SUMMARY
Omicron subvariants continuingly challenge current vaccination strategies. Here, we demonstrate nearly complete escape of the XBB.1.5, CH.1.1, and CA.3.1 variants from neutralizing antibodies stimulated by three doses of mRNA vaccine or by BA.4/5 wave infection, but neutralization is rescued by a BA.5-containing bivalent booster. CH.1.1 and CA.3.1 show strong immune escape from monoclonal antibody S309. Additionally, XBB.1.5, CH.1.1, and CA.3.1 spike proteins exhibit increased fusogenicity and enhanced processing compared with BA.2. Homology modeling reveals the key roles of G252V and F486P in the neutralization resistance of XBB.1.5, with F486P also enhancing receptor binding. Further, K444T/M and L452R in CH.1.1 and CA.3.1 likely drive escape from class II neutralizing antibodies, whereas R346T and G339H mutations could confer the strong neutralization resistance of these two subvariants to S309-like antibodies. Overall, our results support the need for administration of the bivalent mRNA vaccine and continued surveillance of Omicron subvariants.
In brief
Qu et al. show that bivalent booster recipients, compared with monovalent recipients, exhibit higher nAb titers against Omicron subvariants XBB, XBB.1, and XBB.1.5. The CH.1.1 and CA.3.1 variants show more substantial neutralization escape than the XBB variants. Further, structural modeling reveals that the F486P mutation in XBB.1.5 enhances ACE2 binding.
Graphical Abstract
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, continues to circulate across the globe while evolving rapidly. The beginning of 2022 was marked by the emergence of the Omicron BA.1/BA1.1 variant, establishing a turning point in the pandemic with decreased pathogenicity,1–5 increased transmissibility,2 and enhanced immune escape.6–13 During 2022, the prototype Omicron variant has given rise to numerous subvariants, with many displaying even greater immune escape,9,14–22 endangering the efficacy of vaccination efforts.
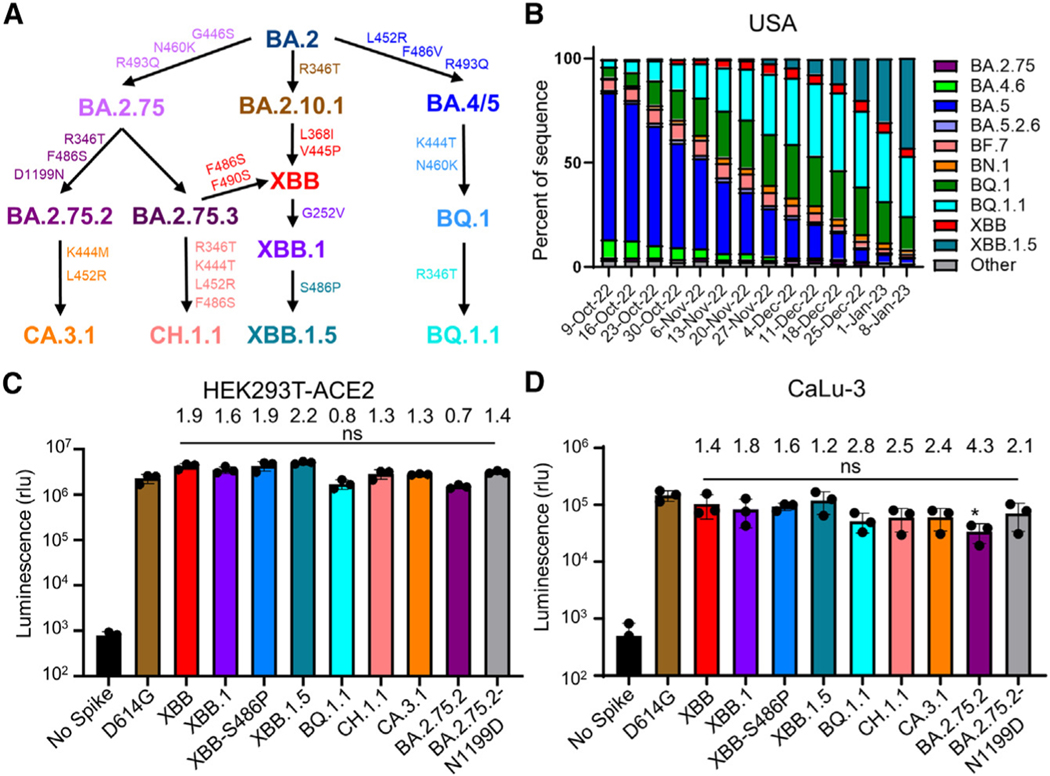
Following a few months of BA.5 dominance in the summer of 2022, a highly immune evasive16,23,24 Omicron subvariant, i.e., BQ.1.1, became the most prevalent in the United States; however, it is now being quickly supplanted by a new subvariant, XBB.1.5.25 The XBB lineage was initially discovered in India in mid-August of 2022, resulting from a recombination event between two BA.2 lineages, BA.2.10.1 and BA.2.75.26 The emergence of this subvariant raised much alarm, as it has brought together a number of mutations in the spike (S) protein with established immune evasion functions, including R346T, G446S, and F486S (Figure 1A).15 Importantly, the efficacies of monoclonal antibody treatments24 and both monovalent23 and bivalent16,27 vaccination strategies, as well as immunity stimulated by infection,23,27 are all reduced against XBB. Recently, XBB has acquired two more mutations in the S protein, including G252V (XBB.1) and G252V + S486P (XBB.1.5) (Figure 1A). The influence of these mutations on XBB.1 and XBB.1.5 is currently unknown, although mutations at residue F486, such as F486V, F486I, and F486S, have been recurring among prior Omicron subvariants,26 representing a critical evolutionary hotspot.28 Given the rapid increased circulation of XBB.1.5 in the United States and other parts of the world (Figures 1B and S1A), it is crucial that we understand its impact on current public health measures.
Figure 1. Distribution and infectivity of emerging Omicron subvariants XBB.1.5, CH.1.1, and CA.3.1.
(A) Schematic depiction of the relationships between different Omicron subvariants, with key lineage-defining amino acid mutations for each displayed.
(B) Distribution of recently emerged Omicron subvariants in the United States starting in early October of 2022 through the beginning of January 2023. Data were collected from the Centers for Disease Control and Prevention25 and plotted using Prism software.
(C and D) Infectivity of pseudotyped lentiviruses carrying each of the indicated S proteins of the Omicron subvariants were determined in (C) HEK293T cells overexpressing human ACE2 and
(D) human lung epithelia-derived CaLu-3 cells. Bars in (C) and (D) represent means ± standard deviation from three biological replicates of one typical experiment. Significance relative to D614G was determined using unpaired two-sided Student’s t tests (n = 3). p values are displayed as ns p > 0.05. The fold change in the mean viral titer of Omicron subvariants was calculated relative to that of D614G.
In addition to BQ.1, BQ.1.1, and XBB subvariants, two other Omicron subvariants derived from BA.2.75, CH.1.1 and CA.3.1, have also drawn attention. CH.1.1 emerged in Southeast Asia in November of 2022 and accounted for more than 25% of infections in some parts of the UK and New Zealand in January 2023; it has caused alarm due to the appearance of the L452R mutation in the S protein,29 which previously appeared in the more pathogenic Delta variant and in the highly transmissible BA.4/ 5 variants.18,30,31 CA.3.1 emerged in the United States in December of 2022 and also carries this critical L452R mutation.29 CA.3.1 is derived from the BA.2.75.2 variant, whereas CH.1.1 is derived from the BA.2.75.3 variant. Notably, BA.2.75.3 lacks the R346T and F486S RBD mutations as well as the D1199N mutation present in BA.2.75.2 (Figure 1A). In contrast, CH.1.1 acquired the R346T and F486S mutations present in BA.2.75.2 as well as harbored the additional K444T and L452R receptor-binding domain (RBD) mutations (Figure 1A). To better assess the impact of these mutations novel to the BA.2.75 clade, we compare CH.1.1 with BA.2.75.2 lacking the D1199N mutation (BA.2.75.2-N1199D) in order to evaluate the effect of the K444T and L452R mutations in the R346T- and F486S-containing background (Figure 1A). In this study, we investigate aspects of S protein biology of XBB.1.5, CH.1.1, and CA.3.1 in comparison with their related variants, including entry into host cells, surface expression, fusogenicity, and processing. Most critically, we determine and compare their sensitivity to the monoclonal antibody (mAb) S309, which has been shown to effectively neutralize some of the previous Omicron subvariants including BQ.1,32,33 as well as neutralizing antibodies stimulated by either bivalent or monovalent mRNA vaccination and previous infection (BA.5 wave), alongside ancestral variants D614G, BA.2, and BA.2.75.2 as well as the previously dominating variant BQ.1.1.
