Abstract
Aim
Interleukin 6 (IL-6) is considered to play a role in the dysbiotic host response in the development of periodontitis. While the inhibition of the IL-6 receptor using monoclonal antibodies is a well-established therapy for some diseases, so far, its potential benefit in patients with periodontitis has not been examined. We tested the association of genetically proxied downregulation of IL-6 signaling with periodontitis to explore whether downregulation of IL-6 signaling could represent a viable treatment target for periodontitis,
Materials and methods
As proxies for IL-6 signaling downregulation, we selected 52 genetic variants in close vicinity of the gene encoding IL-6 receptor that were associated with lower circulating C-reactive protein (CRP) levels in a genome-wide association study (GWAS) of 575 531 participants of European ancestry from the UK Biobank and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Associations with periodontitis were tested with inverse-variance weighted Mendelian randomization in a study of 17 353 cases and 28 210 controls of European descent in the Gene-Lifestyle Interactions in Dental Endpoints (GLIDE) consortium. In addition, the effect of CRP reduction independent of the IL-6 pathway was assessed.
Results
Genetically proxied downregulation of IL-6 signaling was associated with lower odds of periodontitis (odds ratio (OR) = 0.81 per 1-unit decrement in log-CRP levels; 95% confidence interval (CI): [0.66;0.99]; P = 0.0497). Genetically proxied reduction of CRP independent of the IL-6 pathway had a similar effect (OR = 0.81; 95% CI: [0.68; 0.98]; P = 0.0296).
Conclusion
In conclusion, genetically proxied downregulation of IL-6 signaling was associated with lower odds of periodontitis and CRP might be a causal target for the effect of IL-6 on the risk of periodontitis.
Keywords: periodontal disease(s)/periodontitis, cytokine(s), host modulation therapy, epidemiology, cell signaling
Introduction
Periodontal inflammation is initiated by biofilm formation on the tooth surface and further exacerbated by host inflammatory-immune response to dysbiotic microbiome (1, 2). Hence, inflammation control is a central part of periodontitis treatment (3–5). Drugs targeting host response in addition to subgingival scaling and root planing or surgical periodontal therapy are therefore under active investigation (6).
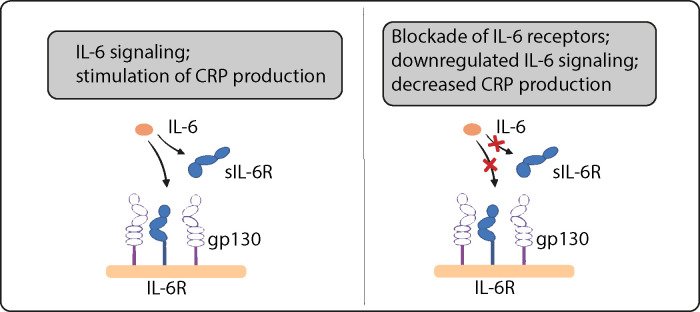
Cytokine balance plays a central role in inflammation (7). Interleukin 6 (IL-6) is a cytokine involved in the body’s innate immune response. It acts on two different signaling pathways: by a specific membrane-bound IL-6 receptor (IL-6R) and by a soluble receptor (sIL-6R), which are called classic and trans-signaling. The inflammatory role of IL-6 is thereby mediated not by the classic pathway, but rather via the trans-signaling pathway where IL-6 binds to sIL-6R and subsequently interacts with cells via the membrane bound signaling molecule gp130 (8, 9). Here, sIL-6R is released from inflammatory cells such as lymphocytes and macrophages by proteolytic cleavage on the cell surface (10).
Inhibition of both membrane and soluble IL-6R using monoclonal antibodies such as tocilizumab, satralizumab, sarilumab or olokizumab is a standard therapy for patients with rheumatoid arthritis or COVID-19 (11, 12). These agents attenuate all forms of IL-6 signaling, reducing serum levels of C-reactive protein (CRP) and other downstream biomarkers.
In this study, we tested the biological effect of IL-6 signaling downregulation on the risk of periodontitis simulating the effect of monoclonal antibodies that target IL-6R by blocking both IL-6 classic and trans-signaling (13–16). Mendelian randomization (MR) is a method to study causal effects on disease outcomes that uses randomly allocated germline genetic variants as proxies for exposures (17). This approach is relatively robust to confounding and reverse causality as these variants stay unchanged after conception. Drug-target MR uses variants in the close vicinity of a gene encoding a protein target as proxies to study causal effects on disease outcomes ( Figure 1 ) (19, 20). Accordingly, genetic variants in the vicinity of the IL6R gene serve as proxies for the targeted pharmacological agent.
Figure 1.
