Abstract
Amyloid-β (Aβ) fibrils in neuritic plaques are a hallmark of Alzheimer’s disease (AD). Since the 42-residue Aβ (Aβ42) fibril is the most pathogenic among different Aβ species, its structural characterization is crucial to our understanding of AD. While several polymorphs have been reported for Aβ40, previous studies of Aβ42 fibrils prepared at neutral pH detected essentially only one structure, with an S-shaped β-sheet arrangement [e.g., Xiao et al., Nat. Struct. Mol. Biol. 2015, 22, 499]. Herein, we demonstrate the feasibility of characterizing the structure of trace amounts of brain-derived and synthetic amyloid fibrils by sensitivity-enhanced 1H-detected solid-state NMR (SSNMR) under ultra-fast magic angle spinning (UFMAS). By taking advantage of the high sensitivity of this technique, we first demonstrate its applicability for the high-throughput screening of trace amounts of selectively 13C- and 15N-labeld Aβ42 fibril prepared with ~0.01% patient-derived amyloid (ca. 4 pmol) as a seed. The comparison of 2D 13C/1H SSNMR data revealed marked structural differences between AD-derived Aβ42 (~40 nmol or ~200 μg) and synthetic fibrils in less than 10 min, confirming the feasibility of assessing the fibril structure from ~1 pmol of brain amyloid seed in ~2.5 h. We also present the first structural characterization of synthetic fully-protonated Aβ42 fibril by 1H-detected 3D and 4D SSNMR. With procedures assisted by automated assignments, main-chain resonance assignments were completed for trace amounts (~42 nmol) of a fully-protonated amyloid fibril in the 1H-detection approach. The results suggest that this Aβ42 fibril exhibits a novel fold or polymorph structure.
Graphical Abstract

Introduction
Alzheimer’s disease (AD) is a fatal neurodegenerative disorder characterized by the accumulation of neuritic plaques and neurofibrillary tangles outside and inside the neurons, respectively.1-3 While neurofibrillary tangles are composed of tau aggregates,4 neuritic plaques mainly consist of fibrillar aggregates of 39–43 residue-long amyloid-β (Aβ) fibrils.5 Among these structures, 40- and 42-residue Aβ (Aβ40 and Aβ42, respectively) fibrils are the predominant isoforms.6-7 Despite their amino acid sequences being different only in two residues located at the C-terminus, these two fibril forms show significantly different characteristics. Aβ42 has been suggested to be more neurotoxic and prone to aggregation compared to Aβ40.8-11 Because the Aβ42 fibril is considered more pathogenic with respect to the AD,11-13 its structural features are the key for developing an AD treatment targeting Aβ aggregation. However, conventional structural tools such as solution NMR and X-ray crystallography are not suitable for characterizing Aβ fibrils, owing to their insoluble and non-crystalline nature. Although cryo-electron microscopy (cryo-EM) has been used to study the structure of non-crystalline amyloid proteins, currently it is only applicable to determine the structure of twisted fibrils with near-atomic resolution.14-16
SSNMR spectroscopy is a powerful technique for elucidating the atomic-level structure of Aβ and other amyloid fibrils.17-43 Although several reports have studied the atomic-level structure of Aβ40 fibril and its mutants by SSNMR,18-31, 44-45 only three high-resolution structural models based on SSNMR are available for Aβ42 fibrils prepared at physiologically relevant neutral pH.27-29 These three structures present a common structural motif, with S-shaped parallel β-sheet folds having similar 13C shifts. Although a recent cryo-EM and SSNMR study suggested another structural motif for an Aβ42 fibril sample prepared at a low (~2) pH,14 relatively little information is available on structural variations of Aβ42 fibrils prepared at physiologically relevant conditions, despite their pathological importance. A major obstacle to the characterization of the Aβ42 fibrils comes from their structural heterogeneity,19, 21, 25, 32, 46 which often prevents the preparation of samples suitable for SSNMR analysis. All atomic models of Aβ42 fibrils were obtained using 13C-detected SSNMR under magic angle spinning (MAS) at low to moderate frequencies (12–20 kHz). Generally, due to its low sensitivity, 13C-detected SSNMR requires several mg of sample to record NMR signals with signal-to-noise (S/N) ratios sufficient for signal assignment or structure determination.25, 27-29, 47 The requirement of large amounts of isotope-labeled amyloid samples for 13C-detected SSNMR analysis is extremely demanding, especially with the difficulties associated with preparing homogeneous amyloid samples. Notably, it has proven impossible to characterize the structures of patient-derived amyloid samples by SSNMR without sacrificing a relatively large section of brain tissue (1–3 g).25
In this study, we evaluate the applicability of 1H-detected SSNMR to characterize homogeneous recombinant/synthetic Aβ42 fibrils and heterogeneous brain-derived Aβ42 fibrils that are available only in pico- to nano-molar amounts. 1H-detected SSNMR is attracting growing interest as a practical tool for studying biological systems.47-56 Especially, ultra-fast MAS (UFMAS) at a frequency of 60 kHz or above offers high 1H resolution for SSNMR, by removing line broadening due to 1H-1H dipolar coupling.47, 50, 54 With a much higher sensitivity per given amount of sample (mass-sensitivity) compared with its traditional 13C-detected approach, 1H-detected SSNMR under UFMAS is well suited for the characterization of trace amounts of biologically relevant compounds.51-55 The sensitivity of 1H-detected SSNMR can be enhanced by incorporating paramagnetic-assisted condensed data collection (PACC),57 drastically reducing the repetition delays and accumulation time of the SSNMR data. By taking advantage of the high sensitivity of 1H-detected SSNMR achieved with these methods, in this work we demonstrate the fast collection of 2D 13Cα/1Hα correlation SSNMR spectra from ~200 μg of in vitro-prepared and brain-derived Aβ42 fibrils that were isotopically labeled at selected residues. Using their SSNMR spectra as fingerprints of amyloid fibril structures, we were able to characterize the structure of amyloid fibril within 33 sec to 8.7 min. As the brain-derived Aβ42 fibril was prepared by growing 13C- and 15N-labeled Aβ42 fibril with approximately 0.010% (~20 ng or ~4 pmol) of amyloid from AD patients as a seed, a minimal amount of patient tissues is required with the present approach. Furthermore, we successfully applied the 1H-detected SSNMR technique under UFMAS at 90 kHz for the structural characterization of a new Aβ42 fibril polymorph prepared from a uniformly 13C- and 15N-labeled sample produced with bacterial protein expression (without brain seed in this case). The excellent sensitivity of 1H-detected SSNMR and the dramatic enhancement of the 1H resolution by UFMAS enabled us to assign most of the resonances in both the protein backbone and side chains using a combination of 3D and 4D SSNMR experiments. Importantly, we were able to record the SSNMR data using only ~42 nmol (~200 μg) of the Aβ42 fibril sample, which is 25–100 times less than the amounts used in earlier studies.14, 27-29, 58-60 We accomplished signal assignment of the chemical shifts based on the 2D–4D SSNMR data with the aid of automated assignment procedures. The comparison of the 13C chemical shifts with those of Aβ42 fibrils with known structures reported in previous studies suggests that our Aβ42 fibril may have a unique structure that was not reported to date. To the best of our knowledge, this is the first 1H-based signal assignment reporting the chemical shifts of both the protein backbone and side chains of amyloid fibrils. Our study will promote the SSNMR-based structural analysis of patient-derived ex vivo amyloid samples that are typically available only in sub-nanomolar amounts.
Results and Discussion
Screening of Picomolar Amounts of Brain-derived Amyloid Fibrils by 1H-detected SSNMR
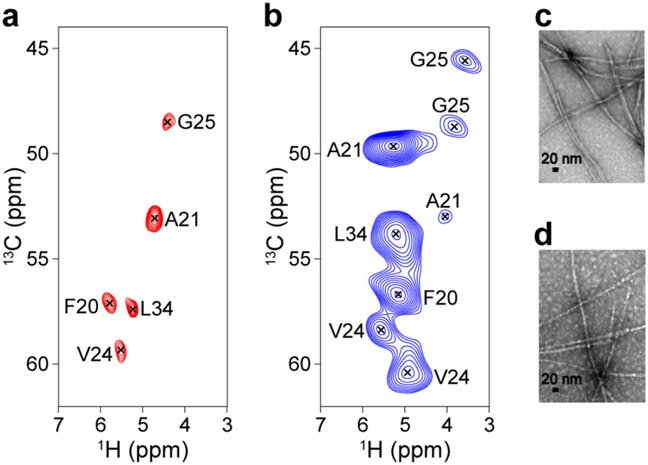
The atomic details of Aβ42 fibrils provide insight into how the amyloid structure is stabilized via molecular contacts and potentially into how therapeutic agents can disrupt toxic aggregates. In addition, very little information is available on the structures of Aβ42 fibrils obtained from AD patients.15, 25-26, 32, 61 Therefore, investigating whether the structure of in vitro-prepared Aβ fibrils actually reflects that of amyloid species in AD brain tissue is a task of great significance. However, the difficulty in achieving mass production of brain-derived Aβ42 fibrils, due to limited sample availability, demands more sensitive and efficient SSNMR techniques to enable the structural comparison of synthetic and AD-derived Aβ fibrils. Herein, we compared the 1H-detected 13Cα/1Hα correlation 2D spectra of in vitro-prepared and brain-derived Aβ42 fibril samples (Figure 1a and b, respectively). Note that the samples were uniformly 13C- and 15N-labeled at selected residues (see caption).
