Abstract
Autophagy plays a crucial role in the development and progression of ischemic acute kidney injury (AKI). However, the function and mechanism of circular RNAs (circRNAs) in the regulation of autophagy in ischemic AKI remain unexplored. Herein, we find that circ-ZNF609, originating from the ZNF609 locus, is highly expressed in the kidney after ischemia/reperfusion injury, and urinary circ-ZNF609 is a moderate predictor for AKI in heart disease patients. Overexpression of circ-ZNF609 can activate AKT3/mTOR signaling and induce autophagy flux impairment and cell apoptosis while inhibiting proliferation in HK-2 cells, which is blocked by silencing circ-ZNF609. Mechanistically, circ-ZNF609 encodes a functional protein consisting of 250 amino acids (aa), termed ZNF609-250aa, the overexpression of which can activate AKT3/mTOR signaling and induce autophagy flux impairment and cell apoptosis in HK-2 cells in vitro and in AKI kidneys in vivo. The blockade of AKT and mTOR signaling with pharmacological inhibitors is capable of reversing ZNF609-250aa-induced autophagy flux impairment and cell apoptosis in HK-2 cells. The present study demonstrates that highly expressed circ-ZNF609-encoded ZNF609-250aa induces cell apoptosis and AKI by impairing the autophagy flux via an AKT/mTOR-dependent mechanism. These findings imply that targeting circ-ZNF609 may be a novel therapy for ischemic AKI.
Keywords: acute kidney injury, renal ischemia/reperfusion, circRNA, protein coding, autophagy
Graphical abstract
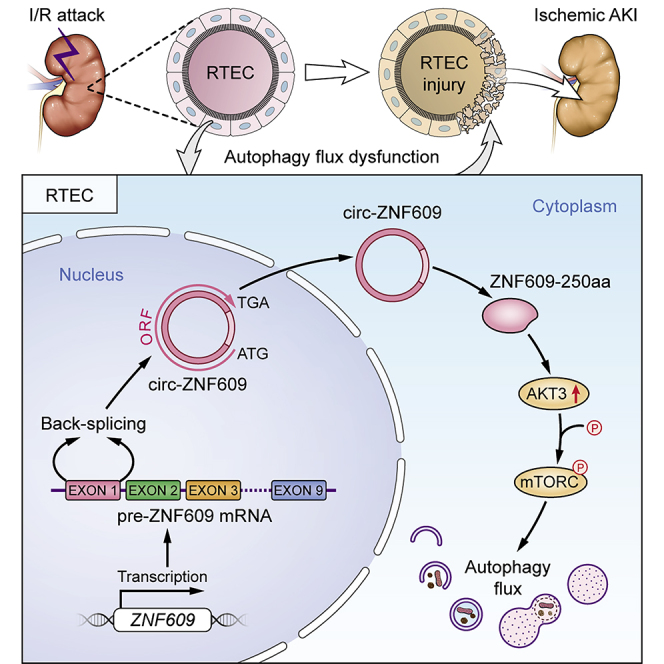
Autophagy regulated by circRNAs is of concern in ischemic acute kidney injury (AKI). Herein, we find that highly expressed circ-ZNF609-encoded ZNF609-250aa induces cell apoptosis and AKI by impairing autophagy flux via the AKT/mTOR signaling pathway. These findings imply that targeting circ-ZNF609 may be a novel therapy for ischemic AKI.
Introduction
Acute kidney injury (AKI) is a common and critical illness worldwide that is characterized by the abrupt loss of renal functions and is induced by renal ischemia/reperfusion (I/R) injury, sepsis, and nephrotoxicity, of which renal I/R injury is a leading cause.1,2 However, the pathogenesis of ischemic AKI has not been fully elucidated, and thus effective treatments remain lacking.
Renal I/R injury has a complex pathophysiology involving activation of cell death programs (e.g., autophagy, apoptosis, and necrosis), oxidative stress, inflammatory responses, mitochondrial damage, and microvascular dysfunction.2,3 Recent studies have confirmed that autophagy, as a self-degradation mechanism responsible for cell death and survival, plays a crucial role in the development and progression of ischemic AKI.4,5,6 Autophagic flux refers to the complete autophagic process, in which the equilibrium between autophagosome formation and clearance by lysosomes is indispensable.7,8 Furthermore, the autophagic process is mainly regulated by the mammalian target of rapamycin (mTOR) pathway.9,10 Cell damage and diseases can occur if there are abnormalities in the autophagic processes and autophagic regulation.
With the rapid innovation of high-throughput sequencing technologies, a growing number of previously unknown transcripts have been identified. Circular RNAs (circRNAs), one of the most concerning types of transcript, are a class of circularly configured RNAs produced from direct backsplicing or exon skipping of precursor mRNA.11 Due to the abundance and stability of circRNAs in eukaryotes, these molecules can widely regulate biological activities. Moreover, evidence has shown that certain circRNAs influence the course of a disease by regulating autophagy, suggesting a critical role for autophagy in the onset and development of various diseases.12 However, the functional role and mechanism of circRNAs in autophagy during the pathogenesis of ischemic AKI remain unexplored. circRNAs usually function as microRNA (miRNA) sponges or regulators of RNA binding proteins to modulate protein-gene transcription.13 An increasing number of circRNAs, such as circ-ZNF609 in myogenesis, circ-E-Cad in glioblastoma, and circMAPK1 in gastric cancer, have been found to encode proteins that are involved in pathophysiological processes.14,15,16 By using high-throughput RNA sequencing, we also find that circ-ZNF609 is markedly upregulated in the ischemic-induced AKI kidney, which is also associated with activation of the PI3K/AKT signaling pathway.17,18 Given that AKT is the primary upstream regulator of mTOR, we speculate that circ-ZNF609 may participate in the pathogenesis of ischemic AKI by regulating autophagy via the AKT/mTOR pathway.
In the present study, we hypothesized that circ-ZNF609 may play a pivotal role in the pathogenesis of ischemic AKI, which was examined in vitro in HK-2 cells and in vivo in I/R-induced AKI by overexpressing or knocking down circ-ZNF609. We found that overexpression of circ-ZNF609 promoted cell death (apoptosis) and AKI by impairing the autophagy flux, which was blocked by silencing circ-ZNF609. Mechanistically, circ-ZNF609 encodes a 250-amino-acid protein to form ZNF609-250aa, which exerts its pathological effect on AKI by impairing the autophagy flux and promoting cell apoptosis via the AKT/mTOR signaling pathway.
Results
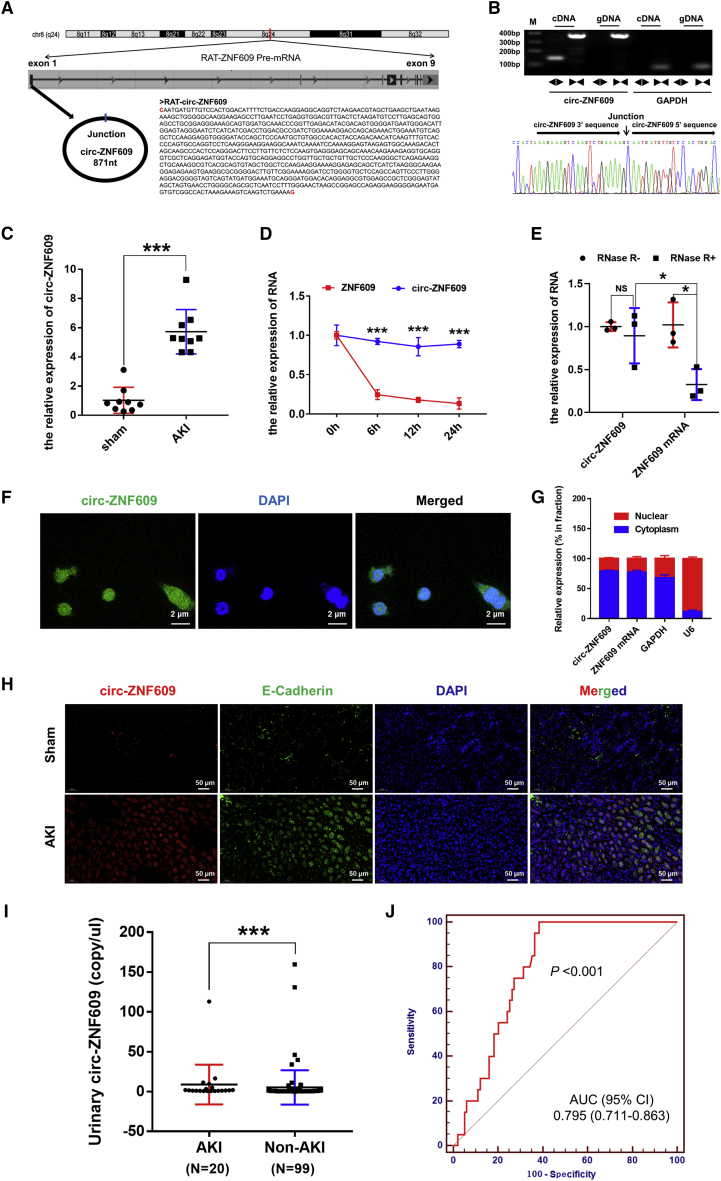
Identification and characterization of circ-ZNF609 in ischemic AKI in animals and clinical specimens
We first characterized circ-ZNF609 in I/R-treated kidney tissues of rats by using two sets of primers (divergent primers and convergent primers) and Sanger sequencing. circ-ZNF609 is derived from the first exon of the ZNF609 gene located on chromosome 8 and has a full length of 871 nt (Figure 1A). The results showed that the divergent primers spanning the circ-ZNF609 junction amplified the predicted PCR products, and Sanger sequencing identified the circular junction (Figures 1B and S1). Quantitative real-time PCR confirmed that the expression of circ-ZNF609 was significantly increased in the kidney tissues of rats with ischemic AKI compared with those of the control group (Figure 1C). Then, conservation analysis of cross-species circ-ZNF609 from humans and rats indicated that circ-ZNF609 sequences are strongly conserved, and the comparison of circ-ZNF609 sequences from rats and humans showed 93% homology (Figure S2). To further confirm the existence of circ-ZNF609, we used an RNase R degradation assay to evaluate the resistance of circ-ZNF609 to RNase R treatment. Figures 1D and 1E show that circ-ZNF609 was resistant to RNase R digestion and did have a longer half-life than linear transcripts of ZNF609. Moreover, fluorescence in situ hybridization (FISH) analysis (Figure 1F) in HK-2 cells revealed that abundant circ-ZNF609 appeared in the cytoplasm of HK-2 cells, as demonstrated by the nuclear mass separation assay (Figure 1G). The FISH analysis in rat kidneys showed that circ-ZNF609 was mainly located in renal tubular epithelial cells (RTECs), and the expression of circ-ZNF609 was low in non-epithelial cells (E-Cadherin and circ-ZNF609 probe as the labels for RTECs and circ-ZNF609, respectively) (Figure 1H). These results indicated that circ-ZNF609 was mainly located and exerted its function in RTECs after renal I/R injury. In vivo and in vitro, the expression of circ-ZNF609 in rat kidneys and HK-2 cells was significantly increased over time after renal I/R injury or cell oxygen-glucose deprivation and reoxygenation (OGD/R), as demonstrated by FISH assay and quantitative real-time PCR (Figure S3). Clinically, we examined the expression of circ-ZNF609 in urinary samples from 119 patients with acute decompensated heart failure (ADHF) or undergoing cardiac surgery under cardiopulmonary bypass (CPB) by droplet digital PCR (ddPCR) to confirm its elevated expression in AKI patients (Figure 1I). Table S1 lists the detailed clinical parameters of the patients mentioned above and shows that the proportion of congestive heart failure and the levels of serum creatine, serum Cys C, and eGFR were significantly different between AKI and non-AKI patients. In addition, we detected the performance of urinary circ-ZNF609 as a predictor of the development of AKI in patients with ADHF or undergoing cardiac surgery under CPB, with a moderate AUC value of 0.795 (95% CI 0.711–0.863) (Figure 1J).