Genomic map for the selected 52 SNPs in the vicinity of the IL6R gene. This figure shows the location of the instrumental SNPs around the IL6R gene on chromosome 1. Variants in the close vicinity of the IL6R gene were used to proxy downregulation of IL-6 signaling and to estimate its genetically proxied effect on periodontitis. Variants associated with CRP based on P<5x10-8 within 300 kilobases of the IL6R gene were selected as genetic proxies of IL-6 signaling downregulation. At the top, chromosome 1 is drawn with the subregion of interest marked in red. The ‘IL6R’ track shows the combined gene model of the alternative transcripts of the IL6R gene. At the bottom, the SNP locations are plotted along the same genomic coordinate. The figure was created using the mapsnp package (18).
Materials and methods
Drug target MR (or cis-MR) is a study design that leverages large genetic data to estimate the effect of drug therapy in the absence of evidence from randomized clinical trials (RCTs). While estimates from other observational study designs are often biased by unmeasured confounding, MR uses genetic instrumental variables to imitate the randomization of RCTs to yield unconfounded effect estimates and has recently been introduced to the field of periodontology (6, 21). For an unbiased effect estimation, three instrumental variable assumptions must be true: The relevance assumption demands a strong association between genetic instruments and the exposure. This assumption can be tested using statistical approaches such as F statistics. The exchangeability assumption demands that genetic instruments are independent of confounders of the exposure-outcome relation. The exclusion restriction assumption finally demands that genetic instruments affect the outcome only via the exposure.
Genetic instruments and study population
Single nucleotide polymorphisms (SNPs) were used to proxy the effect of IL-6 signaling downregulation and to estimate the downstream effect of drugs for blocking IL-6Rs, such as tocilizumab, on periodontitis risk. We identified SNPs within 300 kilobases on either side of the targeted IL6R gene that were associated with CRP serum levels, a reliable downstream biomarker of IL-6 signaling (22) ( Table 1 , Figure 1 ). We assumed that these SNPs directly affect IL6R leading to a modified version or altered quantities of the protein ( Figure 2 ). Variants were robustly associated with CRP in a genome-wide association study (GWAS) meta-analysis of the combined data of 575 531 participants of European descent of the UK Biobank and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium (23). All variants had a p-value <5e-8 for their association with CRP and low linkage disequilibrium (r²<0.2). We estimated the effect of CRP-associated variants on the risk of periodontitis using the GWAS of 17 353 cases and 28 210 controls of European descent from the Gene-Lifestyle Interactions in Dental Endpoints (GLIDE) consortium (24). Periodontitis cases were classified by either the Centers for Disease and Control and Prevention/American Academy of Periodontology (CDC/AAP) or the Community Periodontal Index (CPI) case definition.
Table 1.
Summary of CRP associated variants in IL6R.
| SNP | EA | OA | EAF | BETA | SE | P-value | F statistic |
|---|---|---|---|---|---|---|---|
| rs10796927 | T | C | 0.729 | 0.040 | 0.002 | 2.5e-71 | 330.6 |
| rs111879666 | T | C | 0.039 | -0.055 | 0.005 | 1.5e-28 | 122.3 |
| rs112203594 | A | C | 0.013 | 0.038 | 0.007 | 2.4e-08 | 31.3 |
| rs112505856 | T | C | 0.052 | -0.041 | 0.006 | 1.2e-11 | 46.1 |
| rs11264239 | A | G | 0.057 | -0.038 | 0.005 | 1.3e-16 | 67.9 |
| rs113580743 | A | G | 0.038 | 0.051 | 0.005 | 1.4e-21 | 90.8 |
| rs116037345 | T | C | 0.043 | 0.036 | 0.006 | 2.5e-09 | 35.6 |
| rs116059394 | A | G | 0.951 | -0.046 | 0.004 | 2.5e-24 | 102.7 |
| rs116141616 | A | G | 0.018 | 0.037 | 0.006 | 9.5e-09 | 33.1 |
