Abstract
The human genome is pervasively transcribed, producing a majority of short and long noncoding RNAs (lncRNAs) that can influence cellular programs through a variety of transcriptional and post-transcriptional regulatory mechanisms. The brain houses the richest repertoire of long noncoding transcripts, which function at every stage during central nervous system development and homeostasis. An example of functionally relevant lncRNAs is species involved in spatiotemporal organization of gene expression in different brain regions, which play roles at the nuclear level and in transport, translation, and decay of other transcripts in specific neuronal sites. Research in the field has enabled identification of the contributions of specific lncRNAs to certain brain diseases, including Alzheimer’s disease, Parkinson’s disease, cancer, and neurodevelopmental disorders, resulting in notions of potential therapeutic strategies that target these RNAs to recover the normal phenotype. Here, we summarize the latest mechanistic findings associated with lncRNAs in the brain, focusing on their dysregulation in neurodevelopmental or neurodegenerative disorders, their use as biomarkers for central nervous system (CNS) diseases in vitro and in vivo, and their potential utility for therapeutic strategies.
Keywords: lncRNA, AntagoNAT, neurodevelopment, neurodegeneration, ASO, NEAT1, paraspeckle, RNA-based therapy
Graphical abstract
Srinivas et al. summarize the latest mechanistic findings associated with lncRNAs in the brain, focusing on their dysregulation in neurodevelopmental or neurodegenerative disorders, their use as biomarkers for central nervous system diseases in vitro and in vivo, and their potential utility for therapeutic strategies.
Introduction
Over the last decade, long noncoding RNAs (lncRNAs; with an arbitrary cutoff size of >200 nt) have been the object of intense study, and research addressing their mechanisms of action as well as their contribution to human pathophysiology has multiplied. We now understand the important roles that lncRNAs play in normal cellular function, including growth and death, development and differentiation, and metabolic homeostasis. Characterization of these functions has allowed a greater understanding of the molecular underpinnings of lncRNA action. In the nucleus, lncRNAs can associate with chromatin remodeling complexes, act as co-transcriptional factors, and regulate stages of RNA biogenesis, maturation and localization. In the cytoplasm, they can influence the fate of other RNA or protein molecules by acting as signaling molecules, molecular decoys, scaffolds, cofactors, and targeting agents.
Recent reviews highlight the detailed actions of the best-characterized lncRNAs.1 Briefly, lncRNAs regulate gene expression through multiple mechanisms. In the nucleus, lncRNAs may collaborate in 3D chromatin organization and/or remodeling, chromatin modification, transcription and splicing control, or even mRNA export. In the cytoplasm, they may influence the stability, localization or translation of other RNAs; function as molecular sponges of other RNAs or proteins; or regulate post-translational modifications. This myriad of functions is achieved through the general ability of lncRNAs to interact, through different structured domains, with multiple nucleic acids and proteins, often simultaneously. Most of these functions have been studied in the context of tumorigenesis, but prominent roles have been found in other pathological contexts too, including cardiovascular diseases,2 inflammatory disorders,3 and diabetes.4
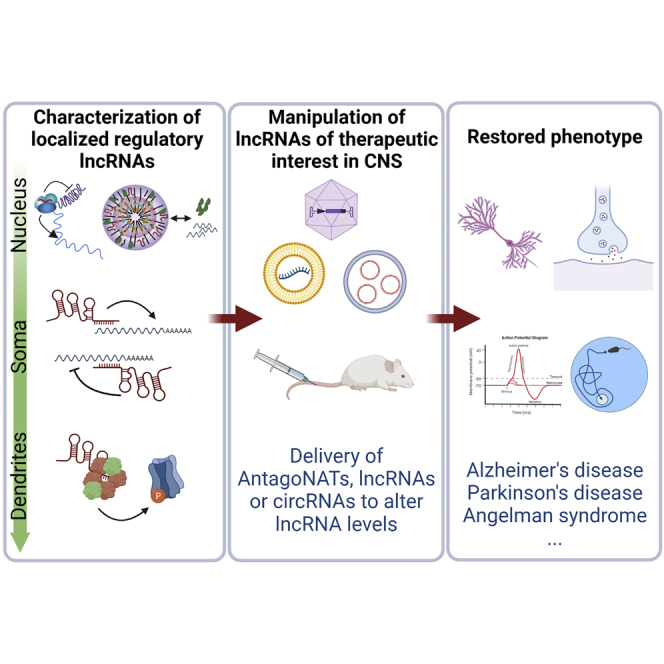
In the nervous system, neuronal maturation, plasticity, and homeostasis are well-orchestrated, complex mechanisms that rely largely on timely regulation of RNA production, localization, and decay. lncRNAs are involved at all stages in these processes; it is therefore unsurprising that the central nervous system (CNS) displays the richest landscape of noncoding RNA species and lncRNA-based regulatory mechanisms, with approximately 40% of all annotated tissue-specific lncRNAs existing in distinct brain regions.5,6 We review the most relevant roles of lncRNAs in the brain with an emphasis on the latest advances, presenting evidence for the value of lncRNAs as biomarkers and perspectives for lncRNAs as bona fide therapeutic targets or tools in the treatment of CNS disorders.
lncRNAs in the brain: Evolution and function
lncRNAs are generally poorly conserved, and despite the fact that approximately one-third of them appear only in the primate lineage,7,8 brain-specific lncRNAs show the highest evolutionary conservation.9 One caveat of these estimations is the fact that structural features, rather than the primary nucleotide sequence, are often the determinants of functionality for lncRNAs. This phenomenon is well illustrated by Xist, a crucial lncRNA for X chromosome inactivation,10,11 and the metastasis associated lung adenocarcinoma transcript 1 MALAT1, a very abundant transcript with highly conserved helices in its functional core.12,13 Importantly, the abundance of lncRNAs produced by an organism correlates with tissue- or organism-specific complexity,11,14 which argues for a contribution of the brain-specific lncRNAs to CNS development. In this regard, prominent examples of relevant lncRNAs in the brain include linc-Brn1b and Dali (which bind epigenetic modifiers and regulate genes required for neocortical development15,16), or Pnky (which is highly expressed in neural stem cells of the developing cortex and interacts with splicing regulators17). These noncoding transcripts represent a common theme in brain-specific lncRNAs: conserved intergenic lncRNAs located adjacent to and co-expressed with protein-coding genes are often involved in large-scale transcriptional programs in CNS development.18 Sometimes, short-range action by lncRNAs is compatible with the regulation of distal sites in trans, often involving binding to chromatin remodeling complexes and reorganization of the 3D genome. For example, Paupar is a CNS-specific lncRNA that acts locally to regulate the expression of the nearby neural transcription factor Pax6 as well as distally by interacting with regulatory elements of multiple genes on different chromosomes.19,20,21 Interestingly, the action of Paupar in trans also involves binding to the PAX6 protein itself to regulate target gene transcription at multiple distal genomic sites, which illustrates how lncRNAs and neural transcription factors can interact to orchestrate cortical differentiation.22 Other examples of nuclear enriched lncRNAs that operate similarly include Evf2 (which interacts with transcription factors and chromatin remodelers to regulate expression of neuronal genes and drive lineage specification)23,24,25 and Gm12371 (which targets multiple genes through regulation of other lncRNAs and is necessary for synaptic transmission in hippocampal neurons).26
Structural lncRNAs in the brain
Some structural lncRNAs with critical roles in brain function deserve special mention: nuclear paraspeckle assembly transcript 1 (NEAT1) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) are two highly conserved and expressed nucleus-retained lncRNAs with structural and regulatory roles.27,28,29,30 Although ubiquitously present in different tissues, they are some of the most abundant lncRNAs in the brain cortex31 and are involved in normal neuronal function as well as in the pathophysiology of neurological disorders.32,33 Indeed, MALAT1 and NEAT1 constitute the major RNA moiety and scaffold of two important subnuclear domains, speckles and paraspeckles, respectively, which regulate the localization and phosphorylation status of splicing factors and other RNA binding proteins, mRNAs, and transcription factors.34 Although prominent roles of MALAT1 in neuropathology are well established,35 MALAT1 function and the impact of its dysregulation have been studied mostly in the context of cancer. By contrast, the impact of NEAT1 function in the CNS deserves special mention when considering the molecular underpinnings of neuropathology. NEAT1 may contribute to phase separation of intrinsically disordered regions, forming a non-membranous ribonucleoprotein milieu that provides a flexible platform for assembly of a number of protein and nucleic acids components. For example, binding to NEAT1 facilitates the condensation propensity of transactive response DNA binding protein of 43 kDa (TDP-43), a DNA and RNA binding protein (RBP) involved in splicing regulation that forms protective nuclear bodies in response to stress, defects in which (e.g., aberrant phosphorylation, cytoplasmic mislocalization, and aggregation) contribute to amyotrophic lateral sclerosis (ALS).36 Alterations in TDP-43 are also typical of other neurological diseases, such as Alzheimer’s disease or frontotemporal lobar degeneration (FTLD).37 Individual-nucleotide resolution cross-linking immunoprecipitation data reveal multiple TDP-43 binding regions at the 5′ and 3′ ends of NEAT1 transcripts, with specific enrichment at UG repeat regions.37,38 This binding preference is consistent with the particular structure of NEAT1 transcripts, where core and structural members of paraspeckles (such as the architectural proteins PSPC1, p54nrb/NONO, and SFPQ) bind along the central region of the transcript,39 whereas auxiliary proteins (e.g., TDP-43) are more present at the periphery (i.e., the paraspeckle shell).40
Despite the existence of a relatively large body of literature about NEAT1 and TDP proteinopathies, the specific mechanism whereby NEAT1 modulates the capacity of TDP-43 and other RBPs for ribonucleoprotein assembly and regulation of transcriptional and post-transcriptional events under physiological conditions remains elusive,41 as does the specific contribution of NEAT1 to TDP-43 mislocalization and the subsequent impact of this mislocalization on mRNA splicing in neurodegeneration.42 Thus, further studies addressing the detailed biophysical properties and roles of NEAT1 and paraspeckles as a hub of RBP function and/or in phase separation are needed for a full understanding of the potential protective role of NEAT1 in these types of proteinopathies (Figure 1).
