Abstract
While macrophages are well-known to polarize across the inflammatory spectrum, neutrophils have only recently been found to activate in a similar fashion in response to pro- or anti-inflammatory stimuli. Matrix metalloproteinase (MMP)-12 mediates neutrophil physiology with direct signaling mechanisms yet to be investigated. We hypothesized MMP-12 may modify neutrophil signaling. Bone marrow neutrophils were stimulated with interleukin (IL-1β; pro-inflammatory), IL-4 (anti-inflammatory), or MMP-12. The secretome was mapped by multi-analyte profiling and intracellular signaling evaluated by array. IL-1β induced a cytokine-mediated inflammatory LPS-like signalome, with upregulation of pro-inflammatory cytokines such as interferon gamma (IFNγ,15.2-fold,p = 0.001), chemokine (C-X-C motif) ligand 1 (CXCL1,8.4-fold,p = 0.005), and tumor necrosis factor alpha (TNFα,11.2-fold,p = 0.004). IL-4 induced strong intracellular signaling with upregulation of mitogen-activated protein kinase kinase (MEK1;1.9-fold,p = 0.0005) and downregulation of signal transducer and activator of transcription 4 (STAT4;0.77-fold,0.001). MMP-12 increased IL-4 secretion 20-fold and induced a robust apoptotic neutrophil signalome with upregulation of forkhead box O1 (FOXO1;1.4-fold,p < 0.0001) and downregulation of WNT signaling with MMP-12 cleavage of the adherens junction components β-catenin, cahderin-3, and catenin-α2. In conclusion, neutrophils shifted phenotype by stimuli, with MMP-12 inducing a unique apoptotic signalome with higher resemblance to the anti-inflammatory signalome.
Keywords: Neutrophil, Matrix metalloproteinase-12, Signalome, Proteomics, Polarization
1. Introduction
Macrophage polarization is a well-studied process, and macrophages transdifferentiate phenotypes based on the type of stimulus present in the environment [1]. For example, interleukin (IL)-1β, interferon (IFN)γ, lipopolysaccharide (LPS), or phorbol 12-myristate 13-acetate (PMA)-stimulated macrophages convert to similar albeit not identical pro-inflammatory phenotypes, also termed classical activation or M1 polarization [2,3]. Similarly, IL-4 or IL-10-stimulated macrophages convert to an anti-inflammatory phenotype, also termed alternative activation or M2 polarization [4,5].
In contrast to macrophages, neutrophils (PMNs) have predominantly been considered a pro-inflammatory cell type, degranulating proteases and other proteins from preformed granules in response to inflammatory or infectious stimuli [6–8]. We have shown that neutrophils actually display a large range of phenotypes over the course of the myocardial infarction (MI) wound healing [9]. Similar results have been shown by various groups in different disease environments [9–16]. Neutrophil phenotypes range from pro-inflammatory early in MI to anti-inflammatory and reparative later [12,17]. The same stimuli that polarize macrophages (e.g., IL-1β and IL-4) likely also polarize neutrophils [18].
Neutrophils are the crucial element of MI resolution as they initiate the early process of necrotic debris removal and later lay ground for extracellular matrix (ECM) remodeling by secreting ECM proteins [19]. The Steffens lab has shown that neutrophil depletion impairs MI wound healing by decreased macrophage differentiation and downregulated debris clearance leading to fibrosis [20]. As neutrophil depletion does not yield favorable results, it is important to know how neutrophils are modulated under different physiological stimuli to target detrimental effects elicited by neutrophils.
Matrix metalloproteinase (MMP)-12 is produced by neutrophils, and neutrophils stimulated with MMP-12 upregulate caspase-3 in vitro [21]. Further, MMP-12 inhibition affects neutrophil physiology in the heart and impairs cardiac wound repair after MI by prolonging neutrophil presence and pro-inflammatory status [21]. Understanding how neutrophils signal in response to MMP-12 stimulation would provide insight into MMP-12 regulation of inflammation.
During the shift from pro-inflammation to anti-inflammation, neutrophils undergo apoptosis to remove the inflammatory signal, and neutrophil apoptosis is a key mechanism for inflammation resolution [17,22]. In MI wound healing, neutrophils undergo apoptosis starting at day 3 of MI in mice and are phagocytosed by macrophages [23]. Compared to necrosis, apoptosis subdues inflammation as it avoids release of danger associated molecular patterns (DAMPs), and further inactivates DAMPs such as high mobility group box protein 1 (HMGB1) [24,25]. Apoptotic cells are inactive but can decoy cytokines and scavenge receptors, aiding in inflammation resolution [26,27]. Likewise, neutrophil apoptosis is delayed when inflammation is prolonged [28].
Therefore, using MI specific polarization stimuli, we evaluated the polarization phenotype and compared neutrophil physiology changes with resolution promoting factor MMP-12 [29]. IL-1β is a major pro-inflammatory signal present at day 1 and 3 of MI, which polarizes neutrophils to pro-inflammatory tissue degrading phenotype by activating them as they enter the infarct site [30,31]. IL-4 is one of the anti-inflammatory signals prevalent later in MI, which is responsible for the change in tissue microenvironment to anti-inflammatory and change in cellular phenotype [23]. We hypothesized that MMP-12 may be a novel modifier of PMN signaling. To address this hypothesis, we mapped the neutrophil protein signalome in response to MMP-12 and compared to stimulation by IL-1β as a classic pro-inflammatory stimulus and IL-4 as a classic anti-inflammatory stimulus.
2. Methods
2.1. Animal use
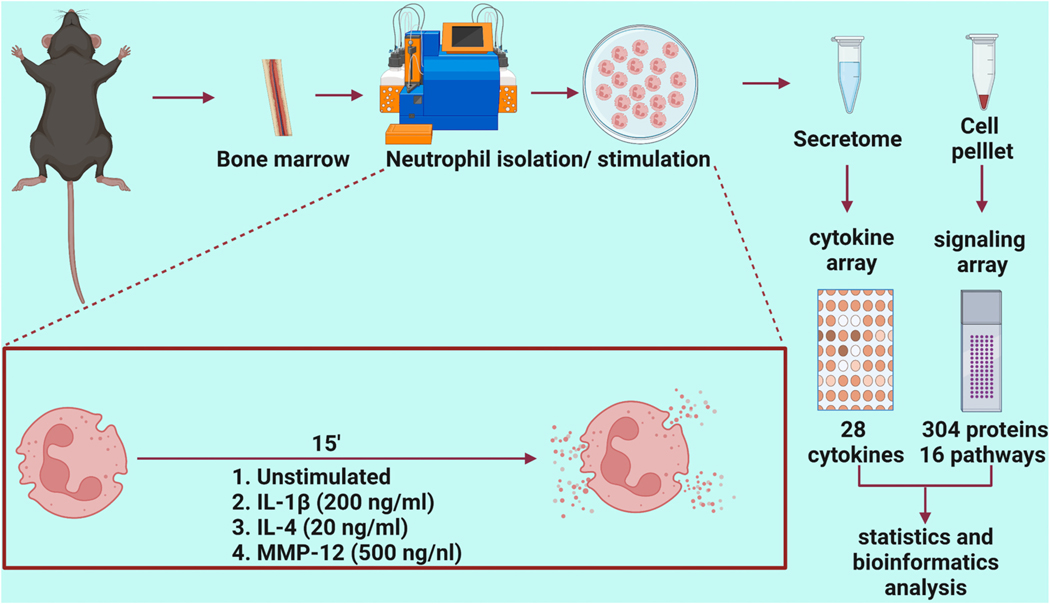
The overall experimental design is detailed in Fig. 1. All animal procedures were performed according to the Guide for the Care and Use of Laboratory Animals, and all protocols were pre-approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Center [32]. C57BL/6 J wild-type mice of both sexes (3–6 months old) were obtained either by purchasing from Jackson Laboratory or from an in-house breeding colony and were housed in the animal facility. All mice were housed together in the same room, under a 12:12 h light-dark cycle and given ad libitum access to standard mouse chow and water.
Fig. 1.
Experimental design. Created with http://BioRender.com.
