Introduction
New advancements in spatial tissue imaging allow for the generation of large datasets that anchor transcriptomic and proteomic expression on histology with high granularity. These highly multiplexed cellular and molecular data provide researchers with an entirely new way to interpret tissue morphology and generate visual clues that may augment existing gold-standard histopathologic interpretation. To best interpret these multimodal data, artificial intelligence methods are indispensable to fuse bright-field histology with diverse spatial -omics methods, including spatial transcriptomics; multiplex fluorescence imaging, including codetection by indexing (CODEX); imaging mass spectrometry; miFISH; and imaging mass cytometry.1–4
Use Case
A 48-year-old White man with hypertension and well-controlled diabetes mellitus but no known kidney disease presented to the emergency department in respiratory distress. He later developed hypotension and pneumonia-related respiratory failure requiring intubation. He cycled through antibiotics before improvement and developed AKI on hospital day 17, requiring KRT. He was discharged on day 31, yet remained dialysis-dependent. A kidney biopsy revealed mild arteriolar hyalinosis and patchy acute tubular necrosis (ATN) with occasional foci of monocytic and lymphocytic infiltrates and occasional mitotic tubular cells. Ten percent of the glomeruli were sclerosed, and <20% of the tubule-interstitium was affected by fibrosis or atrophy. Traces of linear IgG deposits were seen on immunofluorescence. Electron microscopy revealed mild thickening of the glomerular basement membrane without immune complex deposition, consistent with early diabetic changes. After biopsy, the differential diagnosis was (1) pending recovery of ATN, (2) nonrecovery with early signs of CKD, or (3) interstitial nephritis.
The biopsy results did not give insight into recovery or interstitial nephritis and AKI. An approach that integrates molecular analysis may help provide more information for clinicians and pathologists. Specifically, a fused histology and spatial -omics data would identify mitotic tubular cells and their distance to fibrosis, necrosis, and inflammation. CODEX protein immunofluorescence can characterize inflammation in specific immune cells and injury biomarkers in adjacent tubules. Spatial transcriptomics can identify injured tubules and their likelihood of leading to fibrosis. Transcriptomic evidence of receptor ligand interactions can indicate whether damage to epithelial cells arises from nearby immune cells. These technologies provide a comprehensive understanding of biologic processes compared with standard biopsy interpretation.
Select Spatial -Omics Technologies
We focus on spatial transcriptomics (VISIUM) and CODEX as examples of state-of-the-art transcriptomic and multiplex protein imaging modalities because of their ability to register molecular data with bright-field microscopy.
Spatial Transcriptomics
Current spatial transcriptomics technologies localize mRNA expression at the tissue microenvironment, cellular, or subcellular level. Recent advances in multiplexed hybridization technologies, such as merFISH, CosMX, or Xenium, allow single-cell–based transcriptomic signatures of approximately 700–1000 supervised transcripts.5 By contrast, in situ capturing methods, such as VISIUM Spatial Transcriptomics, offer nearly whole transcriptome signatures. This technology uses unique barcodes to localize mRNA expression to a spot in a known location that overlies hematoxylin and eosin histology.1,3 Spots are uniformly distributed across a tissue, allowing for a deep transcriptomic signature. With a robust single-nucleus RNA sequencing atlas, spots may be deconvolved to determine proportions of specific cell types, states, and neighborhoods.6,7
CODEX
CODEX is a fluorescence-based molecular imaging method that facilitates the capture of highly multiplexed images for a high number of protein markers (approximately 40). Researchers have demonstrated the ability to capture cellular diversity in healthy and pathologic kidneys.3,4,8 The key advantage of CODEX is the ability to render equivalent spatial resolution in -omics as with bright-field histology, allowing for one-to-one mapping of cell identity to underlying morphometry. However, incorporation of new targets requires considerable optimization efforts.
Note on Tissue Preparation
Spatial -omics technologies are typically optimized for frozen sections of 7–10 µm thickness. However, formalin-fixed, paraffin-embedded sections with a thickness between 2 and 5 µm are the gold standard for diagnosis. Generating spatial -omics data for thin, formalin-fixed, paraffin-embedded sections is a topic of growing interest.