Figure 1.
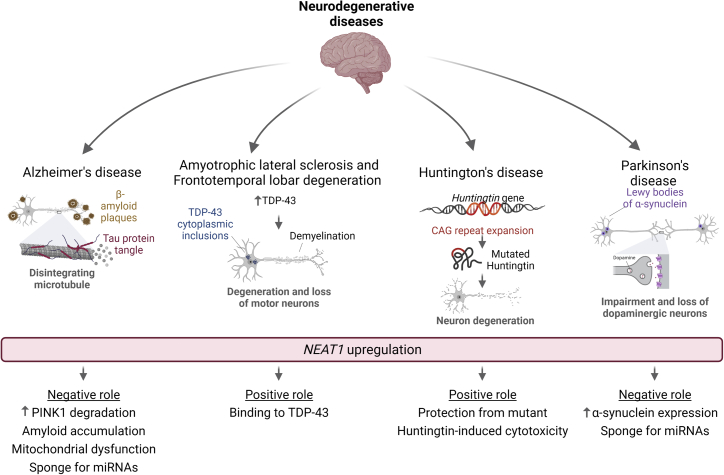
Impact of NEAT1 and paraspeckles on NDs
Alzheimer’s disease (AD) is frequently characterized by accumulation of β-amyloid peptide and disruption of microtubules because of Tau protein tangles. Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) share molecular features, such as an increase of TDP-43 leading to cytoplasmic inclusions and demyelination with consequent loss of motor neurons. Huntington’s disease (HD) is caused by a CAG tri-nucleotide repeat expansion in the Huntingtin (HTT) gene that promotes neuron degeneration. PD disease is characterized by formation of Lewy bodies, a consequence of aggregation of α-synuclein protein, and dysfunction of dopaminergic neurons. The common alteration in all of these diseases is upregulation of NEAT1 with diverse effects. In AD, it has a negative influence, acting as a sponge for miRNAs and promoting amyloid accumulation and mitochondrial dysfunction by PINK1 degradation. In ALS and FTLD, NEAT1 plays a neuroprotective role by binding to TDP-43 and facilitating its phase separation to prevent accumulation of prion-like aggregates. In HD, it prevents cytotoxicity induced by mutant HTT, and in PD, it increases α-synuclein expression and acts as a sponge for miRNAs. This figure was created with BioRender.
In addition, interesting insights into the neurobiology of NEAT1 and paraspeckles arise from the observation that the RNA and protein components of paraspeckles follow circadian rhythms, resulting in nuclear retention of specific transcripts in a paraspeckle-dependent manner that follows rhythmic circadian retention and expression patterns.43 Further research indicates that NEAT1 directly binds to 30% of all transcripts targeted by paraspeckles, suggesting that protein:RNA and RNA:RNA interactions are involved in nuclear retention of mRNAs by paraspeckles.44 Although the detailed mechanisms involved in circadian nuclear retention of mRNAs remains to be determined, this evidence suggests that paraspeckles may play important roles in controlling circadian gene expression at a post-transcriptional level, thereby contributing to the adaptation of organisms to regular changes in their environment. It remains to be investigated whether dysregulation of this circadian control plays roles in human diseases such as neuropsychiatric disorders. Another recently uncovered NEAT1 function with potential impact in brain disorders is its connection with mitochondrial function. The use of genome-wide RNAi screens has revealed genes involved in mitochondrial homeostasis to regulate NEAT1 and paraspeckle formation, suggesting an unexpected link between mitochondria and nuclear bodies. Indeed, mitochondrial stressors alter NEAT1 expression by activating transcription factor 2 (ATF2)-mediated transcriptional upregulation, consequently disrupting paraspeckle morphogenesis. Specifically, mitochondrial stress and altered NEAT1 expression increase the nuclear retention of mRNAs encoding for mitochondrial proteins (so-called mito-mRNAs). Accordingly, treatments that directly alter NEAT1 levels also have an impact on mitochondrial function by changing the retention of some of these mito-mRNAs and causing aberrant mitochondrial homeostasis, pointing to a dynamic cross-talk between mitochondria and paraspeckles in response to changing cellular conditions.45 More generally, the emerging picture is that NEAT1 and paraspeckles act as sensors of a variety of stress signals and mediate important changes in gene expression to allow cellular adaptation to new conditions46 (Figure 2).
Figure 2.
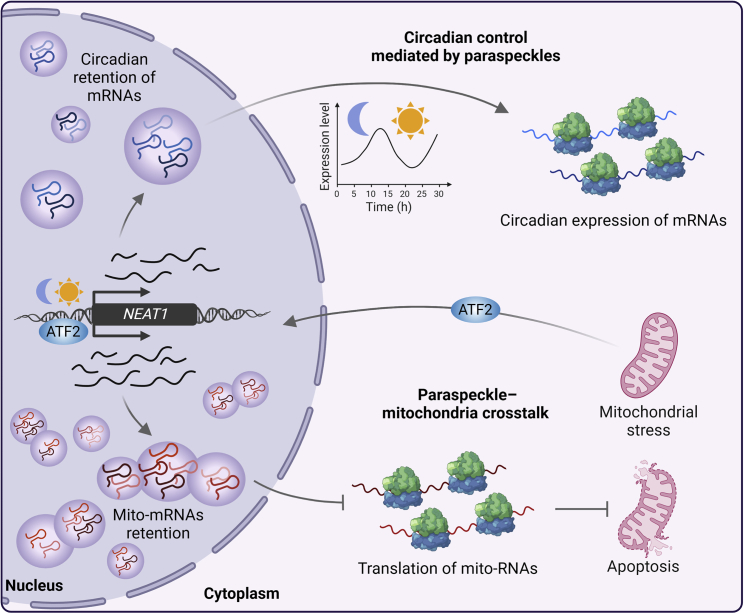
Alternative roles of NEAT1 and paraspeckles: Impact on circadian gene expression and mitochondrial cross-talk
NEAT1, among other components of the paraspeckle, follows a circadian expression pattern. This leads to rhythmic retention of certain mRNAs in the paraspeckle, including some core-clock mRNAs, allowing control of their circadian expression. On the other hand, NEAT1 regulates mitochondrial homeostasis and protects cells from apoptosis through the paraspeckle-mediated function. Mitochondrial stress favors NEAT1 expression by activation of the transcription factor ATF2, which binds to its promoter. NEAT1 upregulation correlates with an increase in the number of paraspeckles and adoption of an elongated morphology, together with enhanced retention of mRNAs. Among them, mRNAs of nucleus-encoded mitochondrial proteins (mito-mRNAs) conform an enriched group, and repression of their translation avoids induction of the mitochondrial pathway of apoptosis. This figure was created with BioRender.
