nAMD receiving anti-VEGF, using a larger dataset of 15,183 paired OCT B-scans from 241 patients [57]. This cGAN model was designed to predict the presence of four abnormal structures on posttreatment OCT, namely the intraretinal fluid, subretinal fluid, pigmented epithelial detachment, and subretinal hyperreflective material, with sensitivity and specificity ranging from 21.2 to 88.2% and 94.6 to 95.1%, respectively. The predictive performance was enhanced after adding fluorescein angiography (FA) and indocyanine green angiography images to the training datasets. As a result of the low sensitivity, the authors concluded that this model is not suitable to be used as a screening tool and further work with a dataset of more variations is warranted. Although GAN synthesized retinal images have overall consistent appearance, generating realistic images with pathological retinal lesions remains as a challenge [58].
Other image modalities
Recently, Tavakkoli et al. proposed a GAN model capable of producing FA from retinal fundus photographs, which was the first DL application to generate images from distinct modalities in ophthalmology [59■■]. Using pairs of FA and fundus images from 59 patients (30 with DR, 29 normal) as the training dataset, they designed a multiscale cGAN comprised of two generators and four discriminators. Their framework was able to produce FA images that are indistinguishable from real ones by three experts and are more accurate than the images produced by another two state-of-the-art cGAN models, as evidenced by significantly lower Fréchet Inception Distance and higher structural similarity measures. This cGAN technique may be a novel alternative to the invasive FA and the expensive OCT angiography with limited field of view. Furthermore, the ability to generate FA based on fundus photographs may improve the efficiency of tele-medicine, in particular during the COVID-19 pandemic when in-clinic examinations becomes challenging [59■■].
OUTCOME MEASURES FOR GENERATIVE ADVERSARIAL NETWORKS
Since GANs are generative models, the evaluation of their outputs – usually images – is not as straightforward as for discriminative models like classifiers, where the predicted label can simply be compared against the ground truth, in a supervised context. For GANs, the evaluation of their outputs may intuitively be based on human judgment of their ‘realness’, which in turn can be broadly considered in two aspects. Firstly, fidelity, in the sense that the generated samples are visually indistinguishable from real samples of the intended class. Fidelity in turn generally depends on various characteristics of the generated images, such as overall quality (being in-focus, etc.), demonstrating plausible object texture and structure, and so forth. Secondly, diversity, where the full range of variation of the intended class is generated. For example, if a GAN had been trained for cars, it would exhibit poor diversity if all images that it generated were of a particular car manufacturer or colour, despite the cars being otherwise realistic. When this happens, possibly due to the GAN’s generator fixating on some particularly plausible output, the GAN is said to undergo mode collapse. It may be possibly to influence the output class distribution of a GAN, by appropriately weighing its training data by class.
Many techniques have been proposed to evaluate GANs outputs, for both qualitative and quantitative measurements [60]. Qualitative methods may involve humans in inspecting and curating the generated images based on graders’ subjective decisions. This may be particularly appropriate in the biomedical domain, since it may not be easily articulated as to why some generated samples are not physiologically plausible. Quantitative methods on the other hand tend not to involve direct human intervention, and the most natural test would perhaps be through classifying the generated samples with a discriminative model trained on real samples. A popular generalization of this basic idea is the Inception Score (IS) [61], which uses an Inception v3 model pretrained on ImageNet, to classify a set of generated images. The IS was claimed by its developers to be highly correlated with human judgment. However, due to its reliance on the ImageNet dataset, it has been noted to be possibly inappropriate when applied to GANs trained on other datasets, being overly sensitive to network weights, and insensitive to prior distribution over labels [62,63].
The Fréchet Inception Distance [41] is perhaps among the most commonly used outcome metrics for GANs, and involves embedding generated images into a feature space expressed by an embedding layer of the Inception v3 model, estimating the mean and covariance for the embeddings of the generated and real data, and computing the Fréchet distance between these two Gaussians. This improves upon IS by being able to quantify mode dropping/collapse, but is still unable to recognize overfitting, as when the GANs reproduce samples from the training data [62]. To address this, qualitative measures such as comparing generated samples to their nearest neighbours in the training data, might be considered [60].