RESULTS
Omicron subvariant XBB.1.5 exhibits an increase in viral infectivity, especially in CaLu-3 cells
First, we determined the infectivity of lentiviruses pseudotyped with each of these subvariant S proteins in HEK293T cells stably expressing human ACE2 (HEK293T-ACE2) and the human lung epithelial cell line CaLu-3. Both XBB and XBB.1.5 exhibited increased infectivity in HEK293T-ACE2 cells, with 1.9 and 2.2 times higher titers compared with D614G, respectively (Figure 1C). The XBB.1.5 variant with lineage-defining mutation S486P and the G225V-containing XBB.1 variant also exhibited an increase in infectivity, with infectivity 1.9 and 1.6 times higher than D614G, respectively (Figure 1C). Of note, the infectivity of XBB.1.5 was not significantly higher than that of XBB (Figure 1C). In contrast to the prototype Omicron BA.1 and subsequent subvariants that showed 3–5 times decreased infectivity in CaLu-3 cells,6,14,17 we found that the infectivity of these XBB subvariants was not significantly different from D614G, with titers only 1.4 and 1.2 times lower for XBB and XBB.1.5, than D614G, respectively (Figure 1D). However, subvariants CH.1.1 and CA.3.1 still exhibited substantial decreases in infectivity, with titers 2.5 and 2.4 times lower than D614G, respectively (Figure 1D). Consistent with previous results, BA.2.75.2 exhibited an infectivity of 4.3 times lower than D614G, the lowest infectivity in CaLu-3 cells compared with D614G among all subvariants tested here (Figure 1D). Together, these results appear to suggest that XBB.1.5, along with XBB, has gained modestly increased infectivity compared with the other Omicron subvariants, including in CaLu-3 cells, although further investigation in primary lung epithelial cells and airway tissue is needed (see discussion).
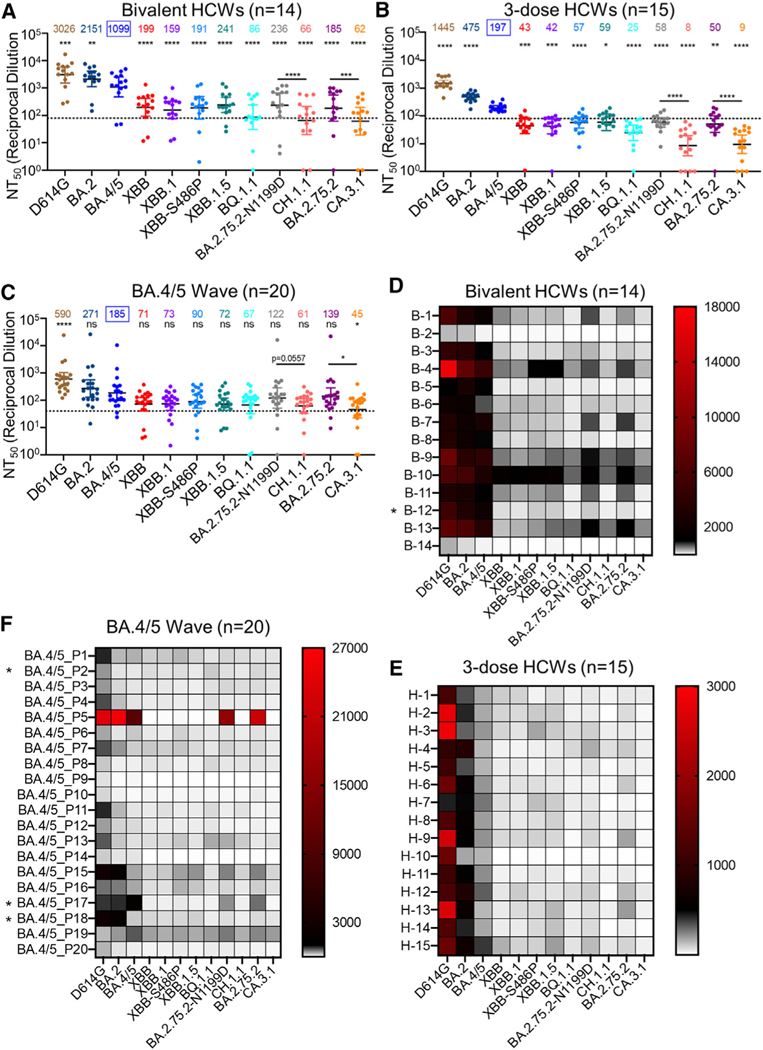
Escape of neutralizing antibodies by XBB.1.5, CH.1.1, and CA.3.1 in bivalent vaccinated sera
In order to investigate neutralization resistance of emerging Omicron subvariants, we used our previously reported pseudotyped lentivirus neutralization assay.34 We first examined their neutralization resistance to sera from 14 healthcare workers (HCWs) who had received a bivalent booster in addition to 2–4 doses of monovalent mRNA vaccine (n = 14, 8 male and 6 female) (Table S1). The bivalent booster formulation included prototype SARS-CoV-2 S and BA.5 S. Among them, 12 HCWs received 3 doses of the monovalent Moderna mRNA-1273 or Pfizer BioNTech BNT162b2 vaccine followed by an additional 1 dose of the bivalent Pfizer or Moderna vaccine, 1 HCW received 2 doses of the monovalent Pfizer BioNTech BNT162b2 vaccine and an additional 1 dose of the bivalent Pfizer vaccine, and 1 HCW received 4 doses of the monovalent Pfizer BioNTech BNT162b2 vaccine plus an additional 1 dose of the bivalent Pfizer vaccine. For COVID-19 status, 4 were negative throughout, 7 became positive during the Omicron wave, and 3 tested positive prior to Omicron. Sera were collected between 23 and 108 days after receiving a bivalent vaccination (median 66 days).
Because of the continued dominance of Omicron subvariants, especially by BA.5 after the summer of 2022, all below comparisons for neutralization were made with BA.4/5 rather than the ancestral D614G. Strong neutralization resistance was exhibited by XBB.1.5, CH.1.1, and CA.3.1, with mean neutralizing antibody (nAb) titers 4.6–7.3 (p < 0.0001), 16.7–20.5 (p < 0.0001), and 17.7–23.2 times (p < 0.0001) lower than BA.4/5, respectively (Figures 2A and 2D). Of note, the nAb titers for the 10 HCWs with breakthrough infection were higher than those of the 4 HCWs without breakthrough infection (Figure S2C), which was consistent with our previous findings,35,36 indicating that breakthrough infection induces higher and broader nAbs. Somewhat surprisingly, XBB.1.5 showed a modest increase in nAb titer compared with the parental XBB variant (Figures 2A, 2D, and S2C). Moreover, neither of the two defining mutations for XBB.1.5 (G252V and S486P) contributed to the enhanced neutralization by bivalent sera, with nAb titers actually 6.9 (p < 0.0001) and 5.8 times (p < 0.0001) lower than BA.4/5, respectively (Figures 2A, 2D, and S2C). Notably, we repeatedly observed that BQ.1.1 exhibited a higher extent of neutralization resistance than all XBB subvariants (see discussion), with a nAb titer 12.8 times lower than BA.4/5 (p < 0.0001) and near the limit of detection (Figures 2A, 2D, and S2C). We found that CH.1.1 had greater immune escape than BA.2.75.2-N1199D, with a 2.7 times reduced nAb titer (p < 0.0001), whereas CA.3.1 exhibited stronger immune escape than its parental BA.2.75.2, with 3 times lower nAb titers (p < 0.0001) (Figures 2A, 2D, and S2C). Overall, we observed comparably strong immune escape among XBB subvariants, including XBB.1.5 compared with BQ.1.1, but much more enhanced neutralization resistance for CH.1.1 and CA.3.1.
Figure 2. Neutralization of Omicron XBB.1.5, CH.1.1, and CA.3.1 subvariants by sera of bivalent or monovalent mRNA vaccinated healthcare workers (HCWs) and BA.4/5 wave infection.
Neutralizing antibody titers were determined using lentiviral pseudotypes carrying each of the indicated S proteins of the Omicron subvariants. They were compared against BA.4/5 and/or respective parental Omicron subvariants as specified in the text. The cohorts included sera from 14 HCWs that received 3 monovalent doses of mRNA vaccine plus a dose of bivalent mRNA vaccine (n = 14) (A and D), 15 sera from HCWs that only received three doses of monovalent mRNA vaccine (B and E), and 20 sera from BA.4/5-wave SARS-CoV-2-infected first responders and household contacts that tested positive during the BA.4/5 wave of infection in Columbus, Ohio (C and F). Bars represent geometric means with 95% confidence intervals. Geometric mean NT50 values are displayed for each subvariant on the top. Statistical significance was determined using log10-transformed NT50 values to better approximate normality. Comparisons between multiple groups were made using a one-way ANOVA with Bonferroni post-test. Comparisons between two groups were performed using a paired, two-tailed Student’s t test with Welch’s correction. Dashed lines indicate the threshold of detection (80 for monovalent and bivalent mRNA vaccinees and 40 for BA.4/5 infection cohort). p values are displayed as ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Heatmaps in (D)–(F) depict neutralizing antibody titers by each individual against each Omicron subvariant tested. Asterisk in (D) indicates the individual infected by SARSCoV-2 within 6 months before the sera sample collection, and asterisk in (F) indicates the individuals who had received three doses of mRNA vaccine before infection.
