Abstract
Targeting different pathways in combinational therapy may lead to synergistic effects with higher drug efficiency. Due to a large number of candidate drugs and the variability in the genomic landscape of the disease, conventional cell culture models have limited success. Three-dimensional (3D) cell culture platforms such as tumoroids not only provide a pathophysiological relevant condition but also allow for low-cost and high-throughput drug screening strategies. Immunostaining of targeted proteins within a tumoroid is challenging as the interior cells are difficult to access via a non-destructive method. Immunohistochemistry (IHC) is an important technique in clinical research to explore the expression of various biomarkers. IHC staining of tumoroids allows non-destructive detection of unstable proteins by direct fixation of cells at the state of tumor microenvironment (TME) context, providing two main advantages. First, the target protein can be fixed without dissociating cells and disintegration of tumoroids into a single-cell suspension. Second, staining the preserved structure of tumoroids helps identify the location of the target proteins as well as the spatial distribution throughout the tumoroid geometry. In this protocol, we describe the detailed methodology of a non-destructive IHC staining of cancer biomarkers which minimizes the manipulation of tumoroids prior to fixation by eliminating multiple centrifugations and shaking steps typically required for removing excess hydrogel and collecting tumoroids. The protocol can be used in studies involving prognostic and predictive biomarker investigations in new anti-tumor drug development strategies.
Keywords: IHC staining, In-vitro tumoroids, Spatial distribution of proteins
Method name: Immunohistochemistry (IHC) staining of in-vitro cancer cell-generated tumoroids
Graphical abstract
Specifications table
| Subject area: | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Immunohistochemistry of tumoroids |
| Name of your protocol: | Immunohistochemistry (IHC) staining of in-vitro cancer cell-generated tumoroids |
| Reagents/tools: |
|
| Experimental design: | Tumoroids were fixed with 10% neutral buffered formalin, washed with DPBS and embedded in 2% agarose solution. Agarose-embedded tumoroids were dehydrated by submerging in ethanol, followed by submerging in Xylene. Agarose blocks were then embedded in melted paraffin wax and placed in cassette for sectioning. Paraffin blocks were sliced using automatic microtome device and deparaffinized. Tumoroids sections were then antibody stained, counterstained with DAPI, and imaged using fluorescent microscopy. |
| Trial registration: | Not applicable |
| Ethics: | Not applicable |
| Value of the Protocol |
|
Description of protocol
Background
Advanced microengineering methods, such as three-dimensional (3D) cell culture platforms and tumoroids, have great potential for a better understanding of complex biological systems [1]. Cancer cell-derived tumoroids are tumor-like organoids, typically generated using microwells, that mimic the 3D evolution of in-vivo tumors. Tumoroids provide the opportunity to resemble complex cell-cell and cell-extracellular matrix (ECM) interactions [2,3]. They also possess the natural diffusion-related characteristics of tumors resulting from their 3D geometry and metabolic-based kinetics reactions. Therefore, they are in-vitro candidates for studies involving biomarkers related to cancer hallmarks, thus enhancing the development of new treatment strategies with improved clinical outcomes [4].
The change in the homeostasis of secreted proteins and small molecules in cancer cells identifies the progression of the tumor. This can be measured by immunostaining of different proteins/biomarkers expressions and distribution at the cellular and subcellular level, using specific antibodies for the target proteins [5]. Immunostaining also enables semi-quantification analysis. In addition, the dynamics of various proteins’ expression necessitate a fast and non-destructive staining procedure for accurate measurements in cells within tumoroids. Proteins that are unstable to small changes in their microenvironment must undergo the least possible manipulation prior to the fixation stage. For instance, hypoxia-inducible factor 1 (HIF-1), one of the key interferent elements in cancer therapies, is among the important tumor-related transcription factors upregulated in hypoxia and hydroxylated in response to oxygen in the order of a few minutes [6]. The immunostaining process of HIF-1 within tumoroids must exclude the cell dissociation step, which is usually carried out using proteolytic enzymes such as trypsin, to avoid HIF hydroxylation.
