Abstract
Inflammation, a hallmark of cancer, has been associated with tumor progression, transition into malignant phenotype and efficacy of the chemotherapeutic agents in cancer. Chronic inflammation provides a favorable environment for tumorigenesis by inducing immunosuppression, whereas acute inflammation prompts tumor suppression by generating anti-tumor immune responses. Inflammatory factors derived from interstitial cells or tumor cells can stimulate cell proliferation and survival by modulating oncogenes and/or tumor suppressors. Recently, a new class of RNAs, i.e., circular RNAs (circRNAs), has been implicated in inflammatory diseases. Although there are reports on circRNAs imparting functions in inflammatory insults, whether these circularized transcripts hold the potential to regulate inflammation-induced cancer or tumor-related inflammation, and modulate the interactions between tumor microenvironment (TME) and the inflammatory stromal/immune cells, awaits further elucidation. Contextually, the current review describes the molecular association between inflammation and cancer, and spotlights the regulatory mechanisms by which circRNAs can moderate TME in response to inflammatory signals/triggers. We also present comprehensive information about the immune cell(s)-specific expression and functions of the circRNAs in TME, modulation of inflammatory signaling pathways to drive tumorigenesis, and their plausible roles in inflammasomes and tumor development. Moreover, the therapeutic potential of these circRNAs in harnessing inflammatory responses in cancer is also discussed.
Keywords: circular RNA, circRNA, inflammation, immunosuppression, tumor microenvironment, inflammatory microenvironment, signal transduction, cancer progression
Graphical abstract
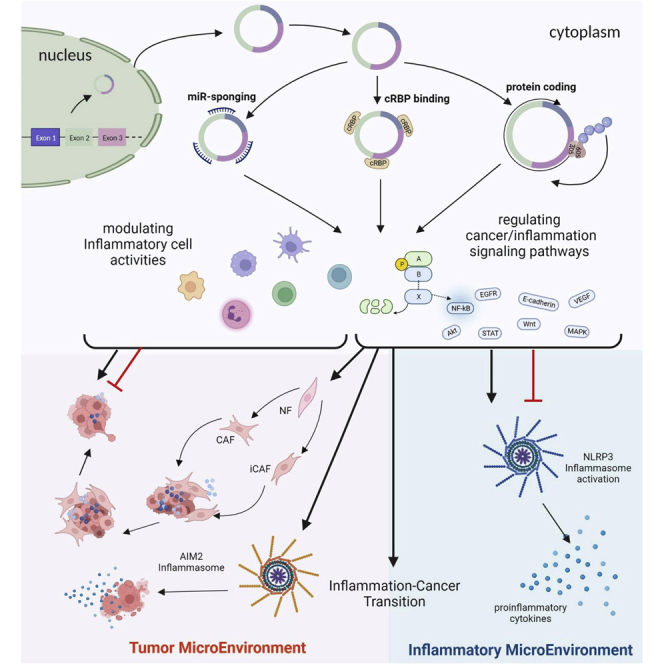
In this review, we describe the molecular association between inflammation and cancer, and spotlight the regulatory mechanisms by which circular RNAs moderate tumor microenvironment in response to inflammatory signals. We present comprehensive information about the expression and functions of the circular RNAs in inflammation-associated tumor microenvironment and tumor development.
Inflammation and cancer
Inflammation, a hallmark of cancer, has been associated with tumor development, promotion, transition into malignant phenotype, and efficacy of chemotherapeutic agents in various forms of cancer.1 Precisely, chronic inflammation provides a favorable environment for tumorigenesis and metastasis by inducing immunosuppression.2 Cancer-inducing niches could be tracked back to chronic inflammation, which entails deep-rooted tissue damage and continuous repair in response to the release of various cytokines and growth factors.3 Concomitantly, tumor eliciting inflammation promotes cancer by stalling anti-tumor immunity, creating a tumor-permissive state by reforming the tumor microenvironment (TME), and carrying out tumor-promoting functions by applying direct signals onto cancer and epithelial cells.4,5 Likewise, anti-cancer therapies may also elicit persistent inflammatory responses, thus sustaining cancer progression by incurring therapeutic resistance.6 On the contrary, acute inflammation prompts a tumor inhibitory environment by generating anti-tumor immune responses by promoting dendritic cell (DC) maturation as well as their function, and the initiation of effector T cells. Besides, acute inflammation can potentially encourage apoptosis/cell death in different types of cancer.7
First proposed by Rudolf Virchow in 1863, the inter-relationship between “cancer” and “inflammation” has been well established, whereby the sites of chronic inflammation can provide a preferred environment for tumor initiation, and tumor biopsies are found to be enriched with inflammatory cells.3 Moreover, persistently unresolved and dysregulated inflammation has been correlated with higher risk of developing malignant phenotypes in various cancer types.8 Both extrinsic and intrinsic inflammation provide a favored situation for tumor development by producing an immunosuppressive TME, through which inflammatory factors derived from the interstitial cells or tumor cells can stimulate cell proliferation and cell survival by either activating oncogenes or inactivating tumor suppressor genes,9,10 as depicted in Figure 1.
Figure 1.
Molecular association between inflammation and cancer
(A) Mechanism of inflammation-induced cancer. Cancer development and progression may, pre-dominantly, be mediated by specific inflammatory cells, called “tumor infiltrating myeloid cells” that are capable of driving carcinogenesis through different schemes of action. Activation of NF-κB and STAT3 in response to the release of various pro-inflammatory cytokines promotes cell-cycle progression and inhibits apoptosis. STAT3 and NF-κB, once activated, can suppress the markers for epithelial differentiation, thereby prompting EMT. STAT3 induction can be hampered by binding of specific ILs to IL-BP. Moreover, secretion of inflammatory cytokines can induce genomic destabilization by activating AID and impair DNA repair by modulating HIF-1α expression, thus advancing tumorigenesis. TNF, tumor necrosis factor; TGF-β, transforming growth factor β; ROS, reactive oxygen species; MYD88, myeloid differentiation primary response 88; ILs, interleukins. (B) Mechanism of cancer-related inflammation. Tumor cells are proficient in engaging T cells, explicitly Tregs, thus exploiting their functional capabilities. These Tregs express specialized CC-chemokine receptors and are recruited to the site of tumor formation through binding of the respective CC-chemokine ligands with these receptors, whereby they encourage angiogenesis by stimulating the release of inflammatory mediators and repress T cell function. Tumor cells may also secrete various chemo-attractants and growth factors that mediate MDSC generation. These MDSCs secrete TGF-β and various other ILs (such as IL-3, IL-4, and IL-10), thus inhibiting the anti-tumor responses by T cells.
In the last two decades, non-coding RNAs have been extensively studied and shown to play important roles in various inflammatory conditions as well as in resolving inflammation.11,12 Quite recently, a new class of such RNAs has been reported to have potential implications in inflammatory diseases, called circular RNAs (circRNAs).13,14,15 circRNAs are largely pre-mRNA derivatives produced as a result of back splicing, unlike linear RNAs that follow the standard canonical splicing process for their formation. Owing to their covalently closed-loop structure and restricted degradation by exonucleases, circRNAs are rather more stable than their linear counterparts.16 circRNAs are generated via intronic complementary sequence-driven, RNA-binding protein (RBP)-driven, or lariat-mediated back splicing facilitated by splicing factors.17 The physiological and pathological functions of circRNAs occur through miRNA sponging, interaction with circRNA-binding proteins (cRBPs), protein translation, or transcriptional regulation.18
Although there are various reports on circRNAs imparting functions in inflammatory insults, whether these circularized transcripts hold the potential to regulate inflammation-induced cancer (extrinsic) or tumor-related inflammation (intrinsic), and modulate the interactions between TME and the inflammatory stromal/immune cells still requires elucidation. In this regard, this review describes the molecular association between inflammation and cancer, and spotlights the regulatory mechanisms by which circRNAs can moderate TME in response to inflammatory signals/triggers. We also present comprehensive information in regard to the immune cell(s)-specific expression and functions of circRNAs in TME, modulation of the inflammatory signaling pathways to drive tumorigenesis, and their plausible roles in inflammasomes and tumor development. Moreover, the therapeutic potential of these related circRNAs in harnessing inflammatory responses in cancer is also discussed.
Role of circRNAs in inflammation-induced carcinogenesis
circRNAs interfere with cancer/inflammation-related signaling pathways
Chronic infections are one of the causes of cancer. Globally, 15.6% and 17.7% of cancer cases reported in 1990 and 1995, respectively, were infection related.19,20 Recently, there has been a surge in studies that link inflammatory diseases to cancer. Tumor initiation, progression, and poor cancer prognosis are associated with dysregulation in both inflammatory and oncogenic signaling pathways.3,21 Upon pathogen sensing, NRLP (NOD-, LRR- and pyrin domain-containing proteins) inflammasomes are assembled to release cytokines, which amplify the inflammatory response by activating subsequent inflammatory signaling pathways, such as NF-κB (Nuclear Factor kappa-light-chain-enhancer of activated B cells) and MAPK (Mitogen-Activated Protein Kinase).22 Inflammation significantly impacts the composition of TME, which consists of fibroblasts, vascular cells, and inflammatory immune cells.6 Based on 8,500 published studies, 41 KEGG pathways were referred to as oncogenic in 49 human cancer types. Of these, the MAPK pathway was associated with up to 40 cancer types. Akt, mTOR, apoptosis, and NF-κB pathways were also involved in at least 34 cancer types, and closely connected to Wnt/β-catenin, STAT (Signal Transducer and Activator of Transcription), and p53 signaling pathways, whereby their synergistic dysregulation may contribute to tumorigenesis.23 These pathways are frequently integrated to form a signaling network between inflammatory diseases and cancer.24 In this regard, circRNAs have been reported to interfere with and regulate such signaling networks as one of their primary biological functions in disease progression.25
Underlying mechanisms for circRNA-mediated regulation of the signaling pathways
circRNAs are implicated in both inflammation and cancers, where they either upregulate or downregulate certain cellular components, thus modulating cancer-related inflammatory signaling pathways through one of the three ways depicted in Figure 2. Many of these circRNAs have been reported to act as a miRNA sponge, thus activating or repressing the signaling pathways involved26 (Table 1). Binding to cRBPs is another proposed mechanism for circRNA function72 (Table 2). In addition, translatable circRNAs encoding novel peptides are also involved in modulating disease-related signaling pathways109,110 (Table 3). Involvement of competing endogenous RNAs in activation or repression of the downstream pathways by sponging miRNA is the primary mechanism of interaction between the circRNA and PI3K/AKT pathway.129 Comprehensive proteogenomic analysis-based characterization of 95 prospectively collected endometrial carcinomas (83 endometrioid and 12 serous tumors) revealed a potential role of circRNAs in regulating epithelial-mesenchymal transition (EMT) through the Wnt/β-catenin pathway.130 Likewise, the mechanistic interplay between circRNAs and the evolutionarily conserved Hippo pathway has also been well studied in the process of carcinogenesis.16
Figure 2.
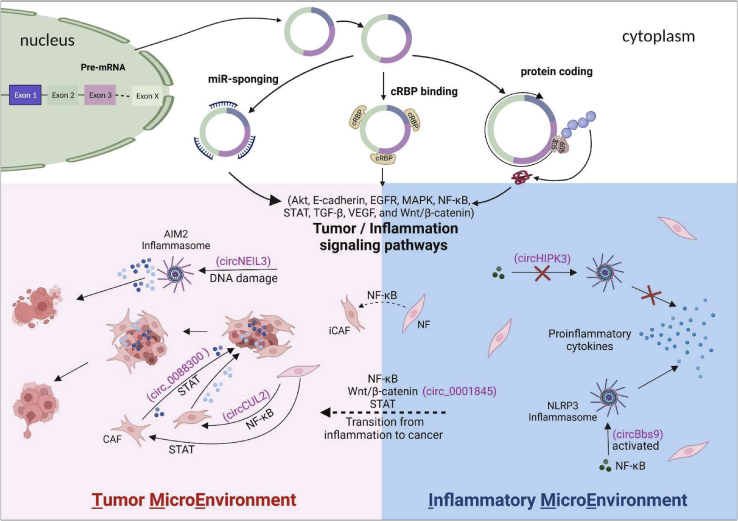
Involvement of circRNAs in tumor- and inflammation-associated microenvironments
circRNAs are generated in the nucleus and released into the cytoplasm. circRNAs regulate tumor/inflammation signaling pathways mainly through miRNA sponging, cRBP binding, and protein translation. The signaling pathways, such as Akt, E-cadherin, EGFR, MAPK, NF-κB, STAT, TGF-β, VEGF, and Wnt/β-catenin, have been found to be involved in both IME and TME. NLRP3 and AIM2 inflammasomes have been identified in IME and TME, respectively. Upon activation of NLRP3/AIM2 inflammasomes by NF-κB/cytosolic DNA, pro-inflammatory cytokines are released into IME/TME to maintain these microenvironments that support the pathogenesis of inflammatory diseases. In IME, sponging of miR-421 by circHIPK3 inhibits the NF-κB pathway and prevents the activation of NLRP3 inflammasome in ischemic injury. circBBS9 promotes NLRP3 inflammasome activation in the PM2.5-induced lung inflammation mice. In TME, sponging miR-1184 by circNEIL3 induces DNA damage and triggers AIM2 inflammasome activation, resulting in pyroptosis upon lung adenocarcinoma radiotherapy. In chronic IME, upregulation of circCUL2 in fibroblasts mediates the conversion of NFs into iCAFs and contributes to CAF heterogeneity in pancreatic ductal adenocarcinoma by inducing the NF-κB signaling pathway. circ_0088300 derived from CAFs regulates STAT signaling by sponging miR-1305 to promote gastric carcinoma. circ_0001845 promotes the transition from colitis to colitis-associated cancer through the Wnt/β-catenin signaling pathway. Furthermore, the interaction between cancer cells and CAF promotes proliferation, migration, and metastasis of cancer cells by releasing cytokines and activating related signaling pathways in TME.
Table 1.