Additionally, generated synthetic images may be evaluated against the GAN training dataset of real images, to ensure that the synthetic images are not merely minimally adapted from the real data. Metrics such as the structural similarity index measure may be used to efficiently compare a sample of the generated data against the real images, in a pairwise manner [64].
ADVANTAGES AND DISADVANTAGES OF GENERATIVE ADVERSARIAL NETWORKS
GANs have gained recognition due to their various advantages over previous generative models. Although other unsupervised generative models share the advantage of not requiring initial human annotation, GANs differ from deep graphical models in that they do not require careful architectural design, and further do not rely on Markov chains for sampling, unlike generative autoencoders [17]. This simplifies and generalizes the modelling process, which may be convenient for researchers primarily interested in medical applications. Empirically, recent GAN architectures have been favoured over alternative generative models for the fidelity of their generated high-resolution images of at least 1024 × 1024 pixels, with higher resolutions likely achievable given advances in hardware and algorithms. Moreover, while GANs do not promise inference capabilities, techniques have been developed to explore their encoded latent space [65]. In the medical domain, GANs have been proposed to augment existing datasets and help preserve patient confidentiality through the generation of additional samples, especially of rarer conditions [66,67].
Nevertheless, GANs do still possess certain disadvantages. Other than the abovementioned possibility of mode collapse, GANs do not explicitly represent the generator’s distribution over the data, which is detrimental to model interpretability. However, this is also the case for other popular generative models. Also, GANs tend to be data intensive, in that the training data needs to sufficiently represent the desired underlying class. To give a concrete example, consider a GAN that is trained to generate ‘abnormal fundus images’, but its training data are almost entirely composed of DR samples, with a few maculopathy samples, and next to no glaucoma examples. In this case, it is very unlikely that the GAN will be able to generate an acceptable diversity of glaucomatous images. This might be addressed to an extent through human-in-the-loop training (Fig. 3), where human guidance is introduced to select acceptable synthetic data generated by the GAN, during the GAN training process. These selected synthetic images can then be used to augment both the training of the discriminator model, and the further fine-tuning of the GAN generator model itself.
FIGURE 3.
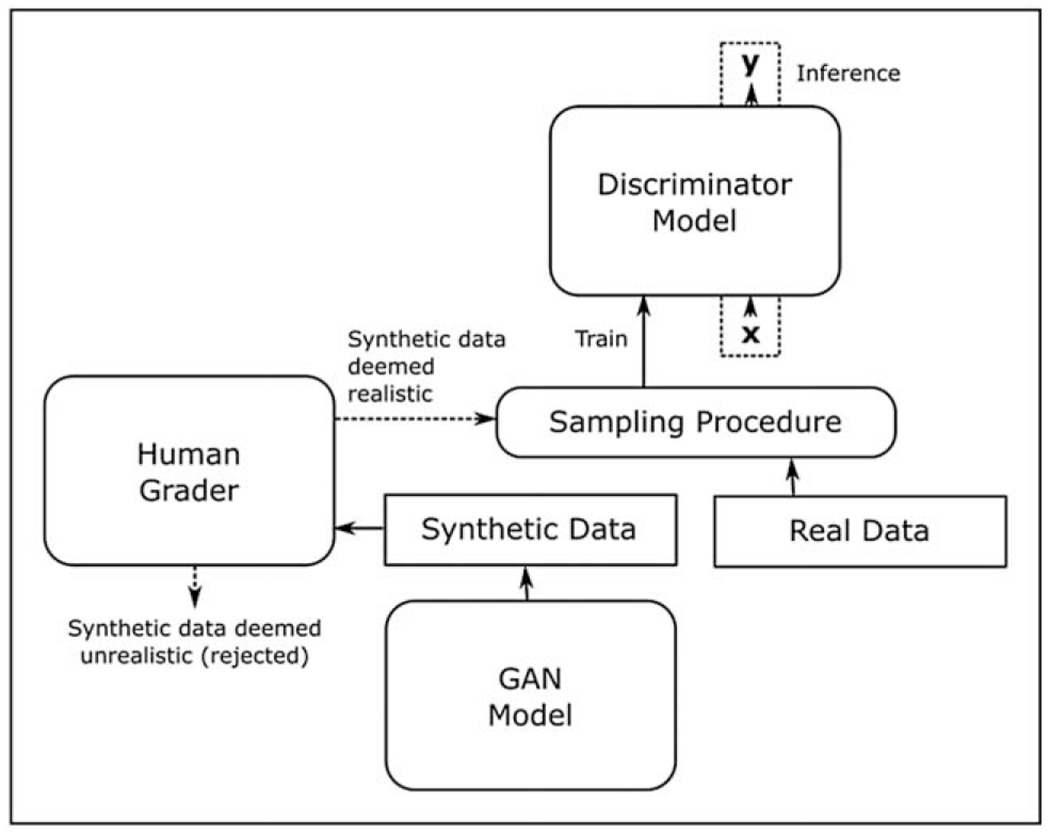
Human-in-the-loop training with a GAN. Human grader(s) arbitrate the generated synthetic data for realism, and the acceptable synthetic data is sampled together with real data to train an improved discriminator model. GAN, generative adversarial network.