The presented protocol details the IHC staining of tumoroids for targeting different cancer biomarkers presented at the state of TME. We utilized EZ-Seed culture platform to generate tumoroids. This platform is made of agarose and allows for culturing monosize tumoroids with the capability of direct embedment in paraffin for easier IHC. Our staining method minimizes the manipulation of tumoroids prior to fixation by eliminating multiple centrifugations and shaking steps typically required for removing excess hydrogel and collecting tumoroids [7,8], providing a non-destructive IHC staining method. It also enables simultaneous IHC staining of multiple uniform organoids under either identical or different treatment conditions. Our approach offers greater control over uniformity, size, and position of the organoids during the staining and ensures consistency in the staining procedure.
Tumoroids fixation
-
1.
Soak tumoroids in 10% neutral buffered formalin for 60 min. If tumoroids are kept in EZ-Seed platform, fill the wells with formalin. If they are placed in a microfluidic chip, inject the formalin into the chamber and make sure the entire camber is covered by formalin.
-
2.
Gently aspire the formalin and wash twice with DPBS. Tumoroids can be properly stored in a fridge before embedding.
Histology mold preparation
Histology mold preparation includes the following steps:
-
1.
Embedding in agarose
Prepare 2% (w/v) agarose solution by dissolving low-melting agarose powder in DPBS. Place a beaker on a hot plate at 100 °C. Stir the solution until it is clear. If needed, cover the beaker with foil to avoid evaporation. Place the fixed and DPBS-washed tumoroids in a 3 cm Petri dish. Let the agarose solution cool down. Before it solidifies, gently pour it into the Petri dish to embed the tumoroids. This step is necessary to keep the tumoroids in place during subsequent steps such as dehydration and Xylene treatment. The height of agarose in the Petri dish must be smaller than the thickness of the cassette (see step 3).
Make sure tumoroids are dispersed within the agarose solution. Let the Petri dish cool down to room temperature (10 min). Pill off the cured agarose block with a blade and cut the block into desired pieces. Make sure each piece has more than one tumoroid and fits inside the cassette.
Note: Do not use hot agarose solution; high temperate may damage the tumoroids and disintegrate the target proteins.
Next steps are optimized for samples with ∼5 mm thickness.
-
2.
Dehydration
To dehydrate the samples, soak the agarose-embedded tumoroids blocks in ethanol according to the following steps,-
▪70% ethanol for 30 min.
-
▪90% ethanol for 30 min.
-
▪100% ethanol for 120 min (3X). Agarose blocks must look white at the end of this step. Repeat this step if there are clear spots in the samples.
To remove ethanol, soak in Xylene according to the following steps,-
▪Xylene for 30 min.
-
▪Xylene for 30 min (clean wash).
-
▪Xylene for 30 min (clean wash). Agarose blocks must become very clear at end of this step. Repeat this step if there are white spots in the samples.
Note: Long exposure to Xylene may damage the samples.
-
▪
-
3.Embedding the agarose blocks in paraffin wax
-
▪Prepare melted paraffin wax in a beaker and keep it on a hotplate at 65 °C.
-
▪Place the samples in cassettes. Label the cassettes according to your experimental conditions.
-
▪Place the cassettes in melted paraffin wax for 120 min
-
▪Refresh the paraffin wax and repeat the previous step.
-
▪
-
4.Preparing histology mold
-
▪Remove the cassette from the wax and remove the sample from the cassette.
-
▪Place the cassette face down on the hot plate and place the agarose blocks in the histology mold on the bench. Each cassette can contain more than one agarose block depending on the number of experimental conditions.
-
▪Pour clean paraffin wax into the mold around the agarose blocks.
-
▪Place the hot cassette on the mold and cover it with wax.
-
▪Let them dry at room temperature for at least 30 min, and transfer to the fridge before slicing.
-
▪
Slicing
Paraffin-embedded tumoroids must be sliced to form monolayer cell distribution (sections) prior to antibody staining as follow,
-
1.
Set the microtome thickness to 10 µm. Note: The slicing velocity can be optimized to user's convenient.
-
2.
Hook the cassettes, containing paraffin blocks, to the clamp. Make sure the cassette is aligned vertically parallel to the blade holder.
-
3.
Load a sharp blade, tighten the blade holder, and make sure the blade's zone of slicing covers the entire paraffin block during the vertical motion.
-
4.
Leave a few millimeters gap between the paraffin block and the blade.
-
5.
Turn on the device. Waite until the blade touches the paraffin block and starts the slicing. Use a tweezer to grab the sections. Gently place the sections on warm water. Continue slicing.
-
6.