Contribution of lncRNAs to normal brain cell physiology
lncRNAs may have important roles in physiological neuronal growth and plasticity, thus impacting cognitive functions. For example, depletion of NEAT1 with small interfering RNAs (siRNAs) or following neuronal potassium chloride stimulation reduces the levels of lysine 9 di-methylation on histone H3 (H3K9me2) at the c-Fos promoter (likely by impairing NEAT1 regulation of the histone methyltransferases EHMT1/2) and consequently upregulates the transcription complex activator protein-1 (AP-1), whose activity is linked to hippocampus-dependent learning tasks.47 In accordance with this link between NEAT1 and neuronal activity, a recent study has provided new insights to understand the impact of knocking out Neat1 in the mouse CNS. Depletion of murine Neat1 expression has a robust phenotype characterized by compromised secretory function and development of critical tissues relating to female reproduction.48,49 While Neat1 knockout mice do not present with an overt neurological phenotype, some deficits may only manifest under certain stress conditions. Indeed, Neat1 knockout (KO) animals suffer under specific stress conditions because of altered neuronal excitability and changes in the alternative splicing patterns of certain genes important for CNS function. These Neat1−/− mice present a distinct behavioral phenotype following a stressful stimulus, which includes hyperlocomotion, decreased anxiety, and impaired sociability, without any apparent neuroinflammation or gross synaptic dysfunction. NEAT1 likely fine-tunes neuronal excitability because Neat1−/− neurons are intrinsically hyperexcitable in culture and have deficiencies in calcium homeostasis. Molecularly, this is concomitant with altered expression of a subset of genes (including the riboflavin kinase [Rfk] gene, which is linked to regulation of circadian rhythms) as well as changes in the splice site choice of a subset of genes, some of which are genetically linked to psychiatric disorders such as schizophrenia and autism spectrum disorder.50
Cytoplasmically/synaptically enriched lncRNAs
The elaborate morphology of neurons confers special features to the subcellular localization of lncRNAs. The intriguing role of NEAT1 in mediating nuclear retention of TDP-43 and its potentially protective roles in certain proteinopathies (Figure 3A) is described above. Additionally, cytoplasmic lncRNAs deserve special mention when considering neuronal function. Different lncRNA entities are finely regulated in their spatiotemporal patterns of expression in the different cytoplasmic subcompartments, suggesting important roles in synaptic plasticity, learning, and memory. As an example, the activity-dependent transported lncRNA (Adeptr) is localized to dendrites by the motor protein Kif2A in a cyclic AMP (cAMP)/protein kinase A (PKA)-dependent manner, where it has a role in maintaining proper localization of the actin-scaffolding regulators ankyrin and spectrin, thus contributing to structural plasticity at the synapse51 (Figure 3B). The brain cytoplasmic 1 (BC1) lncRNA is also specifically transported to dendrites in association with other RBPs, such as Staufen, Translin, and the fragile X syndrome protein (FMRP).52,53,54,55 BC1 was the first ncRNA identified to have a role in mediating de novo protein synthesis at the synapse,56 and its primate functional analog is BC200. BC1/BC200 regulate the initiation phase of translation by directly interacting with eIF4A and eIF4B in dendritic spines, thereby inhibiting formation of the 48S pre-initiation complex.57 However, different ribonucleoprotein complexes with the participation of BC1/BC200 may affect translation differently. For example, in mouse models of Alzheimer’s disease, the BC1-FMRP association has been reported to induce amyloid precursor protein (APP) mRNA translation, and inhibition of BC1 protects against learning and memory deficits58 (Figure 3C). It is unknown to what extent BC200 lncRNA reproduces the functions of the mouse BC1 in human physiopathology, specifically in neurodegenerative disease, and thus its involvement in disease etiology is unclear. Other cytoplasmic lncRNAs are also able to influence translation; the BACE1-AS lncRNA, which is transcribed in the antisense direction relative to the β-secretase 1 (BACE1) coding gene, is stabilized by the neuronal RNA-binding protein HuD and enhances BACE1 mRNA levels and translation through partial base-pairing, with implications for the etiology of neurodegenerative disorders (see below).59
Figure 3.
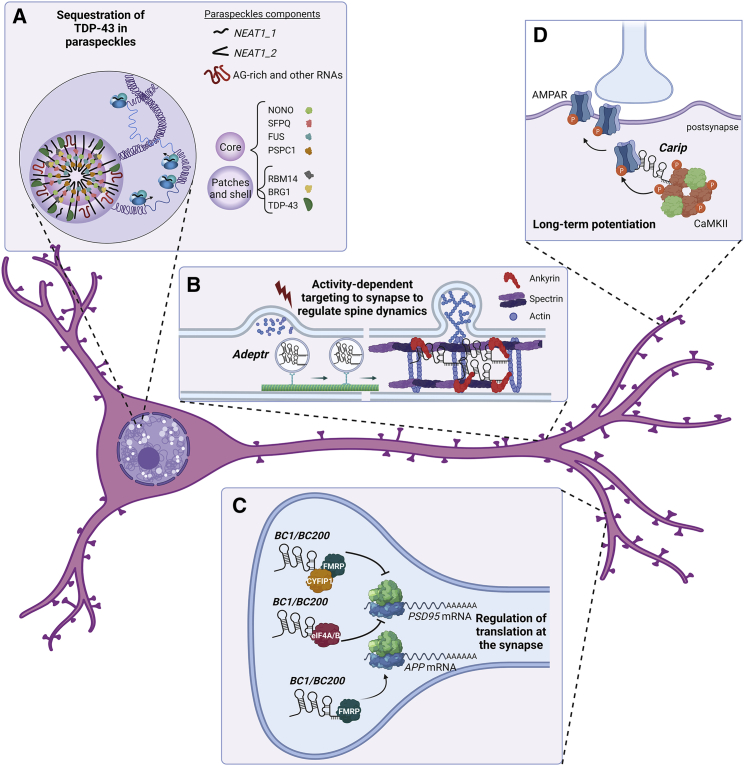
Examples of localized action of lncRNAs in neurons
(A) NEAT1 upregulation may be protective in TDP-43 proteinopathies by mediating nuclear sequestration of TDP-43 in paraspeckles. (B) The lncRNA Adeptr is targeted to dendrites by the motor protein Kif2A upon neuronal activity, where it mediates a cAMP signaling-induced increase in spine density, supporting synaptic activity. Mechanistically, Adeptr acts as a scaffolding agent of at least two regulators of actin polymerization in neuronal dendrites: AnkyrinB and Spectrin. (C) In the wild-type (WT) mouse neocortex, binding of BC1 lncRNA to the FMRP-CYFIP1 complex and to eIF4A/B at the synapse leads to repression of PSD-95 translation, preventing its exaggerated local synthesis. By contrast, in Alzheimer’s mouse models, BC1/BC200-FMRP positively regulates APP mRNA translation. (D) In the hippocampus, binding of the β subunit of the CaMKII complex to the lncRNA Carip enhances phosphorylation of the AMPA receptor and increases conductance of the post-synaptic AMPAR channel, contributing to long-term potentiation and enhancing learning and memory.
lncRNAs in the cytoplasm may also impact post-translational modifications and affect synaptic transmission. In the mouse hippocampus, the Carip lncRNA binds directly to the β subunit of Ca2+/calmodulin-dependent protein kinase II (CaMKII) and regulates post-translational modifications of target proteins (e.g., the excitatory α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptor subunits) in the postsynapse.60,61 Indeed, Carip KO mice display significantly reduced levels of phosphorylated AMPA receptor (AMPAR) and NMDA receptor (NMDAR) subunits, decreased amplitude of postsynaptic excitatory currents, and attenuated long-term potentiation of the hippocampal CA3-CA1 synapses, causing learning and memory deficits in mice. Through dual binding, Carip is suggested to bridge and possibly stabilize the association between the active CaMKII complex and its substrates, the GluA1 and GluA2 subunits of the AMPAR; the mechanistic connection between Carip and NMDAR is less clear (Figure 3D). Although Carip is a conserved transcript also present in humans, it is unknown whether the human ortholog plays a similar role. Modulation of CaMKII might be a shared mechanism among multiple lncRNAs to regulate synaptic plasticity and cognitive processes. For example, upon ischemia, upregulation of the CaMKIIδ and γ subunits facilitates neuronal survival through promotion of IKKα/β signaling and activation of the nuclear factor κB (NF-κB) pathway. This signaling is in part controlled by the intragenic lncRNAs C2dat1 and C2dat2, which overlap with the CAMK2D gene locus in the mouse genome. Downregulation of C2dat1/2 cooperatively reduces CAMK2δ expression through a still unknown mechanism, exacerbating ischemia-induced neuronal death.62,63
Despite remarkable progress in the understanding of lncRNA functions in the brain, there are still large areas to be investigated. For example, few studies consider the stoichiometry of RNA-protein interactions, which are often imbalanced with generally low levels of lncRNA expression. In this regard, the contribution of subcellular condensates to the role of lncRNAs as scaffolds, competitors, or regulators of other molecules is intriguing. Future research can further characterize the diversity of mechanisms used by lncRNAs to modulate neuronal function as well as the contribution of noncoding transcripts to glial cells under physiological and disease conditions.64 Additionally, little is known to date about the impact of lncRNA modifications (especially the most abundant N6-methyladenosine (m6A) mark, a form of methylated adenosine) on their brain functions, although exciting recent advances foresee key roles of modified lncRNAs in neuronal development.65
lncRNAs as biomarkers in disorders of the CNS: Diagnostic relevance
Given the diverse functions of annotated lncRNAs in the CNS, the question of diagnostic and therapeutic utility arises. Here we summarize evidence regarding the use of lncRNAs in CNS disease association and treatment.
Neurodegenerative diseases
Neurodegenerative diseases (NDs) are characterized by progressive loss of nerve cell function and, at the molecular level, accumulation of misfolded proteins and oxidative stress.66 They broadly include Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), dementia, spinocerebellar ataxia (SCA), and ALS. ND patients present with loss of cognitive and/or motor function; however, given that the causes of many NDs remain unknown, there are currently no effective treatments.67
Emerging research reveals that lncRNAs may be more heavily involved in ND pathogenesis than previously assumed. For example, BACE1-AS has been shown to stabilize BACE1 mRNA in vitro in human cells and in vivo in murine brains.68 BACE1-mediated proteolysis of APP leads to formation of hydrophobic β-amyloid peptide aggregates and senile deposits (hallmarks of AD pathophysiology).69,70 These aggregates can increase transcription of BACE1-AS, driving an injurious feedforward APP processing cascade. Furthermore, expression of several lncRNA genes, including BACE1-AS, 51A, and LRP1-AS, is upregulated in AD patient brains.71,72,73 The utility of these studies in developing diagnostic criteria should be considered cautiously because it remains unclear whether and how elevated lncRNA expression mechanistically contributes to the causes rather than simply markers of AD. Additionally, data from large in vivo cohorts controlling for disease severity and, in the case of human samples, comorbidities are underrepresented in these studies.