Figure 1.
Characterization of circ-ZNF609 in ischemic AKI
(A) The origin and sequence of rat circ-ZNF609. (B) The divergent primers detected circ-ZNF609 in cDNA but not in gDNA, and using divergent primers on Sanger sequencing identified the circular junction of circ-ZNF609. (C) The expression of circ-ZNF609 in the kidneys of rats in the sham and ischemic AKI groups was detected by quantitative real-time PCR. n = 9. (D) The relative RNA levels of circ-ZNF609 and linear ZNF609 mRNA at different time points. (E) Quantitative real-time PCR analysis of circ-ZNF609 and linear ZNF609 mRNA after treatment with or without RNase R in HK-2 cells. (F and G) The FISH analysis with junction-specific probes and the nuclear mass separation assay were used to decide the localization of circ-ZNF609 in HK-2 cells. Scale bar: 2 μm. (H) The FISH analysis of rat kidneys was used to decide the localization of circ-ZNF609 in RTECs and non-epithelial cells. E-Cadherin and the circ-ZNF609 probe were the labels for RTECs and circ-ZNF609, respectively. Scale bar: 50 μm. (I) The expression levels of circ-ZNF609 in urinary samples from 119 patients with ADHF or undergoing cardiac surgery under CPB were analyzed by ddPCR. (J) Urinary circ-ZNF609 was a predictor for detecting AKI in patients with ADHF or undergoing cardiac surgery under CPB, with a moderate AUC value of 0.795 (95% CI 0.711–0.863). ADHF, acute decompensated heart failure; CPB, cardiopulmonary bypass; ddPCR, droplet digital PCR. The graph presents the mean ± SD; ∗p < 0.05 and ∗∗∗p < 0.001.
circ-ZNF609 induces AKI, which is associated with autophagy flux impairment and activation of AKT/mTOR signaling in vitro and in vivo
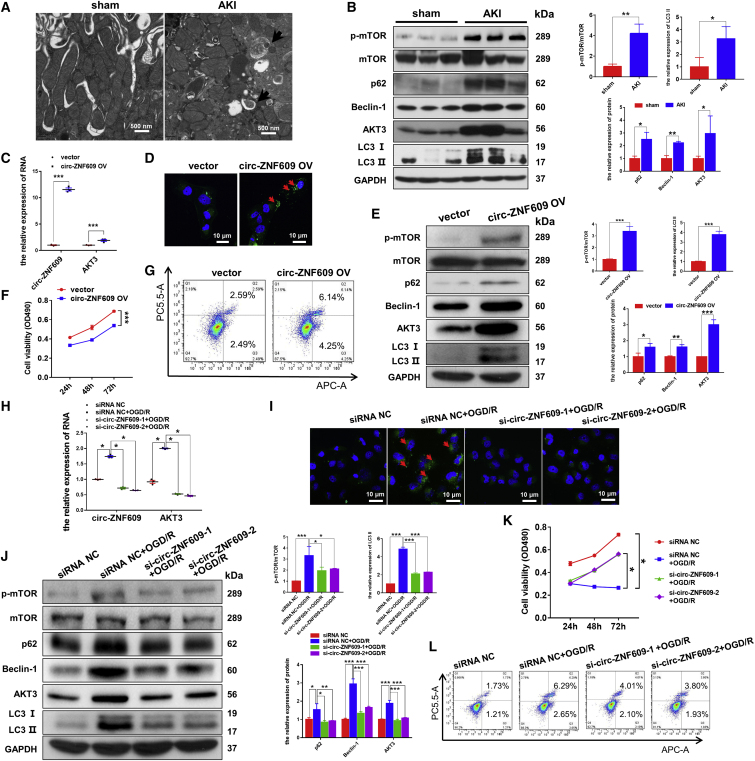
We first investigated the relationships among circ-ZNF609, autophagy, and the AKT/mTOR signaling pathways in vivo and in vitro. By transmission electron microscopy, we found that autolysosomes were accumulated in the ischemic AKI kidney but not in the sham-operated kidneys (Figure 2A). This was associated with an increase in autophagic biomarkers such as p62, Beclin-1, and LC3-II/I and the activation of AKT/mTOR signaling (Figure 2B).
Figure 2.
circ-ZNF609 influences the proliferation and apoptosis of RTECs by regulating autophagic flux via the AKT/mTOR signaling pathway
(A) Transmission electron microscopic images of autophagic structures in the kidney tissues of ischemic AKI and sham rat models. The arrows highlight the main differences in the autophagic structures. Scale bar: 500 nm. (B) Western blotting detected the protein expression of AKT/mTOR/autophagy signaling pathway components in the kidney tissues of rats in the ischemic AKI and sham groups. (C) Quantitative real-time PCR was used to detect the expression of circ-ZNF609 and AKT3 mRNA in the HK-2 cells transfected with the control vector or circ-ZNF609 expression plasmid. (D) Laser confocal microscopy of GFP-LC3 was used to detect autophagosomes in the HK-2 cells transfected with the control vector or circ-ZNF609 expression plasmid. The arrows highlight the main differences in autophagosomes. Scale bar: 10 μm. (E) Western blotting detected the protein expression of AKT/mTOR/autophagy signaling pathway components in the HK-2 cells transfected with the control vector or circ-ZNF609 expression plasmid. (F) MTS assays were used to examine proliferation of the HK-2 cells transfected with the control vector or circ-ZNF609 expression plasmid for 24, 48, and 72 h. (G) Flow cytometry was used to detect apoptosis of the HK-2 cells transfected with the control vector or circ-ZNF609 expression plasmid. (H) Quantitative real-time PCR was used to detect the expression of circ-ZNF609 and AKT3 mRNA in the HK-2 cells treated with circ-ZNF609 siRNA and with or without OGD/R. (I) Laser confocal microscopy of GFP-LC3 was used to detect autophagosomes in the HK-2 cells treated with circ-ZNF609 siRNA and with or without OGD/R. The arrows highlight the main differences in autophagosomes. Scale bar: 10 μm. (J) Western blotting detected the expression of AKT/mTOR/autophagy signaling pathway components in the HK-2 cells treated with circ-ZNF609 siRNA and with or without OGD/R. (K) MTS assays were used to examine cell proliferation of the HK-2 cells treated with circ-ZNF609 siRNA with or without OGD/R for 24, 48, and 72 h. (L) Flow cytometry was used to detect apoptosis of HK-2 cells treated with circ-ZNF609 siRNA with or without OGD/R. OGD/R, oxygen-glucose deprivation and reoxygenation. The graphs present the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Then, we examined the regulatory role of circ-ZNF609 in the autophagy-related cell death pathway and activation of AKT/mTOR signaling in HK-2 cells by overexpressing or silencing circ-ZNF609. As shown in Figure 2C, quantitative real-time PCR detected that circ-ZNF609 and AKT3 mRNAs were highly expressed in the HK-2 cells transfected with the circ-ZNF609 expression plasmid, which was associated with induction of autophagy flux impairment and cell apoptosis as demonstrated by increasing LC3-II, p62, and Beclin-1 and activation of AKT3/mTOR signaling while inhibiting cell proliferation (Figures 2D–2G). In contrast, silencing circ-ZNF609 restored autophagic flux, inhibited the AKT3/mTOR signaling pathway, and protected HK-2 cells from apoptosis while promoting proliferation (Figures 2H–2L). These results collectively suggest that circ-ZNF609 may play a regulatory role in RTEC apoptosis during AKI, possibly by activating AKT/mTOR signaling and causing autophagic flux impairment.