Figure 1.
(a, b) 1H-detected 13C/1H correlation 2D SSNMR spectra of (a) in vitro-prepared and (b) brain-derived Aβ42 fibrils that were uniformly 13C- and 15N-labeled at Phe-20, Ala-21, Val-24, Gly-25, and Leu-34. The signal assignments are shown in the spectra. SSNMR spectra were recorded at a MAS rate of 90 kHz on an 800 MHz Bruker Avance III NMR spectrometer equipped with a JEOL 0.75-mm 1H/13C/15N/2H quad-resonance probe (see Materials and Methods for details). The pulse sequence in Figure S1 was used to collect the data. The experimental time was (a) 33 sec and (b) 8.7 min. (c, d) Transmission electron micrograph (TEM) images of the (c) synthetic and (d) brain-derived Aβ42 fibril samples.
Because of the high sensitivity of our 1H-detected SSNMR approach using UFMAS, the spectrum in Figure 1a was obtained in only 33 s for ~200 μg of the synthetic Aβ42 fibril sample (Figure 1c), which was prepared following the procedure described in ref. 62. The signal assignments of the amino acid-dependent 13Cα chemical shifts indicate that one relatively sharp peak correlated the 1Hα and 13Cα resonances for each labeled residue, suggesting that the sample is made of a single conformer of Aβ; as reported in our previous study, the Aβ42 fibril is known to adopt an S-shaped triple β-sheet arrangement.27 The brain-derived Aβ42 fibril sample (~200 μg) (Figure 1b, d), which was uniformly isotope-labeled at the same residues as those of Figure 1a, was obtained by incubating monomeric 13C- and 15N-labeled Aβ42 with ~0.010% of brain amyloid (~20 ng or ~4 pmol) as seed (see the Materials and Methods section for further details).
To our surprise, the spectrum in Figure 1b shows significantly different chemical shifts from those in Figure 1a. Furthermore, there is more than one peak corresponding to each isotope-labeled residue. Our assignment of Ala-21, for example, suggests the presence of at least one major and one minor peak, indicating two different structures. The major peak does not match the corresponding peak position for Ala-21 in Figure 1a. Similar trends were observed for the Val-24 and Gly-25 residues. These results suggest that the main conformer of the brain-derived Aβ42 fibril has a considerably different structure from the S-shaped β-sheet arrangement of the synthetic Aβ42.27 The presence of the two different conformers limits the sensitivity of Figure 1b compared with that of Figure 1a. It should also be noted that the brain-derived Aβ42 sample contained considerable amounts of brain materials (5.9%) other than Aβ42 fibrils, unlike the synthetic fibril used for Figure 1a. Despite the disadvantages associated with the “native” amyloid sample, we could record a 13C/1H correlation 2D spectrum of the brain-derived Aβ42 fibril within 8.7 min (compared to 33 s for the synthetic Aβ42 fibril). Thus, our analysis could be completed within 2.5 h, even from the same fibril sample of ~10 nmol prepared from ~1 pmol of brain seed. These results show that sensitivity-enhanced 1H-detected SSNMR provides a novel high-throughput route to highlight structural differences between synthetic and patient-derived amyloid fibrils.
For a more precise analysis, we assigned the 13C, 15N, and 1H resonances of the brain-derived Aβ42 fibril sample by recording 1H-detected (H)CCH and (H)CANH 3D SSNMR spectra (Tables S1). Based on our analysis, we identified 3–4 peaks for each isotope-labeled residue, suggesting that the brain-derived Aβ42 sample may contain up to 3–4 different conformers. Then, we compared the 13C secondary chemical shifts of these conformers with those of the previously reported brain-derived and synthetic Aβ42 and Aβ40 fibrils (Table S2).14, 20, 25, 27, 30, 32, 63 The comparison clearly shows that the conformations of the fibrils in our brain-derived Aβ42 sample are distinct from those of the other amyloid fibrils at the compared sites. It should be noted that the brain material used as seeds in preparation of the patient-derived Aβ42 fibrils contained not only Aβ42 seeds (86%), but also limited Aβ40 seeds (14%) and other non-amyloid brain material. Thus, there is a possibility that cross-seeding from Aβ40 fibril might have occurred, simultaneously, although a majority should be self-seeded with brain Aβ42 fibrils considering the inefficiency of the cross-seeding.27
New Polymorph for Aβ42 Fibril
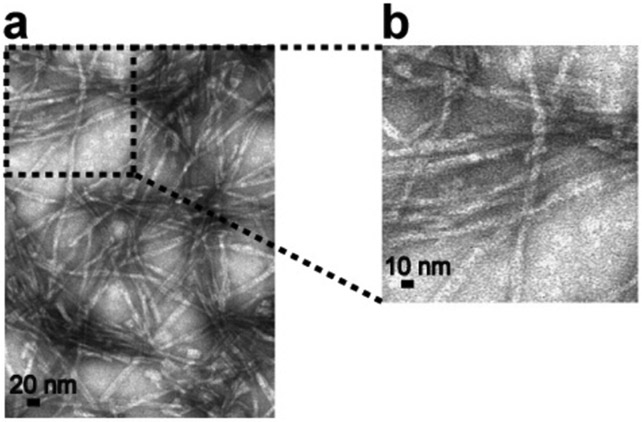
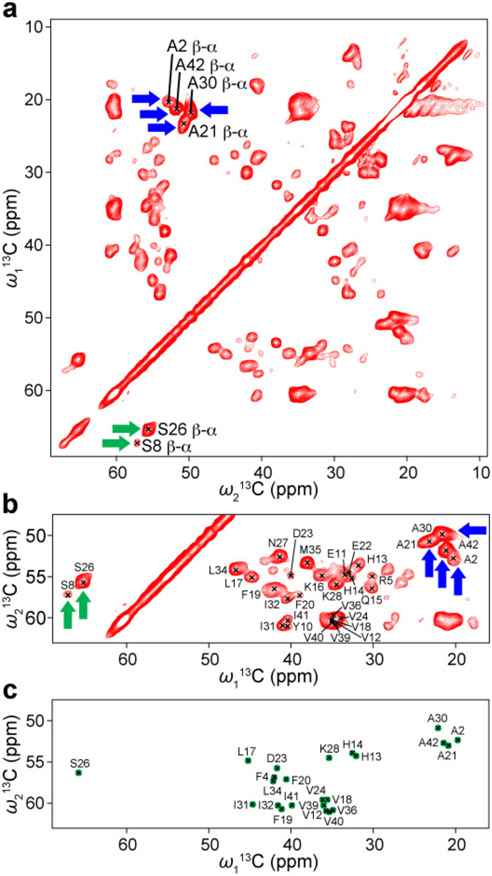
Next, we performed a SSNMR analysis of a structurally homogeneous and uniformly 13C- and 15N-labeled Aβ42 fibril that was newly obtained from a bacteria-expressed sample following a seeding protocol described in a previous study.62 A protein expression system in E. coli BL21 was used to express the Aβ42 peptide using our previously published protocol,64 with minor modifications. Figure 2 shows TEM images of the Aβ42 fibril obtained after three consecutive seeding steps. In the first round of incubation, ~40 μM unlabeled recombinant Aβ42 was incubated without any seeds for ~5 days. In the second and third rounds, 5% (w/w) of seed Aβ42 fibril from the previous round of the incubation was added to a 45–50 μM Aβ42 monomer solution (in the last one with the 13C- and 15N-labeled Aβ42), and we incubated the solution for 3–4 days. The TEM images display bundled fibrils with a diameter of ~10 nm, indicating the presence of a single morphology throughout the sample. Although the resolution is limited, the morphology of the fibrils appears less twisted compared with that observed in our previous studies.27, 62 The conformational homogeneity of the sample was further confirmed by SSNMR analysis, as discussed below. First, we examined the conformational homogeneity of the sample by recording the 1H-detected (H)CCH 3D data, whose 13C/13C 2D projection is shown in Figure 3a, b. We observed only a single set of Cβ-Cα-Hα cross-peaks for each residue type in the spectrum. For example, the amino acid sequence of Aβ42 contained only two Ser residues (Ser-8 and Ser-26), for which we could assign one weak and one strong cross-peak, respectively (green arrows in Figure 3a, b). Similarly, four well-resolved signals were obtained for four Ala residues (Ala-2, Ala-21, Ala-30 and Ala-42) (blue arrows in Figure 3a, b). This finding confirmed that our sample contained only a single structure for the Aβ peptide. The average 13C line width was 1.2 ppm; this relatively narrow value for a fibril sample indicates homogeneous structure. We further confirmed the higher mobility of the N-terminal part of the peptide by examining the intensities of the Cβ-Cα-Hα cross-peaks for Ala and Ser residues. The Ala-2 and Ser-8 residues showed significantly weaker intensities in the 3D spectrum compared with those of the Ala-21, Ala-30, Ala-42, and Ser-26 residues, reflecting more dynamic nature of the N-terminal part of the Aβ42 peptide. Next, we compared the obtained 2D 13C/13C projection with the corresponding spectrum simulated from the known 13Cα/13Cβ assignments for the Aβ42 fibril with S-shaped structure (Figure 3c).27 The comparison clearly highlights a poor match between the two spectra, suggesting that the Aβ42 fibril examined in this study is likely to be a new polymorph species, as further discussed below. This finding was rather surprising, because apart from the source of the Aβ peptide (bacterial expressed vs. chemically synthesized), the incubation protocols were almost identical. Although the origin of the differences is unclear, the isolation of a new polymorph species of homogeneous Aβ42 fibril represents a substantial progress.