XBB.1.5, CH.1.1, and CA.3.1 exhibit an almost complete escape of nAbs in three-dose-vaccinated sera
Next, we investigated neutralization resistance of these new Omicron subvariants in sera from Ohio State University Wexner Medical Center HCWs who had received 3 doses of monovalent mRNA vaccine (n = 15) (Figures 2B, 2E, and S2B). Samples were collected 2–13 weeks after vaccination with a homologous booster dose of the monovalent Pfizer/BioNTech BNT162b2 vaccine (n = 12) or Moderna mRNA-1273 (n = 3). These HCWs included 10 male and 5 female individuals and ranged from 26 to 61 years of age (median 33 years) (Table S1). The average nAb titers of 3-dose mRNA vaccine recipients against D614G, BA.2, and BA.4/5 were about 1.9–5.6 times lower than those of bivalent mRNA vaccines recipients (Figures 2A, 2B, S2A, and S2B); dramatic reductions in neutralization sensitivity were observed for XBB.1.5, CH.1.1, and CA.3.1, which exhibited complete escape from nAbs, with mean nAb titers 3.3–4.5 (p < 0.05), 13.6–24.6 (p < 0.0001), and 15.4–21.9 times (p < 0.0001) lower than BA.4/5, respectively (Figures 2B, 2E, and S2B). Similarly, CH.1.1 and CA.3.1 subvariants also had dramatically lower nAb titers than BA.2.75.2-N1199D and BA.2.75.2, respectively (Figures 2B and 2E). Importantly, the overall trends for each subvariant in the 3-dose mRNA vaccine cohort remained similar to that of bivalent mRNA vaccination (Figures 2A, 2B, 2D, 2E, S2A, and S2B), and this was even more obvious for the subgroup (n = 4) that had relatively high nAb titers (Figure S2D).
Omicron subvariants XBB.1.5, CH.1.1, and CA.3.1 are virtually resistant to neutralization by sera of BA.4/5 infection
We also examined neutralization resistance of XBB.1.5, CH.1.1, and CA.3.1 to sera from a BA.4/5 infection wave among Columbus, Ohio (USA) first responders and household contacts that tested positive for COVID-19 (n = 20) (Figures 2C and 2F). Nasal swab samples were sequenced to identify the specific variant that caused infection, with 4 patients being infected by BA.4 or BA.4-derivative variants, 7 patients being infected with BA.5 or BA.5-derivative variants, and 9 patients being infected with undetermined SARS-CoV-2 variants (Table S1). All sample collection occurred during a BA.4- and BA.5-dominant period in Columbus, Ohio (July 2022 through September 2022). In this cohort, 17 individuals were unvaccinated, and 3 individuals had received 3 doses of either the Pfizer BioNTech BNT162b2 (n = 1) or Moderna mRNA-1272 (n = 2) vaccine (Table S1). Similar to the results shown above for the bivalent and monovalent mRNA vaccines, strong and almost complete neutralization resistance was observed for XBB.1.5, CH.1.1, and CA.3.1, with nAb titers 2.6 (p > 0.05), 3 (p > 0.05), and 4.1 times (p < 0.05) lower than BA.4/5, respectively (Figures 2C, 2F, and S2E). Again, overall trends for each subvariant in this cohort followed the same patterns demonstrated in cohorts described for the bivalent and monovalent mRNA vaccines.
Complete immune escape from mAb S309 by Omicron subvariants CH.1.1 and CA.3.1 but not XBB.1.5 and BQ.1.1
Recent studies show that most therapeutic mAbs lost neutralizing activity either substantially or completely against emerging Omicron subvariants.28,32,33 We examined the resistance of XBB.1.5, CH.1.1, and CA.3.1 along with BQ.1.1 to neutralization by the mAb S309, which is one of the few mAbs still effective against BQ.1 and XBB.32,33 Consistent with published results,28,32 BQ.1.1 and XBB were still effectively neutralized by S309, with half-maximal inhibitory concentration (IC50) values of 0.78 ± 0.06 and 2.50 ± 0.83 μg/mL, respectively (Figures S3A and S3B). The XBB subvariants, including XBB.1, XBB-S486P, and XBB.1.5, showed modestly enhanced sensitivity to S309, with 2.5, 2.1, and 1.3 times lower IC50 than the parental XBB variant, respectively. Interestingly, S309 cannot neutralize CH.1.1, CA.3.1, BA.2.75.2, and BA.2.75.2-N1199D, even at 12 μg/mL, the highest concentration we used.
Fusogenicity, surface expression, and processing of XBB.1.5, CH.1.1, and CA.3.1 S proteins
To investigate the biological function of the S proteins of these new Omicron subvariants, we investigated S fusogenicity, surface expression, and processing. Consistent with our previous reports,15,17,37 all subvariants tested exhibited reduced syncytia formation compared with the ancestral D614G (p < 0.0001) but with a clear increase in fusion relative to BA.2 (Figures 3A and 3B). Like BQ.1.1 and BA.2.75.2,17 subvariants XBB.1.5, CH.1.1, and CA.3.1 also showed enhanced fusogenicity compared with BA.4/5 (Figures 3A and 3B). However, relative to the parental XBB, the XBB.1.5 subvariant and its two single mutants, XBB.1 containing G252V and XBB-S486P, did not demonstrate obvious differences in S fusogenicity (Figures 3A and 3B). The syncytia formation efficiencies of CH.1.1 and CA.3.1 were comparable to that of BA.2.75.2-N1199D but seemed much lower than BA.2.75.2 (Figures 3A and 3B). The apparently higher fusogenicity of BA.2.75.2 compared with BA.2 was consistent with our previous observations.17 Importantly, the increased fusogenicity of these new Omicron subvariants from HEK293T-ACE2 cells were also observed when CaLu3 was used as a target cell (Figures S4A and S4B). Additionally, the differences in fusogenicity of these Omicron subvariants with parental D614G and BA.2 variants could not be attributed to differences in surface expression because comparable levels of fluorescence signal were detected on cells expressing individual S proteins as measured by flow cytometry (Figures 3C and 3D).
Figure 3. Syncytia formation, cell surface expression, and S processing of Omicron XBB.1.5, CH.1.1, and CA.3.1 subvariants.
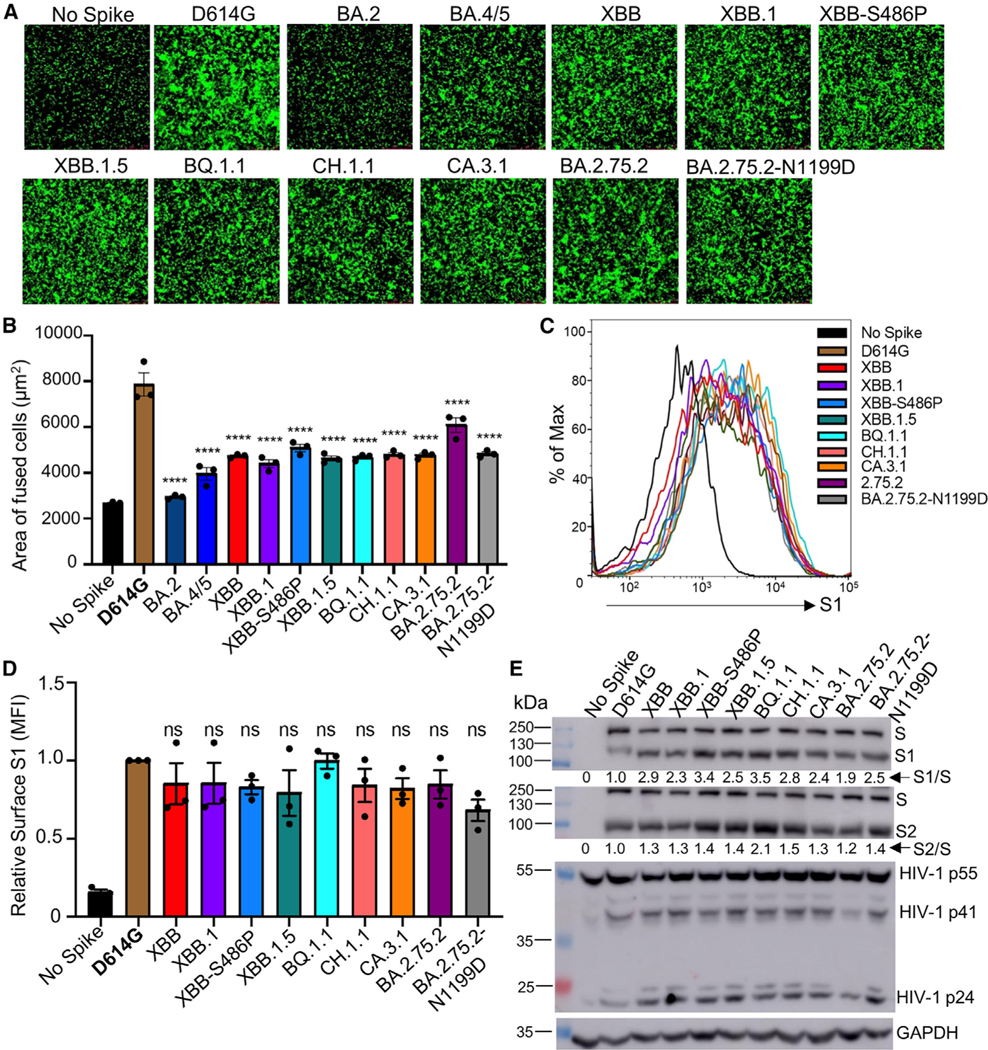
(A and B) Syncytia-forming activity. HEK293T-ACE2 cells were co-transfected with Omicron subvariant S proteins and GFP and incubated for 30 h before (A) imaging and (B) quantifying syncytia. D614G and no S serve as positive and negative controls, respectively. Comparisons in the extent of syncytia for each variant were made against D614G, with p values indicating statistical significance. Similar results were obtained by using CaLu3 cell as target (see data in Figure S4).