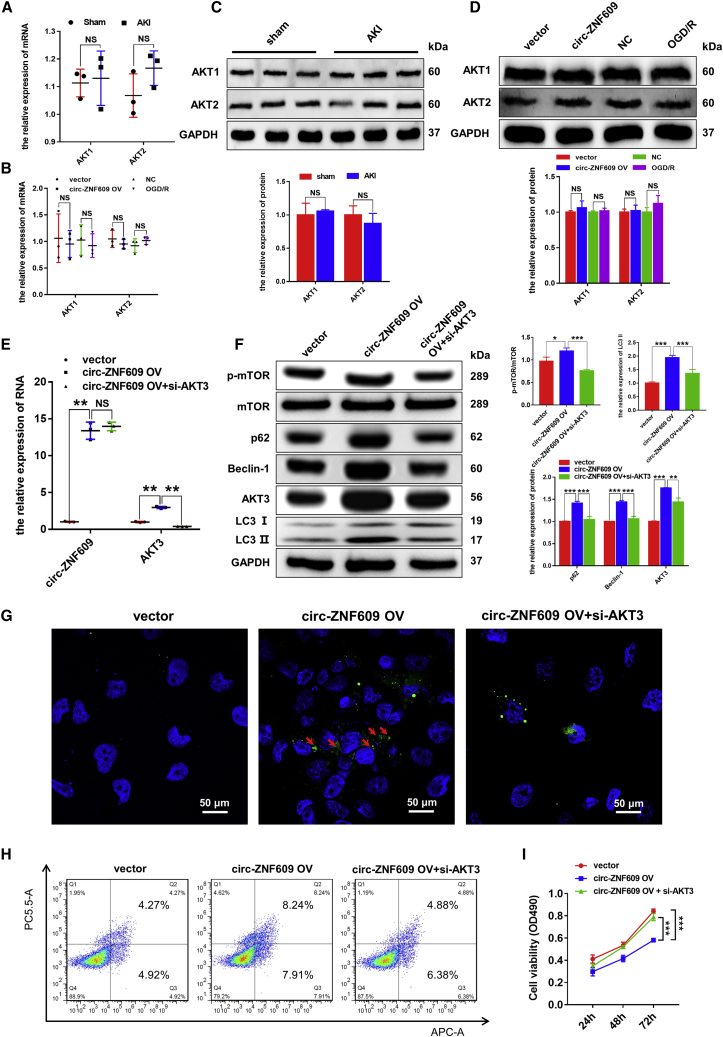
circ-ZNF609 induces autophagy flux impairment and cell apoptosis via an AKT3-dependent mechanism in vitro
We next investigated the mechanisms through which circ-ZNF609 regulates autophagy flux impairment and tubular cell death by targeting AKT3. It is well known that the AKT family has three isoforms (AKT1, AKT2, and AKT3), which share very similar sequences; however, isoform-specific functions have been reported. The quantitative real-time PCR and western blotting results showed that there were no significant differences between AKT1 and AKT2 in the rat kidney tissues with or without I/R injury, HK-2 cells with or without circ-ZNF609 overexpression, and HK-2 cells with or without OGD/R. Therefore, we reasonably believe that AKT3 rather than AKT1 or AKT2 is the target of circ-ZNF609 (Figures 3A–3D). To verify the relationship between circ-ZNF609 and AKT3, we performed circ-ZNF609 overexpression and AKT3 knockdown in HK-2 cells (Figure 3E). Results showed that circ-ZNF609 overexpression significantly enhanced AKT3/mTOR signaling and caused autophagy flux impairment by increasing LC3-II, p62, and Beclin-1, which were blocked by silencing AKT3 with AKT3 small interfering RNAs (siRNAs) (Figure 3F). Similarly, fluorescence imaging of GFP-LC3-II also showed that the increased cytoplasmic autophagosomes induced by overexpressing circ-ZNF609 were also blocked by knocking down AKT3 (Figure 3G). In addition, silencing of AKT3 also blocked circ-ZNF609-induced cell apoptosis and inhibition of cell proliferation in HK-2 cells (Figures 3H and 3I). Thus, circ-ZNF609 induces autophagy flux impairment and promotes cell apoptosis while inhibiting proliferation in HK-2 cells via the AKT3-dependent mechanism.
Figure 3.
circ-ZNF609 suppresses the proliferation, promotes the apoptosis, and induces autophagic flux impairment of RTECs by targeting AKT3 mRNA in vitro
(A and B) Quantitative real-time PCR was performed to detect the RNA expression of AKT1 and AKT2 in the rat kidney tissues with or without I/R injury, HK-2 cells with or without circ-ZNF609 overexpression, and HK-2 cells with or without OGD/R. (C and D) Western blotting was performed to detect the protein expression of AKT1 and AKT2 in the rat kidney tissues with or without I/R injury, HK-2 cells with or without circ-ZNF609 overexpression, and HK-2 cells with or without OGD/R. (E) Quantitative real-time PCR was performed to detect circ-ZNF609 and AKT3 mRNA expression in the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609 plus AKT3 siRNA plasmid. (F) Western blotting was performed to detect the protein expression of AKT/mTOR/autophagy signaling pathway components in the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609 plus AKT3 siRNA plasmid. (G) Laser confocal microscopy of GFP-LC3 in the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609 plus AKT3 siRNA plasmid. The arrows highlight the main differences in autophagosomes. Scale bar: 50 μm. (H) Flow cytometry was used to detect apoptosis of the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609 plus AKT3 siRNA plasmid. (I) MTS assays were used to examine the proliferation of the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609 plus AKT3 siRNA plasmid for 24, 48, and 72 h. OGD/R, oxygen-glucose deprivation and reoxygenation. The graphs present the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
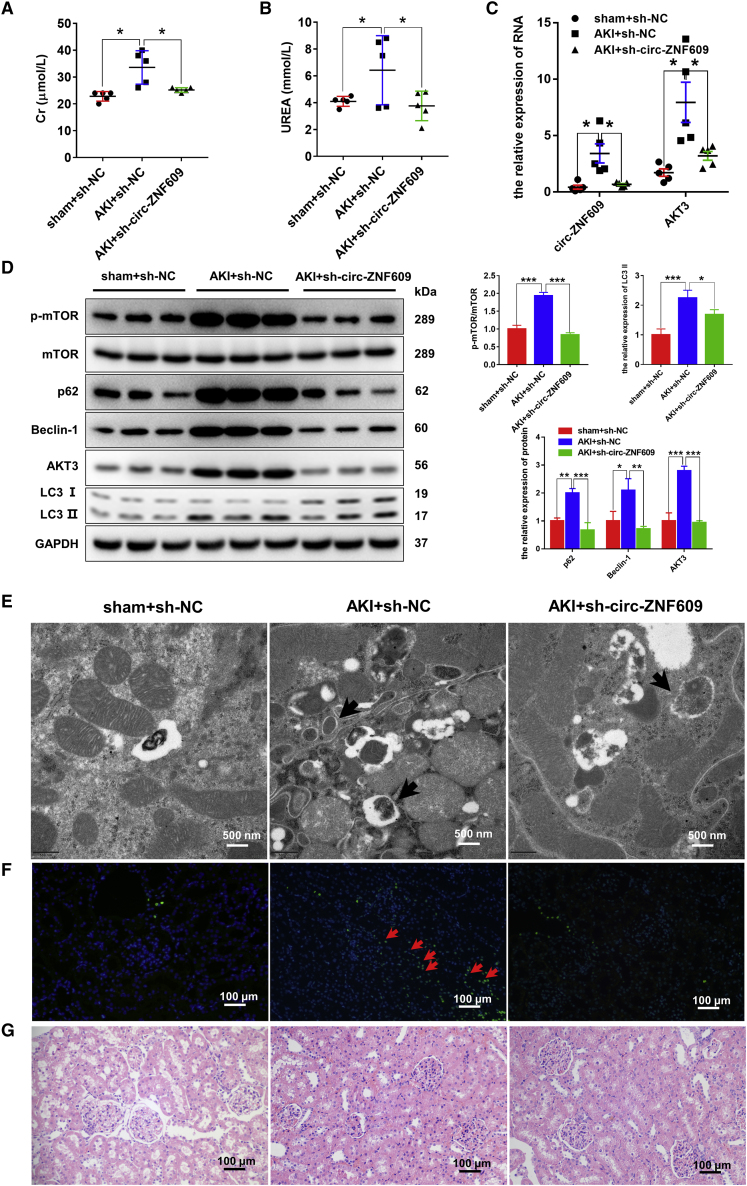
Silencing of circ-ZNF609 is renoprotective by restoring autophagic flux through inhibition of the AKT/mTOR signaling pathway in ischemic-induced AKI kidney
To examine the functional role of circ-ZNF609 in AKI, we knocked down renal circ-ZNF609 by adeno-associated virus (AAV)-mediated sh-circ-ZNF609 before the ischemic AKI model was induced in rats. Compared with the sham-operated animals, AKI rats that received control AAV developed significant AKI with higher levels of serum creatinine and urea, which were largely inhibited in those treated with AAV-sh-circ-ZNF609 (Figures 4A and 4B). Quantitative real-time PCR and western blotting analysis also showed that, compared with control-treated AKI kidney, silencing renal circ-ZNF609 significantly inhibited AKT3/mTOR signaling and improved the autophagy flux impairment as evidenced by inhibition of p62, Beclin-1, and LC3-II expression (Figures 4C and 4D), resulting in blocking of autolysosome accumulation in tubular epithelial cells as detected by transmission electron microscopy (Figure 4E) and apoptosis assessed by TUNEL assay (Figure 4F). Histologically, knockdown of renal circ-ZNF609 also improved ischemic-induced AKI pathology (Figure 4G).
Figure 4.