| rs11811450 | A | G | 0.913 | -0.022 | 0.004 | 2.7e-10 | 39.1 |
| rs12083537 | A | G | 0.807 | 0.066 | 0.002 | 3.1e-159 | 749.4 |
| rs12406117 | A | G | 0.587 | 0.013 | 0.002 | 6.5e-11 | 44.2 |
| rs12726220 | A | G | 0.927 | 0.025 | 0.004 | 1.3e-08 | 32.1 |
| rs12735458 | A | G | 0.987 | 0.077 | 0.009 | 1.1e-18 | 77.2 |
| rs12750774 | A | G | 0.308 | -0.063 | 0.002 | 2.0e-191 | 902.9 |
| rs139364224 | T | C | 0.979 | -0.060 | 0.008 | 5.9e-15 | 61.5 |
| rs139952834 | T | C | 0.029 | 0.064 | 0.009 | 3.3e-12 | 48.1 |
| rs142768042 | T | C | 0.020 | 0.040 | 0.006 | 4.7e-11 | 43.4 |
| rs144671207 | A | G | 0.018 | -0.056 | 0.009 | 1.6e-09 | 36.1 |
| rs145262901 | A | G | 0.018 | -0.057 | 0.010 | 7.0e-09 | 33.4 |
| rs145909430 | T | C | 0.987 | 0.103 | 0.008 | 2.3e-39 | 174.0 |
| rs147830103 | A | G | 0.041 | 0.043 | 0.007 | 7.1e-10 | 38.3 |
| rs16835819 | T | C | 0.989 | 0.080 | 0.009 | 1.4e-20 | 85.6 |
| rs1760798 | T | C | 0.769 | -0.018 | 0.002 | 2.9e-14 | 59.9 |
| rs183641528 | A | G | 0.013 | -0.080 | 0.008 | 7.3e-26 | 111.6 |
| rs1889312 | A | G | 0.428 | -0.022 | 0.002 | 6.5e-28 | 124.3 |
| rs3738028 | A | C | 0.560 | 0.015 | 0.002 | 1.2e-11 | 47.7 |
| rs3766925 | A | T | 0.256 | -0.015 | 0.002 | 6.4e-11 | 44.3 |
| rs3766926 | T | C | 0.805 | -0.046 | 0.002 | 1.0e-78 | 365.8 |
| rs41269913 | T | C | 0.045 | -0.040 | 0.005 | 2.3e-13 | 53.7 |
| rs4133213 | A | C | 0.393 | -0.085 | 0.002 | 1.0e-200 | 1,802.0 |
| rs4509570 | C | G | 0.720 | -0.048 | 0.002 | 9.3e-94 | 437.4 |
| rs4845626 | T | G | 0.187 | 0.052 | 0.003 | 1.4e-85 | 398.5 |
| rs4845663 | T | C | 0.592 | -0.013 | 0.002 | 4.5e-10 | 40.3 |
| rs55676222 | A | T | 0.914 | 0.066 | 0.004 | 2.0e-71 | 313.4 |
| rs56100876 | A | G | 0.018 | -0.106 | 0.008 | 5.4e-39 | 172.5 |
| rs61806853 | T | C | 0.962 | 0.044 | 0.005 | 3.8e-20 | 84.0 |
| rs61811421 | T | C | 0.259 | 0.038 | 0.002 | 2.0e-53 | 245.4 |
| rs67156297 | A | G | 0.250 | 0.031 | 0.002 | 2.7e-43 | 197.3 |
| rs72698179 | A | C | 0.037 | 0.056 | 0.006 | 1.5e-18 | 77.6 |
| rs7523010 | A | T | 0.249 | 0.024 | 0.003 | 1.2e-19 | 85.2 |
| rs7525477 | A | G | 0.480 | 0.029 | 0.002 | 2.6e-43 | 197.3 |
| rs75456865 | A | T | 0.978 | -0.044 | 0.005 | 6.9e-17 | 69.5 |
| rs76289529 | T | C | 0.033 | -0.052 | 0.006 | 6.4e-20 | 83.5 |
| rs77994623 | T | C | 0.136 | 0.046 | 0.003 | 1.6e-61 | 284.0 |
| rs78739139 | A | G | 0.062 | -0.050 | 0.005 | 1.0e-27 | 118.1 |
| rs79438587 | T | C | 0.160 | -0.018 | 0.003 | 1.4e-10 | 39.8 |
| rs79505546 | T | C | 0.011 | -0.059 | 0.009 | 3.9e-11 | 43.3 |
| rs79778789 | A | G | 0.986 | 0.053 | 0.008 | 4.4e-11 | 43.9 |
| rs79794939 | T | C | 0.073 | 0.035 | 0.004 | 9.2e-20 | 81.5 |
| rs8192484 | A | T | 0.973 | -0.041 | 0.007 | 1.3e-08 | 32.6 |
| rs9427092 | T | C | 0.817 | -0.029 | 0.002 | 1.8e-30 | 136.4 |
EA, effect allele. OA, other allele. EAF, effect allele frequency. SE, standard error
Figure 2.
IL-6 signaling. Variants in the IL6R gene act as proxies for downregulation of IL-6 signaling. SNPs in this region that are strongly associated with CRP are assumed to act via modifying IL-6 receptors.
Additional analysis
Our primary analysis includes only variants in the vicinity of the IL6R gene (cis IL6R) acting as a proxy for the target drug effect that contribute to the effect estimate weighted by their effect on CRP. This way we obtain an adequate estimate of IL6R function and its effect on downstream IL-6 signaling. To further examine whether CRP is a potential causal target or merely an otherwise uninvolved marker of IL-6R inhibition we performed an additional analysis using four variants located in the CRP gene (cis CRP) that have previously been selected to fully cover the common variations of this gene in populations of European descent (25). These variants are supposed to alter CRP independent of IL-6 and thus might indicate a causal effect of CRP on the risk of periodontitis.