(C and D) Cell surface expression of S proteins. HEK293T cells used for production of pseudotyped lentiviral vectors bearing S proteins (Figures 1 and 2) from Omicron subvariants were fixed and surface stained for S with an anti-S1 specific antibody T62 followed by flow cytometric analyses. (C) Histogram plots of anti-S1 signals in transfected cells and (D) calculated relative mean fluorescence intensities of each subvariant by setting the value of D614G as 1.
(E) S expression and processing. HEK293T cells used to produce pseudotyped vectors were lysed and probed with anti-S1, anti-S2, anti-GAPDH (loading control), or anti-p24 (HIV capsid, transfection control) antibodies; the signal for anti-GAPDH was from reblotting the membrane of anti-S1, and the signal for anti-S2 was from reblotting the membrane of anti-p24. S processing was quantified using NIH ImageJ used to determine an S1/S or S2/S ratio and normalized to D614G (D614G = 1.0).
Bars in (B) and (D) represent means ± standard error. Dots represent three biological replicates from one typical experiment. Significance relative to D614G was determined using a one-way repeated measures ANOVA with Bonferroni’s multiple testing correction (n = 3). p values are displayed as ns p > 0.05 and ****p < 0.0001.
Next, we investigated the S processing of these Omicron subvariants by immunoblotting using pseudotyped virus producer cell lysates. While the expression levels of these Omicron S proteins were comparable, all XBB subvariants, including XBB.1.5, CH.1.1, and CA.3.1, showed increased S processing compared with D614G; this was evidenced by increased S1/S and S2/S ratios, which was also true for BQ.1.1 and BA.2.75.2, consistent with our previous reports17,38 (Figure 3E). Importantly, S processing for XBB.1.5 remained comparable to that of XBB, though a notable increase in S processing for the XBB-S486P mutant was observed (Figure 3E), which was consistent with its relatively higher cell-cell fusion activity (Figures 3A, 3B, S4A, and S4B). No obvious differences in S processing for CH.1.1 and CA.3.1 were seen compared with the BA.2.75.2 subvariant (Figure 3E).
Homology modeling reveals critical roles of lineage-defining mutations on XBB.1.5, CH.1.1, and CA.3.1 in receptor binding and immune evasion
To determine the impact of mutations S486P and G252V on immune evasion and receptor binding, we modeled the structures of XBB lineage S in complex with either receptor ACE2 or representative nAbs targeting these two residues. Located at the critical RBD-ACE2 contact interface, residue F486 present in the parental BA.2 subvariant is embedded in a hydrophobic groove formed by F28, L79, M82, and Y83 on ACE2; this contrasts with F486S or F486P (present in XBB/XBB.1 or XBB.1.5 subvariants), the side chain of which does not fit into the groove (Figure 4A). In addition, compared with the hydrophilic S486, residue P486 in XBB.1.5 is more hydrophobic, thus forming more energetically favorable interactions with L79 and M82 on ACE2 and enabling better receptor utilization than XBB and XBB.1 (Figure 4A). Moreover, residue F486 is an antigenic hotspot for class I nAb recognition.28 For example, therapeutic mAb AZD8895 focuses its recognition on residue F486, with multiple antibody residues forming a surrounding hydrophobic cage; however, this interaction is abolished by the F486 S/P mutations present in XBB and XBB.1.5 (Figure 4B). Residue G252 is located on the S N-terminal domain (NTD), which is also frequently recognized by nAbs; Figure 4C shows a representative NTD-targeting antibody (COVOX-159) focusing its recognition on residue G252, yet a G252V mutation creates a steric hindrance, abolishing this antibody recognition. Additionally, residues K444 and L452 are located within a common epitope site of class II RBD-targeting nAbs (Figure 4D); however, mutations in these two residues, i.e., K444T/M and L452R present in CH.1.1 and CA.3.1 subvariants, impact these interactions, leading to enhanced escape from established immunity induced by past vaccination or infection.
Figure 4. Homology modeling of key mutations in XBB.1.5, CH.1.1, and CA.3.1.
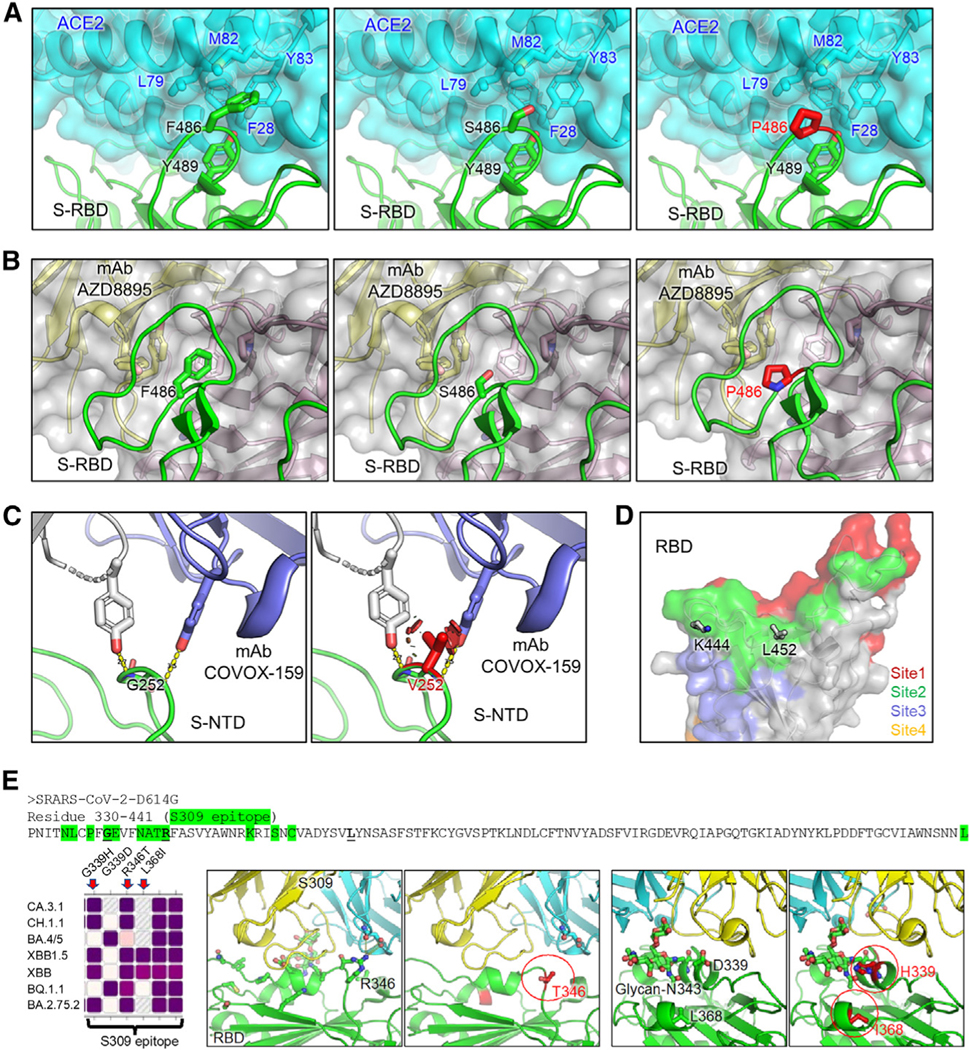
(A) Structures of S receptor-binding domain (RBD)-ACE2 binding interface shown as ribbons.
(B) Structure of RBD with class I antibody AZD8895. The recognition focuses on residue F486, with multiple antibody residues forming a surrounding hydrophobic cage, whereas this interaction is abolished by F486S/P mutation.