Silencing of circ-ZNF609 is renoprotective in rat models of ischemic AKI by restoring autophagic flux through inhibition of the AKT/mTOR signaling pathway
(A and B) The levels of creatinine and urea in the plasma of rat models that underwent renal I/R injury or sham operation with or without sh-circ-ZNF609 by AAV9. n = 5. (C) Quantitative real-time PCR was performed to detect circ-ZNF609 and AKT3 mRNA expression in kidney tissues of ischemic AKI and sham rat models with or without sh-circ-ZNF609 by AAV9. n = 5. (D) Western blotting was used to detect the protein expression of AKT/mTOR/autophagy pathway components in kidney tissues of the ischemic AKI and sham rat models with or without sh-circ-ZNF609 by AAV9. (E) Transmission electron microscopic images of autophagic structures in the kidney tissues of the ischemic AKI and sham rat models with or without sh-circ-ZNF609 by AAV9. The arrows highlight the main differences in autophagic structures. Scale bar: 500 nm. (F) Apoptosis level in rat kidney cells of the ischemic AKI and sham rat models with or without sh-circ-ZNF609 by AAV9 was evaluated by the TUNEL method. The arrows highlight the main differences in cell apoptosis. Scale bar: 100 μm. (G) The histological properties of kidney tissues of the ischemic AKI and sham rat models with or without sh-circ-ZNF609 by AAV9 were observed under a microscope following H&E staining. Scale bar: 100 μm. The graphs present the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
circ-ZNF609 encodes a 250-amino-acid protein, ZNF609-250aa
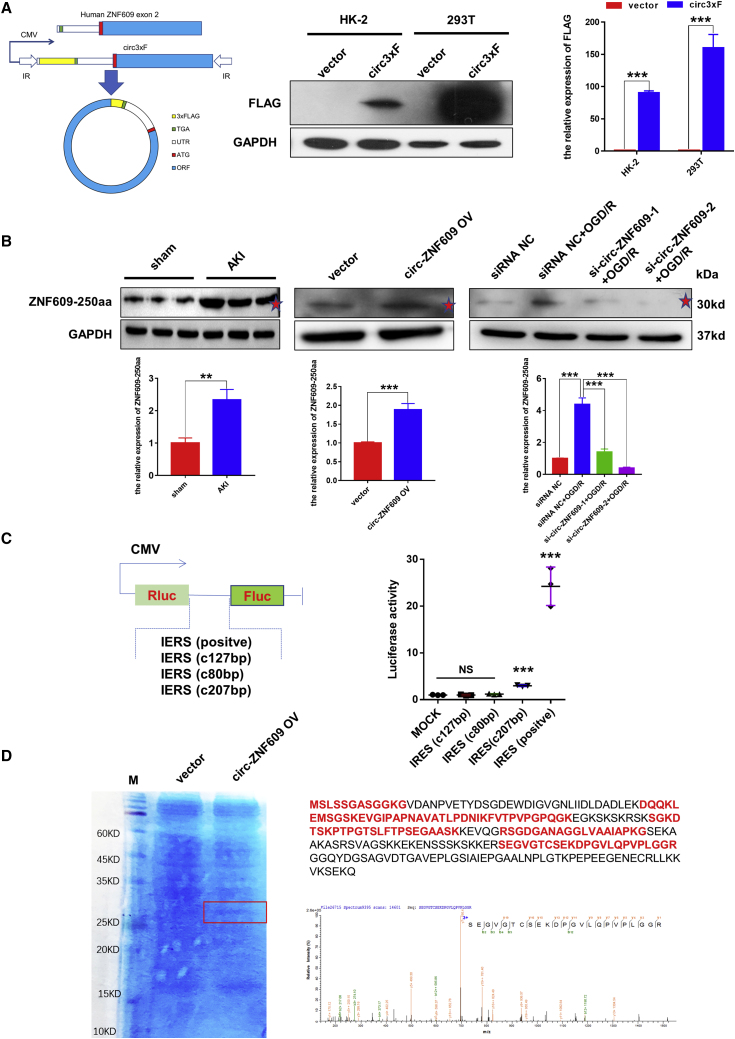
A previous study found that circ-ZNF609 is a circRNA that can be translated and functions in myogenesis.14 In the present study, a series of experiments was performed to show that circ-ZNF609 can be translated and functions in ischemic AKI. According to the prediction results of the online database circRNADb, an open reading frame (ORF) with the initiation codon ATG and an IRES (Internal Ribosome Entry Site) were contained in the sequence of circ-ZNF609. This observation suggests that circ-ZNF609 has the potential to encode a protein of 250 amino acids, which was termed ZNF609-250aa in this study (Figures S4A and S4B). Furthermore, we found that the specific amino acid sequence of the ZNF609-250aa protein can be targeted by Invitrogen’s ZNF609 antibody (Figure S4C). We also compared the full-length protein sequences from ZNF609-250aa and ZNF609 (Figure S4D), circ-ZNF609 ORF sequences from rats and humans (94% homology) (Figure S4E), and the protein sequences encoded by circ-ZNF609 from rats and humans (94% homology) (Figure S4F). These results indicated that the protein-encoding potentials are similar between rat circ-ZNF609 and human circ-ZNF609.
Then, a 3×FLAG tag was inserted upstream of the stop codon to obtain a flagged protein after circularization, namely, circ3×F. Next, exon 2 of the human ZNF609 gene was inserted into the plasmid vector. Western blotting analysis with an anti-FLAG antibody of proteins from the HK-2 and 293T cells transfected with circ3×F showed that in both cell types, in comparison with mock-treated cells, circ3×F gave rise to the flagged proteins (Figure 5A). Furthermore, western blotting with a ZNF609 antibody, which can identify ZNF609 protein and ZNF609-250aa, was performed. The results indicated that the protein level of ZNF609-250aa was increased in kidney tissues of the rats treated with I/R compared with those of the sham group. Similarly, ZNF609-250aa was present in the HK-2 cells treated with circ-ZNF609 overexpression and OGD/R, but circ-ZNF609 knockdown by siRNA reversed the OGD/R-induced ZNF609-250aa increase (Figure 5B). Therefore, to validate the IRES activity of the circ-ZNF609 UTR, a luciferase reporter assay was performed. The CMV promoter drives the transcription of firefly and Renilla luciferase-coding regions, and the spacer in the luciferase bicistronic constructs was substituted with the positive control, an intact 207 bp sequence, or a truncated IRES sequence of circ-ZNF609 (Figure 5C). Notably, the construct with the truncated IRES sequence (127 and 80 bp) could not trigger Fluc activity, indicating that IRES activity of the circ-ZNF609 UTR exists and relies on specific features of the UTR sequence (Figure 5C). Mass spectrometry followed by SDS-PAGE using the circ-ZNF609-overexpressing HK-2 cells further confirmed that ZNF609-250aa was translated from circ-ZNF609, and the peptide sequences had a predicted molecular weight of 31–40 kDa (Figure 5D). Overall, circ-ZNF609 encodes the ZNF609-250aa protein by IRES in RTECs after I/R injury.
Figure 5.
circ-ZNF609 encodes a 250 aa protein termed ZNF609-250aa
(A) Western blotting was used to detect the protein expression of FLAG in the HK-2 and 293T cells treated with control vector or 3×Flag-circ-ZNF609 expression plasmid. (B) Western blotting was used to detect the protein expression of ZNF609-250aa in kidney tissues of the ischemic AKI and sham operation rat models, in the HK-2 cells transfected with control vector or circ-ZNF609 expression plasmid, and in the HK-2 cells treated with or without circ-ZNF609 siRNA and OGD/R. The red stars indicated the ZNF609-250aa and the predicted molecular weight of 31–40 kDa. (C) Luciferase activity of three predictive IRESs in circ-ZNF609. (D) The ZNF609-250aa junction-specific peptide was identified by mass spectrometry. OGD/R, oxygen-glucose deprivation and reoxygenation. The graphs present the mean ± SD; ∗∗p < 0.01 and ∗∗∗p < 0.001.
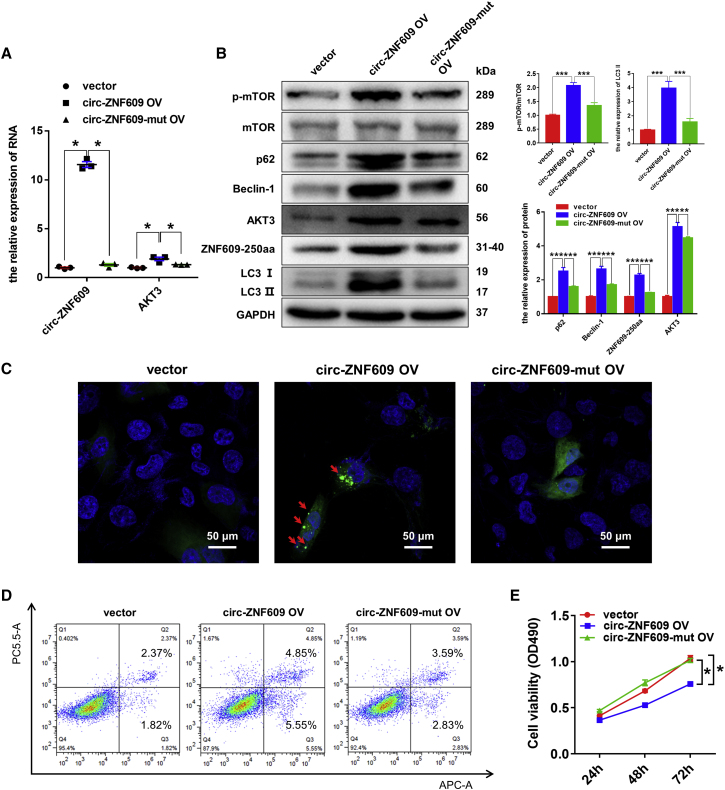
ZNF609-250aa, not circ-ZNF609, induces autophagy flux impairment and promotes the apoptosis while inhibiting proliferation in HK-2 cells
To further explore the biological function of ZNF609-250aa, we first performed a bioinformatics analysis to verify that there is no favorable evidence for circ-ZNF609 to function through regulation of transcription as an miRNA sponge or regulator of AKT3 in ischemic AKI (Document S2. The ceRNA for circ-ZNF609_human, Document S3. The ceRNA for circ-ZNF609_rats, Document S4. The RBP for circ-ZNF609_human, Document S5. The RBP for circ-ZNF609_rats). Then, we transfected the circ-ZNF609 overexpression plasmid and circ-ZNF609 ATG mutant plasmid (the start codon of circ-ZNF609 was mutated) into HK-2 cells. Results shown in Figure 6 show that overexpression of wild-type circ-ZNF609 resulted in enhanced ZNF609-250aa expression, AKT3/mTOR signaling and autophagic flux impairment, and increased cell apoptosis while inhibiting proliferation, as evidenced by quantitative real-time PCR and western blotting (Figures 6A and 6B), laser scanning confocal microscopy (Figure 6C), flow cytometry (Figure 6D), and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) (Figure 6E) assays. In contrast, HK-2 cells transfected with circ-ZNF609 ATG mutant plasmid failed to facilitate ZNF609-250aa expression, to activate AKT3/mTOR signaling, and to induce the autophagy flux impairment (Figures 6B and 6C) and cell apoptosis (Figure 6D) as well as the inhibition of cell proliferation (Figure 6E). Thus, circ-ZNF609 activates AKT3/mTOR signaling to induce autophagic flux impairment and apoptosis while inhibiting cell proliferation by encoding the ZNF609-250aa.