Many other lncRNAs are similarly associated with PD, HD, and ALS and are being investigated in vitro and in vivo.74 For example, recent findings highlight increased NEAT1L (the long isoform of NEAT1) expression in mouse models of polyglutamine diseases like HD, while loss of NEAT1L expression in dopaminergic neuroblastoma cell lines derived from HD patients leads to cellular dysfunction.75 Identifying lncRNAs with a potential role in ND by minimally invasive sampling of patient populations could bolster mechanistic studies and supplement ND diagnosis before onset of severe clinical symptoms, although the sampling rationale and methods would need to be considered carefully in the context of disease. For example, while HD is a monogenic disorder, PD is a heterogeneous disease whose diagnosis could be aided by molecular pathological findings. Given that PD symptoms are marked by selective loss of nigrostriatal dopaminergic neurons,76 it would be important to sample lncRNAs associated with PD (e.g., NEAT1, TUG1, and UCA1, which have not been studied consistently in dopaminergic neurons)74 in this neuronal subpopulation to assess their specificity to the disease pathophysiology. The clinical practicality of neuronal subtype sampling is also an important consideration.
Cancer
Malignant brain tumors include primary and metastatic lesions characterized by uncontrolled cell growth, necrosis, and dynamic angiogenesis that often result in headache, seizures, loss of neurological function, and, ultimately, death.77 While current noninvasive techniques (computed tomography [CT], magnetic resonance imaging [MRI], or positron emission tomography [PET] scans) are useful for identification, structural characterization, and broad localization of these tumors, it is difficult to classify tumor pathology and malignancy by noninvasive procedures alone. Definitive diagnosis by surgical biopsy is complicated by risks of intra- and post-operative functional decline.78 Thus, optimizing noninvasive detection of brain tumor biomarkers in patients of all ages is a key unmet need in neuro-oncology.
lncRNAs have recently emerged as regulators of brain tumor progression and cell stemness.79,80,81 Indeed, lncRNAs such as forkhead box D2 adjacent opposite strand RNA 1 (FOXD2-AS1), homeobox A (HOXA) distal transcript antisense RNA (HOTTIP), and HOX antisense intergenic RNA (HOTAIR) have been shown to promote glioma progression via cell cycle regulation and epigenetic alterations.19,82,83,84 Importantly, certain lncRNAs that cross the blood-brain barrier (BBB) and become enriched in cerebrospinal fluid (CSF) are upregulated in gliomas ex vivo, making them potential biomarkers for brain cancer.85 Although finding a universal marker for brain tumor patients is currently challenging, the ability to assay biomarker lncRNA expression from circulating tumor cells, exosomal noncoding RNAs, and CSF of cancer patients could greatly enhance the efficacy of brain tumor diagnosis. It remains to be seen, however, whether lncRNAs provide added value for brain cancer diagnosis/prognosis or therapy relative to other circulating species such as miRNAs, whose translational potential in cancer is advancing more rapidly.86
Neurodevelopmental disorders
Neurodevelopmental disorders (NDDs) encompass an array of clinical conditions including but not limited to autism spectrum disorder (ASD), fragile X syndrome (FXS), Down syndrome (DS), and Rett syndrome (RTT). They are broadly characterized by behavioral anomalies, intellectual disability, and motor symptoms. Because of their often complex and heterogeneous genetic etiology, NDDs can be difficult to diagnose and target therapeutically, and ongoing research is aimed at uncovering the molecular, genetic, and epigenetic landscape of these disorders.
Many lncRNAs are differentially expressed, mutated (including an increased burden of rare copy number variants of lncRNA loci in ASD), and/or pathogenically implicated in NDDs.87,88,89,90 The lncRNAs AK081227 and AK087060 are significantly upregulated in RTT mouse brains and are associated with downregulation of the gene encoding the gamma-aminobutyric acid receptor Rho 2, a component of inhibitory neurotransmission.91,92 Another class of noncoding RNAs, circular RNAs (circRNAs), shows links to NDD with emerging evidence of protein-regulatory roles in the brain.93 circRNAs are noncoding RNA molecules that undergo intra-transcript back-splicing and covalent linkage to form continuous closed loops lacking 5′ to 3′ polarity.94 Recently, dysregulation of noncoding ultraconserved and circRNAs has been shown to impact the biogenesis of AMPA receptors in RTT models.95 Additionally, brain-derived neurotrophic factor (BDNF), which plays a role in neuronal maturation, survival, and growth,96 is aberrantly diminished in RTT patients, and its antisense transcript, BDNF-AS lncRNA, has been earmarked as a potential RTT therapeutic target.87 When assessing transcriptome diversity in NDD models, it is important to note the caveats associated with techniques like microarray and RNA sequencing (RNA-seq) analysis, which sometimes interfere with RNA integrity during sample processing and often capture expression of abundant RNAs, where lncRNA expression may be relatively lower. Dysregulated lncRNA expression in combination with functional associations with specific NDD etiologies (e.g., for RTT) may, however, help stratify NDD diagnosis.
Psychological and other conditions
circRNAs and lncRNAs are enriched in cells of the CNS97,98 and have been implicated in development of neuropsychiatric and psychological conditions. For example, BDNF-AS was recently found to be elevated in postmortem amygdalae of patients with early-onset alcohol use disorder. BDNF-AS is thought to decrease BDNF expression via recruitment of EZH2, which adds H3K27 trimethyl marks to repress target genes.99 Additionally, multiple circRNAs are dysregulated in whole-blood samples of patients with major depressive disorder, and these circRNAs are enriched in neuronal pathways.100 Building on evidence that several lncRNAs are implicated in inflammation, angiogenesis, and apoptosis in stroke models,101 one ongoing clinical trial is investigating the diagnostic potential of lncRNAs for detection and prognosis of acute ischemic stroke (ClinicalTrials.gov: NCT04175691).
lncRNAs as biomarkers: Empirical evidence
The association of several lncRNAs with ND, cancer, NDD, and psychiatric conditions raises the question of whether they could serve as biomarkers in neuropathological contexts, particularly for diagnosis. A biomarker is specific to the disease in question, can be sampled noninvasively, and is sensitive to early and rapid detection. Biomarkers should help stratify disease risk and prognosis, provide insight into the biological mechanism of disease, and vary in response to treatment.102 Many of the lncRNAs mentioned above have been used as biomarkers under these criteria using minimally invasive methods. In vivo, transcriptomic analysis of plasma-derived cell-free circulating RNAs has revealed BACE1-AS to be a diagnostic marker of AD in patients with varying levels of symptom progression.103 Detection of lncRNA biomarkers from the bloodstream at sites distal to the brain has also been conducted in other NDs with relatively high specificity and sensitivity and, in some cases, has been validated by imaging data and tissue biopsies.104,105,106,107,108,109,110,111,112
While potentially informative, assaying lncRNA expression via the bloodstream requires isolation of serum, plasma, leukocytes, and/or exosomes (because circulating lncRNAs enter the bloodstream packaged in extracellular vesicles or attached to proteins113) and may not be completely representative of molecular changes native to the CNS. Liquid biopsies relying on CSF, which contacts the CNS, have been used to identify lncRNA signatures for diagnosis and preventative surgery in a variety of CNS pathologies.114,115,116 Although these techniques are promising, relying on circulating lncRNAs as biomarkers for CNS disorders is complicated. Canonically, qRT-PCR is used to assay circulating lncRNA expression; however, there is currently no established reference gene set for lncRNAs from different sources (e.g., plasma versus serum versus CSF). Additionally, lncRNAs may be expressed at low levels (and, thus, difficult to sample), depending on the cell type, and the expression of many lncRNAs is not specifically dysregulated in one disorder. For example, NEAT1 expression is aberrant in PD, AD, and ALS.117 For this reason, liquid biopsies for detection of lncRNA biomarkers could be used to supplement existing diagnostic methods rather than as a definitive diagnosis on their own.
lncRNAs as therapeutic targets: Methods, updates, and remaining questions
Beyond their potential use as biomarkers, lncRNAs have been investigated as therapeutic targets in vivo. Many approaches aimed at developing human therapies utilize RNA-based methods that target lncRNA transcripts for degradation or interference. Currently, 11 RNA therapies are approved by the US Food and Drug Administration (FDA) and/or the European Medicines Agency (EMA); of these, only one, nusinersen, aimed at improving spinal muscular atrophy outcomes, is targeted to the CNS via intrathecal administration (the rest target other tissue types86). Most therapies approved for clinical use are either antisense oligonucleotides (ASOs) or siRNAs. ASOs are single-stranded DNA molecules that are designed to be complementary to a target RNA sequence. When bound to the RNA, ASOs inhibit translation, trigger RNase H cleavage of the transcript, and/or interfere with cis-splicing elements, leading to exon exclusion.118 Therefore, the transcript is rendered loss of function. By contrast, siRNAs, which are short single- or double-stranded RNA sequences, take advantage of the endogenous microRNA (miRNA) machinery to cleave target RNAs or recruit repressive proteins. When introduced into the cell, siRNAs load target RNA onto the RNA-induced silencing complex (RISC), which can deadenylate and degrade and/or inhibit translation of the target RNA.