Representative circRNAs that regulate related signaling pathways by sponging miRNAs
| Signaling pathways | circRNAs | miRNAs | Cancer [inflammation] | Ref. |
|---|---|---|---|---|
| Akt | circHIPK3 | miR-29b | [IBD] | 27 |
| circTLK1 | miR-214 | [IRI] | 28 | |
| circANAPC7 | miR-373 | PC | 29 | |
| circHMGCS1 | miR-503-5p | HB | 30 | |
| circVAPA | miR-377-3p/miR-494-3p | SCLC | 31 | |
| circSPON2 | miR-331-3p | PCa | 32 | |
| circSlc8a1 | miR-130b/miR-494 | BC | 33 | |
| E-cadherin | circAKT3 | miR-296-3p | [DN] | 34 |
| ccRCC | 35 | |||
| circHERC4 | miR-556-5p | CRC | 36 | |
| circPRMT5 | miR-30c | UCB | 37 | |
| EGFR | circPRKCB | miR-339-5p | [I/R] | 38 |
| circCDR1AS/ciRS-7 | miR-7 | CRC | 39 | |
| MAPK | circZNF609 | miR-145 | [PU] | 40 |
| circTLK1 | miR-17-5p | [SCM] | 41 | |
| circASAP1 | miR-326/miR-532 | HCC | 42 | |
| circMAPK4 | miR-125a-3p | Gliomas | 43 | |
| circRNA_C190 | miR-142-5p | NSCLC | 44 | |
| circASAP1 | miR-502-5p | GBM | 45 | |
| circFNDC3B | miR-1178-3p | BC | 46 | |
| NLRP3/inflammasome activation | circHipk3 | miR-421 | [Ischemic injury] | 47 |
| circBbs9 | miR-30e-5p | [PM-induced airway inflammation] | 48 | |
| AIM2/inflammasome activation | circNEIL3 | miR-1184 | LUAD | 49 |
| STAT | circ_0007059 | miR-1278 | [LN] | 50 |
| circZNF652 | miR-452-5p | [Asthma] | 51 | |
| circ_0003738 | miR-490-5p | [PsO] | 52 | |
| circSnx5 | miR-544 | [DC-driven immunogenicity] | 53 | |
| circHipk3 | miR124-3p | NSCLC | 54 | |
| circIGHG | miR-142-5p | OSCC | 55 | |
| circ_0088300 | miR-1305 | GC | 56 | |
| circCUL2 | miR-142-3p | 57 | ||
| TGF-β | circTXNRD1 | miR-892a | [PM-induced airway inflammation] | 58 |
| circSlc8a1 | miR-133a | [HF] | 59 | |
| circUCK2 | miR-125b-5p | [IS] | 60 | |
| circCDK14 | miR-125a-5p | [OA] | 61 | |
| circPVT1 | miR-21-5p | [SIONFH] | 62 | |
| circANKS1B | miR-148a-3p/miR-152-3p | BC | 63 | |
| circEHBP1 | miR-130a-3p | 64 | ||
| Wnt/β-catenin | circHIPK3 | miR-30a-3p | [OA] | 65 |
| circ_0025984 | miR-143-3p | [IS] | 66 | |
| circEIF4G3 | miR-4449 | GC | 67 | |
| circSTX6 | miR-449b-5p | PDAC | 68 | |
| YAP/Hippo | circLARP4 | miR-424-5p | GC | 69 |
| circACTN4 | ICC | 70 | ||
| circPVT1 | miR-497-5p | HNSCC | 71 |
Table 2.
Representative circRNAs that regulate related signaling pathways by binding cRBPs
| Signaling pathways | circRNAs | cRBPs | Cancer [inflammation] | Ref. |
|---|---|---|---|---|
| Akt | circCDYL2 | GRB7 | Breast cancer | 73 |
| circKIF18A | FOXC2 | GBMs | 74 | |
| circARHGAP29 | IGF2BP2 | PCa | 75 | |
| circFAM120A | SXT | 76 | ||
| circRHOBTB3 | HuR | CRC | 77,78 | |
| Autophagy/ATG7 | circTICRR | HuR | CC | 79 |
| Hh | circATG7 | HuR | PC | 80 |
| MAPK | hsa_circ_0068631 | EIF4A3 | BC | 81 |
| circPOLR2A | UBE3C/PEBP1 | ccRCC | 82 | |
| circNFIB | MEK1 | ICC | 83 | |
| circVPS13C | RRBP1 | NFPAs | 84 | |
| circPDE5A | WTAP | PCa | 85 | |
| STAT | circSCAR | ATP5B | [NASH] | 86 |
| circSTX6 | CUL2 | PDAC | 68 | |
| circSCMH1 | MeCP2 | [AIS] | 87,88 | |
| mmu_circ_0001109 | STAT3 | [IBD] | 89 | |
| circLRIG3 | EZH2/STAT3 | HCC | 90 | |
| circANRI | PES1 | [AS] | 91 | |
| circPOLR2A | PKR | [SLE] | 92 | |
| circMYBL2 | PTBP1 | AML | 93 | |
| TGF-β | circCDR1as/CIRS-7 | IGF2BP3 | Melanoma | 94,95 |
| circPTEN1 | Smad4 | CRC | 96 | |
| circTHBS1 | HuR | GC | 97 | |
| TNF-α | circDLC1 | HuR | HCC | 98 |
| VEGF | circFndc3b | FUS | [CA] | 99 |
| Wnt/β-catenin | circStag1 | HuR | [PO] | 100 |
| circZNF292 | SDOS | [Anti-inflammation] | 101 | |
| mmu_circ_0001845 | β-catenin | CAC/[IBD] | 89 | |
| circGSK3β | GSK3β | ESCC | 102 | |
| circHuR | CNBP | GC | 103 | |
| circ-CTNNB1 | DDX3 | 104 | ||
| circEIF4G3 | δ-catenin | 67 | ||
| circZKSCAN1 | FMRP | HCC | 105 | |
| circACTN4 | YBX1 | ICC | 70 | |
| circMTCL1 | C1QBP | LSCC | 106 | |
| circRNA_102171 | CTNNBIP1 | PTC | 107 | |
| YAP/Hippo | circYap | eIF4G/PABP | Breast cancer | 108 |
Table 3.
Representative circRNAs that regulate related signaling pathways by encoding proteins
| Signaling pathways | circRNAs | Encoded protein/interacting molecules | Cancer [inflammation] | Ref. |
|---|---|---|---|---|
| Akt | circHER2 | HER2-103/EGFR | TNBC | 111 |
| circGSPT1 | GSPT1-238aa/vimentin/Beclin1/14-3-3 | GC | 112 | |
| circAKT3 | AKT3-174aa/PDK1 | GBM | 113 | |
| EGFR | circEGFR | rtEGFR/EGFR | GBM | 114 |
| circE-Cad | C-E-Cad/EGFR | 115 | ||
| Hh | circSMO | SMO-193a.a./SMO | GBM | 116 |
| MAPK | circNlgn | Nlgn173/H2AX | [HF] | 117,118 |
| circMAPK1 | MAPK1-109aa/MAP2K1 | GC | 119 | |
| circMAPK14 | circMAPK14-175aa/MAP2K6 | CRC | 120 | |
| circASK1 | ASK1-272a.a/Akt1 | LA | 121 | |
| NF-κB | circPLCE1 | circPLCE1-411/RPS3 | CRC | 122 |
| STAT | cGGNBP2 | cGGNBP2- 184aa/STAT3 | ICC | 123 |
| Wnt/β-catenin | circDIDO1 | DIDO1-529aa/PARP1/PRDX2 | GC | 124 |
| circAXIN1 | AXIN1-295aa/APC | 125 | ||
| circβ-catenin | β-catenin-370aa/GSK3β | HCC | 126 | |
| circ-EIF6 | EIF6-224aa/MYH9 | TNBC | 127 | |
| YAP/Hippo | circPPP1R12A | circPPP1R12A-73aa/MST1/LATS1 | CC | 128 |
circRNAs sponge miRNAs to regulate signaling pathways in cancer/inflammation
PubMed search revealed an explosive increase in recent publications on circRNAs regulating the signaling pathways through sponging miRNAs. Representative circRNAs that mediate inflammatory diseases or tumors by sponging miRNAs are listed in Table 1 and part B of Table 4. Involvement of circRNAs in STAT, TGF-β (Transforming Growth Factor β), and NF-κB signaling pathways is related more to inflammation, while Akt, E-cadherin, MAPK, and Hippo-YAP are rather associated with cancer.
Table 4.
NF-kB signaling associated representative circRNAs and circRNAs with multiple functions
| Category | Signaling | circRNAs | Mechanism | Interacting molecules | Cancer [inflammation] | Ref. |
|---|---|---|---|---|---|---|
| A: signaling associated | NF-kB | circPLCE1 | encoding protein | RPS3 | CRC | 122 |
| circPPM1F | cRBP binding | HuR/PPM1F | [T1DM] | 131 | ||
| circDCUN1D4 | HuR/TXNIP | LA | 132 | |||
| circIKBKB | IKKβ | BC | 133 | |||
| mmu_circ_0001109 | p65 | [IBD] | 89 | |||
| circARF3 | miRNA sponging | miR-103 | [Ai] | 134 | ||
| circ_003912 | miR-123/miR-647/miR-31 | [EOLP] | 135 | |||
| circHIPK3 | miR-124 | LC | 136 | |||
| miR-421 | [Ischemic injury] | 47 | ||||
| circSirt1 | miR-132/212 | [VSMC] | 137 | |||
| circNFATC3 | miR-143-3p | BC | 138 | |||
| circCUL2 | miR-203a-5p | PDAC | 139 | |||
| circTLK1 | miR-17-5p | [SCM] | 41 | |||
| miR-214 | [IRI] | 28 | ||||
| circ_0005105 | miR-26a | [OA] | 140,141 | |||
| circ-UQCRC2 | miR-326 | [PP] | 142 | |||
| circRNA104250 | miR-3607-5p | [Airway inflammation] | 143 | |||
| circRNA-000911 | miR-449a | BC | 144 | |||
| circANRIL | miR-622 | [stroke] | 145 | |||
| circGLIS2 | miR-671 | CRC | 146 | |||
| circSHOC2 | miR-7670-3p | [IBI] | 147 | |||
| circLRP6 | miR-205 | [DN] | 148 | |||
| B: multiple functions | Akt | circHIPK3 | miRNA sponging | miR-29b | [IBD] | 27 |
| Wnt/β-catenin | miR-30a-3p | [OA] | 65 | |||
| NF-kB | miR-421 | [Ischemic injury] | 47 | |||
| STAT | miR124-3p | NSCLC | 54 | |||
| MAPK | circTLK1 | miR-136-5p | RCC | 149 | ||
| NF-kB | miR-17-5p | [SCM] | 41 | |||
| miR-214 | [IRI] | 28 | ||||
| E-cadherin | circAKT3 | miR-296-3p | [DN] | 34 | ||
| ccRCC | 35 | |||||
| TGF-β | circPVT1 | miR-21-5p | [SIONFH] | 62 | ||
| YAP/Hippo | miR-497-5p | HNSCC | 71 | |||
| Akt | circSLC8A1 | miR-130b/miR-494 | BC | 33 | ||
| TGF-β | miR-133a | [CH] | 59 | |||
| MAPK | circPOLR2A | cRBP binding | PKR | [SLE] | 92 | |
| STAT | UBE3C | ccRCC | 82 | |||
| YAP/Hippo | circACTN4 | miRNA sponging/cRBP binding | YBX1 | ICC | 70 | |
| Wnt/β-catenin | miR-424-5p | |||||
| EGFR | circCDR1AS/ciRS-7 | IGF2BP3 | melanoma | 94,150 | ||
| TGF-β | miR-7 | CRC | 39 | |||
| VEGF | circFNDC3B | FUS | [CA] | 99 | ||
| MAPK | miR-1178-3p | BC | 46 |
circRNAs bind to cRBPs to regulate signaling pathways in cancer/inflammation
circRNAs can form circRNA-protein complexes to modulate signaling pathways.151 Dysregulation of RBPs has been experimentally verified in different cancer phenotypes, resulting in chronic inflammation and/or autoimmunity.152,153 Among these extensively studied RBPs, human antigen R (HuR) (also named ELAVL1) is the most attractive RBP in the field. HuR is an established regulator of post-transcriptional gene regulation and is overexpressed in many human cancers. HuR drives the activation of a pro-inflammatory phenotype, and is linked to many chronic diseases, such as cardiac hypertrophy, pancreatitis, rheumatoid arthritis, asthma, and cachexia.154 HuR plays essential pathophysiological roles in the development of different liver diseases, including hepatic inflammation, alcoholic liver disease, non-alcoholic fatty liver disease, viral hepatitis, liver fibrosis, and liver cancer.155 LPS (lipopolysaccharide)-induced systemic inflammation causes intimal hyperplasia, and increased expression of Toll-like receptor 4 (TLR4) and HuR.156 Increasing evidence is suggestive of altered signaling pathways as a consequence of interactive crosstalk between circRNAs and HuR in inflammation and cancer. Among the representative circRNAs in Table 3 and part A of Table 4, HuR is the hottest cRBP for circRNAs to interact with. It has been verified that PRKAR2A-derived mmu_circ_0001845 interacts with β-catenin to promote inflammation-to-carcinoma transformation of colitis-associated colorectal carcinoma through the Wnt/β-catenin signaling pathway in vivo and in vitro89 (Figure 2).
circRNAs encode proteins to regulate signaling pathways in cancer/inflammation
circRNAs not only function by miRNA sponging or binding with cRBPs, but can also be translated in a cap-independent manner to encode novel proteins possessing novel physiological roles to either promote or suppress the disease. Some of these circRNA encoded proteins are also involved in signaling pathways to modulate cancer development and progression,109 as listed in Table 4. Of these, only Nlgn173 peptide encoded by circNlgn mediates heart failure by interacting with H2A histone family member X (H2AX) in MAPK signaling,117,118 while others act to regulate cancers by interacting with different proteins associated with Akt, EGFR, Hh, MAPK, NF-κB, STAT, Wnt/β-catenin, and Hippo-YAP pathways.
