Abstract
The RecQ helicase superfamily has been implicated in DNA repair and recombination. At least five human RecQ-related genes exist: RecQ1, BLM, WRN, RecQ4 and RecQ5. Mutations in BLM, WRN and RecQ4 are associated with Bloom, Werner and Rothmund-Thomson syndromes, respectively, involving a predisposition to malignancies and a cellular phenotype that includes increased chromosome instability. RecQ5 is small, containing only a core part of the RecQ helicase, but three isomer transcripts code for small RecQ5α (corresponding to the original RecQ5 with 410 amino acids), new large RecQ5β (991 amino acids) and small RecQ5γ (435 amino acids) proteins that contain the core helicase motifs. By determining the genomic structure, we found that the three isoforms are generated by differential splicing from the RecQ5 gene that contains at least 19 exons. Northern blot analysis using a RecQ5β-specific probe indicates that RecQ5β mRNA is expressed strongly in the testis. Immunocytochemical staining of three N-terminally tagged RecQ5 isomers expressed in 293EBNA cells showed that RecQ5β migrates to the nucleus and exists exclusively in the nucleoplasm, while the small RecQ5α and RecQ5γ proteins stay in the cytoplasm. Immunoprecipitation and an extended cytochemical experiment suggested that the nucleoplasmic RecQ5β, like yeast Sgs1 DNA helicase, binds to topoisomerases 3α and 3β, but not to topoisomerase 1. These results predict that RecQ5β may have an important role in DNA metabolism and may also be related to a distinct genetic disease.
INTRODUCTION
DNA helicases have important roles in cellular DNA events, such as replication, recombination, repair and transcription, by unwinding the duplex DNA (1–3). Multiple DNA helicase families with seven consensus motifs have been found and members within each helicase family also share sequence homologies between motifs (4). The RecQ helicase family includes helicases that have extensive amino acid sequence homologies to the Escherichia coli DNA helicase RecQ implicated in double-strand break repair and suppression of illegitimate recombination (5,6). The human RecQ DNA helicase gene family has five members, RecQ1 (also referred to as RecQL), BLM, WRN, RecQ4 and RecQ5, located on human chromosomes 12p12, 15q26.1, 8p12–11.2, 8q24.3 and 17q25.2–25.3, respectively (7–10). Mutations in BLM, WRN and RecQ4 cause Bloom syndrome (BS), Werner syndrome (WS) and Rothmund-Thomson syndrome (RTS), respectively (8,9,11), genetic disorders that increase the rate of generation of genetic instability in patient cells, resulting in a predisposition to cancer and premature aging (12).
RecQ-type helicases from other organisms are SGS1 of Saccharomyces cerevisiae and rqh1+ of Schizosaccharomyces pombe (13,14). Like E.coli, these unicellular organisms contain a single species of RecQ-type helicase. The Sgs1 protein exists in the nucleolus when the cells are young and migrates to the nucleoplasm as the cells senesce and the nucleolus fragments (15). Mutation of SGS1 suppresses the slow growth phenotype of mutant yeast cells with a mutation in the topoisomerase 3 gene and causes missegregation of the chromosome during meiosis and mitosis (13,16). Biochemically, the Sgs1 protein binds to topoisomerases 2 and 3 (13,16). The rqh1+ gene of S.pombe was cloned by complementation of the UV sensitivity of a rqh1-h2 mutant; rqh1– cells arrested by hydroxyurea in S phase are unrecoverable because of high recombination (14).
Human WRN, BLM and RecQ4 DNA helicases and their disease-causing defective mutations have been extensively characterized by us and others (8,9,11,12). The DNA helicase RecQ5, however, has not been characterized for its gene structure, biochemical nature of the encoded helicase, subcellular localization nor the disease caused by mutation. Our previous phylogenetic study (10) on the sequence of the helicase domain indicated that the product of the RecQ5 gene is evolutionarily close to the small helicase RecQ1, which is characterized as a human progenitor of the RecQ family helicases (7), but has no known relation to human disease. We noted that two size classes of the RecQ helicase family exist, large and small helicases: the large helicases are BLM, WRN and RecQ4 in the nucleus, which contain 1417, 1432 and 1208 amino acids, respectively (8–10); the small helicases are RecQ1 and RecQ5, which contain 649 and 410 amino acids, respectively (7,10).
Extended studies have been mostly with the large helicase members of the RecQ family whose mutation causes disease phenotypes in humans. We and others found that: (i) all three helicases exist in the nucleus (17–19); (ii) their mutation causes the genomes of patient cells to become cytogenetically unstable, but differentially: defective BLM increases the rate of sister chromatid exchange (20), while defective WRN results in variegated translocation mosaicism (21) and defective RecQ4 (RTS) causes trisomerization, although a limited number of case reports exist (22); (iii) notable down-regulation of mRNAs and helicase proteins occurs in patient cells (19,23,24), due perhaps to early termination codon-mediated regulation (25); (iv) only WRN contains exonuclease activity in its N-terminus, whereas its 3′→5′ or 5′→3′ directionality remains to be unequivocally defined (26–29); (v) WRN unwinds DNA/RNA heteroduplexes in addition to DNA (30); (vi) intact BLM and WRN partially suppress hyper-recombination in yeast sgs1 mutant cells (31). These findings suggest that the three disease-causing helicases have different functions in cellular DNA metabolism that cannot be complemented by each other, although the molecular mechanisms behind each helicase reaction need to be clarified.
In this paper we describe the genomic structure of RecQ5, a candidate for the fourth disease-causing RecQ-type helicase gene, regarding new cDNA genes generated by differential splicing and the biochemical nature of proteins coded for by the three gene isoforms. The RecQ5 reported previously by us (10) corresponds to RecQ5α in this paper. Two newly identified isomer genes are referred to here as RecQ5β and RecQ5γ. During the preparation of this manuscript, Sekelsky et al. (32) reported that the Drosophila and human RecQ5 cDNA genes exist in different isoforms generated by alternative splicing. Particularly, three Drosophila isoforms, dRecQ5a–c, which they described in their paper, are consistent in size with the findings of the human isoforms in this study and Drosophila RecQ5a corresponds to human RecQ5β.
