Summary
Background
Retinopathy of prematurity (ROP), a leading cause of childhood blindness, is diagnosed through interval screening by paediatric ophthalmologists. However, improved survival of premature neonates coupled with a scarcity of available experts has raised concerns about the sustainability of this approach. We aimed to develop bespoke and code-free deep learning-based classifiers for plus disease, a hallmark of ROP, in an ethnically diverse population in London, UK, and externally validate them in ethnically, geographically, and socioeconomically diverse populations in four countries and three continents. Code-free deep learning is not reliant on the availability of expertly trained data scientists, thus being of particular potential benefit for low resource health-care settings.
Methods
This retrospective cohort study used retinal images from 1370 neonates admitted to a neonatal unit at Homerton University Hospital NHS Foundation Trust, London, UK, between 2008 and 2018. Images were acquired using a Retcam Version 2 device (Natus Medical, Pleasanton, CA, USA) on all babies who were either born at less than 32 weeks gestational age or had a birthweight of less than 1501 g. Each images was graded by two junior ophthalmologists with disagreements adjudicated by a senior paediatric ophthalmologist. Bespoke and code-free deep learning models (CFDL) were developed for the discrimination of healthy, pre-plus disease, and plus disease. Performance was assessed internally on 200 images with the majority vote of three senior paediatric ophthalmologists as the reference standard. External validation was on 338 retinal images from four separate datasets from the USA, Brazil, and Egypt with images derived from Retcam and the 3nethra neo device (Forus Health, Bengaluru, India).
Findings
Of the 7414 retinal images in the original dataset, 6141 images were used in the final development dataset. For the discrimination of healthy versus pre-plus or plus disease, the bespoke model had an area under the curve (AUC) of 0·986 (95% CI 0·973–0·996) and the CFDL model had an AUC of 0∙989 (0∙979–0∙997) on the internal test set. Both models generalised well to external validation test sets acquired using the Retcam for discriminating healthy from pre-plus or plus disease (bespoke range was 0∙975–1∙000 and CFDL range was 0∙969–0∙995). The CFDL model was inferior to the bespoke model on discriminating pre-plus disease from healthy or plus disease in the USA dataset (CFDL 0·808 [95% CI 0·671–0·909, bespoke 0·942 [0·892–0·982]], p=0·0070). Performance also reduced when tested on the 3nethra neo imaging device (CFDL 0·865 [0·742–0·965] and bespoke 0·891 [0·783–0·977]).
Interpretation
Both bespoke and CFDL models conferred similar performance to senior paediatric ophthalmologists for discriminating healthy retinal images from ones with features of pre-plus or plus disease; however, CFDL models might generalise less well when considering minority classes. Care should be taken when testing on data acquired using alternative imaging devices from that used for the development dataset. Our study justifies further validation of plus disease classifiers in ROP screening and supports a potential role for code-free approaches to help prevent blindness in vulnerable neonates.
Funding
National Institute for Health Research Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and the University College London Institute of Ophthalmology.
Introduction
Retinopathy of prematurity (ROP) is a proliferative retinal vascular disease, typically affecting preterm neonates with low-birthweight. Following landmark findings from the multicentre CRYO-ROP study in 1988, ROP has become a largely treatable disease when sight-threatening features are promptly recognised.1 One such feature is plus disease, a condition characterised by abnormal posterior retinal vessel dilatation and tortuosity.1 Plus disease is highly prognostic of ultimate sight loss and its presence mandates urgent treatment as stipulated by international consensus and guidelines.2
In high-income countries, ROP has traditionally been identified through screening programmes with interval paediatric ophthalmologist-led clinical examination for neonates at risk. For example, the American Academy of Pediatrics recommends screening for all infants with a birthweight of 1500 g or less or a gestational age of 30 weeks or less.3 However, concerns exist over the sustainability of such programmes. Screening ROP examinations are done by clinicians with substantial subspecialty-level experience and improved survival of preterm infants owing to advances in neonatal medicine4 might not be complemented with a corresponding growth in the paediatric ophthalmology workforce. Indeed, over half of neonatologists in a USA-based national survey reported a scarcity of available eye care specialists as a barrier to ROP screening.5 There is growing evidence that telemedicine approaches might provide a more efficient model for care delivery.6 However, such approaches are still limited by the availability of sufficiently trained diagnosticians, a predicament of even greater extent in low-income and middle-income countries (LMICs).7
Automated plus disease identification through deep learning, a subfield of artificial intelligence (AI) inspired by biological neural networks, might be one strategy for improving access to rapid specialist expertise. Since the first report on using deep learning for plus disease detection in 2016,8 the field has matured with the development and validation of several models with high levels of diagnostic accuracy.9–13 However, several challenges remain for such models to progress from high in silico diagnostic accuracy to safe real-world deployment.14 ROP is the most common cause of vision loss in children on a global level, yet the natural history of the disease varies between ethnic groups and there is no harmonisation in guidelines for ROP screening, reinforcing the need for development and validation of models on datasets from areas with the greatest geographical diversity possible. Firstly, most of the literature describing deep learning for ROP thus far has emerged from North America and Asia, reflecting so-called health data poverty where population demographics and health-care infrastructure are such that people from minority ethnic groups and socioeconomically deprived back grounds, might be under-represented in clinical AI development.15 In a condition, such as ROP, whereby natural history of the disease varies between ethnic groups, a mismatch between development and deploy ment populations may lead to poor generalisation upon deployment.16,17 Secondly, guidelines for ROP screening (ie, birthweight and gestational age) are well-established in North America and western Europe; however, most published reports outside these regions have either not articulated the specific criteria for examination or used thresholds that might not be appropriate for population health screening settings; for example, one report involved development data from any baby younger than 37 weeks,13however in the USA screening only occurs for those aged 30 weeks or younger and in the UK the threshold, as of March 2022, is age <31 weeks. Thirdly, models developed using deep learning frameworks frequently generalise poorly when tested on images derived from different devices to the images in the development dataset, and yet only one model for plus disease detection has been validated using images not acquired on the Retcam.18,19 Particularly in the setting of LMICs, the high cost of some devices has been prohibitive to their purchase, use, and wider deployment.20,21
Even when there is greater vigilance to such issues of dataset shift, many institutions still might not have the technical and hardware resources to develop such tools, especially in LMICs, where a predicted increase in ROP rates has led some to refer to sub-Saharan Africa as the new frontier of ROP.22 One potential solution for mitigating dataset shift concerns is local development, validation, and deployment of automated plus disease identification via code-free deep learning (CFDL) tools. Since initially described for medical imaging tasks in 2018, applications of CFDL have been used across a range of retinal diseases, yet comparisons with traditional bespoke deep learning design are few in the medical literature.23,24
In this study, we first aimed to develop and internally validate both bespoke and CFDL plus disease classifiers optimised for a UK population using a real-world training dataset. We used retinal images of neonates eligible for ROP screening according to The Royal College of Ophtha lmologists (RCOphth) criteria from a large single site serving an ethnically and socioeconomically diverse region of London, UK, where health care is free at the point of delivery under the provisions of the National Health Service (NHS). Secondly, we aimed to compare CFDL to bespoke modular architectures, hypothesising that CFDL models would confer similar levels of performance in the detection of plus and pre-plus disease and for assessing imaging quality. Thirdly, we aimed to externally validate both CFDL and bespoke models on four independent datasets from two LMICs (Brazil and Egypt) and the USA, including one composed of images obtained using a different device.
Methods
Dataset and participants
In this retrospective cohort study, consecutive infant retinal images were acquired between Jan 1, 2008, and Jan 31, 2018, as part of routine care, at Homerton University Hospital NHS Foundation Trust, London, UK.