Statistical analysis
The direction of genetic associations between exposure and outcome was harmonized. We performed the MR analysis using the well-documented multiplicative random-effects inverse variance weighted (IVW) method (26). Odds ratios (ORs) were scaled to reflect the equivalent of a 1-unit reduction in serum CRP levels on the log scale. The instrumental variable relevance assumption was tested by the F statistic (17), and F statistics of at least 10 were regarded as an indication of no weak instrument bias. Leave-one-out analysis was used to assess whether the estimate was biased by a single outlier instrument. The exclusion restriction instrumental variable assumption was probed by searching the PhenoScanner database for associations of SNPs with known risk factors of periodontitis, especially tobacco smoking and diabetes (27). Any association between a genetic instrument and a risk factor for periodontitis would hint at horizontal pleiotropy, a violation of the exclusion restriction assumption (17). In this study the risk of violating the exchangeability or exclusion restriction assumption was substantially reduced as the genetic instruments stemmed from the close vicinity of the coding IL6R gene and variants were therefore acting in cis (19, 20). We analyzed the data using R version 4.2.1 (R Foundation for Statistical Computing) using the MendelianRandomization and TwoSampleMR packages and designed the study according to STROBE-MR (28).
Results
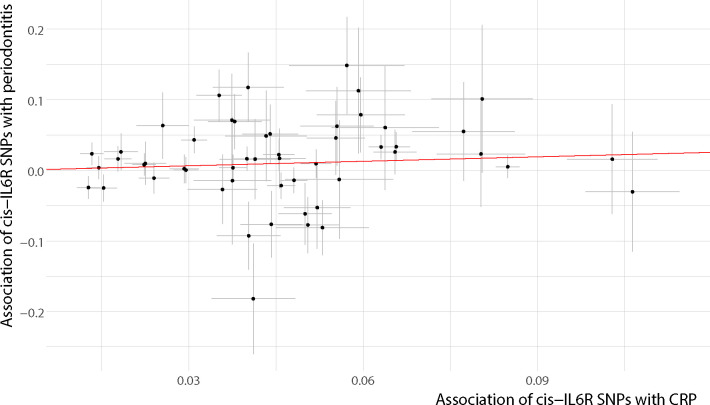
Characteristics of genetic variants in the IL6R gene region used to proxy IL-6R antagonists are presented in Table 1 . In brief, 52 SNPs in the vicinity of IL6R (cis IL6R) were used to proxy IL-6 signaling downregulation. F statistics for the genetic instruments ranged from 31 to 1 802, suggesting no weak instrument bias. Genetically proxied IL-6 signaling downregulation was associated with a reduced odds of periodontitis (OR per 1-unit decrement in log-CRP levels = 0.81; 95% confidence interval (CI): [0.66; 0.99]; P = 0.0497) in the inverse variance weighted (multiplicative random effects) regression ( Table 2 , Figure 3 ). No highly influential leverage points were identified in leave-one-out analyses ( Appendix Table 1 ). Searching for associations of SNPs with known risk factors of periodontitis, we found no evidence for an association of any instrument with smoking or diabetes.
Table 2.
Genetically proxied downregulation of IL-6 signaling and CRP reduction.
| Target | OR | (95% CI) | P-value |
|---|---|---|---|
| cis IL6R | 0.81 | (0.66;0.99) | 0.0497 |
| cis CRP | 0.81 | (0.68;0.98) | 0.0296 |
OR (odds ratio) representing the change in odds of periodontitis per genetically proxied inhibition of drug target equivalent to 1 unit decrease in serum CRP on the log scale; CI, confidence interval; cis IL6R: variants selected from the vicinity of the IL6R gene; cis CRP: variants selected from the vicinity of the CRP gene
Figure 3.
Inverse variance weighted (multiplicative random effects) regression. Instrumental SNPs are plotted with error bars indicating the respective standard errors. The slope of the regression line equals the logarithm of the effect estimate. For better graphical display associations were harmonized to lie on the positive x-axis. SNP, single-nucleotide polymorphism; CRP, c-reactive protein.
The additional analysis, which used variants from the vicinity of the CRP gene (cis CRP), yielded a similar effect of CRP reduction on the risk of periodontitis (OR = 0.81; 95% CI: [0.68; 0.98]; P = 0.0296) ( Table 2 ).
Discussion
In this study, we assessed the potential benefit of inhibiting IL-6 signaling to reduce the risk of periodontitis using the principle of instrumental variable estimation. We used genetic variants in the close vicinity of the IL6R gene to proxy the effect of inhibiting IL-6R. Our results suggest that intervening on this target through pharmacotherapy might support the prevention and treatment of periodontal disease. While we leveraged CRP as a downstream biomarker of IL-6 signaling downregulation, an additional analysis using genetic variants in the CRP gene suggests that CRP might be a substantial causal mediator downstream of IL-6.