MATERIALS AND METHODS
Isolation of human RecQ5 isoform cDNAs
A human testis cDNA library constructed with the pAP3-neo plasmid was purchased from Takara (Kyoto, Japan). DNA from E.coli transformants (~1 × 106 clones) made with recombinant plasmids grown on nitrocellulose filters (Millipore, Japan) was replicated on nylon membranes (Pall, UK) and screened by hybrizidation using a 1.3 kb cDNA fragment covering the open reading frame (ORF) of human RecQ5 as probe. In brief, membranes were hybridized with an [α-32P]dCTP-labeled probe DNA in 5× [0.15 M NaCl, 0.15 M sodium citrate buffer, pH 7.0, (SSC)] containing 10× Denhardt’s solution at 65°C for 20 h. The membranes were washed at 65°C in 0.5× SSC, 0.1% SDS, followed by further washing with 0.1× SSC, 0.1% SDS solution. The membranes were exposed to X-ray films (Kodak) with intensifying screens at –70°C overnight. Colonies on the original nitrocellulose filters corresponding to positive signals were selected. After repeated hybridization, five independent clones were selected that contained the RecQ5α, β and γ cDNAs of almost full size. The sequences of cDNAs were determined by PCR-based cycle sequencing using an automated DNA sequencer (Applied Biosystems).
Determination of the genomic structure of the human RecQ5 gene
Two P1 clones, 15033 and 21570, that partially overlap and cover the contour length of the full-sized RecQ5 gene, were obtained from Genome Systems Inc. (St Louis, MO) after screening using a PCR-based method with two primer sets, 5′-GAGCAGCCTTGTGTTTAGACCTGG-3′/5′-ATCCCCC-ATGTCCAATGTGTCTGG-3′ and 5′-GATATAAGATTGC-GTGGGTTCTGC-3′/5′-CGTGGTCCGCCCAAGAATTAAA-GG-3′, corresponding, respectively, to the 5′- and 3′-untranslated regions of the RecQ5α gene. The exon–intron boundary sequences of the RecQ5 genome were determined by sequencing the P1 DNAs directly using primers prepared from the sequences of the RecQ5 isoform cDNAs.
Northern blot analysis
The expression of RecQ5β mRNA was studied using multiple tissue northern blot analysis (Clontech). The filters were hybridized with an [α-32P]dCTP-labeled RecQ5β-specific EcoRV–XhoI cDNA fragment, corresponding to exons 14–19, prepared from a 6× hemagglutinin (HA)–RecQ5β plasmid at 42°C overnight in 5× SSPE buffer containing 50% formamide, 2% SDS, 10× Denhardt’s solution and 100 µg/ml depurinated salmon sperm DNA. Washing was under highly stringent conditions that were essentially the same as described by Kitao et al. (10): three times with 2× SSC, 0.1% SDS at room temperature and once with 0.2× SSC, 0.1% SDS for 30 min at 65°C. The filters were exposed to X-ray films that were analyzed using a BAS1500 system (Fuji film).
Construction of epitope-tagged expression plasmids
RecQ5α, β or γ cDNA fragments were excised with EcoRI and NotI from pAP3-neo vector DNAs and subcloned into pBluescript II KS+. These plasmids were modified in the 5′-flanking sequences of the first ATG of the RecQ5 gene to isolate the intact ORF using NheI and XhoI digestions. The NheI- and XhoI-digested DNA fragments derived from RecQ5α, β and γ cDNAs were subcloned into the NheI–XhoI sites of the pcDNA3 mammalian expression plasmid containing a 6xHA epitope sequence. The 6xHA epitope was introduced by repeated ligation of oligonucleotides encoding the epitopic amino acid sequence to the 5′ NheI site. To construct the 6xHA–RecQ5βc expression plasmid, the 6xHA–RecQ5β plasmid DNA was digested with NheI to remove the 5′-region upstream of an endogenous NheI site and was self-ligated to code for a C-terminal 246 amino acid polypeptide of RecQ5β.
The cDNAs of human topoisomerases 1, 3α and 3β comprising the full ORF were amplified by PCR using primer sets 5′-TATTCTAGAATGAGTGGGGACCACCTCCACA-ACG-3′/5′-TATGTCGACGCTAAAACTCATAGTCTTCA-TCAGCC-3′ for topoisomerase 1, 5′-TATACTAGTATG-ATCTTTCCTGTCGCCCGCTACG-3′/5′-TATCTCGAGGC-TCATCTGTTCTGAGGACAAAAGG-3′ for topoisomerase 3α and 5′-TATGCTAGCATGAAGACTGTGCTCATGGTTGCTG-3′/5′-TATCTCGAGGGACAGGGTC-ATCATACA-AAGTAGG-3′ for topoisomerase 3β. The amplified cDNA fragments were digested with restriction enzymes whose sites were tagged with primers: XbaI and SalI for topoisomerase 1, SpeI and XhoI for topoisomerase 3α and NheI and XhoI for topoisomerase 3β. The fragments were subcloned downstream of a sequence encoding the N-terminally tagged Flag epitope in a pcDNA3-based vector and were sequenced.
Transfection and expression in 293EBNA cells
Human 293EBNA cells (Invitrogen) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum. The cells were cultured at 37°C in an incubator with 5% CO2. Confluent cells (70%) cultured in 60 mm dishes were transfected with 3 µg of the expression plasmid DNA using Lipofectamine (Gibco BRL) and the transfected cells were grown for 48 h at 37°C.
Immunoprecipitation and western blot analysis
After 48 h transfection in a 60 mm dish, the cells were harvested, washed with phosphate-bufferred saline (PBS) and lysed in 300 µl of lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride). The cell lysate was obtained by centrifugation at 15 000 r.p.m. for 30 min at 4°C. For immunoprecipitation, the cell lysate was incubated with 20 µl of M2 agarose (Eastman Kodak) overnight at 4°C. The protein-bound agarose beads were washed four times with PBS containing 0.05% Tween 20 and the proteins eluted with 20 µl of lysis buffer containing 100 µg/ml of Flag peptide. The proteins were fractionated using SDS–PAGE, transferred to polyvinylidene difluoride membranes and treated with PBS containing 5% (w/v) skimmed milk. The membranes were then incubated for 60 min at 4°C with anti-HA rabbit polyclonal IgG (Santa Cruz) or anti-Flag M2 mouse monoclonal IgG (Eastman Kodak), washed and then incubated for 60 min at 4°C with horse radish peroxidase (HRP)-conjugated anti-rabbit IgG (Amersham) or HRP-conjugated anti-mouse IgG (Dako, Denmark). After incubation, the membranes were washed and developed using ECL™ (Amersham Life Science, UK).