Images were acquired on all babies within the designated study period who fit the 2008 RCOphth screening criteria for ROP (note the RCOphth guidelines were changed in March 2022).25 In brief, any baby who was either born at less than 32 weeks gestational age or has a birthweight of less than 1501 g is recommended to undergo screening for identification of ROP in the 2008 guidelines.
Anonymised retinal images were acquired using the Retcam version 2 device (Natus Medical, Pleasanton, CA, USA) in a range of fixation targets (eg, superior and posterior pole) with a maximum field of view of 130 degrees. Images were manually filtered by one ophthalmologist (MR) for those with capture fixation on the posterior pole. Image pixel resolution was 1600 × 1200 with 24-bit colour depth.
This study received research and development institutional review board approval from the Moorfields Eye Hospital Research and Development Department (BALK1004) and from the UK Health Research Authority (IRAS ID 253237). Because the study is limited to working with data only, the UK Health Research Authority deemed that ethics approval was not required. Consent was not deemed as required because the study was related to research on retrospectively acquired anonymised data. Reporting is in line with the TRIPOD statement.26
Procedures
Image grading was based solely on the retinal image; that is, no additional clinical data (eg, birthweight) were provided during grading. The reference standard was based on a hierarchical grading scheme: each image was independently graded by two junior ophthalmologists with 3 years of ophthalmology experience as either ungradable (opaque media due to corneal opacities, cataract, vitreous opacification or haemorrhage, poor patient fixation limiting the field of view for detection of pathological manifestations, or image artefacts), healthy, pre-plus disease, or plus disease. Two pairs of junior ophthalmologists (JT, SB, CH, and CK) took part in this study. Disagreements between junior ophthalmologists were resolved by a senior paediatric ophthalmologist (GA) with over 25 years of experience, who was masked to the grading of the junior ophthalmologists (appendix 3 p 6).
We exercised two main approaches for model development hypothesising that the architectures (bespoke and CFDL) would confer similar performance. All models were trained firstly for the binary classification of image gradability (defined as sufficient image quality for a clinician to give a confident decision on the absence or presence of plus disease) and secondly for multi-classification into healthy, pre-plus disease, and plus disease. The bespoke model was built on a DenseNet201 for PyTorch, Architecture 20 convolutional neural network architecture that was pre-trained on the ImageNet database (March 11, 2021 update; appendix 3 p 3). Training of the bespoke model was through five-fold cross validation; more precisely, the development dataset was split at a patient level into five folds with four folds for training and one fold for tuning. Iteration using each fold as validation led to the development of five models which were aggregated to an ensemble where the model decision was an average of the output of all five models. To explore the feasibility of code-free deep learning for plus disease detection, we additionally evaluated a CFDL model using the Google Cloud AutoML Vision Application Programming Interface (API), as described previously.23 Models were trained using 40 node hours as recommended by the API.
Performance of the gradability model was evaluated on an internal test set of 308 images of 308 eyes (200 gradable and 108 ungradable) from 155 babies. For the main task of discriminating healthy, pre-plus disease, and plus disease, the internal validation test set consisted of 200 images from 200 eyes of 112 infants. We deliberately oversampled pre-plus and plus disease cases in the internal test set to give more stable estimates of model performance on these classes. Some images were therefore excluded from the development dataset to avoid data leakage (patients were not in both development and test sets). The reference standard was the majority class of three senior paediatric ophthalmologist consultants (A-MH, RH, and HIP; range of experience: 21–45 years). In the case of equal disagreement between the three classes, we took the most severe label. To contextualise the model performance, the internal validation test set was additionally graded by seven other clinicians, consisting of one paediatric ophthalmology consultant, one paediatric ophthalmology fellow, four junior ophthalmology residents, and a neonatal nurse specialist (SH, JT, SB, CH, CK, MR, and JR; appendix 3 p 6). All misclassification errors (defined as those whereby the model output class with the highest probability differed to the reference standard) were visually inspected and are reported. Additionally, to aid model explainability, we developed saliency maps (techniques for attributing model decision-making to specific image pixels) using five techniques (appendix 3 p 3).
Image gradability models were externally validated on a Retcam-based dataset from Alexandria University Hospital, Egypt, consisting of 14 ungradable and 46 gradable images whereby the reference standard was image-based categorisation by a single paediatric ophthalmologist (HK). Models discriminating presence of plus disease were externally validated on four separate datasets: three from LMICs and one from the USA. Three datasets were acquired using the same imaging device as per the development dataset (Retcam) and one using the 3nethra neo device (Forus Health, Bengaluru, India). For the three datasets from the LMICs, the presence of plus disease was graded in a binary fashion—ie, either healthy or pre-plus disease and plus disease. Hence, performance metrics for those three only incorporated a single area under the curve (AUC) metric. The reference standard for all external validation datasets from the LMICs was defined through a combination of both binocular indirect ophthalmoscopy and image-based grading. The Imaging and Informatics in ROP (i-ROP) dataset is a previously described independent USA-based test set from the i-ROP study.27,28 The i-ROP dataset consisted of 100 Retcam images (54 healthy, 31 pre-plus disease, and 15 plus disease) of 70 neonates collected from July 1, 2011, to Dec 31, 2014, with the reference standard diagnosis as previously described. The Brazil dataset consists of 92 images (20 plus or pre-plus disease and 72 healthy) from 46 neonates acquired at the University of São Paulo, Brazil from Jan 1, 2020, to Aug 31, 2022, using the Retcam device. The Egypt Retcam dataset consisted of a total of 45 images (32 plus or pre-plus disease and 13 healthy) from 45 neonates acquired between Jan 1, 2020, and Aug 31, 2022, at the Alexandria University Hospital, Alexandria, Egypt. The Egypt 3nethra dataset consists of 101 images (71 plus or pre-plus disease and 30 healthy) from 33 neonates acquired between April 1, 2018, and August 31, 2022, at the Department of Ophthalmology, Ain Shams University Hospitals and Al Mashreq Eye Centre, Cairo, Egypt. Images were captured using the 3nethra neo, a wide-field camera device providing a maximum 120-degree field of view manufactured by Forus Health. Further information about the external validation datasets, including ethnicity data and reference standard, is provided in appendix 3 (p 7).
Outcomes
The primary outcome was classification accuracy for the diagnosis of plus disease, pre-plus disease, or healthy for the disease detection models in external validations and classification accuracy for the binary classification of image gradability (gradable or ungradable) for the gradability models. Secondary outcomes were inter-reater reliability for the development dataset, internal validation of the disease detection models, misclassfication errors audit, performance comparison between the bespoke and code-free disease detection models, and model explainability through saliancy maps.
Statistical analysis
Inter-rater reliability between two graders was assessed with the quadratic weighted Cohen κ statistic and for more than two graders intraclass correlation coefficient (ICC) was used (appendix 3 p 4). Model performance was estimated through sensitivity, specificity, and AUC using, where needed, a one-versus-all approach and 95% CIs calculated through bootstrapping. To emulate the likely use case of the image gradability and disease detection models, we also provided performance metrics at a set specificity and sensitivity of 1, respectively (ie, 100% of images considered ungradable would be classified as such, and 100% of images with disease would be identified). For visualising the sensitivity and specificity trade-off across different operating points, we generated ROC curves for the bespoke and CFDL models. Bespoke and CFDL models were compared using either the non-parametric approach described by DeLong and colleagues,29 a nonparametric approach for comparing two or more correlated AUCs, or, for test sets which had multiple images per patient, the clustered bootstrap technique to account for clustering.30 All statistical testing was two-tailed with the level of significance set at p<0·05. Analyses were conducted in Python version 3.6.9 and R version 4.1.0.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the manuscript.
