Abstract
We have isolated and determined the structure of the gene encoding the 68 kDa subunit (p68) of the mouse DNA polymerase α–primase complex. The p68 gene consists of four exons and the p68 promoter region lacks TATA and CAAT boxes, but contains a GC-rich sequence, two palindrome sequences and two putative E2F-binding sites. A series of transient expression assays using a luciferase reporter gene indicated that a region from nucleotide position –89 to –30 (–89/–30) with respect to the transcription initiation site is crucial for basal transcription of the p68 gene in proliferating NIH 3T3 cells. In particular, part of the GC-rich sequence (–57/–46) and the palindrome (–81/–62) elements were necessary for promoter activity, both of which share homology with the E-box sequence. Gel mobility shift assays using NIH 3T3 nuclear extracts revealed that the upstream stimulatory factor, known as an E-box-binding protein, binds to these sites. Moreover, we observed binding of E2F to two sites near the transcription initiation site (–11/–3 and +9/+16). A transient luciferase expression assay using synchronized NIH 3T3 cells in G0 phase revealed that these E2F sites are essential for transcription induction of the p68 gene after serum stimulation, but are dispensable for basal transcription. These results indicate that growth-dependent regulation of transcription of the mouse p68 and p180 genes is mediated by a common factor, E2F; however, basal transcription of the genes, interestingly, is regulated by different transcription factors.
INTRODUCTION
The replication of chromosomal DNA is complex and tightly regulated in eukaryotic cells and the multiple replication proteins cooperatively work to replicate the genome during S phase once in a cell cycle (1). In mammalian cells, seven distinct DNA polymerases (α, β, δ, ɛ, γ, ζ and η) have to date been isolated and characterized (2–5). The DNA polymerase α–primase complex (pol α) is one of the major replicative polymerases and is the only DNA polymerase that can initiate DNA synthesis in vivo (6). It consists of four subunits of 180, 68, 54 and 46 kDa, which are highly conserved throughout eukaryotes (2,7). The largest is a catalytic subunit exhibiting DNA polymerase activity (8) and the two small subunits exhibit DNA primase activity (6). The second largest subunit plays a role in regulation of the DNA pol α–primase complex and exhibits significant homologies throughout eukaryotes ranging from yeast to human (9–11). In Saccharomyces cerevisiae the second largest subunit protein is encoded by the POL12 gene and is essential for cell viability and required for both cellular DNA replication and normal cell cycle progression through S phase (12). Recently, we clarified a novel function of p68 using a cDNA expression system in mouse cells (13). We observed that the protein levels of p180, but not the mRNA levels, markedly increased when the p68 gene was co-expressed with the p180 gene, with the result that ectopically generated DNA polymerase activity was dramatically increased. p180 expressed alone was not translocated to the nucleus; however, when p180 and p68 were co-expressed, both proteins were translocated to the nucleus. Thus, p68 plays a significant role in p180 protein synthesis and nuclear translocation.
A number of studies on transcription regulation have shown that several regulatory elements are required for transcription of the genes encoding the proteins involved in DNA replication. Most promoters of these genes lack the TATA box motif and contain Sp1-binding elements, indicating that Sp1 regulates transcription through the GC-box (the human and mouse DNA polymerase α large subunit, human DNA polymerase δ-1, mouse dihydrofolate reductase, human thymidine kinase, mouse Cdc6 and human Cdc7 genes) in the cycling cell (14–19). In addition, it has also been reported that Sp3 activates transcription of the human DNA polymerase δ-1 gene (15). It is known that most of these genes are regulated in the cell cycle, i.e. expression of these genes increases at the G1/S boundary and this transcriptional activation is mediated by E2F (14,15,17,19–21).
We have previously isolated the cDNA of the four subunits of the mouse DNA pol α–primase complex (encoding proteins of 180, 68, 54 and 46 kDa) and have examined the levels of each mRNA species by northern blot hybridization (11). The mRNA levels of all four subunits increased concordantly prior to the peak of DNA synthesis upon inducing proliferation of quiescent cells. Recently, we have characterized the promoter of the p180 gene to investigate its transcriptional regulation. We found that its constitutive transcription is governed by the GA-binding protein and by Sp1, while growth-regulated transcription is regulated by E2F (Izumi et al., unpublished data). Here, we examine the isolation and analysis of the transcriptional regulatory region of the p68 gene in the mouse to see if there is common regulation of the two proteins which form the complex. The p68 gene contains four exons and the 5′-flanking region lacks a TATA box and CAAT box, but contains the consensus sequences for several transcription factors, including ATF, Sp1, E2F, two palindromes and the E-box. A series of transient expression assays revealed that the transcriptional regulatory region was limited to the upstream sequence –89 to –30 from the transcription initiation site. Two sequences (from –81 to –62 and –57 to –46) were found to be critical for transcription. USF (upstream stimulation factor) binds to these sites and is involved in regulation of constitutive transcription of the p68 gene in the cycling cell. Moreover, the p68 gene promoter contains two E2F-binding elements and E2F was shown to bind to these sites and to control growth-dependent transcription.
MATERIALS AND METHODS
Cloning the mouse 68 kDa subunit gene from a genomic DNA library
A mouse genomic DNA library in Charon4A, provided by Dr Matsukage (22), was screened using a 225 bp EcoRI–SmaI fragment of the 68 kDa subunit cDNA (23). To confirm that putative positive clones contained the first exon, we also screened with a 40 bp oligonucleotide (Fig. 1A, exon 1, positions 1–40). We screened 2.3 × 106 plaques of the Charon 4A library and obtained one positive clone which had a 20.1 kb genomic fragment containing exons 1–4. EcoRI digestion of the positive clone yielded four fragments (6E, 6F, 6C and 6B; Fig. 1A); fragment 6E (1719 bp) contained exon 1.
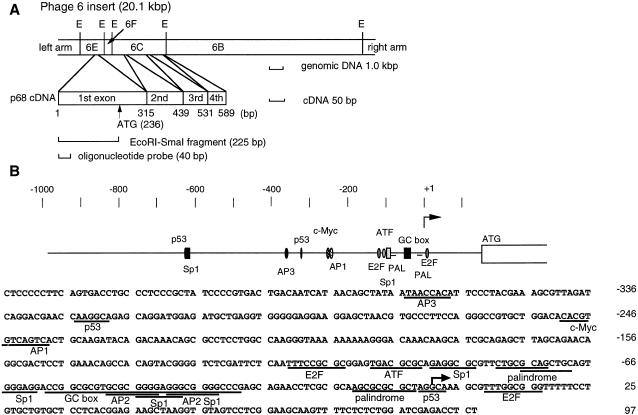
Figure 1.