Figure 6.
ZNF609-250aa, not circ-ZNF609, suppresses the proliferation, promotes the apoptosis, and induces autophagic flux impairment of RTECs by activating the AKT/mTOR signaling pathway in vitro
(A) Quantitative real-time PCR was performed to detect circ-ZNF609 and AKT3 mRNA expression in the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609-mut expression plasmid. (B) Western blotting was used to detect the protein expression of ZNF609-250aa and AKT/mTOR/autophagy signaling pathway components in the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609-mut expression plasmid. (C) Laser confocal microscopy assays of GFP-LC3 in the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609-mut expression plasmid. The arrows highlight the main differences in LC3 autophagosomes. Scale bar: 50 μm. (D) Flow cytometry was used to detect apoptosis of the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609-mut expression plasmid. (E) MTS assays were used to examine cell proliferation of the HK-2 cells transfected with control vector, circ-ZNF609 expression plasmid, or circ-ZNF609-mut expression plasmid for 24, 48, and 72 h. The graphs present the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
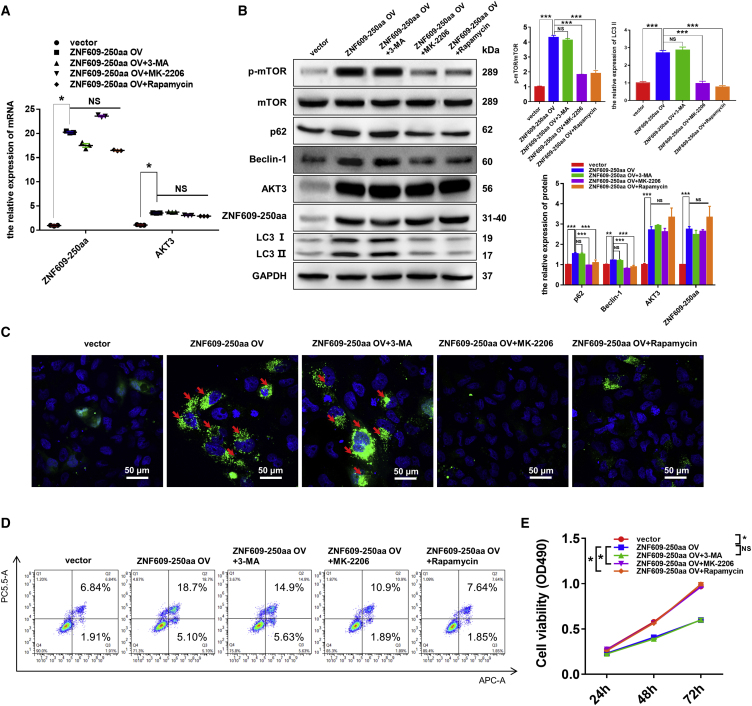
ZNF609-250aa induces autophagy flux impairment via AKT3/mTOR signaling in vitro and in vivo
To investigate the molecular mechanism by which ZNF609-250aa induces ischemic AKI, we transfected the ZNF609-250aa-expressing plasmid into HK-2 cells and supplemented with inhibitors of PI3K (3-MA), AKT (MK-2206), and mTOR (rapamycin) signaling. The overexpression efficiency of ZNF609-250aa mRNA was confirmed by quantitative real-time PCR (Figure 7A). Specifically, the RNA level of AKT3 was significantly increased when ZNF609-250aa was overexpressed in HK-2 cells, and supplementation with 3-MA, MK-2206, and rapamycin did not influence the mRNA expression of AKT3. We further detected the protein levels of ZNF609-250aa, p-mTOR, p62, Beclin-1, AKT3, and LC3-II/I by western blotting (Figure 7B). The results showed that ZNF609-250aa was significantly increased in the HK-2 cells transfected with ZNF609-250aa plasmid compared with the control cells. In addition, the protein levels of p-mTOR, p62, Beclin-1, and LC3-II were markedly increased in the ZNF609-250aa overexpression group compared with the control group, while their expression was significantly reduced after supplementation with MK-2206 and rapamycin. As expected, supplementation with 3-MA showed little impact on autophagy and AKT/mTOR pathway-related proteins. Laser scanning confocal microscopy, flow cytometry, and MTS assays were further performed to explore the impact of ZNF609-250aa and inhibitors on cell autophagy, apoptosis, and viability of HK-2 cells. The results showed that the overexpression of ZNF609-250aa resulted in accumulated autophagosomes (Figure 7C), increased cell apoptosis (Figure 7D), and reduced proliferation (Figure 7E), which were reversed by supplementation with MK-2206 and rapamycin but not 3-MA.
Figure 7.
ZNF609-250aa suppresses the proliferation, promotes the apoptosis, and induces autophagic flux impairment of RTECs by activating the mTOR signaling pathway by targeting AKT3 mRNA in vitro
(A) Quantitative real-time PCR was performed to detect the expression of ZNF609-250aa mRNA and AKT3 mRNA in the HK-2 cells transfected with control vector or ZNF609-250aa expression plasmid with or without 3-MA, MK-2206, or rapamycin. (B) Western blotting was used to detect the protein expression of ZNF609-250aa and AKT/mTOR/autophagy signaling pathway components in the HK-2 cells transfected with control vector or ZNF609-250aa expression plasmid with or without 3-MA, MK-2206, or rapamycin. (C) Laser confocal microscopy assay of GFP-LC3 in the HK-2 cells transfected with control vector or ZNF609-250aa expression plasmid with or without 3-MA, MK-2206, or rapamycin. The arrows highlight the main differences in LC3-autophagosomes. Scale bar: 50 μm. (D) Flow cytometry was used to detect apoptosis of the HK-2 cells transfected with control vector or ZNF609-250aa expression plasmid with or without 3-MA, MK-2206, or rapamycin. (E) MTS assays were used to examine cell proliferation of the HK-2 cells transfected with control vector or ZNF609-250aa expression plasmid with or without 3-MA, MK-2206, or rapamycin for 24, 48, and 72 h. The graphs present the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
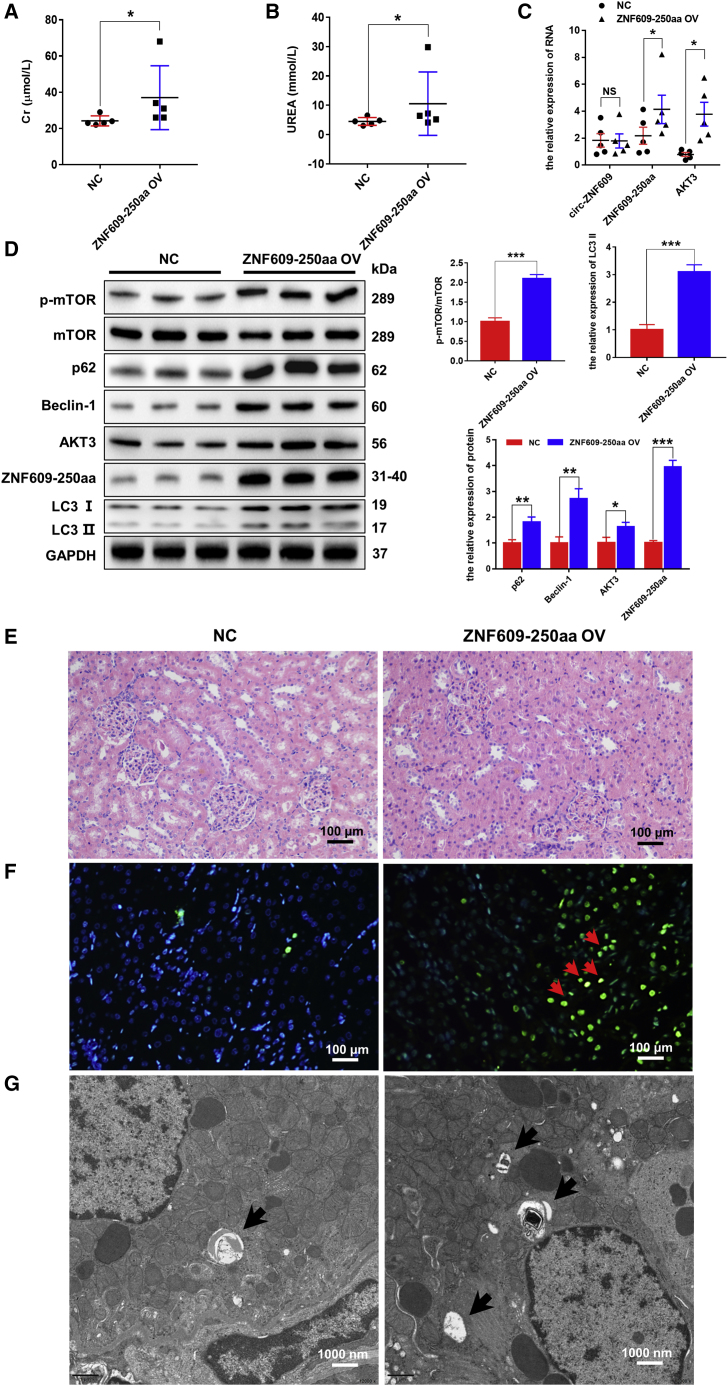
Eventually, we performed in vivo experiments to validate the molecular mechanism by which ZNF609-250aa induces autophagic flux impairment and renal injury by AAV-mediated ov-ZNF609-250aa in rats. The full-length ZNF609-250aa or the control sequence was inserted into a pAV-U6-GFP vector. After the accuracy of the vector was measured by sequencing, AAV9 was packaged, purified, titrated, and injected into the renal veins of rats. The results showed that the overexpression of ZNF609-250aa induced significantly elevated serum creatinine and urea compared with the control group (Figures 8A and 8B). Furthermore, the RNA levels of ZNF609-250aa and AKT3, but not circ-ZNF609, were high in the kidney tissues of the ZNF609-250aa-overexpression group compared with the control group, which indicated that the expression efficiency of ZNF609-250aa in rats is good (Figure 8C). Then, western blotting results showed that p62, Beclin-1, LC3-II/I, AKT3, and p-mTOR were upregulated at the protein level in the kidney tissues of the ZNF609-250aa-overexpression group compared with the control group (Figure 8D). Moreover, the kidney tissues were stained with H&E to determine tissue damage levels under light microscopy. The results showed that ZNF609-250aa overexpression caused dilation, necrosis, and lysis of renal tubules and renal interstitial damage compared with the control group (Figure 8E). Next, cell apoptosis levels were determined by TUNEL assays, and a strong signal was clearly observed in the kidney tissues of the ZNF609-250aa-overexpression group compared with the control group (Figure 8F). Furthermore, autophagic detection via transmission electron microscopy showed that autolysosomes accumulated compared with the control group (Figure 8G).
