Abstract
Understanding the regulation of gene expression requires the identification of cis-acting control elements that modulate gene function. The recent availability of complete genome sequences and profiles of mRNA expression has facilitated the development and utilization of computational methods to identify discrete regulatory elements. We have developed an oligomer counting method that identifies sequences that occur significantly more often in a group of interest relative to other genes in the genome. The use of a second parameter, which measures the frequency of oligomers within the group of interest, allows the detection of false positive signals caused by very infrequent oligomers that would otherwise appear as significant. Applying this method to gene groups that have a common expression pattern or shared function should identify oligomers that comprise cis-acting control elements. As a test of this method, we applied this approach to a set of intron-containing yeast genes, where we easily identified the known splicing signals as control elements. We have used this training set to examine how this method is affected by the length of the oligomer examined, as well as the size and composition of the gene group. These simulations allowed us to identify rules for selecting groups of genes to analyze. Finally, application of this method to nuclear genes encoding proteins targeted to the mitochondria identified a new putative cis-acting sequence in the 3′-untranslated region of this family of genes, which may play a role in mRNA localization or the regulation of mRNA stability or translation.
INTRODUCTION
The expression of genes at the transcriptional and post-transcriptional levels is often controlled by small cis-acting sequence elements in or near the regulated gene. These elements may be recognized by DNA-binding proteins which modulate DNA metabolism (e.g. centromeres and telomeres) or transcription (e.g. promoters) or by RNA-binding proteins which affect RNA processing, RNA localization, translation or RNA degradation. The identification and functional characterization of such cis-acting control elements will be a critical step in developing the tools to interpret and understand complete genomes.
Cis-acting sequences have been identified by a variety of different experimental approaches. Historically, many cis-acting sequences have been identified by mutational analysis of a target gene or suspected regulatory region. In addition, some cis-acting elements have been delineated by the identification of a critical trans-acting regulatory protein, whose binding site is then subsequently determined. Alternatively, cis-acting elements have been identified as shared sequence elements in groups of genes that are co-regulated or undergo similar processing steps. For example, the alignment of several mRNA sequences allowed the identification of a hexanucleotide sequence specifying 3′-end formation and polyadenylation (1). The central logic of this latter approach is that genes that share common regulation or processing should share cis-acting elements that dictate those common events. The recent availability of complete genome sequences (2,3) and expression profiles of most or all of the mRNA species in a population of cells (see for example 4,5) has greatly facilitated the use of these types of computational methods to identify cis-acting elements. For example, by examining elements common to a set of transcripts that are co-regulated, cis-acting transcriptional control elements have been identified (6,7).
We are interested in developing computational methods that can be used to identify cis-acting sequences that modulate post-transcriptional events, such as mRNA splicing, mRNA localization, translation and mRNA degradation. To this end, we have developed a procedure that allows the identification of oligomers that are over-represented in a specific group of co-regulated genes. This approach does not focus exclusively on what sequences are shared in the group of putatively co-regulated genes, but instead identifies oligomers which distinguish the group from the rest of the genome. The results presented here indicate that this method readily detects cis-acting signals involved in mRNA splicing when tested on Saccharomyces cerevisiae genes with coding region introns. Several additional experiments in which the composition of this group was varied to more accurately simulate ‘real world’ experiments indicate that the method performs well even when non-optimal groups of genes are analyzed, indicating that the method should be adequate to identify candidate cis-acting elements in groups of genes which may be co-regulated based on common function or expression pattern. We verified this by identifying a new candidate cis-acting element in the 3′-untranslated region (3′-UTR) of a group of S.cerevisiae genes that encode proteins which are transported into the mitochondrion.
MATERIALS AND METHODS
Sequence files
Chromosome sequences, a FASTA file containing all the spliced coding region sequences and a table listing gene names and functions, chromosome position, number of introns and exon boundaries were downloaded from the Saccharomyces Genome Database (8) on 17 May 1999. A number of repetitive elements were removed from the FASTA file and gene table. These consisted of genes encoded in Ty retrotransposon elements and several highly similar proteins encoded in the subtelomeric regions. Preliminary experiments (data not shown) had indicated that some bias might occur if these repeated sequences were not excluded. Mitochondrially encoded genes were also removed.
Based on data parsed from the gene table file, FASTA files containing the 5′-UTR (nt –100 to –1), 3′-UTR (nt +1 to +150) and unspliced coding region sequences were extracted from the chromosome FASTA files. These files, as well as the Perl source code for the scripts used to extract them, are available at http://www.mcb.arizona.edu/Parker/
Gene group selection
The gene group containing genes with introns was obtained by parsing the gene table file (see above). Genes with a non-zero entry in the introns column were added to the list. The mitochondrial gene group was obtained by a query of the Yeast Protein Database (9). The complex query form was used to retrieve proteins annotated as localized to the mitochondria. Mitochondrially encoded genes were removed by hand and the resulting 281 gene list (available at http://www.mcb.arizona.edu/Parker/ ) was used for the oligomer counting described here.
Oligomer counting method
Complete source code implementing the algorithm described below in Perl is available under the terms of the GNU Public License (GPL, http://www.gnu.org/copyleft/gpl.html ) at http://www.mcb.arizona.edu/Parker/
The oligomer counting method we have described here is dependent on some genome-wide pre-calculation steps (for reasons of computational time reduction). For the purpose of the following description, ‘genome’ indicates a set of genes from a single organism. The gene group of interest is a proper subset of the genome set. The genome may consist of all the genes in an organism (e.g. the S.cerevisiae genome) or only a portion of the genes in an organism (e.g. the genes contained on an oligonucleotide array chip).
The pre-calculation step requires a FASTA file containing the sequences from the region of interest (e.g. 3′-UTR) for every gene in the genome and a list of every oligomer of a given length (e.g. AAAAA to TTTTT for lengths of 5 nt). Two genome reference arrays are determined from this starting point: one which contains the number of times each oligomer is found in the genome and a second which contains the number of different genes each oligomer is found in. Note that the total number of oligomers (used in later steps) can be determined by summing the first array. These two arrays are then written to files, which are then used in all subsequent analyses. This step is somewhat time consuming, but only needs to be performed once for each sequence of interest in the genome.
Once the pre-calculation step is complete, the gene group(s) of interest is scored. The sequence of interest of the genes in the group is counted just as the whole genome was (see above). This generates two arrays specific for the gene group. By subtracting each oligomer from the same oligomer in the whole genome arrays, the two arrays specific for all genes not in the group of interest are obtained. These will be used in the subsequent normalization step.
The representational score RS for a particular oligomer O is calculated as:
RS = (oligomers Ogroup/total oligomersgroup) ÷ (oligomers Oremainder / total oligomersremainder)
where ‘remainder’ indicates the set of all genes not in the group of interest (i.e. ‘remainder’ = whole genome – group of interest). This measure represents how frequently an oligomer occurs in the gene group relative to how frequently it occurs in the genes not in the group.