Patients with periodontitis have been observed with increased levels of IL-6 in serum (29), saliva (29), and gingival crevicular fluid (30). Similarly, a study found increased concentrations of sIL-6R in the gingival crevicular fluid of periodontitis patients and showed in vitro that calprotectin induced sIL-6R production in THP-1 macrophages might be responsible for this association (31). Therefore, patients with periodontitis might be especially susceptible to an excess of IL-6. In contrast, IL-6 secretion in periodontal ligament fibroblasts was suppressed by overexpression of microRNA (miR)-146a in the presence of porphyromonas gingivalis (32) and several studies suggest that expression levels of miR-146a and transglutaminases might similarly be associated with IL-6 expression in inflamed periodontal ligament (33, 34). The continuous observation of IL-6 levels in monkeys with ligature-induced periodontitis revealed that IL-6 occurred as a response to acute initial periodontitis, but remained low in the progression and resolution phases of the disease (35).
Beyond that, the involvement of IL-6 in the development of periodontitis is well recognized (36). IL-6 has pleiotropic effects on lymphocyte promotion and tissue destruction, predominantly mediating B cell activation. Building an IL-6-IL-6R-gp130 complex, IL-6 induces Janus kinase (JAK) to mediate the phosphorylation of signal transducer and activator of transcription 3 (STAT3) and the formation of phosphorylated STAT3 homodimers. Via JAK-STAT3 signaling the expression of IL-6-responsive genes such as suppressor of cytokine signaling 1 (SOCS1) and SOCS3 is upregulated. JAK phosphorylates the cytoplasmic domain of gp130 at tyrosine 759, the binding site of SH2 domain tyrosine phosphatase 2 (SHP2), enforcing the mitogen-activated protein kinase (MAPK) pathway (37). Besides that, IL-6 increased osteocyte-mediated osteoclastic differentiation by activating JAK2 and Receptor activator of nuclear factor-κB ligand (RANKL) in vitro (38).
IL-6R is a popular drug target for the treatment of inflammatory diseases. Blockade of IL-6R with monoclonal antibodies such as tocilizumab is a long-established treatment of rheumatoid arthritis to reduce systemic and articular inflammation. Rheumatoid arthritis and periodontitis are known to be directly associated (39) and a study of 55 patients diagnosed with rheumatoid arthritis and chronic periodontitis first showed improved periodontal conditions after 20 months of medication with Il-6R antagonists (40). Similar studies observed improvements of the periodontal status after only 6 months of treatment (41, 42) suggesting potential benefits of even short term intake of IL-6R antagonists. So far, no study examined the effect of IL-6R antagonists on periodontitis in patients without rheumatoid arthritis. This might be caused by fears of potential adverse effects on immunity, which, however, might be less serious when the drugs are administered locally than when administered systemically (4).
Our analytical approach is novel in dentistry, but well-established in other fields of medicine. Notably, the potential benefit of blocking IL-6R for prevention of coronary heart disease was among the first major applications of the drug target Mendelian randomization approach (43) initiating further research and finally leading to the development of ziltivekimab, a novel IL-6R inhibiting drug specifically for use in atherosclerotic disease (44).
We selected genetic instruments based on their association with CRP instead of circulating IL-6. Blockade of IL-6R would inhibit the transfer of IL-6 into cells and lead to an accumulation of IL-6 in the circulatory system. Therefore, genetic variants in the IL6R gene region that are associated with increased circulating levels of IL-6 would rather indicate reduced IL-6 signaling, leading to opposite directions of association with disease outcomes to those expected based on serum IL-6 measurements. Instead, CRP is a direct indicator of IL-6 signaling (22), and according to our analysis a possible causal mediator for the risk of periodontitis.
CRP is mainly synthesized by hepatocytes in the liver in response to inflammation and tissue damage, but it can also be produced locally by arterial tissue. After binding to polysaccharides, CRP activates the classical complement pathway and prepares ligands for phagocytosis. While CRP serum levels are routinely used to indicate systemic inflammation and nonsurgical periodontal treatment consistently lead to reduced serum CRP levels (45), less is known about the causal role of inflammation markers in general, and CRP in specific, in the pathogenesis of periodontitis (46). While our analyses suggest a causal role of CRP in the pathogenesis of periodontitis, this finding was rather unexpected, and it remains to be seen whether CRP is a suitable target to break the vicious feed-forward loop linking periodontitis to systemic low-grade inflammation.