Results
47158 consecutive infant retinal images were acquired between Jan 1, 2008, and Jan 31, 2018. The final dataset consisted of 7414 posterior pole images of 1370 infants. Individual-level ethnicity and socioeconomic deprivation data were not collected; the aggregate ethnicity of the cohort were: 44% were White, 33% were Black, 13% were south Asian, 4% were other Asian, 5% were Chinese, and 1% were other.
The class distribution across the development, internal validation, and external validation datasets are shown in table 1. Of the original development dataset of 7414 images, 487 (6·6%) were considered ungradable. A further 786 images were removed as they represented images of patients who were in the test set, leaving a final development dataset of 6141 images. Example images of the three classes are shown in appendix 3 (p 9).
Table 1:
Distribution of class for the development, internal test, and external test datasets
| Internal | External | |||||
|---|---|---|---|---|---|---|
| Development (n=6141) | Test (n=200) | i-ROP (n=100) | Brazil (n=92) | Egypt Retcam (n=45) | Egypt 3nethra neo (n=101) | |
| Healthy | 5771 (94·0%) | 111 (55.5%) | 54 (54·0%) | 72 (78.3%) | 13 (28.9%) | 30 (29·7%) |
| Pre-plus disease | 235 (3·8%) | 43 (215%) | 31 (31·0%) | NA | NA | NA |
| Plus disease | 135 (2·2%) | 46 (23·0%) | 15 (15·0%) | NA | NA | NA |
| Pre-plus or plus disease* | NA | NA | NA | 20 (21·7%) | 32 (71·1%) | 71 (70·3%) |
Data are n (%). i-ROP=Imaging and Informatics in Retinopathy of Prematurity. NA=not applicable.
For datasets from Brazil and Egypt the images were labelled as presence of pre-plus or plus disease, or healthy.
Quadratic-weighted κ values between the two pairs of junior ophthalmologists on the development dataset were 0·433 (95% CI 0·404–0·462) and 0·500 (0·461–0·540). Regarding the internal test dataset of 200 images, the ICC between all graders was high at 0·977 (0·972–0·982). The ICC between the three senior paediatric ophthalmologists who formed the reference standard was 0·954 (0·941–0·964), which was higher than resident ophthalmologists (0·801 [0·751–0·842]). Pairwise weighted κ between the ten graders, the majority of three senior paediatric ophthalmologists, and models on the internal validation set are shown in figure 1.
Figure 1: Matrix of pairwise quadratic-weighted κ values.
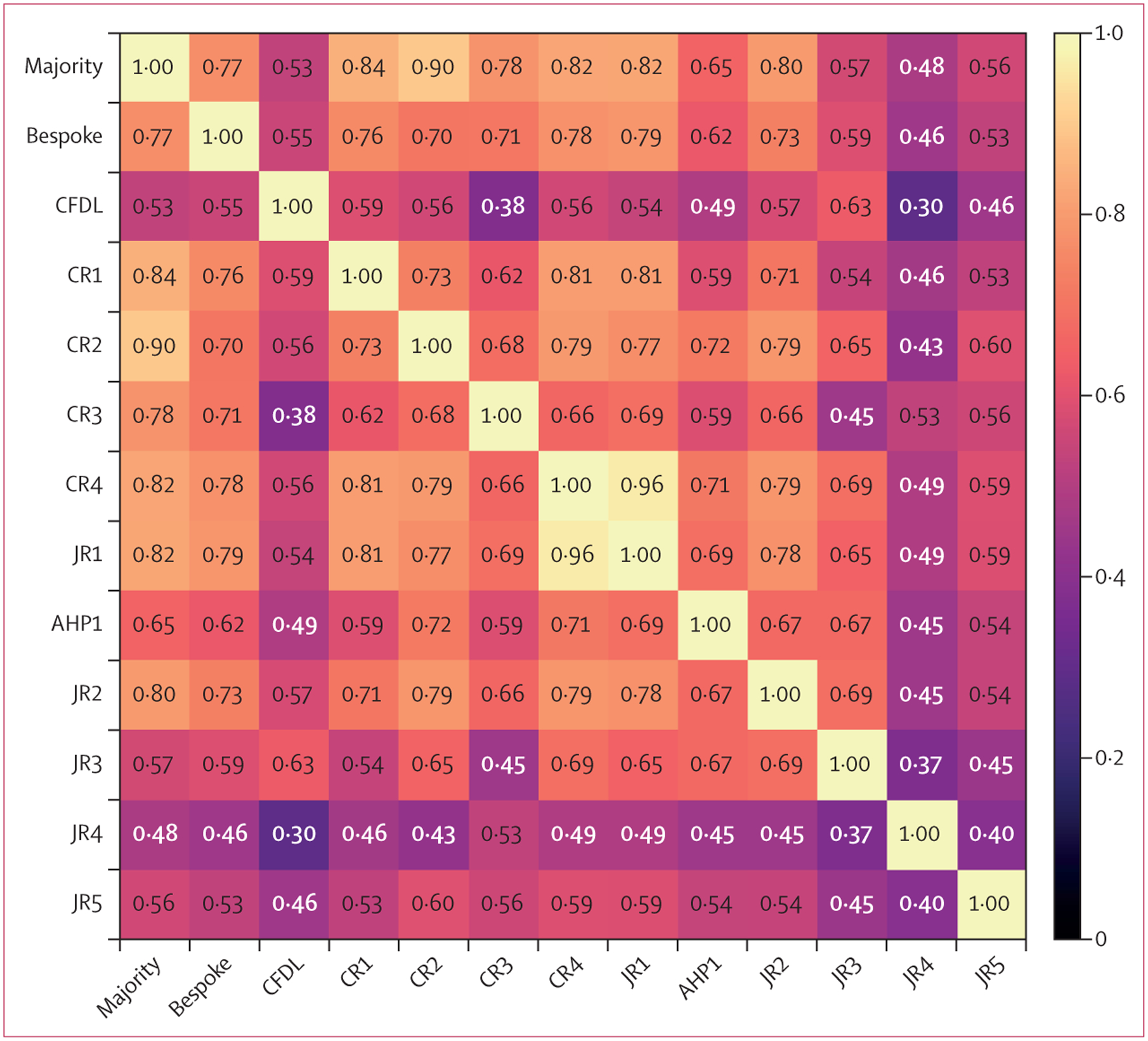
The majority label is based on the majority vote between CR1, CR2, and CR3 so those labels are not independent. CRs are the three senior paediatric ophthalmologists who provided the reference standard. AHP=allied health professional. CFDL=code-free deep learning. CR=consultant rater. JR=junior rater.
On the internal test set, the bespoke model achieved an AUC of 0·979 (95% CI 0·966–0·990) for discriminating gradable from ungradable images. At a specificity of 1, the sensitivity was 0·775 (0·710–0·910). On external validation, the AUC was 0·998 (0·991–1∙000). The bespoke model achieved AUCs of 0·986 (0·973–0·996), 0·927 (0·884–0·962), and 0·974 (0·951–0·991) for the discrimination of healthy, pre-plus disease, and plus disease versus the code-free model and the human expert raters that assessed the internal validation dataset, respectively (figure 2). Performance against health-care profess ionals is shown in table 2. For the detection of healthy versus pre-plus or plus disease, the specificity was 0·691 (95% CI 0·562–0·944) at a set sensitivity of 1. Quadratic-weighted κ between the reference standard and bespoke model was 0·77 (0∙66–0∙87). The bespoke model achieved similar AUCs on the external validation datasets acquired using the Retcam device (0·975 [95% CI 0·942–0·997] for Brazil and 0·976 [0·928–1∙000] for Egypt; table 3) but the performance reduced to an AUC of 0·891 (0·783–0·977) on the Egyptian 3nethra neo dataset.