For subsequent steps, the range of GWO values in the gene group is used to construct a bin series, containing a variable number of fixed size bins. If the range is (low)…(high), the first bin would be (low – bin size) and the last would be (high + bin size). (When the low number would have been <1 gene, it was automatically set to 1 gene.) The representational scores are sorted in these bins. A number (typically 1000) of random gene groups are selected. After ensuring that a similar (±20%) amount of sequence has been chosen (in order to ensure that a similar number of total oligomers are counted), each random group is counted as above. After the representational scores are sorted into bins, the highest score from each bin is saved in a statistical significance array. If no scores for a particular bin are obtained, a value of 0 is placed in the significance array for that bin in that trial. After the trials are complete, the scores in each bin of the significance array are sorted in ascending order. By checking the appropriate row of the sorted array, significance cut-off values are obtained for each bin. The scores from the gene group of interest are then screened for significant oligomers, using the thresholds determined from the random trials. Results are output into text files, imported into Microsoft Excel 98 for post-processing and finally plotted using DeltaGraph 4.5.
Rare element simulations
In order to simulate rare elements, it was necessary to remove a number of randomly selected genes from the genome for each trial. In order to remove the need to re-calculate the genomic reference arrays many times, a modified counting procedure was used. First, a number of genes were randomly selected. Oligomers and genes with oligomer counts were determined for this subset of genes and these values were subtracted from the genome reference arrays to generate the ‘genome’ reference arrays for the reduced genome for that particular trial. Results were then calculated as described above.
Incorrect gene group selection simulations
In order to simulate incorrectly selected gene groups, it was necessary to swap a number of randomly chosen genes from the group of genes with introns for an equal number of randomly chosen genes without introns (i.e. genes not in the training group). For each experiment, the two sets were chosen and then swapped, eliminating any potential back-swapping. Results were then calculated as described above.
RESULTS AND DISCUSSION
General method
The approach we have used is an oligomer counting method, conceptually similar to a method first presented by Staden in 1989 (10), as well as to several other recent methods (11–16, reviewed in 17). The overall goal of this method is to examine the usage of all possible oligomers of a given length in a group of sequences and determine if any are over-represented in those sequences as compared to the oligomer usage in the rest of the genome. The essential steps in the process are as follows (Fig. 1). First, a specific set of genes is chosen to examine. In the test case first discussed below, we have chosen a subset of 225 yeast protein coding genes containing introns within the translated region. Second, for the chosen group of genes, the number of times each oligomer of a particular length occurs is tabulated. For example, for oligomers of 3 nt, the number of occurrences of AAA would be counted, then AAC and so on, up to and including TTT. Counting is done with overlap, so that, for example, the sequence ACGT is counted as one instance of ACG and one of CGT. Third, a similar oligomer count is performed on the remainder of the genome, i.e. all the genes not in the chosen group. Fourth, these oligomer counts are converted into representational scores, referred to as RS. This representational score is the frequency of a given oligomer in the test group (number of occurrences of oligomer O/total number of oligomers of same length) divided by the frequency of the same oligomer in the rest of the genome. An RS value of 1.0 would indicate that the frequency of the oligomer in question is the same in the set of putatively co-regulated genes as it is in the rest of the genome. Since cis-acting elements are expected to be more frequent in the genes where they function, they should have high RS values. To our knowledge, this is the first time a whole eukaryotic genome has been analyzed in this fashion, as opposed to using randomized or Markov model-generated sequences for the purpose of normalization.
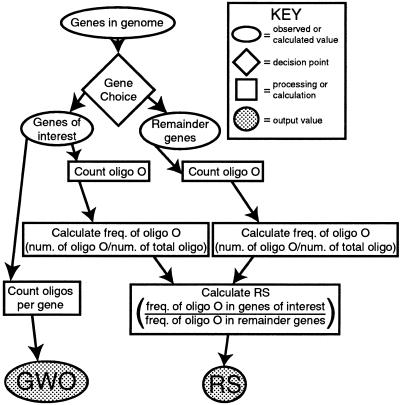
Figure 1.
Flowchart depicting the oligomer counting method. Unshaded ovals indicate values obtained by observation or previously existing data (e.g. genome sequence); the unshaded diamond indicates a decision that must be made; unshaded squares indicate oligomer counting, processing or calculation steps; shaded ovals indicate the output of the method. RS, representational score; GWO, genes with oligomer. See text for details.
The number of genes that contain each oligomer at least once is also simultaneously tabulated. This number, referred to as GWO (genes with oligomer) gives a measure of how many of the genes in the group of interest contain a given oligomer. Cis-acting elements are predicted to be broadly (if not universally) distributed in the groups of genes they regulate and should therefore have high scores for this metric also. Based on this method, strong candidates for cis-acting elements within a group of co-regulated genes will be those oligomers that have both high RS and GWO values. On a two-dimensional scatter plot of these two values, oligomers that contain or that are contained in cis-acting elements would then tend to be found in the upper right quadrant, allowing easy visual identification when results are displayed in this fashion. Use of the GWO score as well as the RS metric allows us to simultaneously quantify oligomer frequency in the gene group of interest relative to all other genes and oligomer abundance within the group.
The statistical significance of the representational scores is determined by a variation of the permutation or resampling statistics often used in quantitative trait mapping (18). Briefly, a large number of random gene groups of the same approximate size as the group of interest (same number of genes, 80–120% of the number of base pairs) are chosen from the entire set of genes in the genome and then counted and scored. The high score from each random trial is recorded. When the high scores from a large number of trials are sorted in ascending order, cut-offs for different levels of significance are found. For example, if the above procedure is used for 1000 random trials, the 950th score (when scores are sorted in a lowest to highest order) would define the P = 0.05 significance level cut-off.
Initial results (data not shown) demonstrated that rare oligomers (those with a high GC content for example) were a source of considerable noise when the above method was utilized to determine statistical significance. Since these rare oligomers will, by definition, have low GWO values, we developed a GWO value-based binning strategy, so that RS values were only compared between oligomers that occurred in a similar number of genes. Briefly, after the group of genes of interest is counted and scored for oligomers of a particular length, the observed range of GWO values is used to define a number of GWO value bins (e.g. 1–10 genes with oligomer, 11–20, etc.) When the subsequent statistical sampling is carried out, the high scores in each bin are recorded and used to determine significance as described above.