Phage 6 insert containing the mouse p68 gene and the nucleotide sequence of the 5′-flanking region of the p68 gene. (A) Physical map of the clone containing mouse p68 genomic DNA. E, EcoRI sites. The positions of the four exons are shown. (B) The 5′-flanking region and the first exon of the mouse p68 gene. (Upper) The putative transcription factor binding sites are indicated. ATG is the translation initiation site. (Lower) The nucleotide sequence is shown from –425 to +97 with respect to the transcription initiation site. The transcription initiation site is marked by an arrow. The consensus sequences for transcription factor binding sites are underlined.
Plasmid construction
The 1.1 kb (EcoRI–XhoI) fragment containing the 5′-untranslated region (995 bp) and part of the first exon (96 bp) of the p68 gene was ligated to XhoI–linker and then ligated to the luciferase reporter vector pGV-B (Toyo Ink., Japan) via the XhoI site [p6E(–995)]. The 5′-end deletion mutants, pS12(–547) and pKL12(–164), derived from p6E(–995), were created using exonuclease III (TaKaRa Shuzou, Japan). The plasmids pKL2(–89), pKCL11(–64), pKLCd(+6) and pKCL19(+55), containing sequence near the promoter region, were constructed using PCR. Positions of the primers were as follows: –89 (pKL2), –64 (pKCL1), +6 (pCLd) and +55 (pKCL19), respectively. The 3′-end deletion mutant (–164/–30) was derived from pKL12(–164). Plasmid pKL12(–164) was digested with ApaI and HindIII, blunt-ended by T4 DNA polymerase and self-ligated. The exact end-points of each construct were confirmed by DNA sequencing.
Site-directed mutagenesis was carried out by a PCR-based method as described previously (24). The mutated plasmids created by the linker scanning method were constructed by a one-step overlap extension PCR method (25). All mutation constructions were derived from pKL12(–164) and the mutation positions of each were confirmed by DNA sequencing.
Cell culture and transient expression assay
Mouse fibroblast NIH 3T3 cells were cultured in DMEM supplemented with 10% calf serum. Transient expression assays using cycling cells were performed with Lipofectamine reagent (Gibco). The cells (2 × 105) were grown in a 35 mm dish for 24 h and transfected with 1.0 µg of test p68 DNA plasmid using Lipofectamine reagent. pRSV-β-gal (0.5 µg) was also included as an internal control to standardize transfection efficiency. Following incubation at 37°C for 5 h, the medium was replaced with fresh growth medium. Forty-eight hours later all cells were harvested in lysate buffer and luciferase activity was measured with a PicaGene assay system (Toyo Ink., Japan). The same extract was used for assay for β-galactosidase activity with chlorophenol red β-d-galactopyranoside (Boehringer Mannheim, Germany) as substrate (26). The luciferase activity was normalized to the β-galactosidase activity. Protein was quantified by the method of Bradford (27). All transient expression assays were carried out in duplicate with at least three independent assays. For transactivation by E2F, 1.0 µg of test plasmid DNA, 0.5 µg of pRSV-β-gal and 0.1 µg of pRC/CMV or pRC/CMV-E2F1 were co-transfected using Lipofectamine reagent and assayed as described above.
Synchronization of transfected cells
Cells (6 × 106) were electroporated (950 µF, 350 µV) with 12.5 µg of reporter plasmid and 7.5 µg of pRSV-β-gal in 0.5 µl of 1× K-PBS (120.7 mM KCl, 30.8 mM NaCl, 8.1 mM Na2HPO4, 1.46 mM KH2PO4, 10 mM MgCl2) at 4°C in a Bio-Rad Gene Pulser apparatus. Electroporated cells were cultured with DMEM containing 0.4% calf serum for 48 h. Cells were released by exchanging the growth medium for medium containing 15% calf serum and cultured at 37°C for 0, 5, 10, 15, 20 or 25 h. To monitor the level of DNA synthesis the cells were grown in the presence of bromodeoxyuridine (BrdU) and BrdU incorporation was quantified by ELISA using BrdU Labeling Detection Kit III (Boehringer Mannheim, Germany).
Mobility gel shift assay
Nuclear extracts were prepared from NIH 3T3 cells as described (28) and dialyzed against buffer containing 300 mM KCl. End-labeled oligonucleotide (0.1 ng) was incubated with 6 µg nuclear extract in 20 µl of reaction mixture (20 mM HEPES–NaOH, pH 7.5, 10% glycerol, 50 mM KCl, 0.05% NP-40, 0.2 mM EDTA, 0.5 mM DTT, 1 µg single-stranded salmon sperm DNA) at 25°C for 20 min. The reaction mixture was run on a 5% acrylamide gel (30:1) in TBE buffer at 4°C (7 V/cm). For the competition assay, 100- or 1000-fold excess amount of competitor was added to the reaction mixture. For supershift experiments, 100 µg of polyclonal rabbit antiserum against E2F-1 (sc-193X; Santa Cruz), E2F-4 (sc-866X; Santa Cruz), USF (sc-862X; Santa Cruz) or c-Myc (sc-764X; Santa Cruz) was incubated in the reaction mixture at 25°C for another 20 min. The following double-stranded oligonucleotides were used as probe or competitor (mutation site underlined): probe 89, 5′-AGGCG-CGTTCTGCGCAGCTGCAGTGGGAGGG-3′; probe 89M, 5′-AGGCGCGTTCTTATCAGCTGCAGTGGG-AGG-3′; probe E, 5′-TGCAGTGGGAGGACCGGCGCGTGC-GCGGG-3′; probe EM, 5′-TGCAGTGGGAGGACCGGATA-GTGCGCGGG-3′; probe 3, 5′-TAGGCAAAGCGTTTGGC-GGGTTTTTCCTG-TGCTGTGCTCC-3′; probe 3M, 5′-TAG-GCAAAGCGTTTGTATGGTTTTTCCTGTGCTGTGCTCC-3′; probe 41, 5′-AG-GGCGGGGCCCGAGCAGAACCTCGCGCAAGCGCGCG-CTAGGCAAAGCGT-3′; probe 41M, 5′-AG-GGCGGGGCC-CGAGCAGAACCTCGCGCAAGCTATCG-CTAGGCAAA-GCGT-3′; probe 41M2, 5′-AGGGCGGG-GCCCGAGCGAA-CCTCTATCAAGCGCGCGCTAGGCA-AAGCGT-3′; Sp1, 5′-ATTCGATCGGGGCGGGGCGAGC-3′ (sc-2503 Santa Cruz); E2F, 5′-ATTTAAGTTTCGCGC-CCTTTCTCAA-3′ (sc-2507; Santa Cruz); E2FM, 5′-AT-TTAAGTTTCGATCCCTTTCTCAA-3′ (sc-2508; Santa Cruz); USF-1, 5′-CA-CCCGGTCACGTGGCCTACACC-3′ (sc-2557; Santa Cruz).
