Summary
In the past decade, the term organoid has moved from obscurity to common use to describe a 3D in vitro cellular model of a tissue that recapitulates structural and functional elements of the in vivo organ it models. The term organoid is now applied to structures formed as a result of two distinct processes: the capacity for adult epithelial stem cells to re-create a tissue niche in vitro and the ability to direct the differentiation of pluripotent stem cells to a 3D self-organizing multicellular model of organogenesis. While these two organoid fields rely upon different stem cell types and recapitulate different processes, both share common challenges around robustness, accuracy, and reproducibility. Critically, organoids are not organs. This commentary serves to discuss these challenges, how they impact genuine utility, and shine a light on the need to improve the standards applied to all organoid approaches.
Associated podcast
For an associated discussion of this work, listen to the latest episode of The Stem Cell Report podcast at https://www.isscr.org/podcast/s2-e10, brought to you by the ISSCR.
While organoids have been heralded as a breakthrough in the modeling of human disease, this review identifies the challenges with such models, provides commentary on risks from overinterpretation, and suggests approaches to improve the validity and hence genuine utility of organoid research outcomes.
Introduction
The modern term organoid refers to cells growing in a defined three-dimensional (3D) environment in vitro to form clusters of cells that self-organize and differentiate into functional cell types, recapitulating the structure and function of an organ in vivo (hence, also called “mini-organs”). Organoids can be derived from either pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), or from tissue-derived stem cells (TSCs) of a given tissue. Pluripotent stem cell-derived organoids form through a process similar to the way in which the organ acquires its distinctive organization and multicellularity during development. By contrast tissue-derived organoids recapitulate the epithelial niche of their tissue of origin. Self-organization within either type of organoid can occur through spatially restricted lineage commitment and/or cell sorting, which requires activation of cell-cell or paracrine signaling pathways with this mediated by both intrinsic cellular components and extrinsic environments such as extracellular matrix (ECM) and media.
Lancaster and Knoblich (2014) defined an organoid as a collection of organ-specific cell types that develops from stem cells or organ progenitors, and self-organizes through cell sorting and spatially restricted lineage commitment in a manner similar to in vivo. They extend the criteria to require organoids to contain multiple organ-specific cell types, recapitulate some type of organ-specific function, and contain cells grouped morphologically in a fashion similar to the organ of origin. As a generic definition of an organoid, this fails to identify the distinctions between pluripotent stem cell and tissue stem cell-derived organoids, which was first given by Huch and Koo (2015). In the case of TSC-derived organoids, it is possible to enforce a state where the cellular complexity is suppressed and the expansion of progenitors displaying specific hallmarks of a regenerative state dominates (Lukonin et al., 2020; Yui et al., 2018). Upon release of stimulus, these cells retain multi-lineage potential. While TSC-derived organoids are regarded as modeling both homeostatic and injury triggered responses, the generation of organoids from PSCs follows a trajectory modeled on organogenesis where there can be intermediary progenitor states but where an organoid is definitively multi-lineage.
While there are clear differences in starting cell type, cellular complexity, and approach, the term organoid is used for both types of 3D cellular models of tissues and both fields have generated significant excitement and interest given the hope that these organotypic models of human origin can model disease. Hence, patient-derived cellular models may substantially improve our ability to model, understand, and potentially treat human disease. However, with hope comes hype. The exponential expansion in interest in using organoid approaches to study human tissues has seen a lot of studies performed with limited rigor, poor model validation, and a lack of unbiased analysis or hypothesis validation. As such, both fields of organoid biology suffer from very similar challenges, and both require substantial consideration with respect to the development of appropriate standards.
As authors, who are stem cell researchers in both the TSC and PSC-derived organoid fields, focused on gut and kidney, respectively, our intention with this report was to introduce both approaches, including the advantages and disadvantages, while providing clear examples of where there is a need for more rigor.
Tissue stem cell-derived organoids
Independent reports from the laboratories of Hans Clevers and Calvin Kuo described how primary intestinal epithelial cells from mouse could be maintained either in Matrigel using defined cell culture components (Sato et al., 2009) or as spheroids in air liquid interface (ALI) cultures on a collagen matrix in the presence of mesenchyme (Ootani et al., 2009) (Figure 1A). In contrast to the ALI cultures, intestinal epithelial cells cultured in Matrigel could be passaged repeatedly even from single Lgr5-expressing stem cells, providing in vitro evidence for their long-term self-renewing potential. Hence, we can define a tissue-derived organoid as a derivative of primary cells that are grown with or without mesenchyme under conditions where they maintain functions characteristic of the tissue of origin.
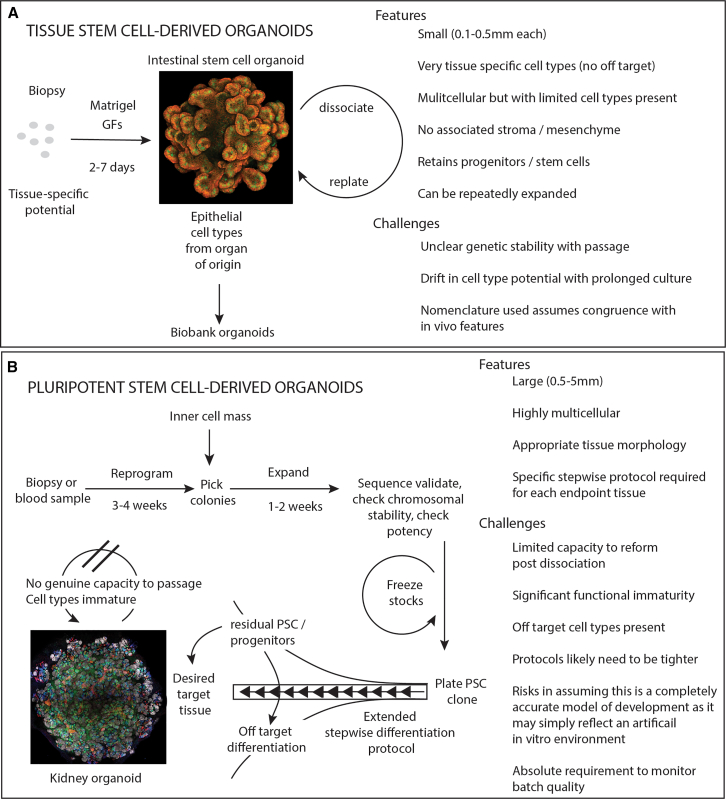
Figure 1.
Generation of organoids highlighting the starting cell source, method of generation, features, and challenges
(A) Tissue stem cell-derived organoids are generated from a patient biopsy, can be repeatedly passaged, and are relatively fast to expand. While multicellular, the self-renewing epithelium includes tissue-specific stem cells, and these ensure that organoids can be propagated upon dissociation and replating. Additional tissue complexity can be engineered via the inclusion of other cell types and engineering of more complex environments/scaffolds. The relevance of this will depend upon the biological question. The image shows a murine intestinal epithelial organoid stained for beta-catenin (red) and Lyzosyme (green).
(B) Pluripotent stem cell-derived organoids require the derivation of a pluripotent stem cell line. This diagram illustrates an induced pluripotent stem cell pathway commencing with a patient biopsy and requiring reprogramming. An alternative is derivation of a new line from the inner cell mass of the blastocyst. All tissue types can be generated from a PSC clone. The resulting organoids are highly multicellular and reflect the target tissue morphology. Organoid formation requires a multi-step differentiation protocol during which there is the risk of off-target cell type formation or the retention of residual undifferentiated stem cells/progenitors. The resulting tissue contains immature cell types and often cannot be passaged. Protocols are being developed to mature component cell types. The image shows a kidney organoid generated from a human iPSC control cell line stained for NEPHRIN (white), LTL (blue), E-CADHERIN (green), and GATA3 (red).