NF-κB signaling bridges the connection between inflammation and cancer
Inflammation, as a body’s protective response, is beneficial to the host in a timely manner. However, chronic inflammation poses high risk by altering the TME, which affects all stages of tumor development.157,158 NF-κB is involved in recruiting inflammatory cells and mediating the release of inflammatory chemokines, thus creating an environment that facilitates cancer progression. Dysregulated NF-κB signaling contributes to the pathogenesis of various inflammatory diseases,159 and is crucial to tumorigenesis as well. As an inducible transcription factor, NF-κB regulates multi-protein inflammasome complexes and mediates the pathogenesis of various inflammatory diseases, such as rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, atherosclerosis, and so on.158 Upon activation of the NLRP3 inflammasomes by NF-κB, pro-inflammatory cytokines are released into the TME or IME (inflammatory microenvironment) to maintain these microenvironments that support the pathogenesis of inflammatory diseases, and to subsequently trigger related signaling pathways in cancer.
Numerous human cancers have constitutive NF-κB activity due to the IME and various oncogenic mutations, which favors tumor growth by remodeling cancer cell metabolism, promoting proliferation, suppressing apoptosis, attracting angiogenesis, and facilitating distant metastasis, especially in solid tumors. Comparably, suppression of the NF-κB pathway leads to tumor regression, and may serve as an effective therapeutic strategy. Therefore, NF-κB is a master regulator in mediating a crosstalk between chronic inflammation and cancer at multiple levels.158,160 In addition, activation of the NF-κB pathway and its downstream pro-inflammatory genes has also been verified in cancer-associated fibroblasts (CAFs) cultured on fibrous polymeric matrices. Moreover, CAFs with NF-κB pathway activation have been observed to increase tumor-promoting proinflammatory properties in breast cancer.161 In addition, mmu_circ_0001109 exacerbates colitis by upregulating STAT and NF-κB signaling pathways, and may play a promoting role during the inflammation or colitis-to-carcinoma transition.89 Therefore, the NF-κB signaling pathway could serve as a bridge that connects the inflammatory cells and inflammation-triggered cancer cells within an immunosuppressive inflammatory TME, thus favoring tumor growth.
As shown in part A of the Table 4, 20 representative circRNAs modulating the NF-κB signaling pathway have been revealed to be associated with various inflammatory diseases (12 circRNAs) and cancers (8 circRNAs). Among these, 15 circRNAs modulate NF-κB signaling by sponging with miRNAs, four of them bind to cRBPs, and circPLCE1 encodes 411 amino acid (circPLCE1-411) peptides that interacts with RPS3 in colorectal cancer.
Multiple functions of circRNAs in signaling pathways related to cancer/inflammation
Among the representative circRNAs listed in the Tables 2, 3, and 4, some circRNAs exhibit multiple functions, which are summarized in part B of Table 4. As shown, most of these circRNAs may have modulatory roles in more than one disease. For instance, circHipk3 sponges four miRNAs to regulate inflammatory bowel disease, osteoarthritis, ischemic injury, and non-small cell lung cancer (NSCLC) through Akt, Wnt/β-catenin, NF-κB, and STAT pathways, respectively. Similarly, circTLK1 regulates two inflammatory diseases and one cancer by sponging three different miRNAs, as indicated.
circRNAs regulate immune cell(s) functions and responses
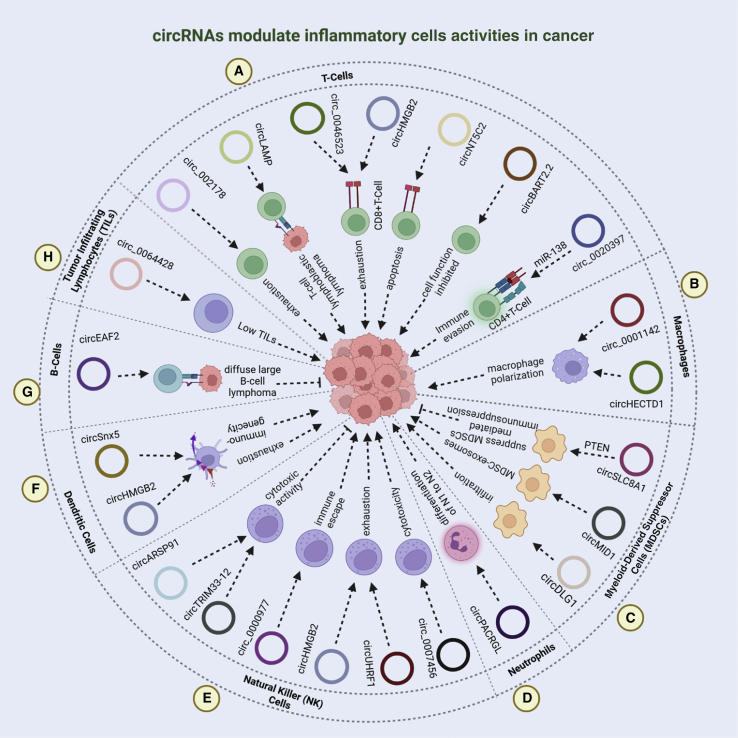
Both immune and tumor cells mutually impact various stages of cancer development and progression, thus adding to the TME intricacy by altering the immune cells to promote evasion, proliferation, and metastasis.3,10 Of these, T lymphocytes, B lymphocytes, natural killer (NK) cells, and antigen-presenting DCs are predominantly involved in tumor suppression, while immunosuppressive functions are imparted by tumor-associated macrophages (TAMs), tumor-associated neutrophils (TANs), CAFs, myeloid-derived suppressor cells (MDSCs), and regulatory T cells (Tregs). Thus far, there are only few studies to report the regulatory and immunomodulatory functions of circRNAs in different immune cells including neutrophils, tumor infiltrating lymphocytes (TILs), DCs, and TAMs in cancer. Nonetheless, certain studies which elucidated the expression and roles of these circRNAs in immune regulation driven by these immune/inflammatory cells are discussed in the subsequent sections and depicted in Figure 3 and Table 5.
Figure 3.
Regulatory functions of circRNAs in inflammatory cell responses during tumorigenesis
(A) In T lymphocytes, circRNA-002178 causes T cell exhaustion by regulating PD1 expression through miR-34 sponging. Similarly, overexpression of hsa_circ_0046523, circHMGB2, and circ-NT5C2 mediates immunosuppression by inducing CD8+ T cell apoptosis and exhaustion by sponging miR-148a-3p, miR-181a-5p/CARM1, and miR-448, respectively, thus promoting tumor growth, invasion, and metastasis. Besides, circ-LAMP1 is upregulated in T cell lymphoblastic lymphoma, whereby it stimulates cell proliferation and alleviates apoptosis through DDR2/RTK signaling dysregulation mediated by miR-615-5p. In nasopharyngeal carcinoma, circBART2.2 binding to RIG-I causes functional inhibition of these cells and immune escape. In CD4+ T cells, hsa_circ_0020397 modulates cell survival, invasion, and apoptosis by sponging miR-138, thus regulating PD-L1 and TERT expression in CRC. (B) In tumor-infiltrating lymphocytes (TILs), hsa_circ_0064428 is downregulated in HCC patients with high TILs and correlated with poor disease prognosis, therefore potentially regulating TIL formation. (C) In B lymphoma cells, upregulation of circEAF2 promotes cell apoptosis and lymphoma cell sensitivity toward epirubicin by targeting Epstein-Barr virus-encoded miR-BART19-3p/APC/β-catenin axis, thus retarding tumor growth by inactivating the Wnt signaling pathway. (D) Pertinent to NK cells, circARSP91 and circTRIM33-12 stimulate NK cell-mediated cytotoxicity, thus producing inhibitory effects on tumor growth and progression. Contrarily, circ_0000977 overexpression alleviates NK cell activity through miR-153 sponging, thus resulting in immune evasion in pancreatic cancer. Likewise, circHMGB2 and tumor-derived exosomal circUHRF1 drive cancer progression by turning off protective immune responses manifested by NK cell exhaustion and resistance to anti-PD1 therapy in NSCLC and HCC, respectively. In addition, hsa_circ_0007456 is revealed to modulate NK cell-mediated cytotoxicity through miR-6852-3p/ICAM-1 axis in HCC. (E) Both circSNX5 and circHMGB2 suppress dendritic cell activity, and promote their functional exhaustion, thus regulating immunogenicity in cancer. (F) circPACRGL has been discovered to bring about differentiation of neutrophils from N1 to N2, and promote cell proliferation, invasion, and migration through the miR-142-3p/miR-506-3p-TGF-β1 axis in CRC. (G) In MDSCs, circSLC8A1 interacts with miR-494, thus suppressing the release of immunosuppressive cytokines in a PTEN-dependent manner. Comparably, MDSC-derived exosomal S100A9 increased circMID1 expression, which functions through miR-506-3p sponging to promote tumorigenesis. Moreover, circDLG1 increases MDSC infiltration, thus promoting gastric cancer. (H) circHECTD1 is highly expressed in macrophages and modulates cell proliferation and migration by sponging miR-320-5p and regulating SLC2A1 activity in glioblastoma multiforme. Besides, exosomal circ_0001142 is overexpressed in breast cancer, whereby it regulates tumor growth, invasion-metastasis, autophagy, and macrophage polarization via miR-361-3p/PIK3CB pathway.
Table 5.
circRNAs associated with the inflammatory diseases that can potentially lead to cancer
| circRNA | Inflammatory disease | Inflammatory/relative cell(s) response | Proposed mechanism | Potential-associated malignancy | Ref. |
|---|---|---|---|---|---|
| circZC3H4 | silicosis | pulmonary macrophage activation | increasing ZC3H4 by miR-212 sponging | lung carcinoma | 210 |
| circHECTD1 | silicosis | macrophage activation and fibroblast proliferation | balancing M1/M2 macrophages by ZC3H12A binding | lung carcinoma | 189 |
| circ-0003528 | bronchitis | macrophage polarization | increasing CTLA4 by miR-224-5p, miR-324-5p and miR-488-5p sponging | lung carcinoma | 211 |
| circATRNL1 | ovarian endometriosis | promoting proliferation, migration, and invasion of Ishikawa cells | increasing YAP1 by miR-141-3p and miR-200a-3p sponging | ovarian carcinoma | 212 |
| circ_0088194 | rheumatoid arthritis | promoting the invasion and migration of rheumatoid arthritis fibroblast-like synoviocytes | increasing MMP2 by miR-766-3p sponging | osteosarcoma | 213 |
| circRNA.33186 | osteoarthritis | promoting proliferation and inhibit apoptosis of IL-1β-treated chondrocytes | increasing MMP-13 by miR-127-5p sponging | osteosarcoma | 214 |
| circFBXW4 | hepatic fibrosis | inhibiting activation, proliferation and induced apoptosis of hepatic stellate cells | increasing FBXW7 by miR-18b-3p sponging | liver cancer | 215 |
| circ_103765 | Crohn’s disease | promoting proliferation and apoptosis of human intestinal epithelial cell | increasing DLL4 by miR-30 family sponging | colorectal carcinoma | 216 |
| circHECTD1 | ulcerative colitis | promoting enterocyte autophagy | increasing HuR by miR-182-5p sponging | colorectal carcinoma | 217 |
| circ_0089172 | Hashimoto’s thyroiditis | N/A | increasing IL-23R by miR-125a-3p sponging | lymphoid tissue lymphoma | 218 |
circRNAs regulate T-, B-, TIL-, and NK-cell-mediated immune responses
T lymphocytes play a significant role in generating responses as part of the adaptive immune system. Tumor-associated antigen can stimulate CD8+ T cell-mediated cytotoxicity, targeting the tumor cells in particular. Nonetheless, the molecular mechanisms involved in tumor promotion may halt their activity due to T cell exhaustion, stemness, senescence, and anergy in humans.162 circRNAs have been reported to incur immunosuppression by activating tumor escape mechanisms and altering T cell-mediated immune response. For instance, high hsa_circ_0046523 expression has been shown to induce apoptosis and CD8+ T cell exhaustion, thus suppressing their function and stimulating the release of immunoinhibitory cytokines, such as TGF-β and IL-10 (interleukin-10). In peripheral blood mononuclear cells, hsa_circ_0046523 inhibited IFN-γ (interferon gamma) and IL-2 secretion, via sponging miR-148a-3p and modulating PD-L1 (programmed death-ligand 1) expression in pancreatic cancer.163 Similarly, circHMGB2 is also reported to repress immune system through miR-181a-5p/CARM1 axis manifested by the exhaustion of CD8+ T cells and resistance to anti-PD-1 in both squamous cell carcinoma and lung adenocarcinoma.164 circRNA-002178 may result in T cell exhaustion by regulating PD1 (programmed death-1) expression through miR-34 sponging.165 circNT5C2 mitigates the immune response by sponging miR-448, thus increasing tumor growth, invasion, and metastasis.166 Besides, circLAMP1 is upregulated in T cell lymphoblastic lymphoma, whereby it stimulates cell proliferation and alleviates apoptosis through sponging miR-615-5p and mediating DDR2/RTK signaling dysregulation.167 Moreover, circ_0020397 has been shown to sponge miR-138, thus activating the immune escape mechanism by altering CD4+ T cells function in tumorigenesis.168 Yet another circRNA encoded by Epstein-Barr virus (EBV) named circBART2.2 has been revealed to mediate immune evasion by inhibiting T cell function in RIG-1 (retinoic acid-inducible gene I)-dependent manner and modulating PD-L1 in nasopharyngeal carcinoma.169 Hence, circRNAs hold functional capabilities to modulate the immune response mediated by lymphocytes in various forms of cancers.