While the primary purpose of the binning of representational scores is to mask the ‘noise’ caused by rare oligomers, the binning also allows a qualitative finer granularity at the high end of the GWO value scale. For example, given a group of 150 genes, it would be unexpected (and quite likely significant) to find a 7 nt oligomer that was present in 145 of these genes. This oligomer could still be interesting even if it had a low RS value (relative to other oligomers with lower GWO scores). Without some kind of binning step in the method, potentially significant oligomers such as these would be overlooked. We chose to use a variable number of bins of a fixed size, so that the range of bin sizes was always based on the number of genes within the gene group of interest. For example, if the range of GWO scores observed in the experimental sample was 1–27 genes and the bin size had been set at 5 genes/bin, then the bin sizes used would be 1–5, 6–10, 11–15, 16–20, 21–25 and 26–30 genes (see Materials and Methods for details). Experiments (see below) demonstrated that selection of bin size (i.e. 5 genes/bin, 10 genes/bin and so on) was not a major factor in the results obtained with our training set. Additionally, before results were plotted, bin boundaries were divided by the number of genes in the group of interest, so that the percentage of genes in a group that contain a particular oligomer could be more easily determined.
Sequence selection
When searching for cis-acting elements, the choice of which region of the genomic sequence to examine is important. Because of our interest in post-transcriptional gene regulation, we chose to focus on sequences likely to be found in mRNAs, which we further subdivided into three groups: 5′-UTR sequences, arbitrarily defined as nucleotides –100 to –1 relative to the ATG of each mRNA; coding region sequences, from (and including) the translation start codon to stop codon of each mRNA; 3′-UTR sequences, arbitrarily defined as nucleotides +1 to +150 relative to the stop codon of each mRNA. It should be emphasized that these choices reflect our particular interests. The method presented here should work equally well on any sequences, provided that the same region of sequence is used for all genes examined and provided an entire genome sequence is available. Partial genome sequences may be sufficient, although using an incomplete genome sequence means that care must be taken to obtain a proper representative sample of genes.
Splicing signals are effectively detected
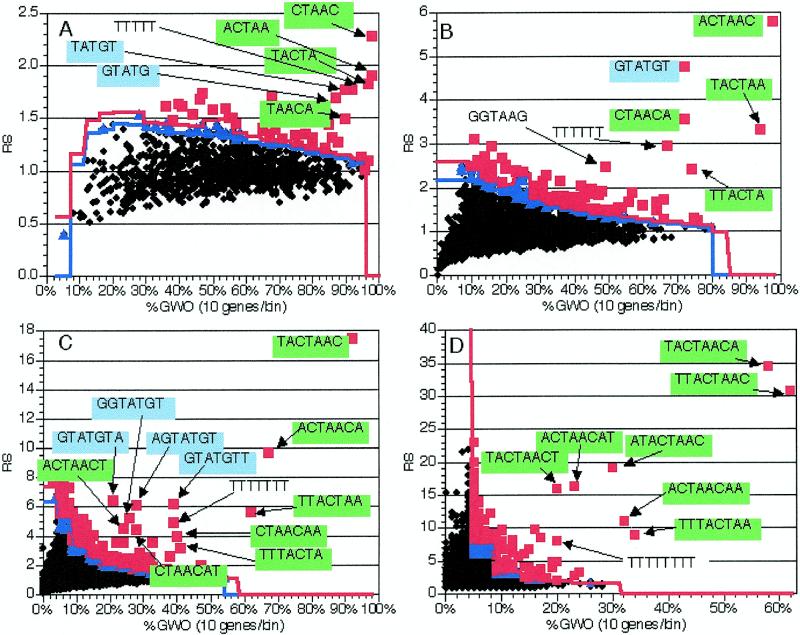
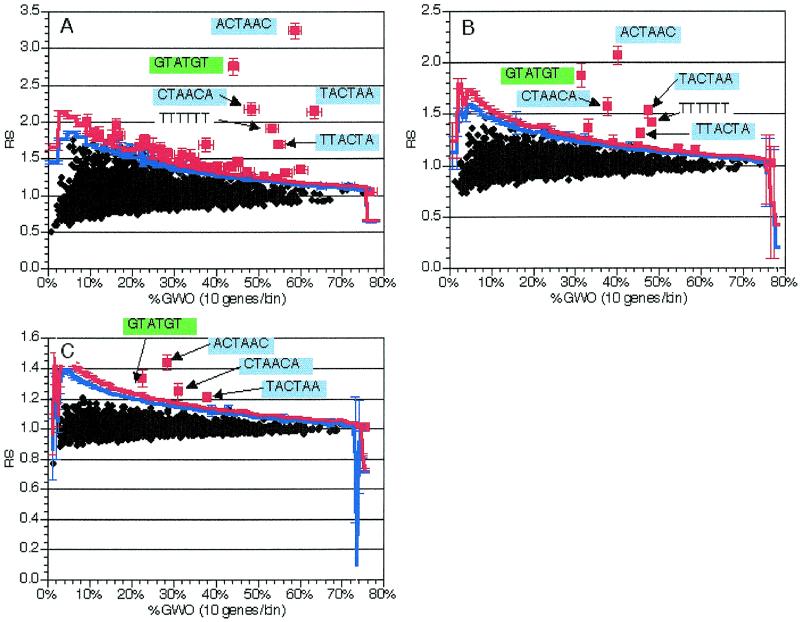
In order to determine the effectiveness of this method, we constructed a training group consisting of the genes containing annotated coding region introns from a recent version of the Saccharomyces Genome Database (8; see Materials and Methods for details; gene names available at http://www.mcb.arizona.edu/Parker ). The sequences between and including the ATG and stop codons (including introns) of these 225 genes were compared to the coding regions of the remainder of the ~6000 yeast genes and the RS and GWO values for all possible 6 nt oligomers were calculated and plotted (Fig. 2B). In this and all other calculations discussed below, 1000 random sets of similar size were also generated and scored for statistical calculation (see above and Materials and Methods). In this calculation, three oligomers particularly stood out on the basis of both RS and GWO values: TACTAA, ACTAAC and GTATGT (Fig. 2B). The first two sequences are the 6 nt oligomers comprising the core of the branch site consensus sequence (TACTAAC) and the third is the consensus 5′ splice site sequence. Two other high scoring oligomers were composed of portions of the TACTAAC branch point sequence, with different bases of the less well conserved flanking regions (e.g. CTAACA and TTACTA). Additionally, TTTTTT was also detected at a significant level, consistent with the observation that homouridine runs are involved in selection of the 3′ splice site (19). Thus, the application of this methodology to a group of genes encoding introns easily and accurately identified the known splicing signals.
Figure 2.
Application of the method to 225 genes with introns. As described in the text, oligomers were counted and analyzed in the coding region (including introns) of 225 genes containing introns. The coding regions of the genes without introns were used for normalization. 1000 random trials were used to determine significance levels; see text for details. Black diamonds, insignificant oligomers; blue triangles, oligomers significant at P = 0.05; red squares, oligomers significant at P = 0.01. Blue line, P = 0.05 significance cut-off in each bin; red line, P = 0.01 cut-off. Text and arrows indicate the sequences of pertinent over-represented oligomers; green backgrounds indicate oligomers comprising or containing TACTAAC, while blue backgrounds indicate the GTATGT 5′ splice site consensus. RS, representational score; %GWO, percent of genes in group with oligomer. %GWO is obtained by dividing GWO values by the number of genes in the group, 225 in this case. (A) Values from analysis of 5 nt oligomers. (B) Values from analysis of 6 nt oligomers. (C) Values from analysis of 7 nt oligomers. (D) Values from analysis of 8 nt oligomers.