Currently, there are no approved RNA therapies for targeting lncRNA molecules in humans. Many approaches exist to target lncRNA expression in the laboratory, including transcriptional or post-transcriptional inhibition, steric hindrance of secondary structure assembly or effector protein interaction, genome editing, and exogenous introduction of lncRNAs. The efficacy of the approach depends in large part on the lncRNA genomic architecture and structure of the transcript.119 One approach that has shown promising results for degradation of natural antisense transcripts (NATs) in the CNS in vivo is use of ASOs. Modified ASOs that decrease NAT expression (AntagoNATs) have been shown to de-repress transcription of the gene encoding BDNF while increasing neuronal outgrowth.120 These ASOs also upregulate wild-type sodium voltage-gated channel alpha subunit 1 (SCN1A) gene expression, which undergoes heterozygous loss-of-function mutations in Dravet syndrome, a rare genetic brain disorder characterized by lifelong epilepsy.121 Similarly, AntagoNAT-mediated reduction of UBE3A-AS, which silences the paternal copy of the ubiquitin protein ligase E3A gene (UBE3A), ameliorates cognitive deficits in murine models of the NDD Angelman syndrome122 (Figures 4A–4C).
Figure 4.
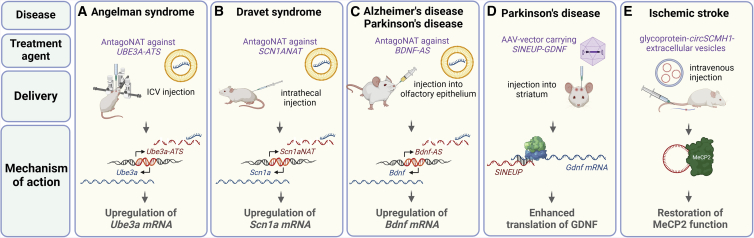
Examples of lncRNAs as potential therapeutic targets in preclinical models of CNS diseases
(A–C) Liposomal delivery of antagoNATs may be used to target lncRNA genes associated with disease. (A) In Angelman syndrome, deficiencies in the maternal allele of the UBE3A gene can be compensated by unsilencing the normally imprinted paternal allele. This is achieved by ASO-mediated downregulation of the antisense UBE3A-ATS transcript, a negative regulator of the sense transcript. In mouse, intracerebroventricular (i.c.v.) injection of ASOs via antagoNAT delivery partially restores UBE3A protein levels and ameliorates cognitive deficits associated with the disease.122 (B) Dravet syndrome is caused by heterozygous mutations in the voltage-gated sodium channel gene SCN1A. Use of antagoNATs against the antisense transcript SCN1ANAT promotes upregulation of SCN1A and improves the seizure phenotype in mice.121 (C) Targeting of the antisense BDNF-AS transcript with antagoNATs produces upregulation of BDNF mRNA, with therapeutic impact on a number of neurodegenerative disorders like AD and PD. Recently, a direct transnasal delivery method has been proposed that delivers the therapeutic agent directly to the olfactory submucosal space, thus enhancing CNS distribution with a minimally invasive strategy.127 (D) Antisense transcripts that partially overlap and promote translation of target mRNAs (SINEUPs) have been employed to upregulate GDNF protein levels and rescue motor deficits and neurodegeneration in mouse models of PD.123 Targeting can be localized via striatal injection. (E) Systemic delivery may also be accomplished intravenously, as in the case of the circRNA SCMH1 in the form of glycoprotein-circSCMH1 extracellular vesicles, which improve functional recovery after ischemic stroke in mice and nonhuman primate ischemic stroke models. Mechanistically, circSCMH1 binds to MeCP2 protein, thereby upregulating transcription of downstream genes involved in brain functions.133 This figure was created with BioRender.
Certain lncRNAs have neuroprotective or restorative effects, and it may be useful to upregulate their expression in disease models. For example, glial cell-derived neurotrophic factor (GDNF) promotes dopaminergic neuron survival and could ameliorate PD symptoms. SINEUPs, a class of antisense lncRNAs containing repetitive short interspersed nuclear elements (SINEs), promote translation of sense coding mRNAs and can be used to activate protein production. Indeed, adeno-associated virus 9 (AAV9)-mediated delivery of GDNF-targeting SINEUPs in the mouse striatum increases endogenous GDNF protein and dopaminergic potentiation while reducing motor deficits and neurodegeneration123 (Figure 4D). SINEUPs were originally identified in the mouse,124 and, to date, the number of functionally validated endogenous SINEUP lncRNAs in human cells is very limited.125 While their presence in the human brain transcriptome is uncertain, SINEUPs have a unique mode of action that may inspire biotechnological advances aimed at recovering the physiological levels of a target protein.126
Although ncRNAs have been considered for therapeutic targeting in several tissue types and diseases, the CNS poses unique challenges. First, the therapy must cross the BBB, which often necessitates direct intrathecal CNS administration or intracerebroventricular injection, both of which can be risky procedures. Recently, minimally invasive nasal depot (MIND), a topical intranasal method, has been shown to successfully deliver antagoNATs against BDNF-AS into the mouse brain127 via the olfactory submucosal space. Additionally, combination of nucleotide therapies with liposomes has been shown to enhance BBB penetration.128 Therapies must also be able to cross the cell membrane, be cell subtype and sequence specific with low off-target effects (at the transcriptome and cellular levels), and show low toxicity and immunogenicity. circRNAs are particularly interesting with respect to the last category because they are less immunogenic than other synthetic molecules129 and can act as miRNA sponges130 that induce continuous loss of miRNA function. Given the abundance of circRNAs in the brain131 and their implications in several CNS-related diseases,132 it will be interesting to follow their use in therapeutic development.
One solution to BBB and cell membrane crossing as well as toxicity and immunogenicity is use of exosomes and other membrane vesicles as therapeutic vehicles. Indeed, delivery of glycoprotein-circSCMH1 via extracellular vesicle intravenous injection has been shown to improve functional recovery and neuronal plasticity in mice with distal middle cerebral artery occlusions with no report of associated toxicity or immune response133 (Figure 4E). Exosomes are also a consideration for PD therapy.134 Combined with novel sequencing methods and emerging functional characterizations of lncRNAs in the CNS, these techniques may bring about substantial advancements in cell- and disease-type-specific ncRNA therapeutic targeting.
Summary
Investigation of lncRNA functions in the CNS is a rapidly expanding field that holds exciting potential for new advances in understanding brain cell physiology and improving disease diagnosis and therapy. This review summarizes important evidence regarding the roles of lncRNAs in brain cellular and molecular processes, highlighting a few lncRNAs that have been studied as biomarkers of CNS disorders involving neurodegeneration, neurodevelopment, and more. Our current understanding is that lncRNAs have a fine-tuning influence in a variety of human diseases and represent a type of molecule that can be easily targeted with high specificity. However, direct involvement of lncRNAs in brain pathologies has been shown for only a reduced number of cases. Given that CNS tissues exhibit large enrichment and diversity of ncRNAs, future research will be key to elucidating the roles of known and still unidentified lncRNAs in the CNS, expanding the potential of these noncodings to serve as diagnostic biomarkers and even targets for treatment of CNS conditions.
Acknowledgments
We thank the CERCA Program/Generalitat de Catalunya and the Josep Carreras Foundation for institutional support. This work was supported by the Ministerio de Ciencia e Innovación (MCI) co-financed by the European Development Regional Fund, “A way to achieve Europe” ERDF, under grant number PID2019-111658RB-I00/AEI/10.13039/501100011033 (to S.G.). T.S. was the recipient of a Fulbright fellowship.
Author contributions
T.S. and S.G. conceived the review topic. T.S., C.M., C.O.-M., and S.G. wrote the entire article and edited it collaboratively.
Declaration of interests
The authors declare no competing interests.