Following the establishment of culture conditions for the small intestine, modifications of the original cell culture conditions have enabled expansion and serial passaging of organoids from essentially all internal organs from both mouse and humans (reviewed in Schweiger and Jensen, 2016). Interestingly, epithelial cells from certain tissues such as liver (cholangiocytes) (Huch et al., 2013), corpus part of the stomach (Stange et al., 2013), and the human fetal and adult small intestine and colon grow like spheroids (Fordham et al., 2013; Huch et al., 2015; Sampaziotis et al., 2017), whereas epithelial cells isolated from salivary gland, pylorus of the stomach (Barker et al., 2010), and mouse small intestine form budding organoids (Maimets et al., 2016; Sato et al., 2009, 2011b). Of note, modification to the original conditions for growing human intestinal organoids transformed them from spheroids into budding organoids (Fujii et al., 2018). Therefore, it remains to be elucidated whether such morphological differences are based on extrinsic or cell intrinsic properties. Altogether, this illustrates that different tissues share common mechanisms for in vitro stem cell self-renewal. The ability to isolate and subsequently expand primary cells as organoids in vitro from even small biopsies (<2 mm3) has facilitated the establishment of biobanks from healthy individuals and patients for studying different types of cancer (Boj et al., 2015; Broutier et al., 2017; Gao et al., 2014; Kawasaki et al., 2020; Sachs et al., 2018; van de Wetering et al., 2015), monogenic disorders such as cystic fibrosis (Dekkers et al., 2013), α1-antitryps in deficiency in the liver (Huch et al., 2015), Alagille (Andersson et al., 2018) syndrome, inflammatory syndromes (Kraiczy et al., 2019; Nishimura et al., 2019), and interactions with microorganisms including Hepatitis B virus (De Crignis et al., 2021), SARS-CoV-2 virus (Lamers et al., 2020), Noro virus (Ettayebi et al., 2016), and genotoxic pks+ E. coli bacteria (Pleguezuelos-Manzano et al., 2020) (Figure 1A). Importantly, such biobanks also represent an entry into the assessment of how genetic variance across populations affects cellular phenotypes. While multicellular, the relative cellular simplicity of these tissue-specific epithelia facilitates simple phenotypic readouts with the challenge of inter-individual variation driven by the genetic heterogeneity between human samples.
One of the best characterized tissue-derived organoid systems to date are organoids derived from the epithelium of the adult small intestine. By recapitulating the in vivo stem cell niche in vitro, single intestinal epithelial stem cells embedded in Matrigel and basal medium supplemented with epidermal growth factor (EGF), the BMP antagonist Noggin, and the Wnt co-factor Rspondin1 will efficiently form organoids from murine tissues (Sato et al., 2009). In the absence of exogenous WNT, growth does depend on the emergence of a secretory Paneth cell as a source of WNT signal (Sato et al., 2011b), and human cultures similarly require addition of exogenous WNT (Sato et al., 2011a, 2011b). Knowledge of stem cell niche components from other tissues has enabled expansion of self-renewing stem cell populations from most human and mouse epithelial tissues (Kim et al., 2020). The formed organoids can be passaged repeatedly, providing the basis for a robust expansion of the primary cell mass. Notably, we still need to understand whether the derived organoid cultures mimic in vivo homeostasis, regeneration, or something completely different. Interestingly, time-resolved analysis of intestinal epithelial cells cultured from single cells revealed that the growth phase of intestinal epithelial organoids can be divided into two phases: 1) a first stage following passaging that recapitulates aspects of tissue regeneration; and 2) a second stage that is intimately linked with the emergence of secretory cells, where the organoids transition into a state mirroring homeostasis (Serra et al., 2019). It will be important to definitively address whether organoids isolated from other tissues follow a similar behavior.
Tissue-derived organoids are derived from tissue biopsies (Figure 1A). Representing the spontaneous expansion and differentiation of an unmanipulated, tissue-specific cell type isolated from a patient organ, tissue-derived organoids represent a model of the epithelial niche of that organ. As such, tissue-derived organoids can only model the organ of origin. Given that most tissue-derived organoid models consist exclusively of epithelial cells, these represent elegant systems for studying how cell fate is regulated in a tissue intrinsic manner. Importantly, it is also possible to add both additional cell types to the cultures of epithelial organoids, such as immune cells, neurons, and fibroblasts to study the cross talk between cellular compartments generating complex coculture systems (Cordero-Espinoza et al., 2021; Kabiri et al., 2014; Lee et al., 2014; Maimets et al., 2022; Ootani et al., 2009). While chromosomal integrity and stability has been a well-debated challenge within the PSC-derived organoid field, this is rarely discussed or investigated in the TSC-derived organoid field. Some analyses across time have suggested substantial genetic stability (Georgakopoulos et al., 2020; Huch et al., 2015; Huch and Koo, 2015), or at least a rate of mutation accumulation similar to what might be anticipated in vivo (Blokzijl et al., 2016). However, observations of drift in replicative and differentiation capacity with time are observed.
Tissues in our body can be divided largely into two categories depending on their ability to undergo tissue renewal. This also specifies the major difference between tissue-derived organoids and those generated via the directed differentiation of PSCs (see below). Whereas tissue-organoids can be derived from essentially all surface epithelia of ectodermal and endodermal origin, this has proven essentially impossible from mesoderm-derived organs such as heart and blood, and neuronal lineages. To study these mesodermal and ectodermal non-epithelial tissues in vitro, it has been necessary to establish directed differentiation protocols that recapitulate signaling events in the developing embryo.
Pluripotent stem cell-derived organoids
The term pluripotency is used to describe an ability to give rise to all lineages of the developing embryo. A transient cellular state during the early embryo, it is possible to support a pluripotent state in vitro either via the isolation of pluripotent cells from the early embryo (referred to as embryonic stem cells; ESCs) or via the reprogramming of adult somatic cells to a pluripotent state (referred to as induced PSCs; iPSCs). An ability to support cells in vitro in a pluripotent state (mouse ESC) was critical for several decades of mouse genetic research via the formation of genetically modified chimeras (Evans and Kaufman, 1981; Martin, 1981). The isolation of the first human ESC (Thomson et al., 1998), however, brought the possibility of recreating human tissues for understanding or treating human disease. As in mouse, the re-expression of key regulators of pluripotency within any adult somatic cell can take that cell back to a state of pluripotency equivalent to the early embryo (Takahashi et al., 2007; Takahashi and Yamanaka, 2006).
The potency of PSC is embodied in their multi-lineage differentiation capacity in teratoma formation and embryoid assays. Initial studies on differentiating these pluripotent cells to a more differentiated endpoint focused on single cell types, but still deployed the knowledge of developmental processes to guide protocol optimization. It was soon observed that directed differentiation of ESCs using developmental principles resulted in the formation of complex self-organizing 3D structures containing multiple cell types arranged as they would be during embryogenesis (Sasai et al., 2012) (Figure 1B). From human PSC, this has included tissues such as the optic cup, retina, all regions of the gastrointestinal tract, the cortex of the brain, kidney, lung, blood vessels, heart, skin, and inner ear (Dye et al., 2015; Eiraku et al., 2008, 2011; Freedman et al., 2015; Hayashi and Yoshino, 2022; Lancaster et al., 2013; Lee et al., 2020; Morizane et al., 2015; Spence et al., 2011; Taguchi et al., 2014; Takasato et al., 2015; Wimmer et al., 2019). Importantly, the formed structures recapitulate cellular interactions between, e.g., neurons, blood lineages, epithelia, and mesenchyme, as the organoids co-evolve to generate tissue-like architecture. This multi-lineage multicellularity made them excellent models of developing organs. The derivation of some organoid types relies on an initial aggregation of PSCs as in embryoid body formation, whereas other organoids are initially guided toward a specific germ layer in 2D cultures, which is then either aggregated to form 3D structures or spontaneously initiate organoid formation (Figure 1B).