Although not as striking as the known splicing signals discussed above, several other oligomers were also detected as being over-represented in this group of genes [shown as red boxes (P < 0.01) in Fig. 2B]. At least one oligomer that was detected as statistically significant (GGTAAG) is not part of any known splicing signal. This oligomer also has a significant representational score when oligomers are counted in coding regions with introns removed (data not shown), so it is likely to be some coding region feature that is over-represented in this set of genes. Examination of the location of the occurrences of this oligomer relative to the start and stop codons and the intron–exon junctions did not show any obvious pattern, although >95% of these oligomers are in-frame (data not shown), encoding Gly-Lys. The over-representation of this oligomer, as well as the high number of other statistically significant oligomers, relative to other gene groups (e.g. compare Figs 2B and 7B) may be due to other biases in this subset of the genes with introns. For example, this intron-containing family of genes is enriched in ribosomal proteins (90/225 genes or 40%), which are generally highly basic and have a high codon bias, which may explain the difference in oligomer distribution.
Figure 7.
Application of the method to 281 genes encoding mitochondrially localized proteins. As described in the text, oligomers were counted and analyzed in the 3′-UTR of 281 genes encoding mitochondrial proteins. Plots and experiments are as in Figure 2, with the exception that %GWO values were determined by dividing by 281, to reflect the change in gene group size, and that oligomer backgrounds are not color coded. (A) Values from analysis of 5 nt oligomers. (B) Values from analysis of 6 nt oligomers. (C) Values from analysis of 7 nt oligomers. (D) Values from analysis of 8 nt oligomers.
Effects of examining different oligomer lengths
We anticipated that the oligomer lengths that were examined would be a critical variable in the results obtained with this method. In order to investigate this issue, we examined oligomers from 3 to 8 nt long in unspliced coding region sequences of the same group of genes with introns as above.
The oligomer length examined affected the results in a variety of manners. First, and as intuitively expected, at oligomer lengths of 3 and 4 nt the components of the splicing signals did not have exceptionally high RS values (data not shown). Second, at an oligomer length of 5 nt the components of the TACTAAC, TTTTTT run and GTATGT (e.g. GTATG and TATGT) elements began to stand out with higher RS and GWO values (Fig. 2A). At an oligomer length of 6 nt the RS value of these elements continued to increase (Fig. 2B). At oligomer lengths of 7 and 8 nt the RS values continued to increase and variants of these elements begin to appear that included more weakly conserved components of an extended splicing signal. For example, at an oligomer length of 7 nt the over-represented oligomers containing the GTATGT 5′ splice site included AGTATGT, GGTATGT, GTATGTT and GTATGTA. Similarly, at oligomer lengths of 8 nt the over-represented oligomers containing the TACTAAC sequence included TACTAACT, TACTAACA ACTAACAT, ACTAACAA, ATACTAAC, TTACTAAC and TTTACTAA.
Based on these types of results, we note two inferences that can be made from the observations on different oligomer lengths. First, the ‘core’, or highly conserved, portion of a cis-acting sequence can be identified. This is because the components of over-represented oligomers are themselves over-represented (usually to a lesser extent). In practical terms, at oligomer lengths shorter than the core element, overlapping oligomers offset by a single nucleotide will be detected. The composite sequence formed by these overlapping oligomers will define the ‘core’ element. For example, GTATG and TATGT are both significant 5 nt oligomers (Fig. 2A); TACTAA and ACTAAC are both significantly over-represented in the 6 nt oligomers (Fig. 2B). Thus, a ‘core’ element will be observed when overlapping over-represented oligomers of length n – 1 coalesce into a single over-represented oligomer of length n.
In addition, information about partially conserved flanking sequences can be found by examining oligomers that are longer than the ‘core’ element. If there is no bias in the flanking sequence, all the ‘core’ element-containing oligomers should be equally over-represented. Conversely, if there are additional partially conserved nucleotides that flank the ‘core’ sequence, one will observe the appearance of multiple over-represented oligomers, which represent the additional partially conserved nucleotides. For example, the highly over-represented oligomers that contain TACTAAC are TACTAACT, TACTAACA, ACTAACAT, ACTAACAA, ATACTAAC, TTACTAAC and TTTACTAA, which is consistent with the results of a recent hidden Markov model-based analysis of S.cerevisiae introns, which defined a WWTACTAACWW extended branch point consensus sequence, where W is A or T (Fig. 2D and fig. 4 in 20). Similarly, the over-represented 7 nt oligomers containing GTATGT are AGTATGT, GGTATGT, GTATGTA and GTATGTT, consistent with the RGTATGTW (where R is A or G and W is A or T) extended 5′ splice site consensus found by the hidden Markov model (Fig. 2C and fig. 4 in 20).
Figure 4.
Effect of small groups on element detection in genes with introns. As described in the text, smaller groups of genes with introns were obtained by discarding randomly selected genes with introns. Plots and experiments are as in Figure 2, with the exception that %GWO is determined by dividing by the appropriate number of genes for each experiment. Sequences are indicated for pertinent oligomers with text and arrows. Error bars indicate standard deviations from 10 independent experiments. All data presented is from analysis of 6 nt oligomers. (A) Average values from analysis of groups of 112 genes with introns. (B) Representative single experiment with a 112 gene group. (C) Average values from analysis of groups of 56 genes with introns. (D) Representative single experiment with a 56 gene group. (E) Average values from analysis of groups of 22 genes with introns. (F) Representative single experiment with a 22 gene group. (G) Average values from analysis of groups of 10 genes with introns. (H) Representative single experiment with a 10 gene group.
