Abstract
NotI linking clones contain sequences flanking NotI recognition sites and were previously shown to be tightly associated with CpG islands and genes. To directly assess the value of NotI clones in genome research, high density grids with 50 000 NotI linking clones originating from six representative NotI linking libraries were constructed. Altogether, these libraries contained nearly 100 times the total number of NotI sites in the human genome. A total of 3437 sequences flanking NotI sites were generated. Analysis of 3265 unique sequences demonstrated that 51% of the clones displayed significant protein similarity to SWISSPROT and TREMBL database proteins based on MSPcrunch filtering with stringent parameters. Of the 3265 sequences, 1868 (57.2%) were new sequences, not present in the EMBL and EST databases (similarity ≤ 90%). Among these new sequences, 795 (24.3%) showed similarity to known proteins and 712 (21.8%) displayed an identity of >75% at the nucleotide level to sequences from EMBL or EST databases. The remaining 361 (11.1%) sequences were completely new, i.e. <75% identical. The work also showed tight, specific association of NotI sites with the first exon and suggest that the so-called 3′ ESTs can actually be generated from 5′-ends of genes that contain NotI sites in their first exon.
INTRODUCTION
Several years ago we proposed an approach combining physical and gene mapping methods, based on NotI linking and jumping clones as framework markers, to define the structure of large regions of human chromosomes (1–3). NotI jumping clones contain DNA sequences adjacent to neighbouring NotI restriction sites and NotI linking clones contain DNA sequences surrounding the same restriction site. The use of clones from linking and jumping libraries for genome mapping offers a promising alternative to the laborious procedures used up to now (1,4).
We constructed a new generation of λ-based phasmid cloning vectors (5). Using these vectors we developed new approaches for construction of linking and jumping libraries. This strategy has several conceptual advantages compared to previous approaches and allows efficient construction of representative libraries, which can also be converted to plasmid form.
In our effort to construct a NotI map of human chromosome 3, we constructed seven chromosome 3-specific (Chr.3-specific) linking and jumping libraries and carefully analysed the Chr.3-specific NotI linking clones obtained from these libraries. We have experimentally confirmed that there is a direct statistical connection between CpG islands (6), NotI sites and expressed sequences in the human genome (3,7,8).
We have partially sequenced more than 1000 NotI linking clones isolated from human Chr.3-specific libraries. Of these clones, 152 were unique Chr.3-specific clones. The clones were precisely mapped using a combination of fluorescence in situ hybridisation (FISH) and hybridisation to somatic cell or radiation hybrids. Two and three colour FISH was used to order the clones that mapped to the same chromosomal region and, in some cases, chromosome jumping was used to resolve ambiguous mapping. When this NotI restriction map was compared to the yeast artificial chromosome (YAC)-based chromosome 3 map, significant differences in several chromosome 3 regions were observed. Thus, this experiment confirmed earlier studies (9,10) that a NotI physical map is more reliable than genetic or radiation hybrid maps.
A search of the EMBL nucleotide database with these sequences revealed homologies (90–100%) to more than 100 different genes or expressed sequence tags (ESTs). Many of these homologies were used to map new genes to chromosome 3. These results suggest that sequencing NotI linking clones, and sequencing CpG islands in general, may complement the EST project and aid in the discovery of all human genes. This method yields information that cannot be obtained by the EST project alone, namely identification of the 5′-ends of genes, including potential promoter/enhancer regions and other regulatory sequences.
Therefore, we propose to use an approach based on sequencing of CpG island clone libraries more widely for identification of human genes, as a complement to the well-established EST sequencing project.
Partially sequenced NotI linking clones can also be used to create new STSs. These STSs can be mapped quickly using PCR and FISH methods and applied to the compilation of a NotI PFGE map of the human genome. This map will be particularly useful for joining orphan contigs and for confirmation of other maps, since this map will be based on native human DNA while other maps are derived from cloned sequences.