2.2. Bone marrow derived neutrophil isolation and stimulation
Neutrophils were isolated from the femur and tibia of mice as previously described [12]. Using a 26-gauge needle and 10 ml syringe filled with RPMI 1640 media supplemented with 1% penicillin/streptomycin and 2 mM EDTA, bone marrow cells were flushed from the bones. The cell suspension was filtered through 30 μm filter to obtain single cell suspension. Single cell suspension was incubated with anti-Ly6G magnetic ultrapure microbeads (Miltenyi Biotec, #130–120–337), and neutrophils were sorted through magnetic columns in the AutoMACS Pro Separator. Purity of Ly6g+ neutrophils using this approach has previously been shown to be >95% by both flow cytometry and immunofluorescence [9]. Neutrophils (1 × 106) were stimulated with IL-1β (200 ng/ml, RnD systems, #401-ML), IL-4 (20 ng/ml, RnD systems, #404-ML), or MMP-12 (500 ng/ml, Enzo Life Sciences #BML-SE138–0010) and incubated for 15 min at 37 °C in 1 ml of RPMI 1640 media. The negative control was unstimulated cells. Samples sizes were n = 3 M and 3 F biological replicates for each stimulus group, performed as a four-group paired stimulation. Following stimulation, cells were centrifuged at 800 xg for 8 min and cell pellets were used for the transcription factor signaling array, while the secretome was used for the cytokine array.
2.3. Signaling protein array
The Cell Signaling Phospho Antibody Array (Full Moon Bio Systems, #PCS300, https://www.fullmoonbio.com/product/cell-signaling-phospho-antibody-array/) measured 304 proteins that represent 16 known signaling pathways, including antibodies against both total and phosphorylated protein. Each glass array slide contained replicate spots for each protein. All procedures were performed according to manufacturer recommendations. Cell pellets (1 × 106 cells) were washed in 1 ml of ice-cold phosphate buffered saline (PBS) for 2 min at 500 xg, repeated twice for a total of 3 washes. The cells were lysed with lysis beads in 50 μl of Extraction Buffer, vortexed for 30 s, placed on ice for 10 min, and the process was repeated 6 times. The samples were centrifuged at 10,000 xg for 5 min at 4 °C, the supernatant transferred to a new tube, centrifuged at 18,000 xg for 15 min at 4 °C and transferred to a new tube. The spin columns were hydrated with 650 μl labeling buffer, and the clear supernatant transferred to a spin column and centrifuged at 750 xg for 2 min. The purified protein in the collection tube was quantified and assessed for quality using a Nanodrop 2000 measuring absorbance at A280. To a new tube, 25 μl of sample, 50 μl labeling buffer, and 3 μl Biotin/DMF solution were added. This mixture was incubated for 1 h at RT with vortexing every 10 min. Stop reagent (35 μl) was added to the tube, and the samples were incubated for 30 min at RT with mixing every 10 min. The samples were stored at −80 °C and used within a month on the arrays.
The antibody arrays were blocked for 30 min at RT and washed. The prepared samples were added to 6 ml of coupling solution and incubated with the arrays on an orbital shaker at 35 rpm for 1 h at RT. The arrays were washed in 30 ml of 1× wash solution at 55 rpm for 10 min 3 times and rinsed in water. To 60 ml detection buffer, 60 μl of Cy-3 streptavidin (GE Healthcare, #PA43001) was added and the array was incubated with 30 ml of the Cy-3 streptavidin mixture for 20 min. The arrays were washed and dried in a centrifuge at 1300 xg for 5 min. The glass slides were packaged and sent to Full Moon BioSystems for analysis using a microarray scanner to image and quantify signal intensity. For each biological replicate, two technical replicates were analyzed. Data were normalized to the median value of the average signal intensity for all antibodies in the array. Values are reported as normalized intensity units and are provided in Supplementary Material 1.
2.4. Multi-analyte cytokine profiling of the secretome
The secretome from the neutrophil stimulation were used for V-PLEX Mouse Cytokine Kit (Mesoscale Discovery, #K15267D), which contained a total of 3 plates, including a 10 protein pro-inflammatory panel, a 9 protein cytokine panel, and a 9 protein TH17 panel, for total of 28 cytokines evaluated. The V-PLEX arrays are validated by Mesoscale Discovery according to the guidelines for fit-for-purpose method development and validation for successful biomarker measurement [33]. Standard cocktail mixture was prepared as per the recommendation in a diluent, diluted to 8 different concentrations for calibration and 50 μl was loaded into the plate in duplicate for each concentration, followed by neutrophil secretome samples (25 μl) in 25 μl of diluent (1:1). The plates were sealed and incubated at RT for 2 h. The plates were washed 3 times with 150 μl of PBS containing 0.05% Tween-20 for each well. Detection antibody solution (25 μl) was added to each well. The plates were sealed and incubated at RT with shaking for 2 h. The plates were washed, and 150 μl of 2× read buffer T was added to each well. The plates were read in the Mesoscale Quikplex SQ 120, and data was processed through the discovery workbench software analysis program (Mesoscale Discovery). Values are reported as normalized intensity units and are provided in Supplementary Material 2.
2.5. MMP-12 substrate analysis
Human recombinant proteins for β-Catenin (Abnova, #H00001499-P01), Cadherin-3 (Abnova, #H00001001-P01), Cadherin-1 (RnD Systems, #8875-EC) and Catenin-α2 (Abnova, #H00001496-P01) were incubated with the active catalytic domain of human MMP-12 (Enzo Life Sciences, #BML-SE138–0010) at a 5:1 ratio (1 μg recombinant protein: 0.2 μg enzyme) at 37 °C in 10 μl of 1× zymogram buffer (Life Technologies, #LC2671) for up to 24 h. Reactions were stopped at each time point by addition of 0.5 μl of 0.5 M EDTA to each 10 μl reaction, which was then stored at 4 °C until the next day and run on SDS-PAGE gels. Timing was adjusted for each protein to observe maximum range of enzymatic degradation. Silver staining was performed using Pierce™ Silver Stain Kit (Thermo Scientific, #24612) as per manufacturer recommendation. The gels were imaged using iBright FL1000. Experiments were run in triplicate, and the data are shown as representative images.
2.6. Statistics and bioinformatics
Statistical analyses were performed using GraphPad Prism 9 according to the guidelines outlined in Statistical Considerations in Reporting Cardiovascular Research [34]. Data are reported as mean ± SEM for group comparisons. The Shapiro-Wilk test was used to test normality, and the data passed. To compare each stimulus group to the unstimulated negative control, a paired Students t-test was used. Raw p values were used for the analysis. Sample size was selected based on power analysis. A sample size of n = 12 (n = 6 in each group) gave a power (1-β) of >0.8, with α = 0.05, minimum desirable effect (MDE) at 30% of the mean and standard deviation set at 20%. A volcano plot was generated to rank based on p value and log-fold change. A proportional Venn diagram was created to visualize inter-group overlap using DeepVenn (http://www.deepvenn.com/) [35]. Enrichment analysis of statistically significant proteins was performed using Enrichr (https://maayanlab.cloud/Enrichr/), a bioinformatics tool developed by the Ma’ayan Laboratory [36]. GO Biological process 2021 was used to determine enriched biological processes. Transcription factor protein-protein interactions were used to identify pathways enriched. A p value <0.05 was considered statistically significant.