Oligomer distribution bin size affects the significance of the results
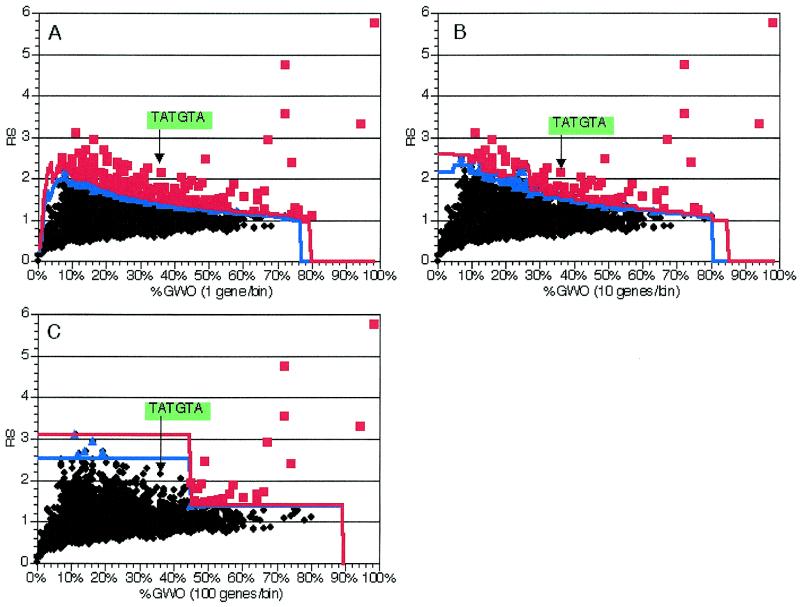
A second parameter that could impact on the results obtained with our method is the size of the bins used in the random trials to determine the significance of over-represented oligomers. In order to determine the effect of this variable, we compared the effects of different bin sizes on the statistical significance in an experiment using 6 nt oligomers in the same training gene group as above. Bin sizes of 1, 2, 5, 10, 25, 50 and 100 genes/bin were examined (Fig. 3 and data not shown). In each case, 1000 independent random sets were generated for each bin size. The smaller bin sizes (1–25 genes/bin, Fig. 3A and B and data not shown) did not have an impact on the significance of the results, as can be seen by the position of the marginally significant TATGTA oligomer (part of the 5′ splice site with one less well conserved 3′-flanking nucleotide) (Fig. 3A and B). With larger bin sizes, this oligomer is no longer detected as significant (Fig. 3C and data not shown). The increased resolution of very small bins (1 or 2 genes/bin) was offset by the observation that their calculation required more computer memory. Subsequent experiments with gene groups of different sizes (see below and data not shown) demonstrated that a bin size that balanced memory usage and statistical resolution was typically one-tenth to one-fifth the number of genes in the group of interest or 10 genes, whichever was smaller.
Figure 3.
Effect of bin size on significant element detection in genes with introns. As described in the text, 225 genes with introns were analyzed using different GWO bin sizes. Plots and experiments are as in Figure 2; the position of an element with differing significance with the different bin sizes is indicated. All data presented are from analysis of 6 nt oligomers. (A) Values from analysis with 1 gene/bin. (B) Values from analysis with 10 genes/bin. (C) Values from analysis with 100 genes/bin.
Variables affecting the choice of the gene group to examine
A critical step in our approach to the identification of cis-acting elements is the choice of the genes in the group that is examined for over-represented oligomers. Critical variables include the size of the selected group and where to draw the distinction between the selected group and the remainder of the genome. For example, consider a case where a number of mRNAs have been identified as increasing in level in response to a particular condition. Is it better to examine a large group containing all the genes encoding the mRNAs that change their level or is it more effective to select a smaller group of genes that undergo larger changes in expression, knowing that some co-regulated genes are likely to be left in the genome control? In order to address these types of issues we manipulated the set of intron-containing genes in various manners and determined the effects on detection of splicing signals as over-represented oligomers.
Effect of the size of the selected gene group. One issue is the number of genes in the selected gene group. In order to examine the effects of group size on the calculation, randomly selected genes with introns were removed from the genome (and hence from the gene group). Six nucleotide oligomers were then counted in unspliced coding region sequences of the remaining genes with introns. Ten independent trials of groups with 112, 56, 22 and 10 genes (40 trials total) were conducted and results were averaged across all trials of a particular size. Averages as well as individual trials were examined for effects on RS and GWO values.
As shown in Figure 4, the average RS values are largely unchanged as the size of the selected gene group decreased. However, the RS values required for a given statistical significance increased substantially as the gene group decreased in size. The smaller gene groups were expected to have a greater number of biased oligomers because of random effects, which would lead to higher significance thresholds, due to the underlying mechanism of our method and statistical calculations. The binning of scores based on oligomer distribution, which normally ameliorates this effect, was not expected to be effective because of the small range of bins available. The results of the small group simulations (Fig. 4) showed that the significance thresholds displayed an inverse correlation with the number of genes per group and that this resulted in the splicing elements not being detected as significant in the smaller gene groups. Nevertheless, it should be noted that we could detect the splicing elements marginally at 10 genes/group and significantly at 22. Based on these simulations, we conclude that this method can effectively detect elements present as few as 20 times in a genome, albeit at a lower significance.
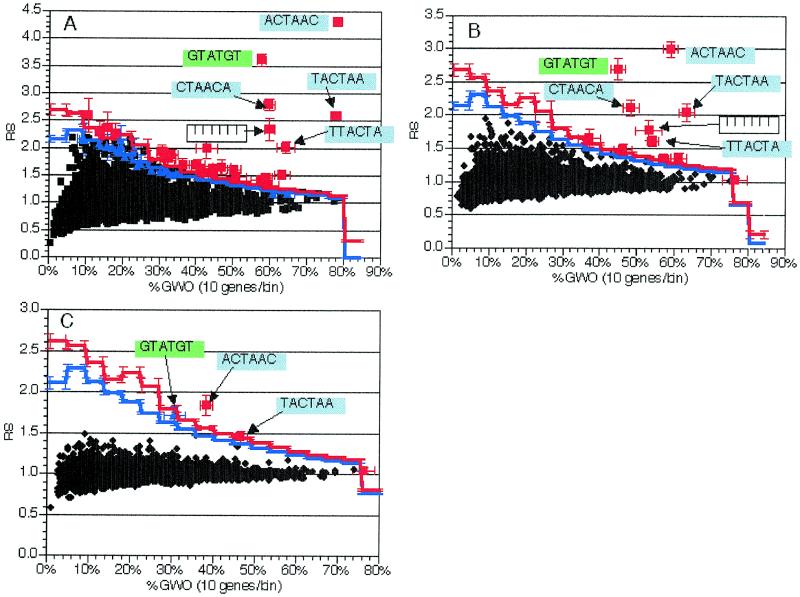
Effects of inappropriate genes in the selected subgroup. Another key component of the selected group of genes is its homogeneity with regard to the common function or regulation being considered. In our test case of genes containing intron sequences, all of the genes in the test group were known to contain introns. This is an ideal case but will often not be met by available data. For example, consider the case of examining all the genes that encode mRNAs that change level in response to an alteration in a mRNA-binding protein that regulates mRNA stability. The population of genes encoding mRNAs that change will include those that are directly affected by the mRNA-binding protein under investigation, but will also include genes encoding mRNAs whose levels change in response to alterations in the directly affected genes. In order to simulate this type of regulatory cascade, we added randomly chosen genes without introns to the training group of genes with introns. In this simulation, the genes with introns represent the primary genes, while the genes without introns represent the secondary, downstream targets of these genes.