Using the previously described procedures we have constructed numerous linking libraries with different restriction enzymes (11) for the purpose of generating representative NotI linking libraries covering the whole human genome. We have also created high density grids containing 50 000 clones and suggested sequencing 15 000–20 000 of these clones in the search for new human genes.
Before starting a large-scale project we decided to perform a pilot study to validate our hypotheses.
The present study provides a direct assessment of the value of NotI linking clones in genomics.
MATERIALS AND METHODS
General methods
All general molecular and microbiology methods were performed according to standard procedures (12). Isolation of plasmid DNA was done using a Biorobot 9600 (Qiagen) with REAL-prep kits according to the instructions. Sequencing gels were run on ABI 377 automated sequencers (Perkin Elmer) according to the manufacturer’s protocols.
GenBank accession nos for the NotI sequences are AQ936570–AQ939834.
NotI linking libraries, clone names and accession numbers
NotI linking libraries were constructed from the CBMI-Ral-Sto cell line as described previously (1,4,11). This cell line was established by immortalisation of human B cells with EBV strain B95-8 and was selected for library construction since it was shown previously that DNA in this cell line is undermethylated (13). A total of 50 000 NotI clones were collected and stored in 384-well plates by GeneScreen Ltd (Dorset, UK).
Nomenclature for the NotI linking libraries (11) and clones used in this study is as follows.
Library name Naming convention
1. HL1NR(A) NR1-NNN, e.g. NR1-135 or NR1-XX (the number of the microplate and the number of the row in the microplate, respectively)-NN, e.g. NR1-AA24
2. HL1NR(B) NR3-NNN, e.g. NR3-055 or NR3-XX (the number of the microplate and the number of the row in the microplate, respectively)-NN, e.g. NR3-BB12
3. HL2NR NR5-NNN, e.g. NR5-002 or NR5-XX (the number of the microplate and the number of the row in the microplate, respectively)-NN, e.g. NR5-CC02
4. HL1NB(A) NB1 (and NB2)-NNN, e.g. NB1-590 or NL1-XX (the number of the microplate and the number of the row in the microplate, respectively)-NN, e.g. NL1-DD24
5. HL1NB(B) NB4-NNN, e.g. NB4-025 or NL4-XX (the number of the microplate and the number of the row in the microplate, respectively)-NN, e.g. NL4-EE04
6. HL2NB NB6-NNN, e.g. NB6-1010 or NL6-XX (the number of the microplate and the number of the row in the microplate, respectively)-NN, e.g. NL6-AA12
Sequencing was done as described previously (2). The sequences of the primers used for the sequencing were as follows: reverse (R), 5′-GGA AAC AGC TAT GAC CAT G-3′; check (C), 5′-GGC AAA GCG CCA TTC GCC ATT-3′; saldel (S), 5′-ATG TAG GTG TTC CAC AGG GTA-3′. Accession numbers for the sequences will be supplied later.
Sequence analysis
Protein and nucleotide similarity searches were performed with the BLAST 1.4 program (14,15). The TREMBL and SwissProt databases were used for protein searches with BLAST parameters H = 0, V = 0. The EMBL and EST databases were used for nucleotide searches with BLAST parameters H = 0, B = 50, V = 50. Similarity data was sorted with MSPcrunch (16) using default (–B 0.8 5 –C upper 75 –C lower 35), stringent (–B 0.8 0 –C upper 80 –C lower 40) and very stringent (–B 0.85 0 –C upper 85 –C lower 45) parameters. Empirical testing suggests that these parameters are effective in removing false matches (17). All short, simple and low complexity repeats were excluded from the analysis.
The search with NotI sequences was performed in September 1999 with EMBL database release 59, SwissProt database release 38, TREMBL database release 11. We used MSPcrunch filtering of the BLAST program for selection of significant matches only and further comparison demonstrated that replacing BLAST 1.4 with BLAST 2.0 does not lead to crucial changes in the output results for these significant matches.