3. Results
3.1. IL-1β induced the neutrophil signalome towards cytokine mediated pro-inflammatory signaling
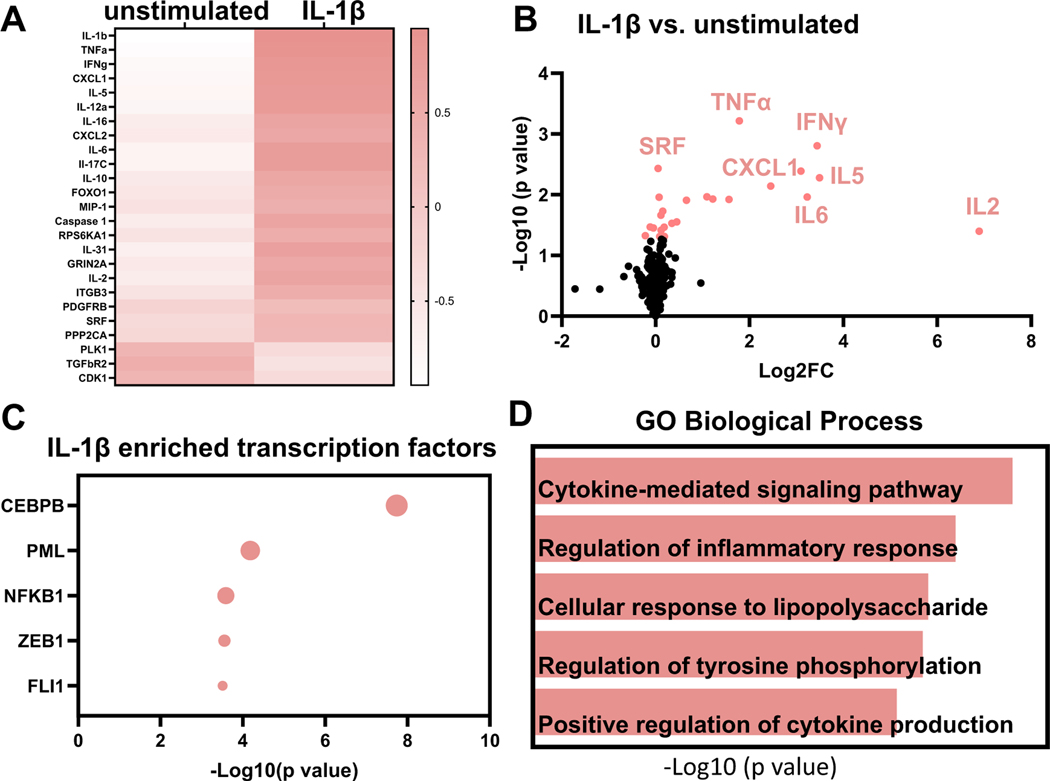
For neutrophils stimulated with IL-1β, there were 25 proteins differentially expressed of the 332 evaluated (7.5%); 22 were upregulated and 3 were downregulated. As a positive control, IL-1β was increased 18,647-fold in the secretome (p < 0.0001). By fold change, the next 4 proteins increased by IL-1β were IL-2 (128-fold, p = 0.03), IFNγ (15-fold, p = 0.001), IL-5 (11-fold, p = 0.004), and IL-6 (8.4-fold, p = 0.01). The heatmap charts the 25 proteins changed in neutrophils with IL-1β treatment (Fig. 2A). By volcano plot that incorporates both fold-change and p value, the top ranked upregulated proteins were TNFα (4-fold, p = 0.0006) and IFNγ (Fig. 2B).
Fig. 2.
IL-1β induced the neutrophil signalome towards cytokine mediated pro-inflammation. A) Heat map shows the top 25 proteins upregulated or downregulated with IL-1β treatment. IL-1β, TNFα, and IFNγ were the top three upregulated proteins based on fold change. B) Volcano plot shows top ranked differentially expressed proteins in neutrophils after IL-1β treatment. C) Enrichment analysis shows top 5 most enriched transcription factors based on p value. CEBPB was the most enriched transcription factor stimulated by IL-1β. D) By enrichment analysis, cytokine-mediated signaling pathway was the most enriched GO biological process in the IL-1β induced neutrophil signalome.
Enrichment analysis showed that CCAAT enhancer binding protein beta (CEBPB) was the most enriched transcription factor represented by the upregulated proteins, followed by promyelocytic leukemia nuclear body scaffold (PML), nuclear factor kappa beta subunit 1 (NFKB1), zinc finger E-Box binding homeobox 1 (ZEB1), and floricaula/leafy-like protein (FL1; Fig. 2C). The most enriched biological pathway by GO biological process was cytokine-mediated signaling (Fig. 2D). IL-1β, therefore, stimulated a strong pro-inflammatory profile, as would be expected. Of note, IL-1β treatment increased IL-10 release in neutrophils. IL-10 is an anti-inflammatory protein, and this finding suggests activation of a negative feedback to limit the maximum pro-inflammatory response. Overall, these results served as a positive control indicative of pro-inflammatory stimulation.
3.2. IL-4 induced the neutrophil signalome towards anti-inflammatory signaling
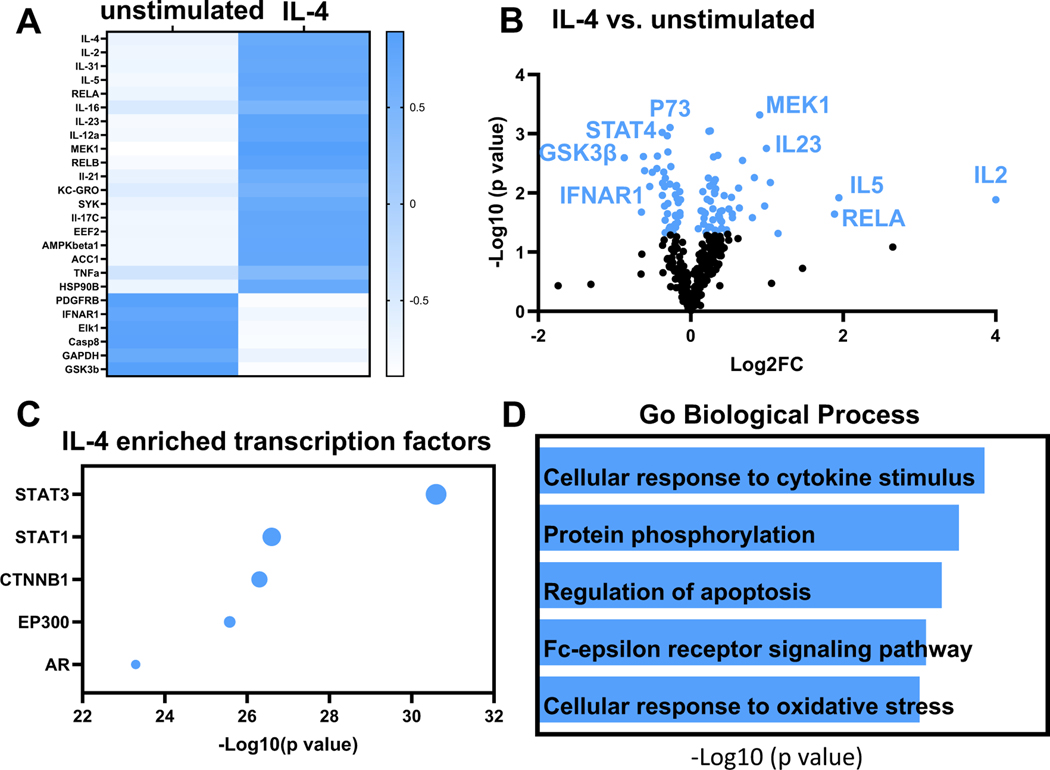
For neutrophils stimulated with IL-4, there were 85 unique proteins differentially expressed out of 332 evaluated (26%); 49 were upregulated and 36 were downregulated. As a positive control, IL-4 was increased 63,420-fold in the secretome (p = 0.02). By fold change, the next 4 factors increased were IL-2 (19-fold, p = 0.01), IL-31 (8-fold, p = 0.006), IL-5 (4-fold, p = 0.01) and RELA/NFkBp65 (3-fold, p = 0.02). While IL-2 was first ranked induced by both IL-1β and IL-4, IL-4 induction of IL-2 was 6.7-fold lower than the amount of IL-2 induced by IL-1β. Induction of IL-2 by both pro- and anti-inflammatory stimuli is consistent with known dual roles of IL-2 in both pathways [37]. The heatmap in Fig. 3A shows the top 25 proteins changed in neutrophils with IL-4 treatment. By volcano plot that incorporates both fold-change and p value, the top ranked upregulated protein induced by IL-4 was mitogen-activated protein kinase kinase (MEK1, 1.9-fold, p = 0.0005). By volcano plot, the top ranked downregulated proteins were tumor protein p73 (P73, 0.83-fold, p = 0.0007) and signal transducer and activator of transcription 4 (STAT4, 0.77-fold, p = 0.001) (Fig. 3B).
Fig. 3.
IL-4 induced the neutrophil signalome towards cytokine release and intracellular signaling response. A) Heat map shows the top 25 proteins upregulated or downregulated with IL-4 treatment. IL-4, IL-2 and IL-31 were the top three upregulated based on fold change. B) Volcano plot shows the top ranked differentially expressed proteins in neutrophils after IL-4 treatment. C) Enrichment analysis shows top 5 most enriched transcription factors based on p value. STAT3 was the most enriched transcription factor stimulated by IL-4. D) By enrichment analysis, cellular response to cytokine stimulus was the most enriched GO biological process in the IL-4 induced neutrophil signalome.