References
- 1.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jusic A., Thomas P.B., Wettinger S.B., Dogan S., Farrugia R., Gaetano C., Tuna B.G., Pinet F., Robinson E.L., Tual-Chalot S., et al. Noncoding RNAs in age-related cardiovascular diseases. Ageing Res. Rev. 2022;77:101610. doi: 10.1016/j.arr.2022.101610. [DOI] [PubMed] [Google Scholar]
- 3.Feng F., Jiao P., Wang J., Li Y., Bao B., Luoreng Z., Wang X. Role of long noncoding RNAs in the regulation of cellular immune response and inflammatory diseases. Cells. 2022;11:3642. doi: 10.3390/cells11223642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C., Wang D., Jiang Z., Gao Y., Sun L., Li R., Chen M., Lin C., Liu D. Non-coding RNAs in diabetes mellitus and diabetic cardiovascular disease. Front. Endocrinol. 2022;13:961802–961816. doi: 10.3389/fendo.2022.961802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer-Bensch G. Emerging roles of long non-coding RNAs as drivers of brain evolution. Cells. 2019;8:1399. doi: 10.3390/cells8111399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washietl S., Kellis M., Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Necsulea A., Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 2014;15:734–748. doi: 10.1038/nrg3802. [DOI] [PubMed] [Google Scholar]
- 9.He Z., Bammann H., Han D., Xie G., Khaitovich P. Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. Rna. 2014;20:1103–1111. doi: 10.1261/rna.043075.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnsson P., Lipovich L., Grandér D., Morris K.V. Evolutionary conservation of long non-coding RNAs; Sequence, structure, function. Biochim. Biophys. Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapusta A., Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30:439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.-D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCown P.J., Wang M.C., Jaeger L., Brown J.A. Secondary structural model of human MALAT1 reveals multiple structure–function relationships. Int. J. Mol. Sci. 2019;20:5610–5619. doi: 10.3390/ijms20225610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G., Mattick J.S., Taft R.J. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle. 2013;12:2061–2072. doi: 10.4161/cc.25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M., et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalei V., Sansom S.N., Kong L., Lee S., Montiel J.F., Vance K.,W., Ponting C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:1–24. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos A.D., Andersen R.E., Liu S.J., Nowakowski T.J., Hong S.J., Gertz C., Salinas R.D., Zarabi H., Kriegstein A.R., Lim D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponjavic J., Oliver P.L., Lunter G., Ponting C.P. Genomic and transcriptional Co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vance K.W., Sansom S.N., Lee S., Chalei V., Kong L., Cooper S.E., Oliver P.L., Ponting C.P. The long non-coding RNA paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlaki I., Alammari F., Sun B., Clark N., Sirey T., Lee S., Woodcock D.J., Ponting C.P., Szele F.G., Vance K.W. The long non-coding RNA Paupar promotes KAP 1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J. 2018;37:e98219. doi: 10.15252/embj.201798219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlaki I., Shapiro M., Pisignano G., Jones S.M.E., Telenius J., Muñoz-Descalzo S., Williams R.J., Hughes J.R., Vance K.W. Chromatin interaction maps identify Wnt responsive cis-regulatory elements coordinating Paupar-Pax6 expression in neuronal cells. PLoS Genet. 2022;18:110102300–110102324. doi: 10.1371/journal.pgen.1010230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Xi J., Wang G., Guo Z., Sun Q., Lu C., Ma L., Wu Y., Jia W., Zhu S., et al. Paupar and pax6 sequentially regulate human embryonic stem cell cortical differentiation. Nucleic Acids Res. 2021;49:1935–1950. doi: 10.1093/nar/gkab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cajigas I., Leib D.E., Cochrane J., Luo H., Swyter K.R., Chen S., Clark B.S., Thompson J., Yates J.R., 3rd, Kingston R.E., Kohtz J.D. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development (Cambridge) 2015;142:2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cajigas I., Chakraborty A., Swyter K.R., Luo H., Bastidas M., Nigro M., Morris E.R., Chen S., VanGompel M.J.W., Leib D., et al. The Evf2 ultraconserved enhancer lncRNA functionally and spatially organizes megabase distant genes in the developing forebrain. Mol. Cell. 2018;71:956–972.e9. doi: 10.1016/j.molcel.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bond A.M., Vangompel M.J.W., Sametsky E.A., Clark M.F., Savage J.C., Disterhoft J.F., Kohtz J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raveendra B.L., Swarnkar S., Avchalumov Y., Liu X.-A., Grinman E., Badal K., Reich A., Pascal B.D., Puthanveettil S.V. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc. Natl. Acad. Sci. USA. 2018;115:E10197–E10205. doi: 10.1073/pnas.1722587115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. Men ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:1–16. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B., Arun G., Mao Y.S., Lazar Z., Hung G., Bhattacharjee G., Xiao X., Booth C.J., Wu J., Zhang C., Spector D.L. The lncRNA malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard D., Prasanth K.V., Tripathi V., Colasse S., Nakamura T., Xuan Z., Zhang M.Q., Sedel F., Jourdren L., Coulpier F., et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barry G., Briggs J.A., Hwang D.W., Nayler S.P., Fortuna P.R.J., Jonkhout N., Dachet F., Maag J.L.V., Mestdagh P., Singh E.M., et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017;7:40127–40211. doi: 10.1038/srep40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirose T., Ninomiya K., Nakagawa S., Yamazaki T. A guide to membraneless organelles and their various roles in gene regulation. Nat. Rev. Mol. Cell Biol. 2022 doi: 10.1038/s41580-022-00558-8. [DOI] [PubMed] [Google Scholar]
- 35.Ghafouri-Fard S., Hussen B.M., Jamali E., Branicki W., Taheri M., Akbari Dilmaghani N. Role of lncRNAs and circRNAs in epilepsy. Ageing Res. Rev. 2022;82:101749. doi: 10.1016/j.arr.2022.101749. [DOI] [PubMed] [Google Scholar]
- 36.Wang C., Duan Y., Duan G., Wang Q., Zhang K., Deng X., Qian B., Gu J., Ma Z., Zhang S., et al. Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle LncRNA NEAT1-mediated liquid-liquid phase separation. Mol. Cell. 2020;79:443–458.e7. doi: 10.1016/j.molcel.2020.06.019. [DOI] [PubMed] [Google Scholar]
- 37.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Zupunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallegger M., Chakrabarti A.M., Lee F.C.Y., Lee B.L., Amalietti A.G., Odeh H.M., Copley K.E., Rubien J.D., Portz B., Kuret K., et al. TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell. 2021;184:4680–4696.e22. doi: 10.1016/j.cell.2021.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao Y.S., Sunwoo H., Zhang B., Spector D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West J.A., Mito M., Kurosaka S., Takumi T., Tanegashima C., Chujo T., Yanaka K., Kingston R.E., Hirose T., Bond C., et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016;214:817–830. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamazaki T., Yamamoto T., Yoshino H., Souquere S., Nakagawa S., Pierron G., Hirose T. Paraspeckles are constructed as block copolymer micelles. EMBO J. 2021;40:11072700–11072719. doi: 10.15252/embj.2020107270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekar D., Tusubira D., Ross K. TDP-43 and NEAT long non-coding RNA: roles in neurodegenerative disease. Front. Cell. Neurosci. 2022;16:954912–954914. doi: 10.3389/fncel.2022.954912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres M., Becquet D., Blanchard M.-P., Guillen S., Boyer B., Moreno M., Franc J.-L., François-Bellan A.M. Circadian RNA expression elicited by 3’-UTR IRAlu-paraspeckle associated elements. Elife. 2016;5:e14837. doi: 10.7554/eLife.14837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacq A., Becquet D., Guillen S., Boyer B., Bello-Goutierrez M.-M., Franc J.-L., François-Bellan A.M. Direct RNA–RNA interaction between Neat1 and RNA targets, as a mechanism for RNAs paraspeckle retention. RNA Biol. 2021;18:2016–2027. doi: 10.1080/15476286.2021.1889253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Hu S.-B., Wang M.-R., Yao R.-W., Wu D., Yang L., Chen L.-L. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018;20:1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- 46.McCluggage F., Fox A.H. Paraspeckle nuclear condensates: global sensors of cell stress? Bioessays. 2021;43:e2000245. doi: 10.1002/bies.202000245. [DOI] [PubMed] [Google Scholar]
- 47.Butler A.A., Johnston D.R., Kaur S., Lubin F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019;12:eaaw9277. doi: 10.1126/scisignal.aaw9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., Fujita Y., Fujimori T., Standaert L., Marine J.-C., Hirose T. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development (Cambridge) 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Standaert L., Adriaens C., Radaelli E., Van Keymeulen A., Blanpain C., Hirose T., Nakagawa S., Marine J.-C. The long noncoding RNA Neat1 is required for mammary gland development and lactation. Rna. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kukharsky M.S., Ninkina N.N., An H., Telezhkin V., Wei W., Meritens C.R.d., Cooper-Knock J., Nakagawa S., Hirose T., Buchman V.L., Shelkovnikova T.A. Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice. Transl. Psychiatry. 2020;10:171–219. doi: 10.1038/s41398-020-0854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grinman E., Nakahata Y., Avchalumov Y., Espadas I., Swarnkar S., Yasuda R., Puthanveettil S.V. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 2021;7:eabf0605. doi: 10.1126/sciadv.abf0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi S., Takashima A., Anzai K. The dendritic translocation of Translin protein in the form of BC1 RNA protein particles in developing rat hippocampal neurons in primary culture. Biochem. Biophy. Res. Commun. 1998;253:448–453. doi: 10.1006/bbrc.1998.9704. [DOI] [PubMed] [Google Scholar]