RESULTS AND DISCUSSION
For the study we generated 3437 sequences flanking NotI sites in human genomic DNA. As expected in the analysis of genomic DNA from EBV-immortalised cells, EBV sequences were identified (51 clone sequences). Sequences were judged to originate from EBV B95-8 if they had at least 90% nucleotide identity over half the length of the sequence. Following EBV sequence removal, redundant sequences (121) were identified. Redundant sequences were defined as sequences having at least 99% identity over half of the length (this safeguard was used to prevent removal of unique sequences that nevertheless have partial overlap due to common repeats or localisation close to NotI, BamHI or EcoRI restriction sites). The remaining 3265 (95%) sequences were classified as unique and were used for further study.
The majority of the clones were sequenced from only one side. These clone sequences covered a total of 1 517 781 bp with an average sequence length of 465 bp. This is more than in our chromosome 3 NotI project (average length 355 bp).
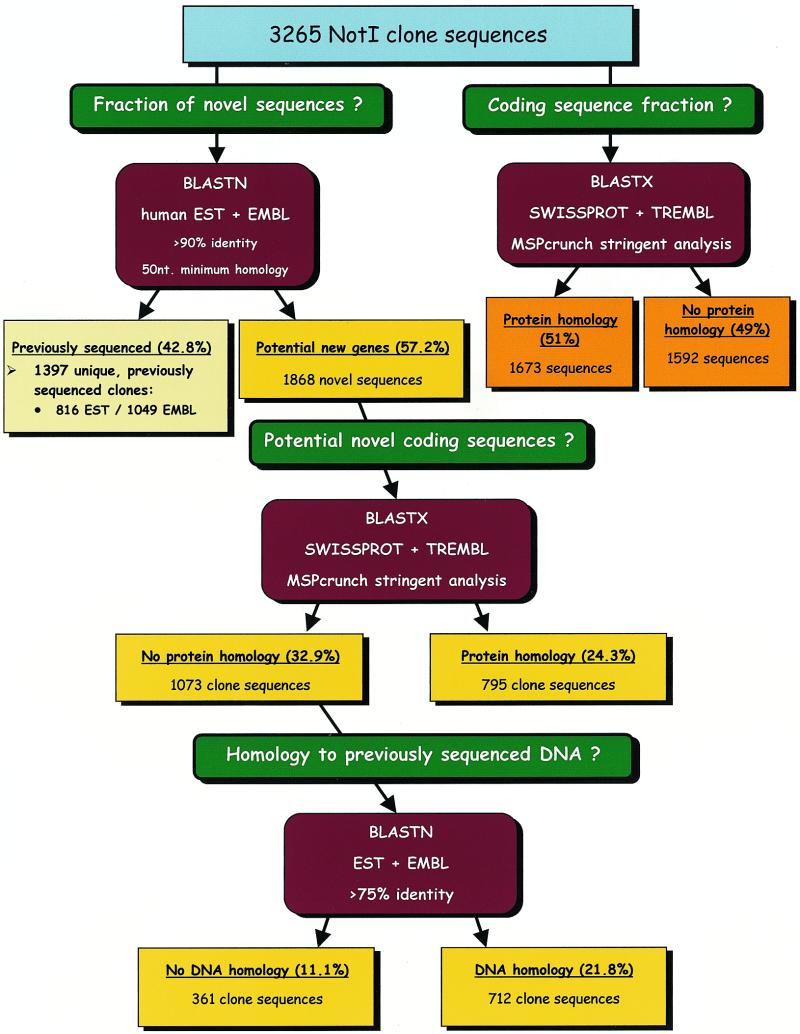
In order to assess the content and usefulness of the data, we devised the simple screening procedure presented in Figure 1. First, we decided to estimate enrichment for protein coding and expressed sequences. A check against the SwissProt and TREMBL databases indicated that 444 823 bp (29.3%) of the total sequence data, representing 51% of the clones, displayed protein similarity based on MSPcrunch output with stringent parameters.
Figure 1.
NotI clone sequence general analysis scheme.