(C) Structures of an immune-dominant region of S N-terminal domain (NTD) with a representative antibody COVOX-159. The nAb recognition on residue G252 is abolished by G252V mutation through creating a steric hindrance (shown as red plates).
(D) Residues K444 and L452 are located within acommon epitope site of class II RBD-targeting neutralizing antibodies represented as green surface.
(E) Antibody S309 epitope and sequence diversity.(Top) S protein sequence (330–441) with residues of antibody S309 epitope highlighted in green, and mutation hotspots in bold font, (left) amino acid variation at residues 339, 346, and 368 among different Omicron subvariants, (middle) R346T abolishes a salt bridge and a hydrogen bond; and (right) G/D339H interferes with the S309 recognition of glycan-N343 while L368I stabilizes it.
The enhanced resistance of CH.1.1 and CA.3.1 to mAb S309 compared with XBB.1.5 (Figures S3A and S3B) was also examined by structural modeling. The epitope of mAb S309 constitutes of 13 residues from 330 to 441, which is highlighted in green of Figure 4E39; notably, the residues R346 and D339 would impact the recognition of SARS-CoV-2S upon certain mutations occurring. For example, the R346T mutation abrogates RBD interactions with the S309 light-chain, including a salt bridge and a hydrogen bond, whereas G339H mutation negatively interferes with the glycan-N343 recognition. In addition, CA.3.1, CH.1.1, and BA.2.75.2 all contain R346T and G339H, which are likely responsible for the strong resistance of these three subvariants to S309. In contrast, L368I, which is only present in all XBB variants, might partially offset the effect of R346T and G339H on S309 binding by stabilizing local conformation near the glycanN343, thus enhancing the glycan recognition (Figure 4E); this could explain why XBB subvariants are still sensitive to neutralization by S309.
DISCUSSION
As SARS-CoV-2 continues to mutate and evolve, it is critical to monitor how the biology of the virus changes and the impact on the efficacy of current vaccines, including the current bivalent mRNA vaccines. In this work, we found that the bivalent mRNA vaccine recipients exhibit approximately 2- to 8-fold higher nAb titers, depending on variants tested, following a 3-dose monovalent mRNA vaccination, and the results are consistent with enhanced vaccine efficacy for the bivalent formula.40 While participants were different in these two cohorts, we did note that one HCW received a third monovalent vaccine dose followed by a fourth dose with the bivalent vaccine plus had a breakthrough infection (testing COVID-19 positive between the third dose and the fourth dose, ~180 days before the bivalent sample collection), and this person exhibited generally 3- to 6-fold higher nAb titers against all variants tested (Table S2). The nearly complete escape of 3-dose monovalent vaccine sera and also BA.4/5 wave infection sera exhibited by all Omicron subvariants, especially XBB subvariants and CH.1.1 and CA.3.1, was remarkable, and this is supported by some recent studies.23,24,27,32,41,42 Notably, XBB.1.5 did not exhibit enhanced neutralization resistance over the recently dominant BQ.1.1 variant, which appears to differ from some recent publications.28,32 However, we would like to point out that the neutralization resistance to sera between XBB and BQ subvariants is probably on comparable levels, especially when the calculated titers between them are near the limit of detection. Nevertheless, it is clear that CH.1.1 and CA.3.1 have a consistently stronger neutralization resistance than XBB, XBB.1, and XBB.1.5, which is astonishing and warrants continuous monitoring and further investigations.
The contribution of breakthrough infection in those vaccinated HCWs indicates that breakthrough infection induces higher and broader nAbs. This is consistent with several reports,43–45 as well as our previous findings.35,36 Notably, it has also been recently demonstrated that SARS-CoV-2 infection before the fourth dose of bivalent vaccine can induce higher nAb titers against Omicron subvariants compared with those who had infection before the fourth dose of monovalent vaccine,44 which seems to support that an infection with an Omicron subvariant before a BA.5 vaccine boost could help overcome immune imprinting. Further study of the impact of breakthrough infection and the durability of immune imprinting are needed.
One curious finding of this study is the modestly but consistently enhanced infectivity of Omicron XBB variants in CaLu-3 cells, especially XBB.1.5, compared with the prototype Omicron BA.1/BA.1.1 and subsequently emerged Omicron subvariants including BQ.1.1 and BA.2.75.2 (Figure 1D). We do not believe that these results are experimental artifacts, as assays were performed side by side at the same time for all variants, and the expression levels of S proteins are also comparable for all variants. One possible explanation is the increased binding of XBB.1.5 to the ACE2 receptor, as recently shown by Richard Cao and colleagues,42 which is also supported by our structural modeling. The initial mutation F486S in XBB is predicted to cause decreased affinity between the S protein and ACE2 due to the introduction of energetically unfavorable contacts between the polar residue and a hydrophobic patch. The subsequent mutation S486P largely reverses this effect, increasing the propensity for hydrophobic interactions with ACE2 and the flexibility of this region of the S protein, thus allowing it to settle further into the binding groove on ACE2 (Figure 4). Consistent with the predicted improvement in ACE2 utilization, we observed a corresponding increase in cell-cell fusion and S processing for XBB subvariants, especially the single point mutant XBB-S486P (Figures 3A, 3B, 3E, S4A, and S4B).
Notably, cell-cell fusion reflects the triggerability of S in response to receptor-mediated conformational changes, and S processing informs the nature of S priming, both of which are related to membrane fusion but are not directly correlated with pathogenesis. However, cell-cell fusion, or syncytia formation, in virus-infected cells has been shown to be associated with the pathogenesis of SARS-CoV-2. In particular, the lower fusogenicity of Omicron has been correlated with a shift in viral tropism toward the upper respiratory tract as opposed to the lower respiratory tract, leading to attenuated diseases.1,4,46 Additionally, it has been established that there is a close association between viral fusogenicity in cultured cells and pathogenicity in hamsters. For example, in contrast to the B.1.1 ancestral variant, the Omicron BA.1 subvariant harboring a less fusogenic S is less pathogenic, whereas the Delta variant, which contains a more fusogenic S, is more pathogenic. Most recently, BA.4/5, which bears a more fusogenic S than BA.2, has been shown to be more pathogenic.4,31,46 Given that the newly emerged Omicron subvariants show increased infectivity, particularly the XBB subvariants, as well as enhanced fusogenicity compared with BA.2,6,9,14,17 which has been associated with pathogenesis for SARS-CoV-2,1,4,31,46 in vivo experiments investigating these aspects of the virus are necessary for the XBB subvariants.
Our structural modeling of the S protein interacting with its receptor and nAbs provide insights for understanding Omicron subvariant evolution. Intriguingly, the structural analysis suggests a sophisticated two-step strategy for the XBB lineage to evade immune recognition and likely outcompete other Omicron subvariants through mutations on the S residue at position 486. F486 has a bulky hydrophobic side chain and is a hotspot for establishing protective immunity against the virus17 (Figure 1A), whereas F486S mutation greatly facilitates evasion of antibody recognition. However, this F486S mutation reduces the efficiency of receptor utilization, which must be counteracted with mutations that promote receptor binding, such as N460K and R493Q, to preserve the viral fitness. Thus, it seems reasonable that once XBB circulation is established, S486 is further mutated into P486, as present in XBB.1.5, thus regaining higher receptor affinity while still maintaining similar immune escape. This combination of enhanced antibody escape and receptor affinity therefore likely enables, and has facilitated, the current dominance of the XBB.1.5 strain. In the cases of subvariants CH.1.1 and CA.3.1, it is clear that these variants have used the same strategy as other Omicron variants including BA.4/5 and BQ.1 by mutating the L452 and K444 sites of vulnerability frequently recognized by class I and II nAbs to evade neutralization, again underscoring the convergent viral evolution. Critically, the conserved mutations R346T and G339H might explain the strong resistance of CA.3.1, CH.1.1, and BA.2.75.2 to the class III mAb S309. We speculate that the potential effect of R346T and G339H mutation on S309 resistance by XBB subvariants could be compensated by the mutation L368I, which is present in all XBB subvariants, by stabilizing the local conformational of S, thus enhancing the glycan-N343 recognition.
Overall, our study highlights the continued waning of 3-dose mRNA vaccine efficacy against newly emerging Omicron subvariants. This effect, plus the reported decay of nAbs over time,36 can be partially rescued by the administration of a bivalent booster plus breakthrough infection, though escape by some subvariants, particularly CH.1.1 and CA.3.1, is still prominent. Hence, continued refinement of current vaccination strategies or investigation of new ones is necessary. The biology of the S protein of Omicron subvariants, notably those of the XBB lineage, also continues to change, emphasizing the importance of continued surveillance and study of emerging variants.