Figure 2: Receiver operating characteristics curves for the bespoke and CFDL models on the internal test set.
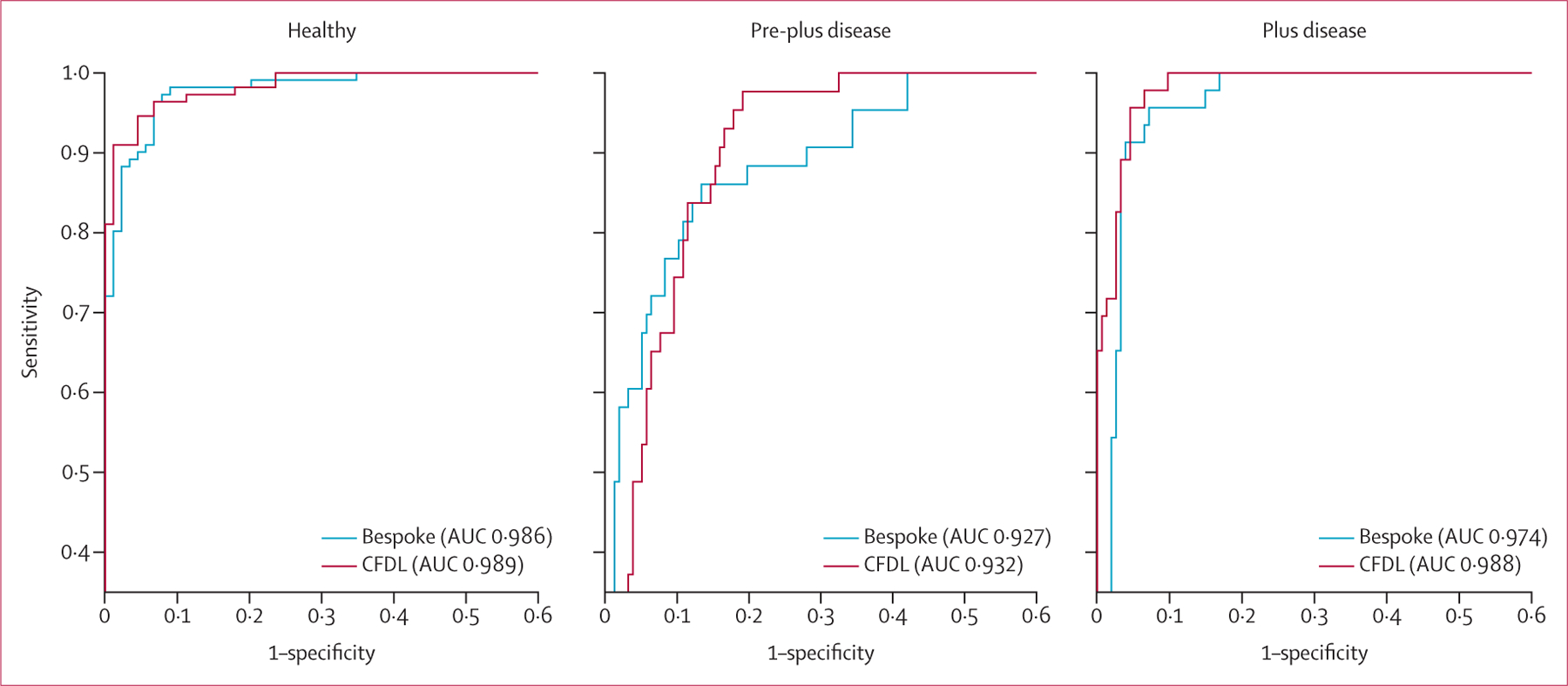
AUC=area under the curve. CFDL=code-free deep learning.
Table 2:
Performance metrics using the majority vote of the three most senior paediatric ophthalmologists (CR1, CR2, and CR3) as reference standard
| Healthy | Pre-plus disease | Plus disease | ||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Bespoke model* | 0.973 | 0.900 (0.640–0.978) | 0.860 | 0.860 (0.612–0.943) | 0.522 | 0.981 (0.948–1.000) |
| CFDL model* | 0.973 | 0.843 (0.700–0.978) | 0.860 | 0.866 (0.796–0.930) | 0.522 | 1.000 (0.994–1.000) |
| CR4 | 0.973 | 0.955 | 0.860 | 0.841 | 0.522 | 0.987 |
| JR1 | 0.964 | 0.955 | 0.860 | 0.873 | 0.652 | 0.987 |
| AHP1 | 0.928 | 0.865 | 0.674 | 0.860 | 0.696 | 0.987 |
| JR2 | 0.964 | 0.921 | 0.744 | 0.930 | 0.826 | 0.968 |
| JR3 | 0.964 | 0.775 | 0.442 | 0.866 | 0.587 | 0.961 |
| JR4 | 0.748 | 0.989 | 0.372 | 0.834 | 0.935 | 0.799 |
| JR5 | 0.901 | 0.843 | 0.558 | 0.796 | 0.522 | 0.961 |
Data are sensitivity, specificity, or specificity (95% CI). CR4 is the consultant rater who was part of the group of seven additional raters for the internal validation of the models but not part of the three consultant raters who provided the reference standard. AHP=allied health professional. CFDL=code-free deep learning. CR=consultant rater. JR=junior rater.
Sensitivity of the bespoke and CFDL models were matched to CR4.
Table 3:
Comparison of performance between bespoke and CFDL approaches across internal and external test sets
| Bespoke model AUC (95% CI) | CFDL model AUC (95% CI) | p value* | |
|---|---|---|---|
| Internal test set | |||
| Gradability | 0.979 (0.966–0.990) | 0.982 (0.970–0.992) | 0.49 |
| Healthy | 0.986 (0.973–0.996) | 0.989 (0.979–0.997) | 0.52 |
| Pre-plus disease | 0.927 (0.884–0.962) | 0.932 (0.896–0.964) | 0.78 |
| Plus disease | 0.974 (0.951–0.991) | 0.988 (0.976–0.996) | 0.089 |
| i-ROP external test set | |||
| Healthy | 1.00 (0.998–1.000) | 0.995 (0.981–1.000) | 0.33 |
| Pre-plus disease | 0.942 (0.892–0.982) | 0.808 (0.671–0.909) | 0.0070† |
| Plus disease | 0.976 (0.938–1.000) | 0.989 (0.967–1.000) | 0.40 |
| Brazil external test set ‡ | |||
| Healthy versus pre-plus or plus disease | 0.975 (0.942–0.997) | 0.969 (0.919–1.000) | 0.73 |
| Egypt external test set (Retcam) ‡ | |||
| Healthy versus pre-plus or plus disease | 0.976 (0.928–1.000) | 0.990 (0.964–1.000) | 0.34 |
| Egypt external test set (3nethra neo) † | |||
| Healthy versus pre-plus or plus disease | 0.891 (0.783–0.977) | 0.865 (0.742–0.965) | 0.31 |
AUC=area under the curve. CFDL=code-free deep learning. i-ROP=Imaging and informatics in Retinopathy of Prematurity.
Hypothesis testing was either through the DeLong test or cluster bootstrapping (for multilevel data).
Considered statistically significant.
For the Brazil and Egypt datasets the reference standard consisted of presence of pre-plus and plus disease versus healthy and hence there is only one binary classification AUC metric.