Immunocytochemistry
The transfected and harvested cells (5 × 104) were washed with PBS. The cells were placed on a glass slide (s8111; Matsunami) using the cytospin method and fixed with 3.7% formaldehyde in PBS. After washing with PBS, the cells were treated with 3% (w/v) skimmed milk in PBS at room temperature and incubated with the first antibody overnight at 4°C. After another washing with PBS, the cells were further incubated with the second antibody for 1 h at room temperature. For subcellular localization of RecQ5 isoforms, 2.5 µg/ml of rabbit anti-HA IgG and biotinylated anti-rabbit IgG were used as first and second antibodies, respectively. To double stain HA- and Flag-tagged proteins, 2.5 µg/ml of rabbit anti-HA IgG and 10 µg/ml of mouse anti-Flag IgG were used as first antibodies and biotinylated anti-rabbit IgG and fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG were used as second antibodies. After washing again with PBS, the cells were incubated with streptavidin–FITC or streptavidin–Texas red for 30 min at room temperature and counterstained with DAPI [5-(N-2,3-dihydroxypropylactamido)-2,4,6-triiodo-N,N′-(bis-2,3- dihydroxypropyl)isophthalamide]. Fluorescent images were visualized using a Nikon Optiphot-2 microscope fitted with a 60× Nikon PlanApo oil immersion objective and a double-pass filter set for fluorescein/DAPI. Images of double labeled cells were produced using a confocal laser scanning microscope (Fluoview, Olympus).
RESULTS
Characterization of the RecQ5 gene and identification of three RecQ5 isomer transcripts generated by differential exon use
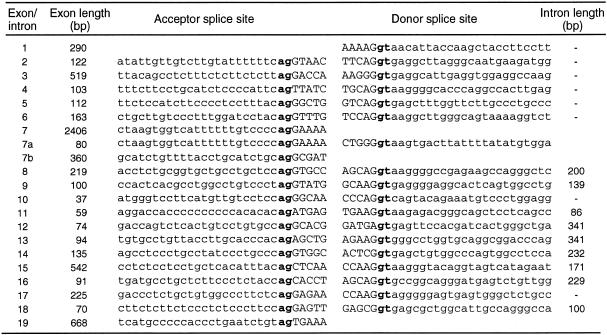
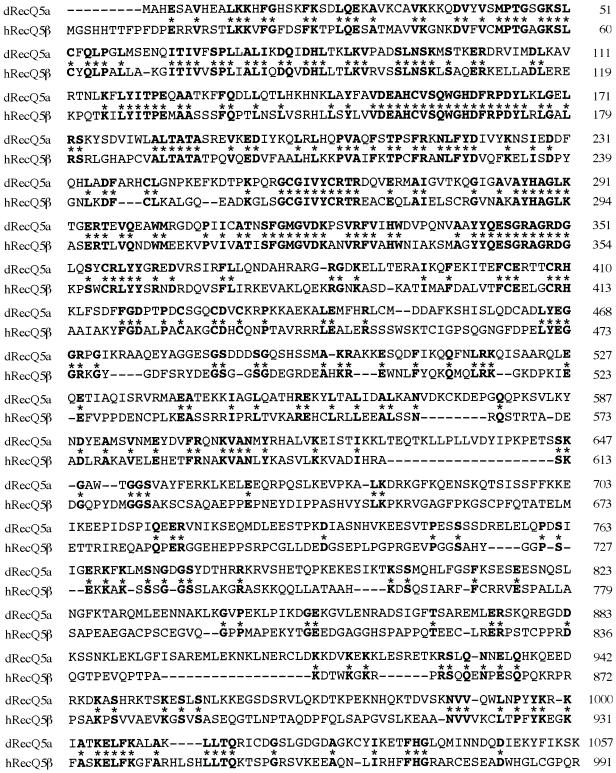
Characterization of the RecQ5 gene using P1 phage DNAs showed that it contained at least 19 exons (Fig. 1A). Table 1 shows the nucleotide sequences at the exon–intron junctions. Previously, we showed that RecQ5 mRNA consists of two transcripts having different sizes of 3.6 and 3.8 kb (10). Our further studies that cloned new RecQ5 cDNAs from human testis mRNAs and determined the sequences, showed that at least three different transcripts code for RecQ5 DNA helicases of different sizes. The original RecQ5 gene (referred to hereafter as RecQ5α) that we reported previously (10) was the largest subset of transcripts (3715 bases) among the three RecQ5 transcript species, consisting of exons 1–7, but encoded a small polypeptide RecQ5α of 410 amino acids (Fig. 1a and b). The second largest transcript, RecQ5β (3703 bases), formed by exons 1–7a and 8–19, coded for the largest polypeptide RecQ5β of 991 amino acids (Fig. 1a and b). The third transcript, RecQ5γ, was the smallest in size (1749 bases), formed by exons 1–7a and exon 7b and coded for the small polypeptide RecQ5γ (Fig. 1a and b). The protein structures of all three polypeptides predicted from the defined genomic sequence share seven motifs conserved for DNA helicases (darkened areas in Fig. 1b). In contrast, the largest polypeptide RecQ5β contained a large C-terminal region that includes a domain homologous to the non-helicase domain of the E.coli RecQ DNA helicase (hatched region in Fig. 1b). Recently, Sekelsky et al. (32) cloned the RecQ5 homolog gene dRecQ5a in Drosophila that encodes a polypeptide of an equivalent size (1057 amino acids) to RecQ5β. A comparison of the amino acids sequences of dRecQ5a and human RecQ5β showed that these two polypeptides share considerable homology, particularly in the N-terminal helicase domain and the C-terminal region (Fig. 2).
Figure 1.