Enrichment analysis showed that signal transducer and activator of transcription (STAT)3 was the most enriched transcription factor for all differentially expressed proteins, followed by STAT1, catenin beta 1 (CTNNB1), EA1 binding protein P300 (EP300), and androgen receptor (AR; Fig. 3C). The most enriched biological process was cellular response to cytokine stimulus (Fig. 3D). IL-4 regulated proteins involved in cell cycle (CDK1, Cyclin D1) and apoptosis (FOXO1, P73). Of note, IL-4 maintained a low baseline of inflammation activation in neutrophils, reflected by increased activation of the NFkB pathway. Overall, these results served as a positive control indicative of anti-inflammatory stimulation.
3.3. MMP-12 induced an IL-4-like neutrophil signalome
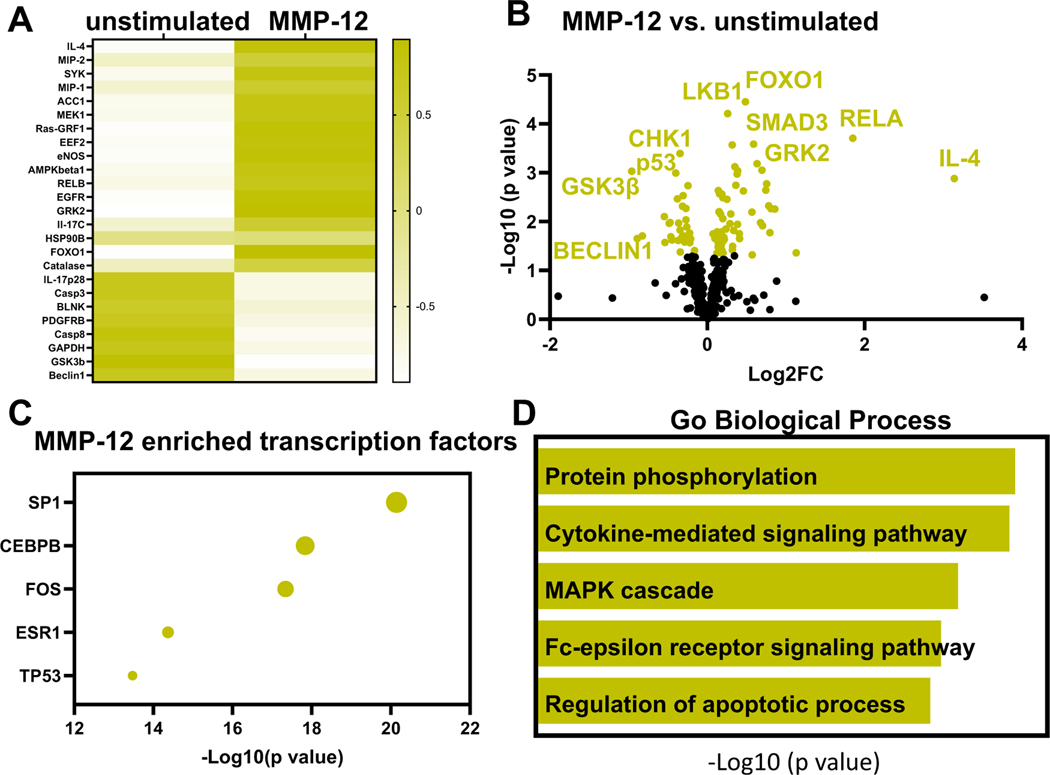
For neutrophils stimulated with MMP-12, there were 81 proteins differentially expressed of the 332 evaluated (24%); 55 were upregulated and 26 were downregulated. MMP-12 stimulated a mixed pro-inflammatory and anti-inflammatory profile. By fold change, IL-4 (20-fold, p = 0.001), RELA/NFkBp65 (4-fold, p = 0.0001), macrophage inflammatory protein (MIP)-2 (2-fold, p = 0.04), SYK (1.8-fold, p = 0.005) and MIP-1 (1.7-fold, p = 0.01) were the highest upregulated proteins. The heatmap in Fig. 4A shows the top 25 proteins changed in neutrophils with MMP-12 treatment. By volcano plot ranking, top upregulated proteins were forkhead box O1 (FOXO1, 1.4-fold, p < 0.0001), liver kinase B1 (LKB1, 1.2-fold, p < 0.0001), RELA proto-oncogene, NFkB subunit (RELA, 3.6-fold, p = 0.0002), G protein coupled receptor kinase 2 (GRK2, 1.5-fold, p = 0.0003), and Smad family member 3 (SMAD3, 1.3-fold, p = 0.003). By volcano plot, the top 3 downregulated proteins were checkpoint kinase 1 (CHK1, 0.79-fold, p = 0.0004), GSK3β (0.50-fold, p = 0.0009) and tumor protein p53 (P53, 0.76-fold, p = 0.0001, Fig. 4B).
Fig. 4.
MMP-12 induced the neutrophil signalome towards apoptosis with a strong intracellular signaling response. A) Heat map shows the top 25 proteins upregulated or downregulated with MMP-12 treatment. IL-4, MIP-2 and SYK were the top three upregulated proteins based on fold change. B) Volcano plot shows top differentially expressed proteins in neutrophils after MMP-12 treatment. C) Enrichment analysis shows top 5 most enriched transcription factors based on p value. SP1 was the most enriched transcription factor stimulated by MMP-12. D) By enrichment analysis, protein phosphorylation was the most enriched GO biological process stimulated by MMP-12.
Enrichment analysis showed that specificity protein 1 (SP1) was the most enriched transcription factor, followed by CEBPB, fos proto-oncogene (FOS), Estrogen receptor 1 (ESR1), and tumor protein 53 (TP53; Fig. 4C). The most enriched biological process by GO biological process was protein phosphorylation, indicating MMP-12 induced strong intracellular signaling (Fig. 4D). MMP-12 stimulated a 3.6-fold increase in RELA indicative of pro-inflammation and a 20-fold increase in IL-4 indicative of anti-inflammation. The top pathways induced by MMP-12 intracellular signaling were associated with the regulation of cell cycle and apoptotic processes.
Out of the 332 proteins evaluated, the Venn diagram in Fig. 5 shows that MMP-12 shared 9 proteins with IL-1β and 45 proteins with IL-4. A total of 10 proteins were unique to IL-β, 33 proteins were unique to IL-4, and 34 proteins were unique to MMP-12. Overall, the neutrophil signalome showed an overall higher resemblance towards neutrophils polarized by IL-4.
Fig. 5.
MMP-12 stimulated the neutrophil signalome towards an IL-4 like response. MMP-12 and IL-4 shared changes in 45 proteins while MMP-12 and IL-1β shared 9 protein changes. Proportional Venn diagram was created using http://deepvenn.com.