MSPcrunch is a BLAST post-processing program applying a number of filtering rules to the output. The filtering ensures that domains with weak but significant hits will not be missed due to other higher scoring domains and ‘junk’ matches with biased composition are eliminated. Higher sensitivity and selectivity are achieved for multiple matching segments in the ‘twilight zone’ thanks to strict consistency criteria (16,17).
Results of a comparison with full-length, human cDNA protein coding sequences from Unigene are shown in Table 1. If we extend the comparison by including not only protein coding but also 5′- and 3′-untranslated regions (UTRs), then the number of matching sequences increased sharply.
Table 1. Similarity between NotI and human Unigene mRNA sequences.
| Similarity region identity | Fraction of NotI clones matching UTR (no. sequences) | Fraction of NotI clones matching coding sequences (no. sequences) | Total |
|---|---|---|---|
| 90% | 4.4% (145) | 7.3% (238) | 11.7% (383) |
| 95% | 3.5% (114) | 4.8% (156) | 8.3% (270) |
| 99% | 1.7% (55) | 1.9% (63) | 3.6% (118) |
The situation changed significantly when we used the EST database for comparison (Table 2). The fraction of NotI clones matching ESTs is much higher than those matching the Unigene full-length cDNAs. The fraction of NotI clones matching human 3′ ESTs is larger than those matching human 5′ ESTs (8) and the sum of the numbers of the sequences matching 5′ and 3′ ESTs is higher than the total number of NotI clone sequences that is likely to be expressed (e.g. for 95% 298 + 575 = 873 >> 779). The reason for this strange sum is that the same NotI sequence can match 5′ as well as 3′ ESTs.
Table 2. Similarity between NotI and expressed human sequences.
| Similarity region identity | Fraction of NotI clones matching human 5′ ESTs (no. sequences) | Fraction of NotI clones matching human 3′ ESTs (no. sequences) | Fraction of NotI clones likely to be expressed based on human EST sequences (no. sequences) |
|---|---|---|---|
| 90% | 13.9% (455) | 23.2% (758) | 30.8% (1005) |
| 95% | 9.1% (298) | 17.6% (575) | 23.9% (779) |
| 99% | 4.0% (129) | 9.3% (305) | 13.2% (430) |
As we can see in Figure 1, from a total of 3265 NotI clone sequences, 1868 (57.2%) are new sequences not represented in the EMBL and EST databases. For these sequences, we checked how many are potential novel coding sequences. Based on MSPcrunch stringent selection we identified 795 such clones (42.6%, or 24.3% from the original 3265 clone sequences). A total of 277 (8.5%) of these passed the very stringent MSPcrunch parameters; the remaining 518 clone sequences should be considered as tentative coding regions. Among the remaining 1073 clone sequences, 66.4% (712 clone sequences or 21.8% of the original sequences) displayed an identity of >75% to sequences in the EMBL and EST databases, as selected by MSPcrunch with stringent parameters, and 361 clones (11.1% of all sequences) are completely novel sequences, without any similarity to previously identified ones.
We have previously shown (18) that NotI sites have a tendency to be located at the 5′-ends of genes. We decided to examine the positions of the NotI sites more specifically. This analysis was based on similarity to a dataset of 5909 complete human cDNA sequences.
When we looked at the distance between the initiating methionine of the cDNA sequence and the beginning of the similarity region, the only visible peak was at the first nucleotide, indicating that the first methionine found in the NotI sequence corresponds in most cases to the first methionine of the cDNA. This result reflects a strong tendency for the NotI site to be close to the first methionine.
From these data we can confirm our previous suggestion that NotI sites are preferentially located close to the beginning of genes. It is therefore possible that the NotI site is part of a consensus motif involved in initiation of transcription or translation. Furthermore, we calculated the distance between the NotI sites and the protein coding regions that were sequenced. The NotI site has a preferred position inside exons, e.g. the first nucleotide of the NotI sequence is inside the protein coding region. In connection with the previous result, this observation looks very interesting and needs further evaluation.