Besides T lymphocytes, B cells are an important module in the TME, whereby they alleviate tumor progression by secreting immunoglobulins (anti-bodies) and modulate other immune cells to produce anti-tumor responses. Pertinently, circRNAs have been reported to modulate B cell-mediated immune responses in various pathological conditions, including cancer. circEAF2 overexpression has been observed to promote apoptosis in EBV-positive B cell lymphoma, thus counteracting cancer progression by regulating the miR-BART19-3p/APC/β-catenin axis in diffuse large B cell lymphoma.170 Pertinent to TILs, downregulation of circ_0064428 has been reported in high TIL patients, and correlated with poor disease prognosis in hepatocellular carcinoma (HCC), therefore potentially regulating TIL formation.171
NK cells have been known to play a critical role as first line defenders against tumor cells by mediating cytotoxic activity in response to chemokine/cytokine secretion. In this regard, specific circRNAs associated with the tumor cells have been shown to impart functions in regulating cytotoxic activity of the NK cells. In HCC, circARSP91 improves the innate immune response by inducing NK cell mediated cytotoxicity.172 Moreover, circTRIM33-12 is observed to have tumor inhibitory effects by increasing NK cell activity through NKG2D expression regulation.173 Contrarily, circ_0000977 overexpression has been reported to alleviate NK cell activity, thus resulting in immune evasion by stimulating HI1FA accumulation through miR-153 sponging in pancreatic cancer.174 Likewise, circHMGB2 and tumor-derived exosomal circUHRF1 drive cancer progression by turning off protective immune responses manifested by NK cell exhaustion and resistance to anti-PD1 therapy in NSCLC and HCC, respectively.164,175 In addition, hsa_circ_0007456 has been shown to modulate NK cell-mediated cytotoxicity through the miR-6852-3p/ICAM-1 axis in HCC.176 Therefore, circRNAs can alter the dynamics of tumor progression by regulating NK cell-mediated cytotoxicity depending on the type of cancer, precisely hinting toward the tissue/organ-specific expression and functions of these circRNAs.
circRNAs regulate immune responses mediated by antigen-presenting DCs
DCs are an important component of the innate immune system that are capable of presenting tumor-associated antigens to naive T cells, thus generating anti-tumor T cell responses.177 Overexpression of circHMGB2 has been associated with exhaustion of DCs in NSCLC, whereby it inhibits the immune response mediated by type 1 IFN. Mechanistically, circHMGB2 increases the expression levels of CARM1 (the downstream effector molecule) by acting as a miR-181a-5p sponge.164 In addition, circSNX5 acts to regulate the activity and function of DCs by sponging miR-544, thus altering the signaling dynamics through suppression of cytokine signaling 1 and impeding PU.1 translocation into the nucleus.53 Comprehensively, both circSNX5 and circHMGB2 suppress DC cell activity, and promote their functional exhaustion, thus regulating DC immunogenicity in cancer. Furthermore, mature DCs have been studied to exhibit high expression of exonic circFSCN1, whereby circFSCN1 knockdown prompts tolerogenic DCs to prevent alloimmune rejection, increase allograft survival, lessen fibrosis, and induce in vivo generation of Tregs.178
circRNAs regulate TANs and circulating neutrophils
Neutrophils (granulocytes) are the most abundant and indispensable component of TME, thus producing significant roles in cancer progression. Both TANs and circulating neutrophils can produce dual roles, i.e., tumor promoting or tumor inhibiting depending on the organ of tumor origin or cancer type. Where circulating neutrophils can enhance the metastatic spread (tumor promoting), TANs are capable to respond either as tumor promoting or inhibiting entities depending on what the TME prompts.179 A specific circRNA population has been implicated in neutrophil function. A circularized exosomal RNA circPACRGL derived from cancer cells has been observed to produce pro-tumorigenic functions and mediate differentiation of neutrophils from N1 to N2 form by modulating the miR-142-3p/miR-506-3p-TGF-β1 axis.180 Therefore, circRNAs can potentially affect the elaborative roles of neutrophils in the TME, and alter the dynamics of cancer progression.
circRNAs regulate MDSC activity
MDSCs compose a big proportion of the cells that are predominantly known to modulate immune response negatively. In tumorigenic conditions, MDSCs are aberrantly activated in the TME, whereby they release immunosuppressive cytokines (iNOS, ROS, and ARG1) that function to repress T cell-mediated cytotoxicity.181 As reported, circSLC8A1 interacts with miR-494 in MDCSs, thus suppressing the release of immunosuppressive cytokines in a PTEN-dependent manner.33 Comparably, MDSC-derived exosomal S100A9 increased circMID1 expression, which functions through miR-506-3p sponging to promote tumorigenesis.182 Moreover, circDLG1 increases MDSC infiltration, thus promoting gastric cancer.183 Therefore, circRNAs hold the potential to control MDSC function and regulate subsequent immune reaction during tumorigenesis.
circRNAs regulate tumor-associated macrophages
Macrophages are the effector cells that are crucial in abridging innate and acquired immune responses through specialized pattern-recognition receptors, i.e., TLRs. These TLRs are capable of recognizing antigens associated with the tumor cells through increased chemokine and cytokine secretion, thus advancing subsequent immune reaction and antigen presentation.184 circRNAs have been reported to perform explicit regulatory functions in modulating such immune responses mediated by macrophages. Relevantly, circHECTD1 is widely expressed in macrophages, and has been described to enhance cell proliferation and migration by modulating miR-320-5p/SLC2A1, miR-1256/USP5, and miR-485-5p/mucin-1 in glioblastoma multiform,185 gastric cancer,186 and HCC, respectively.187
In addition, macrophages are capable of differentiating and polarizing to distinct phenotypes, i.e., M1 and M2, whereby the M1 phenotype is activated primarily by IFN-γ and LPS, and the M2 phenotype is either triggered by the immune complex or some specialized ILs. Intriguingly, both these types of macrophages have been shown to exhibit distinct circRNA expression profiles.188 Henceforth, circRNAs may play critical roles in macrophage polarization and conversion into either of these two forms. In this regard, circHECTD1 has been revealed to have an important role in regulating macrophage polarization.189 Likewise, exosomal circ_0001142, released under endoplasmic reticulum stress has been observed to promote macrophage polarization into M2 type, resulting in breast cancer progression.190
circRNAs regulate the activity of CAFs and inflammatory CAFs
CAFs are one of the components of stromal remodeling and play crucial roles in initiation, progression, metastasis, and therapeutic resistance in most solid tumors.191 CAFs are derived from normal fibroblasts (NFs). As depicted in Figure 2, CAFs exhibit two phenotypes; CAFs and inflammatory CAFs (iCAFs). Evidence suggests that CAFs are driven by TGF-β, while iCAFs are induced by NF-κB signaling in pancreatic cancer.192 CAFs exhibit a matrix-producing contractile phenotype, i.e., myoCAFs relating to myofibroblasts, and iCAFs with immunomodulating secretome-regulating inflammation.193 In HCC, CAFs secrete higher levels of CCL2 and CCL5 than peri-tumor fibroblasts that increase migration through activation of the hedgehog pathway, and higher levels of CCL7 and CXCL16, thus promoting both migration and invasion of HCC cells through the TGF-β pathway.159 Based on the correlation analysis using public datasets containing over 3,000 BC samples, Chen et al. identified a role for iCAFs in tumor progression, which is significantly related to poor prognosis. Single-cell RNA sequencing from 19 different cell types in the bladder carcinoma (BC) microenvironment also indicated iCAFs to be the key factors in BC progression.194 Multiple mechanisms targeting NFs, such as contact signals, extracellular matrix, DNA damage, TGF-β, physiological stress, inflammatory signals, and RTK ligands, activate CAFs in TME.193 CAFs promoted aggressive phenotypes in breast cancer cells through EMT induced by TGF-β1 activation.195 Various inflammatory modulators promote CAF activation through different signaling pathways in a context-dependent manner.193 Interactions between cancer cells and fibroblasts can promote the CAF phenotype in ductal breast carcinoma in situ through the Jagged1/Notch2 signaling pathway.196 Silencing or downregulation of Notch effector CSL in dermal fibroblasts is sufficient for CAF activation and induces senescence of primary fibroblasts from the dermis, oral mucosa, breast, and lung, but not in squamous cell carcinomas.197 IL-1α-induced ELF3/YAP pathway activation also regulates iCAFs differentiation.198 iCAFs represent a major prognosis determining cell type in rectal cancer. Targeted repolarization of CAFs can improve therapeutic response and long-term survival for rectal cancer patients.21 The conversion from NFs to CAFs is mainly mediated by TGF-β and STAT signaling pathways. The crosstalk of CAFs with cancer cells is associated with multiple signaling pathways. CAF-mediated signaling pathways, such as STAT, Wnt/β-catenin, Hippo/YAP, MAPK, EGFR, and NF-κB, are activated by cytokines, chemokines, and growth factors, and are widely involved in cancer cell proliferation, stemness, invasion, migration, metastasis, angiogenesis, EMT process, and therapeutic resistance.24 circRNAs from CAF-derived exosomes contribute to cancer progression.199 Accumulating evidence suggests that abundant miRNAs in CAFs have regulatory roles in tumor progression.24 For example, circHIF1A from CAF exosomes plays an important role in breast cancer by modulating the miR-580-5p/CD44 axis.200 Akt, β-catenin, and MAPK signaling pathways are mediated by CD44.201 As Figure 2 summarizes, CAFs deliver exosomal circ_0088300 to GC tumor cells, and promote proliferation, migration, and invasion of these cells through the STAT signaling pathway by sponging miR-1305.56 Similarly, circCUL2 is specifically expressed in CAFs but not in cancer cells, whereby its upregulation in fibroblasts mediated the conversion of NFs into iCAFs and contributed to CAF heterogeneity in pancreatic ductal adenocarcinoma by inducing the NF-κB signaling pathway. Hence, circCUL2 is critical for inducing and maintaining CAF pro-tumor properties.139 Furthermore, the interaction between cancer cells and CAFs in TME promotes proliferation, migration, and metastasis of cancer cells by releasing cytokines and activating related signaling pathways.
Role of circRNAs in cancer-induced inflammation: Inflammasomes and cancer development
Extensive literature on TME informs that tumor progression is largely driven by the interactive networking between the tumor and the stromal cells. Similarly, the capacity of cancer cells to secrete soluble mediators and the immune response is of utmost importance in mediating tumor progression, whereby cancer cells have the ability to modulate both innate and adaptive immune responses. Pertinently, tumor cells can turn on T cell-mediated responses by modulating Tregs, and can induce macrophage, neutrophil, and MDSC activities as well. Importantly, cancer cells are capable of reprogramming the immune cells into an immuno-inhibitory phenotype through soluble mediators, thus facilitating an anti-tumor immune response.202
Cancer-elicited inflammation is triggered by a variety of immune cells, including macrophages, neutrophils, DCs, NK cells, T lymphocytes, and B lymphocytes.8 Inflammasome formation (a multimeric protein complex) is the pivotal mechanism that drives inflammation in immune cells by activating cysteine protease caspase-1, which subsequently triggers pyroptosis (inflammatory cell death) through the release of inflammatory cytokines.203 In recent years, circRNAs have been thought to play roles in tumor-mediated regulation of the immune system. NLRP3-inflammasome activation by circRNAs, such as circHIPK3 and circBBS9, is related to inflammatory diseases in IME. Pertinently, circBBS9 promotes activation of NLRP3 inflammasome to release pro-inflammatory cytokines into the IME. In contrast, circHIPK3 inhibits activation of NLRP3 inflammasomes (Figure 2). AIM2 (absent in melanoma 2)-inflammasome activation by circRNAs is also associated with cancer development. For example, circNEIL3 activates AIM2 inflammasomes and promotes pyroptosis of lung adenocarcinoma (Figure 2). circRNAs modulate the inflammasome activation in dual ways. circBBS9 promotes NLRP3 inflammasome activation in PM2.5-induced lung inflammation mice.48 circNEIL3 triggers AIM2 inflammasome activation through sponging miR-1184 to induce pyroptosis in lung adenocarcinoma.49 It is known that circHIPK3 can improve blood perfusion in ischemic injury of the skeletal muscles by preventing activation of the NLRP3 inflammasome.47 This is achieved by targeting FOXO3a expression through sponging miR-421, resulting in NF-κB pathway inhibition, and the subsequent release of IL-1β and IL-18 in ischemic injury.
Therapeutic implications of the circRNAs associated with inflammation-related cancers
Attributable to the association between inflammation and tumor progression, exploiting inflammation may plausibly serve as an effective and useful avenue for therapeutic management of different types of cancers. Comparably, IME is also a major attribute that governs the efficacy of various conventional anti-cancer therapies, including chemotherapy, immunotherapy, and radiotherapy. Contextual to this, altered expression profiles of different circRNAs have been related with clinico-pathological parameters that are detectable in various biological specimens, such as biopsies and/or body fluids, and can be exploited to modulate immune response in different cancer types. In this regard, tertiary lymphoid structures have been reported to enhance the efficacy of immunotherapeutic agents in melanoma,204 bladder cancer,205 and gastric cancer.206 However, the exact mechanism might not be fully elucidated thus far; the B cell-driven signaling pathways may sustain better outcomes, explicitly when harnessed in conjunction with the use of circRNAs modulating immune responses in various forms of cancers. For instance, circ0000190 may serve as a potential marker in regard to immunotherapy in lung cancer patients.207 Also, circ_0064428 has been reported to be correlated with poor disease prognosis in high TIL patients diagnosed with HCC.171 Thus, inhibition of the circRNAs that suppress immune system may help in strengthening immune response against different cancers, such as circ-NT5C2, circ_0000977, circLAMP-1, and various others, as depicted in Figure 3.
Besides, circRNAs may be used as potent vaccine adjuvants, thus helping to boost both innate and adaptive immune systems. Transfecting circRNAs in distinct cancer cell lines has been reported to drive the overexpression of specific cytokine-related genes and NF-kB signaling, thus activating an innate immune response.208 Moreover, exogenous circRNAs lacking m6A modification can enhance the expression of genes related with the immune system by stimulating RIG-1.209 Furthermore, DCs may be a promising therapeutic target owing to their ability to serve as a foundation for propagating T cell-mediated anti-tumor responses. In this regard, DCs isolated from mice showed that circFOREIGN could directly stimulate DCs and indirectly mediate CD4+ and CD8+ T cell activities by allowing cross-presentation of the antigen.209 Moreover, melanoma mice vaccinated with circFOREIGN manifested significant increase in overall survival compared with those who received no circFOREIGN injection.209 Tumor-specific DC subsets may exhibit distinct circRNA profiles. Accordingly, exploiting and targeting these DCs through the use of associated circRNAs in an attempt to instigate and promote anti-tumor immune responses can serve as a novel approach for therapeutic management of various types of cancers driven by an inflammatory insult. Therefore, circRNAs can potentially activate immune cells to combat tumors either by functioning as a tumor antigen or being regulated to prompt favorable immune reaction. Nevertheless, if circRNAs can be used in combating anti-cancer therapy-induced inflammation is a niche that still requires rigorous investigation.