Figure 2.
TEM images of seeded Aβ42 fibrils obtained after 144 h incubation of third-generation (G3) fibrils. Scale bars are 20 nm (a) and 10 nm (b).
Figure 3.
(a) 13C/13C 2D projection of a 1H-detected (H)CCH 3D spectrum of 13C- and 15N-labeled Aβ42 fibril. (b) Enlarged view of spectral region extracted from (a), showing the signals corresponding to Cβ-Cα-Hβ correlations, for comparison with the Cβ-Cα assignments of Aβ42 fibrils with known S-shaped structure. (c) A simulated spectrum from the 13Cβ-13Cα assignments of the Aβ42 fibril with S-shaped structure.27
Sequential Assignments and Structural Profiling of the New Polymorph
Next, we demonstrate the effectiveness of the 1H-detected SSNMR approach for signal assignments and structural profiling of the Aβ42 fibril. Previous studies of amyloid fibrils such as HET-s(218–289) by 1H-detected SSNMR required partially deuterated samples,65 which are generally difficult to prepare, for successful signal assignments with enhanced resolution. For the site-specific chemical shift assignment of fully-protonated Aβ42 fibrils, we recorded a combination of 1H-detected 3D and 4D SSNMR data, in addition to the (H)CCH 3D spectrum discussed above. The 13C and 1H resonances from the (H)CCH 3D experiment allowed us to identify the signals based on the residue type in the amino acid sequence of Aβ42 peptide (see Figure S3 in the Supporting Information). Figure 4a shows the representative strip plots of the (H)CA(CON)CAH (green), (H)CANH (red), and (H)CA(CO)NH (blue) 3D spectra, which illustrate the sequential connectivity of the protein backbone from Asn-27–Met-35. In particular, the (H)CA(CON)CAH 3D data (green spectrum in Figure 4a) allowed us to determine the sequential connectivity between Cα(i-1)-Cα(i) resonance pairs for most of the residues (except for the ones having similar 13Cα chemical shifts). Although we initially expected to see only the cross-peaks corresponding to Cα(i-1)-Cα(i)-Hα(i) correlations in the (H)CA(CON)CAH 3D spectrum (marked as black crosses), for most residues we also observed diagonal-peaks (red crosses) corresponding to Cα(i-1)-Cα(i-1)-Hα(i-1) triads via remote Ni-Cα(i-1) magnetization transfer. Thus, most of the 2D (H)CA(CON)CAH strip plots (colored green in Figure 4a) showed two peaks in each strip. Further, Gly-25, Gly-29 and Gly-37 showed cross-peaks at different geminal 1Hα resonances in the (H)CA(CON)CAH 3D spectrum; thus, two strips are displayed in the figure for Gly-29. Moreover, the Asp-1–Gly-9 residues did not show any cross-peaks in the spectrum, presumably due to the dynamics of the N-terminus of the Aβ42 fibril. On the other hand, when two adjacent residues had close 13Cα resonances, the cross- and diagonal-peaks overlapped with each other, making it difficult to separate the exact 13Cα chemical shifts. Thus, we also obtained (H)CANH and (H)CA(CO)NH 3D data, which are traditionally used to establish the sequential connectivity via the 13Cα resonances with respect to 15N and 1HN chemical shifts.51, 66-68 Importantly, this analysis allowed us to determine the 13Cα resonances that could not be distinguished in the (H)CA(CON)CAH 3D spectrum, due to the overlap of the corresponding peaks. However, it should be noted that the traditional approach using (H)CANH and (H)CA(CO)NH 3D experiments alone was not sufficient to establish the signal assignments for the β-sheet-rich amyloid protein, which had very limited spectral dispersion in the 15N/1HN 2D spectrum (Figure S2a). Thus, the novel use of 1Hα-detected (H)CA(CON)CAH 3D data was a crucial element for the successful main-chain assignment on a very small amount of Aβ42 sample. In addition, we carried out a (H)CACONH 4D experiment to assign 13CO resonances (Figure S2b). Since the resolution of the (H)CONH 3D spectrum was not sufficient to assign the signals, we decided to record the 4D data instead. In addition, we also acquired a (H)CX(CA)NH 3D spectrum, which correlates the aliphatic 13C signals with the 15N and 1HN resonances. These data are useful for confirming the accuracy of the sequential connectivity obtained from the analysis of the (H)CANH and (H)CA(CO)NH 3D experiments, because the pattern of the side-chain 13C resonances enables us to assign the peaks with similar 13Cα chemical shifts to their amino acid type. However, the signals of some of the side chains were not detected in the (H)CX(CA)NH 3D spectrum. This may be due to the higher degree of mobility of those side chains. Details on the side-chain assignments can be found in the Figure S3.
Figure 4.
(a) Representative strip plots of (H)CA(CON)CAH (green), (H)CANH (red), and (H)CA(CO)NH (blue) 3D spectra, showing the sequential connectivity from Asn-27 to Met-35. Diagonal peaks corresponding to intra-residue magnetization transfer in the (H)CA(CON)CAH 3D spectrum are marked with red crosses. (b) Graphical representation of successful chemical shift assignments for 13C (green), 15N (blue), and 1H (yellow) resonances in this study. Side-chain 1H and 15N are omitted. Gray squares denote unassigned resonances.
Signal assignments based on a set of the 3D data were performed with the semi-automated assignment program MAGRO NMRView.70 The 13CO assignments were made manually by analyzing the 4D data using the NMRFAM-SPARKY software.71 Chemical shifts obtained by semi-automated methods were manually validated with the 4D data. The assignments were further validated by HIGHLIGHT REDOR experiments on Val-reverse 13C- and 15N-labeled Aβ42 fibril sample (see Supporting Information).51 The completeness of the signal assignment for the Aβ42 fibril is summarized in Figure 4b. Without any deuterated samples, we were able to assign most of the backbone resonances (86% of 13Cα, 76% of 15N, 71% of 13CO, 85% of 1Hα, and 76% of 1HN) and also the majority of the side-chain 13C and 1H resonances (69% of 13C and 54% of 1H). In the N-terminal residues 1–9, the 1HN and 15N signals were largely missing or weak, presumably reflecting the highly dynamic nature of this region. The average 1HN line width was ~0.8 ppm, which is considerably broader than the corresponding line width of 0.2 ppm observed for GB1 microcrystals in the same conditions.51 Nevertheless, our approach enabled the first successful assignment of both protein backbone and side-chain signals from a trace amount (~200 μg, instead of several milligrams) of Aβ42 fibril.