Publicly Available Databases
The Human BioMolecular Atlas Program hosts a database of diverse spatial -omics data for normal reference tissue and organs. The Kidney Precision Medicine Project hosts similar data for patients with CKD and AKI. These data are available through the consortium web portals. Additional clinical metadata are often available on request.
Multi-Omics Data Fusion
The question remains how to best use spatial -omics data to drive digital health. Fusion of multi-omics data with bright-field histology is a growing topic of interest for pathologists and computational researchers alike (Figure 1). The fused dataspace will allow biologists to quickly reference structural and functional relationships of cells in the context of a whole biopsy.
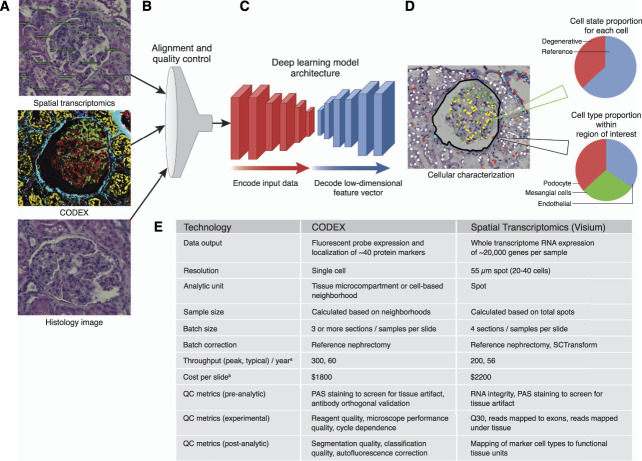
Figure 1.
Deep learning for fusion of two popular spatial technologies. A deep learning model using diverse spatial -omics data as the input and spatial mapping of -omics data on bright-field histology as the output. (A) Glomerulus image from varying modalities including brightfield histology, overlaid spot locations for Visium Spatial Transcriptomics, and Co-Detection by Indexing (CODEX) image. (B) The alignment and quality control step ensures accurate registration of spatial-omics data as well as read quality of transcriptomic or fluorescence data. (C) Representative deep learning model architecture consisting of two sets of convolutional filter banks to first compress input data into a low-dimensional vector and then decode that low-dimensional input and render predictions. (D) Cellular characterization as the output of an ML model, providing insights into both cell-type composition for a given region of interest and an estimate of cellular health (cell state). (E) Comparison of spatial technologies. ML, machine learning; PAS, periodic acid–Schiff; QC, quality control.
A variety of machine learning (ML) approaches are being developed to directly translate bright-field histology images into spatially mapped -omics data. Before input into a ML model, spatial -omics datasets must first be registered to align molecular data with histology. Because these digitally scanned images are exceedingly large (gigapixel area), a patch-based approach is often applied to train models on small portions of slides at a time.9 To ensure that these models are robust to high-dimensional -omics data, it is often necessary to distill the incoming information so that only the most important features are considered. Classical ML methods focused heavily on this dimensionality reduction step to overcome limitations in computational hardware. Currently, vast ML infrastructures (Amazon Web Services Google Collab, Kubeflow, etc.) allow extremely large models to be constructed that are specially equipped to concurrently digest thousands of input values. However, dimensionality reduction of spatial -omics data in general is useful in the context of studying known biological processes. For example, VISIUM data may be better leveraged by translating gene expression into proportions of select cell types within tissue microcompartments (e.g., glomeruli, tubules, vessels).6 Similarly, one can refine CODEX images by isolating particular markers contained within these microcompartments as a better basis of comparison across many individuals. By refining the data dimensionality before injecting to large models, we can reduce the learning gap that these models must overcome and better understand and apply these models to answer important questions.