Conclusion and perspective
Having considered a firmly established relationship between inflammation and cancer, the functional mechanisms as to how circRNAs can regulate the interactive networking between the inflammatory processes and tumor progression remain to be comprehensively elucidated. As presented in this review, more than 90% of the studies recruited have spotlighted miRNA sponging as a mechanism of circRNA function in inflammation and cancer, but much fewer discuss circRNA binding with cRBPs and their translatability into respective peptides as a potential mechanism of circRNA function in either inflammation-induced cancers (extrinsic inflammation) or cancer-related inflammation (intrinsic inflammation). Molecular mechanisms pertinent to the immune cells and the IME that abridge the crosstalk with the tumor cells have been well studied, with STAT3 and NF-κB pathways bridging the two pathological states, i.e., inflammation and cancer. Depending on the tissue-specific distinct expression profiles, as well as tumor-promoting and anti-tumor facets of circRNAs modulating the inflammatory cells functions and the integrated signaling pathways, circRNAs can be exploited to develop novel therapeutic strategies for treating inflammation-induced cancers or cancer-induced inflammation. Knowing that not every organ’s inflammation culminates into tumorigenesis, it would be highly intriguing to investigate if circRNAs have any role in enduring inflammation into tumorigenesis, owing to their tissue-specific expression. Harnessing inflammation through the use of circRNAs can potentially alter the dynamics of tumor development and may prove valuable in devising novel therapeutic options for treating different cancers, together with improving the efficacy of conventional anti-cancerous therapeutic approaches. Nonetheless, it is pivotal to understand the molecular events and signaling cascades behind these bilateral disease mechanisms, i.e., inflammation and tumorigenesis, as well as the functional intricacies of circRNAs, to pave a way for acquiring adequate translational and therapeutic outcomes through the use of circRNA-based anti-cancer therapy for clinical management.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (PJT-155962, and PJT-166107) to B.B.Y.
Author contributions
J.Q. and S.W. designed the structure of the review. J.Q. and S.W. reviewed the literature and collected all essential information. J.Q., S.W., H.Y., and B.B.Y. wrote the paper.
Declaration of interests
There authors declare no competing interests.
References
- 1.Munn L.L. Cancer and inflammation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017;9 doi: 10.1002/wsbm.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Multhoff G., Molls M., Radons J. Chronic inflammation in cancer development. Front. Immunol. 2011;2:98. doi: 10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afify S.M., Hassan G., Seno A., Seno M. Cancer-inducing niche: the force of chronic inflammation. Br. J. Cancer. 2022;127:193–201. doi: 10.1038/s41416-022-01775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGranahan N., Swanton C. Cancer evolution constrained by the immune microenvironment. Cell. 2017;170:825–827. doi: 10.1016/j.cell.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Dong L.Q., Peng L.H., Ma L.J., Liu D.B., Zhang S., Luo S.Z., Rao J.H., Zhu H.W., Yang S.X., Xi S.J., et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J. Hepatol. 2020;72:896–908. doi: 10.1016/j.jhep.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Greten F.R., Grivennikov S.I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piotrowski I., Kulcenty K., Suchorska W. Interplay between inflammation and cancer. Rep. Pract. Oncol. Radiother. 2020;25:422–427. doi: 10.1016/j.rpor.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H., Wu L., Yan G., Chen Y., Zhou M., Wu Y., Li Y. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal. Transduct. Target. Ther. 2021;6:263. doi: 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Hibino S., Kawazoe T., Kasahara H., Itoh S., Ishimoto T., Sakata-Yanagimoto M., Taniguchi K. Inflammation-induced tumorigenesis and metastasis. Int. J. Mol. Sci. 2021;22:5421. doi: 10.3390/ijms22115421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma S., Long T., Huang W.J.M. Noncoding RNAs in inflammation and colorectal cancer. RNA Biol. 2020;17:1628–1635. doi: 10.1080/15476286.2019.1705610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama Y., Fujiu K., Yuki R., Oishi Y., Morioka M.S., Isagawa T., Matsuda J., Oshima T., Matsubara T., Sugita J., et al. A long noncoding RNA regulates inflammation resolution by mouse macrophages through fatty acid oxidation activation. Proc. Natl. Acad. Sci. USA. 2020;117:14365–14375. doi: 10.1073/pnas.2005924117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao X., Cao Y., Guo Z., Wang L., Xiang C. Biological roles and therapeutic potential of circular RNAs in osteoarthritis. Mol. Ther. Nucleic Acids. 2021;24:856–867. doi: 10.1016/j.omtn.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian C., Sun J., Guan W., Zhang L., Zhang X., Yang L., Hu W. Circular RNA circHIPK3 activates macrophage NLRP3 inflammasome and TLR4 pathway in gouty arthritis via sponging miR-561 and miR-192. Inflammation. 2021;44:2065–2077. doi: 10.1007/s10753-021-01483-2. [DOI] [PubMed] [Google Scholar]
- 15.Xu H., Fang M., Li C., Zuo B., Ren J., Zhang Y. BORIS-mediated generation of circular RNAs induces inflammation. Transl. Oncol. 2022;18:101363. doi: 10.1016/j.tranon.2022.101363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qadir J., Li F., Yang B.B. Circular RNAs modulate Hippo-YAP signaling: functional mechanisms in cancer. Theranostics. 2022;12:4269–4287. doi: 10.7150/thno.71708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, F., Lyu, J., Yang, Y., Yang, Q., Santos, C., and Yang, B.B. (2022). An improved model or circular RNA overexpression: using the actin intron reveals high circularization efficiency. Adv. Genet. 2200019. 10.1002/ggn2.202200019 [DOI] [PMC free article] [PubMed]
- 18.Wu N., Qadir J., Yang B.B. CircRNA perspective: new strategies for RNA therapy. Trends Mol. Med. 2022;28:343–344. doi: 10.1016/j.molmed.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Pisani P., Parkin D.M., Muñoz N., Ferlay J. Cancer and infection: estimates of the attributable fraction in 1990. Cancer Epidemiol. Biomarkers Prev. 1997;6:387–400. [PubMed] [Google Scholar]
- 20.Stewart B.W., Kleihues P., editors. World Cancer Report. IARCPress; 2003. [Google Scholar]
- 21.Nicolas A.M., Pesic M., Engel E., Ziegler P.K., Diefenhardt M., Kennel K.B., Buettner F., Conche C., Petrocelli V., Elwakeel E., et al. Inflammatory fibroblasts mediate resistance to neoadjuvant therapy in rectal cancer. Cancer Cell. 2022;40:168–184.e13. doi: 10.1016/j.ccell.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Newton K., Dixit V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012;4:a006049. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F., Wu T., Xu Y., Dong Q., Xiao J., Xu Y., Li Q., Zhang C., Gao J., Liu L., et al. A comprehensive overview of oncogenic pathways in human cancer. Brief. Bioinform. 2020;21:957–969. doi: 10.1093/bib/bbz046. [DOI] [PubMed] [Google Scholar]
- 24.Wu F., Yang J., Liu J., Wang Y., Mu J., Zeng Q., et al. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal. Transduction Targeted Ther. 2021;6:218. doi: 10.1038/s41392-021-00641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 26.Garlapati P., Ling J., Chiao P.J., Fu J. Circular RNAs regulate cancer-related signaling pathways and serve as potential diagnostic biomarkers for human cancers. Cancer Cell Int. 2021;21:317. doi: 10.1186/s12935-021-02017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao L., Ma X.X., Luo J., Chung H.K., Kwon M.S., Yu T.X., Rao J.N., Kozar R., Gorospe M., Wang J.Y. Circular RNA CircHIPK3 promotes homeostasis of the intestinal epithelium by reducing MicroRNA 29b function. Gastroenterology. 2021;161:1303–1317.e3. doi: 10.1053/j.gastro.2021.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Y.F., Zhao L., Wang B.C., Sun J.J., Hu J.L., Zhu X.L., Zhao J., Zheng D.K., Ge Z.W. The circular RNA TLK1 exacerbates myocardial ischemia/reperfusion injury via targeting miR-214/RIPK1 through TNF signaling pathway. Free Radic. Biol. Med. 2020;155:69–80. doi: 10.1016/j.freeradbiomed.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Shi X., Yang J., Liu M., Zhang Y., Zhou Z., Luo W., Fung K.M., Xu C., Bronze M.S., Houchen C.W., Li M. Circular RNA ANAPC7 inhibits tumor growth and muscle wasting via PHLPP2-AKT-TGF-beta signaling Axis in pancreatic cancer. Gastroenterology. 2022;162:2004–2017.e2. doi: 10.1053/j.gastro.2022.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhen N., Gu S., Ma J., Zhu J., Yin M., Xu M., Wang J., Huang N., Cui Z., Bian Z., et al. CircHMGCS1 promotes hepatoblastoma cell proliferation by regulating the IGF signaling pathway and glutaminolysis. Theranostics. 2019;9:900–919. doi: 10.7150/thno.29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua J., Wang X., Ma L., Li J., Cao G., Zhang S., Lin W. CircVAPA promotes small cell lung cancer progression by modulating the miR-377-3p and miR-494-3p/IGF1R/AKT axis. Mol. Cancer. 2022;21:123. doi: 10.1186/s12943-022-01595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao B., Zhu S., Wei X., Chen M.K., Feng Y., Li Z., Xu X., Zhang Y., Wang Y., Zhou J., et al. The circSPON2/miR-331-3p axis regulates PRMT5, an epigenetic regulator of CAMK2N1 transcription and prostate cancer progression. Mol. Cancer. 2022;21:119. doi: 10.1186/s12943-022-01598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Q., Liu T., Feng H., Yang R., Zhao X., Chen W., Jiang B., Qin H., Guo X., Liu M., et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang B., Li W., Ji T.T., Li X.Y., Qu X., Feng L., Bai S. Circ-AKT3 inhibits the accumulation of extracellular matrix of mesangial cells in diabetic nephropathy via modulating miR-296-3p/E-cadherin signals. J. Cell. Mol. Med. 2020;24:8779–8788. doi: 10.1111/jcmm.15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue D., Wang H., Chen Y., Shen D., Lu J., Wang M., Zebibula A., Xu L., Wu H., Li G., Xia L. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol. Cancer. 2019;18:151. doi: 10.1186/s12943-019-1072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He J., Chu Z., Lai W., Lan Q., Zeng Y., Lu D., Jin S., Xu H., Su P., Yin D., et al. Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. J. Hematol. Oncol. 2021;14:194. doi: 10.1186/s13045-021-01210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J., et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 38.Feng D., Wang Z., Zhao Y., Li Y., Liu D., Chen Z., Ning S., Hu Y., Yao J., Tian X. circ-PRKCB acts as a ceRNA to regulate p66Shc-mediated oxidative stress in intestinal ischemia/reperfusion. Theranostics. 2020;10:10680–10696. doi: 10.7150/thno.44250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge R., Gao G. Anti-antioxidant impacts of circZNF609 silence in HaCaT cells through regulating miR-145. Artif. Cells Nanomed. Biotechnol. 2020;48:384–392. doi: 10.1080/21691401.2019.1709863. [DOI] [PubMed] [Google Scholar]
- 41.Qiu Y., Yu Y., Qin X.M., Jiang T., Tan Y.F., Ouyang W.X., Xiao Z.H., Li S.J. CircTLK1 modulates sepsis-induced cardiomyocyte apoptosis via enhancing PARP1/HMGB1 axis-mediated mitochondrial DNA damage by sponging miR-17-5p. J. Cell. Mol. Med. 2021;25:8244–8260. doi: 10.1111/jcmm.16738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Z.Q., Zhou S.L., Li J., Zhou Z.J., Wang P.C., Xin H.Y., Mao L., Luo C.B., Yu S.Y., Huang X.W., et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2020;72:906–922. doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 43.He J., Huang Z., He M., Liao J., Zhang Q., Wang S., Xie L., Ouyang L., Koeffler H.P., Yin D., Liu A. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer. 2020;19:17. doi: 10.1186/s12943-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishola A.A., Chien C.S., Yang Y.P., Chien Y., Yarmishyn A.A., Tsai P.H., Chen J.C.Y., Hsu P.K., Luo Y.H., Chen Y.M., et al. Oncogenic circRNA C190 promotes non-small cell lung cancer via modulation of the EGFR/ERK pathway. Cancer Res. 2022;82:75–89. doi: 10.1158/0008-5472.CAN-21-1473. [DOI] [PubMed] [Google Scholar]
- 45.Wei Y., Lu C., Zhou P., Zhao L., Lyu X., Yin J., Shi Z., You Y. EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro. Oncol. 2021;23:611–624. doi: 10.1093/neuonc/noaa214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H., Bi J., Dong W., Yang M., Shi J., Jiang N., Lin T., Huang J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan B., Zhang Y., Liang C., Liu B., Ding F., Wang Y., Zhu B., Zhao R., Yu X.Y., Li Y. Stem cell-derived exosomes prevent pyroptosis and repair ischemic muscle injury through a novel exosome/circHIPK3/FOXO3a pathway. Theranostics. 2020;10:6728–6742. doi: 10.7150/thno.42259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Penco-Campillo M., Comoglio Y., Feliz Morel Á.J., Hanna R., Durivault J., Leloire M., Mejias B., Pagnuzzi M., Morot A., Burel-Vandenbos F., et al. Author Correction: VEGFC negatively regulates the growth and aggressiveness of medulloblastoma cells. Commun. Biol. 2020;3:758. doi: 10.1038/s42003-020-01502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T., Wu D.M., Luo P.W., Liu T., Han R., Deng S.H., He M., Zhao Y.Y., Xu Y. CircNEIL3 mediates pyroptosis to influence lung adenocarcinoma radiotherapy by upregulating PIF1 through miR-1184 inhibition. Cell Death Dis. 2022;13:167. doi: 10.1038/s41419-022-04561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo P.W., Huang H.T., Ma J., Zuo Y., Huang D., He L.L., Wan Z.M., Chen C., Yang F.F., You Y.W. Circular RNA-0007059 protects cell viability and reduces inflammation in a nephritis cell model by inhibiting microRNA-1278/SHP-1/STAT3 signaling. Mol. Med. 2021;27:113. doi: 10.1186/s10020-021-00372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Xu C., Cai Y., Zou X., Chao Y., Yan Z., Zou C., Wu X., Tang L. CircZNF652 promotes the goblet cell metaplasia by targeting the miR-452-5p/JAK2 signaling pathway in allergic airway epithelia. J. Allergy Clin. Immunol. 2022;150:192–203. doi: 10.1016/j.jaci.2021.10.041. [DOI] [PubMed] [Google Scholar]