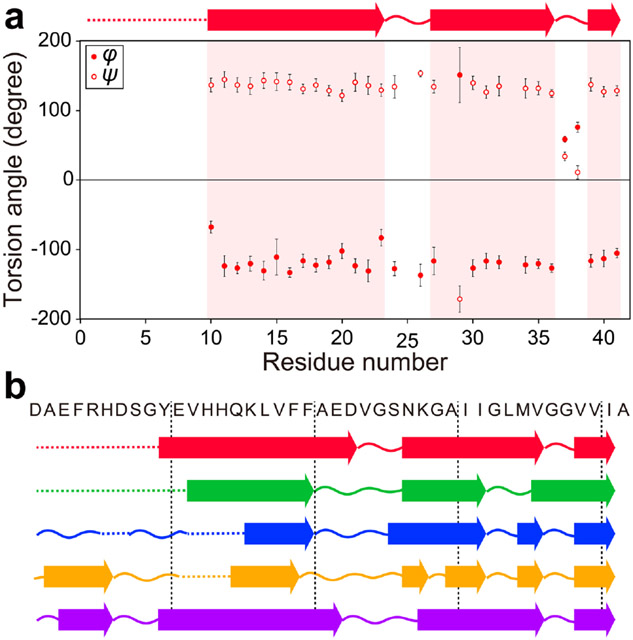
Based on the assigned chemical shifts of the Aβ42 peptide, torsion angles were calculated using the TALOS-N software (Table S6).69 The results indicate that the present Aβ42 fibril is composed of three β-strands spanning the Tyr-10–Asp-23, Asn-27–Vla-36 and Val-39–Ile-41 residues (Figure 5a). Thus, we compared the locations of the β-strands observed in this work with those previously reported for Aβ42 fibril structures (Figure 5b). The present locations of the β-strands are markedly different from those reported in previous studies for S-shaped fibril structures.27-29 This indicates that the Aβ42 fibril examined here possesses a different conformation, compared with Aβ42 fibrils with S-shaped structure. However, the locations of the β-strands obtained in this work show some similarity to the pattern of the β-strands in the fibrils prepared at low pH by Gremer et al.14 Therefore, we further compared chemical shifts of our Aβ42 fibril with those in the previous studies, to investigate any conformational similarities.
Figure 5.
(a) Torsion angles of Aβ42 fibril obtained from TALOS-N analysis.69 Predicted β-strands are shown as arrows on the top of the figure. (b) Comparison of the locations of the β-strands obtained in this study with those of previously reported Aβ42 fibrils. Red arrows represent β-strands predicted in this study. Green, blue, orange, and purple arrows correspond to the β-strands reported by Xiao et al.,27 Colvin et al.,28, 58 Ravotti et al.,29, 59 and Gremer et al.,14 respectively. Curved lines represent non-β-strand regions. Dotted lines represent the regions where there are no predicted secondary structures due to the lack of SSNMR data.
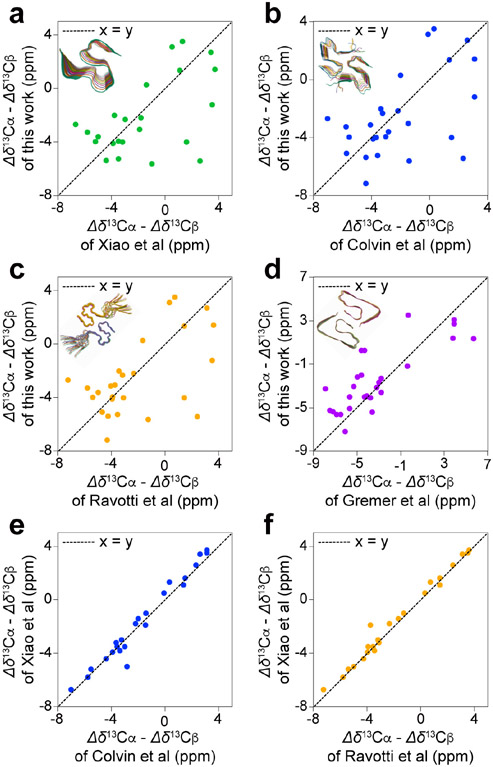
More significant conformational differences emerged when we compared the 13C secondary chemical shifts of the present Aβ42 fibril with those reported in the previous studies mentioned above. Figure 6a-d show the difference between the 13Cα and 13Cβ secondary chemical shifts of the Aβ42 fibril in this study and those of the fibrils reported in other studies for the residues Leu-17–Ala-42. The 13C secondary chemical shifts obtained here are largely inconsistent with those reported in previous studies, with R2 values of (a) 0.29, (b) 0.30, (c) 0.32, and (d) 0.64 denoting poor linear fitting, as shown in Figure 6a-d. For all comparisons, 55%–70% of the residues showed a deviation in the secondary chemical shifts exceeding 1.0 ppm. Similar comparisons of the differences between the 13Cα and 13Cβ secondary chemical shifts of Aβ42 fibrils reported by Xiao et al.,27 Colvin et al.,28, 58 and Ravotti et al.29, 59 show nearly perfect linear fittings, with R2 values of 0.96 and 0.98 for the same residues, as shown in Figure 6e and f, respectively. These three groups reported essentially identical S-shaped structures for Aβ42 fibrils prepared at a neutral pH (pH 7.4–8),27-29, 58-59 as shown in the insets of Figure 6a-c. The 13C chemical shift of the side-chain carboxyl group at Asp-23 (179.3 ppm in the DSS reference) indicated the non-protonated form,72 which is consistent with the neutral pH. These results suggest that the present study involves a conformationally distinct novel polymorph of the Aβ42 fibril, prepared at a physiologically relevant neutral pH. We also confirmed that the 13C chemical shifts of the novel polymorph are notably different from the corresponding chemical shifts for any of the four forms of our brain-derived Aβ42 fibrils (Table S2). At this point, the origin of the different structure of the fibril examined in this work remains unclear.
Figure 6.
Comparison of the 13C secondary chemical shifts obtained in the present work with previous studies of Aβ42 fibrils. The difference between the 13Cα and 13Cβ secondary chemical shifts of the Aβ42 fibril obtained in this study was plotted against that of the fibrils reported by (a) Xiao et al.,27 (b) Colvin et al.,28, 58 (c) Ravotti et al.,29, 59 and (d) Gremer et al.14 The reported structures of the fibrils are shown in the insets; their PDB identifiers are (a) 2MXU , (b) 5KK3, (c) 2NAO and (d) 5OQV. For comparison, the difference between the 13Cα and 13Cβ secondary chemical shifts of the Aβ42 fibril of Xiao et al.27 was plotted against those reported by (e) Colvin et al.28, 58 and (f) Ravotti et al.29, 59 The corresponding plots show excellent linearity, suggesting that the Aβ42 structures of these studies are very similar to each other.
Conclusion
In this study, we demonstrated for the first time that 1H-detected SSNMR is an effective tool for characterizing the structural differences between synthetic and brain-derived Aβ42 fibrils for the samples in a range of nano- to pico-moles. Despite the limited sample amounts available, especially in the case of brain-derived Aβ fibrils, we were able to acquire 2D SSNMR spectra with sufficient S/N ratios within a reasonable experimental time (33 sec to 8.7 min), allowing us to identify the main structural differences between these samples. Our SSNMR data clearly indicate that Aβ42 fibrils in AD brain tissues show different structural features from their synthetic counterparts. Four sets of the cross peaks were identified by 1H-detected 2D–3D SSNMR, suggesting the presence of polymorphs for the brain-derived Aβ42 fibrils. Moreover, the comparison of our SSNMR data for the brain-derived Aβ42 fibrils with those of previously reported Aβ42 and Aβ40 fibrils highlights substantial structural differences.
Furthermore, we report the first (to our knowledge) spectral assignments and structural characterization of fully-protonated Aβ42 fibril using sensitivity-enhanced 1H-detected SSNMR approach under UFMAS. Owing to the excellent sensitivity achieved by the 1H-detection, the analysis could be performed using trace amounts of sample, compared with the larger amounts required by conventional 13C-detected SSNMR. The morphological and conformational homogeneity of the Aβ42 fibril prepared using a seeding protocol allowed us to analyze the sample using 1H-detected SSNMR. Since the resolution of the SSNMR spectra acquired under UFMAS is comparable to that of previous studies, a similar approach could be used for the chemical shift assignment. A combination of 1H-detected 3D and 4D SSNMR spectra was used to perform the assignment of the 13C, 15N, and 1H chemical shifts of both the protein backbone and side chains. In addition, we used a 13C/13C correlation 2D spectrum to assign the 13CO resonance of Ala-42, and some of the aromatic and carbonyl side-chain 13C chemical shifts. In the structured region of the Aβ42 peptide (Tyr-10–Ala-42), our analysis successfully assigned 100% of the 13Cα and 1Hα, 97% of the 15N and 1HN, and 91% of the 13CO resonances of the protein backbone, along with 81% of the 13C and 59% of the 1H resonances of the side chains, within an experimental time of 12.7 days and using only ~200 μg (~42 nmol) of the fibril sample.