When the slicing is done, turn off the device, remove the blade and unhook the paraffin block. Note that paraffin block is still usable if a part of tumoroids remains (stored in the freezer).
-
7.
Label a glass slide, gently hold it underneath one section and grab it. The section sits and attaches on the glass slide. Leave the glass slide in slide holder to remove the excess water. Repeat this step with new glass slides until all the sections are collected. Leave the glass slides at room temperature overnight to dry out. A hot plate can be used for faster dehydration.
Deparaffinization
Deparaffinize by soaking glass slides in,
-
▪
Xylene (2 × 3 min)
-
▪
100% ethanol (2 × 3 min)
-
▪
95% ethanol (3 min)
-
▪
Distilled water (3 min)
Note: Don't let the sections dry out before staining.
Antibody staining
Antibody staining is carried out in the following steps,
-
1.Permeabilization
-
▪Incubate once in TBS + 0.3% Triton-X100 for 10 min at room temperature to permeabilize nuclei (improves intracellular staining)
-
▪
-
2.Blocking
-
▪Add to each section, 3% bovine serum albumin (BSA) and 0.3% Triton X-100 in TBS, and keep it for 20 min at room temperature. Note that all BSA solutions must be 0.2 µm syringe filtered before use.
-
▪
-
3.Primary antibody staining
-
▪Dilute at 1:200 to 1:1000 in 1% BSA, 0.3% Triton X-100 in TBS (follow the antibody instruction if they have specified dilution factors for IHC). Incubate overnight at 4 °C in the humidity chamber. (Alternatively, incubate for 2–4 h at room temperature).
-
▪After the incubation, wash 3x in TBS for 5 min each.
-
▪
-
4.Secondary antibody staining
-
▪Dilute the secondary antibody at 1:500 in 1% BSA, 0.3% Triton X-100 in TBS. (follow the antibody instruction if they have specified dilution factors for IHC). Incubate for 1 hour at room temperature.
-
▪After the incubation, wash 3x in TBS for 5 min each.
-
▪
-
5.Counter-stain with DAPI
-
▪Dilute the 5 mg/mL stock solution down to 1 ug/mL in TBS by diluting by a factor of 5000 (i.e., 10 mL TBS for a 2 µL aliquot).
-
▪Incubate at room temperature for 5 min.
-
▪Wash 3x in TBS for 5 min each.
-
▪
-
6.Mount with Entellan
-
▪Use a plastic dropper to put 3 drops of Entellan on each glass slide. Gently place a rectangular coverslip on top and gently press out any small bubbles.
-
▪Wait at least 30 min to partially dry before imaging.
-
▪
To demonstrate the applicability of the presented protocol, different target proteins were antibody stained.
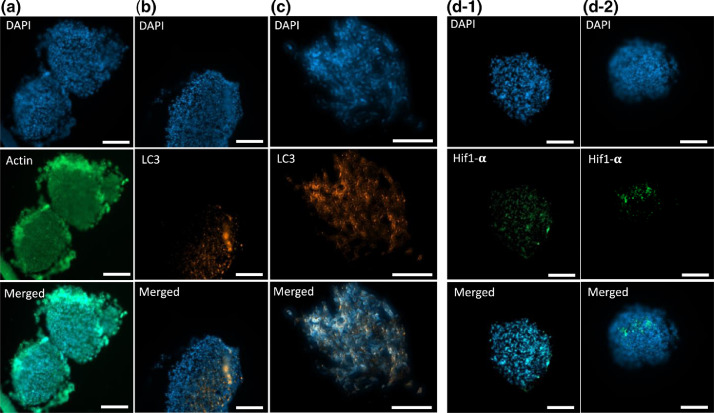
Fig. 1 shows IHC staining of actin, Hif1-α, and LC3 as well as counter-staining with DAPI in human glioblastoma (hGB) U251-derived tumoroids. First, to show the cytoskeleton of cells, U251 tumoroids were IHC stained with actin antibody, shown in Fig. 1(a). The cytoskeleton of a cell regulates different cellular activities such as growth, migration, and structural support, hence, is essential to determine the structure of tumor cells [9].
Fig. 1.
IHC staining of U251 tumoroid cytoskeleton with actin (a), autophagosome with LC3 (b, c), and hypoxia with Hif1-α following the presented protocol (d-1) compared with the tumoroids underwent conventional intermediate steps of washing and centrifugation (d-2). Tumoroids were counterstained with DAPI. Scale bars are 100 µm.