Many different types of PSC-derived organoids do not reach the full functionality characterizing the native tissue but retain fetal properties. Improved maturation can be achieved in some cases with transplantation into animal models (Revah et al., 2022; Subramanian et al., 2019; van den Berg et al., 2018; Watson et al., 2014; Westerling-Bui et al., 2022) or extended time in culture (Quadrato et al., 2017). While some protocols are rapid, with self-organizing structures evident within 2–3 weeks, other protocols are longer and more complicated with evidence in, for example, cortical organoids of continuing cellular complexity and maturation across many months (Camp et al., 2015; Gordon et al., 2021; Velasco et al., 2019). Like tissue-derived organoids, some PSC-derived organoids establish the intrinsic self-renewal capacity of the native tissue and can be serially propagated, such as PSC-derived intestine. In contrast, organoid models from tissue, such as the kidney, cannot be propagated. This reflects the in vivo behavior of these tissues, where progenitor populations responsible for forming the nephrons are present in early kidney organoids but lost with time in culture (Howden et al., 2019). When utilizing such cultures for disease modeling, it is important to take into account this immaturity. In addition, unlike banked tissue-derived organoids, the starting point for disease-modeling experiments generated using PSCs are the pluripotent stem cells themselves, as the differentiated organoids cannot be banked (Figure 1B).
PSC-derived organoids have proven a versatile tool for providing new insight into mechanisms that govern developmental processes in humans and specific disease mechanisms of mutations associated with congenital disorders. These models have enabled insights into how specific cell types contribute to the development of specific tissue structures and have also elucidated how gradients of specific growth factors, cytokines, and morphogens influence cellular identity during organogenesis. Most notable, the ability to study morphogenesis in vitro with PSCs derived from patients, or via introduction of allelic series of mutations associated with congenital disorders, has provided a tool to investigate diseases, including microcephaly, autism spectrum disorder, cystic kidney disease, ciliopathies and glomerulopathies, congenital cardiac anomalies, and many other conditions (Brandão et al., 2020; Cruz et al., 2017; Dorison et al., 2022, 2023; Dvela-Levitt et al., 2019; Forbes et al., 2018; Freedman et al., 2015; Lancaster et al., 2013; Majmundar et al., 2021; Paulsen et al., 2022; Rowe and Daley, 2019; Tran et al., 2022). In addition, PSC-derived organoids have been used to generate tissue models to study microbial interactions including viral infection of SARS-CoV-2 (Vanslambrouck et al., 2022; Yang et al., 2020) and Zika virus (Tang et al., 2016), and bacteria such as Clostridium difficile (Leslie et al., 2015) and Salmonella (Forbester et al., 2015). Finally, injury models, such as cryoinjury or anthracycline-induced cardiotoxicity, have also been modeled (Hofbauer et al., 2021; McOwan et al., 2020; Mills et al., 2017). Collectively, these disease and injury models are hoped to be game changers for the development of new therapeutic strategies for congenital disorders and infectious diseases.
In the case of PSC-derived organoids, any tissue can be generated from the same starting stem cell. By contrast, TSCs can only recapitulate a model of the epithelia of the tissue of origin (Figure 1). This, in principle, avoids the need to obtain multiple tissue biopsies, a process regarded as an invasive procedure whether sampled from a healthy individual or from a patient. While skin biopsy was the most common starting source for deriving human PSCs, this is now highly feasible from small blood samples with simultaneous direct reprogramming and gene editing enabling the rapid derivation of patient and isogenic controls (Howden et al., 2018). As the protocols for directed differentiation of PSCs recapitulate the developmental path of organogenesis, these also represent a unique tool to understand normal developmental processes.
Despite these advantages, the starting cell type for the generation of an iPSC is not present in the patient but is generated in vitro via a process of reprogramming to a pluripotent state. Early approaches to reprogramming also involved genomic integration of the reprogramming genes, with such random integration providing the potential for oncogenic change (Kilpinen et al., 2017). This has been largely mitigated with more recent non-integrating “footprint-free” approaches to reprogramming (Boreström et al., 2014; Ovchinnikov et al., 2015). Nevertheless, the reprogramming process itself is an inefficient process and hence the generation of a PSC line involves a degree of selection. While any two clones selected from the same reprogramming event are theoretically isogenic (contain the same background genome), genetic modifications are known to occur during the reprogramming process, which can provide a selective advantage.
A common need for standards for the application of all organoid models
The ability to generate models of human tissues, whether from pluripotent or tissue stem cells, is a remarkable opportunity to improve our understanding of development, regeneration, and repair in a human context, an opportunity to revolutionize the drug development paradigm and deliver on personalized treatments. Despite this opportunity, any organoid model should be very carefully considered with respect to the validity of the model to the disease in question and the accuracy of any organoid to model the tissue or tissue-state desired. Identifying and acknowledging any limitations is required to ensure appropriate study design, and thereby maximizing the relevance of the results (Table 1). Some key questions can be asked before commencing.
Table 1.
Standards required of organoid experiments depending upon the experimental objective of the study
| Standards required | Experimental objective |
|||
|---|---|---|---|---|
| Novel organoid protocol or refinement of an existing protocol | Confirmation of an ability to model a disease state | Identification of a pathway impacted by a given mutation/disease state | Screening (compounds or genes) | |
| Rigorous validation of the relevance of the protocol, including full characterization of the cell types present and their similarity to in vivo cell types | Required | Required | Equivalence to previous protocol required | Equivalence to previous protocol required |
| Demonstration of transferability of protocol to other lines | Required | Required | ||
| Inclusion of multiple lines carrying mutation/patient variant versus appropriate controls | Required | Required | ||
| Validation of mechanistic hypothesis in alternate lines/differentiations | Required | Required | ||
| Evidence of a robust readout compared with control | Required | Required | ||
| Biological validation in another system | Preferable | Required | ||
Is my organoid an accurate model of my organ of interest?
All tissues are composed of a large number of cell types arranged in a tissue-specific morphology critical for function. The lineage relationships and cellular identities present within many organs are well characterized, but advances in single-cell omics technology is revealing novel component cells/cell states both during development, maturation, and in response to injury. All tissues are vascularized and hence receiving information from and providing information to surrounding organs, and most tissues have an intimate relationship with the immune system of the organism within which they are located. Indeed, most disease states require this level of multisystem complexity for phenotype presentation.
By contrast, few, if any, organoids are a completely accurate model of the tissue they model and hence may not completely recapitulate the disease state of interest. This is the case for both TSC- and PSC-derived organoid models. The growing number of human tissue transcriptional datasets, including developing tissues (Haniffa et al., 2021; Mohenska et al., 2022), are increasingly providing accurate data with which to compare an organoid model with the original tissue. At the same time, advances in bioinformatic approaches for the integration of data are enabling unbiased approaches to the assignment of cellular identity to any cell present within an organoid, together with approaches to harmonize between data from different cell lines, laboratories, and methods (Alquicira-Hernandez et al., 2019; He et al., 2020; Shen et al., 2021). Finally, advances in single-cell proteomics, spatial genomics, and metabolomics approaches, as well as the continuing advances in imaging, will continue to improve our capacity to compare in detail the architecture and cellular complexity of organoids compared with the organ they are to model. The degree to which organoid models genuinely replicate the cellular state of the tissue in question has not always been clearly demonstrated. The use of terms such as bud and crypt when referring to a tissue-derived organoid is not necessarily accurate with respect to the in vivo situation (Guiu and Jensen, 2022; Sugimoto and Sato, 2022). In the case of PSC-derived organoids, the identity of cell types present within these complex structures is often not stringently characterized and the use of antibodies to identify specific cell type equivalents can ignore the presence of inappropriate cell types. There is a tendency to use limited gene sets to declare cellular identity based upon prior literature rather than objective approaches to cell classification, which requires careful benchmarking against available tissue atlases. In both types of organoids, it is likely that the in vitro cellular identity is not completely equivalent to the in vivo tissue as a result of the artificial environment. As in vitro methods used for growing organoids are unlikely to be able to provide cells that exactly match their in vivo counterparts, and different media composition will favor the emergence of specific cellular phenotypes, one also needs to consider what is important for modeling a specific aspect of the tissue or the disease in question.