- 53.Mallardo M., Deitinghoff A., Müller J., Goetze B., Macchi P., Peters C., Kiebler M.A. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Reis S., Oostra B., Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 55.Muslimov I.A., Iacoangeli A., Brosius J., Tiedge H. Spatial codes in dendritic BC1 RNA. J. Cell Biol. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H., Iacoangeli A., Popp S., Muslimov I.A., Imataka H., Sonenberg N., Lomakin I.B., Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Briz V., Restivo L., Pasciuto E., Juczewski K., Mercaldo V., Lo A.C., Baatsen P., Gounko N.V., Borreca A., Girardi T., et al. The non-coding RNA BC1 regulates experience-dependent structural plasticity and learning. Nat. Commun. 2017;8:293. doi: 10.1038/s41467-017-00311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T., Pang P., Fang Z., Guo Y., Li H., Li X., Tian T., Yang X., Chen W., Shu S., et al. Expression of BC1 impairs spatial learning and memory in Alzheimer’s disease via APP translation. Mol. Neurobiol. 2018;55:6007–6020. doi: 10.1007/s12035-017-0820-z. [DOI] [PubMed] [Google Scholar]
- 59.Kang M.J., Abdelmohsen K., Hutchison E.R., Mitchell S.J., Grammatikakis I., Guo R., Noh J.H., Martindale J.L., Yang X., Lee E.K., et al. HuD regulates coding and noncoding RNA to induce APP→Aβ processing. Cell Rep. 2014;7:1401–1409. doi: 10.1016/j.celrep.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lisman J., Yasuda R., Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui X., Zhang R., yang Y., Wu E., Tang Y., Zhao Z., Li C., Yang L., teng X., Ye Y., et al. Identification and characterization of long non-coding RNA Carip in modulating spatial learning and memory. Cell Rep. 2022;38:110398. doi: 10.1016/j.celrep.2022.110398. [DOI] [PubMed] [Google Scholar]
- 62.Xu Q., Deng F., Xing Z., Wu Z., Cen B., Xu S., Zhao Z., Nepomuceno R., Bhuiyan M.I.H., Sun D., et al. Long non-coding RNA C2dat1 regulates CaMKIIδ expression to promote neuronal survival through the NF-κB signaling pathway following cerebral ischemia. Cell Death Dis. 2016;7:e2173. doi: 10.1038/cddis.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye J., Das S., Roy A., Wei W., Huang H., Lorenz-Guertin J.M., Xu Q., Jacob T.C., Wang B., Sun D., Wang Q.J. Ischemic injury-induced CaMKIIδ and CaMKIIγ confer neuroprotection through the NF-κB signaling pathway. Mol. Neurobiol. 2019;56:2123–2136. doi: 10.1007/s12035-018-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen M., Lai X., Wang X., Ying J., Zhang L., Zhou B., Liu X., Zhang J., Wei G., Hua F. Long non-coding RNAs and circular RNAs: insights into microglia and astrocyte mediated neurological diseases. Front. Mol. Neurosci. 2021;14:745066–745118. doi: 10.3389/fnmol.2021.745066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J., Jiang B., Li G.-W., Zheng D., Li M., Xie X., Pan Y., Wei M., Liu X., Jiang X., et al. m6A-modified lincRNA Dubr is required for neuronal development by stabilizing YTHDF1/3 and facilitating mRNA translation. Cell Rep. 2022;41:111693. doi: 10.1016/j.celrep.2022.111693. [DOI] [PubMed] [Google Scholar]
- 66.Gan L., Cookson M.R., Petrucelli L., La Spada A.R. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018;21:1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller J.H., Das V. Potential for treatment of neurodegenerative diseases with natural products or synthetic compounds that stabilize microtubules. Curr. Pharm. Des. 2020;26:4362–4372. doi: 10.2174/1381612826666200621171302. [DOI] [PubMed] [Google Scholar]
- 68.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., 3rd, Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goedert M., Spillantini M.G. A century of Alzheimer’s disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 70.Lacor P.N., Buniel M.C., Furlow P.W., Clemente A.S., Velasco P.T., Wood M., Viola K.L., Klein W.L. Aβ oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer’s disease. J. Neurosci. 2007;27:796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamanaka Y., Faghihi M.A., Magistri M., Alvarez-Garcia O., Lotz M., Wahlestedt C. Antisense RNA controls LRP1 sense transcript expression through interaction with a chromatin-associated protein. Cell Rep. 2015;11:967–976. doi: 10.1016/j.celrep.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Idda M.L., Munk R., Abdelmohsen K., Gorospe M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA. 2018;9:e1463. doi: 10.1002/wrna.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciarlo E., Massone S., Penna I., Nizzari M., Gigoni A., Dieci G., Russo C., Florio T., Cancedda R., Pagano A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 2013;6:424–433. doi: 10.1242/dmm.009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang M., He P., Bian Z. Long noncoding RNAs in neurodegenerative diseases: pathogenesis and potential implications as clinical biomarkers. Front. Mol. Neurosci. 2021;14:685143–685215. doi: 10.3389/fnmol.2021.685143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng C., Spengler R.M., Keiser M.S., Monteys A.M., Rieders J.M., Ramachandran S., Davidson B.L. The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum. Mol. Genet. 2018;27:4303–4314. doi: 10.1093/hmg/ddy331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Funayama M., Nishioka K., Li Y., Hattori N. Molecular genetics of Parkinson’s disease: contributions and global trends. J. Hum. Genet. 2022:1–6. doi: 10.1038/s10038-022-01058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferris S.P., Hofmann J.W., Solomon D.A., Perry A. Characterization of gliomas: from morphology to molecules. Virchows Arch. 2017;471:257–269. doi: 10.1007/s00428-017-2181-4. [DOI] [PubMed] [Google Scholar]
- 78.Malone H., Yang J., Hershman D.L., Wright J.D., Bruce J.N., Neugut A.I. Complications following stereotactic needle biopsy of intracranial tumors. World Neurosurg. 2015;84:1084–1089. doi: 10.1016/j.wneu.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 79.Hart R.P., Goff L.A. Long noncoding RNAs: central to nervous system development. Int. J. Dev. Neurosci. 2016;55:109–116. doi: 10.1016/j.ijdevneu.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M.M., Pletikos M., Meyer K.A., Sedmak G., et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Latowska J., Grabowska A., Zarębska Ż., Kuczyński K., Kuczyńska B., Rolle K. Non-coding RNAs in brain tumors, the contribution of lncRNAs, circRNAs, and snoRNAs to cancer development—their diagnostic and therapeutic potential. Int. J. Mol. Sci. 2020;21:7001–7031. doi: 10.3390/ijms21197001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X., Guo G., Lu Y., Wang S., Zhang Y., Huang Q. Mechanisms and functions of long non coding RNAs in glioma (Review) Oncol. Rep. 2021;45:9. doi: 10.3892/or.2021.7960. [DOI] [PubMed] [Google Scholar]
- 84.Pandey G.K., Mitra S., Subhash S., Hertwig F., Kanduri M., Mishra K., Fransson S., Ganeshram A., Mondal T., Bandaru S., et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 85.Xu K., Jiang X., Ariston Gabriel A.N., Li X., Wang Y., Xu S. Evolving landscape of long non-coding RNAs in cerebrospinal fluid: a key role from diagnosis to therapy in brain tumors. Front. Cell Dev. Biol. 2021;9:737670–737715. doi: 10.3389/fcell.2021.737670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics — challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang S.F., Gao J., Liu C.M. The role of non-coding RNAs in neurodevelopmental disorders. Front. Genet. 2019;10:1033–1110. doi: 10.3389/fgene.2019.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tuncay I.O., Parmalee N.L., Khalil R., Kaur K., Kumar A., Jimale M., Howe J.L., Goodspeed K., Evans P., Alzghoul L., et al. Analysis of recent shared ancestry in a familial cohort identifies coding and noncoding autism spectrum disorder variants. NPJ Genom. Med. 2022;7:13. doi: 10.1038/s41525-022-00284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zarrei M., Burton C.L., Engchuan W., Young E.J., Higginbotham E.J., MacDOnald J.R., Trost B., Chan A.J.S., Walker S., Lamoureux S., et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom. Med. 2019;4:26. doi: 10.1038/s41525-019-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ang C.E., Ma Q., Wapinski O.L., Fan S., Flynn R.A., Lee Q.Y., Coe B., Onoguchi M., Olmos V.H., Do B.T., et al. The novel lncRNA lnc-NR2F1 is proneurogenic and mutated in human neurodevelopmental disorders. Elife. 2019;8:1417700–1417729. doi: 10.7554/eLife.41770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petazzi P., Sandoval J., Szczesna K., Jorge O.C., Roa L., Sayols S., Gomez A., Huertas D., Esteller M. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol. 2013;10:1197–1203. doi: 10.4161/rna.24286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van de Vondervoort I.I.G.M., Gordebeke P.M., Khoshab N., Tiesinga P.H.E., Buitelaar J.K., Kozicz T., Aschrafi A., Glennon J.C. Long non-coding RNAs in neurodevelopmental disorders. Front. Mol. Neurosci. 2013;6:53–59. doi: 10.3389/fnmol.2013.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li M.L., Wang W., Jin Z.B. Circular RNAs in the central nervous system. Front. Mol. Biosci. 2021;8:1–10. doi: 10.3389/fmolb.2021.629593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 95.Siqueira E., Obiols.Guardia A., Jorge-Torres O.C., Oliveira-Mateos C., Soler M., Ramesh-Kumar D., Setién F., van Rossum D., Pascual-Alonso A., Xiol C., et al. Analysis of the circRNA and T-UCR populations identifies convergent pathways in mouse and human models of Rett syndrome. Mol. Ther. Nucleic Acids. 2022;27:621–644. doi: 10.1016/j.omtn.2021.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bathina S., Das U.N. Brain-derived neurotrophic factor and its clinical Implications. Arch. Med. Sci. 2015;11:1164–1178. doi: 10.5114/aoms.2015.56342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hanan M., Soreq H., Kadener S. CircRNAs in the brain. RNA Biol. 2017;14:1028–1034. doi: 10.1080/15476286.2016.1255398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andersen R.E., Lim D.A. Forging our understanding of lncRNAs in the brain. Cell Tissue Res. 2018;371:55–71. doi: 10.1007/s00441-017-2711-z. [DOI] [PubMed] [Google Scholar]