Another interesting question is the association between NotI sites and Alu repeats. We observed one Alu repeat per 11.1 kb (a more realistic number for 90% similarity is one repeat per 27.1 kb). If we assume that the human genome contains 0.5–1 × 106 Alu repeats then the average density for Alu repeats in the human genome will be one repeat per 3–6 kb. Since Giemza-positive and centromeric regions are very deficient in Alu repeats, R-positive T (H3+) bands (19), which contain the vast majority of NotI sites (3,8,10), must be significantly enriched in Alu repeats and the density of these repeats is more than the average in the human genome. Consequently, we can conclude that sequences surrounding NotI sites are significantly deficient in Alu repeats. This is connected and reinforces the previous conclusion that NotI sites are located mainly in the 5′-ends of the genes, as Alu repeats are found preferentially in intergenic and intronic regions.
A total of 165 NotI clone sequences contained regions similar to MER, LINE and retroviral/LTR-like repeats based on a RepeatMasker default minimum Smith–Waterman score of 225 (A.F.A.Smit and P.Green, unpublished results; RepeatMasker at http://repeatmasker.genome.washington.edu/cgi-bin/RM2_reg.pl ).
The data obtained in this pilot experiment are similar to the results of the sequencing of Chr.3-specific NotI clones (8). A comparison of these two sets of data is shown in Table 3.
Table 3. Chromosome 3 and total genome NotI sequence analysis comparison.
| Similarity region identity | Kashuba et al. (8) | This work |
|---|---|---|
| NotI clone sequences 90% identical to human EST/EMBL database sequences | 38% | 42.8% |
| NotI clone sequences 90% identical to 5′ EST | 12.2% | 13.9% |
| NotI clone sequences 90% identical to 3′ EST | 15.8% | 23.2% |
| NotI clone sequences possessing similarity to SwissProt protein sequences | 32.3% | 36.4% |
In this work we again see a surprisingly large fraction of NotI clone sequences with similarity to 3′ ESTs (and ESTs in general). We think that the main reason for this conflict is the presence of internal NotI sites in cDNAs that are cut prior to cloning with NotI and SalI or NotI and EcoRI. On the other hand, such homologies can result from internal priming artefacts created during cDNA library construction, incomplete or alternative splicing or priming from intronic poly(A) tracts (8).
The fraction of sequences with similarity to the databases is higher in the present study than in our previous work (8). There are two obvious reasons for this: the sequences are longer (465 versus 355 bp) and more new sequences have been introduced into the databases. Apart from this, the conclusions of our previous study of Chr.3-specific NotI clones (8) remain valid, i.e. sequencing of NotI clones will result in the isolation of many new genes. In addition, the sequenced and mapped NotI clones will serve as framework markers for verifying contig assemblies and for connecting rough orphan contig sequences into a final, complete sequence of the human genome. Because the general organisation of mammalian genomes is similar we suggest that the conclusions of this paper are valid for other mammals.