We also identified the secondary structural elements in the Aβ42 fibril. The torsion angles calculated by the TALOS-N software69 predicted three β-strands in Tyr-10–Asp-23, Asn-27–Val-36 and Val-39–Ile-41 residues. The comparison of the locations of the β-strands and 13C secondary chemical shifts clearly indicated that the Aβ42 fibril examined in this study is a new polymorph with a different structure from those of previously reported Aβ42 fibril species. The way in which structural differences affect the biological function of the Aβ42 fibril is currently unclear, and further investigations are needed to clarify this aspect. It should be also noted that seeding efficiency of brain-derived amyloid is likely to vary from patients to patients and that the presented data could be a favorable case. Nevertheless, this study demonstrated high propensity of Aβ42 to form multiple forms of fibrils in both in vitro and in vivo (i.e. brain), which was not obvious from the previous studies. We believe that our study can open a new avenue for the analysis of trace amounts of biological systems such as amyloid fibrils, oligomers, and membrane proteins, for which 13C-detected SSNMR might be ineffective.
Materials and Methods
Sample Preparation
The protocol for the preparation of a synthetic Aβ42 fibril sample uniformly 13C- and 15N-labeled at Phe-20, Ala-21, Val-24, Gly-25, and Leu-34 residues can be found in ref. 62. The preparation of the brain-derived Aβ42 fibril sample, which was uniformly 13C- and 15N-labeled at the same residues, is described here. Tissue containing Aβ from the temporal lobe of the AD brain was extracted from a patient (87 year old male) diagnosed with AD and cerebral amyloid angiopathy at The University of Chicago, following the protocol described in ref. 73. The brain tissue material (0.029 mg) was suspended in 70 μL of 10 mM phosphate buffer (pH 7.4) at a concentration of 0.42 mg/mL. This tissue-material sample contained ~3.1 ng/μg and ~0.48 ng/μg of Aβ42 and Aβ40, respectively; the results were suggested by quantitative mass spectroscopy of the C-terminal peptides (VGGVV and VGGVVIA) after cleavage of the Aβ sequence beyond Met-35 with CNBr. To truncate the fibrils and obtain more amyloid seeds, the suspension was sonicated on ice at 65% amplitude of 200 W with 40% duty factor in a 20 sec cycle; the cycle was repeated for 10 min in total with a 5 min rest in the middle. The seeds were introduced into a freshly prepared monomeric Aβ42 solution containing 1 mg of Aβ42 at 40.7 μM in a 10 mM phosphate buffer (pH 7.4). The brain-tissue templated Aβ42 was fibrillized under gentle circular agitation for 2 days at room temperature. Despite the small amount of Aβ seed from the tissues (~0.01%) in the sample, the effectiveness of the seeding was confirmed by thioflavin-T fluorescence (see Figure S5 in SI). The brain-tissue templated fibril sample was pelleted and lyophilized following the same protocol in ref. 27, 62. The use of the human-tissue derived sample for this research was approved by the Tokyo Institute of Technology Human Subjects Research Ethics Review Committee (#2019045, #2020061) and RIKEN Research Ethics Committee (H30-16(2)).
The uniformly 13C- and 15N-labeled Aβ42 fibril sample was prepared as follows. The Aβ42-expression construct was prepared as described in ref. 64. Briefly, the genes for Aβ42 were cloned into the pGEX-2T vector (Sigma) at the BamHI and EcoRI sites, using the N-terminal GST tag and factor Xa cleavage site (IEGR) encoded in the vector. The fusion protein GST-IEGR-Aβ42 was expressed in the BL21-CodonPlus (DE3) bacterial strain. The isotopically labeled protein was expressed in the M9 minimal media containing 5 g/L of 13C6-glucose and 1 g/L of 15N-NH4Cl, with overnight IPTG induction at 27 °C. Then, the cell pellet was collected and the fusion protein was purified by first lysing the cells in a 25 mM Tris-HCl buffer (pH 8.4) containing 0.1% (v/v) of Triton X-100, 0.05% (w/v) of deoxycholic acid, and sodium salt, using a sonic dismembrator (84 W amplitude, 12 cycles of a 10 sec pulse in each minute). The cell debris containing the inclusion bodies were pelleted at 27,000 rpm for 10 min at 4 °C, and then washed in 25 mM Tris-HCl buffer (pH 8.4) containing 0.2% (w/v) of N-lauroylsarcosine (NLS). Eight additional cycles of sonication were incorporated during the wash step followed by centrifugation as described above. The GST-IEGR-Aβ42 fusion protein was dissolved by vortexing in the 25 mM Tris-HCl buffers (pH 8.4) containing 2% of NLS (NLS amount was in a range between 1% to 5% (w/v)). The vortex and centrifugation steps were repeated for multiple rounds until most of the fusion protein was collected in the supernatant. The supernatants containing the GST-IEGR-Aβ42 fusion protein were pooled together and filtered through a 0.8-μm syringe filter.
The fusion protein was cleaved by bovine factor Xa (30–80 units of enzyme per milligram of fusion protein) in 25 mM Tris-HCl (pH 8.4), 0.2 mM CaCl2, and 0.35% NLS, for 16 h at 12 °C quiescently. The cleavage mixture was filtered through a 0.22-μm syringe, and then subjected to gel-filtration by a G-25 column in 10 mM Tris base (pH 10), to desalt the NLS molecules. In the eluate, most of the uncleaved fusion protein and the GST-IEGR tag in dimeric forms were trapped in a spin concentrator (50 kDa molecular weight cutoff, Amicon Ultra-15 UFC905096, EMD Millipore) after centrifuging at 4.7×103 g for 12 min at 4 °C. The flow-through from the centrifugation contained the crude Aβ42 peptide. Subsequent purification and seeded fibrillization were conducted in the same manner as described in ref. 27, 62, except for the injection part for HPLC. The flow-through solution (10 mL) was injected for HPLC purification, and the column was washed at 4 mL/min with mobile phase containing 5% acetonitrile to remove salts for 7 min. Then, we started HPLC purification with acetonitrile gradient, as described in the references. The purity of Aβ42 (> 95%) was verified by MALDI TOF/TOF mass spectroscopy. Briefly, the lyophilized peptide after HPLC purification was weighed and then completely dissolved at 2 mg/mL in an aqueous solution containing 30% acetonitrile (Fisher Scientific) and 0.1% trifluoroacetic acid (TFA; American Bioanalytical) at 4 °C; the solution was subsequently lyophilized again. The lyophilized peptides were stored with drying reagents in a freezer at −20 °C. Before each incubation, the peptide was warmed to room temperature and dissolved in hexafluoroisopropanol (HFIP) (Sigma-Aldrich) at a concentration of ~2 mg/mL; after 1 h, the solution was subsequently lyophilized. This dissolution-lyophilization cycle was repeated twice, according to the previously published protocol. The HFIP-treated peptide was first dissolved in a 10 mM NaOH solution (Fisher Scientific) to 0.6 mM, and then the Aβ solution was diluted to 60 μM at pH 7.4 with a 10 mM phosphate buffer. The fresh Aβ42 peptide solution was filtered by centrifugation with a 50-kDa molecular-mass-cutoff filter (EMD Millipore Amicon Ultra-15 filter, UFC905096) at 4.8 × 103 g for 3 min in order to remove any undissolved peptide or preformed aggregates. The final Aβ monomer concentration was typically ~40 μM. The uniformly 13C- and 15N-labeled Aβ42 fibril was obtained after three consecutive seeding steps. The first two seeding steps were performed with the unlabeled recombinant Aβ42, and the last one with the 13C- and 15N-labeled recombinant Aβ42. The final fibril sample was pelleted by centrifugation; after the buffer was removed, the pelleted sample was lyophilized following the same protocol in ref. 27, 62.