Limitations of the study
Throughout the study, cohorts of relatively small sample size were used to assess nAbs titers against the subvariants. However, previous studies have used cohorts of similar size and generated reliable data that have since been confirmed by other groups. Our cohorts also vary widely in time of sample collection after boosting or infection due to the clinical arrangements around collection of samples. In the bivalent cohort, 10 of 14 HCWs had been infected with SARS-CoV-2, with only one infected within 6 months of sample collection (Figure 2D, B-12 denoted with an asterisk [*]) and 9 infected more than 6 months before sample collection. Given our published results that the mRNA booster vaccine-induced nAb titer drops 17%–20% every 30 days,36 breakthrough infection in this bivalent cohort should not have had a significant impact on their neutralization titers. A small subset of the BA.4/5 convalescent individuals (3 in 20) also received doses of vaccine, though we did not perform subgroup analysis due to the small size of the group. The use of pseudotyped virus instead of live virus for our assays is also a limitation, though we have previously validated our pseudotyped lentiviral system alongside live SARS-CoV-2,34 and pseudotyped vectors are a common system used in the field to evalaute COVID-19 vaccines. Finally, although homology modeling is useful to explain the observations based on functional analyses, it is not as accurate as real structures resolved by crystal tools or cryoelectron microscopy (cryoEM); therefore, the influence of XBB.1.5, CH.1.1, and CA.3.1 signature mutations on ACE2 binding and antibody interaction warrants additional structural and biochemical characterization. Despite these limitations, the dramatic phenotypes of immune evasion by XBB subvariants and CH.1.1 and CA.3.1 are clear, some of which have been corroborated by other studies.23,24,27,32,41,42 Again, our study emphasizes the need for continued surveillance of emerging SARS-CoV-2 variants and investigation of how viral evolution impacts vaccine efficacy and S protein biology.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents and resources can be requested from the lead contact, Dr. Shan-Lu Liu (liu.6244@osu.edu).
Materials availability
Plasmids generated for this study can be made available upon request from the lead contact.
Data and code availability
This paper does not report original code. NT50 values and de-identified patient information will be shared by the lead contact upon request. Any other additional data can be provided for reanalysis if requested from the lead contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Vaccinated and patient cohorts
Three cohorts were utilized in this study, the first being healthcare workers (HCWs) that received 3 homologous doses of mRNA vaccine. These samples were collected under approved IRB protocols 2020H0228, 2020H0527, and 2017H0292. The cohort included 15 HCWs that received homologous doses of either the monovalent Moderna mRNA-1273 (n = 3) or the monovalent Pfizer BioNTech BNT162b2 (n = 12) vaccines. Samples were collected from 14 to 86 days post-booster vaccination (median 40). HCW ages ranged from 26 to 61 (median 33). The cohort included 10 male and 5 female individuals.
The second cohort were HCWs that received a bivalent mRNA booster formulation, including the prototype S and BA.5 S. These samples were collected following the approved IRB protocols 2020H0228, 2020H0527, and 2017H0292. The cohort included 1 HCW that received 2 doses of the monovalent Pfizer BioNTech BNT162b2 vaccines and additional 1 dose of the bivalent Pfizer vaccine, 12 HCWs that received 3 doses of the monovalent Moderna mRNA-1273 or Pfizer BioNTech BNT162b2 vaccines and additional 1 dose of the bivalent Pfizer or Moderna vaccine, and 1 HCW that 4 doses of the monovalent Pfizer BioNTech BNT162b2 vaccines and additional 1 dose of the bivalent Pfizer vaccine. Samples were collected from 23 to 108 days post-bivalent vaccination (median 66). HCW ages ranged from 25 to 48 (median 36). The cohort included 8 male and 6 female individuals.
The third cohort included first responders and their household contacts that tested positive for SARS-CoV-2 infection during the BA.4/5 wave in Columbus, OH. These samples were collected under approved IRB protocols 2020H0527, 2020H0531, and 2020H0240. The cohort included 20 individuals. For each individual, a nasal swab was collected and sequenced to confirm the variant they were infected with. 4 individuals were infected with BA.4 and 7 individuals wereinfectedwith BA.5.The infecting variant could not be determined for the remaining 9 individuals but the dates of collection fall within when BA.4/5 was dominant in Columbus, OH (late July 2022 through late September 2022). Ages ranged from 27 to 58 years (median 44) and the cohort included 4 male and 15 female individuals. The age and gender of one individual are unknown. The cohort included 17 individuals that were unvaccinated and 3 individuals that received 3 homologous does of either the monovalent Pfizer BioNTech BNT162b2 (n = 1) or the monovalent Moderna mRNA-1273(n = 2) vaccines.
Cell lines and maintenance
Human embryonic kidney cell lines HEK293T (ATCC CRL-11268, RRID: CVCL_1926) and HEK293T engineered to overexpress human ACE2 (BEI NR-52511, RRID: CVCL_A7UK) were maintained in DMEM (Gibco, 11965–092) supplemented with 10% FBS (Sigma, F1051) and 0.5% penicillin-streptomycin (HyClone, SV30010). Human adenocarcinoma lung epithelial cell line CaLu-3 (RRID: CVCL_0609) was maintained in EMEM (ATCC, 30–2003) supplemented with 10% FBS and 0.5% penicillin-streptomycin. The cells were incubated at 37°C and 5.0% CO2. Passaging of all cell lines was performed by first washing with Dulbecco’s phosphate buffer saline (Sigma, D5652–10X1L) followed by an incubation in 0.05% Trypsin + 0.53 mM EDTA (Corning, 25–052-CI) until complete cell detachment for splitting.
METHOD DETAILS
Plasmids
Pseudotyped lentiviral vectors were produced as previously described.34 Briefly, vectors are produced through the co-transfection of the HIV-1 vector pNL4–3 with an Env deletion and the SARS-CoV-2 spike of interest. The pNL4–3 vector includes a Gaussia luciferase reporter gene that is secreted by target cells. SARS-CoV-2 spike plasmids were generated in the pcDNA3.1 plasmid backbone either through KpnI and BamHI restriction enzyme cloning by GenScript Biotech (Piscataway, NJ) (D614G, BA.2, BA.4/5, and XBB) or site-directed mutagenesis via PCR (XBB.1, XBB-S486P, XBB.1.5, BQ.1.1, CH.1.1, CA.3.1, BA.2.75.2, and BA.2.75.2-N1199D) and confirmed by Sanger sequencing. All spike constructs include N- and C-terminal FLAG tags.
Pseudotyped lentivirus production and infectivity
Pseudotyped lentiviral vectors were produced as previously described.34 Briefly, HEK293T cells were co-transfected in a 2:1 ration with the pNL4–3-inGluc vector and the spike plasmid of interest using polyethyleneimine transfection (Transporter 5 Transfection Reagent, Polysciences) to produce pseudotyped lentiviral particles. The lentivirus was collected by taking the media of the transfected cells 48 and 72 hours post-transfection. Relative infectivity of the lentivirus was then assessed in both HEK293T-ACE2 and CaLu-3 cells. Gaussia luciferase activity measured at 72 hours post infection for HEK293T and 72 hours for CaLu-3 were used to determine relative infectivity. Gaussia luciferase activity was determined by taking equal volumes of infected cell media and Gaussia luciferase substrate (0.1 M Tris pH 7.4, 0.3 M sodium ascorbate, 10 μM coelenterazine) and combining for an immediate luminescence signal detected by a BioTek Cytation plate reader.
Virus neutralization assay
Pseudotyped lentiviral neutralization assays were performed as previously described.34 First, all serum samples were diluted 4-fold (final dilutions 1:80, 1:320, 1:1280, 1:5120, 1:20480, and no serum control for HCWs samples; final dilutions 1:40, 1:160, 1:640, 1; 2560. 1:10240, and no serum control for HCWs or BA.4/5-Wave samples), whereas mAb S309 were diluted 4-fold from 12 μg/ml. An equal volume of pseudotyped lentivirus was then added to the diluted sera and incubated at 37°C for 1 hour. This neutralized virus mixture was then used to infect HEK292T-ACE2 cells. Gaussia luficerase activity was then determined 48 and 72 hours post infection. 50% neutralization titers (NT50) were determined by least-squares-fit, non-linear regression in GraphPad Prism 9 (San Diego, CA).
Syncytia formation
To measure the extent of cell-cell fusion mediated by the different SARS-CoV-2 spikes, HEK293T cells expressing ACE2 were co-transfected with spike plasmid and GFP.6 Cells were imaged with a Leica DMi8 confocal microscope 30-hours post-transfection. Representative images were selected for presentation. Area of fused cells was determined and quantified using the Leica X Applications Suite, scale bars represent 150 μM.
Cell-cell fusion
Cell-cell fusion assays were carried out as follows. Briefly, HEK293T cells were cotransfected with individual S plus a GFP plasmid; transfected cells were digested next day by trypsin and then cocultured with CaLu-3 cells at a 1:1 ratio. Following 24–48 hrs incubation, the cells were imaged under a Leica DMi8 confocal microscope. The areas of fused cells were quantified using the Leica X Applications Suite.