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Dr Wyeth Wasserman for fruitful discussion, valuable advice and for help in preparing the article. This work was supported by research grants from Pharmacia & Upjohn for the Center for Genomics Research, Swedish Cancer Society, Karolinska Institute and Åke Wiberg Foundation. V.I.K. and V.I.Z. were recipients of fellowships from the Concern Foundation in Los Angeles and the Cancer Research Institute in New York.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed at: CGR and MTC, Karolinska Institute, Box 280, S-171 77 Stockholm, Sweden. Tel: +46 8 728 67 50; Fax: +46 8 31 94 70; Email: eugzab@ki.se AQ936570–AQ939834
REFERENCES
- 1.Zabarovsky E.R., Boldog,F., Erlandsson,R., Allikmets,R.L., Kashuba,V.I., Marcsek,Z., Stanbridge,E., Sumegi,J., Klein G. and Winberg,G. (1991) Genomics, 11, 1030–1039. [DOI] [PubMed] [Google Scholar]
- 2.Zabarovsky E.R., Kashuba,V.I., Zakharyev,V.M., Petrov,N., Pettersson,B., Lebedeva,T., Gizatullin,R., Pokrovskaya,E.S., Bannikov,V.M., Zabarovska,V.I. et al. (1994) Genomics, 21, 495–500. [DOI] [PubMed] [Google Scholar]
- 3.Allikmets R.L., Kashuba,V.I., Pettersson,B., Gizatullin,R., Lebedeva,T., Kholodnyuk,I.D., Bannikov,V.M., Petrov,N., Zakharyev,V.M., Winberg,G. et al. (1994) Genomics, 19, 303–309. [DOI] [PubMed] [Google Scholar]
- 4.Zabarovsky E.R., Kashuba,V.I., Gizatullin,R.Z., Winberg,G., Zabarovska,V.I., Erlandsson,R., Domninsky,D.A., Bannikov,V.M., Pokrovskaya,E., Kholodnyuk,I. et al. (1996) Cancer Detect. Prev., 20, 1–10. [PubMed] [Google Scholar]
- 5.Zabarovsky E.R., Winberg,G. and Klein,G. (1993) Gene, 127, 1–14. [DOI] [PubMed] [Google Scholar]
- 6.Bird A.P. (1987) Trends Genet., 3, 342–347. [Google Scholar]
- 7.Protopopov A.I., Gizatullin,R.Z., Vorobieva,N.V., Protopopova,M.V., Kiss,C., Kashuba,V.I., Klein,G., Kisselev,L.L., Grafodatsky,A.S. and Zabarovsky,E.R. (1996) Chromosome Res., 4, 443–447. [DOI] [PubMed] [Google Scholar]
- 8.Kashuba V.I., Gizatullin,R.Z., Protopopov,A.I., Li,J., Vorobieva,N.V., Fedorova,L., Zabarovska,V.I., Muravenko,O.V., Kost-Alimova,M., Domninsky,D.A. et al. (1999) Gene, 239, 259–271. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa H., Hosoda,F., Arai,Y., Shimizu,K., Ohira,M. and Ohki,M. (1993) Nature Genet., 4, 361–366. [DOI] [PubMed] [Google Scholar]
- 10.Hosoda F., Arai,Y., Kitamura,E., Inazawa,J., Fukushima,M., Tokino,T., Nakamura,Y., Jones,C., Kakazu,N., Abe,T. et al. (1997) Genes Cells, 2, 345–357. [DOI] [PubMed] [Google Scholar]
- 11.Zabarovsky E.R., Allikmets,R., Kholodnyuk,I., Zabarovska,V.I., Paulsson,N., Bannikov,V.M., Kashuba,V.I., Dean,M., Kisselev,L.L. and Klein,G. (1994) Genomics, 20, 312–316. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Ernberg I., Falk,K., Minarovits,J., Busson,P., Tursz,T., Masucci,M.G. and Klein,G. (1989) J. Gen. Virol., 70, 2989–3002. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Gish W. and States,D.J. (1993) Nature Genet., 3, 266–272. [DOI] [PubMed] [Google Scholar]
- 16.Sonnhammer E.L.L. and Durbin,R. (1994) Comput. Appl. Biosci., 10, 301–307. [DOI] [PubMed] [Google Scholar]
- 17.Sonnhammer E.L.L. and Durbin,R. (1997) Genomics, 46, 200–216. [DOI] [PubMed] [Google Scholar]
- 18.Kashuba V.I., Gizatullin,R.G., Protopopov,A.I., Allikmets,R., Korolev,S., Li,J., Boldog,F., Tory,K., Zabarovska,V.I., Marcsek,Z. et al. (1997) FEBS Lett., 419, 181–185. [DOI] [PubMed] [Google Scholar]
- 19.Saccone S., Caccio,S., Kusuda,J., Andreozzi,L. and Bernardi,G. (1996) Gene, 174, 85–94. [DOI] [PubMed] [Google Scholar]