CONCLUSION AND FUTURE DIRECTION
In conclusion, GANs offer a potential innovative solution to address a key challenge in the DL field for ophthalmology, and computational medicine in general: that of limited access to large datasets. Of all the functions of GANs, image synthesis is the most relevant and explored by biomedical research. In ophthalmology, GANs are mainly utilized for synthesizing fundus images and OCT images with and without pathology such as AMD and DR. Although the unmet need of artificial biomedical data generation is of great interest, DL solutions such as GANs still face many challenges in the retinal image synthesis field. First, the development of GANs is data intensive, but how much data is considered sufficient to train GANs remains unknown, and the amount is likely to be task dependent. Second, although Inception Score and Fréchet Inception Distance have been commonly used for quantitative measurement of GANs outputs, current qualitative measurement for the outputs of GANs mainly relies on the subjective judgement of human. Therefore, an objective scale to evaluate the quality of synthetic images such as the realness may be proposed for comparison among different GAN models. Lastly, to test whether GANs could really be the solution to limited access to datasets, future research is warranted to evaluate if GANs generated images could augment the development of DL systems and to test the performance of synthetic image trained DL systems using independent datasets.
KEY POINTS.
There is significant potential to generate a large number of images using GAN, for the training, development, validation and testing of new DL algorithms and systems.
In ophthalmology, GAN has mainly been used to synthesize fundus images and OCT images with and without pathology for the purpose of research.
The development of GAN models is data intensive and there is a lack of consensus on the evaluation of outputs produced by GAN.
Financial support and sponsorship
This work was supported by grants R01EY19474, R01 EY031331, R21 EY031883, and P30 EY10572 from the National Institutes of Health (Bethesda, MD), and by unrestricted departmental funding and a Career Development Award (JPC) from Research to Prevent Blindness (New York, NY).
Footnotes
Conflicts of interest
G.L., T.Y.W. and D.S.W.T. are the co-inventors of a deep learning system for retinal diseases. J.P.C. is a consultant to Boston AI. The remaining authors have no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 2.Abramoff MD, Lou Y, Erginay A, et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Investig Ophthalmol Vis Sci 2016; 57:5200–5206. [DOI] [PubMed] [Google Scholar]
- 3.Ting DSW, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017; 318:2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 2017; 124:962–969. [DOI] [PubMed] [Google Scholar]
- 5.Hood DC, De Moraes CG. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology 2018; 125:1207–1208. [DOI] [PubMed] [Google Scholar]
- 6.Burlina PM, Joshi N, Pekala M, et al. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol 2017; 135:1170–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grassmann F, Mengelkamp J, Brandl C, et al. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology 2018; 125:1410–1420. [DOI] [PubMed] [Google Scholar]
- 8.Brown JM, Campbell JP, Beers A, et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol 2018; 136:803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keel S, Lee PY, Scheetz J, et al. Feasibility and patient acceptability of a novel artificial intelligence-based screening model for diabetic retinopathy at endocrinology outpatient services: a pilot study. Sci Rep 2018; 8:4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhuiyan A, Govindaiah A, Deobhakta A, et al. Development and validation of an automated diabetic retinopathy screening tool for primary care setting. Diabetes Care 2020; 43:e147–e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ting DSW, Lee AY, Wong TY. An ophthalmologist’s guide to deciphering studies in artificial intelligence. Ophthalmology 2019; 126:1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Health Insurance Portability and Accountability Act of 1996 (HIPAA), PUBLIC LAW 104–191 (1996) § 264, 110 Stat.1936. [PubMed]