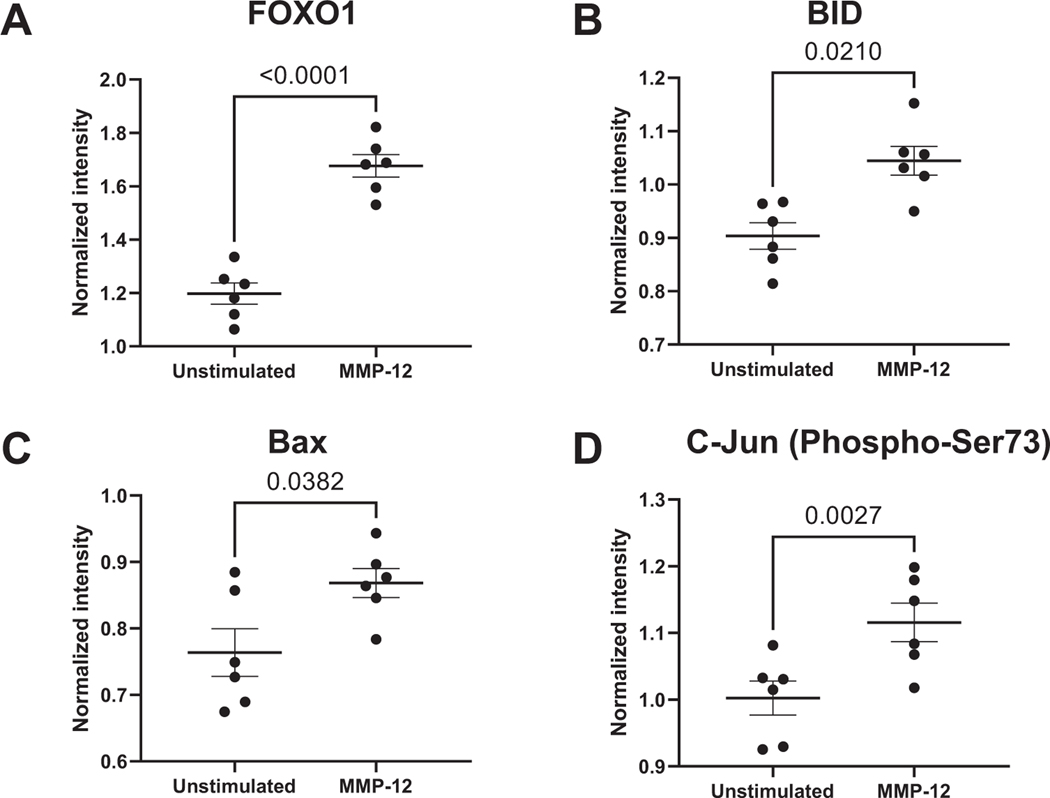
3.4. MMP-12 induced neutrophil apoptosis through upregulation of FOXO1 signaling
As FOXO1 was the highest ranked protein (Fig. 6A) and apoptosis was one of the most-enriched pathways, we evaluated BH3 family protein expression in response to MMP-12. MMP-12 increased the pro-apoptotic proteins Bid (1.7-fold, p = 0.02, Fig. 6B) and Bax (1.1-fold, p = 0.038, Fig. 6C). Of note, IL-1β and IL-4 both also increased FOXO1, but neither stimulated Bid or Bax. Along with FOXO1 signaling, phosphorylation of the proto-oncogene c-Jun (c-Jun) was increased 1.1-fold (p = 0.003, Fig. 6D) in neutrophils reflective of apoptotic signaling with MMP-12 stimulation. The FOXO1 mediated increase in Bcl2-family proteins was responsible for the apoptotic signature induced in the neutrophil signalome.
Fig. 6.
MMP-12 induced neutrophil apoptosis through FOXO1 and c-Jun phosphorylation. MMP-12 increased FOXO1 (A), Bax (B), Bid (C), and c-Jun (D) phosphorylation in neutrophils.
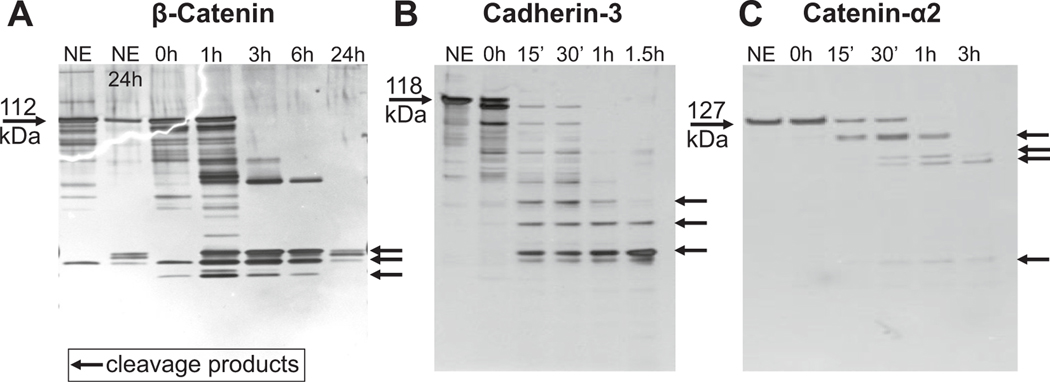
3.5. MMP-12 induced neutrophil adhesion through down regulation of WNT signaling
As GSK3β was downregulated 50%, we looked at WNT signaling components that regulate cell adhesion. β-catenin was not downregulated in MMP-12 stimulated neutrophils (0.84-fold, p = 0.054). To determine if β-catenin was a proteolytic substrate of MMP-12, we performed in vitro substrate analysis by incubating MMP-12 with β-catenin. MMP-12 proteolytically processed β-catenin within 3 h of incubation (Fig. 7A). As β-catenin is associated with adherens junctions and actin-cytoskeleton involved in cellular adhesion, we examined if cadherin-1 (CDH1), cadherin-3 (CDH3), or catenin-α2 (CTNNA2) were also substrates. CDH1 was not cleaved by MMP-12 after 24 h of incubation. MMP-12 cleaved CDH3 (Fig. 7B) and CTNNA2 (Fig. 7C) rapidly within 15 min. Overall, downregulation of WNT signaling through enzymatic cleavage of cadherins and catenin indicated that MMP-12 downregulated proteins involved in cellular adhesion through a post-translational mechanism.
Fig. 7.
MMP-12 downregulated WNT signaling by proteolytically processing catenins and cadherins to reduce cell adhesion. A) MMP-12 cleaved β-catenin within 1 h of incubation. The catalytic domain of MMP-12 was incubated at a 5 (β-catenin): 1(MMP-12) ratio. B) MMP-12 cleaved cadherin-3 within 15 min of incubation. The catalytic domain of MMP-12 was incubated at a 5 (cadherin-3):1 (MMP-12) ratio. C) MMP-12 cleaved catenin-α2 within 15 min of incubation. The catalytic domain of MMP-12 was incubated at a 5 (catenin-α2):1 (MMP-12) ratio.
4. Discussion
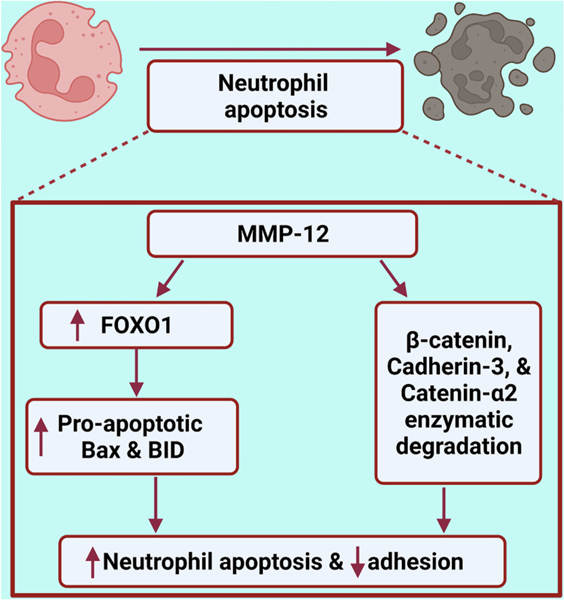
The goal of this project was to use an omics-based approach to identify how the neutrophil signaling proteome responds to a range of stimuli. The major findings were: 1) Neutrophils demonstrated unique polarization profiles in response to specific stimuli; 2) MMP-12 induced a neutrophil signalome that was closer to IL-4 than IL-1β; and 3) MMP-12 polarization shifted the neutrophil signalome towards an apoptotic signature by upregulating FOXO1 and downregulating WNT signaling (Fig. 8). Our results highlight that the neutrophil has more plasticity than previously appreciated. Mapping the unique range of polarization responses in the signalome provides targets to modulate neutrophil cell physiology.
Fig. 8.
MMP-12 induced apoptosis in neutrophils by upregulating FOXO1 and impaired cell adhesion through cleavage of adherent junction proteins. Created with http://BioRender.com.
IL-1β stimulated neutrophils to an expected pro-inflammatory profile, with high cytokine expression (e.g., TNFα and IFNγ) [38]. There was also activation of the inflammasome response with increased caspase 1 to further increase IL-1β production in a positive feedback loop [39]. Enrichment analysis for IL-1β showed strong enrichment for cytokine production, with CEBPB being the most enriched transcription factor. CEBPB is associated with emergency granulopoeisis, and at day 1 after MI neutrophils have increased expression of CEBPB [15]. NFkB and CEBP both regulate the IL-6 promoter, consistent with the >8-fold induction in IL-6 protein [40]. LPS-like response was one of the enriched GO pathways indicating IL-1β induced neutrophil signalome showed a similar pattern to lipopolysaccharide (LPS) stimulation [41,42]. IL-1β induced CXCL1, which induces cellular migration and is also an LPS-like response protein [43]. Overall, IL-1β had robust effect on pro-inflammatory cytokine release.