- 52.Yang L., Zhang C., Bai X., Xiao C., Dang E., Wang G. hsa_circ_0003738 inhibits the suppressive function of Tregs by targeting miR-562/IL-17a and miR-490-5p/IFN-γ signaling pathway. Mol. Ther. Nucleic Acids. 2020;21:1111–1119. doi: 10.1016/j.omtn.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Q., Mang G., Wu J., Sun P., Li T., Zhang H., Wang N., Tong Z., Wang W., Zheng Y., et al. Circular RNA circSnx5 controls immunogenicity of dendritic cells through the miR-544/SOCS1 Axis and PU.1 activity regulation. Mol. Ther. 2020;28:2503–2518. doi: 10.1016/j.ymthe.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X., Mao R., Su W., Yang X., Geng Q., Guo C., Wang Z., Wang J., Kresty L.A., Beer D.G., et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–671. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J., Jiang X., Zou A., Mai Z., Huang Z., Sun L., Zhao J. circIGHG-induced epithelial-to-mesenchymal transition promotes oral squamous cell carcinoma progression via miR-142-5p/IGF2BP3 signaling. Cancer Res. 2021;81:344–355. doi: 10.1158/0008-5472.CAN-20-0554. [DOI] [PubMed] [Google Scholar]
- 56.Shi H., Huang S., Qin M., Xue X., Guo X., Jiang L., Hong H., Fang J., Gao L. Exosomal circ_0088300 derived from cancer-associated fibroblasts acts as a miR-1305 sponge and promotes gastric carcinoma cell tumorigenesis. Front. Cell Dev. Biol. 2021;9:676319. doi: 10.3389/fcell.2021.676319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng L., Sang H., Wei S., Li Y., Jin D., Zhu X., Li X., Dang Y., Zhang G. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol. Cancer. 2020;19:156. doi: 10.1186/s12943-020-01270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Zhu M., Song J., Zeng Y., Xia S., Chen C., Jin M., Song Y. The circular RNA circTXNRD1 promoted ambient particulate matter-induced inflammation in human bronchial epithelial cells by regulating miR-892a/COX-2 axis. Chemosphere. 2022;286:131614. doi: 10.1016/j.chemosphere.2021.131614. [DOI] [PubMed] [Google Scholar]
- 59.Lim T.B., Aliwarga E., Luu T.D.A., Li Y.P., Ng S.L., Annadoray L., Sian S., Ackers-Johnson M.A., Foo R.S.Y. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019;115:1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 60.Chen W., Wang H., Feng J., Chen L. Overexpression of circRNA circUCK2 attenuates cell apoptosis in cerebral ischemia-reperfusion injury via miR-125b-5p/GDF11 signaling. Mol. Ther. Nucleic Acids. 2020;22:673–683. doi: 10.1016/j.omtn.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Shen P., Yang Y., Liu G., Chen W., Chen J., Wang Q., Gao H., Fan S., Shen S., Zhao X. CircCDK14 protects against Osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics. 2020;10:9113–9131. doi: 10.7150/thno.45993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hao Y., Lu C., Zhang B., Xu Z., Guo H., Zhang G. CircPVT1 up-regulation attenuates steroid-induced osteonecrosis of the femoral head through regulating miR-21-5p-mediated Smad7/TGFbeta signalling pathway. J. Cell. Mol. Med. 2021;25:4608–4622. doi: 10.1111/jcmm.16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng K., He B., Yang B.B., Xu T., Chen X., Xu M., Liu X., Sun H., Pan Y., Wang S. The pro-metastasis effect of circANKS1B in breast cancer. Mol. Cancer. 2018;17:160. doi: 10.1186/s12943-018-0914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu J., Luo Y., Zhao Y., Kong Y., Zheng H., Li Y., Gao B., Ai L., Huang H., Huang J., et al. circEHBP1 promotes lymphangiogenesis and lymphatic metastasis of bladder cancer via miR-130a-3p/TGFβR1/VEGF-D signaling. Mol. Ther. 2021;29:1838–1852. doi: 10.1016/j.ymthe.2021.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shang J., Li H., Wu B., Jiang N., Wang B., Wang D., Zhong J., Chen Y., Xu X., Lu H. CircHIPK3 prevents chondrocyte apoptosis and cartilage degradation by sponging miR-30a-3p and promoting PON2. Cell Prolif. 2022;55:e13285. doi: 10.1111/cpr.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou D., Huang Z., Zhu X., Hong T., Zhao Y. Circular RNA 0025984 ameliorates ischemic stroke injury and protects astrocytes through miR-143-3p/TET1/ORP150 pathway. Mol. Neurobiol. 2021;58:5937–5953. doi: 10.1007/s12035-021-02486-8. [DOI] [PubMed] [Google Scholar]
- 67.Zang X., Jiang J., Gu J., Chen Y., Wang M., Zhang Y., Fu M., Shi H., Cai H., Qian H., et al. Circular RNA EIF4G3 suppresses gastric cancer progression through inhibition of β-catenin by promoting δ-catenin ubiquitin degradation and upregulating SIK1. Mol. Cancer. 2022;21:141. doi: 10.1186/s12943-022-01606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng L., Zhang Y., Wu P., Li D., Lu Y., Shen P., Yang T., Shi G., Chen Q., Yuan H., et al. CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2. Mol. Cancer. 2022;21:121. doi: 10.1186/s12943-022-01599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Q., Wang H., Li Z., Li F., Liang L., Zou Y., Shen H., Li J., Xia Y., Cheng Z., et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatol. 2022;76:135–147. doi: 10.1016/j.jhep.2021.08.027. [DOI] [PubMed] [Google Scholar]
- 71.Verduci L., Ferraiuolo M., Sacconi A., Ganci F., Vitale J., Colombo T., Paci P., Strano S., Macino G., Rajewsky N., Blandino G. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18:237. doi: 10.1186/s13059-017-1368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang M.P., Xu W.X., Hou J.C., Xu Q., Wang D.D., Tang J.H. The emerging role of the interactions between circular RNAs and RNA-binding proteins in common human cancers. J. Cancer. 2021;12:5206–5219. doi: 10.7150/jca.58182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ling Y., Liang G., Lin Q., Fang X., Luo Q., Cen Y., Mehrpour M., Hamai A., Liu Z., Shi Y., et al. circCDYL2 promotes trastuzumab resistance via sustaining HER2 downstream signaling in breast cancer. Mol. Cancer. 2022;21:8. doi: 10.1186/s12943-021-01476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang Y., Zhao J., Xu J., Zhang H., Zhou J., Li H., Zhang G., Xu K., Jing Z. Glioblastoma-associated microglia-derived exosomal circKIF18A promotes angiogenesis by targeting FOXC2. Oncogene. 2022;41:3461–3473. doi: 10.1038/s41388-022-02360-4. [DOI] [PubMed] [Google Scholar]
- 75.Jiang X., Guo S., Wang S., Zhang Y., Chen H., Wang Y., Liu R., Niu Y., Xu Y. EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 2022;82:831–845. doi: 10.1158/0008-5472.CAN-21-2988. [DOI] [PubMed] [Google Scholar]
- 76.Natarajan P., Murugesan A.K., Govindan G., Gopalakrishnan A., Kumar R., Duraisamy P., Balaji R., Tanuja, Shyamli P.S., Parida A.K., Parani M. A reference-grade genome identifies salt-tolerance genes from the salt-secreting mangrove species Avicennia marina. Commun. Biol. 2021;4:851. doi: 10.1038/s42003-021-02384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J., Wu Y., Luo X., Jin D., Zhou W., Ju Z., Wang D., Meng Q., Wang H., Fu X., et al. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics. 2021;11:7507–7526. doi: 10.7150/thno.59546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen L., Lei S., Zhang B., Li S., Huang L., Czachor A., Breitzig M., Gao Y., Huang M., Mo X., et al. Skipping of exon 10 in Axl pre-mRNA regulated by PTBP1 mediates invasion and metastasis process of liver cancer cells. Theranostics. 2020;10:5719–5735. doi: 10.7150/thno.42010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu T., Cen Y., Chen Z., Zhang Y., Zhao L., Wang J., Lu W., Xie X., Wang X. Oncogenic circTICRR suppresses autophagy via binding to HuR protein and stabilizing GLUD1 mRNA in cervical cancer. Cell Death Dis. 2022;13:479. doi: 10.1038/s41419-022-04943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He Z., Cai K., Zeng Z., Lei S., Cao W., Li X. Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis. 2022;13:233. doi: 10.1038/s41419-022-04677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X., Chen M., Fang L. hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3. Mol. Ther. Nucleic Acids. 2021;26:122–134. doi: 10.1016/j.omtn.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu Z., Chen S., Liu R., Chen H., Xu B., Xu W., Chen M. Circular RNA circPOLR2A promotes clear cell renal cell carcinoma progression by facilitating the UBE3C-induced ubiquitination of PEBP1 and, thereby, activating the ERK signaling pathway. Mol. Cancer. 2022;21:146. doi: 10.1186/s12943-022-01607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du J., Lan T., Liao H., Feng X., Chen X., Liao W., Hou G., Xu L., Feng Q., Xie K., et al. CircNFIB inhibits tumor growth and metastasis through suppressing MEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol. Cancer. 2022;21:18. doi: 10.1186/s12943-021-01482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang W., Chen S., Du Q., Bian P., Chen Y., Liu Z., Zheng J., Sai K., Mou Y., Chen Z., et al. CircVPS13C promotes pituitary adenoma growth by decreasing the stability of IFITM1 mRNA via interacting with RRBP1. Oncogene. 2022;41:1550–1562. doi: 10.1038/s41388-022-02186-0. [DOI] [PubMed] [Google Scholar]
- 85.Ding L., Wang R., Zheng Q., Shen D., Wang H., Lu Z., Luo W., Xie H., Ren L., Jiang M., et al. circPDE5A regulates prostate cancer metastasis via controlling WTAP-dependent N6-methyladenisine methylation of EIF3C mRNA. J. Exp. Clin. Cancer Res. 2022;41:187. doi: 10.1186/s13046-022-02391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Q., Liu J., Deng H., Ma R., Liao J.Y., Liang H., Hu J., Li J., Guo Z., Cai J., et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183:76–93.e22. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 87.Yang L., Han B., Zhang Z., Wang S., Bai Y., Zhang Y., Tang Y., Du L., Xu L., Wu F., et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 88.Jiang S., Li C., McRae G., Lykken E., Sevilla J., Liu S.Q., Wan Y., Li Q.J. MeCP2 reinforces STAT3 signaling and the generation of effector CD4+ T cells by promoting miR-124-mediated suppression of SOCS5. Sci. Signal. 2014;7:ra25. doi: 10.1126/scisignal.2004824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wan D., Wang S., Xu Z., Zan X., Liu F., Han Y., Jiang M., Wu A., Zhi Q. PRKAR2A-derived circular RNAs promote the malignant transformation of colitis and distinguish patients with colitis-associated colorectal cancer. Clin. Transl. Med. 2022;12:e683. doi: 10.1002/ctm2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun S., Gao J., Zhou S., Li Y., Wang Y., Jin L., Li J., Liu B., Zhang B., Han S., et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 2020;39:252. doi: 10.1186/s13046-020-01779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H., et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 93.Sun Y.M., Wang W.T., Zeng Z.C., Chen T.Q., Han C., Pan Q., Huang W., Fang K., Sun L.Y., Zhou Y.F., et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134:1533–1546. doi: 10.1182/blood.2019000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanniford D., Ulloa-Morales A., Karz A., Berzoti-Coelho M.G., Moubarak R.S., Sánchez-Sendra B., Kloetgen A., Davalos V., Imig J., Wu P., et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70.e15. doi: 10.1016/j.ccell.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen C.L., Tsukamoto H., Liu J.C., Kashiwabara C., Feldman D., Sher L., Dooley S., French S.W., Mishra L., Petrovic L., et al. Reciprocal regulation by TLR4 and TGF-beta in tumor-initiating stem-like cells. J. Clin. Invest. 2013;123:2832–2849. doi: 10.1172/JCI65859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Zheng L., Liang H., Zhang Q., Shen Z., Sun Y., Zhao X., Gong J., Hou Z., Jiang K., Wang Q., et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol. Cancer. 2022;21:41. doi: 10.1186/s12943-022-01495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qiu S., Li B., Xia Y., Xuan Z., Li Z., Xie L., Gu C., Lv J., Lu C., Jiang T., et al. CircTHBS1 drives gastric cancer progression by increasing INHBA mRNA expression and stability in a ceRNA- and RBP-dependent manner. Cell Death Dis. 2022;13:266. doi: 10.1038/s41419-022-04720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu H., Lan T., Li H., Xu L., Chen X., Liao H., Chen X., Du J., Cai Y., Wang J., et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. 2021;11:1396–1411. doi: 10.7150/thno.53227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C., et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen G., Long C., Wang S., Wang Z., Chen X., Tang W., He X., Bao Z., Tan B., Zhao J., et al. Circular RNA circStag1 promotes bone regeneration by interacting with HuR. Bone Res. 2022;10:32. doi: 10.1038/s41413-022-00208-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Heumüller A.W., Jones A.N., Mourão A., Klangwart M., Shi C., Wittig I., Fischer A., Muhly-Reinholz M., Buchmann G.K., Dieterich C., et al. Locus-conserved circular RNA cZNF292 controls endothelial cell flow responses. Circ. Res. 2022;130:67–79. doi: 10.1161/CIRCRESAHA.121.320029. [DOI] [PubMed] [Google Scholar]
- 102.Hu X., Wu D., He X., Zhao H., He Z., Lin J., Wang K., Wang W., Pan Z., Lin H., Wang M. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol. Cancer. 2019;18:160. doi: 10.1186/s12943-019-1095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang F., Hu A., Li D., Wang J., Guo Y., Liu Y., Li H., Chen Y., Wang X., Huang K., et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol. Cancer. 2019;18:158. doi: 10.1186/s12943-019-1094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang F., Fang E., Mei H., Chen Y., Li H., Li D., Song H., Wang J., Hong M., Xiao W., et al. Cis-acting circ-CTNNB1 promotes β-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 2019;79:557–571. doi: 10.1158/0008-5472.CAN-18-1559. [DOI] [PubMed] [Google Scholar]