As noted above, each type of organoid shows specific cellular deficits. Tissue stem cell-derived organoids, while able to re-create the complexity of the epithelium in both a regenerative and a homeostatic postnatal state in vitro, lack the underlying mesenchyme. In many organs, it is well understood that mesenchymal components modulate the epithelial response (Kabiri et al., 2014; McCarthy et al., 2020; Rendl et al., 2005) and hence may well represent a component of a disease state. This lack of endothelial and neuronal elements may limit the capacity to model a given disease state. Conversely, the cellular “simplicity” of tissue-derived organoids means that it is easier to control the signaling environments influencing cell behavior.
PSC-derived organoids model human organogenesis and, as such, frequently fail to reach a stage of maturation beyond a human trimester 1 to 2 fetus. Such organoids are missing many cellular states not yet present in the fetus, limiting the validity of such organoids to model postnatal disease states. For example, while fascinating in their complexity, organoids such as those modeling the cerebral cortex show the presence of early neuronal subtypes that are lost with time, frequently fail to generate critical interneuronal populations, and fail to genuinely display the anatomical layers present in the fetal cortex in vivo (Nowakowski and Salama, 2022; Paulsen et al., 2022; Rosebrock et al., 2022). Similarly, while kidney organoids show a remarkable capacity to self-organize into patterning and segmenting nephrons, individual nephron segments do not display appropriate tubular epithelial maturation (Combes et al., 2019; Wu et al., 2018). This is perhaps most evident within the proximal tubule, which in the postnatal organ is the major site for solute and water reabsorption. While displaying the expression of appropriate transcription factors and the presence of a megalin-cubilin-positive brush border, organoid proximal tubules generally fail to express or fail to appropriately localize key solute transporters required for function. Transplantation studies seem to suggest that further maturation may require apical solute flow (van den Berg et al., 2018). While the culture of kidney organoids in the presence of media flow has been suggested to increase the formation of an endothelial plexus (Homan et al., 2019), no evidence was presented to suggest that this improved proximal tubule maturation. In models of the directed differentiation of PSC to cardiac organoids, the presence of a cardiac fibroblast and endothelial cell population improves the capacity for such structure to commence spontaneous contraction (Giacomelli et al., 2020); however, cardiomyocytes remain proliferative and immature. More research is required here. However, kidney organoid proximal tubule maturation into functionally distinct S1, S2, and S3 segments has now been induced via improved progenitor patterning (Vanslambrouck et al., 2022) while isolation and plating of organoid epithelium has shown upregulation of mature solute channels (Aceves et al., 2022). There is also evidence in both heart and kidney organoid models that maturation can be improved via a change in metabolites, with the inclusion of free fatty acids and a reduction in glucose inducing a more mature phenotype (Mills et al., 2017; Wang et al., 2022).
These limitations must be acknowledged and considered when planning experiments using organoid models. Clearly it is not feasible to deploy a disease model without the presence of the affected target cell type. Similarly, when developing a new protocol for generating or improving organoid models, it is essential to benchmark against the tissue in vivo to demonstrate relevance and/or to have additional functional assays to benchmark against.
How reproducible is my protocol and what are the sources of variation?
Having defined the validity of an organoid protocol (which should be a requirement for all groups recapitulating a published protocol), it is also critical that the protocol is reliable and reproducible and that sources of technical variation are understood. This can include, but is not limited to, plating density, the specific timing of growth factor addition or concentration, and variation in reagents between batch.
All tissue stem cell-derived organoid protocols have been developed as adaptations of the original observation that epithelial stem cell expansion is supported by prolonged Wnt pathway activation. Slight modifications of this original approach have enabled a significant expansion in the tissue models that can be generated from tissue biopsies, and subsequent modifications of the original growth factor combination have enhanced the resemblance between tissues and organoids. Reproducibility among experiments, biological samples, and labs in this field is clearly important. However, as these organoids constitute relatively simple cellular structures in a context where large numbers of organoids can be simultaneously generated, banked/cryopreserved, and analyzed, the impact of technical variation between experiments can be accounted for (Figure 2A). The relatively short duration of any tissue-derived organoid experiment, and the limited number of cell types present, reduces the challenges around reproducibility, but does not remove these altogether. Notably, the simultaneous evaluation of organoids from many individuals will reduce the impact of technical variations on the identification of phenotypic outcomes. It will also dampen the resolution of subtle effects. It is also worth monitoring organoid cultures over time for potential drifts in cellular phenotypes driven by a selection of organoids with enhanced growth potential upon the serial propagation of the cultures. This is particularly important where clonal lines are established following genetic manipulation, such as the introduction of mutations or reporter alleles. Transparency around the initial source of material and whether patient or population diversity exists is therefore critical for study interpretation.
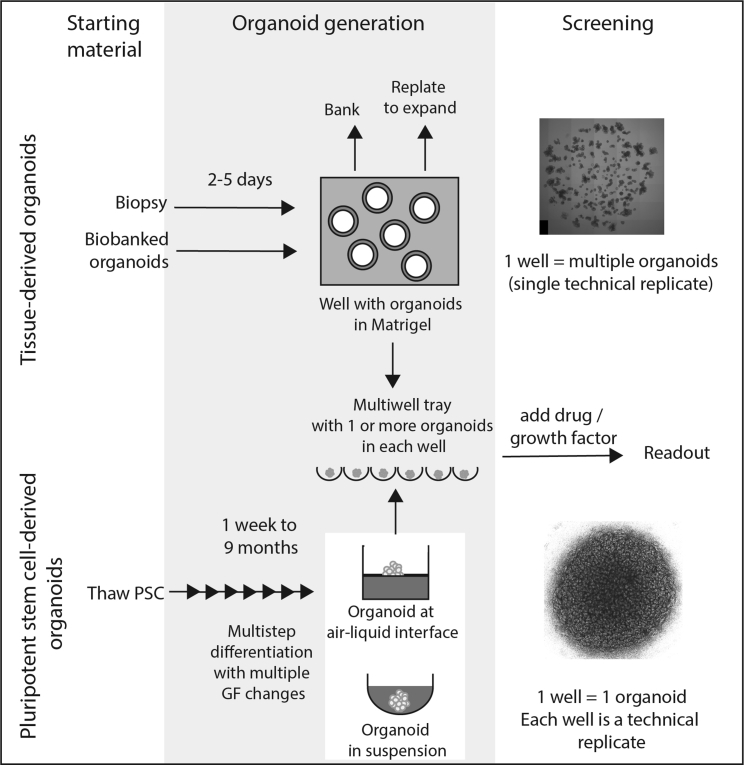
Figure 2.
Overview of organoid generation and experimental design for both tissue-derived and pluripotent stem cell-derived organoids
Tissue-derived organoid experiments can commence using fresh biopsy material but would normally involve an initial expansion and potentially a banking step in order to generate sufficient patient lines for experimental analysis. Features of this type of experiment include a short organoid generation time frame, capacity for extensive passaging, and ready freeze thawing of starting material. Organoids can be generated per well but are frequently cultured via plating into Matrigel with individual cells giving rise to an individual organoid and hence many organoids arising per well. Despite being smaller than PSC-derived organoids, these remain quite large multicellular structures with high-content imaging challenges for developing quantitative phenotypic readouts. PSC-derived organoid experiments all commence with pluripotent stem cells followed by extensive multi-step differentiation protocols, each of which are tailored for a specific tissue endpoint. These are usually larger structures, often with one organoid per well, microwell, or Transwell. Features of this type of experiment include the requirement for extended differentiation prior to experimental analysis. Their size and complexity can make image-based readouts challenging. There is a higher risk of off-target cell types and variability between organoids. Top image shows adult intestinal epithelial organoids derived from the ilium of a healthy individual cultured in a droplet of Matrigel. Bottom image shows a kidney organoid cultured on a Transwell filter.