The CFDL model achieved an AUC of 0·982 (95% CI 0·970–0·992) for discriminating gradable from ungradable images with a sensitivity of 0·850 (0·785–0·915) when setting the specificity to 1. On external validation, the AUC was 0·977 (0·941–0·999). For the task of healthy, pre-plus disease, and plus disease detection versus the bespoke model and expert human raters that assessed the internal validation dataset, the AUCs of the CFDL model on the internal test set were 0·989 (0·979–0·997) for healthy, 0·932 (0·896–0·964) for pre-plus disease, and 0·988 (0·976–0·996) for plus disease (figure 2). At a set sensitivity of 1, the specificity for discrimination of healthy versus pre-plus or plus disease was 0·775 (0·674–0·899). Quadratic-weighted κ between the reference standard and CFDL model was 0∙53 (95% CI 0∙41–0∙66). Performance on the external validation datasets was generally high for datasets where image acquisition was using the Retcam (table 3). A reduction in AUC performance to 0·865 (0·742–0∙965) was noted when tested on the Egypt 3nethra neo dataset.
In general, performances were similar between the bespoke and CFDL models for all tasks on the internal test set and in the external validations from Brazil and Egypt. In the i-ROP external validation test set, there was evidence that the bespoke model had superior diagnostic accuracy to the CFDL model for the detection of pre-plus disease (0·942 [95% CI 0·892–0·982] vs 0·808 [0·671–0·909], p=0·0070; table 3). Some of this difference could be explained by the ensembling approach adopted with the bespoke model; however, even individual model outputs were higher than the CFDL output (appendix 3 p 8).
Figure 3 shows a matrix of cohort labels for the misclassifications in the internal test set. Most misclassifications were when the majority considered the image as pre-plus disease. The CFDL model frequently made a binary decision between plus disease or healthy (appendix 3 p 5). Examples of misclassification errors are shown in appendix 3 (p 10). Examination of saliency maps suggested that pixels involving retinal vasculature, particularly areas of tortuosity and engorgement, influenced the model output (appendix 3 p 11).
Figure 3: Matrix heatmap showing disagreements between the model and graders within the internal test set.
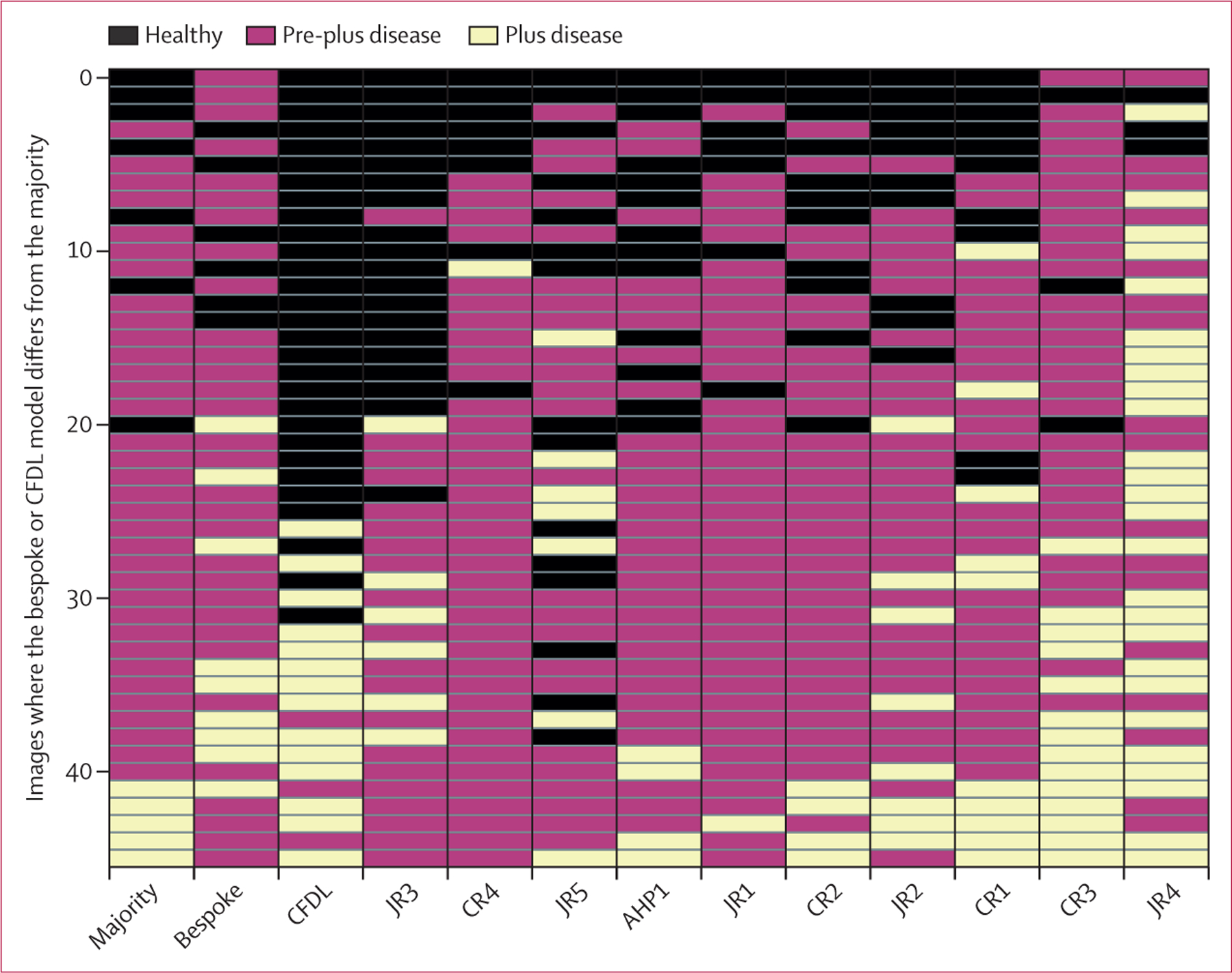
Each row indicates a different observation or image, columns indicate different graders, and colours indicate different classes (healthy, pre-plus disease, and plus disease). Cases are ordered vertically by the mean severity from all ten graders. Horizontally, graders are listed from left to right by sensitivity. All four CRs were included. AHP=allied health professional. CFDL=code-free deep learning. CR=consultant rater. JR=junior rater.
Discussion
In this study, performance of both the bespoke and CFDL models for the discrimination of healthy versus plus or pre-plus disease was similar to senior clinicians currently undertaking ROP screening. Both models generalised well on international external validation datasets using the same imaging device (Retcam); however, the CFDL model performed less well than the bespoke model on the task of detecting pre-plus disease. Both models had a reduction in performance when tested on a separate imaging device, the 3nethra neo. Although deployment of such models requires a thorough evaluation of effectiveness, our diagnostic accuracy results highlight the potential for automated plus disease diagnosis. In the UK, neonatal units are currently leveraging Retcam-based neonatal screening approaches as well as use of CFDL platforms for in-house development of automated deep-learning classification systems for ROP screening.
Previously, we evaluated the feasibility of code-free deep learning methods for medical image classification tasks, reporting similar diagnostic accuracy to contemporary state-of-the-art models.23,31 In the current study, we show similar performance in most tasks between pre-trained bespoke model architectures and an automated deep learning approach (neural architecture search) provided through the Google Cloud. CFDL approaches have limitations in their ability to interrogate and adapt specific details about model architecture. For example, we adopted a cross-validation and ensembling approach for our bespoke models, but these cannot be implemented within CFDL. Indeed, our individual bespoke models performed worse than the ensemble model, however they still exceeded that of the CFDL on the i-ROP external validation dataset (appendix 3 p 7). However, CFDL does provide an alternative option for model development when specialist data science expertise and access to high-performance computing resources (eg, graphics processing units) might be scarce. This scarcity is the case in LMICs, such as in sub-Saharan Africa, where a relative increase in access to specialist neonatal care has led to improved preterm neonatal survival and consequent escalation in the incidence of ROP.32 Moreover, particularly in a disease such as ROP, whereby there is evidence that the natural history might differ between ethnic groups and by socioeconomic status,16,17,33 models developed in one setting might generalise poorly to another due to distributional shift.14 For example, among nine studies in a recent systematic review of deep learning classifiers, no development datasets included neonates from South America or Africa.34,35 Local training facilitated through CFDL approaches might provide some mitigation, allowing neonatal units in poorly resourced regions to develop models optimised to their specific populations.