Val-reverse 13C- and 15N-labeled Aβ42 fibril sample was prepared following a similar protocol used to prepare uniformly 13C- and 15N-labeled Aβ42 fibril with the following additional step; before the IPTG induction, 1.25 mM of unlabeled L-Val was added to the M9 medium.51
For SSNMR experiments, unless otherwise mentioned, the prepared lyophilized Aβ42 fibril samples (~200 μg) were first packed into 0.75-mm JEOL SSNMR MAS rotors, and then ~0.5 μL of 200 mM Cu-EDTA solution was added for paramagnetic doping and incubated for 3 days. For SSNMR experiments of Val-reverse 13C- and 15N-labeled Aβ42 fibril, for which all the residues except for Val and some scrambled Leu were unformly 13C- and 15N-labeled, a sample was packed into a 1-mm (~500 μg) JEOL SSNMR MAS rotor and doped with 200 mM Cu-EDTA solution (~0.5 μL).
SSNMR Experiments
Unless otherwise mentioned, all the SSNMR experiments in this study were recorded at a MAS rate of 90 kHz on a Bruker Avance III 800 MHz NMR spectrometer at RIKEN NMR Facility using a JEOL 0.75-mm 1H/13C/15N/2H quad-resonance probe. During the SSNMR experiments, the temperature of the sample was maintained at ~30 °C by the application of the cooling N2 gas at a flow rate of 400 L/h from a Bruker BCU II unit set to the strong cooling power. SSNMR data for Val-reverse 13C- and 15N-labeled Aβ42 fibril were collected at a MAS rate of 80 kHz on a Bruker Avance III 750 MHz spectrometer at the UIC Center for Structural Biology using a JEOL 1-mm 1H/13C/15N/2H quad-resonance MAS probe while maintaining the sample temperature at ~30 °C by the application of cooling N2 gas at −20 °C. Unless otherwise mentioned, all the SSNMR data were recorded under 1H-decoupling at 10 kHz using WALTZ-16 decoupling during the evolution periods.74 To eliminate 13C-15N J-coupling, a 13C or 15N π-pulse was applied at the middle of the 15N or 13C evolution periods, respectively. During the 1H acquisition, 10 kHz and 5 kHz WALTZ-16 decoupling was used on 13C and 15N channels, respectively. For Val-reverse 13C- and 15N-labeled Aβ42 sample, 1H-decoupling during t1 and t2 evolution was set to 10 kHz using SPINAL-64 scheme. Other experimental parameters used in the SSNMR measurements are summarized in Tables S7-9. All multidimensional SSNMR spectra were processed using the NMRPipe software.75 The 1H and 13C/15N dimensions of the time-domain data were apodized with 45°- and 60°-shifted sine-bell window functions, respectively. All the chemical shifts presented in this work were calibrated based on the DSS standard for 1H and 13C and liquid ammonia for 15N. Secondary chemical shifts were determined based on the random coil chemical shifts given in ref. 76.
Assignment Protocol
Site-specific chemical shift assignments of the 3D SSNMR data collected on uniformly 13C- and 15N-labeled Aβ42 fibril were mainly performed via the MAGRO-NMRView automated assignment software, which is an upgraded version of Kujira, a package of integrated tools for NMR analysis.70 The NMRFAM-SPARKY software71 was used for assignments of 4D SSNMR (H)CACONH data, which were performed manually to assign 13CO resonances. Since the 4D data had a better resolution than (H)CA(CO)NH 3D spectrum, a few resonance assignments from the 3D data that were inconsistent with the 4D data were corrected manually. The chemical shift data for the uniformly 13C- and 15N-labeled Aβ42 fibril were deposited in Biological Magnetic Resonance Bank (BMRB; Entry ID: 26307). Additionally, a comparison of the observed 13Cα, 15N and 1HN resonances with a Val-reverse 13C- and 15N-labeled Aβ42 fibril sample was performed to confirm the accuracy of the assignment (see Supporting Information).
Supplementary Material
ACKNOWLEDGMENT
The structural analysis of the Aβ42 fibrils was mainly supported by a NIH U01 grant (5U01GM098033) to Y. I. The development of the SSNMR analysis for trace amyloid samples was supported by the JST-Mirai Program (grant No. JPMJMI17A2, Japan) to Y. I. The development of high-dimensional SSNMR methods was supported by a JSPS KAKENHI grant (No. JP15K21772, Japan) to Y. I. The preparation of the brain-derived sample was also supported by NIH R01AG048793 (S.C.M.), Alzheimer’s Association Zenith Fellowship Award (S.C.M.), and the Medical Scientist Training Program Grant T32 GM07281 (K.P.S.). The authors thank Prof. Vipin Agarwal at TIFR Hyderabad for introducing CANH TOBSY 3D experiment to them. The authors also thank the RIKEN NMR facility in Yokohama, Japan and its staff members.
Footnotes
Supporting Information
Supporting Information including additional SSNMR data is available free of charge on the ACS Publications website.
Tables showing assigned chemical shifts, experimental parameters used in the measurement of the SSNMR spectra, comparisons of chemical shifts, additional SSNMR spectra, and ThT fluorescence data (PDF).
References
- 1.Finder VH, Alzheimer's Disease: A General Introduction and Pathomechanism. Journal of Alzheimer's Disease 2010, 22, S5–S19. [DOI] [PubMed] [Google Scholar]
- 2.Caraci F; Copani A; Nicoletti F; Drago F, Depression and Alzheimer's disease: Neurobiological links and common pharmacological targets. European Journal of Pharmacology 2010, 626 (1), 64–71. [DOI] [PubMed] [Google Scholar]
- 3.Goedert M; Crowther RA, Amyloid plaques, neurofibrillary tangles and their relevance for the study of Alzheimer's disease. Neurobiol. Aging 1989, 10 (5), 405–406. [DOI] [PubMed] [Google Scholar]
- 4.Brion JP, Neurofibrillary Tangles and Alzheimer’s Disease. European Neurology 1998, 40 (3), 130–140. [DOI] [PubMed] [Google Scholar]
- 5.Cras P; Kawai M; Lowery D; Gonzalez-DeWhitt P; Greenberg B; Perry G, Senile plaque neurites in Alzheimer disease accumulate amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A 1991, 88 (17), 7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta PD; Pirttilä T; Mehta SP; Sersen EA; Aisen PS; Wisniewski HM, Plasma and Cerebrospinal Fluid Levels of Amyloid β Proteins 1-40 and 1-42 in Alzheimer Disease. Arch. Neurol 2000, 57 (1), 100–105. [DOI] [PubMed] [Google Scholar]
- 7.Gravina SA; Ho L; Eckman CB; Long KE; Otvos L; Younkin LH; Suzuki N; Younkin SG, Amyloid β Protein (Aβ) in Alzheimeri's Disease Brain: Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Aβ40 OR Aβ42(43). J. Biol. Chem 1995, 270 (13), 7013–7016. [DOI] [PubMed] [Google Scholar]
- 8.Selkoe DJ, Cell biology of protein misfolding: The examples of Alzheimer's and Parkinson's diseases. Nat. Cell. Biol 2004, 6 (11), 1054–1061. [DOI] [PubMed] [Google Scholar]
- 9.Meisl G; Yang X; Hellstrand E; Frohm B; Kirkegaard JB; Cohen SIA; Dobson CM; Linse S; Knowles TPJ, Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (26), 9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwatsubo T; Odaka A; Suzuki N; Mizusawa H; Nukina N; Ihara Y, Visualization of Aβ42(43) and Aβ40 in senile plaques with end-specific Aβ monoclonals: Evidence that an initially deposited species is Aβ42(43). Neuron 1994, 13 (1), 45–53. [DOI] [PubMed] [Google Scholar]
- 11.Burdick D; Kosmoski J; Knauer MF; Glabe CG, Preferential adsorption, internalization and resistance to degradation of the major isoform of the Alzheimer's amyloid peptide, Aβ1–42, in differentiated PC12 cells. Brain Research 1997, 746 (1), 275–284. [DOI] [PubMed] [Google Scholar]
- 12.Roher AE; Lowenson JD; Clarke S; Woods AS; Cotter RJ; Gowing E; Ball MJ, beta-Amyloid-(1-42) is a major component of cerebrovascular amyloid deposits: implications for the pathology of Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A 1993, 90 (22), 10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarrett JT; Berger EP; Lansbury PT, The carboxy terminus of the .beta. amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer's disease. Biochemistry 1993, 32 (18), 4693–4697. [DOI] [PubMed] [Google Scholar]