Protocol robustness, reproducibility, and sources of variation are a much larger challenge for PSC-derived organoids for a number of reasons. In the first instance, there is the considerably increased cellular complexity of the final organoid when deriving a PSC-derived organoid. Kidney organoids, for example, contain >16 distinct cell types with the relative ratio of each cell type varying between differentiations, even when commencing with the same PSC line (Howden et al., 2019; Phipson et al., 2019). The developmental nature of these organoids also leads to variation in individual cellular maturity between PSC lines, or between biological replicates using the same PSC line. In addition, each experiment commences with a pluripotent stem cell that is then subjected often to an extended period of differentiation to reach an organoid endpoint (Figure 2B). Along that differentiation trajectory, the pluripotent starting cells can generate appropriate endpoint cells as well as off-target cell types. There is also the risk of retention of residual pluripotent cells (Figure 2B). This unwanted heterogeneity is a challenge for quality control and reproducibility between lines and between experiments. An additional challenge is the duration of any differentiation given each experiment starts from a pluripotent stem cell state. A single technical replicate can involve 1 to 9 months to generate. Biological replication by repeating this entire process with the same line multiple times is a slow and an extremely expensive experimental design. Here, parallel technical replicates may suffice, depending upon the question being addressed.
Here also, the derivation of the starting PSC itself introduces a level of variation that is hard to control. While theoretically isogenic, individual iPSCs from a given starting somatic cell are distinct clones and as such do not all differentiate in the same way. This may arise due to genetic variations between clones introduced during reprogramming, clone selection, or expansion. As such, there could be circumstances where a polyclonal pool of PSCs may be better than a clone. The use of CRISPR-Cas9 gene editing to gene correct an identified disease-causing mutation is viewed as an appropriate way to generate a gene-corrected isogenic line to be compared with a mutated patient line. By definition, generation of these isogenic controls will also represent the generation of distinct clones. This is actually not only a problem for PSC-derived organoids, as TSC-derived organoids will similarly arise through clonal selection upon introduction of specific genetic modifications. The challenge is more around the experimental requirement to commence each experiment from a PSC rather than from a banked organoid.
How does this impact the utility of my system?
The major challenge to the field from variations between lines and between experiments is the validity of disease modeling. The direct comparison of any patient versus control organoid, whether generated from a tissue or pluripotent stem cell, will show differences. The challenge is to establish when a difference is biologically relevant and not simply reflective of a technical difference. How this plays out will depend upon the experimental design, the readout, the approach to analysis, and the intended application. For example, there is considerable reliance upon transcriptional profiling, either looking genome wide or around a few target genes, to assess disease-associated differences. In many instances, investigating the impact of a disease gene or perturbation state has involved simply comparing organoids from a single patient with that of either an isogenic or unrelated control line. Such a study design will always identify differences (A versus B comparison looking for differential expression), but these may not be disease relevant. It will therefore be necessary to analyze multiple biological replicates, and/or multiple patient lines. In a recent example, we studied the impact of an allelic series of NPHS2 missense mutations on protein trafficking of the encoded protein, PODOCIN, within the podocytes of kidney organoids (Dorison et al., 2023). This involved transcriptional profiling of age-matched isolated glomeruli from five distinct mutant lines, the parental control together with a patient (homozygous) and parent (heterozygous) line. This entire experiment was done as a biological triplicate, such that all lines were simultaneously differentiated for isolation on three occasions for profiling. While any given mutant/control pair could identify transcriptional changes, there were no common disease-associated transcriptional changes identified (Dorison et al., 2023). While this may appear unsurprising given that the mutated gene does not encode a transcription factor, what this rigorous design reveals is the danger of using differentially expressed genes from less powered experiments as the only readout when performing disease modeling.
Very few studies have specifically assessed the variation between tissue-derived organoids from multiple individuals and across time course studies. The ones assessing potential technical variation do, however, strongly support that adult organoid cultures show less variation both across time series and between individuals (Kraiczy et al., 2019; Mohammadi et al., 2021). The cellular heterogeneity of the organoid derivatives between individuals is still unresolved and it will be important to map this across multiple labs, cell culture conditions, and donors. However, it is worth noting that disease modeling and drug screening using TSC-derived organoids is usually performed using organoids derived from multiple patients rather than a single comparison between patient and control. This is where the power of organoid banking and the capacity for scale out culture formats amenable to high-throughput screening has really come into play (Lukonin et al., 2020; Ostrop et al., 2020; Vlachogiannis et al., 2018). By contrast, high-content format drug screening is in its infancy in the PSC-derived organoid field, although repurposing drug approaches have now been performed (Tran et al., 2022). In both instances, drug screening requires a clear (significant difference between disease state and control) and robust (simple and reliable) readout of phenotype. This has been particularly effective for the screening of response to drugs for cystic fibrosis in which rapid cyst swelling can be readily and rapidly used as a readout of drug efficacy (Dekkers et al., 2016).
How can I mitigate these challenges and what does this mean for standards?
To ensure reproducibility for studies using any type of stem cell-derived organoid, it is essential that certain standards should be met. However, what is essential will depend upon the question being asked or the objective of the study. The establishment of a novel organoid protocol, for example, will need rigorous characterization (cell types present, histological structure, transcriptomic or other omic profiling), analysis of the robustness and transferability of the approach between lines and a thorough comparison to the in vivo tissue being modeled (Table 1). In a study adopting a previously published protocol, evidence must be provided of the successful replication of the original method, even if only as supplementary data. Establishment of the utility of a given protocol to model a disease state requires robust replication of the mutant phenotype in more than one line and the optimal use of comparative controls (isogenic lines are preferable) (Table 1). To test a given biological hypothesis, multiple replicates need to be analyzed, and authors must be transparent with respect to how they have defined their replicates. Stem Cell Reports has included a set of very clear criteria for how authors should define replicates and the distinction between biological and technical replicates. As a standard, hypotheses should be tested in multiple independent TSC and PSC-derived organoid lines to demonstrate robustness. When comparisons are made between different patient lines, it is important to consider that the genetic diversity in human samples is likely to introduce more variance, and the statistical analyses frequently require non-parametric tests using larger sample cohorts (n > 6). In addition to patient-derived lines, more and more studies are reporting on lines that have been engineered to knock out a given gene or insert a specific tag. Here it is important, whenever possible to use multiple genetically engineered clones, or as a minimum, test hypotheses in complementary experiments to provide additional support for the observations (Table 1). By contrast, performing a drug or compound screen is a completely different strategy, as here the objective is to evaluate rapidly as possible many perturbations to a ground state, whether that be control or mutant. Here it is essential that all compounds or gene perturbations tested are performed on organoids all generated at the same time to avoid batch variation. However, all screens focus on identifying hits for subsequent validation, and hence involve different experimental design to developing a new protocol (see Table 1). In principle, these same standards apply to any organoid model. For PSC-derived organoid experiments, the pluripotent status and chromosomal stability of each PSC line must be established. Whether TSC- or PSC-derived, sequencing across any specific disease gene or engineered locus of relevance must be confirmed. These are both already stated requirements for acceptance of studies within Stem Cell Reports.
As the greatest source of variation between organoids is technical batch, any direct comparison between two lines should be performed in parallel within the same experiment. Even distinct differentiations of the same cell line can shift in cell composition and maturation between experiments. For a given protocol, it may be possible to normalize for transcriptional changes previously identified as displaying technical variation for a given protocol (Forbes et al., 2018). This will, however, dampen any ability to identify subtle differences between disease and control. A frequent weakness in the field is the overinterpretation of data from single-cell expression profiling experiments. Single-cell transcription profiling data between individual samples cannot produce a statistically robust evaluation of change in cell type abundance. This can only be achieved via the barcoding and pooling of multiple experiments such that there are replicate experiments being compared (Lawlor et al., 2021). Without an ability to apply such standards to organoid experiments, validation of any claim made must be comprehensively addressed using multiple independent lines of evidence. Indeed, for almost all hypotheses generated using organoids, it is important to validate the assumption in a different model, whether this be clinical or pre-clinical.