- 13.Office for Civil Rights (OCR), HIPAA for Professionals. U.S. Department of Health & Human Services. Available at: https://www.hhs.gov/hipaa/for-professionals/index.html. [Accessed 25 May 2021] [Google Scholar]
- 14.The L. Striking the right balance between privacy and public good. Lancet 2006; 367:275. [DOI] [PubMed] [Google Scholar]
- 15.Office for Civil Rights (OCR), Your Rights Under HIPAA. U.S. Department of Health & Human Services. Available at: https://www.hhs.gov/hipaa/for-individuals/guidance-materials-for-consumers/index.html. [Accessed 18 March 2021] [Google Scholar]
- 16.Informed consent for medical photographs.. Dysmorphology subcommittee of the clinical practice committee, american college of medical genetics. Genet Med 2000; 2:353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodfellow I, Pouget-Abadie J, Mirza M, et al. , editors. Generative Adversarial Networks. ArXiv 2014, abs/1406.2661. [Google Scholar]
- 18.Rezaei M, Yang H, Meinel C. Whole heart and great vessel segmentation with context-aware of generative adversarial networks. In: Maier A, Deserno T, Handels H, Maier-Hein K, Palm C, Tolxdorff T (eds). Bildverarbeitung für die Medizin 2018. Informatik aktuell. Berlin, Heidelberg: Springer Vieweg; 2018. 10.1007/978-3-662-56537-7_89. [DOI] [Google Scholar]
- 19.Madani A, Moradi M, Karargyris A, Syeda-Mahmood T, editors. Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018 4-7 April 2018. [Google Scholar]
- 20.Alex V, Safwan KPM, Chennamsetty SS, Krishnamurthi G. Generative adversarial networks for brain lesion detection. Proc. SPIE 10133, Medical Imaging 2017: Image Processing, 101330G. 10.1117/12.2254487 SPIE Medical Imaging, 2017, Orlando, Florida, United States. [DOI] [Google Scholar]
- 21.Yan P, Xu S, Rastinehad A, Wood B, editors. Adversarial image registration with application for MR and TRUS image fusion. arXiv e-prints 2018; 1804.11024. [Google Scholar]
- 22.Wolterink JM, Leiner T, Viergever MA, Išgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging 2017; 36:2536–2545. [DOI] [PubMed] [Google Scholar]
- 23.Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Analysis 2019; 58:101552. [DOI] [PubMed] [Google Scholar]
- 24.Fujioka T, Mori M, Kubota K, et al. Breast ultrasound image synthesis using deep convolutional generative adversarial networks. Diagnostics (Basel) 2019; 9:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan S, Loew M. Breast cancer detection using synthetic mammograms from generative adversarial networks in convolutional neural networks. J Med Imaging 2019; 6:031411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baydoun A, Xu KE, Heo JU, et al. Synthetic CT generation of the pelvis in patients with cervical cancer: a single input approach using generative adversarial network. IEEE Access 2021; 9:17208–17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Chen H, Loew M, Ko H. COVID-19 CT image synthesis with a conditional generative adversarial network. IEEE J Biomed Health Inform 2021; 25:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Meng L, Zhong J. MAGAN: mask attention generative adversarial network for liver tumor CT image synthesis. J Healthc Eng 2021; 2021:6675259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toda R, Teramoto A, Tsujimoto M, et al. Synthetic CT image generation of shape-controlled lung cancer using semi-conditional InfoGAN and its applicability for type classification. Int J Comput Assist Radiol Surg 2021; 16:241–251. [DOI] [PubMed] [Google Scholar]
- 30.Berṁ dez C, Plassard A, Davis LT, Newton A, Resnick S, Landman B, editors. Learning implicit brain MRI manifolds with deep learning. Medical Imaging; 2018; 10574, doi: 10.1117/12.2293515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine AB, Peng J, Farnell D, et al. Synthesis of diagnostic quality cancer pathology images by generative adversarial networks. J Pathol 2020; 252:178–188. [DOI] [PubMed] [Google Scholar]