Figure 8.
ZNF609-250aa causes renal injury by inducing autophagic flux impairment by activating the AKT/mTOR signaling pathway in vivo
(A and B) The levels of creatinine and urea in plasma of rat models with or without ZNF609-250aa expression by AAV9. n = 5. (C) Quantitative real-time PCR was performed to detect circ-ZNF609, ZNF609-250aa, and AKT3 mRNA expression in kidney tissues of rat models with or without ZNF609-250aa by AAV9. n = 5. (D) Western blotting was used to detect the protein expression of ZNF609-250aa and AKT/mTOR/autophagy pathway components in kidney tissues of rat models with or without ZNF609-250aa expression by AAV9. (E) The histological properties in kidney tissues of rat models with or without ZNF609-250aa expression by AAV9 were observed under a microscope following H&E staining. Scale bar: 100 μm. (F) Apoptosis level in rat kidney cells of rat models with or without ZNF609-250aa expression by AAV9 was evaluated by the TUNEL method. The arrows highlight the main differences in cell apoptosis. Scale bar: 100 μm. (G) Transmission electron microscopic images of autophagic structures in the kidney tissues of rat models with or without ZNF609-250aa expression by AAV9. The arrows highlight the main differences in autophagic structures. Scale bar: 1,000 nm. The graphs present the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Taken together, these results verified that ZNF609-250aa is a crucial contributor to renal injury by inducing autophagic flux impairment via AKT3/mTOR signaling.
Discussion
Autophagy is a conserved sequential step in the degradation of intracellular organelles, proteins, and other macromolecules by the hydrolases of lysosomes.19,20 The normal autophagic process is the cornerstone of maintaining cellular homeostasis, and autophagic flux is proposed to represent the dynamic process between autophagosome formation and clearance by lysosomes. Autophagy is mostly induced in response to renal I/R injury, but both protective and harmful effects are observed, indicating a dual role for autophagy in ischemic AKI.21 In the present study, we found that autophagy is renoprotective when kidneys or RTECs undergo I/R injury. Overexpression of circ-ZNF609 promoted cell apoptosis and AKI by impairing the autophagy flux, which was blocked by silencing circ-ZNF609. Mechanistically, circ-ZNF609 encoded ZNF609-250aa, which exerts its pathological effect on AKI by impairing autophagy flux and promoting cell apoptosis via the AKT/mTOR signaling pathway.
Despite numerous methods for monitoring autophagy,22 we assessed the effects of circ-ZNF609 on autophagy in I/R injury conditions by measuring the classic autophagic markers LC3-I, LC3-II, and p62 in the present study. LC3-I and LC3-II are two forms of LC3, of which LC3-I is cytosolic and formed by the removal of the 22 C-terminal amino acids from LC3, whereas LC3-II is autophagosome membrane bound and formed by the conversion of a fraction of LC3-I into LC3-II.23 LC3-II levels can reflect the extent of autophagosome formation. However, the amount of LC3-II can also increase due to impaired clearance of autophagosomes in lysosomes. Therefore, p62, a protein incorporated into the autophagosome and degraded by autophagy, is frequently employed as a complementary marker of autophagic flux and can accumulate if autophagy is inhibited.24,25 Our results showed that autophagosome, LC3-II, and p62 levels were simultaneously increased in vitro and in vivo after treatment with circ-ZNF609 overexpression or I/R injury, suggesting that defective autophagosome clearance might be responsible for the autophagic flux impairment and that autophagy plays a renoprotective role in ischemic AKI. However, LC3-GFP transgenic mice may be more appropriate for revealing the dynamic variation in autophagic flux if available in further studies.
The involvement of circRNAs in specific types of kidney diseases, including AKI, hypertensive nephropathy, and lupus nephritis, has attracted increased attention.26 Recently, a growing number of studies have shown that most circRNAs function on the target mRNA 3′ UTRs as miRNA sponges in various AKIs. Notably, the elevation or decrease in different circRNAs might exert different effects, including renoprotective effects or enhanced injury. Xu et al. showed that circ-AKT3 could aggravate renal I/R injury by sponging miR-144-5p and activating the Wnt/β-catenin pathway and oxidative stress.27 Ma et al. reported that circ-Ttc3 could relieve inflammation and oxidative stress by regulating Rcan2 expression by binding to and negatively regulating miR-148a in rats with AKI induced by sepsis.28 Cheng et al. systematically explored the circRNA- or lncRNA (long noncoding RNA)-associated competing endogenous RNA mechanisms in a rat model of contrast-induced AKI (CI-AKI) through deep RNA sequencing, providing new insights into the underlying mechanism of CI-AKI.29 Our study showed that circ-ZNF609 is a contributor to ischemic AKI that presented upregulated expression, as confirmed in the various specimens. In terms of functional impact, circ-ZNF609 overexpression suppressed the proliferation and autophagic flux of RTECs and promoted apoptosis, while silencing circ-ZNF609 with siRNAs protected against RTEC injury induced by OGD/R. Notably, our study first reported the positive correlation between circ-ZNF609 and AKI and the effect of circ-ZNF609 on autophagy.
Regarding circ-ZNF609, previous studies have confirmed an influential and pivotal molecule that was significantly differentially expressed in various diseases and functioned by specific mechanisms, including miRNA sponges and encoding proteins. Almost all of the studies focusing on circ-ZNF609 were conducted in malignant diseases and showed that its expression is upregulated and it is a contributor to the development and progression of diseases. For instance, circ-ZNF609 exhibits high expression in lung cancer and promotes lung cancer cell proliferation and migration by acting as a sponge for miR-142-3p and modulating G protein subunit β2.30 He et al. reported that circ-ZNF609 is conspicuously overexpressed in hepatocellular carcinoma and enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of GLI2 expression by targeting miR-15a-5p/15b-5p.31 The evidence mentioned above seems inconsistent with our results that circ-ZNF609 could suppress cell proliferation, promote cell apoptosis, and induce cell injury in ischemic AKI. What is clear, however, is that circ-ZNF609 expression is upregulated in various diseases, including ischemic AKI, and contributes to the development of the pathophysiological progress by influencing cell proliferation and apoptosis. However, there is also evidence about circ-ZNF609 in non-neoplastic lesions, supporting our results. Ge et al. reported that silencing circ-ZNF609 could reveal a protective function in skin oxidative damage by suppressing the loss of cell proliferation, cell apoptosis, and reactive oxygen species (ROS) generation through regulation of the miR-145, JNK, and p38MAPK pathways.32 Liu et al. reported that circ-ZNF609 acts as an endogenous miR-615-5p sponge to sequester and inhibit miR-615-5p activity in vascular endothelial dysfunction, which leads to increased MEF2A expression, reduces cell proliferation, increases cell apoptosis, and suppresses cell migration and tube formation.33 Evidence in non-neoplastic lesions showed that the elevation of circ-ZNF609 also plays a role in inhibiting cell proliferation and aggravates cell or tissue injury by regulating various pathways. Therefore, the discrepancies in the results from different studies may be due to various functions and downstream targets of circ-ZNF609 in different pathogenetic processes.
Notably, Legnini et al. reported the protein-encoding ability of human circ-ZNF609 in myogenesis and found that circ-ZNF609 contains a 753 nt ORF spanning from the putative AUG of the host gene to a STOP codon created 3 nt after the splice junction.14 In addition, the study showed that the protein encoded by circ-ZNF609 might act as a dominant-negative competitor or as a modulator of alternative ZNF609 complex formation. Indeed, our results showed that rat circ-ZNF609 encodes a 250 aa protein with 94% homology in humans from a similar ORF. Furthermore, the protein and mRNA levels of AKT3 and the phosphorylation levels of mTOR were upregulated after renal I/R injury, and both were positively correlated with the expression of circ-ZNF609 and ZNF609-250aa. mTOR, a serine/threonine protein kinase that is mainly involved in regulating cell growth, proliferation, motility, and survival, can be activated by multiple signals, including amino acids, glucose, oxidative stress, and growth factors.34 mTOR complex 1 (mTORC1) is a recognized inhibitor of autophagy, while AKT is a major upstream regulatory factor of mTORC1. In addition, previous studies have shown that the AKT/mTOR pathway is a vitally negative pathway of the autophagic process, as confirmed by our results that autophagic flux of RTECs is impaired when the AKT/mTOR pathway is activated.35,36
Regarding the relationship between the AKT/mTOR signaling pathway and AKI, Zhu et al. reported that αKlotho protein could enhance autophagy by inhibiting the AKT/mTOR signaling pathway, which resulted in suppression of NLRP3 inflammasome-mediated pyroptosis to protect against CI-AKI.37 Hsu et al. found that lactoferrin contributes a renoprotective effect in AKI by inducing autophagy via the activation of AMPK and inhibition of the AKT/mTOR signaling pathway and suppressing oxidative stress-induced cell death and apoptosis via the augmentation of autophagy.38 Similarly, our results indicated that AKT/mTOR signaling pathway inhibition might protect against renal I/R injury by restoring autophagic flux in RTECs. In addition, a recent study reported that circYAP1 could activate the PI3K/AKT/mTOR signaling pathway and protect HK-2 cells from I/R injury by sponging miR-21-5p, indicating that circRNA might play a vital role in ischemic AKI.39 However, in vivo experiments will be crucial for validating the role of circYAP1 in ischemic AKI. In the present study, we confirmed that silencing circ-ZNF609 was renoprotective by inhibiting the AKT/mTOR signaling pathway in rat models that underwent renal I/R injury with or without sh-circ-ZNF609 by AAV9. Furthermore, we employed 3-MA, MK-2206, and rapamycin as inhibitors and mutated circ-ZNF609 to explore the underlying mechanism of circ-ZNF609 in the AKT/mTOR signaling pathway and confirmed that ZNF609-250aa, not circ-ZNF609, activated the mTOR signaling pathway by regulating AKT3 expression and influenced autophagic flux. We first found that circ-ZNF609 plays a crucial role in ischemic AKI by encoding ZNF609-250aa, which could regulate the AKT/mTOR signaling pathway and influence the autophagic flux of RTECs. However, in theory, the best way to gain more precise insights into the molecular mechanism caused by circ-ZNF609 in ischemic AKI is to knock down circ-ZNF609 according to innovative transgenic technology.40 This issue should be addressed in future studies.