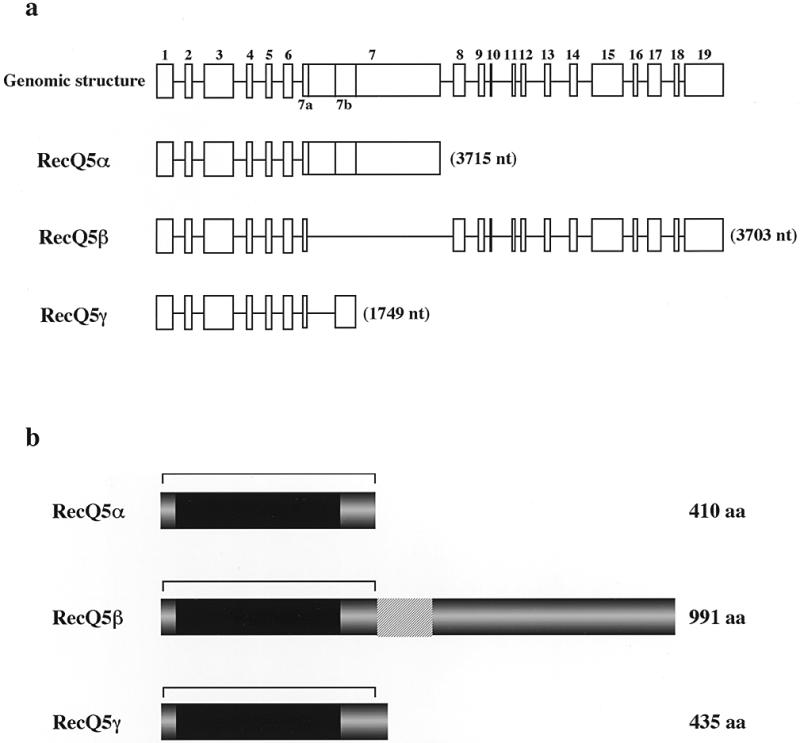
Structures of RecQ5 genes and the encoded RecQ5 proteins. (a) Genomic structure of the RecQ5 gene and the exon uses of three RecQ5 mRNA isomers, RecQ5α, RecQ5β and RecQ5γ. (b) Schematic representation of three RecQ5 helicase isomers, RecQ5α, RecQ5β and RecQ5γ. The sizes of each protein are given on the right. The darkened areas indicate the locations of the helicase domains shared by the RecQ helicase family. The hatched areas indicate the extended homologous region to E.coli RecQ helicase. The regions indicated by bars indicate identical amino acid sequences among the three isoforms.
Table 1. Exon–intron organization of the human RecQ5 gene.
Figure 2.
Comparison of the amino acid sequences of Drosophila melanogaster RecQ5a and human RecQ5β. dRecQ5a, Drosophila melanogaster RecQ5a; hRecQ5β, human RecQ5β. The asterisks denote amino acid residues identical between the two proteins.
Northern blot analysis of the RecQ5β mRNA in human tissues
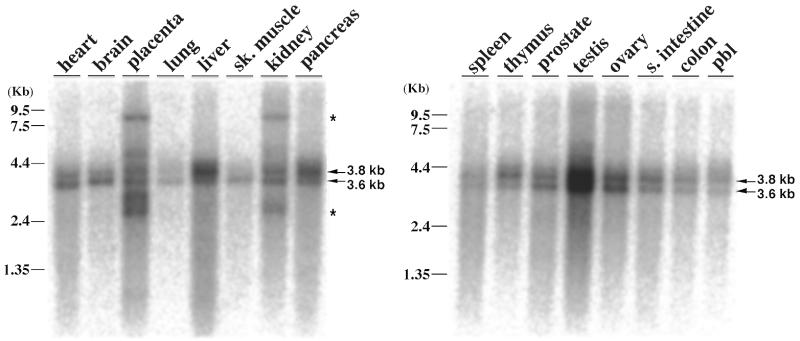
Our previous studies using a DNA probe prepared from the helicase domain of RecQ5α showed that: (i) the profile of RecQ5 expression in human tissues largely resembles that of RecQ1, ubiquitously expressed in all tissues examined, with a notably strong expression in the pancreas and testis, similar to some extent to that of WRN; (ii) at least two major isomer transcripts of 3.6 and 3.8 kb exist (10). To determine whether the expression profile of the new large RecQ5β mRNA differs from that of the original RecQ5 (now renamed RecQ5α), we prepared a DNA probe from the 3′-terminus (exons 14–19) of RecQ5β and carried out northern blot analysis for expression in various tissues under highly stringent conditions (Fig. 3). Again, RecQ5β was expressed in all the tissues and organs examined, with strong expression in the testis. The expression profile was, however, distinct from that of RecQ5α, with reduced expression in the pancreas and increased expression in the kidney. Two major transcripts of 3.6 and 3.8 kb were detected, consistent with our previous findings, but there were other small and large RecQ5β-related RNA species found in the placenta and kidney (indicated by asterisks). These results suggest that the tissue-specific expression of RecQ5β is by selective exon use and perhaps other unidentified RecQ5β-related transcripts exist in the placenta and kidney.
Figure 3.
Multiple tissue northern blot analysis to compare the expression levels of the human RecQ5β gene. A multiple tissue northern blot (Clontech) was prepared with 2 µg/lane poly(A)+ RNA. A DNA fragment containing exons 14–19 of the RecQ5β gene was used for the probe. Arrows show the major species of RecQ5β transcripts of 3.6 and 3.8 kb that were detected in this study and in our previous report (10). Asterisks show the RecQ5β 3′-terminus-related RNA species that were not pursued in this study.