Emerging research in molecular imaging, combined with novel ML approaches, has the potential to provide the medical community with ways to analyze histological data at a depth never before possible. Whether it is with CODEX or spatial transcriptomics, these high-dimensional datasets require the development of complex models to achieve robust, explainable performance for biological and medical applications.8,10 Although challenges abound, there are exciting opportunities for potential developers of data fusion techniques for innovation, developing tools for clinicians to improve patient care. Meeting these challenges requires collaborative efforts of data and image scientists, clinicians, and biologists to formulate the best algorithmic solution in a team science approach.
Acknowledgments
This article is part of the Artificial Intelligence and Machine Learning in Nephrology series, led by series editor Girish N. Nadkarni.
The content of this article reflects the personal experience and views of the author(s) and should not be considered medical advice or recommendation. The content does not reflect the views or opinions of the American Society of Nephrology (ASN) or CJASN. Responsibility for the information and views expressed herein lies entirely with the author(s).
Disclosures
T.M. El-Achkar reports employment with US Department of Veterans Affairs; research funding from NIH-NIDDK and VA Merit; patent: US11053290B2—Modified Tamm-Horsfall Protein and Related Compositions and Methods of Use; and other interests or relationships with the American College of Physicians, the American Physiological Society, and the American Urological Association. S. Jain reports research collaborative agreement with Nanostring (no funds are provided) and research collaborative agreement with Altos Labs (no funds are provided); advisory or leadership roles for NIH—Kidney Precision Medicine Project and NIH—HuBMAP; royalties from Amirsys Inc. for book chapters in Diagnostic Pathology: Kidney Diseases; and other interests or relationships with CZI Human Cell Atlas Project workshop, FASEB-AKI annual workshop, and Nanostring research collaboration. P. Sarder reports research funding from NCI, NIH-NIDDK, and OD; serving as an Associate Editor for PLOS One; and serving as an Editorial Board member for JASN. All remaining authors have nothing to disclose.
Funding
The work is supported by HuBMAP grants OT2 OD033753 from NIH-OD and U54DK134301 from NIDDK and KPMP grants U01 DK133090 and U01DK114933 from NIDDK, as well as P30 DK079312, R01 DK114485, R01 DK131189, R21 DK128668, U01 DK114923, and U2C DK114886 from NIDDK.
Author Contributions
Writing – original draft: Samuel Border.
Writing – review & editing: Samuel Border, Michael T. Eadon, Tarek M. El-Achkar, Sanjay Jain, Nicholas Lucarelli, Pinaki Sarder.
References
- 1.Marx V. Method of the year: spatially resolved transcriptomics. Nat Methods. 2021;18(1):9–14. doi: 10.1038/s41592-020-01033-y [DOI] [PubMed] [Google Scholar]
- 2.Jen K-Y, Murali LK, Lutnick B, et al. In silico multi-compartment detection based on multiplex immunohistochemical staining in renal pathology. Proc SPIE Int Soc Opt Eng. 2021;11603:1160314. doi: 10.1117/12.2581795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann EK, Patterson NH, Rivera ES, et al. Highly multiplexed immunofluorescence of the human kidney using co-detection by indexing. Kidney Int. 2022;101(1):137–143. doi: 10.1016/j.kint.2021.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black S, Phillips D, Hickey JW, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16(8):3802–3835. doi: 10.1038/s41596-021-00556-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14(1):68–18. doi: 10.1186/s13073-022-01075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lake BB, Menon R, Winfree S, et al. An atlas of healthy and injured cell states and niches in the human kidney. bioRxiv. 2021. doi: 10.1101/2021.07.28.454201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo Ferreira R, Sabo AR, Winfree S, et al. Integration of spatial and single-cell transcriptomics localizes epithelial cell–immune cross-talk in kidney injury. JCI Insight. 2021;6(12):e147703. doi: 10.1172/jci.insight.147703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh JN, Matlock MK, Kudose S, et al. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging. 2018;37(12):2718–2728. doi: 10.1109/tmi.2018.2851150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–1309. doi: 10.1038/s41591-019-0508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.House TW. FACT SHEET: Biden-Harris Administration Announces Key Actions to Advance Tech Accountability and Protect the Rights of the American Public; 2022. [Google Scholar]