Inter-observer variability within our study was generally in line with previous reports, however, a novel element of our study was to examine clinicians with a range of experience.27,28,36,37 Senior clinicians (ie, consultant ophthalmologists) showed greater levels of agreement with each other than junior clinicians and agreement varied between the pairs of junior clinicians. Inter-observer agreement in plus disease diagnosis even among senior ophthalmologists is known to be moderate,27,36 which introduces challenges when establishing a rigorous reference standard. Chen and colleagues38 have suggested three means for improving the reproducibility of reference standards based on subjective interpretation: increasing the expertise of graders, increasing the number of graders for each case, and ensuring the disagreement resolution process is unbiased. Our study, which leveraged independent gradings from two junior ophthalmologists with at least 3 years of ophthalmology experience and arbitration by a senior ophthalmologist, aspired to an unbiased resolution process minimising groupthink. However, as the model matures towards real-world deployment, the ground truth is likely to be defined through a larger number of graders with greater experience. Some alternative strategies have been investigated by other groups: the i-ROP Research Consortium combined image labelling from multiple graders with ancillary information from clinical examination.11,39 Such labels are robust but can be pragmatically challenging because they require prospective data collection using standardised protocols for examination. Moreover, work by the same group has also suggested that image-based diagnostic accuracy specifically for plus disease might actually exceed that of ophthalmoscopy examination.40 Another option might be to consider colour fundus photography segmentation tools for automating the extraction of quantitative retinovascular indices, such as venular calibre and arteriolar tortuosity, and use these as surrogates of disease.
Disagreements between graders and the bespoke and CFDL models primarily involved cases labelled as pre-plus disease. For example, almost all misclassifications in the internal test set were where the majority vote was pre-plus disease. Despite a seemingly high AUC of 0∙932 (95% CI 0·896–0·964) for the discrimination of pre-plus disease by the CFDL model, it would usually output either plus disease or healthy. This nuance becomes apparent on (1) external validation in the i-ROP dataset whereby the bespoke model had significantly better performance than the CFDL model for pre-plus disease detection and (2) inspecting the weighted κ values where the agreement between the majority vote and bespoke (κ 0·77) was markedly superior to that between the majority vote and CFDL (κ 0·53). It might be that CFDL is more sensitive to situations with uneven class representation during training. A solution might be to consider that plus disease represents one end of a spectrum of proliferative retinovascular disease and a continuous vascular severity score might be more suitable for modelling, a strategy supported by the latest consensus statement from the International Class-ification of Retinopathy of Prematurity.41 Recent work by the i-ROP Research Consortium has validated a quantitative score and shown use for monitoring disease progression, predicting treatment-requiring ROP, and for post-treatment disease regression.42–44
Several limitations should be considered when interpreting our results. First, due to information governance restrictions, we did not explicitly collect individual-level data on potential confounders of model performance, such as sex and ethnicity, in our development dataset. Although the dataset is derived from a diverse population, we could only report summary information on ethnicity. Formal analyses stratified by these potential confounders would elucidate their contribution in model inference and inform concerns over algorithmic fairness. Second, grading of the dataset was restricted to image quality and the presence of pre-plus or plus disease. Although, in our development dataset the detection of retinal lesions (eg, haemorrhage) in the area captured by the Retcam image was very low, it is conceivable that the models could use these features as shortcut signals. Future work could explicitly annotate images for retinal lesions as well as provide test performance results in groups stratified by their presence or absence. Third, our study leveraged a hierarchical grading approach with the first grader as junior ophthalmologists, yet inter-observer variability of ROP diagnosis among even senior ophthalmologists was moderate. Strategies for improving the ground-truth process through other approaches, as mentioned earlier, could be used for follow up work before deployment. Fourth, grading of image quality and selection of images centred on the optic nerve was manually performed in this study. Although we developed models for image gradability, we did not explicitly assess model performance stratified by levels of image quality or integrate quality assessment into a patient workflow. Before any clinical implementation, further consideration on integrating image quality control and its consequent impact on diagnostic accuracy should be considered, as per other work in diabetic retinopathy and ROP.12,45,46 Fifth, we benefitted from a relatively large dataset for training both our bespoke and CFDL models although other units may not have access to large datasets. It remains unclear how many images constitute a sufficient sample size for modelling plus disease classification; however, most reports have used several thousands of images. There currently is no openly available large dataset of neonatal retinal images; however, one potential solution gaining appeal might be training deep learning models with use of real alongside synthetic neonatal images. This approach has been shown to lead to similar model performance with models trained exclusively on real images in a recent study.47 Finally, our models generalised well when using the same imaging device as the development dataset (Retcam); however, it should be noted that the individual external validation datasets (especially for image gradability) are relatively small. A major impetus for the development of automated deep-learning ROP screening is the substantial variability in neonatal care practice patterns in different health-care settings, whereas all infants in our development dataset had access to UK neonatal care, which could potentially induce some bias. The models’ performance on external validations, however, is indicative of low bias and high generalisability of our models. Further external validation is always helpful in the process of accruing evidence of the models’ broader generalisability. For example, infants with severe ROP in China tend to be larger and older.48
In summary, automated plus disease detection through deep learning provides levels of diagnostic accuracy similar to senior paediatric ophthalmology clinicians in neonates undergoing ROP screening according to the RCOphth criteria. The strong performance of bespoke modular architectures can be emulated by automated deep learning tools, especially in the binary classification task of discriminating referrable from non-referrable disease. Such code-free options might provide an option for LMICs, where an intersection of rising rates of ROP, scarce resources, and few clinicians with the appropriate specialist expertise could result in an increase in rates of childhood blindness. Although further validation and studies of effectiveness across different populations are needed before deployment, deep learning might provide a tool for mitigating the risk of lifelong sight impairment in these young patients.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed, MEDLINE, Scopus, Web of Science, Embase, and arXiv for studies from database inception up to Nov 21, 2022, using the keywords “retinopathy of prematurity”, “plus disease”, “fundus photographs”, “machine learning”, “artificial intelligence (AI)”, and “deep learning”. Only study reports written in English were included into the search and reviewed. Abstracts in English from reports written in other languages were also reviewed. We focused particularly on geographical setting, selection criteria for neonatal examinations, sociodemographic characteristics, reference standard, model development, and comparison with human experts. Diagnostic accuracy was generally high across reports. Almost all studies emerged from North America and Asia. Several studies did not stipulate the inclusion criteria for neonatal examination or used thresholds substantially different from internationally recognised retinopathy of prematurity screening programmes. Only one study using a code-free deep learning platform was identified and no comparisons were seen between code-free and bespoke architectures. Two studies had examined generalisability across different imaging devices, and external validation in a substantially disparate population from the development dataset population was only seen for two models. Comparison with experts was typically limited to few senior paediatric ophthalmologists.