- 14.Gremer L; Schölzel D; Schenk C; Reinartz E; Labahn J; Ravelli RBG; Tusche M; Lopez-Iglesias C; Hoyer W; Heise H; Willbold D; Schröder GF, Fibril structure of amyloid-β(1–42) by cryo–electron microscopy. Science 2017, 358 (6359), 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kollmer M; Close W; Funk L; Rasmussen J; Bsoul A; Schierhorn A; Schmidt M; Sigurdson CJ; Jucker M; Fändrich M, Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nature Communications 2019, 10 (1), 4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh U; Thurber KR; Yau W-M; Tycko R, Molecular structure of a prevalent amyloid-β fibril polymorph from Alzheimer9s disease brain tissue. Proc. Natl. Acad. Sci. U. S. A 2021, 118 (4), e2023089118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tycko R., Solid-state NMR as a probe of amyloid structure. Protein and peptide letters 2006, 13 (3), 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petkova AT; Ishii Y; Balbach JJ; Antzutkin ON; Leapman RD; Delaglio F; Tycko R, A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U. S. A 2002, 99 (26), 16742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petkova AT; Leapman RD; Guo Z; Yau W-M; Mattson MP; Tycko R, Self-Propagating, Molecular-Level Polymorphism in Alzheimer's ß-Amyloid Fibrils. Science 2005, 307 (5707), 262. [DOI] [PubMed] [Google Scholar]
- 20.Petkova AT; Yau W-M; Tycko R, Experimental Constraints on Quaternary Structure in Alzheimer's β-Amyloid Fibrils. Biochemistry 2006, 45 (2), 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paravastu AK; Leapman RD; Yau W-M; Tycko R, Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (47), 18349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertini I; Gonnelli L; Luchinat C; Mao J; Nesi A, A New Structural Model of Aβ40 Fibrils. J. Am. Chem. Soc 2011, 133 (40), 16013–16022. [DOI] [PubMed] [Google Scholar]
- 23.Lopez del Amo JM; Schmidt M; Fink U; Dasari M; Fändrich M; Reif B, An Asymmetric Dimer as the Basic Subunit in Alzheimer’s Disease Amyloid β Fibrils. Angewandte Chemie International Edition 2012, 51 (25), 6136–6139. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z-W; Vugmeyster L; Au DF; Ostrovsky D; Sun Y; Qiang W, Molecular structure of an N-terminal phosphorylated β-amyloid fibril. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (23), 11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J-X; Qiang W; Yau W-M; Schwieters Charles D.; Meredith Stephen C.; Tycko R, Molecular Structure of β-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell 2013, 154 (6), 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paravastu AK; Qahwash I; Leapman RD; Meredith SC; Tycko R, Seeded growth of β-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (18), 7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Y; Ma B; McElheny D; Parthasarathy S; Long F; Hoshi M; Nussinov R; Ishii Y, Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease. Nat. Struct. Mol. Biol 2015, 22 (6), 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colvin MT; Silvers R; Ni QZ; Can TV; Sergeyev I; Rosay M; Donovan KJ; Michael B; Wall J; Linse S; Griffin RG, Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J. Am. Chem. Soc 2016, 138 (30), 9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wälti MA; Ravotti F; Arai H; Glabe CG; Wall JS; Böckmann A; Güntert P; Meier BH; Riek R, Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (34), E4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schütz AK; Vagt T; Huber M; Ovchinnikova OY; Cadalbert R; Wall J; Güntert P; Böckmann A; Glockshuber R; Meier BH, Atomic-Resolution Three-Dimensional Structure of Amyloid β Fibrils Bearing the Osaka Mutation. Angewandte Chemie International Edition 2015, 54 (1), 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber M; Ovchinnikova OY; Schütz AK; Glockshuber R; Meier BH; Böckmann A, Solid-state NMR sequential assignment of Osaka-mutant amyloid-beta (Aβ1–40 E22Δ) fibrils. Biomolecular NMR Assignments 2015, 9 (1), 7–14. [DOI] [PubMed] [Google Scholar]
- 32.Qiang W; Yau W-M; Lu J-X; Collinge J; Tycko R, Structural variation in amyloid-β fibrils from Alzheimer's disease clinical subtypes. Nature 2017, 541 (7636), 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siemer AB; Arnold AA; Ritter C; Westfeld T; Ernst M; Riek R; Meier BH, Observation of Highly Flexible Residues in Amyloid Fibrils of the HET-s Prion. J. Am. Chem. Soc 2006, 128 (40), 13224–13228. [DOI] [PubMed] [Google Scholar]
- 34.Wasmer C; Lange A; Van Melckebeke H; Siemer AB; Riek R; Meier BH, Amyloid Fibrils of the HET-s(218–289) Prion Form a β Solenoid with a Triangular Hydrophobic Core. Science 2008, 319 (5869), 1523. [DOI] [PubMed] [Google Scholar]
- 35.Van Melckebeke H; Wasmer C; Lange A; Ab E; Loquet A; Böckmann A; Meier BH, Atomic-Resolution Three-Dimensional Structure of HET-s(218–289) Amyloid Fibrils by Solid-State NMR Spectroscopy. J. Am. Chem. Soc 2010, 132 (39), 13765–13775. [DOI] [PubMed] [Google Scholar]
- 36.Helmus JJ; Surewicz K; Nadaud PS; Surewicz WK; Jaroniec CP, Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (17), 6284–6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuttle MD; Comellas G; Nieuwkoop AJ; Covell DJ; Berthold DA; Kloepper KD; Courtney JM; Kim JK; Barclay AM; Kendall A; Wan W; Stubbs G; Schwieters CD; Lee VMY; George JM; Rienstra CM, Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat. Struct. Mol. Biol 2016, 23 (5), 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin HK; Boatz JC; Krabbendam IE; Kodali R; Hou ZP; Wetzel R; Dolga AM; Poirier MA; van der Wel PCA, Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nature Communications 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boatz JC; Whitley MJ; Li MY; Gronenborn AM; van der Wel PCA, Cataract-associated P23T gamma D-crystallin retains a native-like fold in amorphous-looking aggregates formed at physiological pH. Nature Communications 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee MC; Yu WC; Shih YH; Chen CY; Guo ZH; Huang SJ; Chan JCC; Chen YR, Zinc ion rapidly induces toxic, off-pathway amyloid-beta oligomers distinct from amyloid-beta derived diffusible ligands in Alzheimer's disease. Scientific Reports 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theint T; Xia YJ; Nadaud PS; Mukhopadhyay D; Schwieters CD; Surewicz K; Surewicz WK; Jaroniec CP, Structural Studies of Amyloid Fibrils by Paramagnetic Solid-State Nuclear Magnetic Resonance Spectroscopy. J. Am. Chem. Soc 2018, 140 (41), 13161–13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuo XF; Wang J; Zhang J; Jiang LL; Hu HY; Lu JX, Solid-State NMR Reveals the Structural Transformation of the TDP-43 Amyloidogenic Region upon Fibrillation. J. Am. Chem. Soc 2020, 142 (7), 3412–3421. [DOI] [PubMed] [Google Scholar]
- 43.Wu XL; Hu H; Dong XQ; Zhang J; Wang J; Schwieters CD; Liu J; Wu GX; Li B; Lin JY; Wang HY; Lu JX, The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis. Nature Communications 2021, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiang W; Yau W-M; Luo Y; Mattson MP; Tycko R, Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (12), 4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elkins MR; Wang T; Nick M; Jo H; Lemmin T; Prusiner SB; DeGrado WF; Stöhr J; Hong M, Structural Polymorphism of Alzheimer’s β-Amyloid Fibrils as Controlled by an E22 Switch: A Solid-State NMR Study. J. Am. Chem. Soc 2016, 138 (31), 9840–9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fändrich M; Meinhardt J; Grigorieff N, Structural polymorphism of Alzheimer Aβ and other amyloid fibrils. Prion 2009, 3 (2), 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishii Y; Wickramasinghe A; Matsuda I; Endo Y; Ishii Y; Nishiyama Y; Nemoto T; Kamihara T, Progress in proton-detected solid-state NMR (SSNMR): Super-fast 2D SSNMR collection for nano-mole-scale proteins. J. Magn. Reson 2018, 286, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishii Y; Tycko R, Sensitivity enhancement in solid state 15N NMR by indirect detection with high-speed magic angle spinning. J. Magn. Reson 2000, 142, 199–204. [DOI] [PubMed] [Google Scholar]