RESULTS
Analysis of the 5′ upstream sequence of the mouse p68 gene
We obtained a clone containing the mouse p68 gene by screening a C57BL-HaeIII partially digested genomic library. Figure 1A shows the genomic DNA construct. The clone (phage 6) contains 20.1 kb of genomic DNA covering the 5′-flanking region and exons 1–4. The four exons were identified by Southern analysis using a cDNA probe and genomic DNA digested with EcoRI, BamHI and HindIII (data not shown). The sizes of the exons are 315 (exon 1), 124 (exon 2), 92 (exon 3) and 58 bp (exon 4), respectively. This clone was digested into four fragments with EcoRI and Southern hybridization showed that one of them (6E fragment) contained exon 1 and the 5′-flanking region. The DNA sequence of the 6E fragment (1719 bp) was obtained by sequencing analysis and exactly corresponded to the cDNA sequence (11).
The transcription initiation site was determined by primer extension analysis and RNase protection assay (Fig. 1B and data not shown). We determined the transcriptional initiation site to be a guanine residue 273 bp upstream from the first base of the start codon.
We determined the nucleotide sequence of clone p6E containing the region –995 to +97 with respect to the transcription initiation site. This promoter does not contain a TATA or CAAT box; the region from –110 to +1 is GC-rich (75%) and contains some putative transcription factor binding sites (Fig. 1B).
Identification of the transcriptional regulatory region
To determine the transcriptional regulatory region, we constructed a plasmid (p6E) which contained the 5′-flanking region of the p68 gene (from –995 to +97) inserted into a luciferase expression vector pGV-B (Fig. 2). Several 5′-end deletion mutants were made using p6E as the starting point. These deletion mutants were transiently transfected into logarithmically growing NIH 3T3 cells and their luciferase activities were measured. To normalize the efficiencies of transfection, an RSV-driven β-gal reporter plasmid was co-transfected as an internal control.
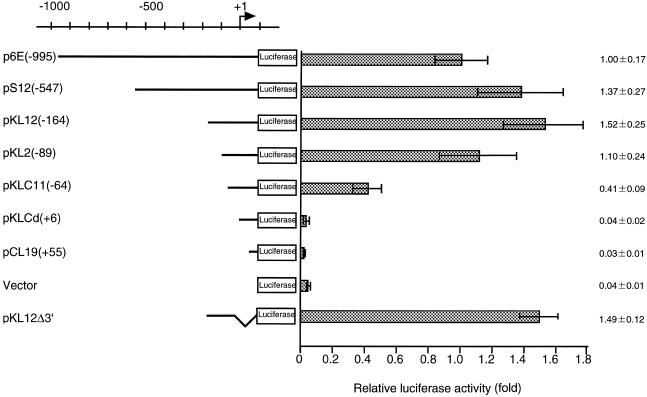
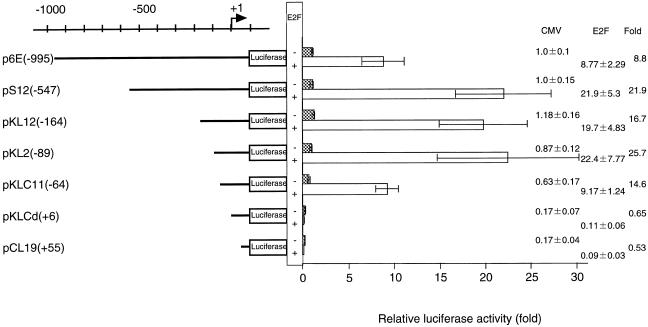
Figure 2.
Transcription activity of the upstream region of the p68 gene. A sequential series of deletion mutants of the p68 gene promoter were made and inserted upstream of the luciferase reporter gene. The number in parentheses indicates the extent of the deletion with respect to the transcription initiation site. Each reporter plasmid (1.0 µg) was independently transfected into NIH 3T3 cells and the luciferase activity determined. Luciferase activity is given as activity relative to that of the full-length construct [i.e. p6E(–995)]. These activities are normalized against β-galactosidase activity. Averages and standard deviations are presented on the right.
Each plasmid construct and its corresponding luciferase activity are shown in Figure 2. Deletion of the sequence from –995 to –164 resulted in a minor increase in promoter activity [pKL12(–164)]. No significant reduction in promoter activity was observed in a plasmid with all 5′ sequence past position –89 deleted; however, deletion up to position –64 decreased promoter activity to 41% of the initial level. Further deletion of the promoter sequence abolished promoter activity. pKL12d3′, which contains the region from –164 to –30, exhibited a similar level of promoter activity to the full-length construct. These results suggest that the region from –89 to –30 is essential for efficient transcription of the p68 gene.