Groups of genes were created that contained the original 225 genes with introns combined with either 225 (50% introns final), 675 (25% introns final) or 2025 (10% introns final) non-intron-containing genes. Ten independent trials of each of the groups totaling 450, 900 and 2250 genes were conducted, where in each trial the additional genes were chosen at random. For each trial, 6 nt oligomers were counted and scored in unspliced coding region sequences. Results for experimental groups were averaged across all trials of a given size, as were the significance thresholds determined from the random trials. The averaged results as well as results from individual trials were examined.
Increases in the group size led to a reduction in the RS value for the oligomers containing the splicing signals (compare Fig. 5A, B and C). However, the oligomers corresponding to the splicing signals were still easily detected as statistically significant in populations where the intron-containing genes were 50 or 25% of the total mix (Fig. 5A and B) and marginally detected even when they constituted 10% of the total genes in the mix (Fig. 5C). It should be noted that the largest group consisted of nearly 40% of the genes in the genome. It is difficult to envision a biologically relevant application of this method where a group with that large a fraction of the genome could not be further subdivided based on additional criteria. Thus, we conclude that this approach is sensitive enough to detect cis-acting elements even in mixed populations where the group of primary targets is as few as 25% of the total mix.
Figure 5.
Effect of regulatory cascade simulation in groups with some intron-containing genes. As described in the text, regulatory cascades were simulated by addition of random numbers of genes without introns to the 225 genes with introns. Plots and experiments are as in Figure 2, with the exception that %GWO is determined by dividing by the appropriate number of genes for each experiment. Sequences are indicated for pertinent oligomers with text and arrows. Error bars indicate standard deviations from 10 independent experiments. All data presented are from analysis of 6 nt oligomers. (A) Average values from analysis of groups with 450 genes [225 (50%) with introns]. (B) Average values from analysis of groups with 900 genes [225 (25%) with introns]. (C) Average values from analysis of groups with 2250 genes [225 (10%) with introns].
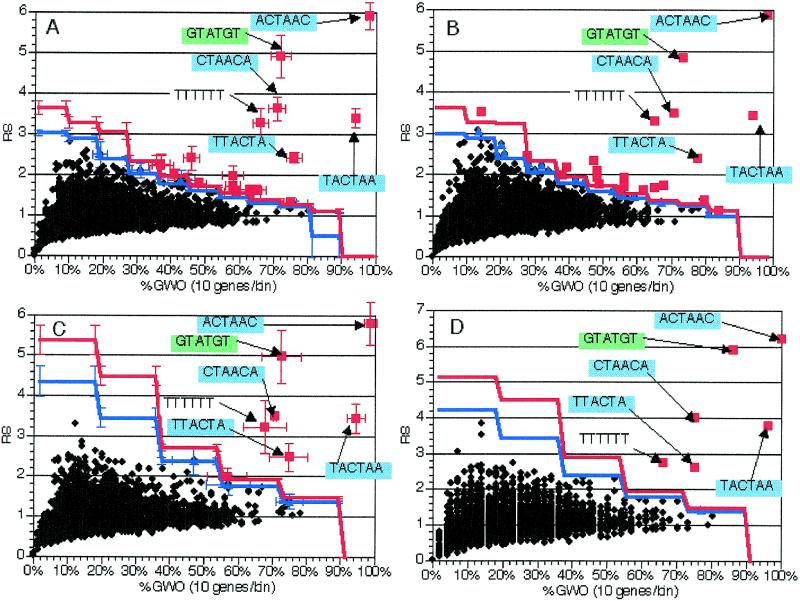
Effects of gene mixing between the selected and control groups. Since the distribution of the putative cis-acting element(s) is unknown when this method is used, the odds of an imperfect gene group selection are quite high. As discussed above, one type of common incorrect selection involves including genes that lack the element of interest, perhaps because they are affected by secondary effects. A second type of incorrect selection could occur if some element-containing genes were not placed in the gene group. The third, and potentially most detrimental, type of potential incorrect gene group selection is a combination of the first and second types, so that the selected gene group contains genes with and without elements, as does the remainder of the genome which is used for normalization. In order to simulate this situation we randomly swapped genes with introns (from the training gene group) and genes without introns (from the rest of the genome). In independent experiments, 56, 112 and 168 genes were swapped between the groups. In these cases the final selected group of 225 genes contained 75, 50 and 25% genes with introns, respectively. The swapped genes were all selected before any genes were exchanged, so no back-swapping occurred. Ten trials of each kind were carried out. In each trial, 6 nt oligomers were counted and scored in unspliced coding region sequences. All scores and significance thresholds were averaged across all trials of a particular size and the results of both the averaged and individual trials were examined.
Since this type of mixing both decreases the numerator and increases the denominator of the ratio leading to the RS value, we anticipated that this type of group selection would have a strong effect on the ability to detect the splicing signals as significant. The effect was predicted to be similar to that observed in the regulation cascade experiment (above), but more severe. As shown in Figure 6, a reduction in representational score occurred that correlated with the number of genes that had been swapped. However, the splicing signals were still statistically significant when the selected group contained 75 or 50% introns. This suggests that the effectiveness of this method is dependent on having at least half of the instances of a given element in the gene group. This, of course, may vary depending on several factors, such as gene group size and element size and distribution. In general, having more instances of a particular element in the group of interest will produce better results.
Figure 6.
Effect of group misselection simulation in groups with some intron-containing genes. As described in the text, selection of inappropriate gene groups was simulated by swapping random numbers of genes without introns with genes with introns. Plots and experiments are as in Figure 2. Sequences are indicated for pertinent oligomers with text and arrows. Error bars indicate standard deviations from 10 independent experiments. All data presented are from analysis of 6 nt oligomers in groups of 225 genes. (A) Average values from analysis of groups with 167 genes with introns and 58 without introns (75% introns in set). (B) Average values from analysis of groups with 113 genes with introns and 112 without introns (50% introns in set). (C) Average values from analysis of groups with 57 genes with introns and 168 without introns (25% introns in set).
Implications for gene group selection
The results detailed above and summarized in Table 1 suggest a strategy for selecting gene groups from the results of an expression profiling experiment. First, since large groups of genes do not appear to impair element detection, at least up to a biologically relevant ceiling value, inclusion of extraneous (element-lacking) genes is not a primary concern. Furthermore, small groups can have a negative effect on element detection and failing to include element-containing genes in the group does have a large detrimental impact. This suggests that an effective strategy would be to analyze the group consisting of those genes having and potentially having the expression profile of interest. If this results in a group that is too large, genes that fit the profile most poorly should be excluded, until the group is of a suitable size.
Table 1. Effects of changes in gene group size and composition on RS and GWO values.
| Change in gene group | Effect on RS | Effect on GWO | Other effects |
|---|---|---|---|
| Reduced number of genes (Fig. 4) | Minimal | None | Increase in statistical cut-off levels |
| Increased number of genes (Fig. 5) | Decrease | Decrease | Increased computing time and memory usage |
| Imperfect gene selection (Fig. 6) | Decrease | Decrease |
This table summarizes the effects of changes in gene groups on RS and GWO values, as described in the text.