- 32.Xu C, Xu L, Ohorodnyk P, et al. Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal GANs. Med Image Anal 2020; 62:101668. [DOI] [PubMed] [Google Scholar]
- 33.Jiao J, Namburete AIL, Papageorghiou AT, Noble JA. Self-supervised ultrasound to MRI fetal brain image synthesis. IEEE Trans Med Imaging 2020; 39:4413–4424. [DOI] [PubMed] [Google Scholar]
- 34.Jin C-B, Kim H, Liu M, et al. Deep CT to MR synthesis using paired and unpaired data. Sensors 2019; 19:2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Han IH, Kim DH, et al. Spine computed tomography to magnetic resonance image synthesis using generative adversarial networks: a preliminary study. J Korean Neurosurg Soc 2020; 63:386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gui J, Sun Z, Wen Y, et al. a review on generative adversarial networks: algorithms, theory, and applications. ArXiv 2020. abs/2001.06937. [Epub ahead of print] [Google Scholar]
- 37.Mirza M, Osindero S. Conditional generative adversarial nets. ArXiv 2014. abs/1411.1784. [Epub ahead of print] [Google Scholar]
- 38.Arjovsky M, Chintala S, Bottou L. Wasserstein Generative Adversarial Networks. In: Doina P, Yee Whye T, editors. Proceedings of the 34th International Conference on Machine Learning. Sydney, Australia: Proceedings of Machine Learning Research: PMLR; 2017. pp. 214–223. [Google Scholar]
- 39.Mao X, Li Q, Xie H, et al. Least squares generative adversarial networks. ArXiv e-prints 2017: 1611.04076 IEEE Int Conf Comput Vis (ICCV) 2017. [Google Scholar]
- 40.Li C, Wand M. Precomputed real-time texture synthesis with Markovian generative adversarial networks. ArXiv 2016. abs/1604.04382. [Epub ahead of print] [Google Scholar]
- 41.Heusel M, Ramsauer H, Unterthiner T, Nessler B, Hochreiter S, editors. GANs trained by a two time-scale update rule converge to a local nash equilibrium. NIPS; 2017. arXiv e-prints:1706.08500. [Google Scholar]
- 42.Karras T, Aila T, Laine S, Lehtinen J. Progressive growing of GANs for improved quality, stability, and variation. ArXiv 2018. abs/1710.10196. [Google Scholar]
- 43.Brock A, Donahue J, Simonyan K. Large scale GAN training for high fidelity natural image synthesis. ArXiv 2019. abs/1809.11096. [Epub ahead of print] [Google Scholar]
- 44.Karras T, Laine S, Aila T. A style-based generator architecture for generative adversarial networks. 2019 IEEE/CVF Conf Comput Vis Pattern Recognit 2019; 4396–4405. [DOI] [PubMed] [Google Scholar]
- 45.Costa P, Galdran A, Meyer MI, et al. End-to-end adversarial retinal image synthesis. IEEE Trans Med Imaging 2018; 37:781–791. [DOI] [PubMed] [Google Scholar]
- 46.Guibas JT, Virdi TS, Li P. Synthetic medical images from dual generative adversarial networks. ArXiv 2017. abs/1709.01872. [Epub ahead of print] [Google Scholar]
- 47.Zhao H, Li H, Maurer-Stroh S, Cheng L. Synthesizing retinal and neuronal images with generative adversarial nets. Med Image Anal 2018; 49:14–26. [DOI] [PubMed] [Google Scholar]
- 48.Yu Z, Xiang Q, Meng J, et al. Retinal image synthesis from multiple-landmarks input with generative adversarial networks. Biomed Eng Online 2019; 18:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burlina PM, Joshi N, Pacheco KD, et al. Assessment of deep generative models for high-resolution synthetic retinal image generation of age-related macular degeneration. JAMA Ophthalmol 2019; 137:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diaz-Pinto A, Colomer A, Naranjo V, et al. Retinal image synthesis and semi-supervised learning for glaucoma assessment. IEEE Trans Med Imaging 2019; 38:2211–2218. [DOI] [PubMed] [Google Scholar]
- 51.■.Zhou Y, Wang B, He X, et al. DR-GAN: conditional generative adversarial network for fine-grained lesion synthesis on diabetic retinopathy images. IEEE J Biomed Health Inform 2020. 1–1. doi: 10.1109/jbhi.2020.304547. [DOI] [PubMed] [Google Scholar]; This study reported a DR fundus photo generator that can be directly deployed to train DR classifier, which is an example of how to incorporate GAN synthesized images into the development of a DL system.