- 99.Bohnsack J.P., Teppen T., Kyzar E.J., Dzitoyeva S., Pandey S.C. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl. Psychiatry. 2019;9:34. doi: 10.1038/s41398-019-0367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang D., Ji Y., Chen X., Chen R., Wei Y., Peng Q., Lin J., Yin J., Li H., Cui L., et al. Peripheral blood circular RNAs as a biomarker for major depressive disorder and prediction of possible pathways. Front. Neurosci. 2022;16:844422–844511. doi: 10.3389/fnins.2022.844422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bao M.H., Szeto V., Yang B.B., Zhu S.-Z., Sun H.-S., Feng Z.-P. Long non-coding RNAs in ischemic stroke review-article. Cell Death Dis. 2018;9:281. doi: 10.1038/s41419-018-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bennett M.R., Devarajan P. Biomarkers of Kidney Disease. Elsevier Inc.; 2011. Characteristics of an ideal biomarker of kidney diseases. [DOI] [Google Scholar]
- 103.Fotuhi S.N., Khalaj-Kondori M., Hoseinpour Feizi M.A., Talebi M. Long non-coding RNA BACE1-AS may serve as an Alzheimer’s disease blood-based biomarker. J. Mol. Neurosci. 2019;69:351–359. doi: 10.1007/s12031-019-01364-2. [DOI] [PubMed] [Google Scholar]
- 104.Wang Q., Han C.-L., Wang K.-L., Sui Y.-P., Li Z.-B., Chen N., Fan S.-Y., Shimabukuro M., Wang F., Meng F.-G. Integrated analysis of exosomal lncRNA and mRNA expression profiles reveals the involvement of lnc-MKRN2-42:1 in the pathogenesis of Parkinson’s disease. CNS Neurosci. Ther. 2020;26:527–537. doi: 10.1111/cns.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang D., Wang P., Bian X., Xu S., Zhou Q., Zhang Y., Ding M., Han M., Huang L., Bi J., et al. Elevated plasma levels of exosomal BACE1 AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Mol. Med. Rep. 2020;22:227–238. doi: 10.3892/mmr.2020.11118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feng L., Liao Y.-T., He J.-C., Xie C.-L., Chen S.-Y., Fan H.-H., Su Z.-P., Wang Z. Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol. 2018;18:4–8. doi: 10.1186/s12883-017-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gagliardi S., Zucca S., Pandini C., Diamanti L., Bordoni M., Sproviero D., Arigoni M., Olivero M., Pansarasa O., Ceroni M., et al. Long non-coding and coding RNAs characterization in peripheral blood mononuclear cells and spinal cord from amyotrophic lateral sclerosis patients. Sci. Rep. 2018;8:2378–2411. doi: 10.1038/s41598-018-20679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cheng J., Duan Y., Zhang F., Shi J., Li H., Wang F., Li H. The role of lncRNA TUG1 in the Parkinson disease and its effect on microglial inflammatory response. Neuromolecular Med. 2021;23:327–334. doi: 10.1007/s12017-020-08626-y. [DOI] [PubMed] [Google Scholar]
- 109.Boros F.A., Maszlag-Török R., Vécsei L., Klivényi P. Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain Res. 2020;1730:146672. doi: 10.1016/j.brainres.2020.146672. [DOI] [PubMed] [Google Scholar]
- 110.Fan Y., Li J., Yang Q., Gong C., Gao H., Mao Z., Yuan X., Zhu S., Xue Z. Dysregulated long non-coding RNAs in Parkinson’s disease contribute to the apoptosis of human neuroblastoma cells. Front. Neurosci. 2019;13:1–12. doi: 10.3389/fnins.2019.01320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu Y., Pang D., Li C., Gu X., Chen Y., Ou R., Wei Q., Shang H. The expression discrepancy and characteristics of long non-coding RNAs in peripheral blood leukocytes from amyotrophic lateral sclerosis patients. Mol. Neurobiol. 2022;59:3678–3689. doi: 10.1007/s12035-022-02789-4. [DOI] [PubMed] [Google Scholar]
- 112.Santoro M., Nociti V., Lucchini M., De Fino C., Losavio F.A., Mirabella M. Expression profile of long non-coding RNAs in serum of patients with multiple sclerosis. J. Mol. Neurosci. 2016;59:18–23. doi: 10.1007/s12031-016-0741-8. [DOI] [PubMed] [Google Scholar]
- 113.Chen F., Wang N., Tan H.-Y., Guo W., Zhang C., Feng Y. The functional roles of exosomes-derived long non-coding RNA in human cancer. Cancer Biol. Ther. 2019;20:583–592. doi: 10.1080/15384047.2018.1564562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Whitlock J.H., Soelter T.M., Williams A.S., Hardigan A.A., Lasseigne B.N. Liquid biopsies in epilepsy: biomarkers for etiology, diagnosis, prognosis, and therapeutics. Hum. Cell. 2022;35:15–22. doi: 10.1007/s13577-021-00624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pan C.Y., Tian M., Zhang L.-L., Tian D., Wang L.-Y., Sun Y.-J., Cui Y.F. lncRNA signature for predicting cerebral vasospasm in patients with SAH: implications for precision neurosurgery. Mol. Ther. Nucleic Acids. 2020;21:983–990. doi: 10.1016/j.omtn.2020.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hossein-Nezhad A., Fatemi R.P., Ahmad R., Peskind E.R., Zabetian C.P., Hu S.-C., Shi M., Wahlestedt C., Zhang J., Faghihi M.A. Transcriptomic profiling of extracellular RNAs present in cerebrospinal fluid identifies differentially expressed transcripts in Parkinson’s disease. J. Parkinsons Dis. 2016;6:109–117. doi: 10.3233/JPD-150737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.An H., Williams N.G., Shelkovnikova T.A. NEAT1 and paraspeckles in neurodegenerative diseases: a missing lnc found? Noncoding. RNA Res. 2018;3:243–252. doi: 10.1016/j.ncrna.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Crooke S.T., Baker B.F., Crooke R.M., Liang X.H. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 2021;20:427–453. doi: 10.1038/s41573-021-00162-z. [DOI] [PubMed] [Google Scholar]
- 119.Goyal A., Myacheva K., Groß M., Klingenberg M., Duran Arqué B., Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45:e12. doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Modarresi F., Faghihi M.A., Lopez-Toledano M.A., Fatemi R.P., Magistri M., Brothers S.P., van der Brug M.P., Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hsiao J., Yuan T.Y., Tsai M.S., Lu C.Y., Lin Y.C., Lee M.L., Lin S.W., Chang F.C., Liu Pimentel H., Olive C., et al. Upregulation of haploinsufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of Dravet syndrome. EBioMedicine. 2016;9:257–277. doi: 10.1016/j.ebiom.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Espinoza S., Scarpato M., Damiani D., Managò F., Mereu M., Contestabile A., Peruzzo O., Carninci P., Santoro C., Papaleo F., et al. SINEUP non-coding RNA targeting GDNF rescues motor deficits and neurodegeneration in a mouse model of Parkinson’s disease. Mol. Ther. 2020;28:642–652. doi: 10.1016/j.ymthe.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 125.Schein A., Zucchelli S., Kauppinen S., Gustincich S., Carninci P. Identification of antisense long noncoding RNAs that function as SINEUPs in human cells. Sci. Rep. 2016;6:33605–33608. doi: 10.1038/srep33605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Espinoza S., Bon C., Valentini P., Pierattini B., Matey A.T., Damiani D., Pulcrano S., Sanges R., Persichetti F., Takahashi H., et al. SINEUPs: a novel toolbox for RNA therapeutics. Essays Biochem. 2021;65:775–789. doi: 10.1042/EBC20200114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Padmakumar S., Jones G., Pawar G., Khorkova O., Hsiao J., Kim J., Amiji M.M., Bleier B.S. Minimally invasive nasal depot (MIND) technique for direct BDNF AntagoNAT delivery to the brain. J. Control Release. 2021;331:176–186. doi: 10.1016/j.jconrel.2021.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grafals-Ruiz N., Rios-Vicil C.I., Lozada-Delgado E.L., Quiñones-Díaz B.I., Noriega-Rivera R.A., Martínez-Zayas G., Santana-Rivera Y., Santiago-Sánchez G.S., Valiyeva F., Vivas-Mejía P.E. Brain targeted gold liposomes improve rnai delivery for glioblastoma. Int. J. Nanomedicine. 2020;15:2809–2828. doi: 10.2147/IJN.S241055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 2019;74:508–520.e4. doi: 10.1016/j.molcel.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 131.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 132.Sekar S., Liang W.S. Circular RNA expression and function in the brain. Noncoding. RNA Res. 2019;4:23–29. doi: 10.1016/j.ncrna.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang L., Han B., Zhang Z., Wang S., Bai Y., Zhang Y., Tang Y., Du L., Xu L., Wu F., et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 134.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., Patel T., Piroyan A., Sokolsky M., Kabanov A.V., Batrakova E.V. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]