IL-4 stimulation induced several signaling changes reflective of an anti-inflammatory and pro-reparative neutrophil. IL-4 induced IL-2, IL-31, and IL-5, all of which are neither strictly pro-inflammatory or anti-inflammatory signals [44–46]. STAT3 was the most enriched transcription factor induced by IL-4, and STAT3 activation is the major signaling pathway activated by IL-10 [47]. STAT3 inactivates IkappaB kinase (IKK) associated with Th1 immune response and is a part of anti-inflammatory signaling [48,49]. In the setting of ischemia reperfusion injury, STAT3 over expression inhibits reactive oxygen species production and offers cardioprotection [50–53]. STAT3 deficient mice had more mature neutrophils in their bone marrow. MEK1 promotes phagocytosis in neutrophils and granulocyte macrophage colony stimulating factor induced delay of apoptosis [54–56]. MEK1 upregulation seen with IL-4 treatment could have protective role in MI as MEK1 impairs MI induced adverse remodeling by increased phosphorylation of STAT3 and downregulation of MMP-9 in the infarct [54,57]. Previously we have shown IL-4 shuts off pro-inflammation in neutrophils induced in vivo by MI [23]. STAT4 downregulation could be responsible for anti-inflammatory action of IL-4 as STAT4 is required for IL-12 induced inflammation in neutrophils [58,59]. Our data indicates that pro-inflammation might be a prerequisite for the full anti-inflammatory action of IL-4 to occur.
MMP-12 stimulation of neutrophils showed a unique apoptotic phenotype. IL-4 is the major cytokine upregulated in neutrophils with MMP-12 treatment followed by macrophage inflammatory proteins 1 and 2 indicating neutrophil to macrophage crosstalk [27,60]. SP1 was the most enriched transcription factor in MMP-12, which when inhibited is protective against myocardial ischemia injury by inhibiting cardiomyocyte cell death [61–63]. CBEPB enrichment was shared between MMP-12 and IL-1β.
FOXO1 was the most upregulated signaling molecule in MMP-12 treated neutrophils, which is associated with apoptosis of various cell types [64,65]. FOXO1 is linked to apoptosis stimulated by increased oxidative stress or DNA damage by upregulating Bcl-2 family proteins [64,66,67]. In MMP-12 stimulated neutrophils, both Bax and Bid were increased. IL-1β or IL-4 both primed neutrophils to undergo apoptosis by upregulating FOXO1, while only MMP-12 completed the signaling. Resveratrol and Curcumin induce p53 independent apoptosis through FOXO1 [65,68,69]. In our cells, p53 was downregulated by MMP-12. FOXO1 also stimulates RELA, which was induced in MMP-12 stimulated neutrophils [70]. MMP-12 also stimulated phosphorylation of other apoptotic proteins such as c-Jun. [71,72]
FOXO1 is a member of WNT signaling to regulate cellular adhesion and migration [73]. For WNT signaling, GSK3β was downregulated by MMP-12. GSK3β phosphorylates β-catenin to degrade it [74]. As MMP-12 enzymatically cleaved β-catenin, downregulation of GSK3β could be a potential negative feedback response. β-catenin is a component of the adherent junction along with other cadherins, catenins and actin filaments [75,76]. MMP-12 enzymatic degradation of adherens junction proteins regulates cellular adhesion and migration, which goes along with its role in apoptosis. The Overall lab has shown that MMP-12 degrades actin-filaments and impairs neutrophil infiltration in arthritis [77]. This corroborates our finding that MMP-12 impaired cell adhesion by degrading adherent junction proteins. Decreased cellular adhesion of neutrophils after MI could limit inflammation in MI.
5. Conclusions
In summary, neutrophils polarize towards a phenotype that reflects the activating stimuli, as revealed by mapping their signalome. There was a strong cytokine-mediated pro-inflammatory LPS-like signalome induced by IL-1β. With IL-4, there was an inflammation resolving signalome. MMP-12 treatment induced a robust apoptotic neutrophil signalome that was more reflective of IL-4 than IL-1β. MMP-12, therefore, may be a candidate resolution promoting factor to curb inflammation through direct actions on neutrophils.
Supplementary Material
Significance:
This study revealed that neutrophils demonstrate unique polarization signaling responses to specific stimuli, with the matrix metalloproteinase (MMP)-12 signalome showing similarity to the IL-4 signalome. MMP-12 polarized neutrophils towards a strong apoptotic signature by upregulating FOXO1 and downregulating WNT signaling. Our results highlight that neutrophils display more plasticity than previously appreciated.
Acknowledgements
Dr. Lindsey is a Stokes-Shackleford Professor at UNMC. We acknowledge funding from National Institutes of Health under Award Numbers HL137319 and U54GM115458, and from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development under Award Numbers 5I01BX000505; the American Cancer Society Research Scholar Grant under award number RSG-19-127-01-CSM; and the Swedish Society for Medical Research under award number P19-0144. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies. All authors have reviewed and approved the article.
Footnotes
Declaration of Competing Interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jprot.2022.104636.
Data availability
Data are provided in supplementary files.
References
- [1].Sica A, Mantovani A, Macrophage plasticity and polarization: in vivo veritas, J. Clin. Invest 122 (3) (2012) 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Orecchioni M, Ghosheh Y, Pramod AB, Ley K, Macrophage polarization: different gene signatures in M1 (LPS+) vs. classically and M2 (LPS–) vs. alternatively activated macrophages, Front. Immunol 10 (2019), 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yu T, Zhao L, Huang X, Ma C, Wang Y, Zhang J, Xuan D, Enhanced activity of the macrophage M1/M2 phenotypes and phenotypic switch to M1 in periodontal infection, J. Periodontol 87 (9) (2016) 1092–1102. [DOI] [PubMed] [Google Scholar]
- [4].Hart PH, Bonder CS, Balogh J, Dickensheets HL, Donnelly RP, Finlay-Jones JJ, Differential responses of human monocytes and macrophages to IL-4 and IL-13, J. Leukoc. Biol 66 (4) (1999) 575–578. [PubMed] [Google Scholar]
- [5].Jung M, Ma Y, Iyer RP, DeLeon-Pennell KY, Yabluchanskiy A, Garrett MR, Lindsey ML, IL-10 improves cardiac remodeling after myocardial infarction by stimulating M2 macrophage polarization and fibroblast activation, Basic Res. Cardiol 112 (3) (2017) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mayadas TN, Cullere X, Lowell CA, The multifaceted functions of neutrophils, Annual Review of Pathology: Mechanisms of Disease. 9 (2014) 181–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Borregaard N, Neutrophils, from marrow to microbes, Immunity. 33 (5) (2010) 657–670. [DOI] [PubMed] [Google Scholar]
- [8].Weigand M, Degroote RL, Amann B, Renner S, Wolf E, Hauck SM, Deeg CA, Proteome profile of neutrophils from a transgenic diabetic pig model shows distinct changes, J. Proteome 224 (2020), 103843. [DOI] [PubMed] [Google Scholar]
- [9].Ma Y, Yabluchanskiy A, Iyer RP, Cannon PL, Flynn ER, Jung M, Henry J, Cates CA, Deleon-Pennell KY, Lindsey ML, Temporal neutrophil polarization following myocardial infarction, Cardiovasc. Res 110 (1) (2016) 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu K, Wang F-S, Xu R, Neutrophils in liver diseases: pathogenesis and therapeutic targets, Cellular & molecular immunology. 18 (1) (2021) 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Giese MA, Hind LE, Huttenlocher A, Neutrophil plasticity in the tumor microenvironment, Blood, The Journal of the American Society of Hematology. 133 (20) (2019) 2159–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Daseke MJ 2nd, Valerio FM, Kalusche WJ, Ma Y, DeLeon-Pennell KY, Lindsey ML, Neutrophil proteome shifts over the myocardial infarction time continuum, Basic Res. Cardiol 114 (5) (2019) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].García-Culebras A, Durán-Laforet V, Peña-Martínez C, Moraga A, Ballesteros I, Cuartero MI, de la Parra J, Palma-Tortosa S, Hidalgo A, Corbí AL, Role of TLR4 (toll-like receptor 4) in N1/N2 neutrophil programming after stroke, Stroke. 50 (10) (2019) 2922–2932. [DOI] [PubMed] [Google Scholar]