The DNA sequence containing the E-box is essential for minimal promoter activity
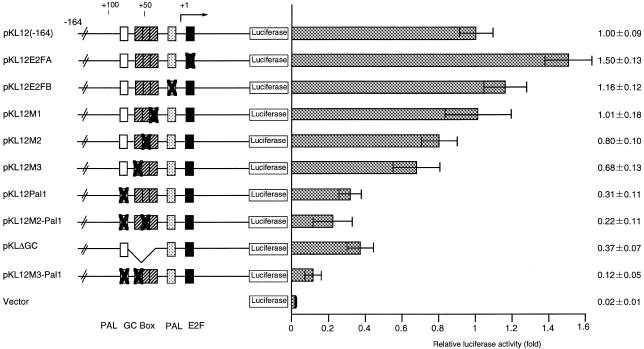
The essential region for promoter activity from –89 to –30 contains the following cis elements that are known to regulate transcription in other systems: Sp1-binding sites, at positions –65 (5′-GGGAGGA-3′), –45 (5′-GGGGAGGG-3′) and –40 (5′-GGGCGGGG-3′); AP2-binding sites, at positions –49 (5′-GCGCGGGGA-3′) and –40 (5′-GGGCGGGG-3′); an E-box, at position –75 (5′-CAGCTG-3′); palindromic sequences at positions –80 (5′-CTGCGCAG-3′) and –76 (5′-GCAGCTGC-3′). To investigate the essential sequence(s) required for promoter activity, we constructed plasmids each containing triple or quadruple contiguous transversion mutations (see Fig. 4) from –78 to –76 (pKL12Pal1), –54 to –52 (pKL12M3), –46 to –44 (pKL12M2) or –37 to –35 (pKL12M1), using pKL12(–164) DNA (Fig. 3). Transient assays using these plasmids were performed. The luciferase activities of pKL12Pal1 and pKLdGC, which had the entire GC-box (at positions –72 to –30) deleted, were significantly decreased. The activity of the doubly mutated clone pKL12M3-Pal1 (positions –78 to –76 and –54 to –52) was reduced to less than 12% of the level of the wild-type promoter (Fig. 3). Interestingly, mutation of the consensus sequence of the Sp1-binding element (pKL12M1 and pKL12M2) did not affect promoter activity.
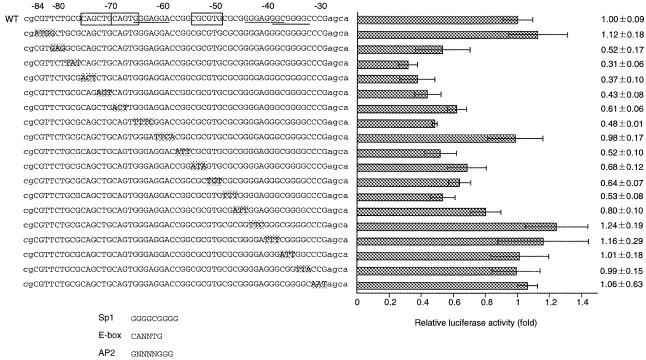
Figure 4.
Determination of the minimal p68 gene promoter. Eighteen mutants were constructed that altered a 3 or 4 bp sequence between positions –84 and –29 of the original construct pKL12(–164). The mutation sites are highlighted on the left. These constructs were transfected into NIH 3T3 cells and luciferase activities were measured. The luciferase activities are indicated as relative activity compared with activity of the wild-type construct (right). WT, wild-type. A consensus sequence for the E-box element and the Sp1-binding element are indicated by the open box and underlining, respectively. Consensus sequences for Sp1, AP2 and the E-box are presented below the construct information.
Figure 3.
Analysis of essential cis-elements for basal transcription of the p68 gene. Mutant constructs are illustrated on the left. Point mutation sites are represented with X. The constructs were transfected into NIH 3T3 cells and luciferase activity was measured. Luciferase activity was normalized against β-galactosidase activity and is given as relative activity compared to that of the wild-type construct pKL12(–164) and are presented on the right. Open and gray boxes represent palindromes and striped and closed boxes represent the GC-box and consensus sequences of E2F-binding elements, respectively.
To determine the minimal promoter region of the p68 gene, 18 mutants were constructed, each of which contains a single 3 or 4 bp mutation between positions –84 and –29 of the pKL12(–164) construct (Fig. 4). The minimal promoter activities of nine mutant plasmids were significantly impaired with mutations within the regions from –81 to –62 and –57 to –46. In particular, when the site at –78 to –76 was mutated, transcription activity was reduced to ~30% of the wild-type promoter. The region from –81 to –62 contains the E-box consensus sequence, which is the binding site (CANNTG) for transcription factors with the basic helix–loop–helix motif (29), and the Sp1-binding element; the region from –57 to –46 contains an E-box-like sequence.
Transcription factor USF binds to the p68 gene promoter
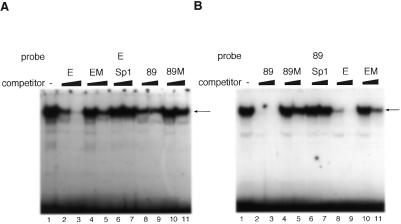
We next determined which proteins bound to these putative binding sites in the promoter using a gel mobility shift assay. We used probe 89 (position –89 to –60) and probe E (position –71 to –43), which cover the required sites for minimal promoter activity. These end-labeled probes were incubated with NIH 3T3 cell extract and the resultant protein–DNA complexes were separated by electrophoresis and visualized by autoradiography. Both of the probes caused a gel mobility shift, indicating that each probe binds the proteins (Fig. 5A and B, lane 1). Since the sequences of these sites are similar, these sites are possibly recognized by the same protein. To test this possibility, excess competitor probe was added to each reaction mixture. In the case of probe E, probe E itself inhibited complex formation (Fig. 5A, lanes 2 and 3). However, probe EM, containing a triple mutation (CGC→ATA) at positions –54 to –52, did not inhibit complex formation (lanes 4 and 5). Probe 89 was able to compete for binding of probe E (Fig. 5A, lanes 8 and 9), while probe 89M, containing the triple mutation GCG→TAT at positions –78 to –76, was unable to compete (Fig. 5A, lanes 10 and 11). The same effects were observed on gel mobility shift assays using end-labeled probe 89. Probes 89 and E inhibited complex formation (Fig. 5B, lanes 2, 3, 8 and 9). However, probes 89M (lanes 4 and 5) and EM (lanes 10 and 11) did not inhibit complex formation. An oligonucleotide containing the Sp1 consensus sequence was not able to compete for binding (Fig. 5A and B, lanes 6 and 7). These results indicate that the same protein binds to these sites.
Figure 5.
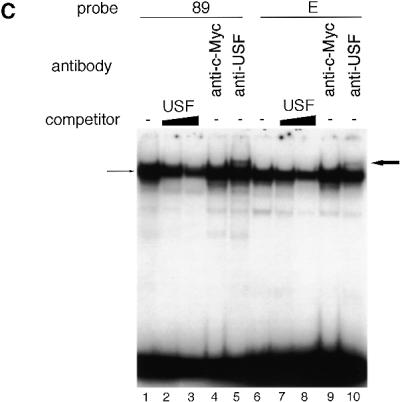
Determination of protein binding to p68 gene promoter sequences. A gel mobility retardation assay was performed using end-labeled probes (89 and E) and nuclear extracts of asynchronous NIH 3T3 cells. An excess (100- or 1000-fold) of unlabeled oligonucleotide was added as competitor. Shifted bands are indicated by arrows. Gel mobility retardation assays used probe E (positions –71 to –43) (A) and probe 89 (positions –89 to –60) (B). These assays were performed in the absence (lane 1) or presence of unlabeled E, EM, Sp1, 89 or 89M oligonucleotide probes. The Sp1 oligonucleotide probe contains the Sp1-binding consensus sequence (GGGGCGGGGC). (C) After complex formation between the nuclear extracts and either probe 89 or E, anti-USF or anti-c-Myc was added to the reaction mixture. Each band was supershifted when anti-USF was added to the reaction mixture (bold arrow). The USF-1 oligonucleotide probe contains the USF-1-binding consensus sequence (CACGTG).