- 49.Ishii Y; Yesinowski JP; Tycko R, Sensitivity enhancement in solid-state C-13 NMR of synthetic polymers and biopolymers by H-1 NMR detection with high-speed magic angle spinning. J. Am. Chem. Soc 2001, 123 (12), 2921–2922. [DOI] [PubMed] [Google Scholar]
- 50.Bertini I; Emsley L; Lelli M; Luchinat C; Mao J; Pintacuda G, Ultrafast MAS Solid-State NMR Permits Extensive C-13 and H-1 Detection in Paramagnetic Metalloproteins. J. Am. Chem. Soc 2010, 132 (16), 5558-+. [DOI] [PubMed] [Google Scholar]
- 51.Wang S; Parthasarathy S; Xiao Y; Nishiyama Y; Long F; Matsuda I; Endo Y; Nemoto T; Yamauchi K; Asakura T; Takeda M; Terauchi T; Kainosho M; Ishii Y, Nano-mole scale sequential signal assignment by 1H-detected protein solid-state NMR. Chem. Commun 2015, 51 (81), 15055–15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwal V; Penzel S; Szekely K; Cadalbert R; Testori E; Oss A; Past J; Samoson A; Ernst M; Böckmann A; Meier BH, De Novo 3D Structure Determination from Sub-milligram Protein Samples by Solid-State 100 kHz MAS NMR Spectroscopy. Angewandte Chemie International Edition 2014, 53 (45), 12253–12256. [DOI] [PubMed] [Google Scholar]
- 53.Wang S; Parthasarathy S; Nishiyama Y; Endo Y; Nemoto T; Yamauchi K; Asakura T; Takeda M; Terauchi T; Kainosho M; Ishii Y, Nano-mole scale side-chain signal assignment by 1H-detected protein solid-state NMR by ultra-fast magic-angle spinning and stereo-array isotope labeling. PloS one 2015, 10 (4), e0122714–e0122714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andreas LB; Jaudzems K; Stanek J; Lalli D; Bertarello A; Le Marchand T; Cala-De Paepe D; Kotelovica S; Akopjana I; Knott B; Wegner S; Engelke F; Lesage A; Emsley L; Tars K; Herrmann T; Pintacuda G, Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (33), 9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanek J; Andreas LB; Jaudzems K; Cala D; Lalli D; Bertarello A; Schubeis T; Akopjana I; Kotelovica S; Tars K; Pica A; Leone S; Picone D; Xu Z-Q; Dixon NE; Martinez D; Berbon M; El Mammeri N; Noubhani A; Saupe S; Habenstein B; Loquet A; Pintacuda G, NMR Spectroscopic Assignment of Backbone and Side-Chain Protons in Fully Protonated Proteins: Microcrystals, Sedimented Assemblies, and Amyloid Fibrils. Angewandte Chemie International Edition 2016, 55 (50), 15504–15509. [DOI] [PubMed] [Google Scholar]
- 56.Stanek J; Schubeis T; Paluch P; Güntert P; Andreas LB; Pintacuda G, Automated Backbone NMR Resonance Assignment of Large Proteins Using Redundant Linking from a Single Simultaneous Acquisition. J. Am. Chem. Soc 2020, 142 (12), 5793–5799. [DOI] [PubMed] [Google Scholar]
- 57.Wickramasinghe NP; Parthasarathy S; Jones CR; Bhardwaj C; Long F; Kotecha M; Mehboob S; Fung LWM; Past J; Samoson A; Ishii Y, Nanomole-scale protein solid-state NMR by breaking intrinsic 1H T1 boundaries. Nature Methods 2009, 6 (3), 215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colvin MT; Silvers R; Frohm B; Su Y; Linse S; Griffin RG, High Resolution Structural Characterization of Aβ42 Amyloid Fibrils by Magic Angle Spinning NMR. J. Am. Chem. Soc 2015, 137 (23), 7509–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravotti F; Wälti MA; Güntert P; Riek R; Böckmann A; Meier BH, Solid-state NMR sequential assignment of an Amyloid-β(1–42) fibril polymorph. Biomolecular NMR Assignments 2016, 10 (2), 269–276. [DOI] [PubMed] [Google Scholar]
- 60.Antzutkin ON; Leapman RD; Balbach JJ; Tycko R, Supramolecular Structural Constraints on Alzheimer's β-Amyloid Fibrils from Electron Microscopy and Solid-State Nuclear Magnetic Resonance. Biochemistry 2002, 41 (51), 15436–15450. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh U; Yau W-M; Tycko R, Coexisting order and disorder within a common 40-residue amyloid-β fibril structure in Alzheimer's disease brain tissue. Chem. Commun 2018, 54 (40), 5070–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao Y; McElheny D; Hoshi M; Ishii Y, Solid-State NMR Studies of Amyloid Materials: A Protocol to Define an Atomic Model of Aβ(1–42) in Amyloid Fibrils. In Peptide Self-Assembly: Methods and Protocols, Nilsson BL; Doran TM, Eds. Springer New York: New York, NY, 2018; pp 407–428. [DOI] [PubMed] [Google Scholar]
- 63.Sgourakis Nikolaos G.; Yau W-M; Qiang W, Modeling an In-Register, Parallel “Iowa” Aβ Fibril Structure Using Solid-State NMR Data from Labeled Samples with Rosetta. Structure 2015, 23 (1), 216–227. [DOI] [PubMed] [Google Scholar]
- 64.Long F; Cho W; Ishii Y, Expression and purification of 15N- and 13C-isotope labeled 40-residue human Alzheimer’s β-amyloid peptide for NMR-based structural analysis. Protein Expression and Purification 2011, 79 (1), 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith AA; Ravotti F; Testori E; Cadalbert R; Ernst M; Böckmann A; Meier BH, Partially-deuterated samples of HET-s(218–289) fibrils: assignment and deuterium isotope effect. J. Biomol. NMR 2017, 67 (2), 109–119. [DOI] [PubMed] [Google Scholar]
- 66.Knight MJ; Webber AL; Pell AJ; Guerry P; Barbet-Massin E; Bertini I; Felli IC; Gonnelli L; Pierattelli R; Emsley L; Lesage A; Herrmann T; Pintacuda G, Fast Resonance Assignment and Fold Determination of Human Superoxide Dismutase by High-Resolution Proton-Detected Solid-State MAS NMR Spectroscopy. Angewandte Chemie International Edition 2011, 50 (49), 11697–11701. [DOI] [PubMed] [Google Scholar]
- 67.Barbet-Massin E; Pell AJ; Retel JS; Andreas LB; Jaudzems K; Franks WT; Nieuwkoop AJ; Hiller M; Higman V; Guerry P; Bertarello A; Knight MJ; Felletti M; Le Marchand T; Kotelovica S; Akopjana I; Tars K; Stoppini M; Bellotti V; Bolognesi M; Ricagno S; Chou JJ; Griffin RG; Oschkinat H; Lesage A; Emsley L; Herrmann T; Pintacuda G, Rapid Proton-Detected NMR Assignment for Proteins with Fast Magic Angle Spinning. J. Am. Chem. Soc 2014, 136 (35), 12489–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou DH; Nieuwkoop AJ; Berthold DA; Comellas G; Sperling LJ; Tang M; Shah GJ; Brea EJ; Lemkau LR; Rienstra CM, Solid-state NMR analysis of membrane proteins and protein aggregates by proton detected spectroscopy. J. Biomol. NMR 2012, 54 (3), 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen Y; Bax A, Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56 (3), 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi N; Iwahara J; Koshiba S; Tomizawa T; Tochio N; Güntert P; Kigawa T; Yokoyama S, KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR 2007, 39 (1), 31–52. [DOI] [PubMed] [Google Scholar]
- 71.Lee W; Tonelli M; Markley JL, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31 (8), 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Platzer G; Okon M; McIntosh LP, pH-dependent random coil H-1, C-13, and N-15 chemical shifts of the ionizable amino acids: a guide for protein pK (a) measurements. J. Biomol. NMR 2014, 60 (2-3), 109–129. [DOI] [PubMed] [Google Scholar]
- 73.Scherpelz KP; Lu J-X; Tycko R; Meredith SC, Preparation of Amyloid Fibrils Seeded from Brain and Meninges. In Protein Amyloid Aggregation: Methods and Protocols, Eliezer D, Ed. Springer New York: New York, NY, 2016; pp 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickramasinghe A; Wang S; Matsuda I; Nishiyama Y; Nemoto T; Endo Y; Ishii Y, Evolution of CPMAS under fast magic-angle-spinning at 100kHz and beyond. Solid State Nucl. Magn. Reson 2015, 72, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A, NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6 (3), 277–293. [DOI] [PubMed] [Google Scholar]
- 76.Wishart DS; Bigam CG; Holm A; Hodges RS; Sykes BD, 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5 (1), 67–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.