In another experiment, to monitor the localization of LC3, an important autophagosome marker in autophagy flux, we IHC stained LC3 in rapamycin/chloroquine-treated tumoroids, depicted in Fig. 1 (b, c).
Hypoxia is another key regulator of cancer cell metabolism that can trigger tumor hallmarks such as invasion and angiogenesis [10]. To induce hypoxia, tumoroids were kept in hypoxic chamber for 24 h and IHC stained with Hif1-α antibody, shown in Fig. 1(d). The instability of Hif1-alpha against oxygen requires a staining procedure with minimum manipulation prior to fixation of cells. Our staining method preserves the expression of hif1-alpha in tumoroid cells by eliminating the challenges of organoid collection and removal of excess hydrogel before fixation. To demonstrate the advantage of eliminating these intermediate steps, we compared the expression of hif1-alpha in tumoroids stained following our protocol, Fig. (d-1), with tumoroids underwent preparation steps, including washing and centrifugation, Fig. (d-2). Results depict a significant decrease in the expression of hif1-alpha due to the intermediate steps, while our approach successfully detected Hif1-alpha expression in tumoroid cells.
Important notes
-
1.
After deparaffinization, do not let the slides dry out! Incubate the primary and secondary antibodies in the airtight glass container with snap lid. Put a wet paper towel and 3–4 mm of water at the bottom. Carefully break a serological pipette into 1/3 segments and place them on the paper towel to hold the slides level and above the water.
-
2.
It is best to perform all steps in the fume hood. Xylene and Entellan fumes are toxic.
-
3.
Using the optimal form of heat induced epitope retrieval (HIER) can help increasing signal intensity. If it is listed in the antibody datasheet.
-
4.
Make sure that the primary antibody is a suitable target for the secondary antibody, i.e., if using a rabbit primary, the secondary should be anti-rabbit. The species the secondary is raised in is irrelevant. i.e., Donkey anti-rabbit, Goat anti-rabbit, and Mouse anti-rabbit would all be suitable secondaries for a Rabbit primary.
CRediT authorship contribution statement
Meitham Amereh: Conceptualization, Methodology, Investigation, Writing – original draft. Mohsen Akbari: Conceptualization, Supervision, Resources, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Mohsen Akbari has conflict of interest in Apricell.
Acknowledgments
Funding: This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). We would like to acknowledge BC cancer Foundation, NSERC, Canada Foundation for Innovation (CFI) and Apricell biotechnology (https://www.apricell.com).
Data availability
Data will be made available on request.
References
- 1.Duval K., et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32(4):266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 2.Kim J., Koo B.-K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21(10):571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubert C.G., et al. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivobrain cancer stem cell organoids. Cancer Res. 2016;76(8):2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang S., Kim D., Lee J., Takayama S., Park J.Y. Engineered microsystems for spheroid and organoid studies. Adv. Healthc. Mater. 2021;10(2) doi: 10.1002/adhm.202001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor C.R. Predictive biomarkers and companion diagnostics. The future of immunohistochemistry:‘in situ proteomics,’ or just a ‘stain’? Appl. Immunohistochem. Molecul. Morphol. 2014;22(8):555–561. doi: 10.1097/PAI.0000000000000126. LWW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muz B., de la Puente P., Azab F., Azab A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Yu L., Chen D., Meng Z., Chen W., Huang W. Protocol for generation of lung adenocarcinoma organoids from clinical samples. STAR Protocol. 2021;2(1) doi: 10.1016/j.xpro.2020.100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiley L.A., Beebe D.C., Mullins R.F., Stone E.M., Tucker B.A. A method for sectioning and immunohistochemical analysis of stem cell–derived 3-D organoids. Curr. Protoc. Stem Cell Biol. 2016;37(1):1C–19. doi: 10.1002/cpsc.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melak M., Plessner M., Grosse R. Actin visualization at a glance. J. Cell Sci. 2017;130(3):525–530. doi: 10.1242/jcs.189068. [DOI] [PubMed] [Google Scholar]
- 10.Abou Khouzam R., et al. Tumor hypoxia regulates immune escape/invasion: influence on angiogenesis and potential impact of hypoxic biomarkers on cancer therapies. Front. Immunol. 2021;11 doi: 10.3389/fimmu.2020.613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.