Added value of this study
We report on the diagnostic accuracy of both bespoke and code-free deep learning models for plus disease across an ethnically and socioeconomically diverse population of premature babies explicitly fulfilling standardised screening criteria in the UK. Bespoke and code-free approaches had similar performance when discriminating healthy from pre-plus disease or plus disease retinal images; however, the code-free model performed inferior to the bespoke model on detection of the minority class pre-plus disease. Overall, performance on the internal test set of both models was similar to senior paediatric ophthalmologists who regularly conduct retinopathy of prematurity screening. Both models generalised well to independent external validation test sets from the USA, Brazil, and Egypt but performance dropped when evaluated on images from a different neonatal camera on the task of discriminating healthy from pre-plus disease or plus disease images.
Implications of all the available evidence
Both bespoke and code-free deep learning approaches confer acceptable performance for the discrimination of healthy and pre-plus disease or plus disease. For resource-limited settings, where hardware and deep learning resources might be scarce, code-free approaches might provide an alternative option for local teams to develop, validate, and potentially deploy in their own populations. This approach might be of particular appeal for retinopathy of prematurity where risk of dataset shift upon generalisation is high—the natural history of the disease differs between ethnic groups, image capture devices might vary between different institutions, and screening criteria are heterogeneous. Further work into the implementation and effectiveness of integrating such classification models into screening programmes are warranted.
Acknowledgments
This research was supported by the National Institute for Health Research Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and the University College London Institute of Ophthalmology. The authors thank the patients, whose data were used as part of this study, as well as their families.
Declaration of interests
SKW is funded through a Medical Research Council Clinical Research Training Fellowship (MR/TR000953/1). MR has received travel fees from Bayer and previously worked as a consultant for IQVIA. PAK is supported by a Moorfields Eye Charity Career Development Award (R190028A) and a UK Research & Innovation Future Leaders Fellowship (MR/T019050/1); receives research support from Apellis; is a consultant for DeepMind, Roche, Novartis, Apellis, and BitFount; is an equity owner in Big Picture Medical; and has received speaker fees from Heidelberg Engineering, Topcon, Allergan, Roche, and Bayer; meeting or travel fees from Novartis and Bayer; and compensation for being on an advisory board from Novartis and Bayer. NP is supported by a National Institute for Health and Care Research AI Award (AI_AWARD02488) and Moorfields Eye Charity Career Development Award (R190031A); co-founder and director of Phenopolis. JPC declares grants or contracts from Genentech and National Institutes of Health; grants from Research to Prevent Blindness; consulting fees from Boston AI; and is an equity owner and chief medical officer for Siloam Vision, a company involved in ROP telemedicine and artificial intelligence. Siloam Vision has no rights or interest in the technology described in this Article and had no part in the design, planning, or conduct of the study. PJP has received speaker fees from Bayer and Roche; meeting or travel fees from Novartis and Bayer; compensation for being on an advisory board from Novartis, Bayer, and Roche; consulting fees from Novartis, Bayer, and Roche; and research support from Bayer. GA declares an institutional grant from Bayer; payment or honoraria from the British and Irish Paediatric and Strabismus Association; and participation on a Data Safety Monitoring Board or Advisory Board for an NHS England Policy Working Group (Ranibizumab for ROP). KB has received speaker fees from Novartis, Bayer, Alimera, Allergan, Roche, and Heidelberg; meeting or travel fees from Novartis and Bayer; compensation for being on an advisory board from Novartis and Bayer; consulting fees from Novartis and Roche; and research support from Apellis, Novartis, and Bayer. All other authors declare no competing interests.
Footnotes
For the Portuguese translation of the abstract see Online for appendix 1
For the Arabic translation of the abstract see Online for appendix 2
For more on DenseNet for PyTorch see https://arxiv.org/abs/1608.06993
For more on ImageNet see https://www.image-net.org/
For more on the Google Cloud AutoML Vision Application Programming Interface see https://cloud.google.com/vision/docs/reference/rest#service:-vision.googleapis.com
Data sharing
Model architecture source code can be accessed at https://github.com/MoorfieldsInnovationLab/Automated-detection-of-plus-disease.
We provide descriptions of the experiments and implementation details in the methods to allow for independent replication. The datasets in this Article are not publicly available; however, collaborations are welcomed. Requests for access to the development dataset and those from Brazil and Egypt are subject to appropriate data sharing and legal provisions and should be directed to the corresponding author at k.balaskas@nhs.net. The USA-based external validation dataset was developed by the Imaging and Informatics in Retinopathy of Prematurity (i-ROP) consortium. Requests for access to the i-ROP dataset should be sent to J Peter Campbell at campbelp@ohsu.edu. More information on i-ROP is available at https://i-rop.github.io/.
References
- 1.Cryotherapy for Retinopathy of Prematurity Cooperative Group. Multicenter trial of cryotherapy for retinopathy of prematurity. Preliminary results. Arch Ophthalmol 1988; 106: 471–79. [DOI] [PubMed] [Google Scholar]
- 2.Early Treatment For Retinopathy Of Prematurity Cooperative Group. Revised indications for the treatment of retinopathy of prematurity: results of the early treatment for retinopathy of prematurity randomized trial. Arch Ophthalmol 2003; 121: 1684–94. [DOI] [PubMed] [Google Scholar]
- 3.Fierson WM, Chiang MF, Good W, et al. Screening examination of premature infants for retinopathy of prematurity. Pediatrics 2018; 142: e20183061. [DOI] [PubMed] [Google Scholar]
- 4.Glass HC, Costarino AT, Stayer SA, Brett CM, Cladis F, Davis PJ. Outcomes for extremely premature infants. Anesth Analg 2015; 120: 1337–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kemper AR, Wallace DK. Neonatologists’ practices and experiences in arranging retinopathy of prematurity screening services. Pediatrics 2007; 120: 527–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SK, Callaway NF, Wallenstein MB, Henderson MT, Leng T, Moshfeghi DM. SUNDROP: six years of screening for retinopathy of prematurity with telemedicine. Can J Ophthalmol 2015; 50: 101–06. [DOI] [PubMed] [Google Scholar]
- 7.Bronsard A, Geneau R, Duke R, et al. Cataract in children in sub-Saharan Africa: an overview. Expert Rev Ophthalmol 2018; 13: 343–50. [Google Scholar]
- 8.Worrall DE, Wilson CM, Brostow GJ. Automated retinopathy of prematurity case detection with convolutional neural networks. In: Carneiro G, Mateus M, Peter L, et al. Deep learning and data labeling for medical applications. Switzerland: Springer Cham, 2016: 68–76. [Google Scholar]
- 9.Tong Y, Lu W, Deng Q-Q, Chen C, Shen Y. Automated identification of retinopathy of prematurity by image-based deep learning. Eye Vis (Lond) 2020; 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Ju R, Chen Y, et al. Automated retinopathy of prematurity screening using deep neural networks. EBioMedicine 2018; 35: 361–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown JM, Campbell JP, Beers A, et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol 2018; 136: 803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Ji J, Zhang M, et al. Automated explainable multidimensional deep learning platform of retinal images for retinopathy of prematurity screening. JAMA Netw Open 2021; 4: e218758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Huang K, Ju R, et al. Evaluation of artificial intelligence-based quantitative analysis to identify clinically significant severe retinopathy of prematurity. Retina 2022; 42: 195–203. [DOI] [PubMed] [Google Scholar]