- 105.Zhu Y.J., Zheng B., Luo G.J., Ma X.K., Lu X.Y., Lin X.M., Yang S., Zhao Q., Wu T., Li Z.X., et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526–3540. doi: 10.7150/thno.32796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Z., Sun A., Yan A., Yao J., Huang H., Gao Z., Han T., Gu J., Li N., Wu H., Li K. Circular RNA MTCL1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1QBP ubiquitin degradation and mediating beta-catenin activation. Mol. Cancer. 2022;21:92. doi: 10.1186/s12943-022-01570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bi W., Huang J., Nie C., Liu B., He G., Han J., Pang R., Ding Z., Xu J., Zhang J. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of beta-catenin pathway. J. Exp. Clin. Cancer Res. 2018;37:275. doi: 10.1186/s13046-018-0936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu N., Yuan Z., Du K.Y., Fang L., Lyu J., Zhang C., He A., Eshaghi E., Zeng K., Ma J., et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019;26:2758–2773. doi: 10.1038/s41418-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wen S.Y., Qadir J., Yang B.B. Circular RNA translation: novel protein isoforms and clinical significance. Trends Mol. Med. 2022;28:405–420. doi: 10.1016/j.molmed.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 110.Khan F.A., Nsengimana B., Khan N.H., Song Z., Ngowi E.E., Wang Y., Zhang W., Ji S. Chimeric peptides/proteins encoded by circRNA: an update on mechanisms and functions in human cancers. Front. Oncol. 2022;12:781270. doi: 10.3389/fonc.2022.781270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li J., Ma M., Yang X., Zhang M., Luo J., Zhou H., Huang N., Xiao F., Lai B., Lv W., Zhang N. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol. Cancer. 2020;19:142. doi: 10.1186/s12943-020-01259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu F., Peng Y., Chang S., Luo X., Yuan Y., Zhu X., et al. Vimentin binds to a novel tumor suppressor protein, GSPT1-238aa, encoded by circGSPT1 with a selective encoding priority to halt autophagy in gastric carcinoma. Cancer Lett. 2022;545:215826. doi: 10.1016/j.canlet.2022.215826. [DOI] [PubMed] [Google Scholar]
- 113.Xia X., Li X., Li F., Wu X., Zhang M., Zhou H., Huang N., Yang X., Xiao F., Liu D., et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu S.S., Liu Y.S., Guo X.Y., Murakami Y., Yang G., Gao X.D., Kinoshita T., Fujita M. A knockout cell library of GPI biosynthetic genes for functional studies of GPI-anchored proteins. Commun. Biol. 2021;4:777. doi: 10.1038/s42003-021-02337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao X., Xia X., Li F., Zhang M., Zhou H., Wu X., Zhong J., Zhao Z., Zhao K., Liu D., et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol. 2021;23:278–291. doi: 10.1038/s41556-021-00639-4. [DOI] [PubMed] [Google Scholar]
- 116.Wu X., Xiao S., Zhang M., Yang L., Zhong J., Li B., Li F., Xia X., Li X., Zhou H., et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021;22:33. doi: 10.1186/s13059-020-02250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Du W.W., Xu J., Yang W., Wu N., Li F., Zhou L., Wang S., Li X., He A.T., Du K.Y., et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ. Res. 2021;129:568–582. doi: 10.1161/CIRCRESAHA.120.318364. [DOI] [PubMed] [Google Scholar]
- 118.Xu J., Du W.W., Wu N., Li F., Li X., Xie Y., Wang S., Yang B.B. The circular RNA circNlgnmediates doxorubicin-inducedcardiac remodeling and fibrosis. Mol. Ther. Nucleic Acids. 2022;28:175–189. doi: 10.1016/j.omtn.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang T., Xia Y., Lv J., Li B., Li Y., Wang S., Xuan Z., Xie L., Qiu S., He Z., et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer. 2021;20:66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang L., Zhou J., Zhang C., Chen R., Sun Q., Yang P., Peng C., Tan Y., Jin C., Wang T., et al. A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6. Clin. Transl. Med. 2021;11:e613. doi: 10.1002/ctm2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang T., Liu Z., She Y., Deng J., Zhong Y., Zhao M., Li S., Xie D., Sun X., Hu X., Chen C. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 2021;520:321–331. doi: 10.1016/j.canlet.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 122.Liang Z.X., Liu H.S., Xiong L., Yang X., Wang F.W., Zeng Z.W., He X.W., Wu X.R., Lan P. A novel NF-kappaB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer. 2021;20:103. doi: 10.1186/s12943-021-01404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li H., Lan T., Liu H., Liu C., Dai J., Xu L., Cai Y., Hou G., Xie K., Liao M., et al. IL-6-induced cGGNBP2 encodes a protein to promote cell growth and metastasis in intrahepatic cholangiocarcinoma. Hepatology. 2022;75:1402–1419. doi: 10.1002/hep.32232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Y., Jiang J., Zhang J., Shen H., Wang M., Guo Z., Zang X., Shi H., Gao J., Cai H., et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol. Cancer. 2021;20:101. doi: 10.1186/s12943-021-01390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peng Y., Xu Y., Zhang X., Deng S., Yuan Y., Luo X., Hossain M.T., Zhu X., Du K., Hu F., et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/beta-catenin signaling pathway to promote gastric cancer progression. Mol. Cancer. 2021;20:158. doi: 10.1186/s12943-021-01457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K.W., Waye M.M.Y., Zhang Q., Fu W.M., Zhang J.F. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y., Wang Z., Su P., Liang Y., Li Z., Zhang H., Song X., Han D., Wang X., Liu Y., et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Mol. Ther. 2022;30:415–430. doi: 10.1016/j.ymthe.2021.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., Wu C., Zhou Q., Hu W., Wu C., Jiang J. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xue C., Li G., Lu J., Li L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal. Transduct. Target. Ther. 2021;6:400. doi: 10.1038/s41392-021-00788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dou Y., Kawaler E.A., Cui Zhou D., Gritsenko M.A., Huang C., Blumenberg L., Karpova A., Petyuk V.A., Savage S.R., Satpathy S., et al. Proteogenomic characterization of endometrial carcinoma. Cell. 2020;180:729–748.e26. doi: 10.1016/j.cell.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang C., Han X., Yang L., Fu J., Sun C., Huang S., Xiao W., Gao Y., Liang Q., Wang X., et al. Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics. 2020;10:10908–10924. doi: 10.7150/thno.48264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liang Y., Wang H., Chen B., Mao Q., Xia W., Zhang T., Song X., Zhang Z., Xu L., Dong G., Jiang F. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol. Ther. Nucleic Acids. 2021;23:355–368. doi: 10.1016/j.omtn.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu Y., Zhang S., Liao X., Li M., Chen S., Li X., Wu X., Yang M., Tang M., Hu Y., et al. Circular RNA circIKBKB promotes breast cancer bone metastasis through sustaining NF-κB/bone remodeling factors signaling. Mol. Cancer. 2021;20:98. doi: 10.1186/s12943-021-01394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Z., Zhang T., Feng R., Huang H., Xia T., Sun C. circARF3 alleviates mitophagy-mediated inflammation by targeting miR-103/TRAF3 in mouse adipose tissue. Mol. Ther. Nucleic Acids. 2019;14:192–203. doi: 10.1016/j.omtn.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang Z., Liu F., Wang W., Ouyang S., Sang T., Huang Z., Liao L., Wu J. Deregulation of circ_003912 contributes to pathogenesis of erosive oral lichen planus by via sponging microRNA-123, -647 and -31 and upregulating FOXP3. Mol. Med. 2021;27:132. doi: 10.1186/s10020-021-00382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kong P., Yu Y., Wang L., Dou Y.Q., Zhang X.H., Cui Y., Wang H.Y., Yong Y.T., Liu Y.B., Hu H.J., et al. circ-Sirt1 controls NF-kappaB activation via sequence-specific interaction and enhancement of SIRT1 expression by binding to miR-132/212 in vascular smooth muscle cells. Nucleic Acids Res. 2019;47:3580–3593. doi: 10.1093/nar/gkz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karedath T., Al-Dasim F.M., Ahmed I., Al-Qurashi A., Raza A., Andrews S.S., Ahmed A.A., Ali Mohamoud Y., Dermime S., Malek J.A. Regulation of circular RNA CircNFATC3 in cancer cells alters proliferation, migration, and oxidative phosphorylation. Front. Cell Dev. Biol. 2021;9:595156. doi: 10.3389/fcell.2021.595156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng S., Hu C., Lin H., Li G., Xia R., Zhang X., Su D., Li Z., Zhou Q., Chen R. circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-kappaB signaling pathway. J. Exp. Clin. Cancer Res. 2022;41:71. doi: 10.1186/s13046-021-02237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wu Y., Zhang Y., Zhang Y., Wang J.J. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol. Int. 2017;41:1283–1289. doi: 10.1002/cbin.10761. [DOI] [PubMed] [Google Scholar]
- 141.Gasparrini M., Mazzola F., Cuccioloni M., Sorci L., Audrito V., Zamporlini F., Fortunato C., Amici A., Cianci M., Deaglio S., et al. Molecular insights into the interaction between human nicotinamide phosphoribosyltransferase and Toll-like receptor 4. J. Biol. Chem. 2022;298:101669. doi: 10.1016/j.jbc.2022.101669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou G., Duan Y., Lu C., Wang W. Knockdown of circ-UQCRC2 ameliorated lipopolysaccharide-induced injury in MRC-5 cells by the miR-326/PDCD4/NF-kappaB pathway. Int. Immunopharmacol. 2021;97:107633. doi: 10.1016/j.intimp.2021.107633. [DOI] [PubMed] [Google Scholar]
- 143.Li X., Jia Y., Nan A., Zhang N., Zhou H., Chen L., Pan X., Qiu M., Zhu J., Zhang H., et al. CircRNA104250 and lncRNAuc001.dgp.1 promote the PM(2.5)-induced inflammatory response by co-targeting miR-3607-5p in BEAS-2B cells. Environ. Pollut. 2020;258:113749. doi: 10.1016/j.envpol.2019.113749. [DOI] [PubMed] [Google Scholar]
- 144.Wang H., Xiao Y., Wu L., Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int. J. Oncol. 2018;52:743–754. doi: 10.3892/ijo.2018.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiang S., Zhao G., Lu J., Jiang M., Wu Z., Huang Y., Huang J., Shi J., Jin J., Xu X., Pu X. Silencing of circular RNA ANRIL attenuates oxygen-glucose deprivation and reoxygenation-induced injury in human brain microvascular endothelial cells by sponging miR-622. Biol. Res. 2020;53:27. doi: 10.1186/s40659-020-00295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen J., Yang X., Liu R., Wen C., Wang H., Huang L., Li W., Zhu Z., Zhu Y., Liu H. Circular RNA GLIS2 promotes colorectal cancer cell motility via activation of the NF-κB pathway. Cell Death Dis. 2020;11:788. doi: 10.1038/s41419-020-02989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen W., Wang H., Zhu Z., Feng J., Chen L. Exosome-shuttled circSHOC2 from IPASs regulates neuronal autophagy and ameliorates ischemic brain injury via the miR-7670-3p/SIRT1 Axis. Mol. Ther. Nucleic Acids. 2020;22:657–672. doi: 10.1016/j.omtn.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen B., Li Y., Liu Y., Xu Z. circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J. Cell. Physiol. 2019;234:21249–21259. doi: 10.1002/jcp.28730. [DOI] [PubMed] [Google Scholar]
- 149.Li J., Huang C., Zou Y., Ye J., Yu J., Gui Y. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol. Cancer. 2020;19:103. doi: 10.1186/s12943-020-01225-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 150.Wu Y., Zhang J., Zhang X., Zhou H., Liu G., Li Q. Cancer stem cells: a potential breakthrough in HCC-targeted therapy. Front. Pharmacol. 2020;11:198. doi: 10.3389/fphar.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu C.X., Chen L.L. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185:2016–2034. doi: 10.1016/j.cell.2022.04.021. [DOI] [PubMed] [Google Scholar]
- 152.Pereira B., Billaud M., Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 153.Akira S., Maeda K. Control of RNA stability in immunity. Annu. Rev. Immunol. 2021;39:481–509. doi: 10.1146/annurev-immunol-101819-075147. [DOI] [PubMed] [Google Scholar]
- 154.Schultz C.W., Preet R., Dhir T., Dixon D.A., Brody J.R. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR) Wiley Interdiscip. Rev. RNA. 2020;11:e1581. doi: 10.1002/wrna.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu R., Wu K., Li Y., Sun R., Li X. Human antigen R: a potential therapeutic target for liver diseases. Pharmacol. Res. 2020;155:104684. doi: 10.1016/j.phrs.2020.104684. [DOI] [PubMed] [Google Scholar]
- 156.Lin F.Y., Chen Y.H., Lin Y.W., Tsai J.S., Chen J.W., Wang H.J., Chen Y.L., Li C.Y., Lin S.J. The role of human antigen R, an RNA-binding protein, in mediating the stabilization of toll-like receptor 4 mRNA induced by endotoxin: a novel mechanism involved in vascular inflammation. Arterioscler. Thromb. Vasc. Biol. 2006;26:2622–2629. doi: 10.1161/01.ATV.0000246779.78003.cf. [DOI] [PubMed] [Google Scholar]
- 157.Jang J.H., Kim D.H., Surh Y.J. Dynamic roles of inflammasomes in inflammatory tumor microenvironment. NPJ Precis. Oncol. 2021;5:18. doi: 10.1038/s41698-021-00154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu T., Zhang L., Joo D., Sun S.C. NF-kappaB signaling in inflammation. Sig. Transduct. Target. Ther. 2017;2:e17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liu J., Chen S., Wang W., Ning B.F., Chen F., Shen W., Ding J., Chen W., Xie W.F., Zhang X. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-beta pathways. Cancer Lett. 2016;379:49–59. doi: 10.1016/j.canlet.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 160.Xia Y., Shen S., Verma I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Balachander G.M., Talukdar P.M., Debnath M., Rangarajan A., Chatterjee K. Inflammatory role of cancer-associated fibroblasts in invasive breast tumors revealed using a fibrous polymer scaffold. ACS Appl. Mater. Inter. 2018;10:33814–33826. doi: 10.1021/acsami.8b07609. [DOI] [PubMed] [Google Scholar]