- 52.Beers A, Brown J, Chang K, et al. High-resolution medical image synthesis using progressively grown generative adversarial networks. ArXiv 2018; 1805.03144. [Google Scholar]
- 53.■.Zheng C, Xie X, Zhou K, et al. Assessment of generative adversarial networks model for synthetic optical coherence tomography images of retinal disorders. Transl Vis Sci Technol 2020; 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that DL framework trained on GAN synthesized OCT images achieved noninferior performance in classifying urgent and nonurgent referrals, comparing to the model trained on real OCT images.
- 54.Ma Y, Chen X, Zhu W, et al. Speckle noise reduction in optical coherence tomography images based on edge-sensitive cGAN. Biomed Opt Express 2018; 9:5129–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kande NA, Dakhane R, Dukkipati A, Yalavarthy PK. SiameseGAN: a generative model for denoising of spectral domain optical coherence tomography images. IEEE Trans Med Imaging 2021; 40:180–192. [DOI] [PubMed] [Google Scholar]
- 56.■.Liu Y, Yang J, Zhou Y, et al. Prediction of OCT images of short-term response to anti-VEGF treatment for neovascular age-related macular degeneration using generative adversarial network. Br J Ophthalmol 2020; 104:1735–1740. [DOI] [PubMed] [Google Scholar]; This study showed that an image-to-image GAN model could predict the posttherapeutic OCT images based on their pretherapeutic ones with satisfactory sensitivity and specificity.
- 57.Lee H, Kim S, Kim MA, et al. Post-treatment prediction of optical coherence tomography using a conditional generative adversarial network in age-related macular degeneration. Retina 2021; 41:572–580. [DOI] [PubMed] [Google Scholar]
- 58.Bellemo V, Burlina P, Yong L, Wong TY, Ting DSW, editors. Generative Adversarial Networks (GANs) for Retinal Fundus Image Synthesis. Cham: Springer International Publishing; 2019. [Google Scholar]
- 59.■■.Tavakkoli A, Kamran SA, Hossain KF, Zuckerbrod SL. A novel deep learning conditional generative adversarial network for producing angiography images from retinal fundus photographs. Sci Rep 2020; 10:21580. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study was the first DL application to generate images from distinct modalities in ophthalmology, which showed that a GAN model was capable of producing fluorescein angiography (FA) from retinal fundus photographs.
- 60.Borji A. Pros and cons of GAN evaluation measures. Comput Vis Image Understand 2019; 179:41–65. [Google Scholar]
- 61.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X, editors. Improved techniques for training GANs. NIPS; 2016. arXiv e-prints: 1606.03498. [Google Scholar]
- 62.Barratt ST, Sharma R. A note on the inception score. ArXiv. 2018; arXiv e-prints 1801.01973. [Google Scholar]
- 63.Lucic M, Kurach K, Michalski M, Gelly S, Bousquet O, editors. Are GANs created equal? A large-scale study. NeurIPS; 2018. ArXiv e-prints:1711.10337. [Google Scholar]
- 64.Zhou W, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004; 13:600–612. [DOI] [PubMed] [Google Scholar]
- 65.Abdal R, Qin Y, Wonka P. Image2StyleGAN: How to Embed Images Into the StyleGAN Latent Space?. Seoul, Korea: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019. 4431–40. [Google Scholar]
- 66.Lim G, Bellemo V, Xie Y, et al. Different fundus imaging modalities and technical factors in AI screening for diabetic retinopathy: a review. Eye and Vis 2020; 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qasim AB, Ezhov I, Shit S, et al. Red-GAN: Attacking class imbalance via conditioned generation. Yet another medical imaging perspective. In: Tal A, Ismail Ben A, Marleen de B, Maxime D, Herve L, Christopher P., editors. Proceedings of the Third Conference on Medical Imaging with Deep Learning; Proceedings of Machine Learning Research: PMLR; 2020. pp. 655–68. [Google Scholar]