S protein surface expression
HEK293T cells used to produce lentiviral vectors were harvested 72-hours post-transfection. Cells were incubated in PBS+5mM EDTA for 7 minutes at 37°C to mediate disassociation. The cells were fixed in 3.7% formaldehyde and stained with anti-SARS-CoV-2 polyclonal S1 antibody (Sino Biological, 40591-T62; RRID: AB_2893171) and secondary antibody anti-Rabbit-IgG-FITC (Sigma, F9887, RRID: AB_259816). S surface expression was measured using a Life Technologies Attune NxT flow cytometer and data was processed using FlowJo v7.6.5 (Ashland, OR).
S protein processing
Lysate from HEK293T cells used to produce lentiviral vectors was collected through a 40-minute incubation on ice in RIPA lysis buffer (50mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, Nonidet P-40, 0.1% SDS) supplemented with protease inhibitor (Sigma, P8340). Samples were run on a 10% acrylamide SDS-PAGE gel and transferred to a PVDF membrane. Membranes were probed with anti-S1 (Sino Biological, 40591-T62; RRID:AB_2893171), anti-S2 (Sino Biological, 40590; RRID:AB_2857932), anti-p24 (NIH HIV Reagent Program, ARP-1513), and anti-GAPDH (Santa Cruz, Cat# sc-47724, RRID: AB_627678). Secondary antibodies included Anti-Rabbit-IgG-HRP (Sigma, A9169; RRID:AB_258434) and Anti-Mouse (Sigma, Cat# A5278, RRID: AB_258232). Blots were imaged using Immobilon Crescendo Western HRP substrate (Millipore, WBLUR0500) and exposed on a GE Amersham Imager 600. Band intensities were quantified using NIH ImageJ analysis software (Bethesda, MD).
Structural modeling and analyses
Structural modeling of XBB spike proteins in complex with either ACE2 receptor or neutralizing antibodies was performed on SWISS-MODEL server using published X-ray crystallography or cryo-EM structures as templates (PDB: 7K8Z, 8DT3, 7L7D, 7XB0, 7XCK, 7YAD, 7NDD). Molecular contacts of XBB mutants were examined and illustrated with PyMOL.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 9 and are described in the figure legends. NT50 values were determined by least-squares fit non-linear regression in GraphPad Prism 9. Error bars in (Figures 1C and 1D) represent means ± standard deviation and in (Figures 3B, 3D, and S4B) represent means ± standard error. Error bars in (Figures 2A–2C and S2) represent geometric means with 95% confidence intervals. Statistical significance was determined using log10 transformed NT50 values to better approximate normality (Figures 2A–2C and S2A–S2E), comparisons between multiple groups were made using a one-way ANOVA with Bonferroni post-test, and a paired, two-tailed Student’s t test with Welch’s correction was used (Figures 2A–2C and S2C–S2E).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-SARS-CoV-2 S1 | Sino Biological | Cat# 40150-T62; RRID:AB_2920715 |
| anti-SARS-CoV-2 S2 | Sino Biological | Cat# 40590-T62; RRID: AB_2857932 |
| anti-p24 | NIH | Cat# ARP-1513; RRID: AB_2832923 |
| anti-GAPDH | Santa Cruz Biotechnology | Cat# sc-47724; RRID: AB_627678 |
| S309 mAb | Pinto et al.39 | N/A |
| anti-Mouse IgG (whole molecule)–Peroxidase | Sigma-Aldrich | Cat# A5278; RRID: AB_258232 |
| anti-Rabbit IgG (whole molecule)–Peroxidase | Sigma-Aldrich | Cat# A9169; RRID: AB_258434 |
| anti-Rabbit IgG (whole molecule)–FITC antibody | Sigma-Aldrich | Cat# F9887; RRID: AB_259816 |
|
| ||
| Biological samples | ||
|
| ||
| 3-dose HCWs Sera | Evans et al.9, Qu et al.14 | N/A |
| 3-dose Bivalent HCWs Sera | This paper | N/A |
| Omicron BA.4/5-wave Infected First Responders and Household Contacts Sera | Qu et al.17 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Transporter 5 Transfection Reagent | Polysciences | Cat# 26008–5 |
| Dulbecco’s Modified Eagles Medium (DMEM) | Sigma-Aldrich | Cat#: 11965–092 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat#: F1051 |
| 0.05% Trypsin + 0.53 mM EDTA | Corning | Cat# 25–052-CI |
| Penicillin-Streptomycin | HyClone | Cat#: SV30010 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Coelenterazine | GoldBio | Cat#: CZ2.5, CAS: 55779–48-1 |
|
| ||
| Deposited data | ||
|
| ||
| NT50 Values and De-identified patient data | SeroNet Coordinating Center, NCI, NIH | N/A |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T | ATCC | Cat#: CRL-11268; RRID: CVCL_1926 |
| HEK293T-ACE2 | BEI Resources | Cat#: NR-52511; RRID: CVCL_A7UK |
| Calu-3 | ATCC | RRID: CVCL_0609 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pNL4–3-inGluc | David Derse, NIH47 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_D614G | GenScript Biotech48 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2 | GenScript Biotech9 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5 | GenScript Biotech9 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75.2 | GenScript Biotech15 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75.2-N1199D | GenScript Biotech15 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BQ.1.1 | GenScript Biotech15 | |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB.1 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB-S486P | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB.1.5 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_CH.1.1 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_CA.3.1 | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism | Version 9.0.0 | GraphPad |
| FlowJo | FlowJo, LLC | https://www.flowjo.com/ |
| ImageJ | Qu et al.15 | https://imagej.nih.gov/ij/ |
| SWISS-MODEL | 49 | https://swissmodel.expasy.org/ |
| PyMOL | Warren DeLano and Sarin Bromberg | https://pymol.org/ |
| Leica Application Suite X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
|
| ||
| Other | ||
|
| ||
| Cytation 5 Imaging Reader | BioTek | N/A |
| Amersham Imager 600 | GE Healthcare Life Sciences | N/A |
Highlights.
Bivalent booster induces 2- to 8-fold higher nAb titer than monovalent against XBB and XBB.1.5
CH.1.1 and CA.3.1 exhibit nearly complete escape of neutralization from bivalent booster
XBB.1.5, CH.1.1, and CA.3.1 show increased fusogenicity compared with BA.2
Homology modeling shows impacts of F486P mutation present in XBB.1.5 on ACE2 binding
ACKNOWLEDGMENTS
We thank the NIH AIDS Reagent Program and BEI Resources for providing important reagents for this work. We also thank the Clinical Research Center/Center for Clinical Research Management of The Ohio State University Wexner Medical Center and The Ohio State University College of Medicine in Columbus, Ohio, specifically Francesca Madiai, Dina McGowan, Breona Edwards, Evan Long, and Trina Wemlinger, for logistics, collection, and processing of samples. In addition, we thank Sarah Karow, Madison So, Preston So, Daniela Farkas, and Finny Johns of the clinical trials team of The Ohio State University for sample collection and other support. This work was supported by a fund provided by an anonymous private donor to OSU. S.-L.L., S.F., D. J., A.R.P., R.J.G., L.J.S., and E.M.O. were supported by the National Cancer Institute of the NIH under award no. U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.P.E. was supported by Glenn Barber Fellowship from The Ohio State University College of Veterinary Medicine. K.X. was supported by The Ohio State University Comprehensive Cancer Center, a Path to K grant through the Ohio State University Center for Clinical & Translational Science, and partially by NIH grants U01 AI173348 and UH2 AI171611. R.J.G. was additionally supported by the Robert J. Anthony Fund for Cardiovascular Research and the JB Cardiovascular Research Fund, and L.J.S. was partially supported by NIH R01 HD095881.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112443.