Both regions contain a sequence similar to the E-box (CANNTG, where N represents any nucleotide residues) (29). Proteins containing helix–loop–helix domains, like USF or c-Myc, are reported to bind to the E-box (30,31). We therefore examined whether USF or c-Myc can bind to these regions by performing supershift assays in the presence of antibodies against these proteins. Antiserum against USF or c-Myc was added to the reaction mixture after each probe was incubated with NIH 3T3 cell extract (Fig. 5C). Supershift bands were observed only in reactions containing either probe in the presence of anti-USF serum, but not in the presence of anti-c-Myc serum (probe 89, Fig. 5C, lane 5; probe E, Fig. 5C, lane 10) Competition assays were also performed. When excess unlabeled oligonucleotides containing USF-1 consensus sequence were added to the reaction mixture, the oligonucleotide competed for binding of probes E and 89 (Fig. 5C, lanes 2, 3, 7 and 8). These results indicate that USF was present in each complex that bound to these regions and also that other factors which have a similar molecular mass may also bind to these sites, since not all signal disappeared upon addition of the competitor oligonucleotides. The molecular weight of the bound protein was determined using a UV crosslinking assay and found to be ~90 kDa (data not shown). USF consists of 43 (USF-1) and 44 kDa (USF-2) subunits and homo- and heterodimers are found (32). Therefore, the molecular weights of USF proteins are 86–88 kDa. These molecular weights match nicely with the molecular weight of the bound protein as determined by the UV crosslinking assay.
Identification of E2F binding sites and effect of E2F on transcription of the p68 gene
The promoter region of the p68 gene contains three E2F-binding site consensus sequences (positions –112 to –104, –19 to –12 and +9 to +16) and one E2F-like palindrome (positions –11 to –3). To investigate whether these sites are really involved in transcriptional regulation by E2F, the E2F expression vector was co-transfected into NIH 3T3 cells together with several 5′-end promoter deletion mutants (Fig. 6). The promoter activity of each deletion mutant was evaluated by luciferase assay, and is presented as relative activity with respect to the activity of p6E(–995) without E2F-1. E2F-1 activated the p6E(–995) promoter 8.8-fold and activated the pS12(–547) promoter 22-fold. It is suggested that the region between –995 and –547 contains a negatively regulated cis element. 5′ deletions involving the sequence between positions –574 and –64 were significantly activated by E2F-1. When the region from –64 to +6 was deleted, transactivation was drastically decreased. These results indicate that E2F activates the p68 gene promoter by interacting with the putative binding sites between –63 and +6.
Figure 6.
Determination of the region which is responsible for transcriptional activation by E2F. A series of 5′ deletion mutants (also used in the experiments presented in Fig. 2) were transfected into NIH 3T3 cells together with the E2F expression vector (pCMV-E2F) or a control plasmid (pCMV). The promoter activity for each construct is indicated as relative activity with respect to the activity of p6E(–995) without E2F-1. Promoter activity with or without E2F-1 is indicated by open or closed bars, respectively. The increase in the relative activity of luciferase with CMV-E2F-1 is presented on the right.
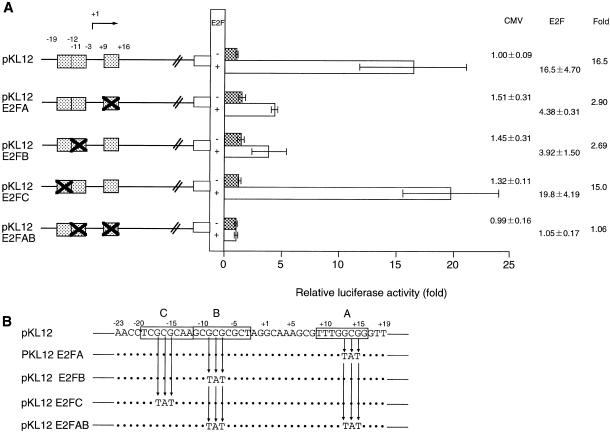
To define the E2F-binding sites, transversion mutations were introduced into three homologous E2F-binding sites (–19/–12, –11/–3 or +9/+16) (Fig. 7). When pKL12 E2FA or pKL12 E2FB was co-transfected with the E2F-1 expression vector into NIH 3T3 cells, transactivation was reduced compared to that observed with pKL12. The activity of pKL12 E2FC was similar to that of the wild-type construct. PKL12 E2FAB, with mutations in both the E2FA and E2FB sites, was not activated even in the presence of E2F-1. These results suggest that both E2FA and E2FB are essential for E2F-dependent activation of the promoter.
Figure 7.
Detection of the sites activated by E2F. (A) Three regions of the p68 gene promoter (+9/+16, –11/–3 and –19/–12) were mutated (the mutations were transversions of each base in the three base stretch and are indicated by X). These sites are represented by hatched boxes. Each mutant plasmid was co-transfected into NIH 3T3 cells with a plasmid carrying CMV-E2F-1 or CMV. The promoter activities are indicated as relative activity with respect to the activity of pKL12 without E2F-1. The promoter activity with and without E2F-1 is represented by open and closed bars, respectively. The increase in the relative activity of luciferase with CMV-E2F-1 is presented on the right. (B) The wild-type sequence and mutation sequence. Open boxes indicate three homologous E2F-binding sites and arrows represent transversion mutations.