The drug target MR approach has some limitations. First, the method models a linear association of genetically proxied IL6R inhibition around the observed mean of CRP and is not able to estimate the effect at extremes of the distribution. Second, the analyses did not account for anti-inflammatory drug use. Third, the result is foremost to be interpreted as a test of the causal association. The actual size of risk lowering achieved through IL-6R inhibitors cannot reliably be predicted in this kind of analysis. Drug target MR effect estimates rather correspond to continuous long-term modulation of drug targets resembling preventive intake. Drug intake as therapy in an acute phase of periodontitis over a relatively short period of time might therefore yield different treatment effects. Fourth, our analyses used data of patients with European ancestry and may not directly be transferable to populations of different heritage. Fifth, the instrumental variable analysis relies on certain unverifiable assumptions, namely exchangeability and exclusion restriction. However, for studies targeting a protein the biasing effect of horizontal pleiotropy should be minimal. Finally, we recognize that effects on health outcomes are mediated by classic and trans signaling in differing ways, and that our genetic instruments may not act in the same way as IL-6R antagonists or other approaches to downregulate IL-6, for example by intervening on miR-146a expression in periodontal tissue.
Conclusion
In conclusion, we found genetic evidence for a reduced risk of periodontitis through downregulation of IL-6 signaling. Our results identified IL-6R as a potential drug target to prevent the development of periodontitis and as a host modulating adjunctive periodontitis therapy. While there is some clinical evidence to support our findings, the effect of IL-6R antagonists should be investigated in more detail. Further genetic studies could help dissect the downstream effect of IL-6 signaling on periodontitis. Beyond that, our study highlights the benefit of leveraging genetic data to investigate drug repurposing and adverse effects in dentistry.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: CRP GWAS data are accessible via the GWAS catalog (https://www.ebi.ac.uk/gwas/) under accession ID GCST90029070. Periodontitis GWAS data are available at https://data.bris.ac.uk/data/dataset/2j2rqgzedxlq02oqbb4vmycnc2.
Ethics statement
The individual studies had previously obtained relevant ethical approval and participant consent. This study complied with all relevant ethical regulations, including the Declaration of Helsinki, and ethical approval for data collection and analysis was obtained by each study from local boards as described in the included GWAS.
Author contributions
MN contributed to conception and design, development of methodology, data acquisition, analysis, interpretation of data, drafted, and critically revised the manuscript. ZA and SR contributed to data analysis and interpretation of data and critically revised the manuscript. TK and BE contributed to interpretation of data and critically revised the manuscript. BH contributed to conception, design, and interpretation of data, and critically revised the manuscript. HB contributed to conception, design, development of methodology, and interpretation of data, and critically revised the manuscript. MG contributed to conception and design, data analysis and interpretation of data, and critically revised the manuscript. SEB contributed to conception, design, development of methodology, analysis, and interpretation of data, and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors acknowledge and thank the investigators of the original GWAS studies for sharing summary data used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1160148/full#supplementary-material
References
- 1. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol (2018) 1612:745–59. doi: 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Dyke TE, Sima C. Understanding resolution of inflammation in periodontal diseases: is chronic inflammatory periodontitis a failure to resolve? Periodontology 2000 (2020) 821:205–13. doi: 10.1111/prd.12317 [DOI] [PubMed] [Google Scholar]
- 3. Balta MG, Papathanasiou E, Blix IJ, van Dyke TE. Host modulation and treatment of periodontal disease. J Dental Res (2021) 1008:798–809. doi: 10.1177/0022034521995157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hajishengallis G, Chavakis T, Lambris JD. Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontology 2000 (2020) 841:14–34. doi: 10.1111/prd.12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Dyke TE. Shifting the paradigm from inhibitors of inflammation to resolvers of inflammation in periodontitis. J Periodontology (2020) 91 Suppl 1:S19–25. doi: 10.1002/JPER.20-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baumeister S-E, Holtfreter B, Reckelkamm SL, Kocher T, Alayash Z, Ehmke B, et al. Genotype-driven NPC1L1 and PCSK9 inhibition and reduced risk of periodontitis. J Clin Periodontol (2022) 50(1):114–20. doi: 10.1111/jcpe.13719 [DOI] [PubMed] [Google Scholar]