Since the issue of gene group selection is of paramount importance in using this method, we have chosen to present a hypothetical situation and explain how the strategy described above could be applied. While we have chosen an example that involves selecting genes from an expression profile, it should be emphasized that these guidelines are also applicable to other situations as well (e.g. selection based on function). Consider an expression profile comparing a wild-type population of yeast cells to a population containing a mutation in an RNA-binding protein that is known to stimulate the degradation of some, but not all, mRNAs. Obviously, the population of mutant cells is predicted to have increased levels of some messages. Further, some of the mRNAs with increased levels are predicted to share a sequence element that directs degradation. Following the strategy outlined above, one approach in this case would be to split the genes into three sets: significantly increased (>2-fold), possibly increased (1.5- to 2-fold) and unaffected genes. The initial gene group would be composed of both significantly and possibly increased genes. If that resulted in a gene group that was too large, the genes with the lowest increases could be removed from the gene group. A second approach would be to group the significantly affected genes for analysis and use only the unaffected genes for normalization purposes, rather than the whole genome. Once putative elements were identified from the significantly affected genes, the possibly affected genes could be searched for instances of the putative elements. A positive correlation between element distribution and/or consensus and the mRNA level increase would be a strong indication that the element had a role in the mRNA degradation process being studied.
Identification of a putative cis-acting element in the 3′-UTR of nuclear genes encoding mitochrondrial proteins
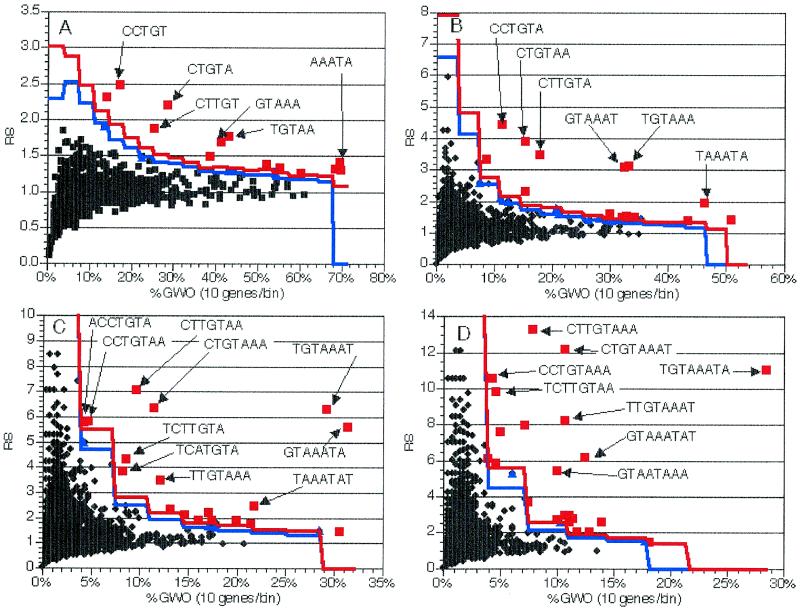
To determine if the method described and characterized above would be effective at identifying unknown potential sequence elements, we performed several experiments on a group of 281 nuclear genes that encode mitochondrial proteins, which were obtained by querying the Yeast Protein Database (9). These genes are of interest because there is experimental data suggesting that the 3′-UTR of these mRNAs may function to target the mRNA to the surface of the mitochondrion and thereby facilitate import of the protein into this organelle (21–25). Examination of the distributions of oligomers from 5 to 8 nt long in the 5′-UTR and coding region sequences (spliced and unspliced) of this group failed to identify any candidate elements (data not shown). However, when the 3′-UTR sequences (+1 to +150 relative to the stop codon) of these genes were examined for the distribution of 6 nt oligomers, a number of partially overlapping sequences were identified as significant (Fig. 7B). Further examination revealed that the components of this composite sequence (CYTGTAAATA, where Y is C or T) also had high RS and GWO values in the 5, 7 and 8 nt oligomer distributions (Fig. 7), making this sequence a strong candidate for being a cis-acting element affecting the function of this class of mRNAs. Additionally, analysis at different oligomer lengths suggests that this 10 nt sequence is the ‘core’ of this element, in terms of detection of overlapping oligomers offset by a single nucleotide. We were not able to detect any significant partially conserved flanking sequence, possibly due to the fact that longer (>8 nt) oligomers were not examined.
The CYTGTAAATA sequence occurs 169 times in the ~14 Mb yeast genome, with 58 of these occurrences being in the ~1 Mb that we termed 3′-UTR sequences (+1 to +150 of each stop codon). When the list of genes containing this sequence is examined, a biased distribution can clearly be detected. The 58 3′-UTR occurrences correspond to 58 genes. Of these 58, 38 are known to have functions in mitochondrial metabolism and four others have sequence features suggestive of such a role (Table 2). Fifteen of the remaining genes have unknown functions. The only gene with a known non-mitochondrial function is CAF17, a component of the CCR4 transcription factor complex (C.Denis, personal communication). Interestingly, mutations of this gene lead to loss of mitochondrial function, suggesting that there is indeed some functional link to mitochondrial function for this gene as well (C.Denis, personal communication). Of the 45 genes with known or putative mitochondrial functions, 35 (77.7%) are involved in post-transcriptional steps of mitochondrial gene expression (primarily translation, but also RNA splicing and degradation and protein complex assembly). Of the 58 genes, only 32 (55.1%) were present in the original selected gene group in this experiment and these 32 comprised only 11.4% of the group of genes analyzed.
Table 2. Genes containing the CYTGTAAATA element.
In the ‘In gene group’ column, Y indicates that the gene was present in the group of 281 genes that were counted and scored, while N indicates that the gene was not present (see Fig. 7). In the ‘Mito function’ column, M indicates known mitochondrial function, P indicates probable mitochondrial function based on sequence similarities and ? indicates unknown function (also see text). In the ‘Gene exp. Function’ column, G indicates proteins that function in some phase of mitochondrial gene expression, such as mRNA splicing, translation, mRNA degradation or protein complex assembly, while a blank indicates that the gene has no known function in gene expression. Gene functions are from the gene table file obtained from the Saccharomyces Genome Database (see Materials and Methods for details) and Moser et al. (26), for YCL036W.
We hypothesized that if this element was bipartite, or composed of two conserved regions separated by a non-conserved region, we would detect the second half of the element as a series of highly significantly over-represented oligomers when the 58 genes were analyzed as a group. However, no new significantly over-represented oligomers were found (data not shown) when oligomer representation in the 3′-UTR sequences of the 58 genes was examined, suggesting that this is not a bipartite element. Possible functions for this element include the coordination of gene expression of this family of genes and/or functioning in the localization of these mRNAs to the mitochondrial surface.