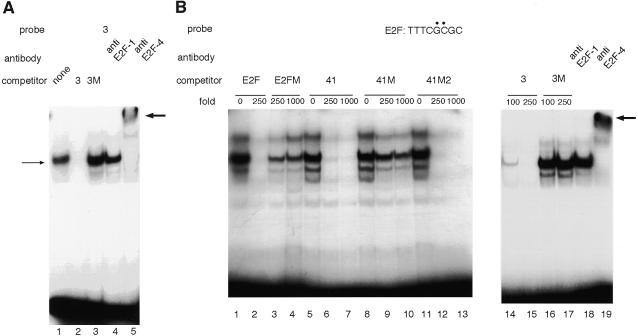
To confirm that cellular E2F binds to these sites, we carried out gel mobility shift assays using probes which contain these sites. Band shifts were observed using the end-labeled probe 3 (positions –3 to +37) (Fig. 8A, lane 1). Probe 3 competed with itself, while probe 3M, containing a mutation (GCG→TAT) at positions +13 to +15 (Fig. 7B, site A), was unable to compete (Fig. 8A, lane 3). Moreover, this complex was supershifted mainly by anti-E2F-4 serum (lanes 6). These results are consistent with the previous reports that E2F-4 accounts for the majority of E2F activity in asynchronous cells (33,34). When we performed the same experiment using probe 41 (positions –41 to +9), we also observed band shifts; however, neither probe 3 nor the E2F-binding consensus sequence competed with these bands. On the other hand, probe 41M, containing a mutation (GCG→TAT) at positions –9 to –7 (Fig. 7B, site B), competed with probe 41 to the same extent as probe 41 itself. Moreover, a supershifted band was not detected by anti-E2F-4 serum (data not shown). These results indicate that the shifted bands with probe 41 represent binding of some other protein factor(s) that is unrelated to E2F. In order to test the possibily that weak binding of E2F to this site (E2FB) is masked by stronger binding of some other protein(s), we carried out gel a mobility shift assay using the E2F-binding consensus sequence (TTTCGCGC) as probe; several shifted bands were detected (Fig. 8B, lane 1). Probe 41 competed with the E2F-binding consensus sequence (Fig. 8B, lanes 6 and 7), while the probe containing a mutation at positions –9 to –7 (probe 41M, Fig. 7B, site B) did not compete (Fig. 8B, lanes 9 and 10). The probe containing a mutation at positions –17 to –15 (probe 41M2, Fig. 7B, site C) competed with the E2F-binding consensus sequence (Fig. 8B, lanes 12 and 13). Probe 3 was able to compete for binding (Fig. 8B, lanes 14 and 15), while probe 3M was not (Fig. 8B, lanes 16 and 17). Moreover, anti-E2F-4 serum supershifted this complex. These results are consistent with those obtained by luciferase assay and demonstrate that E2F binds to positions –11/–3 and +9/+16.
Figure 8.
Identification of the binding site for transcription factor E2F. (A) Gel mobility shift assay was performed using probe 3 in the presence of unlabeled 3 (lane 2) or 3M (lane 3) oligonucleotide as competitor. After incubation of labeled probe 3 and nuclear extract, antibody (anti-E2F-1 or anti-E2F-4) was added to the reaction mixture (lanes 4 and 5). Shifted bands are indicated by thin arrows; supershifted bands are indicated by bold arrows. (B) End-labeled E2F consensus sequence was used as probe and the E2F consensus oligonucleotide E2FM (an oligonucleotide containing a mutation of the E2F-binding sequence), unlabeled 41, 41M, 41M2, 3 or 3M oligonucleotide was added to the reaction mixture as competitor.
Two E2F sites are essential for cell cycle-dependent promoter activation
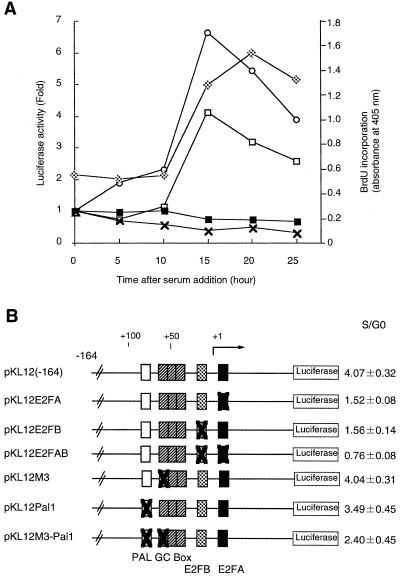
Previously we have shown that the amounts of both the p68 and the p180 subunit mRNAs increased before DNA synthesis (11). What governs cell cycle-dependent p68 promoter activity? Concerning the promoter of the p180 catalytic subunit gene, we have shown that E2F regulates growth-dependent transcription by binding to sequences near the transcription initiation site (Izumi et al., unpublished data). We have examined the possible involvement of E2F in the cell cycle-dependent activation of the p68 promoter. After transfection of pKL12 DNA by electroporation, NIH 3T3 cells were arrested in the G0 phase in the presence of 0.4% serum and then restimulated with 15% serum to proceed into a cell cycle. The stimulated cells were harvested to assay luciferase activity every 5 h. DNA synthesis was monitored by the incorporation of BrdU. As shown in Figure 9A, p68 gene promoter activity started to increase at 5–10 h after serum stimulation and reached its highest level (~4-fold) at 15–20 h after serum stimulation and before DNA synthesis. To define the cis-acting elements as E2F-binding sites, a reporter assay was carried out using pKL12 E2FAB, in which the two E2F-binding sites are mutated. PKL12 E2FAB did not show any promoter activation in response to serum stimulation.
Figure 9.
A transient luciferase expression assay using synchronized NIH 3T3 cells. (A) NIH 3T3 cells were transfected with p68 [KL12(–164)], p180 [pD3(–241)], pKL12AB or pGV-B (control) constructs by electroporation. Open squares, p68 [KL12(–164)]; open circles, p180 [pD3(–241)]; closed squares, pKL12 E2FAB; crosses, control (pGV-B). After culture in DMEM containing 0.4% calf serum, cells were released by growth in medium containing 15% calf serum and assayed every 5 h. Luciferase activity is shown relative to that measured at time 0 prior to serum addition. DNA synthesis was monitored by BrdU incorporation (gray squares). (B) Various mutant constructs were transfected into NIH 3T3 cells and luciferase activity 15 h after serum stimulation was compared to time 0. The promoter activities are indicated as values relative to luciferase activity at time 0.
Moreover, we confirmed that only E2F-binding sites are necessary as cis-acting elements. Various luciferase constructs were transfected into NIH 3T3 cells by electroporation and the cells were harvested at 0 and 15 h after serum addition to compare luciferase activity at each point (Fig. 9B). Mutation in part of the GC-box or upstream palindrome (pKL12M3, pKL12pal1 and pKL12M3-pal1) led to a reduction in promoter activity, but did not affect the growth response. When pKL12E2FA or pKL12 E2FB was transfected into cells, transcriptional activity was not reduced but the growth response was. The promoter whose E2F-binding sites were mutated (pKL12E2FAB) also did not reduce promoter activity but significantly affected the growth response. These results demonstrate that both E2F-binding sites are cis-acting elements necessary for growth-dependent regulation of transcription and only E2F family transcription factors stimulate transcription of the p68 gene promoter at the G1/S boundary.