Nuclear localization of RecQ5β helicase but not of RecQ5α and RecQ5γ helicases
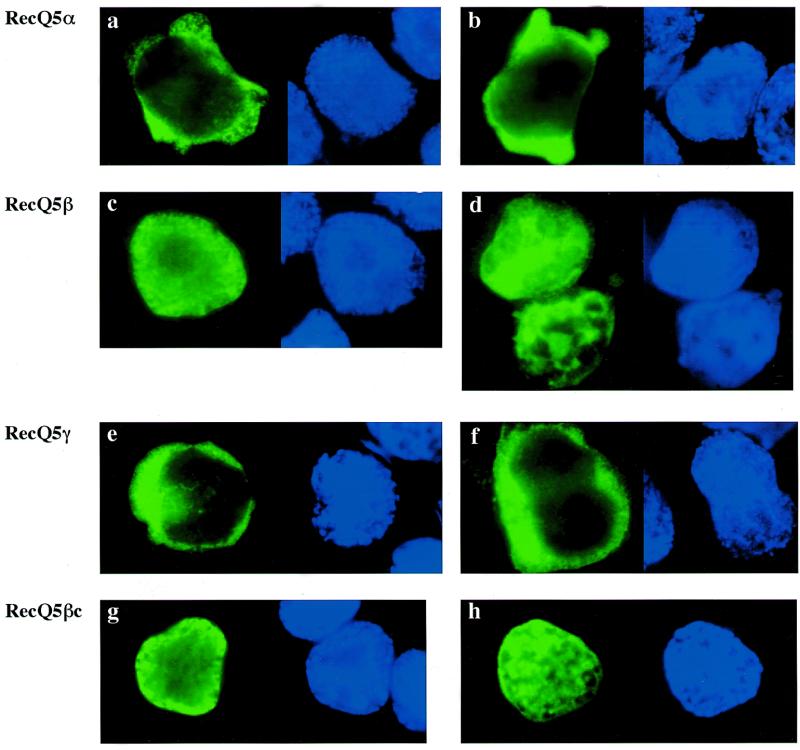
BLM, WRN and RecQ4 helicases migrate to and localize in the nucleus (17–19), while RecQ1 helicase localizes in the nucleus and cytoplasm (33). To understand the subcellular localization of the three RecQ5 helicase isomers, we expressed the genes N-terminally tagged with epitope sequence (YPYDVPDYASL) of influenza virus hemagglutinin (HA) repeated six times in 293EBNA cells. As our preliminary experiments showed that expressed RecQ5β migrates into the nucleus, we also expressed a part of the C-terminus (246 amino acids, referred to as RecQ5βc) to locate the nuclear localization signal (NLS) sequence. Figure 4a shows structures of the expressed proteins. A total of four recombinant proteins were expressed with the sizes confirmed by western blot analysis using a HA-specific antibody (Fig. 4b). The molecular weights of the individual proteins were estimated as 53, 140, 60 and 49 kDa for RecQ5α, RecQ5β, RecQ5γ and RecQ5βc, respectively. The subcellular locations of the expressed proteins were next examined by immunocytochemical staining using the HA-specific antibody and a biotinylated antibody to rabbit immunoglobulin and the expressed proteins were stained green with streptavidin-conjugated FITC (Fig. 5). To locate the position of the nuclei, the same cells were stained with DAPI, which stains duplex DNA blue. The data clearly show that RecQ5α (Fig. 5a and b) and RecQ5γ (Fig. 5e and f) are localized in the cytoplasm. In contrast, RecQ5β migrates to the nucleus (Fig. 5c and d), suggesting that RecQ5β is a nuclear helicase. RecQ5βc also migrated exclusively into the nucleus (Fig. 5g and h), indicating that the NLS of the RecQ5β protein is within this small C-terminal fragment.
Figure 4.
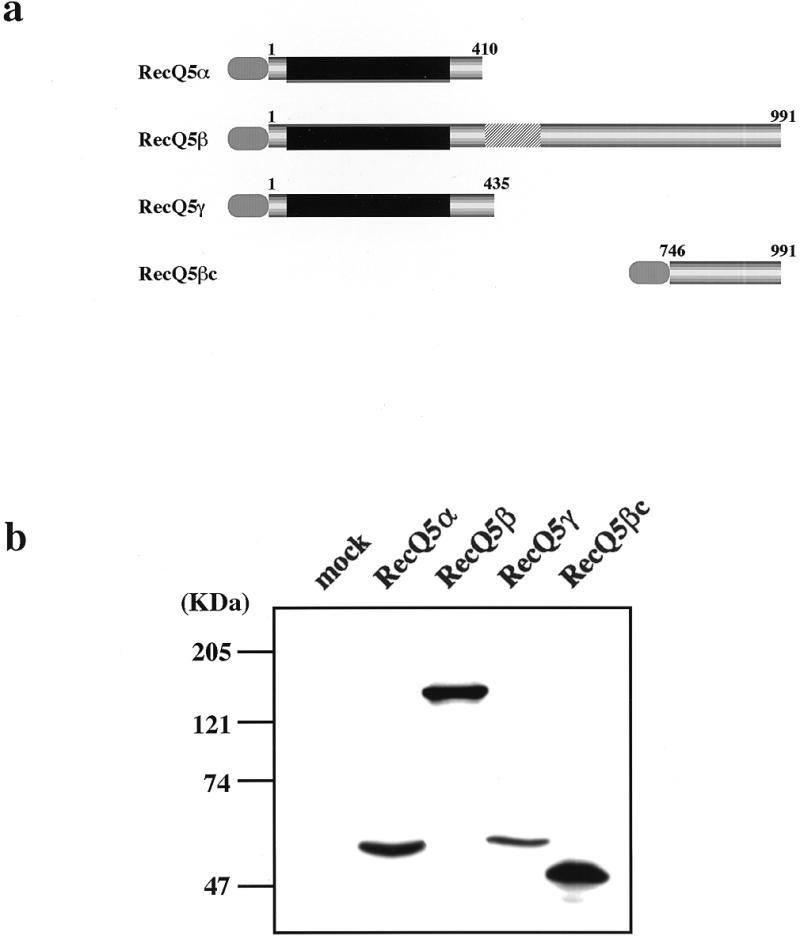
Expression of RecQ5 isoforms in 293EBNA cells. (a) Schematic representations of three recombinant RecQ5 helicase isomers, RecQ5α, RecQ5β and RecQ5γ, all N-terminally tagged with influenza virus 6xHA. RecQ5βc contains an N-terminal 6xHA tag and the C-terminal region of RecQ5β (amino acid residues 746–991). Numbers indicate amino acid residues. (b) Western blot analysis of three RecQ5 helicase isomers and RecQ5βc expressed in 293EBNA cells. Cell lysates prepared from cells transiently transfected with each construct were size fractionated by SDS–PAGE and analyzed by western blot analysis using anti-HA antibody.
Figure 5.
Subcellular localization of RecQ5 isoforms in 293EBNA cells. Each construct was transiently expressed in 293EBNA cells and the cells fixed and stained with anti-HA antibody as shown in Materials and Methods. Localization of the RecQ5 isoform and RecQ5βc proteins are indicated by FITC (green, left panel) and the nuclear positions are shown by DAPI (blue, right panel). (a and b) RecQ5α; (c and d) RecQ5β; (e and f) RecQ5γ; (g and h) RecQ5βc.