In conclusion, our study found a circRNA with upregulated expression, circ-ZNF609, in ischemic AKI tissues and cells. In addition, evidence indicated that circ-ZNF609 encoded a conserved 250 aa peptide, ZNF609-250aa. ZNF609-250aa, not circ-ZNF609 itself, is involved in the I/R injury of RTECs in vitro and in vivo. Furthermore, ZNF609-250aa suppresses the proliferation and promotes the apoptosis of RTECs by inducing autophagic flux impairment through activation of the AKT/mTOR signaling pathway after renal I/R injury. Collectively, our findings intuitively provide valuable insights into the pathogenesis of ischemic AKI. As a novel positive regulator of AKT/mTOR signaling, ZNF609-250aa is a potential diagnostic marker and therapeutic target for ischemic AKI.
Materials and methods
Samples, cell lines, and animals
All urinary samples were collected from patients with ADHF or undergoing cardiac surgery under CPB in Maoming People’s Hospital. The samples in ADHF patients were collected every 24 h for the first 7 days during hospitalization, and the level of circ-ZNF609 on admission and 24 h before the development of AKI was employed as a predictor for non-AKI and AKI, respectively. The samples in CPB patients were collected 24 h before and after the operation. The urinary samples were centrifuged at 3,000 × g for 10 min and the sediment was stored at −80°C. We examined the expression of circ-ZNF609 in urinary samples by ddPCR to confirm its elevated expression in AKI patients. The ddPCR was performed on a MicroDrop-100 ddPCR system (Forevergen, Guangzhou, China) to measure the levels of urinary circ-ZNF609. The ddPCR mixture was prepared with the following: 10 μL of reaction Supermix (Evagreen), 1.8 μL of forward primer, 1.8 μL of reverse primer, and 1.5 μL of template. The mixture was pipetted into the sample wells of a Droplet Generator Cartridge, and droplet generation oil was added into the oil wells. Then, the cartridges were covered with gaskets and placed in a MicroDrop-100A droplet generator to generate droplets, which were then transferred to a 96-well plate and sealed with pierceable foil for PCR amplification. The fluorescence signal of the droplets was recorded with a MicroDrop-100B, and QuantDrop software was used to quantify the expression. This study was approved by the Ethical Committee of Maoming People’s Hospital, and the human materials were obtained with written informed consent.
The human RTEC line HK-2 and the human embryonic kidney cell line 293T were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All these cell lines were maintained in complete medium with 5% CO2 and 95% air to compare the groups. For OGD/R, cells were subjected to glucose-free medium under hypoxic conditions for 4 h. Then, we used standard conditions to maintain the cells in complete medium for 12, 24, 48, and 72 h. In addition, cells cultured in standard medium under normoxic conditions were used as controls. Eight-week-old SD rats weighing 200–250 g were included in this study (Guangdong Medical Laboratory Animal Center) after adaptation to standard laboratory conditions with free access to food and water, except for modeling for more than 1 week at the Forevergen Laboratory Animal Center (Forevergen, Guangzhou, China). All animals were treated following the guidelines of the Committee on Animals of Guangdong Provincial People’s Hospital. SD rats were subjected to I/R surgery to establish ischemic AKI models as described in our previous study.17,18
RNA extraction, gDNA extraction, and quantitative real-time PCR analysis
Total RNA and gDNA were extracted using TRIzol solution (Thermo Fisher Scientific, USA) and the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, K182001), respectively, according to the manufacturer’s protocol. cDNA was generated using a Prime Script RT Master Mix kit (TaKaRa, Japan) by reverse transcription with 500 ng RNA and random primers according to the manufacturer’s protocol. Quantitative real-time PCR was performed using the GoTaq qPCR Master Mix kit according to the manufacturer’s protocol on a StepOnePlus Real-Time PCR System (Applied Biosystems, USA). Primers were synthesized as shown in Table S2. Moreover, the comparative CT method (ΔΔCT) was employed to calculate the relative expression of the gene.
Plasmids, siRNAs, and cell transfection
The sequences of human circ-ZNF609, circ-ZNF609 ATG mutation, circ-ZNF609-250aa, and the wild-type and mutant IRES of circ-ZNF609 were generated through chemical gene synthesis and cloned into the LV003 plasmid (Forevergen Biotechnology, Guangzhou, China) to construct the overexpressing vectors. siRNAs specific to human circ-ZNF609 were purchased from GenePharma (Shanghai, China) and used to transiently silence the expression of circ-ZNF609. According to the manufacturer’s protocol, the vectors and siRNAs mentioned above were transfected with Lipofectamine 2000 transfection reagent (Life Technologies, USA).
Western blotting
The cells and tissues were lysed in RAPI (protein lysis buffer containing protease inhibitor and phosphatase inhibitor) for 30 min on ice. Equal amounts of proteins in cell lysate or tissue lysate were separated by SDS-PAGE gels and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, MA, USA). After incubation with a primary antibody at 4°C overnight, the membranes were hybridized with a secondary antibody at room temperature for 1 h. The immunoreactive signals were visualized using an enhanced chemiluminescence kit (Forevergen Biotechnology, Guangzhou, China). GAPDH was used as an endogenous control to normalize the protein loading. Western blotting was finally developed with ECL solution (Amersham) and exposed to films.
Cell apoptosis analysis
The cell apoptosis level was detected by a flow cytometer using an Annexin V/7-AAD apoptosis detection kit (Biosea, Beijing, China) and the TUNEL method using a One Step TUNEL Apoptosis Assay Kit (C1088; Beyotime, Shanghai, China) according to the manufacturer’s protocol. Flow cytometry was performed as follows. HK-2 cells were resuspended in binding buffer containing Annexin V (BD Biosciences 550474, USA) and 7-AAD (BD Biosciences 559925, USA) at a concentration of 1 × 106 cell/mL and incubated in the dark for 15 min. Then, we detected apoptotic cells under a flow cytometer (BD FACSCalibur, BD Biosciences, USA). The TUNEL assay was performed as follows. Kidney slides were stained with 50 μL of TUNEL solution at 37°C for 1 h in the dark, and apoptotic cells were stained with green fluorescent dye. Then, we detected apoptotic cells by fluorescence microscopy.
Cell viability assay
The MTS assay (Promega, USA) was used to assess cell growth and viability by measuring formazan production following the addition of MTS according to the manufacturer’s protocol. Cells were seeded at a density of 4,000 cells/well in a 96 well plate and incubated in an incubator at 37°C and 5% CO2 for 24, 48, and 72 h. Then, a BioTek ELISA reader (BioTek Instruments) was used to detect the absorbance at a wavelength of 490 nm following cell incubation in a medium containing 10 μL of MTS for 4 h at 37°C.
Luciferase reporter assays
A psicheck2 system was used to amplify the Renilla luciferase (Rluc) and firefly luciferase (Fluc) sequences placed in front and back, respectively. Overlapping PCR generated the full-length sequences of Rluc-Fluc, and the flank sequences were connected to the pCDNA3.1(+) vector by two restriction enzyme sites, Mlu and XhoI. The potential IRES sequences of circ-ZNF609 were amplified and inserted in the middle of Rluc and Fluc by two restriction enzyme sites, BamHI and EcoRI, introduced by primers. The plasmids were transfected into HK-2 and 293T cells with Lipofectamine 2000 transfection reagent, and Rluc and Fluc activities were measured by the Dual Glo luciferase assay (Promega) according to the manufacturer’s protocol.
Transmission electronic microscopy and laser scanning confocal microscopy
Autophagy was detected by transmission electron microscopy and laser scanning confocal microscopy. Transmission electron microscopy was performed as follows. The kidney tissues were fixed with 2.5% glutaraldehyde at 4°C for 2 h and then double fixated, dehydrated, infiltrated, embedded, ultrathin sectioned, stained, and examined under a transmission electron microscope (JEM-1400 Plus, Japan). Laser scanning confocal microscopy was performed as follows. Briefly, HK-2 cells were maintained on glass slides, transfected with LC3-GFP, and then exposed to experimental conditions. Zeiss LMS510 software (EMBL, Germany) was used to record and analyze the cell images visualized by confocal microscopy.
AAV9 transduction of circ-ZNF609
For knockdown and overexpression of circ-ZNF609 and ZNF609-250aa in vivo, full-length sh-circ-ZNF609, ZNF609-250aa, or the control sequence was inserted into a pAV-U6-GFP vector. After the accuracy of the vector was measured by sequencing, AAV serotype 9 (AAV9) was packaged, purified, and titrated by Vigene Biosciences. One hundred microliters of AAV9 (5 × 1013 vg/mL) harboring either sh-circ-ZNF609 or ZNF609-250aa or the control sequence was injected into the renal veins of rats.41 Briefly, the rats were anesthetized; skin, muscle, and fascia were cut; the kidneys were exposed; and then the renal vein was clamped by a microaneurysm clamp. Finally, we injected the AAV particles mentioned above diluted in 100 μL of saline into the renal vein with a 31G needle and removed the clamp 15 min after injection, followed by suturing the incision.
Renal function and histopathology
Rat serum samples were collected and subjected to urea and creatinine measurement to assess renal function by a Roche Cobas C111 analyzer (Roche Diagnostics International, Rotkreuz, Switzerland). Furthermore, paraffin-embedded kidney tissue sections with a thickness of 2 μm were stained with H&E to determine tissue damage levels as previously described.17,18 Signs of dilation, necrosis, and lysis of renal tubules and renal interstitial congestion under light microscopy indicated damage.