DISCUSSION
We have isolated and analyzed the p68 gene promoter region. It lacks both a TATA box and a CAAT box but contains several GC-boxes within a GC-rich region. These characteristics are common to housekeeping genes (35). Most of the TATA-less promoters contain several Sp1-binding sites and/or Initiator (Inr) elements that can act to promote transcriptional initiation in sequences without a TATA box (36,37). The p68 gene promoter has highly conserved consensus binding sequences (i.e. 90% identity) for Sp1 (underlined in Fig. 4); however, alteration of these sites did not significantly affect promoter activity (Figs 3 and 4). A similar result was reported for the ADP-ribosylation factor 3 (ARF3) promoter (38). Although three potential Sp1-binding elements were identified in the ARF3 promoter, Sp1 did not bind to these regions and removal of the GC-rich box did not affect transcriptional activity (38).
The Inr element located near the transcription initiation site exhibits a consensus sequence (i.e. PyPyA+1NT/APyPy, where Py is a pyrimidine) (37). This site is recognized by the basic helix–loop–helix (bHLH) protein TFII-I (transcription factor II-I). Roy et al. (39) found that TFII-I stabilized binding of TBP (TATA box-binding protein) to the adenovirus major late core (adML) promoter and the TBP–TFII-I complex allows formation of a preinitiation complex containing other transcription factors. However, the promoter of the p68 gene does not follow this pattern, since a construct which is deleted from position +96 to –30 can still activate transcription (Fig. 2).
The E-boxes are bound by bHLH proteins (E47, E12, HEB, USF, Myc, etc.) (40–42). These proteins are expressed ubiquitously or cell specifically and regulate the cell cycle and development (40). We observed that USF bound to the p68 gene promoter, but another factor with a similar molecular mass may also bind to these sites, since not all signals disappeared upon addition of competitor oligonucleotide (Fig. 5).
Cell cycle-dependent transcriptional activation of the p68 gene is controlled by E2F, as shown in Figure 9. This is a common feature of transcription of genes encoding DNA replication-related proteins. For example, the 5′-flanking region of the human and mouse polymerase α large subunit, human DNA polymerase δ-1, human thymidine kinase, human Cdc6, human Cdc7, mouse dihydrofolate reductase and human Orc1 genes all contain E2F-binding sites which activate transcription of these genes at the G1/S boundary (14–17,19–21,26). In Drosophila, the cell cycle-dependent transcription of the two largest subunits of DNA polymerase α are regulated by E2F (43,44).
Three putative E2F sites exist in the p68 gene promoter. The most distal E2F site from the transcriptional initiation point (–111/–105) does not affect activation of transcription, even though the sequence of this site shows a higher similarity to the consensus site than do those E2F sites more proximal to the transcriptional initiation site (Fig. 7). Fry et al. (45) generated a series of human dihydrofolate reductase gene promoter mutants involving the E2F site (original position is –10/+2) to examine positional effects of the E2F site on transcriptional activation of the gene. They found that the E2F element confers growth regulation to the promoter only when it is proximal to the transcription initiation site. The p68 promoter may also be regulated by the same mechanism.
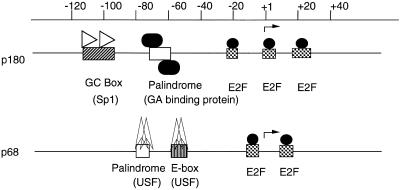
We have previously examined cell cycle-dependent regulation of the mouse p180 gene promoter and found that the E2F sites are essential for cell cycle-dependent activation of transcription of the p180 gene (Izumi et al., unpublished data). The promoter of the p180 gene contains three E2F sites, while the p68 gene promoter contains two E2F sites; all of these sites in both genes exist near the transcriptional initiation site (Fig. 10). The promoter of p180 was activated ~20-fold, while the promoter of p68 was activated ~15-fold by E2F-1 co-transfection in the cycling cell. The promoter activity of the p180 gene in S phase increased about six times compared to that in G0 phase and that of the p68 gene in S phase increased about four times compared to that in G0 phase. These results suggest that the activation rate of transcription might be a reflection of the number of E2F sites. These E2F sites did not affect constitutive transcription activity, but constructs altered at these sites were inactivated for transcriptional activity through G1 to S phase (Fig. 9). These results suggest that the growth-dependent regulation of both promoters is controlled solely by E2F. However, constitutive transcription of the p68 gene is regulated by a different factor. On the other hand, regulation of the p180 gene is controlled by GABP and SP1 (Fig. 10). Regulation of p68 expression may be by a different mechanism since p68 protein controls the translation of p180.
Figure 10.
Transcriptional regulation of the p180 and p68 gene promoters. The upstream sequences of the p180 and p68 genes are presented. The E2F-binding sites of the two promoters are located adjacent to the transcription initiation sites (gray boxes) and E2F activates transcription of both genes in a cell cycle-dependent manner. Basal transcription of the p180 gene is controlled by the Sp1 and GA proteins, whereas basal transcription of the p68 gene is controlled by different factors, including USF.
In conclusion, growth-dependent regulation of p68 and p180 transcription is mediated by a common factor, E2F, although constitutive transcription of these gene is regulated by different transcription factors.
Acknowledgments
ACKNOWLEDGEMENTS
We sincerely thank Dr Shunsuke Ishii for plasmid pRSV-β-gal, Dr Kristian Helin for the E2F-1 cDNAs and Dr Tetsu Akiyama for NIH 3T3 cells. We greatly appreciate critical reading of the manuscript by Drs Toshihiko Eki and Alasdair J.E. Gordon. We also thank Drs Takeshi Mizuno, Chiaki Ohtaka-Maruyama, Kaoru Sugasawa, Kiyoshi Ohtani, Toshihiko Eki and Masamitsu Yamaguchi for helpful comments and discussions and the rest of the Cellular Physiology Laboratory of RIKEN for their helpful suggestions. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan and the Biodesign Research Program of RIKEN (The Institute of Physical and Chemical Research). N.S.N. and M.Y. are special post-doctoral researchers of RIKEN.