- 7. Naruishi K, Nagata T. Biological effects of interleukin-6 on gingival fibroblasts: cytokine regulation in periodontitis. J Cell Physiol (2018) 2339:6393–400. doi: 10.1002/jcp.26521 [DOI] [PubMed] [Google Scholar]
- 8. Rose-John S. Interleukin-6 family cytokines. Cold Spring Harb Perspect Biol (2018) 102. doi: 10.1101/cshperspect.a028415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rose-John S. Blocking only the bad side of IL-6 in inflammation and cancer. Cytokine (2021) 148:155690. doi: 10.1016/j.cyto.2021.155690 [DOI] [PubMed] [Google Scholar]
- 10. Wolf J, Rose-John S, Garbers C. Interleukin-6 and its receptors: a highly regulated and dynamic system. Cytokine (2014) 701:11–20. doi: 10.1016/j.cyto.2014.05.024 [DOI] [PubMed] [Google Scholar]
- 11. Ohta S, Tsuru T, Terao K, Mogi S, Suzaki M, Shono E, et al. Mechanism-based approach using a biomarker response to evaluate tocilizumab subcutaneous injection in patients with rheumatoid arthritis with an inadequate response to synthetic DMARDs (MATSURI study). J Clin Pharmacol (2014) 541:109–19. doi: 10.1002/jcph.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shankar-Hari M, Vale CL, Godolphin PJ, Fisher D, Higgins JPT, Spiga F, et al. Association between administration of IL-6 antagonists and mortality among patients hospitalized for COVID-19: a meta-analysis. JAMA (2021) 3266:499–518. doi: 10.1001/jama.2021.11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Georgakis MK, Malik R, Gill D, Franceschini N, Sudlow CLM, Dichgans M. Interleukin-6 signaling effects on ischemic stroke and other cardiovascular outcomes: a mendelian randomization study. Circ Genom Precis Med (2020) 133:e002872. doi: 10.1161/CIRCGEN.119.002872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Georgakis MK, Malik R, Burgess S, Dichgans M. Additive effects of genetic interleukin-6 signaling downregulation and low-density lipoprotein cholesterol lowering on cardiovascular disease: a 2×2 factorial mendelian randomization analysis. JAHA (2022) 111:e023277. doi: 10.1161/JAHA.121.023277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kappelmann N, Arloth J, Georgakis MK, Czamara D, Rost N, Ligthart S, et al. Dissecting the association between inflammation, metabolic dysregulation, and specific depressive symptoms: a genetic correlation and 2-sample mendelian randomization study. JAMA Psychiatry (2021) 782:161–70. doi: 10.1001/jamapsychiatry.2020.3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khandaker GM, Zuber V, Rees JMB, Carvalho L, Mason AM, Foley CN, et al. Shared mechanisms between coronary heart disease and depression: findings from a large UK general population-based cohort. Mol Psychiatry (2020) 257:1477–86. doi: 10.1038/s41380-019-0395-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Primers (2022) 21. doi: 10.1038/s43586-021-00092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang F. A flexible tool to plot a genomic map for single nucleotide polymorphisms. Source Code Biol Med (2016) 11:5. doi: 10.1186/s13029-016-0052-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmidt AF, Finan C, Gordillo-Marañón M, Asselbergs FW, Freitag DF, Patel RS, et al. Genetic drug target validation using mendelian randomisation. Nat Commun (2020) 111:3255. doi: 10.1038/s41467-020-16969-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holmes MV, Richardson TG, Ference BA, Davies NM, Davey Smith G. Integrating genomics with biomarkers and therapeutic targets to invigorate cardiovascular drug development. Nat Rev Cardiol (2021) 186:435–53. doi: 10.1038/s41569-020-00493-1 [DOI] [PubMed] [Google Scholar]
- 21. Alayash Z, Baumeister S-E, Holtfreter B, Kocher T, Baurecht H, Ehmke B, et al. Inhibition of tumor necrosis factor receptor 1 and the risk of periodontitis. Front Immunol (2023) 14:1094175. doi: 10.3389/fimmu.2023.1094175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plebani M. Why c-reactive protein is one of the most requested tests in clinical laboratories? Clin Chem Lab Med (2023). doi: 10.1515/cclm-2023-0086 [DOI] [PubMed] [Google Scholar]
- 23. Said S, Pazoki R, Karhunen V, Võsa U, Ligthart S, Bodinier B, et al. Genetic analysis of over half a million people characterises c-reactive protein loci. Nat Commun (2022) 131:2198. doi: 10.1038/s41467-022-29650-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shungin D, Haworth S, Divaris K, Agler CS, Kamatani Y, Keun Lee M, et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat Commun (2019) 101:2773. doi: 10.1038/s41467-019-10630-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, Shah T, et al. Association between c reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ (2011) 342:d548. doi: 10.1136/bmj.d548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for mendelian randomization. Stat Methods Med Res (2017) 265:2333–55. doi: 10.1177/0962280215597579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papapanou PN, Demmer RT. Epidemiology of periodontitis. In: Berglundh T, Giannobile WV, Lang NP, Sanz M, editors. Lindhe's clinical periodontology and implant dentistry, 7th ed. Newark: Wiley & Sons; (2022). p. 119–59. [Google Scholar]