These results support several conclusions. First, relying solely on functional annotations in sequence databases is unlikely to isolate all known genes of a particular functional subclass, meaning that additional care must be taken when assembling gene groups based on gene function. Second, the above results indicate that the performance of our method is robust, as predicted by the experiments performed with the genes with introns group (see above and Figs 4–6). Finally, once a putative element has been identified with our method, listing all the genes in the genome that contain it can be a useful check on the ‘correctness’ of the candidate element. Additionally, finding a putative element in a gene with no known function may provide hints as to what processes the encoded protein participates in in the cell.
Future improvements
There are several potential improvements that could be made to our method. The Perl source code for oligomer counting and scoring in this manuscript is available (http://www.mcb.arizona.edu/Parker/ ) and has been released under the GNU Public License (GPL, http://www.gnu.org/copyleft/gpl.html ), which will enable these improvements and other modifications to be made by members of the scientific community as well as our laboratory.
One improvement would be to use a more accurate set of sequences with regards to mRNA structure. Ideally, whole mRNA or promoter sequences (depending on the interests of the researchers) of each gene would be used, to maximize the biological relevance of the search. Unfortunately, given the number of mapped mRNA 5′- and 3′-ends in S.cerevisiae, it is currently easier to check the locations of putative elements versus mapped ends after the putative element has been identified. Another improvement in biological relevance could be achieved by incorporating the results of a wild-type expression profile, so that only the genes being expressed in the relevant transcriptome are searched.
The core oligomer counting algorithm in our method is quite fast. On a modern desktop workstation, the 6 nt oligomers in a set of 100 genes can be counted in well under 1 min. However, repeatedly performing that counting step, selecting appropriate random sets for statistical purposes and counting multiple oligomer lengths means that the entire experiment can take 6 h or more. Reducing this time is a prerequisite if an interactive web-based version of this method is to be deployed. A potentially productive strategy would be to identify several stereotypical gene group sizes (e.g. 50, 100 and 150 genes) and pre-calculate the significance cut-offs for them. This would allow very rapid ‘prototyping’ of gene groups. Those elements identified in this initial step could then be counted with more rigorous statistical methods, which would also be faster since only certain oligomers would need to be counted. Additionally, passing the output oligomers through a contig assembly program could facilitate detection of the longest shared overlapping sequences from a given group of genes. Finally, we would be remiss if we did not acknowledge that many elements are recognized on the basis of secondary structure (e.g. stem–loops). It should be relatively easy to modify the oligomer counting section of our method to instead look for potential stem-forming regions, the distribution of which can then be examined in a fashion analogous to that described here for oligomers.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank the members of the Parker laboratory for support and many helpful discussions. The manuscript was much improved by the critical readings of Drs Carol Dieckmann, David Mount, Bruce Patterson and Sam Ward. J.S.J.A. gratefully thanks Randal L. Schwartz, Tom Christiansen and Larry Wall for the Llama and the Camel. This work was supported by a grant to R.P. from the National Institutes of Health (GM45443).
REFERENCES
- 1.Proudfoot N.J. and Brownlee,G.G. (1976) Nature, 263, 211–214. [DOI] [PubMed] [Google Scholar]
- 2.Cherry J.M., Ball,C., Weng,S., Juvik,G., Schmidt,R., Adler,C., Dunn,B., Dwight,S., Riles,L., Mortimer,R.K. and Botstein,D. (1997) Nature, 387, 67–73. [PMC free article] [PubMed] [Google Scholar]
- 3.The C. elegans Sequencing Consortium (1998) Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 5.Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.,Jr, Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- 6.Tavazoie S., Hughes,J.D., Campbell,M.J., Cho,R.J. and Church,G.M. (1999) Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- 7.Holstege F.C.P., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 8.Cherry J.M., Adler,C., Ball,C., Chervitz,S.A., Dwight,S.S., Hester,E.T., Jia,Y., Juvik,G., Roe,T., Schroeder,M., Weng,S. and Botstein,D. (1998) Nucleic Acids Res., 26, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodges P.E., McKee,A.H.Z., Davis,B.P., Payne,W.E. and Garrels,J.I. (1999) Nucleic Acids Res., 27, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staden R. (1989) CABIOS, 5, 293–298. [DOI] [PubMed] [Google Scholar]
- 11.Ponomarenko M.P., Ponomarenko,J.V., Frolov,A.S., Podkolodny,N.L., Savinkova,L.K., Kolchanov,N.A. and Overton,G.C. (1999) Bioinformatics, 15, 687–703. [DOI] [PubMed] [Google Scholar]
- 12.van Helden J., André,B. and Collado-Vides,J.J. (1998) J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 13.Dandekar T., Beyer,K., Bork,P., Kenealy,M., Pantopolous,K., Hentze,M., Sonntag-Buck,V., Flouriot,G., Gannon,F., Keller,W. and Schreiber,S. (1998) Bioinformatics, 14, 271–278. [DOI] [PubMed] [Google Scholar]
- 14.Thieffry D., Salgado,H., Huerta,A.M. and Collado-Vides,J. (1998) Bioinformatics, 14, 391–400. [DOI] [PubMed] [Google Scholar]
- 15.Wagner A. (1998) Genomics, 50, 293–295. [DOI] [PubMed] [Google Scholar]
- 16.Wolfsberg T.G., Gabrielian,A.E., Campbell,M.J., Cho,R.J., Spouge,J.L. and Landsman,D. (1999) Genome Res., 9, 775–792. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M.Q. (1999) Genome Res., 9, 681–688. [PubMed] [Google Scholar]
- 18.Doerge R.W. and Churchill,G.A. (1996) Genetics, 142, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson B. and Guthrie,C. (1991) Cell, 64, 181–187. [DOI] [PubMed] [Google Scholar]
- 20.Spingola M., Grate,L., Haussler,D. and Ares,M.,Jr (1999) RNA, 5, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoladek T., Vaduva,G., Hunter,L.A., Boguta,M., Go,B.D., Martin,N.C. and Hopper,A.K. (1995) Mol. Cell. Biol., 15, 6884–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lithgow T., Cuezva,J.M. and Silver,P.A. (1997) Trends Biochem. Sci., 22, 110–113. [DOI] [PubMed] [Google Scholar]
- 23.Preiss T. and Lightowlers,R.N. (1993) J. Biol. Chem., 268, 10659–10667. [PubMed] [Google Scholar]
- 24.Preiss T., Hall,A.G. and Lightowlers,R.N. (1993) J. Biol. Chem., 268, 24523–24526. [PubMed] [Google Scholar]
- 25.Lightowlers R.N., Sang,A.E., Preiss,T. and Chrzanowska-Lightowlers,Z.M.A. (1996) Biochem. Soc. Trans., 24, 527–531. [DOI] [PubMed] [Google Scholar]
- 26.Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]