- 162.Davoodzadeh Gholami M., Kardar G.A., Saeedi Y., Heydari S., Garssen J., Falak R. Exhaustion of T lymphocytes in the tumor microenvironment: significance and effective mechanisms. Cell. Immunol. 2017;322:1–14. doi: 10.1016/j.cellimm.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 163.Fu X., Sun G., Tu S., Fang K., Xiong Y., Tu Y., Zha M., Xiao T., Xiao W. Hsa_circ_0046523 mediates an immunosuppressive tumor microenvironment by regulating MiR-148a-3p/PD-L1 Axis in pancreatic cancer. Front. Oncol. 2022;12:877376. doi: 10.3389/fonc.2022.877376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang L.X., Gao J., Long X., Zhang P.F., Yang X., Zhu S.Q., Pei X., Qiu B.Q., Chen S.W., Lu F., et al. The circular RNA circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas via the miR-181a-5p/CARM1 axis. Mol. Cancer. 2022;21:110. doi: 10.1186/s12943-022-01586-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang J., Zhao X., Wang Y., Ren F., Sun D., Yan Y., Kong X., Bu J., Liu M., Xu S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu X., Zhong Y., Li J., Shan A. Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget. 2017;8:114829–114838. doi: 10.18632/oncotarget.22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Deng L., Liu G., Zheng C., Zhang L., Kang Y., Yang F. Circ-LAMP1 promotes T-cell lymphoblastic lymphoma progression via acting as a ceRNA for miR-615-5p to regulate DDR2 expression. Gene. 2019;701:146–151. doi: 10.1016/j.gene.2019.03.052. [DOI] [PubMed] [Google Scholar]
- 168.Zhang X.L., Xu L.L., Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 2017;41:1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 169.Ge J., Wang J., Xiong F., Jiang X., Zhu K., Wang Y., Mo Y., Gong Z., Zhang S., He Y., et al. Epstein-barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81:5074–5088. doi: 10.1158/0008-5472.CAN-20-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao C.X., Yan Z.X., Wen J.J., Fu D., Xu P.P., Wang L., Cheng S., Hu J.d., Zhao W.L. CircEAF2 counteracts Epstein-Barr virus-positive diffuse large B-cell lymphoma progression via miR-BART19-3p/APC/β-catenin axis. Mol. Cancer. 2021;20:153. doi: 10.1186/s12943-021-01458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weng Q., Chen M., Li M., Zheng Y.F., Shao G., Fan W., Xu X.M., Ji J. Global microarray profiling identified. J. Med. Genet. 2019;56:32–38. doi: 10.1136/jmedgenet-2018-105440. [DOI] [PubMed] [Google Scholar]
- 172.Shi L., Yan P., Liang Y., Sun Y., Shen J., Zhou S., Lin H., Liang X., Cai X. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171. doi: 10.1038/cddis.2017.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang P.F., Wei C.Y., Huang X.Y., Peng R., Yang X., Lu J.C., Zhang C., Gao C., Cai J.B., Gao P.T., et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer. 2019;18:105. doi: 10.1186/s12943-019-1031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ou Z.L., Luo Z., Wei W., Liang S., Gao T.L., Lu Y.B. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol. 2019;16:1592–1603. doi: 10.1080/15476286.2019.1649585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang P.F., Gao C., Huang X.Y., Lu J.C., Guo X.J., Shi G.M., Cai J.B., Ke A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shi M., Li Z.Y., Zhang L.M., Wu X.Y., Xiang S.H., Wang Y.G., Zhang Y.Q. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021;12:94. doi: 10.1038/s41419-020-03334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wculek S.K., Cueto F.J., Mujal A.M., Melero I., Krummel M.F., Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020;20:7–24. doi: 10.1038/s41577-019-0210-z. [DOI] [PubMed] [Google Scholar]
- 178.Wang B., Zhou Q., Li A., Li S., Greasley A., Skaro A., Quan D., Min W., Liu K., Zheng X. Preventing alloimmune rejection using circular RNA FSCN1-silenced dendritic cells in heart transplantation. J. Heart Lung Transpl. 2021;40:584–594. doi: 10.1016/j.healun.2021.03.025. [DOI] [PubMed] [Google Scholar]
- 179.Coffelt S.B., Wellenstein M.D., de Visser K.E., KE Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 180.Shang A., Gu C., Wang W., Wang X., Sun J., Zeng B., Chen C., Chang W., Ping Y., Ji P., et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol. Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Veglia F., Sanseviero E., Gabrilovich D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021;21:485–498. doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gao F., Xu Q., Tang Z., Zhang N., Huang Y., Li Z., Dai Y., Yu Q., Zhu J. Exosomes derived from myeloid-derived suppressor cells facilitate castration-resistant prostate cancer progression via S100A9/circMID1/miR-506-3p/MID1. J. Transl. Med. 2022;20:346. doi: 10.1186/s12967-022-03494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen D.L., Sheng H., Zhang D.S., Jin Y., Zhao B.T., Chen N., Song K., Xu R.H. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol. Cancer. 2021;20:166. doi: 10.1186/s12943-021-01475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ayala-Cuellar A.P., Cho J., Choi K.C. Toll-like receptors: a pathway alluding to cancer control. J. Cell. Physiol. 2019;234:21707–21715. doi: 10.1002/jcp.28879. [DOI] [PubMed] [Google Scholar]
- 185.Li W., Wang S., Shan B., Cheng X., He H., Qin J., Tang Y., Zhao H., Tian M., Zhang X., Jin G. CircHECTD1 regulates cell proliferation and migration by the miR-320-5p/SLC2A1 Axis in glioblastoma multiform. Front. Oncol. 2021;11:666391. doi: 10.3389/fonc.2021.666391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cai J., Chen Z., Wang J., Wang J., Chen X., Liang L., Huang M., Zhang Z., Zuo X. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis. 2019;10:576. doi: 10.1038/s41419-019-1814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jiang Q.L., Feng S.J., Yang Z.Y., Xu Q., Wang S.Z. CircHECTD1 up-regulates mucin 1 expression to accelerate hepatocellular carcinoma development by targeting microRNA-485-5p via a competing endogenous RNA mechanism. Chin. Med. J. 2020;133:1774–1785. doi: 10.1097/CM9.0000000000000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Y., Zhang Y., Li X., Zhang M., Lv K. Microarray analysis of circular RNA expression patterns in polarized macrophages. Int. J. Mol. Med. 2017;39:373–379. doi: 10.3892/ijmm.2017.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhou Z., Jiang R., Yang X., Guo H., Fang S., Zhang Y., Cheng Y., Wang J., Yao H., Chao J. circRNA mediates silica-induced macrophage activation via HECTD1/ZC3H12a-dependent ubiquitination. Theranostics. 2018;8:575–592. doi: 10.7150/thno.21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lu C., Shi W., Hu W., Zhao Y., Zhao X., Dong F., Xin Y., Peng T., Liu C. Endoplasmic reticulum stress promotes breast cancer cells to release exosomes circ_0001142 and induces M2 polarization of macrophages to regulate tumor progression. Pharmacol. Res. 2022;177:106098. doi: 10.1016/j.phrs.2022.106098. [DOI] [PubMed] [Google Scholar]
- 191.Barrett R.L., Puré E. Cancer-associated fibroblasts and their influence on tumor immunity and immunotherapy. Elife. 2020;9:e57243. doi: 10.7554/eLife.57243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Biffi G., Oni T.E., Spielman B., Hao Y., Elyada E., Park Y., Preall J., Tuveson D.A. IL1-Induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9:282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sahai E., Astsaturov I., Cukierman E., DeNardo D.G., Egeblad M., Evans R.M., Fearon D., Greten F.R., Hingorani S.R., Hunter T., et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen Z., Zhou L., Liu L., Hou Y., Xiong M., Yang Y., Hu J., Chen K. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020;11:5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yu Y., Xiao C.H., Tan L.D., Wang Q.S., Li X.Q., Feng Y.M. Cancer-associated fibroblasts induce epithelial–mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br. J. Cancer. 2014;110:724–732. doi: 10.1038/bjc.2013.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Strell C., Paulsson J., Jin S.B., Tobin N.P., Mezheyeuski A., Roswall P., Mutgan C., Mitsios N., Johansson H., Wickberg S.M., et al. Impact of epithelial-stromal interactions on peritumoral fibroblasts in ductal carcinoma in situ. J. Natl. Cancer Inst. 2019;111:983–995. doi: 10.1093/jnci/djy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Procopio M.G., Laszlo C., Al Labban D., Kim D.E., Bordignon P., Jo S.H., Goruppi S., Menietti E., Ostano P., Ala U., et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat. Cell Biol. 2015;17:1193–1204. doi: 10.1038/ncb3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tran L.L., Dang T., Thomas R., Rowley D.R. ELF3 mediates IL-1alpha induced differentiation of mesenchymal stem cells to inflammatory iCAFs. Stem Cells. 2021;39:1766–1777. doi: 10.1002/stem.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li C., Teixeira A.F., Zhu H.J., Ten Dijke P. Cancer associated-fibroblast-derived exosomes in cancer progression. Mol. Cancer. 2021;20:154. doi: 10.1186/s12943-021-01463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhan Y., Du J., Min Z., Ma L., Zhang W., Zhu W., Liu Y. Carcinoma-associated fibroblasts derived exosomes modulate breast cancer cell stemness through exonic circHIF1A by miR-580-5p in hypoxic stress. Cell Death Discov. 2021;7:141. doi: 10.1038/s41420-021-00506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hiam-Galvez K.J., Allen B.M., Spitzer M.H. Systemic immunity in cancer. Nat. Rev. Cancer. 2021;21:345–359. doi: 10.1038/s41568-021-00347-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kantono M., Guo B. Inflammasomes and cancer: the dynamic role of the inflammasome in tumor development. Front. Immunol. 2017;8:1132. doi: 10.3389/fimmu.2017.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cabrita R., Lauss M., Sanna A., Donia M., Skaarup Larsen M., Mitra S., Johansson I., Phung B., Harbst K., Vallon-Christersson J., et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 205.Groeneveld C.S., Fontugne J., Cabel L., Bernard-Pierrot I., Radvanyi F., Allory Y., de Reyniès A. Tertiary lymphoid structures marker CXCL13 is associated with better survival for patients with advanced-stage bladder cancer treated with immunotherapy. Eur. J. Cancer. 2021;148:181–189. doi: 10.1016/j.ejca.2021.01.036. [DOI] [PubMed] [Google Scholar]
- 206.Mori T., Tanaka H., Deguchi S., Yamakoshi Y., Miki Y., Yoshii M., Tamura T., Toyokawa T., Lee S., Muguruma K., Ohira M. Clinical efficacy of nivolumab is associated with tertiary lymphoid structures in surgically resected primary tumors of recurrent gastric cancer. PLoS One. 2022;17:e0262455. doi: 10.1371/journal.pone.0262455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Luo Y.H., Yang Y.P., Chien C.S., Yarmishyn A.A., Ishola A.A., Chien Y., Chen Y.M., Huang T.W., Lee K.Y., Huang W.C., et al. Plasma level of circular RNA hsa_circ_0000190 correlates with tumor progression and poor Treatment response in advanced lung cancers. Cancers (Basel) 2020;12:1740. doi: 10.3390/cancers12071740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chen Y.G., Kim M.V., Chen X., Batista P.J., Aoyama S., Wilusz J.E., Iwasaki A., Chang H.Y. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B., et al. N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yang X., Wang J., Zhou Z., Jiang R., Huang J., Chen L., Cao Z., Chu H., Han B., Cheng Y., Chao J. Silica-induced initiation of circular ZC3H4 RNA/ZC3H4 pathway promotes the pulmonary macrophage activation. FASEB J. 2018;32:3264–3277. doi: 10.1096/fj.201701118R. [DOI] [PubMed] [Google Scholar]
- 211.Huang Z., Yao F., Liu J., Xu J., Guo Y., Su R., Luo Q., Li J. Up-regulation of circRNA-0003528 promotes mycobacterium tuberculosis associated macrophage polarization via down-regulating miR-224-5p, miR-324-5p and miR-488-5p and up-regulating CTLA4. Aging (Albany NY) 2020;12:25658–25672. doi: 10.18632/aging.104175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang D., Luo Y., Wang G., Yang Q. CircATRNL1 promotes epithelial-mesenchymal transition in endometriosis by upregulating Yes-associated protein 1 in vitro. Cell Death Dis. 2020;11:594. doi: 10.1038/s41419-020-02784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cai Y., Liang R., Xiao S., Huang Q., Zhu D., Shi G.P., Ouyang Q., Yang M. Circ_0088194 promotes the invasion and migration of rheumatoid arthritis fibroblast-like synoviocytes. Front. Immunol. 2021;12:628654. doi: 10.3389/fimmu.2021.628654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhou Z.B., Huang G.X., Fu Q., Han B., Lu J.J., Chen A.M., Zhu L. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol. Ther. 2019;27:531–541. doi: 10.1016/j.ymthe.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen X., Li H.D., Bu F.T., Li X.F., Chen Y., Zhu S., Wang J.N., Chen S.Y., Sun Y.Y., Pan X.Y., et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics. 2020;10:4851–4870. doi: 10.7150/thno.42423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ye Y., Zhang L., Hu T., Yin J., Xu L., Pang Z., Chen W. CircRNA_103765 acts as a proinflammatory factor via sponging miR-30 family in Crohn's disease. Sci. Rep. 2021;11:565. doi: 10.1038/s41598-020-80663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xu Y., Tian Y., Li F., Wang Y., Yang J., Gong H., Wan X., Ouyang M. Circular RNA HECTD1 mitigates ulcerative colitis by promoting enterocyte autophagy via miR-182-5p/HuR Axis. Inflamm. Bowel Dis. 2022;28:273–288. doi: 10.1093/ibd/izab188. [DOI] [PubMed] [Google Scholar]
- 218.Xiong S., Peng H., Ding X., Wang X., Wang L., Wu C., Wang S., Xu H., Liu Y. Circular RNA expression profiling and the potential role of hsa_circ_0089172 in hashimoto's Thyroiditis via sponging miR125a-3p. Mol. Ther. Nucleic Acids. 2019;17:38–48. doi: 10.1016/j.omtn.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]