Some final words of caution for the field
The purpose of this review was to pause and reflect on not only what has been achieved in the organoid field, but the limitations of the systems. This is not to take away from the remarkable advances coming from human organoid models. The delivery of an organoid-based approach to the personalized evaluation of drug efficacy for the treatment of patients with rare mutations in the CFTR gene (cystic fibrosis), for example, is a major clinical outcome (Dekkers et al., 2016). The application of cell types generated using tissue stem cells and PSCs in organ-on-chip or microphysiological systems will assist in addressing the technical challenges of quality control and throughput but must still be rationally considered based upon their logical biological limitations. However, the relevance of such systems to advancing biological understanding, developing drugs, or delivering cellular therapies will absolutely depend upon a rigorous science. Conversely, overstating the cellular complexity or functionality of such models is undoubtedly damaging to the field.
In a final word of caution, there is a desire to apply PSC-derived organoids to the modeling of late-onset postnatal disease, including conditions such as chronic tissue fibrosis in renal failure and late-onset dementia. Many of these chronic diseases result from long-term ischemic challenges, often including immunological responses. The absence of an immune system or an appropriate metabolic milieu (most tissue culture media are high glucose and organoids are cultured at hyperoxic conditions compared with the tissue in situ) would likely suggest that such models are too simplistic. Conversely, as tissue-derived organoids recapitulate repair and homeostasis of postnatal epithelial niches, these are potentially a more appropriate system to investigate chronic conditions, albeit without the complete complexity of the original tissue. At the very least, it is critical that any data coming from organoid models are appropriately validated in vivo.
Engineering improvements are likely to enable the generation of increased cellular complexity and anatomical accuracy for both organoid types. The proof of concept of this has been shown, for example, in studies in which intestinal epithelial cells originally cultured as organoids were seeded in a microphysiological device recapitulating the structure of the small intestine provided the basis for differentiation into even rare cell types (Nikolaev et al., 2020). The aggregation of organoids representing different tissues—assembloids—is also enabling much more complex human models (Goldrick et al., 2023; Miura et al., 2022). The generation of combination tissue models in which TSC- and PSC-derived cells are combined is also an evolving area of activity. Again, the validity of such models must be established and the challenges acknowledged in the experimental designs (Table 1). Engineering approaches can also assist in the production of large numbers of quality-controlled organoids for high-content screening for application in toxicity and drug screening. However, the accuracy of the readout will continue to depend upon the presence of the appropriate mature cell types crucial to a valid biological readout.
In closing, there is great promise for both types of organoid models. Hence, there is much to look forward to if the caveats of the model are taken into consideration, the readout is a reasonable surrogate of the disease state of interest, and if the field applies appropriate standards to evaluate the validity of data generated from organoid studies. It will be the development and adoption of rigorous standards that will be critical to future progress.
Author contributions
K.B.J., M.H.L.: conceptualization, literature review and analysis, writing, editing, visualization.
Acknowledgments
This work was supported by the Novo Nordisk Foundation Center for Stem Cell Medicine (NNF21CC0073729). M.H.L. holds an NHMRC Senior Principal Research Fellowship (GNT1136085). K.B.J. acknowledges support from Independent Research Fund Denmark (0134-00111B), the Novo Nordisk Foundation (NNF20OC0064376), and from the European Union’s Horizon 2020 research and innovation programme (STEMHEALTH ERCCoG682665). We acknowledge Ker Sin Tan, Jessica Vanslambrouck, Astrid Møller Baattrup, and Robert P. Fordham for the images of organoids.
Conflict of interests
M.H.L. is an inventor on patents in the field of kidney organoid technology.
References
- Aceves J.O., Heja S., Kobayashi K., Robinson S.S., Miyoshi T., Matsumoto T., Schäffers O.J.M., Morizane R., Lewis J.A. 3D proximal tubule-on-chip model derived from kidney organoids with improved drug uptake. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-19293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alquicira-Hernandez J., Sathe A., Ji H.P., Nguyen Q., Powell J.E. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019;20:264. doi: 10.1186/s13059-019-1862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E.R., Chivukula I.V., Hankeova S., Sjöqvist M., Tsoi Y.L., Ramsköld D., Masek J., Elmansuri A., Hoogendoorn A., Vazquez E., et al. Mouse model of Alagille syndrome and mechanisms of Jagged1 missense mutations. Gastroenterology. 2018;154:1080–1095. doi: 10.1053/j.gastro.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M., et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Blokzijl F., de Ligt J., Jager M., Sasselli V., Roerink S., Sasaki N., Huch M., Boymans S., Kuijk E., Prins P., et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj S.F., Hwang C.I., Baker L.A., Chio I.I.C., Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S., et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreström C., Simonsson S., Enochson L., Bigdeli N., Brantsing C., Ellerström C., Hyllner J., Lindahl A. Footprint-free human induced pluripotent stem cells from articular cartilage with redifferentiation capacity: a first step toward a clinical-grade cell source. Stem Cells Transl. Med. 2014;3:433–447. doi: 10.5966/sctm.2013-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão K.O., van den Brink L., Miller D.C., Grandela C., van Meer B.J., Mol M.P.H., de Korte T., Tertoolen L.G.J., Mummery C.L., Sala L., et al. Isogenic sets of hiPSC-CMs harboring distinct KCNH2 mutations differ functionally and in susceptibility to drug-induced arrhythmias. Stem Cell Rep. 2020;15:1127–1139. doi: 10.1016/j.stemcr.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L., Mastrogiovanni G., Verstegen M.M., Francies H.E., Gavarró L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M., et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes A.N., Zappia L., Er P.X., Oshlack A., Little M.H. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 2019;11:3. doi: 10.1186/s13073-019-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Espinoza L., Dowbaj A.M., Kohler T.N., Strauss B., Sarlidou O., Belenguer G., Pacini C., Martins N.P., Dobie R., Wilson-Kanamori J.R., et al. Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell Stem Cell. 2021;28:1907–1921.e8. doi: 10.1016/j.stem.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz N.M., Song X., Czerniecki S.M., Gulieva R.E., Churchill A.J., Kim Y.K., Winston K., Tran L.M., Diaz M.A., Fu H., et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 2017;16:1112–1119. doi: 10.1038/nmat4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Crignis E., Hossain T., Romal S., Carofiglio F., Moulos P., Khalid M.M., Rao S., Bazrafshan A., Verstegen M.M., Pourfarzad F., et al. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife. 2021;10 doi: 10.7554/eLife.60747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., van de Graaf E.A., Nieuwenhuis E.E.S., Houwen R.H.J., et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8:344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W.M., Bijvelds M.J.C., Scholte B.J., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Dorison A., Forbes T.A., Little M.H. What can we learn from kidney organoids? Kidney Int. 2022;102:1013–1029. doi: 10.1016/j.kint.2022.06.032. [DOI] [PubMed] [Google Scholar]