DDBJ/EMBL/GenBank accession no. AB030823
REFERENCES
- 1.Stillman B. (1994) Cell, 78, 725–728. [DOI] [PubMed] [Google Scholar]
- 2.Wang T.S. (1991) Annu. Rev. Biochem., 60, 513–552. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999) EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 6.Foiani M., Lucchini,G. and Plevani,P. (1997) Trends Biochem. Sci., 22, 424–427. [DOI] [PubMed] [Google Scholar]
- 7.Lehman I.R. and Kaguni,L.S. (1989) J. Biol. Chem., 264, 4265–4268. [PubMed] [Google Scholar]
- 8.Wong S.W., Paborsky,L.R., Fisher,P.A., Wang,T.S. and Korn,D. (1986) J. Biol. Chem., 261, 7958–7968. [PubMed] [Google Scholar]
- 9.Collins K.L., Russo,A.A., Tseng,B.Y. and Kelly,T.J. (1993) EMBO J., 12, 4555–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotterill S., Lehman,I.R. and McLachlan,P. (1992) Nucleic Acids Res., 20, 4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazawa H., Izumi,M., Tada,S., Takada,R., Masutani,M., Ui,M. and Hanaoka,F. (1993) J. Biol. Chem., 268, 8111–8122. [PubMed] [Google Scholar]
- 12.Foiani M., Marini,F., Gamba,D., Lucchini,G. and Plevani,P. (1994) Mol. Cell. Biol., 14, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno T., Ito,N., Yokoi,M., Kobayashi,A., Tamai,K., Miyazawa,H. and Hanaoka,F. (1998) Mol. Cell. Biol., 18, 3552–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson B.E., Nasheuer,H.P. and Wang,T.S. (1991) Mol. Cell. Biol., 11, 2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L. and Chang,L.S. (1997) J. Biol. Chem., 272, 4869–4882. [PubMed] [Google Scholar]
- 16.Slansky J.E. and Farnham,P.J. (1996) BioEssays, 18, 55–62. [DOI] [PubMed] [Google Scholar]
- 17.Tommasi S. and Pfeifer,G.P. (1997) J. Biol. Chem., 272, 30483–30490. [DOI] [PubMed] [Google Scholar]
- 18.Hateboer G., Wobst,A., Petersen,B.O., Le Cam,L., Vigo,E., Sardet,C. and Helin,K. (1998) Mol. Cell. Biol., 18, 6679–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J.M., Sato,N., Yamada,M., Arai,K. and Masai,H. (1998) J. Biol. Chem., 273, 23248–23257. [DOI] [PubMed] [Google Scholar]
- 20.Yan Z., DeGregori,J., Shohet,R., Leone,G., Stillman,B., Nevins,J.R. and Williams,R.S. (1998) Proc. Natl Acad. Sci. USA, 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani K., DeGregori,J., Leone,G., Herendeen,D.R., Kelly,T.J. and Nevins,J.R. (1996) Mol. Cell. Biol., 16, 6977–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi M., Hayashi,Y., Hirose,F., Matsuoka,S., Moriuchi,T., Shiroishi,T., Moriwaki,K. and Matsukage,A. (1991) Nucleic Acids Res., 19, 2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chartier F.L., Keer,J.T., Sutcliffe,M.J., Henriques,D.A., Mileham,P. and Brown,S.D. (1992) Nature Genet., 1, 132–136. [DOI] [PubMed] [Google Scholar]
- 24.Ito W., Ishiguro,H. and Kurosawa,Y. (1991) Gene, 102, 67–70. [DOI] [PubMed] [Google Scholar]
- 25.Urban A., Neukirchen,S. and Jaeger,K.E. (1997) Nucleic Acids Res., 25, 2227–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbomel P., Bourachot,B. and Yaniv,M. (1984) Cell, 39, 653–662. [DOI] [PubMed] [Google Scholar]
- 27.Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 28.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl K. (1993) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- 29.Sawadogo M. and Roeder,R.G. (1985) Cell, 43, 165–175. [DOI] [PubMed] [Google Scholar]
- 30.Gregor P.D., Sawadogo,M. and Roeder,R.G. (1990) Genes Dev., 4, 1730–1740. [DOI] [PubMed] [Google Scholar]
- 31.Ayer D.E., Kretzner,L. and Eisenman,R.N. (1993) Cell, 72, 211–222. [DOI] [PubMed] [Google Scholar]
- 32.Sirito M., Lin,Q., Maity,T. and Sawadogo,M. (1994) Nucleic Acids Res., 22, 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tommasi S. and Pfeifer,G.P. (1995) Mol. Cell. Biol., 15, 6901–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moberg K., Starz,M.A. and Lees, J,A. (1996) Mol. Cell. Biol., 16, 1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson L.F. (1992) Curr. Opin. Cell. Biol., 4, 149–154. [DOI] [PubMed] [Google Scholar]
- 36.Pugh B.F. and Tjian,R. (1991) Genes Dev., 5, 1935–1945. [DOI] [PubMed] [Google Scholar]
- 37.Smale S.T. (1997) Biochim. Biophys. Acta, 1351, 73–88. [DOI] [PubMed] [Google Scholar]
- 38.Haun R.S., Moss,J. and Vaughan,M. (1993) J. Biol. Chem., 268, 8793–8800. [PubMed] [Google Scholar]
- 39.Roy A.L., Malik,S., Meisterernst,M. and Roeder,R.G. (1993) Nature, 365, 355–359. [DOI] [PubMed] [Google Scholar]
- 40.Desiderio S. (1995) Curr. Biol., 5, 605–608. [DOI] [PubMed] [Google Scholar]
- 41.Gregor P.D., Sawadogo,M. and Roeder,R.G. (1990) Genes Dev., 4, 1730–1740. [DOI] [PubMed] [Google Scholar]
- 42.Ayer D.E., Kretzner,L. and Eisenman,R.N. (1993) Cell, 72, 211–222. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi M., Hayashi,Y., Hirose,F., Nishimoto,Y. and Matsukage,A. (1997) Nucleic Acids Res., 25, 3847–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi Y., Yamaguchi,M., Hirose,F., Cotterill,S., Kobayashi,J., Miyajima,S. and Matsukage,A. (1996) J. Biol. Chem., 271, 14541–14547. [DOI] [PubMed] [Google Scholar]
- 45.Fry C.J., Slansky,J.E. and Farnham,P.J. (1997) Mol. Cell. Biol., 17, 1966–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]