- 14.Finlayson SG, Subbaswamy A, Singh K, et al. The clinician and dataset shift in artificial intelligence. N Engl J Med 2021; 385: 283–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim H, Liu X, Zariffa N, Morris AD, Denniston AK. Health data poverty: an assailable barrier to equitable digital health care. Lancet Digit Health 2021; 3: e260–65. [DOI] [PubMed] [Google Scholar]
- 16.Eliason KJ, Dane Osborn J, Amsel E, Richards SC. Incidence, progression, and duration of retinopathy of prematurity in Hispanic and White non-Hispanic infants. J AAPOS 2007; 11: 447–51. [DOI] [PubMed] [Google Scholar]
- 17.Aralikatti AKV, Mitra A, Denniston AKO, Haque MS, Ewer AK, Butler L. Is ethnicity a risk factor for severe retinopathy of prematurity? Arch Dis Child Fetal Neonatal Ed 2010; 95: F174–76. [DOI] [PubMed] [Google Scholar]
- 18.Chen JS, Coyner AS, Ostmo S, et al. Deep learning for the diagnosis of stage in retinopathy of prematurity: accuracy and generalizability across populations and cameras. Ophthalmol Retina 2021; 5: 1027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole E, Valikodath NG, Al-Khaled T, et al. Evaluation of an artificial intelligence system for retinopathy of prematurity screening in Nepal and Mongolia. Ophthalmol Sci 2022; 2: 100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinekar A, Rao SV, Murthy S, et al. A novel, low-cost, wide-field, infant retinal camera, “Neo”: technical and safety report for the use on premature infants. Transl Vis Sci Technol 2019; 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinekar A, Jayadev C, Mangalesh S, Shetty B, Vidyasagar D. Role of tele-medicine in retinopathy of prematurity screening in rural outreach centers in India—a report of 20,214 imaging sessions in the KIDROP program. Semin Fetal Neonatal Med 2015; 20: 335–45. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert C, Malik ANJ, Nahar N, et al. Epidemiology of ROP update - Africa is the new frontier. Semin Perinatol 2019; 43: 317–22. [DOI] [PubMed] [Google Scholar]
- 23.Faes L, Wagner SK, Fu DJ, et al. Automated deep learning design for medical image classification by health-care professionals with no coding experience: a feasibility study. Lancet Digit Health 2019; 1: e232–42. [DOI] [PubMed] [Google Scholar]
- 24.Antaki F, Coussa RG, Kahwati G, Hammamji K, Sebag M, Duval R. Accuracy of automated machine learning in classifying retinal pathologies from ultra-widefield pseudocolour fundus images. Br J Ophthalmol 2023; 107: 90–95. [DOI] [PubMed] [Google Scholar]
- 25.British Association of Perinatal Medicine, BLISS. Guideline for the Screening and Treatment of Retinopathy of Prematurity (2008). https://www.bapm.org/resources/37-guideline-for-the-screening-and-treatment-of-retinopathy-ofprematurity-2008 (accessed Dec 11, 2022).
- 26.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Circulation 2015; 131: 211–19. [DOI] [PubMed] [Google Scholar]
- 27.Kalpathy-Cramer J, Campbell JP, Erdogmus D, et al. Plus disease in retinopathy of prematurity: improving diagnosis by ranking disease severity and using quantitative image analysis. Ophthalmology 2016; 123: 2345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell JP, Kalpathy-Cramer J, Erdogmus D, et al. Plus disease in retinopathy of prematurity: a continuous spectrum of vascular abnormality as a basis of diagnostic variability. Ophthalmology 2016; 123: 2338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–45. [PubMed] [Google Scholar]
- 30.Ying G-S, Maguire MG, Glynn RJ, Rosner B. Tutorial on biostatistics: receiver-operating characteristic (ROC) analysis for correlated eye data. Ophthalmic Epidemiol 2022; 29: 117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korot E, Guan Z, Ferraz D, et al. Code-free deep learning for multi-modality medical image classification. Nat Mach Intell 2021; 3: 288–98. [Google Scholar]
- 32.Herrod SK, Adio A, Isenberg SJ, Lambert SR. Blindness secondary to retinopathy of prematurity in sub-Saharan Africa. Ophthalmic Epidemiol 2022; 29: 156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karmouta R, Altendahl M, Romero T, et al. Association between social determinants of health and retinopathy of prematurity outcomes. JAMA Ophthalmol 2022; 140: 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Liu Y, Mitsuhashi T, Matsuo T. Accuracy of deep learning algorithms for the diagnosis of retinopathy of prematurity by fundus images: a systematic review and meta-analysis. J Ophthalmol 2021; 2021: 8883946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan SM, Liu X, Nath S, et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit Health 2021; 3: e51–66. [DOI] [PubMed] [Google Scholar]
- 36.Chiang MF, Jiang L, Gelman R, Du YE, Flynn JT. Interexpert agreement of plus disease diagnosis in retinopathy of prematurity. Arch Ophthalmol 2007; 125: 875–80. [DOI] [PubMed] [Google Scholar]
- 37.Campbell JP, Ryan MC, Lore E, et al. Diagnostic discrepancies in retinopathy of prematurity classification. Ophthalmology 2016; 123: 1795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen PC, Mermel CH, Liu Y. Evaluation of artificial intelligence on a reference standard based on subjective interpretation. Lancet Digit Health 2021; 3: e693–95. [DOI] [PubMed] [Google Scholar]
- 39.Ryan MC, Ostmo S, Jonas K, et al. Development and evaluation of reference standards for image-based telemedicine diagnosis and clinical research studies in ophthalmology. AMIA Annu Symp Proc 2014; 2014: 1902–10. [PMC free article] [PubMed] [Google Scholar]
- 40.Biten H, Redd TK, Moleta C, et al. Diagnostic accuracy of ophthalmoscopy vs telemedicine in examinations for retinopathy of prematurity. JAMA Ophthalmol 2018; 136: 498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang MF, Quinn GE, Fielder AR, et al. International classification of retinopathy of prematurity, third Edition. Ophthalmology 2021; 128: e51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor S, Brown JM, Gupta K, et al. Monitoring disease progression with a quantitative severity scale for retinopathy of prematurity using deep learning. JAMA Ophthalmol 2019; 137: 1022–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta K, Campbell JP, Taylor S, et al. A quantitative severity scale for retinopathy of prematurity using deep learning to monitor disease regression after treatment. JAMA Ophthalmol 2019; 137: 1029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redd TK, Campbell JP, Brown JM, et al. Evaluation of a deep learning image assessment system for detecting severe retinopathy of prematurity. Br J Ophthalmol 2018; published online Nov 23. 10.1136/bjophthalmol-2018-313156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai L, Wu L, Li H, et al. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat Commun 2021; 12: 3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruamviboonsuk P, Tiwari R, Sayres R, et al. Real-time diabetic retinopathy screening by deep learning in a multisite national screening programme: a prospective interventional cohort study. Lancet Digit Health 2022; 4: e235–44. [DOI] [PubMed] [Google Scholar]
- 47.Coyner AS, Chen JS, Chang K, et al. Synthetic medical images for robust, privacy-preserving training of artificial intelligence. Ophthalmol Sci 2022; 2: 100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Zhou X, Zhang Q, et al. Screening for retinopathy of prematurity in China: a neonatal units-based prospective study. Invest Ophthalmol Vis Sci 2013; 54: 8229–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Model architecture source code can be accessed at https://github.com/MoorfieldsInnovationLab/Automated-detection-of-plus-disease.
We provide descriptions of the experiments and implementation details in the methods to allow for independent replication. The datasets in this Article are not publicly available; however, collaborations are welcomed. Requests for access to the development dataset and those from Brazil and Egypt are subject to appropriate data sharing and legal provisions and should be directed to the corresponding author at k.balaskas@nhs.net. The USA-based external validation dataset was developed by the Imaging and Informatics in Retinopathy of Prematurity (i-ROP) consortium. Requests for access to the i-ROP dataset should be sent to J Peter Campbell at campbelp@ohsu.edu. More information on i-ROP is available at https://i-rop.github.io/.