- [14].Bird L, Neutrophil plasticity, Nat. Rev. Immunol 9 (10) (2009) 673. [DOI] [PubMed] [Google Scholar]
- [15].Vafadarnejad E, Rizzo G, Krampert L, Arampatzi P, Arias-Loza A-P, Nazzal Y, Rizakou A, Knochenhauer T, Bandi SR, Nugroho VA, Dynamics of cardiac neutrophil diversity in murine myocardial infarction, Circ. Res 127 (9) (2020) (e232–e49). [DOI] [PubMed] [Google Scholar]
- [16].Calcagno DM, Zhang C, Toomu A, Huang K, Ninh VK, Miyamoto S, Aguirre AD, Fu Z, Heller Brown J, King KR, SiglecF (HI) Marks late-stage neutrophils of the infarcted heart: a single-cell transcriptomic analysis of neutrophil diversification, J. Am. Heart Assoc 10 (4) (2021), e019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Daseke II MJ, Chalise U, Becirovic-Agic M, Salomon JD, Cook LM, Case AJ, Lindsey ML, Neutrophil signaling during myocardial infarction wound repair, Cell. Signal 77 (2021), 109816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ohms M, Möller S, Laskay T, An attempt to polarize human neutrophils toward N1 and N2 phenotypes in vitro, Front. Immunol 11 (2020) 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tenkorang MA, Chalise U, Daseke I, Michael J, Konfrst SR, Lindsey ML, Understanding the mechanisms that determine extracellular matrix remodeling in the infarcted myocardium, Biochem. Soc. Trans 47 (6) (2019) 1679–1687. [DOI] [PubMed] [Google Scholar]
- [20].Horckmans M, Ring L, Duchene J, Santovito D, Schloss MJ, Drechsler M, Weber C, Soehnlein O, Steffens S, Neutrophils orchestrate post-myocardial infarction healing by polarizing macrophages towards a reparative phenotype, Eur. Heart J 38 (3) (2017) 187–197. [DOI] [PubMed] [Google Scholar]
- [21].Iyer RP, Patterson NL, Zouein FA, Ma Y, Dive V, de Castro Brás LE, Lindsey ML, Early matrix metalloproteinase-12 inhibition worsens post-myocardial infarction cardiac dysfunction by delaying inflammation resolution, Int. J. Cardiol 185 (2015) 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fox S, Leitch AE, Duffin R, Haslett C, Rossi AG, Neutrophil apoptosis: relevance to the innate immune response and inflammatory disease, Journal of innate immunity. 2 (3) (2010) 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Daseke II MJ, Tenkorang-Impraim MAA, Ma Y, Chalise U, Konfrst SR, Garrett MR, DeLeon-Pennell KY, Lindsey ML, Exogenous IL-4 shuts off pro-inflammation in neutrophils while stimulating anti-inflammation in macrophages to induce neutrophil phagocytosis following myocardial infarction, J. Mol. Cell. Cardiol 145 (2020) 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Carta S, Castellani P, Delfino L, Tassi S, Vene R, Rubartelli A, DAMPs and inflammatory processes: the role of redox in the different outcomes, J. Leukoc. Biol 86 (3) (2009) 549–555. [DOI] [PubMed] [Google Scholar]
- [25].Luerman GC, Uriarte SM, Rane MJ, McLeish KR, Application of proteomics to neutrophil biology, J. Proteome 73 (3) (2010) 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bonecchi R, Garlanda C, Mantovani A, Riva F, Cytokine decoy and scavenger receptors as key regulators of immunity and inflammation, Cytokine. 87 (2016) 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chalise U, Becirovic-Agic M, Lindsey ML, Neutrophil crosstalk during cardiac wound healing after myocardial infarction, Current Opinion Physiology (2022) 100485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Garlichs C, Eskafi S, Cicha I, Schmeisser A, Walzog B, Raaz D, Stumpf C, Yilmaz A, Bremer J, Ludwig J, Delay of neutrophil apoptosis in acute coronary syndromes, J. Leukoc. Biol 75 (5) (2004) 828–835. [DOI] [PubMed] [Google Scholar]
- [29].Mouton AJ, Gonzalez OJR, Kaminski AR, Moore ET, Lindsey ML, Matrix metalloproteinase-12 as an endogenous resolution promoting factor following myocardial infarction, Pharmacol. Res 137 (2018) 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Toldo S, Van Tassell BW, Abbate A, Interleukin-1 blockade in acute myocardial infarction and heart failure: getting closer and closer, American College of Cardiology Foundation Washington DC (2017) 431–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kain V, Halade GV, Role of neutrophils in ischemic heart failure, Pharmacol. Ther 205 (2020), 107424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Albus U, Guide for the Care and Use of Laboratory Animals (8th Edn), SAGE Publications Sage UK, London, England, 2012. [Google Scholar]
- [33].Lee JW, Devanarayan V, Barrett YC, Weiner R, Allinson J, Fountain S, Keller S, Weinryb I, Green M, Duan L, Fit-for-purpose method development and validation for successful biomarker measurement, Pharm. Res 23 (2) (2006) 312–328. [DOI] [PubMed] [Google Scholar]
- [34].Lindsey ML, Gray GA, Wood SK, Curran-Everett D, Statistical considerations in reporting cardiovascular research, Am. J. Phys. Heart Circ. Phys 315 (2) (2018) (H303–H13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hulsen T, de Vlieg J, Alkema W, BioVenn–a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams, BMC Genomics 9 (1) (2008) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xie Z, Bailey A, Kuleshov MV, Clarke DJ, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Gene set knowledge discovery with Enrichr, Current Protocols. 1 (3) (2021), e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shachar I, Karin N, The dual roles of inflammatory cytokines and chemokines in the regulation of autoimmune diseases and their clinical implications, J. Leukoc. Biol 93 (1) (2013) 51–61. [DOI] [PubMed] [Google Scholar]
- [38].ShPd Oliveira C. Canetti R. Ribeiro F. Cunha, Neutrophil migration induced by IL-1β depends upon LTB 4 released by macrophages and upon TNF-α and IL-1β released by mast cells, Inflammation. 31 (1) (2008) 36–46. [DOI] [PubMed] [Google Scholar]
- [39].Mankan AK, Dau T, Jenne D, Hornung V, The NLRP3/ASC/Caspase-1 axis regulates IL-1β processing in neutrophils, Eur. J. Immunol 42 (3) (2012) 710–715. [DOI] [PubMed] [Google Scholar]
- [40].Xiao W, Hodge DR, Wang L, Yang X, Zhang X, Farrar WL, Co-operative functions between nuclear factors NFκB and CCAT/enhancer-binding protein-β (C/EBP-β) regulate the IL-6 promoter in autocrine human prostate cancer cells, Prostate 61 (4) (2004) 354–370. [DOI] [PubMed] [Google Scholar]
- 41.[] Nick JA, Avdi NJ, Young SK, Lehman LA, McDonald PP, Frasch SC, Billstrom MA, Henson PM, Johnson GL, Worthen GS, Selective activation and functional significance of p38α mitogen-activated protein kinase in lipopolysaccharide-stimulated neutrophils, J. Clin. Invest 103 (6) (1999) 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu J, Li X, Yue Y, Li J, He T, He Y, The inhibitory effect of quercetin on IL-6 production by LPS-stimulated neutrophils, Cell Mol Immunol. 2 (6) (2005) 455–460. [PubMed] [Google Scholar]
- [43].Guijarro-Muñoz I, Compte M, Alvarez-Cienfuegos A, Álvarez-Vallina L, Sanz L, Lipopolysaccharide activates toll-like receptor 4 (TLR4)-mediated NF-κB signaling pathway and proinflammatory response in human pericytes, J. Biol. Chem 289 (4) (2014) 2457–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gorski SA, Lawrence MG, Hinkelman A, Spano MM, Steinke JW, Borish L, Teague WG, Braciale TJ, Expression of IL-5 receptor alpha by murine and human lung neutrophils, PLoS One 14 (8) (2019), e0221113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huang J, Yue H, Jiang T, Gao J, Shi Y, Shi B, Wu X, Gou X, IL-31 plays dual roles in lung inflammation in an OVA-induced murine asthma model, Biology Open. 8 (1) (2019) bio036244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Assier E, Jullien V, Lefort J, Moreau J-L, Di Santo JP, Vargaftig BB, e Silva JRL, Thèze J, NK cells and polymorphonuclear neutrophils are both critical for IL-2-induced pulmonary vascular leak syndrome, J. Immunol 172 (12) (2004) 7661–7668. [DOI] [PubMed] [Google Scholar]