RecQ5β protein interacts with topoisomerases 3α and 3β
The high sequence homology to E.coli RecQ DNA helicase and a clear biochemical nature to localize in the nucleus strongly predict that RecQ5β is most probably a helicase involved in DNA unwinding. Although such a DNA helicase activity of the RecQ5β protein remains to be substantiated using the purified protein, we examined whether the expressed RecQ5β protein interacts with type I topoisomerases that cooperate with helicases. S.cerevisiae Sgs1 DNA helicase, for example, binds to topoisomerase 3 and the two enzymes cooperate to suppress hyper-recombination (13), and sgs1 mutant cells have a poor growth rate in a topoisomerase 1-defective background (34). To determine whether RecQ5β is capable of binding to human type I topoisomerases, we carried out a series of co-expression experiments using human topoisomerases 1, 3α and 3β in 293EBNA cells. In this experiment the RecQ5β protein was expressed as a form N-terminally tagged with influenza virus HA (6xHA–RecQ5β). Topoisomerases were expressed individually as N-terminal Flag-tagged forms (Flag–Top1, Flag–Top3α or Flag–Top3β). After co-expression with individual topoisomerases, RecQ5β protein bound to the topoisomerases and co-immunoprecipitated with anti-Flag antibody was analyzed by western blot analysis using anti-HA antibodies. In these co-expression experiments, 6xHA–RecQ5β and the three topoisomerase proteins were expressed at their expected sizes in 293EBNA cells, as measured by western blot analysis using antibodies specific to HA (Fig. 6a) and to Flag (Fig. 6b). Individual topoisomerases in the cell extracts were then immunoprecipitated by anti-Flag antibody-conjugated agarose beads, eluted by Flag peptide and the 6xHA–RecQ5β protein potentially bound to each topoisomerase was analyzed by western blot analysis using anti-HA antibodies. The results clearly show that 6xHA–RecQ5β was co-immunoprecipitated with Flag–Top3α and Flag–Top3β, but not with Flag–Top1 (Fig. 6c), suggesting that RecQ5β helicase binds specifically to topoisomerases 3α and 3β.
Figure 6.
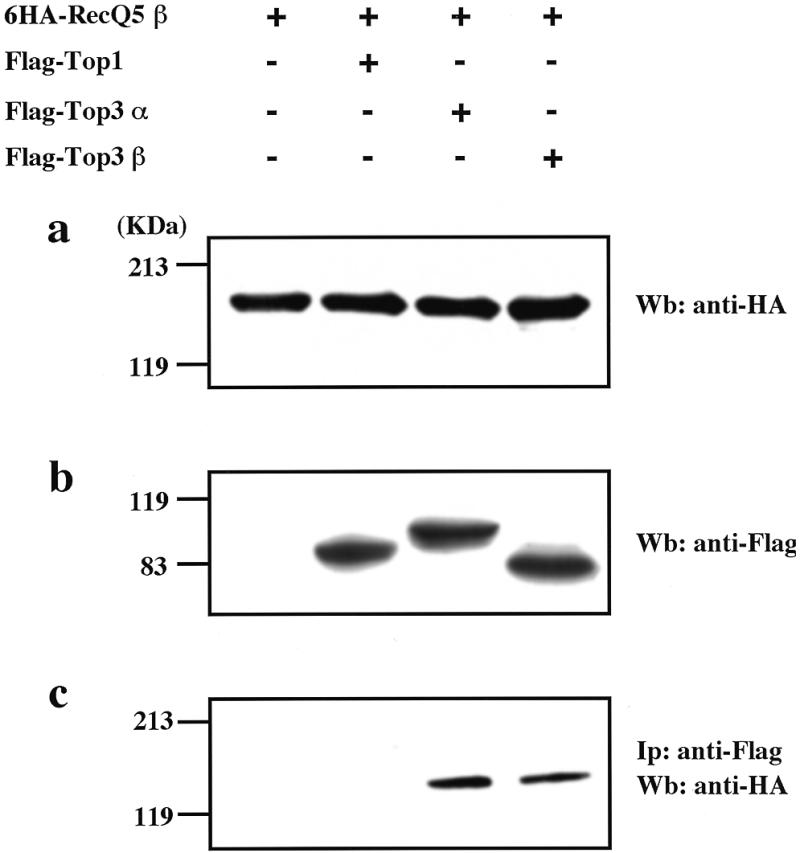
Co-immunoprecipitation of RecQ5β and human topoisomerases 3α and 3β. (a) Expression of 6xHA-tagged RecQ5β protein in 293EBNA cells. 293EBNA cells were transfected with 6xHA–RecQ5β, Flag–Top3α and Flag–Top3β as indicated. Cell lysates were prepared from the cells, size fractionated by SDS–PAGE and analyzed by western blot analysis using anti-HA antibodies. (b) Expression of Flag-tagged type I topoisomerase proteins in 293EBNA cells. Anti-Flag antibodies were used in a western blot analysis. (c) RecQ5β protein co-immunoprecipitated with topoisomerases 3α and 3β. Type I topoisomerases and their binding proteins were precipitated with anti-Flag IgG-conjugated agarose beads and were eluted with Flag peptide. The eluted proteins were size fractionated by SDS–PAGE and analyzed by western blot analysis using anti-HA antibodies.
Co-localization of RecQ5β and topoisomerases 3α and 3β in the nucleoplasm
To confirm that the interactions between RecQ5β and topoisomerases 3α and 3β also occur in cells, we examined the subcellular localization of expressed proteins by immunocytochemical staining using fluorescence-conjugated anti-HA and anti-Flag antibodies. The expressed 6xHA–RecQ5β was visualized as red by staining with Texas red dye and Flag–Top3α and Flag–Top3β proteins were visualized as green by staining with FITC dye (Fig. 7); Figure 7a shows the staining profiles of 6xHA–RecQ5β and Flag–Top3α, and Figure 7b shows those of 6xHA–RecQ5β and Flag–Top3β in the same cells. These data strongly indicate that 6xHA–RecQ5β exists in the nucleoplasm of 293EBNA cells, co-localizing with Flag–Top3α and Flag–Top3β. This apparent co-localization was confirmed by merging the stained profiles of 6xHA–RecQ5β and Flag–Top3α, and 6xHA–RecQ5β and Flag–Top3β, both of which produced matching yellow staining (Fig. 7a and b, right).