- 28. Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ (2021) 375:n2233. doi: 10.1136/bmj.n2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gümüş P, Nizam N, Lappin DF, Buduneli N. Saliva and serum levels of b-cell activating factors and tumor necrosis factor-α in patients with periodontitis. J Periodontology (2014) 852:270–80. doi: 10.1902/jop.2013.130117 [DOI] [PubMed] [Google Scholar]
- 30. Tymkiw KD, Thunell DH, Johnson GK, Joly S, Burnell KK, Cavanaugh JE, et al. Influence of smoking on gingival crevicular fluid cytokines in severe chronic periodontitis. J Clin Periodontol (2011) 383:219–28. doi: 10.1111/j.1600-051X.2010.01684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kajiura Y, Lew J-H, Ikuta T, Nishikawa Y, Kido J-I, Nagata T, et al. Clinical significance of GCF sIL-6R and calprotectin to evaluate the periodontal inflammation. Ann Clin Biochem (2017) 546:664–70. doi: 10.1177/0004563216680232 [DOI] [PubMed] [Google Scholar]
- 32. Tang L, Li X, Bai Y, Wang P, Zhao Y. MicroRNA-146a negatively regulates the inflammatory response to porphyromonas gingivalis in human periodontal ligament fibroblasts via TRAF6/p38 pathway. J Periodontology (2019) 904:391–9. doi: 10.1002/JPER.18-0190 [DOI] [PubMed] [Google Scholar]
- 33. Currò M, Matarese G, Isola G, Caccamo D, Ventura VP, Cornelius C, et al. Differential expression of transglutaminase genes in patients with chronic periodontitis. Oral Dis (2014) 206:616–23. doi: 10.1111/odi.12180 [DOI] [PubMed] [Google Scholar]
- 34. Sattari M, Taheri RA, ArefNezhad R, Motedayyen H. The expression levels of MicroRNA-146a, RANKL and OPG after non-surgical periodontal treatment. BMC Oral Health (2021) 211:523. doi: 10.1186/s12903-021-01883-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebersole JL, Kirakodu S, Novak MJ, Stromberg AJ, Shen S, Orraca L, et al. Cytokine gene expression profiles during initiation, progression and resolution of periodontitis. J Clin Periodontol (2014) 419:853–61. doi: 10.1111/jcpe.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hajishengallis G, Korostoff JM. Revisiting the page & Schroeder model: the good, the bad and the unknowns in the periodontal host response 40 years later. Periodontology 2000 (2017) 751:116–51. doi: 10.1111/prd.12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan W, Wang Q, Chen Q. The cytokine network involved in the host immune response to periodontitis. Int J Oral Sci (2019) 113:30. doi: 10.1038/s41368-019-0064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Q, Zhou X, Huang D, Ji Y, Kang F. IL-6 enhances osteocyte-mediated osteoclastogenesis by promoting JAK2 and RANKL activity in vitro . Cell Physiol Biochem (2017) 414:1360–9. doi: 10.1159/000465455 [DOI] [PubMed] [Google Scholar]
- 39. González-Febles J, Sanz M. Periodontitis and rheumatoid arthritis: what have we learned about their connection and their treatment? Periodontology 2000 (2021) 871:181–203. doi: 10.1111/prd.12385 [DOI] [PubMed] [Google Scholar]
- 40. Kobayashi T, Okada M, Ito S, Kobayashi D, Ishida K, Kojima A, et al. Assessment of interleukin-6 receptor inhibition therapy on periodontal condition in patients with rheumatoid arthritis and chronic periodontitis. J Periodontology (2014) 851:57–67. doi: 10.1902/jop.2013.120696 [DOI] [PubMed] [Google Scholar]
- 41. Kobayashi T, Ito S, Kobayashi D, Kojima A, Shimada A, Narita I, et al. Interleukin-6 receptor inhibitor tocilizumab ameliorates periodontal inflammation in patients with rheumatoid arthritis and periodontitis as well as tumor necrosis factor inhibitors. Clin Exp Dent Res (2015) 12:63–73. doi: 10.1002/cre2.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ancuţa C, Chirieac R, Ancuţa E, Ţănculescu O, Solomon SM, Fătu AM, et al. Exploring the role of interleukin-6 receptor inhibitor tocilizumab in patients with active rheumatoid arthritis and periodontal disease. J Clin Med (2021) 104. doi: 10.3390/jcm10040878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swerdlow DI, Holmes MV, Kuchenbaecker KB, Engmann JEL, Shah T, Sofat R, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet (2012) 3799822:1214–24. doi: 10.1016/S0140-6736(12)60110-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ridker PM, Rane M. Interleukin-6 signaling and anti-Interleukin-6 therapeutics in cardiovascular disease. Circ Res (2021) 12811:1728–46. doi: 10.1161/CIRCRESAHA.121.319077 [DOI] [PubMed] [Google Scholar]
- 45. Machado V, Botelho J, Escalda C, Hussain SB, Luthra S, Mascarenhas P, et al. Serum c-reactive protein and periodontitis: a systematic review and meta-analysis. Front Immunol (2021) 12:706432/full. doi: 10.3389/fimmu.2021.706432/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pink C, Kocher T, Meisel P, Dörr M, Markus MRP, Jablonowski L, et al. Longitudinal effects of systemic inflammation markers on periodontitis. J Clin Periodontol (2015) 4211:988–97. doi: 10.1111/jcpe.12473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: CRP GWAS data are accessible via the GWAS catalog (https://www.ebi.ac.uk/gwas/) under accession ID GCST90029070. Periodontitis GWAS data are available at https://data.bris.ac.uk/data/dataset/2j2rqgzedxlq02oqbb4vmycnc2.