- Dorison A., Ghobrial I., Graham A., Peiris T., Forbes T.A., See M., Das M., Saleem M.A., Quinlan C., Lawlor K.T., et al. Kidney organoids generated using an allelic series of NPHS2 point variants reveal distinct intracellular podocin mistrafficking. J. Am. Soc. Nephrol. 2023;34:88–109. doi: 10.1681/asn.2022060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvela-Levitt M., Kost-Alimova M., Emani M., Kohnert E., Thompson R., Sidhom E.H., Rivadeneira A., Sahakian N., Roignot J., Papagregoriou G., et al. Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell. 2019;178:521–535.e23. doi: 10.1016/j.cell.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A.H., Tsai Y.H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4 doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.L., Qu L., et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Forbes T.A., Howden S.E., Lawlor K., Phipson B., Maksimovic J., Hale L., Wilson S., Quinlan C., Ho G., Holman K., et al. Patient-iPSC-Derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. Am. J. Hum. Genet. 2018;102:816–831. doi: 10.1016/j.ajhg.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbester J.L., Goulding D., Vallier L., Hannan N., Hale C., Pickard D., Mukhopadhyay S., Dougan G. Interaction of Salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 2015;83:2926–2934. doi: 10.1128/iai.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham R.P., Yui S., Hannan N.R.F., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T., et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman B.S., Brooks C.R., Lam A.Q., Fu H., Morizane R., Agrawal V., Saad A.F., Li M.K., Hughes M.R., Werff R.V., et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Matano M., Toshimitsu K., Takano A., Mikami Y., Nishikori S., Sugimoto S., Sato T. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 2018;23:787–793.e6. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Gao D., Vela I., Sboner A., Iaquinta P.J., Karthaus W.R., Gopalan A., Dowling C., Wanjala J.N., Undvall E.A., Arora V.K., et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos N., Prior N., Angres B., Mastrogiovanni G., Cagan A., Harrison D., Hindley C.J., Arnes-Benito R., Liau S.S., Curd A., et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev. Biol. 2020;20:4. doi: 10.1186/s12861-020-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., van Helden R.W.J., Krotenberg Garcia A., Mircea M., Kostidis S., Davis R.P., et al. Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell. 2020;26:862–879.e11. doi: 10.1016/j.stem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrick C., Guri I., Herrera-Oropeza G., O'Brien-Gore C., Roy E., Wojtynska M., Spagnoli F.M. 3D multicellular systems in disease modelling: from organoids to organ-on-chip. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1083175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A., Yoon S.J., Tran S.S., Makinson C.D., Park J.Y., Andersen J., Valencia A.M., Horvath S., Xiao X., Huguenard J.R., et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat. Neurosci. 2021;24:331–342. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiu J., Jensen K.B. In vivo studies should take priority when defining mechanisms of intestinal crypt morphogenesis. Cell. Mol. Gastroenterol. Hepatol. 2022;13:1–3. doi: 10.1016/j.jcmgh.2021.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Taylor D., Linnarsson S., Aronow B.J., Bader G.D., Barker R.A., Camara P.G., Camp J.G., Chédotal A., Copp A., et al. A roadmap for the human developmental cell atlas. Nature. 2021;597:196–205. doi: 10.1038/s41586-021-03620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Yoshino T. Reconstitution of reproductive organ system that produces functional oocytes. Curr. Opin. Genet. Dev. 2022;77 doi: 10.1016/j.gde.2022.101982. [DOI] [PubMed] [Google Scholar]
- He Z., Brazovskaja A., Ebert S., Camp J.G., Treutlein B. CSS: cluster similarity spectrum integration of single-cell genomics data. Genome Biol. 2020;21:224. doi: 10.1186/s13059-020-02147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer P., Jahnel S.M., Papai N., Giesshammer M., Deyett A., Schmidt C., Penc M., Tavernini K., Grdseloff N., Meledeth C., et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299–3317.e22. doi: 10.1016/j.cell.2021.04.034. [DOI] [PubMed] [Google Scholar]
- Homan K.A., Gupta N., Kroll K.T., Kolesky D.B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M.T., Ferrante T., Bonventre J.V., et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S.E., Thomson J.A., Little M.H. Simultaneous reprogramming and gene editing of human fibroblasts. Nat. Protoc. 2018;13:875–898. doi: 10.1038/nprot.2018.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S.E., Vanslambrouck J.M., Wilson S.B., Tan K.S., Little M.H. Reporter-based fate mapping in human kidney organoids confirms nephron lineage relationships and reveals synchronous nephron formation. EMBO Rep. 2019;20 doi: 10.15252/embr.201847483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S.W., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M.A., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Koo B.K. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- Kabiri Z., Greicius G., Madan B., Biechele S., Zhong Z., Zaribafzadeh H., Edison Aliyev J., Aliyev J., Wu Y., Bunte R., et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- Kawasaki K., Toshimitsu K., Matano M., Fujita M., Fujii M., Togasaki K., Ebisudani T., Shimokawa M., Takano A., Takahashi S., et al. An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell. 2020;183:1420–1435.e21. doi: 10.1016/j.cell.2020.10.023. [DOI] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J., et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy J., Nayak K.M., Howell K.J., Ross A., Forbester J., Salvestrini C., Mustata R., Perkins S., Andersson-Rolf A., Leenen E., et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345 doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor K.T., Vanslambrouck J.M., Higgins J.W., Chambon A., Bishard K., Arndt D., Er P.X., Wilson S.B., Howden S.E., Tan K.S., et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater. 2021;20:260–271. doi: 10.1038/s41563-020-00853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Rabbani C.C., Gao H., Steinhart M.R., Woodruff B.M., Pflum Z.E., Kim A., Heller S., Liu Y., Shipchandler T.Z., Koehler K.R. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582:399–404. doi: 10.1038/s41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Bhang D.H., Beede A., Huang T.L., Stripp B.R., Bloch K.D., Wagers A.J., Tseng Y.H., Ryeom S., Kim C.F. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie J.L., Huang S., Opp J.S., Nagy M.S., Kobayashi M., Young V.B., Spence J.R. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 2015;83:138–145. doi: 10.1128/iai.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukonin I., Serra D., Challet Meylan L., Volkmann K., Baaten J., Zhao R., Meeusen S., Colman K., Maurer F., Stadler M.B., et al. Phenotypic landscape of intestinal organoid regeneration. Nature. 2020;586:275–280. doi: 10.1038/s41586-020-2776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimets M., Pedersen M.T., Guiu J., Dreier J., Thodberg M., Antoku Y., Schweiger P.J., Rib L., Bressan R.B., Miao Y., et al. Mesenchymal-epithelial crosstalk shapes intestinal regionalisation via Wnt and Shh signalling. Nat. Commun. 2022;13:715. doi: 10.1038/s41467-022-28369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimets M., Rocchi C., Bron R., Pringle S., Kuipers J., Giepmans B.N.G., Vries R.G.J., Clevers H., de Haan G., van Os R., Coppes R.P. Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals. Stem Cell Rep. 2016;6:150–162. doi: 10.1016/j.stemcr.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar A.J., Buerger F., Forbes T.A., Klämbt V., Schneider R., Deutsch K., Kitzler T.M., Howden S.E., Scurr M., Tan K.S., et al. Recessive NOS1AP variants impair actin remodeling and cause glomerulopathy in humans and mice. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N., Manieri E., Storm E.E., Saadatpour A., Luoma A.M., Kapoor V.N., Madha S., Gaynor L.T., Cox C., Keerthivasan S., et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell. 2020;26:391–402.e5. doi: 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McOwan T.N., Craig L.A., Tripdayonis A., Karavendzas K., Cheung M.M., Porrello E.R., Conyers R., Elliott D.A. Evaluating anthracycline cardiotoxicity associated single nucleotide polymorphisms in a paediatric cohort with early onset cardiomyopathy. Cardiooncology. 2020;6:5. doi: 10.1186/s40959-020-00060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L., et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y., Li M.Y., Revah O., Yoon S.J., Narazaki G., Pașca S.P. Engineering brain assembloids to interrogate human neural circuits. Nat. Protoc. 2022;17:15–35. doi: 10.1038/s41596-021-00632-z. [DOI] [PubMed] [Google Scholar]