Figure 7.
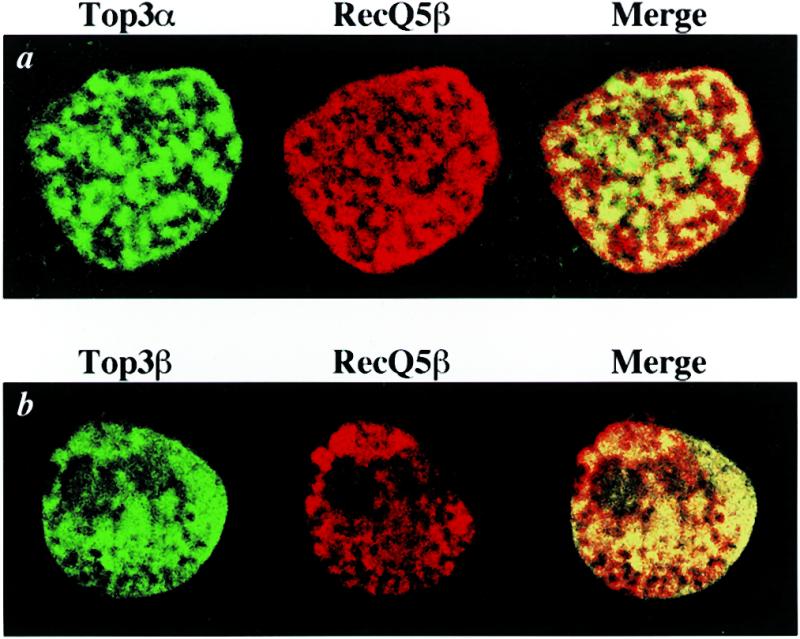
Co-localization of RecQ5β with topoisomerases 3α and 3β. 293EBNA cells were co-transfected with 6×HA–RecQ5β and Flag–Top3α (a) or Flag–Top3β (b), then fixed and stained with anti-HA and anti-Flag antibodies as described in Materials and Methods. Flag–Top3α and Flag–Top3β proteins were detected by FITC (green, left panels) and 6×HA–RecQ5β protein by Texas red (red, center panels). The merged profiles are shown in the right panels. Co-localization of RecQ5β with topoisomerases 3α and 3β is seen in yellow in the right panels.
DISCUSSION
In this paper we have defined the exon–intron structures of the RecQ5 gene, and we show that three RecQ5 helicase isomers, RecQ5α, RecQ5β and RecQ5γ, are generated as a result of differential splicing of the RecQ5 gene transcript. Two of these isomer gene products, RecQ5α and RecQ5γ, are small and localize in the cytoplasm, while RecQ5β migrates into the nucleus and exists in the nucleoplasm, like other human RecQ helicases (Fig. 5). Our amino acid homology search indicated that a potential NLS with the sequence KRPRSQQENPESQPQKRPR exists in the C-terminus (residues 854–872 amino acids) of RecQ5β, which is absent from both RecQ5α and RecQ5γ. A similar bipartite NLS motif comprising two arrays of basic amino acids with a spacer region of any 10 amino acids between them has been found in many nuclear proteins, including nucleoplasmin, p53 and topoisomerases (reviewed in 35). Previously, we identified the NLS as having an array of basic amino acids, RKRKKMPASQRSKRRK (residues 1334–1349) and KRRCFPGSEEICSSSKRSK (residues 1371–1389), in the C-termini of BLM (17) and WRN (18), respectively, and found that most mutations generate truncated helicases lacking the NLS, causing WRN and BLM not to be transported to the nucleus, where the DNA helicases are presumed to function. Importantly, this finding clearly explains why WS and BS patients show a set of similar clinical phenotypes no matter what type of mutation they carry (18). More recently, we showed that WRN helicase can exist not only in the nucleoplasm (36) but also in the nucleolus (19), confirming the previous data of Marciniak et al. (37).
The expression profile of RecQ5β is largely the same as that we previously reported for RecQ5α, although stronger expression of RecQ5β in the testis was noted in this study. These data predict that a mutation in RecQ5β may cause a disease that shows the phenotypes shared by BS, WS and RTS, although whether RecQ5β is a disease-causing gene remains to be investigated. Our studies using immunoprecipitation showed that the expressed RecQ5β binds to topoisomerases 3α and 3β, but not to topoisomerase 1 (Fig. 6). Although the results should be confirmed by the endogenous proteins and their specific antibodies, the data of this study at least support the view that overexpressed RecQ5β shows preferential binding to overexpressed topoisomerases 3α and 3β, similar to yeast Sgs1, which binds to overexpressed topoisomerases 2 and 3 in two-hybrid analyses (13,16). The fact that the expressed RecQ5β failed to bind to overexpressed topoisomerase 1 suggests that the apparent preferential binding to topoisomerases 3α and 3β is unlikely to be mediated simply by DNA. Further immunocytochemical double staining of expressed RecQ5β and topoisomerases 3α or 3β in 293EBNA cells (Fig. 7) was consistent with this conclusion. Gangloff et al. (13) showed that the N-terminus of yeast Sgs1 helicase binds to topoisomerase 3. Whether the interaction between RecQ5β and the two topoisomerases is also mediated by the RecQ5β N-terminus remains to be studied.
Topoisomerases remove positive and/or negative superhelicity from DNA (38). The SV40 T antigen, a viral DNA helicase, binds to topoisomerase 1 (39) and yeast Sgs1 binds to topoisomerases 2 and 3 (13,16), perhaps to cooperatively regulate the superhelicity and relax the distorted DNA structure generated as a result of DNA unwinding. Consistent with this, Harmon et al. (40) recently showed that E.coli RecQ helicase specifically stimulates topoisomerase 3 to fully catenate double-stranded DNA molecules by unwinding a covalently closed DNA, proposing that these two proteins functionally interact and control cellular recombination. In the absence of cooperation between helicase and topoisomerases, an imbalance may occur, yielding potential recombinogenic lesions. It is thus intriguing that RecQ5β is capable of binding to topoisomerases 3α and 3β. This finding strongly supports the view that RecQ5β is probably an active DNA helicase and is involved in nucleoplasmic DNA metabolism, but this remains to be clarified.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by The Organization for Drug ADR Relief (R and D Promotion and Product Review) of the Japanese Government.