- [47].Gaba A, Grivennikov SI, Do MV, Stumpo DJ, Blackshear PJ, Karin M, Cutting edge: IL-10–mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production, J. Immunol 189 (5) (2012) 2089–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yu H, Pardoll D, Jove R, STATs in cancer inflammation and immunity: a leading role for STAT3, Nat. Rev. Cancer 9 (11) (2009) 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murray P, STAT3-mediated Anti-inflammatory Signalling, Portland Press Ltd., 2006. [DOI] [PubMed] [Google Scholar]
- [50].Szczepanek K, Xu A, Hu Y, Thompson J, He J, Larner AC, Salloum FN, Chen Q, Lesnefsky EJ, Cardioprotective function of mitochondrial-targeted and transcriptionally inactive STAT3 against ischemia and reperfusion injury, Basic Res. Cardiol 110 (6) (2015) 1–10. [DOI] [PubMed] [Google Scholar]
- [51].Wu L, Tan J-L, Chen Z-Y, Huang G, Cardioprotection of post-ischemic moderate ROS against ischemia/reperfusion via STAT3-induced the inhibition of MCU opening, Basic Res. Cardiol 114 (5) (2019) 1–15. [DOI] [PubMed] [Google Scholar]
- [52].Harhous Z, Booz GW, Ovize M, Bidaux G, Kurdi M, An update on the multifaceted roles of STAT3 in the heart, Frontiers in cardiovascular medicine. 6 (2019) 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Haghikia A, Ricke-Hoch M, Stapel B, Gorst I, Hilfiker-Kleiner D, STAT3, a key regulator of cell-to-cell communication in the heart, Cardiovasc. Res 102 (2) (2014) 281–289. [DOI] [PubMed] [Google Scholar]
- [54].Lips DJ, Bueno OF, Wilkins BJ, Purcell NH, Kaiser RA, Lorenz JN, Voisin L, Saba-El-Leil MK, Meloche S, Pouysségur J, MEK1-ERK2 signaling pathway protects myocardium from ischemic injury in vivo, Circulation. 109 (16) (2004) 1938–1941. [DOI] [PubMed] [Google Scholar]
- [55].Simard FA, Cloutier A, Ear T, Vardhan H, McDonald PP MEK -independent ERK activation in human neutrophils and its impact on functional responses, J. Leukoc. Biol 98 (4) (2015) 565–573. [DOI] [PubMed] [Google Scholar]
- [56].Downey GP, Butler JR, Tapper H, Fialkow L, Saltiel AR, Rubin BB, Grinstein S, Importance of MEK in neutrophil microbicidal responsiveness, J. Immunol 160 (1) (1998) 434–443. [PubMed] [Google Scholar]
- [57].Yeh CC, Malhotra D, Yang YL, Xu Y, Fan Y, Li H, Mann MJ, MEK1-induced physiological hypertrophy inhibits chronic post-myocardial infarction remodeling in mice, J. Cell. Biochem 114 (1) (2013) 47–55. [DOI] [PubMed] [Google Scholar]
- [58].Mehrpouya-Bahrami P, Moriarty AK, De Melo P, Keeter WC, Alakhras NS, Nelson AS, Hoover M, Barrios MS, Nadler JL, Serezani CH, STAT4 is expressed in neutrophils and promotes antimicrobial immunity, JCI insight. 6 (14) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lentsch AB, Kato A, Davis B, Wang W, Chao C, Edwards MJ, STAT4 and STAT6 regulate systemic inflammation and protect against lethal endotoxemia, J. Clin. Invest 108 (10) (2001) 1475–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chalise U, Daseke MJ, Kalusche WJ, Konfrst SR, Rodriguez-Paar JR, Flynn ER, Cook LM, Becirovic-Agic M, Lindsey ML, Macrophages secrete murinoglobulin-1 and galectin-3 to regulate neutrophil degranulation after myocardial infarction, Molecular Omics. 18 (3) (2022) 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Xu Y, Wang B, Liu X, Deng Y, Zhu Y, Zhu F, Liang Y, Li H, Sp1 targeted PARP1 inhibition protects cardiomyocytes from myocardial ischemia–reperfusion injury via downregulation of autophagy, Frontiers in Cell and Developmental Biology. 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Geng H, Su Y, Huang R, Fan M, Li X, Lu X, Sheng H, Specific protein 1 inhibitor mithramycin a protects cardiomyocytes from myocardial infarction via interacting with PARP, In Vitro Cellular & Developmental Biology-Animal. 57 (3) (2021) 315–323. [DOI] [PubMed] [Google Scholar]
- [63].Li R, Geng H-H, Xiao J, Qin X-T, Wang F, Xing J-H, Xia Y-F, Mao Y, Liang J-W, Ji X-P, miR-7a/b attenuates post-myocardial infarction remodeling and protects H9c2 cardiomyoblast against hypoxia-induced apoptosis involving Sp1 and PARP-1, Sci. Rep 6 (1) (2016) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].McLoughlin TJ, Smith SM, DeLong AD, Wang H, Unterman TG, Esser KA, FoxO1 induces apoptosis in skeletal myotubes in a DNA-binding-dependent manner, Am. J. Phys. Cell Phys 297 (3) (2009) (C548–C55). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Liu X, Zhao H, Jin Q, You W, Cheng H, Liu Y, Song E, Liu G, Tan X, Zhang X, Resveratrol induces apoptosis and inhibits adipogenesis by stimulating the SIRT1-AMPKα-FOXO1 signalling pathway in bovine intramuscular adipocytes, Mol. Cell. Biochem 439 (1) (2018) 213–223. [DOI] [PubMed] [Google Scholar]
- [66].Shen M, Lin F, Zhang J, Tang Y, Chen W-K, Liu H, Involvement of the upregulated FoxO1 expression in follicular granulosa cell apoptosis induced by oxidative stress, J. Biol. Chem 287 (31) (2012) 25727–25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Shao D, Zhai P, Del Re DP, Sciarretta S, Yabuta N, Nojima H, Lim D-S, Pan D, Sadoshima J, A functional interaction between hippo-YAP signalling and FoxO1 mediates the oxidative stress response, Nat. Commun 5 (1) (2014) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhao Z, Li C, Xi H, Gao Y, Xu D, Curcumin induces apoptosis in pancreatic cancer cells through the induction of forkhead box O1 and inhibition of the PI3K/Akt pathway, Mol. Med. Rep 12 (4) (2015) 5415–5422. [DOI] [PubMed] [Google Scholar]
- [69].Hori YS, Kuno A, Hosoda R, Horio Y, Regulation of FOXOs and p53 by SIRT1 modulators under oxidative stress, PLoS One 8 (9) (2013), e73875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kim DH, Kim JY, Yu BP, Chung HY, The activation of NF-κB through Akt-induced FOXO1 phosphorylation during aging and its modulation by calorie restriction, Biogerontology. 9 (1) (2008) 33–47. [DOI] [PubMed] [Google Scholar]
- [71].Watson A, Eilers A, Lallemand D, Kyriakis J, Rubin LL, Ham J, Phosphorylation of c-Jun is necessary for apoptosis induced by survival signal withdrawal in cerebellar granule neurons, J. Neurosci 18 (2) (1998) 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Behrens A, Sibilia M, Wagner EF, Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation, Nat. Genet 21 (3) (1999) 326–329. [DOI] [PubMed] [Google Scholar]
- [73].Dejana E, The role of wnt signaling in physiological and pathological angiogenesis, Circ. Res 107 (8) (2010) 943–952. [DOI] [PubMed] [Google Scholar]
- [74].Huang H, He X, Wnt/β-catenin signaling: new (and old) players and new insights, Curr. Opin. Cell Biol 20 (2) (2008) 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Niessen CM, Gottardi CJ, Molecular components of the adherens junction, Biochimica et Biophysica Acta (BBA)-Biomembranes. 1778 (3) (2008) 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Meng W, Takeichi M, Adherens junction: molecular architecture and regulation, Cold Spring Harb. Perspect. Biol 1 (6) (2009), a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bellac CL, Dufour A, Krisinger MJ, Loonchanta A, Starr AE, Keller U, Lange PF, Goebeler V, Kappelhoff R, Butler GS, Macrophage matrix metalloproteinase-12 dampens inflammation and neutrophil influx in arthritis, Cell Rep. 9 (2) (2014) 618–632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are provided in supplementary files.