Statistical analysis
Continuous variables are reported as the mean ± standard deviation from three independent experiments. Student’s t test and Mann-Whitney U test were used for parametric data and nonparametric data, respectively. All statistical tests were performed using GraphPad Prism software v.7.0 (GraphPad Prism, San Diego, CA, USA) and SPSS 24.0 software (SPSS, Chicago, IL, USA). The p values were two-tailed and were considered statistically significant if less than 0.05.
Acknowledgments
We thank the Maoming People’s Hospital staff for collecting samples and Yiyu Deng, Chunyu Deng, and Maolei Zhang for experimental assistance. C.C. is currently receiving a grant from the major program of Summit Project, Guangdong Province High-Level Hospital Construction Project of Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences (DFJH2020028) and a grant from the program of Outstanding Young Medical Talents in Guangdong Province (KJ012019445).
Author contributions
C.C., O.X., and H.Z. conceived and designed the experiments. H.Z., F.H., Z.H., G.F., H.T., H.Y., W.Q., D.J., X.J., and W.Y. performed the experiments. H.L. and O.X. processed the data. O.X. and F.H. wrote the manuscript. Y.H. and Y.Q. provided expertise.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.09.007.
Supplemental information
Data availability
The authors declare that all relevant data of this study are available within the article or from the corresponding author on reasonable request.
References
- 1.Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron. Clin. Pract. 2012;120:c179–c184. doi: 10.1159/000339789. [DOI] [PubMed] [Google Scholar]
- 2.Bonventre J.V., Yang L. Cellular pathophysiology of ischemic acute kidney injury. J. Clin. Invest. 2011;121:4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvadori M., Rosso G., Bertoni E. Update on ischemia-reperfusion injury in kidney transplantation: pathogenesis and treatment. World J. Transpl. 2015;5:52–67. doi: 10.5500/wjt.v5.i2.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Chen Y., Lai B., Chen Y.X., Xu C.Y., Liu Y.F. Overexpression of SP1 restores autophagy to alleviate acute renal injury induced by ischemia-reperfusion through the miR-205/PTEN/Akt pathway. J. Inflamm. 2021;18:7. doi: 10.1186/s12950-021-00270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian R., Wang P., Huang L., Li C., Lu Z., Lu Z., Wu A., Bao K., Mao W., Huang Q., Xu P. Sanqi oral solution ameliorates renal ischemia/reperfusion injury via reducing apoptosis and enhancing autophagy: involvement of ERK/mTOR pathways. Front. Pharmacol. 2020;11:537147. doi: 10.3389/fphar.2020.537147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu S., Chen X., Pang Y., Zhang Z. Lipocalin-2 protects against renal ischemia/reperfusion injury in mice through autophagy activation mediated by HIF1α and NF-κb crosstalk. Biomed. Pharmacother. 2018;108:244–253. doi: 10.1016/j.biopha.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X.J., Chen S., Huang K.X., Le W.D. Why should autophagic flux be assessed? Acta Pharmacol. Sin. 2013;34:595–599. doi: 10.1038/aps.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlop E.A., Tee A.R. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin. Cell Dev. Biol. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Rubinsztein D.C., Mariño G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Mo Y., Peng M., Zhang S., Gong Z., Yan Q., Tang Y., He Y., Liao Q., Li X., et al. The influence of circular RNAs on autophagy and disease progression. Autophagy. 2021;18:240–253. doi: 10.1080/15548627.2021.1917131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao S., Wu S., Huang S., Liu M., Gao B. Advances in the identification of circular RNAs and research into circRNAs in human diseases. Front. Genet. 2021;12:665233. doi: 10.3389/fgene.2021.665233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X., Xia X., Li F., Zhang M., Zhou H., Wu X., Zhong J., Zhao Z., Zhao K., Liu D., et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol. 2021;23:278–291. doi: 10.1038/s41556-021-00639-4. [DOI] [PubMed] [Google Scholar]
- 16.Jiang T., Xia Y., Lv J., Li B., Li Y., Wang S., Xuan Z., Xie L., Qiu S., He Z., et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer. 2021;20:66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang M., Liu S., Zhou Y., Deng Y., Yin Q., Hu L., Ouyang X., Hou Y., Chen C. Circular RNA involved in the protective effect of losartan on ischemia and reperfusion induced acute kidney injury in rat model. Am. J. Transl. Res. 2019;11:1129–1144. [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y., Peng W., Wei R., Zhou Y., Fang M., Liu S., Deng Y., Yin Q., Ouyang X., Hu L., et al. Rat mRNA expression profiles associated with inhibition of ischemic acute kidney injury by losartan. Biosci. Rep. 2019;39 doi: 10.1042/BSR20181774. BSR20181774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y., He D., Yao Z., Klionsky D.J. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decuypere J.P., Ceulemans L.J., Agostinis P., Monbaliu D., Naesens M., Pirenne J., Jochmans I. Autophagy and the kidney: implications for ischemia-reperfusion injury and therapy. Am. J. Kidney Dis. 2015;66:699–709. doi: 10.1053/j.ajkd.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu M., Kageyama S., Ichimura Y. p62/SQSTM1/A170: physiology and pathology. Pharmacol. Res. 2012;66:457–462. doi: 10.1016/j.phrs.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Jiang P., Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015;75:13–18. doi: 10.1016/j.ymeth.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Ren G.L., Zhu J., Li J., Meng X.M. Noncoding RNAs in acute kidney injury. J. Cell. Physiol. 2019;234:2266–2276. doi: 10.1002/jcp.27203. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y., Jiang W., Zhong L., Li H., Bai L., Chen X., Lin Y., Zheng D. circ-AKT3 aggravates renal ischaemia-reperfusion injury via regulating miR-144-5p/Wnt/β-catenin pathway and oxidative stress. J. Cell Mol. Med. 2020 doi: 10.1111/jcmm.160722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma X., Zhu G., Jiao T., Shao F. Effects of circular RNA Ttc3/miR-148a/Rcan2 axis on inflammation and oxidative stress in rats with acute kidney injury induced by sepsis. Life Sci. 2021;272:119233. doi: 10.1016/j.lfs.2021.119233. [DOI] [PubMed] [Google Scholar]
- 29.Cheng W., Li X.W., Xiao Y.Q., Duan S.B. Non-coding RNA-associated ceRNA networks in a new contrast-induced acute kidney injury rat model. Mol. Ther. Nucleic Acids. 2019;17:102–112. doi: 10.1016/j.omtn.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S., Yang N., Jiang X., Wang J., Dong J., Gao Y. FUS-induced circular RNA ZNF609 promotes tumorigenesis and progression via sponging miR-142-3p in lung cancer. J. Cell. Physiol. 2021;236:79–92. doi: 10.1002/jcp.29481. [DOI] [PubMed] [Google Scholar]
- 31.He Y., Huang H., Jin L., Zhang F., Zeng M., Wei L., Tang S., Chen D., Wang W. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis. 2020;11:358. doi: 10.1038/s41419-020-2441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge R., Gao G. Anti-antioxidant impacts of circZNF609 silence in HaCaT cells through regulating miR-145. Artif. Cells Nanomed. Biotechnol. 2020;48:384–392. doi: 10.1080/21691401.2019.1709863. [DOI] [PubMed] [Google Scholar]
- 33.Liu C., Yao M.D., Li C.P., Shan K., Yang H., Wang J.J., Liu B., Li X.M., Yao J., Jiang Q., Yan B. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buss S.J., Riffel J.H., Katus H.A., Hardt S.E. Augmentation of autophagy by mTOR-inhibition in myocardial infarction: when size matters. Autophagy. 2010;6:304–306. doi: 10.4161/auto.6.2.11135. [DOI] [PubMed] [Google Scholar]
- 35.Heras-Sandoval D., Pérez-Rojas J.M., Hernández-Damián J., Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell. Signal. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Suhara T., Baba Y., Shimada B.K., Higa J.K., Matsui T. The mTOR signaling pathway in myocardial dysfunction in type 2 diabetes mellitus. Curr. Diab. Rep. 2017;17:38. doi: 10.1007/s11892-017-0865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Li S., Lin Q., Shao X., Wu J., Zhang W., Cai H., Zhou W., Jiang N., Zhang Z., et al. αKlotho protein has therapeutic activity in contrast-induced acute kidney injury by limiting NLRP3 inflammasome-mediated pyroptosis and promoting autophagy. Pharmacol. Res. 2021;167:105531. doi: 10.1016/j.phrs.2021.105531. [DOI] [PubMed] [Google Scholar]
- 38.Hsu Y.H., Chiu I.J., Lin Y.F., Chen Y.J., Lee Y.H., Chiu H.W. Lactoferrin contributes a renoprotective effect in acute kidney injury and early renal fibrosis. Pharmaceutics. 2020;12:E434. doi: 10.3390/pharmaceutics12050434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang T., Cao Y., Wang H., Wang Q., Ji J., Sun X., Dong Z. Circular RNA YAP1 acts as the sponge of microRNA-21-5p to secure HK-2 cells from ischaemia/reperfusion-induced injury. J. Cell. Mol. Med. 2020;24:4707–4715. doi: 10.1111/jcmm.15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suenkel C., Cavalli D., Massalini S., Calegari F., Rajewsky N. A highly conserved circular RNA is required to keep neural cells in a progenitor state in the mammalian brain. Cell Rep. 2020;30:2170–2179.e5. doi: 10.1016/j.celrep.2020.01.083. [DOI] [PubMed] [Google Scholar]
- 41.Wang P., Luo M.L., Song E., Zhou Z., Ma T., Wang J., Jia N., Wang G., Nie S., Liu Y., Hou F. Long noncoding RNA lnc-TSI inhibits renal fibrogenesis by negatively regulating the TGF-β/Smad3 pathway. Sci. Transl. Med. 2018;10:eaat2039. doi: 10.1126/scitranslmed.aat2039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all relevant data of this study are available within the article or from the corresponding author on reasonable request.