- Mohammadi S., Morell-Perez C., Wright C.W., Wyche T.P., White C.H., Sana T.R., Lieberman L.A. Assessing donor-to-donor variability in human intestinal organoid cultures. Stem Cell Rep. 2021;16:2364–2378. doi: 10.1016/j.stemcr.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohenska M., Tan N.M., Tokolyi A., Furtado M.B., Costa M.W., Perry A.J., Hatwell-Humble J., van Duijvenboden K., Nim H.T., Ji Y.M.M., et al. 3D-cardiomics: a spatial transcriptional atlas of the mammalian heart. J. Mol. Cell. Cardiol. 2022;163:20–32. doi: 10.1016/j.yjmcc.2021.09.011. [DOI] [PubMed] [Google Scholar]
- Morizane R., Lam A.Q., Freedman B.S., Kishi S., Valerius M.T., Bonventre J.V. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015;33:1193–1200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev M., Mitrofanova O., Broguiere N., Geraldo S., Dutta D., Tabata Y., Elci B., Brandenberg N., Kolotuev I., Gjorevski N., et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- Nishimura R., Shirasaki T., Tsuchiya K., Miyake Y., Watanabe Y., Hibiya S., Watanabe S., Nakamura T., Watanabe M. Establishment of a system to evaluate the therapeutic effect and the dynamics of an investigational drug on ulcerative colitis using human colonic organoids. J. Gastroenterol. 2019;54:608–620. doi: 10.1007/s00535-018-01540-y. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Salama S.R. Cerebral organoids as an experimental platform for human neurogenomics. Cells. 2022;11 doi: 10.3390/cells11182803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., Kuo C.J. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrop J., Zwiggelaar R.T., Terndrup Pedersen M., Gerbe F., Bösl K., Lindholm H.T., Díez-Sánchez A., Parmar N., Radetzki S., von Kries J.P., et al. A semi-automated organoid screening method demonstrates epigenetic control of intestinal epithelial differentiation. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.618552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov D.A., Sun J., Wolvetang E.J. Generation of footprint-free induced pluripotent stem cells from human fibroblasts using episomal plasmid vectors. Methods Mol. Biol. 2015;1330:37–45. doi: 10.1007/978-1-4939-2848-4_4. [DOI] [PubMed] [Google Scholar]
- Paulsen B., Velasco S., Kedaigle A.J., Pigoni M., Quadrato G., Deo A.J., Adiconis X., Uzquiano A., Sartore R., Yang S.M., et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602:268–273. doi: 10.1038/s41586-021-04358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B., Er P.X., Combes A.N., Forbes T.A., Howden S.E., Zappia L., Yen H.J., Lawlor K.T., Hale L.J., Sun J., Wolvetang E. Evaluation of variability in human kidney organoids. Nat. Methods. 2019;16:79–87. doi: 10.1038/s41592-018-0253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C., Puschhof J., Rosendahl Huber A., van Hoeck A., Wood H.M., Nomburg J., Gurjao C., Manders F., Dalmasso G., Stege P.B., et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P., et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M., Lewis L., Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revah O., Gore F., Kelley K.W., Andersen J., Sakai N., Chen X., Li M.Y., Birey F., Yang X., Saw N.L., et al. Maturation and circuit integration of transplanted human cortical organoids. Nature. 2022;610:319–326. doi: 10.1038/s41586-022-05277-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock D., Arora S., Mutukula N., Volkman R., Gralinska E., Balaskas A., Aragonés Hernández A., Buschow R., Brändl B., Müller F.J., et al. Enhanced cortical neural stem cell identity through short SMAD and WNT inhibition in human cerebral organoids facilitates emergence of outer radial glial cells. Nat. Cell Biol. 2022;24:981–995. doi: 10.1038/s41556-022-00929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe R.G., Daley G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019;20:377–388. doi: 10.1038/s41576-019-0100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H., et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Sampaziotis F., Justin A.W., Tysoe O.C., Sawiak S., Godfrey E.M., Upponi S.S., Gieseck R.L., 3rd, de Brito M.C., Berntsen N.L., Gómez-Vázquez M.J., et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- Sasai Y., Eiraku M., Suga H. In vitro organogenesis in three dimensions: self-organising stem cells. Development. 2012;139:4111–4121. doi: 10.1242/dev.079590. [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schweiger P.J., Jensen K.B. Modeling human disease using organotypic cultures. Curr. Opin. Cell Biol. 2016;43:22–29. doi: 10.1016/j.ceb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Serra D., Mayr U., Boni A., Lukonin I., Rempfler M., Challet Meylan L., Stadler M.B., Strnad P., Papasaikas P., Vischi D., et al. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Sun Y., Matsumoto M., Shim W.J., Sinniah E., Wilson S.B., Werner T., Wu Z., Bradford S.T., Hudson J., et al. Integrating single-cell genomics pipelines to discover mechanisms of stem cell differentiation. Trends Mol. Med. 2021;27:1135–1158. doi: 10.1016/j.molmed.2021.09.006. [DOI] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange D.E., Koo B.K., Huch M., Sibbel G., Basak O., Lyubimova A., Kujala P., Bartfeld S., Koster J., Geahlen J.H., et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Sidhom E.H., Emani M., Vernon K., Sahakian N., Zhou Y., Kost-Alimova M., Slyper M., Waldman J., Dionne D., et al. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat. Commun. 2019;10:5462. doi: 10.1038/s41467-019-13382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S., Sato T. Organoid vs in vivo mouse model: which is better research tool to understand the biologic mechanisms of intestinal epithelium? Cell. Mol. Gastroenterol. Hepatol. 2022;13:195–197. doi: 10.1016/j.jcmgh.2021.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., Chuva de Sousa Lopes S.M., Little M.H. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M., et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tran T., Song C.J., Nguyen T., Cheng S.Y., McMahon J.A., Yang R., Guo Q., Der B., Lindström N.O., Lin D.C.H., McMahon A.P. A scalable organoid model of human autosomal dominant polycystic kidney disease for disease mechanism and drug discovery. Cell Stem Cell. 2022;29:1083–1101.e7. doi: 10.1016/j.stem.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L., et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C.W., Ritsma L., Avramut M.C., Wiersma L.E., van den Berg B.M., Leuning D.G., Lievers E., Koning M., Vanslambrouck J.M., Koster A.J., et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 2018;10:751–765. doi: 10.1016/j.stemcr.2018.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanslambrouck J.M., Wilson S.B., Tan K.S., Groenewegen E., Rudraraju R., Neil J., Lawlor K.T., Mah S., Scurr M., Howden S.E., et al. Enhanced metanephric specification to functional proximal tubule enables toxicity screening and infectious disease modelling in kidney organoids. Nat. Commun. 2022;13:5943. doi: 10.1038/s41467-022-33623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernández-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R., et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Heijs B., Kostidis S., Rietjens R.G.J., Koning M., Yuan L., Tiemeier G.L., Mahfouz A., Dumas S.J., Giera M., et al. Spatial dynamic metabolomics identifies metabolic cell fate trajectories in human kidney differentiation. Cell Stem Cell. 2022;29:1580–1593.e7. doi: 10.1016/j.stem.2022.10.008. [DOI] [PubMed] [Google Scholar]
- Watson C.L., Mahe M.M., Múnera J., Howell J.C., Sundaram N., Poling H.M., Schweitzer J.I., Vallance J.E., Mayhew C.N., Sun Y., et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerling-Bui A.D., Fast E.M., Soare T.W., Venkatachalan S., DeRan M., Fanelli A.B., Kyrychenko S., Hoang H., Corriea G.M., Zhang W., et al. Transplanted organoids empower human preclinical assessment of drug candidate for the clinic. Sci. Adv. 2022;8:eabj5633. doi: 10.1126/sciadv.abj5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer R.A., Leopoldi A., Aichinger M., Wick N., Hantusch B., Novatchkova M., Taubenschmid J., Hämmerle M., Esk C., Bagley J.A., et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Uchimura K., Donnelly E.L., Kirita Y., Morris S.A., Humphreys B.D. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23:869–881.e8. doi: 10.1016/j.stem.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Azzolin L., Maimets M., Pedersen M.T., Fordham R.P., Hansen S.L., Larsen H.L., Guiu J., Alves M.R.P., Rundsten C.F., et al. YAP/TAZ-Dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell. 2018;22:35–49.e7. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]