REFERENCES
- 1.Shuai H, Chan JFW, Hu B, Chai Y, Yuen TTT, Yin F, Huang X,Yoon C, Hu JC, Liu H, et al. (2022). Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699. 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 2.Yuan S, Ye Z-W, Liang R, Tang K, Zhang AJ, Lu G, Ong CP, Man Poon VK, Chan CCS, Mok BWY, et al. (2022). Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters.Science 377, 428–433. [DOI] [PubMed] [Google Scholar]
- 3.Hui KPY, Ho JCW, Cheung MC, Ng KC, Ching RHH, Lai KL,Kam TT, Gu H, Sit KY, Hsin MKY, et al. (2022). SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603, 715–720. 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, et al. (2022). Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603, 700–705. 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A,Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, et al. (2022). Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714. 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng C, Evans JP, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, et al. (2021). Neutralization and stability of SARS-CoV-2 omicron variant. Preprint at bioRxiv. 10.1101/2021.12.16.472934. [DOI] [Google Scholar]
- 7.Schmidt F, Muecksch F, Weisblum Y, da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, et al. (2022). Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J.Med 386, 599–601. 10.1056/nejmc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C,Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, et al. (2022). Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675. 10.1038/s41586-02104389-z. [DOI] [PubMed] [Google Scholar]
- 9.Evans JP, Zeng C, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, et al. (2022). Neutralization of SARS-CoV-2 omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe 30, 1093–1102.e3. 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, de la Cruz E, Jorge A, et al. (2022). Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med 28, 481–485. 10.1038/s41591-02201705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia H, Zou J, Kurhade C, Cai H, Yang Q, Cutler M, Cooper D,Muik A, Jansen KU, Xie X, et al. (2022). Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe 30, 485–488.e3. 10.1016/j.chom.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen H, Strengert M, Maaß H, Ynga Durand MA, Katzmarzyk M, Kessel B, Harries M, Rand U, Abassi L, Kim Y, et al. (2022). Diminished neutralization responses towards SARS-CoV-2 Omicron VoC after mRNA or vector-based COVID-19 vaccinations. Sci. Rep 12, 19858. 10.1038/s41598-022-22552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Zhao X, Song J, Wu J, Zhu Y, Li M, Cui Y, Chen Y, Yang L, Liu J, et al. (2022). Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg. Microbes Infect 11, 477–481. 10.1080/22221751.2022.2030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu P, Faraone J, Evans JP, Zou X, Zheng Y-M, Carlin C, Bednash JS, Lozanski G, Mallampalli RK, Saif LJ, et al. (2022). Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med 386, 2526–2528. 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu P, Evans JP, Zheng Y-M, Carlin C, Saif LJ, Oltz EM, Xu K,Gumina RJ, and Liu S-L (2022). Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe 30, 1518–1526.e4. 10.1016/j.chom.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis-Gardner ME, Lai L, Wali B, Samaha H, Solis D, Lee M, Porter-Morrison A, Hentenaar IT, Yamamoto F, Godbole S, et al. (2023). Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. N. Engl. J. Med. Overseas. Ed 388, 183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu P, Evans JP, Faraone J, Zheng Y-M, Carlin C, Anghelina M,Stevens P, Fernandez S, Jones D, Lozanski G, et al. (2022). Enhanced neutralization resistance of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. Cell Host Microbe 31, 9–17.e3. 10.1016/j.chom.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, et al. (2022). BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608, 593–602. 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, Wang M, Yu J,Bowen AD, Chang JY, et al. (2022). Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608, 603–608. 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM,Selvaraj M, Liu C, Mentzer AJ, Supasa P, Duyvesteyn HME, et al. (2022). Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from.vaccine and BA.1 serum. Cell 185, 2422–2433.e13. 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora P, Zhang L, Krüger N, Rocha C, Sidarovich A, Schulz S, Kempf A, Graichen L, Moldenhauer AS, Cossmann A, et al. (2022). SARS-CoV-2 Omicron sublineages show comparable cell entry but differential neutralization by therapeutic antibodies. Cell Host Microbe 30, 1103–1111.e6. 10.1016/j.chom.2022.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hachmann NP, Miller J, Collier ARY, Ventura JD, Yu J, Rowe M,Bondzie EA, Powers O, Surve N, Hall K, and Barouch DH (2022). Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med 387, 86–88. 10.1056/nejmc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uraki R, Ito M, Furusawa Y, Yamayoshi S, Iwatsuki-Horimoto K,Adachi E, Saito M, Koga M, Tsutsumi T, Yamamoto S, et al. (2023). Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis 23, 30–32. 10.1016/S14733099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai M, Ito M, Kiso M, Yamayoshi S, Uraki R, Fukushi S, Watanabe S, Suzuki T, Maeda K, Sakai-Tagawa Y, et al. (2023). Efficacy of a fourth dose of covid-19 mRNA vaccine against omicron subvariants BQ.1.1 and XBB. N. Engl. J. Med, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention (2022). Variant Proportions(COVID Data Tracker). https://covid.cdc.gov/covid-data-tracker/#variant-proportions. [Google Scholar]
- 26.Chen C, Nadeau S, Yared M, Voinov P, Xie N, Roemer C, and Stadler T. (2022). CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics 38, 1735–1737. 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurhade C, Zou J, Xia H, Liu M, Chang HC, Ren P, Xie X, and Shi PY (2023). Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med 29, 344–347. 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 28.Cao Y, Jian F, Wang J, Yu Y, Song W, Yisimayi A, Wang J, An R,Chen X, Zhang N, et al. (2022). Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614, 521–529. 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangavarapu K, Latif AA, Mullen J, Alkuzweny M, Hufbauer E,Tsueng G, Haag E, Zeller M, Aceves CM, Zaiets K, et al. (2023). SARS-CoV-2 (hCoV-19) Mutation Reports. https://outbreak.info/situation-reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing Y-H, Ni W, Wu Q, Li W-J, Li G-J, Wang W-D, Tong J-N, Song X-F, Wong GW-K, and Xing Q-S (2021). SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura I, Yamasoba D, Tamura T, Tanaka S, and Sato K. (2022). Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, et al. (2023). Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286.e8. 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou T, Wang L, Misasi J, Pegu A, Zhang Y, Harris DR, Olia AS,Talana CA, Yang ES, Chen M, et al. (2022). Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 376, eabn8897. 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng C, Evans JP, Pearson R, Qu P, Zheng YM, Robinson RT,Hall-Stoodley L, Yount J, Pannu S, Mallampalli RK, et al. (2020). Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight 5, e143213. 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans JP, Zeng C, Carlin C, Lozanski G, Saif LJ, Oltz EM, Gumina RJ, and Liu SL (2022). Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med 14, eabn8057. 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu P, Faraone JN, Evans JP, Zheng Y-M, Yu L, Ma Q, Carlin C, Lozanski G, Saif LJ, Oltz EM, et al. (2022). Durability of booster mRNA vaccine against SARS-CoV-2 BA.2.12.1, BA.4, and BA.5 subvariants. N. Engl. J. Med. Overseas. Ed 387, 1329–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu P, Faraone JN, Evans JP, Zou X, Zheng Y-M, Carlin C, Bednash JS, Lozanski G, Mallampalli RK, Saif LJ, et al. (2022). Differential evasion of Delta and omicron immunity and enhanced fusogenicity of SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. Preprint at bioRxiv. 10.1101/2022.05.16.492158. [DOI] [Google Scholar]
- 38.Qu P, Evans JP, Faraone J, Zheng Y-M, Carlin C, Anghelina M,Stevens P, Fernandez S, Jones D, Lozanski G, et al. (2022). Distinct neutralizing antibody escape of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. Preprint at bioRxiv. 10.1101/2022.10.19.512891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. (2020). Crossneutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 40.Arbel R, Peretz A, Sergienko R, Friger M, Beckenstein T, Yaron S, Hammerman A, Bilenko N, and Netzer D. (2023). Effectiveness of the bivalent mRNA vaccine in preventing severe covid-19 outcomes: an observational cohort study. Preprints with The Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Chen L-L, Ip JD, Chan W-M, Hung IF-N, Yuen K-Y, Li X, and To KK-W (2022). Omicron sublineage recombinant XBB evades neutralising antibodies in recipients of BNT162b2 or CoronaVac vaccines.Lancet Microbe, 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue C, Song W, Wang L, Jian F, Chen X, Gao F, Wang X, and Cao Y. (2023). Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. Preprint at bioRxiv. 10.1101/2023.01.03.522427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abu-Raddad LJ, Chemaitelly H, Ayoub HH, Yassine HM, Benslimane FM, Al Khatib HA, Tang P, Hasan MR, Coyle P, Al Kanaani Z, et al. (2021). Association of prior SARS-CoV-2 infection with risk of breakthrough infection following mRNA vaccination in Qatar. JAMA 326, 1930–1939. 10.1001/jama.2021.19623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klotz SA, Yates S, Smith SL, Dudley S, Schmidt JO, Shirazi FM,Cooper D, Yang Q, Cai H, Muik A, et al. (2023). Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with bivalent vaccine. N. Engl. J. Med. Overseas. Ed 388, 853–854. 10.1056/NEJMc2029813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muik A, Lui BG, Bacher M, Wallish A-K, Toker A, Finlayson A, Kruger K, Ozhelvaci O, Grikschiet K, Hoehl S, et al. (2022). Omicron BA.2 breakthrough infection enhances cross-neutralization of BA.2.12.1 and BA.4/BA.5. Sci. Immunol 7, eade2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, Kosugi Y,Shirakawa K, Sadamasu K, Kimura I, et al. (2022). Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306. 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, and Derse D. (2010). Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 6, e1000788. 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng C, Evans JP, Faraone JN, Qu P, Zheng YM, Saif L, Oltz EM, Lozanski G, Gumina RJ, and Liu SL (2021). Neutralization of SARS-CoV-2 variants of concern harboring Q677H. mBio 12. 10.1128/mBio.02510-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guex N, Peitsch MC, and Schwede T. (2009). Automated comparativeprotein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 (Suppl 1 ), S162–S173. 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code. NT50 values and de-identified patient information will be shared by the lead contact upon request. Any other additional data can be provided for reanalysis if requested from the lead contact.