REFERENCES
- 1.Waga S. and Stillman,B. (1998) Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen A. and Lucchesi,J.C. (1998) Bioessays, 20, 634–641. [DOI] [PubMed] [Google Scholar]
- 4.Gorbalenya A.E., Koonin,E.V., Donchenko,A.P. and Blinov,V.M. (1989) Nucleic Acids Res., 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kusano K., Sunohara,Y., Takahashi,N., Yoshikura,H. and Kobayashi,I. (1994) Proc. Natl Acad. Sci. USA, 91, 1173–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanada K., Ukita,T., Kohno,Y., Saito,K., Kato,J. and Ikeda,H. (1997) Proc. Natl Acad. Sci. USA, 94, 3860–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seki M., Miyazawa,H., Tada,S., Yanagisawa,J., Yamaoka,T., Hoshino,S., Ozawa,K., Eki,T., Nogami,M., Okumura,K., Taguchi,H., Hanaoka,F. and Enomoto,T. (1994) Nucleic Acids Res., 22, 4566–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 9.Yu C.-E., Oshima,J., Fu,Y.-H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 10.Kitao S., Ohsugi,I., Ichikawa,K., Goto,M., Furuichi,Y. and Shimamoto,A. (1998) Genomics, 54, 443–452. [DOI] [PubMed] [Google Scholar]
- 11.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 12.Lindor N.M., Furuichi,Y., Kitao,S., Shimamoto,A., Arndt,C. and Jalal,S. (2000) Am. J. Med. Genet., 90, 223–228. [DOI] [PubMed] [Google Scholar]
- 13.Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart E., Chapman,C.R., Al-Khodairy,F., Carr,A.M. and Enoch,T. (1997) EMBO J., 16, 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair D.A., Mills,K. and Guarente,L. (1997) Science, 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 16.Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko H., Orii,K.O., Matsui,E., Shimozawa,N., Fukao,T., Matsumoto,T., Shimamoto,A., Furuichi,Y., Hayakawa,S., Kasahara,K. and Kondo,N. (1997) Biochem. Biophys. Res. Commun., 240, 348–353. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto T., Shimamoto,A., Goto,M. and Furuichi,Y. (1997) Nature Genet., 16, 335–336. [DOI] [PubMed] [Google Scholar]
- 19.Kitao S., Lindor,N., Shiratori,M., Furuichi,Y. and Shimamoto,A. (1999) Genomics, 61, 268–276. [DOI] [PubMed] [Google Scholar]
- 20.German J. (1993) Medicine, 72, 393–406. [PubMed] [Google Scholar]
- 21.Martin G.M. (1978) Birth Defects, 14, 5–39. [PubMed] [Google Scholar]
- 22.Lindor N.M., Devries,E.M.G., Michel,V.V., Schad,C.R., Jalal,S.M., Donovan,K.M., Smithson,W.A., Kvols,L.K., Thibodeau,S.N. and Dewald,G.W. (1996) Clin. Genet., 49, 124–129. [DOI] [PubMed] [Google Scholar]
- 23.Yamabe Y., Sugimoto,M., Satoh,M., Suzuki,N., Sugawara,M., Goto,M. and Furuichi,Y. (1997) Biochem. Biophys. Res. Commun., 236, 151–154. [DOI] [PubMed] [Google Scholar]
- 24.Goto M., Yamabe,Y., Shiratori,M., Okada,M., Kawabe,T., Matsumoto,T., Sugimoto,M. and Furuichi,Y. (1999) Hum. Genet., 105, 301–307. [DOI] [PubMed] [Google Scholar]
- 25.Maquat L.E. (1995) RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S., Gray,M.D., Oshima,J., Mian,I.S. and Campisi,J. (1998) Nature Genet., 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen J.C., Gray,M.D., Oshima,J., Kamath-Loeb,A.S., Fry,M. and Loeb,L.A. (1998) J. Biol. Chem., 273, 34139–34144. [DOI] [PubMed] [Google Scholar]
- 28.Kamath-Loeb A.S., Shen,J.C., Loeb,L.A. and Fry,M. (1998) J. Biol. Chem., 273, 34145–34150. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki N., Shiratori,M., Goto,M. and Furuichi,Y. (1999) Nucleic Acids Res., 27, 2361–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki N., Shimamoto,A., Imamura,O., Kuromitsu,J., Kitao,S., Goto,M. and Furuichi,Y. (1997) Nucleic Acids Res., 25, 2973–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamagata K., Kato,J., Shimamoto,A., Goto,M., Furuichi,Y. and Ikeda,H. (1998) Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekelsky J.J., Brodsky,M.H., Rubin,G.M. and Hawley,R.S. (1999) Nucleic Acids Res., 15, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W.S., Seki,M., Yamaoka,T., Seki,T., Tada,S., Katada,T., Fujimoto,H. and Enomoto,T. (1998) Biochim. Biophys. Acta, 1443, 198–202. [DOI] [PubMed] [Google Scholar]
- 34.Lu J., Mullen,J.R., Brill,S., Kleff,S., Romeo,A.M. and Sternglanz,R. (1996) Nature 383, 678–679. [DOI] [PubMed] [Google Scholar]
- 35.Dingwall C. and Laskey,R.A. (1991) Trends Biochem. Sci., 16, 478–481. [DOI] [PubMed] [Google Scholar]
- 36.Shiratori M., Sakamoto,S., Suzuki,N., Tokutake,Y., Kawabe,Y., Enomoto,T., Sugimoto,M., Goto,M., Matsumoto,T. and Furuichi,Y. (1998) J. Cell Biol., 144, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcianiak R.A., Lombard,D.B., Johnson,F.B. and Guarente,L. (1998) Proc. Natl Acad. Sci. USA, 95, 6887–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gangloff S., Lieber,M.R. and Rothstein,R. (1994) Experientia, 50, 261–269. [DOI] [PubMed] [Google Scholar]
- 39.Simmons D.T., Melendy,T., Usher,D. and Stillman,B. (1996) Virology, 222, 365–374. [DOI] [PubMed] [Google Scholar]
- 40.Harmon F.G., DiGate,R.J. and Kowalczykowski,S.C. (1999) Mol. Cell, 3, 611–620. [DOI] [PubMed] [Google Scholar]